- 1Department of Marine Science, Incheon National University, Incheon, South Korea
- 2Marine Environmental Research Division, National Institute of Fisheries Science, Busan, South Korea
- 3Department of Aquaculture and Aquatic Science, Kunsan National University, Gunsan, South Korea
Although environmental drivers are known to shape the abundance and distribution of bacterial communities in the East Sea, the effects of physical processes have not been directly studied. Here, we aimed to examine the influences of water mass mixing (summer) and eddy circulation (winter) on the surface bacterial communities of the Ulleung Basin (UB), East Sea, based on the metagenomic approach. Overall, 490,087 operational taxonomic units (OTUs) were identified from five stations, and prokaryotic abundance was dominant at all stations in both seasons. Among the prokaryotes, most OTUs were affiliated with Proteobacteria, Cyanobacteria, Flavobacteria, and Actinobacteria during summer and winter. Bacterial communities were found to differ with water masses (Changjiang, Tsushima, and North Korea surface water) and eddy circulation, and were strongly correlated with environmental variables, suggesting specific bacterial community responses with specific seasonal physicochemical parameters. Our investigation indicates that together with distance and environment, advection shapes the UB bacterial community composition, helping us better understand the physical cues related to biological composition in the East Sea. However, further studies are needed to ascertain the role of microbial functional genes along with the advection of oceanographic processes in the East Sea to better understand the regional biogeochemical processes.
Introduction
Marine microbes are responsible for nearly half of the global net primary productivity (Falkowski et al., 1998), and are important drivers of ocean biogeochemical cycling (Arrigo, 2005). Bacterial communities play an inevitable role as they decompose organic matter and transfer energy and nutrients to higher trophic levels (Zehr and Kudela, 2011). Due to its characteristics, each bacterium acts as a sensitive indicator of the surrounding environment. Specifically, bacterial composition and abundance in the ocean environment are dependent on temperature, salinity, and nutrient concentration (Henriques et al., 2006; Shade et al., 2007; Winter et al., 2007).
Physical processes (water mass mixing and eddy circulation) in the marine environment are also significant factors that determine the bacterial structure and composition (Mann and Lazier, 2006; Dinasquet et al., 2017; Venkatachalam et al., 2017). In particular, the mixing of water masses causes changes in their physiological processes (respiration, degradation, and production) by mediating the influences of temperature, dissolved oxygen, and nutrient concentrations to determine their community structure composition (Shade et al., 2008; Laque et al., 2010; Celussi et al., 2010; Galand et al., 2010). Eddy circulation imports and/or exports new bacterial species and influences the functional processes of ecosystems (Fuhrman et al., 2015; Lu et al., 2021). Previous studies have observed that eddy variability causes nutrient fluctuation, consequently, altering the bacterial composition (Pinhassi et al., 2006; Zhang et al., 2011) as their production and biomass inside the eddy were changed compared to the surrounding waters (Thyssen et al., 2005; Zhang et al., 2009).
The East Sea (ES) is a semi-enclosed marginal sea consisting of three deep basins (Ulleung, Yamato, and Japan basins) connected to the North Pacific Ocean through shallow straits (Korea, Soya, and Tsugaru Strait) (Figure 1A). The ES is a physically dynamic region owing to its occurrence in many physical processes, such as deep-water formation, subpolar front, thermohaline circulation, coastal upwelling, and eddy circulation (Ichiye, 1984; Kim et al., 1996; Lee et al., 2009). Among the ES basins, Ulleung Basin (UB), located in the southwestern region (Figure 1B) is a biological “hot spot” as its annual-mean biological productivity is nearly 40% higher compared to other basins (Joo et al., 2014). During summer, the low saline Changjiang Diluted Water (CDW) is transported from the East China Sea (ECS) to the UB through the Korea Strait (KS) with high temperature and salinity features of Tsushima Warm Water (TWW), which constitutes the Tsushima Warm Current (TWC) (Chen et al., 1994; Senjyu et al., 2006). In winter, the Ulleung warm eddy (UWE) from the East Korean Warm Current (EKWC) flowing northward along the Korean coast (Cho and Kim, 1996), occurs in the center of the UB (Shin et al., 2019), transporting nutrient rich coastal waters into the center of the UB and subsequently enhancing the microbial production (Hyun et al., 2009; Kim et al., 2012). These seasonally different physical features of water mass mixing in summer and eddy circulation in winter are key factors determining the UB bacterial structure and composition.
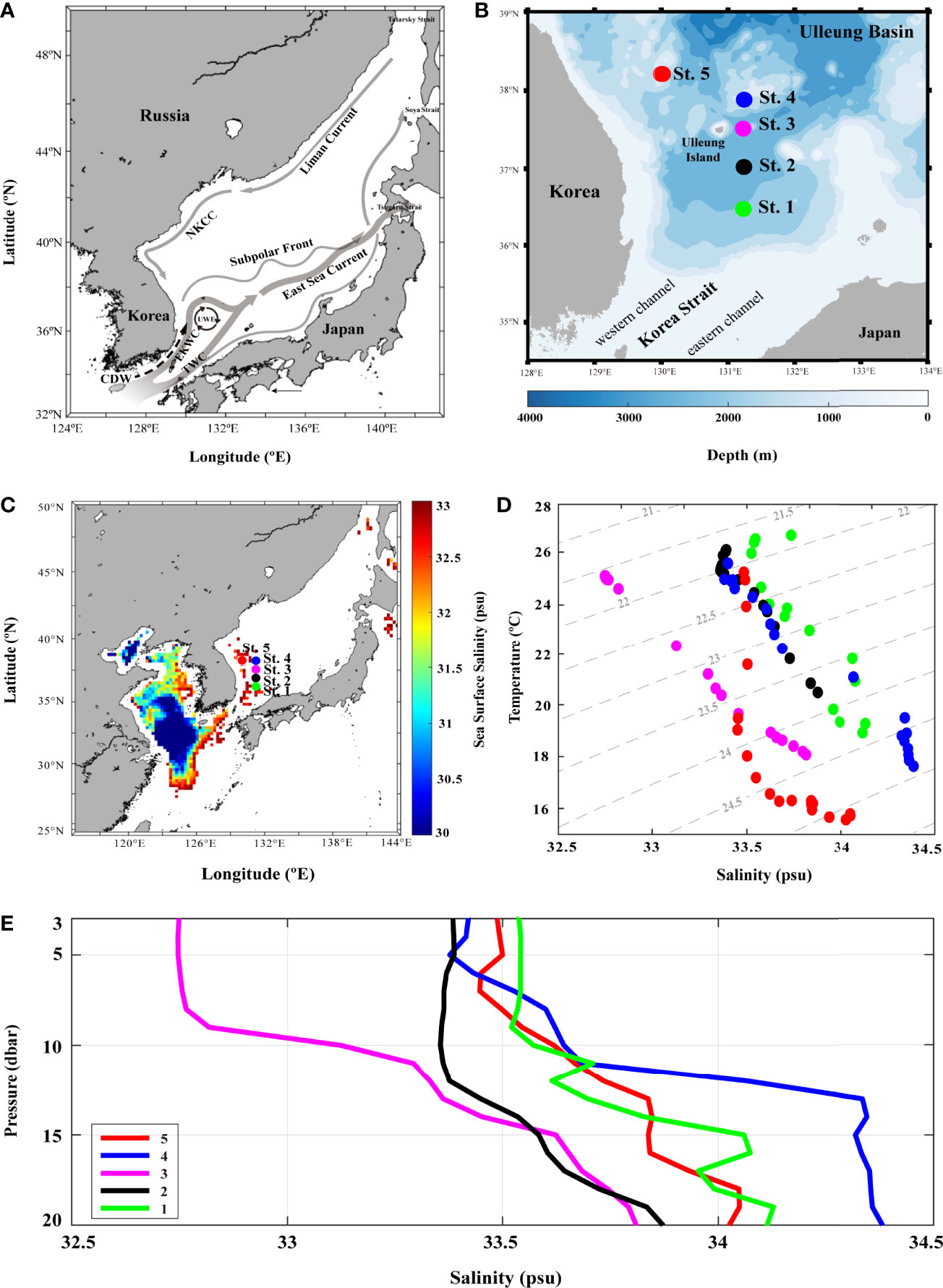
Figure 1 The surface currents identified in the East Sea and five sampling stations in Ulleung Basin (UB). (A) map with arrows represent major surface currents and gyre identified in the East Sea: Changjiang diluted water (CDW) flows from Changjiang river in summer, Tsushima warm current (TWC) enters into the East Sea through Korea strait (KS), and East Korean warm current (EKWC) flows along the eastern coast of Korea and enters UB. Subpolar front (~40°N) divides the East Sea into two regions, southern large eddy kinetic energy region and northern with small eddy kinetic energy region. East Sea current is a broad current that EKWC and Eastern Branch form after leaving the respective coasts of Korea and Japan. North Korea cold current (NKCC) is a year-round mean current along the coast of North Korea. Liman Current (LC), which flows southward along the Siberian coast down to Vladivostok. Ulleung warm eddy (UWE), originating from EKWC, northward branch of the warm and saline TWC, is a well-known physical phenomenon in the center of the southwest East Sea; (B) map of the five sampling stations in UB using Tamgu 3 during August and December, 2019; (C) sea surface salinity gridding map using August (summer) monthly data from Ocean Color; (D) temperature–salinity diagram of 0–20-m depth seawaters at the five stations; and (E) vertical distribution of salinity in 0–20 dbr pressure measured with CTD at each station.
Recently, Lee and Choi (2021) demonstrated the confluence of two water masses (TWW and ES water) in the UB region and suggested that these currents may lead to increases in biological activities. Similarly, Ichinomiya et al. (2019) found that microbial abundance and species composition are significantly influenced by water currents, which are low in the Tsushima Warm Current in northern Japan. In addition, in the upper epipelagic layer (0-200m), eddies known to enhance nutrient inputs to the surface ocean and increasing new production (Falkowsky et al., 1991; Morán et al., 2001) effects on the prokaryotic community are apparent, as indicated by higher prokaryotic abundance, prokaryotic heterotrophic activity, and percentage of active cells within eddies than in the eddy far-field (Baltar et al., 2010). However, fewer studies have been conducted in this region regarding the metagenomic approach, focusing on physically static sedimentary environments with a microbial population and vertical structure influenced by methane gas presence (Jeong et al., 2010; Lee et al., 2013; Shin et al., 2014; Cho et al., 2016). Despite the importance of physical processes in the UB region, little is known about how these processes determine bacterial community composition. To this end, the present study aimed to investigate (i) how water mass mixing (in summer) and eddy circulation (in winter), and (ii) their driving environmental factors determine the spatial distribution of bacterial structure and compositions in the UB region.
Materials and Methods
Sampling and Measuring Physical and Biogeochemical Parameters
Samples were collected from five stations located in the UB aboard R/V Tamgu 3, in August (summer) and December (winter) 2019 (Figure 1B). At each station during the two seasons, temperature, salinity, and dissolved oxygen (DO) were determined using an attached sensor system (Sea-Bird SBE 911 conductivity–temperature–depth) on a rosette sampler (Sea-Bird Electronics, USA). The CTD was calibrated by the manufacturer before we proceed with the measurements at stations and the accuracies of temperature, conductivity, oxygen, and pressure sensors were ± 0.001°C, ± 0.0003 S m -1, ± 2 % and ± 0.015% of the full-scale range, respectively. To determine the nutrients [nitrite (), nitrate (), phosphate (, denoted as PO4), and silicate (SiO2)], seawater was collected in 15 ml pre-acid rinsed plastic bottles and analyzed onboard using a continuous flow auto-analyzer (QuAAtro 39, Seal Analytical, UK). The analytical precision of the nutrients was better than 1%. The nutrient measurements were performed using same method as the used by Hydes et al. (2010) in the GO-SHIP protocol and reference materials (Kanso RMNS) was used to track the performance of the system and the consistency of nutrient data. The nitrite + nitrate observed in this study is defined as dissolved inorganic nitrogen (DIN). To determine chlorophyll-a (Chl-a), 500 ml of seawater was filtered through 25 mm Whatman GF/F filters and frozen at -80°C. In the laboratory, pigments were extracted using 90% acetone (Parsons et al., 1984) and the Chl-a concentrations were measured using a Trilogy fluorometer with an analytical precision of ± 0.05 µg L-1 (CHL NA, Model #7200-002; Turner designs, USA). For Chl-a measurement, EPA 445.0 was used as the standard operating procedure for fluorometric determination. At all stations (5 stations × 2 seasons = 10 samples), 2L of surface seawater was filtered through 0.22 µm cellulose ester membrane filters (Millipore, Ireland) to capture bacterial cells. Filtered samples were immediately frozen and stored at −80°C until DNA extraction.
DNA Isolation and Sequencing of 16S rRNA
Total DNA was extracted from the obtained samples following a standard protocol using a DNA isolation kit (Qiagen, Germany) and the quality of the extracted DNA was cross-checked using NanoDrop (Thermo Scientific, USA). After quality control (QC), the 16s rRNA sequencing library was constructed according to the 16S metagenomic sequencing library preparation protocol (Illumina, San Diego, CA, USA) targeting the V3 and V4 hypervariable regions of the 16S rRNA genes. PCR was performed using the 16S amplicon PCR forward primer: 5′- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and reverse primer: 5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′. The extracted DNA samples were amplified and analyzed using Illumina MiSeq (MiSeq Control Software v2.4.1.3) and Real-Time Analysis (RTA v1.18.54.0). Subsequently, 16S tagging of DNA fragmentation (100–300 bp) was performed using the quantitative kit for sample DNA. Primers were prepared for all DNA fragments, amplified using an adapter, and subjected to library QC using a bioanalyzer, followed by clustering. Sequencing data were processed using QIIME1.9.1 to assemble paired-end reads into tags according to their overlapping relationships. In the pre-processing step, the primer was removed and then demultiplexing and quality filtering (Phred ≥20) were applied. USEARCH7 was used to perform denoising and chimera detection/filtering to group operational taxonomic units (OTUs). The Silva132 and National Center for Biotechnology Information (NCBI) databases were used to determine OTUs with 97% similarity using UCLUST and the close-reference analysis. The OTU table was normalized by dividing each OTU by its 16S copy number abundance. After filtering the generated OTU table using the Biological Observation Matrix format, the resulting sequences were clustered into OTUs based on a similarity threshold of ≥97% using Python Nearest Alignment Space Termination (PyNAST) (Caporaso et al., 2010). Using RDP classifiers, we performed comparative OTU assignments with the database regarding phylum, class, order, family, genus, and species. Permutational multivariate analysis of variance (PERMANOVA) was used to test for significant differences among the groups. The relative abundance of phyla, classes, orders, families, genera, and species was estimated using the STAMP program. Multivariate analysis was performed using principal component analysis, whereas univariate analysis was performed by ANOVA. Raw sequence data from this study were deposited in the NCBI with Sequence Read Archive (accession number: PRJNA778576).
Data Visualization and Statistical Analysis
The surface salinity data for August and surface temperature data for December used in this study were obtained from NASA Ocean Color (https://oceancolor.gsfc.nasa.gov/). Multiple statistical and similarity analyses (ANOSIM; a non-parametric permutation procedure that tests whether differences in dissimilarity between groups exceed differences within groups) were conducted with bray-curtis method using the R program (v.3.6.2, https://www.R-project.org) with gplots, ggplot2, Heatplus, and vegan packages. Non-metric multidimensional scaling (NMDS) analysis was performed using the unweighted Unifrac method to indicate surface bacterial dissimilarity. Shannon diversity indices and Chao1 species abundance indices were used to estimate the alpha diversity of bacterial communities. Distance-based redundancy analysis (db-RDA) was performed to understand the relationships between the bacterial communities and the surrounding physicochemical parameters. Spearman’s rank-order correlation coefficients were used for non-normally distributed data (i.e., between the relative abundance of bacterial taxa and physical and biogeochemical factors) to define the degree of dependence among the variables, giving a value between -1 and +1 (1 represents a total positive correlation, 0 represents no correlation, and -1 represents a negative correlation) using R software.
Results and Discussion
Large Influence of Water Mass Mixing on Surface Bacterial Composition During Summer
The environmental parameters obtained during the investigation showed variable results at each station (Figure 1 and Table 1). In particular, the summer (August) sea surface salinity data showed lower salinity CDW (<31 PSU) widely distributed from Changjiang River mouth to the ECS (Figure 1C). A large discharge of CDW extends to near 28°N, and in turn, the salinity of CDW increases (32–33 PSU) because of the mixing of warm and saline TWW (>20°C and <33.8 PSU) via the Tsushima Current (Park, 1978; Yang et al., 1991; Cho et al., 1997; Chang and Isobe, 2003; Moon et al., 2009). TWW and CDW flowed into the UB through the western channel of the KS (Figures 1B, C), the main path of fresh CDW resulting from strong halocline stratification (Senjyu et al., 2006; Kim et al., 2014). Consequently, the conducting temperature-salinity diagram explains the distribution of the three different water masses during summer (Figure 1D). The surface temperature and salinity observed at St-3 were 25.01°C and 32.74 PSU, respectively, with relatively high PO4 (0.02 µM) and SiO2 (5.63 µM) concentrations (Table 1). The vertical salinity profile also showed that CDW distinctly affected St-3 at pressures of 0–8 dbr (Figure 1E). The mean surface temperature and salinity at Sts-1, 2 and 4 were 25.8°C (range: 24.9–26.46°C) and 33.45 PSU (33.54–33.38 PSU), which were higher than those at St-3, while mean nutrient concentrations were relatively lower (DIN, 0.39 µM; PO4, 0.01 µM; and SiO2, 3.40 µM) (Table 1 and Figure 1D), indicating the influences of TWW. Relatively low temperature (21.55°C) and high DO (8.21 mg L−1) and DIN concentration (0.86 µM) were observed at St-5 (Table 1), located at the highest latitude among other stations with North Korea Surface Water (NKSW), originating from the northern ES and is characterized by low temperature and salinity near 40°N in the UB (Kim and Lee, 2004). Based on these observations, it is evident that the physical mixing of three different water masses (TWW, CDW, and NKSW) influences the UB region during summer. We categorized these observed stations into three groups: TWW (Sts-1, 2, and 4), CDW (St-3), and NKSW group (St-5).
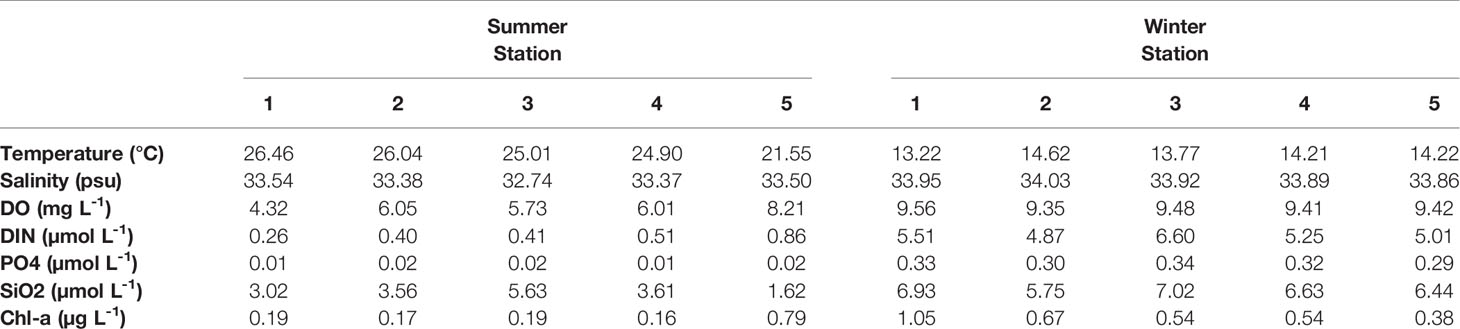
Table 1 Surface physicochemical parameter data including temperature (°C), salinity (PSU), dissolved oxygen (mg L−1), dissolved inorganic nitrogen (µmol L−1), phosphate (µmol L−1), silicate (µmol L−1), and Chl-a (µg L−1) in summer and winter at the five stations in Ulleung Basin.
In summer, metagenome samples collected from five stations and a total of 490,087 operational taxonomic units (OTUs) were identified, belonging to seven phyla, 10 classes, 23 orders, and 37 family levels in the surface bacterial communities (Supplementary Table S1A–D). To determine the influence of physical mixing on bacterial composition, we performed alpha (Shannon diversity and Chao1 species richness indices) and beta diversity (clustering dendrogram, NMDS, dbRDA, and heatmap) analyses (Figures 2 and 3). Clustering analysis computed by Bray–Curtis dissimilarity based on OTUs revealed that stations affected by the same water mass were clustered first: Sts-1, 2, and 4; St-3; and St-5, in sequence (r = 0.12, P < 0.5) (Figure 2A). The CDW group was clustered with the TWW group before the NKSW group (Figure 2A) as the CDW was transported from the ECS to UB by the Tsushima current system (Figure 1A). The NMDS plot represents the dissimilarity of the surface bacterial community with distances in the configuration (Shepard, 1962; Kruskal, 1964; Holland, 2008), showing remarkable dissimilarity of St-3 (Figure 2B) during summer. This was further confirmed by the Shannon diversity and Chao1 species abundance indices with lower values (4.84 and 422.15, respectively) at St-3 (Figures 2C, D). Moreover, the Shannon and Chao1 indices across Sts-1, 2, and 4 were consistently high (mean value: 5.37 and 497.64, respectively). Commonly, at higher latitudes, lower values of Shannon and Chao1 were discovered within the TWW group, whereas St-5 showed a contradictory pattern with relatively high Shannon value but low Chao1 values of 5.38 and 433, respectively (Figures 2C, D). The Chao1 value of St. 4 which belongs to the TWW group, was low in the TWW group; however, it was relatively high compared to that of Sts-3 and 5. As St. 4 is close to St. 5, the St. 4 Chao1 value seems to stand out within the same group.
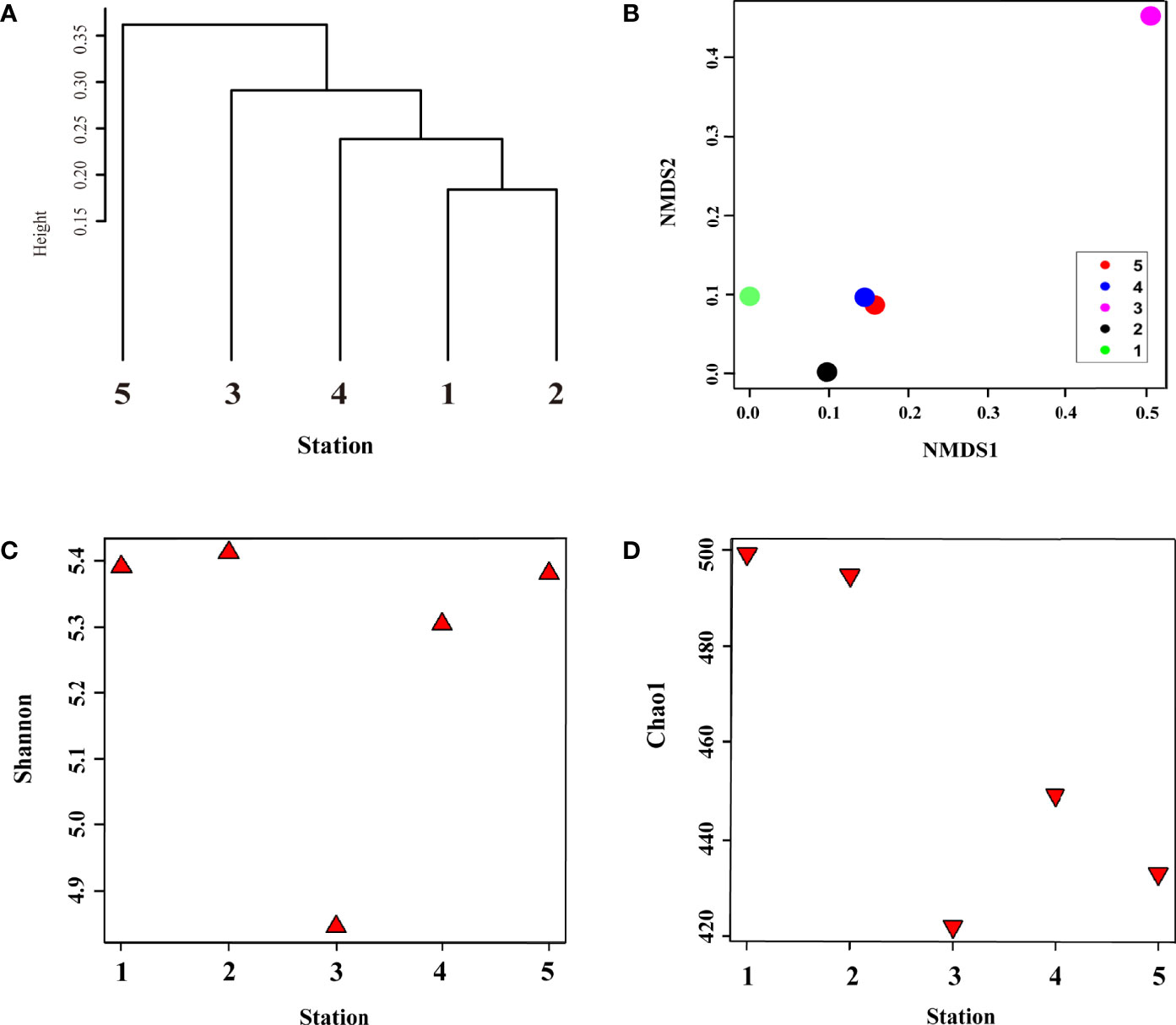
Figure 2 Surface bacterial community composition similarity and differences in diversity and species richness in summer. (A) clustering of samples at the operational taxonomic unit (OTU) level based on Bray–Curtis distance; (B) dissimilarity of bacterial community composition using non-metric dimension scale (NMDS) plot based on unweighted-unifrac method; (C) diversity and (D) species richness represent the alpha diversity of surface bacterial community composition using Shannon and Chao1 indices, respectively.
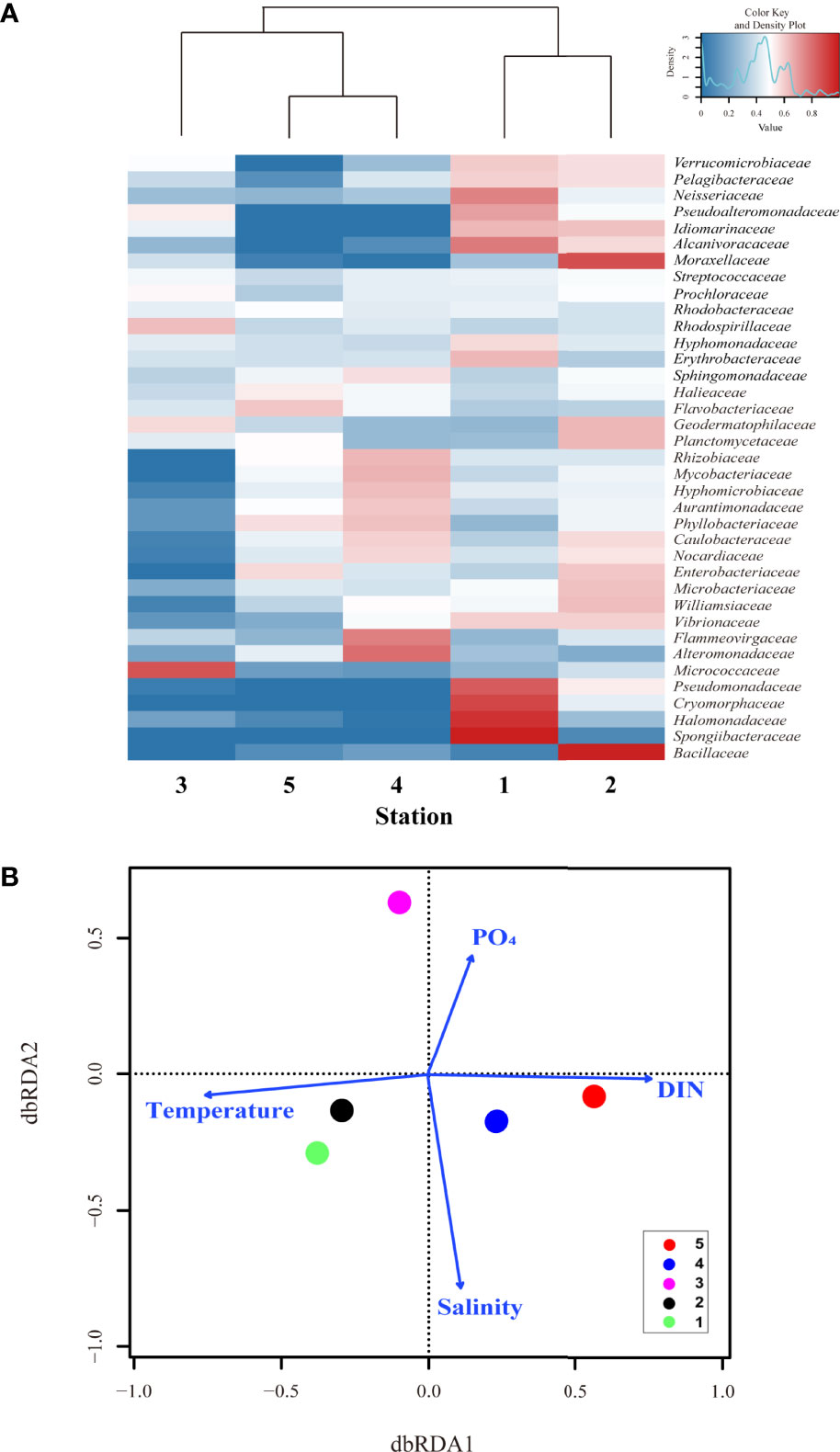
Figure 3 The distance-based redundancy analysis (dbrda) ordination plot and heatmap revealed that environmental conditions controlled the surface bacterial community composition in summertime of UB. (A) hierarchical heatmap showing the relative abundance 37 genera at family level. Color code indicates the relative abundance, ranging from blue (low relative abundance) to red (high relative abundance); and (B) the dbrda ordination diagram of bacterial communities based on OTU at family level and related physical and biogeochemical factors, including temperature, salinity, dissolved inorganic nitrogen (DIN), and phosphate (PO4) concentration.
Visual inspection of a heatmap revealed differences in the relative abundance at the family level (top 37 genera, relative abundance >0.01%) of surface bacterial communities from five stations during summer (Figure 3A). Even within the same TWW group, some bacteria showed differences in relative abundance by station. The relative abundances of Spongiibacteraceae, Halomonadaceae, Cryomorphaceae, Pseudomonadaceae, Neisseriaceae, Pseudoalteromonadaceae, Hyphomonadaceae, and Erythrobacteraceae were high at St. 1 whereas those of Bacillaceae, Williamsiaceae, Microbacteriaceae, Enterobacteriaceae, Planctomycetaceae, Geodermatophilaceae, and Moraxellaceae were high at St. 2. In addition, Alteromonadaceae, Flammeovirgaceae, Rhizobiaceae, Mycobacteriaceae and Hyphomicrobiaceae showed high relative abundances at St. 4. However, Vibrionaceae, Alcanivoracaceae, Idiomarinaceae, Pelagibacteraceae, and Verrucomicrobiaceae were abundant in Sts-1 and 2. In St-3, the relative abundance of Micrococcaceae (belonging to the class Actinobacteria) and Rhodospirillaceae (Alphaproteobacteria) were higher, whereas that of Flavobacteriaceae (Flavobacteriia) and Halieaceae, were high at St-5, even if it was a low value. As shown in Figures 2C, D, which showed high diversity and species abundance, high relative abundance species were more diverse in the TWW group (Sts- 1, 2 and 4) than in other stations. The db-RDA analysis was conducted to identify the environmental factors shaping the bacterial community composition at the family level (Figure 3B; with relative abundance >0.01% in at least one of the samples). Sts-1 and 2 (affected by warm and saline TWW) were closely linked to temperature and salinity. Additionally, owing to the influence of high temperature and saline TWW and its geographical features closely located to St. 5, St. 4 appears to be associated with salinity and DIN (Figure 3B). St-3 (affected by CDW with relatively high PO4 and SiO2 concentration) and St-5 (affected by NKSW with high DO and DIN concentration) were associated with PO4 and DIN, respectively (St. 3 is not closely matched with salinity because the dbRDA plot has shown that high values are connected). These results suggest that the bacterial structure and composition at each station were closely related to the physicochemical characteristics (P < 0.05) that were influenced by different water masses.
Family level bacteria with high relative abundance at one or two stations in each three group (i.e., TWW [Sts-1,2, and 4], CDW [St-3], and NKSW [St-5]) were identified (20 genera). Consequently, they showing considerable differences between the three groups with a correlation value of ≥ 0.5 with the physical and biogeochemical factors of influencing water masses (Figure 4). Verrucomicrobiaceae (belonging to class Verrucomicrobiae), Alcanivoracaceae, Pseudomonadaceae, Spongiibacteraceae (Gammaproteobacteria), and Cryomorphaceae (Flavobacteriia) were highly abundant at Sts-1, 2, and 4 [TWW], influenced by high temperature, salinity, and poor-nutrients depicting a strong positive correlation with temperature (r = 0.97, 1.0, 0.97, 0.89, and 0.89, respectively), and salinity (r = 0.2, 0.3, 0.41, 0.67, and 0.67, respectively) and a negative correlation with DIN concentration (r = -0.97). In addition, Nocardiaceae, Williamsiaceae (belonging to class Actinobacteria), and Vibrionaceae, Spongiibacteraceae (Gammaproteobacteria) were highly dominant in Sts-1, 2, and 4, while Pseudoalteromonadaceae (Gammaproteobacteria), Hyphomonadaceae, Erythrobacteraceae (Alphaproteobacteria), and Cryomorphaceae (Flavobacteriia) at the family level showed positive and negative correlations with temperature and DIN. At St-3 [CDW], the relative abundance of Micrococcaceae, belonging to class Actinobacteria which is mainly distributed in the Changjiang River (Chung et al., 2015), and Rhodospirillaceae (Alphaproteobacteria) were higher and showed a positive relationship with PO4 (r = 0.58 and 0.4, respectively) and SiO2 concentrations (r = 0.2 and 0.98, respectively) (Figure 4). Flavobacteriaceae (belonging to class Bacteroidetes) reflects specific substrate conditions related to the phytoplankton community (Rink et al., 2011) and showed high relative abundance at St-5 [NKSW] (Figure 3A), with a positive correlation with DIN (r = 0.9; P < 0.001), DO and Chl-a concentration (r = 0.7 and 0.21). Based on these results, environmental conditions with specific water mass (TWW, CDW, and NKSW) characteristics (Figure 4) are related to a specific group of bacterial relative abundances (Figure 3) during summer.
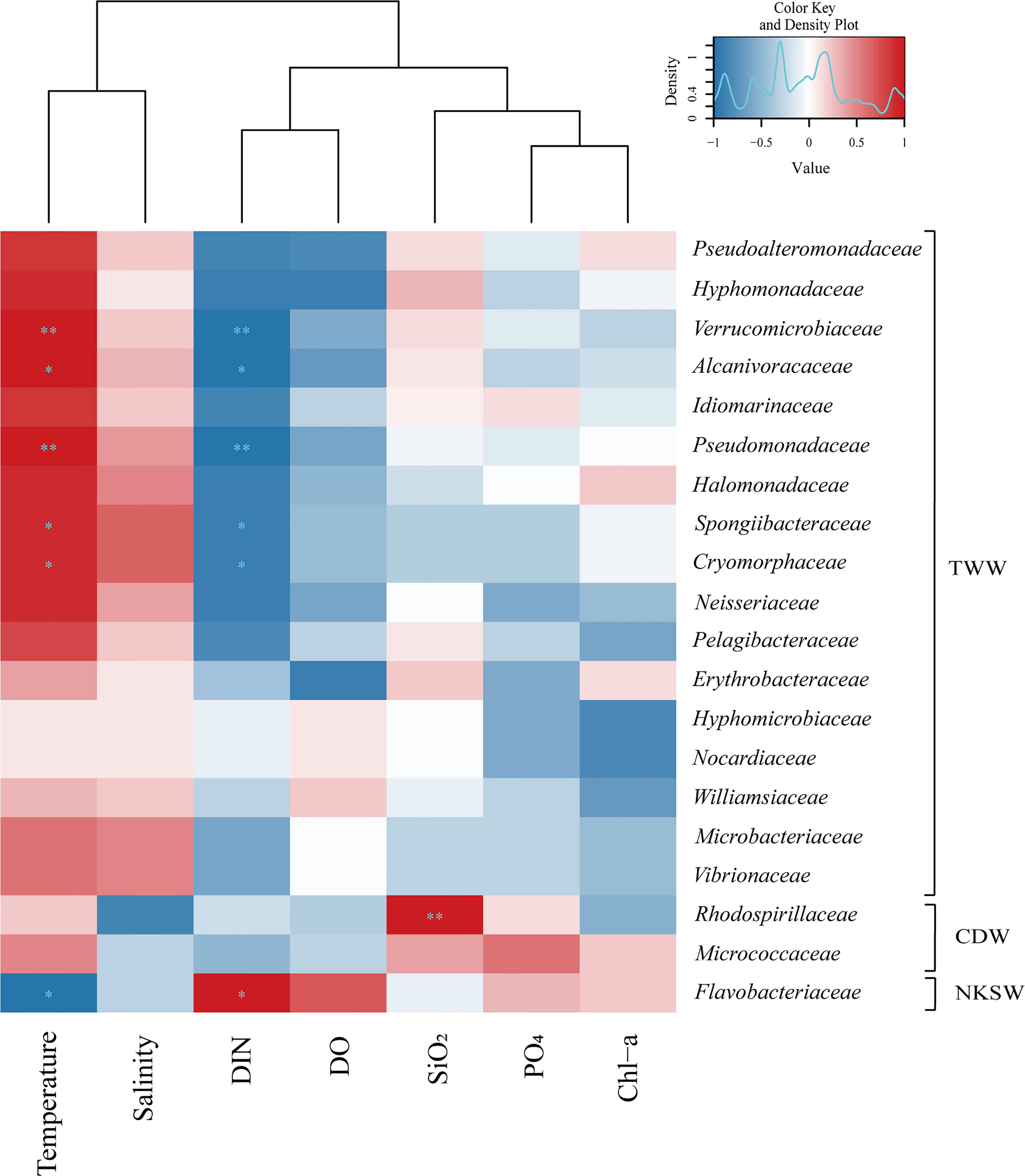
Figure 4 The 20 genera in family level of bacteria which showed significant differences in relative abundance within three groups (i.e., TWW, CDW, and NKSW). Hierarchical heatmap using Spearman’s rank order correlation coefficient showed correlation between the relative abundance of bacteria at the 37 genera family level and physical and biogeochemical factors of temperature, salinity, PO4, SiO2, DO, DIN, and Chl-a concentration in summer. The variations of 20 genera family level relative abundance showed strong correlation to hydrographic characteristics of each water masses. The symbols of * and ** indicate p value less than 0.05 and 0.01, respectively.
Spatial patterns in the marine bacterial structure and community composition have been reported across oceanic water masses with distinct hydrographic properties (Baldwin et al., 2005; Giebel et al., 2009). However, no studies have shown the influence of physical process on bacterial composition in ES, where complex physical mixing of water masses could cause variation of bacterial community composition. In the Kuroshio and Oyashio water masses from the western North Pacific Ocean, Kataoka et al. (2009) reported that different hydrographic conditions, that is temperature, salinity, and nutrient concentrations influence the elevation of bacterial composition, and variation in their phylotype structure in each water mass. Yokokawa et al. (2010) revealed the influence of water masses’ on the spatial variations in bacterial composition abundance, similarity, and activity in the eastern Mediterranean Sea, reflecting their responses to the surrounding physicochemical characteristics. Along with this physiological evidence, recent metagenome studies from the Western Subarctic Pacific Ocean (Li et al., 2018) and the Western Pacific Ocean (Zhang et al., 2018) have shown that surrounding physicochemical parameters could elevate the fundamental metabolic activity of bacterial communities and their composition. Similar to these observations, water masses were classified in this study according to their physicochemical properties, which can elevate the surrounding bacterial communities and their metabolic activities during summer. This assumption was further confirmed by alpha diversity of the bacterial community (Shannon and Chao1 indices) and a positive correlation with the features of water masses associated with physicochemical parameters (Figure 2).
Similar to the present study area (semi-closed marginal sea and occurrence of seasonal physical mixing) multiple investigations have been conducted in the Baltic Sea (Herlemann et al., 2011; Koskinen et al., 2011), demonstrating that the salinity gradient influenced the bacterial composition. However, these studies did not show a clear pattern of richness or diversity contrary to the observations in the present study, which proved a clear distinction between the three water masses (TWW, CDW, and NKSW). Specifically, a bacterial family of 20 genera, among 37 genera showed evident relative abundance differences by physically mixing the three water masses (Supplementary Table S2). For example, 17 genera were dominant at Sts-1, 2, and 4 (influenced by TWW), strongly correlated with temperature and salinity, and negatively correlated with DIN concentration (Figure 4). In addition, two genera, Micrococcaceae and Rhodospirillaceae showed remarkably high relative abundances in St-3 (influenced by CDW) with a high correlation to PO4 and SiO2 concentrations. The highly abundant Flavobacteriaceae at St-5 (affected by NKSW) strongly correlated with the DIN, DO, and Chl-a concentrations. These results suggest that our observation of hydrographic characteristics was formed by three different physical mixing processes that influenced surface bacterial communities and composition during summer in the UB.
A Major Regulatory Factor of UWE on the Surface Bacterial Community Composition During Winter
Sea surface temperature data obtained from satellite observations and environmental data collected from field observations during winter (December) showed distinct physicochemical parameters (Figure 5 and Table 1). This variation could be reflected in the influence of the clockwise UWE (Figure 5A) during this investigation, which is identifiable as a 2°C lower cold-core (16°C) wrapped by the warm filament (18°C). The surface temperature data further depict the EKWC (northward branch of warm and saline Tsushima current that flows along the Korean coast) curved between 37 and 38°N and entrained high Chl-a concentration in coastal waters that formed the UWE ring, supplying highly productive coastal waters to the center of the UB (Hyun et al., 2009; Kim et al., 2012). With the occurrence of UWE in UB during winter, the physicochemical characteristics at St-1 (located at the UWE ring; Figure 5A) showed a lower temperature (13.22°C) but higher Chl-a concentration (1.05 µg L−1) compared to those outside of the eddy ring at St-2 temperature (14.62°C) and Chl-a concentration (0.67 µg L−1) (Figure 5B and Table 1). St-3 (located between Ulleung Island and Dokdo) environmental parameters (Figure 5A) showed a fluctuating environment, which could be the reason for the strong wind (Yamada et al., 2004; Baek et al., 2018) characterized by the highest DIN (6.60 µmol L−1), PO4 (0.34 µmol L−1), and SiO2 (7.02 µmol L−1) concentrations (Table 1). Temperature and salinity in Sts-4 and 5 were similar at 14.21–14.22°C and 33.86–33.89 PSU, respectively, with the lowest Chl-a concentration (0.38–0.54 µg L-1), indicating that these stations were influenced with surface water mass (Figure 5B, Table 1). The temperature-salinity diagram (Figure 5B) confirmed that the occurrence of UWE resulted in differences in physicochemical data from five stations and was categorized similarly within three groups: the UWE ring group (Sts-1 and 2, varied between inside and outside of the UWE ring), Ulleung Island and Dokdo (UID) station (St-3, affected by the geographical features between UID), and surface water group (Sts-4 and 5, affected by wintertime surface water).
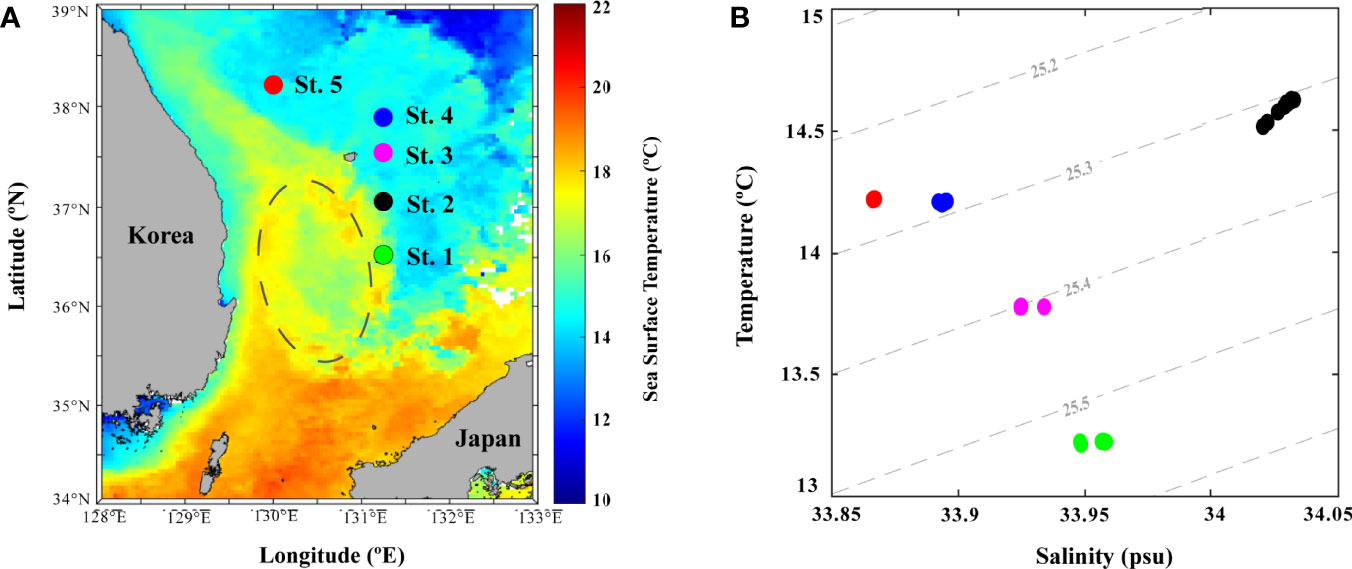
Figure 5 Physicochemical properties of the sampling stations in winter at Ulleung Basin (UB). (A) sea surface temperature gridding map using December monthly data obtained from Ocean Color; and (B) temperature–salinity diagram of 0–20-m depth seawater at five stations.
In total, 254,712 OTUs were identified in winter belonging to 10 genera in the phylum, 11 genera in the class, 23 genera in the order, and 33 genera at the family level (Supplementary Table S3A–D). Cluster analysis of the five samples with Bray–Curtis dissimilarity showed (Figure 6A) that bacterial communities were divided into three clusters with UWE ring group (Sts-1 and 2), surface water group (Sts-4 and 5), and UID station (St. 3). The NMDS plot, describing the dissimilarity of the surface bacterial community (Figure 6B), showed that Sts-1 and 2 were geographically distant from the other three stations, indicating that the bacterial community dissimilarity at Sts-1 and 2 were higher. The Shannon diversity indices of the five stations (Figure 6C) displayed that the lowest Shannon index with a value of 3.92 was observed at St-1, located at the UWE ring. A study reported that these low trophic level organisms (heterotrophic bacteria) dominate in high-nutrient UWE rings formed with coastal upwelling water (Hyun et al., 2009). In addition, Shannon diversity indices at Sts-1 and 2 (Figure 6C) showed a reverse pattern with the Chao1 richness index (Figure 6D) as the Shannon diversity index in St-1 was lower than that at St-2 (3.92 and 4.04, respectively), and the Chao1 species abundance index at St-1 was higher than that at St-2 (327.5 and 308.16, respectively). However, lower Shannon and Chao1 values were observed at St-4 (3.99 and 311.96) than at St-5 (4.24 and 367), unlike Sts-1 and 2 (Figures 6C, D). The highest Shannon and relatively high Chao1 index values (4.24 and 344.88) were observed at St-3, characterized by the highest DIN, PO4, and SiO2 concentrations (Table 1), indicating that this result is associated with nutrient concentration, which significantly affects bacterial niche structure (Cullen, 1991), shaping bacterial communities (Dai et al., 2022) with changes in community diversity and an increase in species abundance as nutrients increased (Gilbert et al., 2012).
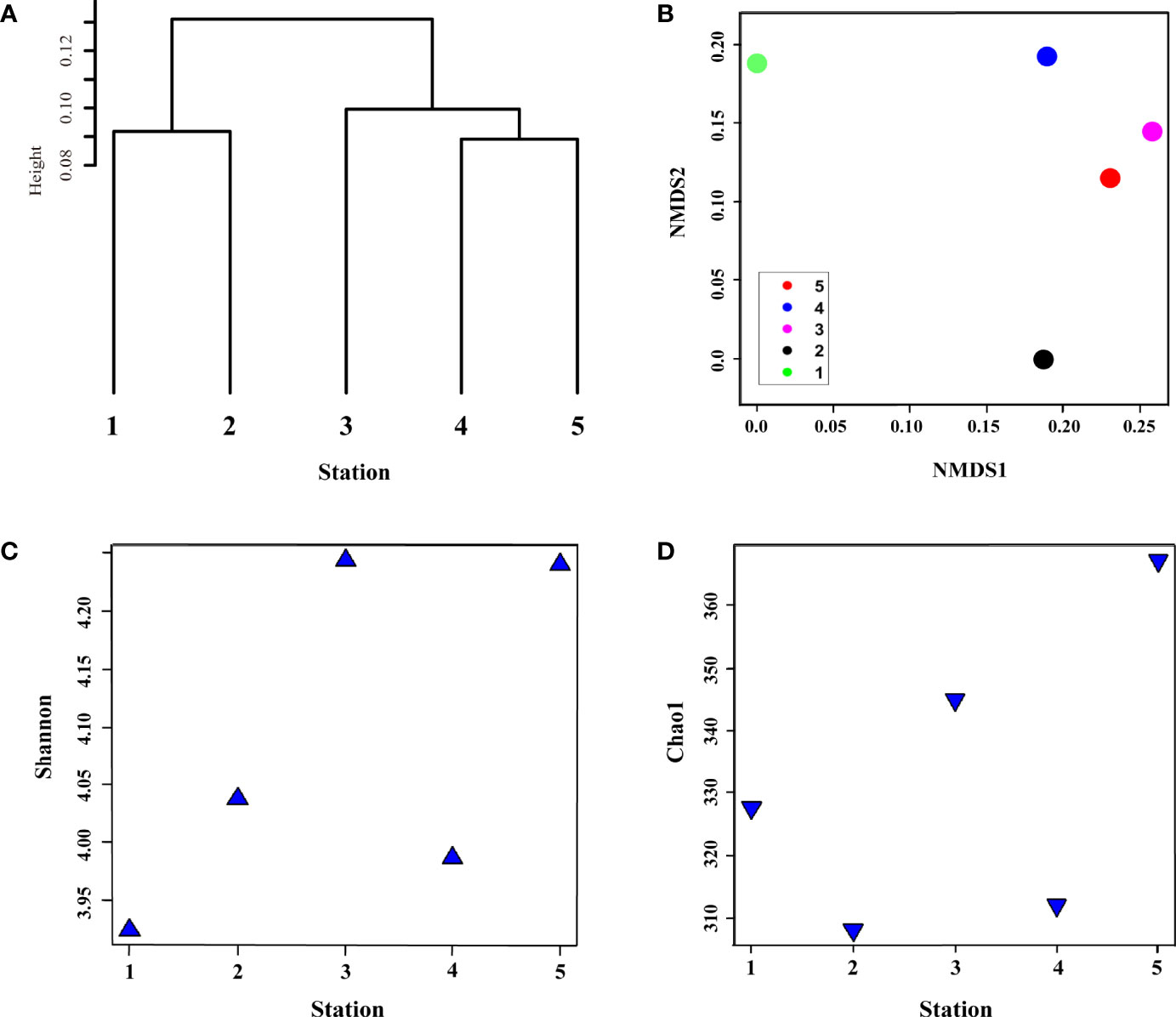
Figure 6 Surface bacterial community composition similarity and difference in abundance and diversity during winter. (A) clustering of samples at the operational taxonomic unit (OTU) level based on Bray–Curtis distance; (B) dissimilarity of bacterial community composition using unweighted-unifrac method, the alpha diversity of surface bacterial community; (C) diversity and (D) species richness using Shannon and Chao1 indices, respectively.
Along with these similarities and diversities (Figure 6), the relative abundance of the top 33 family level bacteria is shown in the heatmap (Figure 7A), and the relative abundance differences in each group were different. Specifically, the relative abundance of Enterococcaceae, Prochloraceae, Flavobacteriaceae, and Porticoccaceae was higher at St. 1, whereas Pseudoalteromonadaceae was abundant at St-2 (Figure 7A). However, Verrucomicrobiaceae showed a high relative abundance at Sts-1 and 2. In addition, Staphylococcaceae, Propionibacteriaceae, and Streptococcaceae were highly abundant at St. 4, whereas Planctomycetaceae, Vibrionaceae, Mycobacteriaceae, Rhodobiaceae, and Alcanivoracaceae were highly abundant at St-5 (Shewanellaceae was abundant in Sts-4 and 5, surface water group). However, there was a physicochemical difference between Sts-1 and 2 due to the occurrence of the UWE ring (Figure 5B), clustering (Figure 6A) and heatmap (Figure 7A) that showed that community composition similarity was high, and bacteria with high relative abundance in common at Sts-1 and 2 are similar. This is considered to be due to other biological factors (i.e., interactions with other microorganisms and phytoplankton community) not covered in the present study potentially influencing the bacterial community structure of the UWE ring group. However, eight bacteria, Hyphomonadaceae, Sphingomonadaceae, Balneolaceae, Lactobacillaceae, Alteromonadaceae, Bifidobacteriaceae, Methylobacteriaceae, and Nocardiaceae showed high relative abundance at St-3, consistent with the result of the high Shannon diversity index (Figure 6C). The db-RDA analysis revealed the physicochemical factors that played a determinant role in shaping surface bacterial community composition at the family level (relative abundance >0.01% in at least one of the samples; Figure 7B). In addition, Sts-1 and 2 were closely linked to Chl-a and salinity St-3 was related to DIN concentration, and Sts-4 and 5 were intertwined with temperature (Figure 7B). These results revealed significant differences in relative abundance (at the family level) in each of the three groups of bacterial communities, which were greatly influenced by environmental features developed by the occurrence of UWE.
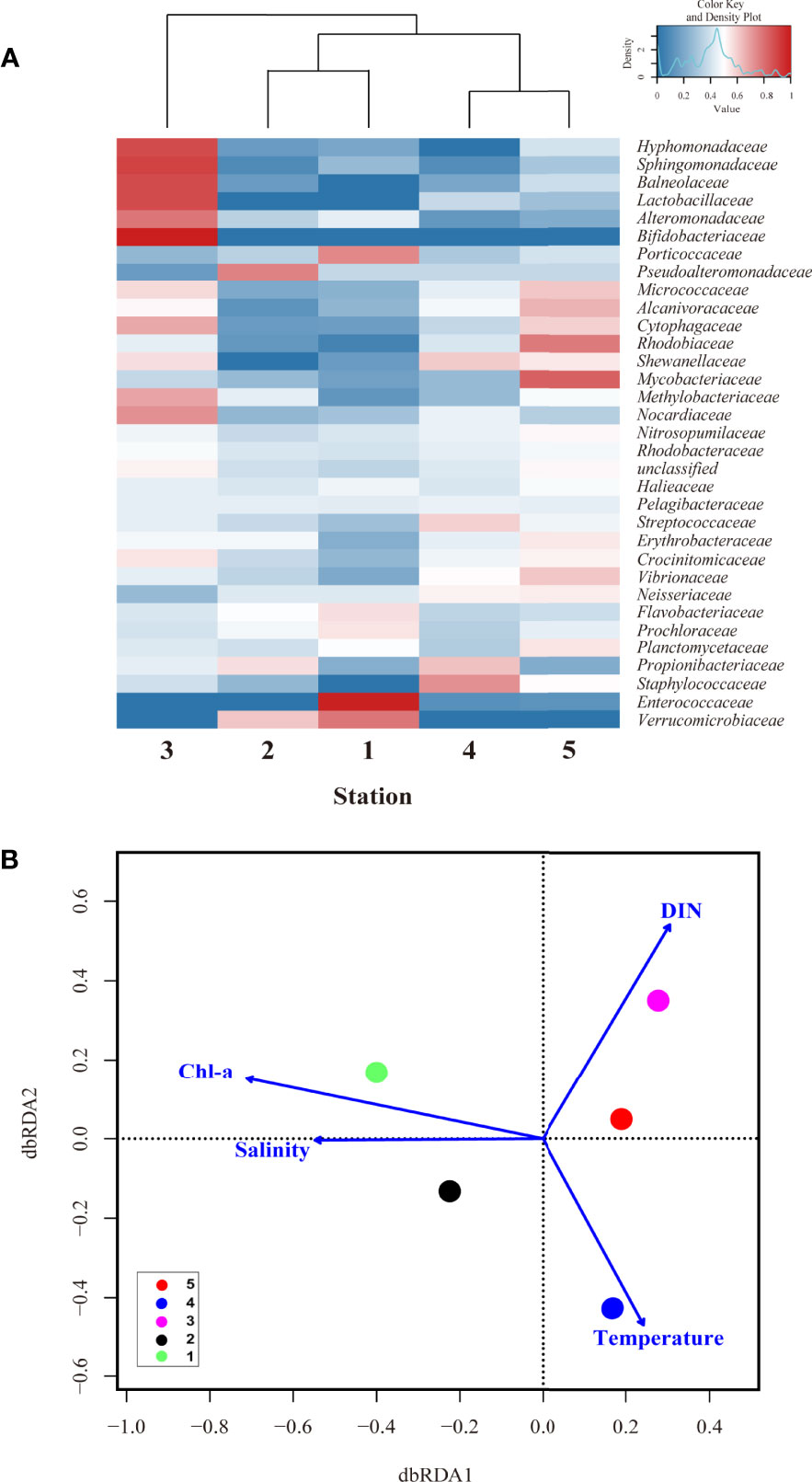
Figure 7 The distance-based redundancy analysis (dbrda) ordination plot and heatmap revealed that environmental conditions controlled the surface bacterial community composition in wintertime of UB. (A) hierarchical heatmap showing the relative abundance at family level (33 genera) in five stations. Color codes indicate the relative abundance, ranging from blue (low relative abundance) to red (high relative abundance); and (B) the dbrda ordination diagram of bacterial communities based on OTU at family level with related physical and biogeochemical factors, including temperature, salinity, dissolved inorganic nitrogen (DIN), and phosphate (PO4) concentration.
The specific influences of environmental characteristics on the bacteria at the family level (20 genera) showed evident differences in the relative abundance of the three groups (UWE, UID, and surface water) (Figure 8). Flavobacteriaceae (member of the order Flavobacteriales), observed to be dominant at Sts-1 and 2, was particularly highly abundant at St-1, located in a high Chl-a concentration UWE ring with a positive correlation with DO and DIN concentrations (r = 0.3 and 0.1). This genus was previously reported to be associated with phytoplankton and responds rapidly to nutrient-rich eddy circulation (Fuhrman et al., 2015). Similarly, a high abundance of Flavobacteriales in euphotic eddy-influenced samples has been observed in the northeastern South China Sea (McGillicuddy et al., 2007). Moreover, Prochloraceae and Verrucomicrobiaceae abundant at Sts-1 and 2 showed high correlations with salinity (r = 0.60) and Chl-a concentration (r = 0.67). In particular, Prochloraceae belonging to the genus Prochlorococcus are significantly influenced by warm or cold eddies (Jing and Liu, 2012; Wang et al., 2016). However, Sphingomonadaceae, Nocardiaceae, Lactobacillaceae, Hyphomonadaceae, Bifidobacteriaceae, Balneolaceae, and Methylobacteriaceae were positively correlated with high DIN, PO4, and SiO2 concentrations, which were abundant at St-3. Vibrionaceae, Erythrobacteraceae, Staphylococcaceae, Streptococcaceae, and Mycobacteriaceae were abundant at Sts-4 and 5 (affected by low-temperature surface water) and showed a positive correlation between temperature and relative abundance.
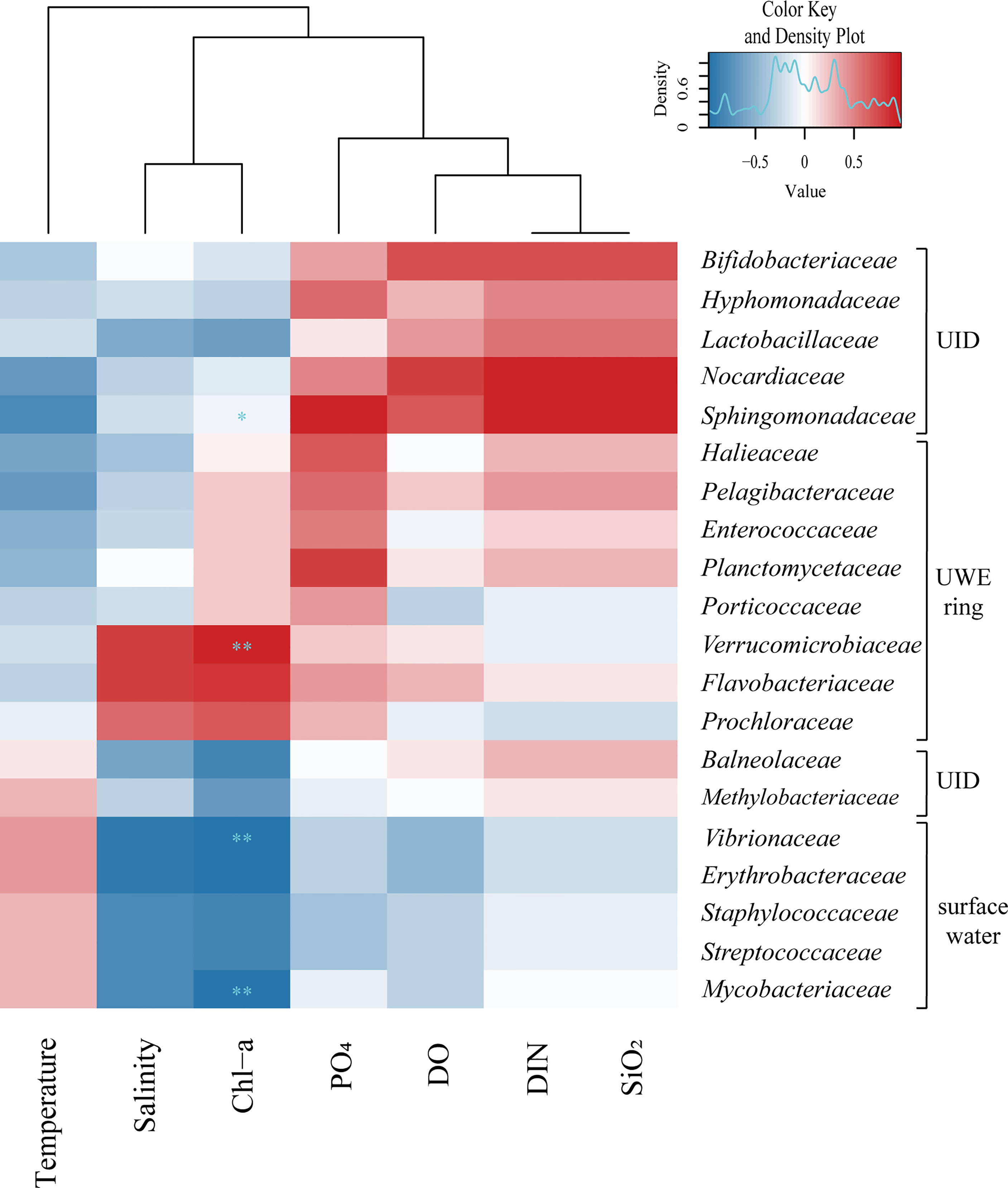
Figure 8 The 20 genera family level bacteria showed marked differences in relative abundance within three groups (i.e., UWE ring, UID, and surface water) classified as the occurrence of Ulleung warm eddy (UWE) during winter. Hierarchical heatmap using Spearman’s correlation coefficient revealed interrelation between the relative abundance of bacterial community (at the family level) and the physical and biogeochemical factors. The variations of surface bacterial community composition with 20 genera family level showed strong correlation to characteristics of environmental parameters created by physical eddy circulation. The symbols of * and ** indicate p value less than 0.05 and 0.01, respectively.
Eddies exert a major control on the phytoplankton productivity by nutrient flux (Huang et al., 2010; He et al., 2016; He et al., 2017; Liu et al., 2017), and on the associated bacterial community structure, diversity, and function (Harke et al., 2021). Eddy offshore shifts from large to small phytoplankton cells and higher to lower autotrophic biomass, has a complex and variable system regarding its hydrological structure and distribution of biological properties and has the highest organic matter concentration (Arístegui et al., 2004). Bacterial communities were also observed to be influenced by cyclonic and anticyclonic eddy circulation. For example, earlier studies noted that bacterial responses to mesoscale eddies (approximately 200km diameter) elevated their biomass and production inside a cold-core eddy compared to the surrounding waters (Thyssen et al., 2005; Zhang et al., 2009; Baltar et al., 2010). More specifically, a recent study by Devresse et al. (2021) on oligotrophic oceans observed that eddies influence the elevation of bacterial abundances and their metabolic activities, that is respiration rate, biomass production, and growth efficiency. Similar to these studies, present study results showed that the difference in bacterial composition at the UWE influenced (Sts-1 and 2) and not influenced stations. In the Sargasso Sea, prokaryotic heterotrophic production increases at the perimeter of the cyclonic eddy relative to the eddy center (Ewart et al., 2008). Likewise, considerable spatial heterogeneity in bacterial community composition was apparent inside and outside the cold-core cyclonic eddies of the subtropical marginal South China Sea (Zhang et al., 2011). Moreover, Li et al. (2014) demonstrated that bacterial communities derived from anticyclonic eddy sites displayed abnormal compositions compared to those of adjacent sites in the Northern South China Sea.
The five stations during winter were categorized based on their environmental features: the UWE ring group (Sts-1 and 2), distinguished from the Chl-a concentration that classifies the effects of the UWE ring with relatively high salinity values; the UID station (St-3), which showed high DIN, PO4, and SiO2 concentrations; and surface water group (Sts-4 and 5), influenced by wintertime surface water. These physicochemical features played a decisive role in the similarity, diversity, and distribution of the surface bacterial composition at each station related to the relative abundance of bacteria (Supplementary Table S4). Interestingly, we observed a clear distinction in the bacterial communities at the family-level Pelagibacteraceae, Halieaceae, Verrucomicrobiaceae, Porticoccaceae (belonging to Proteobacteria), Planctomycetaceae (belonging to Planctomycetes), Enterococcaceae (belonging to Firmicutes), Flavobacteriaceae (belonging to Bacteroidetes), and Prochloraceae (belonging to Cyanobacteria), which were more predominant in Sts-1 and 2. In contrast, Sphingomonadaceae, Hyphomonadaceae, Methylobacteriaceae (belonging to Alphaproteobacteria), Lactobacillaceae (belonging to Firmicutes), Bifidobacteriaceae, Nocardiaceae (belonging to Actinobacteria), and Balneolaceae (belonging to Balneolaeota) were predominantly found in St-3. Meanwhile, the strongest indicator species for Sts-4 and 5 were Vibrionaceae, Erythrobacteriaceae (belonging to Proteobacteria), Mycobacteriaceae (belonging to Actinobacteria), Staphylococcaceae, and Streptococcaceae (belonging to Firmicutes). Collectively, our data indicate that the physical circulation of the UWE during winter was influences the physicochemical conditions of the surrounding water and consequently alters the surface bacterial composition in the UB, ES.
Conclusions
In this study, bacterial communities in surface seawater at five stations in the UB of the ES were studied using a metagenomics approach, to elucidate the specific environmental drivers determining the bacterial diversity, abundance, and composition in this highly dynamic region. The results showed that seasonal physical processes, water mass mixing (in-summer), and eddy circulation (in-winter) led to the development unique physicochemical drivers, which significantly influenced the surface bacterial composition. Our results also indicated the relative abundances of bacteria showed specific responses to specific environmental drivers in summer and winter. To the best of our knowledge, this is the first attempt to elucidate the overall interpretation of the environmental characteristics and bacterial composition of this undertaken UB region, providing evidence for the bacterial community composition shaped by ecological proclivity. Overall, this study extends our knowledge of the bacterial community structure in ES, focusing on their seasonal differences with different ocean physical processes. However, future investigations should investigate more intensive bacterial community composition including their metabolic responses related to advection processes and other parameters known to greatly influence the microbial community (e.g., interaction with other diatoms, competition or symbiotic relationships, phytoplankton community, and day length) to understand the role of the microbial ecosystem in related to biogeochemical cycling in the East Sea.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA778576].
Author Contributions
H-RK, J-HL, J-HK, ST, and I-NK developed the concept and design of the article. H-RK, ST, and I-NK wrote the manuscript. All authors discussed the results and commented on the article. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by a grant from the National Institute of Fisheries Science (R2022035). This work was also supported by the National Research Foundation of Korea (NRF) grand funded by the Korea governments (MSIT) (No. 2022R1A2C100847).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the captain and crew members of the R/V Tamgu 3 for their endless support during the 2019 summer and winter cruises.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.841492/full#supplementary-material
References
Arístegui J., Barton E. D., Tett P., Montero M. F., García-Muñoz M., Basterretxea G., et al. (2004). Variability in Plankton Community Structure, Metabolism, and Vertical Carbon Fluxes Along an Upwelling Filament (Cape Juby, NW Africa). Prog. Oceanogr. 62, 95–113. doi: 10.1016/j.pocean.2004.07.004
Arrigo K. (2005). Marine Microorganisms and Global Nutrient Cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Baek S. H., Lee M., Kim Y. B. (2018). Spring Phytoplankton Community Response to an Episodic Windstorm Event in Oligotrophic Waters Offshore From the Ulleungdo and Dokdo Islands. J. Sea Res. 132, 1–14. doi: 10.1016/j.seares.2017.11.003
Baldwin A. J., Moss J. A., Pakulski J. D., Catala P., Joux F., Jeffrey W. H. (2005). Microbial Diversity in a Pacific Ocean Transect From the Arctic to Antarctic Circles. Aquat. Microb. Ecol. 41, 91–102. doi: 10.3354/ame041091
Baltar F., Aristegui J., Gasol J. M., Lekunberri I., Herndl G. J. (2010). Mesoscale Eddies: Hotspots of Prokaryotic Activity and Differential Community Structure in the Ocean. ISME J. 4, 975–988. doi: 10.1038/ismej.2010.33
Caporaso J.G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R PyNAST: A Flexible Tool for Aligning Sequences to a Template Alignment Bioinformatics (2010), 26 (2), 266–267. doi: 10.1093/bioinformatics/btp636
Celussi M., Bergamasco A., Cataletto B., Umani S. F., Negro P. D. (2010). Water Masses’ Bacterial Community Structure and Microbial Activities in the Ross Sea, Antarctica. Antarct. Sci. 22, 361–370. doi: 10.1017/S0954102010000192
Chang P. H., Isobe A. (2003). A Numerical Study on the Changjiang Diluted Water in the Yellow and East China Seas. J. Geophys. Res. Oceans 108(C9), 3299. doi: 10.1029/2002JC001749
Chen C., Beardsley R. C., Limeburner R., Kim K. (1994). Comparison of Winter and Summer Hydrographic Observations in the Yellow and East China Seas and Adjacent Kuroshio During 1986. Cont. Shelf Res. 14, 909–929. doi: 10.1016/0278-4343(94)90079-5
Cho H., Hyun J. H., You O. R., Kim M., Kim S. H., Choi D. L., et al. (2016). Microbial Community Structure Associated With Biogeochemical Processes in the Sulfate-Methane Transition Zone (SMTZ) of Gas-Hydrate-Bearing Sediment of the Ulleung Basin, East Sea. Geomicrobiol. J. 34 (3), 207–219. doi: 10.1080/01490451.2016.1159767
Cho Y. K., Kim K. (1996). Seasonal Variation of the East Korea Warm Current and its Relation With the Cold Water. La Mer 34, 172–182. doi: 10.1029/2006gl027625
Cho H. J., Moon C. H., Yang H. S., Kang W. B., Lee K. W. (1997). Regeneration Processes of Nutrients in the Polar Front Area of the East Sea III. Distribution Patterns of Water Masses and Nutrients in the Middle-Northern Last Sea of Korea in Octobe. J. Korean Fish. Soc 30, 393–407.
Chung C. C., Gong G. C., Huang C. Y., Lin J. Y., Lin Y. C. (2015). Changes in the Synechococcus Assemblage Composition at the Surface of the East China Sea Due to Flooding of the Changjiang River. Microb. Ecol. 70, 677–688. doi: 10.1007/s00248-015-0608-5
Cullen J. J. (1991). Hypothesis to Explain High-Nutrient Conditions in the Open Sea. Limnol. Oceanogr. 36, 1578–1590. doi: 10.4319/lo.1991.36.8.1578
Dai T., Wen D., Bates C. T., Wu L., Guo X., Liu S., et al. (2022). Nutrient Supply Controls the Linkage Between Species Abundance and Ecological Interactions in Marine Bacterial Communities. Nat. Commun. 13, 175. doi: 10.1038/s41467-021-27857-6
Devresse Q., Becker K. W., Engel A. (2021). Organic Matter Transport by Cyclonic Eddies Formed in Eastern Boundary Upwelling System Supports Heterotrophy in the Open Oligotrophic Ocean. EGU 21-9818. EGU General Assembly 2021
Dinasquet J., Richert I., Logares R., Yager P., Bertilsson S., Riemann L. (2017). Mixing of Water Masses Caused by a Drifting Iceberg Affects Bacterial Activity, Community Composition and Substrate Utilization Capability in the Southern Ocean. Environ. Microl. 19, 2453–2467. doi: 10.1111/1462-2920.13769
Ewart C. S., Meyers M. K., Wallner E. R., McGillicuddy D. C., Carlson C. A. (2008). Microbial Dynamics in Cyclonic and Anticyclonic Mode-Water Eddies in the Northwestern Sargasso Sea. Deep-Sea Res. II 55, 1334–1347. doi: 10.1016/j.dsr2.2008.02.013
Falkowski P. G., Barber R. T., Smetacek V. (1998). Biogeochemical Controls and Feedbacks on Ocean Primary Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science 200, 200–206. doi: 10.1126/science.281.5374.200
Falkowsky P. G., Ziemann D. A., Kolber D. A., Bienfang P. K. (1991). Role of Eddy Pumping in Enhancing Primary Production in the Ocean. Nature 352, 55–58. doi: 10.1038/352055a0
Fuhrman J., Cram J., Needham D. (2015). Marine Microbial Community Dynamics and Their Ecological Interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Galand P. E., Potvin M., Casamayor E. O., Lovejoy C. (2010). Hydrography Shapes Bacterial Biogeography of the Deep Arctic Ocean. ISME J. 4, 564–576. doi: 10.1038/ismej.2009.134
Giebel H. A., Brinkhoff T., Zwisler W., Selje N., Simon M. (2009). Distribution of Roseobacter RCA and SAR11 Lineages and Distinct Bacterial Communities From the Subtropics to the Southern Ocean. Environ. Microbiol. 11, 2164–2178. doi: 10.1111/j.1462-2920.2009.01942.x
Gilbert J. A., Steele J. A., Caporaso J. G., Steinbrück L., Reeder J., Temperton B., et al. (2012). Defining Seasonal Marine Microbial Community Dynamics. ISME J. 6, 298–308. doi: 10.1038/ismej.2011.107
Harke M. J., Frischkorn K. R., Hennon G. M., Haley S. T., Barone B., Karl D. M., et al. (2021). Microbial Community Transcriptional Patterns Vary in Response to Mesoscale Forcing in the North Pacific Subtropical Gyre. Environ. Microbiol. 23, 4807–4822. doi: 10.1111/1462-2920.15677
Henriques I. S., Alves A., Tacão M., Almeida A., Cunha A., Correia A. (2006). Seasonal and Spatial Variability of Free-Living Bacterial Community Composition Along an Estuarine Gradient (Ria De Aveiro, Portugal). Estuar. Coast. Shelf Sci. 68, 139–148. doi: 10.1016/j.ecss.2006.01.015
Herlemann D. P. R., Labrenz M., Jürgens K., Bertilsson S., Waniek J. J., Andersson A. F. (2011). Transitions in Bacterial Communities Along the 2000km Salinity Gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
He Q., Zhan H., Cai S., Li Z. (2016). Eddy Effects on Surface Chlorophyll in the Northern South China Sea: Mechanism Investigation and Temporal Variability Analysis. Deep-Sea Res. I: Oceanogr. Res. Pap. 112, 25–36. doi: 10.1016/j.dsr.2016.03.004
He Q., Zhan H., Shuai Y., Cai S., Li Q. P., Huang G., et al. (2017). Phytoplankton Bloom Triggered by an Anticyclonic Eddy: The Combined Effect of Eddy-Ekman Pumping and Winter Mixing. J. Geophys. Res.: Oceans 112, 4886–4907. doi: 10.1002/2017JC012763
Holland S. M. (2008) Non-Metric Multidimensional Scaling (MDS). Available at: http://strata.uga.edu/software/pdf/mdsTutorial.pdf.
Huang B., Hu J., Xu H., Cao Z., Wang D. (2010). Phytoplankton Community at Warm Eddies in the Northern South China Sea in Winter 2003/2004. Deep-Sea Res. II: Top. Stud. Oceanogr. 57, 1792–1798. doi: 10.1016/j.dsr2.2010.04.005
Hydes D. J., Aoyama M., Aminot A., Bakker K., Becker S., Coverly S., et al. (2010). “Determination of Dissolved Nutrients (N, P, Si) in Seawater With High Precision and Inter-Comparability Using Gas-Segmented Continuous Flow Analysers,” in IOCCP Report No. 14, Version 1. UNESCO/IOC.
Hyun J. H., Kim D., Shin C. W., Noh J. H., Yang E. J., Mok J. S., et al. (2009). Enhanced Phytoplankton and Bacterioplankton Production Coupled to Coastal Upwelling and an Anticyclonic Eddy in the Ulleung Basin, East Sea. Aquat. Microb. Ecol. 54, 45–54. doi: 10.3354/ame01280
Ichinomiya M., Yamada K., Nakagawa Y., Nishino Y., Kasai H., Kuwata A. (2019). Parmales Abundance and Species Composition in the Waters Surrounding Hokkaido, North Japan. Polar Sci. 19, 130–136. doi: 10.1016/j.polar.2018.08.001
Ichiye T. (1984). Some Problems of Circulation and Hydrography of the Japan Sea and the Tsushima Current. Elsevier Oceanogr. Ser. 39, 15–54. doi: 10.1016/S0422-9894(08)70289-7
Jeong I. S., Cho J. C., Bahk J. J., Hyun S. M., Kwon K. K., Lee J. H., et al. (2010). “Vertical Profile of Bacterial Community in the Sediment of UB: Implication of the Presence of Methane-Driven Community,” in Microorganisms in Industry and Environment. 219–226. World Scientific, Lisbon, Portugal. doi: 10.1142/9789814322119_0049
Jing H. M., Liu H. B. (2012). Phylogenetic Composition of Prochlorococcus and Synechococcus in Cold Eddies of the South China Sea. Aquat. Microb. Ecol. 65, 207–209. doi: 10.3354/ame01546
Joo H. T., Park J. W., Son S. H., Noh J. H., Jeong J. Y., Kwak J. H., et al. (2014). Long-Term Annual Primary Production in the Ulleung Basin as a Biological Hot Spot in the East/Japan Sea. J. Geophys. Res. Oceans 119, 3002–3011. doi: 10.1002/2014JC009862
Kataoka T., Hodoki Y., Suzuki K., Saito H., Higashi S. (2009). Tempo-Spatial Patterns of Bacterial Community Composition in the Western North Pacific Ocean. J. Mar. Syst. 77, 197–207. doi: 10.1016/j.jmarsys.2008.12.006
Kim C. S., Cho Y. K., Seo G. H., Choi B. J., Jung K. T., Lee B. G. (2014). Interannual Variation of Freshwater Transport and its Causes in the Korea Strait: A Modeling Study. J. Mar. Syst. 132, 66–74. doi: 10.1016/j.jmarsys.2014.01.007
Kim K., Kim K. R., Kim Y. G., Cho Y. K., Chung J. Y., Choi B. H., et al. (1996). New Findings From CREAMS Observations: Water Masses and Eddies in the ES. J. Korean Fish. Soc 31, 155–163.
Kim I. N., Lee T. S. (2004). Summer Hydrographic Features of the East Sea Analyzed by the Optimum Multiparameter Method. Ocean Polar Res. 26, 581–594. doi: 10.4217/OPR.2004.26.4.581
Kim D., Yang E. J., Kim K. H., Shin C.-W., Park J., Yoo S., et al. (2012). Impact of an Anticyclonic Eddy on the Summer Nutrient and Chlorophyll a Distributions in the Ulleung Basin, East Sea (Japan Sea). ICES Mar. Sci. Symp. 69, 23–29. doi: 10.1093/icesjms/fsr178
Koskinen K., Hultman J., Paulin L., Auvinen P., Kankaanpää H. (2011). Spatially Differing Bacterial Communities in Water Columns of the Northern Baltic Sea. FEMS Microbiol. Ecol. 75, 99–110. doi: 10.1111/j.1574-6941.2010.00987.x
Kruskal J. B. (1964). Multidimensional Scaling by Optimizing Goodness of Fit to a Nonmetric Hypothesis. Psychometrika 29, 1–27. doi: 10.1007/BF02289565
Laque T., Farjalla V. F., Rosado A. S., Esteves F. A. (2010). Spatiotemporal Variation of Bacterial Community Composition and Possible Controlling Factors in Tropical Shallow Lagoons. Microb. Ecol. 59, 819–829. doi: 10.1007/s00248-010-9642-5
Lee J. K., Choi K. H. (2021). Bacterial Communities From the Water Column and the Surface Sediments Along a Transect in the East Sea. J. Korean Mar. Life Sci. 6, 9–22. doi: 10.23005/KSMLS.2012.6.1.9
Lee J. Y., Kang D. J., Kim I. N., Rho T., Lee T., Kang C. K., et al. (2009). Spatial and Temporal Variability in the Pelagic Ecosystem of the East Sea (Sea of Japan): A Review. J. Mar. Syst. 78, 288–300. doi: 10.1016/j.jmarsys.2009.02.013
Lee J. W., Kwon K. K., Azizi A., Oh H. M., Kim W., Bahk J. J., et al. (2013). Microbial Community Structures of Methane Hydrate-Bearing Sediments in the Ulleung Basin, East Sea of Korea. Mar. Pet. Geol. 47, 136–146. doi: 10.1016/j.marpetgeo.2013.06.002
Li Y., Jing H., Xia X., Cheung S., Suzuki K., Liu H. (2018). Metagenomic Insights Into the Microbial Community and Nutrient Cycling in the Western Subarctic Pacific Ocean. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00623
Li J., Li N., Li F., Zou T., Yu S., Wang Y., et al. (2014). Spatial Diversity of Bacterioplankton Communities in Surface Water of Northern South China Sea. PloS One 9 (11), e113014. doi: 10.1371/journal.pone.0113014
Liu F., Tang S., Huang R. X., Yin K. (2017). The Asymmetric Distribution of Phytoplankton in Anticyclonic Eddies in the Western South China Sea. Deep-Sea Res. I: Oceanogr. Res. Pap. 120, 29–38. doi: 10.1016/j.dsr.2016.12.010
Lu Y., Zhang Y., Wang J., Zhang M., Wu Y., Xiao X., et al. (2021). Dynamics in Bacterial Community Affected by Mesoscale Eddies in the Northern Slope of the South China Sea. Microb. Ecol 83, 823–36. doi: 10.1007/s00248-021-01816-6
Mann K. H., Lazier J. R. N. (2006). Dynamics of Marine Ecosystems: Biological-Physical Interactions in the Oceans, 3rd ed. John Wiley and Sons.
McGillicuddy D. J., Anderson L. A., Bates N. R., Bibby T., Buesseler K. O., Carlson C. A., et al. (2007). Eddy/wind Interactions Stimulate Extraordinary Mid-Ocean Plankton Blooms. Science 316, 1021–1026. doi: 10.1126/science.1136256
Moon J. H., Pang I. C., Yoon J. H. (2009). Response of the Changjiang Diluted Water Around Jeju Island to External Forcings: A Modeling Study of 2002 and 2006. Cont. Shelf Res. 29, 1549–1564. doi: 10.1016/j.csr.2009.04.007
Morán X. A. G., Taupier-Letage I., Vázquez-Domínguez E., Ruiz S., Arin L., Raimbault P., et al. (2001). Physical-Biological Coupling in the Algerian Basin (SW Mediterranean): Influence of Mesoscale Instabilities on the Biomass and Production of Phytoplankton and Bacterioplankton. Deep Sea Res. I 48, 405–437. doi: 10.1016/S0967-0637(00)00042-X
Park C. K. (1978). Chemical Oceanographic Aspect of the Cole Water Mass in Offshore of the East Coast of Korea. J. Korean Fish. Soc 11, 49–54.
Parsons T. R., Maika Y., Lalli C. M. (1984). A Manual of Chemical and Biological Methods for Seawater Analysis Vol. 173 (Pergamon Press, Elsevier).
Pinhassi J., Gomez-Consarnau L., Alonso-Saez L., Sala M. M., Vidal M., Pedrós-Alió C., et al. (2006). Seasonal Changes in Bacterioplankton Nutrient Limitation and Their Effects on Bacterial Community Composition in the NW Mediterranean Sea. Aquat. Microb. Ecol. 44, 241–252. doi: 10.3354/ame044241
Rink B., Gruner N., Brinkhoff T., Ziegelmuller K., Simon M. (2011). Regional Patterns of Bacterial Community Composition and Biogeochemical Properties in the Southern North Sea. Aquat. Microb. Ecol. 63, 207–222. doi: 10.3354/ame01493
Senjyu T., Enomoto H., Matsuno T., Matsui S. (2006). Interannual Salinity Variations in the Tsushima Strait and its Relation to the Changjiang Discharge. J. Oceanogr. 62, 681–692. doi: 10.1007/s10872-006-0086-y
Shade A., Jones S. E., McMahon K. (2008). The Influence of Habitat Heterogeneity on Freshwater Bacterial Community Composition and Dynamics. Environ. Microbiol. 10, 1057–1067. doi: 10.1111/j.1462-2920.2007.01527.x
Shade A., Kent A. D., Jones S. E., Newton R. J., Triplett E. W., McMahon K. (2007). Interannual Dynamics and Phenology of Bacterial Communities in a Eutrophic Lake. Limnol. Oceanol. 52, 487–494. doi: 10.4319/lo.2007.52.2.0487
Shepard R. N. (1962). The Analysis Proximities: Multidimensional Scaling With an Unknown Distance Function, I. Psychometrika 27, 125–140. doi: 10.1007/BF02289630
Shin H. R., Kim I., Kim D., Kim C. H., Kang B., Lee E. (2019). Physical Characteristics and Classification of the Ulleung Warm Eddy in the East Sea (Japan Sea). J. Korean Soc. Oceanogr. 24, 298–317. doi: 10.7850/jkso.2019.24.2.298
Shin J. H., Nam J. H., Lee J. W., Lee D. H. (2014). Characteristics of Microbial Community Structures of the Methane Hydrate Sediments in the Ulleung Basin, East Sea of Korea. Korean J. Microbiol. 50, 191–200. doi: 10.7845/kjm.2014.4033
Thyssen M., Lefevre D., Caniaux G., Ras J., Fernandex C. I., Denis M. (2005). Spatial Distribution of Heterotrophic Bacteria in the Northeast Atlantic (POMME Study Area) During Spring 2001. J. Geophys. Res. 110, C07S16. doi: 10.1029/2004JC002670
Venkatachalam S., Ansorge I. J., Mendes A., Melato L. I., Matcher G. F., Dorrington R. A. (2017). A Pivotal Role for Ocean Eddies in the Distribution of Microbial Communities Across the Antarctic Circumpolar Current. PloS One 12 (8), e0183400. doi: 10.1371/journal.pone.0183400
Wang J., Tan Y., Huang L., Ke Z., Tan J., Hu Z., et al. (2016). Response of Picophytoplankton to a Warm Eddy in the Northern South China Sea. Oceanol. Hydrobiol. Stud. 45, 145–158. doi: 10.1515/ohs-2016-0014
Winter C., Hein T., Kavka G., Mach R. L., Farnleitner A. H. (2007). Longitudinal Changes in the Bacterial Community Composition of the Danube River: A Whole-River Approach. ASM J. 73, 421–431. doi: 10.1128/AEM.01849-06
Yamada K., Ishizak J., Yoo S., Kim H., Chiba S. (2004). Seasonal and Interannual Variability of Sea Surface Chlorophyll a Concentration in the Japan/East Sea (JES). Prog. Oceanogr. 61, 193–211. doi: 10.1016/j.pocean.2004.06.001
Yang H. S., Kim S. S., Kang C. G., Cho K. D. (1991). A Study on Sea Water and Ocean Current in the Sea Adjacent to Korea Peninsula -III. Chemical Characteristics of Water Masses in the Polar Front Area of the Central Korean East Sea-. J. Korean Fish. Soc 24, 185–192.
Yokokawa T., Corte D. D., Sintes E., Herndl G. J. (2010). Spatial Patterns of Bacterial Abundance, Activity and Community Composition in Relation to Water Masses in the Eastern Mediterranean Sea. Aquat. Microb. Ecol. 59, 185–195. doi: 10.3354/ame01393
Zehr J. P., Kudela R. M. (2011). Nitrogen Cycle of the Open Ocean: From Genes to Ecosystems. Ann. Rev. Mar. Sci. 3, 197–225. doi: 10.1146/annurev-marine-120709-142819
Zhang Y., Jiao N., Sun Z., Hu A., Zheng Q. (2011). Phylogenetic Diversity of Bacterial Communities in South China Sea Mesoscale Cyclonic Eddy Perturbations. Res. Microbiol. 162, 320–329. doi: 10.1016/j.resmic.2010.12.006
Zhang Y., Sintes E., Chen J. N., Zhang Y., Dai M., Jiao N., et al. (2009). Role of Mesoscale Cyclonic Eddies in the Distribution and Activity of Archaea and Bacteria in the South China Sea. Aquat. Microb. Ecol. 56, 65–79. doi: 10.3354/ame01324
Keywords: physical process, water mass mixing, Eddy circulation, metagenomics, bacterial community composition, Ulleung Basin, East Sea
Citation: Kim H-R, Lim J-H, Kim J-H, Thangaraj S and Kim I-N (2022) Physical Process Controlling the Surface Bacterial Community Composition in the Ulleung Basin of East Sea. Front. Mar. Sci. 9:841492. doi: 10.3389/fmars.2022.841492
Received: 22 December 2021; Accepted: 09 May 2022;
Published: 09 June 2022.
Edited by:
Jie Xu, University of Macau, ChinaReviewed by:
Hangun Kim, Sunchon National University, South KoreaZuriñe Baña, University of the Basque Country, Spain
Copyright © 2022 Kim, Lim, Kim, Thangaraj and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Il-Nam Kim, aWxuYW1raW1AaW51LmFjLmty
 Hyo-Ryeon Kim
Hyo-Ryeon Kim Jae-Hyun Lim
Jae-Hyun Lim Ju-Hyoung Kim
Ju-Hyoung Kim Satheeswaran Thangaraj
Satheeswaran Thangaraj Il-Nam Kim
Il-Nam Kim