- 1Natural Sciences and Science Education, National Institute of Education, Nanyang Technological University, Singapore, Singapore
- 2Department of Biological Sciences, National University of Singapore, Singapore, Singapore
Giant clams harbor dinoflagellates generally of the three genera (Symbiodinium, Cladocopium, and Durusdinium) of phototrophic Symbiodiniaceae. Coccoid dinoflagellates (alias zooxanthellae) are found mainly inside zooxanthellal tubules located in the colorful outer mantle. The symbionts need to obtain carbon, nitrogen and phosphorus from the host for growth and metabolism. The host can absorb exogenous ammonia through the ctenidium and assimilate it into glutamine. Although the host does not normally excrete ammonia, its hemolymph contains only low concentrations of ammonia, indicating that the symbionts can absorb and recycle the ammonia produced metabolically by the host. In this study, we had obtained from the outer mantle of the giant clam, Tridacna squamosa, three major ammonium transporter 2 (AMT2) sequences, one each for Symbiodinium spp. (Symb-AMT2), Cladocopium spp. (Clad-AMT2), and Durusdinium spp. (Duru-AMT2), which comprised 1341 bp, 1308 bp, and 1296 bp, respectively. The respective deduced amino acid sequences contained 447 (~ 46.5 kDa), 436 (~ 45.5 kDa), and 432 (~ 45.0 kDa) residues. Phenogramic and sequence similarity analyses confirmed that these sequences were derived from dinoflagellates. Zooxanthellae-AMT2 (Zoox-AMT2), which represented comprehensively AMT2 of Symbiodinium spp., Cladocopium spp., and Durusdinium spp. was localized at the dinoflagellates’ plasma membranes, indicating that it could partake in the absorption of ammonia from the luminal fluid of the zooxanthellal tubules. Zoox-AMT2 expression was detected in the outer mantle, inner mantle, foot muscle, hepatopancreas and ctenidium of T. squamosa, indicating that the coccoid dinoflagellates residing in all five organs had the potential of ammonia absorption. The outer mantle had the highest transcript level of Zoox-AMT2, and illumination upregulated the protein abundance of Zoox-AMT2 therein. Therefore, it can be deduced that the coccoid dinoflagellates residing in the outer mantle could augment the potential of ammonia absorption in alignment with photosynthesis as the assimilation of ammonia required an increased supply of carbon chains.
Introduction
Nitrogen (N) is crucial to all living organisms, as it is a basic element of amino acids, proteins and nucleic acids. The degradation of nitrogenous compounds in aqueous media produces ammonia comprising molecular NH3 and cationic NH4+, the proportion of which varies according to pH. As the reaction, NH4+ + H2O ↔ NH3 + H3O+, has a pK of ~9.5, NH4+ is the major species of dissolved ammonia at physiological pH (~ 7.2-7.4; Ip and Chew, 2010). In this report, ammonia refers to both NH3 and cationic NH4+. In general, ammonia is the major nitrogenous waste excreted by aquatic animals because ammonia is toxic (Ip and Chew, 2010; Chew and Ip, 2014). However, many reef animals in the tropics, including symbiotic sea anemones, scleractinian corals and giant clams, absorb ammonia from the ambient seawater, particularly during illumination (Muscatine et al., 1979; Wilkerson and Muscatine, 1984; Wilkerson and Trench, 1986; Miller and Yellowlees, 1989). This is because they need to supply N to the Symbiodineaceae living inside their bodies. Tropical waters are usually oligotrophic especially in atolls due to the lack of upwelling. To resolve the problems concerning nutrient availability, many tropical reef animals are phototrophic as they harbor coccoid Symbiodineaceae as symbionts (also called zooxanthellae).
Giant clams (Genus: Tridacna or Hippopus) can be found in Indo-Pacific coral reefs. They commonly harbor three genera of Symbiodineaceae, Symbiodinium spp., Cladocopium spp., and Durusdinium spp. although Gerakladium spp. has also been reported for Tridacna maxima from the French Polynesia (Hernawan, 2008; DeBoer et al., 2012; Ikeda et al., 2017; Lim et al., 2019; Pochon et al., 2019; Guibert et al., 2020). Symbiodineaceae can exist as motile flagellates or coccoid symbionts. The coccoid dinoflagellates reside as symbionts extracellularly inside the lumen of a branched tubular system that originates from the host’s digestive tract (Norton et al., 1992). The majority of symbionts are found inside the tertiary zooxanthellal tubules located in the colorful and extensible outer mantle unique to giant clams. The outer mantle contains iridocytes that can absorb harmful UV radiation (Rossbach et al., 2020) and deflect light of relevant wavelength to the photosynthesizing symbionts (Holt et al., 2014).
Symbiodineaceae express form II ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and conduct C3 photosynthesis (Streamer et al., 1993; Rowan et al., 1996; Mayfield et al., 2014). Thus, the phototrophic potential of coccoid Symbiodineaceae can be estimated based on the transcript level of form II RuBisCO (rbcII). Recently, Poo et al. (2020) designed a set of comprehensive quantitative real-time PCR primers to determine the transcript level of Zoox-rbcII, which consisted of rbcII of Symbiodinium spp. (Symb-rbcII), Cladocopium spp. (Clad-rbcII), and Durusdinium spp. (Duru-rbcII). They used Zoox-rbcII as a molecular indicator to compare the phototrophic potential of five organs in the fluted giant clam, Tridacna squamosa, and reported the highest phototrophic potential in the outer mantle as compared with the inner mantle, foot muscle, hepatopancreas and ctenidium. Photosynthesizing symbionts in the outer mantle release large quantities of photosynthate to the host so as to support the host’s energy and nutritional needs (Fisher et al., 1985; Klumpp et al., 1992), including light-enhanced shell formation (Ip et al., 2017a; Rossbach et al., 2019; Ip and Chew, 2021). In return, the host must supply nutrients containing essential elements (carbon, N and phosphorus) to the symbionts that have no direct access to the ambient seawater. N is needed by the coccoid dinoflagellates to synthesize important nitrogenous compounds including amino acids. Besides catering for their own needs, coccoid dinoflagellates must supply amino acids to the host for muscle synthesis, growth and reproduction. Notably, the host is incapable of synthesizing essential amino acids (Wang and Douglas, 1999) and must obtain them from the symbionts or through filter feeding. With light as the sole energy source, giant clams can grow in Millipore-filtered seawater for more than 10 months (Fitt and Trench, 1981). This confirms that the clam host can obtain all the nutrients needed for growth and survival, including essential amino acids, from the coccoid dinoflagellates (Klumpp and Griffiths, 1994).
Seawater contains NH4+ and NO3− as the major dissolved inorganic N, as well as dissolved organic N (Johansson and Wedborg, 1980; Bronk et al., 2007). The host can absorb inorganic (Fitt et al., 1993a; Ip et al., 2020a) and organic N (Chan et al., 2018; Chan et al., 2019) from the ambient seawater and supply them to the symbionts. As a result, the supplementation of seawater with ammonia augments the growth of juvenile giant clams (Summons et al., 1986; Belda et al., 1993; Fitt et al., 1993a) and enhances the division rate and density of symbionts (Belda et al., 1993; Fitt et al., 1993a). Giant clams absorb exogenous ammonia in light faster than in darkness (Wilkerson and Trench, 1986; Fitt et al., 1993a; Fitt et al., 1993b). The absorption of ammonia occurs mainly in the ctenidium (Rees et al., 1994; Shepherd et al., 1999), and the absorbed ammonia is assimilated into glutamine catalyzed by a ctenidial glutamine synthetase (GS) that displays light-enhanced gene and protein expression levels (Hiong et al., 2017). Hence, the host supplies exogenous ammonia in the form of glutamine-N to the symbionts. Nonetheless, the host constantly produces endogenous ammonia through the degradation of nitrogenous compounds. Yet, despite the lack of ammonia excretion (Muscatine and D’Elia, 1978), the ammonia concentrations in the hemolymph of giant clams remain low, and fluctuate diurnally with lower concentrations during illumination than in darkness (Fitt et al., 1995). Therefore, metabolic ammonia produced endogenously by the host must be effectively absorbed and assimilated by the coccoid dinoflagellates, especially during illumination. In fact, the coccoid dinoflagellates of T. squamosa can assimilate ammonia into glutamine because they express a novel cytosolic glutamine synthetase 1 (GS1) sequence that contains an extra segment characteristic of nucleotide diphosphate kinase (Fam et al., 2018). Notably, the gene and protein expression levels of GS1/GS1 are enhanced during illumination (Teh et al., 2021). Hence, it is logical to deduce that coccoid dinoflagellates could increase the uptake of ammonia from the host during illumination through a transporter that has not been identified.
Members of the ammonium transporter/methylamine permease/Rhesus family can be found in all domains of life, and they mediate the transport of ammonia across membranes (McDonald et al., 2012). Ammonium transporters (AMTs) are ubiquitous in plant, fungi and eubacteria, involving specifically in ammonia absorption (Peng and Huang, 2006). Based on their phylogenetic relationships, plant AMTs can be grouped into two subfamilies, AMT1 and AMT2 (Loqué and von Wirén, 2004; McDonald et al., 2012). AMT2 is more closely related to the AMT of prokaryotes than to plant AMT1 (Loqué and von Wirén, 2004; McDonald et al., 2012). Members of the AMT1 subfamily transport ionic NH4+ as NH3 + H+, and play important roles in high-affinity ammonia uptake at the micromolar level (Yuan et al., 2007; McDonald et al., 2012; Song et al., 2017). By contrast, members of the AMT2 subfamily transport molecular NH3 despite having high affinity for ammonia (Ganz et al., 2020). In Arabidopsis, AMT2;1 (AtAMT2;1) also contributes to low-affinity ammonia transport in the millimolar range (Giehl et al., 2017). Although the concentration of ammonia in the luminal fluid of the zooxanthellal tubules of giant clams is unknown, the ammonia concentration in the hemolymph ranges from ~ 10 µM to ~ 0.25 mM (Fitt et al., 1995). Therefore, the objective of this study was to obtain the full coding cDNA sequence of AMT2 from the coccoid dinoflagellates residing in the outer mantle of T. squamosa. While multiple AMT2 sequences were obtained because of the presence of multiple phylotypes of Symbiodinium, Cladocopium, and Durusdinium, only one major AMT2 sequence for each genus of dinoflagellate, named Symbiodinium-AMT2 (Symb-AMT2), Cladocopium-AMT2 (Clad-AMT2) and Durusdinium-AMT2 (Duru-AMT2), was presented in this report. Molecular characterization and phenogramic analysis of the three deduced amino acid sequences were conducted to verify their identity and dinoflagellate-origin. It was essential to confirm that the comprehensive AMT2 expressed by all three genera of dinoflagellates, named zooxanthellae-AMT2 (Zoox-AMT2), was expressed in the symbiont’s plasma membrane. Hence, an antibody that could react comprehensively with Zoox-AMT2 was designed and custom-made in order to conduct immunofluorescence microscopy. In addition, efforts were made to test the hypothesis that illumination could lead to increases in the gene expression level of Zoox-AMT2 and protein abundance of Zoox-AMT2 in the outer mantle so that ammonia absorption could be augmented in the symbionts during photosynthesis.
Besides containing substantial quantities of coccoid dinoflagellates, giant clams can expel viable dinoflagellates (Morishima et al., 2019) that can repopulate other bleached Symbiodiniaceae-bearing hosts such as scleractinian corals (Umeki et al., 2020). It is therefore important to understand the nutritional aspect of the giant clam-zooxanthellae association, which might provide insights into ways to enhance the growth of giant clams in the wild or in culture. In particular, the clam host absorbs multiple exogenous N-containing compounds, including ammonia, urea and NO3−, to benefit the symbionts, but the transporters involved in the absorption of these nitrogenous substrates in the coccoid dinoflagellates have not been elucidated. Results obtained on dinoflagellate-derived AMT2 would set a foundation for future studies on such transporters in coccoid dinoflagellates of giant clams and other symbiotic animals.
Materials and Methods
Giant Clams and Maintenance Conditions
Adult T. squamosa (575 ± 135 g) were imported directly from Vietnam through Xanh Tuoi Tropical Fish Co., Ltd (Ho Chi Minh City, Vietnam). They were packed individually in plastic bags and shipped to Singapore through air. The giant clams (n = 22) were housed in three glass tanks (L 92 cm × W 62 cm × H 62 cm) containing approximately 320 l of seawater in the Animal Holding Room of the National Institute of Education (Singapore). The salinity, pH, hardness, concentrations of ammonia, calcium, nitrate, and phosphate and the temperature of the seawater were maintained as described by Pang et al. (2021). Briefly, the water had a salinity of 30-32 with a temperature of 25.5-26.5°C. The pH was 8.2-8.4 while the water hardness was at 143-179 ppm. The total ammonia, calcium, nitrate, and phosphate contents were maintained at 0 ppm, 380-420 ppm, 0 ppm, and less than 0.28 ppm, respectively. As we had no information on the conditions of the giant clams’ natural habitat in Vietnam, the giant clams were illuminated with a light intensity that mimicked the irradiance received by T. squamosa in the Red Sea as reported by Jantzen et al. (2008). They were given a month to acclimatize to the above-mentioned conditions under a 12 h dark: 12 h light regimen without any food supply before any experiments were performed. Exemption from approval by the Nanyang Technological University Institutional Animal Care and Use Committee was granted for the use of giant clams in this study.
Experimental Conditions and Tissues Collection
After one month of acclimation to the 12 h dark: 12 h light regimen, four individuals of T. squamosa (control) were killed at the end of the 12-h period of darkness. Tissues samples of the five targeted organs namely the outer mantle, inner mantle, foot muscle, hepatopancreas and ctenidium (gill) were collected. These five organs were chosen as a previous study had confirmed that they contained a certain quantity of coccoid dinoflagellates based on transcript levels of Zoox-rbcII (Poo et al., 2020). In addition, tissue samples of the outer mantle were collected from another 12 individuals (n = 4 for each time point) that had been exposed to light for 3, 6 or 12 h during the 12 h light regimen. Prior to tissue sampling, giant clams were anaesthetized with 0.2% phenoxyethanol. Then the shell-valves were forced open and the adductor muscle was severed in order to excise the tissues. After excision, tissue samples were freeze-clamped in liquid N2 and stored in an ultralow freezer at −80°C until analysis. For immunofluorescence microscopy, tissue samples were dissected from the central region of the outer mantle of three separate individuals after exposure to darkness for 12 h. They (n = 3) were fixed in paraformaldehyde (3.7% in seawater). In addition, three individuals that had been exposed to darkness for 12 h were killed to obtain the outer mantle tissues for the isolation of coccoid dinoflagellates, which was used for the identification of the targeted band in the western blotting experiments.
Total RNA Extraction, cDNA Synthesis, PCR, Cloning, and RACE-PCR
The extraction of total RNA and the synthesis of cDNA were performed according to the methods described by Pang et al. (2021). A set of genus-comprehensive PCR primers (forward: 5’- CTTCTTCTATGGTGGCCTGGT -3’; reverse: 5’- GAAAGCCACACCAGCCAG -3’) was designed based on a total of nine AMT2 sequences with three from Symbiodinium databases (Shoguchi et al., 2018; Chen et al., 2020; González-Pech et al., 2021), three from Cladocopium databases (Davies et al., 2018; Shoguchi et al., 2018; Chen et al., 2020), and three from Durusdinium databases (Ladner et al., 2012; Rosic et al., 2015; Shoguchi et al., 2021) (Supplementary Table S1). They were used to obtain the partial sequences of AMT2 from Symbiodinium, Cladocopium, and Durusdinium in the outer mantle of T. squamosa. PCR was conducted following the method of Pang et al. (2021) using a 9902 Veriti 96-well thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA). The PCR product obtained was cloned with the pGEM®-T Easy Vector system II (Promega, Madison, WI, USA). Partial sequences were obtained from sixty clones that were picked randomly, and their identities were confirmed by comparison with AMT2 sequences available in multiple dinoflagellate databases. Using the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Mountain View, CA, USA), 5’ and 3’ RACE-PCR were performed to obtain the full coding sequence of the major Symb-AMT2, Clad-AMT2 and Duru-AMT2. The RACE primer set designed for Symb-AMT2 was 5’ RACE: 5’-GAAGGCGAGACCTGGAAGCGTGTCCC-3’, 3’ RACE: 5’- GGGACACGCTTCCAGGTCTCGCCTTC-3’. For Clad-AMT2, the RACE primer set consisted of 5’ RACE: 5’-CCAACGAACATGCCGAATCCAGTGGAAC-3’; 3’ RACE: 5’- GTGACTCCGGCCTCCCAGCTTTGG -3’. The RACE primer set for Duru-AMT2 comprised 5’ RACE: 5’-GTCATCTGGAAAGCTGCAAAACAAAGAC-3’; 3’ RACE: 5’- ACTCGGGGCTCCCTGGTCTTTGTT-3’.
Deduced Amino Acid Sequences
The ExPASy Proteomic server (http://web.expasy.org/translate/) was used to translate the three nucleotide sequences into amino acid sequences of Symb-AMT2, Clad-AMT2, and Duru-AMT2. The transmembrane regions (TM) and pore lining amino acid residues were deduced using TMpred provided by Expasy (https://embnet.vital-it.ch/software/TMPRED_form.html).
Phenogramic Analysis
A phenogramic analysis of Symb-AMT2, Clad-AMT2, and Duru-AMT2 together with AMT2 sequences obtained from various databases was performed to confirm their identities. Maximum Likelihood analysis using the program RaxML 8.2.5 was applied to generate the phenogram with 2000 bootstraps (Stamatakis, 2014). An analysis by the ModelGenerator v0.85 revealed LG + G + F as the best-fitting evolutionary model for AMT2 (Whelan and Goldman, 2001; Keane et al., 2006).
Quantitative Polymerase Chain Reaction
To quantify the transcript level of Zoox-AMT2, a set of forward (5’-GGGTTGTCTTTGGGTTCTC-3’) and reverse: (5’-CCCACATCTTCATGTCCA-3’) qPCR primers, which could react comprehensively with Zoox-AMT2, was designed based on a total of nine AMT2 sequences with three from Symbiodinium databases (Shoguchi et al., 2018; Chen et al., 2020; González-Pech et al., 2021), another three from Cladocopium databases (Levin et al., 2016; Davies et al., 2018; Shoguchi et al., 2018), and the final three from Durusdinium databases (Ladner et al., 2012; Rosic et al., 2015; Bellantuono et al., 2019) (Supplementary Table S2). For absolute quantification of the transcripts, plasmid clones of the amplicon were generated following the method of Hiong et al. (2017). The transcript level of Zoox-rbcII was quantified using the set of genus-comprehensive Zoox-rbcII qPCR primers designed by Poo et al. (2020). The amplification efficiencies of the qPCR primers for Zoox-AMT2 and Zoox-rbcII were 91.1% and 93.8%, respectively. qPCR was performed according to the methods of Pang et al. (2021) using a 96-well StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific). Two standard curves were constructed using two different plasmid clones, one generated for Zoox-AMT2 and another one for Zoox-rbcII, to determine the absolute copies of transcript. Results were expressed as copies of transcripts per ng of total RNA (dinoflagellate + host).
Although the transcript level of a gene in the tissue samples of non-symbiotic organisms is commonly expressed with reference to the total RNA in the same tissue of that organism, we expressed the transcript levels of Zoox-AMT2 with reference to the total RNA (dinoflagellate + host) in the outer mantle of T. squamosa in this study. As explained by Mani et al. (2021) and Teh et al. (2021), T. squamosa is a symbiotic animal representing an animal-dinoflagellate association, and its tissue samples, particularly the outer mantle, naturally consisted of a mixture of host tissues and coccoid dinoflagellates. Furthermore, the quantity of symbionts per gram tissue sample, and hence the quantity of dinoflagellate RNA, varies between the same organ of different individuals or between different organs of the same individual (Poo et al., 2020). As our objectives were to compare the expression levels of Zoox-AMT2 in five organs of T. squamosa as well as in the outer mantle of different individuals exposed to various periods of illumination, it would be inappropriate to express results of absolute quantification as copy number of Zoox-AMT2 transcripts per ng of total dinoflagellate RNA. Moreover, it was technically impracticable to quantify the dinoflagellate RNA separated from the host RNA, because changes in the transcript level (and also protein abundance) might occur during the isolation of dinoflagellates from the host tissues before RNA (and protein) extraction. To resolve the problem of variability in quantities of coccoid dinoflagellates, we quantified the transcript levels of Zoox-rbcII, which was expressed only by the coccoid dinoflagellates, in order to calculate the transcript ratio of Zoox-AMT2/Zoox-rbcII. As this ratio is independent of the total RNA (dinoflagellate + host), direct comparisons could be made between different organs concerning the potential of ammonia transport with reference to the phototrophic potential. Of note, results could not be presented according to the 2ΔΔCT method of relative quantification (Livak and Schmittgen, 2001), because the transcript levels of Zoox-rbcII varied among different organs of the same individual or in the outer mantle of different individuals exposed to experimental conditions (Poo et al., 2020; Poo et al., 2021) and did not satisfy the criteria of a reference gene.
Antibodies
As a sequence of AMT derived from the host has been obtained from the ctenidium of T. squamosa previously (Boo et al., 2018), it was essential to design a custom-made antibody that could differentiate the dinoflagellate AMT from the host AMT. Thus, we aligned multiple AMT sequences obtained from various Symbiodiniaceae databases (Supplementary Table S3) to select the highly conserved epitope sequence of YSFWTNLDMKNWD for the production of the genus-comprehensive anti-Zoox-AMT2 antibody outsourced to Genscript (Piscataway, NJ, USA). This epitope sequence corresponded to residues 103–115 of Symb-AMT2, 90–102 of Clad-AMT2 and Duru-AMT2 with similarity of 92.3%, 69.2%, and 69.2%, respectively. To confirm that the anti-Zoox-AMT2 antibody could specifically detect AMT2s of different phylotypes of Symbiodiniaceae, a NCBI BLAST was performed on the epitope sequence. The top 15 candidates obtained from the NCBI BLAST were all dinoflagellate AMTs, some of which had very small E-value (Table 1). Hence, results of immunofluorescence microscopy and western blotting obtained using this antibody should reflect specifically the cellular/subcellular localization and protein abundance of AMT2, respectively.
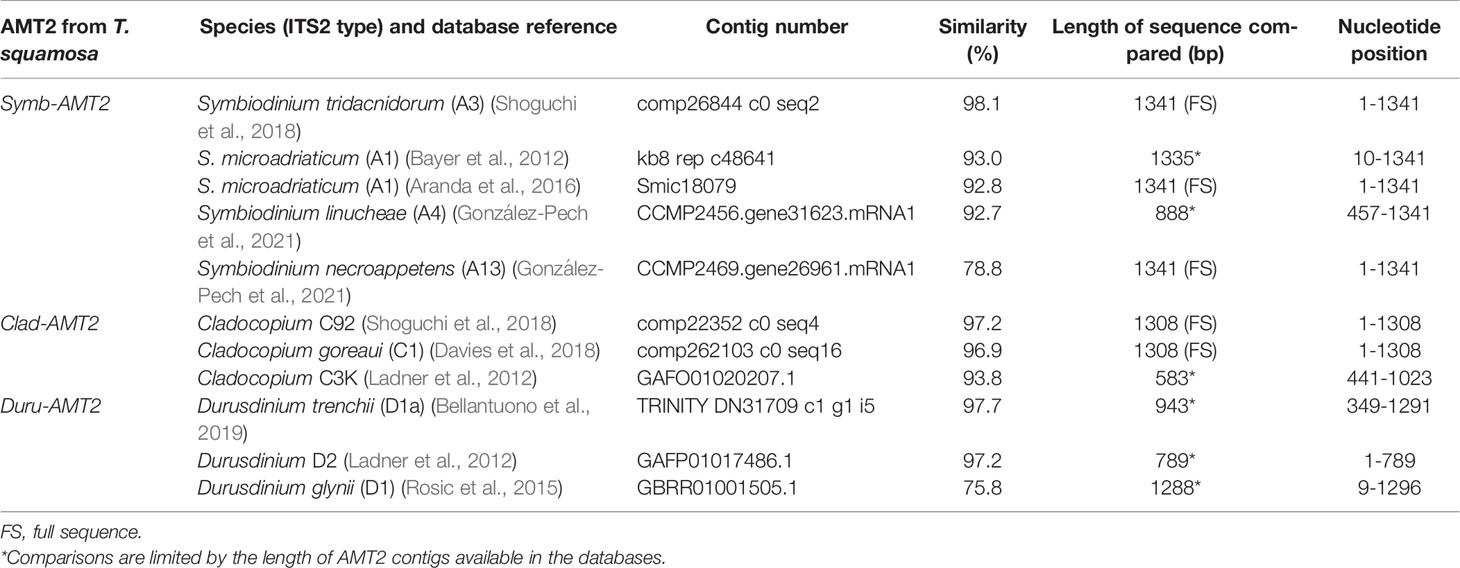
Table 1 A comparison of the nucleotide sequences of ammonium transporter 2 (AMT2) derived from Symbiodinium (Symb-AMT2), Cladocopium (Clad-AMT2) and Durusdinium (Duru-AMT2) of Tridacna squamosa with selected AMT2 contigs obtained from various dinoflagellate databases.
In addition, α-tubulin was selected as the reference protein, and an anti-α-tubulin antibody custom-made by Genscript based on the epitope of PKDVNAAVATIKTK was used to produce the anti- α-tubulin antibody. Mani et al. (2021) and Teh et al. (2021) had reported that this antibody could theoretically react with α-tubulin derived from both the clam host and coccoid dinoflagellates. As T. squamosa represents an animal-dinoflagellate association and its organs naturally consist of a mixture of host’s tissues and symbionts, it would be inappropriate to used dinoflagellate α-tubulin as the reference protein.
SDS-PAGE Electrophoresis and Western Blotting
In order to identify the band of interest and its dinoflagellate-origin, dinoflagellates were isolated from the outer mantle following the method of Poo et al. (2021). The outer mantle tissues from three individuals exposed to 12 h of darkness were excised and cut into smaller pieces. The tissue samples were homogenized twice, at 3200 rpm for 20 s, in 10 volumes of filtered seawater using an Ultra-Turrax T25 homogenizer fitted with an 18G shaft (Ika-Labortechnik, Staufen, Germany). The homogenate was centrifuged at 60 xg for 5 min at 25°C to separate the released dinoflagellates into the supernatant from the residual outer mantle tissues in the pellet. To obtain the remaining dinoflagellates from the pellet, the residual outer mantle tissues were resuspended in 10 volumes of filtered seawater and the homogenization and centrifugation process was repeated. The supernatants from the two rounds of homogenization were combined followed with centrifugation at 60 xg for 5 min at 25°C to obtain the dinoflagellate pellet.
The proteins from the outer mantle of T. squamosa or from the isolated dinoflagellates were extracted and their concentrations were determined according to the methods of Hiong et al. (2017) and Bradford (1976), respectively. The proteins were mixed with Laemmli’s reagent (Laemmli, 1970) and heated at 70°C for 15 min. An aliquot of sample from the outer mantle containing 100 μg protein was separated by SDS-PAGE using step gradient gels (4% stacking, 8% resolving). To confirm that the band of interest was derived from the symbiont and not the host, 20 µg of protein from the isolated dinoflagellates was also separated by SDS-PAGE. The separated proteins were then transferred onto Amersham™ Protran® nitrocellulose membranes (Cytiva, Marlborough, MA, USA). The Pierce Fast Western Blot kit, SuperSignal® West Pico Substrate (Thermo Fisher Scientific) was used to conduct western blotting. The membranes were incubated with the anti-Zoox-AMT2 antibody (1.25 µg ml-1) at 25°C for 1 h, and all subsequent steps were carried out according to the manufacturer’s instructions. Chemiluminescence was captured using the ChemiDoc XRS+ (Bio-Rad Laboratories, Hercules, CA, USA). ImageJ (version 1.50, NIH) was used to quantify the optical densities of the bands of interest. Calibration of the optical densities was carried out with the Stouffer R3705-1C, 37 step reflection scanner scale with increments of 0.05 (Stouffer Industries Inc., Mishawaka, IN, USA). The optical density of the Zoox-AMT2 band was normalized to that of the α-tubulin band in order to express the relative protein abundance of Zoox-AMT2 in the outer mantle. As the protein sample obtained from freshly isolated dinoflagellates was only used to confirm the origin of the Zoox-AMT2 band and not for quantification, the determination of reference protein was omitted.
Immunofluorescence Microscopy
The methods of Pang et al. (2021) were adopted to process the outer mantle samples for immunofluorescence microscopy. Briefly, samples were dehydrated and embedded in Paraplast Plus (Sigma-Aldrich Co.) before sectioning to 5 µm with a Leica RM2125 RTS microtome (Leica, Wetzlar, Germany). The sections were then mounted onto Menzel Gläser SuperFrost Plus Adhesion slides (Thermo Fisher Scientific). Sections were then deparaffinized prior to antigen retrieval by treatment with citraconic anhydrase (Nacalai Tesque, Kyoto, Japan) at 95°C for 5 min, followed by 1% SDS solution at 25°C for 10 min. The sections were then washed with TPBS (0.05% Tween-20, 10 mmol l-1 Na2HPO4, 1.8 mmol l-1 KH2PO4, 137 mmol l-1 NaCl, and 1.8 mmol l-1 KCl at pH 7.4). Sudan Black B (0.1%; Sigma-Aldrich Co.) was applied to the sections in order to reduce autofluorescence. Bovine serum albumin (1%) was used as the blocking agent. As stated in Pang et al. (2021), Signal Enhancer HIKARI Kit obtained from Nacalai Tesque was used to enhance the immunofluorescence. The concentration of the anti-Zoox-AMT2 antibody used for staining was 2.5 μg ml-1. An Olympus BX60 fluorescence microscope equipped with a DP73 CCD digital camera was used to examine the mounted sections according to the procedure of Pang et al. (2021). Adobe Photoshop CC (Adobe Systems, San Jose, CA) was used to process and overlay the photo-images.
Data Analysis
Statistical analyses were performed by the SPSS Statistics software v26 (IBM Corporation, Armonk, NY, USA), with results reported as means + SEM. Comparison among the means and the evaluation of homogeneity of the variance were analyzed by One-way Analysis of Variance (ANOVA) and Levene’s test, respectively. When equal variance prevailed, Tukey’s test was used as the post-hoc test. For unequal variance, the Dunnet’s T3 test was used. With a p-value of < 0.05, the difference between the means was regarded as significant. Results expressed as ratios were analyzed by the non-parametric Friedman test followed by the Wilcoxon Signed-Rank Test. Differences obtained through non-parametric analysis were regarded as statistically significant when the p-value was < 0.01 after Bonferroni adjustment.
Results
Nucleotide and Translated Amino Acid Sequences
The coding cDNA sequences of Symb-AMT2, Clad-AMT2, and Duru-AMT2 obtained from T. squamosa had been deposited into GenBank, and they were assigned the accession numbers of MW023800, MW023801 and MW023802, respectively. The respective lengths of the nucleotide sequences were 1341 bp, 1308 bp, and 1296 bp. Symb-AMT2 had the greatest sequence similarity (98.1%) with AMT2 of Symbiodinium tridacnidorum (ITS2 type A3; Shoguchi et al., 2018) while Clad-AMT2 shared the highest similarity (97.2%) with the AMT2 sequence of Cladocopium ITS2 type C92 (Shoguchi et al., 2018) (Table 1). On the other hand, Duru-AMT2 had the greatest similarity (97.7%) with the AMT2 sequence of Durusdinium trenchii (ITS2 type D1a; Bellantuono et al., 2019) (Table 1).
There were 447 (~ 46.5 kDa), 436 (~ 45.5 kDa) and 432 (~ 45.0 kDa) amino acid residues in the deduced amino acid sequences of Symb-AMT2, Clad-AMT2, and Duru-AMT2, respectively. Each amino acid sequence had an extracellular N-terminus and an intracellular C-terminus, as well as 11 predicted TM (TM1 − TM11; Figure 1). All three amino acid sequences contained the conserved amino acid residues needed for NH4+ binding, the phenylalanine gate, histidine residues, and the AMT signature sequence (Figure 1). Notably, they also consisted of the AMT2 signature sequence DYSGGYVIHLSSGVAGFTAAYWVGPR as defined by Couturier et al. (2007). AMTs have been proposed to function as homotrimers, whereby each monomer contains a central hydrophobic channel through which NH3 is transported (Khademi et al., 2004; Pantoja, 2012). Gly456 is needed to establish trans-interactions between monomers and to activate the transporters. Hence, the conservation of this residue in Symb-AMT2 (Gly411), Clad-AMT2 (Gly400), and Duru-AMT2 (Gly400) indicated that they could undergo oligomerization to form trimers (Pantoja, 2012). A phenogramic analysis confirmed that these three sequences had a dinoflagellate origin, as they were distinct from the AMTs of bivalves (Figure 2).
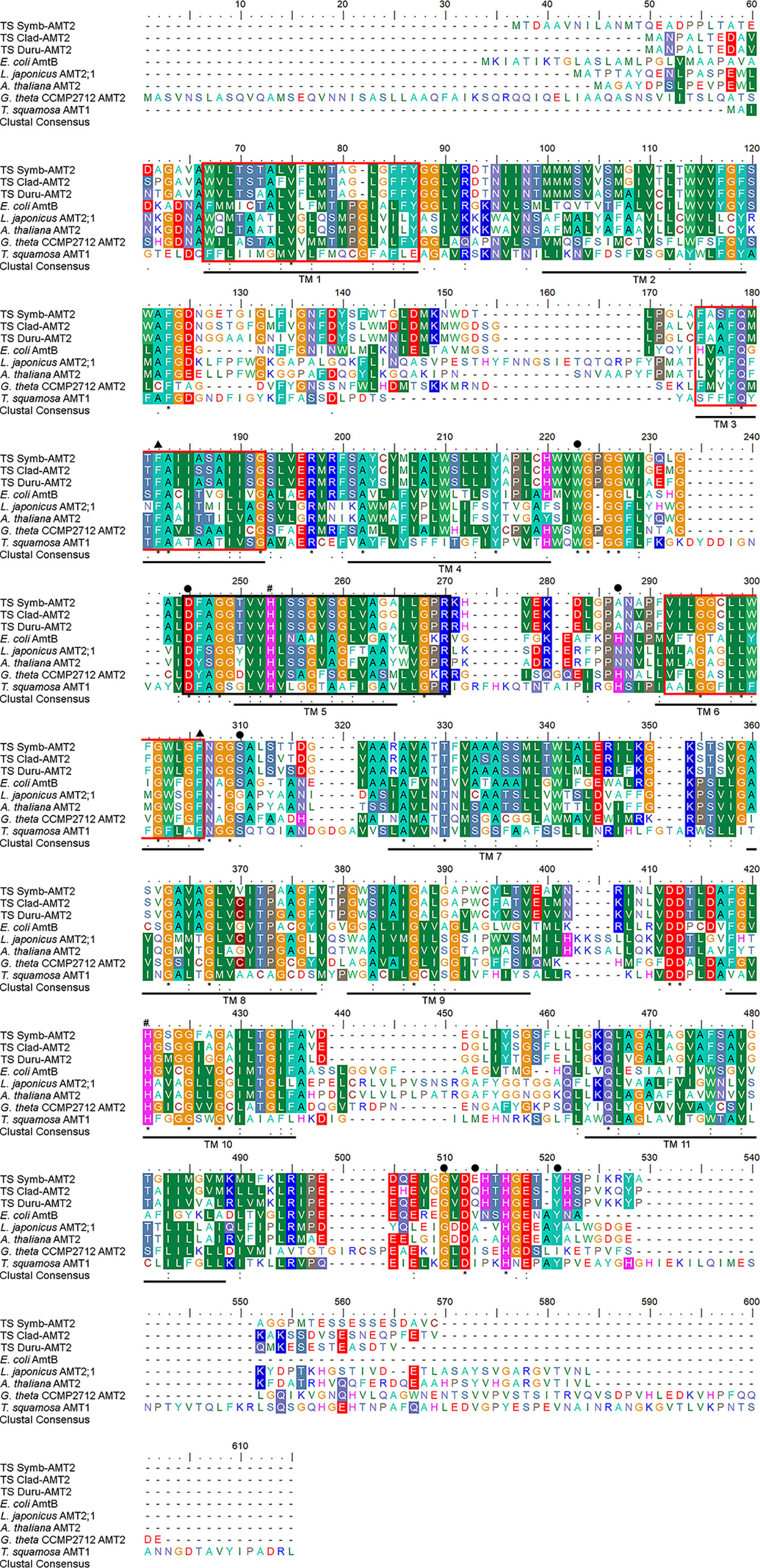
Figure 1 Molecular characterization of the deduced amino acid sequences of ammonium transporter 2 (AMT2) from Symbiodinium (Symb-AMT2), Cladocopium (Clad-AMT2), and Durusdinium (Duru-AMT2) obtained from the outer mantle of Tridacna squamosa. Multiple alignment of Symb-AMT2 (MW023800), Clad-AMT2 (MW023801), and Duru-AMT2 (MW023802) with Escherichia coli AmtB (AAD14837.1), Lotus japonicus AMT2;1 (AAL08212.1), Arabidopsis thaliana AMT2 (AEC09519.1), Guillardia theta CCMP2712 AMT2 (EKX43695.1), and T. squamosa AMT1 (ATC20499). Identical or similar amino acid residues are shaded. Asterisks, colons and periods denote identical amino acid residues, strongly similar amino acids and weakly similar amino acids, respectively. The eleven predicted transmembrane regions (TM 1-TM 11) predicted using MEMSAT3 are underlined in black. The pore lining amino acid residues predicted MEMSAT-SVM were indicated with red boxes. The conserved amino acid residues involved in the binding of NH4+ and the phenylalanine gate are marked with closed circles and triangles, respectively. The conserved histidine residues involved in the deprotonation of NH4+ are demarcated by the pound sign. The AMT signature pattern (D160-[FYWS]-[AS]-G-[GSC]-x(2)-[IV]-x(3)-[SAG](2)-x(2)-[SAG]-[LIVM-F]-x(3)-[LIV-MFYWA](2)-x-[GK]-x-R) indicated with a black box was predicted using ScanProsite tool.
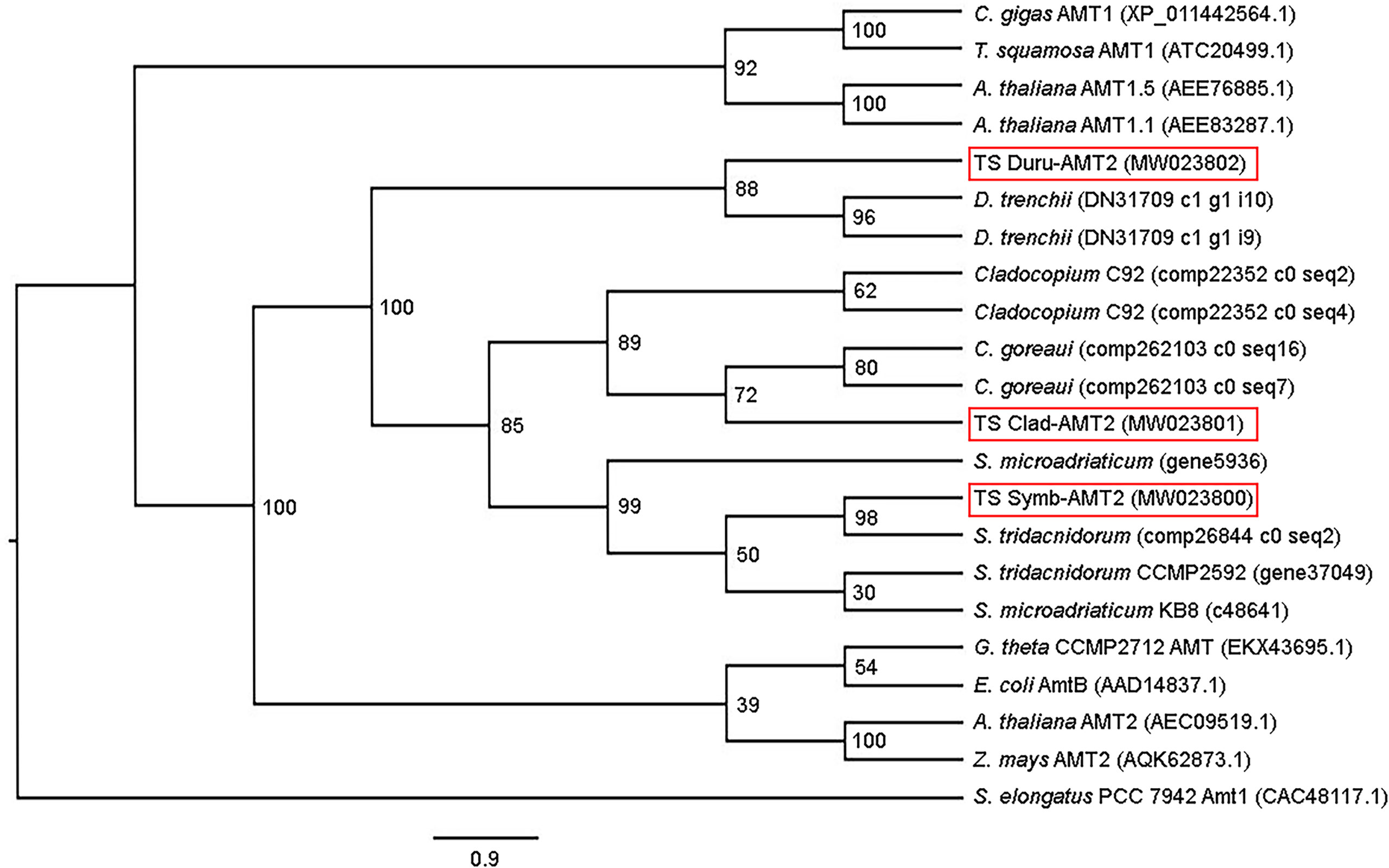
Figure 2 Phenogramic analysis of ammonium transporter 2 (AMT2) from Symbiodinium (Symb-AMT2), Cladocopium (Clad-AMT2), and Durusdinium (Duru-AMT2) obtained from the outer mantle of Tridacna squamosa. Numbers at each branch point represent bootstrap values from 2000 replicates. Amt1 from Synechococcus elongatus is used as the outgroup for comparison. Amino acid sequences of ammonia transporters from the bacterium (Escherichia coli), algae (Cladocopium C92, C. goreaui, Durusdinium trenchii, Guillardia theta, Symbiodinium tridacnidorum, and S. microadriaticum), higher plants (Arabidopsis thaliana and Zea mays), bivalves (Crassostrea gigas and T. squamosa), were obtained from Genbank or various dinoflagellate databases with their accession or contig numbers given in parentheses.
Transcript Levels of Zoox-AMT2 and Zoox-rbcII, and Transcript Ratios of Zoox-AMT2 to Zoox-rbcII in Five Organs
The outer mantle had the highest transcript level of Zoox-AMT2, which was significantly higher than those in the inner mantle (by ~ 4.6-fold), foot muscle (by ~ 16.2-fold), hepatopancreas (by ~ 18.2-fold) and ctenidium (by ~ 136-fold) (one-way ANOVA, F4,15 = 32.446, p-value < 0.05; Figure 3A). Likewise, the transcript level of Zoox-rbcII in the outer mantle was significantly higher than those in the inner mantle (by ~ 5.4-fold), foot muscle (by ~ 17.6-fold), hepatopancreas (by ~ 14.9-fold), and ctenidium (by ~ 158-fold) (one-way ANOVA, F4,15 = 54.541, p-value < 0.05; Figure 3B). The transcript ratios of Zoox-AMT2 to Zoox-rbcII were smaller than one (0.52 to 0.75; Figure 3C) in all the five organs, because the transcript levels of Zoox-rbcII were slightly higher than those of Zoox-AMT2. Overall, the transcript ratios of Zoox-AMT2 to Zoox-rbcII were comparable among the five organs (Figure 3C).
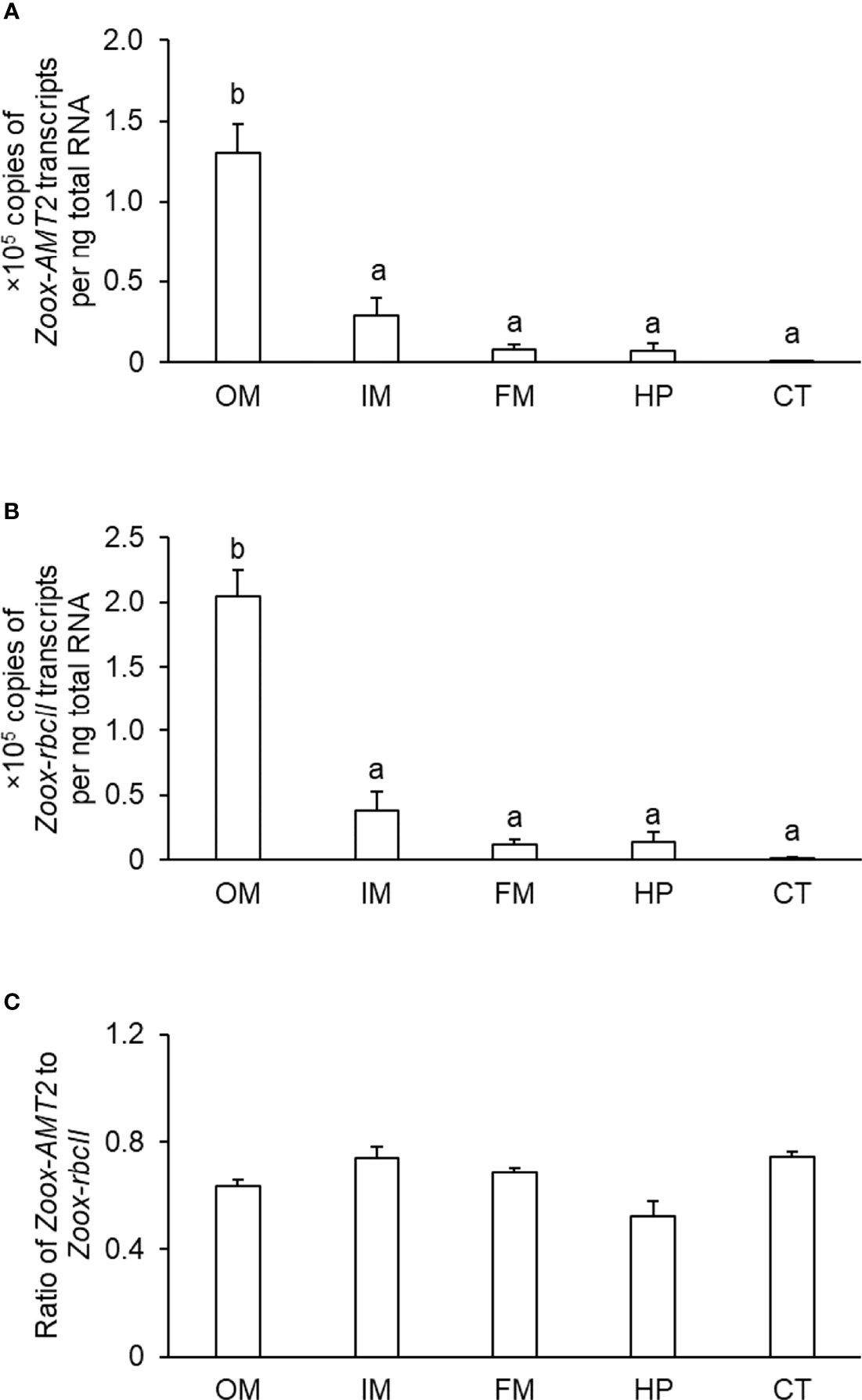
Figure 3 Transcript levels (×105 copies of transcript per ng total RNA) of (A) zooxanthellae-ammonium transporter 2 (Zoox-AMT2) and (B) zooxanthellae-form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Zoox-rbcII), and the (C) transcript ratio of Zoox-AMT2/Zoox-rbcII in the outer mantle (OM), inner mantle (IM), foot muscle (FM), hepatopancreas (HP) and ctenidium (CT) of Tridacna squamosa. Results represent means + SEM. (n = 4). Means not sharing the same letter are significantly different (p-value < 0.05 for transcript levels using ANOVA or p-value < 0.01 for transcript ratios).
Immunolocalization of Zoox-AMT2 in Coccoid Dinoflagellates
Zoox-AMT2 was immuno-localized at the plasma membranes of dinoflagellates in the outer mantle of T. squamosa (Figure 4).
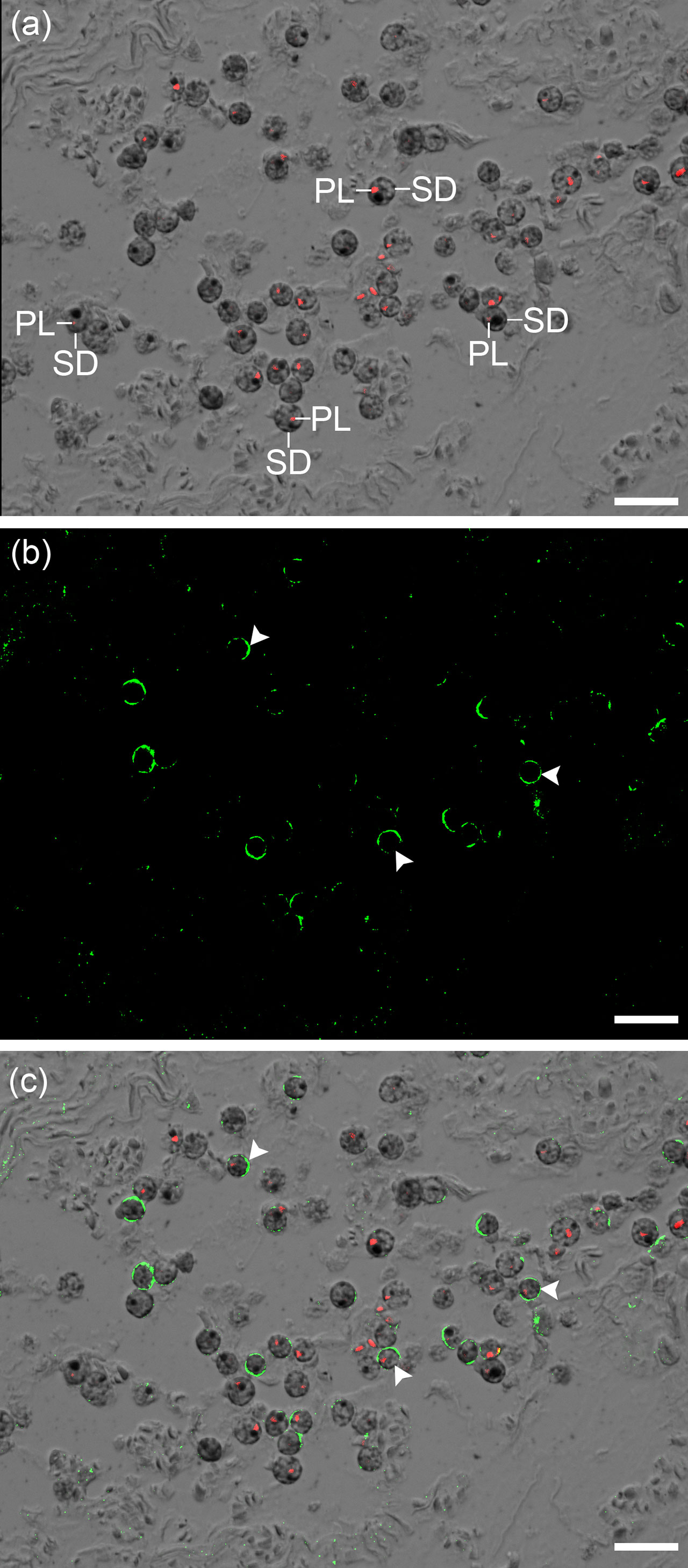
Figure 4 Immunofluorescent labelling of zooxanthellae-ammonium transporter 2 (Zoox-AMT2) in the outer mantle of Tridacna squamosa. Zoox-AMT2 comprises AMT2 of Symbiodinium, Cladocopium and Durusdinium. (A) The combined image of differential interference contrast (DIC) microscopy and the red channel showing autofluorescence of the plastids (PL) in the coccoid dinoflagellates (SD). (B) The image showing green immunofluorescence of Zoox-AMT2. (C) The DIC image overlaid with the red channel and green channel. Arrowheads indicate the localization of Zoox-AMT2 (green) at the plasma membrane of SD. Scale bar: 20 μm. Replicable results were obtained from T. squamosa (n = 3).
Effects of Illumination on the Transcript Level of Zoox-AMT2 and the Protein Abundance of Zoox-AMT2 in the Outer Mantle
Exposure to light had no significant effects on the transcript level of Zoox-AMT2 in the outer mantle of T. squamosa, as compared with the control kept in darkness for 12 h (one-way ANOVA, F3,12 = 0.119, p-value < 0.05; Figure 5).
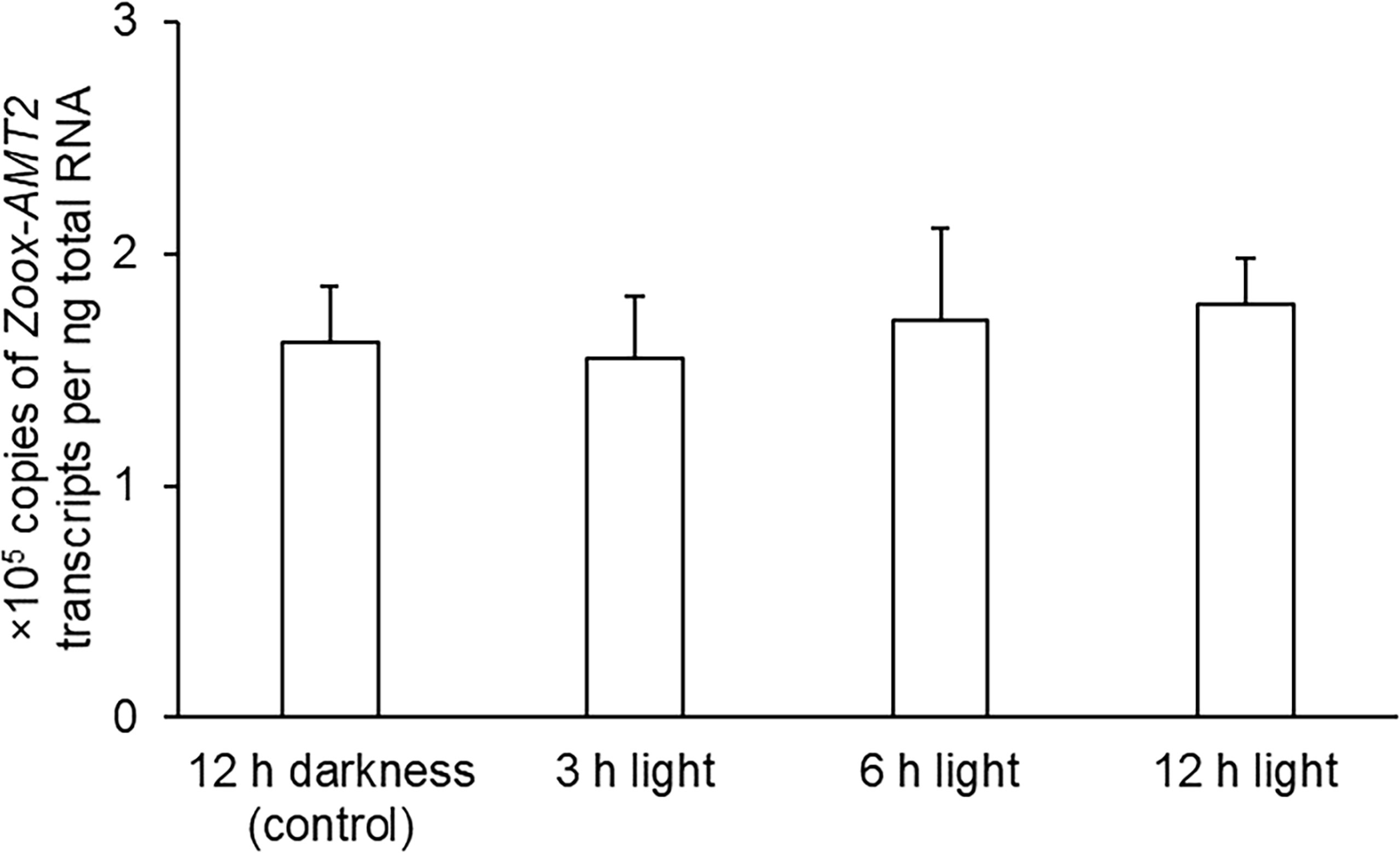
Figure 5 Effects of light on the transcript levels (×105 copies of transcripts per ng total RNA) of zooxanthellae-ammonium transporter 2 (Zoox-AMT2) in the outer mantle of Tridacna squamosa exposed to darkness for 12 h (control) or to light for 3 h, 6 h, or 12 h. Results represent means + SEM (n = 4). Means not sharing the same letter are significantly different (p-value < 0.05).
While the predicted molecular mass of Zoox-AMT2 was ~45 kDa, western blotting revealed a band at ~ 100 kDa (Figure 6A). Nonetheless, the ~ 100 kDa band could be the Zoox-AMT2 band because of three reasons. Firstly, the anti-Zoox-AMT2 was custom-made for this study based on carefully selection of an epitope that was highly conserved among dinoflagellate AMTs. The specificity of this antibody had been confirmed by a BLAST analysis on the epitope sequence, and the top 10 BLAST results, judging by query cover, E-value and percentage identity, were AMTs derived from Symbiodiniaceae (Table 2). Secondly, only one protein band was obtained from dinoflagellates isolated from the outer mantle of T. squamosa (Figure 6A). When taken together with the first reason, this band was highly likely to be Zoox-AMT2. Thirdly, AMTs are known to function as trimers (Khademi et al., 2004; Pantoja, 2012). Therefore, the relatively high molecular mass of ~100 kDa could be due to the formation of Zoox-AMT2 trimer, and trimers of AMTs are known to be resistant to dissociation by SDS/PAGE (Blakey et al., 2002). The fact that transmembrane proteins are known to run at a lower molecular mass than expected due to incomplete unfolding in the presence of SDS (Gaillard et al., 1996) or altered detergent binding (Rath et al., 2009) can explain why the molecular mass of ~ 100 kDa obtained through western blotting was lower than the ~ 135 kDa of the trimer based on calculation. After exposure to light for 12 h, the relative normalized protein abundance of Zoox-AMT2 increased significantly by ~ 4.1-fold in the outer mantle as compared with the control (one-way ANOVA, F3,12 = 5.282, p-value < 0.05; Figure 6B).
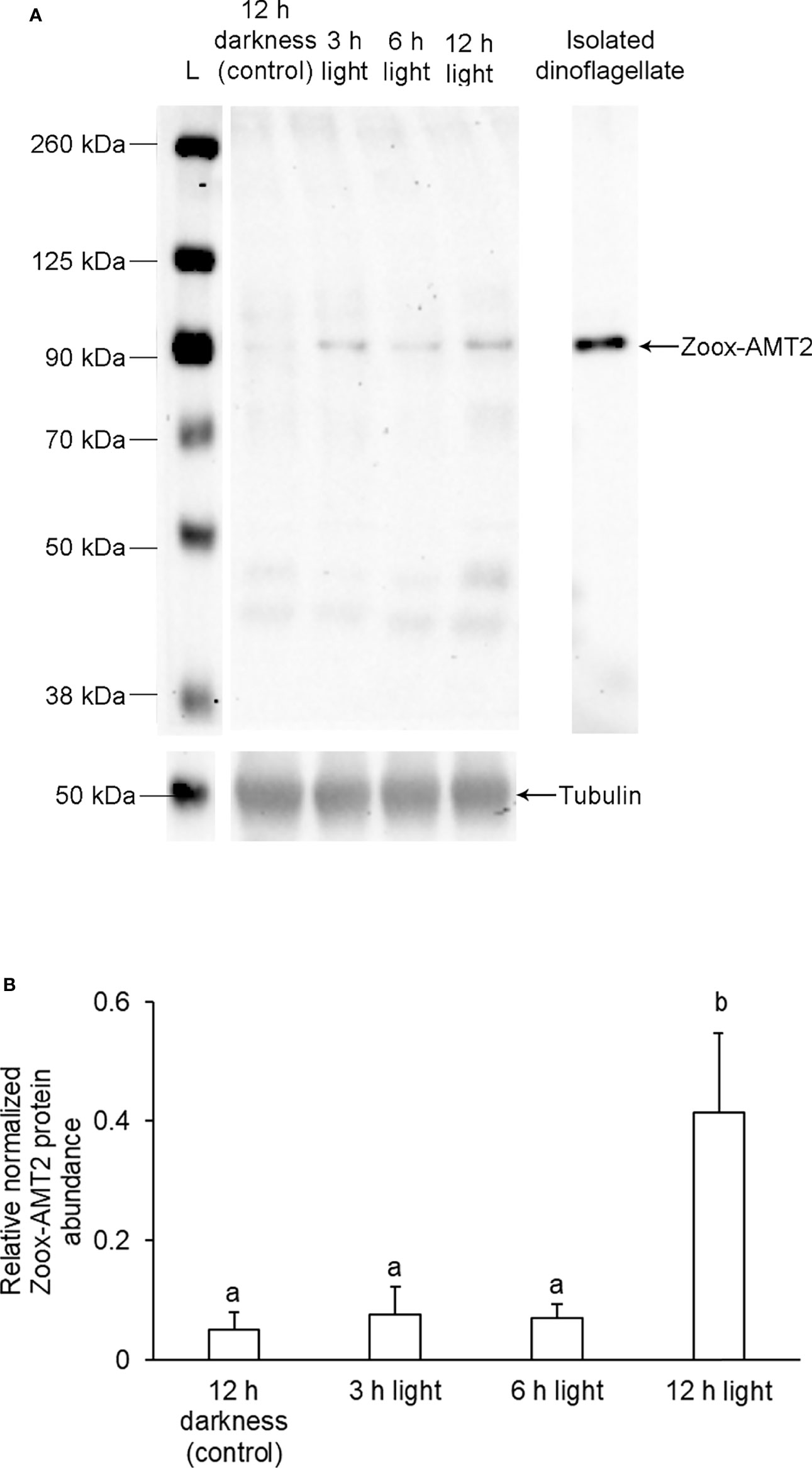
Figure 6 Effects of light exposure on the protein abundance of zooxanthellae-ammonium transporter 2 (Zoox-AMT2) in the outer mantle of Tridacna squamosa. (A) A representative immunoblot of Zoox-AMT2 from the outer mantle and dinoflagellates isolated from the outer mantle of T. squamosa. α-tubulin was used as the reference protein for Zoox-AMT2 from the outer mantle. L represents the ladder. (B) The relative protein abundance of Zoox-AMT2, expressed as arbitrary densitometric units (a.u.), from the outer mantle of T. squamosa exposed to 12 h of darkness (control) or 3, 6, or 12 h of light. Results represent means + SEM. (n = 4). Means that are significantly different from each other are represented by different letters (p-value < 0.05).
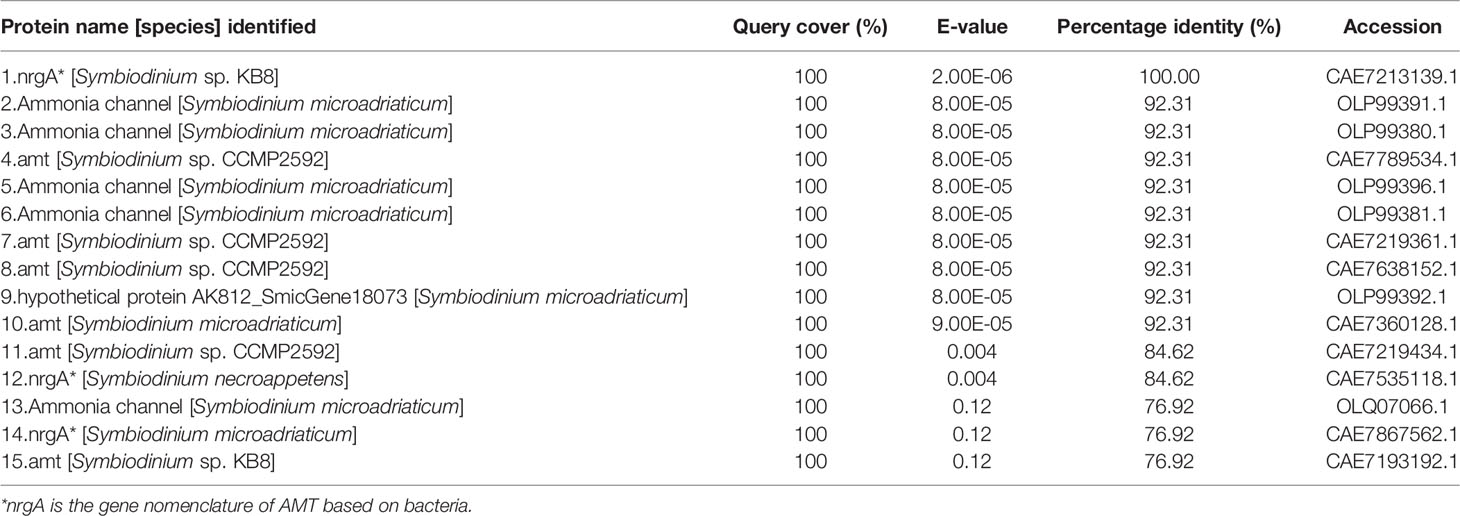
Table 2 BLAST analysis of the epitope sequence (YSFWTNLDMKNWD) of the genus-comprehensive anti-Zoox-AMT2 antibody.
Discussion
Giant clams harbor coccoid dinoflagellates inside zooxanthellal tubules that are located mainly in the colorful outer mantle. The clam host absorbs inorganic (Fitt et al., 1993a; Ip et al., 2020a) and organic N (Chan et al., 2018; Chan et al., 2019) from the ambient seawater through the ctenidium and supplies them to the outer mantle where the majority of symbionts reside. Despite the lack of ammonia excretion and the continuous production of endogenous ammonia, the host’s hemolymph contains only low concentrations of ammonia. Hence, it is logical to deduce that the coccoid dinoflagellates can effectively absorb and assimilate the metabolic ammonia produced endogenously by the host. Indeed, we had obtained from the outer mantle of T. squamosa three major AMT2 nucleotide sequences derived from Symbiodinium spp., Cladocopium spp., and Durusdinium spp. (Symb-AMT2, Clad-AMT2 and Duru-AMT2, respectively). The transcript level of Zoox-AMT2 was the highest in the outer mantle. The coccoid dinoflagellates expressed Zoox-AMT2 proteins at the plasma membranes, denoting that Zoox-AMT2 was positioned to take part in ammonia absorption. Furthermore, the protein abundance of Zoox-AMT2 was upregulated in the outer mantle during illumination. Therefore, it can be deduced that the photosynthesizing coccoid dinoflagellates in the outer mantle augmented the potential of ammonia absorption probably to increase the capacity of amino acid production.
Molecular Characterization of Symb-AMT2, Clad-AMT2, and Duru-AMT2
The deduced Symb-AMT2, Clad-AMT2, and Duru-AMT2 sequences have high similarity with Escherichia coli AmtB (EcAmtB), which is known to be involved in the uptake of ammonia in the bacterium. They contain the conserved amino acid residues needed for NH4+ binding, the phenylalanine gate, and the twin-histidine motif. In the monomer of EcAmtB, the putative NH4+ binding site comprises three amino acid residues, Phe107, Trp148 and Ser219 (Javelle et al., 2008). These three residues are conserved in Symb-AMT2 (Phe129, Trp148 and Ser241), Clad-AMT2 (Phe118, Trp159 and Ser230), and Duru-AMT2 (Phe118, Trp159 and Ser230) obtained from T. squamosa. Molecular dynamic simulations have implicated that the phenylalanine gate is involved in the entry of NH3 from the periplasmic/extracellular side into the pore (Akgun and Khademi, 2011). At the same time, the phenylalanine gate serves to prevent the permeation of ions of similar radii as NH4+ (Nygaard et al., 2006; Javelle et al., 2008). In EcAmtB, the phenylalanine gate comprises Phe107 and Phe215, which correspond to Phe129 and Phe237 of Symb-AMT2, Phe118 and Phe226 of Clad-AMT2, and Phe118 and Phe226 of Duru-AMT2.
It has been proposed that NH4+ could be deprotonated by a pair of conserved histidine residues located close to the centre of the conducting pore of AMT. Then, NH3 is transported down its concentration gradient through the hydrophobic core while the H+ is transferred to the cytoplasm via a polar conduction route (Williamson et al., 2020). In EcAmtB, these two residues are His168 and His318 (Zheng et al., 2004; Javelle et al., 2006; Pantoja, 2012; Hall and Yan, 2013). These two histidine residues are conserved in AtAMT1;2 and AtAMT2 (Ganz et al., 2020), as well as in Symb-AMT2 (His191 and His341), Clad-AMT2 (His180 and His330) and Duru-AMT2 (His180 and His330). The conserved histidine residues also function as a filter to prevent K+ from passing though the pore, allowing for high substrate affinity and selectivity (Hall and Yan, 2013; Williamson et al., 2020).
Zoox-AMT2 Is Associated With the Dinoflagellates’ Plasma Membrane
Zoox-AMT2 was localized at the plasma membrane of the coccoid dinoflagellates of T. squamosa, which is similar to AMTs of Arabidopsis thaliana and Oryza sativa (Ye et al., 2016). Hence, Zoox-AMT2 was positioned in the coccoid dinoflagellates to absorb ammonia from the surrounding luminal fluid. Notably, the ammonia present in the hemolymph must be transported across the tubular epithelial cells through their basolateral and apical membranes into the luminal fluid of the tubules; only then, would ammonia become accessible to the extracellular symbionts. Logically, the tubular epithelial cells would possess transporters to facilitate and regulate the movement of H+ and ammonia. Hence, there could be differences in pH as well as the concentration of ammonia between the hemolymph and the luminal fluid. It has been established that the outer mantle of the clam host possesses a light-enhanced carbon concentration mechanism (CCM), which augments the transport of inorganic carbon from the hemolymph into the luminal fluid. This host-mediated CCM involves vacuolar-type H+-ATPase (VHA; Ip et al., 2018) of the iridocytes, as well as homologs of carbonic anhydrase 2 (CA2; Ip et al., 2017b), and VHA (Ip et al., 2018) of the tubular epithelial cells. Specifically, VHA of the iridocytes and tubular epithelial cells could secrete H+ into the hemolymph and the luminal fluid, respectively. In addition, the symbionts also possess a light-enhanced external CCM to promote and regulate the uptake of inorganic carbon (Mani et al., 2021). They express VHA in intracellular vesicles that can align and merge with the plasma membrane to augment the excretion of H+ into the surrounding luminal fluid during illumination. With the secretion of H+ through the host’s and symbionts’ VHA, NH4+ is probably the major species of dissolved ammonia in the luminal fluid as the pK of NH4+ dissociation is ~9.5. Molecular characterization of Symb-AMT2, Clad-AMT2, and Duru-AMT2 revealed that they could bind with the luminal NH4+ through the putative NH4+ binding sites, deprotonate it through the two conserved histidine residues, and transport the molecular NH3 into the symbionts.
The Implications of Light-Enhanced Expression Level of Zoox-AMT2 in the Outer Mantle
Illumination did not affect the transcript level of Zoox-AMT2 in the outer mantle of T. squamosa. This was probably attributable to the relatively high transcript level of Zoox-AMT2 (× 105 copies of transcript per ng total RNA) in the outer mantle of the control individuals, which implies that it was non-essential to upregulate the transcript level of Zoox-AMT2 in order to increase the protein abundance of Zoox-AMT2 during light exposure.
Western blotting revealed a band of interest (Zoox-AMT2) at ~ 100 kDa, which corroborates the fact that AMT2s act as trimers (Khademi et al., 2004; Pantoja, 2012), and that AMT trimers are resistant to dissociation during SDS/PAGE electrophoresis (Blakey et al., 2002). The apparent discrepancy between the estimated molecular mass (~ 100 kDa; obtained through western blotting) and the calculated molecular mass of the Zoox-AMT2 trimer (~135.0 kDa) could result from the incomplete unfolding of the membrane protein in the presence of SDS (Gaillard et al., 1996) or altered detergent binding (Rath et al., 2009). In support of our results and proposition, the molecular masses of EcAmtB (Blakey et al., 2002) and Archaeoglobus fulgidus Amt1 (Andrade et al., 2005) obtained through western blotting are ~90 kDa, which are lower than the calculated molecular masses of the trimers (133.5 kDa and 120.3 kDa, respectively) based on those of the monomers (44.5 kDa and 40.1 kDa, respectively).
Exposure to light for 12 h augmented the protein abundance of Zoox-AMT2 in the outer mantle of T. squamosa, indicating that the expression of Zoox-AMT2 was regulated at the translational level in the dinoflagellates therein. A similar phenomenon of an increase in protein abundance not being accompanied by an increase in transcript level has been reported for Zoox-rbcII/Zoox-RBCII (Poo et al., 2020), Zoox-GS1/Zoox-GS1 (Teh et al., 2021), and Zooxanthellae-vacuolar H+-ATPase subunit B (Zoox-VHA-B; Mani et al., 2021) in T. squamosa. The light-dependent increase in the potential of ammonia uptake in these dinoflagellates explains why the ammonia concentration of the hemolymph decreases during illumination (Fitt et al., 1995) and why giant clams do not excrete ammonia when exposed to light (Muscatine and D’Elia, 1978).
Besides inorganic carbon fixation (Yellowlees et al., 1993), coccoid dinoflagellates play a crucial role in N assimilation in the giant clam-dinoflagellate holobiont (Rees et al., 1994). As the assimilation of N needs a supply of carbon chains, there must be coupling between these two metabolic processes. In plants, ammonia uptake and AMT expression can be augmented by light (Camañes et al., 2007) independent of circadian rhythm (Couturier et al., 2007; Haydon et al., 2011; Song et al., 2011; Ranathunge et al., 2014; Li et al., 2016). The gene expression of AMT2 of the field mustard, Brassica campestris, (BcAMT2) is strongly affected by diurnal changes, and carbohydrates play a major role in the diurnal changes of its gene expression (Bläsing et al., 2005; Haydon et al., 2011). The highest transcript level of BcAMT2 is detected in the leaves of B. campestris at the end of the light period (Couturier et al., 2007), which coincides with the high demand of carbohydrates for the assimilation of ammonia. Hence, it has been concluded that AMT2 expression is regulated by the availability of carbohydrates (Gazzarrini et al., 1999; Couturier et al., 2007; Li et al., 2016). This can also be applied to coccoid Symbiodiniaceae, as the syntheses of nitrogenous compounds essential for growth and development require carbon-chains and therefore need to occur concurrently with carbon fixation. Indeed, the protein abundances of Zoox-AMT2 (this study), zooxanthellae-URE (Ip et al., 2020b), zooxanthellae-GS1 (Zoox-GS1; Teh et al., 2021), Zoox-RBCII (Poo et al., 2020) and Zoox-VHA-B (Mani et al., 2021) are upregulated in the outer mantle of T. squamosa during illumination. These results indicate that illumination augments the potential of ammonia absorption (by Zoox-AMT2) and the potential of ammonia production through urea degradation (by Zoox-URE) in the coccoid dinoflagellates residing in the outer mantle. The excess ammonia can be converted to glutamine (by Zoox-GS1), which can act as a key substrate for amino acid synthesis (Vander Heiden and DeBerardinis, 2017) using the carbon-chains produced through photosynthesis (by Zoox-RBCII). Alternatively, the excess ammonia can be converted into glutamate by glutamine oxoglutarate aminotransferase (Fam et al., 2018). Importantly, glutamate and glutamine are key substrates for the syntheses of many nitrogenous compounds needed for growth and development, including non-essential amino acids, chlorophyll, nucleic acids, glutathione, cofactors, and secondary metabolites (Newsholme et al., 2003; Moller, 2005).
The Potentials of Ammonia Absorption in Five Organs
Using Zoox-AMT2 as a molecular indicator, we made an attempt to estimate the potential of ammonia transport in coccoid dinoflagellates in the outer mantle, inner mantle, foot muscle, hepatopancreas and ctenidium of T. squamosa. The quantity of dinoflagellates naturally varies among different individuals of T. squamosa, and among different organs of the same individual. Hence, considerable variation in the transcript level of Zoox-AMT2 could be detected in similar organs of different individual giant clams or different organs of the same giant clam. To resolve this, it was essential to determine the transcript levels of Zoox-rbcII using the genera-comprehensive qPCR primers designed by Poo et al. (2020). With this, the transcript ratios of Zoox-AMT2/Zoox-rbcII could be calculated so that the potential of ammonia transport could be evaluated with reference to the phototrophic potential independent of the quantity of symbionts in these host organs. As the transcript levels of Zoox-AMT2 was slightly lower than those of Zoox-rbcII in the five organs, the ratios of Zoox-AMT2/Zoox-rbcII was somewhat smaller than one. Nonetheless, these results indicate that the coccoid dinoflagellates in these five organs had the ability to absorb ammonia from the host. Similar to the potentials of ammonia assimilation based on Zoox-GS1/Zoox-rbcII (Teh et al., 2021), the potentials of ammonia transport relative to the phototrophic potential were grossly similar among the outer mantle, inner mantle, foot muscle, and ctenidium.
In general, the coccoid dinoflagellates residing in the outer mantle of T. squamosa can play a major role in the uptake and assimilation of metabolic ammonia produced by the host because of two reasons. Firstly, the outer mantle had the largest quantity of dinoflagellates and the highest transcript level of Zoox-AMT2 among the five organs studied. Secondly, illumination augments the expression levels of Zoox-AMT2 (this study) and Zoox-RBCII (Poo et al., 2020) in the outer mantle of T. squamosa, indicating an alignment of ammonia absorption with photosynthesis. On the other hand, the whitish inner mantle of T. squamosa is in direct contact with the extrapallial fluid and participates in light-enhanced shell-formation (Boo et al., 2017; Ip et al., 2017a; Boo et al., 2019; Cao-Pham et al., 2019; Chew et al., 2019; Chan et al., 2021). It contains a population of coccoid dinoflagellates at a special region of the inner mantle near the hinge of the shell-valves (Poo et al., 2020). Our results indicate that these symbionts can absorb ammonia from the host for the production of amino acids. The donation of amino acids from the symbionts to the host could support shell formation (Teh et al., 2021), which needs to synthesize a proteinaceous matrix for CaCO3 deposition (Greenfield et al., 1984). The foot muscle is instrumental to lateral movement (Stasek, 1962; Huang et al., 2007), and its tip contains a concentrated population of coccoid dinoflagellates (Poo et al., 2020). Ammonia absorbed by symbionts in the foot muscle can be assimilated into amino acids, which can be shared with the host for muscle production (Teh et al., 2021). The ctenidium consists primarily of epithelial tissues, but its epithelial cells express a large variety of protein catalysts including transporters, channels and enzymes, which are crucial for the absorption of exogenous nutrients. It is therefore important for the symbionts residing in the ctenidium to have the ability to absorb and assimilate ammonia into amino acids to be shared with the host. In comparison, the hepatopancreas had the lowest Zoox-AMT2/Zoox-rbcII among the five organs studied. The reason for this is unclear at present, but it may indicate that symbionts residing in the hepatopancreas might prefer to absorb and assimilate some other types of N-containing compounds.
Conclusion
The recycling of N between the host and its symbionts contribute substantially to the success of giant clams in nutrient-poor waters in the tropics. N-recycling consists of two important elements: firstly, the tightly regulated supply of N from the host to the symbionts, and secondly, the release of essential and non-essential amino acids by the symbionts to the host. Our results confirmed for the first time that coccoid dinoflagellates of T. squamosa express AMT2 in the plasma membranes, corroborating the proposition that these symbionts can absorb and assimilate metabolic ammonia produced by the host. The uptake of ammonia is aligned apparently with photosynthesis, as illumination increases the expression level of Zoox-AMT2 in the outer mantle of T. squamosa. Besides, the host can also increase the absorption of exogenous ammonia (Wilkerson and Trench, 1986; Fitt et al., 1993a; Fitt et al., 1993b), NO3− (Ip et al., 2020a) and urea (Chan et al., 2018; Chan et al., 2019) from the ambient seawater during illumination. Therefore, effort should be made in the future to identify the transporters involved in the absorption of various types of N-containing compounds in coccoid Symbiodiniaceae and to elucidate whether symbionts residing in different organs of T. squamosa would have distinct preference of N-substrates for N metabolism. As the rates of NO3− (Ip et al., 2020a) and urea (Chan et al., 2018; Chan et al., 2019) absorption in T. squamosa are augmented during illumination, the quantity of exogenous N absorbed could be much higher than that of endogenous ammonia produced by the host. Hence, the clam host may not be able to regulate the symbiont population by simply controlling the availability of endogenous ammonia to them. As the symbionts are located extracellularly inside the zooxanthellal tubules, it would be crucial to elucidate the specific transport mechanisms involved in the translocation of ammonia, NO3− and urea from the hemolymph across the epithelial cells of the zooxanthellal tubules into the luminal fluid. Importantly, it is through these transport mechanisms that the host can regulate the supply of N-containing compounds to the symbionts. Additionally, it would be essential to elucidate whether the tubular epithelial cells in the outer mantle of giant clams possess a light-enhanced N-concentration mechanism, similar to the CCM.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: MW023800, MW023801 and MW023802].
Author Contributions
SC and YI designed the experiments. CP and MB performed the experiments and analyzed the data. SC, CP and YI wrote the manuscript. SC, CP and MB participated in animal subjection and sample collection. SC and YI were involved in the analysis of data and approval of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Singapore Ministry of Education administered as grants to SC: (1) National Institute of Education (NIE) Academic Research Fund RI 3/19 CSF, and (2) the NIE Research Support for Senior Academic Administrator Grant RS 1/21 CSF. It was also supported by the Singapore National Research Foundation through a grant (NRF-MSRDP-P22) administered by the Marine Science Research and Development Programme (MSRDP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.835574/full#supplementary-material
References
Akgun U., Khademi S. (2011). Periplasmic Vestibule Plays an Important Role for Solute Recruitment, Selectivity, and Gating in the Rh/Amt/MEP Superfamily. Proc. Natl. Acad. Sci. 108, 3970–3975. doi: 10.1073/pnas.1007240108
Andrade S. L. A., Dickmanns A., Ficner R., Einsle O. (2005). Expression, Purification and Crystallization of the Ammonium Transporter Amt-1 From Archaeoglobus Fulgidus. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 61, 861–863. doi: 10.1107/S1744309105027004
Aranda M., Li Y., Liew Y. J., Baumgarten S., Simakov O., Wilson M. C., et al. (2016). Genomes of Coral Dinoflagellate Symbionts Highlight Evolutionary Adaptations Conducive to a Symbiotic Lifestyle. Sci. Rep. 6, 39734. doi: 10.1038/srep39734
Bayer T., Aranda M., Sunagawa S., Yum L. K., DeSalvo M. K., Lindquist E., et al. (2012). Symbiodinium Transcriptomes: Genome Insights Into the Dinoflagellate Symbionts of Reef-Building Corals. PloS One 7, e35269. doi: 10.1371/journal.pone.0035269
Belda C. A., Lucas J. S., Yellowlees D. (1993). Nutrient Limitation in the Giant Clam- Zooxanthellae Symbiosis: Effects of Nutrient Supplements on Growth of the Symbiotic Partners. Mar. Biol. 117, 655–664. doi: 10.1007/BF00349778
Bellantuono A., Dougan K., Granados-Cifuentes C., Rodriguez-Lanetty M. (2019). Free-Living and Symbiotic Lifestyles of a Thermotolerant Coral Endosymbiont Display Profoundly Distinct Transcriptomes Under Both Stable and Heat Stress Conditions. Mol. Ecol. 28, 5265–5281. doi: 10.5061/dryad.12j173m
Blakey D., Leech A., Thomas G. H., Coutts G., Findlay K., Merrick M. (2002). Purification of the Escherichia Coli Ammonium Transporter AmtB Reveals a Trimeric Stoichiometry. Biochem. J. 364, 527–535. doi: 10.1042/BJ20011761
Bläsing O. E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., et al. (2005). Sugars and Circadian Regulation Make Major Contributions to the Global Regulation of Diurnal Gene Expression in Arabidopsis. Plant Cell 17, 3257–3281. doi: 10.1105/tpc.105.035261
Boo M. V., Hiong K. C., Choo C. Y. L., Cao-Pham A. H., Wong W. P., Chew S. F., et al. (2017). The Inner Mantle of the Giant Clam, Tridacna Squamosa, Expresses a Basolateral Na+/K+-ATPase α-Subunit, Which Displays Light-Dependent Gene and Protein Expression Along the Shell-Facing Epithelium. PloS One 12, e0186865. doi: 10.1371/journal.pone.0186865
Boo M. V., Hiong K. C., Goh E. J. K., Choo C. Y. L., Wong W. P., Chew S. F., et al. (2018). The Ctenidium of the Giant Clam, Tridacna Squamosa, Expresses an Ammonium Transporter 1 That Displays Light-Suppressed Gene and Protein Expression and may be Involved in Ammonia Excretion. J. Comp. Physiol. B 188, 765–777. doi: 10.1007/s00360-018-1161-6
Boo M. V., Hiong K. C., Wong W. P., Chew S. F., Ip Y. K. (2019). Shell Formation in the Giant Clam, Tridacna Squamosa, may Involve an Apical Na+/Ca2+ Exchanger 3 Homolog in the Shell-Facing Epithelium of the Whitish Inner Mantle, Which Displays Light-Enhanced Gene and Protein Expression. Coral Reefs 38, 1173–1186. doi: 10.1007/s00338-019-01848-y
Bradford M. M. (1976). A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bronk D. A., See J. H., Bradley P., Killberg L. (2007). DON as a Source of Bioavailable Nitrogen for Phytoplankton. Biogeosciences 4, 283–296. doi: 10.5194/bg-4-283-2007
Camañes G., Cerezo M., Primo-Millo E., Gojon A., García-Agustín P. (2007). Ammonium Transport and CitAMT1 Expression are Regulated by Light and Sucrose in Citrus Plants. J. Exp. Bot. 58, 2811–2825. doi: 10.1093/jxb/erm135
Cao-Pham A. H., Hiong K. C., Boo M. V., Choo C. Y. L., Pang C. Z., Wong W. P., et al. (2019). Molecular Characterization, Cellular Localization, and Light-Enhanced Expression of Beta-Na+/H+ Exchanger-Like in the Whitish Inner Mantle of the Giant Clam, Tridacna Squamosa, Denote its Role in Light-Enhanced Shell Formation. Gene 695, 102–112. doi: 10.1016/j.gene.2019.02.009
Chan J. W. J., Boo M. V., Wong W. P., Chew S. F., Ip Y. K. (2021). Illumination Enhances the Protein Abundance of Sarcoplasmic Reticulum Ca2+-ATPases-Like Transporter in the Ctenidium and Whitish Inner Mantle of the Giant Clam, Tridacna Squamosa, to Augment Exogenous Ca2+ Uptake and Shell Formation, Respectively. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 251, 110811. doi: 10.1016/j.cbpa.2020.110811
Chan C. Y. L., Hiong K. C., Boo M. V., Choo C. Y. L., Wong W. P., Chew S. F., et al. (2018). Light Exposure Enhances Urea Absorption in the Fluted Giant Clam, Tridacna Squamosa, and Up-Regulates the Protein Abundance of a Light-Dependent Urea Active Transporter, DUR3-Like, in its Ctenidium. J. Exp. Biol. 221, jeb176313. doi: 10.1242/jeb.176313
Chan C. Y. L., Hiong K. C., Choo C. Y. L., Boo M. V., Wong W. P., Chew S. F., et al. (2019). With Illumination, the Fluted Giant Clam, Tridacna Squamosa, Upregulates the Protein Abundance of an Apical Na+: Glucose Cotransporter 1 Homolog in its Ctenidium, and Increases Exogenous Glucose Absorption That can be Impeded by Urea. J. Exp. Biol. 222, jeb195644. doi: 10.1242/jeb.195644
Chen Y., González-Pech R. A., Stephens T. G., Bhattacharya D., Chan C. X. (2020). Evidence That Inconsistent Gene Prediction can Mislead Analysis of Dinoflagellate Genomes. J. Phycol. 56, 6–10. doi: 10.1111/jpy.12947
Chew S. F., Ip Y. K. (2014). Excretory Nitrogen Metabolism and Defense Against Ammonia Toxicity in Air-Breathing Fishes. J. Fish Biol. 84, 603–638. doi: 10.1111/jfb.12279
Chew S. F., Koh C. Z. Y., Hiong K. C., Choo C. Y. L., Wong W. P., Neo M. L., et al. (2019). Light-Enhanced Expression of Carbonic Anhydrase 4-Like Supports Shell Formation in the Fluted Giant Clam Tridacna Squamosa. Gene 683, 101–112. doi: 10.1016/j.gene.2018.10.023
Couturier J., Montanini B., Martin F., Brun A., Blaudez D., Chalot M. (2007). The Expanded Family of Ammonium Transporters in the Perennial Poplar Plant. New Phytol. 174, 137–150. doi: 10.1111/j.1469-8137.2007.01992.x
Davies S. W., Ries J. B., Marchetti A., Castillo K. D. (2018). Symbiodinium Functional Diversity in the Coral Siderastrea Siderea is Influenced by Thermal Stress and Reef Environment, But Not Ocean Acidification. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00150
DeBoer T. S., Baker A. C., Erdmann M. V., Jones P. R., Barber P. H. (2012). Patterns of Symbiodinium Distribution in Three Giant Clam Species Across the Biodiverse Bird’s Head Region of Indonesia. Mar. Ecol. Prog. Ser. 444, 117–132. doi: 10.3354/meps09413
Fam R. R. S., Hiong K. C., Choo C. Y. L., Wong W. P., Chew S. F., Ip Y. K. (2018). Molecular Characterization of a Novel Algal Glutamine Synthetase (GS) and an Algal Glutamate Synthase (GOGAT) From the Colorful Outer Mantle of the Giant Clam, Tridacna Squamosa, and the Putative GS-GOGAT Cycle in its Symbiotic Zooxanthellae. Gene 656, 40–52. doi: 10.1016/j.gene.2018.02.062
Fisher C. R., Fitt W. K., Trench R. K. (1985). Photosynthesis and Respiration in Tridacna Gigas as a Function of Irradiance and Size. Biol. Bull. 169, 230–245. doi: 10.2307/1541400
Fitt W. K., Heslinga G. A., Watson T. C. (1993a). Utilization of Dissolved Inorganic Nutrients in Growth and Mariculture of the Tridacnid Clam Tridacna Derasa. Aquaculture 109, 27–38. doi: 10.1016/0044-8486(93)90483-f
Fitt W. K., Rees T. A. V., Braley R. D., Lucas J. S., Yellowlees D. (1993b). Nitrogen Flux in Giant Clams: Size-Dependency and Relationship to Zooxanthellae Density and Clam Biomass in the Uptake of Dissolved Inorganic Nitrogen. Mar. Biol. 117, 381–386. doi: 10.1007/BF00349313
Fitt W. K., Rees T. A. V., Yellowlees D. (1995). Relationship Between pH and the Availability of Dissolved Inorganic Nitrogen in the Zooxanthella-Giant Clam Symbiosis. Limnol. Oceanogr. 40, 976–982. doi: 10.4319/lo.1995.40.5.0976
Fitt W. K., Trench R. K. (1981). Spawning, Development, and Acquisition of Zooxanthellae by Tridacna Squamosa (Mollusca, Bivalvia). Biol. Bull. 161, 213–235. doi: 10.2307/1540800
Gaillard I., Slotboom D. J., Knol J., Lolkema J. S., Konings W. N. (1996). Purification and Reconstitution of the Glutamate Carrier GltT of the Thermophilic Bacterium Bacillus Stearothermophilus. Biochem. 35, 6150–6156. doi: 10.1021/bi953005v
Ganz P., Ijato T., Porras-Murrilo R., Stührwohldt N., Ludewig U., Neuhäuser B. (2020). A Twin Histidine Motif is the Core Structure for High-Affinity Substrate Selection in Plant Ammonium Transporters. J. Biol. Chem. 295, 3362–3370. doi: 10.1074/jbc.RA119.010891
Gazzarrini S., Lejay L., Gojon A., Ninnemann O., Frommer W. B., von Wirén N. (1999). Three Functional Transporters for Constitutive, Diurnally Regulated, and Starvation-Induced Uptake of Ammonium Into Arabidopsis Roots. Plant Cell 11, 937–947. doi: 10.1105/tpc.11.5.937
Giehl R. F. H., Laginha A. M., Duan F. Y., Rentsch D., Yuan L. X., von Wirén N. (2017). A Critical Role of AMT2;1 in Root-to-Shoot Translocation of Ammonium in Arabidopsis. Mol. Plant 10, 1449–1460. doi: 10.1016/j.molp.2017.10.001
González-Pech R. A., Stephens T. G., Chen Y., Mohamed A. R., Cheng Y., Shah S., et al. (2021). Comparison of 15 Dinoflagellate Genomes Reveals Extensive Sequence and Structural Divergence in Family Symbiodiniaceae and Genus Symbiodinium. BMC Biol. 19, 73. doi: 10.1186/s12915-021-00994-6
Greenfield E. M., Wilson D. C., Crenshaw M. A. (1984). Ionotropic Nucleation of Calcium Carbonate by Molluscan Matrix. Am. Zool. 24, 925–932. doi: 10.1093/icb/24.4.925
Guibert I., Lecellier G., Torda G., Ponchon X., Berteaux-Lecellier V. (2020). Metabarcoding Reveals Distinct Microbiotypes in the Giant Clam Tridacna Maxima. Microbiome 8, 57. doi: 10.1186/s40168-020-00835-8
Hall J., Yan D. (2013). The Molecular Basis of K+ Exclusion by the Escherichia Coli Ammonium Channel AmtB. J. Biol. Chem. 288, 14080–14086. doi: 10.1074/jbc.M113.457952
Haydon M. J., Bell L. J., Webb A. A. R. (2011). Interactions Between Plant Circadian Clocks and Solute Transport. J. Exp. Bot. 62, 2333–2348. doi: 10.1093/jxb/err040
Hernawan U. E. (2008). Symbiosis Between the Giant Clams (Bivalvia: Cardiidae) and Zooxanthellae (Dinophyceae). Biodiversitas 9, 53–58. doi: 10.13057/biodiv/d090113
Hiong K. C., Choo C. Y. L., Boo M. V., Ching B., Wong W. P., Chew S. F., et al. (2017). A Light-Dependent Ammonia-Assimilating Mechanism in the Ctenidia of a Giant Clam. Coral Reefs 36, 311–323. doi: 10.1007/s00338-016-1502-4
Holt A. L., Vahidinia S., Gagnon Y. L., Morse D. E., Sweeney A. M. (2014). Photosymbiotic Giant Clams are Transformers of Solar Flux. J. R. Soc Interface 11, 20140678. doi: 10.1098/rsif.2014.0678
Huang D., Todd P. A., Guest J. R. (2007). Movement and Aggregation in the Fluted Giant Clam (Tridacna Squamosa L.). J. Exp. Mar. Biol. Ecol. 342, 269–281. doi: 10.1016/j.jembe.2006.10.051
Ikeda S., Yamashita H., Kondo S. N., Inoue K., Morishima S. Y., Koike K. (2017). Zooxanthellal Genetic Varieties in Giant Clams are Partially Determined by Species-Intrinsic and Growth-Related Characteristics. PloS One 12, e0172285. doi: 10.1371/journal.pone.0172285
Ip Y. K., Chew S. F. (2010). Ammonia Production, Excretion, Toxicity, and Defence in Fish: A Review. Front. Physiol. 1. doi: 10.3389/fphys.2010.00134
Ip Y. K., Chew S. F. (2021). Light-Dependent Phenomena and Related Molecular Mechanisms in Giant Clam-Dinoflagellate Associations: A Review. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.627722
Ip Y. K., Hiong K. C., Goh E. J., Boo M. V., Choo C. Y. L., Ching B., et al. (2017a). The Whitish Inner Mantle of the Giant Clam, Tridacna Squamosa, Expresses an Apical Plasma Membrane Ca2+-ATPase (PMCA) Which Displays Light-Dependent Gene and Protein Expressions. Front. Physiol. 8. doi: 10.3389/fphys.2017.00781
Ip Y. K., Hiong K. C., Lim L. J. Y., Choo C. Y. L., Boo M. V., Wong W. P., et al. (2018). Molecular Characterization, Light-Dependent Expression, and Cellular Localization of a Host Vacuolar-Type H+-ATPase (VHA) Subunit A in the Giant Clam, Tridacna Squamosa, Indicate the Involvement of the Host VHA in the Uptake of Inorganic Carbon and its Supply to the Symbiotic Zooxanthellae. Gene 659, 137–148. doi: 10.1016/j.gene.2018.03.054
Ip Y. K., Hiong K. C., Teng J. H. Q., Boo M. V., Choo C. Y. L., Wong W. P., et al. (2020a). The Fluted Giant Clam (Tridacna Squamosa) Increases Nitrate Absorption and Upregulates the Expression of a Homolog of SIALIN (H+:2NO3– Cotransporter) in the Ctenidium During Light Exposure. Coral Reefs 39, 451–465. doi: 10.1007/s00338-020-01907-9
Ip Y. K., Koh C. Z. Y., Hiong K. C., Choo C. Y. L., Boo M. V., Wong W. P., et al. (2017b). Carbonic Anhydrase 2-Like in the Giant Clam, Tridacna Squamosa: Characterization, Localization, Response to Light, and Possible Role in the Transport of Inorganic Carbon From the Host to its Symbionts. Physiol. Rep. 5 (23), e13494. doi: 10.14814/phy2.13494
Ip Y. K., Teng G. C. Y., Boo M. V., Poo J. S. T., Hiong K. C., Kim H., et al. (2020b). Symbiodiniaceae Dinoflagellates Express Urease in Three Subcellular Compartments and Upregulate its Expression Levels in Situ in Three Organs of a Giant Clam (Tridacna Squamosa) During Illumination. J. Phycol. 56, 1696–1711. doi: 10.1111/jpy.13053
Jantzen C., Wild C., El-Zibdah M., Roa-Quiaoit H. A., Haacke C., Richter C. (2008). Photosynthetic Performance of Giant Clams, Tridacna Maxima and T. Squamosa, Red Sea. Mar. Biol. 155, 211–221. doi: 10.1007/s00227-008-1019-7
Javelle A., Lupo D., Ripoche P., Fulford T., Merrick M., Winkler F. (2008). Substrate Binding, Deprotonation, and Selectivity at the Periplasmic Entrance of the Escherichia Coli Ammonia Channel AmtB. Proc. Natl. Acad. Sci. 105, 5040–5045. doi: 10.1073/pnas.0711742105
Javelle A., Lupo D., Zheng L., Li X., Winkler F., Merrick M. (2006). An Unusual Twin-His Arrangement in the Pore of Ammonia Channels is Essential for Substrate Conductance. J. Biol. Chem. 281, 39492–39498. doi: 10.1074/jbc.M608325200
Johansson O., Wedborg M. (1980). The Ammonia-Ammonium Equilibrium in Seawater at Temperatures Between 5 and 25 °C. J. Solution Chem. 9, 37–44. doi: 10.1007/BF00650135
Keane T. M., Creevey C. J., Pentony M. M., Naughton T. J., Mclnerney J. O. (2006). Assessment of Methods for Amino Acid Matrix Selection and Their Use on Empirical Data Shows That Ad Hoc Assumptions for Choice of Matrix are Not Justified. BMC Evol. Biol. 6, 29. doi: 10.1186/1471-2148-6-29
Khademi S., O’Cornell J., Remis J., Robles-Colmenares Y., Miercke L. J. W., Stroud R. M. (2004). Mechanism of Ammonia Transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science 305, 1587–1594. doi: 10.1126/science.1101952
Klumpp D. W., Bayne B. L., Hawkins A. J. S. (1992). Nutrition of the Giant Clam Tridacna Gigas (L.) I. Contribution of Filter Feeding and Photosynthates to Respiration and Growth. J. Exp. Mar. Biol. Ecol. 155, 105–122. doi: 10.1016/0022-0981(92)90030-E
Klumpp D. W., Griffiths C. L. (1994). Contributions of Phototrophic and Heterotrophic Nutrition to the Metabolic and Growth Requirements of Four Species of Giant Clam (Tridacnidae). Mar. Ecol. Progr. Ser. 115, 103–115. doi: 10.3354/MEPS115103
Ladner J. T., Barshis D. J., Palumbi S. R. (2012). Protein Evolution in Two Co-Occurring Types of Symbiodinium: An Exploration Into the Genetic Basis of Thermal Tolerance in Symbiodinium Clade D. BMC Evol. Biol. 12, 217. doi: 10.1186/1471-2148-12-217
Laemmli U. K. (1970). Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Levin R. A., Beltran V. H., Hill R., Kjelleberg S., McDougald D., Steinberg P. D., et al. (2016). Sex, Scavengers, and Chaperones: Transcriptome Secrets of Divergent Symbiodinium Thermal Tolerances. Mol. Biol. Evol. 33, 2201–2215. doi: 10.1093/molbev/msw119
Li H., Cong Y., Chang Y. H., Lin J. (2016). Two AMT2-Type Ammonium Transporters From Pyrus Betulaefolia Demonstrate Distinct Expression Characteristics. Plant Mol. Biol. Rep. 34, 707–719. doi: 10.1007/s11105-015-0957-8
Lim S. S. Q., Huang D., Soong K., Neo M. L. (2019). Diversity of Endosymbiotic Symbiodiniaceae in Giant Clams at Dongsha Atoll, Northern South China Sea. Symbiosis 78, 251–262. doi: 10.1007/s13199-019-00615-5
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loqué D., von Wirén N. (2004). Regulatory Levels for the Transport of Ammonium in Plant Roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Mani R., Boo M. V., Poo J. S. T., Ng S. Y., Chew S. F., Ip Y. K. (2021). Molecular Characterization, Cellular Localization and Light-Dependent Expression of Dinoflagellate Vacuolar-Type H+-ATPase (VHA) Subunit B in the Colourful Outer Mantle of the Giant Clam, Tridacna Squamosa, Indicate the Involvement of VHA in CO2 Uptake in the Photosynthesizing Symbionts. Plant Gene 28, 100328. doi: 10.1016/j.plgene.2021.100328
Mayfield A. B., Hsiao Y. Y., Chen H. K., Chen C. S. (2014). Rubisco Expression in the Dinoflagellate Symbiodinium Sp. Is Influenced by Both Photoperiod and Endosymbiotic Lifestyle. Mar. Biotechnol. 16, 371–384. doi: 10.1007/s10126-014-9558-z
McDonald T. R., Dietrich F. S., Lutzoni F. (2012). Multiple Horizontal Gene Transfers of Ammonium Transporters/Ammonia Permeases From Prokaryotes to Eukaryotes: Toward a New Functional and Evolutionary Classification. Mol. Biol. Evol. 29, 51–60. doi: 10.1093/molbev/msr123
Miller D. J., Yellowlees D. (1989). Inorganic Nitrogen Uptake by Symbiotic Marine Cnidarians: A Critical Review. P. R. Soc Lond. B. Bio. 237, 109–125. doi: 10.1098/rspb.1989.0040
Moller S. G. (2005). Plastids: Annual Plant Reviews Volume Thirteen Vol. pp (Boca Raton: CRC Press), 344. doi: 10.1093/aob/mcl018
Morishima S., Yamashita H., O-hara S., Nakamura Y., Quek V. Z., Yamauchi M., et al. (2019). Study on Expelled But Viable Zooxanthellae From Giant Clams, With an Emphasis on Their Potential as Subsequent Symbiont Sources. PloS One 14, e0220141. doi: 10.1371/journal.pone.0220141
Muscatine L., D’Elia C. E. (1978). The Uptake, Retention and Release of Ammonium by Reef Corals. Limnol. Oceanogr. 23, 725–734. doi: 10.4319/lo.1978.23.4.0725
Muscatine L., Masuda H., Burnap R. (1979). Ammonium Uptake by Symbiotic and Aposymbiotic Reef Corals. Bull. Mar. Sci. 29, 572–575.
Newsholme P., Lima M. M. R., Procopio J., Pithon-Curi T. C., Bazotte R. B., Curi R. (2003). Glutamine and Glutamate as Vital Metabolites. Braz. J. Med. Biol. Res. 36, 153–163. doi: 10.1590/s0100-879x2003000200002
Norton J. H., Shepherd M. A., Long H. M., Fitt W. K. (1992). The Zooxanthellal Tubular System in the Giant Clam. Biol. Bull. 183, 503–506. doi: 10.2307/1542028
Nygaard T. P., Rovira C., Peters G. H., Jensen M. (2006). Ammonium Recruitment and Ammonia Transport by E. Coli Ammonia Channel AmtB. Biophys. J. 91, 4401–4412. doi: 10.1529/biophysj.106.089714
Pang C. Z., Ip Y. K., Chew S. F. (2021). Using Transcript Levels of Nitrate Transporter 2 as Molecular Indicators to Estimate the Potentials of Nitrate Transport in Symbiodinium, Cladocopium and Durusdinium of the Fluted Giant Clam, Tridacna Squamosa. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.784662
Pantoja O. (2012). High Affinity Ammonium Transporters: Molecular Mechanism of Action. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00034
Peng J., Huang C. H. (2006). Rh Proteins vs Amt Proteins: An Organismal and Phylogenetic Perspective on CO2 and NH3 Gas Channels. Transfus. Clin. Biol. 13, 85–94. doi: 10.1016/j.tracli.2006.02.006
Pochon X., Wecker P., Stat M., Berteaux-Lecellier V., Lecellier G. (2019). Towards an in-Depth Characterization of Symbiodiniaceae in Tropical Giant Clams via Metabarcoding of Pooled Multi-Gene Amplicons. PeerJ 7, e6898. doi: 10.7717/peerj.6898
Poo J. S. T., Boo M. V., Chew S. F., Ip Y. K. (2021). Using Form II Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase to Estimate the Phototrophic Potentials of Symbiodinium, Cladocopium and Durusdinium in Various Organs of the Fluted Giant Clam, Tridacna Squamosa, and to Evaluate Their Responses to Light Upon Isolation From the Host. Coral Reefs 40, 233–250. doi: 10.1007/s00338-020-02031-4
Poo J. S. T., Choo C. Y. L., Hiong K. C., Boo M. V., Wong W. P., Chew S. F., et al. (2020). Phototrophic Potential and Form II Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Expression in Five Organs of the Fluted Giant Clam, Tridacna Squamosa. Coral Reefs 39, 361–374. doi: 10.1007/s00338-020-01898-7
Ranathunge K., El-kereamy A., Gidda S., Bi Y. M., Rothstein S. J. (2014). AMT1;1 Transgenic Rice Plants With Enhanced NH4+ Permeability Show Superior Growth and Higher Yield Under Optimal and Suboptimal NH4+ Conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009). Detergent Binding Explains Anomalous SDS-PAGE Migration of Membrane Proteins. Proc. Natl. Acad. Sci. 106, 1760–1765. doi: 10.1073/pnas.0813167106
Rees T. A. V., Fitt W. K., Yellowlees D. (1994). Host Glutamine Synthetase Activities in the Giant Clam - Zooxanthellae Symbiosis: Effects of Clam Size, Elevated Ammonia and Continuous Darkness. Mar. Biol. 118, 681–685. doi: 10.1007/bf00347516
Rosic N., Yew E. S. L., Chan C. K. K., Lee H. C., Kaniewska P., Edwards D., et al. (2015). Unfolding the Secrets of Coral-Algal Symbiosis. ISME J. 9, 844–856. doi: 10.1038/ismej.2014.182
Rossbach S., Saderne V., Anton Gamazo A., Duarte C. M. (2019). Light-Dependent Calcification in Red Sea Giant Clam Tridacna Maxima. Biogeosciences 16, 2635–2650. doi: 10.5194/bg-16-2635-2019
Rossbach S., Subedi R. C., Ng T. K., Ooi B. S., Duarte C. M. (2020). Iridocytes Mediate Photonic Cooperation Between Giant Clams (Tridacninae) and Their Photosynthetic Symbionts. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00465
Rowan R., Whitney S. M., Fowler A., Yellowlees D. (1996). Rubisco in Marine Symbiotic Dinoflagellates: Form II Enzymes in Eukaryotic Oxygenic Phototrophs Encoded by a Nuclear Multigene Family. Plant Cell 8, 539–553. doi: 10.1105/tpc.8.3.539
Shepherd D., Leggat W., Rees T. A. V., Yellowlees D. (1999). Ammonium, But Not Nitrate, Stimulates an Increase in Glutamine Concentration in the Haemolymph of Tridacna Gigas. Mar. Biol. 133, 45–53. doi: 10.1007/s002270050441
Shoguchi E., Beedessee G., Hisata K., Tada I., Narisoko H., Satoh N., et al. (2021). A New Dinoflagellate Genome Illuminates a Conserved Gene Cluster Involved in Sunscreen Biosynthesis. Genome Biol. Evol. 13, 2. doi: 10.1093/gbe/evaa235
Shoguchi E., Beedessee G., Tada I., Hisata K., Takeshi K., Takeshi T., et al. (2018). Two Divergent Symbiodinium Genomes Reveal Conservation of a Gene Cluster for Sunscreen Biosynthesis and Recently Lost Genes. BMC Genomics 19, 458. doi: 10.1186/s12864-018-4857-9
Song T., Gao Q., Xu Z., Song R. T. (2011). The Cloning and Characterization of Two Ammonium Transporters in the Salt-Resistant Green Alga, Dunaliella Viridis. Mol. Biol. Rep. 38, 4797–4804. doi: 10.1007/s11033-010-0621-1
Song S. W., He Z. H., Huang X. M., Zhong L. H., Liu H. C., Sun G. W., et al. (2017). Cloning and Characterization of the Ammonium Transporter Genes BaAMT1;1 and BaAMT1;3 From Chinese Kale. Hortic. Environ. Biote. 58, 178–186. doi: 10.1007/s13580-017-0168-3
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stasek C. R. (1962). The Form, Growth and Evolution of the Tridacnidae (Giant Clams). Arch. Zool. Exp. Gen. 101, 1–40.
Streamer M., McNeil Y. R., Yellowless D. (1993). Photosynthetic Carbon Dioxide Fixation in Zooxanthellae. Mar. Biol. 115, 195–198. doi: 10.1007/BF00346335
Summons R. E., Boag T. S., Osmond C. B. (1986). The Effect of Ammonium on Photosynthesis and the Pathway of Ammonium Assimilation in Gymnodinium Microadriaticum In Vitro and in Symbiosis With Tridacnid Clams and Corals. P. R. Soc Lond. B. Bio. 227, 147–159. doi: 10.1098/rspb.1986.0016
Teh L., Poo J. S. T., Boo M. V., Chew S. F., Ip Y. K. (2021). Using Glutamine Synthetase 1 to Evaluate the Symbionts’ Potential of Ammonia Assimilation and Their Responses to Illumination in Five Organs of the Giant Clam, Tridacna Squamosa. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 255, 110914. doi: 10.1016/j.cbpa.2021.110914
Umeki M., Yamashita H., Suzuki G., Sato T., Ohara S., Koike K. (2020). Fecal Pellets of Giant Clams as a Route for Transporting Symbiodiniaceae to Corals. PloS One 15, e0243087. doi: 10.1371/journal.pone.0243087
Vander Heiden M. G., DeBerardinis R. J. (2017). Understanding the Intersections Between Metabolism and Cancer Biology. Cell 168, 657–669. doi: 10.1016/j.cell.2016.12.039
Wang J. T., Douglas A. E. (1999). Essential Amino Acid Synthesis and Nitrogen Recycling in an Alga–Invertebrate Symbiosis. Mar. Biol. 135, 219–222. doi: 10.1007/s002270050619
Whelan S., Goldman N. (2001). A General Empirical Model of Protein Evolution Derived From Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Wilkerson F. P., Muscatine L. (1984). Uptake and Assimilation of Dissolved Inorganic Nitrogen by a Symbiotic Sea Anemone. P. R. Soc Lond. B. Bio. 221, 71–86. doi: 10.1098/rspb.1984.0023
Wilkerson F. P., Trench R. K. (1986). Uptake of Dissolved Inorganic Nitrogen by the Symbiotic Clam Tridacna Gigas and the Coral Acropora sp. Mar. Biol. 93, 237–246. doi: 10.1007/bf00508261
Williamson G., Dias Mirandela G. D., Tamburrino G., Boeckstaens M., Bizior A., Terras E., et al. (2020). A Two-Lane Mechanism for Selective Biological Ammonium Transport. Access Microbiol. 2, 7. doi: 10.7554/eLife.57183
Yellowlees D., Dionisio-Sese M. L., Masuda K., Maruyama T., Abe T., Baillie B., et al. (1993). Role of Carbonic Anhydrase in the Supply of Inorganic Carbon to the Giant Clam-Zooxanthellae Symbiosis. Mar. Biol. 115, 605–611. doi: 10.1007/BF00349368
Ye X., Sun Y., Liu P., Lee I. (2016). Evolutionary Analysis of AMT (Ammonium Transporters) Family in Arabidopsis Thaliana and Oryza Sativa. Mol. Soil Biol. 7, 1–7. doi: 10.5376/msb.2016.07.0011
Yuan L. X., Loqué D., Kojima S., Rauch S., Ishiyama K., Inoue E., et al. (2007). The Organization of High-Affinity Ammonium Uptake in Arabidopsis Roots Depends on the Spatial Arrangement and Biochemical Properties of AMT1-Type Transporters. Plant Cell 19, 2636–2652. doi: 10.1105/tpc.107.052134
Keywords: amino acids, glutamine, coral reefs, photosynthate, symbiosis, zooxanthellae
Citation: Pang CZ, Boo MV, Ip YK and Chew SF (2022) Symbiotic Dinoflagellates of the Giant Clam, Tridacna squamosa, Express Ammonium Transporter 2 at the Plasma Membrane and Increase Its Expression Levels During Illumination. Front. Mar. Sci. 9:835574. doi: 10.3389/fmars.2022.835574
Received: 14 December 2021; Accepted: 28 March 2022;
Published: 21 April 2022.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Ziniu Yu, South China Sea Institute of Oceanology (CAS), ChinaShashank Keshavmurthy, Academia Sinica, Taiwan
Copyright © 2022 Pang, Boo, Ip and Chew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shit F. Chew, c2Z1bi5jaGV3QG5pZS5lZHUuc2c=
 Caryn Z. Pang
Caryn Z. Pang Mel V. Boo
Mel V. Boo Yuen K. Ip
Yuen K. Ip Shit F. Chew
Shit F. Chew