
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 02 February 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.834337
Bacteria play a crucial role in marine biogeochemistry by releasing, consuming and transforming organic matter. Far from being isolated entities, bacteria are involved in numerous cell–cell interactions. Among such interactions, quorum sensing (QS) allows bacteria to operate in unison, synchronizing their actions through chemical communication. This review aims to explore and synthesize our current knowledge of the involvement of QS in the regulation of bacterial processes that ultimately impact marine biogeochemical cycles. We first describe the principles of QS communication and the renewed interest in its study in marine environments. Second, we highlight that the microniches where QS is most likely to occur due to their high bacterial densities are also hotspots of bacterially mediated biogeochemical transformations. Many bacterial groups colonizing these microniches harbor various QS systems. Thereafter, we review relevant QS-regulated bacterial processes in marine environments, building on research performed in both complex marine assemblages and isolated marine bacteria. QS pathways have been shown to directly regulate organic matter degradation, carbon allocation and nutrient acquisition but also to structure the community composition by mediating colonization processes and microbial interactions. Finally, we discuss current limitations and future perspectives to better characterize the link between QS expression and the bacterial mediation of biogeochemical cycles. The picture drawn by this review highlights QS as one of the pivotal mechanisms impacting microbial composition and functions in the oceans, paving the way for future research to better constrain its impact on marine biogeochemical cycles.
Microorganisms dominate life in the ocean. With an estimated 1030 cells in the ocean (Whitman et al., 1998), prokaryotes outnumber any other life form and support the whole marine trophic web. More specifically, heterotrophic prokaryotes degrade up to 50% of the carbon (C) fixed by marine primary producers (Azam et al., 1983) and allow its reinjection to higher trophic levels through the microbial loop (Pomeroy, 1974; Azam et al., 1983). In doing so, they also remineralize nutrients that support the growth of primary producers (Azam and Malfatti, 2007; Pomeroy, 2007). As such, microorganisms are involved in virtually all elemental cycles, including those of C, nitrogen (N), phosphorus (P), sulfur (S) or iron (Fe) (Azam and Malfatti, 2007; Pomeroy, 2007).
Organic matter (OM) is operationally defined as dissolved (DOM) or particulate organic matter (POM). DOM is essentially consumed by free-living (FL) bacteria, which constitute the bulk of the prokaryotic community (Pomeroy, 1974; Azam et al., 1983). As DOM contains compounds ranging from low to high molecular weight, bacteria implement different strategies to exploit them. Some, such as Rhodobacterales, specialize in the uptake of small compounds that can be directly incorporated into the cells (Landa et al., 2017; Ferrer-González et al., 2021). Other species, such as members of the order Flavobacteriales, target high molecular weight substrates (Elifantz et al., 2005; Fernàndez-Gómez et al., 2013; Ferrer-González et al., 2021), which must be processed before they are metabolized. The prevailing model thus far is that heterotrophic prokaryotes use extracellular hydrolytic enzymes to cleave complex molecules into freely available compounds (Arnosti, 2011). This hydrolysis step is considered a crucial rate-limiting step in bacterially mediated OM transformations (Arnosti, 2011). Scavenging bacteria can consume the released substrates without participating in hydrolytic enzyme production (Enke et al., 2019; Reintjes et al., 2019). A third widespread strategy was recently discovered: “selfish” bacteria can bind polysaccharides on their cell surface and uptake the partially degraded products into their periplasmic space, resulting in a minimal diffusive loss of the substrates to other cells (Reintjes et al., 2017, 2019).
In parallel, some bacteria use chemotaxis and motility to locate and colonize microniches, including marine particles, the microalgal phycosphere and macro- and microplastics (Azam, 1998; Pomeroy, 2007; Stocker et al., 2008; Grossart, 2010). These bacteria, referred to as particle-associated (PA) bacteria, are often phylogenetically different (Bižic-Ionescu et al., 2015; Rieck et al., 2015) and possess higher and more diversified metabolic activity than their FL counterparts (Smith et al., 1992; Grossart et al., 2007; Lyons and Dobbs, 2012; Rieck et al., 2015). Consequently, even though they represent a small proportion of the community, they disproportionately impact biogeochemical cycles by degrading and consuming POM, leading to DOM and nutrient release (Smith et al., 1992; Azam and Long, 2001; Grossart, 2010).
Bacteria were long considered unique, distinct entities whose behavior was driven by resource availability and abiotic factors (Shapiro, 1998; Strom, 2008; Schmidt et al., 2019). However, we have since realized the importance of social interactions, giving rise to the field of chemical ecology. Marine bacteria operate in unison through a complex web of social networks, which, intertwined with other environmental and abiotic cues, shape community structure and function (Atkinson and Williams, 2009; Schmidt et al., 2019). These communications occur via chemical signaling, i.e., the exchange of chemical signals between organisms, inducing an effect on the behavior and physiology of the participants (Schmidt et al., 2019). Chemical signaling includes quorum sensing (QS), a common mode of bacterial communication. One key question in environmental chemical ecology is the role of QS signals in the regulation of microbial activities, including degradation and mineralization processes. Together, these microscale interactions may ultimately impact large-scale biogeochemical processes. As such, it is essential to understand how they contribute to ecosystem functioning.
This review aims to explore and synthesize our current knowledge of how QS regulates bacterial processes that ultimately impact global biogeochemical cycles in marine environments. First, we describe QS communication principles and the renewed interest QS study in marine environments. We then review the microniches where QS most likely occurs and the main bacterial taxa colonizing them. These main groups are described, presenting their ecological importance and their various QS systems. Thereafter, we summarize how QS regulates bacterial functions that may directly or indirectly impact biogeochemical cycles. Finally, we outline current limitations and future perspectives on the subject.
Marine bacteria constantly compete for ecological niches, C and nutrients, giving rise to many biotic interactions. These interactions can be chemically mediated by numerous bioactive metabolites, including QS molecules. QS is a density-based microbial language used to collectively regulate the expression of various genes involved in processes such as bioluminescence, biofilm formation or extracellular metabolite production (Miller and Bassler, 2001; Waters and Bassler, 2005). It depends on the production of small pheromone-like autoinducers (AIs) that are continuously produced and sensed by the cells. QS primarily relies on three components: (i) the AI synthase, which produces the signal molecule; (ii) the signal molecule itself; and (iii) the AI receptor, which initiates the transcriptional regulation of target genes when AIs reach their threshold concentration (Miller and Bassler, 2001; Waters and Bassler, 2005). AIs include N-acylhomoserine lactones (AHLs), furanosyl-borate diester (AI-2) and 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal, PQS) (Figure 1), although a much greater diversity of signaling molecules exists (Miller and Bassler, 2001; Waters and Bassler, 2005). AHLs are the best-studied AIs in marine environments, as they led to the discovery of QS (Nealson et al., 1970; Eberhard, 1972) and are widely used among Gram-negative bacteria (Miller and Bassler, 2001). AHLs are produced by an AHL synthase (luxI, ainS or hdtS family) and released to the surrounding environment through either passive diffusion (short-chain AHLs) or active transport (long-chain AHLs) (Kaplan and Greenberg, 1985; Pearson et al., 1999). More recently, it was discovered that hydrophobic AIs (including long-chain AHLs, CAI-1 and PQS) could also be transported through outer membrane vesicles (Li et al., 2016; Toyofuku et al., 2017; Brameyer et al., 2018; Zhao Z. et al., 2021), facilitating their solubilization and specific delivery (Toyofuku et al., 2017; Brameyer et al., 2018). AHL receptors include cytoplasmic LuxR-type and membrane-bound LuxN-type receptors. Interestingly, both AHL synthases and receptors exist as solos, without an accompanying receptor or synthase, respectively (Fuqua, 2006; Buchan et al., 2016). As many as ∼75% of luxR genes are solo receptors (Hudaiberdiev et al., 2015; Subramoni et al., 2015), pointing to the existence of QS-cheaters, i.e., cells that sense AIs without producing them. In addition, QS communications can be shut down by quorum quenching (QQ), adding a layer of complexity to those cell–cell interactions (Grandclément et al., 2016). QQ encompasses different mechanisms, including the enzymatic degradation of AIs (by AHL-lactonase and acylase, for instance) or the blockage of AI reception or production through QS inhibitors (QSIs) (Grandclément et al., 2016; Saurav et al., 2017).
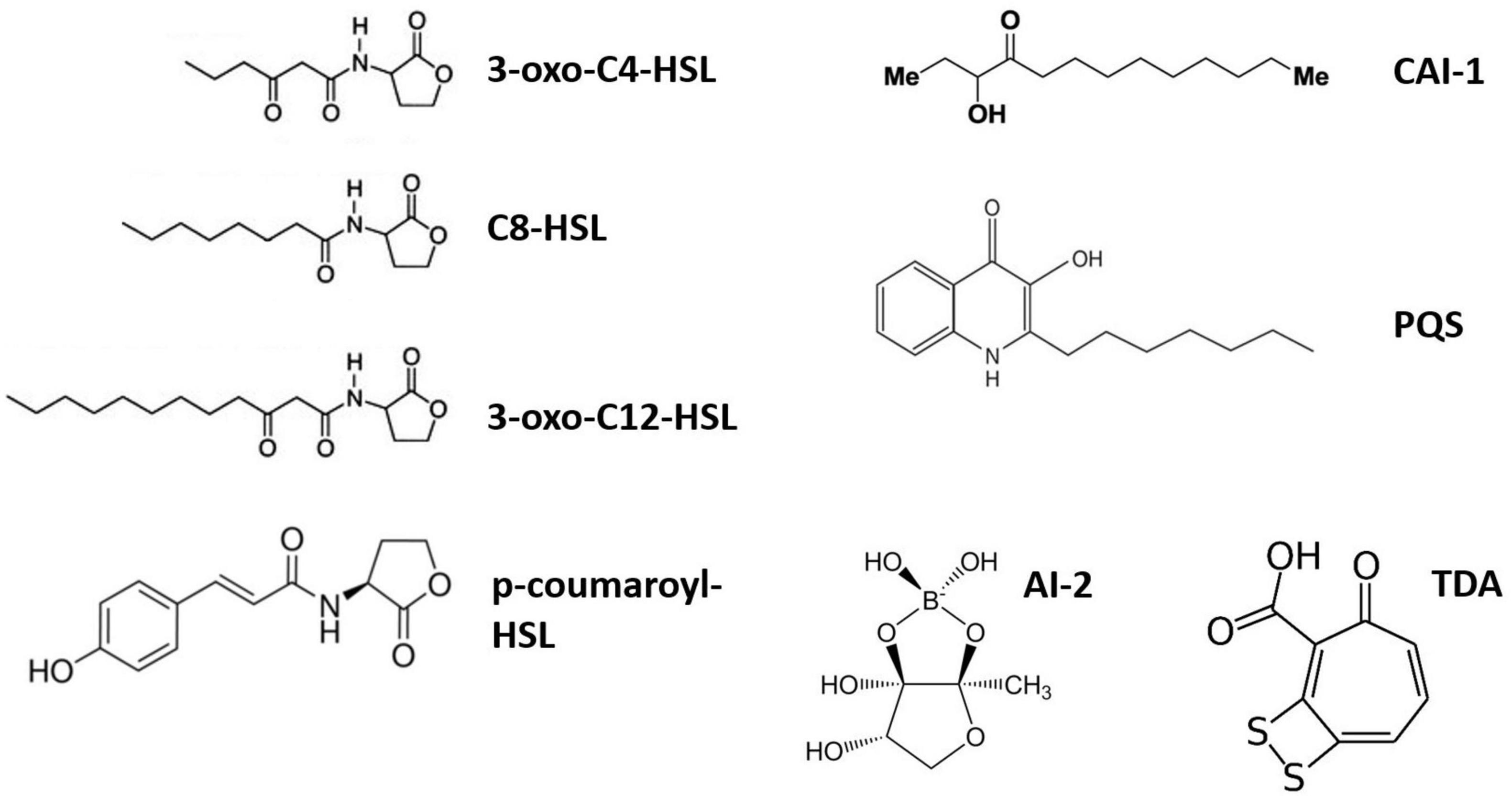
Figure 1. Diversity of AI molecules produced by marine bacteria. Only three AHL (3-oxo- C4-, C8-, and 3-oxo-C12-HSL) structures are illustrated. 3-oxo-C4-HSL, N-3-oxo-butyryl-L-homoserine lactone; C8-HSL, N-octanoyl-L-homoserine lactone; 3-oxo-C12-HSL, N-3-oxo-dodecanoyl-L-homoserine lactone; p-coumaroyl-HSL, N-(p-coumaroyl)-L-homoserine lactone; CAI-1, cholera autoinducer-1 [(S)-3-hydroxytridecan-4-one]; PQS, Pseudomonas quinolone signal (2-heptyl-3-hydroxy-4-quinolone); AI-2, furanosyl-borate diester; TDA, tropodithietic acid.
While the concept of QS was elaborated by studying the marine bacterium Vibrio fischeri (Nealson et al., 1970; Eberhard, 1972; Fuqua et al., 1994), it was subsequently studied mostly through a medical or agronomic lens (Lami, 2019). Little attention was given to QS from an ecological perspective before the 2000s, when studies from McLean et al. (1997) and Bachofen and Schenk (1998) suggested its importance in natural environments. Given the initial definition of QS (i.e., a cell-density-based system), the focus was almost immediately placed on bacteria colonizing particles living among dense biofilms. One of the first hypotheses was that bacteria colonizing marine snow used QS to synchronize the expression of extracellular hydrolytic enzymes, which are crucial for POM mineralization processes (Gram et al., 2002; Kiørboe et al., 2002). Gram et al. (2002) first reported the production of AHLs within bacterial strains isolated from marine snow, supporting this hypothesis. Since then, the advent of omics techniques has allowed us to better appreciate the importance of QS and QQ in marine genomes and metagenomes (Romero et al., 2012; Doberva et al., 2015; Su et al., 2021), more generally raising the question of the importance of QS in the regulation of marine biogeochemical processes.
In marine environments, QS communications are favored in specific microniches corresponding to densely colonized, biofilm-covered, biotic and abiotic surfaces (Hmelo and Van Mooy, 2009; Hmelo, 2017). Indeed, these microniches contain high OM concentrations, favoring a high cellular density. This allows AI accumulation and exacerbates the potential for bacterial interactions and communications (Moons et al., 2009; Dang and Lovell, 2016). In contrast, QS communication are thought to be limited in FL communities due to the spatial separation between the cells and the limited distance of QS signals diffusion (Hmelo and Van Mooy, 2009; Hmelo, 2017). These high cell-density microniches incidentally correspond to hotspots of bacterially mediated OM transformation and include, among others, marine aggregates, microbial mats, various microbiomes, and anthropogenic contaminants.
Marine aggregates, including marine snow particles, are rich in organic substrates, and are a well-known hotspot of bacterial mineralization mediated by extracellular hydrolytic enzymes (Smith et al., 1992; Dang and Lovell, 2016). These degradation processes are key in the ocean’s biogeochemistry, as they solubilize aggregates and thereby modify the flux of sinking POM and its subsequent storage in deep ocean sediments (Smith et al., 1992; Azam and Long, 2001). The marine communities colonizing aggregates can reach densities of 108–109 cells mL–1 (Azam and Long, 2001; Simon et al., 2002). They are especially enriched in Gammaproteobacteria (e.g., Oceanospirillales, Alteromonadales, Pseudomonadales) and Bacteroidetes (e.g., Flavobacteriales, Sphingobacteriales) but also in Alphaproteobacteria (e.g., Rhodobacterales, Rhizobiales, Rickettsiales), Betaproteobacteria (e.g., Burkholderiales) and Planctomycetes (e.g., Planctomycetales) (Rink et al., 2008; Bižic-Ionescu et al., 2015; Mestre et al., 2017, 2020; Li et al., 2021).
Microbial mats develop at liquid–solid interfaces, such as the water-sediment interface. They are thick, vertically layered biofilm structures experiencing sharp physicochemical gradients driving niche differentiation (van Gemerden, 1993; Visscher and Stolz, 2005). Microbial mats are characterized by some of the highest metabolic rates on earth and usually exhibit tight coupling between autotrophic and heterotrophic processes, resulting in the rapid cycling of elements (Visscher and Stolz, 2005). They are composed of dense microbial communities, whose densities may reach 109 cells g–1 (Carreira et al., 2015). Coastal microbial mats have been found to mainly host Alphaproteobacteria (e.g., Rhodobacterales and Sphingomonadales), Gammaproteobacteria (e.g., Chromatiales), Deltaproteobacteria (e.g., Desulfobacteriales and Desulfovibrionales), Bacteroidetes (e.g., Flavobacteriales and Sphingobacteriales), Actinobacteria but also Cyanobacteria and Archaea (Bolhuis and Stal, 2011).
The phycosphere is the diffusive boundary layer immediately surrounding phytoplankton cells. It is enriched in DOM and nutrients due to phytoplankton metabolic activity (Seymour et al., 2017). It also contains compounds such as dimethylsulfoniopropionate that act as chemoattractants for chemotactic bacteria (Stocker, 2012; Seymour et al., 2017). Dimethylsulfoniopropionate is an ecologically relevant S-containing metabolite produced by various marine algae and is an important C and S source for microorganisms (Yoch, 2002). Consequently, the phycosphere fosters a dense community of heterotrophic bacteria that process and transform this phytoplankton-derived OM, reaching densities of 108–1011 cells mL–1 (Sheridan et al., 2002). This coupling increases the rate and efficiency of OM transformation (Stocker, 2012; Seymour et al., 2017) and exerts an ecosystem-scale influence on biogeochemistry processes (Seymour et al., 2017). The typical bacteria often found in association with the phycosphere are dominated by Alphaproteobacteria (e.g., Rhodobacterales), Gammaproteobacteria (e.g., Vibrionales and Oceanospiralles) and Bacteroidetes (e.g., Flavobacteriales) (Goecke et al., 2013; Buchan et al., 2014).
Other relevant microbiomes include coral-, macroalga- or vertebrate-associated communities, among others. Macroalgae are especially important in coastal ecosystems, where they account for a large fraction of C burial (Duarte et al., 2005). Macroalgal biomass recycling is mainly mediated by colonizing epiphytic bacteria that form dense biofilms comprising 103–109 cells cm–2 (Martin et al., 2014; Lage and Graça, 2016). Alphaproteobacteria (e.g., Rhodobacterales and Sphingomonadales), Gammaproteobacteria (e.g., Alteromonadales, Pseudomonadales, Vibrionales), Bacteroidetes (e.g., Cytophagales and Flavobacteriales), Firmicutes (e.g., Bacillales), Actinobacteria (e.g., Actinomycetales) or Planctomycetes (e.g., Planctomycetales) are the major components of these communities (Hollants et al., 2013; Martin et al., 2014; Lage and Graça, 2016; Oh et al., 2021).
Finally, anthropogenic contaminants such as hydrocarbons or plastics are emerging microniches with biogeochemical implications. Hydrocarbons, such as alkanes or polycyclic aromatic compounds (PAHs), are solely composed of C and hydrogen. They represent one of the main sources of marine pollution and can persist for years in the environment, where bacteria play a key role in their biodegradation (Duran and Cravo-laureau, 2016). Hydrocarbonoclastic bacteria mostly belong to Gammaproteobacteria-affiliated genera, including specialized genera such as Cycloclasticus and Alcanivorax (Harayama et al., 2004; Chernikova et al., 2020), but also several other non-specialist bacterial genera such as Pseudomonas, Vibrio and Marinobacter (Dashti et al., 2015; Chernikova et al., 2020; Kumari et al., 2020). The plastisphere (the microbial community colonizing macro- and microplastics) is another niche of emerging relevance in marine biogeochemistry. Every year, tons of plastic waste enter the ocean, and an estimated five trillion plastic particles are currently floating in the seas (Eriksen et al., 2014). Immerged plastics are rapidly colonized by biofilm-forming bacteria, which can reach densities of 108 cells cm–2 (Schlundt et al., 2020), offering new hotspots for bacterial colonization and activity (Zettler et al., 2013). The plastisphere communities are enriched in bacteria belonging to Bacteroidetes (e.g., Flavobacteriales and Sphingobacteriales), Alphaproteobacteria (e.g., Rhodobacterales and Sphingomonadales), Betaproteobacteria (e.g., Burkholderiales) or Gammaproteobacteria (e.g., Oceanospirillales, Alteromonadales, Vibrionales) (Zettler et al., 2013; Schlundt et al., 2020; Wright et al., 2021).
The bacterial hotspots highlighted above are colonized by highly diverse bacteria, dominated by a few reoccurring groups, belonging to the Alphaproteobacteria (e.g., Rhodobacterales and Sphingomonadales), Gammaproteobacteria (e.g., Alteromonadales, Pseudomonadales, and Vibrionales) or Bacteroidetes (e.g., Flavobacteriales). They are ecologically relevant given their abundance and their involvement in all major biogeochemical cycles (Figure 2). These groups are notably largely involved in the consumption and transformation of algal-derived organic substrates that constitute the bulk of DOM (Buchan et al., 2016; Teeling et al., 2016). Interestingly, there is overlap between the taxa colonizing the highlighted biogeochemical hotspots and the taxa known to harbor QS systems. Indeed, the AHL-based QS and QQ genes found in marine metagenomics studies (Romero et al., 2012; Doberva et al., 2015; Huang et al., 2018; Muras et al., 2018; Su et al., 2021) are mostly affiliated with the roseobacter group (Rhodobacterales and Alphaproteobacteria), Gammaproteobacteria genera (such as Pseudomonadales, Alteromonadales or Vibrionales) and Flavobacteriales (Bacteroidetes) (Supplementary Table 1). In addition, several strains affiliated with these groups have been shown to produce AHLs and AI-2 during cultivation (Figure 2 and Supplementary Tables 2, 3). They also encode a variety of other communication systems, although they are much less investigated.
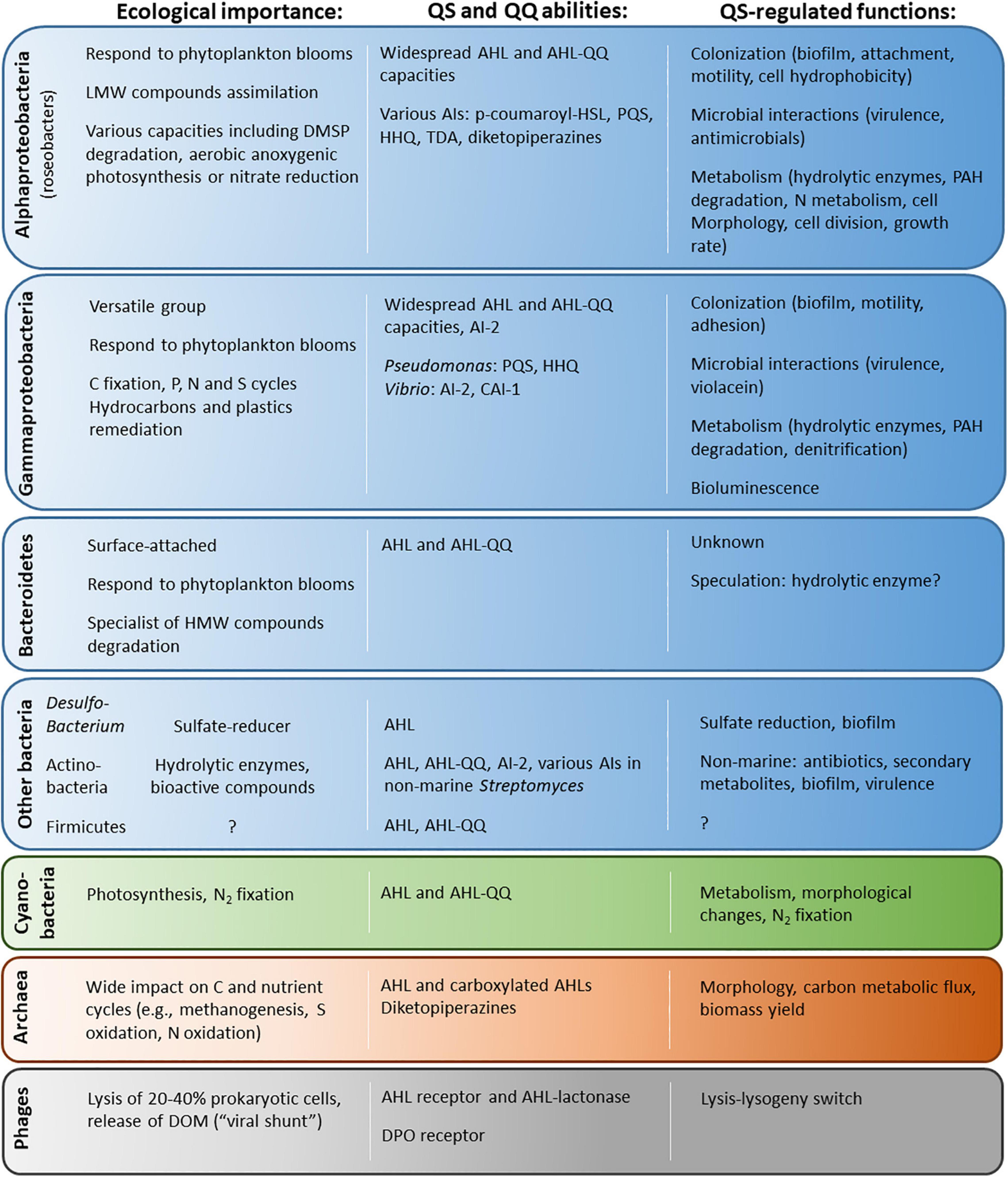
Figure 2. Ecological importance, QS pathways and QS-regulated functions in the main prokaryotic groups (as well as phages) presented in this review. The relevant references can be found in the main text as well as in Supplementary Table 4 concerning QS circuits.
The Alphaproteobacteria class comprises numerous, diverse orders, including Rhodobacterales, Pelagibacterales, and Sphingomonadales. Within the Rhodobacterales order, the roseobacter group is of particular interest (Buchan et al., 2005; Geng and Belas, 2010a). Indeed, it is one of the most readily cultivated and extensively studied marine lineages (Wagner-Döbler and Biebl, 2006; Buchan et al., 2014). In addition, roseobacter members (hereinafter roseobacters) are present in all major marine habitats, including living freely in bulk seawater, in association with phytoplankton, marine snow, and plastics or within sediments and microbial mats (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Luo and Moran, 2014). They can constitute up to 25% of the bacterial communities at the ocean surface (Buchan et al., 2005; Luo and Moran, 2014) and have been reported to be major primary colonizers in coastal waters (Dang et al., 2008).
Alphaproteobacteria are thought to primarily consume small organic molecules such as sugar monomers, amino acids, dimethylsulfoniopropionate or phosphonates (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Luo and Moran, 2014). They possess versatile metabolic capacities with important implications for the C, N and S cycles: they are able to perform aerobic anoxygenic photosynthesis, degrade sulfur compounds (including dimethylsulfoniopropionate) and aromatics, perform assimilatory and dissimilatory nitrate reduction, and produce multiple bioactive secondary metabolites (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Luo and Moran, 2014). They play an especially important role in the microalgal phycosphere, where they are well adapted to form symbioses (Geng and Belas, 2010a).
Alphaproteobacteria, particularly roseobacters, are widely involved in AHL-based QS communication in marine environments. They constitute the majority of the AHL-producing bacteria isolated from various environments, including marine snow, coastal biofilms, or marine seagrasses (Gram et al., 2002; Wagner-Döbler et al., 2005; Huang et al., 2008; Blanchet et al., 2017; Su et al., 2019; Supplementary Table 2). In addition, Rhodobacterales are among the main contributors to AHL-related genes in marine metagenomics studies (Doberva et al., 2015; Huang et al., 2018; Su et al., 2021; Supplementary Table 1). The roseobacter group is perhaps the most frequently investigated group in AHL-based QS studies, and this communication system is widespread among its members (Wagner-Döbler and Biebl, 2006; Buchan et al., 2016). It has been shown that 82% of roseobacters possess at least one luxRI gene cassette and that 28% possess two (Buchan et al., 2016). Additionally, roseobacters often possess solo luxI genes (i.e., genes that are not located near a tandem luxR receptor gene), although their ecological role is unclear thus far (Cude and Buchan, 2013; Buchan et al., 2016). Solo luxI are especially common among the Sulfitobacter, Ruegeria and Phaeobacter genera (Buchan et al., 2016). Among roseobacters, AHL-based QS seems to be involved in the production of antimicrobials (e.g., indigoidine and tropodiethietic acid), polysaccharide production, gene transfer, biofilm formation and motility, among others (Wagner-Döbler and Biebl, 2006; Buchan et al., 2016).
The marine members of the Alphaproteobacteria also use other QS systems (Figure 2 and Supplementary Table 3), including systems based on tropodiethietic acid, p-coumaroyl-homoserine lactone (p-coumaroyl-HSL) or AI-2. Tropodiethietic acid is a sulfur-containing molecule whose production is common among roseobacters (Buchan et al., 2016). It plays dual roles, serving both as an antibiotic and an AI signal at subinhibitory concentrations (Geng and Belas, 2010b; Beyersmann et al., 2017). Similar to AHLs, tropodiethietic acid modulates motility, biofilm and antibiotic production in the marine bacterium Phaeobacter inhibens, which are important traits during algal colonization (Beyersmann et al., 2017).
In addition, p-coumaroyl-HSL production has been demonstrated in the marine strains Silicibacter pomeroyi DSS-3 (Schaefer et al., 2008) and Ruegeria sp. KLH11 (Zan et al., 2015). Schaefer et al. (2008) illustrated that an AHL synthase could use a non-fatty acyl substrate to synthesize aryl-HSL derivatives such as p-coumaroyl-HSL, suggesting that a large pool of fatty acids could be used to generate a number of as-yet-undiscovered AI signals. This could be beneficial to organisms using QS to control functions that are useful only under a particular set of conditions (Schaefer et al., 2008), including during algal/bacterial interactions (Schaefer et al., 2008).
Finally, Alphaproteobacteria also seem to use QS systems based on AI-2. This boron-containing molecule is often considered a universal AI mediating interspecies communication (Xavier and Bassler, 2003). However, the function of AI-2 as a QS signal has been debated, since it may also represent a metabolic side-product in a number of bacteria (Diggle et al., 2007; Rezzonico and Duffy, 2008). In any case, AI-2-producing (Blanchet et al., 2017; Tourneroche et al., 2019) and AI-2-sensing (Pereira et al., 2008; Su et al., 2019) marine and soil Alphaproteobacteria have been reported.
Alphaproteobacteria also encode AHL-QQ mechanisms. They represent a large number of marine bacterial isolates with QQ capacities (Romero et al., 2011a; Blanchet et al., 2017; Rehman and Leiknes, 2018; Su et al., 2019; Supplementary Tables 2, 3). Zhao et al. (2019) reported that marine Alphaproteobacteria constituted 21% of marine isolates with potential AHL-QQ capacities (across 19 reviewed studies). Quite interestingly, Alphaproteobacteria are not the main contributors of enzymatic QQ-related sequences (AHL lactonases and acylases) according to metagenomics studies (Huang et al., 2018; Su et al., 2021). This could suggest that they mainly employ QSI-based QQ, although this could also result from a cultivation bias.
Gammaproteobacteria is one of the most genus-rich prokaryotic taxa. This class includes the well-studied Vibrionales order as well as other easily culturable bacterial genera, such as Alteromonas, Pseudoalteromonas, Marinomas, or Oceanospirillum (Fuhrman and Hagström, 2008). Consequently, the Gammaproteobacteria is a diverse group and its members can thrive under various conditions. They can constitute up to 40% of the bacterial community in different oceans (Sunagawa et al., 2015), sediments (Zinger et al., 2011; Dyksma et al., 2016), or marine snow (Mestre et al., 2017).
This phylogenetic diversity translates into a variety of metabolic functions and an important contribution to biogeochemical-related processes. Gammaproteobacteria generally have high potential growth rates and have been shown to respond rapidly to OM concentration increases (Fuhrman and Hagström, 2008). As such, they participate in C mineralization by processing phytoplankton-derived compounds during blooms with a lifestyle that overlaps with those of Bacteroidetes and Alphaproteobacteria, consuming both high and low molecular weight molecules (Ferrer-González et al., 2021; Francis et al., 2021). Gammaproteobacteria also play a major role in dark C fixation in coastal sediments, representing an important C sink (Dyksma et al., 2016). In addition, they widely participate in P, N, and S nutrient cycles in different habitats (Villarreal-Chiu et al., 2012; Baker et al., 2015; Zhou et al., 2020). Finally, Gammaproteobacteria play an important role in the bioremediation of anthropogenic pollutants such as hydrocarbons or plastic particles. For instance, bacteria of the Alcanivorax genus (Oceanospirillales) are able to degrade petroleum hydrocarbons and are among the first responders to oil spills (Harayama et al., 2004). Several other Gammaproteobacteria genera have been shown to degrade hydrocarbons such as Marinobacter, Oleiphilus, and Oleispira but also the more generalists Vibrio, Pseudoalteromonas, or Marinomonas (Harayama et al., 2004; Zhou et al., 2020). Likewise, plastics are hydrocarbon-derived polymers that can be biodegraded by marine Gammaproteobacteria, notably Pseudomonas- or Vibrio-affiliated strains (Jacquin et al., 2019; Roager and Sonnenschein, 2019).
The Gammaproteobacteria class probably contains bacteria with the best-characterized AHL-based QS systems, such as members of the Vibrio, Pseudomonas, and Acinetobacter genera. They are also among the most common AHL producers isolated from various marine habitats (Cuadrado-Silva et al., 2013; Jatt et al., 2015; Freckelton et al., 2018; Charlesworth et al., 2019; Reen et al., 2019; Su et al., 2019; Urvoy et al., 2021a; Supplementary Table 2). In addition, they constitute the main class contributing to AHL production and sensing genes in marine metagenomics surveys, especially members of Vibrionales, Pseudomonadales, or Alteromonadales (Doberva et al., 2015; Huang et al., 2018; Su et al., 2021; Supplementary Table 1). Gammaproteobacteria AHL pathways have been shown to control various phenotypes with potential biogeochemical relevance, such as biofilm formation, hydrolytic enzyme (including metalloproteases, chitinase, caseinase, amylase, alkaline phosphatase, and DNAse) production, siderophore production and motility (reviewed in Torres et al., 2019).
In addition to AHL signals, marine Gammaproteobacteria possess a range of diverse, interconnected QS pathways involved in inter- and intraspecies communication, and based on AI-2, PQS or CAI-1 signals, among others (Figure 2 and Supplementary Table 3). As noted previously, the function of AI-2 as a QS signal has been debated but has been demonstrated in several Vibrio and Shewanella strains (Sun et al., 2004; Bodor et al., 2008; Rezzonico and Duffy, 2008). AI-2 can be sensed by the periplasmic binding receptors luxPQ (In Vibrionaceae), LsrB and probably other uncharacterized receptors (Chen et al., 2002; Zhang et al., 2020). The AI-2 system has notably been demonstrated to regulate bioluminescence, biofilm formation, motility, chemotaxis, proteases and virulence factor production in various strains, such as the marine V. harveyi (reviewed in Pereira et al., 2013).
Pseudomonas quinolone signal, as suggested by its name, is a QS signal that is considered to be specific to the Pseudomonas genus. The production of PQS is encoded by the pqsABCDE operon, and its immediate precursor is HHQ (4-hydroxy-2-heptylquinoline), which also acts as an AI. The PQS system has mostly been studied in clinical Pseudomonas strains, where it regulates virulence factors (such as biofilm, elastase, rhamnolipid or pyocyanin) production and promotes infection (as reviewed in García-Reyes et al., 2020). However, the synthesis of PQS and HHQ has been discovered in other genera, such as Burkholderia and a marine Pseudoalteromonas bacterium (Vial et al., 2008; Paulsen et al., 2020). In addition, it has been shown that PQS acts as a signal molecule in several Gram-negative and Gram-positive strains (including marine isolates) (Reen et al., 2011) as well as in multispecies marine communities (Wang et al., 2020), highlighting its role as a possible interspecies signal.
Finally, the CAI-1 signaling molecule is an example of an intraspecies-specific AI that is used among the Vibrio genus and synthesized by the CqsA protein (Henke and Bassler, 2004). This AI could regulate virulence factors operating through the CqsS receptor (Henke and Bassler, 2004). Its relevance in the ocean still has to be studied, especially in niches where Vibrio are dominant, such as on certain plastic particles (Zettler et al., 2013) or macroalgae (Hollants et al., 2013).
Marine Gammaproteobacteria are also actively engaged in AHL-QQ mechanisms involving both QQ enzymes and QSIs. Several studies have reported that isolated marine Gammaproteobacteria strains possess QQ capacities, in which they were often the most represented taxon (Torres et al., 2016; Freckelton et al., 2018; Su et al., 2019; Urvoy et al., 2021a; Supplementary Table 2). In their review, Zhao et al. (2019) reported that 27% of marine bacterial isolates with QQ capacities were Gammaproteobacteria. In addition to the common AHL acylase and lactonase, marine strains have been reported to produce QSIs such as cyclic peptides (Marinobacter sp., Photobacterium halotolerans) or phenol derivatives (Vibrio alginolyticus) (as reviewed in Saurav et al., 2017 or Zhao et al., 2019). In marine metagenomics, Gammaproteobacteria often constitute the majority of AHL-QQ enzyme sequences (Huang et al., 2018; Su et al., 2021; Supplementary Table 1).
Bacteroidetes are thought to be among the most abundant groups of marine bacteria with the Proteobacteria and Cyanobacteria (Kirchman, 2002), contributing up to 50% of the total bacteria in some marine ecosystems (Cottrell and Kirchman, 2000; Acinas et al., 2015). The Bacteroidetes phylum comprises the Bacteroidia, Flavobacteria, Sphingobacteria, and Cytophagia classes. Bacteroidetes mostly live attached to surfaces such as aggregates and micro- or macroalgae (Kirchman, 2002). They can constitute one of the most abundant phyla detected on marine and estuarine particles (Delong et al., 1993; Ploug and Grossart, 1999), which was linked to their capacities to colonize surfaces and degrade complex, recalcitrant polymers (Kirchman, 2002). Indeed, Bacteroidetes (particularly Flavobacteriales) are largely considered specialists in polymers degradation and comparative genomics has demonstrated that they contain many peptidases and carbohydrate-active enzymes (Cottrell and Kirchman, 2000; Thomas et al., 2011; Fernàndez-Gómez et al., 2013). Bacteroidetes have evolved a unique degradation system known as polysaccharide utilization loci, in which all relevant genes involved in the cleavage of a particular carbohydrate are grouped together (Grondin et al., 2017). They often include energy-dependent TonB-dependent transporters that allow the sequestration of large molecules in the periplasmic space prior to further degradation, protecting them from scavenging bacteria (Grondin et al., 2017).
Despite their importance in colonization and OM degradation, Bacteroidetes are not well studied in terms of QS pathways. However, several marine Bacteroidetes strains have been shown to produce AHLs during cultivation, such as Flammeovirga sp., Muricauda sp., Tenacibaculum spp., Maribacter sp. and Aquimarina sp. (Huang et al., 2008; Romero et al., 2010; Abdul Malik et al., 2020; Figure 2 and Supplementary Tables 2, 3). In addition, genes related to AHL sensing (Su et al., 2021) and AI-2 synthesis (Huang et al., 2018) have been affiliated with Bacteroidetes during metagenomic surveys (Supplementary Table 1). It seems that marine Bacteroidetes show widespread QQ capacities, as highlighted by several studies (Romero et al., 2010, 2011a,2012; Su et al., 2019; Supplementary Table 2). Zhao et al. (2019) found that Bacteroidetes represented 9% of QQ-capable marine bacterial isolates across 19 studies. Su et al. (2021) also reported that Flavobacteriales contributed to the lactonase and acylase AHL-QQ gene pool during a marine metagenomic study. An interesting finding has been made by Barriuso and Martínez (2018) concerning QS pathways in environmental Bacteroidetes. They examined the QS-related genes (AHLs and AI-2 synthases and receptors) present in 22 metagenomes from natural and artificial biofilm habitats and found that all sequences related to Bacteroidetes corresponded to LuxR-encoding genes. The imbalanced ratio between AHL receptors and synthases points toward the existence of AHL-QS cheaters in this group. This also seems to be reflected by marine metagenomics studies, as Su et al. (2021) showed that Flavobacteriales contributes to AHL-sensing and AHL-QQ genes, although they do not contribute to the AHL-synthase gene pool. Given the lack of data on this phylum, the physiological processes under QS control in Bacteroidetes are far from elucidated, especially concerning the potential role of QS in OM degradation.
Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes represent the majority of isolated bacteria with QS or QQ capacities and are the main contributors of QS genes in marine metagenomic datasets. However, some data show that other ecologically relevant microorganisms may use QS communications, notably members of Actinobacteria, Firmicutes, Cyanobacteria, Archaea and even bacteriophages.
Actinobacteria (Gram-positive) are ubiquitous in the ocean realm (Giovannoni and Stingl, 2005) and can reach high abundance in deep-sea sediments (Zinger et al., 2011; Chen et al., 2016), marine aggregates (Grossart et al., 2004) and macroalgae (Oh et al., 2021). Although their metabolism and roles are largely unknown, these bacteria possess various extracellular hydrolytic enzymes that are able to degrade recalcitrant OM (Lam, 2006; Chen et al., 2016). They are also recognized for their production of diverse bioactive compounds (Lam, 2006; Chen et al., 2016). In terms of QS, several marine AHL-producing strains have been isolated from marine snow (Su et al., 2019), microbial mats (Charlesworth et al., 2019) or marine sponges (Bose et al., 2017; Supplementary Tables 2, 3). In addition, AHL-quenching strains affiliated with Actinobacteria have been isolated from various coastal communities (Romero et al., 2011a), the Mediterranean Sea (Muras et al., 2018), the Colombian Caribbean Sea (Betancur et al., 2017) and marine snow (Su et al., 2019; Supplementary Table 2). These bacteria are mostly affiliated with genera such as Streptomyces, Salinispora or Microbacterium. Actinobacteria also use other QS systems based on AIs such as gamma-butyrolactone or AI-2 (reviewed in Polkade et al., 2016), although this has not been well explored in marine bacteria.
Firmicutes members are mostly Gram-positive bacteria. Their role and prevalence in the oceans are poorly characterized, but they can constitute an important fraction of the epiphytic macroalgal community (Hollants et al., 2013; Oh et al., 2021), microbial mats (Bolhuis and Stal, 2011) and sediments (Zinger et al., 2011). A few studies have reported Firmicutes-affiliated strains isolated from marine environments with AHL-based QS or QQ capacities (Supplementary Table 2). AHL production has been reported in genera such as Bacillus spp. (Freckelton et al., 2018; Charlesworth et al., 2019), Exiguobacterium sp. (Biswa and Doble, 2013), Virgibacillus sp. (Charlesworth et al., 2019) and Halobacillus sp. (Charlesworth et al., 2019). In addition, strains such as Bacillus spp. (Kanagasabhapathy et al., 2009; Romero et al., 2011a; Teasdale et al., 2011), Planomicrobium chinense (Muras et al., 2018) and Oceanobacillus sp. (Romero et al., 2011a) have showed QQ capacities.
As obligate phototrophs, Cyanobacteria are commonly found in the photic zone of all oceans and can account for up to 90% of primary production in oligotrophic waters (Stockner, 1988). Cyanobacteria play a crucial role in N2 fixation, which significantly contributes to global N budgets (Fuhrman and Hagström, 2008). Although QS is not well studied in cyanobacteria, at least two reports have demonstrated AHL production in non-marine axenic cultures of cyanobacteria. Sharif et al. (2008) demonstrated that the epilithic colonial cyanobacterium Gloeothece PCC6909 (and its sheathless mutant PCC6909/1) produced C8-HSL, which seemed involved in carbohydrate and amino acid metabolism. Zhai et al. (2012) reported that a lacustrine Microcystis aeruginosa PCC7820 produced three putative AHLs, although their structure was not fully determined. The authors suggested that QS might regulate morphological changes. In addition, several marine cyanobacteria have been shown to produce AHL-QSIs (as reviewed in Saurav et al., 2017) and AHL-QQ enzymes (Romero et al., 2008). For example, the marine nitrogen-fixing bacterium Anabaena sp. PCC7120 produces the AHL-lactonase AiiC which has a wide range of AHL degradation capacities (Romero et al., 2008).
Although archaea were initially considered extremophiles, they have since been shown to be present in nearly every marine environment, representing an estimated number of ∼1028 cells in the oceans (Karner et al., 2001). They are particularly abundant in the deep ocean, where they can constitute up to 39% of the total picoplanktonic community (Karner et al., 2001). Archaea perform important functions such as methanogenesis, S oxidation and ammonia oxidation (Jarrell et al., 2011; Santoro et al., 2019). Few papers have addressed QS communications in archaea thus far, especially in marine environments. To our knowledge, only Charlesworth et al. (2020) identified marine archaeal strains that induce an AHL biosensor. In addition, Su et al. (2021) highlighted AHL-lactonase sequences affiliated with archaea during a marine metagenomics survey. Additional studies were performed in non-marine environments, suggesting that QS could be an overlooked mechanism in marine archaea. For instance, several archaeal strains isolated from various lakes activated AHL biosensors (Paggi et al., 2003; Liao et al., 2016; Charlesworth et al., 2020), although the underlying compounds were not analytically determined. The Methanosaeta harundinacea 6Ac strain, isolated from an anaerobic digester, produces carboxylated AHLs that modulates cell morphology and C metabolic fluxes (Zhang et al., 2012). In addition, the authors identified the first putative archaeal AHL synthase, which was different from any known bacterial synthase (Zhang et al., 2012). Rajput and Kumar (2017) identified numerous solo luxR genes by mining the Interpro archaeal genomes. Finally, outside of the AHL system, the lacustrine halophilic Haloterrigena hispanica was shown to use diketopiperazines as a QS signal, although their role remains unclear (Tommonaro et al., 2012). Archaeal enzymes and overall metabolism vastly differ from bacteria. As such, the presence of bacterial QS systems in Archaea could point to a punctual adaptation (e.g., lateral gene transfer) to their environment, although this should be further investigated.
Viruses are by far the most abundant entities in the oceans: although they represent only 5% of the marine bacterial biomass, they constitute up to 94% of marine microorganisms (Suttle, 2007). By lysing 20–40% of prokaryotes in surface waters each day, bacteriophages shape the bacterial community composition (BCC) and release copious amounts of labile DOM (“viral shunt”), thus playing a major role in marine biogeochemistry (Suttle, 2005, 2007). Recent studies highlighted that phages could harbor genes related to bacterial QS. Silpe et al. (2020) reported that the vibriophage VP882, infecting Vibrio cholera, carries a gene encoding a DPO-binding (3,5-dimethylpyrazin-2-ol, an AI used by V. cholera) protein driving the lysis-lysogeny decision. In addition, several phages seem to encode AHL-receptor luxR genes, including two phages infecting Aeromonas spp. (Silpe and Bassler, 2019) and marine flavobacterial phages (Bartlau et al., 2021). Finally, Su et al. (2021) reported phage-affiliated AHL lactonases during a metagenomics study.
The numerous QS circuits present among marine bacteria could directly impact biogeochemical cycles through the transcriptional regulation of genes involved in degradation, C allocation and nutrient acquisition, among others. QS may also indirectly modulate those phenotypes by altering bacterial community structure and functions, which has been highlighted in both marine bacterial strains and complex bacterial communities (Figure 3 and Supplementary Tables 3, 4).
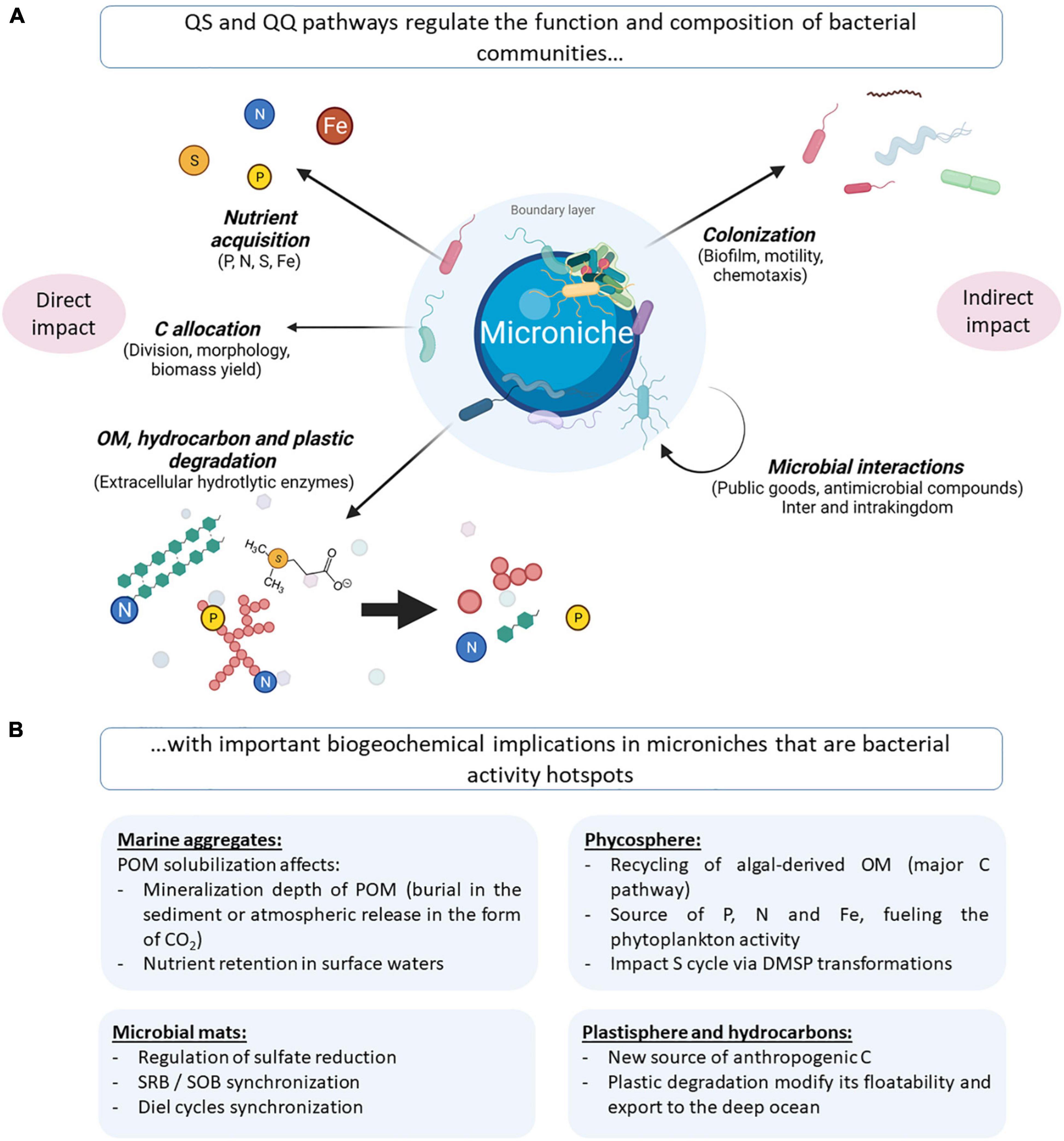
Figure 3. (A) Illustration of QS-regulated processes that potentially impact biogeochemical cycles, either directly through the modulation of OM degradation and nutrient acquisition (left) or indirectly through the modulation of the microbial composition (right). (B) Illustration of the repercussions of the highlighted QS-regulated processes in microniches that are bacterial activity hotspots. OM, organic matter; POM, particulate organic matter; DMSP, dimethylsulfoniopropionate; SRB, sulfate-reducing bacteria; SOB, sulfide-oxidizing bacteria. Created with BioRender.com.
One of the best-studied functions of QS expression in the marine environment is the regulation of extracellular hydrolytic enzyme production, including cell-bound and dissolved (i.e., cell-free) enzymes. The bacterial degradation of OM by hydrolytic enzymes is essential to marine life, as it allows heterotrophic bacteria to grow and initiates the microbial loop (Arnosti, 2011). The possible role of QS in synchronizing hydrolase expression in bacteria would impact C, P, and N cycles through the degradation of various polymers. Several studies have examined the implications of QS in hydrolase expression in either isolated strains (Jatt et al., 2015; Su et al., 2019; Urvoy et al., 2021a) or natural bacterial communities from various environments (Hmelo et al., 2011; Van Mooy et al., 2012; Krupke et al., 2016; Su et al., 2019; Urvoy et al., 2021b; Supplementary Table 4). These studies, mainly performed in the context of AHL-based QS, have largely demonstrated the complexity of QS regulation, as no clear pattern has emerged.
Hmelo et al. (2011) were the first to demonstrate that the addition of AHL signals (3-oxo-C6-HSL or 3-oxo-C8-HSL) to bacterial communities colonizing marine snow particles increased their extracellular aminopeptidase, lipase and phosphatase activities (total activity, including cell-bound and cell-free activities). These findings were later reinforced by Krupke et al. (2016) and Su et al. (2019). Krupke et al. (2016) found that AHL amendment (3-oxo-C8-HSL or a cocktail of C10-, C12- and C14-HSL) caused as much inhibition as stimulation of extracellular aminopeptidase, lipase and phosphatase (total activity) across five locations in the Atlantic and Pacific oceans. They particularly highlighted the complexity of these cell–cell mechanisms, as the responses varied depending on the sampling location, AHL concentration, and time scale. Su et al. (2019) demonstrated that the addition of 3-oxo-C8-HSL to marine snow collected in the Yellow Sea upregulated galactosaminidase and β-xylosidase but downregulated β-glucosidase and mannosidase (cell-free activity) extending QS involvement to glucosidases. Van Mooy et al. (2012) made similar observations in epibiontic bacteria colonizing Trichodesmium colonies, demonstrating that the addition of C8-, 3-oxo-C8-HSL and long-chain AHLs (a cocktail of C10-, C12- and C14-HSL) induced a doubling of extracellular alkaline phosphatase activity (total activity). Finally, Urvoy et al. (2021b) showed that the addition of low concentrations of C4-, C6-, 3-oxo- C8-, C12- or C16-HSL to coastal bacterioplankton communities sampled in two contrasting stages of the spring phytoplankton bloom differently modified their extracellular leucine aminopeptidase, N-acetyl glucosaminidase, lipase and β-glucuronidase activities (total activity), further highlighting the complexity of QS regulation.
A few studies on isolated strains have illustrated the implications of AHL-based QS in hydrolytic enzyme regulation. Jatt et al. (2015) showed that in Pantoea ananatis B9 (Enterobacteriales, Gammaproteobacteria) isolated from marine snow, extracellular (cell-free) phosphatase activity was increased by C10-HSL amendment and decreased by the AHL-lactonase AiiA. Su et al. (2019) highlighted that lipase (cell-free) production was increased by several AHLs and decreased by the AHL-lactonase MomL in Ruegeria mobilis Rm01 (Rhodobacterales, Alphaproteobacteria) isolated from marine snow. Finally, Urvoy et al. (2021a) tested the effect of endogenous AHL disruption on both cell-free and cell-bound β-glucosidase and leucine aminopeptidase in 28 estuarine strains affiliated with Gammaproteobacteria (Pseudomonadales, Vibrionales and Enterobacterales) and Bacteroidetes (Sphingobacteriales) using a lactonase preparation. AHL disruption affected the activity of at least one enzyme in 23 strains, resulting in both increased and decreased activities. Interestingly, AHL disruption could differentially regulate cell-bound and cell-free activities in some strains, highlighting the need to study the two pools of extracellular enzymes separately. In addition to these widespread hydrolases, QS was shown to regulate more specific enzymes degrading anthropogenic compounds in several marine strains. For instance, Mangwani et al. (2015) showed that the marine bacterium Pseudomonas aeruginosa N6P6 (Gammaproteobacteria) used AHL-based QS to regulate PAH (phenanthrene and pyrene) degradation, which was reduced in the presence of an AHL QSI. Kumari et al. (2020) later showed that the same strain mediated naphthalene degradation through QS-regulated naphthalene dioxygenase gene expression. In addition, Yu et al. (2020) used deletion mutants to highlight that the strain Croceicoccus naphthovorans PQ-2 (Sphingomonadales, Alphaproteobacteria), isolated from a marine biofilm, could degrade various PAHs and that AHL-based QS regulated several of its PAH-degrading genes. Finally, Huang et al. (2013) highlighted the evident overlap between PAH degradation and AHL-related genes in bacteria and supported this hypothesis by mining the NCBI genome database, suggesting that AHL-based regulation could play a pivotal role in the bioremediation of hydrocarbons and PAHs.
Much less is known about the impact of non-AHL QS signals on hydrolytic enzymes, which could either exacerbate or mitigate AHL-based regulation. Indeed, Van Mooy et al. (2012) showed that the addition of (S)-4,5-dihydroxy-2,3-pentanedione (DPD, which spontaneously transforms into AI-2 in seawater) decreased extracellular phosphatase production by the epibiont of Trichodesmium colonies, while AHL addition increased phosphatase activity. In contrast, Su et al. (2019) demonstrated that AI-2 addition increased lipase production in R. mobilis Rm01 (Alphaproteobacteria), similar to the effect of AHL amendment.
It is clear that QS is an important mechanism involved in hydrolytic enzyme regulation with potentially strong ecological implications in all the mentioned microniches. In aggregates and marine snow, the QS-based regulation of hydrolases could impact the solubilization of sinking POM, retaining particles and nutrients in surface waters (Hmelo et al., 2011). In the phycosphere, the degradation of organic P by QS-regulated-alkaline phosphatase may impact the productivity of the broader phytoplanktonic community and P cycling at the ocean surface (Van Mooy et al., 2012). The regulation of enzymes degrading hydrocarbons could play a pivotal role in their bioremediation, generating new C sources for marine bacteria and affecting the transport of those compounds to the deep ocean. However, these regulation mechanisms seem to be extremely complex, as no clear trend has emerged: QS-based regulation impact different sets of activities in both isolated strains and natural communities, and AI amendment or disruption can provoke both an increase or decrease in activity, respectively. In addition, these studies highlight the need to investigate both cell-bound and cell-free activities separately.
As the main component of life, it is of utmost importance to understand the mechanisms driving the marine C cycle. In addition to regulating the degradation of C-containing substrates, QS could influence the marine C cycle in more subtle ways by altering phenotypes involved in cell morphology and biomass yield. Using transcriptomic and mutant construction techniques, Patzelt et al. (2013) demonstrated that QS induced morphological heterogeneity and controlled cell division in the marine bacterium Dinoroseobacter shibae (roseobacter, Alphaproteobacteria). Indeed, the wild-type (AHL-producing) strain exhibited a heterogeneous cell morphology, with the coexistence of ovoid and rod-shaped cells reaching up to 10 μm. The elongated cells divided by forming one smaller daughter cell (polar growth), while small ovoid cells divided by binary fission, yielding two equally sized cells. In contrast, the luxI knockout mutant (non-AHL-producing) grew faster, was homogeneous in terms of size and morphology (small ovoid rods) and replicated only via binary fission. As such, by controlling morphology, cell division and growth rates, QS ultimately impacts the utilization and cellular allocation of C. This induced morphological heterogeneity could be beneficial under changing conditions, such as in the phycosphere during a bloom. Other reports in non-marine bacteria further suggest QS involvement in such processes. For instance, in Escherichia coli, AI-2-based QS was reported to modify cell division, cell shape and morphology (DeLisa et al., 2001; Sperandio et al., 2001). Zhang et al. (2012) reported that in the archaeon Methanosaeta harundinacea 6Ac (isolated from an anaerobic digester), carboxylated AHLs facilitated the transition from a short cell morphology to filamentous growth. This change in morphology was accompanied by important changes in metabolism, with an altered carbon metabolic flux that reduced the cellular biomass yield.
Phosphorus is an important element involved in energy storage, cell structure, signal relay and genetic material (White and Dyhrman, 2013). In the ocean, P is essentially available to microorganisms in its phosphate form, which can be released from dissolved and particulate organic P using alkaline phosphatases (Chróst, 1990). As outlined above, AHLs and AI-2 were shown to impact alkaline phosphatase activities (Hmelo et al., 2011; Van Mooy et al., 2012; Krupke et al., 2016). However, QS could be involved in many more mechanisms impacting the fate of this nutrient, as recently demonstrated by Xu et al. (2021) in a lacustrine community. AHL addition to a limnic epiphytic biofilm showed that QS mediated both intra- and extracellular P entrapment, i.e., P storage in polyphosphate-accumulating bacteria or extracellular polymeric substances (EPS) (Xu et al., 2021). QS promoted intracellular P entrapment via the upregulation of 43% of the genes identified as being involved in inorganic and organic P accumulation (e.g., alkaline phosphatases phoA, phoX, and phoD), P uptake and transport (e.g., phosphate-specific transport operon pst), and P regulation (e.g., phoB, phoR, and phoU), as determined via metatranscriptomics. In addition, it promoted extracellular P entrapment via the upregulation of EPS production and the modification of their composition. EPS impact P entrapment by complexing with phosphate through the amine groups present in their polysaccharidic and proteic fractions (Xu et al., 2020). Although performed in a lacustrine biofilm, we hypothesize that similar mechanisms occur in marine environments. QS-regulated P acquisition could be important in the phycosphere, especially in P-limited environments where bacteria could supply (or compete for) available P to phytoplankton cells.
Nitrogen is an important macronutrient and one of the main factors limiting primary production in the oceans (Pajares and Ramos, 2019). It mainly occurs as dinitrogen (N2), only available to nitrogen-fixing microorganisms (mostly cyanobacteria) that convert it into more available forms, such as ammonium, thereby playing a critical role in the N cycle (Voss et al., 2013; Pajares and Ramos, 2019). In addition, processes such as nitrification and denitrification play a crucial role in the maintenance of the global N cycle in the ocean (Pajares and Ramos, 2019). A very limited number of studies have investigated whether QS mediates N transformation in marine (and, more widely, aquatic) environments. For instance, there is strong evidence that QS pathways and denitrification are interconnected in the marine strain D. shibae, as illustrated by Koppenhöfer and Lang (2020). These authors re-analyzed several transcriptomic datasets and showed that the redox regulators controlling denitrification (among others) also interacts with the strain QS pathways. One luxI-knockout mutant also had significantly overexpressed denitrification genes. In addition, Romero et al. (2011b) showed that AHL amendment to the cyanobacterium Anabaena sp. PCCC7120 strongly inhibited its N2 fixation ability, possibly via the post-transcriptional regulation of nitrogenase activity. The regulation of the bacterially mediated transformation of N compounds by QS pathways has also been suggested in several non-marine studies. QS was notably shown to impact processes such as anaerobic ammonium oxidation, ammonia oxidation and N2 fixation in soil and wastewater treatment systems (reviewed in Wang et al., 2021; Zhao Z. C. et al., 2021). In addition, AHLs and PQS were shown to regulate denitrification in the clinical isolate Pseudomonas aeruginosa PAO1 (Gammaproteobacteria) (Toyofuku et al., 2008). The implications of QS-regulated N transformations in marine environments need to be further investigated. They could have strong implications for the phycosphere, where the phytoplankton cells may benefit from the bacterially mediated fixation of N2 into available forms.
Sulfur is a key element for life and primarily occurs in proteins, coenzymes and ligands binding to proteins. It goes through a complex biogeochemical cycle driven by microbial transformation (Sievert et al., 2007; Muyzer and Stams, 2008). Indeed, marine bacteria can use S-containing compounds as electron acceptors or donors to generate energy (Sievert et al., 2007; Muyzer and Stams, 2008). Sulfate-reducing bacteria (SRB) use sulfate as an electron acceptor for OM degradation, playing an important role in anoxic environments such as sediments and bacterial mats (Muyzer and Stams, 2008). Sivakumar et al. (2019) highlighted the link between QS and sulfate reduction in two SRB: the soil bacterium Desulfovibrio vulgaris Hildenborough (Deltaproteobacteria) and the marine bacterium Desulfobacterium corrodens (Deltaproteobacteria). The authors measured the effect of three QSIs on sulfate reduction, specific growth rates and biofilm formation. QSI addition significantly decreased the specific growth rates, the biofilm formation and sulfate reduction abilities of the two strains, even at subinhibitory concentrations. In addition, SRB can work in concert with sulfide-oxidizing bacteria (SOB) that use reduced S as an electron donor (Baumgartner et al., 2006). As such, it has been hypothesized that QS could contribute to this synergy by coordinating their metabolism (Decho et al., 2009, 2010; Montgomery et al., 2013), although experimental data are currently lacking. Consequently, QS-mediated S transformations could be especially important in sediments and microbial mats.
Iron is a vital micronutrient for all living organisms, (co)limiting the growth of microorganisms in nearly all open-ocean waters (Morel et al., 1991). It is involved in numerous metabolic processes, including photosynthesis, respiration and N cycling (Butler, 2005). In addition, Fe is a necessary cofactor for certain alkaline phosphatases, used to access organic P pools (Browning et al., 2017). Bacterial Fe uptake is achieved using siderophores, i.e., Fe-scavenging molecules (Butler, 2005). AHL- and PQS-based QS-regulated siderophore production has been well documented in a few clinical Pseudomonas (Gammaproteobacteria) strains (Ren et al., 2005; Popat et al., 2017) and in several marine Vibrio strains (Wang et al., 2007; Wen et al., 2012; McRose et al., 2018). McRose et al. (2018) notably showed that depending on its cell density, the marine bacterium V. harveyi could produce either cell-associated or soluble siderophores, which likely serve different purposes depending on the environment and growth contexts. Interestingly, several AIs have been shown to possess siderophore activity (Schertzer et al., 2009), suggesting dual roles of these molecules.
Quorum sensing and QQ can also indirectly impact bacterial processes involved in biogeochemistry by altering the BCC and, consequently, its associated functions. Indeed, different bacterial communities may possess different gene contents related to OM degradation and nutrient cycling, depending on the presence of different functional groups (Zimmerman et al., 2013; Arnosti, 2014). QS pathways could play a pivotal role in the structuration of bacterial communities, although few studies have experimentally demonstrated their impact on marine communities thus far (Supplementary Table 4). The mechanisms driving changes in the BCC are unclear and probably multiple. We hypothesize that QS- and QQ-induced changes in the BCC could result from (i) their impact on colonization processes and (ii) their regulation of microbial interactions.
Autoinducer diffusing away from AI-producing cells in microniches (e.g., in the marine snow plume or the phycosphere) could mediate the surface colonization of nearby FL bacteria. In this sense, Dobretsov et al. (2007) demonstrated that different QSIs induced changes in the marine bacterial community colonizing polystyrene Petri dishes. The most affected groups were Alphaproteobacteria, Gammaproteobacteria, and Cytophagales (Bacteroidetes). More recently, Huang et al. (2019) demonstrated that AHL disruption in marine communities colonizing immersed steel coupons coated with the QQ AHL-lactonase SsoPox W263I significantly altered their composition. The lactonase-based degradation of AHLs greatly reduced the abundances of Burkholderiales (Betaproteobacteria), Pseudomonadales (Gammaproteobacteria), and Rhodospirillales (Alphaproteobacteria), among others. The authors simultaneously demonstrated that lactonase coating resulted in a change in the metabolism of colonizing bacteria, reducing steel coupons biocorrosion.
Quorum sensing could impact the composition of colonizing bacterial communities by regulating various phenotypes, such as motility (e.g., swimming, gliding, swarming, and chemotaxis) and adhesion (e.g., biofilm production and cell hydrophobicity), which are known to drive colonization (Dang and Lovell, 2016; Raina et al., 2019). Numerous examples of QS involvement in these phenotypes in marine strains have been reported. For instance, Fei et al. (2020) demonstrated that QS regulates motility and attachment in two Rhodobacterales-affiliated strains (Sulfitobacter pseudonitzschiae F5 and Phaeobacter sp. F10). Gardiner et al. (2015) showed that a luxR knockout of marine Nautella italica R11 (roseobacter) could not form biofilms or attach to the red alga Delisea pulchra. Yu et al. (2020) showed that QS regulated cell hydrophobicity in marine C. naphthovorans PQ-2, driving its adhesion to PAHs. QS widely regulates motility in marine strains affiliated with Vibrio (Lupp and Ruby, 2005; Tian et al., 2008) and Pseudoalteromonas (Ayé et al., 2015). Finally, several studies have implicated QS in biofilm formation among marine and estuarine strains, including Vibrio sp. (Liu et al., 2017; Urvoy et al., 2021a), Rhodobacterales (Su et al., 2019; Fei et al., 2020), Pseudomonadales (Mangwani et al., 2015; Urvoy et al., 2021a) and Pseudoalteromonas sp. (Ayé et al., 2015). As such, QS mediation of colonization through motility, chemotaxis and biofilm formation could impact the colonization of microbial communities (thus, their composition) in all the highlighted microniches.
Once bacterial cells settle into relevant microniches, QS regulates a myriad of phenotypes that could further alter the BCC. Thus far, four studies have demonstrated a direct influence of AI amendment on the composition of marine planktonic communities (Whalen et al., 2019; Urvoy et al., 2021b) and established marine and lacustrine biofilm communities (Wang et al., 2020; Xu et al., 2021). Whalen et al. (2019) showed that HHQ addition to microbial communities sampled along a simulated phytoplanktonic bloom altered the BCC of both PA and FL heterotrophic bacteria, with highly variable effects depending on the bloom stage. Overall, HHQ exposure favored Gammaproteobacteria (including the HHQ producers Pseudoalteromas spp.) and Alphaproteobacteria, while the relative abundance of Bacteroidetes decreased. Whalen et al. (2019) also found that HHQ exposure reduced the growth of eukaryotes (especially nanoeukaryotes) without altering their composition. Urvoy et al. (2021b) showed that amendment with low concentrations of different AHLs (50 nM, C4- to C16-HSL) altered the composition of planktonic bacterial communities sampled at two stages of coastal phytoplanktonic spring bloom as well as their hydrolytic activities. Similar to Whalen et al. (2019), the AHL effect widely differed depending on the bloom stage. The AHL treatment mostly affected Bacteroidetes (only Flavobacteriales), Gammaproteobacteria (mainly Vibrionales, Alteromonadales, and Oceanospirillales) and Alphaproteobacteria (only Rhodobacterales). Wang et al. (2020) tested the effect of AHLs (C6- and C12-HSL), PQS and cyclic di-guanosine monophosphate (c-di-GMP) on the taxonomic structure and functional profiles of 9-day-old biofilms developed on Petri dishes in the coastal subtidal zone of Hong Kong Port. They showed that AI amendments altered the taxonomy and structure of the biofilm communities, especially the C12-HSL and PQS treatments. The PQS treatment specifically increased the relative abundance of groups belonging to Alphaproteobacteria (e.g., Sulfitobacter, Thalassospira, Ruegeria, Erythrobacter) and Gammaproteobacteria (e.g., Alteromonas). The PQS and C12-HSL treatments also increased the relative abundance of genes involved in membrane transport, motility, chemotaxis and cell signaling. Finally, Xu et al. (2021) performed AHL amendments to limnic biofilms (as described above) and showed that it induced a shift in the community composition by increasing the abundance of polyphosphate-accumulating organisms.
The mechanisms driving changes in BCC in these experiments are largely unclear. We can speculate that, in addition to regulating colonization processes, QS further drives these changes by mediating both intra- and interkingdom microbial interactions, for instance, through the regulation of public goods production and prophage lysis. Public goods are compounds providing a collective benefit, including hydrolytic enzymes, siderophores, exopolysaccharides, antibiotics and algaecides (Smith and Schuster, 2019). QS is known to regulate the production of a number of public goods (Whiteley et al., 2017; Mukherjee and Bassler, 2019). In the highlighted articles, QS altered hydrolytic enzymes (Urvoy et al., 2021b) and polysaccharide production (Xu et al., 2021), which could drive changes in the BCC. In the study by Urvoy et al. (2021b), the changes in hydrolytic enzymes and bacterial diversity were correlated, supporting this hypothesis. Additionally, Whalen et al. (2019) indicated that the alteration of phytoplankton growth rates following HHQ exposure suggested that HHQ could mediate interkingdom microbial interactions. Finally, QS could also modulate BCC by triggering the induction of prophages, leading to cell lysis and the release of phage particles. As highlighted above, several phages, including marine phages, have been shown to carry bacterial QS receptors. One of them was shown to regulate the lysis-lysogeny decision in the vibriophage VP882 infecting V. cholerae (Silpe et al., 2020). In addition, AHL amendments induced phage lysis in natural soil and groundwater communities (Ghosh et al., 2009; Liang et al., 2020), resulting in the alteration of the BCC in one study (Liang et al., 2020). To our knowledge, the direct link between QS and prophage induction has yet to be demonstrated in marine communities.
Quorum sensing regulates several microbial processes, ultimately impacting marine biogeochemistry. However, most of the studies to date were performed on isolated bacteria in dense liquid monocultures, and few of them have investigated the role of QS under environmentally relevant conditions. This can be explained by the complexity of studying cell–cell communication in natural environments, requiring multidisciplinary skills to overcome technical limitations, and by the inherent complexity of QS communications (Whiteley et al., 2017; Schmidt et al., 2019; Figure 4).
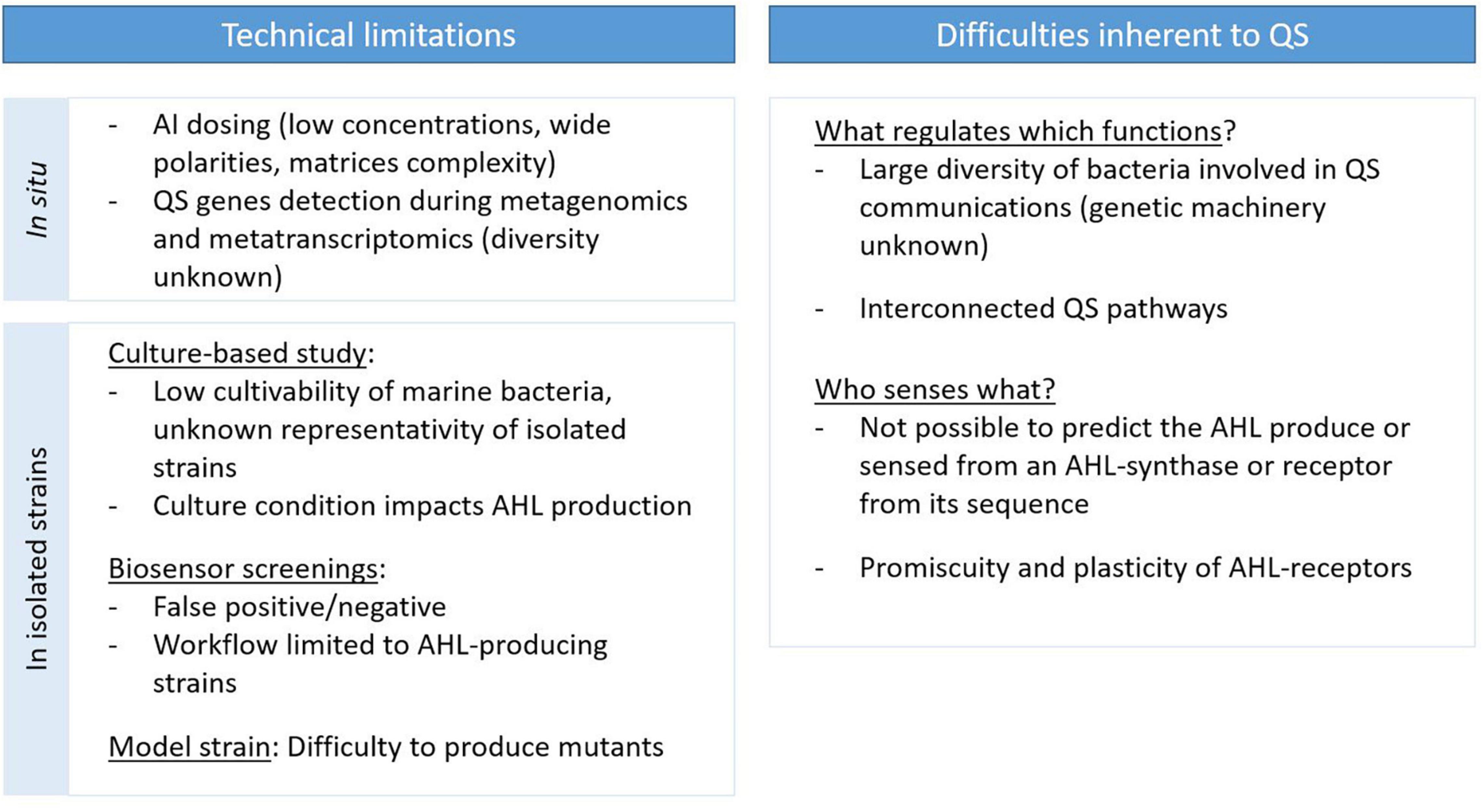
Figure 4. Current limitations and difficulties restricting our ability to investigate QS communications.
Technical limitations are a major constraint restricting our capacity to study QS roles and functions in natural environments, whether it concerns the dosage of AIs or the monitoring of the presence and expression of QS genes. The dosing of AI in natural environments presents a challenge because of their low concentrations, their wide range of polarities and the inner complexity of their environmental matrices (Stock et al., 2021). A few studies have achieved AHL dosing in dense environments such as marine snow particles (Hmelo et al., 2011; Jatt et al., 2015), the phycosphere (Van Mooy et al., 2012), microbial mats (Decho et al., 2009), biofilms (Huang et al., 2009; Xu et al., 2021) and sediments (Stock et al., 2021). These reports remain limited, and no study has yet achieved AHL dosing directly in the water column. There is also a lack of quantitative measurements of other AIs in marine environments. Only Wang et al. (2018) reported AI-2 dosing, with a quantification method that required large sample amounts and a concentration step for both seawater and marine biofilms. Pollara et al. (2021) reported the dosing of HHQ in marine waters via a method involving an adsorption step on polystyrene resin, but it required the integration of 400–600 L of seawater. Although AI dosing is necessary, it does not allow the identification of the AI-producing organisms. Metagenomic (or metatranscriptomic) techniques offer the possibility of surveying the presence (or expression) of genes involved in QS and QQ. However, the diversity of the genetic machinery encoding these systems remains under characterized and those genes can be difficult to detect in such datasets. This is especially true for AHL synthase and receptor genes, which are highly variable: known luxR and luxI genes only share 18–25% and 28–35% similarity, respectively, despite the relatively conserved structure of AHLs (Fuqua et al., 1996). As such, there are most likely new AHL-related gene families to discover. The use of bacterial genes to query archaeal (Zhang et al., 2012) and viral (Silpe and Bassler, 2019) genomes probably contributes to the limited success in the identification of their QS genes thus far.
To circumvent the limitations associated with natural environments, a common method is the isolation and cultivation of bacteria, followed by the screening of their production of QS or QQ compounds using biosensor reporter strains (Supplementary Table 2). Biosensor strains are bacteria that have been engineered to produce a detectable response upon AI sensing (production of bioluminescence, fluorescence or pigments). They are often used as a screening tool due to their sensitivity, easy implementation and low cost. Various biosensors have been developed to sense different AIs, including AHLs (Riedel et al., 2001; Steidle et al., 2001), AI-2 (Miller et al., 2004) and PQS and HHQ (Fletcher et al., 2007). However, this workflow also has some disadvantages. First, only a minor fraction of marine bacteria are cultivable. This limits our study capacity to a few bacterial groups whose representativeness may be debatable. Second, biosensors are prone to produce both false-negative and false-positive results. Indeed, AI receptors can bind non-native ligands that acts as agonists (false-positive) or antagonists (false-negative). Such effects have been demonstrated for various receptors with natural and synthetic molecules (Holden et al., 1999; Geske et al., 2008; Swem et al., 2008). Given these drawbacks, biosensors should be used in concert with analytical methods to confirm the presence of AIs and to identify and quantify them. These analytical methods often include liquid (LC) or gas chromatography (GC) coupled to mass spectrometry (MS) or tandem MS (MS/MS) (Supplementary Table 2). Another limit of this workflow is the dependence upon culture conditions, as factors such as temperature, medium composition (C sources and nutrients) and shaking can influence AI production (Medina-Martínez et al., 2006; Shrout et al., 2006; Boyle et al., 2015). In addition, the use of pure cultures could limit the expression of QS genes (Albataineh et al., 2021). As such, QS systems may be more prevalent than what has been reported to date. The workflow based on bacterial isolation and the detection of AI production de facto restricts QS studies to strains that produce AIs. Consequently, marine QS cheaters are not as actively studied as AHL-producing strains despite their well-established prevalence.
Finally, the lack of marine model strains with adequate molecular tools also limits our ability to study the role of QS in ecologically relevant strains. The construction of deletion or recombinant mutants is crucial to identify QS circuits and their regulated functions. However, our capacity to adapt genetic tools to produce mutants is limited, as this process is time-consuming and expensive, and can be especially difficult outside of a few well-managed groups such as in the Vibrio genus and in roseobacters (Lami et al., 2021). Consequently, only a few marine bacterial QS models currently exist (Lami et al., 2021), including the marine bacteria Vibrio fischeri (in which QS was discovered), Ruegeria sp. KLH11 (a model for studying QS in marine sponge symbionts) (Zan et al., 2015) and Dinoroseobacter shibae (an algal symbiont characterized by a pronounced pleomorphism) (Patzelt et al., 2013; Koppenhöfer et al., 2019).
Several difficulties in the study of QS pathways are linked to their inherent complexity. QS systems occur in a large diversity of bacteria, and the diversity of the genetic machinery encoding these systems is not well known. A single bacterium can produce a multitude of AIs, and single strains producing more than 20 AHLs have been reported (Ortori et al., 2007; Doberva et al., 2017). In addition, QS pathways based on different AI systems are often interconnected, with different hierarchies between them (Long et al., 2009; Lee and Zhang, 2015). Another difficulty concerning AHL-based QS is the impossibility of predicting the AHL synthesized by an AHL synthase or fixed by an AHL receptor from their genetic sequence, due to their low identity (Fuqua et al., 1996; Wellington Miranda et al., 2021). AHL receptors may exhibit some degree of promiscuity; that is to say that they can fix non-AHL ligands, expanding the range of QS crosstalk and responses (Wellington and Greenberg, 2019; Prescott and Decho, 2020). Very little is known about the factors driving QS system selectivity (Wellington Miranda et al., 2021). They also seem highly plastic and can rapidly evolve to adapt their ligand preference (as reviewed in Prescott and Decho, 2020). Consequently, it is difficult to determine the diversity of bacteria producing or targeted by a given AHL.
Several techniques and improvements may alleviate the current constraints and provide new perspectives in environmental QS studies. More specifically, these techniques could help characterize the diversity of QS systems in marine environments as well as their roles and effective ranges. This would allow us to better appreciate the role of QS as a regulatory mechanism involved in bacterial processes that ultimately affect marine biogeochemistry, especially outside of the few well-characterized bacterial groups, such as Vibrio and roseobacters. QS studies can be performed at three scales: (i) in isolated strains, (ii) in simplified bacterial communities, and (iii) in complex natural communities (Figure 5).
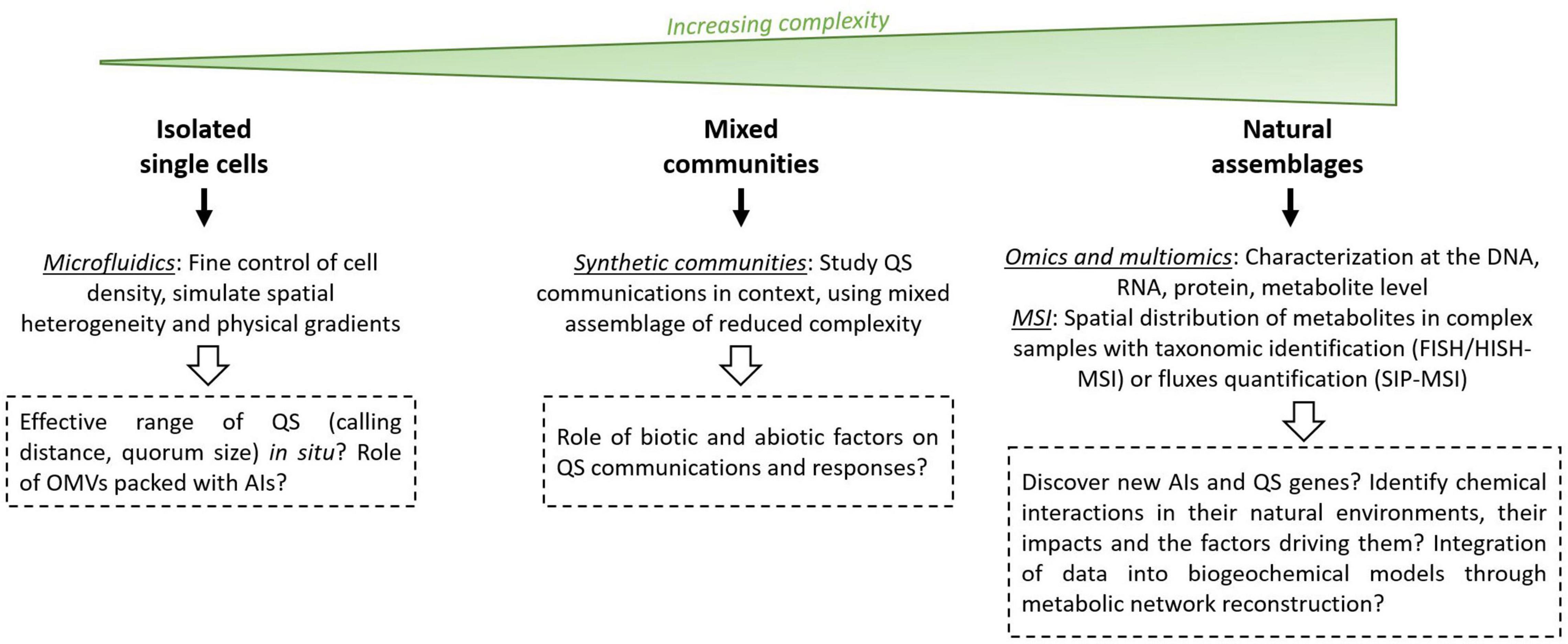
Figure 5. Future perspectives and new tools in QS studies. AI, autoinducer; OMVs, outer membrane vesicles; MSI, mass spectrometry imaging; FISH, fluorescence in situ hybridization; HISH, halogen in situ hybridization; SIP, stable isotope probing.
A first step would be to move from the initial understanding of QS, based on dense, homogeneous liquid monocultures, to a more environmentally relevant perspective. Indeed, the principles of QS were originally formulated using dense monocultures (Nealson et al., 1970; Eberhard, 1972). However, QS regulation most likely results from the integration of various factors that are not accounted for in such models. Consequently, several attempts have been made to redefine QS as a means of sensing: (i) the mass transfer properties of the surrounding medium (“diffusion sensing,” Redfield, 2002); (ii) the combination of cell density, mass transfer properties and the spatial distribution of cells, indistinguishably (“efficiency sensing,” Hense et al., 2007); and (iii) a combination of social (cell density) and physical (mass transfer) properties, which can be distinguished by the integration of several QS pathways (“combinatorial sensing,” Cornforth et al., 2014). These studies highlighted the crucial importance of considering the spatial and physical heterogeneity faced by microorganisms in their environment at the microscale. Environmental conditions could be better simulated through the use of microfluidics (Aleklett et al., 2018). Microfluidics refers to the manipulation of fluids at the microscale, allowing the fine control of various parameters, including flows, chemical gradients and bacterial density (Whitesides, 2006). Microfluidics has already been successfully used to explore the effect of cell-density (down to a few cells) on QS induction (Boedicker et al., 2009), the effect of spatial heterogeneity on QS dynamics (reviewed in Mukherjee and Bassler, 2019) and the effect of AI molecules as chemoattractants (Nagy et al., 2015). However, many unanswered questions remain, such as questions concerning the effective range of AIs in situ (calling distance and quorum size) and the role of outer membrane vesicles-packed AIs. To our knowledge, calling distances (i.e., the distance over which bacteria can communicate) have not been characterized in marine environments, although they have been estimated to 1–100 μm in the tomato rhizosphere (Gantner et al., 2006) and up to 176 μm for clinical P. aeruginosa aggregates (Darch et al., 2018), suggesting the possibility of interaggregates signaling. A few reports have yielded questions regarding the bacterial quorum size (the number of cells required for QS induction), which could be reduced to a few cell clusters under appropriate conditions (Dulla and Lindow, 2008). In addition, the recent discovery of AIs packed in outer membrane vesicles could allow the targeted delivery of hydrophobic AIs over long distances in marine environments and induce single cells (Li et al., 2016; Toyofuku et al., 2017). These discoveries reshuffle the cards concerning the mode and range of action of QS in the environment and need to be addressed to better constrain its impact on bacterial processes and marine biogeochemistry.
Second, as QS is a social trait, the total implications of this mechanism can only be addressed when studied in mixed bacterial communities comprising several bacterial types. Synthetic communities are artificial communities created by coculturing two or more species (Großkopf and Soyer, 2014). They exhibit some of the key features of the mimicked communities with reduced complexity. Such cocultures have mostly been used to study clinically relevant strains thus far (reviewed in Abisado et al., 2018). The more widespread use of synthetic communities coupled with other techniques would allow to study QS in communities more similar to natural assemblages. This could particularly help us assess the roles of biotic and abiotic factors in shaping QS interactions and responses, for instance, in the case of symbiotic associations between a microalga and different synthetic microalgal microbiomes.
Finally, when working on complex natural assemblages, several techniques can be implemented. The use of omics (metagenomics, metatranscriptomics, metabolomics or metaproteomics) and multiomics (the integrated use of several omics approaches) would allow a better depiction of the diversity of QS systems, the functions they regulate and their impact on natural communities. Although omics techniques are increasingly being used, few studies implementing them in the context of marine QS studies have been published thus far. They could notably aid in the discovery of new AIs and their encoding genes, especially in understudied microbial groups. For instance, functional metagenomics (i.e., the construction and screening of libraries containing DNA cloned from environmental metagenomes) is an excellent tool for revealing new QS and QQ genes (Williamson et al., 2005; Weiland-Bräuer et al., 2016), while comparative proteomics or metabolomics could help to discover new AIs (Prado et al., 2016). Multiomics could also allow the identification of chemical communications in their natural environments and a better understanding of the impact of QS-regulated functions in key processes, such as the colonization of marine aggregates by mixed communities. These approaches could help unravel the genetic and molecular bases of chemical communications (e.g., their biosynthesis and transportation pathways) and how biotic and abiotic factors trigger and modulate them. Studies carried out at several levels (i.e., DNA > RNA > metabolites, proteins) would offer a more holistic view of bacterial interactions in situ. Multiomics integrations can lead to metabolic network reconstructions (at the genome or community level) (Prado et al., 2016), allowing the prediction of QS communication impacts on cellular metabolism under changing conditions, ultimately integrating such networks within global biogeochemical flux models.
Another promising emergent tool is the use of mass spectrometry imaging (MSI). MSI is an untargeted approach used to visualize the spatial distribution of molecules. In MSI, the sample surface is swept by a focused ionizing beam. The resulting ions released to the gas phase are analyzed by MS. Several ionization techniques can be used, including matrix-assisted laser desorption/ionization, desorption electrospray ionization and secondary ion mass spectrometry, each offering different advantages (as reviewed in several articles, such as Dunham et al., 2016). MSI provides new opportunities to study QS in its ecological context, as it enables the spatial resolution of metabolites without the need for extraction steps, making it possible to track minute quantities of AIs in complex environments. In addition, MSI can be coupled with other techniques to further expand its abilities, for instance, to allow the identification of the studied bacteria (using fluorescence or halogen in situ hybridization) and to trace the uptake and incorporation of labeled substrates (using stable isotope probing). This could allow the study of the impact of QS on microbial metabolism and organic matter incorporation at the cellular level in a mixed population.
Future research will need to address several questions concerning the prevalence and diversity of QS systems, as well as their role and effective range in marine environments:
• What is the global diversity of AHL-related QS genes, and do unknown families remain to be discovered? What is the prevalence of solo luxI genes and promiscuous AHL receptors? How widespread are non-AHL-based QS systems in the ocean?
• Which other traits are QS-regulated in marine bacteria, and how do they affect biogeochemical cycles? By which mechanisms does QS shape the composition of microbial groups? To what extent does QS contribute to the selection of initial colonizers? How do these interactions drive the evolution of QS systems? What are the role and ecological implications of QQ? What is the role of QS in terms of interkingdom crosstalk and especially in the induction of prophage lysis?
• What is the effective range (calling distance, quorum size) of QS communications in marine environments? Does QS occur outside of biofilms? What are the effective range and the function of outer membrane vesicles-packed AIs in the environment?
Cell–cell communication systems are increasingly being studied in marine environments, where they allow prokaryotes to cooperate with or compete against each other, especially in dense microniches such as marine aggregates, microbial mats and the microalgal phycosphere. Together, these microscale interactions have global, large-scale repercussions. Indeed, QS-based regulations may be important for marine biogeochemical cycles, as they are involved in many ecologically relevant bacterial processes. In addition, this communication system is prevalent among groups such as Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes, which are abundant in marine bacterial hotspots and are involved in all major cycles. QS may directly regulate phenotypes involved in OM degradation, C allocation and nutrient acquisition and may indirectly structure marine communities. However, much remains to be revealed about QS prevalence and function in marine environments. Although an increasing number of studies have described the production of AHLs and QQ compounds in marine strains, other QS systems have been much less described. Beyond the depiction of QS system prevalence and diversity, there are still many unknowns concerning their ranges and roles in marine environments. In the future, we will need to simultaneously (i) improve culture-based studies through the development of new marine model strains, the use of synthetic communities and the better simulation of microenvironment heterogeneity; and (ii) improve studies on natural communities, with better integration of omics and MSI techniques, and the quantification of QS impacts on the fluxes of C and nutrients. Overall, the real extent of bacterial diversity involved in QS and its ecological and biogeochemical consequences remain to be characterized in marine environments.
All authors participated in the manuscript design. MU wrote the first draft. All authors substantially edited, proofread, and approved the final manuscript.
This work was supported by the French National program EC2CO (Ecosphère Continentale et Côtière) and by an ISblue (Interdisciplinary graduate school for the blue planet) project (ANR-17-EURE-0015), co-funded by a grant from the French government under the program ‘Investissements d’Avenir.’ This work was also carried out in conjunction with the European Marine Biological Resource Centre (EMBRC-ERIC) and EMBRC-France. French state funds are managed by the ANR within the Investments of the Future program under the grant number ANR-10-INSB-02.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.834337/full#supplementary-material
Abdul Malik, S. A., Bazire, A., Gamboa-Muñoz, A., Bedoux, G., Robledo, D., García-Maldonado, J. Q., et al. (2020). Screening of surface-associated bacteria from the mexican red alga Halymenia floresii for quorum sensing activity. Microbiology 89, 778–788. doi: 10.1134/S0026261720060132
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9:e02331-17.
Acinas, S. G., Ferrera, I., Sarmento, H., Díez-Vives, C., Forn, I., Ruiz-González, C., et al. (2015). Validation of a new catalysed reporter deposition- fluorescence in situ hybridization probe for the accurate quantification of marine Bacteroidetes populations. Environ. Microbiol. 17, 3557–3569. doi: 10.1111/1462-2920.12517
Albataineh, H., Duke, M., Misra, S. K., Sharp, J. S., and Stevens, D. C. (2021). Identification of a solo acylhomoserine lactone synthase from the myxobacterium Archangium gephyra. Sci. Rep. 11:3018. doi: 10.1038/s41598-021-82480-1
Aleklett, K., Kiers, E. T., Ohlsson, P., Shimizu, T. S., Caldas, V. E., and Hammer, E. C. (2018). Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 12, 312–319. doi: 10.1038/ismej.2017.184
Arnosti, C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Arnosti, C. (2014). Patterns of microbially driven carbon cycling in the ocean: links between extracellular enzymes and microbial communities. Adv. Oceanogr. 2014:706082. doi: 10.1155/2014/706082
Atkinson, S., and Williams, P. (2009). Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 6, 959–978. doi: 10.1098/rsif.2009.0203
Ayé, A. M., Bonnin-Jusserand, M., Brian-Jaisson, F., Ortalo-Magné, A., Culioli, G., Koffi, Nevry R, et al. (2015). Modulation of violacein production and phenotypes associated with biofilm by exogenous quorum sensing N-acylhomoserine lactones in the marine bacterium Pseudoalteromonas ulvae TC14. Microbiology 161, 2039–2052. doi: 10.1099/mic.0.000147
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Azam, F., Fenchel, T., Field, J., Gray, J., Meyer-Reil, L., and Thingstad, F. (1983). The ecological role of water-column microbes in the Sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Bachofen, R., and Schenk, A. (1998). Quorum sensing autoinducers: do they play a role in natural microbial habitats? Microbiol. Res. 153, 61–63. doi: 10.1016/S0944-5013(98)80022-0
Baker, B. J., Lazar, C. S., Teske, A. P., and Dick, G. J. (2015). Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. doi: 10.1186/s40168-015-0077-6
Barriuso, J., and Martínez, M. J. (2018). In Silico analysis of the Quorum Sensing metagenome in environmental biofilm samples. Front. Microbiol. 9:1243. doi: 10.3389/fmicb.2018.01243
Bartlau, N., Wichels, A., Krohne, G., and Adriaenssens, E. M. (2021). Highly diverse flavobacterial phages isolated from North Sea spring blooms. ISME J. [Epub ahead of print]. doi: 10.1038/s41396-021-01097-4
Baumgartner, L. K., Reid, R. P., Dupraz, C., Decho, A. W., Buckley, D. H., Spear, J. R., et al. (2006). Sulfate reducing bacteria in microbial mats: changing paradigms, new discoveries. Sediment. Geol. 185, 131–145. doi: 10.1016/j.sedgeo.2005.12.008
Betancur, L. A., Naranjo-Gaybor, S. J., Vinchira-Villarraga, D. M., Moreno-Sarmiento, N. C., Maldonado, L. A., Suarez-Moreno, Z. R., et al. (2017). Marine Actinobacteria as a source of compounds for phytopathogen control: an integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS One 12:e0170148. doi: 10.1371/journal.pone.0170148
Beyersmann, P. G., Tomasch, J., Son, K., Stocker, R., Göker, M., Wagner-Döbler, I., et al. (2017). Dual function of tropodithietic acid as antibiotic and signaling molecule in global gene regulation of the probiotic bacterium Phaeobacter inhibens. Sci. Rep. 7:730. doi: 10.1038/s41598-017-00784-7
Biswa, P., and Doble, M. (2013). Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett. 343, 34–41. doi: 10.1111/1574-6968.12123
Bižic-Ionescu, M., Zeder, M., Ionescu, D., Orlic, S., Fuchs, B. M., Grossart, H., et al. (2015). Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization. Environ. Microbiol. 17, 3500–3514. doi: 10.1111/1462-2920.12466
Blanchet, E., Prado, S., Stien, D., Oliveira da Silva, J., Ferandin, Y., Batailler, N., et al. (2017). Quorum sensing and quorum quenching in the Mediterranean Seagrass Posidonia Oceanica microbiota. Front. Mar. Sci. 4:218. doi: 10.3389/fmars.2017.00218
Bodor, A., Elxnat, B., Thiel, V., Schulz, S., and Wagner-Döbler, I. (2008). Potential for luxS related signalling in marine bacteria and production of autoinducer-2 in the genus Shewanella. BMC Microbiol. 8:13. doi: 10.1186/1471-2180-8-13
Boedicker, J. Q., Vincent, M. E., and Ismagilov, R. F. (2009). Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chemie Int. Ed. 48, 5908–5911. doi: 10.1002/anie.200901550
Bolhuis, H., and Stal, L. J. (2011). Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. ISME J. 5, 1701–1712. doi: 10.1038/ismej.2011.52
Bose, U., Ortori, C. A., Sarmad, S., Barrett, D. A., Hewavitharana, A. K., Hodson, M. P., et al. (2017). Production of N-acyl homoserine lactones by the sponge-associated marine actinobacteria Salinispora arenicola and Salinispora pacifica. FEMS Microbiol. Lett. 364, 1–7. doi: 10.1093/femsle/fnx002
Boyle, K. E., Monaco, H., van Ditmarsch, D., Deforet, M., and Xavier, J. B. (2015). Integration of metabolic and quorum sensing signals governing the decision to cooperate in a bacterial social trait. PLoS Comput. Biol. 11:e1004279. doi: 10.1371/journal.pcbi.1004279
Brameyer, S., Plener, L., Müller, A., Klingl, A., Wanner, G., and Jung, K. (2018). Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI-1 between Vibrio harveyi cells. J. Bacteriol. 200: e000740-17.
Browning, T. J., Achterberg, E. P., Yong, J. C., Rapp, I., Utermann, C., Engel, A., et al. (2017). Iron limitation of microbial phosphorus acquisition in the tropical North Atlantic. Nat. Commun. 8:15465. doi: 10.1038/ncomms15465
Buchan, A., Gonzalez, J. M., and Moran, M. A. (2005). Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665
Buchan, A., Lecleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Buchan, A., Mitchell, A., Cude, W. N., and Campagna, S. (2016). “Acyl-homoserine lactone-based quorum sensing in members of the marine bacterial Roseobacter clade: complex cell-to-cell communication controls multiple physiologies,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, ed. F. J. De Bruijn (Hoboken, NJ: John Wiley & Sons), 225–233.
Butler, A. (2005). Marine siderophores and microbial iron mobilization. BioMetals 18, 369–374. doi: 10.1007/s10534-005-3711-0
Carreira, C., Staal, M., Middelboe, M., and Brussaard, C. P. D. (2015). Counting viruses and bacteria in photosynthetic microbial mats. Appl. Environ. Microbiol. 81, 2149–2155. doi: 10.1128/AEM.02863-14
Charlesworth, J., Kimyon, O., Manefield, M., Beloe, C. J., and Burns, B. P. (2020). Archaea join the conversation: detection of AHL-like activity across a range of archaeal isolates. FEMS Microbiol. Lett. 367, 1–9. doi: 10.1093/femsle/fnaa123
Charlesworth, J. C., Watters, C., Wong, H. L., Visscher, P. T., and Burns, B. P. (2019). Isolation of novel quorum-sensing active bacteria from microbial mats in Shark Bay Australia. FEMS Microbiol. Ecol. 95, 1–14. doi: 10.1093/femsec/fiz035
Chen, P., Zhang, L., Guo, X., Dai, X., Liu, L., Xi, L., et al. (2016). Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the southwest Indian ridge. Front. Microbiol. 7:1340. doi: 10.3389/fmicb.2016.01340
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Chernikova, T. N., Bargiela, R., Toshchakov, S. V., Shivaraman, V., Lunev, E. A., Yakimov, M. M., et al. (2020). Hydrocarbon-degrading bacteria alcanivorax and marinobacter associated with microalgae Pavlova lutheri and Nannochloropsis oculata. Front. Microbiol. 11:572931. doi: 10.3389/fmicb.2020.572931
Chróst, R. J. (1990). “Microbial ectoenzymes in aquatic environments,” in Aquatic Microbial Ecology: Biochemical and Molecular Approaches, eds J. Overbeck and R. J. Chróst (Berlin: Springer), 47–77.
Cornforth, D. M., Popat, R., McNally, L., Gurney, J., Scott-Phillips, T. C., Ivens, A., et al. (2014). Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc. Natl. Acad. Sci. U.S.A. 111, 4280–4284. doi: 10.1073/pnas.1319175111
Cottrell, M. T., and Kirchman, D. L. (2000). Natural assemblages of marine Proteobacteria and members of the Cytophaga-flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000
Cuadrado-Silva, C. T., Castellanos, L., Arévalo-Ferro, C., and Osorno, O. E. (2013). Detection of quorum sensing systems of bacteria isolated from fouled marine organisms. Biochem. Syst. Ecol. 46, 101–107. doi: 10.1016/j.bse.2012.09.010
Cude, W. N., and Buchan, A. (2013). Acyl-homoserine lactone-based quorum sensing in the Roseobacter clade: complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 4:336. doi: 10.3389/fmicb.2013.00336
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15.Address
Darch, S. E., Simoska, O., Fitzpatrick, M., Barraza, J. P., Stevenson, K. J., Bonnecaze, R. T., et al. (2018). Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl. Acad. Sci. U.S.A. 115, 4779–4784. doi: 10.1073/pnas.1719317115
Dashti, N., Ali, N., Eliyas, M., Khanafer, M., Sorkhoh, N. A., and Radwan, S. S. (2015). Most hydrocarbonoclastic bacteria in the total environment are diazotrophic, which highlights their value in the bioremediation of hydrocarbon contaminants. Microbes Environ. 30, 70–75. doi: 10.1264/jsme2.ME14090
Decho, A. W., Norman, R. S., and Visscher, P. T. (2010). Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 18, 73–80. doi: 10.1016/j.tim.2009.12.008
Decho, A. W., Visscher, P. T., Ferry, J., Kawaguchi, T., He, L., Przekop, K. M., et al. (2009). Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11, 409–420. doi: 10.1111/j.1462-2920.2008.01780.x
DeLisa, M. P., Wu, C., Wang, L., Valdes, J. J., and Bentley, W. E. (2001). DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183, 5239–5247. doi: 10.1128/JB.183.18.5239
Delong, E. F., Franks, D. G., and Alldredge, A. L. (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934.
Diggle, S. P., Gardner, A., West, S. A., and Griffin, A. S. (2007). Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos. Trans. R. Soc. B Biol. Sci. 362, 1241–1249. doi: 10.1098/rstb.2007.2049
Doberva, M., Sanchez-Ferandin, S., Toulza, E., Lebaron, P., and Lami, R. (2015). Diversity of quorum sensing autoinducer synthases in the Global Ocean Sampling metagenomic database. Aquat. Microb. Ecol. 74, 107–119. doi: 10.3354/ame01734
Doberva, M., Stien, D., Sorres, J., Hue, N., Sanchez-Ferandin, S., Eparvier, V., et al. (2017). Large diversity and original structures of acyl-homoserine lactones in strain MOLA 401, a marine rhodobacteraceae bacterium. Front. Microbiol. 8:1152. doi: 10.3389/fmicb.2017.01152
Dobretsov, S., Dahms, H. U., Yili, H., Wahl, M., and Qian, P. Y. (2007). The effect of quorum-sensing blockers on the formation of marine microbial communities and larval attachment. FEMS Microbiol. Ecol. 60, 177–188. doi: 10.1111/j.1574-6941.2007.00285.x
Duarte, C. M., Middelburg, J. J., and Caraco, N. (2005). Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2, 1–8.
Dulla, G., and Lindow, S. E. (2008). Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. U.S.A. 105, 3082–3087. doi: 10.1073/pnas.0711723105
Dunham, S. J. B., Ellis, J. F., Li, B., and Sweedler, J. V. (2016). Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res. 50, 96–104. doi: 10.1021/acs.accounts.6b00503
Duran, R., and Cravo-laureau, C. (2016). Role of environmental factors and microorganisms in determining the fate of polycyclic aromatic hydrocarbons in the marine environment. FEMS Microbiol. Rev. 40, 814–830. doi: 10.1093/femsre/fuw031
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., et al. (2016). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Eberhard, A. (1972). Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 109, 1101–1105.
Elifantz, H., Malmstrom, R. R., Cottrell, M. T., and Kirchman, D. L. (2005). Assimilation of polysaccharides and glucose by major bacterial groups in the delaware estuary. Appl. Environ. Microbiol. 71, 7799–7805. doi: 10.1128/AEM.71.12.7799
Enke, T. N., Datta, M. S., Schwartzman, J., Cermak, N., Schmitz, D., Barrere, J., et al. (2019). Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. Rep. 29, 1528–1535. doi: 10.1016/j.cub.2019.03.047
Eriksen, M., Lebreton, L. C. M., Carson, H. S., Thiel, M., Moore, C. J., Borerro, J. C., et al. (2014). Plastic pollution in the world’s oceans: more than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS One 9:e111913. doi: 10.1371/journal.pone.0111913
Fei, C., Ochsenkühn, M. A., Shibl, A. A., Isaac, A., Wang, C., and Amin, S. A. (2020). Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ. Microbiol. 22, 4761–4778. doi: 10.1111/1462-2920.15228
Fernàndez-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., Gonzàles, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Ferrer-González, F. X., Widner, B., Holderman, N. R., Glushka, J., Edison, A. S., Kujawinski, E. B., et al. (2021). Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 15, 762–773. doi: 10.1038/s41396-020-00811-y
Fletcher, M. P., Diggle, S. P., Cámara, M., and Williams, P. (2007). Biosensor-based assays for PQS, HHQ and related 2-alkyl-4-quinolone quorum sensing signal molecules. Nat. Protoc. 2, 1254–1262. doi: 10.1038/nprot.2007.158
Francis, B., Urich, T., Mikolasch, A., Teeling, H., and Amann, R. (2021). North Sea spring bloom-associated Gammaproteobacteria fill diverse heterotrophic niches. Environ. Microbiomes 16:15. doi: 10.1186/s40793-021-00385-y
Freckelton, M. L., Høj, L., and Bowden, B. F. (2018). Quorum sensing interference and structural variation of Quorum Sensing mimics in Australian soft coral. Front. Mar. Sci. 5:198. doi: 10.3389/fmars.2018.00198
Fuhrman, J. A., and Hagström, Å (2008). “Bacterial and archaeal community structure and its patterns,” in Microbial Ecology of the Oceans, ed. D. L. Kirchman (Hoboken, NJ: Wiley).
Fuqua, C. (2006). The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3161–3171. doi: 10.1128/JB.188.9.3169-3171.2006
Fuqua, C., Winans, S. C., and Greenberg, P. E. (1996). Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751.
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density- responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gantner, S., Schmid, M., Christine, D., Schuhegger, R., Steidle, A., Hutzler, P., et al. (2006). In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56, 188–194. doi: 10.1111/j.1574-6941.2005.00037.x
García-Reyes, S., Soberón-Chávez, G., and Cocotl-Yanez, M. (2020). The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein. J. Med. Microbiol. 69, 25–34. doi: 10.1099/jmm.0.001116
Gardiner, M., Fernandes, N. D., Nowakowski, D., Raftery, M., Kjelleberg, S., Zhong, L., et al. (2015). VarR controls colonization and virulence in the marine macroalgal pathogen Nautella italica R11. Front. Microbiol. 6:1130. doi: 10.3389/fmicb.2015.01130
Geng, H., and Belas, R. (2010a). Molecular mechanisms underlying roseobacter–phytoplankton symbioses. Curr. Opin. Biotechnol. 21, 332–338. doi: 10.1016/j.copbio.2010.03.013
Geng, H., and Belas, R. (2010b). Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J. Bacteriol. 192, 4377–4387. doi: 10.1128/JB.00410-10
Geske, G. D., O’Neill, J. C., Miller, D. M., Wezeman, R. J., Mattmann, M. E., Lin, Q., et al. (2008). Comparative analyses of N-acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. ChemBioChem 9, 389–400. doi: 10.1002/cbic.200700551.Comparative
Ghosh, D., Roy, K., Williamson, K. E., Srinivasiah, S., Wommack, K. E., and Radosevich, M. (2009). Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75, 7142–7152. doi: 10.1128/AEM.00950-09
Giovannoni, S. J., and Stingl, U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437, 343–348.
Goecke, F., Thiel, V., Wiese, J., Labes, A., and Imhoff, J. F. (2013). Algae as an important environment for bacteria - Phylogenetic relationships among new bacterial species isolated from algae. Phycologia 52, 14–24. doi: 10.2216/12-24.1
Gram, L., Grossart, H. P., Schlingloff, A., and Kiørboe, T. (2002). Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68, 4111–4116. doi: 10.1128/AEM.68.8.4111-4116.2002
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., and Faure, D. (2016). Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Grondin, J. M., Tamura, K., Déjean, G., Abbott, D. W., and Brumer, H. (2017). Polysaccharide utilization loci: fuelling microbial communities. J. Bacteriol. 199:e00860-16. doi: 10.1128/JB.00860-16
Grossart, H. P. (2010). Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ. Microbiol. Rep. 2, 706–714. doi: 10.1111/j.1758-2229.2010.00179.x
Grossart, H. P., Schlingloff, A., Bernhard, M., Simon, M., and Brinkhoff, T. (2004). Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 47, 387–396. doi: 10.1016/S0168-6496(03)00305-2
Grossart, H. P., Tang, K. W., Kiørboe, T., and Ploug, H. (2007). Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol. Lett. 266, 194–200. doi: 10.1111/j.1574-6968.2006.00520.x
Großkopf, T., and Soyer, O. S. (2014). Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77. doi: 10.1016/j.mib.2014.02.002
Harayama, S., Kasai, Y., and Hara, A. (2004). Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15, 205–214. doi: 10.1016/j.copbio.2004.04.002
Henke, J. M., and Bassler, B. L. (2004). Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186, 6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004
Hense, B. A., Kuttler, C., Müller, J., Rothballer, M., Hartmann, A., and Kreft, J. U. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239. doi: 10.1038/nrmicro1600
Hmelo, L., and Van Mooy, B. A. S. (2009). Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 54, 127–133. doi: 10.3354/ame01261
Hmelo, L. R. (2017). Quorum sensing in marine microbial environments. Ann. Rev. Mar. Sci. 9, 257–281. doi: 10.1146/annurev-marine-010816-060656
Hmelo, L. R., Mincer, T. J., and Van Mooy, B. A. S. (2011). Possible influence of bacterial quorum sensing on the hydrolysis of sinking particulate organic carbon in marine environments. Environ. Microbiol. Rep. 3, 682–688. doi: 10.1111/j.1758-2229.2011.00281.x
Holden, M. T. G., Chhabra, S. R., De Nys, R., Stead, P., Bainton, N. J., Hill, P. J., et al. (1999). Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x
Hollants, J., Leliaert, F., De Clerck, O., and Willems, A. (2013). What we can learn from sushi: a review on seaweed-bacterial associations. FEMS Microbiol. Ecol. 83, 1–16. doi: 10.1111/j.1574-6941.2012.01446.x
Huang, S., Bergonzi, C., Schwab, M., Elias, M., and Hicks, R. E. (2019). Evaluation of biological and enzymatic quorum quencher coating additives to reduce biocorrosion of steel. PLoS One 14:0217059. doi: 10.1371/journal.pone.0217059
Huang, X., Zhu, J., Cai, Z., Lao, Y., Jin, H., Yu, K., et al. (2018). Profiles of quorum sensing (QS)-related sequences in phycospheric microorganisms during a marine dinoflagellate bloom, as determined by a metagenomic approach. Microbiol. Res. 217, 1–13. doi: 10.1016/j.micres.2018.08.015
Huang, Y., Ki, J., Lee, O. O., and Qian, P. (2009). Evidence for the dynamics of Acyl homoserine lactone and AHL-producing bacteria during subtidal biofilm formation. ISME J. 3, 296–304. doi: 10.1038/ismej.2008.105
Huang, Y., Zeng, Y., Yu, Z., Zhang, J., Feng, H., and Lin, X. (2013). In silico and experimental methods revealed highly diverse bacteria with quorum sensing and aromatics biodegradation systems - A potential broad application on bioremediation. Bioresour. Technol. 148, 311–316. doi: 10.1016/j.biortech.2013.08.155
Huang, Y. L., Ki, J. S., Case, R. J., and Qian, P. Y. (2008). Diversity and acyl-homoserine lactone production among subtidal biofilm-forming bacteria. Aquat. Microb. Ecol. 52, 185–193. doi: 10.3354/ame01215
Hudaiberdiev, S., Choudhary, K. S., Alvarez, R. V., Ligeti, B., Lamba, D., and Pongor, S. (2015). Census of solo LuxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol. 5:20. doi: 10.3389/fcimb.2015.00020
Jacquin, J., Cheng, J., Odobel, C., Pandin, C., Conan, P., Pujo-Pay, M., et al. (2019). Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “plastisphere.”. Front. Microbiol. 10:856. doi: 10.3389/fmicb.2019.00865
Jarrell, K. F., Walters, A. D., Bochiwal, C., Borgia, J. M., Dickinson, T., and Chong, J. P. J. (2011). Major players on the microbial stage: why Archaea are important. Microbiology 157, 919–936. doi: 10.1099/mic.0.047837-0
Jatt, A. N., Tang, K., Liu, J., Zhang, Z., and Zhang, X. H. (2015). Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol. Ecol. 91, 1–13. doi: 10.1093/femsec/fiu030
Kanagasabhapathy, M., Yamazaki, G., Ishida, A., Sasaki, H., and Nagata, S. (2009). Presence of quorum-sensing inhibitor-like compounds from bacteria isolated from the brown alga Colpomenia sinuosa. Lett. Appl. Microbiol. 49, 573–579. doi: 10.1111/j.1472-765X.2009.02712.x
Kaplan, H. B., and Greenberg, E. P. (1985). Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163, 1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985
Karner, M. B., Delong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kiørboe, T., Grossart, H. P., Ploug, H., and Tang, K. (2002). Mechanisms and rates of colonisation of sinking aggregates. Appl. Environ. Microbiol. 68, 3996–4006. doi: 10.1128/AEM.68.8.3996
Kirchman, D. L. (2002). The ecology of Cytophaga-flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100. doi: 10.1016/S0168-6496(01)00206-9
Koppenhöfer, S., and Lang, A. S. (2020). Interactions among redox regulators and the CtrA phosphorelay in Dinoroseobacter shibae and Rhodobacter capsulatus. Microorganisms 8:562. doi: 10.3390/microorganisms8040562
Koppenhöfer, S., Wang, H., Scharfe, M., Kaever, V., Wagner-Döbler, I., and Tomasch, J. (2019). Integrated transcriptional regulatory network of quorum sensing, replication control, and SOS response in dinoroseobacter shibae. Front. Microbiol. 10:803. doi: 10.3389/fmicb.2019.00803
Krupke, A., Hmelo, L. R., Ossolinski, J. E., Mincer, T. J., and Van Mooy, B. A. S. (2016). Quorum Sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front. Mar. Sci. 3:55. doi: 10.3389/fmars.2016.00055
Kumari, S., Mangwani, N., and Das, S. (2020). Naphthalene catabolism by biofilm forming marine bacterium Pseudomonas aeruginosa N6P6 and the role of quorum sensing in regulation of dioxygenase gene. J. Appl. Microbiol. 130, 1217–1231. doi: 10.1111/jam.14867
Lage, O. M., and Graça, A. P. (2016). “Biofilms: an extra coat on macroalgae,” in Algae - Organisms for Imminent Biotechnology, eds N. Thajuddin and D. Dhanasekaran (London: InTech).
Lam, K. S. (2006). Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9, 245–251. doi: 10.1016/j.mib.2006.03.004
Lami, R. (2019). “Quorum sensing in marine biofilms and environments,” in Quorum Sensing: Molecular Mechanism and Biotechnological Application, ed. G. Tommonaro (Cambridge, MA: Academic Press).
Lami, R., Grimaud, R., Sanchez-brosseau, S., Six, C., Thomas, F., West, N. J., et al. (2021). “Marine bacterial models for experimental biology,” in Handbook of Marine Model Organisms in Experimental Biology, eds A. Boutet and B. Schierwater (Boca Raton, FL: CRC Press).
Landa, M., Burns, A. S., Roth, S. J., and Moran, M. A. (2017). Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690. doi: 10.1038/ismej.2017.117
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Li, J., Azam, F., Zhang, S., Biology, M., Diego, S., and Jolla, L. (2016). Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ. Microbiol. 18, 3850–3866. doi: 10.1111/1462-2920.13344
Li, J., Gu, L., Bai, S., Wang, J., Su, L., Wei, B., et al. (2021). Characterization of particle-associated and free-living bacterial and archaeal communities along the water columns of the South China Sea. Biogeosciences 18, 113–133. doi: 10.5194/bg-2020-115
Liang, X., Wagner, R. E., Li, B., Zhang, N., Radosevich, M., and Emerson, J. B. (2020). Quorum sensing signals alter in vitro soil virus abundance and bacterial community composition. Front. Microbiol. 11:1287. doi: 10.3389/fmicb.2020.01287
Liao, Y., Williams, T. J., Ye, J., Charlesworth, J., Burns, B. P., Poljak, A., et al. (2016). Morphological and proteomic analysis of biofilms from the Antarctic archaeon, Halorubrum lacusprofundi. Sci. Rep. 6:37454. doi: 10.1038/srep37454
Liu, J., Fu, K., Wang, Y., Wu, C., Li, F., Shi, L., et al. (2017). Detection of diverse N-Acyl-homoserine lactones in Vibrio alginolyticus and regulation of biofilm formation by lactone in vitro. Front. Microbiol. 8:1097. doi: 10.3389/fmicb.2017.01097
Long, T., Tu, K. C., Wang, Y., Mehta, P., Ong, N. P., Bassler, B. L., et al. (2009). Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 7:e1000068. doi: 10.1371/journal.pbio.1000068
Luo, H., and Moran, M. A. (2014). Evolutionary ecology of the marine roseobacter clade. Microbiol. Mol. Biol. Rev. 78, 573–587. doi: 10.1128/MMBR.00020-14
Lupp, C., and Ruby, E. G. (2005). Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187, 3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005
Lyons, M. M., and Dobbs, F. C. (2012). Differential utilization of carbon substrates by aggregate-associated and water-associated heterotrophic bacterial communities. Hydrobiologia 686, 181–193. doi: 10.1007/s10750-012-1010-7
Mangwani, N., Kumari, S., and Das, S. (2015). Involvement of quorum sensing genes in biofilm development and degradation of polycyclic aromatic hydrocarbons by a marine bacterium Pseudomonas aeruginosa N6P6. Appl. Microbiol. Biotechnol. 99, 10283–10297. doi: 10.1007/s00253-015-6868-7
Martin, M., Portetelle, D., Michel, G., and Vandenbol, M. (2014). Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 98, 2917–2935. doi: 10.1007/s00253-014-5557-2
McLean, R. J. C., Whiteley, M., Stickler, D. J., and Fuqua, W. C. (1997). Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154, 259–263. doi: 10.1016/S0378-1097(97)00336-4
McRose, D. L., Baars, O., Seyedsayamdost, M. R., and Morel, F. M. M. (2018). Quorum sensing and iron regulate a two-for-one siderophore gene cluster in Vibrio harveyi. Proc. Natl. Acad. Sci. U.S.A. 115, 7511–7586. doi: 10.1073/pnas.1805791115
Medina-Martínez, M. S., Uyttendaele, M., Demolder, V., and Debevere, J. (2006). Effect of temperature and glucose concentration on the N-butanoyl-L- homoserine lactone production by Aeromonas hydrophila. Food Microbiol. 23, 534–540. doi: 10.1016/j.fm.2005.09.010
Mestre, M., Ferrera, I., Borrull, E., Ortega-Retuerta, E., Mbedi, S., Grossart, H. P., et al. (2017). Spatial variability of marine bacterial and archaeal communities along the particulate matter continuum. Mol. Ecol. 26, 6827–6840. doi: 10.1111/mec.14421
Mestre, M., Höfer, J., Sala, M. M., and Gasol, J. M. (2020). Seasonal variation of bacterial diversity along the marine particulate matter continuum. Front. Microbiol. 11:1590. doi: 10.3389/fmicb.2020.01590
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L., et al. (2004). Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15, 677–687. doi: 10.1016/j.molcel.2004.07.020
Montgomery, K., Charlesworth, J., LeBard, R., Visscher, P. T., and Burns, B. P. (2013). Quorum sensing in extreme environments. Life 3, 131–148. doi: 10.3390/life3010131
Moons, P., Michiels, C. W., Aertsen, A., Moons, P., Michiels, C. W., and Aertsen, A. (2009). Bacterial interactions in biofilms. Crit. Rev. Microbiol. 35, 157–168. doi: 10.1080/10408410902809431
Morel, F., Rueter, J., and Price, N. (1991). Iron nutrition of phytoplankton and its possible importance in the ecology of ocean regions with high nutrient and low biomass. Oceanography 4, 56–61. doi: 10.5670/oceanog.1991.03
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Muras, A., López-Pérez, M., Mayer, C., Parga, A., Amaro-Blanco, J., and Otero, A. (2018). High prevalence of quorum-sensing and quorum-quenching activity among cultivable bacteria and metagenomic sequences in the mediterranean sea. Genes 9:100. doi: 10.3390/genes9020100
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Nagy, K., Sipos, O., Valkai, S., Gombai, É, Hodula, O., Kerényi, Á, et al. (2015). Microfluidic study of the chemotactic response of Escherichia coli to amino acids, signaling molecules and secondary metabolites. Biomicrofluidics 9:e044105. doi: 10.1063/1.4926981
Nealson, K. H., Platt, T., and Hastings, J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322.
Oh, R. M., Bollati, E., Maithani, P., Huang, D., and Wainwright, B. J. (2021). The microbiome of the reef macroalga sargassum ilicifolium in Singapore. Microorganisms 9:898. doi: 10.3390/microorganisms9050898
Ortori, C. A., Atkinson, S., Chhabra, S. R., Cámara, M., Williams, P., and Barrett, D. A. (2007). Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole–linear ion trap mass spectrometry. Anal. Bioanal. Chem. 387, 497–511. doi: 10.1007/s00216-006-0710-0
Paggi, R. A., Martone, C. B., Fuqua, C., and De Castro, R. E. (2003). Detection of quorum sensing signals in the haloalkaliphilic archaeon Natronococcus occultus. FEMS Microbiol. Lett. 221, 49–52. doi: 10.1016/S0378-1097(03)00174-5
Pajares, S., and Ramos, R. (2019). Processes and microorganisms involved in the marine nitrogen cycle: knowledge and gaps. Front. Mar. Sci. 6:739. doi: 10.3389/fmars.2019.00739
Patzelt, D., Wang, H., Buchholz, I., Rohde, M., Gröbe, L., Pradella, S., et al. (2013). You are what you talk: quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. ISME J. 7, 2274–2286. doi: 10.1038/ismej.2013.107
Paulsen, S. S., Isbrandt, T., Kirkegaard, M., Buijs, Y., Strube, M. L., Sonnenschein, E. C., et al. (2020). Production of the antimicrobial compound tetrabromopyrrole and the Pseudomonas quinolone system precursor, 2-heptyl-4-quinolone, by a novel marine species Pseudoalteromonas galatheae sp. nov. Sci. Rep. 10:21630. doi: 10.1038/s41598-020-78439-3
Pearson, J. P., Van Delden, C., and Iglewski, B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181, 1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999
Pereira, C. S., Mcauley, J. R., Taga, M. E., Xavier, K. B., and Miller, S. T. (2008). Sinorhizobium meliloti, a bacterium lacking the supplied by other bacteria. Mol. Microbiol. 70, 1223–1235. doi: 10.1111/j.1365-2958.2008.06477.x
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Ploug, H., and Grossart, H. P. (1999). Bacterial production and respiration in suspended aggregates - A matter of the incubation method. Aquat. Microb. Ecol. 20, 21–29. doi: 10.3354/ame020021
Polkade, A. V., Mantri, S. S., Patwekar, U. J., and Jangid, K. (2016). Quorum sensing: an under-explored phenomenon in the phylum Actinobacteria. Front. Microbiol. 7:131. doi: 10.3389/fmicb.2016.00131
Pollara, S. B., Becker, J. W., Nunn, B. L., Boiteau, R., Repeta, D., Mudge, M. C., et al. (2021). Bacterial quorum-sensing signal arrests phytoplankton cell division and impacts virus-induced mortality. mSphere 6:e00009-21.
Pomeroy, L. R. (1974). The ocean’s food web, a changing paradigm. Bioscience 24, 499–504. doi: 10.2307/1296885
Popat, R., Harrison, F., Silva, A. C., Easton, S. A. S., Mcnally, L., Williams, P., et al. (2017). Environmental modification via a quorum sensing molecule influences the social landscape of siderophore production. Proc. R. Soc. B Biol. Sci. 284:20170200.
Prado, S., Leblanc, C., and Rebuffat, S. (2016). “Microbiota and chemical ecology,” in Chemical Ecology, eds A.-G. Bagnères and M. Hossaert-Mckey (Hoboken, NJ: John Wiley & Sons, Inc).
Prescott, R. D., and Decho, A. W. (2020). Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 28, 436–444. doi: 10.1016/j.tim.2019.12.004
Raina, J. B., Fernandez, V., Lambert, B., Stocker, R., and Seymour, J. R. (2019). The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294. doi: 10.1038/s41579-019-0182-9
Rajput, A., and Kumar, M. (2017). Computational exploration of putative LuxR solos in Archaea and their functional implications in quorum sensing. Front. Microbiol. 8:798. doi: 10.3389/fmicb.2017.00798
Redfield, R. J. (2002). Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10, 365–370. doi: 10.1016/S0966-842X(02)02400-9
Reen, F. J., Gutiérrez-Barranquero, J. A., McCarthy, R. R., Woods, D. F., Scarciglia, S., Adams, C., et al. (2019). Quorum sensing signaling alters virulence potential and population dynamics in complex microbiome-host interactomes. Front. Microbiol. 10:2131. doi: 10.3389/fmicb.2019.02131
Reen, F. J., Mooij, M. J., Holcombe, L. J., Mcsweeney, C. M., Mcglacken, G. P., Morrissey, J. P., et al. (2011). The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 77, 413–428. doi: 10.1111/j.1574-6941.2011.01121.x
Rehman, Z. U., and Leiknes, T. O. (2018). Quorum-quenching bacteria isolated from red sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Front. Microbiol. 9:1354. doi: 10.3389/fmicb.2018.01354
Reintjes, G., Arnosti, C., Fuchs, B., and Amann, R. (2019). Selfish, sharing and scavenging bacteria in the Atlantic Ocean: a biogeographical study of bacterial substrate utilisation. ISME J. 13, 1119–1132. doi: 10.1038/s41396-018-0326-3
Reintjes, G., Arnosti, C., Fuchs, B. M., and Amann, R. (2017). An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 11, 1640–1650. doi: 10.1038/ismej.2017.26
Ren, D., Zuo, R., and Wood, T. K. (2005). Quorum-sensing antagonist influences siderophore biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 66, 689–695. doi: 10.1007/s00253-004-1691-6
Rezzonico, F., and Duffy, B. (2008). Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. doi: 10.1186/1471-2180-8-154
Rieck, A., Herlemann, D. P. R., Jürgens, K., and Grossart, H. (2015). Particle-associated differ from free-living bacteria in surface waters of the baltic sea particle-associated differ from free-living bacteria in surface waters of the baltic sea. Front. Microbiol. 6:1297. doi: 10.3389/fmicb.2015.01297
Riedel, K., Hentzer, M., Geisenberger, O., Huber, B., Steidle, A., Wu, H., et al. (2001). N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262. doi: 10.1099/00221287-147-12-3249
Rink, B., Martens, T., Fischer, D., Lemke, A., Grossart, H. P., Simon, M., et al. (2008). Short-term dynamics of bacterial communities in a tidally affected coastal ecosystem. FEMS Microbiol. Ecol. 66, 306–319. doi: 10.1111/j.1574-6941.2008.00573.x
Roager, L., and Sonnenschein, E. C. (2019). Bacterial candidates for colonization and degradation of marine plastic debris. Environ. Sci. Technol. 53, 11636–11643. doi: 10.1021/acs.est.9b02212
Romero, M., Avendaño-Herrera, R., Magariños, B., Cámara, M., and Otero, A. (2010). Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol. Lett. 304, 131–139. doi: 10.1111/j.1574-6968.2009.01889.x
Romero, M., Diggle, S. P., Heeb, S., Cámara, M., and Otero, A. (2008). Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 280, 73–80. doi: 10.1111/j.1574-6968.2007.01046.x
Romero, M., Martin-Cuadrado, A. B., and Otero, A. (2012). Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 78, 6345–6348. doi: 10.1128/AEM.01266-12
Romero, M., Martin-Cuadrado, A. B., Roca-Rivada, A., Cabello, A. M., and Otero, A. (2011a). Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 75, 205–217. doi: 10.1111/j.1574-6941.2010.01011.x
Romero, M., Muro-Pastor, A. M., and Otero, A. (2011b). Quorum sensing N-acylhomoserine lactone signals affect nitrogen fixation in the cyanobacterium Anabaena sp. PCC7120. FEMS Microbiol. Lett. 315, 101–108. doi: 10.1111/j.1574-6968.2010.02175.x
Santoro, A. E., Richter, R. A., and Dupont, C. L. (2019). Planktonic marine archaea. Ann. Rev. Mar. Sci. 11, 131–158. doi: 10.1146/annurev-marine-121916-063141
Saurav, K., Costantino, V., Venturi, V., and Steindler, L. (2017). Quorum sensing inhibitors from the sea discovered using bacterial N -acyl-homoserine lactone-based biosensors. Mar. Drugs 15:53. doi: 10.3390/md15030053
Schaefer, A. L., Greenberg, E. P., Oliver, C. M., Oda, Y., Huang, J. J., Bittan-Banin, G., et al. (2008). A new class of homoserine lactone quorum-sensing signals. Nature 454, 595–600. doi: 10.1038/nature07088
Schertzer, J. W., Boulette, M. L., and Whiteley, M. (2009). More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol. 17, 189–195. doi: 10.1016/j.tim.2009.02.001
Schlundt, C., Mark, J. L., Anna, W., and Amaral-zettler, E. R. Z. L. A. (2020). Spatial structure in the “Plastisphere”: molecular resources for imaging microscopic communities on plastic marine debris. Mol. Ecol. Resour. 20, 620–634. doi: 10.1111/1755-0998.13119
Schmidt, R., Ulanova, D., Wick, L. Y., Bode, H. B., and Garbeva, P. (2019). Microbe-driven chemical ecology: past, present and future. ISME J. 13, 2656–2663. doi: 10.1038/s41396-019-0469-x
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Shapiro, J. A. (1998). Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104. doi: 10.1146/annurev.micro.52.1.81
Sharif, D. I., Gallon, J., Smith, C. J., and Dudley, E. (2008). Quorum sensing in Cyanobacteria: N-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium Gloeothece PCC6909. ISME J. 2, 1171–1182. doi: 10.1038/ismej.2008.68
Sheridan, C. C., Steinberg, D. K., and Kling, G. W. (2002). The microbial and metazoan community associated with colonies of Trichodesmium spp.: a quantitative survey. J. Plankton Res. 24, 913–922. doi: 10.1093/plankt/24.9.913
Shrout, J. D., Chopp, D. L., Just, C. L., Hentzer, M., Givskov, M., and Parsek, M. R. (2006). The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62, 1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x
Sievert, S. M., Kiene, R. P., and Schulz-vogt, H. N. (2007). The sulfur cycle. Oceanography 20, 117–123.
Silpe, J. E., and Bassler, B. L. (2019). Phage-encoded LuxR-type receptors responsive to host- produced bacterial quorum-sensing autoinducers. mBio 10: e00638-19.
Silpe, J. E., Bridges, A. A., Huang, X., Coronado, D. R., Duddy, O. P., and Bassler, B. L. (2020). Separating functions of the phage-encoded Quorum-Sensing-activated antirepressor Qtip. Cell Host Microbe 27, 629.e4–641.e4. doi: 10.1016/j.chom.2020.01.024
Simon, M., Grossart, H. P., Schweitzer, B., and Ploug, H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
Sivakumar, K., Scarascia, G., Zaouri, N., Wang, T., Kaksonen, A. H., and Hong, P. Y. (2019). Salinity-mediated increment in sulfate reduction, biofilm formation, and quorum sensing: a potential connection between quorum sensing and sulfate reduction? Front. Microbiol. 10:188. doi: 10.3389/fmicb.2019.00188
Smith, D. C., Simon, M., Alldredge, A. L., and Azam, F. (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359, 139–142. doi: 10.1038/355242a0
Smith, P., and Schuster, M. (2019). Public goods and cheating in microbes. Curr. Biol. 29, R442–R447. doi: 10.1016/j.cub.2019.03.001
Sperandio, V., Torres, A. G., Giro, J. A., and Kaper, J. B. (2001). Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183, 5187–5197. doi: 10.1128/JB.183.17.5187
Steidle, A., Sigl, K., Schuhegger, R., Ihring, A., Schmid, M., Gantner, S., et al. (2001). Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67, 5761–5770. doi: 10.1128/AEM.67.12.5761-5770.2001
Stock, F., Cirri, E., Nuwanthi, S. G. L. I., Stock, W., Ueberschaar, N., Mangelinckx, S., et al. (2021). Sampling, separation, and quantification of N-acyl homoserine lactones from marine intertidal sediments. Limnol. Oceanogr. Methods 19, 145–157. doi: 10.1002/lom3.10412
Stocker, R. (2012). Marine microbes see a sea of gradients. Science 338, 628–633. doi: 10.1126/science.1208929
Stocker, R., Seymour, J. R., Samadani, A., Hunt, D. E., and Polz, M. F. (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U.S.A. 105, 4209–4214. doi: 10.1073/pnas.0709765105
Stockner, J. G. (1988). Phototrophic picoplankton: an overview from marine and freshwater ecosystems. Limnol. Oceanogr. 33, 765–775. doi: 10.4319/lo.1988.33.4part2.0765
Strom, S. L. (2008). Microbial ecology of ocean biogeochemistry: a community perspective. Science 320, 1043–1045. doi: 10.1126/science.1153527
Su, Y., Tang, K., Liu, J., Wang, Y., Zheng, Y., and Zhang, X. H. (2019). Quorum sensing system of Ruegeria mobilis Rm01 controls lipase and biofilm formation. Front. Microbiol. 10:3304. doi: 10.3389/fmicb.2018.03304
Su, Y., Yang, Y., Zhu, X. Y., Zhang, X. H., and Yu, M. (2021). Metagenomic insights into the microbial assemblage capable of quorum sensing and quorum quenching in particulate organic matter in the yellow sea. Front. Microbiol. 11:602010. doi: 10.3389/fmicb.2020.602010
Subramoni, S., Salcedo, D. V. F., and Suarez-Moreno, Z. R. (2015). A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front. Cell. Infect. Microbiol. 5:16. doi: 10.3389/fcimb.2015.00016
Sun, J., Daniel, R., Wagner-Döbler, I., and Zeng, A. P. (2004). Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. doi: 10.1186/1471-2148-4-36
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Suttle, C. A. (2007). Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Swem, L. R., Swem, D. L., Wingreen, N. S., and Bassler, B. L. (2008). Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134, 461–473. doi: 10.1016/j.cell.2008.06.023
Teasdale, M. E., Donovan, K. A., Forschner-Dancause, S. R., and Rowley, D. C. (2011). Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 13, 722–732. doi: 10.1007/s10126-010-9334-7
Teeling, H., Fuchs, B. M., Bennke, C. M., Krüger, K., Chafee, M., Kappelmann, L., et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. elife 5:e11888. doi: 10.7554/eLife.11888
Thomas, F., Hehemann, J. H., Rebuffet, E., Czjzek, M., and Michel, G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2:93. doi: 10.3389/fmicb.2011.00093
Tian, Y., Wang, Q., Liu, Q., Ma, Y., Cao, X., Guan, L., et al. (2008). Involvement of LuxS in the regulation of motility and flagella biogenesis in Vibrio alginolyticus. Biosci. Biotechnol. Biochem. 72, 1063–1071. doi: 10.1271/bbb.70812
Tommonaro, G., Abbamondi, G. R., Iodice, C., Tait, K., and De Rosa, S. (2012). Diketopiperazines produced by the halophilic archaeon, haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol. 63, 490–495. doi: 10.1007/s00248-011-9980-y
Torres, M., Dessaux, Y., and Llamas, I. (2019). Saline environments as a source of potential quorum sensing disruptors to control bacterial infections: a review. Mar. Drugs 17, 1–28. doi: 10.3390/md17030191
Torres, M., Rubio-Portillo, E., Antón, J., Ramos-Esplá, A. A., Quesada, E., and Llamas, I. (2016). Selection of the N-acylhomoserine lactone-degrading bacterium Alteromonas stellipolaris PQQ-42 and of its potential for biocontrol in aquaculture. Front. Microbiol. 7:646. doi: 10.3389/fmicb.2016.00646
Tourneroche, A., Lami, R., Hubas, C., Blanchet, E., Vallet, M., Escoubeyrou, K., et al. (2019). Bacterial-fungal interactions in the kelp endomicrobiota drive autoinducer-2 quorum sensing. Front. Microbiol. 10:1693. doi: 10.3389/fmicb.2019.01693
Toyofuku, M., Morinaga, K., Hashimoto, Y., Uhl, J., Shimamura, H., Inaba, H., et al. (2017). Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509. doi: 10.1038/ismej.2017.13
Toyofuku, M., Nomura, N., Kuno, E., Tashiro, Y., Nakajima, T., and Uchiyama, H. (2008). Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 190, 7947–7956. doi: 10.1128/JB.00968-08
Urvoy, M., Lami, R., Dreanno, C., Daudé, D., Rodrigues, A. M. S., Gourmelon, M., et al. (2021a). Quorum sensing disruption regulates hydrolytic enzyme and biofilm production in estuarine bacteria. Environ. Microbiol. 23, 7183–7200. doi: 10.1111/1462-2920.15775
Urvoy, M., Lami, R., Dreanno, C., Delmas, D., L’Helguen, S., and Labry, C. (2021b). Quorum sensing regulates the hydrolytic enzyme production and community composition of heterotrophic bacteria in coastal waters. Front. Microbiol. 12:780759. doi: 10.3389/fmicb.2021.780759
van Gemerden, H. (1993). Microbial mats: a joint venture. Mar. Geol. 113, 3–25. doi: 10.1016/0025-3227(93)90146-M
Van Mooy, B. A. S., Hmelo, L. R., Sofen, L. E., Campagna, S. R., May, A. L., Dyhrman, S. T., et al. (2012). Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 6, 422–429. doi: 10.1038/ismej.2011.115
Vial, L., Lépine, F., Milot, S., Groleau, M. C., Dekimpe, V., Woods, D. E., et al. (2008). Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J. Bacteriol. 190, 5339–5352. doi: 10.1128/JB.00400-08
Villarreal-Chiu, J. F., Quinn, J. P., and McGrath, J. W. (2012). The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 3:19. doi: 10.3389/fmicb.2012.00019
Visscher, P. T., and Stolz, J. F. (2005). Microbial mats as bioreactors: populations, processes, and products. Palaeogeogr. Palaeoclimatol. Palaeoecol. 219, 87–100. doi: 10.1016/j.palaeo.2004.10.016
Voss, M., Bange, H. W., Dippner, J. W., Middelburg, J. J., Montoya, J. P., and Ward, B. (2013). The marine nitrogen cycle: recent discoveries, uncertainties and the potential relevance of climate change. Philos. Trans. R. Soc. Lond. B 368:20130121.
Wagner-Döbler, I., and Biebl, H. (2006). Environmental biology of the marine roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Wagner-Döbler, I., Thiel, V., Eberl, L., Allgaier, M., Bodor, A., Meyer, S., et al. (2005). Discovery of complex mixtures of novel long- chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. ChemBioChem 6, 2195–2206. doi: 10.1002/cbic.200500189
Wang, N., Gao, J., Liu, Y., Wang, Q., Zhuang, X., and Zhuang, G. (2021). Realizing the role of N-acyl-homoserine lactone-mediated quorum sensing in nitrification and denitrification: a review. Chemosphere 274:129970. doi: 10.1016/j.chemosphere.2021.129970
Wang, Q., Liu, Q., Ma, Y., Rui, H., and Zhang, Y. (2007). LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen Vibrio alginolyticus. J. Appl. Microbiol. 103, 1525–1534. doi: 10.1111/j.1365-2672.2007.03380.x
Wang, R., Ding, W., Long, L., Lan, Y., Tong, H., Saha, S., et al. (2020). Exploring the influence of signal molecules on marine biofilms development. Front. Microbiol. 11:571400. doi: 10.3389/fmicb.2020.571400
Wang, T.-N., Kaksonen, A. H., and Hong, P.-Y. (2018). Evaluation of two autoinducer-2 quantification methods for application in marine environments. J. Appl. Microbiol. 124, 1469–1479. doi: 10.1111/jam.13725
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Weiland-Bräuer, N., Kisch, M. J., Pinnow, N., Liese, A., and Schmitz, R. A. (2016). Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 7:1098. doi: 10.3389/fmicb.2016.01098
Wellington, S., and Greenberg, E. P. (2019). Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio 10:e00146-19.
Wellington Miranda, S., Cong, Q., Schaefer, A. L., Macleod, E. K., Zimenko, A., Baker, D., et al. (2021). A covariation analysis reveals elements of selectivity in quorum sensing systems. eLife 10:e69169.
Wen, Y. I, Kim, H., Son, J., Lee, B., and Kim, K. (2012). Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J. Biol. Chem. 287, 26727–26739. doi: 10.1074/jbc.M112.374165
Whalen, K. E., Becker, J. W., Schrecengost, A. M., Gao, Y., Giannetti, N., and Harvey, E. L. (2019). Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7:93. doi: 10.1186/s40168-019-0711-9
White, A., and Dyhrman, S. (2013). The marine phosphorus cycle. Front. Microbiol. 4:105. doi: 10.3389/fmicb.2013.00105
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442, 368–373. doi: 10.1038/nature05058
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Williamson, L. L., Borlee, B. R., Schloss, P. D., Guan, C., Allen, H. K., and Handelsman, J. (2005). Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 71, 6335–6344. doi: 10.1128/AEM.71.10.6335
Wright, R. J., Langille, M. G. I., Walker, T. R., and Wright, R. J. (2021). Food or just a free ride? A meta-analysis reveals the global diversity of the Plastisphere. ISME J. 15, 789–806. doi: 10.1038/s41396-020-00814-9
Xavier, K. B., and Bassler, B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6, 191–197. doi: 10.1016/S1369-5274(03)00028-6
Xu, Y., Curtis, T., Dolfing, J., Wu, Y., and Rittmann, B. E. (2021). N-acyl-homoserine-lactones signaling as a critical control point for phosphorus entrapment by multi-species microbial aggregates. Water Res. 204:117627. doi: 10.1016/j.watres.2021.117627
Xu, Y., Wu, Y., Esquivel-Elizondo, S., Dolfing, J., and Rittmann, B. E. (2020). Using microbial aggregates to entrap aqueous phosphorus. Trends Biotechnol. 38, 1292–1303. doi: 10.1016/j.tibtech.2020.03.012
Yoch, D. C. (2002). Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68, 5804–5815. doi: 10.1128/AEM.68.12.5804-5815.2002
Yu, Z., Hu, Z., Xu, Q., Zhang, M., Yuan, N., Liu, J., et al. (2020). The luxI/luxR-type quorum sensing system regulates degradation of polycyclic aromatic hydrocarbons via two mechanisms. Int. J. Mol. Sci. 21:5548. doi: 10.3390/ijms21155548
Zan, J., Choi, O., Meharena, H., Uhlson, C. L., Churchill, M. E. A., Hill, R. T., et al. (2015). A solo luxI -type gene directs acylhomoserine lactone synthesis and contributes to motility control in the marine sponge symbiont Ruegeria sp. KLH11. Microbiology 161, 50–56. doi: 10.1099/mic.0.083956-0
Zettler, E. R., Mincer, T. J., and Amaral-zettler, L. A. (2013). Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146.
Zhai, C., Zhang, P., Shen, F., Zhou, C., and Liu, C. (2012). Does Microcystis aeruginosa have quorum sensing? FEMS Microbiol. Lett. 336, 38–44. doi: 10.1111/j.1574-6968.2012.02650.x
Zhang, G., Zhang, F., Ding, G., Li, J., Guo, X., Zhu, J., et al. (2012). Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344. doi: 10.1038/ismej.2011.203
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11:5371. doi: 10.1038/s41467-020-19243-5
Zhao, J., Li, X., Hou, X., Quan, C., and Chen, M. (2019). Widespread existence of quorum sensing inhibitors in marine bacteria: potential drugs to combat pathogens with novel strategies. Mar. Drugs 17:275. doi: 10.3390/md17050275
Zhao, Z., Wang, L., Miao, J., Zhang, Z., Ruan, J., Xu, L., et al. (2021). Regulation of the formation and structure of biofilms by quorum sensing signal molecules packaged in outer membrane vesicles. Sci. Total Environ. 806:151403. doi: 10.1016/j.scitotenv.2021.151403
Zhao, Z. C., Xie, G. J., Liu, B. F., Xing, D. F., Ding, J., Han, H. J., et al. (2021). A review of quorum sensing improving partial nitritation-anammox process: functions, mechanisms and prospects. Sci. Total Environ. 765:142703. doi: 10.1016/j.scitotenv.2020.142703
Zhou, Z., Liu, Y., Pan, J., Toner, B. M., Anantharaman, K., Breier, J. A., et al. (2020). Gammaproteobacteria mediating utilization of methyl-, sulfur- and petroleum organic compounds in deep ocean hydrothermal plumes. ISME J. 14, 3136–3148. doi: 10.1038/s41396-020-00745-5
Zimmerman, A. E., Martiny, A. C., and Allison, S. D. (2013). Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J. 7, 1187–1199. doi: 10.1038/ismej.2012.176
Keywords: marine bacteria, quorum sensing, AHL (N-acyl-homoserine lactone), biogeochemical cycle, hydrolytic enzyme, organic matter degradation, nutrient acquisition
Citation: Urvoy M, Labry C, L’Helguen S and Lami R (2022) Quorum Sensing Regulates Bacterial Processes That Play a Major Role in Marine Biogeochemical Cycles. Front. Mar. Sci. 9:834337. doi: 10.3389/fmars.2022.834337
Received: 13 December 2021; Accepted: 11 January 2022;
Published: 02 February 2022.
Edited by:
Gordon T. Taylor, Stony Brook University, United StatesReviewed by:
Elena Yakubovskaya, Stony Brook University, United StatesCopyright © 2022 Urvoy, Labry, L’Helguen and Lami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marion Urvoy, bWFyaW9uLnVydm95QG91dGxvb2suZnI=; Raphaël Lami, cmFwaGFlbC5sYW1pQG9icy1iYW55dWxzLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.