- 1School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 3College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 4Key Laboratory for Sustainable Development of Marine Fisheries, Ministry of Agriculture and Rural Affairs, Qingdao, China
- 5Shandong Key Laboratory of Marine Fisheries Biotechnology and Genetic Breeding, Qingdao, China
In mammals, the mutation of nipped-B-like protein (nipbl) leads to Cornelia de Lange Syndrome (CdLS), characterized by low birth weight, short stature, and structural abnormalities of the skeleton, heart, and gut. In Chinese tongue sole (Cynoglossus semilaevis), a typical marine fish exhibiting sexual size dimorphism, the nipbl homolog gene (nipped-B-like protein A (nipbla)) was also screened with female higher expression level by somatotropic and reproductive tissues’ transcriptomic analysis. In this study, two nipbla genes, namely, nipbla-w and nipbla-z, were identified from the W and Z chromosomes of C. semilaevis, respectively. Similar to other mammalian and fish species nipbl, the two homolog proteins of C. semilaevis contained two conserved domains, namely, cohesion_HEAT and Nipped-B_C. The phylogenetic tree analysis showed that these two nipbla gametolog proteins were first clustered together and then grouped with other fish species. At least two types of alternative splicing sites were observed in exon 12 of the nipbla-z gene, which produced nipbla-z-tv1 and nipbla-z-tv2. Also, the sex-biased expression patterns of different nipbla-w and nipbla-z transcripts in female and male tissues were revealed by quantitative PCR (qPCR). The highest expression level of nipbla-w was observed in female gonad. While nipbla-z-tv1 exhibited relatively high expression in the muscle, liver, gonad, and brain, nipbla-z-tv2 only showed its expression superiority in the muscle of male individuals. The promoter regions of nipbla genes were amplified, and their transcription activity was successfully verified by a dual-luciferase reporter system. After nipbla-w and nipbla-z knockdown in the brain cell lines by RNA interference, a series of growth-related genes were influenced, including Bone Morphogenetic Protein 4 (bmp4), Wnt Family Member 11 (wnt11), and Sprouty Related EVH1 Domain Containing 2 (spred2). The prediction of transcription factors suggested that c-Jun, sex-determining region Y (SRY), POU Class 1 Homeobox 1 (POU1F1a), myogenic differentiation antigen (MyoD), signal transducer and activator of transcription 5a (STAT5A), and nuclear factor I C (Nfic) might be the putative upstream regulatory factors for nipbla; among them, c-Jun has been verified to effectively regulate the transcriptional activity of nipbla. The identification of two nipbla genes provided important data for interpreting the sexual size dimorphism in C. semilaevis.
Introduction
Sexual dimorphism in the sizes, shapes, and colors between female and male individuals has been broadly described in mammals, reptiles, birds, and fishes (Parker, 1992; Foellmer and Moya-Larano, 2007; Lindenfors et al., 2007; Székely et al., 2007; Mei and Gui, 2015). The sex-biased gene expression was a common phenomenon in various animals (Ranz et al., 2003; Yang et al., 2006; Mank et al., 2007, 2010) and has been linked to the sexual body size of wild turkey (Meleagris gallopavo) (Pointer et al., 2013). Thus, the study of sex-biased gene expression is crucial for understanding the molecular mechanism of sexual dimorphism.
In our previous transcriptomic study, thousands of sex-biased genes have been identified from the somatotropic and reproductive tissues of Chinese tongue sole (Cynoglossus semilaevis) (Wang et al., 2018), which is a female heterogamete (ZW/ZZ) fish species with typical female-biased sexual size dimorphism. One female-biased gene, i.e., nipped-B-like protein A (nipbla), and also one hub gene derived from the positive-related module (Wang et al., 2018) have caused our interest. Importantly, the mutation or disruption of nipbl in mammals leads to transcription dysregulation, which accounts for 60% of Cornelia de Lange syndrome, a disorder characterized by growth delay, limb reduction defects, and structural defects of multiple organs (Krantz et al., 2004; Liu et al., 2009; Mannini et al., 2013; Lindgren et al., 2014). Furthermore, five single-nucleotide polymorphisms (SNPs) of human nipbl based on the GWAS Catalog1 have been identified to be associated with body height and weight (p < 1 × 10–5) (MacArthur et al., 2017). As the homolog gene of the Drosophila melanogaster Nipped-B and fungal Scc2-type sister chromatid cohesion proteins, nipbl in mammals not only plays a vital role in the loading of the cohesin complex onto chromatin by forming a heterodimeric complex with MAU2/SCC4 but also involves in cohesin loading at sites of DNA damage (Muto et al., 2014; Rhodes et al., 2017). In teleost, nipbl has been reported to regulate expression of Hox cluster genes as an upstream factor; thereinto, the hoxA and D clusters are crucial for limb/fin development (Zakany and Duboule, 2007; Muto et al., 2014). More studies show that nipbl deficiency may be the multifactorial origins of heart and gut defects in teleost (Chien et al., 2011). In zebrafish, nipbl is believed to control the cell cycle, which downregulates the canonical Wnt pathway genes observed in patient-specific fibroblasts during neural development (Pistocchi et al., 2013). Regarding embryo development in zebrafish, nipped-B-like protein B (nipblb) loss-of-function has a role of dysregulation in hematological malignancies (Spreafico et al., 2020).
Interestingly, by chromosome location analysis, two allele genes located on chromosomes W and Z of C. semilaevis were found and named as nipbla-w and nipbla-z, respectively. Based on their different expression patterns in RNA-seq, this study aimed to reveal their different chromosome location and genomic structure information, examine their expression in more wide tissues by quantitative PCR (qPCR), survey the transcription activity by promoter region identification and analysis, and assay their knockdown effect by RNA interference.
Materials and Methods
Ethics Statement
The sampling and treatment of the C. semilaevis in this study were approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences (Approval number: YSFRI−2021024, dated 8 December 2021), and all the experimental procedures were carried out in line with the guidelines for the Care and Use of Laboratory Animals at the Chinese Academy of Fishery Sciences.
Fish and Cells
Ten different tissues including brain, liver, testis, ovary, muscle, intestine, spleen, kidney, heart, and skin were isolated from three female and three male individuals of 3-year-old C. semilaevis cultivated in Haiyang Yellow Sea Aquatic Product Co., Ltd (Shandong, China) and stored for subsequent RNA extraction. The gonads, were stored in the alcohol for DNA extraction by TIANamp Marine Animals DNA Kit (Tiangen). The genetic sex identification was performed by the primers sex-F (CCTAAATGATGGATGTAGATTCTGTC) and sex-R (GATCCAGAGAAAATAAACCCAGG) described in the previous study (Liu et al., 2014). Human embryonal kidney (HEK) 293T cells were cultured in DME/F-12 medium (HyClone, UT, United States) supplemented with 10% fetal bovine serum (FBS; Gibco, Australia) and maintained in 5% CO2 at 37°C. C. semilaevis brain cells (CSB) were maintained in L-15 medium supplemented with 10% FBS and 10 ng/ml basic fibroblast growth factor (bFGF; Invitrogen, MD, United States) at 24°C (Wang et al., 2015). Before transfection, cells were cultured in 24-well or 12-well plates with 60–80% density. The Lipofectamine 2000™ (Invitrogen) and riboFECT™ CP Transfection Kit (RiboBio) were separately used for plasmids and siRNA transfection.
The Characterization of Nipbla-w and Nipbla-z in Different Chromosomes
Based on the information of RNA-seq in our previous study, two nipbla homolog genes (GenBank number: 103397102 and 103398179) were found in chromosomes W and Z, respectively. To clarify the two genes, we denoted these two genes as nipbla-w and nipbla-z. The two genes’ locus information in chromosomes W and Z were further analyzed based on the genomic data of C. semilaevis. Also, the nipbla location information in other fish species including zebrafish (Danio rerio), marine medaka (Oryzias melastigma), Senegalese sole (Solea senegalensis), and turbot (Scophthalmus maximus) were exhibited.
The Phylogenetic Analysis of Nipbla Proteins
To identify the ortholog of nipbla of C. semilaevis, a phylogenetic analysis was carried out using MEGA 7.0 and EvolView. In brief, a list of nipbla or nipbl proteins were screened from UniProt by BLAST tools and submitted to MEGA 7.0 for the construction of a phylogenetic tree by the neighbor-joining (NJ) method (Kumar et al., 2016). Furthermore, the Newick file from MEGA 7.0 was uploaded into the EvolView software2 for the phylogenetic relationship presentation and domain description of nipbla/nipbl proteins.
Identification of Alternative Splicing and qPCR Analysis of Nipbla in Female and Male Individual Tissues
First, two primers nipbla-F and nipbla-R, respectively, located in exon 11 and exon 15, were designed to detect the splicing variants. To measure the expression patterns of nipbla-w and nipbla-z in different tissues of female and male individuals, primers nipbla-w-F/R, nipbla-z-tv1-F/R, and nipbla-z-tv2-F/R (Table 1) were first designed based on the mRNA sequences of nipbla-w, nipbla-z-tv1, and nipbla-z-tv2. The qPCR was further carried out, and β-actin was used for the internal reference. In brief, ten tissues, including brain, testis, ovary, heart, intestine, kidney, liver, muscle, skin, and spleen, were isolated from three female and three male individuals of 3-year-old C. semilaevis. The total RNAs of these samples were extracted by TRIzol Reagent and used for cDNA preparation with PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Japan). The 20 μl qPCR reactions were conducted by 95oC for 10 s, followed by 40 cycles of 95oC for 5 s and 60oC for 34 s on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, United States). The 2–ΔΔCt method was employed for evaluating the relative expression levels of genes (Livak and Schmittgen, 2001). The data obtained from qPCR were performed using the OmicShare tools for the least significant difference (LSD) analysis3.
The Promoter Activity Analysis for Two Genes
The putative promoter regions of nipbl-w and nipbl-z were first amplified by the primers (i.e., nipbw+s, nipbw-x, nipbz+s, and nipbz-x in the Table 1) and subcloned into pGL3-Basic to construct the recombinant plasmids pGL3-nipbla-wp and pGL3-nipbla-zp by the restriction enzymes SacI and XhoI. Then, pGL3-nipbla-wp, pGL3-nipbla-zp, pGL3-control, and pGL3-Basic were respectively transfected into HEK 293T cells by Lipofectamine 2000™ following the protocol with 800 ng/well in 24-well plates. The pRL-TK plasmid was used as the internal reference with 40 ng/well. At 48 h, cells transfected by plasmids were detected by a Dual-Luciferase Reporter Gene Assay Kit (Beyotime, Shanghai, China) with a Varioskan Flash spectral scanning multimode reader (Thermo, Vantaa, Finland) for the activities of firefly and Renilla luciferase. Each experiment was performed in triplicate. The abovementioned data were performed using the OmicShare tools for the LSD analysis (see text footnote 3).
Transcription Factor Prediction and Verification
To screen the putative transcription factors, the promoter sequences of nipbla-w and nipbla-z were submitted to the online tool PROMO4 and JASPAR5. To analyze the binding activity of transcription factors, the coding region of a c-Jun homolog in C. semilaevis (XM_025052424.1) was amplified by primers (jun+H and jun-E in Table 1) and ligated with pcDNA3.1 by HindIII and EcoRI to construct pcDNA3.1-c-Jun. Also, the mutation vector of pGL3-nipbl-wp (M) and pGL3-nipbla-zp (M) for c-Jun were constructed by the primers (i.e., wp-junMuF, wp-junMuR, zp-junMuF, and zp-junMuR in Table 1) according to the protocol of the Fast Site-Directed Mutagenesis Kit (Tiangen, China). The promoter and transcription factor plasmids were co-transfected into 293T cells to examine the luciferase activity.
The DNA Methylation Status of Two Gametologs in Different Sexes
To understand the influence of nipbla methylation on gene transcript formation, we analyzed the methylation profiles of the nipbla in genomic sequences of female, male, and pseudo-male individuals. Based on our methylome from C. semilaevis female, male, and pseudo-male gonad (Wang et al., 2021), the methylation levels of nipbla-w and nipbla-z in different sexual gonad tissues were demonstrated by R software. The methylation level analysis, which included the nipbla-w and nipbla-z upstream of 2,000 bp, gene body, and downstream of 2,000 bp regions, was employed for the examination of three sexual genotypes in Chinese tongue sole.
RNAi-Mediated Knockdown for Two Nipbla Gametologs in C. semilaevis
The nipbla small-interfering RNAs (i.e., si-nipbla-w and si-nipbla-z in Table 1) were designed and synthesized by RiboBio Co., Ltd. (Guangzhou, Guangdong Province, China). Even more, a non-specific siRNA negative was executed as a control [negative control (NC)] during the cell transfection process. Three replicates of si-nipbla-w, si-nipbla-z, and NC groups were conducted at 20 nM. After transfection for 48 h, the total RNA was extracted from the cells, and cDNA was synthesized according to the abovementioned methods. The relative expression levels of the genes related to growths, such as Bone Morphogenetic Protein 4 (bmp4), Wnt Family Member 11 (wnt11), and Sprouty Related EVH1 Domain Containing 2 (spred2), and FAT Atypical Cadherin 4 (fat4) were evaluated by qPCR.
Results
Chromosome Locus Information and Gene Structure of Nipbla-w and Nipbla-z
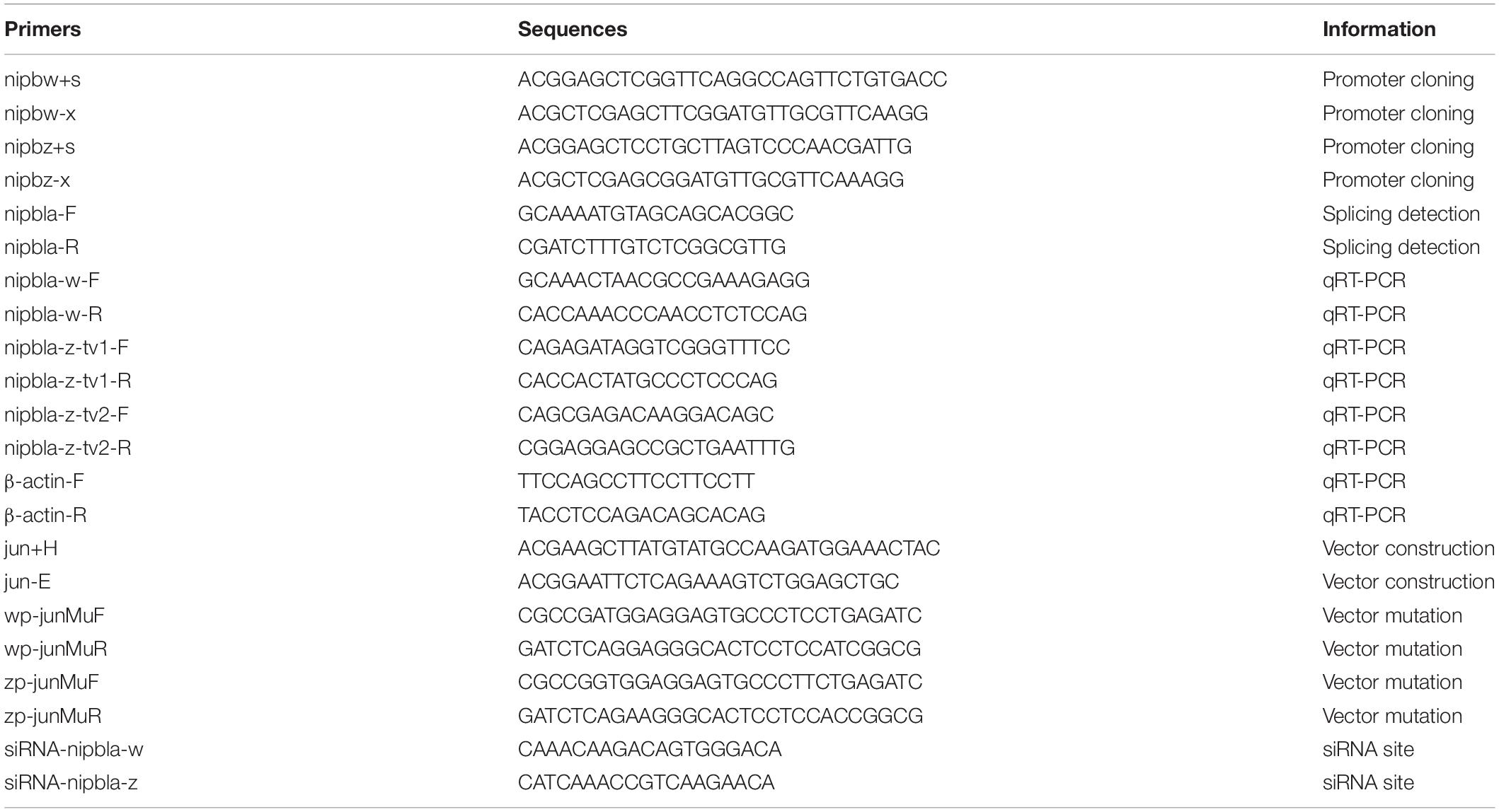
The nipbla-w (GenBank number: 103397102) and nipbla-z (GenBank number: 103398179) genes were separately located in the W and Z chromosomes (Figure 1). Also, no obvious syntenic arrangement was found in these two chromosomes. While the syntenic arrangement relationship of chromosome Z is similar to D. rerio, O. melastigma, S. senegalensis, and S. maximus nipbla. The similarity of mRNA (XM_008335345.2 and XM_017042947.2) and proteins (XP_008333567.1 and XP_016898436.1) derived from NCBI database between nipbla-w and nipbla-z were 88.4% and 88.7%, respectively.
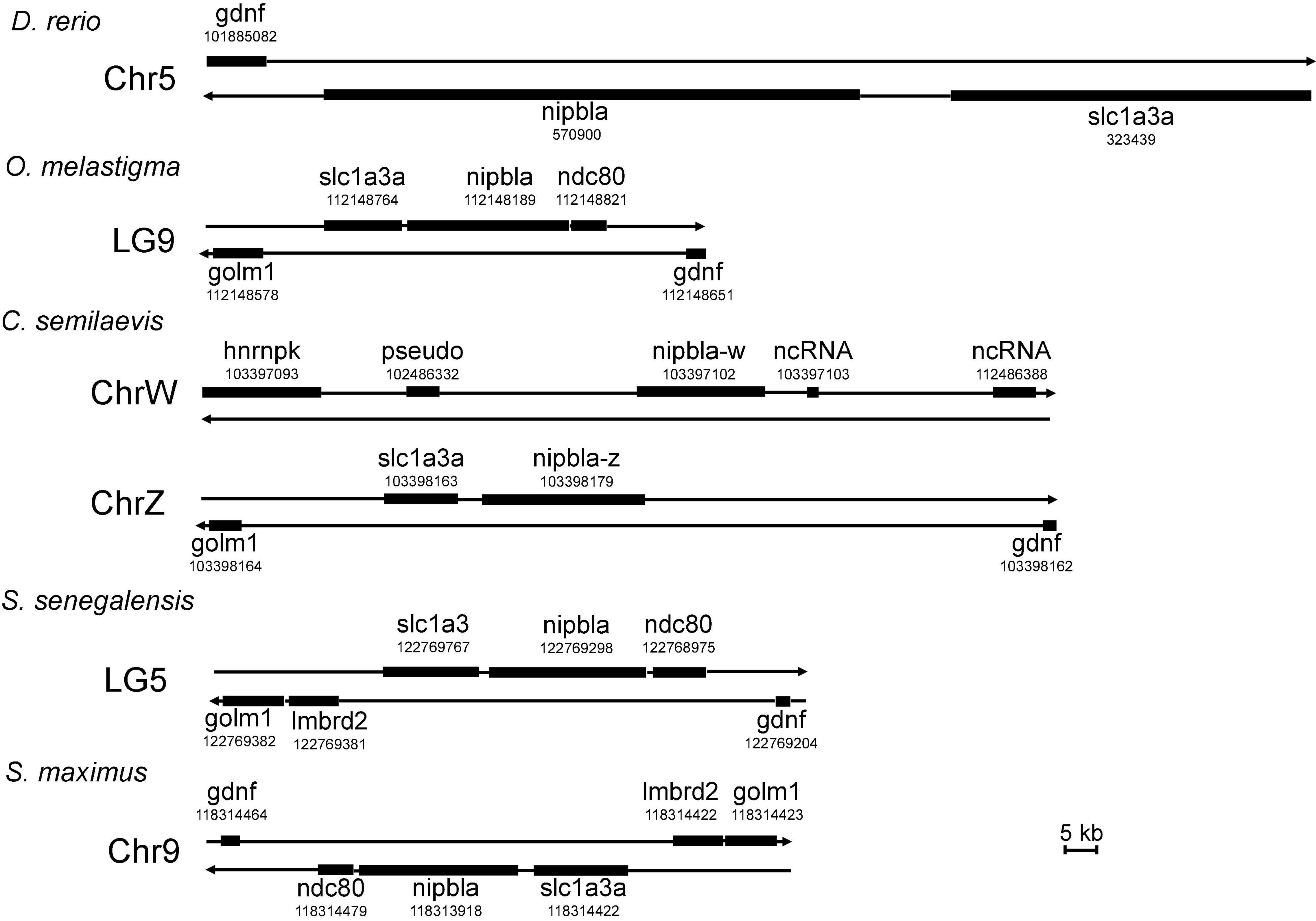
Figure 1. The chromosome location information of nipbla-w and nipbla-z. Danio rerio chromosome 5_7,986,080–8,171,694, Oryzias melastigma LG9_15,171,132–15,252,085, Cynoglossus semilaevis chromosome w_8,015,650-8,133,838, C. semilaevis chromosome z_16,746,071-16,882,207, Solea senegalensis LG5_13,731,413–13,824,383, and Scophthalmus maximus chromosome 9_15,599,751–15,691,435 were illustrated with nipbla and other genes location. hnrnpk, heterogeneous nuclear ribonucleoprotein K; golm1, golgi membrane protein 1; slc1a3a, solute carrier family 1 member 3a; gdnf, glial cell line-derived neurotrophic factor; ndc80, NDC80 kinetochore complex component; ncRNA, non-coding RNA.
The Polygenetic Analysis of Nipbl Proteins
The other nipbla or nipbl proteins were screened by searching the UniProt database using two kinds of nipbla proteins in C. semilaevis. A total of 32 nipbl or nipbla proteins including fruit fly (D. melanogaster), human (Homo sapiens), mouse (Mus musculus), pig (Sus scrofa), chicken (Gallus gallus), zebrafish (D. rerio), medaka (Oryzias latipes), turbot (S. maximus), Nile tilapia (Oreochromis niloticus), channel catfish (Ictalurus punetaus) blind cave fish (Astyanax mexicanus), and so on. The amino acid sequences using the NJ method were submitted to the construction of a polygenetic tree by MEGA 7.0. The subsequent phylogenetic relationship presentation and domain description (Figure 2) by EvolView software indicate that two nipbla homolog proteins in C. semilaevis were first clustered together and then clustered with other fish including medaka and marine medaka. Similar to other mammalian and fish species nipbl, the two homolog proteins of C. semilaevis contained two conserved domains, namely, cohesion_HEAT and Nipped-B_C.
Figure 2. The evolutionary relationships of nipbl proteins in different species. A total of 32 nipbl proteins were used for the phylogenetic tree construction using the neighbor-joining method in MEGA 7.0. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Notably, 32 nipbl proteins are listed as follows: A0A4W6GBV1_LATCA (Lates calcarifer, barramundi perch), A0A3B4VIZ8_SERDU (Seriola dumerili, greater amberjack), A0A3Q0RAT2_AMPCI (Amphilophus citrinellus, midas cichlid), I3KCI3_ORENI (Oreochromis niloticus, Nile tilapia), A0A3Q1G7Y4_9TELE (Acanthochromis polyacanthus, spiny chromis), A0A4U5UV84_COLLU (Collichthys lucidus, big head croaker), A0A3Q3WWU6_MOLML (Mola mola, ocean sunfish), A0A2U9BSM3_SCOMX (S. maximus, turbot), A0A484DBC0_PERFV (Perca flavescens, American yellow perch), G3NY23_GASAC (Gasterosteus aculeatus, three-spined stickleback), XP_008333567.1 (nipbla-w in C. semilaevis), XP_016898436.1 (nipbla-z in C. semilaevis), A0A2I4C8B6_9TELE (Austrofundulus limnaeus, annual killifish), H2LLE4_ORYLA (Oryzias latipes, medaka), A0A3B3WE17_9TELE (Poecilia mexicana, Atlantic molly), A0A2D0SNS8_ICTPU (Ictalurus punctatus, Channel catfish), A0A1S3L2Z4_SALSA (Salmo salar, Atlantic salmon), NIPBL_MOUSE (Mus musculus, mouse), J9NZL2_CANLF (Canis lupus familiaris, Dog), M3W142_FELCA (Felis catus, cat), H2R5I6_PANTR (Pan troglodytes, chimpanzee), NIPBL_HUMAN (Homo sapiens, human), A0A060VZN4_ONCMY (Oncorhynchus mykiss, rainbow trout), A0A3Q2UAU6_CHICK (Gallus gallus, chicken), F7EE79_XENTR (Xenopus tropicalis, western clawed frog), W5KHL5_ASTMX (Astyanax mexicanus, blind cave fish), A0A287BF39_PIG (Sus scrofa, Pig), E1BKC4_BOVIN (Bos taurus, bovine), A0A0G2K0J4_RAT (Rattus norvegicus, rat), NIPB_DROME (Drosophila melanogaster, fruit fly), A0A0R4IGM2_DANRE (D. rerio, zebrafish), and A0A3B3E184_ORYME (O. melastigma, marine medaka). Black square and triangle represent nipbla-w and nipbla-z proteins, respectively.
The Expression Patterns of Two Genes in Female and Male Tissues
By using primers nipbla-F and nipbla-R, respectively, located in the exon 11 and exon 15, the alternative 5’ splicing was detected in the exon 12 of nipbla-z, which produced nipbla-z-tv1 and nipbla-z-tv2 (Supplementary Figure 1). Their sequences are also shown in Supplementary Figure 1.
The qPCR analysis revealed that nipbla-w, nipbla-z-tv1, and nipbla-z-tv2 exhibited sex-biased expression patterns in female and male individuals (Figure 3). In the heart, skin, kidney, intestine, spleen, gonad, liver, brain, and muscle, nipbla-w exhibited generally higher expression levels in female than male individuals and showed significantly highest expression level in female gonad. Concretely for different transcripts, nipbla-z-tv1 and nipbla-z-tv2 showed slight expression difference, and nipbla-z-tv1 demonstrated significantly higher expression levels in the muscle, liver, brain, and gonad of male individuals. But nipbla-z-tv2 exhibited the highest in the muscle of male individuals. The nipbla-w transcripts were gradually increased with gonadal development (Figure 4). Due to its low expression in testes, nipbla-w was not observed in the transcription level of changes during different developmental stages of testes. For the results of expression differences in stages of gonads development, both nipbla-z-tv1 and nipbla-z-tv2 showed a prominent expression in male individuals from 40 dph to 1.5 years, respectively. Furthermore, nipbla-z-tv1 reached a peak transcription level at 6 months, but nipbla-z-tv2, at 1.5 years (Figure 4).
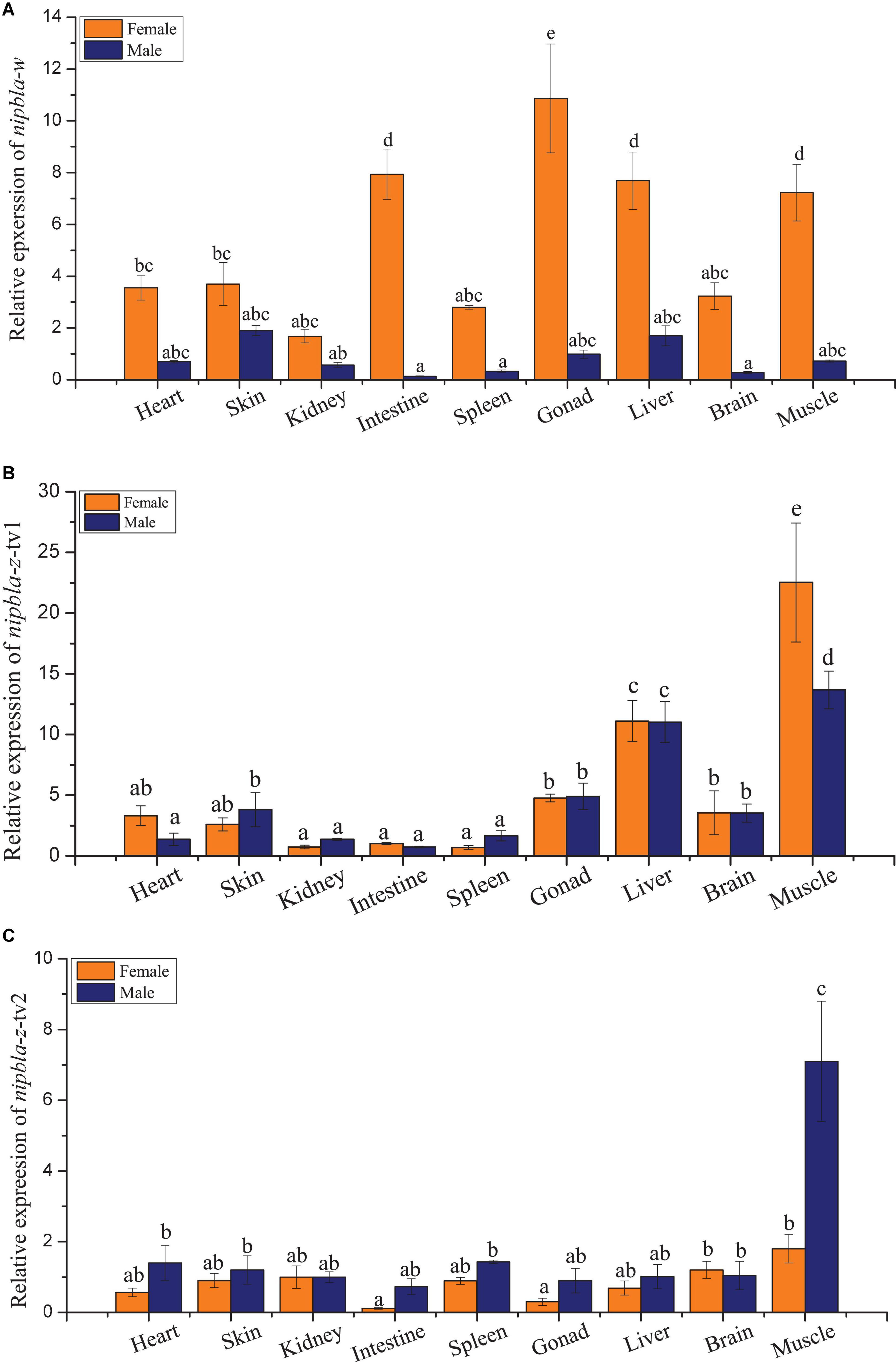
Figure 3. The relative expression patterns of nipbla-w and nipbla-z in female and male tissues. The relative repression levels of nipbla-w and nipbla-z in brain, gonad, spleen, heart, intestine, kidney, liver, muscle, skin, and spleen and of female and male individuals are shown in parts (A–C), respectively. Different letters represent significant differences.
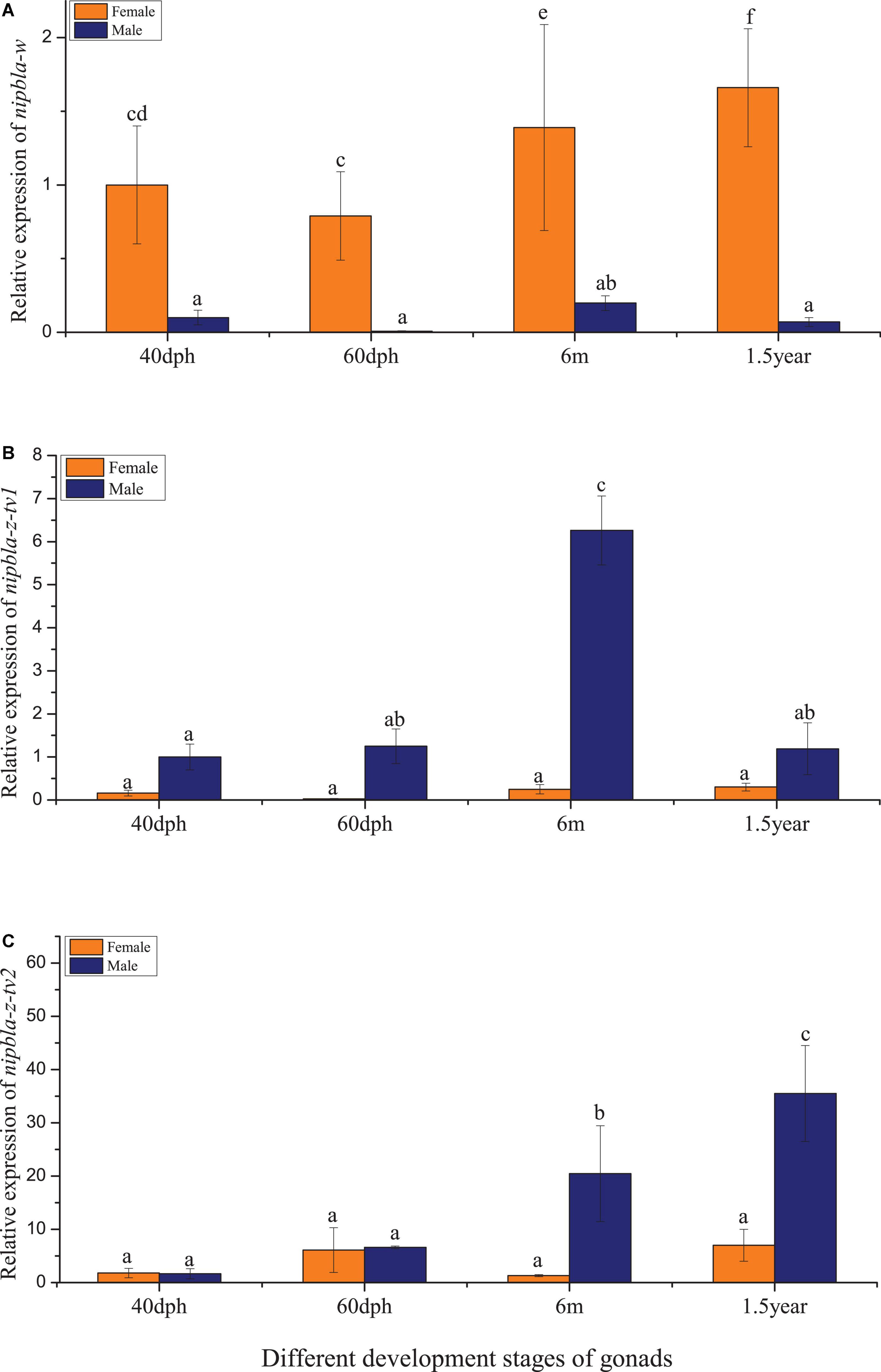
Figure 4. The relative expression patterns of nipbla-w and nipbla-z in female and male individuals in the development of gonads. (A) The nipbla-w transcription in the gonads at different developmental stages. (B) The nipbla-z-tv1 transcription in the gonads at different developmental stages. (C) The nipbla-z-tv2 transcription in the gonads at different developmental stages. Different letters represent significant differences.
The Amplification and Activity Detection of the Putative Promoters
By designing the primers based on the C. semilaevis genome, the putative 1,636 bp and 1,724 bp promoter regions were amplified from the upstream of nipbla-w and nipbla-z genes, respectively, and their GenBank numbers are MW349831 and MW349832, respectively. Their sequences shared 67.6% similarity, and their structure information were illustrated in Supplementary Figure 2. Analyzed by PROMO and JASPAR, several transcription factors including c-Jun, sex-determining region Y (SRY), POU Class 1 Homeobox 1 (POU1F1a), myogenic differentiation antigen (MyoD), signal transducer and activator of transcription 5a (STAT5A), and nuclear factor I C (Nfic) were predicted, and their binding sites were indicated in Supplementary Figure 2. The recombinant plasmids based on these putative promoters were constructed and named as pGL3-nipbla-wp and pGL3-nipbla-zp, respectively. After the transfection with pRL-TK and the detection of dual-luciferase activities (Figure 5A), the pGL3-nipbla-wp and pGL3-nipbla-zp transfected cells manifested significantly higher firefly/Renilla luciferase activity than cells transfected with pGL3-Basic and no transfection cells. This suggested that nipbla-wp and nipbla-zp could successfully drive the expression of luciferase in 293T cells. Also, the activity of nipbla-zp was significantly higher than nipbla-wp.
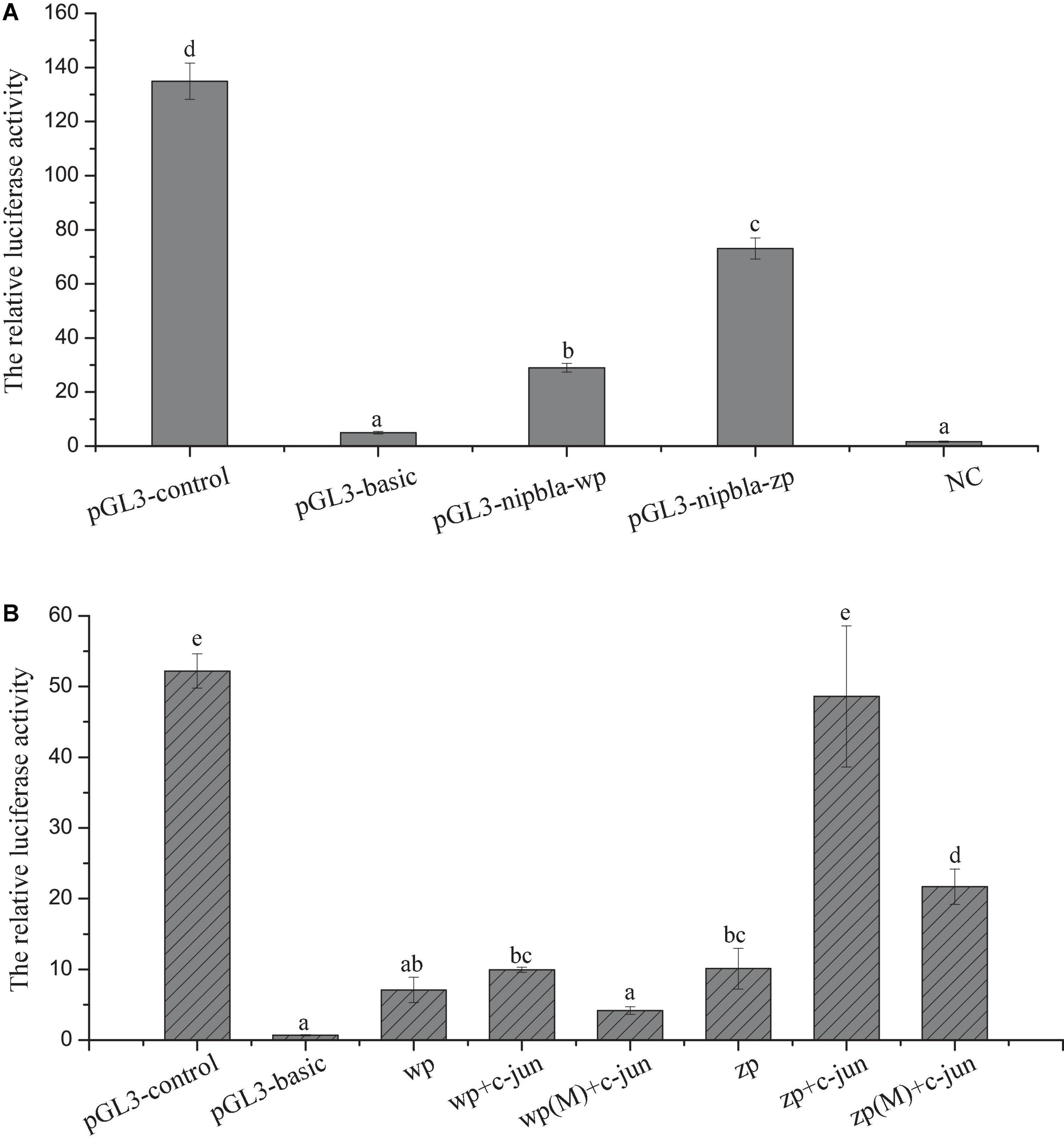
Figure 5. The analysis of nipbla-w and nipbla-z transcriptional activity and the effect of c-Jun on transcriptional activity of nipbla-w and nipbla-z. (A) The analysis of nipbla-w and nipbla-z transcriptional activity. (B) The effect of c-Jun on transcriptional activity of nipbla-w and nipbla-z promoter. Different letters represent significant differences.
To further analyze the activity of key transcription factors, the transfection in 293T cells and the dual-luciferase detection assay were observed that c-Jun could make a significant effect on the transcriptional activity of nipbla-zp, but for nipbla-wp, there was no discernible effect (Figure 5B).
The Epigenetic Modification of Two Genes and the Relationship Between DNA Methylation and Transcription Activity
The analysis of approximately 2 kb up- and downstream flanking regions of the nipbla-w and nipbla-z genomic sequence from the female ovary, male and pseudo-male testes are shown in Figure 6. Interestingly, as shown, there was a clear difference in the gene body, especially located at first exon region, which exhibited difference between female and pseudo-male individuals in nipbla-w and also exhibited difference between female, male, and pseudo-male individuals in nipbla-z.
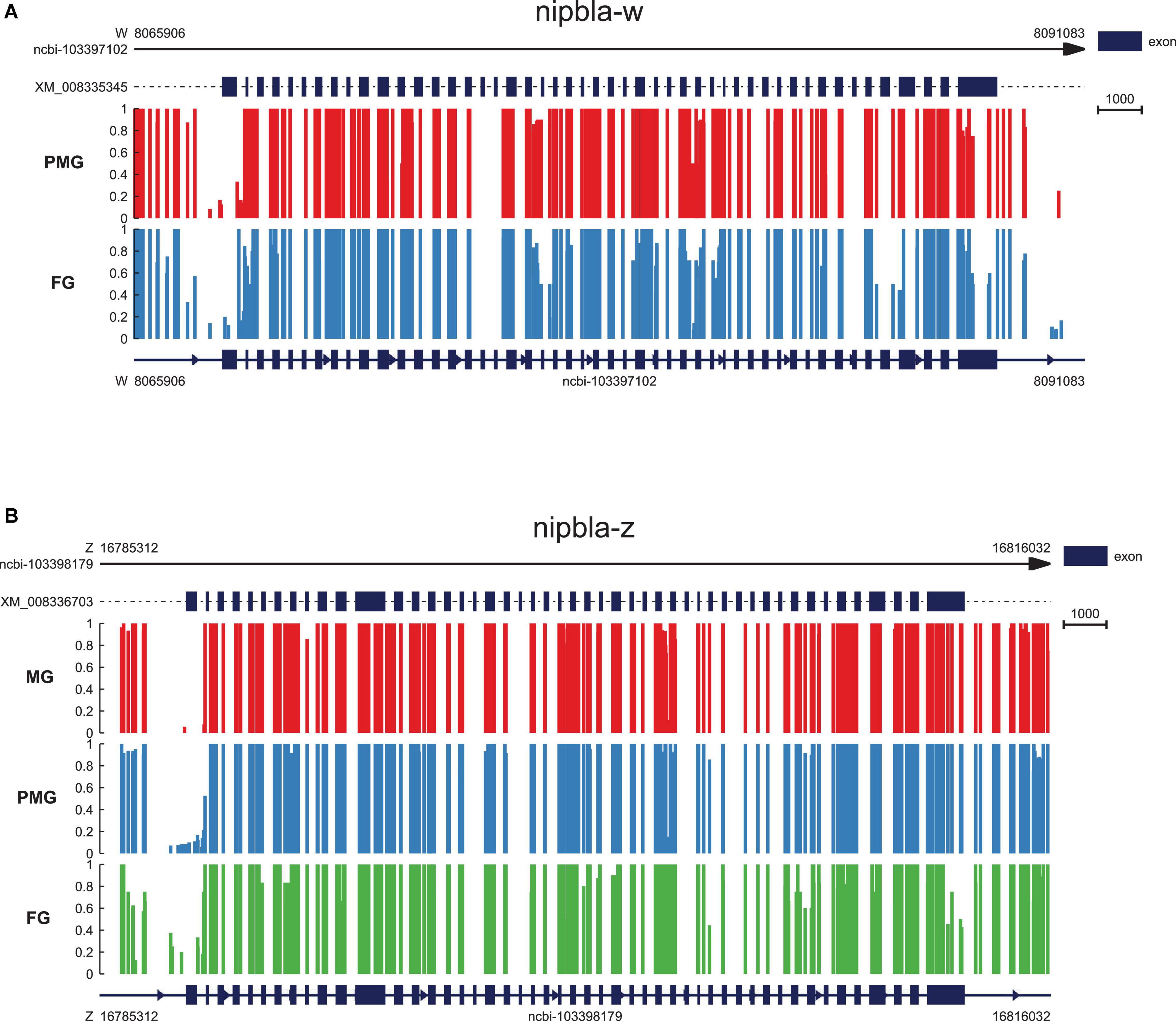
Figure 6. The epigenetic regulation of nipbla-w and nipbla-z in the gonad. (A) The nipbla-w genomic sequence with gene body, 2kb upstream and downstream regions; the methylation level of cytosine is shown with red vertical lines in pseudo-male individuals and blue in male individuals. (B) The nipbla-z genomic sequence with gene body, 2kb upstream and downstream regions; the methylation level of cytosine is shown with red vertical lines in male individuals, blue in pseudo-male individuals, and green in female individuals, respectively.
RNAi of Nipbla and in vitro Growth-Related Gene Expression
The RNAi-mediated knockdown in vitro was assisted for nipbla in the Chinese tongue sole brain cell line (i.e., CSB). The silencing effects of RNAi between nipbla-w and nipbla-z expression in CSB were detected by qPCR after siRNA was transfected into cells. The results revealed that the silencing efficiencies of nipbla-w and nipbla-z were approximately 46% and 55.1%, respectively. Compared with the control, the mRNA levels of neurogenesis, vascular development, and osteogenesis genes, i.e., bmp4, Wnt pathway gene wnt11, fat4, and spred2, were detected after RNAi. As shown in Figure 7, the expression levels of bmp4, wnt11, and spred2 showed strongly alterative after nipbla-w and nipbla-z RNAi, including the upward trend of spred2 and the downregulation expression trends of bmp4 and wnt11.
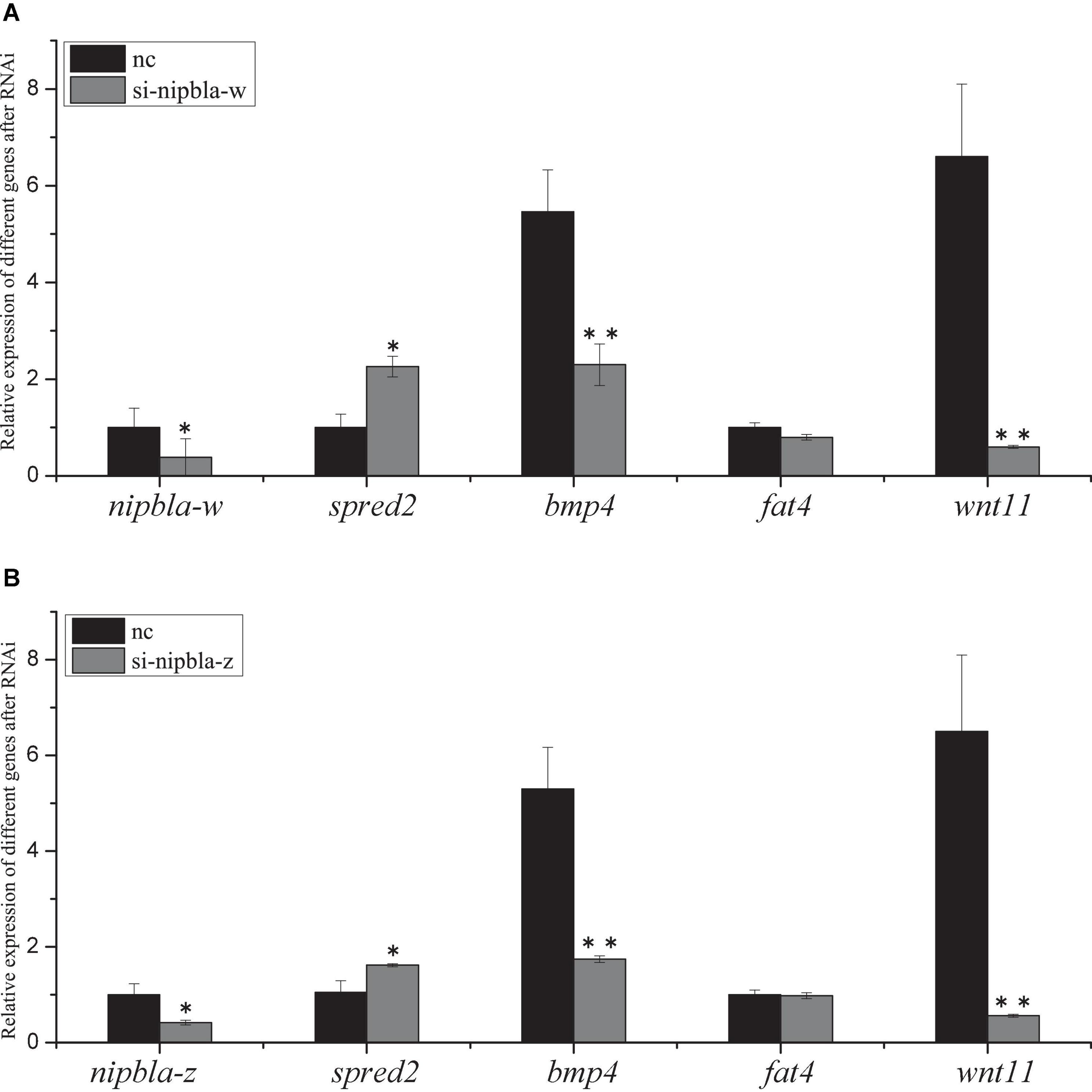
Figure 7. The siRNA effects of nipbla in C. semilaevis brain cells. The analysis of nipbla-w, nipbla-z (tv1 and tv2), Sprouty Related EVH1 Domain Containing 2 (spred2), Bone Morphogenetic Protein 4 (bmp4), FAT Atypical Cadherin 4 (fat4), and wnt11 expression in cultured testis cells after RNAi. (A) Expression of nipbla-w, spred2, bmp4, fat4, and wnt11 after transfection of si-nipbla-w RNA at 48 h. (B) Expression of nipbla-z-tv1, spred2, bmp4, fat4, and wnt11 after transfection of si-nipbla-z siRNA at 48 h. NC indicates the testis cells transfected with the siRNA of the negative control. Asterisks indicate significant differences (p < 0.05) between the treated group and the control group.
Discussion
The phenomenon of sexual size dimorphism that existed in many fish species has severely hindered the sustainable development of aquaculture. In C. semilaevis, a series of genes located in the W and Z chromosomes cause our interest due to their opposite expression pattern. The nipbl loads cohesin onto chromatin, and cohesin mediates sister chromatid cohesion important for mitosis (Dorsett, 2011). Nipbl also involved in specific enhancer-promoter communication (Strachan, 2005). Some reports remind that nipbl co-localizes with the mediator complex at promoters/enhancers of actively transcribed genes in mouse embryonic stem cells (Kagey et al., 2010). The functional range of the nipbl mutations in this gene results in Cornelia de Lange syndrome, a disorder characterized by dysmorphic facial features, growth delay, limb reduction defects, and cognitive disability in most mammals (Garcia et al., 2021). Similarly, nipbl-deficient individuals of teleost also produce observed developmental defects in heat, gut, and limb (Chien et al., 2011; Muto et al., 2014). In fish, nipbl and mediator cooperatively regulate gene (e.g., fgfs, shha, hand2, and Hox) expression to control limb development which led to size reductions and patterning defects (Muto et al., 2014). Thus, we speculated that nipbla was a typical candidate gene for the transcriptional regulation pathway involved in explaining the sexual size dimorphism in C. semilaevis. But the effect of nipbl gene on growth differences caused by sex dimorphism has not been reported in teleost.
Interestingly, two different nipbla, namely, nipbla-w, and nipbla-z, were identified in the C. semilaevis genome of the W and Z chromosomes, respectively, which were the two kinds of homologs of nipbla from our previous transcriptomic study. As shown in Figure 1, the chromosome syntenic locus analysis of nipbla on genome location in C. semilaevis, which compared O. melastigma, S. senegalensis, D. rerio, and S. maximus, obviously reveals that the syntenic arrangement of nipbla in chromosome Z of C. semilaevis was more identical to other teleost species of nipbla. Obviously, the distribution of pseudogenes or non-coding RNA (ncRNA) in chromosome W leads to the inconsistent gene syntenic arrangement, which further confirms the higher gene degeneration in chromosome W than in chromosome Z over a period of evolutionary times (Moghadam et al., 2012; Chen et al., 2014). Interestingly, these relatively conservative linear genes have shown a similar function to nipbla and have given concrete examples about it: slc1a3a and gdnf participate in the regulation of neuronal differentiation and development in teleost (Feng, 2009; Wong et al., 2021). Since nipbl is required for embryonic growth in both fish and mice (Kawauchi et al., 2009; Muto et al., 2011), even more, nipbl is one of the important regulatory factors in the maintenance of 3D genome organization and genome-controlled gene expression through interacts with cohesion (Gao et al., 2019). Interestingly, the alternative splicing sites in exon 12 of nipbla-z in tongue sole attribute to alternative 5’ splicing, which produced two transcript genes nipbla-z-tv1 and nipbla-z-tv2. Similar events related to the sex-specific alternative splicing, which is also discovered in sex differential genes, e.g., dmrt1 and cyp19a1 in Lates calcarifer and factor in the germ line alpha (figla) in C. semilaevis (Shao et al., 2014; Domingos et al., 2018). Regarding growth traits, there is also myostatin (mstn) gene concerned with the alternative splicing phenomenon (Huang et al., 2011). Also, our RNA-seq analysis found that more phenomena of alternative splicing were observed in the male C. semilaevis (unpublished data). Undoubtedly, nipbla as potential gametologs in the W and Z chromosomes, were involved in the growth of sexual dimorphism in the Chinese tongue sole.
The expression analysis revealed that nipbla-w and nipbla-z showed significant expression differences of most tissues between female and male individuals, where nipbla-w occupied a dominant in female individuals, compared with nipbla-z in male individuals. Particularly, even nipbla-z showed superiority in male individuals, especially at different developmental stages, the two transcripts of nipbla-z (i.e., nipbla-z-tv1 and nipbla-z-tv2) showed a consistent expression trend. While, in the 3-year matured male individuals, they revealed the differential expression with emphasis, e.g., nipbla-z-tv1 observed in the muscle, liver, gonad, and brain, but nipbla-z-tv2 preferentially existed in the muscle. It reveals that nipbla-w and nipbla-z play a role in the growth of female and male gonads, respectively, but different transcripts of nipbla-z (nipbla-z-tv1 and nipbla-z-tv2) have expression bias in the muscle, even related to gonad and liver associated with nipbla-z-tv2.
After the in vitro knockdown of nipbla-w and nipbla-z in the cultured brain cells, qPCR showed the mRNA levels of the growth-related genes, e.g., bmp4, wnt11, and spred2, including the upward trend of spred2 and the downregulation expression trends of bmp4 and wnt11. Meanwhile, spred2, which inhibits FGF, probably influences FGF-mediated phosphorylation, and wnt11 all depends on the Wnt signaling pathway (Katoh and Katoh, 2006). Even more, spred2 plays a critical role in cell proliferation and differentiation of the zebrafish brain (Lim et al., 2017). The bmp4 is also influenced after nipbla RNAi; although the mechanisms remain elusive, interestingly, it is reported as a key element to activate the Wnt pathway to promote primordial germ cell formation in chicken (Zuo et al., 2021). Even more, the knockdown of nipbl expression led to size reductions and patterning defects that were observed by dysregulated expression of key early limb development genes, including fgfs, shha, hand2, and multiple Hox genes (Muto et al., 2014). Therefore, we speculated that nipbla may be involved in the growth of sexual dimorphism through the Wnt signaling pathway. Comparatively, nipblb loss-of-function affects DNA replication, ribosome biogenesis, cell cycle, and results involved in pathways related to an overall activation of the Wnt canonical pathway in the early developmental stages of zebrafish embryos (Spreafico et al., 2020). Otherwise, in addition to the Wnt pathway, nipbl can take function through other pathways, and concretely, it influenced the expression of the gene involved in endodermal differentiation (sox32, sox17, foxa2, and gata5) and left-right patterning (spaw, lefty2, and dnah9), which is altered in early embryonic development (Muto et al., 2011). It suggested that nipbl may fulfill functions that are mainly involved in the Wnt signaling pathway and play a role in gene regulation in fish development, while the mechanisms about it need further identification analysis.
The promoter transcription activities of two nipbla genes in C. semilaevis were both successfully verified by the dual-luciferase reporter system in HEK 293T cells. The results reveal that the activity of nipbla-zp was much stronger than that of nipbla-wp. The prediction of several transcription factors including SRY, POU1F1a, MyoD, c-Jun, STAT5A, and Nfic from the promoters of nipbla has provided insight into the possible transcription regulation of nipbla. The transcription factors mentioned above are known to bind with growth-related genes to positively regulate gene expression and accelerate growth rate in mammals and teleosts (Li et al., 1990; Yu et al., 2001; Zhu et al., 2019). Further, we focused on c-Jun because it is reported to involve in cell proliferation and differentiation at the cellular level (Hilberg et al., 1993), and it is also proved as a crucial effector to induce normal growth (Castellazzi et al., 1991). There is a common sense among mammals and fish that c-Jun requires its DNA-binding domain and interacts with estrogen receptor α (ERα) and pituitary homeobox-1 (Pitx1) and then mediates GnRH to effect on early growth (Melamed et al., 2006). In C. semilaevis, c-Jun transcripts are both downregulated in the liver and gonad based on the transcriptomic analysis (Wang et al., 2018), and it effectively affected the transcriptional activity of the promoter of nipbla-z in this study. According to the abovementioned analysis, it is not difficult to infer that nipbla make potential role in growth process regulated by the putative transcription factors, especially c-Jun still needs more evidence to exact.
Furthermore, our study revealed that nipbla-w and nipbla-z exhibited a difference in methylation level in gene body associated with three sexual genotypes in the gonad. There is proof that the alternative splicing of mRNA is linked to the DNA methylation regulation (Shukla et al., 2011). Whether this epigenetic modification would have an effect on the different types of alternative splicing needed to be further investigated. The genome-wide DNA methylation of the liver reveals that nipbl promoter and gene body could be the important regulators for the long-term effects of EE2 in the liver of adult fish (Voisin et al., 2021). This provides that the expression of nipbla may be influenced by methylation patterns.
In summary, two nipbla gametolog genes were separately identified from C. semilaevis chromosomes W and Z. These two genes were found to demonstrate the sex-biased expression pattern in female and male tissues or even involved in different developmental stages of the gonads. The identification, activity verification, and prediction of transcription factors for nipbla promoters provide clues for the transcription regulation of nipbla in C. semilaevis. The identification of two nipbla genes provided important data for interpreting the sexual size dimorphism in C. semilaevis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences.
Author Contributions
NW conceived and designed the experiments, and carried out the promoter activity experiment. YZ, QY, and RS performed fish tissue sampling. YZ, QY, RS, and MZ carried out RNAi interference and the qPCR experiment. YZ and NW analyzed the data and wrote the manuscript. SC provided valuable suggestions to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31873037 and 31730099), National Key R&D Program of China (2018YFD0900205), Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD20), and Taishan Scholar Project of Shandong Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.833070/full#supplementary-material
Footnotes
- ^ www.ebi.ac.uk/gwas
- ^ https://www.evolgenius.info/evolview/#login
- ^ http://www.omicshare.com/tools
- ^ http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3
- ^ http://jaspar.genereg.net/
References
Castellazzi, M., Spyrou, G., La Vista, N., Dangy, J.-P., Piu, F., Yaniv, M., et al. (1991). Overexpression of c-jun, junB, or junD affects cell growth differently. Proc. Natl. Acad. Sci. U.S.A. 88, 8890–8894.
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260. doi: 10.1038/ng.2890
Chien, R., Zeng, W., Kawauchi, S., Bender, M., Santos, R., Gregson, H. C., et al. (2011). Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 286, 17870–17878. doi: 10.1074/jbc.M110.207365
Domingos, J. A., Budd, A. M., Banh, Q. Q., Goldsbury, J. A., Zenger, K. R., and Jerry, D. R. (2018). Sex-specific dmrt1 and cyp19a1 methylation and alternative splicing in gonads of the protandrous hermaphrodite barramundi. PLoS One 13:e0204182. doi: 10.1371/journal.pone.0204182
Dorsett, D. (2011). Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 21, 199–206. doi: 10.1016/j.gde.2011.01.018
Feng, L. (2009). Regulation of Retinoic Acid in Early Zebrafish Development. Washington, DC: University of Washington.
Foellmer, M. W., and Moya-Larano, J. (2007). “Sexual size dimorphism in spiders: patterns and processes,” in Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism, eds D. J. Fairbairn, W. U. Blanckenhorn, and T. Székely (Oxford: Oxford University Press), 71–81. doi: 10.1093/acprof:oso/9780199208784.003.0008
Gao, D., Zhu, B., Cao, X., Zhang, M., and Wang, X. (2019). “Roles of NIPBL in maintenance of genome stability. Semin. Cell Dev. Biol. 90, 181–186. doi: 10.1016/j.semcdb.2018.08.005
Garcia, P., Fernandez-Hernandez, R., Cuadrado, A., Coca, I., Gomez, A., Maqueda, M., et al. (2021). Disruption of NIPBL/Scc2 in Cornelia de Lange Syndrome provokes cohesin genome-wide redistribution with an impact in the transcriptome. Nat. Commun. 12, 1–15. doi: 10.1038/s41467-021-24808-z
Hilberg, F., Aguzzi, A., Howells, N., and Wagner, E. F. (1993). c-Jun is essential for normal mouse development and hepatogenesis. Nature 365, 179–181. doi: 10.1038/365179a0
Huang, K. L., Wang, J. W., Han, C. C., Liu, H. H., Li, L., Dai, F., et al. (2011). Developmental expression and alternative splicing of the duck myostatin gene. Comp. Biochem. Physiol. Part D Genomics Proteomics 6, 238–243. doi: 10.1016/j.cbd.2011.04.002
Kagey, M. H., Newman, J. J., Bilodeau, S., Zhan, Y., Orlando, D. A., Van Berkum, N. L., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. doi: 10.1038/nature09380
Katoh, Y., and Katoh, M. (2006). FGF signaling inhibitor, SPRY4, is evolutionarily conserved target of WNT signaling pathway in progenitor cells. Int. J. Mol. Med. 17, 529–532.
Kawauchi, S., Calof, A. L., Santos, R., Lopez-Burks, M. E., Young, C. M., Hoang, M. P., et al. (2009). Multiple organ system defects and transcriptional dysregulation in the Nipbl+/- mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 5:e1000650. doi: 10.1371/journal.pgen.1000650
Krantz, I. D., Mccallum, J., Descipio, C., Kaur, M., Gillis, L. A., Yaeger, D., et al. (2004). Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36, 631–635. doi: 10.1038/ng1364
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, S., Crenshaw, E. B. 3rd., Rawson, E. J., Simmons, D. M., Swanson, L. W., and Rosenfeld, M. G. (1990). Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature 347, 528–533. doi: 10.1038/347528a0
Lim, F. T., Ogawa, S., Smith, A. I., and Parhar, I. S. (2017). Proteomics identification of potential candidates involved in cell proliferation for early stage of brain regeneration in the adult zebrafish. Zebrafish 14, 10–22. doi: 10.1089/zeb.2016.1319
Lindenfors, P., Gittleman, J. L., and Jones, K. E. (2007). “Sexual size dimorphism in mammals,” in Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism, eds D. J. Fairbairn, W. U. Blanckenhorn, and T. Székely (Oxford: Oxford University Press), 16–26.
Lindgren, E., Hägg, S., Giordano, F., Björkegren, J., and Ström, L. (2014). Inactivation of the budding yeast cohesin loader Scc2 alters gene expression both globally and in response to a single DNA double strand break. Cell Cycle 13, 3645–3658. doi: 10.4161/15384101.2014.964108
Liu, J., Zhang, Z., Bando, M., Itoh, T., Deardorff, M. A., Clark, D., et al. (2009). Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7:e1000119. doi: 10.1371/journal.pbio.1000119
Liu, Y., Chen, S., Gao, F., Meng, L., Hu, Q., Song, W., et al. (2014). SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semilaevis). J. Agric. Biotechnol. 22, 787–792.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
MacArthur, J., Bowler, E., Cerezo, M., Gil, L., Hall, P., Hastings, E., et al. (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901. doi: 10.1093/nar/gkw1133
Mank, J. E., Hultin-Rosenberg, L., Axelsson, E., and Ellegren, H. (2007). Rapid evolution of female-biased, but not male-biased, genes expressed in the avian brain. Mol. Biol. Evol. 24, 2698–2706. doi: 10.1093/molbev/msm208
Mank, J. E., Nam, K., Brunström, B., and Ellegren, H. (2010). Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol. Biol. Evol. 27, 1570–1578. doi: 10.1093/molbev/msq042
Mannini, L., Cucco, F., Quarantotti, V., Krantz, I. D., and Musio, A. (2013). Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 34, 1589–1596. doi: 10.1002/humu.22430
Mei, J., and Gui, J. F. (2015). Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 58, 124–136.
Melamed, P., Zhu, Y., Tan, S. H., Xie, M., and Koh, M. (2006). Gonadotropin-releasing hormone activation of C-jun, but not early growth response factor-1, stimulates transcription of a luteinizing hormone β-subunit gene. Endocrinology 147, 3598–3605.
Moghadam, H. K., Pointer, M. A., Wright, A. E., Berlin, S., and Mank, J. E. (2012). W chromosome expression responds to female-specific selection. Proc. Natl. Acad. Sci. U.S.A. 109, 8207–8211. doi: 10.1073/pnas.1202721109
Muto, A., Calof, A. L., Lander, A. D., and Schilling, T. F. (2011). Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. 9:e1001181. doi: 10.1371/journal.pbio.1001181
Muto, A., Ikeda, S., Lopez-Burks, M. E., Kikuchi, Y., Calof, A. L., Lander, A. D., et al. (2014). Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet 10:e1004671. doi: 10.1371/journal.pgen.1004671
Pistocchi, A., Fazio, G., Cereda, A., Ferrari, L., Bettini, L., Messina, G., et al. (2013). Cornelia de Lange Syndrome: nipbl haploinsufficiency downregulates canonical Wnt pathway in zebrafish embryos and patients fibroblasts. Cell Death Dis. 4, e866–e866. doi: 10.1038/cddis.2013.371
Pointer, M. A., Harrison, P. W., Wright, A. E., and Mank, J. E. (2013). Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genet. 9:e1003697. doi: 10.1371/journal.pgen.1003697
Ranz, J. M., Castillo-Davis, C. I., Meiklejohn, C. D., and Hartl, D. L. (2003). Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742–1745. doi: 10.1126/science.1085881
Rhodes, J., Mazza, D., Nasmyth, K., and Uphoff, S. (2017). Scc2/Nipbl hops between chromosomal cohesin rings after loading. Elife 6:e30000. doi: 10.7554/eLife.30000
Shao, C., Li, Q., Chen, S., Zhang, P., Lian, J., Hu, Q., et al. (2014). Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604–615. doi: 10.1101/gr.162172.113
Shukla, S., Kavak, E., Gregory, M., Imashimizu, M., Shutinoski, B., Kashlev, M., et al. (2011). CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79. doi: 10.1038/nature10442
Spreafico, M., Mangano, E., Mazzola, M., Consolandi, C., Bordoni, R., Battaglia, C., et al. (2020). The Genome-Wide impact of Nipblb loss-of-function on Zebrafish gene expression. Int. J. Mol. Sci. 21:9719. doi: 10.3390/ijms21249719
Strachan, T. (2005). Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr. Opin. Genet. Dev. 15, 258–264. doi: 10.1016/j.gde.2005.04.005
Székely, T., Lislevand, T., and Figuerola, J. (2007). “Sexual size dimorphism in birds,” in Sex, Size and Gender roles: Evolutionary Studies of Sexual Size Dimorphism, eds D. J. Fairbairn, W. U. Blanckenhorn, and T. Székely (Oxford: Oxford University Press), 27–37.
Voisin, A.-S., Suarez Ulloa, V., Stockwell, P., Chatterjee, A., and Silvestre, F. (2021). Genome-wide DNA methylation of the liver reveals delayed effects of early-life exposure to 17-α-ethinylestradiol in the self-fertilizing mangrove rivulus. Epigenetics [Online ahead of print] 1–25. doi: 10.1080/15592294.2021.1921337
Wang, N., Wang, R., Wang, R., and Chen, S. (2018). Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integrat. Genomics 18, 327–339. doi: 10.1007/s10142-018-0595-y
Wang, N., Yang, Q., Wang, J., Shi, R., Li, M., Gao, J., et al. (2021). Integration of transcriptome and methylome highlights the roles of cell cycle and hippo signaling pathway in flatfish sexual size dimorphism. Front. Cell Dev. Biol. 9:743722. doi: 10.3389/fcell.2021.743722
Wang, T. Z., Sun, A., Wang, N., Cui, Z. K., Chen, S. L., and Sha, Z. X. (2015). Establishment and characterization of an astroglial cell line derived from the brain of half-smooth tongue sole (Cynoglossus semilaevis). Dongwuxue Yanjiu 36, 305–310. doi: 10.13918/j.issn.2095-8137.2015.5.305
Wong, C. E. D., Hua, K., Monis, S., Saxena, V., Norazit, A., Noor, S. M., et al. (2021). gdnf affects early diencephalic dopaminergic neuron development through regulation of differentiation-associated transcription factors in zebrafish. J. Neurochem. 156, 481–498. doi: 10.1111/jnc.15108
Yang, X., Schadt, E. E., Wang, S., Wang, H., Arnold, A. P., Ingram-Drake, L., et al. (2006). Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16, 995–1004. doi: 10.1101/gr.5217506
Yu, T. P., Sun, H. S., Wahls, S., Sanchez-Serrano, I., Rothschild, M. F., and Tuggle, C. K. (2001). Cloning of the full length pig PIT1 (POU1F1) CDNA and a novel alternative PIT1 transcript, and functional studies of their encoded proteins. Anim. Biotechnol. 12, 1–19. doi: 10.1081/ABIO-100102975
Zakany, J., and Duboule, D. (2007). The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17, 359–366. doi: 10.1016/j.gde.2007.05.011
Zhu, H., Zhang, Y., Bai, Y., Yang, H., Yan, H., Liu, J., et al. (2019). Relationship between SNPs of POU1F1 gene and litter size and growth traits in shaanbei white cashmere goats. Animals (Basel) 9:114. doi: 10.3390/ani9030114
Keywords: nipbla, sex-biased expression, Chinese tongue sole (Cynoglossus semilaevis), sexual size dimorphism, gametolog
Citation: Zhu Y, Shi R, Yang Q, Zhang M, Chen S and Wang N (2022) Two Nipped-B-Like Protein A (Nipbla) Gametologs in Chinese Tongue Sole (Cynoglossus semilaevis): The Identification of Alternative Splicing, Expression Pattern, and Promoter Activity Analysis. Front. Mar. Sci. 9:833070. doi: 10.3389/fmars.2022.833070
Received: 10 December 2021; Accepted: 31 January 2022;
Published: 18 March 2022.
Edited by:
Alexander Chong Shu-Chien, Universiti Sains Malaysia (USM), MalaysiaReviewed by:
Alejandro S. Mechaly, CONICET Instituto de Investigaciones en Biodiversidad y Biotecnología (INBIOTEC), ArgentinaZhenkui Qin, Ocean University of China, China
Copyright © 2022 Zhu, Shi, Yang, Zhang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Na Wang, d2FuZ25hQHlzZnJpLmFjLmNu
 Ying Zhu
Ying Zhu Rui Shi
Rui Shi Qian Yang
Qian Yang Mengqian Zhang
Mengqian Zhang Songlin Chen
Songlin Chen Na Wang
Na Wang