- 1Department of Marine Biology and Environmental Science, Faculty of Agriculture, University of Miyazaki, Miyazaki, Japan
- 2Center for Earth Surface System Dynamics, Atmosphere and Ocean Research Institute, University of Tokyo, Kashiwa, Japan
- 3Australia Institute of Marine Science, Townsville, QLD, Australia
- 4Aquaculture Research, Extension, Training & Technology Development, College of Micronesia Land Grant Program (NIFA, USDA), Palikir, Micronesia
- 5ENTROPIE UMR 9220, IRD, Université de La Réunion, CNRS, IFREMER, Université de la Nouvelle-Calédonie, Perpignan, France
- 6CRIOBE - EPHE-UPVD-CNRS & Laboratoire d’Excellence “CORAIL”, PSL Université Paris, Perpignan, France
- 7Marine Science Institute CS, University of the Philippines, Diliman, Quezon City, Philippines
- 8 President Office, University of Ryukyus, Nishihara, Japan
- 9Cooperative Research & Extension Department, Career & Technical Education Center, College of Micronesia-FSM, Palikir, Micronesia
- 10Marine Science Institute, University of the Philippines, Diliman, Quezon City, Philippines
- 11Coral Conservation Genetics & Restoration, Hawai’i Institute of Marine Biology, University of Hawai’i, Mānoa, HI, United States
- 12DNA Sequencing Section, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
- 13Marine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
Recurring outbreaks of crown-of-thorns starfish (COTS) severely damage healthy corals, especially in the Western Pacific Ocean. To obtain a better understanding of population genetics of COTS and historical colonization across the Pacific Ocean, complete mitochondrial genomes were sequenced from 243 individuals collected in 11 reef regions. Our results indicate that Pacific COTS (Acanthaster cf. solaris) comprise two major clades, an East-Central Pacific (ECP) clade and a Pan-Pacific (PP) clade, separation of which was supported by high bootstrap value. The ECP clade consists of COTS from French Polynesia, Fiji, Vanuatu and the Great Barrier Reef (GBR). The Hawaii population is unique within this clade, while California COTS are included in EPC clade. On the other hand, the PP clade comprises multiple lineages that contain COTS from Vietnam, the Philippines, Japan, Papua New Guinea, Micronesia, the Marshall Islands, GBR, Vanuatu, Fiji and French Polynesia. For example, a lineage of the PP clade, which has the largest geographic distribution, includes COTS from all of these locations. These results suggest two alternative histories of current geographic distributions of COTS in the Pacific Ocean, an ECP clade ancestry or Western Pacific clade ancestry. Although further questions remain to be explored, this discovery provides an evolutionary context for the interpretation of COTS population structure which will aid future coral reef research in the Pacific Ocean, and ultimately improve reef management of COTS.
Introduction
Coral reefs are the most biodiverse marine ecosystems, and because they nurture edible marine species, furnish biochemicals and novel pharmaceutical leads, provide coastal protection and employment, and contribute to regional cultures, invested marine managers, communities and governments are calling for their preservation (De’ath et al., 2012; Selmoni et al., 2020). However, many coral reefs are currently experiencing severe, cumulative disturbances, including coral bleaching (Hughes et al., 2017), cyclones/typhoons (Harmelin-Vivien, 1994), and massive outbreaks of crown-of-thorns starfish (COTS) (Birkeland and Lucas, 1990; Yasuda et al., 2009; Timmers et al., 2012; Hughes et al., 2014; Yasuda, 2018).
COTS are considered the major and most destructive predators of reef-building corals in the Indo-Pacific (Birkeland, 1990). In this study, we use the following terminology to describe COTS: Acanthaster planci, sensu lato to designate COTS that exist in both the Indian and Pacific Oceans, A. planci to refer to those in the Indian Ocean and A. cf. solaris to indicate COTS in the Pacific Ocean (Haszprunar and Spies, 2014). Although COTS are highly fecund (Birkeland and Lucas, 1990), under normal, undisturbed conditions COTS populations remain relatively constant and their impacts on coral communities negligible (Fabricius et al., 2010) (Figure 1A). However, in the past several decades anthropogenic activities have adversely affected the marine environment, two major impacts being an increased discharge of nutrients (Fabricius et al., 2010) and climate change (Uthicke et al., 2013). These factors have been linked to increased COTS pelagic larval duration (PLD) (Yamaguchi, 1973; Lucas, 1982), and this relatively long PLD, up to several weeks, is strongly implicated in enlargement of the original population and an expansion into new habitats with comparatively homogeneous populations across widespread localities (Birkeland and Lucas, 1990; Vogler et al., 2013). In association with strong ocean currents, the lengthy PLD is also hypothesized to cause successive secondary population outbreaks of COTS (Figure 1B), especially in the Great Barrier Reef (GBR) of Australia and in the Ryukyu Archipelago (RA) of Japan (Birkeland and Lucas, 1990; Benzie and Stoddart, 1992; Kenchington, 1997; Yasuda et al., 2009; Pratchett et al., 2017). These outbreaks can cause substantial loss of coral cover, thereby diminishing the integrity and resilience of reef ecosystems (Timmers et al., 2012; Hughes et al., 2014). In the GBR, 42% of coral reef damage is attributed to COTS predation (De’ath et al., 2012). Similarly, in the RA of Japan, at least two waves of chronic and successive outbreaks spanning 60 years have decimated corals (Yasuda, 2018). Since 2000, over 980,000 COTS have been removed from reefs of Amami Island and the Ryukyus (Nakamura et al., 2014; Yasuda, 2018) (website http://www.churaumi.net/onihitode/onihitode1.html), and from 2011 well over 300,000 COTS have been culled in the GBR (website http://www.environment.gov.au/marine/gbr/case-studies/crown-of-thorns) with further expansion of this program in 2018 removing an additional ~300,000 (https://www.gbrmpa.gov.au/our-work/our-programs-and-projects/crown-of-thorns-starfish-management/crown-of-thorns-starfish-project-dashboard). These programs highlight the protracted nature and high cost of maintaining healthy coral reefs. COTS larval dispersal, therefore, has intensively been studied in the Pacific Ocean (Benzie, 1992; Yasuda et al., 2009; Yasuda et al., 2014; Harrison et al., 2017; Pratchett et al., 2017).
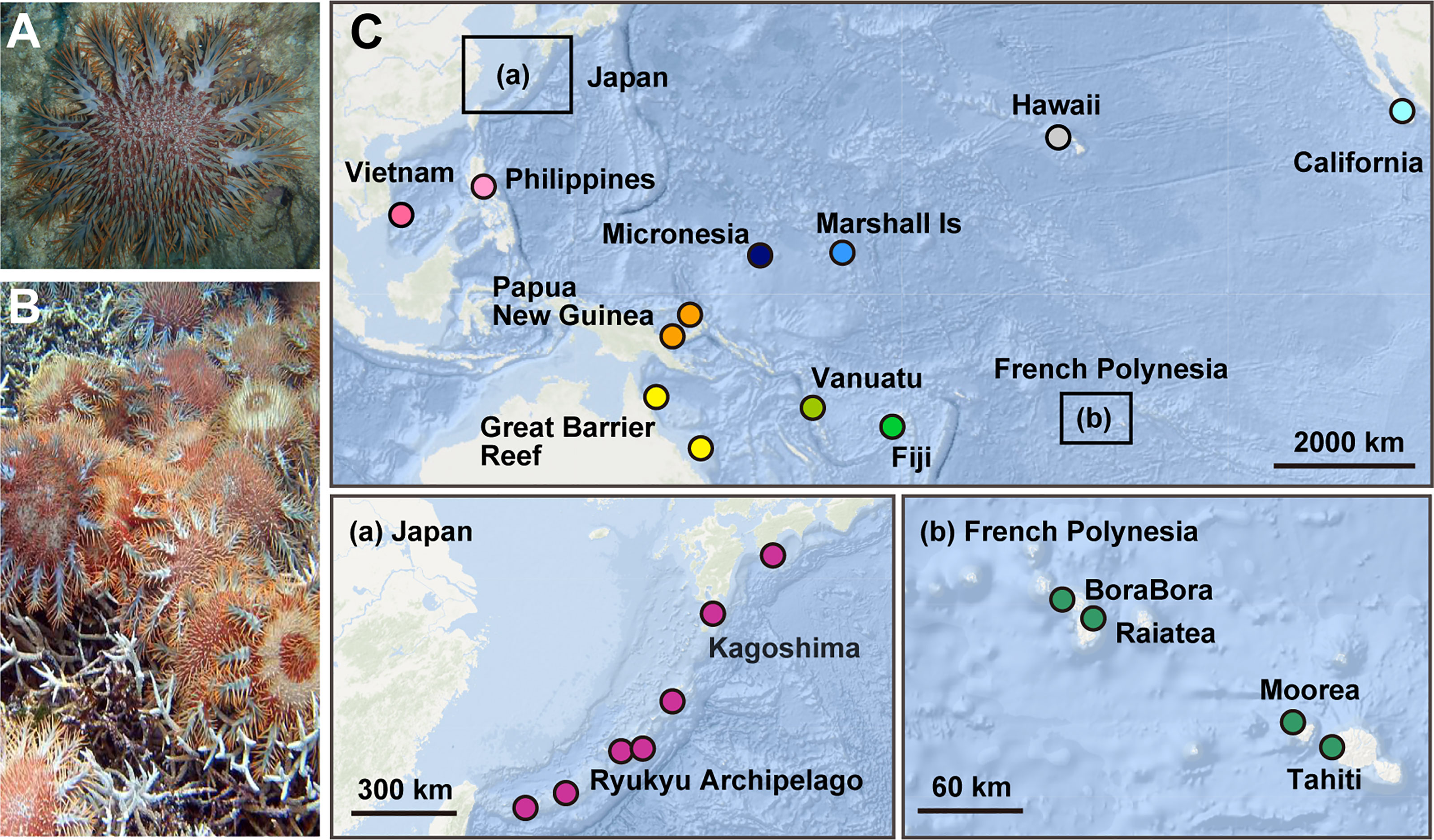
Figure 1 (A) An adult of crown-of-thorns starfish (COTS) on reef-building corals. (B) An outbreak of COTS covering and eating scleractinian corals, causing severe damage to the reef. (C) Collection sites of COTS in the Pacific Ocean. 243 COTS were collected at 23 locations in 14 countries, representing 11 reef regions: French Polynesia (Bora Bora, Moorea, Raiatea and Tahiti), Fiji, Vanuatu, Great Barrier Reef of Australia (GBR; Clack and Shell Reefs), Papua New Guinea (PNG), the Philippines, Vietnam, Japan, Micronesia, the Marshall Islands, and USA (Hawaii and California). Locations in Japan and French Polynesia are enlarged in (a, b).
Mitochondrial DNA (mtDNA) strictly inherits maternally, lacks recombination, and has a low effective population size (1/4th of nuclear genes) with a high mutation rate, and thus the comparison of mtDNA sequences has strong advantages in phylogeographic analysis (Avise, 2000). Based on partial mitochondrial gene [cytochrome oxidase subunit I (COI)] sequences, Vogler et al. (2008) reported the presence of four geographically distributed COTS lineages in the Indo-Pacific, two in the Indian Ocean (Northern and Southern), one in the Red Sea, and one in the Pacific Ocean. In addition, studies using either genes of mitochondrial COI, II and III or nuclear microsatellite locus heterozygosity, or both, have demonstrated a genetically homogenous pattern of A. cf. solaris in the Western Pacific (Vogler et al., 2013; Tusso et al., 2016), as well as in regions associated with western boundary currents (Yasuda et al., 2009), the Hawaiian Islands (Timmers et al., 2011), French Polynesia (Yasuda et al., 2015), and the GBR (Harrison et al., 2017). However, no reports explored population structures of COTS in the entire Pacific Ocean.
Our previous study determined the complete mtDNA sequences of 10 individuals of the three starfish, Asterias amurensis (16,421 bp), Linckia laevigata (16,229 bp) and Acanthaster cf. solaris (16,235 bp) each to infer their population genetics, and we showed this method is more informative than partial sequence comparison for starfish population genetics (Inoue et al., 2020). Here, we sequenced the complete mtDNA of 243 COTS specimens collected from 11 representative localities of the Pacific and conducted molecular phylogenetic analyses to reconstruct detailed genetic geography of Pacific COTS. With this information we considered the question: Are COTS populations in the Pacific genetically homogenous or do they exist as separated lineages as in the case of A. planci in the Indian Ocean? Such information might provide clues essential for future management of massive outbreaks of COTS in the Pacific Ocean.
Materials and Methods
Acanthaster cf. solaris
A total of 243 adult COTS (Acanthaster cf. solaris) were collected between 2006 and 2018 at reefs in the Pacific Ocean (Figure 1C). Their average size was 20 cm in diameter. Fifty-three specimens were collected in French Polynesia, including 13 specimens from Bora-Bora, 16 from Moorea, 9 from Raiatea, and 15 from Tahiti (Supplementary Table 1). Ten specimens were collected from Fiji, 31 from Vanuatu, and 20 from the GBR (10 each from Clack and Shell Reefs). We collected 9 specimens from Papua New Guinea, 4 from the Philippines, 10 from Vietnam, 48 from the Ryukyu Archipelago of Japan and 29 from the Kagoshima Islands of Japan (Supplementary Table 1). A further 9 specimens from Micronesia, 8 from the Marshall Islands, and 12 from the USA (9 from Hawaii and 3 from California) were also collected. Collection sites and sample numbers are reported in Supplementary Table 1.
DNA Sequencing and Assembly of Mitochondria Genomes
Tube feet of adult COTS were dissected with scissors and fixed in 99.5% ethanol. Specimens were kept at 4°C until use for DNA sequencing. Nuclear and mtDNA were extracted using the automated Nextractor® NX-48S system. Extraction was performed following the manufacturer’s protocol using an NX-48 Tissue DNA kit (Genolution Inc., Seoul, Korea). Tube foot tissue was incubated in lysis buffer overnight and extracted DNA was purified with Agencourt AMPure XP magnetic beads immediately before library preparation. DNA concentration was determined with Qubit dsDNA broad range (Thermo Scientific Inc., USA), and the quality of high molecular-weight DNA was checked using an Agilent 4150 TapeStation (Agilent, USA). PCR-free shotgun libraries were constructed using NEBNext® Ultra™ II FS DNA Library Prep Kits for Illumina (New England BioLabs Inc, UK), following the manufacturer’s protocols. Sequencing was performed using an Illumina NovaSeq 6000 sequencer (Illumina Inc., USA).
Approximately 10X coverage of nuclear genome DNA sequences were obtained. Paired-end reads were assembled using NOVOPlasty 2.6.3 (Dierckxsens et al., 2017) with the default settings. Sequencing errors were checked by comparing published sequences of A. planci (A. cf. solaris) (accession number, NC_007788.1) and A. brevispinus (NC_007789.1) (Yasuda et al., 2006). Many of the longer scaffolds analyzed using MitoAnnotator (Iwasaki et al., 2013) showed that they formed the circular structure of the mitochondrial genome. Specifically, the genome consists of a gene set of cytochrome oxidase subunits I, II and III (COI, COII and COIII), cytochrome b (Cyt b), NADH dehydrogenase subunits 1-6 and 4L (ND1-6 and 4L), ATPase subunits 6 and 8 (ATPase6 and 8), two rRNAs, and 22 transfer RNAs (tRNA) (see Figure 1 of Inoue et al., 2020). We used sequences of only circulated mtDNAs for further analyses.
As mentioned above, we used 243 individuals representing 11 coral reef regions of the Pacific Ocean (Figure 1C) and determined the complete mtDNA sequences (16,210~16,246 bp, depending on the individual) of all specimens. Genome sequencing coverage per individual was 1,827X on average, ranging from 34X to 136,220X, indicating the data robustness from each specimen. We unambiguously aligned 16,218 bp sequences, including 1,822 variable sites, which were used for an unrooted tree analysis (Figure 2), a rooted tree analysis (Figure 3) and principal component analysis (PCA) (Figure 4).
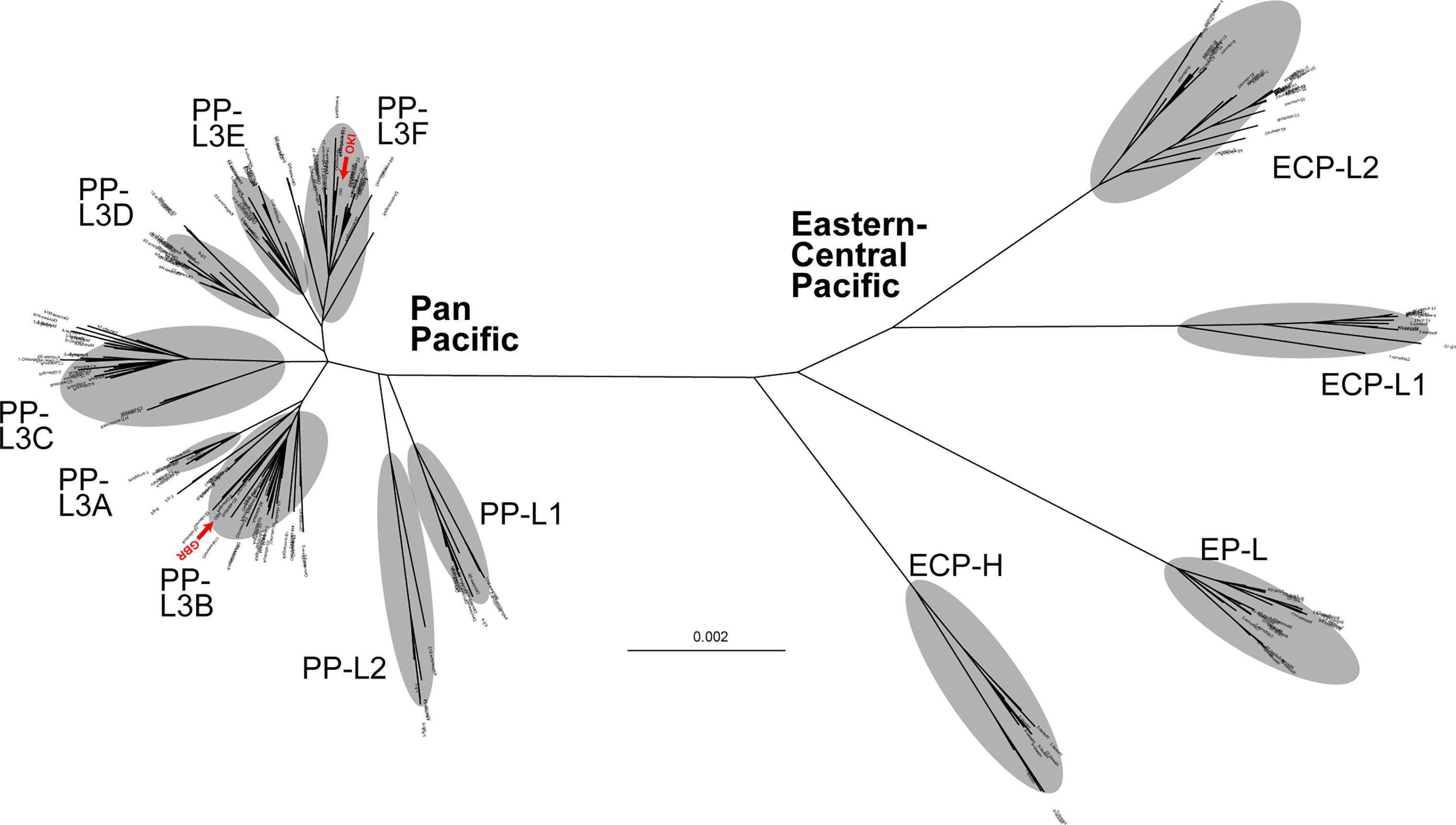
Figure 2 An unrooted phylogenetic tree of individual COTS using the maximum-likelihood (ML) method, based on entire mitochondrial genome sequences (16,218 bp, including 1,822 variable sites), indicating the separation of two clades, Eastern-Central Pacific and Pan Pacific. Red arrowheads indicate sequences (OKI and GBR) decoded in the genome paper (Hall et al., 2017).
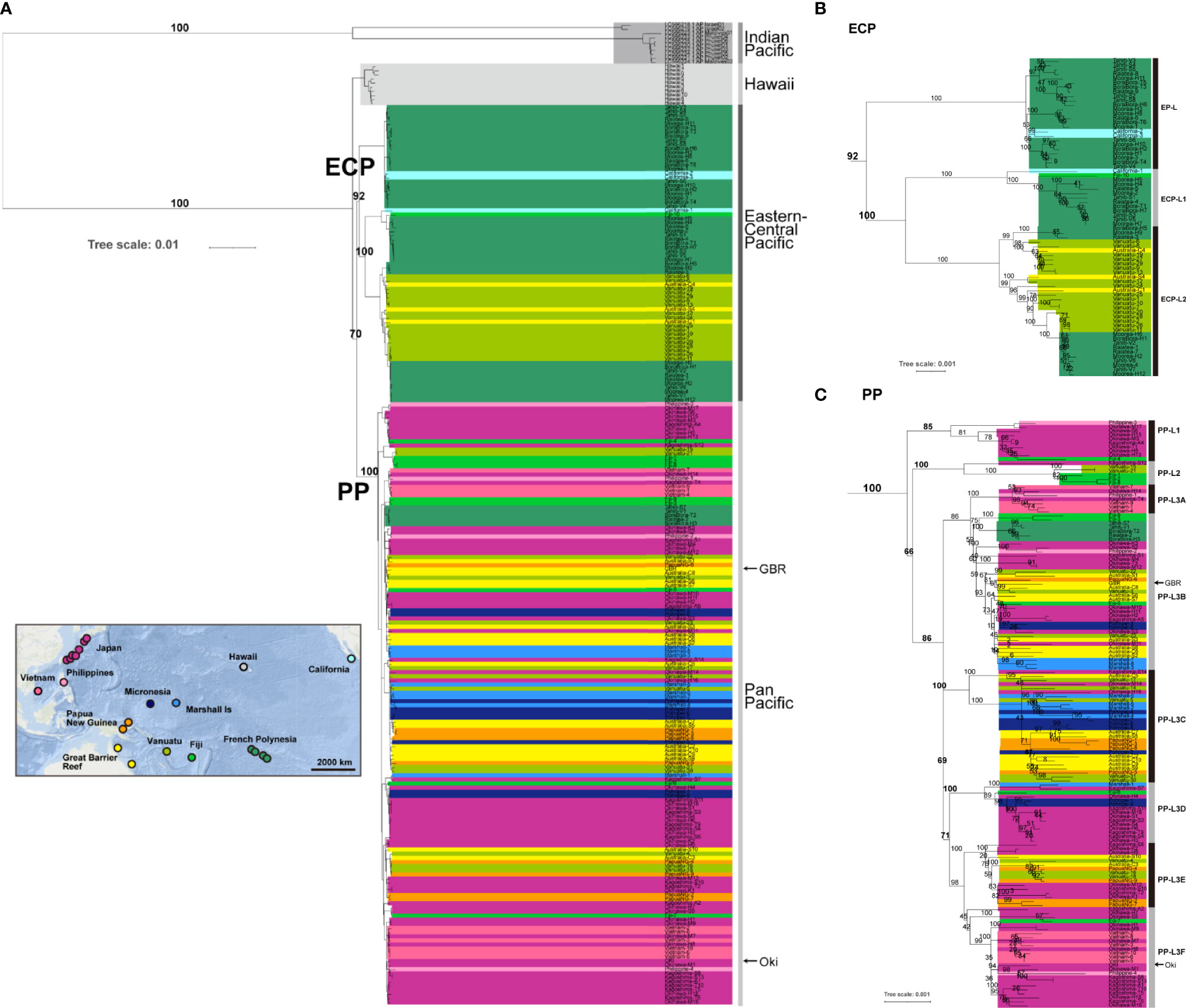
Figure 3 A rooted phylogenetic tree of Acanthaster cf. solaris using the maximum-likelihood (ML) method, based on entire mitochondrial genome sequences (16,219 bp, including 1,822 variable sites). The A. planci of the Indian Ocean was used for rooting. Numbers at some nodes indicate bootstrap values based on 1000 replicates for internal branch support. Arrowheads at the right indicate sequences (OKI and GBR) decoded in the genome paper (Hall et al., 2017). (A) indicates the entire phylogeny, (B) is an enlargement of ECP clade, and (C) is for PP clade.
Phylogenetic Analysis
An unrooted tree was constructed as follows. Whole mtDNA sequences were aligned using MAFFT (Katoh et al., 2005) with the default settings. Multiple sequence alignments were trimmed by removing poorly aligned regions using TRIMAL 1.2 (Capella-Gutiérrez et al., 2009) with the option “gappyout.” The maximum likelihood (ML) trees were created using RAxML 8.2.6 (Stamatakis, 2014). Trees were estimated with the “-f a” option, which invokes rapid bootstrap analysis with 1000 replicates. Trees were displayed using iTOL (Letunic and Bork, 2021).
For a rooted tree, we selected A. planci of the Indian Ocean as an outgroup. Whole mtDNA sequences of A. planci, two individuals from Eilat South Reef (Red Sea) (LC566218.1 and LC566219.1), two from Maldives (LC566223.1 and LC566224.1) and five from Racha Yai Island (Thailand) (LC566230.1, LC566231.1, LC566232.1, LC566233.1, and LC566234.1) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/bioproject/751773). The tree was constructed using the same method as in the case of the unrooted tree.
Principal Component Analysis (PCA)
Population structures were analyzed using model-free approaches. Based on complete mtDNA sequences, PCA was performed on all individuals, using PLINK 1.9 (Purcell and Chang, 2015). Pairwise genetic distances among localities were estimated with Weir and Cockerham’s FST (Weir and Cockerham, 1984) and Nei’s genetic distance (Nei, 1972) using StAMPP (Pembleton et al., 2013).
Divergence Time Estimation
Among the class Asteroidea, the genus Luidia belongs to the order Paxillosida and the genera Linckia and Acanthaster to the order Valvatida. O’Hara et al. (2014) estimated that the divergence of the two orders occurred approximately 185 million years ago (MYA). Using this divergence date and by comparing complete mtDNA sequences, we previously estimated the divergence time between A. brevispinus and A. planci (A. cf. solaris) (Lucas and Jones, 1976) as 21 MYA (Figure 5A) (Inoue et al., 2020). Since in this study mtDNA sequence of A. brevispinus (accession number, NC_007789.1) was added as an outgroup species, thus unambiguously aligned sequences of 16,219 bp, including 3,159 variable sites, were used for rooted tree analyses. Referring to Inoue et al. (2020), a time calibrated tree of populations was roughly estimated using RelTime (Tamura et al., 2012), as implemented in MEGA X (Kumar et al., 2018).
Results
We compared all mtDNA sequences of 243 individuals of COTS and obtained unambiguously aligned 16,218 bp sequences including 1,822 variable sites (Supplementary Figure 1). The variable sites were not evenly distributed in the genome (Supplementary Table 2). Higher rates of variation appeared in the non-coding D-loop region (0.36% on average), as well as in the protein coding sequences (0.13% on average) and rRNA sequences (0.11% on average) (Supplementary Table 2). tRNA sequences showed less variability (0.06% on average). All information was used for phylogenetic analyses.
Two Major COTS Clades Exist in the Pacific Ocean
An unrooted tree analysis
An unrooted tree analysis indicated that COTS populations in the Pacific represent two major clades, tentatively called the East-Central Pacific (ECP) clade and Pan-Pacific (PP) clade (Figure 2). Diversification of the clades was evident by a long branch distance between the two in the tree. That is, the two clades are separated by 0.004 mtDNA sequence substitutions per site (Figure 2). However, no differences in the mitochondrial genome architecture were detected between the two clades.
The ECP clade consists of four sub lineages, EP (Eastern Pacific)-L (lineage), ECP-L1, ECP-L2, and ECP-H (Hawaii) (Figure 2). Although the Hawaii population appeared more independent, we here included it in the ECP clade. The PP clade comprises three major lineages, PP-L1, PP-L2, and PP-L3, the latter of which was further divided into six sub lineages, PP-L3A, PP-L3B, PP-L3C, PP-L3D, PP-L3E, and PP-L3F (Figure 2). An individual collected on the GBR, from which the nuclear genome was sequenced (Hall et al., 2017), belonged to PP-L3B while a genome-decoded OKI individual belonged to PP-L3F (shown by red in Figure 2).
A Rooted Tree Analysis
A rooted tree analysis using Indian Ocean A. planci as an outgroup also revealed two clades of Pacific COTS (Figure 3). The ECP clade was supported by 92% of bootstrap 1000-times replications and PP clade by 100%.
Among the ECP COTS, ECP-H diverged first, then EP-L and ECP-L1/ECP-L2 (Figures 3A, B). The grouping of ECP-H, EP-L, ECP-L1 and ECP-L2 was supported by 100% bootstrap value, respectively. Three lineages of the PP clade, PP-L1, PP-L2, and PP-L3 were supported by 85, 100 and 86%, respectively (Figures 3A, C).
The rooted tree established the Indian Ocean COTS from the Red Sea and those from the Southern Indian Ocean belonged to the same clade with 100% bootstrap support (Figure 3A). An analysis by Vogler et al. (2008) of the four geographically distributed COTS lineages in the Indo-Pacific showed that COTS of the Northern and Southern Indian Oceans form a clade (more than 90% bootstrap support) while Red Sea COTS form another clade with Pacific COTS, although bootstrap support value was low (less than 70%). However, the present comparison of the complete set of mtDNA sequences showed that Red Sea COTS were more closely aligned with Indian Ocean COTS with 100% bootstrap support (Figure 3A). This supports the notion of their being two clades of A. planci, sensu lato, that is A. planci in the Indian Ocean plus Red Sea, and A. cf. solaris in the Pacific Ocean.
Secondly, the difference of branch length between populations of A. planci and that of A. cf. solaris was extensive (Figure 3A). Compared with the longer branch length of Indian Ocean COTS, the length of Pacific Ocean COTS was quite short, suggesting the time interval required for local colonization of this COTS across the entire Pacific Ocean was comparatively short.
Principal Component Analysis (PCA)
PCA of specimens from all sampling locations (Supplementary Table 1) supported the results of molecular phylogenetic analyses (Figure 4). PCA resulted in five unique groups, corresponding to EP-L, ECP-L1, EPC-L2, ECP-H, and PP-L, respectively. Notably, a mixture of COTS from all locations across the Pacific was evident in PP-L (Figure 4, upper right corner). When compared to molecular phylogeny results (Figures 2, 3), grouping of EP-L, ECP-L1 and ECP-H was more strongly demonstrated in PCA (Figure 4). PCA also suggested an affinity of ECP-L2 with PP-L, although this was not as strong (Figure 4).
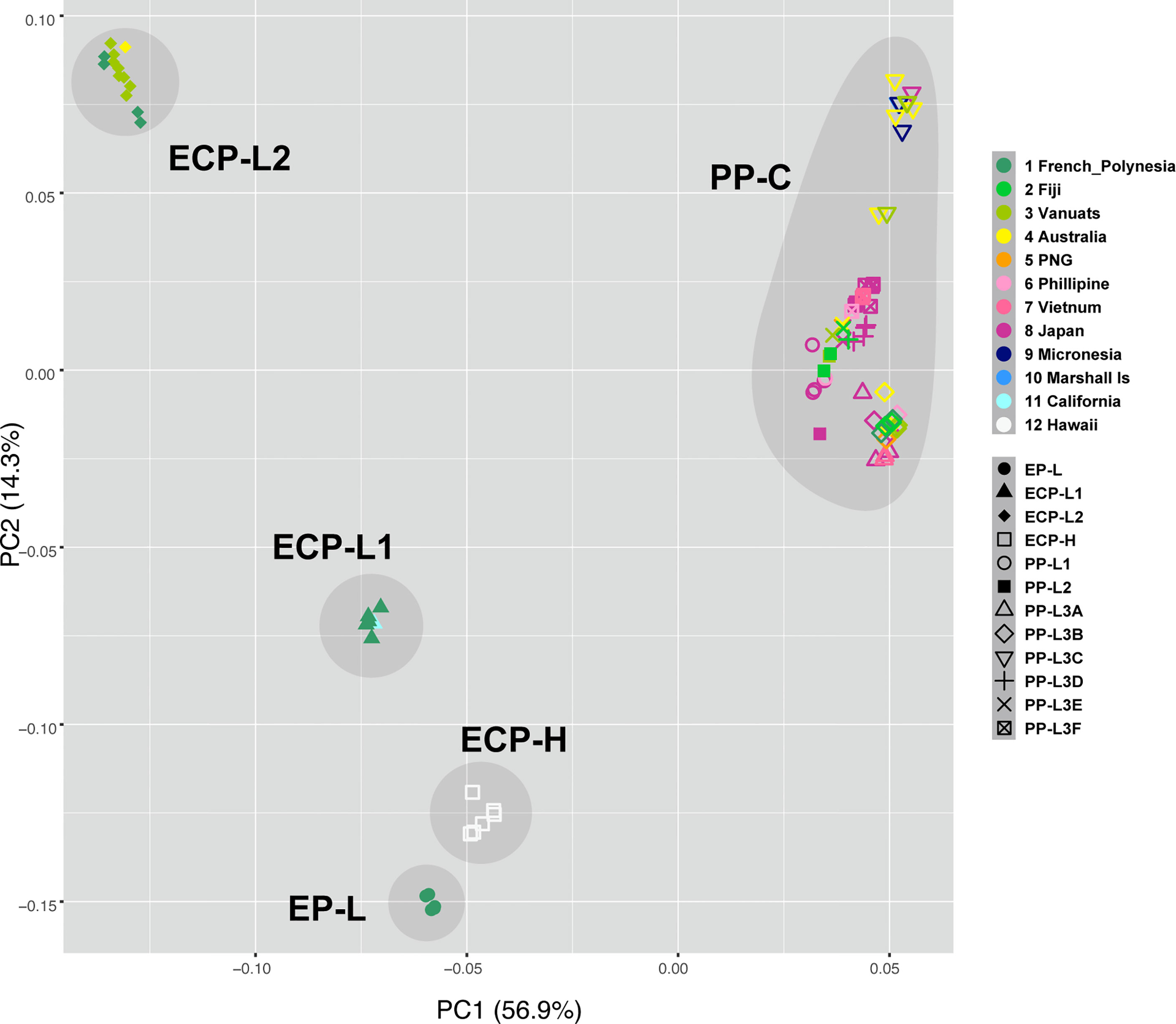
Figure 4 Principle Component Analysis identifies five COTS populations, PP-C, and four lineages of ECP-C, EP-L, ECP-L1, ECP-L2, and ECP-H. PP-C contains COTS collected from all the Pan Pacific countries except for USA, suggesting that these populations are expanded from a common ancestor population. Color codes are shown at the right side.
Population Structure of the East-Central Pacific Clade
The molecular phylogenetic analyses presented above showed that the ECP COTS clade consists of four sub-lineages, EP-L, ECP-L1, ECP-L2, and ECP-H (Figures 2, 3). The EP-L consists of COTS from French Polynesia (Tahiti, Bora Bora, Moorea, and Raiatea) and two individuals from California as one separate sub-clade (Figure 3B). The ECP-L1 contains COTS from French Polynesia, plus an individual from California and Fiji each (Figure 3B). On the other hand, the ECP-L2 comprises two subgroups, but both include COTS of French Polynesia, Fiji, Vanuatu, and GBR (Clack and Shell Reefs). The ECP-H was composed of individuals only from Hawaii (Figures 2, 3).
Two results of particular interest are related to COTS from California and those from Hawaii. First, the two California COTS pertain to ECP-C, one belonging to EP-L and the other to ECP-L1 (Figure 3B). Second, ten Hawaiian COTS formed a discrete group (ECP-H) near the root position and did not include individuals from any other Pacific reefs. This genetic isolation is exceptional but is supported by 100% bootstrap (Figure 3A).
Population Structure of the Pan Pacific Clade
In addition to the four lineages of ECP-C, all of which are comparatively well separated or isolated, two of the three lineages or subgroups of PP-C, that is, PP-L1, PP-L2 and PP-L3, were supported by higher bootstrap values, returning 85%, 100%, and 74%, respectively (Figure 3C). PP-L1 included COTS from the Philippines and Japan, and PP-L2 comprised starfish from Fiji, Vanuatu, and Japan, both branched earlier and were separated from the PP-L3 lineages (Figures 2, 3C).
PP-L3 was a very large group, including not only Western Pacific COTS, but also Eastern Pacific populations from French Polynesia, Fiji, Vanuatu, GBR, Papua New Guinea, the Philippines, Japan, Micronesia, and the Marshall Islands. It consists of six lineages (PP-L3A to PP-L3F) that are not strictly geographically defined, in that each subgroup comprises individuals from several of these areas, except for PP-L3A that was comprised of COTS of the Philippines, Vietnam and Japan (Figure 3C). Of special interest is PP-L3B, which had the largest geographic distribution, including COTS from all locations of French Polynesia, Fiji, Vanuatu, GBR, Papua New Guinea, the Philippines, Vietnam, Japan, Micronesia, and the Marshall Islands. PP-L3C also includes COTS from various locations including Vanuatu, GBR, Papua New Guinea, Japan, Micronesia, and the Marshall Islands (Figure 3C). PP-L3D consists of COTS not only from Japan, Micronesia and the Marshall Islands, but also Fiji. PP-L3E includes COTS from Japan, GBR, Vanuatu, and Micronesia. On the other hand, PP-L3F appears to be a lineage more specific to East Asia, comprising COTS populations in the Philippines, Vietnam, and Japan (Figure 3C).
Divergence Time Estimation
Vogler et al. (2008) estimated that the four clades of A. planci, sensu lato have diverged in the Pliocene-Early Pleistocene, 1.92-3.65 MYA. Using the entire mtDNA sequence of A. brevispinus as an outgroup (see Method), we estimated the divergence time of A. cf. solaris (Figure 5B). Our results indicated that the divergence of PP-C from a common ancestor with ECP-C occurred 2.9 (confidence interval: 2.2–3.7) MYA (Figure 5B). Divergence of PP-L1 and PP-L2/3 occurred around 1.1 (0.8–1.5) MYA and that of PP-L2 and PP-L3 1.0 (0.7–1.5) MYA. Divergence of six sub-lineages of PP-L3, (L3A to L3F) apparently occurred approximately 0.7 MYA, with a range from 0.8 (0.5–1.1) to 0.6 (0.4–0.8) MYA (Figure 5B). This suggests that COTS of PP-C diverged more recently and coincidently compared to those of ECP-C.
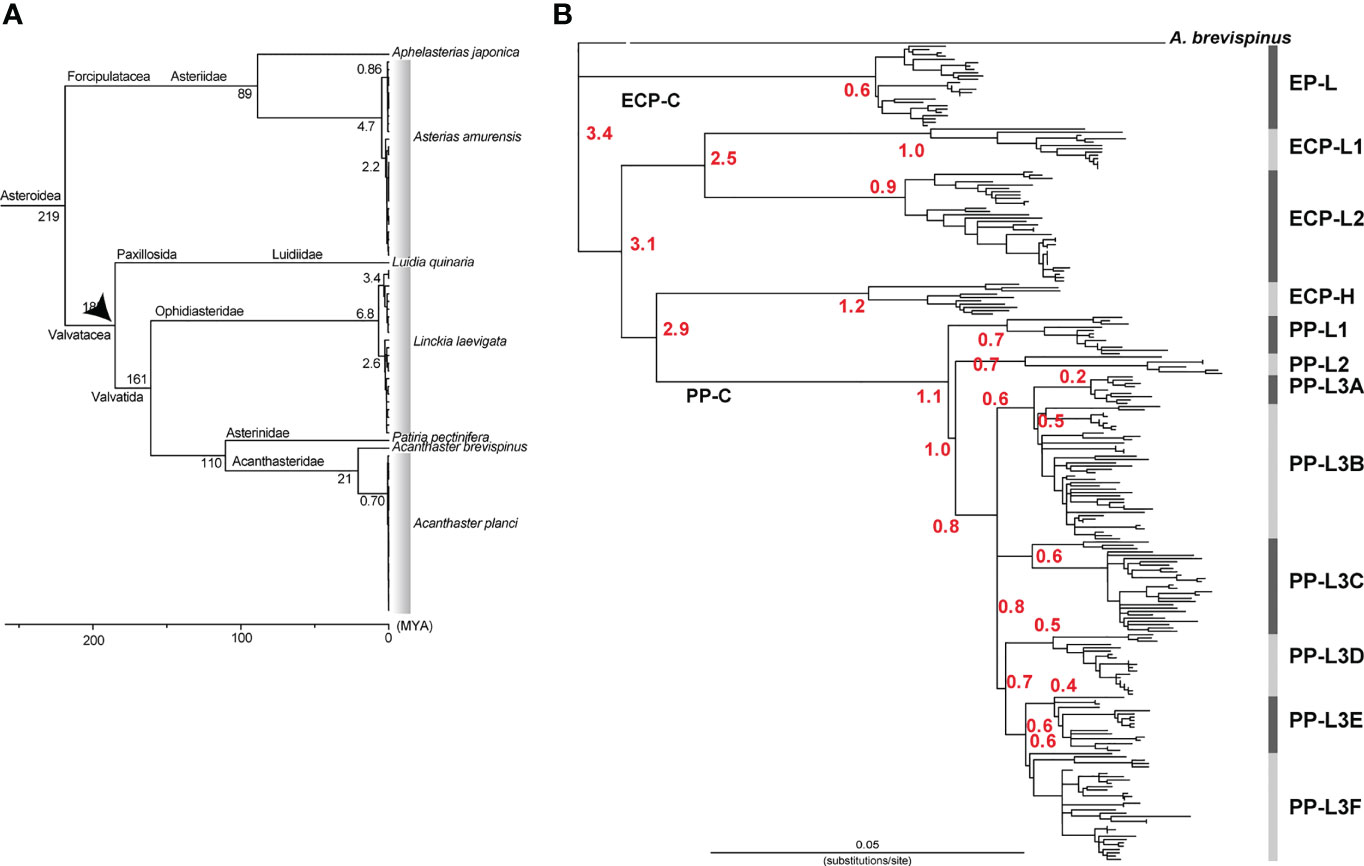
Figure 5 Estimated divergence times (A) between different species of starfish based on Inoue et al (2020) and (B) those between different lineages of COTS (shown by red numerals). The estimation was based on the RelTime method using 21 MYA as the diversification time between of Acanthaster brevispinus and A. cf. solaris.
Discussion
COTS Population Structures in the Pacific Ocean
The present study broadly analyzed phylogeography of COTS in the Pacific Ocean. Based on comparisons of complete mtDNA sequences, COTS in the Pacific are genetically subdivided into two major clades, ECP-C and PP-C (Figures 2, 3). The ECP-C comprises of COTS of French Polynesia, Fiji, Vanuatu, and GBR. In addition, COTS of California and Hawaii are included in this clade, the latter forming a discrete population as ECP-H. On the other hand, PP-C comprises of COTS from all collection sites across the Pacific. These results agree well with previous studies, suggesting that North Central Pacific COTS, including those from Hawaii, form a distinct clade among Pacific COTS (Timmers et al., 2012; Vogler et al., 2013).
The two California COTS pertain to ECP-C, one belonging to EP-L and the other to ECP-L1 (Figure 3). The external morphology of the California COTS is significantly different from counterparts in other areas of the Pacific. Specifically, they tend to have shorter arms, and were initially classified as a separate species, Acanthaster elichii (Birkeland and Lucas, 1990). However, allozyme analysis revealed them to have stronger affinity to COTS of the Western Pacific than to their closest geographical neighbors, the Hawaiian COTS, and were therefore reconfirmed as A. planci (Nishida and Lucas, 1988). This suggests a common ancestry for Eastern Pacific COTS and California COTS. Accordingly, Haszprunar and Spies (2014) classified all Pacific COTS as A. cf. solaris. Therefore, the present result supports the notion that California COTS share an ancestor with those in the Central Pacific, and thus their assignment as A. cf. solaris.
The Hawaiian COTS population is discrete from other populations (Timmers et al., 2012), being phylogenetically categorized as ECP-H. Nishida and Lucas (1988) had previously established this by allozyme analysis and postulated that the independency of this population was likely due to i) the absence of eastward currents at higher latitudes capable of transporting larvae between the Hawaiian Islands and the eastern Pacific, and ii) the North Equatorial Countercurrent that could transport larvae eastward from the Line Islands to the eastern Pacific. Given the distinct genetic distance and long-term isolation, ECP-H might be a cryptic COTS species and future nuclear genomic and ecological studies are warranted to confirm this possibility.
Possible Scenarios of Historical COTS Distribution in the Pacific Ocean
Phylogenetic categorization offers an opportunity to speculate on the oceanic mechanisms that may have been involved in establishing these distinct COTS populations in the Pacific and the Indian Ocean (Figure 6).
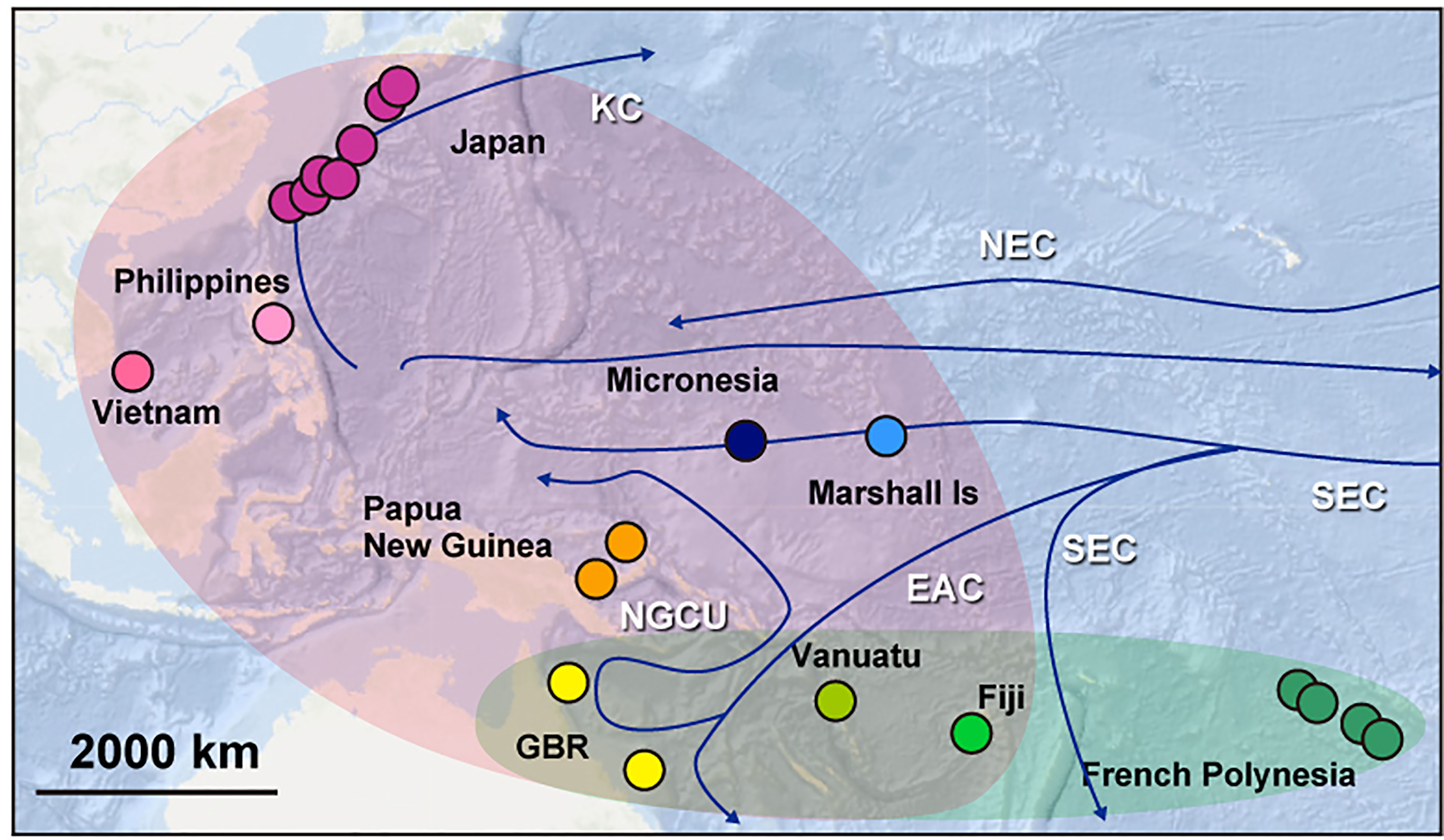
Figure 6 A summary diagram to show a possible historical colonization of COTS with main seawater currents in the Pacific. The Pacific hosts two major groups of COTS. The East-Central Pacific group comprises COTS from French Polynesia, Fiji, Vanuatu, and the GBR (blue). The Pan Pacific group contains COTS from the entire Western Pacific (purple). Blue arrows show the main currents in the Pacific Ocean. KC, Kuroshio Current; CC, California Current; NEC, North Equatorial Current; ECC, Equatorial Countercurrent; SEC, South Equatorial Current; EAC, East Australian Current and NGCU, New Guinea Coastal Undercurrent.
History of the East-Central Pacific (ECP) COTS
The ECP clade was divided into four sub-groups, EP-L, ECP-L1, ECP-L2 and ECP-H. The former three are distinguishable by their geographic distributions, COTS of California and Hawaii having already been discussed above. EP-L is confined to four locations of French Polynesia + California, ECP-L1 encompasses French Polynesia + California + Fiji, and ECP-L2 is confined to French Polynesia, Vanuatu, and GBR. This sub-grouping suggests two possible scenarios relative to their distributional history in the Eastern and Central Pacific. One is the EP-L ancestry hypothesis, whereby COTS that originated in French Polynesia experienced a bottleneck-like founder effect resulting in a comparatively homogenous genetic background (Yasuda et al., 2014). Then they expanded into the central and western regions, ultimately reaching the GBR. In contrast, in the ECP-L2 ancestry hypothesis, a comparatively broad region encompassing GBR, Vanuatu, Fiji and French Polynesia is the original source of COTS, from which EP-L and ECP-L1 became established as separate, independent lineages long ago. The early divergence of EP-L suggests the former scenario is the more plausible. If this is the case, COTS of EPC originated from EP-L and expanded toward the Western Pacific.
Irrespective of these discussions, the fact that ECP COTS are confined to the Eastern and Central Pacific and are less affected by anthropogenic factors strongly suggests that they are not prone to major outbreaks, even though they show local outbreaks (Birkeland, 1990). The smaller size of coral reefs and lower density of suitable coral prey may also be limiting factors at times of outbreak.
History of the Western Pacific COTS
In contrast to the ECP clade, the PP clade comprises COTS from across the entire Pacific, including more the highly populated regions where massive expansions might have occurred repeatedly (Figures 3C, 4, 6). PP-L contains COTS from almost all regions of the Pacific, including French Polynesia, Fiji, Vanuatu, GBR, Papua New Guinea, Vietnam, the Philippines, Japan, Micronesia, and the Marshall Islands. The two PP-L subgroups, PP-L3B and PP-L3C, both contain COTS from all these localities. It is highly likely that this population genetic profile reflects the trajectory of repeated population expansions across the entire Pacific Ocean, with the exception of the U.S. population. One possible explanation is that dispersal of long-lived COTS larvae spawned in the central Pacific is facilitated by the South Equatorial Current, which flows at an average velocity of 20 nautical miles per day from Fiji and Vanuatu toward the GBR, where it bifurcates into the New Guinea Coastal Undercurrent (Figure 6) (Sokolov and Rintoul, 2000; Treml et al., 2008). In combination with the North Equatorial Current, which originates from the Californian Current, it bifurcates into the strong Kuroshio Current that flows from the northeastern Philippines toward Japan (Qiu and Lukas, 1996) (Figure 6). An earlier divergence of PP-L2, which includes COTS from Fiji and Vanuatu, suggests a contribution of these COTS with western Pacific populations via the southernmost branches of the South Equatorial Current (Figure 6).
Another interesting observation here is that COTS of Micronesia and the Marshall Islands are not members of EP-L, but belong in PP-L. This suggests that the westward flow of the South Equatorial Current into the Coral Sea may become disrupted by complex topography, carrying larvae to the intersection of the Equatorial Counter Current, which is an eastward flowing, wind-driven current, thereby transporting Eastern-Central COTS larvae toward California (Wyrtki, 1967) (Figure 6).
Further support linking repeated dispersal among the PP-L3 population comes from comparisons of the entire ~384-Mb genome sequences of two COTS, one from the GBR and the other from Okinawa (OKI), separated by over 5,000 km (Hall et al., 2017). An unexpected result of the genome study was the exceptionally low heterozygosity of the genomes, 0.88% and 0.92% for the GBR and OKI populations, respectively. In addition, reciprocal BLAST analysis of scaffolds longer than 10 kb showed 98.8% nucleotide identity between the GBR and OKI genomes, evidence of the great similarity of their nuclear DNA sequences. Together, the very strong resemblance of the nuclear genomes of the GBR and OKI individuals and their categorization as belonging to the two closely related PP subclades, supports a common ancestor.
These results raise yet another possibility with respect to the geographical extent of the distribution of various COTS lineages. Most of the COTS that belong to ECP-L1 are from Vanuatu. However, other Vanuatu COTS belong to PP-L2, PP-L3B, or PP-L3E. Both lineages of COTS coexist in Vanuatu, one with the capacity for large expansions and the other without. An objective of future population genomics studies will be to sequence and compare complete genomes of both ECP-L and PP-L COTS to try and discover the genetic and genomic features that encode the capacity for outbreaks. Although we postulate that the oldest lineages of extant Pacific COTS originated from a broad Pacific region, including Fuji, Vanuatu and GBR, all scenarios should be examined by future studies using entire nuclear genome sequences.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJDB10499].
Author Contributions
NY, JI, MH, CM, and NS designed the research. NY, MH, MNa, MA, MF, MNi, NT, RR-G, and ZF collected samples. TS and RK sequenced COTS mitochondrial DNA. NY, JI, KH, CM, and NS analyzed data. NY, JI, CM, and NS wrote the manuscript with input from all authors. All authors gave final approval for publication.
Funding
This study was supported by OIST funds (POC5 Project) to the Marine Genomics Unit, and by the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (17H04996 to NY and 18K06396 and 21H04922 to JI.) This research was also supported in part by the Ms. Sumiko Imano Memorial Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the following people for their help with sample collection: Dr. Hugh Sweatman and the AIMS Bioresources Library for GBR samples, Dr. Molly Timmers for Hawaiian samples, Geoff Jones and Jeff Kinch for Papua New Guinean samples, Monal Lal for Fijian samples, Christina Shaw for Vanuatu samples, Hoang Dinh Chieu for Vietnamese samples, and Hiromitsu Ueno for Japanese samples. The DNA Sequencing Section and IT section of OIST are acknowledged for its expert help with genome sequencing and bioinfomatic analysis. Last, but not least, we acknowledge the traditional owners of the lands and sea country on which this research took place.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.831240/full#supplementary-material
References
Avise J. C. (2000). Phylogeography: The History and Formation of Species (Massachusetts: Harvard university press).
Benzie J. (1992). Review of the Genetics, Dispersal and Recruitment of Crown-of-Thorns Starfish (Acanthaster Planci). Marine Freshwater Res. 43, 597–610. doi: 10.1071/MF9920597
Benzie J., Stoddart J. (1992). Genetic Structure of Outbreaking and Non-Outbreaking Crown-of-Thorns Starfish (Acanthaster planci) populations on the Great Barrier Reef. Marine Biol. 112, 119–130. doi: 10.1007/BF00349735
Birkeland C. (1990). “Acanthaster planci,” in Major Management Problem of Coral Reefs (Boca Raton: CRC Press). Charles E. Birkeland, John S. Lucas.
Birkeland C., Lucas J. (1990). “Acanthaster planci,” in Major Management Problem of Coral Reefs (Florida, Boca Raton: CRC Press).
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
De’ath G., Fabricius K. E., Sweatman H., Puotinen M. (2012). The 27–year Decline of Coral Cover on the Great Barrier Reef and Its Causes. Proc. Natl. Acad. Sci. 109, 17995–17999. doi: 10.1073/pnas.1208909109
Dierckxsens N., Mardulyn P., Smits G. (2017). NOVOPlasty: De Novo Assembly of Organelle Genomes From Whole Genome Data. Nucleic Acids Res. 45, e18–e18.
Fabricius K. E., Okaji K., De’ath G. (2010). Three Lines of Evidence to Link Outbreaks of the Crown-of-Thorns Seastar Acanthaster planci to the Release of Larval Food Limitation. Coral Reefs 29, 593–605. doi: 10.1007/s00338-010-0628-z
Hall M. R., Kocot K. M., Baughman K. W., Fernandez-Valverde S. L., Gauthier M. E., Hatleberg W. L., et al. (2017). The Crown-of-Thorns Starfish Genome as a Guide for Biocontrol of this Coral Reef Pest. Nature 544, 231–23436. doi: 10.1038/nature22033
Harmelin-Vivien M. L. (1994). The Effects of Storms and Cyclones on Coral Reefs: A Review. J. Coastal Res., 12, 211–231. Available at: https://www.jstor.org/stable/25735600.
Harrison H. B., Pratchett M. S., Messmer V., Saenz-Agudelo P., Berumen M. L. (2017). Microsatellites Reveal Genetic Homogeneity Among Outbreak Populations of Crown-of-Thorns Starfish (Acanthaster cf. solaris) on Australia’s Great Barrier Reef. Diversity 9, 16.
Haszprunar G., Spies M. (2014). An Integrative Approach to the Taxonomy of the Crown-of-Thorns Starfish Species Group (Asteroidea: Acanthaster): A Review of Names and Comparison to Recent Molecular Data. Zootaxa 25, 271–284. doi: 10.11646/zootaxa.3841.2.6
Hughes R. N., Hughes D. J., Smith I. P. (2014). Limits to Understanding and Managing Outbreaks of Crown-of-Thorns Starfish (Acanthaster spp.). Oceanography Marine Biology: Annu. Rev. 52, 133–200.
Hughes T. P., Kerry J. T., Álvarez-Noriega M., Álvarez-Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global Warming and Recurrent Mass Bleaching of Corals. Nature 543, 373–377. doi: 10.1038/nature21707
Inoue J., Hisata K., Yasuda N., Satoh N. (2020). An Investigation Into the Genetic History of Japanese Populations of Three Starfish, Acanthaster planci, Linckia laevigata, and Asterias amurensis, Based on Complete Mitochondrial DNA Sequences. G3: Genes Genomes Genet. 10, 2519–2528. doi: 10.1534/g3.120.401155
Iwasaki W., Fukunaga T., Isagozawa R., Yamada K., Maeda Y., Satoh T. P., et al. (2013). MitoFish and MitoAnnotator: A Mitochondrial Genome Database of Fish With an Accurate and Automatic Annotation Pipeline. Mol. Biol. Evol. 30, 2531–2540. doi: 10.1093/molbev/mst141
Katoh K., Kuma K.-i., Toh H., Miyata T. (2005). MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kenchington R. (1977). Growth and Recruitment of Acanthaster planci (L.) on the Great Barrier Reef. Biol. Conserv 11, 103 – 118. doi: 10.1016/0006-3207(77)90032-5
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic I., Bork T. (2021). Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 49 W293–W296. doi: 10.1093/nar/gkab301
Lucas J. S. (1982). Quantitative Studies of Feeding and Nutrition During Larval Development of the Coral Reef Asteroid Acanthaster planci (L.). J. Exp. Marine Biol. Ecol. 65, 173–193. doi: 10.1016/0022-0981(82)90043-0
Lucas J. S., Jones M. M. (1976). Hybrid Crown-of-Thorns Starfish (Acanthaster planci x A. brevispinus) Reared to Maturity in the Laboratory. Nature 263, 409–412. doi: 10.1038/263409a0
Nakamura M., Okaji K., Higa Y., Yamakawa E., Mitarai S. (2014). Spatial and Temporal Population Dynamics of the Crown-of-Thorns Starfish, Acanthaster planci, Over a 24-year Period Along the Central West Coast of Okinawa Island, Japan. Marine Biol. 161, 2521–2530. doi: 10.1007/s00227-014-2524-5
Nishida M., Lucas J. S. (1988). Genetic Differences Between Geographic Populations of the Crown-of-Thorns Starfish Throughout the Pacific Region. Marine Biol. 98 (3), 359–368. doi: 10.1007/BF00391112
O’Hara T. D., Hugall A. F., Thuy B., Moussalli A. (2014). Phylogenomic Resolution of the Class Ophiuoidea Unlocks a Global Microfossil Record. Curr. Biol. 24, 1874–1879. doi: 10.1016/j.cub.2014.06.060
Pembleton L. W., Cogan N. O., Forster J. W. (2013). St AMPP: An R Package for Calculation of Genetic Differentiation and Structure of Mixed-Ploidy Level Populations. Mol. Ecol. Resour. 13, 946–952. doi: 10.1111/1755-0998.12129
Pratchett M. S., Caballes C. F., Wilmes J. C., Matthews S., Mellin C., Sweatman H., et al. (2017). Thirty Years of Research on Crown-of-Thorns Starfish, (1986–2016): Scientific Advances and Emerging Opportunities. Diversity 9, 41. doi: 10.3390/d9040041
Purcell S., Chang C. (2015) PLINK 1.9. Available at: www.cog-genomics.org/plink/1.9.
Qiu B., Lukas R. (1996). Seasonal and Interannual Variability of the North Equatorial Current, the Mindanao Current, and the Kuroshio Along the Pacific Western Boundary. J. Geophysical Research: Oceans 101, 12315–12330. doi: 10.1029/95JC03204
Selmoni O., Lecellier G., Ainley L., Collin A., Doucet R., Dubousquet V., et al. (2020). Using Modern Conservation Tools for Innovative Management of Coral Reefs: The MANACO Consortium Journal: Frontiers in Marine Science, Section Marine Conservation and Sustainability. Front. Marine Sci. 7, 609. doi: 10.3389/fmars.2020.00609
Sokolov S., Rintoul S. (2000). Circulation and Water Masses of the Southwest Pacific: WOCE Section P11, Papua New Guinea to Tasmania. J. marine Res. 58, 223–268. doi: 10.1357/002224000321511151
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura K., Battistuzzi F. U., Billing-Ross P., Murillo O., Filipski A., Kumar S. (2012). Estimating Divergence Times in Large Molecular Phylogenies. Proc. Natl. Acad. Sci. U.S.A. 109, 19333–19338. doi: 10.1073/pnas.1213199109
Timmers M. A., Andrews K. R., Bird C. E., deMaintenton M. J., Brainard R. E., Toonen R. J. (2011). Widespread Dispersal of the Crown-of-Thorns Sea Star, Acanthaster planci, Across the Hawaiian Archipelago and Johnston Atoll. J. Marine Biol. 2011, 10. doi: 10.1155/2011/934269
Timmers M., Bird C., Skillings D., Smouse P., Toonen R. (2012). There's No Place Like Home: Crown-of-Thorns Outbreaks in the Central Pacific Are Regionally Derived and Independent Events. PloS One 7, e31159. doi: 10.1371/journal.pone.0031159
Treml E. A., Halpin P. N., Urban D. L., Pratson L. F. (2008). Modeling Population Connectivity by Ocean Currents, a Graph-Theoretic Approach for Marine Conservation. Landscape Ecol. 23, 19–36. doi: 10.1007/s10980-007-9138-y
Tusso S., Morcinek K., Vogler C., Schupp P. J., Caballes C. F., Vargas S., et al. (2016). Genetic Structure of the Crown-of-Thorns Seastar in the Pacific Ocean, with focus on Guam. PeerJ 4, e1970. doi: 10.7717/peerj.1970
Uthicke S., Pecorino D., Albright R., Negri A. P., Cantin N., Liddy M., et al. (2013). Impacts of Ocean Acidification on Early Life-History Stages and Settlement of the Coral-Eating Sea Star Acanthaster planci. PloS One 8, e82938. doi: 10.1371/journal.pone.0082938
Vogler C., Benzie J., Lessios H., Barber P., Worheide G. (2008). A Threat to Coral Reefs Multiplied? Four Species of Crown-of-Thorns Starfish. Biol. Lett. 4, 696–699. doi: 10.1098/rsbl.2008.0454
Vogler C., Benzie J. A. H., Tenggardjaja K., Ambariyanto, Barber P. H., Wörheide G. (2013). Phylogeography of the Crown-of-Thorns Starfish: Genetic Structure Within the Pacific Species. Coral Reefs 32, 515–525. doi: 10.1007/s00338-012-1003-z
Weir B. S., Cockerham C. C. (1984). Estimating F-Statistics for the Analysis of Population Structure. Evolution 38, 1358–1370.
Yamaguchi M. (1973). Early Life History of Coral Reef Asteroids, with Special Reference to Acanthaster planci (L.). Biol. Geology Coral Reefs Biol. 1 II, 369 – 389. doi: 10.1016/B978-0-12-395526-5.50019-5
Yasuda N. (2018). “Distribution Expansion and Historical Population Outbreak Patterns of Crown-of-Thorns Starfish, Acanthaster Planci Sensu Lato, in Japan from 1912 to 2015,” in Coral Reef Studies of Japan. Eds. Iguchi A., Hongo C. (Singapore: Springer Singapore), 125–148.
Yasuda N., Hamaguchi M., Sasaki M., Nagai S., Saba M., Nadaoka K. (2006). Complete Mitochondrial Genome Sequences for Crown-of-Thorns Starfish Acanthaster planci and Acanthaster brevispinus. BMC Genomics 7 (1), 17. doi: 10.1186/1471-2164-7-17
Yasuda N., Nagai S., Hamaguchi M., Okaji K., Gérard K., Nadaoka K. (2009). Gene Flow of Acanthaster planci (L.) in Relation to Ocean Currents Revealed by Microsatellite Analysis. Mol. Ecol. 18, 1574–1590. doi: 10.1111/j.1365-294X.2009.04133.x
Yasuda N., Taquet C., Nagai S., Yoshida T., Adjeroud M. (2014). Genetic Connectivity of the Coral-Eating Sea Star Acanthaster planci during the severe outbreak of 2006–2009 in the Society Islands, French Polynesia. Marine Ecol. 36, 668–678. doi: 10.1111/maec.12175
Keywords: crown-of-thorns starfish (COTS), Pacific Ocean, population genetics, complete mitochondrial genome sequences, two hidden clades
Citation: Yasuda N, Inoue J, Hall MR, Nair MR, Adjeroud M, Fortes MD, Nishida M, Tuivavalagi N, Ravago-Gotanco R, Forsman ZH, Soliman T, Koyanagi R, Hisata K, Motti CA and Satoh N (2022) Two Hidden mtDNA-Clades of Crown-of-Thorns Starfish in the Pacific Ocean. Front. Mar. Sci. 9:831240. doi: 10.3389/fmars.2022.831240
Received: 08 December 2021; Accepted: 21 March 2022;
Published: 27 April 2022.
Edited by:
Jin Sun, Ocean University of China, ChinaReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, United StatesJuan D. Gaitan-Espitia, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2022 Yasuda, Inoue, Hall, Nair, Adjeroud, Fortes, Nishida, Tuivavalagi, Ravago-Gotanco, Forsman, Soliman, Koyanagi, Hisata, Motti and Satoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noriyuki Satoh, norisky@oist.jp; orcid.org/0000-0002-4480-3572
†These authors have contributed equally to this work
 Nina Yasuda1†
Nina Yasuda1†