- 1Guangzhou Key Laboratory of Subtropical Biodiversity and Biomonitoring, Guangdong Provincial Key Laboratory for Healthy and Safe Aquaculture, Guangdong Provincial Engineering Technology Research Center for Environmentally Friendly Aquaculture, School of Life Sciences, South China Normal University, Guangzhou, China
- 2Institute of Life and Environmental Sciences, University of Iceland, Reykjavík, Iceland
Background
The White Cloud Mountain minnow (Tanichthys albonubes Lin, 1932) is a native freshwater fish of Southern China and Northern Vietnam that belongs to the family Tanichthyidae, under the order of Cypriniformes (Mayden and Chen, 2010). This colorful species was selected as pet fish shortly after its discovery at the brooks of White Cloud Mountain (Guangzhou city, Guangdong province) in 1932 and soon became a worldwide popular aquarium fish species due to its low demand for care and captive breeding (Li et al., 2020a). While being introduced to many countries and has established stable hatchery populations by ornamental aquaculture, the White Cloud Mountain minnow was considered to be extinct in its natural distribution as a consequence of habitat loss since the 1960s and hence the species is classified as a second-class state-protected animal in China (Harris, 2008). Fortunately, wild populations were rediscovered in Guangdong (Yi et al., 2004), Guangxi (Li and Li, 2011) and on Hainan Island (Chan and Chen, 2009) in the last two decades, which marks the milestone of its conservation and address the urgent necessity of conducting scientific researches on this endangered species.
To date, most of the attention from the academic world has been given to the phylogenetic status of the White Cloud Mountain minnow and its sister species. The White Cloud Mountain minnow was considered as a member of the subfamily Danioninae (Chen, 1998), and used to be the only species in the genus Tanichthys for more than 50 years, until two new species T. micagemmae (Freyhof and Herder, 2001) and T. thacbaensis (Nguyen and Ngo, 2001) were reported from Middle and North Vietnam in 2001. Based on molecular evidence, Mayden and Chen proposed that the genus Tanichthys should be elevated to the family level (Mayden and Chen, 2010) and this was then confirmed by Tan and Armbruster (2018). Subsequently in 2019, the fourth valid species T. kuehnei was described by Bohlen et al. (2019).
Besides discussions on the phylogenetic status of this species, cryptic species were also found from its previous identified populations. A recent study examined the genetic relationship of eight wild populations crossing its range using multilocus molecular (one mitochondrial gene, two nuclear genes and 13 microsatellite loci) and morphological data (Li et al., 2020b). Li et al. revealed deep hidden genetic divergence among these populations with subtle morphological disparity, which was mainly in the number of branched anal-fin rays. Therefore they proposed the existence of at least seven cryptic species in the T. albonubes complex and highlighted the need for further evaluation of the species complex using genomic tools (Li et al., 2020b).
Although several studies have investigated the population genetics and gene expression of T. albonubes (Chen et al., 2009; Liu et al., 2011; Jing et al., 2013; Luo et al., 2015; Zhao et al., 2018), there are not any genomic-level resources available for this species. In the present study, we aimed to obtain the full-length transcriptome data for White Cloud Mountain minnow using long-read Pacific Biosciences (PacBio) isoform sequencing (Iso-seq) technology. This method is ideal for generating a reference transcriptome for species without whole-genome sequencing data (Workman et al., 2018). In addition, full-length transcriptomic method can obtain more accurate sequences of transcripts compared with common RNA-seq. We believe that the full-length transcriptome data reported here could be helpful for future studies related to various topics, including conservation biology, comparative transcriptome and phylogeny studies related to White Cloud Mountain minnow.
Data Description
Sample Collection
Fish of the White Cloud Mountain minnow (Tanichthys albonubes) were collected from Guangzhou Conghua Nature Reserve for T. albonubes with approval from its management council (Liangkou, Conghua, Guangzhou, Guangdong, China). One juvenile (sex information unknown) and two adult individuals (one female and one male) were caught on May 5 2020 using a hand net. Body weight of these fish ranged from 0.083 to 0.169 g with body length from 1.75-2.27 cm. Their physical conditions were examined by eye to avoid any body damage and disease.
RNA Extraction and Single-Molecule Real-Time Sequencing
All experimental protocols and animal welfare were approved by the Animal Care and Use Committee of South China Normal University. The five tissues (liver, muscles, skin, brain and gills) were frozen in liquid nitrogen immediately after collection, and stored at −80°C in a refrigerator before further use. We mixed the frozen tissues and RNA was isolated using Trizol (Invitrogen, CA, USA) following the manufacturer’s protocols. One μg of total RNA extracted from each of the three individuals was equally pooled together. The RIN value of the extracted RNA was ≥ 8.0. The extracted RNA was used for cDNA synthesis using a SMARTer PCR cDNA Synthesis kit (Clontech, USA). PCR cycles were optimized by setting 10-14 cycles and 12 PCR cycles were finally selected as the cycle parameter for large-scale PCR amplification using PrimeSTAR GXL DNA Polymerase (Clontech, USA). PCR products were purified using 1* and 0.4* AMPure PB Beads. Corresponding PCR products were mixed equally for downstream library preparation. After purification, the cDNA products were then subjected to the construction of SMRTbell template libraries using the SMRTbell Template Prep Kit 1.0 (PacBio). DNA damage and end of cDNA were repaired, which followed by ligation of adapters. Finally, SMRT cells were sequenced on a PacBio Sequel platform using sequencing kit 2.1 with 10 h movie recordings.
Full-Length Transcriptome Assembly
Raw sequencing reads were first processed using the SMRTlink version 6.0 (PacifificBiosciences, Menlo Park, CA, USA) and CCSs (circular consensus sequences) were generated from subread BAM files with the following parameters: pbccs.task_options.max_length=20000, pbccs.task_options.min_length=300. The CCSs were then trimmed and clustered to obtain FLNC (full-length non-chimeric) reads. These reads were further classified into high-quality FLNC reads (predicted accuracy > 99% and supported by at least two full-length reads) and low-quality FLNC reads. Finally, Unigenes were obtained by clustering isoform sequences (identify=98%) using CD-HIT version 4.6.7 (parameters: -c 0.98 -T 6 -G 0 -aL 0.90 -AL 100 -aS 0.98 -AS 30 -M 0 -d 0 -p 1). The dataset of Unigenes (full-length transcripts) was used for the downstream analysis. The completeness of the assembly was assessed using BUSCO version 3.0.1 (Simão et al., 2015) with default parameters.
Functional Annotation
To annotate the full-length transcriptome, the Unigenes sequences were searched in six public databases, including the NCBI non-redundant protein (NR) (http://www.ncbi.nlm.nih.gov), the Swiss-Prot protein (http://www.expasy.ch/sprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg), the Clusters of Orthologous Groups (COG)/EuKaryotic Orthologous Groups (KOG) (http://www.ncbi.nlm.nih.gov/COG) and the evolutionary genealogy of genes: Non-supervised Orthologous Groups database (http://eggnog.embl.de/version_3.0/) and Gene Ontology (GO) (http://geneontology.org/) using the Diamond program version 0.9.7 (Buchfink et al., 2015) (parameters: –more-sensitive -k 10 -e 1e-5). We set an E-value threshold of 1e-5 to match the protein sequences with the highest similarity in the databases.
Prediction of Protein-Coding sequences and Long Non-Coding RNA
The protein-coding sequence (CDSs) of the full-length transcriptome was predicted by BLAST (parameters: -max_target_seqs 1 -evalue 1e-10) search against the NR, KOG and the SwissProt protein databases one by one. If transcripts could be mapped to any one of these three databases, then they will be translated to protein sequences according to standard genetic codes. For those transcripts that failed to match any sequences in the three databases, we used the ESTscan program version 3.0.3 to predict their protein-coding regions (Iseli et al., 1999) with default parameters. To predict long non-coding RNA (lncRNA), CNCI version 2.0 (parameters: -m ve) was used to identify lncRNA and CPC2 version 0.92r2 with default parameters was used to access the protein-coding potential of transcripts. Transcripts with protein-coding potential will then be excluded. PLEK version 1.2 and Pfam version v2015-06-02 (parameters: -e_seq 0.001) were also performed to facilitate exclusion of protein-coding transcripts. Transcripts that were not predicted as CDSs and had length > 200bp were used in the lncRNA analysis. Transcription factors (TFs) and TF families were annotated by comparing transcripts against the Animal Transcription Factor Database version 2.0 (Zhang et al., 2015). To facilitate the use of the full-length transcriptome, we also annotated microsatellites (Simple Sequence Repeats, SSRs) in the transcriptome using the MIcroSAtellite server (http://pgrc.ipk-gatersleben.de/misa/) (MISA, version 1.0) with default parameters. Primer3 version 2.4.0 (parameters: -default_version=2 -io_version=4) was used to design primer pairs for subsequent SSRs validation (Rozen and Skaletsky, 2000).
Results
PacBio Sequencing Data and Full-Length Transcriptome Assembly
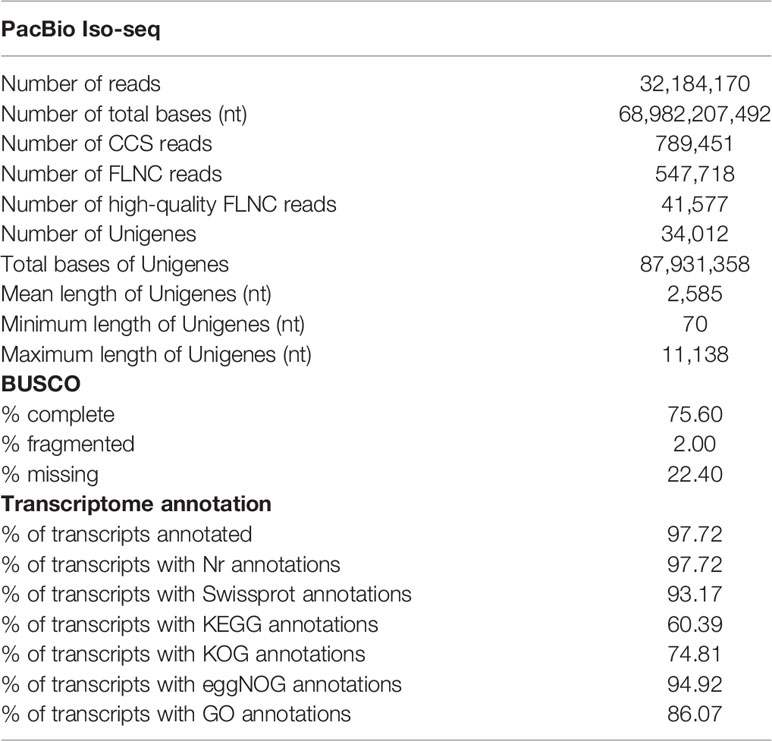
We obtained a total of 32,184,170 reads (68,982,207,492 nt in total) from the PacBio Sequel platform, read length ranging from 51 nt to 238,624 nt with mean length of 2,143 nt (Supplementary Figure 1A). Raw sequencing data have been submitted to the NCBI under the project number PRJNA786952. All subreads were then transferred and 789,451 CCS reads were generated (Table 1). Of these CCS reads, 547,718 were filtered as FLNC reads and finally 41,577 CCS reads were obtained as high-quality FLNC reads. After clustering, 34,012 high-quality FLNC reads (87.93 Mb of nucleotides) were retained and considered as Unigenes, which had a size ranging from 70 nt to 11,138 nt with a mean length of 2,585 nt (Supplementary Figure 1B). BUSCO analysis showed that the completeness of the White Cloud Mountain minnow full-length transcriptome was 75.60%, i.e., of the 4,584 conserved eukaryotic genes searched, 3,462 were found as “complete” in the transcriptome (Supplementary Figure 2). Our full-length transcriptome had a higher proportion of “missing” orthologs (22.40%) than those reported in other full-length transcriptomes from Cypriniformes fishes (e.g., 10.02% in Ictiobus cyprinellus, Ge et al., 2021a; 11.25% in Myxocyprinus asiaticus, Ge et al., 2021b).
Functional Annotations
The full-length transcriptome of the White Cloud Mountain minnow was annotated in six databases. 33,238 full length transcripts (97.72%) were successfully annotated (Table 1 and Supplementary Table 1). Of these annotated transcripts, 31,690 (93.17%), 20,539 (60.39%), 25,443 (74.81%), 32,283(94.92%) and 29,273(86.07%) transcripts were found in the Swissprot, KEGG, KOG, eggNOG and GO databases, respectively (Supplementary Table 1). In the NR database annotation, the top ten species of the annotated results were Sinocyclocheilus rhinocerous (6,776, 20.49%), Sinocyclocheilus anshuiensis (6,181, 18.69%), Carassius auratus (4,929, 14.91%), Cyprinus carpio (4,806, 14.5%), Danio rerio (3,930, 11.89%), Sinocyclocheilus grahami (3,670, 11.1%), Ctenopharyngodon idella (556, 1.68%), Tanichthys albonubes (193, 0.58%), Pygocentrus nattereri (184, 0.56%) and Megalobrama amblycephala (167, 0.51%) (Supplementary Figure 3A).
In the KOG database annotations, transcripts annotated (25,443) were classified into 25 categories, of which “General function prediction only” showed the highest proportion and “Cell motility” had the lowest percentage (Supplementary Figure 3B). Function annotations in the eggNOG database revealed that the majority of transcripts were functionally unknown and three categories had no annotation results (Supplementary Figure 3C).
GO function annotations showed that 25,396, 28,100 and 25,033 transcripts were divided into Biological Process, Cellular Component and Molecular Function, respectively. Among the Biological Process category, cellular process accounts for the highest annotations (Figure 1A). Annotation results from the KEGG database could be divided into five main classes, including 34 secondary classes (Figure 1B). Most of the transcripts assigned were involved in signal transduction.
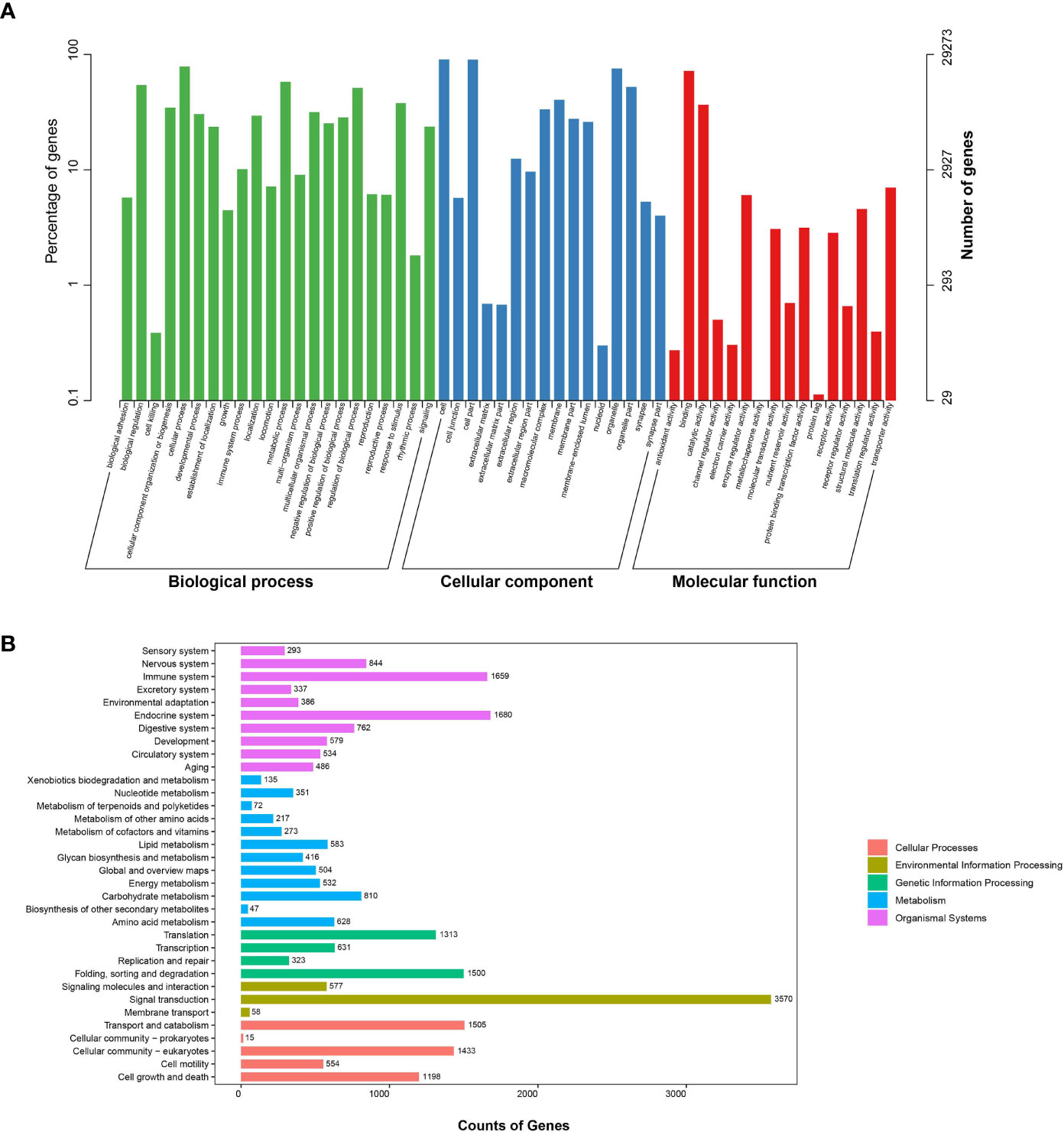
Figure 1 Functional classification of Tanichthys albonubes transcriptome. (A) Two level of GO functional annotations of full-length transcripts. (B) KEGG classification of full-length transcripts.
Prediction of Protein-Coding sequences (CDSs) and Long Non-Coding RNA
33,724 transcripts were predicted as CDSs, of which 33,244 transcripts were predicted by BLAST-based method and 480 transcripts were predicted by ESTscan program (Supplementary Figure 4). Analysis of lnRNA based on four software showed 317 transcripts were lnRNA (Supplementary Figure 5A), with a length range from 203 nt to 5524 nt (mean: 899 nt) (Supplementary Figure 5B) and GC content frequency distribution around 40 (Supplementary Figure 5C). A total of 4,878 TFs and 66 TFs families were obtained in the annotation (Supplementary Figure 6). The top species annotated to was Danio rerio (1,853, 37.99%) (Supplementary Figure 6A) and the first three TFs families were zf-C2H2 (844), Homeobox (401), bHLH (361) (Supplementary Figure 6B). The results of TFs may be useful in researches analyzing gene regulations. Analyses of SSRs revealed that there were six SSRs types in the full-length transcriptome of the White Cloud Mountain minnow (Supplementary Figure 7). The most frequent type was the one unite size and the number of SSRs contained more than two unite size was 16,968. This kind of information will guide marker selection in future population genetic studies of the White Cloud Mountain minnow.
Reuse Potential
We reported the first full-length transcriptome for the White Cloud Mountain minnow (Tanichthys albonubes) using PacBio long-read single molecule real-time (SMRT) sequencing. Our data of assembly and annotation results could benefit future studies regarding comparative transcriptome and whole-genome sequencing of the White Cloud Mountain minnow and its relatives. In addition, this study provides transcriptomic resources of species in the family Tanichthyidae for the first time. Therefore, phylogeny inferences involving this important fish taxon will greatly benefit from the full-length transcriptome we reported. Further studies of gene expression of this species such as analyzing the events of alternative splicing (AS) isoforms could also be achieved by reusing our data.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI [accession: PRJNA786952].
Ethics Statement
The animal study was reviewed and approved by School of Life Sciences, South China Normal University.
Author Contributions
JZ and CL contributed to the study design. CL, SZ, and JJ contributed to the sample preparation and molecular experiments. CL and HX performed the bioinformatic analyses. CL wrote the draft of the manuscript with assistance from JW and HX. All authors read and approved the final manuscript.
Funding
This work was supported financially by the National Natural Science Foundation of China (No. 31772430, No. 31372178) and the Key Project of Science-Technology Basic Condition Platform from the Ministry of Science and Technology of the People’s Republic of China (No. 2005DKA21402).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Shuying Jiang, Mr Chen and Yang at Guangzhou Conghua Nature Reserve for Tanichthys albonubes for assistance in the sample collection. We thank Shanghai Oebiotech Co., LTD for advice on data analysis and technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.831148/full#supplementary-material
Supplementary Data | All the assembly and annotation results are deposited to FigShare: https://doi.org/10.6084/m9.figshare.17109305.v3.
Supplementary Table 1 | Annotation results of the White Cloud Mountain minnow full-length transcriptome based on six databases.
Supplementary Figure 1 | (A) Length distribution of raw reads from PacBio Sequel platform. (B) Length distribution of high-quality FLNC reads.
Supplementary Figure 2 | BUSCO assessment results of the White Cloud Mountain minnow full-length transcriptome.
Supplementary Figure 3 | Annotation of the White Cloud Mountain minnow full-length transcriptome in three databases. (A) Top ten species distribution annotated in NR database. (B) Statistics of KOG function classification. (C) Barplot of eggNOG function classification.
Supplementary Figure 4 | Sequence length distribution of protein-coding sequences. (A) Sequence length distribution of CDSs based on BLAST method. (B) Sequence length distribution of CDSs based on ESTscan program.
Supplementary Figure 5 | Analysis of lnRNA. (A) Venn diagrams of non-coding RNA predicted by four software. (B) Length distribution of lncRNA. (C) GC content frequency distribution.
Supplementary Figure 6 | Analysis of Transcription factors (TFs) and TF families. (A) Top ten species distribution of TFs annotated. (B) TFs families distribution.
Supplementary Figure 7 | SSR type statistics.
References
Bohlen J., Dvor ák T., Thang H. N., Šlechtov á V. (2019). Tanichthys Kuehnei, New Species, From Central Vietnam (Cypriniformes: Cyprinidae). Ichthyol. Explor. Freshw. IEF-1081, 1–10. doi: 10.23788/IEF-1081
Buchfink B., Xie C., Huson H. D. (2015). Fast and Sensitive Protein Alignment Using Diamond. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chan B., Chen X. (2009). Discovery of Tanichthys albonubes Lin 1932 (Cyprinidae) on Hainan Island and Notes on its Ecology. Zool. Res. 30, 209–214. doi: 10.3724/SP.J.1141.2009.02209
Chen M., Bai J. J., He X. P., Jian Q., Ye X., Wu L. X., et al. (2009). Study on the Genetic Analysis and Biology of Transgenic Tanichthys Albonubes Expressing the Red Fluorescent Protein Gene. J. Shanghai Ocean Univ. 18 (6), 649–654.
Freyhof J., Herder F. (2001). Tanichthys micagemmae, A New Miniature Cyprinid Fish From Central Vietnam (Cypriniformes: Cyprinidae). Ichthyol. Explor. Freshw. 12, 215–220.
Ge H., Zhang H., Yang L., Wang H., Tu L., Jiang Z., et al. (2021a). Full-Length Transcriptome Sequencing From the Longest-Lived Freshwater Bony Fish of the World: Bigmouth Buffalo (Ictiobus Cyprinellus). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.736188
Ge H., Zhang H., Zhao Q., Li F., Gu H., Liu S., et al. (2021b). Construction of a Full-Length Transcriptome Resource for the Chinese Sucker (Myxocyprinus Asiaticus), a Rare Protected Fish, Based on Isoform Sequencing (Iso-Seq). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.699504
Harris R. B. (2008). Wildlife Conservation in China: Preserving the Habitat of China's Wild West (New York, NY: M. E. Sharpe).
Iseli C., Jongeneel C. V., Bucher P. (1999). “Estscan: A Program for Detecting, Evaluating, and Reconstructing Potential Coding Regions in EST Sequences,” in International Conference on Intelligent Systems for Molecular Biology, 138–148.
Jing J., Liu H. C., Chen H. H., Hu S. F., Xiao K., Ma X. F. (2013). Acute Effect of Copper and Cadmium Exposure on the Expression of Heat Shock Protein 70 in the Cyprinidae Fish Tanichthys Albonubes. Chemosphere 91 (8), 1113–1122. doi: 10.1016/j.chemosphere.2013.01.014
Li C., Jiang S., Schneider K., Jin J. J., Lin H. D., Wang J., et al. (2020b). Cryptic Species in White Cloud Mountain Minnow, Tanichthys albonubes: Taxonomic and Conservation Implications. Mol. Phylogenet. Evol. 153 (12), 106950. doi: 10.1016/j.ympev.2020.106950
Li C., Jin J. J., Luo J. Z., Wang C. H., Wang J. J., Zhao J. (2020a). Genetic Relationships of Hatchery Populations and Wild Populations of Tanichthys albonubes Near Guangzhou. Biodivers. Sci. 28 (4), 474–484. doi: 10.17520/biods.2019290
Li J., Li X. H. (2011). A New Record of Fish Tanichthys Albonubes (Cypriniformes: Cyprinidae) in Guangxi, China. Chin. J. Zool. 46, 136–140. doi: 10.13859/j.cjz.2011.03.022
Lin S. Y. (1932). New Cyprinid Fishes From White Cloud Mountain, Canton. Lingnan Sci. J. 11, 379–383.
Liu H. C., Chen H. H., Qin J. H., Ma X. F. (2011). Cloning and Expression of Heat Shock Protein 60 cDNA of Tanichthys Albonubes. J. Huazhong Agric. Univ. 30 (5), 635–639. doi: 10.13300/j.cnki.hnlkxb.2011.05.005
Luo J. Z., Lin H. D., Yang F., Yi Z. S., Chan B. P., Zhao J. (2015). Population Genetic Structure in Wild and Hatchery Populations of White Cloud Mountain Minnow (Tanichthys albonubes): For Conservation. Biochem. Syst. Ecol. 62, 142–150. doi: 10.1016/j.bse.2015.08.008
Mayden R. L., Chen W. J. (2010). The World's Smallest Vertebrate Species of the Genus Paedocypris: A New Family of Freshwater Fishes and the Sister Group to the World's Most Diverse Clade of Freshwater Fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 57 (1), 152–175. doi: 10.1016/j.ympev.2010.04.008
Nguyen V. H., Ngo S. V. (2001). Freshwater Fishes of Vietnam. Volume I. Family Cyprinidae (Hanoi: Aquaculture Publishing House), 622 pp.
Rozen S., Skaletsky H. (2000). Primer3 on the WWW for General Users and for Biologist Programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: Assessing Genome Assembly and Annotation Completeness With Single-Copy Orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Tan M., Armbruster J. W. (2018). Phylogenetic Classification of Extant Genera of Fishes of the Order Cypriniformes (Teleostei: Ostariophysi). Zootaxa 4476 (1), 6–39. doi: 10.11646/zootaxa.4476.1.4
Workman R. E., Myrka A. M., Wong G. W., Tseng E., Welch K. C. Jr., Timp W. (2018). Single-Molecule, Full-Length Transcript Sequencing Provides Insight Into the Extreme Metabolism of the Ruby-Throated Hummingbird Archilochus colubris. GigaScience 7, giy009. doi: 10.1093/gigascience/giy009
Yi Z. S., Chen X. L., Wu J. X., Yu S. C., Huang C. E. (2004). Rediscovering the Wild Population of White Cloud Mountain Minnows (Tanichthys albonubes Lin) on Guangdong Province. Zool. Res. 25, 551–555.
Zhang H. M., Liu T., Liu C. J., Song S., Zhang X., Liu W., et al. (2015). AnimalTFDB 2.0: A Resource for Expression, Prediction and Functional Study of Animal Transcription Factors. Nucleic Acids Res. 43, D76–D81. doi: 10.1093/nar/gku887
Keywords: White Cloud Mountain minnow, Tanichthys albonubes, protected fishes, full-length transcriptome, ISO-seq
Citation: Li C, Zhong S, Jin J, Xiao H, Wang J and Zhao J (2022) Full-Length Transcriptome Data for the White Cloud Mountain Minnow (Tanichthys albonubes) From a Wild Population Based on Isoform Sequencing. Front. Mar. Sci. 9:831148. doi: 10.3389/fmars.2022.831148
Received: 08 December 2021; Accepted: 28 March 2022;
Published: 22 April 2022.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Juan Andrés López, University of Alaska Fairbanks, United StatesZhongneng Xu, The University of Tokyo, Japan
Copyright © 2022 Li, Zhong, Jin, Xiao, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zhao, emhhb2p1bkBzY251LmVkdS5jbg==
 Chao Li
Chao Li Shenhang Zhong
Shenhang Zhong Jinjin Jin
Jinjin Jin Han Xiao
Han Xiao Junjie Wang1
Junjie Wang1 Jun Zhao
Jun Zhao