- 1South China Sea Environmental Monitoring Center, State Oceanic Administration, Guangzhou, China
- 2Nansha Islands Coral Reef Ecosystem National Observation and Research Station (NSICR), Guangzhou, China
By far, very few studies have dealt with the ichthyoplankton diversity in Guanghai Bay (China), which is a potential spawning ground for many important fish species. In this study, fish eggs collected in Guanghai Bay were identified through molecular method combined with visual taxonomic method. We employed two mitochondrial gene regions of cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S rRNA) as genetic markers for species identification. Through sequence identification at NCBI, 121 eggs with overlapping size range and easy to be confused were discriminated as four economically important species: seven as silver sillago Sillago sihama, 48 as black-banded sillago Sillago nigrofasciata, 38 as yellow drum Nibea albiflora, and 28 as Pacific seabream Acanthopagrus pacificus. Phylogenetic analyses showed that these 121 eggs clustered in four groups with strong support. To testify the validity of these identification results, species identification through five BarcodingR package methods was also carried out using sequences of 33 fish specimens as a reference library covering four target species. Finally, a highly consensus of species assignment results was achieved across different methods. Morphological characteristics and detailed photographs for eggs from these four species were supplied here. Eggs of each species are pelagic, round, have a smooth chorion and one single oil globule. Embryonic pigment patterns vary as eggs develop and can be used for species distinguishing. Eggs from S. nigrofasciata and A. pacificus were described for the first time in this study. One simple and accurate method for identifying N. albiflora eggs was additionally provided. Moreover, the morphological differences between two Sillago eggs offered supportive evidence for the recent separation of S. nigrofasciata as a new species from S. sihama. All these results would be critical for the discrimination of eggs from these four species and the estimation for their spawning areas. Meanwhile, our study would contribute to the stock assessment and fishery management in Guanghai Bay.
1 Introduction
Precise knowledge of spawning grounds and migration routes for fish species is of particular importance to fishery resource conservation and policies formulating (Fuiman and Werner, 2002; Fox et al., 2005). A traditional way to determine spawning spots and to estimate stock biomass sizes is based on direct observations of adult aggregation through trawl surveys. However, this method is not only time- and labor-consuming, and also ecologically impacting, particularly in the case of low stock abundance (Reeves and Pastoors, 2007; Goodsir et al., 2008). A fishery-independent approach using ichthyoplankton data to infer spawning grounds can reduce the bias caused by misreporting (Watson and Pauly, 2001; Karaiskou et al., 2007), as well as bring minor damage to the structure of fish populations (Lelievre et al., 2010). In contrast with larvae, fish eggs provide more accurate and direct information on the spawning spots because of their passive dispersal capacity in the water column (Petitgas et al., 2006; Richardson et al., 2009). Thus, an accurate identification of fish eggs is a prerequisite for the proper applications of this alternative approach.
The most commonly used approach for fish eggs identification is the direct observation of morphological characters, including egg size, number of oil globules, chorion structure, and width of the perivitelline space (Ikeda et al., 2014). However, it is usually challenging to assign fish eggs to species level through this method because of the paucity of egg morphology knowledge for many species and insufficient diagnostic traits for eggs in early developmental stages (Lelievre et al., 2010; Lewis et al., 2016; Leyva-Cruz et al., 2016).
Molecular analysis aiming at species identification through mitochondrial genetic markers, such as cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S rRNA), has been successfully applied on fish eggs (Kawakami et al., 2010; Lewis et al., 2016; Leyva-Cruz et al., 2016; Hofmann et al., 2017; Choi et al., 2018; Hou et al., 2020; Mateos-Rivera et al., 2020; Hou et al., 2021; Zheng et al., 2021). The most popular method is DNA barcoding based on COI gene analysis, which had been verified of the effectiveness in the taxonomic identification system for fish species (Ward et al., 2005; Ward et al., 2009; Hou et al., 2018). However, the species covered by previous studies were limited, and some of these studies only focused on the feasibility of analytical methods instead of the detailed morphology of fish eggs.
In addition, visual taxonomic features acquired only through DNA-based methods sometimes may not be correct, especially for oil globule numbers. Ethanol is often used as a sample fixative in molecular identification methods because of its high efficiency in preserving DNA. Nonetheless, some morphological changes such as oil globule dissolution, chorion shrinkage, and opaque yolk sac are frequently observed in ethanol-fixed eggs (Lewis et al., 2016; Leyva-Cruz et al., 2016; Zheng et al., 2021). During the last years, successful identification of formalin-fixed eggs through DNA-based methods had been reported (Akimoto et al., 2002; Perez et al., 2005; Karaiskou et al., 2007; Lelievre et al., 2010; Hofmann et al., 2017). However, the efficacy rate varied greatly from less than 20% to 98% among these studies, and the amplification length of some targeted genes was often limited to a small fragment (Akimoto et al., 2002; Perez et al., 2005). Besides, the lengthy procedure and high experimental costs hindered the application of these methods. An alternative solution was to employ formalin as a reference fixative to achieve precise morphological knowledge for fish eggs (Zheng et al., 2021).
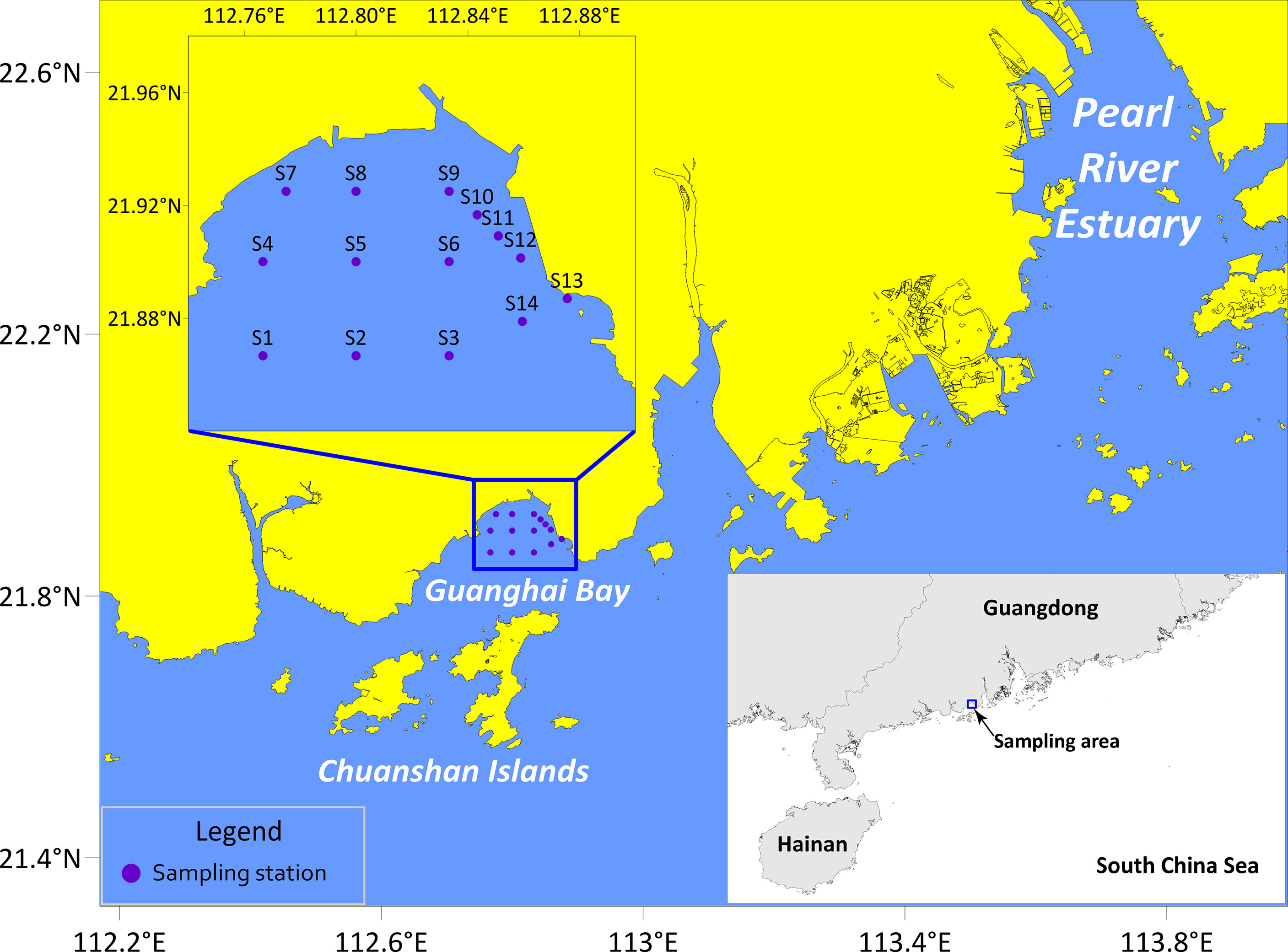
Guanghai Bay (Jiangmen City, Guangdong Province, China) is a semi-enclosed bay with a coastal line of 92.8 km. It is adjacent to the Chuanshan Islands and located to the west of the Pearl River Estuary (Figure 1). The mouth of the bay faces the South China Sea, the east and west banks are framed by mountainous terrains, and the north connects to the inland plain. Most parts of this bay are less than five meters deep, and the maximum water depth is six meters (Chen, 1999). As indicated in Guangdong Economy (2014) report, the Greater Guanghai Bay Economic Zone will be built into a major platform for Guangdong-Hong Kong-Macao cooperation on the west bank of the Pearl River by 2030. With the construction of cargo ports and coastal power plants, and the rapid development of marine aquaculture, transportation, and coastal tourism, human activities have adversely affected the aquatic environment of Guanghai Bay.
The previous report suggested that many economically important fishes spawn in this sea area (Guo et al., 1994). Our surveys on the ichthyoplankton in this area in the winter season revealed that this bay might be the spawning ground of Japanese gizzard shad Nematalosa japonica Regan, 1917, dotted gizzard shad Konosirus punctatus (Temminck and Schlegel 1846), black porgy Acanthopagrus schlegelii (Bleeker 1854), and ovate sole Solea ovata Richardson 1846, as well as an essential nursery habitat for yellowfin seabream Acanthopagrus latus (Houttuyn 1782) (unpublished data). However, very few studies have dealt with fish eggs identification in Guanghai Bay during the last fifteen years except for previous investigations on Commerson’s anchovy Stolephorus commersonii Lacepède 1803 (Tian et al., 2017), S. ovata and A. schlegelii (Zheng et al., 2021). Hence, survey on the ichthyoplankton in this area is urgently important for the estimation of stock loss and fishery resource conservation.
The main objectives of this study are (i) to identify fish eggs of economically important species collected in Guanghai Bay in the winter season using molecular analyses based on mitochondrial COI gene and 16S rRNA gene, (ii) to discriminate formalin-fixed eggs according to the morphological features displayed by ethanol-fixed eggs, (iii) to generalize visual taxonomic characteristics and provide egg photographs based on ethanol- and formalin-fixed eggs.
2 Materials and Methods
2.1 Sample Collection
All fish eggs used in this study were sampled in Guanghai Bay (21°52ʹ00˝-21°55ʹ30˝ N, 112°46ʹ00˝-112°52ʹ32˝ E) during winter surveys in February 2019, December 2019, and January 2020 (Figure 1). Sampling effort was carried out on fourteen stations with surface trawling at a speed of 3.7 km h-1 (2 knots) for 10 min per station. A zooplankton net of a 505 μm mesh size with a mouth diameter of 80 cm and net length of 2.8 m was used. Samples were randomly fixed with 99% ethanol for molecular analyses and morphological observations or with 4% formalin for morphological reference.
For the reference library, fish specimens of the target species and congeneric species were obtained from fishermen and seafood markets around the Guanghai Bay. All samples were labeled with internal code, photographed, and stored at -20°C.
2.2 DNA Extraction and PCR Conditions
Total genomic DNA was extracted from each of the ethanol-fixed eggs and muscle tissue of each adult specimen using 180 µl of NaOH solution (50 mM). After being heated at 95°C for 10 min, 20 µl Tris-HCl buffer (1 M, pH 8.0) was added to the mixture, followed by centrifugation at 13,362 g (12,000 rpm) for 10 min. The supernatant was further used as a DNA template for Polymerase Chain Reactions (PCR). A ca. 650 base pair (bp) fragment of the COI gene was amplified using primers described in Ward et al. (2005). The forward PCR primer was a mixture of primer FishF1 (5′-TCAACCAACCACAAAGACATTGGCAC-3′) and primer FishF2 (5′-TCGACTAATCATAAAGATATCGGCAC-3′). The mixture of primer FishR1 (5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′) and primer FishR2 (5′-ACTTCAGGGTGACCGAAGAATCAGAA-3′) was used as reverse PCR primer. The final concentration of each primer in the mix is 10μM. A ca. 570 bp fragment of the 16S rRNA gene was amplified using primers 16SAR (5′- CGCCTGTTTATCAAAAACAT -3′) and 16SBR (5′- CCGGTCTGAACTCAGATCACG -3′), according to Palumbi et al. (2002). The polymerase chain reactions (PCR) were carried out in a total volume of 50 μl, containing 25 μl of 2 × PCR Buffer, 10 μl of dNTPs (2 mM), 1.5 μl of each primer (10 μM each), 1 μl KOD FX PCR enzyme (TOYOBO, Japan), 2 μl of DNA template and 9 μl of ultrapure water.
The PCR conditions were designed according to the product manual provided by the manufacturer (TOYOBO, Japan). The thermal regime for COI amplification consisted of an initial denaturation at 94°C for 2 min, followed by 38 cycles of denaturation at 98°C for 15 sec, annealing at 55°C for 30 sec and extension at 68°C for 50 sec, followed in turn by a final extension at 68°C for 10 min. The PCR conditions for 16S rRNA were: 94°C for 2 min, 36 cycles of 98°C for 15 sec, 52°C for 30 sec and 68°C for 50 sec, and a final extension at 68°C for 10 min. The purification and bi-directional sequencing of PCR products were performed by Tsingke Biological Technology Inc. (Guangzhou, China).
To construct a reference library, both COI and 16S rRNA gene sequences were amplified for all fish specimens. Most of the early-stage eggs were identified only through 16S rRNA gene. Middle- and late-stage eggs were all identified by using COI sequence and part of them were additionally discriminated through 16S rRNA gene. See Supplementary Table 1 for the developmental stage of and mitochondrial genetic markers used for each egg.
2.3 Data Analysis and Species Identification
Sequences were assembled from forward and reverse reads by using Sequencher® (https://www.genecodes.com/). After trimming of the primer sequences, the final sequences were submitted to the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Supplementary Tables 1, 2). Sequences, trace files, and primer details of fish specimens were also submitted to the Barcode of Life Data systems (BOLD; http://www.boldsystems.org) (Ratnasingham and Hebert, 2007; Ratnasingham and Hebert, 2013) under the project RLSCS.
All egg sequences were differentiated through the Basic Local Alignment Search Tool (BLAST) at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi/). The NCBI sequences that had high similarity (≥99%) with the query sequences were regarded as candidate species (Hubert et al., 2015). The identifications were determined based on the distribution range and the current opinions on the taxonomic status of candidate species.
Sequences of candidate species, congeneric or sympatric species to the egg sample were retrieved from the GenBank (Supplementary Table 3). These downloaded sequences together with egg sequences were used to construct Bayesian inference (BI) trees with MrBayes v3.2.0 (Huelsenbeck and Ronquist, 2001). The family Serranidae was chosen as the outgroup taxa. Sequence alignments were carried out in BioEdit 7.05.3 (Hall, 1999) through Clustal W. Ambiguous gaps at both ends were removed from the alignments. The substitution models of TPM2f+I+G and TIM2e+G were selected respectively for the Bayesian procedure of COI and 16S rRNA by ModelFinder in IQ-TREE (Kalyaanamoorthy et al., 2017). The model TPM2f and TIM2e, which were not implemented in MrBayes, were replaced by the GTR model (Lecocq et al., 2013). Four Markov chains were run for 10,000,000 generations with a sampling frequency of every 1,000 generations. The first 2,500 trees (25%) were discarded as burn-in, and the remaining 7,500 trees were used to construct the 50% majority-rule consensus tree and to estimate the Bayesian posterior probabilities (PP). The BI trees were visualized using iTOL (Letunic and Bork, 2021).
To testify the validity of the identification results through BLAST, all egg sequences were additionally identified through R package BarcodingR version 1.0-3 (Zhang et al., 2017) in R 3.4.0 (R Core Team, 2017). All sequences of fish specimens we collected were used as reference library. The COI sequences were identified through three methods: (i) bpNewTraining (BP) (Zhang et al., 2008), (ii) fuzzyId (FZ) (Zhang et al., 2012), and (iii) Bayesian (BI) (Jin et al., 2013). The 16S rRNA sequences were identified through two methods applicable to non-protein-coding barcodes: BBSIK (parameters: kmer =10, UseBuiltModel = TRUE) (Zhang et al., 2008) and fuzzy-set method (FZKMER, parameters: kmer =10, optimization = TRUE) (Zhang et al., 2012). The 100% consensus of species identified (sp.Identified) through all methods was accepted as the species finalized.
Phylogenetic trees for sequences of eggs and fish specimens were constructed using the same method as described above. TPM2f+G and TIM2e+G were chosen as the best- fitting substitution models for COI and 16S rRNA sequences, respectively.
Sequence analyses for all egg and fish samples of target species were performed by using DnaSP v6 (Rozas et al., 2017), including number of invariable sites (i), variable sites (v), singleton sites (s), and parsimony informative sites (p), as well as haplotypes (h), haplotype diversity (Hd), and nucleotide diversity (π). Sequences alignments were implemented in MEGA X using Clustal W with default settings (Kumar et al., 2018). Genetic distances were calculated based on the Kimura 2-parameter model (K2P; Kimura, 1980) in MEGA X, and the “complete deletion” option was selected for the treatment of gaps and missing data (Nei and Kumar, 2000; Kumar et al., 2018).
2.4 Morphological Observations
Morphometric examinations on fish specimens were carried out following Hubbs and Lagler (2004). Species identifications were based on McKay (1985); McKay (1992); Iwatsuki et al. (2010); Kaga (2013); Chen and Zhang (2015), and Xiao et al. (2021). Common names and scientific names of fishes were, respectively, following FishBase (Froese and Pauly, 2022) and Eschmeyer’s Catalog of Fishes (Fricke et al., 2022).
Photographing and morphometric analyses of ethanol- and formalin-fixed eggs were carried out with a digital camera (Axiocam 512 color, Zeiss, Munich, Germany) connected to a stereoscopic microscope (Discovery V20, Zeiss, Jena, Germany). The ethanol-fixed eggs were rehydrated in distilled water for analysis in this step to eliminate the chorion shrinkage effect caused by ethanol.
The main characteristics used for initial sorting and identification included egg size, shape (spherical, ellipsoidal), chorion (ornamentation, texture), number of oil globules, pigment patterns, and perivitelline space according to Ikeda et al. (2014). Criteria from Ahlstrom and Moser (1980) were used to determine the three developmental stages of fish eggs: early (fertilization to blastopore closure), middle (from blastopore closure to the time that the separating tail begins to curve laterally away from the embryonic axis), and late (from the time that the tail is curved away from the embryonic axis to the time of hatching).
According to morphological features displayed by ethanol-preserved eggs, we identified the formalin-preserved eggs. Oil globule numbers of post-early-stage eggs preserved in ethanol were taken as a criterion because those of early-stage eggs were usually indiscernible. When the oil globule number was inconsistent among post-early-stage eggs preserved in ethanol, embryonic pigment patterns and egg size were the main characters used to discriminate formalin-fixed eggs. The oil globule number was finally determined through comparisons between ethanol- and formalin-fixed eggs. Oil globule characters of congeneric species previously reported were also taken into account. Visual taxonomic features were generalized based on morphological characteristics displayed by both formalin- and ethanol-fixed specimens.
3 Results
3.1 Species Identification
Thirty-three fish specimens were collected as a reference library and assigned to five species based on morphometric examinations: 19 as silver sillago Sillago sihama (Fabricius 1775), eight as black-banded sillago S. nigrofasciata Xiao, Yu, Song and Gao 2021, one as yellow drum Nibea albiflora (Richardson 1846), three as Pacific seabream Acanthopagrus pacificus Iwatsuki, Kume and Yoshino 2010, and two as A. schlegelii (Bleeker 1854) (Supplementary Figures 3–5). Photos of all fish specimens had been deposited in the BOLD under the project RLSCS.
A total of 733 eggs were collected throughout the three winter voyages, among which 391 eggs were preserved in ethanol and 342 were in formalin. Of the ethanol-fixed eggs, 121 eggs with the overlapping size range and easy to be confused were identified as four economically important species. We selected them for further investigation in this study. The other 270 ethanol-fixed eggs were identified as Stolephorus commersonii, A. schlegelii, Solea ovata, speckled tonguesole Cynoglossus puncticeps (Richardson 1846), and Liza sp. through BLAST in NCBI.
Totally, 168 sequences were obtained for the 121 ethanol-fixed eggs, including 61 sequences of COI and 107 sequences of 16S rRNA. Sixty-six sequences with 33 of COI and 33 of 16S rRNA genes were generated for fish specimens. After trimming of the primer sequences, a final length of 655 bp for the COI gene was obtained. The final length for the 16S rRNA gene varied from 576 bp to 578 bp among species. All sequences had no insertions/deletions, heterozygous sites, or stop codons.
Egg sequences differentiation via BLAST (analyses performed on 17 August 2021 and sequences of fish specimens not included) showed that among the 61 egg sequences of COI, five matched 99.69-100% with Sillago sihama, 30 matched 99.85-100% with Nibea albiflora, 18 matched 100% with Acanthopagrus pacificus and 99.85-100% with A. berda, and the last eight matched 100% with both S. asiatica and S. sihama.
Tree topology based on Bayesian inference analysis for COI gene revealed that these 61 egg sequences clustered into four groups with strong support values (PP=1.00) (Figure 2). Five sequences from fish eggs with two in early stage, one in middle stage and two in late stage, grouped in the clade of S. sihama in BI tree. Another 30 specimens (early: 4; middle: 23; late: 3) clustered in the clade of N. albiflora. These 35 eggs can be assigned to species level. Eighteen sequences (middle: 16; late: 2) grouped in one clade with sequences of A. pacificus (GenBank accession number [GBAN]: KY855483, MN520736) and A. berda (GBAN: KJ767796, KJ012251). According to reports from Iwatsuki et al. (2010), Iwatsuki and Heemstra (2010), Hsu et al. (2011), and Hasan et al. (2020), goldsilk seabream A. berda (Fabricius 1775) is distributed in the Indian Ocean, east to Malaysia (Penang Island and Langkawi Island, near Singapore). The fish species occurring in the Western Pacific Ocean and once identified as A. berda was a misidentification. Iwatsuki et al. (2010) had re-described it as a new species, A. pacificus. Those two specimens of A. berda (GBAN: KJ767796, KJ012251) may be inaccurately identified, and one of them (GBAN: KJ767796) had been corrected as A. pacificus in the study by Hasan et al. (2020). The last eight sequences of eggs (early: 3; late: 5) formed one lineage with sequences of S. sihama (GBAN: KU051808) and S. asiatica (GBAN: KU943693), which was due to the discordance with the identification of this species. According to the reports from Chang et al. (2017), these COI individual sequences were assigned to Asian sillago S. asiatica McKay, 1982, whereas Cheng et al. (2021) assigned them to S. sihama. Recently, Xiao et al. (2021) re-described it as a new species, S. nigrofasciata. On the other hand, Xiao et al. (2016) and Cheng et al. (2021) identified a different species as S. asiatica, and the COI sequences from this S. asiatica (GBAN: KM350211, KU051912) did not cluster with that from the former S. asiatica (GBAN: KU943693) identified by Chang et al. (2017) (Figure 2). We assigned these eight egg samples to species level as S. nigrofasciata according to the study from Xiao et al. (2021).
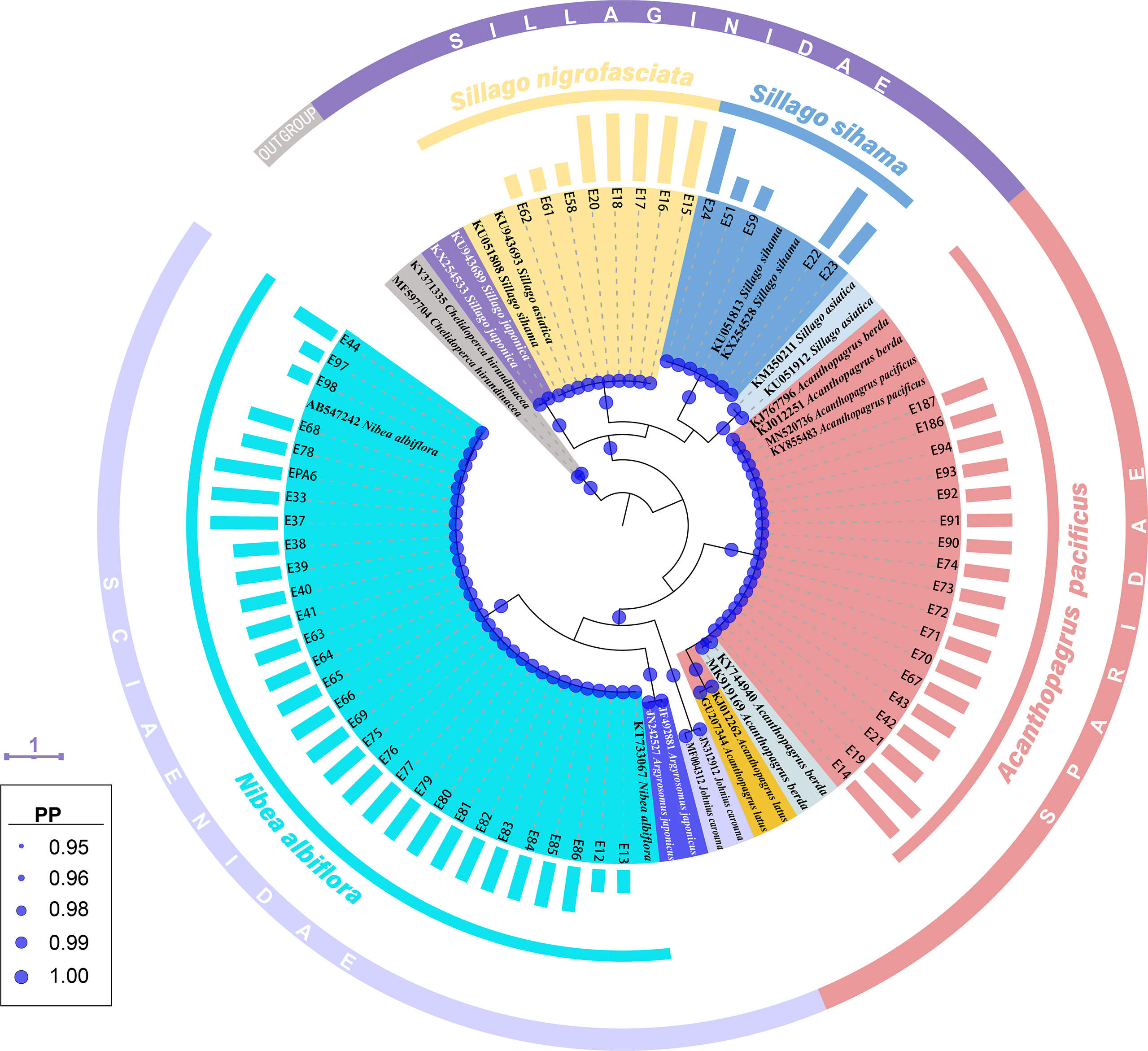
Figure 2 A Bayesian inference (BI) tree based on COI gene of 61 egg sequences combined with 24 sequences downloaded from the GenBank. The final alignments of 518 positions were used to construct this BI tree. The column bars have three sizes (short, medium, long) and represent three developmental stages (early, middle, late) respectively. The scale bar indicates the average number of nucleotide substitutions per site. Values of posterior probability (PP) >=0.95 are shown in circle.
Among the 107 sequences of 16S rRNA obtained for eggs, four sequences with a length of 577 bp matched 100% with S. sihama, 28 with a length of 576 bp matched 99.83-100% with N. albiflora, 27 with a length of 578 bp matched 99.82%~100% with A. pacificus, and the last 48 with a length of 577 bp matched 99.83-100% with S. sihama.
The same grouping results were present on the BI tree for 16S rRNA, where 107 fish egg sequences diverged into four clades with strong support values (PP=1.00) (Figure 3). Four sequences of early-stage eggs clustered in the clade of S. sihama, 27 (early: 10, middle: 15, late: 2) in the clade of A. pacificus, and 28 (early: 10, middle: 18) in the clade of N. albiflora. Referring to the identification results from COI sequence, these 59 eggs were identified to species level. Although the last 48 sequences (early: 43, late: 5) formed a clade with the sequence of S. sihama (GBAN: MF572035), they were assigned to S. nigrofasciata based on the report from Xiao et al. (2021).
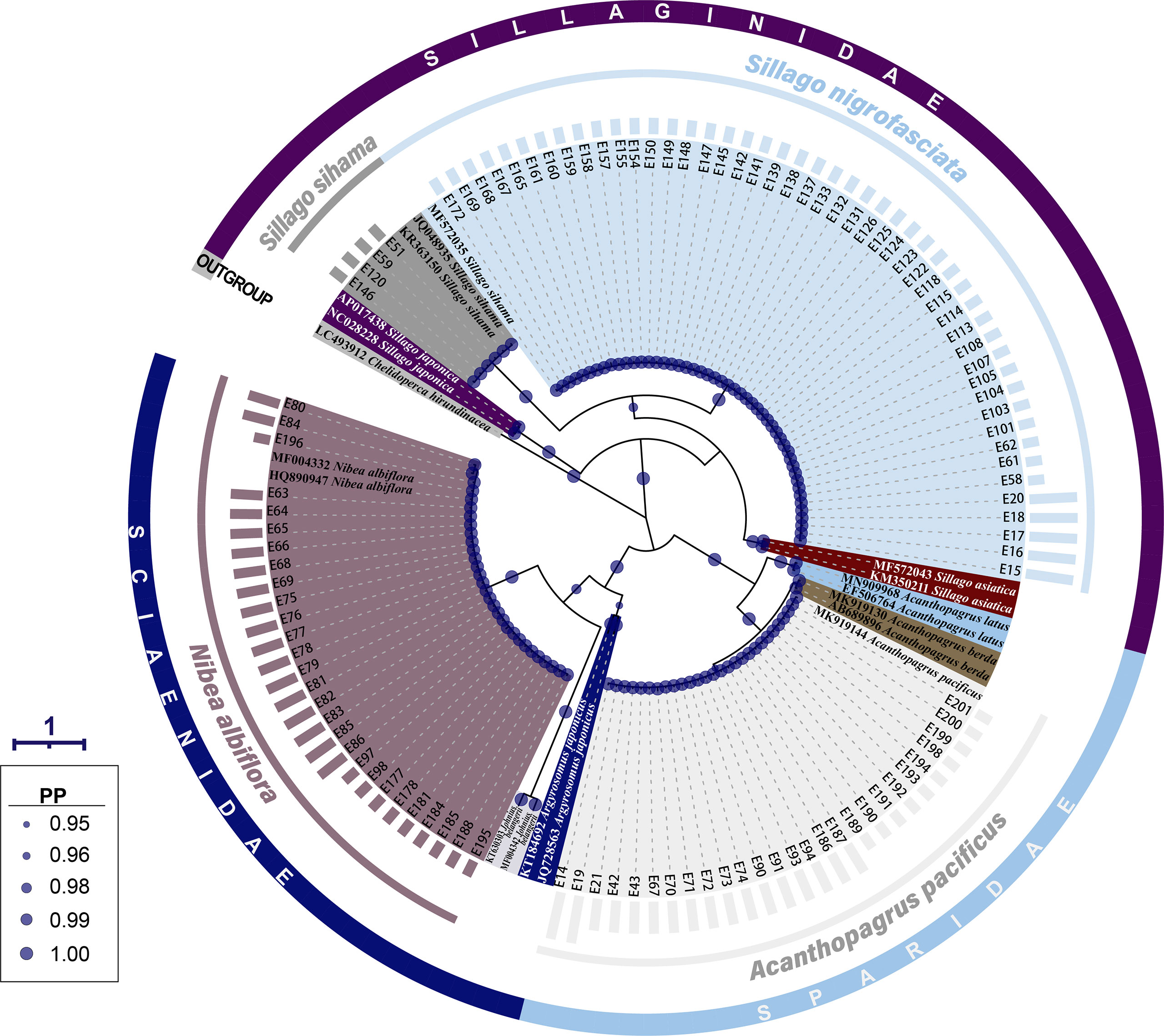
Figure 3 A Bayesian inference (BI) tree based on 16S rRNA gene of 107 egg sequences combined with 19 sequences downloaded from the GenBank. The final alignments of 574 positions were used to construct this BI tree. The column bars have three sizes (short, medium, long) and represent three developmental stages (early, middle, late) respectively. The scale bar indicates the average number of nucleotide substitutions per site. Values of posterior probability (PP) >=0.95 are shown in circle.
Using sequences of 33 fish specimens as a reference library, the identification results of eggs based on COI and 16S genes were highly consistent across different methods in BarcodingR (Table 1, Supplementary Tables 4, 5). The 121 ethanol-fixed eggs were classified as belonging to four species (S. sihama, S. nigrofasciata, N. albiflora, and A. pacificus). The assignment probability (or FMF) of each sequence ranged from a minimum of 0.887 (FZKMER method for egg E70) to a maximum of 1. BI method yielded the most successful assignment results with the probability value of 1 for all query sequences. The success rate values of the other four methods (BP, FZ, BBSIK, and FZKMER) were all 100%. Molecular phylogenetic trees based on COI and 16S rRNA sequences of these 121 eggs and 33 fishes showed congruent grouping results that all egg samples clustered in four clades with high support (PP >0.88) (Supplementary Figures 1, 2). Combing the morphometric and anatomic examinations on fish specimens (Supplementary Figures 3–5), we finally assigned these 121 eggs to four species: S. sihama, S. nigrofasciata, N. albiflora, and A. pacificus.
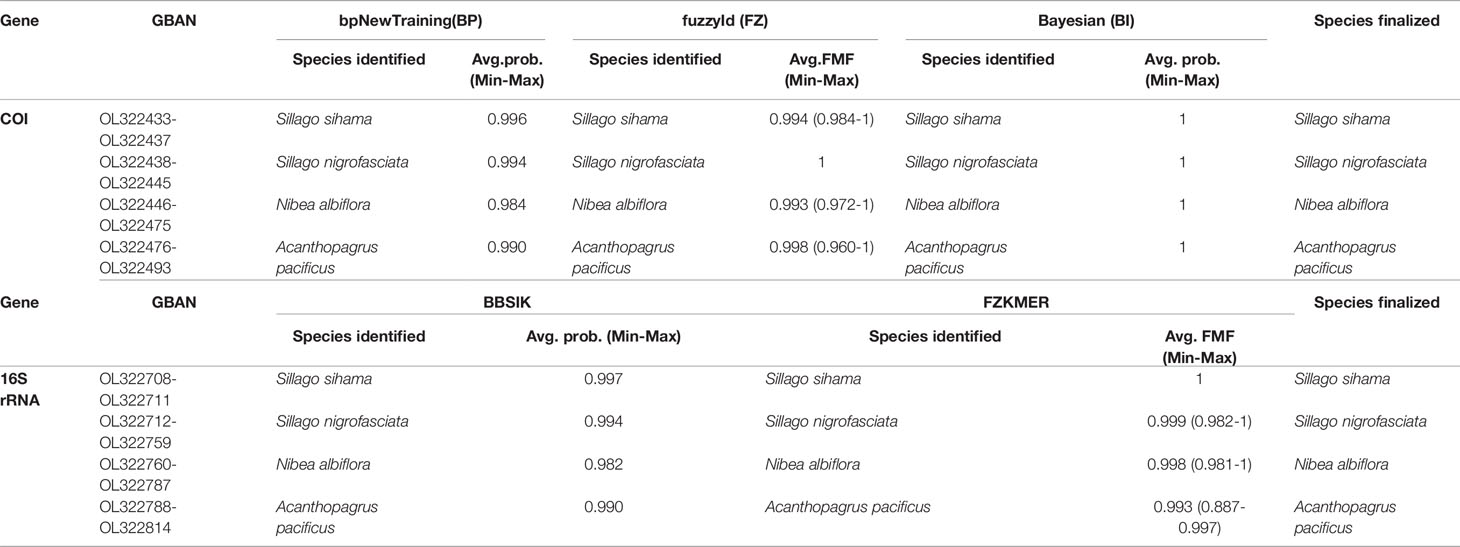
Table 1 Species identification through BarcodingR package with the assignment probabilities (average, minimum and maxium).
N. albiflora displayed high haplotype diversity in the COI gene with an Hd value of 0.641 (Table 2). In contrast, the other three species all attributed Hd values lower than 0.5 for both COI and 16S rRNA sequences. Smaller haplotype and nucleotide diversities were observed in the 16S rRNA sequences than COI sequences, except for A. pacificus. The mean intraspecific genetic distances of the four target species ranged from 0.01% to 0.14% for COI and from 0 to 0.04% for 16S rRNA. The mean pairwise divergence between species for COI ranged from 17.47% (between S. sihama and S. nigrofasciata) to 27.41% (between S. sihama and N. albiflora), and for 16S rRNA from 9.15% (between S. sihama and S. nigrofasciata) to 21.39% (between S. sihama and N. albiflora) (Table 3). All pairwise distances between species were greater than 8.94%, corresponding to the recommended threshold of >2.2% interspecific genetic divergence by Ratnasingham and Hebert (2013).
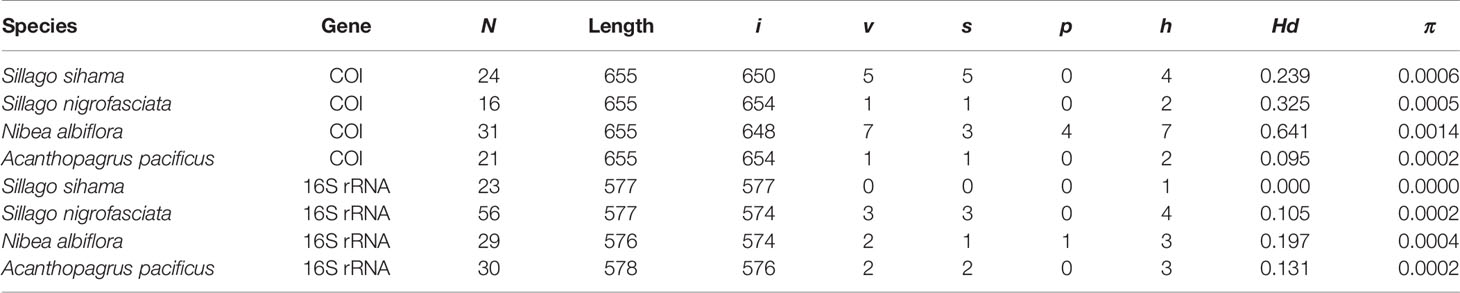
Table 2 Summary statistics of sequence analysis for eggs and fish specimens from four species (N, number of sequences; i, v, s, p, number of invariable sites, variable sites, singleton sites, and parsimony informative sites, respectively; h, number of haplotypes; Hd, haplotype diversity; π, nucleotide diversity).

Table 3 Interspecific divergence based on K2P distance (%) using COI sequences (lower matrix) and 16S rRNA sequences (upper matrix) of eggs and fish specimens from four species.
3.2 Visual Taxonomic Features of the Species
Eggs of these four target species are pelagic and spherical, generally with a smooth chorion and one single oil globule. Among the 121 ethanol-fixed eggs, 69 were in early stage, 40 in middle stage, and twelve in late stage. All the early-stage eggs displayed opaque yolk sac with no discernable oil globules. The absence of oil globule was also observed on 38.89% (seven out of 18) of A. pacificus eggs in post early stage, led by the dissolution effect from ethanol (Table 4 and Supplementary Table 1). According to the morphological features displayed by ethanol-fixed eggs, one late-stage egg of S. nigrofasciata, seven middle- and two late-stage eggs of N. albiflora, six middle- and three late-stage eggs of A. pacificus were discriminated from the formalin-fixed eggs. Early-stage eggs fixed in formalin cannot be guaranteed accurate identification. Visual taxonomic features of each species were acquired according to the morphological traits displayed by both ethanol- and formalin-fixed eggs. See Supplementary Table 1 for diameters of egg and oil globule of each sample.
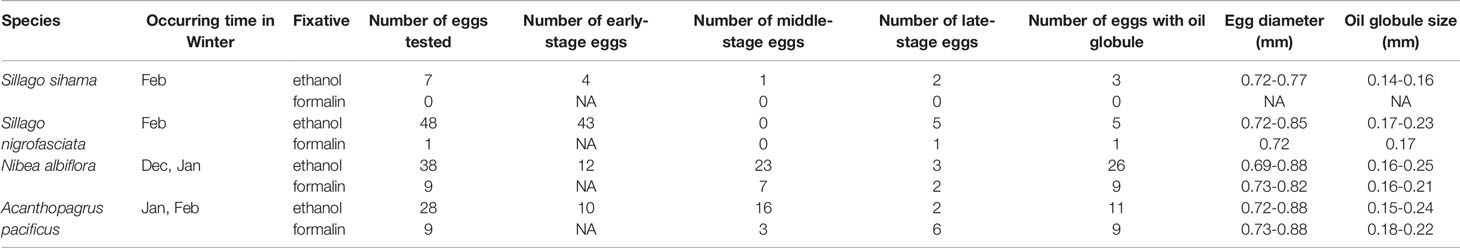
Table 4 Characters of eggs from four economically important fish species in Guanghai Bay, South China Sea (NA indicates no results achieved for a particular category).
3.2.1 Sillago sihama
Eggs of S. sihama range in diameter from 0.72 to 0.77 mm with one single oil globule measuring from 0.14 to 0.16 mm. In middle- and late-stage eggs, stellate melanophores are uniformly present on the oil globule and the dorsal surface of the embryo (Figures 4A–C). The embryos are initially straight with a blunt caudal end (Figure 4C), and curve as developing, with the tail gradually extending and approaching the oil globule (Figures 4A, B). The oil globule in some ethanol-fixed eggs may be invisibly pigmented due to the dissolution (Figures 4B, C). The spawning of this species in Guanghai Bay begins in February, much earlier than the time (April to May) recorded for this species in the South China Sea by Zhang et al.(1985). The egg size obtained in this study is also slightly larger than the dimensions (egg: 0.59-0.68; oil globule: 0.13-0.14) reported by Zhang et al.(1985), but corresponds closely with the described values (egg: 0.68-0.76; oil globule: 0.14-0.18) by Wan (1996). Although S. sihama eggs had been morphologically described with line drawings in previous reports, photographs for middle- and late-stage eggs were first provided in this study, contributing to egg identification of this species.
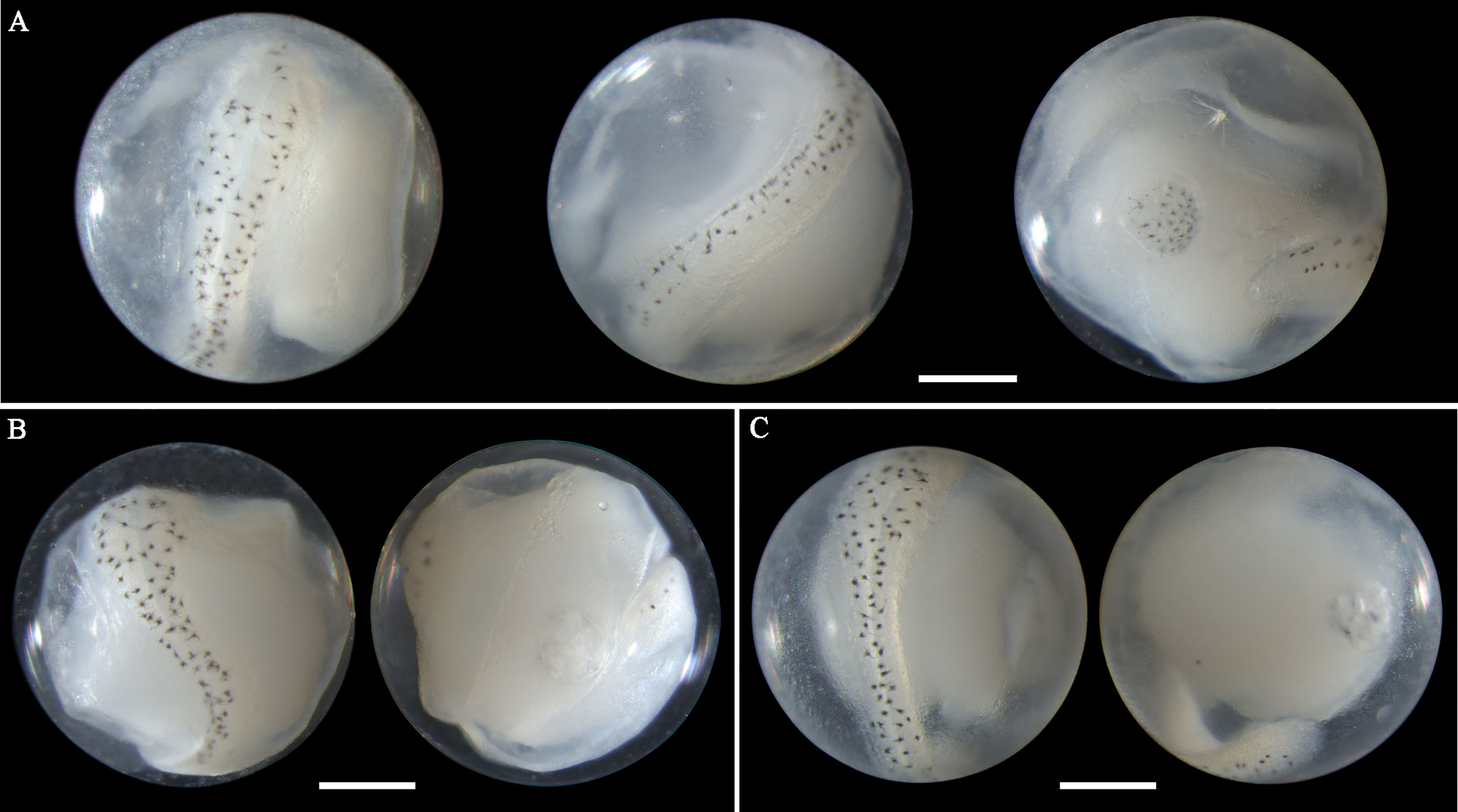
Figure 4 Images of ethanol-fixed eggs of Sillago sihama in (A) late stage with single pigmented oil globule, (B) late stage with single less-pigmented oil globule, and (C) middle stage with single less-pigmented oil globule. Images within each panel represent the same egg photographed from different observation views. Scale bar for images =200μm.
3.2.2 Sillago nigrofasciata
The morphology of eggs from this species has not been described in the literature. The egg diameter of this species ranges from 0.72 to 0.85 mm. The single pigmented oil globule measures 0.17-0.23 mm in size. In late-stage eggs, the embryonic melanophores extend from large stellate-shaped (Figure 5A) to branch-shaped (Figures 5B, C). Meanwhile, the bodies are more densely pigmented. When the embryos develop with finfolds, a series of branched pigments appear on the ventral midline of the trunk and tail (Figure 5C), which is a striking feature of eggs from this species. As the embryos extend and curve, the relative position of the oil globule varies from immediately backward of the caudal end (Figure 5A), through to ventrally close to the surface of the tail (Figure 5B), and to laterally near the trunk or tail (Figure 5C). This species in Guanghai Bay began to spawn in February.
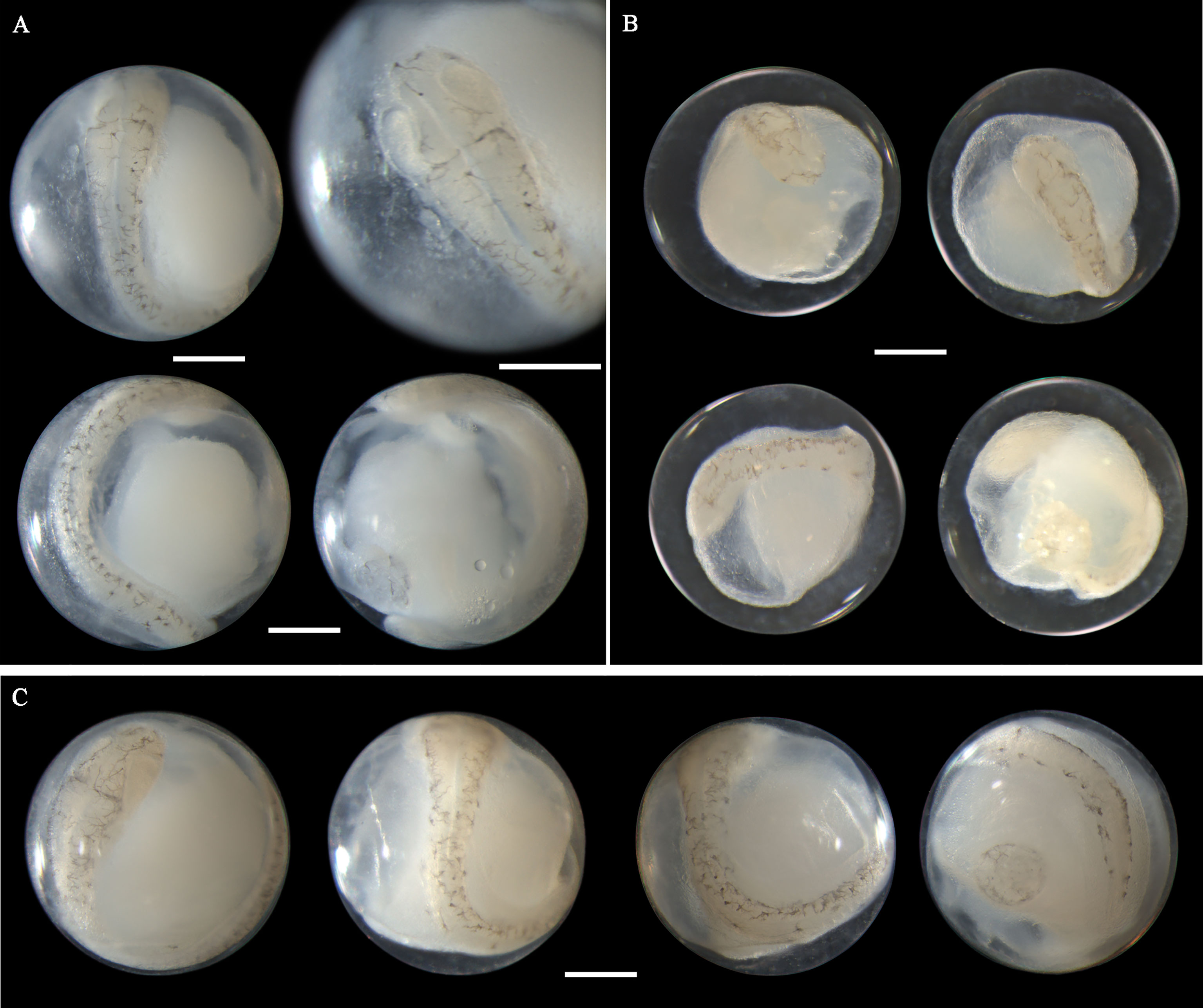
Figure 5 Images of late-stage eggs of Sillago nigrofasciata preserved in (A) ethanol, (B) formalin, and (C) ethanol. Images within each panel represent the same egg photographed from different observation views. Scale bar for images =200μm.
3.2.3 Nibea albiflora
Eggs of N. albiflora range in diameter from 0.69 to 0.88 mm with one single pigmented oil globule 0.16-0.25 mm. In early middle-stage eggs, punctate melanophores are restricted on the dorsal surface of the trunk and slightly packed on about the middle part (Figure 6A). As eggs develop, the embryonic melanophores enlarge and are present towards the head and tail region (Figures 6B, C). The pigment packing region appears more pronounced at this stage, and some melanophores on this region have changed to stellate-shaped. Similar to the former two species, the tail in developing eggs extends towards the oil globule. As eggs develop into late stage, stellate chromatophores appear continuous along the dorsal side of the embryo, and the melanophore clustering area is more heavily pigmented and more striking (Figures 6D, E). This unique morphological trait that pigments clustered in trunk area can conveniently distinguish this species from others. However, according to the reports from Lei and Fan (1981) and Zhang et al.(1985), four pigment-clustering areas are present on the embryo in live eggs of this species. This disparity may be due to the loss of natural pigments in the course of sample fixation. The egg size obtained at the present study closely agreed with dimensions (0.69-0.83mm in diameter) reported by Ikeda et al. (2014). Eggs of N. albiflora occurred in Guanghai Bay from December to January during the winter season.
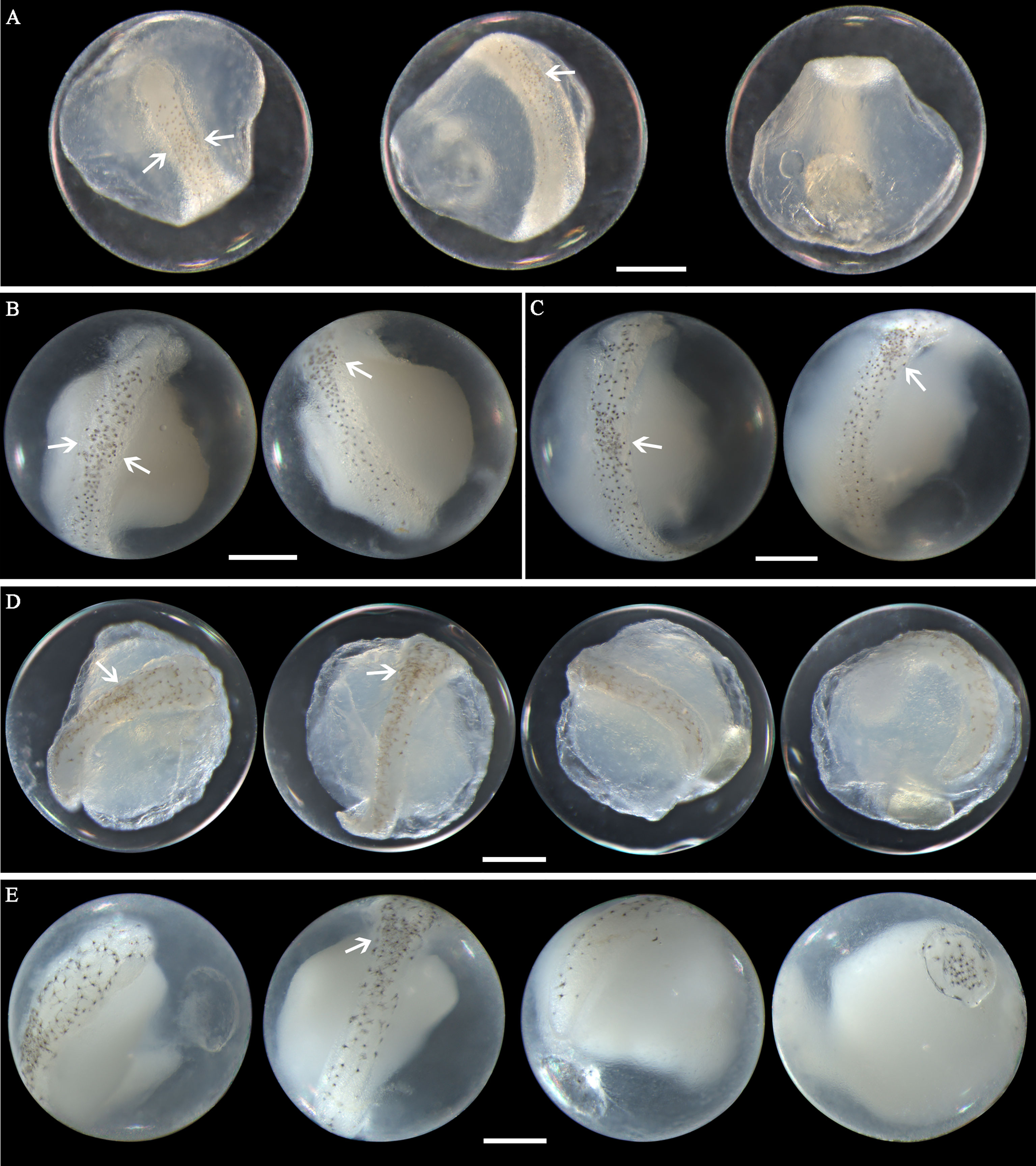
Figure 6 Images of Nibea albiflora eggs (A) fixed in formalin at early middle stage, (B) fixed in ethanol at middle stage, (C) fixed in ethanol at middle stage, (D) fixed in formalin at late stage, and (E) fixed in ethanol at late stage. Arrows indicate pigment clustering area on embryo. Images within each panel represent the same egg photographed from different observation views. Scale bar for images =200μm.
3.2.4 Acanthopagrus pacificus
Eggs from this species have not been morphologically described before. A. pacificus eggs range in diameter from 0.72 to 0.88 mm with one single pigmented oil globule 0.15-0.24 mm. In middle-stage eggs, punctate melanophores are distributed uniformly on the dorsal surface of the embryo (Figures 7A–D). The more developed eggs (Figure 7D) have larger and more widely scattered melanophores than eggs in the early middle stage (Figure 7A). Meanwhile, the tail in developing eggs extends towards the oil globule. As eggs develop to late stage, the melanophores change to branch-shaped and scatter all over the dorsal side of the embryo (Figures 7E–G). In eggs with finfolds, a few discontinuous melanophores ventrally appear on the tail and middle part of the trunk (Figure 7G). No pigmentation clustering area is present on the embryo throughout the developmental stages. Evenly scattered melanophores on the embryo are typical of eggs from this species. A. pacificus spawn in Guanghai Bay from January to February during the winter season.
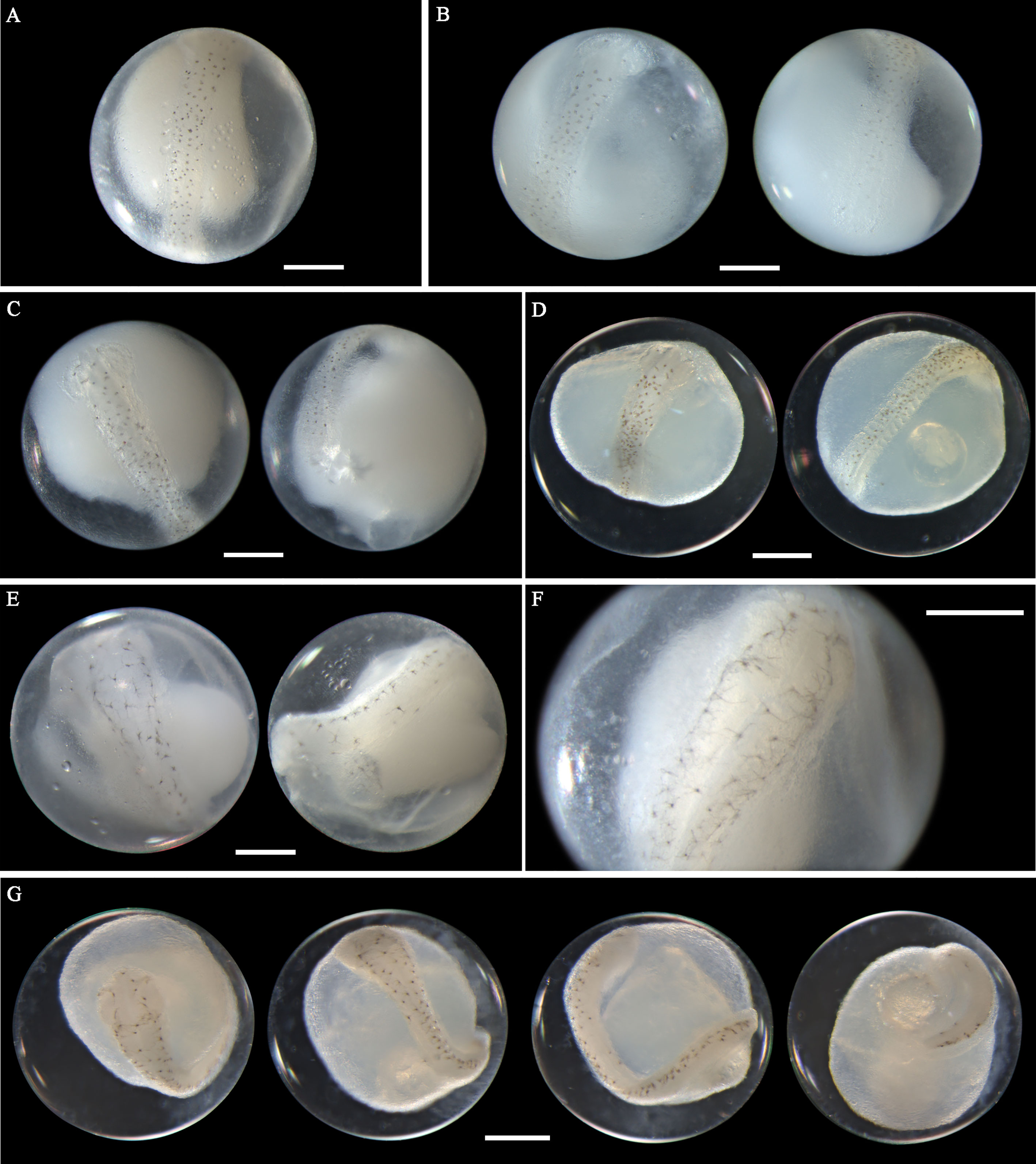
Figure 7 Images of Acanthopagrus pacificus eggs (A–C) fixed in ethanol at middle stage, (D) fixed in formalin at middle stage, (E, F) fixed in ethanol at late stage, and (G) fixed in formalin at late stage. Images within each panel represent the same egg photographed from different observation views. Scale bar for images =200μm.
4 Discussion
4.1 Characters for Egg Discrimination Between Morphologically Similar Species
Eggs from the four species in this study could be confused due to the overlapping range of their diameter and similar oil globule size (Table 4). Usually, myomere number is an essential characteristic in larvae identification, but this is neither practical nor convenient for eggs because of the insufficient myomere number and less discernable myomere boundary. Accordingly, pigment patterns at different developmental stages are the main features used to differentiate these four species in this study.
4.1.1 Sillago sihama versus Sillago nigrofasciata
These two species have very similar egg sizes and spawning times. For eggs in early late stage, the embryonic melanophores of S. sihama are stellate-shaped (Figure 4B), but in the latter, they are branch-shaped (Figure 5A). Additionally, S. sihama eggs have smaller oil globules (Table 4). The morphological disparity in eggs between these two species provided another supportive evidence for the recent separation of S. nigrofasciata as a new species from S. sihama by Xiao et al. (2021).
4.1.2 Sillago nigrofasciata versus Acanthopagrus pacificus
Early late-stage eggs of S. nigrofasciata (Figure 5A) can be confused with late-stage eggs of A. pacificus (Figures 7E–G) due to the very similar melanophore shapes. They can be differentiated via the position of the oil globule. In S. nigrofasciata eggs, the oil globule in this stage is positioned close to the caudal end (Figure 5A), whereas, in the latter, it is laterally located near the trunk or tail (Figures 7E–G). When the oil globule in S. nigrofasciata eggs is positioned at this location, the embryo is more heavily pigmented with densely branched melanophores, especially on the ventral side (Figure 5C). The difference in the spawning time of these two species can be used for egg discriminations (Table 4).
4.1.3 Nibea albiflora versus Acanthopagrus pacificus
Middle-stage eggs from these two species (Figure 6B versus Figure 7D) can be confused because of the similarly pigmented embryos. Although in N. albiflora eggs, the pigment clustering on the middle part of the trunk is not very striking in this stage, the melanophores on this area are slightly larger than those on the other parts of the trunk (Figure 6B). However, in A. pacificus eggs, pigments on the trunk are of similar shape and size, and are evenly scattered with no clustering (Figure 7D). When the melanophores appear on the head region, the pigment clustering area in N. albiflora has been pronounced (Figure 6C), and eggs of these two species can be easily distinguished. Additionally, the difference in the occurring time of these two eggs can be helpful for the differentiation (Table 4).
4.2 Importance of Reference Library of Accurately Identified Fishes
In this study, two mitochondrial gene regions of COI and 16S rRNA consistently differentiate fish eggs into four species. However, when compared with sequences in the GenBank database, two species (S. nigrofasciata, A. pacificus) both have more than one candidate species showing high similarity (>99%). These two cases in this study were more likely caused by the misidentification rather than by the mitochondrial introgression previously reported in the genus of Scomberomorus and Thunnus (Paine et al., 2007; Kawakami et al., 2010).
The most important character used to identify sillaginids is the shape of the swimbladder. S. asiatica can be distinguished from S. sihama in possessing a swim bladder with one single tapering posterior extension versus two posterior extensions in S. sihama (McKay, 1985; McKay, 1992; Kaga, 2013). Based on anatomic examinations on fish specimens we collected, which shared the same COI sequence with S. asiatica identified by Chang et al. (2017), the swim bladder in this species has two posterior extensions (Supplementary Figure 4B), suggesting that the identification by Chang et al. (2017) was incorrect. Additionally, morphological features displayed by these fish specimens corresponded with those of S. nigrofasciata described by Xiao et al. (2021). In another case of A. pacificus, the straight infraorbital edge (Supplementary Figure 3D) and posteriormost head squamation type (Supplementary Figure 5) shown by one adult fish specimen we collected can distinguish this species from A. berda. (Iwatsuki et al., 2010). All these results indicated that a reference library for accurately-identified fishes is essential for the correct identifications of fish eggs through molecular methods.
4.3 Identification and Preservation Methods for Fish Eggs
For early-stage egg identification, molecular analysis shows its superiority over the visual taxonomic method with respect to accuracy. It is almost impossible to visually get reliable identification results when samples are constituted by a large proportion of early-stage eggs with a similar size from many species. For middle- and late-stage eggs, visual taxonomic methods will be more desirable than molecular techniques on the premise that the rate of successful identification can be assured. A reliable application of this convenient method will inevitably stress the need for a precise knowledge of the morphological features of egg species. By far, studies on fish egg morphology covered only a very small proportion of the fish species. Furthermore, most of these studies provided drawings not photographs for egg morphology, or morphological photos were limited to a certain developmental stage. With the goal to discriminate egg species correctly, detailed photos of more developmental stages and more species need to be complemented. Additionally, random molecular analysis on egg specimens is critical to ascertain the accuracy of morphological identifications.
The selection of the preservatives is another crucial issue for egg identifications. Ethanol is reliable for DNA preserving and molecular analysis (Karaiskou et al., 2007; Goodsir et al., 2008), but morphological distortions are often observed in ethanol-fixed eggs (Lewis et al., 2016; Leyva-Cruz et al., 2016; Zheng et al., 2021). As shown in this study, most of the ethanol-fixed eggs had opaque yolk sac because of protein denaturation, making it difficult to distinguish if the embryonic tailbud had lifted off the yolk sac, which can be used as a criterion for the discrimination of middle- and late-stage eggs (Miller and Kendall, 2009). Additionally, oil globule dissolution was common in ethanol-preserved eggs of A. pacificus in this study. Similar cases were also observed in eggs of Solea ovata and A. schlegelii in our previous study (Zheng et al., 2021), which highlighting the necessity of employing formalin as reference fixative to achieve accurate morphological features. On the other hand, formalin-fixed samples usually have low PCR amplification success resulting from DNA fragmentation and histones cross-linking (Akimoto, 2002; Karaiskou et al., 2007). Consequently, the combined use of both kinds of fixatives in the present study was conducive to avoid these adverse effects. Recently, Bourque et al. (2020) reported that 10% hydantoin solution performed effectively in both retaining morphological features and keeping the DNA integrity for ichthyoplankton, which appeared promising as a solution for egg preserving.
5 Conclusion
For the first time, distinguishing characteristics and detailed photographs were supplied in this study for fish eggs from four economically important species in Guanghai Bay, namely silver sillago Sillago sihama, black-banded sillago Sillago nigrofasciata, yellow drum Nibea albiflora, and Pacific seabream Acanthopagrus pacificus. The results will be essential for the spawning ground estimation for these four species. Future study may not only focus on species identification and morphological descriptions for fish eggs in this area, but also the development of inspection methods suitable for long-term ichthyoplankton monitoring programs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the State Oceanic Administration of China.
Author Contributions
Y-JZ and X-LL participated in field work, experiments, and data analysis. Y-JZ and Y-HH wrote the original draft. X-JP provided guidance on data analysis. W-XC designed the study. Q-SL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by Science and Technology Plan Projects of Guangdong Province (Grant No. 2021B1212050025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are deeply grateful to the various fisherfolk for their extensive and warm assistance with the collection of samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.815473/full#supplementary-material
References
Ahlstrom E. H., Moser H. G. (1980). Characters Useful in Identification of Pelagic Marine Fish Eggs. CalCOFI Rep. XXI, 121–131.
Akimoto S., Kinoshita S., Sezaki K., Mitani I., Watabe S. (2002). Identification of Alfonsino and Related Fish Species Belonging to the Genus Beryx With Mitochondrial 16S rRNA Gene and its Application on Their Pelagic Eggs. Fisheries Sci. 68, 1242–1249. doi: 10.1046/j.1444-2906.2002.00561.x
Bourque D. A., Morey K. C., Bradley D. L., Fost B., Daley J. M., Jacobson N., et al. (2020). Dimethyldimethylhydantoin Hydantoin: An Alternative Fluid for Morphological and Genetic Preservation. Biopreservation Biobanking 18, 283–289. doi: 10.1089/bio.2020.0001
Bleeker P. (1854). Faunae Ichthyologicae Japonicae Species Novae. Natuurkundig Tijdschrift voor Nederlandsch Indië 6, 395–426.
Chang C. H., Shao K. T., Lin H. Y., Chiu Y. C., Lee M. Y., Liu S. H., et al. (2017). DNA Barcodes of the Native Ray-Finned Fishes in Taiwan. Mol. Ecol. Resour. 17, 796–805. doi: 10.1111/1755-0998.12601
Chen S. Z. (1999). The Bay Chronicle of China (the Tenth Fascicle) (Beijing: China: Ocean Press), 1–48.
Cheng J., Xiao J. G., Song N., Saha S., Qin J. G., Nomura H., et al. (2021). Molecular Phylogeny Reveals Cryptic Diversity and Swim Bladder Evolution of Sillaginidae Fishes (Perciformes) Across the Indo-West Pacific Ocean. Diversity Distributions 27, 82–94. doi: 10.1111/ddi.13171
Choi H. Y., Oh J., Kim S. (2018). Genetic Identification of Eggs From Four Species of Ophichthidae and Congridae (Anguilliformes) in the Northern East China Sea. PloS One 13, e0195382. doi: 10.1371/journal.pone.0195382
Fabricius J. C. (1775). In Niebuhr C. (ed.) Descriptiones Animalium Avium, Amphibiorum, Piscium, Insectorum, Vermium; Quae In Itinere Orientali Observavit PetrusforskåL. Post Mortem Auctoris Edidit Carsten Niebuhr. Hauniae, Möller, Denmark. 20+XXXIV+164 pp., 1 map.
Fox C. J., Taylor M. I., Pereyra R., Villasana M. I., Rico C. (2005). TaqMan DNA Technology Confirms Likely Overestimation of Cod (Gadus Morhua L.) Egg Abundance in the Irish Sea: Implications for the Assessment of the Cod Stock and Mapping of Spawning Areas Using Egg-Based Methods. Mol. Ecol. 14, 879–884. doi: 10.1111/j.1365-294X.2005.02439.x
Fricke R., Eschmeyer W. N., van der Laan R. eds. (2022). Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Accessed 7 March 2022).
Froese R., Pauly D. eds. (2022). FishBase. World Wide Web electronic publication. Available at: www.fishbase.org.
Fuiman L. A., Werner R. G. (2002). Fishery Science, The Unique Contribution of Early Life Stages (Oxford: Blackwell Science).
Goodsir F., Armstrong M. J., Witthames P. R., Maxwell D. L., Fox C. J. (2008). The Use of Species-Specific TaqMan Probes for Identifying Early Stage Gadoid Eggs Following Formaldehyde Fixation. ICES J. Mar. Sci. 65, 1573–1577. doi: 10.1093/icesjms/fsn180
Guangdong Economy (2014). A New Platform for Guangdong-Hong Kong-Macau Cooperation: The Greater Guanghai Bay Economic Zone of Jiangmen. Guangdong Economy 1, 90 (in Chinese).
Guo J. F., Li M. Z., Yu M. Y. (1994). Marine Organism and Fishery Resources in Insular Areas of Guangdong Province (Guangzhou: Technological Publishing Press).
Hall T. A. (1999). “BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT,” in Nucleic Acids Symposium Series, Vol. 41. 95–98. doi: 10.14601/Phytopathol_Mediterr-14998u1.29
Hasan M. E., Durand J. D., Iwatsuki Y. (2020). Acanthopagrus Datnia (Hamilton 1822), a Senior Synonym of Acanthopagrus Longkpinnis (Valenciennes 1830) (Perciformes: Sparidae). Zootaxa 4750, 151–181. doi: 10.11646/zootaxa.4750.2.1
Hofmann T., Knebelsberger T., Kloppmann M., Ulleweit J., Raupach M. J. (2017). Egg Identification of Three Economical Important Fish Species Using DNA Barcoding in Comparison to a Morphological Determination. J. Appl. Ichthyology 33, 925–932. doi: 10.1111/jai.13389
Hou G., Chen W. T., Lu H. S., Cheng F., Xie S. G. (2018). Developing a DNA Barcode Library for Perciform Fishes in the South China Sea: Species Identification, Accuracy and Cryptic Diversity. Mol. Ecol. Resour. 18, 137–146. doi: 10.1111/1755-0998.12718
Hou G., Wang J. R., Chen Z. Z., Zhou J. L., Huang W. S., Zhang H. (2020). Molecular and Morphological Identification and Seasonal Distribution of Eggs of Four Decapterus Fish Species in the Northern South China Sea: A Key to Conservation of Spawning Ground. Front. Marine Sci. 7. doi: 10.3389/fmars.2020.590564
Hou G., Xu Y. W., Chen Z. Z., Zhang K., Huang W. S., Wang J. R., et al. (2021). Identification of Eggs and Spawning Zones of Hairtail Fishes Trichiurus (Pisces: Trichiuridae) in Northern South China Sea, Using DNA Barcoding. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.703029
Houttuyn M. (1782). Beschryving Van Eenige Japanese Visschen, En Andere Zee-schepzelen. Verhandelingen der Hollandsche Maatschappij der Wetenschappen, Haarlem, 20, 311–350.
Hsu T. H., Madrid A. G. G., Burridge C. P., Cheng H. Y., Gwo J. C. (2011). Resolution of the Acanthopagrus Black Seabream Complex Based on Mitochondrial and Amplified Fragment-Length Polymorphism Analyses. J. Fish Biol. 79, 1182–1192. doi: 10.1111/j.1095-8649.2011.03100.x
Hubbs C. L., Lagler K. F. (2004). Fishes of the Great Lakes Region (Ann Arbor, Michigan: The University of Michigan Press). doi: 10.3998/mpub.17658
Hubert N., Espiau B., Meyer C., Planes S. (2015). Identifying the Ichthyoplankton of a Coral Reef Using DNA Barcodes. Mol. Ecol. Resour. 15, 57–67. doi: 10.1111/1755-0998.12293
Huelsenbeck J. P., Ronquist F. (2001). Mrbayes: Bayesian Inference of Phylogenetic Trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Ikeda T., Hirai A., Tabata S., Onishi Y., Mito S. (2014). “Key to Fish Eggs in Japan,” in An Atlas of Early Stage Fishes in Japan. Ed. Okiyama M. (Hadano-shi, Kanagawa: Tokai University Press), 1–108.
Iwatsuki Y., Heemstra P. C. (2010). Taxonomic Review of the Western Indian Ocean Species of the Genus Acanthopagrus Peter(Perciformes: Sparidae), With Description of a New Species From Oman. Copeia, 123–136. doi: 10.1643/ci-08-073
Iwatsuki Y., Kume M., Yoshino T. (2010). A New Species, Acanthopagrus Pacificus From the Western Pacific (Pisces, Sparidae). Bull. Natl. Mus. Nat. Sci. Ser. A 36, 115–130.
Jin Q., Han H., Hu X., Li X., Zhu C., Ho S. Y. W., et al. (2013). Quantifying Species Diversity With a DNA Barcoding-Based Method: Tibetan Moth Species (Noctuidae) on the Qinghai-Tibetan Plateau. PloS One 8, e64428. doi: 10.1371/journal.pone.0064428
Kaga T. (2013). Phylogenetic Systematics of the Family Sillaginidae (Percomorpha: Order Perciformes). Zootaxa 3642, 1–105. doi: 10.11646/zootaxa.3642.1.1
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). Modelfinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karaiskou N., Triantafyllidis A., Alvarez P., Lopes P., Garcia-Vazquez E., Triantaphyllidis C. (2007). Horse Mackerel Egg Identification Using DNA Methodology. Marine Ecol. 28, 429–434. doi: 10.1111/j.1439-0485.2007.00190.x
Kawakami T., Aoyama J., Tsukamoto K. (2010). Morphology of Pelagic Fish Eggs Identified Using Mitochondrial DNA and Their Distribution in Waters West of the Mariana Islands. Environ. Biol. Fish. 87, 221–235. doi: 10.1007/s10641-010-9592-2
Kimura M. (1980). A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotide-Sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/bf01731581
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lecocq T., Vereecken N. J., Michez D., Dellicour S., Lhomme P., Valterova I., et al. (2013). Patterns of Genetic and Reproductive Traits Differentiation in Mainland vs. Corsican Populations of Bumblebees. PloS One 8, e65642. doi: 10.1371/journal.pone.0065642
Lei J., Fan N. (1981). A Preliminary Observation on the Morphological Characteristics of Embryos, Larvae and Juveniles of Nibea Albiflora (Richardson). Marine Fisheries Res. 4, 77–84.
Lelievre S., Verrez-Bagnis V., Jerome M., Vaz S. (2010). PCR-RFLP Analyses of Formalin-Fixed Fish Eggs for the Mapping of Spawning Areas in the Eastern Channel and Southern North Sea. J. Plankton Res. 32, 1527–1539. doi: 10.1093/plankt/fbq067
Letunic I., Bork P. (2021). Interactive Tree of Life (iTOL) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lewis L. A., Richardson D. E., Zakharov E. V., Hanner R. (2016). Integrating DNA Barcoding of Fish Eggs Into Ichthyoplankton Monitoring Programs. Fishery Bull. 114, 153–165. doi: 10.7755/fb.114.2.3
Leyva-Cruz E., Vasquez-Yeomans L., Carrillo L., Valdez-Moreno M. (2016). Identifying Pelagic Fish Eggs in the Southeast Yucatan Peninsula Using DNA Barcodes. Genome 59, 1117–1129. doi: 10.1139/gen-2015-0151
Mateos-Rivera A., Skern-Mauritzen R., Dahle G., Sundby S., Mozfar B., Thorsen A., et al. (2020). Comparison of Visual and Molecular Taxonomic Methods to Identify Ichthyoplankton in the North Sea. Limnology Oceanography-Methods 18, 599–605. doi: 10.1002/lom3.10387
McKay R. J. (1982) In Dutt S., Sujatha K. On A New Species Of Sillago Cuvier, 1817 (Teleostei: Sillaginidae) From India. Proceedings of the Indian National Science Academy Part B Biological Sciences 48, 611–614.
McKay R. J. (1985). A Revision of the Fishes of the Family Sillaginidae. Memoirs Queensland Museum 23, 1–73.
McKay R. J. (1992). “FAO Species Catalogue.Volume14 Sillaginid Fishes of the World (Family Sillaginidae),” in An Annotated and Illustrated Catalogue of the Sillago, Smelt or Indo-Pacific Whiting Species Known to Date (Rome: FAO).
Miller B. S., Kendall A. W. Jr (2009). Development of Eggs and Larvae In Early Life History of Marine Fishes (Berkeley & Los Angeles, CA: University of California Press), 39–45.
Nei M., Kumar S. (2000). “Chapter 3 Evolutionary Change of DNA Sequences,” in Molecular Evolution and Phylogenetics (New York, NY: Oxford University Press), 33–50.
Paine M. A., McDowell J. R., Graves J. E. (2007). Specific Identification of Western Atlantic Ocean Scombrids Using Mitochondrial DNA Cytochrome C Oxidase Subunit I (COI) Gene Region Sequences. Bull. Marine Sci. 80, 353–367.
Palumbi S. R., Martin A., Romano S., McMillan W. O., Stice L., Grabowski G. (2002). The Simple Fool’s Guide to PCR.version 2.0 (Honolulu: University of Hawaii Press).
Perez J., A´lvarez P., Martinez J. L., Garcia-Vazquez E. (2005). Genetic Identification of Hake and Megrim Eggs in Formaldehyde-Fixed Plankton Samples. ICES J. Mar. Sci. 62, 908–914. doi: 10.1016/j.icesjms.2005.04.001
Petitgas P., Magri S., Lazure P. (2006). One-Dimensional Biophysical Modelling of Fish Egg Vertical Distributions in Shelf Seas. Fisheries Oceanography 15, 413–428. doi: 10.1111/j.1365-2419.2006.00409.x
Ratnasingham S., Hebert P. D. (2007). Bold: The Barcode of Life Data System. Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Ratnasingham S., Hebert P. D. N. (2013). A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PloS One 8, e66213. doi: 10.1371/journal.pone.0066213
R Core Team (2017). R: A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: http://www.R-project.org/.
Reeves S. A., Pastoors M. A. (2007). Evaluating the Science Behind the Management Advice for North Sea Cod. ICES J. Mar. Sci. 64, 671–678. doi: 10.1093/icesjms/fsl048
Regan C. T. (1917). A revision of the clupeoid fishes of the genera Pomolobus, Brevoortia and Dorosoma and their allies. Annals and Magazine of Natural History (Series 8). 19, 297–316. doi: 10.1093/icesjms/fsl048
Richardson D. E., Llopiz J. K., Leaman K. D., Vertes P. S., Muller-Karger F. E., Cowen R. K. (2009). Sailfish (Istiophorus Platypterus) Spawning and Larval Environment in a Florida Current Frontal Eddy. Prog. Oceanography 82, 252–264. doi: 10.1016/j.pocean.2009.07.003
Richardson J. (1846). Report on the Ichthyology of the Seas of China and Japan. Report of the British Association for the Advancement of Science 15th meeting [1845], 187–320.
Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DNASP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Tian F. G., Zheng Y. J., Xiao Y. Z., Fang H. D. (2017). Spawning Season and Variation of the Eggs of Stolephorus Commersonii in Guanghaiwan Bay. J. Appl. Oceanography 36, 395–402. doi: 10.3969/J.ISSN.2095-4972.2017.03.013 (In Chinese).
Temminck C. J., Schlegel H. (1846). Pisces. In: Siebold P. F. de (ed.): Fauna Japonica, Sive Descriptio Animalium, Quae In Itinere Per Japoniam...suscepto Annis 1823-1830 Collegit, Notis, Observationibus Et Adumbrationibus Illustravit Ph. Fr. De Siebold. Parts 10-14:173–269.
Wan R. J. (1996). Morphology of Early Development of Sillago Sihama (Forsskål). Marine Fisheries Res. 17, 35–41.
Ward R. D., Hanner R., Hebert P. D. N. (2009). The Campaign to DNA Barcode All Fishes, FISH-BOL. J. Fish Biol. 74, 329–356. doi: 10.1111/j.1095-8649.2008.02080.x
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. N. (2005). DNA Barcoding Australia’s Fish Species. Phil. Trans. R. Soc B 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Watson R., Pauly D. (2001). Systematic Distortions in World Fisheries Catch Trends. Nature 414, 534–536. doi: 10.1038/35107050
Xiao J. G., Song N., Gao T. X., Zhao Y. H. (2016). The Complete Mitochondrial Genome of Sillago Asiatica (Perciformes: Sillaginidae). Mitochondrial DNA Part A 27, 1644–1645. doi: 10.3109/19401736.2014.958709
Xiao J. G., Yu Z. S., Song N., Gao T. X. (2021). Description of a New Species, Sillago Nigrofasciata Sp. Nov. (Perciformes, Sillaginidae) From the Southern Coast of China. Zookeys 1011, 85–100. doi: 10.3897/zookeys.1011.57302
Zhang A. B., Hao M. D., Yang C. Q., Shi Z. Y. (2017). BarcodingR:An Integrated R Package for Species Identifcation Using DNA Barcodes. Methods Ecol. Evol. 8, 627–634. doi: 10.1111/2041-210X.12682
Zhang R. Z., Lu S. F., Zhao C. Y., Chen L. F., Zang Z. J., Jiang, et al. (1985). Fish Eggs and Larvae in the Offshore Waters of China (Shanghai: Shanghai Scientific and Technological Press).
Zhang A. B., Muster C., Liang H. B., Zhu C. D., Crozier R., Wan P., et al. (2012). A Fuzzy-Set-Theory-Based Approach to Analyse Species Membership in DNA Barcoding. Mol. Ecol. 21, 1848–1863. doi: 10.1111/j.1365-294X.2011.05235.x
Zhang A. B., Sikes D. S., Muster C., Li S. Q. (2008). Inferring Species Membership Using DNA Sequences With Back-Propagation Neural Networks. Systematic Biol. 57, 202–215. doi: 10.1080/10635150802032982
Keywords: fish eggs, molecular identification, morphological characteristics, Guanghai Bay, visual taxonomic method
Citation: Zheng Y-J, Peng X-J, Cai W-X, Lou Q-S, Lyu X-L and Huang Y-H (2022) The Identification of Fish Eggs From Four Economically Important Species in Guanghai Bay (China). Front. Mar. Sci. 9:815473. doi: 10.3389/fmars.2022.815473
Received: 15 November 2021; Accepted: 06 April 2022;
Published: 19 May 2022.
Edited by:
Liping Liu, Shanghai Ocean University, ChinaReviewed by:
Zhihao Wu, Institute of Oceanology (CAS), ChinaNicolas Michel Hubert, Institut de Recherche pour le Développement, France
Copyright © 2022 Zheng, Peng, Cai, Lou, Lyu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Li Lyu, dnR3b3Zfbjc4QDE2My5jb20=; Ying-Hua Huang, eWhoX3Njc0AxNjMuY29t
 Yan-Jing Zheng1
Yan-Jing Zheng1 Xiang-Li Lyu
Xiang-Li Lyu