
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 31 January 2022
Sec. Marine Molecular Biology and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.811284
Outside-host parasite survival is a key parameter to better understand disease mechanisms, especially for marine pathogens transmitted from one host to another through an environmental stage. For non-cultivable micro-parasites like Marteilia refringens, a protozoan parasite infecting the flat oyster Ostrea edulis, investigating this parameter requires innovative approaches. In the present study, we have developed an Environmental DNA (eDNA)-based method allowing detecting and quantifying up to 25 and 10 parasites DNA in seawater and sediment, respectively. This method was used in combination with light and transmission electron microscopy (TEM) to study experimentally parasite survival in seawater and flat oyster faeces after its release from naturally infected oysters. M. refringens DNA could be detected up to 20 days, in both seawater and oyster faeces with a more stable detection over time in the latter. Light and transmission microscopy confirm that parasites leaving flat oysters are sporangia. We also observed a membrane dissolution of the sporangia over time that could indicate the release of parasite spore. This study not only improves our understanding of M. refringens ecology but also highlights the interest to combine molecular and microscopical analysis to study non-cultivable micro-parasites life cycle outside their host.
The native European oyster, Ostrea edulis (Linnaeus, 1758) is naturally found from the Atlantic Ocean and North Sea to the Mediterranean, Adriatic and Black Seas. This oyster species has been consumed since Roman times. However, O. edulis populations showed first signs of overfishing in the XVIIIth century. The subsequent improvement in fishing efficiency and transport technologies in the XXth century intensified the decline in oyster landings in Europe, falling from ∼30,000 tons in 1961 to ∼3,000 tons in 2010 (Gercken and Schmidt, 2014). In addition to overfishing, the demise of O. edulis in Europe is also explained by the emergence of epizootic diseases, including marteiliois (Buestel et al., 2009; Pogoda et al., 2019).
Marteilia refringens is the causative agent of marteiliosis and was first described in 1968 in the context of flat oyster mortality, in Aber Wrac’h, Brittany (France) (Comps, 1970; Grizel et al., 1974). Since its first description in O. edulis, several other bivalve species have been found to be susceptible to M. refringens including the dwarf oyster Ostrea stentina (Elgharsalli et al., 2013), the mussels Mytilus edulis (Le Roux et al., 2001) and Mytilus galloprovincialis (López-Flores et al., 2004), the razor clam Solen marginatus (López-Flores et al., 2008a), the clam Chamelea gallina (López-Flores et al., 2008b), and the dwarf mussel Xenostrobus securis (Pascual et al., 2010). Aquaculture movements have contributed to spread the parasite in almost all European countries and along the Mediterranean basin (Pogoda et al., 2019; OIE, 2021). Because of its impact on natural and farmed flat oyster populations, detection of this infection requires mandatory notification to the World Organization for Animal Health (OIE, 2021).
Previous experimental transmission studies consisting of either inoculating M. refringens cells directly into molluscs or cohabitation of infected with non-infected molluscs failed, suggesting that an intermediate host or a process of parasite maturation might be required in the life cycle (Berthe et al., 2004). The detection of the parasite by PCR and in situ hybridization in copepods Paracartia grani (Audemard et al., 2002; Boyer et al., 2013) or in the congeneric species Paracartia latisetosa (Arzul et al., 2014) supports their involvement in M. refringens cycle. Nevertheless, although it has been possible to infect experimentally copepods P. grani by cohabitation with M. refringens infected flat oysters, trials aiming to infect flat oysters from infected copepods have never been successful so far (Carrasco et al., 2008), leaving the implication of the copepods in M. refringens life-cycle unclear. Additionally, the detection of parasite DNA in other zooplankton species as well as the cnidarian Cereus pedunculatus (Audemard et al., 2002; Carrasco et al., 2007a,b) raises question regarding the role of these species in the transmission of M. refringens.
In the flat oyster O. edulis and other bivalve species, M. refringens infects the digestive system. It develops along the digestive tract by endogenous divisions with young stages mainly found in the stomach and more advanced stages in digestive diverticula. Young stages of the parasite appear as primary cells containing one or more secondary cells within their cytoplasm. Within the digestive diverticula, secondary cells divide to generate four tertiary cells. As the tertiary cells evolve into spores, the size of the primary cell, called sporangiosorus, increases with the appearance of refringent inclusions in the secondary cells or sporangia. These advanced stages of the parasite are released into the lumen of the digestive diverticula. Finally, after the rupture of the sporangiosorus membrane, sporangia are released in flat oyster faeces (Grizel et al., 1974; Perkins, 1976; Berthe et al., 2004; Carrasco et al., 2015).
The observation of marteiliosis in areas where oyster farming has been suspended for several years suggests that the parasite is able to survive outside flat oysters probably under resistant stages such as spores (Grizel, 1985). Nevertheless, the survival and behaviour of the parasite once released from infected oysters have never been formally investigated. However, understanding parasite life cycle and ecology is crucial to understand disease development and identify adapted management measures. In this context, parasite survival outside its host is an important parameter to estimate as it can directly influence disease spreading between oyster populations through water currents, as suggested for Bonamia ostreae (Arzul and Carnegie, 2015).
Environmental DNA (eDNA) based approaches show many advantages to study non-cultivable micro-organisms (Taberlet et al., 2012; Bass et al., 2015) such as M. refringens. Indeed, eDNA approaches allow rapid, non-invasive and efficient monitoring of micro-organisms (Harper et al., 2019). However, DNA detection does not provide any information about the status (active/inactive) or form of the targeted organism. Complementary techniques such as RNA detection (Mérou et al., 2020) or microscopy are required to characterize micro-organisms.
In this context, an eDNA-based approach based on the real-time PCR analysis of artificially spiked seawater and sediment samples with purified parasites has been developed. This approach has been used to monitor the presence of M. refringens in seawater and flat oyster faeces during up to 20 days after its release from naturally infected animals incubated during 24 h in filtered seawater. Complementary light and transmission electron microscopy (TEM) examinations have allowed describing released parasites providing new insights into the cycle of M. refringens outside its bivalve hosts.
Three years old flat oysters O. edulis originated from a hatchery in La Rochelle (France) and maintained in claire ponds known to be infected with M. refringens on Oleron Island (Nouvelle-Aquitaine, France) (Audemard et al., 2001) from April 2018 to April 2019 were maintained individually during 1,5 week at room temperature in 2 L aquaria containing decanted seawater: every three days, faeces samples were collected from each oyster and examined under light microscopy using Malassez-cell haemocytometer to check the presence of M. refringens.
Flat oysters found infected following light microscopy analysis were selected for the next step to monitor the presence of the parasite in seawater and flat oyster faeces.
Efficiency, limit of detection (LOD), and limit of quantification (LOQ) of the real-time PCR approach were established using –20°C frozen pellet of M. refringens isolated from highly infected flat oysters according to Mialhe et al. (1985). A stock parasite suspension was made by resuspending pellet in 0.22 μm filtered seawater before performing 1:10 serial dilutions. Each dilution was filtered on a 1 μm pore size 47 mm diameter polycarbonate membrane (Whatman® Nuclepore™ Track-Etched Membranes) or mixed with 0.25 g of sediment sample known to be negative for M. refringens. In the present study, we used sediment to mimic flat oyster faeces within the development of the real-time PCR approach: for the rest of the paper, “sediment/faeces” will refer to the development of the real-time PCR approach whereas “faeces” will refer to the experimental procedure carried out to monitor M. refringens presence in flat oyster faeces after release. Samples were stored at –80°C until being used for DNA extraction and analysed by real-time PCR as described below.
A group of 10 flat oysters found infected with M. refringens was maintained for 24 h in 2 L of 1 μm filtered seawater at room temperature. After 24 h, oysters were removed, seawater was transferred into a 2 L glass bottle while faeces were transferred into a 50 mL glass bottle with the smallest amount of seawater (just enough to keep it wet). To minimise bacteria proliferation, seawater and faeces suspensions were complemented with Streptomycin-Penicillin G (final concentration: 2.4512 U/mL and 11.0353 U/mL, respectively) to cover a broad bacteria spectrum.
This approach was repeated three times using the same batch of 10 flat oysters. In total, four seawater and faeces suspensions were obtained and stored at 20°C for 20 days.
Samples of seawater and faeces were collected every 1–5 days after homogenizing suspensions by turning them up and down several times, and tested regarding the presence and quantity of M. refringens by Real Time PCR, light microscopy, and Transmission Electron Microscopy (TEM only for faeces samples).
Real-time PCR analyses were carried out from 100 mL and 500 μL of seawater and faeces suspensions, respectively. Seawater samples (100 mL) were filtered through a 1 μm pore size 47 mm diameter polycarbonate membrane (Whatman, Inc., Buckinghamshire, United Kingdom) using a vacuum pump. Each membrane was then cut in four quarters and stored at –80°C until being processed for DNA extraction in 1.5 mL Safe Lock Tubes (Eppendorf, Inc., Hamburg, Germany). For each sample, one quarter membrane was used for DNA extraction, and the three other quarters were stored to redo analysis if needed. Faeces samples (500 μl) were centrifuged at 4000 g for 1 min to remove supernatant. Dry faeces pellets were stored at -80°C in 1.5 mL Safe Lock Tubes (Eppendorf, Inc., Hamburg, Germany) until being processed for DNA extraction.
Light microscopy examination was done from 200 μL of seawater suspension or 200 μl of 1/10 diluted faeces suspension. Samples were cytocentrifuged at 28 g for 5 min on microscopic slides (Polysine Adhesion Slides, J2800AMNT, Thermo Scientific™). Supernatant was removed and microscopic slides were dried before being stained with Hemacolor® (Merck-Millipore, Inc., Burlington, MA, United States).
Transmission electron microscopy examination was performed from 200 μl of faeces suspension. Samples were centrifuged at 22,4 g for 5 min before removing supernatant and fixing dry pellets with 4% glutaraldehyde for at least 24 h at 4°C. Then, samples were washed in 0.2 M cacodylate buffer (3 × 30 min) and post-fixed for 1 h in 1% osmium tetroxide (OsO4) in 0.2 M cacodylate buffer. After post-fixation, samples were cleared in propylene oxide, and embedded in epon resin.
For seawater samples, total DNA was extracted from quarter membranes using the DNeasy® PowerWater® Kit (Qiagen, Inc., Hilden, Germany) according to the manufacturer’s protocol with some modifications as described in Mérou et al. (2020). After 10 min at 65°C, a mechanical cell lysis was carried out using the Precellys®24 bead beater (Bertin Technologies, Inc., Montigny-le-Bretonneux, France), and the following program: 4 lysis cycles of 20 s at 5000 rpm, with 5 s of pause between each cycle.
For sediment or faeces samples, total DNA was extracted using the DNeasy® PowerSoil® Kit (Qiagen, Inc., Hilden, Germany) according to the manufacturer’s protocol with some modifications. After 10 min at 70°C, a mechanical cell lysis was carried out using the Precellys®24 bead beater (Bertin Technologies, Inc., Montigny-le-Bretonneux, France), and the following program: 8 lysis cycles of 20 s at 5000 rpm, with 5 s of pause between each cycle.
For both extraction protocols, DNA was eluted in 50 μL of the elution solution provided in the kit and samples were stored at 4°C until being tested by real-time PCR.
For the detection of M. refringens 18S rDNA from dilution range or experimental samples, amplification reactions were carried out using the M. refringens simplex version of the multiplex real-time PCR described in Canier et al. (2020).
Each sample was analysed in duplicate by real-time PCR in 96-microwell plates using the Mx3000p™ thermocycler sequence detector (Stratagene, Inc., La Jolla, CA, United States). Positive and negative controls as well as standard dilution range were included in each PCR run. Positive controls consisted in DNA extracted from known infected samples. Negative controls consisting in 5 μL of bi-distilled water used in the extraction and real-time PCR steps were added to each PCR plate.
Microscopic slides were stained with Hemacolor® (Merck-Millipore, Inc., Burlington, MA, United States) and screened by light microscopy at 60 times magnification. For seawater suspensions, all M. refringens-like cells were counted in a total 10 cytocentrifugation sections. For faeces suspensions all M. refringens-like cells were counted in a total five cytocentrifugation sections.
Ultrathin sections were obtained from samples embedded in epon resin using copper grids. Sections were double stained with 5% uranyl acetate and 5% lead citrate before being examined at 120 kV on a JEOL 1110 transmission electron microscope associated to a Morada digital camera and iTEM imaging software (Soft Imaging System, Olympus, Shinjuku, Japan).
Real-Time PCR efficiency, LOD, and LOQ were determined for both seawater and sediment/faeces approaches from the analysis of parasite dilution series. Data were analysed using R 4.0.3 (2020-10-10) (R Core Team, 2020) from quantification cycle (Cq) values. When no amplification was obtained, “No Cq” value was replaced by 40, corresponding to the maximum number of cycles. For each tested amount of parasite, Cq values were used as soon as the Coefficient of Variation (CV) computed on all replicates was less than 5%. If CV was higher than 5%, only the furthest replicate from the average Cq was removed per tested amount of parasite.
Following MIQE recommendations (Bustin et al., 2009), LOD was defined as the lowest detected number of parasites in at least 95% of tested replicates and LOQ was defined as the last amount of parasites detected in the linear dynamic range of the standard curve.
After a decimal log conversion of the number of parasites included in the dilution series, PCR efficiency was computed for both matrix on the linear dynamic range of the standard curve using the following formula:
where “a” is the slope of the linear regression performed on Cq ∼ log10 value.
The Cq ∼ log10 value linear regression allowed building standard curves to quantify parasites in each matrix using the following formula:
95% confidence and prediction intervals were calculated using “predict” in Cornillon et al. (2012).
Parasite presence was monitored by testing samples collected from seawater or faeces suspensions containing freshly released parasites in real-time PCR.
“Loess” regression was built on Cq ∼ sampling day and difference between Cq value of each replicate was computed: when difference was higher than 2, furthest Cq value from “loess” regression was removed. When difference was lower than 2, average Cq was computed between replicates.
Number of detected parasites was calculated for day post-release using the following equation obtained from standard curve:
Correlation between the number of detected parasites at t = 0 day and at the last detection day above the detection limit in seawater and faeces was investigated with Spearman’s rank correlation test (rho).
Real-Time PCR limit of detection (LOD), limit of quantification (LOQ), and efficiency were determined following analysis of DNA extracted from 1:10 dilution series of purified M. refringens applied on 1 μm polycarbonate membranes or mixed with 0.25 g of sediment.
After removing outliers Cq values for each tested amount of parasites and matrix as described above, an average coefficient of variation (CV) of 1.77% (±1.80%) and 2.06% (±0.87%) were found for M. refringens DNA detection approach in seawater and sediment/faeces, respectively, suggesting a good repeatability.
In seawater, the smallest amount of parasites (2.5 parasites) was never detected in the four tested replicates whereas 25 parasites were detected four times out of four. Thus, the LOD for M. refringens DNA detection approach in seawater was set at 25 parasites per quarter membrane. In sediment (used to mimic flat oyster faeces to develop the approach), the smallest amount of parasites (1 parasite) was detected one time out of four whereas 10 parasites were detected four times out of four. Thus, the LOD for M. refringens DNA detection approach in sediment/faeces was set at 10 parasites in 0.25 g.
As the LOD of both approaches were also the last amounts in the dynamic linear range of the standard curve, LOQ were set at 25 and 10 parasites for seawater and sediment/faeces approaches, respectively.
From DNA extraction to real-time PCR analysis the efficiency of the method was 88.50% in seawater and 95.96% in sediment/faeces.
The developed approach has allowed the detection and quantification from 25 to 125,000 parasite per quarter membrane using the following linear regression between Cq and decimal logarithm of number of detected parasites (N):
Cq = −3.63*log(N) + 38.70(R2=0.98) (Figure 1A).
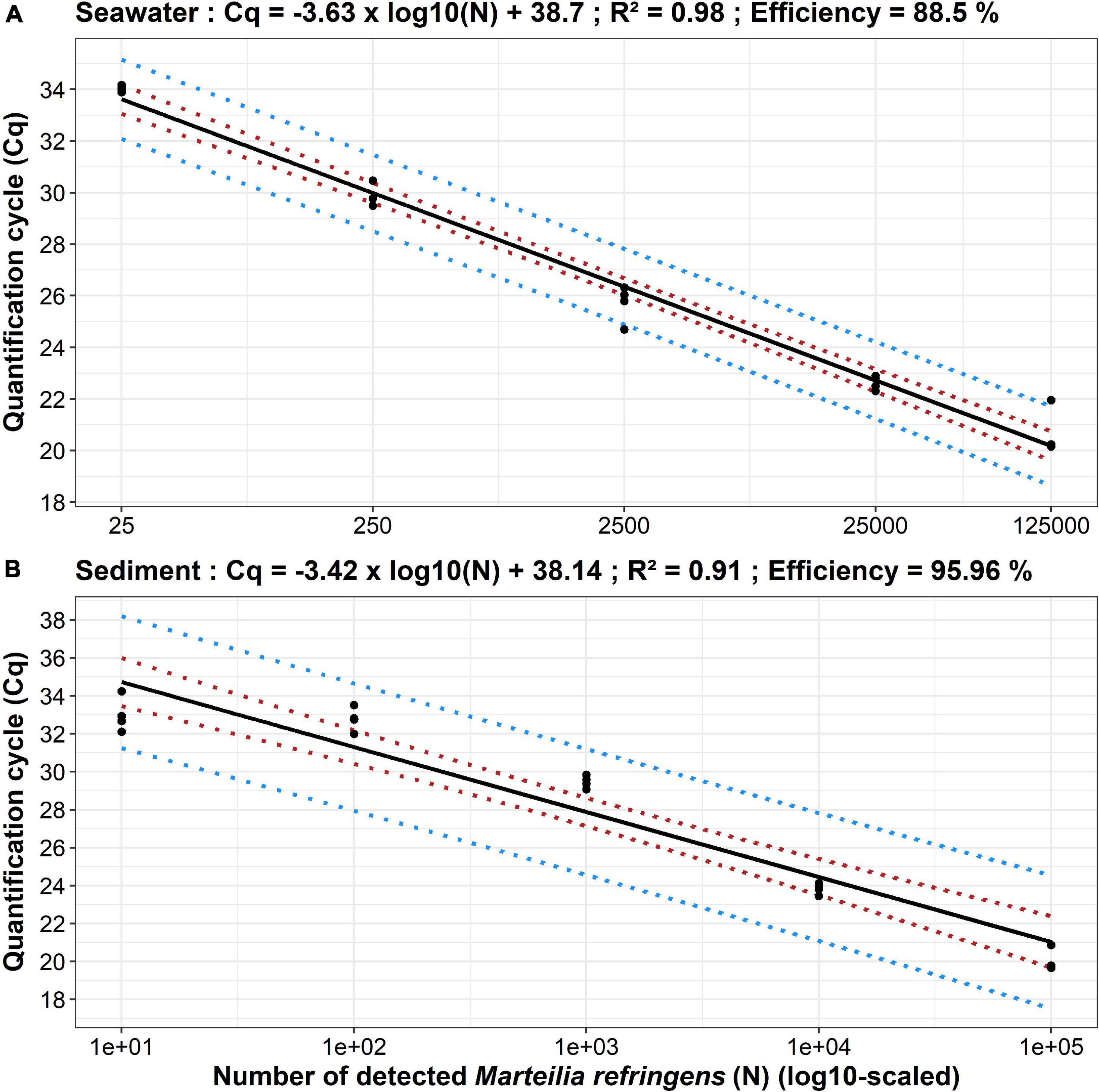
Figure 1. Standard curves established from seawater (A) and sediment (B) real-time PCR analyses of Marteilia refringens dilution series. Dots represent experimental data, solid lines represent linear regression, and dotted coloured lines represent 95% confidence (red) and 95% prediction (blue) intervals. For each parasite amount, n = 4 except for 125,000 parasites (seawater) and 100,000 parasites (sediment) where n = 3.
The developed approach has allowed the detection and quantification from 10 to 100,000 parasite per 0.25 g of sediment/faeces using the following linear regression between Cq and decimal logarithm of number of detected parasites (N):
Cq = −3.42*log(N) + 38.14(R2=0.91) (Figure 1B).
Marteilia refringens presence was monitored for up to 20 days in four seawater and faeces suspensions (A–D) containing parasites freshly released from infected flat oysters.
At t = 0 day, depending on the considered suspension, from 1.02 * 103 to 8.57 * 105 parasites were detected in seawater and from 4.44 * 103 to 1.40 * 106 parasites were detected in faeces (Supplementary Table 1). Whatever the parasite suspension, number of detected parasites was always higher in faeces than in seawater (from 1.63 times higher for suspension A to 9.49 times higher for suspension C) (Figure 2).
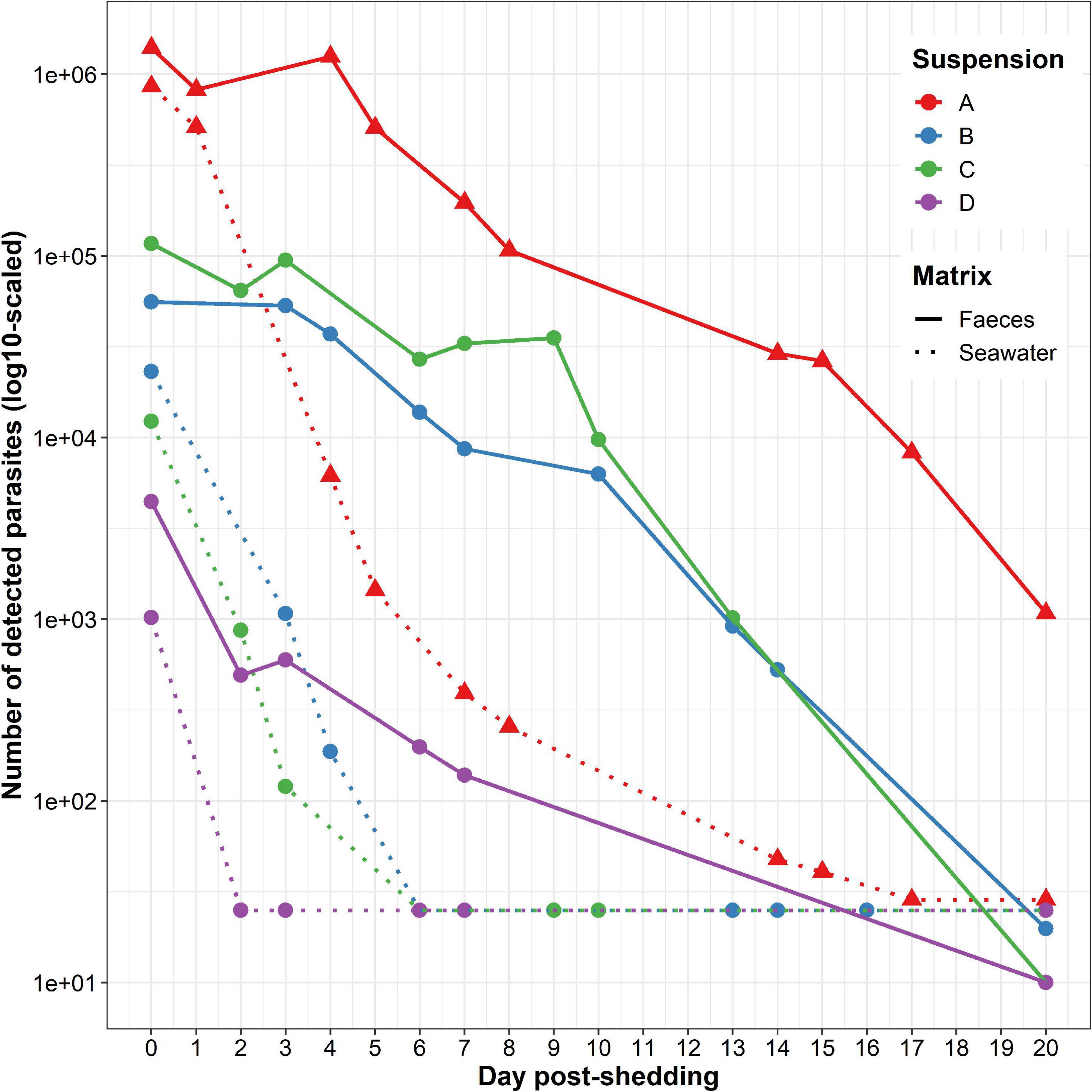
Figure 2. Marteilia refringens DNA detection in the four tested seawater (dotted lines) and faeces (solid lines) suspensions (A–D). Samples of seawater and faeces were collected every 1–5 days during 20 days after removing flat oysters.
At t = 20 days, depending on the considered suspension, between 25 and 29 parasites remained detectable in seawater whereas 10 to 1.08 * 103 parasites were detectable in faeces (Figure 2 and Supplementary Table 1).
Whatever the parasite suspension or matrix, the number of detected parasites globally decreased over time. The initial number of detected parasites appeared positively and strongly correlated with the number of detected parasites at t = 3 or 4 days post-release, depending on the considered suspension. Nevertheless, this observation was not significant (rho = 0.83, p-value = 5.83 * 10–2, Spearman’s rank correlation test rho). For this test, only amounts of parasites above the real-time PCR detection limit were taken into account. Therefore, suspension D was not included in this analysis because the number of detected M. refringens falls under the detection limit of the approach after the first sampling at t = 0 day post-release.
Considering each matrix separately, the number of detected parasites in suspension A was always higher than in other suspensions (Figure 2). In addition, we also observed a rapid decrease between t = 0 day and t = 5 days and a gentler decrease between t = 5 and t = 20 days in seawater. On the contrary, the decrease of parasite detection in faeces appeared stable throughout the whole experiment.
90% of the initial number of parasites was no longer detectable after 2–4 days post-release in seawater and after 6–13 days post-release in faeces.
Marteilia refringens presence was monitored by light microscopy in the different seawater and faeces suspensions. However, parasites were only observed in suspension A samples and thus only results obtained from seawater and faeces suspensions A are reported below.
Marteilia refringens cells were observed between t = 0 and t = 8 days post-release. These cells have been classified in three different “types” depending on the presence of a cell membrane surrounding spores and granules (Figures 3A–F). Types 1 and 2 (Figures 3A,B,D,E) consisted in secondary cells measuring about 20 μm in diameter and including four 3–4 μm spores and between three and five 2–3 μm granules. No secondary cell nucleus was observed. Spores generally showed two strongly basophilic nuclei and a less basophilic cytoplasm. Granules were usually observed close to spores and showed a strong basophilic coloration. Whereas spores and granules were enclosed in a secondary cell whose membrane appeared more or less visible in type 1 or type 2 (Figures 3A,D and 3B, respectively), type 3 consisted only in the 4 free spores associated to 3–5 granules (Figure 3C). Nevertheless, group of spores and granules corresponded to type 3 were counted as one cell, to conserve the same counting method from one cell type to another.
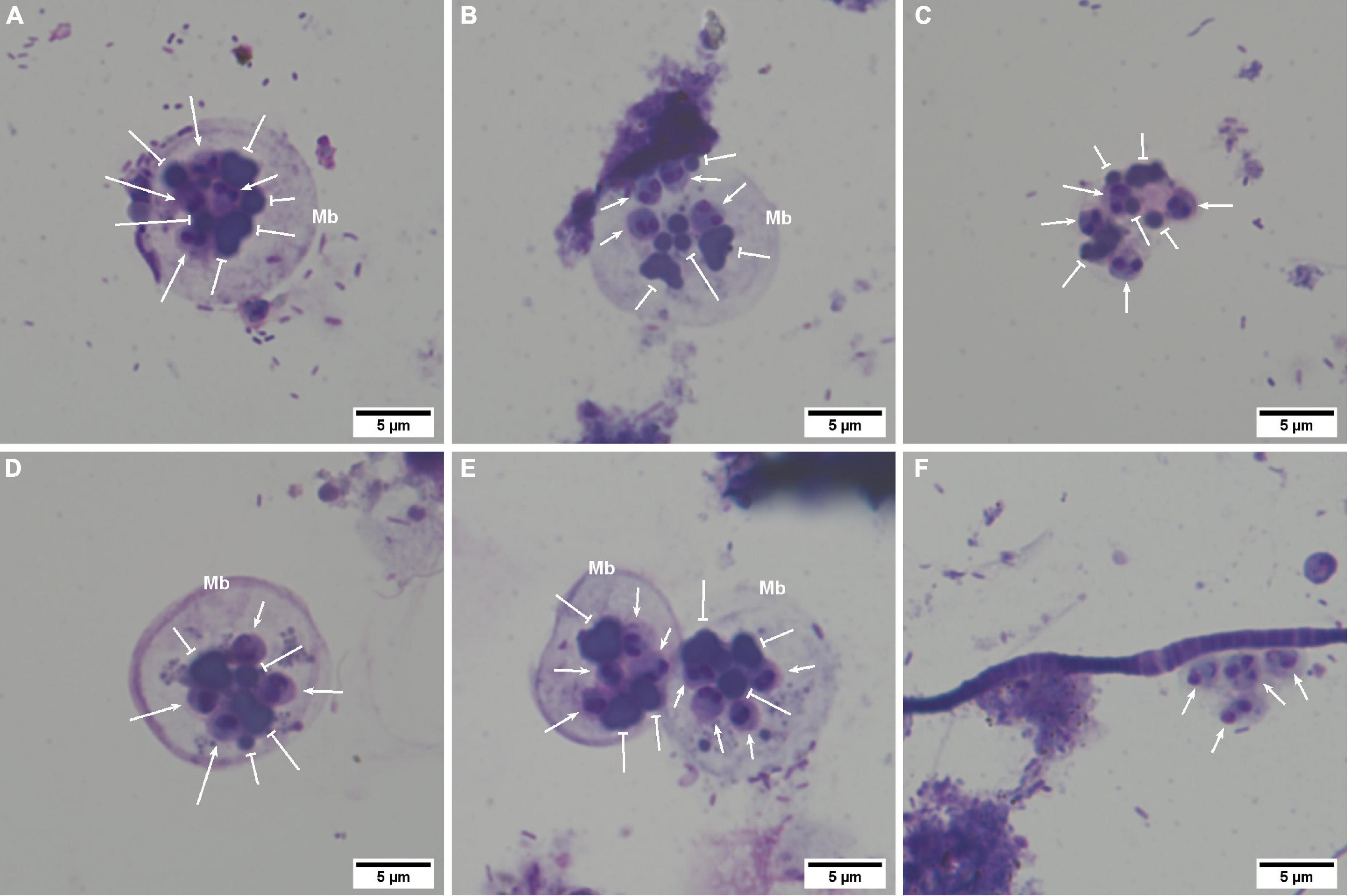
Figure 3. Marteilia refringens cells observed by light microscopy on seawater and faeces cytocentrifugations (Hémacolor® stained). (A) “Type-1 cell” containing at least four spores (notched-head arrow) and five refringent bodies (bar-head arrow). (B) “Type-2 cell” with a less visible membrane (Mb) than in type-1 containing at least four spores and six refringent bodies. (C) “Type-3 cell” with no visible cell membrane and showing at least four spores and four visible refringent bodies. (D) Thick membrane observed sometimes in type-1 cells. (E) Agglomerated secondary-cells. (F) Free spores non-associated with refringent bodies.
The number of observed secondary cells decreased over time (Figure 4). Moreover, the number of parasites in faeces was always higher than in seawater, despite the dilution performed before cytocentrifugation and the lower number of examined sections. As shown in Figure 4, type 1 was mostly observed at t = 0 day, type 2 at t = 0 and t = 1 day and type 3 after t = 1 day. Free spores non-associated with granules (Figure 3F) were also observed in faeces whatever the day and in seawater at t = 0 and t = 1 day.
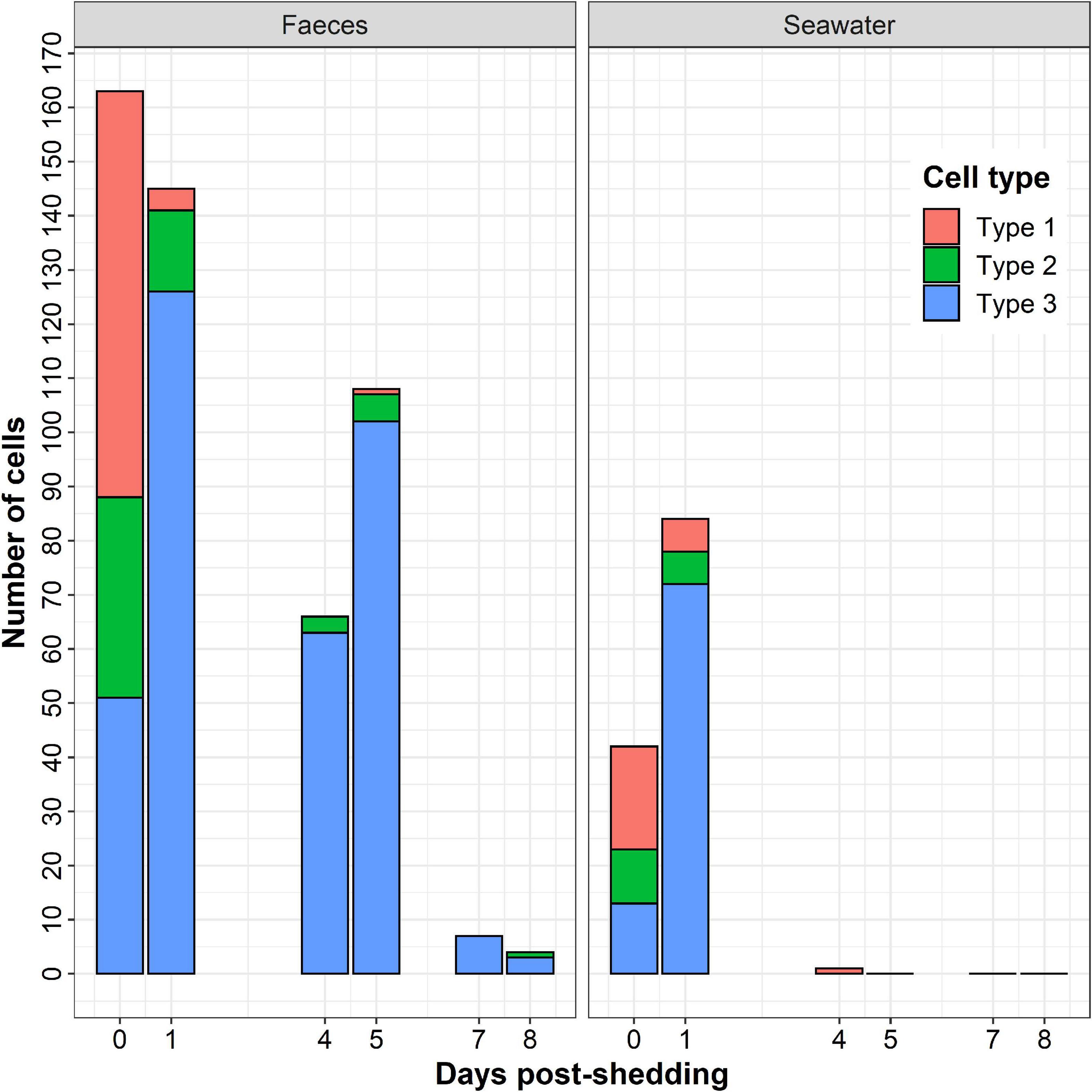
Figure 4. Number of Marteilia refringens cell-types (Type-1: cell with visible membrane containing spores and refringent bodies, Type-2: cell with less visible membrane than in type 1 containing spores and refringent bodies, Type 3: cell without visible membrane containing spores and refringent bodies) observed per day by light microscopy on seawater and faeces cytocentrifugations (Hémacolor® stained).
Finally, secondary cells appeared sometimes agglomerated by 2–4 (Figure 3E) in faeces between t = 0 day and t = 5 days and in seawater at t = 0 day, respectively.
Transmission electron microscopy examination could only be carried out on faeces suspension A at t = 0 day. All measures carried out to describe cell components are referred in Table 1.

Table 1. Measurements of Marteilia refringens sporangia and cell components observed by electron transmission microscopy.
As in light microscopy, sporangia (secondary cell containing matures spores) were surrounded by a more or less visible membrane (Figures 5A,B). Groups of spores surrounded by an undetermined matrix were also observed (Figure 5C). These different pictures might correspond to “types” 1, 2, and 3 described in the previous section. These forms appeared circular/oblong and included 1–3 rounded spores. Spores were associated with 0–5 oblong electron-dense bodies interpreted as refringent bodies.
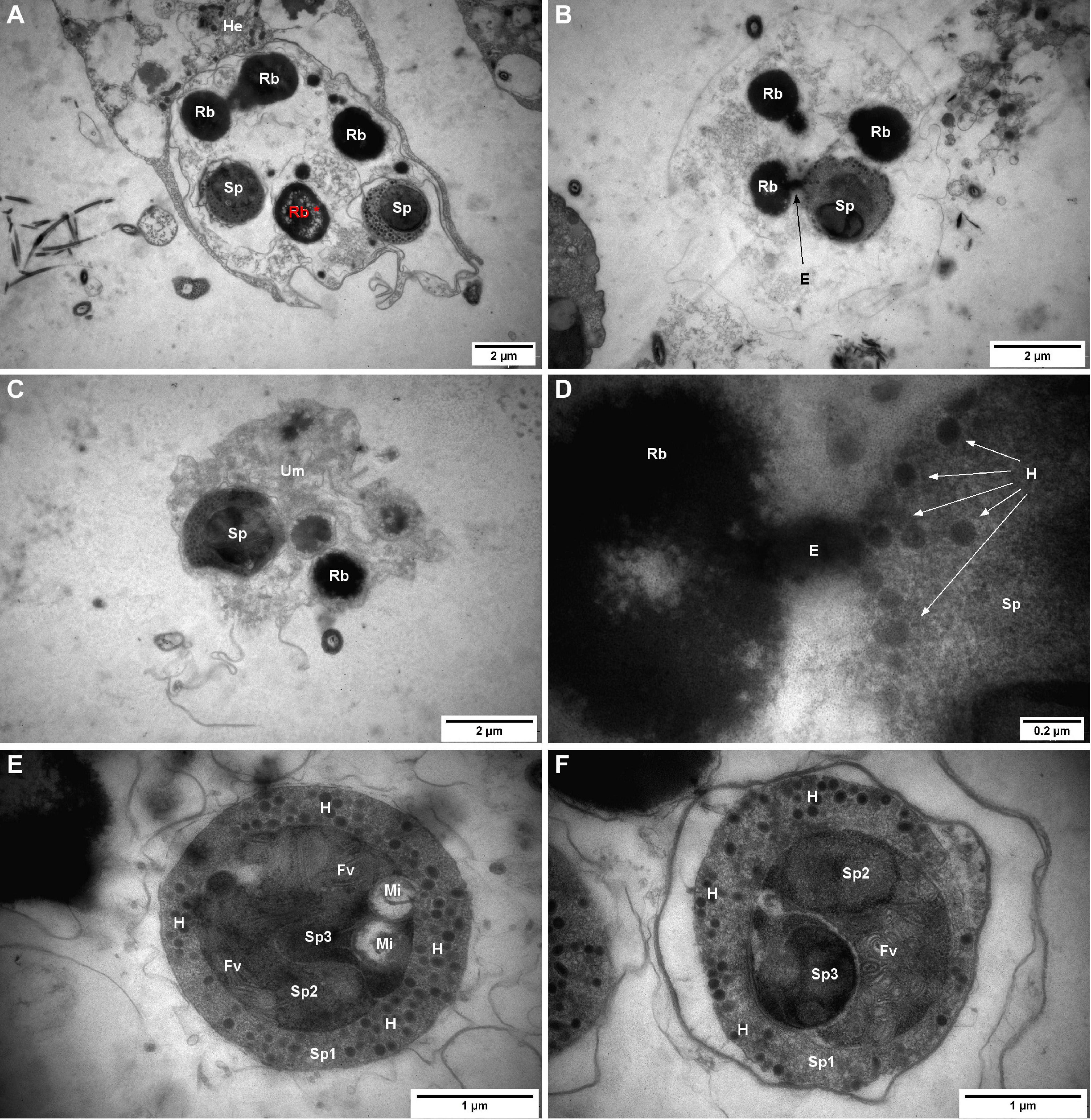
Figure 5. Marteilia refringens cells observed by transmission electron microscopy (TEM) in flat oyster faeces. (A) “Type-1 like cell” containing two matures spores (Sp) and four refringent bodies (Rb) whose show several empty zones (Rb*, red) and surrounded by flat oyster haemocyte (He). (B) “Type-2 like cell” containing one mature spore (Sp) and three refringent bodies (Rb) and showing an excrescence (E) between one refringent body and the mature spore. (C) “Type-3 like cell” containing one mature spore (Sp) and one visible refringent body (Rb) and surrounded by an undetermined matrix (Um). (D) Excrescence between refringent body (Rb) and mature spore (Sp) visible in panel (B) and showing a possible synthesis of haplosporosomes (H). (E) Mature spore containing haplosporosomes (H), mitochondria (Mi), flattened vesicles (Fv), and showing three sporoplasms levels: outermost (Sp1), intermediate (Sp2), and innermost (Sp3). (F) Mature spore containing haplosporosomes (H), flattened vesicles (Fv), and showing three sporoplasms levels: outermost (Sp1), intermediate (Sp2), and innermost (Sp3).
No nucleus or cell organelles besides spores and refringent bodies were observed or identified in sporangia. Among all the observed refringent bodies, 18% (7/39) showed a surrounding membrane and 13% showed a heterogeneous content including like “empty zones” (Figure 5A*). Moreover, in two sporangia, refringent bodies showed excrescence towards spores (Figures 5B,D).
In 71% of observed spores (20/28), it was possible to distinguish three sporoplasm levels (Figures 5E,F): a rounded outermost sporoplasm containing circular haplosporosomes, and an intermediate sporoplasm, containing itself one oblong innermost sporoplasm.
A mass of flattened vesicules or tubules was observed in 80% (12/15) of the observed spores and probably located in the intermediate sporoplasm (Figures 5E,F). Mitochondria with DNA fibrils were observed in some spores and appeared located within the sporoplasm (Figure 5E). In 73% (11/15) of observed outermost sporoplasms, space was observed between spore membrane and spore cytoplasm.
Studying and monitoring non-cultivable small parasites such as M. refringens present some challenges because of their small size, the difficulty to capture them from the environment and the impossibility to isolate them on culture media (Bass et al., 2015). By allowing the detection of cryptic or elusive organisms, large scale sampling, rapid, non-invasive and cost-efficiency monitoring, environmental DNA (eDNA) based-approaches overcome most difficulties met when investigating non-cultivable small parasites (Harper et al., 2019).
In order to study M. refringens survival once released from the flat oyster, we have developed an eDNA based approach allowing both the detection and quantification of parasite DNA in water and in oyster faeces. The method has been developed and optimised by testing serial dilutions water and sediment artificially spiked with parasites isolated from infected flat oysters. Sediment was used to mimic flat oyster faeces during the development of the method because of the similarity between these two matrices (Taberlet et al., 2012; Thomsen and Willerslev, 2015).
In seawater, our method allowed to detect and quantify from 2.5 * 101 to 1.25 * 105 parasites and showed a global efficiency of 88.50%. The performance of our method appears close to the performance of the eDNA based approach developed to detect and quantify the flat oyster parasite B. ostreae (Mérou et al., 2020). In sediment/faeces, the method showed similar results allowing the detection of 101 to 105 parasites with a global efficiency of 95.96%. These approaches both show satisfactory efficiencies, although the detection threshold appears lower in the sediment, which could be due to the presence of inhibitors in seawater or uneven recovery of cells.
The main limitation of targeting DNA to monitor micro-organism presence in environmental samples is that DNA detection does not allow to distinguish between metabolically active forms and inactive forms, such as cysts, spores, or eggs or even free extracellular DNA from degraded or dead cells (Bass et al., 2015). Although DNA persistence in seawater could be very short (as low 10 h) (Dell’Anno and Corinaldesi, 2004), suggesting that its detection can be associated to the recent presence of targeted microorganisms, other approaches such as microscopical ones are needed, especially to describe parasite stages.
The new developed eDNA based method was combined with light and transmission electron microscopy observations to study the survival of M. refringens freshly released from infected flat oysters in both seawater and flat oyster faeces during 20 days.
In real-time PCR, the number of detected parasites decreased in both seawater and flat oyster faeces over time. However, it has been possible to detect M. refringens DNA in both matrices until 20 days after release, suggesting that the parasite is able to persist at least 20 days in seawater and oyster faeces. In comparison, B. ostreae DNA was no longer detected after 7 days post-release in seawater (Mérou et al., 2020). The monitoring carried out in this study was stopped 20 days after parasite release from the flat oysters, however, the parasite may be able to persist longer in the environment. This hypothesis is supported by the apparent ability of M. refringens to persist for several years in a bay whereas the oyster production has been stopped (Grizel et al., 1974; Perkins, 1976), which could be due to the persistence of the parasite under spore forms.
In our study, whatever the considered suspension, the number of detected parasites was always higher in flat oyster faeces than in seawater. Moreover, more than 90% of initially detected parasites were no longer detected after 2–4 days post-release in seawater, against 6–13 days in flat oyster faeces. These results suggest that M. refringens mortality is higher in seawater than in oyster faeces and, in contrast, parasite persistence is higher in flat oyster faeces than in seawater. As the parasite is released from the flat oyster through faeces (Grizel et al., 1974; Perkins, 1976), this matrix might favour its survival, which might explain the difference observed between oyster faeces and seawater. In addition, light microscopy observations revealed that membrane of secondary cells persist during a longer time in faeces than in seawater, which could also support the hypothesis of a higher persistence of the parasites in this matrix. However, the experiment was carried out under controlled conditions (constant temperature, antibiotics, filtered seawater) that might favour or disadvantage parasite survival, depending on its abiotic optimums and its interactions with other microorganisms. As Arzul et al. (2009) did on B. ostreae, it could be interested to investigate abiotic variations on M. refringens survival in seawater and faeces, in order to better understand its survival outside the flat oyster.
In light microscopy, the parasite in seawater and faeces showed the same morphology as described in flat oysters and appeared as 20 μm secondary cells containing four 3–4 μm spores associated with three to five 2–3 μm granules (Longshaw et al., 2001; Elgharsalli et al., 2013; Itoh et al., 2014; Darriba et al., 2020). Moreover, the number of parasites decreased over time and was always higher in flat oyster faeces than in seawater, confirming results obtained by real-time PCR. Different cell-types were observed and might reflect the transformation of the parasite after its release from the flat oyster. Indeed, most of initially observed parasites consisted in secondary cells surrounded by a cell membrane and containing spores and granules. As soon as one day after release, most of parasites do not show any visible membrane around spores and granules.
These results together with the observation of sporangia in the lumen of the digestive diverticula, intestine, and rectum (Grizel et al., 1974; Perkins, 1976) indicate that the parasite is released through the digestive system under secondary cell forms containing spores and granules. The secondary cell membrane is then rapidly degraded, leaving free spores in the environment.
The detection and observation of the parasites preferentially in oyster faeces but also in seawater suggest that soon after its release from the flat oysters the parasite can be spread by currents but probably mostly joins the sediment.
In a recent study on Marteilia cochillia, a parasite of the cockle Cerastoderma edule (Darriba et al., 2020), have shown that this parasite is released from its host in seawater through white thread-like pellets consisting of millions of tightly packed M. cochillia sporangia. In our experiment, we did not observe white thread-like pellets which might be due to the burrowing behaviour of the cockle.
Transmission Electron Microscopy analysis was carried out on faeces samples at t = 0 day. The ultrastructure of the parasite was in accordance with previous descriptions of M. refringens in bivalves (Longshaw et al., 2001; Elgharsalli et al., 2013) as similar cell components and size were observed. However, in our study, different parasite forms could be observed and might correspond to the three cell-types described in light microscopy. When the membrane of the secondary cell was not visible anymore, spores and granules appeared included in an undetermined matrix rather than completely free as suggested in light microscopy.
It is noteworthy that all the secondary cells showed spores with haplosporosomes in the outermost sporoplasm. This observation suggests that only secondary cells with mature spores are released from the flat oysters which is in agreement with previous studies (Grizel et al., 1974; Perkins, 1976). In addition to haplosporosomes, spores displayed mitochondria with DNA fibrils and in the intermediate sporoplasm flattened vesicules or tubules as reported by Perkins (1976). However, in contrast with this study, spores observed herein had darker sporoplasms rich in ribosomes and did not show a clear spore wall. Indeed, when visible, the membrane around the outermost sporoplasm appeared thinner and disconnected from the cell. Although this picture can be an artefact linked to the preparation of the sample, it might also reflect the transformation of the spore when maturing outside its host.
We also noticed excrescences between inclusion bodies and spores, suggesting that refringent bodies might play a role of reserve material used in the maturation of the spores. In comparison with results from Perkins (1976), inclusions were smaller and had a less homogeneous content. As we have not observed a clear spore wall, further spore maturation event might occur and complementary investigation is needed to understand the role of spore organelles including refringent bodies and haplosporosomes as well as the role of the different sporoplasm layers.
To our knowledge, this is the first study investigating M. refringens outside its host O. edulis. The combination of eDNA-based and microscopical approaches have allowed us not only to detect but also quantify and describe the parasite in seawater and flat oyster faeces. Our results suggest that the parasite can survive at least 20 days in both matrices. However, oyster faeces seem to favour parasite survival as compared to seawater. If the parasite can be spread through currents it seems more probable that it mostly joins the sediment which appears as an interesting compartment to investigate regarding the cycle of M. refringens. We confirm that sporangia containing mature spores are released through the digestive tract. Shortly after their release, parasites loose the membrane of the secondary cell releasing spores and refringent bodies. Although transmission electron microscopy has allowed describing the ultrastructure of the spores one day after sporangia release, complementary examination at later stage would be required to better describe the resistant form of the parasite and investigate its potential maturation process. By suggesting that M. refringens can be spread at a metapopulation scale through water currents and can persist for a long time in sediment under spore forms, our results indicate that it may be difficult to completely eradicate M. refringens from the environment highlighting the importance of prevention measures in marteiliosis management.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
NM, SP, and IA conceived the ideas, designed the methodology, collected the data, analysed the data, and wrote the manuscript. CL helped to collect the data, performed confirmation analysis from biological samples, and helped with data analysis. TB collected the data. BC and IA performed transmission electron microscopy analyses. All authors read and edited the manuscript, and gave final approval for publication.
This work was supported by the French Research Institute for Exploitation of the Sea (IFREMER) and Région Nouvelle-Aquitaine. This work received financial support from the European Union Reference Laboratory for Molluscs Diseases, the French National Research Agency (ANR) project ENVICOPAS (15-CE35-0004) and the European project VIVALDI (H2020 n°678589).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.811284/full#supplementary-material
Arzul, I., and Carnegie, R. B. (2015). New perspective on the Haplosporidian parasites of molluscs. J. Invertebr. Pathol. 131, 32–42. doi: 10.1016/j.jip.2015.07.014
Arzul, I., Chollet, B., Boyer, S., Bonnet, D., Gaillard, J., Baldi, Y., et al. (2014). Contribution to the understanding of the cycle of the protozoan parasite Marteilia refringens. Parasitology 141, 227–240. doi: 10.1017/S0031182013001418
Arzul, I., Gagnaire, B., Bond, C., Chollet, B., Morga, B., Ferrand, S., et al. (2009). Effects of temperature and salinity on the survival of Bonamia ostreae, a parasite infecting flat oysters Ostrea edulis. Dis. Aquat. Org. 85, 67–75. doi: 10.3354/dao02047
Audemard, C., Barnaud, A., Collins, C. M., Le Roux, F., Sauriau, P. G., Coustau, C., et al. (2001). Claire ponds as an experimental model for Marteilia refringens life-cycle studies: new perspectives. J. Exp. Mar. Biol. Ecol. 257, 87–108. doi: 10.1016/s0022-0981(00)00330-0
Audemard, C., Le Roux, F., Barnaud, A., Collins, C., Sautour, B., Sauriau, P. G., et al. (2002). Needle in a haystack: involvement of the copepod Paracartia grani in the life-cycle of the oyster pathogen Marteilia refringens. Parasitology 124, 315–323. doi: 10.1017/s0031182001001111
Bass, D., Stentiford, G. D., Littlewood, D. T. J., and Hartikainen, H. (2015). Diverse applications of environmental DNA methods in parasitology. Trends Parasitol. 31, 499–513. doi: 10.1016/j.pt.2015.06.013
Berthe, F. C. J., Le Roux, F., Adlard, R. D., and Figueras, A. (2004). Marteiliosis in molluscs: a review. Aquat. Living Resour. 17, 433–448. doi: 10.1051/alr:2004051
Boyer, S., Chollet, B., Bonnet, D., and Arzul, I. (2013). New evidence for the involvement of Paracartia grani (Copepoda, Calanoida) in the life cycle of Marteilia refringens (Paramyxea). Int. J. Parasitol. 43, 1089–1099. doi: 10.1016/j.ijpara.2013.07.008
Buestel, D., Ropert, M., Prou, J., and Goulletquer, P. (2009). History, status, and future of oyster culture in France. J. Shellfish Res. 28, 813–820. doi: 10.2983/035.028.0410
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Canier, L., Dubreuil, C., Noyer, M., Serpin, D., Chollet, B., Garcia, C., et al. (2020). A new multiplex real-time PCR assay to improve the diagnosis of shellfish regulated parasites of the genus Marteilia and Bonamia. Prev. Vet. Med. 183:105126. doi: 10.1016/j.prevetmed.2020.105126
Carrasco, N., Arzul, I., Chollet, B., Robert, M., Joly, J. P., Furones, M. D., et al. (2008). Comparative experimental infection of the copepod Paracartia grani with Marteilia refringens and Marteilia maurini. J. Fish Dis. 31, 497–504. doi: 10.1111/j.1365-2761.2008.00910.x
Carrasco, N., Green, T., and Itoh, N. (2015). Marteilia spp. parasites in bivalves: a revision of recent studies. J. Invertebr. Pathol. 131, 43–57. doi: 10.1016/j.jip.2015.07.016
Carrasco, N., López-Flores, I., Alcaraz, M., Furones, M. D., Berthe, F. C. J., and Arzul, I. (2007b). First record of a Marteilia parasite (Paramyxea) in zooplankton populations from a natural estuarine environment. Aquaculture 269, 63–70. doi: 10.1016/j.aquaculture.2007.03.030
Carrasco, N., López-Flores, I., Alcaraz, M., Furones, M. D., Berthe, F. C. J., and Arzul, I. (2007a). Dynamics of the parasite Marteilia refringens (Paramyxea) in Mytilus galloprovincialis and zooplankton populations in Alfacs Bay (Catalonia, Spain). Parasitology 134, 1541–1550. doi: 10.1017/S0031182007003009
Comps, M. (1970). Observations sur les causes d’une mortalite anormale des huitres plates dans la Bassin de Marennes. Rev. Trav. Inst. Pêches Marit. 34, 317–326.
Cornillon, P.-A., Husson, F., Jégou, N., Matzner-Lober, E., Josse, J., Guyader, A., et al. (2012). Statistiques Avec R. Rennes: Presses universitaires de Rennes.
Darriba, S., Iglesias, D., and Carballal, M. J. (2020). Marteilia cochillia is released into seawater via cockle Cerastoderma edule faeces. J. Invertebr. Pathol. 172:107364. doi: 10.1016/j.jip.2020.107364
Dell’Anno, A., and Corinaldesi, C. (2004). Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 70, 4384–4386. doi: 10.1128/AEM.70.7.4384-4386.2004
Elgharsalli, R., Aloui-Bejaoui, N., Salah, H., Chollet, B., Joly, J. P., Robert, M., et al. (2013). Characterization of the protozoan parasite Marteilia refringens infecting the dwarf oyster Ostrea stentina in Tunisia. J. Invertebr. Pathol. 112, 175–183. doi: 10.1016/j.jip.2012.11.004
Gercken, J., and Schmidt, A. (2014). Current Status of the European Oyster (Ostrea edulis) and Possibilities for Restoration in the German North Sea. Bonn: Bundesamt für Naturschutz BfN, 1–96.
Grizel, H. (1985). Etude des Recentes Epizooties de L’huitre Plate Ostrea Edulis Linne et de Leur Impact sur L’ostreiculture Bretonne. Ph.D. thesis. Montpellier: Universite des sciences et techniques du Languedoc.
Grizel, H., Comps, M., Bonami, J. R., Cousserans, F., Duthoit, J. L., and Pennec, M. A. (1974). Recherche sur l’agent de la maladie de la glande digestive de Ostrea edulis Linne. Sci. Pech. Bull. Inst. Pêches Marit. 240, 7–30.
Harper, L. R., Buxton, A. S., Rees, H. C., Bruce, K., Brys, R., Halfmaerten, D., et al. (2019). Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds. Hydrobiologia 826, 25–41. doi: 10.1007/s10750-018-3750-5
Itoh, N., Yamamoto, T., Kang, H. S., Choi, K. S., Green, T. J., Carrasco, N., et al. (2014). A novel paramyxean parasite, Marteilia granula sp. nov. (Cercozoa), from the digestive gland of Manila clam Ruditapes philippinarum in Japan. Fish Pathol. 49, 181–193. doi: 10.1016/j.jip.2019.03.006
Le Roux, F., Lorenzo, G., Peyret, P., Audemard, C., Figueras, A., Vivarès, C., et al. (2001). Molecular evidence for the existence of two species of Marteilia in Europe. J. Eukaryot. Microbiol. 48, 449–454. doi: 10.1111/j.1550-7408.2001.tb00178.x
Linnaeus, C. (1758). Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis, Vol. 1, 10th Edn. Holmiae: Laurentius Salvius, 824. Available online at: https://biodiversitylibrary.org/page/726886
Longshaw, M., Feist, S. W., Matthews, R. A., and Figueras, A. (2001). Ultrastructural characterisation of Marteilia species (Paramyxea) from Ostrea edulis, Mytilus edulis and Mytilus galloprovincialis in Europe. Dis. Aquat. Org. 44, 137–142. doi: 10.3354/dao044137
López-Flores, I., De La Herrán, R., Garrido-Ramos, M. A., Navas, J. I., Ruiz-Rejón, C., and Ruiz-Rejón, M. (2004). The molecular diagnosis of Marteilia refringens and differentiation between Marteilia strains infecting oysters and mussels based on the rDNA IGS sequence. Parasitology 129, 411–419. doi: 10.1017/s0031182004005827
López-Flores, I., Garrido-Ramos, M. A., de la Herran, R., Ruiz-Rejón, C., Ruiz-Rejón, M., and Navas, J. I (2008a). Identification of Marteilia refringens infecting the razor clam Solen marginatus by PCR and in situ hybridization. Mol. Cell. Probes 22, 151–155. doi: 10.1016/j.mcp.2008.01.002
López-Flores, I., Robles, F., Valencia, J. M., Grau, A., Villalba, A., De La Herrán, R., et al. (2008b). Detection of Marteilia refringens using nested PCR and in situ hybridisation in Chamelea gallina from the Balearic Islands (Spain). Dis. Aquat. Org. 82, 79–87. doi: 10.3354/dao01966
Mérou, N., Lecadet, C., Pouvreau, S., and Arzul, I. (2020). An eDNA/eRNA-based approach to investigate the life cycle of non-cultivable shellfish micro-parasites: the case of Bonamia ostreae, a parasite of the European flat oyster Ostrea edulis. Microb. Biotechnol. 13, 1807–1818. doi: 10.1111/1751-7915.13617
Mialhe, E., Bachere, E., Le Bec, C., and Grizel, H. (1985). Isolement et purification de Marteilia (Protozoa: Ascetospora) parasites de bivalves marins. C. R. Acad. Sci. Sér. 3 Sci. Vie 301, 137–142.
Pascual, S., Villalba, A., Abollo, E., Garci, M., González, A. F., Nombela, M., et al. (2010). The mussel Xenostrobus securis: a well-established alien invader in the Ria de Vigo (Spain, NE Atlantic). Biol. Invasions 12, 2091–2103. doi: 10.1007/s10530-009-9611-4
Perkins, F. O. (1976). Ultrastructure of sporulation in the European flat oyster pathogen, Marteilia refringens—taxonomic implications. J. Protozool. 23, 64–74. doi: 10.1111/j.1550-7408.1976.tb05247.x
Pogoda, B., Brown, J., Hancock, B., Preston, J., Pouvreau, S., Kamermans, P., et al. (2019). The native oyster restoration alliance (NORA) and the Berlin oyster recommendation: bringing back a key ecosystem engineer by developing and supporting best practice in Europe. Aquat. Living Resour. 32:13. doi: 10.1051/alr/2019012
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Taberlet, P., Coissac, E., Hajibabaei, M., and Rieseberg, L. H. (2012). Environmental DNA. Mol. Ecol. 21, 1789–1793.
Keywords: Marteilia refringens, Ostrea edulis, survival, protozoan, parasite, shellfish, eDNA, transmission electron microscopy
Citation: Mérou N, Lecadet C, Billon T, Chollet B, Pouvreau S and Arzul I (2022) Investigating the Environmental Survival of Marteilia refringens, a Marine Protozoan Parasite of the Flat Oyster Ostrea edulis, Through an Environmental DNA and Microscopy-Based Approach. Front. Mar. Sci. 9:811284. doi: 10.3389/fmars.2022.811284
Received: 08 November 2021; Accepted: 07 January 2022;
Published: 31 January 2022.
Edited by:
Ciro Rico, Institute of Marine Sciences of Andalusia, Spanish National Research Council (CSIC), SpainReviewed by:
Gorka Bidegain, University of the Basque Country, SpainCopyright © 2022 Mérou, Lecadet, Billon, Chollet, Pouvreau and Arzul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabelle Arzul, SXNhYmVsbGUuQXJ6dWxAaWZyZW1lci5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.