- 1Departamento de Botánica, Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 2Instituto de Ecología, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 3UBIPRO, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 4Instituto de Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, Mexico City, Mexico
Deep-sea sediments are vast microbial habitats that cover almost two-thirds of the planet’s surface. Particularly, sediments in hydrothermal vents and oxygen minimum zones (OMZ) represent emblematic, and poorly understood extreme niches that pose strong selective pressures to life, representing untapped reservoirs of a unique microbial diversity. Nonetheless, the mycobiota in these systems remains relatively unknown. Here, we explored fungal diversity and community structure in deep-sea sediments collected from low- and high-temperature vent systems (Pescadero Basin, Pescadero Transform Fault, and Alarcón Rise) and an OMZ (Alfonso Basin) in the southern Gulf of California, by using high-throughput Illumina sequencing of the ITS1 region. We identified 102 fungal amplicon sequence variants (ASVs), principally affiliated to the Ascomycota and Basidiomycota. Our results also evidenced a high proportion of widely distributed, uncultured phylotypes (e.g., unknown clades closely related to the Basidiomycota such as Fungi sp. 18 and 19 formerly reported from deep-sea), evidencing the copious occurrence of novel fungal lineages with limited distribution to deep-sea sediments. Overall, the largest number of ASVs was recovered from high-temperature vent systems, corroborating these as diversity hotspots. In addition, clustering patterns across samples and the FUNGuild analysis revealed characteristic assemblages in each ecosystem, which could be linked to site-specific processes. This study provides baseline data paving the way for a better understanding of deep-sea fungal diversity, evidencing its potential importance in ecosystem functioning.
Introduction
Deep-sea seafloor (below 200 m depth) covers around 95% of Earth’s surface, representing the largest biome (Danovaro et al., 2014, 2017). This biome is characterized by the absence of light (leading to severe organic nutrient limitations), high hydrostatic pressures, and average temperatures of 4°C (Gage and Tyler, 1991). It sustains some of the most extreme and unique ecosystems on Earth, such as hydrothermal vents and oxygen minimum zones (OMZs), yet remains as one of the least studied because of its remoteness and the technological challenges for its investigation (Ramirez-Llodra and Billett, 2006).
On the one hand, hydrothermal vents are extreme environments where fluids that are geothermally heated and enriched in reduced compounds and trace metals exit to the seafloor reaching temperatures from 330 to 400°C (Van Dover, 2014). They host complex biological communities where primary production is fundamentally driven by sulfur-based chemosynthesis, as fluids provide free-living and endosymbiotic bacteria with chemical energy. These fascinating environments are globally distributed along mid-ocean spreading centers, back-arc spreading centers, and at some seamounts (Van Dover et al., 2002; revised in Dick, 2019). On the other hand, OMZ are steady-state low-oxygen layers at mid-water depths (Diaz et al., 2013) that are widespread in the northeast subarctic Pacific, the northern Indian Ocean, and the Eastern Tropical Pacific (Paulmier and Ruiz-Pino, 2009). They are formed in areas of high primary production in the surface waters and poor circulation, where the biological degradation of the sinking organic matter results in oxygen depletion (Rogers, 2000; Levin, 2003). The species that live in these areas have low oxygen tolerance thresholds, morphological adaptations, specialist rather than opportunistic lifestyles, and the potential to utilize chemosynthesis-based nutritional pathways (Levin, 2003). Globally, about 1,150,000 km2 of the seafloor is exposed to oxygen concentrations <0.7 mg O2/l from OMZs (Helly and Levin, 2004).
Our knowledge of deep-sea biodiversity only hints at thousands of undiscovered organisms and their potential benefits (Barbier et al., 2014). Despite its incipient exploration (Grossart et al., 2019), in recent years, assorted deep-sea ecosystems have been recognized as important fungal habitats, including subseafloor igneous crust (e.g., Ivarsson et al., 2016), seafloor sediments (e.g., Vargas-Gastélum et al., 2019), hydrothermal vents (e.g., Burgaud et al., 2009), OMZs (e.g., Orsi and Edgcomb, 2013; Manohar et al., 2014), whale falls (Nagano et al., 2020), among others. Furthermore, increasing evidence supports fungal active involvement in marine biogeochemical processes (Stoeck et al., 2007; Alexander et al., 2009; Stock et al., 2009; Edgcomb et al., 2011; Manohar et al., 2015; Quemener et al., 2020), yet our understanding of deep-sea fungal diversity remains in its infancy.
To date, culture-dependent and independent studies hint at the Ascomycota and Basidiomycota as the most abundant groups in hydrothermal vents (Burgaud et al., 2009; Xu et al., 2017, 2018), as well as Chytridiomycota (Le Calvez et al., 2009). In addition to a fungal lineage formed by environmental 18S rDNA sequences reported exclusively from submarine and continental hydrothermal systems (López-García et al., 2007). Similarly, fungal communities in OMZ seem to be dominated by Basidiomycota, Ascomycota, and early diverging phyla (Jebaraj et al., 2010; Manohar et al., 2015; Peng and Valentine, 2021). Nevertheless, our knowledge of deep-sea fungal diversity in hydrothermal vents and OMZ remains limited, resulting in a rudimentary understanding of ecosystem functioning and resilience.
Since their recent discovery, the biological diversity and ecology of the hydrothermal vents of Pescadero Basin (PB), Pescadero Transform Fault (PTF), and Alarcón Rise (AR), as well as the OMZ of Alfonso Basin (AB) has been increasingly investigated (Goffredi et al., 2017, 2021; Salcedo et al., 2019, 2021; Espinosa-Asuar et al., 2020). Nevertheless, their fungal diversity remained unknown, hampering the robust view of ecosystem functioning. In this work, we report the first ITS rRNA survey of fungal diversity in deep-sea sediments collected in PB, PTF, AR, and AB, in the southern Gulf of California, Mexico. Our results evidenced that a diverse and functionally multifaceted microfungal community inhabits these deep-sea extreme ecosystems, revealing that the conserved core of fungal taxa across ecosystems comprised barely 3.9% of the total community. A lot of work remains to be done in order to obtain a full picture of fungal diversity in this region, and to understand compositional differences across ecosystems. Therefore, major efforts must be directed at further systematic samplings associated to DNA barcoding and culturing to validate our observations.
Materials and Methods
Sampling Sites
Sediment samples were collected in four localities (PB, PTF, AR, and AB), distributed along the SW margin of the Gulf of California and the central portion of the gulf’s mouth (Supplementary Figure 1). This semi-enclosed, dynamic basin is located at the NE Pacific Ocean, between 22°–32°N, and 105°–107°W. Detailed information of the biotic and abiotic setting of the study sites is available in Goffredi et al. (2017); Clague et al. (2018), Paduan et al. (2018), and Sánchez et al. (2018). Briefly, the PB is a sedimented basin located at the entrance of the Gulf of California near 24° N. It includes a series of large carbonate mounds and chimney edifices, where venting fluids reach up to 290°C, and contain high concentrations of hydrocarbons, hydrogen and hydrogen sulfide (Goffredi et al., 2017). The PTF hosts discrete low-temperature hydrothermal vents (fluids exiting at 5°C), with substrate composed of volcanic rubble and sediment (Clague et al., 2018; Salcedo et al., 2019). The AR vent system is located at the SW end of the Gulf of California, and is bounded by the 60 km long Tamayo Transform Fault, which connects it to the 21°N segment of the East Pacific Rise (Paduan et al., 2018). This basalt-hosted system is characterized by surface lava flows; thick sediments are evident only along the transform faults (Clague et al., 2018). It hosts black smokers and chimneys composed of iron, copper, and zinc sulfides. Their end-member fluids reach a maximum temperature of 360°C and contain high concentrations of hydrogen sulfide and metal-sulfides (Paduan et al., 2018). Lastly, the AB is located within La Paz Bay, where the deepwater that enters the basin is drown from low-oxygen intermediate waters; and below 200 m, the water is suboxic to anoxic (González-Yajimovich et al., 2005). Sediments in this system are characterized by terrigenous mud as the main component (Douglas et al., 2002).
Sample Collection
Seven sediment cores were collected by the articulating manipulator arm of the remotely operated vehicle Doc Ricketts, during the expedition Vents and Seeps (2015) by Monterey Bay Aquarium Research Institute, on the R/V Western Flyer under the permit numbers SRE: #CTC/01700/15 and SAGARPA: DGOPA-02919/14 issued by the Mexican government. Onboard, a subsample from the base of each core was obtained (10-cm length section by using a sterile PVC cylinder), sealed, and deep-frozen at −80°C. The material was collected from accessible, randomly chosen sites in the following locations: (1) PB, high-temperature hydrothermal vent fields (three cores); (2) AR, high-temperature hydrothermal vent fields (one core); (3) PTF, low-temperature hydrothermal vents (one core), and (4) AB, OMZ (two cores) (Table 1). The disparity in the number of samples from each locality depended on the limited exploration time and the geological settings of every site.
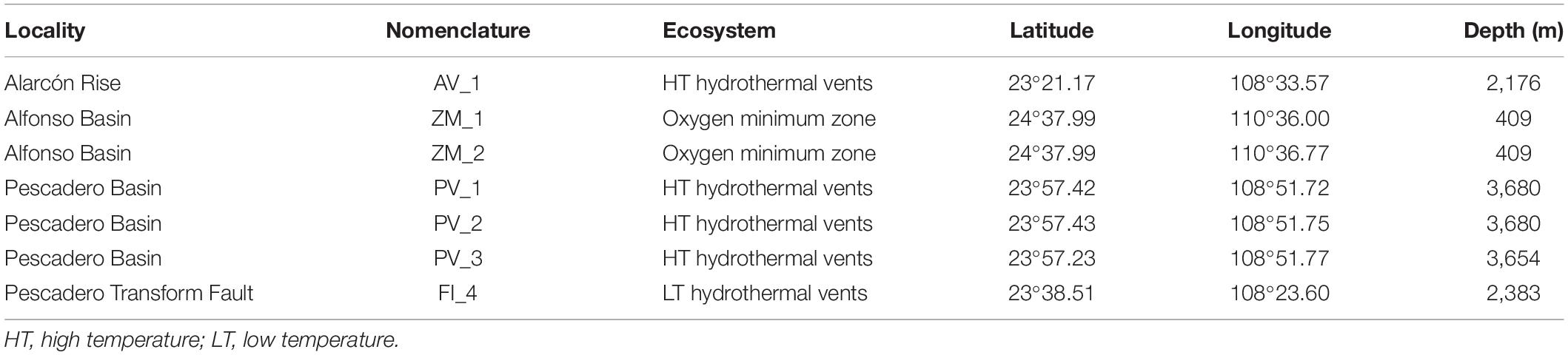
Table 1. Sampling stations where sediment cores were collected in the southern Gulf of California, Mexico.
DNA Extraction, Amplification, and Sequencing
Using the DNeasy PowerMax Soil Kit (Qiagen, Hilden, Germany), total DNA was extracted from 5 g of each sediment sample [removing the exterior layer of the core, as described in Velez et al. (2020)], following the procedures indicated by the manufacturer. The DNA samples were quantified using Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States) and preserved at −20°C until used. All of the steps that involved sample handling and extracting nucleic acids were processed within a laminar flow hood. Illumina MiSeq paired-end (2 × 300) sequencing was performed by the commercial service (amplification, library preparation, and sequencing) provided by the Genomic Services Laboratory (LANGEBIO, Irapuato, Guanajuato, Mexico), targeting the ITS1 region of the ribosomal RNA gene cluster by using primers set ITS1-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′; Gardes and Bruns, 1993) and ITS2 (5′- GCTGCGTTCTTCATCGATGC-3′; White et al., 1990), yielding to around 25,000 reads per subsample (data are available at NCBI GenBank under the BioProject ID PRJNA793088).
Bioinformatics
Raw reads were processed using the fungal ITS-specific workflow proposed by Callahan et al. (2016) using the package dada2 v1.13.1 in R v4.0.2 (R Core Team, 2020). Briefly, we used cutadapt v3.2 (Martin, 2011) to detect and remove primer and adapter contamination from reads. Then, we used the function filterAndTrim to exclude reads with ambiguous bases (maxN = 0) and each read was required to have <1 expected errors based on their quality scores [maxEE = c (1,1), truncQ = 3]. The quality profiles of filtered reads were inspected using plotQualityProfile function. Amplicon sequence variants (ASVs) were inferred using the parametric error model implemented by dada2 for the forward and reverse reads independently, using the function learnErrors (Callahan et al., 2016). As recommended by the authors, the estimated sample-specific error rates were visually inspected using plotErrors. To assemble paired-end reads, we used the function mergePairs considering a minimum of 50 pb of overlap and excluding reads with any mismatch in the overlapping region. Chimeras were removed using the consensus method of removeBimeraDenovo. To discard diversity bias due to differences in the number amplicons per site, we conducted rarefaction and extrapolation curves with the package iNEXT v 2.0.20 (Hsieh et al., 2020).
The taxonomic assignment was performed using the DECIPHER v2.16.1 Bioconductor package (Wright, 2016) against the UNITE_v2020_February2020 database (Abarenkov et al., 2020) implementing the trained classifier available on DECIPHER website.1 We used the IdTaxa function with default settings. To avoid diversity overestimation due to over-split components, all of the of ASVs with a taxonomic assignment higher to genus level (for example, Basidiomycota sp.) were clustered using Cd-Hit (Li and Godzik, 2006) with a threshold of 97%. Particularly, the ASVs classified as “Fungi spp.” were verified and further curated using the BLAST tool of NCBI Genbank non-redundant (nr) database to obtain the top 10 most similar hits. As a further corroboration we used a phylogenetic approach (recommended in Lücking et al., 2020) to explore the taxonomic affinities of our sequences vs. reference material retrieved from NCBI. So, sequences were aligned with MUSCLE v3.8.1551 (Edgar, 2004), and a Neighbor-Joining tree was built using MEGAX (Kumar et al., 2018).
Alpha and Beta Diversity Estimates
To explore the relative abundance and shared taxa among sites, we created a heat map and intersection plots using the R packages ggplot2 (Wickham, 2016) and UpSetR (Conway et al., 2017). We used iNEXT to estimate the rarefied Shannon and Simpson diversities along with their bootstrap 95% confidence intervals (1,000 replicates). To evaluate the equitability of the taxa frequency distribution (also interpreted as dominance), we estimated the Shannon Evenness using the package vegan v2.5-6 (Oksanen et al., 2019). We estimated the differences in taxonomic composition between sampling points through a Bray-Curtis (Abundance-based), and Sorensen dissimilarity (presence-absence) matrices. Both dissimilarity matrices were plotted using the Ward clustering criterion. To represent the differences among sites communities, we performed a principal component analysis (PCA) using a Hellinger-transformed abundance table by using the vegan package.
Functional Guilds
Despite functional categorization provides useful guides to navigate the complex fungal kingdom, confirming the effect of functional diversity on ecosystem processes (Crowther et al., 2013); the ability to assign trophic strategies to individual taxonomic units remains a broad challenge in the field of microbial ecology. Milestone proposals have showed that fungal functional guilds can be approached based on taxonomic affinity, introducing FUNGuild, an open annotation tool for parsing fungal community datasets by ecological guild (Nguyen et al., 2016). We used this approach (FUNGuild v1.1 database2) to assign ecological functions (trophic modes) to our ASVs. It should be noted that unfortunately precise community-wide conclusions are still unattainable since: (1) many genera contain more than one trophic strategy; (2) insufficient guild data are available for several fungal groups (Nilsson et al., 2019); and (3) the lifestyles of marine fungal taxa remain largely unknown. Nonetheless, this invaluable information base should pave the way for further improved work on deep-sea fungi.
Results
Sequence Analysis
Overall we obtained 184,466 reads from the analyzed sediment samples collected in high-temperature hydrothermal vent fields in the Pescadero Basin (47,917 reads) and the Alarcón Rise (32,941 reads), low-temperature hydrothermal vent fields in the Pescadero Transform Fault (61,106 reads), and OMZ in Alfonso Basin (42,502 reads) (Supplementary Table 1). After quality control, a total of 143,244 sequences were used for analysis. These reads were classified into 940 ASVs out of which 102 represented fungal taxa (65,780 reads). All of the rarefaction and extrapolation curves of the fungal assemblages in sediment samples from the different ecosystems reached the asymptote (Figure 1), indicating that the data provided a good description of the diversity.
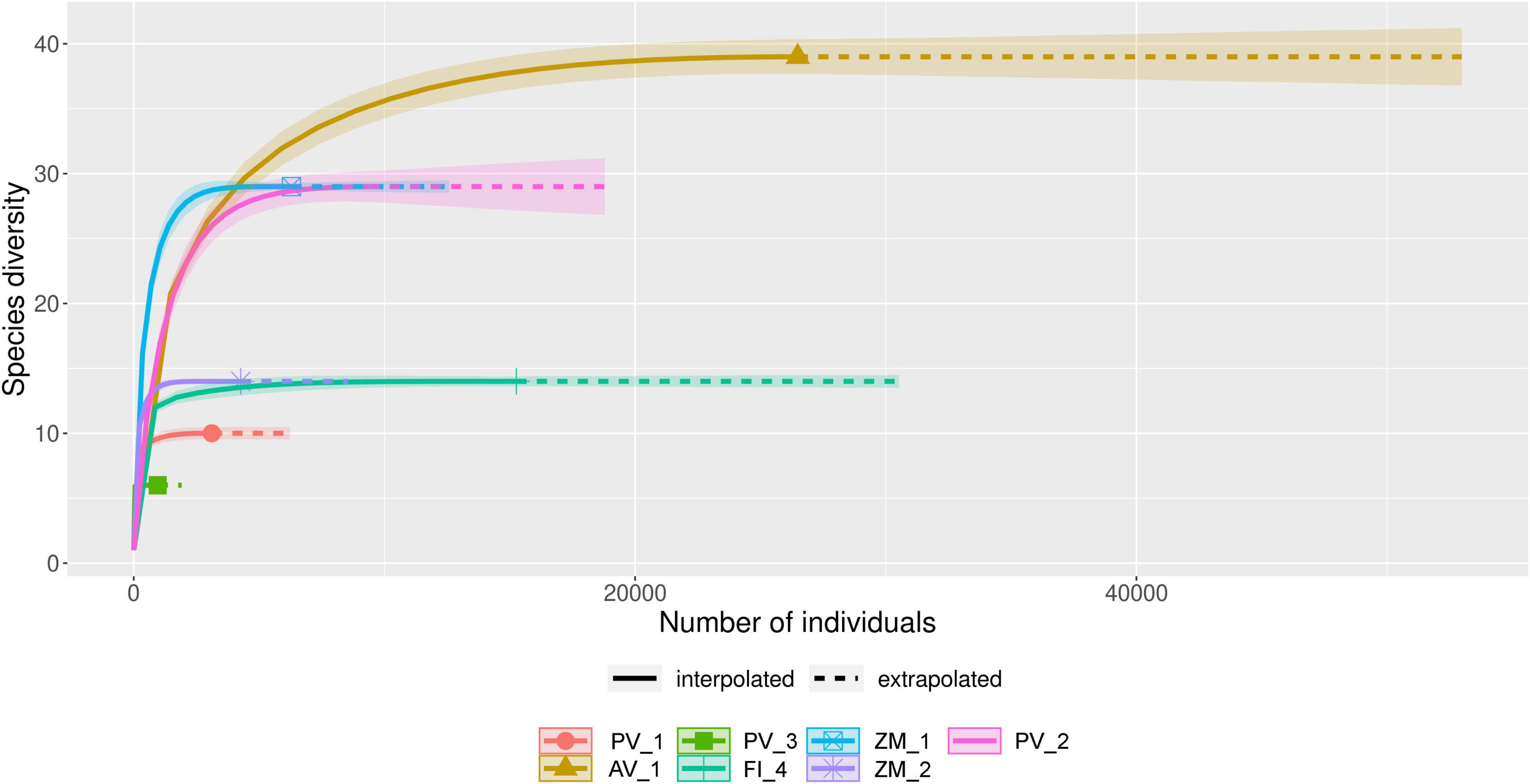
Figure 1. Rarefaction and extrapolation curves of fungal amplicon sequence variants (ASVs) obtained from deep-sea sediment samples collected in the Pescadero Basin (PV_1, PV_2, PV_3), Pescadero Transform Fault (FI_4), Alarcón Rise (AV_1), and Alfonso Basin (ZM_1, ZM_2).
Fungal Taxonomic Diversity
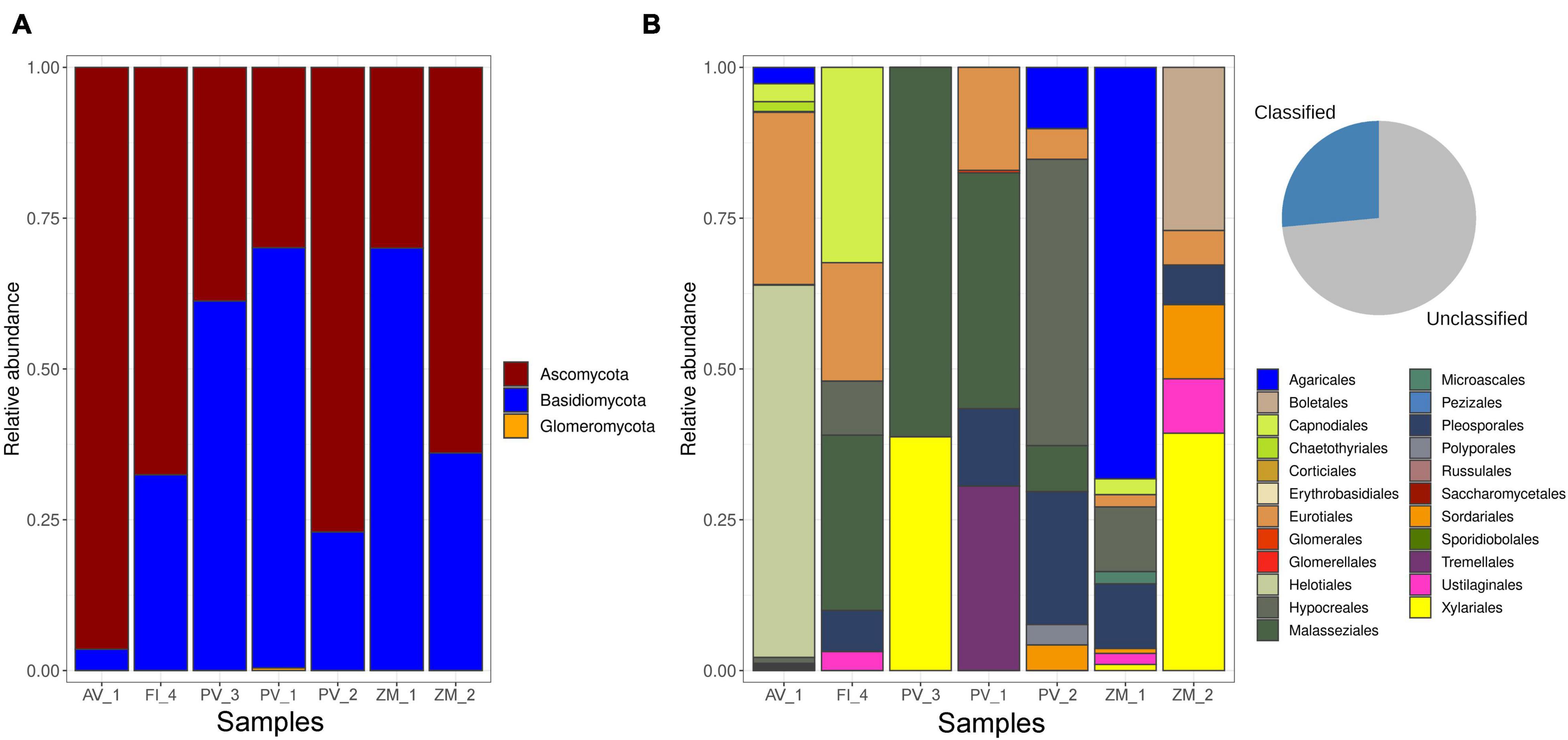
Out of 143,244 reads, 65,780 were taxonomically assigned. In all the studied sites, the fungal community was dominated by the Ascomycota, followed by Basidiomycota. Glomeromycota were present as a negligible proportion of the community at a single site in the Pescadero Basin (Figure 2A). At the order level, dominance patterns varied across ecosystems. For instance in the Alarcón Rise Helotiales dominated the fungal community, whereas Malasseziales occurred abundantly in the Pescadero Transform Fault and Pescadero Basin. Sites in the Alfonso Basin were dominated by Agaricales and Xylariales, respectively (Figure 2B). Overall, at the species level, Fungi sp. 4 and 18 represented the dominant ASVs (Figure 3). These taxa are affiliated to the Pezizomycotina and uncultured Basidiomycota clades from deep-sea, respectively, and could potentially represent currently unknown taxa with limited distribution to deep-sea sediments. Moreover, we detected the abundant occurrence (n = 21) of ASVs that could only be taxonomically assigned at the kingdom level as Fungi sp. These taxa were also corresponded to mostly uncultured fungal clades (Figure 4).
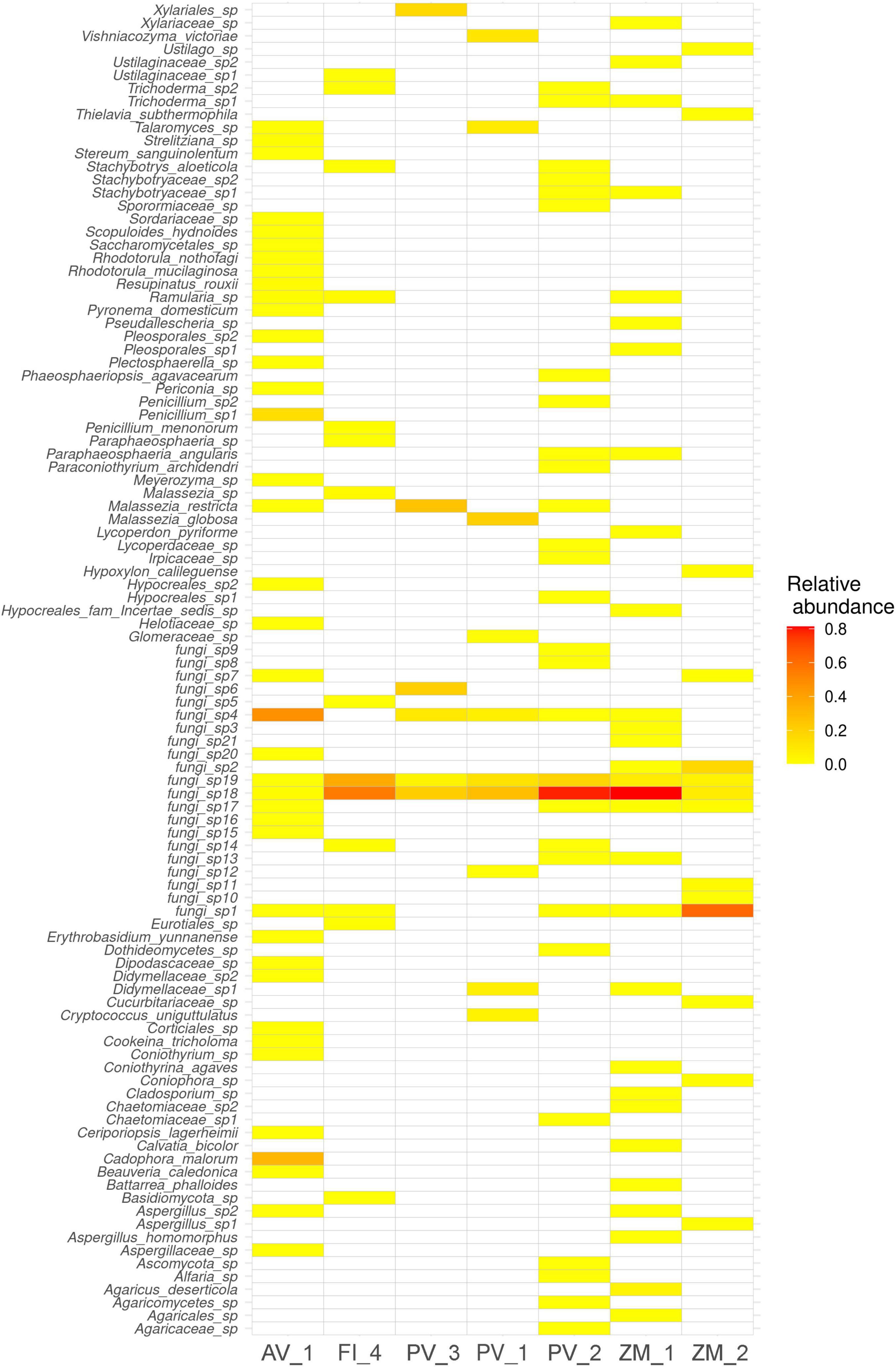
Figure 3. Heatmap (red-white) showing sediment fungal richness and relative abundance at the species level across samples. The color intensity is proportional to fungal relative abundance, with red indicating higher values.
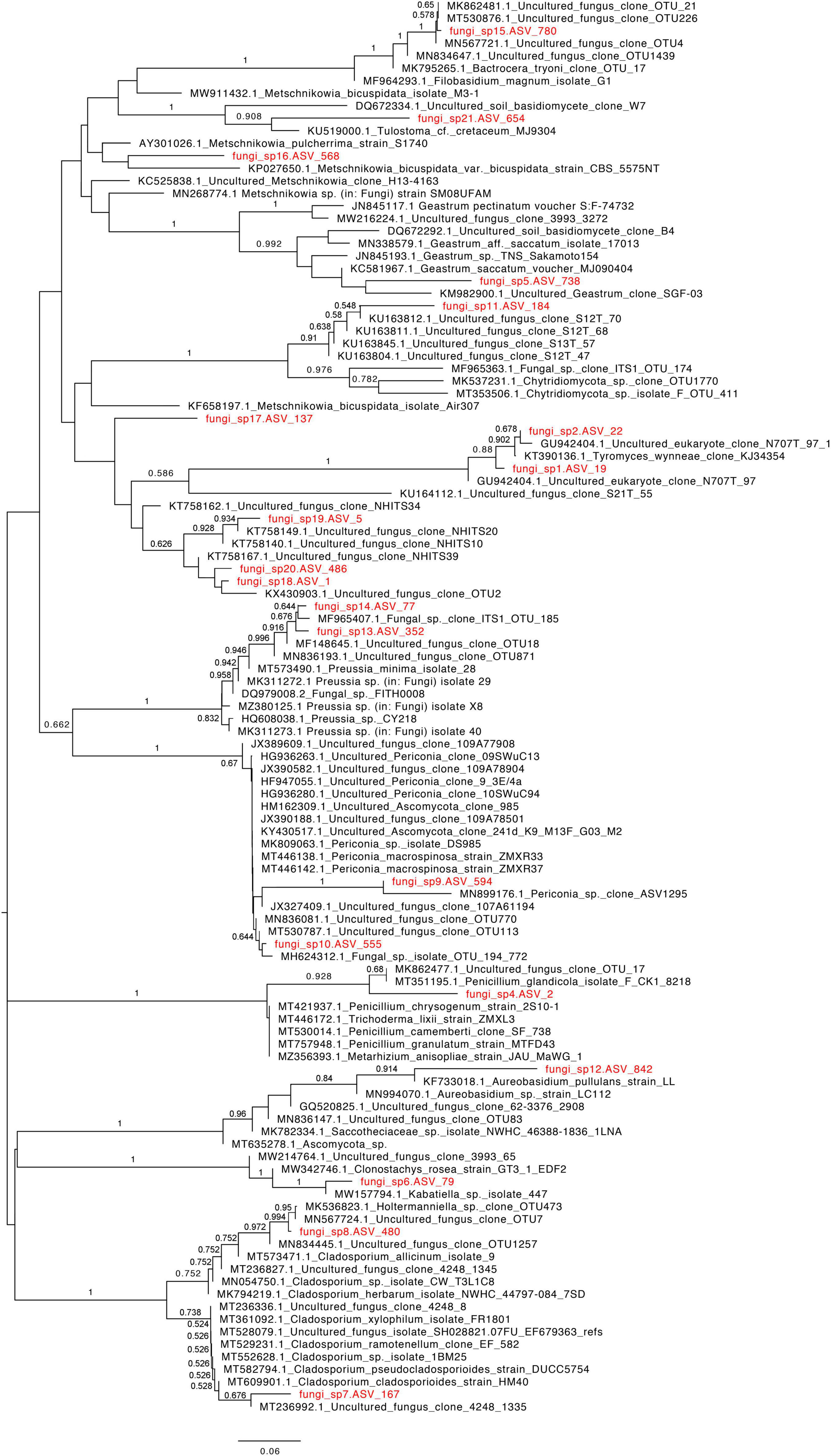
Figure 4. Phylogenetic relationship of deep-sea derived ASVs that were taxonomically assigned at the kingdom level (indicated in red font) with close species based on ITS1 rRNA sequences using the neighbor-joining algorithm. Bootstrap values > 50% are indicated above the branches.
Fungal Alpha and Beta Diversity Indexes
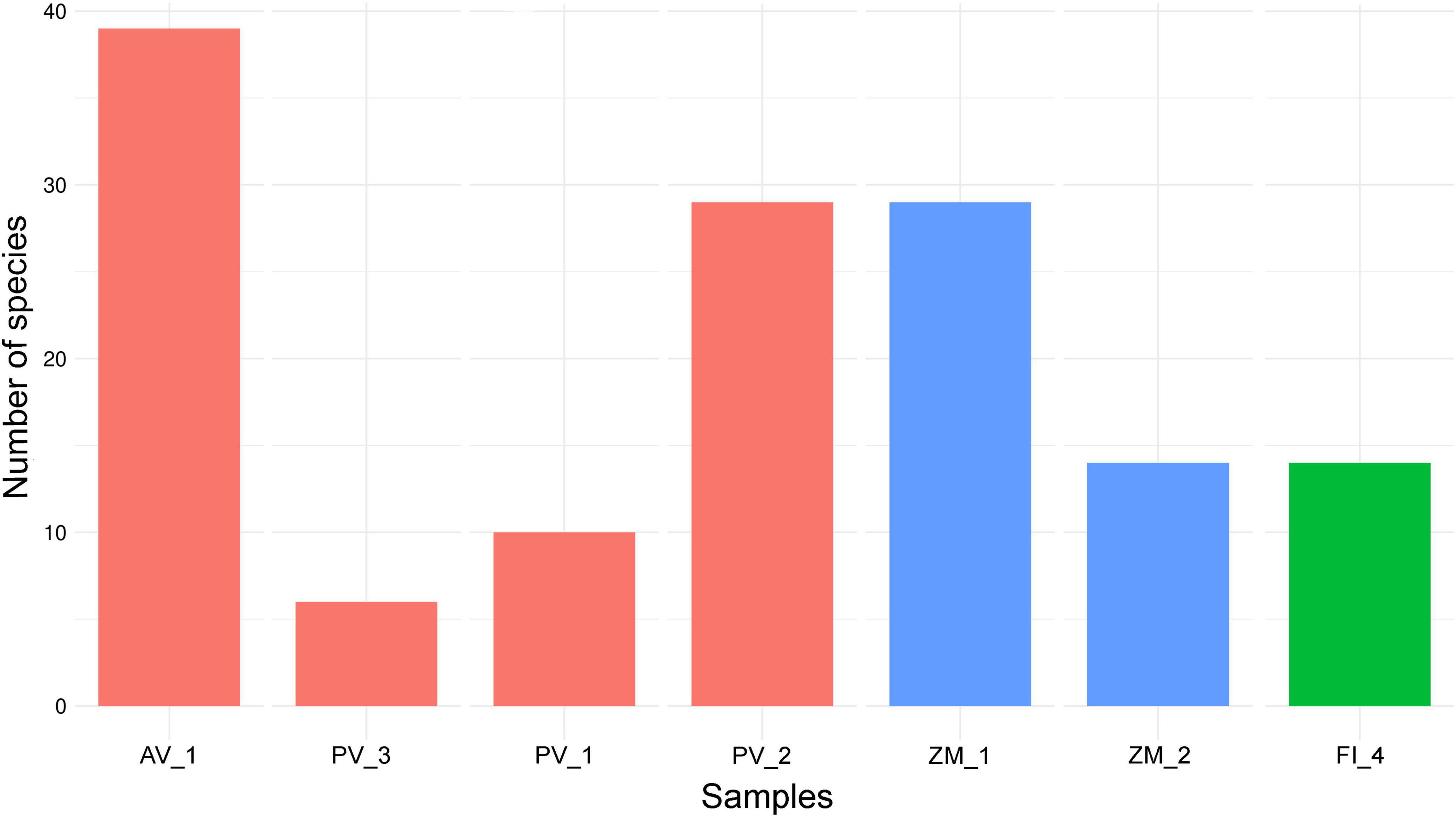
Based on our limited sampling, we observed a heterogeneous number of fungal ASVs across ecosystems, with the highest richness values in the high-temperature vent system of Alarcón Rise (AV_1); whereas the lowest values corresponded to the Pescadero Transform Fault (PV_3; Figure 5). Overall, our results hint that high-temperature vent systems represent the richest ecosystem for fungi (highest number of ASVs) followed by OMZ, which should be confirmed in further studies.
Alpha diversity indexes showed variations in and between ecosystems. For instance, the rarefied Shannon and Simpson metrics agreed that Pescadero Basin showed both, the highest diversity in PV_1 and the lowest diversity in PV_2 (Figures 6A,B and Supplementary Table 1). In addition, the highest evenness was observed in the Pescadero Transform Fault in PV_3 and Pescadero Basin in PV_1, whereas the lowest values corresponded to the Pescadero Basin in PV_2, Alfonso Basin in ZM_1, and the Alarcón Rise in AV_1 (Figure 6C).
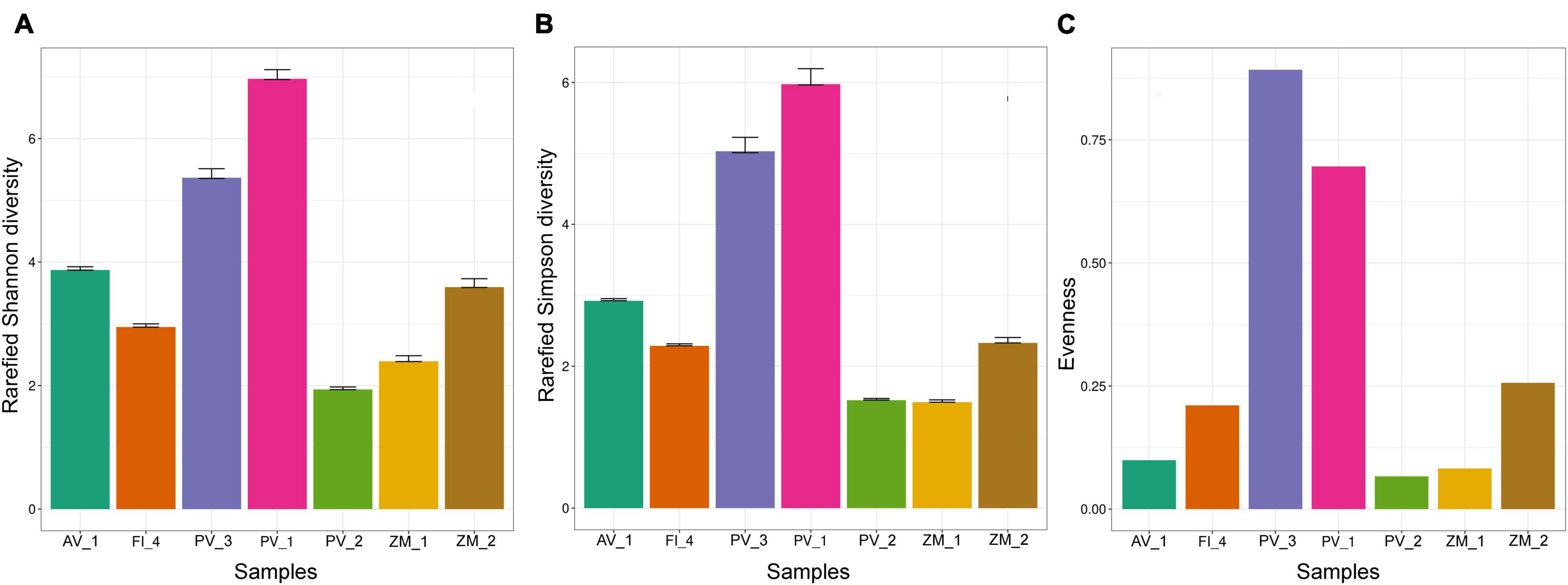
Figure 6. Alpha diversity metrics of fungal ASVs in sediment samples from deep-sea low- and high-temperature hydrothermal vents, and oxygen minimum zones, depicting rarefied Shannon (A) and Simpson (B) indexes, and evenness (C).
Bray-Curtis and Sorensen dissimilarities showed sparse clustering patters. However, both metrics consistently agreed that vent systems in the Alarcón Rise differed from Pescadero Basin (Figure 7). Moreover, the PCA showed sparse clustering patterns among samples collected in the same deep-sea ecosystem with AB and AR samples differing the most (Figure 8). Our results indicated that a large proportion (81.37%) of the ASVs occurred at a single site, whereas only 2 ASVs were shared among the seven samples (Figure 9A). At the system level, the Venn diagram analysis revealed that 3.92% of the ASVs were shared among low- and high-temperature hydrothermal vents, and the OMZ, while 52.94% of the ASVs were specific to high-temperature vents (PB and AR), 24.50% to the OMZ (AB), and 6.86 to the low-temperature vent site (PTF) (Figure 9B). Lastly, no ASVs were shared between the OMZ and the low-temperature vent site.
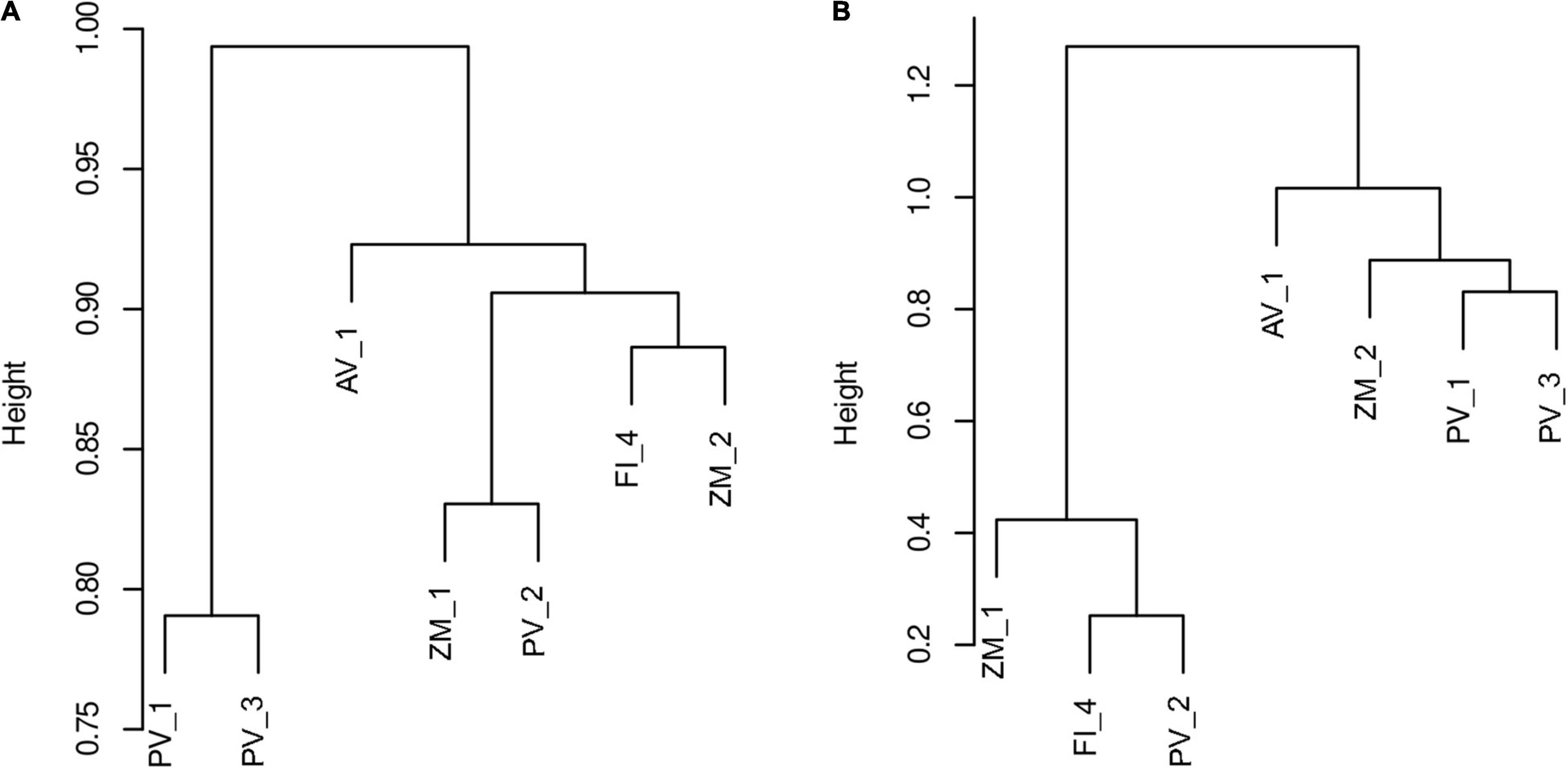
Figure 7. Dendrogram generated using Ward’s clustering criterion showing dissimilarities of fungal communities in sediment samples from deep-sea low- and high-temperature hydrothermal vents, and an oxygen minimum zone based on Sorensen (A) and Bray-Curtis (B) metrics. Height indicates distance given the dissimilarity metrics.
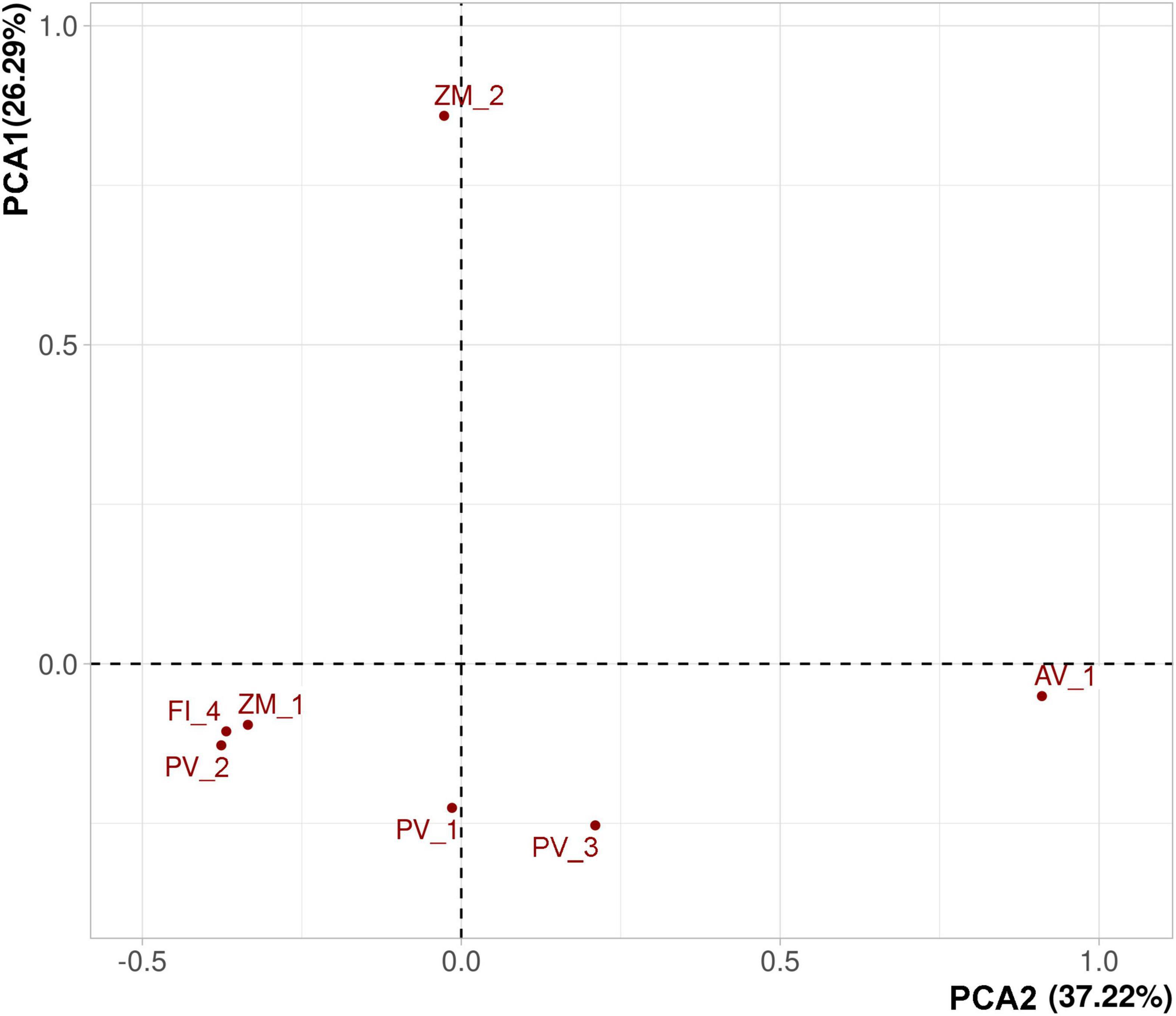
Figure 8. Principal component analysis (PCA) on Hellinger-transformed abundance data showing clustering patterns in fungal ASVs from deep-sea low- and high-temperature hydrothermal vents, and an oxygen minimum zone.
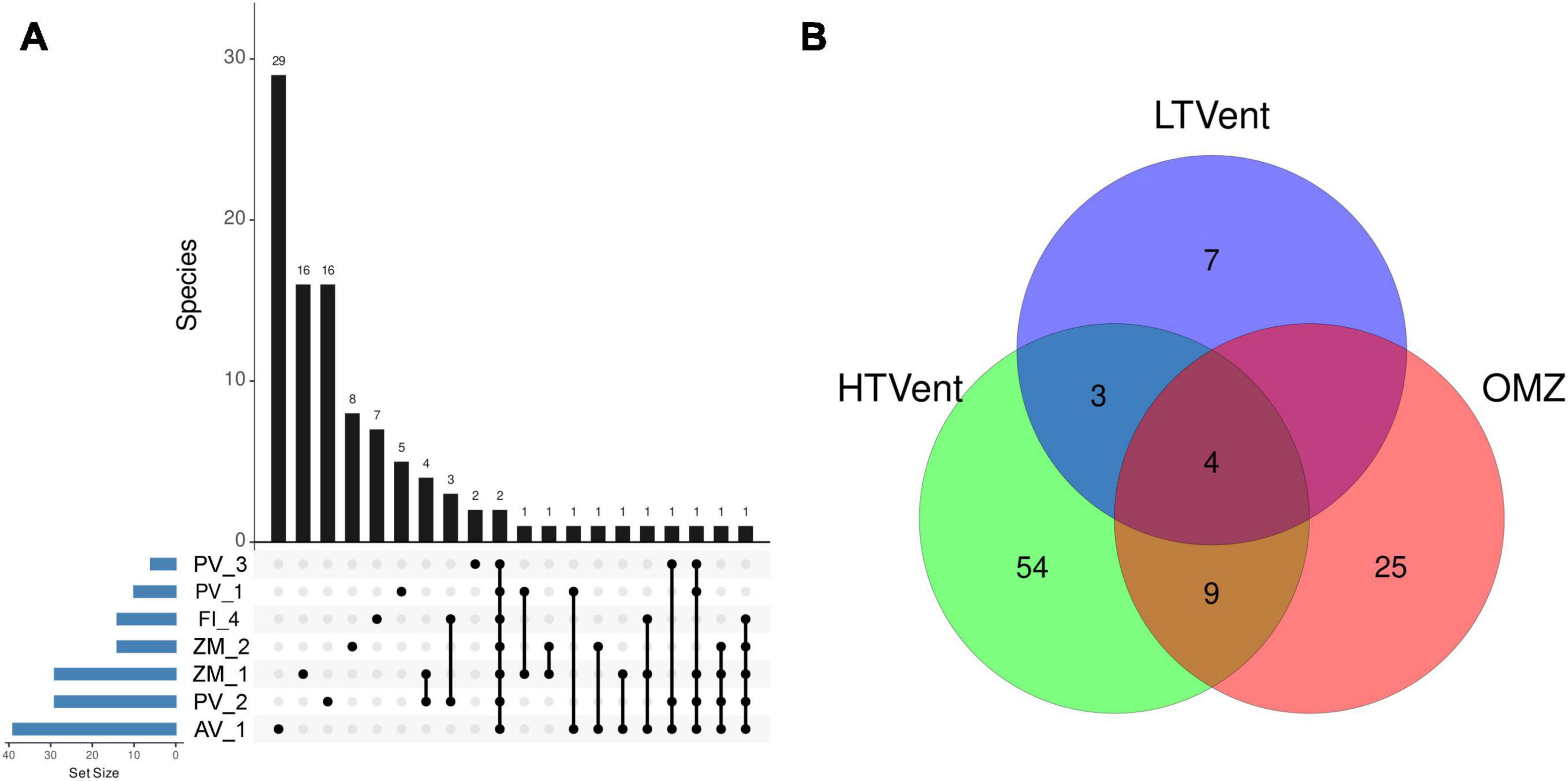
Figure 9. Upset plot indicating overlapping and unique fungal AVSs across the seven sampling sites stacked in the x-axis (A), where the set size represents the overall number of species for each sample; and Venn diagram (B) showing the total number of shared and uniquely retrieved fungal AVSs in deep-sea low-temperature hydrothermal vents, high-temperature hydrothermal vents, and an oxygen minimum zone. HTVent, high-temperature vents; LTVent, low-temperature vent; OMZ, oxygen minimum zone.
Functional Guilds
FUNGuild analysis was performed to predict functions of fungal communities, accounting for the determination of 16.57% of total reads (pie chart Figure 10). No hits were obtained for the site PV_3 in the Pescadero Transform Fault. The analysis revealed characteristic functional guilds in each ecosystem, with the dominance of undefined saprotrophs in the Pescadero Basin, endophytes in the Alarcón Rise, and dung-saprotroph-endophyte-plant pathogen-undefined saprotrophs in Alfonso Basin. The site AV_1 in the Alarcón Rise showed the richest functional diversity (including animal endosymbiont-animal pathogen-endophyte-plant pathogen-undefined saprotrophs, animal pathogens, dung saprotroph-plant saprotrophs, ectomycorrhizal-fungal parasite-plant pathogen-wood saprotrophs, endophytes, plant pathogens, plant pathogen-undefined saprotrophs, plant pathogen-wood saprotrophs, undefined saprotrophs, and wood saprotrophs), whereas PV_1 in the Pescadero Basin the lowest (Figure 10).
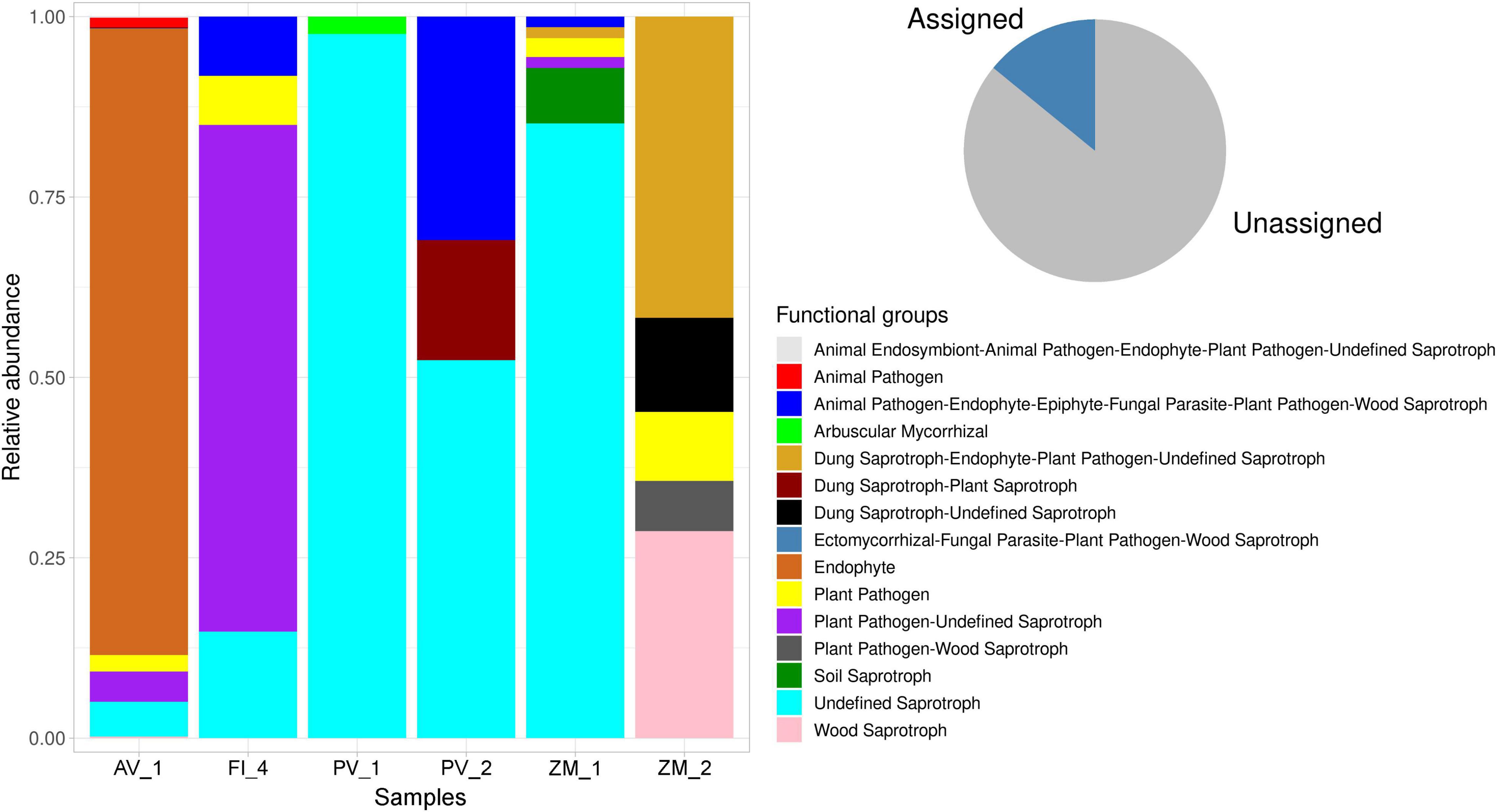
Figure 10. Stacked bar plot showing the relative proportion (y-axis) of the assigned fungal functional guilds (shown in red in the pie chart) across sediment samples from deep-sea low- and high-temperature hydrothermal vents, and an oxygen minimum zone (x-axis).
Discussion
Even though microorganisms dominate the deep marine biosphere, most exploration efforts have solely focused in the investigation of prokaryotes, neglecting a key and copious eukaryotic component of microbial communities: fungi (Nagano and Nagahama, 2012). The present work represents the first report addressing this knowledge gap for three deep-sea extreme ecosystems in the southern Gulf of California: the Pescadero Basin and the Alarcón Rise (high-temperature hydrothermal vents), the Pescadero Transform Fault (low-temperature hydrothermal vents), and the Alfonso Basin (OMZ), unveiling a total of 102 fungal ASVs and characteristic diversity patterns across ecosystems.
Fungal Taxonomic Diversity
Our findings on the dominant occurrence of Ascomycota, followed by Basidiomycota in all the studied sites, agrees with a plethora of investigations in hadal trenches (Xu et al., 2019; Gao et al., 2020), deep-sea sediments (Damare et al., 2006; Singh et al., 2012; Zhang et al., 2014, 2016), OMZs (Jebaraj et al., 2010), methane hydrate-bearing deep-sea marine sediments (Lai et al., 2007), hydrothermal vents (Burgaud et al., 2009; Le Calvez et al., 2009), and chemosynthetic whale falls (Nagano et al., 2020). Furthermore, the occurrence of Glomeromycota as a minor proportion of the community in the Pescadero Transform Fault (PV_1), resembles former findings reporting this phylum as a component of fungal vent communities (Le Calvez et al., 2009; Nagano et al., 2010; Nagahama et al., 2011; Luo et al., 2020). Largely, these results suggest that despite distinctive extreme conditions of the investigated systems, they host a considerable fungal diversity in higher taxonomic categories.
Less than 0.0001% of the deep ocean has been investigated, representing the least explored biome of Earth (Danovaro et al., 2017). Unsurprisingly, 20.58% of our ASVs were identified only to the kingdom level, which sums to accumulating evidence on the rich occurrence of formally undescribed, uncultured fungal phylotypes from deep-sea (revised in Nagano and Nagahama, 2012; Nilsson et al., 2019). Interestingly, the most widely distributed (occurring in all the ecosystems) and top dominant ASV, Fungi sp. 18, showed affinity to previously reported deep-sea phylotypes such as the DSF-group1 (Ascomycota; AB507843, AB507839). This uncultured fungal group is believed to have a restricted distribution to oxygen-depleted deep-sea environments, such as methane cold-seeps, anoxic bacterial mats, and deep-sea sediments (Bass et al., 2007; Takishita et al., 2007; Nagano et al., 2010), occurring perhaps as a parasite of planktonic animals (Nagano and Nagahama, 2012). So, our results validate the wide distribution of this clade across deep-sea extreme systems, including low- and high-temperature hydrothermal vents.
Alpha Diversity Estimates
Our findings on vent systems as the richest ecosystem for fungal ASVs corroborates former postulates on hydrothermal vents as a deep-sea oasis for marine life (Carney, 1994). This has been correlated with high productivity and dynamism at fluid-influenced sites, which support energetically expensive diversity levels (Alfaro-Lucas et al., 2020); in contrast to other relatively homogeneous and stable deep-sea benthic habitats (Snelgrove and Smith, 2002) where communities thrive on the organic matter produced in shallower waters (Ruhl and Smith, 2004). So, it is reasonable to believe that fungi, as heterotrophic microorganisms, take advantage of nutrient pulses in deep-sea extreme ecosystems. In addition, our results revealed opposite diversity patterns to those reported for bacteria from the same samples, where high-temperature vents (PV_1 and PV_3) were the least diverse localities (Espinosa-Asuar et al., 2020), denoting cross-kingdom antagonistic processes that shall be further confirmed.
Pescadero Basin
In terms of taxonomic diversity, the Malasseziales occurred abundantly in the high-temperature vents of PB, evidencing their copious occurrence in this ecosystem (Amend, 2014). Nonetheless, we documented differences across sites in terms of the rarefied diversity indexes (Simpson and Shannon), evenness metrics, and community composition. On the one hand, the highest diversity values were observed in PV_1, where the fungal community was dominated by Fungi sp. 5, 18, and 19 (similar to Geastrum, and uncultured lineages from hydrothermal vents in the Mid-Atlantic and deep-sea sediments in South China, respectively), Malassezia globosa, Vishniacozyma victoriae, and Cryptococcus uniguttulatus. On the other hand, this system also included the site with the lowest diversity values (PV_2), characterized by the abundant occurrence of Fungi sp. 18 and 19. These results confirm strong substrate heterogeneity in PB vents owing to the variety of ecological settings in this locality and differences in fluid regimes and chemical composition (Mullineaux et al., 2018; Paduan et al., 2018). In agreement, Salcedo et al. (2019) proposed the existence of a greater variety of niches in PB than in PTF and AR, inferred by a wide range in the carbon isotopic signatures of sedimentivore consumers, implying the occurrence of numerous free-living microorganisms such as fungi.
Pescadero Transform Fault
Low-temperature vent site of the PTF, showed moderate evenness and diversity values, depicting strong dominance patterns of a few taxa (Fungi sp. 18 and 19) in a species-poor community. At large, the community was dominated by the Dothideomycetes and Malasseziales, suggesting these fungi represent a characteristic element in this locality. However, in contrast to sites in the PB, Ramularia sp., Malassezia sp., and Fungi sp. 14 (similar to Preussia spp., and uncultured fungal phylotypes from ponds) occurred abundantly, denoting distinctive local assemblages. Interestingly, the low fungal diversity in PTF vents is consistent with the low faunal diversity of 16 taxa identified by Goffredi et al. (2017), and the low diversity in trophic guilds and the less complex food web observed by Salcedo et al. (2019). This may be related to the apparent simplicity of this vent site, which is mainly composed of dense clusters of siboglinid worms Escarpia spicata and Lamellibrachia barhami surrounded by uplifted sediment hills and characterized by a discrete low-temperature seepage (Goffredi et al., 2017).
Alarcón Rise
Considering that different and independent ecological processes determine species richness and evenness (Ma, 2005), it is feasible that our results on the highest richness of ASVs and low evenness values for the high-temperature vent system of the Alarcón Rise (site AV_1) may reflect an unstable system where site-specific conditions and spatial patchiness of resources account for a highly rich fungal community that varies in resource utilization strategies (confirmed by the FUNGuild analysis). Moreover, we observed distinctive fungal assemblages with the Helotiales, mostly Cadophora malorum, which has been previously isolated from the endemic shrimp Rimicaris exoculata from the Rainbow hydrothermal site by Rédou et al. (2016) as a dominant element. This finding also resembles former reports on the dominance of this order in deep-sea animal-associated fungi from hydrothermal vents in the Pacific Ocean (Burgaud et al., 2009, 2010), fungi associated with the sponge Stelletta normani in the Atlantic Ocean (Batista-García et al., 2017), and deep-sea coral gardens in the Atlantic Ocean (Marchese et al., 2021), suggesting the animal-related lifestyle of this fungus in AV_1. The highest fungal richness observed in AR vents is consistent with the highest invertebrate richness previously identified since 43 taxa were identified in this locality in comparison to 27 taxa in PB and 16 taxa in PTF (Goffredi et al., 2017; Salcedo et al., 2021).
Alfonso Basin
Although OMZs are considered as inhospitable areas to aerobically respiring organisms, our data evidenced that the analyzed OMZ regions in the AB support thriving fungal communities (including unidentified, uncultured deep-sea fungal phylotypes) that are associated with diverse functional guilds (e.g., undefined saprotrophs, wood saprotrophs, dung saprotroph-endophyte-plant pathogen-undefined saprotroph, animal pathogen-endophyte-epiphyte-fungal parasite-plant pathogen-wood saprotroph, among others). Our results on contrasting richness values, and marked differences in fungal assemblages, even at higher taxonomic levels, evidenced the spatial and environmental patchiness in this deep-sea ecosystem (Dalsgaard et al., 2012), where microbial diversity is known to vary along the oxycline (Wright et al., 2012). Furthermore, our data revealed that around one-fourth of the ASVs (24.5%) were exclusive to the AB, supporting the possibility of fungal ecotypes selection. These results are consistent with previous observations demonstrating fungal taxa metabolic adaptations to utilize nitrate and (or) nitrite as an alternative for oxygen (Shoun et al., 1992), hinting on their potential to participate in anaerobic denitrification processes in the analyzed OMZs.
Beta Diversity Estimates
No single factor determines the global distribution and abundance of all fungi (Nilsson et al., 2019). Our results evidenced the differentiation of fungal communities among an OMZ, low- and high-temperature vent systems, sharing only five ASVs. Particularly, the OMZ site ZM_2 and the vent system site AV_1 were the most distinct sites, in accordance to previous reports on bacterial diversity (Espinosa-Asuar et al., 2020). These results suggest that fungal assemblages in each ecosystem (e.g., AR vs. PB vents) might be explained by the diversity of the surrounding physicochemical conditions (i.e., gradients of temperature, pH, etc.), as well as by the degree of connectivity (Grossart et al., 2019). For instance, the vent field of the PB is composed of calcite, venting fluids are rich in aromatic hydrocarbons, hydrogen, methane, and hydrogen sulfide at a pH of ∼6.5, and has a faunal composition dominated by the siboglinid tubeworm Oasisia aff. alvinae (Goffredi et al., 2017). In contrast, the AR hosts black smokers chimneys constructed of polymetallic sulfides, emanating fluids with high concentrations of H2S and metal-sulfides, where the siboglinid Riftia pachyptila proliferates (Goffredi et al., 2017; Paduan et al., 2018). These marked physical, chemical, and biological differences (presence of exclusive species) might be responsible for high fungal turnover between surrounding sites, yet the specific environmental factors driving these differences remain to be explored.
Our beta diversity results based on Bray-Curtis metrics also resembled former reports for bacteria, evidencing the clustering of the site PV_2 (high-temperature vent in the PB) with FI_4 (low-temperature vent in the PTF; Espinosa-Asuar et al., 2020). This may be associated to the geographical distribution and connectivity of sampling sites within the Gulf of California. Since PV_2 represents the most easterly site within PB, perhaps receiving a low influence on the sediments from venting fluids within the PB. Nonetheless, this might also reflect the occurrence of many singleton species in the dataset, leading to sparse matrices predominantly filled up with zeros (Legendre and Gallagher, 2001), overriding the agreement in presence of species (Ricotta and Podani, 2017).
Functional Guilds
Although true fungal diversity and its role in deep-sea environments are still unclear, facultative parasitic or opportunistic (e.g., Van Dover, 2007; Burgaud et al., 2009), and symbiotic interactions with endemic deep-sea animals have been repeatedly documented. The FUNGuild analysis evidenced characteristic functional fungal signatures across the investigated deep-sea extreme systems. For instance, the dominance of undefined saprotrophs in the Pescadero Basin suggests that the fungi might be involved in important ecological functions in nutrients turnover. Whereas, in the Alarcón Rise and Alfonso Basin potentially symbiotic fungal guilds represented the most common trophic guilds, hinting at animal-associated lifestyles. It is worth mentioning that despite our results indicated that Malassezia restricta (putative opportunistic human pathogen) occurred abundantly in site PV_3, this record was not reflected in the FUNGuild results, representing a significant bias. This highlights the urgent need for additional studies and the development of reference databases to fully elucidate fungal functional roles in deep-sea ecosystems.
Conclusion
This work evidenced a high fungal diversity -at both taxonomic and functional levels- in the sediments collected from distinct vent systems (including low- and high-temperature systems associated with carbonate mounds and black smokers) and an OMZ in the southern Gulf of California. This fungal diversity was characterized by the plentiful occurrence of deep-sea uncultured clades that could potentially represent unknown taxa with limited distribution to deep-sea sediments, hinting at the magnitude of the still unknown species occurring in the oceans. The fungal diversity herein detected included species with different ecological roles suggesting that despite the extreme conditions characterizing deep-sea extreme systems, they host a complex fungal community. Nonetheless, we stress on the importance of conducting further exploration works that include an extensive sampling design, accounting for intra-sample variability.
Our results uncovered distinctive fungal assemblages in each ecosystem, which could be linked to site-specific processes that should be further evaluated. Interestingly, these fungal diversity patterns are consistent with the faunistic data reported in previous studies, hinting chemical and physical heterogeneity as the main drivers of the community structure at microscopic and macroscopic levels. This preliminary work opens the way to new studies of the diversity and ecology of fungi in deep benthic ecosystems. So, additional studies are needed to corroborate in detail fungal ecology in these extreme environments, and to elucidate fungal trophic roles, as well as their potential utilization in bioprospection and bioremediation.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA793088.
Author Contributions
PV conceived the presented idea, verified the analytical methods, conceived and planned the experiments, wrote the manuscript, and supervised the project. DS conceived the presented idea and conducted the sampling. LE-A and AH-M conceived and planned the experiments. JG-P performed the computations and wrote the manuscript. LS conceived the presented idea and supervised the project. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Funding
This research work was financially supported by the DGAPA-PAPIIT-UNAM IN200921, and part of the hydrothermal vent ecology program financed by the Instituto de Ciencias del Mar y Limnología, UNAM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We greatly appreciate the invaluable support of R. Vrijenhoek. We thank reviewers for valuable comments on earlier versions of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.802634/full#supplementary-material
Footnotes
References
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H., et al. (2020). UNITE General FASTA Release for Fungi. Version 04.02.2020. UNITE Community. doi: 10.15156/BIO/786368
Alexander, E., Stock, A., Breiner, H. W., Behnke, A., Bunge, J., Yakimov, M. M., et al. (2009). Microbial eukaryotes in the hypersaline anoxic L’Atalante deep-sea basin. Environ. Microbiol. 11, 360–381. doi: 10.1111/j.1462-2920.2008.01777.x
Alfaro-Lucas, J. M., Pradillon, F., Zeppilli, D., Michel, L. N., Martinez-Arbizu, P., Tanaka, H., et al. (2020). High environmental stress and productivity increase functional diversity along a deep-sea hydrothermal vent gradient. Ecology 101:e03144. doi: 10.1002/ecy.3144
Amend, A. (2014). From dandruff to deep-sea vents: malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog. 10:e1004277. doi: 10.1371/journal.ppat.1004277
Barbier, E. B., Moreno-Mateos, D., Rogers, A. D., Aronson, J. L., Pendleton, L., Danovaro, R., et al. (2014). Ecology: protect the deep sea. Nature 505, 475–477. doi: 10.1038/505475a
Bass, D., Howe, A., Brown, N., Barton, H., Demidova, M., Michelle, H., et al. (2007). Yeast forms dominate fungal diversity in the deep oceans. Proc. Biol. Soc. 274, 3069–3077. doi: 10.1098/rspb.2007.1067
Batista-García, R. A., Sutton, T., Jackson, S. A., Tovar-Herrera, O. E., Balcázar-López, E., Sánchez-Carbente, M. R., et al. (2017). Characterization of lignocellulolytic activities from fungi isolated from the deep-sea sponge Stelleta normani. PLoS One 12:e0173750. doi: 10.1371/journal.pone.0173750
Burgaud, G., Arzur, D., Durand, L., Cambon-Bonavita, M., and Barbier, G. (2010). Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol. Ecol. 73, 121–133. doi: 10.1111/j.1574-6941.2010.00881.x
Burgaud, G., Le Calvez, T., Arzur, D., Vandenkoornhuyse, P., and Barbier, G. (2009). Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ. Microbiol. 11, 1588–1600. doi: 10.1111/j.1462-2920.2009.01886.x
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carney, R. S. (1994). Consideration of the oasis analogy for chemosynthetic communities at Gulf of Mexico hydrocarbon vents. Geo Mar. Lett. 14, 149–159. doi: 10.1007/BF01203726
Clague, D. A., Caress, D. W., Dreyer, B. M., Lundsten, L., Paduan, J. B., Portner, R. A., et al. (2018). Geology of the Alarcón Rise, Southern Gulf of California. Geochem. Geophys. Geosyst. 19, 807–837. doi: 10.1002/2017GC007348
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364
Crowther, T. W., Stanton, D. W. G., Thomas, S. M., A’Bear, A. D., Hiscox, J., Jones, T. H., et al. (2013). Top-down control of soil fungal community composition by a globally distributed keystone consumer. Ecology 94, 2518–2528.
Dalsgaard, T., Thamdrup, B., Farías, L., and Revsbech, N. P. (2012). Anammox and denitrification in the oxygen minimum zone of the eastern South Pacific. Limnol. Oceanogr. 57, 1331–1346. doi: 10.4319/lo.2012.57.5.1331
Damare, S., Raghukumar, C., and Raghukumar, S. (2006). Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res. Part I Oceanogr. Res. Pap. 53, 14–27. doi: 10.1016/j.dsr.2005.09.005
Danovaro, R., Corinaldesi, C., Dell’Anno, A., and Snelgrove, P. V. (2017). The deep-sea under global change. Curr. Biol. 27, R461–R465. doi: 10.1016/j.cub.2017.02.046
Danovaro, R., Snelgrove, P. V. R., and Tyler, P. (2014). Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475. doi: 10.1016/j.tree.2014.06.002
Diaz, R. J., Eriksson-Hägg, H., and Rosenberg, R. (2013). “Chapter 4: Hypoxia,” in Managing Ocean Environments in a Changing Climate, eds K. J. Noone, U. Rashid, and R. J. Diaz (Amsterdam: Elsevier), 67–96. doi: 10.1016/B978-0-12-407668-6.00004-5
Dick, G. J. (2019). The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat. Rev. Microbiol. 17, 271–283.
Douglas, R. G., Gorsline, D., Grippo, A., Granados, I., and González-Yajimovich, O. (2002). “Holocene ocean-climate variations in Alfonso Basin, Gulf of California, Mexico,” in Proceedings of the Eighteen PACLIM Workshop, eds G. J. West and L. D. Buffaloe (Pacific Grove, CA: Asilomar), 7–20.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgcomb, V. P., Beaudoin, D., Gast, R., Biddle, J. F., and Teske, A. (2011). Marine subsurface eukaryotes: the fungal majority. Environ. Microbiol. 13, 172–183. doi: 10.1111/j.1462-2920.2010.02318.x
Espinosa-Asuar, L., Soto, L. A., Salcedo, D. L., Hernández-Monroy, A., Eguiarte, L. E., Souza, V., et al. (2020). “Chapter 7, Bacterial communities from deep hydrothermal systems: the Southern Gulf of California as an example or primeval environments,” in Astrobiology and Cuatro Ciénegas Basin as an Analog of Early Earth, eds V. Souza, A. Segura, and J. S. Foster (Cham: Springer), 149–163. doi: 10.1007/978-3-030-46087-7_7
Gage, J. D., and Tyler, P. A. (1991). Deep-Sea Biology: A Natural History of Organisms at the Deep Sea Floor. Cambridge: Cambridge University Press.
Gao, Y., Du, X., Xu, W., Fan, R., Zhang, X., Yang, S., et al. (2020). Fungal diversity in deep sea sediments from east yap trench and their denitrification potential. Geomicrobiol. J. 37, 848–858. doi: 10.1080/01490451.2020.1789778
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes – Application to identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Goffredi, S. K., Johnson, S., Tunnicliffe, V., Caress, D., Clague, D., Escobar, E., et al. (2017). Hydrothermal vent fields discovered in the southern Gulf of California clarify the role of habitat in augmenting regional diversity. Proc. R. Soc. B Biol. Sci. 284:20170817. doi: 10.1098/rspb.2017.0817
Goffredi, S. K., Motooka, C., Fike, D. A., Gusmao, L. C., Tilic, E., Rouse, G. W., et al. (2021). Mixotrophic chemosynthesis in a deep-sea anemone from hydrothermal vents in the Pescadero Basin, Gulf of Claifornia. BMC Biol. 19:8. doi: 10.1186/s12915-020-00921-1
González-Yajimovich, O., Douglas, R. G., and Gorsline, D. S. (2005). The preserved carbonate record in Holocene sediments of the Alfonso and Pescadero basins, Gulf of California, Mexico. Proc. Geol.′Assoc. 116, 315–330. doi: 10.1016/S0016-7878(05)80050-1
Grossart, H. P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354.
Helly, J. J., and Levin, L. A. (2004). Global distribution of naturally occurring marine hypoxia on continental margins. Deep Sea Res. I Oceanogr. Res. Pap. 51, 1159–1168. doi: 10.1016/j.dsr.2004.03.009
Hsieh, T. C., Ma, K. H., and Chao, A. (2020). iNEXT: iNterpolation and EXTrapolation for Species Diversity. R Package Version 2.0.20. Available online at: http://chao.stat.nthu.edu.tw/wordpress/software-download/ (accessed July, 2021).
Ivarsson, M., Bengston, S., and Neubeck, A. (2016). The igneous oceanic crust – Earth’s largest fungal habitat? Fungal Ecol. 20, 249–255. doi: 10.1016/j.funeco.2016.01.009
Jebaraj, C. S., Raghukumar, C., Behnke, A., and Stoeck, T. (2010). Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol. Ecol. 71, 399–412. doi: 10.1111/j.1574-6941.2009.00804.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai, X., Cao, L., Tan, H., Fang, S., Huagn, Y., and Zhou, S. (2007). Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 1, 756–762. doi: 10.1038/ismej.2007.51
Le Calvez, T., Burgaud, G., Mahé, S., Barbier, G., and Vandenkoornhuyse, P. (2009). Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 75, 6415–6421. doi: 10.1128/AEM.00653-09
Legendre, P., and Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280.
Levin, C. L. (2003). Oxygen minimum zone benthos: adaptation and community response to hypoxia. Oceanogr. Mar. Biol. 41, 1–45.
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
López-García, P., Vereshchaka, A., and Moreira, D. (2007). Eukaryotic diversity associated with carbonates and fluid-seawater interface in Lost City hydrothermal field. Environ. Microbiol. 9, 546–554. doi: 10.1111/j.1462-2920.2006.01158.x
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Ariyawansa, H. A., Aoki, T., et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA fungus, 11, 1–32. doi: 10.1186/s43008-020-00033-z
Luo, Y., Wei, X., Yang, S., Gao, Y. H., and Luo, Z. H. (2020). Fungal diversity in deep-sea sediments from the Magellan seamounts as revealed by a metabarcoding approach targeting the ITS2 regions. Mycology 11, 214–229. doi: 10.1080/21501203.2020.1799878
Ma, M. (2005). Species richness vs evenness: independent relationship and different responses to edaphic factors. Oikos 111, 192–198. doi: 10.1111/j.0030-1299.2005.13049.x
Manohar, C. S., Boekhout, T., Müller, W. H., and Stoeck, T. (2014). Tritirachium candoliense sp. nov., a novel basidiomycetous fungus isolated from the anoxic zone of the Arabian Sea. Fungal Biol. 118, 139–149. doi: 10.1016/j.funbio.2013.10.010
Manohar, C. S., Menezes, L. D., Ramasamy, K. P., and Meena, R. M. (2015). Phylogenetic analyses and nitrate-reducing activity of fungal cultures isolated from the permanent, oceanic oxygen minimum zone of the Arabian Sea. Can. J. Microbiol. 61, 217–226. doi: 10.1139/cjm-2014-0507
Marchese, P., Garzo, L., Young, R., Allcock, L., Barry, F., Tuohy, M., et al. (2021). Fungi populate deep-sea coral gardens as well as marine sediments in the Irish Atlantic Ocean. Environ. Microbiol. 23, 4168–4184. doi: 10.1111/1462-2920.15560
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mullineaux, L. S., Metaxas, A., Beaulieu, S. E., Bright, M., Gollner, S., Grupe, B. M., et al. (2018). Exploring the ecology of deep-sea hydrothermal vents in a metacommunity framework. Front. Mar. Sci. 5:49. doi: 10.3389/fmars.2018.00049
Nagahama, T., Takahashi, E., Nagano, Y., Abdel-Wahab, M. A., and Miyazaki, M. (2011). Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ. Microbiol. 13, 2359–2370. doi: 10.1111/j.1462-2920.2011.02507.x
Nagano, Y., Miura, T., Tsubouchi, T., Lima, A. O., Kawato, M., Fujiwara, Y., et al. (2020). Cryptic fungal diversity revealed in deep-sea sediments associated with whale-fall chemosynthetic ecosystems. Mycology 11, 263–278. doi: 10.1080/21501203.2020.1799879
Nagano, Y., and Nagahama, T. (2012). Fungal diversity in deep-sea extreme environments. Fungal Ecol. 5, 463–471. doi: 10.1016/j.funeco.2012.01.004
Nagano, Y., Nagahama, T., Hatada, Y., Nunoura, T., Takami, H., Miyazaki, J., et al. (2010). Fungal diversity in deep-sea sediments—the presence of novel fungal groups. Fungal Ecol. 3, 316–325. doi: 10.1016/j.funeco.2010.01.002
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109.
Oksanen, J. F., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). Vegan: Community Ecology Package. R Package Version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan (accessed July, 2021).
Orsi, W. D., and Edgcomb, V. P. (2013). Microbial eukaryotes in marine oxygen minimum zones. Polyextremophiles 27, 485–497.
Paduan, J. B., Zierenberg, R. A., Clague, D. A., Spelz, R. M., Caress, D. W., Troni, G., et al. (2018). Discovery of hydrothermal vent fields on Alarcón Rise and in Southern Pescadero Basin, Gulf of California. Geochem. Geophys. Geosyst. 19, 4788–4819. doi: 10.1029/2018GC007771
Paulmier, A., and Ruiz-Pino, D. (2009). Oxygen minimum zones (OMZs) in the modern ocean. Prog. Oceanogr. 80, 113–128. doi: 10.1016/j.pocean.2008.08.001
Peng, X., and Valentine, D. L. (2021). Diversity and N2O production potential of fungi in an oceanic oxygen minimum zone. J. Fungi 7:218. doi: 10.3390/jof7030218
Quemener, M., Mara, P., Schubotz, F., Beaudoin, D., Li, W., Pachiadaki, M., et al. (2020). Meta-omics highlights the diversity, activity and adaptations of fungi in deep oceanic crust. Environ. Microbiol. 22, 3950–3967. doi: 10.1111/1462-2920.15181
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramirez-Llodra, E., and Billett, D. S. M. (2006). “Deep-sea ecosystems: pristine biodiversity reservoir and technological challenges,” in The Exploration of Marine Biodiversity: Scientific and Technological Challenges, ed. C. M. Duarte (Bilbao: Fundación BBVA), 63–92.
Rédou, V., Kumar, A., Hainaut, M., Henrissat, B., Record, E., Barbier, G., et al. (2016). Draft genome sequence of the deep-sea ascomycetous filamentous fungus Cadophora malorum Mo12 from the Mid-Atlantic Ridge reveals its biotechnological potential. Genome Announc. 4, 467–516. doi: 10.1128/genomeA.00467-16
Ricotta, C., and Podani, J. (2017). On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complex. 31, 201–205.
Rogers, A. D. (2000). The role of the oceanic oxygen minima in generating biodiversity in the deep sea. Deep Sea Res. II 47, 119–148. doi: 10.1016/S0967-0645(99)00107-1
Ruhl, H. A., and Smith, K. L. (2004). Shifts in deep-sea community structure linked to climate and food supply. Science 305, 513–515.
Salcedo, D. L., Soto, L. A., and Paduan, J. B. (2019). Trophic structure of the macrofauna associated to deep-vents of the southern Gulf of California: pescadero Basin and Pescadero transform fault. PLoS One 14:e0224698. doi: 10.1371/journal.pone.0224698
Salcedo, D. L., Soto, L. A., and Paduan, J. B. (2021). Trophic interactions among the macrofauna of the deep-sea hydrothermal vents of Alarcón Rise, Southern Gulf of California. Deep Sea Res. Part I 176:133609. doi: 10.1016/j.dsr.2021.103609
Sánchez, A., Rodríguez-Figueroa, G., Shumilin, E., and Ortiz-Hernández, M. C. (2018). Evidence of intense suboxia and anoxia in a tropical bay of the Gulf of California. Continent. Shelf Res. 168, 21–27.
Shoun, H., Kim, D. H., Uchiyama, H., and Sugiyama, J. (1992). Dentrification by fungi. FEMS Microbiol. Lett. 94, 277–281. doi: 10.1111/j.1574-6968.1992.tb05331.x
Singh, P., Raghukumar, C., Murti, R., Verma, P., and Shouche, Y. (2012). Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 5, 543–553. doi: 10.1016/j.funeco.2012.01.001
Snelgrove, P., and Smith, C. R. (2002). “A riot of species in an environmental calm: the paradox of the species-rich deep-sea floor,” in Oceanography and Marine Biology, An Annual Review 40, eds R. N. Gibson, M. Barnes, and R. J. A. Atkinson (London: CRC Press), 311–342.
Stock, A., Bunge, J., Jurgens, K., and Stoeck, T. (2009). Protistan diversity in the suboxic and anoxic waters of the Gotland Deep (Baltic Sea) as revealed by 18S rRNA clone libraries. Aquat. Microb. Ecol. 55, 267–284. doi: 10.3354/ame01301
Stoeck, T., Zuendorf, A., Breiner, H. W., and Behnke, A. (2007). A molecular approach to identify active microbes in environmental eukaryote clone libraries. Microb. Ecol. 53, 328–339. doi: 10.1007/s00248-006-9166-1
Takishita, K., Tsuchiya, M., Kawato, M., Oguri, K., Kitazato, H., and Maruyama, T. (2007). Genetic diversity of micro- bial eukaryotes in anoxic sediment of the saline meromictic lake Namako-ike (Japan): on the detection of anaerobic or anoxic-tolerant lineages of eukaryotes. Protist 158, 51–64. doi: 10.1016/j.protis.2006.07.003
Van Dover, C. L. (2007). “The biological environment of polymetallic sulphides deposits, the potential impact of exploration and mining on this environment, and data required to establish environmental baselines in exploration areas,” in Proceedings of the International Seabed Authority’s Workshop. Polymetallic Sulphides and Cobalt-rich Ferromanganese Crusts Deposits: Establishment of Environmental Baselines and an Associated Monitoring Programme during Exploration, (Kingston: International Seabed Authority), 169–190.
Van Dover, C. L. (2014). Impacts of anthropogenic disturbances at deep-sea hydrothermal vent ecosystem: a review. Mar. Environ. Res. 102, 59–72. doi: 10.1016/j.marenvres.2014.03.008
Van Dover, C. L., German, C. R., Speer, K. G., Parson, L. M., and Vrijenhoek, R. C. (2002). Evolution and biogeography of Deep-Sea vent and seep invertebrates. Science 295, 1253–1257. doi: 10.1126/science.1067361
Vargas-Gastélum, L., Chong-Robles, J., Lago-Lestón, A., Darcy, J. L., Amend, A. S., and Riquelme, M. (2019). Targeted ITS1 sequencing unravels the mycodiversity of deep-sea sediments from the Gulf of Mexico. Environ. Microbiol. 21, 4046–4061. doi: 10.1111/1462-2920.14754
Velez, P., Gasca-Pineda, J., and Riquelme, M. (2020). Cultivable fungi from deep-sea oil reserves in the Gulf of Mexico: genetic signatures in response to hydrocarbons. Mar. Environ. Res. 153:104816. doi: 10.1016/j.marenvres.2019.104816
White, T. J., Bruns, T., Lee, S. J. W. T., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Applic. 18, 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wright, E. S. (2016). Using DECIPHER v2.0 to Analyze Big biological sequence data in R. R J. 8, 352–359. doi: 10.32614/RJ-2016-025
Wright, J. J., Konwar, K. M., and Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Xu, W., Gao, Y. H., Gong, L. F., Li, M., Pang, K. L., and Luo, Z. H. (2019). Fungal diversity in the deep-sea hadal sediments of the Yap Trench by cultivation and high throughput sequencing methods based on ITS rRNA gene. Deep Sea Res. Part I 145, 125–136. doi: 10.1016/j.dsr.2019.02.001
Xu, W., Gong, L. F., Pang, K. L., and Luo, Z. H. (2018). Fungal diversity in deep-sea sediments of a hydrothermal vent system in the Southwest Indian Ridge. Deep Sea Res. Part I Oceanogr. Res. Pap. 131, 16–26. doi: 10.1016/j.dsr.2017.11.001
Xu, W., Guo, S., Pang, K. L., and Luo, Z. H. (2017). Fungi associated with chimney and sulfide samples from a South Mid-Atlantic ridge hydrothermal site: distribution, diversity and abundance. Deep Sea Rese. Part I Oceanogr. Res. Pap. 123, 48–55. doi: 10.1016/j.dsr.2017.03.004
Zhang, X., Tang, G., Xu, X., Nong, X., and Qi, S. (2014). Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One 9:e109118. doi: 10.1371/journal.pone.0109118
Keywords: deep-sea microeukaryotes, extreme environments, ITS metabarcoding, functional diversity, marine mycodiversity
Citation: Velez P, Salcedo DL, Espinosa-Asuar L, Gasca-Pineda J, Hernandez-Monroy A and Soto LA (2022) Fungal Diversity in Sediments From Deep-Sea Extreme Ecosystems: Insights Into Low- and High-Temperature Hydrothermal Vents, and an Oxygen Minimum Zone in the Southern Gulf of California, Mexico. Front. Mar. Sci. 9:802634. doi: 10.3389/fmars.2022.802634
Received: 27 October 2021; Accepted: 26 January 2022;
Published: 17 February 2022.
Edited by:
Paris Vasileios Stefanoudis, University of Oxford, United KingdomReviewed by:
Stefano Varrella, Marche Polytechnic University, ItalyLluvia Vargas Gastelum, Center for Scientific Research and Higher Education in Ensenada (CICESE), Mexico
Copyright © 2022 Velez, Salcedo, Espinosa-Asuar, Gasca-Pineda, Hernandez-Monroy and Soto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia Velez, cHZlbGV6QGliLnVuYW0ubXg=; Diana L. Salcedo, ZGlsZXNhb3JAZ21haWwuY29t
†Retired
 Patricia Velez
Patricia Velez Diana L. Salcedo
Diana L. Salcedo Laura Espinosa-Asuar
Laura Espinosa-Asuar Jaime Gasca-Pineda
Jaime Gasca-Pineda Abril Hernandez-Monroy
Abril Hernandez-Monroy Luis A. Soto4†
Luis A. Soto4†