- 1Key Laboratory of Marine Biotechnology of Zhejiang Province, School of Marine Sciences, Ningbo University, Ningbo, China
- 2Xiangshan Xuwen Seaweed Development Co., Ltd., Ningbo, China
Ulva prolifera, the main causative species of green tide, has drawn much attention. Spore reproduction was one of the main reproduction strategies that could be induced by changing environmental factors, but the mechanism of spore formation remains obscure. Here, we cultured U. prolifera (segments) under the optimal sporulation condition, and four states in total from vegetative to reproductive were defined. Additionally, the chlorophyll fluorescence and transcriptome analysis were determined at these four states. The results showed that: (1) Compared with state I, the photosynthetic capacity (Fv/Fm, Fv′/Fm′, rETRmax) and chlorophyll content (Chl a, Chl b, carotenoids) were enhanced in state II, whereas it decreased in state III and IV (the spore formation period); (2) a total of 41,058 unigenes were expressed during the spore formation process; (3) compared with state I, the genes related with photosynthesis, terpenoid backbone biosynthesis, and carotenoid biosynthesis were significantly upregulated in states II, III, and IV whereas glycolysis was downregulated in state I; (4) some genes of the transcription factors families, such as the C3H family, may be one of the key factors that regulate genes in the spore formation; (5) 574 of the differentially expressed genes (DEGs) associated with flagella biosynthesis were annotated according to Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Orthology, including 20 unigenes encoding intraflagellar transport proteins (IFTs) which had not been reported in previous transcriptome analysis in U. prolifera. This study provides a new perspective of spore formation at the gene transcriptional level, although the detailed transcription regulatory spore formation network remains to be unveiled.
Introduction
Ulva spp., the most ubiquitous and important primary producers around the world’s oceans (Raffaelli et al., 1998), can be used as animal feed in aquaculture or an important protein food source for humans because they are highly rich in proteins (He et al., 2021). They were also the domain species of the green tide, especially for the Ulva prolifera, which is the main causative species for their rapid growth rate and complex reproductive abilities (Cui et al., 2018; Wang et al., 2018a). The reproductive strategies of U. prolifera include parthenogenesis, sexual and asexual, and the asexual spores, sexual male and female gametes (Lin et al., 2008). Recently, many studies about the effects of ecological factors, such as temperature, light intensity on the growth, physiological and biochemical characteristics of U. prolifera have been reported (Cui et al., 2015; Wu et al., 2018a; Wang et al., 2020a). However, the life history of U. prolifera is still very controversial, and whether gametophytes or sporophytes dominate the life cycle has not been determined (Wang et al., 2020b).
For macroalgae, the spore reproduction could be affected by biotic and abiotic factors, such as the thallus fragmentation, temperature, and light intensity (Han et al., 2008; Gao et al., 2010, 2017). Previous studies have shown the following: about 0.5 mm diameter of U. prolifera fragments were almost entirely converted to sporangia (Gao et al., 2010), abrupt temperature changes could stimulate the reproduction of U. rigida (Gao et al., 2017), and the minimum light intensity required for the spore release was 5–10 μmol m–2 s–1 in U. pertusa (Han et al., 2008). The spore release of U. pseudocurvata was also spontaneously triggered as soon as the light cycle began (Smith, 1947). Moreover, to understand better the changes in the reproductive process of Ulva, some studies divided the reproductive progress, from the transformation of vegetative cells into reproductive cells, into two to four stages (Wichard and Oertel, 2010; Carl et al., 2014; Wang et al., 2016; Chavez-Sanchez et al., 2018). Although the previous study showed that spore formation was regulated by the redox status of the plastoquinone pool that could be affected by photosynthetic electron transport chain (PET) (Wang et al., 2016), the regulatory mechanism of spore formation of U. prolifera is not yet clear.
RNA-sequencing, as a revolutionary tool for transcriptomics, has been wildly used for biological inquiries (Wang et al., 2009; Du et al., 2021). The previous study of transcriptome analysis showed some of the potential molecular mechanisms of growth have been revealed, such as the specific photoprotective mechanism associated with LhcSR and PsbS (Zhang et al., 2012). Expressed sequence tags analysis found that some sequences code for proteins involved in the formation of flagella (axonemal-type dynein light chain), the length of the flagella (LF4), and the movement of the flagella (CAM kinase and PF16) (Stanley et al., 2005). Flagella was the key for spores to swimm in the water to survive and proliferate under different environmental conditions (Silflow and Lefebvre, 2001). In U. prolifera and U. linza, the existence of C4-type metabolism pathway also has been confirmed by transcriptome analysis (Xu et al., 2012, 2013). Compared with the other three Ulva species, U. prolifera demonstrated noticeably larger numbers of total transcription factors (TFs) which were prominent in many TF types, and several growth-related genes were enriched (Wang et al., 2019). However, few studies were related to spore formation.
So, in this study, the global mRNA expression patterns of U. prolifera at multiple sampling time points from vegetative cells to matured spores were examined by using RNA-seq technology to understand better the morphological conversion from vegetative cells to matured spores of U. prolifera.
Materials and Methods
Species and Culture Conditions
Ulva prolifera seeding (3 mm) was obtained from Xiangshan Xuwen Seaweed Development Co. Ltd., Xiangshan County, Zhejiang Province, China (29°05′065N, 121°56′153E), and it was identified through morphology and genotyped by sequencing the 18S and ITS rRNA genes. They were cultured at 20°C, 100 μmol m–2 s–1 condition until ca. ≥18 cm length and were used in the following experiments. No specific permits were required for the using this species.
Based on the results of our preexperiment, the adult thalli were cut into 2–3 mm segments (optimal length) and then cultured in sterile artificial seawater (salinity 25) enriched with f/2 medium (without Si) (Guillard and Ryther, 1962) in an illuminated plant incubator (GXZ, Jiangnan Instrument Factory, Ningbo, China) at 25°C, 400 μmol m–2 s–1, 12:12 light/cycle with aerated (350 mL min–1) ambient outdoor air. GeO2 (0.3 mg L–1) was added to remove the diatom (Cui et al., 2015) for the following main experiment. Salinity was measured with a visual hand-held refractometer (Index Instruments, Ramsey, United Kingdom).
Morphology Observation
The process of spore formation and release was observed by using the optical microscope with digital camera (ToupCam™, Touptek, China). According to Carl et al. (2014) and Chavez-Sanchez et al. (2018), four states of thalli segments, from vegetative to reproductive, were defined. Moreover, the synchronization of different thalli was defined as the number of more than 50% of different thalli that changed significantly. The thalli with a high synchronization than 90%, were cultured in the laboratory after being cut into small segments. So, when the state was confirmed, the samples were harvested for transcriptome analysis.
Chlorophyll Fluorescence Parameters Measurement
After defining the state, a PSI fluorometer (AquaPen-C, Photo System Instruments, Drasov, Czechia) was used to determine the effective quantum yield of PSII (Fv′/Fm′), the non-photochemical quenching (NPQ), and the rapid light curves (RLC). After adapting to dark conditions for 15 min, NPQ and Fv′/Fm′ were measured with the cultivated light intensity as the actinic light. RLC was fitted as rETR = I/(aI2 + bI + c) (Eilers and Peeters, 1988), where I is the photon flux density of activity light (μmol m–2 s–1), and a, b, c are the adjustment parameters. The initial slop (α), the maximum electron transport rate (rETRmax), and the light saturation parameter (Ek) were expressed as a function of the parameters a, b, and c as follows: α = 1/c; rETRmax = 1/(b + 2(a × c)1/2); Ek = c/(b + 2(a × c)1/2).
Chlorophyll Content Determination
Chlorophyll a (Chl a), Chlorophyll b (Chl b), and carotenoid (Car) are important photosynthetic chlorophyll in green macroalgae. Approximately 0.01 g freshly weighed thalli were extracted in 3 mL methanol at 4°C in darkness overnight, and then the absorption spectrum of the supernatant was obtained by scanning the sample from 250 to 750 nm with a scanning spectrophotometer (Yuanxi Instrument Co., Ltd, Shanghai, China). The chlorophyll concentration, including Chl a, Chl b. and Car, was calculated according to Wellburn (1994).
RNA Isolation and cDNA Library Construction
Total RNA of the samples was extracted with an E.Z.N.A.® Plant RNA Kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s instructions. The concentration and the quality of RNA were determined using a NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, United States) and Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, United States).
RNA samples and a pooled RNA from these 12 samples (three biological replicates, four states) were submitted for differential expression analysis and de novo transcriptome analysis, respectively. The 13 cDNA libraries were constructed using mRNA library preparation kit (MGIEasy™, MGI, Shenzhen, China). The mRNA with ploy(A) structure was enriched with oligo magnetic adsorption. The enriched mRNAs were fragmented and then reversed into a double-strand cDNA (dscDNA) with an N6 random primer. Sequencing adaptors were linked to the end of the purified dscDNA, and the ligation product was amplified by PCR with specific primers. The PCR product was thermally denatured into a single strand, and then the single-stranded DNA was circularized with a bridge primer to obtain a single-stranded circular DNA library.
RNA-Sequence, de novo Assembly of Transcriptome, and Gene Identification
The library for de novo transcriptome was sequenced by 150 bp double ends sequencing method while the other 12 libraries for differential expression analysis were sequenced by 50-bp single end sequencing method on a DNBSEQ platform, performed by Wuhan Genomic Institution (BGI, Shenzhen, China). Additionally, although the genome of U. prolifera was reported (the accession number in NCBI was SDUY00000000.1; He et al., 2021), the annotation of this genome has not been found in the NCBI database. So, the de novo transcriptome analyzed was chosen in this study. The details were similar to Hu et al. (2019).
Differentially Expressed Genes and Their Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analyses
Differentially expressed genes (DEGs) were identified using the DESeq method based on the negative binomial distribution (Wang et al., 2010). There are two standards to select DEGs, namely, using the Q value < 0.001 and the absolute value of fold change ≥ 2 as two threshold values to screen out DEGs in different comparison groups. The Q value was a corrected p-value calculated by using KOBAS 2.0. method (Mao et al., 2005). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analysis of DEGs were performed using the KEGG database and GO database (He et al., 2019a). A Q value cutoff of 0.05 was set for the identification of remarkably enriched GO and KEGG pathways for DEGs.
Expression Validation by Quantitative Real-Time PCR Analysis
Ten DEGs were randomly selected for quantitative real-time PCR (qRT-PCR) to validate the RNAseq results. The single-strand cDNAs were synthesized from 1 μg of the total RNA using HiScript II 1st strand cDNA synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. qRT-PCR was performed on a Mastercycler Realplex2 realtime PCR instrument (Eppendorf, Germany) using a ChamQTM Universal SYBR qPCR Master Mix Kit (Vazyme, China). The qRT-PCR program was as follows: 95°C for 30 s, 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 20 s. The primers were designed using Primer Premier 5 software (Premier Biosofe International, San Francisco, CA, United States) and are listed in Supplementary Table 1. Four technical replicates were performed for each sample. The relative expression levels of the genes were normalized by comparison with 18S rDNA expression and calculated using the 2–ΔΔCt relative quantitative method (He et al., 2019a).
Statistical Analysis
Three replicates for each treatment were used for all the treatments. The data were shown as means ± SD. The data were analyzed using SPSS software (version 22.0 USA) and assessed by one-way analysis of variance (ANOVA). A turkey post hoc test (Turkey HSD) was performed to show the difference between different states, such as chlorophyll content and the gene expression levels obtained from qRT-PCR among treatments. The significant levels were set as p < 0.05.
Results and Discussion
Morphology Change of Ulva prolifera in the Spore Formation
According to previous studies (Pettett, 2009; Carl et al., 2014; Park, 2020) and based on the observations, the reproductive process of U. prolifera was divided into four states (Figure 1). In detail, first, initially, the chloroplasts were visible to the interior of the green vegetative cell (state I; Figure 1A). Second, the chloroplasts were prepared to displace to one side of the cell (state II; Figure 1B). The spores filled the interior of the cell (state III; Figure 1C) and were matured with a clear shape and an eyespot (state IV; Figure 1D). Previous studies showed that sporulation can be identified in Ulva thalli through a change in coloration (Pettett, 2009; Carl et al., 2014; Park, 2020). During this study, the color of the thalli segments converted to yellow from green, which was significantly observed in states III and IV, indicating the spore formatted in these two states. Moreover, an eyespot was also observed in the states III and IV, and especially in state IV, the eyespot has a clear shape. Therefore, we speculated that states III and IV were the crucial periods of spore formation.
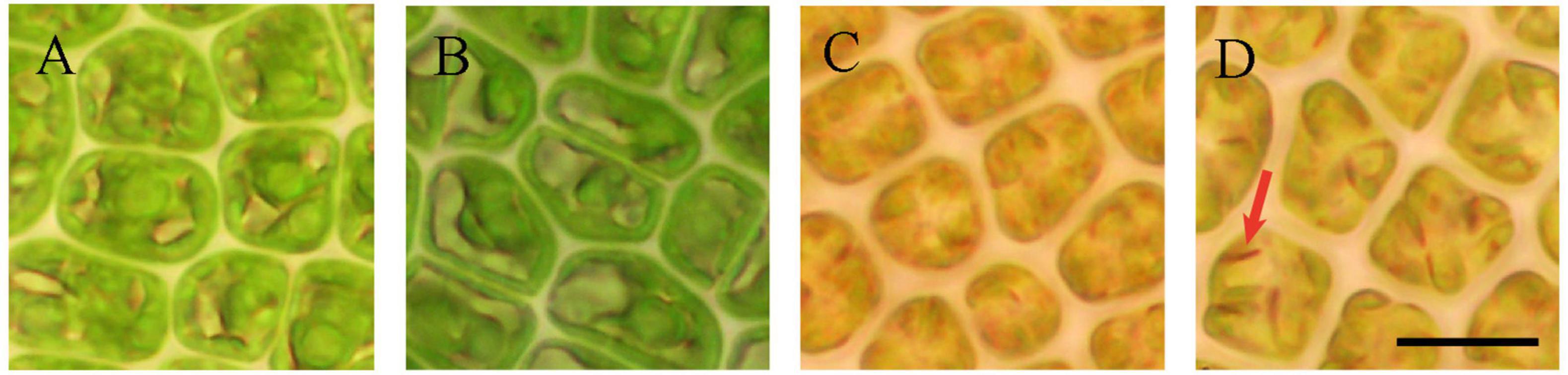
Figure 1. The four states of induced sporogenesis in U. prolifera. (A) vegetative and (B–D) sporogenesis process. The scale bars represent 10 μm. Red arrow means eyespot of spore.
Chlorophyll Fluorescence Parameters and Chlorophyll Content
With the change of algae cell morphology, photosynthesis changed significantly at the same time. Compared with state I, the relative electron transport rate (rETR) and effective quantum yield (Fv’/Fm’) were higher in the states II and III, and state III has higher NPQ than I and II (Figures 2D–F). NPQ is one of the adaptive mechanisms employed by plants and algae for coping with complex environmental change that works by two independent mechanisms, with the faster-activated quenching catalyzed by the monomeric light-harvesting complex (LHCII) proteins and the slowly activated quenching catalyzed by LHCII trimers, both processes depending on zeaxanthin but to a different extent (Gao et al., 2020). Previous study showed that the induction of NPQ was regulated by the generation of zeaxanthin (Zhang et al., 2015). In this study, state I was observed at the end of the light period whereas state II was observed at the end of the dark period, the genes encoding zeaxanthin synthesis were upregulated from state I to state II, which was consistent with the previous study, showing that the content of zeaxanthin could be enhanced by high light (Eismann et al., 2020). But compared to state II, the expression of genes encoding for violaxanthin de-epoxidase (VDE; EC: 1.23.5.1) decreased by 2–4 folds, whereas the expression of genes encoding for beta-ring hydroxylase (LUT) and zeaxanthin epoxidase (ZEP; EC: 1.14.15.21) was upregulated (Supplementary Table 2) in state III. The NPQ in state III was higher than that in state II, but because we did not measure the content of zeaxanthin, the relationship between NPQ and zeaxanthin, as well as the role of zeaxanthin during the sporulation should be further studied. The other chlorophyll fluorescence parameters (Fv/Fm, Fv’/Fm’, NPQ, rETRmax, and α) were significantly decreased in state IV (Figure 2). The rETRmax and α provide an approximation of the rate of electron flow via PS II into the photosynthetic electron transport chain and the efficiency of energy transfer from the light-harvesting antenna to PSII reaction centers, respectively, and these values also reflect the overall photosynthetic capacity of algae (Serodio et al., 2006).
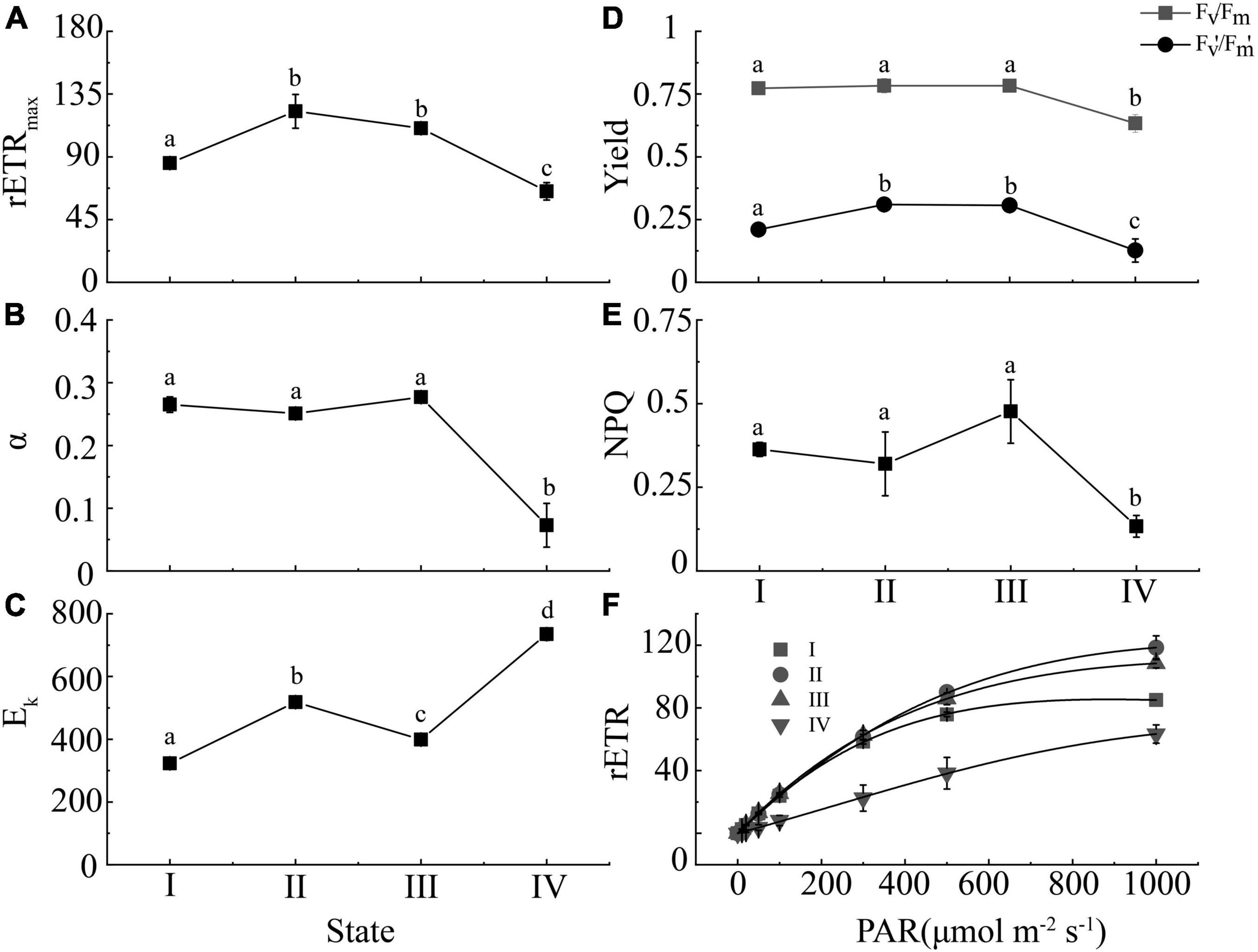
Figure 2. The changed of chlorophyll fluorescence parameters (A: rETRmax; B: α; C: Ek; D: Fv/Fm and Fv’/Fm’; E: NPQ; F: rETR curves) in the four states of spore formation in U. prolifera. Different letters indicate significant differences (p < 0.05) between different states.
Moreover, the highest chlorophyll content (Chl a, Chl b, and Car) was observed in the state II cells (Figure 3A), but gradually decreased in the state III and IV. Although there were no significant differences in the ratio of Chl a to Car or Chl b to Car between state III and IV (Figure 3B), the state IV cells showed the lowest chlorophyll content but highest Car percentage (23%) in the total chlorophyll content (Chl a + Chl b + Car) (Figure 3A). This indicated that the proportion of carotenoids was synchronized with the formation of spores, which was consistent with the previous study (Park, 2020). This may be the reason why the algae color was yellow in states III and IV. Additionally, carotenoid as a type of photoprotection pigment, which indicated higher antistress ability of algae (Siefermann-Harms, 1987), can protect the membranes from oxidants, such as reactive oxygen species (ROS), generated in the spore formation process of U. prolifera.
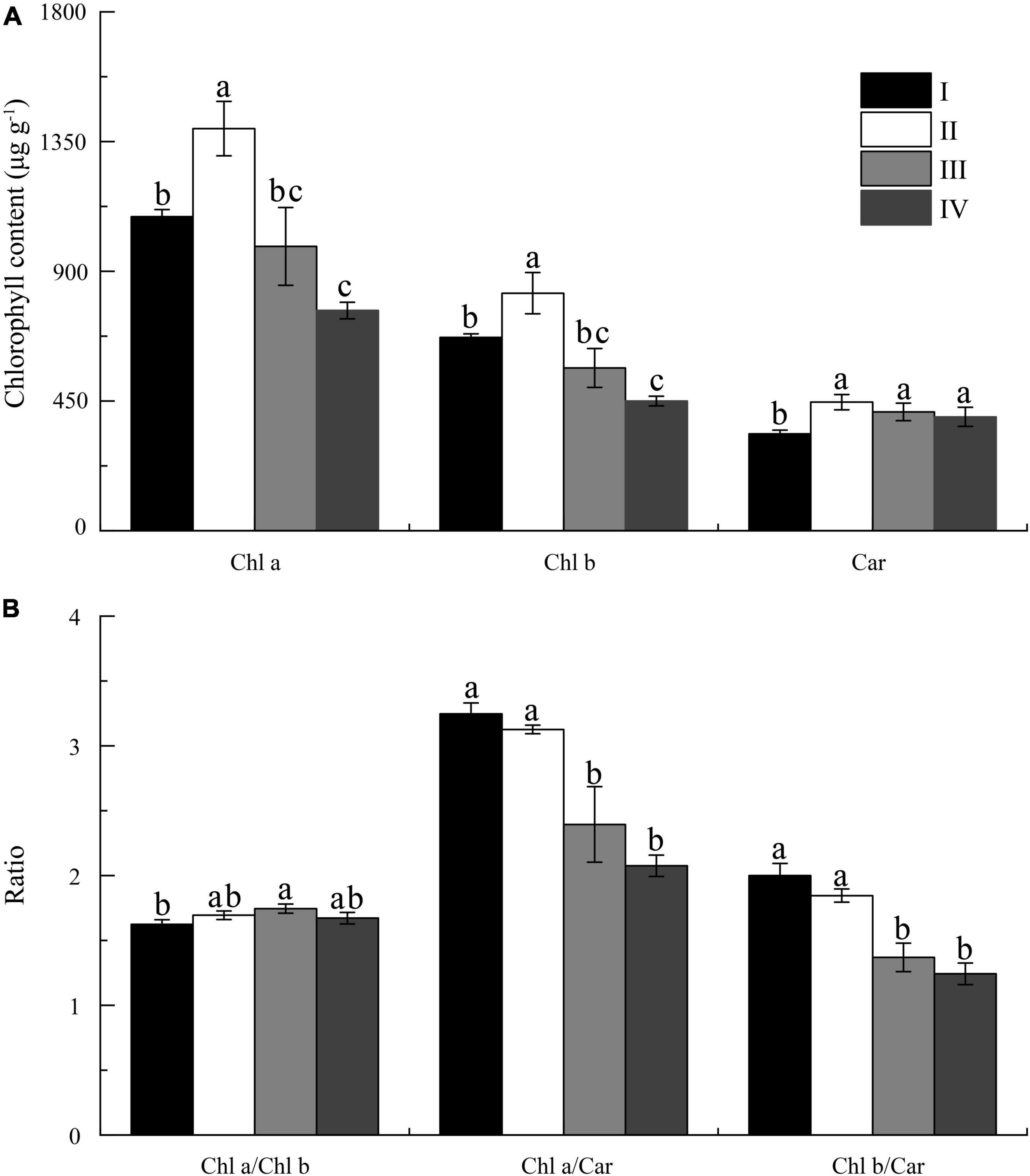
Figure 3. The content (A) and ratio (B) of the Chl a, Chl b, and carotenoids (Car) in four states of spore formation of U. prolifera. Different letters above the columns indicate significant differences (p < 0.05) between different states for the same name along the x-axis.
De novo Assembly of Transcriptome and Gene Functional Annotation
RNA-sequencing using the DNBSEQ platform generated a final 10.56 Gb of clean data. De novo assembly obtained 41,779 unigenes with an N 50 length of 2,619 bp by the Trinity method. The total length was 69,331,130 bp, and the average length was 1,659 bp. After BLASTx searches (E-value ≤ 10–5) against seven public databases, including the NCBI non-redundant protein sequences (Nr), NCBI Nucleotide Sequence Database (Nt), EuKaryotic Orthologous Groups database (KOG), Swiss Prot database (Swiss Prot), Pfam database (Pfam), KEGG, and GO (Supplementary Table 3), the transcriptome expression profile analysis of U. prolifera was performed using 12 cDNA samples, produced 21.68M clean reads each sample in average, and the average mapping rate to the transcriptome described above was 92.54%, and a total of 41,058 unigenes were identified.
To validate the reliability and reproducibility of the RNA-seq analysis, ten unigenes were randomly selected from different metabolism pathways for qRT-PCR validation. The expression patterns of these selected unigenes in our transcriptome data were highly consistent with the qRT-PCR results (Supplementary Figure 1), indicating that the RNA-seq data were credible and sufficient for subsequent analysis.
Principal Component Analysis and Correlation Analysis
Principal component analysis (PCA) was performed to get an overview of the transcriptome difference between different states (Supplementary Figure 2A). The first principal component (PC1), accounting for 34.5% of the total variance, clearly separated samples of II, III, and IV states from those of state I, indicating that spore formation could significantly change the transcriptional component of algal cells. Likewise, correlation analysis results was consistent with that of PCA. Correlation coefficients among samples of the three biological replicates were all greater than 0.9 (Supplementary Figure 2B).
Transcriptomic Changes of Thalli in Different Spore Formation States
The transcriptomic profiles of state II with I, III with II, and IV with III were compared. In detail, compared with state I, a total of 21,149 unigenes, including 11,645 upregulated unigenes (URGs) and 9,504 downregulated unigenes (DRGs) were founded in state II (Supplementary Figure 2C). The enrich result of URGs and DRGs are shown in Supplementary Figures 3, 4. The ribosome would encode amino acids to form polypeptide chain by utilizing mRNA as the model and produce protein for microtubule protein, transporter, cell skeleton protein, and so on. The upregulation of genes in the photosynthesis pathway explained the increased photosynthetic activity in state II with I (Figure 2). Photosynthesis also could provide several stress-resistant mechanisms to affect photosynthesis activity and provide power for other metabolism pathways. A previous study found that ubiquinone and other terpenoid–quinone biosynthesis pathways could provide precursors for carotenoid biosynthesis (Lohr et al., 2012), and the carotenoid could affect photosynthesis (Eismann et al., 2020).
Compared with state II, a total of 20,856 DEGs, including 9,539 URGs and 11,317 DRGs were found in state III (Supplementary Figure 2C). Purine metabolism may be associated with the repair of PS II because defective purine biosynthesis impairs chloroplast biogenesis and alters the induction of high light stress pathways in Arabidopsis (Woo et al., 2011).
Compared with state III, a total of 15,185 DEGs, including 6,473 URGs and 8,712 DRGs were found in state IV (Supplementary Figure 2C). Hormones play essential roles in the germination, growth, and development of plants by signaling pathways (Li et al., 2016), such as abscisic acid (ABA), jasmonate (JA), ethylene, and cytokinin (CK) (Nakashima and Yamaguchishinozaki, 2013).
Differentially Expressed Genes During the Sporulation Process
Photosynthesis
Photosynthesis products are the main energy sources for algae that include light and dark reactions, each of which consists of a series of redox reactions related to energy capture and conversion (Liu, 2016). At the initial step of photosynthesis, light energy is captured by the peripheral light-harvesting complexes, and then the synthesis of ATP and NADPH was affected by regulated Fv′/Fm′ (Yang et al., 2020). Compared with state I, the genes related to photosynthesis participating in the constitution of photosystem I (PS I) (PsaA, PsaB, PsaD, etc.) (Figure 4A, PS I), photosystem II (PS II) (PsbD, PsbC, PsbB, etc.) (Figure 4A, PSII), cytochrome b6/f complex (cytob6/f) (PetA, PetC, PetN, etc.), ATP synthase (F-type ATPase), and photosynthethic electron transport (PET) were upregulated in the state II but were downregulated in state III (Figure 4A and Supplementary Table 4). A previous study showed PsaB encodes P700 chlorophyll A2 apoproteins, one of PS I reaction, and the upregulation of this gene could significantly increase photosynthetic electron transport (Qian et al., 2010), resulting in an increase in photosynthesis (Figure 2). However, the gene expression of photosynthesis was also upregulated in the state D while the chlorophyll fluorescence parameters were decreased (Figure 2). We speculated that the polar growth of algae segment maybe one reason, but it needed to be further studied. Additionally, compared with state I, the gene encoding for ferredoxin NADP reductase (FNR) of NADPH synthesis was upregulated 5. 3-, 1. 6-, and 2-folds in the state II, III, and IV, respectively, indicating higher NADPH and photosynthesis electron transport of PS I in state II, but lower in III and IV. NADPH could enhance CO2 assimilation by providing power (Sui et al., 2015).
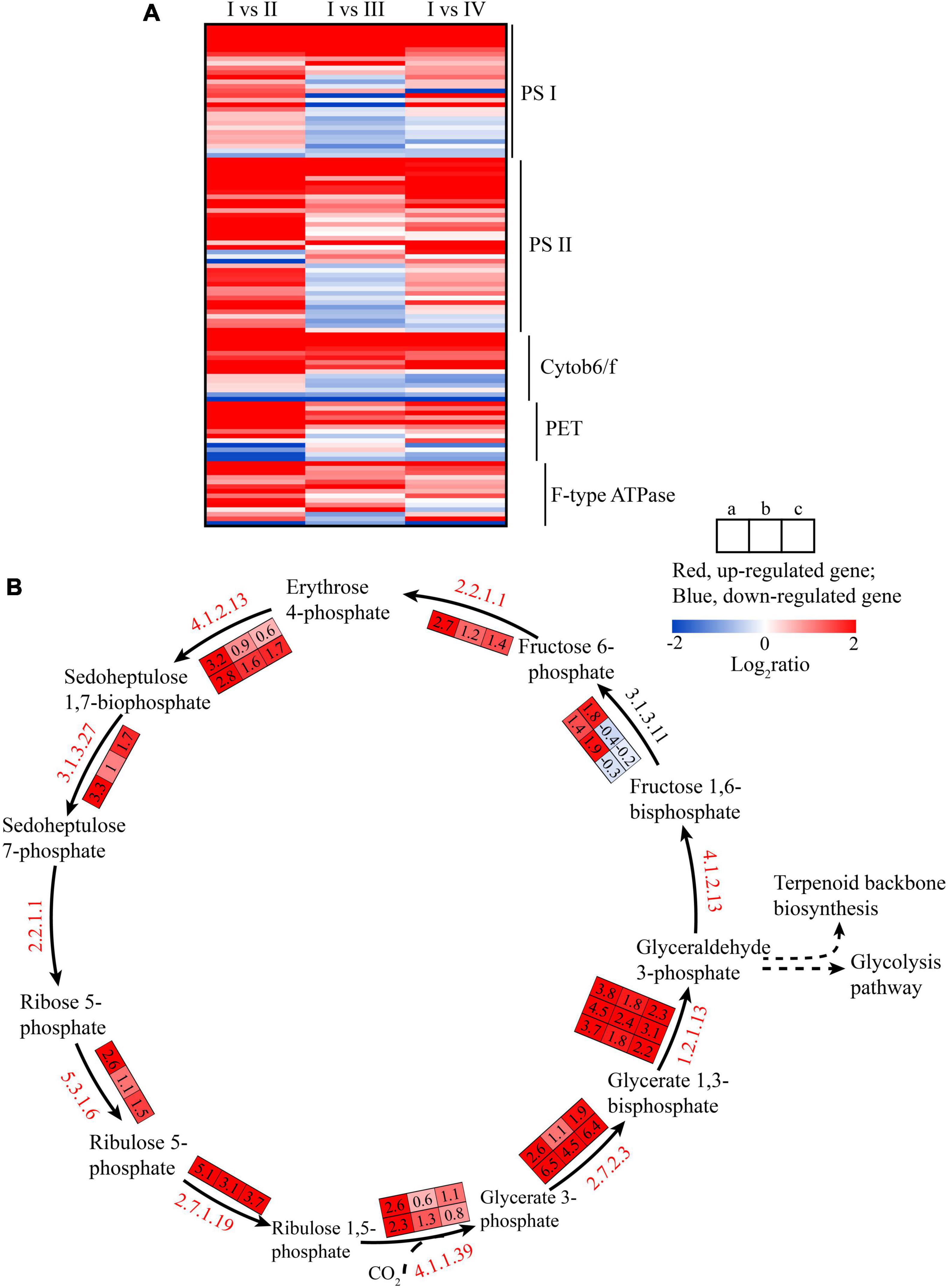
Figure 4. The heatmap of transcriptional changes of genes associated with photosynthesis (A) and carbon fixation pathway (B) in the spore formation of U. prolifera between different states. Column a, I vs. II; Column b, I vs. III; Column c, I vs. IV. Log2 of fold changes are given by shades of red or blue colors according to the scale bar, the larger the value, the darker the color.
The DEGs among four states in carbon fixation pathway are shown in Figure 4B. The rbcl gene, the key gene in carbon fixation, encoding ribulose-bisphosphate carboxylase (Rubisco), was upregulated (Figure 4B), indicating the increase in photosynthesis in U. prolifera (Qian et al., 2010). The rbcl gene could utilize CO2 to catalyze ribulose-1,5-bisphosphate (RuBP) to 3-phosphoglycerate (3-PGA), and during this process, the role of carbonic anhydrases (CA), C4 enzymes [phosphoenolpyruvate carboxylase (PEPC; EC: 4.1.1.31), and phosphoenolpyruvate carboxykinase (PEPCK; EC: 4.1.1.49)] cannot be ignored (Liu et al., 2020). In detail, atmospheric CO2 could be transferred into bundle sheath cells by PEPC and PEPCK, ensuring efficient transfer of CO2 to Rubisco to participate in 3-PGA synthesis (Hatch, 1987), and finally generate 3-phosphoglyceraldehyde (3-PGAld) (Figure 4B). However, the gene expression of PEPCK insignificantly changed from state II to III, whereas the rbcl gene expression was downregulated 2–4 folds, indicating that redundant CO2 existed in the cell, which could decrease the pH of the thylakoid membrane, and which could produce NPQ (Zhang et al., 2015). Moreover, oxaloacetate also could be catalyzed by malate dehydrogenase (MDH; EC: 1.1.1.37) to form malic acid which could be catalyzed by NADP–malate dehydrogenase (NADP-ME; EC: 1.1.1.40) to provide CO2 to Rubisco (Yang et al., 2020). The expression of the gene encoding for MDH was upregulated 2.6 folds while NADP-ME was downregulated 2.1 folds from state II to state III (Supplementary Table 5), indicating the malic acid synthesis increased. Malic acid is an organic acid that could help cells to transduce signal in the stress condition by upregulating gene expression (Kumar et al., 2016) and stimulate spore formation (He et al., 2019b).
Additionally, compared with state II, rbcl gene was downregulated in state III, which might decrease 3-PGA synthetic and inhibit the electrons transport of Cytochrome b6/f complex and PET, but the Fv’/Fm’ has no change, which was measured from PS II, and the PsaB expression was upregulated 2.1 folds. The PsaB is the key gene of PS II (Qian et al., 2010), that might enhance the oxidation of the plastoquinone (PQ), which was caused by plastid terminal oxidase (PTOX). We also found the expression of gene encoding PTOX was upregulated from state II to state III. The role of PTOX was transferring electrons from PQH2 to oxygen during the oxidation of PQ pool to enhance the oxidation state of the plastoquinone pool (Nawrocki et al., 2015). The enhanced oxidation of the plastoquinone pool could promote the formation of spores (Wang et al., 2016) and catalyze NADP to NADPH to provide power for MDH catalyze reaction (Chen et al., 2020). Then, we speculated that PTOX was one of the key genes for regulating spore formation.
The 3-PGAld was a photosynthetic product that could be converted to starch in the chloroplast stroma or enter the cytoplasm via triose phosphate translocator and synthesize sucrose (Yang et al., 2020). It was also the raw material for glycolysis and terpenoid backbone biosynthesis. A previous study showed that glucose-1-phosphate adenylyltransferase (APG; EC: 2.7.7.27), starch synthase (EC: 2.4.1.21), and 1,4-alpha-glucan branching enzyme (GBE; EC: 2.4.1.18) were involved in starch synthesis (Stark et al., 1992), and in this study, the genes responsible for these enzymes were upregulated in state II (Figure 5A). Additionally, the synthesis of 3-PGA was increased in carbon fixation pathway in state II (Figure 4B), which was consistent with the previous study, that is the APG could be regulated by 3-PGA and Pi as positive and negative effectors, respectively (Preiss, 1991). Glucose 1-phosphate (G1P) as the substrate of ADP–glucose (ADPG) was also increased, catalyzed by fructose-bisphosphate aldolase (EC: 4.2.1.13), fructose-1,6-bisphosphatase I (EC: 3.1.3.11), glucose-6-phosphate isomerase (EC: 5.3.1.9), and phosphoglucomutase (EC: 5.4.2.2). The expression of genes of these enzymes were upregulated by 7–9.2, 2.6–3.5, 2.8, and 3.5 folds, respectively. The starch catabolism genes, alpha-amylase (EC: 3.2.1.1) and glycogen phosphorylase (EC: 2.4.1.1), were downregulated 3.2–4.6, 2.1–36.8 folds, implying that the starch content might increase, and the enzymes related to starch synthesis also could be used to synthesize sucrose (Figure 5B). Starch, as the major carbohydrate, could provide power for spore formation (Wang et al., 2016). Notedly, the chloroplast disappeared in state III, but the spore would produce chromatophore at the anterior end (Chen et al., 2011). Chromatophore was the product of primary endosymbiotic integration of a cyanobacterium into a eukaryotic (Reyes-Prieto et al., 2007). Therefore, we suggested that chromatophores have the photosynthesis power as chloroplast and that polar growth occurred in state IV, which may be the reason why the gene expression of starch synthase was downregulated in state II to III and III to IV, but APG was upregulated in the state III to IV, which might prepare the precursor for starch synthesis for the growth of later seedlings.
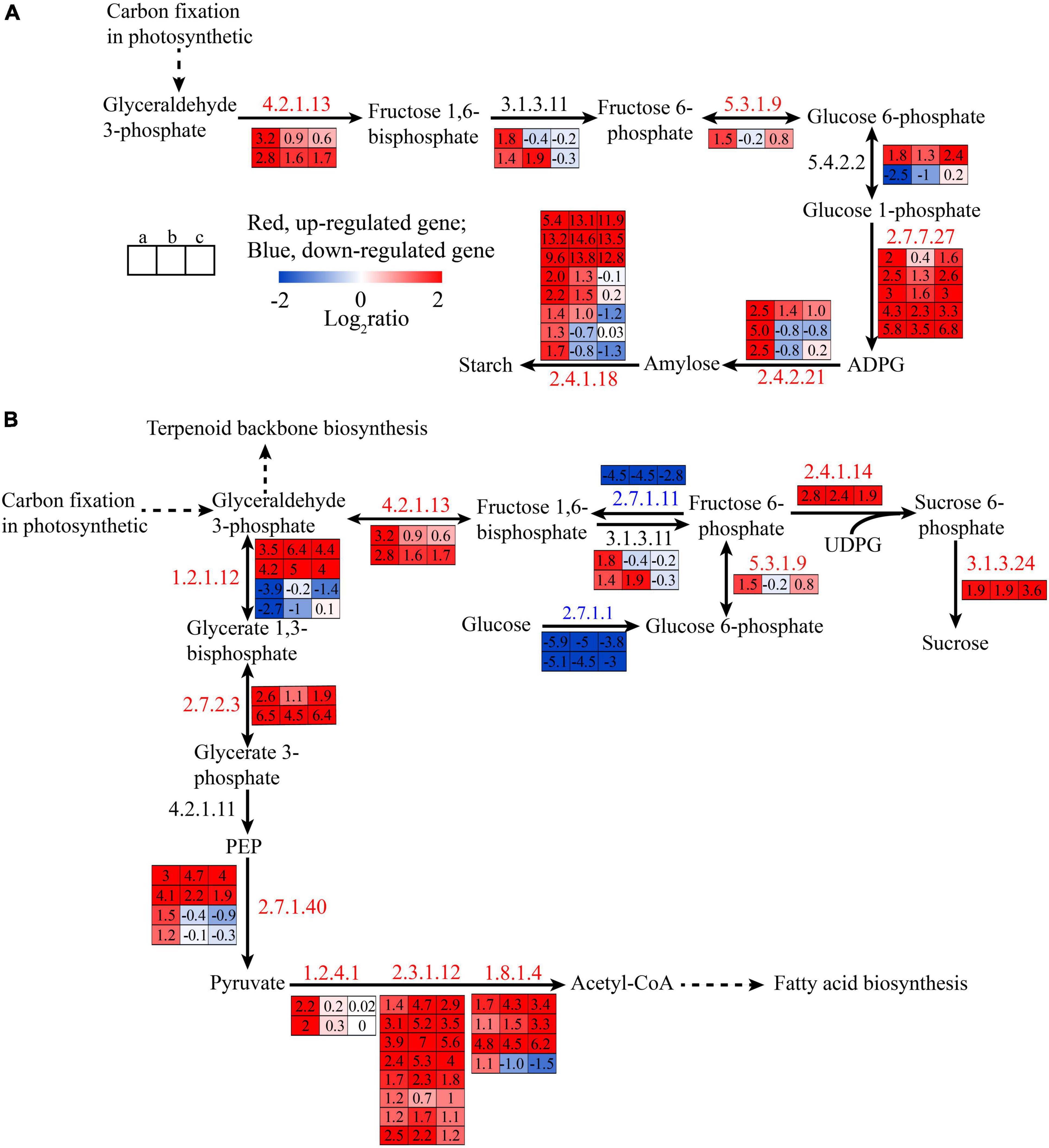
Figure 5. The transcriptional changes of genes associated with starch synthetic (A) and glycolysis pathway (B) in the spore formation of U. prolifera between different states.
Glycolysis
Glycolysis is a fundamental metabolic pathway that breaks down glucose into two three-carbon metabolites and generates energy in the cytoplasm. The DEGs in glycolysis are shown in Figure 5B. Compared with state I, the genes encoding for hexokinase (EC: 2.7.1.1) and 6-phosphofructokinase 1 (EC: 2.7.1.11), which are the rate-limiting enzymes in glycolysis pathway (El-Gharbawy and Koeberl, 2014), were downregulated in state II while the expressions of genes encoding fructose-bisphosphate aldolase, glyceraldehyde 3-phosphate dehydrogenase (EC: 1.2.1.13), and phosphoglycerate kinase (EC: 2.7.2.3) were upregulated. So, we speculated that the 3-PGA increased in the glycolysis pathway because the algae can transfer 3-PGA from carbon fixation pathway to glycolysis via triose phosphate translocator. The increase of 3-PGA could replace glucose as the raw material downstream for glycolysis to produce pyruvate and produce a negative effect for upstream, causing downregulation of the gene expression of hexokinase and 6-phosphofructokinase and upregulation of pyruvate kinase (EC: 2.7.1.40), and all this is shown in the Figure 5B. These results also showed that fructose-6 -phosphate would increase, which was the substrate of sucrose, and the gene expression of sucrose-phosphate synthase (SPS; EC: 2.4.1.14) and sucrose-6-phosphatase (SPP; EC: 3.1.3.24) was upregulated in state II, indicating the glycolysis pathway was downregulated to prepare energy for spore formation in states III and IV, which also was observed in previous study (Wang et al., 2016).
Pyruvate and 3-PGAld from glycolysis and carbon fixation pathway also could be used as the substrate of the terpenoid backbone synthesis (Ganjewala et al., 2009), which also was observed in Figure 6A, and provide precursor for the carotenoids. Pyruvate also could be catalyzed by pyruvate dehydrogenase complex, including three main enzymes: pyruvate dehydrogenase (E1; EC: 1.2.4.1), dihydrolipoamide S-acetyltransferase (E2; EC: 2.3.1.12), and dihydrolipoamide dehydrogenase (E3; EC: 1.8.1.4) (Figure 5B). The gene expression of the three enzymes were upregulated 4–4.6, 2.3–14.9, and 2.1–27.9 folds from state I to II, respectively, and they generated acetyl-CoA, which was the substrate of tricarboxylic acid (TCA) cycle and fatty acid biosynthesis. However, the expressions of genes encoding for TCA cycle were downregulated, including isocitrate dehydrogenase (EC: 1.1.1.42), 2-oxoglutarate dehydrogenase (E1; EC: 1.2.4.2), and dihydrolipoamide succinyl-transferase (E2; EC: 2.3.1.61), indicating that the biosynthesis of fatty acid might increase.
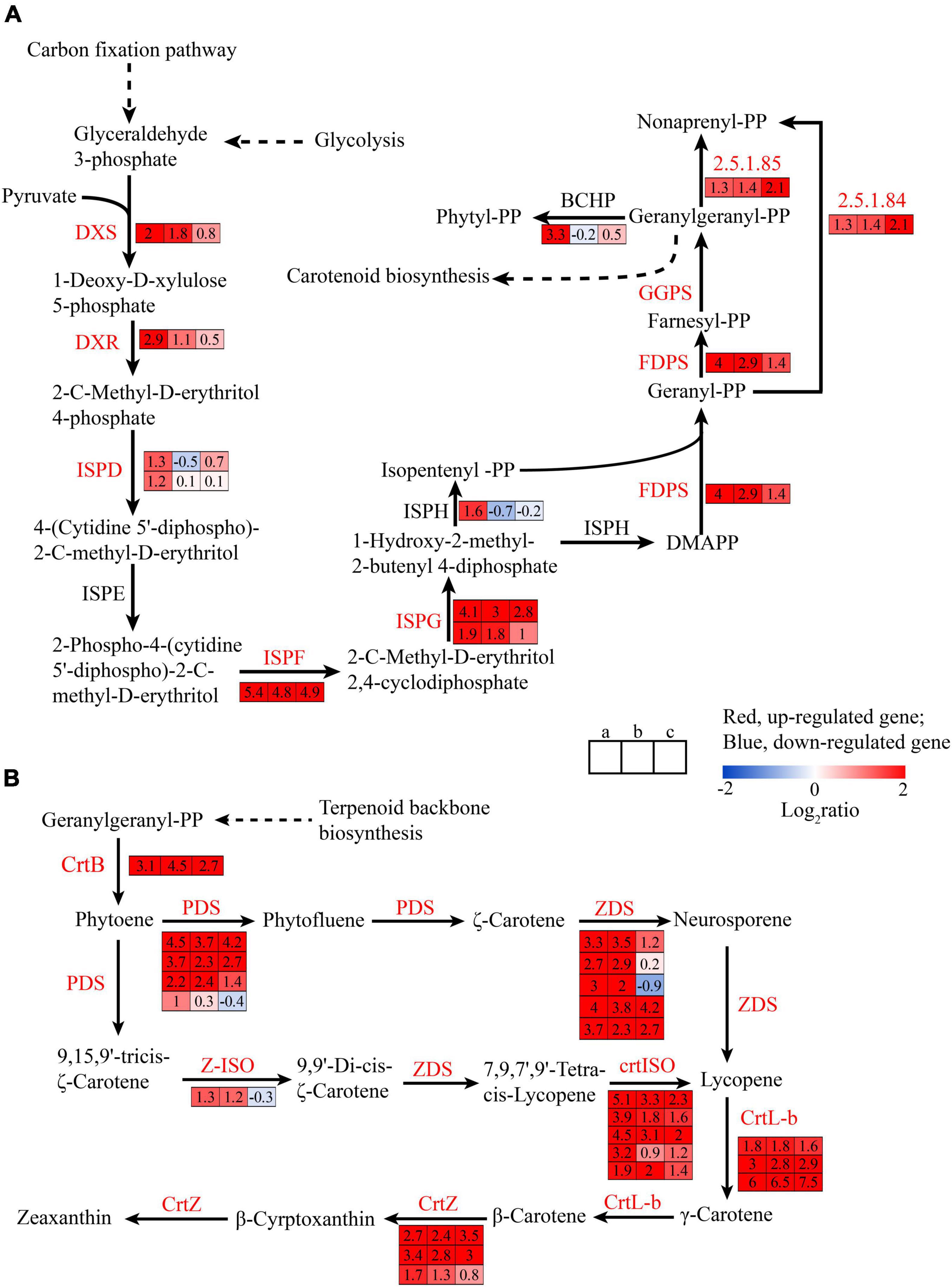
Figure 6. The transcriptional changes of genes associated with terpenoid backbone biosynthesis (A) and carotenoid biosynthesis pathway (B) in the spore formation of U. prolifera between different states.
Terpenoid Backbone Biosynthesis Pathway
Terpenoid backbone biosynthesis pathway was involved in 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in plastids and mevalonate (MVA) pathway in the cytosol (Eismann et al., 2020). In green algae, the MVA pathway is lost and the MEP pathway is the sole pathway (Grauvogel and Petersen, 2007). The DEGs in terpenoid backbone biosynthesis pathway are shown in Figure 6A. The expression of genes encoding for 1-deoxy-D-xylulose 5-phosphate synthase (DXS) and plastid 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) were upregulated to 4 and 7.5 folds, respectively. DXS and DXR catabolizing the first steps in terpenoid backbone biosynthesis pathway are two of the rate-limiting enzymes in terpenoid backbone biosynthesis pathway (Lichtenthaler et al., 1997), which could catalyze 3-PGAld to generate 1-deoxy-D-xylulose 5-phosphate (DOXP) and 2-C-methyl-D-erythritol 4-phosphate (MEP), indicating the synthesis of terpenoid backbone increased. The expression of genes encoding 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (ISPD) and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (ISPH) was upregulated 2.3- to 3-folds and those of genes encoding 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (ISPF), (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ISPG), and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (ISPH) were upregulated 42.2, 3.7–17.1, and 3-folds, respectively, indicating the synthesis of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) was increase which was the precursors for terpenoid synthesis (He et al., 2018). The gene expression of farnesyl diphosphate synthase (FDPS) and geranylgeranyl diphosphate synthase, type II (GGPS), was upregulated 3.7–17.1 folds in state II, indicating that the conversion of 1-hydroxy-2-methyl-2-butenyl 4-diphosphate (HMB-PP) to geranylgeranyl diphosphate (GGPP) was enhanced. GGPP was the precursor for carotenoid biosynthesis and other important terpenoids, such as chlorophylls, tocopherols, phylloquinone, plastoquinone, and gibberellins (Lohr et al., 2012; Egeland, 2016).
Carotenoid Biosynthesis Pathway
Moreover, the expressions of genes encoding for Car biosynthesis pathway were significantly upregulated in three states (II, III, and IV) (Figure 6B), as compared with those in state I, such as phytoene synthase (CrtB), phytoene desaturase (PDS), zeta-carotene desaturase (ZDS), zeta-carotene isomerase (Z-ISO), prolycopene isomerase (crtISO), lycopene beta-cyclase (CrtL-b), and beta-carotene 3-hydroxylase (CrtZ) were upregulated 8.6, 2–22.6, 6.5–16, 2.5, 3.7–34.2, 3.5–64, and 3.2–10.6 folds, respectively (Figure 6B). CrtB is the limiting enzyme for carotenoid biosynthesis which could catalyze two GGPPs to form phytoene (Eismann et al., 2020), and compared with state I, the gene expression was upregulated 8.6, 22.6, 6.5 folds in states II, III, and IV, respectively. These results verified the changes in Car content, which was observed in Figure 3, and Car, in addition to photosynthesis, protects membranes from excess of photon energy by producing NPQ by xanthophyll cycle (Gao et al., 2020) and affects spore formation (Park, 2020). Moreover, some studies showed that carotenoid biosynthesis is related to the accumulation of astaxanthin in Haematococcus pluvialis and Chlorella zofingiensis (Zhang et al., 2016; Du et al., 2021), whereas the expression of CrtZ related to the astaxanthin synthesis pathway was upregulated during the sporulation process (Figure 6B). However, it is still uncertain whether astaxanthin exists in U. prolifera, and the further research is needed.
Transcriptional Factors
Transcription factors are the main regulators that control the expression of genes involved in plant growth, development, and responses to external stimuli (Ahammed et al., 2020). They activate or inhibit genes by binding to promoter functional elements or interacting with other proteins (Ng et al., 2018). In our study, a total of 219 of unigenes involved in 29 TF families were detected during the process of sporulation. Among them, many genes were involved in the AP2-EREBP family, the MYB family, the G2-like family, and the WRKY family whereas the ARF family, Alfin-like family, bHLH family, and bZIP family have only one gene (Figure 7A). The identified 42 of the DEGs, covering 19 families, were constantly significantly upregulated during the process of the spore formation (Figure 7B, label 1), including four G2-like genes, two C2C2-CO-like genes, two PLATZ genes, one bHLH gene, six WRKY genes, seven AP2-EREBP genes, etc. (Supplementary Table 6), whereas 23 of the DEGs were significantly downregulated (Figure 7B, label 2). Additionally, most of the orthologs for the 29 URGs in other species were hypothetical or predicted proteins. Five TFs unigenes which involved four families seemed to be unique in U. prolifera because no orthologs could be found in other species (Supplementary Table 7), and the expressions of CL2970.Contig1_mix12 and Unigene3886_mix12 were significantly downregulated in the process of spore formation. The expressions of Unigene4015_mix12 and Unigene17383_mix12 were significantly upregulated in states III and IV, respectively, as compared with A (Supplementary Table 7). Notedly, states III and IV were the crucial periods of spore formation according to Figure 1. Therefore, we speculated that those two genes would affect spore formation at different times, but need to be further studied.
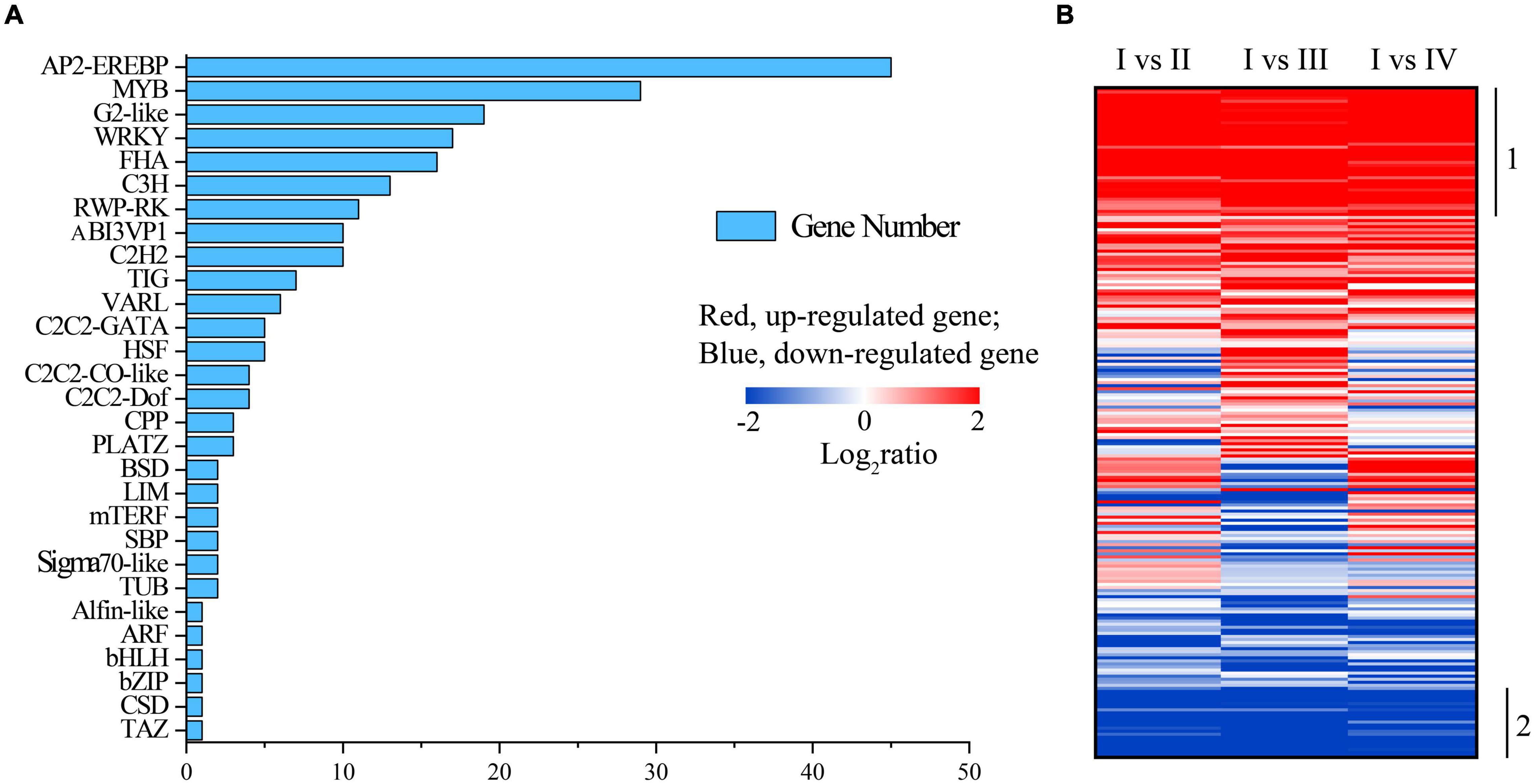
Figure 7. The quantity of transcriptional factors (TFs) (A) and the heatmap of transcriptional changes of genes associated with TF (B) in spore formation of U. prolifera.
The previous study showed that the bZIP TF is involved in regulating adventitious shoot development in Paulownia kawakamii (Low et al., 2001). In our study, the expression of Unigene14540_mix12, which belonged to bZIP family, was highly upregulated in the state IV as compared with other states (including state I, II, and III), indicating that polar growth may have occurred in state IV, and this also was consistent with the results of the light saturation parameter (Ek), which increased in state IV (Figure 2C). The TFs of RWP-RK family have profound and broad influence on the early transcriptome response to nitrate signaling and assimilation (Marchive et al., 2013). Moreover, some TFs of RWP-RK family regulate the storage of starch by controlling carbon partition in Chlamydomonas (Chardin et al., 2014). MYB and WRKY TFs also played important roles in regulatory signal transduction and responses to biotic and abiotic stresses (Smita et al., 2015; Wu et al., 2018b).
The functions of the remaining TFs can be divided into three primary GO categories according to GO annotation, including biological process, cellular component, and molecular function (Supplementary Figure 5). Notedly, the functions of CL1966.Contig2_mix12 of C3H family in the biological process was reproduction and reproduction process, in which the gene expression was upregulated in states II and III whereas it was downregulated in stage IV (Supplementary Figure 6). Moreover, the expression of this gene was the highest in stage III, which is also the main stage of spore formation. All of these results indicated that the main process of reproduction occurred in state III and ended in state IV, which also verified the result of Figure 1 and our speculation that the polar growth might occur in state IV. These results implied that the CL1966.Contig2_mix12 may be one of the key genes in the spore formation. However, the involvement of specific functions of these TFs in the spore formation of U. prolifera merits further in-depth investigations. Additionally, Unigene5846_mix12 has an identity above 40% with diguanylate cyclase, and its overexpression could decrease the sporulation frequency and early sporulation gene transcription of Clostridioides difficile (Edwards et al., 2021), but it did not significantly change in this study, indicating that the spore formation was not inhibited.
Flagella Biosynthesis
The spore of U. prolifera has four flagella (Zhang et al., 2013), and flagella are one of the most ancient cellular organelles, constituted by skeleton protein, providing motility for living in water which is vital for algal cells to survive and proliferate under different environmental conditions (Silflow and Lefebvre, 2001). In this study, 574 of the DEGs associated with flagella biosynthesis in U. prolifera were annotated according to GO annotation (Supplementary Table 8). To understand the expression of genes associated with flagella synthesis in different states for better, we arranged and divided these genes into different groups, noting labels as shown in Figure 8A. Among these genes, 37 of the DEGs were constantly upregulated in the process of spore formation (Figure 8A, label 1), but 12 of the DEGs were constantly downregulated (Figure 8A, label 6); 20 of the DEGs were upregulated in II state but downregulated in states III and IV (Figure 8A, label 3). Although the flagella did not synthesize in state II (Figure 1), the ribosome pathway was significantly upregulated in state II (Supplementary Figure 4A). Therefore, these genes may prepare material for flagella synthesis in the later period, such as protein and cell skeleton, and assemble only the 146 DEGs that were upregulated in state III to produce flagella (Figure 8A, label 4) because the spore was observed in state III according to morphology (Figure 1). Only 44 of the total DEGs that were upregulated in state IV may be associated with spore movement (Figure 8A, label 5), the previous study showed that spore could keep rotating depending on the flagella with much higher speed and collided with the cytocyst wall, and a hole was formed at the top of the cytocyst where germ cells were released one by one in a kind of amoeboid movement (Zhang et al., 2013). Moreover, 243 of the DEGs were upregulated in states II and III state while they were downregulated in IV may have the preparation and assembly effects to flagella synthesis (Figure 8A, label 2). We also found that including 20 of the DEGs encoding intraflagellar transport proteins (IFTs) has the highest expression level in state III, but decreased in IV (Figure 8B). The assembly and maintenance of flagella require the involvement of IFTs, which are also closely related to flagella movement, signal reception, and signaling (Croft et al., 2018; Wang et al., 2018b). However, those genes were annotated in Chlamydomonas reinhardtii and were not reported in previous transcriptome analysis of U. prolifera (He et al., 2019a; Wang et al., 2019; Table 1). Combining the phenomenon of state III, we suggest that state III is a critical period for sporulation and provides an observable expression gene for subsequent exploration of how environmental factors affect sporulation.
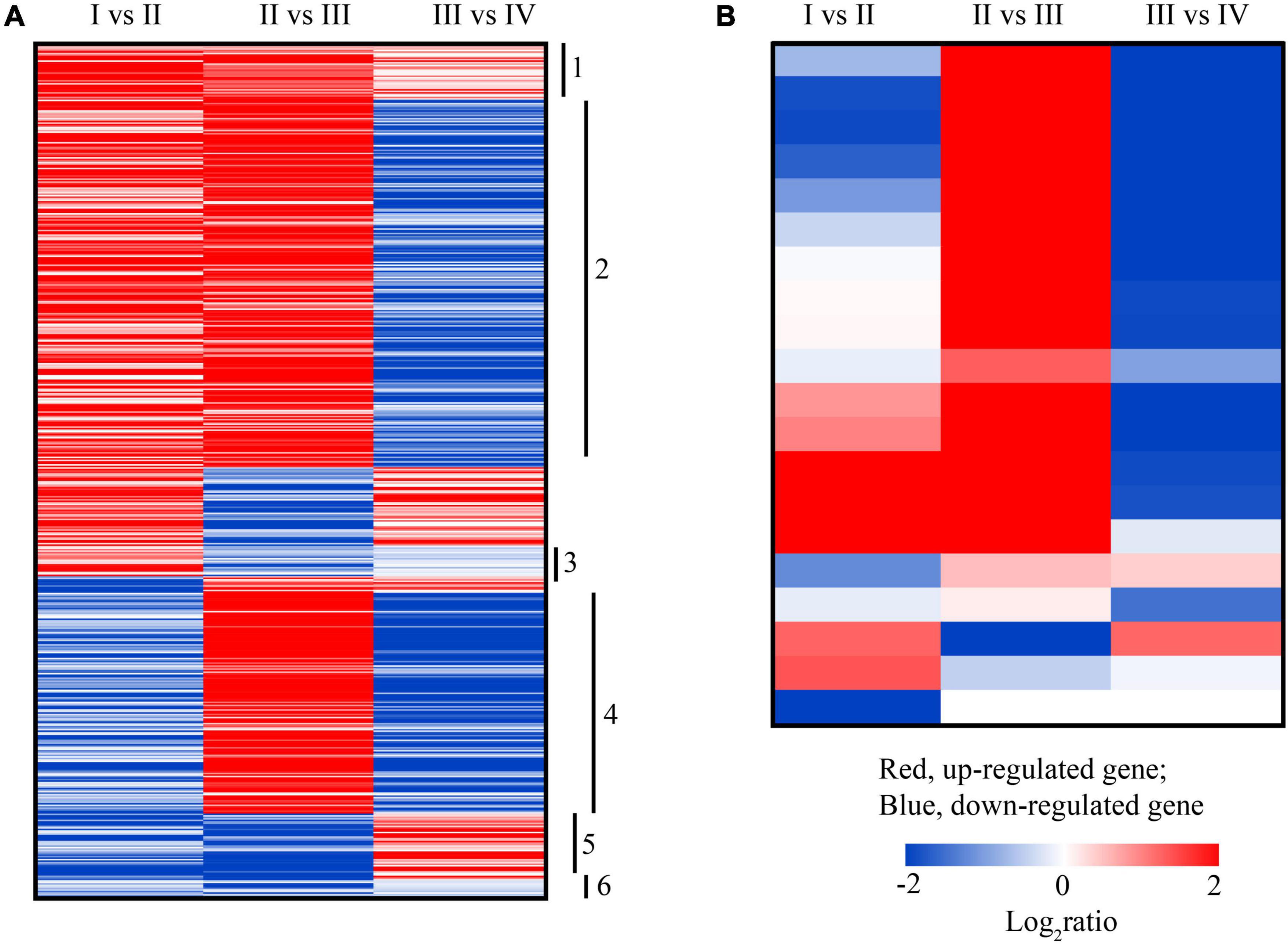
Figure 8. The heatmap of transcriptional changes of genes associated with flagella biosynthesis (A) and intraflagellar transport proteins (IFTs) (B) in the spore formation of U. prolifera between different states.

Table 1. The 20 genes associated with intraflagellar transport proteins (IFTs) expression in spore formation of U. prolifera.
Conclusion
The spore formation of U. prolifera was divided into 4 states, cells in the state II accumulated energy by enhancing photosynthesis and starch synthesis and inhibiting catabolism, and provide power for the spore production in the state III and IV. The synthesis of carotenoid had a high degree correlation with spore formation. According to the expression of gene associated with flagella and intraflagellar transport proteins biosynthesis in U. prolifera, state III is a critical period for sporulation. Moreover, some genes of TF families, such as C3H family, may be one of the key regulating genes in spore formation. These results provide a new perspective of spore formation at the gene transcriptional level although the detailed transcription regulatory spore formation network remains to be unveiled.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA794305.
Author Contributions
YL and NX: conceptualization, funding acquisition, supervision, and writing-review and editing. JJ: data curation and writing-original draft. JJ, YL, CH, WZ, and NX: formal analysis. JJ and YL: visualization. All authors: contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (41876181), the Natural Science Foundation of Zhejiang Province (LY19D060002), General Scientific Research Project of Zhejiang Provincial Department of Education (Y202045555), Graduate Research and Innovation Fund Project (IF2020148), and was also sponsored by K. C. Wong Magna Fund in Ningbo University.
Conflict of Interest
WZ was employed by the company Xiangshan Xuwen Seaweed Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.802006/full#supplementary-material
References
Ahammed, G. J., Li, X., Mao, Q., Wan, H. J., Zhou, G. Z., and Cheng, Y. (2020). The SlWRKY81 transcription factor inhibits stomatal closure by attenuating nitric oxide accumulation in the guard cells of tomato under drought. Physiol. Plantarum. 172, 885–895. doi: 10.1111/ppl.13243
Carl, C., de Nys, R., Lawton, R. J., and Paul, N. A. (2014). Methods for the induction of reproduction in a tropical species of filamentous Ulva. PLoS One 9:e97396. doi: 10.1371/journal.pone.0097396
Chardin, C., Girin, T., Roudier, F., Meyer, C., and Krapp, A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587. doi: 10.1093/jxb/eru261
Chavez-Sanchez, T., Pinon-Gimate, A., Serviere-Zaragoza, E., Lopez-Bautista, J. M., and Casas-Valdez, M. (2018). Ulva blooms in the southwestern Gulf of California: reproduction and biomass. Estuar. Coast. Shelf S. 200, 202–211. doi: 10.1016/j.ecss.2017.11.007
Chen, Q. F., He, P. M., Feng, Z. H., Tang, W. Z., Li, X. Y., Zhang, T., et al. (2011). Reproduction of spores/gametes of floating green tide algae Ulva prolifera. J. Fishery Sci. China 18, 1069–1076. doi: 10.3724/SP.J.1011.2011.00187
Chen, Z., Chen, J., Liu, J. H., Li, L. M., Qin, S., and Huang, Q. (2020). Transcriptomic and metabolic analysis of an astaxanthin-hyperproducing Haematococcus pluvialis mutant obtained by low-temperature plasma (LTP) mutagenesis under high light irradiation. Algae Res. 45:101746. doi: 10.1016/j.algal.2019.101746
Croft, J. T., Zabeo, D., Subramanian, R., and Höög, J. L. (2018). Composition, structure and function of the eukaryotic flagellum distal tip. Essays Biochem. 62, 815–828. doi: 10.1042/EBC20180032
Cui, J. J., Shi, J. T., Zhang, J. H., Wang, L. T., Fan, S. Y., Xu, Z. Y., et al. (2018). Rapid expansion of Ulva blooms in the Yellow Sea, China through sexual reproduction and vegetative growth. Mar. Pollut. Bull. 130, 223–228. doi: 10.1016/j.marpolbul.2018.03.036
Cui, J. J., Zhang, J. H., Huo, Y. Z., Zhu, L. J., Wu, Q., Chen, L. P., et al. (2015). Adaptability of free-floating green tide algae in the Yellow Sea to variable temperature and light intensity. Mar. Pollut. Bull. 101, 660—666. doi: 10.1016/j.marpolbul.2015.10.033
Du, F. C., Hu, C. Y., Sun, X., and Xu, N. J. (2021). Transcriptome analysis reveals pathways responsible for the promoting effect of sucrose on astaxanthin accumulation in Haematococcus pluvialis under high light condition. Aquaculture 530:735757. doi: 10.1016/j.aquaculture.2020.735757
Edwards, A. N., Willams, C. L., Pareek, P., McBride, S. M., and Tamayo, R. (2021). c-di-GMP inhibits early sporulation in Clostridioides difficile. mSphere 6:e00919-21. doi: 10.1128/msphere.00919-21
Egeland, E. S. (2016). “Carotenoids,” in The Physiology of Microalgae. Developments in Applied Phycology, eds M. Borowitzka, J. Beardall, and J. Raven (Cham: Springer).
Eilers, P. H. C., and Peeters, J. C. H. (1988). A model for the relationship between light intensity and the rate of photosynthesis in phytoplankton. Ecol. Model 42, 199–215. doi: 10.1016/0304-3800(88)90057-9
Eismann, A. I., Reis, R. P., da Silva, A. F., and Cavalcanti, D. N. (2020). Ulva spp. carotenoids: responses to environmental conditions. Algal Res. 48:101916. doi: 10.1016/j.algal.2020.101916
El-Gharbawy, A., and Koeberl, D. (2014). Basic biochemical pathway, clinical presentation, diagnosis, and treatment. Inborn. Errors. Metabol. 64, 119–133.
Ganjewala, D., Kumar, S., and Luthra, R. (2009). An account of cloned genes of methylerythritol-4-phosphate pathway of isoprenoid biosynthesis in plants. Curr. Issues Mol. Biol. 11, i35–i45. doi: 10.21775/cimb.011.i35
Gao, G., Clare, A. S., Rose, C., and Caldwell, G. S. (2017). Intrinsic and extrinsic control of reproduction in the green tide-forming alga, Ulva rigida. Environ. Exp. Bot. 139, 14–22. doi: 10.1016/j.envexpbot.2017.03.016
Gao, S., Chen, X. Y., Yi, Q. Q., Wang, G. C., Pan, G. H., Lin, A. P., et al. (2010). A strategy for the proliferation of Ulva prolifera, main causative species of green tides, with formation of sporangia by fragmentation. PLoS One 5:e8571. doi: 10.1371/journal.pone.0008571
Gao, S., Zheng, Z. B., Wang, J., and Wang, G. C. (2020). Slow zeaxanthin accumulation and the enhancement of CP26 collectively contribute to an atypical non-photochemical quenching in macroalga Ulva prolifera under high light. J. Phycol. 56, 393–403. doi: 10.1111/jpy.12958
Grauvogel, C., and Petersen, J. (2007). Isoprenoid biosynthesis authenticates the classification of the green alga Mesostigma viride as an ancient streptophyte. Gene 396, 125–133. doi: 10.1016/j.gene.2007.02.020
Guillard, R. R., and Ryther, J. H. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Han, T., Han, Y. S., Park, C. Y., Jun, Y. S., Kwon, M. J., Kang, S. H., et al. (2008). Spore release by the green alga Ulva: a quantitative assay to evaluate aquatic toxicants. Environ. Pollut. 153, 699–705. doi: 10.1016/j.envpol.2007.09.001
Hatch, M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta Rev. Bioenerg. 895, 81–106. doi: 10.1016/S0304-4173(87)80009-5
He, Y., Ao, Y., Yin, Y., Yuan, A. X., Che, T. J., Li, L., et al. (2019a). Comparative transcriptome analysis between floating and attached Ulva prolifera in studying green tides in the Yellow Sea. Algal Res. 44:101712. doi: 10.1016/j.algal.2019.101712
He, Y. L., Wang, Y. H., and Hu, C. Y. (2019b). Dynamic metabolic profiles of the marine macroalga Ulva prolifera during fragmentation-induced proliferation. PLoS One 14:e0214491. doi: 10.1371/journal.pone.0214491
He, Y., Ma, Y. F., Du, Y., and Shen, S. D. (2018). Differential gene expression for carotenoid biosynthesis in a green alga Ulva prolifera based on transcriptome analysis. BMC Genomics 19:916. doi: 10.1186/s12864-018-5337-y
He, Y., Shen, S. D., Yu, D. C., Wang, Y. H., Yin, J., Wang, Z. L., et al. (2021). The Ulva prolifera genome reveals the mechanism of green tides. J. Oceanol. Limnol. 39, 1458–1470. doi: 10.1007/s00343-020-0212-5
Hu, C. Y., Cui, D. D., Sun, X. J., Shi, J., Song, L. L., Li, Y. H., et al. (2019). Transcriptomic analysis unveils survival strategies of autotrophic Haematococcus pluvialis against high light stress. Aquaculture 513:734430. doi: 10.1016/j.aquaculture.2019.734430
Kumar, M., Kuzhiumparambil, U., Pernice, M., Jiang, Z., and Ralph, P. J. (2016). Metabolomics: an emerging frontier of systems biology in marine macrophytes. Algal Res. 16, 76–92. doi: 10.1016/j.algal.2016.02.033
Li, W. Q., Yamaguchi, S., Khan, M. A., An, P., Liu, X. J., and Tran, L. S. P. (2016). Roles of gibberellins and abscisic acid in regulating germination of Suaeda salsa dimorphic seeds under salt stress. Front. Plant Sci. 6:1235. doi: 10.3389/fpls.2015.01235
Lichtenthaler, H. M., Rohmer, M., and Schwender, J. (1997). Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol. Plantarum 101, 643–652. doi: 10.1034/j.1399-3054.1997.1010327.x
Lin, A. P., Shen, S. D., Wang, J. W., and Yan, B. L. (2008). Reproduction diversity of Enteromorpha prolifera. J. Integr. Plant Biol. 50, 622–629. doi: 10.1111/j.1744-7909.2008.00647.x
Liu, D. Y., Ma, Q., Valiela, I., Anderson, M. D., Kessing, J. K., Gao, K. S., et al. (2020). Role of C4 carbon fixation in Ulva prolifera, the macroalga responsible for the world’s largest green tides. Commun. Biol. 3:494. doi: 10.1038/s42003-020-01225-4
Liu, L. N. (2016). Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. BBA Bioenergetics 1857, 256–265. doi: 10.1016/j.bbabio.2015.11.010
Lohr, M., Schwender, J., and Polle, J. E. (2012). Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae. Plant Sci. 185, 9–22. doi: 10.1016/j.plantsci.2011.07.018
Low, R. K., Prakash, A. P., Swarup, S., Goh, C.-J., and Kumar, P. (2001). A differentially expressed bZIP gene is associated with adventitious shoot regeneration in leaf cultures of Paulownia kawakamii. Plant Cell Rep. 20, 696–700. doi: 10.1007/s00299-001-0401-4
Mao, X. Z., Tao, C., Olyarchuk, J. G., and Wei, L. P. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.2307/1592215
Marchive, C., Roudier, F., Castaings, L., Brehaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. doi: 10.1038/ncomms2650
Nakashima, K., and Yamaguchishinozaki, K. (2013). ABA signaling in stress response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nawrocki, W. J., Tourasse, N. J., Taly, A., Rappaport, F., and Wollman, F. A. (2015). The plastid terminal oxidase: its elusive function points to multiple contributions to plastid physiology. Annu. Rev. Plant Biol. 66, 49–74. doi: 10.1146/annurev-arplant-043014-114744
Ng, D. W., Abeysinghe, J. K., and Kamali, M. (2018). Regulating the regulators: the control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19:3737. doi: 10.3390/ijms19123737
Park, J. (2020). Photosynthetic and biochemical traits change in the green-tide-forming macroalga Ulva pertusa during sporulation. J. Phycol. 56, 549–557. doi: 10.1111/jpy.12969
Pettett, P. (2009). Preliminary Investigation Into the Induction of Reproduction in Ulva spp. in Southeast Queensland for Mass Cultivation Purposes. Master thesis, Downs QLD: University of the Sunshine Coast.
Preiss, J. (1991). Oxford Survey of Plant Molecular and Cellular Biology, Vol. 7. Oxford: Oxford University Press, 59–114.
Qian, H. F., Li, J. J., Pan, X. J., Jiang, H. Y., Sun, L. W., and Fu, Z. W. (2010). Photoperiod and temperature influence cadmium’s effects on photosynthesis-related gene transcription in Chlorella vulgaris. Ecotox. Environ. Safe 73, 1202–1206. doi: 10.1016/j.ecoenv.2010.07.006
Raffaelli, D. G., Raven, J. A., and Poole, L. J. (1998). Ecological impact of green macroalgal blooms. Oceanogr. Mar. Biol. 36, 97–125.
Reyes-Prieto, A., Weber, A. P., and Bhattacharya, D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168. doi: 10.1146/annurev.genet.41.110306.130134
Serodio, J., Vieira, S., Cruz, S., and Coelho, H. (2006). Rapid light-response curves of chlorophyll fluorescence in microalgae: relationship to steady-state light curves and non-photochemical quenching in benthic diatom-dominated assemblages. Photosynth. Res. 2006, 29–43. doi: 10.1007/s11120-006-9105-5
Siefermann-Harms, D. (1987). The light-harvesting and protective function of carotenoids in photosynthetic membranes. Physiol. Plantarum 69, 561–568. doi: 10.1111/j.1399-3054.1987.tb09240.x
Silflow, C. D., and Lefebvre, P. A. (2001). Assembly and motility of eukaryotic cilia and flagella. lessons from Chlamydomonas reinhardtii. Plant Physiol. 127, 1500–1507. doi: 10.1104/pp.127.4.1500
Smita, S., Katiyar, A., Chinnusamy, V., Pandey, D. M., and Bansal, K. C. (2015). Transcriptional regulatory network analysis of MYB transcription factor family genes in Rice. Front. Plant Sci. 6:1157. doi: 10.3389/fpls.2015.01157
Smith, G. M. (1947). On the reproduction of some Pacific coast species of Ulva. Am. J. Bot. 34, 80–87. doi: 10.1002/j.1537-2197.1947.tb12961.x
Stanley, M. S., Perry, R. M., and Callow, J. A. (2005). Analysis of expressed sequence tags from the green alga Ulva linza (Chlorophyta). J. Phycol. 41, 1219–1226. doi: 10.1111/j.1529-8817.2005.00138.x
Stark, D. M., Timmerman, K. P., Barry, G. F., Preiss, J., and Kishore, G. M. (1992). Regulation of the amount of starch in plant tissues by adp glucose pyrophosphorylase. Science 258, 287–292. doi: 10.1126/science.258.5080.287
Sui, N., Yang, Z., Liu, M. L., and Wang, B. S. (2015). Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16:534. doi: 10.1186/s12864-015-1760-5
Wang, C. Y., Jiao, X. M., Zhang, Y., Zhang, L., and Xu, H. (2020a). A light-limited growth model considering the nutrient effect for improved understanding and prevention of macroalgae bloom. Environ. Sci. Pollut. Res. 27, 12405–12413. doi: 10.1007/s11356-020-07822-4
Wang, G. C., Wang, H., Gao, S., Huan, L., Wang, X. L., Gu, W. H., et al. (2020b). Study on the biological mechanism of green tide. Oceanol. Limnol. Sinica 51, 789–808. doi: 10.11693/hyhz20200300078
Wang, H., Lin, A. P., Gu, W. H., Huan, L., Gao, S., and Wang, G. C. (2016). The sporulation of the green alga Ulva prolifera is controlled by changes in photosynthetic electron transport chain. Sci. Rep. 6:24923. doi: 10.1038/srep24923
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., and Zhang, X. G. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, S. Y., Huo, Y. Z., Zhang, J. H., Cui, J. J., Wang, Y., Yang, L. L., et al. (2018a). Variations of dominant free-floating Ulva species in the source area for the world’s largest macroalgal blooms, China: differences of ecological tolerance. Harmful Algae 74, 58–66. doi: 10.1016/j.hal.2018.03.007
Wang, Q. M., Taschner, M., Ganzinger, K. A., Kelley, C., Villasenor, A., Heymann, M., et al. (2018b). Membrane association and remodeling by intraflagellar transport protein IFT172. Nat. Commun. 9:4684. doi: 10.1038/s41467-018-07037-9
Wang, Y., Liu, F., Liu, X. F., Shi, S. T., Bi, Y. P., and Moejes, F. W. (2019). Comparative transcriptome analysis of four cooccurring Ulva species for understanding the dominance of Ulva prolifera in the Yellow Sea green tides. J. Appl. Phycol. 31, 3303–3316. doi: 10.1007/s10811-019-01810-z
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wellburn, A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Wichard, T., and Oertel, W. (2010). Gametogenesis and gamete release of Ulva mutabilis and Ulva lactuca (Chlorophyta): regulatory effects and chemical characterization of the Bswarming inhibitor. J. Phycol. 46, 248–259. doi: 10.1111/j.1529-8817.2010.00816.x
Woo, N. S., Gordon, M. J., Graham, S. R., Rossel, J. B., Badger, M. R., and Pogson, B. J. (2011). A mutation in the purine biosynthetic enzyme ATASE2 impacts high light signalling and acclimation responses in green and chlorotic sectors of Arabidopsis leaves. Funct. Plant Biol. 38, 401–419. doi: 10.1071/FP10218
Wu, H. L., Gao, G., Zhong, Z. H., Li, X. S., and Xu, J. T. (2018a). Physiological acclimation of the green tidal alga Ulva prolifera to a fast-changing environment. Mar. Environ. Res. 137, 1–7. doi: 10.1016/j.marenvres.2018.02.018
Wu, B., Li, M. Y., Xu, Z. S., Wang, F., and Xiong, A. S. (2018b). Genome-wide analysis of WRKY transcription factors and their response to abiotic stress in celery (Apium graveolens L). Biotechnol. Biotec. Eq. 32, 293–302. doi: 10.1080/13102818.2017.1413954
Xu, J. F., Fan, X., Zhang, X. W., Xu, D., Mou, S. L., Cao, S. N., et al. (2012). Evidence of coexistence of C3 and C4 photosynthetic pathways in a green-tide-forming alga, Ulva prolifera. PLoS One 7:e37438. doi: 10.1371/journal.pone.0037438
Xu, J. F., Zhang, X. W., Ye, N. H., Zheng, Z. B., Mou, S. L., Dong, M. T., et al. (2013). Activities of principal photosynthetic enzymes in green macroalga Ulva linza: functional implication of C4 pathway in CO2 assimilation. Sci. China Life Sci. 56, 571–580. doi: 10.1007/s11427-013-4489-x
Yang, Z., Li, J. L., Liu, L. N., Xie, Q., and Sui, N. (2020). Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet sorghum. Front. Plant Sci. 10:1722. doi: 10.3389/fpls.2019.01722
Zhang, J. H., Huo, Y. Z., Yu, K. F., Chen, Q. F., He, Q., Han, W., et al. (2013). Growth characteristics and reproductive capability of green tide algae in Rudong coast, China. J. Appl. Phycol. 25, 795–803. doi: 10.1007/s10811-012-9972-4
Zhang, X. W., Mou, S. L., Cao, S. N., Fan, X., Xu, D., and Ye, N. H. (2015). Roles of the transthylakoid proton gradient and xanthophyll cycle in the non-photochemical quenching of the green alga Ulva linza. Estuar. Coast. Shelf S. 163, 69–74. doi: 10.1016/j.ecss.2014.09.006
Zhang, X. W., Ye, N. H., Liang, C. W., Mou, S. L., Fan, X., Xu, J. F., et al. (2012). De novo sequencing and analysis of the Ulva linza transcriptome to discover putative mechanisms associated with its successful colonization of coastal ecosystems. BMC Genomics 13:565. doi: 10.1186/1471-2164-13-565
Keywords: Ulva prolifera, spore formation, photosynthesis, transcriptome, flagella
Citation: Jiang J, Li Y, Hu C, Zhu W and Xu N (2022) Transcriptomic Analysis Reveals the Regulation Mechanism of Sporogenesis in Ulva prolifera. Front. Mar. Sci. 9:802006. doi: 10.3389/fmars.2022.802006
Received: 26 October 2021; Accepted: 20 January 2022;
Published: 02 March 2022.
Edited by:
Menghong Hu, Shanghai Ocean University, ChinaReviewed by:
Fanna Kong, Ocean University of China, ChinaZhi-Gang Zhou, Shanghai Ocean University, China
Copyright © 2022 Jiang, Li, Hu, Zhu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yahe Li, liyahe@nbu.edu.cn; Nianjun Xu, xunianjun@nbu.edu.cn
 Jianan Jiang1
Jianan Jiang1 Yahe Li
Yahe Li Nianjun Xu
Nianjun Xu