- 1Shenzhen Key Laboratory of Marine Bioresource and Eco-Environmental Sciences, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2State Environmental Protection Key Laboratory of Drinking Water Source Management and Technology, Shenzhen Research Academy of Environmental Sciences, Shenzhen, China
- 3School of Business Administration, Heyuan Polytechnic, Heyuan, China
- 4School of Materials and Environmental Engineering, Shenzhen Polytechnic, Shenzhen, China
Akashiwo sanguinea is a marine algal species associated with harmful algal blooms (HABs). We found that Vibrio brasiliensis H115, isolated from the seawater of Dameisha Bay (Shenzhen), China, can lyse A. sanguinea. At bacteria-to-algae cell ratios of 7,000:1 and 8,000:1, 73.9 ± 1.8 and 81.4 ± 2.8%, respectively, of A. sanguinea were lysed in just 10 min. V. brasiliensis H115 attacks A. sanguinea by secreting extracellular active compounds. Single-factor experiments showed that the optimum growth conditions for strain H115 were medium 2216E with extra sorbitol (1.0%) and peptone (2.0%), pH 7.5, a shaking speed of 200 rpm, and incubated at 40°C for 54 h. Under these optimized conditions, the algicidal efficiency of strain H115 against A. sanguinea improved by 69.4%. Large biomasses were obtained when strain H115 was incubated in 5- and 100-L bioreactors, with algicidal efficiencies against A. sanguinea reaching 94.2 ± 0.5 and 97.1 ± 0.6%, respectively, in 4 h of co-incubation. These results suggest that strain H115 and its active algicidal compounds may be viable for controlling the outbreaks of A. sanguinea.
Introduction
Red tides caused by harmful algal blooms (HABs) have attracted worldwide attention because they significantly threaten fishery resources, marine ecosystems, and human health (Anderson, 2009). Physical, chemical, and biological methods have been proposed for controlling the red tide outbreaks (Elder and Horne, 1978; Sengco and Anderson, 2004; Li et al., 2016). Compared with physical and chemical methods, biological methods have the advantages of low cost, high efficiency, and eco-friendliness (Li et al., 2016) and thus have been widely investigated.
Algicidal bacteria, which can lyse algal cells or inhibit their growth, have attracted much attention in recent decades. Algicidal bacteria isolated from various eutrophic water environments show a wide distribution (Tian et al., 2012; Lu et al., 2016; Zheng et al., 2018). Most identified algicidal bacteria belong to the Cytophaga/Flavobacterium/Bacteroidetes group or the γ-proteobacteria group (Wang M. et al., 2020). Most Gram-negative genera, including Cytophaga, Alteromonas, Pseudoalteromonas, and Pseudomonas, and several Gram-positive genera, including Bacillus, Planomicrobium, and Micrococcus, have been shown to exert algicidal effects (Zhang et al., 2020). Algicidal bacteria are typically species-specific and play important roles in regulating the dynamics of HABs (Yang et al., 2012).
Direct and indirect attacks are the two modes of interaction proposed between the algicidal bacteria and the target algae. The former refers to direct contact between the algicidal bacteria and the targeted algal cells, which leads to the rupture/death of those cells. The latter mode is typically characterized by the release of extracellular active compounds from algicidal bacteria, which inhibit or lyse the algal cells (Azam, 1998; Wang et al., 2011). Approximately 70% of algicidal bacteria utilize an indirect mode of attack (Wang M. et al., 2020). The active compounds released from algicidal bacteria include β-cyano-L-alanine (Yoshikawa et al., 2000), the proteinaceous compound P7 (Wang et al., 2011), benzoic acid (Lu et al., 2016), and novel L-amino acid oxidase (Chen et al., 2011). It has been reported that benzoic acid kills the algal cells by entering into them and acidifying the intracellular reserve, thereby inhibiting respiratory enzyme activity and blocking the acetyl coenzyme-A condensation reaction (Lu et al., 2016). Some active algicidal substances inhibit algal growth by destroying photosynthetic pigments, blocking the respiratory chain, and reducing assimilation products (Tilney et al., 2014; Zhang F. et al., 2018).
Culture conditions are essential for improving the biomass of algicidal bacteria and the production of active algicidal compounds. The components of the culture medium, pH, shaking speed, incubation time, and volume have been proposed as key factors that influence the growth of algicidal bacteria (Fu et al., 2011; Kristyanto et al., 2017). Thus, exploring the optimum culture conditions for a specific algicidal bacterium is the foundation for effectively producing active algicidal compounds and controlling HAB. Previous studies of algicidal bacteria have mainly focused on the algal host specificity, attack mode, and active algicidal compounds, and several studies have sought to improve the biomass of specific algicidal bacteria and the production of active algicidal compounds (Liao and Liu, 2014; Liao et al., 2014; Lyu et al., 2019). For example, the optimization of bacterial culture conditions has successfully improved the algicidal activity of Pseudoalteromonas SP48 against Alexandrium tamarense (Lyu et al., 2019). Three minerals (i.e., KNO3, MnSO4⋅H2O, and K2HPO4) were identified as key growth factors of liquid media that enhance the cell densities of the algicidal bacterium Enterobacter sp. NP23 (Liao et al., 2015a,b). When applied as part of water management, some reports also indicated that the initial concentrations of bacterial cells significantly affected the algicidal activity and the algicidal rate (Tian et al., 2012; Liao and Liu, 2014). Therefore, obtaining a higher cell density of algicidal bacteria is key for the biocontrol of algal blooms.
The dinoflagellate Akashiwo sanguinea is a dominant HAB-forming species (Menden-Deuer and Montalbano, 2015) associated with the death of fish, shellfish, and seabirds (Botes et al., 2003; Jessup et al., 2017). Blooms of this species have been reported in estuaries and coastal areas worldwide (Koening et al., 2014; White et al., 2014; Chen et al., 2019). However, few studies have investigated the isolation of algicidal bacteria against A. sanguinea. At present, only Pseudoalteromonas S1 (P. S1), Bacillus sp. AB-4, and Pseudoalteromonas haloplanktis AFMB-008041 have been shown to exert an algicidal action against A. sanguinea (Kim J.-D. et al., 2009; Kim Y. S. et al., 2009; Sun et al., 2016). Considering the wide distribution of algicidal bacteria, the number of highly efficient algicidal bacteria targeting A. sanguinea remains limited.
Since 1998, A. sanguinea has frequently caused red tides along the coast of China. A. sanguinea blooms were the highest in Guangdong Province around the South China Sea in 1998–2017 (Chen et al., 2019). In this study, we searched for an efficient algicidal bacterium in Dameisha Bay in Shenzhen, Guangdong Province, China. A new algicidal bacterium capable of lysing A. sanguinea was isolated from the seawater of Dameisha Bay during an A. sanguinea bloom in May 2017. The isolate was identified, and its mode of attack on A. sanguinea cells was studied. To obtain a high cell density, growth factors were investigated using single-factor effect experiments. The optimum growth conditions for the bacterial strain were also investigated to further improve algicidal efficiency. Finally, the growth of the bacterial strain was tested in two enlarged systems (5 and 100 L) under optimum conditions to improve the production of biomass and active algicidal compounds. This study was conducted to generate a new indigenous algicidal strain for the potential biocontrol of A. sanguinea and to provide useful information for future HAB management in the marine environment at either the laboratory or field scale.
Materials and Methods
Algal Culture
The Akashiwo sanguinea used in this study was provided by the Algal Culture Collection of the Institute of Hydrobiology at Jinan University (Guangzhou, China). The culture was maintained and incubated in modified f/2 medium (Lananan et al., 2013; Zhang et al., 2014) at 20 ± 2°C with a 12:12 h light/dark cycle (3,500 l×; Sun et al., 2016).
Isolation and Identification of Algicidal Bacteria
Surface seawater samples were collected from Dameisha Bay in Shenzhen, China, in May 2017, where an A. sanguinea bloom occurred. The samples were transferred to the laboratory within 2 h and stored at 4°C until use. Seawater samples were first filtered through 0.45-μm membranes to remove algal cells and then passed through 0.22-μm membranes. The 0.22-μm membrane was placed into 50 ml of preincubated A. sanguinea culture and incubated at 20 ± 2°C under a 12:12 h light/dark cycle (3,500 l×). After 7 days of incubation, 200 μl of each 10-fold serially diluted suspension was streaked onto 2216E agar plates (5 g L–1 of peptone, 1 g L–1 of yeast extract, 0.1 g L–1 of ferric phosphate, 10–12 g L–1 of agar powder, and pH 7.6–7.8) and incubated for 24 h (30°C). The isolates were then streaked onto 2216E plates twice to obtain pure isolates. The pure isolate was inoculated into 2216E medium and stored in 25% glycerol solution at −80°C.
To screen for bacteria with algicidal activity against A. sanguinea, pure cultures of all bacterial isolates were incubated in 100 ml sterilized 2216E medium in a 250-ml flask for 24 h (25°C, 200 rpm) to achieve exponential growth. The bacterial cultures were then added to 50 ml of preincubated algal culture (3,413 ± 176 cells ml–1) at a ratio of 1% (v/v). Morphological changes in the algal cells were monitored every 1 h using a microscope (Olympus IX51, Tokyo, Japan), and algicidal activity against A. sanguinea was calculated. Bacteria that could lyse A. sanguinea cells were considered as algicidal bacteria, and the strain with the strongest effects was selected for further analysis.
Genomic DNA was extracted from the selected strain using an E.Z.N.A. bacterial DNA kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s instructions. A TaKaRa PCR Kit (Sigma, Japan) was used to amplify the 16S rRNA gene in the selected strain using the primers F27 (5′-AGAGTTTGATCMTGGCTCAG-3′) and R1492 (5′-TTGGYTACCTTGTTACGACT-3′). The amplified fragments were sequenced by BGI Company (Shenzhen, China). The obtained sequences were analyzed with standard databases using BLAST from NCBI. A phylogenetic tree of the strain and other reported strains were generated in MEGA 7 software (Kumar et al., 2016) using the neighbor-joining method with 1,000 bootstrap replicates.
Algicidal Activity of Strain H115 and Mode of Action of Algicidal Metabolites
Strain H115 (2 ml) was inoculated into 100 ml sterilized 2216E medium in a 250-ml flask and incubated for 24 h (25°C, 200 rpm) to achieve exponential growth. Bacterial cultures were then added to 50 ml of preincubated algal culture (3,413 ± 176 cells ml–1) at a ratio of 1% (v/v). In the control, 50 ml of sterile 2216E medium was co-incubated with 50 ml of A. sanguinea. Samples were collected at 0, 1, 2.5, and 4 h, and the A. sanguinea cells were counted under a microscope (Olympus IX51, Tokyo, Japan) with a hemocytometer, after staining with Lugol’s reagent (Wang Y. et al., 2020). Algicidal efficiency was calculated as described by Zhang F. et al. (2018):
where N0 and Nt represent the cell number in algal cultures measured at baseline and at different sampling times (t), respectively. All experiments were conducted in triplicate. Chlorophyll fluorescence was detected using a chlorophyll fluorometer (Water-PAM, WALZ, Effeltrich, Germany). The morphological characteristics of algal cells at different times were examined using a scanning electron microscope (SEM; APREO S, Thermo Fisher Scientific, Waltham, MA, United States).
To determine the threshold concentration of strain H115 with algicidal effects against A. sanguinea, strain H115 was added to 1 ml of A. sanguinea cultures with different cell ratios (bacteria-to-algae cell ratios of 1,000:1, 3,000:1, 5,000:1, 7,000:1, and 8,000:1). The A. sanguinea culture (1 ml) without the strain added was used as control. Samples were collected at 10 min and 2, 6, 12, 24, and 48 h to detect algicidal activity. The spread plate method was used to count the number of bacterial cells. Briefly, 1 ml of bacterial culture with a known OD600 was serially diluted, and 200 μl of each 10-fold serially diluted sample was spread on a 2216E agar plate and incubated at 30°C for 24 h to prepare for counting the bacterial cells. During the experiments, the absorbance of the bacteria at 600 nm was adjusted to obtain the desired bacterial cell density.
To investigate the algicidal mode-of-action of strain H115, different fractions of the strain H115 culture, specifically the cell-free supernatant and washed cells, were prepared prior to co-incubation with A. sanguinea. Briefly, 50 ml of bacterial cultures in the exponential growth phase were collected and centrifuged at 10,000g for 10 min, and the supernatant was collected and filtered through 0.22-μm membranes to obtain the cell-free supernatant. The remaining cell pellets were washed with sterilized f/2 medium and resuspended in sterilized f/2 medium (50 ml). The same amounts of bacterial culture and sterilized 2216E medium were used as controls. The different fractions were added to 100 ml of A. sanguinea culture at a bacteria-to-algae cell ratio of 7,000:1. Samples were collected at 12, 24, 36, and 48 h to assess the algicidal efficiency of these fractions against A. sanguinea. The morphology of algal cells co-incubated with cell-free supernatant was examined using a light microscope (Olympus IX51) to confirm the algicidal effect.
Effect of Different Factors on the Growth and Algicidal Activity of Strain H115
The characteristics of different species of algicidal bacteria varied significantly. To further improve the algicidal efficiency of strain H115 and understand its characteristics and possible control mechanisms, the effect of different factors on the biomass of strain H115 and its algicidal efficiency were evaluated. It has been reported that suitable nutrient conditions, pH, and temperature enable the growth of Vibrio sp. and thus stimulate the secretion of active compounds into the extracellular medium (Jellouli et al., 2009; Zhang H. et al., 2018). To identify the factors that influence the growth of strain H115 and its algicidal activity, single-factor experiments were conducted with different extra carbon sources (e.g., fructose, sucrose, lactose, mannitol, sorbitol, and glucose), extra nitrogen sources (e.g., peptone, beef extract, yeast extract, and urea), pH (e.g., 5, 6, 7, 7.5, 8, 9, and 10), temperature (e.g., 20, 25, 30, 35, and 40°C), incubation time (e.g., 12, 24, 36, 48, and 60 h), shaking speeds (e.g., 50, 100, 150, 200, 250, and 300 rpm), and incubation volume (e.g., 30, 60, 90, 120, and 150 ml) using 2216E medium as the basal medium. Strain H115 cultured with 2216E basal medium (pH 7.5) and incubated for 24 h (30°C) was used as the control. The incubation conditions for strain H115 were the same as described in the “Algicidal Activity of Strain H115 and Mode of Action of Algicidal Metabolites” section except for the variables. NaOH or HCl (1 M) was added to the medium to adjust the pH. After 24 h of incubation, cell-free supernatants of strain H115 under different treatments were collected and added to the A. sanguinea culture at a cell ratio of 7,000:1. Samples were collected at 0 and 10 min to determine the algicidal efficiency of strain H115 against A. sanguinea.
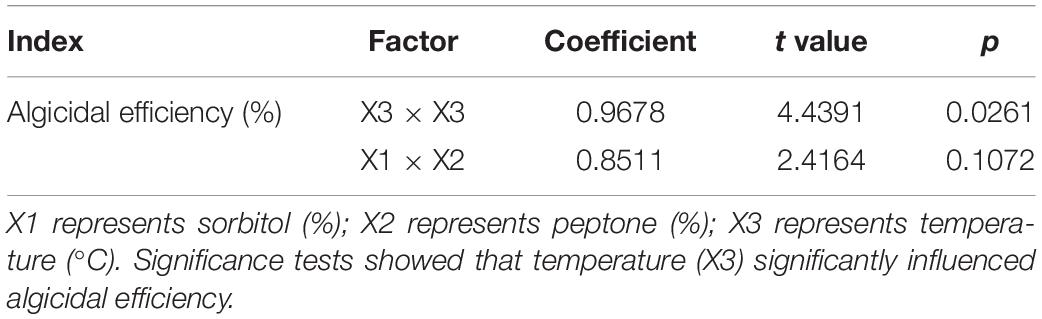
A uniform design was used to optimize the growth conditions of strain H115. A uniform design table U6 (64; Table 1) was adopted with four factors, including the content of sorbitol, peptone, temperature, and incubation time, and two indices, including OD600 and algicidal efficiency, to screen for the optimum growth conditions. The algicidal efficiency of strain H115 against A. sanguinea was further tested under the optimized conditions with a cell ratio of 600:1 (bacteria-to-algae cell ratio), as described earlier, with strain H115 cultured in 2216E medium as a control. Algal cells were counted at 4, 8, 12, 16, and 24 h, and algicidal efficiency was tested, as described earlier.
Scaling Up of Bacterial Production
Under optimized conditions (pH 7.5, 40°C, 200 rpm, 0.3% defoamers, 3 L min–1 flow rate), the incubation system was increased to 5- and 100-L fermentation cylinders to evaluate whether more biomass and active algicidal compounds of strain H115 could be obtained. After 54 h of incubation, 10 ml of the bacterial culture was collected and OD600 measured. Samples were centrifuged at 10,000g for 10 min, and the supernatant was collected and filtered through 0.22-μm membranes to obtain the cell-free supernatant. According to the OD600 and bacterial cell density of the bacterial culture, 41 (for the 5 L system) and 14 μl (for the 100 L system) of cell-free supernatants were added to 10 ml of A. sanguinea to obtain a bacteria-to-algae cell ratio of 600:1. Algal cells were counted at 4, 8, 12, 16, and 24 h, and algicidal efficiency was tested, as described earlier.
Results
Isolation and Characterization of Strain H115
Twenty-three strains of bacteria were isolated from the water samples, among which strain H115 showed the strongest algicidal activity against A. sanguinea. The colony of strain H115 was round, smooth, wet, and gray-white with regular edges and a diameter of 2–3 mm. PCR amplification of the 16S rRNA gene (1,428 bp) and sequencing showed that the H115 sequence exhibited the greatest similarity (95%) to that of Vibrio brasiliensis IS014 (accession number KR186076). Phylogenetic analysis further indicated that strain H115 belonged to V. brasiliensis (Figure 1).
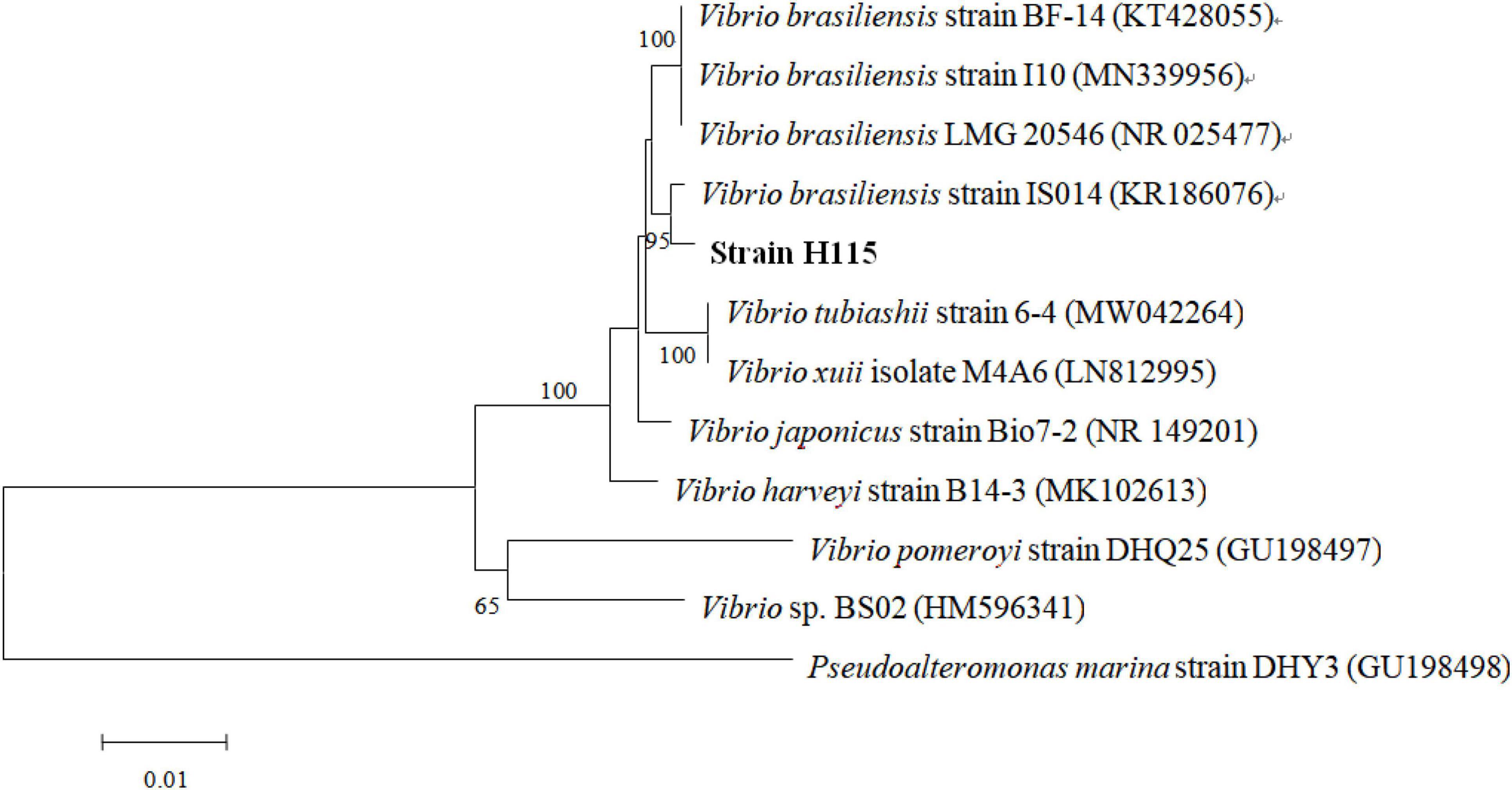
Figure 1. Neighbor-joining tree of 16S rRNA gene sequences of the strain H115 and other reported strains. Bootstrap values over 50% were shown at each node. The scale bar represents a 1% sequence divergence.
Algicidal Activity of Strain H115
After the addition of strain H115, the number of algal cells decreased from 3,413 ± 176 to 250 ± 101 cells ml–1 within 1 h, and 92.7% of the algae were lysed (Figure 2A). After co-incubation for 4 h, 99.2% of cells were lysed. These results indicate the efficient algicidal activity of strain H115 against A. sanguinea. A significant decrease (t-test, p < 0.05) in the activity of chloroplast photoreactive centers in algal cells was observed when compared to the control, which was reflected by the decline in the photosynthetic quantum yield (QY; Figure 2B). The SEM images (Figure 3) further demonstrated the decomposition of A. sanguinea during co-incubation with strain H115.
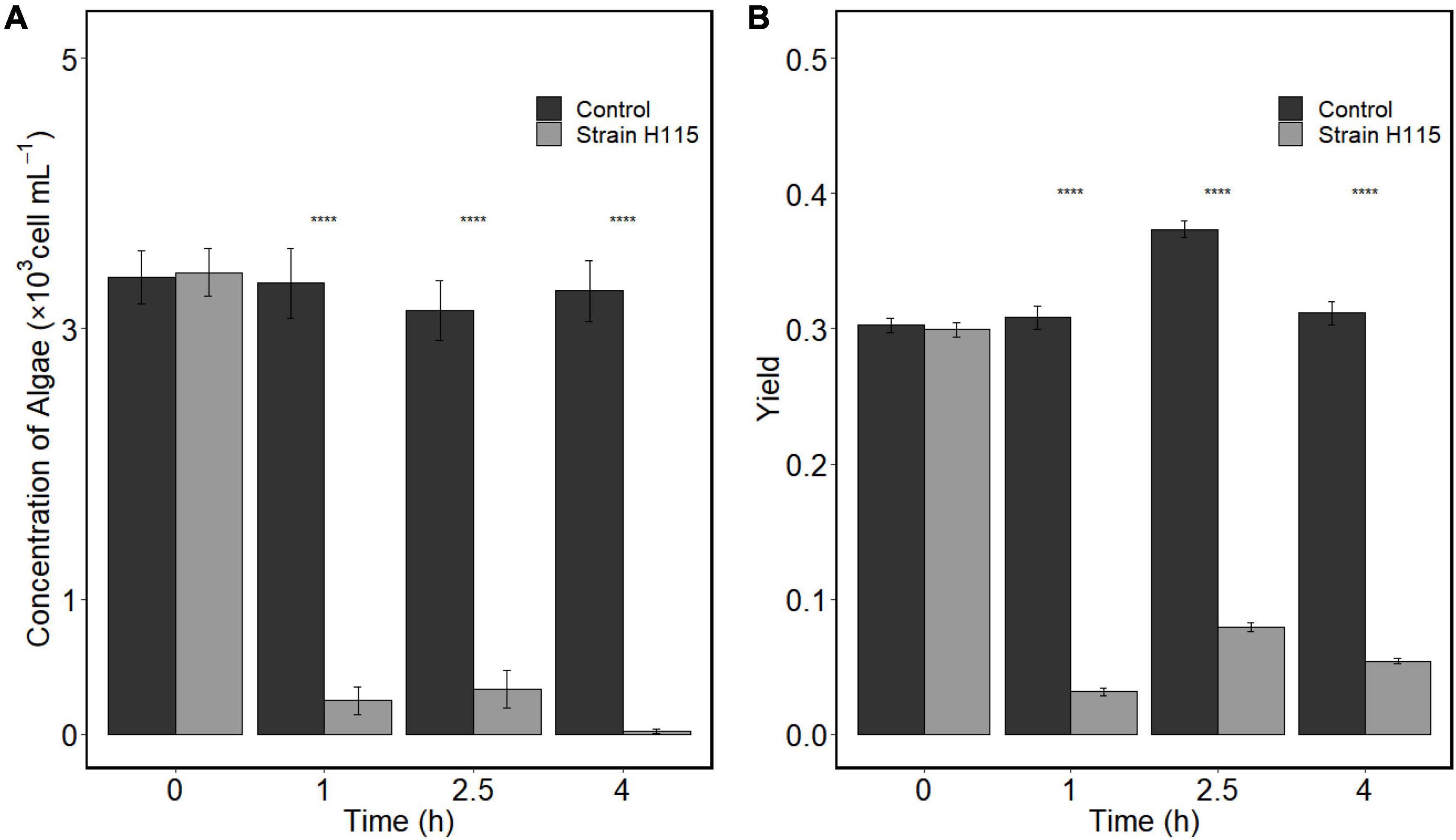
Figure 2. Algicidal effects of strain H115 on Akashiwo sanguinea cells based on (A) algal cell concentration and (B) photosynthetic maximum quantum yield. Paired t-test was conducted to test the difference between experimental group and the control. ****p < 0.05 (data are mean ± SE, n = 3).
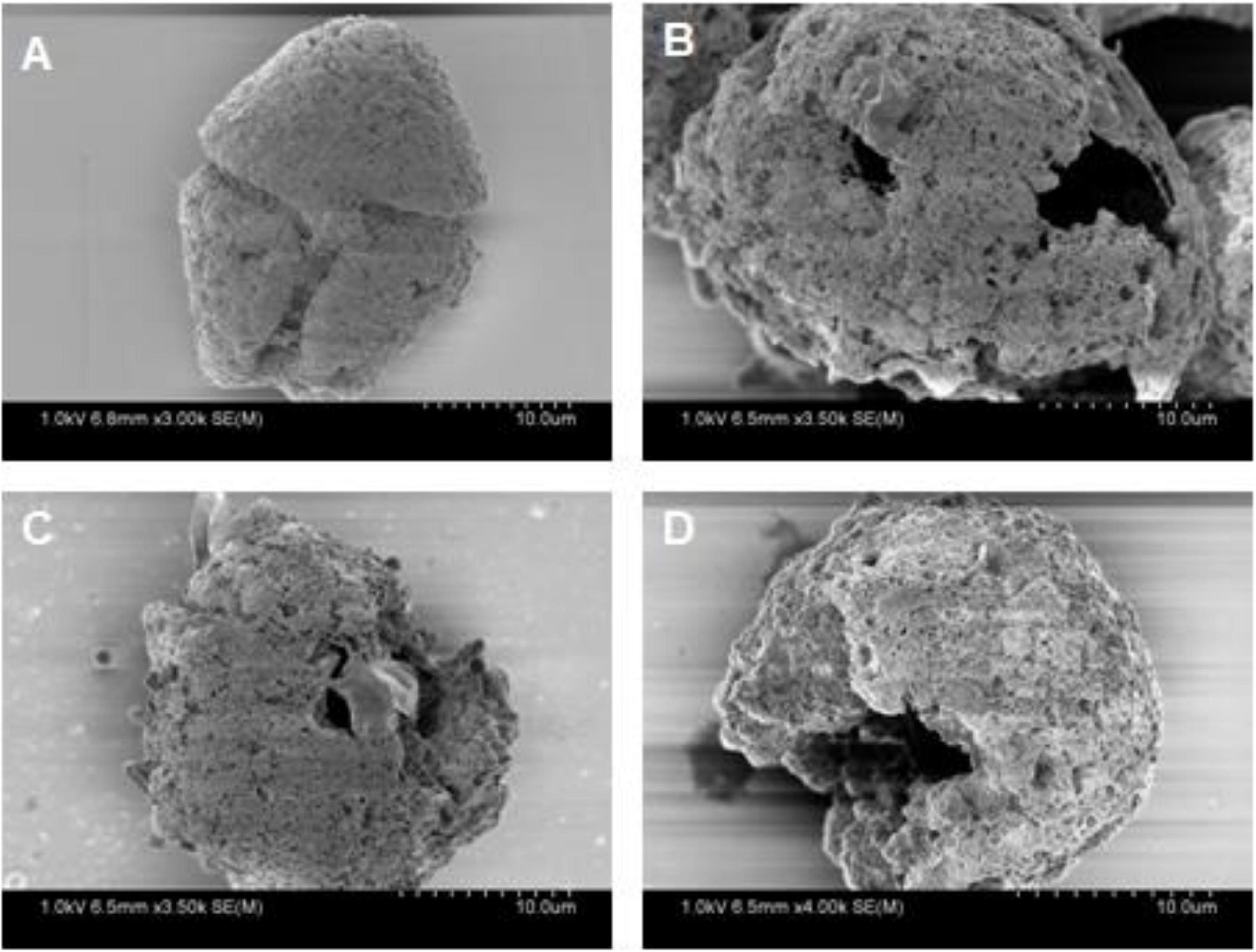
Figure 3. Scanning electron microscope (SEM) images of Akashiwo sanguinea when co-incubated with strain H115 at (A) 0 h, (B) 1 h, (C) 2.5 h, and (D) 4 h.
The algicidal performance of strain H115 against A. sanguinea at different cell ratios was tested to evaluate the optimum bacteria-to-algae cell ratio for further experiments. Algal growth was not inhibited at low cell ratios (1,000:1 and 3,000:1), showing the same trend as the control (Figure 4). When the cell ratio reached 5,000:1, a decrease in the algal cell concentration was observed after 10 min (Figure 4), indicating rapid inhibition of algal cells. The algicidal efficiency increased with the addition of more bacteria. The algicidal efficiencies (10 min) were 73.9 ± 1.8 and 81.4 ± 2.8% at the cell ratios of 7,000:1 and 8,000:1, respectively. After 48 h of incubation, the algicidal efficiency reached 80.8 ± 6.7 and 96.5 ± 0.4%, respectively. In terms of the dosage and treatment time, a cell ratio of 7,000:1 and 10 min of co-incubation were used for subsequent single-factor optimization experiments.
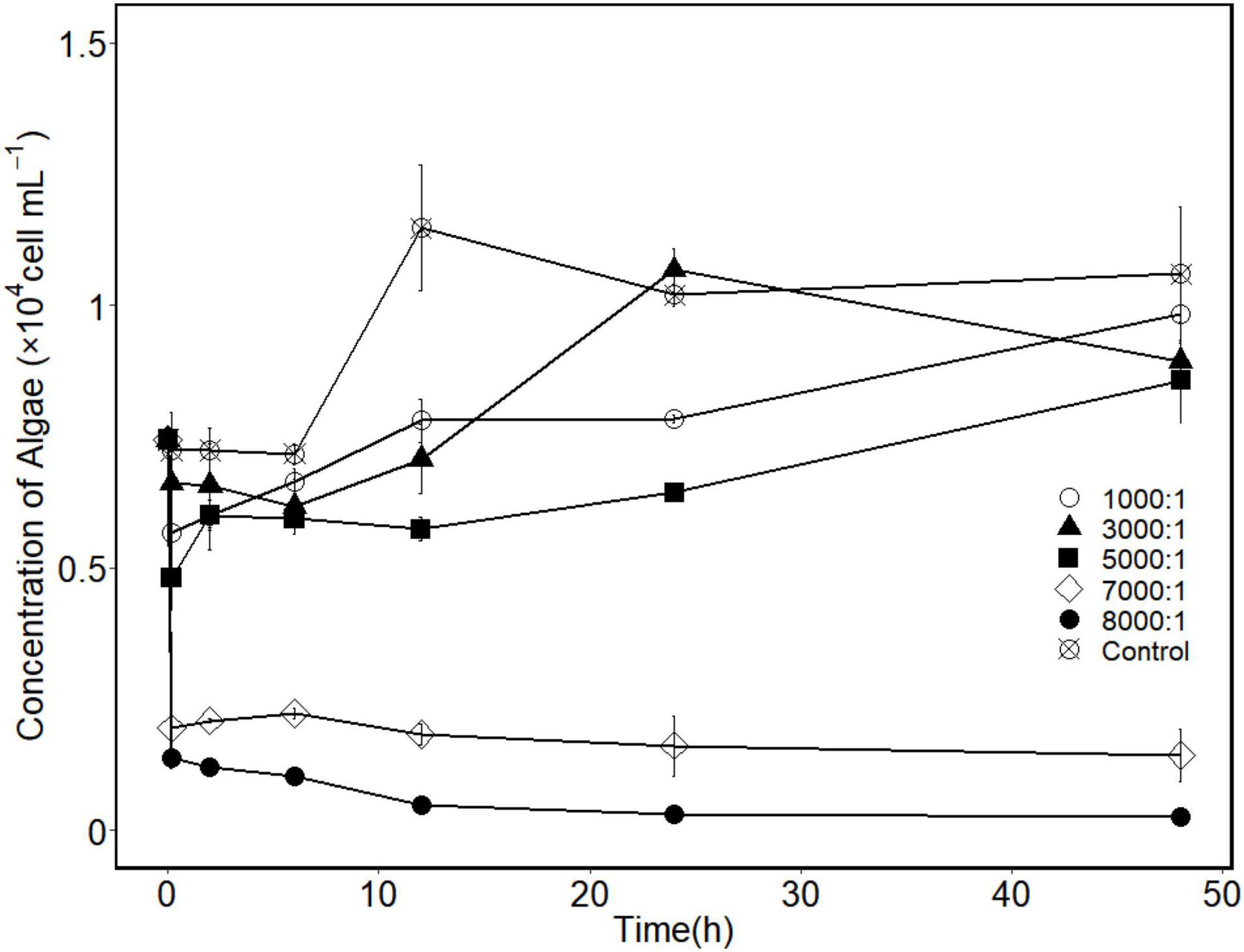
Figure 4. Changes in algal cell concentrations when Akashiwo sanguinea co-incubated with strain H115 under different bacteria-to-algae cell ratios (control means bacteria-to-algae of 0, and data are mean ± SE, n = 3).
Algicidal Activity of Cell-Free Supernatants of Strain H115
Different fractions of bacterial culture were added to A. sanguinea cultures separately to examine the algicidal mode of strain H115. The cell-free supernatant showed an algicidal efficiency of 39.7 ± 1% after treatment for 12 h, which was slightly higher than that of the strain culture (34.6 ± 1.4%, t-test, p = 0.04; Figure 5). In contrast, no algicidal activity was observed in washed bacterial cells. The growth rate of the algal culture co-incubated with washed bacterial cells was lower than that of the control (t-test, p = 0.003). These results suggest that strain H115 lysed A. sanguinea indirectly, and the active compounds causing algal cell death were present in the cell-free supernatant. Light microscopy imaging confirmed that A. sanguinea cells were lysed within 72 h of co-incubation with the cell-free supernatant (Figure 6), confirming the potent algicidal activity of this fraction.
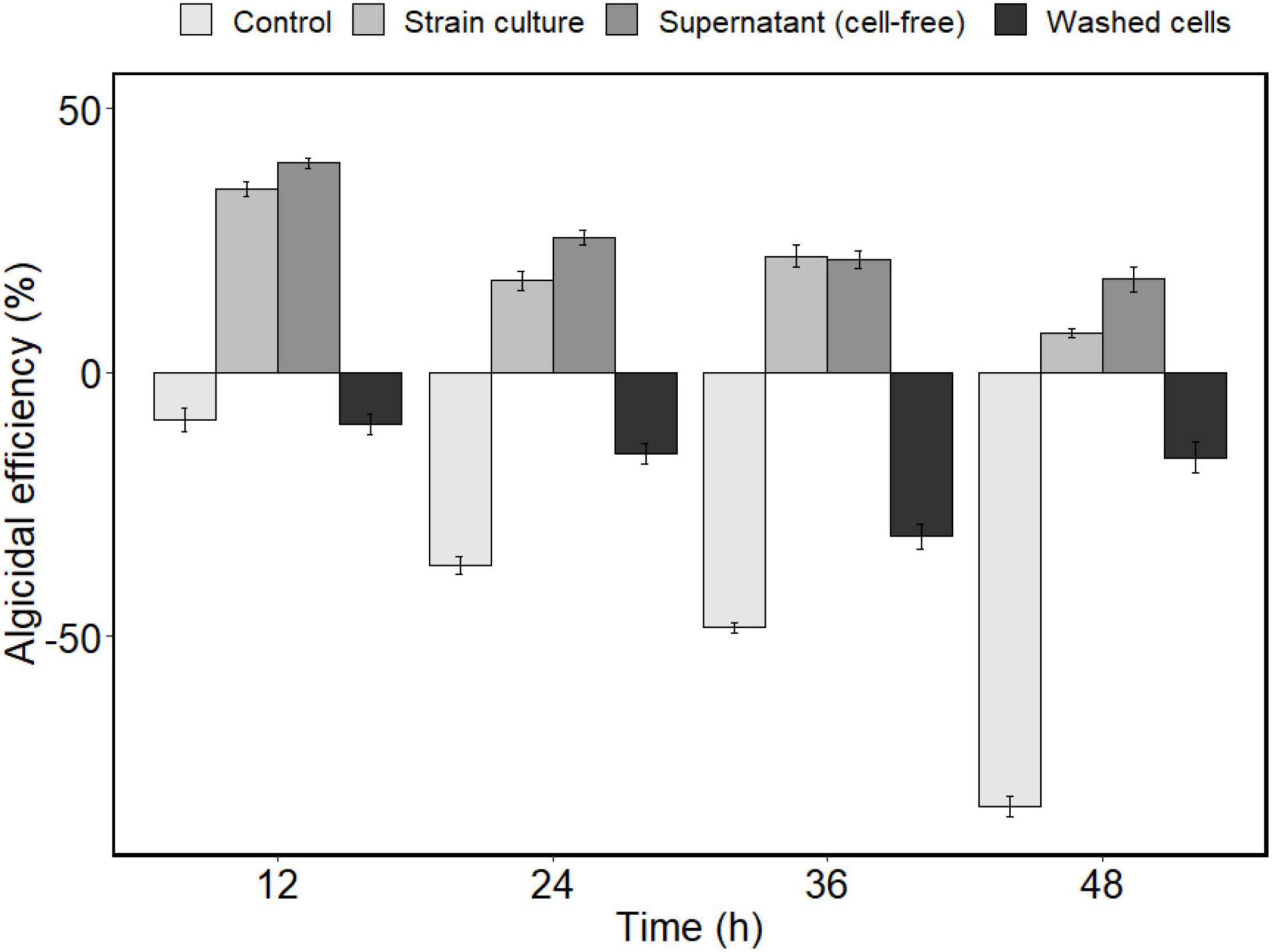
Figure 5. Algicidal efficiency of different fractions of strain H115 culture against Akashiwo sanguinea (control means A. sanguinea co-incubated with 2216E medium. Data are mean ± SE, n = 3).
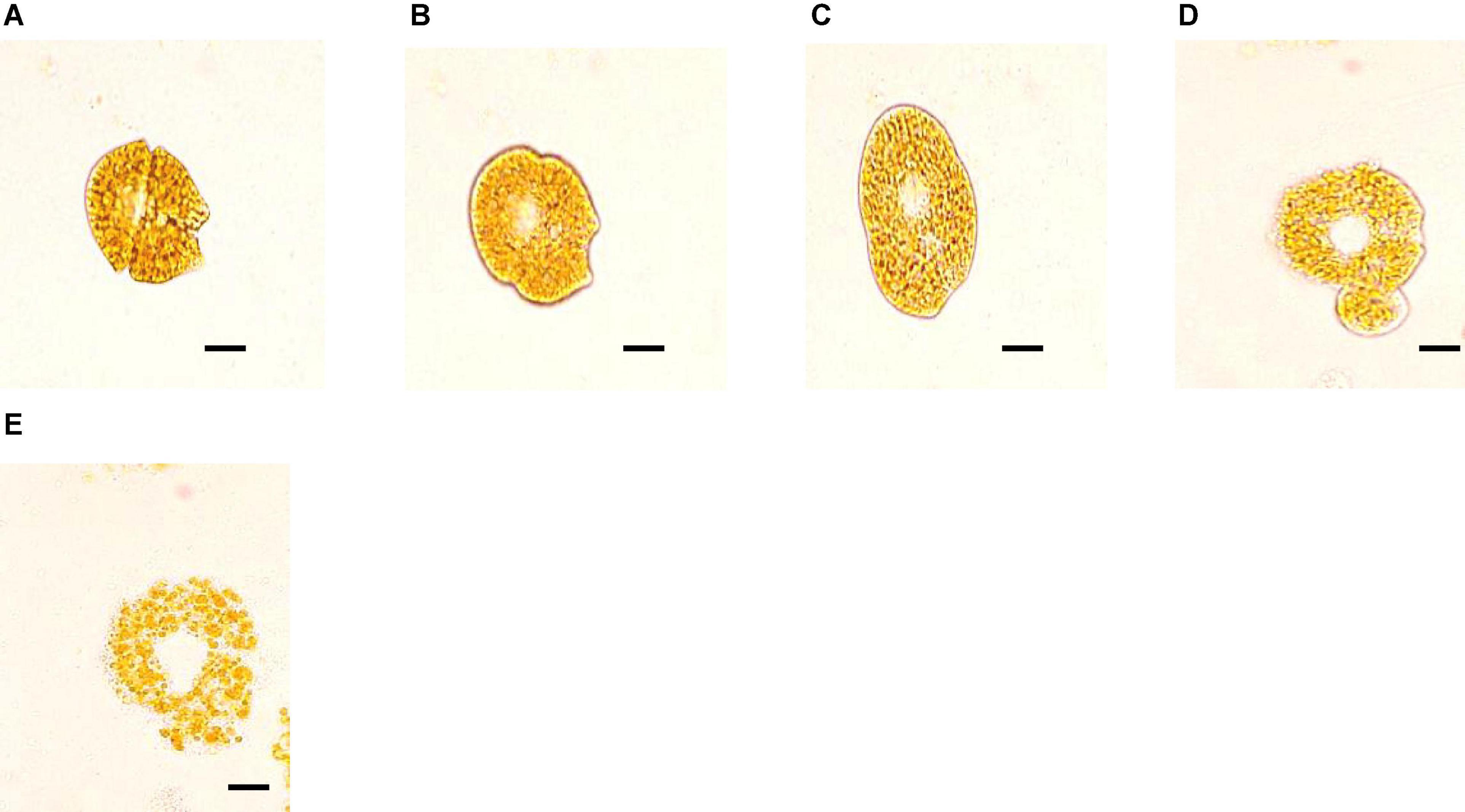
Figure 6. Algicidal effects of cell-free supernatant of strain H115 on Akashiwo sanguinea under a light microscope: (A) normal cell; (B,C) deformed cell; (D) release of cellular contents; and (E) broken thecae. Scale bars represent 5 μm.
Effects of Different Factors on the Growth and Algicidal Activity of Strain H115
Carbon Sources
Different additional carbon sources were added to the 2216E medium to test the effect of an extra carbon source on the growth of strain H115. The cell-free supernatant showed the highest algicidal efficiency in the presence of sorbitol (88.9 ± 0.5%; Table 2), which was significantly higher than that of the control (2216E, t-test, p = 0.02). The other tested carbon sources showed lower algicidal efficiencies than the control. Therefore, sorbitol was selected as the optimum carbon source for strain H115.
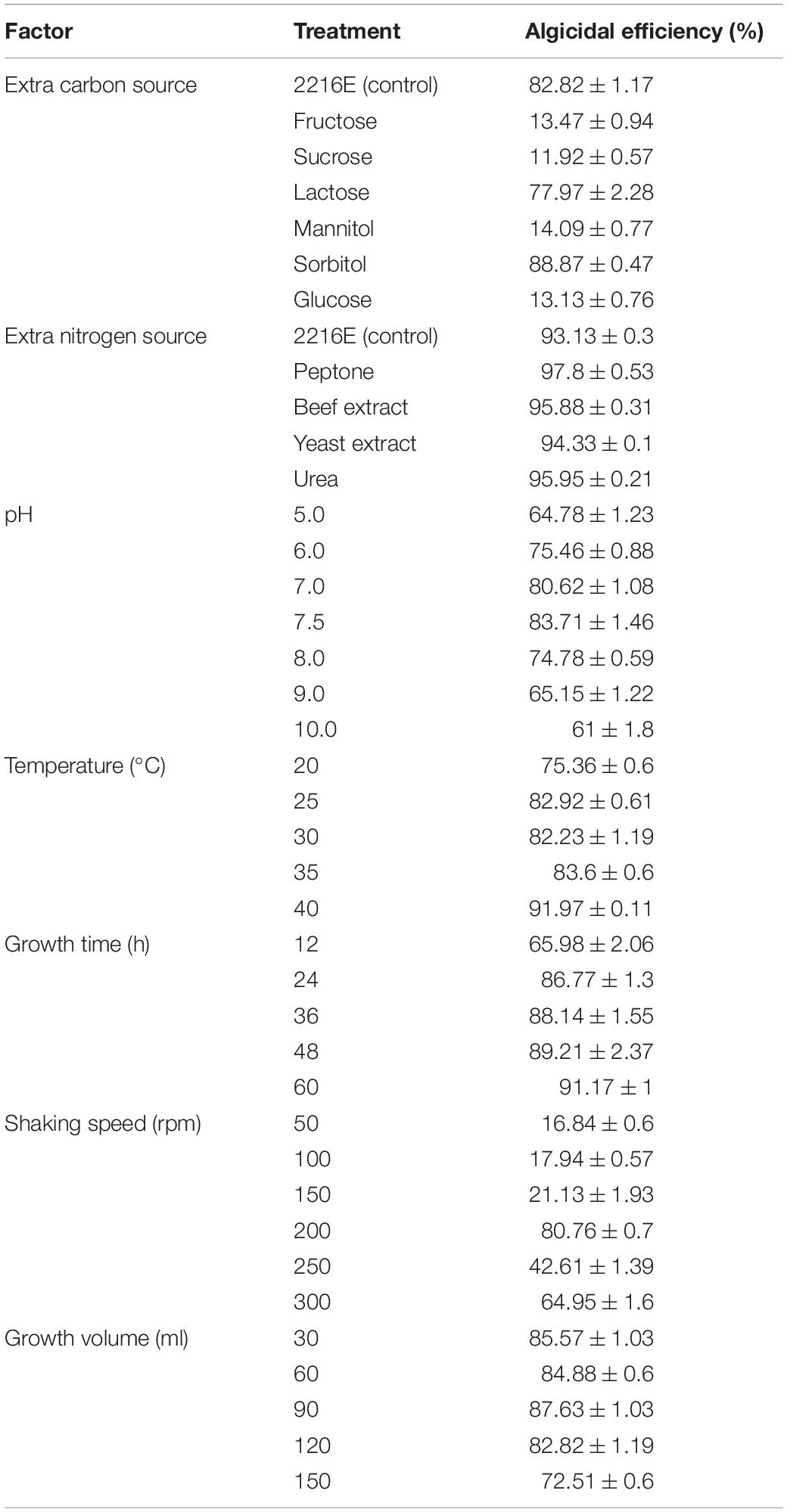
Table 2. Effects of growth conditions of bacterial strain H115 on the algicidal efficiencies against Akashiwo sanguinea.
Nitrogen Source
Strain H115 cultured with extra nitrogen sources (e.g., peptone, urea, beef extract, and yeast extract) showed slightly higher algicidal efficiencies compared to the control (2216E, t-test, p = 0.002). Strain H115 cultured with extra peptone showed the highest algicidal efficiency (97.8 ± 0.5%) against A. sanguinea, followed by urea (95.9 ± 0.2%; Table 2). Thus, peptone was selected as the best nitrogen source in the H115 culture medium.
Initial pH
The effect of pH on the algicidal activity of strain H115 was tested in the pH range of 5–10. At pH 5–7.5, the algicidal efficiency increased with increasing pH, whereas the opposite trend was observed with a further increase in pH (Table 2). Strain H115 cultured at pH 7.5 showed the highest algicidal efficiency (83.7 ± 1.5%), followed by pH 7 (80.6 ± 1.1%), indicating that a slightly alkaline culture was beneficial for the growth of strain H115.
Temperature
Strain H115 was cultured at different temperatures (25–40°C) to determine the effect of temperature on its growth. In general, algicidal efficiency increased with increasing temperature, and the cell-free supernatants showed the highest algicidal efficiency (92.0 ± 0.1%) at 40°C (Table 2).
Incubation Time
Algicidal efficiency increased with increasing incubation time and reached a maximum (91.2 ± 1%) at 60 h, after which algicidal efficiency decreased (Table 2).
Incubation Volume
Strain H115 was inoculated into different volumes of medium to test the effect of culture volume on the growth of the strain. Strain H115 incubated in 90 ml medium showed the highest algicidal efficiency (87.6 ± 1.0%). The algicidal efficiency of strain H115 decreased with further increase in the culture volume (Table 2). Thus, 90 ml was chosen as the optimum incubation volume for the growth of strain H115.
Shaking Speed
The algicidal efficiency of strain H115 increased from 16.8 ± 0.6 to 80.8 ± 0.7% as the shaking speed increased from 50 to 200 rpm. As the shaking speed was further increased to 250 and 300 rpm, algicidal efficiency decreased (Table 2). Thus, the optimum shaking speed for the growth of strain H115 was 200 rpm.
Optimum Growth Conditions for Strain H115 Based on Uniform Design
A uniform design was used to further optimize the growth conditions for strain H115; the results are listed in Table 1. Under the tested culture conditions, the OD600 was 3.03–4.76, and algicidal efficiency was 19–97%. A stepwise regression based on a quadratic polynomial was used to obtain the regression equation for the response variance Y2 (algicidal efficiency):
where X2, X3, and X4 represent the content of peptone (%), temperature (°C), and incubation time (h), respectively. The analysis of variance demonstrated that 92.97% of the total variation was attributed to the independent variables, and the relationship between the algicidal efficiency and the tested factors is explained by the model (F = 9.5576, p = 0.0261). The tests of significance showed that temperature (X3) significantly influenced algicidal efficiency (Y2; Table 3). The highest algicidal efficiency (97%) was obtained under the culture conditions of N2, with the following major factors: 1.0% sorbitol, 2.0% peptone, and incubation for 54 h at 40°C (Table 1). These results are consistent with those obtained in single-factor experiments. However, the maximum OD600 of strain H115 was obtained under the culture conditions of N3 (1.5% sorbitol, 3.0% peptone, and incubation for 48 h at 24°C), demonstrating that N3 medium was suitable for the growth of strain H115.
Under the optimized culture conditions (N2), the algicidal efficiency of strain H115 against A. sanguinea was further tested. Compared with that of the control (2216E medium), the OD600 increased from 1.81 to 3.97. At a bacteria-to-algae cell ratio of 600:1, algicidal efficiency improved from 28.2 ± 7.9% (control) to 97.7 ± 0.1% (N2) after 12 h (Figure 7A).
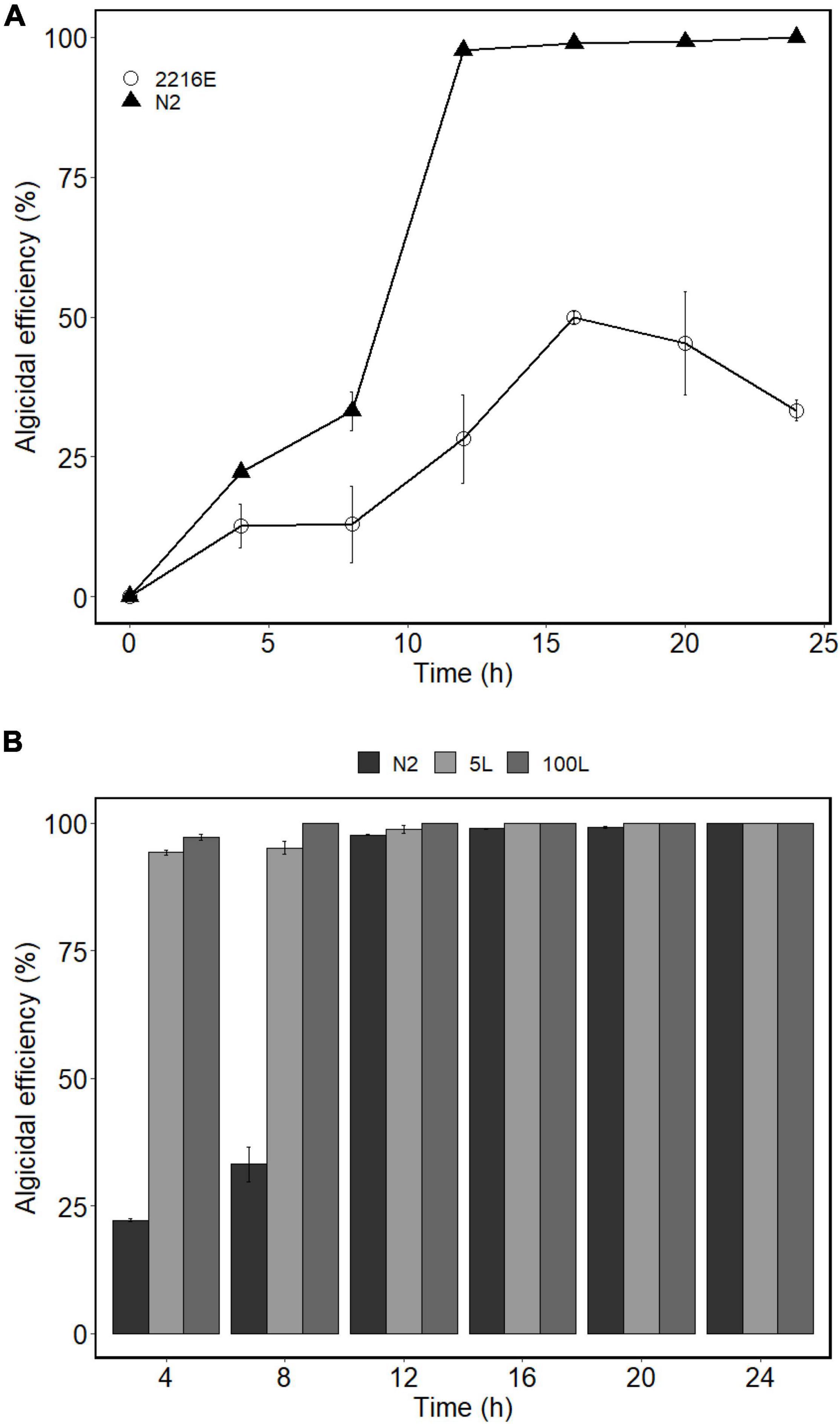
Figure 7. (A) Algicidal efficiencies of strain H115 cultured under different medium against A. sanguinea [2216E medium and optimized medium (N2)]. (B) Algicidal efficiencies of strain H115 cultured under different fermentation cylinders against Akashiwo sanguinea (5 and 100 L). Data are mean ± SE, n = 3.
Growth Under Enlarged Bioreactors
The growth of strain H115 was tested in 5- and 100-L bioreactors under optimized conditions to obtain a higher biomass and more active algicidal compounds. When the incubation system was enlarged to 5 and 100 L, the OD600 reached 5.10 and 30.95, respectively. The algicidal efficiency of strain H115 cultured at 5 and 100 L reached 94.2 ± 0.5 and 97.1 ± 0.6% when co-incubated with A. sanguinea for 4 h, respectively, which was higher than that in flasks (22.2 ± 0.2%; Figure 7B).
Discussion
Recently, various algicidal bacteria have been isolated from lakes, estuaries, and seas (Tian et al., 2012; Lu et al., 2016; Zheng et al., 2018). However, few studies have been conducted to isolate and identify algicidal bacteria that target A. sanguinea. P. S1 isolated from Dapeng Bay in China (Sun et al., 2016), P. haloplanktis AFMB-008041 from Masan Bay in Korea (Kim J.-D. et al., 2009), and Bacillus sp. AB-4 from the coastal waters of Uljin, Korea (Kim Y. S. et al., 2009) were reported to be algicidal against A. sanguinea. In this study, an algicidal bacterium, V. brasiliensis H115, was isolated from the seawater of Dameisha Bay. Reports on algicidal bacteria belonging to Vibrio spp. are rare compared with other commonly recognized algicidal genera (Wang M. et al., 2020). Strains of Vibrio sp. are algicidal against marine algal species, such as A. tamarense (Wang B. X. et al., 2010; Fu et al., 2011; Su et al., 2011), Oscillatoria amphibia (Yoshikawa et al., 2000), and Heterosigma akashiwo and Gymnodinium catenatum (Yoshinaga et al., 1995). The Vibrio sp. co-culture (containing V. brasiliensis and Vibrio tubiashii) was recently confirmed to be algicidal against A. sanguinea (Wang Y. et al., 2020). However, a V. brasiliensis isolate capable of lysing A. sanguinea has not previously been reported.
Similar to the findings for other algicidal bacteria (Tian et al., 2012), the algicidal activity of strain H115 depended on initial bacterial cell density, with a threshold bacteria-to-algae cell ratio of 5,000:1 (Figure 4). At a bacteria-to-algae cell ratio of 7,000:1, algicidal efficiency reached 73.90% after 10 min. Strain H115 showed obvious advantages compared with P. S1, as a shorter time was required to achieve high algicidal efficiency (10 min vs. 4 h; Sun et al., 2016). The algicidal efficiencies of Bacillus sp. AB-4 (Kim Y. S. et al., 2009) and P. haloplanktis AFMB-008041 (Kim J.-D. et al., 2009) against A. sanguinea were only 6.8 and 47.6%, respectively. As strain H115 can effectively lyse A. sanguinea, it may be useful as a preventive agent against A. sanguinea blooms.
Different fractions of the strain culture were tested for their algicidal activity against A. sanguinea. The cell-free supernatant exhibited an algicidal efficiency of 39.70% after treatment for 12 h (Figure 5), suggesting an indirect mode of attack. This is consistent with other identified algicidal bacteria belonging to Vibrio spp. (Wang B. X. et al., 2010; Fu et al., 2011; Su et al., 2011). Vibrio sp. lyses algae by releasing active extracellular algicidal compounds. Unknown algicidal compounds may be metabolic byproducts of strain H115, e.g., β-cyano-L-alanine, a proteinaceous compound P7, and a non-proteinaceous substance were reported as the active algicidal compounds in Vibrio sp. C-979, Vibrio sp. DHQ25, and Vibrio sp. BS02, respectively (Yoshikawa et al., 2000; Fu et al., 2011; Wang et al., 2011). In each case, they breach the cell wall and degrade the algal cells. During the lysis of A. sanguinea, the photosynthetic maximum QY significantly decreased (t-test, p < 0.05; Figure 2B). The SEM (Figure 3) and light microscopy images (Figure 6) confirmed that A. sanguinea cells were disrupted and lysed during co-incubation with the strain and the cell-free supernatant. Some active algicidal substances were reported to inhibit algal growth by destroying photosynthetic pigments, blocking the respiratory chain, and reducing assimilation products (Tilney et al., 2014; Zhang F. et al., 2018). Some algicidal bacteria have been demonstrated to lyse algal cells through cell wall polysaccharide degradation (Kim J.-D. et al., 2009; Wang X. et al., 2010). Hydrolytic enzymes, including beta-glucosidases, amylases, cellulases, and xylanases, were reported to be responsible for the polysaccharide degradation (Kim J.-D. et al., 2009; Chen et al., 2013; Shi et al., 2018). Given the cellulose makeup of dinoflagellate thecal plates, the polysaccharide degradation pathway of cell lysis might explain why some algicidal bacteria showed higher lysis rates on armored dinoflagellates than on athecate dinoflagellates (Shi et al., 2018). Strain H115 may secrete a variety of extracellular bioactive agents that affect the photosynthetic apparatus and the transmission of photosynthetic electrons in A. sanguinea (Zhang et al., 2020). Strain H115 may restrain the growth of A. sanguinea by destroying the cell membrane and inhibiting the antioxidant system (Wang Y. et al., 2020).
Studies aimed at enhancing the cell density of algicidal bacteria are rare because the analysis of the medium content is typically complex (Liao and Liu, 2014; Liao et al., 2014). However, optimizing the medium is key to enhancing the cell density of algicidal bacteria when applied to a control algal bloom. Single-factor effect and uniform design experiments suggested that the highest algicidal efficiency was achieved by adding sorbitol and peptone as carbon and nitrogen sources, respectively. The marine algicidal bacteria Bacillus sp. AB-4 and Marinomonas algicida sp. nov. Yeongu 1-4T were reported to use sorbitol as a carbon source (Kim Y. S. et al., 2009; Kristyanto et al., 2017). Suitable nutrient conditions and appropriate pH and temperature enable the growth of Vibrio sp. and thus stimulate the secretion of active compounds into the extracellular medium (Jellouli et al., 2009; Zhang H. et al., 2018). Peptone is the optimum nitrogen source for strain H115, which is consistent with other algicidal bacteria (Wang et al., 2011; Zheng et al., 2018). However, tryptone has been reported as the optimum nitrogen source for Pseudoalteromonas sp. SP48 (Lyu et al., 2019). For strain H115, the optimum pH was 7.5, which is consistent with that of Vibrio sp. BS02 (7.6–7.8; Fu et al., 2011). However, the optimum temperature was 40°C, which is significantly different from the optimum temperature for the growth of Vibrio sp. BS02 (25–28°C; Fu et al., 2011). Under the optimized growth condition (N2), the algicidal efficiency of strain H115 increased by 69.43%, indicating that the yield of the algicidal compounds was improved after optimization. Moreover, because strain H115 maintained relatively high algicidal efficiency against A. sanguinea (75.4 ± 0.6 to 92.0 ± 0.1%) within 20–40°C (Table 2), this strain and its active algicidal compounds may be stable under natural conditions. Larger biomass and active algicidal compounds were obtained when the incubation system was enlarged to 5- and 100-L bioreactors. We have tested the toxicity of strain H115 to zebra fish and brine shrimp and confirmed the safety of strain H115 for these two species. Further experiments regarding the security evaluation of strain H115 to other HAB species, live-food organisms, and the marine environment should be undertaken. These results demonstrate the potential for applying strain H115 as an algicidal substance against A. sanguinea blooms. Further studies targeting the collection and identification of active algicidal compounds, as well as the evaluations of safety and industrial production, should be conducted.
Conclusion
The marine algicidal bacterium V. brasiliensis H115 was isolated from seawater obtained from Dameisha Bay (Shenzhen, China) and exhibited high algicidal activity against A. sanguinea via an indirect mode of attack. Under optimized conditions, the biomass and algicidal efficiency of strain H115 were significantly improved and further enhanced as the incubation system was increased to 5- and 100-L bioreactors. These results provide a foundation for the potential application of strain H115 and its active algicidal compounds for controlling A. sanguinea blooms.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SL: conceptualization, project administration, funding acquisition, supervision, and writing—original draft. SW: investigation and methodology. LX: conceptualization. YL: investigation, methodology, and formal analysis. HC: conceptualization and supervision. JF: investigation. LO: conceptualization, formal analysis, writing—original draft, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2020YFD0901003), Shenzhen Sustainable Development Science and Technology Project (KCXFZ20201221173404012), the Scientific Research Startup Fund for Shenzhen High-Caliber Personnel of SZPT (No. 6022310040K), and Post-doctoral Later-stage Foundation Project of Shenzhen Polytechnic (6021271015K1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson, D. M. (2009). Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast. Manage. 52, 342–347. doi: 10.1016/j.ocecoaman.2009.04.006
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Botes, L., Smit, A. J., and Cook, P. A. (2003). The potential threat of algal blooms to the abalone (Haliotis midae) mariculture industry situated around the South African coast. Harmful Algae 2, 247–259. doi: 10.1016/s1568-9883(03)00044-1
Chen, B., Kang, W., and Hui, L. (2019). Akashiwo sanguinea blooms in Chinese waters in 1998–2017. Mar. Pollut. Bull. 149:110652. doi: 10.1016/j.marpolbul.2019.110652
Chen, C. Y., Bai, M. D., and Chang, J. S. (2013). Improving microalgal oil collecting efficiency by pretreating the microalgal cell wall with destructive bacteria. Biochem. Eng. J. 81, 170–176. doi: 10.1016/j.bej.2013.10.014
Chen, W. M., Sheu, F. S., and Sheu, S. Y. (2011). Novel L-amino acid oxidase with algicidal activity against toxic cyanobacterium Microcystis aeruginosa synthesized by a bacterium Aquimarina sp. Enzyme Microb. Technol. 49, 372–379. doi: 10.1016/j.enzmictec.2011.06.016
Elder, J. F., and Horne, A. J. (1978). Copper cycles and CuSO4 algicidal capacity in two California lakes. Environ. Manage. 2, 17–30. doi: 10.1007/bf01866443
Fu, L., An, X., Li, D., Zhou, L., Tian, Y., and Zheng, T. (2011). Isolation and alga-inhibiting characterization of Vibrio sp. BS02 against Alexandrium tamarense. World J. Microbiol. Biotechnol. 27, 2949–2956. doi: 10.1007/s11274-011-0778-3
Jellouli, K., Bougatef, A., Manni, L., Agrebi, R., Siala, R., Younes, I., et al. (2009). Molecular and biochemical characterization of an extracellular serine-protease from Vibrio metschnikovii J1. J. Ind. Microbiol. Biotechnol. 36, 939–948. doi: 10.1007/s10295-009-0572-5
Jessup, D. A., Miller, M. A., Ryan, J. P., Nevins, H. M., Kerkering, H. A., Mekebri, A., et al. (2017). Mass stranding of marine birds caused by a surfactant-producing red tide. PLoS One 4:e4550. doi: 10.1371/journal.pone.0004550
Kim, J.-D., Kim, J.-Y., Park, J.-K., and Lee, C.-G. (2009). Selective control of the prorocentrum minimum harmful algal blooms by a novel algal-lytic bacterium Pseudoalteromonas haloplanktis AFMB-008041. Mar. Biotechnol. 11, 463–472. doi: 10.1007/s10126-008-9167-9
Kim, Y. S., Lee, D.-S., Jeong, S.-Y., Lee, W. J., and Lee, M.-S. (2009). Isolation and characterization of a marine algicidal bacterium against the harmful raphidophyceae Chattonella marina. J. Microbiol. 47, 9–18. doi: 10.1007/s12275-008-0141-z
Koening, M., Flores Montes, M., Eskinazi Leça, E., and Tiburcio, A. (2014). New record of Akashiwo sanguinea(Dinophyta) in the tropical estuarine waters of Northeastern Brazil (Western Atlantic). Braz. J. Biol. 74, 191–198. doi: 10.1590/1519-6984.03512
Kristyanto, S., Chaudhary, D. K., Lee, S.-S., and Kim, J. (2017). Characterization of Marinomonas algicida sp nov., a novel algicidal marine bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 67, 4777–4784. doi: 10.1099/ijsem.0.002374
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lananan, F., Jusoh, A., Ali, N., Lam, S. S., and Endut, A. (2013). Effect of Conway Medium and f/2 medium on the growth of six genera of South China Sea marine microalgae. Bioresour. Technol. 141, 75–82. doi: 10.1016/j.biortech.2013.03.006
Li, Y., Liu, L., Xu, Y., Guan, C., Lei, X., Zheng, W., et al. (2016). First evidence of Altererythrobacter sp LY02 with indirect algicidal activity on the toxic dinoflagellate, Alexandrium tamarense. Curr. Microbiol. 73, 550–560. doi: 10.1007/s00284-016-1093-x
Liao, C. L., and Liu, X. B. (2014). High-cell-density cultivation and algicidal activity assays of a novel algicidal bacterium to control algal bloom caused by water eutrophication. Water Air Soil Pollut. 225:2120.
Liao, C. L., Liu, X. B., Liu, R. F., and Shan, L. N. (2015a). Characterization and effects of two algicidal isolates on antioxidase activities of Chlorella pyrenoidosa. Environ. Prog. Sustain. Energy 34, 1647–1651. doi: 10.1002/ep.12170
Liao, C. L., Liu, X. B., Liu, R. F., and Shan, L. N. (2015b). Two novel algicidal isolates kill Chlorella pyrenoidosa by inhibiting their host antioxidase activities. Appl. Biochem. Biotechnol. 177, 567–576. doi: 10.1007/s12010-015-1749-1
Liao, C. L., Liu, X. B., and Shan, L. N. (2014). Optimization of liquid media and biosafety assessment for algae-lysing bacterium NP23. Can. J. Microbiol. 60, 593–597. doi: 10.1139/cjm-2014-0322
Lu, X., Zhou, B., Xu, L., Liu, L., Wang, G., Liu, X., et al. (2016). A marine algicidal Thalassospira and its active substance against the harmful algal bloom species Karenia mikimotoi. Appl. Microbiol. Biotechnol. 100, 5131–5139. doi: 10.1007/s00253-016-7352-8
Lyu, Y.-H., Zhou, Y.-X., Li, Y., Zhou, J., and Xu, Y.-X. (2019). Optimized culturing conditions for an algicidal bacterium Pseudoalteromonas sp. SP48 on harmful algal blooms caused by Alexandrium tamarense. Microbiologyopen 8:e803. doi: 10.1002/mbo3.803
Menden-Deuer, S., and Montalbano, A. L. (2015). Bloom formation potential in the harmful dinoflagellate Akashiwo sanguinea : clues from movement behaviors and growth characteristics. Harmful Algae 47, 75–85. doi: 10.1016/j.hal.2015.06.001
Sengco, M. R., and Anderson, D. M. (2004). Controlling harmful algal blooms through clay flocculation. J. Eukaryot. Microbiol. 51, 169–172. doi: 10.1111/j.1550-7408.2004.tb00541.x
Shi, X., Liu, L., Li, Y., Xiao, Y., Ding, G., Lin, S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal-bloom dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Su, J., Yang, X., Zhou, Y., and Zheng, T. (2011). Marine bacteria antagonistic to the harmful algal bloom species Alexandrium tamarense (Dinophyceae). Biol. Control 56, 132–138. doi: 10.1016/j.biocontrol.2010.10.004
Sun, H.-Y., Zhang, Y., Chen, H.-R., Hu, C.-X., Li, H., and Hu, Z.-L. (2016). Isolation and characterization of the marine algicidal bacterium Pseudoalteromonas S1 against the harmful alga Akashiwo sanguinea. Mar. Biol. 163, 1–8. doi: 10.1080/09593330.2022.2036250
Tian, C., Liu, X., Tan, J., Lin, S., Li, D., and Yang, H. (2012). Isolation, identification and characterization of an algicidal bacterium from Lake Taihu and preliminary studies on its algicidal compounds. J. Environ. Sci. 24, 1823–1831. doi: 10.1016/s1001-0742(11)60983-2
Tilney, C. L., Pokrzywinski, K. L., Coyne, K. J., and Warner, M. E. (2014). Growth, death, and photobiology of dinoflagellates (Dinophyceae) under bacterial-algicide control. J. Appl. Phycol. 26, 2117–2127. doi: 10.1007/s10811-014-0248-z
Wang, B., Yang, X., Lu, J., Zhou, Y., Su, J., Tian, Y., et al. (2011). A marine bacterium producing protein with algicidal activity against Alexandrium tamarense. Harmful Algae 13, 83–88. doi: 10.1016/j.hal.2011.10.006
Wang, B. X., Zhou, Y. Y., Bai, S. J., Su, J. Q., Tian, Y., Zheng, T. L., et al. (2010). A novel marine bacterium algicidal to the toxic dinoflagellate Alexandrium tamarense. Lett. Appl. Microbiol. 51, 552–557. doi: 10.1111/j.1472-765X.2010.02936.x
Wang, M., Chen, S., Zhou, W., Yuan, W., and Wang, D. (2020). Algal cell lysis by bacteria: a review and comparison to conventional methods. Algal Res. 46:101794. doi: 10.1016/j.algal.2020.101794
Wang, X., Li, Z. J., Su, J. Q., Tian, Y., Ning, X. R., Hong, H. S., et al. (2010). Lysis of a red-tide causing alga, Alexandrium tamarense, caused by bacteria from its phycosphere. Biol. Control 52, 123–130. doi: 10.1016/j.biocontrol.2009.10.004
Wang, Y., Li, S., Liu, G., Li, X., Yang, Q., Xu, Y., et al. (2020). Continuous production of algicidal compounds against Akashiwo sanguinea via a Vibrio sp. co-culture. Bioresour. Technol. 295:122246. doi: 10.1016/j.biortech.2019.122246
White, A. E., Watkins-Brandt, K. S., McKibben, S. M., Wood, A. M., Hunter, M., Forster, Z., et al. (2014). Large-scale bloom of Akashiwo sanguinea in the Northern California current system in 2009. Harmful Algae 37, 38–46. doi: 10.1016/j.hal.2014.05.004
Yang, C., Li, Y., Zhou, Y., Zheng, W., Tian, Y., and Zheng, T. (2012). Bacterial community dynamics during a bloom caused by Akashiwo sanguinea in the Xiamen sea area, China. Harmful Algae 20, 132–141. doi: 10.1016/j.hal.2012.09.002
Yoshikawa, K., Adachi, K., Nishijima, M., Takadera, T., Tamaki, S., Harada, K., et al. (2000). Beta-cyanoalanine production by marine bacteria on cyanide-free medium and its specific inhibitory activity toward cyanobacteria. Appl. Environ. Microbiol. 66, 718–722. doi: 10.1128/AEM.66.2.718-722.2000
Yoshinaga, I., Kawai, T., Takeuchi, T., and Ishida, Y. (1995). Distribution and fluctuation of bacteria inhibiting the growth of a marine red tide phytoplankton Gymnodinium mikimotoi in Tanabe Bay (Wakayama Pref., Japan). Fish. Sci. 61, 780–786. doi: 10.2331/fishsci.61.780
Zhang, B., Cai, G., Wang, H., Li, D., Yang, X., An, X., et al. (2014). Streptomyces alboflavus RPS and its novel and high algicidal activity against harmful algal bloom species Phaeocystis globosa. PLoS One 9:e92907. doi: 10.1371/journal.pone.0092907
Zhang, F., Ye, Q., Chen, Q., Yang, K., Zhang, D., Chen, Z., et al. (2018). Algicidal activity of novel marine bacterium Paracoccus sp strain Y42 against a harmful algal-bloom-causing dinoflagellate, Prorocentrum donghaiense. Appl. Environ. Microbiol. 84, e01015–e01018. doi: 10.1128/AEM.01015-18
Zhang, H., Li, H., Lang, D. A., Xu, H., and Zhu, H. (2018). Purification, characterization and potential applications of a novel halotolerant metalloprotease from marine bacterium Vibrio sp LA-05. J. Chem. Technol. Biotechnol. 93, 3627–3637. doi: 10.1002/jctb.5744
Zhang, S.-J., Du, X.-P., Zhu, J.-M., Meng, C.-X., Zhou, J., and Zuo, P. (2020). The complete genome sequence of the algicidal bacterium Bacillus subtilis strain JA and the use of quorum sensing to evaluate its antialgal ability. Biotechnol. Rep. 25:e00421. doi: 10.1016/j.btre.2020.e00421
Keywords: red tides, algicidal bacteria, Akashiwo sanguinea, optimum growth conditions, algicidal effect
Citation: Li S, Wang S, Xie L, Liu Y, Chen H, Feng J and Ouyang L (2022) Identification and Optimization of the Algicidal Activity of a Novel Marine Bacterium Against Akashiwo sanguinea. Front. Mar. Sci. 9:798544. doi: 10.3389/fmars.2022.798544
Received: 15 November 2021; Accepted: 28 February 2022;
Published: 13 April 2022.
Edited by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research (Nofima), NorwayReviewed by:
Xiaobo Liu, Nanjing University of Science and Technology, ChinaDeo Florence Onda, University of the Philippines Diliman, Philippines
Copyright © 2022 Li, Wang, Xie, Liu, Chen, Feng and Ouyang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liao Ouyang, b3V5YW5nbGlhb0Bob3RtYWlsLmNvbQ==
 Shuangfei Li
Shuangfei Li Shilin Wang
Shilin Wang Linshen Xie2
Linshen Xie2 Liao Ouyang
Liao Ouyang