
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 24 February 2022
Sec. Marine Biogeochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.773548
 Fulin Sun1,2,3,4
Fulin Sun1,2,3,4 Xiaomin Xia3,4,5
Xiaomin Xia3,4,5 Meinhard Simon6
Meinhard Simon6 Youshao Wang1,2,3,4
Youshao Wang1,2,3,4 Hui Zhao7
Hui Zhao7 Cuici Sun1,2,3,4
Cuici Sun1,2,3,4 Hao Cheng1,3,4
Hao Cheng1,3,4 Yutu Wang1,2,3,4
Yutu Wang1,2,3,4 Shuibo Hu8
Shuibo Hu8 Jiao Fei1,3,4
Jiao Fei1,3,4 Meilin Wu1,3,4*
Meilin Wu1,3,4*Mesoscale eddies play an important role in ocean energy and material transport. However, the effect of eddies on bacterial and eukaryotic community and their ecological effects remains unclear, regarding anticyclonic eddies (ACE). In this study, bacterial and eukaryotic community composition was examined across an ACE in the South China Sea, using high-throughput sequencing of the 16S rRNA and 18S rRNA gene. Environmental variables reflected the hydrographic characteristics of the ACE, which enhanced bacterial diversity and eukaryotic diversity in most water layers, relative to adjacent regions. Principal component analysis (PCoA) showed that bacterial and eukaryotic communities had certain different compositions between inside and outside the eddy above 75 m water. An obvious effect of the ACE was the increase in abundance and depth distribution of small photosynthetic and heterotrophic bacteria, such as SAR11, Prochlorococcus, Rhodospirillales and Oceanospirillales. While ACE decreased the relative abundance of nutrient-rich phytoplankton (Bacillariophyta and Mamiellophyceae), resulted in more growth space for other eukaryotes that prefer oligotrophic environment (especially Fungi, Dictyochophyceae, and Synurophyceae). Canonical correlation analysis (CCA) showed temperature, salinity, nitrate, phosphate and nitrite had significantly affected on microbial community. The special environment of ACE (especially temperature) shaped the composition of its specific microbe. This study shed important light on the effect of ACEs on environmental conditions to impact marine ecosystem structure.
Mesoscale eddies are a ubiquitous phenomenon in the ocean. They range from tens to hundreds of kilometers in spatial scale, and have a temporal range of days to months (Chelton et al., 2011). Many studies have shown that mesoscale eddies play an important role in biogeochemical cycles, which they influence nutrient dynamics in the euphotic zone (Martin and Pondaven, 2003; Klein and Lapeyre, 2009). Some eddies are characterized by significant particulate carbon, particulate nitrogen and biogenic silica export, others show only enhanced biogenic Si export, or little to no enhancement of elemental particle fluxes (Bidigare et al., 2003; Benitez-Nelson et al., 2007; Zhou et al., 2020). Currently, there exists a relatively clear understanding of the effects of cyclonic eddy (CE) on ecosystems, with CE potentially increasing nutrient concentration, chlorophyll concentration, photosynthetic efficiency and productivity (Sweeney et al., 2003; Chen et al., 2007; Zhang et al., 2009, 2011). However, the effect of anticyclonic eddy on marine ecosystem is not well understood.
The opposite physical processes of anticyclonic eddy (ACE) could result in different environmental changes and nutrient responses (McGillicuddy et al., 1998; Oschlies and Garçon, 1998). Environment changes could lead to changes in the microbial community with different nutrient requirements, and the composition of primary producers in water bodies. The taxonomic diversity of the microeukaryotes significantly differed by eddy polarity (cyclonic versus anticyclonic) and between sampling seasons in the North Pacific Subtropical Gyre (Harke et al., 2021). Low heterotrophic biomass in the microbial food web exists in the center of ACE; in contrast, increases in microbial biomass and high chlorophyll a concentrations are observed at the ACE edge (Mizobata et al., 2002; Christaki et al., 2011; Xiu and Chai, 2011; Wang et al., 2018). Some previous studies have reported that ACE could cause changes in the abundance of some dominant microbial taxa, such as Prochlorococcus, diatoms and dinoflagellates (Zhou et al., 2013; Nishino et al., 2018; Mohan and Biju, 2020). Environmental conditions (especially nutrient availability) in the ACE waters have been prejudicial to phytoplankton, mainly triggered by downwelling, resulted in high cell mortality, which forced photosynthesized carbon to fuel the dissolved pool (Lasternas et al., 2013). Despite the observed differences in microbial distributions in the ACE, as only a few algae taxa were studied, the biological features appeared insufficient to independently characterize ACE and provide information on their microbial community dynamics.
Over the past decade, high-throughput sequencing (HTS) has highlighted the phylogenetic diversity of microbial communities through high-resolution. HTS techniques have been applied to understand the diversity of bacterial communities in the marine environment (Ladau et al., 2013; Sun et al., 2020b; Lu et al., 2021), and provided detailed studies of bacterial community composition and dynamics, which helps to determine relationships and potential interactions between microbial community and environment (Fuhrman et al., 2015). HTS methods can provide detailed information on the composition of phytoplankton communities, revealing the vast unknown diversity of eukaryotes, and have been successfully used in phytoplankton and harmful algal blooms studies (Dasilva et al., 2013; Sunagawa et al., 2015; Stefanidou et al., 2018; Sun et al., 2020a). In general, HTS methods could have the ability to assess microbial diversity and determine its ecological significance in the ocean ecosystem.
Mesoscale eddies are found throughout the SCS, especially the anticyclonic eddies shed from the Kuroshio can occur all year round (Wang et al., 2015). Previous studies have found that the complex topography and the kuroshio eddy shedding process are the two main reasons of eddy in the northern SCS, which are shed from Kuroshio intrusions into the SCS and formed anticyclonic loop (Li et al., 1998; Sangra et al., 2005). The Kuroshio carries quantity of heat and salt from low latitude to midlatitude into the northern SCS, affecting their physical and biogeochemical properties (Yang et al., 2019). Figure 1 showed the effect of Kuroshio on mesoscale eddies in the SCS during our investigation. However, the potential ecological effects of these eddies are not well understood. Further study of this scientific issue is helpful to understand the impact of ACE on the global marine biogeochemical cycle. In this study, we hypothesized that ACE not only affect specific microbial taxa, but also cause changes in the entire microbial community, as well as potential ecological functions. To meet this purpose, an anticyclone eddy was investigated to explore its characteristics of physicochemical properties and microbial community in the South China Sea. Through intensive sampling and high throughput sequencing of the 16S rRNA and 18 rRNA gene, the composition of the bacterial and eukaryotic community was identified, as well as the potential ecosystem effects, further elucidated the role of ACEs in the tropical oceans.
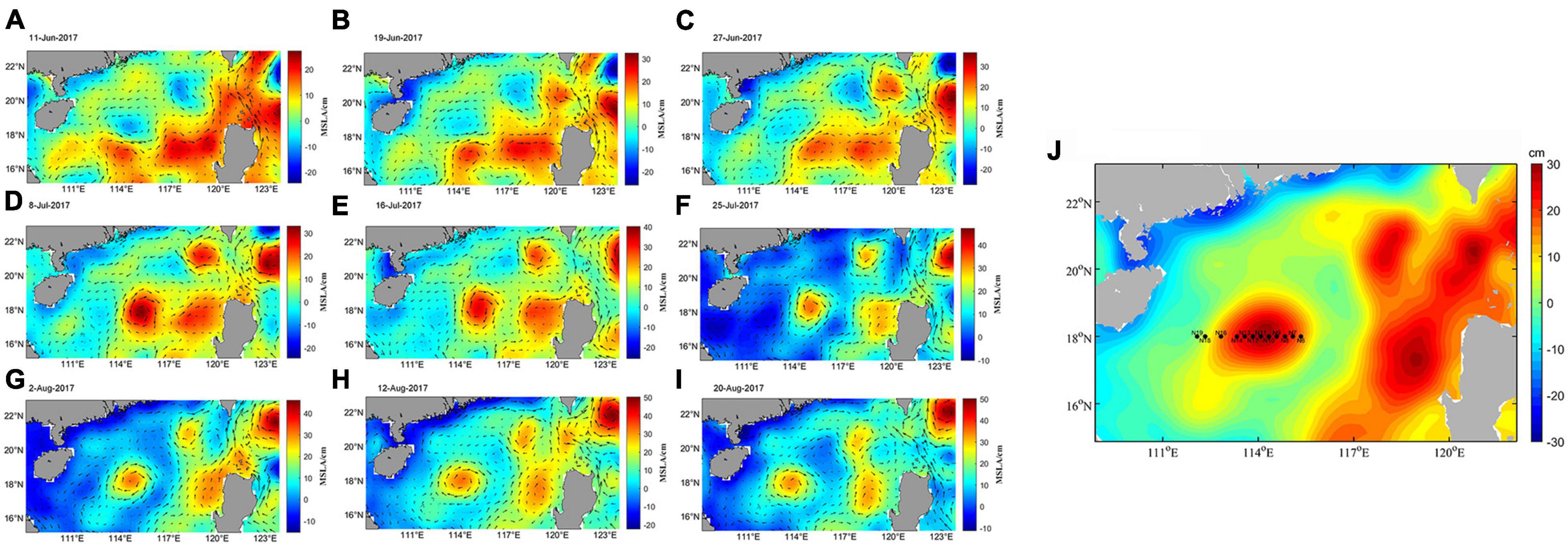
Figure 1. Evolution of the ACE detected by satellite sea surface height and geostrophic current for (A) 11 June, (B) 19 June, (C) 27 June, (D) 8 July, (E) 16 July, (F) 25 July, (G) 2 August, (H) 12 August, and (I) 20 August. (J) Sampling station of the ACE in 10–13 August, 2017; N6-N16 are located in ACE. N9, N10, N11, and N12 are located in the physical center of the eddy. N18 and N19 are located outside the eddy.
A survey cruise was undertaken in the South China Sea on the Shiyan 1 in 10–13 August, 2017 (Figure 1). Samples were collected along the ACE transect from 112 to 114 oE and 18 oN based on sea level anomalies. A conductivity-temperature-depth (CTD) system (SeaBird SBE-911 Plus, United States) was deployed to acquire hydrographic parameters. Seawater samples (12 stations) were collected at eight water depths (5, 25, 50, 75, 100, 150, 200, and 300 m) with CTD 12-L Niskin bottles (General Oceanics, Inc., Miami, FL, United States). After each sample was collected, 2,000 mL of water was filtered for bacterial community analysis and eukaryotic community analysis using polycarbonate membranes (EMD Millipore, United States) with a pore size of 0.22 μm. DNA from filtered membranes were extracted using the PowerWater DNA Isolation Kit (MoBio, United States) according to the DNA extraction protocol under sterile conditions to avoid contamination. Seawater for nutrient analysis (500 mL) was filtered through polycarbonate filters with a pore size of 0.45 μm, and immediately frozen (−20°C) until analysis. Nitrate (NO3), nitrite (NO2), ammonium (NH4), phosphate (PO4) and silicate (SiO4) concentrations were measured on a QuAAtro39 Auto-Analyzer (SEAL Analytical Ltd., United Kingdom). The analytical detection limited was 0.03, 0.05, 0.02, 0.02, and 0.45 μmol L–1 for NH4, NO3, NO2, PO4, and SiO4, respectively.
Sea-surface height anomalies (SSHA) were used to describe and quantify the position and characteristics of mesoscale features. SSHA fields consist of delayed-time reference series that are available online from AVISO1. These have been mapped to global Mercator grids with (1/4) × (1/4)° spatial resolution. The corresponding map for sampling was analyzed using Matlab R2015a (The MathWorks Inc., MA, United States) to create the SSHA map. Stations N6-N16 are located inside the ACE, and N9, N10, N11, and N12 are located in the physical center of the eddy. N18 and N19 are located outside the eddy.
The v3 and v4 regions of 16S rRNA were amplified by polymerase chain reaction (PCR) using the following primers: 5′- CCTACGGRRBGCASCAGKVRVGAAT -3′ (forward) and 5′- GGACTACNVGGGTWTCTAATCC -3′) (reverse) (Li et al., 2019). The PCR comprised a 20 μL reaction volume containing 10 ng of DNA template, 4 μL of 5 × FastPfu buffer, 2 μL of 2.5 mmol/L dNTPs, 0.5 μL of 5 U/μL FastPfu polymerase, 0.5 μL of primer (5.0 μmol/L). The remaining volume was made up with DNA-free water. The PCR reaction conditions were as follows: 95°C for 3 min, 30 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and 72°C for 10 min. 18S rRNA were amplified using the eukaryotic-specific V9 primers 1380F (5′-CCCTGCCHTTTGTACACAC-3′) and 1510R (5′-CCTTCYGCAGGTTCACCTAC-3′) (Amaral-Zettler et al., 2009). The PCR comprised a 20 μL reaction volume containing 4 μL of 5 × FastPfu buffer, 2 μL of 2.5 mmol/L dNTPs, 0.5 μL of 5 U/μL FastPfu polymerase, 0.5 μL of primer (5.0 μmol/L), and 10 ng DNA templates. PCR reaction conditions were as follows: 95°C for 3 min, 30 cycles at 95°C for 30 s, 57°C for 30 s, 72°C for 45 s, and 72°C for 10 min. A positive control and non-template control samples were run to validate PCR. Amplified PCR products were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States). A total of 124 DNA libraries were validated using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), and quantified using a Qubit 2.0 Fluorometer. DNA libraries were multiplexed with equimolar amounts and loaded on an Illumina MiSeq instrument following the manufacturer’s instructions (Illumina, San Diego, CA, United States). Sequencing was performed using a 2 × 300 paired-end configuration, and image analysis and base calling were conducted using the MiSeq Control Software embedded in the MiSeq instrument.
For pair-ended reads obtained by Illumina MiSeq sequencing, barcodes and primers were trimmed from paired-end sequences and then assembled using FLASH (Magoč and Salzberg, 2011). Low-quality reads (qaverage < 20, maxambig > 0 and length < 200 bp) were removed, and Chimera Check was used to remove chimeric sequences. High quality sequences were grouped into operational taxonomic units (OTUs) using the clustering program VSEARCH (version 1.9.6) at a 97% sequence identity. The Ribosomal Database Program (RDP) classifier was used to assign a taxonomic category to all OTUs at a confidence threshold of 0.8.
The representative sequence of each OTU (most abundant) was assigned using BLAST against Silva_123 16S rRNA database2 and the V9_PR2 18S rRNA database (Guillou et al., 2012). 16S rRNA OTUs identified as belonging to either chloroplasts or mitochondria, or which were unknown, were removed. A 18S rRNA OTUs assigned to Metazoan, Archaea and Bacteria were removed from the dataset.
Sequence was first rarefied, then calculated the alpha and beta diversity statistics with QIIME (version 1.9.1) after normalization of the lowest number of sequences. ANOSIM analysis were used to assess for significant differences (p < 0.05) in bacterial and eukaryotic OTUs between samples. Environmental, bacterial and eukaryotic community data representation was performed with ocean data view version 4.7.2 using the DIVA gridding function (Ocean Data View, 20153) to display the location of ACEs. Beta diversity was calculated using weighted UniFrac, principal coordinate analysis (PCoA) was performed with R software (version 3.3.1). Canonical correlation analysis (CCA) was generated using the R software (version 3.3.1) vegan package, and statistical significances of CCA were judged by performing PERMUTEST analysis of variance.
Satellite altimetry can be used to identify the location of ACEs in the study area. The SSHAs of ACEs are positive, whereas those of CEs are negative. Sea level anomalies showed the location of the ACE during this investigation (Figure 1). The vertical distribution of temperature, salinity, NO3, NO2, NH4, PO4, and SiO4 concentrations along the transect were shown in Figure 2. There was a trend of increased temperature, NH4 and SiO4 concentration inside the eddy, while PO4, NO3 and NO2 decreased in comparison to outside the eddy. The salinity in the center of eddy was 0.1 higher than that of the surrounding seawater.
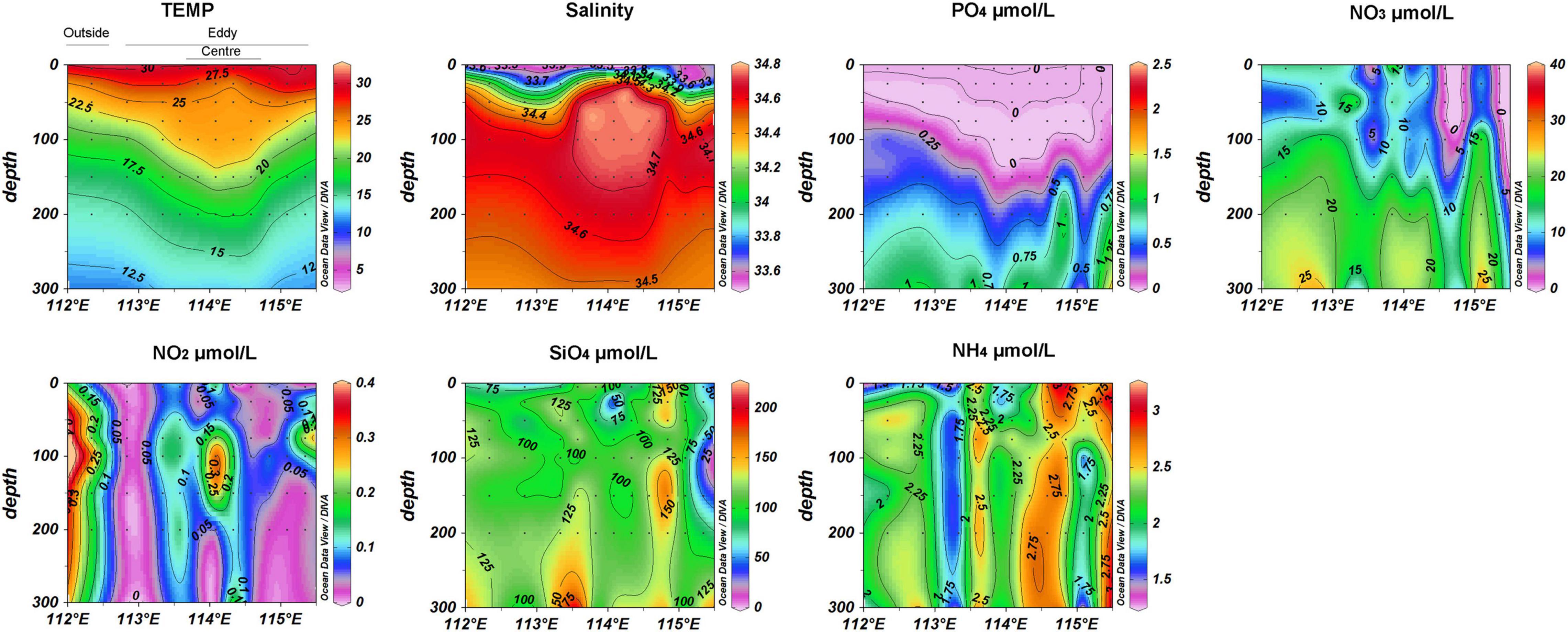
Figure 2. Temperature, salinity and inorganic nutrients (phosphate, nitrate, nitrite, silicate) assessed in a transect over the ACE. Black dots represent samples collected from different sites and depths. The sequence of station positions from right to left in each figure is as follows: N6, N7, N8, N9, N10, N11, N12, N13, N14, N16, N18, and N19.
The diversity of the bacterial and eukaryotic communities was evaluated using Chao and Shannon index (Figure 3). At 5, 25, and 50 m water layers, the diversity of bacterial community was significantly higher in the eddy than outside eddy. Whereas, in other water layers, the bacterial community diversity in the eddy was lower than that outside the eddy. Similar to the bacterial community, the diversity of eukaryotic communities was also significantly higher in the eddy than outside eddy at 5, 25, and 50 m water layers. Except these, the Chao index of eukaryotic communities was higher in the eddy than outside eddy at 100, 150, and 200 m depth. Overall, ACE increased the diversity of bacterial and eukaryotic communities in shallow water of ACE.
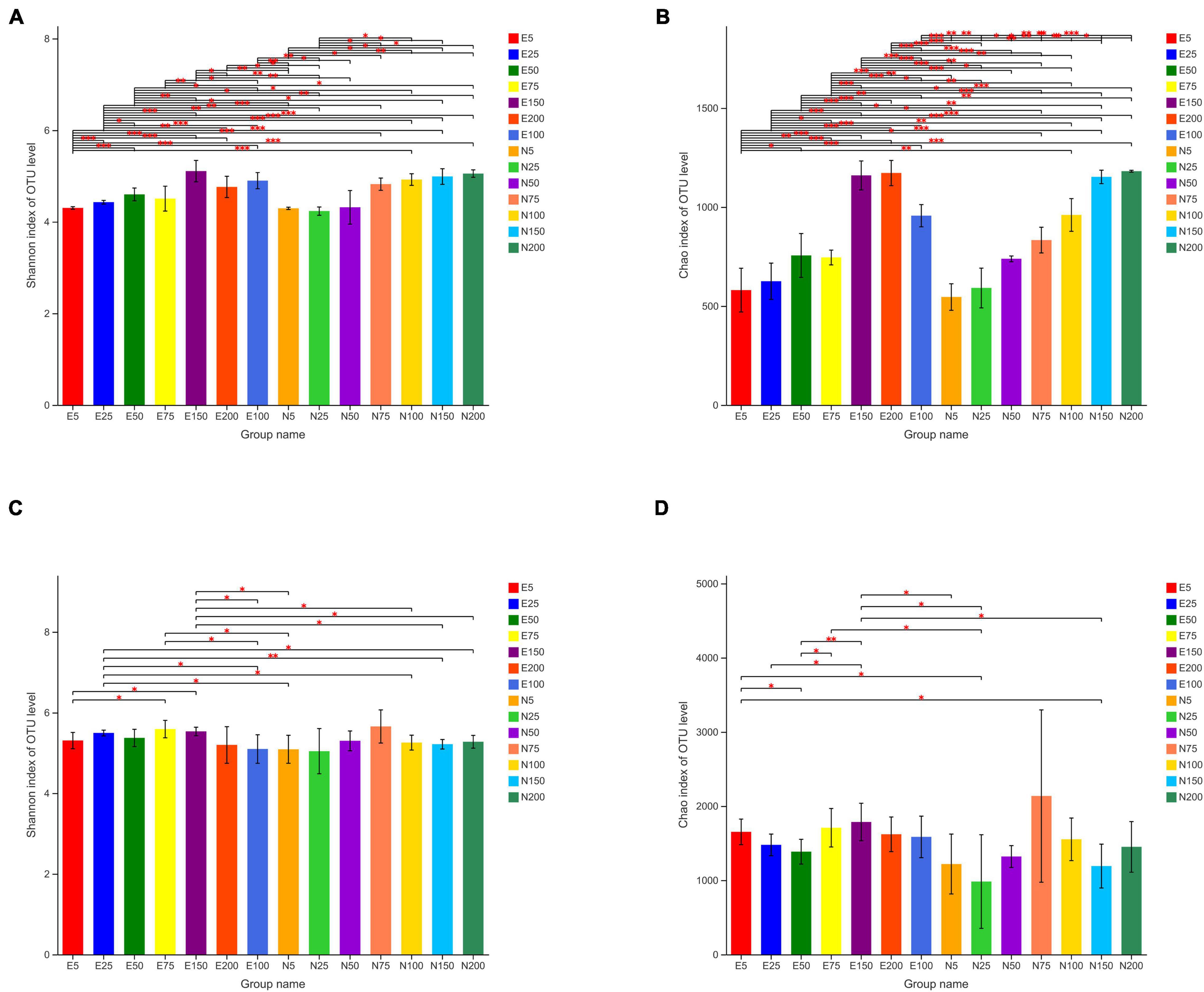
Figure 3. Shannon and Chao index of bacterial and eukaryotic OTUs inside and outside eddies. (A) Shannon index of bacterial OTUs; (B) Chao index of bacterial OTUs; (C) Shannon index of eukaryotic OTUs; (D) Chao index of eukaryotic OTUs; E5-E200 represents the water layer from 5 to 200 m within the eddy; N5-N200 represents the water layer from 5 to 200 m outside the eddy.
Principal component analysis (PCoA) analysis based on OTUs showed that microbial community of ACE was certain different from that of outside eddy (Figure 4). ANOSIM significance analysis showed that there were significant differences in bacterial communities (R = 0.834, p = 0.001) and eukaryotic communities (R = 0.565, p = 0.001) between the eddy and outside eddy in different water layers. As for bacteria, there were obvious differences of community among different water layers. There are obvious distances between the same water layers inside and outside eddy, especially 25, 50, 75, and 100 m. Compared with bacterial community, eukaryotic community is relatively far apart from each other in the same water layer. The differences of eukaryotic communities between eddy and outside eddy were not obvious as the bacteria, but obvious at 25, 50, and 75 m between the eddy and outside eddy.
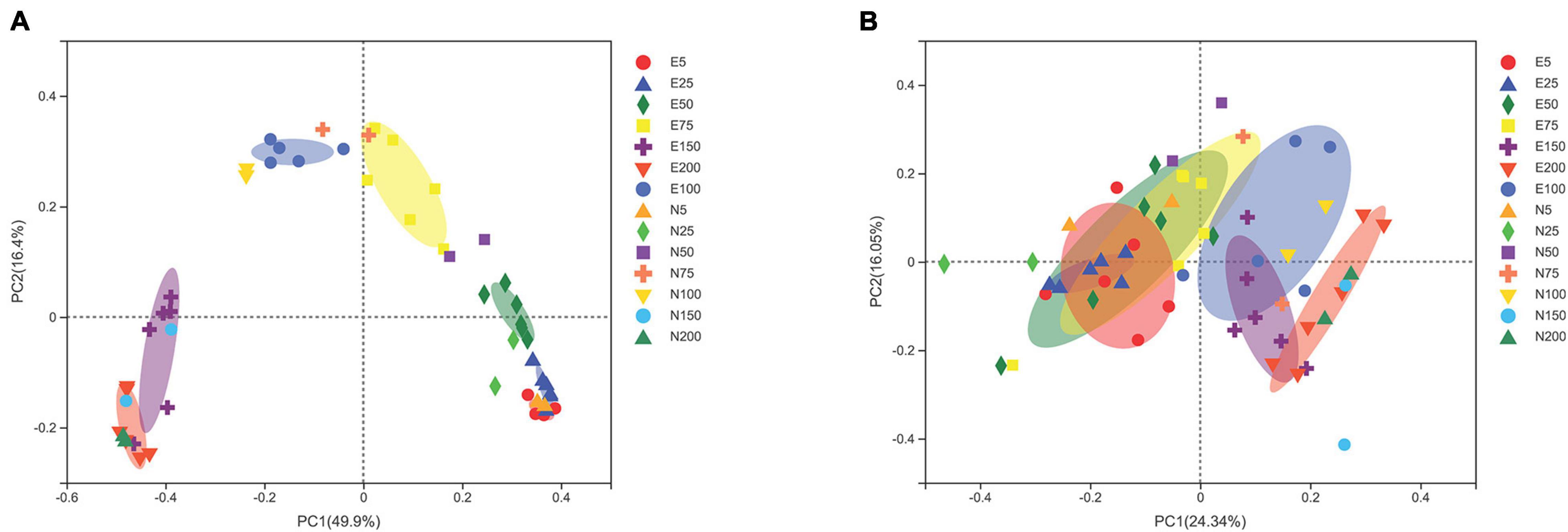
Figure 4. PCoA analysis of bacterial (A) and eukaryotic (B) community structure; E5-E200 represents the water layer from 5 to 200 m within the eddy; N5-N200 represents the water layer from 5 to 200 m outside the eddy.
Figure 4 showed the variation of bacterial communities inside and outside the eddy. Abundances of Alphaproteobacteria and Cyanobacteria were higher inside the ACE than outside the eddy and mainly occurred in waters above 150 m (Figure 5 and Supplementary Material 1). The relative abundance of Gammaproteobacteria and Chloroflexi were also higher inside the ACE than outside the eddy at depths greater than 100 m. However, relative abundance of Bacteroidetes and Actinobacteria had a decreasing trend inside, as opposed to outside the ACE. At the order level (Figure 5 and Supplementary Material 2), the most obvious change produced by the ACE was the increase in the relative abundance and vertical distribution of the SAR11 clade, Prochlorococcus, Oceanospirillales, Rhodospirillales, and Rhodobacterales. These bacteria were mainly recorded above 100 m. Vibrionales and Alteromonadales were also abundant at depths of 100–300 m of the ACE, but exhibited extremely low relative abundances outside the eddy. In contrast, Synechococcus and Flavobacteriales had a lower relative abundance inside the ACE compared with waters outside the eddy.
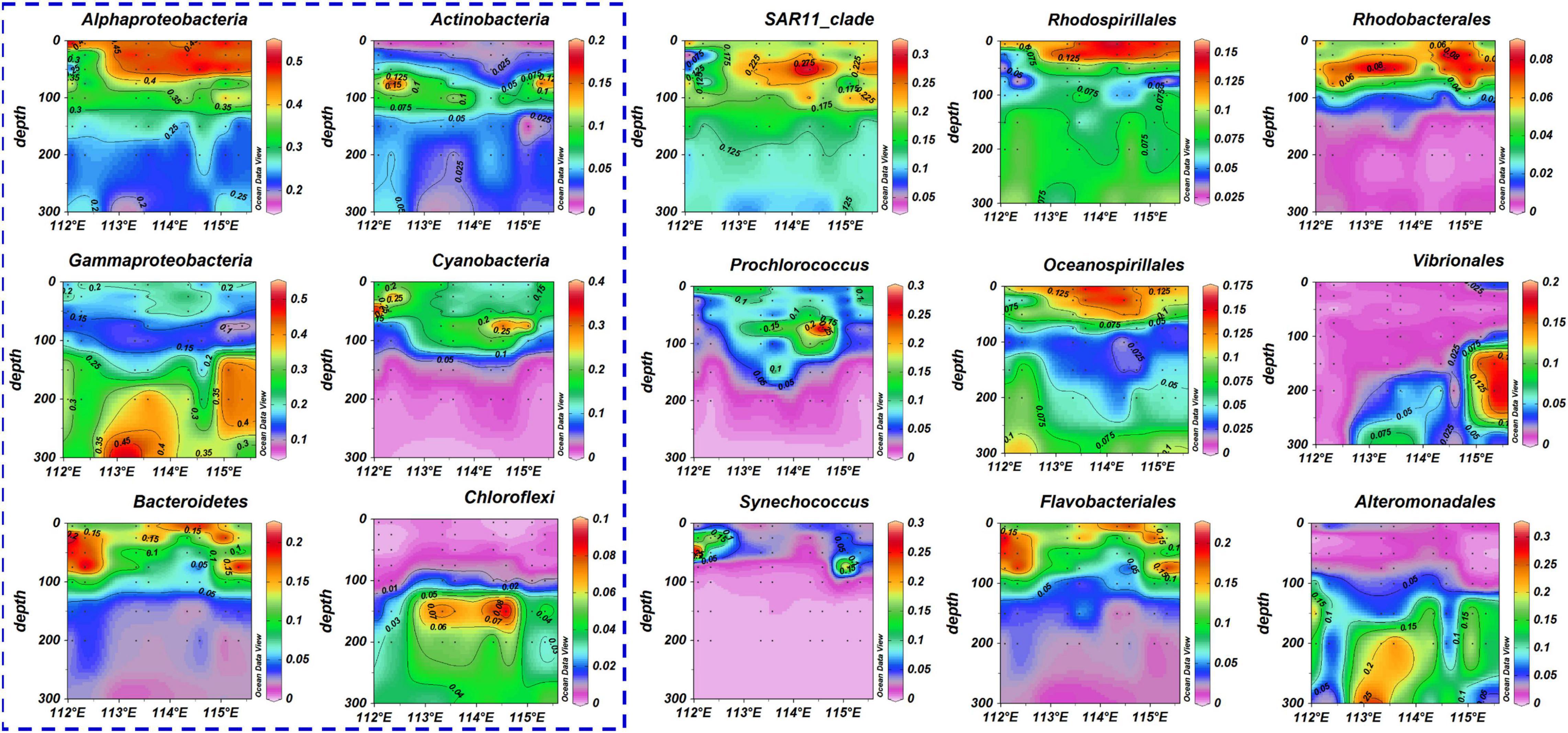
Figure 5. The distribution of the bacterial community at the phylum and order level (blue dotted boxes represent phylum classification). The values in the color bar represent the relative abundance of bacterial taxa and black dots represent samples collected from different stations and depths. The sequence of station positions from right to left in each figure is as follows: N6, N7, N9, N10, N13, N14, N18, and N19.
The dominant taxa Chlorophyta, Ochrophyta, and Haptophyta were more abundant inside than outside the ACE (Figure 6 and Supplementary Material 3). The highest values for dinoflagellates were found at 25 m depth of outside eddy station. Cryptophyta and Fungi were only detected in the center of the eddy, where they were present at high relative abundances. Stramenopiles_x was present at significantly lower relative abundances inside the ACE than in waters outside the eddy.
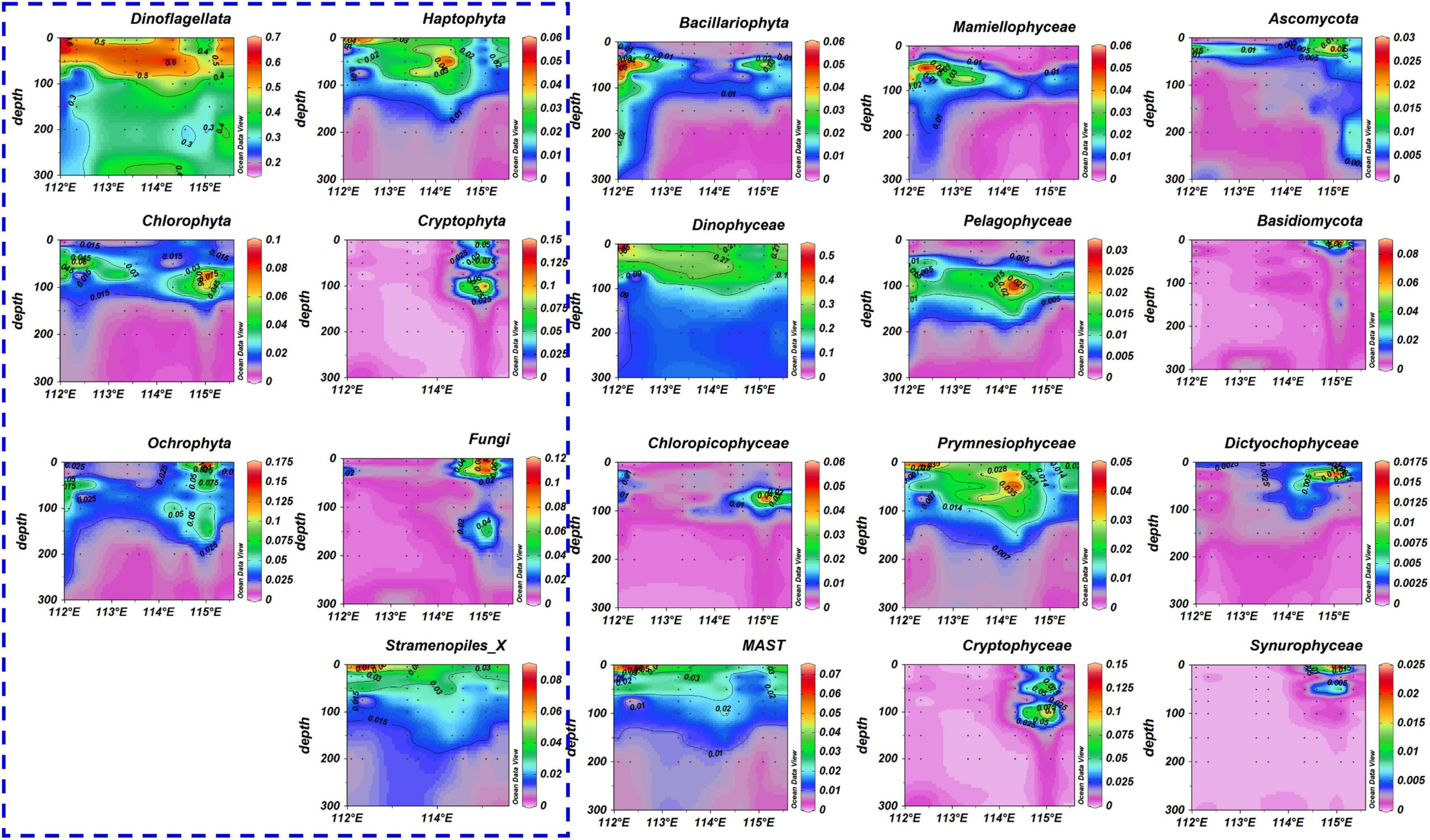
Figure 6. Distribution of the eukaryotic community at the phylum and order level (blue dotted boxes represent phylum classification). The values in the color bar represent the relative abundance of eukaryotic taxa and black dots represents samples collected from different stations and depths. The sequence of station positions from right to left in each figure is as follows: N6, N7, N9, N10, N13, N14, N18, and N19.
At the order level, the most obvious characteristic was the high relative abundance of eukaryotic taxa in the center of the eddy, including Chloropicophyceae, Ascomycota, Basidiomycota, Dictyochophyceae, and Synurophyceae, which were rarely detected in other areas (Figure 6 and Supplementary Material 4). Dinophyceae, MAST, and Prymnesiophyceae had a higher relative abundance outside the ACE than inside the eddy at 5 m depth. However, these taxa increased in relative abundance from 25 to150 m inside the eddy, in comparison to waters outside the eddy. Pelagophyceae had a higher abundance from 50 to150 m inside the eddy, compared with outside the eddy. The dominant taxa, Bacillariophyta and Mamiellophyceae exhibited a gradual decrease in relative abundance (5 – 300 m) from the outside of the eddy to the center of the eddy.
Canonical Correlation Analysis analysis was conducted to explore the relationship between environmental factors and microbial community (Figure 7). For bacteria (Figure 7A), temperature was significantly positively correlated (R2 = 0.76, p = 0.001) with the community in the ACE shallow layer (5, 25, and 50 m). The bacterial community was positively correlated with salinity, nitrate and phosphate (R2 = 0.38∼0.56, p = 0.001) below 100 m water. There was a significantly positive correlation between nitrite and bacterial community at 75 m water (R2 = 0.31, p = 0.004). Similar to bacteria community, temperature also exhibited positive correlation (r2 = 0.67, p = 0.001) with eukaryotic community (Figure 7B) in the ACE shallow layer (5, 25, and 50 m). Eukaryotic communities were positively correlated with salinity, nitrate and phosphate (p = 0.001) below 75 m water.
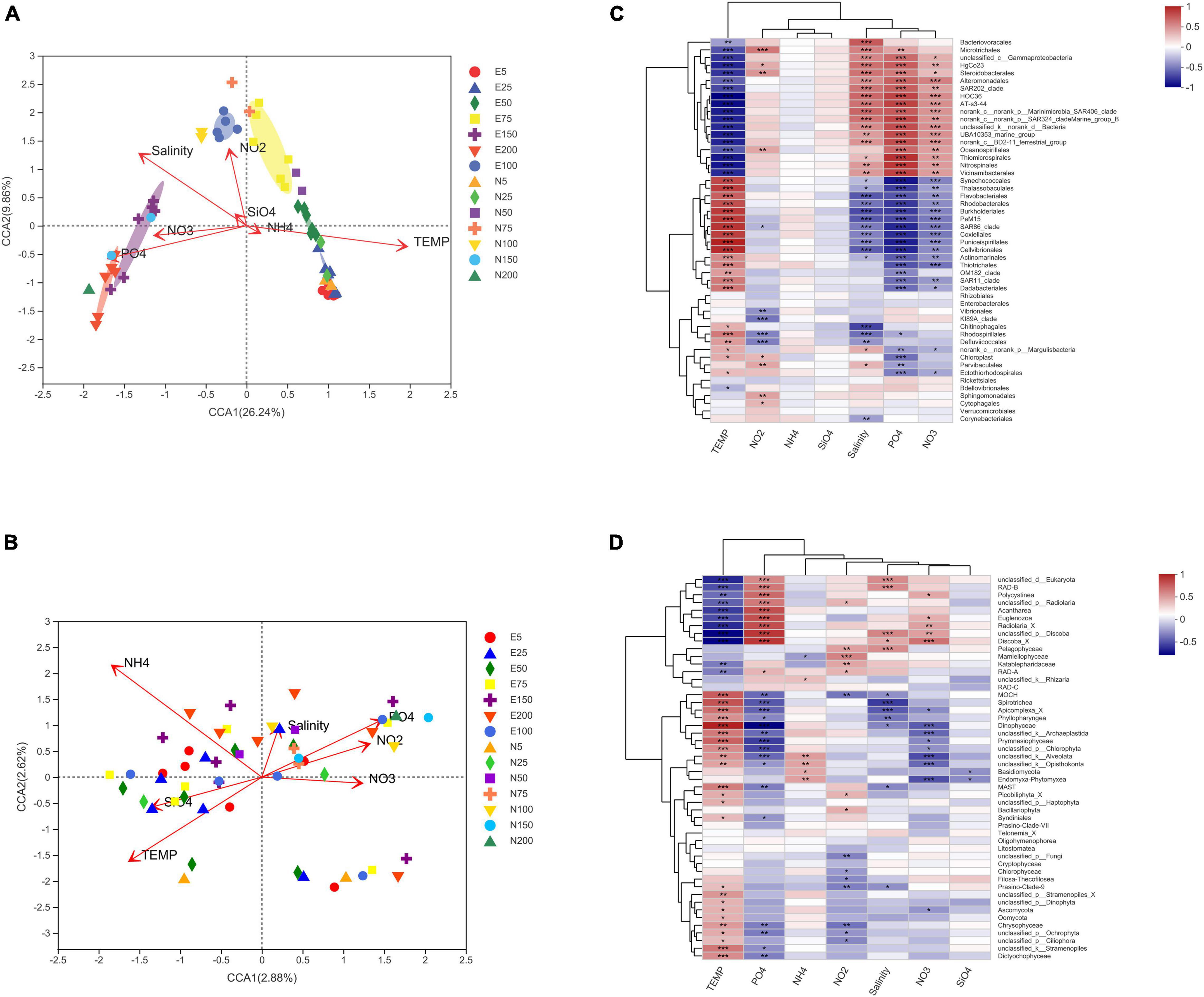
Figure 7. Canonical correlation analysis of environmental factors and bacterial (A) and eukaryotic (B) community; Correlation analysis of environmental factors and bacterial (C) and eukaryotic (D) community; E5-E200 represents the water layer from 5 to 200 m within the eddy; N5-N200 represents the water layer from 5 to 200 m outside the eddy.
Spearman correlation analysis reflects the ecological niche of dominant taxa and their correlation with environmental parameters. Bacterial taxa can be divided into two clusters (Figure 7C). Bacteria in the first cluster was enriched in the shallow layer of mesoscale eddy, such as Flavobacteriales, Rhodobacterales, SAR11 Clade, Prochlorococcus, Cellvibrionales and Rhodospirillales, which were mainly positively correlated with temperature. The second cluster of dominant taxa, which were distributed in relatively deep water, such as Alteromonadales, Nitrospinales, Salinisphaerales, Thermoplasmatales, SAR406 clade, SAR202 clade and Vibrionales, were negatively correlated with temperature, but positively correlated with salinity, phosphate and nitrate (p < 0.01).
For eukaryote, the effect of temperature on dominant taxa was more obvious (Figure 7D). Dominant eukaryotic taxa, such as MAST, Prymnesiophyceae, Dinophyceae, Prymnesiophyceae, Dictyochophyceae, and Ascomycota, which had significantly positive correlation with temperature (p < 0.01 or p < 0.05). Phosphate, nitrate and salinity (especially phosphate) were significantly correlated with eukaryotic taxa (p < 0.01 or p < 0.05). Phosphate was positively correlated with RAD-B, Polycystinea, Acantharea, Radiolaria_X, etc. Salinity was significantly positively correlated with RAD-B and Pelagophyceae. Interestingly, some dominant taxa, such as Basidiomycota, Cryptophyceae, Chloropicophyceae, and Synurophyceae were mainly found in the ACE center, had a low positive correlation with temperature. There is no significant correlation between these taxa and nutrient concentration (p > 0.05).
This study suggested a closely link between the ACE and the response of environmental parameters, bacterial and eukaryotic communities in the South China Sea. ACE has a great influence on the water environment, which leads to the changes of the microbial community composition. Temperature, salinity, nitrate, and phosphate were the main factors on microbial community in ACE. Environmental changes that drive by ACE increased the diversity of microbial community, induced the microbial community to differentiate, finally produce large impacts on ocean ecosystems and biogeochemical cycling.
Anticyclonic eddies has a great influence on marine environmental parameters, such as temperature, salinity and nutrients, which can be used as indicators of the existence of an anticyclonic eddy (Figure 2). Unlike CE, ACE can transport seawater from the surface to the deep ocean, increasing the temperature, and lowering the salinity and nutrient concentration (phosphate, nitrate, and nitrite) of deeper waters, as reported by previous studies (McGillicuddy et al., 1998; Woodward and Rees, 2001; Fong et al., 2008; Shin et al., 2011). The nitrite + nitrate is more than 1 μmol L–1 in surface layer in the South China Sea (Chow et al., 2021). Nitrate was about 5 μmol L–1 in surface layer in the South China Sea (Lu et al., 2021). Nitrate was 16 mmol m–3 in the near-surface values with the lowest values evident in Southern Ocean (Korb and Whitehouse, 2004).
Principal component analysis based on OTUs showed significant differences in bacterial and eukaryotic communities between eddy and outside eddy, indicating the important role of ACE in the change of microbial community. In terms of the variability in bacterial and eukaryotic community, temperature exhibited the most variation among environmental factors. CCA analysis indicated that the high temperature and oligotrophic environment in ACE were responsible for this difference (Figure 7). Temperature was characterized as a strong factor to drive bacterial community structure over time (El-Swais et al., 2015), eukaryotic community structure (Dasilva et al., 2013), as well as a global scale (Ladau et al., 2013; Sunagawa et al., 2015). Salinity could also act as a barrier to separate bacterial community, and has a significant impact on bacterial community structure (Pommier et al., 2007). Previous studies have shown that phosphorus availability plays an important role in the growth and activity of plankton, as required by heterotrophic bacteria in nutrient-poor marine environments (Fuller et al., 2005; Van Mooy et al., 2006). The current study indicated that ACEs change environmental parameters by downwelling, which reduces the differences in environmental parameters between water layers, thus increasing the similarity of biological community structure.
Another significant effect of ACE was increased the diversity of bacterial and eukaryotic communities above 50 m water layers (Figure 5). Due to the lack of relevant studies, we could speculate that the special environment, such as high temperature and low nutrient in ACE, might result in high diversity of microbial community. There are two possible reasons for this phenomenon. One of the mechanisms was that microbial communities were more active and diverse at higher temperature, which favoring different phylogenetic types when subjected to temperature. On the other hand, the ACE could inhibit the growth of relatively nutrient-rich phytoplankton, and resulted in more growth space for other algae and bacteria that prefer oligotrophic environment.
According to the results of microbial taxonomy, bacterial and eukaryotic cells tended to be miniaturized under the influence of ACE. Environment factors in ACE, especially temperature in this study, was identified as the most important driver on controlling the composition of the bacterial and eukaryotic community (Figure 7). In this study, the most significant changes in microbial communities occurred in shallow water above 100 m with temperatures above 22.5°C. The result of CCA indicate temperature is an important loading of variables. Temperature was reported as a key variable of taxonomic in the Red Sea (Thompson et al., 2017), and explained the most variability among significant environmental factors in the South China Sea (Zhang et al., 2018). Regardless of differences in inorganic nutrient loading, temperature was reported as the main impacting factor, enhancing graze on larger phytoplankton species and changing the outcome of coexistence and competition between different phytoplankton species, which induce a shift toward smaller cell sizes (Lewandowska and Sommer, 2010; Morán et al., 2010), and increase picoplanktonic Chlorophyceae and nanoplanktonic Haptophyceae, while decreasing large diatoms and dinoflagellates (Stefanidou et al., 2018). In this study, the high abundant eukaryotic taxa in the ACE, such as MAST, Prymnesiophyceae, Dinophyceae, Dictyochophyceae, Ascomycota are potential beneficiaries of the ACE physical-chemical changes.
For bacteria, it is obvious phenomena that the increase in the relative abundance and the depth distribution of small photosynthetic bacteria and heterotrophic bacteria above the 100 m water layer (Figure 5). These changes in bacterial communities will inevitably produce significant ecological effects. It has been reported that ACEs can reduce primary production by suppressing nutrient lines, thus, facilitating the development of an ecosystem with low biomass and small cells (Landry et al., 1998; Fernández et al., 2008). Some studies have reported a positive response of Prochlorococcus to the ACEs (Fernández et al., 2008; Baltar et al., 2010; Lasternas et al., 2013; Wang et al., 2018; Sun et al., 2021), indicating that Prochlorococcus may benefit from warmer and nutrient poor waters in the ACE. The minute size of Prochlorococcus and SAR11 allows them to maximize their uptake per unit biomass and to extract nutrients at the minuscule bulk concentrations characteristic of the ocean (Lauro et al., 2009). Given the high abundances and contributions of Prochlorococcus to primary production, these changes may have large impacts on ocean ecosystems and biogeochemical cycling (Flombaum et al., 2013).
Anticyclonic eddies increase the relative abundance of bacteria performing marine sulfur metabolism. SAR11 clade and Rhodobacterales, two groups associated with carbon and sulfur biogeochemical cycling (Malmstrom et al., 2004; Brinkhoff et al., 2008; Billerbeck et al., 2016), showed high abundance in ACE, suggesting that they were superior to other groups under warming conditions. In addition, SAR11 and Rhodobacterales are the main participants in the uptake and metabolism of carbon and sulfur compounds in algae, which are significantly correlated with many algae. The increase in the abundance of these algae may stimulate the growth of SAR11 and Rhodobacterales through produced dimethylsulfoniopropionate (DMSP), an important sulfur compound, which will inevitably lead to the enhancement of marine sulfur metabolism.
Phosphate could be the main limiting nutrient to microbial community in the ACE water. Compared with other nutrients, phosphates have a more significant deficiency in the eddy. ACE lead to phosphorus depletion, further resulting in phytoplankton species succession from diatoms to dinoflagellates and cyanobacteria (Ning et al., 2004). ACE enable microorganisms to differentiate according to their nutrient level requirements. Indeed, nutrients concentration in the deeper layer decreased, which resulted in more growth space for algae and bacteria that prefer oligotrophic environment. Nutrient levels have been shown to explain variability of microbial communities in the ACE waters (Figure 7), and the relationship between the upper eddy microbe, and deep-water microbe.
Physical process of downwelling in ACE could also play an important role in the evolution of microbial community. Other studies also reported that downwelling processes in ACE have been significantly prejudicial to phytoplankton, as indicated by high phytoplankton cell mortality of diatoms, and strongly induced a large POC or DOC release in the middle of ACE (Lasternas et al., 2013), which may be the reason for the increase in abundance of heterotrophs (bacteria and fungi) in the center of ACE. It has been reported previously that diatoms were transported laterally suffered from diminishing nutrient availability from the edge to the center of the eddy (Ning et al., 2004). Conversely, pico-phytoplankton can take advantage of its low nutritional needs and low mortality rate for sustainable growth (Baltar et al., 2010; Lasternas et al., 2013).
This study deepened the understanding of the effects of ACE on bacterial and eukaryotic community. As a common phenomenon in the global ocean, ACE provides evidence of the bacterial and eukaryotic communities in the center-outside and upper-deep layers of the eddy. This study provided insights into the internal mechanism of the changes of microbial community structure induced by ACE, which further promoted the understanding of the ecological effect of ACE. However, the changes of microbial functions need further study to better understand the impact of anticyclonic eddy on global marine ecosystem.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA540096 and https://www.ncbi.nlm.nih.gov/genbank/, PRJNA540355.
FS and MW contributed to conception and design of the study. FS wrote the first draft of the manuscript. FS, XX, MS, YSW, MW, SH, and CS performed data analysis and revised the manuscript. HZ, YTW, HC, and JF conducted survey and sampling analysis. All authors contributed to the article and approved the submitted version.
This study was supported financially by the Hainan Province Science and Technology Special Fund (ZDYF2021XDNY131), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0303 and GML2019ZD0305), the National Natural Science Foundation of China (31971480, 41406130, U1901211, 41890852, and 42073078), the National Key Research and Development Plan (2017FY100700), and the Project of Guangdong Science and Technology Department (2017A020216008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dong-Xiao Wang for organizing the cruise “Shiyan 1.”
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.773548/full#supplementary-material
Supplementary Material 1 | The distribution of the bacterial community at the phylum level in a table.
Supplementary Material 2 | The distribution of the bacterial community at the order level in a table.
Supplementary Material 3 | The distribution of the eukaryotic community at the phylum level in a table.
Supplementary Material 4 | The distribution of the eukaryotic community at the order level in a table.
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. doi: 10.1371/journal.pone.0006372
Baltar, F., Arístegui, J., Gasol, J. M., Lekunberri, I., and Herndl, G. J. (2010). Mesoscale eddies: hotspots of prokaryotic activity and differential community structure in the ocean. ISME J. 4, 975–988. doi: 10.1038/ismej.2010.33
Benitez-Nelson, C. R., Bidigare, R. R., Dickey, T. D., Landry, M. R., Leonard, C. L., Brown, S. L., et al. (2007). Mesoscale eddies drive increased silica export in the subtropical Pacific Ocean. Science 316, 1017–1021. doi: 10.1126/science.1136221
Bidigare, R. R., Benitez-Nelson, C., Leonard, C. L., Quay, P. D., Parsons, M. L., Foley, D. G., et al. (2003). Influence of a cyclonic eddy on microheterotroph biomass and carbon export in the lee of Hawaii. Geophys. Res. Lett. 30:1318. doi: 10.1029/2002gl016393
Billerbeck, S., Wemheuer, B., Voget, S., Poehlein, A., Giebel, H.-A., Brinkhoff, T., et al. (2016). Biogeography and environmental genomics of the Roseobacter-affiliated pelagic CHAB-I-5 lineage. Nat. Microbiol. 1:16063. doi: 10.1038/nmicrobiol.2016.63
Brinkhoff, T., Giebel, H.-A., and Simon, M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189, 531–539. doi: 10.1007/s00203-008-0353-y
Chelton, D. B., Schlax, M. G., and Samelson, R. M. (2011). Global observations of nonlinear mesoscale eddies. Prog. Oceanogr. 91, 167–216. doi: 10.1016/j.pocean.2011.01.002
Chen, Y.-L. L., Chen, H.-Y., Lin, I. I., Lee, M.-A., and Chang, J. (2007). Effects of cold eddy on Phytoplankton production and assemblages in Luzon Strait bordering the South China Sea. J. Oceanogr. 63, 671–683. doi: 10.1007/s10872-007-0059-9
Chow, C. H., Shih, Y.-Y., Chien, Y.-T., Chen, J. Y., Fan, N., Wu, W.-C., et al. (2021). The wind effect on biogeochemistry in eddy cores in the Northern South China Sea. Front. Mar. Sci. 8:717576. doi: 10.3389/fmars.2021.717576
Christaki, U., Van Wambeke, F., Lefevre, D., Lagaria, A., Prieur, L., Pujo-Pay, M., et al. (2011). The impact of anticyclonic mesoscale structures on microbial food webs in the Mediterranean Sea. Biogeosci. Discuss. 8, 185–220.
Dasilva, C. R., Li, W. K. W., and Lovejoy, C. (2013). Phylogenetic diversity of eukaryotic marine microbial plankton on the Scotian Shelf Northwestern Atlantic Ocean. J. Plankton Res. 36, 344–363. doi: 10.1093/plankt/fbt123
El-Swais, H., Dunn, K. A., Bielawski, J. P., Li, W. K. W., and Walsh, D. A. (2015). Seasonal assemblages and short-lived blooms in coastal north-west Atlantic Ocean bacterioplankton. Environ. Microbiol. 17, 3642–3661. doi: 10.1111/1462-2920.12629
Fernández, C., Thyssen, M., and Denis, M. (2008). Microbial community structure along 18°W (39°N–44.5°N) in the NE Atlantic in late summer 2001 (POMME programme). J. Mar. Syst. 71, 46–62. doi: 10.1016/j.jmarsys.2007.06.003
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N., et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Fong, A. A., Karl, D. M., Lukas, R., Letelier, R. M., Zehr, J. P., and Church, M. J. (2008). Nitrogen fixation in an anticyclonic eddy in the oligotrophic North Pacific Ocean. ISME J. 2, 663–676. doi: 10.1038/ismej.2008.22
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Fuller, N. J., West, N. J., Marie, D., Yallop, M., Rivlin, T., Post, A. F., et al. (2005). Dynamics of community structure and phosphate status of picocyanobacterial populations in the Gulf of Aqaba Red Sea. Limnol. Oceanogr. 50, 363–375. doi: 10.4319/lo.2005.50.1.0363
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., et al. (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Harke, M. J., Frischkorn, K. R., Hennon, G. M. M., Haley, S. T., Barone, B., Karl, D. M., et al. (2021). Microbial community transcriptional patterns vary in response to mesoscale forcing in the North Pacific Subtropical Gyre. Environ. Microbiol. 23, 4807–4822. doi: 10.1111/1462-2920.15677
Klein, P., and Lapeyre, G. (2009). The oceanic vertical pump induced by mesoscale and submesoscale turbulence. Annu. Rev. Mar. Sci. 1, 351–375. doi: 10.1146/annurev.marine.010908.163704
Korb, R. E., and Whitehouse, M. (2004). Contrasting primary production regimes around South Georgia, Southern Ocean: large blooms versus high nutrient, low chlorophyll waters. Deep Sea Res. Part I Oceanogr. Res. Pap. 51, 721–738. doi: 10.1016/j.dsr.2004.02.006
Ladau, J., Sharpton, T. J., Finucane, M. M., Jospin, G., Kembel, S. W., O’Dwyer, J., et al. (2013). Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 7, 1669–1677. doi: 10.1038/ismej.2013.37
Landry, M. R., Brown, S. L., Campbell, L., Constantinou, J., and Liu, H. (1998). Spatial patterns in phytoplankton growth and microzooplankton grazing in the Arabian Sea during monsoon forcing. Deep Sea Res. PT II Top. Stud. Oceanogr. 45, 2353–2368. doi: 10.1016/S0967-0645(98)00074-5
Lasternas, S., Piedeleu, M., Sangrà, P., Duarte, C. M., and Agustí, S. (2013). Forcing of dissolved organic carbon release by phytoplankton by anticyclonic mesoscale eddies in the subtropical NE Atlantic Ocean. Biogeosciences 10, 2129–2143. doi: 10.5194/bg-10-2129-2013
Lauro, F. M., McDougald, D., Thomas, T., Williams, T. J., Egan, S., Rice, S., et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533. doi: 10.1073/pnas.0903507106
Lewandowska, A., and Sommer, U. (2010). Climate change and the spring bloom: a mesocosm study on the influence of light and temperature on phytoplankton and mesozooplankton. Mar. Ecol. Prog. Ser. 405, 101–111.
Li, L., Nowlin, W. D., and Jilan, S. (1998). Anticyclonic rings from the Kuroshio in the South China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 45, 1469–1482. doi: 10.1016/S0967-0637(98)00026-0
Li, S., Chen, M., Chen, Y., Tong, J., Wang, L., Xu, Y., et al. (2019). Epibiotic bacterial community composition in red-tide dinoflagellate Akashiwo sanguinea culture under various growth conditions. FEMS Microbiol. Ecol. 95:fiz057. doi: 10.1093/femsec/fiz057
Lu, Y., Zhang, Y., Wang, J., Zhang, M., Wu, Y., Xiao, X., et al. (2021). Dynamics in Bacterial Community affected by Mesoscale Eddies in the Northern Slope of the South China Sea. Microb. Ecol. doi: 10.1007/s00248-021-01816-6
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Malmstrom, R. R., Kiene, R. P., Cottrell, M. T., and Kirchman, D. L. (2004). Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70, 4129–4135. doi: 10.1128/aem.70.7.4129-4135.2004
Martin, A. P., and Pondaven, P. (2003). On estimates for the vertical nitrate flux due to eddy pumping. J. Geophys. Res. Oceans 108:3359. doi: 10.1029/2003JC001841
McGillicuddy, D. J. Jr., Robinson, A. R., Siegel, D. A., Jannasch, H. W., Johnson, R., Dickey, T. D., et al. (1998). Influence of mesoscale eddies on new production in the Sargasso Sea. Nature 394:263. doi: 10.1038/28367
Mizobata, K., Saitoh, S. I., Shiomoto, A., Miyamura, T., Shiga, N., Imai, K., et al. (2002). Bering Sea cyclonic and anticyclonic eddies observed during summer 2000 and 2001. Prog. Oceanogr. 55, 65–75. doi: 10.1016/S0079-6611(02)00070-8
Mohan, A. P., and Biju, A. (2020). Influence of warm core eddy on the vertical distribution of autotrophic pico- and nanoplankton in the Bay of Bengal. Mar. Biol. Res. 16, 683–694. doi: 10.1080/17451000.2021.1872795
Morán, X. A. G., López-Urrutia, Á, Calvo-Díaz, A., and Li, W. K. W. (2010). Increasing importance of small phytoplankton in a warmer ocean. Glob. Change Biol. 16, 1137–1144. doi: 10.1111/j.1365-2486.2009.01960.x
Ning, X., Chai, F., Xue, H., Cai, Y., Liu, C., and Shi, J. (2004). Physical-biological oceanographic coupling influencing phytoplankton and primary production in the South China Sea. J. Geophys. Res. Oceans 109:C10005. doi: 10.1029/2004jc002365
Nishino, S., Kawaguchi, Y., Fujiwara, A., Shiozaki, T., Aoyama, M., Harada, N., et al. (2018). Biogeochemical anatomy of a Cyclonic Warm-Core Eddy in the Arctic Ocean. Geophys. Res. Lett. 45, 11284–11292. doi: 10.1029/2018gl079659
Oschlies, A., and Garçon, V. (1998). Eddy-induced enhancement of primary production in a model of the North Atlantic Ocean. Nature 394, 266–269. doi: 10.1038/28373
Pommier, T., Canback, B., Riemann, L., Bostrom, K. H., Simu, K., Lundberg, P., et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16, 867–880. doi: 10.1111/j.1365-294X.2006.03189.x
Sangra, P., Pelegri, J. L., Hernandez-Guerra, A., Arregui, I., Martin, J. M., Marrero-Diaz, A., et al. (2005). Life history of an anticyclonic eddy. J. Geophys. Res. Oceans 110:C002526. doi: 10.1029/2004jc002526
Shin, C.-W., Kim, D., Kim, K. H., Yoo, S., Yang, E. J., Park, J., et al. (2011). Impact of an anticyclonic eddy on the summer nutrient and chlorophyll a distributions in the Ulleung Basin, East Sea (Japan Sea). ICES J. Mar. Sci. 69, 23–29. doi: 10.1093/icesjms/fsr178
Stefanidou, N., Genitsaris, S., Lopez-Bautista, J., Sommer, U., and Moustaka-Gouni, M. (2018). Unicellular eukaryotic community response to temperature and salinity variation in mesocosm experiments. Front. Microbiol. 9:2444. doi: 10.3389/fmicb.2018.02444
Sun, F., Wang, C., Wang, Y., Tu, K., Zheng, Z., and Lin, X. (2020a). Diatom red tide significantly drive the changes of microbiome in mariculture ecosystem. Aquaculture 520:734742. doi: 10.1016/j.aquaculture.2019.734742
Sun, F., Wu, M., Wang, Y., Sun, C., and Xu, Z. (2020b). Diversity and potential function of bacterial communities in different upwelling systems. Estuar. Coast. Shelf. Sci. 237:106698. doi: 10.1016/j.ecss.2020.106698
Sun, F. L., Wang, Y. S., Wu, M. L., Sun, C. C., Jiang, Z. Y., Cheng, H., et al. (2021). Bacterial community variations in the South China Sea driven by different chemical conditions. Ecotoxicology 30, 1808–1815. doi: 10.1007/s10646-021-02455-w
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Sweeney, E. N., McGillicuddy, D. J., and Buesseler, K. O. (2003). Biogeochemical impacts due to mesoscale eddy activity in the Sargasso Sea as measured at the Bermuda Atlantic Time-series Study (BATS). Deep Sea Res. Part Ii Top. Stud. Oceanogr. 50, 3017–3039. doi: 10.1016/j.dsr2.2003.07.008
Thompson, L. R., Williams, G. J., Haroon, M. F., Shibl, A., Larsen, P., Shorenstein, J., et al. (2017). Metagenomic covariation along densely sampled environmental gradients in the Red Sea. ISME J. 11, 138–151. doi: 10.1038/ismej.2016.99
Van Mooy, B. A. S., Rocap, G., Fredricks, H. F., Evans, C. T., and Devol, A. H. (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc. Natl. Acad. Sci. U.S.A. 103, 8607–8612. doi: 10.1073/pnas.0600540103
Wang, L., Huang, B., Laws, E. A., Zhou, K., Liu, X., Xie, Y., et al. (2018). Anticyclonic eddy edge effects on phytoplankton communities and particle export in the northern South China Sea. J. Geophys. Res. Ocean 123, 7632–7650.
Wang, Q., Zeng, L., Zhou, W., Xie, Q., Cai, S., Yao, J., et al. (2015). Mesoscale eddies cases study at Xisha waters in the South China Sea in 2009/2010. J. Geophys. Res. Oceans 120, 517–532. doi: 10.1002/2014jc009814
Woodward, E. M. S., and Rees, A. P. (2001). Nutrient distributions in an anticyclonic eddy in the northeast Atlantic Ocean, with reference to nanomolar ammonium concentrations. Deep Sea Res. Pt II Top. Stud. Oceanogr. 48, 775–793. doi: 10.1016/S0967-0645(00)00097-7
Xiu, P., and Chai, F. (2011). Modeled biogeochemical responses to mesoscale eddies in the South China Sea. J. Geophys. Res. Ocean 116:C10006. doi: 10.1029/2010jc006800
Yang, Y., Wang, D., Wang, Q., Zeng, L., Xing, T., He, Y., et al. (2019). Eddy-induced transport of saline Kuroshio Water Into the Northern South China Sea. J. Geophys. Res. Oceans 124, 6673–6687. doi: 10.1029/2018JC014847
Zhang, Y., Jiao, N., Sun, Z., Hu, A., and Zheng, Q. (2011). Phylogenetic diversity of bacterial communities in South China Sea mesoscale cyclonic eddy perturbations. Res. Microbiol. 162, 320–329. doi: 10.1016/j.resmic.2010.12.006
Zhang, Y., Li, J., Cheng, X., Luo, Y., Mai, Z., and Zhang, S. (2018). Community differentiation of bacterioplankton in the epipelagic layer in the South China Sea. Ecol. Evol. 8, 4932–4948. doi: 10.1002/ece3.4064
Zhang, Y., Sintes, E., Chen, M., Zhang, Y., Dai, M., Jiao, N., et al. (2009). Role of mesoscale cyclonic eddies in the distribution and activity of Archaea and Bacteria in the South China Sea. Aquat. Microb. Ecol. 56, 65–79. doi: 10.3354/ame01324
Zhou, K., Dai, M., Kao, S.-J., Wang, L., Xiu, P., Chai, F., et al. (2013). Apparent enhancement of Th-234-based particle export associated with anticyclonic eddies. Earth Planet. Sci. Lett. 381, 198–209. doi: 10.1016/j.epsl.2013.07.039
Keywords: anticyclonic eddy, bacterial community, eukaryotic community, South China Sea, high throughput sequencing
Citation: Sun F, Xia X, Simon M, Wang Y, Zhao H, Sun C, Cheng H, Wang Y, Hu S, Fei J and Wu M (2022) Anticyclonic Eddy Driving Significant Changes in Prokaryotic and Eukaryotic Communities in the South China Sea. Front. Mar. Sci. 9:773548. doi: 10.3389/fmars.2022.773548
Received: 10 September 2021; Accepted: 14 January 2022;
Published: 24 February 2022.
Edited by:
Jun Sun, China University of Geosciences Wuhan, ChinaReviewed by:
Helga Do Rosario Gomes, Lamont-Doherty Earth Observatory (LDEO), United StatesCopyright © 2022 Sun, Xia, Simon, Wang, Zhao, Sun, Cheng, Wang, Hu, Fei and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meilin Wu, bWx3dUBzY3Npby5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.