- 1Fisheries and Oceans Canada, Nanaimo, BC, Canada
- 2Institute of Life and Earth Sciences, School of Energy, Geoscience, Infrastructure and Society, Heriot-Watt University, Edinburgh, United Kingdom
- 3NatureScot, Edinburgh, United Kingdom
- 4The School of Biological Sciences, University of Aberdeen, Aberdeen, United Kingdom
- 5Division of Molecular and Clinical Medicine, University of Dundee, Dundee, United Kingdom
- 6International Centre for Island Technology, Heriot Watt University, Stromness, United Kingdom
- 7Department of Life Sciences, Natural History Museum London, London, United Kingdom
Modiolus modiolus (horse mussel) reefs are an example of marine biodiversity hotspots of high conservation importance. Due to historical destruction and slow rates of recovery, the habitat is considered threatened and/or declining under the OSPAR Convention for the Protection of the Marine Environment of the North-East Atlantic 1992, and therefore incorporated into the conservation legislation of several countries. An analysis of genetic connectivity and diversity of nine M. modiolus reefs across Scotland, both within and outside of Marine Protected Areas (MPAs), was undertaken using 12 newly developed microsatellite markers. Analyses indicated moderate to high levels of genetic connectivity between all populations and significantly low genetic variance among populations. Generally, a lack of spatial genetic structure was determined though several populations were highlighted as potentially genetically separated. Structure and connectivity results were largely corroborated by network visualization which additionally identified several potentially key populations. All populations showed departure from Hardy–Weinberg Equilibrium (HWE) and positive inbreeding coefficients, suggesting reduced genetic diversity and/or reflecting the high frequency of null alleles observed across populations. However, allelic richness was uniformly high across all reefs compared to previously reported results for the habitat. Results broadly suggest that an open system of M. modiolus populations exists in Scottish waters and align with conclusions from prior larval dispersal modeling. Findings highlight that neither M. modiolus populations nor the MPAs where they are found should be considered discrete, independent entities and support the protection of features within MPAs in concert with non-designated areas and across varying spatial scales. It is proposed that potential for greater protection exists if all relevant Scottish MPAs, i.e., both those where M. modiolus reefs are a designated feature and those that host M. modiolus reefs, had statutory restrictions on all activities that cause damage to the sea bed. Such protection may facilitate the support of vulnerable populations by more resilient populations, particularly under climate change. Furthermore, given that a large number of unprotected M. modiolus populations may be important components in the interdependent system of reef populations, supplementary genetics studies informed by larval dispersal modeling are recommended to identify further key populations for safeguarding.
Introduction
Effective management is critical for ensuring environmental conservation, particularly for species or habitats already under threat or in decline due to current or historical human activity. Moreover, where such habitats provide ecosystem services, the need for sound management measures is magnified both in terms of the potential extent of impact and the urgency of response required. Marine biogenic habitats such as coral reefs, maerl beds, and shellfish reefs, for example, are species-rich hotspots (Rees et al., 2008; Sanderson et al., 2008; Ragnarsson and Burgos, 2012; Henry and Roberts, 2017; Riosmena-Rodríguez, 2017) that provide a range of ecosystem services including the potential to mitigate increases in atmospheric CO2 concentrations by acting as a sink for carbon in their skeletal structure (Ragnarsson and Burgos, 2012; Burrows et al., 2014, 2017) and associated sediments (Kent F. E. et al., 2017; Lee et al., 2020). Fragmentation and reduction of these habitats due to historical damage (Roberts et al., 2011; Cook et al., 2013; Bernard et al., 2019) coupled with the threat of climate change stressors (Gormley et al., 2013; Gormley K. S. et al., 2015), means that these habitats and their respective services will be severely compromised if sufficient protection, appropriate management measures and effective climate change mitigation are not implemented.
The marine bivalve, Modiolus modiolus (Linnaeus, 1,758) (horse mussel) is a reef-forming Arctic-Boreal species with a distribution range that extends from the seas around Scandinavia and Iceland southward to the Bay of Biscay (Rees, 2009), and along Pacific and Atlantic coasts (e.g., Bay of Fundy) in North America (Brown, 1984). M. modiolus is predominantly a sublittoral semi-infaunal species though intertidal populations are occasionally encountered in shallow pools and damp cervices along the lower reaches of the intertidal region (Seed and Brown, 1977). The species is generally long-lived (e.g., 50+ years in several North-East Atlantic populations) and has maximum shell lengths varying from 65 to 210 mm depending on location (e.g., Anwar et al., 1990; Dinesen and Morton, 2014; Brash et al., 2018; Fariñas-Franco and Roberts, 2018). M. modiolus reefs are subtidal, typically occurring at 10–35 m depth (Mair et al., 2000, 2010; Hirst et al., 2012) in fully saline conditions and tide-swept areas, though a smaller proportion can also be found in sheltered bays, fjords, or lochs (Rees, 2009), where temperature and salinity regimes may vary. Likewise, considerable temperature variation exists across the latitudinal range with sea surface and bottom temperatures ranging from 4°C to as high as 19°C over the course of the year (Reid et al., 2010).
M. modiolus reefs are characterized by high species diversity (Rees et al., 2008; Sanderson et al., 2008; Ragnarsson and Burgos, 2012; Buzeta, 2014), and habitat creation by M. modiolus has substantial effects on the composition and abundance of mega-faunal benthic organisms in coastal waters (Ragnarsson and Burgos, 2012; Robinson et al., 2012). As with other bivalve-dominated habitats, M. modiolus reefs also contribute to a number of ecosystem services including carbon storage (Burrows et al., 2017), benthic-pelagic coupling (Kent F. E. et al., 2017), water filtration (Navarro and Thompson, 1996) and habitat provision for commercially important species (Rees, 2009; Kent et al., 2016; Kent F. E. A. et al., 2017). Consequently, M. modiolus biogenic reefs are a conservation priority under Scottish [e.g., Marine (Scotland) Act, 2010], United Kingdom (e.g., United Kingdom Post-2010 Biodiversity Framework) and international (e.g., OSPAR) legislation (OSPAR, 2021). North American populations are yet to be classified under conservation measures, though Bay of Fundy reefs in Canada are currently being considered for protection under the country’s Sensitive Benthic Area (SBA) designation (Buzeta, 2014; DFO, 2015).
M. modiolus reefs are sensitive to physical impacts (Cook et al., 2013) and historical dredging and trawling of seabed areas have caused widespread and long-term damage to reefs in Europe (e.g., Rees, 2009; Strain et al., 2012) and North America (e.g., Bay of Fundy) (Kenchington et al., 2007; Sameoto et al., 2021). The combination of physical damage and slow recovery of the habitat (Mazik et al., 2015) has resulted in decline in the extent of reefs at a number of places across the species’ range (e.g., Rees et al., 2008; Strain et al., 2012; Cook et al., 2013; Wilson et al., 2021). Consequently, M. modiolus is listed as threatened and/or in decline across all OSPAR regions of Europe under the OSPAR Convention for the Protection of the Marine Environment of the North-east Atlantic 1992 (Rees et al., 2008). Further loss of M. modiolus reef habitat is predicted over the next 100 years due to warming of the ocean under climate change (Gormley et al., 2013; Gormley K. S. et al., 2015).
Connectivity, or the linking of spatially discrete populations via the exchange of genetic material, organisms, nutrients and/or energy (Balbar and Metaxas, 2019), is a key consideration for management of separate populations across different geographic areas. This is particularly relevant when identifying where Marine Protected Areas (MPAs) are required in order to ensure adequate inter-connectivity between populations through larval dispersal and for applying appropriate management measures within or outside of individual MPAs. Clarification of the relationship between reefs can aid in determining the extent to which a population within an MPA acts independently or is dependent on populations in other MPAs or outside protected areas (Juffe-Bignoli et al., 2014). The NE Atlantic European states are committed to creating an ‘ecologically coherent network’ of MPAs in accordance with the OSPAR Convention, which highlights connectivity as a key element to assist the interpretation of ecological coherence. Likewise, connectivity has been considered a fundamental element in the development of the Scottish MPA network (Marine Scotland, 2011; MCCIP, 2018). However, whilst regularly touted as a vital component of effective MPA networks, in practice connectivity has been largely ignored, risking that many MPA networks may be ineffective in providing the necessary level of connectivity for them to function as true ecologically coherent networks (Balbar and Metaxas, 2019).
The current study focuses on the genetic aspect of connectivity, where genetic connectivity is defined as “the extent to which populations in different parts of a species’ range are linked by the exchange of larvae, recruits, juveniles or adults” (Palumbi, 2003; i.e., vectors of genetic material). Genetic connectivity has become a major factor in determining and defining the implications of threats to marine biodiversity and is crucial to the MPA network design and decision-making process (Weersing and Toonen, 2009; Schunter et al., 2011). Terrestrial studies have also shown that high gene flow can mitigate against loss of genetic diversity, even in degraded landscapes (Lowe et al., 2005). Consequently, it is an especially critical component for achieving effective protection and management of species of high conservation importance (Lowe and Allendorf, 2010; Carr et al., 2017).
Marine environments have the potential added advantage of large-scale water movements that support relatively high connectivity between populations compared to terrestrial systems. However, where gene flow is not facilitated by adult life stages but rather by larval dispersal, information on larval behavior, longevity and mortality is required to refine estimates of the scale at which populations are connected (Cowen et al., 2000). Marine biogenic habitats such as M. modiolus reefs comprising conglomerations of sessile individuals, for example, are maintained through the physical dispersal of larvae which dictates levels of genetic connectivity and the genetic structure of populations across space and time. The species is predominantly dioecious with occasional hermaphrodites (Wiborg, 1946; Jasim and Brand, 1989; Fariñas-Franco and Roberts, 2018) and age to sexual maturity has been shown to vary between Norwegian, Canadian, Northern Irish and Manx studies from 3 to 8 years of age (see Dinesen and Morton, 2014). Variation in spawning appears to depend on latitude and site-specific conditions with seasonal events, inactive years, prolonged ‘trickle-spawning’ and synchronized epidemic spawning all reported (see De Schweinitz and Lutz, 1976; Dinesen and Morton, 2014 and references therein). During spawning events, adults release eggs and sperm into the water, and the resulting planktotrophic larvae disperse and mature over approximately 30 days to a stage where they are competent to settle (De Schweinitz and Lutz, 1976; Dinesen and Morton, 2014).
For species with a planktotrophic larval stage, gene flow occurs when larvae from a ‘source’ population are dispersed and then recruited into other ‘sink’ populations (Elsäßer et al., 2013). The degree to which individual populations self-recruit or act as larval sources or sinks has critical consequences for maintenance of ecological processes that shape population structure and support population resilience (Cowen and Sponaugle, 2009). Additionally, populations may act as stepping-stone ‘sink and source’ populations which provide important genetic linkages between more remote/distant populations, thus ensuring the connection of populations to each other via larval exchange over multiple generations (Fowler et al., 2019), which is an essential requirement of an effective MPA network (Carr et al., 2017). Indeed, the marine stepping-stone effect has been highlighted as an important mechanism with regards shaping the distribution of a number of marine species with pelagic larval dispersal (Adams et al., 2014; Coolen et al., 2020). Ultimately, whether populations are self-recruiting, require external input of larvae or exist in a stepping-stone array, understanding the dynamics of larval movement and connectivity patterns is critical to MPA network design (Palumbi, 2003; Carr et al., 2017).
There is a tendency toward an assumption of ‘open’ systems of population connectivity, i.e., that larval connectivity is assumed and that there are sufficient ocean currents and mixing. However, there may be an overestimation of connectivity potential between populations. Indeed, modeling that has included the effect of larval mortality and diffusion has illustrated marked decreases in transport potential and highlighted the importance of local retention in maintaining populations (Cowen et al., 2000). Likewise, there is ample evidence of high levels of self-recruitment in marine invertebrate species, even when barriers to dispersal are seemingly absent (Becker et al., 2007; Teske et al., 2016). Bearing this in mind, determining the open versus closed nature of populations is essential to effective marine spatial management, especially with regards to selection of MPAs and wider environmental management measures.
Scotland supports more than 80% of all M. modiolus reefs in the United Kingdom with populations existing in various regions of Scotland’s coastal waters, though most notably along the west coast and running north to the Orkney Islands and Shetland Isles. The Scottish coastline is complex with a bathymetry ranging from deep sea to shallow inshore sea lochs, and circulation patterns that vary over time and space. Large-scale circulation patterns are residually unidirectional (south to north, clockwise) and highly variable, with notable areas of high tidal flow between the mainland and islands. Likewise, small scale flow regimes vary substantially between regions (Gallego et al., 2013; Millar et al., 2019; Rabe et al., 2020). This complex hydrography of the Scottish coastal waters likely promotes varying levels of connectivity or isolation across reef populations.
In Scotland, M. modiolus reef habitat is classified as a Priority Marine Feature (PMF) due to its functional importance, sensitivity and low recovery potential (Tyler-Walters et al., 2016), and a number of reefs fall within the Scottish network of MPAs which comprises Special Areas of Conservation (SACs), Nature Conservation MPAs (NC MPAs), marine components of Sites of Special Scientific Interest (SSSIs), Special Protection Areas (SPAs), and Ramsar sites (Scottish Natural Heritage [SNH] and The Joint Nature Conservation Committee [JNCC], 2012). Across the MPA network, M. modiolus reefs are a protected feature of a number of NC MPAs, designated under the Marine (Scotland) Act 2010 to protect one or more habitats and/or species found on the Scottish Government’s list of PMFs. A number of M. modiolus reefs are also protected as ‘biogenic reefs’ in several SACs. However, it is important to note that protection is not afforded in full as bottom fishing (e.g., scallop dredging, creeling, hand-diving for scallops) is still permitted in MPAs where consultations on fishing activity have not yet been completed. Additionally, where consultations have been undertaken some forms of fishing are still permitted (e.g., creeling, hand-diving for scallops).
The aim of this study was to gain a greater understanding of the nature of the interrelationship and connectedness of the system of Scottish M. modiolus reefs (e.g., open versus closed) and to clarify the extent and potential role of MPAs in maintaining populations. The key objectives were: (i) to determine the genetic structure of M. modiolus reef populations situated in coastal waters off the mainland and islands of Scotland, including selected MPAs; and (ii) to determine levels (e.g., low, moderate, and high) of genetic connectivity between such populations. Results are considered in relation to recent M. modiolus larval dispersal modeling for a similar geographical area (Millar et al., 2019). The findings are designed to provide an indication of whether reefs falling within MPAs (if properly protected) are adequate to contribute to the long-term survival of other reefs (i.e., outside of MPAs) and will highlight where potential changes to the current MPA network may be warranted, especially under the context of climate change. This will support the development of effective management of MPAs toward an ecologically coherent network with increased resilience to climate change.
Materials and Methods
Site Selection
Nine M. modiolus reef sites were selected for genetic screening based on their known presence, accessibility for hand collection using scuba (depth) and geographical coverage (i.e., covering a range of latitudes within Scotland as well as east and west coast populations). Sample sites were chosen within three distinct geographical regions, namely the Shetland Isles, the North-east and Orkney Islands, and the West Coast, and included open coast sites and enclosed sea lochs to allow investigation at differing spatial scales and varying oceanographic conditions (Figure 1 and Table 1). Within each geographical region, M. modiolus reef sites ranged from as little as 10 km apart (e.g., Loch Creran to Port Appin) to much greater distances (e.g., Karlsruhe to Dornoch Firth: 140 km) (Figure 1). Sample sites across the entire study area were up to 700 km apart (e.g., Loch Creran, West Coast to Hascosay Sound, Shetland Isles). Sampling also targeted populations within and outside of MPAs with at least one protected site with M. modiolus reef as a designated feature in each region. While widely dispersed individual M. modiolus may contribute toward genetic connectivity, they were not included in the sampling strategy as reefs are considered key recruitment and settlement sites (Fariñas-Franco and Roberts, 2018). In addition to examination of genetic connectivity, the Noss Head site (Figure 1) was also selected for consideration of self-recruitment potential as it is the largest (3.85 km2) and most abundant known M. modiolus reef in Scotland (Hirst et al., 2012).
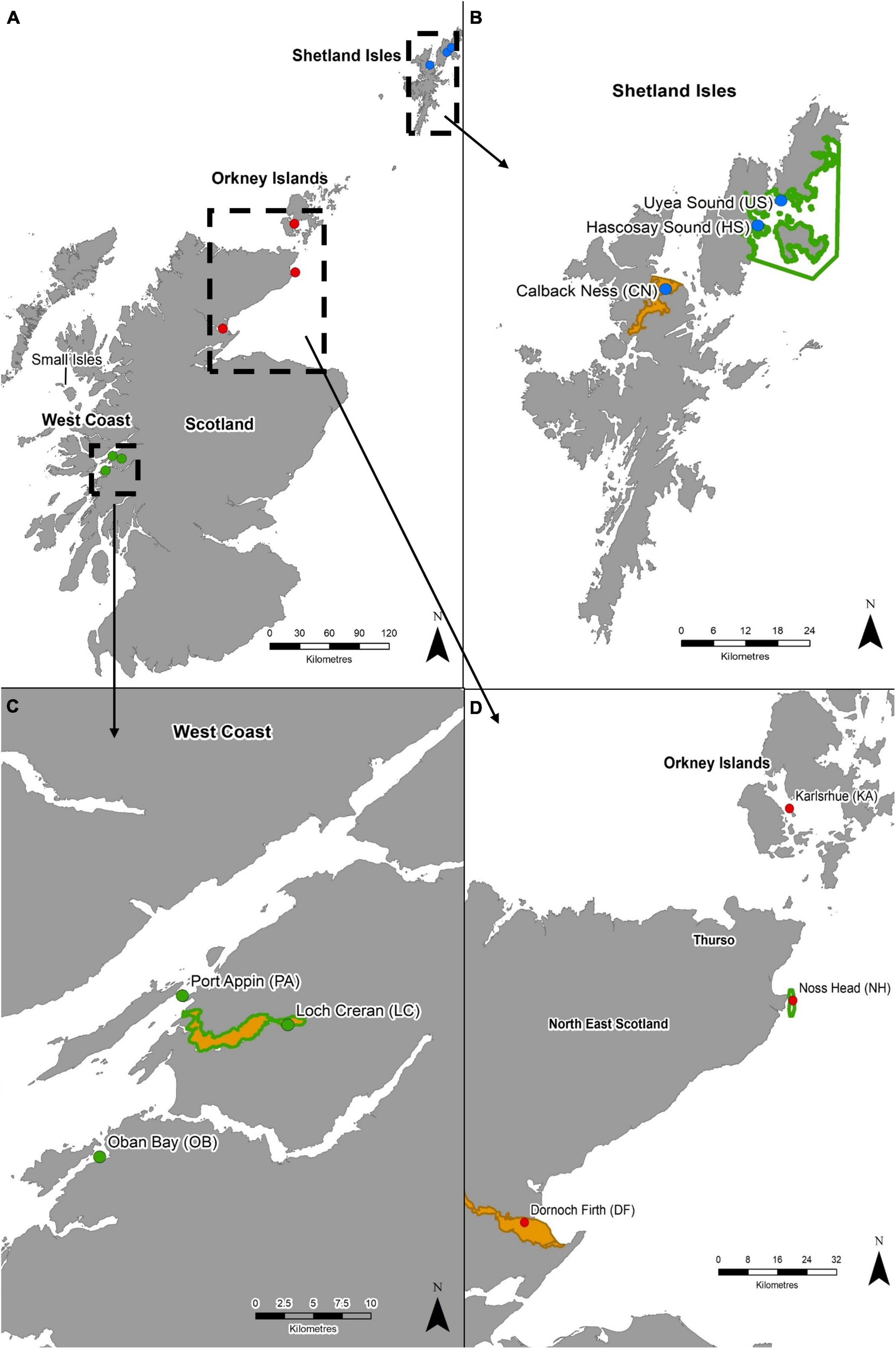
Figure 1. (A) Modiolus modiolus reef populations of Scotland included in genetic analyses. Dashed boxes indicate sampling regions: (B) Shetland Isles, (C) West Coast, and (D) North-east and Orkney Islands. Protected areas are shown as solid orange polygons (SACs) and green polygons (NC MPAs). Coastline Crown © and database right (2021). Ordnance Survey License number 100017908.

Table 1. Names and locations (latitude and longitude) of Modiolus modiolus reefs included in genetic connectivity analyses for three areas in Scotland: (a) Shetland Isles, (b) West Coast, and (c) North-east and Orkney Islands.
Sample Collection
Adult (>80 mm shell length) M. modiolus (n = 50) were hand collected by the Heriot-Watt Scientific Dive Team from selected sites between 2011 and 2016 (Table 1). Additionally, juvenile (<40 mm shell length, as per Anwar et al., 1990) M. modiolus (n = 50) were collected from Noss Head in order to examine the influence of life-stage to genetic connectivity and diversity. Adductor muscle tissue was dissected from each individual, cut into several small pieces, and preserved in approximately 15–20 ml 96% ethanol solution in 25 ml specimen tubes. Samples were stored at 4°C prior to DNA extraction.
Microsatellite Screening
DNA extractions were carried out with Qiagen DNeasy Blood & Tissue Kits (Qiagen, Manchester, United Kingdom) according to the Qiagen protocol (Qiagen, 2006). Presence of high-quality DNA was confirmed by gel electrophoresis. Following biophotometric quantification, samples were diluted to 20 ng/μl with Tris-EDTA buffer solution (pH 8.0) for running in multiplex PCRs. Multiplex Manager software (Holleley and Geerts, 2009) was used to design and optimize multiplex PCRs. Microsatellite markers included four newly developed by Gormley K. S. G. et al. (2015) and eight newly developed by Preziosi and Rowntree Labs, Faculty of Life Sciences, Manchester University (Table 2). Three multiplex PCR reactions were designed with four sets of microsatellite markers used per reaction. All primer sets (Eurofins Genomics; 0.05 μmol, salt-free, lyophilized) were made up to 100 μM in Tris-EDTA buffer solution (pH 8.0) and stored at −20°C until used. Forward markers were labeled with unique fluorescent dyes (NED, HEX, FAM, and PET dyes) to distinguish products within a given set.
Table 2. Microsatellite markers used in genetic analyses including forward and reverse primer sequences, amplicon size range, repeat motif, annealing temperature and marker developer.
Multiplex PCR reactions were carried out using the Qiagen Type-it Microsatellite PCR Kit (Qiagen, Manchester, United Kingdom) and protocol, but prepared to a final reaction volume of 5 μl to extend consumables, comprising 2.5 μl Type-IT Master Mix, 1.5 μl H2O (molecular grade), 0.5 μl primer mix, and 0.5 μl normalized DNA template. All reactions were run on a G-Storm thermocycler (Kapa Biosystems, Wilmington, NC, United States) with a heating period of 95°C for 5 min followed by 35 cycles of: 95°C for 30 s; 60°C for 90 s; 72°C for 30 s; and a final step at 60°C for 30 min. A sub-sample (10%) of each set of PCR samples was run on PCR gel to confirm successful amplification. Genotyping of PCR product was carried out at the University of Dundee where fragments were run alongside a ROX500 size standard in an ABI 3130 Genetic Analyser. Output files were received in FSA file format and Peak Scanner (Version 2; Applied Biosystems, Foster City, CA, United States) software was used to score peaks.
Data Analyses
Genetic Connectivity
Peak data output files were applied to FreeNA (Chapuis and Estoup, 2007) to determine genetic differentiation [FST of Weir (1996)]. Pairwise FST values were calculated with and without ENA correction as described in Chapuis and Estoup (2007). Additionally, pairwise significance tests of differentiation were carried out in FSTAT software (V2.9.3) (Goudet, 1995) and analyses of molecular variance (AMOVAs) (Excoffier et al., 1992) were conducted in ARLEQUIN 3.5.1.3 (Excoffier and Lischer, 2010) with 1,000 permutations and a significance level of p < 0.05. AMOVAs were carried out with all populations (ungrouped) and with populations grouped according to geographical region (N = 3) to examine difference among groups (FCT) and differences among populations within groups (FSC).
Deviation from neutrality of loci was tested to identify markers potentially under selection and examine their influence to differentiation and network connectivity. Outlier analyses was carried out via a coalescent simulation approach using ARLEQUIN 3.5.1.3 (Excoffier and Lischer, 2010). Analyses was carried out using a finite island model with 20,000 simulations, 100 demes, and minimum and maximum heterozygosities of 0 and 1, respectively. Where outlier markers were identified, the above analyses was re-run with neutral markers only to examine whether population differentiation and molecular variance were altered.
Level of differentiation is reported according to pairwise FST values with ENA correction, and defined according to Wright (1978) and Hartl and Clark (1997) where a value lying in the range 0–0.05 indicates low genetic differentiation; a value between 0.05 and 0.15, moderate differentiation; a value between 0.15 and 0.25, great differentiation; and values above 0.25, very great genetic differentiation. These categories have been applied in previous research where a microsatellite approach was applied to examination of genetic connectivity in bivalve populations (Nie et al., 2017; De Souza et al., 2018). However, it should be highlighted that no specific levels of FST divergence are universally accepted as designating particular levels of differentiation, and categorizations are likely to vary according to species and life histories. Consequently, the interpretation of differentiation values via pre-set categories should be made with some degree of caution.
EDENetworks v.2.18 (Kivelä et al., 2015) was utilized to visualize population connectivity across all sites and identify potential source populations. Analyses was carried out via the distance-thresholding approach with sampling-site-based FST distance measures using neutral markers only. An automatic threshold value of 0.0453 was detected and used for primary visualization. Networks were also visualized at manual thresholds of 0.03, 0.04, and 0.06. For all visualizations, network nodes (i.e., populations) were sized according to betweenness values and color-coded according to region, and edges were weighted to illustrate degree of connectivity. Component metrics including components (and populations therein), degrees and cluster coefficients were determined at each threshold value.
Genetic Structure
Population genetic structure was analyzed using two statistical approaches. First, the Bayesian clustering method was implemented in STRUCTURE (version 2.3.1; Pritchard et al., 2000; Falush et al., 2003, 2007). STRUCTURE parameters were set to allow for admixture and correlated allele frequencies with a burn-in period of 100,000 followed by 500,000 iterations. Outputs of five iterations of K = 1–8 were run in STRUCTURE Harvester web v0.6.92 (Earl and vonHoldt, 2012) to determine the uppermost true (estimated) number of genetic units (K). Evaluation of the DeltaK and L(K) plots with the Evanno Method (Evanno et al., 2005) identified the uppermost K-value. Secondly, discriminant analysis of principal components (DAPC) was carried out in ADEGENET (Jombart, 2008; Jombart et al., 2010) for R (R Development Core Team, 2012) with individuals grouped a priori by sampling locations. DAPC maximizes genetic separation among groups, while minimizing variation within groups (Jombart et al., 2010).
Genetic Diversity
Genetic variation of populations was assessed through estimation of allelic frequencies, observed heterogeneity (Ho) and expected heterogeneity (He), and inbreeding coefficients (Fis). FSTAT software (V2.9.3) (Goudet, 1995) was used to calculate Fis, allelic frequency, and number of alleles. Deviations of genotype frequencies from the Hardy–Weinberg Equilibrium (HWE) were tested in ARLEQUIN 3.5 (Excoffier and Lischer, 2010). FreeNA software (Chapuis and Estoup, 2007) was used to calculate allelic richness, frequencies of null alleles and Estimation of Null Allele (ENA) correction. Additionally, contemporary (i.e., estimates apply to the time period encompassed by the samples) effective population size (Ne) was estimated using the linkage disequilibrium method of Hill (1981); Waples (2006) and Waples and Do (2010), as implemented in NeEstimator V2.1 (Do et al., 2014). Pcrit was set to 0.02 (i.e., alleles with frequencies < 0.02 excluded) and confidence intervals were based on the jackknife method (Jones et al., 2016).
Results
Genetic Connectivity
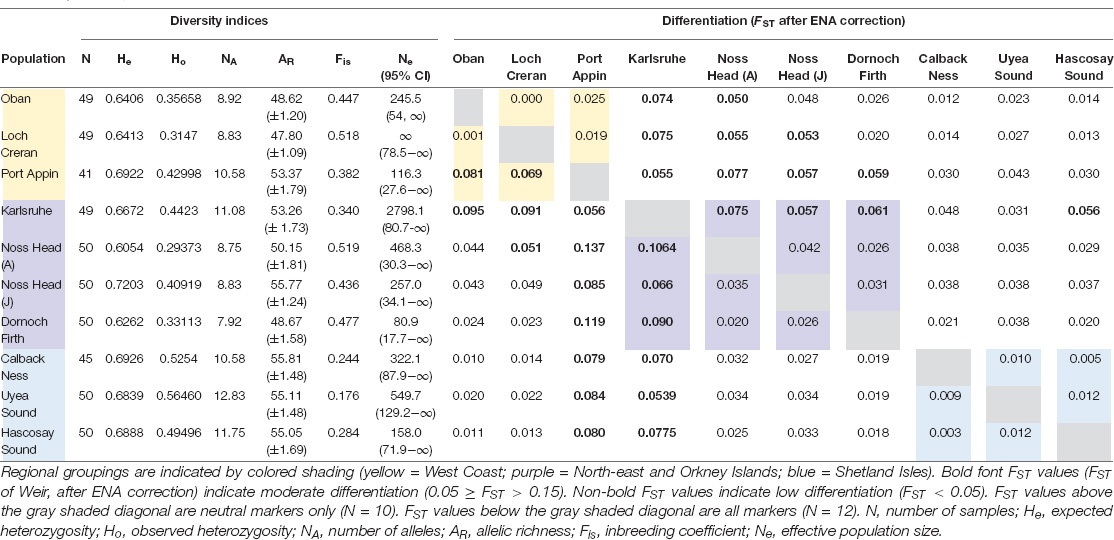
Comparison of analyses with all markers versus neutral markers showed minimal discrepancy between FST values and significance levels across all populations (Tables 3, 4). Overall, all FST values indicated low-moderate differentiation across Scottish M. modiolus reefs (Figure 2 and Tables 3, 4). No differentiation values were considered great or very great in either analyses. However, the exclusion of outliers did generally slightly reduce differentiation values, with the most noticeable effects (i.e., increases) to intra-regional connectivity of West Coast populations and inter-regional connectivity between West Coast and Shetland Isles populations, and Orkney Islands (Karlsruhe) and Shetland Isles populations (Table 4). Consequently, the following genetic connectivity results and discussion are with regards the neutral marker data analyses.
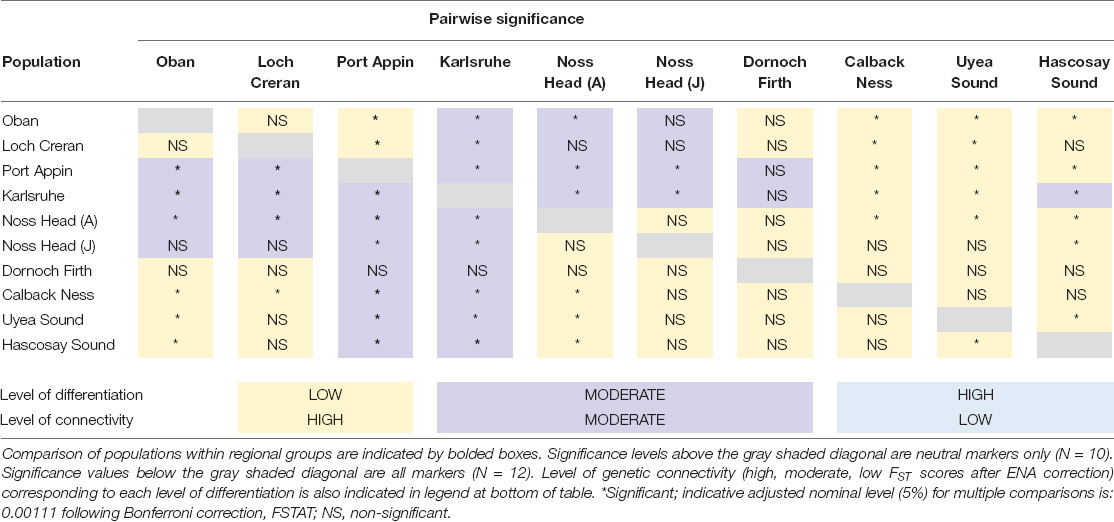
Table 4. Level (low, moderate, and high) and significance of genetic differentiation values for Scottish M. modiolus reefs.
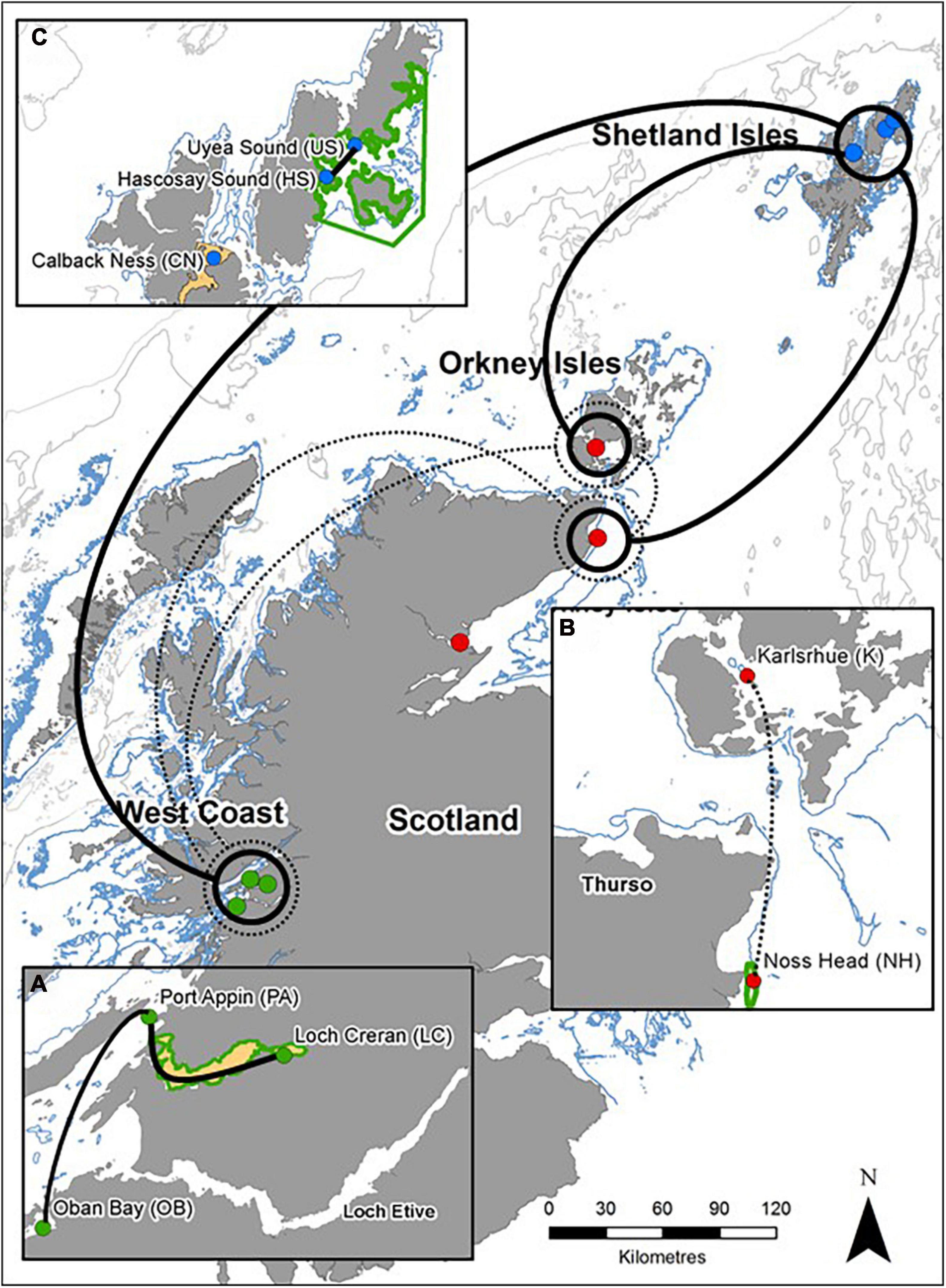
Figure 2. General patterns of genetic connectivity of M. modiolus reef populations of Scotland. Solid lines indicate high genetic connectivity (low differentiation) and dashed lines indicate moderate genetic connectivity (moderate differentiation). Circles contain any/all population(s) that is/are connected to adjoining population(s). Smaller inset maps indicate intra-regional genetic connectivity: (A) West Coast, (B) North-east and Orkney Islands, and (C) Shetland Isles. Coastline Crown © and database right (2021). Ordnance Survey License number 100017908.
Comparison of intra-regional connectivity suggested higher levels of differentiation within populations of North-east and Orkney Islands regions relative to those of the West Coast and Shetland Isles regions. Within the West Coast region, Port Appin showed significant (p < 0.05) low differentiation from both Loch Creran and Oban populations, while low differentiation between Oban and Loch Creran was not significant. Within the North-east and Orkney Islands region, Karlsruhe showed significant moderate differentiation from the Noss Head populations [Noss Head: FST = 0.075 (adults), FST = 0.057 (juveniles)]. Results also suggest potentially low genetic differentiation between Noss Head adults and juveniles, but this result was not statistically significant. Within the Shetland Isles region, all populations showed low differentiation, but genetic differentiation was only statistically significantly different between Hascosay Sound and Uyea Sound (FST = 0.012; Tables 3, 4). AMOVA analyses also indicated low genetic variance among populations (4.35%, p < 0.001). Likewise, when populations were grouped according to geographic area, percentage variation among groups was low (1.39%, p = 0.002) and variance among populations within groups (3.30%, p < 0.001) was similar to that across all populations.
Across all populations, several key relationships were detected (Tables 3, 4). Firstly, West Coast populations (particularly Port Appin) showed significantly moderate differentiation from populations of the North-east and Orkney Islands region but significantly low differentiation from populations of the Shetland Isles’ region. North-east populations of the Scottish mainland (i.e., Noss Head and Dornoch Firth) also showed low differentiation from Shetland Isles populations, though results were only found to be significant between Noss Head adults and all Shetland Isles’ populations, and Noss Head juveniles and a single population (Hascosay Sound). High genetic differentiation (i.e., low genetic connectivity) was not detected for any pair of populations in the current study.
Network visualization (Figure 3) and component data (Table 5) at the automatic threshold (0.0453) indicated two network components with grouping of all populations except Noss Head juveniles in the larger component. Within this main component, the Calback Ness (Shetland Isles) and Oban Bay (West Coast) populations had the highest degree (i.e., highest number of connections in the network, =5) (Table 5). Similarly, Oban Bay showed a high betweenness centrality (Figure 3), suggesting strong connectivity of this population within the M. modiolus system of reefs. Results suggest that these two populations may play key roles in maintaining network connectivity. In contrast, Karlsruhe and Noss Head (adults) showed the lowest degree metrics (Table 5) with corresponding low betweenness centralities (Figure 3). Visualization at lower threshold levels indicated potential sub-structuring of Shetland Isles’ populations (0.03), and of Noss Head (adults) and Dornoch Firth (0.03, 0.04). At a higher threshold level (0.06), network visualization showed Oban Bay (West Coast) with a high betweenness centrality, again highlighting it as a potentially key population.
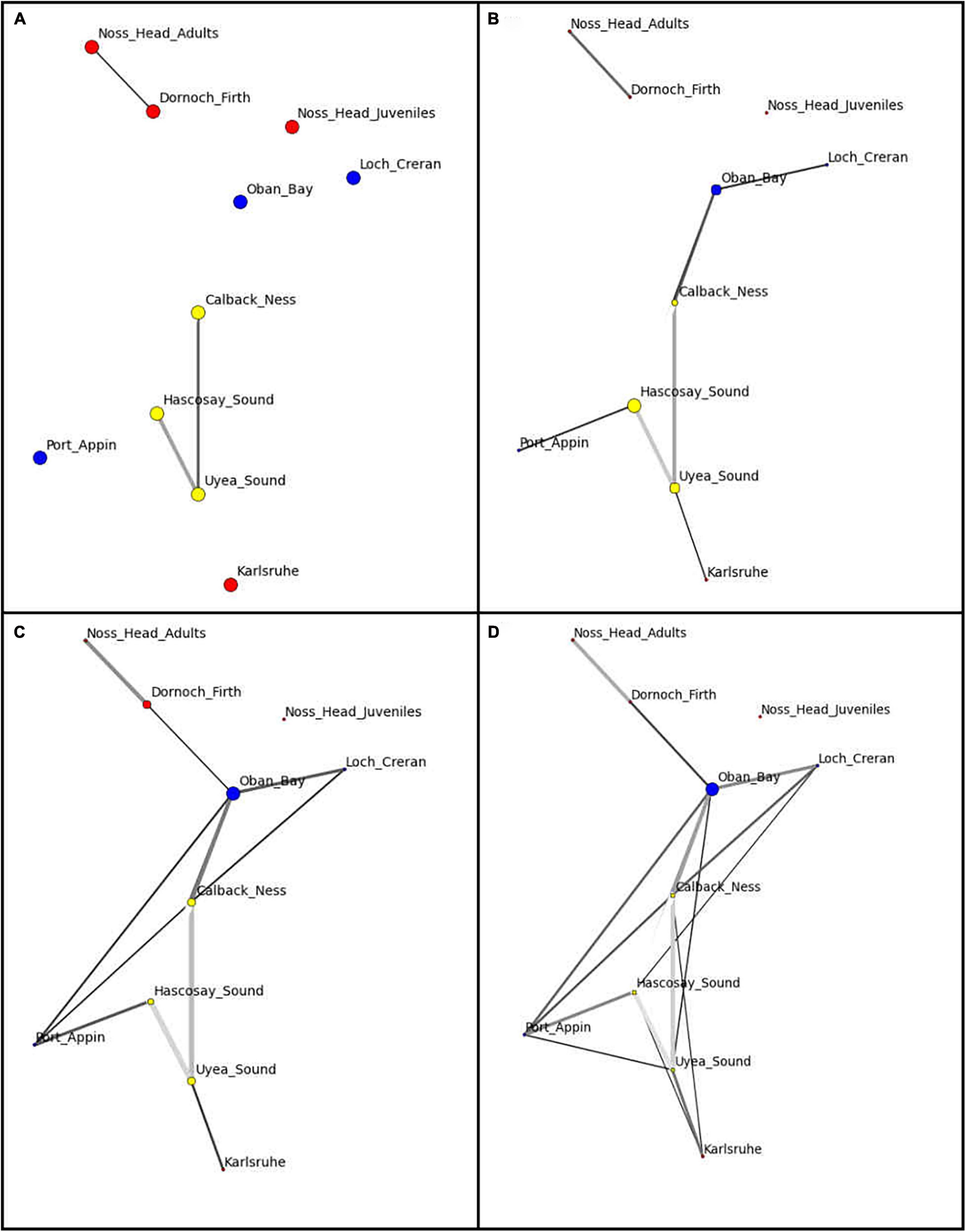
Figure 3. Network visualization of Scottish M. modiolus populations at threshold values of (A) 0.03, (B) 0.04, (C) 0.05 (automatic threshold) and (D) 0.06. Network nodes indicate populations with node color indicating region (yellow: Shetland Isles, blue: West Coast; red: North-East and Orkney Islands) and node size indicating betweenness value. Nodes with a higher number of connecting edges have stronger connectivity to neighboring nodes. Strength of a genetic relationship between two nodes is visualized according to edge weight (darker = stronger).
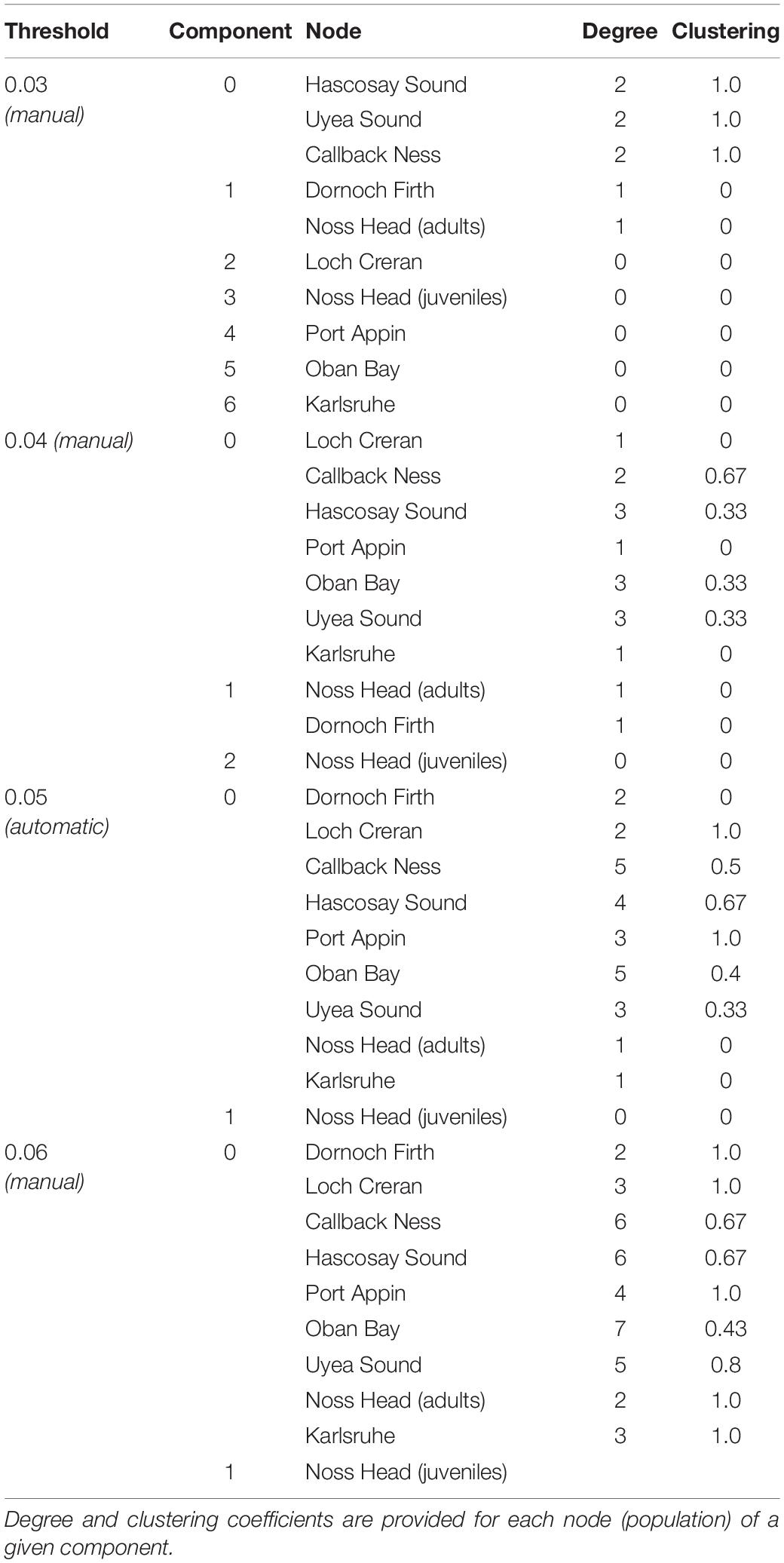
Table 5. Component summary data for network visualization of Scottish M. modiolus populations at an automatic threshold value of 0.0453 (0.05) and at manual threshold values of 0.03, 0.04 and 0.06.
Genetic Structure
STRUCTURE and STRUCTURE Harvester results found K = 2 to be the most likely K-value (Figure 4), indicating there are two main genetic groups across all currently sampled Scottish populations. Group 1 comprised Loch Creran, Oban, Noss Head (adults), Dornoch Firth, Hascosay Sound, Uyea Sound and Callback Ness. Group 2 comprised only the Port Appin and Karlsruhe populations. Noss Head (juveniles) appeared to be part of both groups though associated more strongly with Group 1. DAPC also suggested that Karlsruhe and Port Appin populations are genetically separate from all other populations (Figure 5).

Figure 4. STRUCTURE output with K = 2–5 for M. modiolus reef populations across Scottish regions. Population codes provided along x-axis. Populations showing the same color are part of the same genetic group (K). Population codes as per Table 1.
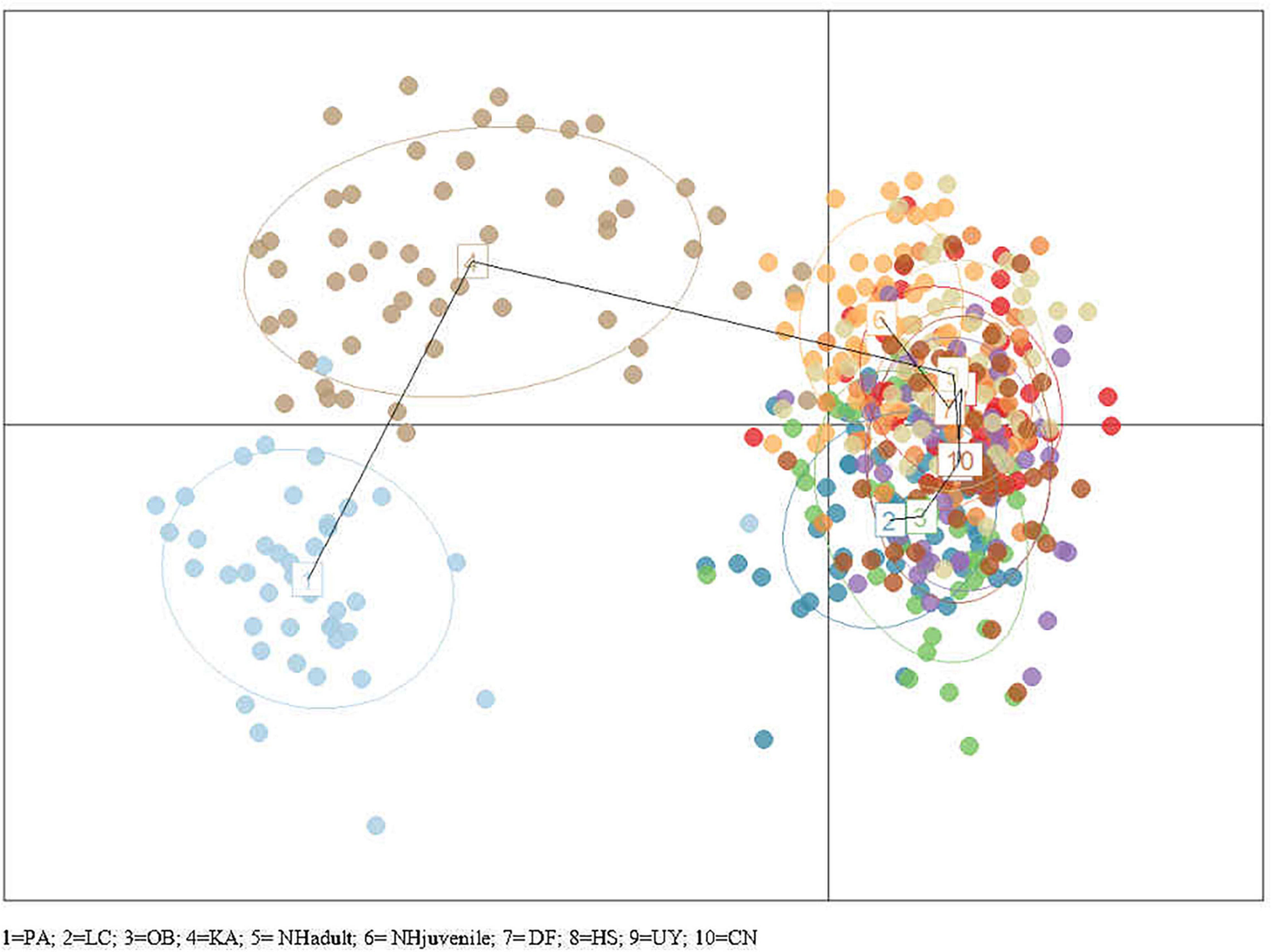
Figure 5. Plot of DAPC for groups defined by sampling locations. A single dot represents an individual. Population centers are indicated by population number (1–10) and are connected by a minimum spanning tree.
Genetic Diversity
Local inbreeding coefficients (Fis scores) of Scottish populations ranged from 0.176 (Uyea Sound) to 0.519 (Noss Head – adults), indicating a heterozygote deficit across all populations (Table 3). Likewise, all Ho values (ranging from 0.2937 in Noss Head adults to 0.5646 in Uyea Sound) were lower than He values (ranging from 0.6054 in Noss Head adults to 0.7203 in Noss Head juveniles), further indicating a heterozygote deficiency (Table 3). All populations deviated from Hardy–Weinberg Equilibrium (HWE). Results suggest a lack of genetic variation, possibly due to population sub-structuring or varied reproductive success, or caused by the preponderance of null alleles. Null alleles were detected across all markers and populations (r > 0.05).
Allelic richness and number of alleles were similar for all populations (Table 3). Across all populations, Uyea Sound had the greatest number of alleles, highest Ho, lowest Fis, and a high allelic richness score (only Callback Ness and Noss Head juveniles had higher allelic richness scores), suggesting higher genetic diversity in this population relative to the other populations. Conversely, Noss Head (adults) had the lowest Ho and highest Fis suggesting relatively low genetic diversity and inbreeding in this population. Regionally, the Shetland Isles appeared to have the greatest genetic diversity while genetic diversity values were similar between the other regions. The Loch Creran, Noss Head (adults) and Noss Head (juveniles) populations showed the greatest deviation from HWE. Contemporary estimates of Ne were variable across sampling sites, ranging from 80.9 (Dornoch Firth) to 2798.1 (Karlsruhe) (Table 3), suggesting considerable variation in genetic drift across populations, regions and life-stages (Noss Head). However, an Ne value of infinity (∞) was obtained for Loch Creran and upper confidence limits for all populations were unbounded (i.e., ∞).
Discussion
Genetic Connectivity and Structure
The low-moderate FST scores (i.e., low-moderate differentiation) determined by the current study suggest ‘moderate-strong’ genetic connectivity of the M. modiolus populations across the study area as a whole and illustrate that genetic connectivity of M. modiolus reef populations can exist over distances >500 km. Distance does not appear to be a key driver of genetic connectivity for the Scottish M. modiolus reef populations included in this study, with examples of moderate differentiation across smaller spatial scales and minimal differentiation across larger spatial scales as well as the converse. Network visualization and component data of M. modiolus reefs also suggest generally strong connectivity between most populations with the potential exceptions of Karlsruhe, Noss Head (adults) and Dornoch Firth. Visualization also indicated that Noss Head juveniles are separate from the main network though this result may be a consequence of various factors (see below for further discussion). Importantly, network visualization identified Oban Bay and Calback Ness populations as potentially critical to maintaining network connectivity, and thus it may be argued that these populations should be earmarked for future (Oban Bay) or continued (Callback Ness) protection.
STRUCTURE analyses indicated two clear genetic groups with no obvious spatial divide. It should be noted that a STRUCTURE result of K = 2 can be a false solution if a hierarchical analysis to fully explore population subdivision is not carried out (Janes et al., 2017), as was the case in the current study. Co-consideration of STRUCTURE and DAPC analyses, however, does suggest similar patterns of genetic structure across all populations, i.e., a lack of genetic structure between all populations except Karlsruhe and Port Appin, with minimal correlation to distance. Additionally, network visualization and component data generally support STRUCTURE and DAPC results.
Variation in the complexity and magnitude of local and regional hydrodynamics is a major influence with regards promoting or hindering larval dispersal to varying degrees and thus directly advancing or limiting gene flow between sites (Robins et al., 2013). Consequently, timing, direction and success of larval dispersal from and recruitment to M. modiolus reef populations is very much location-dependent (Coscia et al., 2012; Gormley K. S. G. et al., 2015; Millar et al., 2019). For example, while reefs situated in open areas of high flow that are exposed to large scale water movement (e.g., headlands and exposed sites) may have greater dispersal potential, long-range dispersal ‘success’ may also lead to dispersal dilution and increased larval mortality rates resulting in lowered recruitment success to other populations (Cowen and Sponaugle, 2009; Adams et al., 2014; Chapman et al., 2021). This could partially explain why the exposed Noss Head reef, located in an area of high spring tide current flows (up to 1.20 ms–1; ABPmer, 2008), showed moderate genetic differentiation from nearby reef sites (e.g., Karlsruhe population in the Orkney Islands). In contrast, significant low genetic differentiation was determined between the Noss Head adults and Shetland Isles populations. Given the predominantly clockwise direction of residual flow in Scotland (Baxter et al., 2008), it is therefore suggested that these more distant northerly populations could be acting as a key larval source for the Noss Head population.
Exposed sites with high flow rates may also experience problems with retention of larvae compared to enclosed bays (Adams et al., 2014). If larvae are dispersed too quickly, there may be reduced opportunity for recruitment to the parent site (i.e., low self-recruitment). Variation in genetic structure between adults and juveniles at the Noss Head reef, for example, could be partially explained by the high flow conditions of the site leading to low levels of larval retention (and therefore decreased potential for self-recruitment) within the population. Larval dispersal modeling from the site has also suggested low rates of retention and low self-recruitment potential (0.4%) (Millar et al., 2019).
Results also suggest that reefs situated within semi-enclosed water bodies, such as Loch Creran, need not act as closed systems reliant on self-recruitment. The complex geography of Scotland’s west coast coupled with high flow rates between islands and sea loch ‘narrows’ may create hydrodynamic conditions that instead aid larval dispersal from enclosed areas (Robins et al., 2013; Adams et al., 2014). Recent modeling studies have confirmed that the residual particle transport direction for plankton (e.g., larvae and sea lice) is predominantly clockwise around the Scottish coastline (Millar et al., 2019; Rabe et al., 2020) and thus south-western M. modiolus reefs such as Loch Creran may play an important role as a direct source or indirect stepping-stone source of larvae for more northern populations via as yet un-sampled populations (Millar et al., 2019). Given that the current study found reefs on the West Coast (e.g., Oban, Loch Creran, and Port Appin) to be collectively well-connected (significant low-moderate genetic differentiation) to all other reef populations except Dornoch Firth, and identified Oban Bay as a potential important source population, a recommendation for their collective protection could be made (Martens et al., 2018). Likewise, additional West Coast M. modiolus reef populations situated north of Oban, Loch Creran and Port Appin should be identified and considered for protection because they may also act as key intermediates in terms of maintaining the open system of populations.
Genetic Diversity
All M. modiolus populations of the current study were shown to be in departure from HWE with positive Fis values, typically equated with inbreeding and increased risk of adaptive decline (Lowe et al., 2004). However, such observations are highly characteristic of bivalve species (Addison and Hart, 2005) and likely to be artifacts of a broadcast spawning reproductive strategy. This approach demands the production of high numbers of gametes leading to increased mutation rates and a subsequent preponderance of null alleles with parallel decreases in heterozygosity (Reece et al., 2004; Addison and Hart, 2005; Coscia et al., 2012; Mariani et al., 2012). Similar results were reported by Gormley K. S. G. et al. (2015) for Irish Sea M. modiolus populations where departure from HWE and positive Fis values were also observed in all populations. Fis results are also comparable to those reported by Addison and Hart (2005) and Olsen et al. (2020) for sessile/sedentary marine invertebrates and those with planktonic larval dispersal, respectively. The ambiguity with regards to whether departure from HWE and high Fis values indicate reduced genetic diversity in M. modiolus reefs of the current study or are merely representative of bivalves is acknowledged and is a known limitation of adopting a microsatellite approach to study natural bivalve populations (Hedgecock et al., 2004).
In contrast, allelic richness was substantially (i.e., approximately fivefold) higher in M. modiolus populations in the current study than of Irish Sea populations (Gormley K. S. G. et al., 2015) suggesting greater genetic diversity in Scottish populations. Allelic richness was also similar across populations (mean 52.36 ± 3.22 SD) likely due to the moderate to high levels of genetic connectivity between reef populations (i.e., an open system). Contemporary estimates of Ne were somewhat uninformative due to large CIs (i.e., upper bounds = ∞) across all populations. While it is thus difficult to make any strong inferences, relative comparison across populations alludes to variable levels of genetic drift over the M. modiolus reef system, likely as a result of variation in sex ratios, reproductive success, age structure and/or migration rates. The lowest point estimate was at Dornoch Firth, potentially suggesting reduced genetic diversity and a higher incidence of inbreeding in this population (Waples et al., 2016).
The variation in genetic diversity and genetic structure observed between juvenile and adult cohorts at Noss Head reef highlights an additional consideration. The genetic dissimilarity of the two groups suggests the population does not rely on self-recruitment, potentially due to poor larval retention. Deviation in genetic structure could also be a consequence of inter-site and inter-annual variation in the scale and timing of key physical, chemical and biological factors that control dispersal, recruitment, growth and mortality, resulting in markedly different patterns of inter-annual recruitment and survival, as has been observed in other reef-forming shellfish populations (Casas et al., 2015). Site-specific selective pressures (e.g., temperature and flow rates) at the Noss Head site and phenotypic differences (e.g., growth rate and shell shape) between source populations could also favor juveniles from particular source populations leading to reduced genetic diversity in adult year classes as a smaller group of out-competing juveniles matures to form the reproductive cohort of the population. This “phenotype-environment mismatch” (DeWitt et al., 1998), where phenotypes of recruits from external source populations are less suited to the local environment and thus have increased mortality before reaching reproductive maturity, is an important consideration as such incompatibilities pose biological barriers to connectivity in marine environments (Marshall et al., 2010). Additionally, sweepstakes recruitment [i.e., where the matching of successful reproductive activity with oceanographic conditions that are favorable to gamete maturation, fertilization, larval development, settlement and recruitment to the adult population is down to chance (Hedgecock and Pudovkin, 2011)], largely characterized by marked variance in reproductive success and chaotic genetic patchiness (CGP) (Vendrami et al., 2021), may also play a role. Consequently, to fully understand the recruitment dynamics at additional reef sites and their potential reliance on larvae from external sources, future genetic analyses should include both juveniles and adults in order to compare genetic signatures between different year classes. Identifying those reefs which are recipients of recruits from different source populations and/or have temporally variable recruitment could also further complicate the interdependency of populations.
Comparison With Modeling Studies
Past studies have combined hydrodynamic modeling with genetic techniques to provide improved determination of connectivity for species with a planktonic larval life-stage (Schunter et al., 2011; Selkoe and Toonen, 2011; Coscia et al., 2012; Gormley K. S. G. et al., 2015; Millar et al., 2019). Co-consideration of both aspects, if possible, is increasingly regarded as best practice. Millar et al. (2019) carried out coupled particle tracking and hydrodynamic modeling (Scottish Shelf Model) for M. modiolus reefs (n = 42) across Scotland, including many of the present study sites [e.g., Port Appin, Loch Creran, Noss Head, Dornoch Firth, and Scapa Flow (Orkney)]. A Lagrangian particle tracking model [FVCOM I-State Configuration Model (FISCM)] was applied to track virtual particles (representing larvae) forced by Scottish Shelf Model hydrodynamic model outputs. Particles were assumed to be passive (i.e., no behavior elements were applied) and were subject to advection by currents and diffusion (Millar et al., 2019). Results from the Millar et al. (2019) and the present study both point toward an open system of M. modiolus reefs in Scottish waters, however, there are some discrepancies between the two approaches.
Genetic results indicate that the Karlsruhe population is moderately differentiated from West Coast populations, whereas the larval dispersal model based on domains of the Scottish Shelf hydrodynamics model deemed reef populations in the Orkney region (Scapa Flow) to be potentially well-connected to those of the West Coast region (Millar et al., 2019). This difference in outcome could be explained by the greater number of Orkney and West Coast release sites included in the larval dispersal model, thereby incorporating more northerly sites along the West Coast that could act as stepping-stone populations between southern and northern regions. Indeed, Millar et al. (2019) demonstrated that M. modiolus reefs in the south-west region of the West Coast (e.g., Port Appin and Loch Creran) generally export larvae south and west while those in the north-west region of the West Coast (e.g., Loch Laxford) generally export larvae west and north. Millar et al. (2019) also proposed that a M. modiolus reef in the Small Isles (not included in the current genetic analyses) may act as a stepping-stone population linking the more southern reef populations on the West Coast and those located further north. Similarly, it is possible that more northerly west coast populations not included in the current study likely serve as additional source or stepping-stone populations to more northwestern populations and thus are integral to maintaining the greater open system of reef populations.
Genetic results also indicate high genetic connectivity between reefs in the Shetland Isles and those of the West Coast and North-east and Orkney Islands regions, whilst Millar et al. (2019) concluded that the Shetland Isles populations are predominantly isolated from all other Scottish reefs. However, when a higher resolution integrated modeling approach was applied to specific sea loch areas of the North-West coast, dispersal outputs indicated improved connectivity between a number of larval release sites (e.g., Loch Laxford SAC; Lochs Duich, Long and Alsh Reefs SAC) and the Shetland Isles (Millar et al., 2019).
Building Resilience to Climate Change
Knowledge of genetic connectivity and diversity of M. modiolus populations may inform the assessment of climate change vulnerability of individual reef populations and aid in clarifying the interdependency of all populations. In general, the moderate to high levels of gene flow between M. modiolus populations included in the current study suggest that adaptive capacity is generally limited within populations at a site level (i.e., reduced potential to adapt to site-specific stress conditions) (Whitlock, 2008; Sanford and Kelly, 2011). Alternatively, it has been suggested that gene flow may in fact increase local adaptive potential by improving genetic diversity, particularly in populations that are exposed to temporal variation in stress levels (Baskett et al., 2010). Consequently, the generally open nature of the system of M. modiolus reefs may result in a diverse gene pool, as suggested by allelic richness results, from which future adaptations may arise (Harley et al., 2006). Moreover, where populations largely rely on external inputs of genetic material for maintaining genetic diversity (e.g., remote/sink populations, populations in decline), it is critically important that connectivity with source populations be maintained, particularly when under threat of increasing magnitudes of environmental stress. An argument for protection may also be made for sink populations in receipt of genetic inputs (i.e., larvae) from multiple source populations, as they may serve as important stores of genetic diversity for application to future restoration efforts (e.g., transplantation for reef restoration) (Almany et al., 2009). Conversely, where reef populations are lost, decreases in genetic diversity may follow (Almany et al., 2009).
The open nature of the Scottish M. modiolus reef system may also facilitate the support of vulnerable populations by more resilient populations. There is the potential that as sea temperatures increase toward the upper thermal limit for M. modiolus (Gormley et al., 2013), reef populations from lower latitudes or shallower (and therefore warmer) locations could provide increasingly resilient larvae to northern/deeper populations and thereby improve adaptive capacity at those sites. Conversely, reefs located in more southern or shallower locations may struggle to adapt to warming conditions, resulting in the future loss of larval supply (with potentially parallel losses in genetic diversity) to more northern or deeper populations. Such losses could result in a domino-effect, as reefs historically dependent on lost reefs for recruitment are pushed into decline, thereby instigating the deterioration of other reefs.
More northern populations of M. modiolus may also serve as sources of larvae to populations located further south along eastern Scottish coasts so that northern (i.e., less thermally-stressed sites) populations may function as both a demographic and adaptive source for higher thermally-stressed locations. Thus, the positive effect of enhanced recruitment (i.e., increased recruitment potential due to external supply) may offset the adverse effect of recruits that are poorly adapted to local conditions. This effect has been documented in coral reef populations where evidence on the connectivity of reef structures is more advanced (Baskett et al., 2010). It is also worth noting that the relatively high allelic richness of populations in the current study as compared to more southern populations of the Irish Sea (Gormley K. S. G. et al., 2015) may indicate a higher adaptive potential (Greenbaum et al., 2014) which could translate to enhanced resilience of M. modiolus populations of Scotland’s reef system as compared to more southern reefs.
In general, an improved understanding of the interplay between isolated/connected, southern/northern and shallow/deep populations in terms of climate adaptation is needed to understand how future conditions may alter connectivity and diversity of populations. Given the likelihood that some reef populations may be lost or pushed into decline under current and/or future conditions, expansion or redesign of the current network to one that is truly “ecologically coherent” may be necessary.
Conservation Recommendations
Given the known distribution of M. modiolus reefs (Figure 6), it is clear that a good number are designated features within the Scottish MPA network, with additional populations, although not specifically designated features, occurring within other MPAs (i.e., hosted populations). Furthermore, the status of M. modiolus as a Priority Marine Feature (PMF) affords a degree of wider protection. Based on the findings of this study together with the potential larval dispersion described by Millar et al. (2019), it is clear that M. modiolus populations nor the MPAs where they are found should be considered discrete, independent entities (Cicin-Sain and Belfiore, 2005). Rather, the protection of features within MPAs must occur in concert with non-designated areas and across varying spatial scales (Cicin-Sain and Belfiore, 2005).
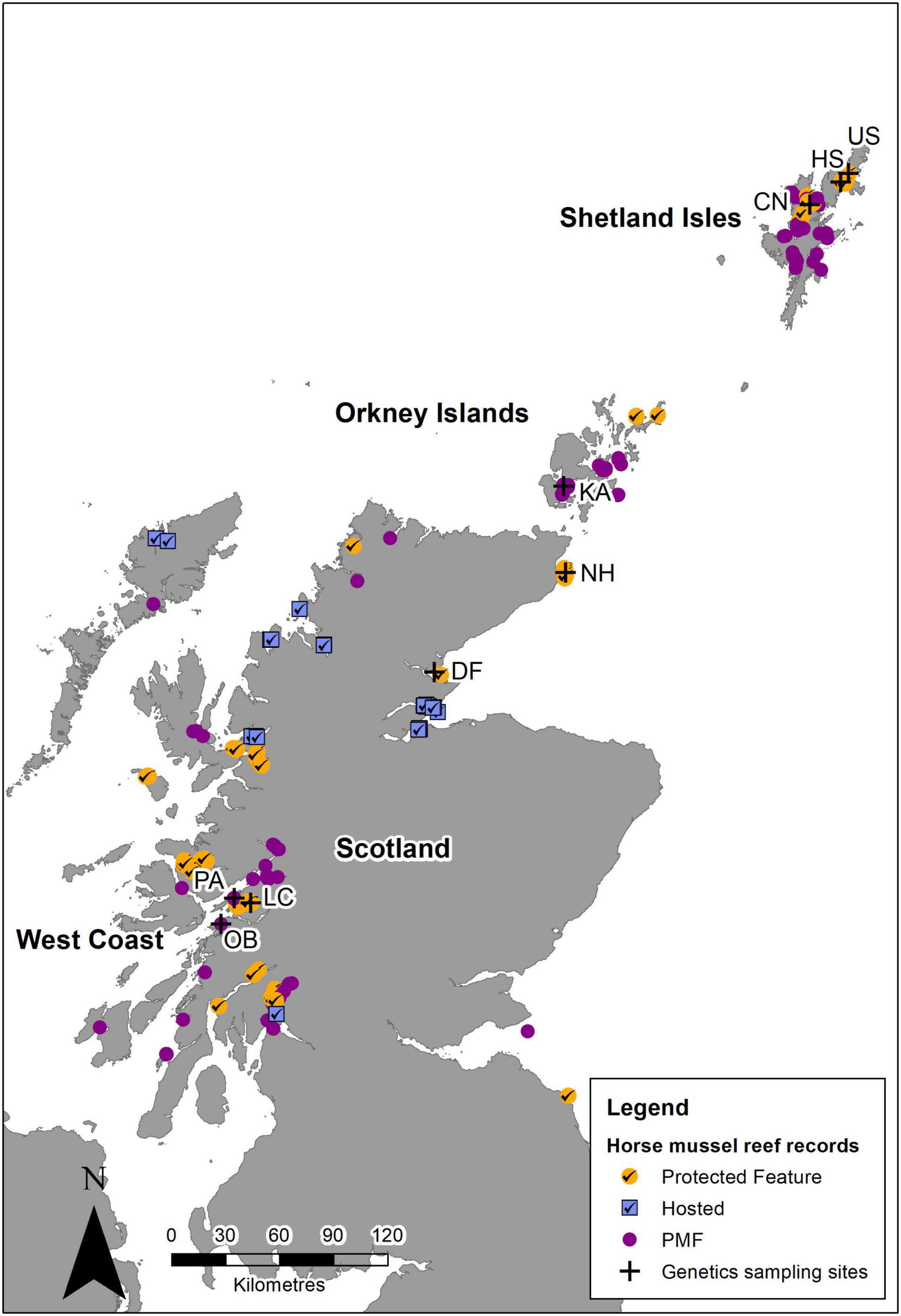
Figure 6. Horse mussel (M. modiolus) reef records based on the Geodatabase for Marine Habitats and Species adjacent to Scotland Coastline (GeMS) showing records within protected areas where they are a designated feature (orange circle with tick), reefs that are hosted in closed area for which the presence of M. modiolus reefs was not the reason for designation (blue square with tick), and M. modiolus reef outside of protected areas and closed areas where they are a Priority Marine Feature (purple circle). Sites where samples were collected for genetic analysis also show (cross) for context. Crown © and database right (2021). Ordnance Survey License number 100017908.
Currently, there are 12 MPAs in Scotland that have M. modiolus reefs as a designated feature (Figure 6). Ten of these have specific management measures in place to prevent any damaging activities1. A further two MPAs host M. modiolus populations that are not designated features of these sites. The potential for greater protection exists if all relevant Scottish MPAs, i.e., both those where M. modiolus reefs are a designated feature and those that host M. modiolus reefs, had statutory restrictions on all activities that cause damage to the sea bed. With such measures in place, the potential for genetic connectivity through larval dispersal would be greatly enhanced. There would remain, however, a large number of M. modiolus populations not protected in this way that may serve as important components in the overall interdependent system of populations. Thus, further genetics studies of additional populations, informed by larval dispersal modeling, is required to identify key locations in need of protection.
A comprehensive program of sampling of both adult and juvenile components of all relevant M. modiolus populations would help determine the relative scale of self-recruitment versus remote recruitment. Based on these findings, a more complete picture of the genetic connectivity of all M. modiolus populations (within and outside of MPAs) could be constructed. Ideally, such findings would then be considered alongside the results of larval distribution models to identify likely gaps in the current network of MPAs. Likewise, those M. modiolus reefs identified through larval modeling as potential key nodes of the network could also be sampled for their genetic connectivity and as necessary designated. As a matter of priority, those reefs included in this study but which are not within an MPA or which are not a designated feature of an MPA should be given full protection through designation and implementation of the necessary management measures. Similarly, ongoing monitoring of levels of genetic diversity across all populations should be factored in to ensure that management strategies are working to improve the allelic richness of populations and to avoid the consequences of genetic erosion (Hoban et al., 2021).
The protection afforded M. modiolus reefs through the designation of MPAs will increase their resilience which is critical in the face of climate change but which to date has rarely been considered in the development of MPA networks (Frost et al., 2016). Whilst more southerly populations are likely to be better adapted to warmer sea water temperatures and thus an important potential source of larvae and ultimately recruits to more northern populations, they are also likely to be some of the first to be lost due to temperature rise (Gormley et al., 2013). If such populations fall outside MPAs, they will be more vulnerable to additional stressors and so there is an increased chance of a vital source of temperature-adapted larvae being lost. Moreover, given the predicted shifts in the extent and distribution of M. modiolus reefs under climate change (Gormley et al., 2013; Gormley K. S. et al., 2015), changes or disruptions to genetic connectivity and diversity are to be expected, highlighting the need for an adaptive management approach (Gormley K. S. et al., 2015).
This study has begun to reveal the complexities and scale of connectivity in M. modiolus reef populations and the challenges and issues that need to be taken into account when designing and establishing an ecologically-coherent MPA network. While results suggest that M. modiolus reef populations are genetically related across both inter- and intra-regional extents (i.e., an open system), a deeper consideration is needed as to whether the current MPA network is “fit for purpose.” If the aim of a network of MPAs is more than the protection of a representative collection of sites/populations, then further consideration is required of the optimization of MPA protection and the requirement for wider environmental management measures. Without an integrated protected network there is a risk that any level of protection within an MPA will be redundant if key source populations outside of the MPA network are damaged or lost. Proper consideration and implementation of these aspects will allow full appreciation of the scale of the protected area network that is required and advance the development of an effective MPA network.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
CM, FK, JB, WS, and JP contributed to the conception and design of the study and wrote sections of the manuscript. FK, JP, and WS carried out collection from sub-tidal sites. CM, KG, and AC carried out the microsatellite screening and genetic analyses. CM, FK, JB, and JP wrote the first draft of the manuscript. All authors contributed to the manuscript revision and approved the submitted version.
Funding
This work was largely funded by Heriot-Watt University (James Watt Scholarship) and NatureScot (formerly Scottish Natural Heritage). Additional funding was received from the MASTS pooling initiative (The Marine Alliance for Science and Technology for Scotland) and their support is gratefully acknowledged. MASTS was funded by the Scottish Funding Council (grant reference HR09011) and contributing institutions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://spatialdata.gov.scot/geonetwork/srv/eng/catalog.search#/metadata/Marine_Scotland_FishDAC_1630
References
ABPmer (2008). Atlas of UK Marine Renewable Resources. ABPmer. Available Online at: http://www.renewables-atlas.info/ (accessed January, 2017).
Adams, T. P., Aleynik, D., and Burrows, M. T. (2014). Larval dispersal of intertidal organisms and the influence of coastline geography. Ecography 37, 698–710. doi: 10.1111/j.1600-0587.2013.00259.x
Addison, J. A., and Hart, M. W. (2005). Spawning, copulation and inbreeding coefficients in marine invertebrates. Biol. Lett. 1, 450–453. doi: 10.1098/rsbl.2005.0353
Almany, G. R., Connolly, S. R., Heath, D. D., Hogan, J. D., Jones, G. P., McCook, L. J., et al. (2009). Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs 28, 339–351. doi: 10.1007/s00338-009-0484-x
Anwar, N. A., Richardson, C. A., and Seed, R. (1990). Age determination, growth rate and population structure of the horse mussel Modiolus modiolus. J. Mar. Biol. Assoc. 70, 441–457. doi: 10.1017/S0025315400035529
Balbar, A. C., and Metaxas, A. (2019). The current application of ecological connectivity in the design of marine protected areas. Glob. Ecol. Conserv. 17:e00569. doi: 10.1016/j.gecco.2019.e00569
Baskett, M. L., Nisbet, R. M., Kappel, C. V., Mumby, P. J., and Gaines, S. D. (2010). Conservation management approaches to protecting the capacity for corals to respond to climate change: a theoretical comparison. Glob. Change Biol. 16, 1229–1246. doi: 10.1111/j.1365-2486.2009.02062.x
Baxter, J. M., Boyd, I. L., Cox, M., Cunningham, L., Holmes, P., and Moffat, C. F. (2008). Scotland’s Seas: Towards Understanding Their State. Aberdeen: Fisheries Research Services.
Becker, B. J., Levin, L. A., Fodrie, F. J., and McMillan, P. A. (2007). Complex larval connectivity patterns among marine invertebrate populations. Proc. Nat. Acad. Sci. U.S.A. 104, 3267–3272. doi: 10.1073/pnas.0611651104
Bernard, G., Romero-Ramirez, A., Tauran, A., Pantalos, M., Deflandre, B., Grall, J., et al. (2019). Declining maerl vitality and habitat complexity across a dredging gradient: insights from in situ sediment profile imagery (SPI). Sci. Rep. 9:16463. doi: 10.1038/s41598-019-52586-8
Brash, J. M., Cook, R. L., Mackenzie, C. L., and Sanderson, W. G. (2018). The demographics and morphometries of biogenic reefs: important considerations in conservation management. J. Mar. Biol. Assoc. U. K. 98, 1231–1240. doi: 10.1017/s0025315417000479
Brown, R. A. (1984). Geographical variations in the reproduction of the horse mussel, Modiolus modiolus (Mollusca: Bivalvia). J. Mar. Biol. Assoc. U. K. 64, 751–770. doi: 10.1017/S0025315400047214
Burrows, M. T., Hughes, D. J., Austin, W. E. N., Smeaton, C., Hicks, N., Howe, J. A., et al. (2017). Assessment of Blue Carbon Resources in Scotland’s Inshore Marine Protected Area Network. Scottish Natural Heritage Commissioned Report No. 957. Edinburgh: Scottish Natural Heritage.
Burrows, M. T., Kamenos, N. A., Hughes, D. J., Stahl, H., Howe, J. A., and Tett, P. (2014). Assessment of Carbon Budgets and Potential Blue Carbon Stores in Scotland’s Coastal and Marine Environment. Scottish Natural Heritage Commissioned Report No. 761. Edinburgh: Scottish Natural Heritage.
Buzeta, M. I. (2014). Identification and Review of Ecologically and Biologically Significant Areas in the Bay of Fundy. DFO Can. Sci. Advis. Sec. Res. Doc. 2013/065.
Carr, M. H., Robinson, S. P., Wahle, C., Davis, G., Kroll, S., Murray, S., et al. (2017). The central importance of ecological spatial connectivity to effective coastal marine protected areas and to meeting the challenges of climate change in the marine environment. Aquat. Conserv. 27, 6–29. doi: 10.1016/j.marpolbul.2021.112244
Casas, S. M., La Peyre, J., and La Peyre, M. K. (2015). Restoration of oyster reefs in an estuarine lake: population dynamics and shell accretion. Mar. Ecol. Prog. Ser. 524, 171–184. doi: 10.3354/meps11198
Chapman, E. C., Rodriguez-Perez, A., Hugh-Jones, T., Bromley, C., James, M. A., Diele, K., et al. (2021). Optimising recruitment in habitat creation for the native European oyster (Ostrea edulis): implications of temporal and spatial variability in larval abundance. Mar. Pollut. Bull. 170:112579. doi: 10.1016/j.marpolbul.2021.112579
Chapuis, M.-P., and Estoup, A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631. doi: 10.1093/molbev/msl191
Cicin-Sain, B., and Belfiore, S. (2005). Linking marine protected areas to integrated coastal and ocean management: a review of theory and practice. Ocean Coast. Manage. 48, 847–868. doi: 10.1016/j.ocecoaman.2006.01.001
Cook, R., Fariñas-Franco, J. M., Gell, F. R., Holt, R. H. F., Holt, T., Lindenbaum, C., et al. (2013). The substantial first impact of bottom fishing on rare biodiversity hotspots: a dilemma for evidence-based conservation. PLoS One 8:e69904. doi: 10.1371/journal.pone.0069904
Coolen, J. W., Boon, A. R., Crooijmans, R., Van Pelt, H., Kleissen, F., Gerla, D., et al. (2020). Marine stepping-stones: connectivity of Mytilus edulis populations between offshore energy installations. Mol. Ecol. 29, 686–703. doi: 10.1534/genetics.114.161380
Coscia, I., Robins, P. E., Porter, J. S., Malham, S. K., and Ironside, J. E. (2012). Modelled larval dispersal and measured gene flow: seascape genetics of the common cockle Cerastoderma edule in the southern Irish Sea. Conserv. Genet. 14, 451–466. doi: 10.1007/s10592-012-0404-4
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annu. Rev. Mar. Sci. 1, 443–466. doi: 10.1146/annurev.marine.010908.163757
Cowen, R. K., Lwiza, K. M., Sponaugle, S., Paris, C. B., and Olson, D. B. (2000). Connectivity of marine populations: open or closed? Science 287, 857–859. doi: 10.1126/science.287.5454.857
De Schweinitz, E. H., and Lutz, R. A. (1976). Larval development of the northern horse mussel, Modiolus modiolus (L.), including a comparison with the larvae of Mytilus edulis L. as an aid in planktonic identification. Biol. Bull. 150, 348–360. doi: 10.2307/1540677
De Souza, F. P., Ruas, C. D. F., Urrea-Rojas, A. M., De Lima, E. C. S., Povh, J. A., Ribeiro, R. P., et al. (2018). Novel microsatellite markers for the invasive golden mussel Limnoperna fortunei. J. Shell. Res. 37, 485–489.
DeWitt, T. J., Sih, A., and Wilson, D. S. (1998). Costs and limits of phenotypic plasticity. Trends Ecol. Ecol. 13, 77–81. doi: 10.1016/S0169-5347(97)01274-3
DFO (2015). Information on Potential Sensitive Benthic Areas in the Bay of Fundy: Head Harbour/West Isles/Passages and the Modiolus Reefs, Nova Scotia Shore. DFO Can. Sci. Advis. Sec. Sci. Resp. 2014/044.
Dinesen, G. E., and Morton, B. (2014). Review of the functional morphology, biology and perturbation impacts on the boreal, habitat-forming horse mussel Modiolus modiolus (Bivalvia: Mytilidae: Modiolinae). Mar. Biol. Res. 10, 845–870. doi: 10.1080/17451000.2013.866250
Do, C., Waples, R. S., Peel, D., Macbeth, G. M., Tillett, B. J., and Ovenden, J. R. (2014). NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 14, 209–214. doi: 10.1111/1755-0998.12157
Earl, D. A., and vonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Elsäßer, B., Fariñas-Franco, J. M., Wilson, C. D., Kregting, L., and Roberts, D. (2013). Identifying optimal sites for natural recovery and restoration of impacted biogenic habitats in a special area of conservation using hydrodynamic and habitat suitability modelling. J. Sea. Res. 77, 11–21. doi: 10.1016/j.seares.2012.12.006
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. doi: 10.1093/genetics/131.2.479
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/164.4.1567
Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578. doi: 10.1111/j.1755-0998.2009.02591.x
Fariñas-Franco, J. M., and Roberts, D. (2018). The relevance of reproduction and recruitment to the conservation and restoration of keystone marine invertebrates: a case study of sublittoral Modiolus modiolus reefs impacted by demersal fishing. Aquat. Conserv. 28, 672–689. doi: 10.1002/AQC.2882
Fowler, A. M., Jørgensen, A.-M., Coolen, J. W. P., Jones, D. O. B., Svendsen, J. C., Brabant, R., et al. (2019). The ecology of infrastructure decommissioning in the North Sea: what we need to know and how to achieve it. ICES J. Mar. Sci. 77, 1109–1126. doi: 10.1093/icesjms/fsz143
Frost, M., Bayliss-Brown, G., Buckley, P., Cox, M., Dye, S. R., Sanderson, W. G., et al. (2016). A review of climate change and the implementation of marine biodiversity legislation in the United Kingdom. Aquat. Conserv. 26, 576–595. doi: 10.1002/aqc.2628
Gallego, A., Gibb, F. M., Tulett, D., and Wright, P. J. (2013). Connectivity of benthic priority marine species within the Scottish MPA network. Mar. Scotland Sci. Scott. Mar. Freshw. Sci. 4, 1–51. doi: 10.7489/1512-1
Gormley, K. S. G., Mackenzie, C. L., Robins, P., Coscia, I., Cassidy, A., and James, et al. (2015). Connectivity and dispersal patterns of protected biogenic beds: implications for the conservation of Horse mussels (L.) in the Irish Sea. PLoS One 10:e0143337. doi: 10.1371/journal.pone.0143337
Gormley, K. S., Hull, A. D., Porter, J. S., Bell, M. C., and Sanderson, W. G. (2015). Adaptive management, international co-operation and planning for marine conservation hotspots in a changing climate. Mar. Pol. 53, 54–66. doi: 10.1016/j.marpol.2014.11.017
Gormley, K. S., Porter, J. S., Bell, M. C., Hull, A. D., and Sanderson, W. G. (2013). Predictive habitat modelling as a tool to assess the change in distribution and extent of an OSPAR priority habitat under an increased ocean temperature scenario: consequences for marine protected area networks and management. PLoS One 8:e68263. doi: 10.1371/journal.pone.0068263
Goudet, J. (1995). FSTAT (Version 1.2): a computer program to calculate F-Statistics. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Greenbaum, G., Templeton, A. R., Zarmi, Y., and Bar-David, S. (2014). Allelic richness following population founding events–a stochastic modeling framework incorporating gene flow and genetic drift. PloS One 9:e115203. doi: 10.1371/journal.pone.0115203
Harley, C. D. G., Randall Hughes, A., Hultgren, K. M., Miner, B. G., Sorte, C. J. B., Thornber, C. S., et al. (2006). The impacts of climate change in coastal marine systems. Ecol. Lett. 9, 228–241. doi: 10.1111/j.1461-0248.2005.00871.x
Hartl, D. L., and Clark, A. G. (1997). Principles of Population Genetics, 3rd Edn. Sunderland, MA: Sinauer Associates.
Hedgecock, D., and Pudovkin, A. I. (2011). Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87, 971–1002. doi: 10.5343/bms.2010.1051
Hedgecock, D., Li, G., Hubert, S., Bucklin, K., and Ribes, V. (2004). Widespread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the Pacific oyster, Crassostrea gigas. J. Shellfish Res. 23, 379–386.
Henry, L. A., and Roberts, J. M. (2017). “Global biodiversity in cold-water coral reef ecosystems,” in Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspot, eds S. Rossi, L. Bramanti, A. Gori, and C. Orejas Saco del Valle (Cham: Spring International Publishing). doi: 10.11646/zootaxa.4878.3.2
Hill, W. G. (1981). Estimation of effective population size from data on linkage disequilibrium. Genet. Res. 38, 209–216. doi: 10.1017/S0016672300020553
Hirst, N., Clark, L., and Sanderson, W. G. (2012). The Distribution of Selected MPA Search Features and Priority Marine Features off the NE Coast of Scotland. Scottish Natural Heritage Commissioned Report No.500. Edinburgh: Scottish Natural Heritage.
Hoban, S., Bruford, M. W., Funk, W. C., Galbusera, P., Griffith, M. P., Grueber, C. E., et al. (2021). Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. BioScience 71, 964–976. doi: 10.1093/biosci/biab054
Holleley, C. E., and Geerts, P. G. (2009). Multiplex Manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. Biotechniques 46, 511–517. doi: 10.2144/000113156
Janes, J. K., Miller, J. M., Dupuis, J. R., Malenfant, R. M., Gorrell, J. C., Cullingham, C. I., et al. (2017). The K= 2 conundrum. Mol. Ecol. 26, 3594–3602. doi: 10.1111/mec.14187
Jasim, A. K., and Brand, A. R. (1989). Observations on the reproduction of Modiolus modiolus in Isle of Man waters. J. Mar. Biol. Assoc. U. K. 69, 373–385.
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Jones, A. T., Ovenden, J. R., and Wang, Y. G. (2016). Improved confidence intervals for the linkage disequilibrium method for estimating effective population size. Heredity 117, 217–223. doi: 10.1038/hdy.2016.19
Juffe-Bignoli, D., Burgess, N. D., Bingham, H., Belle, E. M. S., De Lima, M. G., and Deguignet, M. (2014). Protected Planet Report 2014. Cambridge: UNEP-WCMC.
Kenchington, E. L., Kenchington, T. J., Henry, L. A., Fuller, S., and Gonzalez, P. (2007). Multi-decadal changes in the megabenthos of the Bay of Fundy: the effects of fishing. J. Sea Res. 58, 220–240. doi: 10.1016/j.seares.2007.04.001
Kent, F. E. A., Mair, J. M., Newton, J., Lindenbaum, C., Porter, J. S., and Sanderson, W. G. (2017). Commercially important species associated with horse mussel (Modiolus modiolus) biogenic reefs: a priority habitat for nature conservation and fisheries benefits. Mar. Pollut. Bull. 118, 71–78. doi: 10.1016/j.marpolbul.2017.02.051
Kent, F. E., Gray, M. J., Last, K. S., and Sanderson, W. G. (2016). Horse mussel reef ecosystem services: evidence for a whelk nursery habitat supporting a shellfishery. Int. J. Biodivers. Sci. Ecosyst. Serv. Manag. 12, 172–180.
Kent, F. E., Last, K. S., Harries, D. B., and Sanderson, W. G. (2017). In situ biodeposition measurements on a Modiolus modiolus (horse mussel) reef provide insights into ecosystem services. Estuar. Coast. Shelf. 84, 151–157.
Kivelä, M., Arnaud-Haond, S., and Saramäki, J. (2015). EDENetworks: a user-friendly software to build and analyse networks in biogeography, ecology and population genetics. Mol. Ecol. Res. 15, 117–122. doi: 10.1111/1755-0998.12290
Lee, H. Z., Davies, I. M., Baxter, J. M., Diele, K., and Sanderson, W. G. (2020). Missing the full story: first estimates of carbon deposition rates for the European flat oyster, Ostrea edulis. Aquat. Conserv. 30, 2076–2086.
Lowe, A. J., Boshier, D., Ward, M., Bacles, C. F. E., and Navarro, C. (2005). Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Hered 95, 255–273. doi: 10.1038/sj.hdy.6800725
Lowe, A., Harris, S., and Ashton, P. (2004). Ecological Genetics: Design, Analysis, and Application. Oxford: Blackwell Science Ltd, 326.
Lowe, W. H., and Allendorf, F. W. (2010). What can genetics tell us about population connectivity? Mol. Ecol. 19, 3038–3051. doi: 10.1111/j.1365-294X.2010.04688.x
Mair, J. M., Moore, C. G., Kingston, P. F., and Harries, D. B. (2000). A Review of the Status, Ecology and Conservation of Horse Mussel Modiolus Modiolus Beds in Scotland. Scottish Natural Heritage Commissioned Report F99PA08. Edinburgh: Scottish Natural Heritage.
Mair, J. M., Moore, C. G., Lyndon, A. R., and Sotheran, I. (2010). Biotope Mapping and Survey of Sullom Voe Special Area of Conservation. A Report for Scottish Natural Heritage. Heriot-Watt University: Edinburgh.
Mariani, S., Peijnenburg, K. T. C. A., and Weetman, D. (2012). Independence of neutral and adaptive divergence in a low dispersal marine mollusc. Mar. Ecol. Prog. Ser. 446, 173–187. doi: 10.3354/meps09507
Marine Scotland (2011). Marine Protected Areas in Scotland’s Seas Guidelines on the Selection of MPAs and Development of the MPA Network. Edinburgh: The Scottish Government.
Marshall, D. J., Monro, K., Bode, M., Keough, M. J., and Swearer, S. (2010). Phenotype–environment mismatches reduce connectivity in the sea. Ecol. Lett. 13, 128–140. doi: 10.1111/j.1461-0248.2009.01408.x
Martens, L. E. A., Treml, E. A., and von der Heyden, S. (2018). Genetic and biophysical models help define marine conservation focus sreas. Front. Mar. Sci. 5:268. doi: 10.3389/fmars.2018.00268
Mazik, K., Strong, J., Little, S., Bhatia, N., Mander, L., Barnard, S., et al. (2015). A Review of the Recovery Potential and Influencing Factors of Relevance to the Management of Habitats and Species Within Marine Protected Areas Around Scotland. Scottish Natural Heritage Commissioned Report No. 771. Edinburgh: Scottish Natural Heritage.
MCCIP (2018). Climate Change and Marine Conservation: Horse Mussel Beds, eds M. Smedley et al. Lowestoft: MCCIP. doi: 10.14465.2018.ccmco.002-hom
Millar, H., O’Hara Murray, R., Gallego, A., Gormley, K., and Kent, F. (2019). Connectivity of Selected Priority Marine Features Within and Outwith the Scottish MPA Network. Scottish Natural Heritage Research Report No. 1048. Edinburgh: Scottish Natural Heritage.
Navarro, J. M., and Thompson, R. J. (1996). Physiological energetics of the horse mussel Modiolus modiolus in a cold ocean environment. Mar. Ecol. Prog. Ser. 138, 135–148. doi: 10.3354/meps138135
Nie, H., Huo, Z., Li, J., Guo, W., and Yan, X. (2017). Genetic variation and differentiation in wild and selected manila clam inferred from microsatellite loci. J. Shell. Res. 36, 559–565. doi: 10.2983/035.036.0303
Olsen, K. C., Ryan, W. H., Winn, A. A., Kosman, E. T., Moscoso, J. A., Krueger-Hadfield, S. A., et al. (2020). Inbreeding shapes the evolution of marine invertebrates. Evolution 74, 871–882. doi: 10.1111/evo.13951
OSPAR (2021). Protecting and Conserving the North-East Atlantic and Its Resources. Available Online at: https://www.ospar.org/ (accessed June, 2021).
Palumbi, S. R. (2003). Population genetics, demographic connectivity, and the design of marine reserves. Ecol. Appl. 13, S146–S158.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Qiagen (2006). DNeasy Blood and Tissue Handbook. Available Online at: www.QIAGEN.com (accessed September, 2014).
R Development Core Team (2012). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rabe, B., Gallego, A., Wolf, J., O’Hara Murray, R., Stuiver, C., Price, D., et al. (2020). Applied connectivity modelling at local to regional scale: the potential for sea lice transmission between Scottish finfish aquaculture management areas. Estuar. Coast. Shelf. 238:106716. doi: 10.1016/j.ecss.2020.106716
Ragnarsson, S. A., and Burgos, J. M. (2012). Separating the effects of a habitat modifier, Horse mussels and substrate properties on the associated megafauna. J. Sea Res. 72, 55–63. doi: 10.3390/ijms12010456
Reece, K. S., Ribeiro, W. L., Gaffney, P. M., Carnegie, R. B., and Allen, S. K. (2004). Microsatellite marker development and analysis in the eastern oyster (Crassostrea virginica): confirmation of null alleles and non-mendelian segregation ratios. J. Hered. 95, 346–352. doi: 10.1093/jhered/esh058
Rees, E. I. S. (2009). Assessment of Horse Mussels Beds in the OSPAR Area. Peterborough: Joint Nature Conservation Committee, 22.
Rees, E. I. S., Sanderson, W. G., Mackie, A. S. Y., and Holt, R. H. F. (2008). Small-scale variation within a Horse mussels (Mollusca: Bivalvia) reef in the Irish Sea. III. Crevice, sediment infauna and epifauna from targeted cores. J. Mar. Biol Assoc. U.K. 88, 151–156. doi: 10.1017/S0025315408000052
Reid, P. C., Edwards, M., McQuatters-Gollop, A., Beaugrand, G., Bresnan, E., Brierley, A., et al. (2010). “Charting progress 2: healthy and biological diverse seas feeder report: section 3.3: plankton,” in UKMMAS (2010) Charting Progress 2 Healthy and Biological Diverse Seas Feeder Report, eds M. Frost and J. Hawkridge (London: Department for Environment Food and Rural Affairs on behalf of UKMMAS), 286–377.
Riosmena-Rodríguez, R. (2017). “Natural History of Rhodolith/Maërl Beds: their role in near-shore biodiversity and management,” in Rhodolith/Maërl Beds: A Global Perspective, eds R. Riosmena-Rodríguez, W. Nelson, and J. Aguirre (Cham: Spring International Publishing).
Roberts, D., Allcock, A. L., Fariñas-Franco, J. M., Gorman, E., Maggs, C., Mahon, A. M., et al. (2011). Modiolus Restoration Research Project: Final Report and Recommendations. Belfast: Queen’s University Belfast.
Robins, P. E., Neill, S. P., Gimenez, L., Jenkins, S. R., and Malham, S. K. (2013). Physical and biological controls on larval dispersal and connectivity in a highly energetic shelf sea. Limnol. Oceanogr. 58, 1–21. doi: 10.4319/lo.2013.58.2.0505
Robinson, K. A., Mackie, A. S., Lindenbaum, C., Darbyshire, T., van Landeghem, K. J., and Sanderson, W. G. (2012). “Seabed habitats of the southern Irish Sea,” in Seafloor Geomorphology as Benthic Habitat, eds P. T. Harris and E. K. Baker (Amsterdam: Elsevier).
Sameoto, J. A., Hall, K., Gass, S. E., Keith, D., Kirchhoff, S., and Brown, C. J. (2021). Conservation implications of demographic changes in the horse mussel Modiolus modiolus population of the inner Bay of Fundy. Mar. Ecol. Prog. Series 670, 93–104.
Sanderson, W. G., Holt, R. H. F., Kay, L., Ramsay, K., Perrins, J., McMath, A. J., et al. (2008). Small-scale variation within a horse mussel (Mollusca: Bivalvia) reef in the Irish Sea. II. Epifauna recorded by divers and cameras. J. Mar. Biol Assoc. U. K. 88, 143–149. doi: 10.1017/S0025315408000040
Sanford, E., and Kelly, M. W. (2011). Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 3, 509–535. doi: 10.1146/annurev-marine-120709-142756
Schunter, C., Carreras-Carbonell, J., Macpherson, E., Tintoré, J., Vidal-Vijande, E., Pascual, A., et al. (2011). Matching genetics with oceanography: directional gene flow in a Mediterranean fish species. Mol. Ecol. 20, 5167–5181. doi: 10.1111/j.1365-294X.2011.05355.x
Scottish Natural Heritage [SNH], and The Joint Nature Conservation Committee [JNCC] (2012). Advice to the Scottish Government on the selection of Nature Conservation Marine Protected Areas (MPAs) for the development of the Scottish MPA network. Scottish Natural Heritage Commissioned Report No. 547. Edinburgh: Scottish Natural Heritage.
Seed, R., and Brown, R. A. (1977). A comparison of the reproductive cycles of Modiolus modiolus (L.), Cerastoderma (=Cardium) edule (L.), and Mytilus edulis L. in Strangford Lough, Northern Ireland. Oecologia 30, 173–188. doi: 10.1007/BF00345419
Selkoe, K. A., and Toonen, R. J. (2011). Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 436, 291–305. doi: 10.3354/meps09238
Strain, E. M. A., Allcock, A. L., Goodwin, C. E., Maggs, C. A., Picton, B. E., and Roberts, D. (2012). The long-term impacts of fisheries on epifaunal assemblage function and structure, in a Special Area of Conservation. J. Sea Res. 67, 58–68. doi: 10.1016/j.seares.2011.10.001
Teske, P. R., Sandoval-Castillo, J., Van Sebille, E., Waters, J., and Beheregaray, L. B. (2016). Oceanography promotes self-recruitment in a planktonic larval disperser. Sci. Rep. 6:34205. doi: 10.1038/srep34205
Tyler-Walters, H., James, B., Carruthers, M., Wilding, C., Durkin, O., and Lacey, C., et al. (eds) (2016). Descriptions of Scottish Priority Marine Features (PMFs). Scottish Natural Heritage Commissioned Report No. 406. Edinburgh: Scottish Natural Heritage.
Vendrami, D. L. J., Peck, L. S., Clark, M. S., Eldon, B., Meredith, M., and Hoffman, J. I. (2021). Sweepstake reproductive success and collective dispersal produce chaotic genetic patchiness in a broadcast spawner. Sci. Adv. 7:eabj4713. doi: 10.1126/sciadv.abj4713
Waples, R. S. (2006). A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv. Genet. 7:167. doi: 10.1007/s10592-005-9100-y
Waples, R. S., and Do, C. H. I. (2010). Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol. Appl. 3, 244–262. doi: 10.1111/j.1752-4571.2009.00104.x
Waples, R., Larson, W., and Waples, R. (2016). Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity 117, 233–240. doi: 10.1038/hdy.2016.60
Weersing, K., and Toonen, R. J. (2009). Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 393, 1–12. doi: 10.3354/meps08287
Weir, B. S. (1996). Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer.
Whitlock, M. C. (2008). Evolutionary inference from QST. Mol. Ecol. 17, 1885–1896. doi: 10.1111/j.1365-294X.2008.03712.x
Wiborg, K. (1946). Undersøkelser Overoskjellet (Modiola modiolus L.). I. Alrninnelig Biologi, Vekst og Økonomisk Betydning. Norwegian Fishery and Marine Investigation Reports, VIII, 85. Bergen: Directory of Fisheries.
Wilson, B. R., Brown, C. J., Sameoto, J. A., Lacharité, M., Redden, A. M., and Gazzola, V. (2021). Mapping seafloor habitats in the bay of fundy to assess megafaunal assemblages associated with Modiolus modiolus beds. Estuar. Coast. Shelf Sci. 252:107294. doi: 10.1016/j.ecss.2021.107294
Keywords: genetic connectivity, genetic diversity, horse mussel reefs, marine management, marine protected area (MPA), Modiolus modiolus
Citation: Mackenzie CL, Kent FEA, Baxter JM, Gormley KSG, Cassidy AJ, Sanderson WG and Porter JS (2022) Genetic Connectivity and Diversity of a Protected, Habitat-Forming Species: Evidence Demonstrating the Need for Wider Environmental Protection and Integration of the Marine Protected Area Network. Front. Mar. Sci. 9:772259. doi: 10.3389/fmars.2022.772259
Received: 07 September 2021; Accepted: 25 January 2022;
Published: 03 March 2022.
Edited by:
Sophie von der Heyden, Stellenbosch University, South AfricaReviewed by:
Andrew M. Fischer, University of Tasmania, AustraliaJonathan Paul Aden Gardner, Victoria University of Wellington, New Zealand
Copyright © 2022 Mackenzie, Kent, Baxter, Gormley, Cassidy, Sanderson and Porter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanne S. Porter, Si5TLlBvcnRlckBody5hYy51aw==
 Clara L. Mackenzie
Clara L. Mackenzie Flora E. A. Kent
Flora E. A. Kent John M. Baxter
John M. Baxter Kate S. G. Gormley
Kate S. G. Gormley Andrew J. Cassidy
Andrew J. Cassidy William G. Sanderson
William G. Sanderson Joanne S. Porter
Joanne S. Porter