
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mar. Sci., 10 May 2022
Sec. Marine Molecular Biology and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.757921
This article is part of the Research TopicICYMARE - Early Career Researchers in Marine ScienceView all 32 articles
 Tessa M. Page1,2,3,4*†
Tessa M. Page1,2,3,4*† Jonathan W. Lawley1,2,3*†
Jonathan W. Lawley1,2,3*†As we enter the United Nations Decade of Ocean Science for Sustainable Development, extensive efforts to reverse the decline of ocean health are taking place. Moreover, the need to utilize innovative and integrative approaches to aid in these efforts and address marine ecological questions are urgent. Transcriptomic technologies provide tools to further our understanding of an organism’s biology by allowing researchers to rapidly gain information on the genetic variation of populations and the regulation of cellular processes and pathways through gene presence, absence, and expression. Here, we review the application of transcriptomics in the field of marine ecology over the last decade, following a systematic literature review approach. We found 478 articles that fit our search criteria of using transcriptomic approaches to address ecological hypotheses, with 70% of these studies occurring within the last 5 years. Among the analysed articles, 51.7% involved a type of stressor, 16.6% used transcriptomics to study adaptation, and another 15.9% researched ecological interactions. Most articles investigated species from kingdom Animalia, with a high representation from both molluscs (19.5%) and chordates (13.3%), and only 22% of studies had a fieldwork component. Our review demonstrates how the use of transcriptomic techniques in the field of marine ecology is increasing and how they are being applied. Although there are still challenges researchers experience using such techniques, particularly when annotating genes in non-model species and those with no prior genomic resources, these innovative technologies are extremely valuable in investigating differential gene expression, molecular pathways, and generating genomic resources.
Transcriptomics is the study of the transcriptome, or the total set of RNA molecules in a given sample, such as a cell, tissue, or organism, at a given time (Milward et al., 2016). Transcriptomics provides insight into the regulation of cellular processes, pathways, and mechanisms through specific gene expression (Manzoni et al., 2018). Transcriptomic technologies have greatly advanced since the first attempts to study the transcriptome began in the 1990s (Lowe et al., 2017), with next generation sequencing (NGS) arising in the early 2000s (Lowe et al., 2017). NGS quickly replaced previous technologies because it allowed for large scale differential gene expression studies and detection of novel changes that were previously unobtainable (Manzoni et al., 2018). With NGS came the introduction of RNA-Seq, which opened the door to a broader use of transcriptomics on new systems and disciplines. RNA-Seq enabled a surge in research and transcriptome data on non-model organisms, as well as organisms without prior genomic resources (Todd et al., 2016; Matz, 2018; Gleason, 2019). Transcriptomics has the power to reveal details of an organism’s biology by revealing gene expression changes that cannot be discerned through more targeted approaches or traditional phenotypic and physiological response measurements. For example, Niemisto et al. (2021) have demonstrated through differential gene expression that postlarval responses to ocean warming and acidification of the American lobster, Homarus americanus, are more pronounced than found previously in other physiological and morphometric studies. Basu et al. (2017) found diatom-specific genes that were differentially expressed during the sexual phase that were consistent with the unique diatom life cycle, which has evolved species-specific mechanisms to attract the right mating partner. The aforementioned studies demonstrate how transcriptomic approaches can be effective in supplementing traditional phenotypic observations and in addressing independent hypotheses related to underlying biological processes.
In ecology (sensu Jørgensen and Fath, 2008), transcriptomic techniques have become more prevalent, with reviews addressing technologies such as RNA-Seq as important research tools (Todd et al., 2016; Matz, 2018; Gleason, 2019). In marine ecology, transcriptomics enables researchers to investigate ecologically important traits of species such as resistance to challenging environments (Marin-Guirao et al., 2017), disease or infection resistance (Rosenwasser et al., 2019; Soo et al., 2019), life-history (Qiu et al., 2015), development (Liu et al., 2017; Johnson et al., 2019), and plasticity (Wong et al., 2018; Pargana et al., 2020; Strader et al., 2020) through differences in gene expression. The use of transcriptomics, independently and in combination with other types of analyses, can also shed light on adaptation of species through the discovery and identification of novel genes (Huang et al., 2019) or through revealing genetic variability amongst populations, like in Ribeiro et al., 2019 who paired transcriptomics with single-nucleotide polymorphism analysis. Furthermore, studies pairing transcriptomic technologies with other physiological or biological measurements allow for a multidimensional framework to address ecological hypotheses (e.g., Krol et al., 2020; Liu et al., 2020; Strader et al., 2020).
Reviews on 'omics (e.g., genomics, metabolomics, transcriptomics) techniques across a variety of fields have been published (Stillman and Armstrong, 2015; Todd et al., 2016; Chandhini and Kumar, 2019; Gleason, 2019), highlighting how these tools can be used to answer fundamental questions in conservation (Connon et al., 2018), aquaculture (Chandhini and Kumar, 2019), and ecology and evolution (Dupont et al., 2007; Stillman and Armstrong, 2015; Todd et al., 2016; Gleason, 2019). Previous reviews on ‘omics, and more specifically transcriptomics, have examined the possibilities of applying these types of approaches in the field of marine ecology and how they can provide insight into mechanisms underlying tolerance, acclimatisation, and resilience of species (Dupont et al., 2007; Hofmann and Place, 2007). There is a paucity of recent reviews that layout how the use of transcriptomic technologies has developed and grown in marine ecology, and how transcriptomic applications can further progress this discipline. Herein, we provide an update on how transcriptomics is being used in marine ecology, focusing on the geographical spread of research, the aims of these studies, common methodologies, and organisms studied. We also discuss current gaps in knowledge and propose future directions for the use of transcriptomics in marine ecology as we enter the United Nations Decade of Ocean Science for Sustainable Development (https://www.oceandecade.org).
A systematic literature review was conducted to compile a database of marine ecological studies that have used transcriptomic techniques. We used the Web of Science research engine (https://www.webofknowledge.com) to search only articles published between 2010 and 2020 (including early access from 2021). The following search terms were used: “transcriptom*”, “marine”, and (“ecolog*” OR “environment*”), excluding “freshwater*”, “lake*”, or “river*”, within the title, keywords, and/or abstract of publications. These search criteria resulted in 687 publications (Supplementary Table 1), which were manually reviewed and excluded from downstream use if they: (1) did not use transcriptomic tools to generate original data; (2) did not have an associated ecological hypothesis (i.e., related to the interrelations between living organisms and their environment; Jørgensen and Fath, 2008); (3) did not include marine organisms; or (4) were not peer-reviewed, research articles, but instead reviews or methodological briefs.
The remaining 478 publications were assessed, and the following information was added to a database (Supplementary Table 2): (1) author keywords; (2) location of author institutions; (3) location of fieldwork (if applicable), (4) year of publication; (5) taxonomic information of studied organisms (kingdom, phylum, genus, and species; classification based on World Register of Marine Species - WoRMS, https://www.marinespecies.org); (6) type of study (i.e., laboratory- and/or field-based, the latter considered as a study that did not have an experimental ex situ component); (7) main transcriptomics methodology used [expressed sequence tags by Sanger sequencing (EST-SangerSeq), microarrays, RNA-Seq, reverse transcription quantitative polymerase chain reaction (RT-qPCR), and/or suppression subtractive hybridisation (SSH)]; (8) additional methodologies [differential expression of genes analysis (DEG), RT-qPCR validation, and other integrated data from morphology, physiology, histology, proteomics, metabolomics, and/or other chemical analyses]; and (9) the topic(s) of the research article. Up to two topics were assigned to all articles according to the aim of the research, classified as: (1) anthropogenic stress, which includes any study involving global change (i.e., ocean warming, ocean acidification, hypoxia, and freshening) and pollution (i.e., nutrients, heavy metals, artificial light, microplastics, and chemicals); (2) environmental stress (i.e., non-anthropogenic, biotic and abiotic, usually involving temperature, salinity, starvation, injury, and toxins); (3) development; (4) metabolism; (5) harmful algal blooms; (6) reproduction; (7) adaptation, which includes studies involving evolutionary ecology, species and population comparisons that did not fall within another primary category, epigenetics, acclimation, and acclimatisation; and (8) ecological interactions (e.g., symbiosis, parasitism, commensalism, predation, competition, host-pathogen interactions, and trophic interactions).
The database including information manually assigned to each article was used in downstream analyses and the construction of figures, which were accomplished using the statistical computing software R version 4 (R Core Team, 2017). When necessary, further adjustments to figures were done in Adobe Photoshop 2021. iWantHue (https://medialab.github.io/iwanthue/) was used to search for colour blind friendly colours for all figures. Additionally, author keywords were used to construct a word cloud (https://www.wordclouds.com).
Over the last decade, transcriptomics-based studies in marine ecology have increased. Of the 478 articles published between 2010 and 2020 that were herein analysed, 145 were published between 2010 and 2015, and over double that amount from 2016 onward (Figure 1). Transcriptomic technologies have also evolved over the years, with the use of RNA-Seq increasing and becoming the dominant technology used to study transcriptomes (Figure 1). Around 85% of all articles that used RNA-Seq were based on Illumina systems, most frequently on HiSeq platforms (73% of Illumina-based sequencing), which are now being replaced by NovaSeq and NextSeq. The next top RNA-Seq method was pyrosequencing by 454 Life Sciences at only 8% of studies that used RNA-Seq (Supplementary Table 2), however this platform has now been discontinued. Other sequencing platforms with less than 2% representation include SOLiD and Ion Torrent, as well as the emerging Oxford Nanopore, PacBio SMRT, and BGI/MGI-based sequencing.
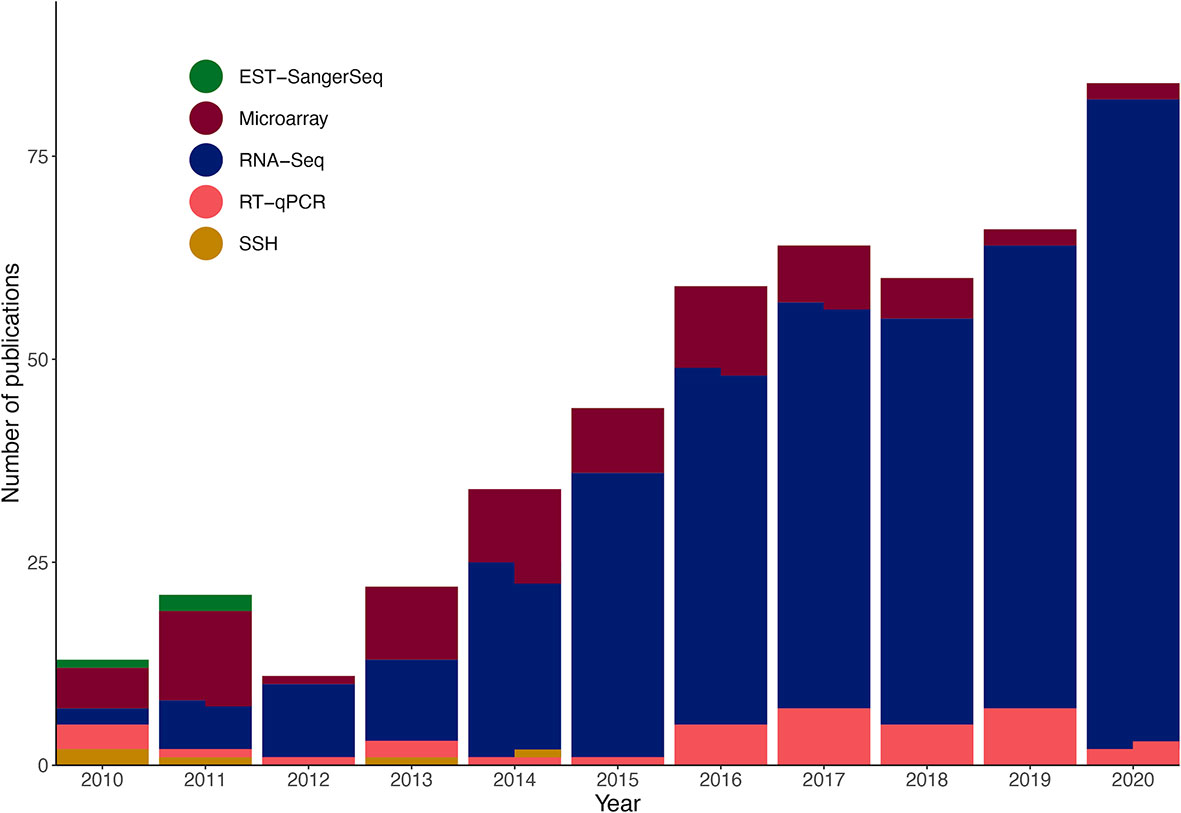
Figure 1 Transcriptomics methodologies used in the marine ecological studies analysed between 2010 and 2020. Some publications used two methods, indicated by split sections in some columns. See Supplementary Tables 1, 2 for details on publications analysed and their methods.
Regarding additional methodologies, most studies involved DEG analysis (75.31%) and 33.05% used RT-qPCR validation (Supplementary Table 2). The use of RT-qPCR validation has decreased within the last 5 years. From 2010 to 2015, 44.43% of studies used RT-qPCR to validate gene expression data, whereas only 27.12% of studies used RT-qPCR for validation between 2016 and 2020 (Supplementary Table 2). Integrative studies (i.e., studies that integrated data from morphology, physiology, histology, proteomics, metabolomics, and/or other chemical analyses) represented 44.98% of articles analysed, of which 71.16% were published after 2015 (Supplementary Table 2).
Of the analysed articles published from 2010 to 2020, the most frequent author-defined keywords, such as “stress” and “adaptation”(Figure 2A), were also among the most frequent manually assigned topics (Figure 2B). The most frequently assigned topics were anthropogenic stress (31.50%), with 17.10% of these investigating the effects of pollution and 14.40% relating to global change effects (Figure 2B). Studies relating to environmental stress (20.20%), adaptation (16.60%), and ecological interactions (15.90%) made up over half of the remaining assigned topics (Figure 2B).
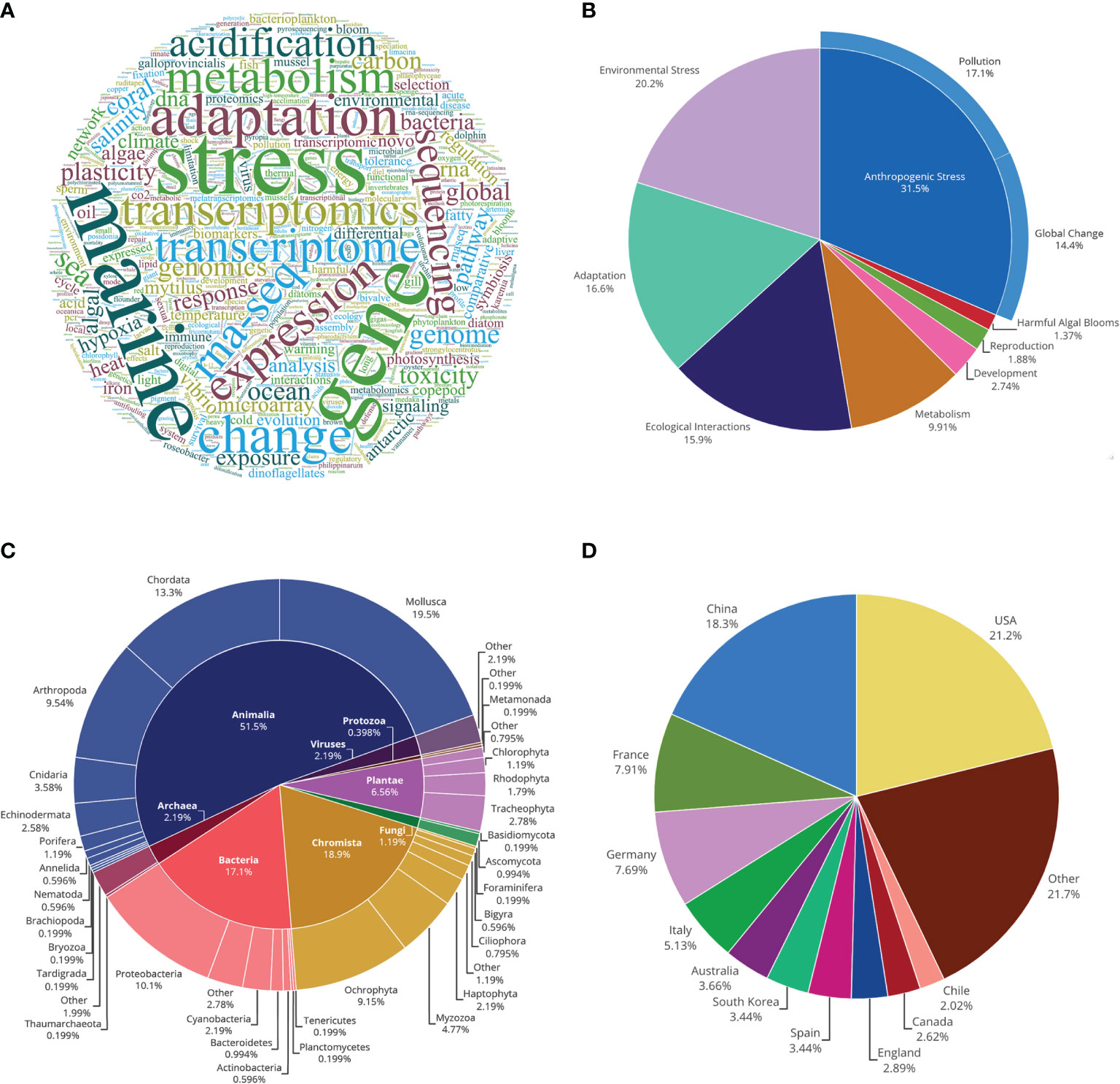
Figure 2 Trends in marine ecological studies using transcriptomics. (A) Word cloud of author keywords, where size of words is proportional to their frequency; (B) Frequency of manually assigned topics; (C) Taxonomic classification of studied organisms, with the inner circle representing kingdom and the outer circle phylum (‘Other’ represents cases when the phylum could not be identified based on information given, such as studies involving organisms from multiple kingdoms); (D) Geographical spread of the research based on countries of author institutions (‘Other’ represents countries with less than 2% representation). See Supplementary Tables 1, 2 for more details.
More than half of all organisms studied belonged to kingdom Animalia (51.50%), 18.90% to Chromista, 17.10% to Bacteria, 6.56% to Plantae, and the remaining 6% to Viruses, Archaea, Fungi, and Protozoa (Figure 2C). Among animals studied, most were molluscs, of which around 60% were bivalves, such as mussels and oysters; or chordates, of which 78% were ray-finned fishes (Actinopterygii) (Supplementary Table 2). All arthropods studied were part of subphylum Crustacea (Supplementary Table 2). Most of the bacteria studied were Proteobacteria or Cyanobacteria, while the Chromista were predominantly Ochrophyta and Myzozoa (Figure 2C), mostly related to unicellular algae. Organisms studied within Plantae were mainly tracheophytes and rhodophytes (Figure 2C), mostly related to seagrasses and macroalgae.
By comparing the proportion of species within the articles analysed to the proportion of accepted marine species, animals appeared as the only underrepresented kingdom, representing 85.38% of accepted species but only 58.39% of the studied species (based on data from WoRMS; Supplementary Table 3). Chordates, however, were among the few overrepresented animal phyla, while rhodophytes (part of Plantae) and Foraminifera (part of Chromista) were the only underrepresented phyla outside Animalia (Supplementary Table 3). Nevertheless, the most species-rich marine invertebrate phyla (Arthropoda and Mollusca, respectively) were also the most well represented (Supplementary Table 3). Overall, the diversity of phyla studied per year in the last decade has increased (Supplementary Table 3).
Lastly, we found almost half of author institutions conducting research in marine ecology using transcriptomics were from the USA (21.2%) or China (18.3%), followed by France with 7.91% (Figure 2D). Only 22% of the analysed articles were considered as primarily field-based (i.e., studies that did not have an experimental laboratory component; Supplementary Table 2). Among field-based studies, 23.15% conducted fieldwork in a location outside the country of any author’s institution, with some of these conducted in international waters and others within the exclusive economic zone of another country (Supplementary Table 2). Therefore, 12.96% of field-based studies did not have at least one author from one or more of the sampling locality countries (Supplementary Table 2).
Transcriptomics provides access to transcriptome-wide gene expression data that allows for characterisation of an organism’s limitations and capacities for traits such as resistance, acclimatisation, plasticity, and development. Our review demonstrates the increasing use of transcriptomics over the years in the field of marine ecology, and how the use of technologies has shifted. Next generation sequencing (NGS) was introduced in the early 2000s (Lowe et al., 2017) however, it took several more years, until 2007-2008, for the first transcriptome studies on non-genome enabled species to be published (Ekblom and Galindo, 2011). As NGS approaches have developed, time to process and overall costs have decreased, making NGS more attractive and obtainable to scientists (Stillman and Armstrong, 2015). In the current review, we show how the use of technologies has shifted from using primarily microarrays and RT-qPCR to RNA-Seq, the latter a NGS approach. Although both RT-qPCR and microarrays are less costly than RNA-Seq, they are limited because they are based on a priori knowledge, whereas NGS allows detection of novel changes and can be used to sequence transcriptomes for the first time (de novo). Furthermore, a study comparing microarrays and RNA-Seq found the latter technology was more accurate than the former (Bradford et al., 2010). Due to the reliability of RNA-Seq methods and our developed knowledge on designing experiments for RNA-Seq considering biological replicates and robust statistical analysis (Coenye, 2021), future differential gene expression results from transcriptomic studies using RNA-Seq should not have to use RT-qPCR for validation. In fact, previous studies indicate that RNA-Seq and RT-qPCR data are highly correlated (Shi and He, 2014; Wu et al., 2014). Methods in transcriptomics are adapting as technologies advance and become more accessible, therefore, enabling researchers to better understand mechanisms that underlay biological processes or traits, indicating transcriptomics to be an impactful tool in marine ecological research.
Transcriptomics is now being used on a wide range of topics in marine ecology, including the investigation of ecological traits such as development and reproduction, as well as broader ecological attributes such as adaptation, interactions, and response to stressors. In general, transcriptomics is most prevalently used to address ecological hypotheses relating to the effects of stressors on marine organisms (e.g., Yum et al., 2015; Shama et al., 2016; Boyen et al., 2020; Niemisto et al., 2021). This highlights the current need in marine ecology, and various other disciplines, to further our understanding of how organisms will fare under future conditions driven by changes in their environment due to anthropogenic stressors. Experimental laboratory studies can help us understand how species respond to these current and predicted stressors. On the other hand, field-based experiments or sampling can give us insight into ecologically important parameters such as, but not limited to, genetic variation within and across populations of species (Bernal et al., 2019; Peng et al., 2020) and insights into adaptation of species to their current environments (Zhang et al., 2016; Heras and Aguilar, 2019), which also can provide insight into their potential for adaptation and/or acclimatisation to predicted future conditions (Gan et al., 2020; Leder et al., 2021). Future studies should therefore aim for both field and laboratory components when using transcriptomics in marine ecology.
Most of the organisms in the studies herein analysed are animals, yet ironically this is the only underrepresented kingdom when the proportion of accepted marine species is considered. Chordates appear as one of the few overrepresented animal phyla. Previous reviews on taxonomic bias in ecology and conservation biology have found similar results, in which vertebrates (part of Chordata) are overrepresented, while invertebrate groups are underrepresented (Clark and May, 2002; Donaldson et al., 2017; Troudet et al., 2017). Considering that morphological and physiological data are more easily acquired for macroorganisms, it is not surprising that there are fewer ecological studies using transcriptomic tools on animals compared to other kingdoms, which, except for Plantae, mostly include microorganisms. Yet, transcriptomic studies have proven invaluable to macroorganisms and microorganisms alike, in some cases providing important information otherwise not obtained through traditional morphological and physiological analyses (e.g., Strader et al., 2020; Niemisto et al., 2021). Integrating these traditional analyses with ‘omics approaches such as transcriptomics, can lead us to more fully understand organisms’ biology and their potential for adaptation and acclimatisation, as suggested by Gleason (2019). Of the studies analysed herein that integrated other data, most were published after 2015, suggesting this type of integrative approach is gaining momentum in marine ecological research and should be considered in future studies when using transcriptomics.
Over the last decade the diversity of phyla studied per year has also increased, yet there is a lack of functional genetic studies for many of these phyla. In some cases, around 40% of sequenced genes are unannotated (e.g., Kong et al., 2014), and in others over 50% of genes could not be associated to a gene ontology (GO) term or Kyoto encyclopedia of genes and genomes (KEGG) pathway (e.g., Ip et al., 2016). This likely occurs due to lack of annotated genetic data from closely related organisms and can lead to vague conclusions for the hypotheses at hand and/or uncertainties when analysing the differential expression of genes (Matz, 2018). The lack of annotated genetic data for non-model organisms is a challenge faced during bioinformatic analysis and when using transcriptomics in marine ecology. Further expanding functional genetics in underrepresented phyla can likely improve these analyses.
Our review also highlights trends in the geographical locations of author institutions and where fieldwork was conducted. Most of the countries with over 2% representation of author institutions (78.3% of all institutions listed), are high-income countries according to the World Bank (https://www.worldbank.org). In some cases, researchers from high-income countries have conducted fieldwork in a location within the exclusive economic zone of another country, often a low-income country, without working or giving credit to local scientists. This practice is called “parachute” or “helicopter” science and it has been highlighted across various fields (Adame, 2021; Stefanoudis et al., 2021). In the current review, we found 12.96% of field-based studies qualified as parachute or helicopter research, largely by scientists from wealthier nations conducting research in low-income countries (albeit not true in all cases). Marine ecologists should strive to develop local meaningful collaborations, which often leads to opportunities for local researchers as well as benefits to the surrounding community.
As we enter the United Nations Decade of Ocean Science for Sustainable Development, there is an urgent need to continue with innovative and integrative research in marine ecology, to help understand marine organisms and their interactions with the environment to drive conservation efforts in a changing ocean. Diversifying the taxa studied and expanding functional research, especially for underrepresented groups such as invertebrate phyla, can improve genetic databases as well as gene annotations, leading to a better understanding of gene function and the molecular processes involved. Field-based studies should avoid “helicopter” or “parachute” science by collaborating with local researchers and benefiting the community of their sampling localities. As RNA-Seq is more reliable today, we suggest that the use of RT-qPCR for validation of well-designed RNA-Seq experiments that are statistically robust is not always necessary. Our review demonstrates how transcriptomics can be used to not only supplement traditional marine ecological studies, allowing us to quantify links between genotype and phenotype particularly when pairing transcriptomics with other ‘omics approaches like metabolomics, but also answer marine ecological questions that may have been previously unanswerable or at best only hypothesised.
All authors conceived of the idea, conducted the literature review, performed the analyses, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
We would like to thank the state of Bremen for kindly supporting the publication of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Dr. Carmel McDougall for reviewing this manuscript and for her helpful comments and edits. This study was conceptualised after we hosted the session “The next generation is here – genomic approaches in marine science” in the International Conference for Young Marine Researchers (ICYMARE) 2020. Therefore, we would like to thank all participants who collaborated in that session, as well as conference organisers Dr. Simon Jungblut, Dr. Viola Liebich, and Jan Boelmann for their support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.757921/full#supplementary-material
Adame F. (2021). Meaningful Collaborations Can End ‘Helicopter Research’. Nature. doi: 10.1038/d41586-021-01795-1
Basu S., Patil S., Mapleson D., Russo M., Vitale L., Fevola C., et al. (2017). Finding a Partner in the Ocean: Molecular and Evolutionary Bases of the Response to Sexual Cues in a Planktonic Diatom. N. Phytol. 215, 140–156. doi: 10.1111/nph.14557
Bernal M., Dixon G., Matz M., Rocha L. (2019). Comparative Transcriptomics of Sympatric Species of Coral Reef Fishes (Genus: Haemulon). PEERJ 7. doi: 10.7717/peerj.6541
Boyen J., Fink P., Mensens C., Hablutzel P., De Troche M. (2020). Fatty Acid Bioconversion in Harpacticoid Copepods in a Changing Environment: A Transcriptomic Approach. Philos. Trans. R. Soc. B-Biol. Sci. 375. doi: 10.1098/rstb.2019.0645
Bradford J. R., Hey Y., Yates T., Li Y., Pepper S. D., Miller C. J. (2010). A Comparison of Massively Parallel Nucleotide Sequencing With Oligonucleotide Microarrays for Global Transcription Profiling. BMC Genomics 11, 1–12. doi: 10.1186/1471-2164-11-282
Chandhini S., Kumar V. J. R. (2019). Transcriptomics in Aquaculture: Current Status and Applications. Rev. Aquac. 11, 1379–1397. doi: 10.1111/raq.12298
Clark J. A., May R. M. (2002). Taxonomic Bias in Conservation Research. Science 297, 191–192. doi: 10.1126/science.297.5579.191b
Coenye T. (2021). Do Results Obtained With RNA-Sequencing Require Independent Verification? Biofilm 3, 100043. doi: 10.1016/j.bioflm.2021.100043
Connon R. E., Jeffries K. M., Komoroske L. M., Todgham A. E., Fangue N. A. (2018). The Utility of Transcriptomics in Fish Conservation. J. Exp. Biol. 221. doi: 10.1242/jeb.148833
Donaldson M. R., Burnett N. J., Braun D. C., Suski C. D., Hinch S. G., Cooke S. J., et al. (2017). Taxonomic Bias and International Biodiversity Conservation Research. FACETS 1, 105–113. doi: 10.1139/facets-2016-0011
Dupont S., Wilson K., Obst M., Sköld H., Nakano H., Thorndyke M. (2007). Marine Ecological Genomics: When Genomics Meets Marine Ecology. Mar. Ecol. Prog. Ser. 332, 257–273. doi: 10.3354/meps332257
Ekblom R., Galindo J. (2011). Applications of Next Generation Sequencing in Molecular Ecology of Non-Model Organisms. Heredity 107, 1–15. doi: 10.1038/hdy.2010.152
Gan Z., Yuan J., Liu X., Dong D., Li F., Li X. (2020). Comparative Transcriptomic Analysis of Deep- and Shallow-Water Barnacle Species (Cirripedia, Poecilasmatidae) Provides Insights Into Deep-Sea Adaptation of Sessile Crustaceans. BMC Genomics 21. doi: 10.1186/s12864-020-6642-9
Gleason L. U. (2019). Applications and Future Directions for Population Transcriptomics in Marine Invertebrates. Curr. Mol. Biol. Rep. 5, 116–127. doi: 10.1007/s40610-019-00121-z
Heras J., Aguilar A. (2019). Comparative Transcriptomics Reveals Patterns of Adaptive Evolution Associated With Depth and Age Within Marine Rockfishes (Sebastes). J. Hered. 110, 340–350. doi: 10.1093/jhered/esy070
Hofmann G., Place S. (2007). Genomics-Enabled Research in Marine Ecology: Challenges, Risks and Pay-Offs. Mar. Ecol. Prog. Ser. 332, 249–255. doi: 10.3354/meps332249
Huang H., Li Z., Chen M. (2019). De Novo Assembly and Characterization of the Transcriptome of the Northern Mauxia Shrimp Acetes Chinensis. Mar. Genomics 47. doi: 10.1016/j.margen.2019.03.007
Ip J., Leung P., Ho K., Qiu J., Leung K. (2016). De Novo Transcriptome Assembly of the Marine Gastropod Reishia Clavigera for Supporting Toxic Mechanism Studies. Aquat. Toxicol. 178, 39–48. doi: 10.1016/j.aquatox.2016.07.006
Jørgensen S. E., Fath B. D. (2008). “Preface,” in Encyclopedia of Ecology. Eds. Jørgensen S. E., Fath B. D. (Oxford: Academic Press), xxix–xxxx. doi: 10.1016/B978-008045405-4.09005-4
Johnson K., Wong J., Hoshijima U., Sugano C., Hofmann G. (2019). Seasonal Transcriptomes of the Antarctic Pteropod, Limacina Helicina Antarctica. Mar. Environ. Res. 143, 49–59. doi: 10.1016/j.marenvres.2018.10.006
Kong F., Li H., Sun P., Zhou Y., Mao Y. (2014). De Novo Assembly and Characterization of the Transcriptome of Seagrass Zostera Marina Using Illumina Paired-End Sequencing. PloS One 9. doi: 10.1371/journal.pone.0112245
Krol E., Noguera P., Shaw S., Costelloe E., Gajardo K., Valdenegro V., et al. (2020). Integration of Transcriptome, Gross Morphology and Histopathology in the Gill of Sea Farmed Atlantic Salmon (Salmo Salar): Lessons From Multi-Site Sampling. Front. Genet. 11. doi: 10.3389/fgene.2020.00610
Leder E., Andr C., Le Moan A., Topel M., Blomberg A., Havenhand J., et al. (2021). Post-Glacial Establishment of Locally Adapted Fish Populations Over a Steep Salinity Gradient. J. Evol. Biol. 34, 138–156. doi: 10.1111/jeb.13668
Liu S., Li J., Wu Y., Ren Y., Liu Q., Wang Q., et al. (2017). De Novo Transcriptome Sequencing of Marine-Derived Aspergillus Glaucus and Comparative Analysis of Metabolic and Developmental Variations in Response to Salt Stress. Genes Genomics 39, 317–329. doi: 10.1007/s13258-016-0497-0
Liu Q., Tang X., Jian X., Yang Y., Ma W., Wang Y., et al. (2020). Toxic Effect and Mechanism of Tris (1,3-Dichloro-2-Propyl)Phosphate (TDCPP) on the Marine Alga Phaeodactylum Tricornutum. Chemosphere 252. doi: 10.1016/j.chemosphere.2020.126467
Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. (2017). Transcriptomics Technologies. PloS Comput. Biol. 13, e1005457. doi: 10.1371/journal.pcbi.1005457
Manzoni C., Kia D. A., Vandrovcova J., Hardy J., Wood N. W., Lewis P. A., et al. (2018). Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 19, 286–302. doi: 10.1093/bib/bbw114
Marin-Guirao L., Entrambasaguas L., Dattolo E., Ruiz J., Procaccini G. (2017). Molecular Mechanisms Behind the Physiological Resistance to Intense Transient Warming in an Iconic Marine Plant. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01142
Matz M. V. (2018). Fantastic Beasts and How To Sequence Them: Ecological Genomics for Obscure Model Organisms. Trends Genet. 34, 121–132. doi: 10.1016/j.tig.2017.11.002
Milward E. A., Shahandeh A., Heidari M., Johnstone D. M., Daneshi N., Hondermarck H. (2016). “Transcriptomics,” in Encyclopedia of Cell Biology. Eds. Bradshaw R. A., Stahl P. D. (Waltham: Academic Press), 160–165. doi: 10.1016/B978-0-12-394447-4.40029-5
Niemisto M., Fields D., Clark K., Waller J., Greenwood S., Wahle R. (2021). American Lobster Postlarvae Alter Gene Regulation in Response to Ocean Warming and Acidification. Ecol. Evol. 11, 806–819. doi: 10.1002/ece3.7083
Pargana A., Musacchia F., Sanges R., Russo M., Ferrante M., Bowler C., et al. (2020). Intraspecific Diversity in the Cold Stress Response of Transposable Elements in the Diatom Leptocylindrus Aporus. Genes 11. doi: 10.3390/genes11010009
Peng C., Ren J., Deng C., Jiang D., Wang J., Qu J., et al. (2020). The Genome of Shaw’s Sea Snake (Hydrophis Curtus) Reveals Secondary Adaptation to Its Marine Environment. Mol. Biol. Evol. 37, 1744–1760. doi: 10.1093/molbev/msaa043
Qiu F., Ding S., Ou H., Wang D., Chen J., Miyamoto M. (2015). Transcriptome Changes During the Life Cycle of the Red Sponge, Mycale Phyllophila (Porifera, Demospongiae, Poecilosclerida). Genes 6, 1023–1052. doi: 10.3390/genes6041023
R Core Team (2017). R: A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Ribeiro A., Canchaya C., Penaloza F., Galindo J., da Fonseca R. (2019). Population Genomic Footprints of Environmental Pollution Pressure in Natural Populations of the Mediterranean Mussel. Mar. Genomics 45, 11–15. doi: 10.1016/j.margen.2018.10.009
Rosenwasser S., Sheyn U., Frada M., Pilzer D., Rotkopf R., Vardi A. (2019). Unmasking Cellular Response of a Bloom-Forming Alga to Viral Infection by Resolving Expression Profiles at a Single-Cell Level. PloS Pathog. 15. doi: 10.1371/journal.ppat.1007708
Shama L., Mark F., Strobel A., Lokmer A., John U., Wegner K. (2016). Transgenerational Effects Persist Down the Maternal Line in Marine Sticklebacks: Gene Expression Matches Physiology in a Warming Ocean. Evol. Appl. 9, 1096–1111. doi: 10.1111/eva.12370
Shi Y., He M. (2014). Differential Gene Expression Identified by RNA-Seq and qPCR in Two Sizes of Pearl Oyster (Pinctada Fucata). Gene 538, 313–322. doi: 10.1016/j.gene.2014.01.031
Soo T., Devadas S., Din M., Bhassu S. (2019). Differential Transcriptome Analysis of the Disease Tolerant Madagascar-Malaysia Crossbred Black Tiger Shrimp, Penaeus Monodon Hepatopancreas in Response to Acute Hepatopancreatic Necrosis Disease (AHPND) Infection: Inference on Immune Gene Response and Interaction. Gut Pathog. 11. doi: 10.1186/s13099-019-0319-4
Stefanoudis P. V., Licuanan W. Y., Morrison T. H., Talma S., Veitayaki J., Woodall L. C. (2021). Turning the Tide of Parachute Science. Curr. Biol. 31, R184–R185. doi: 10.1016/j.cub.2021.01.029
Stillman J. H., Armstrong E. (2015). Genomics are Transforming Our Understanding of Responses to Climate Change. BioScience 65, 237–246. doi: 10.1093/biosci/biu219
Strader M., Kozal L., Leach T., Wong J., Chamorro J., Housh M., et al. (2020). Examining the Role of DNA Methylation in Transcriptomic Plasticity of Early Stage Sea Urchins: Developmental and Maternal Effects in a Kelp Forest Herbivore. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00205
Todd E. V., Black M. A., Gemmell N. J. (2016). The Power and Promise of RNA-Seq in Ecology and Evolution. Mol. Ecol. 25, 1224–1241. doi: 10.1111/mec.13526
Troudet J., Grandcolas P., Blin A., Vignes-Lebbe R., Legendre F. (2017). Taxonomic Bias in Biodiversity Data and Societal Preferences. Sci. Rep. 7, 1–14. doi: 10.1038/s41598-017-09084-6
Wong J., Johnson K., Kelly M., Hofmann G. (2018). Transcriptomics Reveal Transgenerational Effects in Purple Sea Urchin Embryos: Adult Acclimation to Upwelling Conditions Alters the Response of Their Progeny to Differential pCO(2) Levels. Mol. Ecol. 27, 1120–1137. doi: 10.1111/mec.14503
Wu S., Sun J., Chi S., Wang L., Wang X., Liu C., et al. (2014). Transcriptome Sequencing of Essential Marine Brown and Red Algal Species in China and Its Significance in Algal Biology and Phylogeny. Acta Oceanol. Sin. 33, 1–12. doi: 10.1007/s13131-014-0435-4
Yum S., Jo Y., Woo S. (2015). Metabolic Changes in Marine Medaka Fish (Oryzias Javanicus) in Response to Acute 4-Nonlyphenol Toxicity. Biochip. J. 9, 322–331. doi: 10.1007/s13206-015-9408-8
Keywords: transcriptomics, marine ecology, next generation sequencing, gene expression, RNA-Seq, microarrays, marine molecular ecology
Citation: Page TM and Lawley JW (2022) The Next Generation Is Here: A Review of Transcriptomic Approaches in Marine Ecology. Front. Mar. Sci. 9:757921. doi: 10.3389/fmars.2022.757921
Received: 13 August 2021; Accepted: 14 April 2022;
Published: 10 May 2022.
Edited by:
Simon Jungblut, University of Bremen, GermanyReviewed by:
Lani Gleason, California State University, Sacramento, United StatesCopyright © 2022 Page and Lawley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tessa M. Page, dGVzc2EucGFnZUBncmlmZml0aC5lZHUuYXU=; Jonathan W. Lawley, am9uYXRoYW4ud2FuZGVybGV5bGF3bGV5QGdyaWZmaXRodW5pLmVkdS5hdQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.