
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 January 2023
Sec. Marine Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1110587
This article is part of the Research Topic Integration of Development, Physiology and Responses to Environmental Change in Aquatic Invertebrates View all 13 articles
Sexual differentiation and gonad development are important biological processes for bivalve species. The clam Cyclina sinensis is an important cultured marine bivalve widely distributed along with the coastal areas of China. However, the information related to sexual determination/differentiation and gonadal development of C. sinensis almost has no reported. To study the molecular mechanisms of its sexual determination/differentiation and gonadal development, transcriptome analysis was performed in the gonad of C. sinensis, and the potential biological functions of reproduction-related gene were also investigated in this study. The results showed that 1 013 and 427 genes were differentially expressed in the ovary and testis, respectively. KEGG enrichment analysis showed that the DEGs in the gonad were significantly clustered in progesterone-mediated oocyte maturation, cell cycle and oocyte meiosis. Further analysis showed that 23 genes were mainly involved in sex determination/differentiation, including Dmrt1, Sox2/4/9, Foxl2, β-catenin and GATA-type zinc finger protein 1-like. Twenty key genes were mainly involved in the process of spermatogenesis, and five genes encode steroid biosynthesis and metabolism. Fifteen genes related to ovarian development were also identified in this study, such as Vitellogenin, MAM and LDL-receptor class A domain-containing protein 1 and Cell division cycle protein 20 homolog. Moreover, 50µg/L estradiol treatment significantly up-regulated the expression levels of CsVg in the ovary and hepatopancreas. These results highlight the genes involved in sexual determination/differentiation and gonadal development, which enhance our understanding for further studies of reproduction and breeding of C. sinensis and other marine bivalves.
● 23 genes involved in sex determination/differentiation of C. sinensis were identified.
● Twenty key genes played important role in the process of spermatogenesis of C. sinensis.
● 15 key genes played important role in the ovarian development of C. sinensis.
● 50µg/L estradiol treatment significantly up-regulated the expression levels of CsVg.
Sexual reproduction is one of the most universal phenomena in the animal kingdom, and it includes sexual determination/differentiation and gonadal development (Li et al., 2016a; Liu et al., 2016). Sexual determination/differentiation refers to the event that triggers the bipotential primordia to make the fate decision to become testes or ovaries, and it is very flexible in fish, with several species showing a chromosomal basis and others with influencing environmental factors (Devlin and Nagahama, 2002; Sandra and Norma, 2010). Previous studies showed that sex-determining/−differentiating pathways share common genes such as SRY-related HMG box 9 (SOX9), wingless-type MMTV integration site family member-4 (WNT4), R-spondin1 (R-SPO1) and transcription factor forkhead box L subfamily member 2 (FOXL2), among vertebrates and invertebrates, (Bertho et al., 2016; Major et al., 2019; Estermann et al., 2020; Broquard et al., 2021). Among those genes, Sox9 has been identified and proved its crucial roles in male sex determination, male gonad development, and Sertoli cell development and differentiation (Vidal et al., 2002; Chaboissier et al., 2004; Barrionuevo et al., 2009).
WNT4 is a key regulator of ovarian development in mammals, and R-spondin1 tips the balance in sex determination (Capel, 2006; Farhadi et al., 2021). Ning et al. (2021) reported that the transcription factor FoxL2 is an evolutionarily conserved gene playing pivotal roles in regulation of early ovarian differentiation and maintenance in animals. Similarly, as the primary reproductive organs, the ovary and testis play keys role in gametogenesis and steroid regulation of vertebrates and invertebrates (Nagaraju, 2011; Senthilkumaran and Kar, 2021).
As the second largest phylum of invertebrates, mollusks exhibit different reproductive strategies: they are dioecious, hermaphrodites or exhibit sex reversal (Li et al., 2018). Therefore, the studies on reproduction of mollusk, especially commercial mollusk species has received significant research attention in recent years (Wang et al., 2020; Broquard et al., 2021; Shangguan et al., 2022). For example, Santerre et al. (2014) reported that SoxE and β-catenin are involved in early gonadic differentiation of Pacific oyster Crassostrea gigas. Shangguan et al. (2022) reported that 17-alpha-hydroxylase/17,20 lyase (cyp17a1) plays an important role in gonadal development of Hyriopsis cumingii. Meanwhile, several gonadal transcriptome analyses identified candidate genes (SoxH, Sox9, Foxl2, Doublesex- and mab-3-related transcription factor 1 (Dmrt1), sex determining protein Fem-1 (Fem-1), Beta-catenin, wnt4, cyp17a1, Estradiol 17-beta-dehydrogenase 2 (17β-hsd2) and Vitellogenin (Vg)) that participate in gender determination/differentiation and gonadal maturation pathway of mollusks, including Crassostrea hongkongensis, C. gigas, Patinopecten yessoensis, Sinonovacula constricta, Pinctada margaritifera, Tegillarca granosa, Chlamys nobilis and Hyriopsis schlegelii (Teaniniuraitemoana et al., 2014; Zhang et al., 2014; Tong et al., 2015; Li et al., 2016a; Chen et al., 2017; Yue et al., 2018; Broquard et al., 2021; Yao et al., 2021; Zeng et al., 2022). Despites the identification of some candidate genes, studies on sex determination/differentiation and gonadal maturation genes in bivalves and other molluscs were still limited, and more sex and gonad related genes and their functions need to be studied.
The clam, Cyclina sinensis, is one of the important cultured marine bivalves found widely along the coastal areas of China, Korea, Japan, and Southeast Asia (Wei et al., 2020; Ge et al., 2021; Liao et al., 2022). To date, the research on C. sinensis mainly focuses on genetic evaluation, breeding techniques, nutritional value and physiological responses (Wei et al., 2020; Dong et al., 2021; Ge et al., 2021; Liao et al., 2022). However, the information related to sexual determination/differentiation and gonadal development of C. sinensis is limited. Transcriptome sequencing provides an effective way to obtain large amounts of sequence data, has been widely applied to model and non-model species (Qian et al., 2014). The availability of the C. sinensis genome and the reduced cost of next-generation sequencing provide the opportunity for enhance our understanding of the complex regulation mechanisms of reproduction (Wei et al., 2020; Dong et al., 2021; Ge et al., 2021; Liao et al., 2022). Therefore, the present study sequenced C. sinensis gonad transcriptomes and investigated the potential biological functions of reproduction-related genes. The results of this study will help us understand the general underlying molecular mechanisms of bivalve reproduction and provide a scientific basis for the sexual control and breeding of shellfish.
All the study design and animal experiments were conducted in accordance with guidelines of Jiangsu Ocean University’s Animal Care and Use Committee.
The female and male C. sinensis used in this experiment were purchased from a commercial farmer in Qinzhou, Guangxi, China. After being transferred to the laboratory, the clams were acclimated in cement tanks (length×width× depth=100cm×100cm×80cm) filled with natural seawater (salinity 24 ± 1 ppt). All clams were fed daily at 17:00 with a mixture of Nannochloropsis oceanica and Chaetoceros mulleri quantitatively. After acclimation for seven days, the clams were placed in an ice bath for anesthetisation, and the ovary or testis was dissected. The sampled ovary or testis was divided into two pieces: one piece was immediately frozen in liquid nitrogen for total RNA extraction, and the other piece was fixed in 4% paraformaldehyde for histological examination. Based on the histological features of previously reported by Yan (2009), the gonadal development was classified into five stages: proliferation stage (stage I), growing stage (stage II), maturation stage (stage III), spawning stage (stage IV), and spent stage (stage V). Based on the staging results, the ovary or testis tissues from three individuals at growing stage were selected for transcriptome analysis.
Total RNA was extracted from each sample using TRIzol reagent (Invitrogen, USA) according the manufacturer’s instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKara). The integrity and purity of the total RNA quality were determined by 2100 Bioanalyser (Agilent Technologies) and quantified using the ND-2000 (NanoDrop Technologies).
RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions (Illumina, San Diego, CA). The transcriptome library was prepared following TruSeqTM RNA sample preparation kit from Illumina (San Diego, CA) using 1μg total RNA. Then, the synthesized cDNA was subjected to end-repair, phosphorylation and ‘A’ base addition following Illumina’s library construction protocol. After quantification by TBS380, paired-end RNA sequencing library was sequenced with the Illumina NovaSeq 6000 sequencer (2 × 150 bp read length).
The raw paired-end reads were trimmed and quality controlled by fastp with default parameters (Chen et al., 2018). Then, clean data from the samples were used to perform de-novo assembly with Trinity (Grabherr et al., 2011). The assembled transcripts were assessed and optimized with BUSCO (Manni et al., 2021), TransRate (Smith-Unna et al., 2016) and CD-HIT (Fu et al., 2012). All the assembled transcripts were searched against the National Center for Biotechnology Information (NCBI) protein non-redundant (Nr), Swiss-Prot, Pfam, Clusters of Orthologous Groups of proteins, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTX to identify proteins that had the highest sequence similarity with the given transcripts to retrieve their function annotations, and a typical cut-off E-values less than 1.0×10−5 was set.
The expression level of each transcript was calculated using the fragments per kilobase of exon per million mapped reads (FPKM) method (Trapnell et al., 2010). To identify differentially expressed genes (DEGs) across samples, we considered DEGs with |log2 (foldchange)| ≥ 1 and P-value ≤ 0.05 to be significantly different expressed genes. To identify DEGs that were significantly enriched in GO terms and metabolic pathways at P-value ≤0.05, we applied Goatools and KOBAS were applied for GO functional enrichment and KEGG pathway analysis, respectively (Xie et al., 2011).
Eight genes involved in gonadal development and sexual determination/differentiation were randomly selected for validation using qPCR. A reverse first-strand cDNA synthesis kit (RR036A, Takara Bio, Japan) was used to synthesize first-strand cDNA. Primer pairs were designed using Primer 6.0, and all the primer sequences are listed in Table 1. β-actin was used as the internal control to normalize target gene expression. qPCR was carried out with a StepOnePlus™ real-time PCR system using SYBR1 Green I (TakaRa, Japan) according to the manufacturer’s instructions. The volume of qPCR reaction system was 20 μl containing 2 μl of cDNA template, 0.4 μl of each primer (10 mM), 10 μl of SYBR premix, 0.4 μl of ROX Reference Dye (50×) and 6.8 μl of water. PCR conditions were as follows: 95°C for 30s; 40 cycles of 95°C for 5s, 60/60.2°C for 30 s, and 72°C for 30 s. The relative expression level of each gene was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Based on the results of transcriptome sequencing, the higher expression level of CsVg was found in the ovary of C. sinensis, the CsVg was selected for the further analysis in this study. The amino acid sequence and open reading frame (ORF) of CsVg were predicted by the ORF finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The conserved domains of CsVg and other species were analyzed using the online software Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Subsequently, ClustalX and MEGA 5.0 software were used to carry out multiple sequence alignment and phylogenetic tree construction of neighbor-joining (NJ) system. The amino acid sequences of other species used in these analyses were obtained from GenBank, including Xenopus laevis (AB092605.1), Oryzias latipes (AB092605.1), Caenorhabditis elegans (X56213.1), Portunus trituberculatus (DQ000638.1), Scylla paramamosain (KU987908.1), Litopenaeus vannamei (AY321153.2), Haliotis discus hannai (AB360714.1), Scapharca broughtonii (MG580782.1), Tegillarca granosa (JQ266265.2), Crassostrea angulate (JX218047.2), Crassostrea gigas (AB084783.1), Saccostrea glomerata (KU194475.1) and Ostrea edulis (XM_048903027.1).
To assay the gene expression of CsVg in different tissues, several tissues (ovary, hepatopancreas, adductor, pipe, gill, foot, mantle) were dissected from the clams in the gonadal growing stage. In this experiment, different tissues from three individual C. sinensis were used for tissue-specific expression analysis. Meanwhile, the ovary from female clams at each gonadal developmental stage were also collected to determine the abundance of CsVg transcripts. A total of 6-8 clams were used for sampling in each stage. Total RNA isolation, cDNA synthesis and PCR reactions were all performed as described above.
To analyze the transcriptional response of CsVg to estradiol, we performed an in vivo exposure experiment using C. sinensis at the gonadal growing stage was performed. Specifically, 120 healthy clams were randomly divided into three groups of 30. The clams were exposed for 21 days to various nominal estradiol concentration (0+ethanol µg/L, 5 µg/L and 50 µg/L) in accordance with previous literature (Wu et al., 2019). During exposure, the clams were maintained in a 20 L plastic container, and uneaten food and feces were removed before water renewal. Throughout the experiment, the seawater with estradiol was changed daily to ensure that the concentration of estradiol in each group remained invariable during the experiment. At the end of the estradiol exposure, the ovary and hepatopancreas were dissected and stored at -80°C for RNA extraction.
Data are presented as the mean ± standard error (SE). Statistical analyses were performed using SPSS Statistics V22.0. P value of < 0.05 was considered statistically significant. When normal distribution and/or homogeneity of variances were not achieved, data were subjected to the Kruskal-Wallis H nonparametric test, followed by the Games-Howell nonparametric multiple comparison test.
After dissection, the structure of the ovary and testis can be easily distinguished by microscopic observation of gonadal sections. Histological results showed that the ovaries and testes of C. sinensis used for RNA sequencing were at growing stage (stage II) of gonadal maturation (Figure 1).
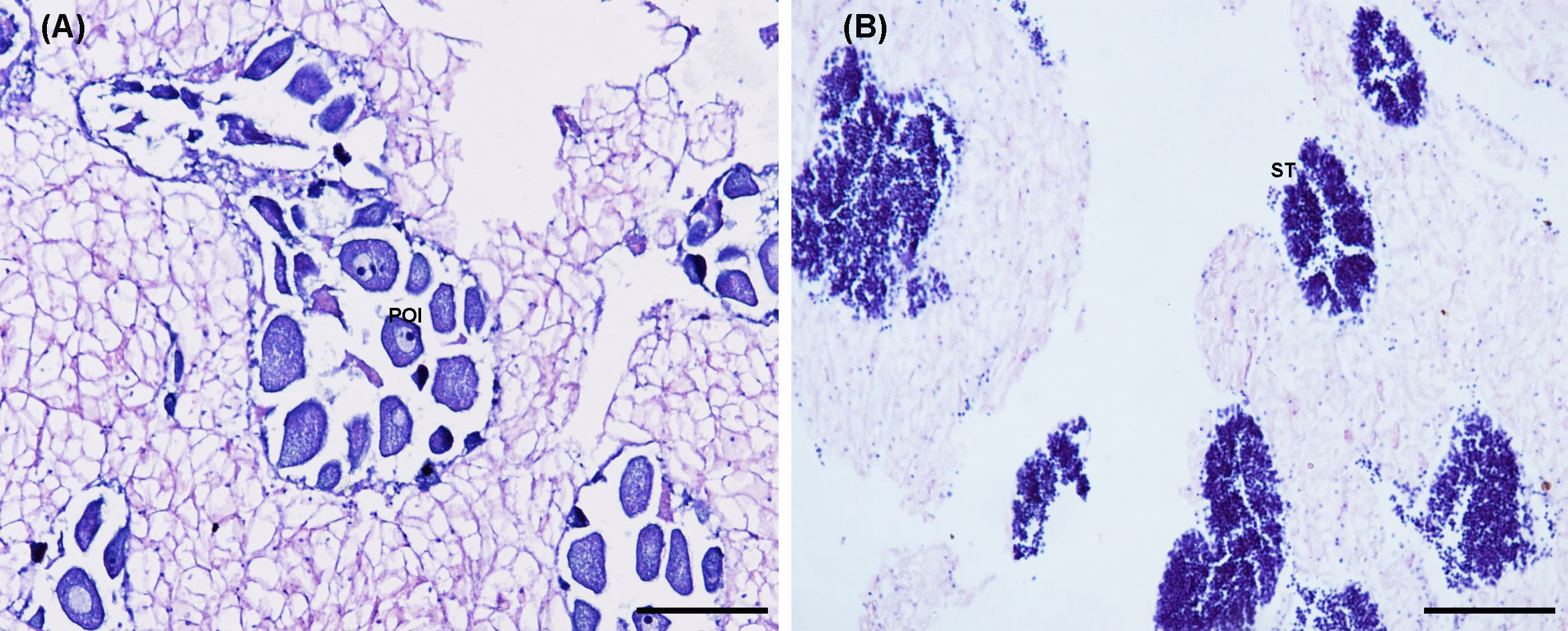
Figure 1 Histological characteristics of the ovary (A) and testis (B) of the C sinensis for transcriptome sequencing. POI, pre-vitellogenesis oocyte; ST: spermatid. Scale bars, 100 μm.
Two cDNA libraries were constructed from the ovary and testis of C. sinensis. After adaptor sequences and low-quality reads were removed, 278 million clean reads, including 140 109 554 reads from the ovary libraries and 137 762 234 reads from the testis libraries, were obtained from the ovary and testis of the C. sinensis transcriptome (Table 2). Among these clean reads, the Q30 in each sample was above 90%. All reads were submitted to the website of NCBI (PRJNA906186).
All clean reads were assembled into 134 124 unigenes, and the minimum, longest, and average lengths of unigenes were 201, 29 212 and 1 095 bp, respectively. Among the unigenes, 77 305 unigenes (57.64%) were successfully annotated by searching against the Nr, Swissport, Pfam, COG, GO and KEGG databases. Especially, 22 095 (28.59%) unigenes were annotated to the Nr database, whereas 15 142 (19.59%) unigenes can be fully annotated to the SwissProt database (Figure 2). In addition, the species most represented in BLASTx searches included Pecten maximus (18.5%), Mizuhopecten yessoensis (17.23%) and Crassostrea gigas (4%) (Figure 3).
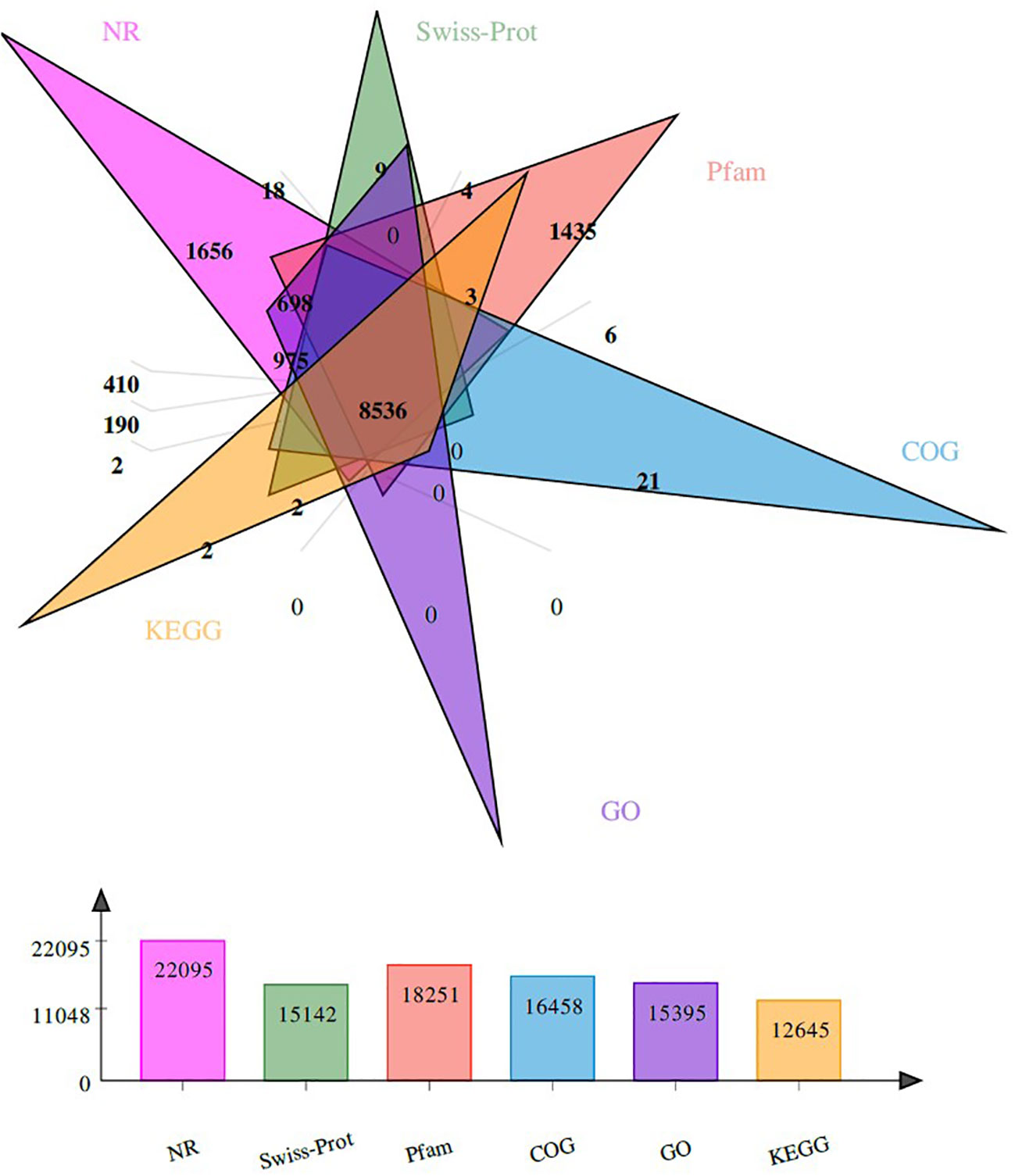
Figure 2 The distribution of unigenes in six databases. The colors show different databases. The number represents the total number of unigenes.
By comparing the databases of ovary and testis, 1 440 DEGs were detected in the transcriptome, of which 1 013 and 427 DEGs were upregulated and downregulated, respectively. To analyze the functions of these DGEs, we conducted GO and KEGG analyses. GO annotation analysis indicated that these DEGs were annotated to three functional ontologies, i.e. molecular function (399 transcripts), cellular component (504 transcripts), and biological process (493 transcripts) (Figure 4). In the biological process category, the metabolic process (GO:0008152) and cellular process (GO:0009987) level 2 terms were the most abundant terms. In the cellular component category, the membrane part (GO:0044425) and cell part (GO:0044464) level 2 terms were the most abundant terms. In the cellular component category, the binding (GO:0005488) and catalytic activity (GO:0003824) level 2 terms were the most abundant terms. KEGG enrichment analysis showed that these DEGs were enriched in 220 specific KEGG metabolic pathways. Several DEGs were mapped to several pathways related with reproduction, such as progesterone-mediated oocyte maturation, cell cycle, cAMP signaling pathway and oocyte meiosis. The top 20 most significantly enriched metabolic pathways are represented in Figure 5.
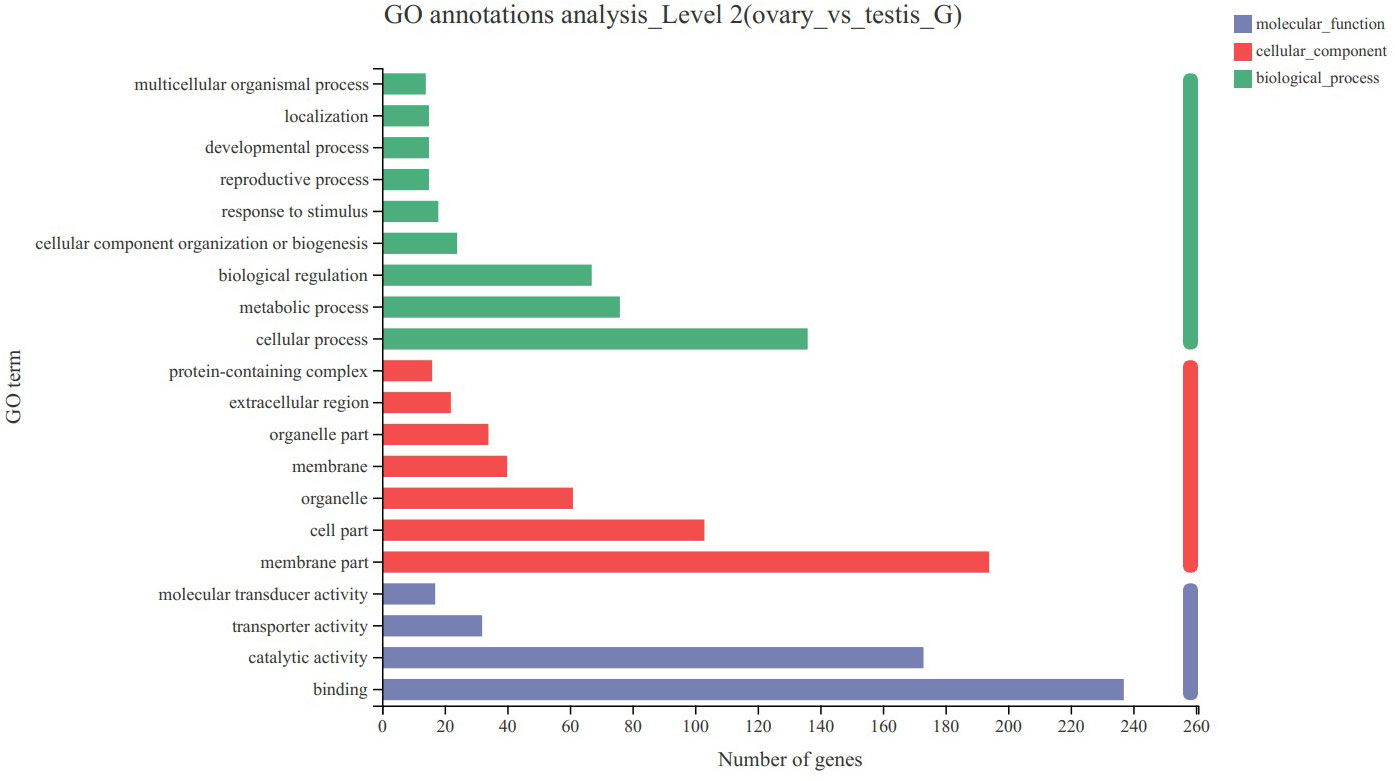
Figure 4 GO annotation analyses of differentially expressed genes (DEGs) in C. sinensis transcriptome. The left vertical coordinate indicates the second level of GO classification. The horizontal coordinate indicates the number of unigene/transcript included in this secondary classification, and the three colors on the right indicate the three major branches of GO.
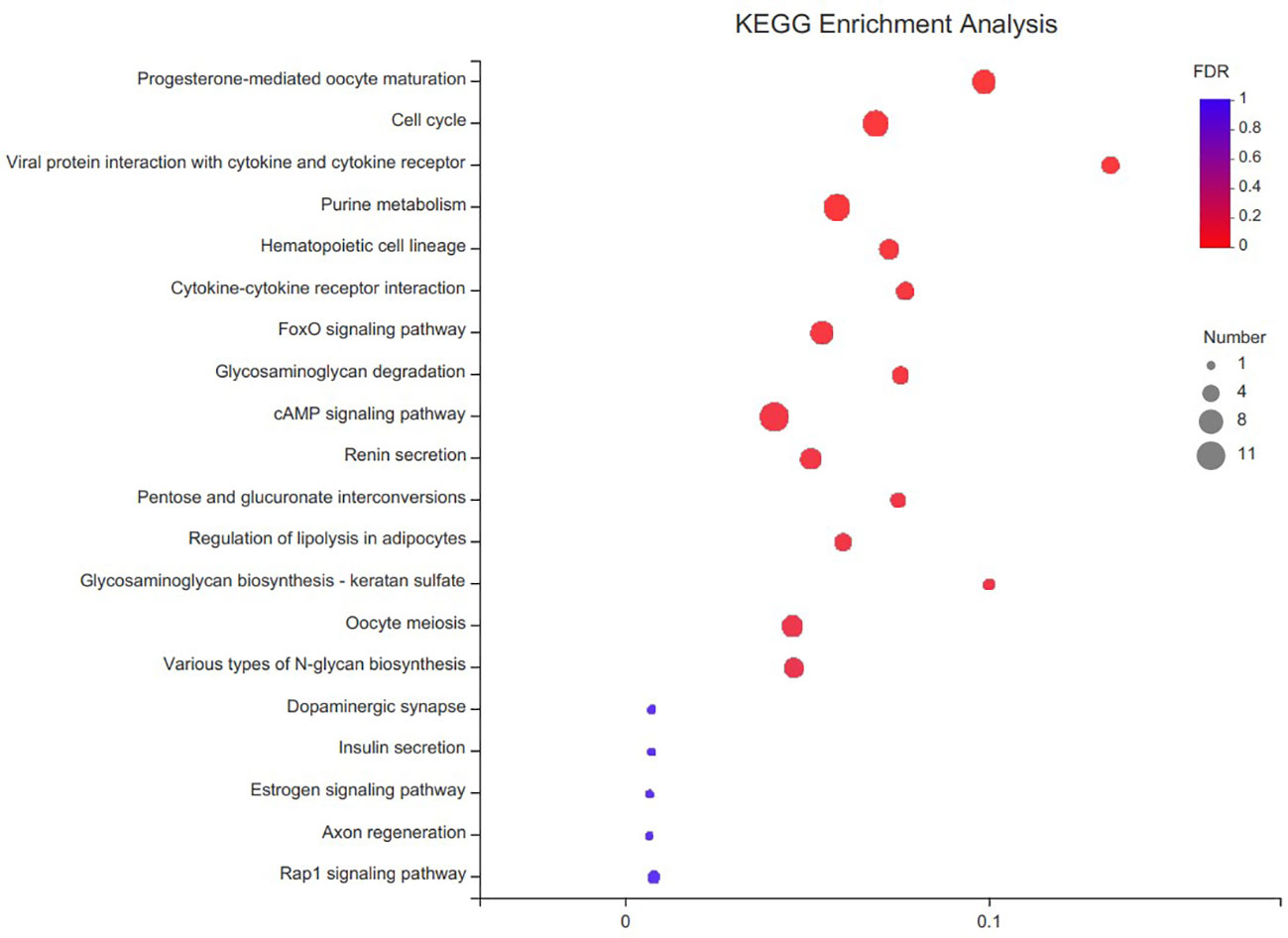
Figure 5 KEGG enrichment analyses of differentially expressed genes (DEGs) in C. sinensis transcriptome. The vertical coordinate is the name of the KEGG metabolic pathway and the horizontal coordinate is the number of transcript/unigene annotated to the pathway.
Through the analysis of the overall gene expression profiles of gonads, several candidate genes involved in various processes of sex determination/differentiation and gonadal development were identified (Table 3). Among the candidate genes, twenty-three key genes were mainly involved in sexual determination/differentiation. Specifically, the transcript levels of Foxl2 and GATA-type zinc finger protein 1-like in the ovary were significantly higher than those in the testis. The expression levels of Dmrt1, Sox9, Fem-1, Follistatin, Transformer-2 protein homolog alpha isoform X5, Zinc finger Y-chromosomal protein 1-like, E3 ubiquitin-protein ligase MARCHF3 isoform X1, Synaptonemal complex protein 1 isoform X2, Transcription factor Runt and Protein dpy-30 in the testis were higher than those in the ovary. Twenty key genes were mainly involved in the process of spermatogenesis, and the higher expression levels of Spermatogenesis-associated protein 17-like isoform X1, Meiotic recombination protein SPO11-like isoform X1, Meiotic recombination protein REC8 homolog isoform X2, Ropporin-1-like protein, Sperm-tail PG-rich repeat-containing protein 2-like (STPG1), Testis-specific serine/threonine-protein kinase 1, Testis-specific serine/threonine-protein kinase 3-like, Sperm-associated antigen 6, Kelch-like protein 10, Armadillo repeat-containing protein 4-like isoform X1 and G2/mitotic-specific cyclin-B3-like were found in the testis. Moreover, fifteen genes related to ovarian development and five genes encoding steroid biosynthesis/metabolism were identified in the ovary and testis.
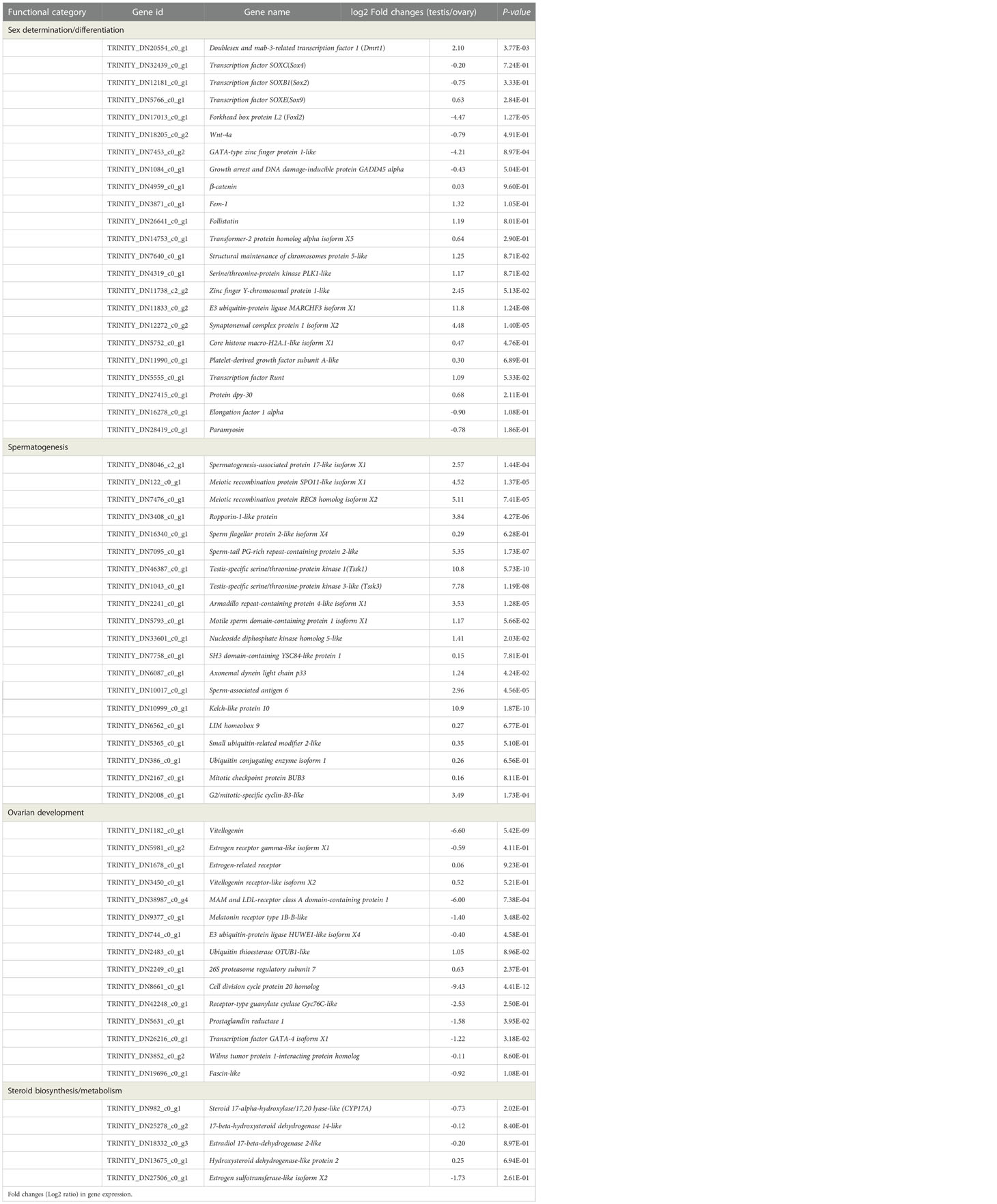
Table 3 Summary of differential and non-differentially expressed genes related to sex determination/differentiation and gonadal development in the transcriptome of C. sinensis.
Eight selected important genes related to sexual determination/differentiation and gonadal development of C. sinensis were determined using qPCR to validate the RNA-seq results. qPCR results showed that Dmrt1, Testis-specific serine/threonine-protein kinase 1 and Sperm-associated antigen 6 had significantly higher expression levels in male gonads (Figure 6), whereas Foxl2, Vg, MAM and LDL-receptor class A domain-containing protein 1, Cell division cycle protein 20 homolog and Prostaglandin reductase 1 had significantly higher expression levels in female gonads (Figure 6). The qPCR results showed that the relative expression patterns of these eight genes were consistent with the RNA-seq results (Figure 6).
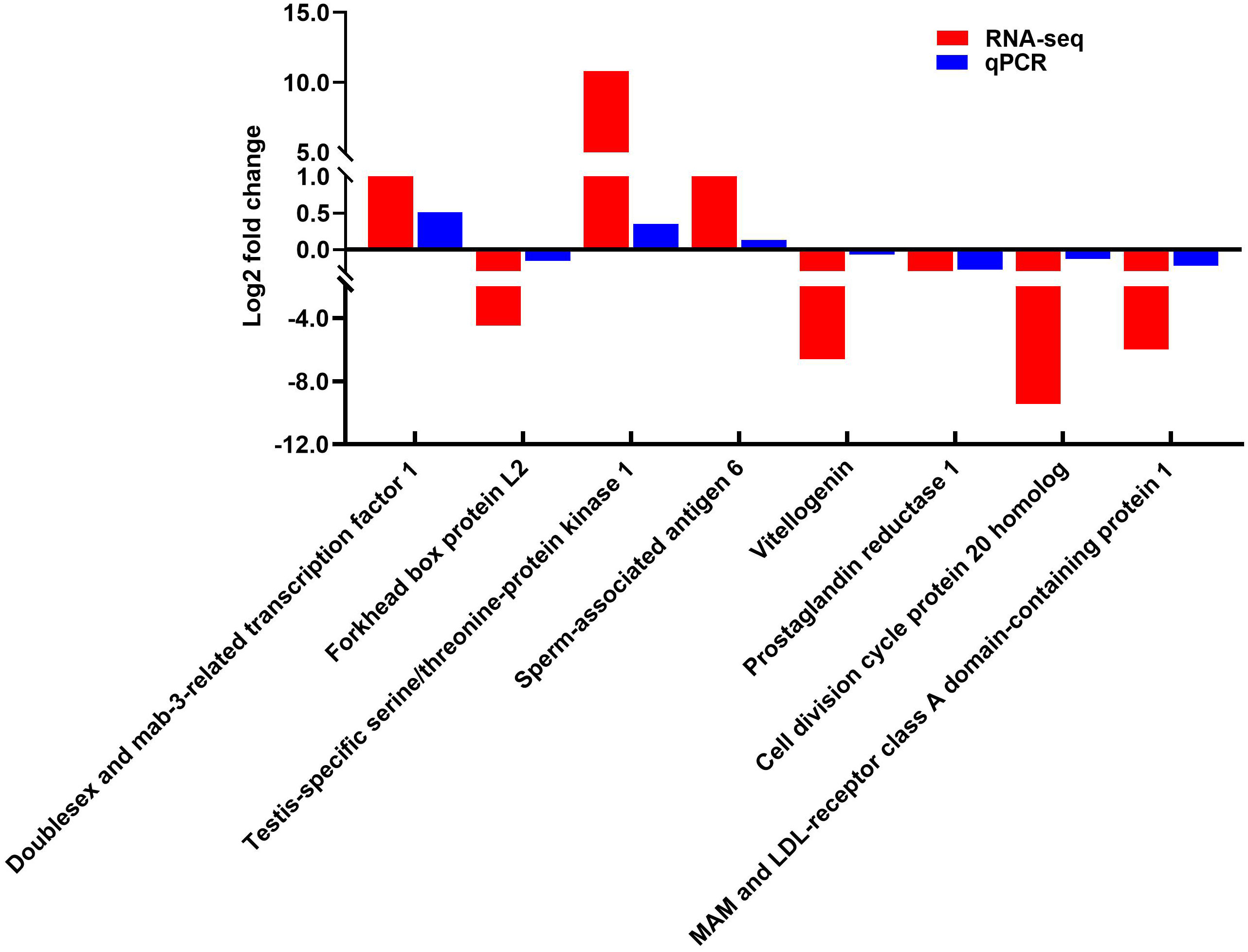
Figure 6 Expression levels of eight selected DEGs normalized to β-actin of C. sinensis was determined by qPCR.
The CsVg sequence contains a 5584 bp open reading frame encoding 1860 amino acids. Comparison of the amino acid sequence encoded by the CsVg with the homologues from other species showed that CsVg contains a conserved DUF1943 and VWD domains (Figure 7A). However, the typical lipoprotein N-terminal domain of known lipid transport proteins was not found in the amino acid sequence of CsVg. Finally, the phylogenetic tree obtained by NJ method revealed that the CsVg from bivalves was clustered in a separate clade from the CsVg from vertebrates and invertebrates (Figure 7B). Evolutionarily, the protein sequence of CsVg is more similar to that of the Pacific abalone Haliotis discus hannai.
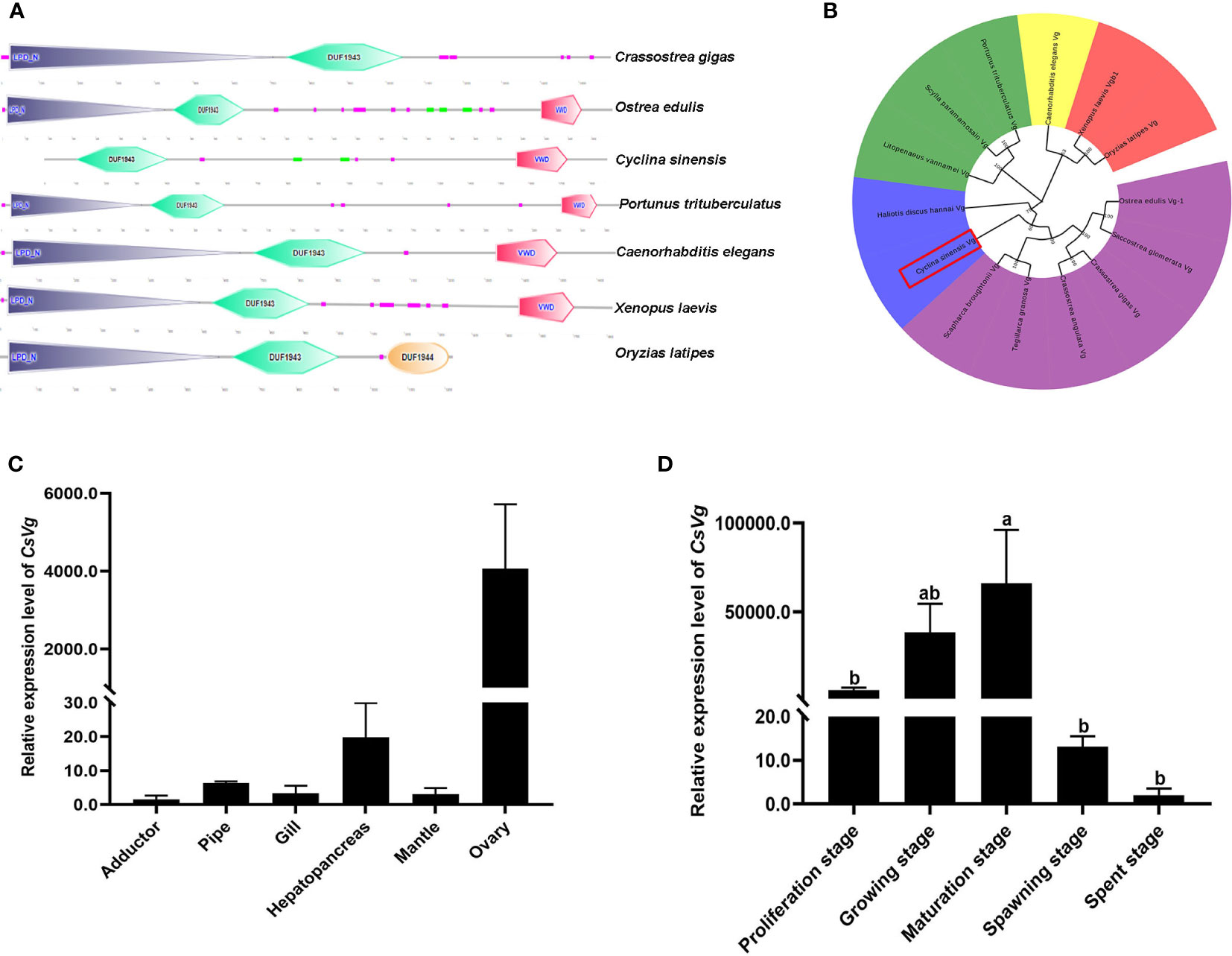
Figure 7 Sequence analysis of CsVg and its expression profile in tissues of C sinensis. (A) Domain structures of CsVg and other selected species. (B) Phylogenetic tree based on amino acid sequences of Vg gene (C) Relative expression of CsVg in different tissues (D) Relative expression of CsVg in different ovarian development stages. The columns with different letter indicate significant difference (P < 0.05).
Tissue distribution analysis demonstrated that CsVg was expressed in various tissues of female C. sinensis (Figure 7C). Remarkably, CsVg had significantly high expression level in the ovary and hepatopancreas. To test the correlation of CsVg expression level with the gonadal development stages, the relative abundance of CsVg transcripts was detected in different stages of ovarian development by qPCR. As shown in Figure 7D, the expression level of CsVg in the ovary increased continuously from proliferation stage to maturation stage of C. sinensis, and it reached the peak level at maturation stage. However, the expression level of CsVg in the ovary decreased from spawning stage to spent stage.
The expression level of CsVg in the ovary of C. sinensis exposure with estradiol is shown in Figure 8A. Compared with the control treatment, the expression level of CsVg in the ovary increased in a dose-dependent manner by estradiol treatments. Moreover, the expression levels of CsVg in the ovary of C. sinensis significantly increased in the 50µg/L estradiol treatment. In the hepatopancreas, estradiol treatments (5µg/L, 50µg/L) also significantly up-regulated the expression levels of CsVg compared with the control (Figure 8B).
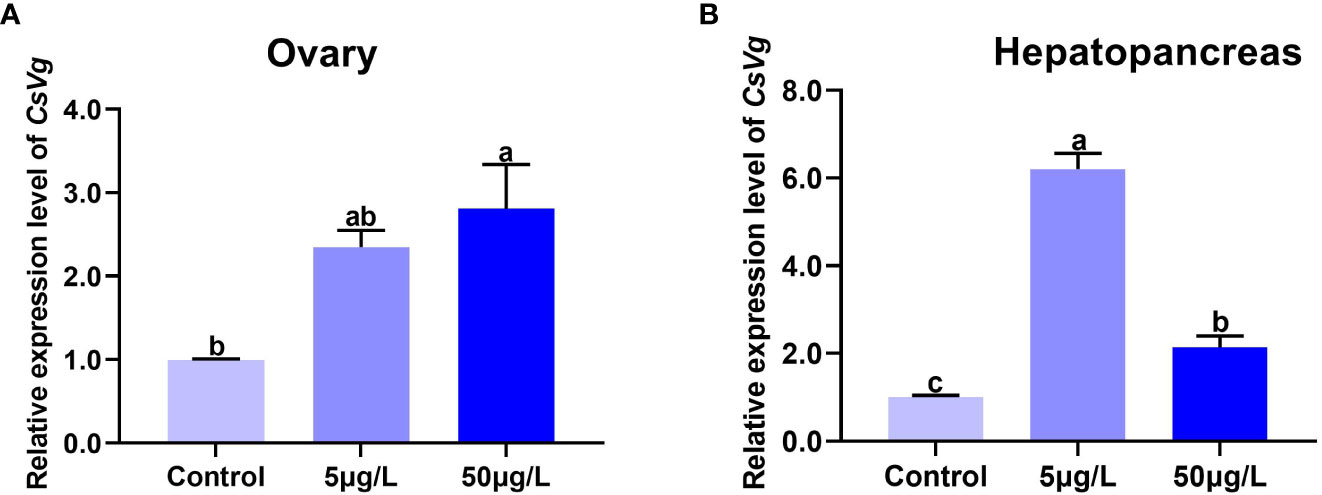
Figure 8 The expression level of CsVg in the ovary (A) and hepatopancreas (B) of C sinensis by different concentrations of estradiol. The columns with different letter indicate significant difference (P < 0.05).
The reproduction of marine invertebrates demonstrates a wide range of sexual reproduction traits, which often manifests through free spawning (Ostrovsky, 2021; Picard et al., 2021). As a typical buried shellfish, the research on the molecular mechanism of C. sinensis reproduction is a key basis work for its genetic selection. Therefore, the present study firstly sequenced C. sinensis gonad transcriptomes, screened out several candidate genes involved in the process of sexual determination/differentiation and gonadal development, and then analyzed the responses of several genes to estradiol. The results of this study will help us understand the general underlying molecular mechanisms of bivalve reproduction and provide a scientific basis for the sexual control and breeding of shellfish.
In this study, serveral DEGs were mapped to several pathways related with to reproduction, including progesterone-mediated oocyte maturation, cell cycle, cAMP signaling pathway, oocyte meiosis and estrogen signaling pathway, which suggests the significance of signal transduction systems and endocrine regulation in gonadal development of C. sinensis. Importantly, twenty-three potential sexual determination/differentiation genes were identified by analysis of the female and male gonad transcriptomes of C. sinensis, these genes included Dmrt1, Sox9, Wnt-4a, Foxl2, GATA-type zinc finger protein 1-like, Elongation factor 1 alpha (EF-1-alpha), Transformer-2 protein homolog alpha isoform X5, Follistatin and Fem-1. The discovery of these reported genes suggests that, similar to other bivalves and mammalian, these genes also play an important role in sexual determination/differentiation of C. sinensis (Li et al., 2016a; Yang et al., 2016; Yao et al., 2021). Robinson et al. (2022) reported that Dmrt are key genes for male and female development of mollusk by the analysis of RNAseq data from eight phylogenetically diverse bivalve species. Adzigbli et al. (2019) reported that Sox9 and Gata-type zinc finger protein 1 are involved in genetic sex determination of pearl oysters. The signalling molecule Wnt-4 is not only crucial for female sexual development, but its signalling is also well implicated in mammalian testis development (Vainio et al., 1999; Naillat et al., 2015). Foxl2 plays a major role in initiating ovarian differentiation in fish and bivalves (Jin et al., 2022; Sun et al., 2022). Follistatin1 not only acts as an inhibitory binding protein of activin in the regulation of oocyte maturation in adult females but also plays a potential role in the masculinization of juveniles (Jiang et al., 2012). EF-1-alpha has been reported to be expressed in male and female germ cells, and it may contribute to the massive protein synthesis required for egg production (Kinoshita et al., 2000; Zhou et al., 2002). Wang et al. (2021a) reported that Transformer-2 plays a potential regulatory role in embryonic sex determination and early gonadal development of Freshwater Pearl Mussel Hyriopsis cumingii. Differently to what reported in H. cumingii, the present study showed the higher expression level of Fem-1 in the testis than in the ovary of C. sinensis (Wang et al., 2021b). The opposite result can potentially be explained by the different functions of various Fem-1 isoforms in distinct species. Tan et al. (2001) reported that Fem-1 plays pivotal roles in sex determination of Caenorhabditis elegans, whereas Fem-2 has a role in apoptosis signaling. Therefore, further research is required in C. sinensis regarding what these genes target and how they function to determine sex.
Spermatogenesis is a complicated process of proliferation and division and involves numerous genes (Yu et al., 2009; Yue et al., 2018). Previous study reported that spermatogenesis-associated protein 17 is a testis-specific apoptosis genes and plays important roles in the gonadogenesis and testis development (Nie et al., 2011). Meiotic recombination protein REC8 is a prominent component of the meiotic prophase chromosome axis, whereas Meiotic recombination protein SPO11 initiates meiotic recombination by generating DNA double-strand breaks (Yoon et al., 2016; Paiano et al., 2020). Ropporin is a spermatogenic cell-specific protein and may be involved in sperm maturation (Chen et al., 2009). In this study, the spermatogenesis-associated protein 17-like isoform X1, meiotic recombination protein SPO11-like isoform X1, meiotic recombination protein REC8 homolog isoform X2 and Ropporin-1-like protein were also identified in the testis, where all of them had higher transcript levels in the testis, demonstrating that the spermatogenesis-associated protein 17-like isoform X1, meiotic recombination protein SPO11-like isoform X1 meiotic recombination protein REC8 homolog isoform X2 and Ropporin-1-like protein are involved in the spermatogenesis of C. sinensis. Notably, members of testis-specific serine/threonine kinases (Tssk) family are required for male fertility in mammals, and targeted deletion of Tssk1 and Tssk2 results in dysregulation of spermiogenesis (Shang et al., 2013; Wang et al., 2022). Several studies reported that Tssk1/2, Tssk3, Tssk4 and Tssk5 play a functional role during spermatogenesis of several mollusks, including the pen shell Atrina pectinate, abalone Haliotis discus hannai and Bay Scallop Argopecten irradians (Li et al., 2016b; Kim et al., 2019; Xue et al., 2021). The present study showed the higher expression level of Tssk1 and Tssk3 in the testis of C. sinensis, indicating that Tssk family may have a role in sperm differentiation in the testis and/or fertilization. Moreover, Murray and Hobbs (2022) reported that Kelch-like protein homolog 10 and Armadillo repeat-containing protein 4 isoform X2 are involved in spermatogenesis and spermatid development/maturation of male blue mussel Mytilus edulis, respectively. Similarly, the Kelch-like protein homolog 10 and Armadillo repeat-containing protein 4 isoform X2 have previously been identified in serveral bivalve species including Mytilus galloprovincialis, Nodipecten subnodosus, Pinctada margaritifera and C. gigas (Craft et al., 2010; Llera-Herrera et al., 2013; Teaniniuraitemoana et al., 2014; Gallardi et al., 2021). In agreement with previous results, the higher expression level of Kelch-like protein 10 and Armadillo repeat-containing protein 4-like isoform X1 was found in the testis, illustrating that these genes play a key role in the sperm development of C. sinensis. Sperm-associated antigen 6 (SPAG6) is an important flagellar protein required for normal flagellar and cilia motility (Jarrell et al., 2020). De Sousa et al. (2014) reported that the mRNA levels of SPAG6 showed a significant correlation with the gonad area of European Clam Ruditapes decussatus. The present study revealed the higher transcript levels of SPAG6 in the testis, suggesting that SPAG6 is essential for the spermatogenesis of C. sinensis.
Ovary are the primary reproductive organs, and their normal development is crucial for bivalve reproduction (Yang et al., 2016; Zhao et al., 2022). Notably, vitellogenesis is involved in the accumulation of the major yolk protein vitellin (Vn), which is important in the development and maturation of oocytes (Saavedra et al., 2012; Kang et al., 2014). As a synthetic precursor of Vn, vitellogenin (Vg) is considered a biological marker of ovarian development in vertebrates and invertebrates (Marin and Matozzo, 2004; Porte et al., 2006). In this study, sequence analysis results showed that CsVg has no the typical lipoprotein N-terminal domain region compared with other known species, which may be attributed to the involvement of different mechanisms of Vg receptor binding during endocytosis (Xie et al., 2009). Previous studies showed that the lipoprotein N-terminal domain is responsible for lipid binding (Morandin et al., 2014). Tissue distribution results showed that CsVg was expressed in various tissues of female C. sinensis, which is consistent with the results on Fujian oyster Crassostrea angulate and C. gigas (Matsumoto et al., 2003; Ni et al., 2014). Remarkably, qPCR data showed that CsVg was abundantly expressed in the ovary, indicating that CsVg plays crucial roles in the ovarian development of C. sinensis. To search for more clues regarding the interaction between CsVg and ovarian development, the temporal expression patterns of CsVg in the ovary was analyzed during the ovarian development of C. sinensis. A relatively abundant expression of CsVg in proliferation stage to maturation stage and a significantly low expression in spent stage further demonstrated that CsVg is involved in the ovarian development of C. sinensis. These results coincide with those of previous studies on bivalve species (Ni et al., 2014; Yang et al., 2016). Vitellogenesis in bivalves is under the control of sex steroid hormone as well as other oviparous animals, and the estradiol levels of bivalve exhibit a seasonal change associated with the reproductive cycle (Osada et al., 2003; Wang and Croll, 2003; Osada et al., 2004). Therefore, to obtain further gain insights into the endocrine regulatory mechanism of C. sinensis reproduction, the regulation of CsVg mRNA expression by estradiol were analyzed in the ovary and hepatopancreas. The present results showed that estradiol treatments significantly upregulated the mRNA expression of CsVg in the ovary, suggesting that estradiol is a primary promoter of Vg mRNA transcription in C. sinensis. Similar estradiol inducibility of Vg was demonstrated in other bivalve species (Osada et al., 2003; Andrew et al., 2010; Qin et al., 2012; Ni et al., 2014). Moreover, the CsVg mRNA transcription in the hepatopancreas was induced by estradiol, which implies that hepatopancreas may also be involved in the vitellogenesis of C. sinensis. Hepatopancreas is another important site of vitellogenesis in crustaceans (Feng et al., 2022). Nonetheless, the regulation mechanism of estradiol in the vitellogenesis of bivalve is largely unknown. In vertebrate, it is well known that estrogen regulates Vg gene transcription mainly through binding to estrogen receptors (ER) on the target organ (Nelson and Habibi, 2013). In this study, the Estrogen receptor gamma-like isoform X1 (ERγ) and Estrogen-related receptor (ERR) were identified in the ovary, indicating that the estradiol-ERγ/ERR-Vg signalling pathway may play an important role the regulation mechanism of estradiol on the vitellogenesis in bivalve. The above hypothesis was also proposed for scallop, mussels and oyster (Wang and Croll, 2003; Ciocan et al., 2010; Ni et al., 2014).
Except for Vg, the present study also discovered other genes associated with ovarian development and steroid biosynthesis/metabolism in the transcriptome data. Specifically, the higher expression levels of MAM and LDL-receptor class A domain-containing protein 1, Melatonin receptor type 1B-B-like, Cell division cycle protein 20 homolog, Prostaglandin reductase 1 and Transcription factor GATA-4 isoform X1 were detected in the ovary, indicating that those genes participate in the ovarian development, steroidogenesis, folliculogenesis and oocyte maturation of C. sinensis. A previous study showed that MAM and LDL-receptor class A domain-containing protein 1 may serve as oogonia or oocyte membrane protein mediating diverse signal transduction to regulate oocyte development (Yue et al., 2018). Takahashi and Ogiwara (2021) reported that melatonin can directly regulate ovarian physiology, including steroidogenesis, folliculogenesis, oocyte maturation and ovulation, by binding to melatonin receptors. Cell division cycle protein 20 is required for spindle assembly and chromosomal segregation during oocyte maturation (Yang et al., 2014). Prostaglandin reductase 1 plays a negative physiological role in the development of oocytes and ovaries (Prasertlux et al., 2011). Transcription factor GATA-4 can be a downstream effector of cAMP/PKA pathway in the regulation of CYP19 gene during folliculogenesis and luteinization (Monga et al., 2012). In addition, five genes related to steroid biosynthesis/metabolism were identified in the transcriptome data. CYP17Ahas been viewed as a critical enzyme for the biosynthesis of sexual steroid, and it performs the 17-alpha-hydroxylation of progesterone and pregnenolone to 17-hydroxyprogesterone (17OHP) and 17-hydroxypreg nenolone, respectively (Athanasoulia et al., 2013). 17 beta-Hydroxysteroid dehydrogenases (17 beta-HSDs) plays a key role in estrogen and androgen steroid metabolism by catalyzing the final steps of steroid biosynthesis (Marchais-Oberwinkler et al., 2011). The present study showed that the CYP17A and three genes related to beta hydroxysteroid dehydrogenase enzymes (17-beta-hydroxysteroid dehydrogenase 14-like, Estradiol 17-beta-dehydrogenase 2-like, Hydroxysteroid dehydrogenase-like protein 2) were identified in the gonad, suggesting that these genes are involved in the biosynthesis of sexual steroids in the C. sinensis. Moreover, the Estrogen sulfotransferase-like isoform X2 was also found in the gonad, which indicates the presence ofa balance system of steroid hormone synthesis in clam gonads. A previous study reported that Estrogen sulfotransferase is a cytosolic enzyme that sulphatesestrogens to inactivate them and regulate their homeostasis (Yi et al., 2021).
In this study, transcriptome sequencing was used to explore the expression level of genes associated with sexual determination/differentiation and gonadal development of C. sinensis. The present study identified 23 genes (Dmrt1, Sox2/4/9, Foxl2, β-catenin and GATA-type zinc finger protein 1-like) involved in sexual determination/differentiation, 20 genes (Spermatogenesis-associated protein 17-like isoform X1, Meiotic recombination protein SPO11-like isoform X1, Meiotic recombination protein REC8 homolog isoform X2, Testis-specific serine/threonine-protein kinase 1, Testis-specific serine/threonine-protein kinase 3-like, Sperm-associated antigen 6 and Kelch-like protein 10) related to spermatogenesis, 15 genes (Vitellogenin, MAM and LDL-receptor class A domain-containing protein 1 and Cell division cycle protein 20 homolog) associated with the ovarian development and 5 genes involved in steroid biosynthesis/metabolism, respectively. Theresults will provide new insights into the mechanisms of sexual determination/differentiation and gonadal development in marine bivalves. In the future, the roles of these genes need to be elucidated to provide a scientific basis for the sexual control and breeding of shellfish.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI - PRJNA906186.
ML: Experimental design, Writing - Original Draft, Data Curation. HN: Formal analysis, Data Curation. ZR: Data Curation, Validation. ZW: Formal analysis, Visualization. SY: Data Curation, Visualization. XL: Experimental design, Formal analysis. ZD: Writing-Editing, Funding acquisition. All authors contributed to the article and approved the submitted version.
This study was funded by the projects (No.42106088) from the Natural Science Foundation of China, the projects (CARS-49) from Modern Agro-industry Technology Research System; the projects (JBGS[2021]034) from the ‘JBGS’ Project of Seed Industry Revitalization in Jiangsu Province; an Open-end Funds (SH20201205) of Jiangsu Key Laboratory of Marine Bioresources and Environment, two practice innovation training program projects (No. 202111641127Y and No. SY202257X) for the Jiangsu College students. Infrastructure costs were partially supported by the Project of Jiangsu Fisheries Science and Technology (SZ-LYG202029).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adzigbli L., Wang Z., Lai Z., Li J., Deng Y. (2019). Sex determination in pearl oyster: A mini review. Aquacult. Rep. 15, 100214. doi: 10.1016/j.aqrep.2019.100214
Andrew M., O'Connor W., Dunstan R., Macfarlane G. (2010). Exposure to 17α-ethynylestradiol causes dose and temporally dependent changes in intersex, females and vitellogenin production in the Sydney rock oyster. Ecotoxicology 19 (8), 1440–1451. doi: 10.1007/s10646-010-0529-5
Athanasoulia A., Auer M., Riepe F., Stalla G. (2013). Rare missense P450c17 (CYP17A1) mutation in exon 1 as a cause of 46, XY disorder of sexual development: Implications of breast tissue 'Unresponsiveness' despite adequate estradiol substitution. Sex Dev. 7 (4), 212–215. doi: 10.1159/000348301
Barrionuevo F., Georg I., Scherthan H., Lecureuil C., Guillou F., Wegner M., et al. (2009). Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev. Biol. 327 (2), 301–312. doi: 10.1016/j.ydbio.2008.12.011
Bertho S., Pasquier J., Pan Q., Le Trionnaire G., Bobe J., Postlethwait J., et al. (2016). Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex Dev. 10 (3), 111–129. doi: 10.1159/000447611
Broquard C., Saowaros S., Lepoittevin M., Degremont L., Lamy J., Morga B., et al. (2021). Gonadal transcriptomes associated with sex phenotypes provide potential Male and female candidate genes of sex determination or early differentiation in Crassostrea gigas, a sequential hermaphrodite mollusc. BMC Genomics 22 (1), 609. doi: 10.1186/s12864-021-07838-1
Capel B. (2006). R-spondin1 tips the balance in sex determination. Nat. Genet. 38 (11), 1233–1234. doi: 10.1038/ng1106-1233
Chaboissier M., Kobayashi A., Vidal V., Lutzkendorf S., Van de Kant H., Wegner M., et al. (2004). Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131 (9), 1891–1901. doi: 10.1242/dev.01087
Chen J., Cai Z., Gui Y. (2009). Advances in the researches of spermatogenic protein, ropporin. Natl. J. Androl. 15 (9), 833–835.
Chen H. P., Xiao G., Chai X., Lin X., Fang J., Teng S. (2017). Transcriptome analysis of sex-related genes in the blood clam Tegillarca granosa. PloS One 12 (9), e0184584. doi: 10.1371/journal.pone.0184584
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), 884–890. doi: 10.1093/bioinformatics/bty560
Ciocan C., Cubero-Leon E., Puinean A., Hill E., Minier C., Osada M., et al. (2010). Effects of estrogen exposure in mussels, Mytilus edulis, at different stages of gametogenesis. Environ. pollut. 158 (9), 2977–2984. doi: 10.1016/j.envpol.2010.05.025
Craft J., Gilbert J., Temperton B., Dempsey K., Ashelford K., Tiwari B., et al. (2010). Pyrosequencing of Mytilus galloprovincialis cDNAs: Tissue-specific expression patterns. PloS One 5 (1), e8875. doi: 10.1371/journal.pone.0008875
De Sousa J., Milan M., Bargelloni L., Pauletto M., Matias D., Joaquim S., et al. (2014). A microarray-based analysis of gametogenesis in two Portuguese populations of the European clam Ruditapes decussatus. PloS One 9 (3), e92202. doi: 10.1371/journal.pone.0092202
Devlin R., Nagahama Y. (2002). Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208 (3-4), 191–364. doi: 10.1016/S0044-8486(02)00057-1
Dong Z., Duan H., Zheng H., Ge H., Wei M., Liu M., et al. (2021). Research progress in genetic resources assessment, culture technique and exploration utilization of Cyclina sinensis. J. Fish. China 45 (12), 2083–2098. doi: 10.11964/jfc.20201212545
Estermann M., Major A., Smith C. (2020). Gonadal sex differentiation: Supporting versus steroidogenic cell lineage specification in mammals and birds. Front. Cell Dev. Biol. 8, 616387. doi: 10.3389/fcell.2020.616387
Farhadi A., Fang S., Zhang Y., Cui W., Fang H., Ikhwanuddin M., et al. (2021). The significant sex-biased expression pattern of sp-Wnt4 provides novel insights into the ovarian development of mud crab (Scylla paramamosain). Int. J. Biol. Macromol. 183, 490–501. doi: 10.1016/j.ijbiomac.2021.04.186
Feng Q., Liu M., Cheng Y., Wu X. (2022). Comparative transcriptome analysis reveals the process of ovarian development and nutrition metabolism in Chinese mitten crab. Eriocheir Sinensis. Front. Genet. 13, 910682. doi: 10.3389/fgene.2022.910682
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28 (23), 3150–3152. doi: 10.1093/bioinformatics/bts565
Gallardi D., Xue X., Mercier E., Mills T., Lefebvre F., Rise M., et al. (2021). RNA-Seq analysis of the mantle transcriptome from Mytilus edulis during a seasonal spawning event in deep and shallow water culture sites on the northeast coast of Newfoundland, Canada. Mar. Genomics 60, 100865. doi: 10.1016/j.margen.2021.100865
Ge H., Liang X., Liu J., Cui Z., Guo L., Li L., et al. (2021). Effects of acute ammonia exposure on antioxidant and detoxification metabolism in clam Cyclina sinensis. Ecotox. Environ. Safe. 211, 111895. doi: 10.1016/j.ecoenv.2021.111895
Grabherr M., Haas B., Yassour M., Levin J., Thompson D., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–U130. doi: 10.1038/nbt.1883
Jarrell Z., Ahammad M., Sweeney K., Wilson J., Benson A. (2020). Characterization of sperm-associated antigen 6 expression in the reproductive tract of the domestic rooster (Gallus domesticus) and its impact on sperm mobility. Poultry Sci. 99 (11), 6188–6195. doi: 10.1016/j.psj.2020.08.009
Jiang N., Jin X., He J., Yin Z. (2012). The roles of follistatin 1 in regulation of zebrafish fecundity and sexual differentiation. Biol. Reprod. 87 (3), 099689. doi: 10.1095/biolreprod.112.099689
Jin L., Sun W., Bao H., Liang X., Li P., Shi S., et al. (2022). The forkhead factor Foxl2 participates in the ovarian differentiation of Chinese soft-shelled turtle Pelodiscus sinensis. Dev. Biol. 492, 101–110. doi: 10.1016/j.ydbio.2022.10.001
Kang B., Okutsu T., Tsutsui N., Shinji J., Bae S., Wilder M. (2014). Dynamics of vitellogenin and vitellogenesis-inhibiting hormone levels in adult and subadult whiteleg shrimp, Litopenaeus vannamei: Relation to molting and eyestalk ablation. Biol. Reprod. 90 (1), 112243. doi: 10.1095/biolreprod.113.112243
Kim E., Kim S., Park C., Nam Y. (2019). Characterization of testis-specific serine/threonine kinase 1-like (TSSK1-like) gene and expression patterns in diploid and triploid pacific abalone (Haliotis discus hannai; gastropoda; Mollusca) males. PloS One 14 (12), 0226022. doi: 10.1371/journal.pone.0226022
Kinoshita M., Kani S., Ozato K., Wakamatsu Y. (2000). Activity of the medaka translation elongation factor 1 alpha-a promoter examined using the GFP gene as a reporter. Dev. Growth Differ. 42 (5), 469–478. doi: 10.1046/j.1440-169x.2000.00530.x
Liao X., Sun Z., Cui Z., Yan S., Fan S., Xia Q., et al. (2022). Effects of different sources of diet on the growth, survival, biochemical somposition and physiological metabolism of clam (Cyclina sinensis). Aquac. Res. 53 (10), 3797–3806. doi: 10.1111/are.15886
Li H., Kong L., Yu R., Li Q. (2016b). Characterization, expression, and functional analysis of testis-specific serine/threonine kinase 1 (Tssk1) in the pen shell Atrina pectinata. Invertebr. Reprod. Dev. 60 (2), 118–125. doi: 10.1080/07924259.2016.1161667
Liu X., Li Y., Liu J., Cui L., Zhang Z. (2016). Gonadogenesis in scallop Chlamys farreri and cf-foxl2 expression pattern during gonadal sex differentiation. Aquac. Res. 47 (5), 1605–1611. doi: 10.1111/are.12621
Livak K., Schmittgen T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta c) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li R., Zhang L., Li W., Zhang Y., Li Y., Zhang M., et al. (2018). FOXL2 and DMRT1L are yin and yang genes for determining timing of sex differentiation in the bivalve mollusk Patinopecten yessoensis. Front. Physiol. 9, 01166. doi: 10.3389/fphys.2018.01166
Li Y., Zhang L., Sun Y., Ma X., Wang J., Li R., et al. (2016a). Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop Patinopecten yessoensis. Mar. Biotechnol. 18 (4), 453–465. doi: 10.1007/s10126-016-9706-8
Llera-Herrera R., Garcia-Gasca A., Abreu-Goodger C., Huvet A., Ibarra A. (2013). Identification of Male gametogenesis expressed genes from the scallop Nodipecten subnodosus by suppressive subtraction hybridization and pyrosequencing. PloS One 8 (9), 0073176. doi: 10.1371/journal.pone.0073176
Major A., Ayers K., Chue J., Roeszler K., Smith C. (2019). FOXL2 antagonises the Male developmental pathway in embryonic chicken gonads. J. Endocrinol. 243 (3), 211–228. doi: 10.1530/JOE-19-0277
Manni M., Berkeley M., Seppey M., Simao F., Zdobnov E. (2021). BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38 (10), 4647–4654. doi: 10.1093/molbev/msab199
Marchais-Oberwinkler S., Henn C., Moeller G., Klein T., Negri M., Oster A., et al. (2011). 17 beta-hydroxysteroid dehydrogenases (17 beta-HSDs) as therapeutic targets: Protein structures, functions, and recent progress in inhibitor development. J. Steroid Biochem. Mol. Biol. 125 (1-2), 66–82. doi: 10.1016/j.jsbmb.2010.12.013
Marin M., Matozzo V. (2004). Vitellogenin induction as a biomarker of exposure to estrogenic compounds in aquatic environments. Mar. pollut. Bull. 48 (9-10), 835–839. doi: 10.1016/j.marpolbul.2004.02.037
Matsumoto T., Nakamura A., Mori K., Kayano T. (2003). Molecular characterization of a cDNA encoding putative vitellogenin from the pacific oyster Crassostrea gigas. Zool. Sci. 20 (1), 37–42. doi: 10.2108/zsj.20.37
Monga R., Ghai S., Datta T., Singh D. (2012). Involvement of transcription factor GATA-4 in regulation of CYP19 gene during folliculogenesis and luteinization in buffalo ovary. J. Steroid Biochem. Mol. Biol. 130 (1-2), 45–56. doi: 10.1016/j.jsbmb.2011.12.010
Morandin C., Havukainen H., Kulmuni J., Dhaygude K., Trontti K., Helantera H. (2014). Not only for egg yolk-functional and evolutionary insights from expression, selection, and structural analyses of Formica ant vitellogenins. Mol. Biol. Evol. 31 (8), 2181–2193. doi: 10.1093/molbev/msu171
Murray H., Hobbs K. (2022). Spatial expression patterning of kelch-like protein homolog 10 (KLHL10), armadillo repeat-containing protein 4 isoform x2 (ARMC4), and a gamete-specific mitochondrial cytochrome c oxidase (MT-CO1) during Spermatogenesis/Spermiogenesis in the mantle of Male blue mussel (Mytilus edulis). J. Shellfish Res. 41 (1), 109–117. doi: 10.2983/035.041.0108
Nagaraju G. (2011). Reproductive regulators in decapod crustaceans: An overview. J. Exp. Biol. 214 (1), 3–16. doi: 10.1242/jeb.047183
Naillat F., Yan W., Karjalainen R., Liakhovitskaia A., Samoylenko A., Xu Q., et al. (2015). Identification of the genes regulated by wnt-4, a critical sSignal for commitment of the ovary. Exp. Cell Res. 332 (2), 163–178. doi: 10.1016/j.yexcr.2015.01.010
Nelson E., Habibi H. (2013). Estrogen receptor function and regulation in fish and other vertebrates. Gen. Comp. Endocr. 192, 15–24. doi: 10.1016/j.ygcen.2013.03.032
Nie D., Liu Y., Xiang Y. (2011). Overexpression a novel zebra fish spermatogenesis-associated gene 17 (SPATA17) induces apoptosis in GC-1 cells. Mol. Biol. Rep. 38 (6), 3945–3952. doi: 10.1007/s11033-010-0511-6
Ning J., Cao W., Lu X., Chen M., Liu B., Wang C. (2021). Identification and functional analysis of a sex-biased transcriptional factor Foxl2 in the bay scallop Argopecten irradians irradians. Comp. Biochem. Phys. B 256, 110638. doi: 10.1016/j.cbpb.2021.110638
Ni J., Zeng Z., Kong D., Hou L., Huang H., Ke C. (2014). Vitellogenin of fujian oyster, Crassostrea angulata: Synthesized in the ovary and controlled by estradiol-17 beta. Gen. Comp. Endocr. 202, 35–43. doi: 10.1016/j.ygcen.2014.03.034
Osada M., Harata M., Kishida M., Kijima A. (2004). Molecular cloning and expression analysis of vitellogenin in scallop, Patinopecten yessoensis (Bivalvia, Mollusca). Mol. Reprod. Dev. 67 (3), 273–281. doi: 10.1002/mrd.20020
Osada M., Takamura T., Sato H., Mori K. (2003). Vitellogenin synthesis in the ovary of scallop, Patinopecten yessoensis: Control by estradiol-17 beta and the central nervous system. J. Exp. Zool. A Comp. Exp. Biol. 299 (2), 172–179. doi: 10.1002/jez.a.10276
Ostrovsky A. (2021). Reproductive strategies and patterns in marine invertebrates: Diversity and evolution. Paleontological J. 55 (7), 803–810. doi: 10.1134/S003103012107008X
Paiano J., Wu W., Yamada S., Sciascia N., Callen E., Cotrim A., et al. (2020). ATM And PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat. Commun. 11 (1), 857. doi: 10.1038/s41467-020-14654-w
Picard M., Vicoso B., Bertrand S., Escriva H. (2021). Diversity of modes of reproduction and sex determination systems in invertebrates, and the putative contribution of genetic conflict. Genes 12 (8), 1136. doi: 10.3390/genes12081136
Porte C., Janer G., Lorusso L., Ortiz-Zarragoitia M., Cajaraville M., Fossi M., et al. (2006). Endocrine disruptors in marine organisms: Approaches and perspectives. Comp. Biochem. Phys. C. 143 (3), 303–315. doi: 10.1016/j.cbpc.2006.03.004
Prasertlux S., Sittikankaew K., Chumtong P., Khamnamtong B., Klinbunga S. (2011). Molecular characterization and expression of the prostaglandin reductase 1 gene and protein during ovarian development of the giant tiger shrimp Penaeus monodon. Aquaculture 322, 134–141. doi: 10.1016/j.aquaculture.2011.09.037
Qian X., Ba Y., Zhuang Q., Zhong G. (2014). RNA-Seq technology and its application in fish transcriptomics. Omics 18 (2), 98–110. doi: 10.1089/omi.2013.0110
Qin Z., Li Y., Sun D., Shao M., Zhang Z. (2012). Cloning and expression analysis of the vitellogenin gene in the scallop Chlamys farreri and the effects of estradiol-17 beta on its synthesis. Invertebr. Biol. 131 (4), 312–321. doi: 10.1111/ivb.12006
Robinson W., Robinson W., Krick K., Murray H., Poynton H. (2022). Comparative phylotranscriptomics reveals putative sex differentiating genes across eight diverse bivalve species. Comp 41, 100952.
Saavedra L., Leonardi M., Morin V., Quinones R. (2012). Induction of vitellogenin-like lipoproteins in the mussel Aulacomya ater under exposure to 17 beta-estradiol. Rev. Biol. Mar. Oceanog. 47 (3), 429–438. doi: 10.4067/S0718-19572012000300006
Sandra G., Norma M. (2010). Sexual determination and differentiation in teleost fish. Rev. Fish Biol. Fisher. 20 (1), 101–121. doi: 10.1007/s11160-009-9123-4
Santerre C., Sourdaine P., Adeline B., Martinez A. (2014). Cg-SoxE and cg-beta-catenin, two new potential actors of the sex-determining pathway in a hermaphrodite lophotrochozoan, the pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. A. 167, 68–76. doi: 10.1016/j.cbpa.2013.09.018
Senthilkumaran B., Kar S. (2021). Advances in reproductive endocrinology and neuroendocrine research using catfish models. Cells 10 (11) 2807. doi: 10.3390/cells10112807
Shangguan X., Mao Y., Wang X., Liu M., Wang Y., Wang G., et al. (2022). Cyp17a effected by endocrine disruptors and its function in gonadal development of Hyriopsis cumingii. Gen. Comp. Endocr. 323, 114028. doi: 10.1016/j.ygcen.2022.114028
Shang P., Hoogerbrugge J., Baarends W., Grootegoed J. (2013). Evolution of testis-specific kinases TSSK1B and TSSK2 in primates. Andrology 1 (1), 160–168. doi: 10.1111/j.2047-2927.2012.00021.x
Smith-Unna R., Boursnell C., Patro R., Hibberd J., Kelly S. (2016). TransRate: reference-free quality assessment of De novo transcriptome assemblies. Genome Res. 26 (8), 1134–1144. doi: 10.1101/gr.196469.115
Sun D., Yu H., Li Q. (2022). Examination of the roles of Foxl2 and Dmrt1 in sex differentiation and gonadal development of oysters by using RNA interference. Aquaculture 548 (2), 737732. doi: 10.1016/j.aquaculture.2021.737732
Takahashi T., Ogiwara K. (2021). Roles of melatonin in the teleost ovary: A review of the current status. Comp. Biochem. Physiol. A. 254, 110907. doi: 10.1016/j.cbpa.2021.110907
Tan K., Chan S., Tan K., Yu V. (2001). The Caenorhabditis elegans sex-determining protein FEM-2 and its human homologue, hFEM-2, are Ca (2+)/calmodulin-dependent protein kinase phosphatases that promote apoptosis. J. Biol. Chem. 276 (47), 44193–44202. doi: 10.1074/jbc.M105880200
Teaniniuraitemoana V., Huvet A., Levy P., Klopp C., Lhuillier E., Gaertner-Mazouni N., et al. (2014). Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: Identification of potential sex differentiation and sex determining genes. BMC Genomics 15, 491. doi: 10.1186/1471-2164-15-491
Tong Y., Zhang Y., Huang J., Xiao S., Zhang Y., Li J., et al. (2015). Transcriptomics analysis of Crassostrea hongkongensis for the discovery of reproduction-related genes. PloS One 10 (8), e0134280. doi: 10.1371/journal.pone.0134280
Trapnell C., Williams B., Pertea G., Mortazavi A., Kwan G., van Baren M., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–U174. doi: 10.1038/nbt.1621
Vainio S., Heikkila M., Kispert A., Chin N., McMahon A. (1999). Female development in mammals is regulated by wnt-4 signalling. Nature 397 (6718), 405–409. doi: 10.1051/medsci/2002182149
Vidal V., Chaboissier M., Schedl A. (2002). Sox9 gene induces testis development XX transgenic mice. M S-Med. Sci. 18 (2), 49–151.
Wang C., Croll R. (2003). Effects of sex steroids on in vitro gamete release in the Sea scallop, Placopecten magellanicus. Invertebr. Reprod. Dev. 44 (2-3), 89–100. doi: 10.1080/07924259.2003.9652559
Wang G., Dong S., Guo P., Cui X., Duan S., Li J. (2020). Identification of Foxl2 in freshwater mussel Hyriopsis cumingii and its involvement in sex differentiation. Gene 754, 144853. doi: 10.1016/j.gene.2020.144853
Wang Y., Duan S., Dong S., Cui X., Wang G., Li J. (2021b). Comparative proteomic study on fem-1b in female and Male gonads in Hyriopsis cumingii. Aquacult. Int. 29 (1), 1–18. doi: 10.1007/s10499-020-00605-1
Wang X., Pei J., Xiong L., Guo S., Cao M., Kang Y., et al. (2022). Identification of the TSSK4 alternative spliceosomes and analysis of the function of the TSSK4 protein in yak (Bos grunniens). Animals 12 (11) 1380. doi: 10.3390/ani12111380
Wang Y., Wang X., Ge J., Wang G., Li J. (2021a). Identification and functional analysis of the sex-determiner transformer-2 homologue in the freshwater pearl mussel, Hyriopsis cumingii. Front. Physiol. 12, 704548. doi: 10.3389/fphys.2021.704548
Wei M., Ge H., Shao C., Yan X., Nie H., Duan H., et al. (2020). Chromosome-level clam genome helps elucidate the molecular basis of adaptation to a buried lifestyle. Iscience 23 (6), 101148. doi: 10.1016/j.isci.2020.101148
Wu Q., Tan Y., Wang J., Xie Q., Huo Z., Fang L., et al. (2019). Effect of estradiol stimulation on dmrt gene expression in Manila clam ruditapes philippinarum. J. Dalian Ocean Univ. 34 (3), 362–369. doi: 10.16535/j.cnki.dlhyxb.2019.03.009
Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xie S., Sun L., Liu F., Dong B. (2009). Molecular characterization and mRNA transcript profile of vitellogenin in Chinese shrimp, Fenneropenaeus chinensis. Mol. Biol. Rep. 36 (2), 389–397. doi: 10.1007/s11033-007-9192-1
Xue X., Zhang L., Li Y., Wei H., Wu S., Liu T., et al. (2021). Expression of the testis-specific Serine/Threonine kinases suggests their role in spermiogenesis of bay scallop Argopecten irradians. Front. Physiol. 12, 657559. doi: 10.3389/fphys.2021.657559
Yan H. (2009). Studies on reproductive physiology of sinonovacula constricta and cyclina sinensis. master thesis (Ocean University of China). doi: 10.7666/d.y1502788
Yang W., Li J., An P., Lei A. (2014). CDC20 downregulation impairs spindle morphology and causes reduced first polar body emission during bovine oocyte maturation. Theriogenology 81 (4), 535–544. doi: 10.1016/j.theriogenology.2013.11.005
Yang D., Yin C., Chang Y., Dou Y., Hao Z., Ding J. (2016). Transcriptome analysis of Male and female mature gonads of Japanese scallop Patinopecten yessonsis. Genes Genom. 38 (11), 1041–1052. doi: 10.1007/s13258-016-0449-8
Yao H., Lin Z., Dong Y., Kong X., He L., Xue L. (2021). Gonad transcriptome analysis of the razor clam (Sinonovacula constricta) revealed potential sex-related genes. Front. Mar. Sci. 8, 725430. doi: 10.3389/fmars.2021.725430
Yi M., Negishi M., Lee S. C. (2021). Estrogen sulfotransferase (SULT1E1): Its molecular regulation, polymorphisms, and clinical perspectives. J. Pers. Med. 11 (3), 194. doi: 10.3390/jpm11030194
Yoon S., Lee M., Xaver M., Zhang L., Hong S., Kong Y., et al. (2016). Meiotic prophase roles of Rec8 in crossover recombination and chromosome structure. Nucleic Acids Res. 44 (19), 9296–9314. doi: 10.1093/nar/gkw682
Yue C., Li Q., Yu H. (2018). Gonad transcriptome analysis of the pacific oyster Crassostrea gigas identifies potential genes regulating the sex determination and differentiation process. Mar. Biotechnol. 20 (2), 206–219. doi: 10.1007/s10126-018-9798-4
Yu K., Hou L., Zhu J., Ying X., Yang W. (2009). KIFC1 participates in acrosomal biogenesis, with discussion of its importance for the perforatorium in the Chinese mitten crab Eriocheir sinensis. Cell Tissue Res. 337 (1), 113–123. doi: 10.1007/s00441-009-0800-3
Zeng Q., Hu B., Blanco A., Zhang W., Zhao D., Martinez P., et al. (2022). Full-length transcriptome sequences provide insight into hermaphroditism of freshwater pearl mussel Hyriopsis schlegelii. Front. Genet. 13, 868742. doi: 10.3389/fgene.2022.868742
Zhang N., Xu F., Guo X. (2014). Genomic analysis of the pacific oyster (Crassostrea gigas) reveals possible conservation of vertebrate sex determination in a mollusc. G3-Genes Genom. Genet. 4 (11), 2207–2217. doi: 10.1534/g3.114.013904
Zhao W., Li Y., Wang Y., Luo X., Wu Q., Huo Z., et al. (2022). Gonadal development and the reproductive cycle of the geoduck clam (Panopea japonica) in the Sea of Japan. Aquaculture 548, 737606. doi: 10.1016/j.aquaculture.2021.737606
Keywords: Cyclina sinensis, sex-differentially expressed genes, spermatogenesis, ovarian development, vitellogenin
Citation: Liu M, Ni H, Rong Z, Wang Z, Yan S, Liao X and Dong Z (2023) Gonad transcriptome analysis reveals the differences in gene expression related to sex-biased and reproduction of clam Cyclina sinensis. Front. Mar. Sci. 9:1110587. doi: 10.3389/fmars.2022.1110587
Received: 29 November 2022; Accepted: 29 December 2022;
Published: 16 January 2023.
Edited by:
Bo Liu, Qingdao Agricultural University, ChinaReviewed by:
Ping Liu, Yellow Sea Fisheries Research Institute (CAFS), ChinaCopyright © 2023 Liu, Ni, Rong, Wang, Yan, Liao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiguo Dong, ZHpnNzcxMkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.