- 1Marine Science and Technology College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
- 2School of Petrochemical Engineering and Environment, Zhejiang Ocean University, Zhoushan, Zhejiang, China
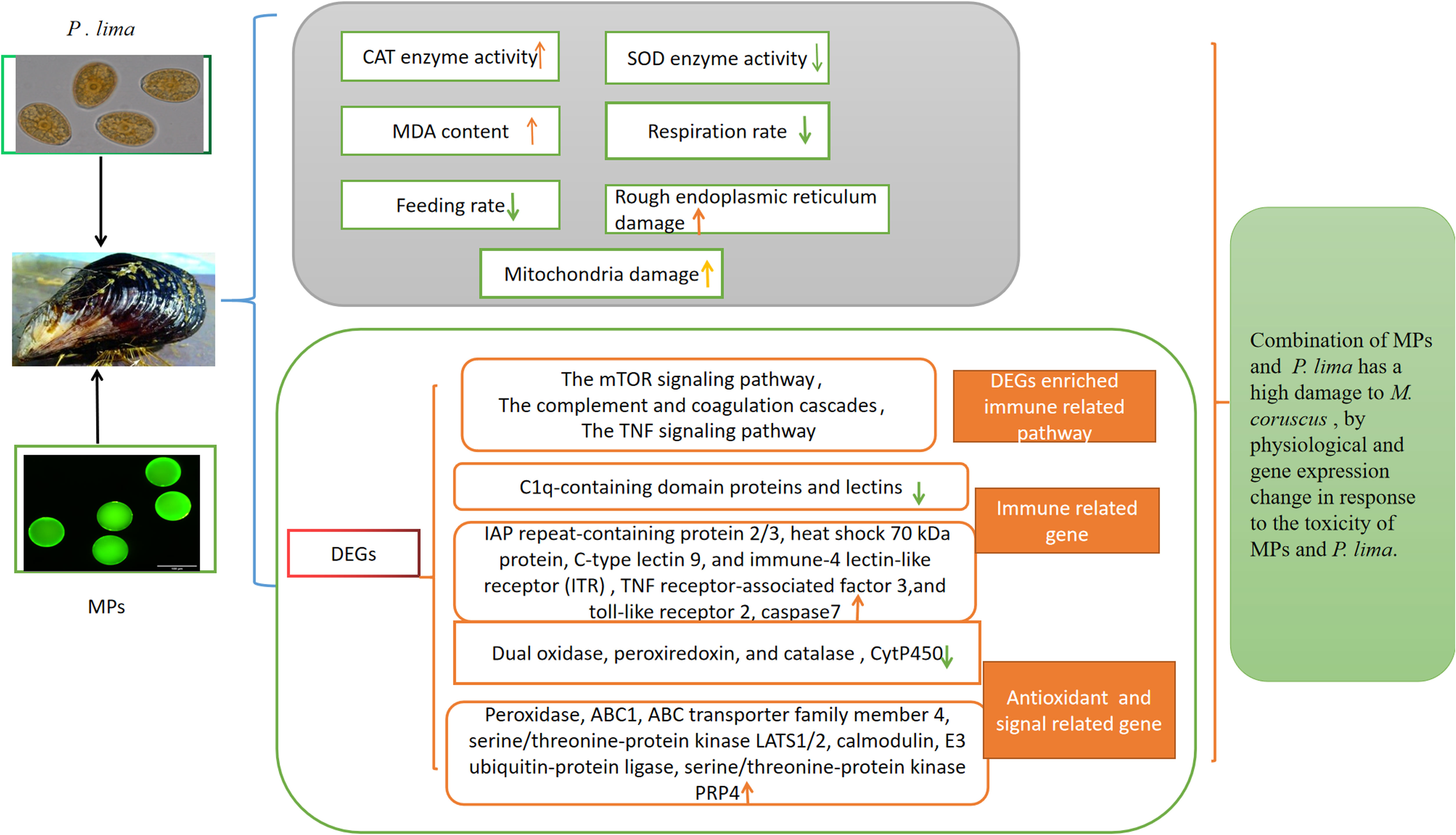
Nowadays, diarrheic shellfish toxicity (DSP) toxin and microplastics (MPs) are commonly found in coastal waters worldwide. Due to their widespread use, their persistence and toxicity, they may induce adverse effects on Mytilus coruscus. However, the underlying toxic mechanisms of DSP and MPs on M. coruscus remain unclear. This study explored the physiological index and transcriptome change of the digestive gland of adult M. coruscus exposed for 3 days to polystyrene (PS) MPs (0.2 mg/L, 90-100 μm) and Prorocentrum lima alone or in combination. The results showed that the CAT activity and MDA content significantly increased, respiration rate and feeding rate significantly decreased. The combination of MPs and P. lima caused more structural damage to the rough surface endoplasmic reticulum and mitochondria in the digestive glands of M. coruscus. The transcriptome analysis showed that 485 and 220 genes were up- and down-regulated, respectively, after exposure to P. lima; 1,989 up-regulated DEGs and 1,098 down-regulated DEGs were identified after exposure to MP treatment, and 1,004 up-regulated DEGs and 664 down-regulated DEGs were identified after exposure to the combination of P. lima and MPs. The DEGs were mainly enriched in the lysosome, mRNA surveillance pathway, carbon metabolism, the mTOR signaling pathway, the complement and coagulation cascades, and the TNF signaling pathway. The MP, P. lima exposure mainly induced the expression of RNA-binding protein musashi, serine/arginine repetitive matrix protein 1, low affinity immunoglobulin epsilon Fc receptor, toll-like receptor 2, caspase 7, calmodulin, E3 ubiquitin-protein ligase, serine/threonine-protein kinase PRP4, glutathione S-transferase, and heat shock 70 kDa protein. MPs and P. lima poison mainly influence the expression of RNA transport, immune related gene, apoptosis, signal related gene, and antioxidant gene change. The combination of MPs and P. lima has a synergistic toxic effect. This study provides a new insights into its physiological and molecular responses of M. coruscus to MPs and P. lima toxic exposure.
Introduction
In recent years, plastic pollution has attracted increasing attention from the public and has become a popular researched topic. Microplastic pollution has been a major ecological and societal issue, specifically for threatening marine ecosystems (Lebreton et al., 2017). The definition of Microplastics (MPs) is an upper size limit of 5 mm plastic particles (Li et al., 2016) that have the characteristics of small size, easy diffusion, and chemical stability. Therefore, MPs are ubiquitous in daily life and easily transported in the marine environment (Scheurer and Bigalke, 2018). Given their small size and easy diffusion, MPs can be ingested easily by marine animals. Previous studies have shown that MPs, mostly microspheres, can cause adverse health impacts (Vital et al., 2021), including growth (Jiang et al., 2022), reproduction (Xu et al., 2020), enzyme activity (Trestrail et al., 2021), energy balance (Weber et al., 2021), and survival (Rist et al., 2016). At the molecular level, MPs ingested by marine animals affect many pathways involved in immune and stress responses, including the apoptotic processe, lysosomal response, antioxidant system (Capolupo et al., 2018), detoxification, and complement system (Li et al., 2021). The filtration rate of Mytilus galloprovincialis was significantly reduced with increasing the concentrations of polyethylene MP (PE-MP). PE-MPs induced oxidative damage and inactivated the antioxidative system (CAT and GST) in the digestive glands at 100 and 1000 μg/L PE-MP treatment (Abidli et al., 2021).
Due to some factors, the intensity and frequency of global warming (Wells et al., 2020), potentially microplastic pollution (Casabianca et al., 2019), and harmful algal blooms (HABs) are increasing worldwide. HABs have become a hot topic due to their increasing frequency and severity (Valdiglesias et al., 2013). The marine phytoplankton P. lima, P. concavum, and P. maculosumis are important HAB microalgae that can produce diarrhetic shellfish poisoning (DSP) and accumulate in bivalve mollusks, causing various toxicities (Chi et al., 2018). As reported, DSP exposure may cause damage to bivalve tissues, change the immune system and induce genotoxicity and cytotoxicity (Chi et al., 2018). Bivalves exposed to P. lima showed genotoxic effects on the gene expression involved in the immune system and cell cycle regulation. Previous studies also showed that some genes, such as glutathione S-transferase (GST), cytochrome P450, and antioxidant system enzymes, may be involved in the detoxification in shellfish (Wei et al., 2019). P. viridis might primarily rely on glutathione S-transferase (GST) and ABC transporters to counteract DSP toxins during short-term exposure. For more prolonged exposure could activate the inhibitors of apoptosis protein (IAP) and the Nrf2 signaling pathway, which in turn reduced the damage of DSP toxins to the mussel (Dou et al., 2020). However, the underlying mechanisms remain unclear after M. coruscus exposure to P. lima.
Mussels are marine bivalve shellfish, and they have a wide geographical distribution and a filter-feeding lifestyle. M. coruscus is often employed as a potential candidate in environmental monitoring, ecotoxicological investigations and assessments of chemical contaminants. M. coruscus is also one of the main cultured shellfish in the East China Sea. In recent years, the presence of microplastics in shellfish tissues, which were obtained either by sampling from marine environments or markets, has been reported in many studies (Hermabessiere et al., 2019). Moreover, the influence of DSP toxins on marine bivalve also worsened. Studies on the influence of DSP in shellfish have shown that DSP not only affects some significant enzyme activities but also affects the expression of genes related to the apoptosis, cytoskeleton, complement system, and immune system (He et al., 2019; Neves et al., 2019; Dou et al., 2020; Huang et al., 2021). To the best of our knowledge, outbreaks of P. lima are often accompanied by the occurrence of microplastics (Do Prado Leite et al., 2022), but the toxic effect mechanisms of the P. lima and microplastic combination on M. coruscus are unclear and require further research. The present study was focused on the changes in the antioxidant enzyme activity, respiration rate, feeding rate, subcellular structure and transcriptome of M. coruscus upon exposure to the DSP-producing dinoflagellate P. lima in combination with microplastics. The results of this study were intended to elucidate the toxic effects of HABs in combination with MPs on marine organisms and provide a better understanding of the interactions mechanisms of complex pollutants in the marine environment.
Materials and methods
Materials
Adult mussels (70–80 mm in length) were collected from a local market in Zhoushan city in the East China Sea. Then, each mussel was scrubbed and rinsed to remove any adhering material (including epiphytic biomass and environmental MPs) and acclimated in an aquarium in aerated artificial seawater (ASW, 26 ± 1) at 18 ± 1°C for four days. The artificial seawater was filtered using a 0.22 μm filter membrane before use to maintain a microplastic-free environment.
The dinoflagellate P. lima was purchased from Shanghai Guangyu Biological Technology Co., Ltd. The P. lima were cultured in f/2 medium and grown in an artificial climate incubator at 20 ± 1°C (light intensity:40 μmol m−2 s−1, 12h/12 h light/dark cycle).
Polystyrene (PS) is one of the most commonly manufactured plastic polymers and is frequently found as microplastic in the marine environment worldwide. Lumisphere uniform green fluorescent microspheres in emulsion at nominal sizes of 90-100 μm were purchased from Tianjin BaseLine ChromTech Research Center, Tianjin, China. These spheres were characterized as having excitation and emission wavelengths of 488 nm and 518 nm, and a density of 10 mg/mL. Images of the MPs obtained from the fluorescence microscope are given in Supplementary Figure 1.
Experimental design
The experimental mussels were divided into four experimental groups, including mussels raised in a tank with clean artificial seawater (ASW) as the control group (denoted CK), an exposure to P. lima (denoted LM), exposure to 0.2 mg/L polystyrene MPs (denoted MPs1), and P. lima and MP co-exposure groups (denoted LM_MPs1). According to previous studies, 0.2 mg/L polystyrene MP, which is close to the concentration of microplastics in realistic pollution scenarios, was chosen (Lasee et al., 2017). Seventy-six mussels were randomly selected to be equally divided into 4 experimental tanks filled with 10 L of membrane-filtered seawater with CK, LM, MP, and LM_MP treatments change to MPs1, and LM_MPs1 treatments. The exposure lasted for 72 h, during which the MPs (0.2 mg/L) and P. lima microalgae (2×103 cells mL−1 determined by LUNA-II™ Automated Cell Counter) were added twice a day, meanwhile the ASW was renewed, and no mussels died during the exposure. After 72 h, the mussels were transferred to 12 tanks (3 repeats for each treatment, 1 mussel for each repeat) for respiration rate measurements. Another 12 tanks with mussels (3 repeats for each treatment, 1 mussel for each repeat) were used for the feeding rate measurement. Following exposure for 72 h, each mussel shell surface was rinsed with sterile seawater. The gills, digestive glands, and kidneys from 3 mussels for each group were snap-frozen under liquid nitrogen and maintained at -80 ℃ until the measurement of biological indices and RNA extraction. Digestive gland tissues from 3 mussel individuals within the same treatment were pooled together as one sample for transcriptome analysis, with 3 repeats for each group. After 72 hours of exposure, one mussel was randomly selected from each tank, dissected to extract the digestive glands (size: 1 mm³), and placed in centrifuge tubes filled with 4% glutaraldehyde. These individuals were used for transmission electron microscopy (TEM) observation.
Enzyme activity
The 100 mg tissues were ground into powder under liquid nitrogen and mixed adding nine times (W/V) the volume of saline. After centrifugation at 3000 rpm for 10 min, the resulting supernatant was used for enzyme activities assay. The SOD and CAT activities and the MDA content were assayed using the Kits (Jian Cheng Bioengineering Research Institute, Nanjing, China), respectively, according to the instructions. Protein concentrations were measured by an enhanced BCA protein assay kit (Beyotime, Shanghai city China) according to the manufacturer’s instructions.
Energy budget
After 72 h of exposure, the energy budget was measured, including the respiration and feeding rates. The respiration rate (mg O2 h−1) determined was according to the dissolved oxygen variations in the ASW. The dissolved oxygen concentration (DO) (mg O2 L−1) was determined by YSI (YSI Pro Plus, Yellow Springs, USA) at the beginning (when moved to the tanks containing new ASW) and 1 h later. The respiration rate was calculated from the difference in DO at the start and 1 h later. After the mussels were moved to the new tanks, we added Isochrysis galbana (the common bait algae, 5×106 cells mL−1 determined by LUNA-II™ Automated Cell Counter) as mussel food. The feeding rate was measured by tracking the changes in hourly algal concentrations in ASW (Wang et al., 2021).
Samples preparation for transmission electron microscopy
The digestive glands (size: 1 mm³) were placed in centrifuge tubes filled with 2.5% glutaraldehyde and incubated overnight at 4°C. The stationary liquid was removed, and rinsed the samples with 0.1 M phosphate buffer (pH 7.0) three times for 15 min each and then 1% osmic acid solution was added, leaving the samples to stand for 2 h. The excess osmic acid liquid was removed, and rinsed the samples with 0.1 M phosphate buffer (pH 7.0) three times for 15 min each. The samples were dehydrated with ethanol solution of graded concentrations (30%, 50%, 70%, 80%, 90%, and 95%). Each concentration was applied for 15 min, with the addition of 100% ethanol and 100% acetone for 20 min. The samples were treated with a 1:1 (v/v) mixture of Spurr’s embedding agent and acetone for 1 h and then a 3:1 (v/v) mixture of Spurr’s embedding agent and acetone for 3 h. After permeation, the samples were embedded and heated overnight at 70 ℃ to obtain the embedded samples. The sections of 70–90 nm samples slices were obtained by a Leica EM UC7 ultrathin slicer. The sections were stained with lead citrate solution and 50% uranium acetate saturation solution for 5–10 min. After being dried, the sliced samples were observed under a Hitachi H-7650 transmission electron microscope.
RNA Extraction
The M. coruscus digestive gland tissue RNA extraction using TRIzol® reagent (Invitrogen), and the genomic DNA was removed using DNaseI (TaKaRa) according to the manufacturer’s instructions. Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit) and agarose gel electrophoresis have used the assay the integrity and purity of the extracted RNA. The high-quality RNA samples (OD260/280 = 1.8-2.2, OD260/230 ≥2.0, 28S:18S ≥ 1.0, >1 μg, RIN ≥ 6.5) were used to construct the cDNA library.
Transcriptome data analysis
Twelve libraries were prepared and sequenced using the DNBSEQ platform by Beijing Genomics Institute (Wuhan, China). The transcriptome raw data presented in the study are deposited in the SRA repository, accession number PRJNA904525. After the raw reads were filtered, we used HISAT to align clean reads to reference genome sequences. The functional annotation of all the unigenes was established by performing a BLAST search against M. coruscus with an e-value cutoff of 1e-5.
Differentially expressed gene analysis
The high-quality reads were mapped to the reference unigene sequences using Bowtie2 v2.2.5 software, and then the gene expression levels were calculated using RSEM (Li and Dewey, 2011). DEGs with a |log2FC|≥1 and Q value ≤ 0.05 were considered significantly differentially expressed genes (Love et al., 2014).
GO enrichment and KEGG pathway enrichment analysis of DEGs
GO and KEGG functional enrichment analyses were applied to identify DEGs significantly enriched in GO terms and metabolic pathways compared with the whole-transcriptome background, with Q-value (adjusted P value) ≤ 0.05. The GO and KEGG pathway enrichment analyses were performed using Goatools (https://github.com/tanghaibao/Goatools), KOBAS (http://kobas.cbi.pku.edu.cn), and the R-function phyper (Zhao et al., 2019).
qRT-PCR validation of gene expression
The isolated RNA sequencing samples were also used to perform real-time quantitative (qRT-PCR) analysis. From the DEGs, 5 related genes were selected to verify the reliability of the transcriptome by real-time fluorescence quantitative PCR. Total RNA was reverse transcribed to single-stranded cDNA using TransScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (one-step gDNA removal, PrimeScriptTM RT Reagent Kit (TaKaRa)) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems) using SYBR Green PCR kits (TaKaRa, China) according to the manufacturer’s instructions. In the present study, β-actin was used as the control gene, and the PCR primers, which were designed using Primer Premier 5 software, are listed in Supplementary Table 1. The qPCR cycling conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Then, the melting curve stage was 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The relative expression was calculated using the 2−ΔΔCT method. All the reactions were performed in triplicate, and each treatment had three biological repeats.
Statistical analyses
One-way ANOVA were performed using SPSS (version 25.0) and two-way ANOVA was performed using GraphPad Prism 8. Three replicates of each experiment and means were separated using Tukey’s multiple range test and Sidak’s multiple comparisons test with a significance threshold of adjusted p value < 0.05.
Results
Biological index analysis
In the present study, the SOD, CAT, MDA content, respiration rate and feeding rate were detected. The oxidative responses in the different tissue and energy budget of mussels are shown in Figure 1.
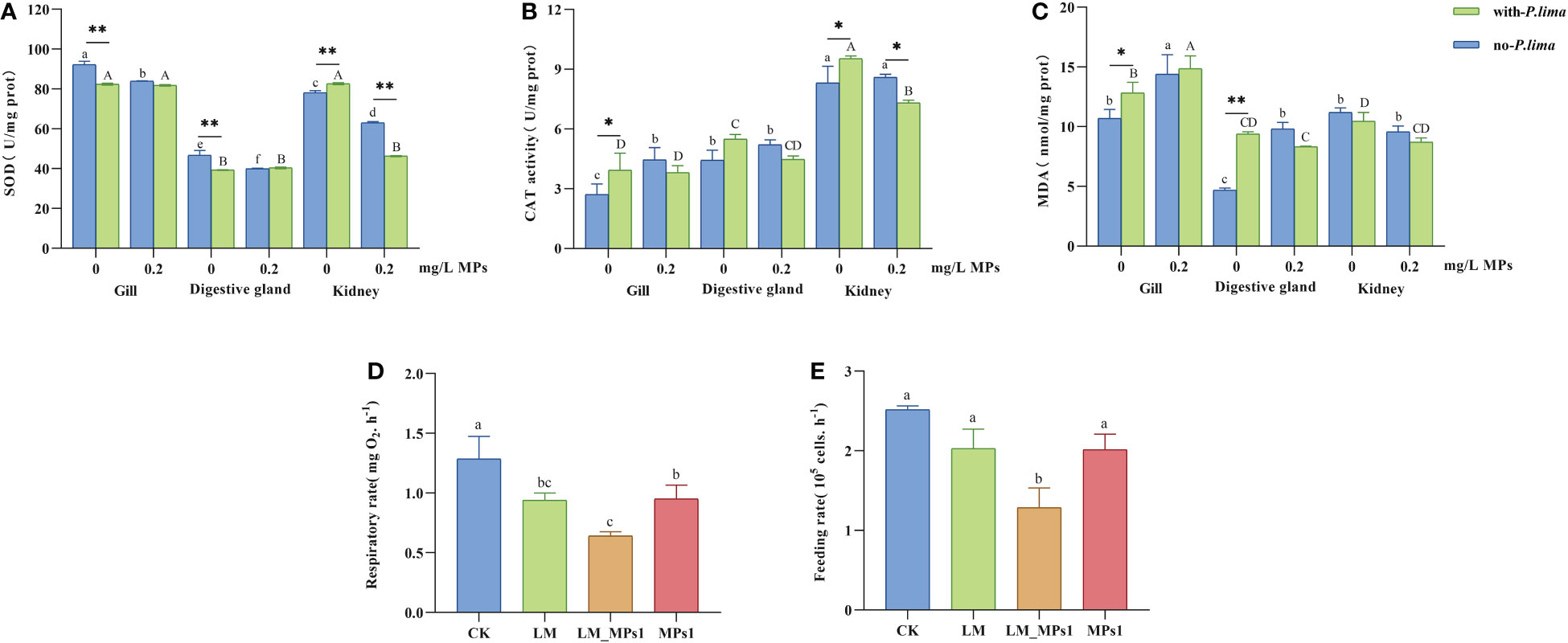
Figure 1 Physiology index change of M. coruscus exposure to MPs and P. lima. (A) SOD activities, (B) CAT activities, (C) MDA content, (D) respiration rate, (E) feeding rate. Different letters indicate significant differences between different tissues in different concentrations of MPs in the presence or absence of P. lima in (A, B, C) (p < 0.05), and different letters indicate significant differences in different groups in (D, E) (p < 0.05), and asterisks indicate significant differences between the presence and absence of P. lima within each fixed concentration of MPs between different tissues. (*p < 0.05; **p < 0.01).
In the no-P. lima treatments, the SOD activity was significantly decreased by MP exposure (p < 0.05). In MP-free treatments, P. lima exposure significantly reduced the SOD activity in the gills and digestive glands but significantly increased SOD activity in the kidney tissue (p < 0.01). Combined exposure to P. lima and MPs significantly decreased SOD activity at high concentrations of MPs (p < 0.01). The SOD activity in the digestive glands was lower than that in the kidneys and gills (Figure 1A).
The CAT activity significantly increased after exposure to single MPs in the gills (p < 0.05), whereas the CAT activity significantly decreased after combined exposure in the kidney (p < 0.05) (Figure 1B). P. lima significantly increased the CAT activity by MP-free treatments in the gills and kidneys (p < 0.05) but significantly decreased the CAT activity by MP exposure in the kidney (p < 0.05). CAT activity is higher in the kidneys of mussels than in the gills and digestive glands (Figure 1B).
The accumulation of MDA increased significantly (p < 0.05) due to MP exposure to the no-P. lima group in the gills and kidneys and the P. lima-exposure group in the gills (Figure 1C). P. lima significantly enhanced MDA concentrations without MPs in the gills (p < 0.05) and digestive gland (p < 0.01) but had no effect at 0.2 mg/L MPs (Figure 1C). The level of lipid peroxidation in the gill was higher than that in the two other tissues (Figure 1C).
The respiration rate was significantly decreased by the three exposure methods in the MPs1, LM, and LM_MPs1 groups. It was significantly lower under the combined exposure than under the single exposure (Figure 1D). For the feeding rate, there were no significant differences among treatments under single exposure in MPs1 and LM but significant decreases after the combined exposure of P. lima and MPs (Figure 1E).
Changes in subcellular ultrastructure in the digestive gland cell of M. coruscus under MP and P. lima exposure
To consolidate the subcellular organizatio change of M. coruscus exposure to MPs and P. lima with the subcellular organization, we observed digestive gland cells using transmission electron microscopy (TEM). The ultrastructural changes in bivalves in response to MPs or P. lima (Vasanthi et al., 2021). Consistently, MPs and harmful algae greatly altered the subcellular structure of the digestive gland cell. TEM results showed a normal cell structure in the control group with numerous ribosomes present at the rough-surfaced endoplasmic reticulum. The mitochondria (M) were normal in shape and size with abundant cristae (Figure 2A). In the P. lima exposure treatments, the rough-surfaced endoplasmic reticulum was significantly swollen, but the ribosomes were still abundant (Figure 2B). In the 0.2 mg/L MP exposure treatment, the ribosomes on the rough-surfaced endoplasmic reticulum were detached, and obvious endoplasmic reticulum membrane dissolutions also appeared (Figure 2C). Swelling of the rough-surfaced endoplasmic reticulum and mitochondria was clear in the combined P. lima and 0.2 mg/L MP exposure group, and the cristae of is the mitochondria were reduced significantly (Figure 2D). TEM analysis revealed that the presence of MPs and P. lima can cause some damage to the digestive gland organelles of M. coruscus, and the combination of MPs and P. lima intensified the toxic effect on the digestive gland.
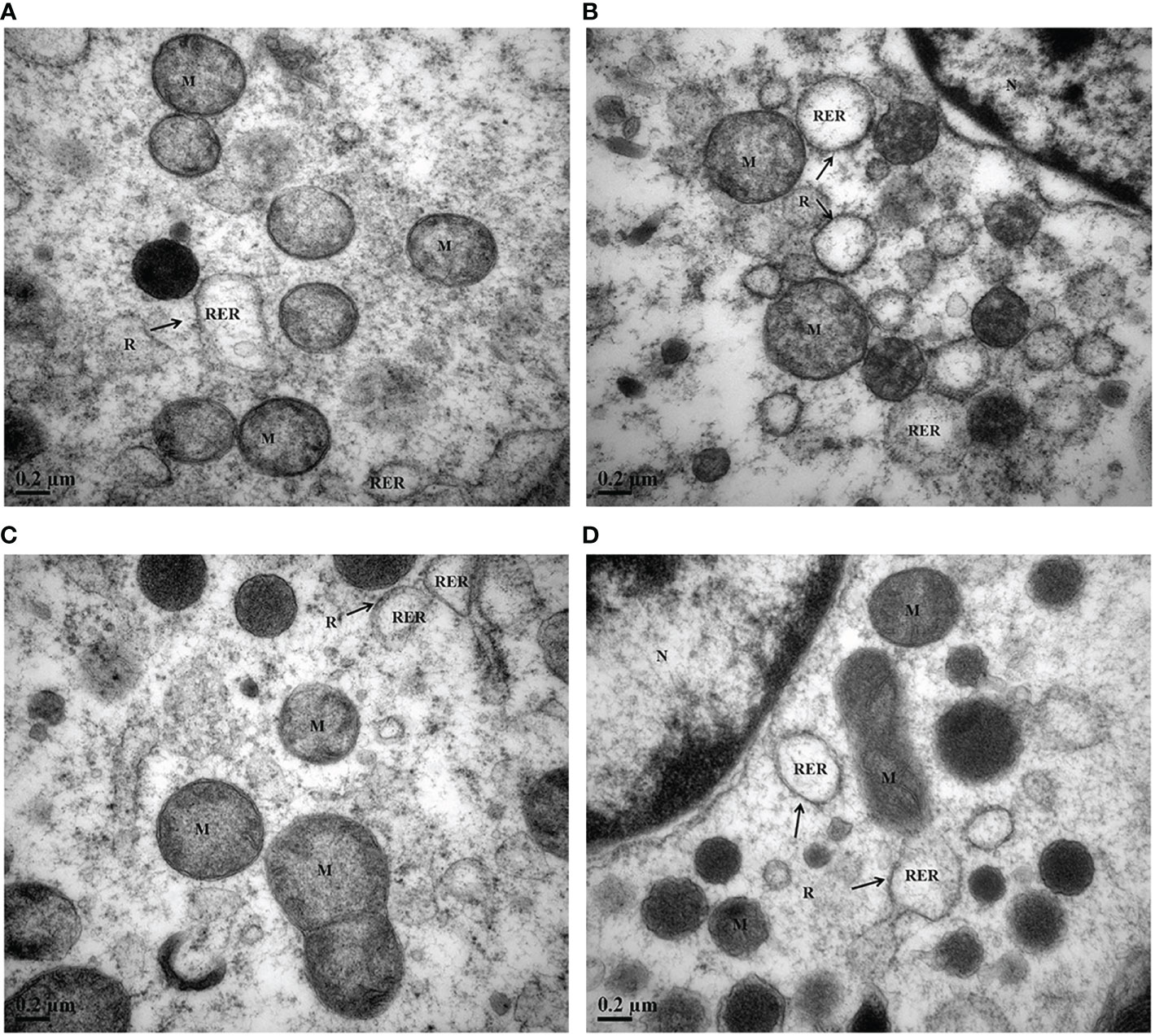
Figure 2 Transmission electron micrographs of the digestive gland of M. coruscus after exposure to MPs and P. lima for 72 h. (A) Control, (B) P. lima exposure, (C) 0.2 mg/L MPs exposure, (D) P. lima and 0.2 mg/L MPs exposure. M, mitochondria; RER, rough-surfaced endoplasmic reticulum; R, ribosome; Mag, ×50000, Scale bar: 0.2μm.
Transcriptome sequencing and gene expression quantity analysis
Twelve cDNA libraries were obtained from the digestive gland of mussel samples, including the control group, P. lima exposure group, MP exposure group, and combined P. lima and MP group, and three biological repeats of each group. Twelve cDNA libraries were sequenced with the DNBSEQ platform, and the sequencing statistics data are outlined in Supplementary Table 2. Each sample average produced approximately 6.36 Gb of data and Q30 > 92% for each library.
The clean reads were mapped to the M. coruscus genome through HISAT2, and in total, 38.53% -74.12% of the alignment was mapped to the genome. The filtered high-quality clean reads were assembled into 27,971 novel transcripts; of these, 21,497 were new alternative splicing subtypes of known protein-coding genes, and 6,474 were transcripts of new protein-coding genes. The size distribution of all the assembled transcripts is shown in Supplementary Figure 2, and most assembled transcripts are have a length of 601-900 bp. As shown in Supplementary Figure 3, a total of 23,426 genes were identified from the digestive glands with FPKM >1, including 16,853, 18,718, 16,745, and 17,661 expressed genes in CK, MPs1, LM, and LM_MPs1, respectively. The numbers of genes crossed in P. lima and the control group, 0.2 mg/L MPs treatment and control groups, and P. lima and 0.2 mg/L MP treatment and control groups were 14,198, 13,936, and 14,255, respectively. A Venn diagram showed the 12,607 genes shared by all the samples (Supplementary Figure 3).
Differentially expressed gene analysis
According to the gene expression levels of each sample, the DEGs were analyzed by DEseq2. Among the transcripts mentioned above, 485 DEGs were up-regulated and 220 DEGs were down-regulated after exposure to P. lima for 72 h compared with the control group conditions, while 1,004 DEGs were up-regulated and 664 DEGs were down-regulated after exposure to P. lima and 0.2 mg/L MPs for 72 h compared with the control group (Figure 3). In addition, 1,989 up-regulated and 1,098 down-regulated DEGs were obtained under MPs treatment compared with the control group conditions, while 78 up-regulated DEGs and 62 down-regulated DEGs were obtained under P. lima and 0.2 mg/L MPs treatment groups compared with P. lima treatment conditions, and 55 up-regulated DEGs and 44 down-regulated DEGs were obtained under P. lima and 0.2 mg/L MPs treatment groups compared with 0.2 mg/L MP exposure conditions (Figure 3). We noted that the most abundant DEGs were present in the 0.2 mg/L MP treatment and P. lima, and 0.2 mg/L MP treatment groups compared with the control group. These results indicated that the digestive gland showed a stronger response to MPs. A Venn diagram (Supplementary Figure 4) was used to elucidate the relationship of the differentially expressed genes between the four treatments in each sample.
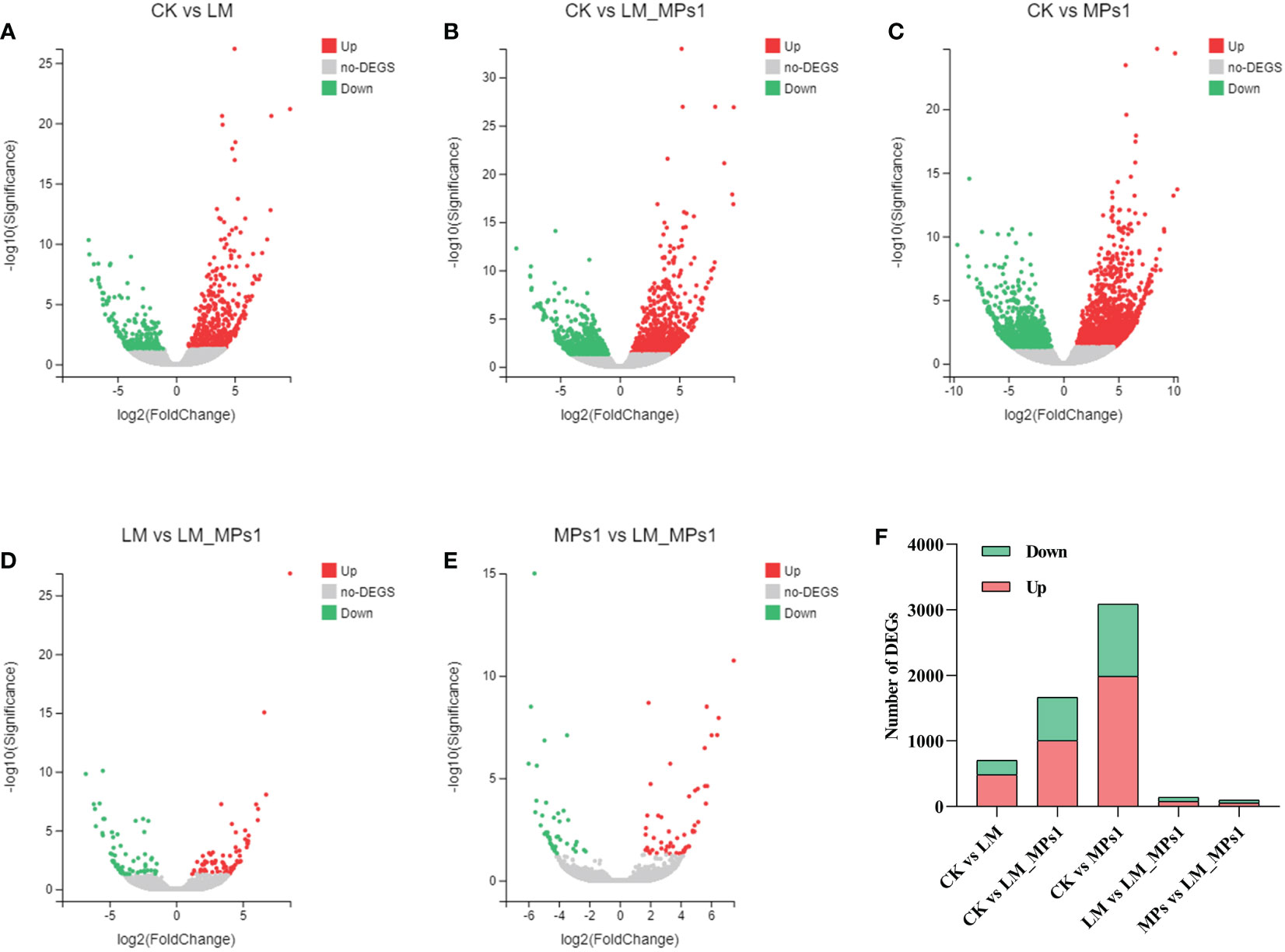
Figure 3 The volcano maps of DEGs in different pairwise comparisons of the digestive gland (A-E). Number of DEGs from pairwise comparisons among the five groups of the digestive gland (F). CK, control; LM, P. lima exposure; LM_MPs1, P. lima & 0.2 mg/L MPs exposure; MPs1: 0.2 mg/L MPs exposure.
To uncover the differentially expressed genes that were activated by the single or combined exposure of P. lima and MPs, the transcripts that passed the critical value (fold change >2, Q value <0.05) were further analyzed, and the results are shown in Table 1. Compared with the control group, the up-regulated genes were leucine-rich repeats and death domain-containing protein, dual serine/threonine and tyrosine protein kinase in the LM group, complement C1q tumor necrosis factor-related protein 6, serine/threonine-protein kinase LATS1/2 in the LM_MPs1 group, dual serine/threonine and tyrosine protein kinase, 2’-5’-oligoadenylate synthetase in the MPs1 group. Compared with the control group, the down-regulated gene was mucin-5 in the LM group, leucine-rich repeat-containing protein 16 in the LM_MPs1 group, and poly [ADP-ribose] polymerase 10/14/15 in the MPs1 group. Under the LM_MPs1 group compared with the LM group conditions, the up-regulated genes were caspase 7 and poly [ADP-ribose] polymerase, and the most downregulated gene was dual serine/threonine and tyrosine protein kinase. Some up- or down-regulated DEGs that are directly involved in the innate immune system and signal transduction, including calmodulin, E3 ubiquitin-protein ligase, toll-like receptor, complement c1q-like protein, and heat shock 70 kDa protein. Some antioxidant-related DEGs, including peroxidase and glutathione S-transferase, are also shown in Table 1.
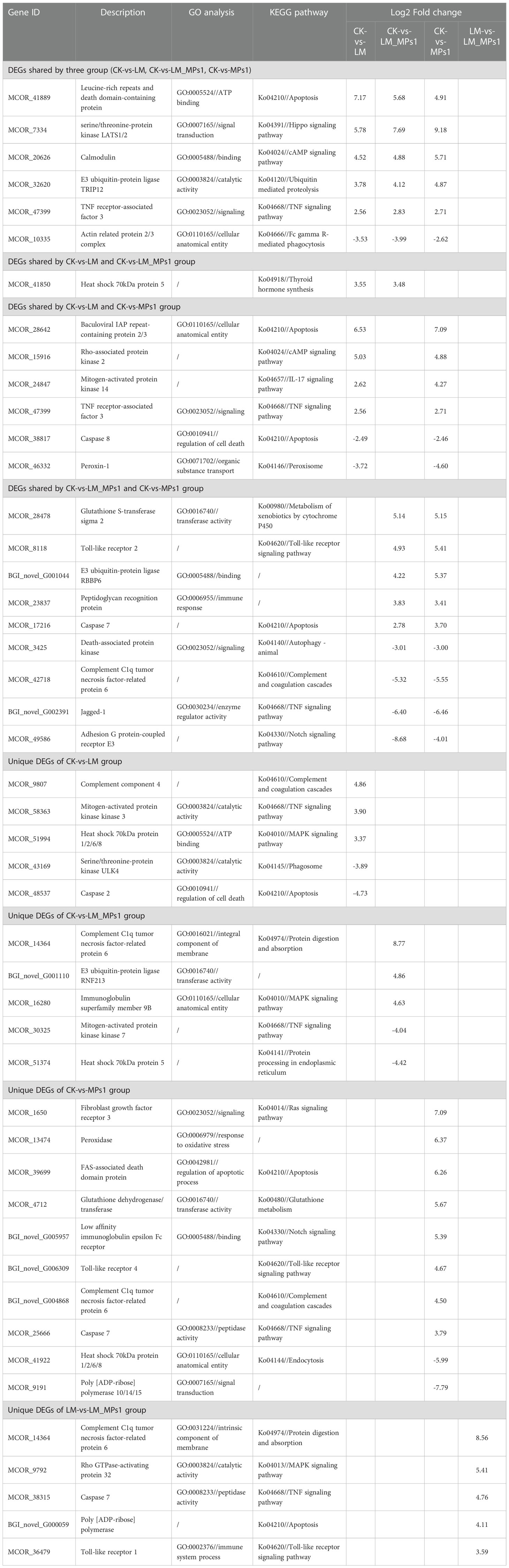
Table 1 The shared and unique DEGs of the digestive gland in M. coruscus among the different comparison groups.
GO annotation and enrichment of DEGs
We classified the 7,845 DEGs with GO terms according to their functions as “biological processes”, “molecular functions” and “cellular components” (Supplementary Figure 5). In the five compared groups, the functional distribution annotated in GO was roughly the same. In the biological process category, the main annotations were cellular process, metabolic process, biological regulation, response to stimulus and signaling. DEGs involved in cellular components were primarily annotated to the intracellular, cellular anatomical entity, and protein-containing complex. In the molecular function category, DEGs were primarily annotated to binding, transporter activity, and catalytic activity. In addition, antioxidant activity, signal transducer activity, and immune system processes are also noted in Supplementary Figure 5. The top twenty GO terms (Q value ≤ 0.05) from the DEG enrichment analysis of each group are shown in Figure 4. The results showed that the most enriched GO terms in the CK vs. LM group were intracellular (GO:0043232) and organelle (GO:0043226). In the CK vs. LM_MPs1 group, intracellular organelles (GO:0043229) and non-membrane-bound organelles (GO:0043228) were the most enriched. In the CK vs. MPs1 group, the most enriched GO terms were protein binding (GO:0005515), protein-containing complex (GO:0032991), and intracellular (GO:0043232). In the MPs1 vs. LM_MPs1 group, the GO terms of positive regulation of cytokine production (GO:0001819), activation of innate immune response (GO:0002218), and activation of immune response (GO:0002253) were the most enriched. Details of all the GO enrichments are provided in Supplementary Table 3. These transcripts may be involved in the response to damage to M. coruscus by MPs and P. lima. The results indicated that organelle damage may be influenced by P. lima and MP exposure.
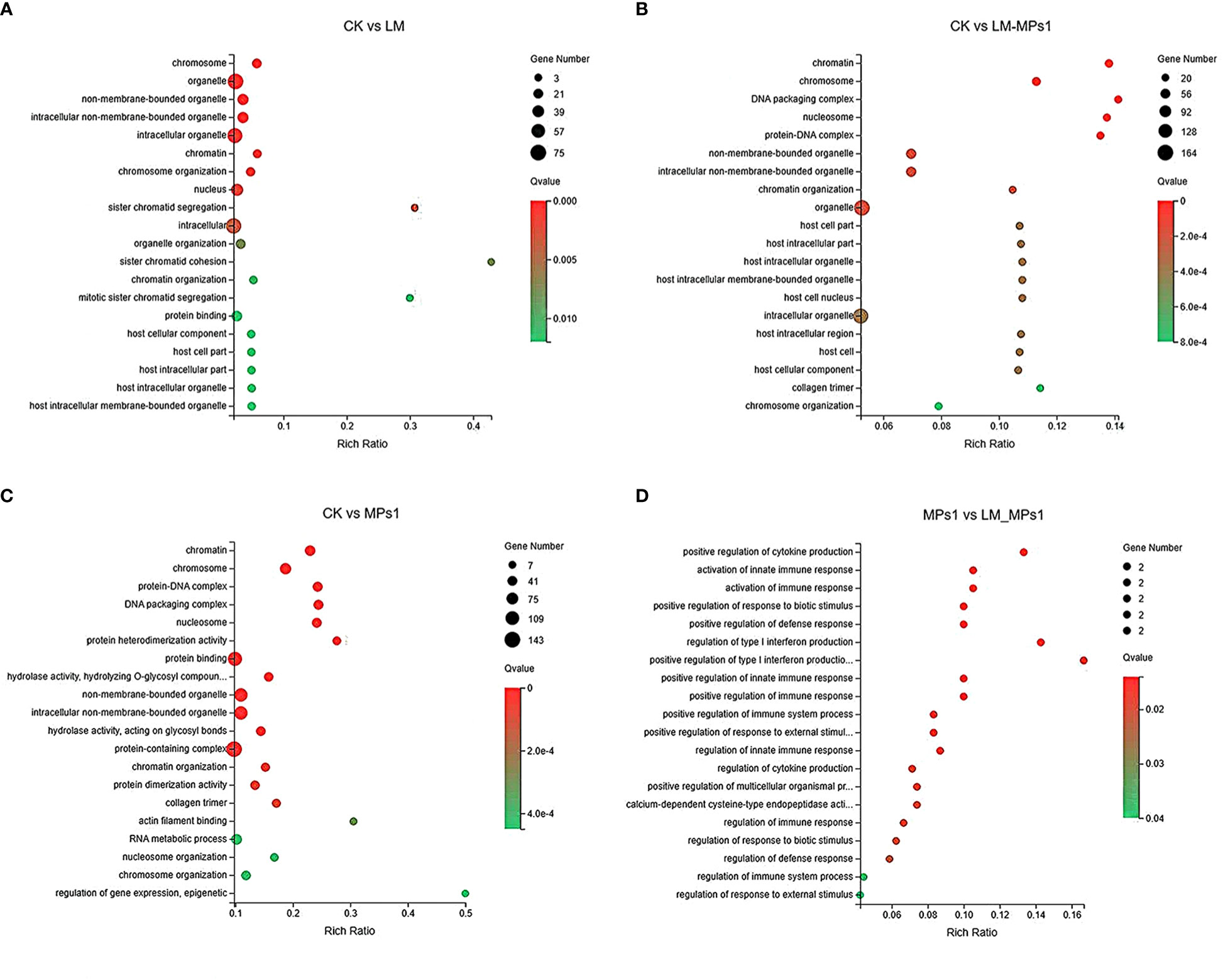
Figure 4 Bubble diagram of top 20 GO terms enriched by DEGs in different pairwise comparisons of the digestive gland (A-D).
KEGG enrichment analyses of DEGs
A KEGG enrichment analysis of DEGs is shown in Figure 5. Totals of 4, 10, and 11 pathways were significantly enriched in the LM, LM_MPs1, and MPs1 groups, respectively, compared with the CK group. The pathway enrichment analysis showed that the most significantly enriched pathways were mRNA surveillance pathway (ko03015), RNA transport (ko03013), protein processing in endoplasmic reticulum (ko04141), the TNF signaling pathway (ko04668) in the CK vs. LM group. In addition, the pathway enriched in lysosome (ko04142), RNA transport (ko03013), carbon metabolism (ko01200), mRNA surveillance pathway (ko03015), mTOR signaling pathway (ko04150), thyroid hormone signaling pathway (ko04919), and penicillin and cephalosporin biosynthesis (ko00311) in the CK vs. LM_MPs1 group. The pathway enriched in protein digestion and absorption (ko04974), thyroid hormone signaling pathway (ko04919), relaxin signaling pathway (ko04926), RNA transport (ko03013), ECM-receptor interaction (ko04512), complement and coagulation cascades (ko04610), lysosome (ko04142), and Fc gamma R-mediated phagocytosis (ko04666) in the CK vs. MPs1 group (Figure 5; Supplementary Table 4). The results indicated that M. coruscus has immune responses and signal change to P. lima and MP exposure.
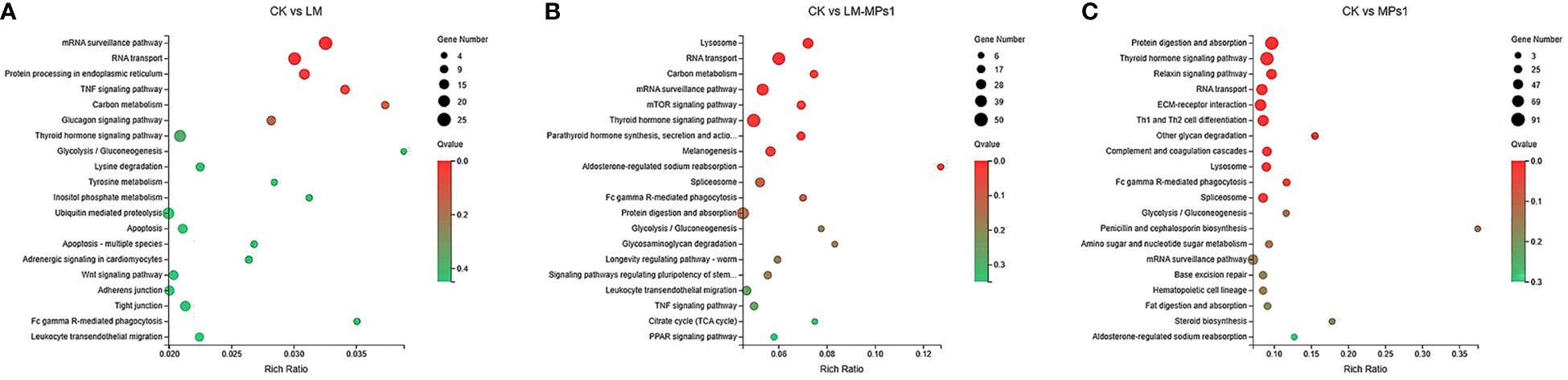
Figure 5 Bubble diagram of top 20 KEGG pathways enrichment of DEGs in different pairwise comparisons of the digestive gland. (A), CK vs. LM; (B), CK vs. LM_MPs1; (C), CK vs. MPs1.
qPCR analysis
The six DEGs with high expression related to immunology, detoxification, and antioxidant ability genes were selected. Two of these genes were suppressed, and the others were induced. Glutathione-S-transferase (GST), heat shock 70 kDa protein (HSP70), toll-like receptor 2 (TLR2), and putative C1q domain containing protein (MgC1q16) were significantly up-regulated (Supplementary Table 5 and Figure 6). Additionally, the detoxifying gene cytochrome P450 family 20 subfamily A (CYP20A) and antioxidant ability-related gene dual oxidase (DUOX) were down-regulated (Supplementary Table 5 and Figure 6). The results indicated that the majority of assayed genes showed almost the similar expression trends as those detected in the transcriptome data.
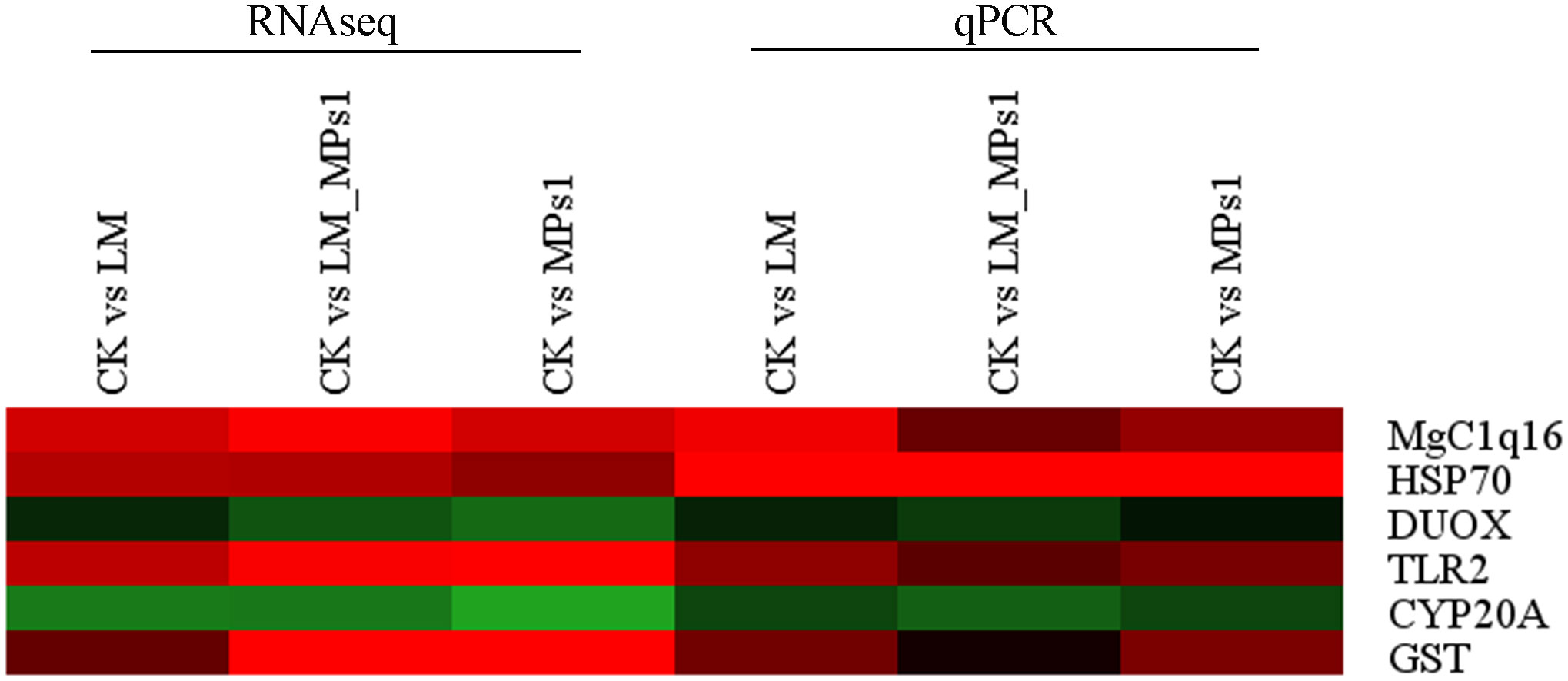
Figure 6 Heatmap of DEGs verify in transcriptome sequencing and qPCR, GST, glutathione-S-transferase; HSP70, heat shock 70kDa protein; CYP20A, cytochrome P450 family 20 subfamily A; TLR2, toll-like receptor 2; MgC1q16, putative C1q domain containing protein; DUOX, dual oxidase. The color panel represents the changes in gene expression levels from down-regulated (green) to up-regulated (red).
Discussion
The study of interactions between organisms and marine pollutants, such as MPs and HABs, is very significant to improving our understanding of the toxicity of those environmental pollutants. In previous studies, exposure to single MPs or P. lima induced physiological stress and immune responses in mussels (Qiu et al., 2020; Trestrail et al., 2021). However, the combined effects of these two environmental pollutants in the context of mussel exposure have not been reported. In our study, the results showed that single and combined exposures influenced oxidative responses and induced the immune response. To investigated the molecular mechanisms of the M. coruscus response to MPs and P. lima and to elucidate the difference in the mussel response of the single or combined group, the immune- and detoxification-related pathways and genes that were differentially expressed in each group were analyzed.
Biological index and TEM analysis
Previous studies indicated that the CAT activity and the MDA concentration in the digestive gland of M. coruscus significantly increased after exposure to 10 µm MPs (Wang et al., 2021). After being exposed to a high density of P. lima for 96 h, the MDA content was increased significantly in the gill tissues of Perna viridis (He et al., 2019). However, in our study, the combined exposure to MPs and P. lima had a significant impact on the CAT activity in the kidney but had no significant impact on the gill and digestive gland. Compared to the single exposure, the MDA level was significantly increased in the combined exposure in the gills but had no significant impact on the digestive gland and kidney. We surmised that digestive gland are insensitive to oxidative reactions. CAT and SOD, the combined scavenger of active oxygen, are considered to be the first line of defense against oxygen free radicals (Wei et al., 2021). In the present study, all the single or combined exposures to MPs or P. lima significantly increased the CAT activity and induced the production of MDA in the tissues of the mussels compared to the control group, implying that those two pollutions caused oxidative damage. SOD activity showed an overall decreasing trend, indicating that MPs and P. lima inhibit SOD activity in the tissues (gills, digestive glands, and kidneys) of mussels. This result was consistent with a previous study showing that the SOD concentrations and SOD gene expression in blue mussels declined after exposure to microplastics (Hamm and Lenz, 2021). The results indicated that antioxidant enzymes may play an important role in reducing microplastic particles and P. lima-induced oxidative stress, and CAT was increased to scavenge ROS (Trestrail et al., 2021).
Interestingly, the enzyme activity results showed that the combined impact of MPs and P. lima was not significant in the digestive gland. However, from the cellular perspective, the TEM results showed that combined exposure caused greater damage to digestive gland cell structures than single exposure. According to (Javadov et al., 2018), mitochondrial swelling is due to damage to the membrane system, ion barrier function of the damaged membrane, and water influx into the mitochondrial matrix. Persistent swelling of mitochondria can eventually lead to apoptosis or necrosis, while mild swelling might be a normal physiological regulation. In the present study, the mitochondria were significantly swollen, and the cristae were shortened and dissolved in the combined group, but in the single treatment, the mitochondria swelled mildly, indicating that the combination of MPs and P. lima causes greater damage to the digestive gland cell structures of mussels.
The respiration and feeding rates of mussels are most affected by the movement of the valves. In our study, the respiration and feeding rates were decreased by the combined exposure of P. lima and MPs. Therefore, combined exposure to P. lima and MPs may affect the valve opening and closure of M. coruscus. It was reported that after exposure to high concentrations (21.6 mg L-1, 216 mg L-1, and 2160 mg L-1) of MPs (1-50 µm), the respiration of mussels was impaired (Rist et al., 2016). The reduction in feeding rate means the less efficient absorption of energy and may affect the growth of individuals (Pedersen et al., 2020). Our results indicated that the energy budget of mussels may be damaged by P. lima and MP exposure. However, the combined exposure causes greater damage to M. coruscus.
Expression analysis of immune-related genes
In our study, the results showed that all the single and combined treatment groups induced some immune response in M. coruscus. It indicated that the combined group had a greater impact on M. coruscus than the P. lima treatment group, but the MPs showed the greatest effect. There are many molecular motifs known as pathogen recognition receptors (PRRs) in mussels, and they can identify potential danger and activate intracellular signaling pathways. PRRs are generally made up of three subfamilies, which are C1q-containing domain proteins and lectins, toll-like receptors and peptidoglycan recognition receptors (PGRPs), and all serve as functional proteins in immune recognition in bivalves (Bouallegui, 2019).
The C1q protein, which contains the globular C1q (gC1q) domain, is involved in the innate immune response, and is found abundantly in the shell, and it participates in the shell formation. The C1q genes, such as MgC1q83 and MgC1q29, were significantly down-regulated in the digestive gland of the mussel Perna viridis following 6 h of exposure to the dinoflagellate P. lima (Dou et al., 2020). Compared with theCK group, we found 6 differentially expressed transcripts, of which 5 DEGs (MgC1q3, MgC1q16, CTRP6, and C1qC) were up-regulated and MgC1q23 was down-regulated after P. lima exposure. In addition, the differential expression of C1q genes also appeared in the MPs1 group (3 up-regulated and 18 down-regulated DEGs) and the combined group (5 up-regulated and 21 down-regulated DEGs). A similar result was also reported in a previous study (Domínguez-Pérez et al., 2021). We speculated that the reason for the differential expression of the C1q family gene may be due to their complex isoforms and distribution. A majority of C1q genes were significantly downregulated in single and combined exposure to MPs and P. lima (Figure 7A), suggesting that DSP toxins and MPs might have a co-negative influence on the complement system of M. coruscus.
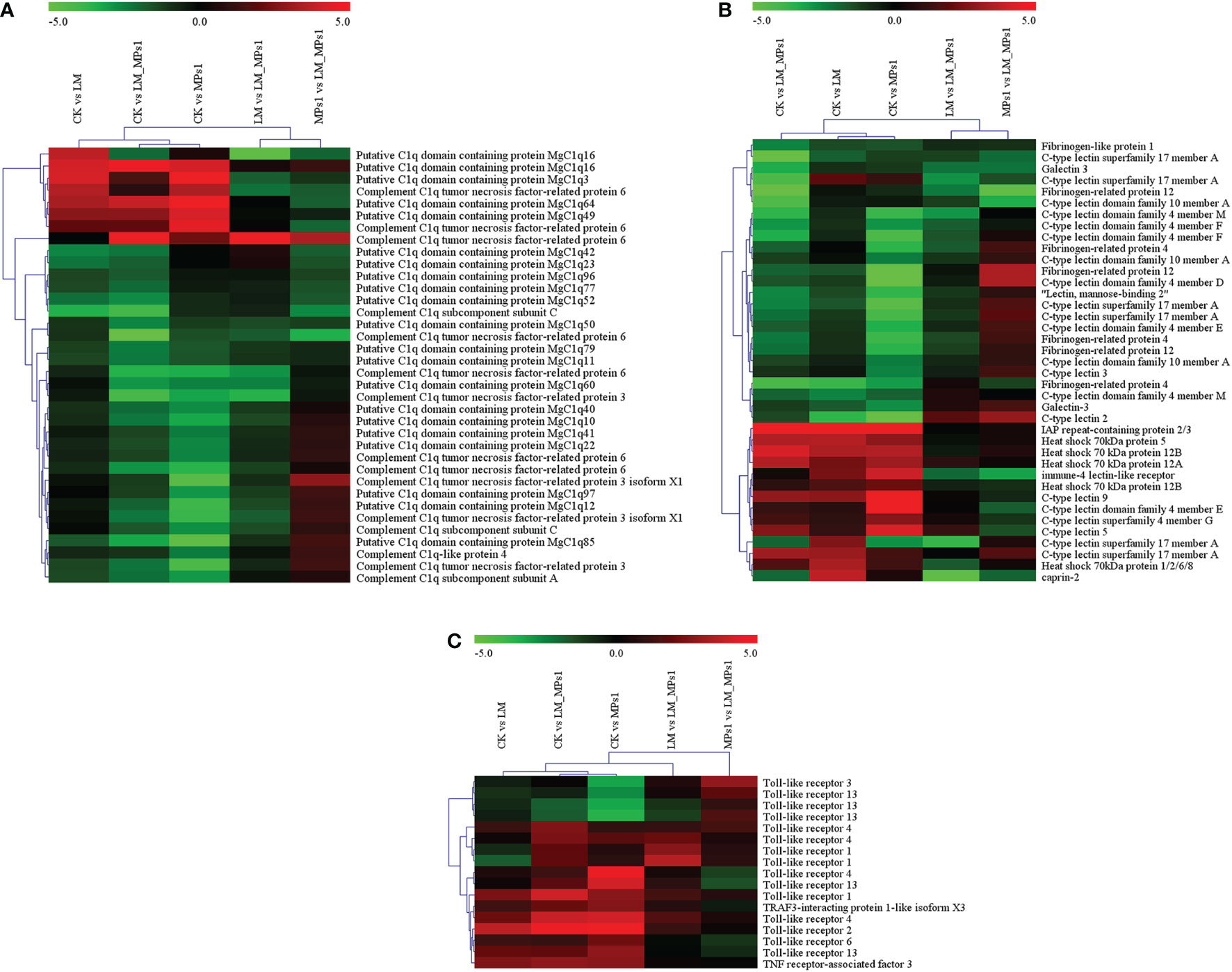
Figure 7 Heatmap analysis of immune-related gene expression in different groups at the transcriptome level. (A), C1q domain protein; (B), Some immune-related genes; (C), Toll-like receptor and TRAF3 genes. The color panel represents the changes in gene expression levels from down-regulated (green) to up-regulated (red).
The C-type lectin (CLT), heat shock protein 70 (Hsp70), fibrinogen-related protein (FREP), and caprine genes are immune-related genes (Wang et al., 2013; Prego-Faraldo et al., 2018). In the present study, we found that the expression of some immune-related genes exhibited highly dynamic changes in response to MPs and P. lima (Figure 7B). Compared with the CK group, IAP repeat-containing protein 2/3 (IAP2/3), heat shock 70 kDa protein 5 (HSP70-5), C-type lectin 9 (CLT-9), and immune-4 lectin-like receptor (ITR) were significantly up-regulated. Fibrinogen-related protein 12 (FREP-12), galectin 3 (Gal-3), and fibrinogen-like protein 1 (FREP-1) were significantly down-regulated in the single and combined groups. High expression changes were observed in the combined group in immune-related genes, such as fibrinogen-like protein 1 (FREP-1), C-type lectin superfamily 17 member A (CLT-17A), heat shock 70 kDa protein 12A (HSP70-12A), and galectin 3 (Gal-3). This corroborated that the combined exposure of MPs and P. lima may have an important effect on the mussel immune system.
Toll-like receptors are type I integral membrane glycoprotein proteins that recognize pathogen-associated molecular profiles (PAMPs). They play a key role in initiating and activating the innate immune system. In the present study, compared with the CK group, 7 TLR DEGs (6 upregulated and 1 downregulated) in the combined group, 6 TLR gene expression levels were significantly different (4 upregulated and 2 downregulated) in the MPs group, and only 1 TLR gene was significantly down-regulated in the P. lima group, which showed that TLR2 and TLR4 were all up-regulated among the TLR genes (Figure 7C). TLR4 has a crucial function in response to viral and bacterial infection (Li et al., 2022), and we found that the TLR4 genes may have the same functions when exposed to MPs and toxic algae. The results were consistent with a previous study showing that under MP exposure, the expression of two toll-like receptors (TLR2 and TLR3) was up-regulated (Gardon et al., 2020). TLRs may play a key role in the induction of the immune response to P. lima and MPs. TRAF3 is a multifunctional immune molecule involved in regulating many immunological responses. In our present study, the expression of TRAF3 in M. coruscus was induced under MP and P. lima exposure (Figure 7C), and the same result was reported in Anodonta woodiana (Qu et al., 2017). The change in the expression of the TLR and TRAF3 genes shows that the severe impact of MPs and P. lima on the immune system of mussels.
Expression analysis of cell apoptosis-related genes
Cell death could be considered a fundamental response to immune and external stimulation stress (Bouallegui, 2019). Apoptosis is an important and multifunctional process that is often considered a signal for pollution monitoring. Caspases, which are related to the mitochondrial apoptotic pathway, are important molecular components of apoptotic cascades in M. galloprovincialis (Estévez-Calvar et al., 2013). In the present study, compared with the CK group, the expression levels of caspase-7 and caspase-6 were significantly up-regulated, caspase-2 and caspase-8 were significantly down-regulated in the single and combined groups (Figure 8). A previous study indicated that caspase-8 in molluscans could be triggered by many biological events, such as chemical exposure, temperature stress (Zhang et al., 2014), and pathogenic infection. In the present study, we obtained the opposite result. Caspase-7 is downstream of mitochondrial-initiated apoptosis, and its function is to commit the cell to apoptosis. The result was consistent with the finding that caspase-7 is upregulated by toxin exposure in oysters (Medhioub et al., 2013). In summary, the results indicated that P. lima and MP exposure induced cell apoptosis in M. coruscus.
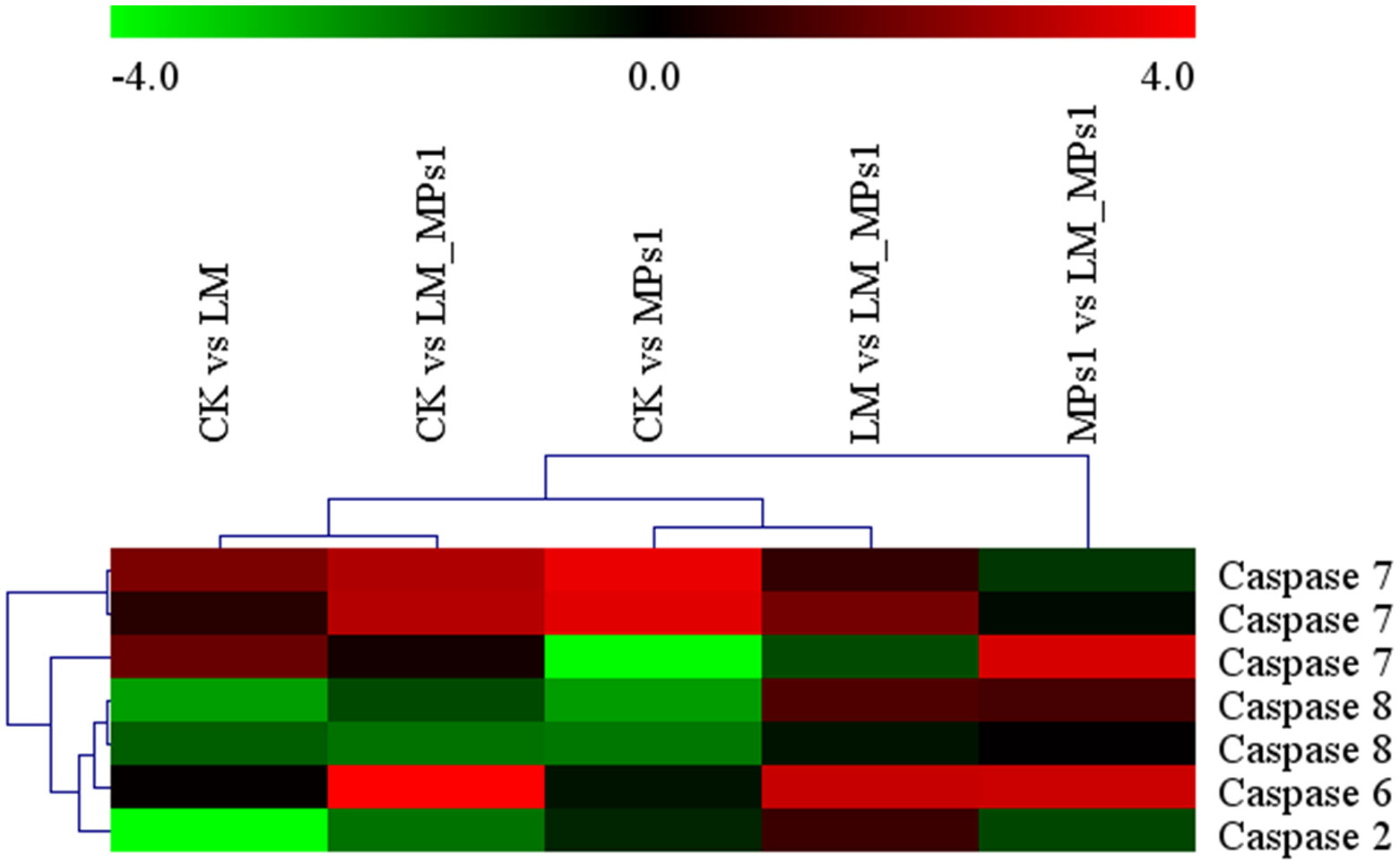
Figure 8 Heatmap analysis of apoptosis-related gene (caspase) expression in different groups. The small panel with a color gradient represents the changes in gene expression levels from down-regulated (green) to up-regulated (red).
Antioxidant and detoxification-related gene expression analysis
In the present study, the detoxification and oxidative responses of M. coruscus were activated after exposure to single and combined MPs and P. lima. Transcriptome analysis indicated that antioxidant-related genes, such as dual oxidase, peroxiredoxin, and catalase, were significantly decreased, while peroxidase was significantly upregulated under single and combined MP and P. lima exposure. The result were consistant with the previous study that the expression of peroxidase in blue mussels (M. edulis) and M. galloprovincialis were increased after exposure to microplastics and P. lima, respectively (Prego-Faraldo et al., 2017; Magara et al., 2019). Glutathione S-transferase (GST), the cytochrome P450 family (CYP450), and ABC transporters play crucial roles in detoxification (Dou et al., 2020). In our study, the expression of GST, and ABC1 was significantly increased under single and combined exposures to MPs and P. lima, with the aim of alleviating the toxic effect of MPs and P. lima on bivalves (Supplementary Figure 6). The results indicated that M. coruscus may mitigate the toxic effects of microplastics and P. lima through the expression of antioxidant and detoxification genes.
Conclusions
In conclusion, the present study revealed that the toxic algae P. lima and MPs induced the immune response and influenced the physiological mechanisms of bivalve species. Single and combined exposures of P. lima and MPs changed the antioxidative enzyme activity and increased the level of lipid peroxidation, damaged the cell sub-microstructure of the digestive gland, and the combined treatment had a high toxic effect on M. coruscus. Regarding the transcriptome, mussels adopt a series of molecular compensatory responses to resist P. lima and MPs, and numerous immune pathways were induced. The MP, P. lima exposure mainly induced the expression of leucine-rich repeats and death domain-containing protein, caspase 7, serine/threonine-protein kinase LATS1/2, serine/threonine-protein kinase PRP4, TNF receptor-associated factor 3, E3 ubiquitin-protein ligase, calmodulin, glutathione S-transferase, and heat shock 70 kDa protein (Figure 9). This study sheds light on the effects of P. lima and MPs on mussels and their immune responses. It will improve our understanding of the future of the mussel aquaculture industry and coastal ecosystems.
Data availability statement
The transcriptome raw data presented in the study are deposited in the SRA repository, accession number PRJNA904525.
Author contributions
M-HF and X-WT contributed to experimental design, statistical analysis, and manuscript preparation. X-WT, RY, C-YX, and Y-KX carried out the experiments. M-HF, ZY, and X-LZ contributed to experiment preparation and data analysis. J-XW, X-JY, ZL, ZY, and M-HF contributed to revision of the manuscript. M-HF, X-JY contributed to funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY20D060006), National College Students’ innovation and entrepreneurship training program (202010340005), and National Natural Science Foundation of China, key international cooperation and exchange project (42020104009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1087667/full#supplementary-material
Supplementary Figure 1 | Images of microplastics (MPs) in the fluorescence microscope.
Supplementary Figure 2 | The number of sequence of different sizes.
Supplementary Figure 3 | Venn diagram of gene number in different treatment.
Supplementary Figure 4 | Venny maps of the DEGs in M. coruscus after MP and P. lima exposure.
Supplementary Figure 5 | The GO functional classification of DEGs in M. coruscus after MP and P. lima exposure.
Supplementary Figure 6 | Heatmap analysis of antioxidant and detoxification-related gene expression analysis in different groups. CK: control; LM: P. lima exposure; LM_MPs1: P. lima and 0.2 mg/L MPs exposure; MPs1: 0.2 mg/L MPs exposure. The small panel with a color gradient represents the changes in gene expression levels from down-regulated (green) to up-regulated (red).
References
Abidli S., Pinheiro M., Lahbib Y., Neuparth T., Santos M. M., Menif N. T. E., et al (2021). Effects of environmentally relevant levels of polyethylene microplastic on Mytilus galloprovincialis (Mollusca: Bivalvia): filtration rate and oxidative stress. Environ. Sci. Pollut. Res. 28, 26643–26552. doi: 10.1007/s11356-021-12506-8
Bouallegui Y. (2019). Immunity in mussels: an overview of molecular components and mechanisms with a focus on the functional defenses. Fish. Shellfish. Immun. 89, 158–169. doi: 10.1016/j.fsi.2019.03.057
Capolupo M., Franzellitti S., Valbonesi P., Lanzas C. S., Fabbri E. (2018). Uptake and transcriptional effects of polystyrene microplastics in larval stages of the Mediterranean mussel Mytilus galloprovincialis. Environ. pollut. 241, 1038–1047. doi: 10.1016/j.envpol.2018.06.035
Casabianca S., Capellacci S., Giacobbe M. G., Dell'Aversano C., Tartaglione L., Varriale F., et al. (2019). Plastic-associated harmful microalgal assemblages in marine environment. Environ. pollut. 244, 617–626. doi: 10.1016/j.envpol.2018.09.110
Chi C., Giri S. S., Jun J. W., Kim S. W., Kim H. J., Kang J. W., et al. (2018). Detoxification- and immune-related transcriptomic analysis of gills from bay scallops (Argopecten irradians) in response to algal toxin okadaic acid. Toxins. 10 (8), 308. doi: 10.3390/toxins10080308
Domínguez-Pérez D., Martins J. C., Almeida D., Costa P. R., Vasconcelos V., Campos A. (2021). Transcriptomic profile of the cockle cerastoderma edule exposed to seasonal diarrhetic shellfish toxin contamination. Toxins. 13 (11), 784(1–7824). doi: 10.3390/toxins13110784
Do Prado Leite I., Menegotto A., Da Cunha Lana P., Júnior L. L. M. (2022). A new look at the potential role of marine plastic debris as a global vector of toxic benthic algae. Sci. Total. Environ. 838 (Pt 3), 156262. doi: 10.1016/j.scitotenv.2022.156262
Dou M., Jiao Y. H., Zheng J. W., Zhang G., Li H. Y., Liu J. S., et al. (2020). De novo transcriptome analysis of the mussel Perna viridis after exposure to the toxic dinoflagellate Prorocentrum lima. Ecotoxicol. Environ. Saf. 192, 110265. doi: 10.1016/j.ecoenv.2020.110265
Estévez-Calvar N., Romero A., Figueras A., Novoa B. (2013). Genes of the mitochondrial apoptotic pathway in Mytilus galloprovincialis. PloS One 8 (4), e61502. doi: 10.1371/journal.pone.0061502
Gardon T., Morvan L., Huvet A., Quillien V., Soyez C., Le Moullac G., et al. (2020). Microplastics induce dose-specific transcriptomic disruptions in energy metabolism and immunity of the pearl oyster Pinctada margaritifera. Environ. pollut. 266 (Pt 3), 115180. doi: 10.1016/j.envpol.2020.115180
Hamm T., Lenz M. (2021). Negative impacts of realistic doses of spherical and irregular microplastics emerged late during a 42 weeks-long exposure experiment with blue mussels. Sci. Total. Environ. 778, 146088. doi: 10.1016/j.scitotenv.2021.146088
He Z. B., Duan G. F., Liang C. Y., Li H. Y., Liu J. S., Yang W. D. (2019). Up-regulation of Nrf2-dependent antioxidant defenses in Perna viridis after exposed to Prorocentrum lima. Fish. Shellfish. Immunol. 90, 173–179. doi: 10.1016/j.fsi.2019.05.003
Hermabessiere L., Paul-Pont I., Cassone A. L., Himber C., Receveur J., Jezequel R., et al. (2019). Microplastic contamination and pollutant levels in mussels and cockles collected along the channel coasts. Environ. pollut. 250, 807–819. doi: 10.1016/j.envpol.2019.04.051
Huang J. H., Jiao Y. H., Li L., Li D. W., Li H. Y., Yang W. D. (2021). Small RNA analysis of Perna viridis after exposure to Prorocentrum lima, a DSP toxins-producing dinoflagellate. Aquat. Toxicol. 239, 105950. doi: 10.1016/j.aquatox.2021.105950
Javadov S., Chapa-Dubocq X., Makarov V. (2018). Different approaches to modeling analysis of mitochondrial swelling. Mitochondrion. 38, 58–70. doi: 10.1016/j.mito.2017.08.004
Jiang W., Fang J., Du M., Gao Y., Fang J., Jiang Z. (2022). Microplastics influence physiological processes, growth and reproduction in the Manila clam, Ruditapes philippinarum. Environ. pollut. 293, 118502. doi: 10.1016/j.envpol.2021.118502
Lasee S., Mauricio J., Thompson W. A., Karnjanapiboonwong A., Kasumba J., Subbiah S., et al. (2017). Microplastics in a freshwater environment receiving treated wastewater effluent. Integr. Environ. Assess. Manage. 13 (3), 528–532. doi: 10.1002/ieam.1915
Lebreton L. C. M., Zwet J., Damsteeg J. W., Slat B., Anthony A., Reisser J. (2017). River plastic emissions to the world’s oceans. Nat. Commun. 8 (1), 15611. doi: 10.1038/ncomms15611
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC. Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li Z., Feng C., Pang W., Tian C., Zhao Y. (2021). Nanoplastic-induced genotoxicity and intestinal damage in freshwater benthic clams (Corbicula fluminea): comparison with microplastics. ACS. Nano 15 (6), 9469–9481. doi: 10.1021/acsnano.1c02407
Li L., Liu W., Fan N., Li F., Huang B., Liu Q., et al. (2022). Scallop IKK1 responds to bacterial and virus-related pathogen stimulation and interacts with MyD88 adaptor of toll-like receptor pathway signaling. Front. Immunol. 13. doi: 10.3389/fimmu.2022.869845
Li W. C., Tse H. F., Fok L. (2016). Plastic waste in the marine environment: A review of sources, occurrence and effects. Sci. Total. Environ. 566-567, 333–349. doi: 10.1016/j.scitotenv.2016.05.084
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Magara G., Khan F. R., Pinti M., Syberg K., Inzirillo A., Elia A. C. (2019). Effects of combined exposures of fluoranthene and polyethylene or polyhydroxybutyrate microplastics on oxidative stress biomarkers in the blue mussel (Mytilus edulis). J. Toxicol. Environ. Health 82 (10), 616–625. doi: 10.1080/15287394.2019.1633451
Medhioub W., Ramondenc S., Vanhove A. S., Vergnes A., Masseret E., Savar V., et al. (2013). Exposure to the neurotoxic dinoflagellate, Alexandrium catenella, induces apoptosis of the hemocytes of the oyster, Crassostrea gigas. Mar. Drugs 11 (12), 4799–4814. doi: 10.3390/md11124799
Neves R. A. F., Santiago T. C., Carvalho W. F., Silva E. D. S., Silva P. M., Nascimento S. M. (2019). Impacts of the toxic benthic dinoflagellate Prorocentrum lima on the brown mussel Perna perna: shell-valve closure response, immunology, and histopathology. Mar. Environ. Res. 146, 35–45. doi: 10.1016/j.marenvres.2019.03.006
Pedersen A. F., Gopalakrishnan K., Boegehold A. G., Peraino N. J., Westrick J. A., Kashian D. R. (2020). Microplastic ingestion by quagga mussels, Dreissena bugensis, and its effects on physiological processes. Environ. pollut. 260, 113964. doi: 10.1016/j.envpol.2020.113964
Prego-Faraldo M. V., Martinez L., Mendez J. (2018). RNA-Seq analysis for assessing the early response to DSP toxins in Mytilus galloprovincialis digestive gland and gill. Toxins. 10 (10), 417. doi: 10.3390/toxins10100417
Prego-Faraldo M. V., Vieira L. R., Eirin-Lopez J. M., Mendez J., Guilhermino L. (2017). Transcriptional and biochemical analysis of antioxidant enzymes in the mussel Mytilus galloprovincialis during experimental exposures to the toxic dinoflagellate Prorocentrum lima. Mar. Environ. Res. 129, 304–315. doi: 10.1016/j.marenvres.2017.06.009
Qiu J., Ji Y., Fang Y., Zhao M., Wang S., Ai Q., et al. (2020). Response of fatty acids and lipid metabolism enzymes during accumulation, depuration and esterification of diarrhetic shellfish toxins in mussels (Mytilus galloprovincialis). Ecotoxicol. Environ. Saf. 206, 111223. doi: 10.1016/j.ecoenv.2020.111223
Qu F., Xiang Z., Zhou Y., Qin Y., Yu Z. (2017). Tumor necrosis factor receptor-associated factor 3 from anodonta woodiana is an important factor in bivalve immune response to pathogen infection. Fish. Shellfish. Immunol. 71, 151–159. doi: 10.1016/j.fsi.2017.10.004
Rist S. E., Assidqi K., Zamani N. P., Appel D., Perschke M., Huhn M. (2016). Suspended micro-sized PVC particles impair the performance and decrease survival in the Asian green mussel Perna viridis. Mar. pollut. Bull. 111 (1-2), 213–220. doi: 10.1016/j.marpolbul.2016.07.006
Scheurer M., Bigalke M. (2018). Microplastics in swiss floodplain soils. Environ. Sci. Technol. 52 (6), 3591–3598. doi: 10.1021/acs.est.7b06003
Trestrail C., Walpitagama M., Miranda A., Nugegoda D., Shimeta J. (2021). Microplastics alter digestive enzyme activities in the marine bivalve, Mytilus galloprovincialis. Sci. Total. Environ. 779, 146418. doi: 10.1016/j.scitotenv.2021.146418
Valdiglesias V., Prego-Faraldo M. V., Pásaro E., Méndez J., Laffon B. (2013). Okadaic acid: more than a diarrheic toxin. Mar. Drugs 11 (11), 4328–4349. doi: 10.3390/md11114328
Vasanthi R. L., Arulvasu C., Kumar P., Srinivasan P. (2021). Ingestion of microplastics and its potential for causing structural alterations and oxidative stress in Indian green mussel Perna viridis- a multiple biomarker approach. Chemosphere. 283, 130979. doi: 10.1016/j.chemosphere.2021.130979
Vital S. A., Cardoso C., Avio C., Pittura L., Regoli F., Bebianno M. J. (2021). Do microplastic contaminated seafood consumption pose a potential risk to human health. Mar. pollut. Bull. 171, 112769. doi: 10.1016/j.marpolbul.2021.112769
Wang J. J., Wang L. L., Yang C. Y., Jiang Q. F., Zhang H., Yue F. (2013). The response of mRNA expression upon secondary challenge with Vibrio anguillarum suggests the involvement of c-lectins in the immune priming of scallop chlamys farreri. Dev. Comp. Immunol. 40 (2), 142–147. doi: 10.1016/j.dci.2013.02.003
Wang S., Zhong Z., Li Z., Wang X., Gu H., Huang W. (2021). Physiological effects of plastic particles on mussels are mediated by food presence. J. Hazard. Mater 404 (Pt A), 124136. doi: 10.1016/j.jhazmat.2020.124136
Weber A., Jeckel N., Weil C., Umbach S., Brennholt N., Reifferscheid G., et al. (2021). Ingestion and toxicity of polystyrene microplastics in freshwater bivalves. Environ. Toxicol. Chem. 40 (8), 2247–2260. doi: 10.1002/etc.5076
Wei Q., Hu C. Y., Zhang R. R., Gu Y. Y., Sun A. L., Zhang Z. M., et al. (2021). Comparative evaluation of high-density polyethylene and polystyrene microplastics pollutants: Uptake, elimination and effects in mussel. Mar. Environ. Res. 169, 105329. doi: 10.1016/j.marenvres.2021.105329
Wei X. M., Lu M. Y., Duan G. F., Li H. Y., Liu J. S., Yang W. D. (2019). Responses of CYP450 in the mussel Perna viridis after short-term exposure to the DSP toxins-producing dinoflagellate Prorocentrum lima. Ecotox. Environ. Safe. 176, 178–185. doi: 10.1016/j.ecoenv.2019.03.073
Wells M. L., Karlson B., Wulff A., Kudela R., Trick C., Asnaghi V., et al. (2020). Future HAB science: directions and challenges in a changing climate. Harmful. Algae. 91, 101632. doi: 10.1016/j.hal.2019.101632
Xu S., Ma J., Ji R., Pan K., Miao A. J. (2020). Microplastics in aquatic environments: occurrence, accumulation, and biological effects. Sci. Total. Environ. 703, 134699. doi: 10.1016/j.scitotenv.2019.134699
Zhang D., Wang H. W., Yao C. L. (2014). Molecular and acute temperature stress response characterizations of caspase-8 gene in two mussels, Mytilus coruscus and Mytilus galloprovincialis. comp. biochem. physiol. b: Biochem. Mol. Biol. 177-178, 10–20. doi: 10.1016/j.cbpb.2014.08.002
Keywords: Mytilus coruscus, microplastics, Prorocentrum lima, transcriptome, physiological response
Citation: Tang X-w, Yu R, Fan M-h, Yang Z, Liao Z, Yang Z-x, Xie C-y, Xuan Y-k, Wang J-x, Zhang X-l and Yan X-j (2022) Physiological and transcriptome analysis of Mytilus coruscus in response to Prorocentrum lima and microplastics. Front. Mar. Sci. 9:1087667. doi: 10.3389/fmars.2022.1087667
Received: 02 November 2022; Accepted: 09 December 2022;
Published: 23 December 2022.
Edited by:
Laura Canesi, University of Genoa, ItalyReviewed by:
Ruiwen Cao, Qilu University of Technology, ChinaXinguo Zhao, Yellow Sea Fisheries Research Institute (CAFS), China
Copyright © 2022 Tang, Yu, Fan, Yang, Liao, Yang, Xie, Xuan, Wang, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei-hua Fan, ZGluZ2VyNTAzQDE2My5jb20=; Xiao-jun Yan, eWFueGpAempvdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiao-wen Tang
Xiao-wen Tang Rong Yu2†
Rong Yu2† Mei-hua Fan
Mei-hua Fan Zhi Liao
Zhi Liao Jian-xin Wang
Jian-xin Wang Xiao-lin Zhang
Xiao-lin Zhang Xiao-jun Yan
Xiao-jun Yan