- 1Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Fisheries College, Jimei University, Xiamen, Fujian, China
- 2Key Laboratory of Cultivation and High-value Utilization of Marine Organisms in Fujian Province, Fujian Collaborative Innovation Center for Exploitation and Utilization of Marine Biological Resource, Fisheries Research Institute of Fujian, Xiamen, China
Large yellow croaker Larimichthys crocea (Richardson) is an important member in family Sciaenidae, and one of most productive mariculture fishes in China. Fluorescence in situ hybridization is a useful tool for cytogenetics and genomics research. Here, we demonstrated that bacterial artificial chromosome (BAC) clones could be used to identify individual chromosomes in large yellow croaker, and also to study chromosome evolution in the related species. By using BAC paired-end sequencing and sequence alignment, 435 BAC clones were anchored to 24 pseudochromosomes of large yellow croaker genome. Among them, 72 BAC clones with low repeat content were selected and passed PCR test, and then grouped by chromosome for FISH test. As a results, there were 67 BAC clones, 2 to 3 BAC clones per chromosome, generated specific and stable signal at expected position. Then, a dual-color FISH probe cocktail composed of 48 of these BAC clones was formulated and used to hybridize metaphase chromosome spreads, resulting in distinct signal patterns on each chromosome pair, which help to distinguish all chromosomes in the metaphase spreads of large yellow croaker. The chromosome-specific BAC-FISH probes were also applied to a close relative of large yellow croaker, Collichthys lucidus, demonstrating that its Y chromosome originated from the fusion of Chr.1 and Chr.7. Thus, our study provides the first set of chromosome-specific FISH probes in family Sciaenidae, which will play an important role in cytogenetics and genomics research in the family.
1 Introduction
Sciaenidae, the largest family in the order Perciformes, includes approximately 283 species in 67 genera worldwide (Nelson et al., 2016). Sciaenid fishes often represent and important commercial fishery resource and aquaculture species. So far, cytogenetic data of Sciaenid fishes have been reported for approximately 40 species, in which only 7 species have been analyzed by Fluorescence in situ hybridization (FISH) with repetitive sequences (Arai, 2011; Zheng et al., 2016; Liao et al., 2017; Xu et al., 2017). The karyotypes of Sciaenid fishes showed outstanding stability, for most of them have a karyotype composing of 48 acrocentric chromosomes (Accioly and Molina., 2008; Arai, 2011). By contrast, high variation in the number and the location of 5S rDNA loci were revealed by FISH (Liao et al., 2017; Xu et al., 2017). In addition, a special karyotype, 2n=48 = 48a for females and 2n=47 = 46a+1m for male, was found in spinyhead croaker Collichthys lucidus, suggesting a multiple sex chromosome system (♀X1X1X2X2/♂X1X2Y), in which the Y chromosome derived from a fusion of two chromosomes (Zhang et al., 2018). Thus, it can be expected that more chromosomal variation could be identified in Sciaenid fishes by using more cytogenetic makers in the context of karyotypic stability.
FISH with chromosome-specific probes can provide abundant and reliable markers for cytogenetic researches (Stein et al., 2001; Yang et al., 2019), including cytogenetic map (Pinkel et al., 1986; Phillips et al., 2006b), chromosome identification (Stein et al., 2001; Yang et al., 2019), chromosome rearrangement (Phillips et al., 2003; Phillips et al., 2013; Bielski et al., 2020), and chromosome evolution across species (Phillips et al., 2001; Ross et al., 2009; do Vale Martins et al., 2021). Chromosome-specific FISH probes have been developed by several methods, such as DOP-PCR after microdissection or flow sorting (Telenius et al., 1992; Vandewoestyne et al., 2009), and screening BACs corresponding to specific linkage groups from a constructed library (Wang et al., 2007). In recent years, assembled genome-based methods have greatly improved the efficiency of developing chromosome-specific FISH probes development, both for BAC screening and bulk oligonucleotide synthesis (Han et al., 2015; Dong et al., 2018).
Large yellow croaker, Larimichthys crocea Richardson, is a member of the family Sciaenidae distributing along the coast of East Asia. The croaker is one of most important mariculture fish species with the highest farmed production in China, and has received intensive studies (Chen et al., 2018). Currently, genetic maps and chromosome-level reference genomes of large yellow croaker are available, and have been applied to support the researches on population genetics and genetic improvement of economic traits (Ning et al., 2007; Wu et al., 2014; Mu et al., 2018; Kong et al., 2019). The karyotype of large yellow croaker was revealed as 2n = 48a with variation in some population (Liao et al., 2017; Xu et al., 2017). Banding and rDNA-FISH were carried out to provide markers for identifying individual chromosomes of large yellow croaker (Liao et al., 2017). However, there is still no way to identify all individual chromosomes in the large yellow croaker. Therefore, the objectives of this study were: (i) to develop a set of chromosome-specific BACs covering all chromosomes in the large yellow croaker; (ii) to develop a dual-color FISH probe cocktail to assist chromosome identification and paring; and (iii) to preliminary test the cross-species applicability of these cytogenetic probes, and to explore the origin of the Y chromosome in spinyhead croaker.
2 Materials and methods
2.1 Sample collection and metaphase chromosome preparations
Samples of large yellow croaker for BAC library construction and somatic chromosome preparation were collected from the breeding section of Jimei university in Ningde, Fujian, China. Spinyhead croaker for somatic chromosome preparation were previously collected from the Sansha Bay (26°42′33″N, 119°46′49″E), Ningde city, Fujian province, China (Zhang et al., 2018). The information of the samples was shown in Supplementary Table 1. Metaphase chromosomes were prepared by using head kidneys according to the methods described previously (Gold et al., 1990). In addition, part of the fin from each fish was fixed in absolute ethyl alcohol to extract genomic DNA.
2.2 BAC library construction and quality evaluation
The BAC library of large yellow croaker was constructed following the methods previously published (Luo and Wing, 2003; Luo et al., 2006). The main steps of the procedure included: i) extracting high molecular weight genomic DNA from a pooled sample of brain tissue from 2 male individuals; ii) partial digesting DNA with restriction enzyme HindIII (0.8 U/μL) (Fermentas); iii) recovering DNA fragments with 110 kb - 220 kb and 220 kb - 300 kb from gel after being separated with pulsed-field gel electrophoresis (PFGE) with a CHEF Mapper (Bio-Rad) for two times; iv) ligating the size-selected DNA fragments with HindIII-digested pHZAUBAC1 vector (Eight Star Bio-tech Co., Ltd., Wuhan, China); v) transforming the ligation products into Escherichia coli strain DH10B T1 Phage-Resistant (Invitrogen); vi) cultivating the transformants on LB plates with appropriate chloramphenicol, X-gal, and IPTG; vii) picking clones and arraying them into 384-well plates containing LB media; viii) incubating the clones in plates for 16 h and freezing them at -80°C.
Eighty clones were randomly selected, and the BAC plasmids were extracted by conventional alkaline lysis method and digested with I-Sce I enzyme (Fermentas) as described previously (Shi et al., 2011). The size of the inserted fragments and the nulling rate were examined by pulsed-field gel electrophoresis. The genome coverage of the BAC library was estimated based on the size of the genome, the number of clones in the library, the average insert size and the null rate.
2.3 Identification of candidate chromosome-specific BAC clones
Totally, 590 BAC clones were selected randomly from the BAC library, and paired end sequenced by Sanger sequencing with Applied Biosystems 3730 DNA Analyzer. The quality of BAC end sequences (BESs) was evaluated with Phred (http://www.phrap.org/) after removing the vector sequence by Seqclean (https://sourceforge.net/projects/seqclean). The resulted high-quality BESs were aligned to the reference genome of large yellow croaker (GenBank assembly accession: GCA_003711585.2) by Blast (Altschul et al., 1997), with an E-value threshold of 1 × 10−5. The high-quality BESs were also aligned to the assembled genome of spinyhead croaker [SCMI00000000.1 (Cai et al., 2019)]. The results of in silico mapping were visualized with Mapchart v2.30.
The insert sequences of mapped BAC clones were deduced from the genomic sequences between the paired BESs, and subjected to estimate the size and the repetitive sequence content. The repetitive sequences were found by using Repeatmasker (http://www.repeatmasker.org/RepeatMasker/). For each chromosome, 3 candidate chromosome-specific BAC clones were selected for further validation according to the following criteria: 1) with insert size around the estimated average; 2) containing as few repetitive sequences as possible.
2.4 Validation of candidate chromosome-specific BAC clones
The correspondence between the selected BAC clones and pseudochromosomes was verified by amplifying fragments of the expected insert sequence. A pair of primers for each BAC clone was designed using Primer Premier 5 software according to the deduced sequence of BAC insert. The primers were synthesized by BGI Genomics Inc. PCR was carried out with a commercial kit (Tiangen Biotech) according to the manual. The conditions of PCR amplification were as following: pre-denaturation at 94 ℃ for 3 min; 40 cycles of denaturation at 94 ℃ for 30 s, annealing at Tm for 30 s, and extension at 72 ℃ for 30 s. The number and the size of PCR amplification products were checked with 1% agarose gel electrophoresis.
After PCR validation, the candidate chromosome specific BAC clones were further tested by FISH. The manipulation of FISH was essentially the same as previously described (Zhang et al., 2018). BAC plasmid DNA was extracted from Escherichia coli using a BAC/PAC DNA extraction kit (Omega). Probes labeled with biotin-11-dUTP or digoxigenin-11-dUTP were prepared with the BAC plasmid DNA by using nick translation kit (Roche). The labeled probe was added into hybridization solution (50% formamide deionized/2 × SSC) and denatured. Chromosome preparation slides were denatured and dehydrated in gradient ethanol solution. The probe solution hybridized with denatured chromosome on the slides at 37°C overnight. After stringent wash, biotin-labeled probes were detected with Avidin-Alexa fluor-488 (Invitrogen), and digoxigenin-labeled probes were detected with Anti-digoxi-Rhodamine (Invitrogen). And then, slide was dyed by DAPI (4’,6-diamidino-2-phenylindole). Metaphase spreads were observed using an Olympus BX53 epifluorescence microscope. Gray-scale images were captured for each color channel with a digital image capture system (Olympus DP 80), and then merged with Cellsens digital image software (Olympus). Background subtraction and image feature intensification were conducted by using the “curve” and “layer overlay” command in Adobe photoshop CC 2017. Chromosome paring and alignment were performed by using the “cut and paste” and “rotation” commands in Adobe photoshop CC 2017.
2.5 Application of chromosome-specific probes
After PCR and FISH validation, 1 to 3 chromosome-specific BAC clones for each pseudochromosome were selected to formulate a probe cocktail. All selected BAC clones were divided into two groups labeled with biotin-11-dUTP or digoxigenin-11-dUTP, and then pooled together for dual-color FISH on metaphase chromosome spreads of large yellow croaker. The chromosomes were identified according to signal pattern, length, and morphology the chromosomes. Chromosome lengths were measured by using “measurement” tool in Adobe photoshop and used to estimate the relative lengths.
Spinyhead croaker is a close relative of large yellow croaker with a fused Y chromosome, which was speculated to originate from the fusion of Chr. 1 and Chr. 7 by a bioinformatic method (Xiao et al., 2020). To explore the Y origin of spinyhead croaker and to test cross-species adaptability of the developed chromosome-specific BAC-FISH probes of large yellow croaker, two BAC clones corresponding to Chr.7 and 18S rDNA corresponding to Chr.1 were labeled with biotin-11-dUTP and digoxigenin-11-dUTP, respectively; and then pooled together for a dual-color FISH on metaphase chromosome spreads of spinyhead croaker.
3 Results
3.1 Construction of BAC Library in large yellow croaker
We constructed a BAC library of large yellow croaker with genomic DNA extracted from brain tissues. The library contained a total of 41,472 BAC clones. To evaluate the quality of library, eighty clones were randomly selected and examined with pulsed field gel electrophoresis (Supplementary Figure 1). Among the eighty clones, there was only one empty clone, indicating an empty load rate of 1.25%. The size of the insert fragments ranged from 120 kb to 150 kb, with an average of 133 kb (Supplementary Figure 2). Based on the genome size, the number of clones in the library, the average size of inserted fragments, and the rate of empty load, the library was estimated to cover the genome of large yellow croaker for 8.1 times.
3.2 Identification of candidate chromosome-specific BAC clones
A total of 590 BAC clones were randomly selected from the constructed BAC library for paired end sequencing. After processing, 1100 high-quality BESs were obtained, covering 0.13% of genome (Table 1). By using BLAST, there were a total of 435 BAC clones with the paired BESs aligned to the same pseudochromosome. The genomic positions of the BES were recorded and used to estimate the expected length of the BAC insert. A total of 342 BAC clones with putative insert sizes around the average (133 kb ± 30 kb) were subjected to further analysis (Supplementary Table 2). Thus, 11 to 32 candidate chromosome-specific BAC clones were obtained for each chromosome, with an average of 18 (Figure 1).
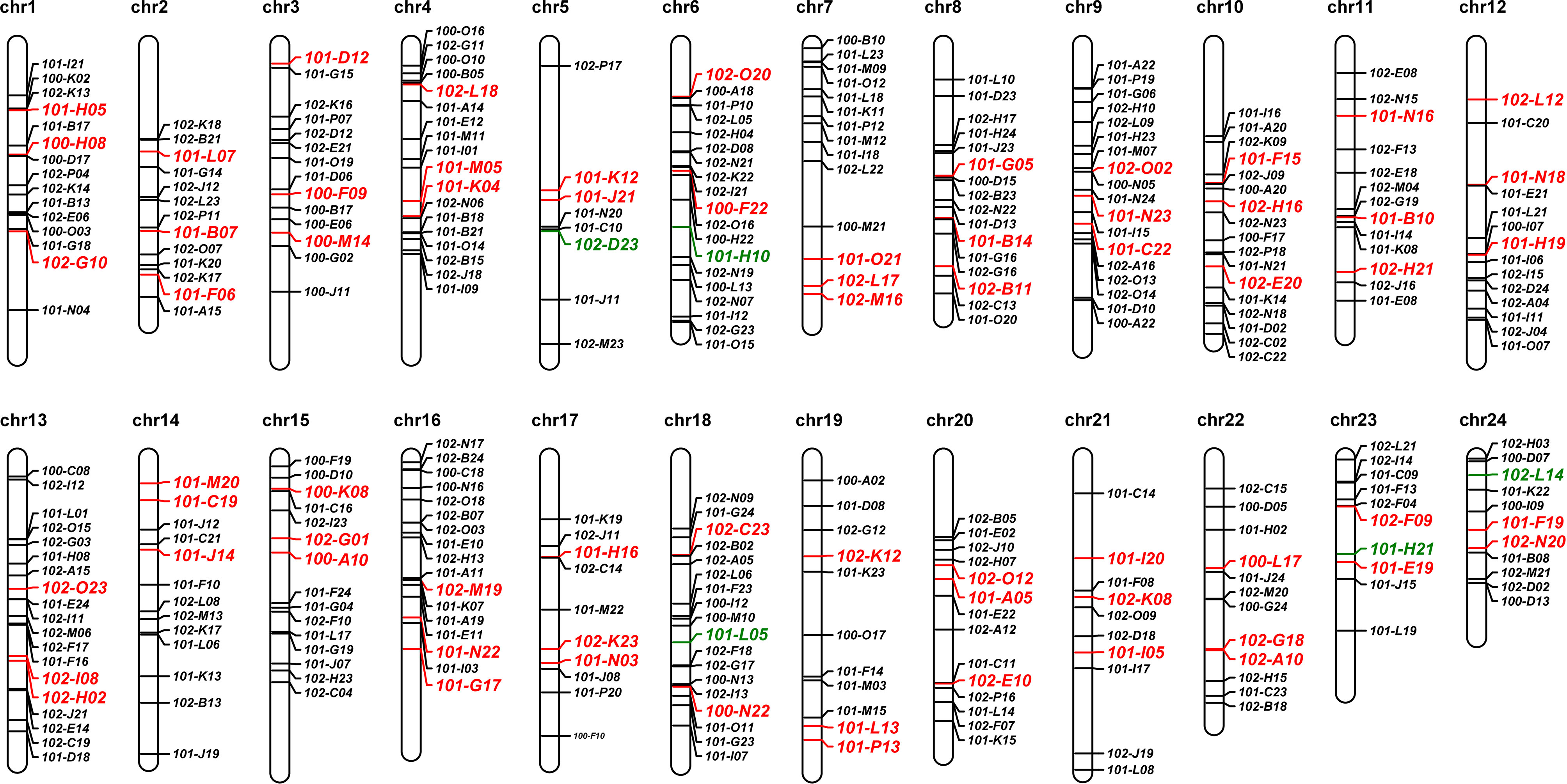
Figure 1 Genomic localization of BAC clones by aligning the end sequences of BACs to the reference genome of L. crocea. The BAC clones with color letters were selected for further FISH verification, red and greed indicating passed and unpassed, respectively.
The repeat content of BAC insertions was estimated to range from 4.24% to 93.5%, with an average of 18.8% ± 10.9% (Supplementary Table 2). Three BAC clones for each pseudochromosome, totally 72 BAC clones, with low content of repeat sequence were selected for further PCR and FISH validaton, which were highlight in color in the ideogram of pseudochromosomes (Figure 1; Supplementary Table 2).
3.3 PCR and FISH verification of chromosome-specific BAC clones of L. crocea
For PCR validation, primers were designed according to putative sequence of the insertions (Supplementary Table 3). The PCR results showed that single product with expected length was amplified with each primer pairs. The representative results of PCR were shown in Supplementary Figure 3. Thus, the accuracy of in silico location of 72 selected BAC clones was preliminarily proved with PCR.
For FISH validation, 72 selected BAC clones were labeled with biotin or digoxigenin. Probes from the same pseudochromosome were pooled for dual-color FISH on metaphase chromosome spreads to examine their location and specificity (Supplementary Figure 4). Figure 2 summarized the FISH results for each pseudochromosome. For 19 chromosome, all 3 selected BAC clones passed FISH validation as they generated 3 specific FISH signals with similar collinearity to the pseudochromosomes. For the other 5 chromosomes including Chr.5, Chr.6, Chr.18, Chr.23 and Chr.24, 2 out of 3 selected BAC clones passed FISH validation. Totally, 67 BAC clones could produce specific signals at the expected positions (Supplementary Table 4). Thus, all chromosomes of large yellow croaker could be identified individually with chromosome-specific FISH probes (Figure 2; Supplementary Figure 4).
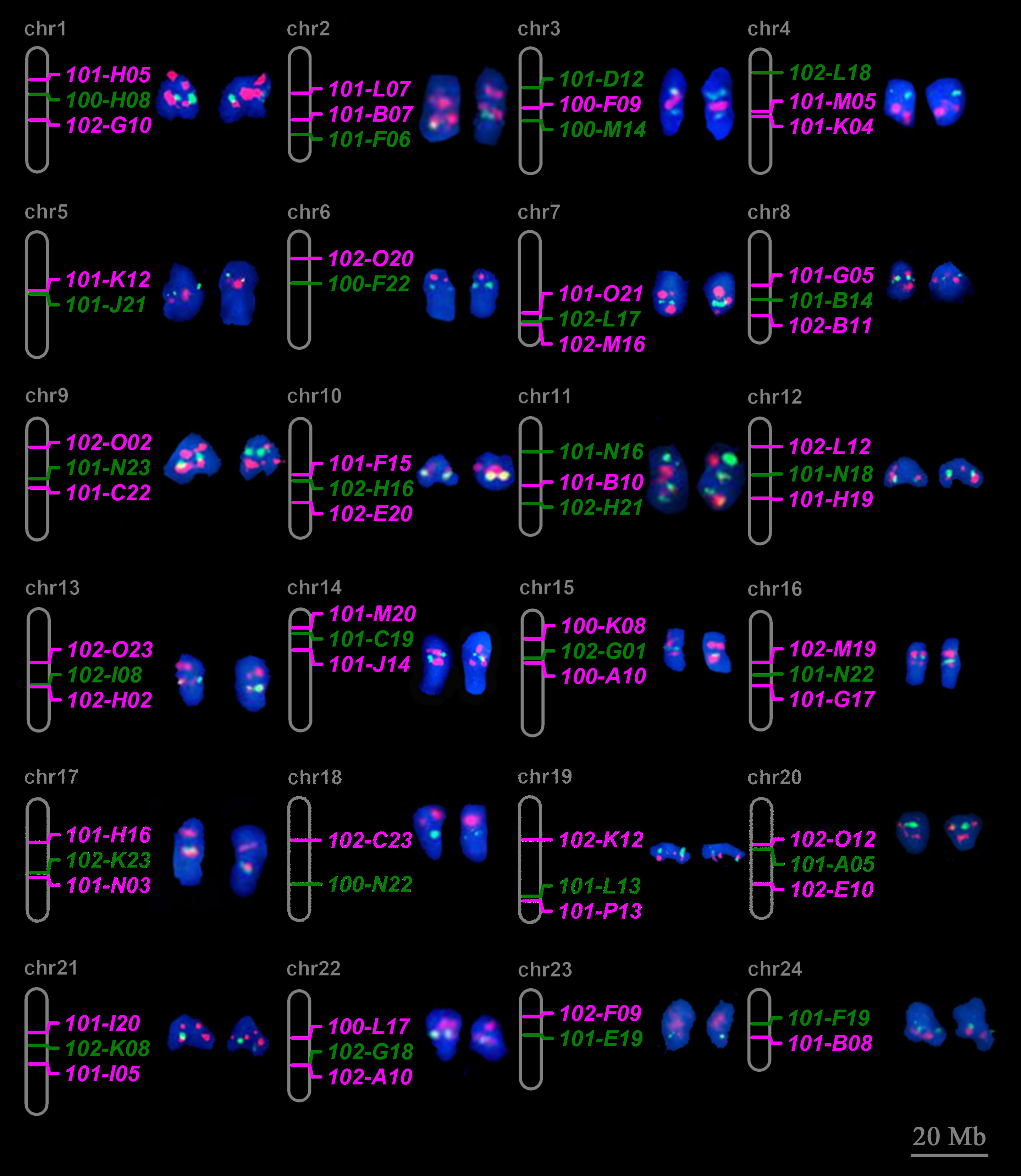
Figure 2 Twenty-four individual chromosomes of large yellow croaker with dual-color FISH signals derived from chromosome-specific BAC clones. The expected chromosomal locations of BAC clones according to alignment of paired BAC end sequence to the reference genome of large yellow croaker are shown on the right side. The corresponding metaphases seen in Supplementary Figure 4. Red and green signals correspond to digoxigenin- and biotin- labeled probes. Scale bar represents 20 Mb for psedochrosome.
FISH results also showed that the distance between FISH signals on chromosome was related to its distribution on pseudochromosome. For example, on Chr.17, 102-K23 and 101-N03 were located in adjacent positions on both pseudochromosome and metaphase chromosome, while 101-H16 was distributed at a more distant position (Figure 2).
3.4 Linking metaphase chromosome to pseudochromosome
On the basis of chromosome specific BACs, we developed a probe cocktail for dual-color FISH to identify the whole set of chromosomes in metaphase cells of large yellow croaker. For each chromosome, 1 to 3 chromosome-specific BACs were selected to produce probes and mixed into two pools according to the scheme shown in Figure 3A. After FISH with the cocktails, red and green signals were generated and formed distinct patterns for each chromosome. Combining FISH signal patterns and chromosome morphology, chromosomes corresponding to pseudochromosomes were all identified (Figure 3B).
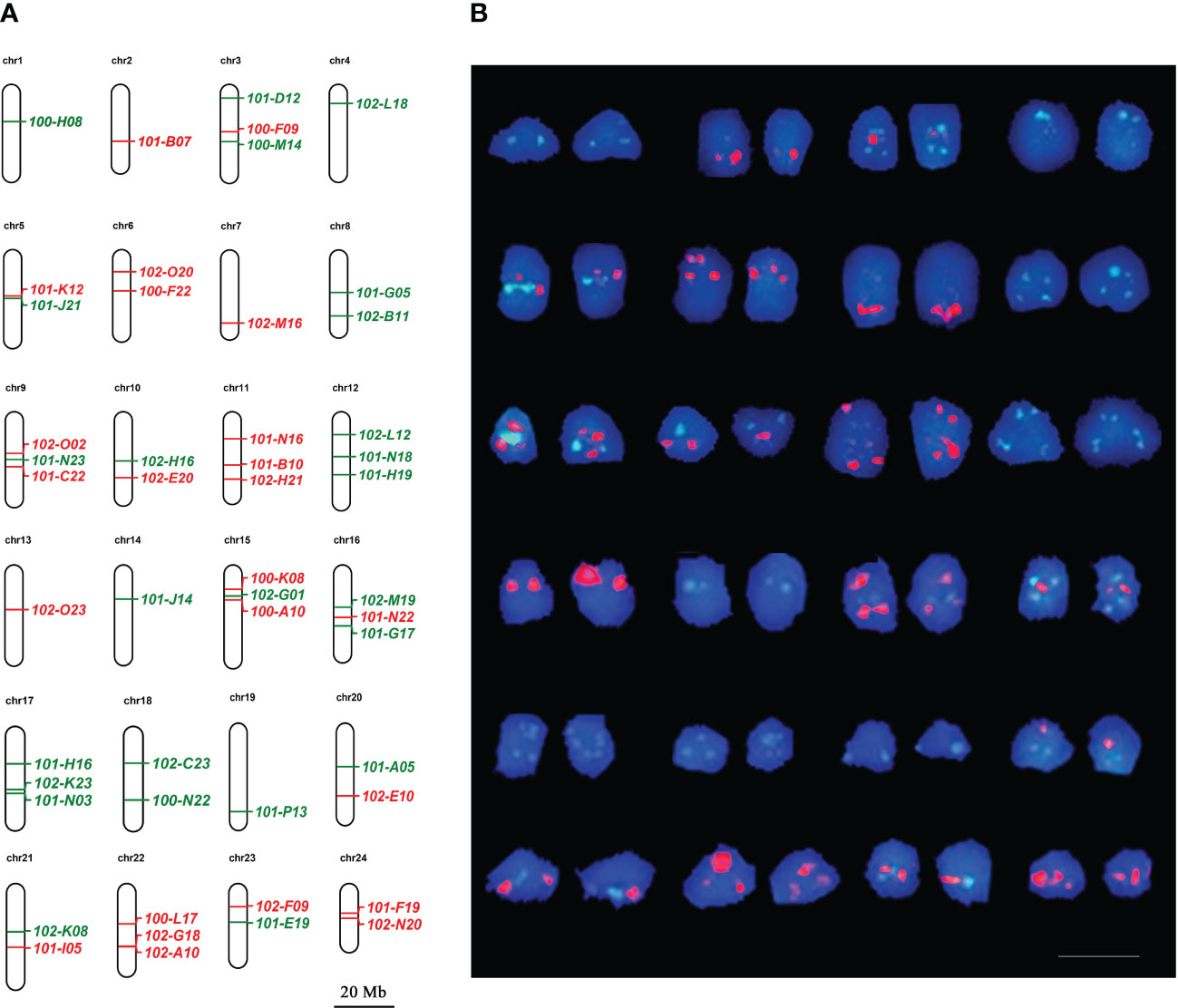
Figure 3 Correspondence between metaphase chromosomes and pseudochromosomes in large yellow croaker. (A) Pseudochromosomes with BAC clones showing the putative locations and signal colors. Bar represents 20 Mb for psedochromosomes. (B) Metaphase chromosomes arranged according to pseudochromsome base on FISH signal pattern. Red and green correspond to digoxigenin- and biotin-labeled probes, respectively. Bar represents 10 μm for metaphase chromosome.
Relative lengths of chromosome were estimated and compared them to the lengths of pseudochromosomes (Supplementary Table 5). The results of correlation analysis showed that they were only weakly correlated (Supplementary Figure 5). Karyotyping was performed by arranging chromosomes in descending order of relative length, linking chromosomes in classic karyotyping and pseudochromosomes in assembled genome (Supplementary Figure 6; Supplementary Table 5).
3.5 Identification of sex chromosomes in spinyhead croaker
By using BLAST, 218 BAC clones were anchored to pseudochromosomes of spinyhead croaker genome (Supplementary Table 6; Supplementary Figure 7). Two BAC clones corresponding to Chr.7 were selected to prepare probes for a dual-color FISH on metaphase spreads of spinyhead croaker, along with probes derived from 18S rDNA that corresponds to Chr.1 (Zhang et al., 2018). In females, Chr.7-specific BAC clones and 18S rDNA were located on two pairs of chromosomes, respectively. While in males, they were located on three chromosomes including a male-specific metacentric chromosome, where Chr. 7-specific BAC clones and 18S rDNA were located, respectively (Figure 4). These results provided cytogenetic evidence that the Y chromosome of spinyhead croaker originated from the fusion of Chr. 1 and Chr. 7.
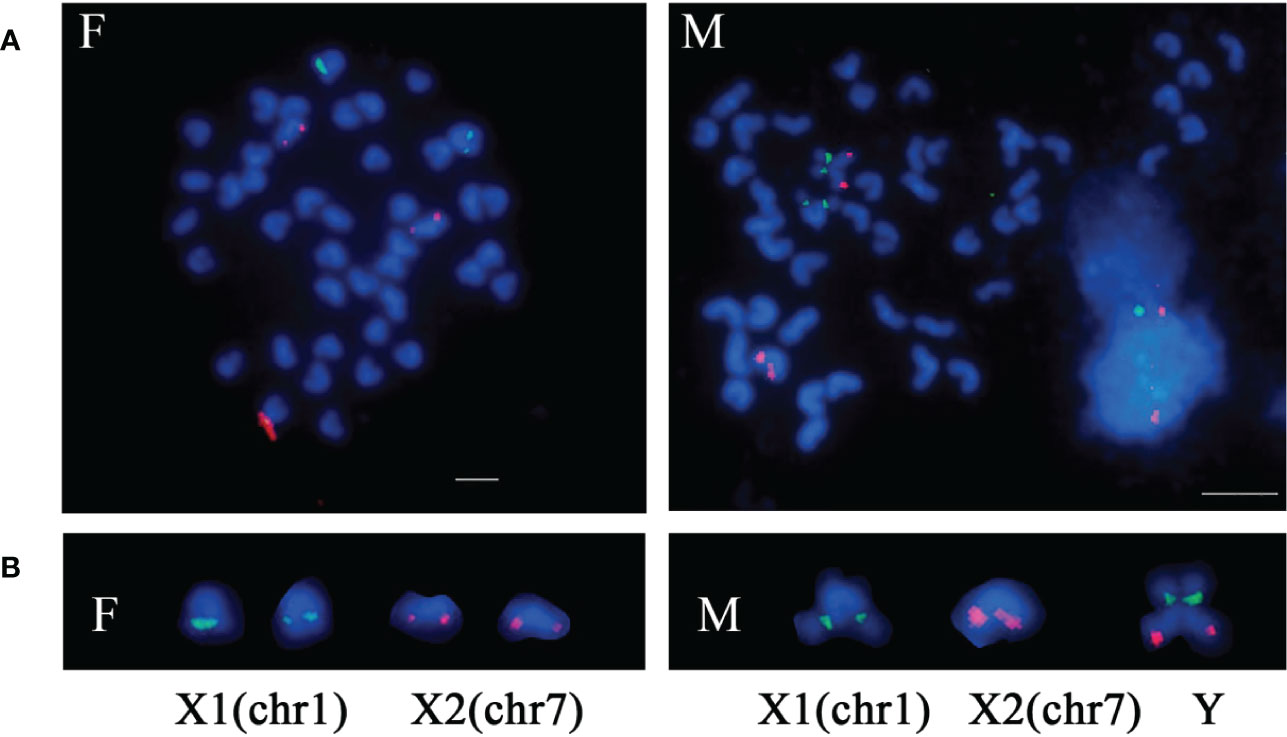
Figure 4 Sex chromosome fusion in spinyhead croaker. (A) Metaphase spreads with FISH signal. (B) The sex chromosomes. The red signals were derived from BAC 101-J21 and BAC 101-K12 corresponding to Chr.7, and the green signals were derived from 18S rDNA corresponding to Chr.1. F indicates female. M indicates male. Bar represents 5 μm.
4 Discussion
Most marine fishes shared a conservative karyotype (2n=48a) (Arai, 2011). However, detailed observations on the chromosomes of marine fish had revealed cryptic chromosome rearrangements (Junior et al., 2006; Ross and Peichel, 2008; da Motta-Neto et al., 2019). In Sciaenidae, karyotypes have been reported in about 40 species, most of which had a conservative formula (2n=48a), but exceptions were also observed (Liao et al., 2018; Zhang et al., 2018). For instances, a multiple sex chromosome system (X1X2Y) was uncovered in C. lucidu (Zhang et al., 2018; Xiao et al., 2020); subtle changes in karyotypes were observed in both L. crocea and L. polyactis, (Xu et al., 2017). In addition, the number and position of 5S rDNA variation among of Sciaenid fishes were revealed by FISH (Liao et al., 2017; Liao et al., 2018). Hence, chromosome evolution in Sciaenidae may be more dynamic beyond the previous knowledge. More cytogenetic researches should be carried out to reveal chromosome rearrangements in Sciaenidae and even in marine fishes, to explore the reasons and biological significance for the macro-karyotype stability coupled with cryptic chromosome rearrangement.
Currently, sequence-based methods have become powerful tools in the studies on chromosome evolution, but cytogenetic methods still played irreplaceable roles (Deakin et al., 2019; Gaffaroglu et al., 2020). In addition, more and more attentions were paid to link DNA sequence and chromosome structure to get comprehensive understanding in dynamic nature and evolution of chromosomes (Iannucci et al., 2021). The BAC clones corresponding to specific chromosomes were the excellent bridges between DNA and chromosomes (Schubert et al., 2001; Wang et al., 2007). In previous studies, chromosome specific BAC clones were developed in other fish species, such as zebrafish, turbot and Atlantic salmon (Phillips et al., 2006a; Phillips et al., 2009; Taboada et al., 2014). In these cases, chromosome-specific BACs were screened out from BAC library by using 3D-PCR according to DNA markers in linkage or physical maps (Taboada et al., 2014). However, the traditional method was very inefficient and tedious (Trifonov et al., 2009).
In present study, bioinformatics methods were used to screen the chromosome specific BACs based on the assembled genome in large yellow croaker. Totally, 435 candidate chromosome specific BAC clones were identified, in which 67 out of 72 selected BAC clone passed PCR and FISH validation, covering all chromosomes of large yellow croaker. In recent years, chromosome-level genomes had been assembled in about 870 fish species (https://www.ncbi.nlm.nih.gov/genome/browse/#!/overview/fish), and BAC libraries were also constructed in many fish species to support the research on genome and genes (Matsuda et al., 2001; Watanabe et al., 2003; Shao et al., 2010; Li et al., 2011). In this context, the bioinformatics method would be a promising way to screen chromosome-specific BACs for chromosome painting.
Teleost fish represent one of the most diverse animal groups in terms of sex determination and differentiation besides outstanding species diversity (Sember et al., 2021; Gong et al., 2022). Fish sex chromosomes include standard constitutions (♀XX/♂XY; ♂ZZ/♀ZW), Y or W chromosome loss-derived systems (♀XX/♂X0, ♂ZZ/♀Z0), and multiple sex chromosome systems (♀X1X1X2X2/♂X1X2Y, ♀XX/♂XY1Y2, ♀X1X1X2X2/♂X1Y1X2Y2, ♂ZZ/♀ZW1W2, ♂Z1Z1Z2Z2/♀Z1W1Z2W2) (Iturra et al., 1997; Kikuchi and Hamaguchi, 2013; Sember et al., 2021). In the Sciaenid fishes, heteromorphic sex chromosomes were found only in spinyhead croaker, which has a multiple sex chromosome system ♀X1X1X2X2/♂X1X2Y (Zhang et al., 2018). One of the two X chromosomes was proved to be Chr.1 corresponding to 18S rDNA by FISH with probes of repetitive sequence (Zhang et al., 2018). That the other X chromosome was Chr.7 was confirmed by BAC-FISH in this study, which was presumed according to sex-dimorphic SNP distribution in our previous study (Xiao et al., 2020). The genome-wide distribution of the sex-dimorphic SNPs also preliminarily revealed the variation of multiple chromosomes among populations of spinyhead croaer (Chen et al., 2022). The studies on population genetics revealed that northern and southern group of spinyhead croaker had a strong differentiation (Cheng et al., 2012; Song et al., 2014). It can be expected that further detailed researches on the sex chromosome of spinyhead croaker will help to understand genetic differentiation among populations, and also provide evidences for understanding the mechanism of sex chromosome fusion.
5 Conclusion
In this study, a set of chromosome-specific BAC-FISH probes covering all chromosomes were developed in large yellow croaker. A dual-color FISH cocktail consisting of 48 chromosome-specific BAC-FISH probes greatly improved chromosome identification in metaphase spreads, allowing to link metaphase chromosomes to pseudochromosomes of the assembled genome. Partial cross-species applicability of the developed chromosome-specific BAC-FISH probes in spinyhead croaker was demonstrated, and cytogenetic evidence that its Y chromosome originates from the fusion of Chr.1 and Chr.7 was provided. Thus, the first set chromosome-specific BAC-FISH probes of Sciaenidae were successfully developed, which would enhance the ability to perform cytological studies for Sciaenid fishes, and then would provide new insight into genome structure and evolution in Sciaenidae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of the Fisheries College of Jimei University Jimei University.
Author contributions
Conceptualization, JiaZ. software, JiaZ. Formal analysis, JiaZ. Investigation, JiaZ and MC. Resources, MC and ZW. Data curation, MC. Writing – original draft preparation, JiaZ and JX. Writing – review & editing, ZW, YW, YJ, JinZ, BL and MC. Supervision, JinZ. Project administration, MC. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China under contract No.41706157 and 31872553, the Seed Industry Innovation and Industrialization Project of Fujian Province under contract 2021FJSCZY01, and the Natural Science Foundation of Fujian Province under contract No.2021J01824.
Acknowledgments
We would like to thank Mengxiang Liao and Songyu Luo for their assistance in BAC library construction of large yellow croaker. We are also grateful to the reviewers for reviewing our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1078110/full#supplementary-material
References
Accioly I. V., Molina W. F.. (2008). Cytogenetic studies in Brazilian marine Sciaenidae and Sparidae fishes (Perciformes). Genet Mol Res. 7 (2), 358–370. doi: 10.4238/vol7-2gmr427
Altschul S. F., Madden T. L., Schaffer A. A., Zhang J. H., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Arai R. (2011). Fish karyotypes: a check list (New York: Springer Science & Business Media). doi: 10.1007/978-4-431-53877-6
Bielski W., Książkiewicz M., Šimoníková D., Hřibová E., Susek K., Naganowska B. (2020). The puzzling fate of a lupin chromosome revealed by reciprocal oligo-FISH and BAC-FISH mapping. Genes 11 (12), 1489. doi: 10.3390/genes11121489
Cai M. Y., Zou Y., Xiao S. J., Li W. B., Han Z. F., Han F., et al. (2019). Chromosome assembly of Collichthys lucidus, a fish of sciaenidae with a multiple sex chromosome system. Sci. Data 6 (1), 132. doi: 10.1038/s41597-019-0139-x
Cheng J., Ma G. Q., Miao Z. Q., Shui B. N., Gao T. X. (2012). Complete mitochondrial genome sequence of the spinyhead croaker Collichthys lucidus (Perciformes, Sciaenidae) with phylogenetic considerations. Mol. Biol. Rep. 39 (4), 4249–4259. doi: 10.1007/s11033-011-1211-6
Chen J. N., Shao X. B., Ye S., Xiao J. Z., Zou Y., Ye K., et al. (2022). Development of sex-specific markers in spinyhead croaker, Collichthys lucidus. Aquaculture 547, 737424. doi: 10.1016/j.aquaculture.2021.737424
Chen S., Su Y., Hong W. (2018). Aquaculture of the large yellow croaker. Aquacult. China 10, 297–308. doi: 10.1002/9781119120759.ch3_10
da Motta-Neto C. C., Cioffi M. D., da Costa G. W. W. F., Amorim K. D. J., Bertollo L. A. C., Artoni R. F., et al. (2019). Overview on karyotype stasis in Atlantic grunts (Eupercaria, Haemulidae) and the evolutionary extensions for other marine fish groups. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00628
Deakin J. E., Potter S., O'Neill R., Ruiz-Herrera A., Cioffi M. B., Eldridge M. D. B., et al. (2019). Chromosomics: Bridging the gap between genomes and chromosomes. Genes 10 (8), 627. doi: 10.3390/genes10080627
Dong G. R., Shen J., Zhang Q., Wang J. P., Yu Q. Y., Ming R., et al. (2018). Development and applications of chromosome-specific cytogenetic BAC-FISH probes in S. spontaneum. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00218
do Vale Martins L., de Oliveira Bustamante F., da Silva Oliveira A. R., da Costa A. F., de Lima Feitoza L., Liang Q., et al. (2021). BAC-and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 130 (2), 133–147. doi: 10.1007/s00412-021-00758-9
Gaffaroglu M., Majtanova Z., Symonova R., Pelikanova S., Unal S., Lajbner Z., et al. (2020). Present and future salmonid cytogenetics. Genes 11 (12), 1462. doi: 10.3390/genes11121462
Gold J. R., Li Y. C., Shipley N. S., Powers P. K. (1990). Improved methods for working with fish chromosomes with a review of metaphase chromosome-banding. J. Fish. Biol. 37 (4), 563–575. doi: 10.1111/j.1095-8649.1990.tb05889.x
Gong G., Xiong Y., Xiao S., Li X.-Y., Huang P., Liao Q., et al. (2022). Origin and chromatin remodelling of young X/Y sex chromosomes in catfish with sexual plasticity. Natl. Sci. Rev., nwac239. doi: 10.1093/nsr/nwac239
Han Y. H., Zhang T., Thammapichai P., Weng Y. Q., Jiang J. M. (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200, 771–779. doi: 10.1534/genetics.115.177642
Iannucci A., Makunin A. I., Lisachov A. P., Ciofi C., Stanyon R., Svartman M., et al. (2021). Bridging the gap between vertebrate cytogenetics and genomics with single-chromosome sequencing (chromSeq). Genes 12 (1), 124. doi: 10.3390/genes12010124
Iturra P., Medrano J. F., Bagley M., Lam N., Vergara N., Marin J. C. (1997). Identification of sex chromosome molecular markers using RAPDs and fluorescent in situ hybridization in rainbow trout. Genetica 101 (3), 209–213. doi: 10.1023/A:1018371623919
Junior P., Molina W. F., Affonso P., Aguilar C. T. (2006). Assessing genetic diversity of Brazilian reef fishes by chromosomal and DNA markers. Genetica 126, 161–177. doi: 10.1007/s10709-005-1446-z
Kikuchi K., Hamaguchi S. (2013). Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dynamics. 242 (4), 339–353. doi: 10.1002/dvdy.23927
Kong S. N., Ke Q. Z., Chen L., Zhou Z. X., Pu F., Zhao J., et al. (2019). Constructing a high-density genetic linkage map for large yellow croaker (Larimichthys crocea) and mapping resistance trait against ciliate parasite cryptocaryon irritans. Mar. Biotechnol. 21, 262–275. doi: 10.1007/s10126-019-09878-x
Liao M. X., Zheng J., Wang Z. Y., Wang Y. L., Zhang J., Cai M. Y. (2018). Molecular cytogenetic of the amoy croaker, Argyrosomus amoyensis (Teleostei, Sciaenidae). J. Oceanol. Limnol. 36, 842–849. doi: 10.1007/s00343-018-6272-0
Liao M. X., Zheng J., Wang Z. Y., Zhang J., Liu X. D., Cai M. Y. (2017). Comparison of chromosome mapping of rDNA between Argyrosomus amoyensis and Larimichthys crocea. J. Fish. China 9, 13–19. doi: 10.11964/jfc.20160910560
Li Y., Xu P., Zhao Z., Wang J., Zhang Y., Sun X. W. (2011). Construction and characterization of the BAC library for common carp cyprinus carpio l. and establishment of microsynteny with zebrafish Danio rerio. Mar. Biotechnol. 13 (6), 706–712. doi: 10.1007/s10126-011-9404-5
Luo M. Z., Kim H. R., Kudrna D., Sisneros N. B., Lee S. J., Mueller C., et al. (2006). Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics 7 (1), 106. doi: 10.1186/1471-2164-7-106
Luo M. Z., Wing R. A. (2003). An improved method for plant BAC library construction. Methods Mol. Biol. 236, 3–20. doi: 10.1385/1-59259-413-1:3
Matsuda M., Kawato N., Asakawa S., Shimizu N., Nagahama Y., Hamaguchi S., et al. (2001). Construction of a BAC library derived from the inbred hd-rR strain of the teleost fish, oryzias latipes. Genes Genet. Syst. 76 (1), 61–63. doi: 10.1266/ggs.76.61
Mu Y. N., Huo J. Y., Guan Y. Y., Fan D. D., Xiao X. Q., Wei J. G., et al. (2018). An improved genome assembly for Larimichthys crocea reveals hepcidin gene expansion with diversified regulation and function. Commun. Biol. 91, 459. doi: 10.1016/j.fsi.2019.04.256
Nelson J. S., Grande T. C., Wilson M. V. (2016). Fishes of the World (John Wiley & Sons). doi: 10.1002/9781119174844
Ning Y., Liu X. D., Wang Z. Y., Guo W., Li Y. Y., Xie F. J. (2007). A genetic map of large yellow croaker Pseudosciaena crocea. Aquaculture 264, 16–26. doi: 10.1016/j.aquaculture.2006.12.042
Phillips R. B., Amores A., Morasch M. R., Wilson C., Postlethwait J. H. (2006a). Assignment of zebrafish genetic linkage groups to chromosomes. Cytogenet. Genome Res. 114, 155–162. doi: 10.1159/000093332
Phillips R. B., Keatley K. A., Morasch M. R., Ventura A. B., Lubieniecki K. P., Koop B. F., et al. (2009). Assignment of Atlantic salmon (Salmo salar) linkage groups to specific chromosomes: Conservation of large syntenic blocks corresponding to whole chromosome arms in rainbow trout (Oncorhynchus mykiss). BMC Genet. 10 (1), 46. doi: 10.1186/1471-2156-10-46
Phillips R. B., Nichols K. M., DeKoning J. J., Morasch M. R., Keadey K. A., Rexroad C., et al. (2006b). Assignment of rainbow trout linkage groups to specific chromosomes. Genetics 174, 1661–1670. doi: 10.1534/genetics.105.055269
Phillips R. B., Park L. K., Naish K. A. (2013). Assignment of chinook salmon (Oncorhynchus tshawytscha) linkage groups to specific chromosomes reveals a karyotype with multiple rearrangements of the chromosome arms of rainbow trout (Oncorhynchus mykiss). G3-Genes. Genom. Genet. 3, 2289–2295. doi: 10.1534/g3.113.008078
Phillips R., Rab P. (2001). Chromosome evolution in the salmonidae (Pisces): An update. Biol. Rev. 76, 1–25. doi: 10.1111/j.1469-185X.2000.tb00057.x
Phillips R. B., Zimmerman A., Noakes M. A., Palti Y., Morasch M. R. W., Eiben L., et al. (2003). Physical and genetic mapping of the rainbow trout major histocompatibility regions: Evidence for duplication of the class I region. Immunogenetics 55, 561–569. doi: 10.1007/s00251-003-0615-4
Pinkel D., Straume T., Gray J. W. (1986). Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Natl. Acad. Sci. 83 (9), 2934–2938. doi: 10.1073/pnas.83.9.2934
Ross J. A., Peichel C. L. (2008). Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 179 (4), 2173–2182. doi: 10.1534/genetics.108.088559
Ross J. A., Urton J. R., Boland J., Shapiro M. D., Peichel C. L. (2009). Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PloS Genet. 5 (2), e1000391. doi: 10.1371/journal.pgen.1000391
Schubert I., Fransz P. F., Fuchs J., Jong J. (2001). Chromosome painting in plants. Methods Cell Sci. 23 (1), 57–69. doi: 10.1023/A:1013137415093
Sember A., Nguyen P., Perez M. F., Altmanová M., Ráb P., Cioffi M. (2021). Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: state of the art and future challenges. Philos. Trans. R. Soc. B. 376 (1833), 20200098. doi: 10.1098/rstb.2020.0098
Shao C. W., Chen S. L., Scheuring C. F., Xu J. Y., Sha Z. X., Dong X. L., et al. (2010). Construction of two BAC libraries from half-smooth tongue sole cynoglossus semilaevis and identification of clones containing candidate sex-determination genes. Mar. Biotechnol. 12, 558–568. doi: 10.1007/s10126-009-9242-x
Shi X., Zeng H., Xue Y., Luo M. (2011). A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant Methods 7 (1), 33. doi: 10.1186/1746-4811-7-33
Song N., Ma G. Q., Zhang X. M., Gao T. X., Sun D. R. (2014). Genetic structure and historical demography of Collichthys lucidus inferred from mtDNA sequence analysis. Environ. Biol. Fish. 97 (1), 69–77. doi: 10.1007/s10641-013-0124-8
Stein J., Phillips R. B., Devlin R. H. (2001). Identification of the y chromosome in chinook salmon (Oncorhynchus tshawytscha). Cytogen. Genome Res. 92 (1-2), 108–110. doi: 10.1159/000056878
Taboada X., Pansonato-Alves J. C., Foresti F., Martinez P., Vinas A., Pardo B. G., et al. (2014). Consolidation of the genetic and cytogenetic maps of turbot (Scophthalmus maximus) using FISH with BAC clones. Chromosoma 123 (3), 281–291. doi: 10.1007/s00412-014-0452-2
Telenius H., Pelmear A. H., Tunnacliffe A., Carter N. P., Behmel A., Fergusonsmith M. A., et al. (1992). Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Gene Chromosome Canc. 4 (3), 257–263. doi: 10.1002/gcc.2870040311
Trifonov V. A., Vorobieva N. N., Rens W. (2009). “FISH with and without COT1 DNA,” in Fluorescence in situ hybridization (FISH) — application guide, vol. 2009. (Berlin, Heidelberg: Springer Press), 99–109. doi: 10.1007/978-3-540-70581-9
Vandewoestyne M., Van Hoofstat D., Van Nieuwerburgh F., Deforce D. (2009). Suspension fluorescence in situ hybridization (S-FISH) combined with automatic detection and laser microdissection for STR profiling of male cells in male/female mixtures. Int. J. Legal. Med. 123 (5), 441–447. doi: 10.1007/s00414-009-0341-z
Wang K., Guo W. Z., Zhang T. Z. (2007). Development of one set of chromosome-specific microsatellite-containing BACs and their physical mapping in Gossypium hirsutum l. Theor. Appl. Genet. 115, 675–682. doi: 10.1007/s00122-007-0598-x
Watanabe M., Kobayashi N., Fujiyama A., Okada N. (2003). Construction of a BAC library for Haplochromis chilotes, a cichlid fish from lake Victoria. Genes Genet. Syst. 78 (1), 103–105. doi: 10.1266/ggs.78.103
Wu C. W., Zhang D., Kan M. Y., Lv Z. M., Zhu A. Y., Su Y. Q., et al. (2014). The draft genome of the large yellow croaker reveals well-developed innate immunity. Nat. Commun. 5, 5227. doi: 10.1038/ncomms6227
Xiao J. Z., Zou Y., Xiao S. J., Chen J. N., Wang Z. Y., Wang Y. L., et al. (2020). Development of a PCR-based genetic sex identification method in spinyhead croaker (Collichthys lucidus). Aquaculture 522, 735130. doi: 10.1016/j.aquaculture.2020.735130
Xu D. D., Molina W. F., Yano C. F., Zhang Y. R., de Oliveira E. A., Lou B., et al. (2017). Comparative cytogenetics in three Sciaenid species (Teleostei, perciformes): Evidence of interspecific chromosomal diversification. Mol. Cytogen. 10 (1), 37. doi: 10.1186/s13039-017-0338-0
Yang Z. J., Li X., Liao H., Hu L. P., Peng C., Wang S. H., et al. (2019). A molecular cytogenetic map of scallop (Patinopecten yessoensis). Mar. Biotechnol. 21 (6), 731–742. doi: 10.1007/s10126-019-09918-6
Zhang S. K., Zheng J., Zhang J., Wang Z. Y., Wang Y. L., Cai M. Y. (2018). Cytogenetic characterization and description of an X1X1X2X2/X1X2Y sex chromosome system in Collichthys lucidus (Richardson 1844). Acta Oceanolog. Sin. 37 (4), 34–39. doi: 10.1007/s13131-018-1152-1
Keywords: Larimichthys crocea, chromosome identification, fluorescence in situ hybridization (FISH), bacterial artificial chromosome (BAC), sex chromosome
Citation: Zhang J, Xie J, Liu B, Wang Z, Wang Y, Jiang Y, Zhang J and Cai M (2022) Development and applications of chromosome-specific cytogenetic BAC-FISH probes in Larimichthys crocea. Front. Mar. Sci. 9:1078110. doi: 10.3389/fmars.2022.1078110
Received: 24 October 2022; Accepted: 02 December 2022;
Published: 22 December 2022.
Edited by:
Gongliang Yu, Institute of Hydrobiology (CAS), ChinaReviewed by:
Dong-Neng Jiang, Guangdong Ocean University, ChinaJie Mei, Huazhong Agricultural University, China
Copyright © 2022 Zhang, Xie, Liu, Wang, Wang, Jiang, Zhang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingyi Cai, bXlpY2FpQGptdS5lZHUuY24=; Jing Zhang, emhqaW5nQGptdS5lZHUuY24=
 Jianpeng Zhang
Jianpeng Zhang Jingyi Xie1
Jingyi Xie1 Zhiyong Wang
Zhiyong Wang Yilei Wang
Yilei Wang Mingyi Cai
Mingyi Cai