- 1State Environmental Protection Key Laboratory of Coastal Ecosystem, National Marine Environmental Monitoring Center, Dalian, China
- 2College of Marine Technology and Environment, Dalian Ocean University, Dalian, China
- 3College of Marine Ecology and Environment, Shanghai Ocean University, Shanghai, China
Vibrio as one of the main pathogens of shellfish diseases can cause serious human seafoodborne gastroenteritis and even death. In this study, we analyzed the bacterial communities from the clam, and compared the resistance phenotypes and genotypes of Vibrio spp. from Meretrix meretrix at different growth stages. High-throughput sequencing analysis revealed the predominance of Proteobacteria (50%) in the bacterial community and Vibrio was one of the dominant genera in the clam hepatopancreas in the summer. Vibrio abundance in Meretrix meretrix positively correlated with the water temperature (p<0.05). A total of 73 Vibrio isolates from Meretrix meretrix were classified into 19 species and the dominant strains included V. mediterranei (19%) and V. harveyi (11%), V. algolyticus (10%), and V. parahaemolyticus (8%). The species and abundance of Vibrio spp. were the highest in the 3-year-old of Meretrix meretrix compared with clams of other ages in the summer. Among the 73 isolates, 68 Vibrio strains were resistant to other 15 antibiotics except for sulfamethoxazole-trimethoprim with 57 resistant phenotypes. The most prevalent resistance was toward clindamycin (76%), followed by amikacin (63%), ampicillin (62%), rifampicin (62%), vancomycin (57%), and amoxicillin (50%). The ARI values of Vibrio spp. in different ages ranged from 0.13 to 0.18, and ARI values of 3-year-old (ARI=0.18) clams are higher than that of other ages clam. Approximately 72% of the resistant isolates showed multidrug-resistant phenotypes with maximum resistance to 15 antibiotics. Tolerance to heavy metals including Cd, Zn, and Cu was detected in the majority of antibiotic resistant isolates. In addition to the co-resistance to the same class of antibiotics, resistance to cephalosporin (CFP, CEP, CZ) were significantly correlated with penicillins (AMP, AMC) (p< 0.01), tetracycline (p < 0.001), sulfanilamide (SXT) (p< 0.01) and quinolone (CIP) (p< 0.01). The heavy metal resistance genes copB and nccA were significantly correlated with the clindamycin resistance phenotype (p<0.01). This study revealed that the habitat of Meretrix meretrix is in low exposure to antibiotics, and a link between heavy metal resistance genes and antibiotic resistance.
1. Introduction
Shellfish mainly inhabit coastal and estuarine environments. Due to the nature of their habitats, shellfish contain a variety of bacterial microbiota, including the Vibrio spp. (Romalde et al., 2014). Vibrio is a Gram-negative bacterium with genetic and metabolic diversity and is an integral part of the global marine ecosystem (Thompson et al., 2004). Vibrio seriously affects shellfish farming. Moreover, it poses a potential danger to humans due to its high detection rate in shellfish farming. Twenty different pathogenic Vibrio species can cause large-scale death of shellfish. Vibrio infection-related diseases usually include gastrointestinal disorders with symptoms like diarrhea, abdominal cramps, nausea, vomiting, and fever (CDC, 2019b). These symptoms may occur within 24 h of ingestion and last for three days. Patients with low immunity or underlying diseases are at a higher risk of death (CDC, 2019a).
Presently, more than 120 species of Vibrio have been found; at least 12 species are known to cause human diseases. The list includes Vibrio alginolyticus, Vibrio cincinnatiensis, Vibrio damsela, Vibrio fluvialis, Vibrio furnisii, Vibrio metschnikovii, Vibrio mimicus, Vibrio cholerae, Vibrio parahaemolyticus, etc. (Oliver, 2015; Economopoulou et al., 2017; Huang et al., 2017; Zago et al., 2017). Antibiotics are often used to prevent and cure aquaculture diseases in recent years. However, the aquaculture industry lacks relevant drug regulations leading to the overuse of antibiotics. Consequently, the Vibrio develop antibiotic resistance, increasing the difficulty of treating human infections (Alsalem et al., 2018; Miranda et al., 2018). Vibrio spp. also showed resistant to the most clinically used antibiotics (Mala et al., 2014; Letchumanan et al., 2015). The incidence of human Vibrio infection and the drug resistance rate of drug-resistant bacteria are also increasing (Kitaoka et al., 2011). Therefore, it is imperative to detect pathogenic Vibrio in aquatic products and prevent food poisoning. The sudden outbreak of Vibrio will seriously affect marine biomass and cause severe economic loss to aquaculture (Moffitt and Cajas-Cano, 2014).
China remains the first major producer of fisheries and aquaculture in the world, with a 35 percent share of the total (FAO (Food and Agriculture Organization, 2022). In 2020, China’s mariculture area covered 1996 thousand hectares. The shellfish occupied 1197 thousand hectares, accounting for 59.99% of the mariculture area. Meretrix meretrix is a beach-buried shellfish widely distributed in both the north and south coastal regions of China, especially in the estuary and tidal flat, such as Liaoning, Shandong, Jiangsu, and Guangxi. Meretrix meretrix grows in a wide range of temperatures and salinity and mainly feeds on planktonic and benthic diatoms. In 2020, the output of clam mariculture from the Liaoning Province was 1.353 million tons, and the area encompassed was 1,53,000 hectares, ranking first in China and far exceeding that of the other provinces (Ministry of Agriculture and Rural Affairs Fisheries Bureau et al., 2021). Meretrix meretrix is currently an important economic shellfish in the coastal mudflat culture in China. However, due to intensified aquaculture and the increasing eutrophication of coastal mudflats, Vibrio spp. as one of the primary pathogen have caused serious harm to the clam aquaculture industry and food safety (Li et al., 2018).
Geligang, as one of the important producing areas of clams Meretrix meretrix in northern China, is located in the east of Liaohe estuary, with an area of about 10000 ha. The annual output of Meretrix meretrix in Geligang area is more than 1000 t. In this study, we investigate the differences in the diversity and abundance of Vibrio isolated from Meretrix meretrix, and analyze antibiotic resistance and heavy metal resistance of Vibrio spp. in Meretrix meretrix at different ages in Geligang, Liaohe estuary in China. This study will support the need for food safety risk assessment of aquatic products.
2. Materials and methods
2.1. Sample collection
Meretrix meretrix samples of different ages were collected from the Geligang aquaculture area in Liaohe estuary in the spring (April), summer (July, August), and autumn (November) of 2019. The 1-year-old and 2-year-old clams were collected in spring. The 1-year-old, 2-year-old, 3-year-old, and 5-year-old of clams were collected in the summer. The 1-year-old, 2-year-old, 3-year-old, and 5-year-old clams were collected in autumn. The collected samples were stored in sterile plastic bags at 4°C and transported to the laboratory for analysis within 24 h. Water temperature and salinity were measured in situ by the YSI ProQuatro Handheld Multiparameter Instrument (YSI, Xylem Inc., NY, USA).
2.2. Isolation and identification of the Vibrio strains
The samples of Meretrix meretrix were divided according to different growth cycles. The hepatopancreas samples of the same age were mixed by three independent samples, and 2–5 groups of parallel samples were set aside for preservation. The clam samples were washed with sterile saline. The clam hepatopancreas (200 g) samples were homogenized in phosphate-buffered saline (PBS; 2.5 mM KH2PO4; pH-7.2) with a blender and subsequently serially diluted with PBS. The homogenate diluents were further plated on Thiosulfate-Citrate-Bile Salts-Sucrose Agar (TCBS Agar; Oxoid, Thermo Fischer Scientific, UK) and incubated at 37°C for 24 h. The presumptive colonies on the TCBS agar plates were re-streaked on Tryptone Soy Agar (Oxoid, Thermo Fischer Scientific, UK) supplemented with 3% Sodium Chloride (TSA + 3% NaCl) and incubated at 37°C for 24 h to achieve a pure isolate (Sujeewa et al., 2009). Vibrio isolates were identified by PCR and sequencing of16S rDNA. Chromosomal DNA from the Vibrio cells were extracted using a QIAGEN DNA extraction kit following the manufacturer’s instructions. 16S rRNA gene amplification and DNA purification were determined as described previously (Park et al., 2018) and sequenced by Sangon Biotech (Shanghai) Co., Ltd. (China). The obtained consensus sequences were subjected to BLAST search at NCBI (http://www.ncbi.nlm.nih.gov/pubmed) for sequence alignment analysis.
2.3. High-throughput sequencing
The genomic DNA was extracted from the clam hepatopancreas using the DNeasy Power Water Total DNA Isolation Kit (QIAGEN, Germany). Bacterial communities were identified using the 16S rRNA gene sequencing technology from Shanghai Personalbio Technology Co., Ltd. (China). The V3-V4 regions of the 16S rRNA gene were amplified using barcodes. The 175 primer sets with the forward primer 341F (5’-CCTACGGGNGGCWGCAG-3’) and the reverse primer 765R with 176 primer sets (5’-GACTACNVGGGTATCTAAT-3’) were used for sequencing. The PCR amplification was performed using the Pfu high-fidelity DNA polymerase (TransGen Biotech), and purification and recovery were managed with magnetic beads. The fluorescence of the PCR amplification product was quantified on the Fluorescence Microplate reader (BioTek, FLx800). The fluorescent reagent was used from the Quant-iT PicoGreen dsDNA Assay Kit. The sequencing library was prepared using the TruSeq Nano DNA LT Library Prep Kit (Illumina, Inc. (USA)). The community DNA fragments were sequenced using the Paired-end Illumina MiSeq platform.
2.4. Antimicrobial susceptibility and heavy metal resistance tests of the Vibrio isolates
The antimicrobial susceptibility of the Vibrio isolates was determined from the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines (Clinical Laboratory Standard Institute, 2016). The isolates were tested for susceptibility toward 16 antimicrobials: Cefazolin (CZ, 30 µg), Cephalotin (CEP, 30 µg), Cefoperazone (CFP, 75 µg), Cefuroxime (CXM, 30 µ g), Ampicillin (AMP, 10 µg), Amoxicillin (AMC, 10 µg), Streptomycin (STR, 10 µg), Gentamicin (GM, 10 µg), Amikacin (AN, 30 µ g), Sulfamethoxazole-Trimethoprim (SXT, 23.75–1.25 µg), Ciprofloxacin (CIP, 5 µg), Clindamycin (DA, 2 µg), Vancomycin (VA, 30 µg), Tetracycline (TCY, 30 µg), Erythromycin (ERY, 15 µg), and Rifampicin (RFP, 5 µg). The results were interpreted as susceptible (S), intermediate (I), and resistant (R) after 12 h of incubation at 30°C using the CLSI standards. Multiple Antibiotic Resistant (MAR) strain is defined as a bacterium resistant to three or more antibiotics (Manjusha et al., 2005). Antibacterial Resistance Index (ARI) was used to analyze the prevalence of resistant isolates from clam and calculated for the same age (Mohanta and Goel, 2014).
The heavy metal resistance of the Vibrio isolates was determined according to a previous method (Kang et al., 2016). The minimal inhibitory concentration (MIC) of the tested heavy metals against the isolates was measured using broth dilution testing (Clinical Laboratory Standard Institute, 2016). The heavy metals used in this study were CdCl2, ZnCl2, and CuCl2.
2.5. Detection of antibiotic resistance genes and heavy metal resistance genes
From the results of the antibiotic and heavy metal resistance phenotypes of Vibrio, 13 resistant genes were selected. The six antibiotic resistance genes included the penicillins resistance gene(ampR), aminoglycosides resistance gene (aadA, strA, and strB) and glycopeptides resistance gene(vanM). The heavy metal resistance genes included copA, copB, and copC genes for Cu2+, nccA and cadD genes for Cd2+, and zntA and zntB resistance genes for Zn2+. The primers and PCR conditions are presented in Table S1 (Sambrook, 2001; Kamika and Momba, 2013; Jiang et al., 2014; Liu et al., 2014; Liu, 2016; Wu et al., 2019; Yang et al., 2020).
2.6. Statistical analyses
The statistical analyses of alpha diversity, beta diversity and differentially abundant taxa were carried out using QIIME2 2019.4. Pearson’s correlation analysis was used to evaluate the relationship between the antibiotic resistant phenotypes and antibiotic resistant genotypes with SPSS version 25 (IBM, Armonk, NY, USA). p value < 0.05 was considered statistically significant.
3. Results
3.1. Microbial community analysis
3.1.1. Microbial alpha diversity
The alpha diversity was determined for each treatment and different seasons. The Good’s Coverage index of 24 clam samples of different ages was >0.99, which means more than 99% of the species diversity was detected with a high coverage (Table S2). The Chao1, Shannon, and Simpson indices showed a trend of first increasing and then decreasing. The highest value was reached in the 3-year-old clams collected in August, and those collected in autumn contributed the lowest. The Chao1 index between the samples in spring and autumn (p <0.05) differed significantly; the Shannon and Simpson indices did not show a seasonal difference. The changing trend of the Good’s Coverage index was autumn>spring>summer, indicating the difference in the sequencing coverage of samples in the four months. The samples in spring and summer were significantly different from those in autumn (p<0.05). Therefore, significant differences existed in the community richness but not in community diversity among the clam samples collected during different seasons (see Table 1; Figure 1).
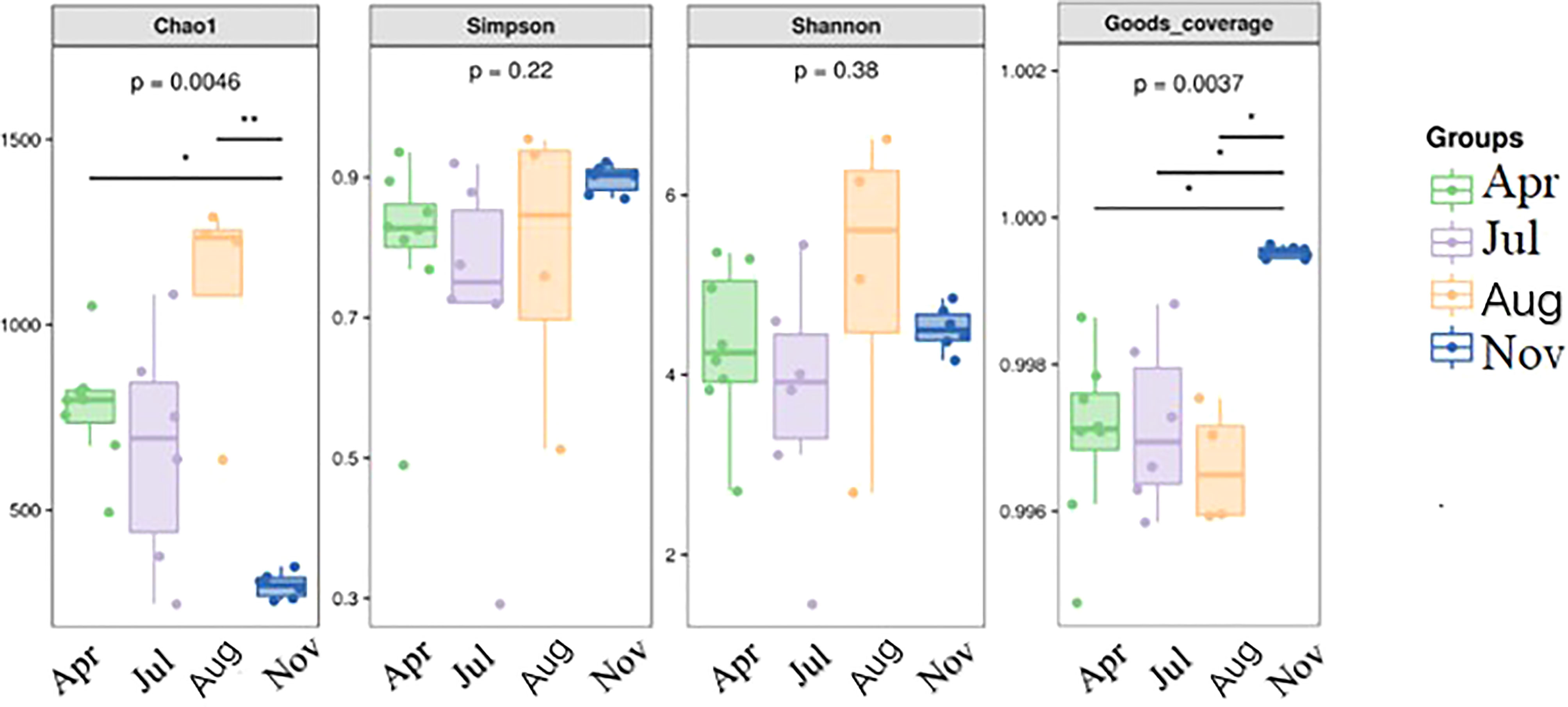
Figure 1 Differences in the Alpha Diversity Index of Meretrix meretrix Samples during different seasons.
3.1.2. Beta diversity
Beta diversity was used to analyze the similarities and differences in the structure of two or more communities. Only the 1-year-old and 2-year-old of Meretrix meretrix were detected in three seasons. Hence, the beta diversity analysis was conducted only for the community structures of the 1-year-old and 2-year-old of Meretrix meretrix. Principal Coordinate Analysis (PCoA) revealed that the dissimilarities in community structure existed in different ages of Meretrix meretrix. Seasonal differences were prevalent in the bacterial community of the clam at the same age (see Figure 2).
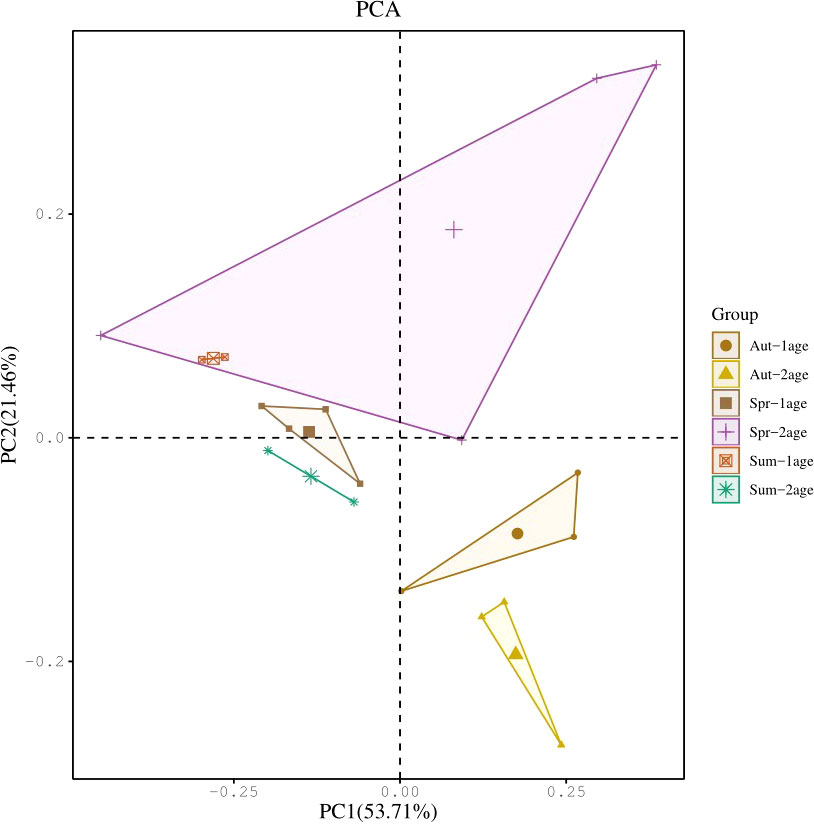
Figure 2 Comparison of the microbial community structures of Meretrix meretrix samples of the same age. Principal Coordinate Analysis (PCoA) of the 16S rRNA gene sequences using Bray-Curtis difference grouped by treatment.
3.1.3. Changes in the microbial community structure at different ages
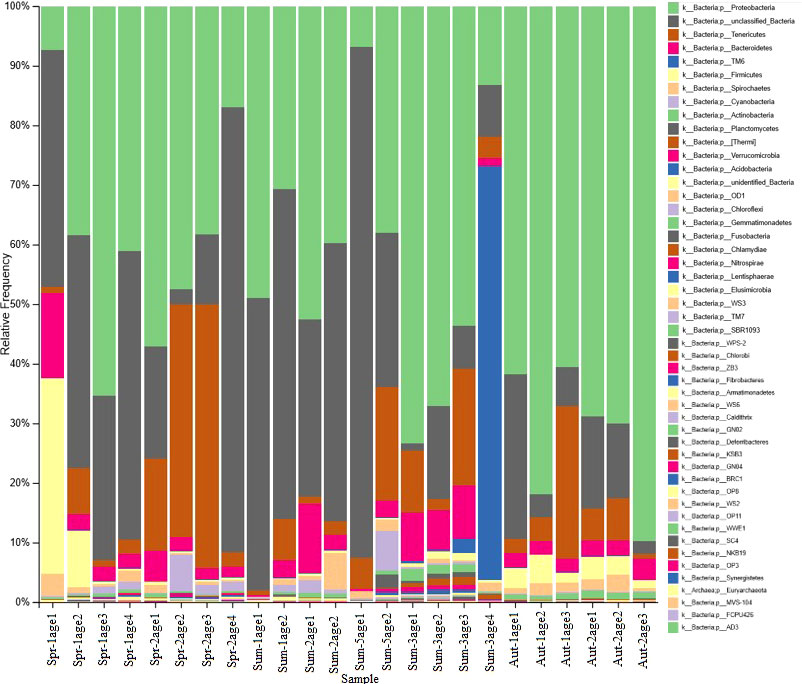
Based on the high-throughput sequencing results, the dominant flora in the Meretrix meretrix included Proteobacteria (50%), Firmicutes (11%), Bacteroides (4%), Spirochaetes (1%), and Cyanobacteria (1%). Proteobacteria mainly included α- Proteobacteria (19%), γ- proteobacteria (19%), β- Proteobacteria (9%), and δ- Proteobacteria (0.5%).
The clam samples collected in the spring mainly comprised Halomonas, Devosia, Cuprum, Lactobacillus, Paleobacterium (1%), Rhizobium (1%), and Sphingomonas (1%). The dominant bacteria in the clam samples collected in the summer (July) included Vibrio, Halomonas, Cuprum, Devosia, Palebacterium, Acinetobacter, Sphingomonas, and Seminibacterium. The clam samples collected in mid-summer (August) mainly had Vibrio and Palebacterium. The dominant bacteria in the clam samples collected in autumn included Ralstonia solanacearum, Devosia, Halomonas, Palebacterium, Pelomonas, Actinobacteria, Enterobacter, Staphylococcus, sediment Bacilli, Rhizobium, and Pseudomonas (see Figure 3).
Vibrio accounted for 0.003%, 8%, 11%, and 0.1% of the bacterial community composition in the spring (April), summer (July), summer (August), and autumn (November), respectively. Thus, the Vibrio abundance in the clam samples increased first and then decreased with change in the season; it was the highest in summer. The relative abundance of Vibrio was the highest in the 3-year-old Meretrix meretrix in summer. Ralstonia caused the significant differences in the species abundance among the Meretrix meretrix during different seasons.
3.2. The abundance and species of Vibrio
The concentration of the culturable bacteria in the clam hepatopancreas ranged from 2.83 × 103 - 1.18 × 105 CFU/g in the spring, summer, and autumn. The abundance of the culturable bacteria increased first and then decreased with the season. The bacterial abundance was the highest in summer, 1–2 orders of magnitude higher than in spring. In the same season, the abundance of the culturable bacteria in the hepatopancreas of the clams of different ages was different. The quantity of the culturable bacteria in the 1-year-old and 2-year-old of Meretrix meretrix collected during the spring, summer, and autumn differed by 2 and 1 orders of magnitude, respectively. Thus, the abundance of the culturable bacteria in the younger Meretrix meretrix was more susceptible to seasonal changes. There was a significant positive correlation between the abundance of the culturable bacteria and temperature (p<0.05).
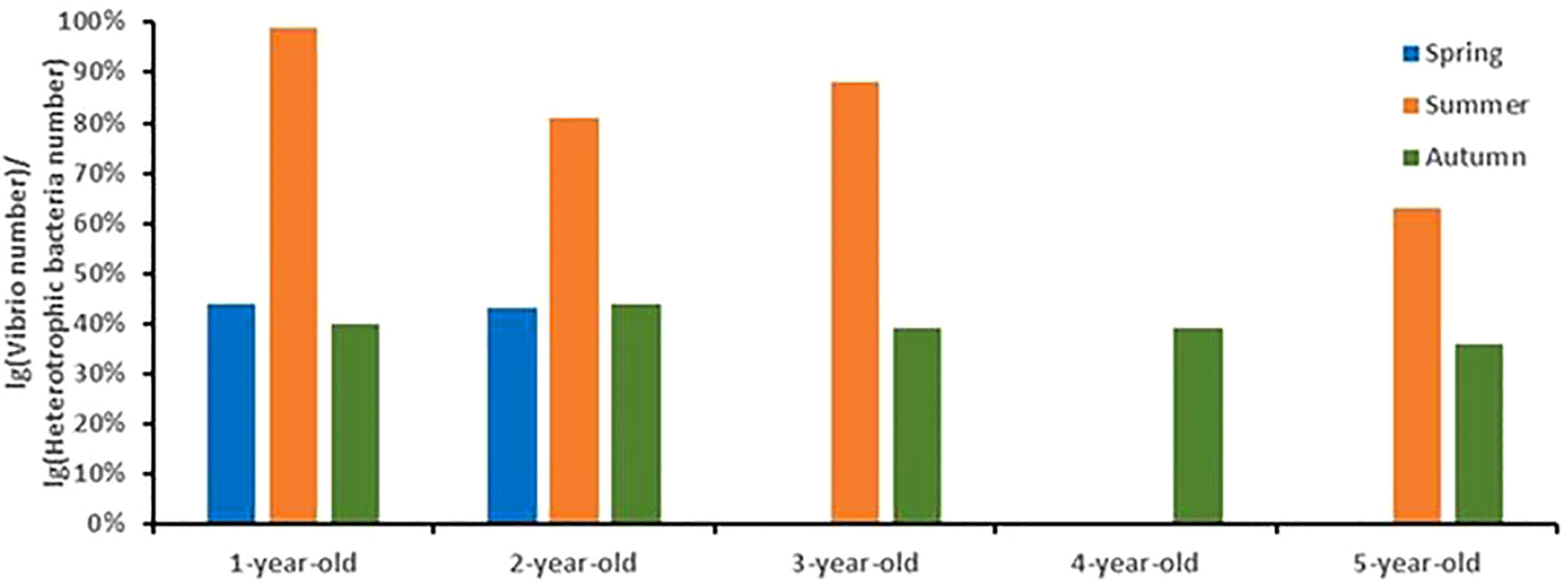
Vibrio abundance in the clam hepatopancreas ranged from 33 to 1.10 × 105 CFU/g in the spring, summer, and autumn. The abundance of the Vibrio and culturable bacteria in the clam hepatopancreas showed the same seasonal variation trend, reaching the highest in summer. The ratio of the logarithmic abundance of Vibrio to culturable bacteria also increased first and then decreased with the season, ranging from 0.36 – 0.99. Moreover, the ratio reached the maximum in summer, ranging from 0.63 – 0.99, indicating that Vibrio was the dominant bacteria in the clam hepatopancreas in summer (see Figure 4).
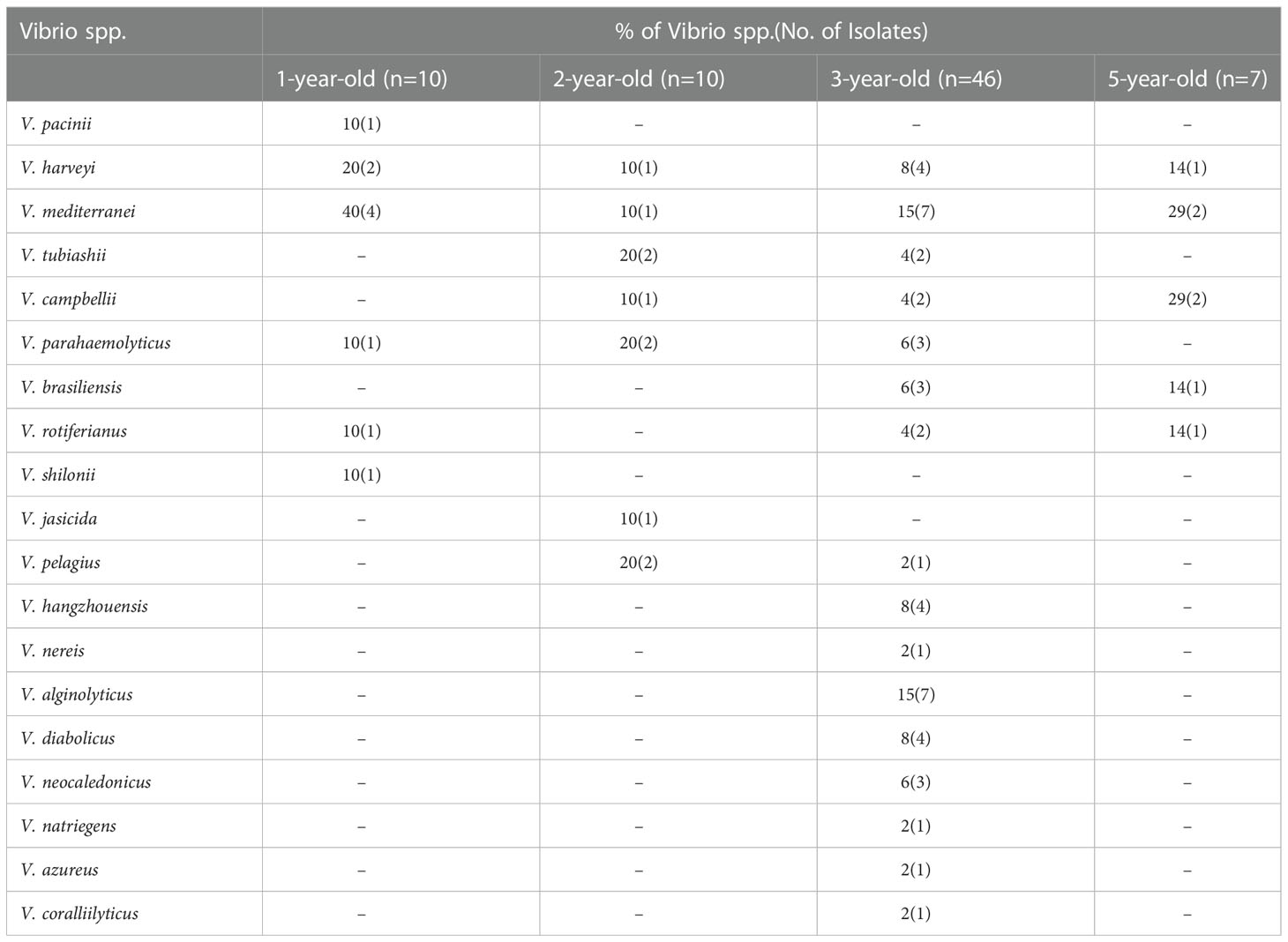
The 16S rRNA gene sequencing technology identified 170 bacterial isolates, and a total of 73 Vibrio isolates of 19 species were obtained. Vibrio mediterranei (19%), V. harveyi (11%), V. algolyticus (10%), and V. parahaemolyticus (8%) were the dominant species. Among them, the 1-year-old, 2-year-old, 3-year-old, and 5-year-old of Meretrix meretrix contained 6, 7, 16, and 5 species of Vibrio, respectively. The species and number of Vibrio isolates in the 3-year-old of Meretrix meretrix were the highest (63%).
3.3. Resistant phenotype analysis
All Vibrio isolates(n=73) were sensitive to sulfamethoxazole-trimethoprim. Among the 73 isolates, 68 Vibrio strains were resistant to other 15 antibiotics with 57 resistant phenotypes. Vibrio showed relatively high resistance to clindamycin (76%), amikacin (63%), ampicillin (62%), rifampicin (62%), vancomycin (57%) and amoxicillin (50%), respectively(Table 2). The ARI values of Vibrio spp. in different ages ranged from 0.13 to 0.18, and ARI values of 2-year-old (ARI=0.17) and 3-year-old (ARI=0.18) clams are higher than that of 1-year-old and 5-year-old clams. Forty-nine MAR Vibrio has been detected in the clams of different ages, and the proportion of MAR Vibrio in the clams at the 3-year-old is the highest (91%) compared with samples of other ages (Figure 5). Among the 49 multiple antibiotic-resistant isolates from the clam, 43 isolates were resistant to three to ten antibiotics and 1 isolate was resistant up to 15 antibiotics(Table S3). Sixteen isolates (24%) and fifteen isolates (22%) from clam samples showed resistance to four antibiotics and two antibiotics with high prevalence, respectively.
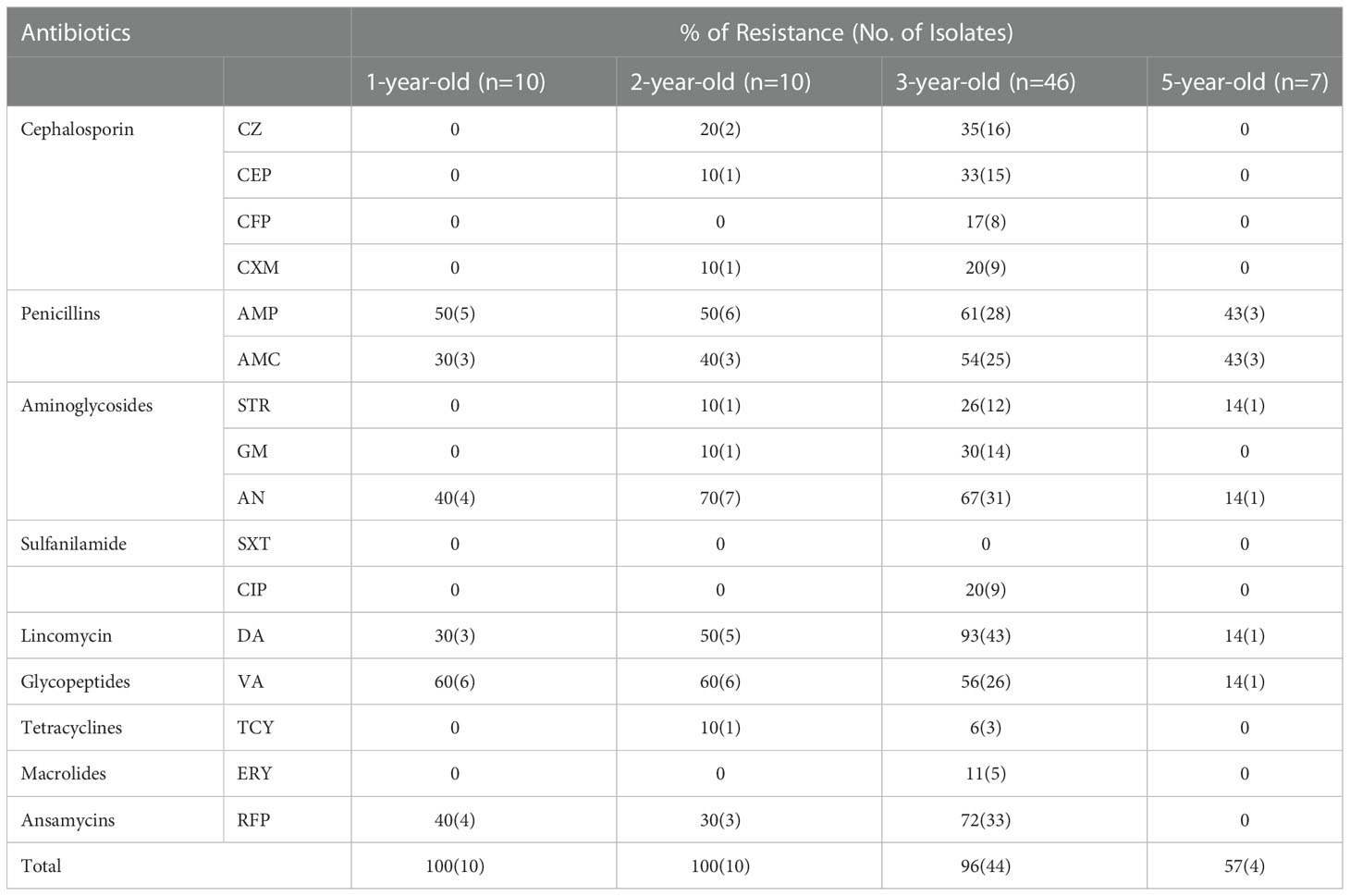
Table 2 Antibiotic resistance phenotypes of Vibrio isolates from Meretrix meretrix in different age.
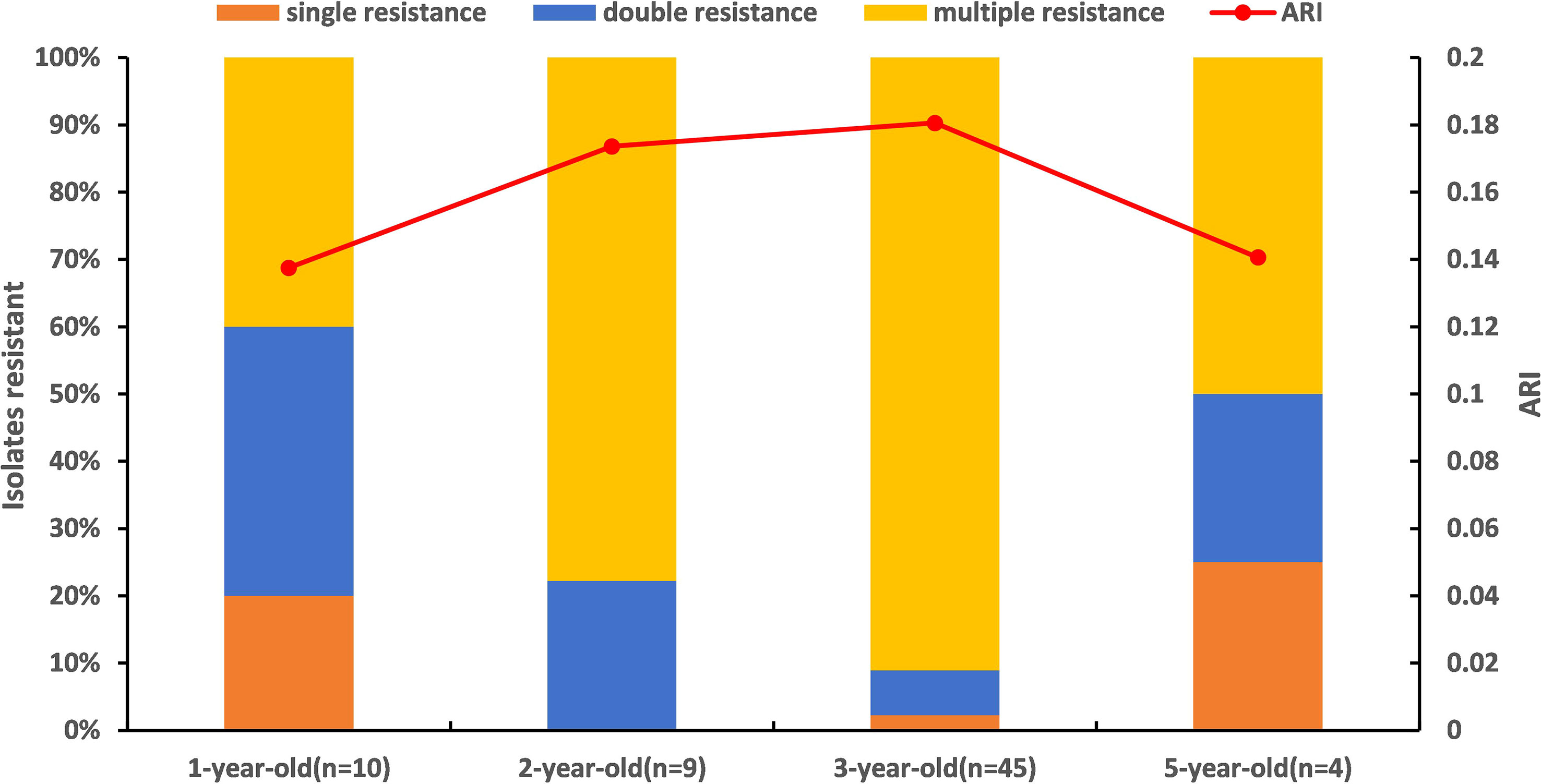
Figure 5 Prevalence of resistant Vibrio spp. and antibiotic resistance index (ARI) from clam Meretrix meretrix in different ages. Single resistance: resistance to one antibiotic; Double resistance: resistance to two antibiotics; Multiple resistance: resistance to three or more antibiotics.
The MIC values of the 73 Vibrio strains for Cd2+, Cu2+, and Zn2+ metal ions were 25 – 125 mg/L, 25 - 300 mg/L, and 50 - 400 mg/L, respectively. The maximum MIC value of Vibrio showed the order of Cd2+ < Cu2+ < Zn2+. The tolerance of Vibrio to Zn2+ increased with age (Table S4).
3.4. Resistant genotype analysis
Based on the results of the antibiotic-resistant phenotypes and heavy metal-resistant phenotypes of the 73 Vibrio strains, 13 associated resistance genes were selected for detection. The resistance genes with a higher detection rate were nccA (14%), aadA (14%), and copB (12%) (Table S5). The copB (20.00%) and nccA (20.00%) genes were the frequently detected resistant genes in the 1-year-old clam. The 2-year-old clam showed more resistance toward the copA (30.00%), ampR (20.00%), and strA (20.00%) genes. The 5-year-old clam showed more resistance toward the copA (14%), nccA (14%), and zntB (14%) genes.
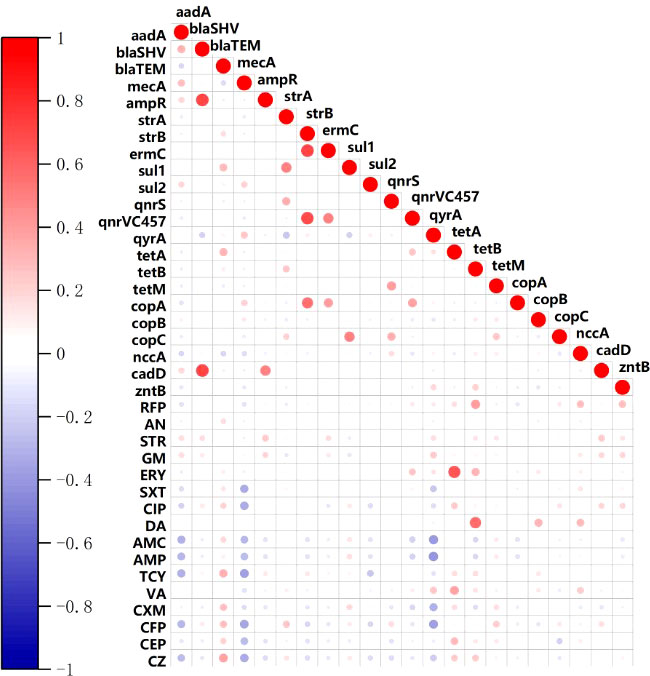
In addition to the co-resistance to the same class of antibiotics, resistance to cephalosporin (CFP, CEP, CZ) were significantly correlated with penicillins (AMP, AMC) (p< 0.01), tetracycline (p < 0.001), sulfanilamide (SXT) (p< 0.01) and quinolone (CIP) (p< 0.01). Aminoglycoside (GM) resistance phenotype is closely correlated with quinolone (CIP) resistance phenotype (p < 0.001). The blaSHV gene was positively correlated with aadA, ampR and cadD gene (p<0.01). The strA gene was positively correlated with sul1(p<0.01), qnrS (p<0.01) and tetB (p<0.05), and negatively correlated with qyrA (p<0.05). The copA was significantly correlated with strB, ermC and qnrVC457 (p<0.01). The heavy metal resistance genes copB and nccA were significantly correlated with the clindamycin resistance phenotype (p<0.01) (Figure 6).
Discussion
Temperature predicts Vibrio abundance quite well (Thompson et al., 2004). The prevalence of V. parahaemolyticus, Vibrio vulnificus, and V. mimicus in the environment positively correlated with temperature (León Robles et al., 2013). In this study, the temperature was higher in the summer (July and August), and lower in the spring (April) and autumn (November). The abundance of Vibrio in Meretrix meretrix in the summer was higher than in the spring and autumn. Vibrio was the dominant bacteria in the clam hepatopancreas in the summer, and there was a significant positive correlation between Vibrio abundance and temperature (p<0.05). Our results were consistent with a previous study demonstrating the increase in Vibrio abundance upon an increase in the temperature within a certain temperature range (Cruz et al., 2016).
Some studies found the greater prevalence of V. alginolyticus, V. cholerae, Vibrio communis, and V. parahemolyticus among all the Vibrio spp. isolated from the clams (Yücel and Balci, 2010; Adebayo-Tayo et al., 2011; Amalina et al., 2019; Abdalla et al., 2022). However, our study indicated the predominance of V. mediterranei in the Vibrio spp. of the clams. The difference in the dominant Vibrio species may be due to different sites or locations of shellfish collection. Vibrio is an indigenous marine bacterium (Hsiao and Zhu, 2020), and not all Vibrio variants are pathogenic (Song et al., 2017). However, three other dominant pathogenic Vibrio, including V. harveyi, V. algolyticus, and V. parahaemolyticus, were also detected in this study. V. parahaemolyticus is the major Vibrio species causing human illness and has emerged as a severe global threat to human health through the consumption of raw or undercooked seafood (Yue et al., 2010; Yue et al., 2011; Silva et al., 2018; Park et al., 2019). V. harveyi is reported to be the primary pathogen of cultured prawns (Stalin and Srinivasan, 2016).
The antibiotic resistance of Vibrio in marine resources is a major global concern for human health (Yang et al., 2017; Silva et al., 2018). The current results showed that 73 Vibrio strains were resistant to 15 other antibiotics except for sulfamethoxazole-trimethoprim. This observation was consistent with a previous study (Abdalla et al., 2022). Moreover, clindamycin (76%), amikacin (63%), ampicillin (62%), rifampicin (62%), vancomycin (57%) and amoxicillin (50%) resistance were very prevalent among the Vibrio isolates in this study. High prevalence of resistances to ampicillin and rifampicin have also been reported in Vibrio isolated from many aquatic products in different regions of the world (Obaidat et al., 2017), which indicating the existence of intrinsic resistance to these antibiotics (Su and Chen, 2020).
ARI is usually used to analyze the prevalence of bacterial resistance in a given population at a specific sites (Mohanta and Goel, 2014). When ARI value >0.2,it means that the isolates are exposed to contamination where antibiotics are often used, and when ARI ≤0.2,it means that antibiotics are seldom or never used (Krumperman, 1983). The ARI value of Vibrio in Meretrix meretrix at all ages was less than 0.2, indicating that the antibiotic level in the meretrix was low. A higher ARI value is detected in the 2-year-old and 3-year-old clams compared to the other ages, indicating the 2-year-old and 3-year-old clams may be more susceptible to antibiotic contamination.
Due to the toxicity, non-biodegradability and bioaccumulation of heavy metals in the food chain, heavy metal pollution is considered as a serious threat to aquatic ecosystems and human health (Diagomanolin et al., 2004). Geligang is reported to have suffered from heavy metal pollution, including zinc, copper and cadmium (Zhang et al., 2016). In this study, our results showed that the heavy metals Cd2+, Cu2+, and Zn2+ were tolerated in the majority of the Vibrio isolates, which may be explained by the existence of these three heavy metal ions in Geligang. Moreover, the concentration of these three heavy metal ions in the clam may be higher due to the enrichment of the clam itself. About 70% of the Vibrio isolates with conjugative elements (ICEs) derived from Yangtze River Estuary displayed strong resistance to Hg (≥1 mM) and Cr (≥10 mM), and the heavy metal contamination is relatively serious in Yangtze River Estuary in China (Song et al., 2013). The tolerance to heavy metals was also found to be prevalent in the V. parahaemolyticus strains with more than two antibiotic resistance phenotypes (Kang et al., 2018), which is consistent with our results. Indeed, the co-resistance between antibiotic resistance and heavy metal resistance has been well confirmed in many studies (Zhao et al., 2012; Kang et al., 2018).
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA901517.
Author contributions
JF: conceptualization. JS and YZ: experimental operation. JS, YZ and TH: manuscript writing. JF and JS: review and acquisition of funding. HM and TS: field sampling. YX and YJ: data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Dalian High Level Talent Innovation Support Plan (2021RD04); Open Project of National Key Laboratory of Environmental Monitoring Quality Control for Environmental Protection (KF202209); the National Key R & D Program of China (Grant 2020 YFA0607601); Millions of Talent Projects of Liaoning Province, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1071371/full#supplementary-material
References
Abdalla T., Al-Rumaithi H., Osaili T. M., Hasan F., Obaid R. S., Abushelaibi A., et al. (2022). Prevalence, antibiotic-resistance, and growth profile of vibrio spp. isolated from fish and shellfish in subtropical-arid area. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.861547
Adebayo-Tayo B. C., Okonko I. O., Esen C. U., Odu N. N., Onoh C. C., Igwiloh N. J. P. (2011). Incidence of potentially pathogenic Vibrio spp. in fresh seafood from itu creek in uyo, akwa ibom state, Nigeria. World Appl. Sci. J. 15 (7), 985–991. doi: 10.1016/j.jip.2009.11.008
Alsalem Z., Elhadi N., Aljeldah M., Alzahrani F., Nishibuchi M. (2018). Characterization of Vibrio vulnificus isolated from the coastal areas in the Eastern province of Saudi Arabia. J. Pure Appl. Microbiol. 12, 1355–1364. doi: 10.22207/JPAM.12.3.38
Amalina N. Z., Santha S., Zulperi D., Amal M. N. A., Yusof M. T., Zamri-Saad M., et al. (2019). Prevalence, antimicrobial susceptibility and plasmid profiling of vibrio spp. isolated from cultured groupers in peninsular Malaysia. BMC Microbiol. 19, 251. doi: 10.1186/s12866-019-1624-2
CDC (2019a) Vibrio species causing vibriosis–people at risk. Available at: www.cdc.gov/vibrio/people-at-risk.html (Accessed October 15 2021).
CDC (2019b) Vibrio species causing vibriosis–symptoms. Available at: https://www.cdc.gov/vibrio/symptoms.html (Accessed October 15 2021).
Clinical Laboratory Standard Institute (2016). Performance standards for antimicobial susceptibility testing: Seventeenth informational supplement M100-S17 (PA, USA: Wayne).
Cruz C. D., Chycka M., Hedderley D., Fletcher G. C. (2016). Prevalence, characteristics and ecology of Vibrio vulnificus found in new Zealand shellfish. J. Appl. Microbiol. 120 (4), 1100–1107. doi: 10.1111/jam.13064
Diagomanolin V., Farhang M., Ghazi-Khansari M., Jafarzadeh N. (2004). Heavy metals (Ni, cr, Cu) in the karoon waterway river, Iran. Toxicol. Lett. 151 (1), 63–68. doi: 10.1016/j.toxlet.2004.02.018
Economopoulou A., Chochlakis D., Almpan M. A., Sandalakis V., Maraki S., Tselentis Y., et al. (2017). Environmental investigation for the presence of Vibrio species following a case of severe gastroenteritis in a touristic island. Environ. Sci. pollut. Res. 24 (5), 4835–4840. doi: 10.1007/s11356-016-8231-7
Food and Agriculture Organization (2022). The state of world fisheries and aquaculture 2022. towards blue Transformation0 (Rome: FAO).
Hsiao A., Zhu J. (2020). Pathogenicity and virulence regulation of Vibrio cholerae at the interface of host-gut microbiome interactions. Virulence 11 (1), 1582–1599. doi: 10.1080/21505594.2020.1845039
Huang Y., Du P., Zhao M., Liu W., Du Y., Diao B., et al. (2017). Functional characterization and conditional regulation of the type VI secretion system in Vibrio fluvialis. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00528
Jiang Y., Yao L., Zhai Y., Wang L. (2014). Studies on molecular mechanism of a multi-drug resistant Vibrio parahaemolyticus strain. J Food Saf. &Quality 5 (1), 77–82. doi: 10.19812/j.cnki.jfsq11-5956/ts.2014.01.014
Kamika I., Momba M. N. B. (2013). Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 13, 28. doi: 10.1186/1471-2180-13-28
Kang C. H., Shin Y., Kim W., Kim Y., Song K., Oh E.-G., et al. (2016). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from oysters in Korea. Environ. Sci. pollut. Res. 23 (1), 918–926. doi: 10.1007/s11356-015-5650-9
Kang C. H., Shin Y., Yu H., Kim S., So J. S. (2018). Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from oysters in Korea. Mar. pollut. Bull. 135, 69–74. doi: 10.1016/j.marpolbul.2018.07.007
Kitaoka M., Miyata S. T., Unterweger D., Pukatzki S. (2011). Antibiotic resistance mechanisms of Vibrio cholerae. J. Med. Microbiol. 60 (4), 397–407. doi: 10.1099/jmm.0.023051-0
Krumperman P. H. (1983). Multiple antibiotic resistance indexing of escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46 (1), 165–170. doi: 10.1128/aem.46.1.165-170.1983
León Robles A., Acedo Félix E., Gomez-Gil B., Quiñones Ramírez E. I., Nevárez-Martínez M., Noriega-Orozco L. (2013). Relationship of aquatic environmental factors with the abundance of Vibrio cholerae, Vibrio parahaemolyticus, Vibrio mimicus and Vibrio vulnificus in the coastal area of guaymas, Sonora, Mexico. J. Water Health 11 (4), 700–712. doi: 10.2166/wh.2013.160
Letchumanan V., Yin W.-F., Lee L.-H., Chan K.-G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticu s isolated from retail shrimps in Malaysia. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00033
Liu X. (2016). Investigation on Antimicrobial Resistance and the Prevalence of qnrVC Gene in Vibrios from Mariculture Sources (Shanghai: Shanghai Ocean University).
Liu X., Sun H., Li Y. (2014). Analysis on enterococcal clinical resistance and detection of new resistance gene vanM in enterococcus faecium. J. Beihua University(Natural Science) 15 (4), 482–485. doi: 10.11713/j.issn.1009-4882.2014.04.014
Li M., Zhao L., Ma J., Zhao N., Luo J., Wang C., et al. (2018). Vibrio vulnificus in aquariums is a novel threat to marine mammals and public health. Transboundary Emerging Dis. 65 (6), 1863–1871. doi: 10.1111/tbed.12967
Mala E., Oberoi A., Alexander V. S. (2014). Vibrio isolates from cases of acute diarrhea and their antimicrobial susceptibility pattern in a tertiary care hospital. Int. J. Basic Appl. Sci. 3 (1), 35–37. doi: 10.14419/ijbas.v3i1.1735
Manjusha S., Sarita G. B., Elyas K. K., Chandrasekaran M. (2005). Multiple antibiotic resistances of Vibrio isolates from coastal and brackish water areas. Am. J. Biochem. Biotechnol. 1 (4), 193–198. doi: 10.3844/ajbbsp.2005.193.198
Ministry of Agriculture and Rural Affairs Fisheries Bureau, National Fisheries Technology Extension Center, China Society of Fisheries (2021). China Fishery statistics yearbook (Beijing: China Agriculture Press).
Miranda C. D., Godoy F. A., Lee M. R. (2018). Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01284
Moffitt C. M., Cajas-Cano L. (2014). Blue growth: the 2014 FAO state of world fisheries and aquaculture. Fisheries (Bethesda) 39 (11), 552–553. doi: 10.1080/03632415.2014.966265
Mohanta T., Goel S. (2014). Prevalence of antibiotic-resistant bacteria in three different aquatic environments over three seasons. Environ. Monit. Assess. 186 (8), 5089–5100. doi: 10.1007/s10661-014-3762-1
Obaidat M. M., Salman A. E. B., Roess A. A. (2017). Virulence and antibiotic resistance of vibrio parahaemolyticus isolates from seafood from three developing countries and of worldwide environmental, seafood, and clinical isolates from 2000 to 2017. J. Food Prot. 80 (12), 2060–2067. doi: 10.4315/0362-028X.JFP-17-156
Oliver J. D. (2015). The biology of Vibrio vulnificus. Microbiol. Spectr. 3 (3), 1–10. doi: 10.1128/microbiolspec.VE-0001-2014
Park Y. G., Lee M. S., Lee D. S., Lee J. M., Yim M. J. (2018). Antibiotic resistance of symbiotic marine bacteria isolated from marine organisms in jeju island of south Korea. J. Oceanogr. Mar. Res. 6 (2), 181. doi: 10.4172/2572-3103.1000181
Park K., Mok J. S., Kwon J. Y., Ryu A. R., Shim K. B. (2019). Seasonal and spatial variation of pathogenic Vibrio species isolated from seawater and shellfish off the gyeongnam coast of Korea in 2013-2016. Korean J. Fish. Aquat.Sci. 52 (1), 27–34. doi: 5657/KFAS.2019.0027
Romalde J. L., Diéguez A. L., Lasa A., Balboa S. (2014). New Vibrio species associated to molluscan microbiota: a review. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00413
Sambrook J. (2001). Molecular Cloning: A Laboratory Manual (New York: Cold Spring Harbor Laboratory Press).
Silva I. P., de Souza Carneiro C., Saraiva M. A. F., de Oliveira T. A. S., de Sousa O. V., Evangelista-Barreto N. S. (2018). Antimicrobial resistance and potential virulence of Vibrio parahaemolyticus isolated from water and bivalve mollusks from bahia, Brazil. Mar. pollut. Bull. 131, 757–762. doi: 10.1016/j.marpolbul.2018.05.007
Song X., Ma Y., Fu J., Zhao A., Guo Z., Malakar P. K., et al. (2017). Effect of temperature on pathogenic and non-pathogenic Vibrio parahaemolyticus biofilm formation. Food Control 73, 485–491. doi: 10.1016/j.foodcont.2016.08.041
Song Y. Z., Yu P., Li B. L., Pan Y. J., Zhang X. J., Cong J., et al. (2013). The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze river estuary, China. BMC Microbiol. 13, 214. doi: 10.1186/1471-2180-13-214
Stalin N., Srinivasan P. (2016). Molecular characterization of antibiotic resistant Vibrio harveyi isolated from shrimp aquaculture environment in the south east coast of India. Microb. Pathogen. 97, 110–118. doi: 10.1016/j.micpath.2016.05.021
Su C. L., Chen L. M. (2020). Virulence, resistance, and genetic diversity of vibrio parahaemolyticus recovered from commonly consumed aquatic products in shanghai, China. Mar. pollut. Bull. 160, 111554. doi: 10.1016/j.marpolbul.2020.111554
Sujeewa A. K. W., Norrakiah A. S., Laina M. (2009). Prevalence of toxic genes of Vibrio parahaemolyticus in shrimps (Penaeus monodon) and culture environment. Int. Food Res. J. 16 (1), 89–95. doi: 10.20431/2349-0365.0406003
Thompson J. R., Randa M. A., Marcelino L. A., Tomita-Mitchell A., Lim E., Polz M. F. (2004). Diversity and dynamics of a north Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70 (7), 4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004
Wu L., Liao J., Pang M., Bao H., Zhou Y., Sun L., et al. (2019). Virulence and antimicrobial resistance of Vibrio harveyi in large yellow croaker. J. Food Saf. Qual. 10 (8), 2111–2119. doi: 10.3969/j.issn.2095-0381.2019.08.005
Yang S., Deng W., Liu S., Yu X., Mustafa G. R., Chen S., et al. (2020). Presence of heavy metal resistance genes in escherichia coli and salmonella isolates and analysis of resistance gene structure in e. coli E308. J. Global Antimicrob. Resist. 21, 420–426. doi: 10.1016/j.jgar.2020.01.009
Yang J. H., Mok J. S., Jung Y. J., Lee K. J., Kwon J. Y., Park K., et al. (2017). Distribution and antimicrobial susceptibility of Vibrio species associated with zooplankton in coastal area of Korea. Mar. pollut. Bull. 125 (1-2), 39–44. doi: 10.1016/j.marpolbul.2018.05.007
Yücel N., Balci Ş. (2010). Prevalence of listeria, aeromonas, and Vibrio species in fish used for human consumption in Turkey. J. Food Prot. 73 (2), 380–384. doi: 10.4315/0362-028X-73.2.380
Yue X., Liu B., Sun L. (2011). Isolation and characterization of a virulent Vibrio spp. bacterium from clams (Meretrix meretrix) with mass mortality. J. Invertebr. Pathol. 106 (2), 242–249. doi: 10.1016/j.jip.2010.10.006
Yue X., Liu B., Xiang J., Jia J. (2010). Identification and characterization of the pathogenic effect of a Vibrio parahaemolyticus-related bacterium isolated from clam meretrix meretrix with mass mortality. J. Invertebr. Pathol. 103 (2), 109–115. doi: 10.1016/j.jip.2009.11.008
Zago V., Zambon M., Civettini M., Zaltum O., Manfrin A. (2017). Virulence-associated factors in Vibrio cholerae non-O1/non-O139 and v. mimicus strains isolated in ornamental fish species. J. Fish. Dis. 40 (12), 1857–1868. doi: 10.1111/jfd.12659
Zhang A., Shao S., Zhao K., Li J. (2016). Nutritional composition and edible safety analysis of two estuarine bivalve clams,Meretrix meretrix and mactra veneriformis in shuangtaizi estuary. J. Fish. China 40 (9), 1497–1504. doi: 10.11964/jfc.20160610422
Keywords: prevalence, antibiotic resistance, heavy metal resistance, Vibrio spp., Meretrix meretrix
Citation: Su J, Zhang Y, Hu T, Ming H, Xie Y, Jin Y, Shi T and Fan J (2022) Prevalence, antibiotic and heavy metal resistance of Vibrio spp. isolated from the clam Meretrix meretrix at different ages in Geligang, Liaohe estuary in China. Front. Mar. Sci. 9:1071371. doi: 10.3389/fmars.2022.1071371
Received: 16 October 2022; Accepted: 06 December 2022;
Published: 20 December 2022.
Edited by:
Songzhe Fu, Dalian Ocean University, ChinaReviewed by:
Xiaojun Zhang, Yangzhou University, ChinaDuochun Wang, National Institute for Communicable Disease Control and Prevention (China CDC), China
Copyright © 2022 Su, Zhang, Hu, Ming, Xie, Jin, Shi and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingfeng Fan, amZmYW5Abm1lbWMub3JnLmNu
 Jie Su1
Jie Su1 Yuan Jin
Yuan Jin Jingfeng Fan
Jingfeng Fan