
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 18 November 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1057814
Dual specificity mitogen-activated protein kinase kinase 1 (MEK1) has been found to regulate aging through the Ras/Raf/MEK/ERK cascade in terrestrial animals. However, few reports have focused on MEK1 promoting aging in marine bivalves. In this study, we aimed to examine the potential roles of MEK1 in the regulation of aging in two Argopecten scallops, the bay scallop A. irradians and the Peruvian scallop A. purpuratus, which are closely related but with distinct lifespan. The complete ORFs of AiiMEK1 and ApuMEK1 were both 1209 bp, encoding 403 identical amino acids but with 41 synonymous SNPs, which may have contributed to the different activities of MEK1 in two scallops. Nutrient restriction, one of the most effective non-genetic means of promoting life span, significantly inhibit the expression of AiiMEK1 and ApuMEK1. The response in hepatopancreas of A. irradians to nutrient restriction was more persistently than that in A. purpuratus. RNAi of AiiMEK1 significantly increased the expression of its downstream genes known to favor longevity, such as FoxO and SOD, while decreased the expression of ERK1/2 and the key genes in the mTOR signaling pathway, as well as the β-GAL activity (a marker for senescence). These results indicated that AiiMEK1 may play a negative role in longevity through Ras/Raf/MEK/ERK pathway. Our results may provide new perspective for understanding of the conservative functions of MEK1 in regulation of aging in animals and benefit the genetic selection of scallops.
Aging is an inevitable physiological phenomenon of any organism, and this process is affected by both genetic and environmental factors, such as temperature, nutrient level, and exogenous stimuli. Multiple pathways have been revealed to be involved in the regulation of aging and lifespan in animals, including the insulin/IGF1-like growth factor signaling pathway, mTOR signaling pathway, JNK pathway and AMPK pathway. Ras/Raf/MEK/ERK multi-layered protein kinase cascade that regulates gene expression by transmitting extracellular signals to the nucleus, may also participate the regulation of longevity. In mice, overexpression of miR-31 drives skin aging by enhancing the Ras/Raf/MEK/ERK signaling, and inhibiting MEK1 kinase activity of this pathway can protect the skin from radiation-induced premature aging (Yu et al., 2021). The Ras/Raf/MEK/ERK pathway is often drawn in a linear manner such that Ras activates Raf, which in turn activates MEK and subsequently ERK (Rouquette-Jazdanian et al., 2012). Mitogen-activated protein kinase kinase1 (MEK1), as a core component of the conserved Ras/Raf/MEK/ERK cascade, plays a central role in these events (Degirmenci et al., 2020; Jin et al., 2020; Qu et al., 2020; Wu et al., 2020). MEK1 is a member of the dual specificity protein kinase family and its most prominent function is to mediate the phosphorylation of threonine and tyrosine in ERK1/2 which in turn regulates various physiological functions through the phosphorylation level of transcription factors of the key genes (Roskoski, 2012; Breitenbach et al., 2019). As this pathway can often be effectively silenced by MEK1 inhibitors, resulting in profound effects on cell growth and longevity, the aging-promoting action of MEK1 has attracted much attention in recent years.
Insulin/IGF-1 signaling (IIS), an evolutionarily conserved signaling pathway which has been known to play a key role in determination of animal lifespan for nearly three decades (Kenyon et al., 1993; Newell Stamper et al., 2018; An et al., 2019; Zhao et al., 2021), is one of the major pathways mediated by MEK1 in lifespan regulation. Numerous studies have demonstrated the importance of the PI3K/AKT/FoxO branch of IIS, and we have found it played a vital role in extending lifespan in Argopecten scallops (Wang et al., 2022; Xu et al., 2022). Previous studies also have identified an equally important role for Ras/Raf/MEK/ERK signaling cascade in IIS-dependent lifespan extension (LeRoith et al., 1995; Luckhart and Riehle, 2007). In Drosophila, the use of the MEK1 kinase inhibitors against the Ras/Raf/MEK/ERK cascade can reduce insulin/IGF-1 (IIS) signaling and extend lifespan (Slack et al., 2015). Caloric restriction remains the most effective non-genetic means of increasing life span in different species (Goldberg et al., 2015; Rojas-Morales et al., 2020) and can downregulate IIS pathway activity (Fontana et al., 2010). In rat heart, the Ras/Raf/MEK/ERK branch of IIS pathway are hyper-activated with aging, and caloric restriction treatment restored ERK1/2, the only substrate of MEK1, activation to near-young values (Castello et al., 2011). In human, under nutritional restriction,the phosphorylation level and activity of ERK1/2 was also reduced along with a decrease in insulin signaling (Arcidiacono et al., 2021). FoxO, a key transcription factor in the insulin/IGF-1 (IIS) signaling pathway, whose overexpression in model organisms was found to be associated with longevity (Webb and Brunet, 2013; Tia et al., 2018), can also be phosphorylated by MEK1 at its serine residues and result in its ubiquitination and subsequent proteasomal degradation (Asada et al., 2007; Yang et al., 2008). In pancreatic cancer cells, the use of MEK1 inhibitor can help block the phosphorylation of FoxO by ERK, eventually leading to cycle arrest and apoptosis in cancer cell (Roy et al., 2010). Conserved mTOR pathway, another important part of aging regulatory network, is also known to interact with the Raf/RAS/MEK/ERK cascade (Fan et al., 2020). In T-cells of mice, MEK1, as an upstream activator, promotes mTORC1 activation that has been reported to be inversely associated with increased lifespan (Gorentla et al., 2011; Baar et al., 2016; Ito et al., 2021). In addition, the role of MEK1 in promoting senescence has been reported in plants. In Arabidopsis thaliana, studies have shown that the overexpression of WRKY53 can lead to early senescence, MEK1 can phosphorylate the WRKY53 transcription factor, but also bind to the promoter region of the WRKY53 gene to increase its relative expression (Miao et al., 2007; Benhamman et al., 2017).
Up to now, most of the studies on lifespan were carried out in terrestrial model organisms. The ocean was considered as the origin of life and rich in species resources. Marine animals, especially marine bivalves with diverse lifestyles and different lifespans, such as Argopecten scallops, could be more suitable for aging and longevity research (Ridgway et al., 2011; Lian et al., 2019). Argopecten scallops are the most cultured scallop species in the world due to their fast growth rates. The bay scallop (A. irradians) originated from the Atlantic coasts of United States and mainly cultured in China, and the Peruvian scallop (A. purpuratus) originated from the Pacific coasts and mainly cultured in Chile and Peru (Zhang et al., 2000; González et al., 2002). Although both belong to the same genus of Argopecten, they have evolved distinct lifespans after long-term adaptation to different natural environments. The bay scallops normally have a shorter lifespan, usually less than 2 years (Ungvari et al., 2013), and die after spawning in the spring of the following year. The Peruvian scallops, on the contrary, can live up to 7-9 years and continue to grow even after sexual maturity (Waller, 1969; González et al., 2002; Ungvari et al., 2013). Introduction of the Peruvian scallops into China and hybridization of the two Argopecten scallops renders us a unique opportunity to explore the regulatory mechanisms of longevity in bivalves. In this study, we attempted to examine the roles of MEK1 in the regulation of aging in these two closely-related bivalves in hope to further our understanding of the mechanisms underlying animal longevity.
Peruvian scallops and bay scallops were cultured in the open sea in the Yangma Island area of Yantai, Shandong Province, China. They were brought into the scallop hatchery of Yantai Spring-Sea AquaSeed Co., Ltd. located in Laizhou, Yantai, Shandong Province, China in early spring and conditioned to mature. They were induced to spawn individually in beakers of 1 L after exposure to air for 30 min. The spawning scallops were watched carefully to collect eggs and sperm separately. Eggs or sperm from different individuals of the same species were pooled together. Then sperm were mixed with eggs of the same species to obtain fertilized eggs for seed production. Approximately 10 days after fertilization, when approximately 50% of larvae developed eyespots, they were placed on plastic collectors. After metamorphosis, the juveniles were moved to a shrimp pond for nursery for a period of one month and then to the open sea for another month before they were dispersed into lantern nets for grow-out. After approximately 9 months of cultivation, the scallops were collected for subsequent experiments.
Total RNA of each experimental sample was separately extracted by the RNA Easy Fast Tissue/Cell Kit (TIANGEN, China), and genomic DNA was removed using RNase-free DNase I (TaKaRa, Dalian, China). 1.2% agarose gel electrophoresis was used to test the RNA integrity and Nanophotometer spectrophotometer was applied to detect RNA purity (odds of OD260/280 and OD260/230); Qubit 2.0 Fluorometer was used for accurate quantification of RNA concentration. Subsequently, the HiScriptIII RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, China) was applied to reverse transcribe total RNA into cDNA, and store at -20°C for later PCR The MEK1 sequences of A.irradians and A.purpuratus from transcriptome data were used to design specific primers by Primer-Blast (https://www.ncbi.nlm.nih.gov/tools/primer-last) for cloning the partial cDNA fragment of these two MEK1 sequences by PCR. PCR reaction systems were carried out in 25μl-volume reaction contains 12.5μl 2X PCR Mix (Takala, Dalian, China), 9.5μl ddH2O, 1μl of forward and reverse primers and 1μl cDNA template. The thermal cycling sequence of PCR was set following parameters: 1cycle of 94°C for 3 min, 30 cycles of 94°C for 30 s, 56°C for 45 s, 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR reaction product was detected by 1.5% agarose gel electrophoresis, the target fragment was recovered and purified, and then ligated with pTOPO-TA vector (Aidlab, Beijing, China) and sequenced by Sangon Biotech Co., Ltd. (Qingdao, China). The ligation product was transformed into DH5α competent cells, and the positive clones were sequenced with M13 primers and sequenced by Sangon Biotech Co., Ltd. (Qingdao, China).
Analysis of the cDNA sequences was conducted to query known sequences in GenBank using the BLASTX search program provided by NCBI (https://www.ncbi.nlm.nih.gov/BLAST/). The open reading frame (ORF) of AiiMEK1 and ApuMEK1 were confirmed using the NCBI ORF Finder tools(http://www.ncbi.nlm.nih.gov) and sequences were translated into amino acid sequences using DNAMAN software. MEGA 7.0 was used to investigate the number of base substitutions per site between AiiMEK1 and ApuMEK1. The functional sites and domains in the deduced amino acid sequence were predicted with SMART programs (https://www.smart.emblheidelbergde/). Multiple sequence alignments of MEK1 proteins were performed using the Clustal W program packaged in DNAMAN 8.0 software. The protein parameters (molecular mass and isoelectric point) of MEK1 were predicted by using the Protparam (http://www.expasy.org/tools/protparam.html), SignalP (https://services.healthtech.dtu.dk/service.php?SignalP-4.1) was used to predict the cleavage site of the signal peptide, the phosphorylation sites were predicted by NetPhos (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1). The secondary and three-dimensional structures of the MEK1 were predicted by SOPMA (http://nhjy.hzau.edu.cn/kech/swxxx/jakj/dianzi/Bioinf7/Expasy/Expasy8.htm) and SWISS-MODEL (https://www.swissmodel.expasy.org/interactive), respectively.
The amino acid sequences of MEK1 from other species were obtained from GenBank database, and multiple sequence alignment of the MEK1 sequences from 11 different species was conducted using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). For phylogenetic tree analysis, MEGA 7.0 software was used with 1,000 replicates to construct a NJ phylogenetic tree based on the amino acid sequences of all the above 36 species.
A.purpuratus and A.irradians irradians at 5 and 12 months of age were collected to examine the expression profiles of MEK1 in various tissues at different developmental stages with 3 replicates for each stage. The sampled tissues including adductor muscles, gills, gonads, hepatopancreases, and mantles were used to extract total RNA and the first-strand cDNA was synthesized using a Reverse transcription kit ReverTra Ace® qPCR RT Master Mix With gDNA Remover (Toyobo, Japan). The expression level of MEK1 in two scallop species were evaluated by quantitative real-time PCR (qRT-PCR) using specific qRT-PCR primers for MEK1 as listed in Table 1. The elongation factor EF-1α was chosen as the reference for internal standardization according to the previous study (Xu et al., 2022). Reactions were carried out on the QuantStudioTM 5 Real-Time PCR Instrument (Applied Biosystems) using SYBR green as fluorescent dye. A complete reaction system for PCR reaction contained 5 μl of Taq Pro Universal SYBR qPCR Master Mix (Vazyme, China), 1 μl of the diluted cDNA, 0.2 μl of each primer (10 μM), and 3.6 μl of sterile distilled H2O. The PCR parameters were set at 95°C for 60 s, followed by 40 cycles of 95°C for 5 s and 60°C for 15 s. All reactions were performed in triplicates. The relative level of genes expression was analyzed using the2−ΔΔCt methods (Livak and Schmittgen, 2001).
Healthy and vigorous 7-month-old bay scallops and Peruvian scallops were randomly selected to explore the differences in the MEK1 gene expression between the two scallops under nutritional restriction. After having been acclimated in filtered and aerated seawater (salinity 30, temperature 22 ± 0.5 °C) for 1 week, each of the two scallops species were equally divided into a nutritional restricted group and a control group, respectively. During the experiment, the control group was fed with 3×104 Chaetoceros sp. per scallop a day while the nutritional restricted group were received only 50% diet of the control group. The hepatopancreas were collected from 3 healthy scallops in each group on 30 and day 56 day and quickly frozen in liquid nitrogen and stored at −80°C for RNA extraction and MEK1 expression analysis.
The sequence-specific AiiMEK1-siRNA was synthesized using the T7 promoter in vitro transcription kit (Takara, China) according to the manufacturer’s instruction. The AiiMEK1-siRNA were dissolved in sterilized 0.1% DEPC water to reach a final concentration of 2μg/μl, and then 1.0% agarose gel electrophoresis and Nanodrop 2000 (Thermo Scientific, USA) were used to detect the fragment integrity and concentration of siRNA. About 120 vigorous 12-month-old A. irradians (33.59 ± 4.19g in whole weight) from the same batch were randomly selected and divided into two groups, the AiiMEK1-siRNA injection group, and the control group. The experiment was performed according to the standard of 1μg (siRNA):1g (wet weight) (Ning et al., 2021). Eighteen microliter of AiiMEK1-siRNA, and PBS were injected into the cardiocoelom of each scallop in the AiiMEK1-siRNA injection group, and control group, respectively, using a micro-syringe. At 6h, 12h,and 24h after injection, the hepatopancreas of five scallops were separately collected and dissected. The hepatopancreas were frozen in liquid nitrogen immediately for RNA extraction and cDNA synthesis. The expression profiles of MEK1 mRNA were examined by real-time quantitative PCR as described above.
After knocking down the expression of AiiMEK1 gene by RNAi, qRT-PCR was used to detect the expression profiles of 9 aging-related genes in hepatopancreases of bay scallop, including the only downstream target gene ERK1/2 (Extracellular Signal-Regulated Kinase 1/2), antioxidative and DNA damage repair genes SOD (superoxide dismutase), CAT (Catalase), FoxO (Forkhead box class O), autophagy-related gene ULK1 (Unc-51 like autophagy activating kinase 1) and nutrient sensing pathway-related genes mTORC1, S6K1 (Ribosomal Protein S6 Kinase B1), 4EBP1 (eukaryotic translation initiation factor 4E binding protein 1) and IF4E (eukaryotic translation initiation factor 4E). The gene-specific primers used for qRT-PCR analysis were shown in Table 1.
Hepatopancreas were collected from three animals in the control and RNAi group 6 h after injection, and β-galactosidase activity was detected using the β-galactosidase assay kit (Microplate method) (purchased from Geruisi Biotechnology, Suzhou, China) in accordance with the manufacturer’s instructions. β-GAL can convert p-nitrobenzene-β-D-galactoside into p-nitrophenol, which has an absorption peak at 405 nm. The β-Gal activity was determined by calculating the rate of increase in absorbance at 405 nm. Tissues were homogenized in 9 volumes of 0.86% cold saline and centrifuged at 12,000 rpm for 10 min at 4°C, then the supernatants were taken and diluted with cold saline and protein concentration was determined using a BCA protein assay kit (Shanghai Epizyme Biotechnology Co., Ltd., Shanghai, China).
Statistical analyses were performed using SPSS 26.0 software. Data obtained from this study were presented as the mean ± standard deviation (S.D.) and analyzed by t-test. Differences were considered statistically significant at P < 0.05 (*) and P < 0.01(**).
The cDNA sequences of AiiMEK1 and ApuMEK1 were cloned and identified respectively, and the complete ORF sequences are deposited in the NCBI GenBank repository, accession numberOP490349 and OP490348. The complete ORFs of AiiMEK1 and ApuMEK1 were both 1209 bp, encoding 403 identical amino acids and the deduced protein molecular weight and theoretical isoelectric point were 44.502 kDa and 6.35, respectively. Forty-one synonymous SNPs were found in AiiMEK1 compared with the ApuMEK1 (Figure 1), including 29 transition and 12 transversion mutations, and transition-to-transversion ratio was 2.42. Among all the conversions of AiiMEK1 sequence, the transition from cytosine (C) to thymine (T) (or G to A on the complementary chain) was more prominent, with 18 loci accounting for 62% of all transition. No potential functional sites such as sequence signal peptide and glycosylation were found in the AiiMEK1 and ApuMEK1 amino acid sequence, but 40 phosphorylation sites were predicted in both sequences (Figure 2A).
An analysis of conserved domains using SMART revealed that both AiiMEK1 and ApuMEK1 contained a conserved domain of serine/threonine protein kinases (S-TKc) with a length of 300 amino acids (Figure 2B). Further analysis of AiiMEK1 and ApuMEK1, sequence revealed that there was an ATP binding site (84-107aa) and a catalytic activity site (196-208aa) within the central domain, which was crucial for its kinase activity (Han et al., 2021). Thirty-two (78%) SNPs in the conserved S-TKc domains and 2 SNPs in the catalytic active site were found between AiiMEK1 and ApuMEK1, with one SNP locating in the near upstream and downstream sequence of the ATP binding site, respectively (Figure 3).
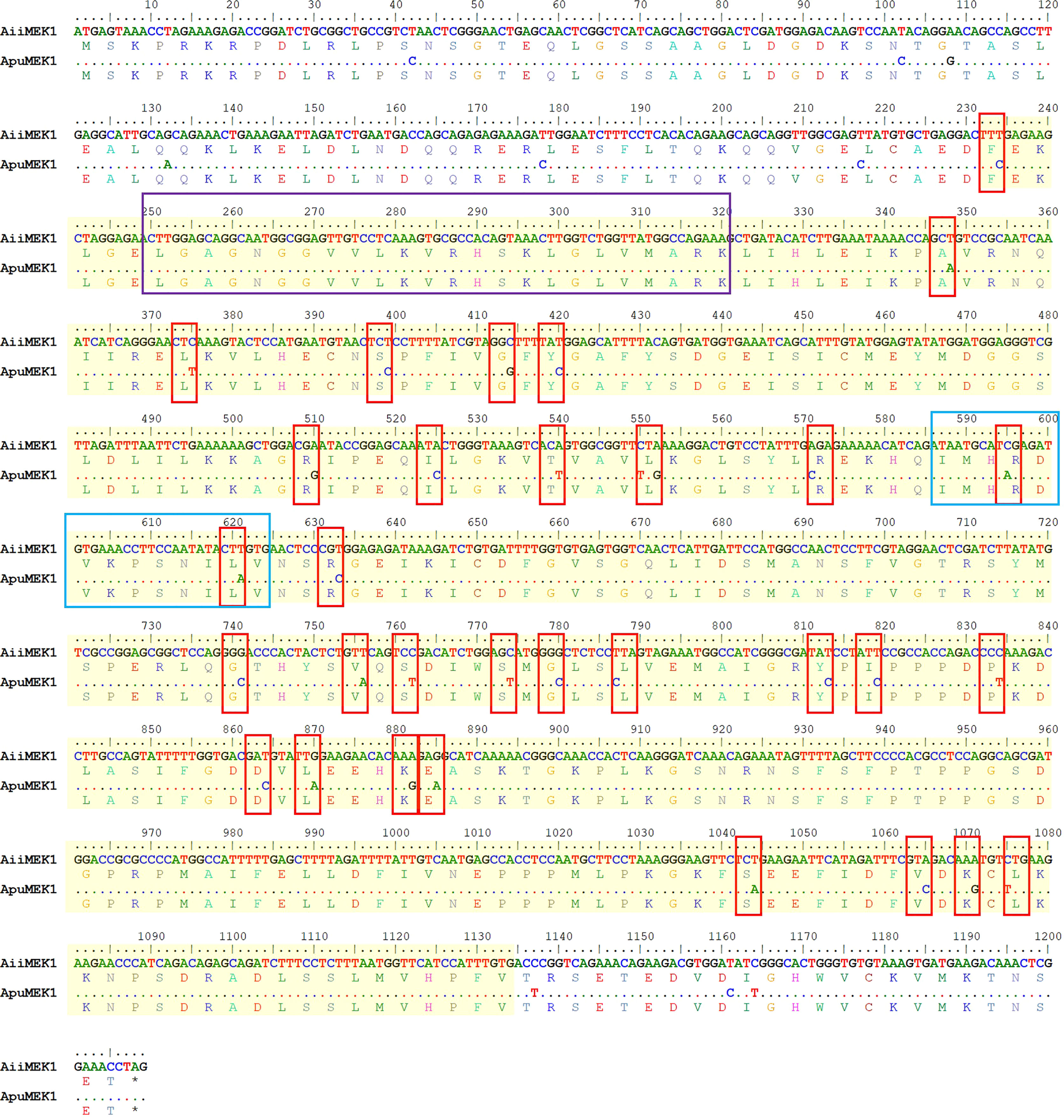
Figure 3 The SNPS in the conserved domains of AiiMEK1 and ApuMEK1. Sequences were highlighted with a yellow shading indicates the region of S_TKc in AiiMEK1 and ApuMEK1. The purple box indicates the ATP-binding site, and blue box indicates the activation site of S_TKc.
Secondary structure prediction of AiiMEK1 and ApuMEK1 demonstrated that the amino acid sequences contained 46.02% random coil, 40.80% α-helix, 9.9% extension chain fraction and 3.23% β-turn (Figure 4A). The tertiary structures of AiiMEK1 and ApuMEK1 were further predicted by the Swiss-Model online tool and the results showed that both AiiMEK1 and ApuMEK1 were mapped to the crystal structure of 6pp9.1.B (BRAF : MEK1 complex) with identities of 70.87% (Figure 4B).
Multiple sequence alignment of AiiMEK1 and ApuMEK1 with other homologous sequences indicated high similarity in protein kinase family characteristics but large evolutionary differences in ATP binding site between invertebrates and vertebrates. AiiMEK1 and ApuMEK1 shared similarity with the homologous sequence in the Mizuhopecten yessoensi (with 99.00% identity), followed by Pecten maximus (with 98.26% identity), Crassostreagigas (with 86.50% identity) and Tegillarca granosa (with 83.98% identity) (Figure 5). However, at the nucleotide level, ApuMEK1 shared high similarity with the homologous sequence in the long-lived species such as Pecten maximus (XM_033895226.1, with 89.44% identity) and Mizuhopecten yessoensi (XM_021516679.1, with 87.66% identity), while AiiMEK1, compared to ApuMEK1, represented lower similarity with MEK1 in Pecten maximus (with 89.19% identity) and Mizuhopecten yessoensi (with 87.51% identity). The topological structure of phylogeny tree showed that the MEK1 of Argopecten scallops were first clustered into a branch, and then clustered into a mollusk cluster with closely related Mizuhopecten yessoensis and Pecten Maximus. The MEK1 of mammals, birds, and fish were clustered separately to form a large branch of MEK1 in vertebrates (Figure 6).

Figure 5 Multiple alignment of amino acid sequences. The red box indicates the region of the S_TKc domain, the yellow box indicates the ATP binding site of this domain and the blue box indicates the active site of S_TKc.
To explore the spatiotemporal expression profile of MEK1 gene in Argopecten scallops, the transcriptional levels of MEK1 gene were detected in the adductor muscles, mantles, gills, gonads, and hepatopancreases of 5-month-old and 12-month-old A. irradians and A. purpuratus, respectively. In 5-month-old A.irradians, the expression of MEK1 was the highest in gill, followed by mantle, gonad and hepatopancreas, and the lowest in adductor muscle (Figure 7A). In 5-month-old A. purpuratus, the expression of MEK1 was the highest in the mantle, followed by the hepatopancreas and gills, and the adductor muscle was also the tissue with the lowest expression of MEK1 (Figure 7B). Compared with 5-month-old scallops, the expression of AiiMEK1 was significantly increased in all tested tissues, especially in adductor muscle, gills and the hepatopancreases were extremely significant in 12-month-old A. irradians(p < 0.01). Compared with 5-month-old scallops, the ApuMEK1 expression was significantly increased in the gills, gonad (p <0.01) and adductor muscle (p < 0.05) but not in mantle and hepatopancreas (p > 0.05) in the 12-month-old scallops (Figure 7B).
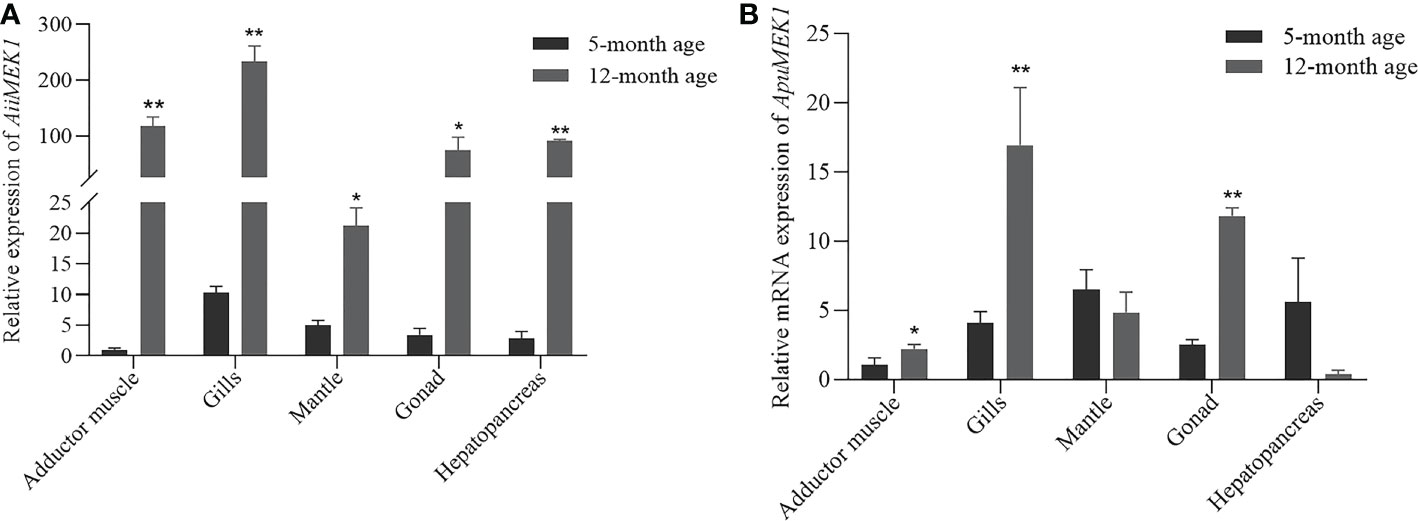
Figure 7 Expression of AiiMEK1 (A) and ApuMEK1 (B) at 5th and 12th month (* p < 0.05 and ** p < 0.01).
The relative mRNA expressions of MEK1 under nutrient restriction were examined in A. irradians and A. purpuratus. In A. irradians, AiiMEK1 expression was significantly decreased in hepatopancreas under nutritional restriction on Day 30 (p < 0.05), while it was significantly increased in the adductor muscle and mantle than that of the control group (Figure 8A); on day 56, the expression of AiiMEK1 in the hepatopancreas of the nutritional restriction group was still significantly lower than that of the control group, but no difference was detected in other tissues (Figure 8B). In A. purpuratus, ApuMEK1 expression level in hepatopancreas under nutritional restriction on Day 30 was also significantly lower (p < 0.05) than that in the control group, whereas the expressions in other tissues in nutritional restriction group were maintained at levels close to that of the control group (Figure 8C); the expression level of ApuMEK1 in the gills and mantle of the nutritional restriction group on day 56 were significantly lower (p < 0.05) than that in the control group on day 56 (Figure 8D).
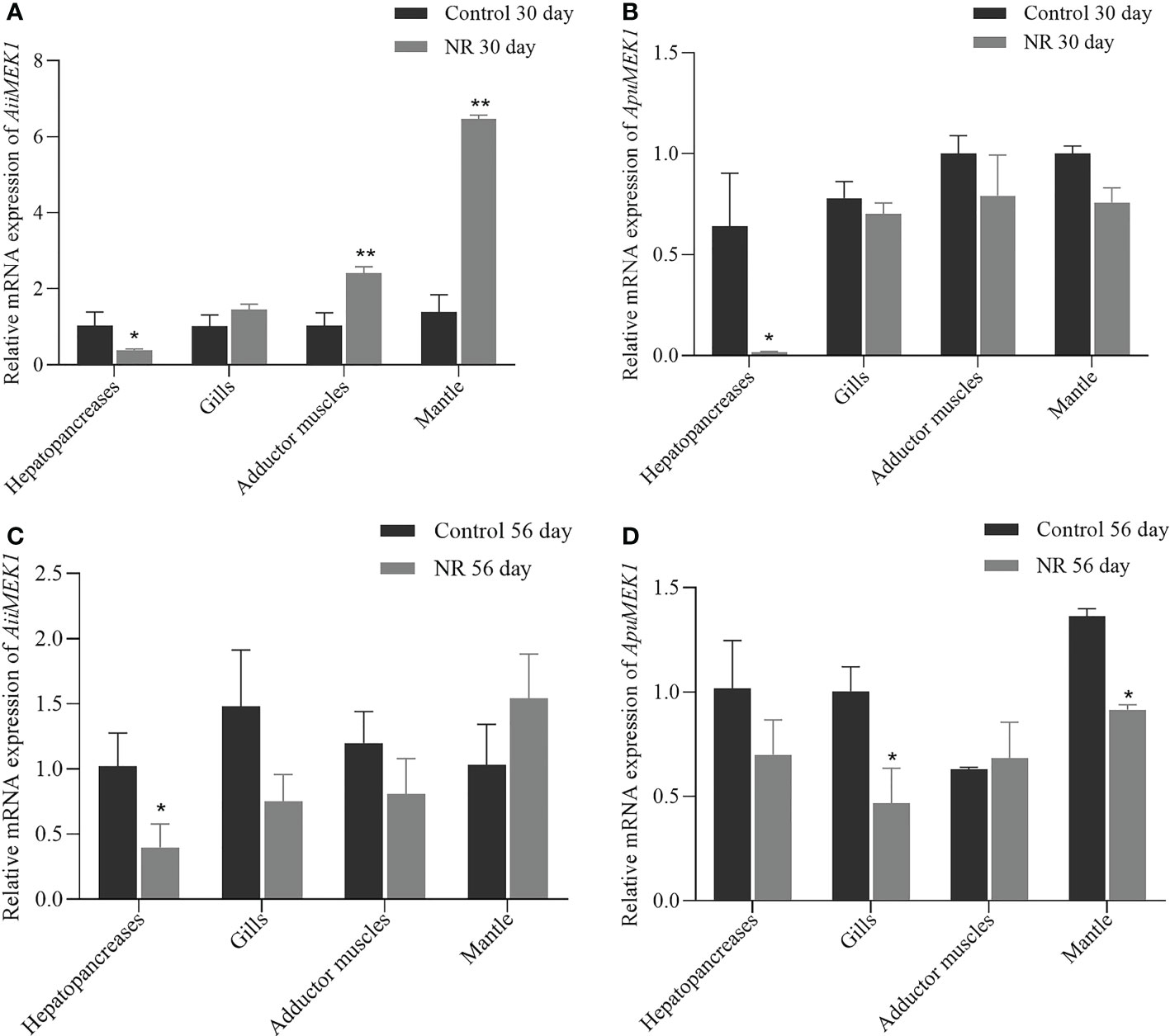
Figure 8 Expression of MEK1 on Day 30 and Day 56 in the dietary restriction group and the control group in Airradians (A, B) and A purpuratus (C, D) (*p < 0.05 and ** p<0.01).
The interference efficiency of the synthesized siRNA on the MEK1 gene in the hepatopancreas of 12-month-old A. irradians is shown in Figure 9. Compared with the control group, the expression of MEK1 gene was significantly inhibited by the interference for 6h, with an interference efficiency of 66.37%. Therefore, hepatopancreas tissue injected with AiiMEK1-siRNA for 6h was selected for subsequent experiment.
β-GAL is a widely used marker of cellular senescence, and its activity increases with age. At 6h after RNAi of MEK1, the β-GAL activity in the experimental group was significantly lower than that in the control group (p < 0.05) (Figure 10).
The expression pattern of aging-related genes in hepatopancreas tissue after RNAi of AiiMEK1 in A. irradians is shown in Figure 11. The results showed that the expressions of antioxidant and DNA damage repair genes SOD and FoxO were significantly up-regulated, while the expression of ERK1/2, the only target gene downstream to MEK1, and CAT and autophagy-related gene ULK1, as well as the nutrient-sensing pathway-related genes including mTORC1, S6K1, 4EBP1 and IF4E were significantly down-regulated.
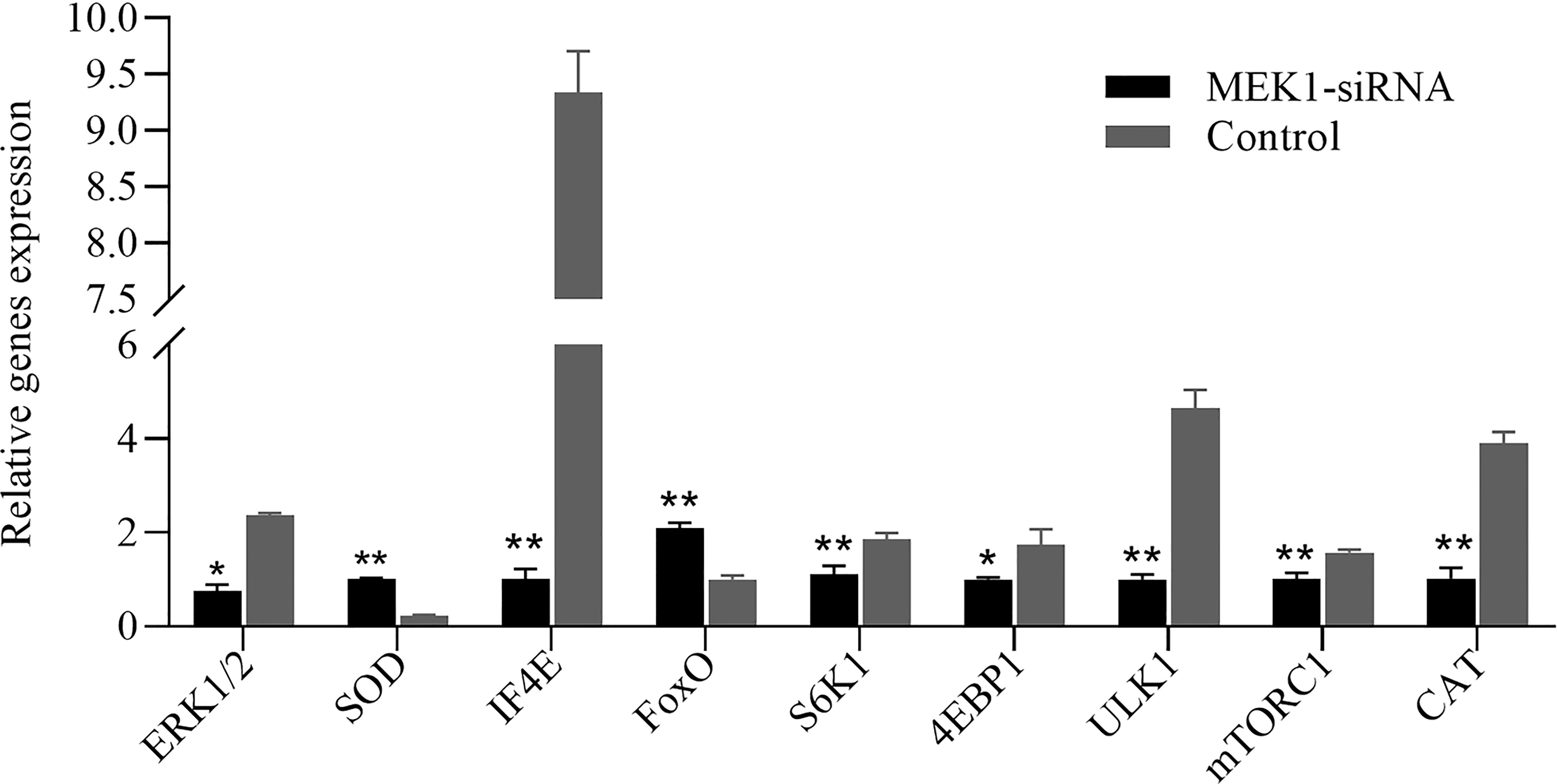
Figure 11 Relative expression levels of other aging-related genes after knockdown of AiiMEK1 gene expression (* p < 0.05 and ** p < 0.01).
In this study, we analyzed the variations in the sequences of MEK1, and examined the expression of MEK1 in response to nutrient restriction in Peruvian scallops and bay scallops, as well as the expression of its downstream target genes and β-GAL activity (a marker for senescence) after RNAi of MEK1. Our results firstly suggested that MEK1 may play a central role on aging in Argopecten scallops.
Sequences of MEK1 with very high similarity were obtained in the two scallops, and in total 41 synonymous SNPs (including 29 transitions and 12 transversion) were discovered between AiiMEK1 and ApuMEK1. It was reported that the transitions occurred more frequently than transversions in gene mutation (Wang et al., 2015; Storz et al., 2019; Mohanta et al., 2020), and in especial for synonymous substitutions (Lim et al., 2017). Previous study has shown the prevalence of cycline (C) to tyrosine (T) transition tends to increase the hydrophobicity of the amino acid and the accumulation of peptides with altered function, may ultimately leading to the premature aging phenotype of Polg mutant mice (Ni et al., 2015). In AiiMEK1, the transition is also dominated by cycline (C) to tyrosine (T), this result implying that high frequency cycline (C) to tyrosine (T) transition may affect the function of AiiMEK1 protein in regulating scallop senescence. In addition, massive literature has reported that synonymous and non-synonymous substitutions can significantly change protein functions through multiple mechanisms, such as protein level, translation accuracy, secretion efficiency, and post-translational modification (Tuller et al., 2010; Walsh et al., 2020). The synonymous SNPs in AiiMEK1 and ApuMEK1 may lead to significant differences in the protein structure and conformation, suggesting that MEK1 in the two scallops may have evolved into different activities during their adaptation to different environments (Drummond et al., 2008). However, the MEK1 nucleotide sequence of other species with long lifespans (such as Pecten maximus, Mizuhopecten yessoensis) share higher similarity with ApuMEK1 than AiiMEK1, indicating that this feature sequence might contribute to longevity in these animals. The deduced amino acid sequences of both AiiMEK1 and ApuMEK1 contained the highly conserved S_TKc domain (300aa) including the ATP binding sites and catalytic segment, which were crucial for MEK1 to function as a protein kinase (Lu et al., 2014; Abdelfatah et al., 2019). This S_TKc domain was identical in amino acid sequence in two scallops and was highly homologous to the S_TKc domain of other invertebrates and vertebrates, indicating the function of MEK1 in different species was evolutionarily conserved. Mutations that occur in MEK1 catalytic core cause constitutive activation of the MEK1 kinase, and the research showed that more than 3/4 of synonymous mutations were harmful and could change the level of gene and protein expression (Shen et al., 2022). However, a total of 32 (78%) SNPs were found between ApuMEK1 and AiiMEK1 in this region, which might lead to different catalytic activity of MEK1 in the two species. Besides, phylogenetic tree analysis showed that MEK1 was obviously clustered with other bivalve MEK1, suggesting that MEK1 was a member of the dual specificity protein kinase family.
As an evolutionary conserved signal transduction factor, the widespread distribution of MEK1 in various tissues of mammals and teleosts has been extensively reported. Likewise, in this study, MEK1 was expressed in all examined tissues, indicating that it was not a tissue-specific gene and may play roles in various physiological processes. With the aging of the bay scallop, the expression of AiiMEK1 was significantly upregulated in all tested tissue especially in adductor muscle and gills. These results suggested that the adductor muscle and gills were aging tissues in the scallops. It has reported that gonad, hepatopancreas and gills of bivalves were the vital organs susceptible to environmental stress (Cannuel et al., 2009; Sun et al., 2014; Dell'Acqua et al., 2019). The increased expression of MEK1 in these tissues at 12 months scallops might have resulted from the complex environmental factors, suggesting that MEK1 may perform the same function under the adverse effects of environmental stress. Inhibiting MEK1 can upregulate the activity of SOD, an important protective enzyme against oxidative damage and thus neutralizing the adverse effects of ROS generated during aging (Subramanian and James, 2010; Mossa et al., 2015; Shi et al., 2021). Research carried out in another closely related short-lived scallop (Argopecten ventricosus, maximum lifespan potential only 2 years) revealed a decrease of SOD and CAT activities in tissues (such as adductor muscle, mantle, and gills) in the elderly population (Guerra et al., 2012). Therefore, we speculated that MEK1 may have promoted tissue senescence by affecting the antioxidant level of tissues in bay scallop.
Prolonging effects of nutritional restriction on animal lifespan has been well established in many experimental animals such as yeast, nematodes, flies, rodents, and primates since its first discovery in healthy rats in 1935 (McCay et al., 1989; Chiang et al., 2012; Laye et al., 2015; Lien et al., 2020). In this study, the expression of MEK1 can be inhibited by nutrient restriction in both species, leading to the upregulation of key genes such as antioxidant genes and those involved in DNA repair (Cho et al., 2016), while inhibiting the activity of the mTOR pathway (Gorentla et al., 2011; Yang et al., 2016). It seems that MEK1 might play a crucial role in the physiological response of the scallops to nutrient availability. MEK1 expression was consistently low in the hepatopancreas of the bay scallop at 30 and 56 days of nutritional restriction, possibly because the more sensitive to starvation. In addition, the expression level of MEK1 in adductor muscle and mantle increased with nutritional restriction on day 30 in A. irradians may be explained by the time of nutritional restriction was too short to derepress MEK1 expressions in mantle and adductor muscle. The decreased expression of MEK1 in gills and mantle rather than hepatopancreas of A. purpuratus on Day 56 of nutrient restriction might be a reflection of the fact that A. purpuratus usually maintain a relatively low level of metabolism in cold water high latitudes, and thus the hepatopancreas were less responsive to food limitations than gills and mantle. Hence, we speculate that MEK1 might also have the conserved function in aging by responding to nutrient availability in A. irradians and A. purpuratus. Research also reported that cold water temperatures and presumably food limitations might be favorable for longevity in bivalves by reducing generation of ROS (Estabrooks, 2007).
To further determine the functions of MEK1 on aging, the expressions of ERK1/2, FoxO, SOD, CAT, ULK1, mTORC1, S6K1, 4EBP1 and IF4E were detected after AiiMEK1 was knocked down by RNAi technology in A. irradians. The results showed that ERK1/2, the only downstream target gene of MEK1 (Olsen et al., 2014), which is known to affect a variety of physiological functions by regulating the phosphorylation level of various transcription factors, was significantly down regulated (Eapen et al., 2011). Rapid and almost complete downregulation of ERK1/2 activity by the inhibition of MEK1 has also been found in other studies (Chen et al., 2014; Lake et al., 2016). The FoxO gene encodes a key regulator of the IIS pathway, and its high expression helps improve the ability to fight oxidative stress in vitro, which in turn increases lifespan (Sun et al., 2017; Hou et al., 2020). The inhibited expression of AiiMEK1 resulted in a very significant increase in the transcription level of FoxO, suggesting that the effect of FoxO in prolonging lifespan may also be conserved in marine animals. Mitochondria of senescent cells produce ROS that can cause protein and lipid damage, lead to telomere shortening and activate DNA damage response. Inhibition of AiiMEK1 expression also significantly increased the relative expression of SOD, probably because FoxO protects quiescent cells from oxidative stress by inducing the expression of SOD (Kops et al., 2002; Liu et al., 2016). However, the relative expression of CAT was significantly downregulated, suggesting that protection of tissues from oxidative damage may be mainly undertaken by SOD in A. irradians (Chi et al., 2016). Studies have shown that mTOR signaling pathway can stimulate protein synthesis through S6K1 kinase and 4EBP1 (Laplante and Sabatini, 2012; Adamson et al., 2014), and inhibition of the mTOR signaling pathway by gene knockout, rapamycin treatment, or dietary restriction can delay aging (Lee et al., 2013; Chang et al., 2015; Dominick et al., 2017). In this study, the expression of mTORC1, S6K1 and 4EBP1 was also significantly decreased with the downregulation of AiiMEK1, implying that mTOR regulatory signals can be reduced by inhibition of AiiMEK1 and postpone the senescence of A. irradians. The physiological process of autophagy can remove toxic and easily aggregated proteins to maintain the normal function of cells and thus prolong life (Melendez et al., 2003; Simonsen et al., 2008; Park and Kim, 2019; Wang et al., 2021). In this study, ULK1, a key gene in autophagy, was significantly lower in the MEK1 knock-down group than that in the control group, might suggest that autophagy is not largely involved in the bay scallops. β-Gal, a widely used marker of cellular aging, is the gold standard for identifying senescence, and its activity increases with age (Shin et al., 2020). In this study, the β-Gal activity of the interference group with low expression of MEK1 was significantly lower than that of the control group, suggesting that MEK1 is tightly associated with antiaging in scallops. Based on the above results, it is thus hypothesized that MEK1 regulates the senescence of A. irradians by up-regulating the activity of mTOR pathway and down-regulating the expression of antioxidant and DNA damage repair genes through Raf/MEK/ERK cascade.
The three-layered protein kinase cascade Raf/MEK1/ERK, with MEK1 as a core member is widely involved in the regulation of many aging and life-span pathways such as mTOR and IIS. In terrestrial animals, it has been confirmed that inhibiting MEK1 can slow down cell aging by inhibiting ERK1/2, but whether MEK1 has similar function in marine invertebrates is not clear. In this study, we cloned and identified MEK1 gene from two closely related scallops with distinct lifespan. We further examined the expression profile of this gene in different tissues at different ages in both species and found that MEK1 expression increased with aging in all tissues. Nutritional restriction significantly decreased the expression of MEK1. In addition, knockdown of MEK1 expression by RNA interference could significantly affect the expression of aging-related genes and inhibit the activity of β-GAL. It is thus speculated that MEK1 is involved in regulating the senescence and life span of Argopecten scallops.
The original contributions presented in the study are publicly available. This data can be found here: GenBank, accession numbers OP490348 and OP490349.
XL and CW designed this study and contributed to the original concept of the project. KY performed the experiments. KY, XL, and WC analyzed the data and wrote the paper. JN, MC, QW, GL, XX, and HX provided the samples. All authors contributed to the article and approved the submitted version.
This project was financially supported by the National Natural Science Foundation of China (No. 31972791 and 42276147); the Earmarked Fund for Agriculture Seed Improvement Project of Shandong Province (No. 2020LZGC016); the Scientific and Technological Project of Yantai, Shandong Province (No. 2022XCZX083); the Earmarked Fund for Shandong Modern Agro-Industry Technology Research System (No. SDAIT-14); the Science and technology smes innovation capacity improvement project in Shandong Province (2021TSGC1229).
GL, XX, and HX were employed by CW. Authors GL, XX and CW were employed by Yantai Spring-Sea AquaSeed, Ltd. Author HX were employed by Jiangsu Baoyuan Biotechnology Co., Ltd.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelfatah S., Berg A., Bockers M., Efferth T. (2019). A selective inhibitor of the polo-box domain of polo-like kinase 1 identified by virtual screening. J. Adv. Res. 16, 145–156. doi: 10.1016/j.jare.2018.10.002
Adamson A. L., Le B. T., Siedenburg B. D. (2014). Inhibition of mTORC1 inhibits lytic replication of Epstein-Barr virus in a cell-type specific manner. Virol. J. 11, 110. doi: 10.1186/1743-422X-11-110
An S. W. A., Choi E. S., Hwang W., Son H. G., Yang J. S., Seo K., et al. (2019). KIN-4/MAST kinase promotes PTEN-mediated longevity of caenorhabditis elegans via binding through a PDZ domain. Aging Cell 18, e12906. doi: 10.1111/acel.12906
Arcidiacono D., Zaramella A., Fabris F., Sanchez-Rodriguez R., Nucci D., Fassan M., et al. (2021). Insulin/IGF-1 signaling is downregulated in barrett's esophagus patients undergoing a moderate calorie and protein restriction program: a randomized 2-year trial. Nutrients 13, 3638. doi: 10.3390/nu13103638
Asada S., Daitoku H., Matsuzaki H., Saito T., Sudo T., Mukai H., et al. (2007). Mitogen-activated protein kinases, erk and p38, phosphorylate and regulate Foxo1. Cell Signal 19, 519–527. doi: 10.1016/j.cellsig.2006.08.015
Baar E. L., Carbajal K. A., Ong I. M., Lamming D. W. (2016). Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice. Aging Cell 15, 155–166. doi: 10.1111/acel.12425
Benhamman R., Bai F., Drory S. B., Loubert-Hudon A., Ellis B., Matton D. P. (2017). The Arabidopsis mitogen-activated protein kinase kinase kinase 20 (MKKK20) acts upstream of MKK3 and MPK18 in two separate signaling pathways involved in root microtubule functions. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01352
Breitenbach T., Lorenz K., Dandekar T. (2019). How to steer and control ERK and the ERK signaling cascade exemplified by looking at cardiac insufficiency. Int. J. Mol. Sci. 20, 2179. doi: 10.3390/ijms20092179
Cannuel R., Beninger P. G., McCombie H., Boudry P. (2009). Gill development and its functional and evolutionary implications in the blue mussel Mytilus edulis (Bivalvia: Mytilidae). Biol. Bull. 217, 173–188. doi: 10.1086/BBLv217n2p173
Castello L., Maina M., Testa G., Cavallini G., Biasi F., Donati A., et al. (2011). Alternate-day fasting reverses the age-associated hypertrophy phenotype in rat heart by influencing the ERK and PI3K signaling pathways. Mech. Ageing Dev. 132, 305–314. doi: 10.1016/j.mad.2011.06.006
Chang G. R., Chiu Y. S., Wu Y. Y., Lin Y. C., Hou P. H., Mao F. C. (2015). Rapamycin impairs HPD-induced beneficial effects on glucose homeostasis. Br. J. Pharmacol. 172, 3793–3804. doi: 10.1111/bph.13168
Chen P. C., Yin J., Yu H. W., Yuan T., Fernandez M., Yung C. K., et al. (2014). Next-generation sequencing identifies rare variants associated with noonan syndrome. Proc. Natl. Acad. Sci. U.S.A. 111, 11473–11478. doi: 10.1073/pnas.1324128111
Chiang W. C., Ching T. T., Lee H. C., Mousigian C., Hsu A. L. (2012). HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148, 322–334. doi: 10.1016/j.cell.2011.12.019
Chi C., Giri S. S., Jun J. W., Kim H. J., Yun S., Kim S. G., et al. (2016). Marine toxin okadaic acid affects the immune function of bay scallop (Argopecten irradians). Molecules 21, 1108. doi: 10.3390/molecules21091108
Cho H. R., Kong Y. J., Hong S. G., Kim K. P. (2016). Hop2 and Sae3 are required for Dmc1-mediated double-strand break repair via homolog bias during meiosis. Mol. Cells 39, 550–556. doi: 10.14348/molcells.2016.0069
Degirmenci U., Wang M., Hu J. (2020). Targeting aberrant RAS/RAF/MEK/ERK signaling for cancer therapy. Cells 9, 198. doi: 10.3390/cells9010198
Dell'Acqua O., Ferrando S., Chiantore M., Asnaghi V. (2019). The impact of ocean acidification on the gonads of three key Antarctic benthic macroinvertebrates. Aquat. Toxicol. 210, 19–29. doi: 10.1016/j.aquatox.2019.02.012
Dominick G., Bowman J., Li X., Miller R. A., Garcia G. G. (2017). mTOR regulates the expression of DNA damage response enzymes in long-lived Snell dwarf, GHRKO, and PAPPA-KO mice. Aging Cell 16, 52–60. doi: 10.1111/acel.12525
Drummond M. J., Dreyer H. C., Pennings B., Fry C. S., Dhanani S., Dillon E. L., et al. (2008). Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J. Appl. Physiol. (1985) 104, 1452–1461. doi: 10.1152/japplphysiol.00021.2008
Eapen A., Ramachandran A., Pratap J., George A. (2011). Activation of the ERK1/2 mitogen-activated protein kinase cascade by dentin matrix protein 1 promotes osteoblast differentiation. Cells Tissues Organs 194, 255–260. doi: 10.1159/000324258
Estabrooks S. L. (2007). The possible role of telomeres in the short life span of the bay scallop, Argopecten irradians irradians (Lamarck 1819). J. Shellfish Res. 26, 307–313. doi: 10.2983/0730-8000(2007)26[307:Tproti]2.0.Co;2
Fan Q., Wang Q., Cai R., Yuan H., Xu M. (2020). The ubiquitin system: orchestrating cellular signals in non-small-cell lung cancer. Cell Mol. Biol. Lett. 25, 1. doi: 10.1186/s11658-019-0193-6
Fontana L., Partridge L., Longo V. D. (2010). Extending healthy life span–from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
Goldberg E. L., Romero-Aleshire M. J., Renkema K. R., Ventevogel M. S., Chew W. M., Uhrlaub J. L., et al. (2015). Lifespan-extending caloric restriction or mTOR inhibition impair adaptive immunity of old mice by distinct mechanisms. Aging Cell 14, 130–138. doi: 10.1111/acel.12280
González M. L., Pérez M. C., López D. A. (2002). Breeding cycle of the northern scallop, Argopecten purpuratus (Lamarck 1819) in southern Chile. Aquacult. Res. 33, 847–852. doi: 10.1046/j.1365-2109.2002.00721.x
Gorentla B. K., Wan C. K., Zhong X. P. (2011). Negative regulation of mTOR activation by diacylglycerol kinases. Blood 117, 4022–4031. doi: 10.1182/blood-2010-08-300731
Guerra C., Zenteno-Savin T., Maeda-Martinez A. N., Philipp E. E., Abele D. (2012). Changes in oxidative stress parameters in relation to age, growth and reproduction in the short-lived catarina scallop argopecten ventricosus reared in its natural environment. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 162, 421–430. doi: 10.1016/j.cbpa.2012.04.018
Han J., Liu Y., Yang S., Wu X., Li H., Wang Q. (2021). MEK inhibitors for the treatment of non-small cell lung cancer. J. Hematol. Oncol. 14, 1. doi: 10.1186/s13045-020-01025-7
Hou W., Pei J., Wang Y., Zhang J., Zheng H., Cui R. (2020). Anti-ageing effects of red ginseng on female Drosophila melanogaster. J. Cell Mol. Med. 24, 3751–3755. doi: 10.1111/jcmm.15029
Ito M., Yurube T., Kanda Y., Kakiuchi Y., Takeoka Y., Takada T., et al. (2021). Inhibition of autophagy at different stages by ATG5 knockdown and chloroquine supplementation enhances consistent human disc cellular apoptosis and senescence induction rather than extracellular matrix catabolism. Int. J. Mol. Sci. 22, 3965. doi: 10.3390/ijms22083965
Jin S., Gao J., Qi Y., Hao Y., Li X., Liu Q., et al. (2020). TGF-beta1 fucosylation enhances the autophagy and mitophagy via PI3K/Akt and ras-Raf-MEK-ERK in ovarian carcinoma. Biochem. Biophys. Res. Commun. 524, 970–976. doi: 10.1016/j.bbrc.2020.02.028
Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. doi: 10.1038/366461a0
Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., et al. (2002). Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321. doi: 10.1038/nature01036
Lake D., Correa S. A., Muller J. (2016). Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol. Life Sci. 73, 4397–4413. doi: 10.1007/s00018-016-2297-8
Laplante M., Sabatini D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
Laye M. J., Tran V., Jones D. P., Kapahi P., Promislow D. E. (2015). The effects of age and dietary restriction on the tissue-specific metabolome of Drosophila. Aging Cell 14, 797–808. doi: 10.1111/acel.12358
Lee K. M., Hwang S. K., Lee J. A. (2013). Neuronal autophagy and neurodevelopmental disorders. Exp. Neurobiol. 22, 133–142. doi: 10.5607/en.2013.22.3.133
LeRoith D., Werner H., Beitner-Johnson D., Roberts C. T. Jr. (1995). Molecular and cellular aspects of the insulin-like growth factor I receptor. Endocr. Rev. 16, 143–163. doi: 10.1210/edrv-16-2-143
Lian S., Wang J., Zhang L., Xing Q., Hu N., Liu S., et al. (2019). Iness itegration of biochemical, cellular, and genetic indicators for understanding the aging procn a bivalve mollusk Chlamys farreri. Mar. Biotechnol. (NY) 21, 718–730. doi: 10.1007/s10126-019-09917-7
Lien W. Y., Chen Y. T., Li Y. J., Wu J. K., Huang K. L., Lin J. R., et al. (2020). Lifespan regulation in alpha/beta posterior neurons of the fly mushroom bodies by Rab27. Aging Cell 19, e13179. doi: 10.1111/acel.13179
Lim S. L., D'Agui H. M., Enright N. J., He T. (2017). Characterization of leaf transcriptome in banksia hookeriana. Genomics Proteomics Bioinf. 15, 49–56. doi: 10.1016/j.gpb.2016.11.001
Liu X., Cai X., Hu B., Mei Z., Zhang D., Ouyang G., et al. (2016). Forkhead transcription factor 3a (FOXO3a) modulates hypoxia signaling via up-regulation of the von hippel-lindau gene (VHL). J. Biol. Chem. 291, 25692–25705. doi: 10.1074/jbc.M116.745471
Livak K. J., Schmittgen T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luckhart S., Riehle M. A. (2007). The insulin signaling cascade from nematodes to mammals: insights into innate immunity of Anopheles mosquitoes to malaria parasite infection. Dev. Comp. Immunol. 31, 647–656. doi: 10.1016/j.dci.2006.10.005
Lu S., Huang W., Wang Q., Shen Q., Li S., Nussinov R., et al. (2014). The structural basis of ATP as an allosteric modulator. PloS Comput. Biol. 10, e1003831. doi: 10.1371/journal.pcbi.1003831
McCay C. M., Crowell M. F., Maynard L. A. (1989). The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition 5, 155–171. doi: 10.1093/JN10.1.63
Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391. doi: 10.1126/science.1087782
Miao Y., Laun T. M., Smykowski A., Zentgraf U. (2007). Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. doi: 10.1007/s11103-007-9198-z
Mohanta T. K., Yadav D., Khan A., Hashem A., Tabassum B., Khan A. L., et al. (2020). Genomics, molecular and evolutionary perspective of NAC transcription factors. PloS One 15, e0231425. doi: 10.1371/journal.pone.0231425
Mossa A. T., Heikal T. M., Belaiba M., Raoelison E. G., Ferhout H., Bouajila J. (2015). Antioxidant activity and hepatoprotective potential of cedrelopsis grevei on cypermethrin induced oxidative stress and liver damage in male mice. BMC Complement Altern. Med. 15, 251. doi: 10.1186/s12906-015-0740-2
Newell Stamper B. L., Cypser J. R., Kechris K., Kitzenberg D. A., Tedesco P. M., Johnson T. E. (2018). Movement decline across lifespan of Caenorhabditis elegans mutants in the insulin/insulin-like signaling pathway. Aging Cell 17, e12704. doi: 10.1111/acel.12704
Ning J., Cao W., Lu X., Chen M., Liu B., Wang C. (2021). Identification and functional analysis of a sex-biased transcriptional factor Foxl2 in the bay scallop argopecten irradians irradians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 256, 110638. doi: 10.1016/j.cbpb.2021.110638
Ni T., Wei G., Shen T., Han M., Lian Y., Fu H., et al. (2015). MitoRCA-seq reveals unbalanced cytocine to thymine transition in polg mutant mice. Sci. Rep. 5, 12049. doi: 10.1038/srep12049
Olsen K. C., Epa A. P., Kulkarni A. A., Kottmann R. M., McCarthy C. E., Johnson G. V., et al. (2014). Inhibition of transglutaminase 2, a novel target for pulmonary fibrosis, by two small electrophilic molecules. Am. J. Respir. Cell Mol. Biol. 50, 737–747. doi: 10.1165/rcmb.2013-0092OC
Park H. J., Kim M. M. (2019). Amentoflavone induces autophagy and modulates p53. Cell J. 21, 27–34. doi: 10.22074/cellj.2019.5717
Qu Z., Ji S., Zheng S. (2020). BRAF controls the effects of metformin on neuroblast cell divisions in C. elegans. Int. J. Mol. Sci. 22, 178. doi: 10.3390/ijms22010178
Ridgway I. D., Richardson C. A., Austad S. N. (2011). Maximum shell size, growth rate, and maturation age correlate with longevity in bivalve molluscs. J. Gerontol. A Biol. Sci. Med. Sci. 66, 183–190. doi: 10.1093/gerona/glq172
Rojas-Morales P., Pedraza-Chaverri J., Tapia E. (2020). Ketone bodies, stress response, and redox homeostasis. Redox Biol. 29, 101395. doi: 10.1016/j.redox.2019.101395
Roskoski R. Jr. (2012). MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem. Biophys. Res. Commun. 417, 5–10. doi: 10.1016/j.bbrc.2011.11.145
Rouquette-Jazdanian A. K., Sommers C. L., Kortum R. L., Morrison D. K., Samelson L. E. (2012). LAT-independent erk activation via Bam32-PLC-gamma1-Pak1 complexes: GTPase-independent Pak1 activation. Mol. Cell 48, 298–312. doi: 10.1016/j.molcel.2012.08.011
Roy S. K., Srivastava R. K., Shankar S. (2010). Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J. Mol. Signal 5, 10. doi: 10.1186/1750-2187-5-10
Shen X., Song S., Li C., Zhang J. (2022). Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature 606, 7915. doi: 10.1038/s41586-022-04823-w
Shi Q., Lang W., Wang S., Li G., Bai X., Yan X., et al. (2021). Echinacea polysaccharide attenuates lipopolysaccharideinduced acute kidney injury via inhibiting inflammation, oxidative stress and the MAPK signaling pathway. Int. J. Mol. Med. 47, 243–255. doi: 10.3892/ijmm.2020.4769
Shin E. Y., Park J. H., You S. T., Lee C. S., Won S. Y., Park J. J., et al. (2020). Integrin-mediated adhesions in regulation of cellular senescence. Sci. Adv. 6, eaay3909. doi: 10.1126/sciadv.aay3909
Simonsen A., Cumming R. C., Brech A., Isakson P., Schubert D. R., Finley K. D. (2008). Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. doi: 10.4161/auto.5269
Slack C., Alic N., Foley A., Cabecinha M., Hoddinott M. P., Partridge L. (2015). The ras-Erk-ETS-signaling pathway is a drug target for longevity. Cell 162, 72–83. doi: 10.1016/j.cell.2015.06.023
Storz J. F., Natarajan C., Signore A. V., Witt C. C., McCandlish D. M., Stoltzfus A. (2019). The role of mutation bias in adaptive molecular evolution: insights from convergent changes in protein function. Philos. Trans. R Soc. Lond B Biol. Sci. 374, 20180238. doi: 10.1098/rstb.2018.0238
Subramanian M. V., James T. J. (2010). Age-related protective effect of deprenyl on changes in the levels of diagnostic marker enzymes and antioxidant defense enzymes activities in cerebellar tissue in wistar rats. Cell Stress Chaperones 15, 743–751. doi: 10.1007/s12192-010-0177-y
Sun X., Chen W. D., Wang Y. D. (2017). DAF-16/FOXO transcription factor in aging and longevity. Front. Pharmacol. 8. doi: 10.3389/fphar.2017.00548
Sun Z., Yang C., Wang L., Wang X., Wang J., Yue F., et al. (2014). The protein expression profile in hepatopancreas of scallop Chlamys farreri under heat stress and Vibrio anguillarum challenge. Fish Shellfish Immunol. 36, 252–260. doi: 10.1016/j.fsi.2013.11.008
Tia N., Singh A. K., Pandey P., Azad C. S., Chaudhary P., Gambhir I. S. (2018). Role of forkhead box O (FOXO) transcription factor in aging and diseases. Gene 648, 97–105. doi: 10.1016/j.gene.2018.01.051
Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., et al. (2010). An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354. doi: 10.1016/j.cell.2010.03.031
Ungvari Z., Csiszar A., Sosnowska D., Philipp E. E., Campbell C. M., McQuary P. R., et al. (2013). Testing predictions of the oxidative stress hypothesis of aging using a novel invertebrate model of longevity: the giant clam (Tridacna derasa). J. Gerontol. A Biol. Sci. Med. Sci. 68, 359–367. doi: 10.1093/gerona/gls159
Waller T. R. (1969). The evolution of the Argopecten gibbus stock (Mollusca: Bivalvia), with emphasis on the tertiary and quaternary species of eastern north America. J. Paleontol. 43, 1–125. doi: 10.1017/S0022336000062466
Walsh I. M., Bowman M. A., Soto Santarriaga I. F., Rodriguez A., Clark P. L. (2020). Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. U.S.A. 117, 3528–3534. doi: 10.1073/pnas.1907126117
Wang Y., Lu X., Wang C., Ning J., Chen M., Yuan K. (2022). Potential roles of FoxO in promoting longevity in larger Argopecten scallops. Aquaculture 561, 738690. doi: 10.1016/j.aquaculture.2022.738690
Wang Y., Pan Y., Liu Y., Disasa D., Akira M., Xiang L., et al. (2021). A new geniposidic acid derivative exerts antiaging effects through antioxidative stress and autophagy induction. Antioxid. (Basel) 10, 987. doi: 10.3390/antiox10060987
Wang J., Raskin L., Samuels D. C., Shyr Y., Guo Y. (2015). Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics 31, 318–323. doi: 10.1093/bioinformatics/btu668
Webb A. E., Brunet A. (2013). FOXO flips the longevity SWItch. Nat. Cell Biol. 15, 444–446. doi: 10.1038/ncb2749
Wu P. K., Becker A., Park J. I. (2020). Growth inhibitory signaling of the Raf/MEK/ERK pathway. Int. J. Mol. Sci. 21, 5436. doi: 10.3390/ijms21155436
Xu H., Lu X., Wang C., Ning J., Chen M., Wang Y., et al. (2022). Potential roles of pten on longevity in two closely related Argopecten scallops with distinct lifespans. Front. Physiol. 13. doi: 10.3389/fphys.2022.872562
Yang J., Zhang P., Krishna S., Wang J., Lin X., Huang H., et al. (2016). Unexpected positive control of NFkappaB and miR-155 by DGKalpha and zeta ensures effector and memory CD8+ T cell differentiation. Oncotarget 7, 33744–33764. doi: 10.18632/oncotarget.8164
Yang J. Y., Zong C. S., Xia W., Yamaguchi H., Ding Q., Xie X., et al. (2008). ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 10, 138–148. doi: 10.1038/ncb1676
Yu Y., Zhang X., Liu F., Zhu P., Zhang L., Peng Y., et al. (2021). A stress-induced miR-31–CLOCK–ERK pathway is a key driver and therapeutic target for skin aging. Nat. Aging 1, 795–809. doi: 10.1038/s43587-021-00094-8
Zhang F., He Y., Yang H. (2000). Introduction engineering of bay scallop and its comprehensive effects. Eng. Sci. 2, 30–35.
Keywords: bivalve, Argopecten scallops, MEK1, aging, nutrition restriction
Citation: Yuan K, Lu X, Ning J, Chen M, Wang Q, Liu G, Xu X, Xu H and Wang C (2022) Potential roles of MEK1 on aging in Argopecten scallops. Front. Mar. Sci. 9:1057814. doi: 10.3389/fmars.2022.1057814
Received: 30 September 2022; Accepted: 31 October 2022;
Published: 18 November 2022.
Edited by:
Hongyu Ma, Shantou University, ChinaCopyright © 2022 Yuan, Lu, Ning, Chen, Wang, Liu, Xu, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Lu, eGlhbHVAeWljLmFjLmNu; Chunde Wang, Y2hlbmRld2FuZzIwMDdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.