
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 13 December 2022
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1050386
 Jiehong Wei1,2,3
Jiehong Wei1,2,3 Renxie Wu4
Renxie Wu4 Laith A. Jawad5
Laith A. Jawad5 Yongshuang Xiao1,2
Yongshuang Xiao1,2 Kar-Hoe Loh6
Kar-Hoe Loh6 Angel Herrera-Ulloa7
Angel Herrera-Ulloa7 Yajun Wang8
Yajun Wang8 Jing Liu1,2*
Jing Liu1,2* Kuidong Xu1,2*
Kuidong Xu1,2*Due to the highly similar external morphology of the Pampus species, misidentifications frequently occur and hinder the understanding of the taxonomy and species distributions of the genus. In this study, we generated 271 mitochondrial sequences and obtained 1,226 sequences from the public databases to understand the species diversity and distributions of the genus Pampus. Most phylogenetic analyses and species delimitations congruently concluded seven valid species within the genus Pampus (i.e., P. argenteus, P. candidus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus). Sequences of P. argenteus are reported through the northern South China Sea to the Japan Archipelago, which covered the type locality of P. echinogaster. Sequences of P. echinogaster in the public databases are all identified as P. argenteus, suggesting that the species should be synonymized with P. argenteus. Furthermore, sequences of P. griseus were identical to our P. cinereus data and, therefore, should be treated as a synonym of the latter. Inference on divergence time and ancestral distribution implied that the genus Pampus originated in the central Indo-Pacific region around 8.35–11.33 million years ago (the late Miocene), associated with the rise of the Indonesian–Australian Archipelago biodiversity hotspot. The divergence between P. cinereus and P. liuorum dated back to 1.20–1.72 million years ago, which might be related to glacial isolation during the Mid-Pleistocene transition.
The genus Pampus Bonaparte, 1834, of the family Stromateidae Rafinesque, 1810, is distinct in having a compressed, silvery body, elongated pelvic bone, absence of pelvic fins, and more than five blade-like spines before their falcate dorsal and anal fins [but spine is absent in Pampus chinensis (Euphrasen, 1788), Liu et al., 2002; Liu et al., 2013a]. Members of the genus Pampus are commercially important fishes in the Indo-West Pacific area (Liu et al., 2002; Radhakrishnan et al., 2019; Roul et al., 2021). There have been concerted efforts to study their reproductive biology, evolution, and population genetics of the Pampus species (e.g., Dadzie et al., 2000; Gupta, 2020; Fan et al., 2022). Nevertheless, the chaotic taxonomies and misidentifications have hindered our understanding of the biology and evolution of this genus (Liu et al., 2013a; Radhakrishnan et al., 2019; Wei et al., 2021).
Due to the highly similar external morphology of the Pampus species, misidentifications frequently occur and greatly affect the understanding of the taxonomy and species distributions of the genus (Wei et al., 2021). Haedrich (1967) recognized three species in the genus Pampus, i.e., Pampus argenteus (Euphrasen, 1788), Pampus echinogaster (Basilewsky, 1855), and P. chinensis, and assigned many species names as synonyms of the three species. With recent efforts, Pampus cinereus (Bloch, 1795), Pampus candidus (Cuvier, 1829), and Pampus punctatissimus (Temminck & Schlegel, 1845) are resurrected as valid species; Pampus liuorum Liu & Li, 2013, and Pampus minor Liu and Li, 1998, are published as new species (Liu and Li, 1998a; Liu and Li, 1998b; Liu et al., 2013a; Liu et al., 2013b; Liu and Li, 2013), demonstrating that the genus Pampus is actually a more diverse group. Our previous study, Wei et al. (2021), confirmed the validities of seven species in the genus Pampus, viz., P. argenteus, P. candidus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus. However, validities of several species names, e.g., P. echinogaster and Pampus nozawae (Ishikawa, 1904), and Pampus griseus (Cuvier, 1833), remained unclear. Haedrich (1967) considered P. echinogaster as a species only found in China, Korea, and Japan, which is distinct from P. argenteus widely distributed in the Indo-West Pacific. The idea is widely used by taxonomic works, and for decades, the two species have been treated as different valid species (e.g., Lindberg and Krasyukova, 1989; Liu et al., 2002; Nakabo, 2002). Although morphological comparison indicated that P. echinogaster is nearly identical to the P. argenteus neotype except in a few metrical characters, the two species are still treated as different valid species by Liu et al. (2013a) because of the proposed north–south difference in their distributions: P. argenteus was considered distributed in offshore Guangdong and probably from Southeast Asia (Liu et al., 2013a); P. echinogaster was considered never found southward to the Taiwan Strait (Nakabo, 2002). Li et al. (2017) also proposed P. echinogaster as a valid species in the Northwest Pacific, but the fin ray, gill raker, and vertebral counts of their specimens overlap with the neotype of P. argenteus designated by Liu et al. (2013a). P. nozawae is published based on a lost type specimen from Hokkaido, northern Japan. The name has been used by several taxonomic works and is recently validated by Jawad and Liu (2017) based on a comparison of the axial skeleton. Although Jawad and Liu (2017) stated several differences between P. nozawae and its congeners, the examined specimens are collected from Southeast Asia instead of Japan, which presumed P. nozawae as a species broadly distributed through Southeast Asia to Japan. Roul et al. (2021) resurrected P. griseus (Cuvier 1833) and assumed that P. griseus inhabits the Indian Ocean, whereas its close relative, P. cinereus, only inhabits the western Pacific, although it is incongruent with the statement by Bloch and Schneider (1801) that P. cinereus inhabits waters around Tranquebar (i.e., Tharangambadi, India). Obviously, taxonomic issues of these Pampus species are accompanied by potential confusion on species distributions. A thorough review of global species diversity and distribution of the genus Pampus is necessary.
Molecular tools have been utilized to distinguish the Pampus species and studied their phylogeny and population genetics (e.g., Sun et al., 2012; Li et al., 2019; Roul et al., 2021). Over 1,500 DNA sequences of the genus Pampus have been deposited on GenBank and the Barcoding of Life Database (BOLD) with well-documented sampling localities in the databases and published studies, which provided a suitable dataset for studying species diversity and distribution of the genus Pampus. However, misidentification of the Pampus sequences frequently occurs (Roul et al., 2021; Wei et al., 2021), which greatly impedes the progress to clarify their taxonomy and distributions. In our previous study, Wei et al. (2021) provided reliable barcodes of cytochrome oxidase unit I (COXI) and cytochrome b (Cytb) from 74 specimens of seven Pampus species (i.e., P. argenteus, P. candidus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus). The specimens have been properly identified morphologically based on taxonomic works of the genus (i.e., Liu and Li, 1998a; Liu et al., 2013b; Liu et al., 2013a; Liu et al., 2013b; Radhakrishnan et al., 2019). A comparison of these barcodes with the global sequence data could provide valuable insight into the species diversity and distribution of the genus Pampus. We performed genetic comparisons on the mitochondrial sequences of the Pampus species from this study, Wei et al. (2021), GenBank, and BOLD to clarify the global species diversity and distribution of the genus Pampus. Inference on divergence times and ancestral distribution were also performed to provide a new perspective on the origin of the Pampus species.
A workflow for clarifying species diversity and distributions of the genus Pampus is illustrated in Figure 1. In brief, we used the properly identified sequences from this study and Wei et al. (2021) to identify the public sequence data and explore the global species diversity of the genus Pampus. Sampling localities of the sequences identified to species level were plotted on a map to illustrate the distributions of the Pampus species.
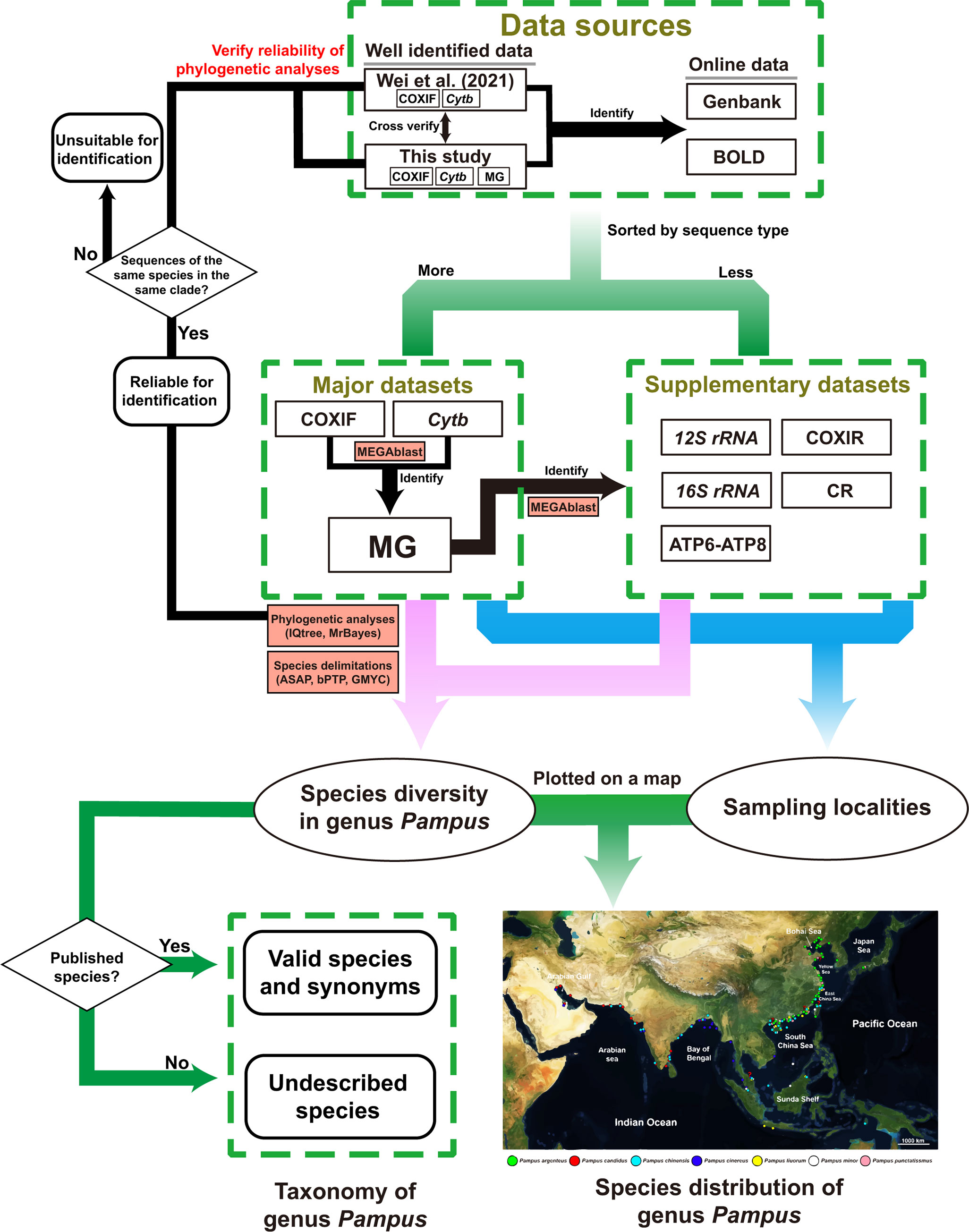
Figure 1 A workflow for exploring the species diversity and distributions of genus Pampus using the global mitochondrial data.
From 2009 to 2021, a muscle sample inventory of the Pampus species containing 209 muscle tissue samples from China, Malaysia, India, Iraq, Kuwait, and Bahrain was established in the Institute of Oceanology, Chinese Academy of Sciences (IOCAS). Most of the samples have either voucher specimens deposited in IOCAS or specimen photos taken upon collection (e.g., Figure 2). Among the 209 samples, 74 had been used in our previous study (Wei et al., 2021). COXI and Cytb sequences of the other 135 samples were obtained in this study using Sanger sequencing. Total genomic DNA was extracted from the muscle samples using TaKaRa MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa Inc., Mountain View, CA, USA) following the product manual. Polymerase chain reactions (PCRs) and Sanger sequencing were performed by Tsingke Ltd. (Qingdao, China). In addition to the 135 samples, mitogenome (MG) sequences were also obtained from muscle tissues of five additional specimens of P. argenteus (specimen numbers: 20120541 and 20181206004), P. liuorum (specimen numbers: 20120541 and 20181206004), and P. punctatissimus (specimen number: 20181206007). The total genomic DNA of the muscle tissues was also extracted using TaKaRa MiniBEST Universal Genomic DNA Extraction Kit. The extractions were sent to SCGene Ltd. (Guangzhou, China) for next-generation sequencing. All the details regarding DNA extraction, primers, polymerase chain reaction, sequencing methods, and assembly of the COXI, Cytb, and mitogenomes are given in Appendix S1. All the samples were identified by J. Liu and J. Wei based on our previous taxonomic works (Liu and Li, 1998a; Liu and Li, 1998b; Liu et al., 2013a; Liu et al., 2013b; Liu and Li, 2013). The obtained sequences were compared with Wei et al. (2021) to ascertain correct identification. The well-identified local data was used as the main reference data to identify the online mitochondrial data of the Pampus species.

Figure 2 Photographs of the seven valid Pampus species. (A) Pampus argenteus (20120469). (B) Pampus candidus (IOCASF000292). (C) Pampus chinensis (201006001). (D) Pampus cinereus (20120481). (E) Pampus liuorum (HBRF03-036). (F) Pampus minor (20120502). (G) Pampus punctatissimus (IOCASF000289).
Mitochondrial sequences from GenBank and BOLD were collected by searching with the keyword “Pampus” in combination with the names of mitochondrial markers, e.g., “cytochrome c oxidase subunit I”, “cytochrome b”, “control region”, and “mitochondrion”. Sequences of the other stromateids (i.e., genera Stromateus Linnaeus, 1758 and Peprilus Cuvier, 1829) were also collected from GenBank as outgroups for molecular analyses.
All the sequences were sorted into datasets by sequence types in Geneious Prime® v2022.1.1 (Biomatters Ltd., Auckland, New Zealand). Because the COXI dataset contained sequences located in different gene regions, we split the COXI dataset by their regions, namely, the COXIF (the forward region of COXI) and COXIR (the rear region of COXI) datasets. Due to extensive unaligned regions and insertions in non-coding genes [e.g., 12S ribosomal RNA (12S rRNA) and 16S ribosomal RNA (16S rRNA)] and control region (CR) of the mitogenomes, we only used their protein-coding genes to form the MG dataset for molecular analyses, i.e., ATP synthase membrane subunit 6 (ATP6), ATP synthase membrane subunit 8 (ATP8), COXI, cytochrome c oxidase subunit II (COXII), cytochrome c oxidase subunit III (COXIII), Cytb, and NADH-ubiquinone oxidoreductase chain 1–6 and 4L (ND1–6 and ND4L). The protein-coding gene sequences of each mitogenome were extracted in the Geneious Prime and concatenated in the SequenceMatrix v1.7.8 (Vaidya et al., 2011). Information on the datasets is summarized in Table 1 and detailed with accession number, sampling localities, and corresponding references in Appendix S2. The major datasets (i.e., the COXIF, Cytb, and MG datasets) contained more sequences, and sampling localities were used for the molecular analyses and studying the distributions of the Pampus species, while the supplementary datasets with unique sampling localities were retained for studying species distributions (Table 1). Sampling localities corresponding to the sequences were plotted on a map to illustrate the distributions of the Pampus species (Figure 3).
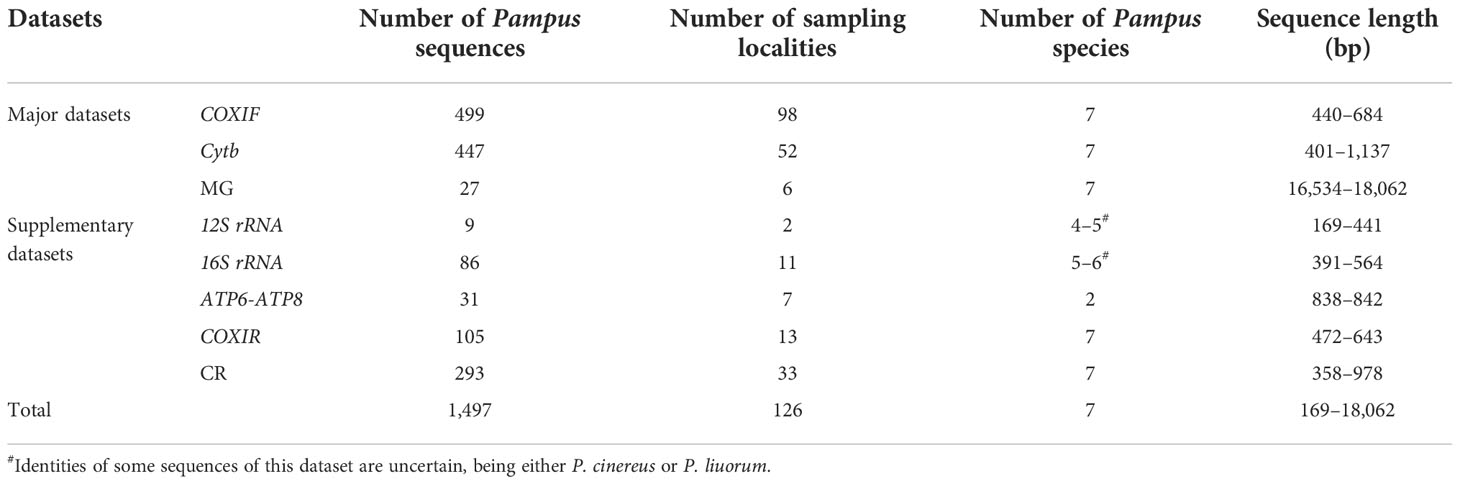
Table 1 Information from the eight datasets collated from 1,497 mitochondrial sequences of genus Pampus used in this study.
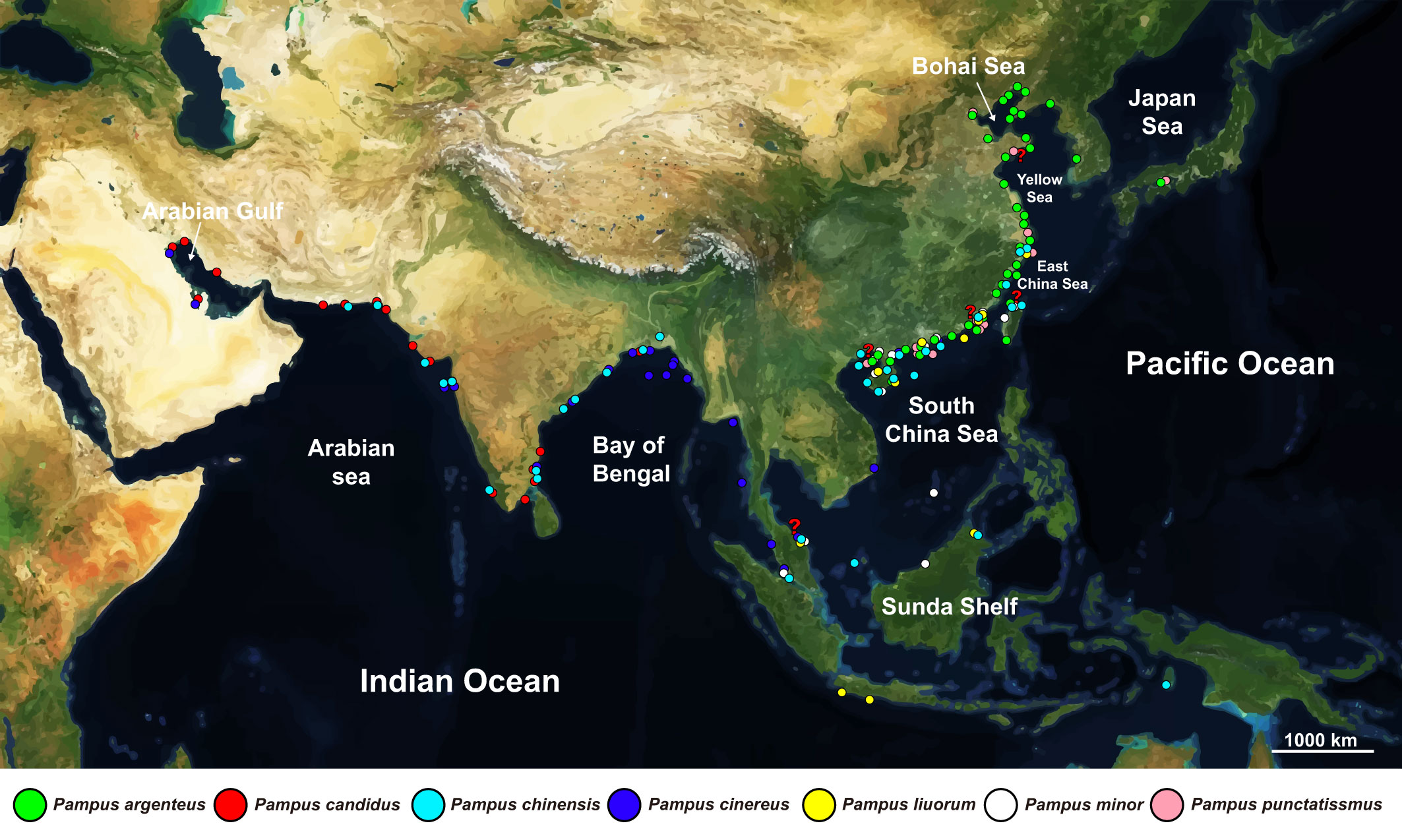
Figure 3 Distributions of the Pampus species based on sampling localities of the eight mitochondrial datasets used in this study. The basic map is generated in Mapbox (https://www.mapbox.com/). Spots on map denote sampling localities of the seven valid species. Geographically close localities are merged into one spot. Red question marks denote that Pampus candidus might not distribute in these sampling localities.
Alignments of the datasets were performed using the MAFFT v7.388 (Katoh and Stanley, 2013) plugin in Geneious Prime with default settings; external gaps were treated as missing data. Any protein-coding gene sequences with reading frameshift, extensive regions of indels, or unaligned nucleotides were excluded from the datasets. The datasets were then compared with GenBank sequences using the MegaBLAST (Morgulis et al., 2008) plugin in Geneious Prime® (Biomatters Ltd.); the first 20 hits with over 98% identical sites were used for preliminary identification. Sequences of non-stromateid species were excluded from the datasets.
Species identifications were mainly performed using the MegaBLAST plugin to compare the sequences with a local database set up in Geneious Prime. The first 20 hits with over 98% identical sites were used for identification. The local database was mainly formed by the well-identified COXIF, Cytb, and mitogenome sequences from this study and Wei et al. (2021). The mitogenomes from public data were also added to the local database after proper identification. Due to the lack of corresponding reference sequences from this study and Wei et al. (2021), sequences of the supplementary datasets were identified based on the mitogenomic data of the local database. Wei et al. (2021) indicated that COXIF sequences of P. cinereus and P. liuorum have very few differences, which might affect the ability of MegaBLAST to identify the two species. Therefore, COXIF sequences of the two species were identified based on the maximum likelihood (ML) tree.
Phylogenetic analyses and species delimitations were performed on the major datasets (Table 1). Whenever possible, ML trees of the supplementary datasets were also generated to eliminate potential misidentification by MegaBLAST. The ML trees are given in Appendix S3. Sequence variation indices of the major datasets (i.e., number of polymorphic sites, parsimony informative sites, and indels) were calculated using DnaSP v6 (Rozas et al., 2017). In the subsequently mentioned analyses, each gene of the MG dataset was treated as a partition. Substitution saturation of the datasets was examined on the three-codon position using the method in DAMBE v7.2.25 of Xia et al. (2003) (Xia, 2018). Best-fit substitution models were selected using jModelTest v2.1.10 (Darriba et al., 2012) based on the Bayesian information criterion (Schwarz, 1978). The results of saturation and model tests are given in Appendix S4. ML trees were inferred in IQtrees v1.6.12 (Nguyen et al., 2015), with 1,000 bootstrap (BS) replicates to estimate the BS values of nodes. For Bayesian inference (BI) trees, two independent Markov chain Monte Carlo (MCMC) runs were performed in MrBayes v3.2 (Ronquist et al., 2012), with four chains for 500,000 generations, sampling every 100 generations. The maximum clade credibility tree of the two runs was generated in MrBayes with the first 25% of samples discarded as burn-in.
Species delimitations were performed using the Assemble Species by Automatic Partitioning (ASAP; Puillandre et al., 2021), single threshold Bayesian Poisson tree processes (bPTP; Zhang et al., 2013), and generalized mixed Yule coalescent (GMYC; Pons et al., 2006; Fontaneto et al., 2007; Fujisawa and Barraclough, 2013) methods in the itaxoTools package v6.2.1 (Vences et al., 2021). All the analyses were performed with the default setting in itaxoTools following the instruction manual (Vences et al., 2021), except that genetic distances were calculated with Kimura’s two-parameter model (Kimura, 1980) instead of the default Jukes–Cantor model distance (Jukes and Cantor, 1967) in the ASAP. Notably, the ASAP of the MG dataset was deployed to each gene partition of the MG dataset, while the bPTP and GMYC were performed using trees generated with the whole MG dataset. The ML trees of the major datasets (Table 1) were used as phylogram input for the bPTP. Ultrametric trees for the GMYC of the major datasets were generated in BEAST v1.10.4 (Drummond and Rambaut, 2007). Two MCMC runs for 10,000,000 generations of each dataset were conducted with the best-fit substitution model and sampling every 1,000 generations. The constant population size model was applied following the instruction of the GMYC (Fujisawa and Barraclough, 2013). Sufficient convergences and burn-in of the runs were checked in Tracer v1.7 (http://tree.bio.ed.ac.uk/software/tracer), ascertaining that the effective sampling sizes of all the parameters were greater than 200. A maximum clade credibility tree (i.e., ultrametric tree) was generated using TreeAnnotator v1.10.4, discarding the first 10% sample as burn-in.
A recent phylogenetic study recovered the sister relationship of the genera Pampus and Stromateus (Arcila et al., 2021). The two genera distribute on either side of the Arabian Peninsula, where the ancient Tethys Seaway was situated. The only fossil genus of the Stromateidae, Pinichthys Bannikov, 1985, was also discovered around the Tethys Seaway (i.e., the Paratethys, Bannikov, 1985). These studies indicate that the stromateids might have a Tethyan origin, and the closure of the Tethys Seaway might have resulted in the isolation and divergence of Stromateus and Pampus. Therefore, we used the closure time of the Tethys Seaway [14.00 million years ago (Mya), Hamon et al., 2013] as the lower limit and the age of the oldest Pinichthys fossil (i.e., Pinichthys pulcher Bannikov, 1988, 32.00–33.70 Mya, the age of its sampling layer, Bannikov, 1988; Popov et al., 2019) as the upper limits to calibrate the time to most recent common ancestor (tMRCA) of Stromateus and Pampus. Due to the lack of mitogenome of the Stromateus species, we estimated the calibrated COXI substitution rate using an additional COXI dataset (the COXIA dataset, Appendix S3) including representative sequences of the Stromateus and Pampus species. The calibrated COXI rate was used for divergence inference. For substitution rate calibration, the random local clock and birth-death process model was selected as the best-fit clock and tree models based on the comparison of Bayes factors, estimated by the path sampling method in BEAST (Baele et al., 2012, Appendix S5). The fossil calibration was applied to the tMRCA of Stromateus and Pampus with an exponential distribution [mean = 5.34, offset = 14.00, 95% highest posterior density interval (HPD95): 14.14–33.7 Mya]. Two independent MCMC runs were conducted with 10,000,000 generations in BEAST v1.10.4 (Drummond and Rambaut, 2007), sampling every 1,000 generations. Sufficient convergence and burn-in of runs were checked with Tracer v1.7 and effective sampling size (ESS) values (above 200). The log file of the two runs was combined in LogCombiner v1.10.4 of the BEAST package, discarding the first 10% of samples as burn-in. The probability density of the calibrated COXI substitution rate was estimated from the combined log file using Tracer v1.7.
To save computational time, a representative MG sequence of each identified Pampus species and the outgroup Peprilus triacanthus (Peck, 1804) was selected to form the reduced MG dataset for divergence time inference. Following substitution rate calibration, the random local clock and the birth-death process model were also applied. The calibrated COXI substitution rate was applied using a normal distribution (mean = 0.0019, standard deviation = 0.0028), covering HPD95 of the rate. Substitution rates of other genes were estimated in BEAST through the MCMC runs. The ML tree of the reduced MG dataset was used as the input tree for BEAST, and the topology was fixed through the MCMC runs. Two independent MCMC runs were conducted with 100,000,000 generations, sampling every 1,000 generations. Sufficient convergence and burn-in of runs were checked with Tracer v1.7 and ESS (above 200). The tree files of the two runs were combined in the LogCombiner v1.10.4 in the BEAST package. The maximum clade credibility tree was generated in TreeAnnotator v1.10.4 of the BEAST package with the first 10% of samples discarded as burn-in.
The ancestral distribution of the genus Pampus was inferred in RASP v4.2 (Yu et al., 2015). The ultrametric-tree file retrieved in divergence time inference was used as the input for RASP after the removal of the outgroup. The consensus tree was generated in RASP. The Indo-Pacific region was divided into three areas with different species diversities: the Indian Ocean (IO; i.e., the Arabian Sea, the Bay of Bengal, and adjacent waters), the central Indo-Pacific [CIP; i.e., the East China Sea, South China Sea, Sunda Shelf, and adjacent waters], and the Northeast Asian waters (NAW; i.e., the Japan Archipelago, Yellow Sea, Bohai Sea, and the adjacent waters). The best-fit model for ancestral reconstruction was determined using BioGeoBEARS (Matzke, 2014) in RASP, and the Statistical Dispersal-Extinction-Cladogenesis model was selected (Beaulieu et al., 2013). Therefore, the ancestral distribution was inferred using the Statistical Dispersal-Extinction-Cladogenesis model allowing six candidate states on nodes: IO, CIP, NAW, IO + CIP, CIP + NAW, and IO + CIP + NAW.
A total of 1,497 mitochondrial sequences of the Pampus species, comprised of 271 newly obtained, 1,120 published, and 106 unpublished sequences from GenBank and BOLD were collected. The COXIF, Cytb, and MG sequences newly obtained in this study were deposited in GenBank (accession numbers: COXIF, OP035984–OP036119; Cytb, OP066061–OP066192; MG, OP096421–OP096425). All the sequences were sorted into eight datasets by genes, i.e., 12S rRNA, 16S rRNA, ATP6-ATP8, COXIF, COXIR, CR, Cytb, and MG datasets (Table 1; Appendix S2). The COXIF dataset contained 499 sequences of the Pampus species and a single sequence of Peprilus medius (Peter, 1869), and their sequence length ranged from 440 to 646 bp. The dataset was aligned into a 690-bp alignment with 143 polymorphic sites, 105 parsimony informative sites, and 10.13% missing data (i.e., external gaps). The Cytb dataset encompassed 447 sequences of the Pampus species and a single sequence of P. medius (outgroup) (401–1137 bp), with 130 polymorphic sites, 102 parsimony informative sites, and 9.70% missing data in its 1,152-bp alignment. The 11,424-bp MG dataset included 28 mitogenomes of the Pampus species and an outgroup P. triacanthus and consisted of 13 protein-coding genes: ATP6 (683 bp), ATP8 (168 bp), COXI (1,551 bp), COXII (691 bp), COXIII (785 bp), Cytb (1,137 bp), ND1 (975 bp), ND2 (1,046 bp), ND3 (349 bp), ND4 (1,381 bp), ND4L (297 bp), ND5 (1,839 bp), and ND6 (522 bp), which summed up to 4,293 polymorphic sites and 3,569 parsimony informative sites. The MG dataset was reduced into having eight MG sequences for divergence time inference, i.e., the outgroup P. triacanthus and seven Pampus species identified in the major datasets (see the results below). No saturated codon position was identified in the three datasets.
All sequences can be identified as the seven Pampus species using MegaBLAST, viz., P. argenteus, P. candidus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus, except that some 12S rRNA and 16S rRNA sequences were uncertainly identified as either P. cinereus or P. liuorum due to their short sequence lengths and the low numbers of mutations of the two genes. All the ML trees reconstructed from the major datasets (i.e., the COXIF, Cytb, and MG datasets, Figure 4) identified seven clades in the genus Pampus. All the sequences of this study and Wei et al. (2021) were assigned to correct clades corresponding to their morphological identities in the ML trees, which suggests the capabilities of these trees to identify the Pampus species (Figure 4; Appendix S3). The correct identities of all the analyzed sequences are summarized in Appendix S2. Cytb sequences of P. argenteus sensu Sun et al. (2012) encompassed three species, i.e., P. liuorum from China, P. cinereus from Myanmar and Thailand, and P. candidus from India and Pakistan. All the sequences of P. echinogaster were identified as P. argenteus (Appendix S2). COXIF sequences of P. echinogaster sensu Li et al. (2017) were nested within the P. argenteus clade (Appendix S2), while those of P. argenteus sensu Li et al. (2013) were nested within the P. candidus clade (Appendix S2). Both COXIF and Cytb sequences of P. griseus from Roul et al. (2021) were identified as P. cinereus, while P. cinereus sensu Roul et al. (2021) were P. liuorum (Appendix S2). Sequences annotated as P. nozawae were identified as either P. liuorum or P. minor (Appendix S2).
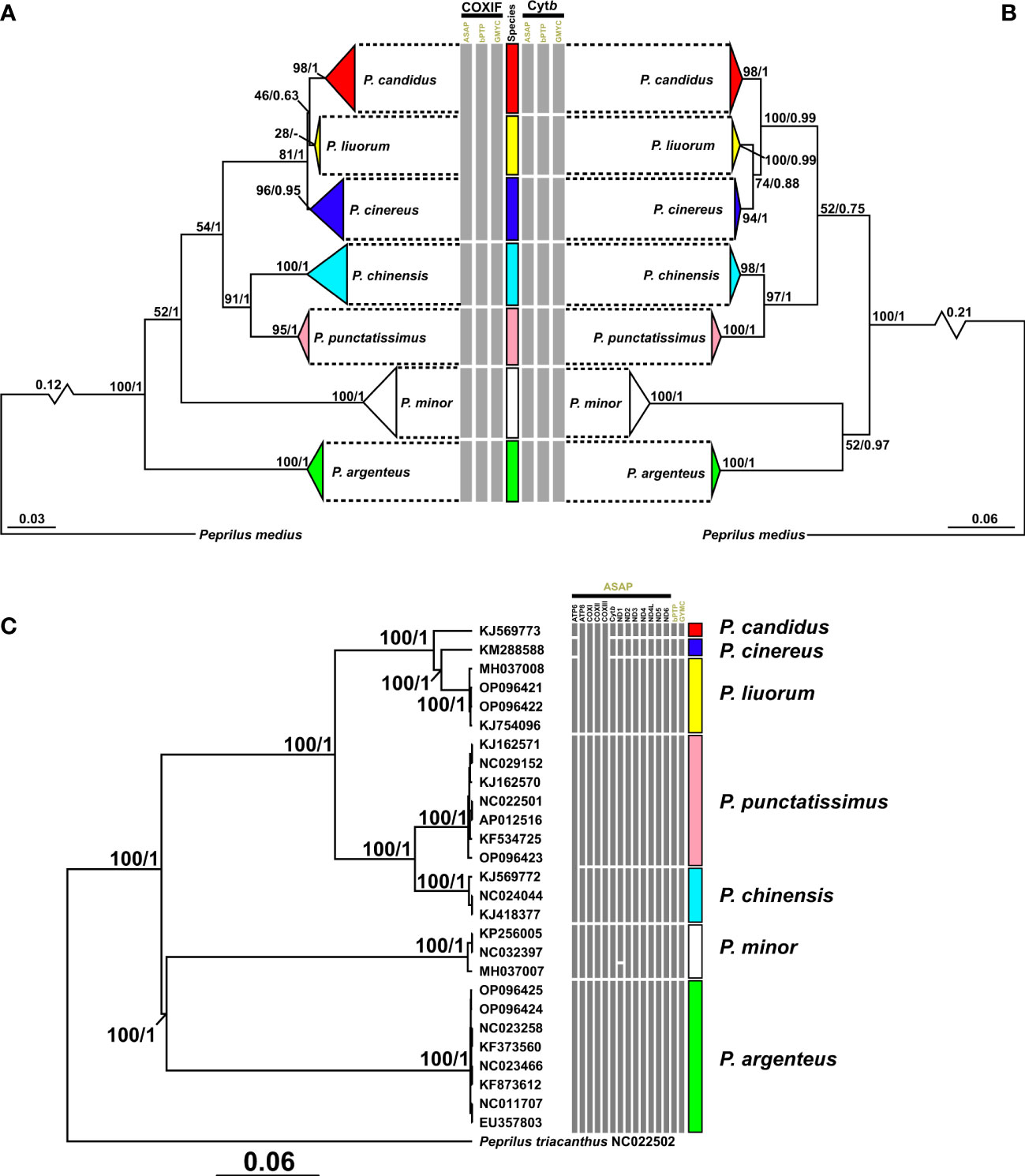
Figure 4 ML trees of the COXIF (A), Cytb (B), and MG (C) datasets. Bootstrap values and posterior probabilities obtained in ML and BI analyses are shown on each node. The broken lines indicate that these branches are condensed, and the original branch lengths are shown on these branches. ML, maximum likelihood; BI, Bayesian inference.
The ML trees of the COXIF and Cytb datasets (Figures 4A, B) recovered different topologies. The monophyly of most species was well supported by BS and posterior probability (PP) values over 75 and 0.95, in phylogenetic trees of the major dataset (Figure 4). However, the monophyly of P. liuorum was either poorly supported by a low BS value of 28 in the ML tree or not indicated in the BI tree of the COXIF dataset (Figure 4A). The COXIF trees resolved P. argenteus as the outermost species sister to all of its congeners. P. minor was resolved as the second outermost species, but the support values of such a relationship were low (COXIF: BS = 52, PP = 1, Figure 4A). The Cytb trees resolved the sister relationship of P. argenteus and P. minor (BS = 52, PP = 0.97). Although the monophyly of P. candidus, P. cinereus, and P. liuorum was well supported in the COXIF and Cytb trees (COXIF: BS = 81, PP = 1; Cytb: BS = 100, PP = 0.99, Figures 4A, B), relationships of the three species were different in the phylogenetic trees. The COXIF tree resolved P. liuorum as the sister species of P. candidus (BS = 46, PP = 0.63, Figure 4A), but the Cytb tree recovered its closer relationship with P. cinereus (BS = 74, PP = 0.88, Figure 4B). P. chinensis was well supported as the sister species of P. punctatissimus (COXIF: BS = 91, PP = 1; Cytb: BS = 98, PP = 1, Figures 4A, B); these two species were sister species to P. candidus, P. cinereus, and P. liuorum (COXIF: BS = 54, PP = 1; Cytb: BS = 66, PP = 0.75). The phylogenetic relationship of the seven species in the MG trees was identical to that in the Cytb trees. The topologies of phylogenetic trees of the MG dataset mostly agreed with those of the Cytb trees. All nodes in the MG trees were well supported by BS and PP values of 100 and 1 (Figure 4C), respectively. Support values for the monophyly of P. cinereus and P. candidus were not available, as the two species have only one mitogenome. The ML tree of the reduced MG dataset showed completely identical relationships among the seven Pampus species to those in the MG dataset (Figure 5B), which was thus used as the input tree for divergence time inference in BEAST.
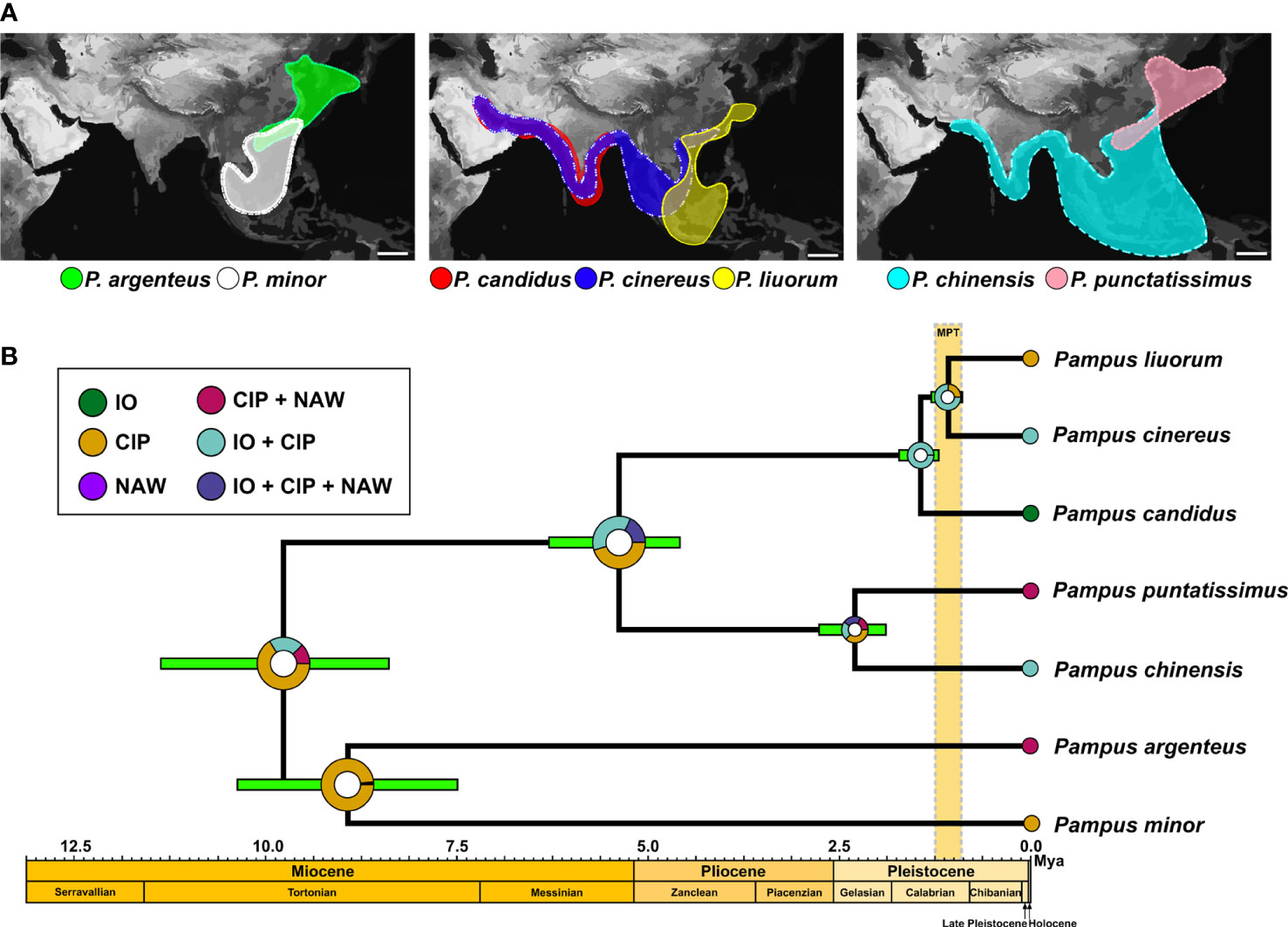
Figure 5 Divergence time inference and ancestral reconstruction of the seven Pampus species. (A) Distributions of each lineage of genus Pampus based on mitochondrial data; scale bars at lower-right corner of the maps indicate 1,000 km. (B) Divergence time inference and ancestral state reconstruction in BEAST v1.10.4 and RASP v4.2. IO, Indian Ocean; CIP, Central Indo-Pacific; NAW, Northeast Asian waters. The intervals of the geographic epochs are plotted based on Steinthorsdottir et al. (2021). The interval of the Mid-Pleistocene transition (MPT) is plotted based on Petrick et al. (2019).
Most species delimitation analyses favored two solutions for operational taxonomic units (OTUs) within the genus Pampus (Figure 4): 1) in the five-OTU solution, P. candidus, P. cinereus, and P. liuorum were delimitated as a single OTU, with other species as different OTUs; 2) in the seven-OTU solution, all the seven species were considered as OTUs severally. The ASAP, bPTP, and GMYC of the COXIF datasets favored the five-OTU solution, while those of the Cytb dataset indicated the seven-OTU solution. For the MG dataset, the bPTP and GMYC agreed on the seven-OTU solution, but the ASAP on its gene partitions indicated different results (Figure 4C): 1) ATP8, COXI, COXII, and COXIII favored the five-OTU solution; 2) Cytb, ND2–6, and ND4L favored the seven-OTU solution; 3) ATP6, P. candidus, P. cinereus, and P. liuorum were delineated as different OTUs, but P. chinensis and P. punctatissimus were merged as a single OTU (i.e., six-OTU solution); 4) ND1 postulated eight OTUs, and an MG sequence of P. minor (accession number, MH037007) was delineated as an additional OTU to the seven OTUs of the valid species.
The sampling localities and correct identities of the mitochondrial sequences were annotated in Figure 3. The COXIF and Cytb datasets contained the highest number of sampling sites among the analyzed datasets (Table 1), while the sampling sites from other datasets were mostly included in those of the two datasets (Appendix S2). The distributions of the seven identified species were similar in both datasets. In brief, all the mitochondrial sequences of the genus Pampus derived from the Indo-West Pacific region, through the Arabian Gulf to coastal Japan (Figure 3). P. argenteus and P. punctatissimus were reported from Japan to the northern South China Sea. The distribution of P. chinensis extended from the Arabian Sea to the East China Sea, south to the Changjiang River estuary. P. cinereus was found through the Arabian Gulf (Bahrain and Kuwait, four specimens, Appendix S2) to the northern South China Sea. P. candidus were mostly found in the IO, from the Arabian Gulf to the Bay of Bengal, but occurrences of the species were sparsely reported in China and Malaysia. P. liuorum and P. minor were endemic to the CIP and not reported in the IO, with their westernmost sequences reported in the Malacca Strait (Figure 3). The distribution range of the genus Pampus encompassed three areas with obviously different species diversities, i.e., the IO (hosted three species, P. candidus, P. chinensis, and P. cinereus), the CIP (hosted six species, P. argenteus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus), and the NAW (hosted two species, P. argenteus and P. punctatissimus; Figure 3). The three areas were thus used as three states for analyses in RASP.
The mean calibrated substitution rate of COXI was 1.19%/million years (HPD95: 0.60–1.75%/million years). The tMRCA of the genus Pampus was 9.76 Mya (HPD95: 8.35–11.33 Mya). The divergence between P. minor and P. argenteus was dated back to 8.9 Mya (HPD95: 7.5–10.37 Mya). The tMRCA of P. candidus, P. cinereus, and P. liuorum was 1.46 Mya (HPD95: 1.20–1.72 Mya), and P. liuorum differentiated from P. cinereus at 1.07 Mya (HPD95: 0.88–1.28 Mya). The divergence time of P. chinensis and P. punctatissimus was 2.33 Mya (HPD95: 1.92–2.79 Mya), while the two species separated from P. candidus, P. cinereus, and P. liuorum in 5.4 Mya (HPD95: 4.57–6.28 Mya).
The Statistical Dispersal-Extinction-Cladogenesis model suggests three ancestral states for the most recent common ancestor (MRCA) of the genus Pampus (CIP, IO + CIP, and CIP + NAW), and the occurrence of these ranges was 66.18%, 21.34%, and 12.47%, respectively (Figure 5B). Therefore, it postulated a CIP origin for the genus Pampus. The MRCA of P. argenteus and P. minor was also assumed to distribute in CIP (98.24%), while the MRCA of P. candidus, P. cinereus, and P. liuorum might distribute in IO + CIP (99.75%, Figure 5B). The model also favored an IO + CIP (75.80%) origin for the MRCA of P. liuorum and P. cinereus. Although the MRCA of P. punctatissimus and P. chinensis had a higher probability (37.01%) for an IO + CIP origin, the probabilities for other states were also high and varied from 18.05% to 22.72% (Figure 5B).
Although a large number of mitochondrial sequences of the genus Pampus have been published today, only seven species could be delimitated (Table 1, Figure 4). It agrees with most conclusions of the recent mitochondrial studies, i.e., Li et al. (2019); Radhakrishnan et al. (2019); Wei et al. (2021), and Roul et al. (2021). In contrast, Yin et al. (2019), based on phylogenetic analyses with a large number of nuclear loci, proposed only five species within the genus Pampus. P. liuorum was synonymized with P. cinereus because the two species formed a mixed clade in the phylogenetic tree of Yin et al. (2019). However, it is questioned that the mixed clade of P. liuorum and P. cinereus in Yin et al. (2019) might actually contain three well-supported subclades, corresponding to three Pampus species (i.e., P. candidus, P. cinereus and P. liuorum, Wei et al., 2021; Roul et al., 2021). In our results, species delimitations with COXIF datasets and ATP8, COXI, COXII, and COXIII partitions of the MG dataset also delineate P. candidus, P. cinereus, and P. liuorum as a single OTU, which is different from most of the delimitation results (Figure 3). The ASAP of ATP6 also delineates P. chinensis and P. punctatissimus as a single OTU, even though the two species are morphologically distinct (Li et al., 2019; Roul et al., 2021; Wei et al., 2021). Their conflicts with most of the delimitation results might be explained by a low level of differentiation among some species in these genes. For example, Wei et al. (2021) indicated that there is only 2–4 bp of difference between the COXIF sequences of P. liuorum and P. cinereus. The low levels of differentiation in these genes might affect the accuracy of species delimitations, and the congruences among different molecular markers should be accounted for. Based on most delimitation results, we conclude seven species in the genus Pampus, which, following Wei et al. (2021), are P. argenteus, P. candidus, P. chinensis, P. cinereus, P. liuorum, P. minor, and P. punctatissimus (Figure 2).
Many studies (e.g., Liu et al., 2013a; Li et al., 2017; Li et al., 2019) hypothesized P. echinogaster and P. argenteus as two species despite their morphological similarities (e.g., see comparisons in Liu et al., 2013a), which overlook their different distributions recorded by earlier works (e.g., Haedrich, 1967; Lindberg and Krasyukova, 1989; Nakabo, 2002). Our results clearly indicated that P. argenteus redescribed by Liu et al. (2013a) was widely distributed through the northern South China Sea to the Bohai Sea and coastal Japan (Figure 3), and all the sequences previously noted as P. echinogaster (e.g., Li et al., 2017) were confidently identified as P. argenteus (Figure 4; Appendix S2). Furthermore, the validity of P. echinogaster proposed by Li et al. (2017) was based on its genetic difference with P. argenteus sensu Li et al. (2013), whereas the latter was identified as P. candidus in our analysis (Figure 4; Appendix S2). Therefore, the validity of P. echinogaster proposed by Li et al. (2017) is incorrect and based on comparisons with misidentified “P. argenteus”. In addition, only P. punctatissimus and P. argenteus were found inhabiting the type locality of P. echinogaster (i.e., the Bohai Sea, Figure 3), supporting that P. echinogaster should be either of these two species. Given that P. echinogaster has been frequently used to represent northern populations of P. argenteus (e.g., Liu et al., 2013a; Li et al., 2017), we propose the species as a junior synonym of P. argenteus.
P. cinereus was frequently reported in several Chinese waters (e.g., Liu et al., 2002; Cui et al., 2010; Li et al., 2019), but “P. cinereus” found in China actually contains two different species. Due to the loss of type specimen, a neotype is designated for P. cinereus, and P. liuorum is proposed to represent the other “P. cinereus” in China (Liu et al., 2013b; Liu and Li, 2013). The validities of the two species have been confirmed by Wei et al. (2021). Roul et al. (2021) assumed that P. cinereus was endemic to West Pacific and resurrected P. griseus, which dominates the Bay of Bengal. Nevertheless, P. griseus and P. cinereus sensu Roul et al. (2021) were identified as P. cinereus and P. liuorum in our results, respectively (Figure 3; Appendix S2), so the resurrection of P. griseus was incorrect due to the misidentification of P. cinereus and P. liuorum. We uncovered that P. cinereus has a broader distribution in the IO and CIP, which also refuted the suggestion of Roul et al. (2021) on the distribution of P. cinereus. Although the exact type locality of P. cinereus was not given in Bloch (1795), the types of P. cinereus might in fact come from waters around India. Bloch (1795) quoted Mr. Christoph Samuel John’s words that the local people made karawade with P. cinereus. The word karawade means dried fish in the Tamil language, which is spoken in Tamil Nadu (southern India). The specimen provider, Mr. John, had been based in Tranquebar (i.e., Tharangambadi, Tamil Nadu) since 1771 (Paepke, 1999), which was also stated as the only known habitat for P. cinereus in Bloch’s later work, i.e., Bloch and Schneider (1801). Our result indicates that P. cinereus redescribed by Liu et al. (2013b) is broadly distributed through the Arabian Gulf to the South China Sea, whereas P. liuorum is not present in the Indian Ocean (Figure 3). Therefore, the neotype designation of P. cinereus by Liu et al. (2013b) appears appropriate, whereas P. liuorum should not be treated as P. cinereus due to its absence in the Indian Ocean (Figure 3).
P. nozawae (Ishikawa, 1904) was treated as valid species in several studies (e.g., Huang et al., 2016; Jawad and Liu, 2017; Hou et al., 2018). However, all the “P. nozawae” sequences were identified as either P. cinereus or P. minor, and none of them were collected from the type locality of P. nozawae (i.e., Hokkaido, Japan). Sequences of P. argenteus and P. punctatissimus have been reported from coastal Japan (Figure 4; Appendix S2), and only these two Pampus species have been persistently recorded in coastal Japan and Russia (e.g., Lindberg and Krasyukova, 1989; Nakabo, 2002). P. argenteus is a species with short dorsal and anal fin lobes (Liu et al., 2013a), while Ishikawa (1904) clearly stated those of P. nozawae “reaching to the base of caudal if laid backward”. The description only suits the morphology of P. punctatissimus (Liu and Li, 1998b). Therefore, P. nozawae might be a synonym of P. punctatissimus. Nevertheless, the validity of P. nozawae could not be eliminated due to a lack of sequence data from coastal Japan and Russia (Figure 3).
Our study revealed that the CIP is a biodiversity hotspot for the genus Pampus. Six of the seven valid species are co-distributed in the CIP region, whereas only three and two species inhabit the IO and NAW regions, respectively (Figure 3). The Indonesian–Australian Archipelago within the CIP hosts over 2,500 fish species (Parravicini et al., 2014; Maxwell et al., 2022) and has been implicated as the center of origin for many marine fish taxa (e.g., Lutjanidae, Rincon-Sandoval et al., 2020; Mulloidei, Santaquiteria et al., 2021). Our RASP analysis supports a CIP origin for the genus Pampus (Figure 5B), which agrees with the conclusion of Fan et al. (2022). The inference on divergence time dated tMRCA of the genus Pampus back to the late Miocene (Figure 5B; mean tMRCA: 9.76 Mya; HPD95: 8.35–11.33 Mya, Steinthorsdottir et al., 2021), which agrees with the late Miocene origins of many modern coral reef fish lineages in the Indonesian–Australian Archipelago (Renema et al., 2008; Bellwood et al., 2017; Steinthorsdottir et al., 2021). These results imply that the origin of the genus Pampus might be related to the development of the Indonesian–Australian Archipelago biodiversity hotspot. The distributions of P. cinereus and P. liuorum overlap near the Sunda Shelf (Figure 3), while Roul et al. (2021) proposed that the species separately dominates the Bay of Bengal and CIP. The divergences among P. candidus, P. cinereus, and P. liuorum were considered to be related to isolations by the Indian Subcontinent and Sunda Shelf (Sun et al., 2012; Fan et al., 2022). In this study, the divergence time of P. cinereus and P. liuorum (mean tMRCA: 1.07; HPD95: 0.88–1.28 Mya) fell in the Mid-Pleistocene transition (0.60–1.25 Mya, Figure 5B), a major transition in global climate (Petrick et al., 2019). The periodicity of the Earth’s glacial-interglacial cycles shifted from 41,000 years to quasi-100,000 years at the Mid-Pleistocene transition, and the glaciations and resultant sea-level changes became more intensive and significant, which accounted for the weakening of the Indonesian Throughflow and exposure of the Sunda Shelf (Petrick et al., 2019). The channel at the Sunda Shelf that connects the populations in the Indian and Pacific Oceans was frequently interrupted during the successive sea level changes in the Pleistocene (Petrick et al., 2019), which is believed to cause trans-oceanic diversification of marine species between the IO and the western Pacific (Ahti et al., 2016; Waldrop et al., 2016; Fan et al., 2022). Therefore, the differentiation between P. cinereus and P. liuorum might be associated with the exposure of the Sunda Shelf during the glacial cooling at the Mid-Pleistocene transition.
In recent years, gene sequences of marine species are rapidly accumulating to meet the growing need for fishery management and environmental protection. Our analyses of these global sequence data revealed new boundaries of the distributions of the Pampus species. P. argenteus used to be considered widely distributed in the IO and CIP (Haedrich, 1967), only inhabiting waters from the northern South China Sea to coastal Japan (Figures 3, 5A). Roul et al. (2021) assume that P. cinereus (i.e., P. griseus of the study) is restricted to the Bay of Bengal and Southeast Asia, but our study indicates that the species is also presented in the Arabian Sea and Arabian Gulf (i.e., Bahrain, Kuwait, and Mumbai, eastern India, Figure 4). Specially, we collected fresh specimens of P. cinereus at landing sites of Bahrain and Kuwait (Figure 4), indicating that the species inhabits the Arabian Gulf (although not a common species here, L. A. Jawad, personal observation). Yamada et al. (1995) stated that P. punctatissimus is not distributed south to the Taiwan Strait, but numerous wild-caught P. punctatissimus and their sequences have been reported by Zhang and Hanner (2012) from the northern South China Sea (Figure 3; Appendix S2), which clearly rejects Yamada et al. (1995) statement. P. punctatissimus is actually a species widely distributed from the northern South China Sea (Figures 3, 5A) to coastal Japan and Russia (Lindberg and Krasyukova, 1989). Nevertheless, commercial transportation might sometimes affect the accuracy of global molecular data. Extra caution should be taken when dealing with sequences of commercial species. We questioned the presence of P. candidus in China reported by Li et al. (2013) because fresh or wild-caught specimens of this species have never been observed in coastal China (personal observation). In fact, China imports “silver pomfrets” (HS code: 03038930) from South Asian countries (e.g., India, Pakistan, and Myanmar, the Custom Statistics Database of China, http://43.248.49.97/). The exportation of P. candidus from India to China, the Middle East, and the EU was also reported by Roul et al. (2021). As a result, P. candidus can be found in fish markets across China (e.g., IOCAS000292 and IOCAS000293 of this study, purchased from a fish market in Qingdao, Appendix S2). The P. candidus specimens reported by Li et al. (2013) are likely import commodities sold in the fish markets of China. We also found a specimen of P. candidus in Pulau Perhentian, Malaysia (Figure 3, IOCAS000181), likely due to commercial transportation of the species from the Bay of Bengal to the eastern Malay Peninsula.
The data presented in the study are deposited in the NCBI Genbank, accession number OP035984–OP036119, OP066061–OP066192, OP096421–OP096425.
Ethical review and approval was not required for the animal study because the samples used in this study were dead specimens purchased from fish market and landing sites, which does not constitute any ethical issue with alive animals.
JW, JL, and KX conceived the research. JW, JL, RW, LJ, KL, and YW conducted the sampling collection. JW, RW, YX, AH-U, KL, and YW performed the laboratory work. JW, RW, and YX performed data curation and analyses. JW wrote the original draft. KX and JL led the review and editing of the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (Nos. 31872195 and 32270472) and Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB42000000).
We are grateful to Dr. Alexandre F. Bannikov of Borissiak Paleontological Institute, Russian Academy of Sciences, for providing morphological and chronological data of the Pinichthys fossils. We also thank Prof. Ronald Fricke of Staatliches Museum für Naturkunde for providing information on P. cinereus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1050386/full#supplementary-material
Ahti P. A., Coleman R. R., DiBattista J. D., Berumen M. L., Rocha L. A., Bowen B. W. (2016). Phylogeography of indo-pacific reef fishes: sister wrasses Coris gaimard and C. cuvieri in the red Sea, Indian ocean and pacific ocean. J. Biogeogr. 43 (6), 1103–1115. doi: 10.1111/jbi.12712
Arcila D., Hughes L. C., Meléndez-Vazquez B., Baldwin C. C., White W. T., Carpenter K.E., et al. (2021). Testing the utility of alternative metrics of branch support to address the ancient evolutionary radiation of tunas, stromateoids, and allies (Teleostei: Pelagiaria). Syst. Biol. 70 (6), 1123–1144. doi: 10.1093/sysbio/syab018
Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., Alekseyenko A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29 (9), 2157–2167.
Bannikov A. F. (1985). Fossil stromateoid fishes (Teleostei) of the Caucasus. Paleontol. J. 19 (4), 7–14.
Bannikov A. F. (1988). A new species of stromateoid fishes (Perciformes) from the lower oligocene of the Caucasus. Paleontol. J. 22 (4), 107–112.
Beaulieu J. M., Tank D. C., Donoghue M. J. (2013). A southern hemisphere origin for campanulid angiosperms, with traces of the break-up of gondwana. BMC Evol. Biol. 13 (1), 80. doi: 10.1186/1471-2148-13-80
Bellwood D. R., Goatley C. H. R., Bellwood O. (2017). The evolution of fishes and corals on reefs: form, function and interdependence. Biol. Rev. 92 (2), 878–901. doi: 10.1111/brv.12259
Bloch M. E., Schneider J. G. (1801). M. e. blochii, systema ichthyologiae iconibus cx ilustratum (Berlin: Sanderian Library).
Cui Z., Liu Y., Liu J., Luan W. (2010). Molecular identification of Pampus fishes (Perciformes, stromateidae). Ichthyol. Res. 57 (1), 32–39. doi: 10.1007/s10228-009-0119-9
Dadzie B. S., Abou-Seedo F., Al-Qattan E. (2000). The food and feeding habits of the silver pomfret, Pampus argenteus (Euphrasen), in Kuwait waters. J. Appl. Ichthyol. 16 (2), 61–67. doi: 10.1046/j.1439-0426.2000.00150.x
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9 (8), 772. doi: 10.1038/nmeth.2109
Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7 (1), 214. doi: 10.1186/1471-2148-7-214
Fan G., Yin G., Sarker A., Li C. (2022). Diversifying of two Pampus species across the indo–pacific barrier and the origin of the genus. Diversity 14 (3), 180. doi: 10.3390/d14030180
Fontaneto D., Herniou E. A., Boschetti C., Caprioli M., Melone G., Ricci C., et al. (2007). Independently evolving species in asexual bdelloid rotifers. PloS Biol. 5 (4), e87. doi: 10.1371/journal.pbio.0050087
Fujisawa T., Barraclough T. G. (2013). Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: a revised method and evaluation on simulated data sets. Syst. Biol. 62 (5), 707–724. doi: 10.1093/sysbio/syt033
Gupta S. (2020). Reviews on the biology and culture of silver pomfret, Pampus argenteus (Euphrasen, 1788). Int. J. Aquat. Biol. 8, 228–245.
Haedrich R. L. (1967). The stromateoid fishes: systematics and a classification. Bull. Mus. Comp. Zool. 135, 31–139.
Hamon N., Sepulchre P., Lefebvre V., Ramstein G. (2013). The role of eastern tethys seaway closure in the middle Miocene climatic transition (ca. 14 ma). Clim. Past 9, 2687–2702. doi: 10.5194/cp-9-2687-2013
Hou G., Chen W.-T., Lu H.-S., Cheng F., Xie S.-G. (2018). Developing a DNA barcode library for perciform fishes in the south China Sea: Species identification, accuracy and cryptic diversity. Mol. Ecol. Resour. 18 (1), 137–146. doi: 10.1111/1755-0998.12718
Huang W., Xu X., Xie Z., Li Y., Wang D., Wang X., et al. (2016). The complete mitochondrial genome of the Pampus nozawae (Perciformes: Stromateidae). Mitochondrial DNA Part A 27 (2), 988–989. doi: 10.3109/19401736.2014.926518
Ishikawa C. (1904). Notes on some new or little known fishes of japan. part I. Proc. Dept. Nat. Hist. Tokyo Imp. Mus. 1, 11–17. 1–17.
Jawad L. A., Liu J. (2017). Comparative osteology of the axial skeleton of the genus Pampus (Family: Stromateidae, perciformes). J. Mar. Biolog. Assoc. U.K. 97 (2), 277–287. doi: 10.1017/S0025315416000369
Jukes T. H., Cantor C. R. (1969). “Evolution of protein molecules,” in Mammal protein metabolism. Ed. Munro H. N. (New York: academic Press), 21–132.
Katoh K., Stanley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 (2), 111–120. doi: 10.1007/BF01731581
Lindberg G. U., Krasyukova Z. V. (1989). Fishes of the Sea of Japan and the adjacent areas of the Sea of Okhotsk and the yellow sea. part 4 (London: CRC Press).
Liu J., Li C. (1998a). A new pomfret species, Pampus minor sp. nov. (Stromateidae) from Chinese waters. Chin. J. Oceanol. Limnol. 16 (3), 280–285. doi: 10.1007/BF02848735
Liu J., Li C. (1998b). Redescription of a stromateid fish Pampus punctatissimus and comparison with Pampus argenteus from Chinese coastal waters. Chin. J. Oceanol. Limnol. 16 (3), 161–166. doi: 10.1007/BF02845182
Liu J., Li C. (2013). A new species of the genus Pampus (Perciformes, stromateidae) from China. Acta Zootaxonomica Sin. 38 (4), 885–890.
Liu J., Li C., Li X. (2002). Studies on Chinese pomfret fishes of the genus Pampus (Pisces: Stromateidae). Sci. Mar. Sin. 44, 240–252.
Liu J., Li C. S., Ning P. (2013a). Identity of silver pomfret Pampus argenteus (Euphrasen 1788) based on specimens from its type locality, with a neotype designation (Teleostei, stromateidae). Acta Zootaxonomica Sin. 38 (1), 171–177.
Liu J., Li C., Ning P. (2013b). A redescription of grey pomfret Pampus cinereus (Bloch 1795) with the designation of a neotype (Teleostei: Stromateidae). Chin. J. Oceanol. Limnol. 31 (1), 140–145. doi: 10.1007/s00343-013-2039-9
Li Y., Song N., Khan F. S., Yanagimoto T., Gao T. (2013). New evidence of morphological characters and DNA barcoding of Pampus argenteus (Euphrase). J. Fish. China 37 (11), 1601–1608. doi: 10.3724/SP.J.1231.2013.38824
Li Y., Zhang Y., Gao T., Han Z., Lin L., Zhang X. (2017). Morphological characteristics and DNA barcoding of Pampus echinogaster (Basilewsky 1855). Acta Oceanol. Sin. 36 (12), 18–23. doi: 10.1007/s13131-017-1124-x
Li Y., Zhou Y., Li P., Gao T., Lin L. (2019). Species identification and cryptic diversity in Pampus species as inferred from morphological and molecular characteristics. Mar. Biodivers. 49 (6), 2521–2534. doi: 10.1007/s12526-019-00976-6
Matzke N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63 (6), 951–970. doi: 10.1093/sysbio/syu056
Maxwell M. F., Leprieur F., Quimbayo J. P., Floeter S. R., Bender M. G. (2022). Global patterns and drivers of beta diversity facets of reef fish faunas. J. Biogeogr. 49 (5), 954–967. doi: 10.1111/jbi.14349
Morgulis A., Coulouris G., Raytselis Y., Madden T. L., Agarwala R., Schäffer A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24 (16), 1757–1764. doi: 10.1093/bioinformatics/btn322
Nakabo T. (2002). Fishes of Japan: with pictorial keys to the species (Vol. 2) (Tokyo: Tokai University Press).
Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Paepke H.-J. (1999). Bloch's fish collection in the museum für naturkunde der Humboldt universität zu Berlin: an illustrated catalog and historical account Vol. 32 (Ruggell (Liechtenstein: Theses Zoologicae), 1–216.
Parravicini V., Villéger S., McClanahan T. R., Arias-González J. E., Bellwood D. R., Bellwood D. R., et al. (2014). Global mismatch between species richness and vulnerability of reef fish assemblages. Ecol. Lett. 17 (9), 1101–1110. doi: 10.1111/ele.12316
Petrick B., Martínez-García A., Auer G., Reuning L., Auderset A., Deik H., et al. (2019). Glacial Indonesian throughflow weakening across the mid-pleistocene climatic transition. Sci. Rep. 9 (1), 16995. doi: 10.1038/s41598-019-53382-0
Pons J., Barraclough T. G., Gomez-Zurita J., Cardoso A., Duran D. P., Hazell. S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55 (4), 595–609. doi: 10.1080/10635150600852011
Popov S. V., Tabachnikova I. P., Pinchuk T. N., Akhmetiev M. A., Zaporozhets N. I. (2019). The reference section of eocene deposits in the belaya river valley, adygea, Western ciscaucasia. Stratigr. Geol. Correl. 27 (1), 118–132. doi: 10.1134/S0869593819010052
Puillandre N., Brouillet S., Achaz G. (2021). ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 21 (2), 609–620. doi: 10.1111/1755-0998.13281
Radhakrishnan D. P., Kumar R. G., Mohitha C., Shanis C. P. R., Kinattumkara B. K., Saidumohammad B. V., et al. (2019). Resurrection and re-description of Pampus candidus (Cuvier), silver pomfret from the northern Indian ocean. Zool. Stud. 58, e7. doi: 10.6620/ZS.2019.58-07
Renema W., Bellwood D. R., Braga J. C., Bromfield K., Johnson K. G., Hall R., et al. (2008). Hopping hotspots: global shifts in marine biodiversity. Science 321 (5889), 654–657. doi: 10.1126/science.1155674
Rincon-Sandoval M., Duarte-Ribeiro E., Davis A. M., Santaquiteria A., Hughes L. C., Baldwin C. C., et al. (2020). Evolutionary determinism and convergence associated with water-column transitions in marine fishes. PNAS 117 (52), 33396–33403. doi: 10.1073/pnas.2006511117
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Roul S. K., Jeena N. S., Kumar R., Vinothkumar R., Rahangdale S., Rahuman S., et al. (2021). Postulating the modality of integrative taxonomy in describing the cryptic congener Pampus griseus (Cuvier) and systematics of the genus Pampus (Perciformes: Stromateidae). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.778422
Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi: 10.1093/molbev/msx248
Santaquiteria A., Siqueira A. C., Duarte-Ribeiro E., Carnevale G., White W. T., Pogonoski J. J., et al. (2021). Phylogenomics and historical biogeography of seahorses, dragonets, goatfishes, and allies (Teleostei: Syngnatharia): assessing factors driving uncertainty in biogeographic inferences. Syst. Biol. 70 (6), 1145–1162. doi: 10.1093/sysbio/syab028
Schwarz G. (1978). Estimating the dimension of a model. Ann. Stat. 6 (2), 461–464. doi: 10.1214/aos/1176344136
Steinthorsdottir M., Coxall H. K., de Boer A. M., Huber M., Barbolini N., Bradshaw N. J., et al. (2021). The Miocene: The future of the past. Paleoceanogr. Paleoclimatology 36 (4), e2020PA004037. doi: 10.1029/2020pa004037
Sun P., Shi Z., Yin F., Peng S. (2012). Population genetic structure and demographic history of Pampus argenteus in the indo-West pacific inferred from mitochondrial cytochrome b sequences. Biochem. Syst. Ecol. 43, 54–63. doi: 10.1016/j.bse.2012.02.028
Vaidya G., Lohman D. J., Meier R. (2011). SequenceMatrix: concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics 27 (2), 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vences M., Miralles A., Brouillet S., Ducasse J., Fedosov A., Kharchev V., et al. (2021). iTaxoTools 0.1: kickstarting a specimen-based software toolkit for taxonomists. Megataxa 6 (2), 77–92. doi: 10.11646/MEGATAXA.6.2.1
Waldrop E., Hobbs J.-P. A., Randall J. E., DiBattista J. D., Rocha L. A., Kosaki R. K., et al. (2016). Phylogeography, population structure and evolution of coral-eating butterflyfishes (Family chaetodontidae, genus Chaetodon, subgenus Corallochaetodon). J. Biogeogr 43 (6), 1116–1129. doi: 10.1111/jbi.12680
Wei J., Wu R., Xiao Y., Zhang H., Jawad L. A., Wang Y., et al. (2021). Validity of Pampus liuorum Liu & Li, 2013, Revealed by the DNA barcoding of Pampus fishes (Perciformes, stromateidae). Diversity 13 (12), 618. doi: 10.3390/d13120618
Xia X. (2018). DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Xia X., Xie Z., Salemi M., Chen L., Wang Y. (2003). An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26 (1), 1–7. doi: 10.1016/S1055-7903(02)00326-3
Yamada U., Irie T., Tokimura T., Tagawa M. (1995). “Hirenaga-managotsuo” Pampus cinereus (Bloch). Seikaiku Suisan Kenkyusho News 83, 1.
Yin G., Pan Y., Sarker A., Baki M. A., Kim J. K., Wu H., et al. (2019). Molecular systematics of Pampus (Perciformes: Stromateidae) based on thousands of nuclear loci using target-gene enrichment. Mol. Phylogenet. Evol. 140, 106595. doi: 10.1016/j.ympev.2019.106595
Yu Y., Harris A. J., Blair C., He X. (2015). RASP (Reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Zhang J., Hanner R. (2012). Molecular approach to the identification of fish in the south China Sea. PloS One 7, e30621. doi: 10.1371/journal.pone.0030621
Keywords: mitochondrial data, Pampus, species distribution, taxonomy, the Indonesian-Australian Archipelago, the middle Miocene transition
Citation: Wei J, Wu R, Jawad LA, Xiao Y, Loh K-H, Herrera-Ulloa A, Wang Y, Liu J and Xu K (2022) Species diversity and distribution of genus Pampus (Pelagiaria: Stromateidae) based on global mitochondrial data. Front. Mar. Sci. 9:1050386. doi: 10.3389/fmars.2022.1050386
Received: 21 September 2022; Accepted: 22 November 2022;
Published: 13 December 2022.
Edited by:
Baojun Tang, East China Sea Fisheries Research Institute (CAFS), ChinaReviewed by:
Zining Meng, Sun Yat-sen University, ChinaCopyright © 2022 Wei, Wu, Jawad, Xiao, Loh, Herrera-Ulloa, Wang, Liu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, amxpdUBxZGlvLmFjLmNu; Kuidong Xu, a3h1QHFkaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.