
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 11 November 2022
Sec. Marine Molecular Biology and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1050022
 Xiaoyu Xin1,2,3
Xiaoyu Xin1,2,3 Chang Liu1,2,3*
Chang Liu1,2,3* Zhaoqun Liu1,2,3
Zhaoqun Liu1,2,3 Yukun Zhang1,2,3
Yukun Zhang1,2,3 Yuqian Gao1,2,3
Yuqian Gao1,2,3 Ting Zhu1,2,3
Ting Zhu1,2,3 Lingling Wang1,2,3,4
Lingling Wang1,2,3,4 Linsheng Song1,2,3,4*
Linsheng Song1,2,3,4*The biosynthesis of shell is a complicated calcification process in the marine bivalve, which can be severely impacted by ocean acidification (OA). Calmodulin (CaM) is a pivotal calcium regulator and thought to be crucial for calcification. In the present study, a CaM (designated CgCaM) with calcium-binding activity was identified from the Pacific oyster Crassostrea gigas with the objective to understand its possible role in the regulation of calcium homeostasis under acidification treatment. The open reading frame (ORF) of CgCaM was of 474 bp encoding a 17.5 kDa protein with four continuous EF-hand domains. CgCaM shared high similarity with CaMs from other invertebrates and vertebrates. The mRNA transcript of CgCaM was constitutively expressed in all detected tissues with the higher expression level in mantle, especially highest in the middle fold of the three folds of mantle. CgCaM was found to be mainly distributed in the mantle epithelium. When the oysters were exposed to acidified seawater, the expression level of CgCaM in the middle fold of mantle and the content of Ca2+ in this fold both decreased significantly. These results collectively suggested that CgCaM was involved in the regulation of calcium homeostasis in the middle fold of mantle under acidification treatment.
Bivalves are a large taxon of calcifying organisms in the ocean, and they have developed tough shell through biologically controlled mineralization process to protect them against predation, support tissues and preclude desiccation (Simkiss and Wilbur, 1989; Marin et al., 2012). The bivalve shell is a typical calcium carbonate (CaCO3) biomineral consisting of approximately 95% CaCO3 and a low proportion of organic matrix, with aragonite and calcite as the main crystalline polymorphs (Marin et al., 2008). In recent years, increasing amounts of atmospheric CO2 was absorbed by the ocean, and the pH value of the seawater was decreased (Caldeira and Wickett, 2003), which called ocean acidification (OA). According to the previous prediction, the pH of the ocean surface is projected to reduce from 8.10 to 7.80 by the end of 2100. Many evidences have demonstrated that ocean acidification (OA) alters the chemical equilibrium of seawater and hence affects the ion homeostasis such as calcium homeostasis in marine bivalves (Li et al., 2016a; Li et al., 2016b), which results in limitation of their growth, development and reproduction (Ross et al., 2011; Hettinger et al., 2013), and even death (Waldbusser et al., 2013).
OA has been demonstrated to disrupt the calcification process of bivalves by affecting the homeostasis of calcium levels (Bednaršek et al., 2014; Chandra Rajan et al., 2021), and the driving mechanism of calcium homeostasis is crucial for bivalves in response to OA. The calcification rate of marine bivalves is reduced due to the decrease of CaCO3 saturation in seawater under elevated CO2 levels (Richards et al., 2018; Chandra Rajan and Vengatesen, 2020). As the main component of shell, sufficient calcium is required for shell calcification. The calcium needed for calcification is mainly absorbed from the external medium in the form of calcium ion (Ca2+) via gill and mantle, then transported to the mineralization sites across mantle by hemocytes (Mount et al., 2004; Addadi et al., 2006; Marin et al., 2012). As the main organ of bivalves for calcification, mantle positively participates in calcium regulation (Li et al., 2004; Sillanpää et al., 2020; Wang et al., 2022). Mantle is composed of inner muscles, nerve fibers and connective tissues bordered by epithelium cell layers on either side (Audino et al., 2015). According to the functional heterogeneity and morphological difference, the mantle is separated into marginal, submarginal and central zone (Fang et al., 2008a). The marginal zone of the mantle has been demonstrated to be particularly active in calcification, and it is further divided into outer fold (OF), middle fold (MF) and inner fold (IF) (Kádár, 2008). The molecular mechanism involved in calcium regulation of mantle is critical to comprehensively understand the adaptation of marine bivalves to OA.
Calmodulin (CaM) is a well-studied calcium-binding protein with EF-hand domains that are widely distributed in all eukaryotic cells (Stevens, 1983). Upon binding to Ca2+, CaM undergoes activity changes and subsequently interacts with downstream proteins to mediate a variety of Ca2+-induced physiological processes (Clapham, 2007), such as cell proliferation, differentiation, apoptosis, glycogen metabolism, and calcium metabolism (Means and Dedman, 1980; Cheung, 1984; Sée et al., 2001). As a fast Ca2+ buffer and pivotal Ca2+ sensor, CaM plays an important role in calcium metabolism processes including calcium absorption, transportation, secretion and deposition (Schwaller, 2012). In vertebrates, the calcium homeostasis is modulated by CaM, which activates or inactivates functional enzymes, cellular receptors and ion channels involved in Ca2+ transportation to affect intracellular Ca2+ concentration (Zhou et al., 2013). In Stylophora pistillata, the interaction of CaM and plasma-membrane Ca2+-ATPase was reported to regulate calcareous skeleton deposition (Zoccola et al., 2004). Several CaMs have been characterized to participate in calcification in bivalves. The CaM in Hyriopsis schlegelii was found to be highly expressed in pearl sac and promote the pearl sac formation (Peng et al., 2018). In the Pacific oyster Crassostrea gigas, the intracellular calcium levels varied significantly after a short-term (7 days) CO2 exposure. While the long-term (60 days) CO2 exposure activated the CaM related signal pathway, and the intracellular calcium recovered to normal levels (Wang et al., 2020). Accumulating reports suggest that CaM is of vital importance in the regulation of bivalve calcium homeostasis under acidification treatment. However, the exact regulatory mechanisms are still not well understood.
The Pacific oyster C. gigas is an important marine calcifying species and a worldwide aquaculture species with economic and ecological significance to the ecosystem (Hendriks et al., 2015). They are distributed in coastal areas worldwide with calcified shells to protect themselves from diverse environmental fluctuations. In the present study, a novel calmodulin (designated CgCaM) was characterized from C. gigas with the main objectives to (1) investigate its expression and distribution in tissues, (2) determine its calcium-binding activity in vitro, and (3) examine the alteration of its expression and calcium content in the MF of mantle after acidification treatment to understand its possible role in the regulation of calcium homeostasis under OA.
The adult Pacific oysters (C. gigas) with an average shell length of 15.0 cm were obtained from a local breeding farm in Dalian, Liaoning province, China. The oysters were pre-acclimated in normal aerated seawater for one week before processing. The experiments were approved by the animal ethics guidelines approved by the Ethics Committee of Dalian Ocean University.
Twenty-four oysters were randomly divided into two groups. The oysters cultured in normal aerated seawater were designated as the control group (pH 8.10 ± 0.05), while those cultured in the seawater continuously bubbled with the air-CO2 mixtures were designated as CO2 treatment group (pH 7.80 ± 0.05). The pH value was controlled using an acidometer (AiKB, China) according to previous reports (Zhang et al., 2019). The partial pressure of CO2 was about 658.1 ± 11.0 ppm and 1217.3 ± 11.6 ppm in pH 8.10 and pH 7.80 groups. And total alkalinity was determined by end-point titration of 25 mmol/L HCl on 50 mL seawater samples.
Oyster samples were collected from each group on 14th day after acidification treatment. Mantle, gill, adductor muscle, hepatopancreas, hemolymph and labial palp were collected from twelve oysters in the control group and CO2 treatment group respectively. The samples from four individuals were randomly mixed as one replicate, and there were three replicates for each group. The hemolymph was extracted and immediately centrifuged at 800 × g, 4°C for 15 min to harvest the hemocytes. The mantle was separated into IF, MF and OF under the microscope Discovery V8 (ZEISS, Germany). The samples for RNA extraction were kept in 1 mL TRIzol reagent (Invitrogen, USA) and stored at -80°C, while the samples for in situ hybridization were fixed with fixative solution (Servicebio, China) for further processing.
Total mRNA was extracted from tissues with TRIzol reagent (Invitrogen, USA) according to the manual. The synthesis of the first-strand cDNA was carried out based on M-MLV RT Usage information (Promega, USA) using the total mRNA treated with DNase I (Takara, China) as template and oligo (dT)-adaptor as primer. The synthesis reaction was first incubated at 42°C for 1 h, and then terminated by heating at 95°C for 5 min. The cDNA mix was diluted and stored at -80°C for subsequent quantitative real-time PCR.
According to the sequence information from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), the open reading frame (ORF) of CgCaM (XM_011428724) was amplified using the specific primers CgCaM-F and CgCaM-R (Table 1). The PCR product was cloned into pMD19-T simple vector (Takara, China) and confirmed by DNA sequencing.
The deduced amino acid sequence was analyzed with the Expert Protein Analysis System (http://www.expasy.org/). The homology searches of the cDNA sequences and protein sequences were conducted with BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast). The protein domain and signal peptide were predicted with the Simple Modular Architecture Research Tool (SMART, http://www.smart.emblheidelberg.de/). ClustalW multiple alignment program was used to create the multiple sequence alignment (http://www.ebi.ac.uk/Tools/clustalw2/). The phylogenetic tree was constructed with the Mega 7.0 program.
The CgCaM mRNA expression level in different tissues was measured by SYBR Green fluorescent quantitative real-time PCR (qRT-PCR) in an ABI PRISM 7500 Sequence Detection System. Specific primers CgCaM-RT-F and CgCaM-RT-R (Table 1) were used to amplify the fragment of CgCaM. The C. gigas elongation factor (CgEF) fragment amplified with primers CgEF- F and CgEF-R (Table 1) was used as internal control. The qRT-PCR experiment was conducted according to previous description (Lv et al., 2019). The relative expression of genes was determined by 2-ΔΔCt method (Livak and Schmittgen, 2001).
The mantle tissues were embedded in paraffin and cut into 5 μm thick sections according to previous report (Johnson et al., 2000; Zhang et al., 2020). In situ hybridization of CgCaM mRNA was carried out on these sections. Digoxigenin-labeled RNA probes were synthesized using a DIG RNA Labeling Kit (Roche, Germany) and detected by a DIG Nucleic Acid Detection Kit (Roche, Germany). The detailed step was performed following the previous reports (Johnson et al., 2000; Huang et al., 2001).
The recombinant protein of CgCaM (rCgCaM) was produced with the prokaryotic expression system. The coding region of CgCaM was amplified using primers CgCaM-Fe and CgCaM-Re (Table 1) with BamH I and Hind III enzyme cutting sites at their 5’ end, respectively. The PCR products were digested with restriction enzymes BamH I and Hind III, and ligated into predigested expression vector pET-30a (Novagen, Germany). The recombinant expression plasmid pET-30a-CgCaM was transformed into Escherichia coli transetta (DE3) (TransGen, China) and the positive colony was incubated in LB medium at 37°C, 220 rpm. When the OD600 of culture medium reached 0.4-0.6, isopropyl-β-D-thiogalactopyranoside (IPTG) was added into the LB medium at the final concentration of 1 mM to induce the expression of rCgCaM. Subsequently, rCgCaM was purified by His-tag Purification Resin (Beyotime, China). Protein concentration was determined by BCA method and stored at -80°C for further experiments.
The electrophoretic migration assay was performed according to the previous method (Burgess et al., 1980; Senawong et al., 2012). The purified rCgCaM protein was separated by 12% nondenaturing polyacrylamide gel electrophoresis in the presence of 20 mM ethylenediaminetetraacetic acid (EDTA) with 1 mM MgCl2, or 1 mM CaCl2, or without MgCl2 and CaCl2, respectively. Bovin serum albumin (BSA) was used as a control. The electrophoretic migration was observed by AI600 RGB.
The mantle tissues harvested from C. gigas were minced and incubated with Pronase (2 mg mL-1) in Hank’s Balanced Salt Solution (HBSS) (G-CLONE, China) without Ca2+ or Mg2+ with a gentle shaking at 37°C for 4 h. The digested tissues were filtered (50 μm mesh), washed and centrifuged at 500 × g in HBSS for 10 min. The pellets were suspended in HBSS and the cell concentration was adjusted to 106 mL-1. The mantle cells were incubated with Fluo-4 AM (5 μM) at 25°C in dark for 20 min. The median fluorescence intensity of 5000 cells was detected by FACS ImageStreamx Mark II and recorded as relative Ca2+ content (Föller et al., 2010).
All data were presented as mean ± S.D. and analyzed by Statistical Package for Social Sciences (SPSS) 16.0. Significant differences between samples were tested by one-way analysis of variance (ANOVA). The differences were considered as significant at p < 0.05.
The complete ORF of CgCaM was of 474 bp, encoding a polypeptide of 158 amino acids with a predicted molecular mass of 17.5 kDa and a theoretical isoelectric point of 4.22 (Figure 1A). There were four continuous typical EF-hand domains in CgCaM (Figure 1B). The deduced amino acid sequence of CgCaM shared high identity with those CaMs in other species (Figure 1C), such as 96.82%, 76.43%, 71.95%, 74.52%, 75.80%, 73.25%, 57.21% and 73.25% identity with that of Crassostrea virginica, Pinctada fucata, Penaeus vannamei, Danio rerio, Drosophila melanogaster, Gallus gallus, Mus musculus and Homo sapiens, respectively.
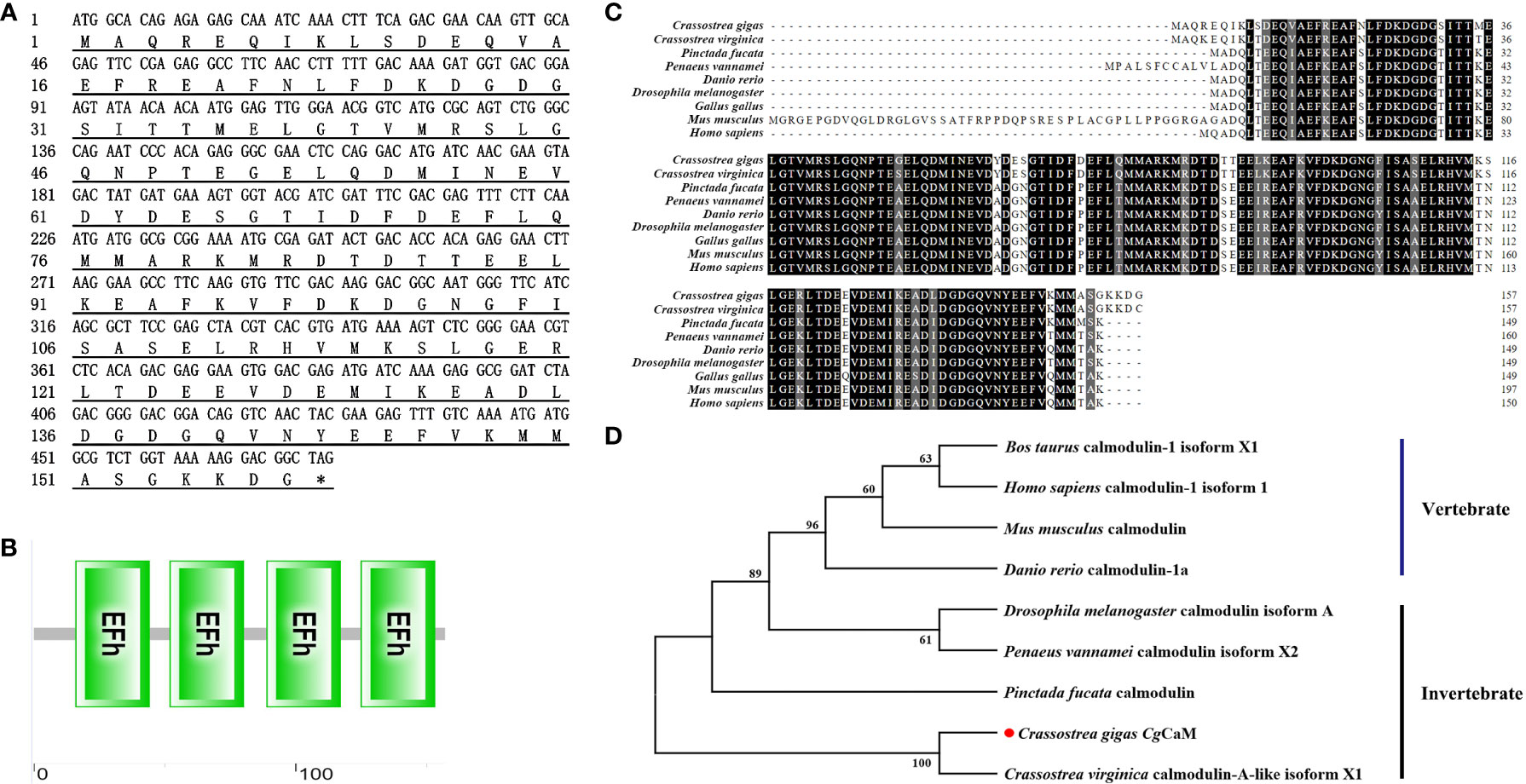
Figure 1 Bioinformatics analysis of CgCaM. (A) Nucleotide and deduced amino acid sequence of CgCaM. (B) Protein domains of CgCaM predicted by SMART analysis. (C) Multiple sequence alignment of CgCaM with known calmodulins from other species, including Crassostrea virginica (XP_022332301.1), Pinctada fucata (AAQ20043.1), Penaeus vannamei (ROT84624.1), Danio rerio (NP_892012.1), Drosophila melanogaster (NP_523710.1), Gallus gallus (NP_990336.1), Mus musculus (NP_001300863.1), Homo sapiens (NP_001350599.1). (D) Phylogenetic relationship of CgCaM. Calmodulin sequences from different species were obtained from GenBank on NCBI, including Bos taurus calmodulin-1 isoform X1 (XP_024853610.1), Homo sapiens calmodulin-1 isoform 1 (NP_001350599.1), Mus musculus calmodulin (AAA66182.1), Danio rerio calmodulin-1a (NP_998516.1), Drosophila melanogaster calmodulin isoform A (NP_523710.1), Penaeus vannamei calmodulin isoform X2 (XP_027237663.1), Pinctada fucata calmodulin (AAQ20043.1), Crassostrea gigas CaM (XP_011427026.1), Crassostrea virginica calmodulin-A-like isoform X1 (XP_022332301.1).
A phylogenetic tree was constructed to analyze the evolutionary relationship of CgCaM with CaMs from other species by neighbor-joining method (Figure 1D). The CgCaM was firstly clustered with calmodulin-A-like protein of C. virginica and then joined into the invertebrate group.
The mRNA transcripts of CgCaM were detected in all the tested tissues, including hemocytes, hepatopancreas, adductor muscle, labial palp, gill and mantle with the highest expression level in mantle, which was 50.67-fold (p < 0.05) of that in hemocytes (Figure 2). CgCaM was also highly expressed in gill and labial palp, which was 36.55-fold (p < 0.05) and 21.94-fold (p < 0.05) of that in hemocytes, respectively. There was no significant difference of CgCaM mRNA expression among hemocytes, hepatopancreas and adductor muscle.
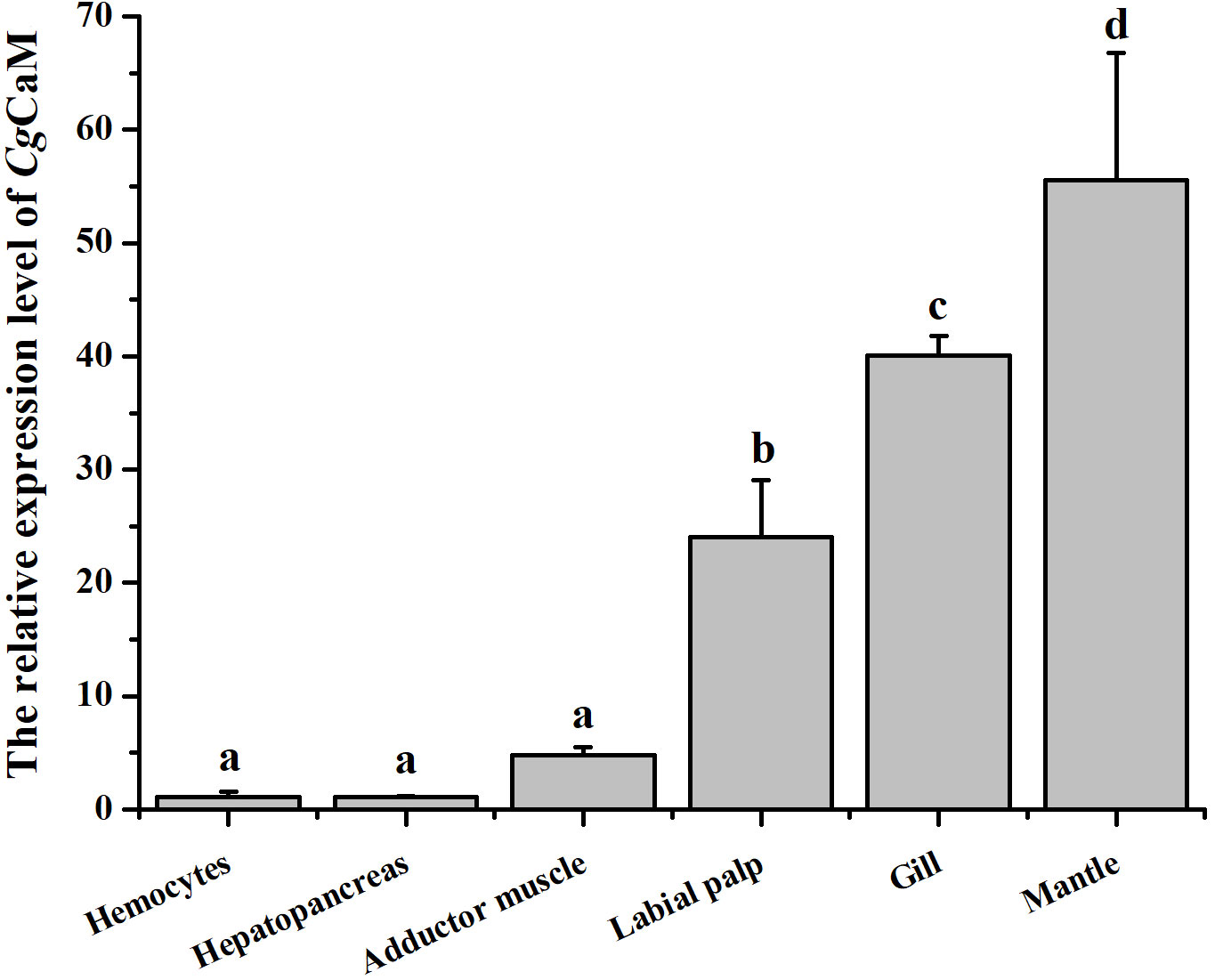
Figure 2 The mRNA expression level of CgCaM in different tissues. Each value was shown as mean ± S.D. (N = 3), and the significant differences (p < 0.05) were marked by different letters (a-d).
The expression level of CgCaM mRNA in IF, MF, OF and the whole mantle of oysters under acidification treatment were determined by quantitative real-time PCR analysis (Figure 3). In the control group, the highest expression level was detected in MF, which was 27.83-fold (p < 0.05) of that in IF. While the mRNA expression of CgCaM in OF and the whole mantle were 11.03-fold (p < 0.05) and 7.75-fold (p < 0.05) of that in IF, respectively. After the acidification treatment for fourteen days, the mRNA expression level of CgCaM in CO2 treatment group was significantly down-regulated in MF, which was 0.13-fold (p < 0.05) of that in the control group. However, the mRNA expression level of CgCaM in OF was up-regulated in CO2 treatment group, which was 1.42-fold (p < 0.05) of that in the control group. There was no significant difference of CgCaM mRNA expression in IF and the whole mantle under acidification treatment.
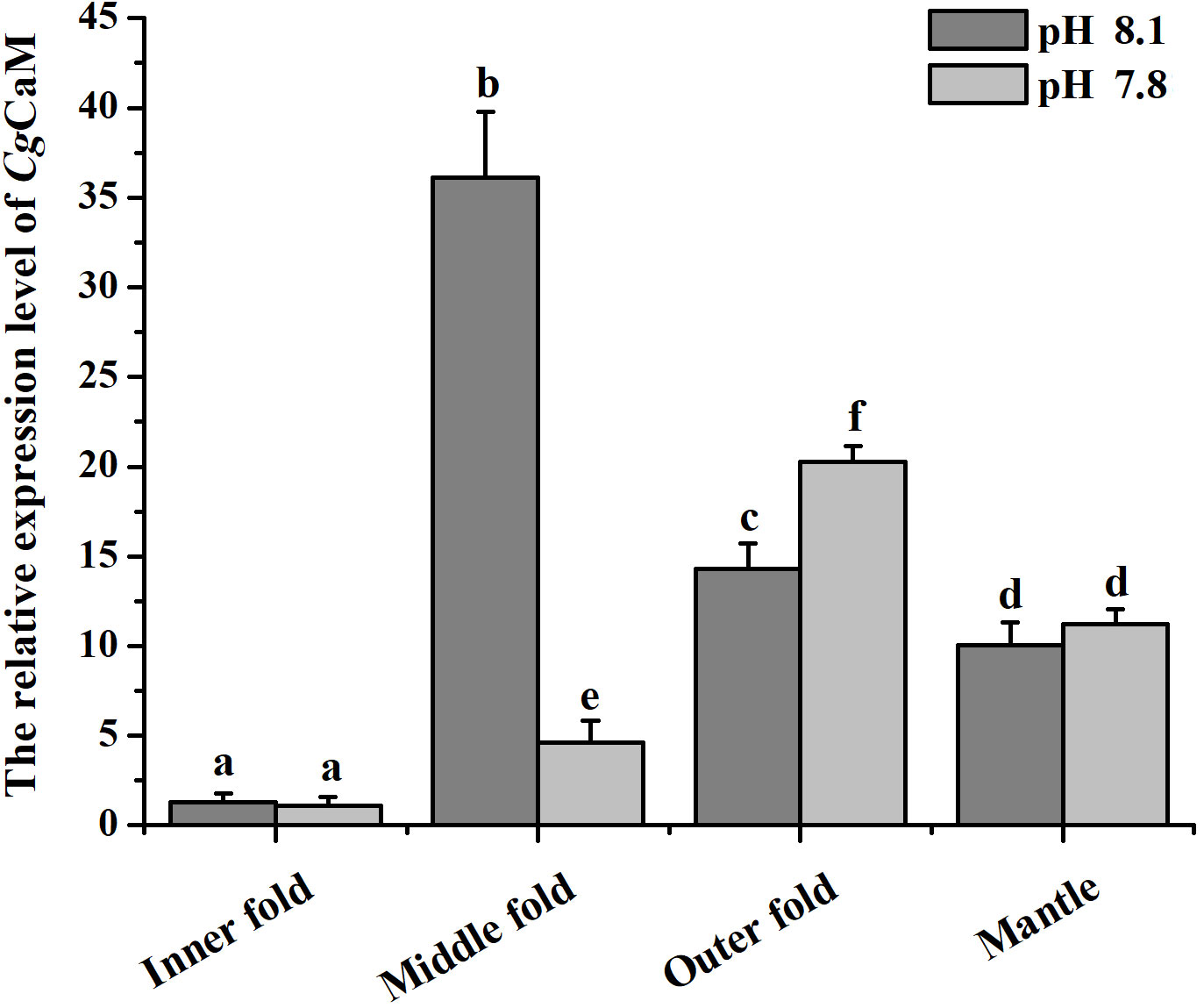
Figure 3 The mRNA expression level of CgCaM in different folds of mantle under acidification treatment. Each value was shown as mean ± S.D. (N = 3), and the significant differences (p < 0.05) were marked by different letters (a-f).
The precise location of CgCaM mRNA in the mantle tissue was analyzed via in situ hybridization. The positive hybridization signals of CgCaM were observed in epithelium of the three mantle folds in the control group (Figure 4B). After acidification treatment, the positive signals in mantle epithelium were weakened, and no obvious positive signals were observed in the outer epithelium of MF (Figure 4D). No signals were detected in negative control group (Figures 4A, C).
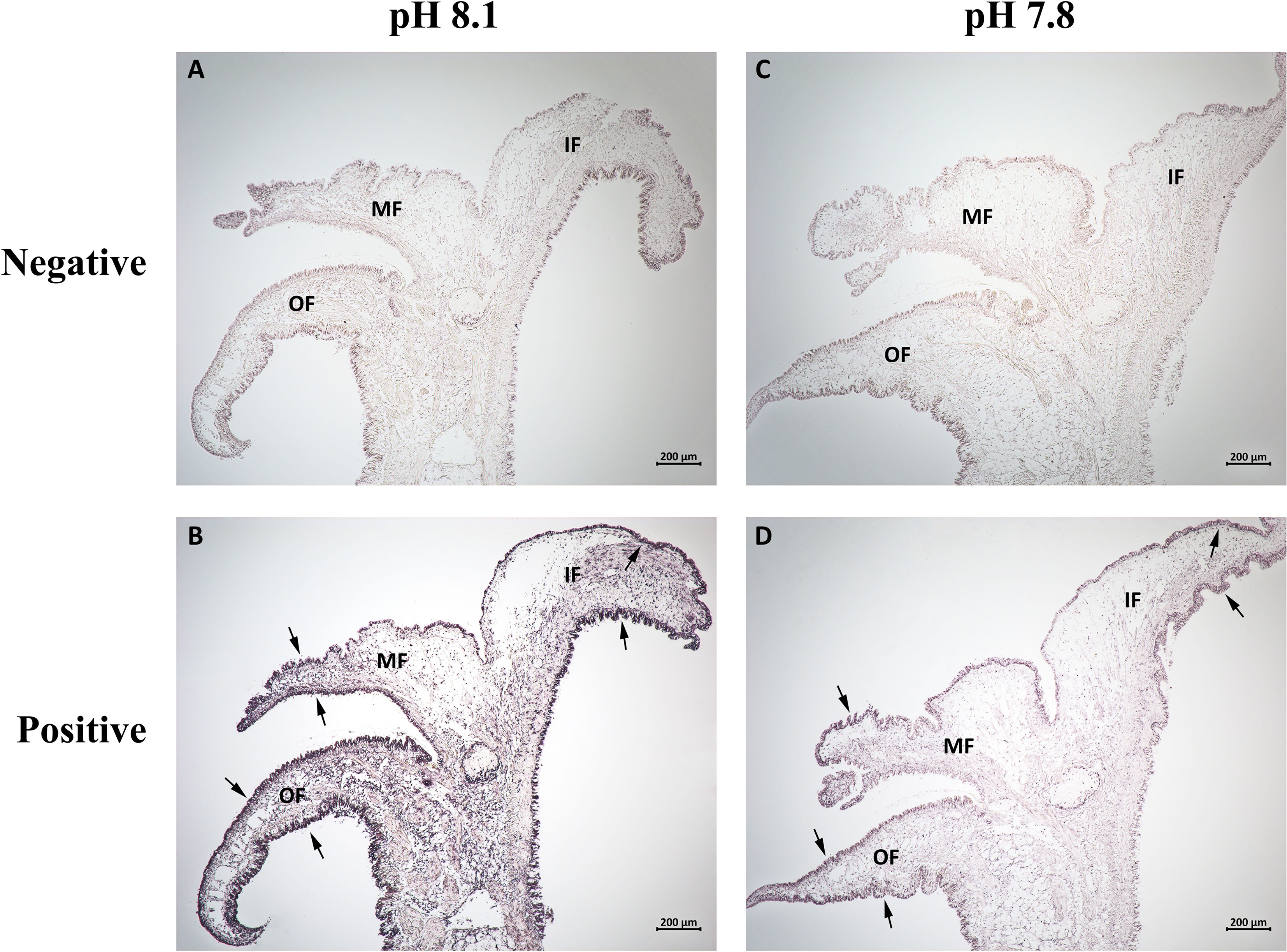
Figure 4 In situ hybridization analysis of CgCaM mRNA in the mantle of oyster C. gigas under acidification treatment. The sections of the oyster mantle were (A, C) hybridized with the sense RNA probes as negative control, while (B, D) hybridized with the anti-sense RNA probes were the positive group. The hybridization positive signals were labeled with arrows. OF, outer fold; MF, middle fold; IF, inner fold.
After IPTG induction, rCgCaM was purified and analyzed by 12% SDS-PAGE. A distinct band was observed with a molecular mass about 25 kDa, which was consistent with the predicted molecular mass of rCgCaM (Figure 5A).
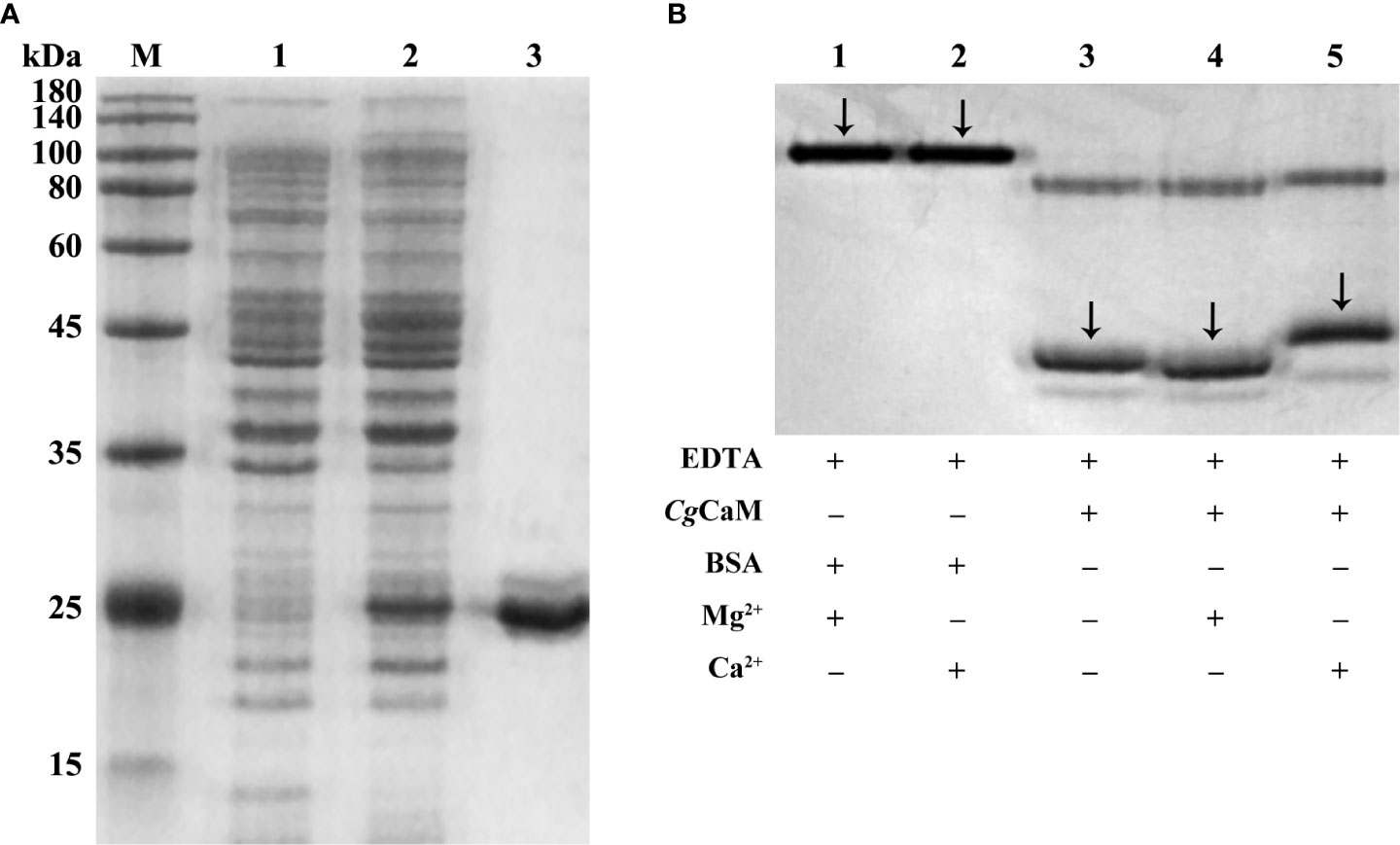
Figure 5 (A) SDS-PAGE analysis of rCgCaM. Lane M: protein molecular standard (kDa); Lane 1: negative control without IPTG induction; Lane 2: positive transformant induced by IPTG; Lane 3: purified rCgCaM. (B) Ca2+-dependent electrophoretic migration analysis of purified rCgCaM. Lane 1: BSA + 20 mM EDTA + 1 mM Mg2+; Lane 2: BSA + 20 mM EDTA + 1 mM Ca2+; Lane 3: rCgCaM + 20 mM EDTA; Lane 4: rCgCaM + 20 mM EDTA + 1 mM Mg2+; Lane 5: rCgCaM + 20 mM EDTA + 1 mM Ca2+.
To further confirm the calcium-binding activity of CgCaM, Ca2+ dependent electrophoretic migration assay was performed. The purified rCgCaM protein with the sample buffer were separated by PAGE (Figure 5B). After addition with 1 mM CaCl2, the band of rCgCaM was observed with obviously retardation effect. No mobility shift was observed in the presence of 1 mM MgCl2. This result suggested that CgCaM protein had calcium-binding activity.
The relative intracellular Ca2+ content in mantle cells of both control group and CO2 treatment group were determined by flow cytometry (Figures 6B, C). In the control group, the Ca2+ content in IF and MF were significantly higher than that in OF, which were both 1.46-fold (p < 0.05) of that in OF. After acidification treatment, the relative intracellular Ca2+ content in CO2 treatment group was significantly increased in IF, which was 1.23-fold (p < 0.05) of that in the control group. While the relative intracellular Ca2+ content in MF and the whole mantle were decreased in CO2 treatment group, which was 0.62-fold (p < 0.05) and 0.81-fold (p < 0.05) of that in the control group, respectively. The results indicated that the Ca2+ content in IF, MF and the whole mantle all changed significantly after acidification treatment. It increased in IF but decreased in MF and whole mantle with showed the most dramatic drop in MF. No significant change of Ca2+ content in OF was observed after the treatment with CO2 (Figure 6A).
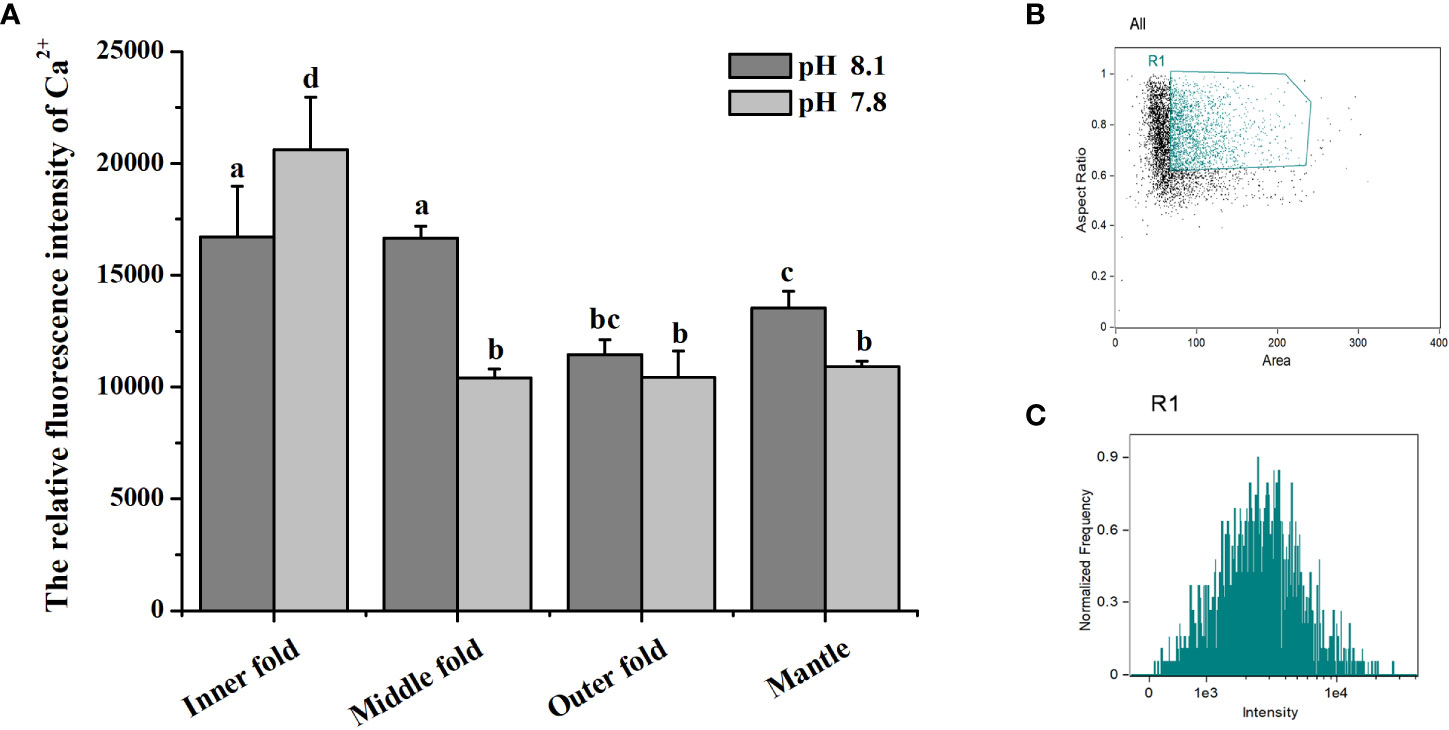
Figure 6 (A) Ca2+ content in oyster mantle cells of control group and CO2 treatment group. Each value was shown as mean ± S.D. (N = 3), and the significant differences (p < 0.05) were marked by different letters (a-d). (B, C) Flow cytometry analysis about the fluorescence intensity of mantle cells incubated with Fluo-4 AM.
Due to the human activities, the emissions of CO2 into the atmosphere is increasing (Field and Barros, 2014), and OA is becoming an irreversible environmental problem with further impacts on marine organisms (Feely et al., 2004; Orr et al., 2005). The calcified shells of bivalves play a vital protective role against environmental stresses, and the reduction of available carbonate saturation in seawater under OA may lead to difficulties in shell biosynthesis (Doney et al., 2009; Thomsen et al., 2015). The shell biosynthesis is a complicated calcification process, calcium homeostasis is crucial for bivalve calcification. CaM is proved to be a pivotal calcium regulator as well as shell formation participant in bivalves (Huang et al., 2007). In the present study, a CaM (designated CgCaM) was identified from C. gigas, and its response against acidification was investigated to explore its possible role in regulation of calcium homeostasis within mantle under OA.
CaM is an important calcium-binding protein with typical EF-hand conformation (Babu et al., 1988). CgCaM identified in the present study contained four continuous typical EF-hand domains (Figures 1A, B) located from 16 to 44 amino acids, 52 to 80 amino acids, 89 to 117 amino acids, and 125 to 153 amino acids. EF-hand is one of the most common calcium-binding domains in both vertebrates and invertebrates, which consists of a loop of twelve residues, and the six residues involved in Ca2+ binding are in positions 1, 3, 5, 7, 9 and 12 (Ikura, 1996; Kretsinger, 1997). As each EF-hand domain is able to bind one Ca2+, the presence of four EF-hand domains in CgCaM enables the mature CgCaM protein to bind four calcium ions. The amino acid sequence of CgCaM was found to share high homology with CaM from other invertebrates and vertebrates (Figure 1C). In the phylogenic tree, CgCaM was clustered with CaM from other bivalves such as C. virginica and P. fucata (Figure 1D). These results suggested that CgCaM belonged to the calcium-binding protein family, which might exhibit the similar functions as its homologues in other invertebrates and vertebrates.
It has been reported that CaM plays an important role in calcification process of marine bivalves. A calmodulin-like protein (CaLP) from pearl oyster P. fucata was reported to modify shell calcite morphology, and the aragonite crystals could be induced when a nacre protein was mixed CaLP (Yan et al., 2007). A mutant of CaLP distributed in mantle epithelium cytoplasm of P. fucata was able to facilitate the mRNA expression of osteopontin (Fang et al., 2008b). Mantle is the most significant tissue involved in calcification of bivalves (Bjärnmark et al., 2016; Yarra et al., 2016). In the present study, the CgCaM was found to be highest expressed in mantle among all examined tissues (Figure 2), suggesting that CgCaM might be involved in the calcification process of oysters. It has been reported that the numbers of EF-hand family members were higher in highly calcified species than that in low calcified species, implying that EF-hand domains were related to biomineralization (Li et al., 2016c). A EF-hand calcium-binding protein EFCBP has been demonstrated to involve in shell formation in P. fucata (Huang et al., 2007). CgCaM was characterized as a calcium-binding protein with four EF-hand domains (Figure 1B), which was further suspected to be involved in calcification. Moreover, the epithelium of mantle is proved to be the exact region regulating the secretion and deposition of shell components (Kocot et al., 2016), and the expression and distribution of CaM in epithelium have also been reported in other bivalves (Li et al., 2004; Fang et al., 2008b). In the present study, CgCaM was found to be mainly distributed in the epithelium of mantle (Figure 4). These results suggested that CgCaM participated in the process of shell calcification in C. gigas.
Mantle is an extremely important organ in bivalves, which plays an essential role in multiple physiological processes (Brake et al., 2004; Abele et al., 2010; Itoh et al., 2010), especially calcification (Zhang et al., 2012). It is divided into three regions with respective function, which have the divergent responses to various physiological processes (Bjärnmark et al., 2016). From the umbo to posterior edge, the three regions of mantle are respectively named as central zone, submarginal zone and marginal zone, while the marginal zone can be further divided into IF, MF and OF. In the shell formation process, the distinct morphogenetic regions are responsible for the synthesis of specific shell layers. The outer periostracum is secreted by epitheliums and basal cells within the periostracal groove between the OF and the MF (Bubel, 1973; Nakayama et al., 2013). The middle prismatic layer is formed by the secretion from columnar epithelial cells in the marginal zone, while the inner nacreous layer is formed by the secretion from the submarginal zone and central zone (Fang et al., 2008a; Gardner et al., 2011; Shi et al., 2012). Most of the previous studies related to the functions of mantle in shell formation focused on the pearl oyster P. fucata. Many shell matrix proteins are mainly distributed in the outer mantle fold or secreted from the epithelium of the outer fold (Nakayama et al., 2013). Besides, the inner mantle fold is also related to shell calcification, it takes up ions required for calcification from the ambient medium (Marin et al., 2008). In Pacific oyster, a previous research proved that engrailed in the middle mantle fold played a major role in shell formation in response to ocean acidification (Zhang et al., 2020). In the present study, the mRNA transcripts of CgCaM were found to be highly expressed in the MF among the three mantle folds of marginal zone (Figure 3), indicating that CgCaM played a major role within the MF during the shell calcification. Calcification is one of the notably affected processes in bivalves in acidified environment (Chandra Rajan et al., 2021; Li et al., 2022). It was reported that the expression of calcification-related genes in mantle were significantly altered under OA to regulate the calcification process (Hüning et al., 2013; Li et al., 2016a). In the present study, the most significant change of CgCaM expression occurred in the MF after acidification treatment (Figure 3). Meanwhile, the positive signal of CgCaM revealed by in situ hybridization was found to disappear only in the outer epithelium of MF (Figure 4). The major function of the mantle MF in shell formation was proved by the previous research (Zhang et al., 2020). These results collectively suggested that CgCaM in the MF of mantle played a major role in the shell calcification of C. gigas in response to OA.
CaM is able to bind Ca2+ and trigger the downstream pathways to mediate a variety of Ca2+-induced physiological processes. As the most important intracellular calcium regulator (Means and Dedman, 1980; Crivici and Ikura, 1995), the no-load CaM does not possess any enzymatic or biological activity. CaM becomes active after it binds with Ca2+ to form Ca2+-CaM complex and undergoes a conformational change to expose the hydrophobic region (Ikura, 1996; Gifford et al., 2007). When extracellular Ca2+ enters the cell, CaM binds Ca2+ more rapidly than other Ca2+-binding proteins (Faas et al., 2011). In this study, CgCaM was certified to display Ca2+-binding activity by Ca2+-dependent electrophoretic shift assay (Figure 5), implying that CgCaM was able to act as an intracellular fast buffer to adjust the free Ca2+ levels and regulate calcium homeostasis. Ca2+ is the most abundant mineral as well as the primary cation in shell formation of C. gigas, and the regulation of calcium levels in the mantle is essential to calcification process (Rousseau et al., 2003). A series of Ca2+-binding proteins, Ca2+-exchangers, Ca2+-channels and Ca2+-ATPases in mantle have been demonstrated to participate in Ca2+ absorption, transport and deposition in order to maintain the calcium homeostasis (Sillanpää et al., 2018; Sillanpää et al., 2020). The expression of these above genes changed greatly under OA stress, which resulted in the calcium homeostasis disequilibrium in marine bivalves (Wang et al., 2020). In the present study, the expression level of CgCaM in the MF of mantle decreased significantly (Figure 3), and the content of Ca2+ was also significantly reduced after C. gigas was exposed to the acidified seawater (Figure 6). Similarly, the significant change of CaM and CaLP expression under long-term OA treatment was also reported to alter the free Ca2+ concentration in mantle cells of P. fucata (Li et al., 2016a). These results inferred that the down-regulated expression of CgCaM under OA resulted in the alteration of Ca2+ content in the mantle MF, which might inhibit calcification process of C. gigas.
In summary, a CgCaM with four continuous EF-hand domains was characterized to exhibit Ca2+-binding activity in oyster C. gigas. The highest mRNA expression level of CgCaM was detected in the MF of mantle. The mRNA expression of CgCaM and Ca2+ content in the MF of mantle decreased significantly after the acidification treatment. These results suggested that CgCaM might affect the calcification process of C. gigas under OA by regulating calcium homeostasis in the MF of mantle.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
XX, CL, and ZL conceived and designed the experiments. XX, YZ, YG, and TZ performed the experiments. XX and CL analyzed the data. ZL, LW, and LS contributed reagents, materials, and analysis tools. XX wrote the original manuscript. CL, LW and LS revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
We are grateful to all the laboratory members for their technical advice and helpful discussions. This research was supported by NSFC grants (Nos. 32072946), National Key R&D Program (2018YFD0900606), the earmarked fund for CARS-49, the Fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research, the Distinguished Professor of Liaoning (XLYC1902012), and Talented Scholars in Dalian Ocean University.
We are grateful to all the laboratory members for their technical advice and helpful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abele D., Kruppe M., Philipp E. E., Brey T. (2010). Mantle cavity water oxygen partial pressure (PO2) in marine molluscs aligns with lifestyle. Can. J. Fish. Aquat. Sci. 67 (6), 977–986. doi: 10.1139/F10-035
Addadi L., Joester D., Nudelman F., Weiner S. (2006). Mollusk shell formation: A source of new concepts for understanding biomineralization processes. Chemistry 12 (4), 980–987. doi: 10.1002/chem.200500980
Audino J. A., Marian J. E., Wanninger A., Lopes S. G. (2015). Mantle margin morphogenesis in Nodipecten nodosus (Mollusca: Bivalvia): New insights into the development and the roles of bivalve pallial folds. BMC Dev. Biol. 15, 22. doi: 10.1186/s12861-015-0074-9
Babu Y. S., Bugg C. E., Cook W. J. (1988). Structure of calmodulin refined at 2.2 a resolution. J. Mol. Biol. 204 (1), 191–204. doi: 10.1016/0022-2836(88)90608-0
Bednaršek N., Feely R. A., Reum J. C., Peterson B., Menkel J., Alin S. R., et al. (2014). Limacina helicina shell dissolution as an indicator of declining habitat suitability owing to ocean acidification in the California current ecosystem. Proc. Biol. Sci. 281 (1785), 20140123. doi: 10.1098/rspb.2014.0123
Bjärnmark N. A., Yarra T., Churcher A. M., Felix R. C., Clark M. S., Power D. M. (2016). Transcriptomics provides insight into Mytilus galloprovincialis (Mollusca: Bivalvia) mantle function and its role in biomineralisation. Mar. Genom. 27, 37–45. doi: 10.1016/j.margen.2016.03.004
Brake J., Evans F., Langdon C. (2004). Evidence for genetic control of pigmentation of shell and mantle edge in selected families of pacific oysters, Crassostrea gigas. Aquaculture 229 (1-4), 89–98. doi: 10.1016/S0044-8486(03)00325-9
Bubel A. (1973). An electron-microscope study of periostracum formation in some marine bivalves. II. The cells lining the periostracal groove. Mar. Biol. 20 (3), 222–234. doi: 10.1007/BF00348988
Burgess W. H., Jemiolo D. K., Kretsinger R. H. (1980). Interaction of calcium and calmodulin in the presence of sodium dodecyl sulfate. Biochim. Biophys. Acta 623 (2), 257–270. doi: 10.1016/0005-2795(80)90254-8
Caldeira K., Wickett M. E. (2003). Oceanography: Anthropogenic carbon and ocean pH. Nature 425 (6956), 365. doi: 10.1038/425365a
Chandra Rajan K., Meng Y., Yu Z., Roberts S. B., Vengatesen T. (2021). Oyster biomineralization under ocean acidification: From genes to shell. Glob. Change Biol. 27 (16), 3779–3797. doi: 10.1111/gcb.15675
Chandra Rajan K., Vengatesen T. (2020). Molecular adaptation of molluscan biomineralisation to high-CO2 oceans - the known and the unknown. Mar. Environ. Res. 155, 104883. doi: 10.1016/j.marenvres.2020.104883
Cheung W. Y. (1984). Calmodulin: Its potential role in cell proliferation and heavy metal toxicity. Fed. Proc. 43 (15), 2995–2999.
Crivici A., Ikura M. (1995). Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24, 85–116. doi: 10.1146/annurev.bb.24.060195.000505
Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A. (2009). Ocean acidification: The other CO2 problem. Ann. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
Faas G. C., Raghavachari S., Lisman J. E., Mody I. (2011). Calmodulin as a direct detector of Ca2+ signals. Nat. Neurosci. 14 (3), 301–304. doi: 10.1038/nn.2746
Fang Z., Feng Q., Chi Y., Xie L., Zhang R. (2008a). Investigation of cell proliferation and differentiation in the mantle of Pinctada fucata (Bivalve, Mollusca). Mar. Biol. 153 (4), 745–754. doi: 10.1007/s00227-007-0851-5
Fang Z., Yan Z., Li S., Wang Q., Cao W., Xu G., et al. (2008bb). Localization of calmodulin and calmodulin-like protein and their functions in biomineralization in P. fucata. Prog. Nat. Sci. 18 (4), 405–412. doi: 10.1016/j.pnsc.2007.11.011
Feely R. A., Sabine C. L., Lee K., Berelson W., Kleypas J., Fabry V. J., et al. (2004). Impact of anthropogenic CO2 on the CaCO3 system in the oceans. Science 305 (5682), 362–366. doi: 10.1126/science.1097329
Field C. B., Barros V. R. (2014). Climate change 2014: impacts, adaptation, and vulnerability. part a: global and sectoral aspects (Cambridge: University Press).
Föller M., Bobbala D., Koka S., Boini K. M., Mahmud H., Kasinathan R. S., et al. (2010). Functional significance of the intermediate conductance Ca2+-activated k+ channel for the short-term survival of injured erythrocytes. Pflugers Arch. 460 (6), 1029–1044. doi: 10.1007/s00424-010-0878-1
Gardner L. D., Mills D., Wiegand A., Leavesley D., Elizur A. (2011). Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyster Pinctada maxima. BMC Genomics 12, 455. doi: 10.1186/1471-2164-12-455
Gifford J. L., Walsh M. P., Vogel H. J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405 (2), 199–221. doi: 10.1042/BJ20070255
Hendriks I. E., Duarte C. M., Olsen Y. S., Steckbauer A., Ramajo L., Moore T. S., et al. (2015). Biological mechanisms supporting adaptation to ocean acidification in coastal ecosystems. Estuar. Coast. Shelf Sci. 152, A1–A8. doi: 10.1016/j.ecss.2014.07.019
Hettinger A., Sanford E., Hill T., Hosfelt J., Russell A., Gaylord B. (2013). The influence of food supply on the response of Olympia oyster larvae to ocean acidification. Biogeosciences 10 (10), 6629–6638. doi: 10.5194/bgd-10-5781-2013
Huang W., Yao B., Sun L., Pu R., Wang L., Zhang R. (2001). Immunohistochemical and in situ hybridization studies of gonadotropin releasing hormone (GnRH) and its receptor in rat digestive tract. Life Sci. 68 (15), 1727–1734. doi: 10.1016/s0024-3205(01)00968-7
Huang J., Zhang C., Ma Z., Xie L., Zhang R. (2007). A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim. Biophys. Acta 1770 (7), 1037–1044. doi: 10.1016/j.bbagen.2007.03.006
Hüning A. K., Melzner F., Thomsen J., Gutowska M. A., Krämer L., Frickenhaus S., et al. (2013). Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: Implications for shell formation and energy metabolism. Mar. Biol. 160 (8), 1845–1861. doi: 10.1007/s00227-012-1930-9
Ikura M. (1996). Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 21 (1), 14–17. doi: 10.1016/S0968-0004(06)80021-6
Itoh N., Okada Y., Takahashi K. G., Osada M. (2010). Presence and characterization of multiple mantle lysozymes in the pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 29 (1), 126–135. doi: 10.1016/j.fsi.2010.02.027
Johnson K. L., Zhen D. K., Bianchi D. W. (2000). The use of fluorescence in situ hybridization (FISH) on paraffin-embedded tissue sections for the study of microchimerism. Biotechniques 29 (6), 1220–1224. doi: 10.2144/00296st01
Kádár E. (2008). Haemocyte response associated with induction of shell regeneration in the deep-sea vent mussel Bathymodiolus azoricus (Bivalvia: Mytilidae). J. Exp. Mar. Biol. Ecol. 362 (2), 71–78. doi: 10.1016/j.jembe.2008.05.014
Kocot K. M., Aguilera F., McDougall C., Jackson D. J., Degnan B. M. (2016). Sea Shell diversity and rapidly evolving secretomes: Insights into the evolution of biomineralization. Front. Zool. 13, 23. doi: 10.1186/s12983-016-0155-z
Kretsinger R. H. (1997). EF-hands embrace. Nat. Struct. Biol. 4 (7), 514–516. doi: 10.1038/nsb0797-514
Li S., Huang J., Liu C., Liu Y., Zheng G., Xie L., et al. (2016a). Interactive effects of seawater acidification and elevated temperature on the transcriptome and biomineralization in the pearl oyster Pinctada fucata. Environ. Sci. Technol. 50 (3), 1157–1165. doi: 10.1021/acs.est.5b05107
Li S., Liu C., Huang J., Liu Y., Zhang S., Zheng G., et al. (2016b). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6, 18943. doi: 10.1038/srep18943
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li S., Xie L., Zhang C., Zhang Y., Gu M., Zhang R. (2004). Cloning and expression of a pivotal calcium metabolism regulator: Calmodulin involved in shell formation from pearl oyster (Pinctada fucata). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 138 (3), 235–243. doi: 10.1016/j.cbpc.2004.03.012
Li X., Yu W., Cai Z., He C., Wei N., Wang X., et al. (2016c). Molecular cloning and characterization of full-length cDNA of calmodulin gene from pacific oyster Crassostrea gigas. Biomed. Res. Int. 2016, 5986519. doi: 10.1155/2016/5986519
Li J., Zhou Y., Qin Y., Wei J., Shigong P., Ma H., et al. (2022). Assessment of the juvenile vulnerability of symbiont-bearing giant clams to ocean acidification. Sci. Total Environ. 812, 152265. doi: 10.1016/j.scitotenv.2021.152265
Lv X., Wang W., Han Z., Liu S., Yang W., Li M., et al. (2019). The dicer from oyster Crassostrea gigas functions as an intracellular recognition molecule and effector in anti-viral immunity. Fish Shellfish Immunol. 95, 584–594. doi: 10.1016/j.fsi.2019.10.067
Marin F., Le Roy N., Marie B. (2012). The formation and mineralization of mollusk shell. Front. Biosci. (Schol. Ed.) 4 (3), 1099–1125. doi: 10.2741/s321
Marin F., Luquet G., Marie B., Medakovic D. (2008). Molluscan shell proteins: primary structure, origin, and evolution. Curr. Top. Dev. Biol. 80, 209–276. doi: 10.1016/S0070-2153(07)80006-8
Means A. R., Dedman J. R. (1980). Calmodulin–an intracellular calcium receptor. Nature 285 (5760), 73–77. doi: 10.1038/285073a0
Mount A. S., Wheeler A. P., Paradkar R. P., Snider D. (2004). Hemocyte-mediated shell mineralization in the eastern oyster. Science 304 (5668), 297–300. doi: 10.1126/science.1090506
Nakayama S., Suzuki M., Endo H., Iimura K., Kinoshita S., Watabe S., et al. (2013). Identification and characterization of a matrix protein (PPP-10) in the periostracum of the pearl oyster, Pinctada fucata. FEBS Open Bio 3, 421–427. doi: 10.1016/j.fob.2013.10.001
Orr J. C., Fabry V. J., Aumont O., Bopp L., Doney S. C., Feely R. A., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437 (7059), 681–686. doi: 10.1038/nature04095
Peng K., Liu F. L., Wang J. H., Hong Y. J. (2018). Calmodulin highly expressed during the formation of pearl sac in freshwater pearl mussel (Hyriopsis schlegelii). Thalassas: an Int. J. Mar. Sci. 34 (1), 219–225. doi: 10.1007/s41208-017-0054-x
Richards M., Xu W., Mallozzi A., Errera R. M., Supan J. (2018). Production of calcium-binding proteins in Crassostrea virginica in response to increased environmental CO2 concentration. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00203
Ross P. M., Parker L., O’Connor W. A., Bailey E. A. (2011). The impact of ocean acidification on reproduction, early development and settlement of marine organisms. Water 3 (4), 1005–1030. doi: 10.3390/w3041005
Rousseau M., Plouguerne E., Wan G., Wan R., Lopez E., Fouchereau-Peron M. (2003). Biomineralisation markers during a phase of active growth in Pinctada margaritifera. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 135 (2), 271–278. doi: 10.1016/s1095-6433(03)00070-9
Schwaller B. (2012). The regulation of a cell's Ca2+ signaling toolkit: the Ca2+ homeostasome. Adv. Exp. Med. Biol. 740, 1–25. doi: 10.1007/978-94-007-2888-2_1
Sée V., Boutillier A. L., Bito H., Loeffler J. P. (2001). Calcium/calmodulin-dependent protein kinase type IV (CaMKIV) inhibits apoptosis induced by potassium deprivation in cerebellar granule neurons. FASEB J. 15 (1), 134–144. doi: 10.1096/fj.00-0106com
Senawong G., Laha T., Loukas A., Brindley P. J., Sripa B. (2012). Cloning, expression, and characterization of a novel Opisthorchis viverrini calcium-binding EF-hand protein. Parasitol. Int. 61 (1), 94–100. doi: 10.1016/j.parint.2011.07.012
Shi S., Jiao Y., Du X., Zhang Y. (2012). Ultrastructure of mantle epithelial cells in Pinctada martensii. Guangdong Agric. Sci. 39, 135–137.
Sillanpää J. K., Cardoso J., Félix R. C., Anjos L., Power D. M., Sundell K. (2020). Dilution of seawater affects the Ca2+ transport in the outer mantle epithelium of Crassostrea gigas. Front. Physiol. 11. doi: 10.3389/fphys.2020.00001
Sillanpää J. K., Sundh H., Sundell K. S. (2018). Calcium transfer across the outer mantle epithelium in the pacific oyster, Crassostrea gigas. Proc. Biol. Sci. 285, 20181676. doi: 10.1098/rspb.2018.1676
Stevens F. C. (1983). Calmodulin: an introduction. Can. J. Biochem. Cell Biol. 61 (8), 906–910. doi: 10.1139/o83-115
Thomsen J., Haynert K., Wegner K. M., Melzner F. (2015). Impact of seawater carbonate chemistry on the calcification of marine bivalves. Biogeosciences 12 (14), 4209–4220. doi: 10.5194/bg-12-4209-2015
Waldbusser G. G., Brunner E. L., Haley B. A., Hales B., Langdon C. J., Prahl F. G. (2013). A developmental and energetic basis linking larval oyster shell formation to acidification sensitivity. Geophys. Res. Lett. 40 (10), 2171–2176. doi: 10.1002/grl.50449
Wang X., Li C., Lv Z., Zhang Z., Qiu L. (2022). A calcification-related calmodulin-like protein in the oyster Crassostrea gigas mediates the enhanced calcium deposition induced by CO2 exposure. Sci. Total Environ. 833, 155114. doi: 10.1016/j.scitotenv.2022.155114
Wang X., Wang M., Wang W., Liu Z., Xu J., Jia Z., et al. (2020). Transcriptional changes of pacific oyster Crassostrea gigas reveal essential role of calcium signal pathway in response to CO2-driven acidification. Sci. Total Environ. 741, 140177. doi: 10.1016/j.scitotenv.2020.140177
Yan Z., Fang Z., Ma Z., Deng J., Li S., Xie L., et al. (2007). Biomineralization: functions of calmodulin-like protein in the shell formation of pearl oyster. Biochim. Biophys. Acta 1770 (9), 1338–1344. doi: 10.1016/j.bbagen.2007.06.018
Yarra T., Gharbi K., Blaxter M., Peck L. S., Clark M. S. (2016). Characterization of the mantle transcriptome in bivalves: Pecten maximus, Mytilus edulis and Crassostrea gigas. Mar. Genom 27, 9–15. doi: 10.1016/j.margen.2016.04.003
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang Y., Liu Z., Song X., Huang S., Wang L., Song L. (2019). The inhibition of ocean acidification on the formation of oyster calcified shell by regulating the expression of Cgchs1 and Cgchit4. Front. Physiol. 10, 1034. doi: 10.3389/fphys.2019.01034
Zhang Y., Liu Z., Zong Y., Zheng Y., Li Y., Han Z., et al. (2020). The increased expression of an engrailed to sustain shell formation in response to ocean acidification. Front. Physiol. 11. doi: 10.3389/fphys.2020.530435
Zhou Y., Xue S., Yang J. J. (2013). Calciomics: integrative studies of Ca2+-binding proteins and their interactomes in biological systems. Metallomics 5 (1), 29–42. doi: 10.1039/c2mt20009k
Keywords: ocean acidification, calmodulin, calcium homeostasis, mantle, Crassostrea gigas
Citation: Xin X, Liu C, Liu Z, Zhang Y, Gao Y, Zhu T, Wang L and Song L (2022) Calmodulin regulates the calcium homeostasis in mantle of Crassostrea gigas under ocean acidification. Front. Mar. Sci. 9:1050022. doi: 10.3389/fmars.2022.1050022
Received: 21 September 2022; Accepted: 26 October 2022;
Published: 11 November 2022.
Edited by:
Taewoo Ryu, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Liqiang Zhao, Guangdong Ocean University, ChinaCopyright © 2022 Xin, Liu, Liu, Zhang, Gao, Zhu, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Liu, bGl1Y2hhbmdAZGxvdS5lZHUuY24=; Linsheng Song, bHNoc29uZ0BkbG91LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.