- 1State Environmental Protection Key Laboratory of Coastal Ecosystem, National Marine Environmental Monitoring Center, Dalian, China
- 2College of Marine Ecology and Environment, Shanghai Ocean University, Shanghai, China
- 3Groundwater Provincial Engineering Research Center for Urban Water Recycling and Environmental Safety, Shenzhen International Graduate School, Tsinghua University, Shenzhen, China
- 4College of Marine Technology and Environment, Dalian Ocean University, Dalian, China
The existence of potentially pathogenic bacteria seriously threatens aquatic animals and human health. Estuaries are closely related to human activities, and the detection of pathogens is important for aquaculture and public health. However, monitoring only indicator microorganisms and pathogens is not enough to accurately and comprehensively estimate water pollution. Here, the diversity of potentially pathogenic bacteria in water samples from the Liaohe estuary was profiled using 16S next-generation sequencing (16S NGS) and quantitative polymerase chain reaction (qPCR) analysis. The results showed that the dominant genera of environmental pathogens were Pseudomonas, Vibrio, Mycobacterium, Acinetobacter, Exiguobacterium, Sphingomonas, and Legionella, and the abundance of enteric pathogens was significantly less than the environmental pathogens, mainly, Citrobacter, Enterococcus, Escherichia-Shigella, Enterobacter, Bacteroides. The qPCR results showed that the 16S rRNA genes of Vibrio were the most abundant, with concentrations between 7.06 and 9.48 lg copies/L, followed by oaa gene, fliC gene, trh gene, and uidA gene, and the temperature and salinity were the main factors affecting its abundance. Variance partitioning analysis (VPA) analysis of spatial factors on the potential pathogen’s distribution (19.6% vs 5.3%) was greater than environmental factors. In addition, the co-occurrence analysis of potential pathogens in the estuary revealed significant co-occurrence among the opportunistic pathogens Testosteronemonas, Brevimonas vesicularis, and Pseudomonas putida. Our findings provide an essential reference for monitoring and occurrence of potentially pathogenic bacteria in estuaries.
Introduction
The estuarine ecosystem is a highly productive and dynamic buffer zone between freshwater and oceans (McLusky and Elliott, 2004). Population growth, urbanization, and industrialization have increased the input of organic and inorganic matter (such as sewage from human activities, agricultural wastewater, and industrial waste) into coastal water (Yeleliere et al., 2018), resulting in eutrophication with a large influx of pathogenic microorganisms into the estuaries and finally into the sea (Mei Liu et al., 2011; Wang et al., 2015). According to reports, at least 80% of the wastewater generated by human activities is discharged into rivers and oceans without any treatment (Unesco, 2017), and the burden of human diseases caused by marine pathogens has been estimated to be 4 million people lost annually (Leal Diego et al., 2013). Therefore, it is vital to study the pathogenic bacteria in estuaries.
Pathogens are divided into two categories: enteric and environmental (Leclerc et al., 2008; Percival, 2014). Enteric pathogens are mainly derived from fecal contamination and do not easily proliferate in-vitro but survive for a long time under favorable conditions (Brookes et al., 2004). In contrast, the distribution of environmental pathogens is related to various environmental factors (Jacobs et al., 2009). Recent studies have shown that few environmental pathogens, such as Legionella pneumophila and Pseudomonas aeruginosa, were responsible for increasing outbreaks (Percival, 2014; Liang et al., 2020). Some enteric pathogens, such as Vibrio, Escherichia coli, and Salmonella spp., are attributed to an increase in seafood disease outbreaks (Ramaiah, 2002; Rodrigues et al., 2011; Khandeparker et al., 2015), particularly Vibrio parahaemolyticus and Vibrio vulnificus (Ruiz-Cayuso et al., 2021). Vibrio-infected oysters caused the first major outbreak in Europe in 1999 (Lozano-León et al., 2003). Although in recent years, studies on estuaries focused mainly on fecal indicator bacteria, such as pathogenic E. coli and Enterococci (Griffith et al., 2009), there is a lack of understanding of potentially pathogenic bacteria. Poor correlations between fecal indicator bacteria (FIB) and pathogens (Boehm et al., 2003; Harwood et al., 2005) suggest that relying solely on FIB levels to indicate fecal contamination is problematic. Extra-intestinal sources of FIB, such as sediments, soil, and aquatic vegetation, can confuse the relationship between pathogens and FIB (Lamb et al., 2017). Furthermore, many environmental factors may affect the abundance of pathogenic bacteria, with eutrophic conditions, salinity, and temperature considered the most important (Campbell and Kirchman, 2013; Wu et al., 2019a). However, there is limited research on the distribution and factors that influence the spatial morphology of estuaries. Therefore, contemporary research is critical to understanding the diversity of the pathogen community and its coexistence patterns in estuaries.
Good management of the quality of potential pathogens in estuarine water and efficient and exhaustive pathogen monitoring is critical to detect the dominant pathogens. In previous studies, the distribution of pathogens (e.g., Vibrio, FIB, or several enteric pathogens) in estuarine waters was studied primarily by culture or quantitative polymerase chain reaction (qPCR) (Baliarsingh et al., 2021; Soueidan et al., 2021). Compared to culture-dependent methods, qPCR has the advantages of specificity and sensitivity and is widely used to detect pathogenic bacteria. Typically, the researcher chooses the target, thus overlooking some key pathogens in complex environments. Recently, next-generation sequencing (NGS) methods of the 16S rRNA gene have been used to study bacterial pathogens in water (Cui et al., 2019). Studies have shown that while this method can detect hundreds of pathogens, the results are incompletely quantified for potential pathogens. In conclusion, there are limitations in monitoring potentially pathogenic bacteria by traditional culturable and single qPCR or 16S NGS methods.
In this study, surface seawater from eutrophication was collected from the Liaohe estuary, and 16S rRNA gene-targeted NGS technology was used to analyze diversity. Our study aimed to 1) characterize the diversity and abundance of potentially pathogenic bacteria in the Liaohe estuary, 2) uncover distribution factors affecting potential pathogenic bacteria, and 3) identify co-occurrence patterns among potentially pathogenic bacteria.
Materials and methods
Sampling and collection
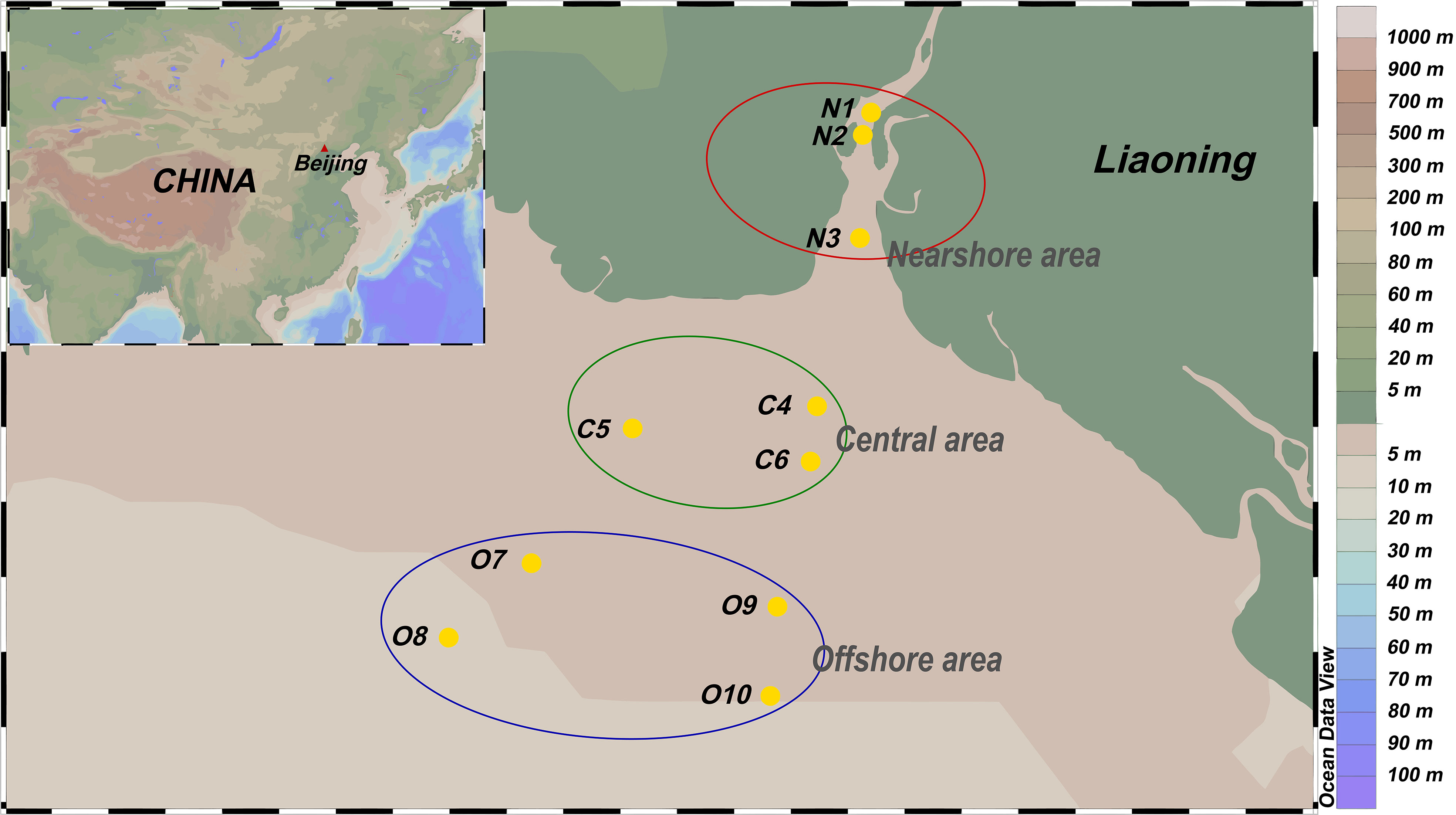
The samples were collected in the Liaohe estuary during the wet season (September 2017). Ten sampling points were established according to salinity and distance from the estuary, which was divided into three areas: near-shore area (N), central area (C) and offshore area (O) (Figure 1). A ZH6547 sampler was used to collect about 4L of seawater from the surface (15-30 cm) and stored in sterile plastic bottles in triplicate. The seawater samples were transported in ice bags (4°C) and delivered to the laboratory within 24 h. Each seawater sample was used for DNA extraction.
Water physicochemical properties
Physicochemical properties for each seawater sample, i.e., seawater depth (Depth), pH, T (Temperature), salinity (Sal), chemical oxygen demand (COD), and dissolved oxygen (DO) were tested using a YSI (Yellow Springs Instrument Company) multi-meter. Phosphate (PO43–) was measured spectrophotometrically (State Oceanic Administration, 2007a), while nitrate (NO3–), nitrite (NO2–), and ammonia (NH4+) concentrations were determined following the marine monitoring specification (State Oceanic Administration, 2007b). Cu, Zn, Pb, Cd, and Cr were measured using flame or flameless atomic absorption spectrophotometry (Cd and Cr) (State Oceanic Administration, 2007c). Suspended solids(SS) were measured by the gravimetric method (National Environmental Protection Agency, 1989).
DNA extraction, sequencing, and data processing
The seawater sample (1L) was filtered through a 0.22 µm filter (Sterivex-GS, Millipore, Billerica, MA, USA), and the DNA was extracted using the Power Seawater® DNA Isolation Kit (MO BIO Inc., Carlsbad, CA, USA). The extracted DNA samples were sent to BOZERON Bioinformatics Institute (Shanghai, China) in triplicate and sequenced using 2 × 300 bp paired-end sequencing through an Illumina MiSeq platform. The extracted DNA was amplified by the V4 and V5 regions of the 16S rRNA gene using the primers 515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 907R: 5′-GGACTACHVGGGTWTCTAAT -3′ by PCR (Ge and Yu, 2017). The raw Illumina sequences were splitting the sequencing data and the construction of barcodes by using the QIIME2 (Bolyen et al., 2019), the sequences were processed by USEARCH(version 7.1) (Edgar, 2010), combined paired-end sequences with the -fasteq_mergepairs command, then the primers were excised (-fastx_truncate), subsequently, the low-quality sequences (-fastq_filter) and redundancy (-fastx_uniques) were filtered and removed, respectively. Then, the operational taxonomic units (OTUs) with 97% similarity were clustered by the -cluster_otus command based on the UPARSE algorithm. Finally, the characteristic sequences were obtained after removing the chimera and the host based on the SILVA v119 database (Yilmaz et al., 2014), and to the OTU table were compared and generated using the -otutab command of USEARCH (-id 0.97).
Accession number
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA855276.
Potential pathogen identification based on 16S NGS
Potential pathogens in the sequencing data were identified from the pathogen databases. A reference database of pathogenic bacteria was created from the literature and the National Marine Environment Monitoring Center pathogen database (https://microbiol.nmemc.org.cn/) and reference lists from other studies, including (59 genera and 177 species) (Wu et al., 2019b) (Table S1). The data included bacterial pathogens from breeding areas, bathing pathogenic microorganisms, and estuarine areas. The representative 16S rRNA gene sequences of the bacterial pathogen species were extracted from NCBI’s Reference Sequence (RefSeq) database. Sequences were annotated to the genus level by SILVA. For further analysis at the species level, unique reads classified for the genera in the reference database of human pathogenic bacteria were screened and aligned with representative sequences of pathogenic species within the same genera using BLAST.
qPCR
A total of five representative potential pathogenic bacterial genes were selected for qPCR amplification, including the uidA gene of E. coli, oaa gene of P. aeruginosa, fliC gene of Vibrio alginolyticus, 16S rRNA gene of Vibrio, trh gene of Vibrio parahaemolyticus because of their prevalence at both the genus and species levels in the screening results by 16S NGS and representative in an estuary. E. coli was considered a fecal contamination indicator and a common enteric waterborne potential pathogen.
Primer information and qPCR conditions are listed in Table S2. The qPCR analysis was performed using an ABI Prism 7500 Sequence Detection System (Applied Biosystems, US) with a 20 µL reaction volume containing 10 µL of TB Green Premix Ex Taq II (Takara, Dalian, China), 0.4 µL of each primer (10 mM), 0.4 µL of ROX II (50X) (Takara, Dalian, China), 2 µL of template DNA and double-distilled water (ddH2O). For the TaqMan qPCR reactions,10 µL consisted of Premix Ex Taq (2X), 0.4 µL of each primer (10 mM), 0.8 µL probe, 0.2 µL ROX Reference Dye (50X), 2 µL of template DNA, and double-distilled water (ddH2O).
The calibration curves based on the appropriate ten-fold dilution series of the corresponding gene standards were run with the test samples and the amplification efficiency was determined to be 80- 105% by the qPCR test, with R2 > 0.99.
Statistical analysis
All statistical analyses were performed using the R program (http://cran.r-project.org/) implemented with various packages. A set of spatial variables was first generated based on the latitude and longitude coordinates of the sampling points, employing a neighborhood matrix principal coordinates (PCNM) analysis method (Borcard and Legendre, 2002). Redundancy Analysis (RDA) was used to analyze the potential pathogen community composition and environmental/spatial variables in the studied estuary. The environmental and spatial variables were screened by the R package ‘vegan’ prior to RDA analysis (Blanchet et al., 2008), using the ‘varpart’ function of the “vegan” package. Only significant (p < 0.05) environmental and spatial variables were selected to perform a Variance partitioning analysis (VPA) to assess the relative impact of environmental and spatial variables on the distribution of potential pathogens. Environmental (E) and spatial (S) variables were explained using pure environmental variables (E|S), pure spatial variables (S|E), and the combined effect of both components (E∩S). The remaining proportion of the differences cannot be explained. Boxplots were drawn using the R package “ggpubr.” A symbiotic network of potentially pathogenic bacteria was inferred from Spearman’s correlation matrix based on estimating abundances of 16S rRNA gene of the potentially pathogenic bacterial species and environmental factors. The “iGraph” package was used for network analysis and drawn using Cytoscape software (version 3.9.0). All statistical tests at p < 0.05 were considered significant.
Results
Physicochemical attributes of the estuary water
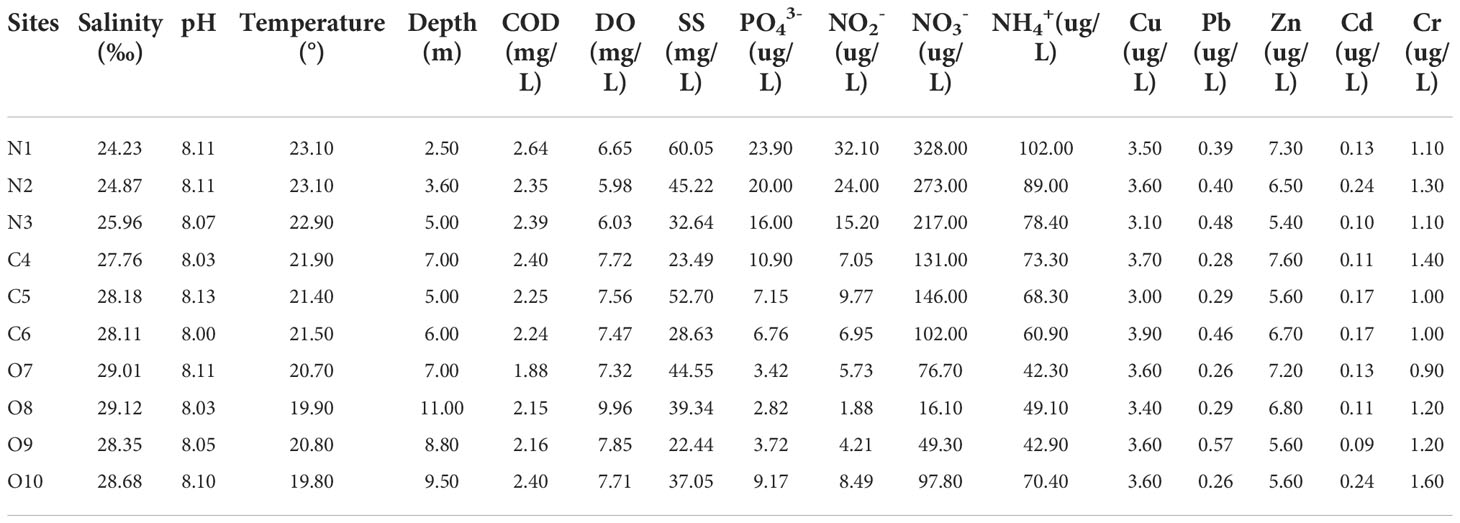
The physicochemical parameters of the samples from ten stations in the Liaohe estuary are shown in Table 1. The surface seawater temperature was between 19.80 and 23.10°C, with the highest recorded in the N area and the lowest in the O area. The salinity gradually increased from the N to the O area, ranging from 24.23-29.12 ‰. Nitrate content was between 16.10-328.00 µg/L, and the ammonia was 42.30-102.00 µg/L. The highest nitrate and ammonia content was found in the N area, the lowest nitrate content was found in the O area, and the lowest ammonia was found in the C area. Phosphate content ranged from 2.82 to 23.90 µg/L, with a similar trend as nitrate. The heavy metal content showed little change between the stations. The concentration ranges were Cu 3.00-3.90 µg/L, Pb 0.26-0.57 µg/L, Zn 5.40-7.60 µg/L, Cd 0.09-0.24 µg/L, Cr 0.90-1.60 µg/L.
Diversity of potential pathogenic bacteria community by 16S NGS
In total, 496,527 qualified sequence reads were obtained from the water samples, averaging 49,653 reads per sample. According to the bacterial database, 15 genera and 31 species were identified as potentially pathogenic bacteria. Five genera of potential enteric pathogens dominated, including Citrobacter, Enterococcus, Escherichia-Shigella, Enterobacter, and Bacteroides, while the potential environmental pathogen genera were more diverse, including Pseudomonas, Vibrio, Mycobacterium, Acinetobacter, Exiguobacterium, Sphingomonas and Legionella, predominated (Figure 2A). The relative abundance and Shannon-Wiener diversity index of dominant potential pathogen species in the estuary are shown in Figure 2B. At the genus level, the relative abundance of Pseudomonas (44.42% and 51.80%) was comparatively high in the N and C areas, while the O area had the highest relative abundance of Vibrio (72.10%). Similarly, Mycobacterium and Acinetobacter were detected in several samples (6 out of 10 samples), and their highest relative abundance was detected in the N area (3.13% and 2.23%) samples. The highest relative abundances of Enterobacter (0.36%) and Bacteroides (0.03%) were mainly concentrated in the C4 site of the C area. Brevundimonas was the most commonly detected potential pathogen in the estuary, with higher relative abundance in the N (0.57%) and O areas (0.24%). Related sequences of Brevundimonas were identified as potential opportunistic pathogens associated with bacteremia.
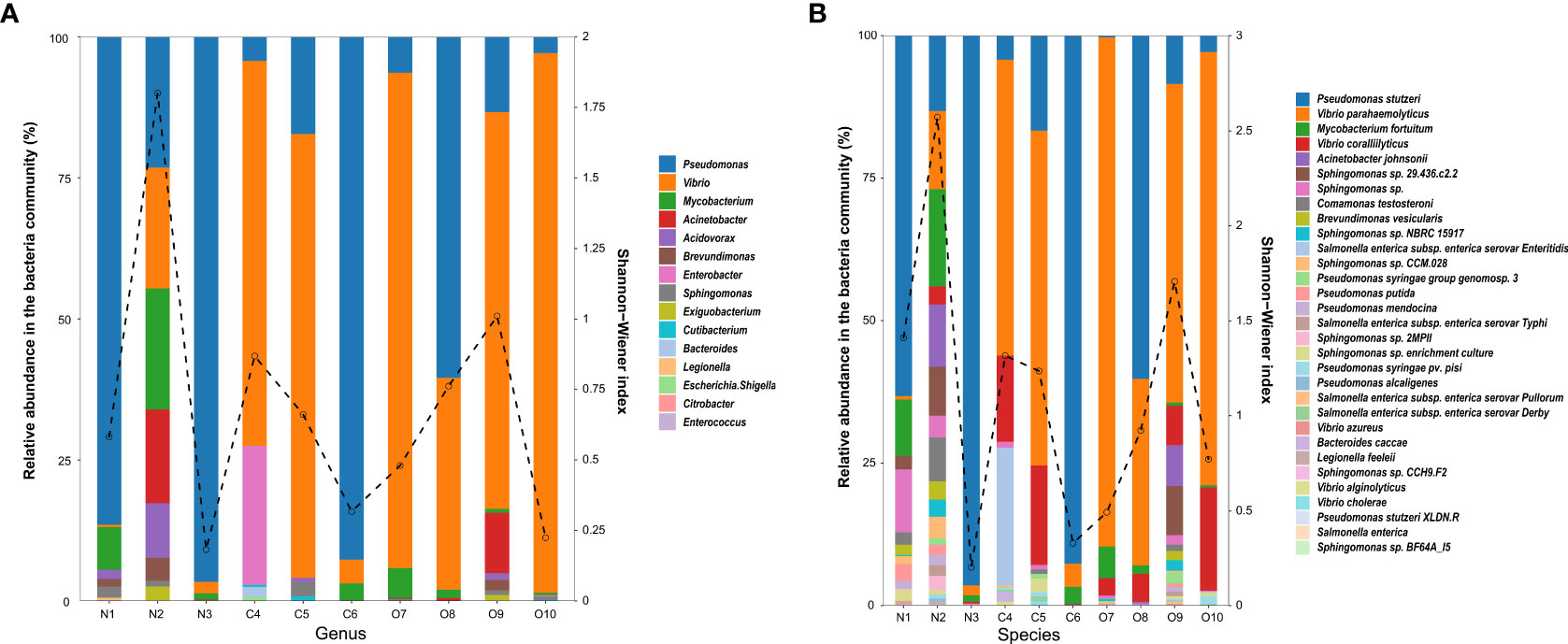
Figure 2 Relative abundance of the potentially pathogenic bacterial genus (A) and species (B) in the estuary. The dashed line indicates the Shannon-Wiener index.
Pseudomonas stutzeri, Mycobacterium fortuitum, and Acinetobacter johnsonii were the main environmental pathogen at the species level. P. stutzeri (51.44%) showed the highest abundance in the C area. V. alginolyticus and V. parahaemolyticus were the main potential pathogenic bacteria in the estuary, and the abundance trend was from the N (13,47% and 7.45%) to the O (38.38% and 65.63%) area, which initially decreased and then increased. Comamonas testosteroni and Brevundimonas vesicularis were potential opportunistic pathogens in the estuary and found most abundant in the N (1.32% and 0.57%) area. Legionella feeleii (0.09%) from the animal and human intestinal tract had the highest abundance in the N area. According to the relative abundance of potential pathogen species and the Shannon-Wiener diversity index, the N2 site in the N area and the O9 site in the O area had the highest proportion of potential pathogen species.
Quantification of typical pathogenic bacteria by qPCR
The abundance of five genes, uidA gene, oaa gene, fliC gene,16S rRNA gene of Vibrio, and trh gene, in the ten water samples were obtained using qPCR (Figure 3). The abundance of the uidA gene was between 0 and 3.80 lg copies/L; however, they were not detected in some sites in N and C areas. The abundance of the 16S rRNA Vibrio gene was 7.06 to 9.48 lg copies/L. The abundance of trh and fliC genes showed the same characteristics. trh gene showed the highest abundance in water from the N area (3.93 and 5.51 lg copies/L), while the abundance of fliC gene was about 102 of the trh gene. The abundance of the oaa gene decreased sequentially from nearshore to distant sea, with the highest value in the N area (7.31 lg copies/L) and the lowest in the O area (5.84 lg copies/L).
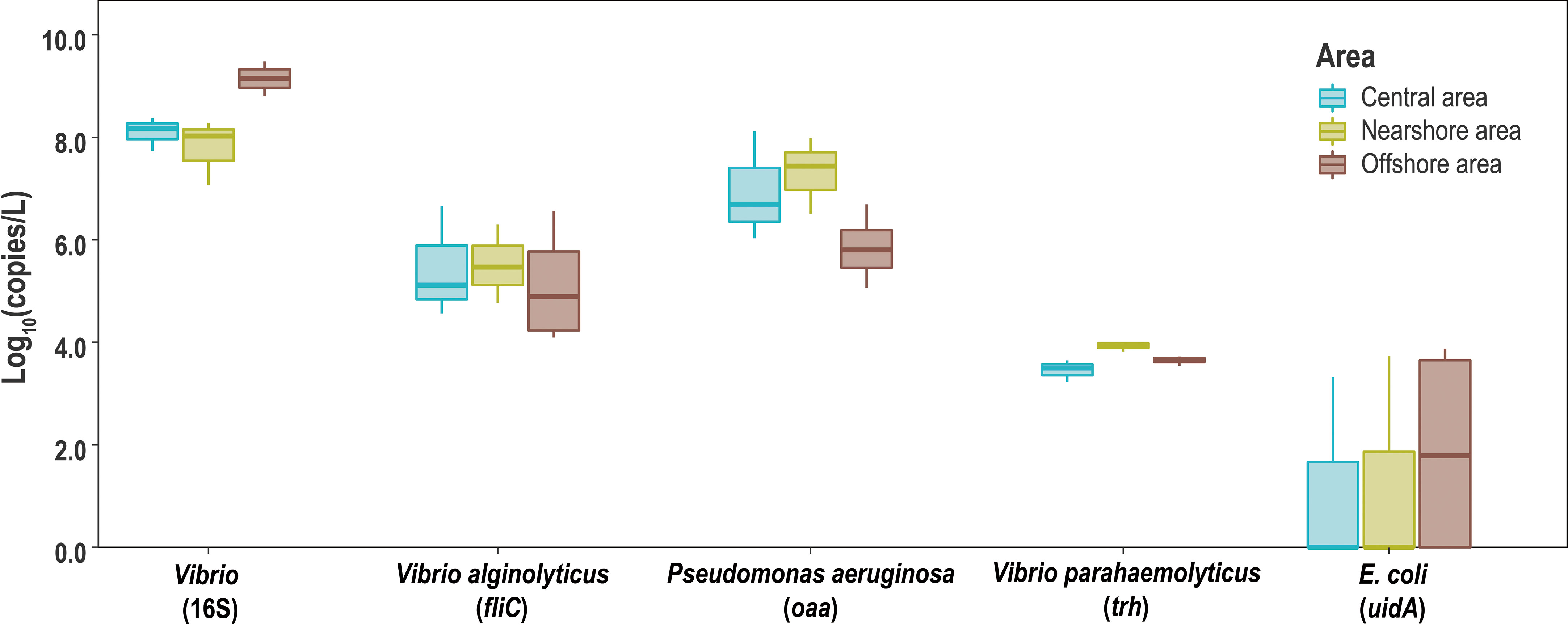
Figure 3 Boxplot of 16S rRNA gene of Vibrio and maker genes of V. alginolyticus, V. parahaemolyticus, P. aeruginosa, and E. coli.
Impact factors in the distribution of potential pathogens
Correlation analysis of quantitative potential pathogenic bacteria with environmental factors was performed to investigate the relationship between environmental factors and potentially pathogenic bacteria. The 16S rRNA gene of Vibrio abundance showed a positive correlation with salinity (Spearman’s correlation, r = 0.806, p<0.01) (Figure 4) and a negative correlation with N, P Nutrients (PO43–, NO3–, NO2–, NH4+) and temperature (r =-0.772, p<0.01). 16S NGS potential pathogenic results were correlated with environmental factors, and it was concluded that P. stutzeri showed a positive correlation with Pb (Spearman’s correlation, r = 0.798, p<0.01) (Figure 5) and Salmonella enterica subsp. enterica serovar enteritidis showed a positive correlation with Cu (Spearman’s correlation, r = 0.697, p<0.01). V. alginolyticus showed a negative correlation with temperature (Spearman’s correlation, r =-0.675, p<0.01) and a positive correlation with salinity (Spearman’s correlation, r = 0.624, p<0.01).
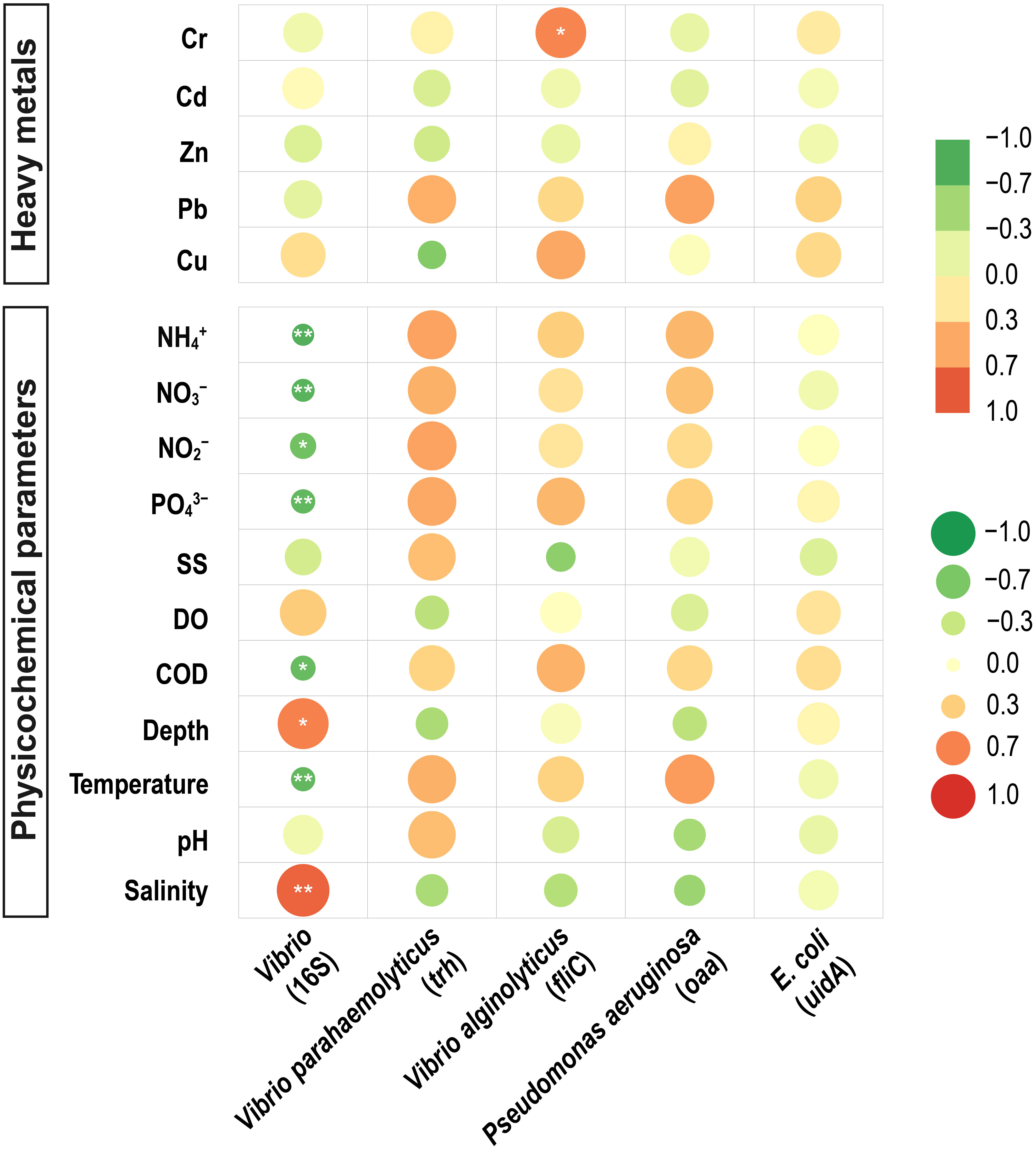
Figure 4 Correlation between abundance, maker genes, and environmental factors. Horizontal axes represent the gene abundance (Vibrio16S rRNA, V. parahaemolyticus trh, V. alginolyticus fliC, P. aeruginosa oaa, E. coli uidA). The vertical axis represents the environmental factors. The circle’s color inside the grid indicates a positive or negative Pearson’s correlation; the circle’s color depth and size denote the correlation’s strength. The deeper the red, the higher the positive correlation between the two factors; the deeper the green, the higher the negative correlation. *significant correlation at 0.05 level, **significant correlation at 0.01 level.
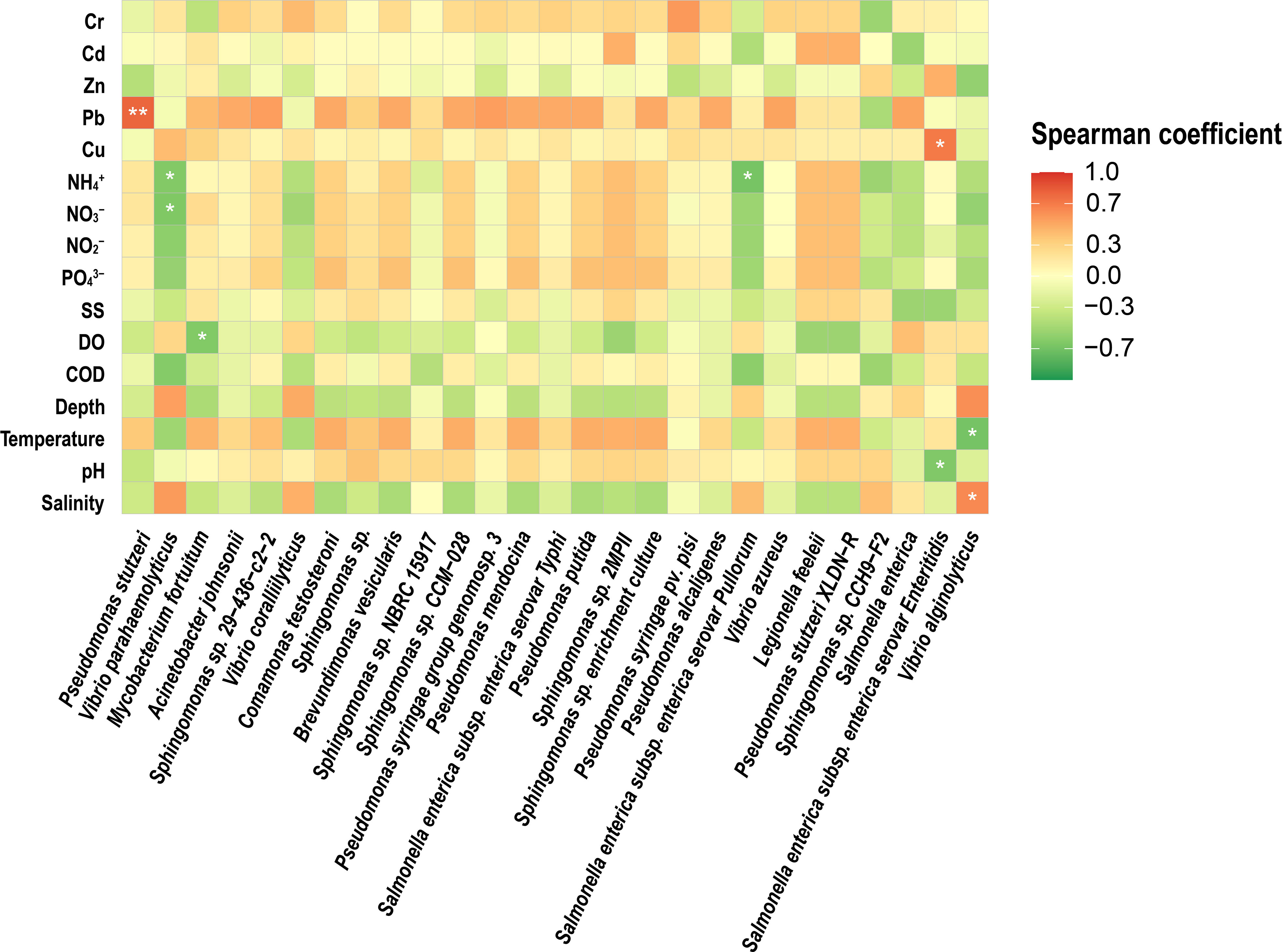
Figure 5 Heatmap showing the Spearman correlations between the 16S rRNA gene abundances of potentially pathogenic bacterial species and environmental variables in the studied estuary samples. *significant correlation at 0.05 level, **significant correlation at 0.01 level.
The effects of spatial and environmental variables on potential pathogenic bacteria in the estuary were explored by statistical analysis, which indicated that four environmental (i.e., pH, COD, Pb, Cu) and two spatial variables (i.e., PCNM1, PCNM2) had a significant (p < 0.05) influence on the distribution of potentially pathogenic bacteria in the water samples (Figure 6). RDA1 and RDA2 contributed 46.42% and 22.55% of the total variation, respectively. The RDA biplot indicated that four environmental (i.e., pH, COD, Pb, Cu) and two spatial variables (i.e., PCNM1, PCNM2) had a major influence on the composition of the bacterial community. VPA showed that environmental and spatial factors explained 5.3% and 19.6% (Figure 7) of the potentially pathogenic bacteria distribution variation in the studied estuary, respectively. However, a large amount of variation (85.3%) remained unexplained.
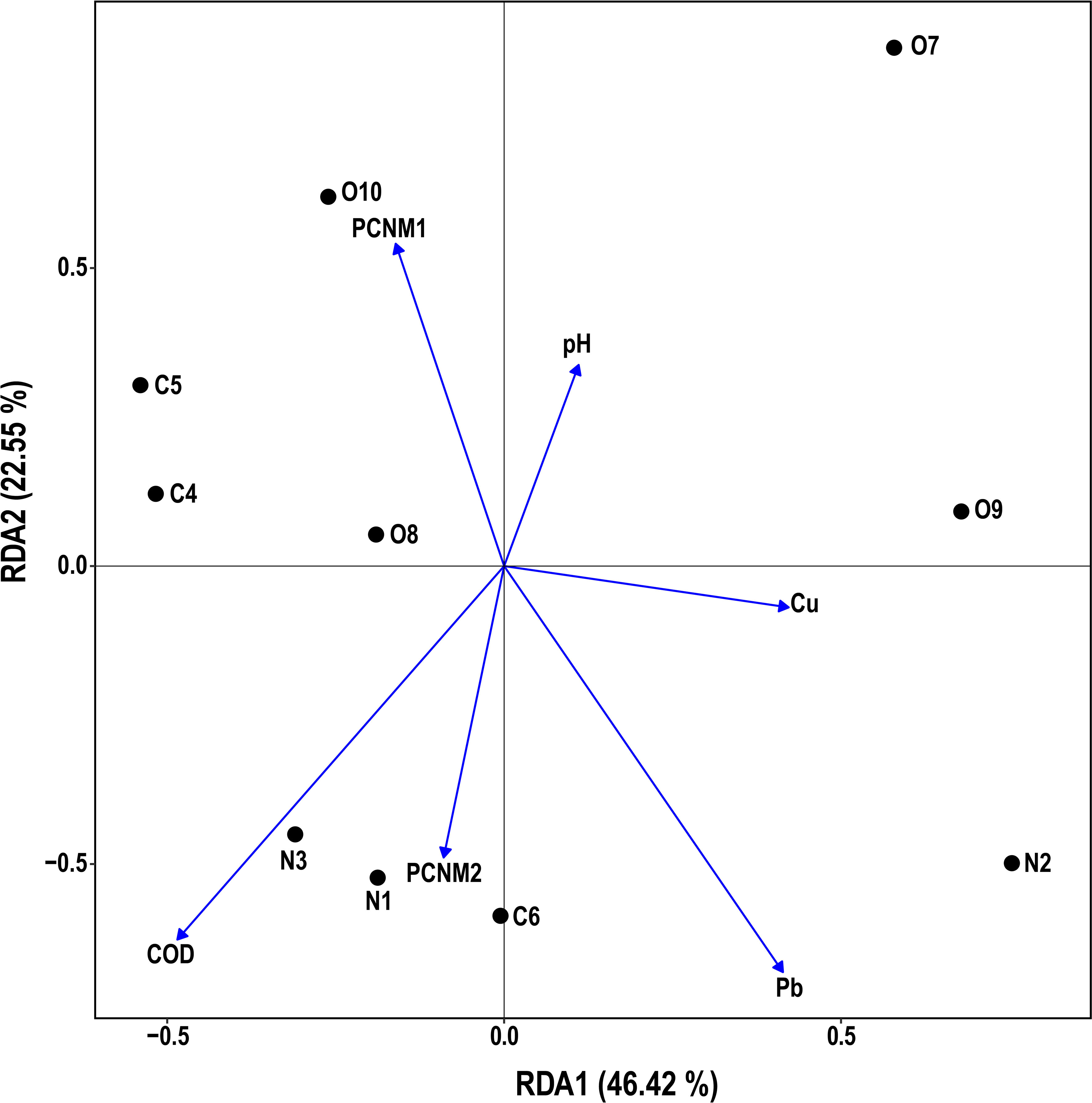
Figure 6 A Redundancy analysis (RDA) of environmental/spatial variables and potentially pathogenic bacterial community structure.
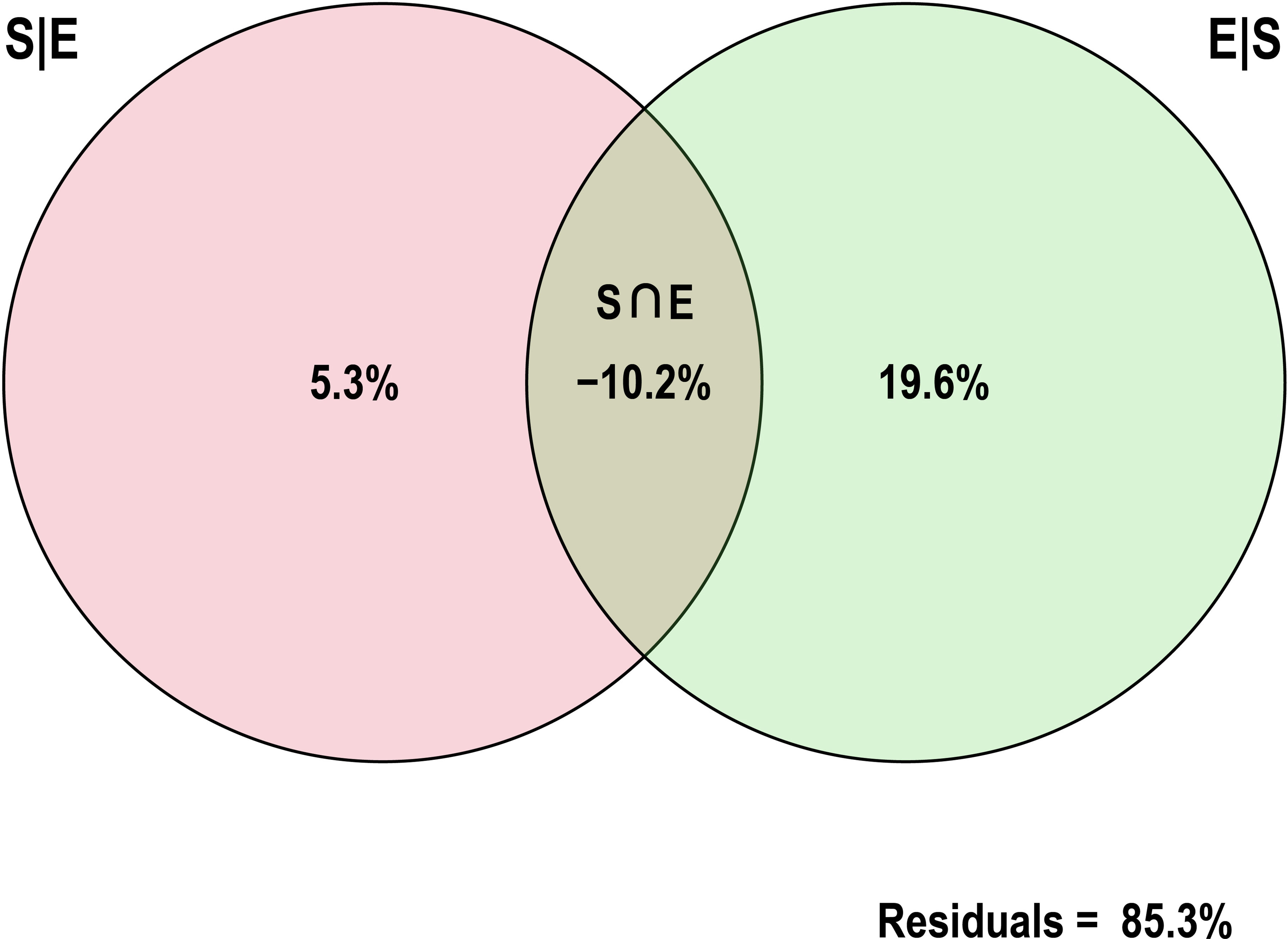
Figure 7 Variation partitioning analysis (VPA) showing the relative influence of spatial and environmental properties on the potentially pathogenic bacterial distribution in the studied estuary samples.
Co-occurrence patterns of potentially pathogenic bacteria in estuary
The co-occurrence characteristics of potentially pathogenic bacteria distribution had 22 network nodes representing potential pathogenic bacteria species and 43 edges representing the correlations between co-occurrence (Figure 8). Our results showed that Pseudomonas putida co-occurred with Sphingomonas spp. and had a significant positive correlation with Sphingomonas sp. CCM-028, Sphingomonas sp. 29-436-c2-2, and Sphingomonas sp. NBRC15917 (p<0.05). Furthermore, three potential opportunistic pathogens, such as C. testosterone, B. vesicularis and P. putida, co-occurred.
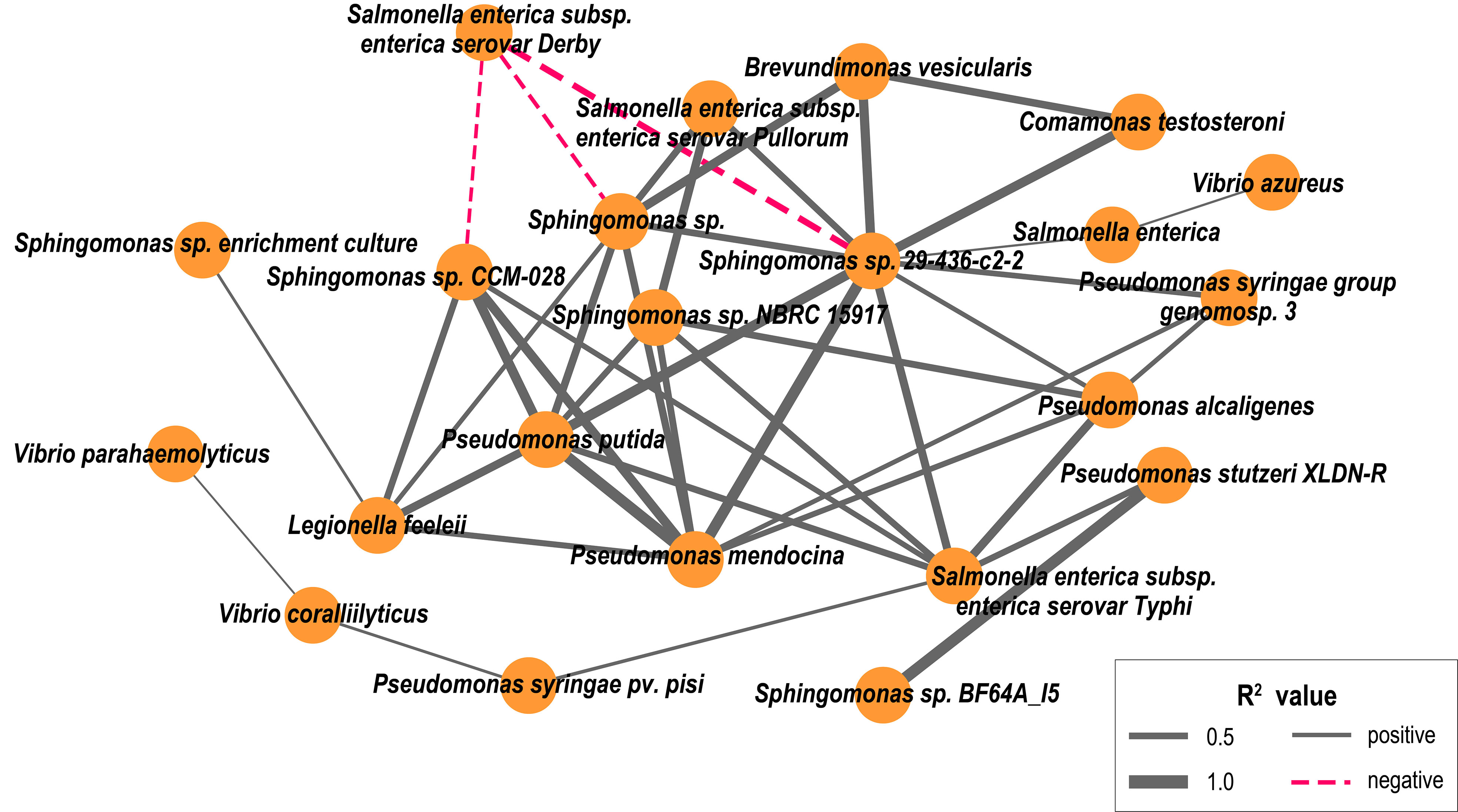
Figure 8 Connections between pathogenic bacteria indicate (p < 0.05) Spearman rank correlations between them. Line thickness reflects the strength of the correlation relationship between pathogenic species (R2 value), and the line color reflects the relationship between pathogenic species, with positive correlations (solid line, gray) and negative correlations (dashed line, red).
Discussion
Co-detection of pathogens by 16S NGS and qPCR
16S rRNA gene sequencing can be used for microbial identification in diagnostic laboratories (Janda and Abbott, 2007). Thus, this method can improve the reference potential pathogen species library by increasing the number of genera and species and avoiding false positives. Potential pathogens that were relatively high in 16S NGS results, such as V. parahaemolyticus and V. alginolyticus, were also detected in our study by qPCR assays (Figure 3). Meanwhile, the potential pathogen targets with low abundances or undetected by 16S NGS, such as Pseudomonas aeruginosa, were more abundant when determined by qPCR than by 16S NGS. This may be due to the lower resolution of 16S NGS, consistent with the results of Cui et al. (Cui et al., 2017). Related studies have shown that increasing reads by at least 500 bp can improve 16S NGS resolution (Janda and Abbott, 2007). We speculate that the cause could be that P. aeruginosa was in low abundance, necessitating more sequencing reads per sample. Therefore, combining 16S NGS with qPCR can be advantageous, and the potential pathogen diversity that 16S NGS can reveal could provide preliminary potential pathogen screening information and relative abundance for comprehensive and further qPCR analysis.
Distribution of potential pathogens in the estuary
The 16S NGS results showed that the distribution of potentially pathogenic bacteria in the Liaohe estuary was mainly abundant in the nearshore N area. According to previous studies (Ming et al., 2020), the estuary had the characteristics of high sewage discharge, high population density, and eutrophication which could be the main reason for the high abundance of potentially pathogenic bacteria in the nearshore N area. Our study detected Vibrio with high abundance using both 16S NGS and qPCR techniques. Vibrio is a symbiotic bacterium that lives in various environments and is an important potential pathogen in estuaries. In this experiment, the abundance of Vibrio in the Liaohe estuary was relatively high compared to the eutrophic estuarine environments such as the Yangtze river estuary (Wang et al., 2020), Sydney harbor (Siboni et al., 2016) and Shandong bay (Xu et al., 2020). This is due to seasonal rainfall or hydrodynamic processes (such as tides and storms) in the Liaohe estuary may have resuspended sediment particles and their attached pathogens into the water column, leading to irregular elevated levels of pollution. Consistent with our hypothesis and influencing factors is the study by Perkins et al. (Perkins et al., 2014), who conducted experiments on the effect of estuarine hydrodynamics on the abundance of pathogenic microorganisms and found that sediment resuspension is an important factor increasing public health risks in seawater.
Interestingly, an unusually high abundance of Vibrio coralliilyticus was observed, suggesting that the symbiotic features benefit both parties in distinct Vibrio spp. Vibrio was found to control virulence factors by density-related synthesis and co-infection with other potential pathogens that produce population sensing molecules (Wendling et al., 2014; Lemire et al., 2015; Tout et al., 2015), indicating that interactions between different bacterial species or the community affect the abundance of V. coralliilyticus. Secondly, the temperature may be one of the important factors. A study revealed that late summer is beneficial for the growth of V. coralliilyticus (Gradoville et al., 2018), which is comparable with the sampling season of our study, suggesting that temperature had a significant impact on the growth of V. coralliilyticus. Third, V. coralliilyticus grew more quickly because of favorable estuarine circumstances. According to studies (Macián Rovira et al., 2000; Froelich et al., 2012), high estuarine salinity promoted the growth of V. coralliilyticus while decreasing the abundance of Vibrio vulnificus to almost zero. A similar phenomenon occurred in our experiment, which confirmed the importance of estuarine hydrological conditions for V. coralliilyticus. Fourth, the higher abundance of or growth of V. corallilyticus may be related to ocean conditions. Some researchers have hypothesized that upwelling may transport high concentrations of V. corallilyticus to the coast (Elston et al., 2008), and that rising water temperatures may also drive the explosive growth of V. corallilyticus (Tout et al., 2015; Amin et al., 2016).
Typical enteric potential pathogenic E. coli mostly enter the estuary from domestic and industrial wastewater (Jeamsripong et al., 2018). Compared to the results of other estuaries (He et al., 2017), our findings suggested that fecal pollution in the Xinhai estuary was more severe, as the abundance of E. coli in the Liaohe estuary was lower than that in the Xinhai estuary. Some studies showed that the lower the salinity, the higher the FIB level, and thus, FIB is more sensitive to high salinity (He et al., 2007; Soueidan et al., 2021). However, in this study, the highest abundance was found in the lower part of the estuary, suggesting that it may be due to rainfall and runoff, where E. coli was washed into distant waters.
Influencing factors on the distribution of potential pathogens in estuary
Our experimental results showed little correlation between most potential pathogens and environmental factors, which demonstrated no association between the majority of possible pathogens and environmental parameters. This conclusion is supported by the findings of López-Hernández et al. (López-Hernández et al., 2015), who evaluated the effect of environmental parameters on the seasonal density of potentially pathogenic V. parahaemolyticus in American oysters by regression analysis, they found that there was no correlation between environmental conditions and V. parahaemolyticus densities, which was thought to be related to changes in water temperature in the study waters. However, our study found a strong association between environmental factors and a common potential pathogen, Vibrio. According to the quantitative qPCR data, temperature and salinity were correlated with Vibrio, and our conclusions are supported by the experimental results of Oberbeckmann et al. and Vezzulli et al. Oberbeckmann et al. (Oberbeckmann et al., 2012) used Spearman’s analysis techniques to model the seasonal dynamics of Vibrio populations offshore the North Sea, where temperature and salinity were identified as the main determinants of Vibrio abundance. A generalized additive model used by Vezzulli et al. (Vezzulli et al., 2016) demonstrated that an increase in sea surface temperature led to a long-term increase in Vibrio abundance, showing the importance of temperature for Vibrio. However, the correlation between temperature and Vibrio abundance is currently controversial. In our study, we found that not all Vibrio spp. were correlated with temperature, but with the span of water temperature in the season in the Liaoning River estuary. For example, Liang et al. (Liang et al., 2019) showed a significant association between temperature and Vibrio abundance in seasons with a broad temperature range, but the correlation was insignificant in the summer, indicating the significance of the temperature range setting. In addition, we found very low abundance of V. vulnificus in 16S NGS results, which may be related to seawater salinity. According to Tamplin et al. (Tamplin, 2001), the abundance of V. vulnificus decreases to a level not related to human health in environments with salinity above 20 ppt, from which it can be inferred that the Liaoning estuary is not suitable for V. vulnificus.
The relationship between temperature and Vibrio virulence genes is a hot research topic. Recent investigations have revealed a strong correlation between temperature and the expression of Vibrio virulence genes, raising the possibility that warm waters could increase its pathogenic potential and endanger public health (Green et al., 2019; Canellas et al., 2021). Previous studies have shown that trh gene is the main virulence gene commonly found in V. parahaemolyticus (López-Hernández et al., 2015; Canellas et al., 2021). In our experiments, trh gene and fliC gene were selected as virulence genes for Vibrio. The fliC gene was chosen as a target gene for the detection of V. alginolyticus because it is an important, highly conserved gene in bacteria and that allows it to swim in highly viscous liquid or semi-solid environments (Tang and Grossart, 2007; Lippolis et al., 2016) as well as aids in seeking nutrients and settling on particles to escape bacterial predation (Matz et al., 2005; Asplund et al., 2011). A similar distribution pattern of trh gene in V. parahaemolyticus and fliC gene in V. alginolyticus confirmed Mookerjee’s findings (Mookerjee et al., 2015), indicating that horizontal gene transfer of these virulence genes occurs in this context. Additionally, because V. alginolyticus is a potential opportunistic pathogen whose virulence is influenced by environmental factors (McDevitt-Irwin et al., 2017), it is possible to infer that the existing environmental factors in the Liaohe estuary were favorable to its proliferation.
Compared to the environmental temperature, salinity is a more compelling critical factor (Takemura et al., 2014). Most experiments showed a significant positive correlation between Vibrio and salinity (Perkins et al., 2014; Krishna et al., 2021), consistent with our results. According to a study, the potential opportunistic pathogenic V. alginolyticus and V. parahaemolyticus are halotolerant (Mookerjee et al., 2015), and thus the 16S NGS results of this experiment showed that their distribution patterns showed dominance at high salinity. In addition, there is a complex relationship between nutrients and Vibrio, such as the abundance of V. parahaemolyticus associated with allochthonous inputs (He et al., 2017; Krishna et al., 2021). In the moderately high saline downstream areas of estuaries, mixing estuarine water with flows such as sewers during the rainy season may be a second peak of Vibrio outbreaks.
The examination of spatial components and analysis of environmental factors also revealed a significant effect on potentially pathogenic bacteria in water (p<0.05). It was shown that one of the key mechanisms of potentially pathogenic bacteria transmission in the ocean is the movement of water masses (Krishna et al., 2021), suggesting that estuarine hydrodynamics may be an important determinant. Additionally, spatial variables can strongly explain potential pathogenic bacteria aggregation patterns and provide a favorable reference for coexistence among potential pathogenic bacteria, such as cooperation or competition in terms of nutrition, space, and material use (Deng et al., 2012). A large proportion (85.3%) of the potentially pathogenic bacteria distribution in this study is unexplained, and many key factors were not identified, which may be explained on the one hand by the fact that most potential bacterial communities have processes of growth, mortality, migration, deposition, and extinction that are jointly determined by spatial scales and environmental gradients. For example, Hanson et al. (Hanson et al., 2012) suggested that microorganisms are mainly influenced by evolutionary and ecological effects, where selection, dispersal, drift, and mutation are the main processes of interaction between microorganisms and the environment, and Berga et al. (Berga et al., 2015) suggested that processes such as viral lysis or predation can be influenced by environmental factors and thus may lead to changes in the microbial community. On the other hand, the unexplained part and the high autocorrelation between spatial and environmental factors may be the difference between VPA and other methods.
Co-occurrence patterns of potentially pathogenic bacteria in the estuary
Microbial network analysis is an important method for studying potential interactions between bacteria. This study found that some potentially pathogenic bacteria were present in the estuaries studied simultaneously, possibly due to potential pathogenic bacteria from a common source of pollution (such as feces and sewage). Salmonella and Legionella originate from the same source of contamination and have co-existed (Pandey et al., 2014), suggesting that the source of contamination is one of the factors affecting the potential pathogen’s coexistence. Furthermore, Brevundimonas vesicularis (Denet et al., 2017), Comamonas testosterone (Orsini et al., 2014), and Pseudomonas putida were found to be potential human pathogens (Fernández et al., 2015), which may be co-existing.
The potential pathogens are affected by heavy metals and water temperature. Many potential pathogens are subjected to environmental stress to carry the commonality of heavy metal resistance genes. Therefore, co-existing potential pathogens may have similar ecological niches, and the co-occurrence of potential opportunistic pathogens in estuarine areas should be treated with caution. We also found that bacterial populations (Pseudomonas and Sphingomonas) correlated highly with pollutant degradation. Several studies reported the degradation of pollutants by bacteria, Pseudomonas and Sphingomonas being the main strain for the degradation of aromatic pollutants (Li et al., 2020), and Pseudomonas stutzeri can contribute to organic pollutants (Gao et al., 2020). Biodegradation processes can also remove heavy metals. Pseudomonas Putida can remediate oil-contaminated soils (Titah et al., 2021), Brevundimonas vesicularis can remove lead from wastewater (Resmi et al., 2010), and Acinetobacter johnsonii can degrade organophosphorus pesticides and are resistant to heavy metals (Xie et al., 2009). Thus, these suggested that the coexistence of potentially pathogenic bacteria is due to functional redundancy, and the functional coherence of potential bacteria can maintain ecological functions in response to rapid environmental perturbations.
Conclusion
This study revealed the distribution and factors that influence the distribution and coexistence of potentially pathogenic bacteria in the Liaohe estuary using 16S NGS and qPCR methods. It was found that most potentially pathogenic bacteria in the estuary area were mainly environmental pathogens, while the proportion of enteric pathogens was small, indicating that sewage discharge was the primary source of estuarine pollution. Secondly, the influence of spatial factors on potential pathogenic bacteria was found to be much greater than that of environmental factors, but temperature and salinity were still critical factors that influenced the typical potential pathogenic bacteria Vibrio. In addition, co-occurrence analysis of potential pathogens showed that some potential opportunistic pathogens might increase the risk of multiple infections. These findings guide the safe monitoring of estuarine waters. More research is needed to identify possible pathogenic bacteria in future estuarine water quality safety monitoring to assess the potential risk of potentially pathogenic bacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HH analyzed all samples and data, contributed to the experimental design and wrote the manuscript. SZ wrote reviews and edited, and contributed to manuscript writing. HM collected all samples and contributed to the experimental design. ML participated in the experiments. JX analyzed the experimental results. YX revised the manuscript. WW participated in the experiment. JF designed the experiments and supported the study. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by People’s Livelihood Science and Technology Project of Liaoning Province, China (2021JH2/10300001), Millions of Talent Projects of Liaoning Province, China, Dalian high level talent innovation support plan (2021RD04), the National Key R & D Program of China (2017YFC1404500).
Acknowledgments
The National Marine Environmental Monitoring Center (China), State Environmental Protection Key Laboratory of Coastal Ecosystem, is acknowledged by the authors for providing the experimental settings.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1044260/full#supplementary-material
References
Amin A. K. M. R., Feng G., Al-saari N., Meirelles P. M., Yamazaki Y., Mino S., et al. (2016). The first temporal and spatial assessment of vibrio diversity of the surrounding seawater of coral reefs in ishigaki, Japan. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01185
Asplund M. E., Rehnstam-Holm A.-S., Atnur V., Raghunath P., Saravanan V., Härnström K., et al. (2011). Water column dynamics of vibrio in relation to phytoplankton community composition and environmental conditions in a tropical coastal area. Environ. Microbiol. 13, 2738–2751. doi: 10.1111/j.1462-2920.2011.02545.x
Baliarsingh S. K., Lotliker A. A., Srichandan S., Basu A., Nair T. M. B., Tripathy S. K. (2021). Effect of tidal cycle on escherichia coli variability in a tropical estuary. Bull. Environ. Contam Toxicol. 106, 622–628. doi: 10.1007/s00128-021-03106-w
Berga M., Östman Ö., Lindström E. S., Langenheder S. (2015). Combined effects of zooplankton grazing and dispersal on the diversity and assembly mechanisms of bacterial metacommunities. Environ. Microbiol. 17, 2275–2287. doi: 10.1111/1462-2920.12688
Blanchet F. G., Legendre P., Borcard D. (2008). Forward Selection of Explanatory Variables. Ecology 89, 2623–2632. doi: 10.1890/07-0986.1
Boehm A. B., Fuhrman J. A., Mrše R. D., Grant S. B. (2003). Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon bay, Catalina island, California. Environ. Sci. Technol. 37, 673–680. doi: 10.1021/es025934x
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Borcard D., Legendre P. (2002). All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153, 51–68. doi: 10.1016/S0304-3800(01)00501-4
Brookes J. D., Antenucci J., Hipsey M., Burch M. D., Ashbolt N. J., Ferguson C. (2004). Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 30, 741–759. doi: 10.1016/j.envint.2003.11.006
Campbell B. J., Kirchman D. L. (2013). Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J. 7, 210–220. doi: 10.1038/ismej.2012.93
Canellas A. L. B., Lopes I. R., Mello M. P., Paranhos R., de Oliveira B. F. R., Laport M. S. (2021). Vibrio species in an urban tropical estuary: Antimicrobial susceptibility, interaction with environmental parameters, and possible public health outcomes. Microorganisms 9, 1007. doi: 10.3390/microorganisms9051007
Cui Q., Fang T., Huang Y., Dong P., Wang H. (2017). Evaluation of bacterial pathogen diversity, abundance and health risks in urban recreational water by amplicon next-generation sequencing and quantitative PCR. J. Environ. Sci. 57, 137–149. doi: 10.1016/j.jes.2016.11.008
Cui Q., Huang Y., Wang H., Fang T. (2019). Diversity and abundance of bacterial pathogens in urban rivers impacted by domestic sewage. Environ. pollut. 249, 24–35. doi: 10.1016/j.envpol.2019.02.094
Denet E., Coupat-Goutaland B., Nazaret S., Pélandakis M., Favre-Bonté S. (2017). Diversity of free-living amoebae in soils and their associated human opportunistic bacteria. Parasitol. Res. 116, 3151–3162. doi: 10.1007/s00436-017-5632-6
Deng Y., Jiang Y.-H., Yang Y., He Z., Luo F., Zhou J. (2012). Molecular ecological network analyses. BMC Bioinf. 13, 113. doi: 10.1186/1471-2105-13-113
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Elston R. A., Hasegawa H., Humphrey K. L., Polyak I. K., Häse C. C. (2008). Re-emergence of vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis. Aquat. Organisms 82, 119–134. doi: 10.3354/dao01982
Fernández M., Porcel M., de la Torre J., Molina-Henares M. A., Daddaoua A., Llamas M. A., et al. (2015). Analysis of the pathogenic potential of nosocomial pseudomonas putida strains. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00871
Froelich B. A., Williams T. C., Noble R. T., Oliver J. D. (2012). Apparent loss of vibrio vulnificus from north Carolina oysters coincides with a drought-induced increase in salinity. Appl. Environ. Microbiol. 78, 3885–3889. doi: 10.1128/AEM.07855-11
Gao J., Zhu T., Liu C., Zhang J., Gao J., Zhang J., et al. (2020). Ammonium removal characteristics of heterotrophic nitrifying bacterium pseudomonas stutzeri GEP-01 with potential for treatment of ammonium-rich wastewater. Bioprocess Biosyst. Eng. 43, 959–969. doi: 10.1007/s00449-020-02292-x
GB11901-89(1989) .Water Quality-Determination of Suspended Substance-Gravimetric Method. (Beijing, China: Ministry of Ecology and Environment).
Ge Y., Yu S. (2017). Next-generation sequencing for investigating the diversity of microorganisms and pathogenic bacteria in a water source. Trop. J. Pharm. Res. 16, 1503. doi: 10.4314/tjpr.v16i7.7
Gradoville M. R., Crump B. C., Häse C. C., White A. E. (2018). Environmental controls of oyster-pathogenic vibrio spp. in Oregon estuaries and a shellfish hatchery. Appl. Environ. Microbiol. 84, e02156–e02117. doi: 10.1128/AEM.02156-17
Green T. J., Siboni N., King W. L., Labbate M., Seymour J. R., Raftos D. (2019). Simulated marine heat wave alters abundance and structure of vibrio populations associated with the pacific oyster resulting in a mass mortality event. Microb. Ecol. 77, 736–747. doi: 10.1007/s00248-018-1242-9
Griffith J. F., Cao Y., McGee C. D., Weisberg S. B. (2009). Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res. 43, 4900–4907. doi: 10.1016/j.watres.2009.09.017
Hanson C. A., Fuhrman J. A., Horner-Devine M. C., Martiny J. B. H. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506. doi: 10.1038/nrmicro2795
Harwood V. J., Levine A. D., Scott T. M., Chivukula V., Lukasik J., Farrah S. R., et al. (2005). Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71, 3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005
He L.-M., Lu J., Shi W. (2007). Variability of fecal indicator bacteria in flowing and ponded waters in southern California: Implications for bacterial TMDL development and implementation. Water Res. 41, 3132–3140. doi: 10.1016/j.watres.2007.04.014
He Y., Sen B., Shang J., He Y., Xie N., Zhang Y., et al. (2017). Seasonal influence of scallop culture on nutrient flux, bacterial pathogens and bacterioplankton diversity across estuaries off the bohai Sea coast of northern China. Mar. pollut. Bull. 124, 411–420. doi: 10.1016/j.marpolbul.2017.07.062
Jacobs J., Rhodes M., Sturgis B., Wood B. (2009). Influence of environmental gradients on the abundance and distribution of mycobacterium spp. in a coastal lagoon estuary. Appl. Environ. Microbiol. 75, 7378–7384. doi: 10.1128/AEM.01900-09
Janda J. M., Abbott S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jeamsripong S., Chuanchuen R., Atwill E. R. (2018). Assessment of bacterial accumulation and environmental factors in sentinel oysters and estuarine water quality from the phang nga estuary area in Thailand. Int J Environ Res Publ Health 15, 1–17. doi: 10.3390/ijerph15091970
Khandeparker L., Anil A. C., Naik S. D., Gaonkar C. C. (2015). Daily variations in pathogenic bacterial populations in a monsoon influenced tropical environment. Mar. pollut. Bull. 96, 337–343. doi: 10.1016/j.marpolbul.2015.04.051
Krishna K., Veettil V. P., Anas A., Nair S. (2021). Hydrological regulation of vibrio dynamics in a tropical monsoonal estuary: a classification and regression tree approach. Environ. Sci. pollut. Res. 28, 724–737. doi: 10.1007/s11356-020-10486-9
Lamb J. B., van de Water J. A. J. M., Bourne D. G., Altier C., Hein M. Y., Fiorenza E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Leal Diego A. G., Dores Ramos A. P., Marques Souza D. S., Durigan M., Greinert-Goulart J. A., Moresco V., et al. (2013). Sanitary quality of edible bivalve mollusks in southeastern Brazil using an UV based depuration system. Ocean Coast. Manage. 72, 93–100. doi: 10.1016/j.ocecoaman.2011.07.010
Leclerc H., Schwartzbrod L., Dei-Cas E. (2008). Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol 28, 371–409. doi: 10.1080/1040-840291046768
Lemire A., Goudenège D., Versigny T., Petton B., Calteau A., Labreuche Y., et al. (2015). Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 9, 1523–1531. doi: 10.1038/ismej.2014.233
Liang J., Liu J., Wang X., Lin H., Liu J., Zhou S., et al. (2019). Spatiotemporal dynamics of free-living and particle-associated vibrio communities in the northern Chinese marginal seas. Appl. Environ. Microbiol. 85, e00217–e00219. doi: 10.1128/AEM.00217-19
Liang Z., Yu Y., Ye Z., Li G., Wang W., An T. (2020). Pollution profiles of antibiotic resistance genes associated with airborne opportunistic pathogens from typical area, pearl river estuary and their exposure risk to human. Environ. Int. 143, 105934. doi: 10.1016/j.envint.2020.105934
Li Q., Huang Y., Wen D., Fu R., Feng L. (2020). Application of alkyl polyglycosides for enhanced bioremediation of petroleum hydrocarbon-contaminated soil using sphingomonas changbaiensis and pseudomonas stutzeri. Sci. Total Environ. 719, 137456. doi: 10.1016/j.scitotenv.2020.137456
Lippolis J. D., Brunelle B. W., Reinhardt T. A., Sacco R. E., Thacker T. C., Looft T. P., et al. (2016). Differential gene expression of three mastitis-causing escherichia coli strains grown under planktonic, swimming, and swarming culture conditions. mSystems 1, e00064–e00016. doi: 10.1128/mSystems.00064-16
López-Hernández K. M., Pardío-Sedas V. T., Lizárraga-Partida L., Williams J., de J., Martínez-Herrera D., et al. (2015). Environmental parameters influence on the dynamics of total and pathogenic vibrio parahaemolyticus densities in crassostrea virginica harvested from mexico’s gulf coast. Mar. pollut. Bull. 91, 317–329. doi: 10.1016/j.marpolbul.2014.11.015
Lozano-León A., Torres J., Osorio C. R., Martínez-Urtaza J. (2003). Identification of tdh-positive vibrio parahaemolyticus from an outbreak associated with raw oyster consumption in Spain. FEMS Microbiol. Lett. 226, 281–284. doi: 10.1016/S0378-1097(03)00604-9
Macián Rovira M. C., Rodríguez Arias C., Aznar Novella R., Garay Auban E., Pujalte Domarco M. J. (2000). Identification of Vibrio spp. (other than V. vulnificus) recovered on CPC agar from marine natural samples. Int. Microbiol. 3, 51–53.
Matz C., McDougald D., Moreno A. M., Yung P. Y., Yildiz F. H., Kjelleberg S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of vibrio cholerae. Proc. Natl. Acad. Sci. 102, 16819–16824. doi: 10.1073/pnas.0505350102
McDevitt-Irwin J. M., Baum J. K., Garren M., Vega Thurber R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00262
McLusky D. S., Elliott M. (2004). The Estuarine Ecosystem: Ecology, Threats and Management third ed. (Oxford: Oxford University Press), 216 pp.
Mei Liu S., Wei Li L., Zhang Z. (2011). Inventory of nutrients in the bohai. Continental Shelf Res. 31, 1790–1797. doi: 10.1016/j.csr.2011.08.004
Ming H., Fan J., Chen Q., Su J., Song J., Yuan J., et al. (2020). Diversity and abundance of denitrifying bacteria in the sediment of a eutrophic estuary. Geomicrobiol J. 38, 199–209. doi: 10.1080/01490451.2020.1822959
Mookerjee S., Batabyal P., Sarkar M. H., Palit A. (2015). Seasonal prevalence of enteropathogenic vibrio and their phages in the riverine estuarine ecosystem of south Bengal. PloS One 10, e0137338. doi: 10.1371/journal.pone.0137338
Oberbeckmann S., Fuchs B. M., Meiners M., Wichels A., Wiltshire K. H., Gerdts G. (2012). Seasonal dynamics and modeling of a vibrio community in coastal waters of the north Sea. Microb. Ecol. 63, 543–551. doi: 10.1007/s00248-011-9990-9
Orsini J., Tam E., Hauser N., Rajayer S. (2014). Polymicrobial bacteremia involving comamonas testosteroni. Case Rep. Med. 2014, e578127. doi: 10.1155/2014/578127
Pandey P. K., Kass P. H., Soupir M. L., Biswas S., Singh V. P. (2014). Contamination of water resources by pathogenic bacteria. AMB Express 4, 51. doi: 10.1186/s13568-014-0051-x
Percival S. L. (2014). Microbiology of waterborne diseases: microbiological aspects and risks. 2nd ed. (Amsterdam: Elsevier, Academic Press).
Perkins T. L., Clements K., Baas J. H., Jago C. F., Jones D. L., Malham S. K., et al. (2014). Sediment composition influences spatial variation in the abundance of human pathogen indicator bacteria within an estuarine environment. PloS One 9, e112951. doi: 10.1371/journal.pone.0112951
Ramaiah N. (2002) Coliform and human pathogenic bacteria in tourism affected water bodies in north goa. Available at: https://drs.nio.org/xmlui/handle/2264/1338.
Resmi G., Thampi S. G., Chandrakaran S. (2010). Brevundimonas vesicularis: A novel bio-sorbent for removal of lead from wastewater. Int. J. Environ. Res. 4, 281–288. doi: 10.22059/IJER.2010.20
Rodrigues V., Ramaiah N., Kakti S., Samant D. (2011). Long-term variations in abundance and distribution of sewage pollution indicator and human pathogenic bacteria along the central west coast of India. Ecol. Indic. 11, 318–327. doi: 10.1016/j.ecolind.2010.05.010
Ruiz-Cayuso J., Trujillo-Soto T., Rodriguez-Iglesias M., Almagro-Moreno S. (2021). Effects of temperature and salinity interaction on vibrio spp and vibrio parahaemolyticus in the intercontinental Euro-African atlantic. doi: 10.21203/rs.3.rs-607386/v1
Siboni N., Balaraju V., Carney R., Labbate M., Seymour J. R. (2016). Spatiotemporal dynamics of vibrio spp. within the Sydney harbour estuary. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00460
Soueidan J., Warren A., Pearson M., Montie E. W. (2021). A changing estuary: Understanding historical patterns in salinity and fecal coliform levels in the may river, SC. Mar. pollut. Bull. 168, 112384. doi: 10.1016/j.marpolbul.2021.112384
State Oceanic Administration (2007a). The Specification for Marine Monitoring–Part 4: Seawater Analysis (GB17378.4-2007). (Beijing: Chinese. Standards Press of China) pp. 117–119.
State Oceanic Administration (2007b). The specification for marine monitoring–part 4: Seawater analysis (GB17378.4-2007). (Beijing: Chinese. Standards Press of China) pp. 109–117.
State Oceanic Administration (2007c). The specification for marine monitoring–part 4: Seawater analysis (GB17378.4-2007). (Beijing: Chinese. Standards Press of China) pp. 10–24.
Takemura A., Chien D., Polz M. (2014). Associations and dynamics of vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00038
Tamplin M. L. (2001). Coastal vibrios: Identifying relationships between environmental condition and human disease. Hum. Ecol. Risk Assessment: Int. J. 7, 1437–1445. doi: 10.1080/20018091095113
Tang K. W., Grossart H.-P. (2007). Iron effects on colonization behavior, motility, and enzymatic activity of marine bacteria. Can. J. Microbiol. 53, 968–974. doi: 10.1139/W07-059
Titah H. S., Pratikno H., Purwanti I. F., Wardhani W. K. (2021). Biodegradation of crude oil spill using bacillus subtilis and pseudomonas putida in sequencing method. J. Ecol. Eng. 22, 157–167. doi: 10.12911/22998993/142913
Tout J., Siboni N., Messer L. F., Garren M., Stocker R., Webster N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural vibrio populations associated with the coral pocillopora damicornis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00432
Vezzulli L., Grande C., Reid P. C., Hélaouët P., Edwards M., Höfle M. G., et al. (2016). Climate influence on vibrio and associated human diseases during the past half-century in the coastal north Atlantic. Proc. Natl. Acad. Sci. 113, E5062–E5071. doi: 10.1073/pnas.1609157113
Wang X., Liu J., Liang J., Sun H., Zhang X.-H. (2020). Spatiotemporal dynamics of the total and active vibrio spp. populations throughout the changjiang estuary in China. Environ. Microbiol. 22, 4438–4455. doi: 10.1111/1462-2920.15152
Wang G., Peng J., Yang D., Zhang D., Li X. (2015). Current levels, composition profiles, source identification and potentially ecological risks of polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs) in the surface sediments from bohai Sea. Mar. pollut. Bull. 101, 834–844. doi: 10.1016/j.marpolbul.2015.11.028
Wendling C. C., Batista F. M., Wegner K. M. (2014). Persistence, seasonal dynamics and pathogenic potential of vibrio communities from pacific oyster hemolymph. PloS One 9, e94256. doi: 10.1371/journal.pone.0094256
Wu D.-M., Dai Q.-P., Liu X.-Z., Fan Y.-P., Wang J.-X. (2019a). Comparison of bacterial community structure and potential functions in hypoxic and non-hypoxic zones of the changjiang estuary. PloS One 14, e0217431. doi: 10.1371/journal.pone.0217431
Wu G., Yang J., Jiang H., Deng Y., Lear G. (2019b). Distribution of potentially pathogenic bacteria in the groundwater of the jianghan plain, central China. Int. Biodeterioration Biodegradation 143, 104711. doi: 10.1016/j.ibiod.2019.05.028
Xie S., Liu J., Li L., Qiao C. (2009). Biodegradation of malathion by acinetobacter johnsonii MA19 and optimization of cometabolism substrates. J. Environ. Sci. 21, 76–82. doi: 10.1016/S1001-0742(09)60014-0
Xu W., Gong L., Yang S., Gao Y., Ma X., Xu L., et al. (2020). Spatiotemporal dynamics of vibrio communities and abundance in dongshan bay, south of China. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.575287
Yeleliere E., Cobbina S. J., Duwiejuah A. B. (2018). Review of ghana’s water resources: the quality and management with particular focus on freshwater resources. Appl. Water Sci. 8, 93. doi: 10.1007/s13201-018-0736-4
Keywords: potential pathogens, spatial distribution, co-occurrence, estuary, quantitative polymerase chain reaction (qPCR), 16S next-generation sequencing (16S NGS)
Citation: Huang H, Zan S, Ming H, Li M, Xu J, Xie Y, Wang W and Fan J (2022) 16S Next-generation sequencing and quantitative PCR reveal the distribution of potential pathogens in the Liaohe Estuary. Front. Mar. Sci. 9:1044260. doi: 10.3389/fmars.2022.1044260
Received: 14 September 2022; Accepted: 22 November 2022;
Published: 09 December 2022.
Edited by:
Juan Feng, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2022 Huang, Zan, Ming, Li, Xu, Xie, Wang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingfeng Fan, jffan@nmemc.org.cn
†These authors have contributed equally to this work and share first authorship
 Huiling Huang
Huiling Huang