- 1Department of Fisheries, Faculty of Marine Science and Technology, University of Hormozgan, Bandar Abbas, Iran
- 2Department of Natural Resources and Environmental Engineering, School of Agriculture, Shiraz University, Shiraz, Iran
- 3Marine Biodiversity Exploitation and Conservation (MARBEC), UMR 9190, Univ. Montpellier, IFREMER, IRD, CNRS, Sète, France
- 4Department of Marine Biology, Faculty of Marine Science and technology, University of Hormozgan, Bandar Abbas, Iran
A few species of mollusks display color variation in their soft tissues. In pearl oysters, the color polymorphism in mantle tissue is associated with the color and radiance of shell and pearl. The study of biomineralization related genes in mantle tissue of pearl oysters can be used as a suitable approach to better identify the molecular mechanisms that influence shell and pearl quality and color variations. In this study, we investigated the expression of biomineralization-related genes in black and orange mantle morphotypes of pearl oyster, Pinctada persica in both warm and cool seasons using quantitative real-time PCR. Our results showed that the genes involved in biomineralization of the prismatic and nacre layer, i.e.; ASP, KRMP, MRNP34, SHELL, SHEM1B, LINKINE, PIF, SHEM5, NACREIN, and in pigmentation (TYR2A) were significantly higher expressed in orange phenotype compared to those of black one. The higher expression of ASP, KRMP, SHEM5, LINKINE and NACREIN in orange phenotype was only observed in warm season, but PIF, SHELL, SHEM1B, and TYR2A were upregulated in both warm and cool seasons. These results suggest the existence of different genetic processes between the two color morphs of P. persica and the more active role of genes in orange morphotype, particularly in warmer season. This study provides better understanding of the molecular mechanisms underlying biomineralization in pearl oysters.
Introduction
Pearl oysters occur around the world with considerable variation in the shell and pearl phenotype in each species (Strack, 2008; Fougerouse et al., 2008). Although the shell color variation in intra and inter species of mollusks is common, color variation in soft tissues has been reported for only few species (Ky et al., 2019). For example, the black-lip pearl oyster (Pinctada margaritifera), a widely distributed Indo-Pacific pearl oyster species (Strack, 2008), consists of three distinct phenotypic color morphs of mantle tissue i.e., black, albino, and orange (Shirai and Nakamura, 1994; Hwang and Okutani, 2003; Wada and Tëmkin, 2008). However, the individuals with black mantle phenotype are the most frequent, and the albino and orange morphotypes are rare mutants (Ky et al., 2019). On the contrary, P. persica which is the most important pearl oyster species of Persian Gulf, displays two mantle color morphotypes, orange and black, but the black phenotype is rare and the orange morphotype is found at higher frequencies (Parvizi et al., 2017). P. persica was previously known as P. margaritifera var. persica (Jameson, 1901; Lal et al., 2017), but recently has been recognized as a new species (Ranjbar et al., 2015). The cross examination on P. margaritifera demonstrated that the variations in mantle color follow the Mendelian inheritance pattern (Ky et al., 2019). Ky et al. (2016) showed that the orange allele of P. margaritifera is recessive to the black one (Ky et al., 2016), and therefore the black mantle color is the dominant morphotype of the species. It is likely that the black phenotype of P. persica is the mutant phenotype, but it should receive more researches attention.
Studies have shown that in pearl oysters, the color polymorphism of mantle tissue is linked to the color of shell and pearl (Ky et al., 2019) dependent of transcriptional activity of biomineralization genes leading to shell and pearl formation (Wada, 1983; Clark et al., 2010; Joubert et al., 2010; Gardner et al., 2011; Shi et al., 2013; Freer et al., 2014; Sun et al., 2015; Wang et al., 2017). The color variation of shell is result of genes involved in different functions from shell structure to pigment synthesis. Differential expression of many genes known to be involved in the formation of the calcitic layer (e.g Aspein, Prismalin, Shematrin and etc), suggesting that variation in shell color could primarily take place in the calcitic layer (Lemer et al., 2015). Moreover, the aragonite binding pigments related to the biomineralization process also play a role in the color of the nacreous layer (Snow et al., 2004). Difference in expression patterns of some genes (like Pif-177, Collagen alpha-1 (XI) chain precursor, Zinc metalloprotease), involved in the formation of the aragonite tablets of the nacre and in the biosynthesis chain of melanin, suggesting that color polymorphism occur at different levels in the shell structure (Lemer et al., 2015). Genes involved in the melanin pathway including tyrosinase which is known as a key biomineralization enzyme (Sun et al., 2015) are among the other potential elements of the shell layers coloration process (Lemer et al., 2015).
The study of biomineralization related genes expression in mantle tissue can be used as a suitable approach to better identify the differentially expressed genes in different morphotypes and their potential role in shell and pearl quality and color variations. Lemer et al. (2015) and Ky et al. (2019) investigated the expression of some candidate genes associated to different mantle morphotypes in P. margaritifera. Using suppressive and subtractive hybridization (SSH) method and RT-qPCR, Lemer et al. (2015) identified differentially expressed genes between black and full albino phenotypes of P. margaritifera (Lemer et al., 2015). Comparing the differential expression of a set of genes in black versus orange mutant phenotypes, both in graft tissue and pearl sac, Ky et al. (2019) also underlined the existence of differential expression of four upon six genes and underlined very different pathways in the orange phenotypes than in the albino ones. Their study also revealed a strong influence of the orange phenotype on the color of pearl, leading to lighter colors that may be an interesting source of diversification for jewellery (Ky et al., 2019). The black phenotype shell has dark color and the orange one exhibit red outer shell phenotype (Ky et al., 2016). The inner shell of orange phenotype, in the contact area with the mantle at the edge of the shell (band color), is paler than that of the black type (Ky et al., 2019).
The pearl oyster P. persica inhabiting the shallow and warm waters of the Persian Gulf are known to produce the most famous pearl qualities in the world. The pearl production in this region and in this species is believed to be one of the first of its kind in the world (Bowen, 1951). Unlike its closely related species (P. margaritifera), P. persica has a distinct phenotypic color morph of mantle tissue, so that the orange mantle color of P. persica is the dominant morphotype compared to the black one.
Altogether the unique morphological characteristic, economic value and decreasing trend of population, this species could also be considered as an interesting pearl oyster species to investigate the molecular mechanisms involved in phenotypic variation, particularly for color traits, and biomineralization processes of shell and pearl formation. Using a deep literature mining across published studies in pearl oysters, we selected a suite of genes involved in biomineralization, shell formation and color polymorphism in pearl oysters, and then evaluated their expression in the mantle tissue of two morphotypes of P. persica using quantitative reverse transcription PCR (qRT-PCR). In molluscan, temperature is one of the important factors influencing physiological processes (Taylor and Venn, 1979; Park, 2001) and a marked effect has been reported on biomineralization (Yao and Somero, 2014). Given the potential influence of the environmental factors, i.e., temperature on transcriptome and biomineralization (Yao and Somero, 2014; Li et al., 2016), we also investigated the expression of biomineralization genes in black and orange morphotypes of P. persica in both warm and cool seasons.
Materials and methods
Thirty-one adult species of P. persica from two different color phenotypes of mantle tissue (21 orange and 10 black; Figures 1A, B) with 100–170 mm in dorso-ventral length were collected from Larak Island, Persian Gulf (26° 53.385’ N. 56 ° 21.061’ E; Figure 1C) by scuba diving during warm (September 2019, 32 °C) and cool (March 2020, 23 °C) seasons. The sample size including six black and 12 orange morphotypes collected in warm season, plus four black and nine orange phenotypes sampled in cool season. The graft part of the mantle tissue was dissected and stored at −80°C until subsequent RNA extraction.

Figure 1 Two common morphotypes of Pinctada persica found in Persian Gulf which were investigated in this study. (A) Orange morphotype, (B) Black morphotype. (C) Sampling site on Larak Island, Persian Gulf, Iran. The map was created using QGIS Desktop (Version 3. 14).
RNA was extracted from 50 mg of the mantle tissue using Trizol® reagent (GeneAll, Korea) in RNase free environment, according to the manufacturer’s recommendations with some modifications to improve the purity of RNAs. The tissues were homogenized in liquid nitrogen and treated in Trizol® reagent, accordingly. RNA was precipitated in isopropanol and washed in 75% ethanol two times and finally eluted in 50 ul DEPC treated water (Biobasic, Canada). RNA was quantified using a Nanodrop spectrophotometer (Thermo Scientific, MA) and the quality of the RNA was checked for degradation using 1% (w/v) agarose gel electrophoresis. Total RNA for each individual was then treated with DNase reagent (Thermo Scientific, MA) to cleanup potential DNA residuals. Afterwards, cDNA was synthesized using 1µg of total RNA as a template, and a cDNA synthesis kit (SMOBio, Taiwan) containing 1 µl dT primer and 0.2 µl random primers, following the manufacturer’s instructions.
In order to identify the relative gene expression profiles of black and orange mantle tissues, 16 candidate genes were targeted based on their functions (Table 1). The selected genes were divided into 5 categories (defined based on Lemer et al. (2015) and Ky et al. (2019) studies) as follow; 1] genes involved in the formation of the prismatic layer: Aspein, KRMP, Peroxidas, Shematrin1, Prismalin, Shematrin5 and Calmodulin (Suzuki et al., 2004; Tsukamoto et al., 2004; Yano et al., 2006; Zhang et al., 2006; Marie et al., 2012; Lemer et al., 2015; Le Luyer et al., 2019); 2] the nacre layer-related genes including MRNP34, Perline, SHELL, Linkine and Pif-177 (Gervis and Sims, 1992; Samata et al., 1999; Suzuki et al., 2009; Marie et al., 2012; Lemer et al., 2015; Le Luyer et al., 2019); 3] the gene Nacrein involved in the formation of both prismatic and nacre layers (Miyamoto et al., 1996; Yano et al., 2006; Marie et al., 2012); 4] the genes involved in the color of the shell, Tyrosinase2 and Chitin (Lemer et al., 2015) and 5] a gene indirectly involved in biomineralization process, SERP (Ky et al., 2019).
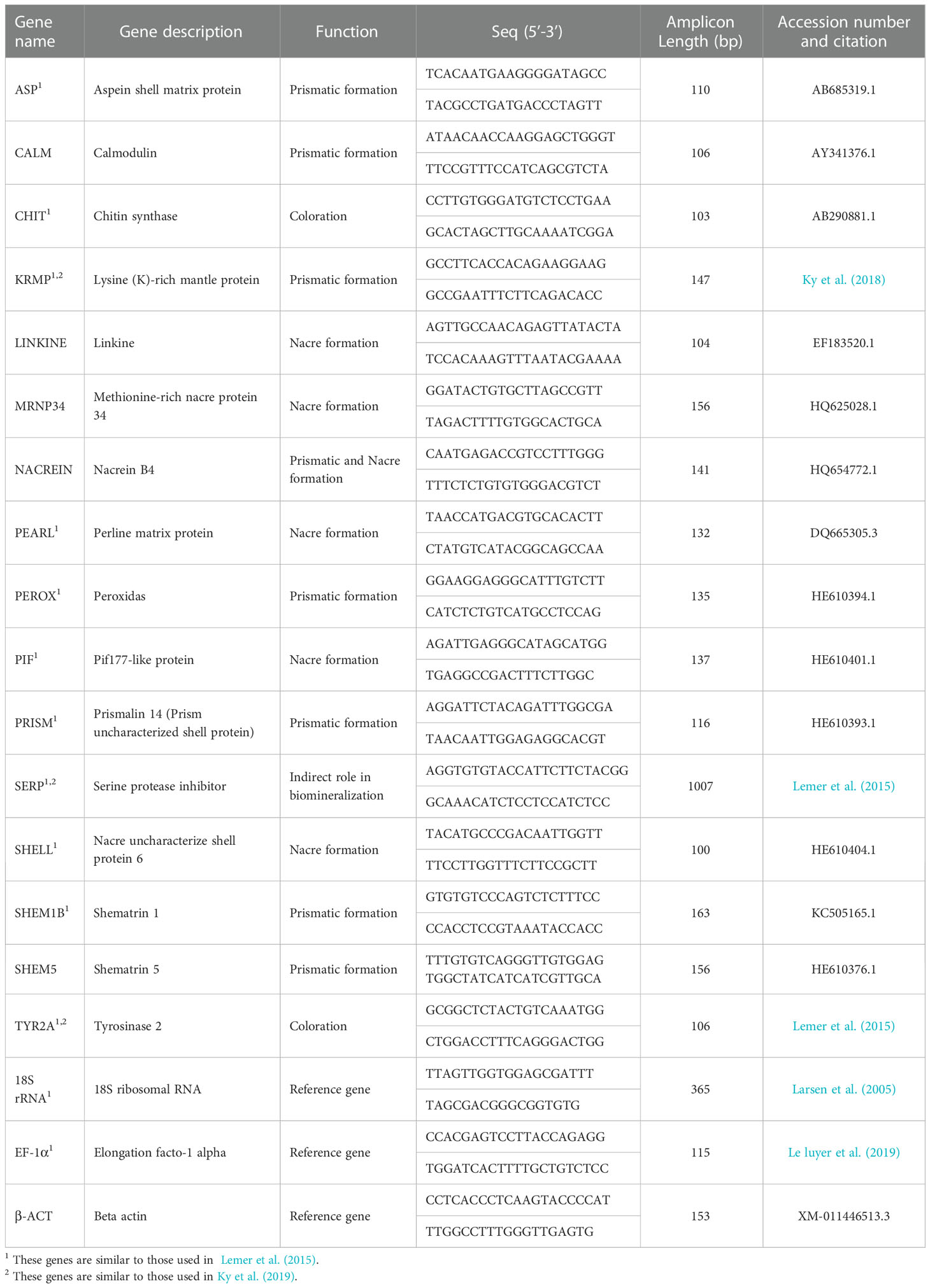
Table 1 Details of the candidate genes and the primer pairs used for gene expression analysis in Pinctada persica in this study.
For normalization of the target genes, various housekeeping genes including 18S ribosomal RNA (18SrRNA), Beta actin (β-actin) and Elongation factor 1 alpha (EF-a1) were initially considered as internal reference genes. The18S gene was shown as the most stable reference gene for qPCR in P. persica (Supplementary Figure 1) and was chosen for normalizing the target genes owing to its constitutive expression patterns in mantle tissue. The primers were designed using the Primer3 software (https://bioinfo.ut.ee/primer3-0.4.0; Primer size: 19-23, Tm: 57-62, Product size ranges: 100-170; Table 1). The specificity of the designed primers was checked by primer blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). In addition, the primers for genes CHIT, KRMP and SERP were adopted from pervious studies (Table 1). Then all the primer pairs (Table 1) were checked on synthesized cDNA from P. persica using normal PCR to ensure their amplification success and consistency.
qRT-PCR amplifications were carried out on a StepOne Real-Time PCR (ThermoFisher Scientific, MA) in final volume of 10 µl included 1 μl cDNA, 5 μl of 2X SYBR green mix (Ampliqon, Denmark) and 0.25 μM of each primer (listed in Table 1). The reaction started with initial denaturation at 94°C for 120 s and then 40 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 20 s followed by gradual thermo scanning (0.3 degree per step) for melting curve analysis. The RT-qPCR reactions were run in duplicate for each sample. The Ct values were exported after baseline normalization using the Real-Time PCR StepOne Software (ThermoFisher Scientific, MA). PCR efficiencies for each gene were calculated using LinRegPCR (Ramakers et al., 2003). The relative expression of target genes was calculated by 2–ΔΔCT method (Livak and Schmittgen, 2001), and then were log2-transformed.
The log2-transformed gene expression data were tested for normality using Shapiro-Wilk test and homogeneity of variance using Leven’s test to meet T-test assumptions. T-student test was used to check the source of significant difference in gene expression between the two morphotypes and the comparison of gene expression between black and orange morphotypes in both warm and cool seasons. For those data failed in normality test, the non-parametric analysis, Mann-Whitney test, was used for analysis. Statistical analysis was conducted using SigmaPlot-v11 (version 11).
The principal components analysis (PCA) was carried out in order to transform the variables and indicated the ordination of target genes against independent variables (color and season) using the fviz_pca function of the factoextra R package (Kassambara and Mundt, 2017). This function provided 95% confidence ellipses for groups within each data set (Houde et al., 2019; Akbarzadeh et al., 2020).
Results
Two color phenotypes of P. persica i.e., orange and black mantle were observed among the specimens sampled in our study in which the orange phenotype was more abundant than the black one. The results of qRT-PCR revealed that a set of genes were differentially expressed between two phenotypes of P. persica. Among the genes involved in the formation of the prismatic layer, the mRNA expression of ASP, KRMP, SHEM1B and SHEM5 were significantly higher in the orange phenotype compared to those of black morphotype (P<0.05). The mRNA expression of PEROX and PRISM showed no significant differences between the two investigated phenotypes (Figure 2A). The results also showed that the gene CALM was only expressed in the orange phenotype, while 90% of individuals with black morphotype showed no CALM mRNA expression.
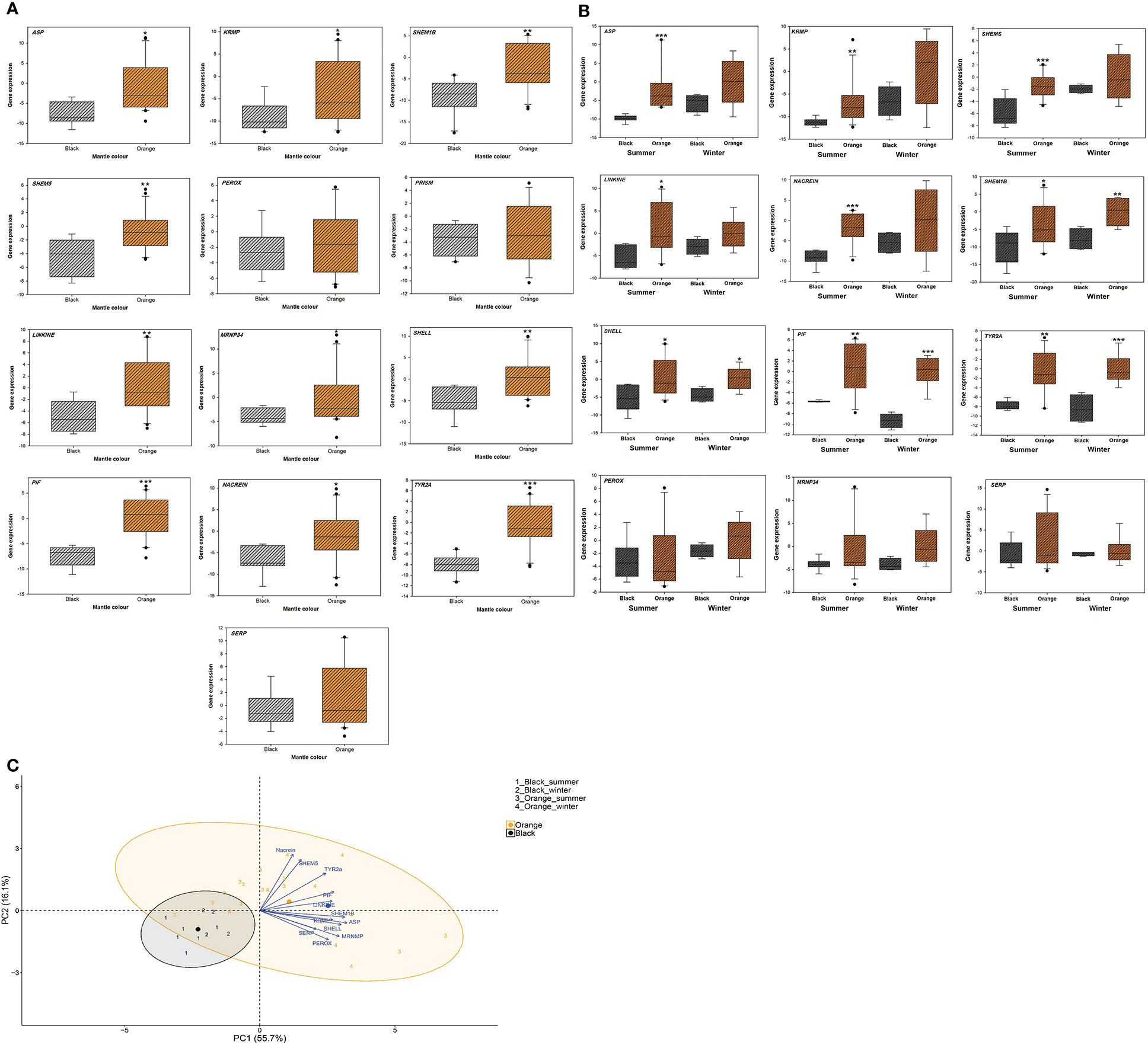
Figure 2 (A) The box plots of the expression of genes involved in biomineralization process in orange and black phenotypes of the pearl oyster Pinctada persica. (B) The box plots of the expression of genes involved in biomineralization process in black and orange phenotypes of the pearl oyster Pinctada persica, in winter (cool) and summer (warm) seasons. Grey bars represent data for the black phenotype and orange bars represent data for the orange phenotype. Y axis is in the logarithmic scale (gene expression). Error bars indicate standard deviations. The genes CALM, PEARL and CHIT were only expressed in the orange phenotype. Statistical differences between the phenotypes are indicated by asterisks: *:P<0.05, **:P<0.01, ***:P<0.001. (C) Canonical plots of the first two principal components of the biomineralization genes for two mantle morphs (black and orange) of the pearl oyster Pinctada persica sampled in winter (cool) and summer (warm) seasons. Principal component analysis (PCA) was performed on the entire dataset. Ellipses represent 95% confidence areas for the groups within treatments. Centroids are represented by the largest point of the same color. Arrows represent loading vectors of the genes.
All the genes related to nacre layer formation (MRNP34, PEARL, SHELL, LINKINE and PIF), showed significantly higher transcript levels in the orange morphotype compared to the black phenotype (P<0.05). The only exception was the PEARL gene in which no transcripts were detected in the individuals with black phenotype, while expression of this gene was detected in 52.3% of individuals with orange mantle type (Figure 2A).
The NACREIN gene which is involved in the formation of both prismatic and nacre layers, also showed significantly higher expression in the mantle of orange morphotype compared to the black morphotype (P<0.05; Figure 2A). Moreover, among the pigmentation related genes, TYR2A showed significantly higher mRNA expression in the orange morphotype compared to the black one (P<0.001; Figure 2A). The CHIT transcripts were not detected in the black morphotype and only 42.8% individuals of orange phenotype showed CHIT transcripts. In addition, no significant differences were observed in the expression of the gene SERP between two morphotypes (P>0.05; Figure 2A).
The variations of the gene transcripts in both phenotypes of P. persica in each season are shown in Figure 2B. In warm season, the mRNA expression of ASP, KRMP, SHEM5, LINKINE and NACREIN was significantly higher in the orange phenotype compared to those of black morphotype (P<0.05; Figure 2B), while no significant differences of these genes were observed between the two phenotypes in winter (Figure 2B). The SHEM1B, SHELL, PIF and TYR2A genes were significantly higher in the orange phenotype compared to those of black one in both warm and cool seasons (P<0.05; Figure 2B).
The PCA analysis revealed that the first cleavage factor is color, with a separation of orange and black groups as shown in PC1 in Figure 2C. The level of variation range in orange group was also more important than that of black phenotype. There was no clear separation between the individuals sampled in two seasons for either color (Figure 2C).
Discussion
In pearl oysters, the color polymorphism of mantle tissue is tightly linked to the color and radiance of shell and pearl (Ky et al., 2019). Therefore, the study of biomineralization and the genes related to this biological process in mantle tissue of pearl oysters can be used as a suitable approach to better understanding the molecular mechanisms that influence shell and pearl quality and color variations. In this study, we investigated the transcription of a panel of genes encoding proteins involved in biomineralization and pigmentation in black and orange mantle morphotypes of pearl oyster, P. persica in both warm and cool seasons using qRT-PCR. Our results showed differential expression of genes between two color morphs of P. persica. The major genes chosen for their involvement in biomineralization of the prismatic and nacre layer, and pigmentation systematically exhibited a significantly higher expression in orange compared to the black phenotypes, suggesting the more active role of these genes in the formation and the color of shell and probably the pearl in the orange morphotype of P. persica.
Shell coloration has been shown to be related to the original pigments and the chemical interactions between the pigments and the shell matrix in mollusks (Williams, 2017). The pigments may be inserted in the outer or other layers of shells (Budd et al., 2014; Williams, 2017). The molecular pathway of the melanin synthesis has an important contribution to the shell and pearl pigmentation (Palumbo, 2003; Lemer et al., 2015). According to the results of the present study, the TYR2A gene that is the most important enzyme in the melanin production (Ding et al., 2021) was higher expressed in the orange phenotype of P. persica compared to that of black mantle phenotype. The tyrosinases play diverse roles in mollusk tissues and shells including shell matrix formation (Degens et al., 1967; Gordon and Carriker, 1980; Waite, 1983; Feng et al., 2017), pigmentation (Nagai et al., 2007; Takgi and Miyashita, 2014; Feng et al., 2017), and producing of the periostracum (Waite and Wilbur, 1976; Waite, 1983; Feng et al., 2017). It is likely that tyrosinases play a different role in the formation of the shell (calcitic) layer and melanin synthesis between two color morphs of P. persica which may result in different amounts of pigments and pigments intensity in different phenotypes (Stenger et al., 2021). The relationship between gene expression and shell pigmentation was reported in the clam Meretrix meretrix with high expression of the genes in the deeper shell color samples (Yue et al., 2015). In addition, the relationship between the shell and mantle pigmentation (Brake et al., 2004) and heritability of shell pigmentation (Evans et al., 2009) were described in Pacific oyster, Crassostrea gigas (Kang et al., 2013). Opposite to the findings of Ky et al. (2019) and Lemer et al. (2015), our results showed significantly differential expression of TYR2A between two morphotypes of P. persica. The high expression of TYR2A in orange phenotype may thus be related to the composition of non-functional melanin protein, a hypothesis reported for other genes such as Zinc metalloprotease (King and Summers, 1988; Oetting and King, 1999; Lemer et al., 2015; Ky et al., 2019).
In addition to tyrosinase, chitin related proteins play prominent roles in the shell formation of bivalve (Feng et al., 2017) and melanin biosynthesis (Lemer et al., 2015), however the expression of CHIT here was only detected in the orange phenotype, suggesting that an alternative pathway might exist in the black phenotype of P. persica. Both CHIT and PEARL genes which are strongly involved in the formation of nacre in pearl oysters (Samata et al., 1999; Miyashita et al., 2000; Kono et al., 2000; Suzuki and Nagasawa, 2007; Yan et al., 2007; Montagnani et al., 2011) and Calmodulin which is prismatic related gene (Marie et al., 2012) only expressed in orange phenotype of P. persica. The calcium binding proteins such as calmodulin, probably play an important role in the regulation of uptake, transport and secretion of calcium and may be essential regulators in the extracellular nacrein construction (Miyamoto et al., 1996; Huang et al., 2007). The MRNP34 may be putatively involved in protein/protein interactions, by analogy with other proteins, like calmodulin, which shows two hydrophobic methionine-rich domains that bind to the target protein (Siivari et al., 1995) and involved in nacre formation (Marie et al., 2012). Therefore, the high expression of these genes in orange morph may confirm their interaction that leads to the enhanced biomineralization and the regulation of microstructure of nacreous layer in this phenotype. Taken together, these results show the important roles of these genes in the calcium metabolism during nacre and prismatic layers formation in orange phenotype of P. persica compared to the black one, which should lead to different rate of calcification, calcium content and the aragonite and calcite crystals arrangements in the shell and pearl. It is likely that other genes from different pathways or the genes that their function is still unknown play similar roles in the black morphotype of P. persica
The peroxidases are formed of two clusters that could be classified as melanin biosynthesis group and the shell formation group (Feng et al., 2017). In this study, we did not observe different expression of PEROX between the two morphotypes, suggesting this gene may not be a key gene of melanin synthesis in P. persica.
Higher expression of PIF in orange phenotype of P. persica than that of black phenotypes confirmed the distinct functions of this gene between two color morphs. Pif-177 is an important component of the organic matrix for the nacreous layer growth of pearl oysters (Suzuki et al., 2009), consists of aragonite-binding domain, chitin-binding domain, and protein-protein interaction domain (Feng et al., 2017). It is believed that PIF is involved in the establishment of color in the shell of P. margaritifera (Lemer et al., 2015). Lemer et al. (2015) argued that the lower expression of PIF in albino than that of black morphotype of P. margaritifera, is involved in their different aragonite crystals arrangement that is associated to lower quality in the colorless nacre layer of albino morphotype (Lemer et al., 2015). Therefore, it can be suggested that the higher expression of PIF in orange phenotype of P. persica might lead to a higher quality of nacre in this phenotype.
The transcripts of two genes encoding shematrin proteins (SHEM1B and SHEM5) were remarkably higher expressed in orange phenotype of P. persica than those of black mantle phenotype. The shematrin proteins are the most common proteins synthesized in the mantle edge of mollusks (Yano et al., 2006; Marie et al., 2012). These proteins are secreted into the prismatic layer of the shell, which is thought to provide a template for calcification and are responsible for the hardness of the shell (Yano et al., 2006). These results show the important roles of the shematrin family proteins in calcification of different pearl oyster morphotypes that can cause different hardness in shell and pearl.
Aspein shell matrix protein is known as the most acidic protein in mollusk shell matrix protein that controls the contribution of calcite (Tsukamoto et al., 2004). Higher expression of ASP and KRMP genes that encode proteins involved in calcite layer formation (Tsukamoto et al., 2004; Zhang et al., 2006) in the orange phenotype of P. persica might indicate that the calcite layer framework and arrangement differ in the orange compared to the black phenotypes. In P. margaritifera, higher expression of ASP and lower expression of KRMP was observed in the black compared to albino phenotypes (Lemer et al., 2015), while no differences in the expression of KRMP were reported between black and orange mantle of P. margaritifera (Ky et al., 2019). Such contrasted expression patterns of genes between closely related species suggest the complex and species specific (and possibly evolutionary convergent) genetic processes are involved in the biomineralization in mantle tissue of pearl oysters. Moreover, both ASP and NACREIN play important roles in pearl quality and color phenotypes (Blay et al., 2018). It has been reported that the expression of ASP is associated with the pearl sac and quality traits (surface defects, lustre and grade) and color in pearl oysters (Blay et al., 2017). Since the high expression of NACREIN and ASP was observed in orange phenotype of P. persica compared to the black one, we expect to observe different quality and color of pearls between two morphotypes.
Matrix proteins regulate the crystal phase, shape, size and nucleation of CaCO3 crystals (Liu et al., 2012). Altogether, differential expression of the nacre and calcite layer-related genes i.e., ASP, KRMP, SHEM1B, SHEM5, LINKINE, MRNP34, SHELL, PIF, NACREIN and coloration (TYR2A) between two morphotypes of P. persica might indicate the different regulation of these genes and its consequences in the rate of calcification, the design of aragonite and calcite crystals in the shell and pearl, the calcium content, the thickness and weight of the prismatic and nacre layers, intensity and pattern of pigmentation, variability of nacre color of shell and pearl color in oysters with different mantle morphotypes in this species. Some studies investigated the gene expression patterns with different phenotypic traits, which showed differential gene expression between different phenotypic groups (Shi and He, 2014; Lemer et al., 2015; Yue et al., 2015; Blay et al., 2017; Blay et al., 2018; Ky et al., 2018; Ky et al., 2019; Stenger et al., 2021). In contrast to our study, the genes encoding the organic matrix proteins produced by the mantle were found to be highly expressed in the black specimens of P. margaritifera. It is stated that the pigmentation-related genes i.e., SHEM4, KRMP, MP8, CHIT and SERP are involved in the black eumelanin synthesis (Lemer et al., 2015). Moreover, Lemer et al. (2015) reported more expression of genes involved in prismatic layer in the black specimens of P. margaritifera and suggested that variation in shell color can primarily occur in the prismatic layer (Lemer et al., 2015). Therefore, the origin of color can result from multiple interactions between both prismatic and nacre layer, and the pigmentation-related genes, which may be conducted by distinct genes in congeneric, even in closely related species.
The beauty and lustre of pearls are related to the surface nacreous layer. The nacre layer is made of CaCO3 aragonite tablets, chitin and matrix proteins (Greígoire, 1957; Watabe, 1965; Wise, 1970). The accurate and orderly assembly of the crystals and organic matrix not only increases lustre by reflecting light uniformly from the layered compartment (Mann, 2001), but also strengthens the nacreous layer to provide fracture resistance (Smith et al., 1999). The highest levels of matrix protein expression is correlated with the ordered deposition of CaCO3 (Liu et al., 2012). According to our result, we might expect a high quality of pearl in orange phenotype of P. persica compared to the black one, however experimental approaches are needed to test this hypothesis. Effective selection based on shell and mantle edge color has been already reported in the Pacific oyster (Kang et al., 2013). The selection programs can improve traits such as pearl size, color, darkness, surface defects and grade (Blay et al., 2018).
Comparing two phenotypes of P. persica in warm and cool seasons showed that the upregulation of ASP, KRMP, SHEM5, LINKINE and NACREIN genes in orange phenotype was only observed in warm season, but PIF, SHELL, SHEM1B, and TYR2A were significantly upregulated in the orange phenotype in both warm and cool seasons. It is likely that warmer season might modulate higher expression of genes involved in biomineralization process in more dominant phenotype of P. persica inhabiting sub-tropical ecosystem of Persian Gulf, and this might affect the quality and color of pearls in orange phenotype of P. persica. However, other environmental factors also might affect the expression of biomineralization genes in mantle tissue of pearl oysters. Previous studies demonstrated the responses of biomineralization-related genes to food, ocean acidification and temperature in pearl oysters (Liu et al., 2012; Moya et al., 2012; Joubert et al., 2014; Li et al., 2016). Various studies have showed the effect of temperature on the relative thickness and growth of the shell in oysters (Pouvreau and Prasil, 2001; Kanazawa and Sato, 2008). In the closely related species, P. margaritifera inhabiting cooler temperatures, exposing to high temperature caused a decrease in the expression biomineralization genes and negatively impacted shell growth [79]. It has been suggested that the thermal thresholds are probably specific to the ecological limits of the pearl oysters [79]. Further experiments might be needed to explore the possible influence of inhabiting warmer temperatures on the nature of nacre and the possible evolution of dominant phenotypes, and possibly the potential for high quality pearl production in natural populations of P. persica.
Conclusion
The goal of present study was to compare the expression of genes involved in biomineralization and pigmentation in the orange and black phenotypes of P. persica. Our results showed different expression profiles of genes between these two color morphs. Major genes involved in biomineralization and pigmentation (except for PEROX and PRISM) were significantly higher expressed in the orange phenotype of P. persica that is more dominant morphotype in Persian Gulf, which is contrary to the results obtained in the closely related species, P. margaritifera with higher expression of biomineralization and pigmentation genes in the black dominant morphotype (Lemer et al., 2015; Ky et al., 2019).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was approved by Qeshm Department of Environment of QFAO.
Author contributions
MR supervised the study and conducted the sampling. MR and AF designed the experiments. FP, AF and MR carried out the molecular genetic studies and experiments in the laboratory and wrote the manuscript. AA, MR, FP and SA-H analyzed the data and contributed to reviewing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by Persian Gulf Biotechnology Park (PGBP), Qeshm Island, Iran grant no. 431.98.
Acknowledgments
Especial thanks go to Kaveh Khosraviani (CellKGP Company, Genetic and Biotechnology laboratory, Karaj, Iran) for laboratory and technical support. We are also grateful to Persian Gulf Biotechnology Park (PGBP) for their generous support to develop this study. The authors also would like to acknowledge the support provided by Eghbal Zobeiri for the map preparation. All individual rights, animal rights and scientific ethics have been observed in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1038692/full#supplementary-material
References
Akbarzadeh A., Houde A. L. S., Sutherland B. J., Günther O. P., Miller K. M. (2020). Identification of hypoxia-specific biomarkers in salmonids using RNA-sequencing and validation using high-throughput qPCR. G3. 10 (9), 3321–3336. doi: 10.1534/g3.120.401487
Blay C., Planes S., Chin-Long K. (2017). Donor and recipient contribution to phenotypic traits and the expression of biomineralisation genes in the pearl oyster model pinctada margaritifera. Sci. Rep. 7 (1), 1–12. doi: 10.1038/s41598-017-02457-x
Blay C., Planes S., Ky C.-L. (2018). Crossing phenotype heritability and candidate gene expression in grafted black-lipped pearl oyster Pinctada margaritifera, an animal chimera. J. Hered. 109 (5), 510–519. doi: 10.1093/jhered/esy015
Bowen R. L. (1951). The pearl fisheries of the Persian gulf. Middle East J. 5 (2), 161–180. Available at: http://www.jstor.org/stable/4322269.
Brake J., Evans F., Langdon C. (2004). Evidence for genetic control of pigmentation of shell and mantle edge in selected families of pacific oysters. Crassostrea gigas. Aquac. 229 (1-4), 89–98. doi: 10.1016/S0044-8486(03)00325-9
Budd A., McDougall C., Green K., Degnan B. M. (2014). Control of shell pigmentation by secretory tubules in the abalone mantle. Front. Zool. 11 (1), 1–9. doi: 10.1186/s12983-014-0062-0
Clark M. S., Thorne M. A., Vieira F. A., Cardoso J. C., Power D. M., Peck L. S. (2010). Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genom. 11 (1), 1–14. doi: 10.1186/1471-2164-11-362
Degens E. T., Spencer D. W., Parker R. H. (1967). Paleobiochemistry of molluscan shell proteins. Comp. Biochem. Physiol. 20 (2), 553–579. doi: 10.1016/0010-406X(67)90269-1
Ding J., Wen Q., Huo Z., Nie H., Qin Y., Yan X. (2021). Identification of shell-color-related microRNAs in the Manila clam Ruditapes philippinarum using high-throughput sequencing of small RNA transcriptomes. Sci. Rep. 11 (1), 1–8. doi: 10.1038/s41598-021-86727-9
Evans S., Camara M. D., Langdon C. J. (2009). Heritability of shell pigmentation in the pacific oyster, crassostrea gigas. Aquac. 286 (3-4), 211–216. doi: 10.1016/j.aquaculture.2008.09.022
Feng D., Li Q., Yu H., Kong L., Du S. (2017). Identification of conserved proteins from diverse shell matrix proteome in Crassostrea gigas: characterization of genetic bases regulating shell formation. Sci. Rep. 7 (1), 1–12. doi: 10.1038/srep45754
Fougerouse A., Rousseau M., Lucas J. S. (2008). “Soft tissue anatomy, shell structure and biomineralisation,” in The pearl oyster. Eds. Southgate P. C., Lucas J. S.. (Amsterdam: The Netherlands: Elsevier), 77–102.
Freer A., Bridgett S., Jiang J., Cusack M. (2014). Biomineral proteins from Mytilus edulis mantle tissue transcriptome. Mar. Biotechnol. 16 (1), 34–45. doi: 10.1007/s10126-013-9516-1
Gardner L. D., Mills D., Wiegand A., Leavesley D., Elizur A. (2011). Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyster pinctada maxima. BMC Genom 12 (1), 1–16. doi: 10.1186/1471-2164-12-455
Gervis M. H., Sims N. A. (1992). The biology and culture of pearl oysters (Bivalvia pteriidae) (London, United Kingdom: Overseas Development Administration (ODA) and International Center for Living Aquatic Resources Management (ICLARM), Manila, Philippines: WorldFish).
Gordon J., Carriker M. (1980). Sclerotized protein in the shell matrix of a bivalve mollusc. Mar. Biol. 57 (4), 251–260. doi: 10.1007/BF00387568
Greígoire C. (1957). Topography of the organic components in mother-of-pearl. J. Cell Biol. 3 (5), 797–808. doi: 10.1083/jcb.3.5.797
Houde A. L. S., Akbarzadeh A., Günther O. P., Li S., Patterson D. A., Farrell A. P., et al. (2019). Salmonid gene expression biomarkers indicative of physiological responses to changes in salinity and temperature, but not dissolved oxygen. J. Exp. Biol. 222 (13), jeb198036. doi: 10.1242/jeb.198036
Huang J., Zhang C., Ma Z., Xie L., Zhang R. (2007). A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim. Biophys. Acta Gen. Subj. 1770 (7), 1037–1044. doi: 10.1016/j.bbagen.2007.03.006
Hwang J. J., Okutani T. (2003). Taxonomy and distribution of the genera pteria and pinctada (Bivalvia: Pteriidae) in Taiwan. JFST. 30 (3), 199–216. Available at: https://www.airitilibrary.com/Publication/alDetailedMesh?docid=03794180-200309-30-3-199-216-a.
Jameson L. H. (1901). On the identity and distribution of the mother-of-pearl oysters; with a revision of the subgenus margaritifera. Proc. Zool. Soc. London 70 (2), 372–394.
Joubert C., Linard C., Le Moullac G., Soyez C., Saulnier D., Teaniniuraitemoana V., et al. (2014). Temperature and food influence shell growth and mantle gene expression of shell matrix proteins in the pearl oyster pinctada margaritifera. PLoS One 9 (8), e103944. doi: 10.1371/journal.pone.0103944
Joubert C., Piquemal D., Marie B., Manchon L., Pierrat F., Zanella-Cléon I., et al. (2010). Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genom. 11 (1), 1–13. doi: 10.1186/1471-2164-11-613
Kanazawa T., Sato Si. (2008). Environmental and physiological controls on shell microgrowth pattern of Ruditapes philippinarum (Bivalvia: Veneridae) from Japan. J. Molluscan Stud. 74 (1), 89–95. doi: 10.1093/mollus/eym049
Kang J.-H., Kang H.-S., Lee J.-M., An C.-M., Kim S.-Y., Lee Y.-M., et al. (2013). Characterizations of shell and mantle edge pigmentation of a pacific oyster, Crassostrea gigas, in Korean peninsula. Asian-australas J. Anim. Sci. 26 (12), 1659. doi: 10.5713/ajas.2013.13562
Kassambara A., Mundt F. (2017). Factoextra: extract and visualize the results of multivariate data analyses. R Package Version. 1 (5), 337–354. Available at: https://CRAN.R-project.org/package=factoextra.
King R. A., Summers C. (1988). Albinism. Dermatol. Clin. 6, 217–228. doi: 10.1016/S0733-8635(18)30668-5
Kono M., Hayashi N., Samata T. (2000). Molecular mechanism of the nacreous layer formation in pinctada maxima. Biochem. Biophys. Res. Commun. 269 (1), 213–218. doi: 10.1006/bbrc.2000.2274
Ky C.-L., Blay C., Broustal F., Koua M. S., Planes S. (2019). Relationship of the orange tissue morphotype with shell and pearl colouration in the mollusc pinctada margaritifera. Sci. Rep. 9 (1), 1–9. doi: 10.1038/s41598-019-41581-8
Ky C. L., Nakasai S., Pommier S., Sham Koua M., Devaux D. (2016). The mendelian inheritance of rare flesh and shell colour variants in the black-lipped pearl oyster (Pinctada margaritifera). Anim. Genet. 47 (5), 610–614. doi: 10.1111/age.12454
Ky C.-L., Quillien V., Broustal F., Soyez C., Devaux D. (2018). Phenome of pearl quality traits in the mollusc transplant model pinctada margaritifera. Sci. Rep. 8 (1), 1–11. doi: 10.1038/s41598-018-20564-1
Lal M. M., Southgate P. C., Jerry D. R., et al. (2017). Swept away: Ocean currents and seascape features influence genetic structure across the 18,000 Km Indo-Pacific distribution of a marine invertebrate, the black-lip pearl oyster Pinctada margaritifera. BMC Genomics 18, 66. doi: 10.1186/s12864-016-3410-y
Larsen J. B., Frischer M. E., Rasmussen L. J., Hansen B. W. (2005). Single-step nested multiplex PCR to differentiate between various bivalve larvae. Mar. Biol. 146 (6), 1119–29.
Le Luyer J., Auffret P., Quillien V., Leclerc N., Reisser C., Vidal-Dupiol J., et al. (2019). Whole transcriptome sequencing and biomineralization gene architecture associated with cultured pearl quality traits in the pearl oyster, pinctada margaritifera. BMC Genom 20 (1), 1–11. doi: 10.1186/s12864-019-5443-5
Lemer S., Saulnier D., Gueguen Y., Planes S. (2015). Identification of genes associated with shell color in the black-lipped pearl oyster, pinctada margaritifera. BMC Genom 16 (1), 1–14. doi: 10.1186/s12864-015-1776-x
Li S., Liu C., Huang J., Liu Y., Zhang S., Zheng G., et al. (2016). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO 2 and temperature. Sci. Rep. 6 (1), 1–10. doi: 10.1038/srep18943
Liu W., Huang X., Lin J., He M. (2012). Seawater acidification and elevated temperature affect gene expression patterns of the pearl oyster pinctada fucata. PLoS One 7 (3), e33679. doi: 10.1371/journal.pone.0033679
Liu X., Li J., Xiang L., Sun J., Zheng G., Zhang G., et al. (2012). The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc. R. Soc. B. 279 (1730), 1000–1007. doi: 10.1098/rspb.2011.1661
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Mann S. (2001). Biomineralization: principles and concepts in bioinorganic materials chemistry (New York, United States: Oxford University Press on Demand).
Marie B., Joubert C., Belliard C., Tayale A., Zanella-Cléon I., Marin F., et al. (2012). Characterization of MRNP34, a novel methionine-rich nacre protein from the pearl oysters. Amino Acids 42 (5), 2009–2017. doi: 10.1007/s00726-011-0932-0
Marie B., Joubert C., Tayalé A., Zanella-Cléon I., Belliard C., Piquemal D., et al. (2012). Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. PNAS. 109 (51), 20986–20991. doi: 10.1073/pnas.1210552109
Miyamoto H., Miyashita T., Okushima M., Nakano S., Morita T., Matsushiro A. (1996). A carbonic anhydrase from the nacreous layer in oyster pearls. PNAS. 93 (18), 9657–9660. doi: 10.1073/pnas.93.18.9657
Miyashita T., Takagi R., Okushima M., Nakano S., Miyamoto H., Nishikawa E., et al. (2000). Complementary DNA cloning and characterization of pearlin, a new class of matrix protein in the nacreous layer of oyster pearls. Mar. Biotechnol. 2 (5), 409–418. doi: 10.1007/PL00021687
Montagnani C., Marie B., Marin F., Belliard C., Riquet F., Tayalé A., et al. (2011). Pmarg-pearlin is a matrix protein involved in nacre framework formation in the pearl oyster pinctada margaritifera. Chembiochem 12 (13), 2033–2043. doi: 10.1002/cbic.201100216
Moya A., Huisman L., Ball E., Hayward D., Grasso L., Chua C., et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol. Ecol. 21 (10), 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Nagai K., Yano M., Morimoto K., Miyamoto H. (2007). Tyrosinase localization in mollusc shells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 146 (2), 207–214. doi: 10.1016/j.cbpb.2006.10.105
Oetting W. S., King R. A. (1999). Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum. Mutat. 13 (2), 99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C
Palumbo A. (2003). Melanogenesis in the ink gland of sepia officinalis. Pigment Cell Res. 16 (5), 517–522. doi: 10.1034/j.1600-0749.2003.00080.x
Park M. S. (2001). Reproductive cycle and biochemical composition of the ark shell Scapharca broughtonii (Schrenck) in a southern coastal bay of Korea. J. Shellfish Res. 20, 177–184. Available at: https://ci.nii.ac.jp/naid/10015421016/.
Parvizi F., Monsefi M., Noori A., Ranjbar M. S. (2017). Mantle histology and histochemistry of three pearl oysters: Pinctada persica, Pinctada radiata and pteria penguin. Molluscan Res. 38 (1), 11–20. doi: 10.1080/13235818.2017.1387039
Pouvreau S., Prasil V. (2001). Growth of the black-lip pearl oyster, Pinctada margaritifera, at nine culture sites of French Polynesia: synthesis of several sampling designs conducted between 1994 and 1999. Aquat Living Resour. 14 (3), 155–163. doi: 10.1016/S0990-7440(01)01120-2
Ramakers C., Ruijter J. M., Lekanne Deprez R. H., Moorman A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Ranjbar M. S., Zolgharnien H., Yavari V., Archangi B., Salari M. A., Arnaud-Haond S., et al. (2015). Rising the Persian gulf black-lip pearl oyster to the species level: fragmented habitat and chaotic genetic patchiness in pinctada persica. Evol. Biol. 43 (1), 131–143. doi: 10.1007/s11692-015-9356-1
Samata T., Hayashi N., Kono M., Hasegawa K., Horita C., Akera S. (1999). A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett. 462 (1-2), 225–229. doi: 10.1016/S0014-5793(99)01387-3
Shi Y., He M. (2014). Differential gene expression identified by RNA-seq and qPCR in two sizes of pearl oyster (Pinctada fucata). Gene. 538 (2), 313–322. doi: 10.1016/j.gene.2014.01.031
Shirai S., Nakamura S. (1994). Pearls and pearl oysters of the world (Okinawa, Japan: Marine planning).
Shi Y., Yu C., Gu Z., Zhan X., Wang Y., Wang A. (2013). Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes. Mar. Biotechnol. 15 (2), 175–187. doi: 10.1007/s10126-012-9476-x
Siivari K., Zhang M., Palmer A. G., Vogel H. J. (1995). NMR studies of the methionine methyl groups in calmodulin. FEBS Lett. 366 (2-3), 104–108. doi: 10.1016/0014-5793(95)00504-3
Smith B. L., Schäffer T. E., Viani M., Thompson J. B., Frederick N. A., Kindt J., et al. (1999). Molecular mechanistic origin of the toughness of natural adhesives, fibres and composites. Nature. 399, 761–763. doi: 10.1038/21607
Snow M. R., Pring A., Self P., Losic D., Shapter J. (2004). The origin of the color of pearls in iridescence from nano-composite structures of the nacre. Am. Min. 89 (10), 1353–1358. doi: 10.2138/am-2004-1001
Stenger P.-L., Ky C.-L., Reisser C., Duboisset J., Dicko H., Durand P., et al. (2021). Molecular pathways and pigments underlying the colors of the pearl oyster Pinctada margaritifera var. cumingii (Linnaeus 1758). Genes. 12 (3), 421. Available at: https://www.mdpi.com/2073-4425/12/3/421.
Strack E. (2008). “Introduction,” in The pearl oyster, vol. p . Eds. Southgate P. C., Lucas J. S. (Amsterdam: The Netherlands: Elsevier), 1–35.
Sun X., Yang A., Wu B., Zhou L., Liu Z. (2015). Characterization of the mantle transcriptome of yesso scallop (Patinopecten yessoensis): identification of genes potentially involved in biomineralization and pigmentation. PLoS One 10 (4), e0122967. doi: 10.1371/journal.pone.0122967
Suzuki M., Murayama E., Inoue H., Ozaki N., Tohse H., Kogure T., et al. (2004). Characterization of prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem. J. 382 (1), 205–213. doi: 10.1042/BJ20040319
Suzuki M., Nagasawa H. (2007). The structure–function relationship analysis of prismalin-14 from the prismatic layer of the Japanese pearl oyster, pinctada fucata. FEBS J. 274 (19), 5158–5166. doi: 10.1111/j.1742-4658.2007.06036.x
Suzuki M., Saruwatari K., Kogure T., Yamamoto Y., Nishimura T., Kato T., et al. (2009). An acidic matrix protein, pif, is a key macromolecule for nacre formation. Science. 325 (5946), 1388–1390. doi: 10.1126/science.1173793
Takgi R., Miyashita T. (2014). A cDNA cloning of a novel alpha-class tyrosinase of Pinctada fucata: its expression analysis and characterization of the expressed protein. Enzyme Res. 2014, 9. doi: 10.1155/2014/780549
Taylor A., Venn T. (1979). Seasonal variation in weight and biochemical composition of the tissues of the queen scallop, Chlamys opercularis, from the Clyde Sea area. J. Mar. Biolog Assoc. 59 (3), 605–621. doi: 10.1017/S0025315400045628
Tsukamoto D., Sarashina I., Endo K. (2004). Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem. Biophys. Res. Commun. 320 (4), 1175–1180. doi: 10.1016/j.bbrc.2004.06.072
Wada K. (1983). White coloration of the prismatic layer in inbred Japanese pearl oyster, pinctada fucata (Yoshokukenho, Japan: Bulletin of National Research Institute of Aquaculture).
Wada K. T., Tëmkin I. (2008). “Taxonomy and phylogeny: Commercial species,” in The pearl oyster. Amsterdam, vol. p . Eds. Southgate P. C., Lucas J. S. (The Netherlands: Elsevier), 37–75.
Waite J. (1983). “Quinone-tanned scleroproteins,” in The Mollusca. Eds. Saleudin A. S. M., Wilbur K. M. (New York: Academic), 467–504.
Waite J. H., Wilbur K. M. (1976). Phenoloxidase in the periostracum of the marine bivalve Modiolus demissus dillwyn. J. Exp. Zool. 195 (3), 359–367. doi: 10.1002/jez.1401950304
Wang X., Liu Z., Wu W. (2017). Transcriptome analysis of the freshwater pearl mussel (Cristaria plicata) mantle unravels genes involved in the formation of shell and pearl. Mol. Genet. Genom. 292 (2), 343–352. doi: 10.1007/s00438-016-1278-9
Watabe N. (1965). Studies on shell formation: XI. crystal–matrix relationships in the inner layers of mollusk shells. J. Ultrastruct Res. 12 (3), 351–370. doi: 10.1016/S0022-5320(65)80104-6
Wise S. W. (1970). Microstructure and mode of formation of nacre (mother-of-pearl) in pelecypods, gastropods, and cephalopods. Eclogae Geol Helv. 63, 775–797. Available at: https://ci.nii.ac.jp/naid/10015599322/.
Yan Z., Jing G., Gong N., Li C., Zhou Y., Xie L., et al. (2007). N40, a novel nonacidic matrix protein from pearl oyster nacre, facilitates nucleation of aragonite in vitro. Biomacromolecules 8 (11), 3597–3601. doi: 10.1021/bm0701494
Yano M., Nagai K., Morimoto K., Miyamoto H. (2006). Shematrin: a family of glycine-rich structural proteins in the shell of the pearl oyster pinctada fucata. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 144 (2), 254–262. doi: 10.1016/j.cbpb.2006.03.004
Yao C.-L., Somero G. N. (2014). The impact of ocean warming on marine organisms. Sci. Bull. 59 (5), 468–479. doi: 10.1007/s11434-014-0113-0
Yue X., Nie Q., Xiao G., Liu B. (2015). Transcriptome analysis of shell color-related genes in the clam meretrix meretrix. Mar. Biotechnol. 17 (3), 364–374. doi: 10.1007/s10126-015-9625-0
Zhang C., Xie L., Huang J., Chen L., Zhang R. (2006). A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem. Biophys. Res. Commun. 342 (2), 632–639. doi: 10.1016/j.bbrc.2006.01.182
Keywords: gene expression, mantle color, biomineralization, qRT-PCR, pearl oyster
Citation: Parvizi F, Akbarzadeh A, Farhadi A, Arnaud-Haond S and Ranjbar MS (2023) Expression pattern of genes involved in biomineralization in black and orange mantle tissues of pearl oyster, Pinctada persica. Front. Mar. Sci. 9:1038692. doi: 10.3389/fmars.2022.1038692
Received: 07 September 2022; Accepted: 23 December 2022;
Published: 19 January 2023.
Edited by:
Hyun Woo Kim, Pukyong National University, Republic of KoreaReviewed by:
Dahui Yu, Beibu Gulf University, ChinaHuayang Guo, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2023 Parvizi, Akbarzadeh, Farhadi, Arnaud-Haond and Ranjbar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Sharif Ranjbar, c2hhcmlmLnJhbmpiYXJAaG9ybW96Z2FuLmFjLmly; Arash Akbarzadeh, YWtiYXJ6YWRlaEBob3Jtb3pnYW4uYWMuaXI=
 Fatemeh Parvizi
Fatemeh Parvizi Arash Akbarzadeh1*
Arash Akbarzadeh1* Sophie Arnaud-Haond
Sophie Arnaud-Haond Mohammad Sharif Ranjbar
Mohammad Sharif Ranjbar