- College of Animal Science and Technology, Yangzhou University, Yangzhou, China
Vibrio mimicus is a global causative agent of vibriosis in a variety of aquatic animals and causes major economic losses to aquaculture. It could survive in water for extended periods of time under environmental stress, but its survival strategy remains unknow. This study described the survival, virulent and gene expression changes of V. mimicus cells undergoing starvation stress. After 4 weeks’ cultivation in media without nutrients, V. mimicus Y4 showed reduced rates of activity with marked changes in morphology and physiological activities. The culturable cell counts declined gradually to 104 CFU/mL and the shape changed from rod-shaped to coccoid with short rods or spherical. The motility of starved cells decreased after starvation and the biofilm production was significantly lower than wild cells. The starved cells still produced β-hemolysis, lecithinase and caseinase, but its infectivity to Macrobrachium nipponense was weakened. To investigate the mechanism behind morphological and physiological changes, we further analyzed differently expressed genes (DEGs) between starved and wild cells at the whole transcriptional level. The RNA-seq analysis demonstrated that large-scale DEGs were involved in transferase, membrane, dehydrogenase, synthase, flagellar, hemolysin, pilus assembly, and starvation, etc. Among them, the well-known virulence-related genes were downregulated significantly, including vmh, pilA, vipA, capB, tadC, huvX, ompA, etc. These data provide a key resource to understand the regulatory mechanisms of V. mimicus to starvation stress.
Introduction
Vibrio mimicus is a gram-negative bacterial pathogen distributing widely in both freshwater and estuarine environments. It is one of the most prevalent pathogens in aquaculture, which could induce high mortality among aquatic animals, impeding healthy and sustainable development of aquaculture (Vieira et al., 2001). For example, the outbreak of white spot disease (WSD) in Penaeus monodon was reported to be related with the infection of V. mimicus (Ramalingam et al., 2017), and it also could infect freshwater catfish and prawns and induce mortality with short disease duration (Geng et al., 2014; Yu et al., 2020; Jiang et al., 2021). Furthermore, consumption of freshwater fish and seafood contaminated with V. mimicus might cause symptoms of gastroenteritis, diarrhea and food poisoning in humans (Chitov et al., 2009; Kay et al., 2012; Tercero-Alburo et al., 2014; Muller et al., 2016). The severe impact of V. mimicus on aquaculture industry and human health attracts people’s attention, and an increasing number of studies focused on its detection and pathogenesis have been conducted (Neogi et al., 2018; Yu et al., 2020).
Bacteria can be subject to multiple environmental stresses during their life, including nutrient limitation, extreme temperature, osmotic concentrations, oxygen, copper stress and organic pollutants (Oliver, 2010; Fu, et al., 2014). Among them, nutrient deficiency is the most common stress which microorganisms routinely encounter in natural ecosystems (Gao et al., 2018). It has been reported that Vibrio spp. can differentiate into the viable but nonculturable (VBNC) state to maintain viability and survive several years in food deprived condition (Abdallah et al., 2009; Kim et al., 2018), which could contribute to the spreading of V. mimicus in the environment and hazarding public health. A common feature of bacteria subjected to starvation is the ‘rounding up’ phenomenon, which cells become rounder and adopt a coccus shape (Carroll et al., 2001). In addition, the growth, survival rate, and pathogenicity were also reported to be reduced compared to the non-starved cells (Eguchi et al., 2000; Amel et al., 2008; Chen et al., 2009). These phenotypic signs induced by starvation were found in Vibrio spp. (Alonso et al., 2002; Citterio et al., 2004), but the response of V. mimicus in survival and pathogenicity under starvation stress are not clear.
This study assessed the potential of V. mimicus to survive under starvation stress and further investigated the changes in morphology, motility, and pathogenicity induced by starvation. Besides, to reveal the molecular mechanism behind phenotypic changes, transcriptomic analyses between starved and wild cells were examined at the whole transcriptional level. These results will shed new light on understanding of the stress response mechanisms of V. mimicus under starvation.
Materials and methods
Bacterial strains and starvation stress
V. mimicus Y4 was isolated from Macrobrachium nipponense with red body disease in Jiangsu province, China (Jiang et al., 2021). It was preserved in 30% glycerol at -80°C in our lab, and the individual colony of the isolate Y4 were incubated in 100 mL of LB liquid medium at 28°C while shaking (180 rpm) for 18 h. V. mimicus cells were harvested by centrifugation at 6,000 g for 10 min and washed twice with sterile saline solution (0.9% w/v NaCl). The cells were resuspended in sterile Erlenmeyer glass flasks containing 100 mL of sterile saline with a final concentration of 1×109 CFU/mL and incubated at 28°C in sterile saline solution without nutrients for 4 weeks. Experiments were performed in triplicates for statistical analysis. The culturable number of bacterial cells was obtained using LB plates. The freshly prepared log-phase V. mimicus cell culture without starvation treatment was used as the control.
Growth curve
The wild and 4-week starved strain was incubated at 28°C to the initial exponential (OD570 = 0.2). The values of OD570 were recorded at 0-24 h. From the OD570 data, growth curves were plotted comparing the wild and 4-week starved strain. Three independent biological replicates were performed for each data point.
Ultrastructural analysis
Morphological changes between 4-week starved and wild cells were monitored under scanning electronic microscope (SEM) as previously described with minor modification (Arias et al., 2012). Briefly, 10 µL of bacteria was fixed in 2.5% glutaraldehyde (v/v) on 8 × 8 mm glass slides at 4°C overnight. Then, samples were dehydrated in a graded ethanol series (50, 70, 90, and 100%), coated with gold palladium alloy in an EMS 550X (Electron Microscopy Science), and examined under Zeiss EVO 50 electronic microscope (Zeiss, Germany).
Bacterial motility test
After 4-week starvation, the motility of V. mimicus strain Y4 was measured as described previously (Xu et al., 2014). Briefly, a puncture inoculated a single colony in the middle of the plates containing 0.4% agar. The diameter of the halo surrounding the punctured portion of the agar media was measured 24 h post-inoculation.
Microtiter plate biofilm production assay
Biofilm formation in microplates was quantified by crystal violet staining according to a previously described method (Dueholm et al., 2013), with minor modifications. Overnight cultures of wild-type and 4-week starved strain were diluted at a 1:1000 ratio in fresh sterile LB, added to 96-well microtiter plates at a volume of 200 μL per well, and incubated at 28°C with shaking for 12-72 h. The wells were aspirated and washed with 0.1 M phosphate buffer (PBS). The biofilms were stained with 250 μL of 1% crystal violet for 15 min, then washed twice with PBS, solubilized the crystal violet with ethanol for 15 min, and measured the absorbance (OD570) using a Synergy HT microplate reader.
Determination of extracellular enzymes and hemolysin
The presence of lecithinase, caseinase, and lipase of the 4-week starved cells were determined by directly culturing the starved cells on LB nutrient agar containing 10% egg yolk (lecithinase test), 10% skim milk (caseinase test), 1% Tween-80 (lipase test), and 7% rabbit erythrocytes (β-hemolytic activity test) as substrate. These plates were incubated at 28°C for 24-48 h, and the presence of the lytic halo around the colonies was observed.
Bacterial virulence assay
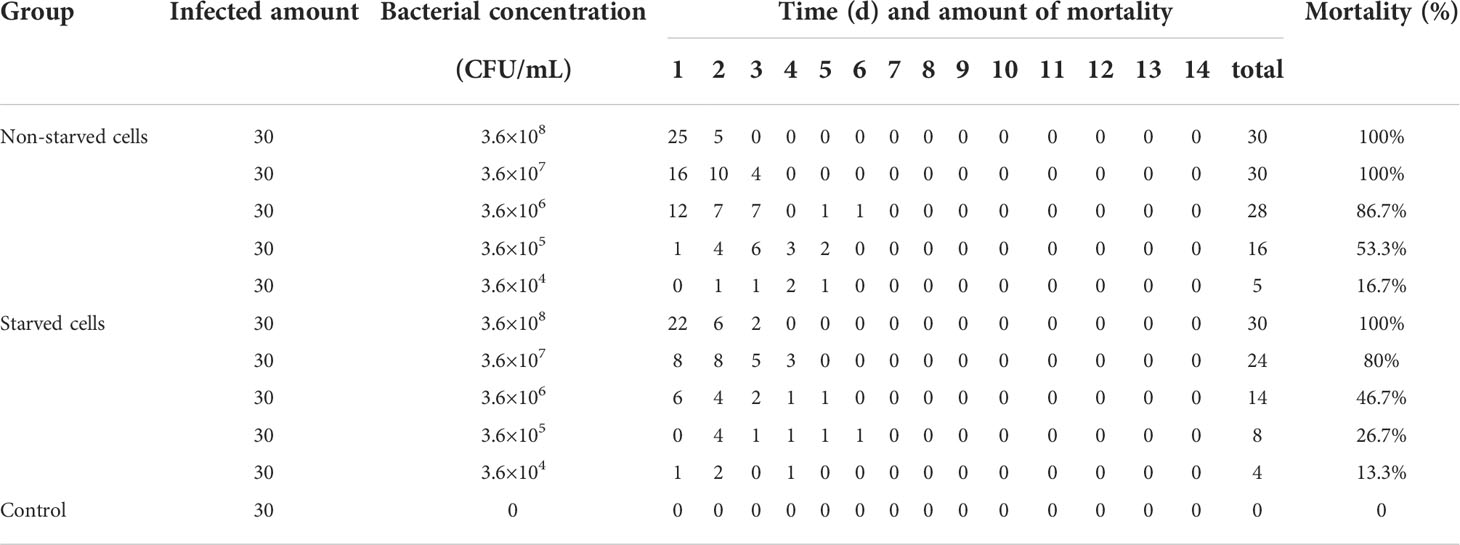
To examine the pathogenicity, healthy M. nipponense was infected by bath immersion with 4-week starved and wild V. mimicus. Experimental M. nipponense (average weight was 1.64 ± 0.6 g) were provided by an aquaculture farm located at Changzhou Country of Jiangsu Province, China. A total of 330 prawns were divided into 11 groups and continuously immersed with five different doses (3.6×108, 3.6×107, 3.6×106, 3.6×105 and 3.6×104 CFU/mL), respectively, and prawns in the rest tank were maintained in freshwater as control group. During the experiment, prawns were maintained in 30-L aquarium at 26 ± 0.5°C water temperature, and the water was aerated with 5.0 mg/L of dissolved oxygen. The mortality of prawns were checked during 7 d post infection. All experiments were repeated three times and the mean value was taken for the calculation of the median lethal dose (LD50) values according to the description of Saganuwan (Saganuwan, 2011) (Supplementary Table 1).
Transcriptome analysis
Samples of 4-week starved and wild cells (n=3) were sent to Beijing Novogene Co., Ltd, and subjected to total RNA isolation and cDNA preparations. The library was sequenced on the Illumina Hiseq 2500 platform. The clean reads of starved and wild cells were filtered from the raw data, and aligned to the reference genome and sequence using Bowtie 2. The DESeq R package was used to determine the differentially expressed genes (DEGs) between the starved and wild cells samples, and the q-value <0.05 and |log2(FoldChange)|>1 was adjusted as the threshold for significantly different expression. These DEGs was identified by GO and KEGG database annotations (Conesa et al., 2005; Minoru et al., 2013). The sequencing data has been successfully deposited in SRA database (PRJNA770024).
Validation of DEGs using qRT-PCR
Fourteen genes were selected to confirm the RNA-seq results by qRT-PCR. Primers were used for qRT-PCR and 16S rRNA gene was used to normalize the expression level (Table 1). The relative gene expressions were determined using 2-ΔΔCt method. Expression of the detected genes was analyzed by one-Way ANOVA using SPSS 16.0. All quantitative data were expressed as means ± SD.
Results
Survival under starvation stress
The survival rate of V. mimicus Y4 under starvation at 28°C was determined over a 4-week period (Figure 1A). Overall, V. mimicus Y4 was still culturable after 4-week starvation. The percentage of the strain Y4 culturable bacterial cells was reduced to 68.13% of their initial level during starvation period with Log10 CFU/mL changing from 10.4028 ± 0.1991 to 7.0878 ± 0.1102. In addition, the viability of starved cells is maintained and no significant change in growth capacity was observed at 4-week-starved cells compared with the wild strain, and the growth curve was shown in Figure 1B.
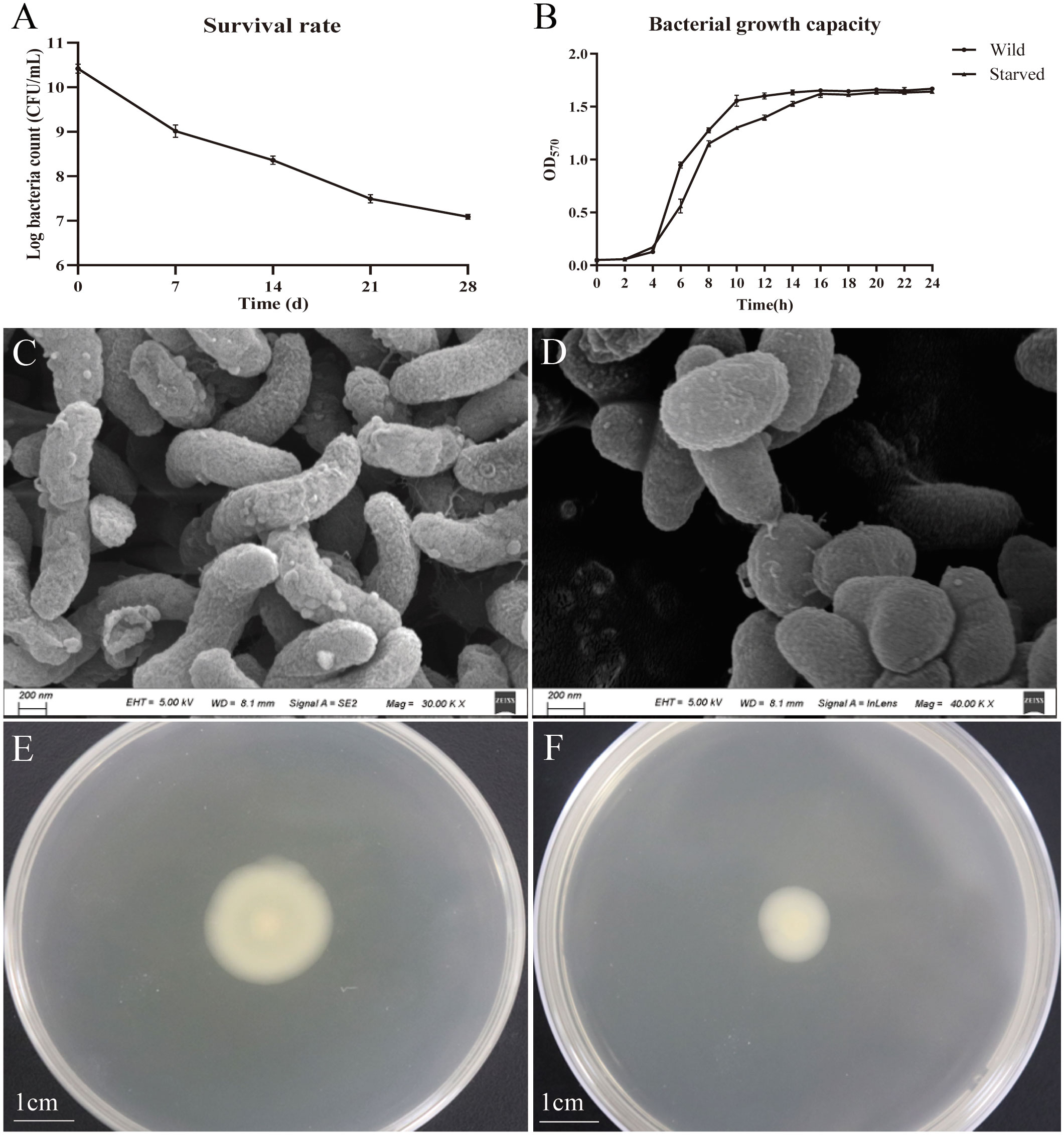
Figure 1 Survival, growth curves, morphological and motility changes of V. mimicus Y4 under starvation stress. Error bars: ± SD. (A) Survival rates; (B) Bacterial growth capacity. *P<0.05, **P<0.01. n=3; (C) morphological character of wild cells; (D) morphological character of 4-week starved cells; (E) motility of wild cells; (F) motility of 4-week starved cells.
Phenotype changes of V. mimicus after starvation stress
The wild V. mimicus cells were changed from rods shape to short rods, and the average cell length of the initial V. mimicus had significantly decreased from 1.6 ± 0.2 µm to 1.0 ± 0.2 µm by using scanning electron micrograph (Figures 1C, D). Furthermore, V. mimicus had significantly decreased in motility (from 2.2 ± 0.2 µm to 1.2 ± 0.2 µm) after starvation (Figures 1E, F). In addition, biofilm production of the starved cells was decreased with 0.24, 0.67, 0.40 and 0.54 fold than wild cells over a period of 12-72 h, respectively (Figure 2). The significant difference was found between biofilm formation at all time points.
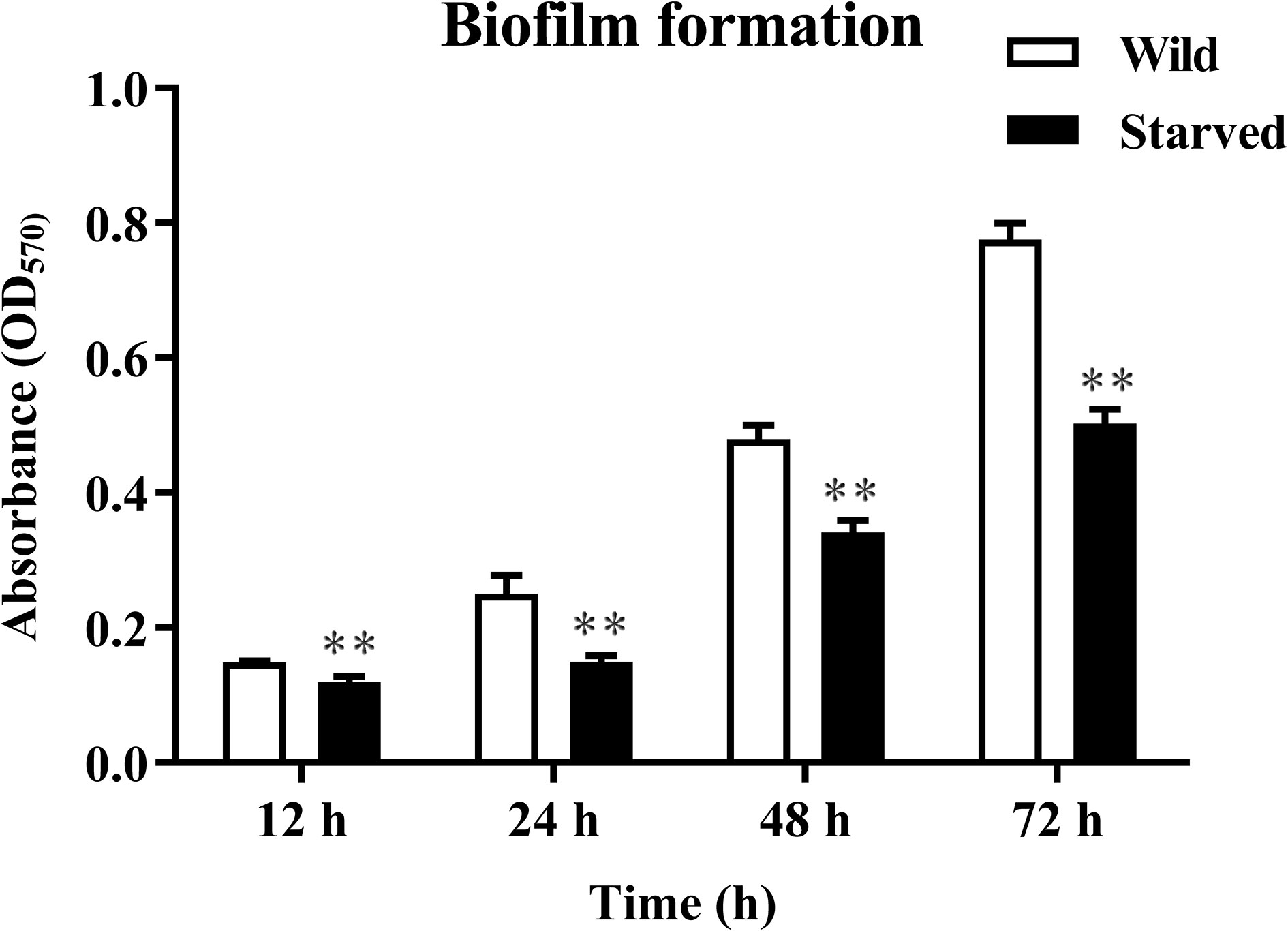
Figure 2 Biofilm production of wild and starved strains incubated at 28°C for 24 h and 48 h. **P<0.01. n=3.
Virulence factors and pathogenicity of the starved strain
Consistent with the wild strain, 4-week-starved V. mimicus still could produce β-hemolysis, lecithinase and caseinase, and did not produce lipase (Figure 3). However, the lytic halo around the colonies reduced in the starved strain. To further examine the pathogenicity changes, the wild and 4-week-starved V. mimicus were used to infect healthy M. nipponense.
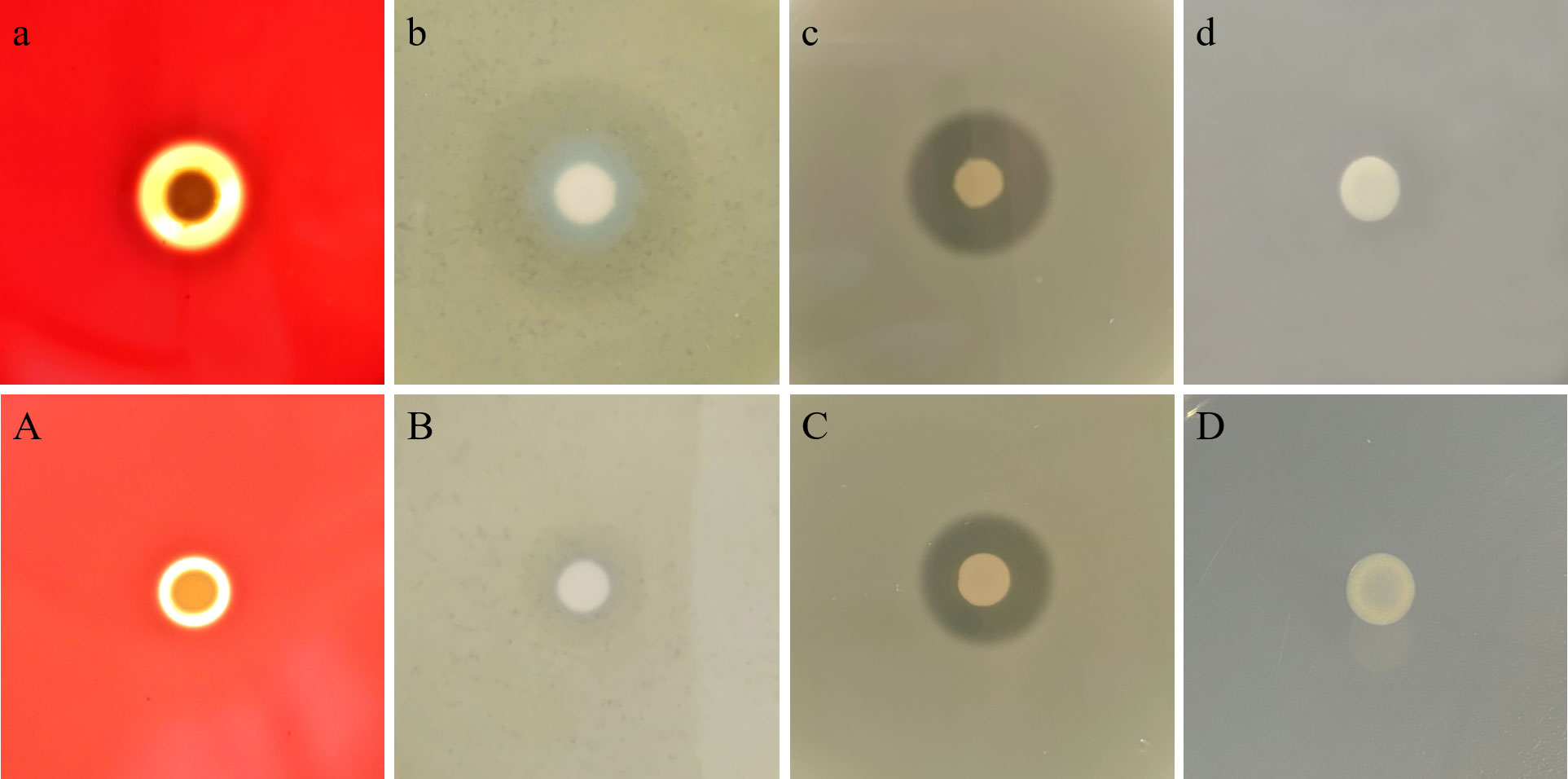
Figure 3 Virulence factors determination of V. mimicus Y4 after starvation stress. (A–D): hemolysin, lecithinase, caseinase, and lipase activities of 4-week starved cells; (a–d): hemolysin, lecithinase, caseinase, and lipase activities of wild cells.
Table 2 showed that the virulence of starved cells decreased with an LD50 value (2.45×106 CFU/mL), compared with wild cells (3.09×105 CFU/mL). These indicated that the LD50 of V. mimicus Y4 decreased by an order of magnitude after starvation stress, suggesting that starvation might affect the virulence of bacteria.
Transcriptional changes during starvation
DEGs analysis
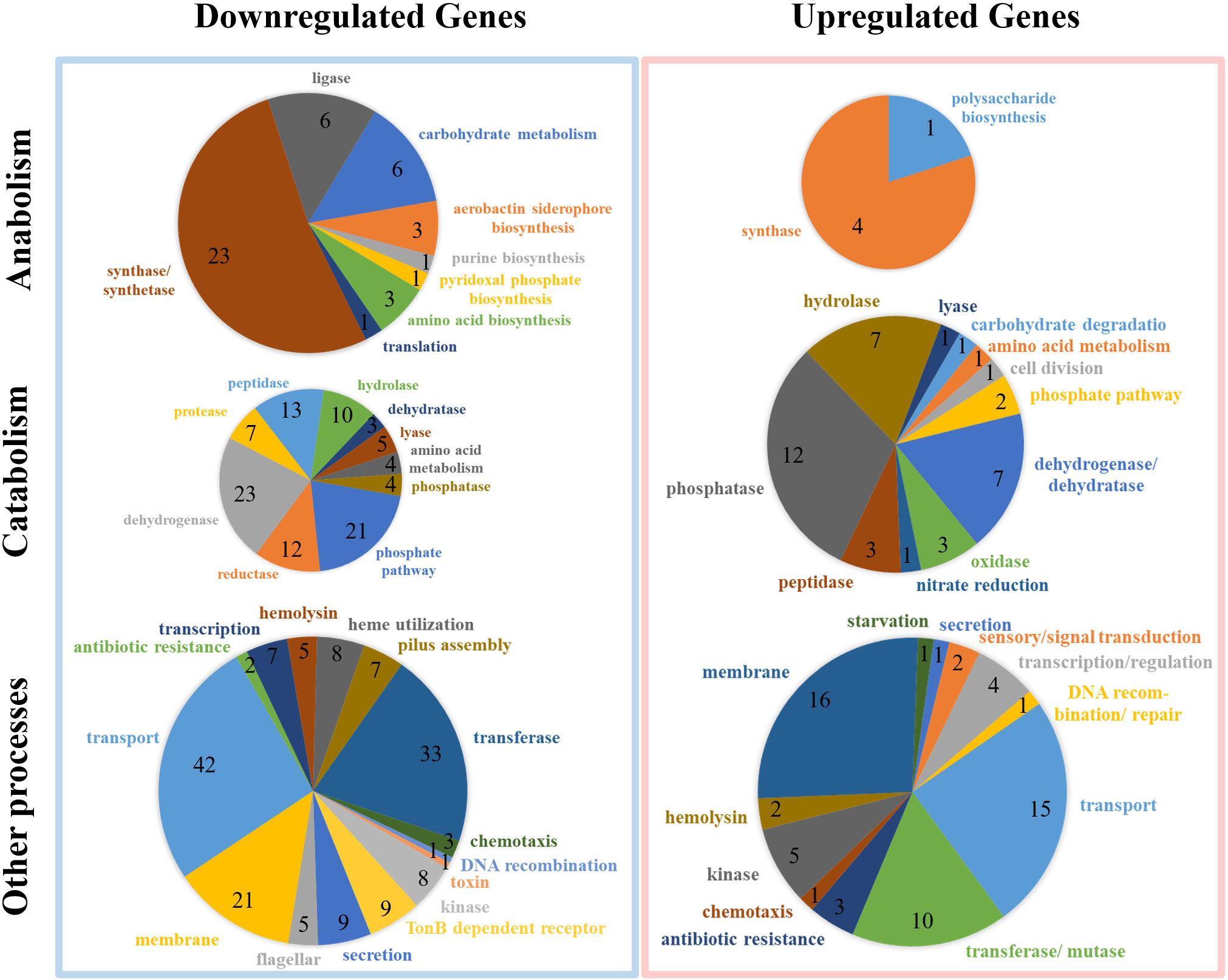
A total of 435 DEGs (119 up-regulated genes and 316 downregulated genes) were identified in the 4-week starved cells by comparison with the wild cells (Supplementary Figure 1). Figure 4 showed DEGs analysis in the wild and starved V. mimicus. Most of the anabolism related genes (44 out of 49, 89.8%) showed decreased expression level under starvation stress, and similar expression changes were found in catabolism related genes with 102 downregulated and 39 up-regulated (Supplementary Table 2). The expression changes of metabolically related gene implied that the V. mimicus stringent response increases the capacity of the cell to respond to a wider range of nutrient sources. Further, most of virulent-related genes involved in hemolysin, hume utilization, pilus assembly, toxin, secretion, etc. were also downregulated, which is consistent with the weakened virulence described above. Remarkably, a large number of flagellar assembly protein-related genes showed significantly downregulated expression after starvation, including flgD gene, flgF gene and fliF gene, etc., which might reduce the flagella synthesis, affect the motility of V. mimicus, and further decrease the bacterial adhesion and invasion ability to the host. Meanwhile, biofilm formation related genes, including rbmA, rbmB, ompU, toxT and luxS, etc., and capsular polysaccharide synthesis enzyme cpsA, cpsC were also downregulated after starvation, which influence to form the biofilm in V. mimicus. In addition, many genes with unknown function were regulated by starvation, which will be further studied.
GO and KEGG enrichment analysis of DEGs
The DEGs of V. mimicus after starvation stress were enriched and classified into three major categories: biological process, cellular component, and molecular function via GO enrichment analysis (Figure 5A; Supplementary Table 3). In the biological process category, small molecule metabolic process was the most highly represented. In the cellular component category, the most abundant category of the DEGs was membrane, which can improve tolerance in stress. Moreover, oxidoreductase activity was the most dominant category in molecular function category, indicative of starvation stress response.
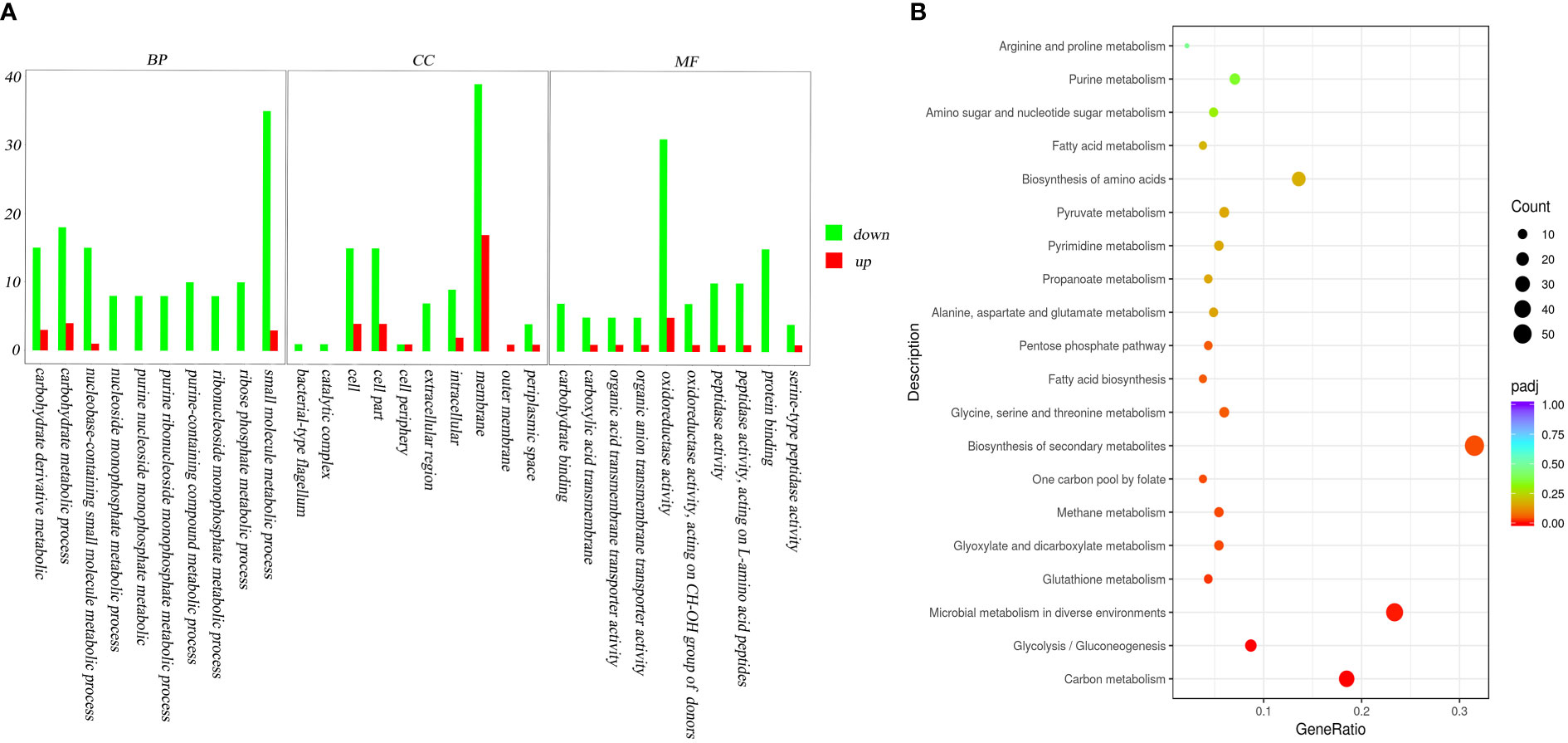
Figure 5 GO analysis and KEGG pathways of DEGs to the initial and the 4-week starved V. mimicus. (A): GO analysis; (B): KEGG pathways The x-axis indicates percentages of DEGs belonging to the corresponding pathway. The left y-axis represents the top 20 pathways. The sizes of the bubbles represent the number of DEGs in the corresponding pathway, and the colors of the bubbles represent the enrichment p-value of the corresponding pathway. KEGG terms with P-value <0.05 were considered significantly enriched by DEGs.
A total of 60 biochemical metabolism and signal pathways were enriched by KEGG enrichment analysis of DEGs. A majority of the DEGs in the enriched KEGG pathways were downregulated with a percentage of 89.6%. The top three enriched KEGG pathways were biosynthesis of secondary metabolites, microbial metabolism in diverse environments and carbon metabolism, which might improve V. mimicus survival by regulating the requirements of energy (Figure 5B).
Verification of the DEGs by qRT-PCR
To further verify the accuracy of sequencing, fourteen DEGs in the transcriptome data were purposefully chosen for relative qRT-PCR. Among them, the expression of flgD, ompA, tonB, luxS, capB, pilA, tadC, huvX, and vmh were down-regulated and the expression of matE, lysE, lysM and dnaJ were up-regulated. The qRT-PCR results showed similar expression trends with transcriptome profiling, confirming the reliability of transcriptome sequencing results (Figure 6).
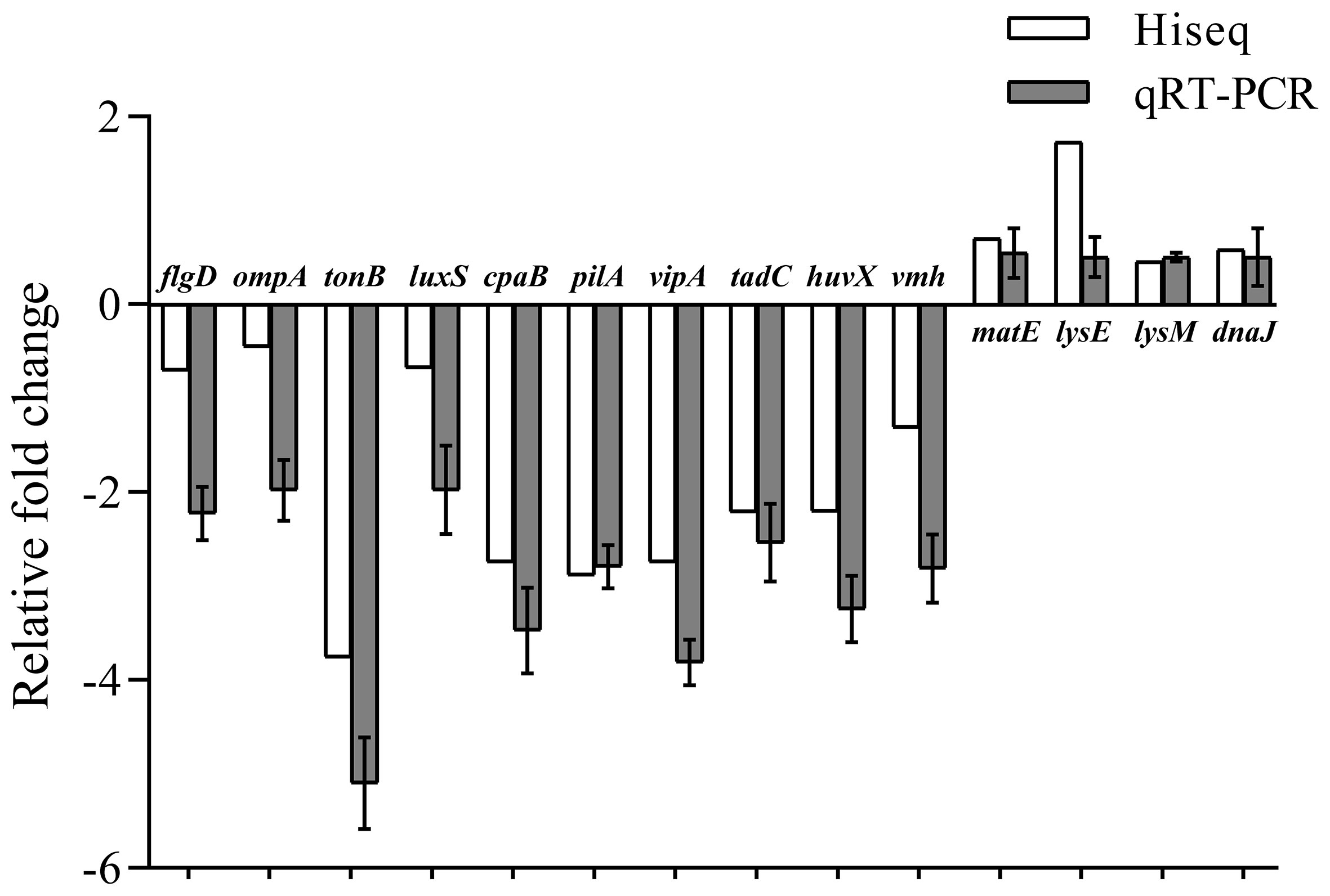
Figure 6 Comparative analysis of qRT-PCR and transcriptome sequencing results of DEGs of V. mimicus before and after starvation stress.
Discussion
Bacteria are usually challenged by various degrees of nutrient deprivation and starvation at some point in their life cycle, and they can sense and respond to this stress through changing their phenotypic and genetic repertoire (Rosen et al., 2006; Gao et al., 2018). In general, starvation-induced activities often differentiate into resistant forms to remain viable in starvation condition (Arias et al., 2012). This study described the ability of V. mimicus to survive under starvation stress for a period of 4 weeks, and further investigate the changes of morphological, viability, culturability, virulence and gene expression changes. Results in this study indicated that V. mimicus undergoes survival and virulent changes in the starvation stress, representing adaptative evolution to new environmental conditions.
The reduction in cell size has been observed in many bacteria after long-term nutrient limitation (Gray et al., 2019), including Vibrio spp. such as V. parahaemolyticus (Chen et al., 2009; Su et al., 2013) and V. fluvialis (Amel et al., 2008). In addition, Vibrio can obtain a better survival ability in their environment by forming biofilm, growing rapidly to a certain population density, or the expression of genes in bacteria responding to environmental stress (Huang et al., 2020); for instance, sigma factors, two-component system, quorum sensing, flagellar assembly, etc. regulate natural competence in V. cholereae (Gao, et al., 2022). In our study, the size reduction in V. mimicus was also found after starvation, which might contribute to reduce the requirements for cell maintenance and to protect bacteria from environmental stress. Meanwhile, morphological changes, swimming motility of V. mimicus also decreased significantly after starvation. The bacterial motility mode is driven by rotating flagella (Berg, 2003; Gao et al., 2018). The RNA-seq results showed that dozens of flagellar assembly related genes, including flgG, fliK, flrC, etc., were downregulated after starvation which well explained the decreased motility. Reportedly, flagellar assembly KEGG pathway was also downregulated in starved cells of V. cholerae (Gao, et al., 2022). Apart from motility, flagella can also influence biofilm formation by affecting bacterial adhesion (Choy et al., 2004). In this study, biofilm formation was reduced in V. mimicus Y4 after starvation which is in consistence with that in Campylobacter jejuni (Svensson et al., 2014). Changes of morphology, swimming motility and biofilm formation are important feature for V. mimicus to increase adaptability.
Like many other food-borne pathogens, V. mimicus can survive long periods in low nutrient conditions on account of the genes used to adapt to environmental stress (Klancnik et al., 2009). A global view of up-regulated and down-regulated genes indicated that a large proportion of DEGs were involved in metabolism, transport and membrane. Metabolism plays an important role in reactivating starved cells in VBNC state (Villarino et al., 2000). A major of DEGs involved in metabolism were downregulated, especially in the anabolism, which indicated V. mimicus Y4 minimized the requirements for cell maintenance to survive starvation. The transport adaptation is the clearest example of starvation response, and some transport-related proteins, including some ABC transporter components and fructose specific PTS enzyme, may contribute to the uptake of alternative sources (Voigt et al., 2007). Membrane systems can change their composition and the concentrations of unsaturated fatty acids, phospholipids, and peptidoglycans to improve tolerance (Margrete et al., 2014). The expression changes of genes involved in metabolism, transport and membrane indicated their role in protecting the cells from stresses, but the function and regulatory mechanism need to be further studied. In addition, previous research found that limitation of nutrients leads to the significant downregulation of genes controlling central carbon metabolism, biosynthesis of lipids, amino acids, and nucleotides (Montánchez et al., 2014). The abundant KEGG pathways of downregulated genes included carbon metabolism, biosynthesis of amino acids, fatty acid biosynthsis, which are similar with the previous research.
Pathogenic bacteria subvert host life through adhering and colonizing the host surface, penetrating and proliferating in the host tissue, producing toxins to destroy cell function, and eventually causing organ infection (Hao et al., 2013). The drastic changes in morphology, swimming motility and biofilm formation induced by starvation might affect the attachment and colonization ability of the pathogen Y4, and results in bacterial virulence assay verified this hypothesis. Although the starved cells could still produce β-hemolysis, lecithinase and caseinase, the lytic halo around the colonies reduced compared with the wild strain. Besides, virulence-related genes showed downregulated expression level and the infectivity of Y4 to M. nipponense also decreased after starvation. Therefore, some virulent genes were downregulated, such as flgD, ompA, luxS, vmh, and pilA, which also contribute to the decrease of pathogenicity. Weakened virulence under starvation stress was also reported in other bacteria. Similarly, V. anguillarum starved cells presented a decline of the ability to cause infection and the expression of virulence genes compared to the non-starved cells (Gao et al., 2018). The downregulated expression levels of virulence regulator gene were found in V. harveyi under starvation stress (Liu et al., 2020). These results indicate that starvation can weaken the virulence of bacteria.
This study demonstrated that V. mimicus cells were still culturable after 4-week starvation, but showed reduction in size, changed morphology from rods to short rods, and weakened pathogenicity. Genes involved in metabolism, virulence and adaptive evolution were differently expressed which might contribute to the survival and virulent changes under starvation. These data provide a key resource to determine the specific response of V. mimicus to starvation stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
All treatments involving animals were carried out under the strict guidelines of Animal Experiment Ethics Committee of Yangzhou University.
Author contributions
ZJ, XG, QJ, and XZ conceived the project and designed the experiments. ZJ, QQ, CW, and YZ performed the experiments. SG, JL, and PJ analyzed the data. ZJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Fund (31972830, 32202982), the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS [2022] 501, JATS [2022] 504), the National Key Research and Development Project (2019YFD0900305), Revitalizing of Seed Industry - the Open Competition Mechanism to Select the Best Candidates Projects (JBGS [2021] 120), Natural Science Foundation of Jiangsu Province (BK20220584).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1035268/full#supplementary-material
Supplementary Figure 1 | Different expressed genes in the wild and 4-weeks starved V. mimicus.
References
Abdallah F. B., Kallel H., Bakhrouf A. (2009). Enzymatic, outer membrane proteins and plasmid alterations of starved Vibrio parahaemolyticus and Vibrio alginolyticus cells in seawater. Arch. Microbiol. 191 (6), 493–500. doi: 10.1007/s00203-009-0477-8
Alonso J. L., Mascellaro S., Moreno Y., Ferrus M. A., Hernandez J. (2002). Double-staining method for differentiation of morphological changes and membrane integrity of Campylobacter coli cells. Appl. Environ. Microbiol. 68 (10), 5151–5154. doi: 10.1128/AEM.68.10.5151-5154.2002
Amel K. N., Amine B., Amina B. (2008). Survival of Vibrio fluvialis in seawater under starvation conditions. Microbiol. Res. 163 (3), 323–328. doi: 10.1016/j.micres.2006.06.006
Arias C. R., LaFrentz S., Cai W., Olivares-Fuster O. (2012). Adaptive response to starvation in the fish pathogenFlavobacterium columnare: cell viability and ultrastructural changes. Bmc. Microbiol. 12 (1), 266. Available at: http://www.biomedcentral.com/1471-2180/12/266
Berg H. C. (2003). The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72 (1), 19. doi: 10.1146/annurev.biochem.72.121801.161737
Carroll J. W., Mateescu M. C., Chava K., Colwell R. R., Bej A. K. (2001). Response and tolerance of toxigenic Vibro cholerae O1 to cold temperatures. Anto. Van. Leeuw. 79 (3-4), 377. doi: 10.1023/A:1012004725373
Chen S. Y., Jane W. N., Chen Y. S., Wong H. C. (2009). Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int. J. Food. Microbiol. 129 (2), 157–165. doi: 10.1016/j.ijfoodmicro.2008.11.009
Chitov T., Kirikaew P., Yungyune P., Ruengprapan N., Sontikun K. (2009). An incidence of large foodborne outbreak associated with Vibrio mimicus. Eur. J. Clin. Microbiol. 28 (4), 421–424. doi: 10.1007/s10096-008-0639-7
Choy W. K., Zhou L., Syn K. C., Zhang L. H., Swarup S. (2004). MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 186 (21), 7221. doi: 10.1128/JB.186.21.7221-7228.2004
Citterio B., Casaroli A. L., Pierfelici L., Battistelli M., Baffone W. (2004). Morphological changes and outer membrane protein patterns in Helicobacter pylori during conversion from bacillary to coccoid form. New Microbiologica 27 (4), 353–360. Available at: http://europepmc.org/article/MED/15646049
Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
Dueholm M. S., Sndergaard M. T., Nilsson M., Christiansen G., Nielsen P. H. (2013). Expression of fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. MicrobiologyOpen. 2 (3), 365–382. doi: 10.1002/mbo3.81
Eguchi M., Fujiwara E., Miyamoto N. (2000). Survival of Vibrio anguillarum in freshwater environments: Adaptation or debilitation. J. Infect. Chemother. 6 (2), 126–129. doi: 10.1007/PL00012152
Fu X., Liang W., Du P., Yan M., Kan B. (2014). Transcript changes in Vibrio cholerae in response to salt stress. Gut. Pathog. 6, 47. doi: 10.1186/s13099-014-0047-8
Gao X., Pi D., Chen N., Li X., Liu X., Yang H., et al. (2018). Survival, virulent characteristics, and transcriptomic analyses of the pathogenic Vibrio anguillarum under starvation stress. Front. Cell. Infect. Mi. 8, 389. doi: 10.3389/fcimb.2018.00389
Gao X., Zhang Z., Qian Q., Chen Q., Gu S., Li J., et al. (2022). Survival characteristics and transcriptomic analyses analyses reveal the adaptive response of the aquatic pathogen Non-O1/O139Vibrio cholerate to starvation stress. Microbiol Spectr. 10(3), 01939–21. doi: 10.1128/spectrum.01939-21
Geng Y., Liu D., Han S., Zhou Y., Wang K. Y., Huang X. L., et al. (2014). Outbreaks of vibriosis associated with vibrio mimicus in freshwater catfish in China. Aquaculture. 433, 82–84. doi: 10.1016/j.aquaculture.2014.05.053
Gray D. A., Dugar G., Gamba P., Strahl H., Jonker M. J., Hamoen L. W. (2019). Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. 10 (1), 890. doi: 10.1038/s41467-019-08719-8
Hao B., Mo Z. L., Xiao P., Pan H. J. (2013). Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl. Microbiol. Biot. 97 (6), 2575–2585. doi: 10.1007/s00253-012-4372-x
Huang Z., Yu K., Fang Y., Dai H., Cai H., Li Z., et al. (2020). Comparative genomics and transcriptomics analyses reveal a unique environmental adaptability of Vibrio fujianensis. Microorganisms. 8, 555. doi: 10.3390/microorganisms8040555
Jiang Z., Gao X., Jiang Q., Zhu X., Zhou Y., Zhang Z., et al. (2021). Genomic characterization and pathogenicity analysis of the Vibrio mimicus Y4 causing red body disease in Macrobrachium nipponense. Aquaculture. 548 (2), 737701. doi: 10.1016/j.aquaculture.2021.737701
Kay M. K., Cartwright E. J., Maceachern D., Mccullough J., Barzilay E., Mintz E., et al. (2012). Vibrio mimicus infection associated with crayfish consumption, Spokane, washingto. J. Food. Protect. 75 (4), 762–764. doi: 10.4315/0362-028X.JFP-11-410
Kim J. S., Chowdhury N., Yamasaki R., Wood T. K. (2018). Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 20 (6). doi: 10.1111/1462-2920.14075
Klancnik A., Guzej B., Jamnik P., Vukovi D., Abram M., Možina S. S. (2009). Stress response and pathogenic potential of Campylobacter jejuni cells exposed to starvation. Res. Microbiol. 160 (5), 345–352. doi: 10.1016/j.resmic.2009.05.002
Liu X., Gao X., Chen N., Zhang Y., Li X., Zhang Y., et al. (2020). Transcriptional responses to starvation of pathogenic Vibrio harveyi strain DY1. J. Oceanol. Limnol. 38 (2), 579–587. doi: 10.1007/s00343-019-8350-3
Margrete S., Leanti L., Thomas M., Snipen L. G., Nes I. F., Anders B. D., et al. (2014). Transcriptomic and functional analysis of NaCl-induced stress in Enterococcus faecalis. PloS One 9 (4), e94571. doi: 10.1371/journal.pone.0094571
Minoru K., Susumu G., Yoko S., Masayuki K., Miho F., Mao T. (2013). Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic. Acids Res. 42 (D1), 199–205. doi: 10.1093/nar/gkt1076
Montánchez I., Arana I., Parada C., Garaizabal I., Orruño M., Barcina I., et al. (2014). Reprogramming of Vibrio harveyi gene expression during adaptation in cold seawater. FEMS. Microbiol. Ecol. 87 (1), 193–203. doi: 10.1111/1574-6941.12216
Muller S., Chau H., Boudreaux K. (2016). Vibrio mimicus as the rare cause of acute diarrheal illness. J. Louisiana State Med. Soc. 168 (6), 192–193. Available at: http://europepmc.org/article/med/28045686
Neogi S. B., Chowdhury N., Awasthi S. P., Asakura M., Yamasaki S. (2018). Novel cholera toxin variant and ToxT regulon in environmental Vibrio mimicus isolates: Potential resources for the evolution of Vibrio cholerae hybrid strains. Appl. Environ. Microb. 85 (3), e01977–e01918. doi: 10.1128/AEM.01977-18
Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS. Microbiol. Rev. 34 (4), 415–425. doi: 10.1111/j.1574-6976.2009.00200.x
Ramalingam A. R., Akshaya P., Debasis D., Sujeet K. (2017). Investigations on white spot disease outbreak in Penaeus monodon (Fabricius 1798) in association with Vibrio mimicus infection in the sunderbans, West Bengal, India. Indian. J. Fish. 64 (1), 56–60. doi: 10.21077/ijf.2017.64.1.54833-09
Rosen R., Matthysse A. G., Becher D., Biran D., Yura T., Hecker M., et al. (2006). Stress-induced proteins of Agrobacterium tumefaciens. Fems. Microbiol. Ecol. 35 (3), 277–285. doi: 10.1111/j.1574-6941.2001.tb00813.x
Saganuwan S. A. (2011). A modified arithmetical method of reed and muench for determination of a relatively ideal median lethal dose (LD50). Afr J. Pharm. Pharmaco. 5 (12), 1543–1546. doi: 10.5897/AJPP11.393
Su C. P., Jane W. N., Wong H. C. (2013). Changes of ultrastructure and stress tolerance of Vibrio parahaemolyticus upon entering viable but nonculturable state. Int. J. Food. Microbiol. 160 (3), 360–366. doi: 10.1016/j.ijfoodmicro.2012.11.012
Svensson S. L., Mark P., Gaynor E. C., Roop R. M. (2014). Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PloS One 9 (8), e106063. doi: 10.1371/journal.pone.0106063
Tercero-Alburo J. J., González-Márquez H., Bonilla-González E., Quiñones-Ramírez E., Vázquez-Salinas C. (2014). Identification of capsule, biofilm, lateral flagellum, and type IV pili in Vibrio mimicus strains. Microb. Pathogenesis. 76, 77–83. doi: 10.1016/j.micpath.2014.09.012
Vieira V. V., Teixeira L. F. M., Vicente A. C. P., Momen H., Salles C. A. (2001). Differentiation of environmental and clinical isolates of Vibrio mimicus from Vibrio cholerae by multilocus enzyme electrophoresis. Appl. Environ. Microb. 67 (5), 2360–2364. doi: 10.1128/AEM.67.5.2360-2364.2001
Villarino A., Bouvet O. M., Regnault B., Martin-Delautre S., Pad G. (2000). Exploring the frontier between life and death in Escherichia coli: evaluation of different viability markers in live and heat- or UV-killed cells. Res. Microbiol. 151 (9), 755–768. doi: 10.1016/S0923-2508(00)01141-4
Voigt B., Hoi L. T., Jürgen B., Albrecht D., Ehrenreich A., Vieth B., et al. (2007). The glucose and nitrogen starvation response of Bacillus licheniformis. PROTEOMICS. 7 (3), 413–423. doi: 10.1002/pmic.200600556
Xu X., Wu Q., Zhang J., Cheng J., Zhang S., Wu K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food. Control. 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Keywords: Vibrio mimicus, starvation stress, survival, virulent changes, transcriptome analysis
Citation: Jiang Z, Gu S, Qian Q, Li J, Ji P, Wu C, Zhang Y, Gao X, Jiang Q and Zhang X (2022) Survival virulent characteristics and transcriptomic analyses of Vibrio mimicus exposed to starvation. Front. Mar. Sci. 9:1035268. doi: 10.3389/fmars.2022.1035268
Received: 02 September 2022; Accepted: 29 September 2022;
Published: 17 October 2022.
Edited by:
Songzhe Fu, Dalian Ocean University, ChinaReviewed by:
Duochun Wang, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaRuijun Li, Dalian Ocean University, China
Pimonsri Mittraparp-arthorn, Prince of Songkla University, Thailand
Copyright © 2022 Jiang, Gu, Qian, Li, Ji, Wu, Zhang, Gao, Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Zhang, enhqOTMwN0AxNjMuY29t
 Ziyan Jiang
Ziyan Jiang Xiaojian Gao
Xiaojian Gao Qun Jiang
Qun Jiang Xiaojun Zhang
Xiaojun Zhang