
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 06 October 2022
Sec. Marine Biotechnology and Bioproducts
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1034945
Two new sulfur-containing benzofurans, talarobenzofurans A and B (1 and 2), one new benzofuran talarobenzofuran C (3) and two new α-pyrones talaropyrones A and B (5 and 6), along with five known compounds eurothiocin A (4), nodulisporipyrone A (7), peniazaphilin B (8), ramulosin (9) and 6-hydroxyramulosin (10) were isolated and identified from the culture extract of the mangrove-derived fungus Talaromyces sp. WHUF0341 guided by OSMAC (one strain-many compounds) strategy. Their structures were established by extensive spectroscopic data analysis, the modified Mosher’s method and electronic circular dichroism (ECD) calculations. Structurally, compounds 1 and 2 possesed the unique thioester moiety derived from benzofuran and 2-hydroxy-3-mercaptopropionic acid, which was rarely-observed in natural products. The α-glucosidase inhibitory and antibacterial activities of the isolated compounds were evaluated. Compounds 3 and 4 exhibited inhibitory effects against α-glucosidase activity with IC50 values of 48.9 ± 2.16 and 8.8 ± 1.08 µM, respectively. The inhibitory type of 3 on α-glucosidase was determined as non-competitive-type inhibition with Ki value of 242.3 μM by using Lineweaver-Burk double reciprocal and Dixon single reciprocal plots. Meanwhile, the binding model of 3 and 4 with α-glucosidase was determined by molecular docking assay. The biosynthetic pathways of compounds 1−4 were also proposed. This study suggested that benzofurans 3 and 4 could be potential lead compounds for the hypoglycemic drugs.
Fungi are considered to be rich sources of biologically active compounds for drug discovery (Newman and Cragg, 2020; Carroll et al., 2021). However, due to the rediscovery of previously described natural products of fungi from traditionally investigated habitats, the attention of researchers has been increasingly attracted to fungi from unique ecological niches such as mangrove-derived fungi (El-Bondkly et al., 2021). Mangroves are a diverse group of higher plants vegetating in the intertidal zones in subtropical and tropical climates (Ellison and Farnsworth, 1992; Nagelkerken et al., 2008). Mangrove-derived fungi, living in these unique environments, are considered as a new resource for diverse secondary metabolites with a wide range of bioactivities, including cytotoxic (Meng et al., 2016; Liu et al., 2021a; Tao et al., 2021), antimicrobial (Hemberger et al., 2013; Valente et al., 2020), enzyme inhibitory (Liu et al., 2014), anti-inflammatory (Liu et al., 2018), antioxidative (Guo et al., 2015) and antiviral activities (Yu et al., 2016).
As a part of our ongoing search for novel bioactive metabolites from mangrove-derived fungi (Liu et al., 2021b; Hou et al., 2021), the strain Talaromyces sp. WHUF0341, isolated from a sample of mangrove soil in Yalong Bay, Hainan Province, People’s Republic of China, was screened out for a detailed chemical investigation. To expand the chemical diversity of secondary metabolites, this strain was cultured in different media (Supplementary Table S1) using the OSMAC (one strain-many compounds) strategy (Pan et al., 2019). HPLC-UV analysis (Supplementary Figure S1) showed that the secondary metabolites in solid rice medium with 3% sea salt are more abundant than those in other culture media. Large scale fermentation on this medium was then performed for 30 days. Chemical investigation of the EtOAc extract of this fungus led to the isolation of five new compounds, talarobenzofurans A−C (1−3) and talaropyrones A and B (5 and 6), along with five known compounds eurothiocin A (4) (Liu et al., 2014), nodulisporipyrone A (7) (Zhao et al., 2015), peniazaphilin B (8) (Zhang et al., 2018), ramulosin (9) (Wang et al., 2014) and 6-hydroxyramulosin (10) (McMullin et al., 2017) (Figure 1). All of the isolated compounds were evaluated for their α-glucosidase inhibitory and antibacterial activities. Herein, we report the isolation, structure elucidation, bioactivities and putative biosynthetic pathways of these compounds.
The ECD spectra were measured on a Chirascan V100 instrument (Applied Photophysics Ltd, Leatherhead, Surrey, UK). Optical rotations were measured with an Anton Paar MCP 200 Automatic Polarimeter (Anton Paar, Graz, Austria). UV data were recorded using a Thermo Genesys-10S UV/Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). IR data were obtained on a Nicolet IS5 FT-IR spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). 1H and 13C NMR data were acquired with Bruker Avance-500 spectrometer (Bruker, Bremen, Germany) using solvent signals (Acetone-d6: δC/H 29.8, 206.1/2.05; CDCl3: δC/H 77.2/7.26; DMSO-d6: δC/H 39.5/2.50) as references. Mass data were performed on an Agilent Accurate-Mass-Q-TOF LC/MS 6520 instrument (Agilent Technologies, Santa Clara, CA, USA). HPLC separations were performed on an Agilent 1260 instrument equipped with a variable-wavelength UV detector with a flow rate of 2.0 mL/min. α-Glucosidase (from Saccharomyces cerevisiae, 33 U/mg), p-nitrophenyl-α-D-glucopyranoside (p-NPG) and acarbose purchased from Shanghai yuanye Bio-Technology Co., Ltd (Shanghai, China).
The sample of mangrove soil was collected from the Yalong Bay, Hainan Province, People’s Republic of China. The fungal strain WHUF0341 was isolated on MEA medium (1.5% sea salt, 1.7% malt extract, 0.3% peptone, 0.2% agar) from this sample and preserved in sterilized 30% glycerol at -80°C. Phylogenetic analysis based on ITS sequence and morphological observation indicated that the strain WHUF0341 should be identified as Talaromyces sp. (GenBank Accession No. ON564542.1). The strain was deposited in the Institute of Microbiology, Chinese Academy of Sciences, Beijing.
Four liquid media and two solid media (each medium contained 3% sea salt as control) were tested to identify the optimum culture conditions (Supplementary Table S1). The fungus Talaromyces sp. WHUF0341 was grown on potato dextrose agar (PDA) at 28°C for 10 days. Several pieces of agar plugs (about 0.5 × 0.5 × 0.5 cm3) were then inoculated into 250 mL Erlenmeyer flasks containing 50 mL of media (0.4% glucose, 1% malt extract, and 0.4% yeast extract) at room temperature on an orbital shaker at 200 rpm for 3 days to produce the seed culture. Finally, 5.0 mL of the spore inoculums obtained from liquid phase cultivation was transferred into 50 × 500 mL Erlenmeyer flasks each containing 80 g rice medium and 120 mL of distilled H2O with 3% sea salt and incubated at 25°C under static condition for 30 days.
The fermented rice material was extracted repeatedly with EtOAc for three times, and the combined filtrate was evaporated to dryness under reduced pressure to afford the crude extract (65.0 g). The crude extract was then fractionated by silica gel vacuum liquid chromatography (VLC) eluted with a gradient of petroleum ether (PE)/EtOAc (from 20:1 to 1:2, v/v) to get seven fractions (Fr. 1−7). The fraction 2 (0.6 g, eluted with PE/EtOAc 4:1) was fractionated by Sephadex LH-20 column chromatography (CC) eluting with CH2Cl2/MeOH 1:1 to afford compounds 4 (7.0 mg) and 9 (6.3 mg). The fraction 3 (5.4 g, eluted with PE/EtOAc 1:1) was subjected to octadecylsilyl column chromatography (ODS CC) with MeOH-H2O gradient elution to yield nine subfractions (Fr. 3-1−3-9). The subfraction 3-2 (49.0 mg, eluted with 60% MeOH-H2O) was purified by semi-preparative RP-HPLC (Agilent Zorbax SB-C18 column; 5 μm; 10 × 250 mm; 34% CH3CN-H2O for 35 min; 2.0 mL/min) to afford compound 1 (5.0 mg, tR 23.2 min). The subfraction 3-5 (44.0 mg, eluted with 65% MeOH-H2O) was further purified by semi-preparative RP-HPLC (40% CH3CN-H2O for 50 min; 2.0 mL/min) to obtain compound 7 (7.3 mg, tR 43.5 min). The fraction 4 (1.4 g, eluted with PE/EtOAc 1:2) was subjected to ODS CC using a MeOH-H2O gradient elution and the subfraction 4-2 (93.0 mg, eluted with 30% MeOH-H2O) was further purified by semi-preparative RP-HPLC (30% CH3CN-H2O for 15 min; 2.0 mL/min) to get compound 8 (6.1 mg, tR 11.6 min). The subfraction 4-4 (52.0 mg, eluted with 40% MeOH-H2O) was also purified by semi-preparative RP-HPLC (50% MeOH-H2O for 25 min; 2.0 mL/min) to afford compound 6 (2.3 mg, tR 13.0 min). The fraction 5 (0.6 g, eluted with PE/EtOAc 3:7) was refractionated by Sephadex LH-20 CC eluting with MeOH and the subfraction 5-1 (102.0 mg) was purified by semi-preparative RP-HPLC (55% MeOH-H2O for 50 min; 2.0 mL/min) to obtain compound 2 (4.8 mg, tR 41.2 min). The fraction 6 (2.9 g, eluted with PE/EtOAc 1:3) was chromatographed on an ODS column using a gradient eluent of MeOH-H2O, the fraction 6-4 (63.0 mg, eluted with 30% MeOH-H2O) was purified by semi-preparative RP-HPLC (28% CH3CN-H2O for 40 min; 2.0 mL/min) to yield compounds 3 (2.2 mg, tR 29.2 min) and 5 (1.4 mg, tR 13.6 min). The fraction 7 (0.2 g, eluted with PE/EtOAc 1:9) was separated by Sephadex LH-20 CC eluting with MeOH to yield three fractions (Fr. 7-1−7-3), the fraction 7-2 (15.3 mg) was purified by semi-preparative RP-HPLC (25% CH3CN-H2O for 22 min; 2.0 mL/min) to afford compound 10 (2.0 mg, tR 15.0 min).
Talarobenzofuran A (1): yellow oil; [α]25D −58.9 (c 0.1, MeOH); UV (MeOH) λmax (log ϵ) 229 (2.12), 275 (2.01) nm; ECD (c 0.54 × 10-3 M, MeOH) λmax (Δϵ) 219 (−1.03), 242 (−0.22), 272 (−1.73), 329 (+0.10) nm; IR (neat) νmax 3420, 2973, 1743, 1635, 1277, 1233, 1148 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 371.1168 [M + H]+ (calcd for C17H23O7S, 371.1159).
Talarobenzofuran B (2): yellow oil; [α]25D −60.9 (c 0.1, MeOH); UV (MeOH) λmax (log ϵ) 229 (2.13), 271 (1.93) nm; ECD (c 0.77 × 10-3 M, MeOH) λmax (Δϵ) 215 (−1.33), 239 (+0.03), 276 (−1.24), 344 (−0.01) nm; IR (neat) νmax 3367, 2975, 1721, 1636, 1233, 1023 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 357.1010 [M + H]+ (calcd for C16H21O7S, 357.1003).
Talarobenzofuran C (3): yellow oil; [α]25D −49.9 (c 0.1, MeOH); UV (MeOH) λmax (log ϵ) 229 (2.15), 267 (1.99) nm; ECD (c 0.79 × 10-3 M, MeOH) λmax (Δϵ) 211 (−2.13), 230 (−0.20), 266 (−2.06), 332 (+0.01) nm; IR (neat) νmax 3420, 2974, 1616, 1358, 1270, 1161, 1089 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 253.1077 [M + H]+ (calcd for C13H17O5, 253.1071).
Talaropyrone A (5): white solid; [α]25D +65.9 (c 0.1, MeOH); UV (MeOH) λmax (log ϵ) 207 (2.20), 280 (2.05) nm; ECD (c 0.78 × 10-3 M, MeOH) λmax (Δϵ) 205 (−7.66), 282 (+4.60), 320 (+0.20) nm; IR (neat) νmax 3391, 2931, 1694, 1567, 1456, 1246 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z 279.1206 [M + Na]+ (calcd for C13H20O5Na, 279.1203).
Talaropyrone B (6): light yellow solid; [α]25D +67.0 (c 0.1, MeOH); UV (MeOH) λmax (log ϵ) 210 (2.02), 280 (2.05) nm; ECD (c 0.74 × 10-3 M, MeOH) λmax (Δϵ) 205 (−1.62), 282 (+0.91), 317 (+0.04) nm; IR (neat) νmax 3350, 2940, 1694, 1627, 1564, 1257 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z 293.1001 [M + Na]+ (calcd for C13H18O6Na, 293.0996).
Compound 1 (0.5 mg for each) was dissolved in pyridine-d5 (100 µL) and transferred into clean NMR tubes, then S-MTPA chloride (10 µL) and R-MTPA chloride (10 µL) were added to each of the NMR tubes, and reaction mixture was stirred at room temperature overnight, which afforded the (R)-MTPA ester and (S)-MTPA ester derivatives of compound 1. 1H NMR data were obtained directly from the reaction NMR tubes. The ΔδH = δH(S)-MTPA-ester – δH(R)-MTPA-ester values around the stereogenic centers of the MTPA esters were determined by 1H NMR spectra.
(R)-MTPA ester of 1 (1a): 1H NMR (pyridine-d5, 500 MHz) δH 6.574 (1H, d, J = 14.8 Hz, H-7), 4.867 (1H, m, H-2), 3.841 (3H, m, H-16), 3.533 (2H, m, H-13), 3.308 (2H, s, H-3), 2.245 (3H, m, H-11), 1.504 (3H, s, H-10), 1.386 (3H, s, H-9).
(S)-MTPA ester of 1 (1b): 1H NMR (pyridine-d5, 500 MHz) δH 6.587 (1H, d, J = 16.9 Hz, H-7), 4.920 (1H, m, H-2), 3.838 (3H, br s, H-16), 3.538 (2H, d, J = 9.7 Hz, H-13), 3.306 (2H, br s, H-3), 2.256 (3H, m, H-11), 1.480 (3H, d, J = 3.9 Hz, H-10), 1.379 (3H, d, J = 5.9 Hz, H-9).
Conformational analysis within an energy window of 3.0 kcal/mol was performed by using the OPLS3 molecular mechanics force field via the MacroModel (Ji and Xu, 2021) panel of Maestro 10.2. The conformers were then further optimized with the software package Gaussian 09 (Frisch et al., 2009) at the B3LYP/6-311G(2d,p) level, and the harmonic vibrational frequencies were also calculated to confirm their stability. Then the 60 lowest electronic transitions for the obtained conformers in vacuum were calculated using time-dependent density functional theory (TDDFT) methods at the B3LYP/6-311G(2d,p) level. ECD spectra of the conformers were simulated using a Gaussian function. The overall theoretical ECD spectra were obtained according to the Boltzmann weighting of each conformers.
The method used for the α-glucosidase inhibitory activity assay was based on previously reported literature (Fan et al., 2020). The reaction mixture consisted of 50 µL of PBS (100 mM, pH 7.0), 20 µL of p-NPG (25 mM) and 20 µL of the test compounds dissolved in DMSO at different concentrations. After incubation at 37°C for 10 min, 10 µL of the enzyme (2 U/mL) was added and the mixture was further incubated at 37°C for 30 min. Finally, the absorbance was measured at 405 nm on an automatic microplate reader. Acarbose was used as the positive control (IC50 = 12.2 ± 0.66 µM). All experiments were carried out in triplicate.
The Lineweaver–Burk plot was used to determine the inhibitory mode of 3 on α-glucosidase. The enzymatic reactions were performed and monitored with different concentrations of 3 (0, 100, 200, and 400 μM) and substrate (3.125, 6.25 and 12.5 mM), with the concentration of the enzyme being constant (0.5 U/mL). And the Dixon plot was used to calculated the inhibition constant (Ki) (He et al., 2021).
The molecular docking simulations of 3 and 4 with α-glucosidase was performed according to the previously reported method (Chen et al., 2022). First, the chemical and MM2 energy minimized 3D structures of compounds 3 and 4 were obtained by using Chemdraw (20.0) and Chem3D (version 20.0), respectively. Meanwhile, the crystal structure of α-glucosidase (PDB ID: 3TOP) was obtained from the RCSB Protein Data Bank (www.rcsb.org). The AutoDock Vina was used to extract co-crystal ligands, determine the docking pocket and score the molecular docking of ligands and receptor protein. Finally, the visible results of possible binding sites were provided using PyMOL version 2.3.4.
The results were expressed as means ± standard deviations (n = 3). All of the data were analyzed by one-way analysis of variance (ANOVA) and following Tukey’s test. GraphPad Prism 9.0 software was used for statistical analysis. A p value < 0.05 was considered statistically significant.
Talarobenzofuran A (1) was isolated as yellow oil with the molecular formula of C17H22O7S deduced from HRESIMS at m/z 371.1168 [M + H]+ (calcd for C17H23O7S 371.1159), corresponding to 7 degrees of unsaturation. The IR spectrum showed hydroxy and carbonyl absorption bands at 3420 cm–1 and 1743 cm–1, respectively. Analysis of the 1H, 13C and HSQC NMR data of 1 (Table 1 and Supplementary Figures S2–S4) displayed the presence of three methyls (δC/H 22.3/2.41, 26.2/1.20, 25.6/1.24), two methylenes (δC/H 34.6/3.37 and 3.52, 28.3/3.14 and 3.08), three methines (δC/H 91.8/4.69, 105.3/6.22, 70.6/4.43), one methoxy group (δC/H 52.6/3.73), three exchangeable protons (δH 9.79, 4.76, 3.70), six aromatic carbons (δC 112.5, 119.5, 139.7, 155.1, 164.9, 105.3), two carbonyl carbons (δC 173.6, 195.7) and one sp3 hybrid carbon (δC 71.6), implying that two rings existed in 1 to match the 7 degrees of unsaturation. The 1H and 13C NMR data of 1 were similar with those of the co-isolated compound eurothiocin A (4) (Liu et al., 2014), indicating the presence of the benzofuran subunit. The HMBC correlations (Figure 2 and Supplementary Figure S6) from H-2 to C-7a (δC 164.9), from H2-3 to C-3a, C-4 and C-7a, and from H-7 (δH 6.22) to C-3a, C-5 and C-7a, combined with the 1H-1H COSY correlations (Figure 2 and Supplementary Figure S5) of H-2/H2-3 confirmed the presence of the benzofuran subunit. In addition, the HMBC correlations from the phenolic hydroxyl OH-4 (δH 9.79) to C-3a, C-4 and C-5, and from the aromatic methyl H3-11 to C-5, C-6 and C-7 located the hydroxy and methyl group at C-4 and C-6, respectively. Furthermore, the HMBC correlations from H-2 to C-8, C-9, and C-10, from OH-8 to C-2, C-8, C-9 and C-10, and from the geminal methyl groups H3-9 and H3-10 to C-2 and C-8 indicated that the 2-hydroxypropan-2-yl group was located at C-2. Meanwhile, the 1H-1H COSY correlations of OH-14/H-14/H2-13, together with the HMBC correlations from the methylene H2-13 (δC/H 34.6/3.52 and 3.37) to the carbonyl carbons C-12 (δC 195.7) and C-15 (δC 173.6), from the oxymethine H-14 to C-15, from OH-14 to C-13, C-14 and C-15, and from H3-16 to C-15 established the methyl lactate (C-13–C-16) moiety with the connection of C-13 and C-12 via the same sulfur atom to form thioester moiety. On the basis of the above results, the planar structure of 1 was elucidated as showed (Figure 1).
The relative configurations of C-2 and C-14 in 1 could not be assigned due to the lack of relevant ROESY correlations (Supplementary Figure S7). As the presence of a secondary alcohol at C-14, the modified Mosher’s method (Ohtani et al., 1995) was performed to determine the absolute configuration of C-14. Treatment of 1 with (S)- or (R)-α-methoxy-α-trifluoromethylphenylacetyl chloride [(S)- or (R)-MTPA-Cl] in the presence of pyridine afforded the R-MTPA ester (1a) and S-MTPA ester (1b), respectively. The chemical shift differences (Δδ = δ(S)-MTPA-ester – δ(R)-MTPA-ester) for the diastereomeric esters 1b and 1a were calculated in order to assign the absolute configuration at C-14. According to Mosher’s rule, the absolute configuration at C-14 of 1 was clearly established as S (Figure 3). Then the absolute configuration of C-2 in 1 was deduced by a comparison of the experimental and calculated electronic circular dichroism (ECD) spectra generated by the time-dependent density functional theory (TDDFT) (Nugroho and Morita, 2014) for two stereoisomers (2R,14S)-1 and (2S,14S)-1 was performed. The experimental ECD curve of 1 matched well with the calculated ECD curve of (2R,14S)-1 (Figure 4A), which allowed the elucidation of the absolute configuration of 1 as 2R,14S.
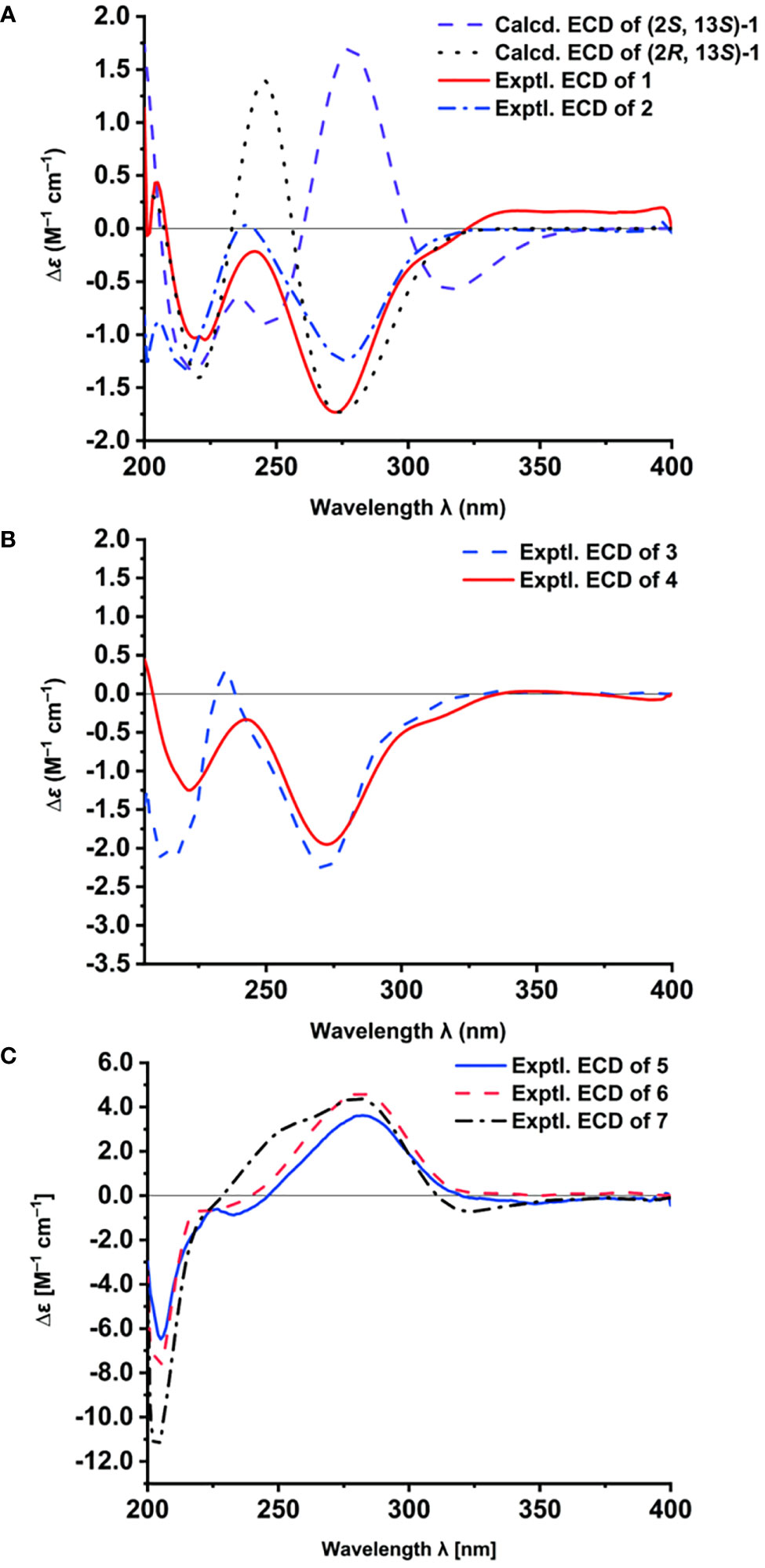
Figure 4 The ECD spectra of 1–7. Calculated and experimental ECD spectra of 1 and experimental ECD spectrum of 2 (A); Experimental ECD spectra of 3 and 4 (B); Experimental ECD spectra of 5–7 (C).
Talarobenzofuran B (2) was obtained as a yellow oil. Its molecular formula was determined as C16H20O7S (7 degrees of unsaturation) by HRESIMS (m/z 357.1010 [M + H]+, calcd for C16H21O7S 357.1003), which is 14 mass units less than that of 1. The NMR spectroscopic data (Table 1) of 2 were similar to those of 1, except that the methoxy group (δC/H 52.6/3.73) in 2 was replaced by a hydroxyl, which was supported by MS data and the HMBC correlations (Figure 2 and Supplementary Figure S12) from H2-13 and H-14 to the carboxylic carbon C-15. The absolute configuration of 2 was deduced to be the same as that of 1 on the basis of biosynthetic consideration, which was further confirmed by the similar Cotton effects in the experimental ECD spectra of 1 and 2 (Figure 4A). Thus, the absolute configuration of compound 2 was designated as 2R,14S.
Talarobenzofuran C (3) was also obtained as a yellow oil. Its molecular formula was determined to be C13H16O5 (6 degrees of unsaturation) by HRESIMS at m/z 253.1077 [M + H]+ (calcd for C13H17O5 253.1071), which is 30 mass units less than that of eurothiocin A (4). Analyses of the 1D (Table 1 and Supplementary Figures S14, S15) and 2D (Figure 2 and Supplementary Figures S16-S18) NMR data of 3 indicated the same structure skeleton to that of 4. The main difference was that the presence of the carboxylic group in 3 instead of the methyl thioester moiety, as evidenced by the MS data. On the basis of these results, the planar structure of 3 was elucidated (Figure 1). The experimental ECD spectrum of 3 (Figure 4B) showed similar Cotton effects with those of eurothiocin A (4) (Liu et al., 2014), indicating the R configuration at C-2. Therefore, the absolute configuration of compound 3 was established as 2R.
Talaropyrone A (5) was obtained as a light yellow solid with the molecular formula of C13H20O5 deduced from the HRESIMS data (m/z 279.1206, [M + Na]+, calcd for C13H20O5Na 279.1203), suggesting four degrees of unsaturation. Analysis of the 1H, 13C and HSQC NMR data of 5 (Table 2 and Supplementary Figures S19–S21) revealed the presence of two exchangeable protons (δH 5.59 and 4.32, respectively), one methoxy group (δC/H 56.3/3.81), six methylenes, one oxymethine (δC/H 68.9/4.23), four olefinic carbons with two protonated carbons and one carbonyl carbon (δC 163.3). Analysis of its 1D NMR data (Table 2 and Supplementary Figures S19, S20) revealed that the structure of compound 5 was closely related to that of compound 7 (Zhao et al., 2015), indicating the presence of the α-pyrone moiety. The noticeable difference was that the absence of NMR signals for a methyl and the presence of a methylene (δC/H 60.7/3.36) group and a hydroxyl (δH 4.23) group in 5. The 1H-1H COSY (Figure 2 and Supplementary Figure S22) correlations of H2-12/H2-13, and the HMBC (Figure 2 and Supplementary Figure S23) correlations from OH-13 to C-12, from H2-13 to C-11 showed that hydroxymethyl group was connected to C-12. The ECD spectrum (Figure 4C) and specific optical rotation of compound 5 were similar to those of nodulisporipyrone A (7) (Zhao et al., 2015), which indicated that the configuration of compound 5 was 7R. Thus, the structure of 5 was defined and named talaropyrone A.
Talaropyrone B (6) was obtained as white solid with the molecular formula C13H18O6 based on HRESIMS data (m/z 293.1001, [M + Na]+, calcd for C13H18O6Na 293.0996), which is 14 mass units more than that of 5. The 1D NMR data (Table 2 and Supplementary Figures S24, S25) revealed that compound 6 had a similar core skeleton to compound 5. The main difference was that the hydroxymethyl group at C-12 in compound 5 was replaced by a carboxyl group in compound 6, which was further supported by HMBC correlations from H2-11 and H2-12 to the carbonyl carbon C-13 (δC 174.7) (Figure 2 and Supplementary Figure S28). The similar positive optical rotation and the similar ECD spectrum of 5 and 6 (Figure 4C) allowed the assignment of its absolute configuration 7R being the same as that of 5. Thus, the structure of talaropyrone B (6) was established as shown.
On the basis of the NMR and MS spectroscopic data comparison with those optical reported in the literature, the structures of the known compounds were identified as eurothiocin A (4) (Liu et al., 2014), nodulisporipyrone A (7) (Zhao et al., 2015), peniazaphilin B (8) (Zhang et al., 2018), ramulosin (9) (Wang et al., 2014) and 6-hydroxyramulosin (10) (McMullin et al., 2017).
Structurally, compounds 1 and 2 possesed the unique thioester moiety derived from benzofuran and 2-hydroxy-3-mercaptopropionic acid, which was rarely-observed in natural products. Although several natural products with a methyl thioester group have been reported such as echinomycin (Watanabe et al., 2006), thiocoraline (Al-Mestarihi et al., 2014), lincomycin A (Spížek and Řezanka, 2017) and collismycin A (Garcia et al., 2012), compounds 1 and 2 possesed previously undescribed lactyl thioester moiety. Compound 3 is a new analogue of the benzofuran 4. While compounds 5 and 6 are new α-pyrone derivatives. Biogenetically, the aromatic polyketide orsellinic acid, which was biosynthesized from one unit of acetyl-CoA and three units of malonyl-CoA (Hashimoto et al., 2014), could be the biosynthetic precursor for compounds 1−4, first via isoprenylation, oxidation and dehydration reactions to form compound 3. Subsequently, compound 3 undergoes a thioesterification reaction to generate compound 4. In addition, compound 3 combines with cysteine via thioesterification, deamination and methylation reactions to form 1 and 2. The proposed biosynthetic pathways of compounds 1–4 was illustrated in Scheme 1.
Compounds 1−10 were also evaluated for antibacterial activities against Staphylococcus aureus (CGMCC 1.2465), Streptococcus pneumoniae (CGMCC 1.1692), Escherichia coli (CGMCC 1.2340) and Bacillus subtilis (ATCC 6633), and none of these compounds showed antibacterial activities toward these bacteria (MIC > 50 µg/mL). Meanwhile, compounds 1−10 were evaluated for the in vitro α-glucosidase inhibitory activities (Table 3). Compounds 3 and 4 showed moderate inhibitory effect with IC50 values of 48.9 ± 2.16 and 8.8 ± 1.08 µM, respectively (acarbose as positive control with IC50 value of 12.2 ± 0.66 µM).
To gain some insight about the inhibitory type of 3 on α-glucosidase, Lineweaver–Burk double reciprocal and Dixon single reciprocal plots were used to analyzed the data and determined the inhibit mode. In the Lineweaver–Burk plot, the Vmax value decreased with the increment of concentration, and the lines intersected on the x-axis (p-NPG concentrations), suggesting that the Km value was unchanged, implying the non-competitive-type inhibition of 3 (Figure 5A). Dixon plot was drawn by plotting the reciprocal of velocity against inhibitory concentrations at varying concentrations of p-NPG and the Ki value of 3 was further calculated as 242.3 μM (Figure 5B).
The molecular docking study was performed to identify the possible orientations and binding interactions of the 3 and 4 in the active site of the α-glucosidase. The molecular docking models of 3 and 4 are illustrated in Figure 6. The results of docking revealed that 3 formed two hydrogen bonds with Arg-1510 and Asp-1526 residues (Figure 6A), 4 formed two hydrogen bonds with Arg-1510 and His-1584 residues (Figure 6B). The different hydrogen bonding sites may result in different inhibitory effects, and the results of this docking experiment may help to understanding the binding mechanism of benzofurans and α-glucosidase.
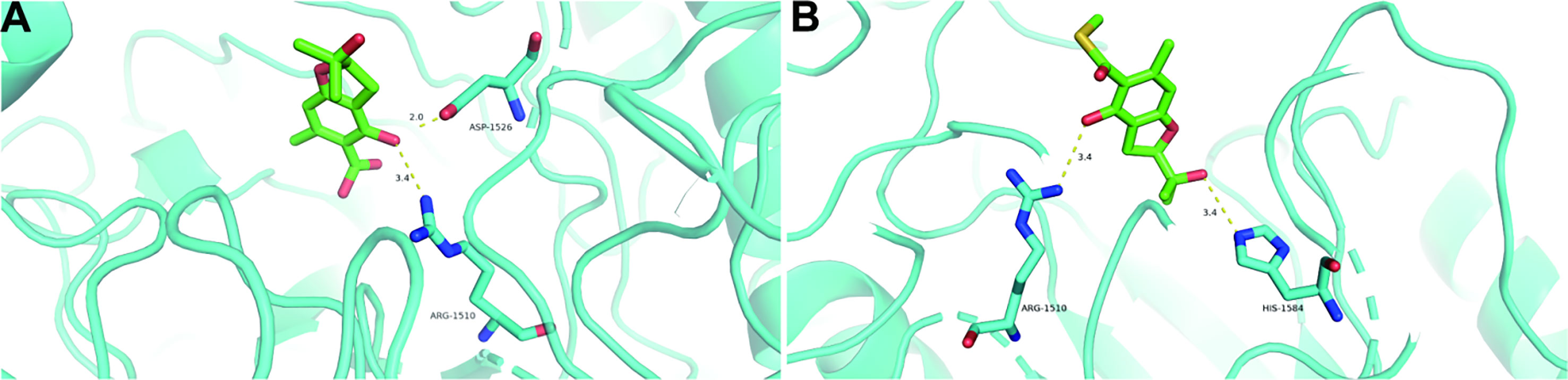
Figure 6 Docking poses and interactions of 3 (A) and 4 (B) with α-glucosidase (PDB ID: 3TOP). Hydrogen bond was denoted with yellow dash line.
In summary, five new compounds including two thionic benzofurans talarobenzofurans A and B (1 and 2), one benzofuran talarobenzofuran C (3) and two α-pyrones talaropyrones A and B (5 and 6), along with five known ones were isolated from the fermentation of the mangrove-derived fungus Talaromyces sp. WHUF0341 guided by OSMAC strategy. Compounds 1 and 2 possesed the unique thioester moiety derived from benzofuran and 2-hydroxy-3-mercaptopropionic acid. Compounds 3 and 4 exhibited inhibitory effects against α-glucosidase activity. The enzyme kinetics assay indicated the non-competitive-type inhibition of 3 towards α-glucosidase. Furthermore, the interactions of 3 and 4 with α-glucosidase were investigated by molecular docking assay. And the biosynthetic pathways of compounds 1–4 was also proposed. These findings not only expanded the chemical diversities of the natural benzofuran and α-pyrone families, but also provided valuable clues for searching new antidiabetic candidates from the mangrove-derived fungi.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conceptualization: LL. Methodology: RZ, JZ and RH. Resources: RZ, YX and KH. Data curation, RZ, JZ, and RH. Writing–original draft preparation: RZ and JZ. Writing–review and editing: RH, KH, and LL. Project administration and funding acquisition: LL. All authors contributed to the article and approved the submitted version.
This research was funded by grants from National Key Research and Development Program of China (2021YFC2100600 and 2018YFC0311000) and the National Natural Science Foundation of China (32022002 and 21977113).
The authors thank the National Special Project and the National Natural Science Foundation for funding. We also appreciate Junnan Shen and Desheng Li (School of Pharmaceutical Sciences, Wuhan University) for their contribution on strain isolation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1034945/full#supplementary-material
Al-Mestarihi A. H., Villamizar G., Fernández J., Zolova O. E., Lombó F., Garneau-Tsodikova S. (2014). Adenylation and S-methylation of cysteine by the bifunctional enzyme TioN in thiocoraline biosynthesis. J. Am. Chem. Soc 136, 17350–17354. doi: 10.1021/ja510489j
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2021). Marine natural products. Nat. Prod. Rep. 38, 362–413. doi: 10.1039/D0NP00089B
Chen S. J., Tian D. M., Wei J. H., Li C., Ma Y. H., Gou X. S., et al. (2022). Citrinin derivatives from Penicillium citrinum Y34 that inhibit α-glucosidase and ATP-citrate lyase. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.961356
El-Bondkly E. A. M., El-Bondkly A. A. M., El-Bondkly A. A. M. (2021). Marine endophytic fungal metabolites: A whole new world of pharmaceutical therapy exploration. Heliyon 7, e06362. doi: 10.1016/j.heliyon.2021.e06362
Ellison A. M., Farnsworth E. J. (1992). The ecology of belizean mangrove-root fouling communities: Patterns of epibiont distribution and abundance, and effects on root growth. Hydrobiologia 247, 87–98. doi: 10.1016/0022-0981(90)90139-4
Fan J. R., Kuang Y., Dong Z. Y., Yi Y., Zhou Y. X., Li B., et al. (2020). Prenylated phenolic compounds from the aerial parts of Glycyrrhiza uralensis as PTP1B and α-glucosidase inhibitors. J. Nat. Prod. 83, 814–824. doi: 10.1021/acs.jnatprod.9b00262
Frisch M. J., Trucks G. W., Schlegel H. B. M., Scuseria G. E., Robb M. A., Cheeseman J. R., et al. (2009). Gaussian 09, revision A.1.
Garcia I., Vior N. M., Brana A. F., Gonzalez-Sabin J., Rohr J., Moris F., et al. (2012). Elucidating the biosynthetic pathway for the polyketide-nonribosomal peptide collismycin a: Mechanism for formation of the 2,2'-bipyridyl ring. Chem. Biol. 19, 399–413. doi: 10.1016/j.chembiol.2012.01.014
Guo W. Q., Kong X. L., Zhu T. J., Gu Q. Q., Li D. H. (2015). Penipyrols A–B and peniamidones A–D from the mangrove derived Penicillium solitum GWQ-143. Arch. Pharm. Res. 38, 1449–1454. doi: 10.1007/s12272-014-0513-3
Hashimoto M., Nonaka T., Fujii I. (2014). Fungal type III polyketide synthases. Nat. Prod. Rep. 31, 1306–1317. doi: 10.1039/C4NP00096J
He X. F., Chen J. J., Huang X. Y., Hu J., Zhang X. K., Guo Y. Q., et al. (2021). The antidiabetic potency of Amomum tsao-ko and its active flavanols, as PTP1B selective and α-glucosidase dual inhibitors. Ind. Crop Prod. 160, 112908. doi: 10.1016/j.indcrop.2020.112908
Hemberger Y., Xu J., Wray V., Proksch P., Wu J., Bringmann G. (2013). Pestalotiopens A and B: stereochemically challenging flexible sesquiterpene-cyclopaldic acid hybrids from Pestalotiopsis sp. Chem. Eur. J. 19, 15556–15564. doi: 10.1002/chem.201302204
Hou B. L., Liu S. S., Huo R. Y., Li Y. Q., Ren J. W., Wang W. Z., et al. (2021). New diterpenoids and isocoumarin derivatives from the mangrove-derived fungus Hypoxylon sp. Mar. Drugs 19, 362. doi: 10.3390/md19070362
Ji C. F., Xu X. Z. (2021). Recent advancements of macrolide hybrids against Staphylococcus aureus. Curr. Top. Med. Chem. 21, 2455–2473. doi: 10.2174/1568026620999201203213733
Liu H. B., Liu Z. M., Chen Y. C., Tan H. B., Li S. N., Li D., et al. (2021a). Cytotoxic diaporindene and tenellone derivatives from the fungus Phomopsis lithocarpus. Chin. J. Nat. Med. 19, 874–880. doi: 10.1016/s1875-5364(21)60095-x
Liu G. R., Niu S. B., Liu L. (2021b). Alterchromanone A, one new chromanone derivative from the mangrove endophytic fungus Alternaria longipes. J. Antibiot. (Tokyo). 74, 152–155. doi: 10.1038/s41429-020-00364-4
Liu S., Su M. Z., Song S. J., Hong J. K., Chung H. Y., Jung J. H. (2018). An anti-inflammatory PPAR-γ agonist from the jellyfish-derived fungus Penicillium chrysogenum J08NF-4. J. Nat. Prod. 81, 356–363. doi: 10.1021/acs.jnatprod.7b00846
Liu Z. M., Xia G. P., Chen S. H., Liu Y. Y., Li H. X., She Z. G. (2014). Eurothiocin A and B, sulfur-containing benzofurans from a soft coral-derived fungus Eurotium rubrum SH-823. Mar. Drugs 12, 3669–3680. doi: 10.3390/md12063669
McMullin D. R., Green B. D., Prince N. C., Tanney J. B., Miller J. D. (2017). Natural products of Picea endophytes from the Acadian forest. J. Nat. Prod. 80, 1475–1483. doi: 10.1021/acs.jnatprod.6b01157
Meng L. H., Wang C. Y., Mándi A., Li X. M., Hu X. Y., Kassack M. U., et al. (2016). Three diketopiperazine alkaloids with spirocyclic skeletons and one bisthiodiketopiperazine derivative from the mangrove-derived endophytic fungus Penicillium brocae MA-231. Org. Lett. 18, 5304–5307. doi: 10.1021/np400614f
Nagelkerken I., Blaber S. J. M., Bouillon S., Green P., Haywood M., Kirton L. G., et al. (2008). The habitat function of mangroves for terrestrial and marine fauna: a review. Aquat. Bot. 89, 155–185. doi: 10.1016/j.aquabot.2007.12.007
Newman D. J., Cragg G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803. doi: 10.1021/acs.jnatprod.9b01285
Nugroho A. E., Morita H. (2014). Circular dichroism calculation for natural products. J. Nat. Med. 68, 1–10. doi: 10.1007/s11418-013-0768-x
Ohtani I. I., Hotta K., Ichikawa Y., Isobe M. (1995). Application of modified mosher’s method to α-aromatic secondary alcohols. exception of the rule and conformational analyses. Chem. Lett. 24, 513–514. doi: 10.1246/cl.1995.513
Pan R., Bai X. L., Chen J. W., Zhang H. W., Wang H. (2019). Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: a literature review. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00294
Spížek J., Řezanka T. (2017). Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 133, 20–28. doi: 10.1016/j.bcp.2016.12.001
Tao L. Y., Zhang J. Y., Liang Y. J., Chen L. M., Zhen L. S., Wang F., et al. (2021). Cytotoxic diaporindene and tenellone derivatives from the fungus Phomopsis lithocarpus. Chin. J. Nat. Med. 19, 874–880. doi: 10.3390/md8041094
Valente S., Cometto A., Piombo E., Meloni G. R., Ballester A. R., González-Candelas L., et al. (2020). Elaborated regulation of griseofulvin biosynthesis in Penicillium griseofulvum and its role on conidiation and virulence. Int. J. Food Microbiol. 328, 108687. doi: 10.1016/j.ijfoodmicro.2020.108687
Wang J. Y., Wang G. P., Zhang Y. L., Zheng B. Q., Zhang C. L., Wang L. W. (2014). Isolation and identification of an endophytic fungus Pezicula sp. in Forsythia viridissima and its secondary metabolites. World. J. Microbiol. Biotechnol. 30, 2639–2644. doi: 10.1007/s11274-014-1686-0
Watanabe K., Hotta K., Praseuth A. P., Koketsu K., Migita A., Boddy C. N., et al. (2006). Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nat. Chem. Biol. 2, 423–428. doi: 10.1038/nchembio803
Yu G. H., Zhou G. L., Zhu M. L., Wang W., Zhu T. J., Gu Q. Q., et al. (2016). Neosartoryadins A and B, fumiquinazoline alkaloids from a mangrove-derived fungus Neosartorya udagawae HDN13-313. Org. Lett. 18, 244–247. doi: 10.1021/acs.orglett.5b02964
Zhang D. W., Zhao J. Y., Wang X. W., Zhao L. L., Liu H. Y., Wei Y. Z., et al. (2018). Peniazaphilin A, a new azaphilone derivative produced by Penicillium sp. CPCC 400786. J. Antibiot. 71, 905–907. doi: 10.1038/s41429-018-0077-4
Keywords: Talaromyces sp., mangrove-derived fungus, thioester, α-glucosidase inhibitory activity, sulfur-containing benzofurans, molecular docking
Citation: Zhang R, Zhang J, Huo R, Xue Y, Hong K and Liu L (2022) Sulfur-containing benzofurans and α-pyrones from the mangrove-derived fungus Talaromyces sp. WHUF0341. Front. Mar. Sci. 9:1034945. doi: 10.3389/fmars.2022.1034945
Received: 02 September 2022; Accepted: 22 September 2022;
Published: 06 October 2022.
Edited by:
Weihua Jiao, Shanghai Jiao Tong University, ChinaReviewed by:
Cheng-shi Jiang, University of Jinan, ChinaCopyright © 2022 Zhang, Zhang, Huo, Xue, Hong and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Liu, bGl1bEBpbS5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.