
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 09 January 2023
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1026742
The bigfin reef squid, Sepioteuthis lessoniana, are a valuable commercial species in East Asian regions such as Taiwan and Japan. A lack of genomic information limits the application of potential aquaculture techniques, especially in breeding when considering the hatching rate of offspring. In some squids and cuttlefishes, symbiotic bacteria are transmitted from the accessory nidamental gland (ANG) to the jelly coat of eggs. In Hawaiian bobtail squid, these parent-delivered mutualistic bacteria play an important role in preventing lethal biofouling of the eggs and accelerating the hatch rate of offspring. The bacterial consortium, which is housed in the female squids ANG, are governed by host selection during female maturation. Immune functions are typically used to explain the regulatory mechanism of symbioses by host selection. In this study, we evaluated the transcripts featured in bacterial selection and maintenance during ANG development using RNA-seq. Different developmental stages of ANGs (stages 1–4) were sequenced. The de novo transcriptome assembly resulted in 524,918 unigenes. Two groups, non-pigmentation group (stage 1 and stage 3) and pigmentation group (stage 4), were clustered by transcriptome-wide expression profile analysis. The gene expression analyses indicated that 9,475 differential expression genes (DEGs) in three different phases and 1,363 (14.3%) DEGs were matched in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Furthermore, KEGG-enriched analysis results suggested that immune responses are a dominant pathway in the non-pigmentation group (stage 1 and stage 3) whereas lipid metabolism and metabolism of flora fermentation are dominant in the pigmentation group (stage 4). Although the host immunity plays an important role during bacterial colonization of the ANG in bigfin reef squid, our results showed that most immune-related genes had a reduced transcriptomic level in the pigmentation group compared with the non-pigmentation group. Therefore, our results provide new insight to understand the regulatory mechanisms of initial bacterial colonization and later bacterial pigmentation in the bigfin reef squid.
Several cephalopods have two specialized-organs, known as light organ and accessory nidamental gland (ANG), which house symbionts. The light organ is a bacteriogenic photophore, which is found in both sexes of certain Sepiolidae and Myopsida species (Lindgren et al., 2012). Unlike the light organ, the ANG is found only in females of some squid (Sepiolidae, Myopsida) and cuttlefish (Sepiida) species and is absent in other cephalopods (Oegopsida, Octopodiformes, Nautiloidea) (Lindgren et al., 2012). In Hawaiian bobtail squid (Euprymna scolopes), it was found that the ANG houses many types of bacteria and it delivers a bacteria-laden coating from the parent to the egg capsule which protects the eggs from fungal fouling (Kerwin et al., 2019). This bacterial consortium within the ANG is populated via horizontal transmission from the marine environment (Kaufman et al., 1998; Li et al., 2019). Although the bacterial consortia of ANGs show variety in different species (Collins et al., 2012; Kerwin and Nyholm, 2017; Yang et al., 2021), its composition shifts gradually during female maturation (Kerwin et al., 2021; Yang et al., 2021). The dynamics of the bacterial consortium in the ANG appear to comprise an intricate process governed by host selection during female maturation. For the light organ, comprehensive mechanisms have been described for bacterial selection and maintenance of the symbiotic luminescent bacterium, Vibrio fischeri (V. fischeri), in Hawaiian bobtail squid (reviewed in Nyholm and McFall-Ngai, 2021). However, symbiont shuffling and regulation in the ANG are not well understood. In order to understand the regulation of cephalopod ANG to the symbiotic bacterial consortium during female maturation, transcriptome analysis was used in this study, especially focused on genes involved in the immune defensive functions of bigfin reef squid ANG during female maturation.
The animal immune system not only defends against microbial infection but also assists in the colonization of symbiotic microorganisms to maintain microbiota–host homeostasis (Cerf-Bensussan and Gaboriau-Routhiau, 2010). In invertebrates, innate immune functions are based on the recognition of non-self patterns and the presence of cellular danger signals (Rosenstiel et al., 2009). Pattern recognition receptors (also called pattern recognition molecules, PRRs/PRMs) are sensor molecules that recognize motifs uniquely present in bacterial cell walls (pathogen/microbe-associated molecular patterns, PAMPs/MAMPs) such as peptidoglycans (most bacteria), lipoteichoic acid (gram-positive bacteria), and lipopolysaccharides (gram-negative bacteria) (Dziarski, 2004; Royet et al., 2011). After receptors bind with PRR/PRM–PAMP/MAMP complexes at the interface of epithelial barriers, they destroy foreign material by induction of defense effectors (Rosenstiel et al., 2009; Royet et al., 2011; Castillo et al., 2015). This innate immune system is known to be involved in the maintenance of commensal bacteria–host homeostasis in different tissues such as gut microbiota in animals (reviewed in Gómez and Balcázar, 2008; Royet et al., 2011), gill symbionts in vent mussel, Bathymodiolus septemdierum (Ikuta et al., 2019), light organ luminescent bacteria, V. fischeri, in Hawaiian bobtail squid (McAnulty and Nyholm, 2017; Nyholm and Graf, 2012), and ANG microbiota in bigfin reef squid (Li et al., 2022). Therefore, the innate immune system serves important biological roles in controlling bacterial assemblages in the ANG of cephalopods.
A single symbiotic bacterial species (V. fischeri) produces bioluminescence in the light organ of the Hawaiian bobtail squid which is controlled by a daily biological rhythm through the quorum-sensing system (Boettcher et al., 1996; Lupp et al., 2003). On the contrary, a complex consortium of microbes synthesizes carotenoids (pigments) which accumulate in the ANG at the time of female maturation (Lum-Kong and Hastings, 1992; Kerwin et al., 2021; Yang et al., 2021). As a result, the color of the ANG changes from white to yellow, orange, and red during sexual maturation in female squids (Bloodgood, 1977; Li et al., 2019; Li et al., 2022). Stressful environments can also lead to carotenoid accumulation as a defense in response to harmful conditions (Asker et al., 2007; Dieser et al., 2010; Mohammadi et al., 2012). Variations in light, temperature, salinity and pH have been reported to trigger carotenogenesis in some bacterial species (reviewed in Ram et al., 2020). Carotenoids also play a role in overall fitness (Johler et al., 2010) and virulence within the host for some pathogenic bacteria (Liu et al., 2005; Mohammadi et al., 2012). Furthermore, carotenoid accumulation (pigment formation) is regulated by a quorum-sensing system in certain bacteria, with high bacterial densities triggering the expression of specific genes (Rutherford and Bassler, 2012). Therefore, the protective antioxidant effects of carotenoids are important during the late phase of bacterial colonization with high bacterial densities. These data also demonstrate that the establishment and maintenance of the complex bacterial consortium in the ANG is an intricate process governed by host selection during initial bacterial colonization and later bacterial pigmentation.
The bigfin reef squid belongs to Myopsida, which possess only the ANG organ; the light organ is absent in this species (Lindgren et al., 2012). The development of the ANGs has been previously studied in female bigfin reef squid (Li et al., 2019) and other coleoid cephalopods (Lum-Kong, 1992; Kaufman et al., 1998; Kerwin et al., 2021). During ANG development, the bacteria are gradually transferred from the outer epithelial cells layers by environmental microbiota; these cell layers are deeply invaginated and form tubule structures in the inner epithelial cell layers for bacterial flora (Lum-Kong, 1992; Kaufman et al., 1998; Li et al., 2019; Kerwin et al., 2021). Four developmental stages of the ANG have been categorized in bigfin reef squid by gonadal and ANG characteristics (Figure 1): juvenile squid (primary oocyte stage) with a colorless ANG without visible bacterial flora (stage 1, bacterial transmission), immature squid (previtellogenic oocyte stage) with a colorless ANG with bacterial flora (stage 2, early phase of bacterial colonization), maturing squid (early vitellogenic oocyte stage) with a white/light-orange ANG and large numbers of bacterial flora (stage 3, late phase of bacterial colonization), and mature squid (mature eggs observe in oviduct) with a pigmented ANG and large numbers of bacterial flora (stage 4, bacterial pigmentation) (Li et al., 2019). Furthermore, our recent study shows that transcripts of one of the PPRs, peptidoglycan recognition protein 2 (Pgrp2), are highly correlated with ANG development and bacterial colonization in bigfin reef squid (Li et al., 2022). Bigfin reef squid Pgrp2 is mainly expressed in the epithelial cells of certain secondary tubules of ANG (Li et al., 2022). Therefore, these data suggest that the host innate immune system may participate in the maintenance of the complex consortium in the ANG of bigfin reef squids.
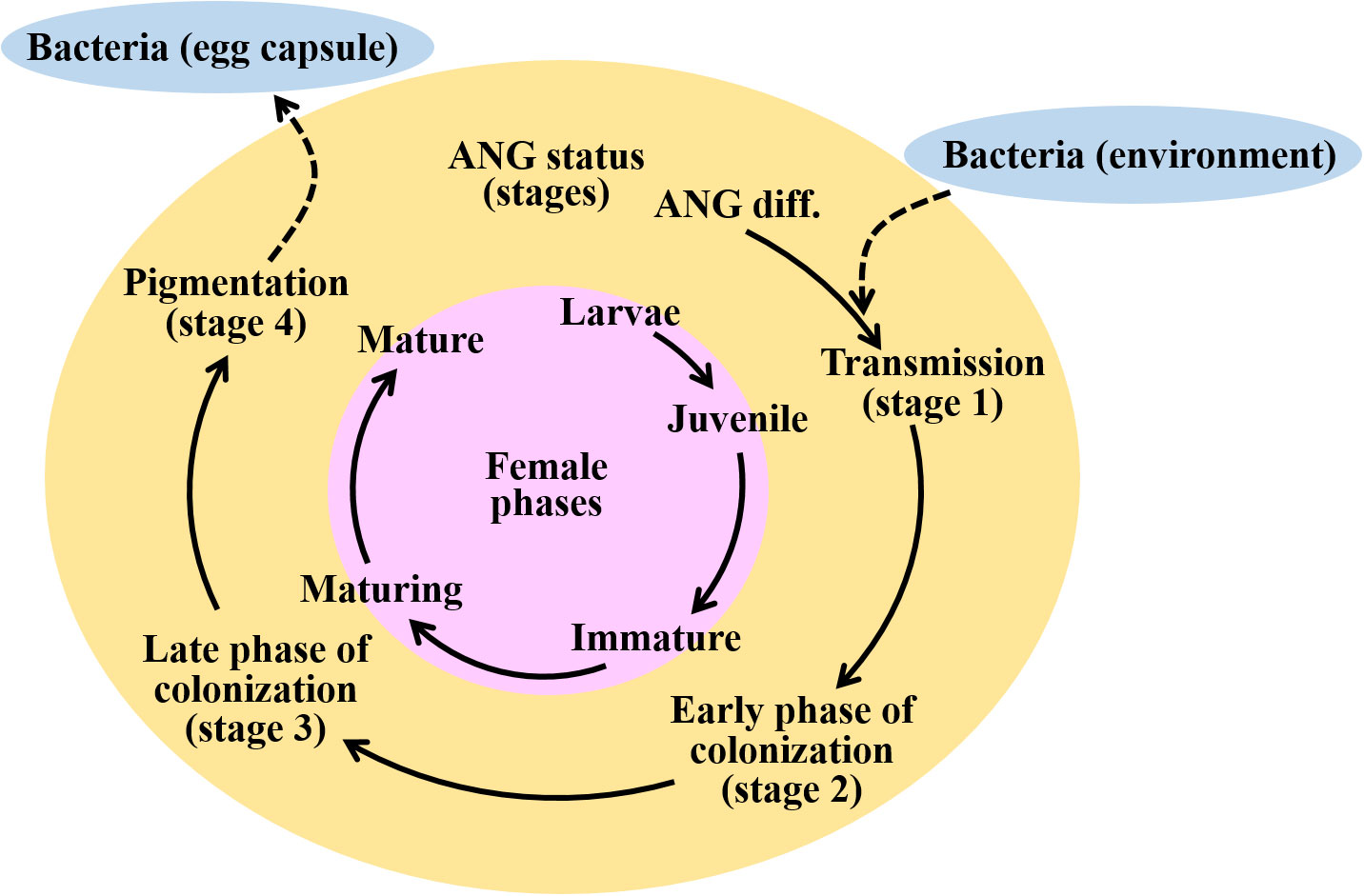
Figure 1 Schematic diagram representing the ANG developmental statues during female maturation in bigfin reef squid. Initially, the bacterial consortium of the ANG is populated by horizontal transmission from the environment. Four developmental stages of the ANG can be identified through gonadal and ANG characteristics. Bacterial transmission (stage 1), juvenile squid with primary oocytes, a colorless ANG, and no visible bacterial flora; early phase of bacterial colonization (stage 2), immature squid with previtellogenic oocytes, a colorless ANG, and bacterial flora; late phase of bacterial colonization (stage 3), maturing squid with early vitellogenic oocyte, a white/light-orange ANG, and a large number of bacterial flora; bacterial pigmentation (stage 4), mature squid with mature eggs observed in oviduct, a pigmented ANG, and large numbers of bacterial flora (stage 4). Finally, these symbiotic bacteria are transmitted to the jelly coat of the egg. Pink, yellow, and green colors indicate the female phases and ANG stages, respectively. Blue color indicates the marine environment. Dash lines indicate the horizontal transmission of bacteria from the marine environment to ANG or bacteria releasing from ANG to egg capsule.
During the last two decades, RNA sequencing has been applied in fundamental research for transcriptome analysis. The transcriptome has been used to study the innate immune system in different tissues of cephalopods as follows: white body (hematopoietic tissue) of the southern bobtail squid Euprymna tasmanica (Salazar et al., 2015), hemocytes of the common octopus Octopus vulgaris (Castellanos-Martínez et al., 2014) and Hawaiian bobtail squid (Collins et al., 2012), light organ of the Hawaiian bobtail squid (Goodson et al., 2005; Belcaid et al., 2019), and ANG of the European cuttlefish Sepia officinalis (Cornet et al., 2015). However, a comprehensive study on the mechanisms of bacterial selection and maintenance in the ANG during female maturation has not yet been completed. In order to understand the regulation of the symbiotic bacterial consortium in the cephalopod ANG during female maturation, transcriptome analysis was used in this study. We presented the first comprehensive ANG transcriptome analysis of the bigfin reef squid by de novo sequencing and annotation of the data generated by Illumina platform. Our data can provide insight into the regulatory mechanism of the ANG to the bacterial consortium during female maturation, especially on genes involved in immune defensive during the sexual development of female bigfin reef squid.
Bigfin reef squids were purchased from a fisherman on Heping Island (Keelung city, Taiwan) from the autumn of 2016 to the spring of 2017. The squids were collected by hand jigging from a boat off the near region of Heping Island (off the northeastern coast of Taiwan). The squid were anesthetized with 5% ethanol/seawater at room temperature and then decapitated after tissue sampling. The reproductive phase of females was determined based on ovarian stage by oocyte size and morphological characteristics (mature eggs in oviduct), as described previously (Chen et al., 2018). The ANG stage was determined based on morphological characteristics and confirmed by histology, in accordance with our previous study (Li et al., 2019). The stage of samples in this study was presented in our recent study (Li et al., 2022), including juvenile squid (stage 1, n = 3), immature squid (stage 2, n = 1), maturing squid (stage 3, n = 4), and mature squid (stage 4, n = 5). The ANGs were further divided into three groups and used for RNA analysis in this study (Figure 2): early transmission phase (stage 1), middle colonization phase (stage 3), and late pigmentation phase (stage 4). The biological characteristics of the sampled squid are shown in Table 1. All procedures and investigations were approved by the National Taiwan Ocean University Institutional Animal Care and Use Committee and were performed in accordance with standard guidelines.
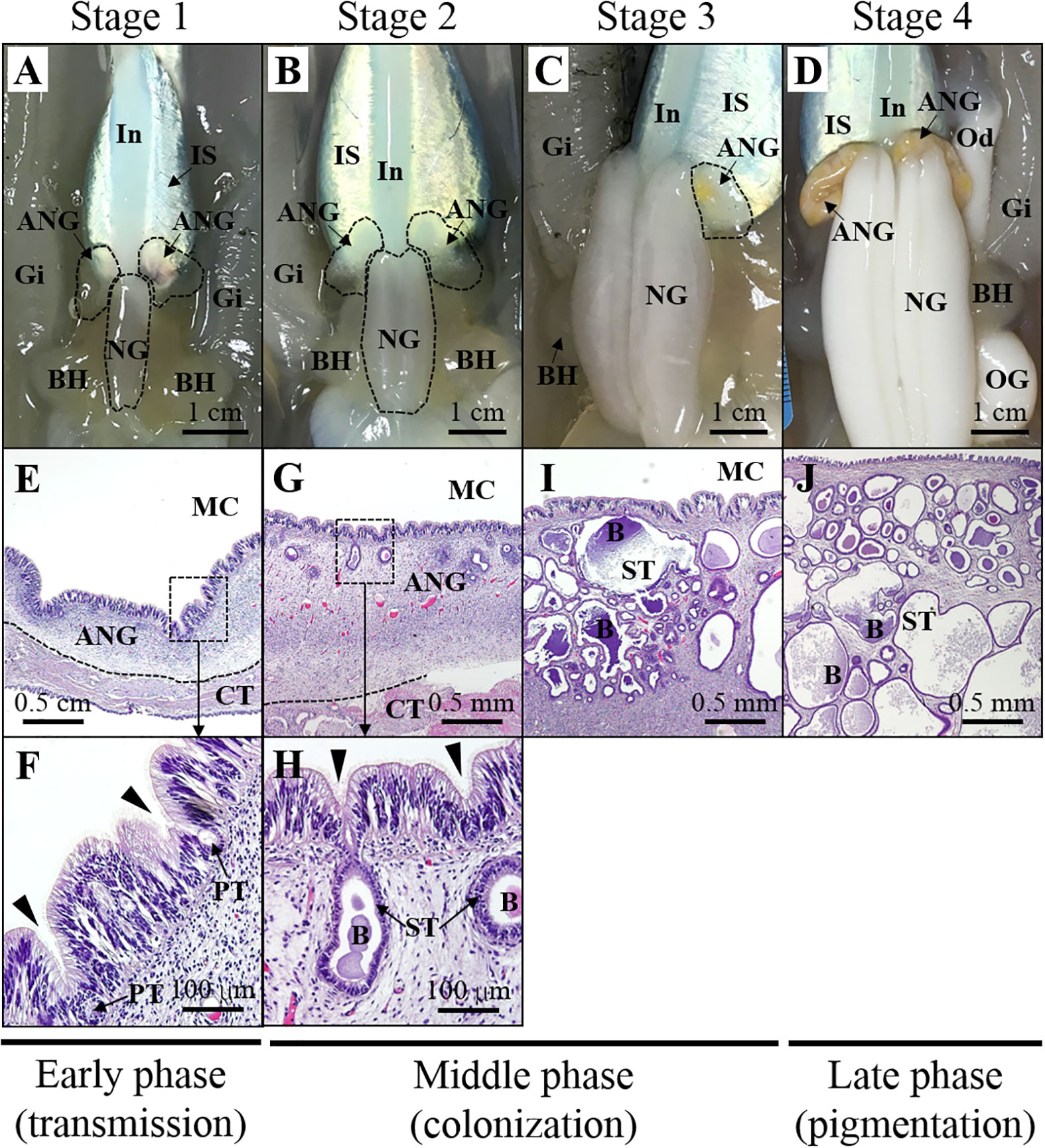
Figure 2 Morphological and histological changes during ANG growth. Relationship between morphology (A–D) and histology (E–J) of ANG during female maturation. Morphological changes in ANG stages are shown as follows: (A) juvenile squid with a colorless ANG (stage 1); (B) immature squid with a colorless ANG with several white spots (stage 2); (C) maturing squid with a white/yellow ANG (stage 3); (D) mature squid with a pigmentary ANG (stage 4). Histological changes in ANG stages are shown as follows:(E, F) juvenile squid without visible bacterial colony in the ANG (stage 1); (G, H) immature squid with several bacterial colonies in the ANG (stage 2); (I, J) bacterial colonies widely spread in the ANG (stage 3 and stage 4). Based on the process of ANG development, three phases are divided in this study, namely, early transmission phase, middle colonization phase, and late pigmentation phase. ANG, accessory nidamental gland; B, bacteria; BH, branchial heart; CT, connective tissue; Gi, gills; In, intestine; IS, ink sac; NG, nidamental gland; OG, oviductal gland; PT, primary tubule; ST, secondary tubule.
Tissue histology was performed according as described in our previous study (Chen et al., 2018). The ANGs and ovaries were fixed with 4% paraformaldehyde in PBS at 4°C for 16 h, dehydrated in methanol, and stored at -20°C. The dehydrated ANGs were serially transferred to ethanol and xylene and then embedded in paraffin. The ANG sections (5-µm thickness) were rehydrated with PBS and subjected to hematoxylin and eosin staining.
Tissues were homogenized in TRIzol reagent (Invitrogen). Extraction of nucleic acids was performed according to the manufacturer’s protocol. The RNA degradation of isolated total RNA was assessed by gel electrophoresis. The quantity of isolated total RNA was determined using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific).
RNA qualification, cDNA library construction, and Illumina sequencing were conducted by Genomics, Inc. (New Taipei City, Taiwan). The concentration and integrity of RNA samples were determined using a Qubit® 2.0 Fluorometer (Life Technologies) and Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. RNA-seq libraries were constructed using the Illumina TruSeq Stranded mRNA Library Prep Kit (Illumina). Paired-end (150-bp) transcriptome sequencing was performed on the Illumina NextSeq 500 platform.
Data processing was modified as described in our previous study (Tseng et al., 2022). The raw reads were deposited in the NCBI Sequence Read Archive database (BioProject ID PRJNA814067). Raw reads were processed using the Galaxy platform (http://usegalaxy.org) to analyze the data. Clean reads were generated from raw reads using the Trimmomatic trimming tool (ILLUMINACLIP = TruSeq3-PE.fa:2:30:10:8, SLIDINGWINDOW = 4:5, LEADING = 5, TRAILING = 5, MINLEN = 25) by removing adapters and low-quality reads. The clean reads of all samples were pooled together for de novo assembly with Trinity v2.9.1. Quality assessment of the de novo assembled transcriptome was performed by computing the ExN50 statistic and BUSCO. To identify the contaminants from holobionts, a DIAMOND tool (Buchfink et al., 2021) analysis of all sequences against the NCBI NR database was used to identify bacterial sequences. Through the above data processing procedure, accurate transcripts were generated as the reference transcripts/genes for subsequent analysis.
To identify putative genes involved in the immune response, the sequences obtained in this study were screened using the GO terms (innate + immune) after annotation of all the sequences. The putative immune-related transcripts were identified by looking for these specific keywords in the hit descriptions of proteins of the present de novo transcriptome database. Furthermore, genes important in the cephalopod immune response, according to the literature (reviewed in Castillo et al., 2015), were searched in the reference bigfin reef squid ANG transcriptome. Full-length cDNA sequences of these immune-related genes in mollusk were retrieved from NCBI GenBank, and local BLAST searches were conducted (BLASTP, cutoff e-value 1e-5) against translated sequences from the reference bigfin reef squid ANG transcriptome.
The clean reads for each sample were mapped to the reference transcripts using the Salmon in the Galaxy platform. The TPM (transcripts per million) values were used to represent the expression level of each sample’s transcripts. Statistically significant differentially expressed genes (DEGs) between the different groups were identified using DESeq2 software, and the filter parameters were | log2 Fold Change| > 1 and adjusted p value < 0.001. These DEGs were clustered using hierarchical clustering of Pearson correlation with the Trimmed Means of M values (TMM). TransDecoder was used to identify putative open reading frames (ORFs) from the DEGs. For functional annotation, predicted peptides of DEGs were annotated using the public database eggNOG v5.0 with eggNOG-mapper v2 (Cantalapiedra et al., 2021). This tool can be used to find clusters of orthologous groups in the eggNOG database and annotate them using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The functional properties of genes and gene products in organisms can be annotated by GO. KEGG pathway analysis was performed to identify pathways for intracellular molecular interactions and their organism-specific variations, not including human disease pathways.
Quantitative real-time PCR (qPCR) was carried out using CFX Connect™ Real-Time PCR Detection System (Bio-Rad Laboratories) with SYBR Green Master Mix (Bio-Rad Laboratories). Elongation factor 1 alpha (Ef1a; GenBank accession no. MG924746) was used as an internal standard to normalize the gene expression levels. Specific qPCR primers for target genes are listed in Table 2. The qPCR specificity was confirmed by a single melting curve in samples and template-containing controls. No signal was detected by qPCR in non-template controls. The reaction efficiency of different genes was evaluated using serially diluted standards. The relative gene expression levels were determined according to the 2-ΔΔCt method, and the highest value of the target genes was defined as 1. Using the same ANGs used for qPCR analysis in our recent studies (Li et al., 2022), the transcript levels of Pgrp2 were analyzed by qPCR in our recent studies (Li et al., 2022).
Data are shown as the mean ± standard deviation (SD). One-way ANOVA followed by a Games–Howell test (homogeneity of variance <0.05) was used to determine the significance of differences among three groups. Statistical Package for the Social Sciences (SPSS) software was used. In all cases, P < 0.05 indicated a significant difference.
To study the regulation of the bacterial consortium by the squid ANG, 13 cDNA libraries were constructed from the different stages of the ANG during female maturation, including stage 1 (3 libraries), stage 2 (1 library), stage 3 (4 libraries), and stage 4 (5 libraries). A total of 213.1 million (64.6 Gb) raw reads was obtained from the 13 libraries that contained 53.3 million reads (16.2 Gb), 81.1 million reads (24.6 Gb), and 78.8 million reads (23.8 Gb) in the early transmission phase, middle colonization phase, and late pigmentation phase, respectively (Table 3). After filtering out the low-quality and ambiguous reads, a total of 60.8-Gb reads (94.1%) was obtained from the early transmission phase (15 Gb, 92.6%), middle colonization phase (23.2 Gb, 94.3%), and late pigmentation phase (22.6 Gb, 95%) (Table 3). These clean reads were used for the subsequent analysis. The de novo assembly generated 524,918 unigenes containing 747,982 transcripts, and the N50 value was 454 bp involving all the contigs (E100) (Table 4). The ExN50 statistic showed that the maximum N50 value was 1,838 bp (E84) (Figure S1). These data only represent the top most highly expressed genes (16,073 unigenes out of the total of 524,918 unigenes) that represent 84% of the total normalized expression data. In total, 2.1% of the transcripts (16,979 transcripts) in the transcriptome were clustered in the non-eukaryotic group, representing only 0.6~1.9% of the total expression data in each library. These data revealed that a small amount of contaminating genes was due to microorganisms. Benchmarking Universal Single-Copy Orthologs (BUSCOs) analysis showed that these unigenes contained 4,592 (86.7%) complete BUSCOs and 148 (2.8%) fragmented BUSCOs, whereas 555 (10.5%) BUSCOs were missing out of 5,295 single-copy orthologs of Mollusca (Table 4).
A threshold of TPM >1 in at least one ANG sample was used to define genes as expressed. In total, 209,719 genes were used for transcriptome-wide expression analysis. The global gene expression patterns of ANGs at different stages were hierarchically clustered in a heatmap using the Pearson correlation coefficient as a distance measure (Figure 3). Two different groups were clearly separated. One consisted of ANGs at the early transmission phase and middle colonization phase (branch 1: stage 1 and stage 3), and the other consisted of ANGs at the later pigmentation phase (branch 2: stage 4). In branch 1 (non-pigmentation group), stage 1 and stage 3 ANGs are clustered together. Global gene expression of ANGs started to change at the later pigmentation phase (stage 4), the period when mature squid have a pigmented ANG with a large numbers of bacterial flora.
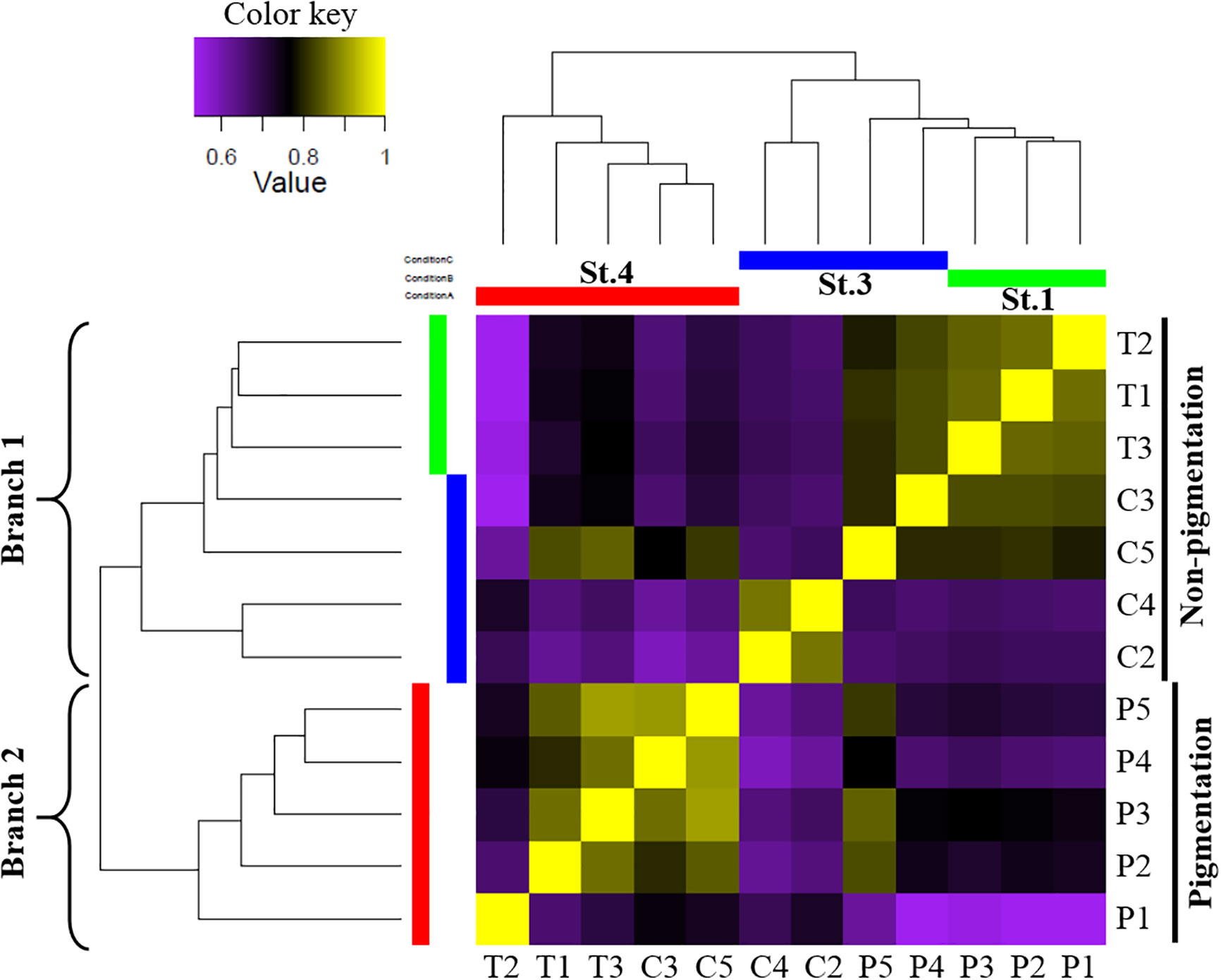
Figure 3 Global gene expression patterns in different ANG stages. Correlation heatmap using the Pearson correlation coefficient as distance measure result. Heatmap indicates that the early transmission phase (stage 1) and middle colonization phase (stage 3) were grouped in branch 1, and the late pigmentation phase (stage 4) was alone in branch 2. The expression patterns are depicted according to the color scale. St, stage; T, transmission phase; C, colonization phase; P, pigmentation phase.
Pairwise comparisons of ANGs from three different stages (stages 1, 3, and 4) identified a total of 9,475 DEGs. Subsequently, expression profiling was performed on these DEGs at three different ANG stages (Figure 4A). Furthermore, according to the temporal characteristics of gene expression across the three ANG stages, these DEGs were divided into five clusters (Figure 4B). In total, 3,162, 1,492, 922, 2,648, and 1,251 unigenes clustered into groups 1, 2, 3, 4, and 5, respectively (Figure 4B). It is worth noting that the gene expression profiles in group 1 and group 4 showed that there were significant changes as the ANGs shifted from the non-pigmentation group (stage 1 and stage 3) to the pigmentation group (stage 4). Furthermore, over half of the DEGs (61.3%) were clustered in these two groups (groups 1 and 4). Combined with global gene expression patterns in ANGs, our results suggested that host physiological characteristics are dramatically altered from the middle colonization phase (stage 3) to the later pigmentation phase (stage 4).
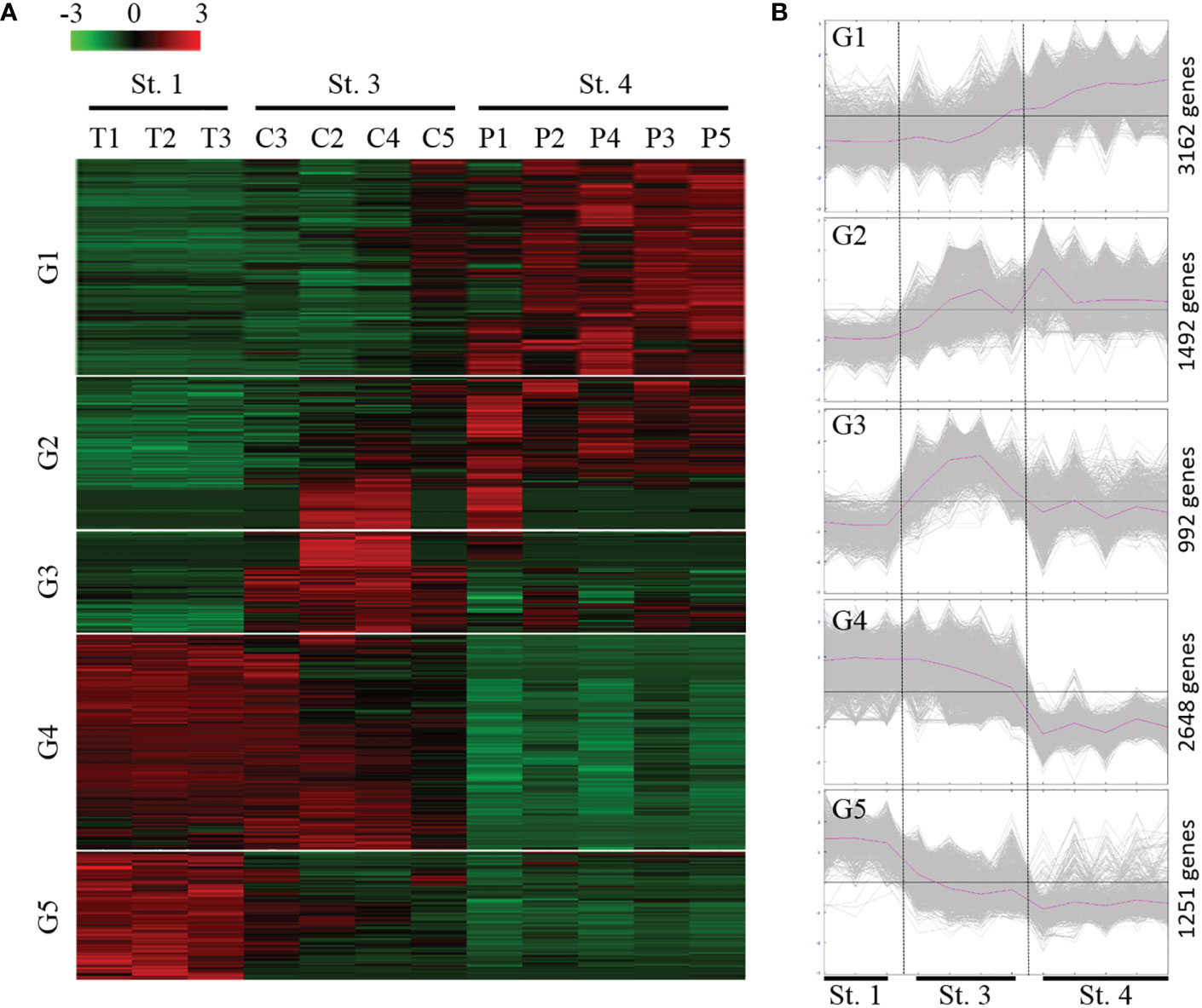
Figure 4 Expression profiles of the DEGs during the ANG growth. (A) Heatmap for cluster analysis of the differentially expressed genes (DEGs) by hierarchical clustering of Pearson correlation with the Trimmed Means of M values. Green and red indicate downregulated and upregulated genes, respectively. (B) MeV cluster analysis of DEGs from the expression profiles during the ANG growth. according to the temporal gene expression across three ANG stages (stage 1, stage 3, and stage 4); these DEGs were divided into five groups. St, stage; G, group.
Among the 9,545 DEGs, 1,223 DEGs (12.8%) and 1,165 DEGs (12.2%) had matches in the GO and KEGG databases, respectively. In total, 8,182 (85.7%) DEGs did not have matches in any databases. GO annotation (level 2) categorized these DEGs into 26 biological processes, two cellular components, and 17 molecular functions (Figure S2). Cellular, metabolic, and biological regulation processes were the most dominant categories of biological processes, whereas cellular anatomical entities and protein-containing complexes were the two cellular component categories. Binding and catalytic activity were the most dominant categories of molecular functions. Furthermore, these DEGs were classified into 32 KEGG subcategories, including 11 metabolism, four genetic information processing, three environmental information processing, four cellular processes, and 10 organismal systems (Figure S3).
Furthermore, the GO term-enriched profiles (p value < 0.05) and KEGG pathway-enriched profiles (p value < 0.05 with five genes in a pathway) in the five main expression profiles (groups) are shown in Figures 5, 6, respectively. According to the GO term-enriched profiles and KEGG-enriched profiles, our data showed temporal characteristics in these five main expression profiles. We suggest that the immune response may be important in the early transmission phase and middle colonization phase (stage 1 and stage 3), whereas lipid metabolism (fatty acid degradation and elongation) and metabolism of flora fermentation (propanate and butanoate metabolism) may be important in the late pigmentation phase (stage 4) (Figure 6). The genes of these dominant pathways in both groups are listed in Table S1. Interestingly, the KEGG pathway map results indicated that immune functions are not the dominant regulatory mechanism for homeostasis in the late pigmentation phase of the ANG during female maturation.

Figure 5 GO enrichment analysis of DEGs. The dominant GO (Gene Ontology) categories of differentially expressed genes (DEGs) in five groups. Temporal expression profiles of these five groups shows in Figure 4 G, group.

Figure 6 KEGG enrichment analysis of DEGs. A) The functionally enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways found in the analysis of differentially expressed genes (DEGs) in five groups. Temporal expression profiles of these five groups shows in Figure 4. G, group.
A screening of the GO terms “innate + immune” allowed us to identify genes with a possible immune function in the present ANG transcriptome database. Furthermore, potential immune-related genes were selected from the present ANG transcriptome database, based on the literature (reviewed in Castillo et al., 2015). Among the transcripts identified, a total of 365 potentially immune-related genes were obtained from the reference bigfin reef squid ANG transcriptome database in this study. We further analyzed the transcriptomic levels of these immune-related genes in different stages. The global immune-related gene expression patterns of ANGs at different stages were hierarchically clustered in a heatmap using the Pearson correlation coefficient as a distance measure (Figure 7). Two different groups were clearly separated. One consisted of ANGs in all samples of the early transmission phase (stage 1, n = 3) and most samples of the middle colonization phase (stage 3, n = 3) (branch 1), and the other consisted of ANGs in a sample of the middle colonization phase and all samples in the later pigmentation phase (stage 4, n = 5) (branch 2). In branch 2 (non-pigmentation group), stage 3 (C5) and stage 4 ANGs (P1–P5) were clustered together. Expression profiling was performed on these immune-related genes at three different ANG stages (Figure 7). According to the temporal characteristics of gene expression across three ANG stages, these genes were divided into five clusters (Figure 7). In total, 27, 75, 42, 12, and 209 genes were clustered into groups 1, 2, 3, 4, and 5, respectively (Figure 7). Global immune-related gene expression of ANGs in group 5 showed that most genes (57.3%) were reduced at the later pigmentation phase (stage 4), the period when mature squid have a pigmented ANG with large numbers of bacterial flora. The DEGs of these immune-related genes in different groups (Figure 4) are listed in Table S2. Thus, our results indicated that innate immunity is also involved in the regulation of the bacterial consortium during ANG growth.
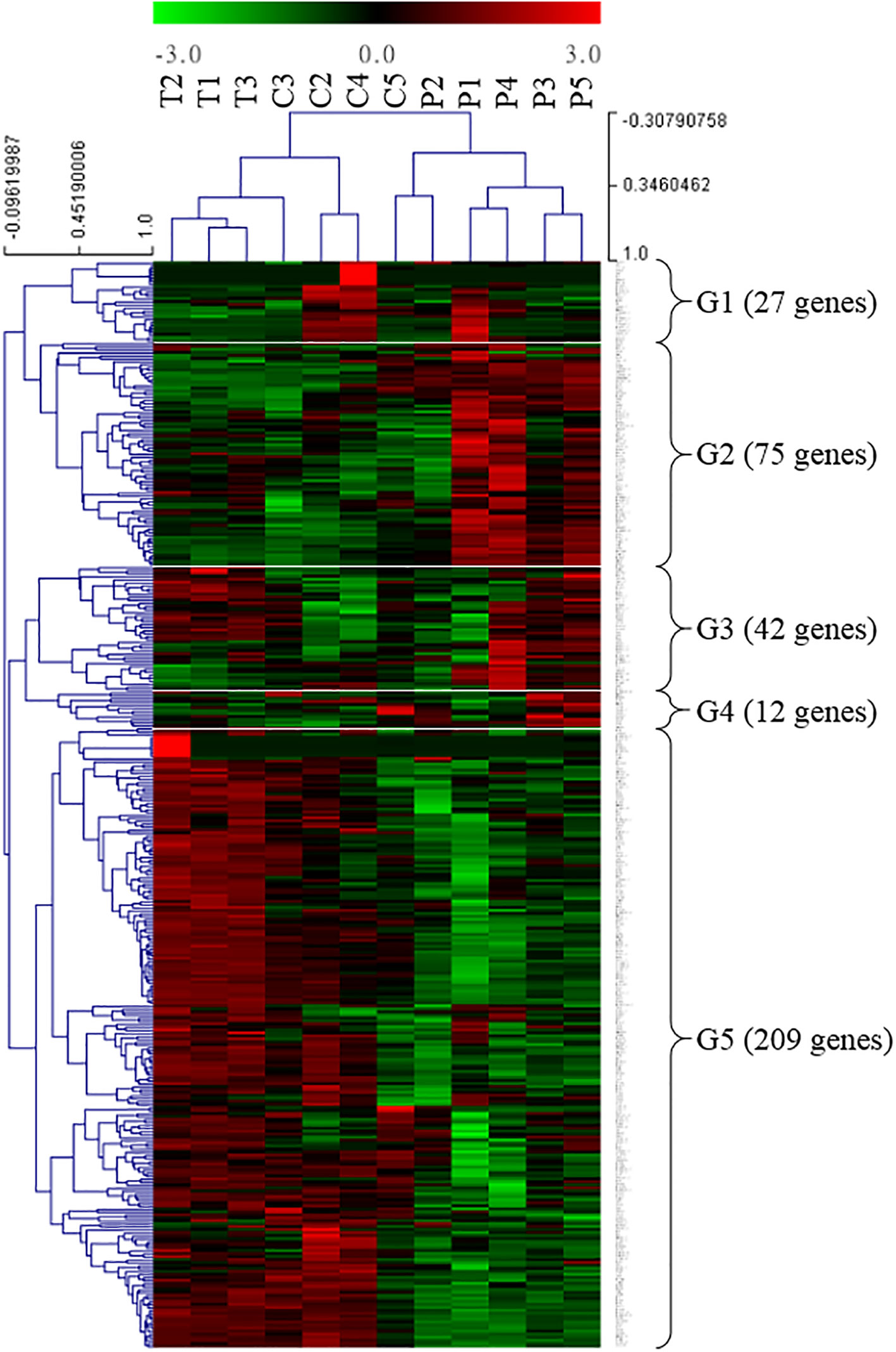
Figure 7 Transcriptomic levels of immune-related genes in different ANG stages. Heatmap for cluster analysis of the immune-related genes by hierarchical clustering of Pearson correlation with the Trimmed Means of M values. According to the gene expression, these DEGs were divided into five groups. Green and red indicate downregulated and upregulated genes, respectively. G, group; T, transmission; C, colonization; P, pigmentation.
In our transcriptome, we showed that the transcriptomic levels of Pgrp2 (group 2 in Figure 7) were similar to our qPCR results (Figure 8). Furthermore, according to the transcriptomic profile, there are several other genes that are changed during bacterial colonization and have similar expression profiles to Pgrp2 transcripts, including the cluster of differentiation 109 (Cd109), nuclear receptor coactivator 7 (Ncoa7), and Pgrp3-like (Pgrp3l) genes. Several genes were randomly selected for comparison of the expression profiles in the non-pigmentation group and pigmentation group, including alkaline phosphatase 1 (Alp1), solute carrier family 26 member 6 (Slc26a6), GTP cyclohydrolase I (Gch1), and carbonic anhydrase 4 (Ca4). The expression of these eight genes was confirmed by qPCR (Figure 8). These results demonstrate that our transcriptomic analysis is reliable and highly correlated with the qPCR analysis results.
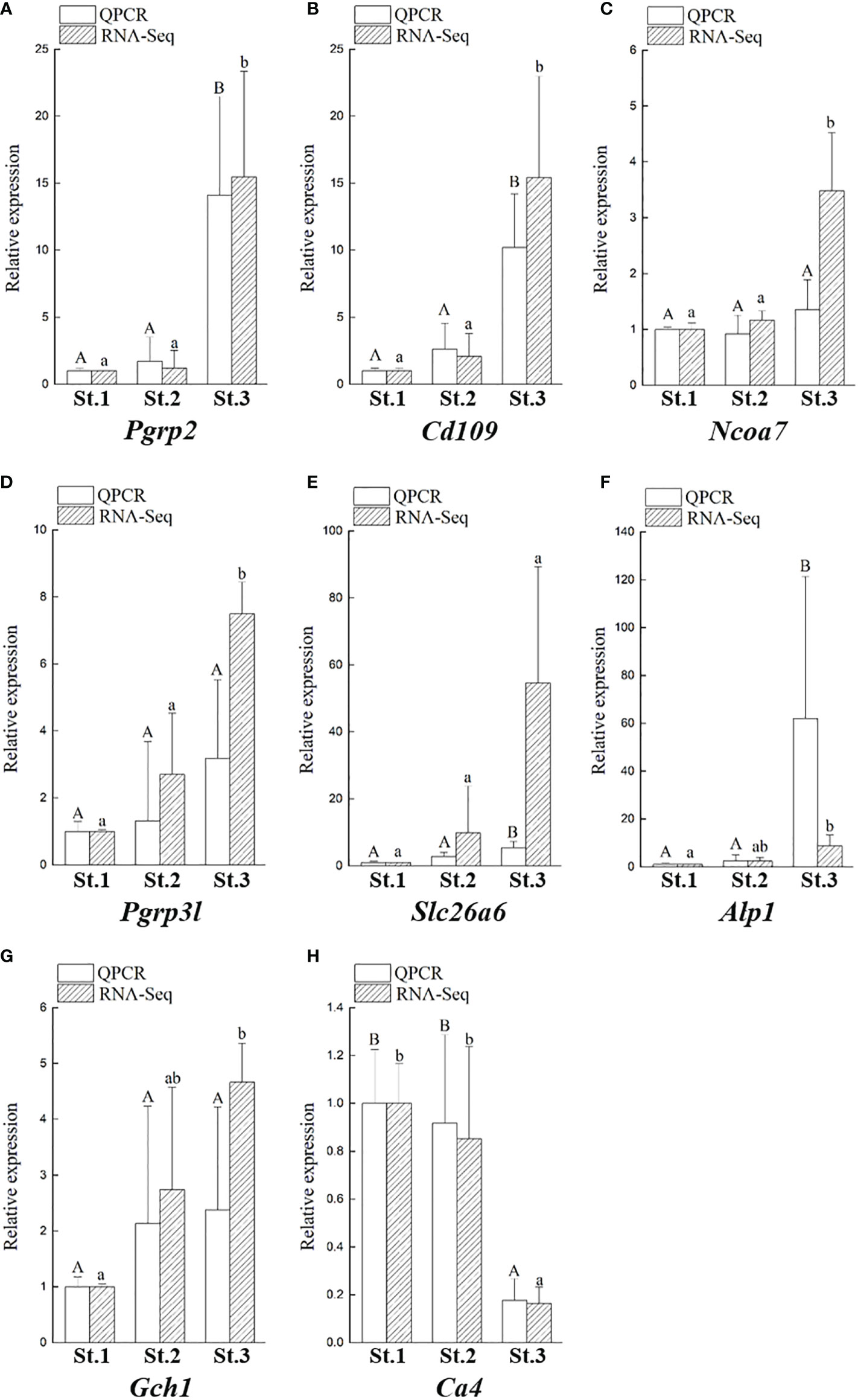
Figure 8 Validation of eight DEGs obtained from the transcriptomic results by qPCR. Comparison of transcriptomic levels, and gene transcripts calculated from qPCR during ANG growth in female squids, including Pgrp2 (A), Cd109 (B), Ncoa7 (C), Pgrp3l (D), Slc26a6 (E), Alp1 (F), Gch1 (G), and Ca4 (H). Data are expressed as the mean ± SD. Different letters indicate significant differences between stages of ANGs (P < 0.05). St, stage; Alp1, alkaline phosphatase 1; Cd109, cluster of Differentiation 109; Gch1, GTP cyclohydrolase 1; Ncoa7, nuclear receptor coactivator 7; Pgrp2, peptidoglycan recognition protein 2; Pgrp3l, peptidoglycan recognition protein 3-like, peptidoglycan recognition protein 3-like; Slc26a6, solute carrier family 26 member 6.
The bigfin reef squid, Sepioteuthis lessoniana, is a commercially valuable species in some East Asian regions such as Taiwan and Japan. Although knowledge of cephalopod culture methods is advanced, commercialization is still constrained by the highly selective feeding habits of cephalopods (Vidal et al., 2014). Aquaculture techniques for this species have been established at the laboratory and aquarium scales but still need to be improved to reach market volumes. The lack of genomic information about bigfin reef squid hinders the application of potential aquaculture techniques, especially those focused on breeding related to the hatching rate of offspring. In Hawaiian bobtail squid, female ANG-delivered commensal bacteria play an important role in preventing lethal biofouling on the egg and accelerating the hatching rate of offspring (Kerwin et al., 2019). Therefore, in the present work, we attempted to understand the immune-related genes involved in bacterial selection and maintenance during ANG development using an RNA-seq approach. This is the first transcriptome analysis of the ANGs in Teuthida cephalopods and the first comprehensive transcriptomic study of the ANG during female maturation in cephalopods. Based on comparisons of the transcriptomic profiles of the different ANG stages during female maturation, the present study provides a potential host regulatory mechanism initial bacterial colonization and late bacterial pigmentation in bigfin reef squid. Similar to a study of Hawaiian bobtail squid, in which the ANG had a high number of ANG-specific transcripts (Belcaid et al., 2019), most DEGs (87.9%) in the ANG of bigfin reef squid were not annotated in any reference database. We further characterized the transcriptomic profiles of innate immune-related genes in the ANG during female maturation. Our data suggest that the immune response is important during bacterial transmission and colonization, whereas lipid metabolism (fatty acids) and metabolism of flora fermentation are important during bacterial pigmentation.
The ANG houses a bacterial consortium that is horizontally transmitted from the environmental community in bigfin reef squid (Li et al., 2019; Li et al., 2022) and other coleoid cephalopods (Kaufman et al., 1998; Kerwin et al., 2021). Furthermore, the microbiota of the ANG undergoes a shift during female maturation of the bigfin reef squid (Yang et al., 2021) and Hawaiian bobtail squid (Kerwin et al., 2021). Our previous study further suggested that the environment affects the microbial compositions of individuals, whereas maturity levels exert progressive effects on microbial dynamics over time (Yang et al., 2021). These host-selected bacteria are transmitted to the egg jelly coat in some squid and cuttlefish (Kerwin and Nyholm, 2017; Huang et al., 2018). Suppression of the bacteria deposited on the eggs by antibiotic treatment led to susceptibility to heavy biofouling and very low hatching rates compared with untreated eggs in Hawaiian bobtail squid (Kerwin et al., 2019). Therefore, such changes in bacterial diversity in the ANG during maturation stages are strongly selected by the host. These host-selected bacteria may have the same role in Hawaiian bobtail squid that maternal-delivery bacteria play an important role in promoting embryo survival by preventing biofouling of the egg capsule.
Host immunity is widely thought to underlie the regulatory mechanism of host–microorganism interactions. Innate immune reactions have been studied in several cephalopod species and comprise two parts: 1) cellular defenses, which refers to the activation of hemocytes that circulate the body and phagocytize encapsulated microbes, and 2) humoral defenses, which refers to the recognition of PAMPs/MAMPs by PPRs/PRMs that activate conserved signaling pathways to release effectors that kill microorganisms (reviewed in Castillo et al., 2015). Furthermore, when host immunity is used to regulate symbiont populations, targeted immune responses that recognize symbionts and suppress but do not eliminate symbiont populations should evolve and be maintained. Unlike the single bacterial population hosted in the light organ of the bobtail squid (Ruby and Asato, 1993; Nyholm et al., 2002), a complex microbial consortium is hosted in the ANG of some squid and cuttlefish (Pichon et al., 2005; Collins et al., 2012; Kerwin and Nyholm, 2017; Yang et al., 2021). PPRs/PRMs are detected in light organs and hemocytes of the Hawaiian bobtail squid, including five peptidoglycan recognition proteins (Pgrps, Pgrp1–5), a lipopolysaccharide-binding protein (Lbps, Lbp1), and three bactericidal permeability-increasing proteins (Bpis, Bpi2–4) (Kremer et al., 2013; Krasity et al., 2015). In the present study, five Pgrps (Pgrp2–5 and Pgrp3l) and two Bpis (Bpi3 and Bpi3l) were identified in the ANG transcriptome of bigfin reef squid. Furthermore, we found that only two Pgrps (Pgrp2 and Pgrp3l) and two Bpis (Bpi3 and Bpi3-like) were correlated with ANG growth and they may be involved in bacterial selection and maintenance during female maturation.
The functions of PGRPs can include both direct bactericidal effects and indirect effects, such as the induction of antimicrobial compound production and the modulation of inflammation and immune responses (Royet et al., 2011). PGRPs are conserved from insects to mammals, and no members of the PGRP family have been found in nematodes (Dziarski and Gupta, 2006; Montaño et al., 2011) or crustaceans (Liu et al., 2011; Bai et al., 2020). Based on their structural characteristics and cellular functions, PGRPs have been divided into three forms: secreted proteins (with a signal peptide), transmembrane receptors, and intracellular proteins (Royet et al., 2011). Unlike insects, which have many Pgrp genes, only a few Pgrp genes have been found in cephalopods (reviewed in Castillo et al., 2015). In our previous study, four Pgrp genes were found in the ANG of bigfin reef squid after construction of a draft transcriptome database (Li et al., 2022). Moreover, a new GPI-anchored Pgrp gene (Pgrp3l) was found in the present study. Taken together, five Pgrp genes have been found in the ANG of bigfin reef squid, including secreted Pgrp2 and Pgrp5, GPI-anchored Pgrp3 and Pgrp3l, and the Pgrp4 surface receptor. In the present study, only the Pgrp2 transcript levels were correlated with ANG growth (Figure 8). Our previous study in bigfin reef squid also showed that Pgrp2 mRNA is predominantly expressed in the ovary and the ANG whereas Pgrp4 and Pgrp5 mRNA are predominantly expressed in hemocytes. In Hawaiian bobtail squid, PGRPs participate in bacterial selection and maintenance of the symbiotic bacterium within the light organ (Nyholm and Graf, 2012; McAnulty and Nyholm, 2016; Nyholm and McFall-Ngai, 2021). Secreted PGRP1 and PGRP2 play an integral role in the recognition of symbiotic bacteria during V. fischeri colonization of the Hawaiian bobtail squid light organ (Troll et al., 2009; Troll et al., 2010). After V. fischeri colonize the crypts of light organs, PGRP2 is secreted into the lumen of the crypts to degrade toxic peptidoglycans secreted by the bacteria, whereas the PGRP1 level decreases dramatically (Troll et al., 2009; Troll et al., 2010). The expression of secreted Pgrp5 transcripts is decreased in hemocytes of Hawaiian bobtail squid upon removal of symbiotic bacteria (Collins et al., 2012). Thus, colonization of the light organ by V. fischeri influences Pgrp gene expression in the Hawaiian bobtail squid (Collins et al., 2012; Nyholm and McFall-Ngai, 2021). Furthermore, PGRP2 showed bactericidal activity in bigfin reef squid (Li et al., 2022) and Hawaiian bobtail squid (Troll et al., 2010). Other secreted PGRPs also showed bactericidal activity in different species, including some bivalves (Kong et al., 2018; Huang et al., 2019; Yang et al., 2019), teleosts (Li et al., 2007; Kim et al., 2010; Li et al., 2020), and mammals (Lu et al., 2006). Taken together, our data suggest that ANG-expressed Pgrp2 and Pgrp3l may participate in bacterial colonization and pigmentation during ANG growth in female bigfin reef squid. The potential roles of hemocyte-expressed Pgrp4 and Pgrp5 should be further examined with regard to bacterial colonization of the ANG.
Like PGRPs, the functions of the LBP/BPI family are diverse. LBPs typically bind to lipopolysaccharide (produced by gram-negative bacteria) to mediate the activation of immune cells triggering the production of inflammatory mediators (Fenton and Golenbock, 1998). In contrast, BPIs are bactericidal (Elsbach, 1998) in mammals. LBPs/BPIs are not conserved in invertebrates since no members of the LBP/BPI family have been found in some animals, such as arthropods and platyhelminths (Irazoqui et al., 2010; Baron et al., 2016). In the present study, two Lbp/Bpi genes were found in the ANG of bigfin reef squid, including secreted Bpi3 and Bpi3-like. Furthermore, these two genes had high transcriptomic levels in the bacterial colonization period, which gradually decreased during final ANG maturation (bacterial pigmentation) (Figure 7). In Hawaiian bobtail squid, a low expression of Lbp1 mRNA was observed in squid without bacterial colonization by the symbiont V. fischeri compared with squid with bacterial colonization (Chun et al., 2006). Furthermore, immunofluorescence has shown that strong signals of LBP1 proteins are present within the crypts that house the colonizing symbionts (Chun et al., 2008). In contrast, both BPI2 and BPI4 are expressed in the epithelia of different tissues facing environmental microbes in Hawaiian bobtail squid (Chen et al., 2017). Other secreted LBPs/BPIs also show bactericidal activity and sensitivity to bacterial/lipopolysaccharide challenge in different species, including sea cucumber (Shao et al., 2015), some bivalves (Mao et al., 2013), and teleosts (Tang et al., 2015; Liu et al., 2021). Taken together, these data suggest that LBP/BPIs may be involved in protecting the surface from biofouling by different microbes but that their roles are not restricted to symbiont homeostasis in cephalopods.
In light organ of Hawaiian bobtail squid, once symbioses are established, the host must regulate symbiotic V. fischeri to prevent excessive replication and overutilization of host resources (Nyholm and McFall-Ngai, 2021). In contrast, the maintenance of the bacterial consortium in the ANG may be not only regulated by host selection but also coordinated by bacterial consortium competition. Therefore, the reduced number of genes encoding PPRs/PRMs and their receptors that participate in the host (ANG) regulatory mechanism may be associated with the high complexity of the bacterial consortium in the ANG compared with the single population of bacteria in the light organ.
Although the host immunity plays an important role during bacterial colonization of the ANG in bigfin reef squid, our results showed that most immune-related genes had a reduced transcriptomic level in the bacterial pigmentation compared with the bacterial colonization (Figures 6, 7). According to the KEGG enrichment results, most of the genes found such as those involved in apoptosis, pattern recognition receptors (Toll and Imd signaling pathway, Toll-like receptor signaling pathway, and RIG-I-like receptor signaling pathway), cytokines (TNF signaling pathway and IL-17 signaling pathway), and immune-mediated transcription factor (NF-kB signaling pathway) (Figure 6). These pathways also play roles in the cephalopod ANG, as exemplified by the immune-related pfam domains in Hawaiian bobtail squid (Belcaid et al., 2019) and the Toll/NF-kB family in cuttlefish (Sepia officinalis) (Cornet et al., 2015). In contrast, the low transcriptomic levels of some immune-related genes during ANG growth may be due to the reduced proportion of the outer layer of the ANG in the whole organ as its development processes. A similar expression pattern is also found in our previous study in transferrin (Tf) (Li et al., 2019). Tf is mainly expressed in the outer layer of ANG during ANG growth, and qPCR analysis showed that Tf expression levels decreased during ANG growth (Li et al., 2019). Thus, these immune-related genes who had reduced expression levels during ANG growth may be localized in the outer layer of ANG. Further studies of distribution of these genes are required for a better understanding of the host–bacteria interaction in bigfin reef squid.
The most dominant KEGG pathway results indicated that lipid metabolism and metabolism of flora fermentation are important for the regulation of bacterial pigmentation in bigfin reef squid (Figure 6). Studies on host control mechanisms in mammals that shape the intestinal (colonic) microbiota to be beneficial suggest that colonic epithelial cells play a central role in the process (reviewed in Litvak et al., 2018). In Hawaiian bobtail squid, acidic pH in symbiotic light organs is a cue that stimulates an anticipatory defensive response during the initiation of symbiosis (Schwartzman et al., 2019). These data are of some interest because immune functions have typically been used to explain symbioses in most previous studies (Gerardo et al., 2020). In these cases, microorganism–host cell and microorganism–microorganism communications are also involved in the regulatory mechanism of homeostasis through non-immune pathways. In mammals, obligate anaerobic bacteria convert food fiber into fermentation products, including the short-chain fatty acids butyrate, propionate, and acetate, to maintain differentiated colonocytes (intestinal epithelial cells) in a certain metabolic state (Alex et al., 2013; Byndloss et al., 2017). The metabolism of these colonocytes is characterized by high oxygen consumption, which maintains epithelial hypoxia to limit the amount of oxygen diffusing from the host into the gut lumen (Byndloss et al., 2017). In contrast, pathogens use their virulence factors to ablate a homeostatic surface of colonocytes and alter colonocyte metabolism to obtain critical resources from host cells to allow them to compete against the resident gut microbiota (reviewed in Litvak et al., 2018). These results suggest that metabolism functions as a control switch for epithelial cells, mediating a shift between homeostatic and dysbiotic communities.
In bigfin reef squid, based on the structural characteristics, several types of epithelial cells are found in the secondary tubules of the ANG during ANG pigmentation (Li et al., 2019). Our recent study (Li et al., 2022) and preliminary data (unpublished data) showed that the mRNA expression of some genes is variable in different color regions of the ANG. Furthermore, our present results indicate that the metabolism of flora fermentation is the dominant KEGG pathway in bacterial pigmentation (Figure 6). Therefore, we suggest that the metabolism of epithelial cells may shape the ANG microbiota in bigfin reef squid. Alternatively, the symbionts could be considered the nutrient source of the host under starvation in some symbioses, such as those involving the deep-sea tubeworm, Riftia pachyptila (Hinzke et al., 2019), and several bivalve species (Caro et al., 2009; König et al., 2015; Ponnudurai et al., 2017). This metabolic interaction includes substrate transfer to the symbiont by abundant host protein degradation-related proteins and nutrient allocation from symbiont to host by symbiont digestion (Ponnudurai et al., 2017; Hinzke et al., 2019). Thus, these protein degradation-related proteins may also be involved in bacterial digestion and further regulate the homeostasis of ANG and its symbiotic microbiota during bacterial pigmentation.
In this study, we examined the transcriptomic profiles of the ANG during female maturation in bigfin reef squid. Although host immunity plays an important role during bacterial colonization of the ANG, our results showed that most immune-related genes had a reduced transcriptomic level in bacterial pigmentation compared with bacterial colonization. Furthermore, our results indicated that lipid (fatty acid) metabolism and metabolism of flora fermentation are important for the regulation of bacterial pigmentation. These data are interesting because immune functions have typically been used to explain the symbioses in most previous studies (Gerardo et al., 2020).
This transcriptomic study provides new data to increase our knowledge of the mechanisms of bacterial selection and maintenance in the ANG association of bigfin reef squid. Additionally, as the non-annotated transcripts correspond to unknown novel transcripts or to unreviewed transcripts, the unannotated DEGs deserve attention in future functional analyses in bigfin reef squid. Future studies are needed to determine the full-length transcriptome of bigfin reef squid to generate a better reference database of the ANG transcripts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ , PRJNA814067.
The animal study was reviewed and approved by National Taiwan Ocean University Institutional Animal Care and Use Committee.
CC and H-WL collected the samples and conducted the histology and RNA analysis. P-WT conducted the transcriptome analysis. Y-CT and C-FC gave the comments. G-CW designed and conducted experiment, analyzed the data, and participated in compiling the main manuscript. All authors reviewed and approved the manuscript.
This work was partly supported by grants from the Ministry of Science and Technology (MOST 104-2923-B-019-001-MY4), National Science and Technology Council (NSTC 111-2326-B-019-001-MY3) and by the Center of Excellence for the Oceans, National Taiwan Ocean University, from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) of Taiwan.
We thank Mr. Cyuan-Hong Syu for preserving bigfin reef squid at Heping Island, Keelung City, Taiwan. An English of draft manuscript was corrected by Ms. Emily Corrigan and further corrected by American Journal Experts.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SY-Y declared a shared affiliation with the authors P-WT and Y-CT to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1026742/full#supplementary-material
Supplementary Table 1 | The DEGs (differentially expressed genes) involved in dominant KEGG pathways.
Supplementary Table 2 | The DEGs (differentially expressed genes) of immune-related genes.
Supplementary Figure 1 | Expression-dependent N50 (ExN50) as calculated against a fraction of the total expressed data (Ex). ExN50 at the point of assembly saturation (84%) and traditional N50 are highlighted.
Supplementary Figure 2 | Functional annotation of DEGs in the GO database. The counts of differentially expressed genes (DEGs) matched in GO (Gene Ontology) categories. The x-axis indicates 45 GO categories. The y-axis indicates the number of DEGs that were matched.
Supplementary Figure 3 | KEGG enrichment analysis of DEGs. A) The 32 functionally enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways found in the analysis of differentially expressed genes (DEGs) in three different stages of ANGs (stage 1, stage 3, and stage 4).
Alex S., Lange K., Amolo T., Grinstead J. S., Haakonsson A. K., Szalowska E., et al. (2013). Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 33, 1303–1316. doi: 10.1128/MCB.00858-12
Asker D., Beppu T., Ueda K. (2007). Unique diversity of carotenoid-producing bacteria isolated from misasa, a radioactive site in Japan. Appl. Microbiol. Biotechnol. 77, 383–392. doi: 10.1007/s00253-007-1157-8
Bai L., Zhou K., Li H., Qin Y., Wang Q., Li W. (2020). Bacteria-induced IMD-Relish-AMPs pathway activation in Chinese mitten crab. Fish Shellfish Immunol. 106, 866–875. doi: 10.1016/j.fsi.2020.08.046
Baron O. L., Deleury E., Reichhart J. M., Coustau C. (2016). The LBP/BPI multigenic family in invertebrates: Evolutionary history and evidences of specialization in mollusks. Dev. Comp. Immunol. 57, 20–30. doi: 10.1016/j.dci.2015.11.006
Belcaid M., Casaburi G., McAnulty S. J., Schmidbaur H., Suria A. M., Moriano-Gutierrez S., et al. (2019). Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc. Natl. Acad. Sci. U. S. A. 116, 3030–3035. doi: 10.1073/pnas.1817322116
Bloodgood R. A. (1977). The squid accessory nidamental gland: Ultrastructure and association with bacteria. Tissue. Cell 9, 197–208. doi: 10.1016/0040-8166(77)90016-7
Boettcher K. J., Ruby E. G., McFall-Ngai M. J. (1996). Bioluminescence in the symbiotic squid Euprymna scolopes Is controlled by a daily biological rhythm. . Comp. Physiol. A 179, 65–73. doi: 10.1007/BF00193435
Buchfink B., Reuter K., Drost H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Byndloss M. X., Olsan E. E., Rivera-Chávez F., Tiffany C. R., Cevallos S. A., Lokken K. L., et al. (2017). Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Sci. 357, 570–575. doi: 10.1126/science.aam9949
Cantalapiedra C. P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Caro A., Got P., Bouvy M., Troussellier M., Gros O. (2009). ). effects of long-term starvation on a host bivalve (Codakia orbicularis, lucinidae) and its symbiont population. Appl. Environ. Microbiol. 75, 3304–3313. doi: 10.1128/AEM.02659-08
Castellanos-Martínez S., Arteta D., Catarino S., Gestal C. (2014). De novo transcriptome sequencing of the Octopus vulgaris hemocytes using illumina RNA-seq technology: Response to the infection by the gastrointestinal parasite Aggregata octopiana. PloS One 9, e107873. doi: 10.1371/journal.pone.0107873
Castillo M. G., Salazar K. A., Joffe N. R. (2015). The immune response of cephalopods from head to foot. Fish Shellfish Immunol. 46, 145–160. doi: 10.1016/j.fsi.2015.05.029
Cerf-Bensussan N., Gaboriau-Routhiau V. (2010). The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 10, 735–744. doi: 10.1038/nri2850
Chen F., Krasity B. C., Peyer S. M., Koehler S., Ruby E. G., Zhang X., et al. (2017). Bactericidal permeability-increasing proteins shape host-microbe interactions. mBio 8, e00040–e00017. doi: 10.1128/mBio.00040-17
Chen C., Li H. W., Ku W. L., Lin C. J., Chang C. F., Wu G. C. (2018). Two distinct vitellogenin genes are similar in function and expression in the bigfin reef squid Sepioteuthis lessoniana. Biol. Reprod. 99, 1034–1044. doi: 10.1093/biolre/ioy131
Chun C. K., Scheetz T. E., Bonaldo ,.M., Brown B., Clemens A., Crookes-Goodson W. J., et al. (2006). An annotated cDNA library of juvenile euprymna scolopes with and without colonization by the symbiont vibrio fischeri. . BMC Genomics 7, 154. doi: 10.1186/1471-2164-7-154
Chun C. K., Troll J. V., Koroleva I., Brown B., Manzella L., Snir E., et al. (2008). Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-Vibrio association. Proc. Natl. Acad. Sci. U. S. A. 105, 11323–11328. doi: 10.1073/pnas.0802369105
Collins A., Schleicher T., Rader B., Nyholm S. (2012). Understanding the role of host hemocytes in a squid/Vibrio symbiosis using transcriptomics and proteomics. Front. Immunol. 3. doi: 10.3389/fimmu.2012.00091
Cornet V., Henry J., Corre E., Le Corguillé G., Zatylny-Gaudin C. (2015). The Toll/NF-κB pathway in cuttlefish symbiotic accessory nidamental gland. Dev. Comp. Immunol. 53, 42–46. doi: 10.1016/j.dci.2015.06.016
Dieser M., Greenwood M., Foreman C. M. (2010). Carotenoid pigmentation in antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 42, 396–405. doi: 10.1657/1938-4246-42.4.396
Dziarski R. (2004). Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 40, 877–886. doi: 10.1016/j.molimm.2003.10.011
Dziarski R., Gupta D. (2006). The peptidoglycan recognition proteins (PGRPs). Genome. Biol. 7, 232. doi: 10.1186/gb-2006-7-8-232
Elsbach P. (1998). The bactericidal/permeability-increasing protein (BPI) in antibacterial host defense. J. Leukoc. Biol. 64, 14–18. doi: 10.1002/jlb.64.1.14
Fenton M. J., Golenbock D. T. (1998). LPS-binding proteins and receptors. J. Leukoc. Biol. 64, 25–32. doi: 10.1002/jlb.64.1.25
Gerardo N. M., Hoang K. L., Stoy K. S. (2020). Evolution of animal immunity in the light of beneficial symbioses. Philos. Trans. R. Soc Lond. B Biol. Sci. 375, 20190601. doi: 10.1098/rstb.2019.0601
Gómez G. D., Balcázar J. L. (2008). A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52, 145–154. doi: 10.1111/j.1574-695X.2007.00343.x
Goodson M. S., Kojadinovic M., Troll J. V., Scheetz T. E., Casavant T. L., Soares M. B., et al. (2005). Identifying components of the NF-kappaB pathway in the beneficial euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl. Environ. Microbiol. 71, 6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005
Hinzke T., Kleiner M., Breusing C., Felbeck H., Häsler R., Sievert S. M., et al. (2019). Host-microbe interactions in the chemosynthetic Riftia pachyptila symbiosis. mBio 10, e02243–e02219. doi: 10.1128/mBio.02243-19
Huang J. D., Lee S. Y., Chiang T. Y., Lu C. C., Lee M. F. (2018). Morphology of reproductive accessory glands in female Sepia pharaonis (Cephalopoda: Sepiidae) sheds light on egg encapsulation. J. Morphol. 279, 1120–1131. doi: 10.1002/jmor.20835
Huang Y., Pan J., Li X., Ren Q., Zhao Z. (2019). Molecular cloning and functional characterization of a short peptidoglycan recognition protein from triangle-shell pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol. 86, 571–580. doi: 10.1016/j.fsi.2018.12.007
Ikuta T., Tame A., Saito M., Aoki Y., Nagai Y., Sugimura M., et al. (2019). Identification of cells expressing two peptidoglycan recognition proteins in the gill of the vent mussel, Bathymodiolus septemdierum. Fish Shellfish Immunol. 93, 815–822. doi: 10.1016/j.fsi.2019.08.022
Irazoqui J. E., Urbach J. M., Ausubel F. M. (2010). Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10, 47–58. doi: 10.1038/nri2689
Johler S., Stephan R., Hartmann I., Kuehner K. A., Lehner A. (2010). Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 76, 1053–1061. doi: 10.1128/AEM.01420-09
Kaufman M. R., Ikeda Y., Patton C., van Dykhuizen G., Epel D. (1998). Bacterial symbionts colonize the accessory nidamental gland of the squid Loligo opalescens via horizontal transmission. Biol. Bull. 194, 36–43. doi: 10.2307/1542511
Kerwin A. H., Gromek S. M., Suria A. M., Samples R. M., Deoss D. J., O'Donnell K., et al. (2019). Shielding the next generation: Symbiotic bacteria from a reproductive organ protect bobtail squid eggs from fungal fouling. mBio 10, 5. doi: 10.1128/mBio.02376-19
Kerwin A. H., McAnulty S. J., Nyholm S. V. (2021). Development of the accessory nidamental gland and associated bacterial community in the Hawaiian bobtail squid, euprymna scolopes. Biol. Bull. 240, 205–218. doi: 10.1086/713965
Kerwin A. H., Nyholm S. V. (2017). Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and developing eggs. Environ. Microbiol. 19, 1463–1475. doi: 10.1111/1462-2920.13665
Kim M. Y., Jang J. H., Lee J. W., Cho J. H. (2010). Molecular cloning and characterization of peptidoglycan recognition proteins from the rockfish, sebastes schlegeli. Fish Shellfish Immunol. 28, 632–639. doi: 10.1016/j.fsi.2009.12.023
Kong X., Liu H., Li Y., Zhang H. (2018). Two novel short peptidoglycan recognition proteins (PGRPs) from the deep sea vesicomyidae clam Archivesica packardana: Identification, recombinant expression and bioactivity. Front. Physiol. 9, 1476. doi: 10.3389/fphys.2018.01476
König S., Le Guyader H., Gros O. (2015). Thioautotrophic bacterial endosymbionts are degraded by enzymatic digestion during starvation: Case study of two lucinids Codakia orbicularis and C. orbiculata. Microsc. Res. Tech. 78, 173–179. doi: 10.1002/jemt.22458
Krasity B. C., Troll J. V., Lehnert E. M., Hackett K. T., Dillard J. P., Apicella M. A., et al. (2015). Structural and functional features of a developmentally regulated lipopolysaccharide-binding protein. mBio 6, e01193–e01115. doi: 10.1128/mBio.01193-15
Kremer N., Philipp E. E., Carpentier M. C., Brennan C. A., Kraemer L., Altura M. A., et al. (2013). Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. . Cell. Host. Microbe 14, 183–194. doi: 10.1016/j.chom.2013.07.006
Li H. W., Chen C., Kuo W. L., Lin C. J., Chang C. F., Wu G. C. (2019). The characteristics and expression profile of transferrin in the accessory nidamental gland of the bigfin reef squid during bacteria transmission. Sci. Rep. 9, 20163. doi: 10.1038/s41598-019-56584-8
Li H. W., Kuo W. L., Chen C., Tseng Y. C., Chang C. F., Wu G. C. (2022). The characteristics and expression profile of peptidoglycan recognition protein 2 in the accessory nidamental gland of the bigfin reef squid during bacterial colonization. Front. Mar. Sci. 9, 825267. doi: 10.3389/fmars.2022.825267
Lindgren A. R., Pankey M. S., Hochberg F. G., Oakley T. H. (2012). Multi-gene phylogeny of Cephalopoda supports convergent morphological evolution in association with multiple habitat shifts in the marine environment. A BMC Evol. Biol. 12, 129–129. doi: 10.1186/1471-2148-12-129
Litvak Y., Byndloss M. X., Bäumler A. J. (2018). Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076. doi: 10.1126/science.aat9076
Liu G. Y., Essex A., Buchanan J. T., Datta V., Hoffman H. M., Bastian J. F., et al. (2005). Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215. doi: 10.1084/jem.20050846
Liu H., Wu C., Matsuda Y., Kawabata S., Lee B. L., Söderhäll K., et al. (2011). Peptidoglycan activation of the proPO-system without a peptidoglycan receptor protein (PGRP)? Dev. Comp. Immunol. 35, 51–61. doi: 10.1016/j.dci.2010.08.005
Liu Y., Zha H., Han X., Yu S., Chai Y., Zhong J., et al. (2021). Molecular characterization and functional analysis of the bactericidal permeability-increasing protein/LPS-binding protein (BPI/LBP) from roughskin sculpin (Trachidermus fasciatus). Dev. Comp. Immunol. 123, 104133. doi: 10.1016/j.dci.2021.104133
Li X., Wang S., Qi J., Echtenkamp S. F., Chatterjee R., Wang M., et al. (2007). Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immun. 27, 518–529. doi: 10.1016/j.immuni.2007.07.020
Li X., Yuan S., Sun Z., Lei L., Wan S., Wang J., et al. (2020). Gene identification and functional analysis of peptidoglycan recognition protein from the spotted sea bass (Lateolabrax maculatus). Fish Shellfish Immunol. 106, 1014–1024. doi: 10.1016/j.fsi.2020.08.041
Lum-Kong A. (1992). A histological study of the accessory reproductive organs of female Loligo forbesi (Cephalopoda: Loliginidae). J. Zool. 226, 469–490. doi: 10.1111/j.1469-7998.1992.tb07493.x
Lum-Kong A., Hastings T. S. (1992). The accessory nidamental glands of Loligo forbesi (Cephalopoda: Loliginidae): characterization of symbiotic bacteria and preliminary experiments to investigate factors controlling sexual maturation. J. Zool. 228, 395–403. doi: 10.1111/j.1469-7998.1992.tb04443.x
Lupp C., Urbanowski M., Greenberg E. P., Ruby E. G. (2003). The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50, 319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x
Lu X., Wang M., Qi J., Wang H., Li X., Gupta D., et al. (2006). Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 281, 5895–5907. doi: 10.1074/jbc.M511631200
Mao Y., Zhou C., Zhu L., Huang Y., Yan T., Fang J., et al. (2013). Identification and expression analysis on bactericidal permeability-increasing protein (BPI)/lipopolysaccharide-binding protein (LBP) of ark shell, scapharca broughtonii. Fish Shellfish Immunol. 35, 642–652. doi: 10.1016/j.fsi.2013.05.025
McAnulty S. J., Nyholm S. V. (2016). The role of hemocytes in the Hawaiian bobtail squid, Euprymna scolopes: A model organism for studying beneficial host-microbe interactions. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.02013
Mohammadi M., Burbank L., Roper M. C. (2012). Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 78, 6859–6865. doi: 10.1128/AEM.01574-12
Montaño A. M., Tsujino F., Takahata N., Satta Y. (2011). Evolutionary origin of peptidoglycan recognition proteins in vertebrate innate immune system. BMC Evol. Biol. 11, 79. doi: 10.1186/1471-2148-11-79
Nyholm S. V., Deplancke B., Gaskins H. R., Apicella M. A., McFall-Ngai M. J. (2002). Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68, 5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002
Nyholm S. V., Graf J. (2012). Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827. doi: 10.1038/nrmicro2894
Nyholm S. V., McFall-Ngai M. J. (2021). A lasting symbiosis: How the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat. Rev. Microbiol. 19, 666–679. doi: 10.1038/s41579-021-00567-y
Pichon D., Gaia V., Norman M. D., Boucher-Rodoni R. (2005). Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (Cephalopoda: Loliginidae and idiosepiidae). Mar. Biol. 147, 1323–1332. doi: 10.1007/s00227-005-0014-5
Ponnudurai R., Kleiner M., Sayavedra L., Petersen J. M., Moche M., Otto A., et al. (2017). Metabolic and physiological interdependencies in the bathymodiolus azoricus symbiosis. ISME J. 11, 463–477. doi: 10.1038/ismej.2016.124
Ram S., Mitra M., Shah F., Tirkey S. R., Mishra S. (2020). Bacteria as an alternate biofactory for carotenoid production: A review of its applications, opportunities and challenges. J. Funct. Foods. 67, 103867. doi: 10.1016/j.jff.2020.103867
Rosenstiel P., Philipp E. E., Schreiber S., Bosch T. C. (2009). Evolution and function of innate immune receptors: insights from marine invertebrates. J. Innate. Immun. 1, 291–300. doi: 10.1159/000211193
Royet J., Gupta D., Dziarski R. (2011). Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat. Rev. Immunol. 11, 837–851. doi: 10.1038/nri3089
Ruby E. G., Asato L. M. (1993). Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159, 160–167. doi: 10.1007/BF00250277
Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold. Spring. Harb. Perspect. Med. 2, a012427. doi: 10.1101/cshperspect.a012427
Salazar K. A., Joffe N. R., Dinguirard N., Houde P., Castillo M. G. (2015). Transcriptome analysis of the white body of the squid euprymna tasmanica with emphasis on immune and hematopoietic gene discovery. PloS One 10, e0119949. doi: 10.1371/journal.pone.0119949
Schwartzman J. A., Lynch J. B., Flores Ramos S., Zhou L., Apicella M. A., Yew J. Y., et al. (2019). Acidic pH promotes lipopolysaccharide modification and alters colonization in a bacteria-animal mutualism. Mol. Microbiol. 112, 1326–1338. doi: 10.1111/mmi.14365
Shao Y., Li C., Che Z., Zhang P., Zhang W., Duan X., et al. (2015). Cloning and characterization of two lipopolysaccharide-binding protein/bactericidal permeability–increasing protein (LBP/BPI) genes from the sea cucumber Apostichopus japonicus with diversified function in modulating ROS production. Dev. Comp. Immunol. 52, 88–97. doi: 10.1016/j.dci.2015.04.015
Tang L., Liang Y., Jiang Y., Liu S., Zhang F., He X., et al. (2015). Identification and expression analysis on bactericidal permeability-increasing protein/lipopolysaccharide-binding protein of blunt snout bream, megalobrama amblycephala. Fish Shellfish Immunol. 45, 630–640. doi: 10.1016/j.fsi.2015.05.013
Troll J. V., Adin D. M., Wier A. M., Paquette N., Silverman N., Goldman W. E., et al. (2009). Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell. Microbiol. 11, 1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x
Troll J. V., Bent E. H., Pacquette N., Wier A. M., Goldman W. E., Silverman N., et al. (2010). Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 12, 2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x
Tseng P. W., Wu G. C., Kuo W. L., Tseng Y. C., Chang C. F. (2022). The ovarian transcriptome at the early stage of testis removal-induced male-to-female sex change in the protandrous black porgy acanthopagrus schlegelii. Front. Genet. 13. doi: 10.3389/fgene.2022.816955
Vidal E. A. G., Villanueva R., Andrade J. P., Gleadall I. G., Iglesias J., Koueta N., et al. (2014). Cephalopod culture: current status of main biological models and research priorities. Adv. Mar. Biol. 67, 1–98. doi: 10.1016/B978-0-12-800287-2.00001-9
Yang S. H., Chen C., Hsieh Y. E., Yang S. Y., Li H. W., Ching T. Y., et al. (2021). Bacterial dynamics in the accessory nidamental gland of Sepioteuthis lessoniana throughout maturation. Microbes Environ. 36, ME21030. doi: 10.1264/jsme2.ME21030
Keywords: ANG, cephalopod, innate immunity, symbiosis, bacterial colonization
Citation: Tseng P-W, Li H-W, Chen C, Tseng Y-C, Chang C-F and Wu G-C (2023) Transcriptomic profile of symbiotic accessory nidamental gland during female maturation in bigfin reef squid. Front. Mar. Sci. 9:1026742. doi: 10.3389/fmars.2022.1026742
Received: 24 August 2022; Accepted: 14 December 2022;
Published: 09 January 2023.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Changkao Mu, Ningbo University, ChinaCopyright © 2023 Tseng, Li, Chen, Tseng, Chang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan-Chung Wu, Z2N3dUBtYWlsLm50b3UuZWR1LnR3
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.