
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 20 October 2022
Sec. Marine Biotechnology and Bioproducts
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1023287
This article is part of the Research Topic Biotechnology of Marine Microorganisms View all 7 articles
 Aikaterini A. Zompra1*†
Aikaterini A. Zompra1*† Styliani A. Chasapi1†
Styliani A. Chasapi1† Matthew S. Twigg2
Matthew S. Twigg2 Karina Salek3
Karina Salek3 Ioannis Anestopoulos4
Ioannis Anestopoulos4 Alex Galanis4
Alex Galanis4 Aglaia Pappa4
Aglaia Pappa4 Tony Gutierrez3
Tony Gutierrez3 Ibrahim M. Banat2
Ibrahim M. Banat2 Roger Marchant2
Roger Marchant2 Stephen R. Euston5,6*
Stephen R. Euston5,6* Mihalis I. Panayiotidis7*
Mihalis I. Panayiotidis7* Georgios A. Spyroulias1*
Georgios A. Spyroulias1*Synthetic surfactants are used in several industries, including manufacturing, pharmaceutical and cosmetic’s, food and feed, agriculture, petroleum and environmental remediation for their ability to adsorb to fluid and solid-water interfaces. However, their widespread use and their synthetic preparation through environmentally unfavorable processes counterbalances the value of this class of reagents. This fact has stimulated new efforts to exploit natural sources of surfactants, such as new classes of bacterial systems or manipulation of existing biological systems, that may produce, through an environmentally friendly process, new biodegradable surfactants and emulsifiers of high commercial value. A downside of microbial production of biobased chemicals such as these types of chemicals, is that their fermentation often yields crude materials consisting of several bioproducts with complex physical and chemical properties. Extraction, identification, and efficient characterization of biosurfactants from a crude mixture of biomolecules requires carefully designed, and detailed analytical processes using state-of-the-art methods. The purpose of this review article is to present the current state-of-the-art and future outlook on the various multidisciplinary biophysical methods applied in the discovery, extraction identification, and in-depth characterization of microbially-produced surface‐active compounds.
Surfactants are surface active agents (SAAs) that can reduce the surface tension between two phases (normally fluid or solid). SAAs facilitate the formation of oil in water and/or water in oil emulsions and promote the creation of both stable gels and foams (Naughton et al., 2019). Surfactants are examples of amphiphiles (from the Greek amphis = both and philia= friendship), compounds with distinct hydrophilic and hydrophobic parts (Ghosh et al., 2020). Their amphiphilic nature allows surfactants to associate at the interface between aqueous and non-aqueous substances, thereby facilitating their application in areas related with solubilization of hydrophobic agents, detergency and foaming, emulsification, wetting and dispersion (Kitamoto et al., 2002; Amaral et al., 2010; Marchant and Banat, 2012). Over the years, synthetic surfactants have been utilized widely by several industries, such as medical manufacturing and pharmaceutical, food, agriculture, petroleum and environmental remediation. The environment is negatively affected by this widespread application of synthetic SAAs and as a result these chemicals are becoming increasingly unpopular with consumers. Environmental concerns and the growing global demand for sustainable technologies have led to a new aim and researchers try to replace synthetic compounds with biosurfactants that are produced from renewal resources, and they are biodegradable (De Almeida et al., 2016; Farias et al., 2021).
Biosurfactants have attracted much attention in recent years and the demand for those that are microbially-produced for use in industrial processes and products is increasing. They are categorized according to their microbial origin, molecular weight, composition, and properties. Compounds with high-molecular-weight (HMW) usually reported as bioemulsifiers, include, proteins, lipoproteins, lipopolysaccharides, and heteropolysaccharides. Compounds with low-molecular-weight (LMW) are usually termed biosurfactants, and include glycolipids (rhamnolipids, trehalose lipids, sophorolipids and mannosylerythritol lipids) and lipopeptides (Rosenberg and Ron, 1999; Mukherjee et al., 2006; Abdel‐Mawgoud et al., 2010; Amaral et al., 2010; Sriram et al., 2011).
The methodologies based on analytical chemistry which are used for the reporting of SAAs detection vary widely from simple thin layer chromatography (TLC) and colorimetric assays that provide an approximate indication of the type of biosurfactant, to advanced methods, such as high-resolution mass and NMR spectroscopic methods that give the necessary information on the molecular masses and detection of structural features. However, there is a lack of consistent methodologies and/or regulatory protocols for detection and/or structural characterization of these molecules according to their molar weight (LMW, HMW) and their classification (rhamnolipids, lipopeptides, lipoproteins, proteins etc.). The increasing demand for environmentally friendly biosurfactants/bioemulsifiers for use in industrial processes, and the absence of accurate analytical methods for their detection, requires further effort in the design of standard procedures for their identification and classification. Also, it has been noted (Twigg et al., 2021) that many reports on microbially-produced SAAs are detrimental to the field as they lack robust experimental evidence generated by validated analytical methodologies (Priji et al., 2017).
In this review article, we present and critically evaluate a wide range of analytical methodologies applied for the characterization of SAAs from microbial sources. Significant efforts have been made to identify those techniques best suited to molecular weight determination, composition, sequencing and highly-resolved structure elucidation.
Lipopeptides (LPS) are a category of microbial secondary metabolites with exceptional biological properties and acting as amphiphilic compounds. They are isolated from cell broth of different kinds of microorganisms, such as from the genera Bacillus, Pseudomonas, and Streptomyces (Raaijmakers et al., 2010). The hydrophobic moieties include lipophilic fatty acids and non-polar amino acids, while their hydrophilic part is mainly consisted of polar amino acids.
Polymeric surfactants are high molecular weight biosurfactants/bio-emulsifiers, produced from different bacteria and contain proteins, lipoproteins, polysaccharides, lipo-polysaccharides, or complex compounds with many of these structural groups and several of them are not fully structural characterized. The most studied polymeric surfactants are Emulsan and Alasan. Emulsan is a lipoheteropolysaccharide isolated from Acinetobacter calcoaceticus RAG-1 (ATCC 31012) with a molecular weight of around 1,000 kDa, whereas Alasan is a complex of a protein with an anionic polysaccharide and its molecular weight also found at 1,000 kDa. This polymeric complex is produced from Acinetobacter radioresistens (Rosenberg et al., 1979; Navon-Venezia et al., 1995). Many other polymeric biosurfactants have been found but several of them are still partially or completely uncharacterized. Surfactin and surfactin-like lipopeptides produced by members of the genus Bacillus are one of the most significant categories of these natural produced SAAs. Surfactin is a cyclic heptapeptide conjugated to a β-hydroxy fatty acid with 13 to15 C atoms via a lactone bond and this lipopeptide is produced by several strains of Bacillus isolated from terrestrial and marine environments. Iturins, another important class of lipopeptides, consisted of a heptapeptide skeleton linked to a C13 to C17 β-amino fatty acid chain. Iturins, show strong in vitro fungitoxicity with their ability to form ion-conducting pores on membranes of fungi and they present different structural characteristics in terms of amino acid sequence and in the length and branching of their fatty acid chain. Fengycin is composed of a β-hydroxy fatty acid chain attached to a decapeptide, also forming a cyclic lactone ring (Aranda et al., 2005) (Ongena and Jacques, 2008). Two major lipopeptides of fengycin group are fengycin A and fengycin B with a Val and an Ala at position 6, respectively (Vater et al., 2002; Sun et al., 2006).
Concerning microbial glycolipids, four classes are the most important and studied: rhamnolipids, sophorolipids, trehalose lipids and mannosylerythritol lipids. These differ in the nature of the hydrophilic sugar head group (rhamnose, sophorose, trehalose or mannopyranosyl-D-erythritol) as well as the number, chain length and degree of unsaturation of the lipid hydrophobic groups (Twigg et al., 2018; Tripathi et al., 2019).
As mentioned, low resolution analytical chemistry methodologies can be used for the analysis of biosurfactants, such as thin layer chromatography (TLC) and colorimetric assays, to more advanced methods, high-resolution mass, and NMR spectroscopic methods, that could give more information on the molecular mass and their structural characteristics. Typical chromatographic techniques used to characterize and quantify molecules, include TLC, preparative TLC (PTLC), column chromatography (CC) or advanced semi-preparative/preparative HPLC. Careful selection of the method should be performed taking in consideration the properties of the target analyte(s) compound, such as its solubility, stability, charge, and molecular size. Thus, the careful selection of chromatographic methods and the stationary phases to be used are crucial for the analysis and/or purification of complex biosurfactant mixtures, such as a mixture of lipopeptides, glycolipids. The analytical tools for the evaluation of identification and structural characterization of lipopeptides, glycolipids and HMW bioemulsifiers are summarized in Anestopoulos et al. (2020).
Thin layer chromatography (TLC) is a fast and simple chromatographic tool and can be used to detect the presence of lipopeptides, rhamnolipids and sophorolipids while preparative TLC (PTLC) is suitable for the purification of small quantities of these chemicals (Symmank et al., 2002). The visualization of the lipopeptides spots on TLC plates can be performed by the following three chromogenic reagents: ninhydrin solution (0.5% ninhydrin in acetone) for the detection of amino acids (amino groups); phenol-sulphuric acid solution (3% phenol and 5% sulphuric acid in absolute ethanol) for the detection of carbohydrates, and iodine vapors for detecting lipids (Razafindralambo et al., 1993; Das et al., 2009). Lipopeptides have specific retention factor (Rf) values depending on solvent or solvent mixture system. Therefore, these specific values can be used in compound identification by comparing the unidentified sample with known lipopeptides or biosurfactants (Smyth et al., 2014). Also, TLC is a simple method for the detection of glycolipids and could assist with providing possible different structural types of the present glycolipids (de Koster et al., 1994). A typical solvent system for developing rhamnolipids is chloroform:methanol:acetic acid (6.5:1.5:0.2, v/v/v) while for sophorolipids the solvent system is chloroform:methanol:water (6.5:1.5:0.2, v/v/v) (Asmer et al., 1988; Déziel et al., 2000). In Table 1, several examples are provided, including TLC and PTLC methodology towards characterization of biosurfactant. Although TLC method is quick and relatively inexpensive, it provides little to no detailed information regarding congener composition, is non-quantitative, and can provide false positive results depending on detection method and purity of the sample.

Table 1 Examples of applied analytical methodologies for the efficient structural characterization of different biosurfactants/bioemulsifiers types.
The isolation and purification of the individual lipopeptide biosurfactant from the production mixture is required for the successful structural characterization. High-performance liquid chromatography (HPLC) is an excellent chromatographic method for the separation of peptides/lipopeptides, and individual isolation (of each peptide) could be achieved using preparative or semi-preparative chromatographic columns (Aguilar, 2004; Tulla-Puche et al., 2015). HPLC and ultra-performance liquid chromatography (UPLC) systems have been also used for quantitative detection of lipopeptides derived from marine microorganisms (Biniarz and Łukaszewicz, 2017). HPLC analysis is usually performed using C8 or C18 reverse-phase columns with a water/acetonitrile (H2O/ACN) gradient elution system. Common detection methods for the samples absorbed onto stationary phase and then eluted from HPLC columns are UV spectroscopy at λ=214-220 nm and 274, 280 nm for detection of peptide bonds and aromatic amino acids, respectively. Furthermore, performing refractive index (RI), fluorescence, electrical conductivity and MS as a detection method could be applied. The application of HPLC method has been reported for the characterization, purification, and even quantification and of several well-known lipopeptides (Table 1) (Janek et al., 2010; Kim et al., 2010).
HPLC can also be applied for the separation of glycolipids when coupled with evaporative light scattering detector (ELSD) or mass spectrometry (MS) and this could provide useful data concerning the identification and quantification of them. Furthermore, coupling HPLC and UV (HPLC-UV) can be applied when the glycolipid biosurfactants have been derivatized to p-bromophenacyl esters (Schenk, 1995; Mata-Sandova et al., 1999). However, the modification that provides the most reliable information for the identification and characterization of biosurfactants, is when an HPLC system has been coupled to a mass spectrometer (MS) or to tandem MS (MS/MS) (Rudden et al., 2015; Smyth et al., 2016).
Size exclusion chromatography (SEC) is often used for the analysis of the molecular weight and structural characterization of macromolecules such as proteins. Also, it is considered as a powerful tool for characterization of polysaccharides. This modern column chromatography method provides information about the whole molecular weight distribution since it can separate molecules according to their hydrodynamic volumes. Thus, molecules are separated because of their exclusion from pores in the column packing material during their migration through the column matrix. First, elute larger molecules since they are too large to be retained in pores on the column and later the smaller ones, since they interact more with the column material. SEC is the most popular approach for molecular weight determination and separation of macromolecules since it has a simple operating procedure, high versatility and there are a wide range of matrices commercially available. Another important advantage of SEC is that analysis and separation of complex samples could be performed under optimized conditions allowing maintenance of stability and activity of the analytes and preserving resolution of analysis. For SEC analysis, the common detectors are Refractive Index (RI) or UV, and the elution volumes could be determined against high molecular weight standards. Using this method has shown that polymeric bioemulsifiers can aggregate according to the ionic strength of the solution they are dissolved in, and this can affect how they are resolved in a SEC column and, in turn, assigned a molecular weight (Gutierrez et al., 2009). SEC (and gel permeation chromatography, GPC) coupled with multi angle light scattering (SEC-MALS) can be used to determine the absolute molecular weight. SEC-MALS is one of the most robust tools for the molecular characterization of polymeric materials (Wyatt, 1993; Tarazona and Saiz, 2003). This sophisticated analysis has proved successful for the characterization associating and micelle-forming polymers such as the microbial polysaccharide, xanthan in dilute solutions (Matsuda et al., 2009). In another example, analysis of the marine derived polysaccharide, carrageenan (kappa- and iota) by GPC-MALS revealed a molecular weight dependence of the coil to helix transition. These conformations (coil and helix) of the degraded and non-degraded carrageenan samples were studied using MALS coupled to GPC, and their weight-average molecular weight, MW. More specifically, the iota-carrageenan presented a Mw, helix at 738 000Da, coil at 332 000Da and the kappa-carrageenan a MW, helix to be 468000 Da and the coil at 324000 Da for the for non-degraded and sonicated samples (Viebke et al., 1995). Another important class of polysaccharides (with surfactant characteristics found in brown marine algae and echinoderms) are the fucoidans, a class of fucose-rich sulfated carbohydrates (Berteau and Mulloy, 2003). Like most polymers, fucoidans are polydisperse, therefore their structure cannot be defined by a unique molecular weight since it is a mixture of molecular weights. Thus, molecular weights are frequently mentioned as a molecular range, average or median peak weight. This complex sample, with different molecular weight polysaccharides, can be more complicated, since fucoidans and other bioemulsifiers are extracted from seaweeds and several other polymeric and branched molecules could be present. Additionally, molecules with similar molecular weight could appear with different branching patterns, as a result of the original polysaccharide being hydrolyzed during the procedure of extraction. In that case, it will be a plethora of molecules, even for apparently the same molecular weight. Table 1 lists additional examples for the characterization of polymeric surfactants and their applied methodologies.
The combination of liquid chromatography and MS - two powerful analytical techniques - offers the capability for rapid, cost-effective and quantitative determinations of peptides/lipopeptides, glycolipids and biosurfactants. Coupling HPLC or UHPLC with MS gives the advantage of defining retention time(s) of a lipopeptide or biosurfactant along with its mass spectral fingerprint, permitting the separation and identification of individual components from complex samples. The first steps of purification are essential to remove impurities of the production/extraction procedure and to isolate the desired fraction and concentrate the sample, especially when the lipopeptides/biosurfactant are present at very low concentrations. The pre-purification protocols and methodologies of biosurfactants are determined by the expected type of compound to be produced by the microorganism being investigated and are extensively discussed elsewhere (Heyd et al., 2008). The most commonly used LC separation method for quantitative LC-MS is reversed phase (RP) separation, a classic methodology in peptide analysis. Thus, structural identification of lipopeptide biosurfactants could be achieved by (LC-MS). The production extracts could be examined for their composition using HPLC and retention times are compared with standard lipopeptides (like surfactin, fengycin etc.) when using mobile and static phases of the chromatography with the same characteristics. It is important to take into consideration several parameters, such as the selection and effects of mobile and stationary phase, flow rate, temperature (chromatography application), type of ionization and detection modes, and other spectrometric conditions contributing to the mass measurement (mass spectrometry application). All the previous parameters contribute to a successful characterization of a lipopeptide/biosurfactant (van den Broek et al., 2008). LC-MS is the most suitable strategy for a discovery-based approach especially when studying lipopeptides (unknown), as many SAAs are readily amenable to LC-MS analysis. Also, performing MS/MS or MSn experiments and the fragmentation data obtained from them, with accurate mass measurement of precursor and/or product ions, offer a tremendous amount of information and validate the proposed lipopeptide or biosurfactant structure. In a study by Rudden et al. (2015), an UHPLC-MS/MS method was developed and validated for the rapid and quantification of individual rhamnolipids congeners (Rudden et al., 2015). The method was validated for linearity, precision (intra- and inter-day) and accuracy according to FDA guidelines.
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) coupled with mass spectrometry (MALDI-TOF-MS) with its high sensitivity and high sample throughput is a powerful method providing information for the molecular masses of intact peptides-lipopeptides and rapid characterization of low molecular weight or high molecular weight biosurfactants. Peptides (lipopeptides or glycopeptides), with molecular masses below 5,000 Da) bearing free amino acid groups that can be protonated easily using the α-cyano-4-hydroxycinnamic acid matrix. These peptidyl-SAAs often afford intense signals (Harvey, 2003). MALDI-TOF is a method that can be applied for the determination of the molecular mass of HMW and lipopeptide-biosurfactants that have been previously purified. The applied matrix for HMW biosurfactants (>10,000) is sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid). Usually, the purified and desalted fractions obtained after gel filtration and/or preparative (or semi-preparative) reversed-phase HPLC are displaced to the MALDI-TOF target after co-crystallization with the appropriate matrix, dried on a platform, onto which the laser is fired to form gaseous ions. Then, the formed ions are separated in a mass spectrometer based on their time of flight (TOF). MALDI can be used to determine the full molecular mass of purified LP/SAAs and the easy sample preparation as well as the recent developments in small bench top MALDI-TOF-MS instruments have made this an attractive method for lipopeptide and biosurfactant molecular mass determination. Well-known lipopeptides, such as surfactins and iturins are detected in the m/z range 1,000–1,100 and fengycins in the m/z range 1,400–1,600 by MALDI-TOF-MS in their single protonated forms or as Na+, K+ adducts in the m/z range 800–2,000 (Kim et al., 2010). Although normally MALDI-TOF requires isolated and HPLC-pure samples, an innovative method for rapid sensitive detection and efficient structural characterization of lipopeptide biosurfactants by using whole microbial cells and crude culture extracts (Vater et al., 2002). Furthermore, MALDI-TOF-MS/MS analysis could define the amino acid sequence of lipopeptides without performing hydrolytic treatment and subsequentially amino acids analysis and, unlike Edman degradation, MS/MS analysis does not need the molecule to be purified to homogeneity since the mass analyzer could select only the ions of interest (Pueyo et al., 2009).
Fourier transform-infrared spectroscopy (FT-IR) is a fast, direct, and low-cost technique for the characterization of the structure of lipopeptides and other biosurfactants, and can assist to identification of functional groups present in these compounds (Romero et al., 2007). This spectroscopy targets to measure the vibrations of a molecule after excitation by IR radiation at a specific wavelength range. The wave number positions of absorbance, intensities and width peaks are useful for functional group characterization, and identification of the type of isolate biosurfactant. There are characteristic absorbance bands of the functional groups of lipopeptides that appear on an IR spectrum (Long, 2004). However, as these functional groups and their characteristic IR bands are not unique to the SAAs and it is possible to be present in other extracellular components of the analysis sample. Thus, the sample is expected to be purified to homogeneity to ensure these specific detections. The process of production and purification of the microbial SAAs with optimized conditions are described extensively elsewhere (Roelants et al., 2016; Twigg et al., 2021).
For example, in a study for characterization of biosurfactant production by Bacillus subtilis isolates for microbial enhanced oil recovery (MEOR) applications (Nikolova et al., 2020), characteristic IR bands of peptides were observed at 3305 cm-1 (typical for NH-stretching mode), 1643 cm-1 (1645 cm-1 CO–N bond, stretching mode); and at 1543cm-1 (deformation mode of the N–H bond with C–N, stretching mode) (Pereira et al., 2013). In addition, also reported the presence of aliphatic chains (–CH3, –CH2–), from the vibration bands between 1465–1368 cm-1, and 2957– 2855 cm-1. The chemical characterization using several more analytical techniques showed that the studied extracts present similar mixtures, containing C13-, C14- and C15-surfactin lipopeptides.
For amino acid analysis of an unknown lipopeptide, the sample should be first heated with appropriate acid to hydrolyze all peptide bonds and then the resulted amino acid mixture is purified and estimated quantitatively, usually with a chromatographic method based on phenyl isothiocyanate derivatization (Cohen et al., 1986). Edman degradation is the classic technique for amino acid sequence identification of peptides/proteins using a chemical procedure and is a suitable method for determination of the amino acids in the peptide portion of purified lipopeptides. This analytical procedure could be used to peptides/proteins, where the N-terminus has not been altered (pGlu or Acetyl group) and requires that lipopeptide is in their correspondent linear peptide and not the cyclic form. Also, it is important for the proper identification of sequence that there is sufficient starting material, and it is of high purity. During Edman sequencing procedure, lipopeptides and proteins should be purified to near homogeneity to avoid mixed results (Zachara and Gooley, 2000). The main disadvantage of this method is that it is relatively slow (45 min/per residue), but the amino acid sequence assignment leads to results of high accuracy without casting doubts, as it happens sometimes with MS/MS approaches. Table 1 lists several examples from literature (Hathout et al., 2000; Gutierrez et al., 2020) in which amino acid analysis was performed towards structural characterization of unknown biosurfactants/bioemulsifiers possessing peptide fragments.
Fatty acid analysis to acquire crucial information for the identification of the structure of them requires a cleavage (hydrolytic) of the bond among the lipid portion and their hydrophilic head group. For lipopeptides, this is a hydrolysis of the bond between the peptide/protein part of the lipopeptide and the lipid portion. Separation and identification of the lipids is achieved by Gas Chromatography (GC) or GC-MS analysis of hydrolysates after derivatization of the fatty acid chains to fatty acid methyl esters (FAME) (by esterification) and additional conversion to trimethylsilyl (TMS) derivatives (Yakimov et al., 1995). For glycolipids, the process starts with a cleavage (hydrolytic) of the bond between the lipid part and carbohydrate and following derivatization of the subsequent fatty acid chains to FAME analyzed also by GC or GC-MS (Brandl et al., 1988).
Many of the methods described above for analysis, identification, and structural characterization of lipopeptides can be applied for the analysis of high molecular weight surfactants/bioemulsifiers. Analytical methods like sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) and/or digestions by proteases are needless for lipopeptide molecules since they do not exhibit the same complexity as proteins. However, analysis of polymeric surfactants and bioemulsifiers can benefit from the techniques since the digested molecules result into smaller molecular fragments/components allowing a more comprehensive analysis. For determining carbohydrate content, lipid composition, and protein content, colorimetric assays like the Bradford assay are commonly used. Determination of the content type and the percentage of the microbial produced and uncharacterized surfactants/bioemulsifiers, is very crucial since it defines which analytical methods should be applied for the structural analysis and characterization of the desired molecule(s) (Smyth et al., 2014). It is usually necessary to divide the initial material into smaller portions to perform a precise structural analysis and ease the determination of the chemical formulation of the unknown biosurfactant/bioemulsifier. Lipid and carbohydrate components can be then removed and characterized by gas chromatography mass spectrometry (GC-MS) after derivatization (Monteiro et al., 2007) The structural analysis of a possible polypeptide segment of a biosurfactant molecule can be performed after its removal by enzymatic treatment (Strader et al., 2006). Edman degradation can be used to determine the individual amino acid sequences of relatively small peptides, but also automated amino acid analysis and/or mass spectrometric techniques (MS/MS fragmentation) as described previously for lipopeptides There are several studies that have used an array of methods to characterize polymeric bioemulsifiers from marine microorganisms (Gutierrez et al., 2007a; Gutierrez et al., 2007b; Gutierrez et al., 2008; Gutierrez et al., 2012; Gutierrez et al., 2020), including the use of SEC (see above), NMR (discussed below) and other analytical and chemical methods.
In order to properly understand and analytically approach a macromolecule, the characteristic of paramount importance is its molecular weight, whether this molecule is synthetically produced or naturally occurring (microbial product) (Tarazona and Saiz, 2003; Gutierrez et al., 2020). Knowing the molecular weight of the polysaccharide is essential since it is indicative of their nature, type, and biotechnological applications. It is critical to have knowledge of the molecular weight scale-range not only for analytical purposes, but also for its application in medical and commercial settings. However, determining a molecule’s molecular weight has proven to be a very difficult task, despite its essential nature (Gil et al., 2022). As an example, polysaccharides exhibit a variety of physicochemical properties in solution which they set to introduce several difficulties when it comes to their determination. The most common native characteristic is their polydispersity which implies that they consist of a variety of different molecular weight structural units. Furthermore, when in solution, they tend to take over/adapt to different conformations proportional to their mass of solute per volume (g/L) (possibility of self-association in case of polysaccharides), making it difficult to determine with accuracy their conformation.
Among the biosurfactants, the chemical class of lipopeptides is of particular significance since their structure is favorable for various industrial applications including cosmetics, and medicine. Differences in their amino-acid sequences and in the number of carbon atoms of the fatty acids aliphatic chains give lipopeptides diverse chemical structures and physicochemical features. A standard structure of lipopeptides consists of a hydrophilic head (which may be either a linear hydrocarbon or a lactone ring-characteristic of cyclic lipopeptides) and a hydrophobic tail, which mostly is composed by a fatty acid. There are some cases where the hydrophobicity might be induced also from the amino acid sequence of hydrophobic amino acids like Gly, Ala, Val, Leu, Ile, Pro, Phe, Met and Trp.
Analysis through specific and suitable NMR experiments can provide information on the structure of both the peptide and the fatty acid structural/chemical part of lipopeptide. A variety of standard two-dimensional NMR experiments exists, which can provide detailed data on the position of linkages between the fatty acid chain and the peptide. Identification and assignment of the amino acid sequence is feasible through a series of NMR experiments including heteronuclear 2D 1H-15N HSQC NMR experiments, since –NH, -NH2, groups are abundant in amino acids like Trp, Lys, Gln and Asn of the peptide moiety. The amidic -NH- group of the peptide backbone appears between 7-10 ppm proton spectral region, since it is adjacent to the carbonyl carbon atom (C=O) of each amino acid in sequence. All representative proton resonances of a lipopeptide molecule are depicted and indicated in the 1H 1D NMR spectrum of a complex bioemulsifier extract in Figure 1.
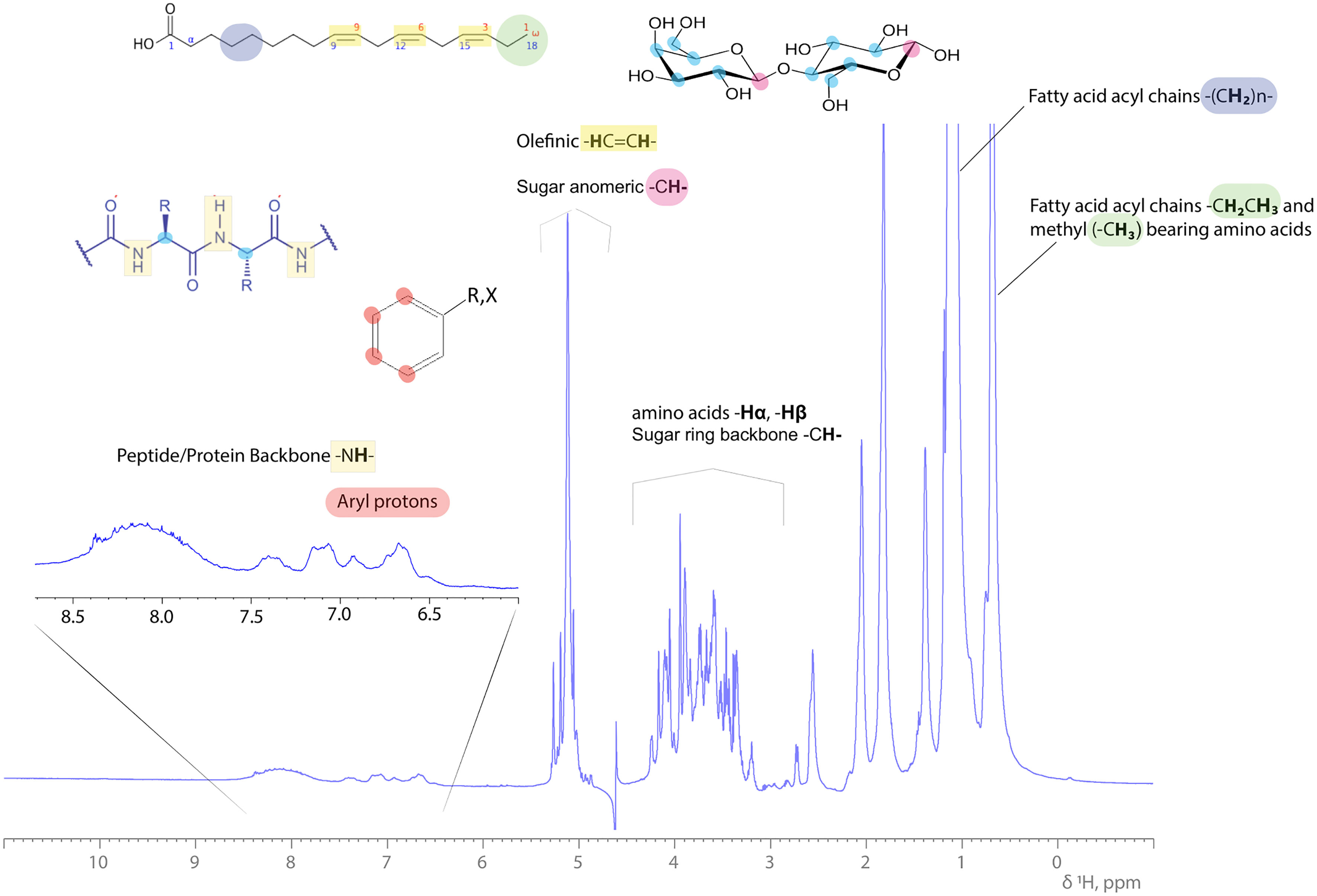
Figure 1 1H 1D NMR spectrum of a complex bioemulsifier extract in an aqueous (H2O:D2O, 90:10%) solution. Characteristic resonances are annotated across the 1H ppm scale, indicating the protein/peptide backbone amide protons, saccharide (sugar ring) and olefinic (double bonds in fatty acid aliphatic chain) protons, amino acid side chain protons (Hα, Hβ and -CH3) and FAs’ methylene and methyl protons arising upfield. Examples of structural blocks are depicted and highlighted along with their indicative 1H signals.
NMR spectroscopy is a powerful tool in obtaining detailed structural information about complex carbohydrates, polysaccharides, and glycolipids. Advanced 1H-NMR methods including homo- and heteronuclear experiments like TOCSY (also known as HOHAHA experiment (two-dimensional experiment homonuclear Hartmann-Hahn)), COSY (correlation spectroscopy), NOESY (Nuclear Overhauser effect spectroscopy) and HMBC (heteronuclear multiple bond correlation). Structural information through bond or through space correlations can be elucidated by applying the above NMR experiments. Two dimensional experiments are also being used for determining the presence, type, and location of saccharide substituents, such as acetyl, methyl, phosphate, and sulfate groups.
Homonuclear two-dimensional (2D) NMR experiments for molecules like lipopeptides/peptides should be handled in a slightly different analytic approach from the one regarding those of organic synthesis compounds. This is because the protons of each amino-acid form a characteristic and distinct pattern of correlation peaks in homonuclear 2D 1H-1H COSY and 2D 1H-1H TOCSY NMR spectra. COSY NMR spectrum illustrates the connectivities between protons belonging to adjacent carbons (connected through one bond), and J coupled nitrogen atoms. To better understand the COSY connectivities, let us assume the chemical structure of a carbohydrate molecule which incorporates the pattern of–Ci-1H-CiH-Ci+1H-; the Hi proton (attached to the Ci) in a COSY spectrum will give a single correlation with the protons Hi-1 and Hi+1. Each one of these connectivities has one-bond distance and in COSY spectrum are illustrated as scalar-coupled correlations of H atoms. A TOCSY NMR spectrum provides correlation peaks between the proton Hi-1 and the proton Hi+1, but also can reveal correlations with protons of atoms that are 3, 4 or 5 bonds distant from each other, as well. In the frame of the NMR assignment workflow, COSY and TOSY experiments are followed by the 2D 1H-1H NOESY NMR spectra which provides information about correlations arising from interactions through space, being at the maximum distance of 5,5 Å. The sequential couplings for peptides/lipopeptides that are presented through the homonuclear 2D 1H-1H correlation spectra, are used for the identification of specific amino acid spin patterns, as well as in NOESY spectra for the evaluation of the sequential couplings along the peptide backbone. Sequential couplings of Hα(i)-NH(i+2,3,4), NH(i)-NH(i ± 1) and Hα(i)-Hβ(i+3) are characteristic when the peptide backbone adopts α-helix structure, while strong correlation peaks are of the type ηα(i)-NH(i+1) and weak correlations NH(i)-NH(i ± 1) are characteristic for β-sheet (Kawano et al., 1990; Ploutno and Carmeli, 2002).
Through an 1D 13C NMR experiment, the presence of the carbonyl carbons can be determined with δC ~ 170-182 ppm, which are attributable to the C=O groups of the amino acids. Additionally, one of the most characteristic carbon chemical shifts is from the amino-acid’s Cα which appear at δC 51-63 ppm, except for the Cα of Glycine which resonates ~ 45 ppm. Signals at δC ~70 and ~80 ppm along with the presence of a pair of doublets at 2.40 ppm which is coupled with multiplet arising from the methine signal of an α- or β-hydroxy fatty acid proton resonate between 4.90-5.07 ppm may be indicative for its coupling with the amide carbonyl signal of an amino acid (Kalinovskaya et al., 2002; Liu et al., 2010). The population and the number of methyl carbons which belong to fatty acid aliphatic chain and/or to amino acids can be determined by both 1D 13C and 2D 1H-13C HSQC NMR spectra. The resonances of these methyl-end groups can be identified through the combination of TOCSY and HMBC correlations. Amino acid-fatty acid sequences of lipopeptides can be constructed by 2D ROESY and HMBC NMR experiments using long range proton-proton and proton-carbon correlations and correlations between amide protons and α-protons. An example is the specific detection of valine through the pattern of isopropyl spin systems. Valine residues in lipopeptides can be determined through COSY and multiplicity edited HSQC correlations. However, their order in the amino acid sequence can be completed by careful analysis of HMBC correlations as described in linear lipopeptides from the marine cyanobacterium Lyngbya majuscula (McPhail et al., 2007).
2D HMBC spectra can provide significant information about lipopeptides, revealing characteristic correlation patterns of these types of molecules. Connectivities and correlations arising for each residue and between residues, can be examined extensively and further functional groups or linkages like ureido groups, methoxy groups, β-hydroxy fatty acids or other aliphatic linkages can be identified (Tareq et al., 2014; Son et al., 2016; Xiu et al., 2017). A common spectral pattern of lipopeptides present in the standard proton 1D NMR spectrum (but also in 2D 1H-13C HSQC spectra) is the region between 0.70-2.00 ppm. Fatty acids’ protons and protons arising from the aliphatic side chain of the hydrophobic amino acids appear with heavily overlapping signals. Several solvent mixtures are being tested to achieve better resolution within this spectral region and to ascertain the distinct number of alkyl-methine (R3C-H), alkyl methylene (R-CH2-R) and alkyl methyl (R-CH3) groups.
Trans- and cis- geometry of aliphatic chains can be characterized from the presence of NOESY correlations between the representative olefinic protons. Such information can be obtained from the splitting patterns and coupling constants. Olefinic cis and trans couplings across double bonds - 3JHH coupling constants - are very reliable indicators of stereochemistry with 3Jtrans > 3Jcis (Alexandri et al., 2017).
Molecular structure elucidation for glycolipids, saccharides, polysaccharides, and lipo-polysaccharides is usually combined with analytical data from mass spectrometry and chromatography. 1H-NMR spectra can be very informative in terms of the general “screening” of such molecules and diagnostic for the presence of contamination in samples of SAAs. Chemical shifts and coupling constants present in NMR spectra from high molecular weight SAAs of this type, are influenced from the high structural similarity, directly affecting the complexity of hetero- and homonuclear 2D NMR spectra.
Until today, most of the NMR-characterized polysaccharides and lipopolysaccharides, are not classified as SAAs (Olsthoorn et al., 2000; Starr et al., 2013). This fact may be due to the high complexity and the high molecular weight they exhibit since NMR analysis is a method limited to molecular weights ≤10000 g/mol due to signal to noise ratio.
SAAs like glycolipids, saccharides, polysaccharides, and lipopolysaccharides (characterized by NMR spectroscopy) unfortunately cannot bear the advantage of being labelled with 13C. Thus, the NMR experiments based on carbon detection need a relatively high amount of SAA material to acquire good quality and the desired dispersion of carbon signals in NMR spectra.
Saccharides have characteristic structural entities and their conformation exhibit some unique parameters, which can be very helpful in structural elucidation through NMR. The anomeric protons and carbons of each saccharide usually have a very specific chemical shift range between δη 5.80-4.20 ppm and δC 95-110 ppm for proton and carbon frequencies, respectively. Significant information can be obtained through the anomeric proton’s chemical shift and coupling constants for the saccharide’s anomeric center geometry. Chemical shifts of the α-anomeric protons are usually located between 4.90-5.80 ppm, while β-anomeric protons between 4.20-4.90 ppm. Chemical shifts of the α-anomeric carbons are usually located between 92-103 ppm, while β-anomeric carbons between 103-110 ppm. From the scalar coupling constants 3JH1, H2, the conformational status of the anomeric proton can be further evaluated/concluded, since for the α-conformation every glucopyranose sugar units appears with a 3-4 Hz coupling, while β-conformation appears with a 7-8 Hz coupling (Agrawal, 1992; Chen et al., 2016; Hadad et al., 2017).
For surfactants that consist of a disaccharide or a relatively small polysaccharide, even the 1D 1H NMR spectrum can be informative about the number of structurally different saccharide units present in the molecule. Integration of the anomeric signals and of their relative ratios provides their ratio in the mixture. Signals arising from methylene (-CH2-) and methine (-CH-) protons belonging to the sugar ring are located between 3.20- 4.20 ppm in aqueous solutions. A typical example of a 1H-13C HSQC NMR spectrum of a rhamnolipid standard mixture purchased from Sigma Aldrich is presented in Figure 2.
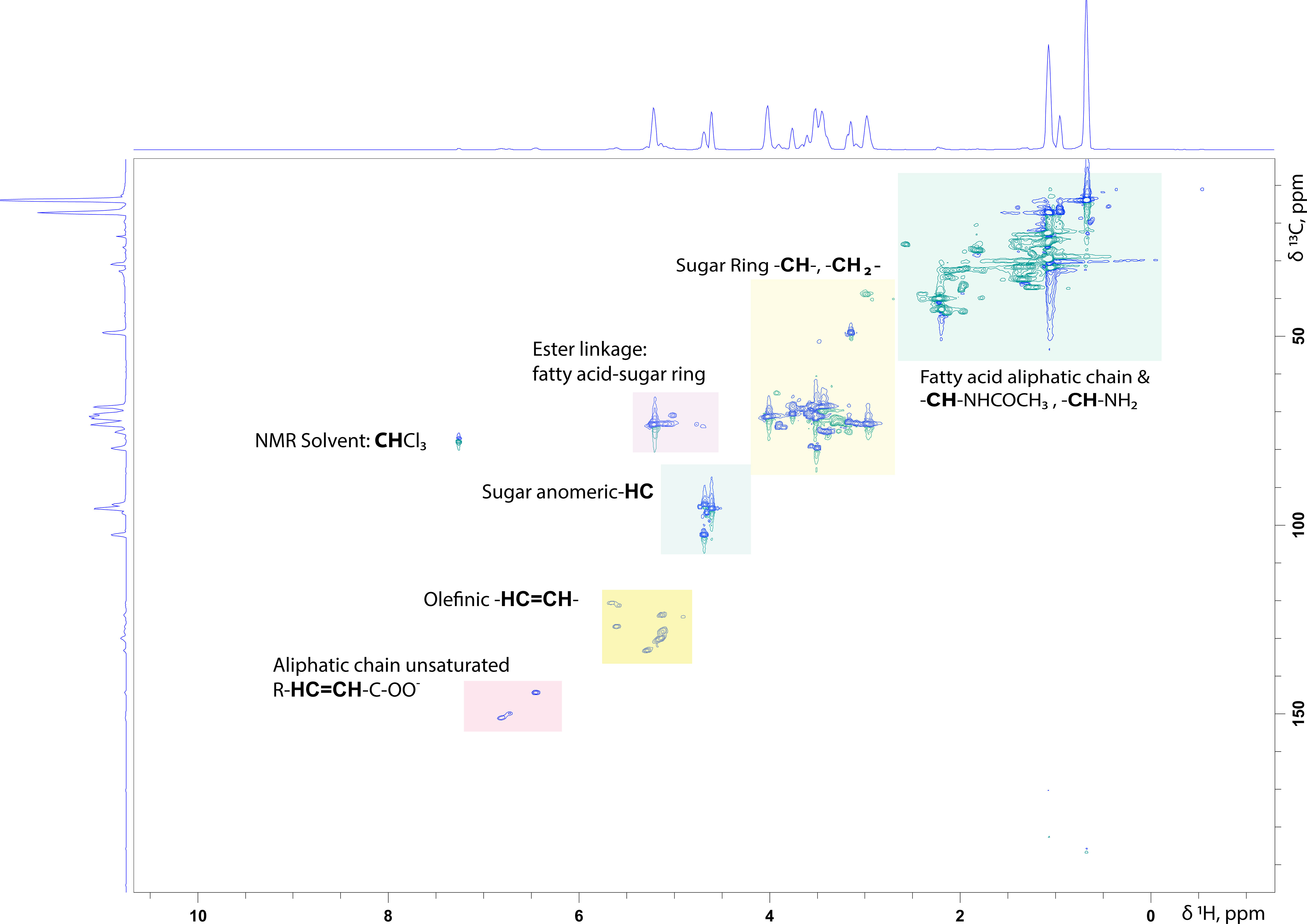
Figure 2 1H-13C HSQC NMR spectrum of a Rhamnolipid standard mixture persuaded from SIGMA Aldrich in CDCl3: MeOD (70:30%, v/v). In the spectrum are highlighted the characteristic structural parts of such molecules.
In cases, where the surfactant under analysis consists of more than one saccharide units, signals tend to overlap, making the assignment for each saccharide unit difficult. Among the most characteristic features of saccharides present in such molecules are, the methyl-end groups which are part of 6-deoxy-saccharides, together with the H-3/3’ from sialic acid that resonate between 2.80-1.20 ppm, while the H-2/2’ axial or equatorial from Ketodeoxyoctonic acids (Kdo) resonate at 3.24-3.29 ppm (Zähringer et al., 2015; Ovchinnikova et al., 2016). The protons adjacent to N-acetyl groups, like N-acetyl-D-glucosamine, also resonate in the same spectral region (Pereira et al., 2015). Furthermore, of great interest the detection and identification of amino sugars which are part of a polysaccharide/lipopolysaccharide. In such a case, the observation of the characteristic protons arising from the adjacent carbon atoms to the N-acetyl or -NH2 groups, along with the resonances of all the –NH-, -NH2 protons which resonate downfield at 7.00-8.50 ppm (sharp doublets, J= 7-8 Hz) are crucial for the amino sugar moiety recognition. Protons of the amino sugars’ amine groups exhibit a unique NOE correlation pattern in the NOESY NMR spectrum, revealing correlations with the polysaccharide’s backbone – such as i+1NH-iH1, i+1NH-iH2 - and in some cases also within the sugar ring (Tian et al., 2018). This characteristic NOESY pattern is of great importance since it is has not been described in the literature so far and it is diagnostic for the type and the composition of saccharide units of the polysaccharide/lipopolysaccharide SAA.
13C chemical shifts of the saccharide backbone carbons resonate between 50-80 ppm. In sugar rings where carbons are substituted with amines or methoxy groups or N-acetylated groups, the carbons resonances are shifted downfield to between 40-60 ppm due to deshielding phenomena, whereas unsubstituted carbons resonate between 60-80 ppm. Carbons of methyl end groups resonate between 15-30 ppm, while carbonyl atoms do so between 160-185 ppm (Tedesco et al., 2016). In the case of saccharides with either disulfated or monosulfated residues as in O-disulfated fucose and 2-O-monosulfated fucose it is very challenging to be distinguished only through the NMR spectra. Both molecules may exhibit a very similar spectral pattern due to the same saccharide backbone, although there might be a few indicative changes due to the position of the substitution usually at the resonances of the H-3 and H-5 signals (Yamada et al., 1992; Cimino et al., 2001). The most significant proton and carbon resonances of SAA’s functional groups which have been already reported and characterized by various experts on the field are summarized in Table 2 (Bubb, 2003; Clément et al., 2010; Post et al., 2012; Tareq et al., 2014; Sarpal et al., 2016; Tan et al., 2016; Zompra et al., 2021).
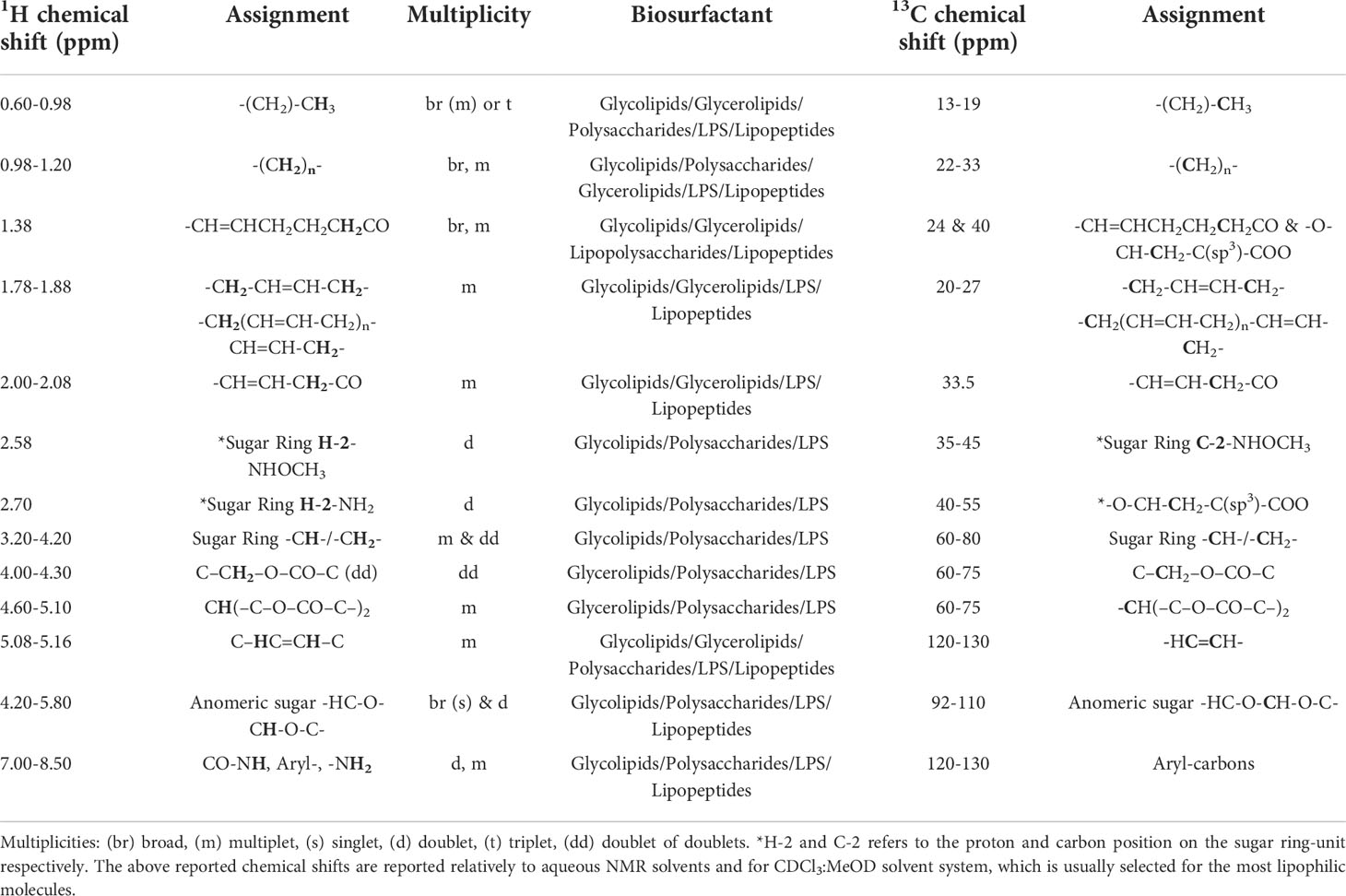
Table 2 Common chemical shifts of characteristic and functional groups of Biosurfactants’ structure.
Multiplicities induced from the existing linkages between monomeric sugar units or for linkages existing between multiple saccharide units or saccharide linked with fatty acids, can increase the complexity of the NMR spectra. Furthermore, linkages among sugar units are formed between the anomeric hydroxyl group of one saccharide unit and any other hydroxyl-bearing carbon in another saccharide unit.
The observation of long-range scalar couplings among anomeric signals and proton/carbon correlation peaks arising from characteristic linkages (ester bonds of linked fatty acids, fatty acid-saccharide, glycerol-fatty acids, glycerol-ethanolamine, glycerol-choline) detected in 2D HMBC spectra along with specific NOE (nuclear Overhauser effect) correlations between two sequential residues can confirm the linkages between polysaccharide surfactants. These correlations provide information about intra- and inter-residue distances, allowing the saccharides’ sequence to be determined and the linkage positions of the residues. Validation of these can be completed through COSY and TOCSY correlations. Glycosidic linkages can be tracked from 2D-NOESY experiments, with the strong NOEs indicating linkages between distant saccharide units or saccharide and fatty acids (Faber et al., 2001).
Regarding the characterization of polysaccharides or lipopolysaccharides, the determination of the structure is based on the determination of the connectivities of each saccharide unit through the identification of glycosidic bonds. Glycosidic linkages might have diverse connections polysaccharides like (16) or (14) linkages and can be confirmed from long-range HMBC 1H-13C coupling between H-1 of the first unit and C-6 or C-4 of the sequential sugar unit. It is noteworthy
that for polysaccharides with linear chain O-glycosidic bonds of either 13, 14 or 16 linkage there is an obvious influence on the chemical shift of the α anomeric protons and carbons (Zähringer et al., 2015).
Evaluation of 2D NOESY spectra can provide useful information about the connectivities for the structure of the surfactant. Where the surfactants have saccharide units without any glycosidic linkage with another saccharide unit, NOESY correlations will appear only for the protons within the same saccharide unit (Müller-Loennies et al., 2002). Correlation NMR experiments, COSY and TOCSY, can be used to assign the complete set of a monosaccharide’s ring protons. Specifically, through TOCSY NMR spectra it is possible to identify the complete series of the C-H of the saccharide backbone, having as a point of reference the anomeric H-1. Usually, the heterogeneity of the samples, does not allow the measurement of coupling constants from the 1D TOCSY experiments.
Glycerol-lipids and liposaccharides have characteristic ester linkages in the lipid component of the saccharide unit or the glycerol backbone which are evident only through long range correlations in 2D 1H-13C HMBC spectra due to the ability of the magnetization to pass through heteroatoms like oxygen and nitrogen. As mentioned, through HMBC, residue connectivities of H1/C3, H1/C4 residue A’ connectivities in position 3 and 4 of residue B’ respectively can be validated. Branching residues can be distinguished from COSY NMR spectra and can be confirmed from the heteronuclear multiple bond correlation HMBC spectrum (Clément et al., 2010).
2D 1H-13C HSQC spectra of those molecules are indicative and may represent their unique NMR signature, however it could be very similar between the same class (furanoses, pyranose) of glycerol-lipid or lipopolysaccharide. The resonances of the saccharide or glycerol backbone are located in a very narrow spectral area of 3.00 - 4.50 with minimum chemical shift variation between the different saccharide units or polyol/glycerol backbone. While the resonances of the lipid part exhibit standard chemical shifts which allow the assignment of structural details, such as the position of double bonds and the number of different double bonds (ω-3, ω-6, ω-9) in the fatty acids, the methylene protons but not the C atom length of the fatty acid aliphatic chain (Salinero et al., 2012; Kügler et al., 2015).
The qualitive information of biosurfactants’ content and as well the precise structural determination of them requires a multidisciplinary approach and use of several analytical methods (Figure 3). Confirmation of biosurfactant/bioemulsifier presence (e.g., identification of orthologs of genes responsible for biosurfactant production) and sufficient purification of them, should be followed by an analytical set of combined methods which can contribute to the efficient structural characterization of biosurfactants.
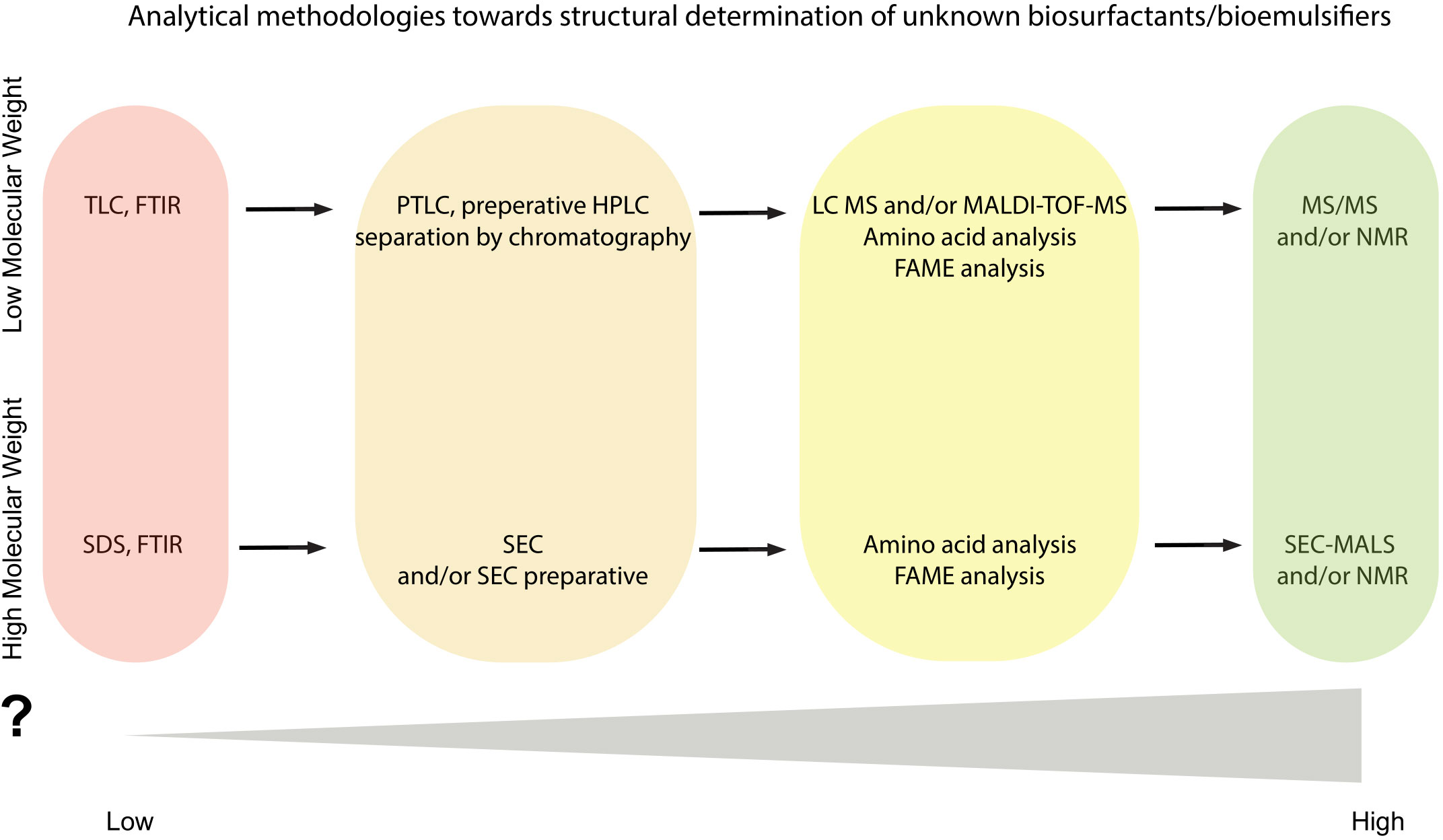
Figure 3 Flowchart of analytical methodologies towards structural determination of unknown biosurfactant(s)/bioemulsifier(s). Color gradient is indicative of the information acquired from each analytical step.
As mentioned, TLC and/or FTIR can be used for the detection of lipopeptides, rhamnolipids and low molecular weight biosurfactants in the initial purified samples but these methods cannot stand alone for sufficient characterization of them and may function as first steps towards the full characterization (Table 1; Figure 3). Application of the HPLC method has been reported for characterization, purification, and even quantification and of several well-known lipopeptides and rhamnolipids but this technique requires at least the use of internal standards. Alternatively, HPLC could assist to separation and isolation of each congener and lead to high purity samples in sufficient yields for further analysis towards structural determination (Figure 3). As an example, for the lipopeptides such as surfactin and fengycin there are several studies reporting the importance of separation/fractionation of the isolate to further structure elucidation (Table 1).
Mass spectrometry analysis requires high purity SAAs but provides information for the molecular weight and tandem MS (MS/MS) method can determine the presence of congeners of glycolipids. The fragmentation data obtained from MS/MS, with accurate mass measurement of precursor and/or product ions could lead to molecular recognition of them and confirm the presence of the biosurfactant with low molecular weight. Therefore, HPLC-MS/MS is considered by far the most accurate method for rhamnolipid identification to date (Heyd et al., 2008; Rudden et al., 2015). If, therefore, a microbial isolate sample considered as a mixture of rhamnolipids, then pseudomolecular ions and fragmentation ion patterns should be detected. This technically means that, in negative operation mode, Rha-C10-C10 with pseudomolecular ion (m/z) 503, will afford the two major fragment ions, Rha-C10 with m/z 333 and m/z 339 from the loss of Rha unit. While for the detection of di-rhamnolipid Rha-Rha-C10-C10, the m/z 649 with fragment ions at m/z 479, 339, 169 (Heyd et al., 2008).
Chemical composition can be tricky when it comes to NMR analysis and atomic level characterization. Lipopolysaccharides, polysaccharides, oligosaccharides, lipopeptides and glycopeptides can be analyzed in depth through standard 2D NMR experiments, although in samples where there is more than one and/or in combination, the analysis might become more challenging. NMR is well suited for the characterization of isolated compounds/molecules, but it can be very informative in case of complex mixtures (NMR application in fraud detection (Sobolev et al., 2019; Dimitrakopoulou et al., 2021) in food and pharmaceutical industry (Remaud et al., 2013), or NMR analysis of biosimilars (Hodgson et al., 2019) through the ease of pattern recognition analysis. Each molecular type of them requires a coupled/parallel analytical technique such as MS, to provide the information of a fully characterized molecule (Figure 3).Additionally, it is worth noting that many microbial biosurfactants face the issue of the presence endotoxin molecules, also known as LPS, in co-production. This fact introduces many problems in downstream processing and can become a steppingstone in characterization, following commercial production and applications (Taguchi et al., 2015). Endotoxin-LPS can be identified via NMR spectroscopy and can be applied as an analytical- validation step in down or upstream process. Regarding NMR key-features, LPS bear the same structural building blocks as a polysaccharide or a liposaccharide molecule, since it is consisting of mainly 3 distinct structural elements, the glycolipid part ‘Lipid A’, the core oligosaccharide and the O-specific oligosaccharide known as ‘antigen-O’ or ‘O-chain’ (Caroff and Karibian, 2003; Leone et al., 2007). These blocks are covalently linked, and their characterization encompass both MALDI MS and NMR analytical techniques. The last decade, there is an upcoming interest in NMR spectrometry application on the structural characterization of LPS-endotoxins or LPS-selective domains from marine Gram-negative bacteria (Vinogradov et al., 2003; Silipo et al., 2005; Nazarenko et al., 2011). Most of the research studies embrace the detailed structural characterization approach on isolated endotoxin-LPS molecules. However, no studies stress out the coexistence of a LPS-endotoxin and the biosurfactant/bioemulsifier molecule of interest. Herein, we suggest that NMR spectroscopy can be very helpful and indicative tool of such a case-scenario. The lipopolysaccharide or the oligosaccharide NMR correlation pattern, along with information about the magnitude (knowledge of the molecular weight) of the molecule(s) under investigation can be used to monitor the composition of the biosurfactant/bioemulsifier, in between the processing steps.
Although extensive efforts have been made in recent years to characterize biosurfactants, a rapid and of high-precision quantification analysis of SAAs remains a challenge. Studies of the properties of SAAs, optimization of the production and application of them in industry, food, drugs, cosmetics, etc., demand fast procedures, accurate and efficacious tools for their identification and quantification. Accurate determination of the bioemulsifier/biosurfactant analogues ratio is intrinsic, since may help in accurately attributing them with a function. In this direction, a variety of analytical methods are described here, including chromatographic and spectroscopic techniques for qualitative and quantitative assessment of SAAs’ content. Combination of these analytical methods is suggested to be the ideal approach for determination of the SAAs towards their structure elucidation.
AZ, methodology, investigation, resources, writing- original draft preparation, and conceptualization. SC, investigation, methodology, writing-original draft preparation, and editing. KS, MT, IA, and AP, writing-review and editing. AG, review and editing. IB, TG, RM, SE, MP, and GS, conceptualization, investigation methodology, supervision, funding acquisition, and resources. All authors contributed to the article and approved the submitted version.
This research was funded by the European Union Framework Programme for Research and Innovation, Horizon 2020, under Grant Agreement No. 635340 MARISURF. This was also supported by the INSPIRED (MIS 5002550) which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure,’ funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund). EU FP7 REGPOT CT-2011-285950 – “SEE-DRUG” project is acknowledged for the purchase of UPAT’s 700 MHz NMR equipment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CC, Column chromatography; NMR, Nuclear magnetic resonance; COSY, COrrelated SpectroscopY; NOESY, Nuclear Overhauser effect spectroscopy; ELSD, evaporative light scattering detector; pGlu, Pyroglutamic acid; FAME, Fatty acid methyl esters; PTLC, preparative thin layer chromatography; FT-IR, Fourier transform- infrared spectroscopy; RI, Refractive index; GC, Gas chromatography; ROESY, Rotating Frame Overhauser Enhancement Spectroscopy; GPC, gel permeating chromatography; MS, Mass spectrometry ; HMW, High molecular weight; RP, reversed phase; HSQC, Heteronuclear Single Quantum Coherence; SAAs, Surface active agents; HPLC, high pressure liquid chromatography; SDS-PAGE, Sodium dodecyl sulfate polyacrylamide-gel electrophoresis; HMBC, heteronuclear multiple bond correlation; SEC, size exclusion chromatography; HR, high resolution; TLC, Thin layer chromatography; LC, liquid chromatography; TMS, trimethylsilyl; LPS, lipopeptides; TOCSY, Total Correlation Spectroscopy; LMW, Low molecular weight; TOF, Time-of-flight; MALDI, Matrix-assisted laser desorption ionization; UPLC, ultra-performance liquid chromatography.
Abdel-Mawgoud A. M., Lépine F., Déziel E. (2010). Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86, 1323–1336. doi: 10.1007/s00253-010-2498-2
Agrawal P. K. (1992). NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 31, 3307–3330. doi: 10.1016/0031-9422(92)83678-R
Alexandri E., Ahmed R., Siddiqui H., Choudhary M., Tsiafoulis C., Gerothanassis I. (2017). High resolution NMR spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules 22, 1663. doi: 10.3390/molecules22101663
Amaral P. F., Coelho M. A. Z., Marrucho I. M., Coutinho J. A. (2010). “Biosurfactants from yeasts: characteristics, production and application,” in Biosurfactants, (New York, NY: Springer)236–249. doi: 10.1007/978-1-4419-5979-9_18
Anestopoulos I., Kiousi D. E., Klavaris A., Maijo M., Serpico A., Suarez A., et al. (2020). Marine-derived surface active agents: Health-promoting properties and blue biotechnology-based applications. Biomolecules 10 (6), 885. doi: 10.3390/biom10060885
Aranda F. J., Teruel J. A., Ortiz A. (2005). Further aspects on the hemolytic activity of the antibiotic lipopeptide iturin a. Biochim. Biophys. Acta (BBA)-Biomembranes 1713, 51–56. doi: 10.1016/j.bbamem.2005.05.003
Asmer H.-J., Lang S., Wagner F., Wray V. (1988). Microbial production, structure elucidation and bioconversion of sophorose lipids. J. Am. Oil Chem. Soc. 65, 1460–1466. doi: 10.1007/BF02898308
Berteau O., Mulloy B. (2003). Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology 13, 29–40. doi: 10.1093/glycob/cwg058
Biniarz P., Łukaszewicz M. (2017). Direct quantification of lipopeptide biosurfactants in biological samples via HPLC and UPLC-MS requires sample modification with an organic solvent. App. Microbiol. Biotechnol. 101, 4747–4759. doi: 10.1007/s00253-017-8272-y
Brandl H., Gross R. A., Lenz R. W., Fuller R. C. (1988). Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. App. Environ. Microbiol. 54, 1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988
Bubb W. A. (2003). NMR spectroscopy in the study of carbohydrates: Characterizing the structural complexity. Concepts Magn. Reson. A: Bridg. Educ. Res. 19 (1), 1–19. doi: 10.1002/cmr.a.10080
Caroff M., Karibian D. (2003). Structure of bacterial lipopolysaccharides. Carbohydr. Res. 338, 2431–2447. doi: 10.1016/j.carres.2003.07.010
Chen J., Song X., Zhang H., Qu Y. (2006). Production, structure elucidation and anticancer properties of sophorolipid from wickerhamiella domercqiae. Enzyme Microb. Technol. 39 (3), 501–506. doi: 10.1016/j.enzmictec.2005.12.022
Chen J., Sun J., Deering R. W., DaSilva N., Seeram N. P., Wang H., et al. (2016). Rhizoleucinoside, a rhamnolipid–amino alcohol hybrid from the rhizobial symbiont bradyrhizobium sp. BTAi1. Org. Lett. 18, 1490–1493. doi: 10.1021/acs.orglett.6b00461
Cimino P., Bifulco G., Casapullo A., Bruno I., Gomez-Paloma L., Riccio R. (2001). Isolation and NMR characterization of rosacelose, a novel sulfated polysaccharide from the sponge mixylla rosacea. Carbohydr. Res. 334, 39–47. doi: 10.1016/S0008-6215(01)00141-0
Clément M.-J., Tissot B., Chevolot L., Adjadj E., Du Y., Curmi P. A., et al. (2010). NMR characterization and molecular modeling of fucoidan showing the importance of oligosaccharide branching in its anticomplementary activity. Glycobiology 20, 883–894. doi: 10.1093/glycob/cwq046
Cohen S. A., Bidlingmeyer B. A., Tarvin T. L. (1986). PITC derivatives in amino acid analysis. Nature 320, 769–770. doi: 10.1038/320769a0
Das P., Mukherjee S., Sen R. (2009). Substrate dependent production of extracellular biosurfactant by a marine bacterium. Bioresour. Technol. 100, 1015–1019. doi: 10.1016/j.biortech.2008.07.015
Daverey A., Pakshirajan K. (2009). Production, characterization, and properties of sophorolipids from the yeast candida bombicola using a low-cost fermentative medium. Appl. Biochem. Biotechnol. 158 (3), 663–674. doi: 10.1007/s12010-008-8449-z
De Almeida D. G., Soares Da Silva R. C., Luna J. M., Rufino R. D., Santos V. A., Banat I. M., et al. (2016). Biosurfactants: Promising molecules for petroleum biotechnology advances. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01718
De Koster C.-G., Heerma W., Pepermans H.-A., Groenewegen A., Peters H., Haverkamp J. (1995). Tandem mass spectrometry and nuclear magnetic resonance spectroscopy studies of candida bombicola sophorolipids and product formed on hydrolysis by cutinase. Anal. Biochem. 230 (1), 135–148. doi: 10.1006/abio.1995.1448
de Koster C. G., Vos B., Versluis C., Heerma W., Haverkamp J. (1994). High-performance thin-layer chromatography/fast atom bombardment (tandem) mass spectrometry of pseudomonas rhamnolipids. Biol. Mass Spectrom 23, 179–185. doi: 10.1002/bms.1200230402
Déziel E., Lépine F., Milot S., Villemur R. (2000). Mass spectrometry monitoring of rhamnolipids from a growing culture of pseudomonas aeruginosa strain 57RP. Biochim. Biophys. Acta (BBA) - Mol. Cell Biol. Lipids 1485, 145–152. doi: 10.1016/S1388-1981(00)00039-1
Dimitrakopoulou M. E., Matzarapi K., Chasapi S., Vantarakis A., Spyroulias G. A. (2021). Nontargeted 1H NMR fingerprinting and multivariate statistical analysis for traceability of Greek PDO vostizza currants. J. Food Sci. 86 (10), 4417–4429. doi: 10.1111/1750-3841.15873
Faber E. J., Kamerling J. P., Vliegenthart J. F. G. (2001). Structure of the extracellular polysaccharide produced by lactobacillus delbrueckii subsp. bulgaricus 291. Carbohydr. Res. 331, 183–194. doi: 10.1016/S0008-6215(01)00012-X
Farias C. B. B., Almeida F. C. G., Silva I. A., Souza T. C., Meira H. M., Soares da Silva R. F., et al. (2021). Production of green surfactants: Market prospects. Electron J. Biotechnol. 51, 28–39. doi: 10.1016/j.ejbt.2021.02.002
Ghosh S., Ray A., Pramanik N. (2020). Self-assembly of surfactants: An overview on general aspects of amphiphiles. Biophys. Chem. 265, 106429. doi: 10.1016/j.bpc.2020.106429
Gil C. V., Rebocho A. T., Esmail A., Sevrin C., Grandfils C., Torres C. A. V., et al. (2022). Characterization of the thermostable biosurfactant produced by burkholderia thailandensis DSM 13276. Polymers 14, 2088. doi: 10.3390/polym14102088
Gupta R., Prabhune A.-A. (2012). Structural determination and chemical esterification of the sophorolipids produced by candida bombicola grown on glucose and α-linolenic acid. Biotechnol. Lett. 34 (4), 701–707. doi: 10.1007/s10529-011-0818-y
Gutierrez T., Biller D., Shimmield T., Green D. H. (2012). Metal binding properties of the EPS produced by halomonas sp. TG39 and its potential in enhancing trace element bioavailability to eukaryotic phytoplankton. BioMetals 25, 1185–1194. doi: 10.1007/s10534-012-9581-3
Gutierrez T., Morris G., Ellis D., Mulloy B., Aitken M. D. (2020). Production and characterization of a marine halomonas surface-active exopolymer. Appl. Microbiol. Biotechnol. 104, 1063–1076. doi: 10.1007/s00253-019-10270-x
Gutierrez T., Morris G., Green D. H. (2009). Yield and physicochemical properties of EPS from halomonas sp. strain TG39 identifies a role for protein and anionic residues (sulfate and phosphate) in emulsification of n-hexadecane. Biotechnol. Bioeng 103, 207–216. doi: 10.1002/bit.22218
Gutierrez T., Mulloy B., Bavington C., Black K., Green D. H. (2007b). Partial purification and chemical characterization of a glycoprotein (putative hydrocolloid) emulsifier produced by a marine antarctobacter species. Appl. Microbiol. Biotechnol. 76, 1017–1026. doi: 10.1007/s00253-007-1091-9
Gutierrez T., Mulloy B., Black K., Green D. H. (2007a). Glycoprotein emulsifiers from two marine halomonas species: chemical and physical characterization. J. Appl. Microbiol. 103, 1716–1727. doi: 10.1111/j.1365-2672.2007.03407.x
Gutierrez T., Shimmield T., Haidon C., Black K., Green D. H. (2008). Emulsifying and metal ion binding activity of a glycoprotein exopolymer produced by pseudoalteromonas species TG12. Appl. Environ. Microbiol. 74, 4867–4876. doi: 10.1128/AEM.00316-08
Hadad M. J., Zhang W., Turney T., Sernau L., Wang X., Woods R. J., et al. (2017). “NMR spin-couplings in saccharides: Relationships between structure, conformation and the magnitudes of JHH, JCH and JCC values,” in NMR in glycoscience and glycotechnology (London: Royal Society of Chemistry), 20–100.
Harvey D. J. (2003). Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates and glycoconjugates. Int. J. Mass Spectrom 226, 1–35. doi: 10.1016/S1387-3806(02)00968-5
Hathout Y., Ho Y.-P., Ryzhov V., Demirev P., Fenselau C. (2000). Kurstakins: a new class of lipopeptides isolated from bacillus thuringiensis. J. Nat. Prod. 63 (11), 1492–1496. doi: 10.1021/np000169q
Häussler S., Nimtz M., Domke T., Wray V., Steinmetz I. (1998). Purification and characterization of a cytotoxic exolipid of burkholderia pseudomallei. Infect. Immun. 66 (4), 1588–1593. doi: 10.1128/IAI.66.4.1588-1593.1998
Heyd M., Kohnert A., Tan T.-H., Nusser M., Kirschhofer F., Brenner-Weiss G., et al. (2008). Development and trends of biosurfactant analysis and purification using rhamnolipids as an example. Anal. Bioanal. Chem. 391, 1579–1590. doi: 10.1007/s00216-007-1828-4
Hodgson D. J., Ghasriani H., Aubin Y. (2019). Assessment of the higher order structure of humira®, remicade®, avastin®, rituxan®, herceptin®, and enbrel® by 2D-NMR fingerprinting. J. Pharm. BioMed. Anal. 163, 144–152. doi: 10.1016/j.jpba.2018.09.056
Hošková M., Schreiberová O., Ježdík R., Chudoba J., Masák J., Sigler K., et al. (2013). Characterization of rhamnolipids produced by non-pathogenic acinetobacter and enterobacter bacteria. Bioresour. Technol. 130, 510–516. doi: 10.1016/j.biortech.2012
Janek T., Łukaszewicz M., Rezanka T., Krasowska A. (2010). Isolation and characterization of two new lipopeptide biosurfactants produced by pseudomonas fluorescens BD5 isolated from water from the Arctic archipelago of Svalbard. Bioresour. Technol. 101, 6118–6123. doi: 10.1016/j.biortech.2010.02.109
Jemil N., Manresa A., Rabanal F., Ayed H. B., Hmidet N., Nasri M. (2017). Structural characterization and identification of cyclic lipopeptides produced by bacillus methylotrophicus DCS1 strain. J. Chrom. B 1060, 374–386. doi: 10.1016/j.jchromb.2017.06.013
Jiang J., Zhang H., Zhang C., Han M., Du J., Yang X., et al. (2021). Production, purification and characterization of ‘Iturin a-2’a lipopeptide with antitumor activity from Chinese sauerkraut bacterium bacillus velezensis T701. Int. J. Pept. Res. Ther. 27 (3), 2135–2147. doi: 10.1007/s10989-021-10241-9
Kalinovskaya N. I., Kuznetsova T. A., Ivanova E. P., Romanenko L. A., Voinov V. G., Huth F., et al. (2002). Characterization of surfactin-like cyclic depsipeptides synthesized by bacillus pumilus from ascidian halocynthia aurantium. Mar. Biotechnol. 4, 179–188. doi: 10.1007/s1012601-0084-4
Kawano K., Yoneya T., Miyata T., Yoshikawa K., Tokunaga F., Terada Y., et al. (1990). Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the beta-sheet structure. J. Biol. Chem. 265, 15365–15367. doi: 10.1016/S0021-9258(18)55402-8
Kim P. I., Ryu J., Kim Y. H., Chi Y. T. (2010). Production of biosurfactant lipopeptides iturin a, fengycin and surfactin a from bacillus subtilis CMB32 for control of colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 20, 138–145. doi: 10.4014/jmb.0905.05007
Kitamoto D., Isoda H., Nakahara T. (2002). Functions and potential applications of glycolipids biosurfactants–from energy–saving materials to gene delivery carriers. J. Biosci. Bioeng 94, 187–201. doi: 10.1016/S1389-1723(02)80149-9
Konishi M., Fukuoka T., Morita T., Imura T., Kitamoto D. (2008). Production of new types of sophorolipids by candida batistae. J. Oleo Sci. 57 (6), 359–369. doi: 10.5650/jos.57.359
Kowall M., Vater J., Kluge B., Stein T., Franke P., Ziessow D., et al (1998). Separation and characterization of surfactin isoforms produced by bacillus subtilis OKB 105. J. Colloid Interface Sci. 204, 1–8. doi: 10.1006/jcis.1998.5558
Kristoffersen V., Rämä T., Isaksson J., Andersen J.-H., Gerwick W.-H., Hansen E. (2018). Characterization of rhamnolipids produced by an Arctic marine bacterium from the pseudomonas fluorescence group. Mar. Drugs 16 (5), 163. doi: 10.3390/md16050163
Kügler J. H., Muhle-Goll C., Hansen S. H., Völp A. R., Kirschhöfer F., Kühl B., et al. (2015). Glycolipids produced by rouxiella sp. DSM 100043 and isolation of the biosurfactants via foam-fractionation. AMB Express 5, 1–11. doi: 10.1186/s13568-015-0167-7
Kügler J.-H., Muhle-Goll C., Kühl B., Kraft A., Heinzler R., Kirschhöfer F., et al. (2014). Trehalose lipid biosurfactants produced by the actinomycetes tsukamurella spumae and t. pseudospumae. Appl. Microbiol. Biotechnol. 98 (21), 8905–8915. doi: 10.1007/s00253-014-5972-4
Leone S., Silipo A., L. Nazarenko E., Lanzetta R., Parrilli M., Molinaro A. (2007). Molecular structure of endotoxins from gram-negative marine bacteria: An update. Mar. Drugs 5, 85–112. doi: 10.3390/md503085
Liu X., Ren B., Chen M., Wang H., Kokare C. R., Zhou X., et al. (2010). Production and characterization of a group of bioemulsifiers from the marine bacillus velezensis strain H3. Appl. Microbiol. Biotechnol. 87, 1881–1893. doi: 10.1007/s00253-010-2653-9
Long D. A. (2004). Infrared and raman characteristic group frequencies. tables and charts George Socrates John Wiley and sons, Ltd, Chichester, third edition 2001. J. Raman Spectrosc. 35, 905–905. doi: 10.1002/jrs.1238
Ma Z., Hu J., Wang X., Wang S. (2014). NMR spectroscopic and MS/MS spectrometric characterization of a new lipopeptide antibiotic bacillopeptin B1 produced by a marine sediment-derived bacillus amyloliquefaciens SH-B74. J. Antibiot. 67, 175–178. doi: 10.1038/ja.2013.89
Marchant R., Banat I. M. (2012). Microbial biosurfactants: Challenges and opportunities for future exploitation. Trends Biotechnol. 30, 558–565. doi: 10.1016/j.tibtech.2012.07.003
Mata-Sandoval J. C., Karns J., Torrents A. (1999). High-performance liquid chromatography method for the characterization of rhamnolipid mixtures produced by pseudomonas aeruginosa UG2 on corn oil. J. Chromatogr. A 864, 211–220. doi: 10.1016/S0021-9673(99)00979-6
Matsuda Y., Biyajima Y., Sato T. (2009). Thermal denaturation, renaturation, and aggregation of a double-helical polysaccharide xanthan in aqueous solution. Polym. J. 41, 526–532. doi: 10.1295/polymj.PJ2008300
McPhail K. L., Correa J., Linington R. G., González J., Ortega-Barría E., Capson T. L., et al. (2007). Antimalarial linear lipopeptides from a Panamanian strain of the marine cyanobacterium lyngbya majuscula. J. Nat. Prod. 70, 984–988. doi: 10.1021/np0700772
Monteiro S. A., Sassaki G. L., de Souza L. M., Meira J. A., de Araújo J. M., Mitchell D. A., et al. (2007). Molecular and structural characterization of the biosurfactant produced by pseudomonas aeruginosa DAUPE 614. Chem. Phys. Lipids 147, 1–13. doi: 10.1016/j.chemphyslip.2007.02.001
Mukherjee S., Das P., Sen R. (2006). Towards commercial production of microbial surfactants. Trends Biotechnol. 24, 509–515. doi: 10.1016/j.tibtech.2006.09.005
Müller-Loennies S., Lindner B., Brade H. (2002). Structural analysis of deacylated lipopolysaccharide of escherichia coli strains 2513 (R4 core-type) and F653 (R3 core-type). FEBS J. 269, 5982–5991. doi: 10.1046/j.1432-1033.2002.03322.x
Nam J., Jung M. Y., Kim P. I., Lee H. B., Kim S. W., Lee C. W. (2015). Structural characterization and temperature-dependent production of C17-fengycin b derived from bacillus amyloliquefaciens subsp. plantarum BC32-1. Biotechnol. Bioproc. Eng. 20 (4), 708–713. doi: 10.1007/s12257-015-0350-3
Naughton P. J., Marchant R., Naughton V., Banat I. M. (2019). Microbial biosurfactants: current trends and applications in agricultural and biomedical industries. J. Appl. Microbiol. 127, 12–28. doi: 10.1111/jam.14243
Navon-Venezia S., Zosim Z., Gottlieb A., Legmann R., Carmeli S., Ron E. Z., et al. (1995). Alasan, a new bioemulsifier from acinetobacter radioresistens. Appl. Environ. Microbiol. 61, 3240–3244. doi: 10.1128/aem.61.9.3240-3244.1995
Nazarenko E. L., Crawford R. J., Ivanova E. P. (2011). The structural diversity of carbohydrate antigens of selected gram-negative marine bacteria. Mar. Drugs 9, 1914–1954. doi: 10.3390/md9101914
Nikolova C., Gutierrez T. (2020). Use of microorganisms in the recovery of oil from recalcitrant oil reservoirs: current state of knowledge, technological advances and future perspectives. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02996
Olsthoorn M. M., Petersen B. O., Duus J., Haverkamp J., Thomas-Oates J. E., Bock K., et al. (2000). The structure of the linkage between the O-specific polysaccharide and the core region of the lipopolysaccharide from salmonella enterica serovar typhimurium revisited. FEBS J. 267, 2014–2027. doi: 10.1046/j.1432-1327.2000.01205.x
Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ovchinnikova O. G., Doyle L., Huang B. S., Kimber M. S., Lowary T. L., Whitfield C. (2016). Biochemical characterization of bifunctional 3-Deoxy-beta-d-manno-oct-2-ulosonic acid (beta-kdo) transferase KpsC from escherichia coli involved in capsule biosynthesis. J. Biol. Chem. 291, 21519–21530. doi: 10.1074/jbc.M116.751115
Pereira J. F. B., Gudiña E. J., Costa R., Vitorino R., Teixeira J. A., Coutinho J. A. P., et al. (2013). Optimization and characterization of biosurfactant production by bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel 111, 259–268. doi: 10.1016/j.fuel.2013.04.040
Pereira A. G. B., Muniz E. C., Hsieh Y.-L. (2015). 1H NMR and 1H–13C HSQC surface characterization of chitosan–chitin sheath-core nanowhiskers. Carbohydr. Polym. 123, 46–52. doi: 10.1016/j.carbpol.2015.01.017
Ploutno A., Carmeli S. (2002). Modified peptides from a water bloom of the cyanobacterium nostoc sp. Tetrahedron 58, 9949–9957. doi: 10.1016/S0040-4020(02)01326-1
Post D. M., Yu L., Krasity B. C., Choudhury B., Mandel M. J., Brennan C. A., et al. (2012). O-Antigen and core carbohydrate of vibrio fischeri lipopolysaccharide: composition and analysis of their role in euprymna scolopes light organ colonization. J. Biol. Chem. 287 (11), 8515–8530. doi: 10.1074/jbc.M111.324012
Price N.-P., Ray K.-J., Vermillion K.-E., Dunlap C.-A., Kurtzman C.-P. (2012). Structural characterization of novel sophorolipid biosurfactants from a newly identified species of candida yeast. Carbohydr. Res. 348, 33–41. doi: 10.1016/j.carres.2011.07.016
Priji P., Sajith S., Unni K. N., Anderson R. C., Benjamin S. (2017). Pseudomonas sp. BUP6, a novel isolate from malabari goat produces an efficient rhamnolipid type biosurfactant. J. Basic Microbiol. 57, 21–33. doi: 10.1002/jobm.201600158
Pueyo M. T., Bloch C. Jr, Carmona-Ribeiro A. M., di Mascio P. (2009). Lipopeptides produced by a soil bacillus megaterium strain. Microb. Ecol. 57, 367–378. doi: 10.1007/s00248-008-9464-x
Raaijmakers J. M., De Bruijn I., Nybroe O., Ongena M. (2010). Natural functions of lipopeptides from bacillus and pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x
Razafindralambo H., Paquot M., Hbid C., Jacques P., Destain J., Thonart P. (1993). Purification of antifungal lipopeptides by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 639, 81–85. doi: 10.1016/0021-9673(93)83091-6
Remaud G. S., Bussy U., Lees M., Thomas F., Desmurs J. R., Jamin E., et al. (2013). NMR spectrometry isotopic fingerprinting: A tool for the manufacturer for tracking active pharmaceutical ingredients from starting materials to final medicines. Eu J. Pharm. Sci. 48 (3), 464–473. doi: 10.1016/j.ejps.2012.12.009
Rezanka T., Siristova L., Sigler K. (2011). Rhamnolipid-producing thermophilic bacteria of species thermus and meiothermus. Extremophiles 15 (6), 697–709. doi: 10.1007/s00792-011-0400-5
Roelants S. L. K. W., Ciesielska K., De Maeseneire S. L., Moens H., Everaert B., Verweire S., et al. (2016). Towards the industrialization of new biosurfactants: Biotechnological opportunities for the lactone esterase gene from starmerella bombicola. Biotechnol. Bioeng 113, 550–559. doi: 10.1002/bit.25815
Romero D., de Vicente A., Rakotoaly R. H., Dufour S. E., Veening J. W., Arrebola E., et al. (2007). The iturin and fengycin families of lipopeptides are key factors in antagonism of bacillus subtilis toward podosphaera fusca. Mol. Plant Microbe Interact. 20, 430–440. doi: 10.1094/MPMI-20-4-0430
Rosenberg E., Perry A., Gibson D. T., Gutnick D. L. (1979). Emulsifier of arthrobacter RAG-1: specificity of hydrocarbon substrate. Appl. Environ. Microbiol. 37, 409–413. doi: 10.1128/aem.37.3.409-413.1979
Rosenberg E., Ron E. Z. (1999). High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 52, 154–162. doi: 10.1007/s002530051502
Rudden M., Tsauosi K., Marchant R., Banat I. M., Smyth T. J. (2015). Development and validation of an ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) method for the quantitative determination of rhamnolipid congeners. Appl. Microbiol. Biotechnol. 99, 9177–9187. doi: 10.1007/s00253-015-6837-1
Salinero C., Feás X., Mansilla J. P., Seijas J. A., Vázquez-Tato M. P., Vela P., et al. (2012). ¹H-nuclear magnetic resonance analysis of the triacylglyceride composition of cold-pressed oil from camellia japonica. Molecules 17, 6716–6727. doi: 10.3390/molecules17066716
Sarpal A. S., Silva P. R. M., Monteiro T. V., Fonseca M., Cunha V. S. (2016). Monitoring of development of certified reference material of biodiesel by high field NMR spectroscopic techniques. J. Sci. Ind. Metrol. 1, 3. doi: 10.1016/0021-9673(94)01127-Z
Schenk T., Schuphan I., Schmidt B.. (2005). High-performance liquid chromatographic determination of the rhamnolipids produced by Pseudomonas aeruginosa. J. Chromatogr. A. 693, 7–13. doi: 10.1016/0021-9673(94)01127-z
Silipo A., Leone S., Molinaro A., Sturiale L., Garozzo D., Nazarenko E. L., et al. (2005). Complete structural elucidation of a novel lipooligosaccharide from the outer membrane of the marine bacterium shewanella pacifica. Eur. J. Org. Chem. 2005, 2281–2291. doi: 10.1002/ejoc.200400882
Smyth T. J., Rudden M., Tsaousi K., Marchant R., Banat I. M. (2014). “Protocols for the isolation and analysis of lipopeptides and bioemulsifiers,” in Hydrocarbon and lipid microbiology protocols (Berlin, Heidelberg: Springer), 3–28. doi: 10.1007/8623_2014_29
Sobolev A. P., Thomas F., Donarski J., Ingallina C., Circi S., Marincola F. C., et al. (2019). Use of NMR applications to tackle future food fraud issues. Trends Food Sci. Technol. 91, 347–353. doi: 10.1016/j.tifs.2019.07.035
Son S., Ko S.-K., Jang M., Kim J., Kim G., Lee J., et al. (2016). New cyclic lipopeptides of the iturin class produced by saltern-derived bacillus sp. KCB14S006. Mar. Drugs 14, 72. doi: 10.3390/md14040072
Sriram M. I., Gayathiri S., Gnanaselvi U., Jenifer P. S., Raj S. M., Gurunathan S. (2011). Novel lipopeptide biosurfactant produced by hydrocarbon degrading and heavy metal tolerant bacterium escherichia fergusonii KLU01 as a potential tool for bioremediation. Bioresour. Technol. 102, 9291–9295. doi: 10.1016/j.biortech.2011.06.094
Starr K. F., Porsch E. A., Heiss C., Black I., Azadi P., Geme J. W. S. III (2013). Characterization of the kingella kingae polysaccharide capsule and exopolysaccharide. PloS One 8, e75409. doi: 10.1371/journal.pone.0075409
Strader M. B., Tabb D. L., Hervey W. J., Pan C., Hurst G. B. (2006). Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal. Chem. 78, 125–134. doi: 10.1021/ac051348l
Sun L., Lu Z., Bie X., Lu F., Yang S. (2006). Isolation and characterization of a co-producer of fengycins and surfactins, endophytic bacillus amyloliquefaciens ES-2, from scutellaria baicalensis georgi. World J. Microbiol. Biotechnol. 22, 1259–1266. doi: 10.1007/s11274-006-9170-0
Symmank H., Franke P., Saenger W., Bernhard F. (2002). Modification of biologically active peptides: production of a novel lipohexapeptide after engineering of bacillus subtilis surfactin synthetase. Protein Eng. 15, 913–921. doi: 10.1093/protein/15.11.913
Taguchi S., Ooi T., Mizuno K., Matsusaki H. (2015). Advances and needs for endotoxin-free production strains. Appl. Microbiol. Biotechnol. 99, 9349–9360. doi: 10.1007/s00253-015-6947-9
Tang J. S., Gao H., Hong K., Yu Y., Jiang M. M., Lin H. P., et al. (2007). Complete assignments of 1H and 13C NMR spectral data of nine surfactin isomers. Magn. Reson. Chem. 45 (9), 792–796. doi: 10.1002/mrc.2048
Tan K. C., Wakimoto T., Abe I. (2016). Sulfoureido lipopeptides from the marine sponge discodermia kiiensis. J. Nat. Prod. 79 (9), 2418–2422. doi: 10.1021/acs.jnatprod.6b00586
Tarazona M. P., Saiz E. (2003). Combination of SEC/MALS experimental procedures and theoretical analysis for studying the solution properties of macromolecules. J. Biochem. Biophys. Methods 56, 95–116. doi: 10.1016/S0165-022X(03)00075-7
Tareq F., Lee M., Lee H.-S., Lee J.-S., Lee Y.-J., Shin H. (2014). Gageostatins a–c, antimicrobial linear lipopeptides from a marine bacillus subtilis. Mar. Drugs 12, 871–885. doi: 10.3390/md12020871
Tedesco P., Maida I., Palma Esposito F., Tortorella E., Subko K., Ezeofor C. C., et al. (2016). Antimicrobial activity of monoramnholipids produced by bacterial strains isolated from the Ross Sea (Antarctica). Mar. Drugs 14, 83. doi: 10.3390/md14050083
Tian G.-Z., Hu J., Zhang H.-X., Rademacher C., Zou X.-P., Zheng H.-N., et al. (2018). Synthesis and conformational analysis of linear homo-and heterooligomers from novel 2-c-branched sugar amino acids (SAAs). Sci. Rep. 8, 1–8. doi: 10.1038/s41598-018-24927-6
Tokumoto Y., Nomura N., Uchiyama H., Imura T., Morita T., Fukuoka T., et al. (2009). Structural characterization and surface-active properties of a succinoyl trehalose lipid produced by rhodococcus sp. SD-74. J. Oleo Sci. 58 (2), 97–102. doi: 10.5650/jos.58.97
Toren A., Orr E., Paitan Y., Ron E. Z., Rosenberg E. (2002). The active component of the bioemulsifier alasan from acinetobacter radioresistens KA53 is an OmpA-like protein. J. Bacteriol. 184 (1), 165–170. doi: 10.1128/JB.184.1.165-170.2002
Tripathi L., Twigg M. S., Zompra A., Salek K., Irorere V. U., Gutierrez T., et al. (2019). Biosynthesis of rhamnolipid by a marinobacter species expands the paradigm of biosurfactant synthesis to a new genus of the marine microflora. Microb. Cell Fact 18, 164. doi: 10.1186/s12934-019-1216-8
Tulla-Puche J., El-Faham A., Galanis A. S., de Oliveira E., Zompra A. A., Albericio F. (2015). “Methods for the peptide synthesis and analysis,” in Peptide chemistry and drug design (Hoboken, NJ, USA: John Wiley & Sons, Inc), 11–73.
Twigg M. S., Baccile N., Banat I. M., Déziel E., Marchant R., Roelants S., et al. (2021). Microbial biosurfactant research: time to improve the rigour in the reporting of synthesis, functional characterization and process development. Microb. Biotechnol. 14, 147–170. doi: 10.1111/1751-7915.13704
Twigg M. S., Tripathi L., Zompra A., Salek K., Irorere V. U., Gutierrez T., et al. (2018). Identification and characterisation of short chain rhamnolipid production in a previously uninvestigated, non-pathogenic marine pseudomonad. Appl. Microbiol. Biotechnol. 102, 8537–8549. doi: 10.1007/s00253-018-9202-3
van den Broek I., Sparidans R. W., Schellens J. H. M., Beijnen J. H. (2008). Quantitative bioanalysis of peptides by liquid chromatography coupled to (tandem) mass spectrometry. J. Chromatogr. B 872, 1–22. doi: 10.1016/j.jchromb.2008.07.021
Vater J., Kablitz B., Wilde C., Franke P., Mehta N., Cameotra S. S. (2002). Matrix-assisted laser desorption ionization-time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of bacillus subtilis c-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 68, 6210–6219. doi: 10.1128/AEM.68.12.6210-6219.2002
Viebke C., Borgström J., Piculell L. (1995). Characterisation of kappa- and iota-carrageenan coils and helices by MALLS/GPC. Carbohydr. Polym. 27, 145–154. doi: 10.1016/0144-8617(95)00017-2
Vinogradov E., Korenevsky A., Beveridge T. J. (2003). The structure of the rough-type lipopolysaccharide from shewanella oneidensis MR-1, containing 8-amino-8-deoxy-Kdo and an open-chain form of 2-acetamido-2-deoxy-D-galactose. Carbohydr. Res. 338, 1991–1997. doi: 10.1016/S0008-6215(03)00325-2
Wyatt P. J. (1993). Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272, l–40. doi: 10.1016/0003-2670(93)80373-S
Xiu P., Liu R., Zhang D., Sun C. (2017). Pumilacidin-like lipopeptides derived from marine bacterium bacillus sp. strain 176 suppress the motility of vibrio alginolyticus. Appl. Environ. Microbiol. 83, e00450–e00417. doi: 10.1128/AEM.00450-17
Yakimov M. M., Timmis K. N., Wray V., Fredrickson H. L. (1995). Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface bacillus licheniformis BAS50. Appl. Environ. Microbiol. 61, 1706–1713. doi: 10.1128/aem.61.5.1706-1713.1995
Yamada S., Yoshida K., Sugiura M., Sugahara K. (1992). One- and two-dimensional 1H-NMR characterization of two series of sulfated disaccharides prepared from chondroitin sulfate and heparan Sulfate/Heparin by bacterial eliminase digestion. J. Biochem. 112, 440–447. doi: 10.1093/oxfordjournals.jbchem.a123919
Zachara N. E., Gooley A. A. (2000). “Identification of glycosylation sites in mucin peptides by edman degradation,” in Glycoprotein methods and protocols: The mucins. Ed. Corfield A. P., (Totowa, New Jersey, United States: Humana Press) 121–128. doi: 10.1385/1-59259-048-9:121
Zähringer U., Ittig S., Lindner B., Moll H., Schombel U., Gisch N., et al. (2015). NMR-based structural analysis of the complete rough-type lipopolysaccharide isolated from capnocytophaga canimorsus. J. Biol. Chem. 290, 25273. doi: 10.1074/jbc.A114.571489
Keywords: biosurfactants, biophysical analysis, NMR, LC-MS, HPLC, SEC
Citation: Zompra AA, Chasapi SA, Twigg MS, Salek K, Anestopoulos I, Galanis A, Pappa A, Gutierrez T, Banat IM, Marchant R, Euston SR, Panayiotidis MI and Spyroulias GA (2022) Multi-method biophysical analysis in discovery, identification, and in-depth characterization of surface‐active compounds. Front. Mar. Sci. 9:1023287. doi: 10.3389/fmars.2022.1023287
Received: 19 August 2022; Accepted: 06 October 2022;
Published: 20 October 2022.
Edited by:
Sophia Letsiou, Agricultural University of Athens, GreeceReviewed by:
Eduardo J. Gudiña, University of Minho, PortugalCopyright © 2022 Zompra, Chasapi, Twigg, Salek, Anestopoulos, Galanis, Pappa, Gutierrez, Banat, Marchant, Euston, Panayiotidis and Spyroulias. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aikaterini A. Zompra, YXpvbXByYUB1cGF0cmFzLmdy; Stephen R. Euston, Uy5SLkV1c3RvbkBody5hYy51aw==; Mihalis I. Panayiotidis, bWloYWxpc3BAY2luZy5hYy5jeQ==; Georgios A. Spyroulias, Ry5BLlNweXJvdWxpYXNAdXBhdHJhcy5ncg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.