- 1School of Agriculture, Ludong University, Yantai, China
- 2College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
Mass mortalities of oysters during the summer have been reported for years. Among the biological factors causing oyster death in summer, Vibrio infection is considered to be the most important factor. Vibrio alginolyticus is one of the main pathogens causing oyster diseases. V. alginolyticus can cause disease and death of fish, shrimp, shellfish, and other aquatic products. Human infection with V. alginolyticus can also cause skin infections, external ear canal infections, and other diseases. Hemocytes in the blood of Crassostrea gigas resemble vertebrate macrophages and play an important role in the immune regulation of organisms. Therefore, it is of great significance to analyze the blood transcriptome to study the anti-pathogen mechanism of the C. gigas. In our study, we performed a transcriptome profile of gene expression on C. gigas infected with V. alginolyticus at 12 and 48 hours, and identified 2,494 and 1,165 differentially expressed genes (DEGs) at two sampling time points. Then, the functions of DEGs were identified using GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) functional enrichment analysis, revealing that there were multiple significantly enriched GO terms and KEGG signaling pathways related to immunity. And a protein-protein interaction (PPI) network was construct using DEGs enriched in significant enriched immune-related KEGG signaling pathways. At last, we screened and validated 12 key genes with multiple protein interaction numbers or involved in more KEGG signaling pathways, and verified their expression changes by quantitative RT-PCR (qRT-PCR). For the first time, we studied the immune defense mechanism of C. gigas blood based on protein-protein interaction network, which explains how C. gigas lacking specific immunity survive in a pathogen-rich environment. This research provides reference for solving the problem of high mortality of C. gigas and other mollusks in summer, and provides reference for the future production of some disease-resistant C. gigas.
Introduction
The innate or nonspecific responses exist in all living organisms, which is a quick response, efficient and ancient immune defense system (Macagno et al., 2010). Some mollusks, such as bivalves, live in estuaries rich in pathogenic microorganisms, and they concentrate these microorganisms when they eat (Riisgard and Larsen, 2010). They lack specific immunity and can only depend on innate immune system to resist foreign microorganisms (Gestal et al., 2008). In invertebrates, innate defense relies on two elements: cells (Hemocytes) and body fluids (antimicrobial peptides, lectins, and lysozymes) (Söderhäll, 2010; Bachère et al., 2015). Hemocytes are considered to be the main effective components in the immune response of invertebrates (Zhang et al., 2014). Hemocytes, similar to vertebrate macrophages, act as an important role in immune protection (Buchmann, 2014).
Pacific oyster (Crassostrea gigas) originated from East Asia, and is widely cultivated in the coastal sea area of Korea, Japan, and northern China at the present stage (Guo, 2009). As a filter-feeding and sessile invertebrate living in estuaries and intertidal areas, C. gigas must cope with and adapt to the dynamic and changeable environment. dynamic and changing environment (Wang et al., 2018). How C. gigas lacking specific immunity survive in this pathogen-rich environment is a question worth exploring. There is growing evidence that over a long process of evolution and adaptation, oysters have developed a very complicated immune system, which can respond to various pathogens and environmental pressures (Huang et al., 2017; Xing et al., 2018; Huang et al., 2019; Chen et al., 2021). Mass mortalities of oysters during the summer have been reported for years (De Decker et al., 2011). Among the biological factors causing oyster death in summer, Vibrio infection is considered to be the most important factor (Meng et al., 2015). In the process of breeding, many oyster diseases are caused by Vibrio. The reported pathogenic Vibrio including Vibrio alginolyticus, Vibrio splendens, Vibrio giganteus, etc (Javier et al., 2017). V. alginolyticus is a gram-negative bacterium that is widespread in oceans and estuaries, and is one of the main Vibrio species that cause oyster disease, and can be isolated from patients, seawater, and aquatic products in a variety of environments (Lajnef et al., 2012; Xie et al., 2020). V. alginolyticus can cause disease and death of fish (Xie et al., 2020), shrimp (Xie et al., 2020), shellfish (Gómez-León et al., 2005; González-Escalona et al., 2006), and other aquatic products, and bring huge economic losses. If humans are infected with V. alginolyticus, they will suffer from skin infections, infections of the external ear canal and other diseases (Reilly et al., 2011; Zhou et al., 2021).
Recent research shows that transcriptome sequencing is a common method to study changes in biological immune regulation. For example, transcriptome analysis can be used to compare the changes of gene expression in different treatment conditions and tissues (Li et al., 2018). This technique is already widely used in bivalves such as Crassostrea angulata (Zeng et al., 2019), Pinctada fucata (Guan et al., 2017), Crassostrea virginica (Zhang et al., 2014). However, the transcriptome of blood of C. gigas infected with V. alginolyticus was not reported.
In our study, transcriptional profiles of oyster blood were examined at 12 and 48 h post-injection of V. alginolyticus. We then performed a bioinformatics analysis, including screening for differentially expressed genes (DEGs), Gene Ontology (GO) functional enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. We established protein interaction networks that is associated with immunity to analyze the role of hub genes in C. gigas immune process. Finally, we screened and verified the expression patterns of 12 key genes by quantitative RT-PCR (qRT-PCR). As a result of this research will provide an important basis for future studies on the immune mechanism of C. gigas infected with V. alginolyticus, and provide an important and valuable resource for the oyster aquaculture industry.
Materials and methods
Oysters and bacteria
All the C. gigas used in this experiment came from Kongtong Island (Yantai, Shandong Province, China). These C. gigas were about half a year old, whose average shell length and average weight were 42.40 cm (range: 40.31 cm - 49.99 cm) and 26.59 g (range: 17.34 g - 39.08 g). These oysters were grown in filtered seawater at 25 ± 1°C and fed with spirulina and replaced with clean seawater daily. V. alginolyticus were presented by researcher Bai Changming’s team. After being retrieved, the bacteria were temporarily stored in at -20 °C refrigerator for use. Before injection, V. alginolyticus was cultured in 2216E liquid medium at 28°C for 10 h to logarithmic proliferation phase.
Sample treatment and RNA preparation
We randomly divided healthy oysters into three groups: 10 oysters for blank control group (BCG), 70 oysters for PBS control group (PCG), 70 oysters for V. alginolyticus experiment group (VEG). The blank group was placed in normal aerated seawater, 50 μL V. alginolyticus (2 × 109 CFU) were injected into adductor muscle of oysters in VEG and oysters in PCG were injected with the same volume of PBS. In blank group, 6 oysters which blood was collected from the blood sinus were randomly selected before injection, then added with 1 mL TRIzol reagent and stored at -80°C for later extraction of blood RNA. At 12 h and 48 h post-injection, blood was collected in the PBS control group and V. alginolyticus experiment group in the same way. The blood of 6 oysters was randomly drawn at each sampling point for RNA extraction: BCG for 0 h (B-0h), PCG for 12 h (P-12h), VEG for 12 h (V-12h), PCG for 48 h (P-48h), and VEG for 48 h (V-48h). For each group, we randomly selected 6 oysters and divided them into 3 groups, and the RNA of the same mole ratio of oysters in each group was combined as templates for constructing RNA-Seq library. We used the remaining blood RNA after library construction for quantitative verification by quantitative real-time PCR.
Library construction and illumina sequencing
The extracted total RNA was used as the raw material for library construction. Total RNA was processed and library construction completed using the method of Li and Ge et al. (Li et al., 2018; Ge et al., 2018). Finally, the library preparation was sequenced on the Illumina NovaSeq platform to generate 150 bp paired reads.
Data processing and differential expression analysis
The raw data needs to be screened to ensure the quality and accuracy of subsequent analyses. It mainly includes removing reads with adapter, removing reads containing unknown base information, and removing inferior quality reads. Then calculate the cleaning data and GC content of Q20 and Q30. The clean reads were mapped to the C. gigas reference genome (Qi et al., 2021) by HISAT2 v2.0.5. Using Feature Counts v1.5.0-p3 count the number of reads mapped to each gene. The FPKM (fragments of transcribed sequence sequenced per million base pairs) that can measure expression levels was then calculated for each gene based on the read counts and the gene length mapped to that gene. Using the DESeq2 R package (1.20.0) screen out differentially expressed genes between different treatment groups. Genes with p-value ≤ 0.05 and |log2fold change| ≥ 1 were designated as DEGs.
Functional annotation and enrichment analysis
We performed GO and KEGG enrichment analysis on the screened DEGs using DAVID v6.8 (Jiao et al., 2012), which enables us to better comprehend the high-level functions and roles of these DEGs in biological systems.
Functional protein association networks construction
The PPI interaction network was established by the default parameter of STRING v11.5 online tool to further study the connections and interactions between genes in immune regulatory pathways.
Gene expression validation by quantitative real-time PCR
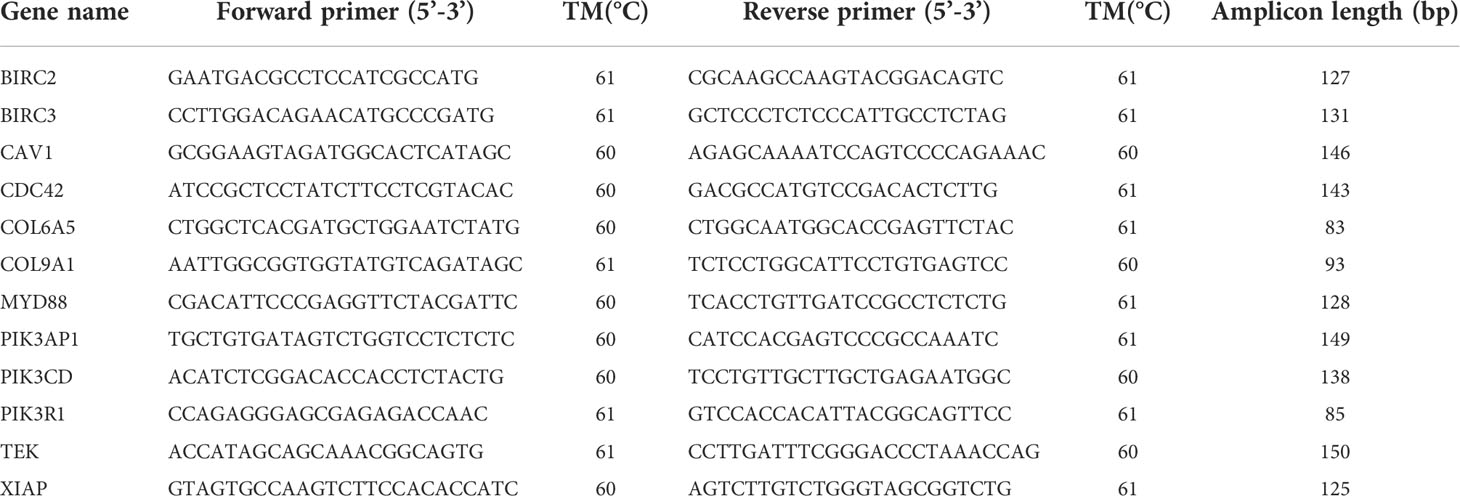
We examined 12 DEGs using quantitative real-time RT-PCR to validate the accuracy of transcriptome sequencing. The experimental material consisted of 6 oysters from each sampling point. Specific primers for selected genes are designed by Primer Premier 5.0. C. gigas β-actin (ACTB) was used as the reference gene because its expression levels were relatively stable at various time points during the test. Quantitative RT-PCR was performed in a total volume of 20 μL containing 1 μL of template cDNA, 0.8 μL of forward and reverse primers, 10 μL of TB Green® Premix Ex Taq™ II (Takara), and 7.4 μL of DEPC H2O by using CFX connect Real-Time PCR Detection System at 95°C for 30 s predeformation, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. We used the 2-ΔΔCT method to determine the relative expression of selected genes at each sampling point. The primer information of the selected 12 genes is shown in Table 1.
Results
Transcriptome sequence and reads mapping
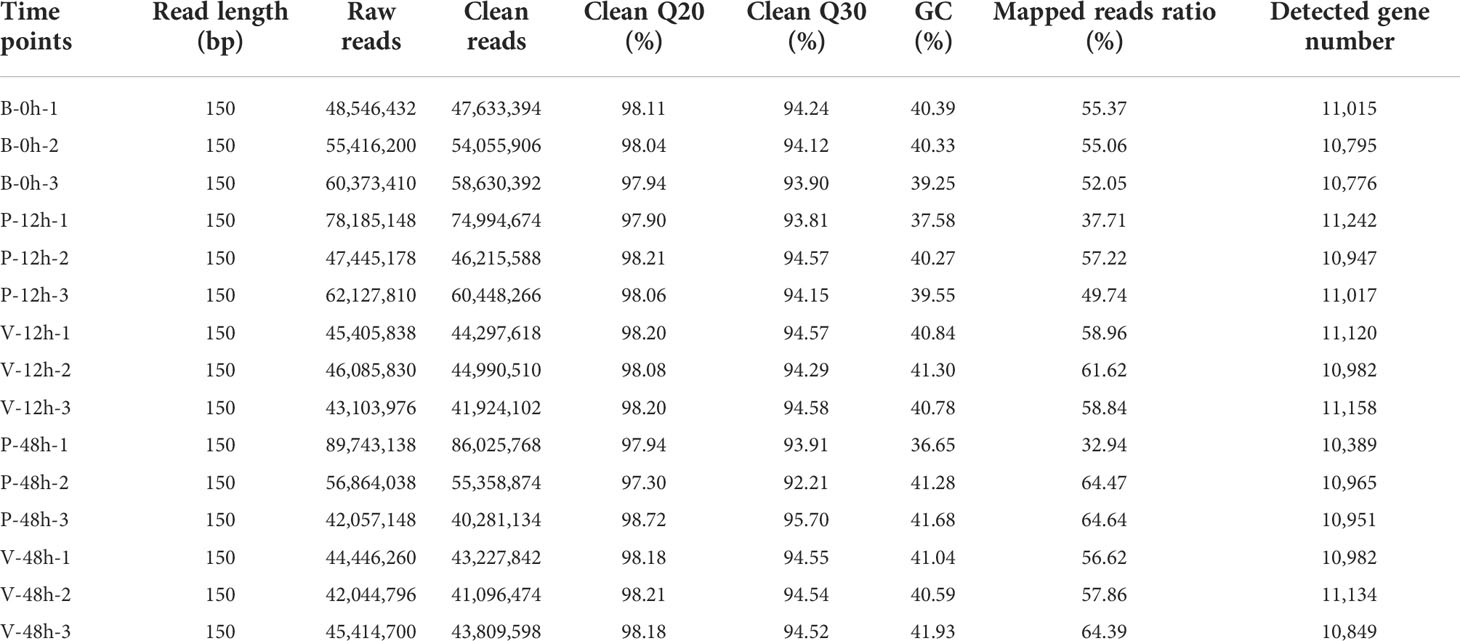
Transcriptome sequencing was performed at five sampling points, with three replicates per group. The RNA sequencing results are shown in Table 2. The screened clean reads were mapped to the reference genome (Qi et al., 2021), where the number of genes detected in each group was approximately 11,000. The resulting genes were then compared with the SwissProt database for further functional annotation.
Differential expression analysis
Differential expression analysis showed that two DEG lists were obtained in the comparison of V-12h versus P-12h and V-48h versus P-48h, generating 2,494 and 1,165 DEGs, respectively. Among these, 1,221 DEGs were up-regulated and 1,273 DEGs were down-regulated at 12h; 693 DEGs were up-regulated and 472 DEGs were down-regulated at 48h (Tables S1, S2, and Figure 1). Figure 2 shows all DEGs of C. gigas after infection with V. alginolyticus. Each of these DEGs may be involved in immunity of C. gigas, so a union of them (3,498) was selected for future analysis (Figure 2 and Table S4). Heatmap (Figure 3 and Table S4) visually presents the expression changes and clustering distribution of these DEGs.
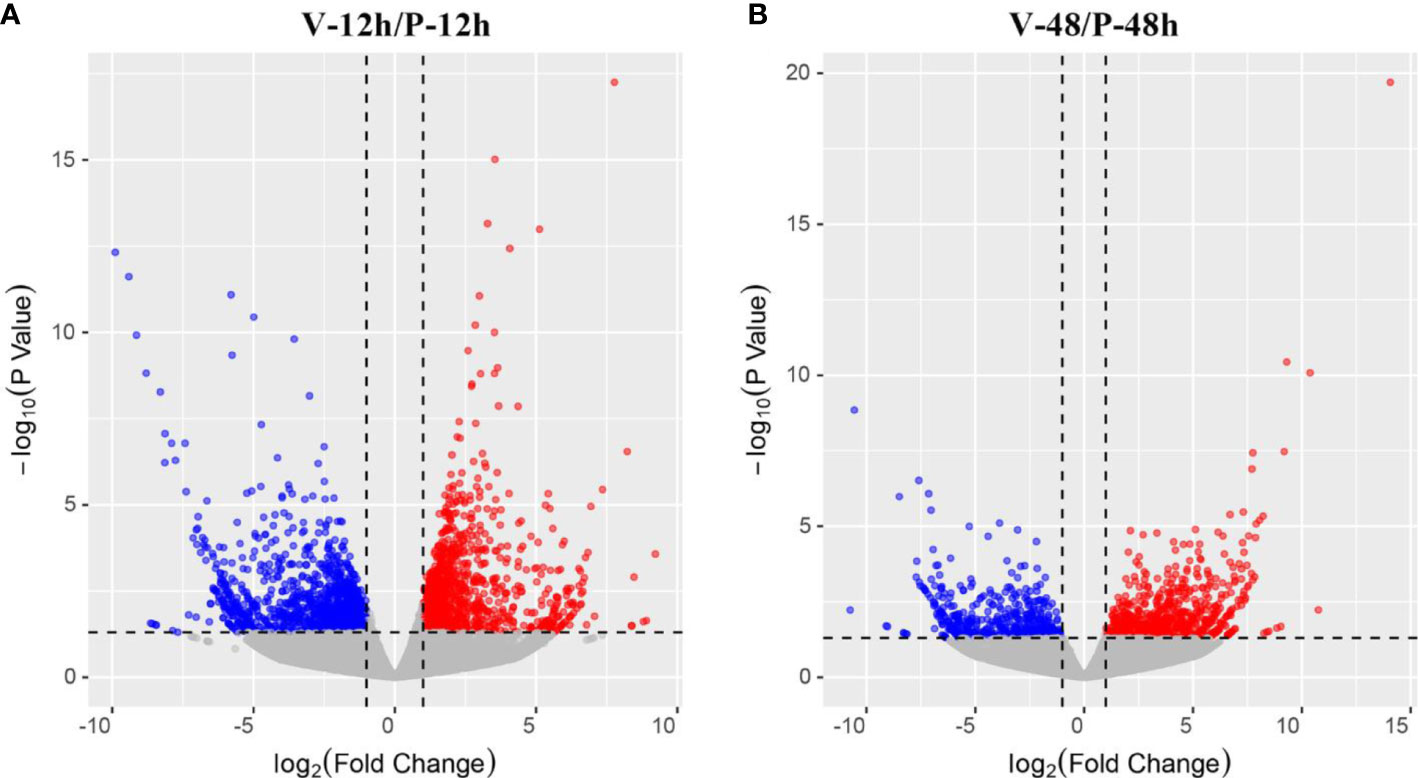
Figure 1 (A) Volcano plot of the distribution trend of DEGs between V-12h and P-12h groups. Each dot in this diagram represents a gene. Red dots represent up-regulated DEGs; blue dots represent down-regulated DEGs, and grey dots are not DEGs. (B) Volcano plot of the distribution trend of DEGs between V-48h and P-48h samples.
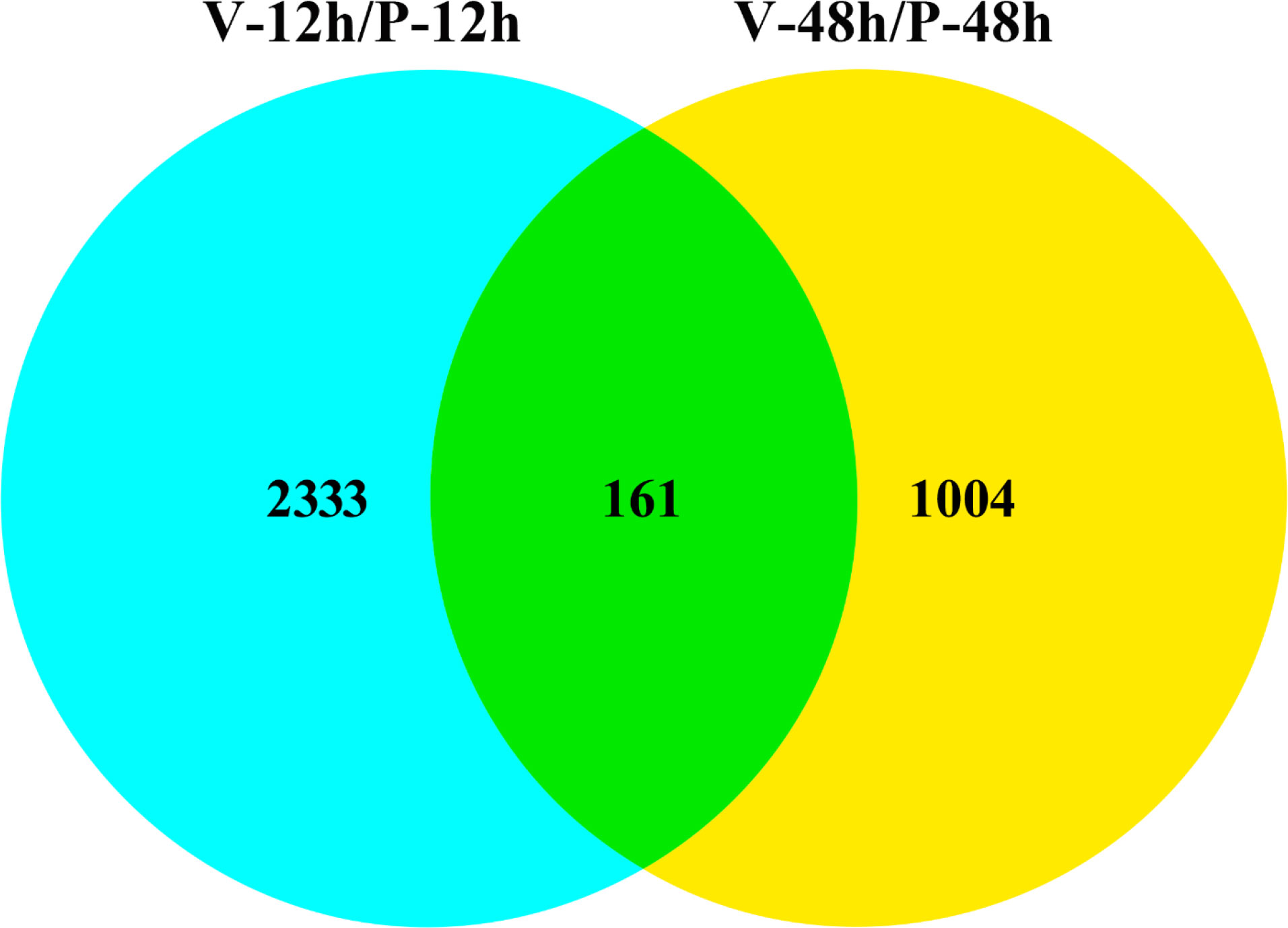
Figure 2 Venn diagram of overlapping DEGs at 12 h post-injection (blue) and 48 h post-injection (yellow). 2,333 DEGs are differentially expressed only at 12 h of infection; 1,004 DEGs are differentially expressed only at 48 h of infection; and 161 DEGs are differentially expressed at both two time points.
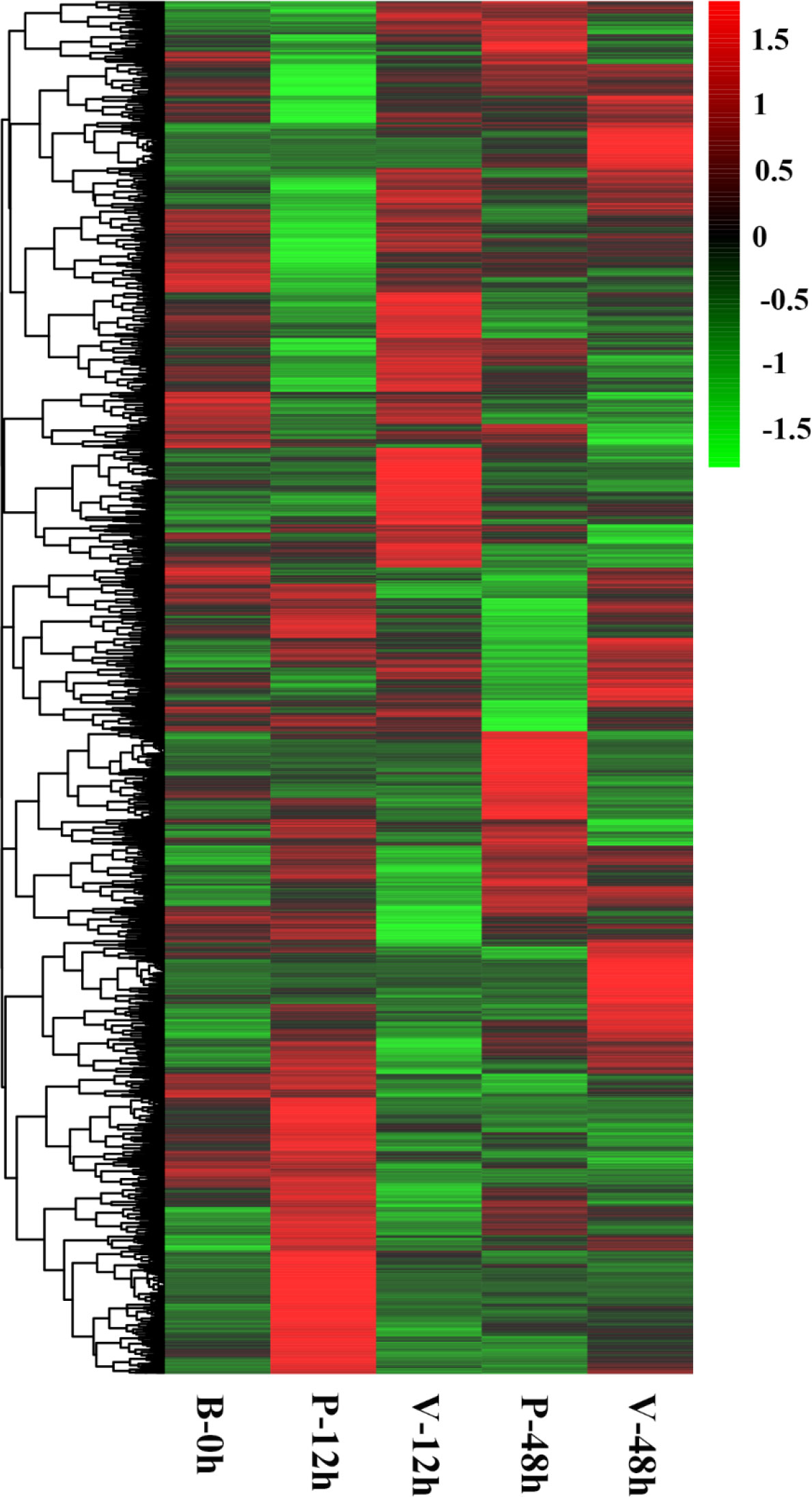
Figure 3 Heatmap analysis of expression patterns of all DEGs at the three time points. In this figure, each row represents a gene, and each column represents a sample group. Green to red indicates the increase of gene expression level.
GO and KEGG pathway enrichment analysis of DEGs
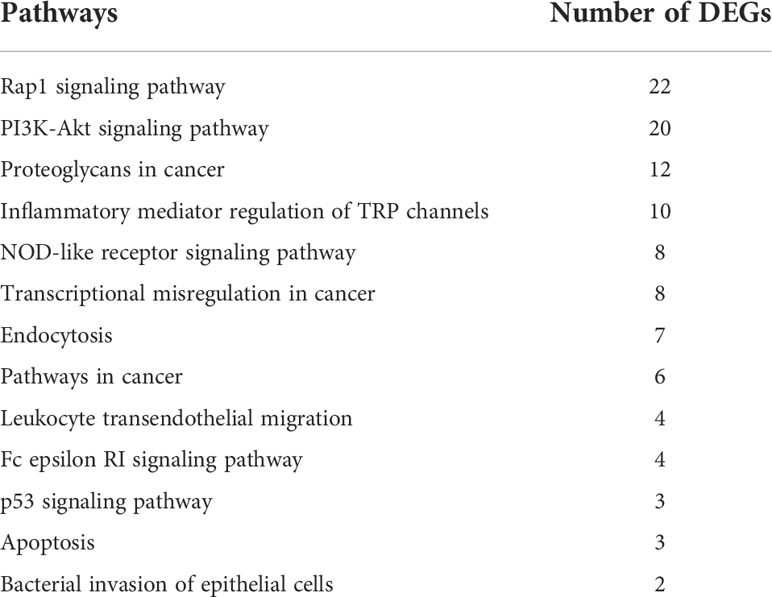
In our research, we selected 3,498 DEGs and performed GO and KEGG functional enrichment analysis on them. GO functional enrichment analysis results were divided into three ontologies. There are 151 third-class subclasses in biological process, 46 third-class subclasses in cellular component and 57 third-class subclasses in molecular function (Table S5). Level-3 GO terms of the top 20 biological process or top 10 cellular component and molecular function are shown in Figure 4. Through KEGG pathway analysis, we can further understand the role of DEGs in biological process, especially immune-related functions. Specifically, out of 3,498 DEGs, there are 339 annotated 156 KEGG signaling pathways (Table S6 and Figure 5). Among these pathways, there were 13 important and significantly enriched signaling pathways that are associated with immunity (Table 3).
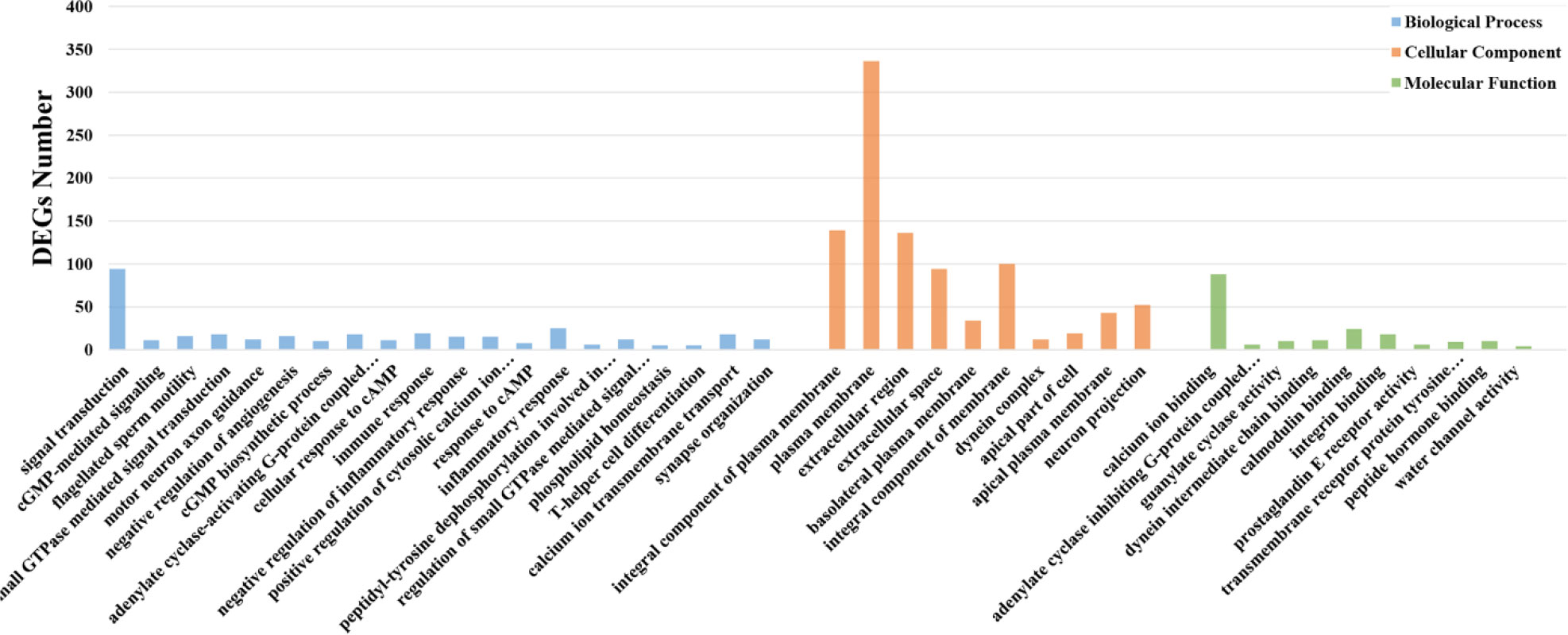
Figure 4 GO enrichment analysis of DEGs. Different distributions of level-3 GO annotation in three ontologies. The abscissa represents the enriched GO term; the ordinate represents DEGs number of the corresponding GO term.
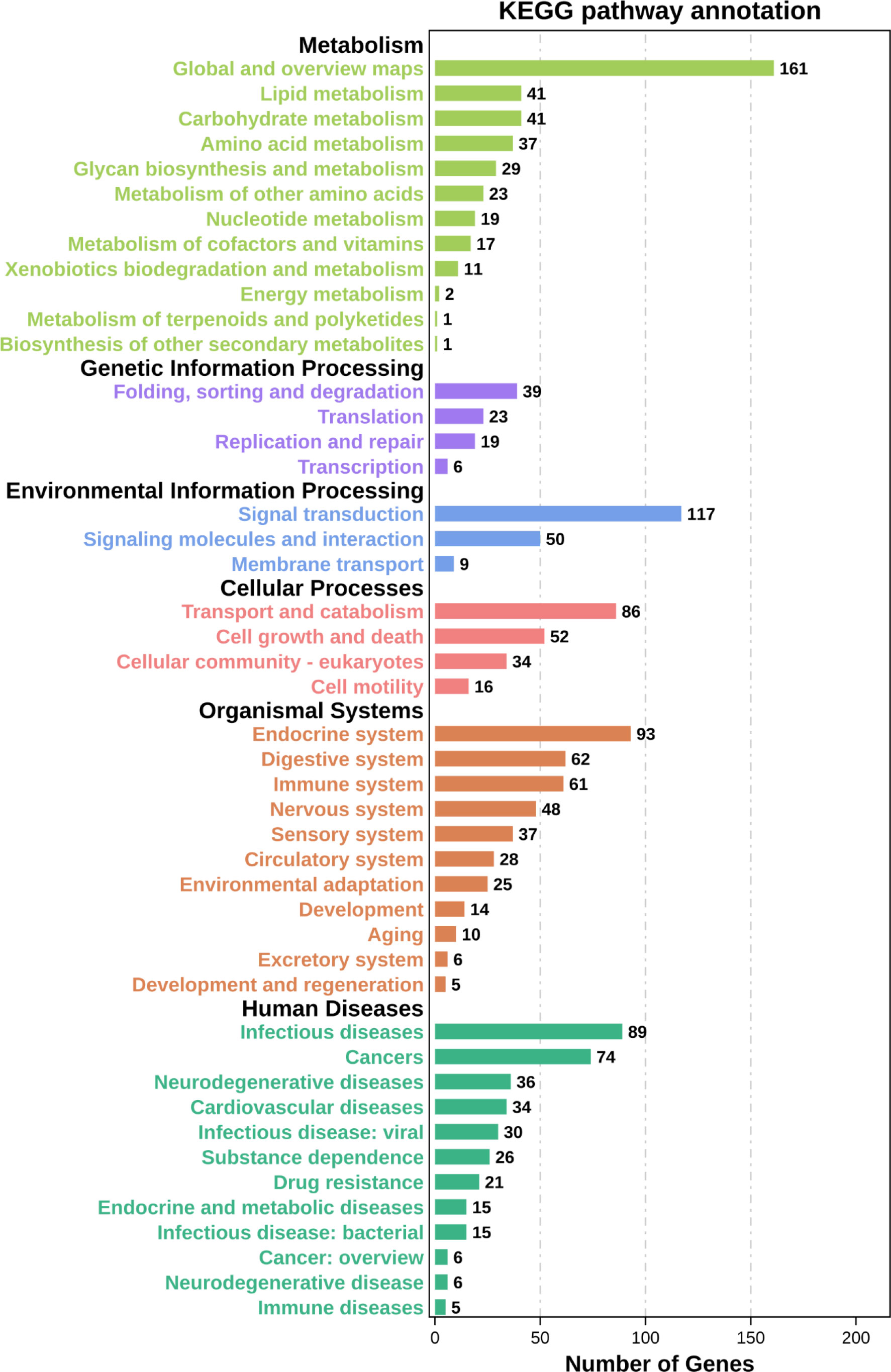
Figure 5 KEGG enrichment analysis of DEGs. The ordinate represents level-2 KEGG classes and the abscissa represents the corresponding DEGs number.
Construction of immune-related protein interaction networks
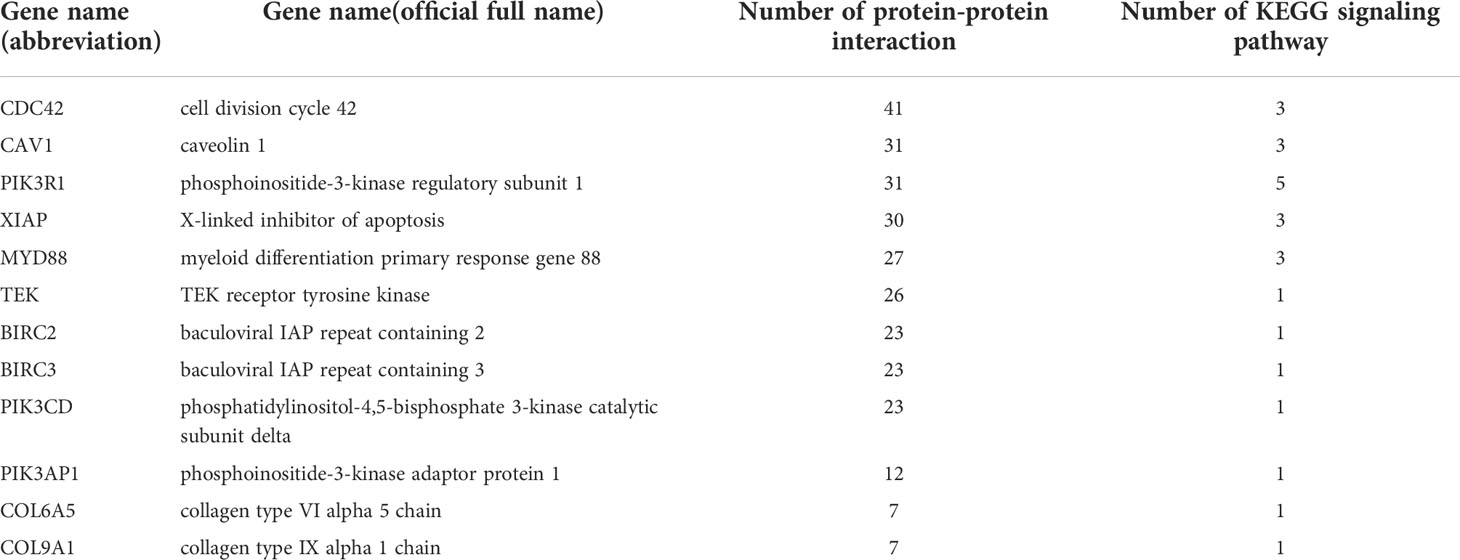
Protein is the material basis for the immune defense function of organisms, and are involved in tissue formation and organ development of the immune system. Constructing protein-protein interaction (PPI) network is helpful to discover important genes in immune regulation. In this study, we used 71 DEGs which were significantly enriched in 13 important immune-related signaling pathways to set up the PPI network (Figure 6). Information about specific network parameters is shown in (Table 4). From this table, we can see that the number of protein interaction edges corresponding to the 43 immune-related DEGs we selected is higher than the expected number of edges, indicating that there is a significant interaction relationship between these genes.
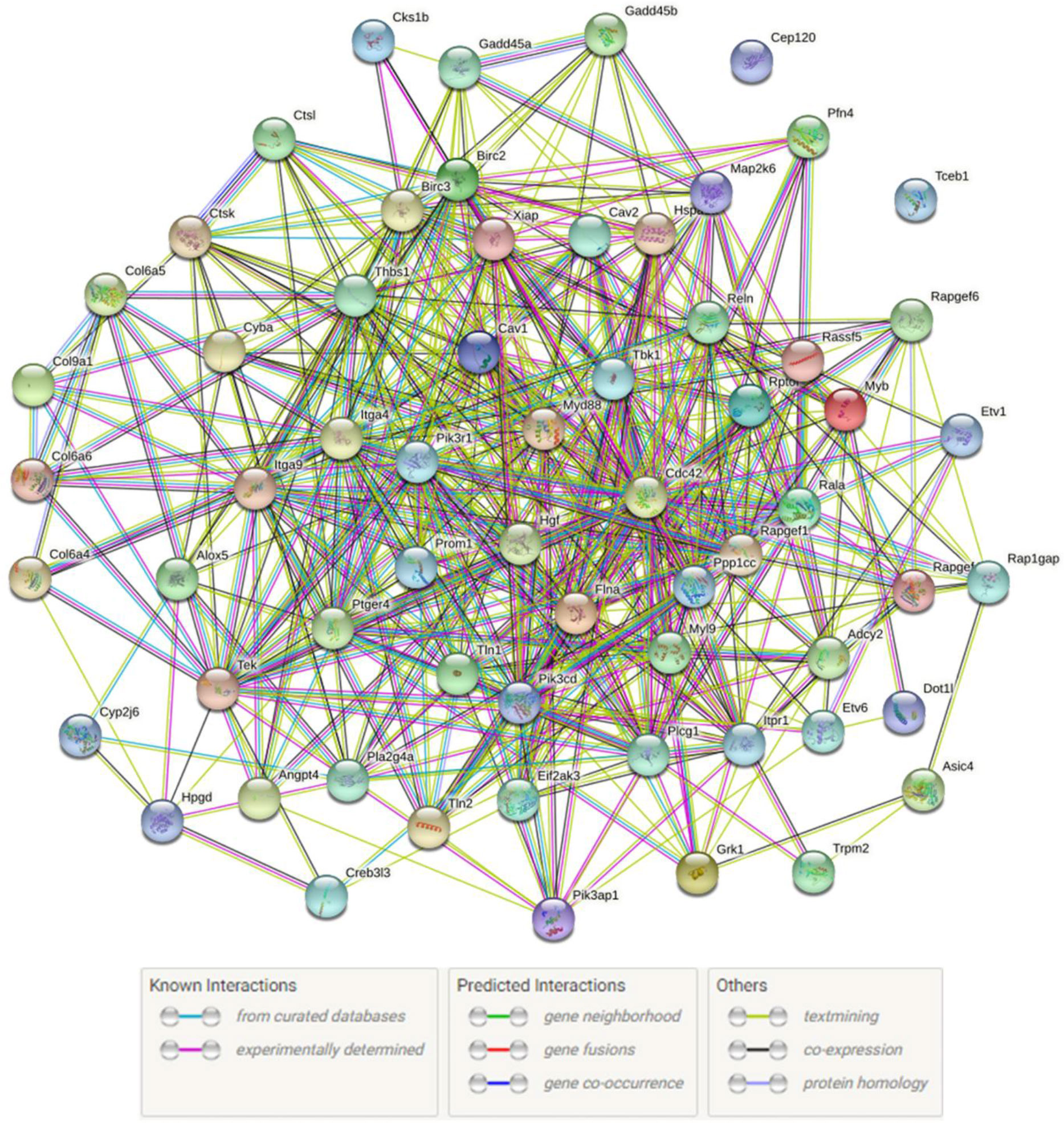
Figure 6 Immune-related PPI networks. Network nodes represents proteins. The legend represents the interaction relationship between the individual proteins.
Analysis of important DEGs associated with immune responses
In our study, we chiefly analyzed the interaction between immune-related DEGs. In total, we validated 12 key DEGs with multiple interactions or involvement in various signaling pathways (Table 5). These 12 key genes are divided into 5 categories: Inhibitor of apoptosis family, Collagen family, PI3K-Akt signaling pathway, NOD-like receptor signaling pathway and other important genes that are involved in various immune processes in oysters. Through these immune-related gene families and pathways, the immune defense mechanism of C. gigas was studied.
Quantitative RT-PCR validation
In order to verify the results of transcriptome analysis, we detected the relative expression changes of 12 immune-related DEGs at 3 time points by using quantitative RT-PCR. The fold change detected by qPCR of selected DEGs was compared with fold change detected by RNA-Seq. As shown in Figure 7, the expression trends of the selected genes at each time point were verified by qRT-PCR to be the same as the sequencing results, indicating that the RNA-Seq sequencing results in this study were accurate.
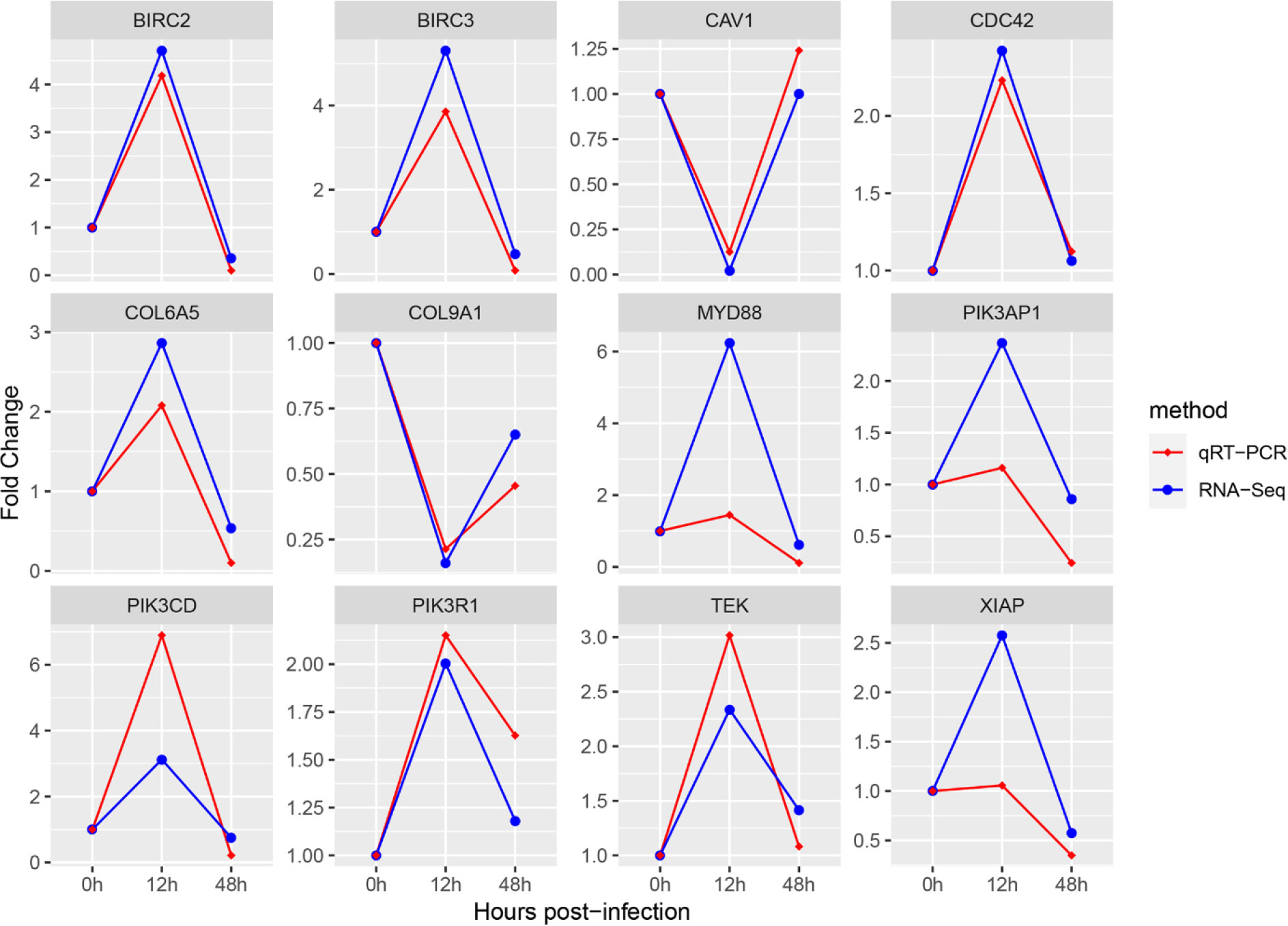
Figure 7 Comparison of 12 hub genes quantitative RT-PCR and RNA-Seq expression changes. The transcriptional expression levels of the selected DEGs were all normalized by the expression levels of the β-actin gene. The abscissa represents the change of sampling time, and the ordinate represents the fold change of experimental group compared with control group.
Discussion
The purpose and significance of transcriptome study
The high mortality of oysters in summer seems to be caused by the complex physiological interaction between host, environment and invasive pathogens (Samain, 2011). It was found that Vibrio infection was an important factor for serious reduction of oyster production in summer (Saulnier et al., 2010). Hemocytes of C. gigas, similar to vertebrate macrophages, play vital function in immune protection. Sequencing the transcriptome of the blood of C. gigas can help us understand preferably how C. gigas adapts to the environment affluent in pathogens. In order to better expound the changes in immune regulation of oyster infected with V. alginolyticus, we identified 2,494 and 1,165 DEGs at 12 h and 48 h post-injection, respectively. Every DEG may be involved in the oyster immune process, so we choose to use their union (3,498 DEGs) for the following analysis.
Enrichment of immune-related GO terms and KEGG pathways
GO enrichment analysis of DEGs after V. alginolyticus stimulation showed that some immune-related terms including immune response, negative regulation of immune response, and immune response were significantly enriched in biological processes (Figure 4). Some pathways significantly enriched in KEGG pathway, such as PI3K-Akt signaling pathway and NOD-like receptor signaling pathway are related to immunity. These results fully indicate that there are affluent immune-related genes in the blood of C. gigas, and various pathways in immune regulation are activated to eliminate the threat posed by V. alginolyticus. In-depth discussion of these enriched GO terms and KEGG Pathways provides us with ideas to explore the immune regulation mechanism of C. gigas infected with V. alginolyticus, find out the reasons for the high mortality of C. gigas in summer and ways to improve them.
Speculation of hub genes
Protein is the material basis of the immune defense function of the organism. Systematic analysis of the interaction between a large number of proteins and genes in the biological system is of great significance to explore the biological signals and immune regulation of C. gigas under special physiological conditions. Therefore, we constructed an immune-related PPI using the 71 DEGs obtained by KEGG enrichment analysis. As can be seen from Table 4, the number of interactions between the selected proteins is greater than expected. These results suggest that proteins interact as a group to regulate biological function. Therefore, proteins with many interactions are defined as immune-related hub proteins, and the genes corresponding to these hub proteins are hub genes that require attention and validation.
Functional analysis of hub genes and KEGG signaling pathways
In this study, the transcriptome analysis of the blood of the C. gigas compared the changes of the immune response at different time points after C. gigas infection with V. alginolyticus, and revealed the special molecular response of the C. gigas to the stimulation of V. alginolyticus. At last, twelve hub genes were identified, which have various protein-protein interactions and participate in multiple immune-related KEGG signaling pathways.
Inhibitor of apoptosis family
Apoptosis is a basic biological phenomenon by which many organisms can clear damaged and infected cells (Gervais et al., 2016). Inhibitor of apoptosis (IAP) protein maintains the stability between cell expansion and cell death and promote the immune responses (Qu et al., 2015). In contrast to necrosis, apoptosis is actively performed by the cell. Typical features of apoptosis have been described in oysters, such as DNA fragmentation, cell shrinks, and rapid blebbing and chromatin condenses (Sunila and Labanca, 2003; Gervais et al., 2016). In this study, XIAP, BIRC2, and BIRC3 were selected as hub genes for functional research. In this study, the expression levels of XIAP, BIRC2 and BIRC3 in oysters infected with V. alginolyticus at 12 h and 48 h increased first and then decreased compared with the control group. XIAP can effectively inhibit endogenous and exogenous pathway stimuli to regulate caspase activation and apoptosis (Kamada, 2013). Previous studies have demonstrated that transcripts encoding IAP proteins are significantly up-regulated during the initial phase of oyster exposure to Vibrio (Tanguy et al., 2018), which consistent with our study. Overexpression of IAP in the blood of C. gigas indicates that apoptosis is activated during the stress period of V. alginolyticus to maintain homeostasis.
NOD-like receptor signaling pathway
NOD-like receptor (NLR) protein is a common pattern recognition receptor that regulate cellular pathways that control growth and immune responses to stimuli, and is pivotal in cellular infection, pathogen recognition and innate immune responses (Shaw et al., 2010; Platnich and Muruve, 2019). Some recent studies have suggested that NLRs also play a rather important role in different immune processes including inflammatory responses (Zhen and Zhang, 2019) and cell death (Heim et al., 2019). Many hub genes are enriched into NOD-like receptor signaling pathway in the blood transcriptome of C. gigas. Genes enriched in pathways, MYD88 and CYBA, were up-regulated at 12 h post-injection and down-regulated at 48 h post-injection. MYD88 is an essential component of the innate immune, and is also pivotal in priming and activating the immune defense system (Du et al., 2013). Many MYD88 isoforms have been found in different mollusks, and the expression of MYD88 isoforms tended to be up-regulated under pathogen attack (Qiu et al., 2007; Guo et al., 2018; Priyathilaka et al., 2018). The CYBA gene encodes P22phox protein that is capable of destroying foreign bacteria and fungi as well as other pathogens (San Jose et al., 2008; Stasia, 2016). The results showed that many genes were enriched in the NOD-like receptor signaling pathway when C. gigas was infected with V. alginolyticus, which indicate that NLR plays a very important role in innate immune response of C. gigas and helps protect it from pathogens.
PI3K-Akt signaling pathway
PI3K-Akt signaling pathway can be activated by a variety of different types of bacteria and toxins, and takes part in the regulation of many basic cellular activities, such as cell growth, proliferation, survival, migration, invasion, apoptosis and metabolism (Cao et al., 2019; Liu et al., 2020). In this study, many hub genes such as phosphoinositide 3-kinases (PI3K) family and collagen family are enriched in this pathway. The genes of the PI3K family were all up-regulated after V. alginolyticus stress compared with the control group. Activation of PI3K can improve the immune defense ability of organism against pathogens and environmental pollutants and coordinate the phagocytosis of mollusk (Sun et al., 2016). The previous researches have shown that collagen and integrin can participate in the migration of oyster hemocytes, allowing hemocytes to penetrate into infected tissues and can also enhance the phagocytosis of hemocytes (Li et al., 2019; Lv et al., 2020). These results show that PI3K-Akt signaling pathway was activated by V. alginolyticus infection of C. gigas, and the upregulated expression of hub immune related genes improved the immune ability of the oyster, so as to cope with the harm caused by V. alginolyticus. The up-regulated expression of many immune-related genes enriched in the pathway enhanced the resistance of oyster to pathogens.
The top three hub genes in the number of interactions
In this research, we focused on the interaction between different immune genes, so as to analyze the changes of immune defense mechanism of C. gigas after infection with V. alginolyticus. We screened and analyzed the top three genes for protein interaction: CDC42, CAV1 and PIK3R1, which are also involved in multiple immune signaling pathways. CDC42, a small GTP-binding protein of the Rho family, is a core protein involved in cell cycle regulation and plays key roles in various signaling and biological processes in many organisms (Gerasimcik et al., 2017). CDC42 as a regulator controls the generation of functional granulocytes in C. gigas, and also regulates phagocytosis of granulocytes (Mao et al., 2020). CAV1 is an important functional protein in the caveolin family (Udayantha et al., 2018), and involve in many common physiological processes such as lipid transport, membrane transport and signal transduction (Liu et al., 2002). CAV1 can regard as the core of immune-related signaling pathways, such as NF-kappa B signaling pathway (Garrean et al., 2006) or Jak-Stat signaling pathway (Guo et al., 2011). PIK3R1 actively regulates the PI3K-Akt signaling pathway, promoting apoptosis and metabolic activity (Fan et al., 2020). Among them, CDC42, PIK3R1 were up-regulated at 12 h post-injection and slightly up-regulated at 48 h post-injection. These results suggest that these two genes may take part in the immune regulation of C. gigas. However, CAV1 was down-regulated at 12 h post-infection, and this result needs further research and verification. These three genes are involved in various immune processes, and have the largest number of protein interactions in our research. Therefore, it is necessary to explore the functions of these three genes in the blood immunity of C. gigas.
Other KEGG signaling pathways and hub genes
In addition to the signaling pathways and hub genes discussed above, we also identified other genes and pathways involved in the C. gigas immune response to V. alginolyticus. For example, inflammatory mediator regulation of TRP, Fc epsilon RI signaling pathway, TEK. These pathways and hub genes are involved in the immune defense of many organisms and have been validated in previous studies (Yano et al., 1997; Klemm and Ruland, 2006; Duitama et al., 2021). The details of the signaling pathways and genes involved in the immune process in the blood of C. gigas infected with V. alginolyticus require us to further explore in future studies.
Conclusion
In this research, we performed the transcriptome analysis of C. gigas blood after infection with V. alginolyticus. and constructed an immune-related PPI network. A total of 12 hub genes were identified, which participate in various KEGG signaling pathways and having various protein interactions with each other. For the first time, we studied the immune defense mechanism of C. gigas blood based on protein–protein interaction network. This research provides abundant reference data for future research on the specific response of C. gigas to V. alginolyticus infection. At the same time, it also provides reference for solving the high mortality of C. gigas and other mollusks in summer and help breed new varieties with stronger antibacterial properties.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA861156&o=acc_s%3Aa
Author contributions
JY, ZL and WW designed and supervised the study. EZ, LD, XB, XY, YL and YF prepared the samples. EZ, XB and ZL analyzed all sequencing data. EZ, ZL and WW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Earmarked Fund for Agriculture Seed Improvement Project of Shandong Province, China (No. 2020LZGC016), fund of central government guiding local for scientific and technological development (No. YDZX2021017).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1017445/full#supplementary-material
References
Bachère E., Rosa R. D., Schmitt P., Poirier A. C., Merou N., Charrière G. M., et al. (2015). The new insights into the oyster antimicrobial defense: Cellular, molecular and genetic view. Fish Shellfish Immunol. 46 (1), 50–64. doi: 10.1016/j.fsi.2015.02.040
Buchmann K. (2014). Evolution of innate immunity: Clues from invertebrates via fish to mammals. Front. Immunol. 5, 459. doi: 10.3389/fimmu.2014.00459
Cao Z., Liao Q., Su M., Huang K., Jin J., Cao D. (2019). AKT and ERK dual inhibitors: The way forward? Cancer Lett. 459, 30–40. doi: 10.1016/j.canlet.2019.05.025
Chen H., Cai X., Qiu H., Fang J., Wu X. (2021). A novel c-type lectin from Crassostrea gigas involved in the innate defense against Vibrio alginolyticus. Biochem. Biophys. Res. Commun. 566, 155–163. doi: 10.1016/j.bbrc.2021.05.092
De Decker S., Normand J., Saulnier D., Pernet F., Castagnet S., Boudry P. (2011). Responses of diploid and triploid pacific oysters Crassostrea gigas to Vibrio infection in relation to their reproductive status. J. Invertebrate Pathol. 106 (2), 179–191. doi: 10.1016/j.jip.2010.09.003
Duitama M., Moreno Y., Santander S. P., Casas Z., Sutachan J. J., Torres Y. P., et al. (2021). TRP channels as molecular targets to relieve cancer pain. Biomolecules 12 (1), 1. doi: 10.3390/biom12010001
Du Y., Zhang L., Huang B., Guan X., Li L., Zhang G., et al. (2013). Characterization, and expression of two myeloid differentiation factor 88 (Myd88) in pacific oyster, Crassostrea gigas. J. World Aquaculture Soc. 44 (6), 759–774. doi: 10.1111/jwas.12077
Fan Z., Liu Y., Shi Z., Deng K., Zhang H., Li Q., et al. (2020). MiR-155 promotes interleukin-1beta-induced chondrocyte apoptosis and catabolic activity by targeting PIK3R1-mediated PI3K/Akt pathway. J. Cell. Mol. Med. 24 (15), 8441–8451. doi: 10.1111/jcmm.15388
Garrean S., Gao X. P., Brovkovych V., Shimizu J., Zhao Y. Y., Vogel S. M., et al. (2006). Caveolin-1 regulates NF-kappaB activation and lung inflammatory response to sepsis induced by lipopolysaccharide. J. Immunol. 177 (7), 4853–4860. doi: 10.4049/jimmunol.177.7.4853
Ge J., Liu C., Tan J., Bian L., Chen S. (2018). Transcriptome analysis of scyphozoan jellyfish Rhopilema esculentum from polyp to medusa identifies potential genes regulating strobilation. Dev. Genes Evol. 228 (6), 243–254. doi: 10.1007/s00427-018-0621-z
Gerasimcik N., He M., Dahlberg C. I. M., Kuznetsov N. V., Severinson E., Westerberg L. S. (2017). The small rho GTPases Rac1 and Rac2 are important for T-cell independent antigen responses and for suppressing switching to IgG2b in mice. Front. Immunol. 8, 1264. doi: 10.3389/fimmu.2017.01264
Gervais O., Chollet B., Renault T., Arzul I. (2016). Flat oyster follows the apoptosis pathway to defend against the protozoan parasite Bonamia ostreae. Fish Shellfish Immunol. 56, 322–329. doi: 10.1016/j.fsi.2016.07.021
Gestal C., Roch P., Renault T., Pallavicini A., Paillard C., Novoa B., et al. (2008). Study of diseases and the immune system of bivalves using molecular biology and genomics. Rev. Fisheries Sci. 16, 133–156. doi: 10.1080/10641260802325518
Gómez-León J., Villamil L., Lemos M. L., Novoa B., Figueras A. (2005). Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ. Microbiol. 71 (1), 98–104. doi: 10.1128/AEM.71.1.98-104.2005
González-Escalona N., Blackstone G. M., DePaola A. (2006). Characterization of a Vibrio alginolyticus strain, isolated from alaskan oysters, carrying a hemolysin gene similar to the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 72 (12), 7925–7929. doi: 10.1128/AEM.01548-06
Guan Y., He M., Wu H. (2017). Differential mantle transcriptomics and characterization of growth-related genes in the diploid and triploid pearl oyster E. Mar. Genomics 33, 31–38. doi: 10.1016/j.margen.2017.01.001
Guo X. (2009). Use and exchange of genetic resources in molluscan aquaculture. Rev. Aquaculture. 1, 251–259. doi: 10.1111/j.1753-5131.2009.01014.x
Guo B., Liu S., Li J., Liao Z., Liu H., Xia H., et al. (2018). Identification and functional characterization of three myeloid differentiation factor 88 (MyD88) isoforms from thick shell mussel Mytilus coruscus. Fish Shellfish Immunol. 83, 123–133. doi: 10.1016/j.fsi.2018.09.027
Guo C. J., Yang X. B., Wu Y. Y., Yang L. S., Mi S., Liu Z. Y., et al. (2011). Involvement of caveolin-1 in the jak-stat signaling pathway and infectious spleen and kidney necrosis virus infection in mandarin fish (Siniperca chuatsi). Mol. Immunol. 48 (8), 992–1000. doi: 10.1016/j.molimm.2011.01.001
Heim V. J., Stafford C. A., Nachbur U. (2019). NOD signaling and cell death. Front. Cell Dev. Biol. 7, 208. doi: 10.3389/fcell.2019.00208
Huang B., Tang X., Zhang L., Li L., Zhang G. (2019). IKKϵ-like plays an important role in the innate immune signaling of the pacific oyster (Crassostrea gigas). Fish Shellfish Immunol. 93, 551–558. doi: 10.1016/j.fsi.2019.07.074
Huang Q., Yu M., Chen H., Zeng M., Chen D. (2017). LRFN (leucine-rich repeat and fibronectin type-III domain-containing protein) recognizes bacteria and promotes hemocytic phagocytosis in the pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 72, 622–628. doi: 10.1016/j.fsi.2017.11.049
Javier D., Barja J. L., Romalde J. L. (2017). New insights into pathogenic vibrios affecting bivalves in hatcheries: Present and future prospects. Front. Microbiol. 8, 762. doi: 10.3389/fmicb.2017.00762
Jiao X. L., Sherman B. T., Huang D. W., Stephens R., Baseler M. W., Lane H. C., et al. (2012). DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 28 (13), 1805–1806. doi: 10.1093/bioinformatics/bts251
Kamada S. (2013). Inhibitor of apoptosis proteins as E3 ligases for ubiquitin and NEDD8. Biomolecular Concepts 4 (2), 161–171. doi: 10.1515/bmc-2012-0036
Klemm S., Ruland J. (2006). Inflammatory signal transduction from the fc epsilon RI to NF-kappa b. Immunobiology 211 (10), 815–820. doi: 10.1016/j.imbio.2006.07.001
Lajnef R., Snoussi M., Romalde J. L., Nozha C., Hassen A. (2012). Comparative study on the antibiotic susceptibility and plasmid profiles of Vibrio alginolyticus strains isolated from four Tunisian marine biotopes. World J. Microbiol. Biotechnol. 28 (12), 3345–3363. doi: 10.1007/s11274-012-1147-6
Li H., Kong N., Sun J., Wang W., Li M., Gong C., et al. (2019). A C1qDC (CgC1qDC-6) with a collagen-like domain mediates hemocyte phagocytosis and migration in oysters. Dev. Comp. Immunol. 98, 157–165. doi: 10.1016/j.dci.2019.04.010
Li Z., Liu X., Liu J., Zhang K., Yu H., He Y., et al. (2018). Transcriptome profiling based on protein-protein interaction networks provides a core set of genes for understanding blood immune response mechanisms against Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 78, 100–113. doi: 10.1016/j.dci.2017.09.013
Liu R., Chen Y., Liu G., Li C., Song Y., Cao Z., et al. (2020). PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 11 (9), 797. doi: 10.1038/s41419-020-02998-6
Liu P., Rudick M., Anderson R. G. W. (2002). Multiple functions of caveolin-1. J. Biol. Chem. 277 (44), 41295–41298. doi: 10.1074/jbc.R200020200
Lv Z., Qiu L., Wang W., Liu Z., Liu Q., Wang L., et al. (2020). The members of the highly diverse Crassostrea gigas integrin family cooperate for the generation of various immune responses. Front. Immunol. 11, 1420. doi: 10.3389/fimmu.2020.01420
Macagno E. R., Gaasterland T., Edsall L., Bafna V., Soares M. B., Scheetz T., et al. (2010). Construction of a medicinal leech transcriptome database and its application to the identification of leech homologs of neural and innate immune genes. BMC Genomics 11, 407. doi: 10.1186/1471-2164-11-407
Mao F., Wong N. K., Lin Y., Zhang X., Liu K., Huang M., et al. (2020). Transcriptomic evidence reveals the molecular basis for functional differentiation of hemocytes in a marine invertebrate, Crassostrea gigas. Front. Immunol. 11, 911. doi: 10.3389/fimmu.2020.00911
Meng J., Zhang L., Huang B., Li L., Zhang G. (2015). Comparative analysis of oyster (Crassostrea gigas) immune responses under challenge by different Vibrio strains and conditions. Molluscan Res. 35 (1), 1–11. doi: 10.1080/13235818.2014.919696
Platnich J. M., Muruve D. A. (2019). NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophysics 670, 4–14. doi: 10.1016/j.abb.2019.02.008
Priyathilaka T. T., Bathige S., Lee S., Lee J. (2018). Molecular identification and functional analysis of two variants of myeloid differentiation factor 88 (MyD88) from disk abalone (Haliotis discus discus). Dev. Comp. Immunol. 79, 113–127. doi: 10.1016/j.dci.2017.10.010
Qi H., Li L., Zhang G. (2021). Construction of a chromosome-level genome and variation map for the pacific oyster Crassostrea gigas. Mol. Ecol. Resour. 21 (5), 1670–1685. doi: 10.1111/1755-0998.13368
Qiu L., Song L., Yu Y., Xu W., Ni D., Zhang Q. (2007). Identification and characterization of a myeloid differentiation factor 88 (MyD88) cDNA from zhikong scallop Chlamys farreri. Fish Shellfish Immunol. 23 (3), 614–623. doi: 10.1016/j.fsi.2007.01.012
Qu T., Zhang L., Wang W., Huang B., Li Y., Zhu Q., et al. (2015). Characterization of an inhibitor of apoptosis protein in Crassostrea gigas clarifies its role in apoptosis and immune defense. Dev. Comp. Immunol. 51 (1), 74–78. doi: 10.1016/j.dci.2015.02.011
Reilly G. D., Reilly C. A., Smith E. G., Baker-Austin C. (2011). Vibrio alginolyticus-associated wound infection acquired in British waters, Guernsey, July 2011. Eurosurveillance 16 (42), 3. doi: 10.2807/ese.16.42.19994-en
Riisgard H. U., Larsen P. S. (2010). Particle-capture mechanisms in suspension-feeding invertebrates. Mar. Ecol. Prog. 413 (18), 255–293. doi: 10.3354/meps08755
Samain J.-F. (2011). Review and perspectives of physiological mechanisms underlying genetically-based resistance of the pacific oyster Crassostrea gigasto summer mortality. Aquat. Living Resour. 24 (3), 227–236. doi: 10.1051/alr/2011144
San Jose G., Fortuno A., Beloqui O., Diez J., Zalba G. (2008). NADPH oxidase CYBA polymorphisms, oxidative stress and cardiovascular diseases. Clin. Sci. 114 (3), 173–182. doi: 10.1042/CS20070130
Saulnier D., De Decker S., Haffner P., Cobret L., Robert M., Garcia C. (2010). A large-scale epidemiological study to identify bacteria pathogenic to pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microbial Ecol. 59 (4), 787–798. doi: 10.1007/s00248-009-9620-y
Shaw P. J., Lamkanfi M., Kanneganti T. D. (2010). NOD-like receptor (NLR) signaling beyond the inflammasome. Eur. J. Immunol. 40 (3), 624–627. doi: 10.1002/eji.200940211
Söderhäll K. (2010). Invertebrate immunity. Adv. Exp. Med. Biol. 708. doi: 10.1007/978-1-4419-8059-5
Stasia M. J. (2016). CYBA encoding p22(phox), the cytochrome b558 alpha polypeptide: Gene structure, expression, role and physiopathology. Gene 586 (1), 27–35. doi: 10.1016/j.gene.2016.03.050
Sunila I., Labanca J. (2003). Apoptosis in the pathogenesis of infectious diseases of the eastern oyster Crassostrea virginica. Dis. Aquat. Organisms 56 (2), 163–170. doi: 10.3354/dao056163
Sun Y., Zhang X., Wang G., Lin S., Zeng X., Wang Y., et al. (2016). PI3K-AKT signaling pathway is involved in hypoxia/thermal-induced immunosuppression of small abalone Haliotis diversicolor. Fish Shellfish Immunol. 59, 492–508. doi: 10.1016/j.fsi.2016.11.011
Tanguy M., Gauthier-Clerc S., Pellerin J., Danger J. M., Siah A. (2018). The immune response of mytilus edulis hemocytes exposed to Vibrio splendidus LGP32 strain: A transcriptomic attempt at identifying molecular actors. Fish Shellfish Immunol. 74, 268–280. doi: 10.1016/j.fsi.2017.12.038
Udayantha H. M. V., Bathige S., Priyathilaka T. T., Lee S., Kim M. J., Lee J. (2018). Identification and characterization of molluscan caveolin-1 ortholog from Haliotis discus discus: Possible involvement in embryogenesis and host defense mechanism against pathogenic stress. Gene Expression Patterns 27, 85–92. doi: 10.1016/j.gep.2017.11.003
Wang L., Song X., Song L. (2018). The oyster immunity. Dev. Comp. Immunol. 80, 99–118. doi: 10.1016/j.dci.2017.05.025
Xie J., Bu L., Jin S., Wang X., Zhao Q., Zhou S., et al. (2020). Outbreak of vibriosis caused by Vibrio harveyi and Vibrio alginolyticus in farmed seahorse Hippocampus kuda in China. Aquaculture 523, 735168. doi: 10.1016/j.aquaculture.2020.735168
Xing D., Li Q., Kong L., Yu H. (2018). Heritability estimate for mantle edge pigmentation and correlation with shell pigmentation in the white-shell strain of pacific oyster, Crassostrea gigas. Aquaculture 482, 73–77. doi: 10.1016/j.aquaculture.2017.09.026
Yano M., Iwama A., Nishio H., Suda J., Takada G., Suda T., et al. (1997). Expression and function of murine receptor tyrosine kinases, TIE and TEK, in hematopoietic stem cells. Blood 89 (12), 4317–4326. doi: 10.1182/blood.v89.12.4317
Zeng Z., Tan Q., Huang Z., Shi B., Ke C. (2019). Differential gene expression related to morphological variation in the adductor muscle tissues of diploid and triploid fujian oysters, Crassostrea angulata. Aquaculture Res. 50 (12), 3567–3578. doi: 10.1111/are.14312
Zhang L., Li L., Zhu Y., Zhang G., Guo X. (2014). Transcriptome analysis reveals a rich gene set related to innate immunity in the Eastern oyster (Crassostrea virginica). Mar. Biotechnol. 16 (1), 17–33. doi: 10.1007/s10126-013-9526-z
Zhang T., Qiu L., Sun Z., Wang L., Zhou Z., Liu R., et al. (2014). The specifically enhanced cellular immune responses in pacific oyster (Crassostrea gigas) against secondary challenge with Vibrio splendidus. Dev. Comp. Immunol. 45 (1), 141–150. doi: 10.1016/j.dci.2014.02.015
Zhen Y., Zhang H. (2019). NLRP3 inflammasome and inflammatory bowel disease. Front. Immunol. 10, 276. doi: 10.3389/fimmu.2019.00276
Keywords: Crassostrea gigas, Vibrio alginolyticus, immune response, protein-protein interaction networks, transcriptome
Citation: Zhang E, Dong L, Bao X, Yang X, Li Y, Feng Y, Yang J, Li Z and Wang W (2022) Transcriptome profiling combined with network analysis deepens the understanding of immune response mechanisms in blood of pacific oyster Crassostrea gigas infected by Vibrio alginolyticus. Front. Mar. Sci. 9:1017445. doi: 10.3389/fmars.2022.1017445
Received: 12 August 2022; Accepted: 05 September 2022;
Published: 16 September 2022.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, (CAS), ChinaReviewed by:
Xubo Wang, Ningbo University, ChinaSergey Ivanovich Maslennikov, National Scientific Center of Marine Biology, Russia
Copyright © 2022 Zhang, Dong, Bao, Yang, Li, Feng, Yang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zan Li, bGl6YW5seG1AMTYzLmNvbQ==; Weijun Wang, d3dqMjUzMDYxNkAxNjMuY29t
 Enshuo Zhang1
Enshuo Zhang1 Jianmin Yang
Jianmin Yang Zan Li
Zan Li Weijun Wang
Weijun Wang