- 1Integrative Science Center of Germplasm Creation in Western China (Chongqing) Science City & Southwest University, Chongqing, China
- 2Jiaxing Key Laboratory for New Germplasm Breeding of Economic Mycology, Jiaxing, Zhejiang, China
The effect of thermal changes on the physiology and behavior of fish is a major research focus in the face of ongoing global warming. There is little information about the effects of temperature increase on fish in the wild. However, the consequences of temperature increase on fish in controlled laboratory conditions can provide insights into what can be expected in the wild. Triplophysa bleekeri, a high-plateau fish, exhibits high sensitivity to high temperatures, suggesting it to be a good model to investigate the impact of temperature increase on fish. In this study, we analyzed the effect of gradual temperature increase on transcriptional and metabolic levels of T. bleekeri subjected to a gradual temperature change of 0.5°C/day until temperatures of 10°C, 13°C, 16°C, and 19°C were reached. Transcriptomics results of the liver, gut, spleen, and trunk kidney showed that metabolic pathways are widely involved in the response to increased temperatures in T. bleekeri. Lipidomics results further indicated that the lipid composition was altered by increased temperatures, and three lipids (PC 14:0e/22:1, PC 18:0e/22:5, and TAG 14:3-21:2-21:2) were identified as potential biomarkers of heat stress in T. bleekeri. Moreover, a decline in unsaturated fatty acid levels was observed in T. bleekeri under high temperatures. These results suggest that high temperatures modify the metabolomic pathways. Overall, our results help improve the understanding of physiological responses in fish to increased temperatures, and provide valuable information predicting the consequences of global warming on fish.
Introduction
Temperature acts as an important abiotic factor that constrains the geographical distribution and controls the physiological and behavioral parameters of fish (Nilsson et al., 2009; Dadras et al., 2017; Forgati et al., 2017). Fish have evolved the ability to adapt to a certain range of temperature variations in nature, such as seasonal variations and diurnal fluctuations (Crozier and Hutchings, 2014). However, the magnitude of temperature change approaching or beyond the thermal tolerance range of fish can lead to severe sublethal disturbances and mortality (Donaldson et al., 2008). In the context of ongoing global warming, threats for over one-third freshwater fish species have increased considerably (Barbarossa et al., 2021). Fish populations, biodiversity, and distribution have been altered by rising temperature coupled with increased extreme climate events around the world, and some fish species even face local extinction (Free et al., 2019; Punzón et al., 2016). Hence, there is a need to better understand the impact of temperature increase on fish physiology for predicting the consequences of global warming on fish.
The capacity of fish to cope with temperature changes is closely related to the degree of phenotypic plasticity (Norin et al., 2016; Keen et al., 2017). Phenotypic plasticity is defined as the ability of a genotype to produce different phenotypes in response to different environmental conditions (Pigliucci et al., 2006). When fishes face a challenge caused by temperature changes, they attempt to alter their physiological and behavioral traits through producing a cascade of plastic responses (e.g., neuroendocrine, metabolic, cellular, and immunological responses), a process termed acclimation, to resist sublethal and lethal effects (Donaldson et al., 2008; Alfonso et al., 2021). Increasing evidence shows that transcriptional variabilities are key contributors to phenotypic plasticity, allowing organisms to adapt to a changing environment (Smith et al., 2013; Wellband and Heath, 2017; Ecker et al., 2018; Li et al., 2021). Understanding the transcriptional variability of fish to altered temperature is critical for assessing the heat/cold acclimation ability in the face of climate change. Coincident with the advancement of sequencing, RNA-seq offers opportunities to explore transcriptional responses as well as molecular mechanisms underlying responses (Oomen and Hutchings, 2017). The transcriptional plasticity to temperature decrease has been described in multiple teleosts using RNA-seq (Long et al., 2012; Zhou et al., 2019; Liu et al., 2020; Long et al., 2020). However, there is little information to explain the transcriptional responses to temperature increase in fishes (Alfonso et al., 2021). Both acute and chronic exposures to high temperature are regarded as a stress by fish, inducing transcriptional changes in processes including stress response, signal transduction, metabolism, and immune response (Smith et al., 2013; Guo et al., 2016; Huang et al., 2018; Chen et al., 2021). With respect to long-term heat stress, the number of differentially expressed genes (DEGs) involved in stress and immune response decreased comparing to that of short-term challenge studies (Narum and Campbell, 2015; Li et al., 2017), which suggests that fishes acclimate to temperature increase after chronic heat stress.
Stenothermal high-plateau fishes are expected to be vulnerable to global warming. However, the mechanism underlying the response of high-plateau fish to temperature increase is still largely unknown. Triplophysa bleekeri, an endemic fish inhabiting the peripheral areas of Qinghai-Tibetan Plateau, is widely distributed at elevations of 200–3,000 m (Wang, 2013). The optimal temperature range of T. bleekeri is 14–15°C, and this fish can spawn even in December (approximately 10°C) (Wang et al., 2013). However, T. bleekeri dies as a result of long-term exposure to temperatures exceeding 21°C. This means that T. bleekeri is very sensitive to high temperatures. Therefore, T. bleekeri is an ideal model for studying the impact on fish due to temperature increase. To gain a comprehensive understanding of transcriptional responses to temperature increases, we detected the transcriptional variations in multiple tissues (liver, gut, trunk kidney, and spleen) under four temperature conditions (10°C, 13°C, 16°C, and 19°C). This approach differs from those in previously reported studies on transcriptional responses to temperature changes in fish in that previous studies used a single or two tissues for RNA-seq. The transcriptional change of genes involved energy metabolism is a critical part of the plasticity response to temperature increases (Veilleux et al., 2015; Jeffries et al., 2016). Hence, we hypothesized that the transcriptional levels of metabolism pathways are affected by temperature increases in T. bleekeri, resulting in changes in metabolites. Liver tissue is a lynchpin in metabolic reprogramming in response to temperature changes (Barat et al., 2016; Lyu et al., 2018; Paul et al., 2021). In this study, we characterized the lipid metabolite changes in the liver of T. bleekeri at high temperatures using a hybrid quadrupole time-of-flight mass spectrometry (UHPLC-QTOF-MS) and gas chromatography-mass spectrometry (GC-MS) analysis. To the best of our knowledge, only a few studies have examined the metabolic profile of fish at high temperatures. Our findings reveal the transcriptional and metabolic response of T. bleekeri exposed to high temperatures, and provide valuable information for understanding how fish biology will be affected by global warming.
Materials and methods
Animals and sampling
Fish were caught from a tributary of the Yangtze River using brails and were then transferred to the laboratory. Fish were maintained in indoor tanks (50 × 40 × 30 cm) with a water-circulating system at 15°C under a 14L:10D lighting cycle. All fish were fed tubificid worms twice daily. Fish were acclimated under these conditions for 3 weeks before the formal experiment. After acclimation, fish (weight 5.6 ± 0.8 g, length 7.1 ± 0.6 cm) were randomly split into four groups, i.e., 10°C group (3 tanks, n = 9/tank), 13°C group (3 tanks, n = 9/tank), 16°C group (3 tanks, n = 9/tank), and 19°C group (3 tanks, n = 9/tank). The temperatures were gradually declined/increased from 15°C to four temperature conditions (10°C, 13°C, 16°C, and 19°C) at a rate of approximately 0.5°C/day using a thermostat. Fish were maintained at aforementioned temperature conditions for 2 weeks. Thereafter, fish were anesthetized by immersion in ice-cold water and were immediately dissected to collect the liver, gut, trunk kidney, and spleen samples. These tissues were quickly frozen in liquid nitrogen for 1 h and then stored at −80°C.
All experimental protocols were approved by Southwest University, and the study was carried out under the protocols of the Institutional Animal Care and Use Committee of Southwest University.
RNA sequencing
Total RNA was extracted from the liver, gut, trunk kidney, and spleen using an Animal Total RNA Isolation Kit (Foregene, China). The quality and quantity of RNA were detected as our previous study (Yuan et al., 2020). Samples with a total RNA concentration ≥ 10 μg and RIN ≥ 8 were used for sequencing. The RNA sequence library was constructed using a paired-end sample preparation kit (Illumina Inc., USA), and the library was sequenced on an Illumina HiSeq 2000 sequencer.
RNA-seq data analysis
A quality control step was conducted on the raw sequencing reads of each sample before bioinformatics analysis. The raw reads were filtered, and then clean reads were aligned to the reference genome of T. Bleekeri (Yuan et al., 2020) using HISAT (Kim et al., 2015). The differentially expressed genes (DEGs) between the low-temperature and high-temperature groups were analyzed using DESeq2 (Love et al., 2014). The DEGs of each group were identified and compared with the other group in T. Bleekeri. Genes with p-value < 0.01 and |log2FC| > 1 were considered as DEGs. GO enrichment and KEGG pathway analyses for DEGs were performed using Tbtools (Chen et al., 2020). R packages ggplot and cytoscape were used for visualization of the Go and KEGG enrichment results. The Short Time-series Expression Miner (STEM) analysis based on FPKM values was conducted to cluster, compare, and visualize gene expression data from the four temperature groups. Genes which had similar expression patterns were clustered into a profile (Ernst and Bar-Joseph, 2006).
Validation of RNA-Seq results using real-time PCR
Total RNA was extracted from the tissue samples, viz., liver, gut, trunk kidney, and spleen, as described above. The synthesis of cDNA was conducted as described in our previous study (Yuan et al., 2021). Real-time PCR (RT-PCR) was performed on the LightCycle 96 (Roche, USA). Primers for the target genes were designed based on the genomic and transcriptomic data of T. bleekeri (Yuan et al., 2020) and are listed in Supplementary Table A.1. β-actin was used as the reference gene, and the relative gene expression was analyzed as described in our previous study (Yuan et al., 2021).
Lipid extraction for UHPLC-QTOF-MS and GC-MS analysis
Lipid extraction was carried out on liver samples, and six liver samples each were sampled from fish of 10°C, 13°C, 16°C, and 19°C groups. Into an EP tube, 25 mg of each liver sample, 200 μL of water, and 480 μL of extracting solution (VMTBE: VMeOH = 5: 1) were added sequentially. This mixture was homogenized (35 Hz, 4 min) and sonicated for 10 min. After repeating this process three times, the mixture was incubated (1 h) at -40°C and centrifuged at 4°C (3,000 rpm, 15 min), and then the supernatant (350 μL) was dried at 37°C. The dried sample was then reconstituted in 50% methanol in DCM (200 μL) by sonication for 10 min. The mixture was centrifuged at 4°C (13,000 rpm, 15min). Subsequently, the 75 μL of supernatant was used for UHPLC-QTOF-MS analysis.
Similarly, each liver sample (50 mg) was extracted with 460 μL of extracting solution (VIsopropanol: Vn-Hexane = 2: 3), and then mixed with 40 μL of Octadecanoic-D35 acid (1 mg/L). The mixture was placed in an ice-water bath for 5 min and then centrifuged at 4°C (12,000 rpm, 15 min). The supernatant was transferred to a fresh tube. Next, the extracting solution (500 μL) was added to the mixture, which was then placed in an ice-water bath and centrifuged for 15 min, as mentioned above. The supernatant was collected and mixed with the previously obtained supernatant. The resulting supernatant (400 μL) was then added to a fresh tube, dried under nitrogen, and mixed with 200 μL of methanol and 100 μL of (trimethylsilyl) diazomethane and maintained for 15 min. This mixture was dried under nitrogen, reconstituted in 160 μL of n-hexane, and then centrifuged at 4°C (12,000 rpm, 5 min). The resulting supernatant was used for GC-MS analysis.
UHPLC-QTOF-MS analysis of lipids
The lipid analysis was performed by an UHPLC System (AB Sciex, USA), coupled with a high-resolution mass spectrometer (Triple TOF 5600, AB Sciex). Lipid species were separated using mobile phases A and B. The mobile phase A consisted of 40% water, 60% ACN, and 10 mmol/L HCOONH4, and the mobile phase B consisted of 10% ACN and 90% IPA. The mobile phase was carried out as described previously (Zhang et al., 2021). The injection volume was 1 μL for the positive ionization (POS) model and negative ionization (NEG) model, respectively. MS/MS spectra were acquired on an information-dependent acquisition (IDA) model using the Triple TOF 5600 mass spectrometer. In the IDA model, the full scan survey MS data was continuously evaluated using analyst TF 1.7 (AB Sciex).
GC-MS analysis of free fatty acids
The free fatty acid (FFA) profiles in the liver samples were analyzed using an Agilent 7890 B gas chromatography system coupled with an Agilent 5977B mass spectrometer and an Agilent DB-FastFAME capillary column (Agilent Technologies, USA). Helium was used as the carrier gas (flow rate: 3.0 mL/min). The temperature program conditions were as follows: the initial temperature was maintained at 75°C for 1 min, raised to 200°C (rate: 50°C/min), and maintained at 200°C for 15 min. The temperature was then increased to 210°C (rate: 2°C/min) for 1 min, finally increased to 230°C (rate: 10°C/min) and maintained at 230°C for 16.5 min. The analytes were ionized by electron impact (EI) at -70 eV. Subsequently, the mass spectrometry data were obtained in Scan/SIM mode with continuous scanning ions from m/z 33 to 400.
Lipid data processing
The format of raw data files (.wiff format) was converted to special format (mzXML) (Holman et al., 2014). Then, the XCMS software (parameters: minfrac=0.5; cutoff=0.3) was used to generate the peak information of MS1, including the retention time (RT), peak area, and m/z values (Tautenhahn et al., 2012). Lipids were identified using the LipidBlast library based on RT and m/z values in the MS/MS spectra (Kind et al., 2013). The principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA) were performed as previous study (Wen et al., 2018), and the VIP was obtained using OPLS-DA. The SPSS 23 was used to analyze data using Student’s t-test. The lipids with p-values < 0.05, and VIP > 1 were considered significantly different. The GraphPad Prism 9 software was used for visualization of lipids. The differential lipids were normalized by Z-score following the equation:
Z = (x-μ)/σ
in which, z is the value of Z-score, x is a value of data, μ is a mean of data, and σ is a number of standard deviation of data. The normalized differential lipids were analyzed by K-means clustering using fpc and cluster package in R to illustrate pattern in lipids with the increasing temperature.
Results
RNA-seq analysis reveals metabolic reprogramming under high temperatures
To comprehensively investigate the transcriptional response to temperature variations, 48 samples from the liver, gut, spleen, and trunk kidney at 10°C, 13°C, 16°C, and 19°C were used for library construction and sequenced. In total, 2,176 Mb of raw reads were obtained. All raw reads were deposited in the NCBI SRA database. After filtering the raw reads, approximately 2,133 Mb clean reads were obtained from each library. The average Q20 and Q30 of the clean reads were 98.14% and 94.24%, respectively. Approximately 92.47% of the clean reads from each library were mapped to the reference genome of T. bleekeri (Supplementary Table A.2).
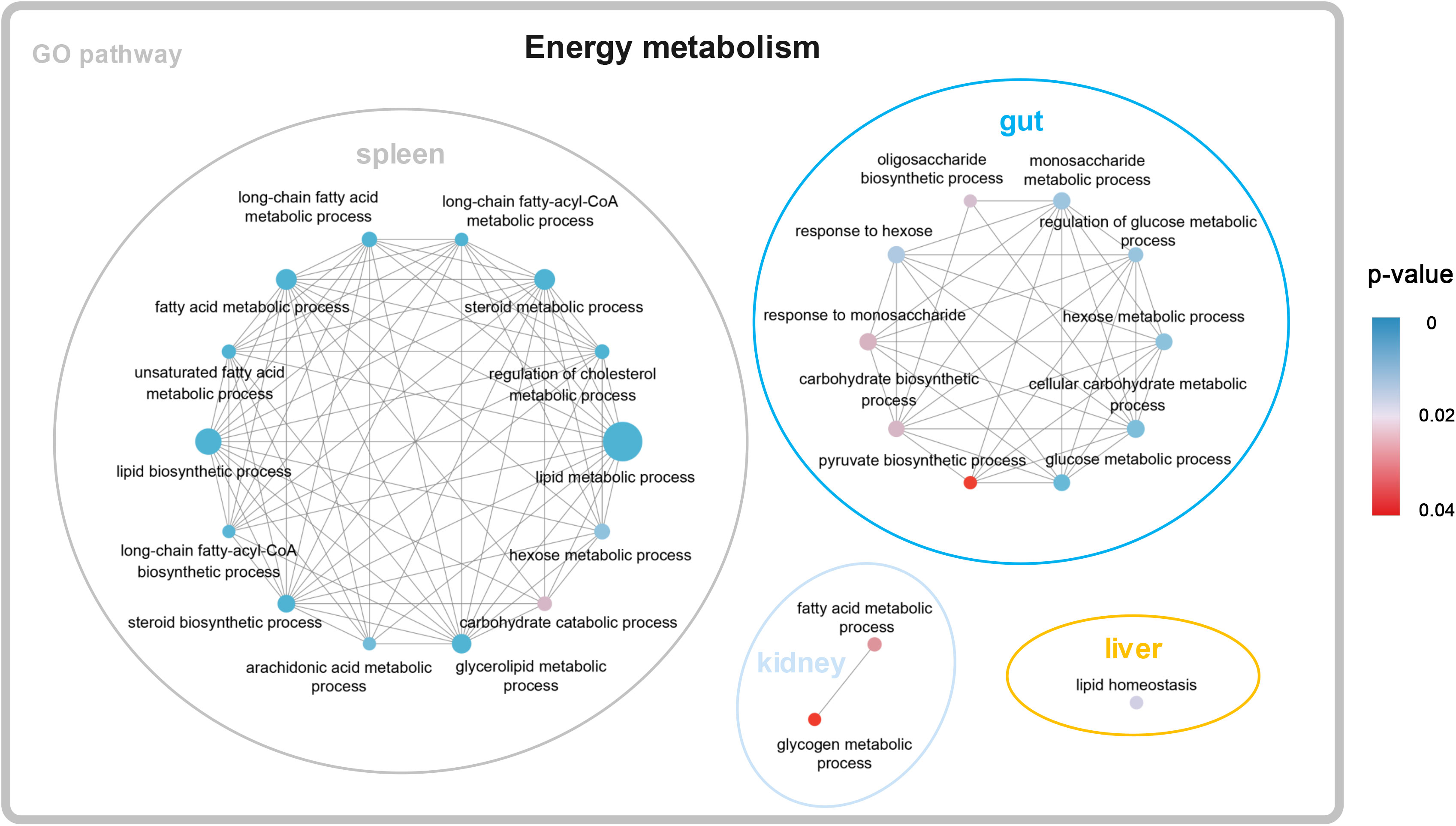
A total of 1,150, 1,709, 1,243, and 510 DEGs were identified in the gut, spleen, trunk kidney, and liver among the four temperature groups (10°C, 13°C, 16°C, and 19°C) through pairwise transcriptome comparison. The largest number of DEGs was observed in the aforementioned tissues between 19°C group and 10°C group (Supplementary Table A.3). The functional enrichment analysis on the KEGG and GO parameters for DEGs was conducted to elucidate molecular pathways involved in transcriptional response to temperature variations. DEGs (19°C group vs. 10°C group) were mainly enriched in categories related to energy metabolism, such as lipid metabolic process, lipid biosynthetic process, steroid metabolic process, fatty acid metabolic process, cellular carbohydrate metabolic process, and glucose metabolic process (Figure 1; Supplementary Table A.4). Besides, DEGs were also significantly enriched in signal transduction, immune response, stress response (Supplementary Table A.4). Carnitine palmitoyltransferase 1A (CPT1A) was shared as a DEG by all four tissues, and was upregulated in the 19°C group (Supplementary Figure A.1; Supplementary Table A.4).
To validate the RNA-seq results, DEGs in the liver, gut, spleen, and trunk kidney of four groups were randomly selected and analyzed by RT-PCR. The expression patterns of DEGs detected using RT-PCR were highly consistent with RNA-seq data (Supplementary Figure A.2).
Clustering analysis reveals dynamic expression profiles of genes with increasing temperature
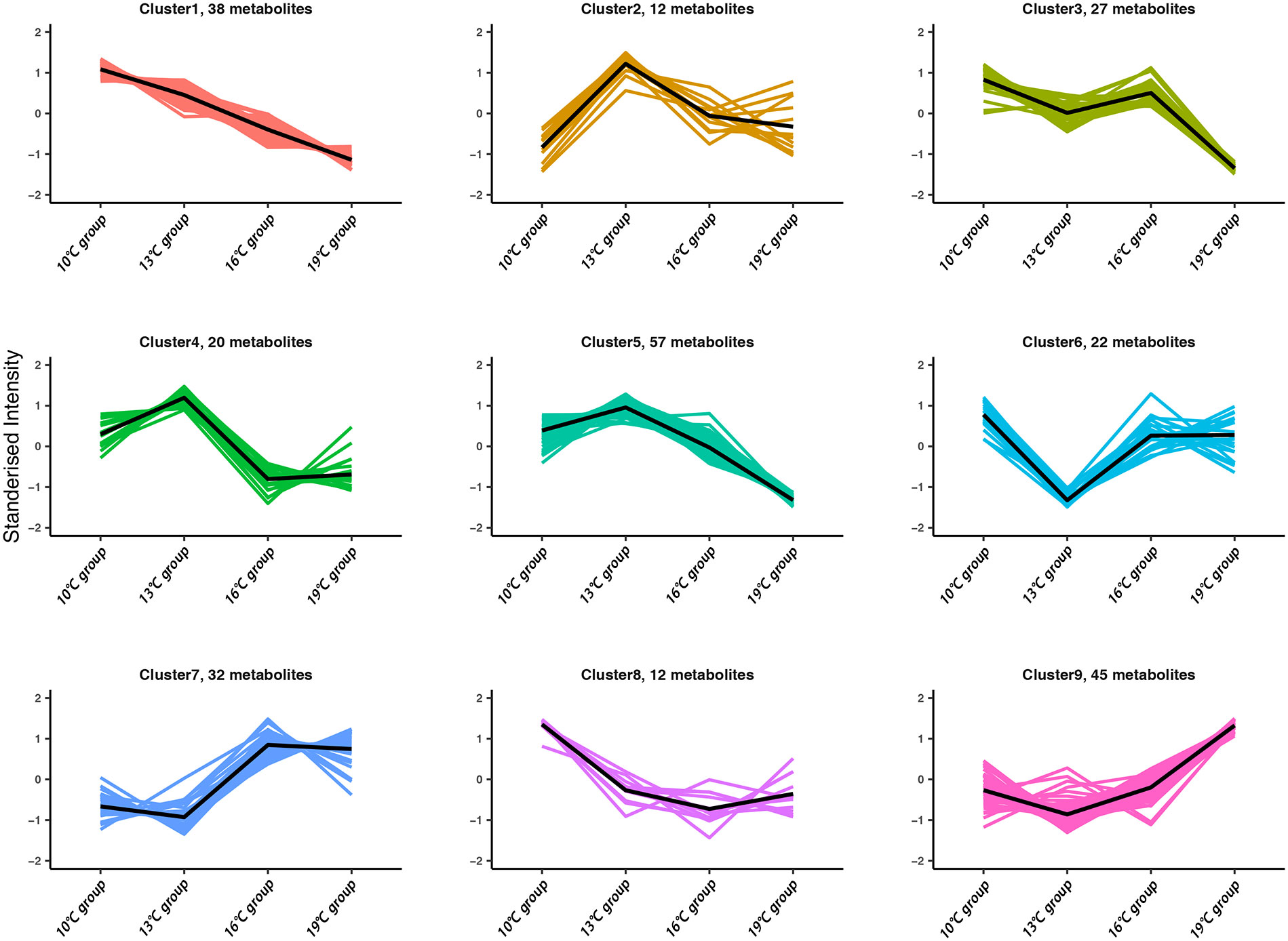
A clustering analysis (STEM) was conducted to investigate the dynamic expression patterns of genes with increasing temperature. A total of 10, 9, 9, and 8 significant expression profiles (P < 0.01) were identified in the gut, spleen, trunk kidney, and liver, respectively (Supplementary Figure A.3). The clustered profile 25 and 0 reflect responsive genes that consistently increase or decrease along with increasing temperature, respectively. The biological functions of genes in profile 25 and 0 (P < 0.01) were further analyzed through GO and KEGG assignments. Genes in profile 25 mainly enriched in arachidonic acid metabolism, Hedgehog signaling pathway, Fanconi anemia pathway, DNA repair and recombination pathway, mTOR signaling pathway, thyroid hormone signaling pathway, insulin signaling pathway, metabolic process, and response to stimulus (Figure 2). The majority of genes in profile 0 were involved in ribosome biogenesis, glycosylphosphatidylinositol (GPI)-anchored proteins, adipocytokine signaling pathway, glucagon signaling pathway, signal transduction, and cellular response to stimulus (Figure 2).
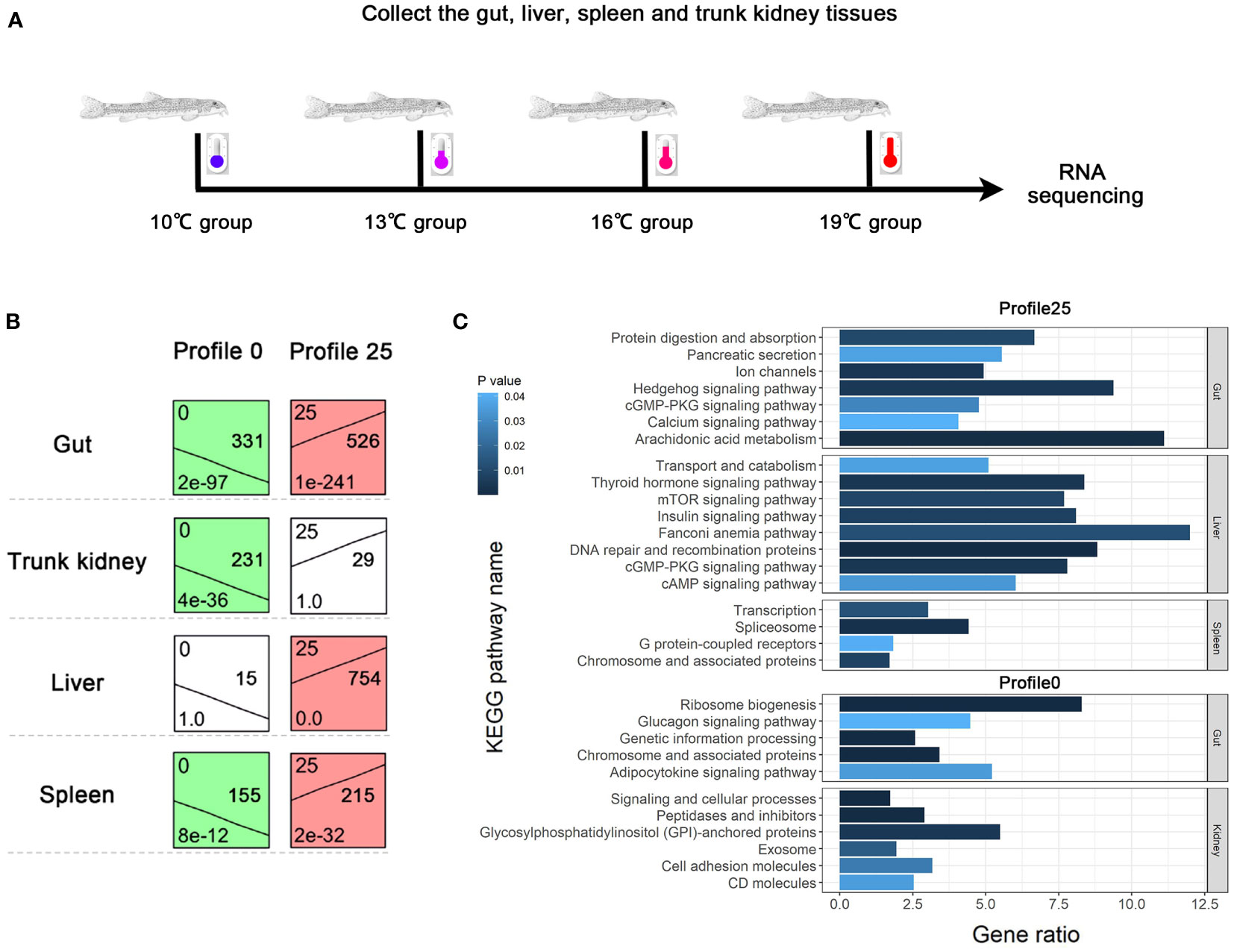
Figure 2 Dynamic expression profiles of genes with increasing temperature. (A) Temperature conditions and sample collections. (B) Expression patterns of consistently increasing and decreasing genes identified by STEM analysis. (C) KEGG enrichment results of genes belonging to profile 0 and 25.
Untargeted lipidomics shows lipid remodeling along with increasing temperature
Twenty-four liver samples from the 10°C, 13°C, 16°C, and 19°C groups were processed for UHPLC-QTOF-MS analysis in the POS and NEG modes. PCA and OPLS-DA analysis indicated that aforementioned groups achieved good separation of lipid extracts (Figure 3; Supplementary Figure A.4). Finally, 2,560 lipids were identified based on existing databases. These detected lipids were further analyzed to identify differential lipids. In total, 265 differential lipids were identified among the four temperature groups. These differential lipids were predominantly attributable to phosphatidylcholine (PC, 12.08%), triacylglycerol (TAG, 10.57%), and phosphatidylethanolamine (PE, 7.55%) (Figure 3). The largest number of differential lipids was observed in the 19°C group vs. 13°C group (142), followed by the 19°C group vs. 10°C group (105) (Supplementary Table A.5). The differential lipids with high fold change in 19°C group vs. 13°C group, and 19°C group vs. 10°C group are showed in the Figure 3.
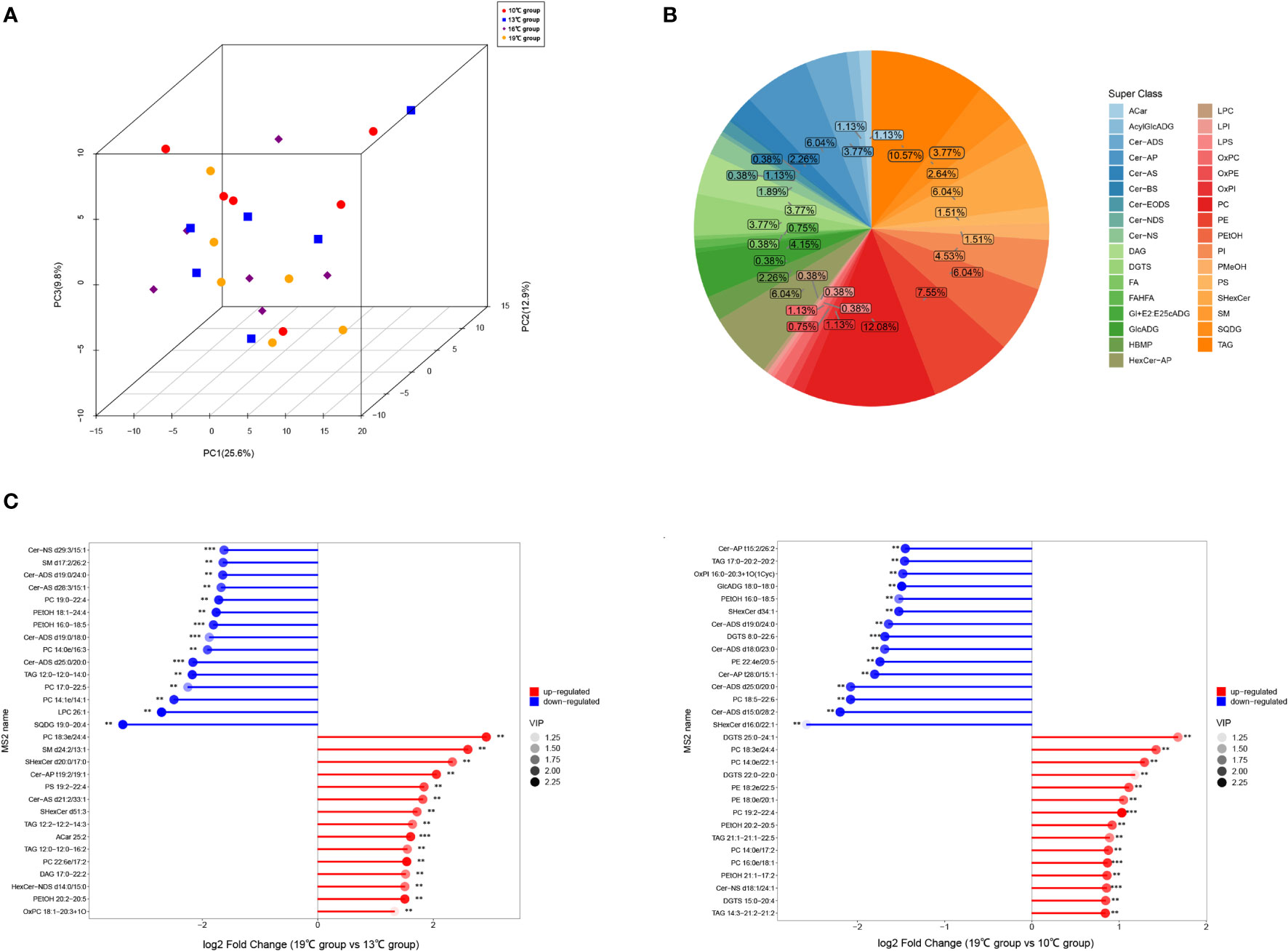
Figure 3 Results of untargeted lipidomics based on UHPLC-QTOF-MS analysis. (A) PCA plots of the samples. (B) Classification of differential lipids among four temperature groups. (C) Differential lipids with high fold change in 19°C group vs. 13°C group, and 19°C group vs. 10°C group.
Clustering analysis reveals potential markers of heat stress
To investigate the dynamic change of differential lipids with increasing temperature, the K-means analysis was conducted. The total of nine clusters was obtained (Figure 4), and the list of differential lipids from each cluster is shown in Supplementary Table A.6. The clusters 1 and 9 contain consistently decreasing and increasing lipids with increasing temperature, respectively. PC 14:0e/22:1, PC 18:0e/22:5, and TAG 14:3-21:2-21:2, which belong cluster 9, have a high content under temperature 19°, suggesting that these three lipids can be used as potential biomarkers of heat stress in T. bleekeri (Figure 4; Supplementary Table A.6).
Targeted lipidomics reveals decreased concentration of unsaturated fatty acids under high temperature
The targeted lipidomics was conducted using a GC-MS platform to evaluate the effect of high temperature on free fatty acids (FFAs). 49 FFAs were detected, and 14 FFAs were significantly altered in 19°C group compared to that of 10°C group (Figure 5). Among these 14 FFAs, only heneicosanoic acid was significantly elevated in the 19°C group. Unsaturated fatty acids, such as 9-elaidic acid, 9-palmitelaidic acid, 10-pentadecenoic acid, 11-octadecenoic acid, 11-eicosenoic acid, 6,9,12-linolenic acid, 7,10,13,16-docosatetraenoic acid, and 4,7,10,13,16-docosapentaenoic acid, were significantly decreased in the 19°C group.
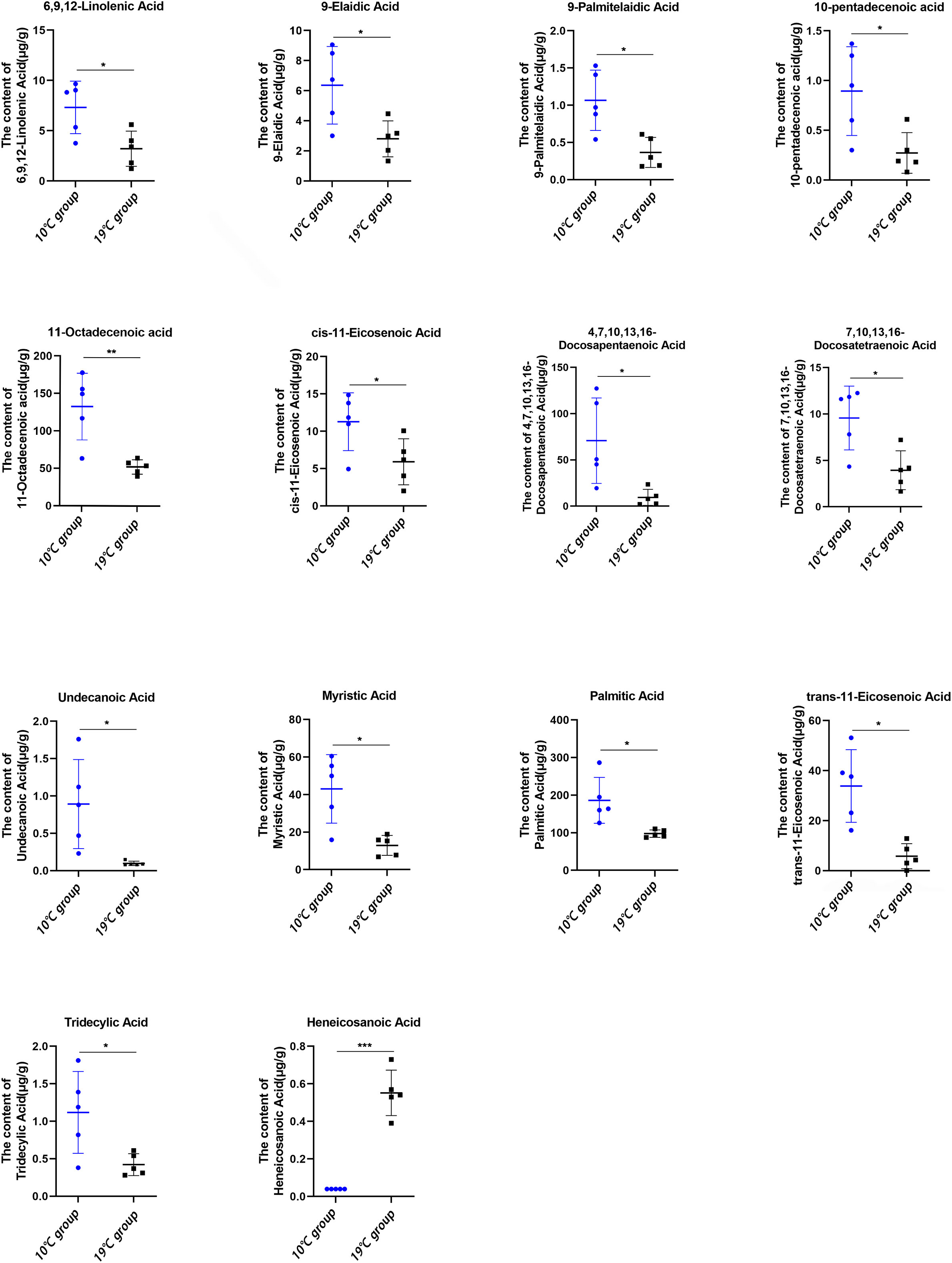
Figure 5 Absolute concentration of free fatty acids in the liver between the 10°C and 19°C groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
In this study, we used two postgenomic approaches, transcriptomics and lipidomics, to reveal the transcriptional and lipid alterations in T. bleekeri with increasing temperature. RNA-seq analysis reveals that metabolic pathways are widely involved in the response to temperature changes. In T. bleekeri, DEGs (19°C vs. 10°C) were significantly enriched in the lipid metabolic process, lipid biosynthetic process, steroid metabolic process, and fatty acid metabolic process. This observation is similar to the results observed in other fish. The expressions of genes involved in lipid metabolism category were significantly changed in response to elevated temperature in Sparus aurata (Balbuena-Pecino et al., 2019), and Scophthalmus maximus (Zhao et al., 2021). In Acanthochromis polyacanthus, a tropical damselfish, lipid metabolism genes were among the most upregulated transgenerationally at elevated temperatures (Veilleux et al., 2015). These results suggest that lipid metabolism has a critical role in heat resistance in fish, which is a conservation mechanism for responding to high temperatures.
To explore the role of lipid metabolism in temperature acclimation of T. bleekeri, mass spectrometry-based lipidomics was used in this study. As the liver is a central player in lipid metabolism, the differences in lipid profiles of the liver among 10°C, 13°C, 16°C, and 19°C groups were investigated. Significant lipid remodeling was observed in T. bleekeri with increasing temperature. The contents of PC, TAG, and PE account for the majority of differential lipids. PC and PE, which act as structural lipids, are an essential component of the cellular membrane (Coskun and Simons, 2011). In fish, the phospholipid composition of the cell membrane was altered by temperature (Farkas et al., 2001; Grim et al., 2010). In rainbow trout (Oncorhynchus mykiss), the levels of PC and lysophosphatidylcholine (LysoPC) in the liver were reduced after exposure to high temperatures (Li et al., 2022). Here, most of the glycerophospholipids containing polyunsaturated fatty acids (PUFAs), such as PC 18:5-22:6 and PE 22:4e-20:5, were downregulated in the 19°C group (Supplementary Table A.5.). It suggests that these PUFAs play an imporatnt role in lipid remodeling in T. bleekeri under high temperatures, since the proportion of saturated and unsaturated acyl chains in membrane lipids is closely related to membrane viscosity and fluidity (Ernst et al., 2016). Fatty acids are also an important component of membrane phospholipids as well as a major energy source that play crucial physiological roles in various tissues (Wakil and Abu-Elheiga, 2009). The previous studies showed that fatty acids are involved in metabolic response to temperature changes. In Antarctic notothenioid fish, high temperature (6°C) induced the saturated fatty acids, and reduced the unsaturated fatty acids (Malekar et al., 2018). Conversely, the content of unsaturated fatty acids increased to maintain membrane fluidity in fish at low temperatures (Grim et al., 2010; He et al., 2015). In this study, PUFAs, such as linolenic acid (C18:3, n-6), docosatetraenoic acid (C22:4, n-6), and docosapentaenoic acid (C22:5, n-3), were significantly decreased in the 19°C group. To sum up, altered lipid saturation in response to temperature change is a major thermal acclimation mechanism. Except above-mentioned n-3/n-6 PUFAs, palmitic acid (C16:0), palmitoleic acid (C16:1), and octadecenoic acid (C18:1) significantly decreased in the 19°C group. Accumulation of fatty acyls containing either n-3 or n-6 indicates the reduced utility of these fatty acids through oxidation, whereas an increase in 16:0, 16:1, 18:0, and 18:1 fatty acids indicates their increased de novo biosynthesis under physiological conditions (Han, 2016). Hence, we speculated that high temperatures decrease lipid biosynthesis but enhance fatty acid oxidation in T. bleekeri. RNA-seq results also support the hypothesis that the genes involved in the biosynthesis of phospholipids and TAG, such as phosphatidate phosphatase-1 (lipin-1) (Bou Khalil et al., 2009), were downregulated in the 19°C group (Supplementary Table A.3). Conversely, phospholipase B1 (PLB1), which is involved in the degradation of phospholipids (Wright et al., 2007), was upregulated in the 19°C group (Supplementary Table A.4). Besides, the expression of CPT1A in the liver, gut, spleen, and trunk kidney was upregulated at 19°C (Supplementary Table A.4). CPT1A, a major rate-limiting enzyme, participates in the fatty acid β-oxidation process and plays an essential role in maintaining energy homeostasis (Schlaepfer and Joshi, 2020). The similar results also observed in Salmo salar that increasing temperature significantly reduced levels of genes (fas, cpla2, and elovl2) related to fatty acid biosynthesis, and increased levels of CPT1A and pparα involved in fatty acid β-oxidation (Norambuena et al., 2015). Hence, lipid biosynthesis were suppressed, whereas fatty acid β-oxidation was enhanced in fish at high temperature.
High temperatures affect lipid metabolism and influence carbohydrate metabolism. A previous study showed that the concentrations of glycogen in Notothenia coriiceps rapidly increased, followed by a decline, in response to varying glucose-6-phosphatase (G6Pase) levels after exposure to high temperatures (Forgati et al., 2017). The acute increase in temperature reduced glycolysis in the heart of Notothenia rossii, while the opposite result was observed in the muscle (Souza et al., 2018). In Scophthalmus maximus, high temperatures increased expression levels of G6Pase, a key enzyme of gluconeogenesis, suggesting that gluconeogenesis was activated to cope with thermal stress (Yang et al., 2020). In this study, our results showed that DEGs were significantly enriched in the glycogen, glucose, hexose, oligosaccharide, and monosaccharide metabolic process. Further, the expression level of glucose-6-phosphate dehydrogenase (G6PD), which catalyzes the first step in the pentose phosphate pathway, was significantly increased, whereas the expression level of 6-phosphofructokinase (Pfk), a critical rate-limiting enzyme in the process of glycolysis, was significantly decreased in the 19°C group. We speculated that high temperatures decrease glycolysis, but enhance the pentose phosphate pathway in T. bleekeri.
Apart from energy metabolism, many key physiological processes, such as immune, stress resistance, and DNA repair systems are strongly affected by high temperatures. In Oreochromis mossambicus, the immune parameters such as leucocyte count, phagocytic activity, and phagocytic index decreased when fish was transferred to high temperatures, suggesting that high temperatures reduced the immune capability of fish (Ndong et al., 2007). In Salmo salar, temperature increase negatively affected complement bacteriolytic activity (Jokinen et al., 2011). In T. Bleekeri, DEGs were significantly enriched in the Hippo signal pathway, NOD-like receptors signal pathway, and RIG-I-like receptor signaling pathway (Supplementary Table A.4). All these pathways play an indispensable role in the innate immune system (Fritz et al., 2006; Zeng et al., 2010; Zhang et al., 2018). Further, genes related to innate immune function, such as interferon regulatory factor 3 (IRF3) and CXC chemokine receptor 1 (CXCR1) were significantly downregulated in the 19°C group. Therefore, we speculated that the innate immunity of T. Bleekeri was diminished by high temperatures. Arachidonic acid (ARA) not only takes part in modulation of lipid metabolism, but also act an esstensial role in stress resistance (Xu et al., 2022). The STEM analysis showed the expression of genes involved in arachidonic acid metabolism significantly increased in T. bleekeri with increasing temperature.The previous study showed that Solea senegalensis prefers to select a diet rich in ARA with the seasonal temperature changes (Norambuena et al., 2012). In Morone saxatilis, higher dietary ARA improved the productive performance under sub-optimal temperatures (Araújo et al., 2021). These results suggest ARA act roles in thermal stress resistance. High temperatures induced the generation of reactive oxygen species (ROS) and antioxidant proteins (Devireddy et al., 2021). In T. bleekeri, we found that the expression level of glutathione peroxidase (GPX) increased under high temperatures. GPX, a key enzymes in the antioxidant system, protect organisms from oxidative stresses (Srikanth et al., 2013). It suggests that T. bleekeri initiates the expression of GPX to reduce the negative effects of oxidative damage induced by high temperature. Furthermore, increased DNA damage was observed in many species of fish exposed to high temperatures (Cheng et al., 2018; Castro et al., 2020). Here, STEM analysis showed that the expression levels of genes involved in Fanconi anemia pathway and DNA repair and recombination pathway constantly increased along with increasing temperature. These pathways all participate in DNA repair (Kim and D’andrea, 2012), which suggests that T. bleekeri may exhibit functional acclimation to increased temperatures.
Conclusion
In this study, transcriptomics and lipidomics approaches were used to comprehensively study effects of high temperatures on the transcriptional and metabolic levels of T. bleekeri. High temperatures induced a series of transcriptional plasticity responses in T. bleekeri, which resulted in altered levels of lipid metabolites. The decreased unsaturated fatty acid levels found in the lipidomics analysis were consistent with the downregulated genes involved in the lipid biosynthesis pathway found by RNA sequencing. Our results thus suggest that lipid metabolism was altered by temperature increase. Further, three lipids (PC 14:0e/22:1, PC 18:0e/22:5, and TAG 14:3-21:2-21:2) which showed rising content with increasing temperature and consequently, high content under high temperatures were identified as potential biomarkers of heat stress in T. bleekeri. However, further studies should be explored to identify whether these lipids can also be used as biomarkers of heat stress in other fishes. Overall, our results help improve the understanding of transcriptional and metabolic variations in fish in response to high temperatures, and provide essential information for forecasting potential impacts of global warming on fish biology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Southwest University.
Author contributions
ZW and DY conceived and designed the study. DY, XC, HG, and SW collected the samples. DY, XL, and JS performed the molecular experiments. HW, DY, and SX performed the bioinformatics analysis. DY, SX, and ZW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (Grant No. SWU-KQ22016), and the National Natural Science Foundation of China (Grant No. 32072980).
Acknowledgments
We gratefully acknowledge the assistance of Biotree Bio-Technique Co., Ltd. (Shanghai, China) in metabolomics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1017142/full#supplementary-material
Supplementary Table 3 | Differentially expressed genes in the liver, gut, spleen, and trunk kidney among four temperature groups.
Supplementary Table 4 | KEGG and GO enrichment results of differentially expressed genes between the 19°C and 10°C groups.
Supplementary Table 5 | The differential lipids in the liver among four temperature groups.
Supplementary Table 6 | The differential lipids of each cluster identified by k-means analysis.
Supplementary Figure 1 | Significantly differentially expressed genes. (A) The shared differentially expressed genes of 4 tissues between the 19°C and 10°C groups. (B) Significantly differentially expressed genes involved in energy metabolism and immune response.
Supplementary Figure 2 | Comparisons between RNA-seq and qPCR results.
Supplementary Figure 3 | Expression patterns of genes identified by STEM analysis.
Supplementary Figure 4 | Score scatter plot of the OPLS-DA model.
References
Alfonso S., Gesto M., Sadoul B. (2021). Temperature increase and its effects on fish stress physiology in the context of global warming. J. Fish. Biol. 98, 1496–1508. doi: 10.1111/jfb.14599
Araújo B. C., Rodriguez M., Honji R. M., Rombenso A. N., Rio-Zaragoza O. B., Cano A. (2021). Arachidonic acid modulated lipid metabolism and improved productive performance of striped bass (Morone saxatilis) juvenile under sub-to optimal temperatures. Aquaculture. 364-365, 198–205. doi: 10.1016/j.aquaculture.2020.735939
Balbuena-Pecino S., Riera-Heredia N., Vélez E. J., Gutiérrez J., Navarro I., Riera-Codina M., et al. (2019). Temperature affects musculoskeletal development and muscle lipid metabolism of gilthead sea bream (Sparus aurata). Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00173
Barat A., Sahoo P. K., Kumar R., Goel C., Singh A. K. (2016). Transcriptional response to heat shock in liver of snow trout (Schizothorax richardsonii)–a vulnerable Himalayan cyprinid fish. Funct. Integr. Genomics 16, 203–213. doi: 10.1007/s10142-016-0477-0
Barbarossa V., Bosmans J., Wanders N., King H., Bierkens M. F. P., Huijbregts M. A. J., et al. (2021). Threats of global warming to the world’s freshwater fishes. Nat. Commun. 12, 1701. doi: 10.1038/s41467-021-21655-w
Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., et al. (2009). The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 50, 47–58. doi: 10.1194/jlr.M800204-JLR200
Castro J. S., Braz-Mota S., Campos D. F., Souza S. S., Val A. L. (2020). High temperature, pH, and hypoxia cause oxidative stress and impair the spermatic performance of the Amazon fish Colossoma macropomum. Front. Physiol. 11. doi: 10.3389/fphys.2020.00772
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng C. H., Guo Z. X., Luo S. W., Wang A. L. (2018). Effects of high temperature on biochemical parameters, oxidative stress, DNA damage and apoptosis of pufferfish (Takifugu obscurus). Ecotoxicol. Environ. Saf. 150, 190–198. doi: 10.1016/j.ecoenv.2017.12.045
Chen Y., Liu E., Li C., Pan C., Zhao X., Wang Y., et al. (2021). Effects of heat stress on histopathology, antioxidant enzymes, and transcriptomic profiles in gills of pikeperch Sander lucioperca. Aquaculture 534, 736277. doi: 10.1016/j.aquaculture.2020.736277
Coskun U., Simons K. (2011). Cell membranes: The lipid perspective. Structure. 19, 1543–1548. doi: 10.1016/j.str.2011.10.010
Crozier L. G., Hutchings J. A. (2014). Plastic and evolutionary responses to climate change in fish. Evol. Appl. 7, 68–87. doi: 10.1111/eva.12135
Dadras H., Dzyuba B., Cosson J., Golpour A., Siddique M. A. M., Linhart O. (2017). Effect of water temperature on the physiology of fish spermatozoon function: a brief review. Aquac. Res. 48, 729–740. doi: 10.1111/are.13049
Devireddy A. R., Tschaplinski T. J., Tuskan G. A., Muchero W., Chen J. G. (2021). Role of reactive oxygen species and hormones in plant responses to temperature changes. Int. J. Mol. Sci. 22 (16), 8843. doi: 10.3390/ijms22168843
Donaldson M. R., Cooke S. J., Patterson D. A., Macdonald J. S. (2008). Cold shock and fish. J. Fish. Biol. 73, 1491–1530. doi: 10.1111/j.1095-8649.2008.02061.x
Ecker S., Pancaldi V., Valencia A., Beck S., Paul D. S. (2018). Epigenetic and transcriptional variability shape phenotypic plasticity. BioEssays 40, 1700148. doi: 10.1002/bies.201700148
Ernst J., Bar-Joseph Z. (2006). STEM: A tool for the analysis of short time series gene expression data. BMC Bioinf. 7, 191. doi: 10.1186/1471-2105-7-191
Ernst R., Ejsing C. S., Antonny B. (2016). Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 428, 4776–4791. doi: 10.1016/j.jmb.2016.08.013
Farkas T., Fodor E., Kitajka K., Halver J. (2001). Response of fish membranes to environmental temperature. Aquac. Res. 32, 645–655. doi: 10.1046/j.1365-2109.2001.00600.x
Forgati M., Kandalski P. K., Herrerias T., Zaleski T., Machado C., Souza M. R. D. P., et al. (2017). Effects of heat stress on the renal and branchial carbohydrate metabolism and antioxidant system of Antarctic fish. J. Comp. Physiol. B. 187, 1137–1154. doi: 10.1007/s00360-017-1088-3
Free C. M., Thorson J. T., Pinsky M. L., Oken K. L., Wiedenmann J., Jensen O. P. (2019). Impacts of historical warming on marine fisheries production. Science. 363, 979–983. doi: 10.1126/science.aau1758
Fritz J. H., Ferrero R. L., Philpott D. J., Girardin S. E. (2006). Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 7, 1250–1257. doi: 10.1038/ni1412
Grim J. M., Miles D. R., Crockett E. L. (2010). Temperature acclimation alters oxidative capacities and composition of membrane lipids without influencing activities of enzymatic antioxidants or susceptibility to lipid peroxidation in fish muscle. J. Exp. Biol. 213, 445–452. doi: 10.1242/jeb.036939
Guo L., Wang Y., Liang S., Lin G., Chen S., Yang G. (2016). Tissue-overlapping response of half-smooth tongue sole (Cynoglossus semilaevis) to thermostressing based on transcriptome profiles. Gene 586, 97–104. doi: 10.1016/j.gene.2016.04.020
Han X. (2016). Lipidomics: Comprehensive mass spectrometry of lipids (Hoboken, NJ: John Wiley & Sons). doi: 10.1002/9781119085263.ch5
He J., Qiang J., Yang H., Xu P., Zhu Z. X., Yang R. Q. (2015). Changes in the fatty acid composition and regulation of antioxidant enzymes and physiology of juvenile genetically improved farmed tilapia Oreochromis niloticus (L.), subjected to short-term low temperature stress. J. Therm. Biol. 53, 90–97. doi: 10.1016/j.jtherbio.2015.08.010
Holman J. D., Tabb D. L., Mallick P. (2014). Employing ProteoWizard to convert raw mass spectrometry data. Curr. Protoc. Bioinf 46, 13.24.1–13.24.9. doi: 10.1002/0471250953.bi1324s46
Huang J., Li Y., Liu Z., Kang Y., Wang J. (2018). Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish. Shellfish. Immunol. 82, 32–40. doi: 10.1016/j.fsi.2018.08.002
Jeffries K. M., Connon R. E., Davis B. E., Komoroske L. M., Britton M. T., Sommer T., et al. (2016). Effects of high temperatures on threatened estuarine fishes during periods of extreme drought. J. Exp. Biol. 219, 1705–1716. doi: 10.1242/jeb.134528
Jokinen I. E., Salo H. M., Markkula E., Rikalainen K., Arts M. T., Browman H. I. (2011). Additive effects of enhanced ambient ultraviolet b radiation and increased temperature on immune function, growth and physiological condition of juvenile (parr) Atlantic salmon, Salmo salar. Fish. Shellfish. Immunol. 30, 102–108. doi: 10.1016/j.fsi.2010.09.017
Keen A. N., Klaiman J. M., Shiels H. A., Gillis T. E. (2017). Temperature-induced cardiac remodelling in fish. J. Exp. Biol. 220, 147–160. doi: 10.1242/jeb.128496
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kind T., Liu K. H., Lee D. Y., DeFelice B., Meissen J. K., Fiehn O. (2013). LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 10, 755–758. doi: 10.1038/nmeth.2551
Li Y., Huang J., Liu Z., Zhou Y., Xia B., Wang Y., et al. (2017). Transcriptome analysis provides insights into hepatic responses to moderate heat stress in the rainbow trout (Oncorhynchus mykiss). Gene 619, 1–9. doi: 10.1016/j.gene.2017.03.041
Li L., Liu Z., Quan J., Lu J., Zhao G., Sun J. (2022). Metabonomics analysis reveals the protective effect of nano−selenium against heat stress of rainbow trout (Oncorhynchus mykiss). J. Proteomics. 259, 104545. doi: 10.1016/j.jprot.2022.104545
Li A., Li L., Zhang Z. Y., Li S. M., Wang W., Guo X. M., et al. (2021). Noncoding variation and transcriptional plasticity promote thermal adaptation in oysters by altering energy metabolism. Mol. Biol. Evol. 38, 5144–5155. doi: 10.1093/molbev/msab241
Liu L., Zhang R., Wang X., Zhu H., Tian Z. (2020). Transcriptome analysis reveals molecular mechanisms responsive to acute cold stress in the tropical stenothermal fish tiger barb (Puntius tetrazona). BMC Genomics 21, 1–14. doi: 10.1186/s12864-020-07139-z
Long Y., Li X., Li F., Ge G., Liu R., Song G., et al. (2020). Transcriptional programs underlying cold acclimation of common carp (Cyprinus carpio l.). Front. Genet. 11. doi: 10.3389/fgene.2020.556418
Long Y., Li L., Li Q., He X., Cui Z. (2012). Transcriptomic characterization of temperature stress responses in larval zebrafish. PLoS. One 7, e37209. doi: 10.1371/journal.pone.0037209
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lyu L., Wen H., Li Y., Li J., Zhao J., Zhang S., et al. (2018). Deep transcriptomic analysis of black rockfish (Sebastes schlegelii) provides new insights on responses to acute temperature stress. Sci. Rep. 8, 9113. doi: 10.1038/s41598-018-27013-z
Malekar V. C., Morton J. D., Hider R. N., Cruickshank R. H., Hodge S., Metcalf V. J. (2018). Effect of elevated temperature on membrane lipid saturation in Antarctic notothenioid fish. PeerJ 6, e4765. doi: 10.7717/peerj.4765
Narum S. R., Campbell N. R. (2015). Transcriptomic response to heat stress among ecologically divergent populations of redband trout. BMC Genomics 16, 103. doi: 10.1186/s12864-015-1246-5
Ndong D., Chen Y. Y., Lin Y. H., Vaseeharan B., Chen J. C. (2007). The immune response of tilapia Oreochromis mossambicus and its susceptibility to Streptococcus iniae under stress in low and high temperatures. Fish. Shellfish. Immunol. 22, 686–694. doi: 10.1016/j.fsi.2006.08.015
Nilsson G. E., Crawley N., Lunde I. G., Munday P. L. (2009). Elevated temperature reduces the respiratory scope of coral reef fishes. Global Change Biol. 15, 1405–1412. doi: 10.1111/j.1365-2486.2008.01767.x
Norambuena F., Estévez A., Sánchez-Vázquez F. J., Carazo I., Duncan N. (2012). Self-selection of diets with different contents of arachidonic acid by Senegalese sole (Solea senegalensis) broodstock. Aquaculture. 364, 198–205. doi: 10.1016/j.aquaculture.2012.08.016
Norambuena F., Morais S., Emery J. A., Turchini G. M. (2015). Arachidonic acid and eicosapentaenoic acid metabolism in juvenile Atlantic salmon as affected by water temperature. PloS One 10, e0143622. doi: 10.1371/journal.pone.0143622
Norin T., Malte H., Clark T. D. (2016). Differential plasticity of metabolic rate phenotypes in a tropical fish facing environmental change. Funct. Ecol. 30, 369–378. doi: 10.1111/1365-2435.12503
Oomen R. A., Hutchings J. A. (2017). Transcriptomic responses to environmental change in fishes: Insights from RNA sequencing. Facets. 2, 610–641. doi: 10.1139/facets-2017-0015
Paul N., Novais S. C., Silva C. S. E., Mendes S., Kunzmann A., Lemos M. F. L. (2021). Global warming overrides physiological anti-predatory mechanisms in intertidal rock pool fish Gobius paganellus. Sci. Total Environ. 776, 145736. doi: 10.1016/j.scitotenv.2021.145736
Pigliucci M., Murren C. J., Schlichting C. D. (2006). Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 209, 2362–2367. doi: 10.1242/jeb.02070
Punzón A., Serrano A., Sánchez F., Velasco F., Preciado I., González-Irusta J. M., et al. (2016). Response of a temperate demersal fish community to global warming. J. Mar. Syst. 161, 1–10. doi: 10.1016/j.jmarsys.2016.05.001
Schlaepfer I. R., Joshi M. (2020). CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 161, bqz046. doi: 10.1210/endocr/bqz046
Smith S., Bernatchez L., Beheregaray L. B. (2013). RNA-Seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genomics 14, 375. doi: 10.1186/1471-2164-14-375
Souza M. R. D. P., Herrerias T., Zaleski T., Forgati M., Kandalski P. K., Machado C., et al. (2018). Heat stress in the heart and muscle of the Antarctic fishes Notothenia rossii and Notothenia coriiceps: carbohydrate metabolism and antioxidant defence. Biochimie 146, 43–55. doi: 10.1016/j.biochi.2017.11.010
Srikanth K., Pereira E., Duarte A. C., Ahmad I. (2013). Glutathione and its dependent enzymes’ modulatory responses to toxic metals and metalloids in fish–a review. Environ. Sci. pollut. Res. 20, 2133–2149. doi: 10.1007/s11356-012-1459-y
Tautenhahn R., Patti G. J., Rinehart D., Siuzdak G. (2012). XCMS online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039. doi: 10.1021/ac300698c
Veilleux H. D., Ryu T., Donelson J. M., van Herwerden L. V., Seridi L., Ghosheh Y., et al. (2015). Molecular processes of transgenerational acclimation to a warming ocean. Nat. Clim. Change. 5, 1074–1078. doi: 10.1038/nclimate2724
Wakil S. J., Abu-Elheiga L. A. (2009). Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50 Supplement, S138–S143. doi: 10.1194/jlr.R800079-JLR200
Wang Z. J. (2013). The reproductive biology and hematology of triplophysa bleekeri from daning river (Southwest university).
Wang Z., Huang J., Zhang Y. (2013). The reproductive traits of Triplophysa bleekeri in the daning river. Freshw. Fish. (Chongqing, China: Southwest University) 43, 8–12. doi: 10.3969/j.issn.1000-6907.2013.05.002
Wellband K. W., Heath D. D. (2017). Plasticity in gene transcription explains the differential performance of two invasive fish species. Evol. Appl. 10, 563–576. doi: 10.1111/eva.12463
Wen B., Jin S. R., Chen Z. Z., Gao J. Z. (2018). Physiological responses to cold stress in the gills of discus fish (Symphysodon aequifasciatus) revealed by conventional biochemical assays and GC-TOF-MS metabolomics. Sci. Total Environ. 640–641, 1372–1381. doi: 10.1016/j.scitotenv.2018.05.401
Wright L. C., Santangelo R. M., Ganendren R., Payne J., Djordjevic J. T., Sorrell T. C. (2007). Cryptococcal lipid metabolism: phospholipase B1 is implicated in transcellular metabolism of macrophage-derived lipids. Eukaryot. Cell. 6, 37–47. doi: 10.1128/EC.00262-06
Xu H., Meng X., Wei Y., Ma Q., Liang M., Turchini G. M. (2022). Arachidonic acid matters. Rev. Aquacult. 1, 1–33. doi: 10.1111/raq.12679
Yang S., Zhao T., Ma A., Huang Z., Liu Z., Cui W., et al. (2020). Metabolic responses in Scophthalmus maximus kidney subjected to thermal stress. Fish. Shellfish. Immunol. 103, 37–46. doi: 10.1016/j.fsi.2020.04.003
Yuan D., Chen X., Gu H., Zou M., Zou Y., Fang J., et al. (2020). Chromosomal genome of Triplophysa bleekeri provides insights into its evolution and environmental adaptation. GigaScience 9, giaa132. doi: 10.1093/gigascience/giaa132
Yuan D. Y., Wang B., Tang T., Lei L., Zhou C. W., Li Z. Q., et al. (2021). Characterization and evaluation of the tissue distribution of CRH, apelin, and GnRH2 reveal responses to feeding states in Schizothorax davidi. fish physiol. Biochem. 47, 421–438. doi: 10.1007/s10695-020-00922-5
Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., et al. (2010). Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330. doi: 10.1016/j.cell.2010.03.029
Zhang X., Liu L., Wang L., Pan Y., Hao X., Zhang G., et al. (2021). Comparative lipidomics analysis of human milk and infant formulas using UHPLC-Q-TOF-MS. J. Agric. Food Chem. 69, 1146–1155. doi: 10.1021/acs.jafc.0c06940
Zhang Y., Zhang H., Zhao B. (2018). Hippo signaling in the immune system. Trends Biochem. Sci. 43, 77–80. doi: 10.1016/j.tibs.2017.11.009
Zhao T., Ma A., Huang Z., Liu Z., Sun Z., Zhu C., et al. (2021). Transcriptome analysis reveals that high temperatures alter modes of lipid metabolism in juvenile turbot (Scophthalmus maximus) liver. Comp. Biochem. Physiol. Part D. Genomics Proteomics. 40, 100887. doi: 10.1016/j.cbd.2021.100887
Keywords: temperature change, heat stress response, lipid metabolism, metabolic reprogramming, high-plateau fish
Citation: Yuan D, Wang H, Liu X, Wang S, Shi J, Cheng X, Gu H, Xiao S and Wang Z (2022) High temperature induced metabolic reprogramming and lipid remodeling in a high-altitude fish species, Triplophysa bleekeri. Front. Mar. Sci. 9:1017142. doi: 10.3389/fmars.2022.1017142
Received: 11 August 2022; Accepted: 04 October 2022;
Published: 18 October 2022.
Edited by:
Hongsheng Yang, Institute of Oceanology (CAS), ChinaReviewed by:
Mohammad Abdul Momin Siddique, Noakhali Science and Technology University, BangladeshTianming Wang, Zhejiang Ocean University, China
Copyright © 2022 Yuan, Wang, Liu, Wang, Shi, Cheng, Gu, Xiao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijun Xiao, c2hpanVuX3hpYW9AMTYzLmNvbQ==; Zhijian Wang, d2FuZ3pqMTk2OUAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
 Dengyue Yuan
Dengyue Yuan Haoyu Wang
Haoyu Wang Xiaoqin Liu
Xiaoqin Liu Siya Wang
Siya Wang Jinfeng Shi
Jinfeng Shi Xinkai Cheng
Xinkai Cheng Haoran Gu
Haoran Gu Shijun Xiao
Shijun Xiao Zhijian Wang
Zhijian Wang