
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 23 September 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1011420
This article is part of the Research TopicMollusk Breeding and Genetic Improvement - Volume IIView all 6 articles
The receptors of serotonin, also known as 5-hydroxytryptamine receptor (5-HTR) can mediate regulatory metamorphosis processes in a variety of mollusks. Studying the mechanisms of metamorphosis of the pearl oyster is significant to elucidate breeding, resource recovery and marine pearl production. In this study, two 5-HTR genes from Pinctada fucata martensii (Pm5-HTR2 and Pm5-HTR4) were cloned. A total of 1623 bp open reading frame was identified in Pm5-HTR2, and a 1185 bp open reading frame was detected in Pm5-HTR4; these open reading frames encoded a 540-residue polypeptide and a 394-residue polypeptide, respectively. We also conducted a domain analysis, which indicated that Pm5-HTR2 and Pm5-HTR4 contained a seven-transmembrane domain and revealed that the receptors had high similarity to Crassostrea gigas 5-HTR2 (54.62%) and 5-HTR4 (66.23%). Sequence analysis demonstrated conserved advanced structure and motifs (the DRY/ERY and NPXXY motifs). The expression pattern analysis revealed high expression levels of Pm5-HTR2 and Pm5-HTR4 during the developmental stages. ISH analysis showed that Pm5-HTR2 was primarily expressed in the FE, B, T, EU, and EL stages and Pm5-HTR4 was mainly expressed in the FE, B, T, D, EU, and EL stages. These results suggest that 5-HTRs may play key roles in P. f. martensii larval metamorphosis.
The neurotransmitter 5-hydroxytryptamine (5-HT), more commonly known as serotonin, is a key signaling molecule in mammals that regulates many physiological processes, such as learning, emotion, sleep, locomotion, reproduction, and pain perception (Weiger, 1997). 5-HT also is one of the oldest and most ubiquitous developmental regulators in vertebrates and invertebrates. In invertebrates, 5-HT plays a crucial role in growth, development, memory, mating, and biological rhythms (Norville et al., 2010; Johnson et al., 2011). In mollusks, 5-HT and 5-HT receptors (5-HTRs) are key components of a variety of physiological activities, including locomotion (Filla et al., 2004), feeding (Kawai et al., 2011), and reproduction (Panasophonkul et al., 2009); researchers have also demonstrated their involvement in memory (Kandel, 2001) and circadian rhythms (Levenson et al., 1999). Furthermore, 5-HT can regulate metamorphosis and larval settlement in invertebrates, including hydroids (McCauley, 1997; Mayorova et al., 2014), ascidians (Zega et al., 2005), barnacles (Yamamoto et al., 1996), and mollusks (Couper and Leise, 1996; Urrutia et al., 2004; Mapara et al., 2008). For example, the larvae of freshwater molluscs and marine polychaetes, endogenous 5-HT released from the neurons of the apical sensory organ (ASO) in response to external stimuli retarded larval development at premetamorphic stages, and accelerated it at metamorphic stages (Glebov et al., 2014). In ascidian larvae, 5-HT signaling was suggested to trigger metamorphosis (Zega et al., 2005). 5-HT was suggested to act via larval nervous system and downstream 5-HT receptors. However, the underlying functions of 5-HT in development of pearly oyster are unclearly.
5-HT regulates a plethora of biological functions by binding to several receptors (Tierney, 2001). Researchers have identified seven 5-HT receptor subtypes (5-HT1–7) in mammals, and all seven have been characterized as G-protein-coupled receptors (GPCRs) according to biochemical and pharmacological criteria (Roth, 2006). In mammals, these receptors are involved in synthesizing second messengers that activate mitogen-activated protein kinase as well as protein kinases A and C (Ghirardi et al., 1992; Li et al., 1995; Martin et al., 1997). By interacting with G-protein-coupled receptors, 5-HT alters the concentration of intracellular cAMP in larvae, thereby participating in larval settlement and metamorphosis (Heyland et al., 2011). Three types of 5-HT receptors (5-HT1-, 5-HT4- and 5-HT7-like) from Helicoma trivolvis are functionally active at premetamorphic (trochophore, veliger) and metamorphic (veliconcha) stages (Glebov et al., 2014).
The lifestyles of most aquatic invertebrates are complex and consist of two phases: a larval (planktonic) phase followed by an adult (benthic) phase. The intricate process of larval development is regulated by several internal and external elements. Metamorphosis, in which the planktonic larvae transition to adult bivalves, is a vital stage in the life cycle of the bivalve and has a direct impact on changes in the population because of the mortality rate of 80%-90% (Yang et al., 2022). Research has demonstrated that 5-HT is a crucial molecule in the transition from the planktonic stage to the adult stage, including larval settlement and metamorphosis. Serotonergic pathways, which involve 5-HT and its precursors (5-hydroxytryptophan and L-tryptophan), help regulate metamorphosis in many invertebrates (Zhao et al., 2003; Urrutia et al., 2004; Yu et al., 2008; Alfaro et al., 2011; Cao et al., 2020). Zhao et al. (2003) reported that 10−3 M serotonin led to the highest rate of larval settlement in Pinctada maxima and did not have any toxic effects on the larva. However, studies on 5-HTR-particularly role of its signaling pathways in larval development and during metamorphosis are scarce.
Pinctada fucata martensii is a subspecies of pearl oyster predominantly distributed in subtropical and tropical marine environments (Li et al., 2018); it is the main species of seawater pearl oysters cultivated in China and is cultured in Guangdong Province, Guangxi Province, and Hainan Province (He et al., 2020). Studying the mechanisms of metamorphosis as well as the larval growth and development patterns of the pearl oyster is essential to elucidate breeding, resource recovery, population changes, and distribution patterns in natural marine areas. Our previous research revealed that the upregulation of 5-HTRs (5-HTRs 1, 2, and 4) promoted larval metamorphosis in P. f. martensii (Zhang et al., 2021). Therefore, in the current study, two 5-HTR genes were cloned and analyzed to explore their functions in larval development.
The animals used in this study were 1.5-year-old P. f. martensii black shell line pearl oysters (Deng et al., 2013) that were reared on pearl farm in Liusha bay (Zhanjiang, Guangdong Province, China). Sperm and eggs were obtained by dissecting the parents, and after artificial insemination, the larvae of P. f. martensii were grown in nursery pools (temperature: 25 ± 1°C; salinity: 30 ± 1‰). Larvae were collected at the following stages: juvenile (J), spat (S), eyed larvae (EL), early umbo larvae (EU), D-stage larvae before feeding (DF), D-stage larvae (D), trochophore (T), early trochophore (ET), gastrula (G), blastula (B), fertilized egg (FE), and egg (E). Liquid nitrogen was used to freeze some samples immediately, and these samples were kept at −80°C until they underwent RNA extraction; 5% MgCl2 was used to anesthetize the other larvae, which were then washed with filtered sterile seawater, fixed in 4% paraformaldehyde containing 1% DEPC, and stored at 4°C for in situ hybridization experiments.
Trizol reagent (Thermo-Fisher Scientific, USA) was used to isolate total RNA from samples at different developmental stages, and gel electrophoresis was used to detect RNA integrity on 1% agarose gel. To measure RNA quantity, the OD260/OD280 ratio was calculated with a NanoDrop 2000 spectrophotometer (Thermo-Fisher Scientific, USA). Subsequently, M-MLV reverse transcriptase obtained from Promega (USA) was employed to synthesize cDNA from 2 μg aliquots of RNA following the protocol of the manufacturer. Partial sequences of Pm5-HTR2 and Pm5-HTR4 were obtained from the P. f. martensii genome (Du et al., 2017) and larval transcriptome (Zhang et al., 2021), and a SMART RACE cDNA Amplification Kit (Clontech, USA) was used to carry out RACE reactions. The 5′ untranslated region (5′UTR) (partial sequence of Pm5-HTR2 and Pm5-HTR4) and 3′ untranslated region (3′UTR) (Pm5-HTR2 and Pm5-HTR4) were isolated via nested polymerase chain reaction (PCR). The 5′UTR of Pm5-HTR2 was obtained using unpublished data on the full-length transcriptome of P. f. martensii. Primers (presented in Table 1) were based on the partial sequences of Pm5-HTR2 and Pm5-HTR4.
BLASTX (http://www.ncbi.nlm.nih.gov/blast) and DNAMAN were used to evaluate and merge the complete sequences of Pm5-HTR4 and Pm5-HTR2. The Open Reading Frame Finder (https://www.ncbi.nlm.nih.gov/orffinder/) was then employed for the prediction of the open reading frame (ORF). Subsequently, the amino acid sequences were identified using Expasy ProtParam (http://web.expasy.org/protparam/), and the transmembrane domain was analyzed by TMHMM Server version 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0). NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to assess the similarity between 5-HTR amino acid sequences from different species, and multiple sequences were aligned with the sequences of 5-HTR2 (Crassostrea virginica, XP_022343922.1; Mytilus galloprovincialis, VDI68244.1; Haliotis rufescens, XP_046365272.1; Crassostrea gigas, XP_011430856.2; Pecten maximus, XP_033761220.1) and 5-HTR4 (C. virginica, XP_022341057.1; C. gigas, XP_011430667.2; P. maximus, XP_033756774.1; Mizuhopecten yessoensis, XP_021369092.1; and Mytilus coruscus, CAC5420242.1) using Clustal W. The advanced structure was determined by Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/protocol) on the basis of sequences from M. galloprovincialis (VDI68244.1) and C. gigas (XP_011430667.2). The amino acid sequences of 5-HTRs from various species (Haliotis discus hannai: QEZ90770.1, QEZ90772.1, QEZ90773.1, QEZ90775.1, and QEZ90776.1; Pinctada fucata: AIW04132.1; Aplysia californica: NP_001240691.1 and XP_005105784.1; M. yessoensis: XP_021369092.1; Homo sapiens: CAA71462.1, AAA92622.1, NP_000863.1, NP_000858.3, and NP_000859.1; Mus musculus: NP_001347227.1; Xenopus laevis: NP_001082744.1; Equus caballus: NP_001157454.1; and Bos taurus: NP_001035575.1) were employed to build a phylogenetic tree with MEGA6 software. To obtain the confidence value for the phylogenetic investigation, the neighbor-joining method was utilized; 1000 bootstrap replications were carried out.
Quantitative real-time RT-PCR (qRT-PCR) was performed with β-actin as the reference gene to determine mRNA expression. Research has showed that β-actin was the most stable reference gene for different tissues and different embryonic developmental stages (Wang and He, 2013) and have been widely used as reference genes for qRT-PCR analysis (Wang et al., 2019; Lai et al., 2021; Chen et al., 2022). qRT-PCR was performed with SYBR Green Master Mix (Thermo-Fisher Scientific, USA) according to the protocol recommended by the manufacturer. All reactions were run in triplicate. Using the complete cDNA sequences from the Pm5-HTR2 and Pm5-HTR4 genes, primers (100 to 300 bp in length) were created with a T7 promoter (GCGTAATACGACTCACTATAGGG) inserted upstream of the forward primer to perform in situ hybridization (ISH). PCR amplification was used to isolate the target fragments, which were then recovered and purified. The recovered products were subsequently transcribed by DIG RNA Labeling Mix and T7 RNA polymerase, ultimately yielding a digoxigenin-labeled RNA probe. D-shaped larvae, umbo larvae, eyed larvae, and juveniles were decalcified with 30% EDTA solution containing 1% DEPC for 1–12 h (the decalcification time was gradually extended as development progressed), and the other stages of larvae were washed with DEPC 3–5 times for 2 min. The treated larvae underwent in situ hybridization using the Enhanced Sensitivity ISH Detection Kit IV (CY3) (Boster) following the instructions from the manufacturer, and observations were recorded by microscopic examination.
SPSS software (IBM, Chicago, IL, USA) was used to perform all statistical analyses in this study. Significant differences (P < 0.05) in expression levels between different stages of development were identified by one-way analysis of variance (ANOVA) followed by Duncan’s multiple comparison test. The expression levels of Pm5-HTR2 and Pm5-HTR4 were analyzed by comparative CT (2−ΔΔCT) method (Livak and Schmittgen, 2001).
The 5′-UTR, 3′-UTR, and the complete sequence of Pm5-HTR2 were 483 bp, 192 bp, and 2298 bp, respectively. The Pm5-HTR2 sequence analysis identified an ORF of 1623 bp; in addition, the gene encoded 540 amino acids (Figure 1A), and its molecular weight was 60.94 kDa. Transmembrane prediction using hidden Markov models (TMHMM) revealed seven transmembrane domains, each consisting of 19–22 residues. According to the predictions of the NetPhos 3.1 server, Pm5-HTR2 contains 106 phosphorylation sites, and threonine, serine, and tyrosine residues serve as potential phosphorylation sites for multiple protein kinases (Supplementary Table 1).
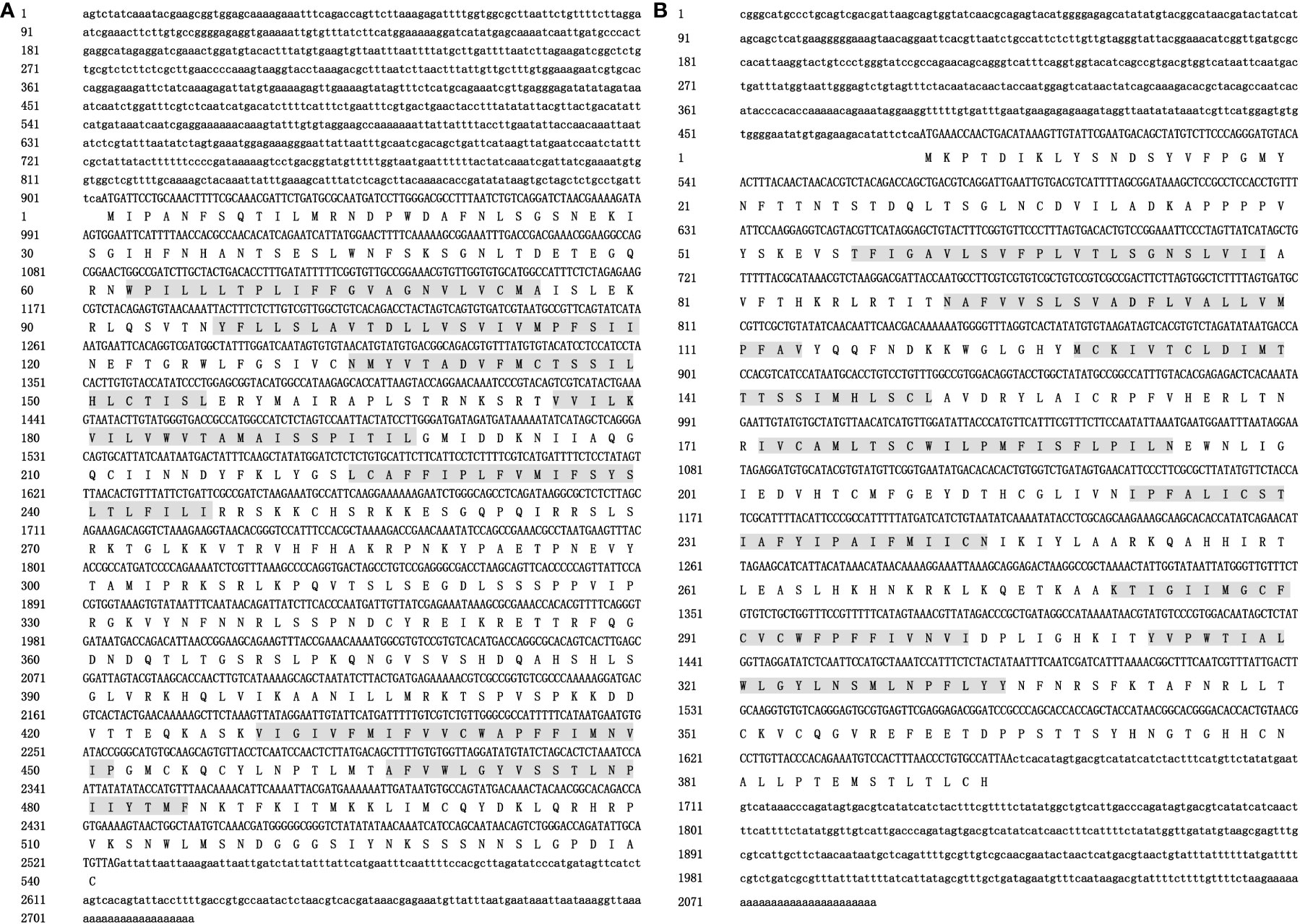
Figure 1 The nucleotide and deduced amino acid sequences of Pm5-HTR2 (A) and Pm5-HTR4 (B). The 5′-UTR and 3′-UTR are indicated by small letters; capital letters represent the ORF and the deduced amino acid (aa) sequences; and the letters with gray background shows the transmembrane domain.
The 5′-UTR, 3′-UTR, and the complete sequence of Pm5-HTR4 were 479 bp, 428 bp, and 2092 bp, respectively. The ORF identified by Pm5-HTR4 sequence analysis was 1185 bp and encoded a total of 394 amino acids (Figure 1B); the gene’s molecular weight was 44.59 kDa. TMHMM analysis revealed seven transmembrane domains, each consisting of 22 residues. The NetPhos 3.1 server predicted the presence of 31 phosphorylation sites and indicated that threonine, serine, or tyrosine residues serve as potential phosphorylation sites for multiple protein kinases (Supplemental Table 2).
The 5-HTR2 and 5-HTR4 sequences from different species were used to construct multiple sequence alignment, which revealed that the 5-HTR amino acids were conserved. In addition, DRY and NPXXY (the receptor activation residues found in the GPCRs) were conserved in the 5-HTR2 and 5-HTR4 structural profiles (Figure 2). BLASTP was used to search the NCBI database, which revealed that Pm5-HTR2 exhibited the highest homology with the C. gigas 5-HTR2 (54.62%), and Pm5-HTR4 exhibited the highest homogony with C. gigas 5-HTR4 (66.23%).

Figure 2 Multiple sequence alignment of Pm5-HTR2 (A) and Pm5-HTR4 (B). The numbers on the right present the total amino acid of each protein. The conserved residues believed to be involved in 5-HT–receptor binding is indicated by asterisks. The rectangular box indicates the conserved residues of GPCRs.
The advanced structure of the P. f. martensii and M. galloprovincialis 5-HTR2s and P. f. martensii and C. gigas 5-HTR4s were predicted by Phyre2, and the results were visualized by Chimera 1.8.1. Multiple alpha helices were identified in 5-HTR2 and 5-HTR4, and helix–loop–helix structures separated these motifs (Figure 3). The advanced structures of the 5-HTR2s of P. f. martensii and M. galloprovincialis were similar, as were the ERY and NPXXY motifs of these species. Correspondingly, the advanced structures of the 5-HTR4s from P. f. martensii and C. gigas were similar, as were the DRY and NPXXY motifs. These findings validated the conservation of the predicted 5-HTR structure. In addition, we observed a short carboxy-terminal domain and a long third cytoplasmic loop in the sequence.
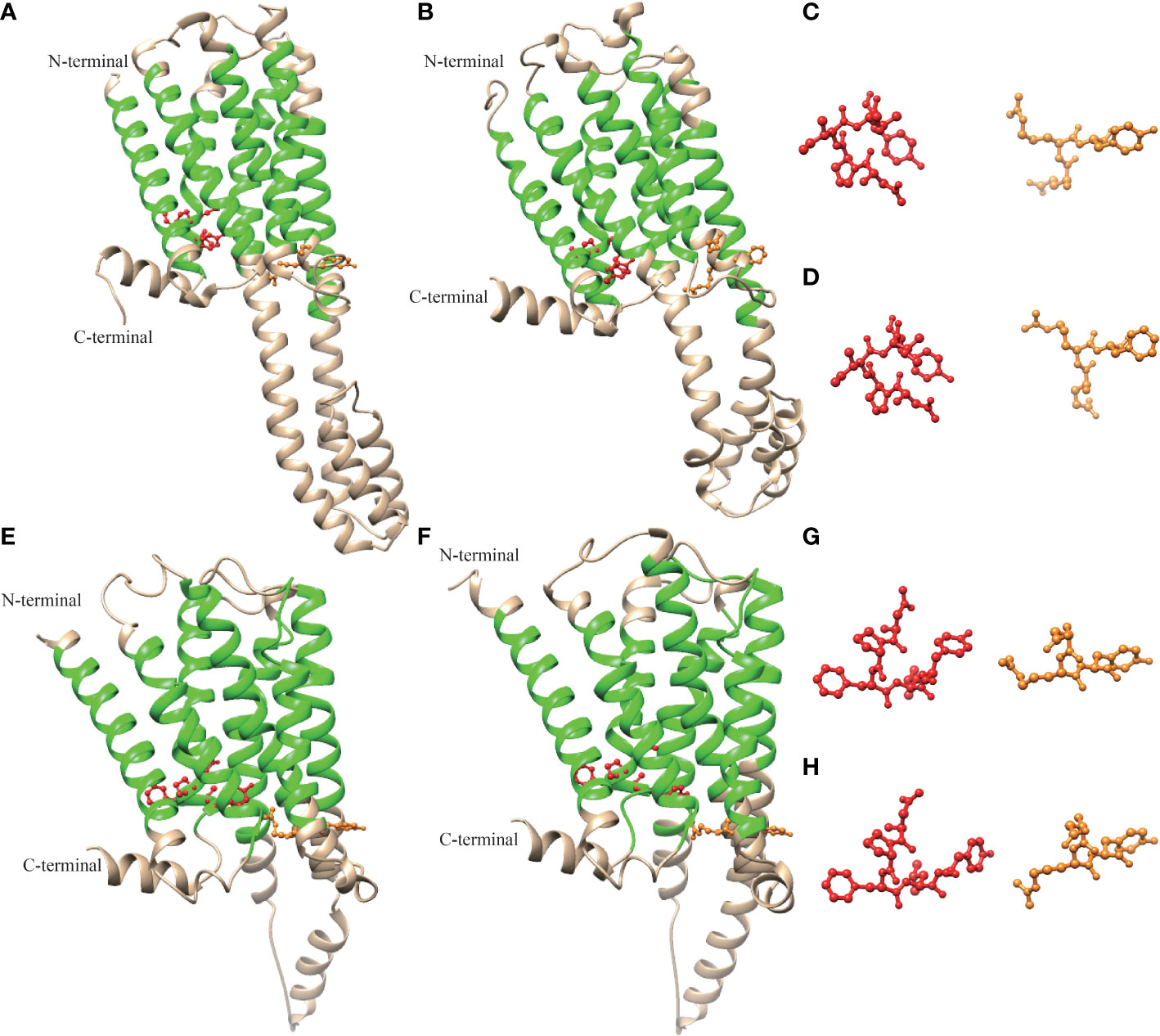
Figure 3 Molecular model of the three-dimensional structure of Pm5-HTR2 (A), Mg-HTR2 (B) (VDI68244.1), Pm5-HTR4 (E), and Cg5-HTR4 (F) (XP_011430667.2). The ERY/DRY and NPXXY motifs are shown in orange and red, respectively. Panels (C, D, G, H) show the ERY/DRY and NPXXY motifs of panels (A, B, E, F), respectively.
To investigate possible evolutionary relationships, the 5-HTRs of various vertebrate and invertebrate species were used to construct a phylogenetic tree. The results presented in Figure 4 show the clustering of the 5-HTRs into various clades. According to the phylogenetic tree analysis, Pm5-HTR2 showed the closest relationship to the 5-HTR2 of H. discus hannai, and the 5-HTR2s of P. f. martensii, H. discus hannai, H. sapiens, and X. laevis were clustered and separated with other 5-HTRs. Pm5-HTR4 clustered with the 5-HTR4 of the bivalve species (H. discus hannai, A. californica, and M. yessoensis). The results of the phylogenetic tree analysis were consistent with those of traditional taxonomy.
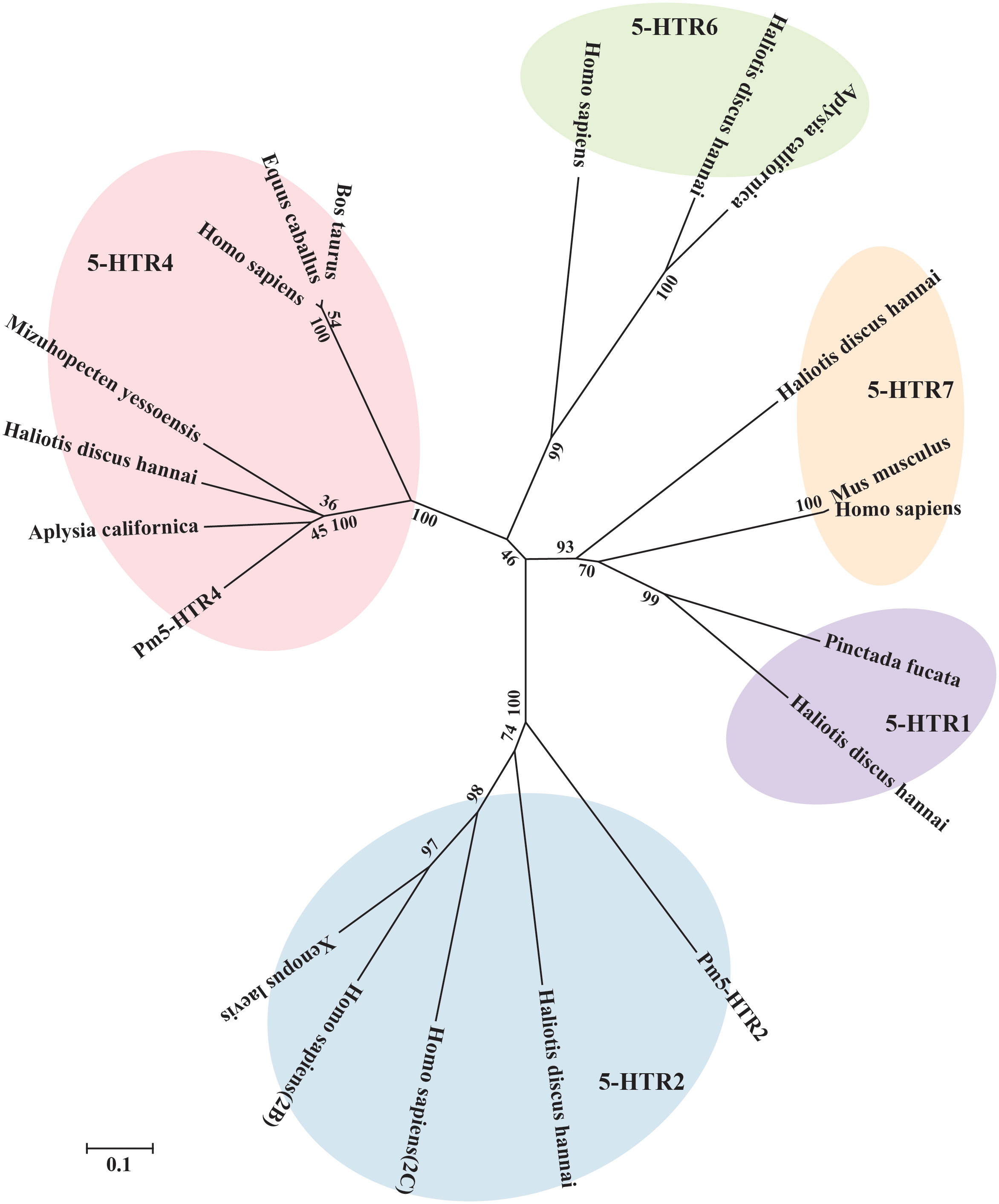
Figure 4 Phylogenetic tree analysis of Pm5-HTR2 and Pm5-HTR4 from different species. The phylogenetic tree was constructed by MEGA software 6.05 using the neighbor-joining method with 1000 bootstrap replications. The scale bar indicates a branch length of 0.1.
To analyze the expression level of Pm5-HTR2 and Pm5-HTR4 at different development stages, qRT-PCR method was utilized. Both Pm5-HTR2 and Pm5-HTR4 were expressed in the following stages: J, S, EL, EU, DF, D, T, ET, G, B, FE, and E (Figure 5). Pm5-HTR2 expression in G was significantly elevated compared with its expression in the other stages (P < 0.05) (Figure 5A). Pm5-HTR4 was highly expressed in the early development stages and the EU stage (Figure 5B). In addition, ISH was used to detect Pm5-HTR2 and Pm5-HTR4 in different developmental stages. The Pm5-HTR2 or Pm5-HTR4 ISH signal was indicated by red fluorescence; the nuclear staining signal (which indicates the integrity of the tissue or individual) was indicated by blue fluorescence. As shown in Figure 6, the ISH results revealed that Pm5-HTR2 was primarily expressed in the FE, B, T, EU, and EL stages; in addition, strong hybridization was identified in the T and EL stages (Figure 6B-a and Figure 6B-c). Pm5-HTR4 was mainly expressed in the FE, B, T, D, EU, and EL stages (Figure 7), and strong hybridization signals were detected in the FE, T, D, and EU stages (Figure 7B-a and Figure 7B-c).
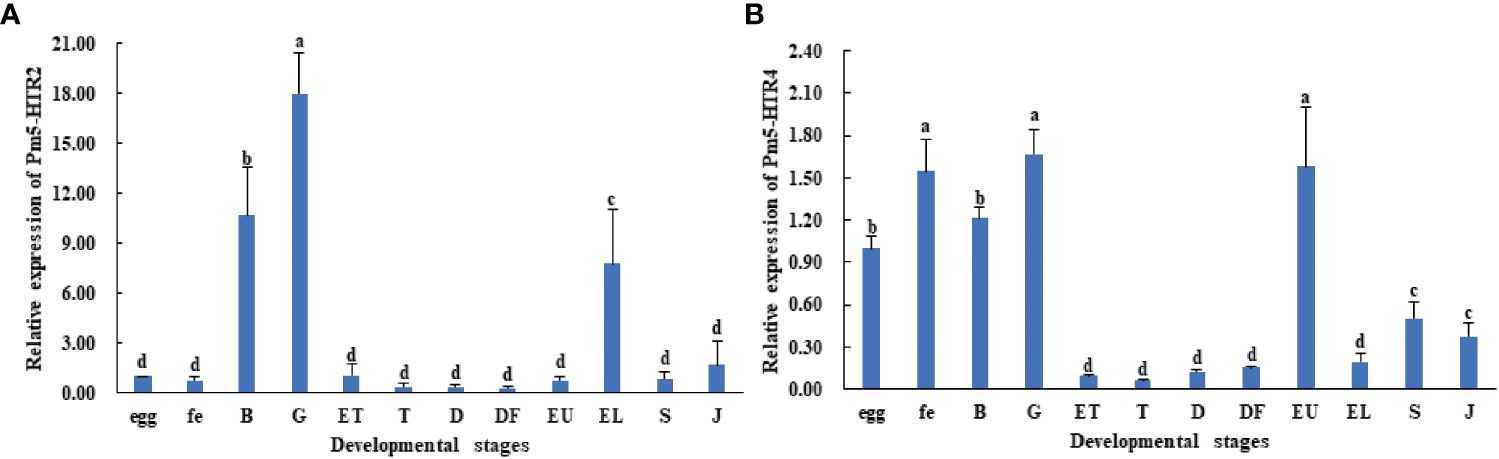
Figure 5 Expression pattern of Pm5-HTR2 (A) and Pm5-HTR4 (B) in development stages. Bars with different superscripts indicate significant differences (P < 0.05). E, egg; FE, fertilized egg; B, blastula; G, gastrula; ET, early trochophore; T, trochophore; D, D-stage larvae; DF, D-stage larvae before feeding; EU, early umbo larvae; EL, eyed larvae; S, spat; J, juvenile.
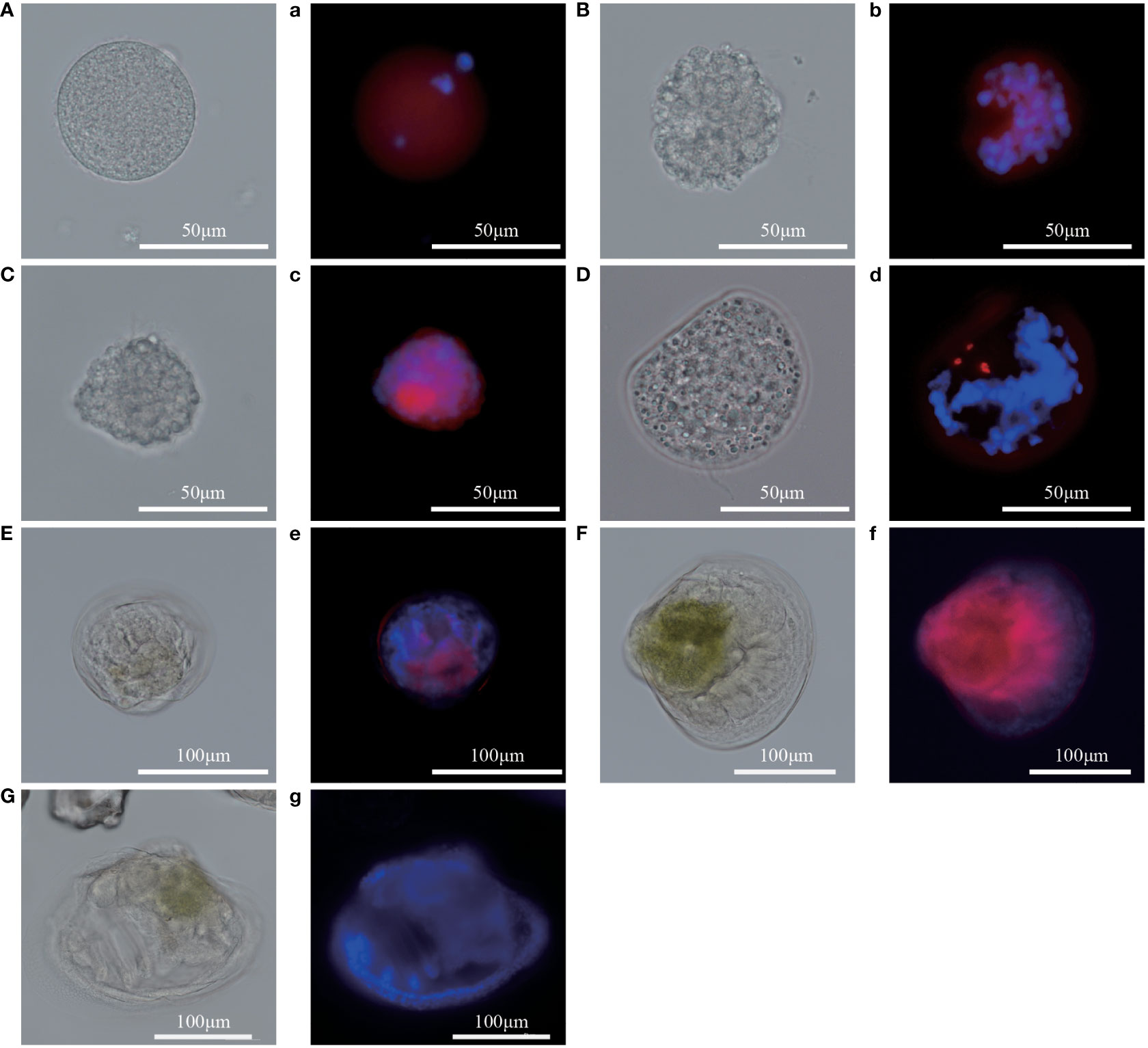
Figure 6 In situ hybridization results of Pm5-HTR2 at the developmental stage. (A–G) photos of the experimental group under white light; (a–g) photos of the experimental group under fluorescent light. (A/a), fertilized ovum; (B/b), blastocyst; (C/c), trochophore; (D/d), D larvae; (E/e), umbo larvae; (F/f), eyed larvae; (G/g), juvenile. Red fluorescence: in situ hybridization signal of the gene; blue fluorescence: nucleus.
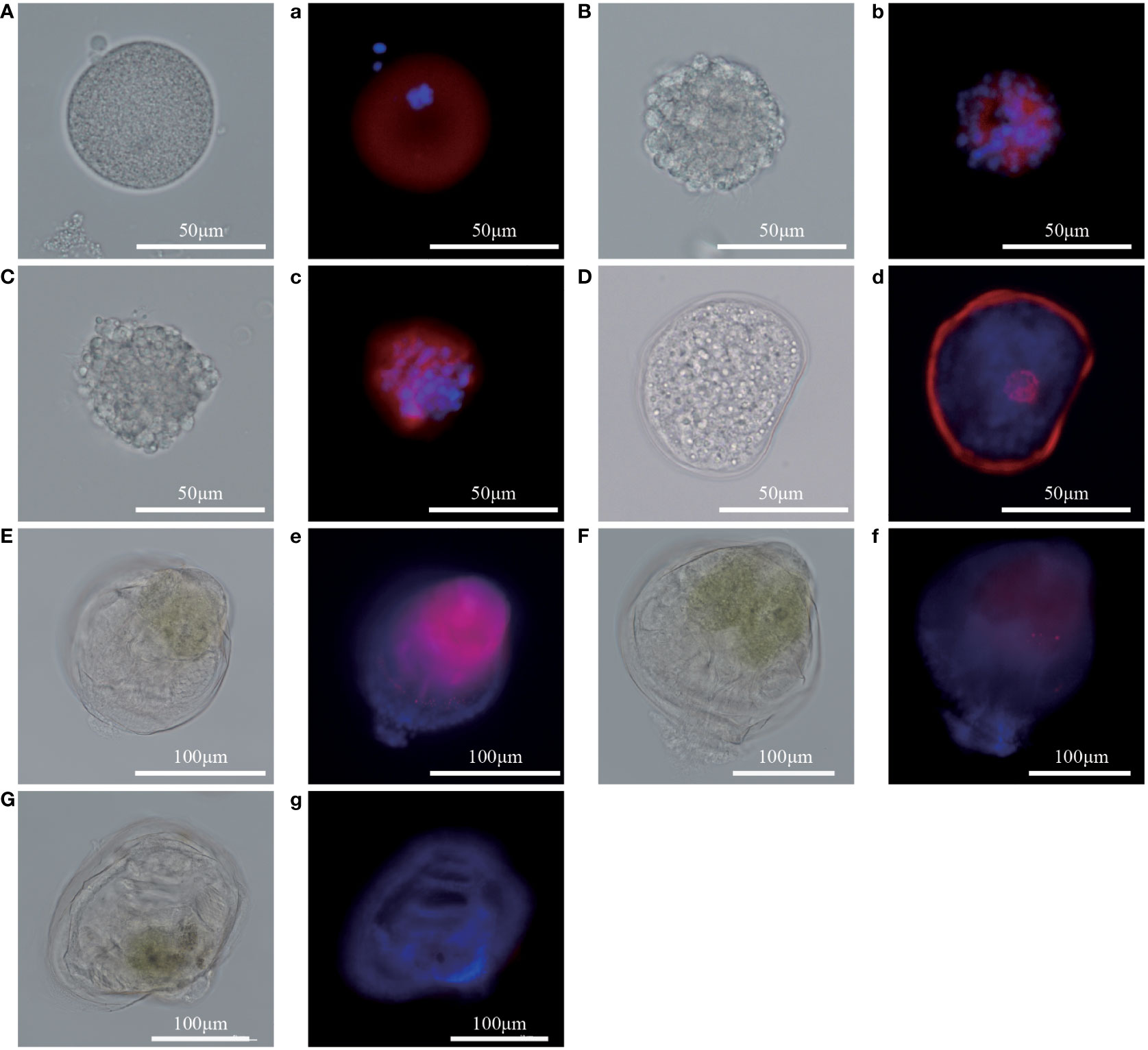
Figure 7 In situ hybridization results of Pm5-HTR4 at the developmental stage. (A–G) photos of the experimental group under white light; a–g: photos of the experimental group under fluorescent light. (A/a), fertilized ovum; (B/b), blastocyst; (C/c), trochophore; (D/d), D larvae; (E/e), umbo larvae; (F/f), eyed larvae; (G/g), juvenile. Red fluorescence: in situ hybridization signal of the gene; blue fluorescence: nucleus.
Research has showed that serotonergic pathway which includes the manoamine neurotransmitter 5-HT, involved in larval development and metamorphosis in H. trivolvis and P. f. martensii (Glebov et al., 2014; Zhang et al., 2021; Yang et al., 2022). In this study, two full-length of 5-HTRs obtained from P. f. martensii were used to explore their potential roles in this species’ development. The protein sequences of both Pm5-HTR2 and Pm5-HTR4 contained seven transmembrane domains; these were identified as typical G-protein-coupled chemoreceptors. Serotonin receptor was characterized with seven hydrophobic transmembrane helical segments with unique sequence patterns (including a short intracellular loop domain and a long third cytoplasmic loop) (Tierney, 2001), which is similar to the result of the present study. These intracellular loops and extracellular loops connect these transmembrane helices (Baldwin, 1993). Receptor activation in the GPCR family is likely attributable to the DRY/ERY and NPXXY motifs of TM-III and TM-VII, respectively (Sharker et al., 2020). Furthermore, the phosphorylation site for protein kinase A or C may be essential in various signal transduction cascades (Ali et al., 2016). According to multiple sequence alignment, the binding of 5-HT with its receptor involves the conserved residues Ala, Phe, and Trp (Mapara et al., 2008). Consistent with these findings, advanced structure analysis demonstrated the conserved structure of the full protein sequence and motif (Sharker et al., 2020). In this study, a phylogenetic tree analysis revealed that Pm5-HTR2 and Pm5-HTR4 clustered with 5-HTR2 and 5-HTR4, respectively, which supports the results of the abovementioned studies.
The signaling of 5-HT2R stimulates the phosphatidylinositol–calcium second messenger system, regulates the action of phosphatidylinositol 3-kinase and its downstream signaling pathways, and induces cells to release intracellular calcium ions. 5-HT4R activates adenylate cyclase activity primarily by coupling with G proteins, thereby elevating the level of the intracellular second messenger cAMP, which activates corresponding functional effects in cells (Claeysen et al., 1996). Furthermore, 5-HT2R is necessary for the release of dopamine and 5-HT, the uptake of 5-HT, and the modulation of extracellular dopamine and 5-HT levels, thus affecting neural activity (Lamiay et al., 2006; Doly et al., 2008) during mammalian development stages. The results of the RT-qPCR and ISH of Pm5-HTR2 and Pm5-HTR4 during P. f. martensii development revealed that both receptors are expressed widely during development especially in the early development stages which indicated their functions in the early development (Zheng et al., 2019; Zhang et al., 2021). The experimental tractability of H. trivolvis embryos revealed that 5-HT, 5-HTR and neurotransmitter plays multiple roles during embryonic development (Christopher et al., 1996; Mapara et al., 2008).
In invertebrates, different types of 5-HT receptors have varying affinities for serotonin; for example, Balanus amphitrite has two receptors with different affinities for serotonin. In one study, 5-HT receptors inhibited larval settlement when larvae were exposed to 0.1 μM serotonin, whereas they promoted larval settlement when larvae were exposed to high concentrations of serotonin (100 μM) (Yamamoto et al., 1996). Different 5-HTR isoforms (5-HTR1, 5-HTR4, and 5-HTR7) were also identified in the genome of the gastropod Helisoma trivolvis; as the gastropod transitioned to the metamorphic stage, researchers observed downregulated 5-HT4-like and 5-HT7-like receptor expression, along with downregulated expression of the downstream G protein. By contrast, they observed that the expression of the 5-HT1-like receptor was upregulated, as was the expression of its downstream G protein (Glebov et al., 2014). However, in some mollusks, such as Phestilla sibogae (Hadfield, 1984) and H. rufescens (Morse et al., 1979), 5-HT did not exhibit inductive activity during larval metamorphosis. In the present study, Pm5-HTR2 was highly expressed in the eyed larvae, whereas 5-HTR4-like was significantly upregulated in the umbo larvae. These results aligned with the results of ISH. Pm5-HTR2 showed highly expression in the eyed larvae and indicated its roles in the larval settlement and metamorphosis. Furthermore, Pm5-HTR4 showed highly expression in the stages before the eyed larvae which may provide the possibility of it in the materials or molecules preparation for the metamorphosis process. These results suggest that these receptors might function during different stages or have different functions in larval settlement and metamorphosis. These studies suggest that different marine invertebrate larvae may have different 5-HT and 5-HTR regulatory mechanisms during metamorphosis which require further investigation.
In the present study, two 5-HTR genes (Pm5-HTR2 and Pm5-HTR4) from P. f. martensii were cloned. Both contained a typical seven-transmembrane domain and demonstrated high identity with C. gigas 5-HTR2 (54.62%) and 5-HTR4 (66.23%). Sequence analysis revealed the conserved advanced structure and DRY/ERY and NPXXY motifs. Pm5-HTR2 and Pm5-HTR4 were expressed widely in the developmental stages, which was consistent with the results of ISH. These findings indicate that 5-HTRs participate in larval metamorphosis; thus, our study provides novel evidence to support the investigation of this process in bivalves.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CY and YD designed the research. SZ, YH, and QX conducted the research. CY and RH analyzed data. SZ, CY, YH, QX, JZ, RH, JL, and YD contributed to the final writing of the paper. All authors have read and approved the final manuscript.
This work was supported by Science and Technology Programof Guangdong Province (Grant No. 2021B0202020003), the Fund of the Key Laboratory of Tropical Marine Ecosystem and Bioresource, MNR (2021QN08), National Natural Science Foundation of China (Grant No. 32102817), Department of Education of Guangdong Province (Grant No. 2020ZDZX1045 and 2021KCXTD026), the earmarked fund for CARS-49, and Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (Grant No. 2022KJ146).
We thank TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1011420/full#supplementary-material
Alfaro A. C., Young T., Ganesan A. M. (2011). Regulatory effects of mussel (Aulacomya maoriana iredale 1915) larval settlement by neuroactive compounds, amino acids and bacterial biofilms. Aquac. 322, 158–168. doi: 10.1016/j.aquaculture.2011.08.038
Ali E. S., Hua J., Wilson C. H., Tallis G. A., Zhou F. H., Rychkov G. Y., et al. (2016). The glucagon-like peptide-1 analogue exendin-4 reverses impaired intracellular Ca2+ signalling in steatotic hepatocytes. BBA - Mol. Cell Res. 1863, 2135–2146. doi: 10.1016/j.bbamcr.2016.05.006
Baldwin M. J. (1993). The probable arrangement of the helices in G protein-coupled receptors. EMBO. J. 12, 1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x
Cao F. J., Zhong F. Z., Yang C. Y., Hao R. J., Wang Q. H., Liao Y. S., et al. (2020). Transcriptomic analysis of differentially expressed genes in the larval settlement and metamorphosis of peanut worm Sipunculus nudus. Aquacult Rep 18, 100475. doi: 10.1016/j.aqrep.2020.100475
Chen Q. Y., Yu W. M., Hao R. J., Yang J. M., Yang C. Y., Deng Y. W., et al. (2022). Molecular characterization and SNPs association with growth-related traits of myosin heavy chains from the pearl oyster Pinctada fucata martensii. Aquac. Res. 53, 2874–2885. doi: 10.1111/are.15802
Christopher K., Chang J., Goldberg J. (1996). Stimulation of cilia beat frequency by serotonin is mediated by a Ca2+ influx in ciliated cells of Helisoma trivolvis embryos. J. Exp. Biol. 199, 1105–1113. doi: 10.1242/jeb.199.5.1105
Claeysen S., Sebben M., Journot L., Bockaert J., Dumuis A. (1996). Cloning, expression and pharmacology of the mouse 5-HT4L receptor. FEBS Lett. 398, 19–25. doi: 10.1016/S0014-5793(96)01132-5
Couper J. M., Leise E. M. (1996). Serotonin injections induce metamorphosis in larvae of the gastropod mollusc Ilyanassa obsoleta. The Biological Bulletin 191, 178–186. doi: 10.2307/1542921
Deng Y. W., Fu S., Lu Y. Z., Du X. D., Wang Q. H., Huang H. L., et al. (2013). Fertilization, hatching, survival, and growth of third-generation colored pearl oyster (Pinctada martensii) stocks. J. Appl. Aquaculture. 25, 113–120. doi: 10.1080/10454438.2013.788311
Doly S., Valjent E., Setola V., Callebert J., Herve D., Launay J. M., et al. (2008). Serotonin 5-HT2B receptors are required for 3, 4-methylenedioxymethamphetamine-induced hyperlocomotion and 5-HT release in vivo and in vitro. J. Neurosci. 28, 2933–2940. doi: 10.1523/JNEUROSCI.5723-07.2008
Du X. D., Fan G. Y., Jiao Y., Zhang H., Guo X. M., Huang R. L., et al. (2017). The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 6 (8), 1–12. doi: 10.1093/gigascience/gix059
Filla A., Hiripi L., Elekes K. (2004). Serotonergic and dopaminergic influence of the duration of embryogenesis and intracapsular locomotion of Lymnaea stagnalis l. Acta Biol. Hung. 55, 315–321. doi: 10.1556/ABiol.55.2004.1-4.37
Ghirardi M., Braha O., Hochner B., Montarolo P. G., Kandel E. R., Dale N. (1992). Roles of PKA and PKC in facilitation of evoked and spontaneous transmitter release at depressed and nondepressed synapses in aplysia sensory neurons. Neuron. 9, 479–489. doi: 10.1016/0896-6273(92)90185-G
Glebov K., Voronezhskaya E. E., Khabarova M. Y., Ivashkin E., Nezlin L. P., Ponimaskin E. G. (2014). Mechanisms underlying dual effects of serotonin during development of Helisoma trivolvis (Mollusca). BMC Dev. Biol. 14, 1–19. doi: 10.1186/1471-213X-14-14
Hadfield M. G. (1984). Settlement requirements of molluscan larvae: new data on chemical and genetic roles. Aquac. 39, 283–298. doi: 10.1016/0044-8486(84)90272-2
He C. Z., Hao R. J., Deng Y. W., Yang C. Y., Du X. D. (2020). Response of pearl oyster Pinctada fucata martensii to allograft-induced stress from lipid metabolism. Fish shellfish Immunol. 98, 1001–1007. doi: 10.1016/j.fsi.2019.11.028
Heyland A., Degnan S., Reitzel A. M. (2011). Emerging patterns in the regulation and evolution of marine invertebrate settlement and metamorphosis Vol. 3 (Oxford, United Kingdom: Oxford University Press), 29–42.
Johnson O., Becnel J., Nichols C. D. (2011). Serotonin receptor activity is necessary for olfactory learning and memory in Drosophila melanogaster. Neuroscience. 192, 372–381. doi: 10.1016/j.neuroscience.2011.06.058
Kandel E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science. 294, 1030–1038. doi: 10.1126/science.1067020
Kawai R., Kobayashi S., Fujito Y., Ito E. (2011). Multiple subtypes of serotonin receptors in the feeding circuit of a pond snail. Zool Sci. 28, 517–525. doi: 10.2108/zsj.28.517
Lai Z. X., Adzigbli L., Chen Q. Y., Hao R. J., Liao Y. S., Deng Y. W., et al. (2021). Identification and allelic variants associated with cold tolerance of PmPIAS in Pinctada fucata martensii. Front. Physiol. 12, 634838. doi: 10.3389/fphys.2021.634838
Lamiay J. M., Schneider B., Loric S., Prada M. D., Kellermann O. (2006). Serotonin transport and serotonin transporter-mediated antidepressant recognition are controlled by 5-HT2B receptor signaling in serotonergic neuronal cells. FASEB. J. 20, 1843–1854. doi: 10.1096/fj.06-5724com
Levenson J., Byrne J. H., Eskin A. (1999). Levels of serotonin in the hemolymph of Aplysia are modulated by light/dark cycles and sensitization training. J. Neurosci. 19, 8094–8103. doi: 10.1523/JNEUROSCI.19-18-08094.1999
Li X. C., Giot J., Kuhl D., Hen R., Kandel E. (1995). Cloning and characterization of two related serotonergic receptors from the brain and the reproductive system of Aplysia that activate phospholipase c. J. Neurosci. 15, 7585–7591. doi: 10.1523/JNEUROSCI.15-11-07585.1995
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Li J. H., Yang C. Y., Wang Q. H., Du X. D., Deng Y. W. (2018). Growth and survival of host pearl oyster Pinctada fucata martensii (Dunker 1880) treated by different biofouling-clean methods in China. Estuarine Coast. Shelf Sci. 207, 104–108. doi: 10.1016/j.ecss.2018.04.009
Mapara S., Parries S., Quarrington C., Ahn K. C., Gallin W. J., Goldberg J. I. (2008). Identification, molecular structure and expression of two cloned serotonin receptors from the pond snail, Helisoma trivolvis. J. Exp. Biol. 211, 900–910. doi: 10.1242/jeb.013953
Martin K. C., Michael D., Rose J. C., Barad M., Casadio A., Zhu H., et al. (1997). MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron. 18, 899–912. doi: 10.1016/S0896-6273(00)80330-X
Mayorova T., Kach J., Kosevich I. (2014). Pattern of serotonin-like immunoreactive cells in scyphozoan and hydrozoan planulae and their relation to settlement. Acta Zool. 95, 272–282. doi: 10.1111/azo.12023
McCauley D. W. (1997). Serotonin plays an early role in the metamorphosis of the hydrozoan Phialidium gregarium. Dev. Biol. 190, 229–240. doi: 10.1006/dbio.1997.8698
Morse D. E., Hooker N., Duncan H., Jensen L. (1979). γ-aminobutyric acid, a neurotransmitter, induces planktonic abalone larvae to settle and begin metamorphosis. Science. 204, 407–410. doi: 10.1126/science.204.4391.407
Norville K., Sweeney S. T., Elliott C. J. H. (2010). Postmating change in physiology of male Drosophila mediated by serotonin (5-HT). J. Neurogenet. 24, 27–32. doi: 10.3109/01677060903477601
Panasophonkul S., Apisawetakan S., Cummins S. F., York P. S., Degnan B. M., Hanna P. J., et al. (2009). Molecular characterization and analysis of a truncated serotonin receptor gene expressed in neural and reproductive tissues of abalone. Histochem Cell Biol. 131, 629–642. doi: 10.1007/s00418-009-0555-7
Roth B. L. (2006). The serotonin receptors: from molecular pharmacology to human therapeutics. (Totowa, NJ, USA: THE HUMANA PRESS).
Sharker M. R., Sukhan Z. P., Kim S. C., Lee W. K., Kho K. H. (2020). Identification, characterization, and expression analysis of a serotonin receptor involved in the reproductive process of the pacific abalone, Haliotis discus hannai. mol. Biol. Rep. 47, 555–567. doi: 10.1007/s11033-019-05162-2
Tierney A. (2001). Structure and function of invertebrate 5-HT receptors: a review. Comp. Biochem. Physiol.-A Mol. Integr. Physiol. 128, 791–804. doi: 10.1016/S1095-6433(00)00320-2
Urrutia P. M., Okamoto K., Fusetani N. (2004). Acetylcholine and serotonin induce larval metamorphosis of the Japanese short-neck clam Ruditapes philippinarum. J. Shellfish. Res. 23, 93–101. doi: 10.1111/j.1365-2761.2004.00537.x
Wang Q., He M. X. (2013). Stability comparison of reference genes of Pinctada fucata by real-time qPCR. South China Fisheries Science. 9 (6), 33–40. doi: 10.3969/j.issn.2095-0780.2013.06.006
Wang Q. H., Wei W. L., Liu Y., Zheng Z., Du X. D., Jiao Y. (2019). HSP40 gene family in pearl oyster Pinctada fucata martensii: Genome-wide identification and function analysis. Fish shellfish Immunol. 93, 904–910. doi: 10.1016/j.fsi.2019.08.029
Weiger W. A. (1997). Serotonergic modulation of behaviour: a phylogenetic overview. Biol. Rev. 72, 61–95. doi: 10.1017/S0006323196004975
Yamamoto H., Tachibana A., Kawaii S., Matsumura K., Fusetani N. (1996). Serotonin involvement in larval settlement of the barnacle, Balanus amphitrite. J. Exp. Zool. 275, 339–345. doi: 10.1002/(SICI)1097-010X(19960801)275:5<339::AID-JEZ2>3.0.CO;2-Q
Yang C. Y., Zhang J. B., Zeng Y. T., Liao Y. S., Deng Y. W., Wang Q. H. (2022). Effect of chemical cues on settlement of pearl oyster Pinctada fucata martensii eyespot larvae. J. Guangdong Ocean Univ. 42, 29–34. doi: 10.3969/j.issn.1673-9159.2022.01.005
Yu X., He W., Gu J. D., He M., Yan Y. (2008). The effect of chemical cues on settlement of pearl oyster Pinctada fucata martensii (Dunker) larvae. Aquac. 277, 83–91. doi: 10.1016/j.aquaculture.2008.02.010
Zega G., Pennati R., Groppelli S., Sotgia C., De Bernardi F. (2005). Dopamine and serotonin modulate the onset of metamorphosis in the ascidian Phallusia mammillata. Dev. Biol. 282, 246–256. doi: 10.1016/j.ydbio.2005.03.012
Zhang J. B., Xiong X. W., Deng Y. W., Zheng Z., Yang C. Y., Du X. D. (2021). Integrated application of transcriptomics and metabolomics provides insights into the larval metamorphosis of pearl oyster (Pinctada fucata martensii). Aquac. 532, 736067. doi: 10.1016/j.aquaculture.2020.736067
Zhao B., Zhang S., Qian P. Y. (2003). Larval settlement of the silver-or goldlip pearl oyster Pinctada maxima (Jameson) in response to natural biofilms and chemical cues. Aquac. 220, 883–901. doi: 10.1016/S0044-8486(02)00567-7
Keywords: Pinctada fucata martensii, Pm5-HTR2, Pm5-HTR4, larval metamorphosis, molecular cloning
Citation: Zhu S, He Y, Xu Q, Zhang J, Yang C, Hao R, Li J and Deng Y (2022) Molecular cloning, characterization, and expression of two 5-HTRs from the pearl oyster Pinctada fucata martensii. Front. Mar. Sci. 9:1011420. doi: 10.3389/fmars.2022.1011420
Received: 04 August 2022; Accepted: 08 September 2022;
Published: 23 September 2022.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, (CAS), ChinaReviewed by:
Zhongming Huo, Dalian Ocean University, ChinaCopyright © 2022 Zhu, He, Xu, Zhang, Yang, Hao, Li and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuangye Yang, eWFuZ2N5QGdkb3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.