- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, China
- 2Collaborative Innovation Center for Zhejiang Marine High-efficiency and Healthy Aquaculture, Ningbo University, Ningbo, China
- 3Department of Genetic Breeding, Zhejiang Mariculture Research Institute, Wenzhou, China
Intertidal macroalgae have adapted to deal with environmental stresses, in particular desiccation. The phytohormone abscisic acid (ABA) plays an important role in the regulation of a suite of critical responses in plants, including desiccation tolerance. The red alga Neoporphyra haitanensis contains a high level of ABA, but its mechanism of action in N. haitanensis is unknown. In this study, the effect of ABA treatment on the relative water content of thalli during the dehydration-rehydration cycle was evaluated and it was identified that ABA treatment resulted in decreased thalli water loss and an accelerated rehydration process. The effects of addition of ABA upon the activation of antioxidant responses, photosynthetic parameters and gene transcription profiles of N. haitanensis were also evaluated. The results revealed that exogenous ABA regulated the activation of the antioxidant system, including by increasing the activity of antioxidant enzymes, increasing the concentrations of ascorbic acid (AsA) and glutathione (GSH), as well as upregulating the expression of genes encoding antioxidant enzymes and enzymes that synthesize AsA and GSH. ABA treatment also inhibited photosynthesis by reducing light capture and down-regulating the Calvin cycle to save energy; ABA supplementation further simultaneously activated repair mechanisms to avoid excessive DNA and protein damage. The results presented herein demonstrate that ABA plays a crucial role in the regulation of stress tolerance in seaweeds, which may be of great importance when examining the adaptation of this organism to the intertidal environment.
Introduction
When the ancestors of plants first inhabited the land, being able to tolerate dryness was an obstacle that was overcome. During the stress of desiccation, the energy not used in carbon fixation will result in the formation of reactive oxygen species (ROS); these species will impact the stability of membranes, the photosystem, and macromolecules in cells (Tafvizi et al., 2020). To tolerate desiccation, plants have evolved complex multi-genic and multi-factorial strategies and mechanisms to prevent oxidative damage and maintain the native structures of macromolecules and membranes. One of these protective mechanisms is the inactivation of the photosystem and cessation of energy transfer from phycobilisomes to PS II during the early stages of desiccation. An additional strategy is the induction of a complex array of antioxidant enzymes and redox molecules, e.g. the antioxidant activity of the glutathione-ascorbate cycle system to support light energy dissipation (Dinakar et al., 2012; Gechev et al., 2012). The marked accumulation of physically protective and osmotically active substances, particularly sucrose, trehalose and proline, also exerts important effects (Lűttge et al., 2011). In addition, protein turnover and DNA-repair are further wide-spread strategies in plants (Gaff and Oliver, 2013). Abscisic acid (ABA) plays a vital role in promoting dehydration tolerance in plants (Guajardo et al., 2016). The reaction of plants to ABA includes the modulation of antioxidant enzymes (Jiang and Zhang, 2002; Hu et al., 2005), the triggering of genetic production of stress proteins, the generation of osmotically active sugars, and growth inhibition to mediate cellular protection during desiccation (Lűttge et al., 2011).
For intertidal seaweeds, especially high-tide seaweeds, dehydration tolerance is crucial (Sahoo et al., 2002; Blouin et al., 2011). Studies demonstrated that when intertidal seaweeds dry out, photosynthesis, antioxidant systems and osmotic regulators, such as floridoside and isofloridoside will all respond to the change in environment (Weiwer et al., 2009; López-Cristoffanini et al., 2015). The response patterns are very similar to those of land plants, indicating that the adaptation strategies of photosynthetic organisms to dryness are evolutionarily conserved.
Knowledge regarding ABA signaling has been reported by the study of land plants. Plants contain ABA in varying concentrations, e.g. Arabidopsis thaliana contains 18.92-491.85 nmol kg-1 FW (fresh weight) of ABA (Lang et al., 1994), while Oryza sativa contains about 300 nmol·kg-1 FW of ABA (Xin et al., 2020). In algae, more than 100 species were identified to contain ABA, with concentrations ranging from 0.9 to 34 nmol kg-1 FW (Mehrotra et al., 2014; Sulochana and Arumugam, 2016). Amongst algae species, Halopithys incurva contains 1.85 nmol·kg-1 of ABA, whereas Gracilaria bursa-pastoris contains 0.92 nmol·kg-1 of ABA (Yaln et al., 2019). Moreover, several homologous ABA biosynthesis genes in higher plants have been identified in algae; Ulva mutabilis possesses zeaxanthin epoxidase, 9-cis-epoxy carotenoid dioxygenase and short-chain alcohol dehydrogenase genes. Of these, it has been speculated that algae may possess an ABA biosynthesis pathway similar to that of higher plants (De Clerck et al., 2018). This led us to hypothesize that ABA provides a similar function in seaweed as it does in land plants. Regarding this, it was identified that ABA altered the photosynthetic efficiency of Chlamydomonas reinhardtii (Al-Hijab et al., 2019). Furthermore, ABA was reported to exert a protective effect against oxidative damage by enhancing the specific activity and genetic expression of antioxidant enzymes such as catalase (CAT) and ascorbateperoxidase (APX) in C. reinhardtii (P.A. Dangeard) (Yoshida et al., 2003; Yoshida et al., 2004). In two Chlorella strains (Chlorella vulgaris and Chlorella pyrenoidesa), the administration of ABA increased lipid accumulation (Wu et al., 2018); in Cyanidioschyzon merolae, the exogenous doses of ABA inhibited the cell cycle G1/S transition (Ando et al., 2016). These results indicate the importance of ABA in regards to stress resistance in seaweeds.
Porphyra sensu lato within the Bangiales order of Rhodophyta are typical high tide zone seaweeds. They can rehydrate normally after losing more than 95% of their water content. Porphyra contain relatively high levels of ABA, Neoporphyra haitanensis contains 10-30 μg·g-1dry weight of ABA, which is 4-7 times higher than the levels identified in seaweeds grown in low tide zones (Zhao et al., 2016). Therefore, to investigate whether ABA is involved in the mechanisms of desiccation tolerance in N. haitanensis, the responses of N. haitanensis to ABA were analyzed in the study herein. As such, we investigated the key physiological and metabolic changes in N. haitanensis after treatment with ABA, the regulation of photosynthetic capacity, the antioxidant system and osmotic pressure regulators were focused on in particular. Meanwhile, a transcriptomic analysis was performed to understand the mechanism by which ABA facilitates the algae when it is responding to stress.
Material and methods
Experimental material
N. haitanensis thalli samples were collected from a coastal farm at Xiangshan Harbor (121°57’5’’E, 29°46’30’’N) within the Zhejiang province of China on October 14, 2019. The formal identification of the plant material was carried out by Prof Qijun Luo. No voucher specimen has been deposited in a genebank and no special permission is needed to study this species. Sampling was permitted by the local government (Xiangshan County Government) and the local department of fisheries (Ningbo Ocean & Fishery Bureau). Healthy thalli measuring 15-20 cm in length were selected before being washed in sterilized seawater (collected from Xiangshan Harbor) prior to the following treatment.
Experimental design
ABA (Shanghai yuanye Bio-Technology Co., Ltd) was dissolved in ethanol. Thalli were grown in seawater at a density of 0.1 mg·mL-1, before being exposed to ABA at concentrations of 10, 100 and 300 μM for 1 and 6 hours. Sterilized seawater without ABA was used as a control. Next, thalli were washed with sterile seawater to remove ABA. To conduct the dehydration and rehydration experiment, the fresh weight of ABA treated thalli was measured before they were hung on strings to simulate dehydration stress; they were stored at an air temperature of 22 ± 2°C, with a relative humidity of 70 ± 5%, a light irradiation level of 40 µmol·m-2·s-1, a photoperiod of 12 h: 12 h (L: D), and an air flow speed of 1.5 ms-1. The dried thalli samples were then immersed in sterile seawater for rehydration. Thalli samples were taken at different time points for weighing (n = 10).
The relative moisture content (RWC) of the thalli was calculated using the formula: RWC (%) = (Fw − Dw)/(FTw − Dw) ×100, where Fw is the weight of thalli after dehydration for a specific amount of time, Dw represents the dry weight after incubation at 80°C for 6 hours, and FTw is the fresh thalli weight (Xu et al., 2018).
For the ABA stimulation study, thalli grown in seawater at a density of 0.1 mg·mL-1 were exposed to ABA at concentrations of 10 or 100 μM for 1 and 6 hours. The thalli were frozen in liquid nitrogen and stored at −80°C prior to transcriptomic analyses. Some samples were taken from each treatment for physiological analysis.
Measurement of malondialdehyde and proline concentration
The malondialdehyde (MDA) and proline concentration of the thalli dehydrated for 0, 15 and 30 minutes were detected. The samples were ground to a powder with liquid nitrogen. 0.1 g of these powdered samples was added to cell lysis buffer solution (Beyotime Biotechnology, Shanghai) for homogenization. The extracted suspension was then centrifuged at 2,500 ×g for 10 minutes and the supernatant was collected to determine the MDA and proline concentration using the corresponding kits according to the manufacturer’s instructions (Nanjing Jiancheng Biotech Inc. MDA, No. A003-1; proline, No. A107-1). In brief, MDA was determined with the classical thiobarbituric acid method (Schmedes and Hølmer, 1989). Proline concentration was determined with a ninhydrin reagent (Lee et al., 2018) wherein one milliliter of extract was combined with 2 mL acid ninhydrin reagent (1.25% ninhydrin in 80% glacial acetic acid) in a water bath at 100°C for 30 min. The reaction was stopped by placing the samples in an ice bath for 10 min. The absorbance of the mixture was finally read at 520 nm.
Chlorophyll fluorescence parameter determination
The chlorophyll fluorescence parameters of photosystem II (PSII) (Fv/Fm) for the thalli samples were recorded using a MAXI IMAGING-PAM system (Walz, Effeltrich, Germany). After 10 minutes of dark adaptation, the fluorescence of chlorophyll a was measured, a minimum fluorescence (F0) and maximal fluorescence (Fm) were determined and the fluorescence of chlorophyll a was calculated as Fv/Fm= (Fm-F0)/Fm (Kitajima and Butler, 2019).
Phycobiliprotein concentration measurement
The phycobiliprotein concentration was measured as previously described, with modifications. In brief, frozen samples (0.1 g in powder form) were homogenized with 8 mL of cold 0.1 M phosphate-buffered saline (PBS) at pH7.0. Ultrasonic extraction was performed for 10 minutes, and then cracked at -20°C for 2 hours. The extract was centrifuged at 10,000 ×g for 5 minutes to separate the supernatant and allow for measurement with absorbance wavelengths of 498 nm, 614 nm and 651 nm. The phycoerythrin (PE), phycocyanin (PC) and allophycocyanin (APC) concentrations were calculated according to the following equations:
Assessment of antioxidant enzyme activities
Treated fresh samples were homogenized in 90% cell lysis buffer (Beyotime Biotechnology, Shanghai) and then centrifuged at 2,500 ×g for 10 minutes (Jia et al., 2011). Supernatants containing the enzymes were collected, and the total protein concentration was determined using the Bradford method (Bradford, 1976). CAT, glutathione peroxidase (GPX), and glutathione reductase (GR) activities, in addition to total glutathione (GSH) and ascorbic acid (AsA) concentrations were measured using Assay kits according to the manufacturer’s instructions (Nanjing Jiancheng Biotech Inc. CAT, No. A007-1; GPX, No. A005-1, GR, No. A062-1; GSH, No. A061-1; and AsA, No. A009-1, respectively).
Measurement of floridoside and isofloridoside concentration
The procedures for extraction of floridoside and isofloridoside from each sample were carried out as previously described (Zhao et al., 2019). Each sample was extracted in ethanol: H2O (7: 3, v/v) before being centrifuged. The supernatants were used to analyze the floridoside and isofloridoside concentration on an Acquity UPLC using a BEH Amide UPLC column (2.1 mm × 100 mm, 1.7 μm, Waters, USA). The constant solvent system was 90% acetonitrile (A)–10% water (10 mM CH3COONH4). The flow rate was 0.4 mL·min-1 and the injection volume was 5 μL. Mass spectrometry was performed on a Xevo TQ-S microMS (Waters, USA) equipped with an electrospray ionization source in the negative mode using multiple reactions monitoring, for quantification. The ionization conditions were as follows: the vaporizer temperature was 500°C, the spray voltage was 0.5 kV, the flow rate of the N2 was 16 L·min-1 and the gas pressure of the collision gas (Ar) was 6×10−6 mbar. The calibration curves for floridoside and isofloridoside were constructed with standard compounds extracted directly from N. haitanensis.
RNA isolation, library construction, and sequencing
Thalli from different treatment groups were collected and snap-frozen in liquid nitrogen to allow for the subsequent extraction of RNA, using a Plant RNA extraction Kit (R6827, OMEGA). Each extracted RNA sample (with a concentration greater than 600 ng·μL-1) was submitted for transcriptomic sequencing (n = 3). RNA-seq libraries were prepared using the Illumina mRNA-seq Library Preparation kit (Illumina) and the samples were sequenced using the Illumina HiSeq 2000 platform.
RNA-seq data analysis
After filtering low-quality reads, clean reads were mapped to the N. haitanensis genome using HISAT2 (Kim et al., 2015). The StringTie program (Pertea et al., 2015) was used to predict novel transcripts, which were combined with genome annotations to obtain the final transcriptome set. Reads were mapped to the transcriptome dataset using Bowtie2 software (Langmead, 2010) and quantified using RSEM (Li and Dewey, 2011). Correlations between samples and replicates were examined using principal component analysis (PCA), performed by the SIMCA-P software package (www.umetrics.com). FPKM was used to calculate all gene transcripts. Differentially expressed genes (DEGs), defined as those with |log2(fold change)| >1 and an FDR significance score (padj) < 0.05, were identified using DEseq2 (v.1.22.2) (Love et al., 2014) and subjected to the Kyoto Encyclopedia of Genes and Genomes(KEGG)-based annotation enrichment using hypergeometric tests (Kanehisa, 2000; Kanehisa, 2019; Kanehisa et al., 2021). FDR ≤ 0.05 was used as the threshold for KEGG pathways with significant enrichment. For transcriptome enrichment analysis, we used the Phyper program within R to calculate P values. Heatmaps were constructed using Euclidian distances and complete linkage clustering with the p heatmap package for R. To compare differences between treatments, the processed transcriptomic datasets were quantile normalized, transformed and clustered using hierarchical clustering. The terms with an abundance of P < 0.05 were visualized using a heatmap constructed based on Euclidian distances using TBTools (https://github.com/CJ-Chen/TBtools/releases) (Chen et al., 2020) and using the heatmap package for R. Statistical significance (P values) for each KEGG pathway was obtained using the hypergeometric test algorithm. Univariate analysis of variance (ANOVA) was performed to determine the significance of differences in relative concentration between the different treatment groups.
Quantitative real-time PCR validation
Total RNA from different groups was extracted using a Plant RNA extraction Kit (R6827, Omega Bio-tek Inc., Norcross, Georgia, USA). mRNA in the extracted total RNA was reversely transcribed into first-strand cDNA according to the protocol supplied with PrimeScript™ RT Master Mix (TaKaRa Biotechnology Co., Ltd., Dalian, China), and qRT-PCR amplification reactions were performed using a LightCycler®96 Real-Time PCR System (Roche, Basel, Switzerland). Primers used are listed in Supplementary Table 1. The qRT-PCR amplification program is: 94°C for 30 s; 94°C for 5 s, 60°C for 15 s, 72°C for 10 s, 40 cycles. Relative expression was calculated using the 2-ΔΔCt method with β-actin as the reference gene.
Statistical analysis
For each experiment, at least three biological replicates were prepared for each group. The results were presented as the mean ± standard deviation (SD). Statistical significance of RWC changes between different groups were analyzed using a post hoc Benjamini-Hochberg test after a one-way ANOVA, further analysis was undertaken using one-way ANOVA tests. Data acquired was analyzed using SPSS version 10.0 (SPSS, Inc., Chicago, IL, USA). Differences between the groups were considered statistically significant P < 0.05.
Results
Effects of ABA on adaptation to drying stress of N. haitanensis
N. haitanensis was exposed to the air and the rate of water loss in the control group was rapid. Following 30 minutes of drying, the RWC of the thalli dropped to 4.58%. ABA treatment slowed down the rate of water loss, however, a concentration trend was not identified. When the concentration of ABA was increased from 10 μM to 100 μM, the protective effect was gradually enhanced. The RWC of thalli subjected to 100 μM ABA for 1 or 6 hours was maintained at 8.19 and 9.62% after 30 minutes of dehydration (P < 0.01, Figures 1A, B; Supplementary Table 2). After that, the dehydrated thalli were immersed in sterile seawater for rehydration. The control group RWC rose to 64.97% after 1 minute and reached 85.38% after 10 minutes. ABA treatment accelerated the rate of rehydration, with the 100 μM treatment group exhibiting the best acceleration effect. After 10 minutes of rehydration, the RWC of thalli exposed to 100 μM ABA for 1 and 6 hours rose to 93.87% and 94.17%, respectively, 8.49% and 8.79% higher, respectively, than the RWC of control group (P < 0.01). However, when the concentration of ABA treatment was 300 μM, its effect in facilitating thalli attenuate water loss and speed recovery was reduced. Therefore, we focused on the concentration of 100 μM to explore the protective mechanism of ABA in depth.
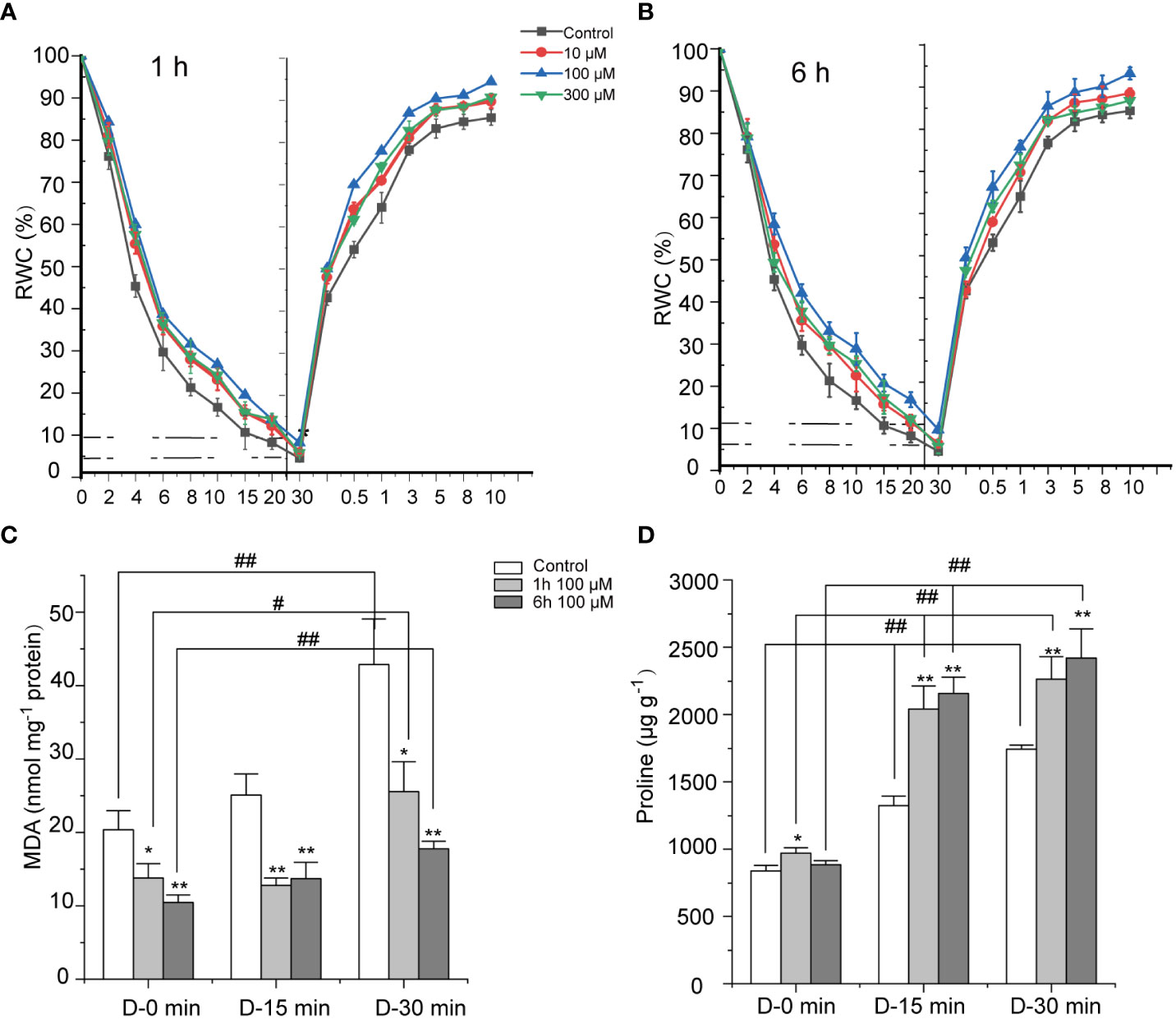
Figure 1 Effects of abscisic acid (ABA) on the dehydration tolerance of Neoporphyra haitanensis. (A, B) Effect of ABA on the relative water content (RWC) of N. haitanensis during the dehydration–rehydration cycle process. The thalli were pre-treated with different concentrations of ABA for 1 or 6 hours, before being exposed to air for 30 minutes to allow for drying, following rehydration in sterile seawater for 10 minutes. The RWC of thalli samples were measured at each time point (n = 5). The statistical significance is shown in Supplementary Table 2. (C, D) Effects of ABA on malondialdehyde (MDA) and proline contents in N. haitanensis during dehydration. The thalli were pre-treated with 100 μM of ABA for 1 or 6 hours, then exposed to air for 15 or 30 minutes to allow for drying. The MDA and proline contents were determined. *P < 0.05, **P < 0.01, compared to the Control group, #P < 0.05, ##P < 0.01, compared to the same treatment group with different drying periods, n = 3.
The effects of 100 μM ABA on the concentration of MDA, a marker for oxidative stress were evaluated (Figure 1C). During dehydration, the MDA concentration in the control group increased significantly as the length of dehydration time increased, whilst the levels of MDA increased 2.1 times after drying for 30 minutes compared to the hydrated sample (P < 0.01, Figure 1B). ABA treatment resulted in a significant decrease in MDA concentration, not only in the dry state, but also in fresh thalli (P < 0.05). After 15 minutes of dehydration, the MDA levels of thalli treated with 100 μM of ABA for 6 hours was 1.83 times lower than that of the control group (P < 0.01). The longer the drying time, the greater the MDA level decreased following ABA treatment, with MDA levels 2.41 times less than those of the control group following 30 minutes of dehydration (P < 0.01).
Proline is an important osmotic regulator in plants (Alvarez et al., 2021). Figure 1D shows that proline increased significantly when N. haitanensis was dehydrated (P < 0.01). When N. haitanensis was dehydrated for 30 minutes, the concentration of proline increased by 2.08 times (P < 0.01) compared to the levels of fresh thalli. ABA treatment further increased the proline concentration in dehydrated thalli, whilst it had little effect on fresh samples. A 100 μM ABA pretreatment for 1 hour improved the level of proline to 2.26 mg·g-1, which was 2.34 times higher than the levels of fresh thalli (P < 0.01) and 1.30 times higher than that of the dried control group (P < 0.01). It was identified the longer the pre-treatment time, the more obvious the proline concentration increase was.
Differentially expressed genes in response to ABA
To explore the transcriptional changes of N. haitanensis in response to ABA treatment, samples treated with 10 and 100 μM of ABA for 1 and 6 hours were selected for sequencing. Using the Illumina HiSeq 2000 platform, a total of 9.7×108 clean reads with pair-end 150 bp were obtained. 72.67% of the clean reads could be mapped to the N. haitanensis reference genome (Supplementary Table 3). The average sequence depth of coverage was 183.65× (Supplementary Table 4). Based on the clean reads, we obtained expression of 10,934 genes, including 9,010 genes from the N. haitanensis reference genome and 1,924 novel genes. We randomly selected nine unigenes to validate the RNA-seq analysis using qRT-PCR. The expression of these genes in the transcriptome data was generally consistent with the qRT-PCR results (Supplementary Figure 1).
PCA was performed to investigate the global responses of N. haitanensis to ABA. Obvious separation was observed among the control group and the two different time-period ABA treatment groups. PCA analyses revealed that treatment period was the main factor that influenced transcription; this was apparent because the two concentration groups were clustered very close to each other when treated for 1 hour, and a similar phenomenon was observed for the 6 hour groups. However, the two groups treated at the same concentration for different time-periods were widely separated (Figure 2A). It is known from the above that 100 μM ABA treatment promoted the best protection against thalli dehydration stress. Therefore, we analyzed the transcriptome data of the thalli treated with 100 μM ABA for 1 and 6 hours in depth. DESeq2 was used to compare the DEGs between the control group and ABA treated groups. DEGs were subjected to KEGG-based annotation enrichment (Supplementary Table 5), and the pathways were sorted by P value. The pathways with a P value < 0.05 were listed in a heatmap (Figure 2B). There were 2,034 DEGs differentially expressed when comparing the control group and the ABA 1 hour treatment group, some of these pathways involved in “Proteasome”, “Endocytosis” and “Fatty acid degradation” were significantly enriched (P < 0.01). Moreover, 2,355 DEGs were enriched when comparing the control group and the 6 hour treatment group, with “DNA replication”, “Photosynthesis-antenna proteins”, “Ribosome” pathways significantly altered (P < 0.01). There were 2,804 DEGs enriched when comparing the 1 and 6 hour ABA treatment groups, including “DNA replication”, “Photosynthesis-antenna proteins”, “Pentose phosphate pathway” and “Carbo fixation in photosynthetic organisms” (P < 0.05).
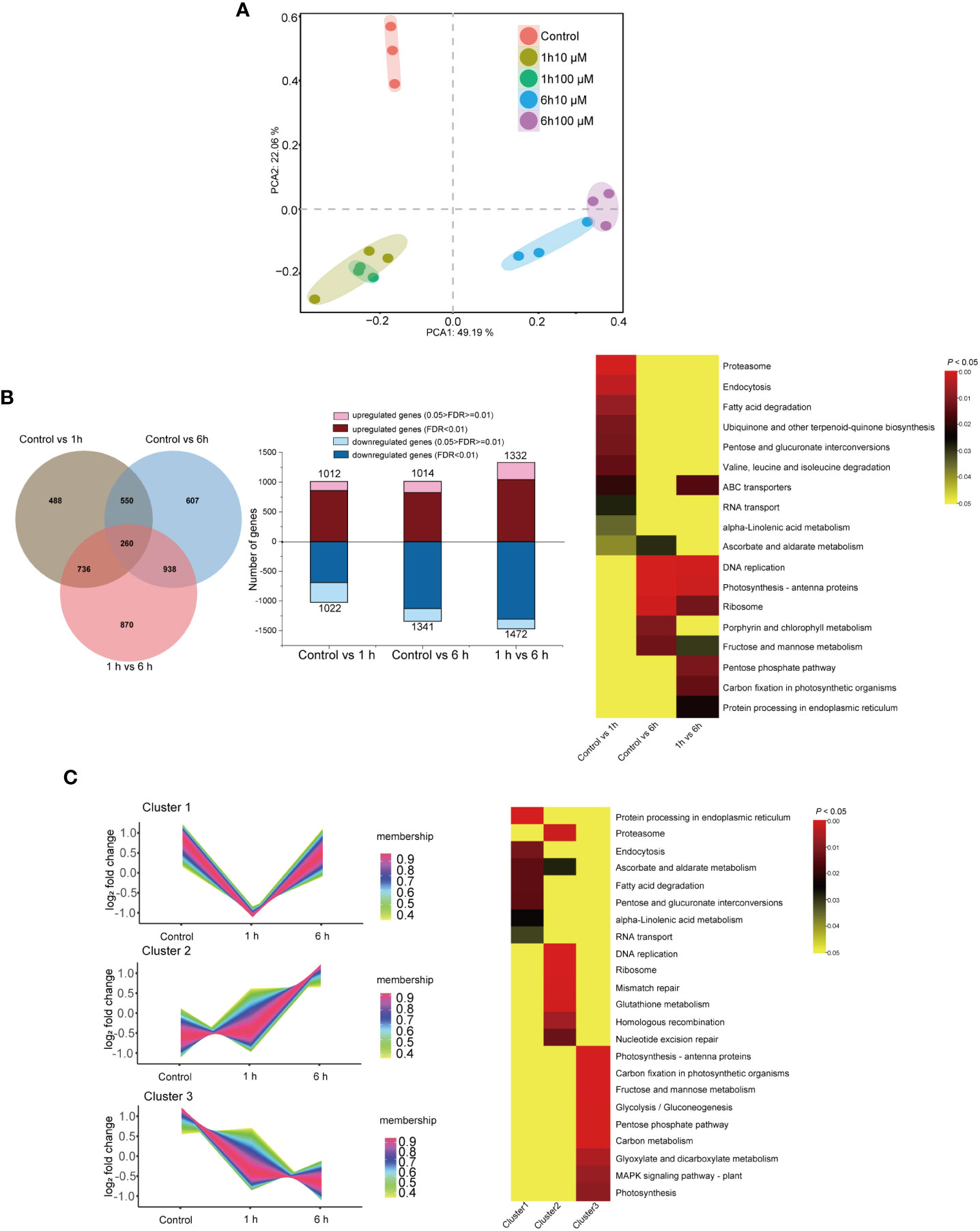
Figure 2 Transcriptional responses of Neoporphyra haitanensis to abscisic acid (ABA) treatment. (A) Principal components analysis (PCA) of the transcriptomic profile of individual traits of N. haitanensis in response to ABA treatment. (B) A Venn diagram of differentially expressed genes (DEGs), the number of DEGs across treatments and a heatmap of significantly enriched pathways for DEGs. (C) Clusters of scaled transcript profiles of N. haitanensis following ABA treatment. Cluster memberships were determined by the similarities of objects measured with respect to subspaces. The corresponding heatmap describes significantly enriched KEGG pathways for each cluster.
Following DEG analyses, pathways with a P value < 0.05 were selected for cluster analysis. As such, 7,193 DEGs were clustered into three major groups. The gene expression levels in Cluster 1 were down-regulated after 1 hour of ABA treatment, however expression of genes in this cluster recovered in the 6 hour group. KEGG enrichment revealed that pathways involved in “Protein processing in endoplasmic reticulum”, “Ascorbate and aldarate metabolism”, “Fatty acid degradation” were differential (P < 0.05, Figure 2C). With the extension of treatment time, the expression of genes in cluster 2 were upregulated. Most of these DEGs were involved in “Nucleotide excision repair”, “Homologous recombination”, “Glutathione metabolism”, “Proteasome”, “DNA replication”, and “Mismatch repair” pathways (P < 0.01). The expression levels of DEGs in cluster 3 were significantly down-regulated when the thalli were treated with ABA. Most of these cluster 3 DEGs were associated with “Photosynthesis”, “Carbon fixation in photosynthetic organisms”, “Photosynthesis-antenna proteins” and carbohydrate metabolic pathways (P < 0.01).
Inhibition of photosynthesis and carbon fixation in response to ABA
ABA had an influence on the maximum photosynthetic efficiency of N. haitanensis. Fv/Fm dropped significantly, by 8.74%, when treated with 100 μM ABA for 1 hour (P < 0.01). In the following 6 hours, the Fv/Fm continued to decline, but the value remained above 0.55, indicating that energy conversion in the PSII reaction center could still continue (Figure 3A). In addition, it was identified that the concentrations of three phycobiliproteins, PE, PC and APC were all decreased. The level of PE was decreased by 17.49% after 1 hour treatment and 34.91% after 6 hours (P < 0.01, Figure 3B); after six hours of treatment, PC levels also decreased by 24.03% (P < 0.01).
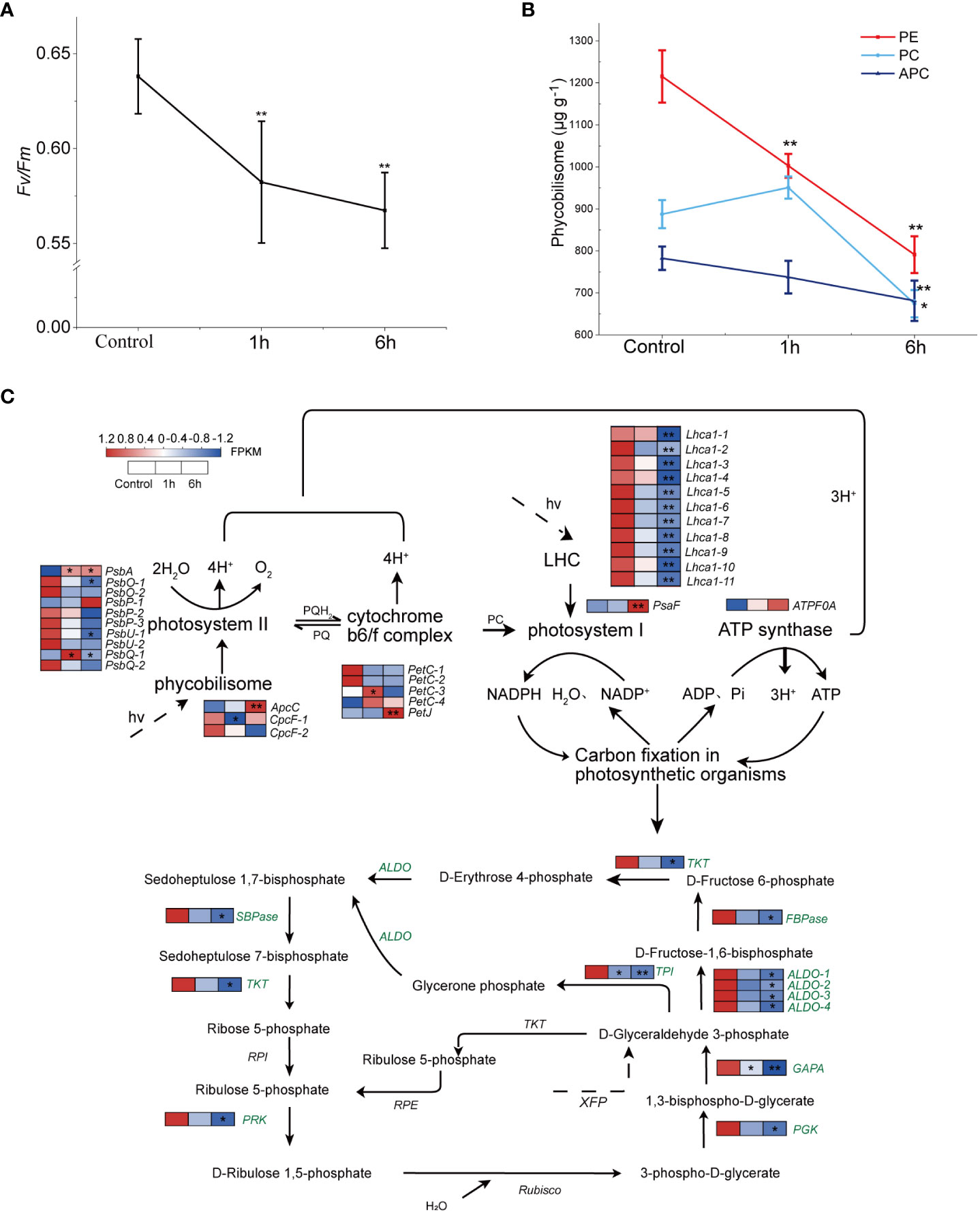
Figure 3 Effects of exogenous abscisic acid (ABA) on photosynthesis in Neoporphyra haitanensis. The thalli were treated with 100 μM of ABA for 1 or 6 hours. (A) Effects of ABA treatment on the photochemical efficiency (Fv/Fm) of thalli. (B) Changes to the content of three phycobiliprotein in response to ABA. (C) Transcriptome expression profiles of the N. haitanensis photosynthetic system and Calvin cycle following ABA treatment. Data for the Control group, the 100 μM ABA treatment groups (both 1 and 6 hours) (from left to right) is shown near the pathway as heatmaps. The heatmaps show gene expression levels (normalized FPKM) (n = 3). For every panel, *P < 0.05 and **P < 0.01, significance was determined when comparing treatment groups to the control group.
In the cluster analysis above, the photosynthesis-antenna proteins, as well as carbon fixation and a range of glycometabolism pathways were all down-regulated. Here, a total of 31 genes were annotated in the pathway of photosynthesis (Figure 3C; Supplementary Table 6). Of these, two cpcF genes were down-regulated, which was consistent with the PC concentration change. It was noticed that several genes of the light harvesting pigment complex (lhca1) and most of the photosystem II core complex protein genes (Psb) were down-regulated by ABA, especially when treated for 6 hours (P < 0.05). However, the ATP synthase gene ATPF0A was up-regulated after ABA treatment.
The down-regulation of the photosynthetic system seems to drive the change of the carbon fixation pathway. After ABA treatment, the genes annotated in the carbon fixation pathway in N. haitanensis were down-regulated as a whole (Figure 3C). There were significant differences in the expression of a sequence genes including fructose-bisphosphate aldolase (ALDO), fructose-1,6-bisphosphatase (FBPase) and sedoheptanose-1,7-bisphosphatase (SBPase) (P < 0.05) after 1 hour of treatment. After 6 hours, FBPase was down regulated 14.69 times (P < 0.05), whilst SBPase was down-regulated 35.13 times (P < 0.05).
Activation of the antioxidant system in response to ABA
32 genes relating to the ‘glutathione metabolism’ pathway were annotated and differentially expressed (Supplementary Table 6). The expression levels of APX, GR and GPX genes were almost all up-regulated following 6 hours of treatment. For example, two APX genes were significantly up-regulated (P < 0.05), APX2 increased 2.46 times; five GR and three GPX genes were also significantly up-regulated (P < 0.05), including GSH-PX1 (3.15 times) (Figure 4A).
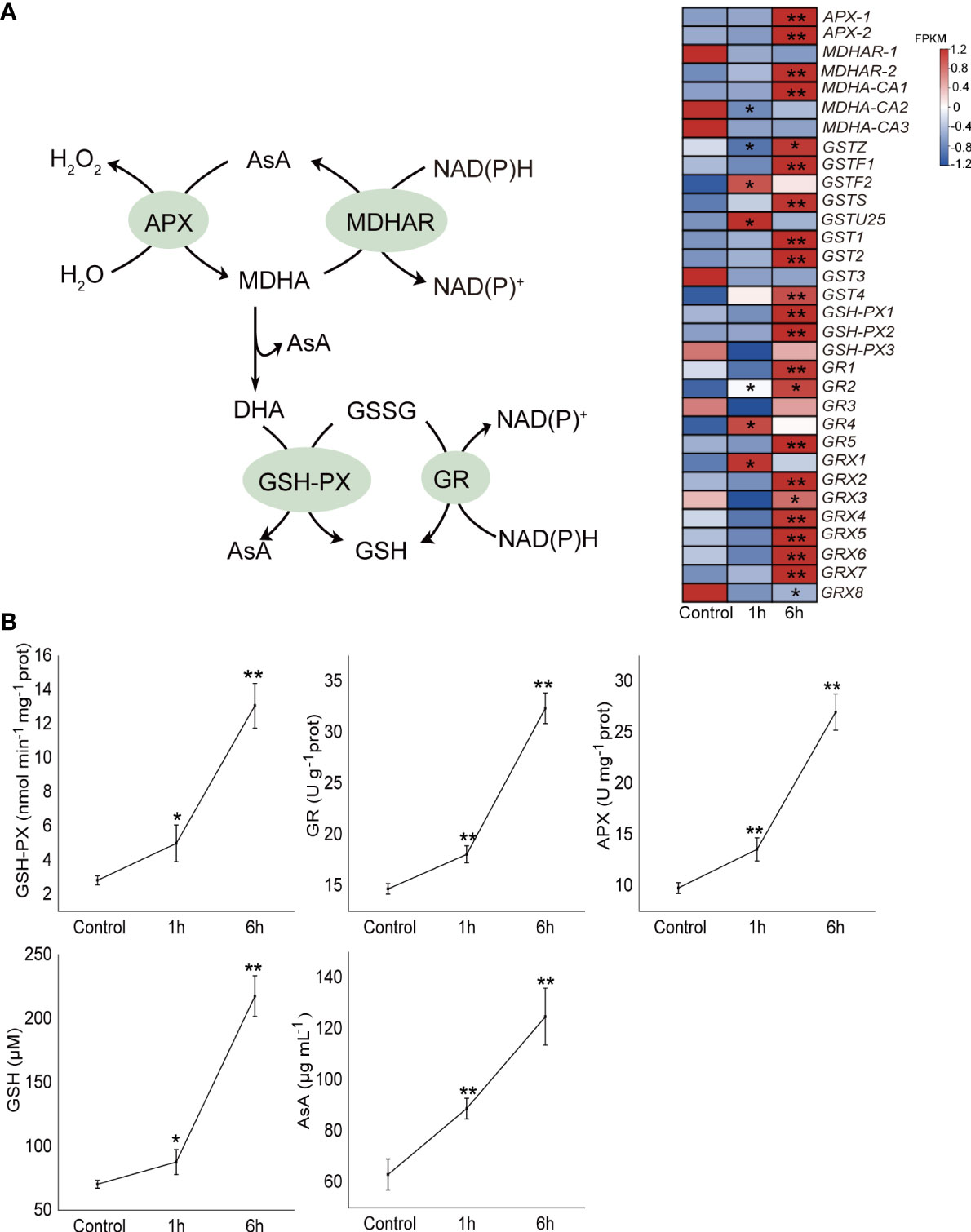
Figure 4 Effect of exogenous abscisic acid on the ascorbate (AsA)-glutathione (GSH) antioxidant system of Neoporphyra haitanensis. (A) Heatmaps of the transcriptional patterns of AsA-GSH antioxidant pathway genes in response to abscisic acid (ABA) treatment. The data shows the transcriptomic profiles from the control, 100 μM ABA 1 and 6 hour groups. FPKM values of genes in different groups were used to construct the heatmaps after a z-score normalization (n = 3). (B) Effect of ABA on the antioxidant enzyme activities and antioxidant concentrations in N. haitanensis. Thalli were treated with 100 μM ABA for 1 and 6 hours, respectively. The samples were collected for enzyme activity analysis. APX, ascorbate peroxidase; GR, glutathione reductase; GPX, glutathione peroxidase. *P < 0.05, **P < 0.01 when compared to control group.
The activities of three important antioxidant enzymes GPX, GR and APX, and the concentrations of two intermediates, GSH and AsA, were further examined (Figure 4B). The activities of the three enzymes in N. haitanensis were significantly up-regulated in response to ABA treatment. Following 6 hours of ABA treatment, the activities of GPX, GR and APX increased by 4.65, 2.20 and 2.76 times respectively in comparison with the control group (P < 0.01). In addition, the concentrations of GSH and AsA were also significantly increased, levels rose by 3.09 and 1.98 times compared to the levels of the control group (P < 0.01).
Abscisic acid enhances the accumulation of isofloridoside
Both responded to ABA treatment, the expression of four putative floridoside-6-phosphate synthases genes (PhTPS1-4) in N. haitanensis, responsible for the synthesis of floridosides and isofloridoside of red algae, increased. PhTPS-1 was up-regulated after 1 hour of treatment, while PhTPS2-4 levels were increased mainly after 6 hours, 1.97, 1.91 and 1.57 times higher than the control, respectively (P < 0.05, Figure 5A; Supplementary Table 6). In addition, the concentrations of floridoside and isofloridoside in N. haitanensis treated with ABA were analyzed (Figure 5B). It was identified that floridoside was not affected by ABA stimulation (P > 0.05); however, isofloridoside levels increased significantly after 1 hour of treatment, up to 2.15 times higher than the control group and this increase was maintained after 6 hours of treatment (P < 0.01), indicating that isofloridoside is the main substance regulated by ABA in N. haitanensis.
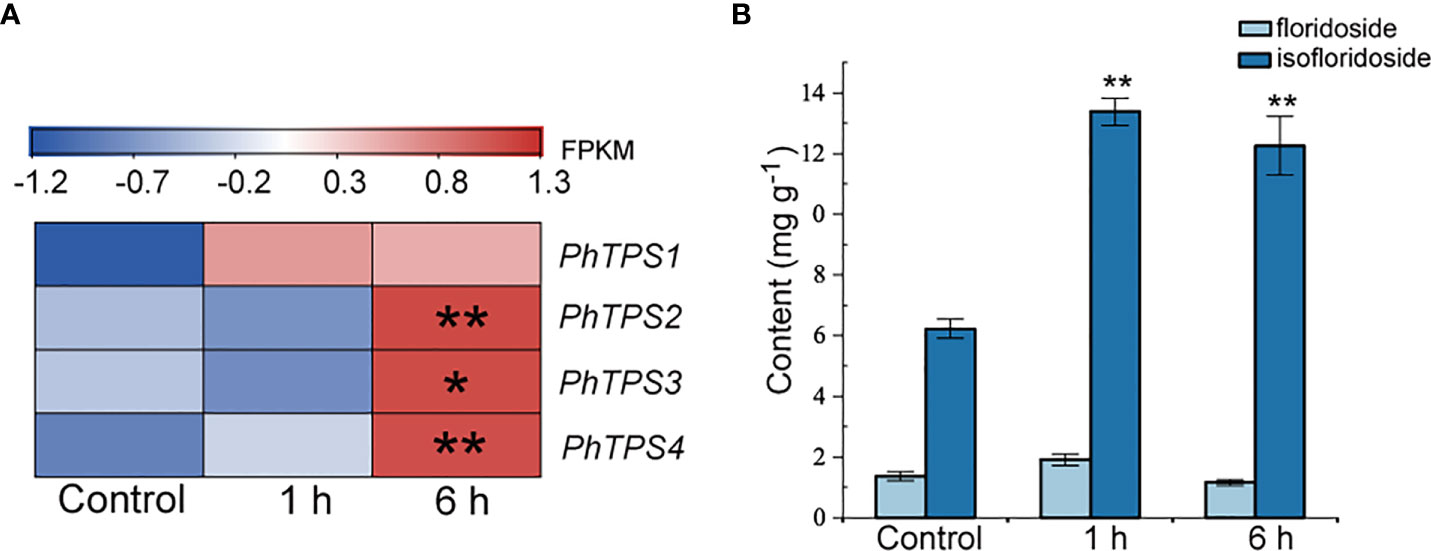
Figure 5 Effect of exogenous abscisic acid (ABA) on the expression of PhTPS1-4 (A), and on the content of floridoside and isofloridoside (B) in Neoporphyra haitanensis. The thalli were treated with 100 μM ABA for 1 or 6 hours. FPKM values of expressed genes in different groups were used to construct the heatmaps after a z-score normalization (n = 3). *P <0.05, **P <0.01 when compared to control group.
Increased DNA repair and protein degradation in response to ABA
The regulation of DNA repair is important for cell survival following exposure to stress. Several major mechanisms contribute to the removal of damaged DNA, including the nucleotide excision repair, mismatch repair, homologous recombination and proteasomes. The 26S proteasome, which consists of a 19S regulatory cap and a 20S catalytic core, degrades polyubiquitinated proteins in eukaryotic cells (Tomko and Hochstrasser, 2011) It was noted that a total of 24 subunit genes of this proteasome were differential expressed. ABA caused most of these proteasome genes to be up-regulated (Figure 6; Supplementary Table 6), Some revealed up-regulation following 1 hour of treatment which was sustained in the 6 hour treatment group; these included RPT1-5; RPN6, 8 and 12. Other genes were up-regulated after 6 hours of treatment, such as RPT 6, RPN3, 6, 9 and 10. In addition, genes involved in DNA mismatch repair, homologous recombination and nucleotide excision repair were also up-regulated during ABA treatment, especially after 6 hours (Figure 6). These results indicated that ABA initiates processes that accelerate the removal of misfolded proteins and the repair of DNA in N. haitanensis.
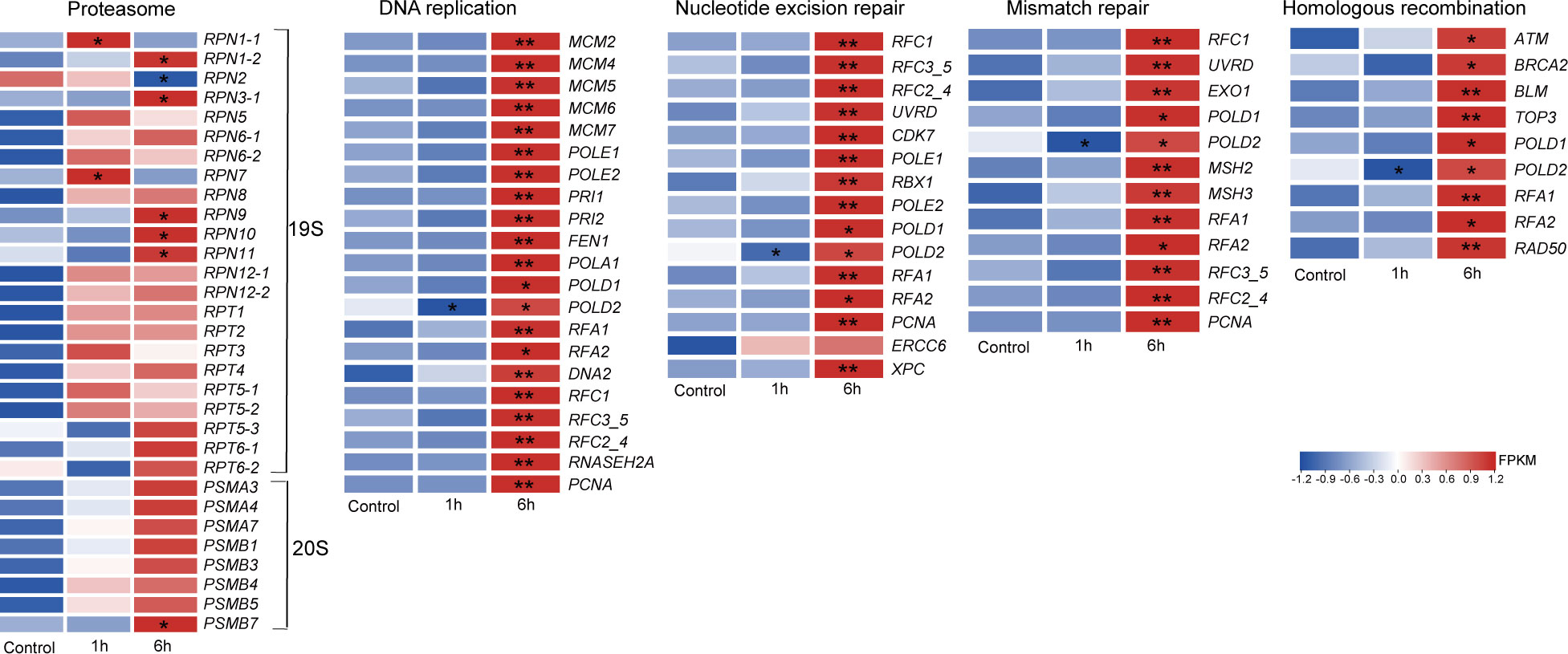
Figure 6 Differential expression patterns of repair systems in Neoporphyra haitanensis. The thalli were treated with 100 μM abscisic acid for 1 or 6 hours. The heatmap reveals the normalized FPKM values of transcripts (n = 3). *P <0.05, **P <0.01 when compared to control group.
Discussion
Plants that grow on land often suffer in response to dryness. ABA plays an important role in the adaptation of these plants to the stress of drought. ABA can regulate drought adaptation in plants in a variety of ways, such as increasing the levels of osmotic pressure regulators, enhancing antioxidant enzyme activity and regulating drought resistance gene expression (Papacek et al., 2017). Due to daily tide cycles, algal species inhabiting the intertidal zone are periodically exposed to air. This condition results in a loss of intracellular water content, commonly known as desiccation. Recently, a combination of multiple omics to elucidate the desiccation tolerance mechanism of N. haitanensis was used and this experiment identified that the photosynthetic system and antioxidant system were both regulated to allow the algae to adapt to desiccation (Chen et al., 2022). The presence of ABA has been reported in approximately 100 species of algae. The concentration of ABA is significantly higher in some algae, such as Pyropia orbicularis (Guajardo et al., 2016). High ABA levels were identified in N. haitanensis and it was speculated as to whether ABA played a role in N. haitanensis’ desiccation tolerance.
In this study, exposure to dry conditions led to a rapid decrease in the RWC of thalli, while ABA pretreatment reduced the rate of RWC decreasing during the drying process. As well as this, pretreatment allowed the thalli to recover more water content during rehydration. Similar to work conducted in angiosperms, suggests that the application of ABA enhances survival in response to extreme periods of water limitation (Mcadam and Sussmilch, 2020). Meanwhile, the protective effect of ABA did not exhibit a concentration trend, with 100 μM promoting the most potent effect, whereas at 300 μM, the effect diminished; these data indicate that excessive concentrations of ABA may have a negative effect on N. haitanensis. Furthermore, we did note that although ABA slowed the rate of water loss during drying, the RWC of thalli still decreased to less than 10%, indicating that when exposed to dry conditions the loss of water from thalli is inevitable to a certain extent. Then, we addressed whether or not drying plays a role in the survival of N. haitanensis. It has previously been demonstrated that a short period of drying is beneficial to the survival of N. haitanensis, as this process can eliminate bacteria on the surface of the algae, keeping it healthy (Wan, 2020). Therefore, ABA may regulate thalli water loss and rehydration by promoting cells to reduce the rate of evaporation, helping cells to retain more water, thus reducing the damage caused by rapid water loss and achieving rapid recovery. However, the propensity of thalli to lose water does not change.
The changes to the transcriptional profile of thalli after ABA treatment provided interesting information. It was observed that the 10 and 100 μM treatment groups were clustered closely together, far from the control group, indicating that ABA induced significant transcriptional changes in N. haitanensis, while the two distinct ABA concentration groups exhibited similar gene transcriptional changes. As such, 100 μM ABA triggered a series of cellular responses in thalli, including the rapid suppression of the photosynthetic system, the activation of the antioxidant system, and the induction of the macromolecular repair system, indicating the initiation of a series of protective mechanisms following ABA treatment.
Photosynthesis and carbon fixation is pivotal for all the plants that are affected by abiotic stresses (Sharma et al., 2020). Studies have reported that exogenous ABA can weaken the ability of higher plants to photosynthesize (Yudina et al., 2020). In this study, Fv/Fm decreased under the influence of ABA, which is consistent with the results of other studies. It was identified that this decrease was related to the reduction of phycobiliproteins and the inhibition of photosynthesis transcription in N. haitanensis by ABA. By decreasing the phycobiliprotein levels and down-regulating the photosynthesis-antenna protein genes (Lhca1), ABA can reduce the ability of N. haitanensis to capture light; however, the photosynthetic system was not completely shut down. It was observed that although the Fv/Fm decreased somewhat, it remained above 0.55, suggesting that the main purpose of ABA may not be to fully switch off normal photochemical efficiency, but merely to reduce photo-oxidation during drying by reducing light capture. The up-regulated levels of ATPF0A also indicated that ABA did not inhibit energy production at a transcriptional level but might utilize the accumulated H+ to promote conversion and energy production to allow the algae to adapt to stress. The overall downregulation of the Calvin cycle may be a protective strategy, which will decrease the assimilation of CO2, consequently reducing the biomass accumulation and decreasing the requirement for ATP in these processes, freeing it up for use in protective processes. This is a commonly seen strategy in plants undergoing abiotic stress (Sharma et al., 2020).
The production of ROS during desiccation is a common phenomenon in organisms. In N. haitanensis, desiccation resulted in the loss of up to 95% of the total water content, significantly enhancing the production of ROS, which was reflected in the increased MDA concentration. ABA has been demonstrated to trigger defense responses to protect plants from oxidative damage during dehydration (Wang et al., 2010). ABA modulates ROS and the antioxidant activity in enzyme such as CAT, APX, and peroxiredoxine, resulting in a greater tolerance to oxidative stress (Guajardo et al., 2016). Here, exposure to exogenous ABA was identified to modulate the activation of the antioxidant system of N. haitanensis. This was achieved through the up-regulation of the expression of genes in the AsA-GSH redox cycle, increasing the specific activity of antioxidant enzymes APX, GPX and GR; this was in addition to enhancing the production of AsA and GSH, which may help to reduce the increase of ROS caused by drought. Regulation by ABA on an enzymatic level has been recorded in microalga C. reinhardtii, where the transcription and activity of the enzymes superoxide dismutase (SOD), APX, and CAT increased after exposure to ABA (Yoshida et al., 2004). Similar results were obtained in P. orbicularis, wherein exposure to exogenous ABA resulted in increased activity of the antioxidant system during hydric stress (Yoshida et al., 2004). These results demonstrate the participation of ABA in attenuating the overproduction of ROS during dehydration.
Drought is a typical osmotic stress. Stress responses to drought are regulated by ABA-dependent signal transduction in plants. Organisms accumulate large amounts of compatible solutes or osmoprotectants. These compounds stabilize membranes and proteins in the dry state in a glass-like structure. Examples of compatible solutes of the encountered in various organisms include non-reducing sugars, polyols and amino acids, such as proline, sucrose or trehalose in plants (Cushman and Oliver, 2011). During the vascular resurrection of plants, ABA triggers the accumulation of osmotically active sugars or proline which mediates cellular protection during desiccation (Cao et al., 2020). In this research, drought stress stimulated proline accumulation which was further increased by exogenous ABA treatment. A putative connection between ABA and osmotically active sugars has recently been put forward (Živanović et al., 2020). Floridoside and isofloridoside, the stable low molecular weight carbohydrates in red algae, represent the main sink for photosynthetic carbon (Arad and Levy-Ontman, 2010). Their functions are similar to sucrose or trehalose in plants. They serve as osmolytes and will accumulate in large quantities under stress conditions such as high salt, dehydration and high temperature (Barbier et al., 2005). However, most red algae use floridoside as an osmotic pressure regulator; for example, Bangia atropurpurea, Gracilaria sordida, and Grateloupia doryphore accumulate floridoside in a hyperosmostic environment. However, there are exceptions, Poterioochromonas malhamensis and Porphyra columbina use isofloridoside as their main osmotic pressure regulator (Yu and Pedersén, 1990; Ekman et al., 1991; Simon-Colin and Bessières, 2002; Eggert and Karsten, 2010). Here, in response to ABA treatment, the concentrations of isofloridoside but not floridoside significantly increased. Since the levels of isofloridoside in N. haitanensis are much higher than that of floridoside, it can be concluded that the incremental changes to a minimal amount of floridoside are not used to control osmotic pressure adjustment. The further accumulation of isofloridoside may effectively stabilize the macromolecular substances and membrane structures of cells, preventing them from being damaged by drying. Moreover, the four putative PhTPS genes were all up-regulated by ABA treatment. PhTPS1, 3, 4 have been validated in vitro to be involved in the biosynthesis of floridoside and isofloridoside, with isofloridoside being the main product (Sun et al., 2019). These results indicated that ABA regulates the osmotic pressure balance of dehydrating N. haitanensis cells by regulating proline and isofloridoside levels.
It was demonstrated that the intact 26S proteasome, or its constituent parts, may serve additional roles in nucleotide excision repair, perhaps as molecular chaperones that promote the proper folding of repair proteins or disassembly of protein complexes (Sweder and Madura, 2002). Furthermore, the 19S proteasome has been shown to have not only a stimulatory role in nucleotide excision repair, but is involved in the repair of DNA double-strand breaks. In this study, 26 proteasome genes were annotated in the transcriptome of N. haitanensis. In response to ABA, most of these genes were up-regulated, indicating that proteasome synthesis was accelerated to degrade abnormal proteins. In addition, the DNA repair system was also massively up-regulated, suggesting ABA ensured the repair and stability of cell macromolecular constructs in N. haitanensis under stress by activating the repair mechanism.
Conclusion
These observations potentially provide insight into the evolutionarily conserved role of ABA in red algae, the early evolutionary stage of photosynthetic eukaryotes. The results presented here suggest that ABA may have evolved as a signal to regulate stress tolerance in seaweed, being crucial for their survival in an intertidal environment. ABA administration slowed down the rate of water loss in thalli and sped up the rate of rehydration; this was achieved through the avoidance photooxidation by reducing light capture and the reduction of carbon fixation to save energy. In addition to this, ABA treatment simultaneously activated repair mechanisms and the antioxidant system to avoid excessive ROS mediated damage and allow for the rapid repair of macromolecular structures. Currently, ABA has been identified to have a number of roles in higher plants. The strategies we identified herein may not only participate in enabling N. haitanensis to adapt to desiccation stress, but may further be involved in other biological processes. It will be of great interest to elucidate the mechanisms by which ABA exerts additional roles in algae in future research.
Data availability statement
The data presented in the study are deposited in the SRA databases repository, accession number PRJNA791973.
Author contribution
CZ performed the majority of the experimental work and analyzed the transcriptome data. JC performed metabolite detection and analysis. RY and QL cultured the seaweed. PZ and TW assisted in the collection and pretreatment of seaweed. HC designed the experiments and wrote the original draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0900305), the National Natural Science Foundation of China (31872540), Major Scientific and Technological Project of Ningbo (2021Z004 and 2021Z103), Major Scientific and Technological Project of Zhejiang Province (2021C02069-9), Zhejiang Province Nature Science Foundation of China (LY22C190002), China Agriculture Research System of MOF and MARA and the K. C. Wong Magna Fund in Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1007193/full#supplementary-material
References
Al-Hijab L., Gregg A., Davies R., Macdonald H., Wilson I. (2019). Abscisic acid induced a negative geotropic response in dark-incubated Chlamydomonas reinhardtii. Sci. Rep. 9, 12063. doi: 10.1038/s41598-019-48632-0
Alvarez M. E., Savouré A., Szabados L. (2021). Proline metabolism as regulatory hub. Trends Plant Sci. 27, 39–55. doi: 10.1016/j.tplants.2021.07.009
Ando H., Tanaka K., Kobayashi Y., Hanaoka M. (2016). Abscisic acid participates in the control of cell cycle initiation through heme homeostasis in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol. 57, 953–960. doi: 10.1093/pcp/pcw054
Arad S. M., Levy-Ontman O. (2010). Red microalgal cell-wall polysaccharides: biotechnological aspects. Curr. Opin. Biotechnol. 21, 358–364. doi: 10.1016/j.copbio.2010.02.008
Barbier G., Larson M. D., Halgren R. G., Wilkerson C., Garavito R. M., Christoph B. (2005). Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137, 460–474. doi: 10.1104/pp.104.051169
Blouin N. A., Brodie J. A., Grossman A. C., Xu P., Brawley S. H. (2011). Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16, 29–37. doi: 10.1016/j.tplants.2010.10.004
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. analytical biochemistry. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cao X., Wu L., Wu M., Zhu C., Zhang J. (2020). Abscisic acid mediated proline biosynthesis and antioxidant ability in roots of two different rice genotypes under hypoxic stress. BMC Plant Biol. 20, 198. doi: 10.1186/s12870-020-02414-3
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen H. M., Chu J. S., Chen J. J., Luo Q. J., Wang H., Lu R., et al. (2022). Insights into the ancient adaptation to intertidal environments by red algae based on a genomic and multiomics investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 39, msab315. doi: 10.1093/molbev/msab315
Cushman J. C., Oliver M. J. (2011). “Understanding vegetative desiccation tolerance using integrated functional genomics approaches within a comparative evolutionary framework,” in Plant desiccation tolerance. Eds. Lűttge U., Beck E., Bartels D. (Berlin, Heidelberg: Springer Press), 307–338.
De Clerck O., Kao S. M., Bogaert K. A., Blomme J., Foflonker F., Kwantes M., et al. (2018). Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol. 28, 2921–2933. doi: 10.1016/j.cub.2018.08.015
Dinakar C., Djilianovb D., Bartels D. (2012). Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 182, 29–41. doi: 10.1016/j.plantsci.2011.01.018
Eggert A., Karsten U. (2010). “Low molecular weight carbohydrates in red algae-an ecophysiological and biochemical perspective,” in Red algae in the genomic age. Eds. Seckbach J., Chapman D. J. (Dordrecht: Springer Press), 443–456.
Ekman P., Yu S., Pedersen M. (1991). Effects of altered salinity, darkness and algal nutrient status on floridoside and starch content, α-galactosidase activity and agar yield of cultivated Gracilaria sordida. Br. Phycol. J. 26, 123–131. doi: 10.1080/00071619100650091
Gaff D. F., Oliver M. (2013). The evolution of desiccation tolerance in angiosperm plants: a rare yet common phenomenon. Funct. Plant Biol. 40, 315–328. doi: 10.1071/FP12321
Gechev T. S., Dinakar C., Benina M., Toneva V., Bartels D. (2012). Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 69, 3175–3186. doi: 10.1007/s00018-012-1088-0
Guajardo E., Correa J. A., Contreras-Porcia L. (2016). Role of abscisic acid (ABA) in activating antioxidant tolerance responses to desiccation stress in intertidal seaweed species. Planta 243, 767–781. doi: 10.1007/s00425-015-2438-6
Hu X., Jiang M., Zhang A., Lu J. (2005). Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 223, 57–68. doi: 10.1007/s00425-005-0068-0
Jia F. X., Dou W., Hu F., Wang J. (2011). Effects of thermal stress on lipid peroxidation and antioxidant enzyme activities of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Fla. Entomol. 94, 956–963. doi: 10.1653/024.094.0432
Jiang M., Zhang J. (2002). Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. doi: 10.1093/jxb/erf090
Kanehisa M. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kanehisa M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. doi: 10.1002/pro.3715
Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. doi: 10.1093/nar/gkaa970
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kitajima M., Butler W. L. (2019). Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. BBA-Bioenergetics 376, 105–115. doi: 10.1016/0005-2728(75)90209-1
Lűttge U., Beck E., Bartels D. (2011). Plant desiccation tolerance (Germany: Springer-Verlag Berlin Heidelberg).
Lang V., Mantyla E., Welin B., Sundberg B., Palva E. T. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104, 1341–1349. doi: 10.1104/pp.104.4.1341
Langmead B. (2010). Aligning short sequencing reads with bowtie. Curr. Protoc. Bioinform. 32, 11–17. doi: 10.1002/0471250953.bi1107s32
Lee M. R., Kim C. S., Park T., Choi Y. S., Lee K. H. (2018). Optimization of the ninhydrin reaction and development of a multiwell plate-based high-throughput proline detection assay. Anal. Biochem. 556, 57–62. doi: 10.1016/j.ab.2018.06.022
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12, 1–16. doi: 10.1186/1471-2105-12-323
López-Cristoffanini C., Zapata J., Gaillard F., Potin P., Correa J. A., Contreras-Porcia L. (2015). Identification of proteins involved in desiccation tolerance in the red seaweed Pyropia orbicularis (Rhodophyta, bangiales). Proteomics 15, 3954–3968. doi: 10.1002/pmic.201400625
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Mcadam S. M., Sussmilch F. C. (2020). The evolving role of abscisic acid in cell function and plant development over geological time. semin. Cell Dev. Biol. 109, 39–45. doi: 10.1016/j.semcdb.2020.06.006
Mehrotra R., Bhalothia P., Bansal P., Basantani M. K., Bharti V., Mehrotra S. (2014). Abscisic acid and abiotic stress tolerance-different tiers of regulation. J. Plant Physiol. 171, 486–496. doi: 10.1016/j.jplph.2013.12.007
Papacek M., Christmann A., Grill E. (2017). Interaction network of ABA receptors in grey poplar. Plant J. Cell Mol. Biol. 92, 199–210. doi: 10.1111/tpj.13646
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Sahoo D., Tang X., Yarish C. (2002). Porphyra: the economic seaweed as a new experimental system. Curr. Sci. 83, 1313–1316. Available at: http://www.jstor.org/stable/24106954
Schmedes A., Hølmer G. (1989). A new thiobarbituric acid (TBA) method for determining free malondialdehyde (MDA) and hydroperoxides selectively as a measure of lipid peroxidation. J. Am. Oil Chem. Soc 66, 813–817. doi: 10.1007/BF02653674
Sharma S., Joshi J., Kataria S., Verma S. K., Chatterjee S., Jain M., et al. (2020). “Regulation of the Calvin cycle under abiotic stresses: an overview” in Plant life changing environment. Eds. Tripathi D. K., Singh V. P., Chauhan D. K., Sharma S., Prasad S. M., Dubey N. K., et al. (London: Academic Press), 681–717.
Simon-Colin C., Bessières M. A. (2002). An alternative HPLC method for the quantification of floridoside in salt-stressed cultures of the red alga Grateloupia doryphora. J. Appl. Phycol. 14, 123–127. doi: 10.1023/A:1019539225950
Sulochana S. B., Arumugam M. (2016). Influence of abscisic acid on growth, biomass and lipid yield of Scenedesmus quadricauda under nitrogen starved condition. Bioresour. Technol. 213, 198–203. doi: 10.1016/j.biortech.2016.02.078
Sun M. X., Zhu Z. J., Chen J. J., Yang R., Luo Q. J., Wu W., et al. (2019). Putative trehalose biosynthesis proteins function as differential floridoside-6-phosphate synthases to participate in the abiotic stress response in the red alga Pyropia haitanensis. BMC Plant Biol. 19, 325. doi: 10.1186/s12870-019-1928-2
Sweder K., Madura K. (2002). Regulation of repair by the 26S proteasome. J. Biomed. Biotechnol. 2, 94–105. doi: 10.1155/S1110724302205033
Tafvizi F., Tafreshi S. A. H., Toluei Z., Toghyani M. A. (2020). Stress responses of the green microalga, Dunaliella salina to PEG-induced drought. J. Mar. Biolog. Assoc. UK 100, 1043–1052. doi: 10.1017/S0025315420000971
Tomko R. J. Jr, Hochstrasser M. (2011). Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 60, 13–20. doi: 10.1007/s12013-011-9178-4
Wan W. M. (2020). Test of seeding net refrigeration technology in the cultivation of Pyropia haitanensis. J. Fish. Res. 42, 635–641. doi: 10.14012/j.cnki.fjsc.2020.06.012
Wang X., Kuang T., He Y. (2010). Conservation between higher plants and the moss Physcomitrella patens in response to the phytohormone abscisic acid: a proteomics analysis. BMC Plant Biol. 10, 192. doi: 10.1186/1471-2229-10-192
Weiwer M., Sherwood T., Linhardt R. J. (2009). Synthesis of floridoside. J. Carbohydr. Chem. 40, 8769–8772. doi: 10.1080/07328300802408843
Wu G., Gao Z., Du H., Lin B. (2018). The effects of abscisic acid, salicylic acid and jasmonic acid on lipid accumulation in two freshwater Chlorella strains. J. Gen. Appl. Microbiol. 64, 42–49. doi: 10.2323/jgam.2017.06.001
Xin P., Guo Q., Li B., Cheng S., Yan J., Chu J. (2020). A tailored high-efficiency sample pretreatment method for simultaneous quantification of 10 classes of known endogenous phytohormones. Plant Commun. 1, 100047. doi: 10.1016/j.xplc.2020.100047
Xu Z., Xin T., Bartels D., Li Y., Gu W., Yao H., et al. (2018). Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Mol. Plant 11, 983–994. doi: 10.1016/j.molp.2018.05.003
Yaln S., Okudan E., Karakaş Z., Önem A. N., Başkan K. S. (2019). Identification and quantification of some phytohormones in seaweeds using UPLC-MS/MS. J. Liq. Chromatogr. Relat. Technol. 42, 475–484. doi: 10.1080/10826076.2019.1625374
Yoshida K., Igarashi E., Mukai M., Hirata K., Miyamoto K. (2003). Induction of tolerance to oxidative stress in the green alga, Chlamydomonas reinhardtii, by abscisic acid. Plant Cell Environ. 26, 451–457. doi: 10.1046/j.1365-3040.2003.00976.x
Yoshida K., Igarashi E., Wakatsuki E., Miyamoto K., Hirata K. (2004). Mitigation of osmotic and salt stresses by abscisic acid through reduction of stress-derived oxidative damage in Chlamydomonas reinhardtii. Plant Sci. 167, 1335–1341. doi: 10.1016/j.plantsci.2004.07.002
Yudina L., Sukhova E., Sherstneva O., Grinberg M., Ladeynova M., Vodeneev V., et al. (2020). Exogenous abscisic acid can influence photosynthetic processes in peas through a decrease in activity of h+-ATPase in the plasma membrane. Biology 9, 324. doi: 10.3390/biology9100324
Yu S., Pedersén M. (1990). The effect of salinity changes on the activity of α-galactosidase of the red algae Gracilaria sordida and G. tenuistipitata. Bot. Mar. 33, 385. doi: 10.1515/botm.1990.33.5.385
Zhao J., Xu P., Chen J. J., Luo Q. J., Yang R., Chen H. M., et al. (2016). Simultaneous analysis of nine phytohormones in Porphyra haitanensis by high performance liquid chromatography coupled with triple quadrupole tandem mass spectrometry. Food Sci. 37, 216–222. doi: 10.7506/spkx1002-6630-201616035
Zhao J., Yang Y., Zhao Q. (2019). Changes of the content of floridosides isomers in Porphyra haitanensis under high temperature stress. J. Nucl. Agric. Sci. 33, 109–117. doi: 10.11869/j.issn.100-8551.2019.01.0103
Keywords: abscisic acid, Neoporphyra haitanensis, dehydration, intertidal environment, adaptation
Citation: Zhang C, Chen J, Yang R, Luo Q, Wang T, Zhang P and Chen H (2022) Abscisic acid activates desiccation tolerance responses in intertidal seaweed Neoporphyra haitanensis. Front. Mar. Sci. 9:1007193. doi: 10.3389/fmars.2022.1007193
Received: 30 July 2022; Accepted: 29 September 2022;
Published: 17 October 2022.
Edited by:
Tifeng Shan, Institute of Oceanology (CAS), ChinaReviewed by:
Octavio R. Salazar, King Abdullah University of Science and Technology, Saudi ArabiaHiroyuki Mizuta, Hokkaido University, Japan
Copyright © 2022 Zhang, Chen, Yang, Luo, Wang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haimin Chen, Y2hlbmhhaW1pbkBuYnUuZWR1LmNu
 Caixia Zhang1
Caixia Zhang1 Rui Yang
Rui Yang Haimin Chen
Haimin Chen