
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 02 February 2022
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.824533
Pearl gentian grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂) is a hybrid fish with high commercial value. It is widely cultured on the Asian coast; however, it is not cold-tolerant. Although we have previously characterized the liver transcriptomic responses of this grouper to cold stress, the roles of miRNAs and transcription factors (TFs) in cold resistance and the underlying regulatory mechanisms are still unclear. In this study, we integrated miRNA and mRNA sequencing data for pearl gentian grouper under cold stress and constructed a miRNA-TF-mRNA regulatory network. Furthermore, we screened seven key miRNAs (i.e., gmo-miR-221-5p, ssa-miR-7132b-5p, ola-let-7c, ssa-miR-25-3-5p, ccr-miR-489, gmo-miR-10545-5p, ccr-miR-122) that regulated target genes (including TF ACSS2, TF PPARD, TF PPP4CB; CYP2J2, EHHADH, RXRs, NR1D2, PPP1CC-A, PPP2R1A, FOXK2, etc.). These miRNAs participated in several important pathways and biological processes by the direct or indirect regulation of target genes, such as antioxidation and membrane fluidity, glucose and lipid metabolism, circadian rhythm, DNA repair, and apoptosis. The key cold-related miRNAs, TFs, and genes and their potential regulatory relationships identified in this study provide a deeper understanding of the complex molecular basis of the response to low-temperature environments in the grouper. In particular, our results provide the first identification for the role of NR1D2 gene in the cold tolerance of fish via the regulation of circadian rhythm. Furthermore, the key miRNAs and genes provide a basis for the molecular breeding of new cold-tolerant varieties of the pearl gentian grouper.
As aquatic ectothermic animals, the survival and growth of fish are closely related to the water temperature (Yoneda and Wright, 2005). When the water temperature reaches or drops below the lower tolerance limit, a cascade of complex physiological, biochemical, and behavior responses occur in fishes (Donaldson et al., 2008). Many physiological, biochemical, and transcriptome studies have showed that in the process of continuous water temperature decline, the metabolic rate of fishes decreases gradually, accompanied by decreases in oxygen uptake and ammonia excretion (Li et al., 2017); however, the blood glucose content could increase via increased glucose and fat metabolism to supplement this energy loss (Cheng et al., 2017). When low-temperature stress exceeds the tolerance range in fish, hydrogen peroxide and free radicals produced by oxidative phosphorylation cannot be eliminated in a timely manner, which could cause irreversible damage to cells and lead to increased DNA repair and even apoptosis (Gabbianelli et al., 2003). These cold-induced responses involve a large number of genes and biological pathways. The underlying causes of transcriptomic changes are closely related to the regulatory roles of both microRNAs (miRNAs) and transcription factors (TFs) (Bartel, 2009). Only a few studies have identified the functional roles of miRNAs and TFs in the cold stress response in fish (Yang et al., 2011; Nie et al., 2019). Moreover, the main pathways involved in the response to cold stress differ substantially among studies.
MiRNAs are a class of small non-coding RNAs (18–25 nt) with pivotal roles in the post-transcriptional regulation of genes (Bushati and Cohen, 2007). The wide application of miRNA microarray and next-generation sequencing technologies has led to miRNA expression profiling studies of many species, especially mammals and plants (Lv et al., 2010). Despite several studies focused on the identification and functional roles of miRNAs in fish development, osmotic pressure regulation, and immune responses (Yan et al., 2012), relatively little is known about the functions of miRNAs in low-temperature stress in fish. Hung et al. (2016) confirmed that dre-miR-29b of zebrafish larvae can accurately regulate the core clock gene PER2 (Period Circadian Regulator 2) under cold shock stress, although weak correlations between cold-affected miRNAs and mRNAs were reported in an earlier study of cold-acclimated zebrafish (Yang et al., 2011). Subsequently, miRNAs associated with enhanced cold tolerance in the turbot Scophthalmus maximus have been shown to be related to circadian pathways and various signaling pathways (Nie et al., 2019). During cold stress in the common carp Cyprinus carpio, miRNAs regulate genes actively involved in energy metabolism and cell membrane components (Sun et al., 2020). Moreover, clear interactions among miRNAs and signaling pathways under cold stress have been revealed in the farmed tilapia Oreochromis niloticus (Qiang et al., 2018a). Collectively, numerous genes are directly and precisely regulated by miRNAs in several important biological processes involved in energy metabolism, antioxidation, the biological clock, and other processes, and miRNAs play key roles in genomic plasticity against low-temperature stress in fishes.
MiRNAs indirectly regulate a number of target genes via the regulation of TFs (Zhou et al., 2008). In particular, TFs can regulate the expression of target genes at a certain time point by combining with a specific sequence upstream of the 5′ end of the gene (Lovering et al., 2021). Numerous studies have demonstrated the biological functions of TFs in the specific physiological responses of a wide variety of animals and plants (Liu et al., 2019). TFs also play essential roles in the regulation of fish embryonic development, cell differentiation, and hypoxia stress (Joseph et al., 2017). However, little is known about the functional roles of fish TFs and their effects under low-temperature conditions. Nie et al. (2019) reported that TFs of the turbot S. maximus are mainly involved the cell membrane, signal transduction, and circadian rhythm pathways via regulatory genes under cold stress. TFs and miRNAs can accurately regulate gene expression pre- and post-transcription, respectively. Thus, the construction of a miRNA-TF-mRNA regulatory network will clarify the complex regulatory relationships involved in the response to low-temperature stress in fish.
Pearl gentian grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂) is an F1 hybrid fish characterized by fast growth and strong disease resistance (Fan et al., 2014). It has become a popular grouper variety with high commercial value in the aquaculture industry along the Asian coast (Shapawi et al., 2014), particularly in southern China (Lin et al., 2017). However, this hybrid grouper is not cold-tolerant. Within the water temperature range of 20–13°C, the feeding and activity of the pearl gentian grouper decrease significantly as the temperature decreases, and its lower critical temperature is 11° (Liang et al., 2013). This obviously limits the breeding scope and environment of this grouper, which is not conducive to the expansion of its aquaculture industry. For this reason, we previously analyzed changes in the liver transcriptome of pearl gentian grouper under cold stress by integrating PacBio SMRT-Seq and Illumina RNA-Seq technologies and successfully screened various cold-related genes and biological pathways; our results indicated that energy homeostasis may play a crucial role in physiological responses to cold stress in the species (Miao et al., 2021). However, the regulatory relationships and dynamics of these genes are still unclear, and the construction of miRNA-TF-mRNA network is necessary to clarify regulatory mechanisms at the pre- and post-transcriptional levels underlying the transient response to low-temperature stress in this grouper and to narrow the screening of cold-related candidate genes. A clear understanding of these molecular mechanisms will contribute to the breeding of cold-tolerant fish varieties by the editing or knockout of cold-sensitive genes (Li et al., 2016).
In this study, we investigated the complex regulatory relationships among miRNAs, TFs, and mRNAs under cold stress in the pearl gentian grouper and constructed a miRNA-TF-mRNA regulatory network. Based on high-quality sequencing data, we first identified miRNAs and screened differentially expressed (DE) miRNAs in cold-stressed fish, followed by analyses of the molecular functions and biological pathways associate with their target genes. Second, we used the STEM (Short Time-series Expression Miner) trend analysis to obtain expression profiles for miRNAs that were consistently expressed with decreasing temperatures. Finally, we integrated all DE miRNAs, DE TFs, and differentially expressed genes (DEGs) to construct a miRNA-TF-mRNA regulatory network and clarify important pathways and biological reactions. Our findings provide new insights into the molecular regulatory mechanism underlying adaptation to low-temperature environments in this grouper and are expected to facilitate the molecular breeding of cold-tolerant varieties.
The experimental animal protocols in the present study were reviewed and approved by the Animal Experimental Ethics Committee of Guangdong Ocean University, China (approval number: 0901-2019). Experiment procedures were performed in accordance with the Provisions and Regulations for the National Experimental Animal Management Regulations (China, July 2013) and the Experimental Animal Policies and Regulations of Guangdong Province (China, October 2010).
The experimental process (Figure 1A) included temporary fish culture, cold stress treatment, liver tissue collection under anesthesia, and total RNA extraction from samples and was performed following the methods described in our previous transcriptome study (Miao et al., 2021). Briefly, 150 healthy juveniles of the pearl gentian grouper hatched from Hainan (northern South China Sea, China) breeding parents were equally divided into three parallel experimental tanks. In the pre-experiment, we found that a considerable number of experimental fishes died at 11°C. Accordingly, we set 12°C as the lowest stress temperature point to obtain rich gene expression information and sufficient sample size under the extreme low-temperature stress. The water temperature in the three experimental tanks was gradually decreased from 25 to 20°C, 15, and 12°C at a constant rate of 1°C/h and then kept at 12°C for 6 h using a cooling-water machine. Three samples were randomly sampled from each tank at each of the five temperature points, for a total of 45 samples. These sampling groups were named CT (control group), LT20, LT15, LT12, and LT12-6. Considering that the liver is an important metabolic organ in the process of fish temperature adaptation (Hazel and Williams, 1990), it was sensitive to the transcriptomic changes of various pathways under temperature stress (Gracey et al., 2004; Sun et al., 2019). So, the liver of pearl gentian grouper was used for the targeted analysis sample in this study. After testing the total RNA quality of all 45 liver samples, 15 better quality RNA samples (taking one sample per parallel tank of all groups) were selected for subsequent small RNA library construction and sequencing.
Small RNA fragments were separately selected from each of the 15 total RNA samples using TruSeq Small RNA Library Preparation Kits (Illumina, San Diego, CA, United States) following the manufacturer’s instructions. Illumina adapters were directly added to the 5′ and 3′ ends of small RNA, and first strand cDNA was synthesized by reverse transcription and purified to obtain a cDNA library of about 150 bp sequences. After quality control using the Agilent 2100, small RNA libraries were sequenced on the Illumina HiSeq 2000 platform. Finally, 50 bp single-end reads were generated and used for subsequent analyses (Figure 1B).
Clean reads were obtained from raw data by a stringent filtering process, including the removal of low-quality reads (containing Ns), 5′ adapter contaminants, reads without 3′ adapter sequences, and reads containing poly-N sequences. Known miRNA, rRNA, tRNA, snRNA, snoRNA sequences were then identified and annotated by mapping small RNA clean reads to miRbase v22 (Kozomara et al., 2019) and Rfam v12.0 (Nawrocki et al., 2015) databases using bowtie2 v2.2.2 (Langmead and Salzberg, 2012). The programs mirEvo v1.3 (Wen et al., 2012) and miRdeep v2.0 (Friedländer et al., 2012) were integrated to predict novel miRNAs by identifying the Dicer cleavage site and secondary structure and calculating the minimum free energy of unannotated miRNA tags.
Expression levels of known and novel miRNAs in each sample were standardized by Transcripts Per Million (TPM). Pearson correlation coefficients (PCC) for expression levels in different samples were evaluated based on a standardized expression data of all miRNAs. Differential expression between groups was evaluated using the DESeq2 v3.18.1 (Love et al., 2014) R package. The DE miRNAs were determined using threshold values of Q < 0.05 and | log2 (Fold Change)| > 1. Subsequently, the DE miRNAs were visualized by volcano diagrams using the HiPlot.1 Functional annotation and hyper-geometric distribution enrichment analyses of the target genes of DE miRNAs were performed using DIAMOND v0.9.24 (Buchfink et al., 2014) based on the Gene Ontology (GO) (Ashburner et al., 2006) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) databases. The GO terms and KEGG pathways were considered significantly enriched when FDR ≤ 0.05.
The STEM (Ernst and Bar-Joseph, 2006) program was used to characterize the expression profiles of miRNA that were consistently expressed with gradient temperature changes in all groups. First, the miRNA expression data standardized by the TPM algorithm were used as the input and then converted into preprocessed data by the log2 algorithm. Second, the maximum unit of change between time nodes and the minimum Pearson correlation coefficient were set to 2 and 0.7, respectively, for the construction of expression profiles. Finally, the miRNA expression profiles with statistical significance (P < 0.05) were screened for visualization and further analysis. The miRNA profiles that were significantly correlated with the temperature gradient were used for subsequent GO and KEGG enrichment analyses conducted using DIAMOND.
All full-length transcripts and DEGs were derived from our previous transcriptome data obtained by Illumina and PacBio sequencing. We applied MiRanda v3.3 (Riffo-Campos et al., 2016) and TargetScan v7.2 (Agarwal et al., 2015) to predict potential target genes of DE miRNAs based on DEG sequences, and overlapping target genes were selected for further analysis. The high-quality full-length transcript sequences were mapped to Animal TFDB v3.0 (Hu et al., 2019) to identify potential TFs using DIAMOND. A differential expression analysis of TFs among groups was performed using the DESeq2 v3.18.1 R package, with P < 0.05 and | log2 (Fold Change)| > 1 as thresholds to determine the DE TFs.
Pairwise relationships between DE miRNAs, DE TFs, and DEGs were evaluated using PCC > 0.7 and P < 0.05 as thresholds to construct a potential miRNA-TF-mRNA regulatory network. A network diagram was drawn using Cytoscape v3.7.2 (Shannon et al., 2003). Nodes with different shapes in the network represent different molecule types (i.e., miRNA, TF, and gene), different colors represent up- and down-regulation, and arcs and arrows indicate regulatory relationships. Further, by integrating pathways involved in each branch of the miRNA-TF-mRNA regulatory network, the cold-induced reaction process was revealed.
To verify the accuracy of sequencing data, five DE miRNAs and six target genes were randomly selected for qRT-PCR to detect expression trends in all groups. Specific primers were designed based on the miRNA and target gene sequences using Primer 5 (Table 1). Total RNA (1 μg) from each sample was used to synthesize cDNA using the TUREscript 1st Stand cDNA Synthesis Kit (AidLab, Beijing, China). Since 18S rRNA was verified by qPCR as stably expressed in all groups, it was used as an internal reference gene for miRNAs and target genes. The qRT-PCR system included 20 μL of reaction mixture consisting of 7.8 μL of RNase-free water, 10 μL of 2 × SYBR® Green Supermix reagent (SYBR, Dalian, Liaoning, China), 1 μL of cDNA, and 0.6 μL of each primer. Amplification was performed using an Analytik Jena QTower 2.2 (Analytik Jena AG, Jena, Germany) under the following conditions: preheating at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. All samples were evaluated in three repeated reactions and a no template control was included for each verified miRNA and target gene. Then, an amplification curve was generated to evaluate the range of cycle threshold (CT) values, and the melting curve was used to determine the specificity of primers and primer dimers. Lastly, the relative expression levels of miRNAs and target genes were calculated using the comparative CT method (2−ΔΔCT).
A total of 4.82 Gbp of clean small RNA reads (Q30 > 97.24%) were obtained after filtering 11.28 Gbp of raw reads. As illustrated in Table 2, the GC content ranged from 49.16 to 49.80%, and the error rate per sample was < 0.01%. Among the 15 samples, the base numbers of clean reads and unique reads ranged from 284,153,887 to 284,153,887 and from 133,080 to 218,551, respectively. Sequence lengths were mainly 18–35 nt. The most frequent lengths were 22 nt (43.97%), 21 nt (21.48%), and 23 nt (15.12%).
The abundances of known miRNAs, novel miRNAs, rRNAs, tRNAs, snRNAs, snoRNAs, and other sequences in each sample are listed in Table 3. Among small RNA reads, the most frequent type was known miRNAs, followed by rRNAs, snoRNAs, snRNAs, and tRNAs.
In total, 668 known miRNAs were identified by mapping small RNA tags to the miRbase database. These miRNAs belonged to various miRNA families, such as mir0124, mir0190, mir0205, mir0218, and mir0455. In an analysis of the first base of mature miRNAs, adenine (A) or uracil (U) was the primary first bases. In addition, 48 novel miRNAs were predicted, of which 12 novel miRNAs were expressed in all samples, including novel_2, novel_7, novel_10, novel_33, novel_34, novel_38, novel_44, novel_47, novel_63, novel_69, novel_77, and novel_85. The secondary structures of these novel miRNAs were drawn to show the detailed stem-loop structure and mature miRNA sequences (indicated by red lines in Figure 2).
Expression data for all 716 miRNAs were standardized by the TPM algorithm and were used for analyses of correlations and differential expression among groups. PCC for pairwise comparisons between samples were in the range of 0.944–0.995 (Figure 3).
We detected 27 (5 up- and 22 down-regulated), 23 (5 up- and 18 down-regulated), 15 (1 up- and 14 down-regulated), and 38 (13 up- and 25 down-regulated) DE miRNAs in the CT vs. LT20, CT vs. LT15, CT vs. LT12, and CT vs. LT12-6 comparisons, respectively (Figure 4 and Supplementary Tables 1–4). Notably, 4 DE miRNAs were shared among the four comparisons. Moreover, 4 (1 up- and 3 down-regulated), 11 (4 up- and 7 down-regulated), 28 (12 up- and 16 down-regulated), 5 (2 up- and 3 down-regulated), 24 (12 up- and 12 down-regulated), and 17 (6 up- and 11 down-regulated) DE miRNAs were observed in the LT20 vs. LT15, LT20 vs. LT12, LT20 vs. LT12-6, LT15 vs. LT12, LT15 vs. LT12-6, and LT12 vs. LT12-6 comparisons, respectively. Obviously, the most DE miRNAs (38) were screened in the comparison between the CT group and the LT12-6 group. In addition, 89 unique miRNAs were screened in the pairwise comparisons among all groups, and 72 unique miRNAs were detected in the pairwise comparisons between the control group and the four low-temperature groups.
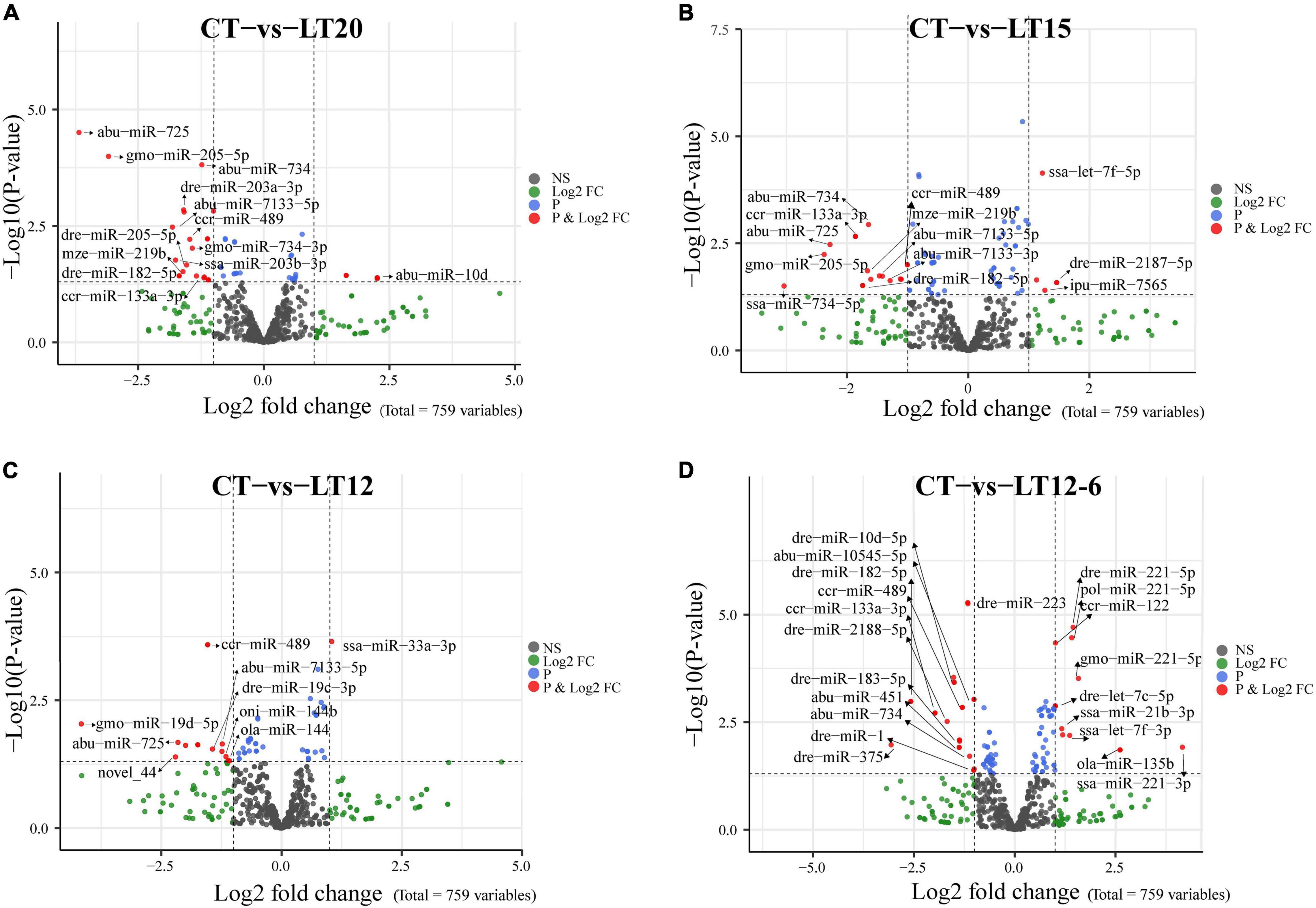
Figure 4. Volcano plot showing the up-regulation or down-regulation of DE miRNAs in pairwise comparisons between the control group and four low-temperature groups. (A) Comparison of CT with LT20 groups. (B) Comparison of CT with LT15 groups. (C) Comparison of CT with LT12 groups. (D) Comparison of CT with LT12-6 groups. Dark gray dots (NS): miRNAs without significant differences between groups. Red dots (P and Log2FC): miRNAs that meet the thresholds of P < 0.05 and log2FC > 1.
GO and KEGG pathway enrichment analyses of the DE miRNAs in the pairwise comparisons among groups were performed to further evaluate the functions of target genes regulated by miRNAs under cold stress. The main GO enrichment terms in all pairwise comparisons were generally similar (Figures 5A,C), including metabolic process, single-organism process, and cellular process in the biological process category, cell, cell part, and membrane in the cellular component category, and binding, catalytic activity, and transporter activity in the molecular function category. Several common pathways, such as vitamin digestion and absorption, biotin metabolism, AMPK signaling pathway, fat digestion and absorption, and phagosome pathways were within the top 20 significantly enriched pathways in the KEGG enrichment analysis (Figures 5B,D).
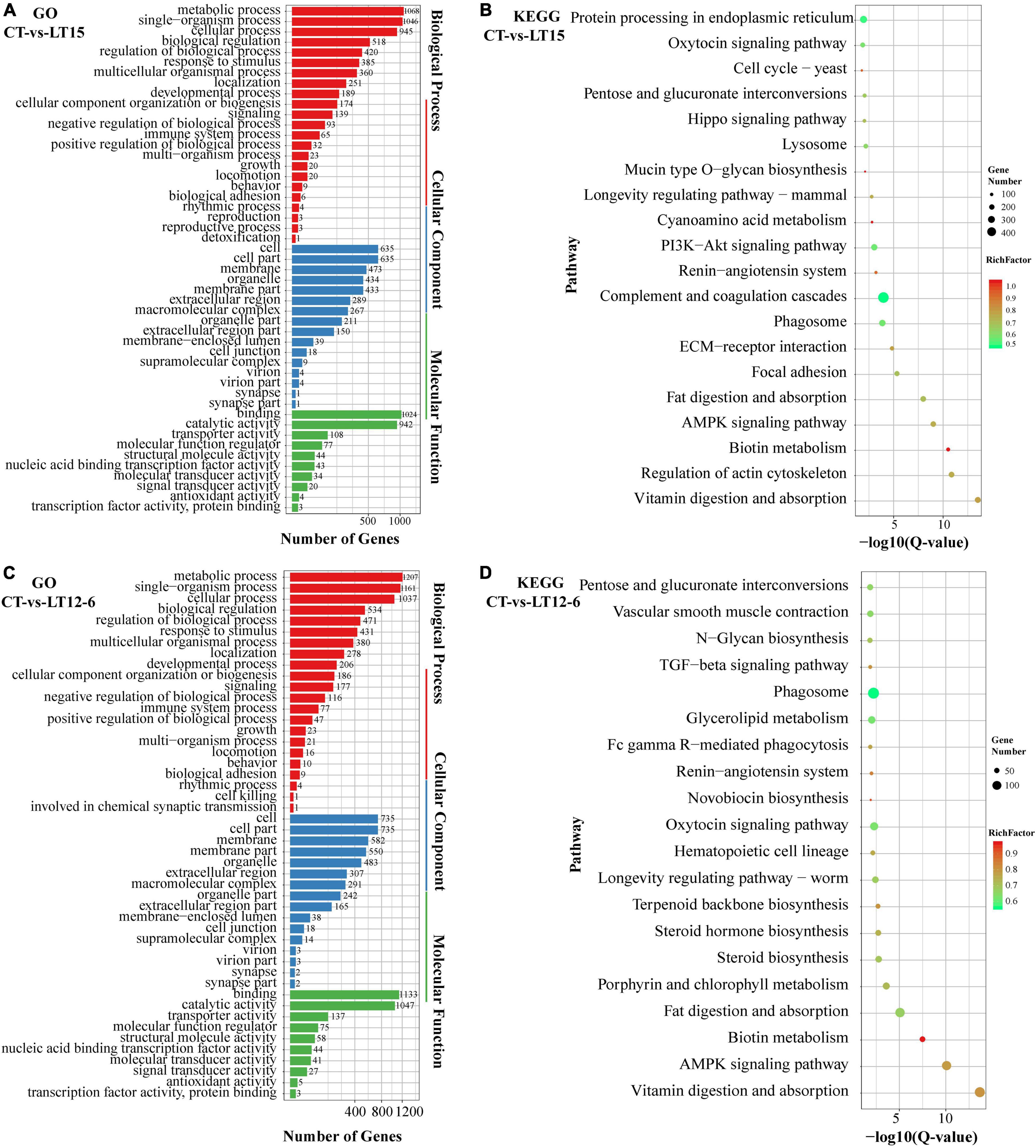
Figure 5. Summary of GO and KEGG pathway enrichment analyses of DE miRNAs in the CT vs. LT15 (A,B) and CT vs. LT12-6 (C,D) comparisons.
Using STEM trend analysis, 20 expression profiles were obtained for all miRNAs in the five groups, and expression changes in profile 0, profile 2, and profile 18 were statistically significant. As illustrated in Figure 6A, profile 0 was characterized by the overall down-regulation of miRNAs and profile 18 involved an overall up-regulation of miRNAs. The expression trend in profile 2 was unclear. In particular, 79 consistently down-regulated miRNAs were assigned to profile 0, most of which showed slight expression changes (Figure 6B). In profile 0, the normalized range of fold change values [log2(V(i)/V(0)] for most miRNAs with down-regulated expression was 0 to –4 [| Fold Change| ∈ (1, 16)], and a few miRNAs had values from –4 to –8 [| Fold Change| ∈ (16, 256)]. In contrast, 32 miRNAs were consistently up-regulated in profile 18 (Figure 6C), most of which were up-regulated in the LT20 group. Expression trends in profile 18 varied. Some miRNAs maintained their original expression levels after an initial down- or up-regulation, while others were continuously down- or up-regulated at all temperatures. The normalized range of values for up-regulated expression for the 32 miRNAs was 0–8 [| Fold Change| ∈(1, 256)].
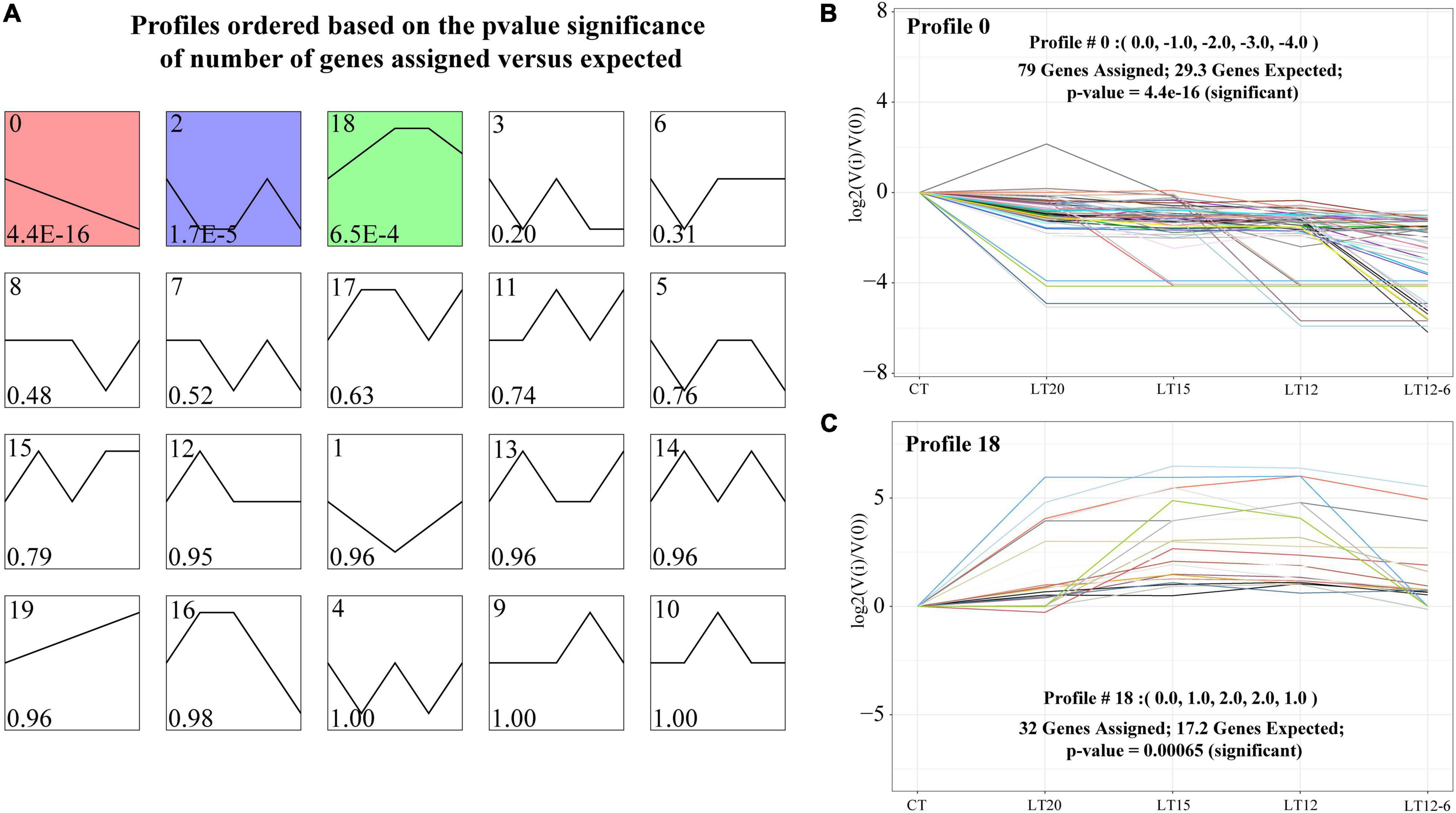
Figure 6. STEM analysis and corresponding miRNA expression profiles. (A) Expression profiles of consistently up- or down-regulated miRNAs were screened (P < 0.05). (B) Profile 0: miRNAs were uniformly down-regulated in all groups. (C) Profile 18: miRNAs were uniformly up-regulated in all groups.
Further, the GO and KEGG enrichment analyses of miRNA target genes assigned to profiles 0 and 18 were performed to reveal the key biological processes and pathways related to low-temperature responses. As demonstrated in Figure 7, the main GO terms associated with profiles 0 and 18 were similar, while the KEGG enrichment pathways involving miRNA target genes varied between the two profiles. In the GO enrichment analysis (Figures 7A,C), the main terms for the two profiles included metabolic process, cellular process, and single-organism process in the biological process category, cell, cell part, and membrane in the cellular component category, and binding, catalytic activity, and molecular function regulator in the molecular function category. Enriched KEGG pathways involving genes in profile 0 mainly included the AMPK signaling pathway, amino sugar and nucleotide sugar metabolism, phagosome, regulation of actin cytoskeleton, and arginine biosynthesis (Figure 7B). Profile 18 was mainly enriched in RNA transport, protein processing in endoplasmic reticulum, terpenoid backbone biosynthesis, platelet activation, and PI3K-Akt signaling pathways (Figure 7D).
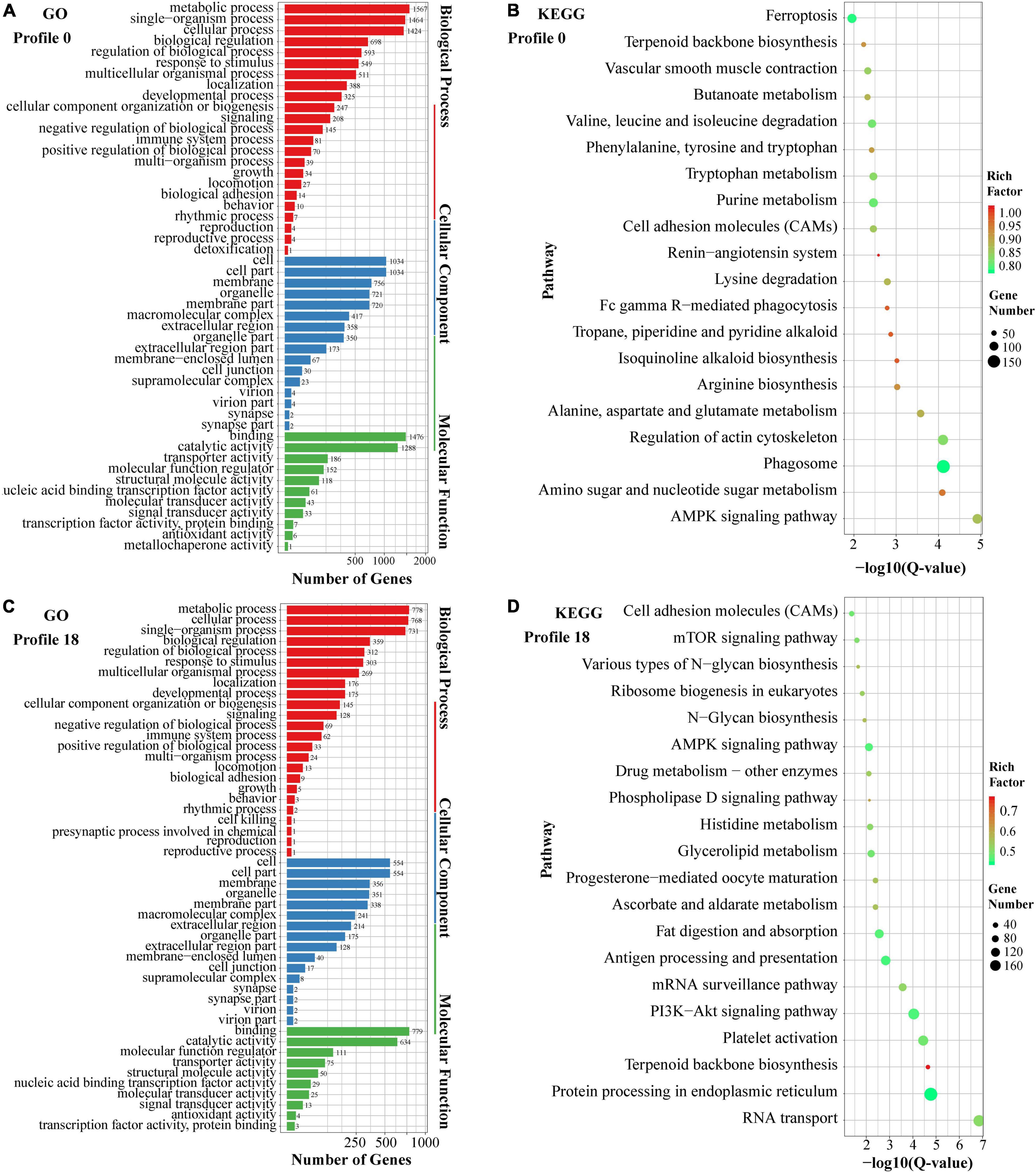
Figure 7. Summary of GO and KEGG pathway enrichment analyses of the miRNA target genes in profiles 0 (A,B) and 18 (C,D).
Predicting target DEGs of all DE miRNAs provides a basis for annotation and enrichment analyses as well as the development of regulatory networks. Based on our 3,271 previously identified DEGs, 89 DE miRNA target genes were predicted in this study, and 9,637 miRNA–mRNA interacting pairs were obtained.
In the present study, 917 TFs were predicted against the Animal TFDB, and these could be assigned to 46 TF families. The top three TF families with the largest number of TFs were TF_bZIP (131 TFs), zf-C2H2 (124 TFs), and bHLH (103 TFs). In total, 63 (40 up- and 23 down-regulated), 138 (84 up- and 54 down-regulated), 148 (110 up- and 38 down-regulated), and 266 (143 up- and 123 down-regulated) TFs were differentially expressed in CT vs. LT20, CT vs. LT15, CT vs. LT12, and CT vs. LT12-6, respectively (Figure 8 and Supplementary Tables 5–8). Moreover, 25 (15 up- and 10 down-regulated), 91 (70 up- and 21 down-regulated), 186 (111 up- and 75 down-regulated), 56 (35 up- and 21 down-regulated), 129 (72 up- and 57 down-regulated), and 90 (35 up- and 55 down-regulated) DE TFs were identified in LT20 vs. LT15, LT20 vs. LT12, LT20 vs. LT12-6, LT15 vs. LT12, LT15 vs. LT12-6, and LT12 vs. LT12-6, respectively. The most DE TFs (270) were screened between the CT group and the LT12-6 group. Further analyses revealed that 368 unique DE TFs were obtained in pairwise comparisons among all groups, most of which (328) were obtained in pairwise comparisons between only the control group and the four low-temperature groups.
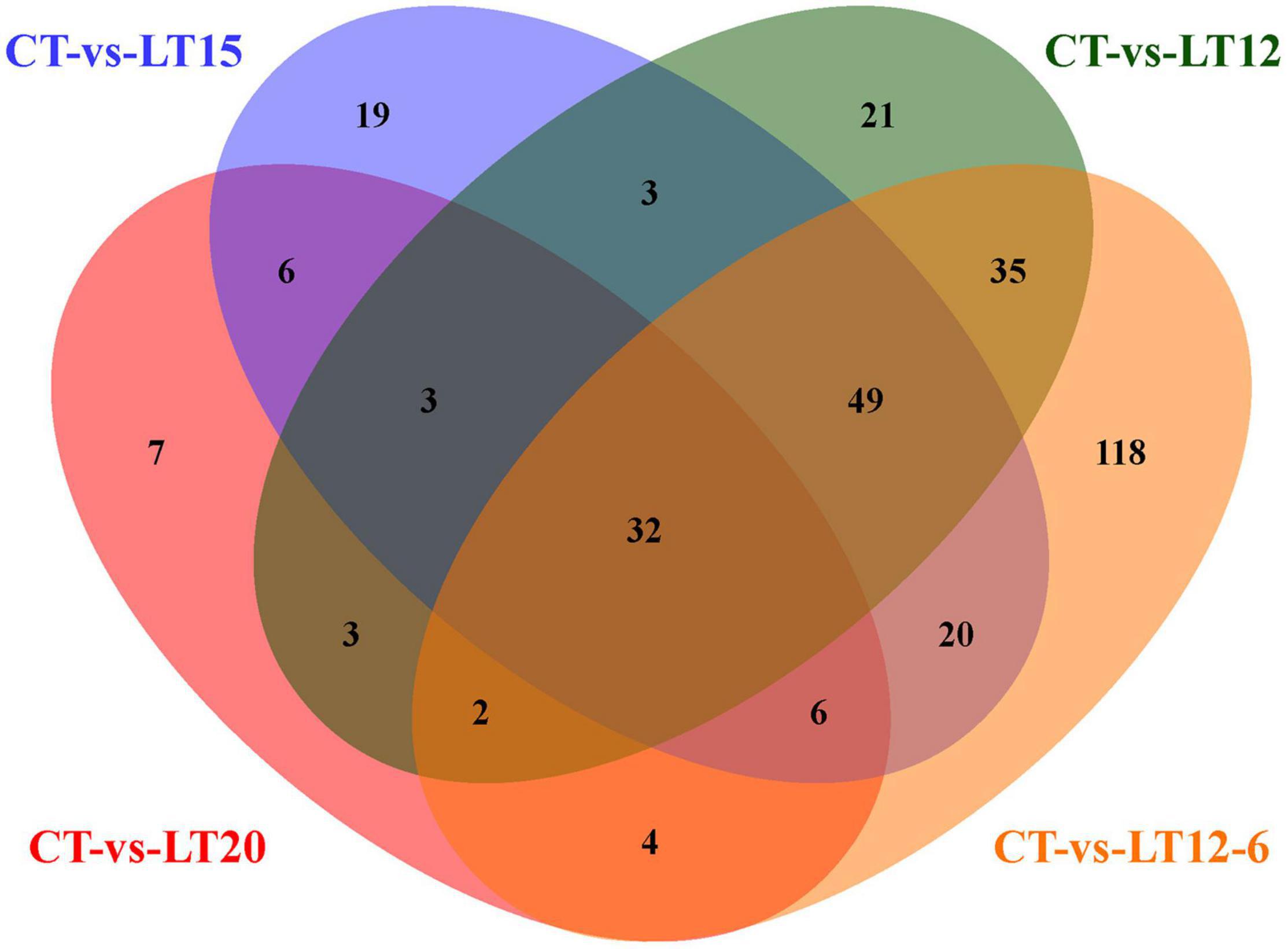
Figure 8. Number and intersection of DE miRNAs in pairwise comparisons between the control group and four low-temperature groups.
The factors related to low-temperature stress were screened by further exploring the interactions and regulatory relationships among DE miRNAs, DE TFs, and DEGs. As a result, 161 regulatory interactions involving the above-described 9637 miRNA-mRNA pairs were selected to construct a potential miRNA-TF-mRNA regulatory network (Figure 9), using thresholds of PCC > 0.7 and P < 0.05. Although there were fewer up-regulated miRNAs than down-regulated miRNAs, the up-regulated miRNAs regulated more target genes and TFs (Figure 9).
The direct regulation of target genes by miRNAs and indirect regulation of genes via TFs were both observed in the miRNA-TF-mRNA regulatory network (Figure 9). For example, gmo-miR-221-5p directly regulated CYP2J2 (Cytochrome P450 2J2), CYP4B1 (Cytochrome P450 4B1), TM7SF3 (Transmembrane 7 Superfamily Member 3), and GYS2 (Glycogen Synthase 2), ssa-miR-25-3-5p regulated TSKU (Tsukushi, Small Leucine Rich Proteoglycan) and NR1D2 (Nuclear Receptor Subfamily 1 Group D Member 2), and ccr-miR-489 and ccr-miR-122 regulated PPP1CC-A (Protein Phosphatase 1 Catalytic Subunit Gamma) and FOXK2 (Forkhead Box K2), respectively. The miRNA ssa-miR-7132b-5p indirectly regulated EHHADH (Enoyl-CoA Hydratase and 3-Hydroxyacyl CoA Dehydrogenase), ACAT2 (Acetyl-CoA Acetyltransferase 2), HADHA (Hydroxyacyl-CoA Dehydrogenase Trifunctional Multienzyme Complex Subunit Alpha), PCK1, PCK2 (Phosphoenolpyruvate Carboxykinase 1, 2) via the TF ACSS2 (Acyl-CoA Synthetase Short Chain Family Member 2), ola-let-7c indirectly regulated RXRAA, RXRBA, RXRGB (Retinoid X Receptor Alpha, Beta, Gamma) by the regulation of the TF PPARD (Peroxisome Proliferator Activated Receptor Delta), and gmo-miR-10545-5p had an indirect effect on PPP2R1A (Protein Phosphatase 2 Regulatory Subunit 1) and PTPA (Protein Phosphatase 2 Phosphatase Activator) by regulating the TF PPP4CB (Protein Phosphatase 4 Catalytic Subunit). In brief, these regulatory mechanisms and relationships may contribute to the response to cold stress in the pearl gentian grouper.
The relative expression levels of five miRNAs and six target genes in all groups were validated by qRT-PCR. As demonstrated in Figure 10A, all five miRNAs (i.e., ola-let-7c, ccr-miR-122, drc-let-7c-5p, gmo-miR-221-5p, and dre-miR-221-5p) were up-regulated. Among the six target genes, NPR2, FOXK2, NIPSNAP1, and MAF1 were down-regulated and PPP1CC-A and C7 were up-regulated. These results for miRNAs and target genes were consistent with those obtained by quantitative RNA-Seq (Figure 10) and regulatory network construction (Figure 9).
Based on an integrated analysis of small RNA sequencing data and previous RNA-Seq data, we obtained 89 DE miRNAs, 368 DE TFs, and 3271 DEGs involved in the response to cold stress in the pearl gentian grouper. A miRNA-TF-mRNA network was constructed based on 23 DE miRNAs, 24 DE TFs, and 91 DEGs (Figure 9). By further exploring the functions and pathways related to these molecules, we finally confirmed that 28 key molecules, including 7 DE miRNAs, 3 DE TFs, and 18 DEGs, were involved in several important pathways and biological reactions associated with cold stress in pearl gentian grouper by both direct and indirect regulatory relationships (Figure 11). The main functions identified in this analysis could be summarized into four general categories (Figure 11): antioxidation and membrane fluidity, glucose and lipid metabolisms, circadian rhythm, and DNA repair and apoptosis.
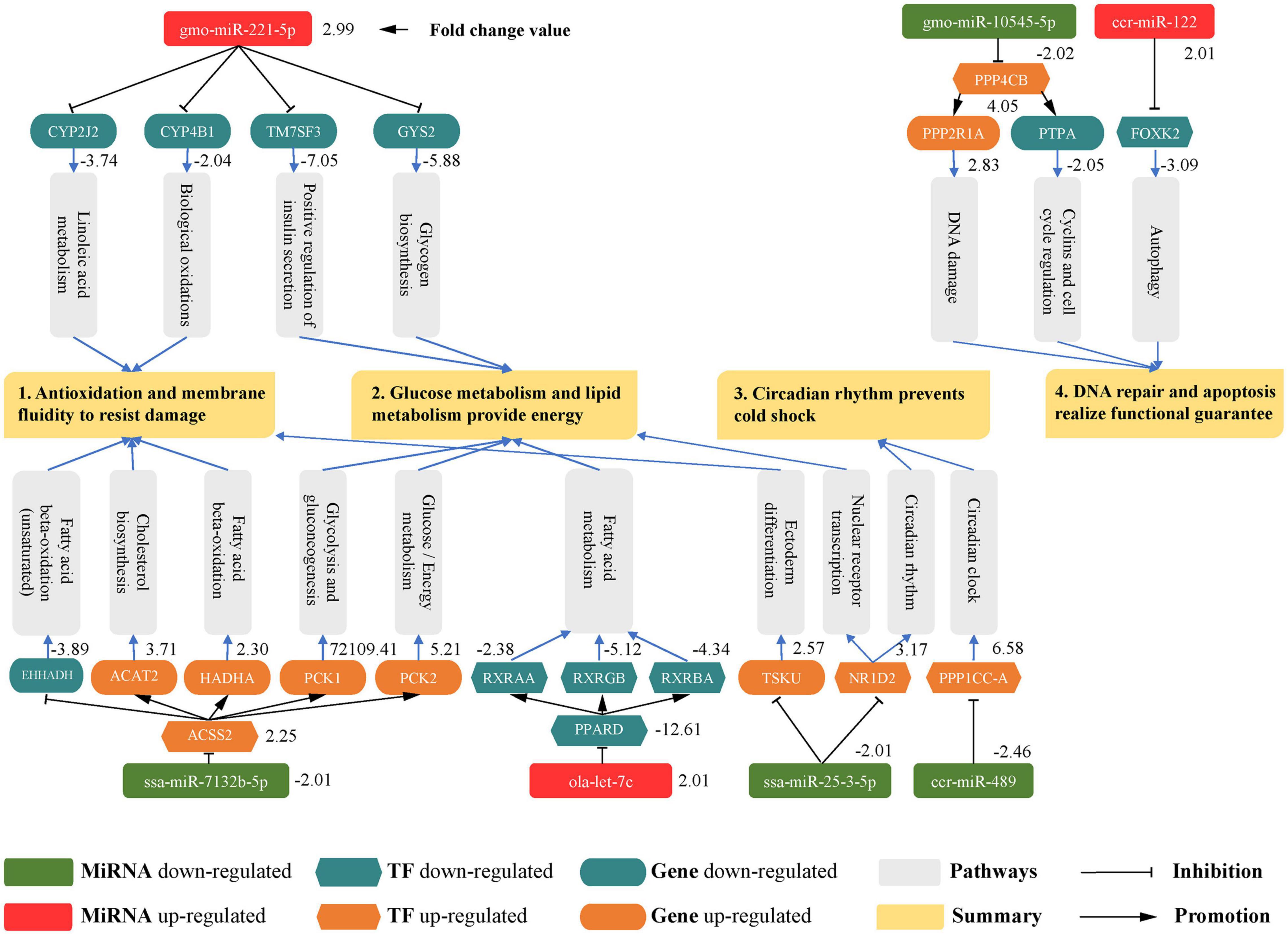
Figure 11. Several important pathways and biological reactions associated with the miRNA-TF-mRNA regulatory network involved in the response to cold stress in pearl gentian grouper.
The oxidation of long-chain fatty acids is closely related to the stability of cell membrane components, and long-chain unsaturated fatty acids can maintain the fluidity of membrane fat and normal physiological functions (Jobin et al., 2013). Low temperatures could induce the abnormal oxidation of lipids and proteins, thus destroying the structure and function of the cell membrane (Malek et al., 2004). Hence, the antioxidant system plays a crucial role in the early stage of cold stress in animals.
In this study, we observed that the up-regulation of gmo-miR-221-5p inhibits the expression of CYP2J2 and CYP4B1; these two genes were involved in linoleic acid metabolism and biological oxidation, respectively. As cytochrome P450 monooxygenases, CYP2J2 and CYP4B1 are involved in the catabolism of polyunsaturated fatty acids (PUFAs) in various tissues (Wu et al., 1996), and the oxidation reaction involving CYP2J2 could reduce the fluidity of the cell membrane (Cheng et al., 2017). Accordingly, the down-regulation of CYP2J2 and CYP4B1 reduced the catabolism and oxidation of unsaturated fatty acids, which was conducive to the maintenance of the fluidity and normal function of cell membrane in the pearl gentian grouper under cold stress. On the other hand, the down-regulation of ssa-miR-7132b-5p weakened the inhibitory effect on the TF ACSS2, which reduced the expression of EHHADH. This gene was mainly involved in the fatty acid β-oxidation (unsaturated) pathway of pearl gentian grouper. As a bifunctional peroxisomal enzyme, EHHADH contains enoyl-CoA hydratase and 3-hydroxyacylcoa dehydrogenase, catalyzing the second and third steps of fatty acid β-oxidation of ultra-long chain fatty acid (Ranea-Robles et al., 2021). Therefore, the down-regulation of EHHADH reduced the oxidation of long-chain unsaturated fatty acids. This is beneficial for the resistance of oxidative damage in the cell membrane under cold stress and for the maintenance of the stability of unsaturated fatty acids. Taken together, these miRNAs and genes may reduce the oxidation of unsaturated fatty acids in the cell membrane.
In addition, we found that the down-regulation of ssa-miR-7132b-5p indirectly up-regulated the expression of ACAT2 and HADHA by regulating the TF ACSS2. The ACAT2 gene encodes cytosolic acetoacetyl-CoA thiolase, which participates in cholesterol biosynthesis (Kursula et al., 2005). Cholesterol is indispensable in animal cell membranes; it can effectively regulate the permeability, molecular order, and elasticity of lipid membranes (Xu et al., 2018). Thus, the up-regulation of the ACAT2 detected in our study could help to maintain the steady state of cell membrane cholesterol and ensure the basic fluidity and biological function of the cell membrane under cold stress. The HADHA gene acylates monomer dicarboxylic acid into cardiolipin, which is a major membrane phospholipid in eukaryotic mitochondria and plays a key role in cell apoptosis and ATP production (Taylor et al., 2012). This suggests that the up-regulation of HADHA was conducive to maintaining mitochondrial membrane function in pearl gentian grouper, thereby ensuring the progression of the electron transport chain in mitochondrial intima and the generation of ATP to provide energy during cold stress. We further discovered that the down-regulation of ssa-miR-25-3-5p weakened the inhibitory effect on TSKU. This gene was mainly involved in the ectoderm differentiation pathway of pearl gentian grouper. TSKU can stabilize the cholesterol content by reducing the circulation of high-density lipoprotein cholesterol, the outflow capacity of cholesterol, and the conversion of cholesterol into bile acids in the liver (Niimori et al., 2014). Therefore, we infer that the up-regulation of TSKU would maintain the stability of the cholesterol content in the cell membrane of pearl gentian grouper. This ensures the fluidity of the cell membrane under cold stress conditions. In brief, these miRNAs and genes may contribute to the fluidity of the cell membrane by maintaining essential lipid components, such as cholesterol and cardiolipin.
Collectively, we conclude that the three miRNAs gmo-miR-221-5p, ssa-miR-25-3-5, and ssa-mir-7132b-5p played pivotal roles in the processes of antioxidation and the maintenance of cell membrane fluidity under cold stress in pearl gentian grouper.
Energy is a necessary and important material source during adaptation to environmental temperature changes. Under continuous low-temperature conditions, a temperature decrease will directly cause the rapid loss of energy and further affect the normal biological and behavioral processes of animals (Sun et al., 2020). Carbohydrate and lipid metabolism are the main metabolic pathways involved in resistance to cold stress in fish (Wen et al., 2018).
In our regulatory network, the up-regulation of gmo-miR-221-5p inhibited the expression of both TM7SF3 and GYS2. These two genes were involved in the positive regulation of insulin secretion and glycogen biosynthesis, respectively. TM7SF3 promotes insulin secretion, thereby contributing to the inhibition of cytokine-induced pancreatic β cell death (Beck et al., 2011). Insulin reduces the blood glucose content to normal levels by reducing the production of glucose in the liver and promoting the storage of glycogen in both the liver and muscle and triglyceride fuel in adipose tissue (Najjar, 2003). By inhibiting TM7SF3, the gmo-miR-221-5p may weaken the hypoglycemic effect of insulin in response to cold stress, thereby stabilizing the blood glucose level in pearl gentian grouper. Glycogen synthase encoded by the GYS2 gene catalyzes glycogen synthesis. This synthase promotes the transfer of glucose molecules from UDP-glucose to the terminal branch of glycogen molecules (Orho et al., 1998). Consequently, the inhibition of GYS2 expression by gmo-miR-221-5p may prevent glycogen synthesis to maintain the blood glucose level and provide necessary energy for pearl gentian grouper under cold stress via glucose metabolism. We further discovered that the down-regulation of ssa-miR-7132b-5p indirectly up-regulated both PCK1 and PCK2 by regulating the TF ACSS2. The PCK1 and PCK2 genes participate in glycolysis and gluconeogenesis and in glucose/energy metabolism, respectively. At low glucose levels, PCK1 catalyzes the conversion of oxaloacetate to phosphoenolpyruvate (PEP) and generates glucose from other precursors derived from the lactic acid and citric acid cycle (Latorre-Muro et al., 2018). PCK2 catalyzes the compensatory transformation from phosphoenolpyruvate to oxaloacetate at high glucose levels (Latorre-Muro et al., 2018). Thus, the indirect up-regulation of both PCK1 and PCK2 could jointly regulate the stability of blood glucose levels, preventing energy loss caused by cold stress in pearl gentian grouper. In summary, the above miRNAs and genes may maintain the stability of blood glucose by participating in glucose metabolism under low-temperature conditions, e.g., by reducing insulin and glycogen synthesis.
We also found that the up-regulation of ola-let-7c inhibited the expression of the TF PPARD, thus decreasing the expression of RXRAA, RXRGB, and RXRBA genes, involved in fatty acid, triacylglycerol, and ketone-body metabolism pathway. The protein encoded by TF PPARD is considered an integratory of transcription inhibition and nuclear receptor signaling, which may inhibit the transcription activities of PPARA/PPARG (Peroxisome Proliferator-Activated Receptors Alpha and Gamma) induced by RXR genes (Van Der Veen et al., 2005). PPARA/PPARG are the key regulators of lipid homeostasis (Kliewer et al., 1997) and have been implicated in the regulation of lipid metabolism and adipocyte differentiation (Gorla-Bajszczak et al., 1999). Hence, the regulatory axis of ola-let-7c, TF PPARD, and three RXR genes promoted the maintenance of PPARA/PPARG activity in pearl gentian grouper, thereby maintaining lipid homeostasis in the liver and adipocyte differentiation in low-temperature environments. Furthermore, our results showed that the down-regulation of ssa-miR-25-3-5p weakened the inhibition of NR1D2 gene, involved in the nuclear receptor transcription pathway. NR1D2 encodes a member of the nuclear hormone receptor family and participates in a lipid metabolic pathway in a heme-dependent manner as a transcriptional inhibitor (Yu et al., 2018). NR1D2 regulates lipid and energy homeostasis in the skeletal muscle by inhibiting genes associated with lipid metabolism and regulates lipid metabolism in the liver by inhibiting APOC3 (Apolipoprotein C3) (Ramakrishnan et al., 2005; Bugge et al., 2012). Therefore, the up-regulation of NR1D2 detected in pearl gentian grouper was beneficial for the regulation of lipid metabolic homeostasis in the muscle and liver and for the maintenance of basic energy to resist cold stress. In short, the miRNAs and genes described are involved in lipid metabolism-related pathways and therefore may provide energy stored in lipids for necessary biological processes.
To sum up, the four miRNAs gmo-miR-221-5p, ssa-miR-7132b-5p, ola-let-7c, and ssa-miR-25-3-5p contributed to the regulation of blood glucose levels and energy in pearl gentian grouper under cold stress by directly or indirectly regulating the expression of their corresponding target genes and TFs.
In addition to its role in lipid metabolism, NR1D2 gene also regulates the circadian rhythm of pearl gentian grouper in a heme-dependent manner. The NR1D2 could directly inhibit the expression of the core clock components ARNTL (Aryl Hydrocarbon Receptor Nuclear Translocator Like) and CRY1 (Cryptochrome Circadian Regulator 1) and form a key negative regulatory branch of the circadian rhythm (Crumbley and Burris, 2011). NR1D2 plays a pivotal role in the regulation of the circadian rhythm in mammals (Laing et al., 2015) and has been a focus of human cancer research (Tong et al., 2020). In studies of fish, NR1D2 has only been shown to play a functional role in the circadian rhythm of muscle cells, which is beneficial for resistance to cell damage (Amaral and Johnston, 2012; Wu et al., 2016). However, this gene has not been reported in studies of fish responses to cold temperatures. In this study, we found that the down-regulation of ssa-miR-25-3-5p reduced the inhibition of NR1D2 in the pearl gentian grouper. Accordingly, we concluded that the up-regulation of NR1D2 may negatively regulate the core clock and its key elements in the circadian rhythm, thereby improving resistance to cold stress damage as well as cold tolerance and preventing cold shock in extremely cold environments. To our knowledge, our results provide the first identification that NR1D2 gene improves the cold tolerance of fish via the regulation of the circadian rhythm.
Moreover, we found that the down-regulation of ccr-miR-489 reduced the inhibition of PPP1CC-A gene. PPP1CC-A cooperates with CSNK1D (Casein Kinase 1 Delta) and CSNK1E (Casein Kinase 1 Epsilon) to jointly regulate the activity and stability of circadian clock elements and to determine the length of the circadian cycle (Song et al., 2015). PPP1CC-A also regulates the normal operation of the circadian clock by regulating PER1 (Period Circadian Regulator 1) and PER2 phosphorylation. Hung et al. (2016) confirmed that the core clock gene PER2 could enhance the cold tolerance of zebrafish larvae via its regulatory association with dre-miR-29b, although this gene was not identified in our study. Therefore, we inferred that PPP1CC-A was involved in the regulation of core clock genes in the circadian rhythm of pearl gentian grouper.
The miRNAs and genes described above may prevent pearl gentian grouper from entering cold shock in the later stage of lower-temperature stress by regulating the circadian rhythm, thereby improving the ability to cope with cold environments.
Cold stress causes a free radical imbalance, and free radical accumulation leads to DNA damage and cell function damage (Qiang et al., 2018b). When low-temperature stress exceeds the tolerance range of cells, cell signaling disruptions, extensive DNA damage, and apoptosis can occur (Elmore, 2007), affecting normal physiological activities and survival.
In this study, the down-regulation of gmo-miR-10545-5p reduced the inhibitory effect on the expression of TF PPP4CB and increased the expression of PPP2R1A gene. As an important protein phosphatase, the TF PPP4CB is involved in DNA repair, DNA damage checkpoint signaling, and apoptosis (Cohen et al., 2005). In particular, it catalyzes the dephosphorylation of RPA2 (Replication Protein A2) to increase expression and mediates the effective recruitment of RAD51 (RAD51 Recombinase) to chromatin, which is an important step in DNA repair (Lee et al., 2010). PPP2R1A encodes the scaffold A subunit of PPP2CA (protein phosphatase 2 catalytic subunit α, also known as PP2A) (Yang et al., 2013). As a substrate of RIDD (IRE1α-dependent decay), PPP2R1A participates in the RIDD catalytic reaction under DNA damage, which could promote DNA repair (Xu et al., 2018). Consequently, the up-regulation of PPP2R1A could provide sufficient substrate for RIDD, conducive to the repair of DNA damage. These results suggest that TF PPP4CB and PPP2R1A gene may protect against DNA instability and damage caused by cold stress in pearl gentian grouper.
Additionally, we found that the down-regulation of gmo-miR-10545-5p indirectly inhibited the expression of PTPA gene by regulating the TF PPP4CB. PTPA is mainly involved in cyclins and cell cycle regulation pathways in pearl gentian grouper. PTPA with peptidyl prolyl isomerase activity can stimulate weak phosphoserine phosphatase activity of the serine/threonine phosphatase PP2A (Chao et al., 2006). Luo et al. (2014) reported that the down-regulation of PTPA reduced the mitochondrial membrane potential, induces Box translocation into mitochondria, and releases Cyt C, which would induce apoptosis. Accordingly, the down-regulation of PTPA detected here may promote apoptosis via mitochondrial pathways, which is useful in maintaining tissue stability in pearl gentian grouper under cold stress. In our study, the up-regulation of ccr-miR-122 was observed to inhibit the expression of FOXK2 gene, involved in apoptosis and autophagy pathway. As a transcription inhibitor of autophagy in muscle cells and fibroblasts, FOXK2 inhibits the induction of autophagy via the TF FOXO3 (Bowman et al., 2014). This suggests that the down-regulation of FOXK2 could reduce the inhibitory effect on the TF FOXO3, which may enhance autophagy (i.e., the clearance of apoptotic cells and harmful substances) of cold stress. Taken together, the above miRNAs and genes may contribute to the stress response and reduce damage in pearl gentian grouper under extreme cold stress by regulating apoptosis and autophagy, thereby maintaining the basic functions of tissues and organs.
In conclusion, miRNA gmo-miR-10545-5p and ccr-miR-122 may play important roles in DNA repair, apoptosis, and autophagy by regulating their corresponding TF and target genes, thereby improving the survival of pearl gentian grouper in the later stage of low-temperature stress.
In this study, DE miRNAs, mRNAs, and TFs in pearl gentian grouper were screened, and a candidate miRNA-TF-mRNA regulatory network related to cold stress was constructed. The network involved two regulatory mechanisms, i.e., four miRNAs directly regulated eight target genes and three miRNAs indirectly regulated ten target genes via the regulation of three TFs. These miRNAs, TFs, and mRNAs played essential roles in several important pathways and biological reactions in pearl gentian grouper under cold stress, such as antioxidant and membrane fluidity, glucose and lipid metabolism, circadian rhythm regulation, DNA repair, and autophagy. In short, the two regulatory mechanisms revealed in pearl gentian grouper protected against damage caused by cold stress and promoted the maintenance of normal physiological activity. Of note, the potential function of NR1D2 gene in cold tolerance in fish via the regulation of the circadian rhythm was identified for the first time in this study. The complex regulatory relationships revealed in the present study not only provide a deeper understanding of the molecular mechanism underlying the adaptation of pearl gentian grouper to low-temperature environments but also provide clear directions for cultivating cold-tolerant varieties in the future.
We detected a number of TFs with important roles as bridges in our regulatory network. However, our prediction analyses revealed only a few TFs involved in miRNA regulation for further pathways analyses. Since the miRNA-mRNA regulatory relationship was the focus of our work, further studies of the detailed regulatory roles of additional TFs are needed, including ATAC-Seq studies of TFs in pearl gentian grouper and analyses of regulatory effects on target genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA779537).
The animal study was reviewed and approved by the Animal Experimental Ethics Committee of Guangdong Ocean University.
B-BM, R-XW, and S-FN: conceptualization, validation, resources, writing original draft preparation, writing review and editing, and visualization. S-FN and B-BM: methodology and software and data curation. B-BM and R-XW: formal analysis. B-BM, Z-BL, and YZ: investigation. R-XW and S-FN: supervision, project administration, and funding acquisition.
This work was supported by the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), China (Grant No. ZJW-2019-06), the Science and Technology Planning Project of Guangdong Province, China (Grant No. 2017A030303077), the National Natural Science Foundation of China (Grant No. 31372532), and the Special Program for Outstanding Young Teachers of Guangdong Ocean University, China (Grant No. HDYQ2017002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Zhanjiang Hist Aquatic Technology Co., Ltd. for providing experimental materials and sites.
The Supplementary Material for this article can be found online at: https://doi.org/10.6084/m9.figshare.17693360.v1
Agarwal, V., Bell, G. W., Nam, J. W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. doi: 10.7554/eLife.05005
Amaral, I. P. G., and Johnston, I. A. (2012). Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302:2011. doi: 10.1152/ajpregu.00367.2011
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2006). Gene ontology: tool for the unification of biology. Nat. Genet. 34, 1535–1540.
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Beck, A., Isaac, R., Lavelin, I., Hart, Y., Volberg, T., Shatz-Azoulay, H., et al. (2011). An siRNA screen identifies transmembrane 7 superfamily member 3 (TM7SF3), a seven transmembrane orphan receptor, as an inhibitor of cytokine-induced death of pancreatic beta cells. Diabetologia 54, 2845–2855. doi: 10.1007/s00125-011-2277-3
Bowman, C. J., Ayer, D. E., and Dynlacht, B. D. (2014). Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat. Cell Biol. 16, 1202–1214. doi: 10.1038/ncb3062
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Bugge, A., Feng, D., Everett, L. J., Briggs, E. R., Mullican, S. E., Wang, F., et al. (2012). Rev-erbα and rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667. doi: 10.1101/gad.186858.112
Bushati, N., and Cohen, S. M. (2007). MicroRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205. doi: 10.1146/annurev.cellbio.23.090506.123406
Chao, Y., Xing, Y., Chen, Y., Xu, Y., Lin, Z., Li, Z., et al. (2006). S structure and mechanism of the phosphotyrosyl phosphatase activator. Mol. Cell 23, 535–546. doi: 10.1016/j.molcel.2006.07.027
Cheng, C. H., Ye, C. X., Guo, Z. X., and Wang, A. L. (2017). Immune and physiological responses of pufferfish (Takifugu obscurus) under cold stress. Fish Shellfish Immunol. 64, 137–145. doi: 10.1016/j.fsi.2017.03.003
Cohen, P. T. W., Philp, A., and Vázquez-Martin, C. (2005). Protein phosphatase 4from obscurity to vital functions. FEBS Lett. 579, 3278–3286. doi: 10.1016/j.febslet.2005.04.070
Crumbley, C., and Burris, T. P. (2011). Direct regulation of CLOCK expression by REV-ERB. PLoS One 6:17290. doi: 10.1371/journal.pone.0017290
Donaldson, M. R., Cooke, S. J., Patterson, D. A., and Macdonald, J. S. (2008). Cold shock and fish. J. Fish Biol. 73, 1491–1530. doi: 10.1111/j.1095-8649.2008.02061.x
Elmore, S. (2007). Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516. doi: 10.1080/01926230701320337
Ernst, J., and Bar-Joseph, Z. (2006). STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:1–11. doi: 10.1186/1471-2105-7-191
Fan, B., Liu, X. C., Meng, Z. N., Tan, B. H., Wang, L., Zhang, H. F., et al. (2014). Cryopreservation of giant grouper Epinephelus lanceolatus (bloch, 1790) sperm. J. Appl. Ichthyol. 30, 334–339. doi: 10.1111/jai.12321
Friedländer, M. R., MacKowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gabbianelli, R., Lupidi, G., Villarini, M., and Falcioni, G. (2003). DNA damage induced by copper on erythrocytes of gilthead sea bream sparus aurata and mollusk scapharca inaequivalvis. Arch. Environ. Contam. Toxicol. 45, 350–356. doi: 10.1007/s00244-003-2171-1
Gorla-Bajszczak, A., Juge-Aubry, C., Pernin, A., Burger, A. G., and Meier, C. A. (1999). Conserved amino acids in the ligand-binding and τ (i) domains of the peroxisome proliferator-activated receptor α are necessary for heterodimerization with RXR. Mol. Cell. Endocrinol. 147, 37–47. doi: 10.1016/S0303-7207(98)00217-2
Gracey, A. Y., Fraser, E. J., Li, W., Fang, Y., Taylor, R. R., Rogers, J., et al. (2004). Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Natl. Acad. Sci. U.S.A. 101, 16970–16975. doi: 10.1073/pnas.0403627101
Hazel, J. R., and Williams, E. E. (1990). The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 29, 167–227. doi: 10.1016/0163-7827(90)90002-3
Hu, H., Miao, Y. R., Jia, L. H., Yu, Q. Y., Zhang, Q., and Guo, A. Y. (2019). AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38. doi: 10.1093/nar/gky822
Hung, I. C., Hsiao, Y. C., Sun, H. S., Chen, T. M., and Lee, S. J. (2016). MicroRNAs regulate gene plasticity during cold shock in zebrafish larvae. BMC Genom. 17:922. doi: 10.1186/s12864-016-3239-4
Jobin, M. L., Bonnafous, P., Temsamani, H., Dole, F., Grélard, A., Dufourc, E. J., et al. (2013). The enhanced membrane interaction and perturbation of a cell penetrating peptide in the presence of anionic lipids: toward an understanding of its selectivity for cancer cells. Biochim. Biophys. Acta Biomembr. 1828, 1457–1470. doi: 10.1016/j.bbamem.2013.02.008
Joseph, S. R., Pálfy, M., Hilbert, L., Kumar, M., Karschau, J., Zaburdaev, V., et al. (2017). Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, 1–31. doi: 10.7554/eLife.23326
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kliewer, S. A., Sundseth, S. S., Jones, S. A., Brown, P. J., Wisely, G. B., Koble, C. S., et al. (1997). Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. U.S.A. 94, 4318–4323. doi: 10.1073/pnas.94.9.4318
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). MiRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kursula, P., Sikkilä, H., Fukao, T., Kondo, N., and Wierenga, R. K. (2005). High resolution crystal structures of human cytosolic thiolase (CT): a comparison of the active sites of human CT, bacterial thiolase, and bacterial KAS I. J. Mol. Biol. 347, 189–201. doi: 10.1016/j.jmb.2005.01.018
Laing, E. E., Johnston, J. D., Möller-Levet, C. S., Bucca, G., Smith, C. P., Dijk, D. J., et al. (2015). Exploiting human and mouse transcriptomic data: identification of circadian genes and pathways influencing health. BioEssays 37, 544–556. doi: 10.1002/bies.201400193
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Latorre-Muro, P., Baeza, J., Armstrong, E. A., Hurtado-Guerrero, R., Corzana, F., Wu, L. E., et al. (2018). Ddynamic acetylation of phosphoenolpyruvate carboxykinase toggles enzyme activity between gluconeogenic and anaplerotic reactions. Mol. Cell 71, 718–732.e9. doi: 10.1016/j.molcel.2018.07.031
Lee, D. H., Pan, Y., Kanner, S., Sung, P., Borowiec, J. A., and Chowdhury, D. (2010). A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 17, 365–372. doi: 10.1038/nsmb.1769
Li, L. J., Zhao, W., Tao, S. S., Li, J., Xu, S. Z., Wang, J. B., et al. (2017). Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell. Immunol. 319, 17–27. doi: 10.1016/j.cellimm.2017.06.004
Li, M., Zhao, L., Page-McCaw, P. S., and Chen, W. (2016). Zebrafish genome genome engineering engineering using using the CRISPR–Cas9 systemsystem. Trends Genet. 32, 815–827. doi: 10.1016/j.tig.2016.10.005
Liang, H. F., Huang, D. K., Wu, Y. H., Wang, C. Q., and Zhong, W. J. (2013). Effects of temperature and salinity on survival and food intake of grouper hybrid (Epinephelus lanceolatus ♀× E. fuscoguttatus ♂). J. Guangdong Ocean Univ. 33, 22–26.
Lin, Y. C., Chao, T. Y., Yeh, C. T., Roffler, S. R., Kannagi, R., and Yang, R. B. (2017). Endothelial SCUBE2 interacts with VEGFR2 and regulates VEGF-induced angiogenesis. Arterioscler. Thromb. Vasc. Biol. 37, 144–155. doi: 10.1161/ATVBAHA.116.308546
Liu, C., Schläppi, M. R., Mao, B., Wang, W., Wang, A., and Chu, C. (2019). The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 17, 1834–1849. doi: 10.1111/pbi.13104
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Lovering, R. C., Gaudet, P., Acencio, M. L., Ignatchenko, A., Jolma, A., Fornes, O., et al. (2021). A GO catalogue of human DNA-binding transcription factors. Biochim. Biophys. Acta Gene Regul. Mech. 1864:194765. doi: 10.1016/j.bbagrm.2021.194765
Luo, S. W., Cai, L., Liu, Y., and Wang, W. N. (2014). Functional analysis of a dietary recombinant fatty acid binding protein 10 (FABP10) on the Epinephelus coioides in response to acute low temperature challenge. Fish Shellfish Immunol. 36, 475–484. doi: 10.1016/j.fsi.2013.12.028
Lv, D. K., Bai, X., Li, Y., Ding, X. D., Ge, Y., Cai, H., et al. (2010). Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459, 39–47. doi: 10.1016/j.gene.2010.03.011
Malek, R. L., Sajadi, H., Abraham, J., Grundy, M. A., and Gerhard, G. S. (2004). The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Arative Biochemistry Biochem. Physiol. C Toxicol. Toxicol. Pharmacol. 138, 363–373. doi: 10.1016/j.cca.2004.08.014
Miao, B. B., Niu, S. F., Wu, R. X., Liang, Z. B., Tang, B. G., Zhai, Y., et al. (2021). Gene expression profile and co-expression network of pearl gentian grouper under cold stress by integrating illumina and pacbio sequences. Animals 11:1745. doi: 10.3390/ani11061745
Najjar, S. (2003). Insulin action: molecular basis of diabetes. eLS 1–10. doi: 10.1038/npg.els.0001402
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. doi: 10.1093/nar/gku1063
Nie, M., Tan, X., Lu, Y., Wu, Z., Li, J., Xu, D., et al. (2019). Network of microRNA-transcriptional factor-mRNA in cold response of turbot Scophthalmus maximus. Fish Physiol. Biochem. 45, 583–597. doi: 10.1007/s10695-019-00611-y
Niimori, D., Kawano, R., Niimori-Kita, K., Ihn, H., and Ohta, K. (2014). Tsukushi is involved in the wound healing by regulating the expression of cytokines and growth factors. J. Cell Commun. Signal. 8, 173–177. doi: 10.1007/s12079-014-0241-y
Orho, M., Bosshard, N. U., Buist, N. R. M., Gitzelmann, R., Aynsley-Green, A., Blümel, P., et al. (1998). Mutations in the liver glycogen synthase gene in children with hypoglycemia due to glycogen storage disease type O. J. Clin. Invest. 102, 507–515. doi: 10.1172/JCI2890
Qiang, J., Cui, Y. T., Tao, F. Y., Bao, W. J., He, J., Li, X. H., et al. (2018a). Physiological response and microRNA expression profiles in head kidney of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to acute cold stress. Sci. Rep. 8, 1–15. doi: 10.1038/s41598-017-18512-6
Qiang, J., Tao, Y. F., Bao, J. W., Chen, D. J., Li, H. X., He, J., et al. (2018b). High fat diet -induced miR - 122 regulates lipid metabolism and fat deposition in genetically improved farmed tilapia (GIFT, Oreochromis niloticus) liver. Front. Physiol. 9:1–11. doi: 10.3389/fphys.2018.01422
Ramakrishnan, S. N., Lau, P., Burke, L. J., and Muscat, G. E. O. (2005). Rev-erbβ regulates the expression of genes involved in lipid absorption in skeletal muscle cells: Evidence for cross-talk between orphan nuclear receptors and myokines. J. Biol. Chem. 280, 8651–8659. doi: 10.1074/jbc.M413949200
Ranea-Robles, P., Violante, S., Argmann, C., Dodatko, T., Bhattacharya, D., Chen, H., et al. (2021). Murine deficiency of peroxisomal l-bifunctional protein (EHHADH) causes medium-chain 3-hydroxydicarboxylic aciduria and perturbs hepatic cholesterol homeostasis. Cell. Mol. Life Sci. 78, 5631–5646. doi: 10.1007/s00018-021-03869-9
Riffo-Campos, ÁL., Riquelme, I., and Brebi-Mieville, P. (2016). Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 17:987. doi: 10.3390/ijms17121987
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shapawi, R., Ebi, I., Yong, A. S. K., and Ng, W. K. (2014). Optimizing the growth performance of brown-marbled grouper, Epinephelus fuscoguttatus (forskal), by varying the proportion of dietary protein and lipid levels. Anim. Feed Sci. Technol. 191, 98–105. doi: 10.1016/j.anifeedsci.2014.01.020
Song, H., Pu, J., Wang, L., Wu, L., Xiao, J., Liu, Q., et al. (2015). ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy 11, 1308–1325. doi: 10.1080/15548627.2015.1060386
Sun, J. L., Zhao, L. L., Wu, H., Lian, W. Q., Cui, C., Du, Z. J., et al. (2019). Analysis of miRNA-seq in the liver of common carp (Cyprinus carpio L.) in response to different environmental temperatures. Funct. Integr. Genomics 19, 265–280. doi: 10.1007/s10142-018-0643-7
Sun, J. L., Zhao, L. L., Wu, H., Liu, Q., Liao, L., Luo, J., et al. (2020). Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci. Total Environ. 713:135157. doi: 10.1016/j.scitotenv.2019.135157
Taylor, W. A., Mejia, E. M., Mitchell, R. W., Choy, P. C., Sparagna, G. C., and Hatch, G. M. (2012). Human Trifunctional trifunctional protein protein alpha alpha links links Cardiolipin cardiolipin remodeling remodeling to betabeta-oxidationoxidation. PLoS One 7:628. doi: 10.1371/journal.pone.0048628
Tong, H., Liu, X., Li, T., Qiu, W., Peng, C., Shen, B., et al. (2020). Nr1d2 accelerates hepatocellular carcinoma progression by driving the epithelial-to-mesenchymal transition. Onco. Targets. Ther. 13, 3931–3942. doi: 10.2147/OTT.S237804
Van Der Veen, J. N., Kruit, J. K., Havinga, R., Baller, J. F. W., Chimini, G., Lestavel, S., et al. (2005). Reduced cholesterol absorption upon PPARδ activation coincides with decreased intestinal expression of NPC1L1. J. Lipid Res. 46, 526–534. doi: 10.1194/jlr.M400400-JLR200
Wen, B., Jin, S. R., Chen, Z. Z., and Gao, J. Z. (2018). Physiological responses to cold stress in the gills of discus fish (Symphysodon aequifasciatus) revealed by conventional biochemical assays and GC-TOF-MS metabolomics. Sci. Total Environ. 640–641, 1372–1381. doi: 10.1016/j.scitotenv.2018.05.401
Wen, M., Shen, Y., Shi, S., and Tang, T. (2012). MiREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 13:140. doi: 10.1186/1471-2105-13-140
Wu, P., Li, Y. L., Cheng, J., Chen, L., Zhu, X., Feng, Z. G., et al. (2016). Daily rhythmicity of clock gene transcript levels in fast and slow muscle fibers from Chinese perch (Siniperca chuatsi). BMC Genom. 17:1008. doi: 10.1186/s12864-016-3373-z
Wu, S., Moomaw, C. R., Tomer, K. B., Falck, J. R., and Zeldin, D. C. (1996). Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J. Biol. Chem. 271, 3460–3468. doi: 10.1074/jbc.271.7.3460
Xu, D., You, Q., Chi, C., Luo, S., Song, H., Lou, B., et al. (2018). Transcriptional response to low temperature in the yellow drum (Nibea albiflora) and identification of genes related to cold stress. Comp. Biochem. Physiol. Part D Genom. Proteom. 28, 80–89. doi: 10.1016/j.cbd.2018.07.003
Yan, B., Zhao, L. H., Guo, J. T., and Zhao, J. L. (2012). MiR-429 regulation of osmotic stress transcription factor 1 (OSTF1) in tilapia during osmotic stress. Biochem. Biophys. Res. Commun. 426, 294–298. doi: 10.1016/j.bbrc.2012.08.029
Yang, R., Dai, Z., Chen, S., and Chen, L. (2011). MicroRNA-mediated gene regulation plays a minor role in the transcriptomic plasticity of cold-acclimated Zebrafish brain tissue. BMC Genom. 12:605. doi: 10.1186/1471-2164-12-605
Yang, R., Yang, L., Qiu, F., Zhang, L., Wang, H., Yang, X., et al. (2013). Functional genetic polymorphisms in PP2A subunit genes confer increased risks of lung cancer in southern and eastern Chinese. PLoS One 8:285. doi: 10.1371/journal.pone.0077285
Yoneda, M., and Wright, P. J. (2005). Effects of varying temperature and food availability on growth and reproduction in first-time spawning female Atlantic cod. J. Fish Biol. 67, 1225–1241. doi: 10.1111/j.1095-8649.2005.00819.x
Yu, M., Li, W., Wang, Q., Wang, Y., and Lu, F. (2018). Circadian regulator NR1D2 regulates glioblastoma cell proliferation and motility. Oncogene 37, 4838–4853. doi: 10.1038/s41388-018-0319-8
Keywords: cold stress, pearl gentian grouper, miRNA-TF-mRNA, regulatory network, miRNA expression profile
Citation: Miao B-B, Niu S-F, Wu R-X, Liang Z-B and Zhai Y (2022) The MicroRNAs-Transcription Factors-mRNA Regulatory Network Plays an Important Role in Resistance to Cold Stress in the Pearl Gentian Grouper. Front. Mar. Sci. 8:824533. doi: 10.3389/fmars.2021.824533
Received: 30 November 2021; Accepted: 27 December 2021;
Published: 02 February 2022.
Edited by:
Jose Fernando Lopez-Olmeda, University of Murcia, SpainCopyright © 2022 Miao, Niu, Wu, Liang and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ren-Xie Wu, d3VyZW54aWVAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.