- 1Department of Biological Sciences, Bowling Green State University, Bowling Green, OH, United States
- 2Division of Marine Science and Conservation, Nicholas School of the Environment, Duke University Marine Laboratory, Duke University, Beaufort, NC, United States
Plastics of various chemistries pollute global water bodies. Toxic chemicals leach with detrimental and often unpredictable impacts on the surrounding ecosystems. We found that seawater leachates of plastic pre-production pellets from 7 recycle categories are acutely toxic to stage II barnacle nauplii; lethal concentration 50 (LC50s) were observed in 24-h leachates from dilutions ranging from 0.007 to 2.1 mg/mL of seawater. Based on previous observations that macro-organismal settlement on fouling management coatings of various toxicities can be used to predict the toxicity of the coating, we hypothesized that interaction of plastic pre-production pellets with emerging microbiomes would exhibit patterns indicative of the chemistry at the pellet surface. We used amplicon sequencing of bacterial 16S ribosomal RNA genes to characterize the microbiomes that developed from 8 through 70 days on pellets exposed to the same flowing ambient seawater. Diversity and composition of the microbiomes colonizing plastic pellets changed over time and varied with plastic type. Microbial taxa belong to taxonomic groups known to consume hydrocarbons, to be prevalent following marine oil spills, or to live on fouling management surfaces. Microbiomes were still distinct between plastic types at Day 70, suggesting that differences in the physicochemical characteristics of the underlying plastics continue to exert variable selection of surface microbial communities. A random forest-based sample classifier correctly predicted 93% of plastic types using microbiome compositions. Surface microbiomes have promise for use in forensically identifying plastic types and potential toxicities.
Introduction
Marine plastic pollution is a dire environmental concern throughout the global oceans, with its scale constantly growing (Jambeck et al., 2015; Brandon et al., 2019; Lau et al., 2020). Between the years 2016 and 2040, an estimated 710 million metric tons of plastic are predicted to enter the environment, even if immediate and drastic action from all stakeholders is taken to curb plastic pollution (Lau et al., 2020). While the term “plastic” conjures the sense of a single inert polymer, plastics contain a “diverse chemical cocktail” (Rochman et al., 2019) of hundreds of molecules (e.g., heavy metals, organic pollutants such as polychlorinated biphenyls and polycyclic aromatic hydrocarbons) introduced during manufacturing or adsorbed during use and disposal that leach out of various polymer backbones (Rochman et al., 2013, 2014, 2019; Li et al., 2016a; Andrady, 2017). Further, environmental forces (e.g., oxidation, UV light, physical weathering) can alter the chemistry of additives and increase the rate of leaching (Teuten et al., 2009; Haider and Karlsson, 2000; Suhrhoff and Scholz-Böttcher, 2016; Walker et al., 2021). Due to these processes, plastics are dynamic, with both their surface chemistries and leachate compositions changing over time and generating novel environments. That plastics are chemically active and dynamic underscores the need to understand their interactions and toxicity in the marine environment (Diana et al., 2020).
Plastic debris in marine and freshwater environments provide new surfaces for colonization by microbes and macro-organisms. Sometimes referred to as “the Plastisphere” (Zettler et al., 2013), plastics and the communities that assemble on their surfaces constitute novel ecosystems unique from that of the surrounding seawater and other marine particles (De Tender et al., 2015; Kirstein et al., 2016; Oberbeckmann et al., 2016; Debroas et al., 2017), with limited overlap of microbial taxa between communities on plastic particles and the surrounding seawater (Zettler et al., 2013). With ever-growing plastic pollution in global water bodies and the recalcitrance of plastics whether at the surface, in the water column or on ocean sediments (Arthur et al., 2009; Jahnke et al., 2017; Worm et al., 2017), the plastisphere ecosystems will continue to increase in area in the future.
Microbiomes contain a wealth of information about their environment and other factors governing their diversity and membership. Microbes colonize every exposed surface in aqueous environments to form diverse biofilm communities of bacteria, diatoms, protozoa, and extracellular polymeric substances (Zobell and Allen, 1935; Dobretsov et al., 2009; Hadfield, 2010). Initial attachment, growth, and ultimate structure of biofilms is influenced by numerous factors, including surface chemistry (Baier, 1970; Meyer et al., 1988; Teughels et al., 2006), presence of toxins, toxicants and compounds detrimental to microbial growth (Ostuni et al., 2001), resource availability, environmental conditions, and biotic interactions such as competition and cooperation/cross-feeding (Shikuma and Hadfield, 2005, 2010; Hadfield, 2010). Accordingly, researchers may examine the dissimilarities between microbial community composition on various plastic types (including changes over time) to potentially inform differences in chemistries at the surface.
Starting with one of the first published reports on the microbiomes of marine plastic debris (Zettler et al., 2013), most plastic microbiome studies have been based on plastics collected from the ocean and other water bodies (e.g., Carson et al., 2013; Zettler et al., 2013; Reisser et al., 2014; De Tender et al., 2015; Debroas et al., 2017; Roager and Sonnenschein, 2019; Amaral-Zettler et al., 2020; Bhagwat et al., 2021). While study sampling locations have spanned much of the world’s oceans, including transects of the Atlantic (Zettler et al., 2013) and Pacific Oceans (Carson et al., 2013), the North Sea (De Tender et al., 2015), Baltic Sea (Roager and Sonnenschein, 2019), Mediterranean Sea (Roager and Sonnenschein, 2019), oceans surrounding Australia (Reisser et al., 2014), and a saltwater lake (Bhagwat et al., 2021) among others (see review by Roager and Sonnenschein, 2019), they have all consistently revealed distinct microbiomes from surrounding seawater. Many studies have focused on plastics harvested from surface waters (e.g., Zettler et al., 2013; Debroas et al., 2017), though others examined the microbiomes of plastic found on beaches (e.g., De Tender et al., 2015; Curren and Leong, 2019). Due to buoyancy, debris loading patterns and ocean currents, the representation of plastic recycling categories [e.g., polyethylene (PE), polypropylene (PP), polyethylene terephthalate (PET), polystyrene (PS)] identified varies across studies, though PE is one of the most collected plastics (Roager and Sonnenschein, 2019), consistent with PE being the most commonly produced non-fiber plastic making up 36% of the total (Geyer et al., 2017). The size of the plastics collected varies between studies. Despite the differences between studies, dominant bacterial families are: Flavobacteriaceae, Saprospiraceae, Hyphomonadaceae, Rhodobacteraceae, Erythrobacteraceae, Sphingomonadaceae, Comamonadaceae, Alcanivoracaceae, Pseudoalteromonadaceae, Oceanospirillaceae, and Vibrionaceae (Roager and Sonnenschein, 2019).
Since most studies of plastic microbiomes are on plastic debris collected from water bodies (Carson et al., 2013; Zettler et al., 2013; Reisser et al., 2014; Amaral-Zettler et al., 2020), “life histories” do not exist and thus we do not know about incubation times, initial entry location, and many other factors. Experimental incubations have been conducted but have been limited to tracking plastic colonization over the first few days (Dang et al., 2008) or for one fixed exposure time (Artham et al., 2009; Oberbeckmann et al., 2015). These snapshots miss the temporal aspect of microbiome development in concordance with plastic leaching and weathering. Additional studies are needed to constrain our understanding of microbiome development on various plastic types from initial colonization to later successional stages.
Here, we have used microbiome techniques to test the hypothesis that microbiomes reflect the nature of the plastic on which they are growing and can therefore be used to forensically determine plastic composition. We base this hypothesis upon a Rittschof and Holm (1997) report that macro-fouling communities reflect the chemical nature of the fouling management coating that they have colonized. Rittschof and Holm (1997) demonstrated that a dominant mechanism of antifouling could be predicted by mapping the macrofouling communities on the coatings with unknown mechanisms of action onto coatings with known mechanisms of action. When this was done, “non-toxic foul release coatings” with highly toxic organo-tin catalysts aligned with highly toxic fouling management coatings and explained part of the mechanism of action of the foul release coatings. Similar to antifouling coatings, plastics have distinctive chemical compositions, surface chemistries and release of toxic leachate molecules, all of which are likely to influence the fouling microbial communities.
Here, we use plastic pre-production pellets to test the hypothesis. Plastic pre-production pellets are the starting material for generating macroplastics such as packaging, containers, utensils, and pipes. The pellets have contaminants from processing and generally trade secret additives that include plasticizers, mold release compounds, UV protection, antioxidants etc., that yield products with desired characteristics. Pre-production pellets are distinctly different from other kinds of similarly named microplastics. The best example is polystyrene microspheres, which are engineered particles that are in multiple micron size ranges, contain anti-aggregation chemistries, and are delivered in solutions with preservatives (Yu and Chan, 2020a). Microspheres are small enough to be consumed by larvae and some can enter tissues of larvae (Yu and Chan, 2020b) and fish. In contrast, pre-production pellets are 2–4 mm in all dimensions, are delivered dry (not in preservatives), are too large to be consumed by larvae and are ingested and egested by larger animals without entering tissues (Allen et al., 2017; Diana et al., 2020).
We found that all 7 recycle categories (resin identification codes) of plastic pellets leach molecules that are acutely toxic to barnacle larvae. To assess the relationships between plastic leaching and the surface microbiomes, we examined the development of microbiomes on plastic pre-production pellets incubated in flowing seawater over 70 days. We sampled the microbiomes at 8, 16, 24, 32, and 70 days, and determined their microbiomes using 16S rRNA gene sequencing. We hypothesized that the microbiomes that initially develop on the pre-production pellets would be different because each plastic type has slightly different surface energy and contains different additives and contaminants, yet when pellets cease leaching molecules their microbiomes should become similar. However, we observed that communities remained different throughout the experiment, even after 70 days of exposure.
Materials and Methods
Plastic Pre-production Pellets
Pre-production pellets were obtained from Chase Plastic Services (Clarkston, Michigan). The seven recycling categories were: high-density polyethylene (HDPE, translucent), low-density polyethylene (LDPE, translucent), polycarbonate (PC, clear), polypropylene (PP, translucent), polyvinyl chloride (PVC, white), polystyrene (PS, clear), and polyethylene terephthalate (PET, blue). Pellets ranged in size from 2 to 4 mm in at least one dimension. There was no substantial variation in pellets’ surface area-to-volume ratio between recycle categories.
Toxicity of Pre-production Pellets
We determined the concentration of 24-h leachate that results in the 50% mortality (LC50) of stage II nauplii of the striped barnacle Amphibalanus amphitrite, following the methods described in Rittschof et al. (1992). From extensive study of fouling management coatings (Rittschof and Holm, 1997) and work with polydimethylsiloxane (PDMS) networks (Holm et al., 2005; Rittschof et al., 2008, 2011), we know that constituents in the polymers leach out rendering coatings toxic for variable periods of time until the leach rates are too low to detect. In these experiments, we leached known weights of each of the seven recycle types of plastic pre-production pellets in filtered, aged seawater (ASW) for 24 h and tested a series of dilutions (0, 0.001, 0.005, 0.01, 0.1, 1.0 mg/mL) to determine the dose-response relationship of naupliar mortality for each recycle type. We added 16–35 nauplii per 3 mL tube. A custom Basic program for probit analysis was used to calculate LC50 values. Determination of death in nauplii is difficult since the only vital sign is movement. Since healthy nauplii swim continuously, our endpoint dead includes immobile larvae and moribund larvae, those that do not swim continuously because earlier work showed these larvae imminently die (Rittschof et al., 1992). All larvae were transferred from test tube to a Bogorov tray and the number of larvae swimming forward and moribund/dead nauplii counted. Larvae with impaired swimming were considered moribund and counted as dead. The percent mortality was determined at each concentration tested for each plastic type and the ASW-only control.
Seawater Incubation of Plastic Pre-production Pellets
Fourteen glass mason jars were used to expose a known number of each of the 7 recycle types of plastic pre-production pellets (two replicates) to seawater, for 70 days, beginning in August 2017. During the first 12 days of the assay, pellets were in 500 mL seawater with air and water was changed every day; after that time, the mason jars were covered with a fiberglass standard U.S. window screen (18 × 16) to prevent pellets from escaping the jars due to water flow. Jars were exposed to approximately 10 mL per minute flows of the same single-pass sand-filtered ambient seawater until the end of the experiment (Day 70). Over the duration of the experiment, the seawater had an ambient temperature between 28 and 24°C (at 0:00 GMT according to the NOAA station 8656483 Beaufort, Duke Marine Lab, NC) and a salinity between 28.2 and 35.6 PSU (courtesy of Piver’s Island Coastal Observatory).
A 2.5 × 5.5 cm glass vial filled to 3 cm high with pellets from each of the 7 recycle types of plastic pre-production pellets was used as a measure. The number of pellets contained in the 3 cm was counted and multiplied by the number of vials that would be added to each mason jars; the total weight of the pellets to be used in the assay was obtained. Four vials for each of the recycle types of plastic pre-production pellets were added to their respective mason jars except for PET, where only one measure of pellets was added due to insufficient pellets. At each sampling interval (8, 16, 24, 32, and 70 days), pellets were removed and stored at −80°C until DNA extraction.
DNA Extraction
Genomic DNA from incubated plastic pre-production pellet samples was extracted in semi-replicated fashion, in which 5 pellets of each type were processed in a first series and then repeated in a second series. DNA extraction was performed using the DNeasy PowerSoil Kit (Qiagen, Germantown, MD, United States) according to manufacturer’s protocol. In brief, pellets were added to individual PowerBead tubes with flame-sterilized forceps. Samples were incubated in sodium dodecyl sulfate (SDS)-containing buffer to lyse microbial cells, followed by vortexing for 10 min. A series of buffer additions was performed to remove cellular debris, PCR inhibitors and other contaminants. Solutions were transferred to spin columns, where nucleic acids bind to silica matrix through a subsequent wash step. In the final step, DNA was eluted with a Tris-based buffer. Extracted DNA was stored at −80°C until sequencing.
Amplicon Sequencing and Sequence Processing
The bacterial community compositions of plastic microbiomes were characterized by 16S rRNA amplicon sequencing at the Duke Center for Genomic and Computational Biology (Durham, NC). Amplicons spanning the V3–V4 variable regions were generated using the Klindworth primer set (Klindworth et al., 2013; forward primer 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG GGNGGCWGCAG; reverse primer 5′ GTCTCGTGGGCTC GGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAA TCC) and unique barcodes for multiplex sequencing, following the Illumina 16S Metagenomic Sequencing Library Preparation protocol (Illumina, 2013). PCR reactions were thermocycled as following: 98 °C for 3 min; and 25 cycles at 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; with a final extension at 72 °C for 5 min. Concentrations of amplicon libraries were accessed using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Q32854) and a Promega GloMax plate reader. Libraries were pooled in equimolar amounts prior to sequencing. Sequencing was performed by the Duke Sequencing and Genomic Technologies (SGT) shared resource on an Illumina MiSeq instrument configured for 300 base-pair (bp) paired-end sequencing runs.
Sequences were demultiplexed and assigned to corresponding samples using CASAVA (Illumina). Sequences were processed using USEARCH v9 (Edgar, 2010) as previously described (Ward et al., 2017). Briefly, low-quality sequence ends were trimmed at Phred quality (Q) of 30 using a 10-bp running window. Paired-end reads were merged when reads had a 10 bp overlap or greater with no mismatches, and the resulting merged sequences were then filtered to remove reads with more than 1 expected error or less than 400 nucleotides in length. Singleton sequences were removed before the remaining sequences were assigned to operational taxonomic units (OTU) of 100% pairwise identity (zero-radius OTU; zOTU) using the UNOISE3 denoising algorithm (Edgar, 2017); chimeric sequences were excluded during this step. This process yielded a total of 7,293 zOTUs and 2,843,481 sequences in the dataset.
Microbiome Characterization and Community Analyses
The processed sequence dataset was imported to R v3.6.2 for community diversity analysis using vegan v2.5-6 (Dixon, 2003) and phyloseq v1.30.0 packages (McMurdie and Holmes, 2013). The taxonomies of 16S rRNA gene sequences were classified using the RDP naïve Bayesian classifier using the Greengenes v13.8 database. Prior to alpha diversity comparisons, libraries were sub-sampled to equal rarefaction depths. Richness (number of zOTUs) and Shannon Diversity Index (SDI) was calculated for all samples. Non-metric dimensional scaling (NMDS) on Bray-Curtis dissimilarities was performed to assess and visualize the temporal dynamics of microbial communities from all plastic categories. A cluster dendrogram was constructed based on Bray-Curtis dissimilarity and K-means clustering (k = 4). For family-level dynamics, the 10 most abundant families across the entire dataset were selected. Their relative abundances were used to generate bar plots of averages and standard error bars, faceted by plastic type and experiment day. The sequence of the abundant diatom chloroplast OTU was queried via Nucleotide BLAST (NCBI; National Library of Medicine, 1988). A machine learning sample classifier was performed in QIIME2 using plastic type and experimental day as the separate predictors. Prior to this step, rare OTUs were removed using a cutoff of 0.1% of the mean sample depth (i.e., 42,000 sequences). The QIIME2 “sample-classifier” function was implemented using the Random Forest Classifier estimator with 100 trees grown.
Results
Naupliar Toxicity
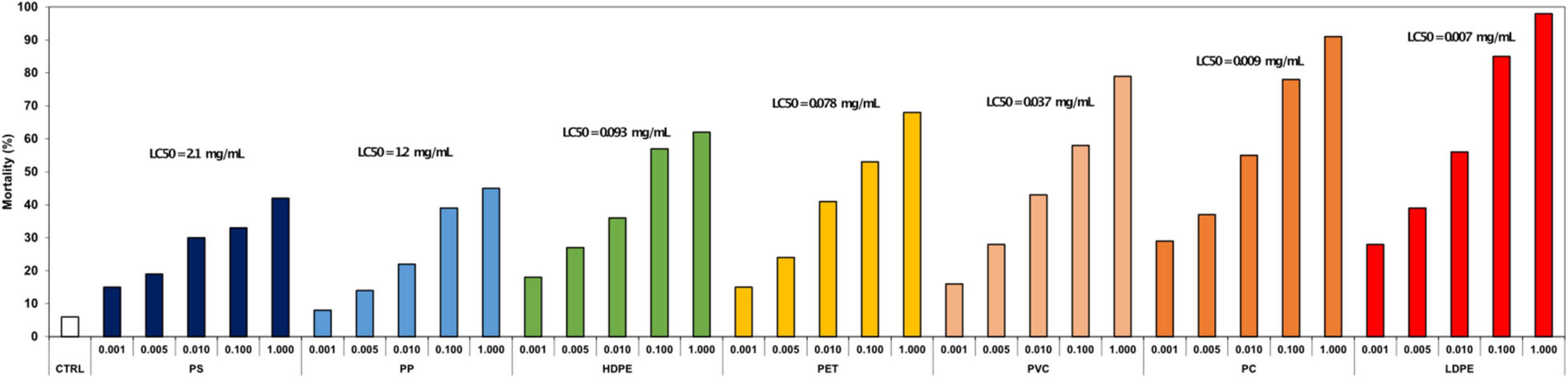
Leachates of plastic pre-production pellets from all recycle types were acutely toxic to barnacle nauplii. Increased mortality was observed at nearly all leachate concentrations from every plastic type, except for PP at 0.001 mg/mL. Calculated LC50 values of the various plastic types ranged 150-fold from the equivalent of 2.1 mg of pellet leached for 24 h in 1 mL of seawater for polystyrene (PS) to as low as the equivalent of 0.007 mg of pellet in 1 mL of water for low-density polyethylene (LDPE) (Figure 1).
Plastic Microbiome Communities
Over 1,000 OTUs were observed in the libraries from microbial communities established on plastic pellets after the first 8 days (Supplementary Figure 1). Over the 70-day study duration, OTU richness and SDI gradually increased on all plastics tested except for PVC, whose alpha diversity remained lower than other plastics throughout the study. A brief dip in richness and SDI was seen at Day 24 for several plastic types (i.e., HDPE, LDPE, PC, PET, PP) due to dominance of Rhodobacteraceae, but diversity returned to previous levels by Day 32.
Non-metric dimensional scaling (NMDS) of the bacterial community compositions indicated that microbiomes on plastics are dynamic, changing substantially over the experimental period (Figure 2A). Further, the trajectories in ordination space of microbiomes on all plastics are oriented in the same general direction, moving downward on axis 2 and to the right on axis 1 over the study period. The microbiomes on PVC are offset from those of the other plastic recycling categories, indicating substantial differences in community compositions, which is supported by average Bray-Curtis dissimilarity of 88.1% (range of 75.2–96.2%) between PVC and other plastics in time-matched communities (Figure 2B). Sample clustering based on community composition, in which PVC libraries at all-time points form their own cluster (Cluster 4), confirmed the distinctiveness of PVC microbiomes at all-time points. Clustering also revealed for microbiomes of the other plastics, that sampling time is a factor that differentiated communities with clusters forming from communities at Days 8 and 70 (Figure 2B).
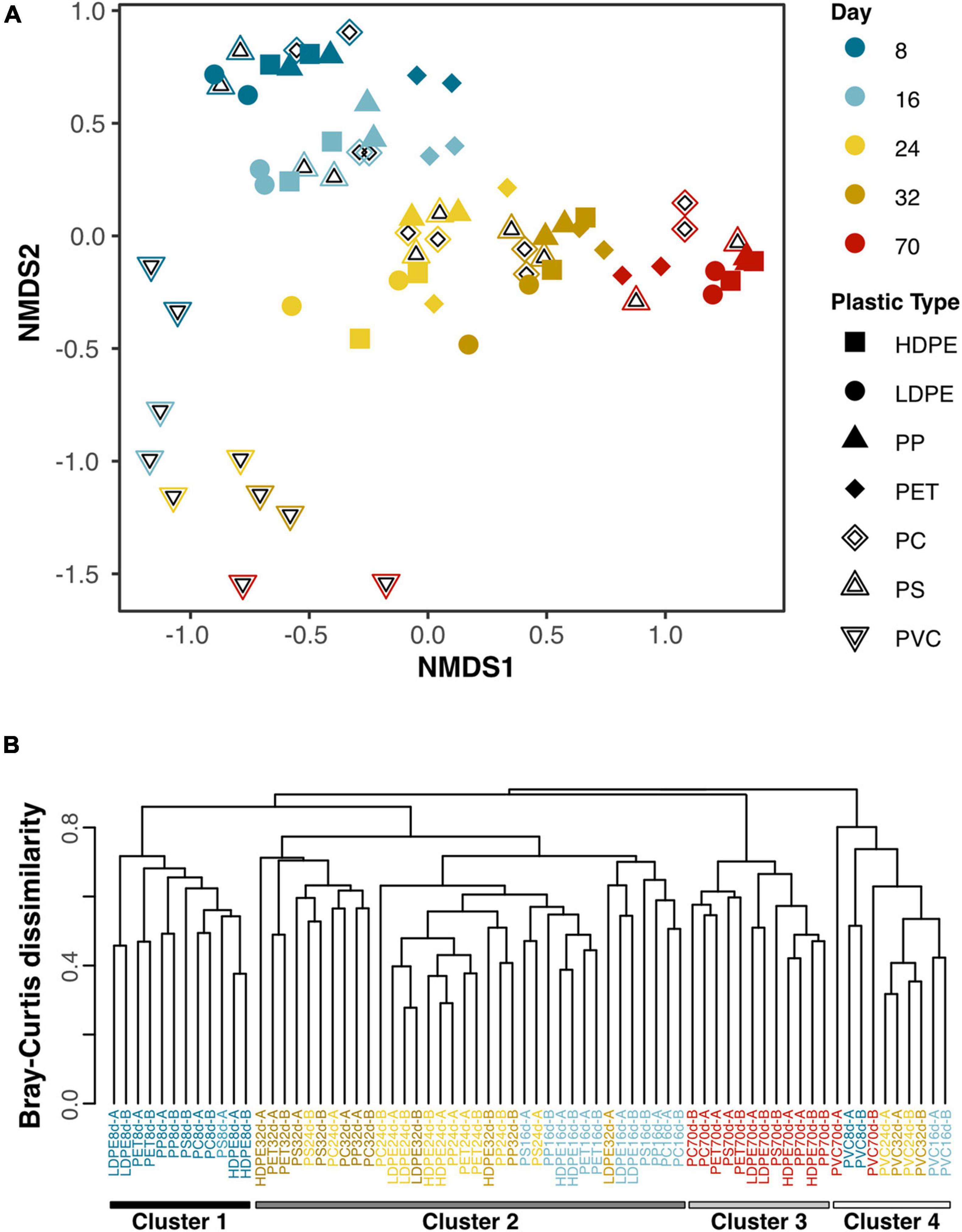
Figure 2. (A) Non-metric dimensional scaling (NMDS) plot of all samples (2 replicates of each plastic recycle category-by-timepoint combination) based on community composition at the OTU level. Colors illustrate the different sampling days and shapes represent plastic types. (B) Dendrogram showing similarities between the microbiomes from the 7 plastic types at the 5 time points. Sample naming conventions are the abbreviation of the plastic recycle category, followed by the sampling day and the replicate (A,B). K-means clustering indicates communities fall into 4 primary clusters. Cluster 4 consists of PVC microbiomes at all 5 time points. Cluster 1 and Cluster 3 are made up of the Day 8 and 70 timepoint microbiomes for the 6 non-PVC plastic types. Cluster 2 is made up of the 6 non-PVC plastic types from the Days 16, 24, and 32 sampling time points.
Throughout the study, microbiomes were dominated by Proteobacteria (30–94% of libraries), followed by Bacteroidetes (1–28%, Supplementary Figure 2). At later time points (Days 24, 32, and 70), phyla Planctomycetes and Cyanobacteria (consisting of diatom chloroplasts) become more abundant, though still in lower relative abundances than the two dominant phyla. The PVC communities contrasted with the other plastics due to the absence of Cyanobacteria, Planctomycetes and Actinobacteria at all time points.
While some differences in the microbiomes between plastic types are evident at the phylum level, patterns at the family level are more informative. The ten most abundant families fell into four categories based on their abundance patterns, reflecting their alignment with community successional stages (Figure 3). Flavobacteriaceae and Oceanospirillaceae make up the Early stage colonizers that were found in high relative abundances (12.3–25.7% and 3.2–26.9%, respectively) at Day 8 and their abundances diminished at later times. While Rhodobacteraceae was also found in comparable abundances at Day 8, it continued increasing with longer incubation prior to Day 70. The Mid stage Hyphomonadaceae and HTCC2089 rose in abundance reaching their peaks on Day 16 (7.0–14.1% and 1.8–7.1%, respectively), while Rhodobacteraceae hit its maximum on Day 24 (28.5–79.0%). In the Late stage (Days 32 and 70), Pirellulaceae and Thalassiosiraceae emerge and reach abundances 4.7–18.5% and 10.7–24.9% on non-PVC plastics after not being present at earlier times. While abundant at earlier times, Saprospiraceae reached its highest abundances (2.9–11.7%) on Day 70.
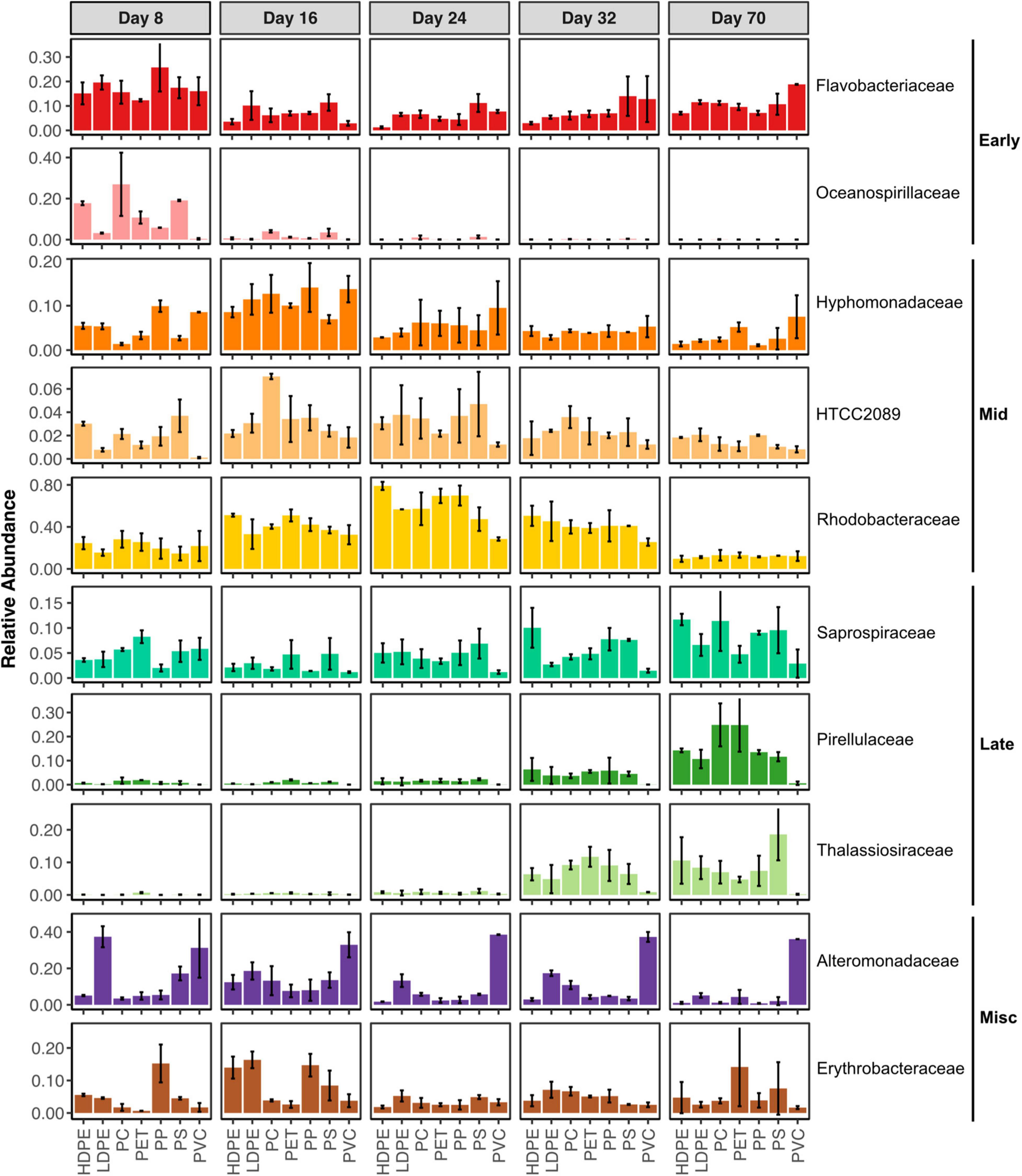
Figure 3. Ten most abundant microbial families at different sampling times. Families have been ordered according to their temporal patterns. Early stage (“Early”) families are most abundant in Day 8. Middle-stage (“Mid”) families are most abundant in Days 16, 24, and 32. Late -stage (“Late”) families are most abundant in Day 70. Miscellaneous (“Misc”) do not have an apparent temporal pattern.
While the majority of the abundant families displayed temporal dynamics, other groups exhibited preference toward or against specific plastic types. Alteromonadaceae was primarily found in PVC microbiomes at 30–40% relative abundances, while mostly being present at lower abundances on other plastics. Families abundant on other plastics were absent on PVC, specifically Oceanospirillaceae, Pirellulaceae, and Thalassiosiraceae. In contrast to the other families, the occurrence of Erythrobacteraceae lacked a clear pattern, as it was sporadically abundant with no apparent preference to plastic type.
Predicting Plastic Type From Microbiome Composition
A sample classifier employing random forests was used to determine if plastic types could be reliably predicted based on the community composition of the samples. The prediction correctly identified plastics with 93% accuracy (13 of 14 samples) (Figure 4). The classifier incorrectly labeled a HDPE sample as LDPE.
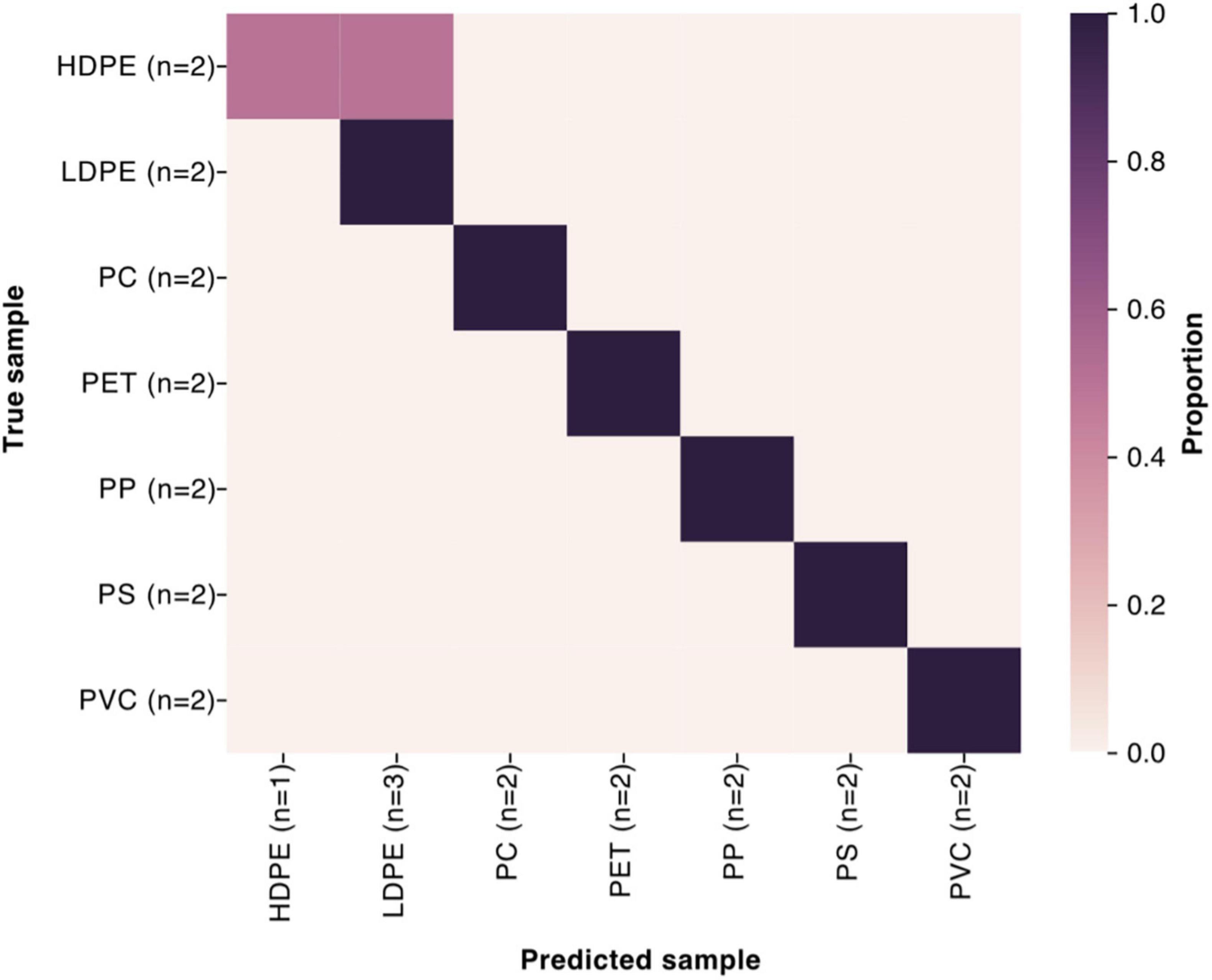
Figure 4. Results of machine learning sample classifier, using the microbial community composition to predict the plastic type with 93% accuracy (13 out of 14 samples). Labels with (n = 2) to indicate that the test set consists of 2 samples from each plastic type, while the rest of the samples constitute the training set to train the sample classifier.
Discussion
Plastics Are Not What You Might Think
Plastic pre-production pellets are dynamic platforms that reflect the chemistry of the polymer, additives and contaminants. Though these plastics are not likely to serve as the sole carbon source in coastal ocean waters, all plastics leach a complex array of molecules. For example, in the case of plastic packaging, a comprehensive review by Groh et al. (2019) reported that at least 906 different chemicals from every aspect of production and manufacture are associated with plastic packaging. Although migration of plastic-associated molecules has been modeled into food (Biryol et al., 2017), we are not aware of similar efforts focusing on predicted impacts toward microbiomes or fouling macro-organisms interacting with plastic surfaces.
Pre-production pellets have complex physical and chemical properties. A material’s surface energy (Zisman, 1964; Baier, 1970; Meyer et al., 1988), influences immediate interactions between biology (e.g., biofilms) and the surface (Zisman, 1964; Baier, 1970; Meyer et al., 1988). These immediate interactions can activate spores, alter motility, disrupt bacterial integrity (Burchard et al., 1990) and trap larvae on the surface (Rittschof and Costlow, 1989; Gerhart et al., 1992; Pechenik et al., 1993). However, the general surface phenomena are short-lived and all low-toxicity surfaces, such as glass and silanized surfaces, have similar surface chemistries after 3–5 days (Meyer et al., 1988). Surface roughness and hydrophobicity (wettability) are other factors that may influence microbial settlement on a surface (Rummel et al., 2017). The dynamic leaching and uptake chemistry and surface energy of plastics set the stage for the naupliar toxicity and microbial community development reported here.
Plastics as Dynamic Platforms
When one considers that the microbiome on the surface of a pre-production pellet is living within the no-slip boundary layer, it follows those semi-labile organic compounds, toxicants, metals, and endocrine disruptors (along with the other chemicals mentioned above) are diffusing from the plastic substrate and bathing the microbiome on the pellet surface. Simultaneously, nutrients, salts, organic compounds, and toxins are diffusing from the water column into the boundary layer. Bacterial and algal exudates and metabolic products add to the mix and diffuse into the plastic and into the water column. Thus, the microbiomes on each type of plastic are exposed to chemical fluxes in both directions and to metabolites generated from within their bacterial community. That the microbiomes on the surfaces of various plastic types remain dissimilar at the final time point (though some microbiomes on LDPE, HDPE and PP become more similar to each other), suggests that the processes imbuing plastic surfaces with their physicochemical activities are still active after 70 days of seawater exposure.
If plastics were inert as is commonly reported, the microbiomes on all plastic types should be highly similar. Our intent with this time series incubation was to pin down when all surfaces reached a common state. Given that microbiome compositions did not fully converge within the 70-day period, strongly indicates that the plastics did not reach a common state, instead retaining differences in their underlying polymer chemistries that continue to impact the communities on the surface. In addition to the community-level differences captured by ordination and clustering (Figures 2A,B), the delayed colonization of plastic surfaces by Thalassiosiraceae and Pirellulaceae until Day 32 for non-PVC plastics and never for PVC (Figure 3) underscores the long-term persistence of a substrate effect on microbiome development.
Pristine Plastic Pellet Leachate Is Acutely Toxic to Nauplii
Although Li et al. (2016a) showed that plastic leachates from finished products (e.g., plates, soda bottles) from all seven plastic recycle categories were variably toxic to stage II barnacle nauplii, to our knowledge this is the first study to test acute naupliar toxicity to leachates of plastic pre-production pellets. Seawater leachates of all seven recycle categories of pre-production pellets are acutely toxic to barnacle nauplii. For the least potent plastic, soaking 1 mg of polystyrene plastic in a milliliter of water generates a solution that kills 42% of barnacle larvae in 24 h. For the most potent plastic, the leachate can be diluted to the equivalent of 7.0 μg of plastic leached in seawater and kill half the larvae. As neither the polymer or its monomer leach, toxicity can be ascribed to the 50 + to several hundred compounds that can be detected by high resolution mass spectrometry (Li et al., 2016a), such as unpolymerized labile organic carbon molecules, petrochemical constituents, heavy metals, organometals with steroidal activity, organic environmental steroids, toxicants and flavors, that can elicit a potent biological response (Al-Malack, 2001; Hahladakis et al., 2018; Groh et al., 2019; Diana et al., 2020). Unlike most other products in which compound classes can indicate relative toxicities to various organisms, the plastic recycle category provides little information about biological activity of molecules impacting microorganisms.
It is likely the barnacle larvae we tested died from multiple insults. Barnacle larvae are swimming in and drinking the complex mixtures of molecules leaching out of pellets. Death could be primarily due to surfactants, endocrine disruptors interfering with molting, organometals, or heavy metals. The toxicity assay, though sensitive (Rittschof et al., 1992), cannot discriminate most of these kinds of differences.
At the macrofouling level, surfaces with different surface energies initially show dramatic differences in larval settlement over the first few days (Roberts et al., 1991; Li et al., 2016b). Over a month time interval, macro-organismal communities on relatively inert surfaces like glass are all similar (Holm et al., 1997). However, on surfaces with additional chemical properties such as generation of reactive oxygen species or degradation, communities remain distinct (Gatley-Montross et al., 2017; Seeley et al., 2020).
Plastic Microbiome Differences Do Not Directly Reflect Acute Toxicity to Barnacle Larvae
We found that there was not a clear relation between acute toxicity to barnacle larvae and the microbiomes that colonize pre-production pellets. Mechanisms of acute toxicity vary widely between barnacle larvae and microbes, so a direct correlation is not expected. While naupliar experiments measure toxin-mediated insults on individuals of a population, microbiome studies capture community response to both toxins and consumable nutrients, such as organic carbon. While we found that PVC had a distinct microbiome from the other plastic recycling categories and inhibited abundances of microbial families prevalent in other plastic types, its leachate was the third most acutely toxic, with a LC50 at 0.037 mg/mL—higher than both PC and LDPE at 0.009 mg/mL and 0.007 mg/mL, respectively. Acute nauplii toxicity was tested with pristine plastic pellets. Since plastics are continuously weathered in marine environments, further larval toxicity studies may consider exposing model animals to plastic pellets that have been weathered in the environment or sampled at various time points over the laboratory seawater exposure, which would be consistent with suggestions by Bucci et al. (2019) to extend the ecological relevance of plastic pollution research.
Selection and Succession on Plastic Surfaces
Microbial community composition is shaped in large part by selection of abiotic factors that determine relative fitness differences between taxa (Martiny et al., 2006; Nemergut et al., 2013). In this study, abiotic factors influencing microbial fitness include both nutrients (e.g., labile and semi-labile organic compounds) and inhibitors (e.g., heavy metals, organic toxicants). The observed compositional dissimilarities between microbiomes of various plastic types that developed while incubated in the same seawater and environmental conditions strongly indicate that distinct surface chemistries of the plastic types were responsible for differences in early colonizing communities at Day 8 and persisted to contribute to the differences in mid- and late-stage communities. Strong selection does not preclude contributions of stochastic ecological processes that might maintain community differences after initial environmental selection are relaxed (Nemergut et al., 2013), though priority effects may have limited impact in this experimental setup with flowing seawater allowing for continuous recruitment of microbial taxa from the coastal ocean microbiome (Fukami, 2015).
As has been previously reported by numerous studies, marine plastic microbiomes were substantially different from free-living microbial communities (Ward et al., 2017) or those attached to natural particles (Yung et al., 2016). The plastic microbiomes contained Hyphomonadaceae, Erythrobacteraceae, HTCC2089, and other lower abundance bacterial families at relative abundances not commonly observed in bacterioplankton of Beaufort Inlet (Yung et al., 2016; Ward et al., 2017), the source community from which recruitment of the plastic microbiomes occurs. In contrast, most families we observed here were also observed in other ocean plastic microbiomes (Zettler et al., 2013; Bryant et al., 2016). The low overlap in microbiome membership between plastics and seawater underscores the highly selective nature of community assembly on surfaces of plastic particles.
While most marine plastisphere studies have mostly focused on polyethylene and polypropylene particles more prevalent in surface waters due to their buoyancies, one of the most striking findings of this study was the drastic divergence of PVC microbiomes from the other plastic types. Recent studies have also observed distinct microbial communities on PVC in comparison to LDPE, HDPE, PP and glass slides after 1 week of incubation in the Northern Adriatic (Pinto et al., 2019). The presence of two plasticizers, di(2-ethylhexyl) phthalate (DEHP) and di-isononyl phthalate (DINP), was hypothesized to cause microbial toxicity, yet microbiomes converged by the end of the 2-month incubation period despite plasticizer concentrations remaining stable. In a separate sediment mesocosm study, microbiomes amended with PVC were distinct after 16 days, with inhibition of nitrification and denitrification processes (Seeley et al., 2020). The variable responses across studies illustrate the complexities inherent in studying plastics in the same recycling category but generated at different times, with different starting materials and additives, and tested in different environments (Li et al., 2016b; Diana et al., 2020; Seeley et al., 2020; Zhu et al., 2020).
Few studies to date investigated the progression of marine plastic surfaces over time, yet our study reveals substantial temporal changes in the microbiomes of all plastic types. While microbiome trajectories preserved differences between plastic types, the temporally variable nature of plastic microbiomes highlights how differences in incubation duration complicate comparisons between studies. Microbial community composition, OTU richness and SDI on all non-PVC plastics changed over the study period. The temporal patterns in microbial family relative abundances are suggestive of successional changes, potentially driven by resource availability, competition and toxicity. Early stage communities were dominated by Rhodobacteraceae and Flavobacteriaceae, common marine bacteria that are consumers of ocean DOM (Buchan et al., 2014) and often primary surface colonizers in coastal marine settings (Dang and Lovell, 2000; Dang et al., 2008; Elifantz et al., 2013). At later time points, Rhodobacteraceae constitute larger relative abundances while Hyphomonadaceae and HTCC2089 increase in abundance. In an examination of community assembly on model organic carbon particles, a consistent successional pattern was revealed in which early colonizers were replaced by primary particle degraders and then secondary consumers that may metabolize further degradation or act as cheaters (Datta et al., 2016). Given that many of the abundant families are associated with hydrocarbon degradation and marine oil spills (discussed below), it is a compelling hypothesis that some early- to mid-stage colonizers may contribute to degradation of plastic leachate. Additionally, we must also consider microbial groups’ resistances to toxic leachates and intermediates in hydrocarbon degradation. For example, the appearance of Thalassiosiraceae and Pirellulaceae at Days 32 and 70 may reflect late-stage succession due to microbial habitat primed by earlier community development or alternatively reduced toxicity due to several weeks worth of leaching.
Differences over time of community compositions reflect that the plastic microbiomes undergo colonization and succession. The relative abundances of major families are indicative of the particular stages of succession.
Plastic as a Harbor for Hydrocarbon-Degrading Bacteria
Plastics are manufactured from petroleum, which is primarily comprised of hydrocarbons (Abrajano et al., 2007; Harding et al., 2007). Marine plastic debris has been previously found to be associated with the hydrocarbon-degrading cyanobacteria Phormidium spp. and Pseudoalteromonas, in contrast to the surrounding seawater (Zettler et al., 2013; Debroas et al., 2017). By extracting petroleum from the ocean and then polluting the marine environment with plastic pollution, researchers posit that marine plastics are creating a natural experiment in the oceans (Amaral-Zettler et al., 2020).
Found within bacterial communities from this study are multiple members that are known or potential hydrocarbon degraders. For instance, we identified HTCC2089 on all plastics primarily during mid-stage period. Several Acinetobacter spp., the dominant genus of HTCC2089 in our study, are capable of n-alkane (C10–C20) degradation, catalyzed by alkane hydroxylase encoded by alkM (Head et al., 2006). Isolated representatives of Acinetobacter spp. have been linked to PAH degradation in pure culture, and Acinetobacter has been found to be enriched in soils following petroleum hydrocarbon contamination (Head et al., 2006). Our study also found Oceanospirillaceae spp. on all plastic types primarily as an early colonizer. Oceanospirillaceae spp. are known to become enriched in seawater containing hydrocarbon compounds (Lamendella et al., 2014; Kleindienst et al., 2016). These species induce expression of various alkane degradation genes upon oil exposure, while also possessing other genes for degrading more resistant compounds such as benzene, toluene, ethylbenzene, xylenes, and polycyclic aromatic hydrocarbons (Lamendella et al., 2014). While Rhodobacteraceae are common marine heterotrophic bacteria and frequent biofilm colonizers (Dang and Lovell, 2000; Dang et al., 2008; Elifantz et al., 2013), Rhodobacteraceae have been reported as a potential hydrocarbon-degrading group and were enriched in bacterial communities in the waters surrounding the Deepwater Horizon Oil Spill as well as the Gulf of Mexico beach sands impacted by the spill (Lamendella et al., 2014). We found Rhodobacteraceae on all plastics throughout the entire study period, consistent with Bhagwat et al. (2021). While others have suggested that the microbial communities observed on marine plastics seem more indicative of a general proclivity for surface attachment, the specific enrichment of bacterial clades with hydrocarbon-degrading attributes not typically associated with natural coastal ocean particles in that environment suggest the specific selection of microbiota by the chemical composition of the plastic substratum itself. To test this hypothesis, a follow-up study employing metatranscriptomics and stable isotope probing is warranted to address whether the microbial taxa on plastic surfaces are utilizing leachates and plastic-bound components, or simply using the plastic surface as a physical substrate and consuming dissolved nutrients from the water column.
Microbial Forensics
Given the massive strides in analytical chemistry and high-resolution mass spectrometry, at first glance it is counterintuitive to think about using microbiomes to learn about surfaces and habitats. However, significant obstacles remain in chemical analysis especially of manmade polymer networks (Hahladakis et al., 2018; Groh et al., 2019; Wiesinger et al., 2021). Further, microbiomes and specific taxa within can present high sensitivities to various stressors (Ward et al., 2019). As an example, key taxa that are only enriched on surfaces leaching organo-tins or other highly toxic compounds can provide evidence that cannot be provided by any other means. Using the organotin example, biological impacts are seen at < 10 ng/l (McClellan-Green et al., 2006), well below the limits of detection of analytical devices especially with complex mixtures.
The sample classifier results underscore how distinct the communities on different plastics are, as machine learning can consistently (93%) determine the plastic type the communities were on. We envision development of analytical tools based upon microbes that can be used to determine the mix of pollutants in a harbor or the nature of the molecules leaching out of a coating or plastic food packaging. These societally relevant compounds could be added intentionally by the unknowing manufacturer or might be contaminants in the materials purchased to make the polymers. With the rapid advances in modern sequencing and reporting technology and dramatic reduction in costs, using microbiome approaches might be the next major advance in assessing environmental water quality. The data support our hypothesis that microbiomes can be used to identify plastics. It remains to be seen if microbiomes can be used to identify particular chemicals leaching from plastics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA780594/, PRJNA780594.
Author Contributions
DR and TS conceived of the study. BO and KK carried out the experiments. CW processed the sequencing data, performed the community analysis, and designed the figures. BO analyzed the nauplii toxicity study. CW, ZD, DR, and BO wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
DR and ZD acknowledged the Oak Foundation for their support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We kindly acknowledge the Piver’s Island Coastal Observatory (PICO) for the salinity data provided in this study. We thank the Duke Center for Genomic and Computational Biology for providing the 16S sequencing services. We would also like to thank Joel Meyer and Richard Di Giulio for assigning a term manuscript that allowed an author to work on the literature review for this manuscript. We would also like to thank the Integrative Bioinformatics for Investigating and Engineering Microbiomes Graduate Traineeship at Duke University for relevant training.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.807327/full#supplementary-material
References
Abrajano, T. A., Yan, B., Song, J., Bopp, R., and O’Malley, V. (2007). “9.13 - High-Molecular-Weight Petrogenic and Pyrogenic Hydrocarbons in Aquatic Environments,” in Treatise on Geochemistry, eds H. D. Holland and K. K. Turekian (Oxford: Pergamon), 1–50. doi: 10.1016/B0-08-043751-6/09055-1
Allen, A. S., Seymour, A. C., and Rittschof, D. (2017). Chemoreception drives plastic consumption in a hard coral. Mar. Pollut. Bull. 124, 198–205. doi: 10.1016/j.marpolbul.2017.07.030
Al-Malack, M. H. (2001). Migration of lead from unplasticized polyvinyl chloride pipes. J. Hazard. Mater. 82, 263–274. doi: 10.1016/s0304-3894(00)00366-6
Amaral-Zettler, L. A., Zettler, E. R., and Mincer, T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Andrady, A. L. (2017). The plastic in microplastics: A review. Mar. Pollut. Bull. 119, 12–22. doi: 10.1016/j.marpolbul.2017.01.082
Artham, T., Sudhakar, M., Venkatesan, R., Nair, C. M., Murty, K., and Doble, M. (2009). Biofouling and stability of synthetic polymers in sea water. Int. Biodeterioration Biodegrad. 63, 884–890. doi: 10.1016/j.ibiod.2009.03.003
Arthur, C., Baker, J., and Bamford, H. (2009). Proceedings of the international workshop on the occurrence, effects, and fate of microplastic marine debris. Washington, D.C: National Oceanic and Atmospheric Administration.
Baier, R. E. (1970). “Surface properties influencing biological adhesion,” in Adhesion in biological systems, ed. R. S. Manley (New York, NY: Academic Press), 15–48.
Bhagwat, G., Zhu, Q., O’Connor, W., Subashchandrabose, S., Grainge, I., Knight, R., et al. (2021). Exploring the Composition and Functions of Plastic Microbiome Using Whole-Genome Sequencing. Environ. Sci. Technol. 55, 4899–4913. doi: 10.1021/acs.est.0c07952
Biryol, D., Nicolas, C. I., Wambaugh, J., Phillips, K., and Isaacs, K. (2017). High-throughput dietary exposure predictions for chemical migrants from food contact substances for use in chemical prioritization. Environ. Int. 108, 185–194. doi: 10.1016/j.envint.2017.08.004
Brandon, J. A., Jones, W., and Ohman, M. D. (2019). Multidecadal increase in plastic particles in coastal ocean sediments. Sci. Adv. 5:aax0587. doi: 10.1126/sciadv.aax0587
Bryant, J. A., Clemente, T. M., Viviani, D. A., Fong, A. A., Thomas, K. A., Kemp, P., et al. (2016). Diversity and Activity of Communities Inhabiting Plastic Debris in the North Pacific Gyre. mSystems. 1, e24–e16. doi: 10.1128/mSystems.00024-16
Bucci, K., Tulio, M., and Rochman, C. M. (2019). What is known and unknown about the effects of plastic pollution: A meta-analysis and systematic review. Ecol. Applicat. 2019:e02044. doi: 10.1002/eap.2044
Buchan, A., LeCleir, G., Gulvik, C., and Gonzalez, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Burchard, R. P., Rittschof, D., and Bonaventura, J. (1990). Adhesion and Motility of Gliding Bacteria on Substrata with Different Surface Free-Energies. Appl. Environ. Microbiol. 56, 2529–2534. doi: 10.1128/aem.56.8.2529-2534.1990
Carson, H. S., Nerheim, M. S., Carroll, K. A., and Eriksen, M. (2013). The plastic-associated microorganisms of the North Pacific Gyre. Mar. Poll. Bull. 75, 126–132. doi: 10.1016/j.marpolbul.2013.07.054
Curren, E., and Leong, S. C. Y. (2019). Profiles of bacterial assemblages from microplastics of tropical coastal environments. Sci. Total Environ. 655, 313–320. doi: 10.1016/j.scitotenv.2018.11.250
Dang, H., and Lovell, C. R. (2000). Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66, 467–475. doi: 10.1128/AEM.66.2.467-475.2000
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Datta, M., Sliwerska, E., Gore, J., Polz, M. F., and Cordero, O. X. (2016). Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7:11965. doi: 10.1038/ncomms11965
De Tender, C. A., Devriese, L. I., Haegeman, A., Maes, S., Ruttink, T., and Dawyndt, P. (2015). Bacterial Community Profiling of Plastic Litter in the Belgian Part of the North Sea. Environ. Sci. Technol. 49, 9629–9638. doi: 10.1021/acs.est.5b01093
Debroas, D., Mone, A., and Ter Halle, A. (2017). Plastics in the North Atlantic garbage patch: A boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 599–600, 1222–1232. doi: 10.1016/j.scitotenv.2017.05.059
Diana, Z., Sawickij, N., Rivera, N. A., Hsu-Kim, H., and Rittschof, D. (2020). Plastic pellets trigger feeding responses in sea anemones. Aquat. Toxicol. 222:105447. doi: 10.1016/j.aquatox.2020.105447
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Vegetation Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dobretsov, S., Teplitski, M., and Paul, V. (2009). Mini-review: quorum sensing in the marine environment and its relationship to biofouling. Biofouling 25, 413–427. doi: 10.1080/08927010902853516
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2017). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv [Preprint]. doi: 10.1101/081257
Elifantz, H., Horn, G., Ayon, M., Cohen, Y., and Minz, D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 85, 348–357. doi: 10.1111/1574-6941.12122
Fukami, T. (2015). Historical Contingency in Community Assembly: Integrating Niches, Species Pools, and Priority Effects. Annu. Rev. Ecol. Evolut. Syst. 46, 1–23. doi: 10.1146/annurev-ecolsys-110411-160340
Gatley-Montross, C. M., Finlay, J. A., Aldred, N., Cassady, H., Destino, J. F., Orihuela, B., et al. (2017). Multivariate analysis of attachment of biofouling organisms in response to material surface characteristics. Biointerphases 12:051003. doi: 10.1116/1.5008988
Gerhart, D. J., Rittschof, D. I, Hooper, R., Eisenman, K., Meyer, A. E., Baier, R. E., et al. (1992). Rapid and inexpensive quantification of the combined polar components of surface wettability: application to biofouling. Biofouling 5, 251–259. doi: 10.1080/08927019209378246
Geyer, R., Jambeck, J. R., and Law, K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3. doi: 10.1126/sciadv.1700782
Groh, K. J., Backhaus, T., Carney-Almroth, B., Geueke, B., Inostroza, P. A., Lennquist, A., et al. (2019). Overview of known plastic packaging-associated chemicals and their hazards. Sci. Total Environ. 651, 3253–3268. doi: 10.1016/j.scitotenv.2018.10.015
Hadfield, M. G. (2010). Biofilms and Marine Invertebrate Larvae: What Bacteria Produce That Larvae Use to Choose Settlement Sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hahladakis, J. N., Velis, C. A., Weber, R., Iacovidou, E., and Purnell, P. (2018). An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 344, 179–199. doi: 10.1016/j.jhazmat.2017.10.014
Haider, N., and Karlsson, S. (2000). Kinetics of migration of antioxidants from polyolefins in natural environments as a basis for bioconversion studies. Biomacromolecules 1, 481–487. doi: 10.1021/bm0000283
Harding, K. G., Dennis, J. S., von Blottnitz, H., and Harrison, S. T. L. (2007). Environmental analysis of plastic production processes: Comparing petroleum-based polypropylene and polyethylene with biologically-based poly-β-hydroxybutyric acid using life cycle analysis. J. Biotechnol. 130, 57–66. doi: 10.1016/j.jbiotec.2007.02.012
Head, I., Jones, D., and Röling, W. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Holm, E. R., Orihuela, B., Kavanagh, C. J., and Rittschof, D. (2005). Variation among families for characteristics of the adhesive plaque in the barnacle Balanus amphitrite. Biofouling 21, 121–126. doi: 10.1080/08927010512331344188
Holm, E. R., Cannon, G., Roberts, D., Schmidt, A. R., Sutherland, J. P., and Rittschof, D. (1997). The influence of initial surface chemistry on development of the fouling community at Beaufort, North Carolina. J. Exp. Mar. Biol. Ecol. 215, 189–203.
Jahnke, A., Arp, H. P. H., Escher, B. I., Gewert, B., Gorokhova, E., Kühnel, D., et al. (2017). Reducing Uncertainty and Confronting Ignorance about the Possible Impacts of Weathering Plastic in the Marine Environment. Environ. Sci. Technol. Lett. 4, 85–90. doi: 10.1021/acs.estlett.7b00008
Jambeck, J. R., Geyer, R., Wilcox, C., Siegler, T. R., Perryman, M., Andrady, A., et al. (2015). Plastic waste inputs from land into the ocean. Science 347, 768–771. doi: 10.1126/science.1260352
Kirstein, I. V., Kirmizi, S., Wichels, A., Garin-Fernandez, A., Erler, R., Löder, M., et al. (2016). Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 120, 1–8. doi: 10.1016/j.marenvres.2016.07.004
Kleindienst, S., Grim, S., Sogin, M., Bracco, A., Crespo-Medina, M., and Joye, S. B. (2016). Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J. 10, 400–415. doi: 10.1038/ismej.2015.121
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1. doi: 10.1093/nar/gks808
Lamendella, R., Strutt, S., Borglin, S. E., Chakraborty, R., Tas, N., Mason, O. U., et al. (2014). Assessment of the Deepwater Horizon oil spill impact on Gulf coast microbial communities. Front. Microbiol. 5:130. doi: 10.3389/fmicb.2014.00130
Lau, W. W. Y., Shiran, Y., Bailey, R. M., Cook, E., Stuchtey, M. R., Koskella, J., et al. (2020). Evaluating scenarios toward zero plastic pollution. Science 369, 1455–1461. doi: 10.1126/science.aba9475
Li, H. X., Getzinger, G. J., Ferguson, P. L., Orihuela, B., Zhu, M., and Rittschof, D. (2016a). Effects of Toxic Leachate from Commercial Plastics on Larval Survival and Settlement of the Barnacle Amphibalanus amphitrite. Environ. Sci. Technol. 50, 924–931.
Li, H.-X., Orihuela, B., Zhu, M., and Rittschof, D. (2016b). Recyclable plastics as substrata for settlement and growth of bryozoans Bugula neritina and barnacles Amphibalanus amphitrite. Environ. Poll. 218, 973–980.
Martiny, J. B. H., Bohannan, B. J. M., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
McClellan-Green, P., Romano, J., and Rittschof, D. (2006). Imposex Induction in the Mud Snail, Ilyanassa obsoleta by Three Tin Compounds. Bull. Environ. Contam. Toxicol. 76, 581–588. doi: 10.1007/s00128-006-0959-1
McMurdie, P. J., and Holmes, S. (2013). phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meyer, A. E., Baier, R. E., and King, R. W. (1988). Initial fouling of nontoxic coatings in fresh, brackish, and sea water. Canad. J. Chem. Engine. 66, 55–62. doi: 10.1002/cjce.5450660108
National Library of Medicine (1988). National Center for Biotechnology Information (NCBI)[Internet]. Bethesda, MD: National Library of Medicine (US). Available online at: https://www.ncbi.nlm.nih.gov/
Nemergut, D. R., Schmidt, S. K., Fukami, T., O’Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Oberbeckmann, S., Löder, M. G., and Labrenz, M. (2015). Marine microplastic-associated biofilms–a review. Environ. Chem. 12, 551–562. doi: 10.1071/en15069
Oberbeckmann, S., Osborn, A. M., and Duhaime, M. B. (2016). Microbes on a Bottle: Substrate, Season and Geography Influence Community Composition of Microbes Colonizing Marine Plastic Debris. PLoS One 11:e0159289. doi: 10.1371/journal.pone.0159289
Ostuni, E., Chapman, R. G., Liang, M. N., Meluleni, G., Pier, G., Ingber, D. E., et al. (2001). Self-Assembled Monolayers That Resist the Adsorption of Proteins and the Adhesion of Bacterial and Mammalian Cells. Langmuir 17, 6336–6343. doi: 10.1021/la010552a
Pechenik, J. A., Rittschof, D., and Schmidt, A. R. (1993). Influence of Delayed Metamorphosis on Survival and Growth of Juvenile Barnacles Balanus-Amphitrite. Mar. Biol. 115, 287–294. doi: 10.1007/bf00346346
Pinto, M., Langer, T. M., Hüffer, T., Hofmann, T., and Herndl, G. J. (2019). The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One 14:e0217165. doi: 10.1371/journal.pone.0217165
Reisser, J., Shaw, J., Hallegraeff, G., Proietti, M., Barnes, D. K. A., Thums, M., et al. (2014). Millimeter-Sized Marine Plastics: A New Pelagic Habitat for Microorganisms and Invertebrates. PLoS One 9:e100289. doi: 10.1371/journal.pone.0100289
Rittschof, D., and Costlow, J. D. (1989). Bryozoan and barnacle settlement in relation to initial surface wettability a comparison of laboratory and field studies. Sci. Mar. 53, 411–416.
Rittschof, D., and Holm, E. R. (1997). “Antifouling and foul-release: A primer. Recent Advances in Marine Biotechnology,” in Endocrinology and Reproduction, Vol. 1, eds M. Fingerman, R. Nagabhushanam, M.-F. Thompson, and N. H. Enfield (Hauppauge, NY: Science Publishers), 497–512.
Rittschof, D., Clare, A. S., Gerhart, D. J., Mary, S. A., and Bonaventura, J. (1992). Barnacle in vitro assays for biologically active substances: Toxicity and Settlement inhibition assays using mass cultured Balanus amphitrite amphitrite darwin. Biofouling 6, 115–122. doi: 10.1080/08927019209386217
Rittschof, D., Orihuela, B., Harder, T., Stafslien, S., Chisholm, B., and Dickinson, G. H. (2011). Compounds from silicones alter enzyme activity in curing barnacle glue and model enzymes. PLoS ONE 6:e16487. doi: 10.1371/journal.pone.0016487
Rittschof, D., Orihuela, B., Stafslien, S., Daniels, J., and Christianson, D. (2008). Barnacle reattachment: a tool for studying barnacle adhesion. Biofouling 24, 1–9. doi: 10.1080/08927010701784920
Roager, L., and Sonnenschein, E. C. (2019). Bacterial candidates for colonization and degradation of marine plastic debris. Environ. Sci. Technol. 53, 11636–11643. doi: 10.1021/acs.est.9b02212
Roberts, D., Rittschof, D., Holm, E., and Schmidt, A. R. (1991). Factors Influencing Initial Larval Settlement - Temporal, Spatial and Surface Molecular-Components. J. Exp. Mar. Biol. Ecol. 150, 203–221. doi: 10.1016/0022-0981(91)90068-8
Rochman, C. M., Brookson, C., Bikker, J., Djuric, N., Earn, A., Bucci, K., et al. (2019). Rethinking microplastics as a diverse contaminant suite. Environ. Toxicol. Chem. 38, 703–711. doi: 10.1002/etc.4371
Rochman, C. M., Hentschel, B. T., and Teh, S. J. (2014). Long-Term Sorption of Metals Is Similar among Plastic Types: Implications for Plastic Debris in Aquatic Environments. PLoS One 9:e85433. doi: 10.1371/journal.pone.0085433
Rochman, C. M., Hoh, E., Hentschel, B. T., and Kaye, S. (2013). Long-Term Field Measurement of Sorption of Organic Contaminants to Five Types of Plastic Pellets: Implications for Plastic Marine Debris. Environ. Sci. Technol. 47, 1646–1654. doi: 10.1021/es303700s
Rummel, C. D., Jahnke, A., Gorokhova, E., Kühnel, D., and Schmitt-Jansen, M. (2017). Impacts of Biofilm Formation on the Fate and Potential Effects of Microplastic in the Aquatic Environment. Environ. Sci. Technol. Lett. 4, 258–267. doi: 10.1021/acs.estlett.7b00164
Seeley, M. E., Song, B., Passie, R., and Hale, R. C. (2020). Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat. Commun. 11, 1–10. doi: 10.1038/s41467-020-16235-3
Shikuma, N. J., and Hadfield, M. G. (2005). Temporal variation of an initial marine biofilm community and its effects on larval settlement and metamorphosis of the tubeworm Hydroides elegans. Biofilms 2, 231–238.
Shikuma, N. J., and Hadfield, M. G. (2010). Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling 26, 39–46. doi: 10.1080/08927010903282814
Suhrhoff, T. J., and Scholz-Böttcher, B. M. (2016). Qualitative impact of salinity, UV radiation and turbulence on leaching of organic plastic additives from four common plastics — A lab experiment. Mar. Poll. Bull. 102, 84–94. doi: 10.1016/j.marpolbul.2015.11.054
Teughels, W., Assche, N. V., Sliepen, I., and Quirynen, M. (2006). Effect of material characteristics and/or surface topography on biofilm development. Clin. Oral Implants Res. 17, 68–81. doi: 10.1111/j.1600-0501.2006.01353.x
Teuten, E. L., Saquing, J. M., Knappe, D. R. U., Barlaz, M. A., Jonsson, S., Björn, A., et al. (2009). Transport and release of chemicals from plastics to the environment and to wildlife. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2027–2045. doi: 10.1098/rstb.2008.0284
Walker, I., Montaño, M. D., Lankone, R. S., Fairbrother, D. H., and Ferguson, P. L. (2021). Influence of CNT loading and environmental stressors on leaching of polymer-associated chemicals from epoxy and polycarbonate nanocomposites. Environ. Chem. 18:131. doi: 10.1071/EN21043
Ward, C. S., Pan, J., Colman, B. P., Wang, Z., Gwin, C., Williams, T. C., et al. (2019). Conserved microbial toxicity responses for acute and chronic silver nanoparticle treatments in wetland mesocosms. Environ. Sci. Technol. 53, 3268–3276. doi: 10.1021/acs.est.8b06654
Ward, C., Yung, C. M., Davis, K., Blinebry, S. K., Williams, T. C., Johnson, Z. I., et al. (2017). Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 11, 1412–1422.
Wiesinger, H., Wang, Z., and Hellweg, S. (2021). Deep dive into plastic monomers, additives, and processing aids. Environ. Sci. Technol. 55, 9339–9351. doi: 10.1021/acs.est.1c00976
Worm, B., Lotze, H. K., Jubinville, I., Wilcox, C., and Jambeck, J. (2017). Plastic as a Persistent Marine Pollutant. Annu. Rev. Environ. Resour. 42, 1–26. doi: 10.1146/annurev-environ-102016-060700
Yu, S.-P., and Chan, B. K. K. (2020a). Effects of polystyrene microplastics on larval development, settlement, and metamorphosis of the intertidal barnacle Amphibalanus amphitrite. Ecotoxicol. Environ. Safety 194:110362. doi: 10.1016/j.ecoenv.2020.110362
Yu, S.-P., and Chan, B. K. K. (2020b). Intergenerational microplastics impact the intertidal barnacle Amphibalanus amphitrite during the planktonic larval and benthic adult stages. Environ. Poll. 267:115560.
Yung, C. M., Ward, C. S., Davis, K. M., Johnson, Z. I., and Hunt, D. E. (2016). Insensitivity of diverse and temporally variable particle-associated microbial communities to bulk seawater environmental parameters. Appl. Environ. Microbiol. 82, 3431–3437. doi: 10.1128/AEM.00395-16
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhu, M., Chernick, M., Rittschof, D., and Hinton, D. E. (2020). Chronic dietary exposure to polystyrene microplastics in maturing Japanese medaka (Oryzias latipes). Aquat. Toxicol. 220:105396. doi: 10.1016/j.aquatox.2019.105396
Zisman, W. A. (1964). Contact Angle, Wettability, and Adhesion. Adv. Chem. 43, 1–51. doi: 10.1021/ba-1964-0043.ch001
Keywords: microbiome, plastic, marine debris, pre-production pellets, toxicity, additives
Citation: Ward CS, Diana Z, Ke KM, Orihuela B, Schultz TP and Rittschof D (2022) Microbiome Development of Seawater-Incubated Pre-production Plastic Pellets Reveals Distinct and Predictive Community Compositions. Front. Mar. Sci. 8:807327. doi: 10.3389/fmars.2021.807327
Received: 02 November 2021; Accepted: 17 December 2021;
Published: 14 January 2022.
Edited by:
Kelli Zargiel Hunsucker, Florida Institute of Technology, United StatesReviewed by:
Nick Aldred, University of Essex, United KingdomHolly Sweat, Smithsonian Marine Station (SMS), United States
Benny K. K. Chan, Academia Sinica, Taiwan
Copyright © 2022 Ward, Diana, Ke, Orihuela, Schultz and Rittschof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher S. Ward, Y2hyd2FyZEBiZ3N1LmVkdQ==; Daniel Rittschof, cml0dEBkdWtlLmVkdQ==
†These authors share first authorship
 Christopher S. Ward
Christopher S. Ward Zoie Diana
Zoie Diana Kate Meicong Ke2
Kate Meicong Ke2 Daniel Rittschof
Daniel Rittschof