- 1Department of Biology, Boston University, Boston, MA, United States
- 2Marine Laboratory, University of Guam, Mangilao, GU, United States
- 3Mote Marine Laboratory, Summerland Key, FL, United States
Reef-building corals form complex relationships with a wide range of microbial partners, including symbiotic algae in the family Symbiodiniaceae and various bacteria. These coral-associated communities can be shaped to varying degrees by environmental context. Sedimentation can structure a coral’s microbial community by altering light availability for symbiotic algae, triggering the coral’s stress response, or serving as a reservoir for both pathogenic and essential bacterial and algal symbionts. To examine the influence of sedimentation on a coral’s microbiome, we used 16S rDNA and ITS-2 amplicon sequencing to characterize the bacterial and algal communities associated with the massive scleractinian coral Porites lobata across pairs of sites along a naturally occurring sedimentation gradient in Fouha Bay, southern Guam. Additionally, we investigate the influence of proximity to sediment on the coral colony scale, by sampling from the edge and center of colonies as well as the nearby sediment. The P. lobata colonies associated with several different genotypes of Cladocopium C15 algal symbionts and often harbored different genotypes within a single colony. However, the different Cladocopium genotypes showed no structuring according to colony position or location along the sedimentation gradient. Bacterial communities were largely consistent across the sedimentation gradient, however, some rarer taxa were differentially abundant across sites. Planococcaceae shows higher abundance closer to the river mouth in coral colonies in both the edge and center of colonies. Peredibacter also shows high abundance near the river mouth but only in sediment and the edges of the colony. We find sediment plays a larger role structuring bacterial communities at the colony scale compared to a coral’s position along the sedimentation gradient. Edge communities look more similar to the sediment compared to the center communities and are also enriched in similar pathways such as those involved in nitrogen fixation. We also find center samples to be dominated by Endozoicomonas compared to the edge, supporting a role for this taxon in structuring bacterial communities and limiting bacterial diversity in coral colonies. Together these results show the differential impact sedimentation can have between sections of the coral colony microhabitat.
Introduction
Reef-building corals form complex relationships with a wide range of microbial partners, including symbiotic algae in the family Symbiodiniaceae and a consortium of bacteria. These relationships can be established vertically (transmission from maternal colonies through egg provisioning), horizontally (acquisition from the local environment) or a combination of both (Byler et al., 2013). These transmission methods can lead to different strategies for responding to changing environmental conditions. For example, some horizontal transmitters replace their dominant algal symbiont with another compatible symbiont species that is more tolerant to a particular stressor (Huang et al., 2020). Once the stressful event is over, the coral host usually recovers its original symbiont species (Sampayo et al., 2016). In comparison, vertical transmitters are limited to either changing the relative abundances of their algal symbionts to alter their community composition (Gates, 1990; Warner et al., 2002; Berkelmans and van Oppen, 2006) or not changing at all (Goulet, 2006, 2007).
While transmission of the bacterial component of the coral microbiome receives comparatively less attention than their algal counterparts, bacterial transmission research has gained recent traction following the revelation of the importance of these partners. Bacteria can provide the coral with a variety of benefits [reviewed in Bourne et al. (2016)] including antibacterial compounds to protect against pathogens (Ritchie, 2006), ultraviolet light–absorbing pigments to reduce the impact of high light conditions (Ravindran et al., 2013) and provisioning important nutrients (Wild et al., 2004). Vertical transmission of bacteria, likely through the mucus layer that coats egg-sperm bundles (Damjanovic et al., 2020b) has been observed for several coral species (Leite et al., 2017; Bernasconi et al., 2019; Damjanovic et al., 2020b). Bacteria can also be transmitted horizontally to the coral from a variety of sources including sediment and the water column but also more charismatic sources such as fireworms (Aeby and Santavy, 2006), echinoderms (Chong-Seng et al., 2011), gastropods (Raymundo et al., 2010; Nicolet et al., 2018), and bites from butterflyfish (Aeby and Santavy, 2006; Raymundo et al., 2010; Chong-Seng et al., 2011; Martin et al., 2018; Nicolet et al., 2018) and parrotfish (Ezzat et al., 2020). While the main source of bacteria in the early life history stages of corals is thought to be the water column (Patten et al., 2008; Apprill et al., 2009, 2012), sediment bacterial communities are among the most taxonomically diverse within coral reef habitats (Schöttner et al., 2012) and bacterial taxa in the coral mucus layer show higher similarity with sediment than with surrounding seawater (Carlos et al., 2013). The plethora of bacterial sources that potentially interact with coral life stages makes understanding bacterial establishment in corals complex, with the exact role of reef sediment in these relationships remaining largely underexplored.
High sediment loads in environments such as reefs near river mouths can dramatically impact the health of a coral, directly affecting coral host physiology and changes in the microbial community. Sedimentation can contribute to compromised coral health (Weber et al., 2012; Sheridan et al., 2014; Pollock et al., 2016) coral species community structure can shift dramatically with proximity to river mouths (West and Van Woesik, 2001). Sediment can negatively impact the coral host directly by removing sperm from seawater surface during reproduction events (Ricardo et al., 2015) and forcing the coral to incur energetic costs through sediment shedding, as the coral would otherwise be smothered (Fabricius and Sea, 2005). Suspended sediments can reduce light availability as silt and clay particles block sunlight, thereby reducing photosynthesis in symbiotic corals (Cortes and Risk, 1985; Fabricius and Wolanski, 2000; Fabricius et al., 2003). Sedimentation further promotes low oxygen environments (Li et al., 2014) and increases the availability of organic material and nutrients on reefs (Haapkylä et al., 2011). These environmental impacts can alter the coral microbiome by changing abiotic conditions, some of which facilitate the growth of harmful bacteria (Erftemeijer et al., 2012). Sediment can also directly deliver harmful bacteria to the coral host, as may be the case with most instances of stony coral tissue loss disease (Rosales et al., 2020). Sediment thus can have a multifold impact on the coral organism including via restructuring of its microbial members.
Given the potential role of sediment as a major force in structuring coral microbial communities, it is possible that within a single colony, a coral polyp’s distance from the boundary between the coral colony edge and the sediment substrate might determine its microbial composition. Corals can exhibit spatial intra-colony variation in their Symbiodiniaceae communities (Rowan et al., 1997; Reimer et al., 2006) and differences between sides of colonies (Garren et al., 2006) can directly correspond to light exposure (van Oppen et al., 2001; Ulstrup and Van Oppen, 2003; Kemp et al., 2015). These spatial differences in Symbiodiniaceae communities can result in within colony variation in bleaching during heat stress events (Kemp et al., 2014). While relatively understudied compared to Symbiodiniaceae, there is also evidence for intra-colony variation in coral bacterial communities. Corals can host discrete bacterial communities between compartments within a polyp (Engelen et al., 2018), between the mucus, tissue and skeleton of a coral (Sweet et al., 2011; Pollock et al., 2018), between different parts of the colony within both the tissue and/or mucus (Rohwer et al., 2002; Hansson et al., 2009; Daniels et al., 2011; Damjanovic et al., 2020a) as well as the skeleton (Marcelino et al., 2018). The ability for a single coral colony to harbor diverse microbial communities suggests that differences in sediment exposure across an individual colony may lead to intra-colony variation in the microbiome and its associated functions.
Here, we tested whether shifts in coral-associated microbial communities depend on proximity to sediment within a coral colony, as well as between sites along an established sedimentation gradient. We tested this hypothesis in Fouha Bay, Guam using the ubiquitous massive scleractinian coral, Porites lobata. In the absence of human activities, natural levels of suspended sediments on reefs are usually less than 5 mg/l (Larcombe et al., 1995; Kleypas, 1996), but on reefs adjacent to degraded watersheds, suspended sediments can reach 1,000 mg/l during periods of heavy rains, as is the case for Fouha Bay (Wolanski et al., 2003; Rongo, 2004). During 1988 a coastal road was constructed within the watershed, which resulted in high sediment loads into the bay and buried and killed many corals from 1988 to 1990 (Richmond, 1993). Since then, roughly 10 major floods annually contribute to large sediment loads which are largely retained within the bay; flushing occurs only about 2–5 times per year by storm-driven swells (Wolanski et al., 2003). There is an established gradient of sediment load in the bay (Figure 1) with sediment loads decreasing with distance from the river mouth (Rongo, 2004). Coral richness declines exponentially with increasing sedimentation rate along this channel (Minton, 2015). However, P. lobata exists at almost all sites along this gradient. We sampled algal and bacterial communities from individual P. lobata colonies across this gradient to assess the role of sediment proximity in structuring the coral microbiome between sites and within coral colonies. We show that coral-associated microbial community composition depends on proximity to sediment within a coral colony. In addition, distance to the river mouth (site) has an effect on coral microbiome community composition, but this is largely limited to the rarer taxa living on the edge of the coral colony.
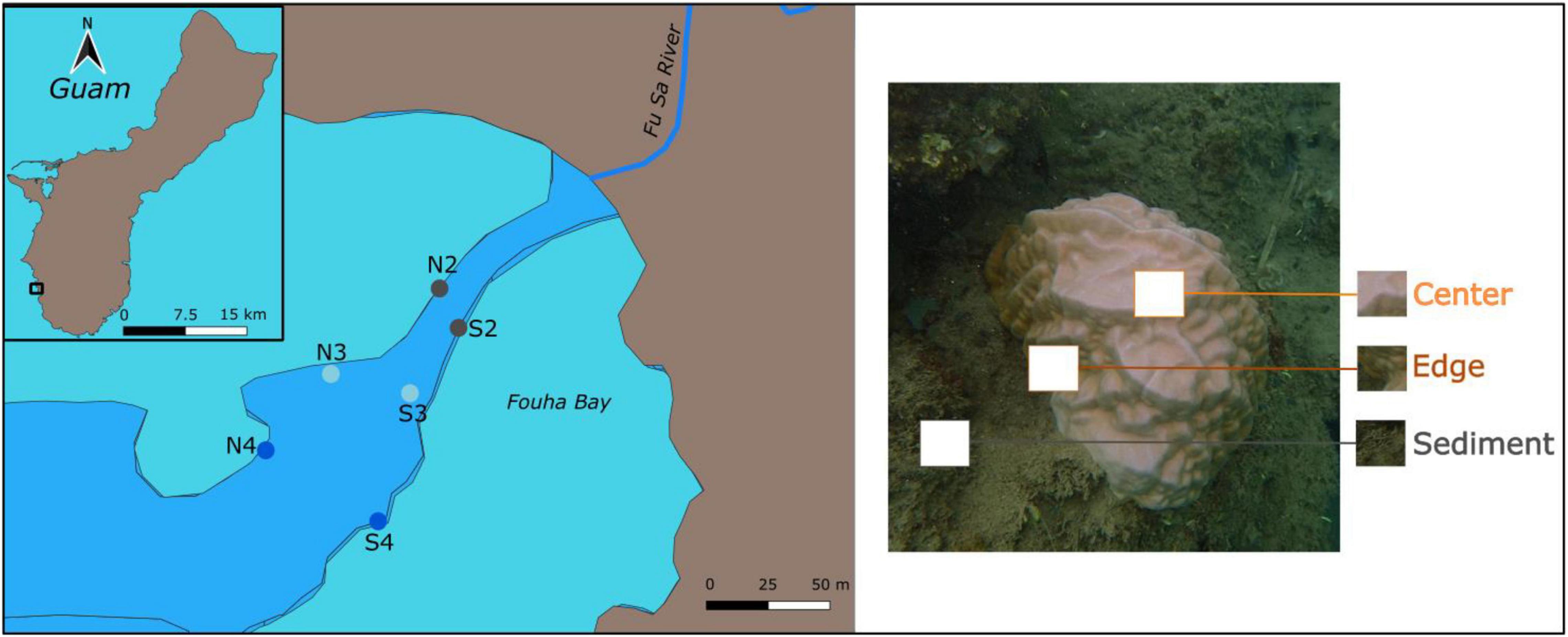
Figure 1. Left panel: Map of Porites lobata coral collection locations (N = 6) in Fouha Bay on the island of Guam (inset). These locations are labeled based on their northern (N) or southern (S) location within the bay and distance from river mouth (2 = closest to mouth, 4 = furthest from mouth), which is a proxy for sedimentation (2 = highest sedimentation, 4 = lowest sedimentation). Right panel: Picture of a P. lobata colony showing the three sampled positions for each coral colony.
Materials and Methods
Coral Colony and Environmental Data Collection
Eight to ten P. lobata colonies were sampled at each of the six locations in Fouha Bay, Guam (Figure 1) via hammer and chisel in April 2019. These locations were further clustered into sites (2–4) that reflect position along the sedimentation gradient as described by Rongo (2004; Figure 1). Site 1 was excluded from this study as it is virtually devoid of coral, with only a few colonies of Leptastrea purpurea remaining. The site closest to the river mouth (site 2) not only experiences higher sedimentation but also lower average light and more frequent decreases in salinity compared to the other two sites. All three sites experience similar temperatures (Supplementary Figure 1). The La Sa Fua River discharges into Fouha Bay through a small canyon cut through the reef flat. This canyon is about 20–30 m wide with depth varying between 0.5 m at the shore and about 7 m at the reef edge. Corals grow along the walls of the canyon with some additional coral outcrops scattered throughout the bay. All specimens collected for this study were growing on the edges of the canyon. All colonies (N = 52) were sampled in the center of the colony, half (N = 23) were also sampled at the edge and sediment (N = 18) was collected adjacent (<0.25 m) to the coral colony. Samples were placed in collection bags and flash frozen in liquid nitrogen upon collection and transferred to −80°C until DNA isolation.
Environmental data were sampled at 30 min intervals over 8 months (April 19, 2018–January 19, 2019) that spanned both dry and wet seasons. Temperature and conductivity were sampled using a HOBO U24-002-C (Onset Computer Corporation, Bourne, MA, United States) saltwater temperature/conductivity logger; conductivity was converted to salinity following calibration using a refractometer. Illuminance was recorded using HOBO UA-002-08 pendant temperature/light data logger.
To supplement the center versus edge intra-colony symbiont structure hypothesis addressed herein, contributions from fifteen P. lobata colonies collected during Klepac and Barshis (2020) were also included. P. lobata is a dominant species of the backreef lagoon pools on Ofu Island in American Samoa. Although these colonies are not influenced by sedimentation, three well-studied pools experience contrasting environmental regimes (high variability, moderate variability, and low variability), which could contribute to site-wide differences in Symbiodiniaceae composition. Five large colonies (6–10 m2) per site were sampled using a core punch (1.27 cm) and rock hammer. From each colony a core was taken from the top centroid and ∼0.5–1 m down (depending on the size of the colony; N = 30 samples) from the top sampling location to determine intra-colony differences in Symbiodiniaceae.
ITS2 and 16S Metabarcoding
DNA was extracted using the DNeasy PowerSoil (Qiagen, Hilden, Germany) kit at the University of Guam Marine Laboratory. Tissue samples were first placed in a bead beater for 30 s (with C1 solution), followed by centrifugation and transfer of the resulting supernatant to a fresh 1.5 ml tube. DNA was purified using a QIAcube automated liquid handler (Qiagen, Hilden, Germany). Bacterial 16S (for center, edge, and sediment samples) and algal ITS2 libraries (for center and edge only) were prepared at Boston University. 16S preps also included a negative control prepped with nuclease-free water. ITS2 libraries were generated following Baumann et al. (2017) and 16S libraries were generated by amplifying the V4/V5 region using modified 515f (Parada et al., 2016) and modified 806r (Apprill et al., 2015). These two library sets were then pooled in 1:2 ITS2:16S and sequenced on an Illumina MiSeqV2 (paired-end 250 bp) at Tufts University Core Facility (TUCF). All samples were prepared for 16S but sediment samples were excluded from ITS2 analysis as P. lobata transmits symbionts vertically (Richmond and Hunter, 1990). Colonies from Ofu Island were prepped for ITS2 as described in Klepac and Barshis (2020).
Symbiodiniaceae Community Analyses
Raw paired-end sequences for each sample were processed using SymPortal (Hume et al., 2019) with default parameters. ITS2 type profiles (representative of putative Symbiodiniaceae genotypes) were predicted and characterized by specific sets of defining intragenomic ITS2 sequence variants (DIVs). Pearson’s Chi-Squared test was performed to compare differences in dominant ITS2 type profiles between sites and intra-colony positions. T-tests were used to compare bacterial beta diversity between dominant ITS2 type profiles that were different between center and edge versus colonies that showed the same profile.
Bacterial Community Analyses
16S Data were pre-processed in bbmap (Chaisson and Tesler, 2012) and cutadapt (Martin, 2013) removed primer sequences. DADA2 (Callahan et al., 2016) truncated reads, calculated error rates, de-duplicated reads, inferred sequence variants, merged paired reads, and removed chimeras (98% not chimeras). In total, 8,663 non-bimeric amplicon sequence variants (ASVs) were assigned taxonomy using the Silva v132 dataset (Glöckner et al., 2017) with a minimum bootstrap confidence of 50. Using phyloseq (McMurdie and Holmes, 2013), ASVs assigning to family “Mitochondria,” order “Chloroplast,” and any with taxonomic designations defined as “Eukaryota” were removed. Using the R package decontam (Davis et al., 2017), ASVs that were determined to be contaminants through the use of the negative control were also removed. ASVs were then mapped against NCBI’s NT database using BLASTN (-e value 1e-5 -max_target_seqs 10) and all ASVs belonging to Eukaryota were removed. The resulting ASV table was rarefied to 5,000 reads (Supplementary Figure 2) using vegan (Oksanen et al., 2020). MCMC.OTU (Matz, 2016) trimmed ASVs representing <0.1% of counts or only present in less than 2% of samples, which retained 1,548 out of 7,654 ASVs. MCMC.OTU also identified putative outlier samples (defined as total counts falling below z-score cutoff of -2.5) and four samples were removed from all downstream 16S analyses (Supplementary Data).
To estimate bacterial diversity, Phyloseq (McMurdie and Holmes, 2013) calculated three alpha diversity metrics on an untrimmed, rarefied dataset: observed ASV richness, Shannon index, and inverse Simpson’s index. Shapiro and Levene’s tests were used to assess normality and heteroskedasticity, respectively. ANOVA, Kruskal-Wallis, or Welch’s t-tests measured significance across intra-colony positions and sites. Beta diversities were compared using Bray-Curtis distances on trimmed datasets transformed to relative abundances. A PERMANOVA using the Adonis function in the vegan package (Oksanen et al., 2020) was run with 1,000 permutations to compare Bray-Curtis distances between intra-colony positions and sites. Post hoc pairwise PERMANOVAs were run as needed.
Bacterial Indicator Species and Overrepresented Pathways
The R package IndicSpecies with the command “multipatt” (De Cáceres and Jansen, 2016) was used to identify bacterial genera whose abundance was significantly associated with particular intra-colony position and site along the sedimentation gradient (p-value threshold = 0.05). PICRUSTV2 (Caicedo et al., 2020) was used to look for overrepresented pathways along the sedimentation gradient and across the coral colony. Copy numbers of gene families were predicted using the EC number database, which were then weighted by the relative abundance of ASVs. MetaCyc pathway abundances were inferred based on weighted EC number abundances and a generalized linear model (GLM) was used to compare pathway enrichment between intra-colony position and sites along the sedimentation gradient via the R package aldex (Gloor et al., 2020).
Results
Algal Communities Associated With Porites lobata
ITS2 Metabarcoding data showed that samples were dominated by Cladocopium C15, but several distinct ITS2 profiles (commonly referred to as types; LaJeunesse, 2002) exist in this population (Figure 2). Individual coral colonies can harbor distinct C15 types between intra-colony positions, although intra-colony position does not predict the Symbiodiniaceae type (X-squared = 26.682, df = 22, p-value = 0.2235). Similarly, colonies from Ofu Island also showed distinct profiles within colonies but intra-colony position did not predict Symbiodiniaceae type (X-squared = 5.5824, df = 6, p-value = 0.4716; Supplementary Figure 3). Within Fouha Bay, site did not predict Symbiodiniaceae type within both center (X-squared = 26.682, df = 22, p-value = 0.2235) and edge samples (X-squared = 15.754, df = 16, p-value = 0.4703). Comparisons of 16S beta diversity between a center-edge pair for a colony that showed intra-colony position-specific differences in dominant ITS2 profiles were no different from the pairwise beta diversity of a center-edge pair for colonies that maintained the same ITS2 profile (t = −1.2871, df = 36, p-value = 0.2063; Supplementary Figure 4).
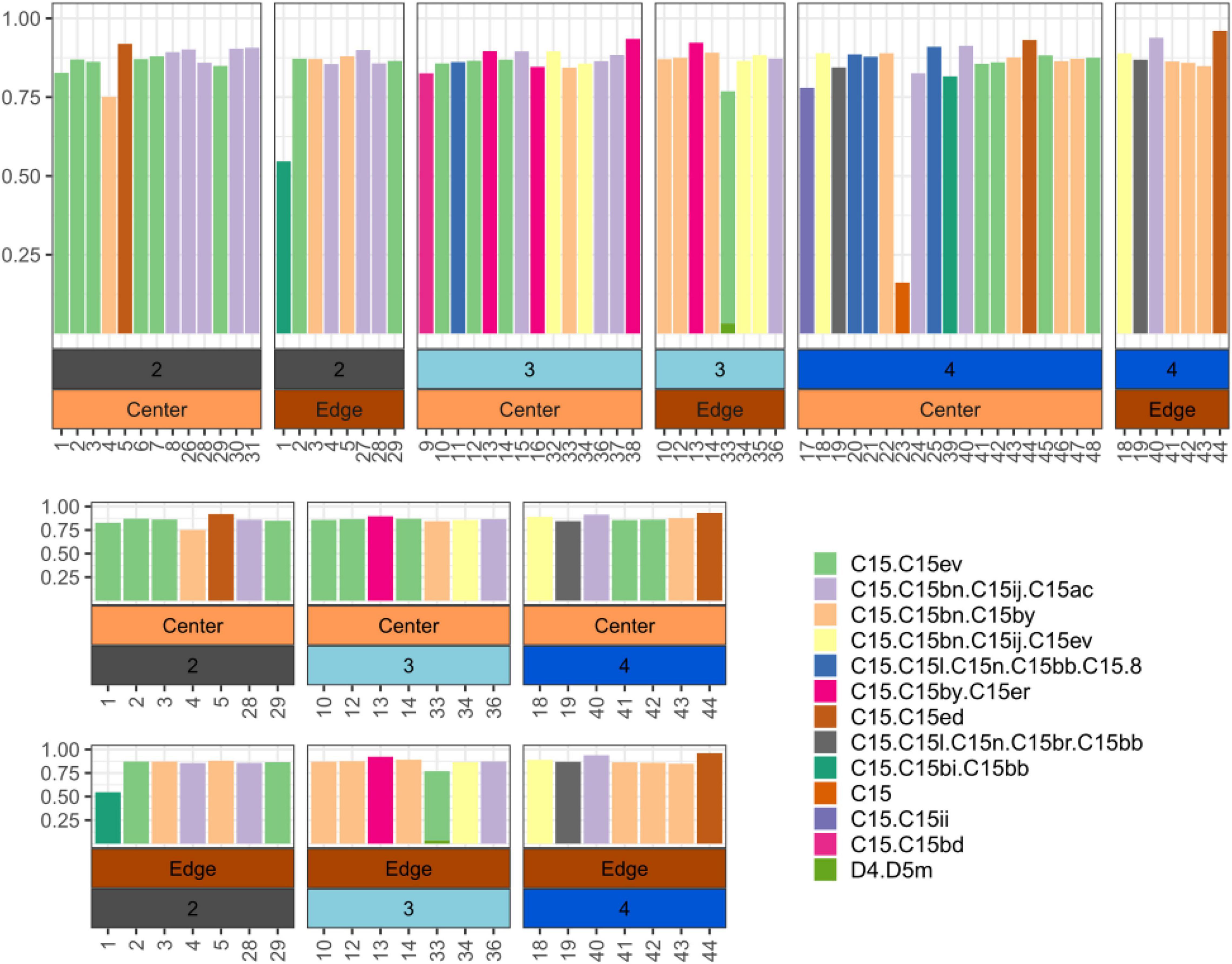
Figure 2. Top: Relative abundance of dominant ITS2 profiles across all Porites lobata colonies along the sedimentation gradient (2: site closest to river mouth - 4: site furthest away from river mouth). Bottom: Relative abundance of dominant ITS2 type profiles across only P. lobata colonies where both edge and center were sampled. Numbers on X-axis correspond to unique coral colonies sampled.
Bacterial Communities Associated With Porites lobata
16S Alpha diversity and beta diversity comparisons for both center and edge communities revealed no significant differences along the sedimentation gradient (Figures 3A,B and Supplementary Figure 5). Alpha diversity for sediment communities was also not significantly different across sites (Supplementary Figure 5). However, beta diversity was significantly different between site 2 (the site with the highest sedimentation) and sites 3 and 4 (Figure 3C). Comparisons of diversity between intra-colony positions revealed striking differences, with edge, center and sediment communities all exhibiting significant differences in both alpha and beta diversity (Figure 4). Sediment bacterial communities were more similar to edge communities than they were to center communities (Supplementary Figure 6). Dominant bacterial families from center samples included Endozoicomonadaceae (mean 42.2% relative abundance), Xenococcaceae (6.3%) and Amoebophilaceae (5%) (Supplementary Figure 7). Dominant bacterial families from edge samples included Endozoicomonadaceae (11.7%) and Amoebophilaceae (6.1%) (Supplementary Figure 8). Dominant bacterial families from sediment samples included Cyclobacteriaceae (5.4%) and Rhodobacteraceae (5.3%) (Supplementary Figure 9).
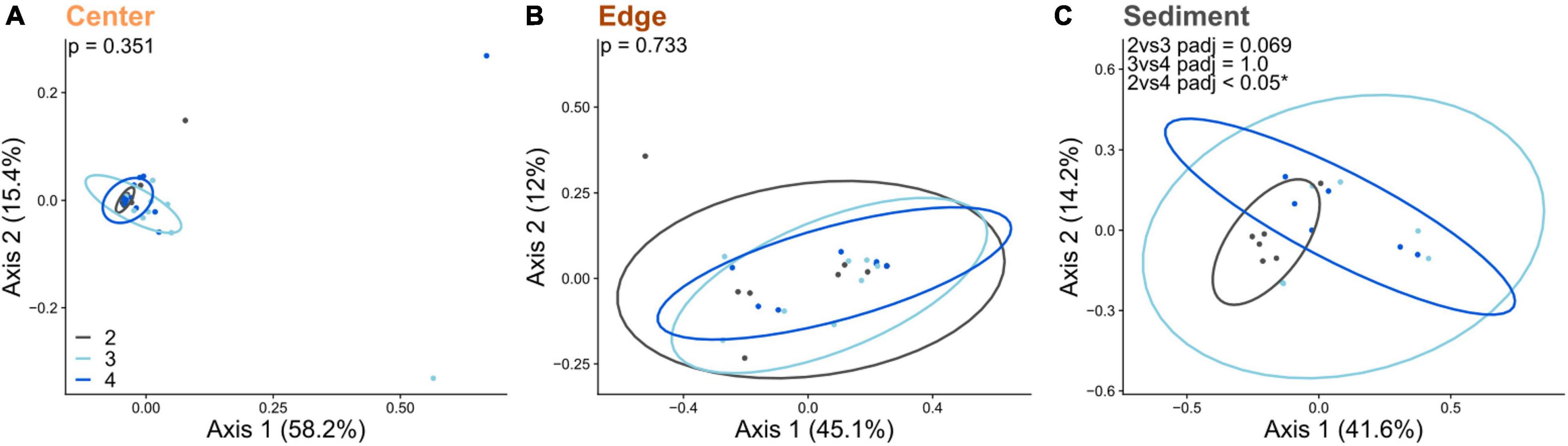
Figure 3. Principal coordinates analysis (PCoA) based on Bray-Curtis dissimilarities of 16S bacterial communities across sites along a sedimentation gradient (2 = high, 3 = moderate, 4 = low sediment load) for (A) center of coral colonies, (B) edge of coral colonies, and (C) in the reef sediment. Percentages on each axis indicate the amount of variation explained by each axis. P-values indicate significant results of PERMANOVA tests for panels (A,B) and pairwise PERMANOVA for panel (C).
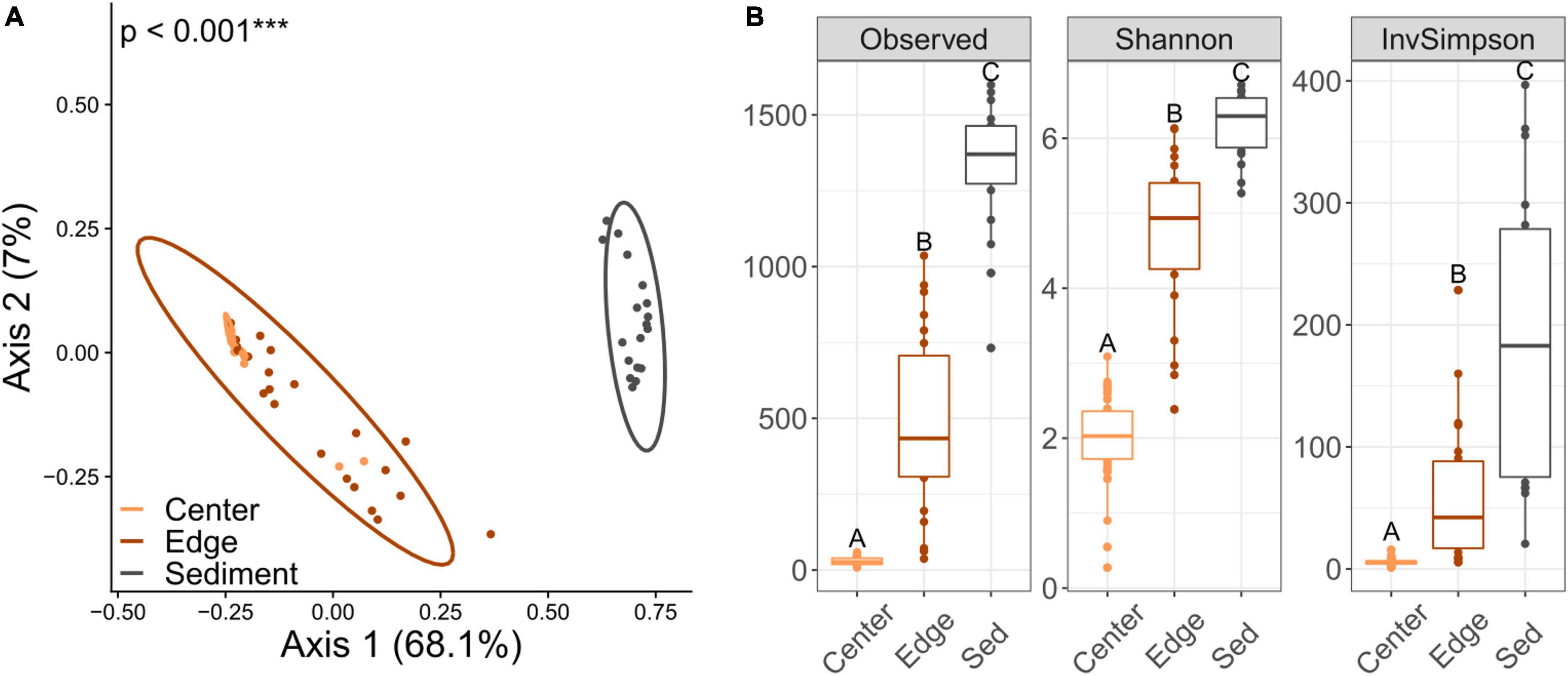
Figure 4. (A) Principal coordinates analysis based on Bray-Curtis dissimilarities of 16S bacterial communities across intra-colony positions regardless of site location. Percentages on each axis indicate the amount of variation explained by each axis. p-values indicate significant results of the PERMANOVA test. (B) Alpha diversity metrics for bacterial communities across intra-colony positions. The box represents the interquartile range (IQR) between the upper and lower quartile. The whiskers maximally extend 1 time beyond the IQR. Letters denote significant (p < 0.001) differences between groups based on Tukey’s post hoc tests of honest significant difference.
Indicator Species Between Bacterial Communities
For edge samples, several ASVs were more abundant at site 2 (the site with the highest sedimentation) or more abundant at sites 2 and 3 compared to site 4 (least sedimentation). These included ASVs belonging to genera Catenococcus, Cohaesibacter, Filomicrobium, Halioglobus, Peredibacter, Pirellula, Rhodopirellula, Woeseia, and Kiloniellaceae (Supplementary Figure 10). For samples collected from the center of the coral colony, several ASVs belonging to the genus Endozoicomonas were more abundant at sites 3 or 4 compared to 2 (i.e., associated with lower sedimentation). One ASV, of the genus Burkholderiaceae, was more abundant at sites 2 and 3 compared to 4 (Supplementary Figure 10). Only one ASV, which belonged to the genus Planococcaceae, was an indicator species for both edge and center colonies across sites. This ASV was most abundant for both intra-colony positions at site 2 (Figure 5A). Additionally, only 1 ASV, belonging to Peredibacter, was an indicator species for both coral (edge only) and sediment across sites. This ASV was also the most abundant for both sample types at site 2 (Figure 5B).
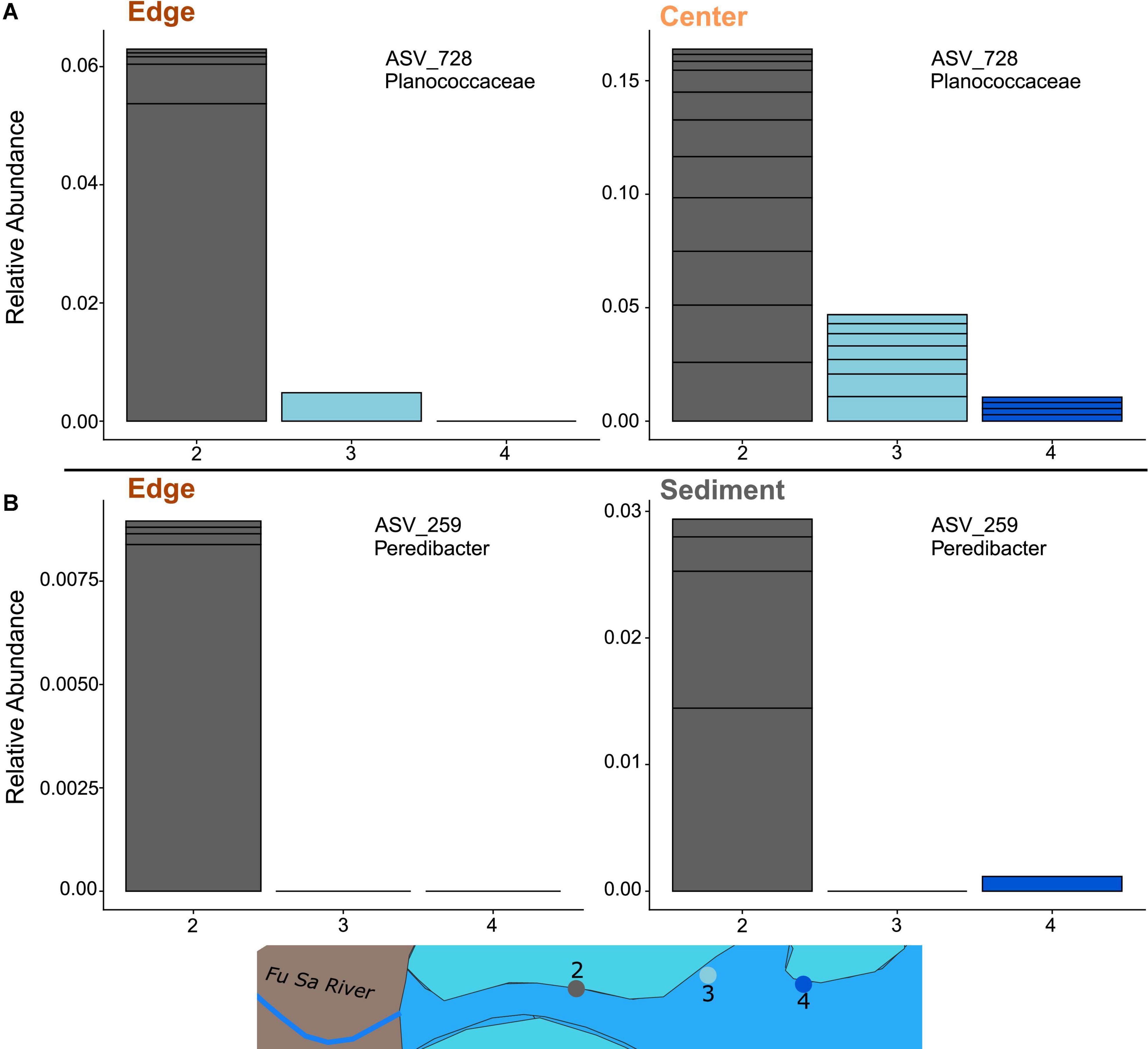
Figure 5. Relative abundance of overlapping indicator species ASVs for (A) overlap of P. lobata colony edge and center and (B) overlap of coral colony edge and reef sediment. Stacked bins represent different ASVs from the same family/genera of bacteria. Bar colors correspond to sampling sites along the sedimentation gradient.
Enrichment of Functional Pathways in Bacterial Communities
Functional pathways significantly differentially enriched when comparing colony edge and center, but not between colony edge and reef sediment, were examined to investigate differences between the center and edge of colonies that were likely mediated by bacteria available in nearby sediment. All pathways that were overrepresented at the edge and sediment compared to the center are involved in cobalamin and amino acid synthesis (Supplementary Figure 11).
Pathways that were significantly enriched when comparing both coral colony edge and center as well as colony edge and reef sediment communities were then examined to investigate differences between the center and the edge of the colony that were not necessarily due to uptake at the edge from local sediment communities. There were many more pathways enriched in this group compared to those that had no differences between edge and sediment. This comparison was further split into three groups. Pathways that were most highly represented in the center, then the edge, and then the sediment included carbohydrate and amino acid degradation and TCA cycle pathways (Supplementary Figure 12). Pathways that were most highly represented in the sediment, then the edge, and then the center involved denitrification and methanogenesis (Supplementary Figure 13). Finally, pathways that were most highly represented in the edge, then sediment, and then center consisted of amino acid metabolism pathways (Supplementary Figure 14).
Discussion
Porites lobata Harbors Distinct Cladocopium C15 Genotypes Within Individual Colonies
We sampled a total of 52 P. lobata corals along a steep sedimentation gradient in Fouha Bay, Guam, and characterized their Symbiodiniaceae algae (ITS2) and bacterial (16S) microbiome communities across the center and edge of coral colonies (ITS2, 16S) and immediately adjacent to sediment (16S only). We observed that P. lobata in Fouha Bay almost exclusively associated with algae in the genus Cladocopium and harbors different putative C15 genotypes even within a single colony. Colonies of P. lobata from Ofu Island also harbored different genotypes of Cladocopium C15 genotypes within a single colony. Massive Porites corals were historically considered to have fixed and non-mixed Symbiodiniaceae associations (Fay and Weber, 2012) with most populations of this coral being associated with a single Cladocopium species, commonly referred to as the C15 lineage (Lajeunesse et al., 2004; Thornhill et al., 2006). These strict associations with a single algal lineage were previously thought to be a result of the vertical transmission during reproduction (Fay and Weber, 2012). However, it has since been discovered that massive Porites corals can harbor mixed Cladocopium and Durusdinium communities (Terraneo et al., 2019; Tan et al., 2020) and application of the within-sample informative intragenomic sequences used by SymPortal (Hume et al., 2019) revealed different colonies within a population of P. lobata can harbor different genotypes of Cladocopium (Gardner et al., 2019; Ziegler et al., 2019; Camp et al., 2020). This study adds to the literature providing evidence against the fixed and non-mixed infection theories by demonstrating intra-colony variation of dominant Cladocopium C15 genotypes within a single colony of P. lobata across small (Fouha Bay) and large (Guam and American Samoa) geographic locations. There are two possible scenarios for explaining mixed genotypes here and they are not mutually exclusive: (1) multiple genotypes are inherited vertically and symbionts “shuffle” (Baker et al., 2004; Berkelmans and van Oppen, 2006); (2) cryptic horizontal transmission of Symbiodiniaceae (Quigley et al., 2018). Further work sequencing ITS2 profiles of the water column, nearby sediment, and P. lobata larvae is needed in order to test these hypotheses.
There was neither a clear pattern of ITS2 profile structuring according to intra-colony position nor a discernable pattern of ITS2 profiles along the sedimentation gradient, suggesting that the ITS2 community composition in P. lobata is not directly driven by sedimentation. Massive Porites can harbor different dominant ITS2 profiles across latitudinal gradients (Terraneo et al., 2019), seasons (Ziegler et al., 2015), and habitats [e.g., higher diversity of ITS2 profiles among colonies in the open ocean compared to those in mangrove habitats; (Camp et al., 2019)]. These previous studies suggest that ITS2 profile structuring for this genus of corals is determined by the local environment. However, we do not observe such structuring here, implying either differences in profiles are random or due to differences in the microenvironment not captured by this study. It is possible resampling following the flooding events might reveal shifts in dominant profile-types. Interestingly, there was a trend where 7/10 colonies that showed differing profiles between the colony center and the colony edge hosted C15.C15bn.C15 on the edge. Furthermore, of these seven, six hosted C15.C15ev at the center. This suggests colonies hosting certain Symbiodiniaceae taxa might be more likely to host divergent populations between the center and edge. However, a greater sample size will be needed to properly investigate this pattern.
Porites lobata-Associated Bacterial Communities Are Dominated by Endozoicomonas
For P. lobata-associated bacterial communities, we observed largely homogenous bacterial communities along the sedimentation gradient. The majority of coral tissue samples from the center of the colony show high homogeneity and were dominated by ASVs belonging to the bacterial genus Endozoicomonas (Proteobacteria: Oceanospirillales). The ecological role of Endozoicomonas in the coral microbiome remains unknown, albeit there are numerous theories, most of which reflect a positive role in coral health [reviewed in Neave et al. (2016)]. Previous work found that Endozoicomonas increased in abundance across several species of coral in response to heat stress and persisted 10 months after heat stress events (Maher et al., 2020). It is therefore possible that the dominance of Endozoicomonas observed here may be a result of the sediment-induced stress associated with the riverine environment of Fouha Bay. As Endozoicomonas abundance did not change across sites within the bay, we theorize that “stress events”–in this case acute large-scale flooding and associated sedimentation events that occur several times a year affecting the entire bay (Rongo, 2004)–play a stronger structuring role than the chronic sedimentation gradient observed for most of the year.
Our results are particularly interesting in light of those reported in Sweet et al. (2019), which found low abundance of Endozoicomonas in massive P. lobata less than 30 km from Fouha Bay. Given that the Guam site from Sweet et al. (2019) experiences little to no sedimentation, it is tempting to speculate that severely sedimented environments are associated with the dominance of Endozoicomonas. Alternatively, given massive Porites is a cryptic species complex (Forsman et al., 2009, 2020), these differences might reflect microbial associations correlating with discrete host lineages. The Fouha Bay P. lobata population might show an absence of community changes along the sedimentation gradient because it belongs to a lineage with an inflexible bacterial microbiome, for which there is also precedent in Endozoicomonas coral dominated colonies (Pogoreutz et al., 2018). Regardless, future work investigating host genetic structure as well as common garden experiments exposing corals to different sediment loads would be the next steps to understanding the forces that structure these microbial communities.
Rarer Taxa Show Differential Abundances in Porites lobata-Associated Bacterial Communities Along a Sedimentation Gradient
Only the rarer bacterial taxa demonstrated abundance patterning according to the site along the sedimentation gradient. The single ASV that was differentially abundant across sites for both center and edge samples belongs to the family Planococcaceae and was most abundant closest to the river mouth, regardless of where on the colony we sampled. This family contains many genera that exhibit diverse functions (Shivaji et al., 2014) making it difficult to disentangle the potential role these microbes are playing. However, this family is well represented in coral samples worldwide (Rodríguez-Gómez et al., 2021) and has also previously been reported to change abundance based on proximity to different pollution sources (Qin et al., 2020). Another ASV which belongs to the genus Peredibacter was differentially abundant across the sedimentation gradient for both edge and sediment samples. This ASV was also more abundant closest to the river mouth and is part of a genus classified as a predatory, Gram-negative, bacteriovorus group ubiquitous in soil and sewage (Davidov and Jurkevitch, 2004) that has previously been found in association with Pocillopora coral hosts (Doering et al., 2021). In edge samples, two other bacteriovorus ASVs–Pirellula and Rhodopirellula–were also in their greatest abundance at sites by the river mouth. The presence of these three bacteriovores in greater abundance near the river mouth at the edge of colonies might be significant as micropredators can drive microbiome changes in corals even at low abundances (Doering et al., 2021).
Two pathogenic bacteria, Catenococcus and Cohaesibacter were also present in highest abundances at the edge of coral colonies closer to the river mouth. Catenococcus is a member of the Vibrionaceae family and has been described as a pathogen in the seaweed Kappaphycus alvarezii in which infection by Catenococcus thiocyli causes bleaching (Zheng et al., 2016), and showed toxic activity in both sponges (Yoghiapiscessa et al., 2016) and clams (Guibert et al., 2020), suggesting that edges of coral colonies may be more susceptible to infection closer to the river mouth. Cohaesibacter has been shown to be enriched in corals exhibiting stony coral tissue loss disease (Rosales et al., 2020; Becker et al., 2021) as well as in white plague-affected corals (Sunagawa et al., 2009; Roder et al., 2014), providing further evidence that colonies at the high sedimentation site might be more at risk for coral disease. Two ASVs from putative denitrifiers Filomicrobium (Asakura et al., 2014) and Kiloniellaceae (Wiese et al., 2020) were also abundant closest to the river mouth. These findings suggest the functions of the bacterial communities are largely the same across sites, consistent with the observed lack of pathway enrichment, but differences in some of the rarer bacteria reflects the potential for differential functional capabilities. Additionally, these site differences in rare bacteria are only seen in edge samples suggesting the edges of coral colonies are more vulnerable to infection by potentially harmful bacteria compared to the center of the coral colony.
We acknowledge that there are limitations with this study in regard to the association between abundance of these rare ASVs and sediment stress. There are other environmental variables that we are unable to decouple across the sediment gradient. For one, we note that the site closest to the river experiences more extreme drops in salinity throughout the year compared to the other two sites, likely due to the freshwater influx during the rainy season. Previous research on the impact of reduced salinity on coral microbiomes is limited, but short-term salinity stress has been shown to increase Vibrio infection of the coral Montipora capitata (Shore-Maggio et al., 2018), suggesting hyposalinity can restructure bacterial communities. The site closest to the river also experiences lower light. While this is likely due to the presence of sediment in the water column, it demonstrates the difficulty of disentangling the exact mechanisms by which higher sedimentation might be impacting microbial communities. Finally, we acknowledge the possibility that the overlap between sediment and coral bacterial communities for these few rare taxa could be a result of coral expulsion of these taxa into the sediment instead of pickup from the sediment into the coral microbiome. Future work should include sampling along a sediment gradient in areas without coral colonies as well as ex situ experiments designed to parse apart the relative impacts of sediment and salinity on the structuring of coral bacterial communities.
Bacterial Communities of Porites lobata Show Greater Structuring According to Intra-colony Position Than Location Along the Sedimentation Gradient
Contrary to the minimal differences in coral-associated bacterial communities observed across the sites along the sedimentation gradient, we find significant differences in bacterial diversity between samples collected from the center and edge of colonies. Similar variation in bacterial communities has previously been shown for massive Porites across the skeleton (Marcelino et al., 2018), but we describe discrete bacterial communities based on intra-colony position within the tissue of massive Porites. High diversity differences between positions of the colony are likely due to proximity of the edge samples to surrounding sediments. This is reflected in the greater similarity between edge and sediment communities versus center and sediment communities. Additionally, many ASVs that were observed in greater abundance at the edge compared to the center were not differentially abundant between the edge and the sediment. One might have also expected to observe higher overlap between edge and sediment communities at sites with greater sedimentation, but this was not a pattern observed in our dataset. However, this study only sampled a single time point during the dry season, making it possible that edge communities exhibit increased similarity with sediment communities during large scale sedimentation events during the rainy season. Additionally, we found that colonies that hosted different Cladocopium genotypes between intra-colony positions did not show any greater differences in their bacterial communities, suggesting the forces structuring microbial communities are different between the algal and bacterial partners.
These differences in bacterial composition between colony edge and center samples also corresponded to substantial differences in functional profiles of the bacterial communities. Edge and sediment communities both showed functional enrichment of pathways relating to cobalamin and amino acid synthesis compared to center communities. Both Symbiodiniaceae and coral cells lack the ability to synthesize certain vitamins, including vitamin B12 (cobalamin), which is important for nucleic acid biosynthesis (Wagle et al., 1958) and is provided to them by bacterial symbionts (Hopkinson and Morel, 2009; Agostini et al., 2012). Greater availability of these important compounds at the edge might sustain increased coral tissue growth rates at the leading edge compared to the center of a coral colony. Edge communities were also enriched for methanogenesis and denitrification pathways compared to the center. As high sedimentation can produce low oxygen environments (Li et al., 2014), enrichment of methanogenesis at the edge might compensate for carbon buildup by providing alternative mechanisms for carbohydrate breakdown at the colony edge that do not require oxygen. Higher denitrification at the edge might be a response to a disruption in the N to P ratio as bacteria that mediate N availability through denitrifying processes can strengthen host tolerance to nutrient replete conditions and help maintain a favorable N to P ratio (Tilstra et al., 2019). However, given Fouha Bay’s proximity to a riverine environment, it seems unlikely that these corals are living under nitrogen limited conditions. Regardless, nitrogen availability impacts the coral-algal symbiont relationship as this association is largely maintained through provision and limitation of N from host to symbiont (Falkowski et al., 1993; Beraud et al., 2013; Tilstra et al., 2019). Intra-colony spatial differences in nitrogen availability suggest possible differences in the stability of this relationship across the colony. At the center, bacterial communities showed greater enrichment in functions relating to carbohydrate metabolism. This same pattern is exhibited by bacterial communities in coral colonies that are more resilient to heat stress (Ziegler et al., 2017). As heat stress can modulate the relative abundance of different sugar compounds in coral mucus (Lee et al., 2016), it is possible sediment stress is acting similarly, resulting in niche differentiation between the edge and center and ultimately discrete bacterial communities that have different metabolic profiles. Further research should investigate the concentration of these relevant compounds across the colony and in coral tissue experiencing different levels of sediment exposure to corroborate these interpretations.
Conclusion
Our findings imply that thorough characterization of a coral’s microbiome and its potential ecological roles requires sampling across a coral colony to capture the variability of its microbiome. More work is needed to elucidate if bacterial communities associated with coral colony edges have any significant bearing on the health of the rest of the colony. The notion that bacterial structuring in corals occurring within a colony can be much greater than structuring between colonies, even across a steep environmental gradient, suggests coral health and resilience at small spatial scales is challenging to predict. Additionally, this study adds to the literature describing Endozoicomonas-dominated coral colonies contributing to a seemingly fixed microbiome. Endozoicomonas species possess relatively large genomes, coding for diverse metabolites that may contribute to coral resilience (Neave et al., 2017). Characterizing the genome and metabolome of Endozoicomonas strains in Fouha Bay will be necessary to begin understanding their role in Porites lobata resilience to sedimentation.
Data Availability Statement
All scripts for analyses and figures can be found at https://github.com/jamesfifer/Fouha. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA774203.
Author Contributions
JF, BB, and SD conceived the project. JB and BB collected the samples from the field and extracted DNA. JF and VB carried out library preparation work. JF analyzed the data with assistance from NK, created the figures, and drafted the manuscript. SD and BB contributed to the writing. SD, BB, NK, and CK provided critical revisions. All authors contributed to the article and approved the submitted version.
Funding
Funding for this project came from startup funds to SD from Boston University. BB and JB were supported by Guam NSF EPSCoR (OIA-1946352). VB was supported by Greater Boston Area Research Opportunities For Young Women (GROW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Hanny Rivera, Dan Wuitchik, Colleen Bove, and Hannah Aichleman for comments on experimental design and figures, Boston University’s SCC for facilitating computational work and Ben Hume for the SymPortal analysis. We wish to thank Melissa Gabriel for providing environmental data for characterization of our study site. We also thank Marine Laboratory captain Jason Miller for facilitating the field work for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.805202/full#supplementary-material
References
Aeby, G. S., and Santavy, D. L. (2006). Factors affecting susceptibility of the coral. Mar. Ecol. Progr. Ser. 318, 103–110.
Agostini, S., Suzuki, Y., Higuchi, T., Casareto, B. E., Yoshinaga, K., Nakano, Y., et al. (2012). Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31, 147–156. doi: 10.1007/s00338-011-0831-6
Apprill, A., Marlow, H. Q., Martindale, M. Q., and Rappé, M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Apprill, A., Marlow, H. Q., Martindale, M. Q., and Rappé, M. S. (2012). Specificity of associations between bacteria and the coral Pocillopora meandrina during early development. Appl. Environ. Microbiol. 78, 7467–7475. doi: 10.1128/AEM.01232-12
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Asakura, T., Date, Y., and Kikuchi, J. (2014). Comparative analysis of chemical and microbial profiles in estuarine sediments sampled from kanto and tohoku regions in Japan. Anal. Chem. 86, 5425–5432. doi: 10.1021/ac5005037
Baker, A. C., Starger, C. J., McClanahan, T. R., and Glynn, P. W. (2004). Corals’ adaptive response to climate change. Nature 430, 741–741. doi: 10.1038/430741a
Baumann, J., Davies, S., Aichelman, H., and Castillo, K. (2017). Coral Symbiodinium community composition across the Belize Mesoamerican Barrier Reef System is driven by host species and environmental variability. bioRxiv [Preprint]. doi: 10.1007/s00248-017-1096-6
Becker, C., Brandt, M., Miller, C., and Apprill, A. (2021). Stony coral tissue loss disease biomarker bacteria identified in corals and overlying waters using a rapid field-based sequencing approach. bioRxiv [Preprint]. doi: 10.1101/2021.02.17.431614
Beraud, E., Gevaert, F., Rottier, C., and Ferrier-Pages, C. (2013). The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674. doi: 10.1242/jeb.085183
Berkelmans, R., and van Oppen, M. J. H. (2006). The role of zooxanthellae in the thermal tolerance of corals: a “nugget of hope” for coral reefs in an era of climate change. Proc. R. Soc. Lond. B Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Bernasconi, R., Stat, M., Koenders, A., Paparini, A., Bunce, M., and Huggett, M. J. (2019). Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 10:1529. doi: 10.3389/fmicb.2019.01529
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Byler, K. A., Carmi-Veal, M., Fine, M., and Goulet, T. L. (2013). Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS One 8:e59596. doi: 10.1371/journal.pone.0059596
Caicedo, H. H., Hashimoto, D. A., Caicedo, J. C., Pentland, A., and Pisano, G. P. (2020). Overcoming barriers to early disease intervention. Nat. Biotechnol. 38, 669–673. doi: 10.1038/s41587-020-0550-z
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., and Johnson, A. J. A, and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. methods. 13, 581–583.
Camp, E. F., Edmondson, J., Doheny, A., Rumney, J., Grima, A. J., Huete, A., et al. (2019). Mangrove lagoons of the Great Barrier Reef support coral populations persisting under extreme environmental conditions. Mar. Ecol. Progr. Ser. 625, 1–14. doi: 10.3354/meps13073
Camp, E. F., Suggett, D. J., Pogoreutz, C., Nitschke, M. R., Houlbreque, F., Hume, B. C. C., et al. (2020). Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39, 701–716. doi: 10.1007/s00338-019-01889-3
Carlos, C., Torres, T. T., and Ottoboni, L. M. M. (2013). Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci. Rep. 3:1624. doi: 10.1038/srep01624
Chaisson, M. J., and Tesler, G. (2012). Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. doi: 10.1186/1471-2105-13-238
Chong-Seng, K. M., Cole, A. J., Pratchett, M. S., and Willis, B. L. (2011). Selective feeding by coral reef fishes on coral lesions associated with brown band and black band disease. Coral Reefs 30, 473–481. doi: 10.1007/s00338-010-0707-1
Cortes, J., and Risk, M. J. (1985). A reef under siltation stress: Cahuita, Costa Rica. Bull. Mar. Sci. 36, 339–356.
Damjanovic, K., Blackall, L. L., Peplow, L. M., and van Oppen, M. J. H. (2020a). Assessment of bacterial community composition within and among Acropora loripes colonies in the wild and in captivity. Coral Reefs 39, 1245–1255. doi: 10.1007/s00338-020-01958-y
Damjanovic, K., Menéndez, P., Blackall, L. L., and van Oppen, M. J. H. (2020b). Early life stages of a common broadcast spawning coral associate with specific bacterial communities despite lack of internalized bacteria. Microb. Ecol. 79, 706–719. doi: 10.1007/s00248-019-01428-1
Daniels, C. A., Zeifman, A., Heym, K., Ritchie, K. B., Watson, C. A., Berzins, I., et al. (2011). Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Progr. Ser. 426, 29–40. doi: 10.3354/meps09024
Davidov, Y., and Jurkevitch, E. (2004). Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54, 1439–1452. doi: 10.1099/ijs.0.02978-0
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2017). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. bioRxiv [Preprint]. doi: 10.1101/221499
De Cáceres, M., and Jansen, F. (2016). Package ‘indicspecies’ for R: Relationship Between Species and Groups of Sites. R Cran. 1–31.
Doering, T., Wall, M., Putchim, L., Rattanawongwan, T., Schroeder, R., Hentschel, U., et al. (2021). Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome 9, 1–16. doi: 10.1186/s40168-021-01053-6
Engelen, A. H., Mota, C., Serrao, E. A., Coelho, M., and Marbà, N. (2018). Differentiation in fitness-related traits in response to elevated temperatures between leading and trailing edge populations of marine macrophytes. PLoS One 13:e0203666. doi: 10.1371/journal.pone.0203666
Erftemeijer, P. L. A., Riegl, B., Hoeksema, B. W., and Todd, P. A. (2012). Environmental impacts of dredging and other sediment disturbances on corals: a review. Mar. Pollut. Bull. 64, 1737–1765. doi: 10.1016/j.marpolbul.2012.05.008
Ezzat, L., Lamy, T., Maher, R. L., Munsterman, K. S., Landfield, K. M., Schmeltzer, E. R., et al. (2020). Parrotfish predation drives distinct microbial communities in reef-building corals. Anim. Microbiome 2:5. doi: 10.1186/s42523-020-0024-0
Fabricius, K. E., and Sea, R. (2005). Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar. Pollut. Bull. 50, 125–146. doi: 10.1016/j.marpolbul.2004.11.028
Fabricius, K. E., and Wolanski, E. (2000). Rapid smothering of coral reef organisms by muddy. Estuar. Coast. Shelf Sci. 50, 115–120.
Fabricius, K. E., Wild, C., Wolanski, E., and Abele, D. (2003). Effects of transparent exopolymer particles and muddy terrigenous sediments on the survival of hard coral recruits. Estuar. Coast. Shelf Sci. 57, 613–621. doi: 10.1016/S0272-7714(02)00400-6
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and McCloskey, L. (1993). Population control in symbiotic corals – Ammonium ions and organic materials maintain the density of zooxanthellae. BioScience 43, 606–611. doi: 10.2307/1312147
Fay, S. A., and Weber, M. X. (2012). The occurrence of mixed infections of Symbiodinium (Dinoflagellata) within individual hosts. J. Phycol. 48, 1306–1316. doi: 10.1111/j.1529-8817.2012.01220.x
Forsman, Z. H., Barshis, D. J., Hunter, C. L., and Toonen, R. J. (2009). Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol. Biol. 9:45. doi: 10.1186/1471-2148-9-45
Forsman, Z. H., Ritson-Williams, R., Tisthammer, K. H., Knapp, I. S. S., and Toonen, R. J. (2020). Host-symbiont coevolution, cryptic structure, and bleaching susceptibility, in a coral species complex (Scleractinia; Poritidae). Sci. Rep. 10:16995. doi: 10.1038/s41598-020-73501-6
Gardner, S. G., Camp, E. F., Smith, D. J., Kahlke, T., Osman, E. O., Gendron, G., et al. (2019). Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol. Evol. 9, 938–956. doi: 10.1002/ece3.4662
Garren, M., Walsh, S. M., Caccone, A., and Knowlton, N. (2006). Patterns of association between Symbiodinium and members of the Montastraea annularis species complex on spatial scales ranging from within colonies to between geographic regions. Coral Reefs 25, 503–512. doi: 10.1007/s00338-006-0146-1
Gates, R. D. (1990). Seawater temerature and sublethal bealching in Jamaica. Coral Reefs 8, 193–197.
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Gloor, A. G., Fernandes, A., Macklaim, J., Albert, A., Links, M., Quinn, T., et al. (2020). Package ‘ALDEx2’.
Goulet, T. L. (2006). Most corals may not change their symbionts. Mar. Ecol. Progr. Ser. 321, 1–7. doi: 10.3354/meps321001
Goulet, T. L. (2007). Most scleractinian corals and octocorals host a single symbiotic zooxanthella clade. Mar. Ecol. Progr. Ser. 335, 243–248. doi: 10.3354/meps335243
Guibert, I., Lecellier, G., Torda, G., Pochon, X., and Berteaux-Lecellier, V. (2020). Metabarcoding reveals distinct microbiotypes in the giant clam Tridacna maxima. Microbiome 8:57. doi: 10.1186/s40168-020-00835-8
Haapkylä, J., Unsworth, R. K. F., Flavell, M., Bourne, D. G., Schaffelke, B., and Willis, B. L. (2011). Seasonal rainfall and runoff promote coral disease on an inshore reef. PLoS One 6:e16893. doi: 10.1371/journal.pone.0016893
Hansson, L., Agis, M., Maier, C., and Weinbauer, M. G. (2009). Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Mar. Ecol. Progr. Ser. 397, 89–102. doi: 10.3354/meps08429
Hopkinson, B. M., and Morel, F. M. M. (2009). The role of siderophores in iron acquisition by photosynthetic marine microorganisms. BioMetals 22, 659–669. doi: 10.1007/s10534-009-9235-2
Huang, Y. Y., Carballo-Bolaños, R., Kuo, C. Y., Keshavmurthy, S., and Chen, C. A. (2020). Leptoria phrygia in Southern Taiwan shuffles and switches symbionts to resist thermal-induced bleaching. Sci. Rep. 10:7808. doi: 10.1038/s41598-020-64749-z
Hume, B. C. C., Smith, E. G., Ziegler, M., Warrington, H. J. M., Burt, J. A., LaJeunesse, T. C., et al. (2019). SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080. doi: 10.1111/1755-0998.13004
Kemp, D. W., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
Kemp, D. W., Thornhill, D. J., Rotjan, R. D., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2015). Spatially distinct and regionally endemic Symbiodinium assemblages in the threatened Caribbean reef-building coral Orbicella faveolata. Coral Reefs 34, 535–547. doi: 10.1007/s00338-015-1277-z
Klepac, C. N., and Barshis, D. J. (2020). Reduced thermal tolerance of massive coral species in a highly variable environment: reduced heat tolerance of massive corals. Proc. R. Soc. Lond. B Biol. Sci. 287, 19–21. doi: 10.1098/rspb.2020.1379rspb20201379
Kleypas, J. A. (1996). Coral reef development under naturally turbid conditions: fringing reefs near Broad Sound, Australia. Coral Reefs 15, 153–167. doi: 10.1007/BF01145886
LaJeunesse, T. C. (2002). Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400. doi: 10.1007/s00227-002-0829-2
Lajeunesse, T. C., Bhagooli, R., Hidaka, M., deVantier, L., Done, T., Schmidt, G., et al. (2004). Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Progr. Ser. 284, 147–161.
Larcombe, P., Ridd, P. V., Prytz, A., and Wilson, B. (1995). Factors controlling suspended sediment on inner-shelf coral reefs, Townsville, Australia. Coral Reefs 14, 163–171. doi: 10.1007/BF00367235
Lee, S. T. M., Davy, S. K., Tang, S. L., and Kench, P. S. (2016). Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 7:371. doi: 10.3389/fmicb.2016.00371
Leite, D. C. A., Leão, P., Garrido, A. G., Lins, U., Santos, H. F., Pires, D. O., et al. (2017). Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front. Microbiol. 8:176. doi: 10.3389/fmicb.2017.00176
Li, J., Chen, Q., Long, L. J., Dong, J. D., Yang, J., and Zhang, S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4:7320. doi: 10.1038/srep07320
Maher, R. L., Schmeltzer, E. R., Meiling, S., McMinds, R., Ezzat, L., Shantz, A. A., et al. (2020). Coral microbiomes demonstrate flexibility and resilience through a reduction in community diversity following a thermal stress event. Front. Ecol. Evol. 8:555698. doi: 10.3389/fevo.2020.555698
Marcelino, V. R., Van Oppen, M. J. H., and Verbruggen, H. (2018). Highly structured prokaryote communities exist within the skeleton of coral colonies. ISME J. 12, 300–303. doi: 10.1038/ismej.2017.164
Martin, M. (2013). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 7, 2803–2809.
Martin, S. B., Sasal, P., Cutmore, S. C., Ward, S., Aeby, G. S., and Cribb, T. H. (2018). Intermediate host switches drive diversification among the largest trematode family: evidence from the Polypipapiliotrematinae n. subf. (Opecoelidae), parasites transmitted to butterflyfishes via predation of coral polyps. Int. J. Parasitol. 48, 1107–1126. doi: 10.1016/j.ijpara.2018.09.003
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Minton, D. (2015). Changes in Coral Reef Structure Along a Sediment Gradient in Fouha Bay. Final Report for National Oceanic and Atmospheric Administration Pacific Islands Regional Office. Honolulu, HI: National Oceanic and Atmospheric Administration Pacific Islands Regional Office, 190.
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2017). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Nicolet, K. J., Chong-Seng, K. M., Pratchett, M. S., Willis, B. L., and Hoogenboom, M. O. (2018). Predation scars may influence host susceptibility to pathogens: evaluating the role of corallivores as vectors of coral disease. Sci. Rep. 8:5258. doi: 10.1038/s41598-018-23361-y
Oksanen, A. J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2020). Package ‘vegan’.
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Patten, N. L., Mitchell, J. G., Middelboe, M., Eyre, B. D., Seuront, L., Harrison, P. L., et al. (2008). Bacterial and viral dynamics during a mass coral spawning period on the Great Barrier Reef. Aquat. Microb. Ecol. 50, 209–220. doi: 10.3354/ame01179
Pogoreutz, C., Rädecker, N., Cárdenas, A., Gärdes, A., Wild, C., and Voolstra, C. R. (2018). Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol. Evol. 8, 2240–2252. doi: 10.1002/ece3.3830
Pollock, F. J., Lamb, J. B., Field, S. N., Heron, S. F., Schaffelke, B., Shedrawi, G., et al. (2016). Correction: sediment and turbidity associated with offshore dredging increase coral disease prevalence on nearby reefs (PLoS ONE (2016) 9:7 (e102498) PLoS One 11:e0165541. doi: 10.1371/journal.pone.0165541
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9:4921. doi: 10.1038/s41467-018-07275-x
Qin, Z., Zhao, Z., Jiao, W., Han, Z., Xia, L., Fang, Y., et al. (2020). Coupled photocatalytic-bacterial degradation of pyrene: removal enhancement and bacterial community responses. Environ. Res. 183:109135. doi: 10.1016/j.envres.2020.109135
Quigley, K. M., Warner, P. A., Bay, L. K., and Willis, B. L. (2018). Unexpected mixed-mode transmission and moderate genetic regulation of Symbiodinium communities in a brooding coral. Heredity 121, 524–536. doi: 10.1038/s41437-018-0059-0
Ravindran, J., Kannapiran, E., Manikandan, B., Francis, K., Arora, S., Karunya, E., et al. (2013). UV-absorbing bacteria in coral mucus and their response to simulated temperature elevations. Coral Reefs 32, 1043–1050. doi: 10.1007/s00338-013-1053-x
Raymundo, L. J., Halford, A. R., Maypa, A. P., and Kerr, A. M. (2010). Functionally diverse reef-fish communities ameliorate coral disease (Proceedings of the National Academy Sciences of United States of America (2009) 106, 40 (17067-17070) Proc. Natl. Acad. Sci. U.S.A. 107:513. doi: 10.1073/pnas.0913173107
Reimer, J. D., Takishita, K., Ono, S., Maruyama, T., and Tsukahara, J. (2006). Latitudinal and intracolony ITS-rDNA sequence variation in the symbiotic dinoflagellate genus Symbiodinium (Dinophyceae) in Zoanthus sansibaricus (Anthozoa: Hexacorallia). Phycol. Res. 54, 122–132. doi: 10.1111/j.1440-1835.2006.00419.x
Ricardo, G. F., Jones, R. J., Clode, P. L., Humanes, A., and Negri, A. P. (2015). Suspended sediments limit coral sperm availability. Sci. Rep. 5:18084. doi: 10.1038/srep18084
Richmond, R. H. (1993). Coral reefs: Present problems and future concerns resulting from anthropogenic disturbance. Integr. Comparat. Biol. 33, 524–536. doi: 10.1093/icb/33.6.524
Richmond, R., and Hunter, C. (1990). Reproduction and recruitment of corals: comparisons among the Caribbean, the Tropical Pacific, and the Red Sea. Mar. Ecol. Progr. Ser. 60, 185–203. doi: 10.3354/meps060185
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Progr. Ser. 322, 1–14. doi: 10.3354/meps322001
Roder, C., Arif, C., Bayer, T., Aranda, M., Daniels, C., Shibl, A., et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8, 31–39. doi: 10.1038/ismej.2013.127
Rodríguez-Gómez, C., Durán-Riveroll, L. M., Okolodkov, Y. B., Oliart-Ros, R. M., García-Casillas, A. M., and Cembella, A. D. (2021). Diversity of bacterioplankton and bacteriobenthos from the veracruz reef system, southwestern gulf of Mexico. Microorganisms 9, 1–26. doi: 10.3390/microorganisms9030619
Rohwer, F. L., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Progr. Ser. 243, 1–10. doi: 10.3354/meps243001
Rongo, T. (2004). Coral Community Change along a Sediment Gradient in Fouha Bay, Guam. Master’s thesis. Mangilao, GU: University of Guam Marine Laboratory.
Rosales, S. M., Clark, A. S., Huebner, L. K., Ruzicka, R. R., and Muller, E. M. (2020). Rhodobacterales and rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 11:681. doi: 10.3389/fmicb.2020.00681
Rowan, R., Knowlton, N., Baker, A., and Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265–269. doi: 10.1038/40843
Sampayo, E. M., Ridgway, T., Franceschinis, L., Roff, G., Hoegh-Guldberg, O., and Dove, S. (2016). Coral symbioses under prolonged environmental change: living near tolerance range limits. Sci. Rep. 6:36271. doi: 10.1038/srep36271
Schöttner, S., Wild, C., Hoffmann, F., Boetius, A., and Ramette, A. (2012). Spatial scales of bacterial diversity in cold-water coral reef ecosystems. PLoS One 7:e32093. doi: 10.1371/journal.pone.0032093
Sheridan, C., Grosjean, P., Leblud, J., Palmer, C. V., Kushmaro, A., and Eeckhaut, I. (2014). Sedimentation rapidly induces an immune response and depletes energy stores in a hard coral. Coral Reefs 33, 1067–1076. doi: 10.1007/s00338-014-1202-x
Shivaji, S., Srinivas, T. N. R., and Reddy, G. S. N. (2014). “The family Planococcaceae,” in The Prokaryotes: Firmicutes and Tenericutes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), doi: 10.1007/978-3-642-30120-9
Shore-Maggio, A., Aeby, G. S., and Callahan, S. M. (2018). Influence of salinity and sedimentation on Vibrio infection of the Hawaiian coral Montipora capitata. Dis. Aquat. Organ. 128, 63–71. doi: 10.3354/dao03213
Sunagawa, S., Desantis, T. Z., Piceno, Y. M., Brodie, E. L., Desalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white Plague disease-associated community changes in the caribbean coral montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Sweet, M. J., Croquer, A., and Bythell, J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52. doi: 10.1007/s00338-010-0695-1
Sweet, M., Burian, A., Fifer, J., Bulling, M., Elliott, D., and Raymundo, L. (2019). Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7:139. doi: 10.1186/s40168-019-0759-6
Tan, Y. T. R., Wainwright, B. J., Afiq-Rosli, L., Ip, Y. C. A., Lee, J. N., Nguyen, N. T. H., et al. (2020). Endosymbiont diversity and community structure in Porites lutea from Southeast Asia are driven by a suite of environmental variables. Symbiosis 80, 269–277. doi: 10.1007/s13199-020-00671-2
Terraneo, T. I., Fusi, M., Hume, B. C. C., Arrigoni, R., Voolstra, C. R., Benzoni, F., et al. (2019). Environmental latitudinal gradients and host-specificity shape Symbiodiniaceae distribution in Red Sea Porites corals. J. Biogeogr. 46, 2323–2335. doi: 10.1111/jbi.13672
Thornhill, D. J., Fitt, W. K., and Schmidt, G. W. (2006). Highly stable symbioses among western Atlantic brooding corals. Coral Reefs 25, 515–519. doi: 10.1007/s00338-006-0157-y
Tilstra, A., El-Khaled, Y. C., Roth, F., Rädecker, N., Pogoreutz, C., Voolstra, C. R., et al. (2019). Denitrification aligns with N2 fixation in Red Sea Corals. Sci. Rep. 9:19460. doi: 10.1038/s41598-019-55408-z
Ulstrup, K. E., and Van Oppen, M. J. H. (2003). Geographic and habitat partitioning of genetically distinct zooxanthellae (Symbiodinium) in Acropora corals on the Great Barrier Reef. Mol. Ecol. 12, 3477–3484. doi: 10.1046/j.1365-294X.2003.01988.x
van Oppen, M. J., Palstra, F. P., Piquet, A. M., and Miller, D. J. (2001). Patterns of coral-dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host-symbiont selectivity. Proc. Biol. Sci. 268, 1759–1767. doi: 10.1098/rspb.2001.1733
Wagle, S. R., Vaughan, D. A., Mistry, S. P., and Johnson, B. C. (1958). Vitamin B12 and nucleic acid biosynthesis. J. Biol. Chem. 239, 917–921.
Warner, M. E., Chilcoat, G. C., McFarland, F. K., and Fitt, W. K. (2002). Seasonal fluctuations in the photosynthetic capacity of photosystem II in symbiotic dinoflagellates in the Caribbean reef-building coral Montastraea. Mar. Biol. 141, 31–38. doi: 10.1007/s00227-002-0807-8
Weber, M., De Beer, D., Lott, C., Polerecky, L., Kohls, K., Abed, R. M. M., et al. (2012). Mechanisms of damage to corals exposed to sedimentation. Proc. Natl. Acad. Sci. U.S.A. 109, E1558–E1567. doi: 10.1073/pnas.1100715109
West, K., and Van Woesik, R. (2001). Spatial and temporal variance of river discharge on Okinawa (Japan): Inferring the temporal impact on adjacent coral reefs. Mar. Pollut. Bull. 42, 864–872. doi: 10.1016/S0025-326X(01)00040-6
Wiese, J., Imhoff, J. F., Horn, H., Borchert, E., Kyrpides, N. C., Göker, M., et al. (2020). Genome analysis of the marine bacterium Kiloniella laminariae and first insights into comparative genomics with related Kiloniella species. Arch. Microbiol. 202, 815–824. doi: 10.1007/s00203-019-01791-0
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y. M., and Jørgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Wolanski, E., Richmond, R. H., Davis, G., and Bonito, V. (2003). Water and fine sediment dynamics in transient river plumes in a small, reef-fringed bay, Guam. Estuar. Coast. Shelf Sci. 56, 1029–1040. doi: 10.1016/S0272-7714(02)00321-9
Yoghiapiscessa, D., Batubara, I., and Wahyudi, A. T. (2016). Antimicrobial and antioxidant activities of bacterial extracts from marine bacteria associated with sponge Stylotella sp. Am. J. Biochem. Biotechnol. 12, 36–46. doi: 10.3844/ajbbsp.2016.36.46
Zheng, Y., Yu, M., Liu, Y., Su, Y., Xu, T., Yu, M., et al. (2016). Comparison of cultivable bacterial communities associated with Pacific white shrimp (Litopenaeus vannamei) larvae at different health statuses and growth stages. Aquaculture 451, 163–169. doi: 10.1016/j.aquaculture.2015.09.020
Ziegler, M., Grupstra, C. G. B., Barreto, M. M., Eaton, M., BaOmar, J., Zubier, K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10:3092. doi: 10.1038/s41467-019-10969-5
Ziegler, M., Roder, C., Bchel, C., and Voolstra, C. R. (2015). Niche acclimatization in Red Sea corals is dependent on flexibility of host-symbiont association. Mar. Ecol. Progr. Ser. 533, 163–176. doi: 10.3354/meps11365
Keywords: coral, bacteria, intra-colony, sediment, Symbiodiniaceae
Citation: Fifer JE, Bui V, Berg JT, Kriefall N, Klepac C, Bentlage B and Davies SW (2022) Microbiome Structuring Within a Coral Colony and Along a Sedimentation Gradient. Front. Mar. Sci. 8:805202. doi: 10.3389/fmars.2021.805202
Received: 30 October 2021; Accepted: 21 December 2021;
Published: 28 January 2022.
Edited by:
Ranjeet Bhagooli, University of Mauritius, MauritiusReviewed by:
Esti Kramarsky-Winter, Ben-Gurion University of the Negev, IsraelAnnette Koenders, Edith Cowan University, Australia
Copyright © 2022 Fifer, Bui, Berg, Kriefall, Klepac, Bentlage and Davies. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James E. Fifer, SmFtZXMuZS5maWZlckBnbWFpbC5jb20=
 James E. Fifer
James E. Fifer Vy Bui1
Vy Bui1 Justin T. Berg
Justin T. Berg Nicola Kriefall
Nicola Kriefall Courtney Klepac
Courtney Klepac Bastian Bentlage
Bastian Bentlage Sarah W. Davies
Sarah W. Davies