
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Mar. Sci. , 10 December 2021
Sec. Coral Reef Research
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.800737
This article is part of the Research Topic Coral Reef Research Methods View all 22 articles
This article is a correction to:
In vitro Symbiosis of Reef-Building Coral Cells With Photosynthetic Dinoflagellates
 Kaz Kawamura1*†‡
Kaz Kawamura1*†‡ Satoko Sekida2‡
Satoko Sekida2‡ Koki Nishitsuji3†
Koki Nishitsuji3† Eiichi Shoguchi3†
Eiichi Shoguchi3† Kanako Hisata3
Kanako Hisata3 Shigeki Fujiwara1†
Shigeki Fujiwara1† Noriyuki Satoh3*†
Noriyuki Satoh3*†A Corrigendum on
In vitro Symbiosis of Reef-Building Coral Cells With Photosynthetic Dinoflagellates
by Kawamura, K., Sekida, S., Nishitsuji, K., Shoguchi, E., Hisata, K., Fujiwara, S., and Satoh, N. (2021). Front. Mar. Sci. 8:706308. doi: 10.3389/fmars.2021.706308
In the original article, in order to show that cells of the IVB5 line are derived from planula larvae of the coral Acropora tenuis, the authors presented transcriptome data from the IVB5 line cells in Table 1, Supplementary Table 1, and BioProjectID, PRJDB11647 (BioSampleID, SAMD00325495; and RawReadsID, DRA012135) as published. However, we later discovered that the medium of our culture system contains not only coral cells, but also other contaminating organisms. The RNA-seq data therefore included sequences of organisms other than Acropora tenuis. As a consequence, the data did not always support the coral origin of IVB5 line cells. Therefore, we have withdrawn these transcriptome-related data. Instead, we present experimental results of immunocytochemistry to show in vitro symbiosis of reef-building coral cells with photosynthetic dinoflagellates more clearly. The following corrections have been made:
1. Materials and Methods, Immunocytochemistry with Antibodies Specific to Acropora tenuis, paragraph one has been replaced with:
“In a previous study, we made two rabbit antibodies against synthetic oligopeptides, one corresponding to a part of A. tenuis Snail protein (a zinc finger transcriptional repressor) and the other to a part of A. tenuis Fat1 protein (a Fat-like cadherin-related tumor suppressor homolog) (for details, see Kawamura et al., 2021). To confirm that cells of the IVB5 line that engulfed algae are of A. tenuis, we carried out immunocytochemistry using the antibodies. Cultured cells suspended in the culture medium were centrifuged at 300 x g for 5 min and resuspended in phosphate-buffered salt solution (PBS). Cells were fixed with 4% paraformaldehyde in PBS for 15 min in an ice bath. After quenching with 200 mM glycine for 2 min and permeabilizing with 0.1% Triton X-100 for 10 min, cells were incubated in a mixture of 0.25% blocking reagent (Roche, Mannheim, Germany) and 5% skim milk in PBS for 30 min. Then, they were reacted with the rabbit primary antibody diluted 400-fold with PBS for 1 h and with goat anti-rabbit secondary antibody labeled with fluorescein isothiocyanate (FITC) (Vector Laboratory, Burlingame, CA, USA) diluted 200-fold with PBS for 30 min. After washing by centrifugation for 5 min twice with PBS, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). They were observed by means of a confocal microscopy system (ECLIPSE C1si, Nikon Co. LTD., Tokyo, Japan).”
2. Results, Acropora tenuis IVB5-Line Cells, paragraphs 1–3 have been replaced with:
“IVB5 is one of 20 cryo-reserved cell lines established from A. tenuis planula larvae in 2020 (Figure 1A). After replating the culture line several times, cells were frozen in 2-mL serum tubes in liquid nitrogen for permanent preservation. A few months later, cells in a tube were melted back into culture medium to proliferate as the original line of cells did (Figure 1A). Although the IVB5 line is polyclonal and contains several types of cells with different morphologies, the majority are dark, flattened, amorphous cells, 20~30 μm in length (Figure 1A). The line also contains a few brilliant cells (Figure 1A) and small elongated cells (Figure 1A). A large vacuole (Figure 1B1, B3) and several small vesicles (Figure 1B1, B2) are found in the cytoplasm of flattened amorphous cells. They extend lamellipodia and sometimes filopodia as well (Figure 1B2, B3) and show moderate locomotor activity (Supplementary Movie 1).
Using immunocytochemistry, we examined dark, flattened, amorphous cells that did not engulf symbiotic dinoflagellates (Supplementary Figure 1A) and cells that did (Supplementary Figures 1B, 1C). Although the former maintained their flattened, amorphous morphology, the latter became spherical (described later). Both showed distinct fluorescent signals in response to A. tenuis-specific antibodies (Supplementary Figures 1A-1C). In the case of anti-AtSnail, signals were restricted to the nucleus (Supplementary Figure 1C), while in the case of anti-AtFat1, fluorescent signals appeared throughout the entire cell bodies (Supplementary Figures 1A, 1B). In contrast, negative control cells that were stained with non-immunized rabbit serum did not exhibit FITC signals, but confirmed DAPI signals (Supplementary Figure 1D). These results indicate that cells of the IVB5 line that engulfed symbiotic algae are of Acropora tenuis.”
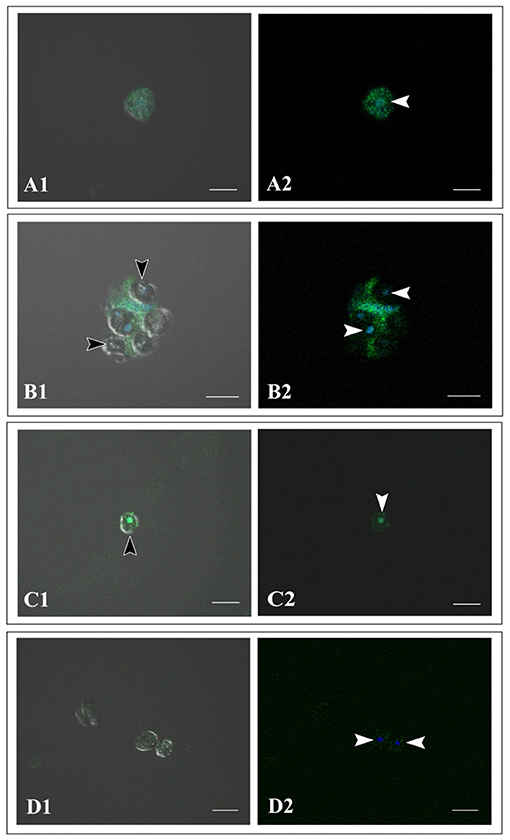
Supplementary Figure 1. Immunocytochemistry of cells of Acropora tenuis IVB5 line with specific antibodies, anti-AtFat (A,B) and anti-AtSnail (C). (D) A negative control stained with non-immunized rabbit serum. (A1–D1) Bright field images and (A2–D2) dark field images. (A) A flattened, amorphous cell that does not contain symbiotic algae. (B–D) Spherical cells that have engulfed symbiotic algae for two weeks or more. Black arrowheads indicate engulfed symbiotic algae and white arrowheads indicate nuclei of coral cells. DAPI stains the nucleus blue. Distinct staining with A. tenuis-specific antibodies indicates that the IVB5 line cells that have engulfed symbiotic algae are coral cells. Scale bar, 20 μm.
3. Results, The Dinoflagellate, Breviolum minutum, paragraph one have been replaced with:
Breviolum minutum “(Figures 1C,D) has been maintained in our laboratories for 8 years. During the proliferation stage, brown cells appear globular, approximately 8 μm in diameter, do not extend flagella, and show no locomotor activity. On the other hand, during steady state, some algal cells extend flagellae and swim in the culture medium. The B. minutum genome has been sequenced (Shoguchi et al., 2013). The identity of B. minutum used in this study was identified by partial genome sequence to be strain ITS2-type B1 (Supplementary Figure 2).”

Supplementary Figure 2. Confirmation of the used Symbiodiniaceae strain by PCR amplified 28S rDNAs sequences (Zardoya et al., 1995). The alignment shows that the 28S rDNAs sequences from PCR amplification corresponded to scaffold 5386 in the draft genome of Breviolum minutum (Shoguchi et al., 2013), confirming that the strain is the B. minutum. The single nucleotide difference between two sequences is due to a variation among 28S rDNAs copies of the clone.
4. Results, Occurrence of in vitro Symbiosis of Coral Cells With Dinoflagellates, paragraphs 1-2 have been replaced with:
“A 200–250-μL drop of culture medium containing B. minutum was added to each well of a 24-well multiplate that contained subconfluent coral cells in approximately 1 mL of growth medium. Most immobilized B. minutum gradually settled at the bottom of the culture plate wells after 5 or 6 min. The first change detected after mixing animal and algal cells was increased locomotor activity of flattened amorphous cells. Immediately after mixing, coral cells developed filopodia and actively extended and retracted pseudopodia (Figures 1C,D,E1, arrows). They crept faster than those in plates that lacked dinoflagellates (Supplementary Movie 1). Interactions between cells of the two taxa occurred shortly after mixing (Figure 1C), followed by coral cell phagocytosis of dinoflagellates (Figure 1D). Coral cells incorporated coccoid cells, but not thecate motile cells. One day after mixing, one or two and sometimes three dinoflagellates were found in the cytoplasm of individual cultured coral cells (Figure 1E1, E2). We repeated experiments more than five times and obtained the same results, indicating that this in vitro symbiosis is reproducible.
We examined whether the culture medium affects the interaction between coral and dinoflagellate cells. Three media, the conditioned cell growth medium that had been used for cell culture for 2 weeks or more, newly prepared cell growth medium, and basic seawater medium, were examined. One day after inoculation in the conditioned medium, 50.2 ± 30.2% (number of observation fields, n = 12, which contained approximately 20 cells) of cultured host cells incorporated algae (Supplementary Figure 3A). In newly prepared growth medium, algal uptake was observed in 45.7 ± 28.8% (n = 14) of all cells (Supplementary Figure 3B), and in seawater medium, 45.5 ± 30.3% (n = 7) (Supplementary Figure 3C). These results indicate that 1 day after inoculation, coral cell symbiosis with algal cells occurred similarly in each of the three media in approximately 50% of the cultured coral cells. Several days after mixing, coral cells did not show further incorporation of dinoflagellates. Even when dinoflagellates were present near or attached to coral cells, they appeared to show no interest in dinoflagellates.”
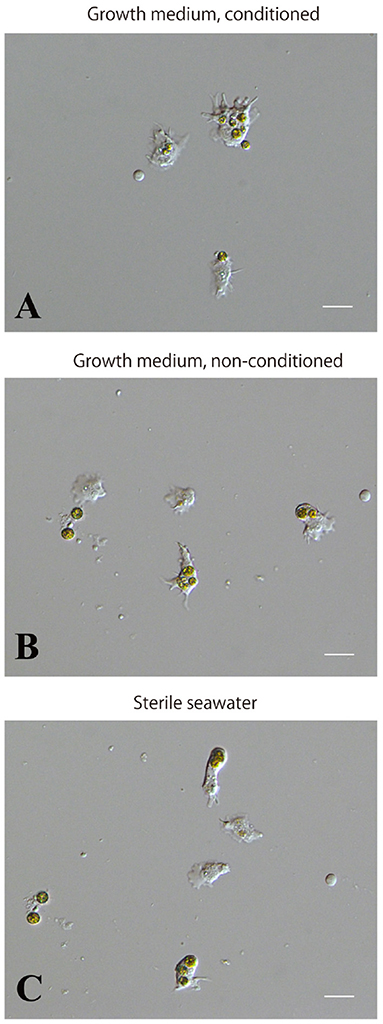
Supplementary Figure 3. Incorporation of symbiotic dinoflagellates by IVB5 coral cells in different culture media. (A) Conditioned, growth medium. (B) Non-conditioned, growth medium. (C) Basic, seawater-based medium. Incorporation of symbiotic dinoflagellates was observed one day after addition of algae. Symbiosis between the two organisms occurred in all three media. Scale bar, 20μm.
5. The Data Availability Statement should have been removed.
6. The Author Contributions statement should have read “KK and NS conceived and designed the study and prepared the manuscript. ES and SS cultured dinoflagellates. KK, SS, KN, KH, SF, and NS carried out analyses. All authors commented on it.”
7. The Acknowledgments should have read “We would like to thank all members of the Marine Genomics Unit at OIST for their support. Steven D. Aird (https://www.sda-technical-editor.org) is acknowledged for editing the manuscript. Professor Virginia Weis, one of the reviewers of this manuscript, kindly noted the mistake in our transcriptome presentation.”
The corrected Supplementary Material is shown at this article and is available online.
The authors apologize for these errors and state that they do not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Kawamura, K., Nishitsuji, K., Shoguchi, E., Fujiwara, S., and Satoh, N. (2021). Establishing sustainable cell lines of a coral, Acropora tenuis. Mar. Biotechnol. 23, 373–388. doi: 10.1007/s10126-021-10031-w
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., Takeuchi, T., Hisata, K., Tanaka, M., Fujiwara, M., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Keywords: In vitro symbiosis, corals, dinoflagellates, Acropora, endoderm, phagocytosis
Citation: Kawamura K, Sekida S, Nishitsuji K, Shoguchi E, Hisata K, Fujiwara S and Satoh N (2021) Corrigendum: In vitro Symbiosis of Reef-Building Coral Cells With Photosynthetic Dinoflagellates. Front. Mar. Sci. 8:800737. doi: 10.3389/fmars.2021.800737
Received: 23 October 2021; Accepted: 19 November 2021;
Published: 10 December 2021.
Edited by:
James Davis Reimer, University of the Ryukyus, JapanReviewed by:
Virginia M. Weis, Oregon State University, United StatesCopyright © 2021 Kawamura, Sekida, Nishitsuji, Shoguchi, Hisata, Fujiwara and Satoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaz Kawamura, a2F6dWtAa29jaGktdS5hYy5qcA==; Noriyuki Satoh, bm9yaXNreUBvaXN0Lmpw
†ORCID: Kaz Kawamura orcid.org/0000-0003-2118-9511
Koki Nishitsuji orcid.org/0000-0002-4015-7139
Eiichi Shoguchi orcid.org/0000-0003-3136-5558
Noriyuki Satoh orcid.org/0000-0002-4480-3572
Shigeki Fujiwara orcid.org/0000-0001-9464-180X
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.