- 1Key Lab of Freshwater Animal Breeding, Ministry of Agriculture, College of Fisheries, Huazhong Agricultural University, Wuhan, China
- 2Department of Aquatic Bioscience, Graduate School of Agricultural and Life Science, The University of Tokyo, Tokyo, Japan
- 3College of Marxism, Shanghai University of Finance and Economics, Shanghai, China
- 4Engineering Research Center of Green Development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt, Ministry of Education/Provincial Engineering Laboratory for Pond Aquaculture, College of Fisheries, Huazhong Agricultural University, Wuhan, China
Introduction
Small non-coding RNAs (sncRNAs) with a length <200 nucleotides (nt) play a crucial role in gene regulation at the transcriptional and post-transcriptional levels, and are mainly classified as miRNAs, piRNAs and siRNAs based on their size and Argonaute partner in biogenesis (Li and Liu, 2011). miRNAs and siRNAs are generated from double-stranded precursors by DICER and are of 20–23 nt in length, whereas piRNAs are generated from single-stranded precursors by PIWI and have a length of 24–31 nt (Huang et al., 2021). Among these, only miRNAs have been extensively investigated in animals, while both piRNA and siRNA have not been explored sufficiently. The well-known function of miRNAs it the regulation of gene expression, which is involved in immune system, differentiation and proliferation, growth and development, tumorigenesis, and cell death (O'Connell et al., 2010). To date, miRNAs have been identified and characterized in several crab species including Chinese mitten crab Eriocheir sinensis (Song et al., 2014; He et al., 2015; Chen et al., 2020; Fu et al., 2021; Luo et al., 2021), mud crab Scylla paramamosain (Li et al., 2013; Jia et al., 2018; Wang et al., 2018; Waiho et al., 2019; Jin et al., 2020; Lai et al., 2020), swimming crab Portunus trituberculatus (Ren et al., 2016; Meng et al., 2018, 2020), and freshwater crab Sinopotamon henanense (Xu et al., 2019). However, most of the studies often evaluated specific tissue expressed miRNAs in crabs.
piRNAs are a widespread strategy to effectively suppress transposable element (TE) activity to safeguard the genome from detrimental insertion mutagenesis (Tóth et al., 2016), a common issue in most animal germlines. Ubiquitously expressed piRNAs were discovered in the soma and germ lines of Annelida, Cnidaria, Echinodermata, Arthropoda, and Mollusca, but underwent tremendous changes in the Chordata (Huang et al., 2021). Interestingly, abundant piRNA-sized reads with a length of 24–31 nt were detected in the gonadal tissues of crabs (He et al., 2015; Meng et al., 2018; Luo et al., 2021), but very few studies have noticed these piRNAs in crabs (Waiho et al., 2020). Crabs, as important aquaculture species, have high commercial value as a food source. Their genetic mechanisms involved in growth, reproduction, and immune response are currently active research areas for aquaculture species. With the development of next-generation sequencing (NGS), it is possible to identify high-throughput miRNAs and piRNAs in non-model organisms by small RNA sequencing (sRNA-seq).
In this data report, we performed a small RNA sequencing analysis and detected abundant miRNAs and piRNAs in three Brachyuran crabs (E. sinensis, E. japonica, and P. trituberculatus). miRNAs were found in all investigated tissues, whereas piRNAs were specifically detected in gonadal tissues in Brachyuran crabs. The quality control of sequencing data was conducted to present a high-quality dataset. This data descriptor provides sufficient information for small RNAs in Brachyuran crabs, which is useful for future miRNAs and piRNAs studies in crabs and related species, and serves as an important reference for studies on genomics and genetic studies in crabs.
Methods
Sample Collections
Three Brachyuran crabs (E. sinensis, E. japonica, and P. trituberculatus) were collected from Tokyo Bay (35°60′N, 140°07′E) and the nearby fishery market (Ameyoko Market, Tokyo) in October 2020. Eight individuals of each species were collected and dissected the hepatopancreas, gill, leg muscle, and gonad (ovary or testis). The harvested tissues were stored in 2 ml tubes with NucleoProtect® RNA solution (TaKaRa, Shiga, Japan) for 12 h at 4°C and then stored at −80°C until further analysis.
RNA Extraction, Library Construction, and Sequencing
Total RNA was extracted from the tissues by using the ReliaPrepTM miRNA Cell and Tissue Miniprep System (Promega, WI, USA) according to the protocols. The Qubit RNA Assay Kit in Qubit 3.0 Fluorometer (Life Technologies, CA, USA) and RNA ScreenTape Assay Kit in Agilent 2200 TapeStation (Agilent Technologies, Waldbronn, Germany) were used to assess the total RNA quantity and quality. For each Brachyuran crabs, two biological replicates were made for each tissue, and each replicate mixed with three equal total RNA from the same tissues. The libraries were constructed by using the SMARTer smRNA-Seq Kit for Illumina (TaKaRa, CA, USA) following the manufacturer's instructions and sequenced on an Illumina HiSeq 2500 platform with a 50-bp single-end module (Macrogen, Tokyo).
Data Pre-processing
To obtain high-quality sRNA reads, the raw data were filtered by removing poly (A) and poly (N) sequences, and low-quality reads with an average quality score ≤ 20. The adapter sequences were trimmed by using the fastx_toolkit, followed by size filtration with a length range of 15–35 nt. The clean reads of these crabs were then filtered by removing genome no-mapping reads or known small non-coding RNAs from Rfam by using bowtie (v1.2.1, -f -k 3 -v2). The genome of E. sinensis (https://www.ncbi.nlm.nih.gov/genome/?term=Eriocheir+sinensis+) was used as reference genome for mitten crabs (E. sinensis and E. japonica), and the genome of P. trituberculatus (https://www.ncbi.nlm.nih.gov/genome/?term=Portunus+trituberculatus) was used as reference genome for swimming crab.
miRNA and piRNA Identification and Annotation
The sRNA reads were pooled from different replicates and tissues for each species, respectively, to perform miRNA prediction with miRBase 22.0 (Kozomara et al., 2019) and miRDeep2 (v.2.0.0.8) (Friedländer et al., 2012) using default parameters. After miRNA profiling, the miRNA reads were removed from the clean reads of gonadal tissues using seqkit (v0.7.3, common module) (Shen et al., 2016), which were considered as putative piRNAs for subsequent analysis. The ping-pong amplification signatures of piRNA reads were calculated by PPmeter (Jehn et al., 2018) and unitas (Gebert et al., 2017), while proTRAC (Rosenkranz and Zischler, 2018) were used to identify the piRNA clusters based on genomic mapping result. The reads have a length of 24–31 nt and significant ping-pong signal were considered as canonical piRNAs in these species.
Downstream Analysis
The total miRNA reads of each library were used for normalization to analyze differentially expressed miRNAs (DEMs) among different tissues. The miRNA counts were normalized according to the reads per million reads (RPM) method. The differential expression analysis of miRNAs was conducted by the DESeq2 package in R software (v4.0.2), and the miRNAs with a |log2 (Fold change) | ≥ 1) and adjusted p < 0.05 were considered as DEMs.
Technical Validation
The quantity and quality of extracted total RNA were validated using Agilent 2200 TapeStation with an RNA ScreenTape Assay Kit (Supplementary Figure 1A). The RIN value > 8.5 of samples can be used for total RNA mixture. The cDNA libraries were validated by using a High Sensitivity D1000 ScreenTape Kit in Agilent 2200 TapeStation (Supplementary Figure 1B). The cleanup cDNA products were purified, and their sizes were selected using AMPure XP beads (Beckman Coulter, CA, USA) in order to obtain the appropriate size (<150 bp). The post-size-selection library was also validated by Agilent 2200 TapeStation (Agilent Technologies) with a High Sensitivity D5000 ScreenTape Kit (Supplementary Figure 1C). In total, we constructed thirty small RNA libraries for the three crabs in which two biological replicates for each tissue.
Results
A total of 132.6 million raw reads were generated by Illumina sRNA-seq with a range of 1.7–8.7 million reads for each library (Supplementary Table 1). After removing the low-quality, genomic no-mapping or outside-of-size range reads, the clean reads were remained for the statistical analysis of sequence length (Supplementary Figure 2), and subsequently mixed for miRNA identification for each species. A total of 200, 161, and 129 miRNAs were identified in E. sinensis, E. japonica, and P. trituberculatus, respectively (Supplementary Table 2). Among these miRNAs, 12 miRNAs were detected in all crabs, namely miR-2a, miR-29a-3p, miR-34-3p, miR-87, miR-125a-5p, miR-181d-3p, miR-184-5p, miR-279, miR-375-5p, miR-680, miR-2424, and miR-7171-5p, whereas another 41 miRNAs were detected in both of E. sinensis and E. japonica (Figure 1A). The expression patterns of these identified miRNAs differed even among tissues of the same species (Figure 1B, Supplementary Figure 3). After identifying and removing the known and novel miRNAs, the remaining reads were considered as putative reads for piRNA processing in gonadal tissues. Abundant piRNA-sized reads with a length of 24–27 nt were both detected in the ovary and testis of these crabs. A significant 10-nt overlap between sense and antisense strands was detected in the ovary and testis samples of these three crabs (Figures 1C–E), which was consistent with piRNAs generated by the ping-pong amplification mechanism.
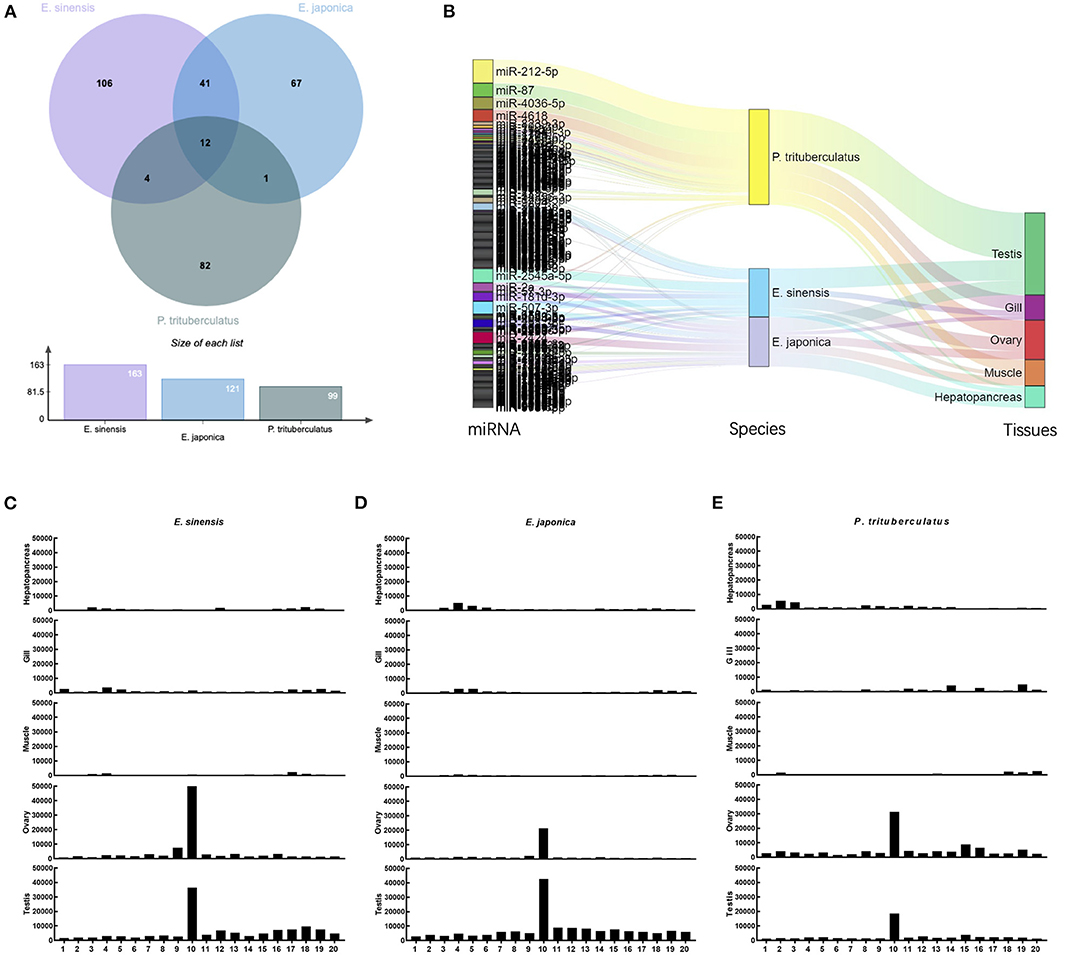
Figure 1. miRNA and piRNA identifications in three crabs. (A) Venn diagram of common and unique known miRNAs identified in these crabs; (B) Sankey diagrams depicting relationships between miRNA, species and tissue patterns; Ping-pong pair reads of putative piRNAs from somatic and gonadal tissues of E. sinensis (C), E. japonica (D), and P. trituberculatus (E).
In conclusion, our research provides a high-quality landscape of small non-coding RNAs in Brachyuran crabs, especially about the novel piRNAs detected in the gonadal tissues. The dataset provides a valuable resource for screening small RNAs in Brachyuran crabs and the related species, which will contribute to improve our understanding of the molecular mechanisms of these small RNAs in Brachyuran crabs.
Code Availability
The software utilized in this study were described in the part of methods and the following list:
1. FastQC version0.11.9 was used for the quality assessment of the FASTQ data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
2. Cutadapt version1.9.1 was used to trim the adapter sequence (http://code.google.com/p/cutadapt/).
3. Fastq_to_fasta.py from fastx_toolkit was used to transfer the sequencing data to fastq format (http://hannonlab.~cshl.edu/fastx_toolkit/index.html).
4. filter_fasta_by_size.pl was used to filter clean read by length (https://github.com/jorvis/biocode/tree/master/fasta).
5. Seqkit version0.7.3 common module was used to remove known RNAs (http://bioinf.shenwei.me/seqkit).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: DDBJ DRA [accession: DRA012855].
Ethics Statement
Ethics review and approval/written informed consent was not required as per local legislation and institutional requirements.
Author Contributions
SH designed and performed the experiments, analyzed the data, and drafted the manuscript. LZ collected the samples and assisted with the data analysis. XC conceived the study, supervised the work, and co-wrote the manuscript. All authors reviewed the manuscript.
Funding
This study was financially supported by National Key R&D Program of China (Project Number: 2018YFD0900200) and the Fundamental Research Funds for the Central Universities of China (Project Number: 2662020SCPY002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.795118/full#supplementary-material
References
Chen, M. Y., Yue, Y. X., He, J. Y., Li, P., Yan, J., Wang, F. F., et al. (2020). Screening and identification of microRNAs during larval metamorphic development of Chinese mitten crab Eriocheir sinensis. Aquac. Res. 51, 2322–2335. doi: 10.1111/are.14576
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Fu, L., Gao, T., Jiang, H., Zhang, Y., and Pan, J. L. (2021). Integrated miRNA-mRNA transcriptomic analysis of hepatopancreas reveals molecular strategies in Chinese mitten crab (Eriocheir sinensis) under acute nitrite stress. Aquacult. Int 29, 1015–1030. doi: 10.1007/s10499-021-00672-y
Gebert, D., Hewel, C., and Rosenkranz, D. (2017). unitas: the universal tool for annotation of small RNAs. BMC Genomics 18:644. doi: 10.1186/s12864-017-4031-9
He, L., Wang, Y. L., Li, Q., Yang, H. D., Duan, Z. L., and Wang, Q. (2015). Profiling microRNAs in the testis during sexual maturation stages in Eriocheir sinensis. Anim. Reprod. Sci. 162, 52–61. doi: 10.1016/j.anireprosci.2015.09.008
Huang, S. Q., Yoshitake, K., and Asakawa, S. A. (2021). A review of discovery profiling of PIWI-interacting RNAs and their diverse functions in metazoans. Int. J. Mol. Sci. 222:1166. doi: 10.3390/ijms222011166
Jehn, J., Gebert, D., Pipilescu, F., Stern, S., Kiefer, J., Hewel, C., et al. (2018). PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun. Biol. 1:137. doi: 10.1038/s42003-018-0141-4
Jia, X., Zhou, M., Zou, Z., Lin, P., Wang, Y., and Zhang, Z. (2018). Identification and comparative analysis of the ovary and testis microRNAome of mud crab Scylla paramamosain. Mol. Reprod. Dev. 85, 519–531. doi: 10.1002/mrd.22989
Jin, Q., Tian, G., Wu, J., Jiang, H. L., and Zhu, F. (2020). Identification and characterization of hemocyte microRNAs in mud crab Scylla paramamosain in response to Vibrio parahemolyticus infection. Aquaculture 524:735288. doi: 10.1016/j.aquaculture.2020.735288
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Lai, Y., Jin, Q., and Zhu, F. (2020). Differential expression of microRNAs in mud crab Scylla paramamosain in response to white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. 105, 1–7. doi: 10.1016/j.fsi.2020.06.055
Li, L., and Liu, Y. (2011). Diverse small non-coding RNAs in RNA interference pathways. Methods Mol. Biol. 764, 169–182. doi: 10.1007/978-1-61779-188-8_11
Li, S., Zhu, S., Li, C., Zhang, Z., Zhou, L., Wang, S., et al. (2013). Characterization of microRNAs in mud crab Scylla paramamosain under Vibrio parahaemolyticus infection. PLoS ONE 8:e73392. doi: 10.1371/journal.pone.0073392
Luo, B. Y., Xiong, X. Y., Liu, X., He, X. Y., and Qiu, G. F. (2021). Identification and characterization of sex-biased and differentially expressed miRNAs in gonadal developments of the Chinese mitten crab, Eriocheir sinensis. Mol. Reprod. Dev. 88, 217–227. doi: 10.1002/mrd.23459
Meng, X., Zhang, M., Gao, B., Lv, J., Li, J., and Liu, P. (2020). Integrative proteomic and microRNA analysis: Insights into mechanisms of eyestalk ablation-induced ovarian maturation in the swimming crab Portunus trituberculatus. Front. Endocrinol. 11:533. doi: 10.3389/fendo.2020.00533
Meng, X., Zhang, X., Li, J., and Liu, P. (2018). Identification and comparative profiling of ovarian and testicular microRNAs in the swimming crab Portunus trituberculatus. Gene 640, 6–13. doi: 10.1016/j.gene.2017.10.026
O'Connell, R. M., Rao, D. S., Chaudhuri, A. A., and Baltimore, D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122. doi: 10.1038/nri2708
Ren, X., Cui, Y., Gao, B., Liu, P., and Li, J. (2016). Identification and profiling of growth-related microRNAs of the swimming crab Portunus trituberculatus by using solexa deep sequencing. Marine Genomics 28, 113–120. doi: 10.1016/j.margen.2016.03.010
Rosenkranz, D., and Zischler, H. (2018). proTRAC: a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinform. 13:5. doi: 10.1186/1471-2105-13-5
Shen, W., Le, S., Li, Y., and Hu, F. (2016). SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11:e0163962. doi: 10.1371/journal.pone.0163962
Song, Y. N., Shi, L. L., Liu, Z. Q., and Qiu, G. F. (2014). Global analysis of the ovarian microRNA transcriptome: implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea:Decapoda). BMC Genomics 15:547. doi: 10.1186/1471-2164-15-547
Tóth, K. F., Pezic, D., Stuwe, E., and Webster, A. (2016). The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol. 886, 51–77. doi: 10.1007/978-94-017-7417-8_4
Waiho, K., Fazhan, H., and Zhang, Y. (2019). Gonadal microRNA expression profiles and their potential role in sex differentiation and gonadal maturation of mud crab Scylla paramamosain. Mar. Biotechnol. 21, 320–334. doi: 10.1007/s10126-019-09882-1
Waiho, K., Fazhan, H., Zhang, Y., Li, S., Zhang, Y., Zheng, H., et al. (2020). Comparative profiling of ovarian and testicular piRNAs in the mud crab Scylla paramamosain. Genomics 112, 323–331. doi: 10.1016/j.ygeno.2019.02.012
Wang, H., Wei, H., Tang, L., Lu, J., Mu, C., and Wang, C. (2018). Identification and characterization of miRNAs in the gills of the mud crab (Scylla paramamosain) in response to a sudden drop in salinity. BMC Genomics 19:609. doi: 10.1186/s12864-018-4981-6
Keywords: miRNA, piRNA, somatic tissue, gonadal tissue, crabs
Citation: Huang S, Zhang L and Cao X (2021) Transcriptional Landscape of Small Non-coding RNAs in Somatic and Gonadal Tissues of Brachyuran Crabs (Genus Eriocheir and Portunus). Front. Mar. Sci. 8:795118. doi: 10.3389/fmars.2021.795118
Received: 14 October 2021; Accepted: 18 November 2021;
Published: 07 December 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Hongyu Ma, Shantou University, ChinaKhor Waiho, University of Malaysia Terengganu, Malaysia
Copyright © 2021 Huang, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojuan Cao, Y2FveGlhb2p1YW5AbWFpbC5oemF1LmVkdS5jbg==
 Songqian Huang1,2
Songqian Huang1,2 Xiaojuan Cao
Xiaojuan Cao