- 1Department of Ocean Science, The Hong Kong University of Science and Technology, Hong Kong, Hong Kong SAR, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Department of Biology, Hong Kong Baptist University, Hong Kong, Hong Kong SAR, China
- 4State Key Laboratory of Marine Environmental Science, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 5Faculty of Biosciences and Aquaculture, Nord University, Bodø, Norway
Sclerolinum annulatus n. sp. (Annelida: Siboglinidae) is described based on specimens collected from soft sediment of the Haima cold seep in the South China Sea. Morphologically, S. annulatus n. sp. is distinct in having a tube with transverse rings and a forepart (i.e., anterior region) containing one arched row of elongated plaques on both sides of the dorsal furrow. Genome skimming, assembly, and annotation produced a nearly complete mitogenome of S. annulatus n. sp. with 15,553 bp nucleotides that encodes 13 protein-coding genes, two rRNA, and 22 tRNA. Phylogenetic analyses based on the mitochondrial cytochrome c oxidase I (cox1) gene and a concatenated dataset comprising the mitochondrial cox1 and 16S rRNA genes along with the nuclear 18S rRNA gene both strongly support the placement of S. annulatus n. sp. in the genus Sclerolinum Southward, 1961. Based on cox1, S. annulatus n. sp. is most closely related to an undescribed siboglinid from off Kushiro in Japan (“Pogonophora” sp. Kushiro-SK-2003). Transmission electron microscopy, microbial 16S rRNA amplicon sequencing, phylogenetic reconstruction, and stable isotope analyses together indicate that S. annulatus n. sp. hosts a single phylotype of sulfur-oxidizing endosymbionts.
Introduction
Siboglinids are annelid tubeworms inhabiting deep-sea chemosynthesis-based ecosystems, including hydrothermal vents, hydrocarbon seeps, decaying organic materials (e.g., sunken wood and whale falls), and reduced sediment (Hilário et al., 2011). Recent morphological and molecular studies have placed these tubeworms, formerly known as the phylum Pogonophora (Southward, 1961) or Vestimentifera (Jones, 1988), into the family Siboglinidae Caullery, 1914 under the phylum Annelida with four evolutionary lineages: Frenulata, Osedax, Sclerolinum, and Vestimentifera (Rouse et al., 2004; Halanych, 2005; Hilário et al., 2011; Li et al., 2015). Adults of siboglinids are mouthless and gutless, and reliant on symbiotic bacteria obtained each generation via horizontal transmission from their surrounding habitats for nutrition (Nussbaumer et al., 2006; Li et al., 2018; Yang et al., 2020). Most frenulates, Sclerolinum, and vestimentiferans harbor chemosynthetic sulfur-oxidizing symbionts within the host cells termed bacteriocytes, located in an organ of their trunk region known as the trophosome (Bright and Giere, 2005; Li et al., 2018). However, methane-oxidizing symbionts have only been confirmed in one species of frenulate (Schmaljohann and Flügel, 1987), and suggested in some other siboglinids (e.g., Pimenov et al., 2000). Osedax is the only group of siboglinids that lacks a discrete trophosome, with females harboring heterotrophic symbionts in their branching roots to penetrate into whale bones for nutrition (Rouse et al., 2004).
Compared to the relatively large vestimentiferans, Sclerolinum Southward, 1961 is a genus of slender tubeworms with a body divided into a forepart (i.e., anterior region) with two tentacles (i.e., palps), a cephalic lobe (i.e., prostomium), a long trunk mostly occupied by the trophosome, and a segmented opisthosoma (i.e., posterior region) that is homologous to those of frenulates and vestimentiferans (Southward et al., 2005; Eichinger et al., 2013). To date, only seven species of Sclerolinum have been described in the global ocean (Figure 1): (1) S. sibogae Southward, 1961 (East Indies; 462–1,914 m water depth); (2) S. brattstromi Webb, 1964 (Norwegian fiords; 90–870 m water depth) (Webb, 1965); (3) S. minor Southward, 1972 (off Honduras, Panama, and Colombia; 470–1,285 m water depth); (4) S. major Southward, 1972 (off Panama and Colombia; 470–1,470 m water depth); (5) S. magdalenae Southward, 1972 (off northeast Colombia near Magdalena River delta; 605–920 m water depth); (6) S. javanicum Ivanov and Selivanova, 1992 (Java Trench; 6,820–6,850 m water depth); and (7) S. contortum Smirnov, 2000 (Haakon Mosby Mud Volcano, Loki’s Castle and Storegga Slide in the Norwegian Sea, Gulf of Mexico, and Antarctic; 745–2,700 m water depth) (Smirnov, 2000; Eichinger et al., 2013; Georgieva et al., 2015). Sclerolinum tubeworms have been reported from a variety of reducing environments. For instance, (1) S. sibogae and S. magdalenae are partly buried in muddy sediment (Southward, 1961, 1972); (2) S. brattstromi, S. minor, S. major, and S. javanicum inhabit decaying wood and other vegetative debris (Webb, 1964; Southward, 1972; Ivanov and Selivanova, 1992); and (3) S. contortum lives in soft mud, decaying wood, and other vegetative debris; besides, S. contortum is also capable of settling on deployed carbonate substrates (Smirnov, 2000; Gaudron et al., 2010). In addition, undescribed Sclerolinum species have been reported from an unknown habitat off Kushiro in Japan (Kojima et al., 1997), seep sediment in the Sea of Okhotsk (Sahling et al., 2003), and vent sediment of the Loihi Seamount in Hawaii (Sahling et al., 2005).
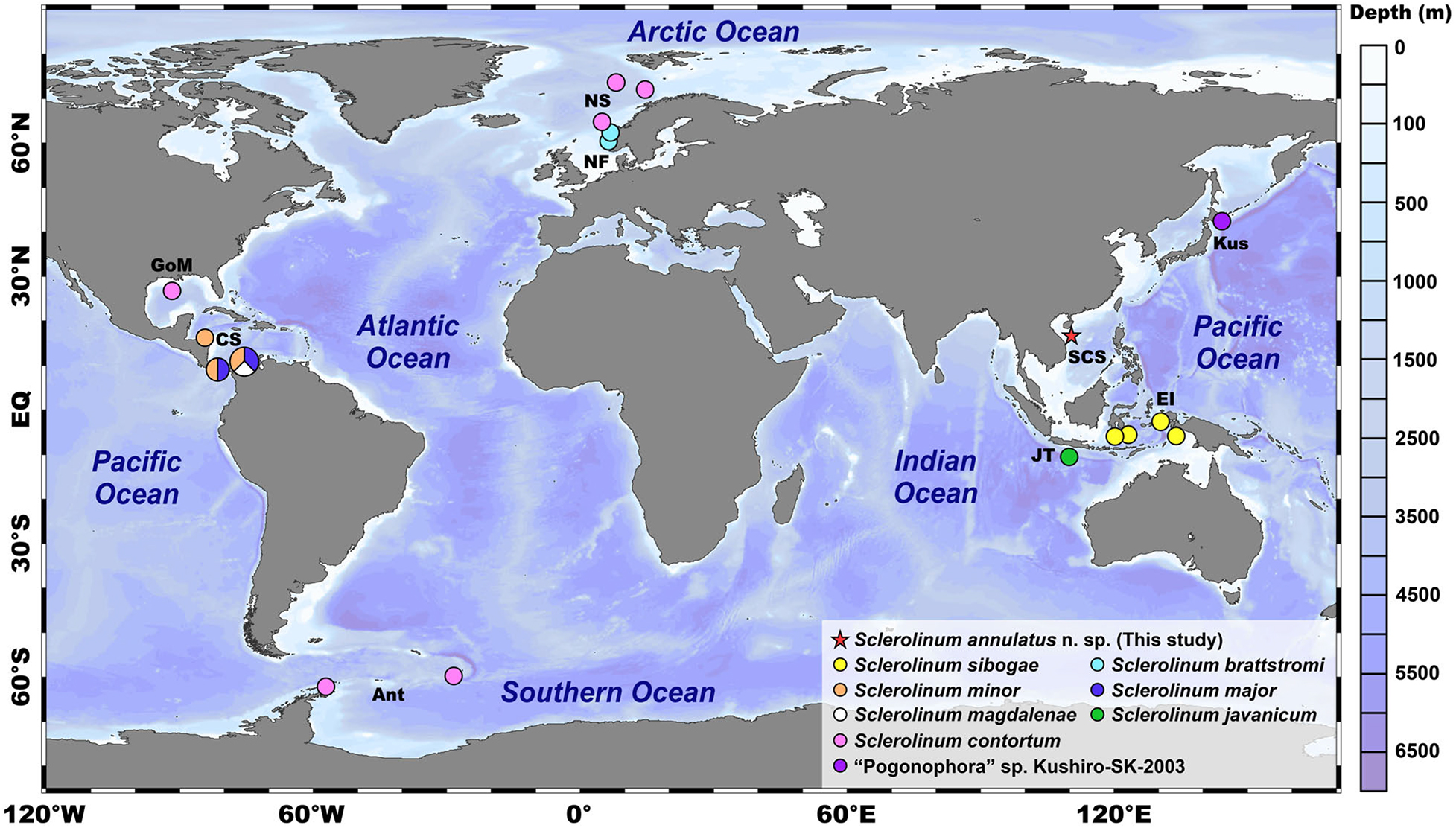
Figure 1. A map showing the known distributions of seven described Sclerolinum species, the type locality of Sclerolinum annulatus n. sp. (i.e., the Haima cold seep in the SCS as indicated by a red asterisk), along with an undescribed species of Sclerolinum “Pogonophora” sp. Kushiro-SK-2003. The map was created using Ocean Data View (ODV) v.5.0 (https://odv.awi.de). Color coding represents the water depth (unit: m). Ant, Antarctic; CS, Caribbean Sea; EI, East Indies; EQ, equator; GoM, Golf of Mexico; JT, Java Trench; Kus, Kushiro; NF, Norwegian fiords; NS, Norwegian Sea; SCS, South China Sea. Notes: the geographic coordinates of Sclerolinum javanicum inhabiting the Java Trench and “Pogonophora” sp. Kushiro-SK-2003 from off Kushiro in Japan are not available, therefore, their distributions are indicative only.
The South China Sea is a marginal sea in the Northwest Pacific with three areas along the northern continental slope known to exhibit active seepage activities, namely the Jiaolong Ridge (also known as Site F or Formosa Ridge) (Suess et al., 2005; Fujikura, 2007; Feng et al., 2018; Zhao et al., 2020), the Four Way Closure Ridge (Klaucke et al., 2016), and the Haima cold seep (Liang et al., 2017). To date, only a few species of symbiont-hosting macrobenthos have been detected to inhabit seep areas in the South China Sea, including four species of bivalves (“Bathymodiolus” aduloides Hashimoto and Okutani, 1994, Gigantidas platifrons (Hashimoto and Okutani, 1994), Archivesica marisinica (Chen et al., 2018), and Gigantidas haimaensis Xu et al., 2019), one species of squat lobster (Shinkaia crosnieri Baba and Williams, 1998), and one species of vestimentiferan (Paraescarpia echinospica Southward et al., 2002) (Yang et al., 2020; Zhao et al., 2020; Ip et al., 2021; Sun et al., 2021).
During a remotely operated vehicle (ROV) cruise in 2019, we discovered dense aggregations of Sclerolinum tubeworms in the Haima cold seep (Figure 2). The objectives of this study were, therefore, to describe the morphological characteristics of this Sclerolinum species, assemble its mitogenome, determine its phylogenetic relationships with other siboglinids, and characterize the composition and trophic mode of its symbiotic bacteria.

Figure 2. An in situ photograph showing an aggregation of Sclerolinum annulatus n. sp. (rough length × width: 10 m × 3 m) mainly surrounded by numerous empty shells of the vesicomyid clam Archivesica marissinica and a few empty shells of the bathymodioline mussel Gigantidas haimaensis. Many other macrobenthos are also present around this aggregation, such as the squat lobster Munidopsis sp. and the brittle star Ophiophthalmus sp. Photo credit: Dr. Jun Tao from Guangzhou Marine Geological Survey (Guangzhou, China).
Materials and Methods
Sample Collection and Preservation
Samples were collected in May 2019 using the ROV Haima on-board the research vessel (R/V) Haiyang 6 of Guangzhou Marine Geological Survey (Guangzhou, China) from the Haima cold seep in the South China Sea (represented by a red asterisk in Figure 1; 16°54.04′N, 110°28.45′E, 1,433 m water depth). All the retrieved samples are incomplete with the anterior and/or trunk regions only. The samples were rinsed several times with chilled seawater to remove the attached sediment. Some individuals (within their tubes) were preserved in 100% ethanol or 10% formalin diluted with Milli-Q water for morphological investigation. Other individuals (also within their tubes) were frozen at −80°C for molecular and stable isotope analyses. Type specimens have been deposited in Tropical Marine Biodiversity Collections of the South China Sea, Chinese Academy of Sciences (Guangzhou, China) as vouchers (Catalog numbers: TMBC030854–TMBC030861).
Morphological Characterization
Morphological features were examined under a Motic SMZ-171 Stereo Microscope (Motic, Xiamen, China), and photographed with a Canon 700D Camera (Canon, Tokyo, Japan) mounted on the microscope. To visualize morphological details, different parts of the tubes and the extracted soft body were carefully dissected for scanning electron microscopy (SEM) in Hong Kong Baptist University. In brief, the dissected tubes and tissues were treated with a gradient of hexamethyldisilazane-ethanol solutions (i.e., 50, 75, and 100%, each for 10 min), dried inside a fume hood, mounted on conductive carbon adhesives, sputter-coated with gold, and observed under a LEO 1530 Field Emission Scanning Electron Microscope (LEO Elektronenmikroskopie GmbH, Oberkochen, Germany).
To examine the symbiont morphology, the trophosome tissue (along with the tube) of one specimen fixed in 10% formalin was cut into small blocks of 2 mm in length, and sent to Electron Microscope Unit of The University of Hong Kong for transmission electron microscopy (TEM). Briefly, the tissue blocks were changed to cacodylate buffer (0.1 M sodium cacodylate-HCl buffer pH 7.4) containing 0.1 M sucrose to remove the fixative, post-fixed with 1% osmium tetroxide in cacodylate buffer for an hour at room temperature, washed in three changes of cacodylate buffer, then dehydrated with a gradient of ethanol solutions (i.e., 50, 70, and 90% all diluted in Milli-Q water, each for 5 min; 100%, three changes with each for 10 min). The samples were afterward exposed to propylene oxide (two changes with each for 5 min), infiltrated in epoxy resin/propylene oxide 1:1 mixture for one and a half hour followed by epoxy resin for an hour both at 37°C, and then embedded in plastic capsules that filled with epoxy resin. After polymerization at 60°C overnight, the embedded tissue blocks were cut into ultrathin sections in 100-nm thickness via a Leica Ultracut UCT Ultramicrotome (Leica, Wetzlar, Germany), mounted on the 150-mesh hexagonal copper grids, stained with 2% aqueous uranyl acetate for 20 min, washed well in running Milli-Q water, stained with Reynold’s lead citrate for 15 min, and thereafter examined under a Philips CM 100 Transmission Electron Microscope (Philips, Amsterdam, Netherlands).
DNA Extraction, Sequencing, and Assembly
Genomic DNA was extracted from individuals T1–T3 (along with their tubes due to the difficulties to extract the soft body from the tube) using the CTAB method (Stewart and Via, 1993). Integrity of DNA was monitored with 1% agarose gel electrophoresis. Purity of DNA was checked using a NanoPhotometer ® Spectrophotometer (IMPLEN, Calabasas, CA, United States). Concentration of DNA was measured using the Qubit ® dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA, United States) together with a Qubit ® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States). One DNA sequencing library with an insert size of approximately 350 bp was constructed with the NEBNext ® Ultra™ DNA Library Prep Kit for Illumina ® (NEB, Ipswich, MA, United States) for each individual, and was then sequenced on an Illumina ® NovaSeq 6000 Sequencer (Illumina, San Diego, CA, United States) in Novogene (Beijing, China) to generate 150 bp paired-end reads. Raw reads were filtered using Trimmomatic v.0.38 (Bolger et al., 2014) to remove adapters and low-quality sequences with the following settings: ILLUMINACLIP: Adapters.fas:2:30:10, LEADING = 10, TRAILING = 10, SLIDINGWINDOW = 4:15, and MINLEN = 40.
Obtained high-quality reads of each individual were de novo assembled using CLC Genomics Workbench v.20.0.4 (QIAGEN Bioinformatics, Aarhus, Denmark) under the default settings. The putative mitochondrial contig and the nuclear 18S rRNA gene were identified by applying the BLASTn option of BLAST + v.2.10.1 (Camacho et al., 2009) to search against the mitogenome and 18S rRNA gene sequences of other siboglinids retrieved from GenBank1 with an E-value threshold of 1e-50. The putative mitochondrial contig of each specimen obtained via CLC Genomics Workbench v.20.0.4 was then served as the bait for its mitogenome assembly using GetOrganelle v.1.7.5 (Jin et al., 2020) under the default settings. Alignment of the obtained mitogenomes of these three specimens was performed using MUSCLE (Edgar, 2004) implemented in MEGA X (Kumar et al., 2018) under the default settings, aiming to elucidate their genetic similarities and finalize the mitogenome with manually revision if necessary.
Mitogenome Annotation
Annotation of the mitogenome was performed using MITOS2 Web Server (Donath et al., 2019) under the default settings, except to change the genetic code to “5 invertebrate”. Resultant boundaries of each protein-coding gene (PCG) and rRNA were further examined and adjusted by aligning with the PCGs and rRNAs extracted from several published mitogenomes of siboglinids, including S. brattstromi (Li et al., 2015), using MUSCLE implemented in MEGA X under the default settings. A gene map of the mitogenome was constructed using CGView Server (Grant and Stothard, 2008). Base constitutions, codon compositions, as well as relative synonymous codon usage (RSCU) patterns were computed using MEGA X. Alignment of each mitochondrial PCG of our specimen with that derived from the mitogenome of S. brattstromi (Li et al., 2015) was carried out using MUSCLE in MEGA X under the default settings. The Kimura-2-parameter (K2P) model (Kimura, 1980) in MEGA X was then adopted to calculate the pairwise genetic distance of each mitochondrial PCG between these two species.
Genetic Distance Estimation and Phylogenetic Analyses of Siboglinids Based on the Cox1 Gene
Sequences of the mitochondrial cytochrome c oxidase I (cox1) gene of 38 siboglinids representing 14 genera and two cirratuliformians (serving as the outgroup) were downloaded from GenBank for genetic distance estimation and phylogenetic analyses, with accession numbers provided in Supplementary Table 1.
To estimate the genetic distances, alignment of the cox1 gene was conducted using MUSCLE in MEGA X under the default settings. Poorly aligned positions were eliminated by Gblocks Server2 under the default settings. The K2P model in MEGA X was then used for genetic distance calculation.
To analyze the phylogenetic relationships, alignment of the cox1 gene was also performed using MUSCLE in MEGA X under the default settings. Nevertheless, to retain as much nucleotide information as possible for phylogenetic reconstruction, poorly aligned positions were removed using Gblocks Server under the relaxed settings, which allow smaller final blocks, gap positions within the final blocks, and less strict flanking positions. Afterward, phylogenetic analyses were carried out using the maximum-likelihood (ML) approach implemented in IQ-TREE v.1.6.12 (Nguyen et al., 2015) and the Bayesian inference (BI) approach implemented in MrBayes v.3.2.7 (Ronquist et al., 2012). The best-fitting nucleotide-substitution model was determined based upon the Bayesian information criterion (BIC) via ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQ-TREE v.1.6.12. If the model selected by ModelFinder was not available in MrBayes v.3.2.7, the closest one available was used instead.
In particular, the ML analysis was performed by applying the GTR + F + I + G4 nucleotide-substitution model to run 1,000 replicates of the Shimodaira-Hasegawa-like (SH-like) approximate likelihood ratio tests (Guindon et al., 2010) together with 1,000 standard non-parametric bootstraps (NB) using IQ-TREE v.1.6.12. The BI analysis was executed by utilizing the GTR + I + G nucleotide-substitution model, running four independent Markov chains for 10 million generations, and sampling every 1,000 generations with the first 25% discarded as the burn-in using MrBayes v.3.2.7.
Phylogenetic Analyses of Siboglinids Based on a Concatenated Dataset of Three Genes
Sequences of the mitochondrial 16S rRNA and nuclear 18S rRNA genes of 26 siboglinids representing 11 genera and two cirratuliformians (serving as the outgroup) were also downloaded from GenBank for phylogenetic analyses based on the concatenated dataset of three genes (i.e., cox1 + 16S rRNA + 18S rRNA), with accession numbers summarized in Supplementary Table 2. Alignment and trimming of each gene followed the same methods detailed in the molecular analyses of cox1. Concatenation of the three gene alignments was achieved using SequenceMatrix v.1.7.8 (Vaidya et al., 2011). Methods for phylogenetic reconstruction of the concatenated three genes were identical to those of cox1, except that the best-fitting nucleotide-substitution model was assessed for each gene alignment and then applied to each gene partition. Particularly, the nucleotide-substitution model applied to the ML analysis was GTR + F + I + G4 for cox1, GTR + F + I + G4 for 16S rRNA, and TIM3e + I + G4 for 18S rRNA, and that applied to the BI analysis was GTR + I + G for cox1, GTR + I + G for 16S rRNA, and GTR + I + G for 18S rRNA.
Microbial 16S rRNA Amplicon Sequencing and Analyses
Genomic DNA was extracted from individuals T4–T6 (along with their tubes due to the difficulties to extract the small soft body from the tube) for sequencing the V3–V4 region of the microbial 16S rRNA gene. Integrity, purity, and concentration of DNA were examined using 1% agarose gel electrophoresis, a NanoPhotometer ® Spectrophotometer (IMPLEN, Calabasas, CA, United States), and the Qubit ® dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA, United States) together with a Qubit ® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States), respectively. One DNA sequencing library was constructed with the NEBNext ® Ultra™ DNA Library Prep Kit for Illumina ® (NEB, Ipswich, MA, United States) with each individual labeled using a unique barcode, and then sequenced on an Illumina ® NovaSeq 6000 Sequencer (Illumina, San Diego, CA, United States) in Novogene (Beijing, China) to generate 250 bp paired-end reads. Data filtering was carried out using the same method depicted in the section of DNA Extraction, Sequencing, and Assembly to remove adapters and low-quality sequences. Thereafter, the microbial operational taxonomic unit (OTU) identification, quantification, and taxonomy prediction were performed using USEARCH v.11.0.667 (Edgar, 2010) according to the following steps: (1) high-quality read pairs were merged using the -fastq_mergepairs command under the default settings; (2) merged reads were further filtered using the -fastq_filter command, with the -fastq_maxee parameter set to 1; (3) unique reads were obtained after removing the replicate and singleton reads with the -fastx_uniques command; (4) OTU assignment was achieved based on the UPARSE algorithm implemented in the -cluster_otus command with a similarity cutoff of 97%, and chimeras detected in this step were also excluded from further analyses; (5) an OTU table was generated using the -otutab command by mapping the merged reads to the obtained OTUs, with the mapping efficiency applied to quantify the relative abundance of each OTU; and (6) taxonomy of theses OTUs was predicted using the SINTAX algorithm (Edgar, 2016) implemented in the -sintax command to research against the Ribosomal Database Project (RDP) 16S rRNA gene training set v.16 with a bootstrap confidence cutoff of 0.80.
Sequences of the symbiont 16S rRNA gene reported from 28 siboglinids representing 12 genera and two Methylococcaceae (serving as the outgroup) were downloaded from GenBank for phylogenetic analyses. Additionally, the gill symbiont 16S rRNA gene reported from eight bivalve species representing eight genera were also included for phylogenetic inferences due to their genetic affinities to the symbionts of siboglinids based on the online BLASTn search3 and/or previous studies (e.g., Eichinger et al., 2014; Reveillaud et al., 2018). Methods for sequence alignment, trimming, phylogenetic reconstruction, and K2P genetic distance estimation were identical to those applied to the molecular analyses of cox1. The nucleotide-substitution model utilized in the ML analysis was GTR + F + R3 and in the BI analysis was GTR.
Stable Isotope Analyses
Individuals T7–T9 (along with their tubes due to the difficulties to extract the small soft body from the tube) and individuals T10–T12 (along with their tubes due to the difficulties to extract the small soft body from the tube) were combined into two samples (i.e., Sample 1: containing individuals T7–T9; Sample 2: containing individuals T10–T12) for stable isotope analyses of carbon (C), nitrogen (N), and sulfur (S) to understand the trophic mode of their symbionts. For the C and N stable isotope analyses, the two samples were freeze-dried, homogenized, weighted using a Mettler Toledo ® Microbalance (Mettler Toledo, Columbus, OH, United States), and then sealed into tin capsules. For the S stable isotope analysis, the two freeze-dried and homogenized samples were further screened with the 200-mesh sieves, dried inside an oven at 60°C, weighted using a Mettler Toledo ® Microbalance (Mettler Toledo, Columbus, OH, United States), mixed with 0.2 mg vanadium pentoxide, and thereafter sealed into tin capsules. These processed samples were analyzed using a Sercon Integra2 Elemental Analyzer Isotope Ratio Mass Spectrometry (Sercon Instruments, Crewe, United Kingdom) in The Third Institute of Oceanography, Ministry of Natural Resources (Xiamen, China). Values were reported in permille (‰) with the δ notation, relative to the standards Vienna Pee Dee Belemnite (VPDB) for C, air N2 for N, and the Canyon Diablo Triolite (CDT) for S. The precision for δ13C, δ15N, and δ34S determinations were all ± 0.2‰.
Results
Systematics
Phylum: Annelida
Family: Siboglinidae Caullery, 1914
Genus: Sclerolinum Southward, 1961
Type species: Sclerolinum sibogae Southward, 1961
Sclerolinum annulatus n. sp. (Figures 2–4)
http://zoobank.org/NomenclaturalActs/6c80eb8a-7550-4c7b-a6f8-2d631463ba34.
Type Locality
The Haima cold seep, 1,433 m water depth, off southern Hainan Island, on the northwestern slope of the South China Sea.
Type Material
Type specimens are incomplete, with the anterior and/or trunk regions but lacking the opisthosomal region. They were collected in May 2019 and are currently deposited in Tropical Marine Biodiversity Collections of the South China Sea, Chinese Academy of Sciences (Guangzhou, China). Holotype (TMBC030854): tube width 0.38 mm, tube length 42.05 cm. Paratypes (TMBC030855–TMBC030861): tube width 0.35–0.53 mm, tube length 31.86–87.85 cm.
Diagnosis
Tube straight, often possessing pronounced, irregularly transverse rings, in anterior and middle regions. Cephalic lobe with a rounded triangular tip pointing anteriorly. Dorsal plaques elongated, three on each side of dorsal furrow, forming a caret shape pointing anteriorly. Ventral plaques oral, present in two irregular rows.
Description
Tubes brownish in color, straight, flexible, and elastic, protruding as dense aggregations from sediment surface to water column (Figure 2). Tube outer surface with clearly defined transverse rings in anterior (Figures 3A,F) and middle regions (Figure 3B); rings sparsely spaced in anterior region (Figures 3A,F), closely spaced in middle region (Figure 3B), and absent in posterior region (Figure 3C). Tube walls composed of multi-layered fibers (Figures 3D,E). Tube diameter 0.35–0.53 mm. Tube thickness 23–41 μm. Tube length up to 87.85 cm. Two wrinkled tentacles (Figures 3F,G, 4A), varying greatly in length from 1.4 to 12.5 mm according to different states of contraction among measured specimens. Forepart 75–190 μm in diameter, with a deep dorsal furrow extending posteriorly from the base of two tentacles (Figure 4A). Epidermal glands scattered on forepart, more densely distributed anterior than posterior to bridle (i.e., thicken cuticular ridge) (Figures 4A,B). Cephalic lobe located on ventral side, with a rounded triangular tip pointing anteriorly (Figure 4D). Bridle located approximately 0.7 mm posterior to cephalic lobe, formed by 15 elongated to oval plaques with diameter ranging from 32 to 52 μm (Figures 4A–E). Dorsal plaques elongated, three on each side of dorsal furrow, forming a caret shape pointing anteriorly (Figures 4A–C). Ventral plaques present in two irregularly rows, each with 4–5 plaques (Figures 4D,E). Transition between the relatively smooth forepart and the highly wrinkled and densely papillated trunk region clear. Oval plaques scattered on top of epidermal papillae in trunk region (Figure 4F).
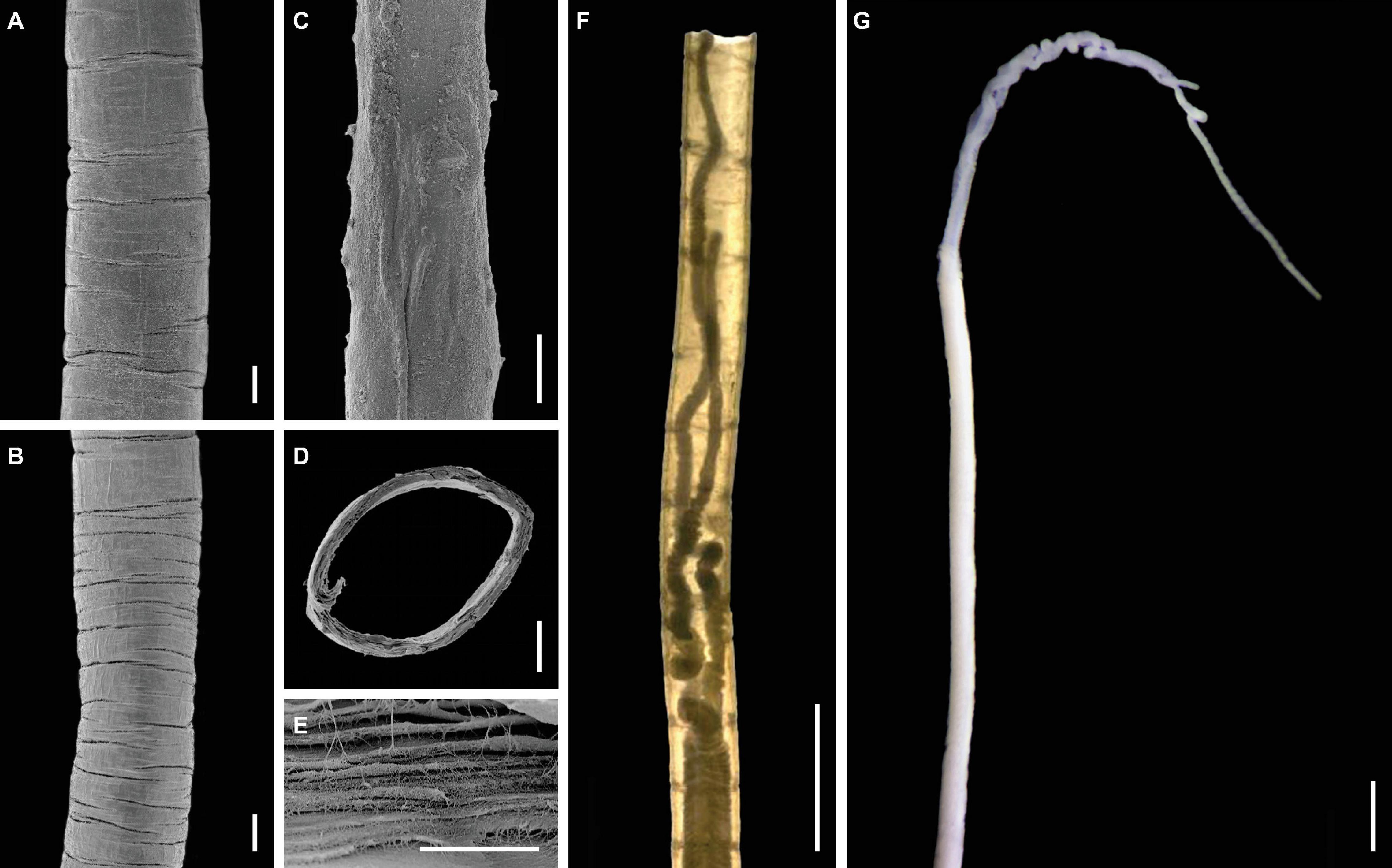
Figure 3. Tube and overall features of Sclerolinum annulatus n. sp. (A) Scanning electron microscopy (SEM) of the anterior region of the tube with sparser transverse rings (holotype TMBC030854). (B) SEM of the middle region of the tube with denser transverse rings (holotype TMBC030854). (C) SEM of the posterior region of the tube, which is rather smooth with no transverse rings (holotype TMBC030854). (D) SEM of the cross section of the tube (holotype TMBC030854). (E) SEM of a close-up section of the tube, showing the multi-layered and fibrous structure (paratype TMBC030855). (F) Light micrograph, showing the sparser transverse rings on the anterior region of the tube and two tentacles of the tubeworm (paratype TMBC030856). (G) Light micrograph in lateral view, showing two tentacles and the forepart. Scale bars: (A–D) 100 μm; (E) 10 μm; (F,G) 1 mm.
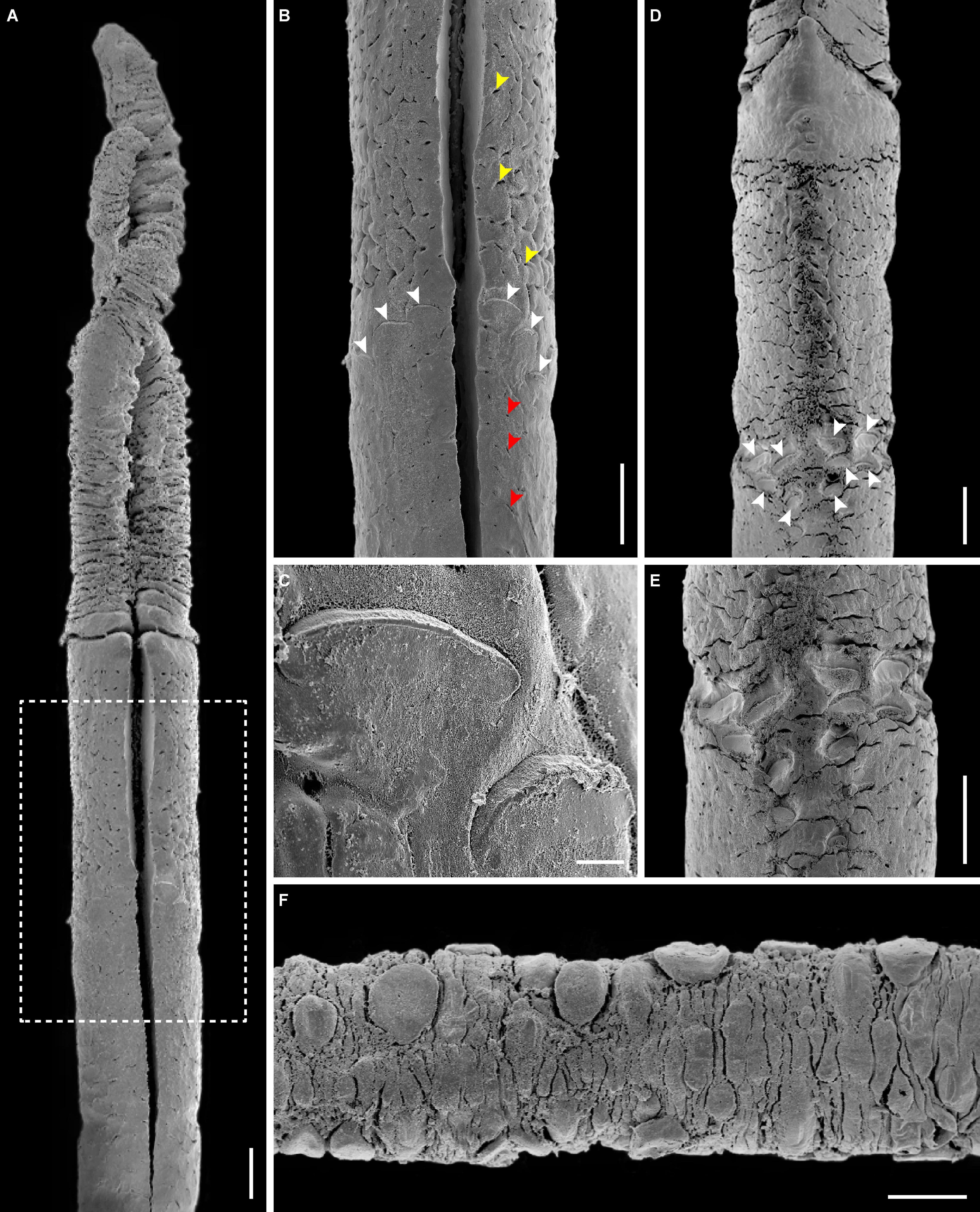
Figure 4. Morphological details of Sclerolinum annulatus n. sp. (A) Scanning electron microscopy (SEM) in dorsal view, showing two tentacles and the forepart (holotype TMBC030854). (B) SEM of a close-up section of the forepart within the white dotted box in (A), showing the epidermal glands (indicated by yellow and red arrows) and the elongated dorsal plaques of the bridle arranged in a caret shape pointing anteriorly (indicated by white arrows). (C) SEM of a close-up section of (B), showing the elongated dorsal plaques. (D) SEM in ventral view, showing the cephalic lobe with a rounded triangular tip pointing anteriorly at the base of two tentacles, the forepart, and the oral ventral plaques arranged in two rows (indicated by white arrows) (paratype TMBC030856). (E) SEM of a close-up section of (D), showing the arrangement of ventral plaques. (F) SEM of the trunk region, showing the scattered oval plaques situated on top of the epidermal papillae (paratype TMBC030857). Scale bars: (A,B,D–F) 100 μm; (C) 10 μm.
Distribution
Within the South China Sea, S. annulatus n. sp. has only been reported from the Haima cold seep—the type locality situated on the northwestern continental slope of the South China Sea.
Etymology
The species epithet “annulatus” refers to “ring” in Latin, which reflects the rings on the tube of this species.
Remarks
Sclerolinum annulatus n. sp. has a relatively large tube (0.35–0.53 mm diameter) among the seven described Sclerolinum species. The only two Sclerolinum species with comparable tube sizes are S. major (0.20–0.84 mm) (Southward, 1972) and S. contortum (0.20–0.61 mm) (Smirnov, 2000; Eichinger et al., 2013). Sclerolinum annulatus n. sp. stands out from all the seven described Sclerolinum species in that its tube is relatively straight and has clearly defined transverse rings in the anterior and middle regions (Figures 3A,B,F), which resembles the tubes of frenulates (Southward, 1972; Smirnov, 2000; Sen et al., 2020). In comparison, the tubes of most other Sclerolinum species are rather smooth (Southward, 1961). Although the tube of S. sibogae was described as deeply wrinkled (Southward, 1961) and those of some S. magdalenae as weakly wrinkled (Southward, 1972), they do not form complete rings. The tube of S. contortum is also different in typically having several bends (although it is smooth in one population) (Eichinger et al., 2013) or having only faint transverse rings (Georgieva et al., 2015). Furthermore, the bridle shape of S. annulatus n. sp. is unique among all the seven described Sclerolinum species in that it is composed of one obliquely arranged row of elongated plaques on each side of the furrow on the dorsum, and two rows of transversely arranged oval plaques on the ventrum (Figures 4A–E).
Key to species of Sclerolinum Southward, 1961
1a. Dorsal furrow present on the forepart …… 2
1b. Dorsal furrow absent on the forepart …… 6
2a. Dorsal plaques scattered around the bridle …… S. major Southward, 1972
2b. Dorsal plaques around the bridle arranged in one row …… 3
3a. Plaques around the bridle arranged in one straight line circling the body …… S. magdalenae Southward, 1972
3b. Plaques around the bridle not arranged in one straight line…… 4
4a. Plaques around the bridle sparsely distributed …… S. minor Southward, 1972
4b. Plaques around the bridle closely distributed …… 5
5a. Tube with bends, smooth, or with faint rings; plaques on the ventral side arranged in one row …… S. contortum Smirnov, 2000
5b. Tube straight, with clear rings; plaques on the ventral side arranged in two rows …… S. annulatus n. sp.
6a. Plaques absent around the girdle, present irregularly in the trunk …… S. sibogae Southward, 1961
6b. Plaques present around the girdle …… 7
7a. Dorsal plaques around the bridle arranged in a caret shape, with the tip pointing anteriorly …… S. brattstromi Webb, 1964
7b. Dorsal plaques around the bridle arranged in a caret shape, with the tip pointing posteriorly…… S. javanicum Ivanov and Selivanova, 1992
General Features of the Mitogenome
The assembled mitogenomes of S. annulatus n. sp. T1–T3 do not form a circle. They vary in length from 15,565 to 15,590 bp, with 15,553 bp being identical. Therefore, only the consensus nucleotides, with an overall base composition of 30.51% for A, 23.55% for C, 12.49% for G, and 33.45% for T, were retained as the mitogenome of S. annulatus n. sp. for downstream analyses. It contains 13 PCGs (i.e., atp6, atp8, cox1–3, cytb, nad1–6, and nad4l), two rRNA, and 22 tRNA which are typical of bilaterian mitogenomes (Boore, 1999), with potential missing nucleotides in the control region that lies between trnR and trnH (Figure 5 and Supplementary Table 3). Gene order of the S. annulatus n. sp. mitogenome is identical to that of other siboglinids reported to date (Li et al., 2015; Sun et al., 2018; Zhou et al., 2020). All 13 mitochondrial PCGs use ATG as the start codon. Most mitochondrial PCGs use TAA as the stop codon, except that cox1 uses TA, cox2 uses TAG, and nad2, nad5, as well as nad6 use a single T.
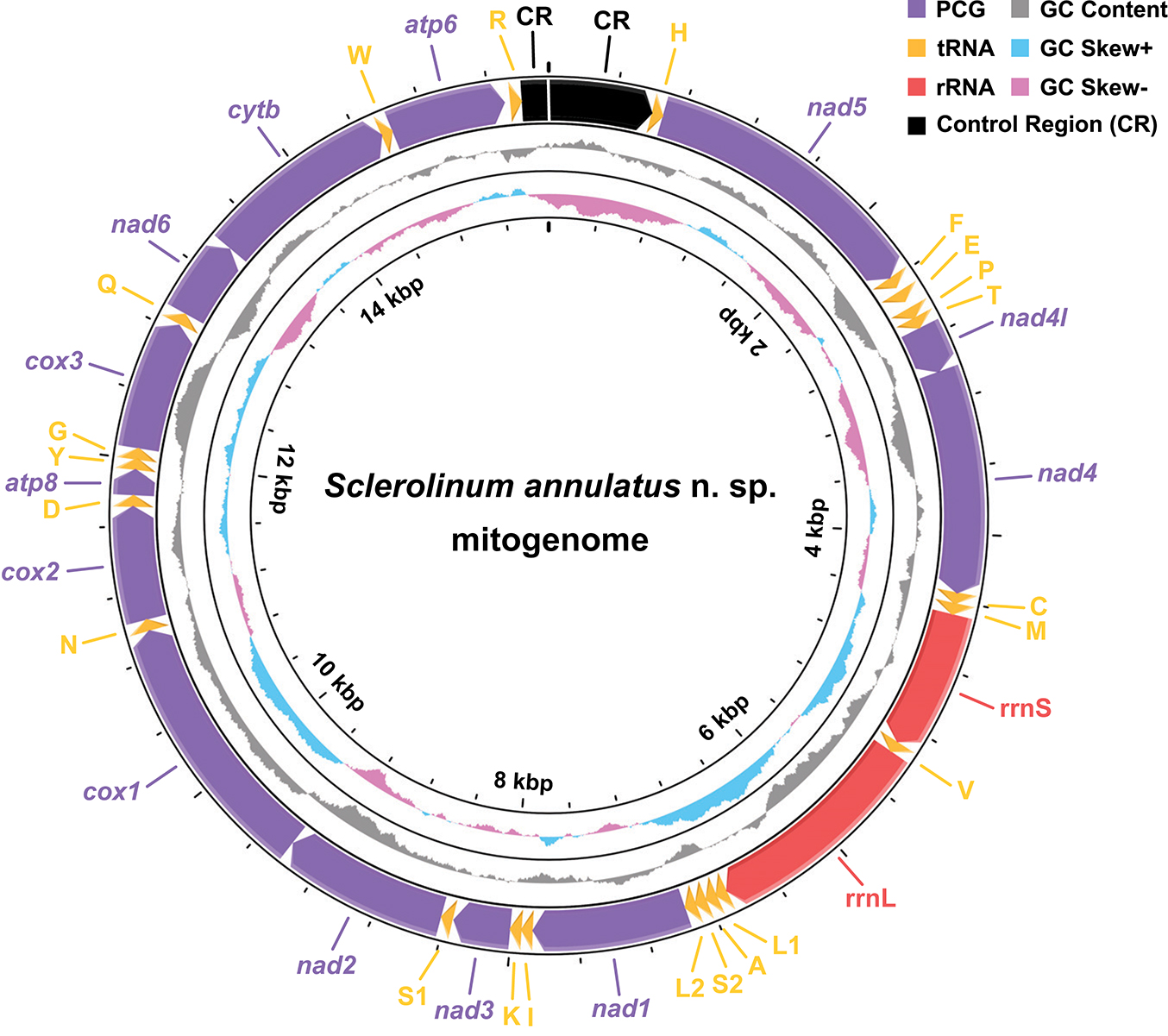
Figure 5. A map of the nearly complete mitogenome of Sclerolinum annulatus n. sp., with the direction of gene transcription indicated by arrows. Protein-coding genes (PCGs) are indicated by purple arrows, tRNA genes by yellow arrows, rRNA genes by red arrows, and the control region (CR) by a black arrow. A gap located between two CR fragments implies probably missing nucleotides. The tRNA genes are represented according to their single-letter IUPAC-IUB abbreviations (i.e., L1, CUN; L2, UUR; S1, AGN; S2, UCN). The GC content is plotted as the deviation from the average GC content of the entire sequence. The GC-skew is plotted as the deviation from the average GC-skew of the entire sequence.
Comparing the 13 mitochondrial PCGs between S. annulatus n. sp. and S. brattstromi uncovered that their K2P genetic distances range from 15.00% (cox2) to 28.66% (atp8), with an average value of 23.75% (Figure 6A). Utilization of the start and stop codons of the 13 mitochondrial PCGs of S. annulatus n. sp. and S. brattstromi is almost consistent, except for the stop codons of nad1 (TAA for S. annulatus n. sp. and TAG for S. brattstromi) and nad6 (a single T for S. annulatus n. sp. and TAA for S. brattstromi). Lengths of 12 out of the 13 mitochondrial PCGs of these two species are identical. The only exception occurs in nad6, with 472 bp for S. annulatus n. sp., whereas 471 bp for S. brattstromi. With the exclusion of the stop codons, the 13 mitochondrial PCGs of S. annulatus n. sp. and S. brattstromi exhibit a total of 3,687 and 3,686 codons, respectively. The three most frequently used codons of these two Sclerolinum mitogenomes are consistent, including Leu1 (CUN), Ile (AUY), and Ser2 (UCN), each with a codons per thousand codons (CDsp T) value > 80 (Figure 6B). Results of the RSCU patterns further illustrate that the predominant synonymous codons in the 13 mitochondrial PCGs of these two Sclerolinum species are also conserved, which generally have an over-usage of A and T at the third codon positions (Figure 6C).
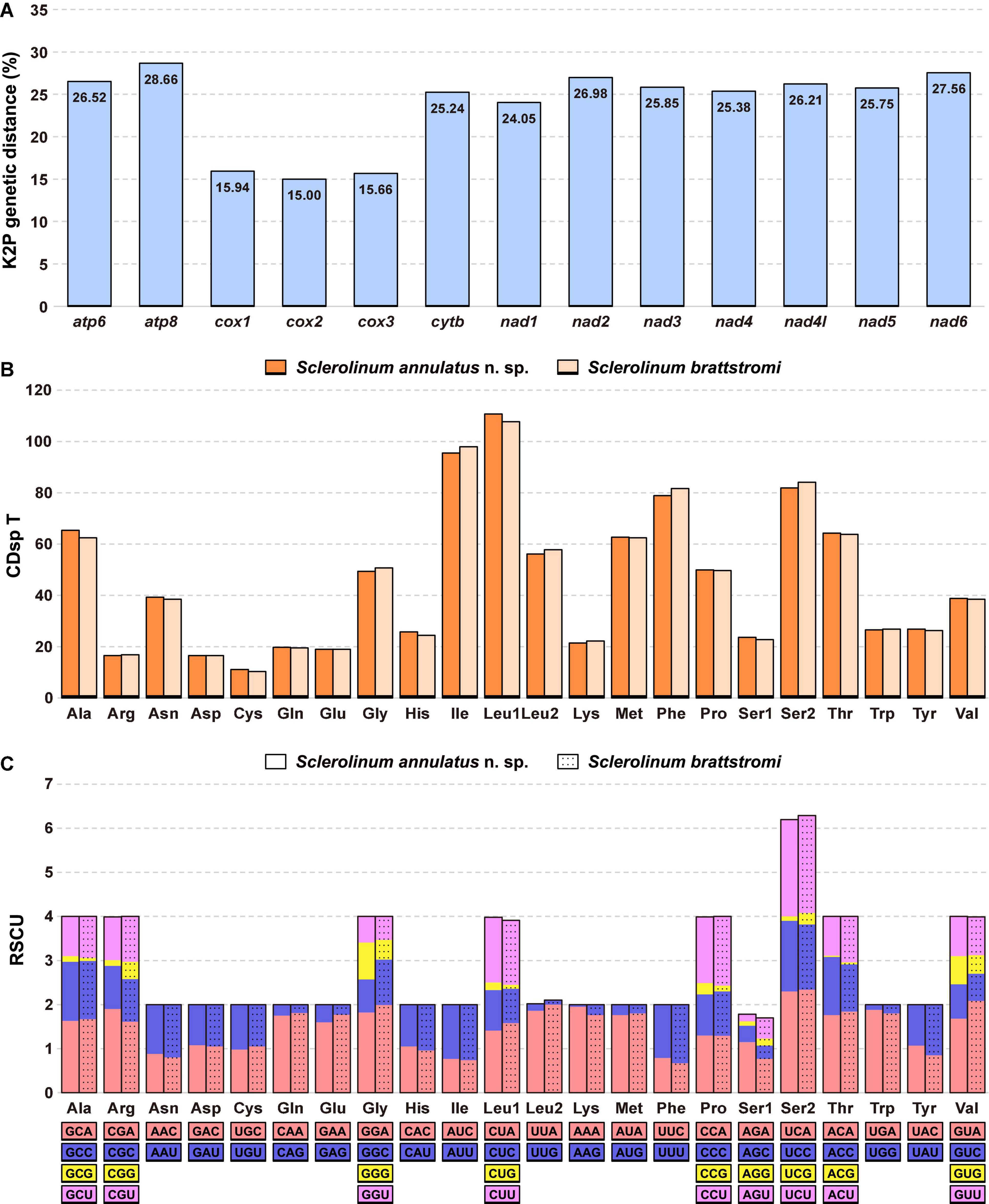
Figure 6. Comparison of the Sclerolinum annulatus n. sp. and Sclerolinum brattstromi mitogenomes. (A) The Kimura-2-parameter (K2P) genetic distance (%) of each mitochondrial protein-coding gene (PCG) between the two species. (B) Codon usage patterns indicated by codons per thousand codons (CDsp T) and (C) relative synonymous codon usage (RSCU) patterns of 13 mitochondrial PCGs.
Genetic Distances of Siboglinids Based on the Cox1 Gene
Alignment using MUSCLE followed by trimming using Gblocks Server (under the default settings) results in a 514-bp alignment of the cox1 gene from 41 siboglinids. The K2P genetic distance of cox1 between the examined frenulates ranges from 18.02% to 24.54%, between the examined Osedax ranges from 11.39% to 24.79%, between the examined vestimentiferans ranges from 0.39% to 21.30%, and between the examined Sclerolinum ranges from 1.98% to 18.78% (Supplementary Table 4). Among them, S. annulatus n. sp. is most closely related to “Pogonophora” sp. Kushiro-SK-2003 collected via deep-sea trawling from an unknown habitat off Kushiro in Japan (1,500 m water depth) (Kojima et al., 1997), with a K2P genetic distance of 1.98%. By contrast, S. annulatus n. sp. is distantly related to S. brattstromi and S. contortum with a K2P genetic distance ranging from 17.16% to 18.78%, respectively. Furthermore, the K2P genetic distance between S. contortum collected from four populations from the Arctic to the Antarctic ranges from 0.19% to 1.38%.
Phylogenetic Reconstruction of Siboglinids
Alignment using MUSCLE followed by trimming using Gblocks Server (under the relaxed settings) resulted in a 1,187-bp alignment of the cox1 gene from 41 siboglinids and two cirratuliformians. Phylogenetic reconstruction led to four major clades of siboglinids with at least two well-supported values (i.e., ML-SH/ML-NB > 80 and/or BI > 0.8), including Frenulata (ML-SH/ML-NB/BI: 92.3/79/1.00), Osedax (ML-SH/ML-NB/BI: 100/99/1.00), Sclerolinum (ML-SH/ML-NB/BI: 99.7/99/1.00), and Vestimentifera (ML-SH/ML-NB/BI: 85.4/64/0.82) (Figure 7). Specifically, in the Sclerolinum clade, S. annulatus n. sp. T1–T3 form a small clade that is sister to “Pogonophora” sp. Kushiro-SK-2003. These four sequences form a well-supported clade (ML-SH/ML-NB/BI: 100/100/1.00) that is sister to another well-supported clade (ML-SH/ML-NB/BI: 89.9/95/0.99) of Sclerolinum, which includes S. brattstromi from Norwegian fiords along with four S. contortum from the Arctic to the Antarctic.
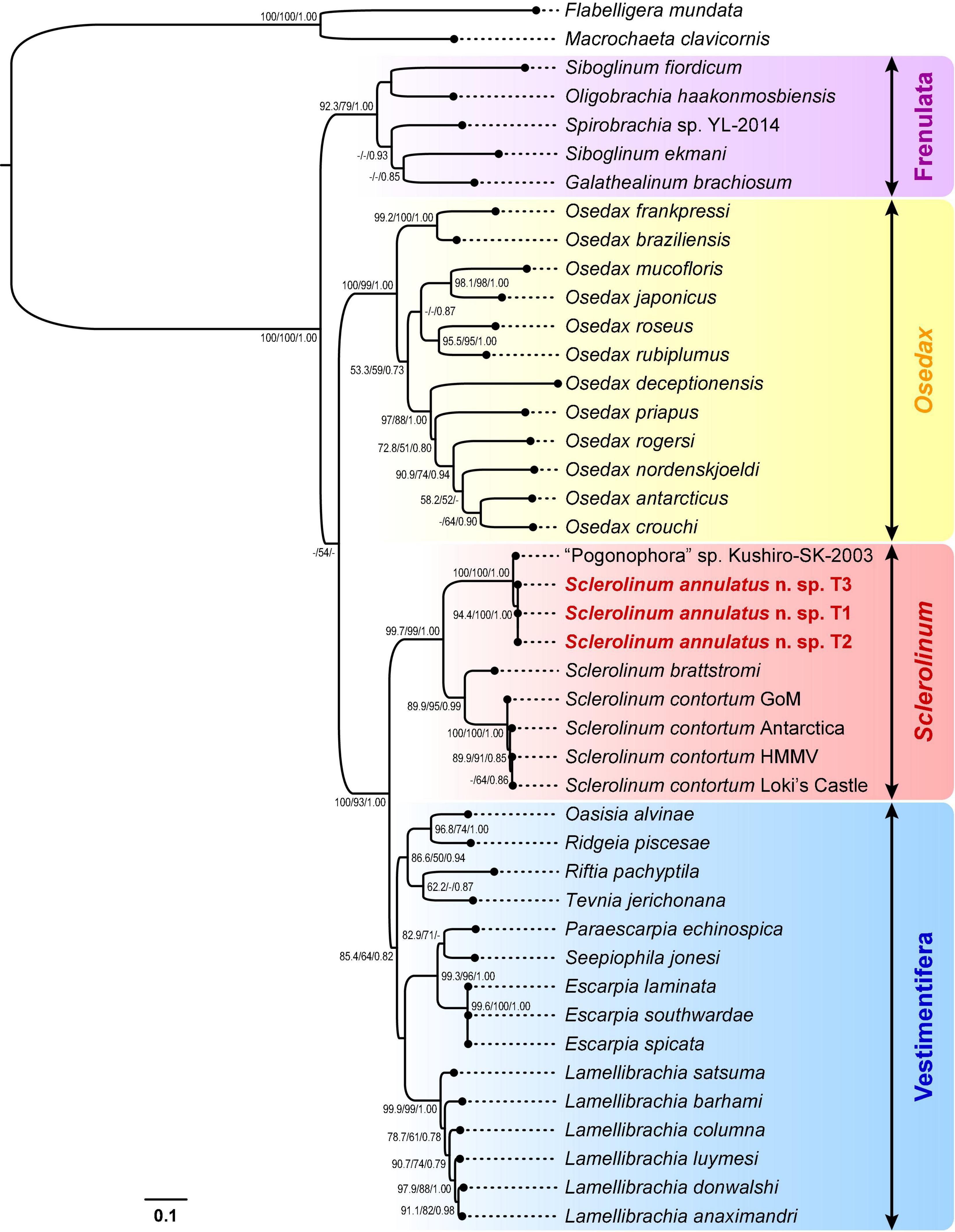
Figure 7. A phylogenetic tree of siboglinid tubeworms based on the maximum-likelihood (ML) topology inferred from a 1,187-bp gap-containing alignment of the cox1 gene. Bootstrap values from the Shimodaira-Hasegawa-like (SH-like) approximate likelihood ratio tests and the standard non-parametric bootstraps (NB) of the ML analysis, as well as the posterior probabilities from the Bayesian inference (BI) analysis, are shown at nodes (ML-SH/ML-NB/BI). Dashes (-) indicate bootstrap values < 50 or posterior probabilities < 0.70. Scale bar: the number of substitutions per site. GoM, Gulf of Mexico; HMMV, Haakon Mosby Mud Volcano.
Alignment using MUSCLE followed by trimming using Gblocks Server (under the relaxed settings) resulted in a 1,211-bp alignment of cox1, a 532-bp alignment of 16S rRNA, and a 1,735-bp alignment 18S rRNA from 29 siboglinids and two cirratuliformians. Phylogenetic reconstruction based on this 3,478-bp concentrated dataset also unveiled four major clades of siboglinids (Figure 8). Among the Sclerolinum clade (ML-SH/ML-NB/BI: 100/100/1.00), S. annulatus n. sp. T1–T3 form a well-supported clade (ML-SH/ML-NB/BI: 100/100/1.00) that is sister to another well-supported Sclerolinum clade (ML-SH/ML-NB/BI: 98.8/100/1.00) comprising S. brattstromi from Norwegian fiords and two S. contortum from the Antarctica and the Haakon Mosby Mud Volcano. Compared to the cox1 tree (Figure 7), the support values of the four major siboglinid clades in the three-gene tree have in general increased (Figure 8). Nevertheless, phylogenetic relationships among the four siboglinid clades remain unsolved, with the support values between Osedax and the other three clades being quite low (Figures 7, 8).
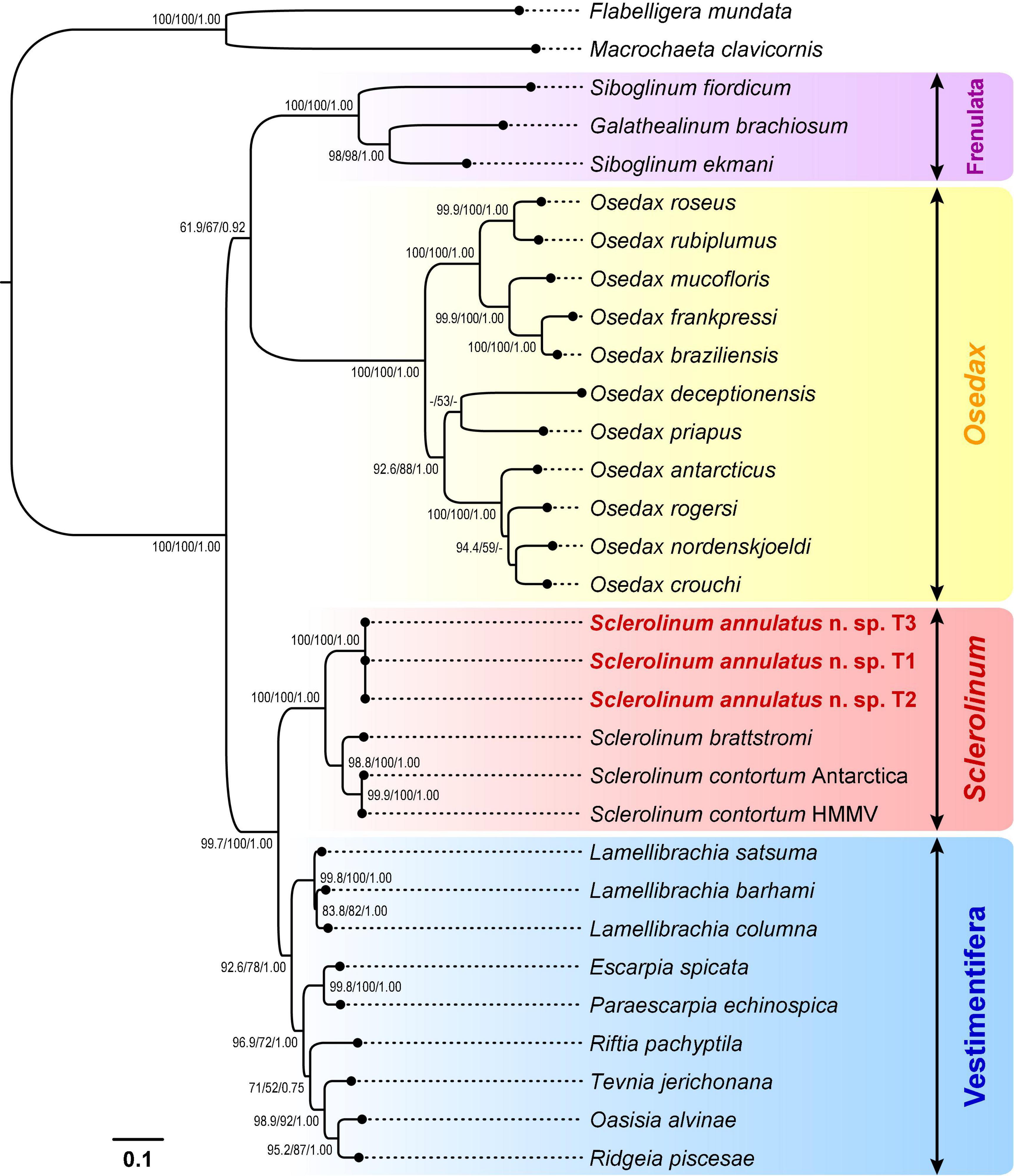
Figure 8. A phylogenetic tree of siboglinid tubeworms based on the maximum-likelihood (ML) topology inferred from a 3,478-bp concatenated alignment of the cox1 (1,211 bp with gaps), 16S rRNA (532 bp with gaps), and 18S rRNA (1,735 bp with gaps) genes. Bootstrap values from the Shimodaira-Hasegawa-like (SH-like) approximate likelihood ratio tests and the standard non-parametric bootstraps (NB) of the ML analysis, as well as the posterior probabilities from the Bayesian inference (BI) analysis, are shown at nodes (ML-SH/ML-NB/BI). Dashes (-) indicate bootstrap values < 50 or posterior probabilities < 0.70. Scale bar: the number of substitutions per site. HMMV, Haakon Mosby Mud Volcano.
Morphology and Phylogeny of Symbionts
Images of TEM revealed intracellular symbionts inside the bacteriocytes within the trophosome of S. annulatus n. sp., which are rod- or coccus-shaped in cross sections, ranging in size from 0.4 to 2.0 μm with no internal stacked membranes (Figures 9A,B). Amplicon sequencing of the microbial 16S rRNA gene and OTU assignment of S. annulatus n. sp. T4–T6 resulted in 337 OTUs in T4 (Supplementary Tables 5, 6), 358 OTUs in T5 (Supplementary Tables 5, 7), and 422 OTUs in T6 (Supplementary Tables 5, 8). Nevertheless, only one OTU (i.e., OTU-1) in each individual has a high relative abundance (i.e., 84.27% in T4, 82.33% in T5, and 78.95% in T6), whereas the other OTUs are all of low abundance (i.e., < 1.50%) (Supplementary Tables 6–8).
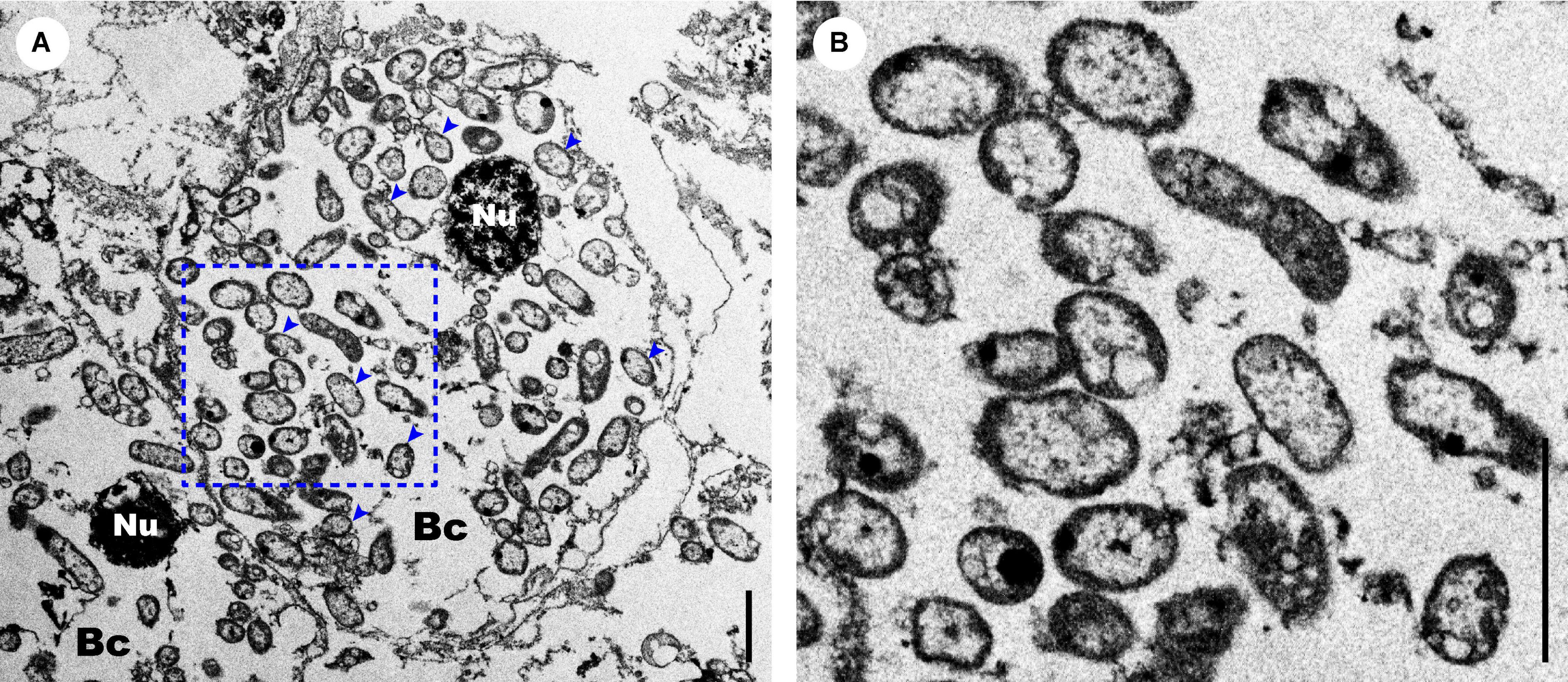
Figure 9. Morphological features of the Sclerolinum annulatus n. sp. endosymbionts. (A) Transmission electron microscopy (TEM) of the trunk region, showing the endosymbionts inside the bacteriocytes are rod or coccus in shape without internal stacked membranes (indicated by blue arrows). (B) TEM of a close-up section of the endosymbionts within the blue dotted box in (A). Scale bars: (A,B) 2 μm. Bc, bacteriocyte; Nu, nucleus.
Alignment using MUSCLE followed by trimming using Gblocks Server (under the relaxed settings) resulted in a 1,434-bp alignment of the microbial 16S rRNA gene from 29 siboglinids, eight bivalves, along with two Methylococcaceae outgroups. Phylogenetic analyses based on the microbial 16S rRNA gene elucidated the symbionts of Sclerolinum as a well-supported clade (ML-SH/ML-NB/BI: 98.4/100/1.00), which contains OTU-1 of S. annulatus n. sp. T4–T6 and two sulfur-oxidizing symbionts hosted by S. contortum from the Gulf of Mexico and the Haakon Mosby Mud Volcano (Figure 10 and Supplementary Figure 1). Similarly, the symbionts of frenulates (ML-SH/ML-NB/BI: 99.9/100/1.00) and Osedax (ML-SH/ML-NB/BI: 100/100/1.00) each form a well-supported clade (Figure 10 and Supplementary Figure 1). However, the symbionts of vestimentiferans are paraphyletic, with 24 forming a clade that is sister to a clade of three bivalve symbionts and four being nested between the bivalve and Sclerolinum symbionts (Figure 10 and Supplementary Figure 1).
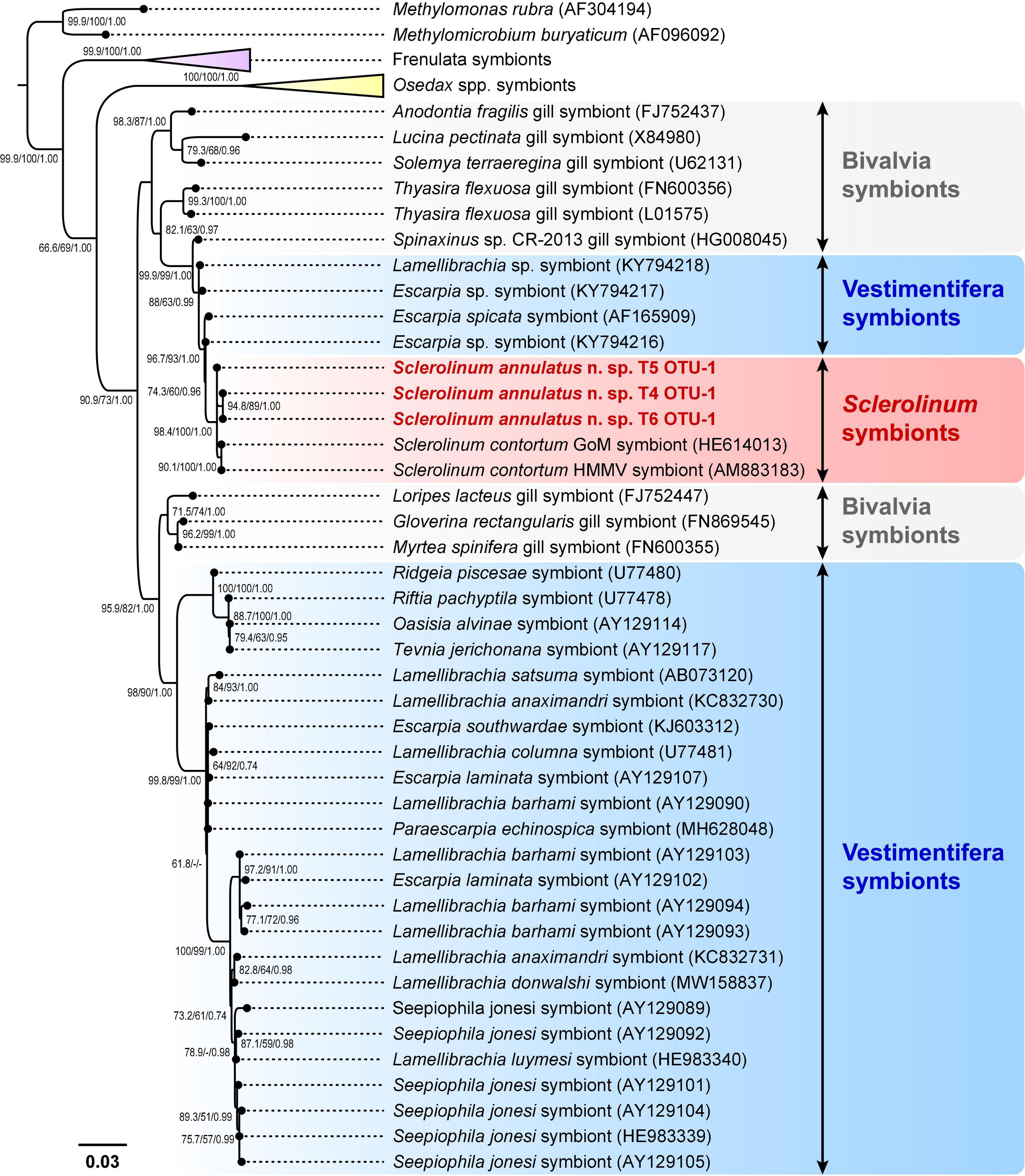
Figure 10. A simplified phylogenetic tree of siboglinid symbionts and their relatives based on the maximum-likelihood (ML) topology inferred from a 1,434-bp gap-containing alignment of the microbial 16S rRNA gene. Bootstrap values from the Shimodaira-Hasegawa-like (SH-like) approximate likelihood ratio tests and the standard non-parametric bootstraps (NB) of the ML analysis, as well as the posterior probabilities from the Bayesian inference (BI) analysis, are shown at nodes (ML-SH/ML-NB/BI). Dashes (-) indicate bootstrap values < 50 or posterior probabilities < 0.70. Scale bar: the number of substitutions per site. GoM, Gulf of Mexico; HMMV, Haakon Mosby Mud Volcano. The full phylogenetic tree refers to Supplementary Figure 1.
Alignment using MUSCLE followed by trimming using Gblocks Server (under the default settings) resulted in a 429-bp alignment of microbial 16S rRNA gene of five Sclerolinum. Among them, OTU-1 of S. annulatus n. sp. T4–T6 exhibit high genetic similarities, with a K2P genetic distance ranging from 0 to 0.47% (Supplementary Table 9). In addition, these OTU-1 are genetically closely related to the sulfur-oxidizing symbionts of S. contortum, with a K2P genetic distance ranging from 0.47% to 0.94% (Supplementary Table 9).
Stable Isotope Composition
The two samples exhibit a similar stable isotope composition. Sample 1 (i.e., containing individuals T7–T9) has a δ13C, δ15N, and δ34S value of −38.57‰, 3.16‰, and −2.40‰, respectively. Sample 2 (i.e., containing individuals T10–T12) has a δ13C, δ15N, and δ34S value of −33.70‰, 2.28‰, and −3.78‰, respectively.
Discussion
In the present study, we described the morphological features of Sclerolinum annulatus n. sp., obtained its nearly complete mitogenome, determined its phylogenetic position in the family Siboglinidae Caullery, 1914, and provided evidence for its endosymbionts inside the trophosome being sulfur oxidizers.
Morphologically, S. annulatus n. sp. is distinguishable from its congeneric species in that its tube has clearly defined transverse rings similar to the rings on the frenulate tubes (Figure 3) and the unique arrangement of plaques in its bridle (Figure 4). Phylogenetic reconstruction inferred from two datasets (Figure 7: cox1 and Figure 8: a concatenated dataset of cox1 + 16S rRNA + 18S rRNA) and the K2P genetic distance estimation based on the cox1 gene (Supplementary Table 4) support the recognition of this new species as three individuals are nested within a clade of two other described Sclerolinum species, while they exhibit substantial interspecific K2P genetic distances with these two species. Besides, phylogenetic analyses based upon cox1 indicate that “Pogonophora” sp. Kushiro-SK-2003 (Kojima et al., 1997) belongs to Sclerolinum (Figure 7). However, it has a K2P genetic distance of 1.98% from S. annulatus n. sp. (Supplementary Table 4). In a large-scale DNA barcoding study that involved amplification of cox1 from 1,876 specimens representing 333 provisional species, Carr et al. (2011) found an average intraspecific K2P genetic distance of 0.38%, and in most species, this distance is much less than 2%. Therefore, it is likely that “Pogonophora” sp. Kushiro-SK-2003 represents an undescribed species of Sclerolinum in the Northwest Pacific.
The complete mitogenomes of siboglinids are circular (Li et al., 2015), whereas that of S. annulatus n. sp. is not, indicating the latter may be incomplete. Nevertheless, given that the mitogenome of S. annulatus n. sp. contains all 37 genes typical in bilaterian mitogenomes (Boore, 1999) with a length close to that of S. brattstromi (15,553 bp for S. annulatus n. sp. vs. 15,383 bp for S. brattstromi), it should be nearly complete, with a small gap probably located in its control region. As having been reported in some siboglinid mitogenomes, the control region is difficult to be assembled using short reads alone due to the occurrence of repetitive sequences (Boore and Brown, 2000; Jennings and Halanych, 2005; Li et al., 2015). Further long-read sequencing based assembly approach would help to address this issue in the future. The mitogenome of S. annulatus n. sp. possesses the same gene order as S. brattstromi, which is also identical to that of other siboglinid mitogenomes (Li et al., 2015; Sun et al., 2018; Zhou et al., 2020). In addition, the codon usage patterns along with RUSC of the S. annulatus n. sp. and S. brattstromi mitogenomes are highly conserved, despite the large K2P genetic distances between all their 13 mitochondrial PCGs (Figure 6).
Compared to the symbionts of other siboglinids, very little is known about the symbionts of Sclerolinum species, with only that of S. contortum having been characterized (Pimenov et al., 1999, 2000; Lösekann et al., 2008; Eichinger et al., 2014). Earlier studies based on radiotracer determination (Pimenov et al., 1999) and TEM (Pimenov et al., 2000) suggested that S. contortum inhabiting seep sediment of the Haakon Mosby Mud Volcano may host methane-oxidizing symbionts in its trophosome. By contrast, more recent phylogenetic inferences, stable isotope analyses, and fluorescent in situ hybridization experiments unveiled a single phylotype of sulfur-oxidizing symbionts inside the trophosome of S. contortum dwelling in seep sediment of the Haakon Mosby Mud Volcano (Lösekann et al., 2008) and the Gulf of Mexico (Eichinger et al., 2014). These different lines of evidence underline that the symbionts of S. contortum reported in Pimenov et al. (1999, 2000) might also be thioautotrophs with an unusually light δ13C signature and an exceptional morphology that are atypical in sulfur-oxidizing bacteria. Alternatively, the S. contortum examined by Pimenov et al. (1999, 2000) might be another siboglinid species that is morphologically similar to S. contortum whereas hosts methanotrophs.
In the present study, the presence of intracellular symbionts inside the trophosome of S. annulatus n. sp. was detected via TEM. These symbionts have no internal stacked membranes (Figure 9), indicating that they are not methane-oxidizing bacteria (Cavanaugh et al., 1987). Stable isotope analyses of the tubeworm fragments detected less negative δ13C values of S. annulatus n. sp. (i.e., −38.57‰ and −33.70‰) than that of S. contortum collected from seep sediment of the Haakon Mosby Mud Volcano (i.e., −47.9‰) (Lösekann et al., 2008), but within the range of δ13C values (i.e., −40‰ to −30‰) of other deep-sea organisms, including some vestimentiferans, that harbor chemoautotrophic sulfur oxidizers (Nelson and Fisher, 1995; Feng et al., 2018). The symbiont morphological characteristics and the δ13C signature thus together illustrate that the symbionts of S. annulatus n. sp. are sulfur-oxidizing bacteria, which may mainly utilize a carbon source less depleted in 13C, such as inorganic carbon (Lösekann et al., 2008). The low δ34S values of S. annulatus n. sp. (i.e., −3.78‰ and −2.40‰) also imply that the assimilation of hydrogen sulfide by its symbionts might be primarily derived from the sulfate-dependent anaerobic oxidation of methane (Feng et al., 2015). Besides, the δ15N values of S. annulatus n. sp. (i.e., 2.28‰ and 3.16‰) are lower than the sediment δ15N values (i.e., 5‰–6‰) in the South China Sea, indicating the assimilation of some locally occurring and isotopically light nitrate or ammonium by the symbionts of S. annulatus n. sp. (Feng et al., 2015).
Application of the microbial 16S rRNA amplicon sequencing resulted in more than 300 OTUs from S. annulatus n. sp. (Supplementary Table 5), nonetheless, only the OTU-1 of each individual is present in high abundance (Supplementary Tables 6–8), suggesting that OTU-1 represents the symbionts, or at least the most dominant symbionts, of S. annulatus n. sp. Co-occurrence of multiple phylotypes of symbionts has been well known for macrobenthos inhabiting chemosynthesis-based ecosystems, including those belonging to different siboglinid lineages (e.g., Kubota et al., 2007; Dubilier et al., 2008; Verna et al., 2010; Zimmermann et al., 2014). In comparison, previous studies of S. contortum revealed only one phylotype of sulfur-oxidizing symbionts inside its trophosome (Lösekann et al., 2008; Eichinger et al., 2014). Although we were unable to separate the trophosome of S. annulatus n. sp. from its tube, given the high abundance of OTU-1 and the low abundance of all the other OTUs identified herein, we consider that S. annulatus n. sp. likely only hosts OTU-1 as its symbionts. Moreover, the close phylogenetic relationships between the symbionts of S. annulatus n. sp. (i.e., OTU-1) and S. contortum (Figure 10 and Supplementary Figure 1) support that OTU-1 belongs to sulfur-oxidizing bacteria. The small genetic distances between the symbionts of S. annulatus n. sp. (i.e., OTU-1) and S. contortum (Supplementary Table 9) further indicate that the symbionts of these two Sclerolinum species may be conspecific or have diverged only recently. Nevertheless, whole-genome sequencing of the siboglinid symbionts should be conducted in future studies to better understand their intra- and inter-specific diversity and differences in metabolic capabilities.
In the Haima cold seep, S. annulatus n. sp. forms huge aggregations consisting of hundreds to thousands of individuals. The posterior of the tube is inserted into sediment, which may allow the tubeworm to gain access to the reducing compounds available in porewater (Freytag et al., 2001). Numerous empty shells of the vesicomyid clam A. marissinica along with a few empty shells of the bathymodioline mussel G. haimaensis have been observed surrounding S. annulatus n. sp. (Figure 2). This phenomenon indicates that the S. annulatus n. sp. aggregations may have developed after the decline of the local chemosymbiotrophic bivalve populations. A similar successional pattern from mollusks to tubeworms has been proposed for cold seep communities in the Gulf of Mexico (Cordes et al., 2009). Additionally, S. annulatus n. sp. aggregations develop a complex three-dimensional habitat for various macrobenthos, such as the squat lobster Munidopsis sp. and the brittle star Ophiophthalmus sp. (Figure 2). A resembling association between Sclerolinum and other macrobenthos has been observed for S. contortum inhabiting the Storegga Slide seep area (Vanreusel et al., 2009) and the Loki’s Castle vent field (Kongsrud and Rapp, 2012) in the Norwegian Sea. It has been suggested that S. contortum might be capable of releasing sulfate and hydrogen ions into sediment to facilitate sustained sulfide production as other siboglinid tubeworms (Dattagupta et al., 2006). Besides, S. contortum may also have the ability to modify its local environments by releasing iron and manganese from sediment to ambient water (Aquilina et al., 2014). These multiple lines of evidence imply Sclerolinum tubeworms as an ecologically important taxon in worldwide chemosynthesis-based ecosystems. Therefore, further in-depth studies are desired to enhance our knowledge on their species diversity, biogeographical distribution, adaptive evolution, as well as symbiotic relationships with sulfur-oxidizing bacteria in the coming future.
Data Availability Statement
Raw Illumina sequencing data of Sclerolinum annulatus n. sp. have been deposited in the National Centre for Biotechnology Information (NCBI) under the BioProject PRJNA771719. Sequences of the mitogenome (accession numbers OL681896–OL681898) and the nuclear 18S rRNA gene (accession numbers OL681870–OL681872) of S. annulatus n. sp. T1–T3 have been deposited in the GenBank. Sequences of the microbial 16S rRNA gene of all the identified OTUs from S. annulatus n. sp. T4–T6 have been summarized in Supplementary Tables 6–8.
Author Contributions
J-WQ, P-YQ, and TX conceived and designed this project. J-WQ and TX collected the samples. YS, ZW, TX, and AS performed morphological examination and description. TX conducted molecular analyses and led manuscript writing. All authors edited the manuscript and approved the submission.
Funding
This project was supported by the National Key R&D Program, Ministry of Science and Technology, China (2018YFC0310005), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0409, SMSEGL20SC01, and SMSEGL20SC02), the Major of Basic and Applied Basic Research Project of Guangdong Province (2019B030302004), and the University Grants Committee of the Hong Kong Special Administrative Region (12101021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Eve Southward for guidance on morphological observation. We thank Guangzhou Marine Geological Survey (Guangzhou, China) for organizing the cruise, the captain and crew of the R/V Haiyang 6 for their support, the chief scientist Dr. Jun Tao, the pilots Mr. Yuankeng Huang and Mr. Yanian Zhang, and all the other members of the ROV Haima team for collecting the samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.793645/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genbank
- ^ http://molevol.cmima.csic.es/castresana/Gblocks_server.html
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
References
Aquilina, A., Homoky, W. B., Hawkes, J. A., Lyons, T. W., and Mills, R. A. (2014). Hydrothermal sediments are a source of water column Fe and Mn in the Bransfield Strait, Antarctica. Geochim. Cosmochim. Acta 137, 64–80. doi: 10.1016/j.gca.2014.04.003
Baba, K., and Williams, A. B. (1998). New Galatheoidea (Crustacea, Decapoda, Anomura) from hydrothermal systems in the West Pacific Ocean Bismarck Archipelago and Okinawa Trough. Zoosystema 20, 143–156.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Boore, J. L., and Brown, W. M. (2000). Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 17, 87–106. doi: 10.1093/oxfordjournals.molbev.a026241
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carr, C. M., Hardy, S. M., Brown, T. M., Macdonald, T. A., and Hebert, P. D. (2011). A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS One 6:e22232. doi: 10.1371/journal.pone.0022232
Caullery, M. (1914). Sur les Siboglinidae, type nouveau d’invertébrés receuillis par l’expédition du Siboga. C. R. Hebd. Séances Acad. Sci. 158, 2014–2017.
Cavanaugh, C. M., Levering, P. R., Maki, J. S., Mitchell, R., and Lidstrom, M. E. (1987). Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325, 346–348. doi: 10.1038/325346a0
Chen, C., Okutani, T., Liang, Q., and Qiu, J. (2018). A noteworthy new species of the family Vesicomyidae from the South China Sea (Bivalvia: Glossoidea). Venus 76, 29–37. doi: 10.18941/venus.76.1-4_29
Cordes, E. E., Bergquist, D. C., and Fisher, C. R. (2009). Macro–ecology of Gulf of Mexico cold seeps. Annu. Rev. Mar. Sci. 1, 143–168. doi: 10.1146/annurev.marine.010908.163912
Dattagupta, S., Miles, L. L., Barnabei, M. S., and Fisher, C. R. (2006). The hydrocarbon seep tubeworm Lamellibrachia luymesi primarily eliminates sulfate and hydrogen ions across its roots to conserve energy and ensure sulfide supply. J. Exp. Biol. 209, 3795–3805. doi: 10.1242/jeb.02413
Donath, A., Jühling, F., Al-Arab, M., Bernhart, S. H., Reinhardt, F., Stadler, P. F., et al. (2019). Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47, 10543–10552. doi: 10.1093/nar/gkz833
Dubilier, N., Bergin, C., and Lott, C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. doi: 10.1038/nrmicro1992
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2016). SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv [Preprint]. doi: 10.1101/074161 bioRxiv 074161,
Eichinger, I., Hourdez, S., and Bright, M. (2013). Morphology, microanatomy and sequence data of Sclerolinum contortum (Siboglindae, Annelida) of the Gulf of Mexico. Org. Divers. Evol. 13, 311–329. doi: 10.1007/s13127-012-0121-3
Eichinger, I., Schmitz-Esser, S., Schmid, M., Fisher, C. R., and Bright, M. (2014). Symbiont-driven sulfur crystal formation in a thiotrophic symbiosis from deep-sea hydrocarbon seeps. Environ. Microbiol. Rep. 6, 364–372. doi: 10.1111/1758-2229.12149
Feng, D., Cheng, M., Kiel, S., Qiu, J. W., Yang, Q., Zhou, H., et al. (2015). Using Bathymodiolus tissue stable carbon, nitrogen and sulfur isotopes to infer biogeochemical process at a cold seep in the South China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 104, 52–59. doi: 10.1016/j.dsr.2015.06.011
Feng, D., Qiu, J. W., Hu, Y., Peckmann, J., Guan, H., Tong, H., et al. (2018). Cold seep systems in the South China Sea: an overview. J. Asian Earth Sci. 168, 3–16. doi: 10.1016/j.jseaes.2018.09.021
Freytag, J. K., Girguis, P. R., Bergquist, D. C., Andras, J. P., Childress, J. J., and Fisher, C. R. (2001). A paradox resolved: sulfide acquisition by roots of seep tubeworms sustains net chemoautotrophy. Proc. Natl. Acad. Sci. U.S.A. 98, 13408–13413. doi: 10.1073/pnas.231589498
Fujikura, K. (2007). “Vent-type chemosynthetic community associated with methane seep at the Formosa Ridge, off southwest Taiwan,” in Proceedings of the International Conference on Gas Hydrate: Energy, Climate and Environment, Taipei.
Gaudron, S. M., Pradillon, F., Pailleret, M., Duperron, S., Le Bris, N., and Gaill, F. (2010). Colonization of organic substrates deployed in deep-sea reducing habitats by symbiotic species and associated fauna. Mar. Environ. Res. 70, 1–12. doi: 10.1016/j.marenvres.2010.02.002
Georgieva, M. N., Wiklund, H., Bell, J. B., Eilertsen, M. H., Mills, R. A., Little, C. T. S., et al. (2015). A chemosynthetic weed: the tubeworm Sclerolinum contortum is a bipolar, cosmopolitan species. BMC Evol. Biol. 15:280. doi: 10.1186/s12862-015-0559-y
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Halanych, K. M. (2005). Molecular phylogeny of siboglinid annelids (a.k.a. pogonophorans): a review. Hydrobiologia 535, 297–307. doi: 10.1007/s10750-004-1437-6
Hashimoto, J., and Okutani, T. (1994). Four new mytilid mussels associated with deepsea chemosynthetic communities around Japan. Venus 53, 61–83. doi: 10.18941/venusjjm.53.2_61
Hilário, A., Capa, M., Dahlgren, T. G., Halanych, K. M., Little, C. T. S., Thornhill, D. J., et al. (2011). New perspectives on the ecology and evolution of siboglinid tubeworms. PLoS One 6:e16309. doi: 10.1371/journal.pone.0016309
Ip, J. C. H., Xu, T., Sun, J., Li, R., Chen, C., Lan, Y., et al. (2021). Host–endosymbiont genome integration in a deep-sea chemosymbiotic clam. Mol. Biol. Evol. 38, 502–518. doi: 10.1093/molbev/msaa241
Ivanov, A. V., and Selivanova, R. V. (1992). Sclerolinum javanicum sp. n., a new pogonophoran living on rotten wood. A contribution to the classification of Pogonophora. Biol. Morya (Vladivostok) 1–2, 27–33.
Jennings, R. M., and Halanych, K. M. (2005). Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): evidence for conserved gene order in Annelida. Mol. Biol. Evol. 22, 210–222. doi: 10.1093/molbev/msi008
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., DePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Jones, M. L. (1988). The Vestimentifera, their biology, systematic and evolutionary patterns. Oceanol. Acta Spec. 8, 69–82.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Klaucke, I., Berndt, C., Crutchley, G., Chi, W. C., Lin, S., and Muff, S. (2016). Fluid venting and seepage at accretionary ridges: the four way closure ridge offshore SW Taiwan. Geo Mar. Lett. 36, 165–174. doi: 10.1007/s00367-015-0431-5
Kojima, S., Segawa, R., Hashimoto, J., and Ohta, S. (1997). Molecular phylogeny of vestimentiferans collected around Japan, revealed by the nucleotide sequences of mitochondrial DNA. Mar. Biol. 127, 507–513. doi: 10.1007/s002270050039
Kongsrud, J. A., and Rapp, H. T. (2012). Nicomache (Loxochona) lokii sp. nov. (Annelida: Polychaeta: Maldanidae) from the Loki’s Castle vent field: an important structure builder in an Arctic vent system. Polar Biol. 35, 161–170. doi: 10.1007/s00300-011-1048-4
Kubota, N., Kanemori, M., Sasayama, Y., Aida, M., and Fukumori, Y. (2007). Identification of endosymbionts in Oligobrachia mashikoi (Siboglinidae, Annelida). Microbes Environ. 22, 136–144. doi: 10.1264/jsme2.22.136
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, Y., Kocot, K. M., Schander, C., Santos, S. R., Thornhill, D. J., and Halanych, K. M. (2015). Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol. Phylogenet. Evol. 85, 221–229. doi: 10.1016/j.ympev.2015.02.008
Li, Y., Liles, M. R., and Halanych, K. M. (2018). Endosymbiont genomes yield clues of tubeworm success. ISME J. 12, 2785–2795. doi: 10.1038/s41396-018-0220-z
Liang, Q., Hu, Y., Feng, D., Peckmann, J., Chen, L., Yang, S., et al. (2017). Authigenic carbonates from newly discovered active cold seeps on the northwestern slope of the South China Sea: constraints on fluid sources, formation environments, and seepage dynamics. Deep Sea Res. Part I Oceanogr. Res. Pap. 124, 31–41. doi: 10.1016/j.dsr.2017.04.015
Lösekann, T., Robador, A., Niemann, H., Knittel, K., Boetius, A., and Dubilier, N. (2008). Endosymbioses between bacteria and deep-sea siboglinid tubeworms from an Arctic cold seep (Haakon Mosby mud volcano, Barents Sea). Environ. Microbiol. 10, 3237–3254. doi: 10.1111/j.1462-2920.2008.01712.x
Nelson, D. C., and Fisher, C. R. (1995). “Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps,” in Deep-Sea Hydrothermal Vents, ed. D. M. Karl (Boca Raton, CA: CRC Press), 125–167.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nussbaumer, A. D., Fisher, C. R., and Bright, M. (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345–348. doi: 10.1038/nature04793
Pimenov, N., Savvichev, A., Rusanov, I., Lein, A., Egorov, A., Gebruk, A., et al. (1999). Microbial processes of carbon cycle as the base of food chain of Haakon Mosby mud volcano benthic community. Geo Mar. Lett. 19, 89–96. doi: 10.1007/s003670050097
Pimenov, N. V., Savvichev, A. S., Rusanov, I. I., Lein, A. Y., and Ivanov, M. V. (2000). Microbiological processes of the carbon and sulfur cycles at cold methane seeps of the North Atlantic. Microbiology 69, 709–720. doi: 10.1023/A:1026666527034
Reveillaud, J., Anderson, R., Reves-Sohn, S., Cavanaugh, C., and Huber, J. A. (2018). Metagenomic investigation of vestimentiferan tubeworm endosymbionts from Mid-Cayman Rise reveals new insights into metabolism and diversity. Microbiome 6, 1–15. doi: 10.1186/s40168-018-0411-x
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rouse, G. W., Goffredi, S. K., and Vrijenhoek, R. C. (2004). Osedax: bone-eating marine worms with dwarf males. Science 305, 668–671. doi: 10.1126/science.1098650
Sahling, H., Galkin, S. V., Salyuk, A., Greinert, J., Foerstel, H., Piepenburg, D., et al. (2003). Depth-related structure and ecological significance of cold-seep communities–a case study from the Sea of Okhotsk. Deep Sea Res. Part I Oceanogr. Res. Pap. 50, 1391–1409. doi: 10.1016/j.dsr.2003.08.004
Sahling, H., Wallmann, K., Dählmann, A., Schmaljohann, R., and Petersen, S. (2005). The physicochemical habitat of Sclerolinum sp. at Hook Ridge hydrothermal vent, Bransfield Strait, Antarctica. Limnol. Oceanogr. 50, 598–606. doi: 10.4319/lo.2005.50.2.0598
Schmaljohann, R., and Flügel, H. J. (1987). Methane-oxidizing bacteria in Pogonophora. Sarsia 72, 91–98. doi: 10.1080/00364827.1987.10419707
Sen, A., Didriksen, A., Hourdez, S., Svenning, M. M., and Rasmussen, T. L. (2020). Frenulate siboglinids at high Arctic methane seeps and insight into high latitude frenulate distribution. Ecol. Evol. 10, 1339–1351. doi: 10.1002/ece3.5988
Smirnov, R. V. (2000). Two new species of Pogonophora from the arctic mud volcano off northwestern Norway. Sarsia 85, 141–150. doi: 10.1080/00364827.2000.10414563
Southward, E. C. (1972). On some Pogonophora from the Caribbean and the Gulf of Mexico. Bull. Mar. Sci. 22, 739–776.
Southward, E. C., Schulze, A., and Gardiner, S. L. (2005). Pogonophora (Annelida): form and function. Hydrobiologia 535, 227–251. doi: 10.1007/s10750-004-4401-6
Southward, E. C., Schulze, A., and Tunnicliffe, V. (2002). Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: a new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin. J. Nat. Hist. 36, 1179–1197. doi: 10.1080/00222930110040402
Stewart, C., and Via, L. E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–751.
Suess, E., Huang, Y., Wu, N., Han, X., and Su, X. (2005). South China Sea Continental Margin: Geological Methane Budget and Environmental Effects of Methane emissions and Gas Hydrates. RV SONNE Cruise Report. Kiel: Leibniz Institute for Science and Mathematics Education at the University of Kiel.
Sun, Y., Liang, Q., Sun, J., Yang, Y., Tao, J., Liang, J., et al. (2018). The mitochondrial genome of the deep-sea tubeworm Paraescarpia echinospica (Siboglinidae, Annelida) and its phylogenetic implications. Mitochondrial DNA Part B Resour. 3, 131–132. doi: 10.1080/23802359.2018.1424576
Sun, Y., Sun, J., Yang, Y., Lan, Y., Ip, J. C. H., Wong, W. C., et al. (2021). Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica. Mol. Biol. Evol. 38:msab203. doi: 10.1093/molbev/msab203
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vanreusel, A., Andersen, A., Boetius, A., Connelly, D., Cunha, M., Decker, C., et al. (2009). Biodiversity of cold seep ecosystems along the European margins. Oceanography 22, 110–127. doi: 10.5670/oceanog.2009.12
Verna, C., Ramette, A., Wiklund, H., Dahlgren, T. G., Glover, A. G., Gaill, F., et al. (2010). High symbiont diversity in the bone-eating worm Osedax mucofloris from shallow whale-falls in the North Atlantic. Environ. Microbiol. 12, 2355–2370. doi: 10.1111/j.1462-2920.2010.02299.x
Webb, M. (1964). A new bitentaculate pogonophoran from Hardangerfjorden, Norway. Sarsia 15, 49–55. doi: 10.1080/00364827.1964.10409528
Webb, M. (1965). Notes on the distribution of Pogonophora in Norwegian fjords. Sarsia 18, 11–15. doi: 10.1080/00364827.1965.10409544
Xu, T., Feng, D., Tao, J., and Qiu, J. W. (2019). A new species of deep-sea mussel (Bivalvia: Mytilidae: Gigantidas) from the South China Sea: morphology, phylogenetic position, and gill-associated microbes. Deep Sea Res. Part I Oceanogr. Res. Pap. 146, 79–90. doi: 10.1016/j.dsr.2019.03.001
Yang, Y., Sun, J., Sun, Y., Kwan, Y. H., Wong, W. C., Zhang, Y., et al. (2020). Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts. ISME J. 14, 135–150. doi: 10.1038/s41396-019-0520-y
Zhao, Y., Xu, T., Law, Y. S., Feng, D., Li, N., Xin, R., et al. (2020). Ecological characterization of cold-seep epifauna in the South China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 163:103361. doi: 10.1016/j.dsr.2020.103361
Zhou, Y., Li, Y., Cheng, H., Halanych, K. M., and Wang, C. (2020). The mitochondrial genome of the bone-eating worm Osedax rubiplumus (Annelida, Siboglinidae). Mitochondrial DNA Part B Resour. 5, 2267–2268. doi: 10.1080/23802359.2020.1772680
Keywords: deep-sea, siboglinid, sulfur-oxidizing bacteria, Haima cold seep, chemosynthesis
Citation: Xu T, Sun Y, Wang Z, Sen A, Qian P-Y and Qiu J-W (2022) The Morphology, Mitogenome, Phylogenetic Position, and Symbiotic Bacteria of a New Species of Sclerolinum (Annelida: Siboglinidae) in the South China Sea. Front. Mar. Sci. 8:793645. doi: 10.3389/fmars.2021.793645
Received: 12 October 2021; Accepted: 24 December 2021;
Published: 08 February 2022.
Edited by:
Gustavo Fonseca, Federal University of São Paulo, BrazilReviewed by:
Katrine Worsaae, University of Copenhagen, DenmarkNadezhda Rimskaya-Korsakova, Lomonosov Moscow State University, Russia
Copyright © 2022 Xu, Sun, Wang, Sen, Qian and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Yuan Qian, Ym9xaWFucHlAdXN0Lmhr; Jian-Wen Qiu, cWl1andAaGtidS5lZHUuaGs=
†Present address: Arunima Sen, Department of Arctic Biology, University Centre in Svalbard, Longyearbyen, Norway
 Ting Xu
Ting Xu Yanan Sun
Yanan Sun Zhi Wang
Zhi Wang Arunima Sen
Arunima Sen Pei-Yuan Qian
Pei-Yuan Qian Jian-Wen Qiu
Jian-Wen Qiu