- 1CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory of Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3College of Marine Science, University of Chinese Academy of Sciences, Beijing, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
- 6Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC, Canada
Thalassiosira is a species-rich genus in Bacillariophyta with important ecological contribution to primary productivity but can also pose negative impact on ecology by developing harmful algal blooms (HABs). However, genomic resources of only a few Thalassiosira species are currently available. Here, we constructed complete chloroplast genomes (cpDNAs) of six Thalassiosira strains (representing six Thalassiosira species T. rotula, T. profunda, T. nordenskioeldii, T. tenera, T. weissflogii, and Thalassiosira sp.), and compared them with published cpDNAs of other diatoms. Comparative analysis revealed that Thalassiosira cpDNAs have generally conventional and conserved quadripartite structure with important exceptions. Gene orders of cpDNAs of Thalassiosira sp. (CNS00561) and T. oceanica were different from that of other Thalassiosira species. Additionally, endosymbiotic gene transfers (EGT) were found to occur in the evolution of Thalassiosira cpDNAs. Furthermore, genomic regions of cpDNAs were found to be highly variable, which could be used to construct molecular markers for distinguishing Thalassiosira species with high resolution and high specificity. This study also demonstrated that Thalassiosira species emerged roughly around 51 MYA and diversified 17–28 MYA. Thalassiosira cpDNAs are not only valuable as super-barcode for phylogenetic analysis, but also important for functional and evolutionary analysis of diatoms.
Introduction
Thalassiosira (Mediophyceae, Bacillariophyta) is a species-rich genus with 287 recorded species, of which 175 have been officially accepted (Guiry and Guiry, 2021). Most of these Thalassiosira species are considered to be cosmopolitan marine species, with only a few found in both fresh and brackish waters (Hasle, 1978; Hasle and Lange, 1989; Round et al., 1990). Thalassiosira species have been identified in major coastal regions in China, including the East China Sea, the South China Sea, the Changjiang Estuary, and the Jiaozhou Bay (Cheng et al., 1993; Dong and Jiao, 1995; Li et al., 2013, 2014). Altogether, 54 Thalassiosira species and six varieties have been recorded (Gao and Cheng, 1992; Li et al., 2008a,b, 2013; Guo et al., 2018a,b).
Thalassiosira species represent a major group of diatoms, which are estimated to perform about one-fifth of global photosynthesis and contribute 40% of the marine primary productivity (Nelson et al., 1995; Gao et al., 2011). Thalassiosira species are not only critically important in ecological habitats, but also widely used as nutritious feed in aquaculture (Emmerson, 1980; Anger et al., 1986; Thompson et al., 1996; Kiatmetha et al., 2011), and for producing biodiesel (Nurachman et al., 2012). Some Thalassiosira species including T. weissflogii have been used as water quality indicators (Morelli et al., 2009; Araujo and Souza-Santos, 2013) and some are often used in nano-materials research (Losic et al., 2007; Pérez-Cabero et al., 2008).
However, many Thalassiosira species can cause harmful algal blooms (HABs) with negative impact on fisheries and tourism (Yu and Chen, 2019). Polyunsaturated short chain aldehydes (PUA) released by Thalassiosira species have been reported to inhibit the development of copepod eggs, affecting the balance and stability of marine ecosystem (Ianora et al., 1996). Eleven Thalassiosira species have been reported to be HAB species, including T. rotula, T. diporocyclus, T. curviseriata, T. weissflogii, T. nordenskioeldii, T. pacifica, T. mala, T. excentrica, T. hyaline, T. subtilis, and T. decipiens (Guo, 2004; Li, 2006; Liang, 2012; Li et al., 2013, 2014, 2018). Many instances of Thalassiosira HABs have been recorded in coastal waters in China over the past 20 years including ten major T. rotula HABs with the maximum area reaching 1000 km2 (Liang, 2012), one T. diporocyclus HAB in 2001–2002, and one T. curviseriata HAB in 2005 (Chen et al., 2004; Xie et al., 2008).
Species identification is a critical step in research on the Thalassiosira species, which was done primarily based on the structural and ultrastructural characteristics under light and electron microscopes. The distinction between Thalassiosira species often depends on cell sizes, valve structural features, areolae array and density, and the number, position shape of strutted and labiate processes (Li et al., 2008b). Typical structural characteristics of areola of Thalassiosira are a foramen of external valve face and cribrum of internal valve face, a single central fultoportula, a ring of marginal fultoportulas, and a single marginal rimoportula (Hasle, 1973; Li, 2009). However, these morphological features are both subtle and diverse (Park and Lee, 2010; Sar et al., 2011) with high taxonomic requirements for researchers, and some morphological features are variable to some degree (Fryxell and Hasle, 1977; McMillan and Johansen, 1988; Guo et al., 2017), which often leads to unclear and inconclusive classification and identification of Thalassiosira species. The application of molecular information greatly improved Thalassiosira species identification at both interspecific and intraspecific levels (Mallatt and Sullivan, 1998; Álvarez, 2003; Bleidorn et al., 2003; Kaczmarska et al., 2006). Whittaker et al. (2012) analyzed the molecular phylogeny of Thalassiosira species with the help of full-length 18S rDNAs, which was adopted by Alverson et al. (2007) and Hoppenrath et al. (2007). Alverson et al. (2007) combined 18S rDNA with psbC and rbcL for analyzing and identifying more Thalassiosira taxa. Whittaker et al. (2012) found that ITS could be used as the molecular marker to probe intraspecific genetic diversity of T. rotula. Nevertheless, the resolution provided by these common molecular markers is often inadequate for distinguishing all closely related Thalassiosira species (Whittaker, 2014). Thus, molecular markers with higher resolution are urgently needed.
With the development of next-generation DNA sequencing technologies, a large number of chloroplast genomes (cpDNAs) have been generated and used as the next-generation of molecular markers because they harbor sufficient variations for discriminating closely related species and strains (Kumar et al., 2009; Parks et al., 2009). Because of the cpDNAs much longer sequence length than the common molecular markers, richer genetic information, and contain a large number of protein-coding genes (PCGs), they are usually called super-barcode (Li et al., 2015; Fu et al., 2018; Ji et al., 2019). To date, 55 cpDNAs of diatom species have been published, ranging in size from ∼110 kbp to ∼200 kbp. The cpDNAs of only three Thalassiosira species have been reported, including the cpDNAs of T. pseudonana (NC_008589) (Oudot-Le Secq et al., 2007), T. oceanica (NC_014808) (Lommer et al., 2010), and T. weissflogii (NC_025314) (Sabir et al., 2014).
In this study, we reported six complete cpDNAs of Thalassiosira species for the first time, which were compared against cpDNAs of other Thalassiosira species and other diatom species. Comparative analysis revealed highly variable regions in the cpDNAs of Thalassiosra species that may serve as molecular markers for species identification. Furthermore, we also evaluated the divergene of Thalassiosira species from other diatom species.
Materials and Methods
Strain Isolation, Culturing and Characterization
Six candidate Thalassiosira strains (CNS00050, CNS00051, CNS00052, CNS00439, CNS00472, and CNS00561) were isolated from water samples collected in various China coastal regions including the Jiaozhou Bay (Vessel “Chuangxin,” March and June 2019), the Yellow Sea (Vessel “Beidou”, April 2019), the Changjiang Estuary (Vessel “Xiang Yang Hong 18”, August 2020), and the East China Sea (Vessel “Beidou”, April 2019) (Figure 1A and Table 1) using single cell capillary method. These strains were cultured using L1 medium (1 ‰ volume fraction Na2SiO3⋅9H2O was added). The culture temperature was maintained at 19°C, and the illumination intensity was kept from 2000 Lx to 3000 Lx at the photoperiod of 12 h light: 12 h darkness. Morphological features of algal cells were observed using Zeiss microscope (Leica Microsystems). Full-length 18S rDNA sequences of the six strains were obtained by Polymerase Chain Reaction (PCR) amplification using forward primer 28F (5′-CGAATTCAACCTGGTTGATCCTGCCAGT-3′) and reverse primer 42R (5′-CCGGATCCTGATCCTTCT GCAGGTTCACCTAC-3′) (Gu, 2007). PCR amplification was programmed with thermal settings of an initial denaturation at 94°C (4 min), followed by 32 cycles of the following 3 steps: 94°C (1 min), 57°C (1 min 50 s), and 72°C (2 min) and finally, an elongation step of 72°C (10 min). Subsequently, the amplicons were sequenced by the methods of Sanger dideoxy. Full-length 18S rDNA sequences were used as molecular markers for identifying species based on their similarities to 18S rDNA sequences of known Thalassiosira species.
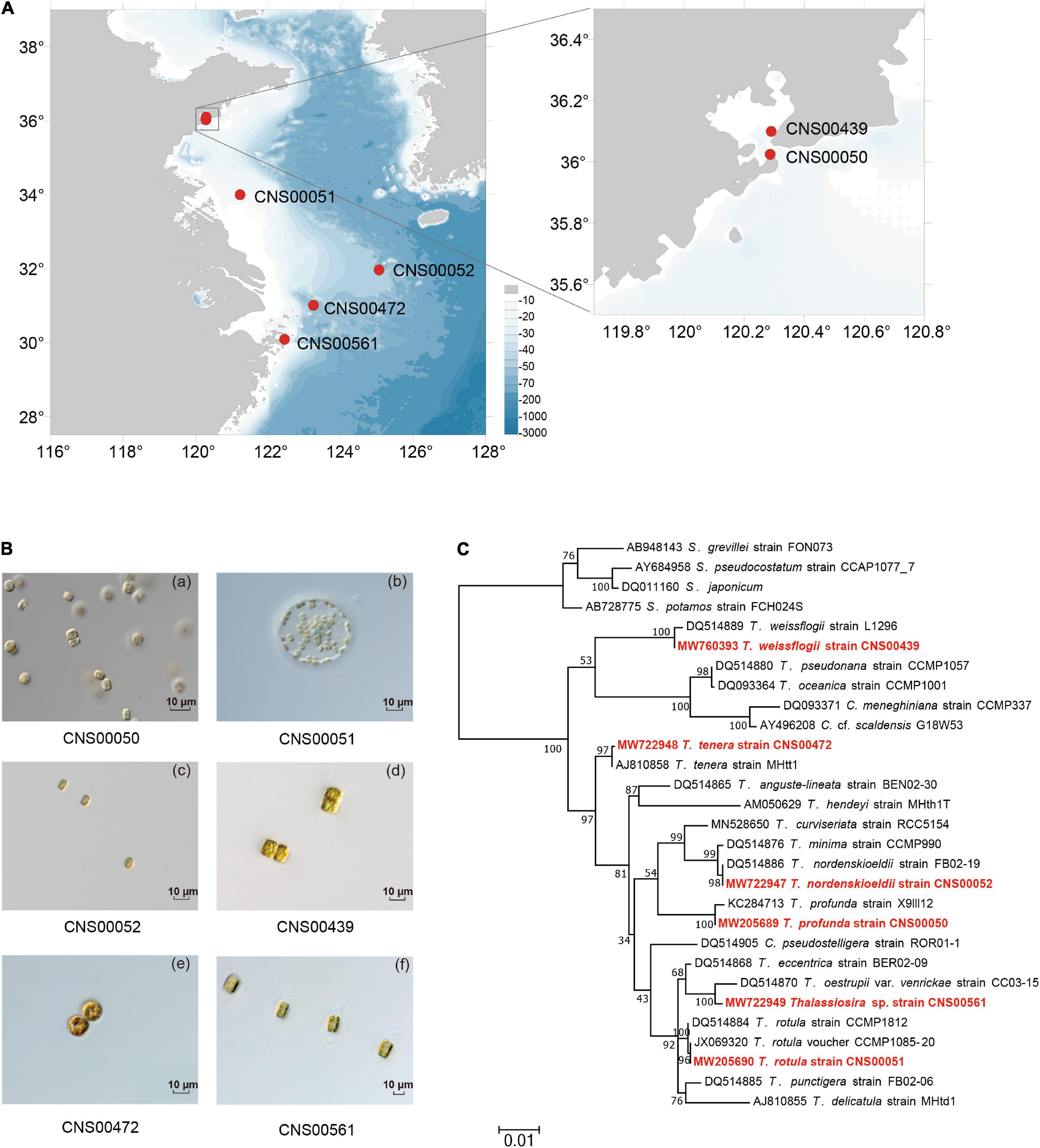
Figure 1. Sampling location and species identification of six Thalassiosira strains in this study. (A) Sampling sites in China coastal waters. (B) Light microscopy of six Thalassiosira strains (CNS00050, CNS00051, CNS00052, CNS00439, CNS00472, and CNS00561). (C) Maximum likelihood (ML) phylogenetic tree based on the nuclear 18S rDNA. The strains highlighted in red were data generated in this study.
DNA Library Preparation and Sequencing
Algal cultures were collected by centrifugation, and the algal muds were stored in liquid nitrogen for subsequent DNA extraction. Total DNA of each sample was extracted using DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). The DNA Library of each sample was prepared using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, United States). After qualification, each DNA library was sequenced using Illumina NovaSeq 6000 platform (Illumina, United States) at Novogene (Beijing, China), yielding about 5 Gb sequencing data of paired-end reads with 150 bp in length.
Assembly and Annotation of Chloroplast DNA
Illumina sequencing results of each strain were assembled using GetOrganelle v1.7.1 (Jin et al., 2020) with ORGANELLE_TYPE: embplant_pt assisted by SPAdes v3.13.2 (Bankevich et al., 2012) and Platanus-allee v2.2.2 (Kajitani et al., 2019). With the cpDNA of T. pseudonana (GenBank accession number: NC_008589) (Oudot-Le Secq et al., 2007) as a reference, we screened the scaffolds of the target cpDNA from the whole genome assembly for cpDNA assembly using BLASTN v2.10.0. The cpDNA assembly of each strain was examined by aligning Illumina reads using the MEM algorithm of BWA v0.7.17 (Li and Durbin, 2010). VarScan v2.3.9 (Koboldt et al., 2009) and IGV v2.8.12 (Robinson et al., 2011) were used to examine mutation sites and to verify the assembly results. Finally, a circular cpDNA was obtained for each strain. Using the same strategy, we also obtained DNA sequences of common molecular markers, including full-length 18S rDNAs, 28S rDNA D1-D2, ITS, 16S rDNA, rbcL, and cox1 for each strain (Supplementary Table 1).
Annotation of cpDNAs was performed using MFannot1 (transl_table = 11) and annotation results were verified using multiple software including Open Reading Frame Finder (ORF finder)2 with genetic code 11 for identifying open reading frames (orfs), tRNAscan-SE 2.0 (Chan and Lowe, 2019) with default setting for identifying tRNAs, MEGA X (Kumar et al., 2018) and BLASTN for identifying rRNAs and introns by sequence comparison with genes from closely related species, and RNAweasel3 for identifying the type of introns. Genome maps were drawn using Organellar Genome DRAW (OGDRAW) (Greiner et al., 2019). Annotations of 55 published diatom cpDNAs were downloaded from NCBI and were verified using the same strategy.
Genome Comparison
Six newly assembled complete cpDNAs of Thalassiosira species were compared against published cpDNAs of Thalassiosira species including T. weissflogii (NC_025314) (Sabir et al., 2014), T. pseudonana (NC_008589) (Oudot-Le Secq et al., 2007), and T. oceanica (NC_014808) (Lommer et al., 2010). Multiple sequence alignments of complete cpDNAs and synteny analysis were performed using Mauve v2.4.0 (Darling et al., 2010) with default setting. Pairwise gene rearrangements of cpDNAs were visualized using CIRCOS v0.69.9 (Krzywinski et al., 2009). The borders of the large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR) regions were identified and analyzed to show the IR expansions and contractions.
Phylogenetic Analysis
Maximum likelihood (ML) phylogenetic tree was constructed using concatenated amino acid sequences of 95 PCGs (Supplementary Table 1) shared by 61 diatom cpDNAs, including six Thalassiosira and 55 other diatoms, with the Ochrophyta species Triparma laevis (AP014625) as the out-group taxon. Sequence alignment, editing and concatenation were completed using MAFFT v7.471 (Katoh and Standley, 2013), trimal v1.2 (Capella-Gutierrez et al., 2009) with default setting, and phyutility (Smith and Dunn, 2008), respectively. Phylogenetic trees were constructed using IQtree v1.6.12 (Trifinopoulos et al., 2016) with data_type: AA, model: edge-linked partition model with 1000 bootstrap alignments.
Nucleotide Divergence Analyses and Mutation Hotspots Identification
We extracted 123 PCGs shared by cpDNAs of nine Thalassiosira strains, which were aligned by MAFFT v7.471 (Katoh and Standley, 2013) and analyzed for nucleotide diversity (Pi) using DnaSP v5.10 (Rozas, 2009). Fragment length differences of the above conserved gene blocks in the nine Thalassiosira cpDNAs were identified using Mauve v2.4.0 (Darling et al., 2010) with a sliding window of 600 bp with the step size of 50 bp. To characterize regions with length differences, Primer Premier 6 (Premier Biosoft International, Palo Alto, CA, United States) was used to design PCR amplification primers (the forward primer: 5′- GTTCTRCATTCRGTACATTCTA -3′ and the reverse primer: 5′- ATCWGGTAGAGCAGTTRTR -3′) for amplifying in the conserved region, located on both sides of the regions with different length. PCR amplification was programmed with thermal settings of an initial denaturation at 94°C (4 min), followed by 32 cycles of the following 3 steps: 94°C (1 min), 50°C (1 min 20 s), and 72°C (2 min) and finally, an elongation step of 72°C (10 min).
Estimation of Divergence Time
We used 127 PCGs (Supplementary Table 2) shared by 23 cpDNAs (including nine Thalassiosira species) to estimate the divergence time at the nucleotide level using MCMCTree in PAML (Yang, 2007). Branch lengths, gradient (g), and Hessian (H) were estimated using maximum likelihood estimates (MLE) (dos Reis et al., 2013, p. 10) and GTR+G substitution model (model = 7) with independent rates clock model (clock = 2). We estimated divergence times by Bayesian analysis. After discarding the first 50,000 (10%) trees as burn-in, twice of 500 million generations were set for the MCMC chains, with sample frequency set to 50. Finally, the tree file was visualized with Figtree v1.4.34.
Age calibration of divergence times was based on the fossil record. First, lower Jurassic diatom fossils were used to define a minimum divergence time of 174 MYA between diatoms and Ectocarpus (5–95% quantiles = 176–202 MYA) (Matari and Blair, 2014). The second calibration point, which corresponded to the prior divergence time of Rhizosolenia, was set at 91.5 MYA (5–95% quantiles = 90–93 MYA) (Sinninghe Damsté et al., 2004), according to the chemical fossil record of C25 highly branched isoprenoid (HBI) alkenes from Upper Turonian. The crown node of Fragilariopsis was constrained with a uniform distribution from 5 to 25 MYA, which estimated by the change or absence of directional movement structures (e.g., terminal fissures) in the fossil (Sims et al., 2019).
Results
Morphological and Molecular Identification of Thalassiosira Species
Six candidate Thalassiosira strains (CNS00050, CNS00051, CNS00052, CNS00472, CNS00439, and CNS00561) were characterized based on their morphological features (Figure 1B). Similarity of their molecular marker sequences to known Thalassiosira species were used for species annotation (Figure 1C and Supplementary Figure 1). For example, the strain CNS00051 was determined to be T. rotula because its cells were 40–45 μm in diameter, which were similar to reported sizes (Hoppenrath et al., 2007; Li et al., 2013), and each cell contained several yellow-brown chloroplasts (Figure 1Bb). Its full-length 18S rDNA sequence (MW205690) clustered closely with those of two T. rotula strains CCMP1085-20 (Whittaker et al., 2012) and CCMP1812 (Alverson et al., 2007) in the 18S rDNA-based phylogenetic tree (Figure 1C) with high percentage identities (Supplementary Table 3). The annotation of the strain CNS00051 as T. rotula was also supported by the high similarities of its 28S rDNA D1–D2, ITS, 16S rDNA, and rbcL to their corresponding reference sequences (Alverson et al., 2007; Kooistra et al., 2008; Supplementary Figure 1).
Similarly, the strains CNS00050, CNS00052, CNS00472, and CNS00439 were annotated as T. profunda, T. nordenskioeldii, T. tenera, and T. weissflogii, respectively (Figure 1C and Supplementary Table 3). The strain CNS00561 could not be annotated with certainty to a specific Thalassiosira species although it had the typical morphology of Thalassiosira species (Figure 1Bf), suggesting that it may represent a new Thalassiosira species, or a known Thalassiosira species whose molecular markers have not be characterized. Thus, we referred this species as Thalassiosira sp. (CNS00561).
Construction and Comparative Analysis of Thalassiosira Chloroplast DNAs
We successfully assembled complete cpDNAs for the above six Thalassiosira strains, each representing one Thalassiosira species (Table 2). The cpDNAs of five Thalassiosira species including T. profunda, T. rotula, T. nordenskioeldii, T. tenera, and Thalassiosira sp. (CNS00561) were assembled for the first time, while the cpDNA of a T. weissflogii strain has been previously published (Sabir et al., 2014). In this study, we assembled a second cpDNA of a different strain of T. weissflogii (CNS00439) that was isolated from the Jiaozhou Bay in China (Table 1). Each of the six cpDNAs was mapped as a single circular DNA, with a typical quadripartite structure consisting of one LSC, one SSC and a pair of inverted repeats (IRa and IRb) (Bendich, 2004; Daniell et al., 2016; Figure 2 and Table 2).
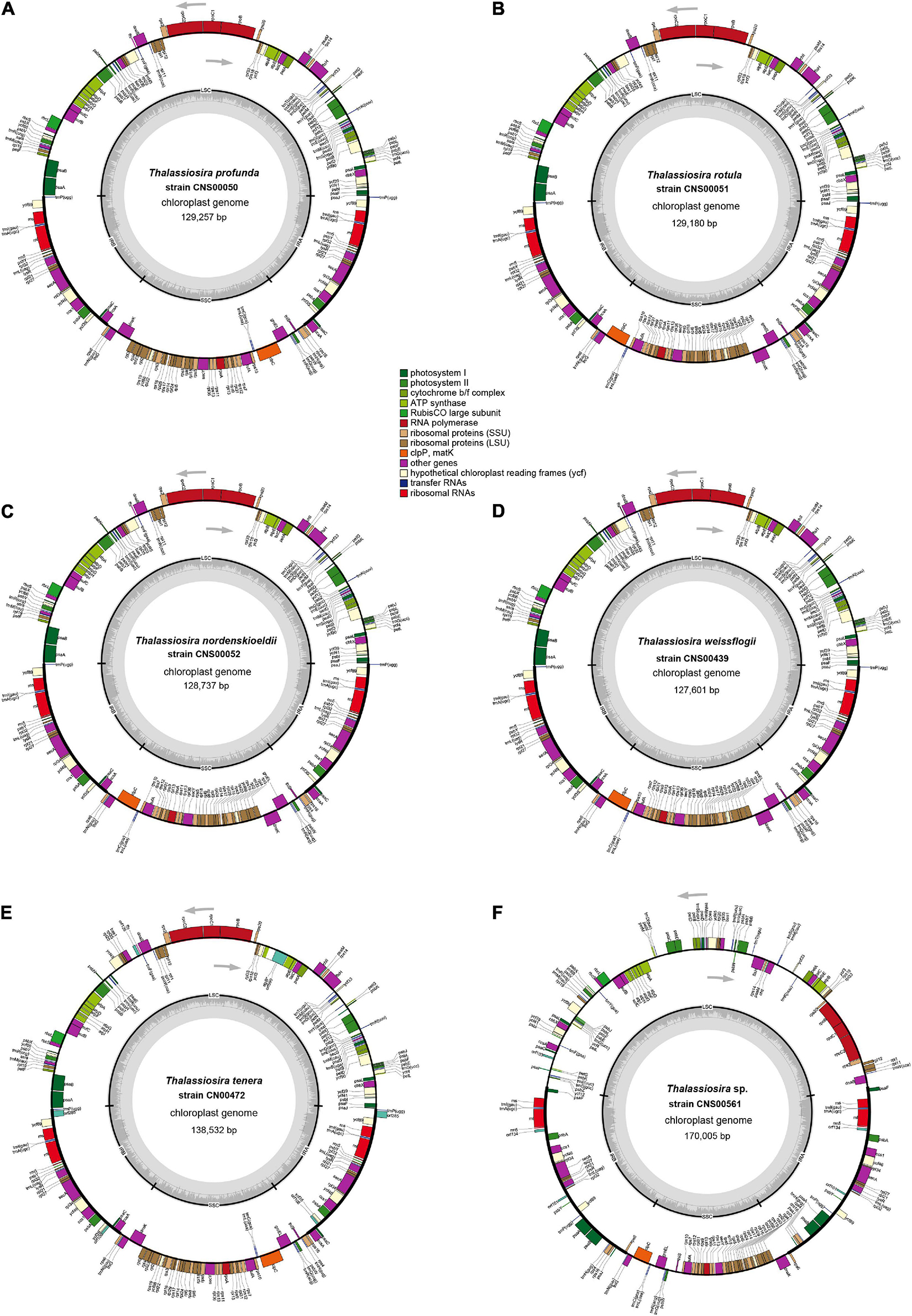
Figure 2. Gene maps of six newly Thalassiosira chloroplast genomes. The strain of (A) T. profunda (CNS00050), (B) T. rotula (CNS00051), (C) T. nordenskioeldii (CNS00052), (D) T. weissflogii (CNS00439), (E) T. tenera (CNS00472), and (F) Thalassiosira sp. (CNS00561). The transcriptional direction inside the ring is clockwise and the transcriptional direction outside the ring is counterclockwise. The color of the gene boxs indicates the type of gene.
While GC contents of the nine cpDNAs, including the six cpDNAs assembled in this study, were quite similar, falling between 29.4% and 30.9%, sizes of these cpDNAs varied substantially, ranging from 127,601 bp in T. weissflogii (CNS00439) to 170,005 bp in Thalassiosira sp. (CNS00561). The larger cpDNA size of Thalassiosira sp. (CNS00561) was primarily due to the larger sizes of the pair of IRs, which accounted for 38.5% of its complete cpDNA size. Twenty-nine genes were found in each of the two IRs in the cpDNA of Thalassiosira sp. (CNS00561), which was substantially more than those found in the IRs of cpDNAs of other Thalassiosira strains. The cpDNA of T. oceanica was the second largest among the nine cpDNAs, with a length of 141,790 bp and a GC content of 30.4%. Its IR region was also relatively long (23,693 bp), accounting for 33.4% of the full length.
Intergenic spacers were small for all nine Thalassiosira cpDNAs described in this project, with the median sizes ranging from 68.5 to 82.0 bp and the average sizes ranging from 100.7 to 296.85 bp, and the largest intergenic spacers being 2676 bp, located between ycf33 and trnK(uuu) in Thalassiosira sp. (CNS00561). No intergenic spacer was found between a pair of genes rpl24-rpl14 in all nine Thalassiosira cpDNAs as expected because it is common in other reported diatoms, except for those in the cpDNA of Nitzschia palea (MH113811), which has a small interval of 1 bp (Crowell et al., 2019). Of all 61 published diatom cpDNAs, the median of intergenic spacers were less than 100 bp and the average value were less than 300 bp (Figure 3). The average length of intergenic spacers in diatom cpDNAs were much smaller than that of green algae, whose average values were roughly between 300 and 1000 bp (Turmel et al., 2017). This is most likely due to the fact that diatoms have larger IR regions (∼20 kbp for combined length of two IR regions), while green algae have smaller IR regions or no IR regions.
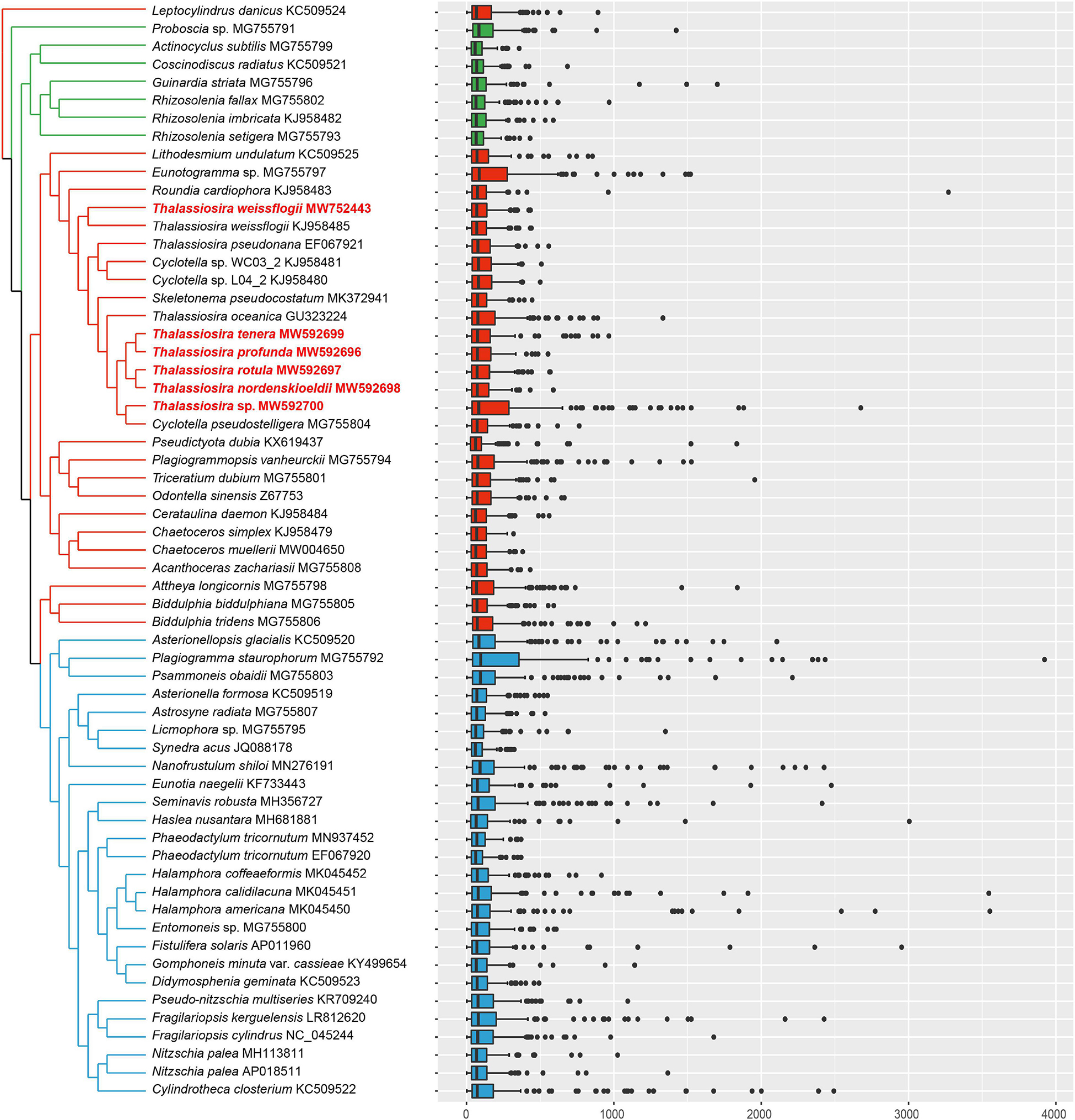
Figure 3. Intergenic spacers of all 61 published diatom cpDNAs with the topology of Maximum likelihood (ML) phylogenetic tree based on 95 shared PCGs of cpDNAs. The strains highlighted in red were data generated in this study.
Nevertheless, a small number of relatively large intergenic spacers were identified in the cpDNAs of diatoms, including the intergenic spacer between rbcS and orf143 in the cpDNA of Plagiogramma staurophorum (MG755792), which was 3923 bp in size, the intergenic spacer between ycf3 and orf124 in the cpDNA of Halamphora americana (MK045450), which was 3522 bp in size, and the intergenic spacer between orf99 and atpB in the cpDNA of Halamphora calidilacuna (MK045451), which was 3545 bp in size.
Four pairs of overlapping genes were found in all nine Thalassiosira cpDNAs. The gene pairs atpD-atpF, sufC-sufB, rpl4-rpl23, and psbD-psbC overlapped by 4, 1, 8, and 53 bp, respectively. Among them, the gene pair psbD-psbC overlapped in all other reported diatoms, whose overlap length was between 44 bp in Entomoneis sp. and 57 bp in Roundia cardiophora. The gene pair rpl4-rpl23 also appeared in almost all diatom cpDNAs, except Pseudo-nitzschia multiseries (KR709240) and Fragilariopsis kerguelensis (LR812620). Similarly, the gene pairs atpD-atpF and sufC-sufB were widely found in most diatom cpDNAs, such as Rhizosolenia fallax (MG755802) of Coscinodiscophyceae, Odontella sinensis (Z67753) of Mediophyceae, and Nitzschia palea (MH113811) of Bacillariophyceae.
The nine Thalassiosira cpDNAs shared 155 genes, including 123 PCGs, 27 tRNA, three rRNA, and two additional RNA genes (Table 2). The proportions of coding sequences of these nine cpDNAs were rather different, ranging from 68.0% of Thalassiosira sp. (CNS00561) cpDNA to 86.1% of T. weissflogii cpDNA. The 27 tRNAs were sufficient to satisfy all the requirements for in organelle protein synthesis. All three shared rRNA genes, including 5S rDNA, 16S rDNA, and 23S rDNA, were present in IRs. Two other RNA genes, transfer-messenger RNA (ssra) and plastid signal recognition particle RNA (ffs), were found in all of these nine cpDNAs of Thalassiosira. In addition, the cpDNA of T. oceanica not only had these two RNA genes, but also encoded an additional flrn (ffs-like RNA) (Lommer et al., 2010).
Introns in Thalassiosira Chloroplast DNAs
A group II intron was identified in atpB of the T. tenera cpDNA. The intron was 2874 bp in size, contributing to the total length of the T. tenera cpDNA. In contrast, no introns were identified in the cpDNAs of all other Thalassiosira species analyzed in this project. This result was not surprising because introns were found in only five other reported diatom cpDNAs including atpB in Seminavis robusta and Proboscia sp., petD in Plagiogramma staurophorum and Halamphora calidilacuna, psaA in Toxarium undulatum, and petB in Halamphora calidilacuna (Table 3), most of which are group II introns in PCGs. A group I intron with 764 bp in size was also found in the rnl gene of Seminavis robusta. In this study, the intron found in the atpB of T. tenera cpDNA contains one orf (orf590) encoding a peptide of 590 amino acids, which was a reverse transcriptase (RT) and an HNH endonuclease with self-splicing activity. Phylogenetic analysis showed that orf590 had high similarity with orf586 within the intron of atpB of the Chlorophyta species Pseudoneochloris marina cpDNA (KY407657) (Supplementary Figure 2; Turmel et al., 2017).
Similar observations were also found in Seminavis robusta of diatom and Ulva of Chlorophyta (Brembu et al., 2014; Cai et al., 2017; Suzuki et al., 2018). orf582 in the intron of atpB gene of diatom Seminavis robusta has a closer phylogenetic relationship with orfs (orf581, orf572, and orf573) in the introns of atpB gene of three Ulva species (Ulva ohnoi (AP018696), Ulva ohnoi (KX579943), and Ulva compressa (MW548841)). Therefore, it is highly possible that this intron was transferred horizontally from the green algae Pseudoneochloris marina or related species of P. marina to T. tenera.
Endosymbiotic Gene Transfer of petF in Thalassiosira
Compared with the cpDNAs of other Thalassiosira species analyzed in this study (e.g., T. weissflogii, T. pseudonana, T. tenera, T. profunda, T. rotula, and T. nordenskioeldii), the cpDNA of T. oceanica missed one PCG petF, and the cpDNA of Thalassiosira sp. (CNS00561) missed four PCGs including petF, psaE, rpl36, and ycf35. Among these genes, petF has been extensively studied for its role in iron uptake (McKay et al., 1997; Roy et al., 2020). The lack of petF in the cpDNA of T. oceanica has been found to be correlated with the transfer of this gene to the host genome (Lommer et al., 2010; Roy et al., 2020) via endosymbiotic gene transfer (EGT) (Timmis et al., 2004). We hypothesized that the lack of petF in the cpDNA of Thalassiosira sp. (CNS00561) was also correlated with the gain of this gene in the nuclear genome (Figure 4). To test this hypothesis, we searched for potential homologs of these four PCGs in the assembled whole genome sequences based on Illumina reads of Thalassiosira sp. (CNS00561), including both nuclear and organelle genomes. A candidate gene orf128 was found in the whole genome assembly of CNS00561 strain. It encoded a peptide with similarity to petF (63.8–76.0%) (Supplementary Table 4), suggesting that the nuclear gene orf128 corresponded to the cpDNA gene petF. Interestingly, a putative petF gene (named PETF) was also found in the nuclear genomes of T. profunda (CNS00050), T. rotula (CNS00051), and T. nordenskioeldii (CNS00052), respectively, suggesting similar EGT events in these Thalassiosira species. The exact locations of these putative PETF genes in the nuclear genomes remained be determined. The peptides encoded by these PETF genes showed high conservation with the peptides encoded by the cpDNA petF genes, suggesting that these nuclear PETF genes were obtained via EGTs (Figure 4B and Supplementary Table 4). All peptides encoded by the PETF genes had 60 aa at the N-termini that showed high similarity to chloroplast transport peptide, which can be found in the LI818 that codes for chlorophyll-binding light-harvesting protein (Armbrust et al., 2004). petF was also missing in the cpDNA of T. oceanica (Lommer et al., 2010).
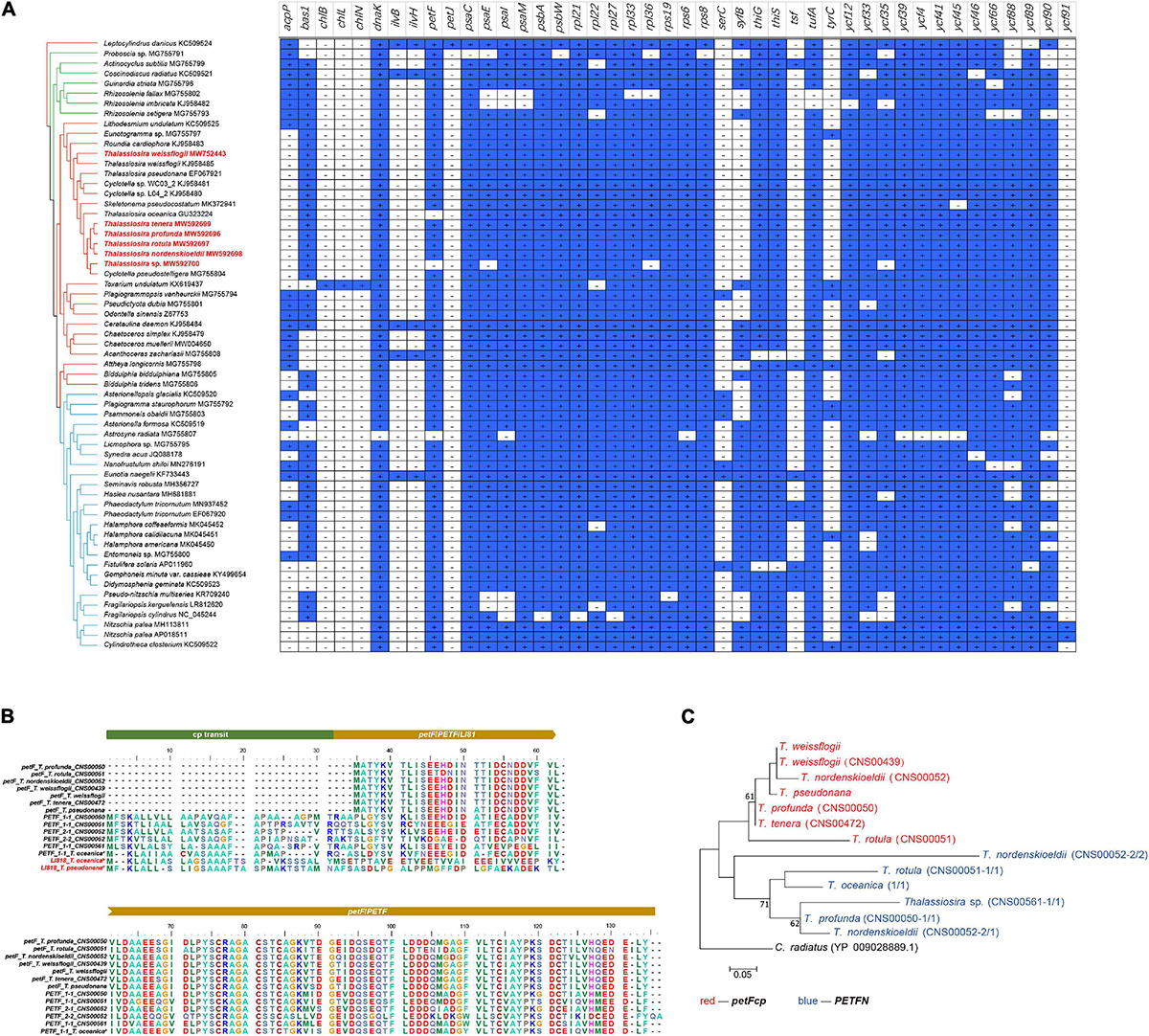
Figure 4. Gene loss and gain events of all 61 published diatom cpDNAs. (A) Presence or absence of 44 PCGs in all 61 published diatom cpDNAs with the topology of Maximum likelihood (ML) phylogenetic tree based on 95 shared PCGs of cpDNAs. Blue squares marked with a plus sign indicate the presence of the gene, and white squares marked with a minus sign indicate the absence of the gene. The strains highlighted in red were data generated in this study. (B) Protein sequence alignment of petF in the cpDNAs and PETF in the nuclear genomes from nine Thalassiosira strains. (C) Phylogenetic tree based on petF in the cpDNAs (red font) and PETF in the nuclear genomes (blue font) from nine Thalassiosira strains.
The loss of three genes psaE, rpl36, and ycf35 from the cpDNA of Thalassiosira sp. (CNS00561) was not correlated with the gain of the corresponding genes in its nuclear genome, suggesting that these genes might have been lost in evolution. Alternatively, these genes could have been missed in the genome assemblies.
The cpDNA of Thalassiosira sp. (CNS00561) encoded three orfs (orf101, orf103, and orf134) that were not found in the cpDNAs of other Thalassiosira species.
Phylogenetic Analysis of Thalassiosira Chloroplast DNAs
To explore the evolutionary positions of these Thalassiosira strains in diatoms, a core set of 95 PCGs (Supplementary Table 1) shared by 61 diatom cpDNAs were used to construct Maximum likelihood (ML) phylogenetic tree (Figure 5). Most of the nodes in the tree had 100% bootstrap (BP) values, indicating strong support. The class Bacillariophyceae was a monophyletic group and sister to Clade 3c of the class Mediophyceae with 100% BP support. The class Coscinodiscophyceae contained two clades, Clade 2a and 2b with Clade 2a containing a single species Proboscia sp.
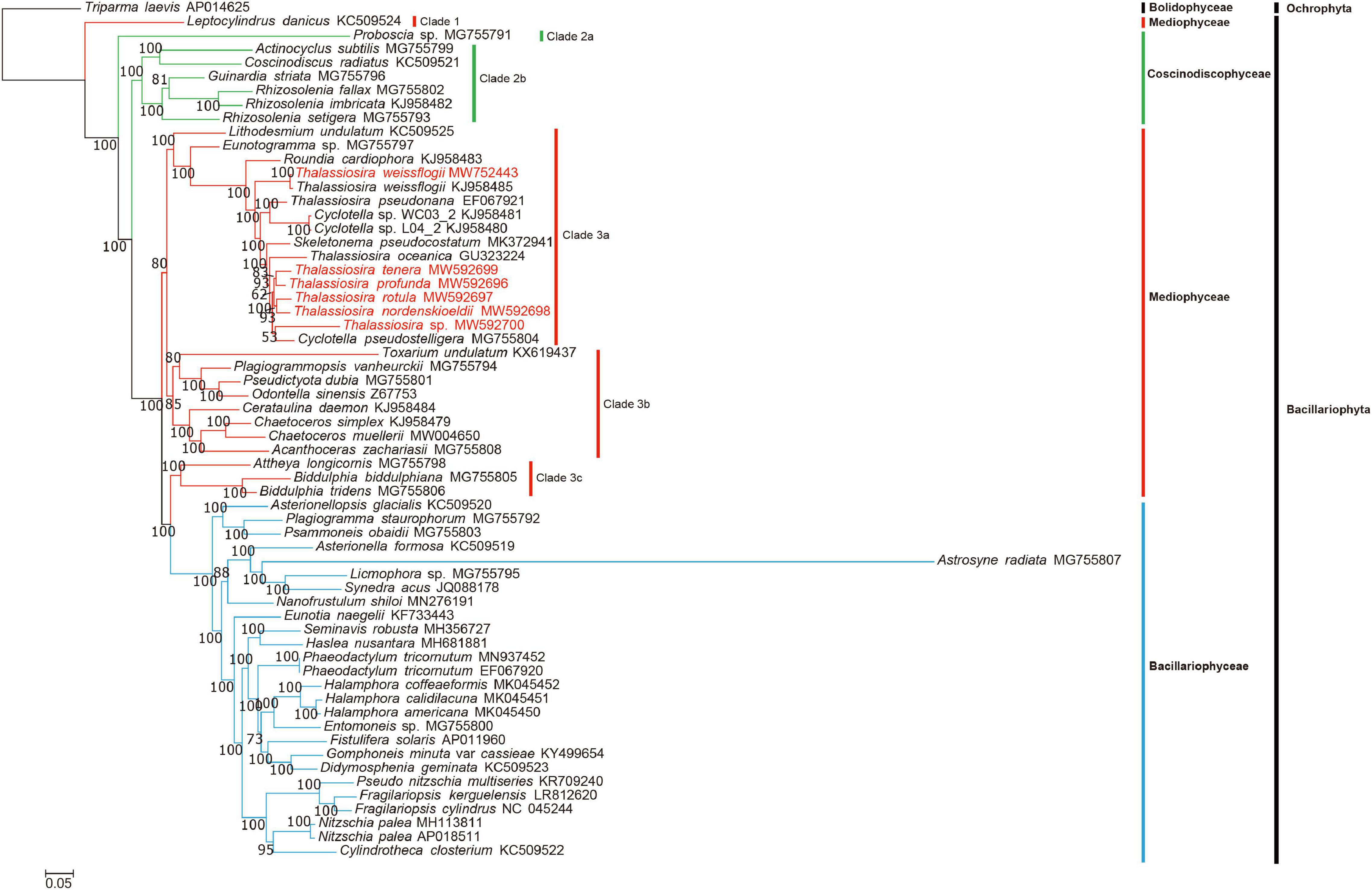
Figure 5. Maximum likelihood (ML) phylogenetic tree based on concatenated amino acid sequences of 95 PCGs shared by 61 diatom cpDNAs, with the Ochrophyta species Triparma laevis (AP014625) as the out-group taxa. The numbers beside branch nodes are the percentage of 1000 bootstrap values. The strains highlighted in red were data generated in this study.
The class Mediophyceae contained four clades (Clade 1, 3a, 3b, and 3c). All Thalassiosira species (red) were contained in Clade 3a. In addition to Thalassiosira species, Clade 3a included some species from other genera, highlighting the complexity of their taxonomical relationships (Yu et al., 2018). T. pseudonana cpDNA clustered with those of species of the genus Cyclotella, which was consistent to previous studies that proposed renaming T. pseudonana (Alverson et al., 2011). Similarly, T. weissflogii might be renamed as well because it did not cluster with other Thalassiosira species (T. oceanica, T. tenera, T. profunda, T. rotula, and T. nordenskioeldii). Clade 3a also included the genus Skeletonema.
Synteny Analysis of Thalassiosira Chloroplast DNAs
Comparative analysis of the cpDNAs of species in the genera Thalassiosira, Skeletonema, and Cyclotella with the cpDNA of Roundia cardiophora (KJ958483) (Sabir et al., 2014) uncovered high collinearity (Figure 6). In particular, the cpDNAs of T. weissflogii, T. pseudonana, T. rotula, T. nordenskioeldii, and C. pseudostelligera were perfectly collinear with that of R. cardiophora cpDNA (Figures 6, 7A), suggesting high conservation of these cpDNAs. Nevertheless, many intra-genus genome rearrangement events were identified among the cpDNAs of Thalassiosira species (Figure 6), suggesting the value of cpDNAs as super-barcodes (Li et al., 2015). An inversion event was found between the cpDNAs of two Cyclotella strains (Cyclotella sp. WC03_2 and Cyclotella sp. L04_2) and T. rotula (Figures 6, 7B). The gene block rpl20-rpl19 ranging from rpl20 to rpl19 contained 27 genes with a length of ∼16,000 bp (Supplementary Table 5). Between the cpDNAs of S. pseudocostatum and T. rotula, an inversion involving the complete SSC (rps6-rps16) involving 45 genes with a length of ∼26,500 bp was found (Figure 7C and Supplementary Table 5). This was due to the presence of two equimolar isomers in the cpDNAs, which differ from another by the relative orientation of the SSC (Linne and Kowallik, 1992). Another inversion event was identified between the T. rotula cpDNA and the cpDNAs of T. tenera and T. profunda (Figures 6, 7D). This inversion involved a gene block (clpC-dnaK) with 34 genes in partial SSC regions of T. tenera (CNS00472) and T. profunda (CNS00050) (Figure 7D and Supplementary Table 5). This block spanned ∼21,200 bp.
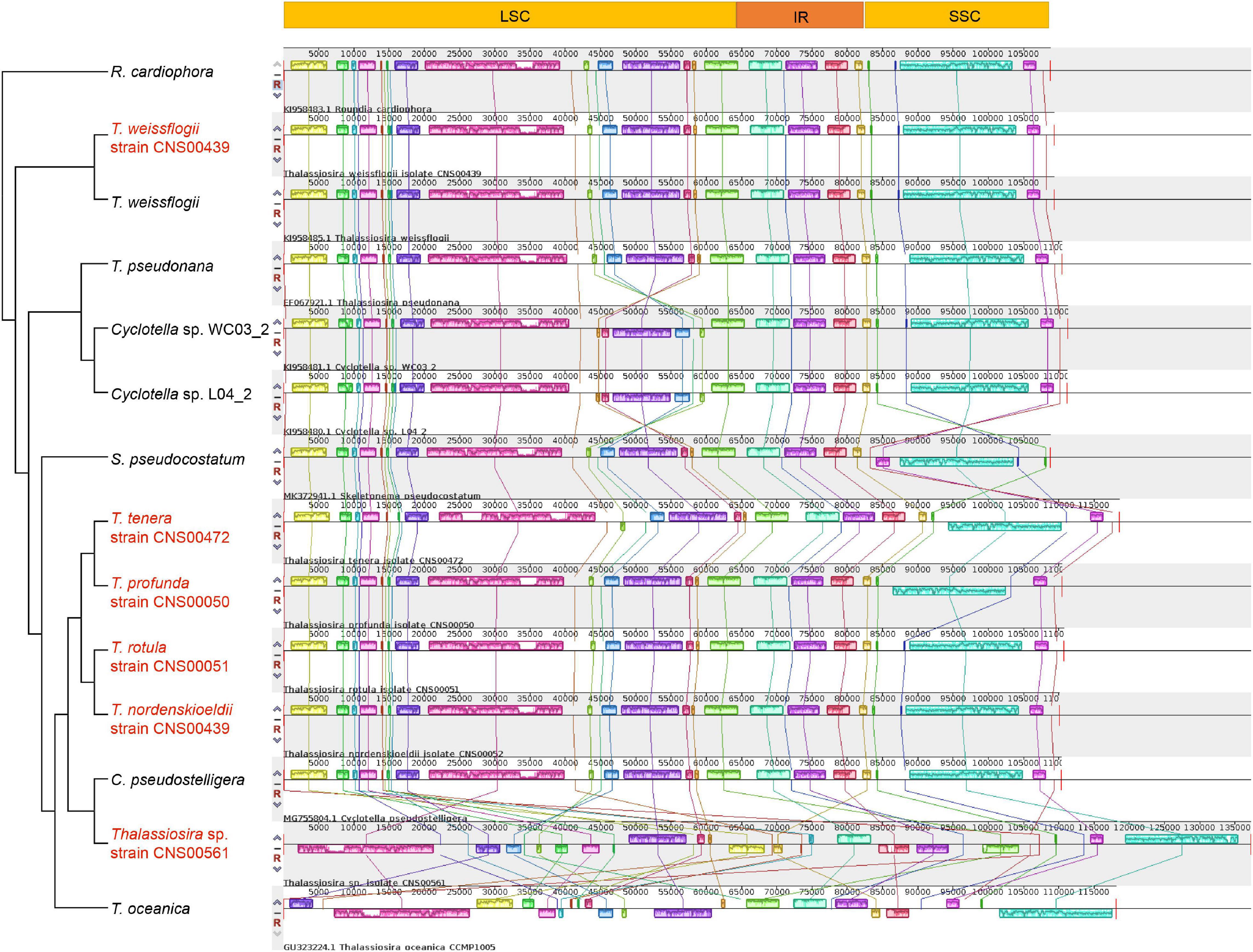
Figure 6. Collinearity analysis of the six newly Thalassiosira cpDNAs for this study and other reported cpDNAs in the three genera (Thalassiosira, Skeletonema, and Cyclotella) using Mauve. Rectangular blocks with the same color indicate collinear regions. The strains highlighted in red were data generated in this study.
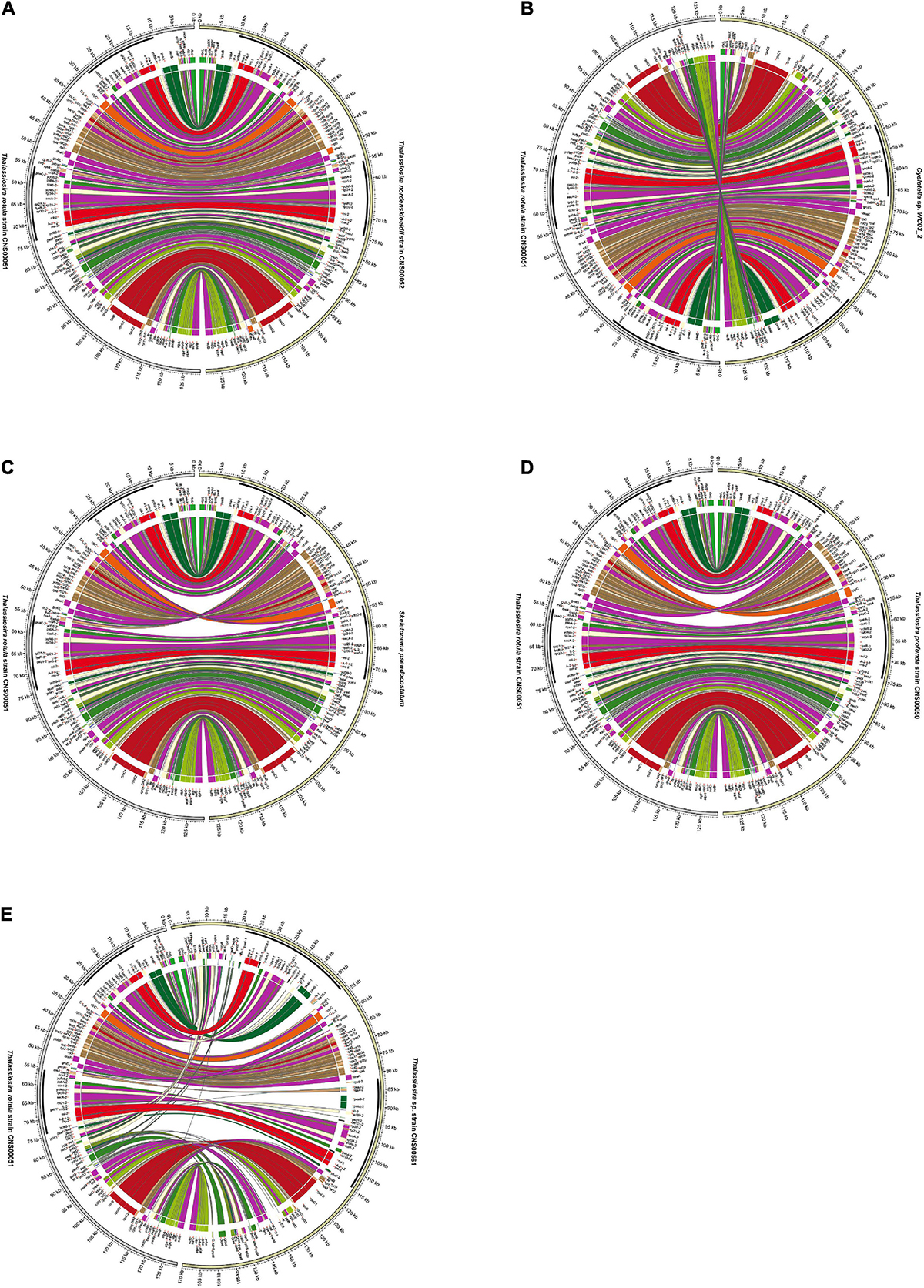
Figure 7. Pairwise collinearity comparison between the cpDNAs of T. rotula (CNS00051) and species in the genera Thalassiosira, Skeletonema, and Cyclotella (A) T. nordenskioeldii (CNS00052), (B) Cyclotella sp. WC03_2, (C) S. pseudocostatum, (D) T. profunda (CNS00050), and (E) Thalassiosira sp. (CNS00561) by the Circos diagram.
The cpDNA of Thalassiosira sp. (CNS00561) was exceptional with extensive rearrangements when it was compared with cpDNAs of other Thalassiosira species. For example, its cpDNA and T. rotula cpDNA showed extensive rearrangements (Figure 7E). Nevertheless, conserved multi-gene blocks could still be identified. For example, the large gene block named petA-trnW(cca) in LSC consisted of 16 genes and its gene order was highly conserved with cpDNAs of species of three genera (Thalassiosira, Skeletonema, and Cyclotella). Another gene block, atpA-sufB, containing eight genes, also showed conserved gene order. In addition, the gene order of gene block rps10-dnaK within SSC was shared by the three genera (Thalassiosira, Skeletonema, and Cyclotella) (Supplementary Table 5).
Inverted Repeat Region Expansions and Contractions
Comparative analysis of the quadripartite structures and corresponding boundaries of cpDNAs of species in the three genera (Thalassiosira, Skeletonema, and Cyclotella) revealed some IR regions experienced expansions or contractions (Figure 8).

Figure 8. Comparative analysis of the boundaries of LSC, SSC, and IR regions among six newly Thalassiosira cpDNAs for this study and other reported cpDNAs in the three genera (Thalassiosira, Skeletonema, and Cyclotella). The strains highlighted in red were data generated in this study.
In cpDNAs of species of the three genera Thalassiosira, Skeletonema, and Cyclotella, the psaA-trnP(ugg) genes were located in the LSC/IRb boundaries. The distances between psaA and the LSC/IRb boundaries ranged from 92 to 142 bp, while the distances between trnP(ugg) and the LSC/IR boundaries were from 69 to 116 bp. And ccsA-rps6 genes were located at the SSC/IRb boundaries. IRb region only extended into the PCG rps6 with 9 bp inside the IRb region of R. cardiophora cpDNA. However, rps6 of the three genera were completely included inside the SSC with 29 to 55 bp to the SSC/IRa boundaries. At the boundaries of LSC/IRa, the distances between ccsA were 229 to 288 bp to the boundaries. However, except for Cyclotella sp. WC03_2 (KJ958481), Cyclotella pseudostelligera (MG755804) and T. profunda, the SSC/IRb boundaries located in the PCG rps16 with a region ranged from 2 to 21 bp. And in all three genera, the psaJ were situated inside the LSC regions, which were 21 to 134 bp from the LSC/IRa boundaries.
While the IR regions of most cpDNAs were generally conserved with minor exceptions, IR regions of the cpDNAs of two species Thalassiosira sp. (CNS00561) and T. oceanica showed substantial cpDNA genome rearrangements relative to other Thalassiosira cpDNAs, which made some genes enter or leave the IR region, resulting in the difference of gene copy number. For example, the psaA and psaB genes were located in the IR region of Thalassiosira sp. (CNS00561), which were often located in the LSC region of other Thalassiosira species, resulted in one more copy of these genes in the cpDNAs of Thalassiosira sp. (CNS00561). For the same reason, the clpC of T. oceanica also had an additional copy (Figure 6). In contrast, the ccsA gene, which was usually located in the IR region, was located in the LSC region of Thalassiosira sp. (CNS00561), so it had one less copy than other Thalassiosira species.
Sequence Variations Among Chloroplast DNAs of Different Thalassiosira Species
Each of the 123 PCGs shared in the nine cpDNAs of the genus Thalassiosira was individually aligned and calculated for nucleotide diversity (Pi value) (Supplementary Figure 3). Pi ranged from 0.0162 (psbN) to 0.2237 (rpl29). Three genes, thiS, clpC and rpl29, have higher nucleotide diversity, with Pi greater than 0.2. The photosystem II protein gene (psb), except psbW, had lower Pi than other genes. Similarly, as an important light-regulated gene, psb often has low nucleotide diversity and high homology and conservation in the cpDNAs of higher plants (Xiong et al., 2020; Yang et al., 2020; Wen et al., 2021).
The analysis of Pi value of a sequence from multiple strains requires consistent collinearity. However, due to genome rearrangements among cpDNAs of the genus Thalassiosira, only three conserved gene blocks (atpA-sufB, petA-trnW(cca), and rps10-dnaK) were selected for mutation hotspots identification (Table 3). The block atpA-sufB was about 6700 bp in length and contained eight genes, corresponding to the region 122,247–128,970 bp of the cpDNAs of T. profunda (CNS00050), the block petA-trnW(cca) was about 19,300 bp in length and contained 16 genes, corresponding to the region 94,965–114,298 bp of the cpDNAs of T. profunda (CNS00050), and the block rps10-dnaK was about 17,800 bp in length and contained 31 genes, corresponding to the region 30,305–48,361 bp of the cpDNAs of T. profunda (CNS00050). With a sliding window of 600 bp with the step size of 50 bp, variation hotspots with length differences among the cpDNAs of nine Thalassiosira strains were found in the above-mentioned three gene blocks. A region about 1000 bp in block petA-trnW(cca) [location: 99,796–100,867 bp in the cpDNAs of T. profunda (CNS00050)], involving rpl33, rps20, and rpoB was identified to have high levels of variations (Figure 9A, red arrow). Figure 9A showed that in the region indicated by the arrow, cpDNAs of nine Thalassiosira strains have different sequence lengths, with the longest in the T. pseudonana cpDNA and the shortest in the Thalassiosira sp. (CNS00561) cpDNA. DNA sequences of this region from the cpDNAs had different lengths in these nine strains of Thalassiosira, ranging from 835 to 1087 bp (Figure 9B). The phylogenetic relationship revealed by this region of nine Thalassiosira strains was similar to but not identical to the phylogenetic relatationship of these strains revealed using cpDNA PCGs (Figure 5). We named this region Thalassiosira chloroplast 1, which was abbreviated as thcp1. Phylogenetic analysis based on ML method showed that eight Thalassiosira species could be adequately separated using thcp1 as a molecular marker (Figure 9B). This indicates that thcp1 can be used as molecular marker for distinguishing Thalassiosira species with high resolution. The primers were used to amplify DNAs extracted from 18 phytoplankton species including nine Bacillariophyta species [three Mediophyceae species (red), three Coscinodiscophyceae species (green), and three Bacillariophyceae species (yellow)] (Figure 9C), two Haptophyta species (Isochrysis galbana and Phaeocystis globosa), four Dinoflagellata species (Amphidinium carterae, Alexandrium tamarense, Karenia mikimotoi, and Prorocentrum donghaiense), and three Ochrophyta species (Heterosigma akashiwo, Aureococcus anophagefferens, and Chattonella marina). PCR amplifications were successful for all Thalassiosira species with amplicons with different sizes. However, PCR amplifications were also successful for five additional species of the classes Mediophyceae and Coscinodiscophyceae. With the abundance of cpDNAs information of more species, it is possible to improve the molecular markers to make up for the lack of specificity.
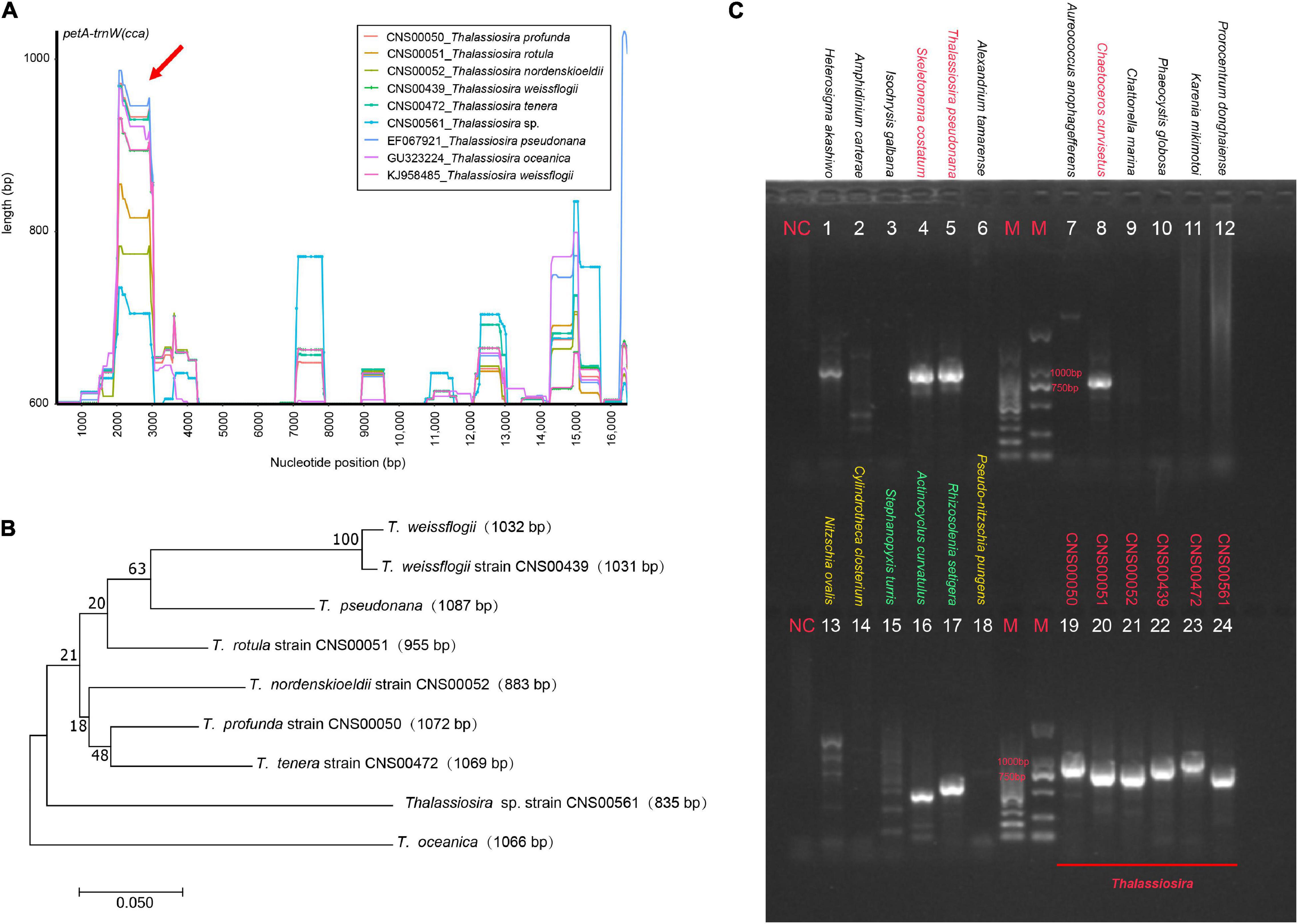
Figure 9. Mutation hotspots among the cpDNAs of nine Thalassiosira strains. (A) Variation hotspots with length differences for collinearly conserved gene blocks petA-trnW(cca). (B) Maximum likelihood (ML) phylogenetic analysis based on molecular marker thcp1. (C) Amplification results of thcp1 from the six newly Thalassiosira and 18 other phytoplankton species belonging to Bacillariophyta, Haptophyta, Dinoflagellata, and Ochrophyta.
Speciation of Thalassiosira Species
The 127 PCGs (Supplementary Table 2) shared in cpDNAs of the 23 species (15 of Mediophyceae, four of Bacillariophyceae, three of Coscinodiscophyceae and one of Phaeophyceae) were subjected to divergence times estimation (Figure 10).
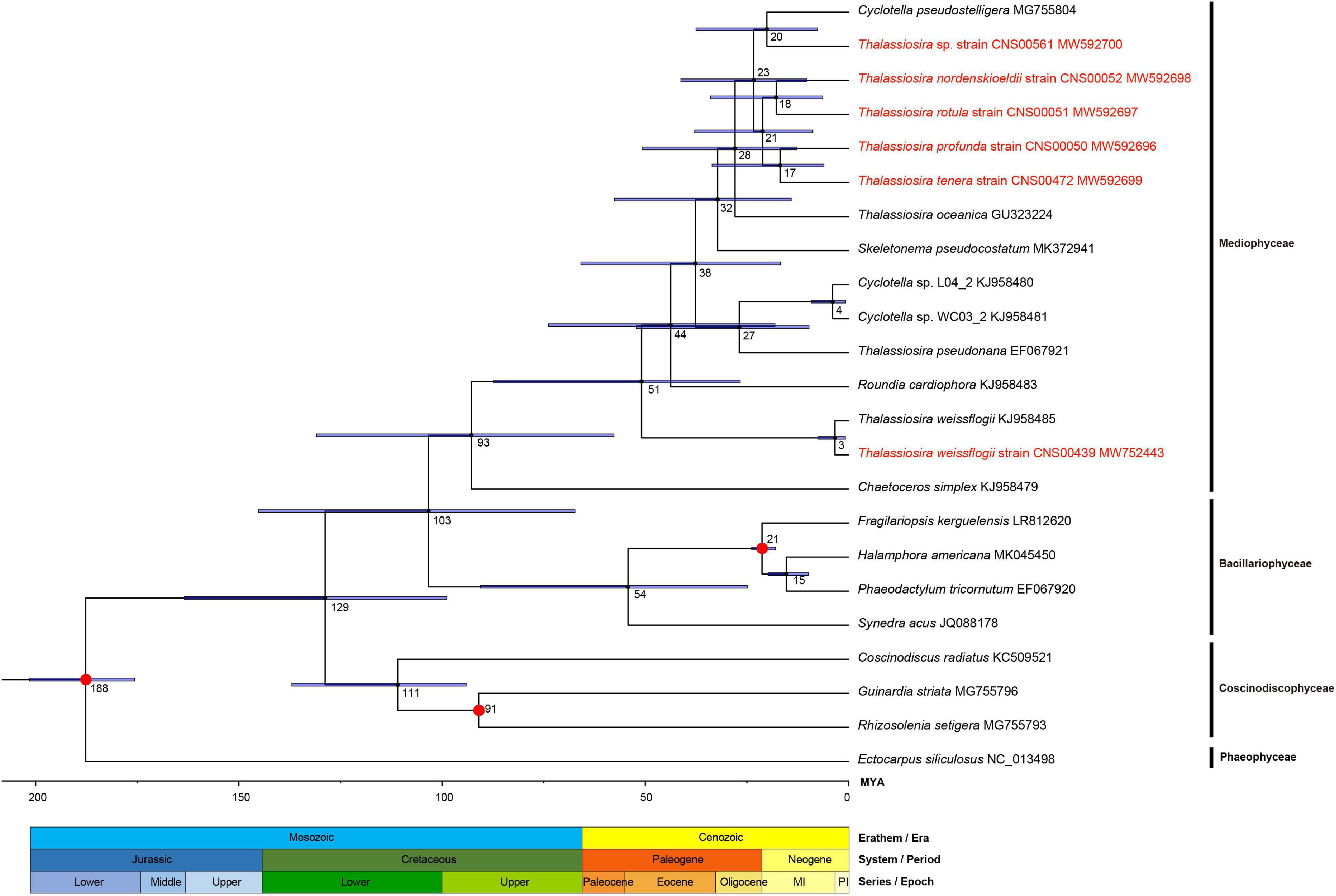
Figure 10. Divergence time estimations for Thalassiosira species by Bayesian analysis based on the amino acid sequence of 127 PCGs. The fossil calibration taxa are indicated with red points on the corresponding nodes. Horizontal bars represent 95% highest posterior density (HPD) values of the estimated divergence time. The strains highlighted in red were data generated in this study.
The results suggested that the first event of diversification within the Phylum Bacillariophyta occurred 129 MYA in lower Cretaceous and the class Coscinodiscophyceae split from it preferentially at 111 MYA. The other two classes, Mediophyceae and Bacillariophyceae were estimated to have separated from each other about 103 MYA. And within Mediophyceae, age estimation for the crown node of Thalassiosira was 51 MYA in the early Eocene. The branching of T. weissflogii in the class Mediophyceae was estimated to have occurred 51 MYA and diverged into different strains at three MYA. Furthermore, the divergence time between the T. pseudonana and Cyclotella was estimated at 27 MYA in Oligocene. The crown node of the remaining Thalassiosira was estimated to have occurred 28 MYA and speciation of them concentrated between 17 and 28 MYA.
Discussion
Thalassiosira Chloroplast DNA Organization
In this study, six complete cpDNAs of Thalassiosira strains were assembled, substantially expanded the total number of cpDNAs of Thalassiosira species from three to nine. Thus, cpDNAs of nine Thalassiosira strains corresponding to eight Thalassiosira species were compared and analyzed. The size of these cpDNAs varied significantly, from 127,601 to 170,005 bp, apparently due to a combination of gene density and IR expansions and contractions (Ravi et al., 2007). Among this, gene density was usually related to the length of intergenic spacers, the presence or absence of introns and gain and loss of genes (Ravi et al., 2007; Bedoya et al., 2019). In the nine cpDNAs, Thalassiosira sp. (CNS00561) had the longest intergenic spacers (median value was 82 bp, maximum value was 2676 bp and average value was 297 bp) and the longest IR region (32,702 bp). Similarly, T. oceanica and T. tenera also had longer intergenic spacers and IR region. In addition, the cpDNA of T. tenera had a group II intron with a length of 2874 bp in atpB. As a result of the above reasons, these three cpDNAs were incompact and had long copy length of IR region, which eventually lead to their larger genome size.
Gene rearrangement and the expansion and contraction of IR region are important characteristics for the analysis of genome evolution (Ravi et al., 2007). Although the organization of plastid is generally stable and conserved, gene rearrangement occurs frequently. The fragmented and large-scale rearrangement the cpDNAs of Thalassiosira sp. (CNS00561) and T. oceanica could be regarded as unique genomic characteristics of the two species, and species classification and identification by cpDNAs has been gradually accepted by taxonomists (Kumar et al., 2009; Wu et al., 2010; Yang et al., 2013; Li et al., 2015). As super-barcodes (Li et al., 2015), cpDNAs can show the differences between Thalassiosira species in terms of genome size, gene gain or loss and gene order, which can greatly improve the resolution of low classification levels (Parks et al., 2009) and help to describe species diversity more accurately.
Gene Loss and Endosymbiotic Gene Transfers in Thallassiosira Chloroplast DNAs
The nine complete cpDNAs of Thalassiosira contained a set of 155 shared genes, including 123 PCGs, 27 tRNA, three rRNA, and two other RNA genes. Chloroplast is considered to be derived from free-living cyanobacteria, and cpDNA is much smaller than that of its predecessors, but it still retains many proteins that are not encoded by itself (Martin, 1998). It is widely accepted that endosymbiotic genes are exported to the nuclear genomes (nDNAs) through EGT (Timmis et al., 2004). Therefore, many primitive PCGs of cpDNAs have been or are being transferred to nDNAs. In addition to the reported missing petF in T. oceanica (Lommer et al., 2010), the cpDNA of Thalassiosira sp. (CNS00561) also missed four PCGs, including petF, ycf35, rpl36, and psaE. The loss of these four genes has also been recorded in the cpDNAs of other species. Among them, petF of Chlorophytes + Streptophytes, Euglenophytes and Alveolates was missing from cpDNAs and transferred to nDNAs (Cui et al., 2006), and the same event also occurred in T. oceanica (Lommer et al., 2010). Similarly, the conserved region of petF can also be found in the non-cpDNA of CNS00561 (PETF), and its front-end amino acid sequence was highly similar to the chloroplast transport peptide of T. oceanica PETF, which may provide the ability for PETF to transfer into chloroplast to play its role. Studies have proposed that the low iron in the sea can be the important factors causing the transfer of petF encoding ferredoxin from cpDNAs to nDNAs, which is an environment-driven evolutionary strategy (Strzepek and Harrison, 2004), which also confirms the remarkable plasticity of chloroplast and nuclear genomes (Lommer et al., 2010). In addition, in three of the six newly constructed cpDNAs of Thalassiosira, including CNS00050, CNS00051, and CNS00052, we found PETF in both cpDNA and non-cpDNA. Such functional PCGs were transferred to the nuclear DNAs, and degenerate copies were often present in organelle genomes, which was considered as an intermediate state of EGT (Brennicke et al., 1993). Until the nuclear copy gene can function normally, the chloroplast copy gene may accumulate some deleterious mutations and be deleted (Ravi et al., 2007).
Except Thalassiosira sp. (CNS00561), psaE was missing in the cpDNAs of five other diatoms, namely, Proboscia sp. (MG755791), Fragilariopsis kerguelensis (LR812620), Pseudo-nitzschia multiseries (KR709240), and Rhizosolenia imbricate (KJ958482), and Rhizosolenia fallax (MG755802). All classes of diatoms, including Mediophyceae, Coscinodiscophyceae, and Bacillariophyceae, have species with psaE loss. It’s worth noting that, the cpDNAs of another species of genus Fragilariopsis, Fragilariopsis cylindrus (NC_045244), and another species of genus Rhizosolenia, Rhizosolenia setigera (MG755793), had complete psaE, suggesting that the loss of psaE in Coscinodiscophyceae occurred at the branch node of Guinardia and the two newly evolved Rhizosolenia. And in class Bacillariophyceae, the loss of psaE occurred at the node where the branch including genus Fragilariopsis and Pseudo-nitzschia was generated, but it was obtained in F. cylindrus again. Similarly, rpl36 was also missing in the above-mentioned Proboscia sp. and R. fallax. However, the loss of ycf35 was more extensive, and it was missing in the cpDNAs of eight species including Proboscia sp., P. multiseries and F. kerguelensis in 61 diatoms. Gene deletion or transfer is a survival mode of adaptation to the environment, which may lead to the diversity of adaptive phenotypes (Roy et al., 2020). However, due to the lack of genome-wide information and the limited sampling of research species, the mechanism and evolutionary relationship of gene loss and transfer are still unclear. In addition, gene gain and loss could be used as a shortcut to distinguish between two species (Hebert et al., 2004), because super-barcodes are more effective than traditional barcodes in detecting genetic loss.
Phylogenetic Analysis and Speciation of Thalassiosira Species
In this study, phylogenetic analysis of 61 diatoms was conducted based on 95 shared PCGs of cpDNAs. The results were consistent with 103 shared PCGs in the cpDNAs of 40 diatoms by Yu et al. (2018), and even expanded the analysis of representative species in each class compared with Yu’s analysis (Yu et al., 2018). This study focused on Clade 3a, including the genus Thalassiosira, which was generated roughly around 93 MYA. The results showed that T. weissflogii first differentiated from the root of all the branches of the genus Thalassiosira around 51 MYA, which supported by the conclusion that the genus Thalassiosira, as mentioned by Sims et al. (2019), evolved around the Eocene-Oligocene boundary. Then, T. pseudonana and two strains of Cyclotella were clustered into one branch with 100% BS, which also supports the view proposed by Alverson (Alverson et al., 2011) that T. pseudonana should be classified in the genus Cyclotella and named as Cyclotella nana. After that, the other six Thalassiosira strains were gradually differentiated at late Miocene to Pliocene around 17–28 MYA. During this period global changes such as the establishment of permanent Antarctic ice-sheets (Barron and Baldauf, 1995), along with changes in ambient temperature, increased the diversity of Thalassiosira species to adapt to the new marine environment (Sims et al., 2019). The genus Thalassiosira was separated by Skeletonema and Cyclotella in phylogeny, which was different from the classification of Thalassiosira based on morphology. The evolutionary status of the three genera may be more accurate when more extensive and balanced sampling analysis of each genus are performed, which may lead to a reclassification of some species.
Conclusion
In this study, we constructed six complete chloroplast genomes of Thalassiosira species, and analyzed them in a comparative framework with published cpDNAs of three Thalassiosira species and 54 diatom species. Gene content of cpDNAs was approximately the same within the Thalassiosira species. petF was missing in the cpDNAs of both T. oceanica and Thalassiosira sp. (CNS00561) but found in their nuclear genomes, which might be an EGT for adapting to low iron environment. A group II intron containing one orf was identified in atpB of the T. tenera cpDNA, which was also found in some diatom species, which was thought to be acquired from green algae through HGT. In addition, complete cpDNAs have been proposed as super-barcodes for discriminating more closely related species with higher resolution. At the same time, mutation hotspots may be developed as molecular markers for species identification. This study can not only provide an in-depth understanding of the cpDNA characteristics of Thalassiosira species, but also provide powerful resources for genetic evolution and speciation analysis of Thalassiosira.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm. nih.gov/genbank/, MW592696, http://www.ncbi.nlm.nih.gov/genbank/, MW592697, https://www.ncbi.nlm.nih.gov/genbank/, MW592698, https://www.ncbi.nlm.nih.gov/genbank/, MW5926 99, https://www.ncbi.nlm.nih.gov/genbank/, MW592700, https://www.ncbi.nlm.nih.gov/genbank/, MW752443, https://www.ncbi.nlm.nih.gov/, PRJNA684688.
Author Contributions
KL and NC designed the research. KL drafted the manuscript. NC revised the manuscript. KL, YC, ZC, and SL collected samples. KL and QX conducted the data analysis. All authors have read and agreed to the submitted version of the manuscript.
Funding
This research was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB42000000), Chinese Academy of Sciences Pioneer Hundred Talents Program (to NC), Taishan Scholar Project Special Fund (to NC), Qingdao Innovation and Creation Plan (Talent Development Program – Fifth Annual Pioneer and Innovator Leadership Award to NC, 19-3-2-16-zhc), and National Key Research and Development Program of China (2017YFC1404300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to all staffs of marine ecological environment genomics research group in Institute of Oceanology, Chinese Academy of Sciences. We are grateful to colleagues from the Jiaozhou Bay Marine Ecosystem Research Station for their help in field sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.788307/full#supplementary-material
Supplementary Figure 1 | Maximum likelihood (ML) phylogenetic tree based on the nuclear 28S rDNA D1–D2 domains for species identification of six Thalassiosira strains in this study. The strains highlighted in red were data generated in this study.
Supplementary Figure 2 | Phylogenetic tree based on ORFs in the introns of T. tenera and five other reported diatom cpDNAs with ORFs in the introns of Rhodophyta and Chlorophyta cpDNAs as reference. The sequence in the red box was the objects of this study. The sequences highlighted in red were red algae, and in green are green algae.
Supplementary Figure 3 | The nucleotide diversity (Pi value) of each of the 123 PCGs shared in the nine Thalassiosira cpDNAs.
Supplementary Table 1 | Gene Set 1 – 95 shared protein-coding genes partitioned by functional groups.
Supplementary Table 2 | Gene Set 2 – 127 shared protein-coding genes partitioned by functional groups.
Supplementary Table 3 | Molecular markers and their significant alignment information.
Supplementary Table 4 | PID after amino acid 60 of petFs and the putative PETFs.
Supplementary Table 5 | Main gene blocks information.
Footnotes
- ^ https://megasun.bch.umontreal.ca/RNAweasel/
- ^ https://www.ncbi.nlm.nih.gov/orffinder
- ^ https://megasun.bch.umontreal.ca/RNAweasel/
- ^ http://tree.bio.ed.ac.uk/software/figtree/
References
Álvarez, I. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/S1055-7903(03)00208-2
Alverson, A. J., Beszteri, B., Julius, M. L., and Theriot, E. C. (2011). The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol. Biol. 11:125. doi: 10.1186/1471-2148-11-125
Alverson, A. J., Jansen, R. K., and Theriot, E. C. (2007). Bridging the Rubicon: phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Mol. Phylogenet. Evol. 45, 193–210. doi: 10.1016/j.ympev.2007.03.024
Anger, K., Anger, V., and Hagmeier, E. (1986). Laboratory studies on larval growth of Polydora ligni, Polydora ciliata, and Pygospio elegans (Polychaeta, Spionidae). Helgolnder Meeresuntersuchungen. 40, 377–395. doi: 10.1007/BF01983819
Araujo, C. F., and Souza-Santos, L. P. (2013). Use of the microalgae Thalassiosira weissflogii to assess water toxicity in the Suape industrial-port complex of Pernambuco, Brazil. Ecotoxicol. Environ. Saf. 89, 212–221. doi: 10.1016/j.ecoenv.2012.11.032
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barron, J. A., and Baldauf, J. G. (1995). Cenozoic marine diatom biostratigraphy and applications to paleoclimatology and paleoceanography. Siliceous Microfossils Paleontol. Soc. Short Courses Paleontol. 8, 107–118. doi: 10.1017/S2475263000001446
Bedoya, A. M., Ruhfel, B. R., Philbrick, C. T., Madrinan, S., Bove, C. P., Mesterhazy, A., et al. (2019). Plastid genomes of five species of riverweeds (podostemaceae): structural organization and comparative analysis in malpighiales. Front. Plant Sci. 10:1035. doi: 10.3389/fpls.2019.01035
Bendich, A. J. (2004). Circular chloroplast chromosomes: the grand illusion. Plant Cell. 16, 1661–1666. doi: 10.1105/tpc.160771
Bleidorn, C., Vogt, L., and Bartolomaeus, T. (2003). New insights into polychaete phylogeny (Annelida) inferred from 18S rDNA sequences. Mol. Phylogenet. Evol. 29, 279–288. doi: 10.1016/S1055-7903(03)00107-6
Brembu, T., Winge, P., Tooming-Klunderud, A., Nederbragt, A. J., Jakobsen, K. S., and Bones, A. M. (2014). The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar Genomics 16, 17–27. doi: 10.1016/j.margen.2013.12.002
Brennicke, A., Grohmann, L., Hiesel, R., Knoop, V., and Schuster, W. (1993). The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBS Lett. 325, 140–145. doi: 10.1016/0014-5793(93)81430-8
Cai, C., Wang, L., Zhou, L., He, P., and Jiao, B. (2017). Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One 12:e0184196. doi: 10.1371/journal.pone.0184196
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, S., Gao, Y., Du, H., Dong, Q., and Huang, C. (2004). First recording of Thalassiosira diporocyclus bloom in the southeast china sea. Oceanol. limnol. Sin. 35, 130–137.
Cheng, Z., Gao, Y., and Liu, S. (1993). Nano-Diatoms In Fujian Coast. Beijing: China Ocean Press, 91.
Crowell, R. M., Nienow, J. A., and Cahoon, A. B. (2019). The complete chloroplast and mitochondrial genomes of the diatom Nitzschia palea (Bacillariophyceae) demonstrate high sequence similarity to the endosymbiont organelles of the dinotom Durinskia baltica. J. Phycol. 55, 352–364. doi: 10.1111/jpy.12824
Cui, L., Veeraraghavan, N., Richter, A., Wall, K., Jansen, R. K., Leebens-Mack, J., et al. (2006). ChloroplastDB: the chloroplast genome database. Nucleic Acids Res. 34, D692–D696. doi: 10.1093/nar/gkj055
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134. doi: 10.1186/s13059-016-1004-2
Darling, A. E., Mau, B., and Perna, N. T. (2010). Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
Dong, J., and Jiao, N. (1995). Ecological Studies On Nanoplanktonic Diatoms In Jiaozhou Bay, China. Beijing: Science Press, 96–102.
dos Reis, M., Álvarez-Carretero, S., and Yang, Z. (2013). MCMCTree Tutorials. Available online at: http://abacus.gene.ucl.ac.uk/software/MCMCtree.Tutorials.pdf (accessed April 28, 2017).
Emmerson, W. (1980). Ingestion, growth and development of Penaeus indicus larvae as a function of Thalassiosira weissflogii cell concentration. Mar. Biol. 58, 65–73. doi: 10.1007/BF00386881
Fryxell, G. A., and Hasle, G. R. (1977). The genus Thalassiosira: some species with a modified ring of central strutted processes. Nova Hedwigia Beih 54, 67–98.
Fu, C., Wu, C., Ye, L., Mo, Z., Jie, L., Chang, Y., et al. (2018). “Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide,” in Proceedings Of The Abstracts Of The 85th Annual Conference Of Chinese Society Of Botany (1993-2018).
Gao, Y., and Cheng, Z. A. (1992). New species and two new varieties of Thalassiosira. J. Xiamen Univ. (Nat. Sci.) 31, 291–294.
Gao, Y., Liang, J., Chen, C., Li, X., Peng, X., and Liu, G. (2011). Studies on biodiversity and ecological importance of marine diatoms. J. Xiamen Univ. (Nat. Sci.) 50, 455–463.
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Gu, H.-F. (2007). The first record of ensiculifera balech and fragilidium balech (Dinophyceae) from Chinese coast. Acta Phytotaxonomica Sin. 45, 828–840. doi: 10.1360/aps07001
Guiry, M. D., and Guiry, G. M. (2021). AlgaeBase. World-wide Electronic Publication. Galway: National University of Ireland.
Guo, H. (2004). Illustrations Of Planktons Responsible For The Blooms In Chinese Coastal Waters (In Chinese). Beijing: Ocean Press.
Guo, X., Guo, Y., and Li, Y. (2018a). Thalassiosira allenii var. Striata as a new variety in the genus Thalassiosira cleve. Acta Hydrobiol. Sin. 42, 824–831.
Guo, X., Guo, Y., and Li, Y. (2018b). Thalassiosira minuscula var. bicustodis, a new variety in the genus Thalassiosira. Plant Sci. J. 36, 508–517.
Guo, Y., Wu, G., and Li, Y. (2017). Re-examination and assessment of the morphological traits of the diatom genus Thalassiosira Cleve, a case study of Thalassiosira allenii Takano. Plant Sci. J. 35, 194–204.
Hamsher, S. E., Keepers, K. G., Pogoda, C. S., Stepanek, J. G., Kane, N. C., and Kociolek, J. P. (2019). Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PLoS One 14:e0217824. doi: 10.1371/journal.pone.0217824
Hasle, G. R. (1973). Some marine plankton genera of the diatom family Thalassiosiraceae. Beihefte Nova Hedwigia. 45, 1–49.
Hasle, G. R. (1978). Some freshwater and brackish water species of the diatom genus Thalassiosira Cleve. Phycologia 17, 263–292. doi: 10.2216/i0031-8884-17-3-263.1
Hasle, G. R., and Lange, C. B. (1989). Freshwater and brackish water Thalassiosira (Bacillariophyceae): taxa with tangentially undulated valves. Phycologia 28, 120–135. doi: 10.2216/i0031-8884-28-1-120.1
Hebert, P., Penton, E. H., Burns, J. M., Janzen, D. H., and Hallwachs, W. (2004). Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A. 101, 14812–14817. doi: 10.1073/pnas.0406166101
Hoppenrath, M., Beszteri, B., Drebes, G., Halliger, H., Van Beusekom, J. E. E., Janisch, S., et al. (2007). Thalassiosiraspecies (Bacillariophyceae, Thalassiosirales) in the north sea at helgoland (German Bight) and Sylt (North Frisian Wadden Sea)–a first approach to assessing diversity. Eur. J. Phycol. 42, 271–288. doi: 10.1080/09670260701352288
Ianora, A., Poulet, S. A., Miralto, A., and Grottoli, R. (1996). The diatom Thalassiosira rotula affects reproductive success in the copepod Acartia clausi. Mar. Biol. 125, 279–286. doi: 10.1007/BF00346308
Ji, Y., Liu, C., Yang, Z., Yang, L., He, Z., Wang, H., et al. (2019). Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol. Ecol. Resour. 19, 1333–1345. doi: 10.1111/1755-0998.13050
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Kaczmarska, I., Beaton, M., Benoit, A. C., and Medlin, L. K. (2006). Molecular phylogeny of selected members of the order Thalassiosirales (Bacillariophyta) and evolution of the fultoportula. J. Phycol. 42, 121–138. doi: 10.1111/j.1529-8817.2006.00161.x
Kajitani, R., Yoshimura, D., Okuno, M., Minakuchi, Y., Kagoshima, H., Fujiyama, A., et al. (2019). Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 10:1702. doi: 10.1038/s41467-019-09575-2
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kiatmetha, P., Siangdang, W., Bunnag, B., Senapin, S., and Withyachumnarnkul, B. (2011). Enhancement of survival and metamorphosis rates of Penaeus monodon larvae by feeding with the diatom Thalassiosira weissflogii. Aquac. Int. 19, 599–609. doi: 10.1007/s10499-010-9375-y
Koboldt, D. C., Chen, K., Wylie, T., Larson, D. E., McLellan, M. D., Mardis, E. R., et al. (2009). VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285. doi: 10.1093/bioinformatics/btp373
Kooistra, W. H., Sarno, D., Balzano, S., Gu, H., Andersen, R. A., and Zingone, A. (2008). Global diversity and biogeography of Skeletonema species (bacillariophyta). Protist 159, 177–193. doi: 10.1016/j.protis.2007.09.004
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Hahn, F. M., McMahan, C. M., Cornish, K., and Whalen, M. C. (2009). Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 9:131. doi: 10.1186/1471-2229-9-131
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Philos. Soc. 90, 157–166. doi: 10.1111/brv.12104
Li, Y. (2006). Ecological Characteristics And Taxonomic Studies On Nano-Diatoms In Coastal Waters of China. Ph.D. thesis. Xiamen: University of Xiamen.
Li, Y. (2009). Morphological characteristics comparisons between Thalassiosira and Coscinodiscus. Bull. Bot. Res. 29, 282–288.
Li, Y., Gao, Y., and Lü, S. (2008a). Newly recorded species of Thalassiosira from China I. J. Xiamen Univ. (Nat. Sci.) 47, 286–290.
Li, Y., Gao, Y., and Lü, S. (2008b). New record species of Thalassiosira for China (II). J. Trop. Oceanogr. 27, 40–44.
Li, Y., Guo, Y. Q., and Guo, X. H. (2018). Morphology and molecular phylogeny of Thalassiosira sinica sp. nov. (Bacillariophyta) with delicate areolae and fultoportulae pattern. Eur. J. Phycol. 53, 122–134. doi: 10.1080/09670262.2017.1386329
Li, Y., Zhao, Q., and Lü, S. (2013). The genus Thalassiosira off the guangdong coast, South China Sea. Bot. Mar. 56, 83–110. doi: 10.1515/bot-2011-0045
Li, Y., Zhao, Q., and Lü, S. (2014). Taxonomy and species diversity of the diatom genus Thalassiosira (Bacillariophyceae) in Zhejiang coastal waters, the East China Sea. Nova Hedwigia 99, 373–402. doi: 10.1127/0029-5035/2014/0170
Liang, Y. (2012). Investigation And Evaluation Of Red Tide Disasters In China (1933-2009). Beijing: OceanPress.
Linne, K. H., and Kowallik, K. V. (1992). Structural organization of the chloroplast genome of the chromophytic alga Vaucheria bursata. Plant Mol. Biol. 18, 83–95. doi: 10.1007/BF00018459
Lommer, M., Roy, A.-S., Schilhabel, M., Schreiber, S., Rosenstiel, P., and LaRoche, J. (2010). Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11:718. doi: 10.1186/1471-2164-11-718
Losic, D., Mitchell, J. G., Lal, R., and Voelcker, N. H. (2007). Rapid fabrication of micro- and nanoscale patterns by replica molding from diatom biosilica. Adv. Funct. Mater. 17, 2439–2446. doi: 10.1002/adfm.200600872
Mallatt, J., and Sullivan, J. (1998). 28S and 18S rDNA sequences support the monophyly of lampreys and hagfishes. Mol. Biol. Evol. 15, 1706–1718. doi: 10.1093/oxfordjournals.molbev.a025897
Martin, W. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118, 9–17. doi: 10.1104/pp.118.1.9
Matari, N. H., and Blair, J. E. (2014). A multilocus timescale for oomycete evolution estimated under three distinct molecular clock models. BMC Evol. Biol. 14:101. doi: 10.1186/1471-2148-14-101
McKay, R. M., Geider, R. J., and LaRoche, J. (1997). Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to fe stress. Plant Physiol. 114, 615–622. doi: 10.1104/pp.114.2.615
McMillan, M., and Johansen, J. R. (1988). Changes in valve morphology of Thalassiosira decipiens (Bacillariophyceae) cultured in media of four different salinities. Br. Phycol. J. 23, 307–316. doi: 10.1080/00071618800650341
Morelli, E., Marangi, M. L., and Fantozzi, L. (2009). A phytochelatin-based bioassay in marine diatoms useful for the assessment of bioavailability of heavy metals released by polluted sediments. Environ. Int. 35, 532–538. doi: 10.1016/j.envint.2008.09.012
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A., and Quéguiner, B. (1995). Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 9, 359–372. doi: 10.1029/95GB01070
Nurachman, Z., Hartati, Anita, S., Anward, E. E., Novirani, G., Mangindaan, B., et al. (2012). Oil productivity of the tropical marine diatom Thalassiosira sp. Bioresour. Technol. 108, 240–244. doi: 10.1016/j.biortech.2011.12.082
Oudot-Le Secq, M. P., Grimwood, J., Shapiro, H., Armbrust, E. V., Bowler, C., and Green, B. R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277, 427–439. doi: 10.1007/s00438-006-0199-4
Park, J.-S., and Lee, J.-H. (2010). A study on the fine structure of marine diatoms in Korean coastal waters: Genus Thalassiosira 5. Algae 25, 121–131. doi: 10.4490/algae.2010.25.3.121
Parks, M., Cronn, R., and Liston, A. (2009). Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84. doi: 10.1186/1741-7007-7-84
Pérez-Cabero, M., Puchol, V., Beltrán, D., and Amorós, P. (2008). Thalassiosira pseudonana diatom as biotemplate to produce a macroporous ordered carbon-rich material. Carbon 46, 297–304. doi: 10.1016/j.carbon.2007.11.017
Ravi, V., Khurana, J. P., Tyagi, A. K., and Khurana, P. (2007). An update on chloroplast genomes. Plant Syst. Evol. 271, 101–122. doi: 10.1007/s00606-007-0608-0
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Round, F. E., Crawford, R. M., and Mann, D. G. (1990). The diatoms: biology & morphology of the genera. Q. Rev. Biol. 167, 110–116.
Roy, A. S., Woehle, C., and LaRoche, J. (2020). The transfer of the ferredoxin gene from the chloroplast to the nuclear genome is ancient within the paraphyletic genus Thalassiosira. Front. Microbiol. 11:523689. doi: 10.3389/fmicb.2020.523689
Rozas, L. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Ruck, E. C., Linard, S. R., Nakov, T., Theriot, E. C., and Alverson, A. J. (2017). Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr. Genet. 63, 499–507. doi: 10.1007/s00294-016-0652-9
Sabir, J. S., Yu, M., Ashworth, M. P., Baeshen, N. A., Baeshen, M. N., Bahieldin, A., et al. (2014). Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One 9:e107854. doi: 10.1371/journal.pone.0107854
Sar, E. A., Sunesen, I., Lavigne, A. S., and Lofeudo, S. (2011). Thalassiosira rotula, a heterotypic synonym of Thalassiosira gravida: morphological evidence. Diatom Res. 26, 109–119. doi: 10.1080/0269249X.2011.573691
Sims, P. A., Mann, D. G., and Medlin, L. K. (2019). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. doi: 10.2216/05-22.1
Sinninghe Damsté, D., Muyzer, G., Abbas, B., Rampen, S. W., Masse, G., Allard, W. G., et al. (2004). The rise of the rhizosolenoid diatoms. Science 304, 584–587. doi: 10.1126/science.1096806
Smith, S. A., and Dunn, C. W. (2008). Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716. doi: 10.1093/bioinformatics/btm619
Strzepek, R. F., and Harrison, P. J. (2004). Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. doi: 10.1038/nature02954
Suzuki, S., Yamaguchi, H., Hiraoka, M., and Kawachi, M. (2018). Mitochondrial and chloroplast genome sequences of Ulva ohnoi, a green-tide-forming macroalga in the Southern coastal regions of Japan. Mitochondrial DNA B Resour. 3, 765–767. doi: 10.1080/23802359.2018.1483778
Thompson, P. A., Guo, M. X., and Harrison, P. J. (1996). Nutritional value of diets that vary in fatty acid composition for larval Pacific oysters (Crassostrea gigas). Aquaculture 143, 379–391. doi: 10.1016/0044-8486(96)01277-X
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Turmel, M., Otis, C., and Lemieux, C. (2017). Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 7:994. doi: 10.1038/s41598-017-01144-1
Wen, F., Wu, X., Li, T., Jia, M., Liu, X., and Liao, L. (2021). The complete chloroplast genome of Stauntonia chinensis and compared analysis revealed adaptive evolution of subfamily Lardizabaloideae species in China. BMC Genomics 22:161. doi: 10.1186/s12864-021-07484-7
Whittaker, K. A. (2014). Biogeography And Nested Patterns Of Genetic Diversity In The Diatom Thalassiosira Rotula. Ph.D. Dissertations. Kingston, RI: University Of Rhode Island.
Whittaker, K. A., Rignanese, D. R., Olson, R. J., and Rynearson, T. A. (2012). Molecular subdivision of the marine diatom Thalassiosira rotula in relation to geographic distribution, genome size, and physiology. BMC Evol. Biol. 12:209. doi: 10.1186/1471-2148-12-209
Wu, F. H., Chan, M. T., Liao, D. C., Hsu, C. T., Lee, Y. W., Daniell, H., et al. (2010). Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10:68. doi: 10.1186/1471-2229-10-68
Xie, W., Yang, L. I., and Gao, Y. (2008). First record of Thalassiosira curviseriata takano (Bacillariophyta) and its bloom in the East China Sea. Acta Oceanol. Sin. 27, 124–132.
Xiong, Y., Xiong, Y., He, J., Yu, Q., Zhao, J., Lei, X., et al. (2020). The complete chloroplast genome of two important annual clover species, Trifolium alexandrinum and T. resupinatum: genome structure, comparative analyses and phylogenetic relationships with relatives in leguminosae. Plants (Basel) 9:478.
Yang, J. B., Tang, M., Li, H. T., Zhang, Z. R., and Li, D. Z. (2013). Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13:84. doi: 10.1186/1471-2148-13-84
Yang, X., Xie, D. F., Chen, J. P., Zhou, S. D., Yu, Y., and He, X. J. (2020). Comparative analysis of the complete chloroplast genomes in allium subgenus cyathophora (Amaryllidaceae): phylogenetic relationship and adaptive evolution. Biomed. Res. Int. 2020:1732586. doi: 10.1155/2020/1732586
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yu, M., Ashworth, M. P., Hajrah, N. H., Khiyami, M. A., Sabir, M. J., Alhebshi, A. M., et al. (2018). Evolution of the plastid genomes in diatoms: plastid genome evolution. Adv. Bot. Res. 85, 129–155. doi: 10.1016/bs.abr.2017.11.009
Keywords: Thalassiosira genus, chloroplast genome, comparative genomics, endosymbiotic gene transfers, super-barcodes, speciation
Citation: Liu K, Chen Y, Cui Z, Liu S, Xu Q and Chen N (2021) Comparative Analysis of Chloroplast Genomes of Thalassiosira Species. Front. Mar. Sci. 8:788307. doi: 10.3389/fmars.2021.788307
Received: 02 October 2021; Accepted: 02 December 2021;
Published: 20 December 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Raúl A. González-Pech, University of South Florida, United StatesArun Prasanna, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2021 Liu, Chen, Cui, Liu, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nansheng Chen, Y2hlbm5AcWRpby5hYy5jbg==; Y2hlbm5Ac2Z1LmNh
 Kuiyan Liu
Kuiyan Liu Yang Chen1,2,3,4
Yang Chen1,2,3,4 Zongmei Cui
Zongmei Cui Qing Xu
Qing Xu Nansheng Chen
Nansheng Chen