- 1Natural Sciences and Science Education, National Institute of Education, Nanyang Technological University, Singapore, Singapore
- 2Department of Biological Sciences, National University of Singapore, Singapore, Singapore
Giant clams are important ecosystem engineers of coral reefs because they harbor large quantities of phototrophic Symbiodiniaceae dinoflagellates of mainly genera Symbiodinium, Cladocopium, and Durusdinium. The coccoid dinoflagellates donate photosynthate and amino acids to the clam host, which in return needs to supply inorganic carbon and nitrogen to them. The host can conduct light-enhanced absorption of nitrate (NO3–), which can only be metabolized by the symbionts. This study aimed to clone nitrate transporter 2 (NRT2) from the symbionts of the fluted giant clam, Tridacna squamosa. Here, we report three major sequences of NRT2 derived from Symbiodinium (Symb-NRT2), Cladocopium (Clad-NRT2) and Durusdinium (Duru-NRT2). Phenogramic analysis and molecular characterization confirmed that these three sequences were NRT2s derived from dinoflagellates. Immunofluorescence microscopy localized NRT2 at the plasma membrane and cytoplasmic vesicles of the symbiotic dinoflagellates, indicating that it could partake in the uptake and transport of NO3–. Therefore, the transcript levels of Symb-NRT2, Clad-NRT2, and Duru-NRT2 could be used as molecular indicators to estimate the potential of NO3– transport in five organs of 13 T. squamosa individuals. The transcript levels of form II ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcII) of Symbiodinium (Symb-rbcII), Cladocopium (Clad-rbcII) and Durusdinium (Duru-rbcII) were also determined in order to calculate the transcript ratios of Symb-NRT2/Symb-rbcII, Clad-NRT2/Clad-rbcII, and Duru-NRT2/Duru-rbcII. These ratios expressed the potentials of NO3– transport with reference to the phototrophic potentials in a certain genus of coccoid dinoflagellate independent of its quantity. Results obtained indicate that Symbiodinium generally had a higher potential of NO3– transport than Cladocopium and Durusdinium at the genus level. Furthermore, some phylotypes (species) of Symbiodinium, particularly those in the colorful outer mantle, had very high Symb-NRT2/Symb-rbcII ratio (7–13), indicating that they specialized in NO3– uptake and nitrogen metabolism. Overall, our results indicate for the first time that different phylotypes of Symbiodiniaceae dinoflagellates could have dissimilar abilities to absorb and assimilate NO3–, alluding to their functional diversity at the genus and species levels.
Introduction
Giant clams (genera Tridacna and Hippopus) are members of reef ecosystems in the tropical Indo-Pacific. They generally live in symbiosis with three genera of phototrophic dinoflagellates (Symbiodinium, Cladocopium, and Durusdinium) belonging to family Symbiodiniaceae (LaJeunesse et al., 2004, 2018; Takabayashi et al., 2004; Hernawan, 2008), although giant clams of the French Polynesia may also contain Gerakladium (Pochon et al., 2019; Guibert et al., 2020). As animal-dinoflagellate associations, giant clams can flourish in nutrient-poor tropical waters where light is adequately available. The life cycle of Symbiodiniaceae dinoflagellates consists of a free-living flagellate stage and a symbiotic coccoid stage. Giant clams harbor the coccoid dinoflagellates (also called zooxanthellae) extracellularly inside a tubular system surrounded by hemolymph. These symbionts are found predominantly in the lumen of tertiary zooxanthellal tubules located in the colorful outer mantle, which can be extended beyond the edge of the shell-valve to receive irradiance needed by the symbionts for photosynthesis (Norton et al., 1992; Ip et al., 2017b). Photosynthesizing symbionts release a large portion of photosynthate to the host to support its energy and nutritional needs (Fisher et al., 1985; Klumpp et al., 1992). As a result, the host can conduct light-enhanced shell formation and grow to large sizes (Ip et al., 2017a; Rossbach et al., 2019; see Ip and Chew, 2021 for a review). In return, the host must supply the symbionts with inorganic carbon, phosphorus and nitrogen as they are separated from the ambient seawater.
Symbiodiniaceae dinoflagellates possess form II ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (Rowan et al., 1996; Mayfield et al., 2014; Poo et al., 2020, 2021), and fix inorganic carbon through C3 photosynthesis (Streamer et al., 1993). Poo et al. (2020) used the transcript level of zooxanthellae-form II RuBisCO (Zoox-rbcII), which comprised rbcII of Symbiodinium (Symb-rbcII), Cladocopium (Clad-rbcII), and Durusdinium (Duru-rbcII), as a molecular indicator to examine the phototrophic potentials of five organs (colorful outer mantle, whitish inner mantle, foot muscle, hepatopancreas and ctenidium) in the fluted giant clam, Tridacna squamosa. They reported that the outer mantle of T. squamosa had a significantly higher transcript level of Zoox-rbcII, and hence a higher phototrophic potential, than the other four organs that are located inside the mantle cavity and shaded from direct irradiance (Poo et al., 2020). Subsequently, Poo et al. (2021) made a pioneering attempt to design three sets of genus-specific primers that could differentiate Symb-rbcII, Clad-rbcII, and Duru-rbcII with the aim of estimating the relative abundances of Symbiodinium, Cladocopium, and Durusdinium harbored by T. squamosa. They reported that Durusdinium was the dominant genus of Symbiodiniaceae dinoflagellates present in individuals of T. squamosa obtained from Vietnam (Poo et al., 2021). They also examined coccoid dinoflagellates freshly isolated from the outer mantle of T. squamosa, and demonstrated that Symb-rbcII, Clad-rbcII, and Duru-rbcII exhibited different responses to light at the transcriptional level (Poo et al., 2021). Thus, they suggested that similar methods could be applied to study other genes of physiological importance in order to elucidate the functional diversity of various phylotypes of Symbiodiniaceae dinoflagellates (Poo et al., 2021).
While inorganic carbon is needed for the production of carbohydrates through photosynthesis, nitrogen is crucial for the formation of amino acids, proteins and nucleic acids. For instance, nitrogen is a vital element for chlorophyll biosynthesis in phototrophic dinoflagellates (Bernhard, 2010). For coccoid dinoflagellates, they have to synthesize amino acids not only for themselves but also for the host, which requires a large supply of amino acids for the production of muscle proteins. The clam host needs muscles to retract the extended colorful outer mantle, to close the shell valves (adductor muscle) and to generate lateral movements (foot muscle). It has been reported that the muscle of T. squamosa contains essential amino acids (Liu et al., 2019) that cannot be synthesized by the clam host (Wang and Douglas, 1999). Yet, T. squamosa can live and grow in Millipore-filtered seawater with light as the sole energy source for more than 10 months (Fitt and Trench, 1981), implying that the host can obtain all their nutrients, including essential amino acids, from its phototrophic symbionts (Klumpp and Griffiths, 1994). However, coccoid dinoflagellates are nitrogen-deficient (Wilkerson and Trench, 1986) and do not have access to the ambient seawater. Therefore, the host must absorb exogenous ammonia (Fitt et al., 1993a), urea (Chan et al., 2018, 2019), and NO3– (Ip et al., 2020) to support nitrogen metabolism in the symbionts. This is unique among aquatic animals, which generally excrete ammonia as the major nitrogenous waste, often together with a small quantity of urea (Ip and Chew, 2010; Chew and Ip, 2014).
In seawater, dissolved inorganic nitrogen is present as NO3– and NH4+, while dissolved organic nitrogen is available as urea and amino acids. The concentration of NO3– in seawater (<500 μmol N 1–1) is much higher than those of NH4+ (2 μmol N 1–1) and urea (25 μmol N 1–1) (Collos and Berges, 2003). Although the environmental toxicity of NO3– is relatively low (Westin, 1974; Tomasso and Carmichael, 1986; Jensen, 1996), nitrite (NO2–) is toxic to most aquatic animals. Hence, unlike algae and plants, aquatic animals generally absorb little NO3– from the environment because the reduction of NO3– to NO2– inside their bodies can lead to NO2– poisoning (Camargo and Alonso, 2006). Instead, aquatic animals excrete small quantities of endogenous NO3– in order to avoid NO3– reduction and the resulting NO2– toxicity. By contrast, T. squamosa absorbs NO3– from the external seawater, and the rate of NO3– absorption is augmented by illumination (Ip et al., 2020). The ctenidium (gill) of T. squamosa expresses a homolog of SIALIN, which functions as an electrogenic H+:2NO3– cotransporter. SIALIN is localized at the apical membrane of the epithelial cells near the tips of ctenidial filaments to absorb exogenous NO3–. Furthermore, illumination leads to significant increases in the transcript level of SIALIN and the protein abundance of SIALIN in the ctenidium, indicating that it can play a role in light-enhanced NO3– absorption. As the clam host cannot assimilate NO3–, the NO3– absorbed through the ctenidium must be dedicated to the symbionts. Indeed, the addition of NO3– to the ambient seawater can increase the growth rate of the host and the density of its symbionts (Fitt et al., 1993b). Hence, coccoid dinoflagellates must be able to absorb NO3– from the luminal fluid of the zooxanthellal tubules through the plasma membrane. As NO3– is an anion that cannot permeate the hydrophobic phospholipid bilayer freely, its transport through the symbiont’s plasma membrane must involve certain types of membrane transporters or channels.
In algae and plants, the transport of NO3– across plasma membranes involves two types of nitrate transporters (NRTs), NRT1s and NRT2s (Dagenais-Bellefeuille and Morse, 2013, 2016), which co-transport NO3– and H+. NRT1s belong to the NRT1/peptide transporter (NPF) family, while NRT2s are members of the nitrate/nitrite porter family (NNP). Both NNP and NPF families are grouped under the major facilitator superfamily (MFS). NRT1s and NRT2s correspond to the earlier defined physiological categories of low- (Léran et al., 2014) and high-affinity (Orsel et al., 2002; Krapp et al., 2014) NO3– transporters, respectively (Crawford and Glass, 1998; Forde, 2000). The high-affinity systems typically operate in the range of 10 – 250 μM NO3–, while the low-affinity systems only become functional important above these concentrations. Free-living dinoflagellates can absorb NO3– from the ambient seawater (Paasche et al., 1984; Fan and Glibert, 2005; Leong et al., 2010), and they are known to express NRT2 (Dagenais-Bellefeuille and Morse, 2016; Pechkovskaya et al., 2020).
As the NO3– concentration in the extracellular fluid of the clam host needs to be low to avoid NO3– reduction leading to NO2– toxicity, it is logical to hypothesize that coccoid Symbiodiniaceae dinoflagellates would possess some types of high-affinity type NO3– transporter. Therefore, this study was undertaken to clone and sequence NRT2 from the symbionts residing in the outer mantle of T. squamosa. Due to the presence of various phylotypes (species) of Symbiodinium, Cladocopium, and Durusdinium, we had obtained multiple NRT2 sequences. However, only one major cDNA coding sequence of NRT2 for each genus of dinoflagellate was presented in this report. These three major sequences were named Symbiodinium-NRT2 (Symb-NRT2), Cladocopium-NRT2 (Clad-NRT2), and Durusdinium-NRT2 (Duru-NRT2). Their identities as NRT2 and origins from dinoflagellates were confirmed through molecular characterization and phenogramic analysis. An antibody that could bind comprehensively with NRT2 derived from all three genera of dinoflagellates, named zooxanthellae-NRT2 (Zoox-NRT2), was custom-made to confirm the localization of Zoox-NRT2 at the plasma membrane by immunofluorescence microscopy. In addition, we made a pioneering attempt to use the transcript levels of Symb-NRT2, Clad-NRT2, and Duru-NRT2 as molecular indicators to estimate the potential of NO3– transport in phylotypes of Symbiodinium, Cladocopium, and Durusdinium. Three sets of genus-specific quantitative real-time polymerase chain reaction (qPCR) primers were designed to determine the transcript levels of Symb-NRT2, Clad-NRT2, and Duru-NRT2 in five organs (colorful outer mantle, whitish inner mantle, foot muscle, ctenidium, and hepatopancreas) of T. squamosa. However, the transcript levels of these three NRT2s could vary considerably among individuals of T. squamosa as they naturally harbor different quantities and proportions of dinoflagellate phylotypes in various organs. To resolve this problem, we also determined the transcript levels of Symb-rbcII, Clad-rbcII, and Duru-rbcII based on the genus-specific qPCR primers designed by Poo et al. (2021). The aim was to calculate the ratios of Symb-NRT2/Symb-rbcII, Clad-NRT2/Clad-rbcII, and Duru-NRT2/Duru-rbcII for a specific organ of each T. squamosa individual, as these ratios could provide information on the potential of NO3– transport with reference to the phototrophic potential for each genus of dinoflagellate independent of its quantity in the tissue sample.
Giant clams are important ecosystem engineers of coral reefs because they harbor large quantities of coccoid Symbiodiniaceae dinoflagellates. They can expel intact and viable dinoflagellates that can repopulate bleached Symbiodiniaceae-bearing hosts including scleractinian corals (Morishima et al., 2019; Umeki et al., 2020). While giant clams are known to harbor multiple phylotypes of Symbiodiniaceae dinoflagellates, the physiological reasons behind it remain enigmatic. Results obtained from this study were expected to furnish novel information on whether different phylotypes of Symbiodinium, Cladocopium, and Durusdinium would have disparate potentials of NO3– transport, and hence different abilities to use NO3– as a substrate to produce essential nitrogenous compounds for themselves and the host. Such information may shed light on the divergent physiological roles of Symbiodiniaceae dinoflagellates at the genus or even the phylotype (species) level, and provide insights into their distinct contributions to the physiological needs of the giant clam-dinoflagellate holobiont.
Materials and Methods
Giant Clam and Maintenance
Sixteen T. squamosa weighing 550 ± 150 g were imported directly from Vietnam through Xanh Tuoi Tropical Fish Co. Ltd. On arrival, the specimens were distributed into three tanks, each with a dimension of 92 cm (L) by 62 cm (W) by 62 cm (H) containing approximately 320 l of seawater at Salinity 30–32 and 26°C. Artificial seawater was prepared with Red Sea salt (Red Sea, Houston, TX, United States). The salinity and temperature of the seawater were monitored using a Pro30 conductivity meter (YSI Incorporated, Yellow Springs, OH, United States). The pH of the seawater was maintained at 8.2–8.4; the hardness at 143–179 ppm; the calcium content at 380–420 ppm; the phosphate content at <0.28 ppm; the total ammonia and NO3– contents at 0 ppm. Each tank was illuminated with two sets of four feet Aquazonic T5 lighting systems, and each system consisted of two white light tubes and two actinic blue light tubes. The underwater light intensity (photosynthetic photon flux density; PPFD) reaching the clams was ∼115 − 125 μmol photons m–2 s–1 (400−700 nm) as determined by a SKP 215 PAR Quantum sensor connected to the SKP 200 display meter (Skye Instruments Ltd, United Kingdom). This level of irradiance mimicked the light intensity received by T. squamosa in its natural habitat at a depth of ∼20 m (Jantzen et al., 2008). No food was supplied to the giant clams during the 1 month of acclimatization under a 12 h: 12 h dark: light regimen. Approval on the use of giant clams in this study was exempted by the Nanyang Technological University Institutional Animal Care and Use Committee.
Exposure to Light Conditions and Collection of Tissues
After 1 month of acclimatization, individuals of T. squamosa (n = 13) that had been exposed to light for 3 h were sampled randomly from the three tanks. Three hours of light exposure is chosen as giant clams are known to display light-enhanced phenomena, and the transcript levels of many transporters and enzymes could be enhanced after 3 – 6 h of illumination (see Ip and Chew, 2021 for a review). To minimize stress on the clams, they were anesthetized in 0.2% phenoxyethanol prior to tissue sampling. The shell valves were forced open to sever the adductor muscle. Samples of the outer mantle, inner mantle, foot muscle, hepatopancreas, and ctenidium (gill) were excised. Excised tissue samples were blotted dry, freeze-clamped in liquid nitrogen, and stored at −80°C until further processing. Separately, tissue samples of the outer mantle were collected from three other individuals of T. squamosa for immunofluorescence microscopy (n = 3). Excised outer mantle tissues were fixed in 3.7% paraformaldehyde prepared with seawater for 18 h at 4°C.
Total RNA Extraction and cDNA Synthesis
The total RNA of a tissue sample was extracted using TRI Reagent™ (Sigma-Aldrich Co., St Louis, MO, United States), and purified with a PureLink™ RNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, United States). The concentration of the purified RNA was determined using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies Inc., Wilmington, DE, United States), and the RNA integrity was checked by agarose gel electrophoresis. A RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific) was used to convert the purified RNA into cDNA.
Polymerase Chain Reaction, Cloning, and Rapid Amplification of cDNA Ends
The partial sequences of NRT2 from Symbiodinium, Cladocopium, and Durusdinium were obtained using a set of genus-comprehensive PCR primers (Forward: 5′-GCACT GTTCAGCAGAATCC-3′; Reverse: 5′-GGCTGTGAGTTGTC CACCA-3′) designed at the homologous regions of nine NRT2 sequences obtained from various dinoflagellate databases (Supplementary Table S1). The PCR reaction was performed using a 9902 Veriti 96-well thermal cycler (Thermo Fisher Scientific) with the cycling conditions: 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 1.5 min and a final extension at 72°C for 10 min. The pGEM®-T Easy Vector system II (Promega, Madison, WI, United States) was used to clone the PCR products obtained. Sixty clones were picked randomly and sequenced. The partial sequences obtained were identified by comparing with NRT sequences available in multiple dinoflagellate databases. A major sequence of NRT2 was identified for each genus of Symbiodinium, Cladocopium, and Durusdinium. To obtain the full coding sequence of the major Symb-NRT2, Clad-NRT2 and Duru-NRT2, 5′ and 3′ Rapid Amplification of cDNA Ends (RACE)-PCR were performed using the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Mountain View, CA, United States). Three sets of RACE primers were designed specifically for Symb-NRT2 (5′ RACE: 5′-AGCTGGAACTGCTGCACAGTCCGTGA-3′, 3′ RACE: 5′-TGTCATCACGGACTGTGCAGCAGTTCCA-3′), Clad-NRT2 (5′ RACE: 5′-GCAGTCCCACTCGCTAAAATGTCC-3′; 3′ RACE: 5′-ACTGCAATCCCGTGCCACAGGACATT-3′) and Duru-NRT2 (5′ RACE: 5′-GTGCAGTAGCATTGTT GGCGATATCGG-3′; 3′ RACE: 5′-TTGAAGTACAAGAA CATTTCCACGACGG-3′). The full coding sequences of Symb-NRT2, Clad-NRT2, and Duru-NRT2 were deposited into Genbank.
Amino Acid Sequences and Phenogramic Analysis
The ExPASy Proteomic server1 was used to deduce the amino acid sequences of Symb-NRT2, Clad-NRT2, and Duru-NRT2 from their respective nucleotide sequences. TMpred provided by Expasy2, was used to identify the transmembrane regions and pore lining amino acid residues. The identities of Symb-NRT2, Clad-NRT2 and Duru-NRT2 were confirmed by conducting a phenogramic analysis together with NRT2 sequences obtained from various databases. The phenogram was generated using Maximum Likelihood analysis using the program RaxML 8.2.5 (Stamatakis, 2014) with 2000 bootstraps. Using ModelGenerator v0.85 (Keane et al., 2006), the best-fitting evolutionary model for NRT2 was determined to be WAG + G + F (Whelan and Goldman, 2001).
Determination of the Transcript Levels by qPCR
Three sets of genus-specific qPCR primers, one each for Symb-NRT2 (forward: 5′-TGAAGACAGGTCTGGAGTA-3′; reverse: 5′-CGCATATGGGCTCTTCT-3′), Clad-NRT2 (forward: 5′-AGAATGATGATACCAATCCCAC-3′; reverse: 5′-CAAA CACAGTCCGCCAG-3′), and Duru-NRT2 (forward: 5′-GAAGTACAAGAACATTTCCACGAC-3′; reverse: 5′-AAAC GCACTTGGACAGCAC-3′) were designed by aligning Symb-NRT2, Clad-NRT2, and Duru-NRT2 with nine NRT2 sequences selected from various dinoflagellate databases (Supplementary Table S2). In order to verify the specificity of the designed Symb-NRT2 primer, Clad-NRT2 primer, and Duru-NRT2 primer, efforts were made to generate three different plasmid clones, each of which contained the insert of the amplicon region of Symb-NRT2, Clad-NRT2, or Duru-NRT2 following the method of Hiong et al. (2017). Then, qPCR was performed using these three plasmid clones as substrates to confirm that each set of genus-specific primers would only react with the plasmid containing the corresponding insert. The amplification efficiencies of the qPCR primers for Symb-NRT2, Clad-NRT2, and Duru-NRT2 were 102.4, 92.9, and 101.9%, respectively. Genus-specific qPCR primers designed by Poo et al. (2021) were adopted to quantify the transcript levels of Symb-rbcII, Clad-rbcII, and Duru-rbcII from T. squamosa. The amplification efficiencies of the primer set for Symb-rbcII, Clad-rbcII, and Duru-rbcII were 95.1, 95.1, and 112.0%, respectively.
qPCR was performed using a 96-well StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific). Each reaction, in a total volume of 10 μl, consisted of 5 μl of qPCRBIO SyGreen Mix Hi-ROX (PCR Biosystems Inc., Wayne, PA, United States), 0.3 μl of forward primer (10 μmol l–1), 0.3 μl of reverse primer (10 μmol l–1), and an appropriate amount of cDNA. The qPCR cycling conditions included a 20 s denaturation and enzyme activation at 95°C, followed with 40 cycles of 95°C for 3 s and a specific temperature for a certain gene for 30 s. The specific temperature that lasted 30 s for Symb-rbcII, Clad-rbcII, and Duru-rbcII were 58, 56, and 55°C, respectively. For Symb-NRT2, Clad-NRT2, and Duru-NRT2, the respective temperature used in the 40 cycles were 57, 60, and 60°C. The dissociation curve obtained after each run was analyzed to verify the homogeneity of the PCR product and the specificity of the PCR reaction. Three standard curves were constructed using the three different plasmid clones as standards for Symb-rbcII, Clad-rbcII, and Duru-rbcII. The transcript levels Symb-rbcII, Clad-rbcII, and Duru-rbcII in a sample were calculated based on these three standard curves and expressed as copies of transcripts per ng of total RNA.
Antibodies
A genus-comprehensive anti-Zoox-NRT2 antibody was custom-made by Genscript (Piscataway, NJ, United States) based on the epitope sequence of MADFKLKVDESNKA, which was selected from a highly conserved region of six NRT2 sequences retrieved from various dinoflagellate databases (Supplementary Table S3). This epitope sequence, corresponding to residues 1−14 of Symb-NRT2, Clad-NRT2, and Duru-NRT2 with similarity of 100, 71.4, and 78.6%, respectively. Thus, anti-Zoox-NRT2 could possibly bind with NRT2s of all phylotypes of Symbiodinium, Cladocopium, and Durusdinium.
Immunofluorescence Microscopy
The fixed outer mantle sample was dehydrated in ethanol and cleared using HistoChoice Clearing Agent (Sigma-Aldrich Co.) before embedding in Paraplast Plus (Sigma-Aldrich Co.). Sections of 5 μm was prepared using a Leica RM2125 RTS microtome (Leica, Wetzlar, Germany) and mounted on Menzel Gläser SuperFrost Plus Adhesion slides (Thermo Fisher Scientific). The deparaffinized section was treated with citraconic anhydrase (Nacalai Tesque, Kyoto, Japan) at 95°C for 5 min, followed with 1% SDS solution at 25°C for 10 min for the retrieval of antigen. The section was then washed with TPBS containing 0.2% Triton-X, 10 mmol l–1 Na2HPO4, 1.8 mmol l–1 KH2PO4, 137 mmol l–1 NaCl, and 1.8 mmol l–1 KCl at pH 7.4. To reduce autofluorescence, the section was treated with 0.1% Sudan Black B (Sigma-Aldrich Co.) in 70% ethanol for 10 min and washed three times with TPBS. Blocking was performed with 1% bovine serum albumin in TPBS at 25°C for 1 h. Thereafter, the section was incubated with the anti-Zoox-NRT2 antibody (2.5 μg ml–1 diluted with Signal Enhancer HIKARI Solution A obtained from Nacalai Tesque) at 25°C for 1 h and rinsed three times with TPBS. The section was incubated with 2.5 μg ml–1 of fluorochrome-coupled goat anti-rabbit gamma globulin diluted with Signal Enhancer HIKARI Solution A (Alexa Fluor 488; Thermo Fisher Scientific) for 1 h at 25°C. Finally, they were mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific Inc.) for microscopy.
The mounted section was examined under a fluorescence microscope (Olympus BX43F; Olympus Corporation, Tokyo, Japan) and the images were acquired using an Olympus DP80 camera and the cellSens Imaging software (Olympus). Differential interference contrast microscopy (DIC) was applied to examine tissue structures and orientation. The red autofluorescence of the plastids of dinoflagellates was examined using the U-MWIG Interference Green Fluorescence Filter with an excitation wavelength of 520–550 nm. The green fluorescence resulting from the staining by the primary antibody and Alexa Fluor 488 was acquired using the Olympus U-WNIBA Blue Fluorescence Filter with an excitation wavelength of 470–490 nm. Overlaying of the images and adjustment of brightness were performed using Adobe Photoshop CC (Adobe Systems, CA, United States).
Data Analysis
Values were reported as means ± SEM unless otherwise stated. The non-parametric Friedman test of differences followed by the Wilcoxon Signed-Rank Test were applied for data expressed as percentages or ratios in Tables 4–8 and Figure 4. Differences obtained among means were considered statistically significant with p-values < 0.017 after Bonferroni adjustment. For Table 3, One-way Analysis of Variance (ANOVA) was used for comparison among the means of the transcript levels of the genus-specific NRT2 or those of genus-specific rbcII in a particular organ while Levene’s test was used to assess the homogeneity of the variance. This was followed by Dunnett’s T3 test as the variance was assumed not to be equal. On the other hand, the paired t-test was used to compare the means between the genus-specific NRT2 and the corresponding genus-specific rbcII in a particular organ in Table 3. The differences between the two means were regarded as significant when the p-value was <0.05. All comparisons were performed with the use of SPSS Statistics software v26 (IBM Corporation, Armonk, NY, United States).
Results
Nucleotide Sequences, Translated Amino Acid Sequences, and Phenogramic Analysis
The complete cDNA coding sequences of Symb-NRT2, Clad NRT2, and Duru-NRT2 obtained from the outer mantle of T. squamosa comprised 1614, 1659, and 1635 bp, respectively. They have been deposited into GenBank with the respective accession numbers of MZ014639, MZ014640, and MZ014641. A comparison of these three NRT2 sequences with NRT2 contigs from multiple dinoflagellate databases revealed that Symb-NRT2 and Clad-NRT2 had the highest similarity (98.6%) to the NRT2 sequence of Symbiodinium tridacnidorum (ITS2 type A3; Shoguchi et al., 2018; 98.6%) and the NRT2 sequence of Cladocopium goreaui (ITS2 type C1; Davies et al., 2018; 99.5%), respectively (Table 1). For Duru-NRT2, it had the highest similarity to the NRT2 sequence of Durusdinium trenchii (ITS2 type D1a; Bellantuono et al., 2019; 99.3%; Table 1).
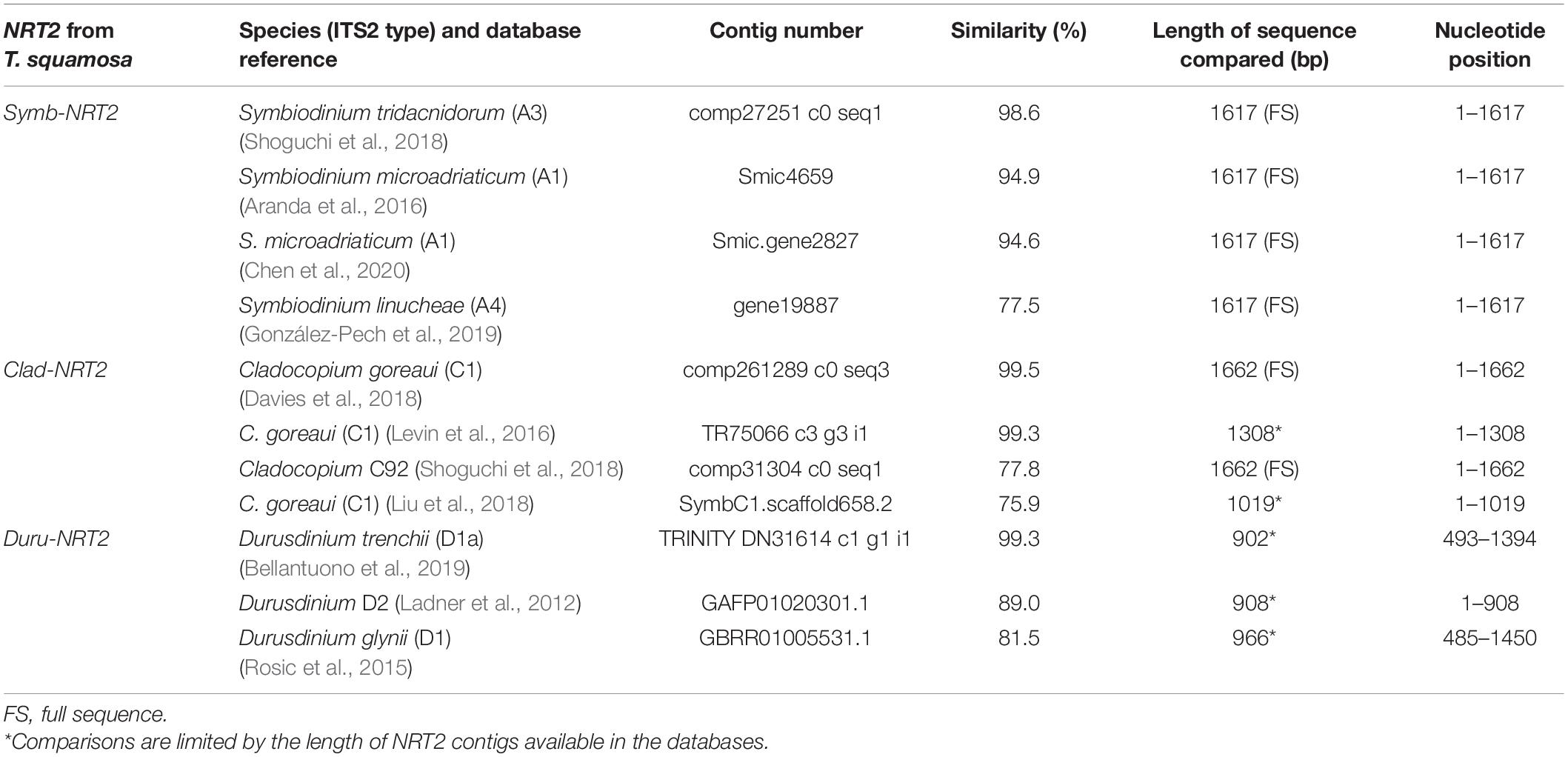
Table 1. A comparison of the nucleotide sequence of nitrate transporter 2 (NRT2) derived from Symbiodinium (Symb-NRT2), Cladocopium (Clad-NRT2), or Durusdinium (Duru-NRT2) of Tridacna squamosa with selected NRT2 contigs obtained from various symbiotic dinoflagellate databases, with information on the species/ITS2 type, database reference, contig number and the length of the sequence for comparison of selected NRT2 contigs.
The deduced amino acid sequences of Symb-NRT2, Clad- NRT2 and Duru-NRT2 contained 538 (∼58.5 kDa), 553 (∼60.2 kDa), and 545 (∼59.4 kDa) residues, respectively. A multiple alignment of Symb-NRT2, Clad-NRT2, and Duru-NRT2 with other NRT sequences from Genbank demonstrated that they consisted of 12 predicted transmembrane regions (TMs), with an intracellular N-terminus and an intracellular C-terminus (TM 1−TM 12; Figure 1). They contained a conserved MFS motif (Forde, 2000) between TM 2 and TM 3 (G-x-x-x-D/N-R/K-x-G-R-R/K) (corresponding to amino acid residues 169–178 in Symb-NRT2, 166–175 in Clad-NRT2, and 170–179 in Duru-NRT2). Two nitrate signature motifs (Trueman et al., 1996; Unkles et al., 2004a,2012) were present in Symb-NRT2 (residues 229–249 and residues 456–476), Clad-NRT2 (residues 226–246 and residues 453–473), and Duru-NRT2 (residues 230–250 and residues 457–477). In addition, the putative substrate-binding site that could form hydrogen bonds with NO3– (Yan et al., 2013) was also present in Symb-NRT2, Clad-NRT2, and Duru-NRT2. This binding site consisted of two positively charged amino acid residues (corresponding to Arg-165 and Arg-382 in Symb-NRT2, Arg-162 and Arg-379 in Clad-NRT2, and Arg-166 and Arg-383 in Duru-NRT2) and two polar residues (corresponding to Asn-246 and Tyr-338 in Symb-NRT2, Asn-243 and Tyr-335 in Clad-NRT2, and Asn-247 and Tyr-339 in Duru-NRT2). The Glu residue involved in the symport of H+ and NO3– (Akhtar et al., 2015; Jacquot et al., 2017) was conserved (corresponding to Glu-345 in Symb-NRT2, Glu-342 in Clad-NRT2, and Glu-346 in Duru-NRT2), implying that Symb-NRT2, Clad-NRT2, and Duru-NRT2 could act as H+-dependent symporters.

Figure 1. Molecular characterization of the deduced amino acid sequences of nitrate transporter 2 from Symbiodinium (Symb-NRT2), Cladocopium (Clad-NRT2), and Durusdinium (Duru-NRT2) obtained from the outer mantle of Tridacna squamosa. Multiple alignment of Symb-NRT2 (MZ014639), Clad-NRT2 (MZ014640), and Duru-NRT2 (MZ014641) with Symbiodinium microadriaticum NRT2.5 (OLQ11495.1), Chlamydomonas reinhardtii NRT (XP_001694496.1), Aspergillus nidulans NrtA (AAA76713.1), and Arabidopsis thaliana NRT2.5 (NP_172754.1). Shaded residues indicate identical or similar amino acid residues. Asterisks denote identical amino acid residues, colons denote strongly similar amino acids, and periods denote weakly similar amino acids. The twelve predicted transmembrane regions (TM 1–TM 12) underlined in black were predicted using TMpred provided by ExPASy Bioinformatics Resource portal. Black boxes denote substrate-binding residues. Hydrophobic residues and polar residues that make up the gate at the substrate binding site are indicated with black triangles and black circles, respectively. The two nitrate signature regions, NS1 and NS2, are marked with red and blue boxes, respectively: [FYK]-X3-[ILQRK]-X-[GA]-X-[VASK]-X-[GASN]-[LIVFQ]-X1,2-G-X-G-[NIM]-X-G-[GVTA].
The identities of Symb-NRT2, Clad-NRT2, and Duru-NRT2 were supported by phenogramic analysis (Figure 2), which grouped them with NRT2 sequences of algae, plants, and dinoflagellates, but distinct from NRT1. Notably, they were clustered with other NRT2 sequences from the same genus of dinoflagellate.
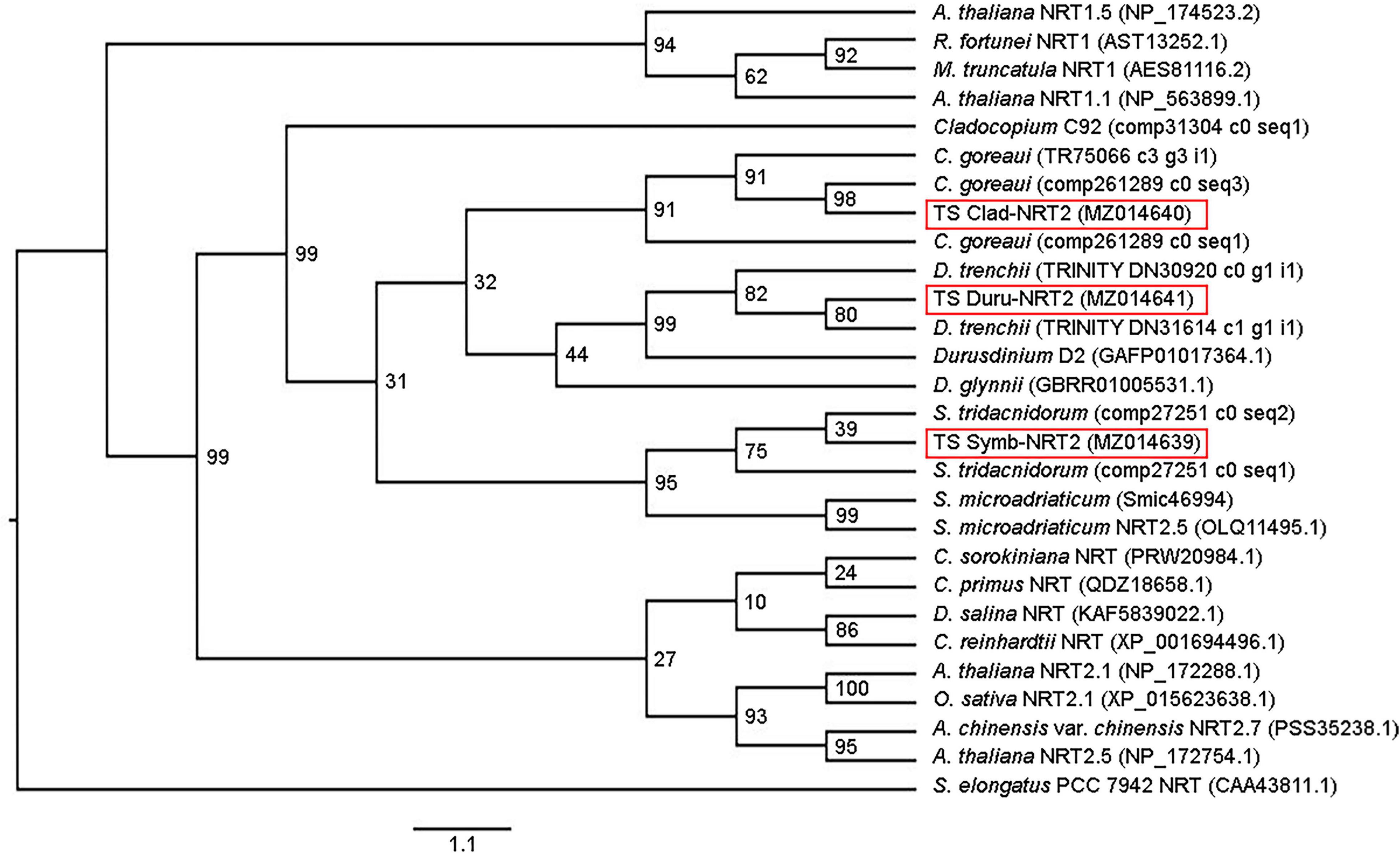
Figure 2. Phenogramic analysis of nitrate transporter 2 from Symbiodinium (Symb-NRT2), Cladocopium (Clad-NRT2), and Durusdinium (Duru-NRT2) from the outer mantle of Tridacna squamosa. Numbers at each branch point represent bootstrap values from 2000 replicates. NRT from Synechococcus elongatus is used as the outgroup. Amino acid sequences of nitrate transporters from algae (Chlamydomonas reinhardtii, Chlorella sorokiniana, Chloropicon primus, Cladocopium C92, C. goreaui, Dunaliella salina, Durusdinium D2, D. glynnii, D. trenchii, Symbiodinium tridacnidorum, and S. microadriaticum) and higher plants (Actinidia chinensis var. chinensis, Arabidopsis thaliana, Medicago truncatula, Oryza sativa, and Rhododendron fortunei) were obtained from Genbank or various dinoflagellate databases with their accession or contig numbers given in parentheses.
Localization of NRT2 by Immunofluorescence Microscopy
NRT2 immuno-labeling was detected at the plasma membrane of the coccoid dinoflagellates in the outer mantle of T. squamosa, in support of a possible role in transporting NO3– across the plasma membrane (Figure 3). NRT2 immunofluorescence was also detected in certain intracellular vesicles inside the cytoplasm of the coccoid dinoflagellates.
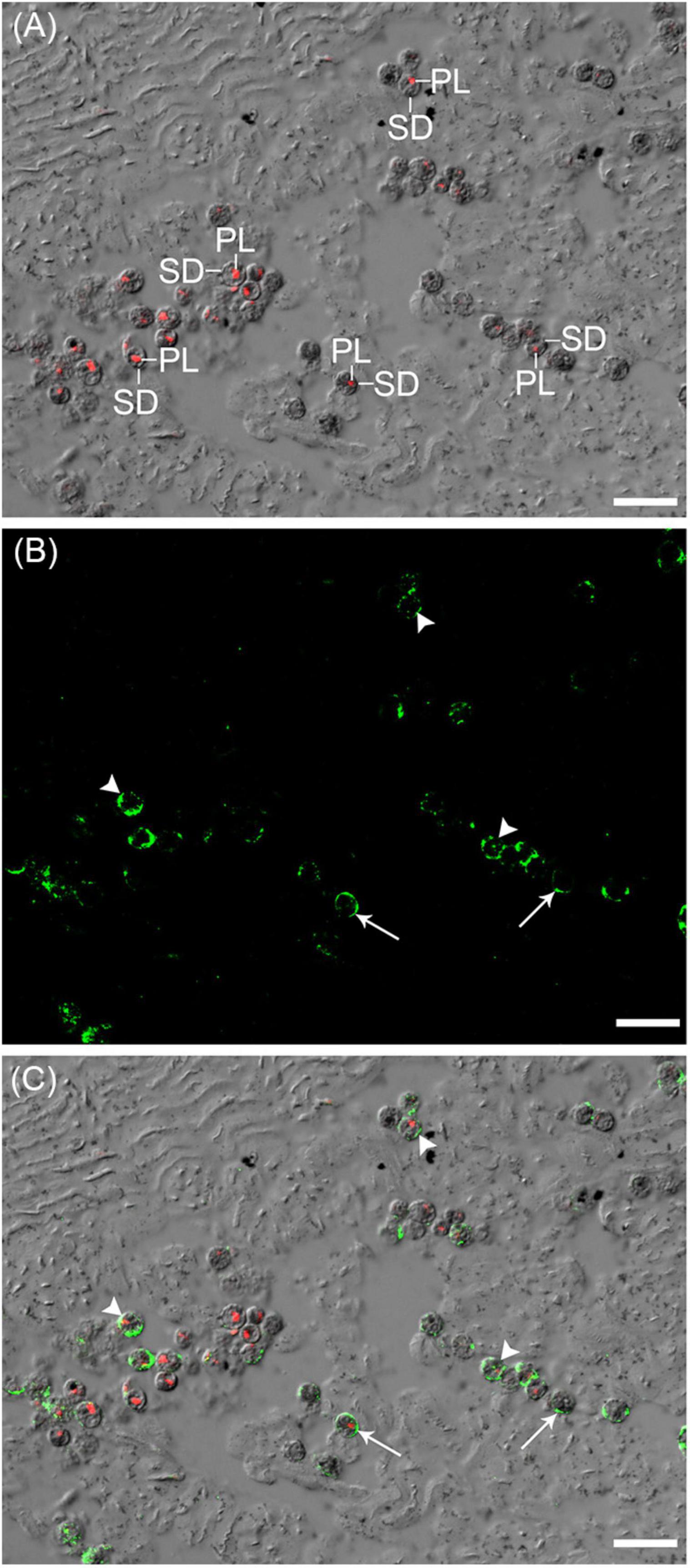
Figure 3. Immunolabeling of zooxanthellae-nitrate transporter 2 (Zoox-NRT2), which comprised NRT2 of Symbiodinium, Cladocopium and Durusdinium, in the outer mantle of Tridacna squamosa. (A) The differential interference contrast (DIC) image is overlaid with the red channel showing autofluorescence of the plastids (PL) of symbiotic dinoflagellates (SD). (B) The green channel showing green immunofluorescence of Zoox-NRT2 labeled with the anti-Zoox-NRT2 antibody. (C) The DIC image overlaid with the red channel and green channel. Arrows indicate Zoox-NRT2 immunolabeling (green) of the plasma membrane of SD. Arrowheads indicate Zoox-NRT2 immunolabeling (green) of intracellular vesicles in SD. Scale bar: 20 μm. Replicable results were obtained from three individuals of T. squamosa.
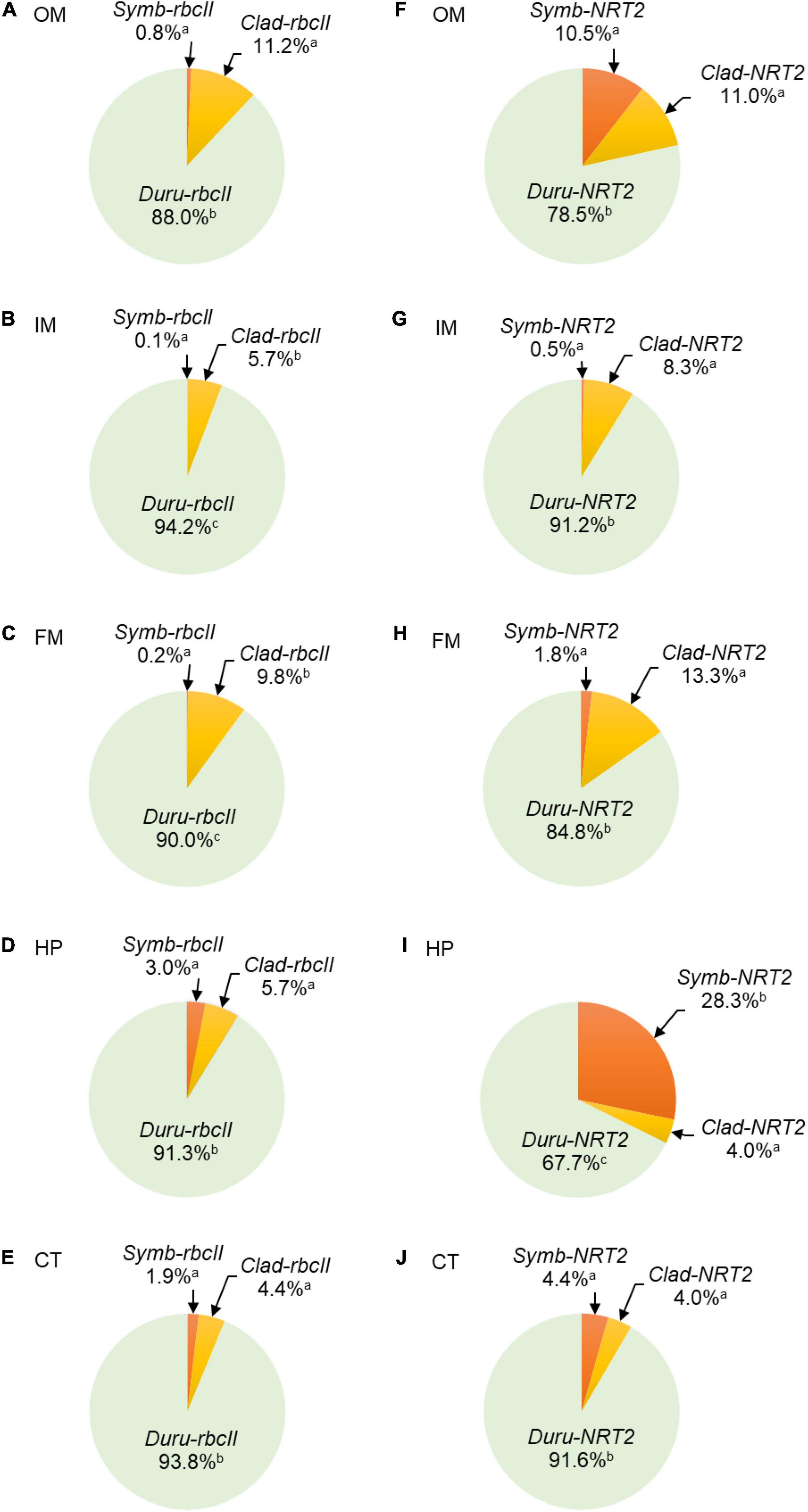
Figure 4. Proportion of transcript levels of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) or transcript levels of nitrate transporter 2 (NRT2) derived from Symbiodinium (Symb-rbcII and Symb-NRT2, respectively), Cladocopium (Clad-rbcII and Clad-NRT2, respectively) and Durusdinium (Duru-rbcII and Duru-NRT2, respectively) in five organs of Tridacna squamosa. Results (n = 13) are expressed as mean percentages in (A,F) the outer mantle (OM), (B,G) the inner mantle (IM), (C,H) the foot muscle (FM), (D,I) the hepatopancreas (HP), and (E,J) the ctenidium (CT). Means that are significantly different from each other are labeled with different letters (p-value < 0.017 after Bonferroni adjustment).
Genus-Specificity of the Three Sets of qPCR Primers
The genus-specificity of the primer sets designed for Symb-NRT2, Clad-NRT2 and Duru-NRT2 were validated with three plasmid clones, each containing the insert of one of the three targeted amplicons. Indeed, a specific set of primer reacted positively only with its related plasmid clone (Table 2). Poo et al. (2021) had verified the genus-specificity of the primer sets designed for Symb-rbcII, Clad-rbcII, and Duru-rbcII previously.
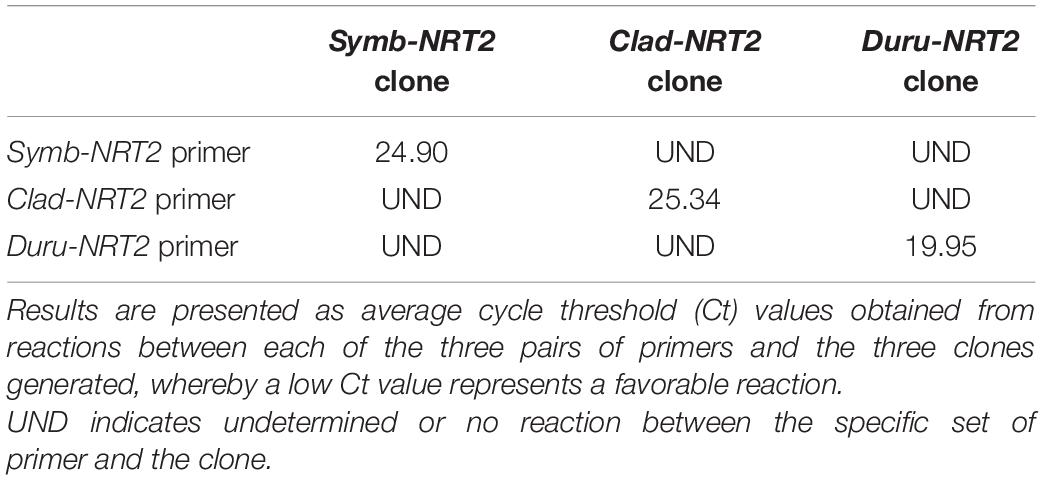
Table 2. Specificity of the three sets of quantitative real-time PCR (qPCR) primers designed for nitrate transporter 2 (NRT2) of Symbiodinium (Symb-NRT2), Cladocopium (Clad-NRT2) and Durusdinium (Duru-NRT2) derived from the outer mantle of Tridacna squamosa, as demonstrated by qPCR using three different plasmid clones with each clone containing specifically the insert of the amplicon region of the Symb-NRT2, Clad-NRT2, or Duru-NRT2 primers.
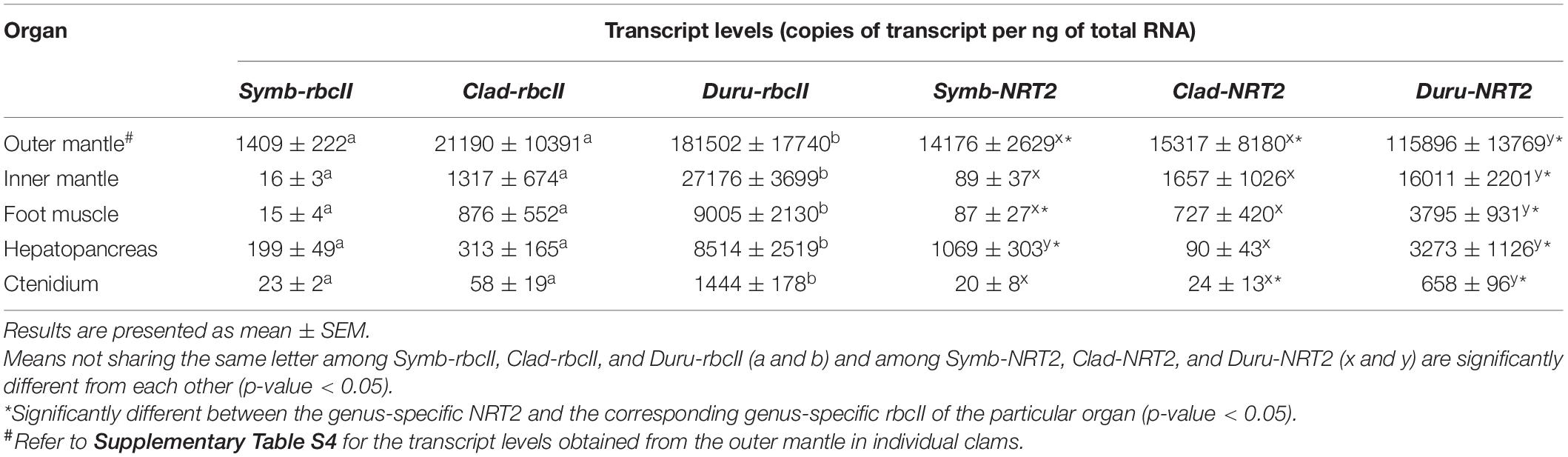
Table 3. Transcript levels (copies of transcript per ng of total RNA) of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) and nitrate transporter 2 (NRT2) derived from Symbiodinium (Symb-rbcII and Symb-NRT2, respectively), Cladocopium (Clad-rbcII and Clad-NRT2, respectively) and Durusdinium (Duru-rbcII and Duru-NRT2, respectively) in the outer mantle, inner mantle, foot muscle, hepatopancreas, and ctenidium (n = 13) of Tridacna squamosa.
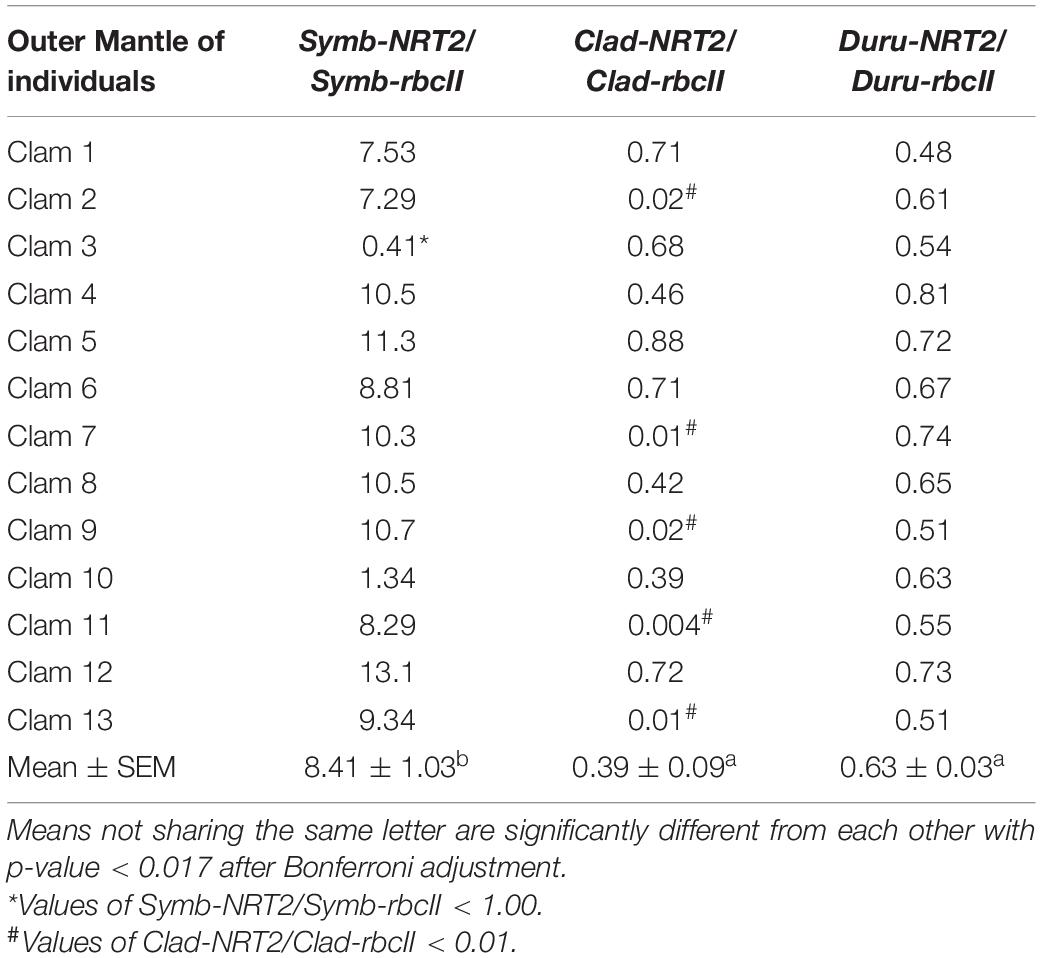
Table 4. Ratios of the transcript levels of nitrate transporter 2 (NRT2) to that of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) from Symbiodinium (Symb-NRT2/Symb-rbcII), Cladocopium (Clad-NRT2/Clad-rbcII), and Durusdinium (Duru-NRT2/Duru-rbcII) obtained from the outer mantle of Tridacna squamosa (n = 13).
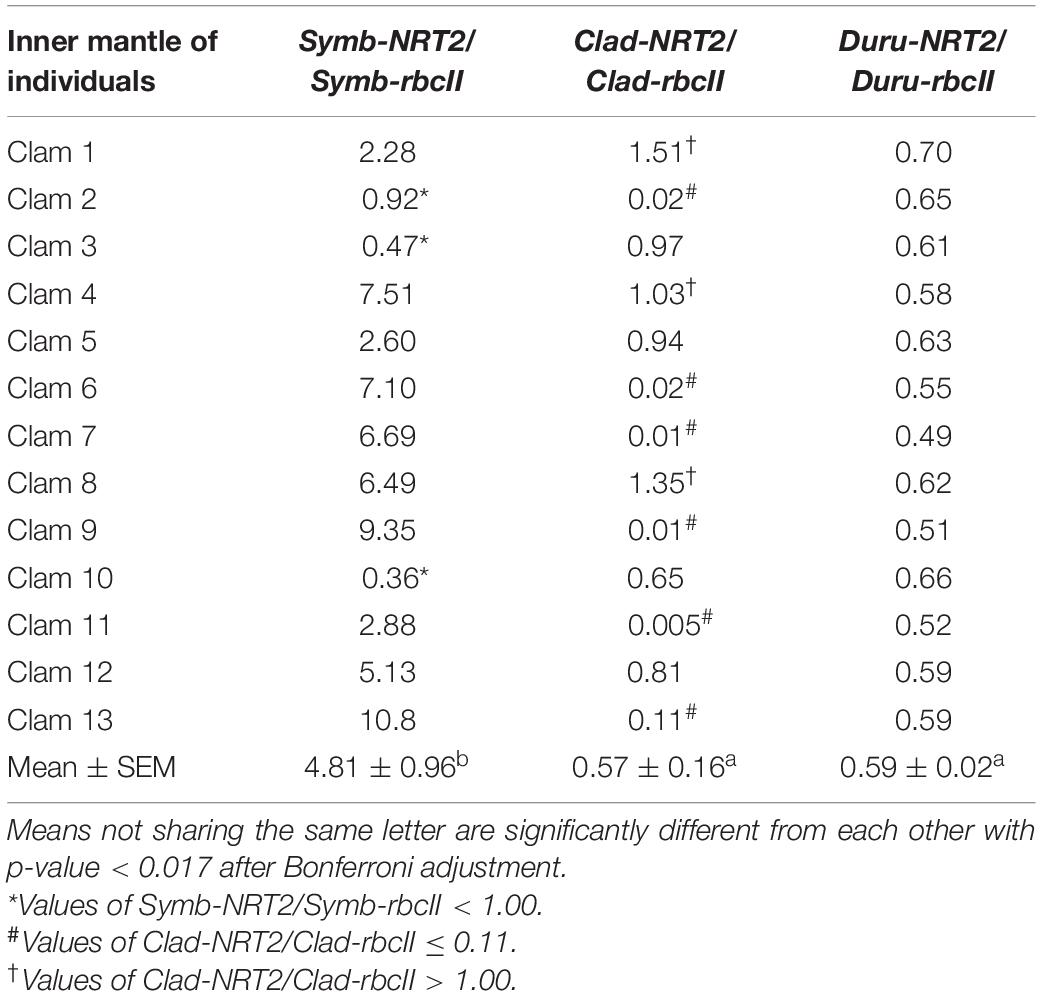
Table 5. Ratios of the transcript levels of nitrate transporter 2 (NRT2) to that of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) from Symbiodinium (Symb-NRT2/Symb-rbcII), Cladocopium (Clad-NRT2/Clad-rbcII), and Durusdinium (Duru-NRT2/Duru-rbcII) obtained from the inner mantle of Tridacna squamosa (n = 13).
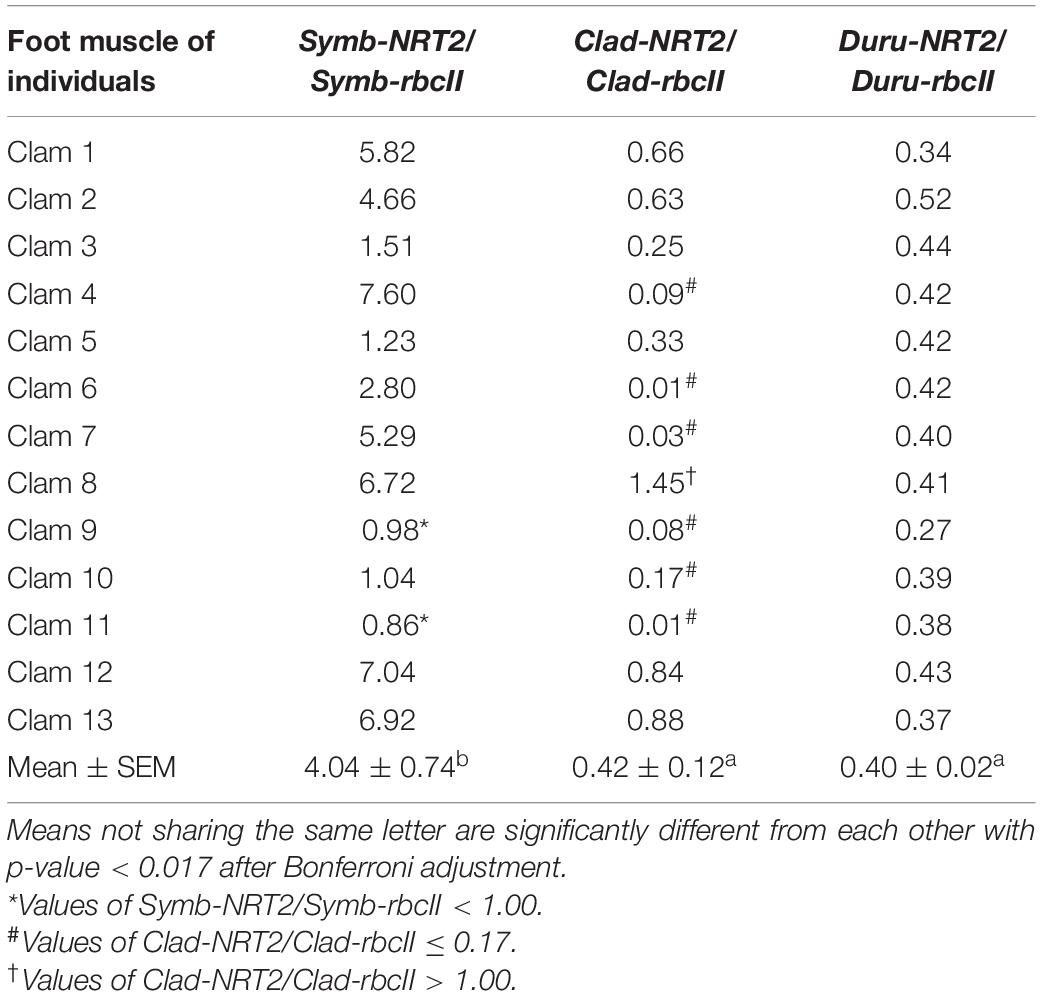
Table 6. Ratios of the transcript levels of nitrate transporter 2 (NRT2) to that of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) from Symbiodinium (Symb-NRT2/Symb-rbcII), Cladocopium (Clad-NRT2/Clad-rbcII), and Durusdinium (Duru-NRT2/Duru-rbcII) obtained from the foot muscle of Tridacna squamosa (n = 13).
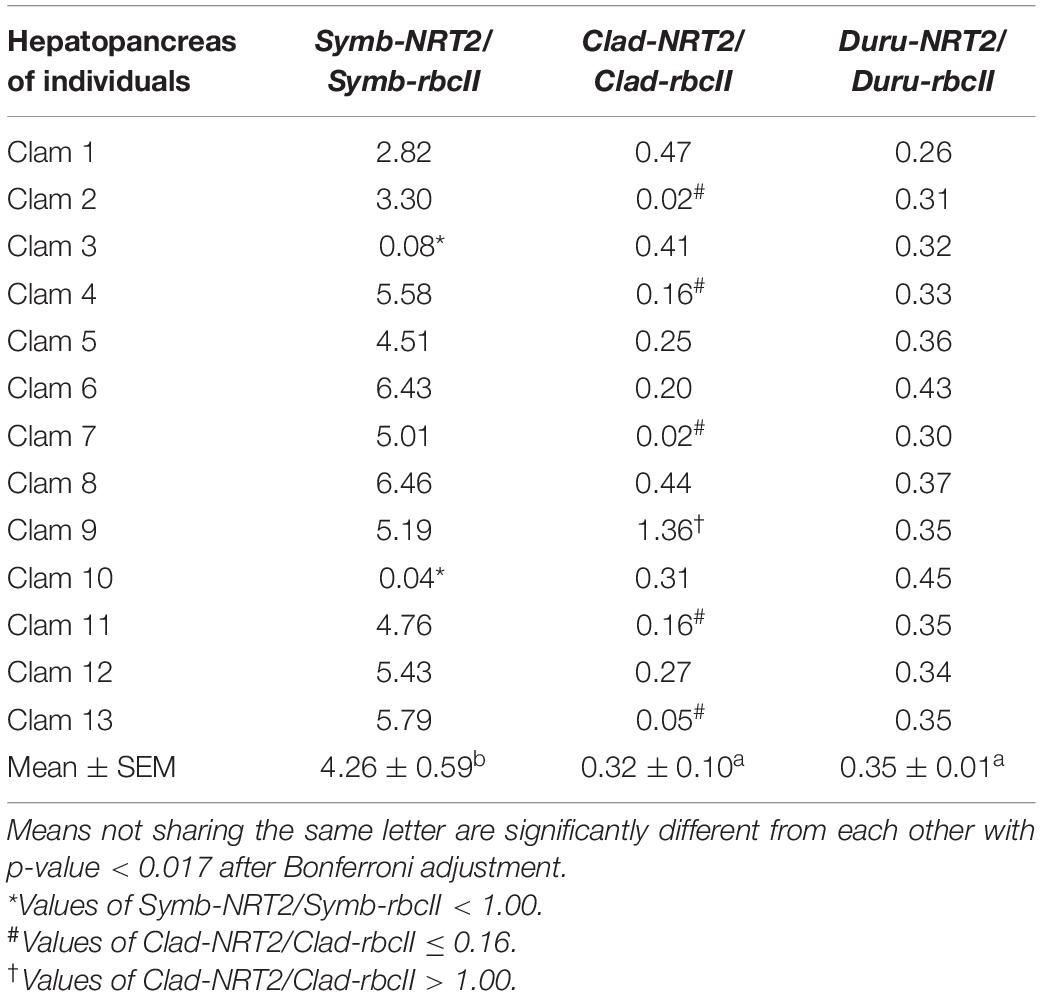
Table 7. Ratios of the transcript levels of nitrate transporter 2 (NRT2) to that of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) from Symbiodinium (Symb-NRT2/Symb-rbcII), Cladocopium (Clad-NRT2/Clad-rbcII), and Durusdinium (Duru-NRT2/Duru-rbcII) obtained from the hepatopancreas of Tridacna squamosa (n = 13).
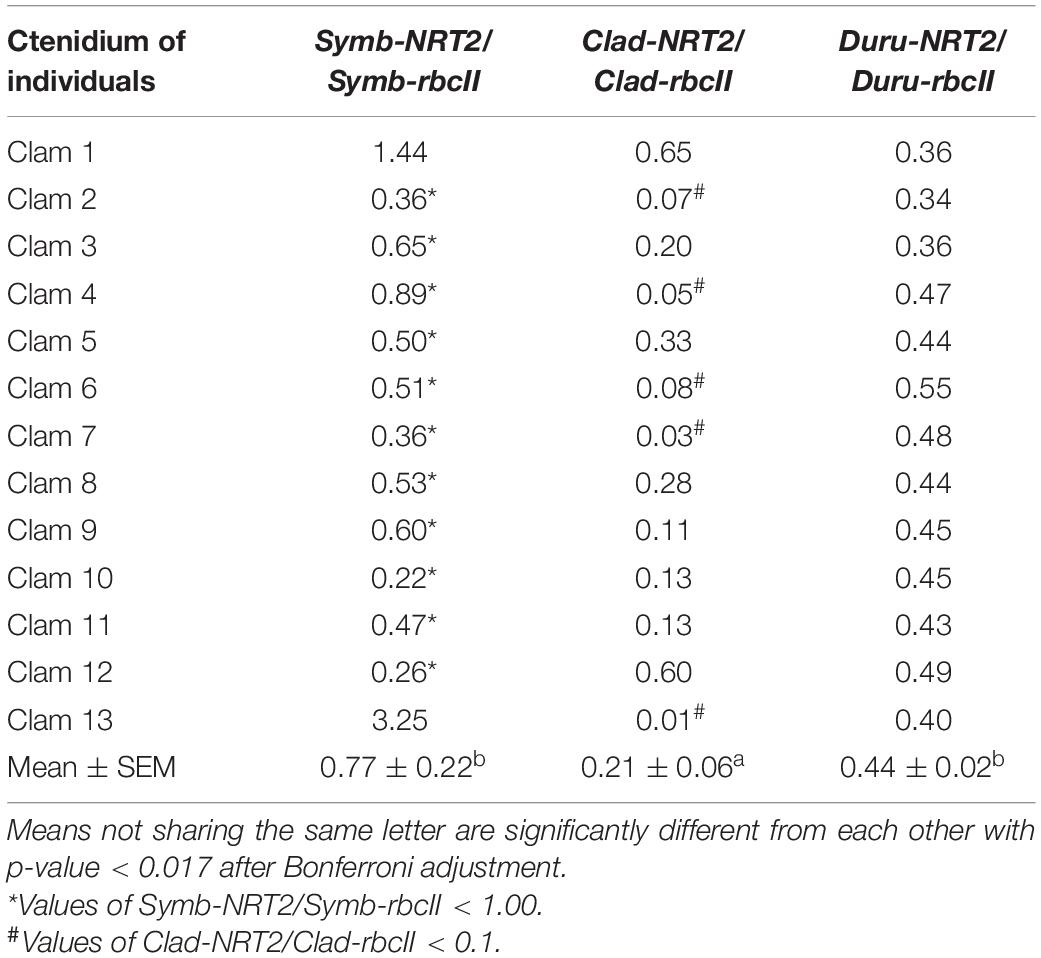
Table 8. Ratios of the transcript levels of nitrate transporter 2 (NRT2) to that of form II ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcII) from Symbiodinium (Symb-NRT2/Symb-rbcII), Cladocopium (Clad-NRT2/Clad-rbcII), and Durusdinium (Duru-NRT2/Duru-rbcII) obtained from the ctenidium of Tridacna squamosa (n = 13).
The Proportions of Genus-Specific NRT2 and rbcII Transcripts in Various Organs
A comparison of genus-specific NRT2 and rbcII in T. squamosa indicates that the mean transcript levels of Symb-NRT2 were significantly higher than those of Symb-rbcII in the outer mantle, foot muscle and hepatopancreas, but they were comparable in the inner mantle and ctenidium (n = 13; Table 3). Of note, the most prominent difference in transcript level of Symb-NRT2 and Symb-rbcII was observed in the outer mantle. By contrast, the mean transcript levels of Clad-NRT2 were either significantly lower than (as in the outer mantle and ctenidium) or comparable to (as in the inner mantle, foot muscle and hepatopancreas) the transcript levels of Clad-rbcII. The transcript levels of Duru-NRT2 were significantly lower than those of Duru-rbcII in these five organs (n = 13; Table 3).
For rbcII, the mean transcript level of Duru-rbcII was significantly higher than those of Symb-rbcII and Clad-rbcII (n = 13; Table 3) in all the five organs studied, indicating that Durusdinium was the major dinoflagellate genus in the individuals of T. squamosa obtained from Vietnam (Figure 4). Based on ANOVA, the transcript levels of Clad-rbcII were not significantly different from those of Symb-rbcII in the five organs because equal variance could not be assumed (n = 13; Table 3). Overall, the percentage proportions of transcript levels of rbcII derived from the three genera of Symbiodiniaceae dinoflagellates in the five organs of these 13 individuals of T. squamosa were Duru-rbcII > > Clad-rbcII = Symb-rbcII (Figure 4).
For NRT2, the mean transcript level of Duru-NRT2 was also significantly higher than those of Symb-NRT2 and Clad-NRT2 in four of the organs studied, except the hepatopancreas (n = 13; Table 3), corroborating the proposition that Durusdinium was the major dinoflagellate genus in these T. squamosa individuals (Figure 4). Furthermore, the mean transcript levels of Clad-NRT2 were statistically comparable to those of Symb-NRT2 in the outer mantle, inner mantle, foot muscle, and ctenidium (Table 3) due to the high variation of transcript levels among different individuals (n = 13). By contrast, the mean transcript level of Symb-NRT2 was significantly higher than that of Clad-NRT2 in the hepatopancreas (n = 13; Table 3). Hence, unlike rbcII, the percentage proportions of transcript levels of NRT2 derived from the three genera of Symbiodiniaceae dinoflagellates varied among the five organs of T. squamosa (Figure 4). For instance, the estimated proportions of Symb-rbcII transcripts in the outer mantle, hepatopancreas and ctenidium were 0.8, 3.0, and 1.9%, respectively, which were considerably lower than the estimated proportions of Symb-NRT2 transcripts in these three organs (outer mantle, 10.5%; hepatopancreas, 28.3%; ctenidium, 4.4%; Figure 4).
Genus-Specific NRT2/rbcII Ratios in the Outer Mantle, Inner Mantle, Foot Muscle, and Hepatopancreas
The mean transcript ratios of Symb-NRT2/Symb-rbcII (N = 13) for the outer mantle (Table 4), inner mantle (Table 5), foot muscle (Table 6), and hepatopancreas (Table 7) were > 1.0 because the transcript levels of Symb-NRT2 were generally higher than those of Symb-rbcII in these organs (Table 3). The mean transcript ratios of Symb-NRT2/Symb-rbcII (n = 13) for the outer mantle (8.4; Table 4) was significantly higher than those for the inner mantle (4.8; Table 5), foot muscle (4.0; Table 6) and hepatopancreas (4.3; Table 7). For each of these four organs, the mean transcript ratios of Symb-NRT2/Symb-rbcII (n = 13) were significantly higher than the mean transcript ratios of Clad-NRT2/Clad-rbcII and Duru-NRT2/Duru-rbcII, which were statistically comparable to each other (Tables 4–7).
For the outer mantle, while 12 of the 13 individuals examined displayed values > 1.0 for Symb-NRT2/Symb-rbcII, clam 3 had a distinctly low Symb-NRT2/Symb-rbcII value of 0.41 (Table 4). Of the 13 individuals examined, eight had ratios of Clad-NRT2/Clad-rbcII ranging between 0.39 and 0.88, but clams 2, 7, 9, 11, and 13 had noticeably lower Clad-NRT2/Clad-rbcII ratios of 0.018, 0.010, 0.022, 0.004, and 0.007, respectively (Table 4). In comparison, the transcript ratios of Duru-NRT2/Duru-rbcII were relatively constant in the outer mantle of these 13 individuals.
For the inner mantle, 10 of the 13 individuals examined displayed Symb-NRT2/Symb-rbcII values > 1.0, but clams 2, 3, and 10 had values < 1.0 (Table 5). Of the 13 individuals examined, three (clams 1, 4, and 8) had Clad-NRT2/Clad-rbcII transcript ratios > 1.0 (ranging from 1.0 to 1.5; Table 5). For the other 10 individuals, six of them (clams 2, 6, 7, 9, 11, and 13) had values close to or below 0.10. The transcript ratios of Duru-NRT2/Duru-rbcII in the inner mantle were again relatively constant among the 13 individuals.
For the foot muscle, out of the 13 individuals examined, 11 of them displayed Symb-NRT2/Symb-rbcII ratios > 1.0, except for clams 9 and 11 (Table 6). One individual (clam 8) had a Clad-NRT2/Clad-rbcII > 1.0 and clams 4, 6, 7, 9, 10 and 11 had values ≤ 0.17 (Table 6). In comparison, the transcript ratios of Duru-NRT2/Duru-rbcII in the foot muscle were relatively constant among the 13 individuals.
For the hepatopancreas, two (clam 3 and 10) of the 13 individuals examined had Symb-NRT2/Symb-rbcII values < 1.0 (Table 7). One individual (clam 9) had a Clad-NRT2/Clad-rbcII > 1.0, and five individuals (clams 2, 4, 7, 11, and 13) had values ≤ 0.16 (Table 7). In comparison, the transcript ratios of Duru-NRT2/Duru-rbcII in the hepatopancreas were relatively constant among the 13 individuals.
Genus-Specific NRT2/rbcII Ratios in the Ctenidium
For the ctenidium, the mean transcript ratio of Symb-NRT2/Symb-rbcII (n = 13; Table 8) was <1.0 because the transcript level of Symb-NRT2 was generally lower than that of Symb-rbcII (Table 3), which was dissimilar to the other four organs. Nevertheless, the mean transcript ratio of Symb-NRT2/Symb-rbcII (n = 13) was significantly higher than that of Clad-NRT2/Clad-rbcII, but comparable to that of Duru-NRT2/Duru-rbcII (Table 8).
Out of the 13 individuals examined, only clams 1 and 13 had Symb-NRT2/Symb-rbcII values > 1.0 (Table 8). As a result, the mean transcript ratio of Symb-NRT2/Symb-rbcII for the ctenidium (0.77; Table 8) was the lowest among the five organs studied (Tables 4–7). The transcript ratios of Clad-NRT2/Clad-rbcII in the ctenidium of all 13 individuals were <1.0, with five of them having particularly low values (<0.10). In comparison, the transcript ratios of Duru-NRT2/Duru-rbcII in the hepatopancreas were relatively constant among the 13 individuals.
Discussion
Giant clams harbor multiple phylotypes of Symbiodiniaceae dinoflagellates in variable proportions. The phylotype composition of dinoflagellates can influence the host’s growth rate (Hernawan, 2008; DeBoer et al., 2012) and affect the host’s ability to withstand environmental changes (Ikeda et al., 2017). Hence, it is logical to deduce that different phylotypes of Symbiodiniaceae dinoflagellates could play distinct physiological roles in the giant clam-dinoflagellate holobiont. One important role of coccoid dinoflagellates is to synthesize essential amino acids and share them with the host, but the host must supply them with nitrogen. Uniquely, the clam host conducts light-enhanced NO3– absorption through the ctenidium to benefit its symbionts. Indeed, our results confirm that Symbiodinium, Cladocopium, and Durusdinium of T. squamosa express NRT2 in the plasma membrane. They also indicate for the first time that different phylotypes of Symbiodiniaceae dinoflagellates in the coccoid stage could have different potentials of NO3– transport and hence different ability to assimilate NO3– into nitrogenous compounds such as amino acids.
Molecular Properties of Symb-NRT2, Clad-NRT2, and Duru-NRT2
Symb-NRT2, Clad-NRT2 and Duru-NRT2 were probably derived from S. tridacnidorum, C. goreaui, and D. trenchii, respectively. Similar to other members of MFS, Symb-NRT2, Clad-NRT2, and Duru-NRT2 had 12 TMs, consisting of two sets of six transmembrane domains linked by a cytosolic loop (see review by Forde, 2000). In addition, Symb-NRT2, Clad-NRT2, and Duru-NRT2 contained a conserved MFS motif between TM 2 and TM 3 (Forde, 2000). A distinguishing feature of the NNP among members of the MFS is the presence of two glycine-rich nitrate signature motifs (Trueman et al., 1996; Unkles et al., 2004a,2012; Yan et al., 2013), and these motifs were present in Symb-NRT2, Clad-NRT2 and Duru-NRT2. By contrast, members of the NPF family have only one signature motif (F-Y-x-x-I-N-x-G-S-L) in TM 5 (Steiner et al., 1995). Symb-NRT2, Clad-NRT2, and Duru-NRT2 could form hydrogen bonds with NO3– because of the presence of the putative NO3– binding site (Yan et al., 2013). The presence of the residue equivalent to Glu-330 in NrtA of Aspergillus nidulans (Unkles et al., 2004a,b), which is known to be involved in the symport of H+ and NO3– (Akhtar et al., 2015; Jacquot et al., 2017), in Symb-NRT2, Clad-NRT2, and Duru-NRT2 implies that they are H+-dependent symporters.
NRT2 Is Localized at the Plasma Membrane of Coccoid Dinoflagellates
NRT2 is localized at the plasma membrane of the coccoid dinoflagellates of T. squamosa, indicating that it can transport NO3– from the luminal fluid of the zooxanthellal tubule into the symbionts. The intracellular vesicles that display NRT2 immunofluorescence could be involved in the transfer of NRT2 from the internal membranes to the plasma membrane as suggested previously for Arabidopsis thaliana (Wirth et al., 2007). Hence, the transcript levels of Symb-NRT2, Clad-NRT2, and Duru-NRT2 can be appropriately used as molecular indicators to estimate the potential of NO3– transport (or uptake) in Symbiodinium, Cladocopium, and Durusdinium, respectively.
Molecular characterization indicates that Symb-NRT2, Clad-NRT2, and Duru-NRT2 probably rely on an inwardly directed [H+] gradient to drive the active absorption of NO3–. Recently, Mani et al. (2021) reported that coccoid dinoflagellates of T. squamosa could excrete H+ into the luminal fluid of zooxanthellal tubules through the merging of intracellular vesicles containing vacuolar-type H+-ATPase (VHA) subunit B (VHA-B) with the plasma membrane. The excreted H+ could augment the dehydration of luminal HCO3– and promote the absorption of CO2 by the photosynthesizing symbionts. The inwardly directed [H+] gradient generated by VHA across the plasma membrane could also drive the transport of NO3– through NRT2 into the symbiont. The absorbed NO3– could be reduced to NH4+. Then, a portion of the carbon fixed during photosynthesis could be utilized to assimilate NH4+ into amino acids catalyzed by glutamate dehydrogenase, glutamine synthetase and glutamine oxoglutarate aminotransferase (Padgett and Leonard, 1996). Some of the amino acids synthesized could be donated to the host in support of its growth and development. Of note, glutamine synthetase and glutamine oxoglutarate aminotransferase have been cloned from dinoflagellates of T. squamosa (Fam et al., 2018; Teh et al., 2021).
Durusdinium Is the Dominant Genus of Dinoflagellates in Tridacna squamosa Obtained From Vietnam
Cladocopium has been reported as the major genus of dinoflagellate found in T. squamosa from Japan (Ikeda et al., 2017), which is temperate and subtropical. However, based on the transcript levels of Symb-rbcII, Clad-rbcII, and Duru-rbcII, Poo et al. (2021) reported that T. squamosa (n = 4) obtained from Vietnam contained mainly Durusdinium (85–95%). Although we also worked with T. squamosa from Vietnam, the individuals involved were different from those examined by Poo et al. (2021). In general, the transcript levels of Symb-rbcII, Clad-rbcII, and Duru-rbcII reported by Poo et al. (2021) were higher than the corresponding results obtained in this study, which could be due to differences in the environmental conditions in the natural habitats of these two batches of T. squamosa. Nonetheless, in agreement with Poo et al. (2021), results of this study (n = 13) also denoted Durusdinium as the dominant genus of dinoflagellates in our experimental animals based on the transcript levels of NRT2 (78–92%) and rbcII (88–94%). This could be related to the tropical environmental conditions of and the availability of Symbiodiniaceae dinoflagellate phylotypes in Vietnam waters. Durusdinium is generally tolerant of a variety of environmental stressors (Brown et al., 2002; van Oppen et al., 2009), including high light intensity that would result in bleaching (Kemp et al., 2014) and turbid reef environments (Ulstrup and Van Oppen, 2003; LaJeunesse et al., 2010; Tonk et al., 2013). Thus, the high abundance of Durusdinium in the outer mantle of T. squamosa from Vietnam could have provided these clams with an advantage under stressful conditions, particularly at elevated temperature in the tropics. Based on the transcript level of Symb-rbcII, individuals of T. squamosa obtained from Vietnam contained relatively small populations of Symbiodinium (1–13%, Poo et al., 2021; 0.1–3%, this study), which could be due to the rarity of Symbiodinium in Indo-Pacific waters as compared to the Red Sea and the Caribbean waters (LaJeunesse, 2002; Baker, 2003).
The Implications of Different Percentage Proportions of Symb-rbcII and Symb-NRT2 Transcripts in Certain Organs of Tridacna squamosa
There are discrepancies in the percentage proportions of Symb-rbcII and Symb-NRT2 transcripts in three organs of the 13 individuals of T. squamosa examined in this study. The percentage proportions of Symb-rbcII in the outer mantle, ctenidium and hepatopancreas (0.8, 1.9, and 3.0%, respectively) were apparently lower than the percentage proportions of Symb-NRT2 in the same organ (10.5, 4.4, and 28.3%, respectively). A logical explanation is that some phylotypes of Symbiodinium in these three organs expressed more transcripts of NRT2 per dinoflagellate cell than phylotypes of Cladocopium and Durusdinium. Hence, our results indicate the presence of different phylotypes of Symbiodinium with different expression levels of NRT2 in the five organs T. squamosa. They also signify that some Symbiodinium phylotypes had a higher potential of NO3– transport than the phototrophic potential. Thus, it was crucial to analyze the transcript ratios of NRT2 to rbcII.
Symbiodinium Generally Have Higher Potential of NO3– Transport Than Cladocopium and Durusdinium
The transcript ratio of NRT2/rbcII estimated for each genus of dinoflagellates (e.g., Symb-NRT2/Symb-rbcII) could indicate whether that genus would have a high or low potential of NO3– transport relative to the potential of phototrophy. Any difference in the transcript ratios among the three genera of dinoflagellates would suggest different potentials of NO3– transport and therefore different abilities to assimilate NO3– for the production of amino acids and other nitrogenous compounds. The mean transcript ratios of Symb-NRT2/Symb-rbcII were significantly higher than those of Clad-NRT2/Clad-rbcII and Duru-NRT2/Duru-rbcII in the outer mantle, inner mantle, foot muscle and hepatopancreas of T. squamosa, and the greatest differences were observed in the outer mantle. Hence, it can be concluded that the phylotypes of Symbiodinium generally had higher potential of NO3– transport than the phylotypes of Cladocopium and Durusdinium in these four organs. For the ctenidium, Symb-NRT2/Symb-rbcII was also significantly higher than Clad-NRT2/Clad-rbcII but comparable to Duru-NRT2/Duru-rbcII. These results imply that the composition of Symbiodinium phylotypes present in the ctenidium of T. squamosa were dissimilar to those present in the other four organs, and particularly distinct from those in the outer mantle.
Some Phylotypes (Species) of Symbiodinium Might Have Higher Potential of NO3– Transport Than the Phototrophic Potential
Symbiodiniaceae dinoflagellates are phototrophic and express high transcript levels of rbcII (Poo et al., 2020, 2021). Yet, the mean transcript levels of Symb-NRT2 in the outer mantle, inner mantle, foot muscle and hepatopancreas of 13 T. squamosa individuals were substantially higher than the corresponding mean transcript levels of Symb-rbcII, resulting in the mean Symb-NRT2/Symb-rbcII ratios of 4.0–8.4 in these four organs. In the outer mantle, the mean Symb-NRT2/Symb-rbcII ratio of 8.4 was much greater than the mean Clad-NRT2/Clad-rbcII ratio of 0.39 and the mean Duru-NRT2/Duru-rbcII ratio of 0.63. These results are unusual as they indicate for the first time that certain phylotypes of Symbiodinium could specialize in NO3– absorption, and hence NO3–assimilation, more so than photosynthesis. They also indicate that the potentials of NO3– transport in these Symbiodinium phylotypes were greater than those in phylotypes of Cladocopium and Durusdinium in general.
Two Physiologically Distinct Phylotypes (Species) of Symbiodinium Based on Symb-NRT2/Symb-rbcII
Of the 13 individuals examined, 11 had ratios of Symb-NRT2/Symb-rbcII that ranged between 7.2 and 13.1 in the outer mantle of T. squamosa, but the ratios for clam 3 (0.41) and clam 10 (1.34) were distinctly < 7. These results indicate that the phylotypes of Symbiodinium in clam 3 and clam 10 had strikingly lower potential of NO3– transport than those in the other 11 clams. Hence, there could be two physiologically distinct phylotypes (species) of Symbiodinium as defined by high or low potential of NO3– transport.
To analyze our results, a Symb-NRT2/Symb-rbcII ratio of 1.0 (i.e., equal transcript levels of Symb-NRT2 and Symb-rbcII) was set arbitrarily to differentiate the Symbiodinium phylotypes with high potential of NO3– transport from those with low potential of NO3–. In the 13 individuals examined in this study, the numbers of clams that displayed a ratio of >1.0 for the outer mantle, inner mantle, foot muscle, hepatopancreas and ctenidium were 12, 10, 11, 11, and 2, respectively. These results indicate that the proportion of phylotypes of Symbiodinium with high potential of NO3– transport varied among the five organs, and that the ctenidium contained mainly Symbiodinium phylotypes with low potential of NO3– transport. It is noteworthy that while muscle tissues can be found in the other four organs, the ctenidium comprises mainly epithelial tissues dedicated for gases exchange, membrane transport and acid-base balance. It could be that the demand for using NO3– to form amino acid is low in the symbionts of the ctenidium in comparison with the symbionts of the other four organs. It could also imply that the symbionts of the ctenidium prefer to use other source of nitrogen, e.g., ammonia, for nitrogen metabolism. On the other hand, the predominant presence of Symbiodinium phylotypes with high potential of NO3– transport in the outer mantle facilitates the effective coupling of NO3– transport and assimilation with photosynthesis. Hence, it can be concluded that the phylotype compositions of Symbiodiniaceae dinoflagellates could vary among organs of T. squamosa based on the host’s physiological needs. Nonetheless, how the relevant phylotypes of Symbiodinium find the way to and establish in different organs of T. squamosa needs to be elucidated in the future.
Cladocopium Phylotypes (Species) Also Display Different Potentials of NO3– Transport
Based on Clad-NRT2/Clad-rbcII, the outer mantle of T. squamosa contained two physiologically distinct phylotypes of Cladocopium. Of the 13 individuals examined, eight had ratios of Clad-NRT2/Clad-rbcII ranging between 0.39 and 0.88. However, clams 2, 7, 9, 11, and 13 had distinctly lower Clad-NRT2/Clad-rbcII ratios (<0.1) and hence lower potential of NO3– transport than the other eight individuals. An examination of Clad-NRT2/Clad-rbcII ratios in the inner mantle, foot muscle, and hepatopancreas indicates the presence of some phylotypes of Cladocopium with Clad-NRT2/Clad-rbcII slightly > 1. The inner mantle, foot muscle, hepatopancreas, and ctenidium apparently contained dissimilar proportions of Cladocopium phylotypes with either high or low potentials of NO3– transport. Overall, Cladocopium phylotypes in the ctenidium, with a mean Clad-NRT2/Clad-rbcII ratio of 0.21 (n = 13), appeared to have lower potentials of NO3– transport than those in the other four organs (0.32–0.57). Again, this might indicate the relative low demand for using NO3– to form amino acids in the ctenidium as compared with the other four organs.
Implications and Perspective
Using NRT2 and rbcII as molecular indicators, our results indicate for the first time that Symbiodiniaceae dinoflagellates harbored by T. squamosa could have different potentials of NO3– transport, and offer insights into the functional diversity among coccoid dinoflagellates at the genus level (Symbiodinium, Cladocopium, and Durusdinium). A corollary of some phylotypes of Symbiodinium having a preference of NO3– uptake and assimilation is that different members of Symbiodiniaceae could be specialized in absorbing and utilizing distinct types of nitrogenous compounds (e.g., ammonia, urea and glutamine) furnished by the host. An important implication is that it would not be possible for the clam host to regulate its general symbiont population by simply reducing the availability of endogenous ammonia to them through the recycling of metabolic ammonia into non-essential amino acids, as has been suggested for scleractinian corals (Falkowski et al., 1993; Cui et al., 2019; Xiang et al., 2020). This is particularly the case for those Symbiodinium phylotypes that can effectively absorb and assimilate NO3–. Rather, the clam host might be able to regulate the population of a specific Symbiodiniaceae phylotype in a certain organ by controlling the translocation of a particular nitrogen-containing compound from the hemolymph through the tubular epithelial cells into the luminal fluid of the zooxanthellal tubules.
Importantly, our results signify that, in the coccoid stage, certain phylotypes (species) of Symbiodinium might specialize in nitrogen metabolism more so than photosynthesis as reflected by their high potential of NO3– transport with transcript ratios of Symb-NRT2/Symb-rbcII much greater than one. This implies that different phylotypes of Symbiodiniaceae dinoflagellates could have different abilities to donate amino acids and carbohydrates to the host. As the host requires ∼20 different types of amino acid for growth and reproduction, it is highly unlikely that these amino acids are donated equally by all the phylotypes of dinoflagellates. Rather, there could be a division of labor among different phylotypes of coccoid dinoflagellates in the supply of nutrients (e.g., specific types of carbohydrate or amino acid) to the host. Therefore, efforts should be made in the future to develop a large variety of molecular indicators (e.g., transporters of ammonia, urea and glutamine, as well as enzymes involved in the syntheses of various amino acids) in order to elucidate the distinct physiological roles of various Symbiodiniaceae phylotypes in the giant clam-dinoflagellate holobiont.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MZ014639, MZ014640, and MZ014641).
Author Contributions
SC and YI designed the experiments, wrote the manuscript, and involved in the analysis of data and approval of the manuscript. CP performed the experiments and analyzed the data. SC and CP participated in animal subjection and sample collection. All the authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Singapore Ministry of Education through grants to SC through National Institute of Education (NIE) Academic Research Fund RI 3/19 CSF and the NIE Research support for Senior Academic Administrator Grant RS 1/21 CSF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.784662/full#supplementary-material
Footnotes
References
Akhtar, N., Karabika, E., Kinghorn, J. R., Glass, A. D. M., Unkles, S. E., and Rouch, D. A. (2015). High-affinity nitrate/nitrite transporters NrtA and NrtB of Aspergillus nidulans exhibit high specificity and different inhibitor sensitivity. Microbiology 161, 1434–1446. doi: 10.1099/mic.0.000088
Aranda, M., Li, Y., Liew, Y. J., Baumgarten, S., Simakov, O., Wilson, M. C., et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6:39734. doi: 10.1038/srep39734
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Bellantuono, A. J., Dougan, K. E., Granados-Cifuentes, C., and Rodriguez-Lanetty, M. (2019). Free-living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol. Ecol. 28:12j173m. doi: 10.5061/dryad.12j173m
Bernhard, A. (2010). The nitrogen cycle: processes, players, and human impact. Nat. Edu. Knowl. 2:12.
Brown, B., Downs, C., Dunne, R., and Gibb, S. (2002). Exploring the basis of thermotolerance in the reef coral Goniastrea aspera. Mar. Ecol. Prog. Ser. 242, 119–129. doi: 10.3354/meps242119
Camargo, J. A., and Alonso, Á (2006). Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ. Int. 32, 831–849. doi: 10.1016/j.envint.2006.05.002
Chan, C. Y. L., Hiong, K. C., Boo, M. V., Choo, C. Y. L., Wong, W. P., Chew, S. F., et al. (2018). Light exposure enhances urea absorption in the fluted giant clam Tridacna squamosa and up-regulates the protein abundance of a light-dependent urea active transporter DUR3-like in its ctenidium. J. Exp. Biol. 221:jeb176313. doi: 10.1242/jeb.176313
Chan, C. Y. L., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2019). Increased apical sodium-dependent glucose transporter abundance in the ctenidium of the giant clam Tridacna squamosa upon illumination. J. Exp. Biol. 222:jeb195644. doi: 10.1242/jeb.195644
Chen, Y., González-Pech, R., Stephens, T., Bhattacharya, D., and Chan, C. (2020). Evidence that inconsistent gene prediction can mislead analysis of dinoflagellate genomes. J. Phycol. 56, 6–10. doi: 10.1111/jpy.12947
Chew, S. F., and Ip, Y. K. (2014). Excretory nitrogen metabolism and defence against ammonia toxicity in air-breathing fishes. J. Fish Biol. 84, 603–638. doi: 10.1111/jfb.12279
Collos, Y., and Berges, J. A. (2003). “Nitrogen Metabolism in phytoplankton,” in Encyclopedia of Life Support Systems, ed. C. M. Duarte (Paris: EOLSS Publishers).
Crawford, N. M., and Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/s1360-1385(98)01311-9
Cui, G., Liew, Y. J., Li, Y., Kharbatia, N., Zahran, N. I., Emwas, A.-H., et al. (2019). Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLoS Genet. 15:e1008189. doi: 10.1371/journal.pgen.1008189
Dagenais-Bellefeuille, S., and Morse, D. (2013). Putting the N in dinoflagellates. Front. Microbiol. 4:00369. doi: 10.3389/fmicb.2013.00369
Dagenais-Bellefeuille, S., and Morse, D. (2016). The main nitrate transporter of the dinoflagellate Lingulodinium polyedrum is constitutively expressed and not responsible for daily variations in nitrate uptake rates. Harmful Algae 55, 272–281. doi: 10.1016/j.hal.2016.03.021
Davies, S., Ries, J., Marchetti, A., and Castillo, K. (2018). Symbiodinium functional diversity in the coral Siderastrea siderea is influenced by thermal stress and reef environment, but not ocean acidification. Front. Mar. Sci. 5:00150. doi: 10.3389/fmars.2018.00150
DeBoer, T. S., Baker, A. C., Erdmann, M. V., Ambariyanto Jones, P. R., and Barber, P. H. (2012). Patterns of Symbiodinium distribution in three giant clam species across the biodiverse Bird’s Head region of Indonesia. Mar. Ecol. Progr. Ser. 444, 117–132. doi: 10.3354/meps09413
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and McCloskey, L. (1993). Population control in symbiotic corals. BioScience 43, 606–611. doi: 10.2307/1312147
Fam, R. R. S., Hiong, K. C., Choo, C. Y. L., Wong, W. P., Chew, S. F., and Ip, Y. K. (2018). Molecular characterization of a novel algal glutamine synthetase (GS) and an algal glutamate synthase (GOGAT) from the colorful outer mantle of the giant clam, Tridacna squamosa, and the putative GS-GOGAT cycle in its symbiotic zooxanthellae. Gene 656, 40–52. doi: 10.1016/j.gene.2018.02.062
Fan, C., and Glibert, P. M. (2005). Effects of light on nitrogen and carbon uptake during a Prorocentrum minimum bloom. Harmful Algae 4, 629–641.
Fisher, C. R., Fitt, W. K., and Trench, R. K. (1985). Photosynthesis and respiration in Tridacna gigas as a function of irradiance and size. Biol. Bull. 169, 230–224. doi: 10.2307/1541400
Fitt, W. K., and Trench, R. K. (1981). Spawning, development and acquisition of zooxanthellae by Tridacna squamosa (Mollusca, Bivalvia). Biol. Bull. 161, 213–235. doi: 10.2307/1540800
Fitt, W. K., Heslinga, G. A., and Watson, T. C. (1993a). Utilization of dissolved inorganic nutrients in growth and mariculture of the tridacnid clam Tridacna derasa. Aquaculture 109, 27–38. doi: 10.1016/0044-8486(93)90483-f
Fitt, W. K., Rees, T. A. V., Braley, R. D., Lucas, J. S., and Yellowlees, D. (1993b). Nitrogen flux in giant clams: size-dependency and relationship to zooxanthellae density and clam biomass in the uptake of dissolved inorganic nitrogen. Mar. Biol. 117, 381–386. doi: 10.1007/BF00349313
Forde, B. G. (2000). Nitrate transporters in plants: structure, function and regulation. BBA Biomembr. 1465, 219–235. doi: 10.1016/s0005-2736(00)00140-1
González-Pech, R., Chen, Y., Stephens, T., Shah, S., Mohamed, A., Lagorce, R., et al. (2019). Genomes of Symbiodiniaceae reveal extensive sequence divergence but conserved functions at family and genus levels. BioRxiv [Preprint]. doi: 10.1101/800482
Guibert, I., Lecellier, G., Torda, G., Pochon, X., and Berteaux-Lecellier, V. (2020). Metabarcoding reveals distinct microbiotypes in the giant clam Tridacna maxima. Microbiome 8:57. doi: 10.1186/s40168-020-00835-8
Hernawan, U. E. (2008). Review: symbiosis between the giant clams (Bivalvia: Cardiidae) and zooxanthellae (Dinophyceae). Biodiversitas 9, 53–58. doi: 10.13057/biodiv/d090113
Hiong, K. C., Choo, C. Y. L., Boo, M. V., Ching, B., Wong, W. P., Chew, S. F., et al. (2017). A light-dependent ammonia-assimilating mechanism in the ctenidia of a giant clam. Coral Reefs 36, 311–323. doi: 10.1007/s00338-016-1502-4
Ikeda, S., Yamashita, H., Kondo, S., Inoue, K., Morishima, S., and Koike, K. (2017). Zooxanthellal genetic varieties in giant clams are partially determined by species-intrinsic and growth-related characteristics. PLoS One 12:e0172285. doi: 10.1371/journal.pone.0172285
Ip, Y. K., and Chew, S. F. (2010). Ammonia production, excretion, toxicity, and defense in fish: a review. Front. Physiol. 1:00134. doi: 10.3389/fphys.2010.00134
Ip, Y. K., and Chew, S. F. (2021). Light-dependent phenomena and related molecular mechanisms in giant clam-dinoflagellate associations: a review. Front. Mar. Sci. 8:627722. doi: 10.3389/fmars.2021.627722
Ip, Y. K., Hiong, K. C., Teng, J. H. Q., Boo, M. V., Choo, C. Y. L., Wong, W. P., et al. (2020). The fluted giant clam (Tridacna squamosa) increases nitrate absorption and upregulates the expression of a homolog of SIALIN (H+:2NO3– cotransporter) in the ctenidium during light exposure. Coral Reefs 39, 451–465. doi: 10.1007/s00338-020-01907-9
Ip, Y. K., Koh, C. Z. Y., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., et al. (2017b). Carbonic anhydrase 2-like in the giant clam, Tridacna squamosa: characterization, localization, response to light, and possible role in the transport of inorganic carbon from the host to its symbionts. Physiol. Rep. 5:e13494. doi: 10.14814/phy2.13494
Ip, Y. K., Hiong, K. C., Goh, E. J. K., Boo, M. V., Choo, C. Y. L., Ching, B., et al. (2017a). The whitish inner mantle of the giant clam, Tridacna squamosa, expresses an apical Plasma Membrane Ca2+-ATPase (PMCA) which displays light-dependent gene and protein expressions. Front. Physiol. 8:781. doi: 10.3389/fphys.2017.00781
Jacquot, A., Li, Z., Gojon, A., Schulze, W., and Lejay, L. (2017). Post-translational regulation of nitrogen transporters in plants and microorganisms. J. Exp. Bot. 68, 2567–2580. doi: 10.1093/jxb/erx073
Jantzen, C., Wild, C., El-Zibdah, M., Roa-Quiaoit, H., Haacke, C., and Richter, C. (2008). Photosynthetic performance of giant clams, Tridacna maxima and T. squamosa, Red Sea. Mar. Biol. 155, 211–221. doi: 10.1007/s00227-008-1019-7
Jensen, F. B. (1996). Uptake, elimination and effects of nitrite and nitrate in freshwater crayfish (Astacus astacus). Aquat. Toxicol. 34, 95–104. doi: 10.1016/0166-445x(95)00030-8
Keane, T. M., Creevey, C. J., Pentony, M. M., Naughton, T. J., and Mclnerney, J. O. (2006). Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6:29. doi: 10.1186/1471-2148-6-29
Kemp, D., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W., and Schmidt, G. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
Klumpp, D. W., and Griffiths, C. L. (1994). Contributions of phototrophic and heterotrophic nutrition to the metabolic and growth requirements of four species of giant clam (Tridacnidae). Mar. Ecol. Prog. Ser. 115, 103–115. doi: 10.3354/MEPS115103
Klumpp, D. W., Bayne, B. L., and Hawkins, A. J. S. (1992). Nutrition of the giant clam Tridacna gigas (L.) I. Contribution of filter feeding and photosynthates to respiration and growth. J. Exp. Mar. Biol. Ecol. 155, 105–122. doi: 10.1016/0022-0981(92)90030-E
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Ladner, J. T., Barshis, D. J., and Palumbi, S. R. (2012). Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol. Biol. 12:217. doi: 10.1186/1471-2148-12-217
LaJeunesse, T. (2002). Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400. doi: 10.1007/s00227-002-0829-2
LaJeunesse, T. C., Bhagooli, R., Hidaka, M., DeVantier, L., Done, T., Schmidt, G. W., et al. (2004). Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Prog. Ser. 284, 147–161. doi: 10.3354/meps284147
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
LaJeunesse, T., Pettay, D., Sampayo, E., Phongsuwan, N., Brown, B., Obura, D., et al. (2010). Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785–800. doi: 10.1111/j.1365-2699.2010.02273.x
Leong, S. C. Y., Maekawa, M., and Taguchi, S. (2010). Carbon and nitrogen acquisition by the toxic dinoflagellate, Alexandrium tamarense in response to different nitrogen sources and supply modes. Harmful Algae 9, 48–58.
Léran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Levin, R. A., Beltran, V. H., Hill, R., Kjelleberg, S., Mcdougald, D., Steinberg, P. D., et al. (2016). Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215. doi: 10.1093/molbev/msw201
Liu, C., Li, T., Liu, E., Li, C., Wang, A., and Gu, Z. (2019). Proximate composition, amino acid content, and fatty acid profile of the adductor muscle and mantle from two species of the giant clams Tridacna crocea and Tridacna squamosa. J. Shellfish Res. 38, 529–534. doi: 10.2983/035.038.0303
Liu, H., Stephens, T., González-Pech, R., Beltran, V., Lapeyre, B., Bongaerts, P., et al. (2018). Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1:95. doi: 10.1038/s42003-018-0098-3
Mani, R., Boo, M. V., Poo, J. S. T., Ng, S. Y., Chew, S. F., and Ip, Y. K. (2021). Molecular characterization, cellular localization and light-dependent expression of dinoflagellate vacuolar-type H+-ATPase (VHA) subunit B in the colourful outer mantle of the giant clam, Tridacna squamosa, indicate the involvement of VHA in CO2 uptake in the photosynthesizing symbionts. Plant Gene 28:100328. doi: 10.1016/j.plgene.2021.100328
Mayfield, A. B., Hsiao, Y. Y., Chen, H. K., and Chen, C. S. (2014). Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotechnol. 16, 371–384. doi: 10.1007/s10126-014-9558-z
Morishima, S., Yamashita, H., O-hara, S., Nakamura, Y., Quek, V. Z., Yamauchi, M., et al. (2019). Study on expelled but viable zooxanthellae from giant clams, with an emphasis on their potential as subsequent symbiont sources. PLoS One 14:e0220141. doi: 10.1371/journal.pone.0220141
Norton, J. H., Shepherd, M. A., Long, H. M., and Fitt, W. K. (1992). The zooxanthellal tubular system in the giant clam. Biol. Bull. 183, 503–506. doi: 10.2307/1542028
Orsel, M., Krapp, A., and Daniel-Vedele, F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. structure and gene expression. Plant Physiol. 129, 886–896. doi: 10.1104/pp.005280
Paasche, E., Bryceson, I., and Tangen, K. (1984). Interspecific variation in dark nitrogen uptake by dinoflagellates. J. Phycol. 20, 394–401.
Padgett, P. E., and Leonard, R. T. (1996). Free amino acid levels and the regulation of nitrate uptake in maize cell suspension cultures. J. Exp. Bot. 47, 871–883. doi: 10.1093/jxb/47.7.871
Pechkovskaya, S. A., Knyazev, N. A., Matantseva, O. V., Emelyanov, A. K., Telesh, I. V., Skarlato, S. O., et al. (2020). Dur3 and nrt2 genes in the bloom-forming dinoflagellate Prorocentrum minimum: Transcriptional responses to available nitrogen sources. Chemosphere 241:125083. doi: 10.1016/j.chemosphere.2019.125083
Pochon, X., Wecker, P., Stat, M., Berteaux-Lecellier, V., and Lecellier, G. (2019). Towards an in-depth characterization of Symbiodiniaceae in tropical giant clams via metabarcoding of pooled multi-gene amplicons. PeerJ 7:e6898. doi: 10.7717/peerj.6898
Poo, J. S. T., Boo, M. V., Chew, S. F., and Ip, Y. K. (2021). Using form II ribulose-1,5-bisphosphate carboxylase/oxygenase to estimate the phototrophic potentials of Symbiodinium, Cladocopium and Durusdinium in various organs of the fluted giant clam, Tridacna squamosa, and to evaluate their responses to light upon isolation from the host. Coral Reefs 40, 233–250. doi: 10.1007/s00338-020-02031-4
Poo, J. S. T., Choo, C. Y. L., Hiong, K. C., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2020). Phototrophic potential and form II ribulose-1,5-bisphosphate carboxylase/oxygenase expression in five organs of the fluted giant clam, Tridacna squamosa. Coral Reefs 39, 361–374. doi: 10.1007/s00338-020-01898-7
Rosic, N., Yew, E. S. L., Chan, C. K. K., Lee, H. C., Kaniewska, P., Edwards, D., et al. (2015). Unfolding the secrets of coral–algal symbiosis. ISME J. 9, 844–856. doi: 10.1038/ismej.2014.182
Rossbach, S., Saderne, V., Anton, A., and Durarte, C. M. (2019). Light-dependent calcification in Red Sea giant clam Tridacna maxima. Biogeosciences 16, 2635–2650. doi: 10.5194/bg-16-2635-2019
Rowan, R., Whitney, S. M., Fowler, A., and Yellowlees, D. (1996). Rubisco in marine symbiotic dinoflagellates: form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 8, 539–553. doi: 10.1105/tpc.8.3.539
Shoguchi, E., Beedessee, G., Tada, I., Hisata, K., Kawashima, T., Takeuchi, T., et al. (2018). Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genom. 19:458. doi: 10.1186/s12864-018-4857-9
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steiner, H., Naider, F., and Becker, J. (1995). The PTR family: a new group of peptide transporters. Mol. Microbiol. 16, 825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x
Streamer, M., McNeil, Y. R., and Yellowless, D. (1993). Photosynthetic carbon dioxide fixation in zooxanthellae. Mar. Biol. 115, 195–198. doi: 10.1007/BF00346335
Takabayashi, M., Santos, S. R., and Cook, C. B. (2004). Mitochondrial DNA phylogeny of the symbiotic dinoflagellates (Symbiodinium, Dinophyta). J. Phycol. 40, 160–164. doi: 10.1111/j.0022-3646.2003.03-097.x
Teh, L., Poo, J. S. T., Boo, M. V., Chew, S. F., and Ip, Y. K. (2021). Using glutamine synthetase 1 to evaluate the symbionts’ potential of ammonia assimilation and their responses to illumination in five organs of the giant clam, Tridacna squamosa. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 255:110914. doi: 10.1016/j.cbpa.2021.110914
Tomasso, J. R., and Carmichael, G. J. (1986). Acute toxicity of ammonia, nitrite, and nitrate to the guadalupe bass, Micropterus treculi. Bull. Environ. Contam. Toxicol. 36, 866–870. doi: 10.1007/bf01623596
Tonk, L., Sampayo, E. M., Weeks, S., Magno-Canto, M., and Hoegh-Guldberg, O. (2013). Host-specific interactions with environmental factors shape the distribution of Symbiodinium across the Great Barrier Reef. PLoS One 8:e68533. doi: 10.1371/journal.pone.0068533
Trueman, L. J., Richardson, A., and Forde, B. G. (1996). Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene 175, 223–231. doi: 10.1016/0378-1119(96)00154-0
Ulstrup, K. E., and Van Oppen, M. J. H. (2003). Geographic and habitat partitioning of genetically distinct zooxanthellae (Symbiodinium) in Acropora corals on the Great Barrier Reef. Mol. Ecol. 12, 3477–3484. doi: 10.1046/j.1365-294x.2003.01988.x
Umeki, M., Yamashita, H., Suzuki, G., Sato, T., Ohara, S., and Koike, K. (2020). Fecal pellets of giant clams as a route for transporting Symbiodiniaceae to corals. PLoS One 15:e0243087. doi: 10.1371/journal.pone.0243087
Unkles, S. E., Karabika, E., Symington, V. F., Cecile, J. L., Rouch, D. A., Akhtar, N., et al. (2012). Alanine scanning mutagenesis of a high-affinity nitrate transporter highlights the requirement for glycine and asparagine residues in the two nitrate signature motifs. Biochem. J. 447, 35–42. doi: 10.1042/BJ20120631
Unkles, S. E., Rouch, D. A., Wang, Y., Siddiqi, M. Y., Glass, A. D. M., and Kinghorn, J. R. (2004a). Two perfectly conserved arginine residues are required for substrate binding in a high-affinity nitrate transporter. PNAS 101, 17549–17554. doi: 10.1073/pnas.0405054101
Unkles, S. E., Wang, R., Wang, Y., Glass, A. D. M., Crawford, N. M., and Kinghorn, J. R. (2004b). Nitrate reductase activity is required for nitrate uptake into fungal but not plant cells. J. Biol. Chem. 279, 28182–28186. doi: 10.1074/jbc.m403974200
van Oppen, M. J. H., Baker, A. C., Coffroth, M. A., and Willis, B. L. (2009). “Bleaching resistance and the role of algal endosymbionts coral bleaching,” in Coral bleaching: Patterns, Processes, Causes and Consequences, eds M. van Oppen and J. Lough (Berlin: Springer), 83–102.
Wang, J. T., and Douglas, A. E. (1999). Essential amino acid synthesis and nitrogen recycling in an alga–invertebrate symbiosis. Mar. Biol. 135, 219–222. doi: 10.1007/s002270050619
Westin, D. T. (1974). Nitrate and nitrite toxicity to salmonoid fishes. Progress Fish Cult. 36, 86–89.
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Wilkerson, F. P., and Trench, R. K. (1986). Uptake of dissolved inorganic nitrogen by the symbiotic clam Tridacna gigas and the coral Acropora sp. Mar. Biol. 93, 237–246. doi: 10.1007/bf00508261
Wirth, J., Chopin, F., Santoni, V., Viennois, G., Tillard, P., Krapp, A., et al. (2007). Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282, 23541–23552. doi: 10.1074/jbc.m700901200
Xiang, T., Lehnert, E., Jinkerson, R. E., Clowez, S., Kim, R. G., DeNofrio, J. C., et al. (2020). Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations. Nat. Comm. 11:108. doi: 10.1038/s41467-019-13963-z
Keywords: amino acids, coral reefs, photosynthate, Symbiodiniaceae dinoflagellates, symbiosis, zooxanthellae, Tridacna squamosa
Citation: Pang CZ, Ip YK and Chew SF (2021) Using Transcript Levels of Nitrate Transporter 2 as Molecular Indicators to Estimate the Potentials of Nitrate Transport in Symbiodinium, Cladocopium, and Durusdinium of the Fluted Giant Clam, Tridacna squamosa. Front. Mar. Sci. 8:784662. doi: 10.3389/fmars.2021.784662
Received: 28 September 2021; Accepted: 15 November 2021;
Published: 06 December 2021.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Fangfang Yang, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaGuoxin Cui, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2021 Pang, Ip and Chew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shit F. Chew, c2Z1bi5jaGV3QG5pZS5lZHUuc2c=
 Caryn Z. Pang
Caryn Z. Pang Yuen K. Ip
Yuen K. Ip Shit F. Chew
Shit F. Chew