- 1The Swire Institute of Marine Science, School of Biological Sciences, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 2Biological Oceanography Division, Council of Scientific and Industrial Research (CSIR)-National Institute of Oceanography, Dona Paula, India
- 3CAS Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou, China
- 4Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
Elevated carbon dioxide levels in ocean waters, an anthropogenic stressor, can alter the chemical equilibrium of seawater through a process called ocean acidification (OA). The resultant reduction of pH can be detrimental during the early developmental stages of the commercially important edible Pacific oyster Crassostrea gigas; the ability of larvae to join a population is likely to be compromised by declining ocean pH. Given this threat, it is important to study the molecular mechanisms that these organisms use to overcome OA stress at the gene expression level. Here, we performed transcriptome profiling in oyster larvae following exposure to ambient (8.1) and reduced (7.4) pH during the pre-settlement growth period (i.e., 18 d post fertilization) using RNA-seq with Illumina sequencing technology. In total, 1,808 differentially expressed genes (DEGs) were identified, 1,410 of which were matched by BLAST against the Swiss-Prot database. Gene ontology classification showed that most of these DEGs were related to ribosomal, calcium ion binding, cell adhesion and apoptotic processes. Pathway enrichment analysis revealed that low pH (7.4) enhanced energy production and organelle biogenesis but prominently suppressed several immune response pathways. Moreover, activation of the MAPK signaling pathway was observed along with inhibition of the Wnt, VEGF, and ErbB pathways, highlighting the fact that the initiation of stress responses is given priority over larval development or shell growth when the larvae cope with low pH. In conclusion, our study demonstrated a unique gene expression profiling approach in studying oyster larval responses to OA, which not only provides comprehensive insights into the mechanisms underlying oyster tolerance to CO2-driven decreases in ocean pH but also supplies a valuable genomic resource for further studies in this species.
Introduction
Carbon dioxide levels in the atmosphere and oceans have been increasing at unprecedented rates, since the beginning of the industrial revolution (Falkowski et al., 2000). One-third of atmospheric anthropogenic CO2 emission dissolves in the ocean, raising acidity of the oceans that in turn leads to changes in the carbonate chemistry, the process called as ocean acidification (OA) (Raven et al., 2005). Addition of excess amount of CO2 in the ocean leads to overall shift in the seawater acid-base chemistry tending toward more acidic condition, reducing pH and saturation state of carbonate minerals (Hurd et al., 2019; Downey-Wall et al., 2020). Subsequent increase in carbonic acid reduces the availability of carbonate ions in oceans, thus affecting marine organisms that depend on ocean carbonate levels to form their calcium carbonate shells (Fabry et al., 2008; Guinotte and Fabry, 2008; Cooley et al., 2009; Thomsen et al., 2018; Hurd et al., 2019). Ocean acidification studies revealed both positive and negative effects on marine organisms (Kroeker et al., 2010, 2011). Though some marine organisms are resilient and can acclimate to changes (Kroeker et al., 2010; Munday et al., 2011; McCulloch et al., 2012), most shell-forming invertebrates, such as sea urchins, corals and mollusks (Parker et al., 2013), find it difficult to calcify in the presence of elevated CO2 levels and reduced pH in seawater (Gazeau et al., 2007, 2010, 2013; Kurihara et al., 2007; Miller et al., 2009; Talmage and Gobler, 2009; Todgham and Hofmann, 2009; Sheppard Brennand et al., 2010; Parker et al., 2012; Doney et al., 2020). Ocean acidification also impacts homeostasis and behavioral response in marine fishes (Esbaugh et al., 2012; Jutfelt et al., 2013; Hamilton et al., 2014) and leads to metabolic suppression to conserve energy for survival and immune responses in many marine organisms (Bibby et al., 2007, 2008; Lannig et al., 2010; Hernroth et al., 2011).
The impact of ocean acidification is generally predicted according to effects on physiological processes such as calcification (Kurihara and Shirayama, 2004; Miller et al., 2009; O’Donnell et al., 2010; Zhao et al., 2017), growth and development (Dupont et al., 2008; Talmage and Gobler, 2009; Sheppard Brennand et al., 2010; Ginger et al., 2013), energy metabolism (Beniash et al., 2010; Lannig et al., 2010; Nakamura et al., 2011), immune response (Castillo et al., 2017; Xing et al., 2019; Zhao et al., 2020) and reproduction (Parker et al., 2009, 2010; Ross et al., 2011). Studies in commercial hatcheries (Talmage and Gobler, 2009; Dineshram et al., 2012, 2013) and wild populations (Miller et al., 2009; Parker et al., 2010, 2011; Waldbusser et al., 2011) have revealed a significant impact of ocean acidification on larval oyster growth.
A thorough understanding of the mechanisms that control the physiological processes is needed to uncover the sensitivity of the organism exposed to OA (Widdicombe and Spicer, 2008). In this context, studies investigating the molecular mechanisms of responses of marine organisms to ocean acidification assume importance, in evaluating whether they can withstand the deleterious effects associated with it. Although currently available transcriptomic analyses and genomic resources are useful in determining mRNA plasticity in response to OA, the availability of these techniques is still limited to only a few species (Todgham and Hofmann, 2009; Todgham and Stillman, 2013). For example, gene expression studies of early developmental stages in coral identified a number of genes affected by OA (Moya et al., 2012; Rivest et al., 2018). Studies on sea urchins also revealed the overall molecular responses of sea urchin larvae exposed to CO2 (Todgham and Hofmann, 2009; Devens et al., 2020). OA is known to interfere with several key physiological process in marine organisms, transcriptomics has proven to be effective method to elucidate the molecular level response and when coupled with phenotypic data confers a better understanding of whole organismal response (Davies et al., 2016; Melzner et al., 2020; Strader et al., 2020).
To explore the adaptation and tolerance mechanisms of oyster larvae to low pH, this study performed transcriptome sequencing of early stage (pediveliger) oysters exposed to seawater at a low pH 7.4 (projected to reflect the atmospheric CO2 emission scenario in 2300, by the Intergovernmental Panel on Climate Change IPCC) (Caldeira and Wickett, 2005), using RNA-seq with Illumina sequencing, to identify underlying adaptation and tolerance mechanisms of oyster larvae coping with environmental pH variations and to provide an integrated view of the cellular pathways affected; to emphasize the physiological responses of ocean acidification.
Materials and Methods
Larval Culture and CO2 Perturbation Set Up
Natural populations of adult pacific oysters were acclimated in natural seawater with salinity 24 psu and a temperature of 24°C for 2 days prior to experimental spawning. D-shaped larvae collected after strip spawning of 8 male and 23 female adult oysters, C. gigas were used for low pH exposure experiments (Breese and Malouf, 1975). Larvae of the C. gigas cultured from the early D-shaped developmental stage (i.e., 12 h post-fertilization) to the pediveliger stage (i.e., 15–17 day post-fertilization) under OA condition (Ko et al., 2014). For detailed information on the culture conditions, please refer to the “Materials and Methods” section of Ko et al. (2014) and Dineshram et al. (2016). The experiment was done in three replicates, using 40-liter tanks for the larval culture. Each larval culture tanks were mixed with air to obtain the ambient CO2 levels for the control (pH 8.1) and CO2 pre-mixed air for the low pH (7.4), which was within environmentally relevant range predicted for the Pacific oysters in the fluctuating coastal marine systems and reflects the futuristic atmospheric CO2 emission scenarios (Zeebe, 2008), for the year 2300 as predicted by the Intergovernmental Panel on Climate Change IPCC (Caldeira and Wickett, 2005). The low pH 7.4 was chosen to reflect the futuristic CO2 emissions as per the IPCC projected scenarios for the study region with respect to decreased pH to represent 2300 years considering far future acidification (Zhai et al., 2013). The CO2 concentration in the experimental tanks was regulated through flowmeters (Cole-Parmer Inc., United States) and pH of the experimental system was constantly monitored thrice a day using a pH meter (Orion Star™, Thermo Electron Co., United States), respectively.
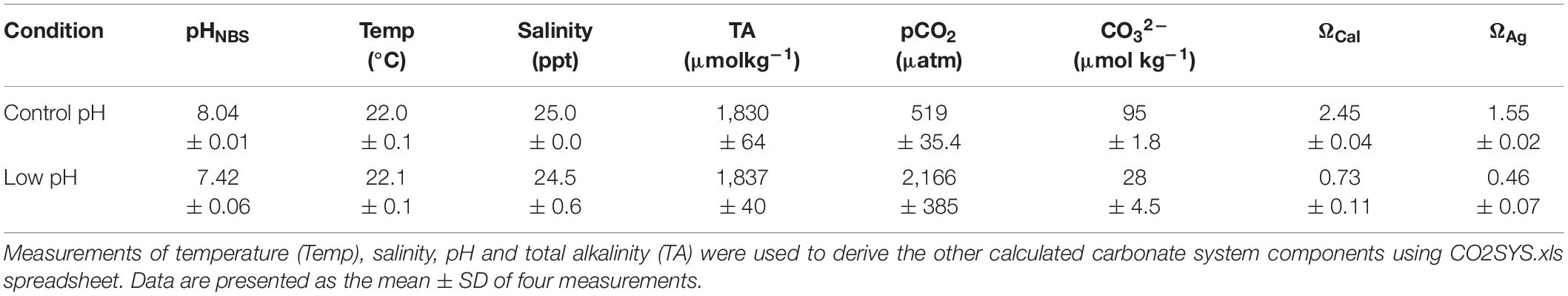
Regular laboratory oyster larval rearing procedures was followed throughout the experiment that includes filtered seawater (0.22μm), ambient temperature: 22–24°C maintained using a continuous flow through system water bath. The carbonate chemistry of seawater used in the experiment was determined by total alkalinity (TA) measurements using Alkalinity Titrator (AC-A2, Apollo SciTech’s Inc., United States), calibrated against certified seawater reference material (Batch 103, A.G. Dickson, Scripps Institution of Oceanography). Using the measured temperature, salinity, pH and TA, other parameters of carbonate chemistry were calculated using co2sys.xls spreadsheet (Pelletier et al., 2007) with the equilibrium constants K1, K2 by Mehrbach (Mehrbach et al., 1973) and KSO4 constants by Dickson (Dickson and Millero, 1987). The carbonate chemistry conditions for the pH levels were described in Table 1. Samples from pediveliger larvae with a size of ∼280 μm were collected after 18 days and stored at –80°C prior to quantitative transcriptome analysis. This study aimed to provide a set of differential uni-genes for the oyster pediveliger larvae exposed to seawater at elevated CO2 levels, low pH 7.4 using next generation high-throughput transcriptome sequencing.
RNA Extraction
Total cellular RNA was extracted from larval samples (each group in triplicates) exposed to pH 8.1 and 7.4 using RNeasy mini kit (Qiagen). Briefly, the experiment started with four biologically independent replicates (replicated culture tanks), however, we have used only three of these replicates for transcriptomic analysis. This pooling of samples was done with purpose, i.e., to reduce biological variability between replicate samples obtained from multiple parents and to deduct the treatment effect without baseline variation (Dineshram et al., 2015, 2016; Griffith et al., 2019). RNA concentration and integrity were measured by NanoDrop (Thermo Fisher Scientific) and Agilent 2100 BioAnalyzer (Agilent Technologies), respectively. After treatment with DNase I, mRNA isolated with Oligotex mRNA Mini Kit (Qiagen) was quantified using NanoDrop 2000 spectrophotometer (Thermo Scientific). The mRNA pooled from triplicate samples in equal quantities was ready for cDNA synthesis.
cDNA Library Construction and RNA-Seq
cDNA libraries were constructed following TruSeq™ RNA sample preparation guide (Illumina). Briefly, the mRNA was fragmented to sizes of 120–200 bp. Then, first and second strand were synthesized using SuperScript II Reverse Transcriptase (Invitrogen) and Second Strand Master Mix, respectively. The double strand (ds) cDNA were isolated using Ampure XP beads. The adapter containing the A-Tailing oligonucleotide was ligated to dsDNA, and then amplified the library using 12 cycles of PCR. After purification, libraries were quantified by Qubit® 2.0 Fluorometer and validated by Agilent 2100 bioanalyzer to confirm the insert size and calculate the mole concentration. Clusters were generated by cBot with the library dilution of 10 pM, and finally were sequenced on the Illumina Genome Analyzer (Hiseq 2500).
Quality Control, Pre-processing, and Genome Mapping
Quality of raw data generated from Illumina sequencing was checked by FastQC.1 The raw reads were preprocessed using the FASTX2 to remove the adapter sequences, low-quality reads, and reads less than 20 bp in length. Then, the clean sequence reads obtained from preprocessing were aligned to the C. gigas genome3 using Tophat 2.0 (Trapnell et al., 2009), allowing maximum 2 mismatch per read with genome sequence. Final results were stored in BAM format files.
Differential Gene Expression Analysis
The mapped reads were counted and then normalized with transcript length using cufflink (Trapnell et al., 2010), resulting in FPKM (Fragments Per Kilo bases per Million reads) for each transcript from two libraries. Thus, FPKM value facilitated to calculate the expression abundance of each gene that was on behalf of one transcript within two libraries. The differentially expressed genes (DEGs) were identified using DEGseq with each pair-wise comparison (Wang et al., 2010). Furthermore, the DEGs were filtered using the threshold of false discovery rate (FDR ≤ 0.1) and the absolute value of log2 (FC) (≥ 1).
Differentially Expressed Gene Identification, Classification, and Pathway Analysis
The DEGs were homology-searched using BLASTX against the Swiss-Prot database with an E-value of 1e–5. Gene names were assigned to each sequence based on the best BLAST hit. To annotate sequences, the Gene Ontology (GO) terms were assigned to query sequences by BLAST2GO (Götz et al., 2008), producing group of genes classified in the three ontology categories, cellular component, biological process and molecular function. Furthermore, pathways were analyzed using the online KEGG Automatic Annotation server (Kyoto encyclopedia of genes and genomes4) with bi-directional best-hit setting. Finally, functional networks of enriched KEGG pathways were visualized using ClueGO.5
Validation of Transcriptomic Profiles by Real Time-Quantitative Polymerase Chain Reaction
Expression of five up-regulated genes such as ADP-ribosylation factor 1, Hemicentin-1, Iron (II)-dependent oxidoreductase EgtB, NADH dehydrogenase and Glutathione S-transferase omega-1 and three down-regulated genes such as Zinc finger protein, FMRF- amide neuropeptides and Calcium/Calmodulin-dependent protein kinase type I revealed as differentially expressed due to low pH exposure by RNA sequencing was determined by RT-qPCR (Table 2). PCR amplifications was performed in triplicate biological samples on a Roche 454 sequencer (United States) in a final volume of 20 μl containing 1μl of template RNA mixed with 10 μl of 2X Master Mix (Roche) and 0.8 μl of each forward and reverse primer in each RT- PCR reaction for cDNA synthesis. Amplifications was carried out for 40 cycles on a 7500 Real Time PCR system (Applied Biosystems) under conditions of 30 s at 95°C for 1 cycle, 5 s at 95°C, 30 s at 60°C for 40 cycles. Thermal denaturing step to generate a dissociation curve to verify amplification efficiency and primer dimer was performed by increasing temperature from 65–99°C with a 5 s hold for every one degree temperature increment. RT-qPCR products were also further checked on 1.5% agarose gels to check the specificity of amplicons. The mRNA expression of each gene was quantified and calculated using their normalized expression level relative to the housekeeping gene by the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Results and Discussion
General Characteristics of Transcriptome Assembly
Two libraries were sequenced with Illumina (GEO accession numbers SRR960964 and SRR967068) and characteristics of transcriptome were summarized below. There were 16,602,870 and 14,795,142 raw reads generated from the control and one low pH library, respectively. After low quality reads removal and adaptor trimming, a total of 15,081,854 effective reads were produced from the control group, while 13,455,384 effective reads were obtained from the treated group, whose effective reads ratio were 90.8 and 90.9%, respectively. These effective reads were then mapped into C. gigas genome (Zhang et al., 2012), revealing 11,515,356 reads in control group and 9,746,368 reads in low pH group were able to be mapped. The mapping ratios for the control and low-pH libraries mapping ratios were 83.4 and 81.1%, respectively. Finally, these mapped reads were collected and used for quantification and annotation analyses.
Differential Expression Genes and Gene Annotation
To investigate the mechanisms underlying oyster larval tolerance to low pH, transcriptomic expression profiling was performed by comparing the control (ambient pH) library and low pH libraries using the FPKM method. Scatter plot showed a total of 1,808 DEGs obtained from the larval oysters exposed to low pH treatment, including 1,176 up-regulated and 632 down-regulated genes, respectively (Figure 1). Among these genes, 1410 DEGs were significantly matched against Swiss-Prot database by BLASTX, accounting for 78% of all DEGs (Supplementary Table 1). After functional annotation of these DEGs, all assigned GO terms were classified into three GO categories: biological process (1,138, 26.4%), cellular component (1,380, 32.1%) and molecular function (1,787, 41.5%) (Figure 2 and Supplementary Table 2). Within molecular function, structural constituents of ribosome (GO: 0003735) and calcium ion binding (GO: 0005509) were highly represented (p < 0.001), highlighting the effect of protein translation and shell formation at low pH. Regarding the cellular component, the most represented (p < 0.001) GO categories were nucleus (GO: 0005634) and extracellular region (GO: 0005576). As in the case of biological process, translation (GO: 0006412) was the most significantly represented (p < 0.001) followed by proteolysis (GO: 0006508), cell adhesion (GO: 0007155) and apoptotic process (GO: 0006915). Almost two-thirds of the differentially regulated genes identified in the current study were up-regulated. The proportion of down-regulated genes was lower, indicating that pH stress does not suppress the activities of major cellular pathways but rather predominantly induces cellular homeostatic mechanisms that up-regulate energy metabolism, cell signaling and ribosomal biogenesis. This gene regulation pattern confirms that low pH alters post-transcriptional regulatory mechanisms irrespective of gene expression.
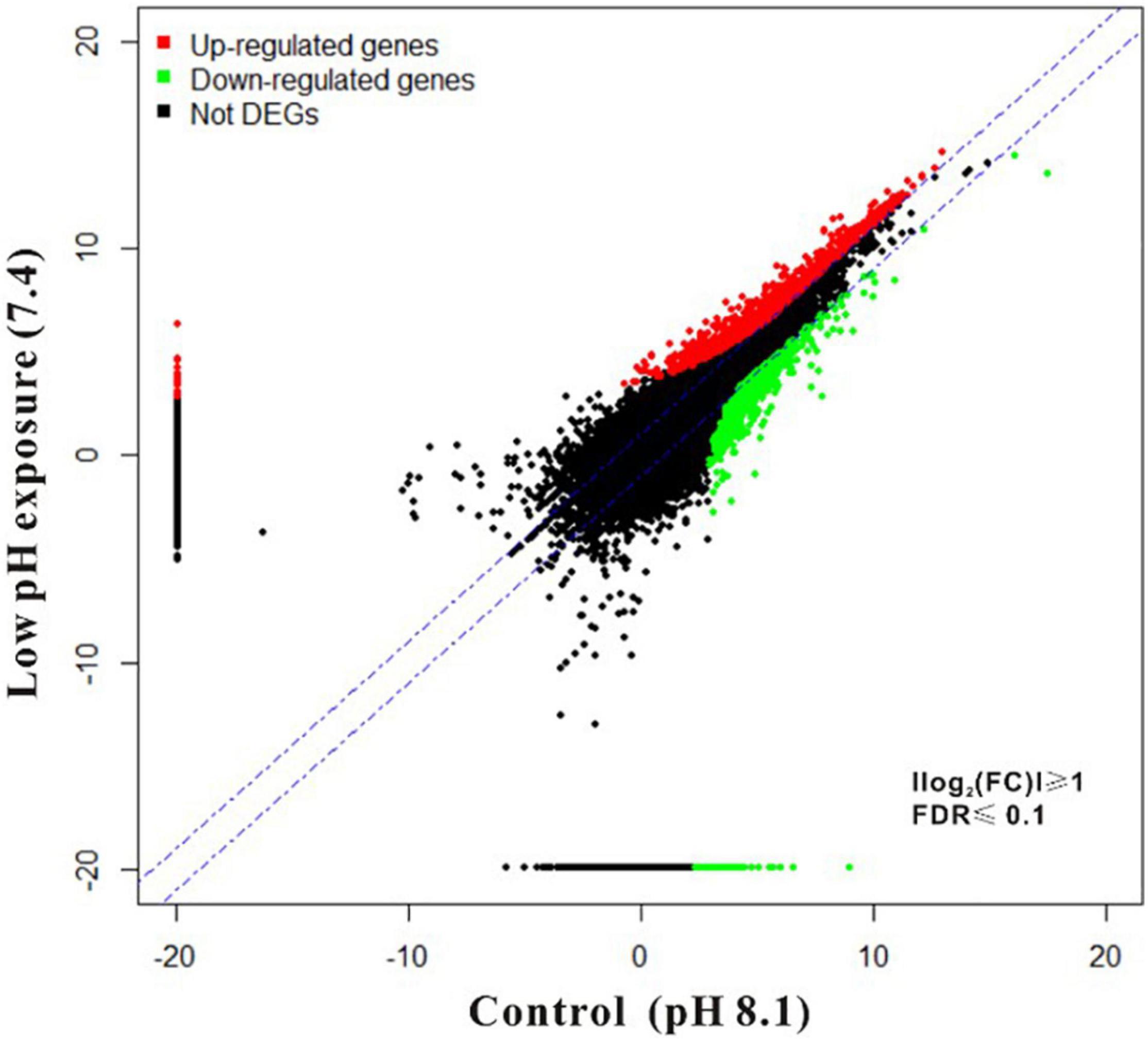
Figure 1. Scatter-plots depicting genomic expression profiles of oyster larvae under low pH exposure. FPKM was quantified based on Illumina RNA-seq analysis and 22,381 representative genes are shown. Red and green indicate genes that were significantly up- and down-regulated in low pH group compared to control group based on filtering by FDR ≤ 0.1 and the absolute value of log2 (FC) (≥ 1). The remaining genes are shown in black.
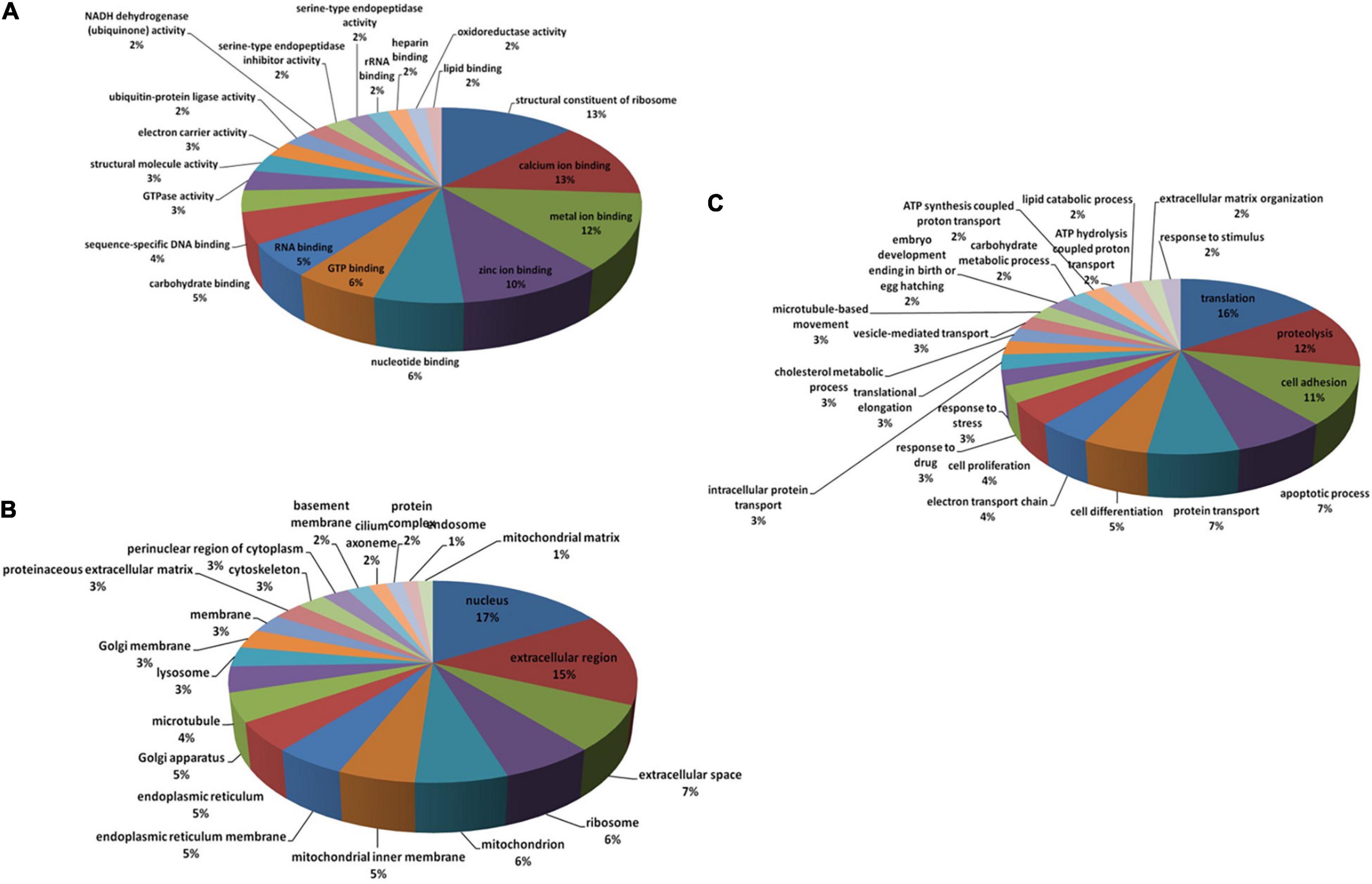
Figure 2. Classification of the annotated DEGs according to GO (Gene Ontology) terms. The distribution of top-level GO terms is depicted as three categories: Molecular function (A), Cellular component (B) and Biological process (C).
Biological Pathways Responsive to Ocean Acidification Acclimation
To explore the possible biological pathways associated with low pH DEGs, this study mapped these DEGs into the KEGG database. Among them, 315 DEGs received KEGG annotations and were assigned to 27 pathways (Figure 3 and Supplementary Table 3). Most of the DEGs were found to be enriched in a few pathways, such as Ribosome (ko03010, 42 DEGs), MAPK signaling pathway (ko04010, 37 DEGs), Viral carcinogenesis (ko05203, 35 DEGs), Peroxisome (ko04146, 34 DEGs) and Oxidative phosphorylation (ko00190, 29 DEGs) with high significance (p < 0.001). Based on the intersection of biological functions and pathways identified in this transcriptomic study, ClueGO Cytoscape pathway analysis was performed to provide integrated and comprehensive insights (Bindea et al., 2009, 2013; Figure 4).
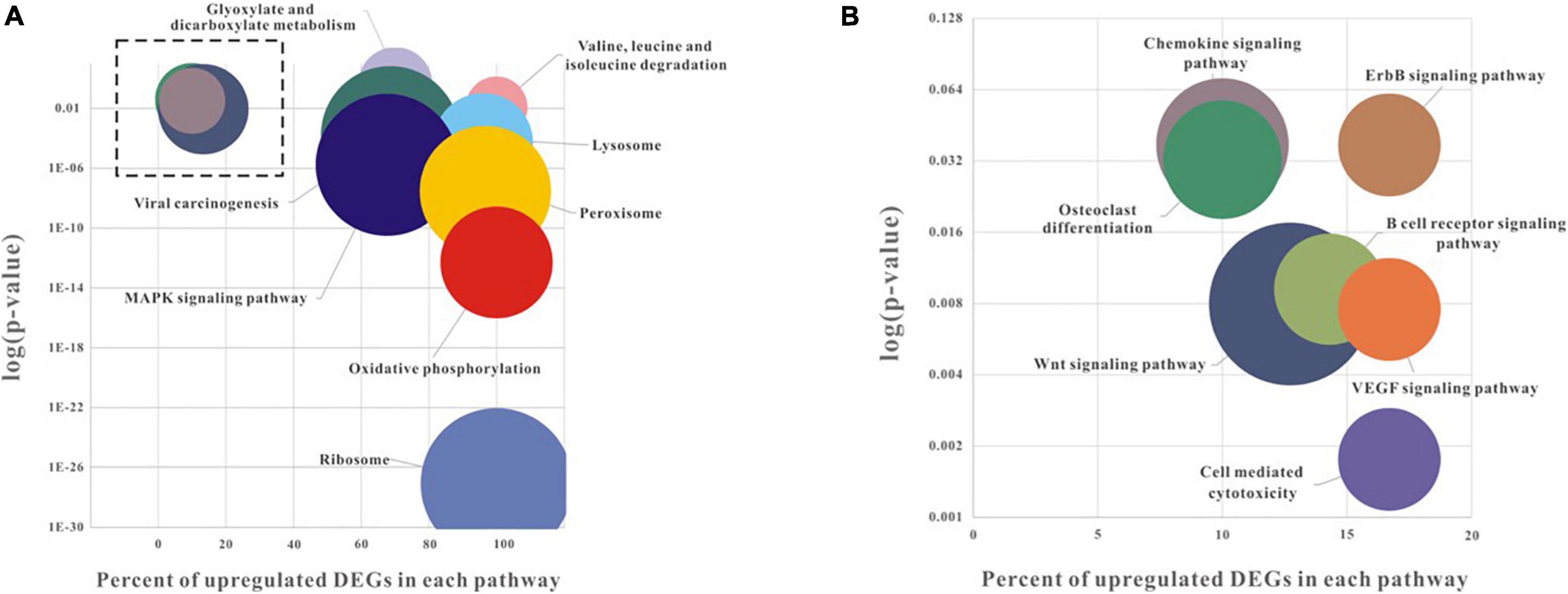
Figure 3. Pathway enrichment analysis of low pH induced DEGs. Higher magnification of the boxed areas within (A) is shown in (B). The longitudinal axis refers to the log10 (p-value), and p-value < 0.05 was used as a threshold to select significant KEGG pathways. The vertical axis represents the percent of up-regulated DEGs in total DEGs. The bubble size indicates the numbers of DEGs enriched in each pathway.
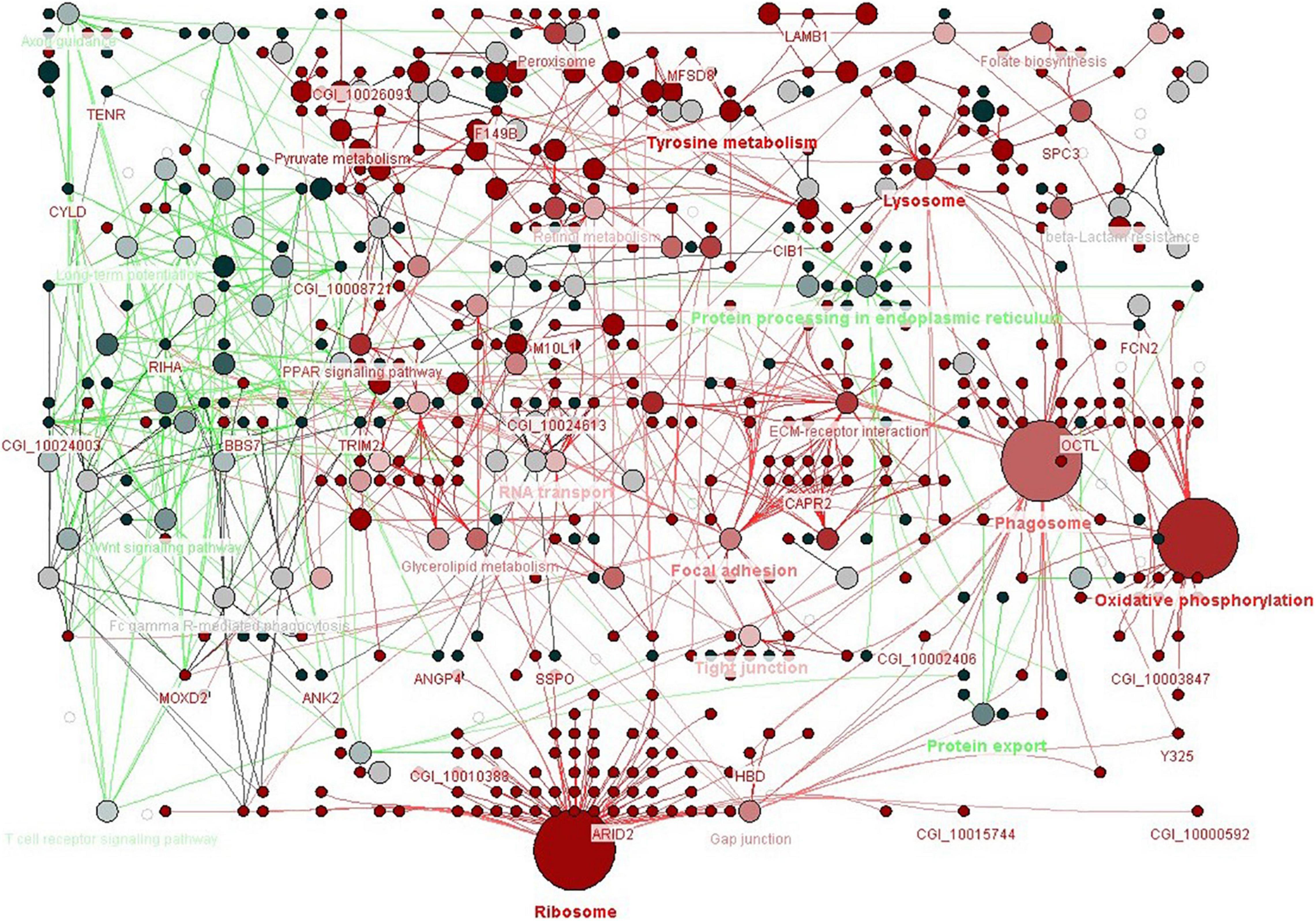
Figure 4. ClueGo pathway analysis of enriched differentially expressed genes. Functional group networks of enriched KEGG pathways induced in oyster larvae to low pH are represented using a cerebral layout. GO terms are shown by nodes and are linked based on their kappa score (> 0.3). The node size represents the term enrichment significance, and the labels shown are the most significant leading group terms. Terms with up-regulated and down-regulated genes are shown in red and green, respectively. The color gradient represents the extent of regulation. Terms and pathways with no significance are shown in gray.
Genes for Energy Production and Cellular Homeostasis
Strikingly, all 23 DEGs assigned to the Oxidative phosphorylation (ko00190) showed to be increased after low pH treatment (two fold change) in genes NADH dehydrogenase, isoforms of ATP synthase, cytochrome c oxidase and V-type ATPase involved in electron transport chain. Oxidative phosphorylation is a metabolic pathway to produce ATP and energy release by the oxidation of nutrients. Thus, oyster larvae likely increase their energy production by up-regulating electron transport chain genes in response to low pH exposure. These results were consistent with previous studies in which adult oysters exposed to elevated temperature exhibited increased mitochondrial respiration (Chamberlin, 2004; Ivanina et al., 2012). In contrast to previous studies that showed metabolic depression in response to low-pH stress in adult blue mussel (Hüning et al., 2013), sea urchins (Padilla-Gamiño et al., 2013) and corals (Todgham and Hofmann, 2009; Kaniewska et al., 2012, 2015; Moya et al., 2012; Rocker et al., 2015), the current study found that genes corresponding to the basal energy metabolic pathways were up-regulated by two fold. The metabolic pathways involved in energy production were significantly down-regulated in PDI stage of pacific oyster larvae (Liu et al., 2020). Also studies on adult pearl oyster Pinctada fucata and razor clams reported metabolic depression along with up-regulation of V-type ATPase and energy producing genes in response to ocean acidification (Li et al., 2016a; Peng et al., 2017) showing that the metabolic depression can also happen without the suppression of energy producing genes. However, other studies in mussels (Thomsen and Melzner, 2010; Fernández-Reiriz et al., 2012), sea-urchins (Stumpp et al., 2011), in arctic pteropods (Comeau et al., 2010) and Corals (Vidal-Dupiol et al., 2013; Davies et al., 2016; González-Pech et al., 2017; Rivest et al., 2018) showed either up-regulation or no change in the expression of energy metabolism genes in response to moderate changes in the ocean acidification. The expression pattern observed in the current study suggests that aerobic pathways are activated to generate more energy to compensate for low pH-induced energy depletion.
Immune Responsive Genes
Immune responses are energy-intensive processes that require large amounts of metabolic energy which involves physiological trade-offs in energy balance to meet energy demands upon activation of the immune response (French et al., 2009). Several pathways involved in immune response were identified by the pathway enrichment analysis, including the Chemokine signaling pathway (ko04062), Natural Killer cell mediated cytotoxicity (ko04650) and B cell receptor signaling pathway (ko04662). Interestingly, all three immune pathways were dramatically inhibited in the low pH group, comprising of 90, 83.3, and 85.7% of the down-regulated DEGs, respectively. Immune related genes in the hemocytes were significantly down-regulated in adult Crassostrea gigas when exposed to ocean acidification (Wang et al., 2016) but it was found to be increased in other oyster species (Goncalves et al., 2016, 2017). Most of the genes corresponding to the immune pathways encoded kinases responsible for immune defense in many bivalves. Kinases are usually activated by stressors such as UV, pollutants, heat and osmotic stress in most bivalves (Canesi et al., 2006). The roles of kinases in defense mechanisms have been illustrated in mussel hemocytes; mussels exposed to a bacterial challenge showed induction of kinase-mediated signal transduction pathways (Canesi et al., 2006). The down-regulation of genes encoding kinases such as mitogen activated protein kinase 3 (–2.8-fold), cAMP-dependent protein kinase catalytic subunit (–3-fold) and glycogen synthase kinase-3 beta (–2.6-fold), in addition to the up-regulation of phosphatidylinositol 3-kinase regulatory subunit alpha (+3.1-fold), clearly suggests that seawater acidification could inhibit immune responses in pediveliger larvae through impaired signal transduction. Notably, down-regulation of fucolectin-1, an immune protein that binds to bacterial surfaces and contributes to defense, was also observed, consistent with the effect of OA on the innate immune system (Honda et al., 2000). Stimulation of TLR pathway and immune factors is known to occur in Crassostrea gigas when exposed to low pH (Cao et al., 2018). In humans, B-cell lymphoma/leukemia 11A (Bcl11a) functions as a key transcriptional factor that controls the normal immune cell development, and in the present study its homolog in pediveliger larvae was also suppressed upon exposure to low pH seawater conditions. Therefore, OA may impair immune cell development and immune signaling in oysters. However, this speculation requires further verification (Liu et al., 2003).
Oxidative Stress and Ribosome Biogenesis Genes
In total, 34 and 18 DEGs involved in formation of Peroxisome (ko04146) and Lysosome (ko04142) formation, respectively, were mapped and shown to be activated, and 32 and 17 of these DEGs were up-regulated at the mRNA level. Additionally, 42 DEGs were assigned to the ribosome pathway (ko03010), and all of the associated DEGs were up-regulated after exposure to low pH. Most of the genes involved in the peroxisomal pathway were oxidases involved in the production of reactive oxygen species (ROS) in cells (Cancio and Cajaraville, 1997). Similarly induction of oxidative stress and subsequent increase in ROS and apoptosis has been observed in the adult Crassostrea gigas (Wang et al., 2016; Cao et al., 2018) while there was no significant increase ROS production in adult Crassostrea gigas (Wang et al., 2020), Saccostrea glomerata (Goncalves et al., 2016, 2017) under long term exposure to low pH. The up-regulation of these genes in low-pH seawater suggests that oxidative stress is induced in mollusks, especially in oysters, by intracellular acidosis and inefficient electron transport (Tomanek et al., 2011). Increased oxidative damage caused by low pH also induced the expression of ubiquitin-related genes in this study, including Ubiquitin-like modifier-activating enzyme 1, Ubiquitin-conjugating enzyme E2, E3 ubiquitin-protein ligase SHPRH, UBR4, HRD1, and de-ubiquitinase BRCC36. On contrary, NADH dehydrogenase ubiquinone and oxidoreductase activity were down-regulated in Crassostrea gigas larvae prior to shell formation (Liu et al., 2020). Considering the crucial role of ubiquitin system in survival against many environmental stressors, including oxidation, activation of ubiquitin system may contribute to host protection from OA-derived stress (Hanna et al., 2007). Activation of ubiquitination protein in Crassostrea hongkongensis (Lim et al., 2021) and up-regulation of oxidoreductase activity was observed in Crassostrea virgininca (Downey-Wall et al., 2020). Lysosomal genes such as Cathepsin L1, F and B, also were found up-regulated (≥2fold) in this study. Studies using different forms of stressors including temperature (Zhang et al., 2006), pollutants (Sheehan and Power, 1999) and salinity (Stickle et al., 1985) have observed lysosomal membrane destabilization leading to the release of lysosomal enzymes into the cytoplasm (Moore et al., 1982). In this study, the oyster larvae displayed lysosomal destabilization in response to low pH stress. Increased activity of lysosomal enzymes plays a vital role in the mobilization of energy reserves to ensure normal metabolic function during periods of stress in bivalves (Tremblay et al., 1998). However, excessive lysosomal activity may cause drastic changes in the cellular activities leading to apoptosis (Zhao et al., 2003; Sokolova, 2009). Similarly, the highly active, autolytic, arylsulfatases, were found to be up-regulated by fourfold in this study. Spinal cord injury in mice causes local tissue acidification and stimulates increased arylsulfatase B activity to improve locomotor function (Yoo et al., 2013). Therefore, it can be assumed that increased catabolism triggered by lysosomal enzymes may help to maintain the cellular metabolism in low pH-stressed oyster larvae. The increased expression of the 40S and 60S ribosomal subunits revealed in the study also appears to suggest an increase in energy requirements during low pH stress.
Signal Transduction and Calcification Regulatory Genes
In total, 79 DEGs were assigned into four main signaling pathways, including the MAPK signaling pathway (ko04010), the Wnt signaling pathway (ko04310), the vascular endothelial growth factor (VEGF) signaling pathway (ko04370) and the ErbB signaling pathway (ko04012). Among these pathways, the MAPK signaling pathway was significantly activated with 67.6% of total mapped DEGs expressed after low pH treatment. Several MAPK genes are highly expressed in the oyster larvae with a strong coverage in the transcriptome dataset.
The activation of MAPK genes may disrupt many signal transduction pathways in cells including immune response, cell death and other kinase-mediated signaling cascades. Induction of MAPK gene pathways has proven to be a useful biomarker of the responses of marine invertebrates to a variety of environmental stressors (Canesi et al., 2006; Hardege et al., 2011), and MAPK pathways could thus provide information on the responses of early stage oyster larvae to low-pH exposure. However, the Wnt, VEGF and ErbB signaling pathways appeared to be suppressed, with 86.7, 83.3, and 83.3% of the constituent genes down-regulated.
Genes related to calcium transport encoding cAMP-dependent protein kinase catalytic subunit, calcium/calmodulin-dependent protein kinase type II subunit delta and calcineurin subunit B type 1 and Wnt genes were found to be down-regulated during acidification stress in the present study. Wnt genes encode highly conserved signaling molecules that play an important role in embryogenesis, cell differentiation and osteogenesis in both vertebrates and invertebrates (Angers and Moon, 2009; De Robertis, 2010; Holstein, 2012; Karako-Lampert et al., 2014). Decreased calcification and retarded growth might result from the differential expression pattern of Wnt genes under ocean acidification as evidenced from the studies in coral holobiont exposed to low-pH conditions (Karako-Lampert et al., 2014). Recent studies reported hypermethylation of Wnt receptor catabolic process, calcium ion binding, calcium dependent phosphorylation in pediveliger larvae of Crassostrea hongkongensis when exposed to ocean acidification (Lim et al., 2021). Pacific oyster larvae at 14–16 h post fertilization, showed a delay in shell formation with an increase in the expression of tyrosinase gene, carbonic anhydrase, calcium and chitin binding proteins when exposed to pH of 7.5 and 7.6 (De Wit et al., 2018). The expression of genes related to key process in shell formation including calcium transportation, bicarbonate transport and organic matrix where significantly inhibited at the stage of initial shell formation (PDI) of pacific oyster larvae, when exposed to low pH 7.4 (Liu et al., 2020) disrupting the mobilization of calcium ions. Supplementary Table 4 provides complete literature of genes and pathways regulated in response to ocean acidification in marine bivalves across life stages. The expression levels of genes related to chitin synthesis was significantly decreased while the expression of genes related to chitin degradation was found to be increased in D shaped larvae of Crassostrea gigas at pH 7.4, negatively effecting chitin biosynthesis and hence biomineralization (Zhang et al., 2019). In adult pacific oyster, a series of calcium binding proteins and calcium signaling pathways were up-regulated in response to OA indicating regulation of calcium homeostasis under OA (Wang et al., 2020), also in larval sea-urchins (Evans et al., 2013) on the contrary, calcium dependent signaling pathway were suppressed in pearl oyster Pinctada fucata under low pH (Li et al., 2016b) but whereas in case of Crassostrea virginica, no significant expression of these genes was observed upon exposure to ocean acidification (Downey-Wall et al., 2020). The stimulation of calcium related genes as reported in some might be helping the larvae to survive and grow under low pH but in a slower rate (De Wit et al., 2018). However, the initial shell formation requires endogenous energy from metabolic process, the metabolic suppression can lead to alteration in the energy allocation to shell formation delaying or inhibiting the initial shell synthesis (Liu et al., 2020).
Similarly, VEGF and ErbB pathway genes involved in invertebrate cell proliferation and angiogenesis were also down-regulated, suggesting that decreased calcification affects building and repair of skeletal tissues (Green et al., 2013). Most molluscan species have evolved to cope with severe environmental conditions by suppressing their metabolic rate. The degree of metabolic suppression is controlled by the down-regulation of phosphorylation inside the cell (Hochachka et al., 1993). Among the ErbB genes, the negative regulation of the serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform in the low-pH environment may affect the relative activities of phosphatases initiating metabolic suppression (Hand and Hardewig, 1996).
Validation of RNA-Seq by RT-qPCR
Eight differentially expressed genes obtained by RNA sequencing in response to OA exposure were selected to confirm the accuracy of gene expression by RT-qPCR approach. Further, the fold changes for the validated genes were shown to have a slight variation in their relative gene expression levels when compared between RNA-Seq and RT-qPCR methods (Figure 5).
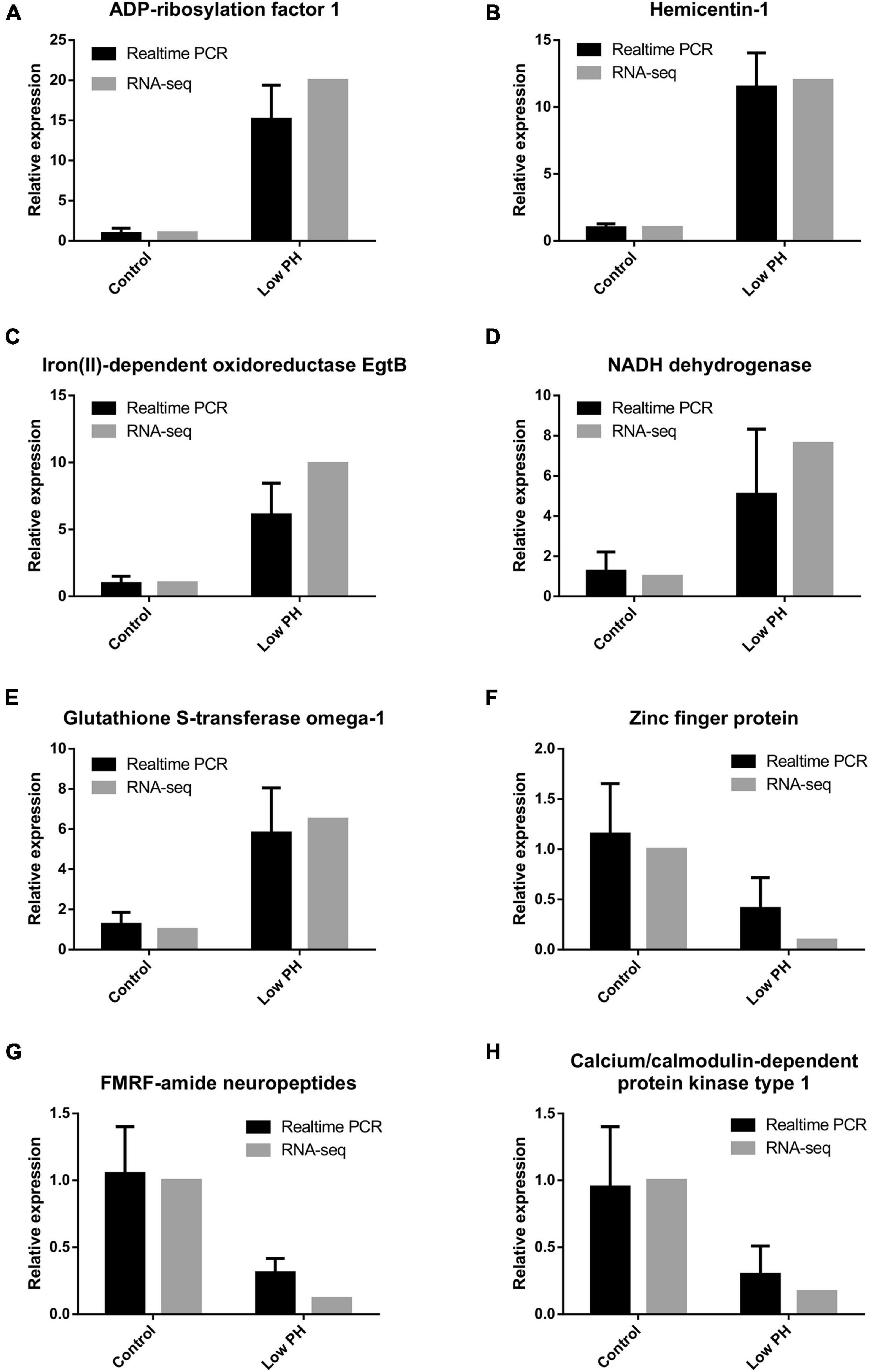
Figure 5. (A–H) Comparison of eight differentially expressed genes analyzed using RNA-seq and RT-qPCR method.
Conclusion
This study described a unique transcriptome profiling analysis of pediveliger larvae exposed to low pH and also provided comprehensive insights into the mechanism underlying their tolerance to extremely decreased pH. We identified a number of OA-responsive genes that were enriched in various pathways involved in controlling basic physiological processes. In particular, our results revealed that oyster larvae may adopt an energy “trade-off” strategy through the suppression of energy-demanding physiological processes, such as immune responses, calcification or shell growth, to overcome the stress induced by OA. In addition, our transcriptome data analyses could also provide a substantial and valuable genomic resource for further studies of this ecologically and economically important aquatic species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: SRR960964 and SRR967068).
Author Contributions
RD and YZ designed the research, analyzed the data, and drafted the final version of the manuscript. RD performed the larval culture experiments with SX, GK, and VT. RD and JL performed transcriptome analysis with the help of YZ and ZY. RD and KS performed the literature review and discussion of the results. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Science Foundation of China (No. 32073002). The larval rearing and ocean acidification facility of this project was funded by a three GRF grants from the HKSAR-RGC (Grant Nos. 17303517, 17304619, and 17120120).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Mr. Tuo Yao for helping in the animal collection from Tsingdao.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.782583/full#supplementary-material
Supplementary Table 1 | List of all BLAST matched DEGs with Swissprot database (XLSX).
Supplementary Table 2 | List of GO classification of all DEGs (XLSX).
Supplementary Table 3 | List of all enriched pathway under seawater acidification (XLSX).
Supplementary Table 4 | Genes and pathways regulated in response to ocean acidification in marine bivalves across life stages (docx).
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ http://hannonlab.cshl.edu/fastx_toolkit/
- ^ http://gigadb.org/dataset/10030
- ^ http://www.genome.jp/kegg/kaas/
- ^ http://apps.cytoscape.org/apps/cluego
References
Angers, S., and Moon, R. T. (2009). Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477.
Beniash, E., Ivanina, A., Lieb, N. S., Kurochkin, I., and Sokolova, I. M. (2010). Elevated level of carbon dioxide affects metabolism and shell formation in oysters Crassostrea virginica. Mar. Ecol. Prog. Ser. 419, 95–108.
Bibby, R., Cleall-Harding, P., Rundle, S., Widdicombe, S., and Spicer, J. (2007). Ocean acidification disrupts induced defences in the intertidal gastropod Littorina littorea. Biol. Lett. 3, 699–701. doi: 10.1098/rsbl.2007.0457
Bibby, R., Widdicombe, S., Parry, H., Spicer, J., and Pipe, R. (2008). Effects of ocean acidification on the immune response of the blue mussel Mytilus edulis. Aquat. Biol. 2, 67–74.
Bindea, G., Galon, J., and Mlecnik, B. (2013). CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663. doi: 10.1093/bioinformatics/btt019
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Breese, W. P., and Malouf, R. E. (1975). Hatchery Manual for the Pacific Oyster. ORESU-H-75-002. Corvallis: Oregon State University Sea Grant College Program.
Caldeira, K., and Wickett, M. E. (2005). Ocean model predictions of chemistry changes from carbon dioxide emissions to the atmosphere and ocean. J. Geophys. Res. Oceans 110:2671.
Cancio, I., and Cajaraville, M. P. (1997). Histochemistry of oxidases in several tissues of bivalve molluscs. Cell Biol. Int. 21, 575–584. doi: 10.1006/cbir.1997.0182
Canesi, L., Betti, M., Ciacci, C., Lorusso, L., Pruzzo, C., and Gallo, G. (2006). Cell signalling in the immune response of mussel hemocytes. Invertebr. Surv. J. 3, 40–49.
Cao, R., Liu, Y., Wang, Q., Zhang, Q., Yang, D., Liu, H., et al. (2018). The impact of ocean acidification and cadmium on the immune responses of Pacific oyster, Crassostrea gigas. Fish Shellf. Immunol. 81, 456–462.
Castillo, N., Saavedra, L. M., Vargas, C. A., Gallardo-Escárate, C., and Détrée, C. (2017). Ocean acidification and pathogen exposure modulate the immune response of the edible mussel Mytilus chilensis. Fish Shellf. Immunol. 70, 149–155. doi: 10.1016/j.fsi.2017.08.047
Chamberlin, M. E. (2004). Top-down control analysis of the effect of temperature on ectotherm oxidative phosphorylation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287, R794–R800. doi: 10.1152/ajpregu.00240.2004
Comeau, S., Jeffree, R., Teyssié, J.-L., and Gattuso, J.-P. (2010). Response of the Arctic pteropod Limacina helicina to projected future environmental conditions. PLoS One 5:e11362. doi: 10.1371/journal.pone.0011362
Cooley, S. R., Kite-Powell, H. L., and Doney, S. C. (2009). Ocean acidification’s potential to alter global marine ecosystem services. Oceanography 22, 172–181. doi: 10.5670/oceanog.2009.106
Davies, S. W., Marchetti, A., Ries, J. B., and Castillo, K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3:112. doi: 10.3389/fmars.2016.00112
De Robertis, E. M. (2010). Wnt signaling in axial patterning and regeneration: lessons from planaria. Sci. Signal. 3:e21. doi: 10.1126/scisignal.3127pe21
De Wit, P., Durland, E., Ventura, A., and Langdon, C. J. (2018). Gene expression correlated with delay in shell formation in larval Pacific oysters (Crassostrea gigas) exposed to experimental ocean acidification provides insights into shell formation mechanisms. BMC Genomics 19:160. doi: 10.1186/s12864-018-4519-y
Devens, H. R., Davidson, P. L., Deaker, D. J., Smith, K. E., Wray, G. A., and Byrne, M. (2020). Ocean acidification induces distinct transcriptomic responses across life history stages of the sea urchin Heliocidaris erythrogramma. Mol. Ecol. 29, 4618–4636. doi: 10.1111/mec.15664
Dickson, A., and Millero, F. J. (1987). A comparison of the equilibrium constants for the dissociation of carbonic acid in seawater media. Deep Sea Res. A Oceanogr. Res. Pap. 34, 1733–1743.
Dineshram, R., Chandramouli, K., Ko, G. W. K., Zhang, H., Qian, P. Y., Ravasi, T., et al. (2016). Quantitative analysis of oyster larval proteome provides new insights into the effects of multiple climate change stressors. Global Change Biol. 22, 2054–2068. doi: 10.1111/gcb.13249
Dineshram, R., Sharma, R., Chandramouli, K., Yalamanchili, H. K., Chu, I., and Thiyagarajan, V. (2015). Comparative and quantitative proteomics reveal the adaptive strategies of oyster larvae to ocean acidification. Proteomics 15, 4120–4134. doi: 10.1002/pmic.201500198
Dineshram, R., Thiyagarajan, V., Lane, A., Ziniu, Y., Xiao, S., and Leung, P. T. (2013). Elevated CO2 alters larval proteome and its phosphorylation status in the commercial oyster, Crassostrea hongkongensis. Mar. Biol. 160, 2189–2205. doi: 10.1007/s00227-013-2176-x
Dineshram, R., Wong, K. K., Xiao, S., Yu, Z., Qian, P. Y., and Thiyagarajan, V. (2012). Analysis of Pacific oyster larval proteome and its response to high-CO2. Mar. Pollut. Bull. 64, 2160–2167. doi: 10.1016/j.marpolbul.2012.07.043
Doney, S. C., Busch, D. S., Cooley, S. R., and Kroeker, K. J. (2020). The impacts of ocean acidification on marine ecosystems and reliant human communities. Annu. Rev. Environ. Resour. 45, 83–112. doi: 10.1146/annurev-environ-012320-083019
Downey-Wall, A. M., Cameron, L. P., Ford, B. M., Mcnally, E. M., Venkataraman, Y. R., Roberts, S. B., et al. (2020). Ocean acidification induces subtle shifts in gene expression and DNA methylation in mantle tissue of the Eastern oyster (Crassostrea virginica). Front. Mar. Sci. 7:566419. doi: 10.3389/fmars.2020.566419
Dupont, S., Havenhand, J., Thorndyke, W., Peck, L., and Thorndyke, M. (2008). Near-future level of CO2-driven ocean acidification radically affects larval survival and development in the brittlestar Ophiothrix fragilis. Mar. Ecol. Prog. Ser. 373, 285–294. doi: 10.3354/meps07800
Esbaugh, A. J., Heuer, R., and Grosell, M. (2012). Impacts of ocean acidification on respiratory gas exchange and acid–base balance in a marine teleost, Opsanus beta. J. Comp. Physiol. B 182, 921–934. doi: 10.1007/s00360-012-0668-5
Evans, T. G., Chan, F., Menge, B. A., and Hofmann, G. E. (2013). Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Mol. Ecol. 22, 1609–1625. doi: 10.1111/mec.12188
Fabry, V., Seibel, B., Feely, R., and Orr, J. (2008). Impacts of ocean acidification on marine fauna and ecosystem processes. ICES J. Mar. Sci. 65:414. doi: 10.1093/icesjms/fsn048
Falkowski, P., Scholes, R. J., Boyle, E. E. A., Canadell, J., Canfield, D., Elser, J., et al. (2000). The global carbon cycle: a test of our knowledge of earth as a system. Science 290, 291–296. doi: 10.1126/science.290.5490.291
Fernández-Reiriz, M. J., Range, P., Álvarez-Salgado, X. A., Espinosa, J., and Labarta, U. (2012). Tolerance of juvenile Mytilus galloprovincialis to experimental seawater acidification. Mar. Ecol. Prog. Ser. 454, 65–74. doi: 10.3354/meps09660
French, S. S., Moore, M. C., and Demas, G. E. (2009). Ecological immunology: the organism in context. Integr. Comp. Biol. 49, 246–253.
Gazeau, F., Gattuso, J.-P., Dawber, C., Pronker, A., Peene, F., Peene, J., et al. (2010). Effect of ocean acidification on the early life stages of the blue mussel Mytilus edulis. Biogeosciences 7, 2051–2060. doi: 10.5194/bg-7-2051-2010
Gazeau, F., Parker, L. M., Comeau, S., Gattuso, J.-P., O’connor, W. A., Martin, S., et al. (2013). Impacts of ocean acidification on marine shelled molluscs. Mar. Biol. 160, 2207–2245. doi: 10.1007/s00227-013-2219-3
Gazeau, F., Quiblier, C., Jansen, J. M., Gattuso, J. P., Middelburg, J. J., and Heip, C. H. (2007). Impact of elevated CO2 on shellfish calcification. Geophys. Res. Lett. 34:e028554.
Ginger, K. W., Vera, C. B., Dennis, C. K., Adela, L. J., Yu, Z., and Thiyagarajan, V. (2013). Larval and post-larval stages of Pacific oyster (Crassostrea gigas) are resistant to elevated CO2. PLoS One 8:e64147. doi: 10.1371/journal.pone.0064147
Goncalves, P., Anderson, K., Thompson, E. L., Melwani, A., Parker, L. M., Ross, P. M., et al. (2016). Rapid transcriptional acclimation following transgenerational exposure of oysters to ocean acidification. Mol. Ecol. 25, 4836–4849. doi: 10.1111/mec.13808
Goncalves, P., Jones, D. B., Thompson, E. L., Parker, L. M., Ross, P. M., and Raftos, D. A. (2017). Transcriptomic profiling of adaptive responses to ocean acidification. Mol. Ecol. 26, 5974–5988. doi: 10.1111/mec.14333
González-Pech, R. A., Vargas, S., Francis, W. R., and Wörheide, G. (2017). Transcriptomic resilience of the Montipora digitata holobiont to low pH. Front. Mar. Sci. 4:403. doi: 10.3389/fmars.2017.00403
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Green, D. W., Padula, M. P., Santos, J., Chou, J., Milthorpe, B., and Ben-Nissan, B. (2013). A therapeutic potential for marine skeletal proteins in bone regeneration. Mar. Drugs 11, 1203–1220. doi: 10.3390/md11041203
Griffith, A. W., Harke, M. J., DePasquale, E., Berry, D. L., and Gobler, C. J. (2019). The harmful algae, Cochlodinium polykrikoides and Aureococcus anophagefferens, elicit stronger transcriptomic and mortality response in larval bivalves (Argopecten irradians) than climate change stressors. Ecol. Evol. 9, 4931–4948. doi: 10.1002/ece3.5100
Guinotte, J. M., and Fabry, V. J. (2008). Ocean acidification and its potential effects on marine ecosystems. Ann. N. Y. Acad. Sci. 1134, 320–342. doi: 10.1196/annals.1439.013
Hamilton, T. J., Holcombe, A., and Tresguerres, M. (2014). CO2-induced ocean acidification increases anxiety in Rockfish via alteration of GABA A receptor functioning. Proc. Biol. Sci. 281:20132509. doi: 10.1098/rspb.2013.2509
Hand, S. C., and Hardewig, I. (1996). Downregulation of cellular metabolism during environmental stress: mechanisms and implications. Annu. Rev. Physiol. 58, 539–563. doi: 10.1146/annurev.ph.58.030196.002543
Hanna, J., Meides, A., Zhang, D. P., and Finley, D. (2007). A ubiquitin stress response induces altered proteasome composition. Cell 129, 747–759. doi: 10.1016/j.cell.2007.03.042
Hardege, J., Rotchell, J., Terschak, J., and Greenway, G. (2011). Analytical challenges and the development of biomarkers to measure and to monitor the effects of ocean acidification. TrAC Trends Anal. Chem. 30, 1320–1326. doi: 10.1016/j.trac.2011.07.004
Hernroth, B., Baden, S., Thorndyke, M., and Dupont, S. (2011). Immune suppression of the echinoderm Asterias rubens (L.) following long-term ocean acidification. Aquat. Toxicol. 103, 222–224. doi: 10.1016/j.aquatox.2011.03.001
Hochachka, P. W., Lutz, P. L., Sick, T. J., and Rosenthal, M. (1993). Surviving Hypoxia: Mechanisms of Control and Adaptation. Boca Raton, FL: CRC Press.
Holstein, T. W. (2012). The evolution of the Wnt pathway. Cold Spring Harb. Perspect. Biol. 4:a007922.
Honda, S., Kashiwagi, M., Miyamoto, K., Takei, Y., and Hirose, S. (2000). Multiplicity, structures, and endocrine and exocrine natures of eel fucose-binding lectins. J. Biol. Chem. 275, 33151–33157. doi: 10.1074/jbc.M002337200
Hüning, A. K., Melzner, F., Thomsen, J., Gutowska, M. A., Krämer, L., Frickenhaus, S., et al. (2013). Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: implications for shell formation and energy metabolism. Mar. Biol. 160, 1845–1861.
Hurd, C. L., Beardall, J., Comeau, S., Cornwall, C. E., Havenhand, J. N., Munday, P. L., et al. (2019). Ocean acidification as a multiple driver: how interactions between changing seawater carbonate parameters affect marine life. Mar. Freshw. Res. 71, 263–274. doi: 10.1111/gcb.15223
Ivanina, A. V., Kurochkin, I. O., Leamy, L., and Sokolova, I. M. (2012). Effects of temperature and cadmium exposure on the mitochondria of oysters (Crassostrea virginica) exposed to hypoxia and subsequent reoxygenation. J. Exp. Biol. 215, 3142–3154. doi: 10.1242/jeb.071357
Jutfelt, F., Bresolin De Souza, K., Vuylsteke, A., and Sturve, J. (2013). Behavioural disturbances in a temperate fish exposed to sustained high-CO2 levels. PLoS One 8:e65825. doi: 10.1371/journal.pone.0065825
Kaniewska, P., Campbell, P. R., Kline, D. I., Rodriguez-Lanetty, M., Miller, D. J., Dove, S., et al. (2012). Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS One 7:e34659. doi: 10.1371/journal.pone.0034659
Kaniewska, P., Chan, C.-K. K., Kline, D., Ling, E. Y. S., Rosic, N., Edwards, D., et al. (2015). Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PLoS One 10:e0139223. doi: 10.1371/journal.pone.0139223
Karako-Lampert, S., Zoccola, D., Salmon-Divon, M., Katzenellenbogen, M., Tambutté, S., Bertucci, A., et al. (2014). Transcriptome analysis of the scleractinian coral Stylophora pistillata. PLoS One 9:e88615. doi: 10.1371/journal.pone.0088615
Ko, G. W., Dineshram, R., Campanati, C., Chan, V. B., Havenhand, J., and Thiyagarajan, V. (2014). Interactive effects of ocean acidification, elevated temperature, and reduced salinity on early-life stages of the Pacific oyster. Environ. Sci. Technol. 48, 10079–10088. doi: 10.1021/es501611u
Kroeker, K. J., Kordas, R. L., Crim, R. N., and Singh, G. G. (2010). Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol. Lett. 13, 1419–1434. doi: 10.1111/j.1461-0248.2010.01518.x
Kroeker, K. J., Micheli, F., Gambi, M. C., and Martz, T. R. (2011). Divergent ecosystem responses within a benthic marine community to ocean acidification. Proc. Natl. Acad. Sci. U.S.A. 108, 14515–14520. doi: 10.1073/pnas.1107789108
Kurihara, H., Kato, S., and Ishimatsu, A. (2007). Effects of increased seawater pCO2 on early development of the oyster Crassostrea gigas. Aquat. Biol. 1, 91–98. doi: 10.3354/ab00009
Kurihara, H., and Shirayama, Y. (2004). Effects of increased atmospheric CO2 on sea urchin early development. Mar. Ecol. Prog. Ser. 274, 161–169.
Lannig, G., Eilers, S., Pörtner, H. O., Sokolova, I. M., and Bock, C. (2010). Impact of ocean acidification on energy metabolism of oyster, Crassostrea gigas—changes in metabolic pathways and thermal response. Mar. Drugs 8, 2318–2339. doi: 10.3390/md8082318
Li, S., Huang, J., Liu, C., Liu, Y., Zheng, G., Xie, L., et al. (2016a). Interactive effects of seawater acidification and elevated temperature on the transcriptome and biomineralization in the pearl oyster Pinctada fucata. Environ. Sci. Technol. 50, 1157–1165. doi: 10.1021/acs.est.5b05107
Li, S., Liu, C., Huang, J., Liu, Y., Zhang, S., Zheng, G., et al. (2016b). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6, 1–10. doi: 10.1038/srep18943
Lim, Y.-K., Cheung, K., Dang, X., Roberts, S. B., Wang, X., and Thiyagarajan, V. (2021). DNA methylation changes in response to ocean acidification at the time of larval metamorphosis in the edible oyster, Crassostrea hongkongensis. Mar. Environ. Res. 163:105214. doi: 10.1016/j.marenvres.2020.105214
Liu, P., Keller, J. R., Ortiz, M., Tessarollo, L., Rachel, R. A., Nakamura, T., et al. (2003). Bcl11a is essential for normal lymphoid development. Nat. Immunol. 4, 525–532. doi: 10.1038/ni925
Liu, Z., Zhang, Y., Zhou, Z., Zong, Y., Zheng, Y., Liu, C., et al. (2020). Metabolomic and transcriptomic profiling reveals the alteration of energy metabolism in oyster larvae during initial shell formation and under experimental ocean acidification. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-62963-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McCulloch, M., Falter, J., Trotter, J., and Montagna, P. (2012). Coral resilience to ocean acidification and global warming through pH up-regulation. Nat. Clim. Change 2, 623–627. doi: 10.1038/srep42405
Mehrbach, C., Culberson, C., Hawley, J., and Pytkowicx, R. (1973). Measurement of the apparent dissociation constants of carbonic acid in seawater at atmospheric pressure 1. Limnol. Oceanogr. 18, 897–907.
Melzner, F., Mark, F. C., Seibel, B. A., and Tomanek, L. (2020). Ocean acidification and coastal marine invertebrates: tracking CO2 effects from seawater to the cell. Annu. Rev. Mar. Sci. 12, 499–523. doi: 10.1146/annurev-marine-010419-010658
Miller, A. W., Reynolds, A. C., Sobrino, C., and Riedel, G. F. (2009). Shellfish face uncertain future in high CO2 world: influence of acidification on oyster larvae calcification and growth in estuaries. PLoS One 4:e5661. doi: 10.1371/journal.pone.0005661
Moore, M. N., Pipe, R. K., and Farrar, S. V. (1982). Lysosomal and microsomal responses to environmental factors in Littorina littorea from Sullom Voe. Mar. Pollut. Bull. 13, 340–345.
Moya, A., Huisman, L., Ball, E., Hayward, D., Grasso, L., Chua, C., et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol. Ecol. 21, 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Munday, P. L., Gagliano, M., Donelson, J. M., Dixson, D. L., and Thorrold, S. R. (2011). Ocean acidification does not affect the early life history development of a tropical marine fish. Mar. Ecol. Prog. Ser. 423, 211–221.
Nakamura, M., Ohki, S., Suzuki, A., and Sakai, K. (2011). Coral larvae under ocean acidification: survival, metabolism, and metamorphosis. PLoS One 6:e14521. doi: 10.1371/journal.pone.0014521
O’Donnell, M. J., Todgham, A. E., Sewell, M. A., Hammond, L. M., Ruggiero, K., Fangue, N. A., et al. (2010). Ocean acidification alters skeletogenesis and gene expression in larval sea urchins. Mar. Ecol. Prog. Ser. 398, 157–171.
Padilla-Gamiño, J. L., Kelly, M. W., Evans, T. G., and Hofmann, G. E. (2013). Temperature and CO2 additively regulate physiology, morphology and genomic responses of larval sea urchins, Strongylocentrotus purpuratus. Proc. R. Soc. B Biol. Sci. 280:20130155. doi: 10.1098/rspb.2013.0155
Parker, L. M., Ross, P. M., and O’connor, W. A. (2009). The effect of ocean acidification and temperature on the fertilization and embryonic development of the Sydney rock oyster Saccostrea glomerata (Gould 1850). Glob. Change Biol. 15, 2123–2136.
Parker, L. M., Ross, P. M., and O’connor, W. A. (2010). Comparing the effect of elevated pCO2 and temperature on the fertilization and early development of two species of oysters. Mar. Biol. 157, 2435–2452.
Parker, L. M., Ross, P. M., and O’connor, W. A. (2011). Populations of the Sydney rock oyster, Saccostrea glomerata, vary in response to ocean acidification. Mar. Biol. 158, 689–697. doi: 10.1007/s00227-010-1592-4
Parker, L. M., Ross, P. M., O’connor, W. A., Borysko, L., Raftos, D. A., and Pörtner, H. O. (2012). Adult exposure influences offspring response to ocean acidification in oysters. Glob. Change Biol. 18, 82–92. doi: 10.1016/j.scitotenv.2021.146704
Parker, L. M., Ross, P. M., O’connor, W. A., Pörtner, H. O., Scanes, E., and Wright, J. M. (2013). Predicting the response of molluscs to the impact of ocean acidification. Biology 2, 651–692.
Pelletier, G., Lewis, E., and Wallace, D. (2007). CO2SYS. XLS: A Calculator for the CO2 System in Seawater for Microsoft Excel/VBA. Washington, DC: Washington State Department of Ecology.
Peng, C., Zhao, X., Liu, S., Shi, W., Han, Y., Guo, C., et al. (2017). Ocean acidification alters the burrowing behaviour, Ca2+/Mg2+-ATPase activity, metabolism, and gene expression of a bivalve species, Sinonovacula constricta. Mar. Ecol. Prog. Ser. 575, 107–117. doi: 10.3354/meps12224
Raven, J., Caldeira, K., Elderfield, H., Hoegh-Guldberg, O., Liss, P., Riebesell, U., et al. (2005). Ocean Acidification Due to Increasing Atmospheric Carbon Dioxide. London: The Royal Society.
Rivest, E. B., Kelly, M. W., Debiasse, M. B., and Hofmann, G. E. (2018). Host and symbionts in Pocillopora damicornis larvae display different transcriptomic responses to ocean acidification and warming. Front. Mar. Sci. 5:186. doi: 10.3389/fmars.2018.00186
Rocker, M. M., Noonan, S., Humphrey, C., Moya, A., Willis, B. L., and Bay, L. K. (2015). Expression of calcification and metabolism-related genes in response to elevated pCO2 and temperature in the reef-building coral Acropora millepora. Mar. Genom. 24, 313–318. doi: 10.1016/j.margen.2015.08.001
Ross, P. M., Parker, L., O’connor, W. A., and Bailey, E. A. (2011). The impact of ocean acidification on reproduction, early development and settlement of marine organisms. Water 3, 1005–1030. doi: 10.1073/pnas.1007273107
Sheehan, D., and Power, A. (1999). Effects of seasonality on xenobiotic and antioxidant defence mechanisms of bivalve molluscs. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 123, 193–199. doi: 10.1016/s0742-8413(99)00033-x
Sheppard Brennand, H., Soars, N., Dworjanyn, S. A., Davis, A. R., and Byrne, M. (2010). Impact of ocean warming and ocean acidification on larval development and calcification in the sea urchin Tripneustes gratilla. PLoS One 5:e11372. doi: 10.1371/journal.pone.0011372
Stickle, W., Moore, M., and Bayne, B. (1985). Effects of temperature, salinity and aerial exposure on predation and lysosomal stability of the dogwhelk Thais (Nucella) lapillus (L.). J. Exp. Mar. Biol. Ecol. 93, 235–258. doi: 10.1016/0022-0981(85)90242-4
Strader, M. E., Wong, J. M., and Hofmann, G. E. (2020). Ocean acidification promotes broad transcriptomic responses in marine metazoans: a literature survey. Front. Zool. 17:7. doi: 10.1186/s12983-020-0350-9
Stumpp, M., Wren, J., Melzner, F., Thorndyke, M., and Dupont, S. (2011). CO2 induced seawater acidification impacts sea urchin larval development I: elevated metabolic rates decrease scope for growth and induce developmental delay. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 160, 331–340. doi: 10.1016/j.cbpa.2011.06.022
Talmage, S. C., and Gobler, C. J. (2009). The effects of elevated carbon dioxide concentrations on the metamorphosis, size, and survival of larval hard clams (Mercenaria mercenaria), bay scallops (Argopecten irradians), and Eastern oysters (Crassostrea virginica). Limnol. Oceanogr. 54, 2072–2080. doi: 10.4319/lo.2009.54.6.2072
Thomsen, J., and Melzner, F. (2010). Moderate seawater acidification does not elicit long-term metabolic depression in the blue mussel Mytilus edulis. Mar. Biol. 157, 2667–2676.
Thomsen, J., Ramesh, K., Sanders, T., Bleich, M., and Melzner, F. (2018). Calcification in a marginal sea–influence of seawater [Ca2 +] and carbonate chemistry on bivalve shell formation. Biogeosciences 15, 1469–1482.
Todgham, A. E., and Hofmann, G. E. (2009). Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. J. Exp. Biol. 212, 2579–2594. doi: 10.1242/jeb.032540
Todgham, A. E., and Stillman, J. H. (2013). Physiological responses to shifts in multiple environmental stressors: relevance in a changing world. Integr. Comp. Biol. 53, 539–544. doi: 10.1093/icb/ict086
Tomanek, L., Zuzow, M. J., Ivanina, A. V., Beniash, E., and Sokolova, I. M. (2011). Proteomic response to elevated pCO2 level in eastern oysters, Crassostrea virginica: evidence for oxidative stress. J. Exp. Biol. 214, 1836–1844. doi: 10.1242/jeb.055475
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tremblay, R., Myrand, B., and Guderley, H. (1998). Temporal variation of lysosomal capacities in relation to susceptibility of mussels, Mytilus edulis, to summer mortality. Mar. Biol. 132, 641–649.
Vidal-Dupiol, J., Zoccola, D., Tambutté, E., Grunau, C., Cosseau, C., Smith, K. M., et al. (2013). Genes related to ion-transport and energy production are upregulated in response to CO2-driven pH decrease in corals: new insights from transcriptome analysis. PLoS One 8:e58652. doi: 10.1371/journal.pone.0058652
Waldbusser, G. G., Voigt, E. P., Bergschneider, H., Green, M. A., and Newell, R. I. (2011). Biocalcification in the eastern oyster (Crassostrea virginica) in relation to long-term trends in Chesapeake Bay pH. Estuar. Coasts 34, 221–231.
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Q., Cao, R., Ning, X., You, L., Mu, C., Wang, C., et al. (2016). Effects of ocean acidification on immune responses of the Pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 49, 24–33. doi: 10.1016/j.fsi.2015.12.025
Wang, X., Wang, M., Wang, W., Liu, Z., Xu, J., Jia, Z., et al. (2020). Transcriptional changes of Pacific oyster Crassostrea gigas reveal essential role of calcium signal pathway in response to CO2-driven acidification. Sci. Total Environ. 741:140177. doi: 10.1016/j.scitotenv.2020.140177
Widdicombe, S., and Spicer, J. I. (2008). Predicting the impact of ocean acidification on benthic biodiversity: what can animal physiology tell us? J. Exp. Mar. Biol. Ecol. 366, 187–197. doi: 10.1016/j.jembe.2008.07.024
Xing, Q., Wang, J., Zhao, Q., Liao, H., Xun, X., Yang, Z., et al. (2019). Alternative splicing, spatiotemporal expression of TEP family genes in Yesso scallop (Patinopecten yessoensis) and their disparity in responses to ocean acidification. Fish Shellfish Immunol. 95, 203–212. doi: 10.1016/j.fsi.2019.10.026
Yoo, M., Khaled, M., Gibbs, K. M., Kim, J., Kowalewski, B., Dierks, T., et al. (2013). Arylsulfatase B improves locomotor function after mouse spinal cord injury. PLoS One 8:e57415. doi: 10.1371/journal.pone.0057415
Zhai, W., Zheng, N., Huo, C., Xu, Y., Zhao, H., Li, Y. W., et al. (2013). Subsurface low pH and carbonate saturation state of aragonite on China side of the North Yellow Sea: combined effects of global atmospheric CO2 increase, regional environmental changes, and local biogeochemical processes. Biogeosciences 10, 3079–3120.
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang, Y., Liu, Z., Song, X., Huang, S., Wang, L., and Song, L. (2019). The inhibition of ocean acidification on the formation of oyster calcified shell by regulating the expression of Cgchs1 and Cgchit4. Front. Physiol. 10:1034. doi: 10.3389/fphys.2019.01034
Zhang, Z., Li, X., Vandepeer, M., and Zhao, W. (2006). Effects of water temperature and air exposure on the lysosomal membrane stability of hemocytes in Pacific oysters, Crassostrea gigas (Thunberg). Aquaculture 256, 502–509.
Zhao, M., Antunes, F., Eaton, J. W., and Brunk, U. T. (2003). Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. Eur. J. Biochem. 270, 3778–3786.
Zhao, X., Han, Y., Chen, B., Xia, B., Qu, K., and Liu, G. (2020). CO2-driven ocean acidification weakens mussel shell defense capacity and induces global molecular compensatory responses. Chemosphere 243:125415. doi: 10.1016/j.chemosphere.2019.125415
Keywords: Crassostrea gigas, pediveliger larvae, ocean acidification, transcriptome, biological pathways
Citation: Dineshram R, Xiao S, Ko GWK, Li J, Smrithi K, Thiyagarajan V, Zhang Y and Yu Z (2021) Ocean Acidification Triggers Cell Signaling, Suppress Immune and Calcification in the Pacific Oyster Larvae. Front. Mar. Sci. 8:782583. doi: 10.3389/fmars.2021.782583
Received: 24 September 2021; Accepted: 17 November 2021;
Published: 13 December 2021.
Edited by:
Pengzhi Qi, Zhejiang Ocean University, ChinaReviewed by:
Jianmin Zhao, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaYongbo Bao, Zhejiang Wanli University, China
Copyright © 2021 Dineshram, Xiao, Ko, Li, Smrithi, Thiyagarajan, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Zhang, eXpoYW5nQHNjc2lvLmFjLmNu; Ziniu Yu, Y2FybHp5dUBzY3Npby5hYy5jbg==
†These authors share first authorship
 Ramadoss Dineshram
Ramadoss Dineshram Shu Xiao
Shu Xiao Ginger Wai Kuen Ko
Ginger Wai Kuen Ko Jun Li
Jun Li Kovilingal Smrithi2
Kovilingal Smrithi2 Vengatesen Thiyagarajan
Vengatesen Thiyagarajan Yang Zhang
Yang Zhang Ziniu Yu
Ziniu Yu