
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 October 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.761628
This article is part of the Research Topic The Physiological and Molecular Response of Aquatic Animals to Environmental Stresses View all 23 articles
Ascidians, particularly those highly invasive ones, are typical fouling organisms to cause significantly negative ecological and economic influence in coastal ecosystems. Stolon, which is the unique structure of some solitary ascidians to complete the essential process of adhesion, possesses extremely high tolerance to environmental stresses during biofouling and invasions. However, the mechanisms underlying environmental tolerance remain largely unknown. Here, we used the quantitative proteomics technology, isobaric tags for relative and absolute quantitation (iTRAQ), to investigate the molecular response to environmental challenges (temperature and salinity) in the stolon of a highly invasive fouling ascidian, Ciona robusta. When compared with the control, a total of 75, 86, 123, and 83 differential abundance proteins were identified under low salinity, high salinity, low temperature, and high temperature stress, respectively. Bioinformatic analyses uncovered the key pathways under both temperature and salinity stresses, including “cytoskeleton,” “signal transduction,” and “posttranslational modification,” which were involved in stolon structure stability, protein synthesis, and stress response activation. Under the low salinity stress, the “extracellular matrix” pathway was identified to play a crucial role by regulating cell signal transduction and protein synthesis. To deal with the high salinity stress, stolon could store more energy by activating “carbohydrate/lipid transport” and “catabolism” pathways. The energy generated by “lipid metabolism” pathway might be beneficial to resist the low temperature stress. The upregulation of “cell cycle” pathway could inhibit cell growth, thus helping stolon conserve more energy against the high temperature stress. Our results here provide valuable references of candidate pathways and associated genes for studying mechanisms of harsh environmental adaptation and developing antifouling strategies in marine and coastal ecosystems.
Biofouling, the undesired adherence of fouling organisms on various submerged surfaces, is one of the most concerned environmental issues in global aquatic ecosystems (Bellard et al., 2013; Ricciardi et al., 2017; Briski et al., 2018). More than 4000 species have been recorded as biofoulers in both marine and freshwater ecosystems (Nakano and Strayer, 2014). Among these diverse organisms, mussels, barnacles, ascidians, sea stars, and tube worms are the most representative taxa (Flammang et al., 2015; Li X. et al., 2021). These taxa usually use their specialized organs/structures, such as mussel byssus (Andrade et al., 2015), sea star and sea urchin tube feet (Santos et al., 2013), ascidian stolon (Li et al., 2019), and barnacle base plate (von Byern and Grunwald, 2010), to firmly adhere to the surfaces of underwater substrates to achieve their successful biofouling. In order to solve the biofouling problem, it is a prerequisite to understanding the adhesive processes of these organs/structures and the associated mechanisms, as well as environment-organism interactions that can affect the adhesive abilities of fouling taxa (Li et al., 2019). In the last decade, numerous studies have confirmed that the underwater adhesion is largely biomacromolecule-mediated, and remarkable adhesive proteins, such as mussel foot proteins (Mfps), barnacle cement proteins, sea star footprint proteins, and sea urchin tube feet cement secreted by the adhesive organs/structures of these fouling organisms, have been identified as the crucial elements for marine biofouling (von Byern and Grunwald, 2010; Hennebert et al., 2012; Santos et al., 2013; Zhang et al., 2017b). Despite the fact that environmental changes also largely determine the successful biofouling (Chen et al., 2021), limited information is available on their influential mechanisms, particularly on the complex interaction mechanisms between environmental factors and adhesion-related proteins.
Studies have illustrated that environmental changes can affect biofouling by altering the expression and functions of adhesion-related proteins. For example, the fouling ability of the barnacle Balanus amphitrite decreased with temperature increased from 15 to 25°C, and such decreased ability of biofouling was owing to the expression changes of cement proteins (Johnston, 2010). The expression of some Mfps reduced significantly in the foot of Mytilus coruscus exposed to higher temperature, thus affecting the byssus production and further weakening biofouling (Li Y. F. et al., 2020). Other environmental stressors, such as pH (ocean acidification), nanoparticles, and microplastics, can also weaken the fouling strength by influencing adhesion-related protein expression (Hu et al., 2015; Scott et al., 2019; Khan et al., 2020; Shi et al., 2020). In addition, studies found that several metabolic pathways associated with adhesion were also involved in the response of the fouling organs/structures of marine organisms to environmental changes. “Osmoregulation” and “cell cycle” were identified as crucial common pathways in the feet of Mytilus galloprovincialis and Mytilus trossulus under salinity stresses (Lockwood and Somero, 2011). The pathways of “metabolic pathway,” “focal adhesion,” and “cytoskeleton” participated in the response to cadmium challenges in the foot of Perna viridis (Zhang et al., 2017c). Available evidence from these studies suggests that anti-stress strategies and associated adhesion-related protein responses should be taxa- or even organ/structure-specific in marine biofoulers, and more efforts are needed to comprehensively investigate common and specific mechanisms underlying environmental challenges.
Ascidians such as Ciona, Styela, Botrylus, and Didemnum are fouling taxa in coastal ecosystems (Dijkstra and Simkanin, 2016). Even worse, many species of these taxa are highly invasive, largely spreading the negative effects of biofouling and further threatening local biological communities and global industries including underwater facilities, shipping, and aquaculture (Cahill et al., 2012; Zhan et al., 2015; Yan et al., 2017; Kim et al., 2019). Among ascidians, C. robusta is a notorious invader for its extremely high biofouling capacity mainly derived from the rootlike fouling structure, the stolon (Li et al., 2019). Stolon can enlarge the binding area between the ascidian body and substrate surface by releasing adhesive proteins to enhance the interfacial adhesion (Pennati and Rothbächer, 2014; Ueki et al., 2018). The stolon can still maintain its structural stability and underwater adhesive ability under various challenges including temperature and salinity stresses, which has been used repeatedly when studying the response mechanisms of ascidians to environmental stresses (Renborg et al., 2014; Hawes et al., 2018; Huang et al., 2019; Li et al., 2019). Former studies have demonstrated that C. robusta body displayed high tolerance to temperature and salinity stresses, which were related to multiple layers of mechanisms such as rapid microevolution and adaptation, phenotypic plasticity, and metabolism trade-off (Renborg et al., 2014; Dijkstra and Simkanin, 2016; Hawes et al., 2018; Huang and Zhan, 2020; Chen et al., 2021). As a special adhesive structure, it remains unknown how C. robusta stolon, particularly the adhesion-related proteins in stolon, actively respond to environmental stressors. Recently, several adhesion-related proteins have been identified from C. robusta stolon using mass spectrometry technologies (Ueki et al., 2018; Li et al., 2019), bringing us an opportunity to reveal the molecular response mechanisms of C. robusta stolon to environmental challenges.
Isobaric tags for relative and absolute quantitation (iTRAQ) has been widely applied in analyzing the effects of environmental changes on fouling organisms at the proteomics level, mainly because such technique has outstanding accuracy, high-throughput, strong sensitivity, and dynamic detection ability (Han et al., 2013; Ji et al., 2014; Zhang et al., 2015; Tang et al., 2020). With this technique, this study aimed to reveal the proteomic response of C. robusta stolon to temperature and salinity challenges. We detected the changes in protein expression in the C. robusta stolon exposed to different temperature and salinity stresses. Subsequently, the functional annotation and network analyses on these stress-related proteins were conducted to reveal the common and differential response mechanisms of this adhesive structure to environmental changes.
Ciona robusta adults (average length of 6 cm) adhered on scallop cages were collected from the Longwangtang Aquaculture Farm, Dalian, Liaoning Province, China (38°49′′N, 121°24′′E) in September 2019. Collected ascidians were acclimatized in the filtered (the size of the filter membrane is 0.45 μm) and aerated seawater at 22 ± 1°C, 30 ± 1 psu, and pH 8.1 ± 0.1 (natural conditions at the collection site) for 1 week. They were fed with the dried algae powder mixture of Spirulina sp. and Chlorella sp. daily. After acclimation, we conducted a preliminary experiment to select suitable exposure time and level for temperature and salinity stresses. The temperature gradient were designed as 5, 10, 30, and 40°C, while the salinity gradient were designed as 12, 20, 40, and 50 psu according to the results obtained from previous investigations (Carver et al., 2006; Bouchemousse et al., 2016; Chen et al., 2018). Ascidian individuals were exposed to the different gradients of environmental stresses for 1 week. The analysis results showed that exposing C. robusta to the levels of 12 psu, 40 psu, 10°C, 30°C for 120 h could represent the low salinity, high salinity, low temperature and high temperature stresses on this species. After preliminary experiment, the healthy ascidians with obvious stimuli responses were randomly assigned to five groups: control (C, 22°C/30 psu), high salinity (HS, 22°C/40 psu), low salinity (LS, 22°C/12 psu), high temperature (HT, 30°C/30 psu), and low temperature (LT, 10°C/30 psu). Each exposure group contained three separate 20 L tanks as replicates and 40 individuals in each replicate tank (Figure 1). After exposure for 120 h, the stolon that was still attached to the substrate surface was dissected from 18 individuals in each tank and pooled together as a mixed biological replicate. A total of 270 stolon tissues (18 individuals × 3 replicates × 5 groups) were collected, and the residual seawater on the sampled stolon surface was removed with the sterilized absorbent paper. The stolon samples were then rapidly frozen with liquid nitrogen and preserved at −80°C until proteomics analysis.
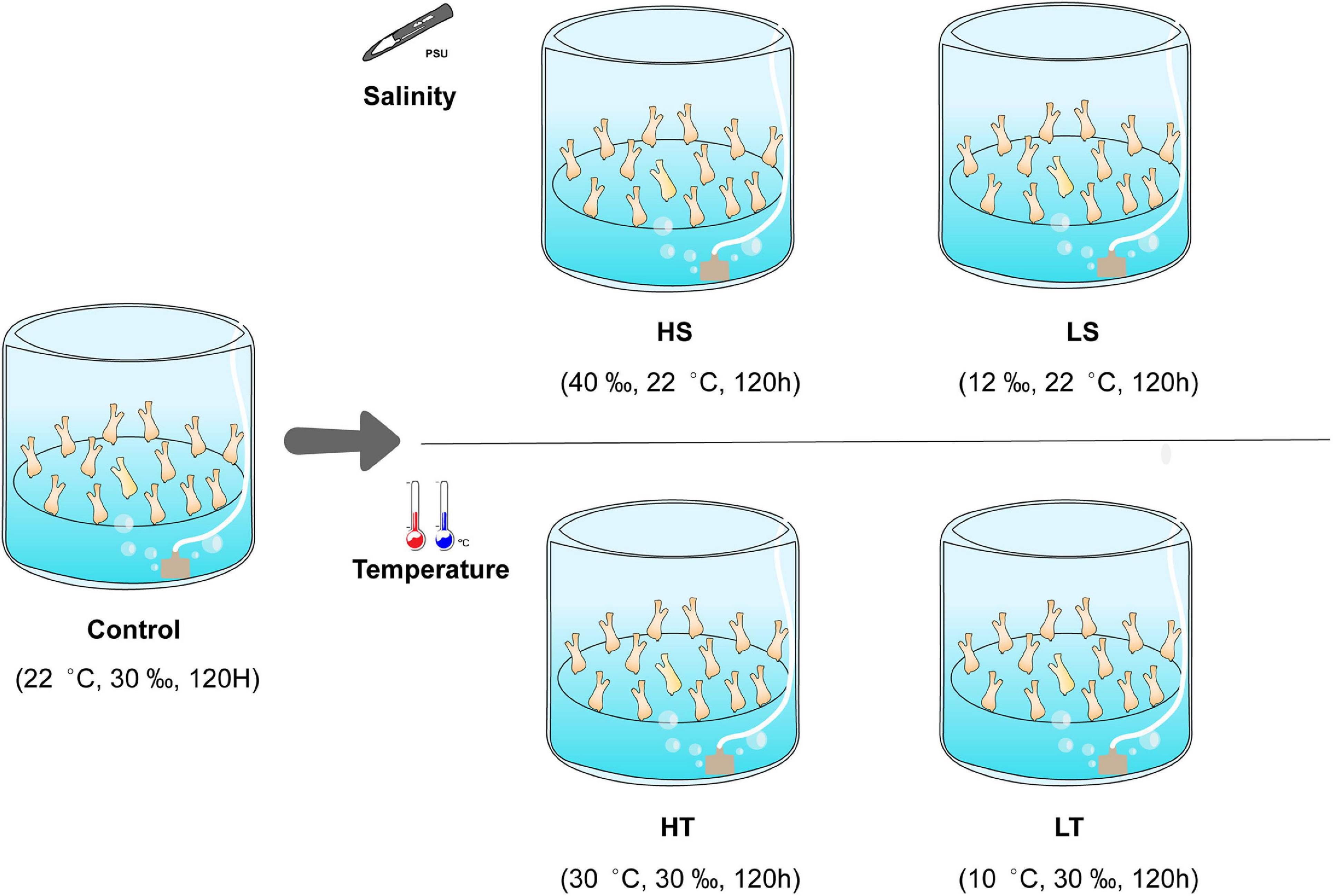
Figure 1. Schematic diagram of temperature and salinity challenges. HS, high salinity; LS, low salinity; HT, high temperature; LT, low temperature; control, natural seawater.
To extract the total proteins, the collected samples were ground into powder with liquid nitrogen. A total of 3 mL radio-immunoprecipitation assay buffer was added to the powder and homogenized with a glass homogenizer. After centrifugation at 16,000 g for 30 min, the supernatant was deposited by adding fourfold volume of cold acetone containing 10 mM DTT at 20°C for about 3 h. Another centrifugation at 20,000 g for 30 min was performed at 4°C and the supernatant was then discarded. The collected precipitate was resuspended with 800 μL of cold lysate containing a solution of 8 M urea, 30 mM 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES), 1 mM PMSF, 2 mM EDTA, and 10 mM DTT, and then incubated at 56°C for 1 h (Lopez et al., 2017). The iodoacetamide was rapidly added into the resuspended sample to obtain the solution with the final concentration of 55 mM by incubating it at room temperature for 1 h in the dark. Following a second round of centrifugation at 20,000 g for 30 min at 4°C, the protein-containing supernatant was collected and stored at −80°C for subsequent analyses.
The purity of extracted stolon proteins was analyzed using the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) method (Li et al., 2019), while the protein concentration was determined using the Bradford method (Bradford, 1976). Subsequently, the filter-aided sample preparation method was used for protein digestion. In brief, a total of 100 μg of stolon proteins was added into the 10 K tube for ultrafiltration. A total of 200 μL of 50% triethylammonium bicarbonate (TEAB) buffer was added to the obtained solution and then centrifuged at 14,000 g for 40 min at 4°C, and this step was repeated once. The protein mixture was then treated with 1 μg/μL of Sequencing Grade Modified trypsin (Promega, Madison, WI, United States) at 37°C for 24 h. The target peptides were lyophilized with a lyophilizer and collected into a new centrifuge tube. Finally, the peptide samples were redissolved in 50% TEAB buffer. Sample labeling was performed using an iTRAQ 8-plex reagent kit (AB Sciex, Framingham, MA, United States) following the manufacturer’s instructions. The isotopes 116, 117, 118, 119, and 121 were selected to label the stolon proteins in the control, LS, HS, LT, and HT groups, respectively.
A set of iTRAQ-labeled samples were combined, desalted, and vacuum-dried. The labeled peptide mixture was dissolved with buffer A (10 mM ammonium formate, pH = 10) and separated with a reversed phase C18 column (75 μm × 10 cm, 5 μm, 300 Å, Agela Technologies) mounted on an ultimate 3000 nano LC system (Dionex, Sunnyvale, CA, United States). Finally, a total of 16 fractions were harvested and freeze-dried. Each fraction was re-dissolved with 5 μL 0.1% formic acid and passed through a Nano LC-MS/MS system at a flow rate of 300 nL/min. The separated compounds were then eluted into an Electrospray Ionization Orbitrap of the Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States), setting in positive ion mode and data-dependent manner with full MS scan at 350–2,000 m/z, full scan resolution at 70,000, and MS/MS scan resolution at 17,500. The minimum signal threshold for MS/MS scan was set at 1E + 5 and the isolation width was set at 2 Da.
The raw data of MS analysis was converted into files with mgf format using the Proteome Discoverer 1.4 software (Thermo Fisher Scientific Inc., Bremen, Germany). The obtained clean data was searched using MASCOT software (Matrix Science, London, United Kingdom; version 2.3.0) to identify proteins. The protein database for C. robusta in UniProt1 was used as the reference database for protein identification as described by a previously published protocol (Li et al., 2019). The summed intensities of the matched spectrum were used for quantizing the relative expression ratios of proteins, and one protein was quantified using at least two spectra. Subsequently, these ratios were transformed to log2 intensities (Kuplik et al., 2019). The protein abundance differences between the exposed groups (LS, HS, LT, and HT) and the control group were evaluated by using t-test method combined with the Benjamini–Hochberg correction (Ji et al., 2014). The DAPs, including the up-regulated and the down-regulated proteins, were identified using p-value < 0.05 and fold change ≥ 1.2 or ≤0.83, respectively. The raw data obtained from LC-MS/MS have been uploaded to the public repository iProX (ID: IPX0002919000).
The functional annotation of the obtained DAPs was performed using OmicsBean online program2, which includes the enrichment functions of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG, Gene and Consortium, 2000; Kanehisa et al., 2008). After removing the invalid values and antilibrary data, a total of 75, 86, 123, and 83 DAPs in the four groups were identified. The hypergeometric test was used to find the significantly enriched GO terms and KEGG pathways with the criterion of p-value < 0.01. Clusters of Orthologous Groups (COG) enrichment was also employed to functionally classify the DAPs based on the orthology concept (Tatusov et al., 2000). The Venn diagram online tool3 was used to analyze the overlapped DAPs among different exposure groups. According to the Venn results, the coexisting DAPs of salinity and temperature stresses were evaluated by protein--protein interactions (PPI). The PPI network analysis was conducted to further clarify the inter-relationships and expression patterns of the DAPs related to salinity/temperature challenges by using the Search Tool for the Retrieval of Interacting Genes4 (version 11.0) and Cytoscape software5 (version 3.7.2) with default parameters.
A total of 33,068 MS/MS counts were generated from the stolon across all five groups (control included). By searching against the UniProt database, 3,930 unique peptides and 1,083 proteins were identified (Supplementary Table 1). A total of 597 (approximately 55.12%) of the identified proteins had at least two unique peptides. Our results illustrated that most of the proteins (approximately 75%) were composed of 100–700 amino acids (Supplementary Figure 1), while 87.63% of the identified proteins contained less than seven peptides (Supplementary Figure 2). The molecular weight of most of the identified proteins was less than100 kDa, accounting for 80% of the total proteins. Moreover, 15 proteins with low molecular weight (<10 kDa) and 135 proteins with high molecular weight (>100 kDa) were identified (Supplementary Figure 3). The distribution of protein coverage illustrated that the coverage with less than 5, 5–15, 15–30, and 30–100%, accounting for 37.65, 36.93, 18.12, and 7.30% of the total proteins in stolon, respectively (Supplementary Figure 4).
For salinity stresses, a total of 75 DAPs including 49 up-regulated and 26 down-regulated proteins were identified from the stolon exposed to LS, while a total of 86 DAPs including 54 up-regulated and 32 down-regulated proteins were identified from the stolon exposed to HS. For temperature stresses, a total of 123 DAPs containing 79 up-regulated and 44 down-regulated proteins were identified from the stolon exposed to LT, while a total of 83 DAPs containing 51 up-regulated and 32 down-regulated proteins were identified from the stolon exposed to HT (Figure 2). Venn analysis showed that a total of 27 DAPs were overlapped in both HS and LS groups, while 48 DAPs were overlapped in both HT and LT groups (Figure 3). The detailed information on these DAPs is shown in Supplementary Table 2.
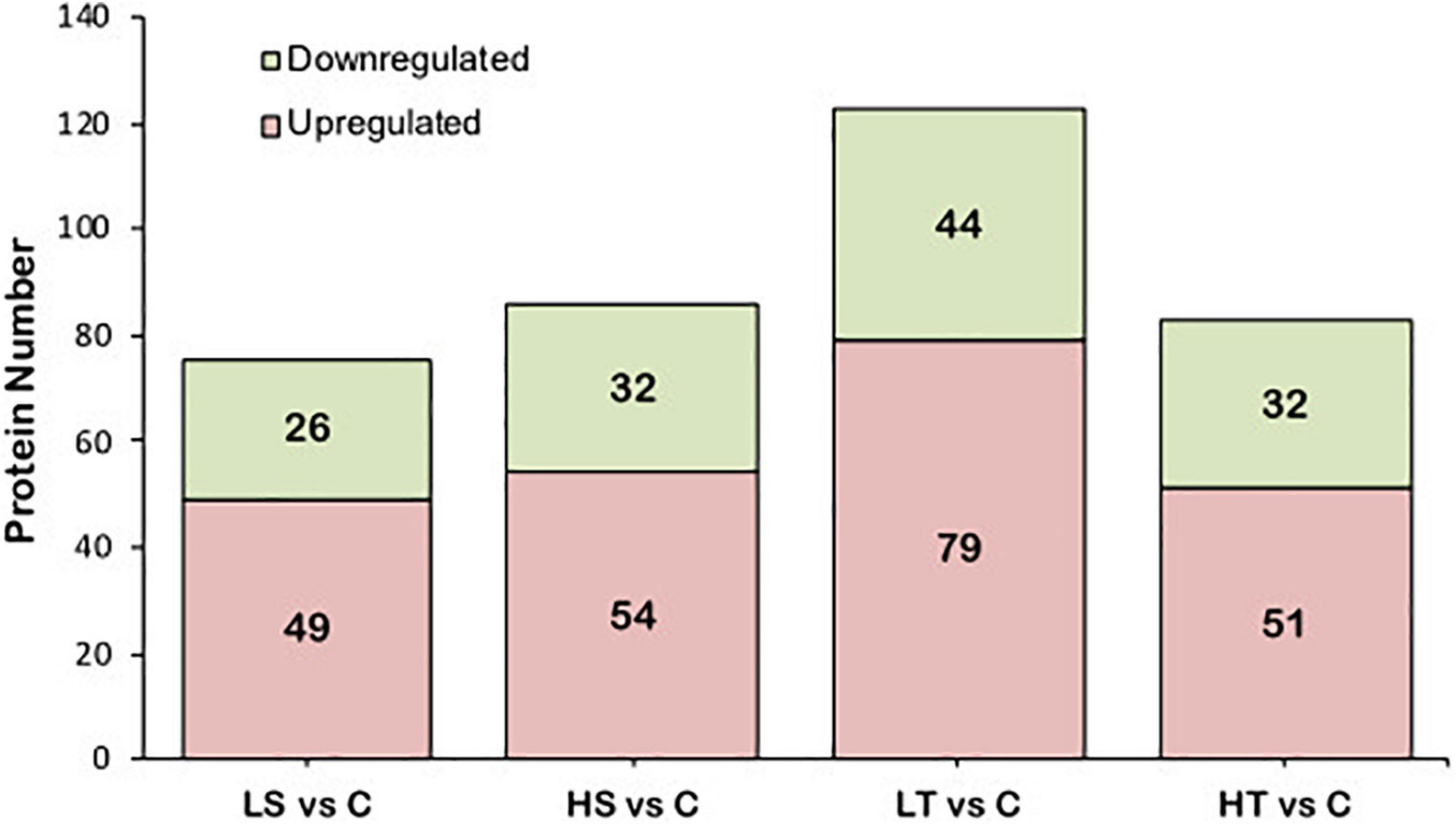
Figure 2. Analysis of the differential abundance proteins in the stolon of Ciona robusta exposed to temperature and salinity stresses. HS, high salinity; LS, low salinity; HT, high temperature; LT, low temperature; C, natural seawater control.
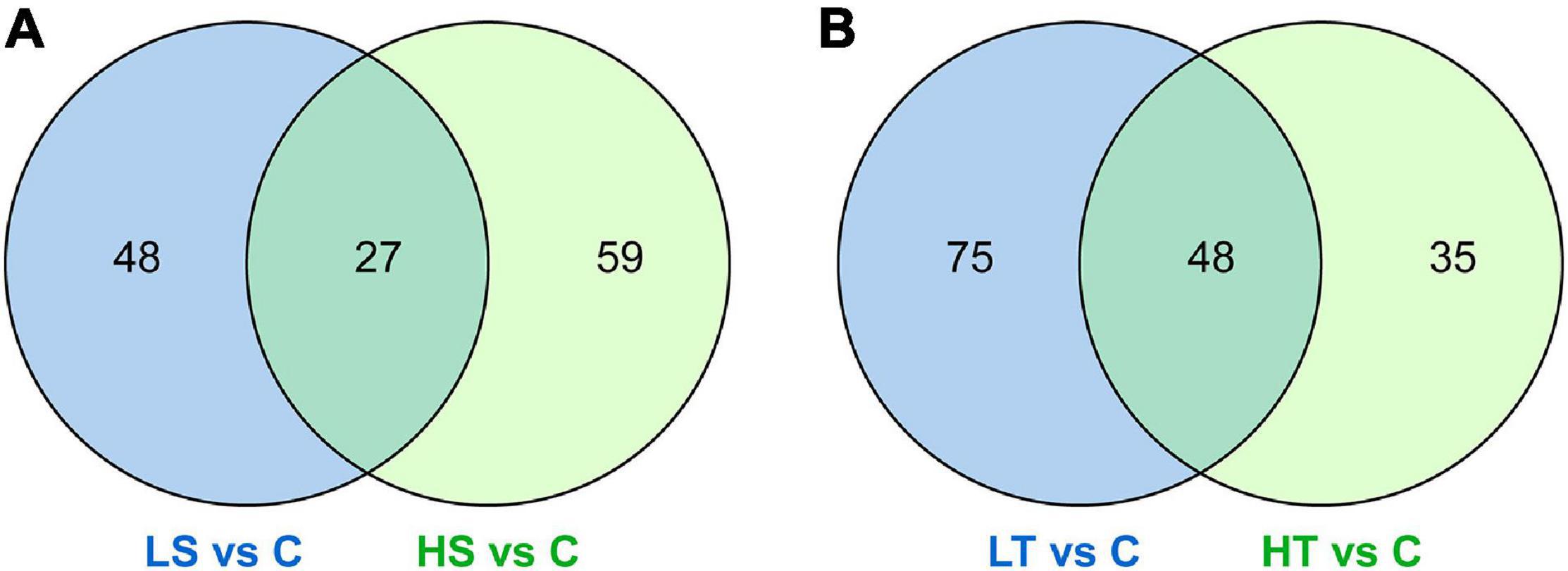
Figure 3. Venn diagrams showing the common differential abundance proteins identified from the stolon of Ciona robusta exposed to salinity and temperature stresses. (A) Salinity stresses and (B) temperature stresses. HS, high salinity; LS, low salinity; HT, high temperature; LT, low temperature; C, natural seawater control.
Gene Ontology enrichment analysis on the DAPs showed that these proteins were enriched into three categories (Figure 3 and Supplementary Tables 3–6): biological process (BP), cell component (CC), and molecular function (MF). The “Cytoskeleton” term was significantly enriched in both HS and LS groups. The terms “extracellular region,” “cellular ion homeostasis,” “cellular cation homeostasis,” and “extracellular matrix structural constituent” were only enriched in the LS group (Figure 4A). The terms “cytoskeletal part,” “cytoskeleton organization,” and “proteolysis” were only enriched in the HS group (Figure 4B). Meanwhile, the terms “cytoskeleton,” “polymeric cytoskeletal fiber,” and “carbohydrate binding” were enriched in both HT and LT groups. The terms “extracellular space,” “calcium ion binding,” and “enzyme inhibitor activity” were enriched in the LT group (Figure 4C). The significantly enriched terms in the HT group were “microtubule,” “cell cycle,” and “nucleoside-triphosphatase activity” (Figure 4D).
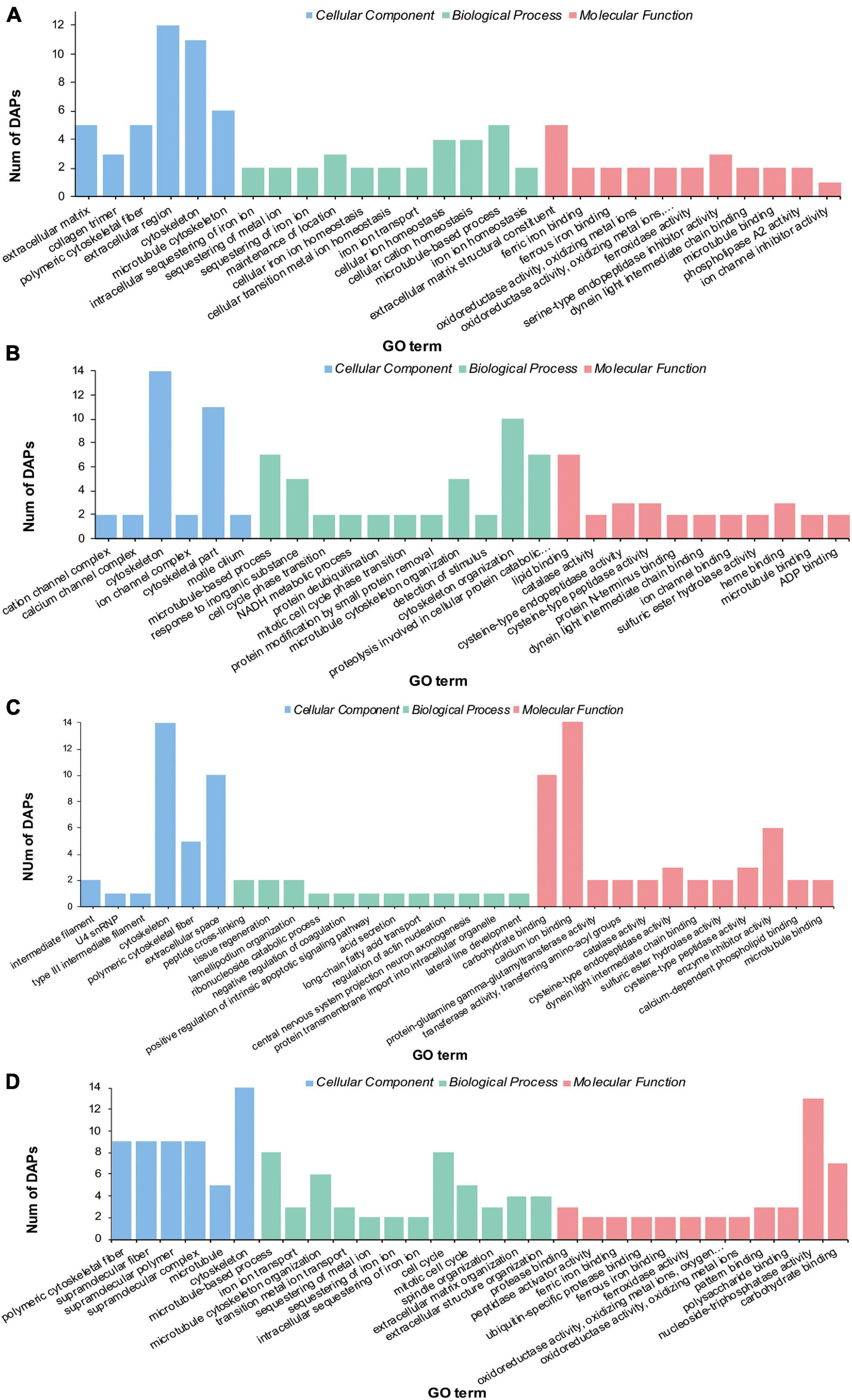
Figure 4. Gene ontology enrichment of differential abundance proteins in the stolon of C. robusta exposed to salinity and temperature stresses. (A) Low salinity, (B) high salinity, (C) low temperature, and (D) high temperature.
By performing KEGG pathway enrichment analyses, the “extracellular matrix (ECM)” pathway was specifically enriched in the stolon exposed to LS (Figure 5A), whereas the “transport and catabolism” and “signal transduction” were the most significant pathways enriched in the stolon exposed to HS (Figure 5B). Furthermore, two pathways, including “signal transduction” and “lipid metabolism,” were significantly enriched in the stolon exposed to LT (Figure 5C), whereas the pathways were “signal transduction” and “cell growth and death” in the HT group (Figure 5D).

Figure 5. Kyoto encyclopedia of genes and genomes enrichment analysis of the differential abundance proteins in the stolon of C. robusta exposed to salinity and temperature stresses. (A) Low salinity, (B) high salinity, (C) low temperature, and (D) high temperature. The number represents the number of proteins enriched to the pathways.
The DAPs identified from the stolon exposed to LS, HS, LT, and HT were enriched into 16, 18, 19, and 17 COG categories, respectively (Figure 6). Among these, the most significant categories in the LS group were “signal transduction mechanisms,” “cytoskeleton,” “extracellular structures,” and “posttranslational modification, protein turnover, chaperones” (Figure 6A), whereas the most significant ones in the HS group were “cytoskeleton,” “signal transduction mechanisms,” and “posttranslational modification, protein turnover, chaperones” (Figure 6B). Meanwhile, the most significant categories in the LT group were “posttranslational modification, protein turnover, chaperones,” “signal transduction mechanisms,” and “cytoskeleton” (Figure 6C), whereas they were “cytoskeleton,” “posttranslational modification, protein turnover, chaperones,” “cell cycle control, cell division, chromosome partitioning,” “inorganic ion transport and metabolism,” and “signal transduction mechanisms” in the HT group (Figure 6D).
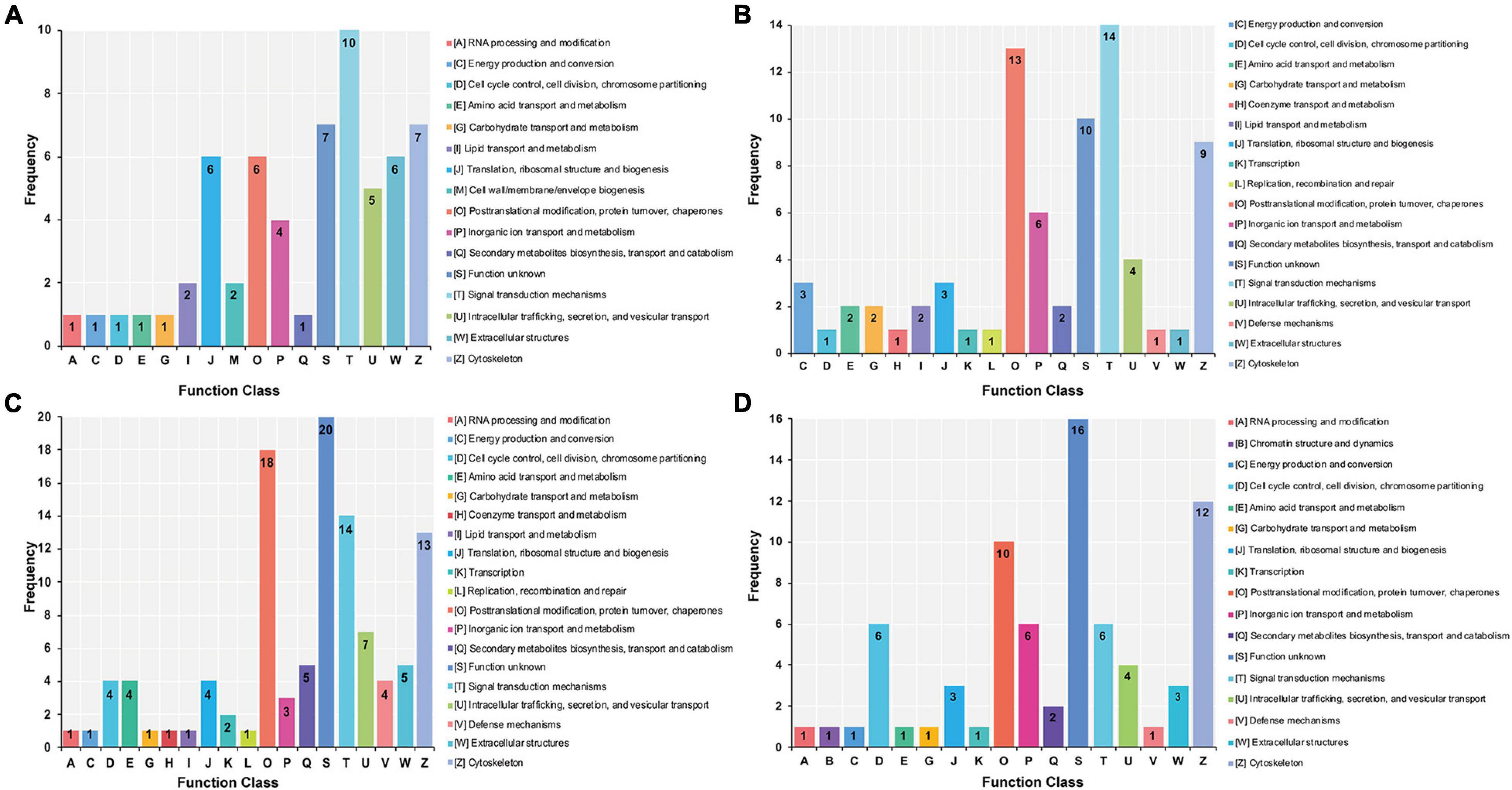
Figure 6. Clusters of orthologous analysis of differential abundance proteins in the stolon of C. robusta exposed to salinity and temperature stresses. (A) Low salinity, (B) high salinity, (C) low temperature, and (D) high temperature. The number represents the number of proteins enriched to the terms.
A total of 27 and 48 DAPs were identified by using the PPI method from the stolon exposed to salinity and temperature stresses, respectively (Figure 7). After hiding the disconnected nodes, only 22 and 29 proteins were correlated to each other in the salinity and temperature networks, respectively. Among these DAPs, one KEGG pathway “ribosome” was significantly enriched under high and low salinity stresses, while two KEGG pathways, “ribosome” and “phagosome,” were significantly enriched under high and low temperature stresses, respectively.
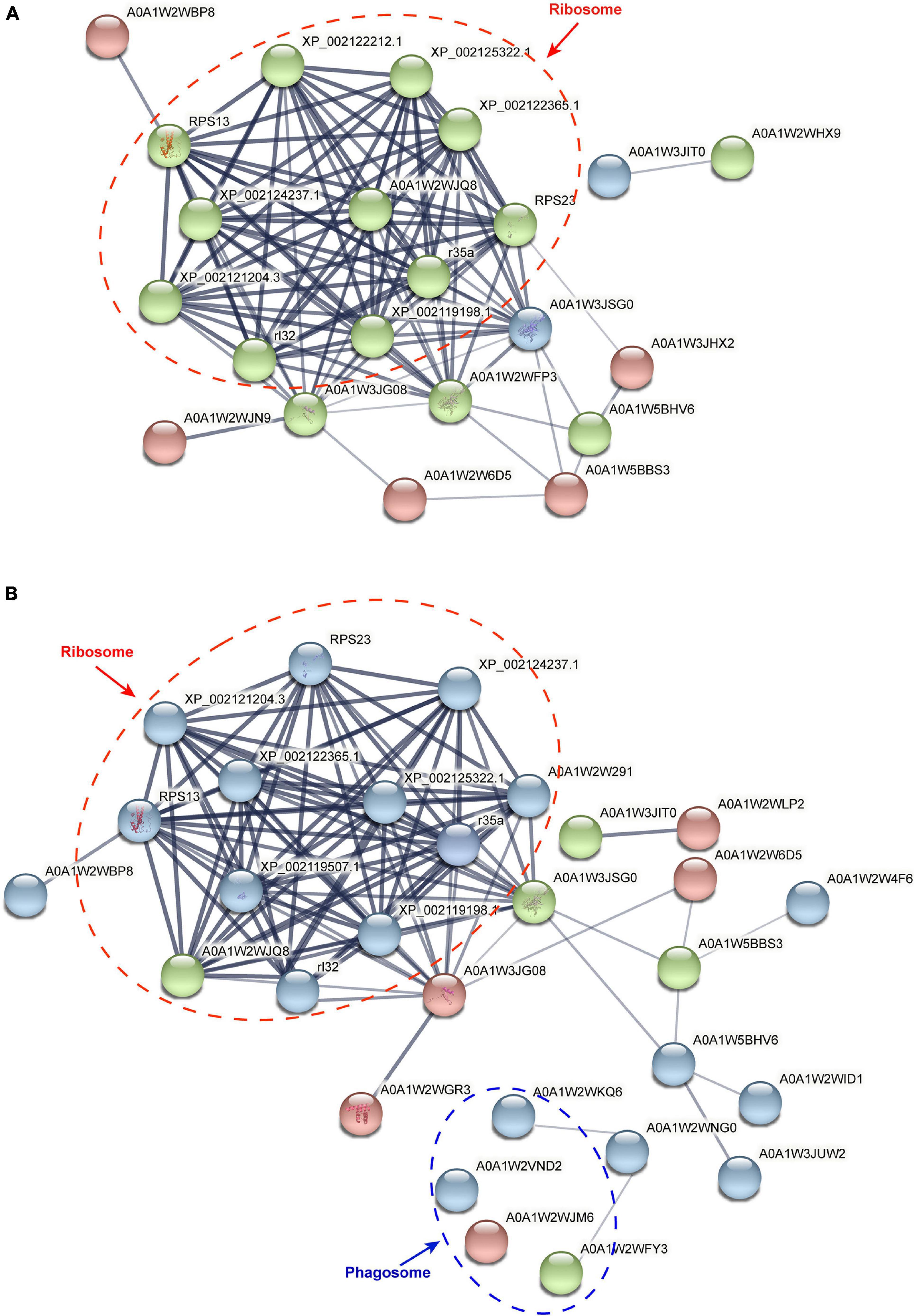
Figure 7. Protein–protein interaction analysis of differential abundance proteins in the stolon of C. robusta exposed to salinity and temperature stresses. (A) Salinity stresses. (B) Temperature stresses. The circles represent differentially abundant proteins (DAPs). Red circle: the DAPs enriched in the stolon exposed to high salt/temperature stresses. Blue circle: the DAPs enriched in the stolon exposed to low salt/temperature stresses. Green circle: the DAPs enriched in the stolon exposed to both high and low salt/temperature stresses. The thickness of the line indicates the strength of data support.
In aquatic species, salinity is a common environmental stressor that can remarkably affect metabolism and osmotic regulation of various organisms (Vargas-Chacoff et al., 2015). Marine animals can maintain their most critical physiological functions through multiple molecular mechanisms when exposed to salinity stresses, despite that there are alterations in osmotic pressure and cytoplasmic composition in their cells (Rautsaw et al., 2020). In our study, the identified DAPs in C. robusta exposed to both low and high salinity stresses were significantly enriched in the pathways “posttranslational modifications (PTMs),” “cytoskeleton,” and “signal transduction,” suggesting the involvements of these pathways in the responses of stolon to salinity stress.
During their synthesis, package, and release, proteins are successively modified. The PTMs, such as phosphorylation, glycosylation, and hydroxylation, are crucial for controlling protein conformation and increasing proteome complexity in organisms (Cloutier and Coulombe, 2013). PTM is indeed a common characteristic of numerous adhesive proteins of marine organisms (Flammang et al., 2015). In these PTMs, protein glycosylation has emerged as an important biochemical process in marine bioadhesion. An example is the marine mussel adhesive protein Pvfp-1, which has extensive threonine O-glycosylation (Zhao et al., 2009). Protein hydroxylation was found in all the plaque proteins (Mfp-1 to Mfp-6) of mussels and cement proteins (Pc-1 and Pc-2) of tubeworms (Waite et al., 2005; Lee et al., 2011). In addition, phosphoproteins have also been detected from the mussel foot proteins such as Mcfp-5 and Mcfp-6 and tubeworm cement proteins such as Pc-3A and Pc-B (Zhao et al., 2005; Zhao and Waite, 2006). Our results here indicated that the response of the C. robusta stolon to salinity stresses might be regulated by PTMs (Figure 6). Such a finding provides new evidence of PTMs to implicate in stolon attachment processes of C. robusta, although the PTMs have not been detected in the adhesive proteins of this species so far.
We found that several DAPs associated with salinity stresses were significantly enriched in the “cytoskeleton” term (Figure 4). Cytoskeletal proteins provide structural organization to cells in eukaryotic organisms (Bogatcheva and Machado, 2020). They are involved in transmitting important regulatory signals during cell activation, keeping the cellular structure stable, and maintaining signal communication between different cells (Bogatcheva and Machado, 2020). Furthermore, the cytoskeletal proteins may have major roles in adjusting biological rhythms by sensing environmental parameters (Artigaud et al., 2014). Therefore, C. robusta may adjust its cytoskeleton management strategy when exposed to salinity stresses, thereby consolidating the cell growth efficiency and further maintaining the structural stability of the stolon. This may be a fact that the stolon can secret some adhesive proteins that permanently glue the tunicate to the substrate under harsh salinity stresses.
Signal transduction pathway is a vital biochemical process in which cells convert extracellular signals from the surrounding environments into intracellular specific reactions (Kling, 1998). The signal transduction is initiated as one of the classic strategies to deal with environmental salinity changes in most aquatic animals (Buckley et al., 2006; Zhang et al., 2015; Dou et al., 2018; Li Y. et al., 2021). In our study, the identified DAPs were significantly enriched in “signal transduction” pathway, indicating the importance of signal transduction in coping with salinity stresses (Figure 5). Indeed, previous studies have confirmed that signal transduction could help aquatic organisms perceive their environmental changes (van der Geer, 2013). Moreover, a study showed that signal transduction-related pathways, such as G protein-coupled receptor, Ras GTPase, and P13K/Akt/mTOR pathways, could regulate the response of the oyster Crassostrea gigas to salinity stimulation (Zhang et al., 2015). Li Y. et al. (2021) reported that the MAPK signaling pathway was activated in the gills of the razor clam Sinonovacula constricta after exposure to salinity stresses (Li Y. et al., 2021). All these findings suggest that signal transduction-related pathways were activated under salinity challenges. Our results here provide one more layer of evidence to support this conclusion, further confirming that the C. robusta stolon could respond to salinity stresses by activating various signal transduction pathways.
The “extracellular matrix (ECM)” term was significantly enriched in the stolon exposed to low salinity. ECM is a complex and dynamic structure that provides the scaffold wherein cells are located. Apart from its function as the principal scaffold of cells, ECM provides the signals regulating cell behaviors and triggers multiple biological activities that are essential for tissue morphological homeostasis (Theocharis et al., 2016). A previous study revealed that Botrylloides nigrum and Botryllus planus ascidians exhibited dramatic morphological response to low salinity stresses, including the expansions of cloacal cavities and distensions of pharyngeal baskets and neural glands (Dijkstra and Simkanin, 2016). Similarly, the changes in the ECM of C. robusta stolon might enhance its defense ability to low salinity stresses by regulating morphological homeostasis of stolon. In addition, the ECM in stolon might not only provide physical scaffolds but also regulate many cellular processes by inducing growth, migration, differentiation, and morphogenesis to respond to salinity changes, and this has been reported in the rough skin sculpin Trachidermus fasciatus and the mud crab Scylla paramamosain (Theocharis et al., 2016; Ma et al., 2018; Zhang et al., 2020).
We also observed the significant enrichment of the DAPs associated with the “transport and catabolism” in the stolon exposed to high salinity stress. An earlier study on the sea cucumber Holothuria leucospilota showed that the protein-dominated catabolism was significantly enhanced under low salinity stresses, whereas the carbohydrate- or lipid-dominated catabolism was significantly enhanced under high salinity stresses (Yu et al., 2013). The proteomic response of C. robusta stolon to higher salinity might be consistent with that of sea cucumber, and the response mechanisms of stolon to hypersaline stresses likely rely on the activation of carbohydrate/lipid transport and catabolism that can produce energy.
Temperature is known to significantly affect marine organisms’ physiology (Pineda et al., 2012; Dong et al., 2019; Irvine et al., 2019). Indeed, temperature changes can affect the adhesive organs/structures of marine organisms. A typical case is byssus adhesion reduction in mussels caused by heat treatment, which has been considered an antifouling strategy for mussels (Perepelizin and Boltovskoy, 2011). Our results here illustrate that the stolon of C. robusta was also influenced by temperature changes at the molecular level. We found the significant enrichment of the term “cytoskeletal proteins” in C. robusta stolon exposed to temperature challenges (Figure 4). Similarly, earlier analyses showed that the changes in the abundance of cytoskeletal proteins in the ascidians Ciona intestinalis and Ciona savignyi were closely associated with thermal stresses (Serafini et al., 2011). Studies on marine teleosts and invertebrates also documented the recombination of the temperature-induced cytoskeletal structure (Vornanen et al., 2005; Buckley et al., 2006; Lockwood et al., 2010; Tomanek and Zuzow, 2010; Jayasundara et al., 2015). Such available evidence suggests that the cytoskeletal proteins in the C. robusta stolon might be involved in temperature stress response through two mechanisms: the alterations in cytoskeletal protein abundance and recombination of the cytoskeletal structure.
Molecular chaperones are involved in the folding or unfolding of proteins and the assembly or disassembly of larger macromolecular complexes (Cloutier and Coulombe, 2013). They are found in all cell types in animals, and their activities are tightly regulated to maintain normal cell functions. Previous studies showed that the higher steady-state levels of molecular chaperones might underlie the capacity of C. intestinalis to out-compete C. savignyi in warm habitats (Serafini et al., 2011). Meanwhile, many molecular chaperones, including heat shock proteins 24, heat shock proteins 90, and calcium-binding protein, were likely to act as chaperones for maintaining the cytoskeletal structure under temperature stresses, preventing the aggregation of the denatured proteins (Lockwood et al., 2010; Lopez et al., 2017). The significant enrichment of chaperoning molecules in our study demonstrates that homeostasis regulation is a crucial mechanism for the C. robusta stolon to respond to temperature stresses. These molecular chaperones likely help the C. robusta stolon achieve stable cytoskeletal structure.
Besides cytoskeleton and molecular chaperones, the term of “calcium-binding proteins” was also significantly enriched in the C. robusta stolon under both low and high temperature stresses. Ca2+ is a highly universal intracellular signal regulating several molecular pathways and cellular processes, including cell proliferation, excitability, exocytosis, and transcription (Berridge et al., 2003; Clapham, 2007). Intracellular calcium-binding proteins can be divided into two categories. One category consists of proteins with Ca2+-binding function, which can transport Ca2+ across cell membranes and thus irreversibly regulate the concentration of Ca2+ in the surrounding environments. Another category can decode Ca2+ signals into functionally specific signals (Carafoli et al., 2001). An investigation showed that the signaling triggered via regulating the intracellular Ca2+ concentration may be involved in the thermal response of the oyster C. gigas (Zhang et al., 2015). We cannot determine the specific functions of calcium-binding proteins in C. robusta stolon, but the enrichment of these proteins in our study suggests that calcium signaling in the stolon should be one of the important molecular mechanisms in response to temperature stresses.
The “phagosome” pathway was enriched in the stolon exposed to both low and high temperature stresses in our study (Figure 7). Phagocytosis is an important congenital defense mechanism for macrophages against bacterial infections (Pradhan et al., 2018). In eukaryotes, phagosomes can engulf potential pathogenic microorganisms and apoptotic cells through binding to the lysosome (Kinchen and Ravichandran, 2008). The enrichment of DAPs in the “phagosome” pathway under temperature stresses implies that temperature might be a remarkable environmental factor leading to the apoptosis of stolon cells in C. robusta. The phagosome is a highly dynamic organelle, even in healthy individuals, and it can phagocytose billions of dead cells per day. However, under stress conditions, they were more inclined to eliminate apoptotic cells (Dean et al., 2019). Therefore, the phagosome may assist in maintaining the homeostasis of stolon cells and the structural stability via phagocytizing apoptotic cells in the C. robusta stolon.
For most organisms, the dynamic equilibrium of lipid metabolism is the fundamental physiological status in maintaining vital activities (Gu et al., 2017). This means that some lipids are constantly oxidized to meet metabolic needs, whereas others are synthesized and stored (Zhang et al., 2017a; Gyamfi et al., 2018). Chilling stress is a common challenge to many aquatic organisms, and responding to chilling stress requires lipid metabolic reprogramming. Eventually, animals usually generate more energy by degrading some lipids to resist cold stresses (Brodte et al., 2008). The enrichment of the “lipid metabolism” pathway has been found in many marine organisms such as Eleginops maclovinus, M. galloprovincialis, and Pelteobagrus vachelli in response to low temperature stresses (Brodte et al., 2008). Our study illustrated that “lipid metabolism” was the only pathway enriched in the C. robusta stolon exposed to low temperature. Mechanistically, lipid metabolism may provide more energy for the C. robusta stolon by suppressing the lipogenesis capacity and enhancing the lipolysis capacity during chilling stresses. Unlike the mechanism by which the stolon responded to low temperature, the “cell cycle” term exhibited a significant enrichment in the stolon in response to high temperature stress. Heat is a common stress during ascidian invasions and failure to adapt to this stressor can result in programmed cell demise and decreased viability (Carafoli et al., 2001). Previous studies found that marine organisms could respond to heat stress via the arrested cell cycle progression. For example, the marine teleost Gillichthys mirabilis could conserve energy through inhibiting body cell growth and proliferation to cope with thermal challenges (Buckley et al., 2006). C. robusta may follow this mechanism and its stolon can inhibit cell growth and eliminate the damaged cells, assisting stolon in preserving energy to defend against high temperature stresses.
Despite the stress-specific pathways observed in this study, the response to both temperature and salinity stresses was not completely independent. Venn diagram analysis revealed that “cytoskeleton,” “signal transduction,” and “posttranslational modification” were the overlapping pathways in both salinity and temperature stresses. The common response was also detected by several studies, showing that ascidians mitigated adverse environmental challenges through similar mechanisms (Serafini et al., 2011; Huang et al., 2019; Li H. et al., 2020; Wei et al., 2020; Chen et al., 2021).
Based on PPI analysis, the DAPs enriched in “ribosome” pathway were changed significantly under both salinity and temperature stresses, indicating that these stresses may affect protein synthesis and metabolism in C. robusta stolon (Figure 7). Marine organisms usually rely on the regulation of proteins related to stress response, such as cytoskeletal proteins and adhesive proteins, to maintain cellular homeostasis and limit the adverse effects of harmful environmental stimuli (Vind et al., 2020). Cytoskeletal proteins, which are used to sustain the stable state of cellular/organ structure and communication, are synthesized by ribosomes (Bashline et al., 2014; Bogatcheva and Machado, 2020). Additionally, the proteins related to adhesive functions in marine fouling organisms are also synthesized by ribosomes (Lafontaine and Tollervey, 2001; Jimenez et al., 2019). Previous studies have indicated the presence of adhesive proteins synthesized by ribosomes in the stolon of C. robusta (Li et al., 2019). The enrichment of “ribosome” pathway in C. robusta may be helpful for the stolon to synthesize some proteins associated with the defense against salinity and temperature stresses.
By using iTRAQ technique, our study revealed the dynamic proteomic response to salinity and temperature stresses in the stolon of the highly fouling ascidian C. robusta. Functional enrichment analysis recovered common and challenge-specific response pathways (Figure 8). In addition to the common pathways such as “cytoskeleton,” “signal transduction,” and “posttranslational modification,” “extracellular matrix,” “carbohydrate/lipid transport and catabolism,” “lipid metabolism,” and “cell cycle” play crucial roles in dealing with the stresses of low salinity, high salinity, low temperature, and high temperature, respectively. The findings here illustrate that C. robusta stolon could respond to environmental challenges by developing complex and diverse molecular mechanisms. The pathways and associated proteins obtained in this study provide candidate references for further studies of molecular mechanisms of marine biofouling, local environmental adaptation during invasions, and formulating antifouling strategies in marine and coastal ecosystems.
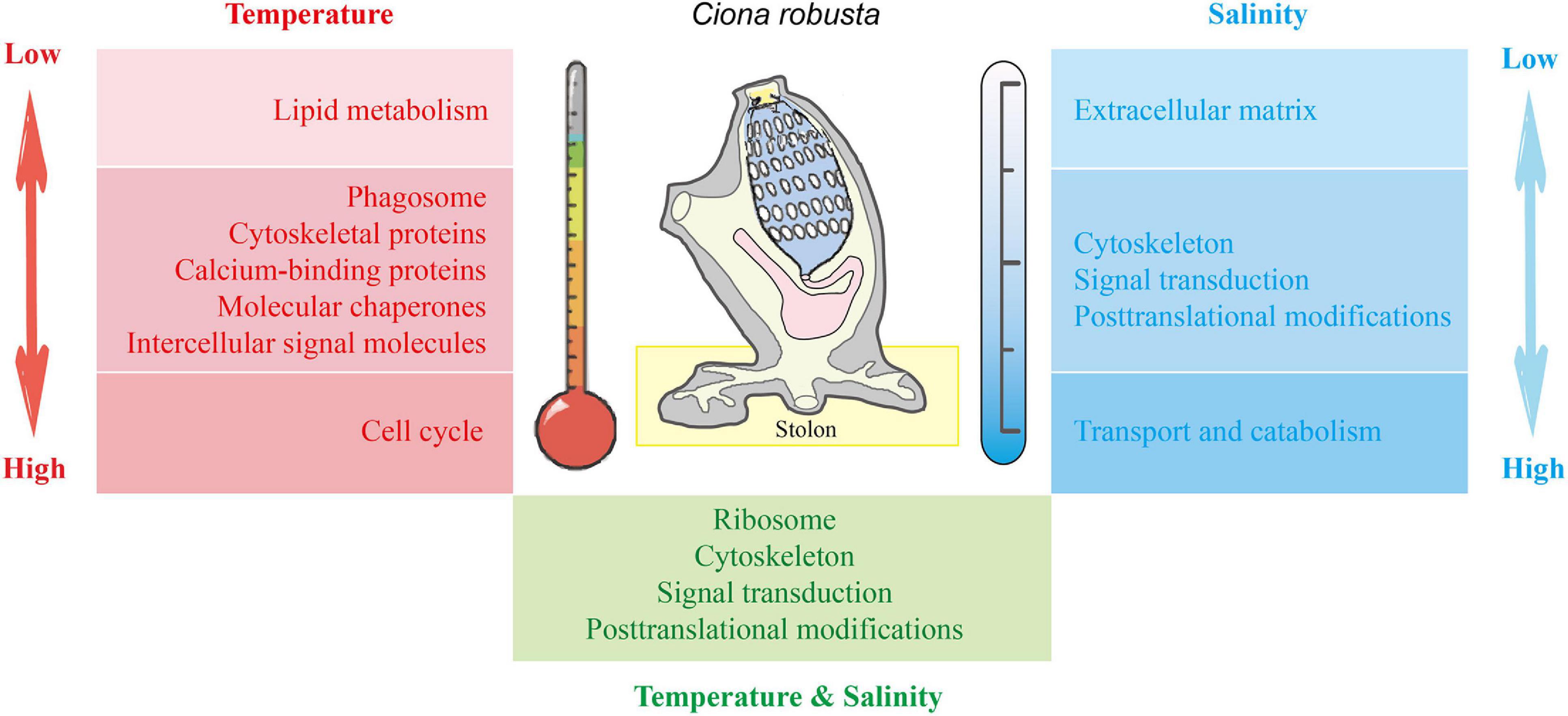
Figure 8. The molecular mechanisms associated with the response of C. robusta stolon to salinity and temperature stresses. Red: temperature stresses. Blue: salinity stresses. Green: interaction of temperature and salinity stresses. The shades of red and blue colors indicate the stress degree of temperature and salinity, respectively. Light color: low salt/temperature stresses. Dark color: high salt/temperature stresses.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD026891.
AZ, SL, and XL conceived the study. XL and SL designed the experiments, conducted the experiments, analyzed the data, and wrote the manuscript. JC and RF analyzed the data. All authors contributed to the revisions of this manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 42076098 and 32061143012), Youth Innovation Promotion Association, Chinese Academy of Sciences (Grant No. 2018054), and Open Project of Key Laboratory of Environmental Biotechnology, Chinese Academy of Sciences (Grant No. kf2018005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Zunchun Zhou and Bei Jiang for the assistance with ascidian collection and providing experimental sites for acclimation and treatments. Great thanks to all editors and reviewers for valuable and constructive comments on early versions of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.761628/full#supplementary-material
Andrade, G. R., Araújo, J. L. F., Nakamura Filho, A., Guañabens, A. C. P., Carvalho, M. D., and Cardoso, A. V. (2015). Functional Surface of the golden mussel’s foot: morphology, structures and the role of cilia on underwater adhesion. Mater. Sci. Eng. C 54, 32–42. doi: 10.1016/j.msec.2015.04.032
Artigaud, S., Lavaud, R., Thébault, J., Jean, F., Strand, O., Strohmeier, T., et al. (2014). Proteomic-based comparison between populations of the great scallop, Pecten maximus. J. Proteomics 105, 164–173. doi: 10.1016/j.jprot.2014.03.026
Bashline, L., Lei, L., Li, S., and Gu, Y. (2014). Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 7, 586–600. doi: 10.1093/mp/ssu018
Bellard, C., Thuiller, W., Leroy, B., Genovesi, P., Bakkenes, M., and Courchamp, F. (2013). Will climate change promote future invasions? Glob. Chang. Biol. 19, 3740–3748. doi: 10.1111/gcb.12344
Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003). Calcium signaling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. doi: 10.1038/nrm1155
Bogatcheva, N. V., and Machado, R. F. (2020). Cytoskeletal Proteins, 2nd Edn. Amsterdam: Elsevier Inc.
Bouchemousse, S., Bishop, J. D. D., and Viard, F. (2016). Contrasting global genetic patterns in two biologically similar, widespread and invasive Ciona species (Tunicata, Ascidiacea). Sci. Rep. 6:24875. doi: 10.1038/srep24875
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Briski, E., Chan, F. T., Darling, J. A., Lauringson, V., MacIsaac, H. J., Zhan, A., et al. (2018). Beyond propagule pressure: importance of selection during the transport stage of biological invasions. Front. Ecol. Environ. 16, 345–353. doi: 10.1002/fee.1820
Brodte, E., Graeve, M., Jacob, U., Knust, R., and Pörtner, H. O. (2008). Temperature-dependent lipid levels and components in polar and temperate eelpout (Zoarcidae). Fish Physiol. Biochem. 34, 261–274. doi: 10.1007/s10695-007-9185-y
Buckley, B. A., Gracey, A. Y., and Somero, G. N. (2006). The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J. Exp. Biol. 209, 2660–2677. doi: 10.1242/jeb.02292
Cahill, P., Heasman, K., Jeffs, A., Kuhajek, J., and Mountfort, D. (2012). Preventing ascidian fouling in aquaculture: screening selected allelochemicals for anti-metamorphic properties in ascidian larvae. Biofouling 28, 39–49. doi: 10.1080/08927014.2011.648624
Carafoli, E., Santella, L., Branca, D., and Brini, M. (2001). Generation, control, and processing of cellular calcium signals. Crit. Rev. Biochem. Mol. Biol. 36, 107–260. doi: 10.1080/20014091074183
Carver, C. E., Mallet, A. L., and Vercaemer, B. (2006). Biological synopsis of the solitary tunicate Ciona intestinalis. Can. Manuscr. Rep. Fish. Aquat. Sci. 2746:55.
Chen, Y., Gao, Y., Huang, X., Li, S., and Zhan, A. (2021). Local environment-driven adaptive evolution in a marine invasive ascidian (Molgula manhattensis). Ecol. Evol. 11, 4252–4266. doi: 10.1002/ece3.7322
Chen, Y., Shenkar, N., Ni, P., Lin, Y., Li, S., and Zhan, A. (2018). Rapid microevolution during recent range expansion to harsh environments. BMC Evol. Biol. 18:187. doi: 10.1186/s12862-018-1311-1
Cloutier, P., and Coulombe, B. (2013). Regulation of molecular chaperones through post-translational modifications: decrypting the chaperone code. Biochim. Biophys. Acta - Gene Regul. Mech. 1829, 443–454. doi: 10.1016/j.bbagrm.2013.02.010
Dean, P., Heunis, T., Härtlova, A., and Trost, M. (2019). Regulation of phagosome functions by post-translational modifications: a new paradigm. Curr. Opin. Chem. Biol. 48, 73–80. doi: 10.1016/j.cbpa.2018.11.001
Dijkstra, J. A., and Simkanin, C. (2016). Intraspecific response of colonial ascidians to variable salinity stress in an era of global change. Mar. Ecol. Prog. Ser. 551, 215–225. doi: 10.3354/meps11719
Dong, H., Yan, D., Deng, L., Guo, H., Wang, K., Wu, M., et al. (2019). Purification effect of oysters based on the analysis of environmental parameters. Stud. Eng. Technol. 6:78. doi: 10.11114/set.v6i1.4299
Dou, L., He, K., Higaki, T., Wang, X., and Mao, T. (2018). Ethylene signaling modulates cortical microtubule reassembly in response to salt stress. Plant Physiol. 176, 2071–2081. doi: 10.1104/pp.17.01124
Flammang, P., Hennebert, E., Maldonado, B., Ladurner, P., and Santos, R. (2015). Experimental strategies for the identification and characterization of adhesive proteins in animals: a review. Interface Focus 5, 1–19. doi: 10.1098/rsfs.2014.0064
Gene, T., and Consortium, O. (2000). Gene ontology: tool for the. Gene Expr. 25, 25–29. doi: 10.1038/75556
Gu, Y., He, L., Zhao, C., Wang, F., Yan, B., Gao, Y., et al. (2017). Biochemical and transcriptional regulation of membrane lipid metabolism in maize leaves under low temperature. Front. Plant Sci. 8:2053. doi: 10.3389/fpls.2017.02053
Gyamfi, D., Awuah, E. O., and Owusu, S. (2018). Lipid Metabolism: An Overview. Amsterdam: Elsevier Inc.
Han, Z., Sun, J., Zhang, Y., He, F., Xu, Y., Matsumura, K., et al. (2013). ITRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J. Proteome Res. 12, 2090–2100. doi: 10.1021/pr301083e
Hawes, N. A., Tremblay, L. A., Pochon, X., Dunphy, B., Fidler, A. E., and Smith, K. F. (2018). Effects of temperature and salinity stress on DNA methylation in a highly invasive marine invertebrate, the colonial ascidian Didemnum vexillum. PeerJ 2018, 1–18. doi: 10.7717/peerj.5003
Hennebert, E., Wattiez, R., Waite, J. H., and Flammang, P. (2012). Characterization of the protein fraction of the temporary adhesive secreted by the tube feet of the sea star Asterias rubens. Biofouling 28, 289–303. doi: 10.1080/08927014.2012.672645
Hu, M., Li, L., Sui, Y., Li, J., Wang, Y., Lu, W., et al. (2015). Effect of pH and temperature on antioxidant responses of the thick shell mussel Mytilus coruscus. Fish Shellfish Immunol. 46, 573–583. doi: 10.1016/j.fsi.2015.07.025
Huang, X., Li, S., and Zhan, A. (2019). Genome-wide identification and evaluation of new refrence genes for gene expression analysis under temperature and salinity stresses in Ciona savignyi. Front. Genet. 10:71. doi: 10.3389/fgene.2019.00071
Huang, X., and Zhan, A. (2020). Highly dynamic transcriptional reprogramming and shorter isoform shifts under acute stresses during biological invasions. RNA Biol. 18, 340–353. doi: 10.1080/15476286.2020.1805904
Irvine, S. Q., McNulty, K. B., Siler, E. M., and Jacobson, R. E. (2019). High temperature limits on developmental canalization in the ascidian Ciona intestinalis. Mech. Dev. 157, 10–21. doi: 10.1016/j.mod.2019.04.002
Jayasundara, N., Tomanek, L., Dowd, W. W., and Somero, G. N. (2015). Proteomic analysis of cardiac response to thermal acclimation in the eurythermal goby fish Gillichthys mirabilis. J. Exp. Biol. 218, 1359–1372. doi: 10.1242/jeb.118760
Ji, C., Wu, H., Wei, L., and Zhao, J. (2014). iTRAQ-based quantitative proteomic analyses on the gender-specific responses in mussel Mytilus galloprovincialis to tetrabromobisphenol A. Aquat. Toxicol. 157, 30–40. doi: 10.1016/j.aquatox.2014.09.008
Jimenez, L., Yu, H., McKenzie, A. J., Franklin, J. L., Patton, J. G., Liu, Q., et al. (2019). Quantitative proteomic analysis of small and large Extracellular Vesicles (EVs) reveals enrichment of adhesion proteins in small EVs. J. Proteome Res. 18, 947–959. doi: 10.1021/acs.jproteome.8b00647
Johnston, L. A. (2010). Temperature Affects Adhesion of the Acorn Barnacle (Balanus amphitrite). Master’s theses Project reports. California, CA: California Polytechnic State University.
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, 480–484. doi: 10.1093/nar/gkm882
Khan, F. U., Hu, M., Kong, H., Shang, Y., Wang, T., Wang, X., et al. (2020). Ocean acidification, hypoxia and warming impair digestive parameters of marine mussels. Chemosphere 256:127096. doi: 10.1016/j.chemosphere.2020.127096
Kim, M. K., Kim, D. H., Park, J., Kim, D. H., Yoon, T. J., Kim, D. G., et al. (2019). Effects of temperature and salinity on the egg development and larval settlement of Ciona robusta (Ascidiacea, Phlebobranchia, Cionidae). Ocean Sci. J. 54, 97–106. doi: 10.1007/s12601-018-0056-5
Kinchen, J. M., and Ravichandran, K. S. (2008). Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9, 781–795. doi: 10.1038/nrm2515
Kuplik, Z., Novak, L., and Shenkar, N. (2019). Proteomic profiling of ascidians as a tool for biomonitoring marine environments. PLoS One 14:e215005. doi: 10.1371/journal.pone.0215005
Lafontaine, D. L. J., and Tollervey, D. (2001). The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2, 514–520. doi: 10.1038/35080045
Lee, B. P., Messersmith, P. B., Israelachvili, J. N., and Waite, J. H. (2011). Mussel-Inspired adhesives and coatings. Annu. Rev. Mater. Res. 41, 99–132. doi: 10.1146/annurev-matsci-062910-100429.Mussel-Inspired
Li, H., Huang, X., and Zhan, A. (2020). Stress memory of recurrent environmental challenges in marine invasive species: ciona robusta as a case study. Front. Physiol. 11:94. doi: 10.3389/fphys.2020.00094
Li, Y. F., Yang, X. Y., Cheng, Z. Y., Wang, L. Y., Wang, W. X., Liang, X., et al. (2020). Near-future levels of ocean temperature weaken the byssus production and performance of the mussel Mytilus coruscus. Sci. Total Environ. 733:139347. doi: 10.1016/j.scitotenv.2020.139347
Li, S., Huang, X., Chen, Y., Li, X., and Zhan, A. (2019). Identification and characterization of proteins involved in stolon adhesion in the highly invasive fouling ascidian Ciona robusta. Biochem. Biophys. Res. Commun. 510, 91–96. doi: 10.1016/j.bbrc.2019.01.053
Li, X., Li, S., Huang, X., Chen, Y., Cheng, J., and Zhan, A. (2021). Protein-mediated bioadhesion in marine organisms: a review. Mar. Environ. Res. 170:105409. doi: 10.1016/j.marenvres.2021.105409
Li, Y., Niu, D., Wu, Y., Dong, Z., and Li, J. (2021). Integrated analysis of transcriptomic and metabolomic data to evaluate responses to hypersalinity stress in the gill of the razor clam (Sinonovacula constricta). Comp. Biochem. Physiol. - Part D Genomics Proteomics 38:100793. doi: 10.1016/j.cbd.2021.100793
Lockwood, B. L., Sanders, J. G., and Somero, G. N. (2010). Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J. Exp. Biol. 213, 3548–3558. doi: 10.1242/jeb.046094
Lockwood, B. L., and Somero, G. N. (2011). Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol. Ecol. 20, 517–529. doi: 10.1111/j.1365-294X.2010.04973.x
Lopez, C. E., Sheehan, H. C., Vierra, D. A., Azzinaro, P. A., Meedel, T. H., Howlett, N. G., et al. (2017). Proteomic responses to elevated ocean temperature in ovaries of the ascidian Ciona intestinalis. Biol. Open 6, 943–955. doi: 10.1242/bio.024786
Ma, J., Chen, T., Wu, S., Yang, C., Bai, M., Shu, K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217. doi: 10.1093/nar/gky869
Ma, Q., Liu, X., Feng, W., Liu, S., and Zhuang, Z. (2018). Analyses of the molecular mechanisms associated with salinity adaption of Trachidermus fasciatus through combined iTRAQ-based proteomics and RNA sequencing-based transcriptomics. Prog. Biophys. Mol. Biol. 136, 40–53. doi: 10.1016/j.pbiomolbio.2018.02.003
Nakano, D., and Strayer, D. L. (2014). Biofouling animals in fresh water: biology, impacts, and ecosystem engineering. Front. Ecol. Environ. 12:167–175. doi: 10.1890/130071
Pennati, R., and Rothbächer, U. (2014). Bioadhesion in ascidians: a developmental and functional genomics perspective. Interface Focus 5, 1–10. doi: 10.1098/rsfs.2014.0061
Perepelizin, P. V., and Boltovskoy, D. (2011). Hot water treatment (chronic upper lethal temperature) mitigates biofouling by the invasive Asian mussel Limnoperna fortunei in industrial installations. Environ. Sci. Technol. 45, 7868–7873. doi: 10.1021/es2014852
Pineda, M. C., Turon, X., and López-Legentil, S. (2012). Stress levels over time in the introduced ascidian Styela plicata: the effects of temperature and salinity variations on hsp70 gene expression. Cell Stress Chaperones 17, 435–444. doi: 10.1007/s12192-012-0321-y
Pradhan, G., Shrivastva, R., and Mukhopadhyay, S. (2018). Mycobacterial PknG targets the Rab7l1 signaling pathway to inhibit phagosome–lysosome fusion. J. Immunol. 201, 1421–1433. doi: 10.4049/jimmunol.1800530
Rautsaw, R. M., Schramer, T. D., Acuña, R., Arick, L. N., DiMeo, M., Mercier, K. P., et al. (2020). Genomic adaptations to salinity resist gene flow in the evolution of floridian watersnakes. Mol. Biol. Evol. 38, 745–760. doi: 10.1093/molbev/msaa266
Renborg, E., Johannesson, K., and Havenhand, J. (2014). Variable salinity tolerance in ascidian larvae is primarily a plastic response to the parental environment. Evol. Ecol. 28, 561–572. doi: 10.1007/s10682-013-9687-2
Ricciardi, A., Blackburn, T. M., Carlton, J. T., Dick, J. T. A., Hulme, P. E., Iacarella, J. C., et al. (2017). Invasion science: a horizon scan of emerging challenges and opportunities. Trends Ecol. Evol. 32, 464–474. doi: 10.1016/j.tree.2017.03.007
Santos, R., Barreto, A., Franco, C., and Coelho, A. V. (2013). Mapping sea urchins tube feet proteome - A unique hydraulic mechano-sensory adhesive organ. J. Proteomics 79, 100–113. doi: 10.1016/j.jprot.2012.12.004
Scott, N., Porter, A., Santillo, D., Simpson, H., Lloyd-Williams, S., and Lewis, C. (2019). Particle characteristics of microplastics contaminating the mussel Mytilus edulis and their surrounding environments. Mar. Pollut. Bull. 146, 125–133. doi: 10.1016/j.marpolbul.2019.05.041
Serafini, L., Hann, J. B., Kültz, D., and Tomanek, L. (2011). The proteomic response of sea squirts (genus Ciona) to acute heat stress: a global perspective on the thermal stability of proteins. Comp. Biochem. Physiol. - Part D Genomics Proteomics 6, 322–334. doi: 10.1016/j.cbd.2011.07.002
Shi, W., Guan, X., Sun, S., Han, Y., Du, X., Tang, Y., et al. (2020). Nanoparticles decrease the byssal attachment strength of the thick shell mussel Mytilus coruscus. Chemosphere 257:127200. doi: 10.1016/j.chemosphere.2020.127200
Tang, Q., Xu, Y., Deng, C., Cheng, C., Dai, Z., Yang, Z., et al. (2020). Differential proteomic analysis to identify proteins associated with apomeiosis in Boehmeria tricuspis (Hance) Makino Using an iTRAQ-Based strategy. J. Proteome Res. 1, 661–669. doi: 10.1021/acs.jproteome.0c00586
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Theocharis, A. D., Skandalis, S. S., Gialeli, C., and Karamanos, N. K. (2016). Extracellular matrix structure. Adv. Drug Deliv. Rev. 97, 4–27. doi: 10.1016/j.addr.2015.11.001
Tomanek, L., and Zuzow, M. J. (2010). The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossulus to acute heat stress: Implications for thermal tolerance limits and metabolic costs of thermal stress. J. Exp. Biol. 213, 3559–3574. doi: 10.1242/jeb.041228
Ueki, T., Koike, K., Fukuba, I., and Yamaguchi, N. (2018). Structural and mass spectrometric imaging analyses of adhered tunic and adhesive projections of solitary ascidians. Zoolog. Sci. 35, 535–547. doi: 10.2108/zs180051
Vargas-Chacoff, L., Saavedra, E., Oyarzún, R., Martínez-Montaño, E., Pontigo, J. P., Yáñez, A., et al. (2015). Effects on the metabolism, growth, digestive capacity and osmoregulation of juvenile of Sub-Antarctic Notothenioid fish Eleginops maclovinus acclimated at different salinities. Fish Physiol. Biochem. 41, 1369–1381. doi: 10.1007/s10695-015-0092-3
Vind, A. C., Genzor, A. V., and Bekker-Jensen, S. (2020). Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res. 48, 10648–10661. doi: 10.1093/nar/gkaa757
von Byern, J., and Grunwald, I. (eds) (2010). Biological Adhesive Systems. Vienna: Grunwald Springer.
Vornanen, M., Hassinen, M., Koskinen, H., and Krasnov, A. (2005). Steady-state effects of temperature acclimation on the transcriptome of the rainbow trout heart. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 289, 1177–1184. doi: 10.1152/ajpregu.00157.2005
Waite, J. H., Andersen, N. H., Jewhurst, S., and Sun, C. (2005). Mussel adhesion: finding the tricks worth mimicking. J. Adhes. 81, 297–317. doi: 10.1080/00218460590944602
Wei, J., Zhang, J., Lu, Q., Ren, P., Guo, X., Wang, J., et al. (2020). Genomic basis of environmental adaptation in the leathery sea squirt (Styela clava). Mol. Ecol. Resour. 20, 1414–1431. doi: 10.1111/1755-0998.13209
Yan, T., Han, S., Wang, J., Lin, H., and Cao, W. (2017). An overview of fouling ascidians. Acta Ecol. Sin. 37, 6647–6655.
Yu, Z., Qi, Z., Hu, C., Liu, W., and Huang, H. (2013). Effects of salinity on ingestion, oxygen consumption and ammonium excretion rates of the sea cucumber Holothuria leucospilota. Aquac. Res. 44, 1760–1767. doi: 10.1111/j.1365-2109.2012.03182.x
Zhan, A., Briski, E., Bock, D. G., Ghabooli, S., and Hugh, J. (2015). Ascidians as models for studying invasion success. Mar. Biol. 162, 2449–2470. doi: 10.1007/s00227-015-2734-5
Zhang, X., Liu, X., Zeng, L., Ye, Z., Hu, B., and Hu, W. (2017b). Barnacle adhesion: research on the substract detection and cement solidification. Prog. Biochem. Biophys. 44, 204–214. doi: 10.16476/j.pibb.2016.0390
Zhang, X., Ruan, Z., You, X., Wang, J., Chen, J., Peng, C., et al. (2017c). De novo assembly and comparative transcriptome analysis of the foot from Chinese green mussel (Perna viridis) in response to cadmium stimulation. PLoS One 12:e176677. doi: 10.1371/journal.pone.0176677
Zhang, G., Zhang, J., Wen, X., Zhao, C., Zhang, H., Li, X., et al. (2017a). Comparative iTRAQ-Based quantitative proteomic analysis of pelteobagrus vachelli liver under acute hypoxia: implications in metabolic responses. Proteomics 17, 1–10. doi: 10.1002/pmic.201700140
Zhang, Y., Sun, J., Mu, H., Li, J., Zhang, Y., Xu, F., et al. (2015). Proteomic basis of stress responses in the gills of the pacific oyster Crassostrea gigas. J. Proteome Res. 14, 304–317. doi: 10.1021/pr500940s
Zhang, Y., Wu, Q., Fang, S., Li, S., Zheng, H., Zhang, Y., et al. (2020). mRNA profile provides novel insights into stress adaptation in mud crab megalopa, Scylla paramamosain after salinity stress. BMC Genomics 21:559. doi: 10.1186/s12864-020-06965-5
Zhao, H., Sagert, J., Hwang, D. S., and Waite, J. H. (2009). Glycosylated hydroxytryptophan in a mussel adhesive protein from Perna viridis. J. Biol. Chem. 284, 23344–23352. doi: 10.1074/jbc.M109.022517
Zhao, H., Sun, C., Stewart, R. J., and Waite, J. H. (2005). Cement proteins of the tube-building polychaete Phragmatopoma californica. J. Biol. Chem. 280, 42938–42944. doi: 10.1074/jbc.M508457200
Keywords: ascidian, biofouling, invasive species, proteomics, salinity, temperature, environmental stress
Citation: Li X, Li S, Cheng J, Fu R and Zhan A (2021) Proteomic Response to Environmental Stresses in the Stolon of a Highly Invasive Fouling Ascidian. Front. Mar. Sci. 8:761628. doi: 10.3389/fmars.2021.761628
Received: 20 August 2021; Accepted: 30 September 2021;
Published: 26 October 2021.
Edited by:
Lingling Wang, Dalian Ocean University, ChinaReviewed by:
Jian-Wen Qiu, Hong Kong Baptist University, Hong Kong, SAR ChinaCopyright © 2021 Li, Li, Cheng, Fu and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aibin Zhan, emhhbmFpYmluQGhvdG1haWwuY29t; YXpoYW5AcmNlZXMuYWMuY24=; Shiguo Li, c2dsaUByY2Vlcy5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.