- 1Department of Biological Sciences, National University of Singapore, Singapore, Singapore
- 2Tropical Marine Science Institute, National University of Singapore, Singapore, Singapore
- 3The Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat Gan, Israel
- 4School of Natural and Physical Sciences, University of Papua New Guinea, Port Moresby, Papua New Guinea
- 5Yale-NUS College, National University of Singapore, Singapore, Singapore
- 6Centre for Nature-Based Climate Solutions, National University of Singapore, Singapore, Singapore
- 7Lee Kong Chian Natural History Museum, National University of Singapore, Singapore, Singapore
Reef-building corals live in symbiosis with the phototrophic dinoflagellate family Symbiodiniaceae, which comprises diverse genera such as Cladocopium and Durusdinium. Pachyseris speciosa, a widely distributed Indo-Pacific coral found in a variety of reef habitats, is known to be associated with these two Symbiodiniaceae genera, but little is known about the biogeographic variability of the endosymbiont communities across the region. In this study, the diversity and dominance patterns of Symbiodiniaceae at the western and eastern areas of the Central Indo-Pacific region were examined. We sampled Pachyseris speciosa colonies at seven and six sites in Singapore and Papua New Guinea, respectively, and genotyped their endosymbionts based on the internal transcribed spacer (ITS) markers using two distinct methods, quantitative polymerase chain reaction (qPCR) and high-throughput sequencing (HTS). Results showed 92% of all colonies in Singapore exhibiting Cladocopium dominance. There was a higher abundance of Durusdinium compared to Cladocopium in certain colonies from one site, Pulau Hantu (mean Durusdinium abundance of 90%, compared to 0–14% among all other sites). In contrast, variation in the endosymbiont communities was generally higher among sites in Papua New Guinea. Cladocopium expectedly dominated most colonies (75%), although colonies from Kimbe Bay (85%) and Kavieng (65%) showed Durusdinium dominance. Between localities, relative genus abundances based on qPCR were not significantly different, but HTS showed that the ratio of Durusdinium over Cladocopium was significantly higher in Papua New Guinea corals. Notably, 6% of colonies from Singapore and 15% from Papua New Guinea showed endosymbiont dominance patterns that were inconsistent between the two methods, underscoring the need for further validation of symbiotic algal quantification based on HTS. The richness of ITS2 type profiles was clearly lower among colonies from the impacted and turbid reefs of Singapore compared to the less urbanized reefs of Papua New Guinea. These coral intraspecific variations of Symbiodiniaceae communities within and among localities suggest that local conditions are important drivers of endosymbiosis and may ultimately influence corals’ resilience against global stressors such as ocean warming.
Introduction
Reef-building stony corals (Cnidaria: Anthozoa: Scleractinia) are associated with the dinoflagellate alga Symbiodiniaceae, forming an ecologically critical symbiotic relationship (Falkowski et al., 1984; Stanley and Fautin, 2001; Baker, 2003). The partnership yields multiple ecological benefits for both the coral host and algal endosymbiont, providing the basis of the tropical coral reef ecosystem and its survival. Symbiotic corals depend on the photosynthetic activities of their mutualistic dinoflagellates (Boilard et al., 2020; LaJeunesse, 2020), or Symbiodiniaceae, to produce the energy required for calcium carbonate skeletal growth of the coral hosts, ultimately driving coral reef development (Al-Horani et al., 2003). In return, in hospite Symbiodiniaceae cells are provided with a light-enriched environment and the by-products of host metabolism (Banin et al., 2003).
These extremely diverse microalgae are currently recognized as several phylogenetically distinct genera (LaJeunesse et al., 2018; Boilard et al., 2020). Reef-building stony corals typically host multiple Symbiodiniaceae species and are most commonly associated with four genera (Symbiodinium, Breviolum, Cladocopium, and Durusdinium) (Rowan and Knowlton, 1995; Baker, 2003; Little et al., 2004; Stat et al., 2008b). The endosymbiont community may grant varying degrees of fitness to the coral host depending on the holobiont’s capability to tolerate environmental stressors and bleaching (Baker, 2001; Stat et al., 2011).
Symbiodiniaceae endosymbiont community composition has been shown to vary between biogeographic regions, host species, reef depth, light accessibility and shifting environmental dynamics, among other factors (Rowan and Knowlton, 1995; Rowan et al., 1997; Stat et al., 2008a; Rouzé et al., 2016; Eckert et al., 2020; Hume et al., 2020). In particular, the diversity of Symbiodiniaceae in various geographic locations and host taxa has been studied extensively (LaJeunesse, 2002; LaJeunesse et al., 2004; Huang et al., 2011; Keshavmurthy et al., 2014; Hume et al., 2015; Leveque et al., 2019; Terraneo et al., 2019; Teschima et al., 2019; Tan et al., 2020). For example, latitudinal variation in Symbiodiniaceae genus dominance has been observed in the Saudi Arabian Red Sea, with shifts from Cladocopium dominance to Durusdinium dominance in Porites colonies along a north-south gradient (Terraneo et al., 2019; see also Chen et al., 2019). Fundamentally, with Cladocopium and Durusdinium known to be the most abundant endosymbionts in tropical reefs, corals hosting Durusdinium were initially thought to be more tolerant against thermal stress and turbidity fluctuations (Stat and Gates, 2011; Silverstein et al., 2015), whereas Cladocopium was generally considered the more stress-sensitive genus (Chen et al., 2003; Stat and Gates, 2011). However, it is now well understood that Cladocopium is found in corals throughout a large depth range and among multiple host species, and even when present in marginal conditions, there may be taxon-specific associations and local adaptations that result in it being the dominant genus (Hume et al., 2015; Rouzé et al., 2016; Innis et al., 2018). For example, corals in the Red Sea exhibit strong species-specific associations and display Cladocopium dominance despite a strong latitudinal gradient and facing high temperatures (Ziegler et al., 2017; Osman et al., 2020). Similarly, corals withstanding extreme temperatures in the Persian Gulf maintain predominant associations with Cladocopium (Howells et al., 2020).
The diversity and functioning of specific coral-Symbiodiniaceae relationships rely on the sensitivity of symbionts to thermal stress, irradiance and/or host-specificity (LaJeunesse et al., 2004). The composition of the symbionts within the host can be influenced by changing environments and driven by various host or symbiont processes (Kemp et al., 2014). Community alterations are achieved via “switching” (changes in the endosymbiont community composition via exchange of symbionts from the environment) or “shuffling” (shifts in the relative abundances of present symbionts) (Baker, 2003). The holobiont’s capacity to undergo these “switching” or “shuffling” processes is vital for its resilience against environmental stress (Baker, 2004; Sampayo et al., 2008; Kemp et al., 2014). More generally, the persistence of coral reefs and the survival of its associated organisms depend greatly on the responses of the coral-Symbiodiniaceae relationship to environmental change (Arif et al., 2014). Seeing as temperature anomalies due to global warming are predicted to occur more frequently (Hughes et al., 2017a,b), studying the diversity of symbionts in different regions is critical for understanding geographic variations in coral reef persistence.
The zooxanthellate coral, Pachyseris speciosa, is one of the most common and abundant corals found in the Central Indo-Pacific region with a wide distribution through various depths (DeVantier and Turak, 2017; Bongaerts et al., 2021). Symbiodiniaceae communities hosted by P. speciosa have been examined in the Great Barrier Reef, Red Sea, French Polynesia, and Singapore (LaJeunesse et al., 2004; Bongaerts et al., 2011; Ziegler et al., 2015b; Tanzil et al., 2016; Goulet et al., 2019; Meistertzheim et al., 2019; Jain et al., 2020; Smith et al., 2020). However, little is known about the biogeographic variabilities of its endosymbionts across the Central Indo-Pacific as comparisons among localities are limited.
Despite chronically high sediment deposition, turbidity and low light levels, the hyper-urbanized shores of Singapore have been home to 255 stony coral species (Huang et al., 2009). Pachyseris speciosa is one of the most common corals dominating the shallow reefs here (Wong et al., 2018; Chow et al., 2019) and is associated with Cladocopium and Durusdinium (Tanzil et al., 2016; Jain et al., 2020; Smith et al., 2020). By contrast, Papua New Guinea, which is generally much less affected by urbanization, has a greater coral diversity and is part of the Coral Triangle (Allen et al., 2003; Veron et al., 2011, 2015). Its reefs hold more than 500 stony species that are of vital importance to the local communities and their self-sustainable lifestyles (Allen et al., 2003; Nicholls, 2004; The Nature Conservancy, 2004). Apart from a study investigating the response of endosymbiont communities to elevated carbon dioxide, little is known about the Symbiodiniaceae communities associated with P. speciosa here (Noonan et al., 2013).
In this study, we characterize the relative abundances of Cladocopium and Durusdinium across various sites in Singapore and Papua New Guinea using two distinct quantification methods, quantitative polymerase chain reaction (qPCR) and high-throughput sequencing (HTS). The universal presence and known association of Pachyseris speciosa with Cladocopium and Durusdinium allow their relative abundances to be examined among coral colonies from the two regions. Here, we quantify genus abundances and dominance by the relative concentrations of the nuclear ribosomal internal transcribed spacer 1 (ITS1) region for Durusdinium and Cladocopium as evaluated by qPCR, and by the relative HTS read abundances of the internal transcribed spacer 2 (ITS2) region for the two genera. In accordance with the SymPortal framework (Hume et al., 2019), we also report on the ITS2 type profiles of P. speciosa that account for intragenomic variation and are representative of putative Symbiodiniaceae species. Our data on the relative abundances of the endosymbiont taxa between the western and eastern Central Indo-Pacific highlight the regional biogeographic drivers of similarities and differences in Symbiodiniaceae diversity. Furthermore, a sympatric species complex representing three distinct lineages of P. speciosa has been discovered recently (Bongaerts et al., 2021; Feldman et al., 2021). Using a cleaved amplified polymorphic sequence (CAPS) assay, we uncover the different P. speciosa lineages from our collections and determine whether they affect Symbiodiniaceae genus dominance. Additionally, we test the interchangeability of using qPCR and HTS to detect relative abundances of Cladocopium and Durusdinium based on a larger and geographically wider set of samples relative to our previous study (Jain et al., 2020).
Materials and Methods
Field Collections
Reefs at Singapore and Papua New Guinea were accessed via SCUBA and Pachyseris speciosa colonies were haphazardly sampled using a hammer and chisel. A total of 266 colonies were sampled, including 146 from seven different sites in Singapore between 25th and 31st May 2018. Between 18 and 25 samples were collected each from Terumbu Pempang Tengah (1°13′45.1″N 103°43′50.5″E), St. John’s Island (01°13′09.0″N 103°50′44.0″E), Pulau Semakau (01°12′11.5″N 103°45′15.0″E), Sisters’ Island (or Pulau Subar Laut; 01°12′49.0″N 103° 49′59.0″E), Pulau Hantu (01°13′36.5″N 103°44′47.0″E), Raffles Lighthouse (or Pulau Satumu; 01°09′36.5″N 103°44′24.0″E) and Kusu Island (or Pulau Tembakul; 01°13′33.0″N 103°51′34.0″E). From Papua New Guinea, 120 samples were taken, comprising 20 colonies from each of six sites – Kimbe Bay (5°09′55.2″S 150°30′00.2″E), Rabaul (4°12′13.9″S 152°09′54.6″E), Kavieng (2°33′02.1″S 150°47′42.2″E), Madang (5°13′54.0″S 145°48′05.3″E), Milne Bay (10°25′50.5″S 150°43′13.4″E) and Port Moresby (9°27′54.0″S 147°08′34.5″E) – between 1st and 28th September 2018 (Figure 1). All sampled fragments were transported to the laboratory in seawater and fixed in 100% molecular grade ethanol, or stored in 20% salt-saturated DMSO immediately following collection.
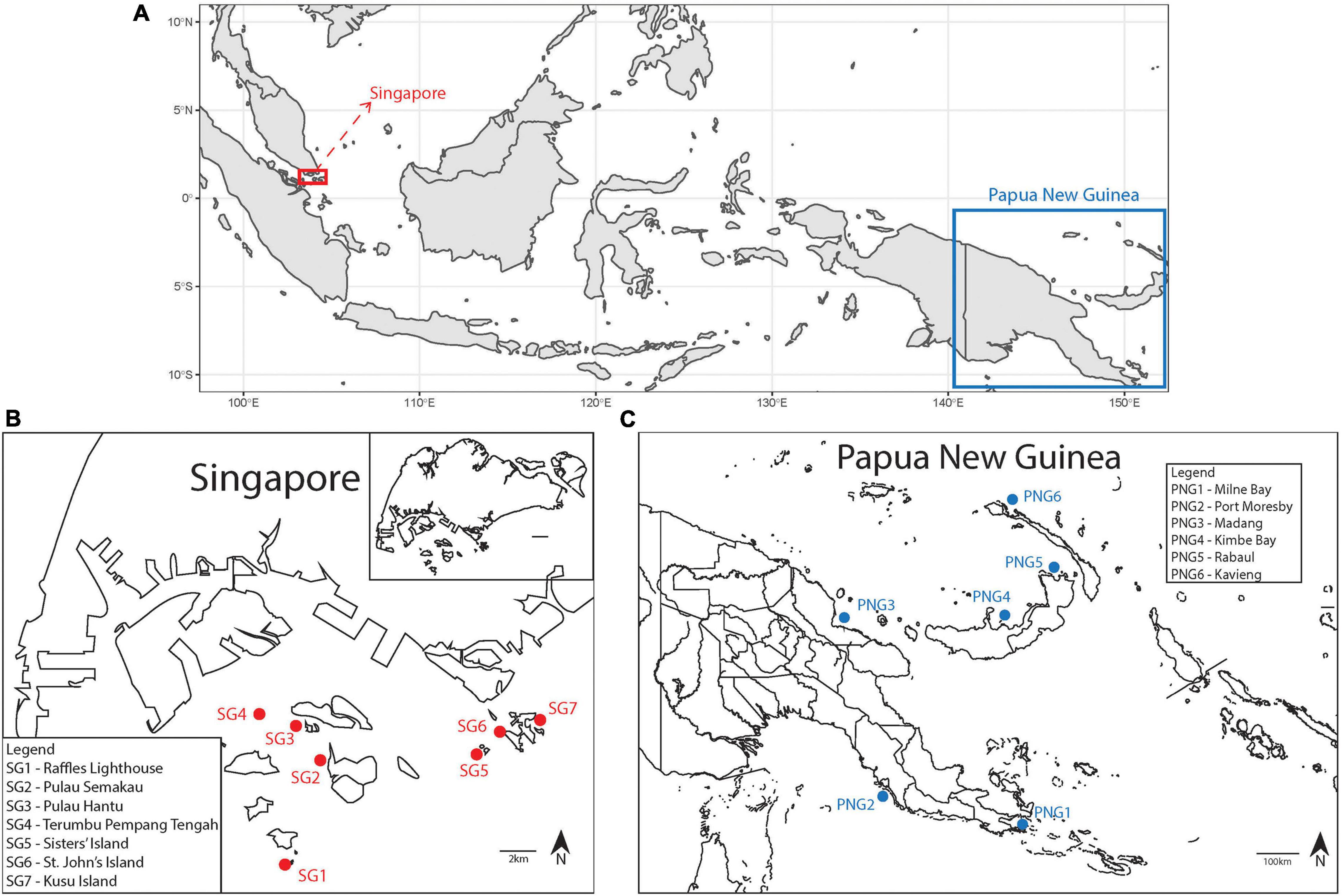
Figure 1. (A) Map showing Singapore and Papua New Guinea. (B) Map of Singapore with study sites (SG1 to SG7) indicated in red (inset: Map of the entire Singapore island for reference). (C) Map of Papua New Guinea with study sites (PNG1 to PNG6) indicated in blue.
DNA Extraction
Total genomic DNA was extracted from all the 146 Singapore samples and 120 Papua New Guinea samples using the DNeasy Blood and Tissue Kit (Qiagen Inc., Hilden, Germany) according to the manufacturer’s protocol. A 2–4 cm2 fragment of coral tissue was used for each sample extraction. Fragments were cut into smaller pieces using sterile forceps and scissors prior to addition of buffers. DNA was eluted in 150 μL nuclease-free water. Extracted DNA samples were stored at −80°C until further processing.
Quantitative Polymerase Chain Reaction
The ITS1 region, located between the 18S and 5.8S rRNA genes, was targeted for the quantification of relative Cladocopium and Durusdinium rDNA copy numbers. Two samples from a previous study were used to generate concentration standards for the ITS1 amplicons of Cladocopium and Durusdinium (Jain et al., 2020). The amplicons were quantified using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, MA, United States) with the Life Technologies Qubit 3.0 Fluorometer (Thermo Fisher Scientific, MA, United States) and then diluted to 50 ng/μL. For amplification of Cladocopium, the universal forward primer [5′–AAG GAG AAG TCG TAA CAA GGT TTC C–3′ (Ulstrup and Van Oppen, 2003)] and Cladocopium-specific reverse primer [5′–AAG CAT CCC TCA CAG CCA AA–3′ (Ulstrup and Van Oppen, 2003)] were used. For Durusdinium, the universal forward primer and the Durusdinium-specific reverse primer [5′–CAC CGT AGT GGT TCA CGT GTA ATA G–3′ (Ulstrup and Van Oppen, 2003)] were used. The amplification profile consisted of an initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 53°C for 45 s, 72°C for 45 s and a 5 min extension step at 72°C. The size of the PCR products (∼100 bp) was confirmed with gel electrophoresis. The products were then purified using SureClean Plus (Bioline Inc., London, United Kingdom) and quantified with the Qubit assay as mentioned above. Amplicon concentration standards were prepared by diluting the purified PCR product to 1 ng/μL, and then serially diluting to get 10 standards (1 to 10–9 ng/μL) each for Cladocopium and Durusdinium. All standards were run alongside each qPCR experiment.
All DNA extracts were quantified and diluted to 1 ng/μL. SsoAdvanced SYBR Green SuperMix (Bio-Rad, CA, United States), the above-mentioned primers (Ulstrup and Van Oppen, 2003) and diluted standardized DNA template were used for qPCR using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, CA, United States). All standards and samples were run in triplicates. The thermocycling profile used consists of an initial denaturation at 98°C for 3 min followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. A melt curve analysis from 65 to 95°C (0.5°C increase per 5 s) was subsequently performed to verify the specificity of the amplification.
CFX Manager Software (Bio-Rad, CA, United States) was used to analyze the efficiency of standards and to enumerate the relative quantities of Cladocopium and Durusdinium (expressed as log-scaled ratio of Durusdinium concentration to Cladocopium concentration) in each sample based on the standard curve, following Jain et al. (2020).
Illumina Library Preparation, Sequencing and Bioinformatics
The ITS2 region, located between the 5.8S and 28S rRNA genes, was targeted for conventional PCR and HTS. All DNA extracts were quantified and diluted to 1 ng/μL. Reagent volumes, PCR cycling conditions and PCR clean-up protocols followed the 16S Metagenomic Sequencing Library Preparation guide for the Illumina MiSeq System (Illumina, 2013). This two-step protocol included an amplicon PCR (1st step) with the ITS2-specific SYM_VAR primer pair [SYM_VAR_5.8S2: 5′–GAATTGCAGAACTCCGTGAACC–3′ (Hume et al., 2015), SYM_VAR_REV: 5′–CGGGTTCWCTTGTYTGACTTCATGC–3′ (Hume et al., 2013)]. The index PCR (2nd step) added unique dual indexes (see Supplementary Table 1) and adapters complementary to the MiSeq flow cell for binding. Indexed PCR products were normalized using the SequalPrep™ Normalization Plate Kit (Thermo Fisher Scientific, MA, United States), pooled and sequenced on the Illumina MiSeq sequencing platform (V3 chemistry; 30% PhiX spike-in) for 300-bp paired-end reads.
Demultiplexed paired fastq.gz files containing Symbiodiniaceae ITS2 reads were analyzed using the SymPortal framework (Hume et al., 2019), run locally. Briefly, sequence filtering and a standardized quality control pipeline were conducted using mothur 1.39.5 (Schloss et al., 2009), the BLAST + suite (Camacho et al., 2009) and minimum entropy decomposition (MED; Eren et al., 2015).
Identifying Distinct Lineages of Pachyseris speciosa
Bongaerts et al. (2021) discovered distinct lineages within Pachyseris speciosa that represented a sympatric species complex and developed a CAPS assay to identify each of the lineages. The three lineages were referred to as “green,” “blue” and “red” (see also Feldman et al., 2021). To determine whether the different lineages influenced Symbiodiniaceae genus dominance in the corals studied here, we performed the CAPS assay on our P. speciosa samples.
DNA extracted from all the colonies from Singapore (n = 146) was assigned to one of the three lineages using the CAPS assay following Bongaerts et al. (2021). As we found no link between host lineage and Symbiodiniaceae genus dominance in the Singapore samples (see below), up to five randomly chosen samples from each site in Papua New Guinea (n = 21) were tested. Briefly, total DNA extracted from each sample was amplified using lineage-specific primers (PsPe-Green, PsPe-Blue and PsPe-Red) and purified using SureClean Plus. Purified products were then digested with the associated restriction enzyme (Green-HhaI, Blue-HaeIII, and Red-Taqα1, respectively) and digestion verified on a 3% gel. Following enzymatic digestion, the “green” or “blue” lineage was determined with positive digestion observed using the associated HhaI and HaeIII restriction enzyme. The “red” lineage was assigned to samples showing no digestion using the Taqα1 restriction enzyme. To verify digestion of PCR amplicons that could not be visualized through gel imaging, the purified PCR products were sequenced using Sanger sequencing. The sequences were analyzed in Geneious Prime 2020.2.4 (Kearse et al., 2012) and a virtual digest was performed to determine the lineages.
Statistical Analyses
Data were analyzed using R 3.6.1 (R Core Team, 2017). Two separate runs of the nested analysis of variance (ANOVA) were performed to assess the differences in ITS1 and ITS2 Durusdinium to Cladocopium ratios among sites in each locality and between the two localities. Specifically, the ratio of Durusdinium to Cladocopium ITS1 concentrations for each sample obtained from the qPCR experiments were compared between localities (Singapore vs. Papua New Guinea) with sites nested within localities. Separately, the ratios of Durusdinium to Cladocopium ITS2 HTS read abundances were also compared between the two localities with sites nested within localities. The Tukey test was further performed to resolve statistical differences between sites in each locality.
To determine if and how the qPCR tests and HTS reads were comparable in the relative Durusdinium vs. Cladocopium levels obtained, we ran a linear model to quantify the relationship between relative (Durusdinium divided by Cladocopium) ITS1 concentrations and ITS2 read abundances at the sample level. Data were log-transformed for normality and homoscedasticity.
To characterize the geographical variation of Symbiodiniaceae communities at different localities and sites, we report the ITS2 type profiles obtained from SymPortal.
Results
Quantitative Polymerase Chain Reaction
The qPCR quantification of the relative abundances of Durusdinium vs. Cladocopium ITS1 in the 146 colonies sampled across Singapore showed limited variation among the seven sites examined (Terumbu Pempang Tengah, St. John’s Island, Pulau Semakau, Sisters’ Island, Pulau Hantu, Raffles Lighthouse and Kusu Island; p = 0.392, df = 6, F = 1.056; Figure 2). Post hoc Tukey tests further revealed insignificant pairwise site differences in the ratios of the relative abundances of Durusdinium vs. Cladocopium (p values ranging from 0.4 to 1).
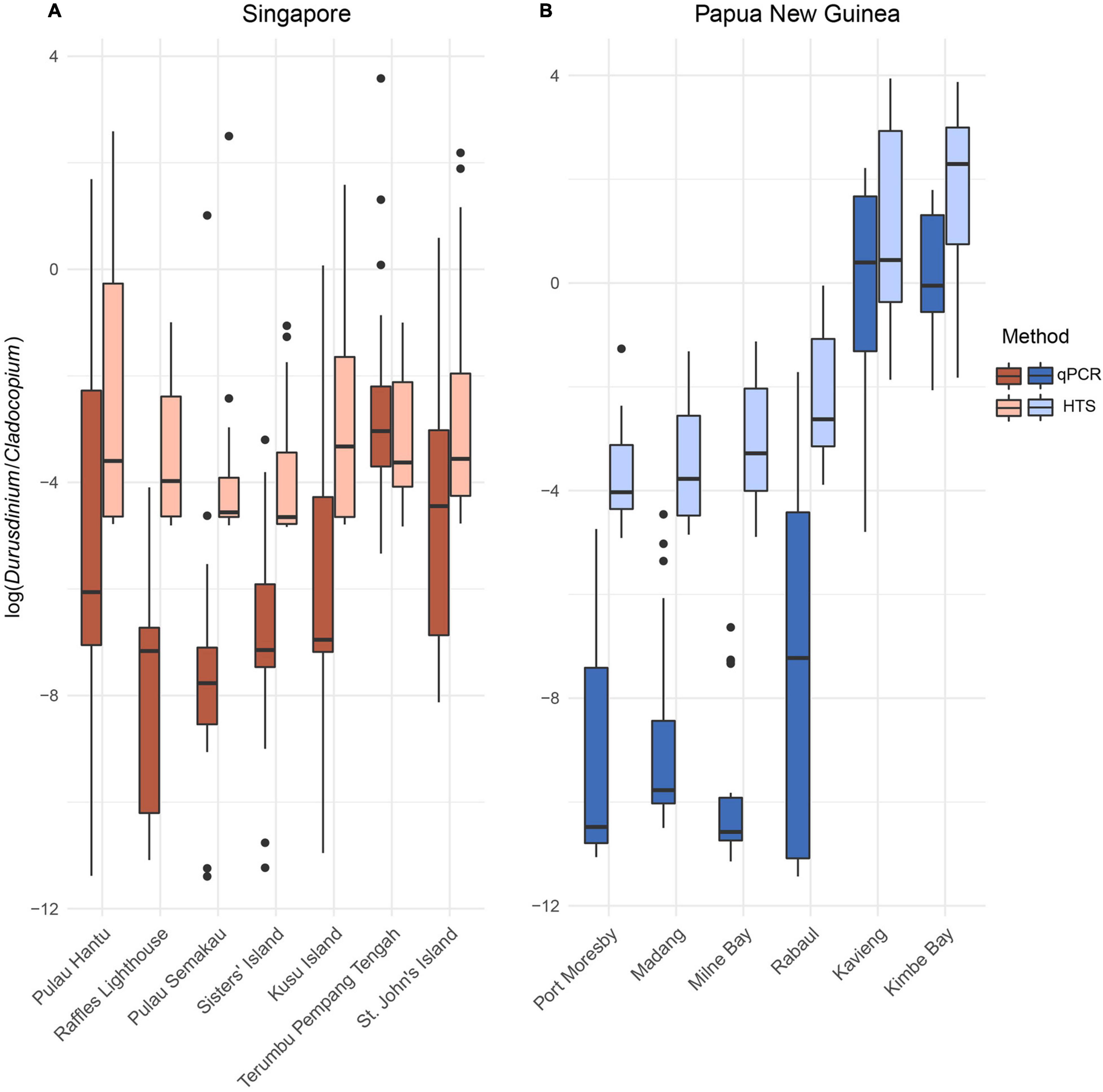
Figure 2. Ratios of Durusdinium vs. Cladocopium relative abundances (log-transformed ratios) in the (A) seven sites across Singapore and (B) six sites across Papua New Guinea obtained via quantitative polymerase chain reaction (qPCR) and high-throughput sequencing (HTS).
Among the 120 Papua New Guinea colonies, the ITS1 ratios were significantly different between sites (Port Moresby, Madang, Milne Bay, Rabaul, Kavieng, and Kimbe Bay; p < 0.001, df = 5, F = 7.136) with post hoc Tukey tests revealing significantly greater proportions of Durusdinium in colonies sampled from Kavieng compared to the other four sites (Madang, Milne Bay, Rabaul and Port Moresby; p < 0.001). There were no significant differences in the relative abundances of Cladocopium and Durusdinium between samples from Madang, Milne Bay, Rabaul and Port Moresby. Overall, ITS1 Durusdinium to Cladocopium ratios between Singapore and Papua New Guinea were not significantly different (p = 0.494, df = 1, F = 0.470).
High-Throughput Sequencing
Illumina sequencing reads of the 146 Pachyseris speciosa colonies sampled across the seven sites in Singapore yielded a total of 19,721,036 raw reads, resulting in 9,860,518 contigs. Following quality control and sequence filtering, a total of 8,113,743 Symbiodiniaceae ITS2 sequences comprising 77,067 unique sequences were retained. The relative abundances of Durusdinium vs. Cladocopium ITS2 reads within the 146 coral colonies were not significantly different between the seven sites in Singapore (p = 0.118, df = 6, F = 1.732; Figure 2). Post hoc Tukey tests indicated higher proportions of Durusdinium in colonies from Pulau Hantu compared to Raffles Lighthouse (p = 0.131), although no statistically significant differences were detected among all sites. These statistical similarities and differences between the sites in Singapore were generally consistent with patterns obtained with ITS1 ratios.
Illumina sequencing reads of the 120 Pachyseris speciosa colonies sampled across the six sites in Papua New Guinea yielded a total of 20,018,361 raw reads, resulting in 10,365,381 raw contigs. Following quality control and sequence filtering, a total of 7,688,010 Symbiodiniaceae ITS2 sequences comprising 93,003 unique sequences were retained. The relative abundances of Durusdinium vs. Cladocopium ITS2 reads were significantly different among the six Papua New Guinea sites (p = 0.0032, df = 5, F = 3.801). Post hoc Tukey test showed larger proportions of Durusdinium detected in colonies from Kavieng compared to those from Madang, Milne Bay, Rabaul and Port Moresby (p < 0.05). There were no significant differences in the relative abundances of Cladocopium and Durusdinium between samples from Madang, Milne Bay, Rabaul and Port Moresby. Colonies from Kimbe Bay and Kavieng had similar proportions of Durusdinium. These results were consistent with the relative abundances derived from qPCR. However, the ITS2 Durusdinium vs. Cladocopium ratios based on HTS showed significant differences between Singapore and Papua New Guinea (p = 0.0015, df = 1, F = 10.352).
Methodological and Lineage Variations
Overall, the patterns of relative Durusdinium vs. Cladocopium abundances were consistent between the qPCR tests and HTS reads (Figure 2). At the sample level, relative ITS1 concentrations determined via qPCR and ITS2 HTS read abundances could be explained by a significant linear relationship at each locality (Singapore: R2 = 0.435, p << 0.001; Papua New Guinea: R2 = 0.778, p << 0.001; Figure 3). However, six and 15 colonies from Singapore and Papua New Guinea, respectively, recorded as having Durusdinium-dominated communities with HTS were actually Cladocopium-dominated according to qPCR. Furthermore, three colonies each from Singapore and Papua New Guinea were recorded as Durusdinium-dominated with qPCR but actually showed Cladocopium dominance using HTS.
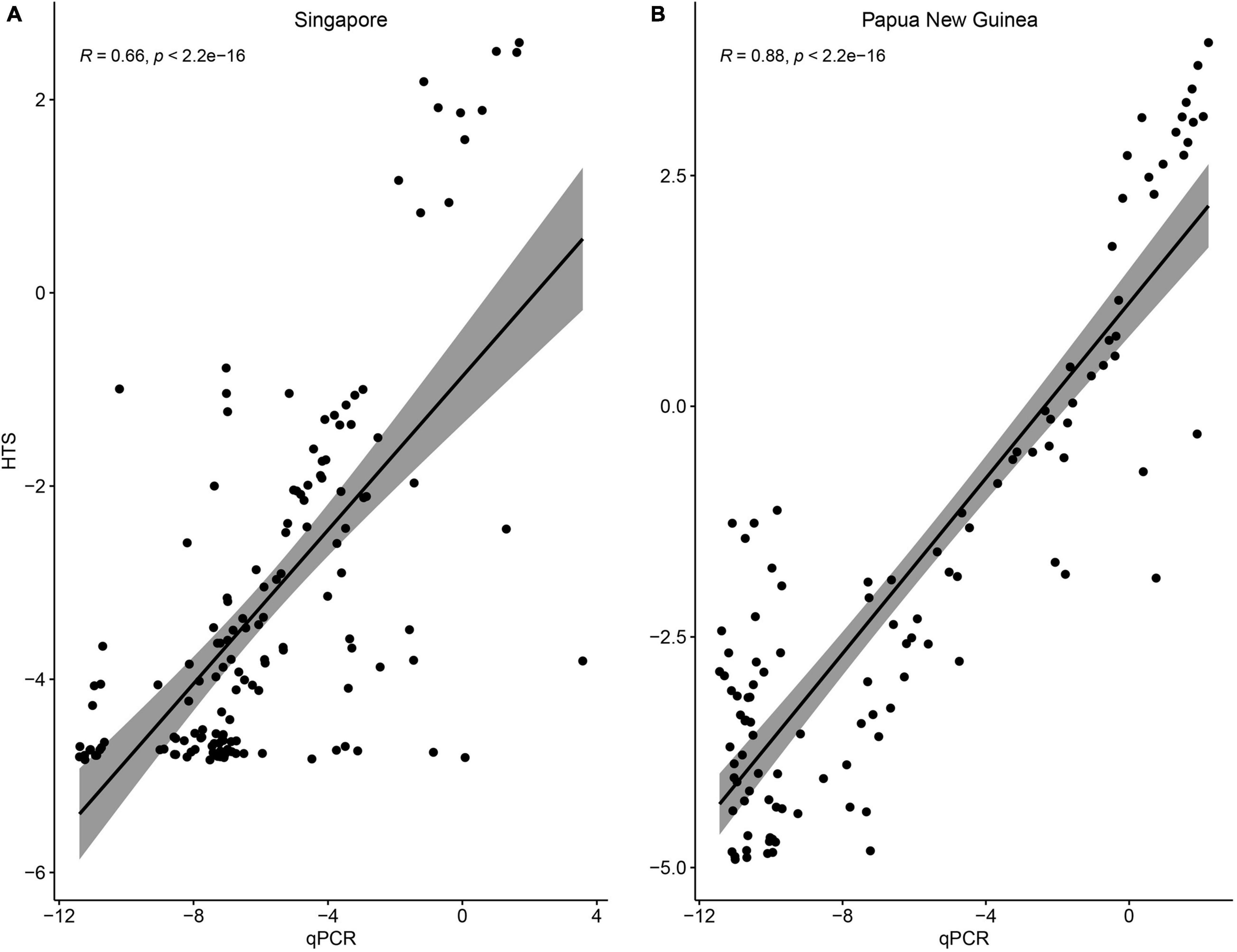
Figure 3. Relationship between the log-transformed relative abundances of Durusdinium vs. Cladocopium ITS1 concentrations (quantitative polymerase chain reaction) and ITS2 read abundances (high-throughput sequencing) on a sample level among colonies from (A) Singapore and (B) Papua New Guinea.
Based on the CAPS assay following Bongaerts et al. (2021), two lineages (“green” and “blue”) occur sympatrically within Singapore and Papua New Guinea. No “red” lineage was identified. To associate the host lineages with the Symbiodiniaceae communities, we compared Cladocopium and Durusdinium dominance in each colony with the assigned lineage (Supplementary Table 2). Colonies that showed a dominance of Durusdinium were assigned to either a “green” or “blue” lineage. There was no clear association of the “green” lineage with dominance of Durusdinium, so we surmised that there was no discernible link between host lineage and endosymbiont community composition.
ITS2 Type Profiles
Based on the SymPortal framework, nine ITS2 type profiles were found in Singapore P. speciosa, including five Cladocopium and four Durusdinium profiles (Figure 4). The five profiles of Cladocopium were associated with eight unique ITS2 sequences and the four profiles of Durusdinium were also associated with eight unique ITS2 sequences. The profile C27-C3-1319-C27a was detected in colonies across all sites except for St. John’s Island where only C27-C3-C27a was detected, suggesting the absence of the Cladocopium sequence “1319” at this particular site. Colonies from Raffles Lighthouse, Terumbu Pempang Tengah and Sisters’ Island showed none of the Durusdinium profiles. Colonies from the other sites contained Durusdinium type profiles, with D1/D4-D1c detected predominantly in colonies from Pulau Hantu and St. John’s Island, and D1-D4-D4c-D4f-D1l detected in colonies from Pulau Semakau and Kusu Island.
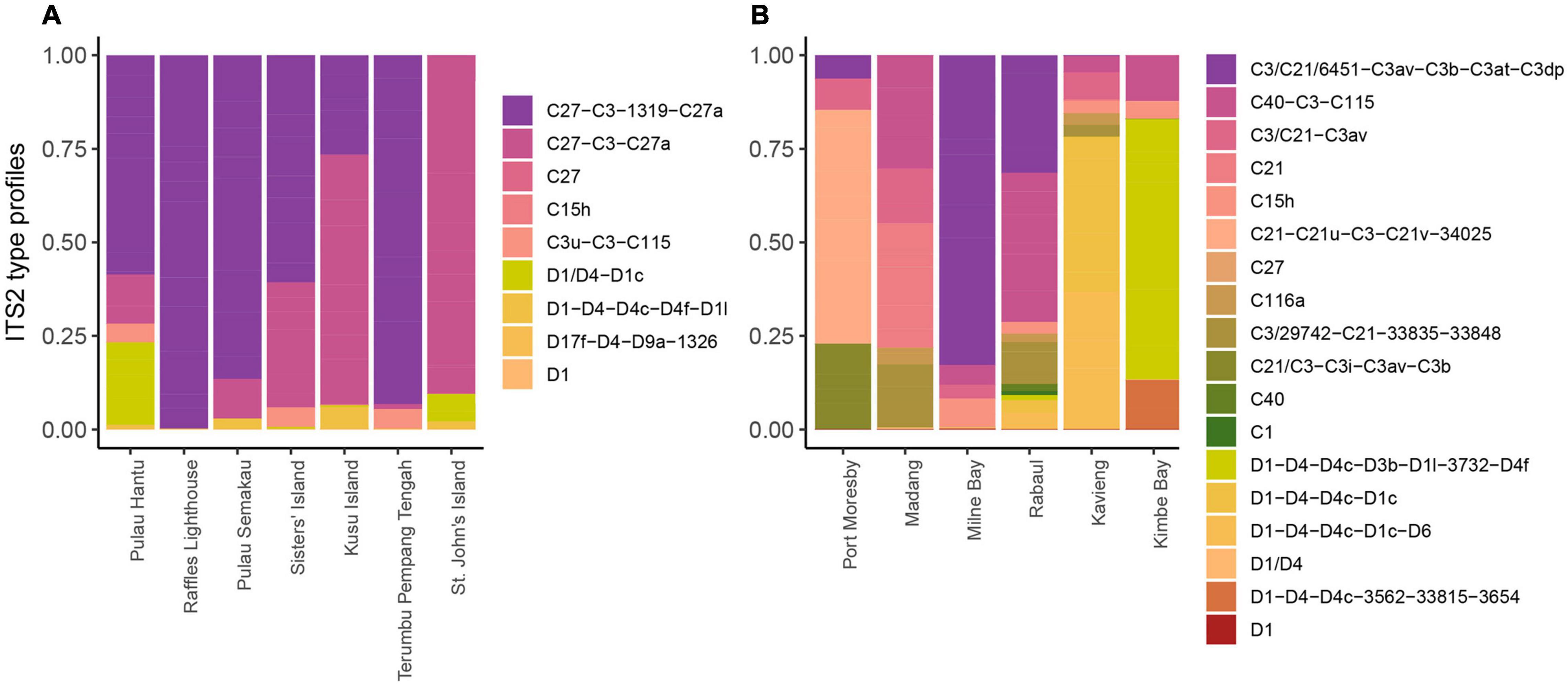
Figure 4. Diversity and relative abundances of Symbiodiniaceae ITS2 type profiles among the sites studied in (A) Singapore and (B) Papua New Guinea.
Colonies from Papua New Guinea showed a higher number of ITS2 type profiles (18) as compared to colonies from Singapore. There were 12 ITS2 type profiles assigned to Cladocopium and six type profiles assigned to Durusdinium across all the six sites sampled (Figure 4). Each site showed dominance of a distinct ITS2 type profile. The dominant type profile detected in colonies from Port Moresby, Madang, Milne Bay and Rabaul belonged to Cladocopium whereas colonies from Kavieng and Kimbe Bay showed dominance by Durusdinium type profiles (Port Moresby: C21-C21u-C3-C21v-34025; Madang: C21; Milne Bay: C3/C21/6451-C3av-C3b-C3at-C3dp; Rabaul: C40-C3-C115; Kavieng: D1-D4-D4c-D1c and Kimbe Bay: D1-D4-D4c-D3b-D1l-3732-D4f).
Discussion
Geographical Variation
In this study, the relative abundances of Symbiodiniaceae genera and ITS2 type profiles hosted by the common reef-building stony coral, Pachyseris speciosa, were compared among colonies from Singapore and Papua New Guinea. Cladocopium C27-C3-1319-C27a was the most abundant ITS2 type profile found in all colonies across sites in Singapore with the exception of St. John’s Island where C27-C3-C27a was detected (Figure 4). This is expected as previous studies have recorded similar results across multiple sites in Singapore (Jain et al., 2020; Smith et al., 2020) with C27 dominating most P. speciosa colonies. However, the type profile C27/C3/C3u-C115 was the most dominant across all six sites examined by Smith et al. (2020). This marginal difference may be the result of 3 years separating the sampling by Smith et al. (2020) in 2015 and the present study in 2018. In particular, a global-scale coral bleaching event (GCBE) occurred here in 2016 (Toh et al., 2018), leading to a shift in the Symbiodiniaceae communities in at least one of the sites, Pulau Hantu (Jain et al., 2020), even as the corals had recovered by 2018 (Ng et al., 2020).
Generally, ITS2 type profiles of Cladocopium were more abundant among colonies in Singapore compared to type profiles of Durusdinium, corroborating the findings at Pulau Hantu by Jain et al. (2020). While there were five Cladocopium and four Durusdinium type profiles, the dominant Cladocopium type profiles included the sequence of C27 and all the Durusdinium type profiles included the sequence of D1. These were also the dominant sequences detected for each of the genera in Jain et al. (2020). High Durusdinium abundance was detected in 26% of colonies from Pulau Hantu (Jain et al., 2020), with fewer colonies from Pulau Semakau (5%), Kusu Island (6%), and St. John’s Island (14%). There were no Durusdinium-dominated colonies at Raffles Lighthouse, Sisters’ Island and Terumbu Pempang Tengah.
The increased relative abundances of Durusdinium in colonies from Pulau Hantu compared to others could be a result of thermal or urbanization-related stress (Jain et al., 2020), given that certain Durusdinium species are commonly associated with resistance to high temperatures and turbidity fluctuations (Baker, 2003; Ulstrup and Van Oppen, 2003; Jones et al., 2008). Durusdinium has been suggested to play an important role in the survival of Singapore’s corals (Poquita-Du et al., 2020). The warm, turbid environment subject corals growing here with challenges not usually ideal for coral health (e.g., close proximity to a major urban center and high sedimentation rates), and furthermore, these reefs are susceptible to major bleaching events (Guest et al., 2016; Ng et al., 2020). Indeed, Durusdinium has mostly been associated with other marginal reefs comparable to those of Singapore (LaJeunesse et al., 2010; Keshavmurthy et al., 2014). Slight changes in the endosymbiont communities are known to be influenced by various environmental stressors (Buddemeier and Fautin, 1993; Baker, 2001). The limited differences in endosymbiont communities between various sites in Singapore suggest the stresses (e.g., thermal, sediment) that P. speciosa is exposed to may be similar among sites.
In colonies from Papua New Guinea, Cladocopium type profiles were the most abundant at all sites except Kimbe Bay and Kavieng, where Durusdinium dominated most colonies. Rather than the C27-C3-1319-C27a dominance exhibited by corals in Singapore, colonies from Papua New Guinea hosted mostly distinct Cladocopium type profiles at different sites. Colonies from Kimbe Bay and Rabaul had the most abundant C40-C3-C115, whereas colonies from Kavieng, Milne Bay, Madang and Port Moresby hosted C3/C21-C3av, C3/C21/6451-C3av-C3b-C3at-C3dp, C21 and C21-C21u-C3-C21v-34025, respectively. These Cladocopium type profiles included sequences of either C3 or C21, or both. Notably, Cladocopium C3, C21, and C40 are regarded as generalists due to their associations with a wide range of reef coral taxa in the tropics at various depths (LaJeunesse et al., 2003, 2004). In fact, the C3 group comprises many divergent lineages that are symbiotic with distinct host taxa (LaJeunesse, 2005; Thornhill et al., 2014). The relatively high variability in the dominant type profile between sites observed at Papua New Guinea suggests a certain level of local adaptation even as Cladocopium C3, C21 and C40 share very similar ITS2 sequences (LaJeunesse et al., 2003).
Pachyseris speciosa colonies from Kimbe Bay and Kavieng were dominated by Durusdinium. The distinction of these two sites based on Durusdinium dominance does not appear to be spatially driven, although Kimbe Bay, Rabaul and Kavieng are on the islands of New Britain and New Ireland off the New Guinea mainland and samples from these sites contained higher Durusdinium levels than the others. It is known that the abundance of Durusdinium may be elevated in corals undergoing bleaching, and may not revert to the pre-bleaching community post-recovery (Tye Pettaya et al., 2015; Boulotte et al., 2016; LaJeunesse et al., 2016). However, these sites do not appear to have bleached in recent years (Mana, R. R., unpublished data), suggesting that local environmental factors such as light intensity, sea surface temperature and currents may promote Durusdinium dominance in Kimbe Bay and Kavieng. In particular, Kavieng faces the Pacific Ocean and is directly exposed to the South Equatorial Current, likely experiencing distinct local conditions compared to the other more sheltered sites to the south. Further tests and environmental measurements are needed to explain the Symbiodiniaceae dominance patterns at Papua New Guinea.
Comparison of the type profiles between Singapore and Papua New Guinea showed highly diverse endosymbiont communities within and between localities (Figure 4). In particular, the SymPortal framework revealed 18 type profiles throughout Papua New Guinea, which is twice the number detected in Singapore. It remains unclear why the endosymbiont compositions between Singapore and Papua New Guinea differ (LaJeunesse et al., 2004; Bongaerts et al., 2011; Cooper et al., 2011; Ziegler et al., 2015b), but these biogeographical patterns could be a consequence of different environmental factors and thermal regimes as discussed above (Tonk et al., 2013; Ziegler et al., 2015a,2017). We note that the spatial distances between sampling sites in Papua New Guinea (∼100 km) are considerably greater than those in Singapore (∼10 km). This could mean greater connectivity and/or more similar environmental conditions among sites in Singapore, and may explain the relatively homogenized Symbiodiniaceae communities within Singapore compared to Papua New Guinea.
Methodological Variation
Clearly there is a need to extend our comparisons of the endosymbiont communities to more localities in the Central Indo-Pacific using complementary approaches. As we have demonstrated, several colonies recorded as having Durusdinium-dominated communities with HTS were actually Cladocopium-dominated according to qPCR (Figure 3). Therefore, it may even be necessary to revisit samples that have been used in the past with HTS to infer that symbiosis with Durusdinium is associated with thermal tolerance (Baker, 2001; Baker et al., 2004; Stat et al., 2013). Similarly, in our previous study which explored the Symbiodiniaceae community patterns in P. speciosa during and after the 2016 GCBE (Jain et al., 2020), three colonies were found to be Durusdinium-dominant based on HTS and Cladocopium-dominant based on qPCR. Nevertheless, overall patterns in the relative abundances of Durusdinium vs. Cladocopium remained consistent.
In this study, differences between localities in the levels of Durusdinium vs. Cladocopium appeared to be magnified with HTS. Our results showed significant differences in the relative abundances of Durusdinium and Cladocopium between Singapore and Papua New Guinea for the HTS results but not for qPCR, likely driven by the higher richness of ITS2 type profiles detected in corals from Papua New Guinea. qPCR is a sensitive method for detecting endosymbionts at a much higher resolution compared to more traditional methods like DGGE. While the technology of HTS has allowed us to detect sequence variants much more easily, it needs to be validated further for the purpose of understanding coral-algal symbiosis as the PCR step may be prone to amplification bias (Neilson et al., 2013). PCR may also pick up free-living Symbiodiniaceae in the corals’ immediate external environment (Santos, 2004; Coffroth et al., 2006; Sze et al., 2018; Fujise et al., 2021). Hence, further exploration of the Symbiodiniaceae community around the coral is important to quantify the coral-algal symbiosis more precisely by accounting for environmental DNA signals. We suggest that water and sediment surrounding the coral colony of interest should be sampled at the same time for Symbiodiniaceae characterization. Overall, since the inferences of Symbiodiniaceae dominance differ at a sample level, we urge caution when examining endosymbiont abundances, and suggest verifying HTS results with qPCR for more precise estimates.
Indeed, precise characterization of the roles of different Symbiodiniaceae taxa would provide insights on the adaptation of coral reefs as global stressors exacerbate. The large colony-level variations of the coral-Symbiodiniaceae relationship even within a small reef area suggest that local adaptation is ongoing that may allow certain colonies to persist despite the accelerating environmental change (McClanahan et al., 2004; Hoogenboom et al., 2017). Such mosaicism of coral fitness at the reef level would be important for forecasting the performance of local reefs amid global climate change.
Data Availability Statement
All sequencing reads associated with this work have been deposited at the National Center for Biotechnology Information under the BioProject Accession PRJNA754701.
Author Contributions
SJ, BW, and DH contributed to conception and design of the study. SJ performed the molecular analyses and analyzed the data. SJ and DH wrote the first draft of the manuscript. All authors contributed to the collection of samples, contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by the National Research Foundation (NRF), Prime Minister’s Office, Singapore and the Israel Science Foundation under their 2nd Joint Grant Call (NRF2017NRF-ISF002-2658; Grant Number 2658/17), as well as the Ocean Park Conservation Foundation, Hong Kong (OT01.1819).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Allen Chen and SK for sharing the initial protocol and discussions, as well as members of the Reef Ecology Laboratory for assistance and support. We are also grateful to Nalini Puniamoorthy, Peter Todd, SK, HH and DT for constructive comments and suggestions. Coral collections were performed under a Singapore National Parks Board research permit (NP/RP16-156) and a Papua New Guinea research permit (AA927408).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.759744/full#supplementary-material
References
Al-Horani, F. A., Al-Moghrabi, S. M., and De Beer, D. (2003). The mechanism of calcification and its relation to photosynthesis and respiration in the scleractinian coral Galaxea fascicularis. Mar. Biol. 142, 419–426. doi: 10.1007/s00227-002-0981-8
Allen, G. R., Kinch, J., McKenna, S. A., and Seeto, P. (2003). A Rapid Marine Biodiversity Assessment of Milne Bay Province, Papua New Guinea—Survey II (2000). RAP Bulletin of Biological Assessment 29. Washington, DC: Conservation International.
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Baker, A. C. (2001). Reef corals bleach to survive change. Nature 411, 765–766. doi: 10.1038/35081151
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Baker, A. C. (2004). “Symbiont diversity on coral reefs and its relationship to bleaching resistance and resilience,” in Coral Health and Disease, eds E. Rosenberg and Y. Loya (Berlin: Springer), 177–194.
Baker, A. C., Starger, C. J., McClanahan, T. R., and Glynn, P. W. (2004). Corals’ adaptive response to climate change. Nature 430, 741–741. doi: 10.1038/430741a
Banin, E., Vassilakos, D., Orr, E., Martinez, R. J., and Rosenberg, E. (2003). Superoxide dismutase is a virulence factor produced by the coral bleaching pathogen Vibrio shiloi. Curr. Microbiol. 46, 418–422. doi: 10.1007/s00284-002-3912-5
Boilard, A., Dubé, C. E., Gruet, C., Mercière, A., Hernandez-Agreda, A., and Derome, N. (2020). Defining coral bleaching as a microbial dysbiosis within the coral holobiont. Microorganisms 8:1682. doi: 10.3390/microorganisms8111682
Bongaerts, P., Cooke, I. R., Ying, H., Wels, D., den Haan, S., Hernandez-Agreda, A., et al. (2021). Morphological stasis masks ecologically divergent coral species on tropical reefs. Curr. Biol. 31, 2286–2298. doi: 10.1016/j.cub.2021.03.028
Bongaerts, P., Sampayo, E. M., Bridge, T. C. L., Ridgway, T., Vermeulen, F., Englebert, N., et al. (2011). Symbiodinium diversity in mesophotic coral communities on the Great Barrier Reef: a first assessment. Mar. Ecol. Prog. Ser. 439, 117–126. doi: 10.3354/meps09315
Boulotte, N. M., Dalton, S. J., Carroll, A. G., Harrison, P. L., Putnam, H. M., Peplow, L. M., et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 10, 2693–2701. doi: 10.1038/ismej.2016.54
Buddemeier, R. W., and Fautin, D. G. (1993). Coral bleaching as an adaptive mechanism. Bioscience 43, 320–326. doi: 10.2307/1312064
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chen, B., Yu, K., Liang, J., Huang, W., Wang, G., Su, H., et al. (2019). Latitudinal variation in the molecular diversity and community composition of Symbiodiniaceae in coral from the South China Sea. Front. Microbiol. 10:1278. doi: 10.3389/fmicb.2019.01278
Chen, C. A., Lam, K. K., Nakano, Y., and Tsai, W. S. (2003). A stable association of the stress-tolerant zooxanthellae, Symbiodinium clade D, with the low-temperature-tolerant coral, Oulastrea crispata (Scleractinia: Faviidae) in subtropical non-reefal coral communities. Zool. Stud. 42, 540–550.
Chow, G. S. E., Chan, Y. K. S., Jain, S. S., and Huang, D. (2019). Light limitation selects for depth generalists in urbanised reef coral communities. Mar. Environ. Res. 147, 101–112. doi: 10.1016/j.marenvres.2019.04.010
Coffroth, M. A., Lewis, C. F., Santos, S. R., and Weaver, J. L. (2006). Environmental populations of symbiotic dinoflagellates in the genus Symbiodinium can initiate symbioses with reef cnidarians. Curr. Biol. 16, 985–987. doi: 10.1016/j.cub.2006.10.049
Cooper, T. F., Ulstrup, K. E., Dandan, S. S., Heyward, A. J., Kühl, M., Muirhead, A., et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc. R. Soc. B Biol. Sci. 278, 1840–1850. doi: 10.1098/rspb.2010.2321
DeVantier, L., and Turak, E. (2017). Species richness and relative abundance of reef-building corals in the Indo-West Pacific. Diversity 9:25. doi: 10.3390/d9030025
Eckert, R. J., Reaume, A. M., Sturm, A. B., Studivan, M. S., and Voss, J. D. (2020). Depth influences Symbiodiniaceae associations among Montastraea cavernosa corals on the Belize Barrier Reef. Front. Microbiol. 11:518. doi: 10.3389/fmicb.2020.00518
Eren, A. M., Morrison, H. G., Lescault, P. J., Reveillaud, J., Vineis, J. H., and Sogin, M. L. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9, 968–979. doi: 10.1038/ismej.2014.195
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and Porter, J. W. (1984). Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709. doi: 10.2307/1309663
Feldman, B., Afiq-Rosli, L., Simon-Blecher, N., Bollati, E., Wainwright, B. J., Bongaerts, P., et al. (2021). Distinct lineages and population structure of the coral Pachyseris speciosa in the small equatorial reef system of Singapore. Coral Reefs. doi: 10.1007/s00338-021-02160-4
Fujise, L., Suggett, D. J., Stat, M., Kahlke, T., Bunce, M., Gardner, S. G., et al. (2021). Unlocking the phylogenetic diversity, primary habitats, and abundances of free-living Symbiodiniaceae on a coral reef. Mol. Ecol. 30, 343–360. doi: 10.1111/mec.15719
Goulet, T. L., Lucas, M. Q., and Schizas, N. V. (2019). “Symbiodiniaceae genetic diversity and symbioses with hosts from shallow to mesophotic coral ecosystems,” in Coral Reefs of the World, eds Y. Loya, K. Puglise, and T. Bridge (Cham: Springer), 537–551.
Guest, J. R., Low, J., Tun, K., Wilson, B., Ng, C., Raingeard, D., et al. (2016). Coral community response to bleaching on a highly disturbed reef. Sci. Rep. 6:20717. doi: 10.1038/srep20717
Hoogenboom, M. O., Frank, G. E., Chase, T. J., Jurriaans, S., Álvarez-Noriega, M., Peterson, K., et al. (2017). Environmental drivers of variation in bleaching severity of Acropora species during an extreme thermal anomaly. Front. Mar. Sci. 4:376. doi: 10.3389/fmars.2017.00376
Howells, E. J., Bauman, A. G., Vaughan, G. O., Hume, B. C. C., Voolstra, C. R., and Burt, J. A. (2020). Corals in the hottest reefs in the world exhibit symbiont fidelity not flexibility. Mol. Ecol. 29, 899–911. doi: 10.1111/mec.15372
Huang, D., Tun, K., Chou, L. M., and Todd, P. A. (2009). An inventory of zooxanthellate scleractinian corals in Singapore, including 33 new records. Raffles Bull. Zool. 22, 69–80.
Huang, H., Dong, Z., Huang, L., Yang, J., Di, B., Li, Y., et al. (2011). Latitudinal variation in algal symbionts within the scleractinian coral Galaxea fascicularis in the South China Sea. Mar. Biol. Res. 7, 208–211. doi: 10.1080/17451000.2010.489616
Hughes, T. P., Barnes, M. L., Bellwood, D. R., Cinner, J. E., Cumming, G. S., Jackson, J. B. C., et al. (2017a). Coral reefs in the Anthropocene. Nature 546, 82–90. doi: 10.1038/nature22901
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017b). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Hume, B. C. C., D’Angelo, C., Burt, J. A., Baker, A. C., Riegl, B., and Wiedenmann, J. (2013). Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar. Pollut. Bull. 72, 313–322. doi: 10.1016/j.marpolbul.2012.11.032
Hume, B. C. C., D’Angelo, C., Smith, E. G., Stevens, J. R., Burt, J. A., and Wiedenmann, J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci. Rep. 5:8562. doi: 10.1038/srep08562
Hume, B. C. C., Mejia-Restrepo, A., Voolstra, C. R., and Berumen, M. L. (2020). Fine-scale delineation of Symbiodiniaceae genotypes on a previously bleached central Red Sea reef system demonstrates a prevalence of coral host-specific associations. Coral Reefs 39, 583–601. doi: 10.1007/s00338-020-01917-7
Hume, B. C. C., Smith, E. G., Ziegler, M., Warrington, H. J. M., Burt, J. A., LaJeunesse, T. C., et al. (2019). SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080. doi: 10.1111/1755-0998.13004
Illumina (2013). 16S Metagenomic Sequencing Library Preparation. Available online at: http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed January 15, 2020).
Innis, T., Cunning, R., Ritson-Williams, R., Wall, C. B., and Gates, R. D. (2018). Coral color and depth drive symbiosis ecology of Montipora capitata in Kāne‘ohe Bay, O‘ahu, Hawai‘i. Coral Reefs 37, 423–430. doi: 10.1007/s00338-018-1667-0
Jain, S. S., Afiq-Rosli, L., Feldman, B., Levy, O., Phua, J. W., Wainwright, B. J., et al. (2020). Homogenization of endosymbiont communities hosted by equatorial corals during the 2016 mass bleaching event. Microorganisms 8:1370. doi: 10.3390/microorganisms8091370
Jones, L., Acolado, P., Cala, Y., Cobi Án, D., Coelho, V., Hernández, A., et al. (2008). “The effects of coral bleaching in the northern Caribbean and western Atlantic,” in Status of Caribbean Coral Reefs after Bleaching and Hurricanes in 2005, eds C. Wilkinson and D. Souter (Townsville, Qld: Global Coral Reef Monitoring Network), 73–84.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kemp, D. W., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
Keshavmurthy, S., Meng, P. J., Wang, J. T., Kuo, C. Y., Yang, S. Y., Hsu, C. M., et al. (2014). Can resistant coral-Symbiodinium associations enable coral communities to survive climate change? A study of a site exposed to long-term hot water input. PeerJ 2:e327. doi: 10.7717/peerj.327
LaJeunesse, T. C. (2002). Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400. doi: 10.1007/s00227-002-0829-2
LaJeunesse, T. C. (2005). “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol. Biol. Evol. 22, 570–581. doi: 10.1093/molbev/msi042
LaJeunesse, T. C. (2020). Zooxanthellae. Curr. Biol. 30, R1110–R1113. doi: 10.1016/j.cub.2020.03.058
LaJeunesse, T. C., Bhagooli, R., Hidaka, M., DeVantier, L., Done, T., Schmidt, G. W., et al. (2004). Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Prog. Ser. 284, 147–161. doi: 10.3354/meps284147
LaJeunesse, T. C., Forsman, Z. H., and Wham, D. C. (2016). An Indo-West Pacific ‘zooxanthella’ invasive to the western Atlantic finds its way to the Eastern Pacific via an introduced Caribbean coral. Coral Reefs 35, 577–582. doi: 10.1007/s00338-015-1388-6
LaJeunesse, T. C., Loh, W. K. W., Van Woesik, R., Hoegh-Guldberg, O., Schmidt, G. W., and Fitt, W. K. (2003). Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48, 2046–2054. doi: 10.4319/lo.2003.48.5.2046
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol 28, 2570.e–2580.e. doi: 10.1016/j.cub.2018.07.008
LaJeunesse, T. C., Pettay, D. T., Sampayo, E. M., Phongsuwan, N., Brown, B., Obura, D. O., et al. (2010). Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785–800. doi: 10.1111/j.1365-2699.2010.02273.x
Leveque, S., Afiq-Rosli, L., Ip, Y. C. A., Jain, S. S., and Huang, D. (2019). Searching for phylogenetic patterns of Symbiodiniaceae community structure among Indo-Pacific Merulinidae corals. PeerJ 7:e7669. doi: 10.7717/peerj.7669
Little, A. F., Van Oppen, M. J. H., and Willis, B. L. (2004). Flexibility in algal endosymbioses shapes growth in reef corals. Science 304, 1492–1494. doi: 10.1126/science.1095733
McClanahan, T. R., Baird, A. H., Marshall, P. A., and Toscano, M. A. (2004). Comparing bleaching and mortality responses of hard corals between southern Kenya and the Great Barrier Reef, Australia. Mar. Pollut. Bull. 48, 327–335. doi: 10.1016/j.marpolbul.2003.08.024
Meistertzheim, A. L., Pochon, X., Wood, S. A., Ghiglione, J. F., and Hédouin, L. (2019). Development of a quantitative PCR–high-resolution melting assay for absolute measurement of coral-Symbiodiniaceae associations and its application to investigating variability at three spatial scales. Mar. Biol. 166:13. doi: 10.1007/s00227-018-3458-0
Neilson, J. W., Jordan, F. L., and Maier, R. M. (2013). Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. J. Microbiol. Methods 92, 256–263. doi: 10.1016/j.mimet.2012.12.021
Ng, C. S. L., Huang, D., Toh, K. B., Sam, S. Q., Kikuzawa, Y. P., Toh, T. C., et al. (2020). Responses of urban reef corals during the 2016 mass bleaching event. Mar. Pollut. Bull. 154:111111. doi: 10.1016/j.marpolbul.2020.111111
Noonan, S. H. C., Fabricius, K. E., and Humphrey, C. (2013). Symbiodinium community composition in scleractinian corals is not affected by life-long exposure to elevated carbon dioxide. PLoS One 8:e63985. doi: 10.1371/journal.pone.0063985
Osman, E. O., Suggett, D. J., Voolstra, C. R., Pettay, D. T., Clark, D. R., Pogoreutz, C., et al. (2020). Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 8:8. doi: 10.1186/s40168-019-0776-5
Poquita-Du, R. C., Huang, D., Chou, L. M., and Todd, P. A. (2020). The contribution of stress-tolerant endosymbiotic dinoflagellate Durusdinium to Pocillopora acuta survival in a highly urbanized reef system. Coral Reefs 39, 745–755. doi: 10.1007/s00338-020-01902-0
R Core Team (2017). R: A Language and Environment for Statistical Computing. Available online at: http://www.R-project.org (accessed March 1, 2021)
Rouzé, H., Lecellier, G., Saulnier, D., and Berteaux-Lecellier, V. (2016). Symbiodinium clades A and D differentially predispose Acropora cytherea to disease and Vibrio spp. colonization. Ecol. Evol. 6, 560–572. doi: 10.1002/ece3.1895
Rowan, R., and Knowlton, N. (1995). Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 92, 2850–2853. doi: 10.1073/pnas.92.7.2850
Rowan, R., Knowlton, N., Baker, A. C., and Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265–269. doi: 10.1038/40843
Sampayo, E. M., Ridgway, T., Bongaerts, P., and Hoegh-Guldberg, O. (2008). Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. U.S.A. 105, 10444–10449. doi: 10.1073/pnas.0708049105
Santos, S. R. (2004). Phylogenetic analysis of a free-living strain of Symbiodinium isolated from Jiaozhou Bay, P.R. China. J. Phycol. 40, 395–397. doi: 10.1111/j.1529-8817.2004.03186.x
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Silverstein, R. N., Cunning, R., and Baker, A. C. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob. Change Biol. 21, 236–249. doi: 10.1111/gcb.12706
Smith, E. G., Gurskaya, A., Hume, B. C. C., Voolstra, C. R., Todd, P. A., Bauman, A. G., et al. (2020). Low Symbiodiniaceae diversity in a turbid marginal reef environment. Coral Reefs 39, 545–553. doi: 10.1007/s00338-020-01956-0
Stanley, G. D., and Fautin, D. G. (2001). The origins of modern corals. Science 291, 1913–1914. doi: 10.1126/science.1056632
Stat, M., Bird, C. E., Pochon, X., Chasqui, L., Chauka, L. J., Concepcion, G. T., et al. (2011). Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS One 6:e15854. doi: 10.1371/journal.pone.0015854
Stat, M., and Gates, R. D. (2011). Clade D Symbiodinium in scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011:730715. doi: 10.1155/2011/730715
Stat, M., Loh, W. K. W., Hoegh-Guldberg, O., and Carter, D. A. (2008a). Symbiont acquisition strategy drives host-symbiont associations in the southern Great Barrier Reef. Coral Reefs 27, 763–772. doi: 10.1007/s00338-008-0412-5
Stat, M., Morris, E., and Gates, R. D. (2008b). Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 105, 9256–9261. doi: 10.1073/pnas.0801328105
Stat, M., Pochon, X., Franklin, E. C., Bruno, J. F., Casey, K. S., Selig, E. R., et al. (2013). The distribution of the thermally tolerant symbiont lineage (Symbiodinium clade D) in corals from Hawaii: correlations with host and the history of ocean thermal stress. Ecol. Evol. 3, 1317–1329. doi: 10.1002/ece3.556
Sze, Y., Miranda, L. N., Sin, T. M., and Huang, D. (2018). Characterising planktonic dinoflagellate diversity in Singapore using DNA metabarcoding. Metabarcoding Metagenom. 2:e25136. doi: 10.3897/mbmg.2.25136
Tan, Y. T. R., Wainwright, B. J., Afiq-Rosli, L., Ip, Y. C. A., Lee, J. N., Nguyen, N. T. H., et al. (2020). Endosymbiont diversity and community structure in Porites lutea from Southeast Asia are driven by a suite of environmental variables. Symbiosis 80, 269–277. doi: 10.1007/s13199-020-00671-2
Tanzil, J. T. I., Ng, A. P. K., Tey, Y. Q., Tan, B. H. Y., Yun, E. Y., and Huang, D. (2016). A preliminary characterisation of Symbiodinium diversity in some common corals from Singapore. Cosmos 12, 15–27. doi: 10.1142/s0219607716500014
Terraneo, T. I., Fusi, M., Hume, B. C. C., Arrigoni, R., Voolstra, C. R., Benzoni, F., et al. (2019). Environmental latitudinal gradients and host-specificity shape Symbiodiniaceae distribution in Red Sea Porites corals. J. Biogeogr. 46, 2323–2335. doi: 10.1111/jbi.13672
Teschima, M. M., Garrido, A., Paris, A., Nunes, F. L. D., and Zilberberg, C. (2019). Biogeography of the endosymbiotic dinoflagellates (Symbiodiniaceae) community associated with the brooding coral Favia gravida in the Atlantic Ocean. PLoS One 14:e0213519. doi: 10.1371/journal.pone.0213519
The Nature Conservancy (2004). Designing a Resilient Network of Marine Protected Areas in Kimbe Bay, West New Britain, Papua New Guinea. Papua New Guinea: The Nature Conservancy.
Thornhill, D. J., Lewis, A. M., Wham, D. C., and LaJeunesse, T. C. (2014). Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution 68, 352–367. doi: 10.1111/evo.12270
Toh, T. C., Huang, D., Tun, K., and Chou, L. M. (2018). “Summary of coral bleaching from 2014 to 2017 in Singapore,” in Status of Coral Reefs in East Asian Seas Region: 2018, eds T. Kimura, K. Tun, and L. M. Chou (Tokyo: Ministry of the Environment of Japan and Japan Wildlife Research Center), 21–23.
Tonk, L., Sampayo, E. M., Weeks, S., Magno-Canto, M., and Hoegh-Guldberg, O. (2013). Host-Specific interactions with environmental factors shape the distribution of Symbiodinium across the Great Barrier Reef. PLoS One 8:e68533. doi: 10.1371/journal.pone.0068533
Tye Pettaya, D., Wham, D. C., Smith, R. T., Iglesias-Prieto, R., and LaJeunesse, T. C. (2015). Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc. Natl. Acad. Sci. U.S.A. 112, 7513–7518. doi: 10.1073/pnas.1502283112
Ulstrup, K. E., and Van Oppen, M. J. H. (2003). Geographic and habitat partitioning of genetically distinct zooxanthellae (Symbiodinium) in Acropora corals on the Great Barrier Reef. Mol. Ecol. 12, 3477–3484. doi: 10.1046/j.1365-294X.2003.01988.x
Veron, J., DeVantier, L., Turak, E., Green, A. L., Kininmonth, S., Stafford-Smith, M., et al. (2011). “The Coral Triangle,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer), 47–55.
Veron, J., Stafford-Smith, M., DeVantier, L., and Turak, E. (2015). Overview of distribution patterns of zooxanthellate Scleractinia. Front. Mar. Sci. 1:81. doi: 10.3389/fmars.2014.00081
Wong, J. S. Y., Chan, Y. K. S., Ng, C. S. L., Tun, K., Darling, E. S., and Huang, D. (2018). Comparing patterns of taxonomic, functional and phylogenetic diversity in reef coral communities. Coral Reefs 37, 737–750. doi: 10.1007/s00338-018-1698-6
Ziegler, M., Arif, C., Burt, J. A., Dobretsov, S., Roder, C. M., LaJeunesse, T. C., et al. (2017). Biogeography and molecular diversity of coral symbionts in the genus Symbiodinium around the Arabian Peninsula. J. Biogeogr. 44, 674–686. doi: 10.1111/jbi.12913
Ziegler, M., Roder, C., Bchel, C., and Voolstra, C. R. (2015a). Niche acclimatization in Red Sea corals is dependent on flexibility of host-symbiont association. Mar. Ecol. Prog. Ser. 533, 163–176. doi: 10.3354/meps11365
Keywords: Central Indo-Pacific, high-throughput sequencing, internal transcribed spacer, qPCR, reef corals, Scleractinia, Symbiodiniaceae
Citation: Jain SS, Afiq-Rosli L, Feldman B, Kunning I, Levy O, Mana RR, Wainwright BJ and Huang D (2021) Endosymbiont Communities in Pachyseris speciosa Highlight Geographical and Methodological Variations. Front. Mar. Sci. 8:759744. doi: 10.3389/fmars.2021.759744
Received: 17 August 2021; Accepted: 18 November 2021;
Published: 22 December 2021.
Edited by:
Shashank Keshavmurthy, Academia Sinica, TaiwanReviewed by:
Hui Huang, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaDaniel J. Thornhill, National Science Foundation (NSF), United States
Copyright © 2021 Jain, Afiq-Rosli, Feldman, Kunning, Levy, Mana, Wainwright and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudhanshi S. Jain, amFpbi5zdWRoYW5zaGlAZ21haWwuY29t; Danwei Huang, aHVhbmdkYW53ZWlAbnVzLmVkdS5zZw==
 Sudhanshi S. Jain
Sudhanshi S. Jain Lutfi Afiq-Rosli1,2
Lutfi Afiq-Rosli1,2 Oren Levy
Oren Levy Benjamin J. Wainwright
Benjamin J. Wainwright