
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mar. Sci., 11 October 2021
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.758694
This article is part of the Research TopicMarine Microbiomes: Towards Standard Methods and Best PracticesView all 10 articles
 Robyn M. Samuel1,2*
Robyn M. Samuel1,2* Raissa Meyer3
Raissa Meyer3 Pier Luigi Buttigieg4
Pier Luigi Buttigieg4 Neil Davies5
Neil Davies5 Nicholas W. Jeffery6
Nicholas W. Jeffery6 Christopher Meyer7
Christopher Meyer7 Christina Pavloudi8
Christina Pavloudi8 Kathleen Johnson Pitz9
Kathleen Johnson Pitz9 Maxime Sweetlove10
Maxime Sweetlove10 Susanna Theroux11
Susanna Theroux11 Jodie van de Kamp12
Jodie van de Kamp12 Alison Watts13
Alison Watts13Biomolecular ocean observing and research is a rapidly evolving field that uses omics approaches to describe biodiversity at its foundational level, giving insight into the structure and function of marine ecosystems over time and space. It is an especially effective approach for investigating the marine microbiome. To mature marine microbiome research and operations within a global ocean biomolecular observing network (OBON) for the UN Decade of Ocean Science for Sustainable Development and beyond, research groups will need a system to effectively share, discover, and compare “omic” practices and protocols. While numerous informatic tools and standards exist, there is currently no global, publicly-supported platform specifically designed for sharing marine omics [or any omics] protocols across the entire value-chain from initiating a study to the publication and use of its results. Toward that goal, we propose the development of the Minimum Information for an Omic Protocol (MIOP), a community-developed guide of curated, standardized metadata tags and categories that will orient protocols in the value-chain for the facilitated, structured, and user-driven discovery of suitable protocol suites on the Ocean Best Practices System. Users can annotate their protocols with these tags, or use them as search criteria to find appropriate protocols. Implementing such a curated repository is an essential step toward establishing best practices. Sharing protocols and encouraging comparisons through this repository will be the first steps toward designing a decision tree to guide users to community endorsed best practices.
The term “omics” generally means studying anything holistically, and here we take a broad view of biomolecular omics that includes, but is not limited to: quantitative target gene amplification (e.g., qPCR, qNASBA etc.), (meta)barcoding, (meta)genomics, (meta)transcriptomics, (meta)proteomics, and metabolomics; and field collection approaches that target organisms or parts thereof, including single-celled organisms (microorganisms), as well as environmental DNA (eDNA). In the marine realm, omic techniques are used to assess and monitor biodiversity, reveal population structure and gene flow, and discover new compounds with applications in medicine and industry. Rapid advances in omic research, and the declining cost of high-throughput sequencing technologies (Wetterstrand, 2020) support the increasing application of omics in marine microbiome research.
The recent expansion in marine omics has led to a proliferation of protocols specific to multiple applications. However, these protocols are rarely shared publicly with sufficient detail to reliably reproduce a study (Dickie et al., 2018). While the omics community has already achieved high standards for sharing sequence data through the International Nucleotide Sequence Database Collaboration, these data often lack sufficient metadata and provenance information on the protocols used (Dickie et al., 2018), undermining efforts to implement the Findable, Accessible, Interoperable and Reusable (FAIR) data principles (Wilkinson et al., 2016). These limitations create challenges for marine microbiome research and operations from individual labs up to global (meta)data analysis efforts such as MGnify (Mitchell et al., 2019), which must identify data collected using comparable methods, in order to integrate and re-use data for meta-analysis (Berry et al., 2020). Moreover, a lack of protocol-sharing impedes the identification of comparable methods needed for global monitoring efforts aiming to understand, and sustainably manage the changing marine ecosystem (Aylagas et al., 2020; Berry et al., 2020; Makiola et al., 2020).
Many projects are looking to develop best practices for omics research: standards organizations, such as the Genomic Standards Consortium’s (GSC) Genomic Biodiversity Interest Group, the Biodiversity Information Standards (TDWG) and the Biocode Commons are working collaboratively toward standards specifications for genomic observatories (Davies et al., 2012, 2014). Large campaigns, such as the Earth Microbiome Project (Gilbert et al., 2014; Thompson et al., 2017), TARA Oceans (Sunagawa et al., 2020), and the Australian Microbiome Initiative (AM; Bissett et al., 2016; Brown et al., 2018; doi: 10.4227/71/561c9bc670099), have already developed standardized practices, and innovative software enterprises, such as protocols.io, are providing powerful solutions for sharing protocols. Yet there is currently no global, publicly-supported infrastructure developed explicitly for encouraging the exchange and harmonization of omic protocols, so these valuable contributions remain fragmented and underutilized.
For marine ecosystems, the Intergovernmental Oceanographic Commission’s Ocean Best Practices System (OBPS) provides a public repository for all ocean research methodological documentation that can interlink protocols, standard specifications, and other guidelines. The OBPS seeks to support continuous convergence of methods as they undergo community refinement to become best practices (Hörstmann et al., 2021). In collaboration with the broader omics community, through the Omic BON initiative (Buttigieg et al., 2019), we propose to develop a best practice system specific to marine omics research, leveraging the framework of the OBPS to curate a global repository for marine omics protocols.
As part of the omics/eDNA session at the 4th OBPS workshop, we discussed recommendations and community needs for an omics/eDNA specific best practices system. Recognizing an urgent need for the ocean omics community to get organized as the UN Decade of Ocean Science for Sustainable Development starts, we identified the demand for publishing protocols into a user-friendly decision tree framework. With such a framework we would aim to support protocol selection, increase protocol findability and improve recognition for protocol developers. In a series of focused follow-up meetings, we identified that an omics decision tree would require a library of constituent parts (the protocols) and framework to: (1) locate where the protocol fits within the entire omics workflow (outlined in section “Ocean Omics Methodology Categories”), and (2) organize protocols using focused descriptive terms (metadata tags), based on what the protocol does and how/why it is used (outlined in section “Essential Metadata for Omics Protocols”).
The typical omics workflow involves a series of protocols, which take a project from ideation, through to publication, and on to societal use. Protocols from each step in the omics workflow hold valuable information for different groups. For example, sample collection protocols may be most relevant to scientists/technicians in the field, whereas local stakeholders and indigenous communities may primarily engage with aspects of how the project and resulting data address and impact important ethical, legal, and societal issues (Nagoya Protocol, 2010; Carroll et al., 2020). Documenting details and provenance for the entire marine omics workflow requires input from multiple parties, as each step of the workflow may be conducted by different individuals or groups. The omics OBPS therefore needs to identify these key methodological categories, to allow protocols and accompanying metadata to be uploaded in modules that link together to form the entire workflow.
We propose twelve protocol categories (Figure 1A) for ocean omics research and operations. Protocols and guidelines are assigned into these categories according to the purpose they serve1. Categories 5–12 outline methodological categories for operational activities used in the AM Initiative (van de Kamp et al., 2019). Categories 1–4 were identified to additionally cover cross-cutting documentation in the omics workflow: (1) Society, (2) Sampling/observational design, (3) Ethics and law, and (4) Data management.
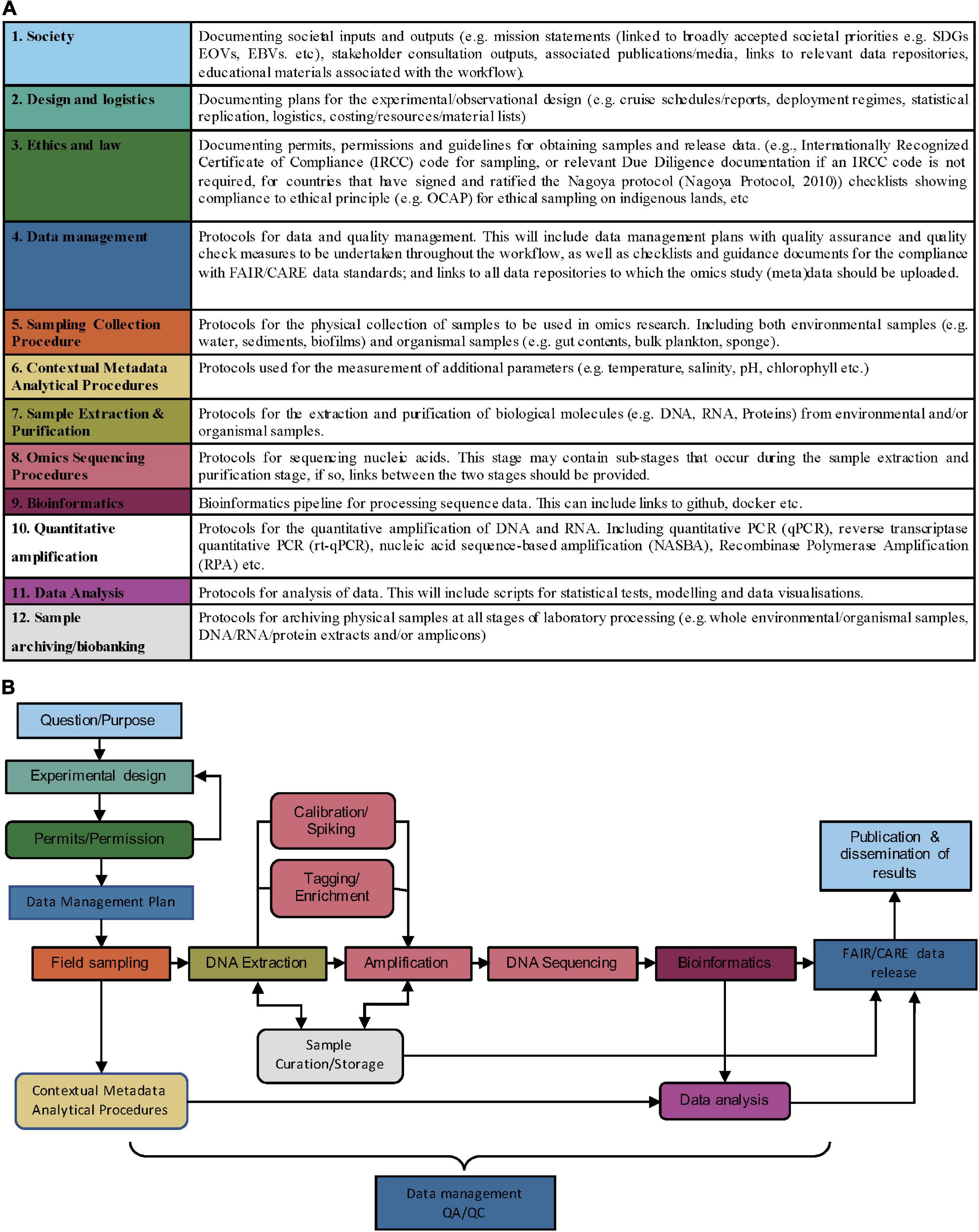
Figure 1. (A) Proposed methodology categories to enhance exchange of ocean omics analysis knowhow. Protocols, guidelines, and other methodologies in some of these categories (such as Sample archiving/biobanking, Data Management, and Society) are cross-cutting and may apply at multiple points in the workflow. (B) Example workflow for a DNA metabarcoding project. Colors correspond to the methodology categories outlined in panel (A) and arrows indicate the order of the workflow. Square boxes show essential steps in a metabarcoding workflow, whereas rounded boxes indicate non-essential steps. Data management and QA/QC are required throughout the entire workflow.
1. Society—All workflows should begin and end with society; societal needs inform the question or purpose behind the research, and societal impacts show the value in the research once it has been completed.
2. Design and logistics—This category covers the practical logistics for implementing ocean omics research and operations, including the experimental/observational design formulated to address the societal priorities outlined in 1.
3. Ethics and law—A survey of workshop participants highlighted a need for guidance on sharing data and complying with important ethical and legal requirements (Simpson et al., 2021). This category will include information on permits and permission required to obtain samples and release data. Collating and publishing this information will firstly provide examples for how previous projects have adhered to legal requirements/ethical principles and secondly stimulate discussion on how to facilitate adherence to these requirements and principles, perhaps through checklists, templates, or training materials.
4. Data management—The data management plan (DMP) is designed to support all the downstream steps according to the ethics, legalities and societal needs identified in (1–3), while making sure that the (meta)data flows to the right stakeholders in society that we need to interface with. DMPs should be drafted prior to data collection and referred to throughout the workflow to ensure that quality assurance and quality checks take place, and that detailed information on (meta)data requirements for both short and long-term (meta)data storage is given. There is a growing body of tools and best practices surrounding DMPs, including principles for making them more machine-actionable, that should be leveraged in omic protocols and associated infrastructure (see Miksa et al., 2019). Publishing documentation on omics specific DMPs will increase transparency for funders by providing direct links to the protocols they refer to. Furthermore, collating examples of omics specific DMPs will provide insight into what the community needs from omics specific data management tools.
In Figure 1B, we give an example of a DNA metabarcoding workflow, where the colour of each step corresponds to a methodology category in Figure 1A. Protocols uploaded to OBPS can be assigned (tagged) to the relevant omics categories. The granularity of protocols uploaded to the OBPS may include individual uploads for sub-stages (i.e., Tagging/Enrichment within 4, Omics sequencing procedures), or single documents spanning multiple methodology categories (i.e., 7, Sample extraction and purification, through to 9, Bioinformatics). To accommodate these levels of granularity, each upload could be tagged with single or multiple methodology category and linked to those protocols pre- and succeeding it. The granular use of methodology categories will increase modularity within the omics workflow and facilitate the mixing and matching of methods from various projects.
The interplay between the activities within and across the steps within a workflow—and how they bring value to the community and society—is complex and beyond the scope of this article; however, we have provided an initial perspective on this using the Porter’s value chain approach (Porter, 1985; Supplementary Figure 1).
The targeted discovery and reuse of protocols can be improved if protocols are effectively described using standardized metadata terms on upload to OBPS and other platforms. Terms and checklists to standardize metadata about primary sequence or biodiversity data already exist [GSC’s Minimum Information about any (x) Sequence checklist (MIxS; Yilmaz et al., 2011) and TDWG’s Darwin Core standard (Wieczorek et al., 2012)]; however, no such standards have thus far been published for metadata about omics protocols.
Here we present initial suggestions for the Minimum Information for an Omic Protocol (MIOP), a set of ten metadata categories which could correspond to ten key decision tree questions asked to identify the relevant protocol for any project. The ten MIOP categories (Table 1) consist of five novel categories (methodology category, purpose, resources, analysis, target) and five categories already used in the GSC’s MIxS (project, geographic location, broad-scale environmental context, local environmental context, and environmental medium). Each category is linked to a set of predefined keywords (metadata terms) from existing vocabularies or ontologies; except for the “project” category, which contains project names, affiliations, and contact details and the “methodology category” outlined in section “Ocean Omics Methodology Categories” (Figure 1A). Omics users would then select the most appropriate keywords for each category, assigning the terms as metadata for the protocol. This will improve the FAIRness of our protocol data, by allowing consequent users to search the protocol database using the same set of keywords; thereby, limiting the proliferation of descriptive keywords (e.g., mapping synonyms) and increasing the findability of protocols.
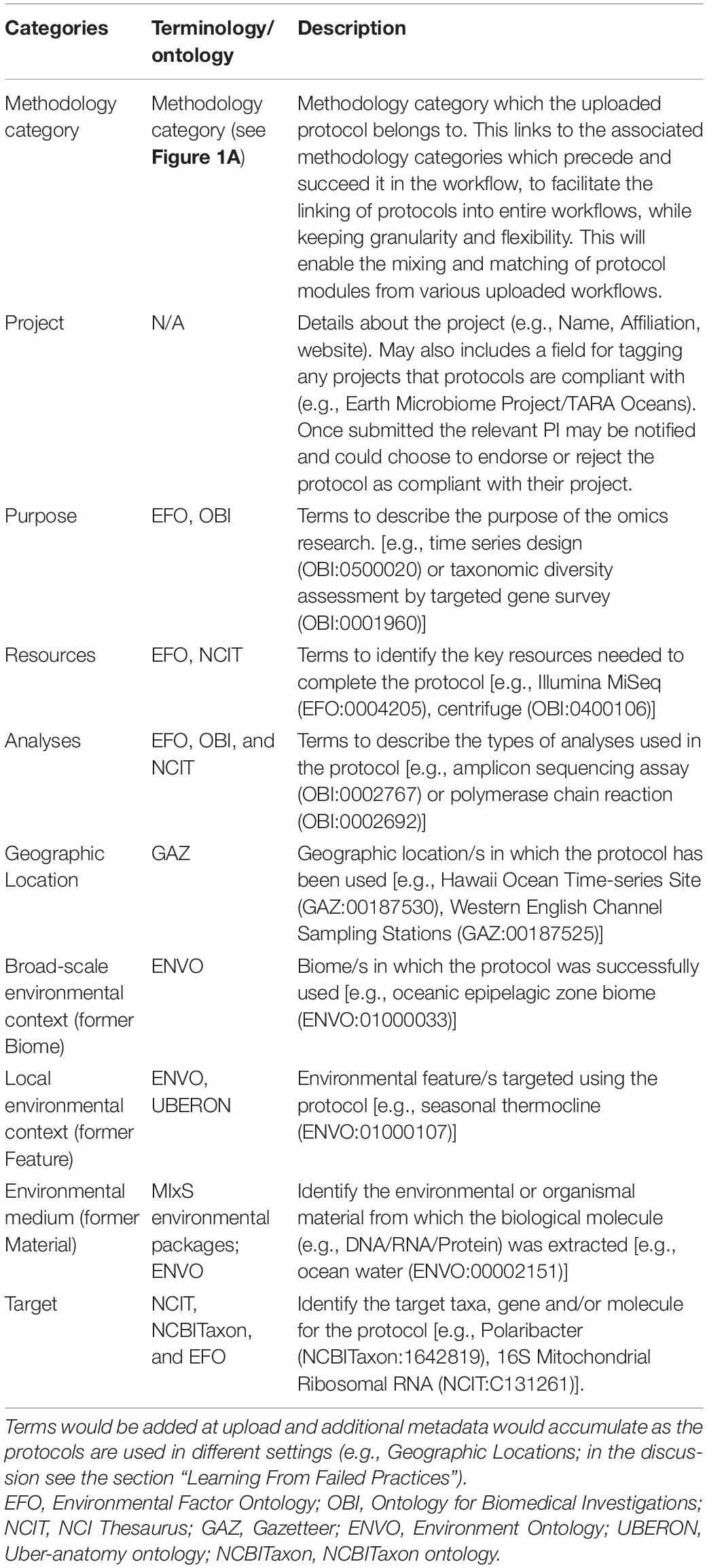
Table 1. Description of keyword categories for protocol metadata and the terminologies (controlled vocabularies, thesauri, and ontologies) containing the relevant keywords.
Ocean Best Practices System provides a neutral, global public repository for ocean community practices. It is a stable and persistent foundation that can host protocols themselves, or link to other protocol tools and functionalities that can (and should) continue to be developed by other organizations including the private sector. The primary function of Omics OBPS would be to publish and archive omics protocols to enhance their global visibility and discoverability, and provide stable links to the entire workflow of protocols. Expanding and improving the functionality of the OBPS for omics protocols will help the community mature by providing a structured system in which context-based best practices can be discovered and identified. A transparent and structured process for handling our omics protocols will be an essential step toward operationalizing omics observing.
Increasing protocol transparency, through detailed publication on OBPS, also means that simple cited protocol strings can become a core component of methods sections in publications. Those strings can then be harvested by machines to generate a graph of “what came before” and “what came after.” When used with the decision tree recommendations this process could point out the most recent protocol development to users and would essentially provide the decision-tree resource we are aiming for. Such an approach enables “practices” (which might be defined as “protocol strings”) to emerge from how protocols are actually being used in the community. Assessment of which of these practices represent a “best” practice in a given context is a distinct challenge, but not a unique one in knowledge sectors. Peer endorsement and citation metrics are two commonly employed ranking mechanisms that could also be applied here.
Community-use metrics offer a way to capture the community’s preference for certain protocols. We suggest that metrics such as times cited, user upvotes, and number of associated data records all be recorded and used to rank lists of relevant protocols. Combined with the MIOP-based grouping into methodology categories, this process will help accelerate the identification of potential best practices within each category. Narrowing down the list of relevant protocols will additionally provide the basis for more targeted and rigorous scientific comparisons between multiple potential best practices for a given scientific endeavor. Outputs of such comparisons may offer further information about the superiority of certain protocols, and could be considered in addition to the more general community-use metrics2. Furthermore, focusing on these community driven best practices will help to reveal protocols that are effective and convenient for a broad range of research facilities. This in turn can reduce literature biases toward novel state of the art practices, which may not be feasible for mainstream use.
During the initial workshop, participants outlined a desire for a best practice system to include “failed practices” and flag when a protocol may limit or eliminate a range of downstream applications. While this type of functionality would not be immediately addressed by implementing MIOP metadata, there would be potential for users to provide feedback for protocols using MIOP metadata and Boolean operators. For example, if a protocol, originally designed for seawater, was used with freshwater samples, the user could upload additional MIOP metadata using “AND freshwater” if the protocol was successful or “NOT freshwater” if unsuccessful. Thereby, broadening the findability of successful protocols and documenting potential limitations to be aware of. Documenting these failed attempts has the potential to save both time and resources.
Minimum Information for an Omic Protocol may additionally promote collaboration between groups. For example, the “Project” category is an administrative metadata field that will describe the project (study or program) for which the protocol was developed, including contact details and affiliated institution. To create links between similar projects and facilitate collaboration, it would be possible to introduce an option to tag a protocol as compliant with pre-existing projects. In such cases, a notification could be sent to the PI of the lead project, allowing them to add or reject the protocol to their list of compliant protocols. Protocols linked this way could form overarching protocol concepts, which may contain a variety of versions and accepted, cross-comparable protocols that include minor adaptations to make them suitable in different circumstances.
An endorsement process for a global observation network has already been developed by Global Ocean Observing System (GOOS) in cooperation with OBPS, to encourage standardized methods for global observations and for reporting on GOOS’ Essential Ocean Variables (EOVs) (Miloslavich et al., 2018; Hermes, 2020). To gain this endorsement, protocols will have to undergo a rigorous community review process that will be strengthened if there is a large source of omics protocols to compare with on the OBPS. Standardized practices and official endorsements are likely to become increasingly valuable as countries begin to use legislation to make biodiversity targets legally binding. Any omic method used to measure biodiversity impacts will need to undergo legal scrutiny if it is used as evidence of a country/organization meeting or failing to meet biodiversity targets. Therefore, protocols officially endorsed through international programmes, such as GOOS, are likely to hold more sway legally. Broad participation from the omics community in open sharing and reviewing of protocols on the OBPS will help to ensure that community endorsed best practices are representative of the wider community needs and not only focused on expensive state of the art methodologies.
Machine readable tracking of protocol versions presents an opportunity to visually map the progression of protocols by linking all versions to a “concept,” as implemented in Zenodo and GitHub. Like software, omic protocols may be updated, corrected, and improved necessitating forms of version control and tracking, such as the use of semantic versioning (Hörstmann et al., 2020; Preston-Werner, 2021). Implementing this would help to increase recognition for the scientists/technicians/students involved in protocol development through citable documentation of their contributions.
Machine-readable and machine-actionable protocols are becoming more important as autonomous technologies evolve. Devices such as the Environmental Sample Processor (ESP) and the Robotic Cartridge Sampling Instrument (RoCSI) are currently being used and developed for autonomous collection, preservation, and in situ analysis of omics samples (Yamahara et al., 2019; National Oceanography Centre, 2021). Eventually, smart sensing platforms using these technologies will be able integrate data from various sensors and satellites to implement adaptive sampling regimes or extraction protocols based on real-time environmental observations (Whitt et al., 2020). To reach this goal a variety of protocols will need to be translated into a machine actionable format using common workflow language. A systematic review of protocols will help to devise such machine actionable formats and protocol templates may help to bridge the gap between lab-based protocol development and in situ autonomous use.
Multiple groups within the omics community are actively developing best practices for their field. To ensure that all these efforts are effectively utilized, a concerted and community wide effort will be needed to gather and organize these practices. By harnessing the OBPS infrastructure and further developing the MIOP metadata we can: (1) allow protocols to be searched for within a decision tree framework; (2) establish a system that encourages the systematic review of protocols; and (3) reveal community preferences through the accumulation of community use data. Taking these steps toward a structured and global public repository of omics protocols will increase transparency and streamline biomolecular ocean observing research to foster the collaborative networks needed to achieve global scale biodiversity observations.
This manuscript has been released as a preprint at https://zenodo.org/record/5482852#.YVNeo55KjAM (Samuel et al., 2021).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AW, CM, and RS constructed the main text figure with input from all authors. RM, PB, and RS developed the supplementary figure. All authors contributed to the discussion and wrote the manuscript.
RS was supported by the Natural Environment Research Council (NERC) NEXUSS Studentship (grant number NE/N012070/1). RM’s contributions were supported by the European Union’s Horizon 2020 Research and Innovation Programmes under grant agreement N° 862923, project AtlantECO (Atlantic ECOsystem assessment, forecasting and sustainability), and grant agreement N° 862626, project EuroSea (Improving and Integrating European Ocean Observing and Forecasting Systems for Sustainable use of the Oceans). NJ was supported by the Fisheries and Oceans Canada Marine Conservation Targets programme. CP was supported by BIOIMAGING-GR (MIS 5002755) implemented under “Action for Strengthening Research and Innovation Infrastructures” funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund), ECCO (project ID 343) funded by the 1st call for the support of Post-doctoral Researchers by the Hellenic Foundation for Research and Innovation (HFRI) and FutureMARES (grant agreement N° 869300) funded by the European Union’s Horizon 2020 Research and Innovation Programme. In addition, CP’s research is co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Programme «Human Resources Development, Education and Lifelong Learning» in the context of the project “Reinforcement of Postdoctoral Researchers—2nd Cycle” (MIS-5033021), implemented by the State Scholarships Foundation (IKY). JK was supported by the CSIRO Environomics Future Science Platform and the Australian Microbiome Initiative. The Australian Microbiome initiative was supported by funding from Parks Australia through the Bush Blitz program funded by the Australian Government and BHP, Bioplatforms Australia and the Integrated Marine Observing System (IMOS) through the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS), and the CSIRO. KP was supported by the David and Lucile Packard Foundation and the National Aeronautics and Space Administration (NASA) grants 80NSSC20M0001 and 80NSSX21M003.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank participants and organisers from the OBPS workshop IV omics/eDNA working group (Simpson et al., 2021), who have contributed to discussions on how to create an ocean best practice system to serve the biomolecular omics community.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.758694/full#supplementary-material
Aylagas, E., Borja, A., Pochon, X., Zaiko, A., Keeley, N., Bruce, K., et al. (2020). Translational molecular ecology in practice: linking DNA-based methods to actionable marine environmental management. Sci. Tot. Environ. 744:140780. doi: 10.1016/j.scitotenv.2020.140780
Berry, O., Jarman, S., Bissett, A., Hope, M., Paeper, C., Bessey, C., et al. (2020). Making environmental DNA (eDNA) biodiversity records globally accessible. Environ. DNA 3, 699–705. doi: 10.1002/edn3.173
Bissett, A., Fitzgerald, A., Meintjes, T., Mele, P., Reith, F., Dennis, P., et al. (2016). Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. GigaScience 5:21. doi: 10.1186/s13742-016-0126-5
Brown, M., van de Kamp, J., Ostrowski, M., Seymour, J., Ingleton, T., Messer, L., et al. (2018). Systematic, continental scale temporal monitoring of marine pelagic microbiota by the australian marine microbial biodiversity initiative. Sci. Data 5:180130. doi: 10.1038/sdata.2018.130
Buttigieg, P., Appeltans, W., Canonico, G., Chavez, F. P., Deck, J., Glöckner, F. O., et al. (2019). “Building a strategy towards an omic biodiversity observation network (Omic BON),” in Proceedings of the OceanObs’19 conference, Honolulu.
Carroll, S., Garba, I., Figueroa-Rodríguez, O., Holbrook, J., Lovett, R., Materechera, S., et al. (2020). The CARE Principles for indigenous data governance. Data Sci. J. 19:43. doi: 10.5334/dsj-2020-043
Davies, N., Field, D., Amaral-Zettler, L., Clark, M., Deck, J., Drummond, A., et al. (2014). The founding charter of the genomic observatories network. GigaScience 3:2. doi: 10.1186/2047-217x-3-2
Davies, N., Meyer, C., Gilbert, J., Amaral-Zettler, L., Deck, J., Bicak, M., et al. (2012). A call for an international network of genomic observatories (GOs). GigaScience 1:5. doi: 10.1186/2047-217x-1-5
Dickie, I., Boyer, S., Buckley, H., Duncan, R., Gardner, P., Hogg, I., et al. (2018). Towards robust and repeatable sampling methods in eDNA -based studies. Mol. Ecol. Resour. 18, 940–952. doi: 10.1111/1755-0998.12907
Gilbert, J., Jansson, J., and Knight, R. (2014). The Earth Microbiome project: successes and aspirations. BMC Biol. 12:69. doi: 10.1186/s12915-014-0069-1
Hermes, J. (2020). GOOS Best Practices Endorsement Process. Version 1. Paris: Global Ocean Observing System.
Hörstmann, C., Buttigieg, P., Simpson, P., Pearlman, J., and Waite, A. (2021). Perspectives on documenting methods to create ocean best practices. Front. Mar. Sci. 7:556234. doi: 10.3389/fmars.2020.556234
Hörstmann, C., Buttigieg, P. L., Simpson, P., Pearlman, J., Karstensen, J., and Waite, A. M. (2020). Towards a Best Practice for Developing Best Practices in Ocean Observation (BP4BP): Supporting Methodological Evolution through Actionable Documentation. Paris: UNESCO.
Makiola, A., Compson, Z., Baird, D., Barnes, M., Boerlijst, S., Bouchez, A., et al. (2020). Key questions for next-generation biomonitoring. Front. Environ. Sci. 7:197. doi: 10.3389/fenvs.2019.00197
Miksa, T., Simms, S., Mietchen, D., and Jones, S. (2019). Ten principles for machine-actionable data management plans. PLoS Comput. Biol. 15:e1006750. doi: 10.1371/journal.pcbi.1006750
Miloslavich, P., Bax, N., Simmons, S., Klein, E., Appeltans, W., Aburto-Oropeza, O., et al. (2018). Essential ocean variables for global sustained observations of biodiversity and ecosystem changes. Glob. Change Biol. 24, 2416–2433. doi: 10.1111/gcb.14108
Mitchell, A., Almeida, A., Beracochea, M., Boland, M., Burgin, J., Cochrane, G., et al. (2019). MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 48, D570–D578. doi: 10.1093/nar/gkz1035
Nagoya Protocol (2010). Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity: (Chapter XXVII: Environment). Nagoya: United Nations Treaty Collection.
National Oceanography Centre (2021). Auto-Sampler Device and Method of Autosampling. Paris: National Oceanography Centre.
Porter, M. (1985). Competitive Advantage: Creating and Sustaining Superior Performance. New York, NY: Free Press.
Preston-Werner, T. (2021). Semantic Versioning 2.0.0. Semantic Versioning. Available online at: https://semver.org/ (accessed July 28, 2021).
Samuel, R. M., Meyer, R., Buttigieg, P. L., Davies, N., Jeffery, N. W., Meyer, C., et al. (2021). Towards a global public repository of community protocols to encourage best practices in biomolecular ocean observing and research. Zenodo [Preprint]. doi: 10.5281/zenodo.5482852
Simpson, P., Pearlman, F., and Pearlman, J. (eds) (2021). Evolving and Sustaining Ocean Best Practices Workshop IV, 18; 21-25 & 30 Sep 2020 [Online]: Proceedings, Volumes 1 & 2. IOC Workshop Report No. 294. Paris: UNESCO.
Sunagawa, S., Acinas, S., Bork, P., Bowler, C., Eveillard, D., Gorsky, G., et al. (2020). Tara Oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445. doi: 10.1038/s41579-020-0364-5
Thompson, L., Sanders, J., McDonald, D., Amir, A., Ladau, J., Locey, K., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
van de Kamp, J., Brown, M., Ostrowski, M., Seymour, J., Smith, M., Raes, E., et al. (2019). The Australian Microbiome Initiative-towards microbial omics at the continental scale. Geophys. Res. Abstr. 21:88.
Wetterstrand, K. (2020). The Cost of Sequencing a Human Genome [WWW Document]. Available online at: www.genome.gov/about -genomics/fact-sheets/Sequencing-Human-Genome-cost (accessed March 28, 2021).
Whitt, C., Pearlman, J., Polagye, B., Caimi, F., Muller-Karger, F., Copping, A., et al. (2020). Future vision for autonomous ocean observations. Front. Mar. Sci. 7:697. doi: 10.3389/fmars.2020.00697
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
Wilkinson, M., Dumontier, M., Aalbersberg, I., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
Yamahara, K., Preston, C., Birch, J., Walz, K., Marin, R., Jensen, S., et al. (2019). In situ autonomous acquisition and preservation of marine environmental DNA using an autonomous underwater vehicle. Front. Mar. Sci. 6:373. doi: 10.3389/fmars.2019.00373
Keywords: omics, eDNA, ocean best practices, ocean observations, metadata, protocol management, methods
Citation: Samuel RM, Meyer R, Buttigieg PL, Davies N, Jeffery NW, Meyer C, Pavloudi C, Pitz KJ, Sweetlove M, Theroux S, van de Kamp J and Watts A (2021) Toward a Global Public Repository of Community Protocols to Encourage Best Practices in Biomolecular Ocean Observing and Research. Front. Mar. Sci. 8:758694. doi: 10.3389/fmars.2021.758694
Received: 14 August 2021; Accepted: 21 September 2021;
Published: 11 October 2021.
Edited by:
Stéphane Pesant, European Bioinformatics Institute (EMBL-EBI), United KingdomReviewed by:
Gerhard Josef Herndl, University of Vienna, AustriaCopyright © 2021 Samuel, Meyer, Buttigieg, Davies, Jeffery, Meyer, Pavloudi, Pitz, Sweetlove, Theroux, van de Kamp and Watts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robyn M. Samuel, ci5tLnNhbXVlbEBzb3Rvbi5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.