- 1Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Japan
- 2Fisheries Resources Institute, Japan Fisheries Research and Education Agency, Nagasaki, Japan
Euphausiids are abundant micronekton and important links between higher and lower trophic levels in marine ecosystems; however, their detailed diets cannot be fully understood by conventional microscopy, especially in subtropical areas. Here, we report the euphausiid community structure in the California Current (CC) area and the eastern/western North Pacific subtropical gyre (ESG and WSG) and detail the feeding ecology of the dominant species (Euphausia pacifica, E. brevis, and E. hemigibba) in each region using a combined approach of gut content analysis via 18S V9 metabarcoding and stable carbon and nitrogen isotope analysis. A pronounced omnivorous feeding of all studied euphausiid species was supported by both methods: phytoplanktonic taxonomic groups (Dinophyta, Stramenopiles, and Archaeplastida), Copepoda, and Hydrozoa were detected in the gut contents; all the three euphausiid species displayed an intermediate trophic position between the net plankton (0.2–1.0 mm) and the myctophid fish (15.2–85.5 mm). However, Hydrozoa found in euphausiid gut contents likely derived from a potential cod-end feeding, based on isotope analysis. E. pacifica in the CC province ingested more autotrophic prey, including pelagophyte and green algae, due to a greater abundance of Stramenopiles and Archaeplastida in shallow layers of CC water. On the other hand, non-autotrophic prey such as mixotrophic Kareniaceae dinoflagellates, Pontellidae and Clausocalanidae copepods, and Sphaerozoidae rhizarian contributed more to the diets of E. brevis and E. hemigibba because of a lower chlorophyll a concentration or potentially a scarcity of autotrophic prey availability in ESG and WSG. The feeding patterns of dominant euphausiid species conducting filter feeding were thus largely determined by phytoplankton prey availability in the environments. Dietary difference across three species was also indicated by stable isotope analysis, with a lower mean trophic level of E. pacifica (2.32) than E. brevis (2.48) and E. hemigibba (2.57). These results verify direct trophic interactions between euphausiids and primary production and suggest that the omnivorous feeding habit is a favorable character for dominant Euphausia species.
Introduction
Euphausiids are key micronekton that are widely distributed across ocean basins of the world. Their standing stock constitutes a significant portion of the total biomass of zooplankton and micronekton in marine pelagic ecosystems (Nicol, 2003). Some euphausiid species perform diel vertical migration and present vertical distributions between 200 and 500 m during the day and above 100 m at night (Brinton, 1967, 1979; Taki, 2008; Werner and Buchholz, 2013), accelerating the transportation of organic matter from the euphotic to the mesopelagic layer. Euphausiids generally prey on phytoplankton by filter-feeding or raptorially feed on larger particles, such as zooplankton (Dilling et al., 1998; Park et al., 2011). Concomitantly, they are a major source of food for commercially important fishes, marine mammals, and birds (Kinsey and Hopkins, 1994; Hipfner, 2009; Sogawa et al., 2017; Kim et al., 2019). Thus, euphausiids are important links between lower and higher trophic levels and investigating their feeding ecology would lead to the further understanding oceanic food web structure and energy flows in marine ecosystems.
Gut content analysis is a widely used method to reveal the natural feeding ecology of an organism. By direct observation of the gut contents under the microscope, prior studies confirmed a diatom-rich feeding pattern dominated by the Fragilariopsis spp. in Euphausia vallentini (Gurney et al., 2001); using gut pigments analysis, omnivory (including diatoms, dinoflagellates, and copepod nauplii) and a decreasing feeding activity during daytime in E. pacifica were verified (Nakagawa et al., 2003). However, the microscopical gut content analysis presents several limitations. For example, euphausiids tend to grind prey with their mandibles, which usually increases the difficulty in identifying the detailed prey taxa; food items with soft bodies are easier to be digested and might be overlooked due to a trace amount of remains in the guts (Sarda and Valladares, 1990; Clarke et al., 2020). Metabarcoding based on the high-throughput DNA sequencing technologies can overcome this limitation and has become an effective tool for gut content analysis with a high taxonomic resolution (Pompanon et al., 2012; Berry et al., 2015; Albaina et al., 2016). 18S metabarcoding has been applied to the Antarctic krill to investigate the different preferences of the diatom Chaetoceros spp. in fjords and open waters (Cleary et al., 2018). Nonetheless, it should be noted that the snapshots provided by the gut content analysis, independent of the method used, only recover a temporary diet of the organisms at a certain time before capture.
Stable isotope analysis could provide time-integrated information of trophic relationships to complement the limitations of gut content analysis, even though this method lacks sufficient information on detailed prey (Tverin et al., 2019). The stable nitrogen isotope ratio is a suitable tracer for studying predator-prey relationships and trophic positions of organisms because predators in higher trophic levels exhibit 15N enrichments relative to their diet (DeNiro and Epstein, 1981; Minagawa and Wada, 1984; Fry, 1988). Likewise, the stable carbon isotope ratio is suitable for inferring dietary sources in the food web. Using these two biomarkers, E. pacifica in the Yellow Sea cold water mass was found to feed on relatively large zooplankton, such as Calanus sinicus (Kim et al., 2019), and an ontogenetic diet shift of E. pacifica was confirmed (Park et al., 2011). Therefore, combining gut content analysis and stable isotope analysis in feeding ecology investigation is more precise than previous studies conducted only with one method (Whitaker et al., 2019; Nakamura et al., 2020).
The North Pacific Ocean covers a large area of pelagic ecosystems with complex distributions of water masses, including the subarctic water, transition region of the California Current (CC), eastern and western regions of the North Pacific central (subtropical) water, and Pacific equatorial water (Sverdrup et al., 1942). Prior biogeographical studies reported a total of 59 euphausiid species in the Pacific Ocean with their vertical and horizontal distributions, biomasses, and life history information, and revealed a comparative distribution pattern of euphausiid species in analogous water masses (Boden et al., 1955; Brinton, 1962). However, feeding habits of North Pacific euphausiids, which might be one of the factors determining their community structures, were mostly studied on the numerically predominant E. pacifica or the larger Thysanoessa spp. that inhabited in the subarctic North Pacific (e.g., T. inspinata; Kim et al., 2010). Only a few comparative feeding ecology investigations of euphausiid species have been conducted in low-latitude ecosystems, such as the oligotrophic North Pacific subtropical gyre.
In this study, we first investigate the community structures of euphausiids from the CC region and the eastern and western subtropical gyres (ESG and WSG) in the North Pacific. Furthermore, we used a combined approach of 18S V9 metabarcoding and stable isotope analysis to reveal detailed prey information and trophic positions of the dominant euphausiid species in these three provinces and clarified the ecological importance of euphausiids in the food web. Finally, we compared the feeding ecology among euphausiid species and provided more insights into feeding habit determining the community structure of euphausiids. We also explored the merits and limitations of DNA metabarcoding as a tool for feeding ecology investigations.
Materials and Methods
Field Collections
Samplings were carried out during the KH-17-4 cruise on the RV Hakuho-Maru (Japan Agency for Marine-Earth Science and Technology) at three stations in the North Pacific Ocean between August 13 and October 2, 2017. Stations 1, 2, and 3 were located in the California Current (CC) region, the eastern subtropical gyre (ESG) region, and the western subtropical gyre (WSG) region, respectively (Figure 1A). Vertical profiles of water temperature and salinity between 0 and 500 m were recorded with a conductivity, temperature, and depth (CTD) system (SBE-911 plus; Sea-Bird Electronics). Chlorophyll a (chl a) was extracted with N, N-dimethylformamide, and the concentration was analyzed using a Turner fluorometer (Turner Designs).
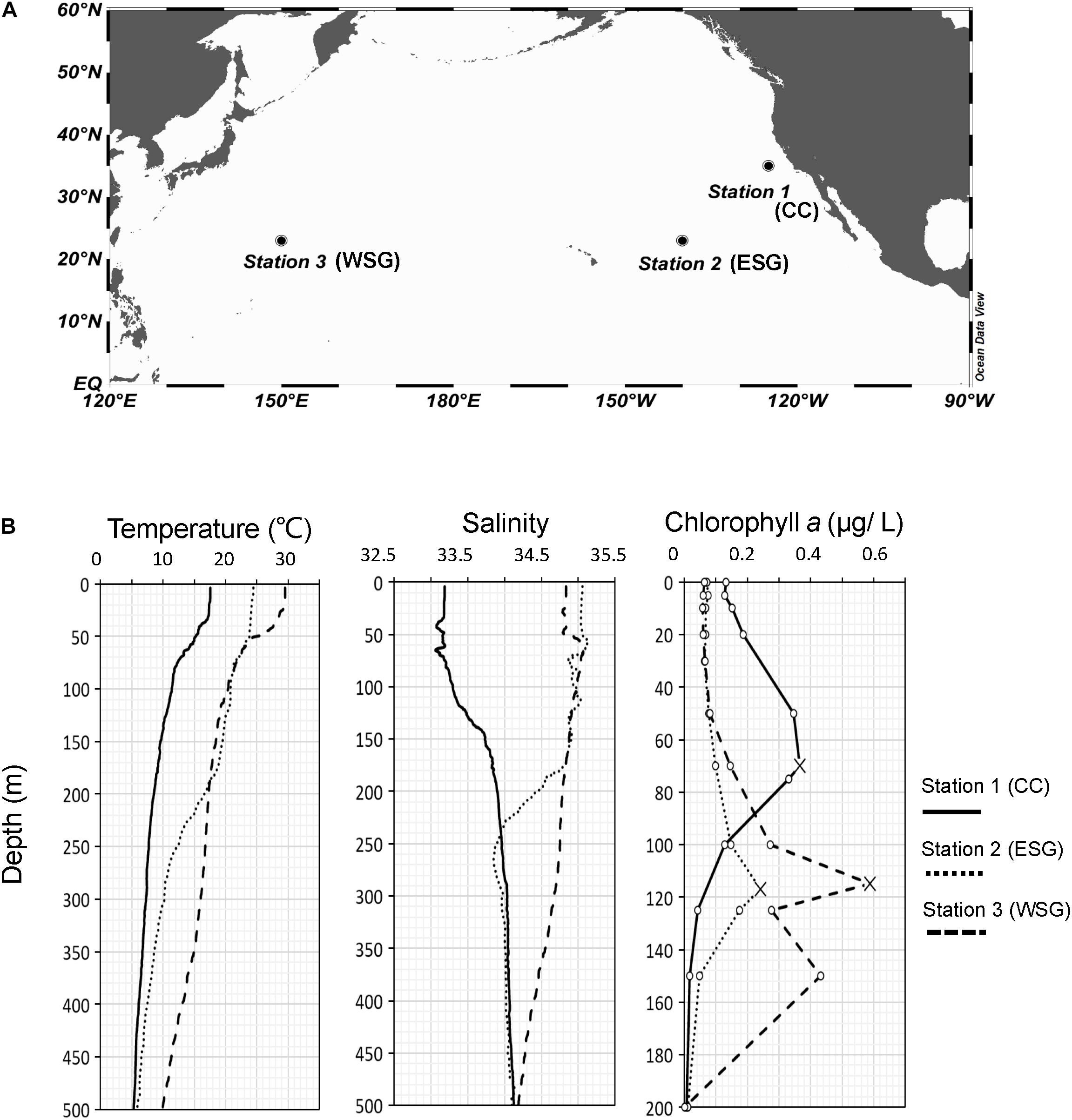
Figure 1. (A) Sampling stations in North Pacific Ocean. (B) Vertical profiles of water temperature and salinity at 0–500 m, and vertical distribution of chlorophyll a concentration at 0–200 m at station 1, 2, and 3. Crosses marked the depths of the subsurface chlorophyll maximum (SCM) in each station.
Euphausiids were collected at night by oblique tows of Matsuda-Oozeki-Hu Trawl (MOHT; Oozeki et al., 2007) with a 2 m2 mouth opening and a 1.95 mm mesh size from approximately 500 m depth to the surface (Table 1). A flow meter was attached to the net to measure the water volumes filtered. Samples were split on board promptly after net landing, and one-fourth of aliquots of the specimens were placed into a −80°C deep freezer immediately after splitting and kept frozen before subsequent laboratory operations for feeding ecology analysis. Another one-eight or one-fourth of aliquots of samples were fixed with 5% buffered formaldehyde for community structure analysis. To characterize the environmental communities as potential prey of euphausiids, seawater was collected at the surface (0 m) using an acid-cleaned bucket and at 10 m, the subsurface chlorophyll maximum (SCM), and at 100 m using acid-cleaned Teflon-coated Niskin bottles attached to the CTD. A 5 L aliquot of the water sample at each depth was immediately filtered on a Nuclepore polycarbonate membrane filter (Whatman, NJ, United States) with a 3.0 μm pore size, and the filters were also strictly preserved at −80°C. However, the seawater samples from station 3 were probably not processed timely, resulting in a large proportion of bacteria in the environmental communities according to the metabarcoding results. Thus, the environmental community data at station 3 was excluded from this study.
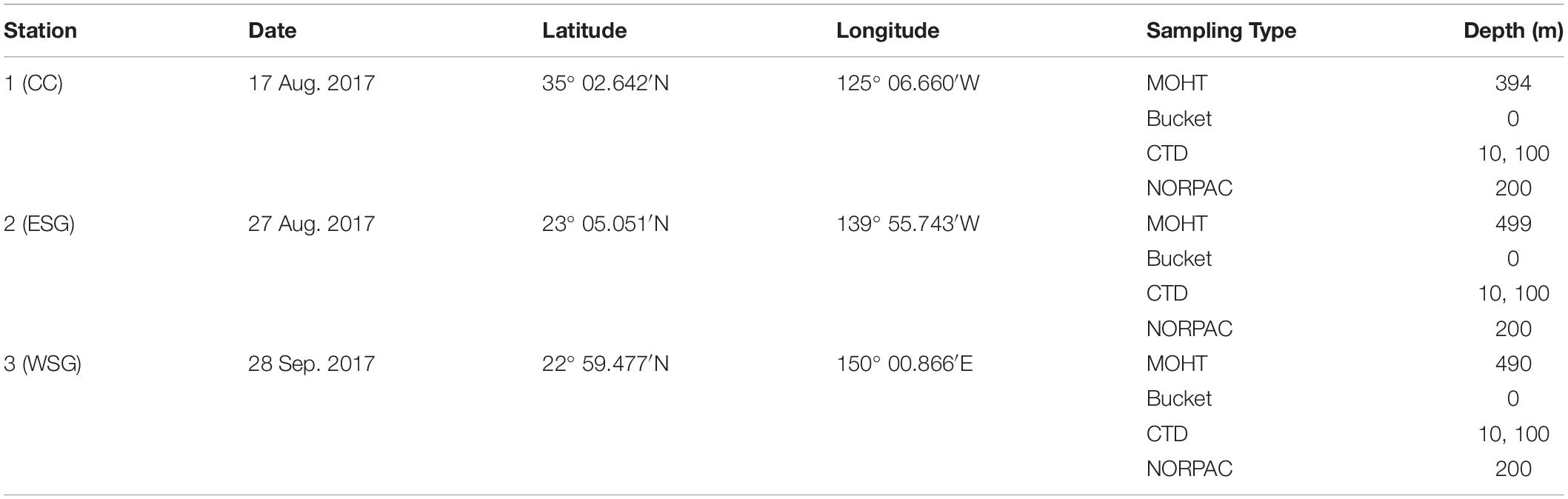
Table 1. Sampling information of station 1, 2, and 3 in different water masses of the North Pacific Ocean.
For the isotopic analysis, suspended particulate organic matter (POM) at 0 m was used as data of primary producers. In addition to the euphausiid samples, we analyzed net-plankton (size ranged from 0.2 to 1.0 mm) and myctophid (size ranged from 15.2 to 85.5 mm) in each station. For a sampling of POM, aliquots of 3–5 L surface seawater (0 m) collected using acid-cleaned buckets were filtered onto pre-combusted GF/F filters via gentle vacuum after pre-filtration with a 200 μm nylon mesh. Net-plankton samples were collected from a 200 m depth from the surface using vertical tows of a NORPAC net (45 cm mouth diameter, 100 μm mesh size), and then immediately size-fractionated into small (0.2–0.5 mm) and large (0.5–1.0 mm) fractions. Myctophids were selected from the micronekton samples collected by the same tow using a MOHT net for the euphausiid samplings. All samples were frozen at −20°C until laboratory processes.
Community Structures Analysis
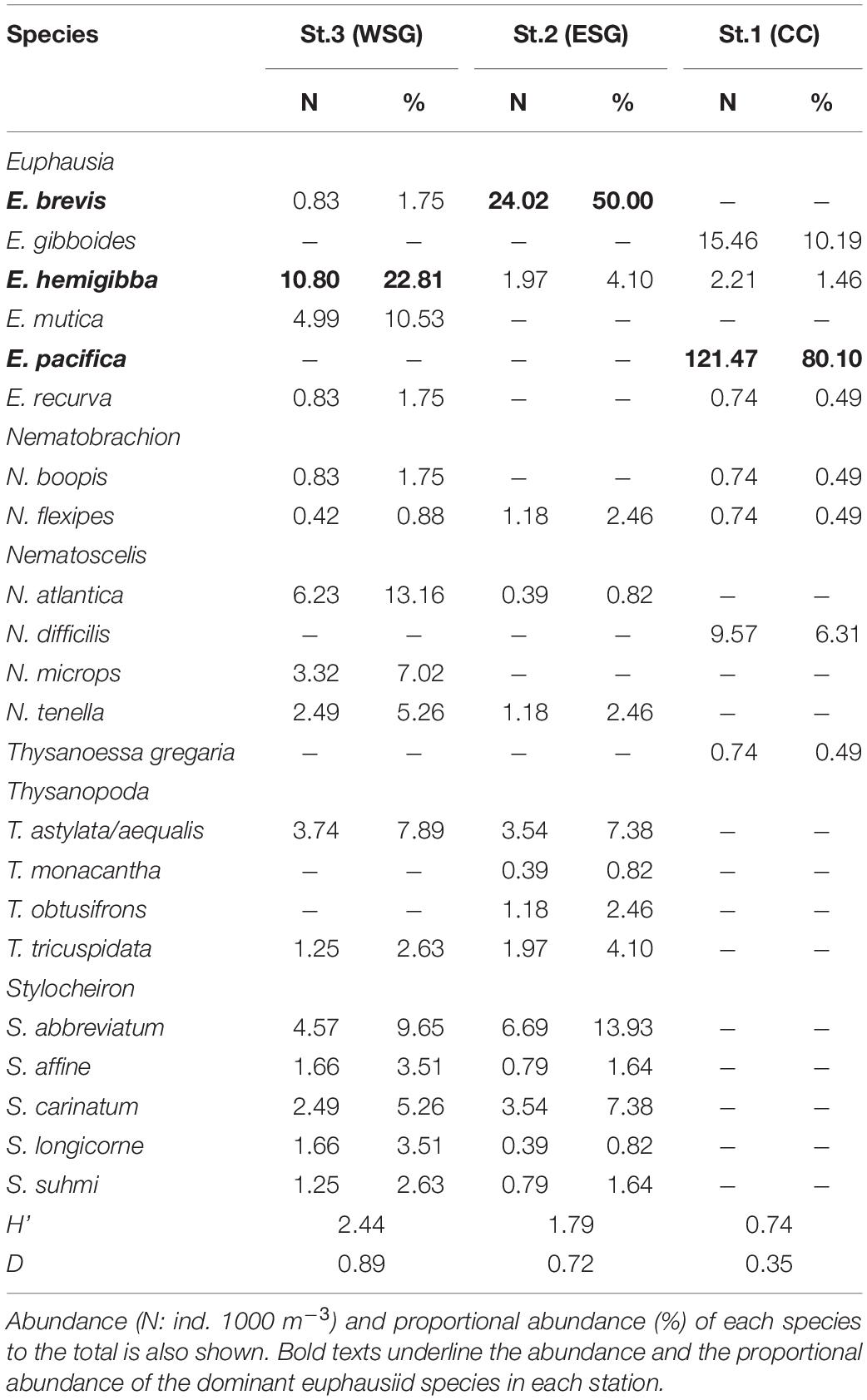
All adult euphausiids at each station were identified to the lowest possible taxonomic level according to Baker et al. (1990) under a stereomicroscope (Nikon SMZ-745T), and the abundance for each species was standardized as individuals (ind.) 1,000 m–3. The Shannon index (H’; Shannon, 1948) and the Simpson’s diversity index (D; Simpson, 1949) were calculated to compare the euphausiid species diversity among the stations.
Euphausiid Processes and High Throughput Sequencing
The feeding ecology of the adults of the dominant euphausiid species at each station was further analyzed. Frozen samples were naturally thawed on ice, and 20–30 euphausiid individuals of the target species were randomly selected during the thawing. In the dissection process, the cephalothorax section was firstly excised to avoid the potential net feeding influence from the ingested food in the stomach of a euphausiid. To minimize DNA degradation, the whole gut was quickly removed and immediately transferred into a 30 μL of precooled 5% Chelex buffer (BIO-RAD) in a 1.5 mL Eppendorf tube kept on ice. Eyes of one of the selected individuals of each species were used as negative controls and treated in the same method as the gut contents. The remains of the euphausiid tissues were preserved back to −80°C until the stable isotope analysis. All the dissection facilities were sterilized every time before touching a different part of one euphausiid individual and dissecting another individual to avoid cross-contamination.
Total DNA of the gut contents and eyes were extracted according to the methods of Nagai et al. (2012). For the environmental samples, total DNA extraction was carried out with the MagAttract PowerWater DNA/RNA Kit (QIAGEN). Polymerase chain reaction (PCR) amplifications were conducted for all samples using KOD Plus version 2 (Toyobo), with the eukaryotic universal primers 1389F and 1510R (Amaral-Zettler et al., 2009) according to the methods of Hirai et al. (2017). Adaptors used to differentiate samples during sequencing on Illumina MiSeq and dual-index target sequences were added in the 2nd and 3rd rounds of PCR. The PCR products of every round were diluted 20 times with diethylpyrocarbonate (DEPC)-treated water and used as templates for the next round. The final PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN). Concentrations of the final PCR products were measured using a Qubit 3.0 Fluorometer (Invitrogen), and 100 ng of each purified PCR product was pooled. A sequencing run on Illumina MiSeq was carried out using MiSeq Reagent Kit v2 (FASMAC Co), and raw data of 2 × 250 bp paired-end sequence reads were obtained.
Bioinformatics and Statistics
All raw sequence reads were quality-filtered using Trimmomatic (Bolger et al., 2014). Short- and low-quality fragments were eliminated, sequencing adaptors were clipped, and all technical sequence reads were trimmed into appropriate length using the following settings: CROP: 135; MINLEN: 50; LEADING: 20; TRAILING: 20; and SLIDINGWINDOW: 30:30. Paired-end sequences were merged, and further quality-filtrating was performed using MOTHUR (Schloss et al., 2009) according to the methods of Hirai et al. (2017). Briefly, the reads with more than six homopolymers and uncertain bases were removed, and qualified sequences were aligned against SILVA 138.1 database (Quast et al., 2013). Aligned sequences were single-linkage pre-clustered and screened to remove potential chimeric sequences with a reference dataset in UCHIME (Edgar et al., 2011). In this study, the reference database V9_PR2 from the Tara Oceans project (de Vargas et al., 2015) was used to classify the remaining high-quality sequences into different taxonomic groups using a naive Bayesian classifier (Wang et al., 2007), with a threshold greater than 80%. Only the Eukaryotic sequences were preserved for further analysis, and sequences classified into “Malacostraca” and “Arthropoda_unclassified” were excluded as a possible read from the euphausiid host. Mammalian, fungi, and parasitic taxa, which were determined according to Cleary and Durbin (2016), were also removed. Finally, preserved sequences were clustered into operational taxonomic units (OTUs) at a 99% similarity threshold.
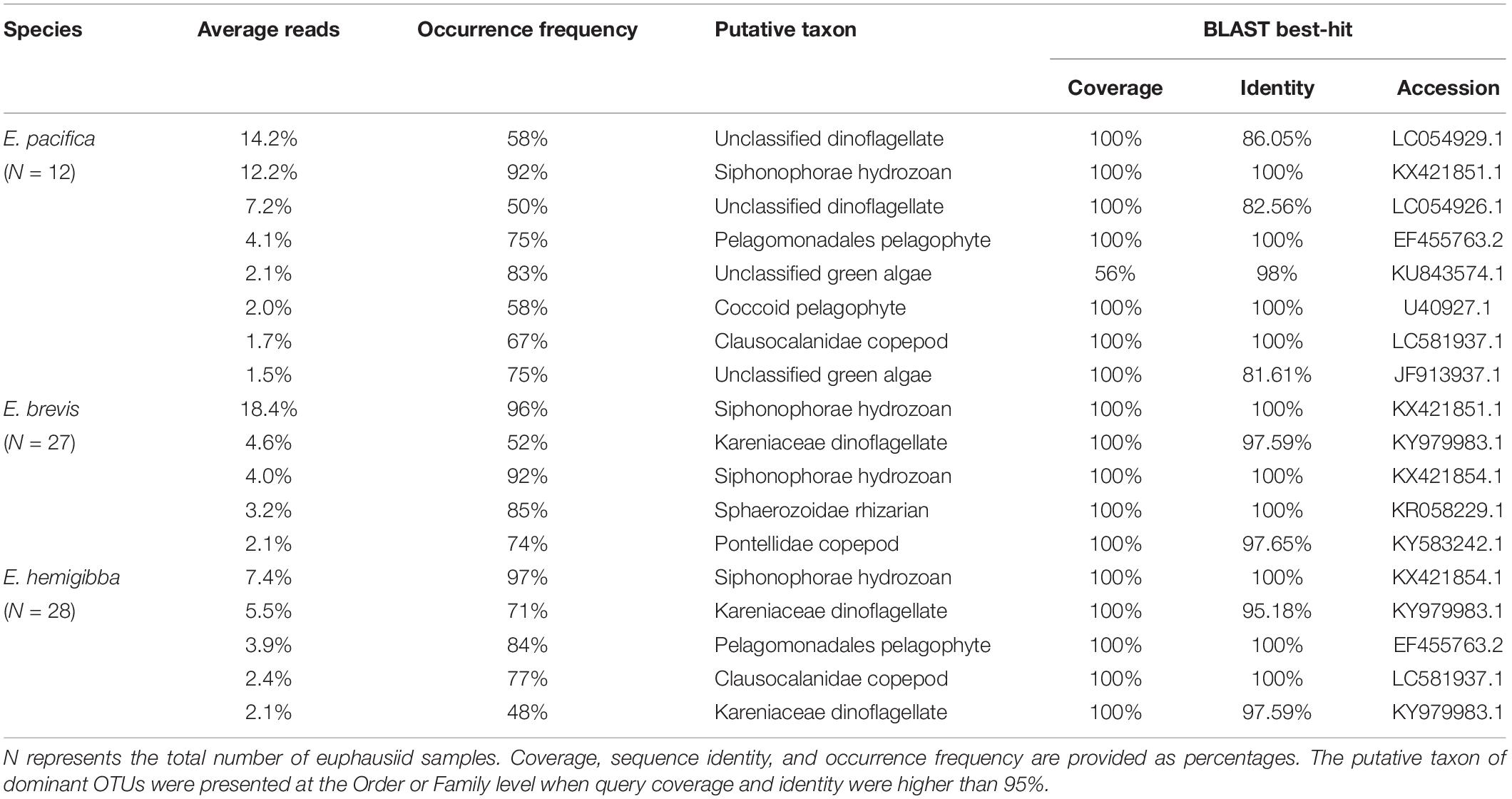
After quality controlling and taxonomic classification, to present the gut content composition of euphausiid individuals and the environmental community structure of different stations, the relative sequence read abundances of all taxonomic supergroups in Eukaryota were measured for each euphausiid and environmental sample. A Kruskal-Wallis test with Dunn post hoc test was conducted on the relative abundances of main supergroups in each sample within and across different euphausiid species using IBM SPSS statistics 23 (Zar, 1999). Moreover, to evaluate the gut content composition differences among different euphausiid species, the Bray-Curtis similarity index was first calculated on euphausiid gut content composition of square root-transformed data, and the homogeneity of dispersion was evaluated using permutational analysis of multivariate dispersions (PERMDISP). Differences among species were then tested using permutational analysis of variance (PERMANOVA) and a posterior pairwise test. PERMANOVA was conducted using Type III sum of squares and the unrestricted model, and number of permutations was 9999 for all permutation-based tests. To detect those prey supergroups responsible for the possible observed differences between euphausiid diets, a similarity percentages (SIMPER) analysis was also performed. All these analyses were conducted in PRIMER version 7 with the add-on package PERMANOVA+ (Anderson et al., 2008). Furthermore, to clarify main preys of all the euphausiid species with detailed taxonomy information, the percentage of each prey OTU in each sample was computed and then averaged among all euphausiid individuals at each station. For all three euphausiid species, detected prey OTUs comprising more than 1.5% of the total gut content reads and with occurrence frequency higher than 40% were deemed as dominant prey OTUs (Harms-Tuohy et al., 2016; Deagle et al., 2019). Every dominant prey OTU was identified to the lowest possible taxonomic level by BLASTN searches against the NCBI “nt” database.
Stable Isotope Analysis
Particulate organic matter (POM), net-plankton, and euphausiid samples for stable isotope analysis were oven-dried at 50°C overnight. Filter samples of POM were exposed to HCl fumes for 2 h to remove the carbonate and dried again at 50°C. All euphausiid individuals and 1–10 mg of the net-plankton samples in the two fractions were homogenized, and approximately 0.5–1.5 mg of each homogenized sample was measured (two euphausiid individuals were pooled when the sample weight was insufficient). For the myctophid samples, the dorsal muscles of 11–14 specimens from each station were dried and homogenized. Half of the sample was lipid-extracted using chloroform/methanol (2:1, v/v) for the measurement of δ13C (Folch et al., 1957).
Stable nitrogen and carbon isotope ratios were measured using a DELTA V advantage mass spectrometer (Thermo Fisher Scientific Inc.) connected to an elemental analyzer. Amino acids with different isotopic values (alanine, histidine, and glycine), which were certified using the standard sample provided by the International Atomic Energy Agency, were used to obtain the calibration curves. Alanine (δ15N = −1.2‰, δ13C = −19.9‰) was measured once every 10 samples for correction of a shift of the baseline. As for the trophic position analysis, Minagawa and Wada (1984) reported an average stepwise increase in δ15N of 3.4 ± 1.1‰ per trophic level across a wide range of habitats, including marine habitats. Thus, the trophic levels of the analyzed samples were calculated as:
Differences in the trophic levels at the different sampling stations were analyzed by one-way analysis of variance (ANOVA) with Fisher’s least significant difference (LSD) post hoc test using IBM SPSS Statistics 23.
Results
Environmental Conditions
A lower temperature and salinity were observed at station 1 in the CC compared to the other two stations in the subtropical gyre (Figure 1B). At station 1, the temperature decreased gradually with increasing depth, from 17.6°C on the surface to 5.3°C at 500 m, and salinity increased slightly from 33.2 to 34.1. Stations 2 and 3 in the subtropical gyre shared more similar environmental conditions, especially in the epipelagic layer: the water temperature of station 3 was higher at the surface and below 200 m, but was nearly the same as station 2 at 50–200 m (average of 20.2°C in station 2 and 19.7°C in station 3); the salinity of these two stations were in common (average of 34.9) in the upper 200 m. A higher chl a concentration (0.13 μgL–1) at the surface (0 m depth) was observed at station 1 than at station 2 (0.07 μgL–1) and 3 (0.06 μgL–1). Deeper SCM depths were observed at station 2 (117 m) and station 3 (115 m), compared with station 1 (70 m). The chl a concentration at the SCM depth of each region was the highest at station 3 (0.59 μgL–1), followed by station 1 (0.37 μgL–1) and station 2 (0.24 μgL–1).
Euphausiid Community Structure
The total abundance of euphausiids was the highest at station 1 (152 ind. 1,000 m–3) and the lowest at station 3 (48 ind. 1,000 m–3 at station 2, 47 ind. 1,000 m–3 at station 3), and 22 euphausiid species belonging to six genera were identified (Figure 2 and Table 2). E. pacifica was the most abundant species at station 1, contributing approximately 80.0% of the abundance (121 ind. 1,000 m–3) of the euphausiid community. Other euphausiids at station 1 were mainly attributed to E. gibboides and Nematoscelis difficilis. E. brevis and E. hemigibba were the dominant euphausiid species at stations 2 and 3, respectively. However, in comparison with E. pacifica at station 1, dominance of these two subtropical species was considerably low, 24 ind. 1,000 m–3 for E. brevis and 11 ind. 1,000 m–3 for E. hemigibba, accounting for 50.0% and 22.8% abundance at stations 2 and 3, respectively. The euphausiid community compositions at stations 2 and 3 were similar at the genus level. Euphausiid species in the genera Stylocheiron and Thysanopoda were detected frequently at both stations 2 and 3 but not at station 1. Abundances of these genera were higher at station 2 than at 3. In contrast to the species abundance, H’ and D were higher in the subtropical gyre (stations 2 and 3) than at station 1 of CC area. The highest species diversity was observed at station 3 (Table 2).
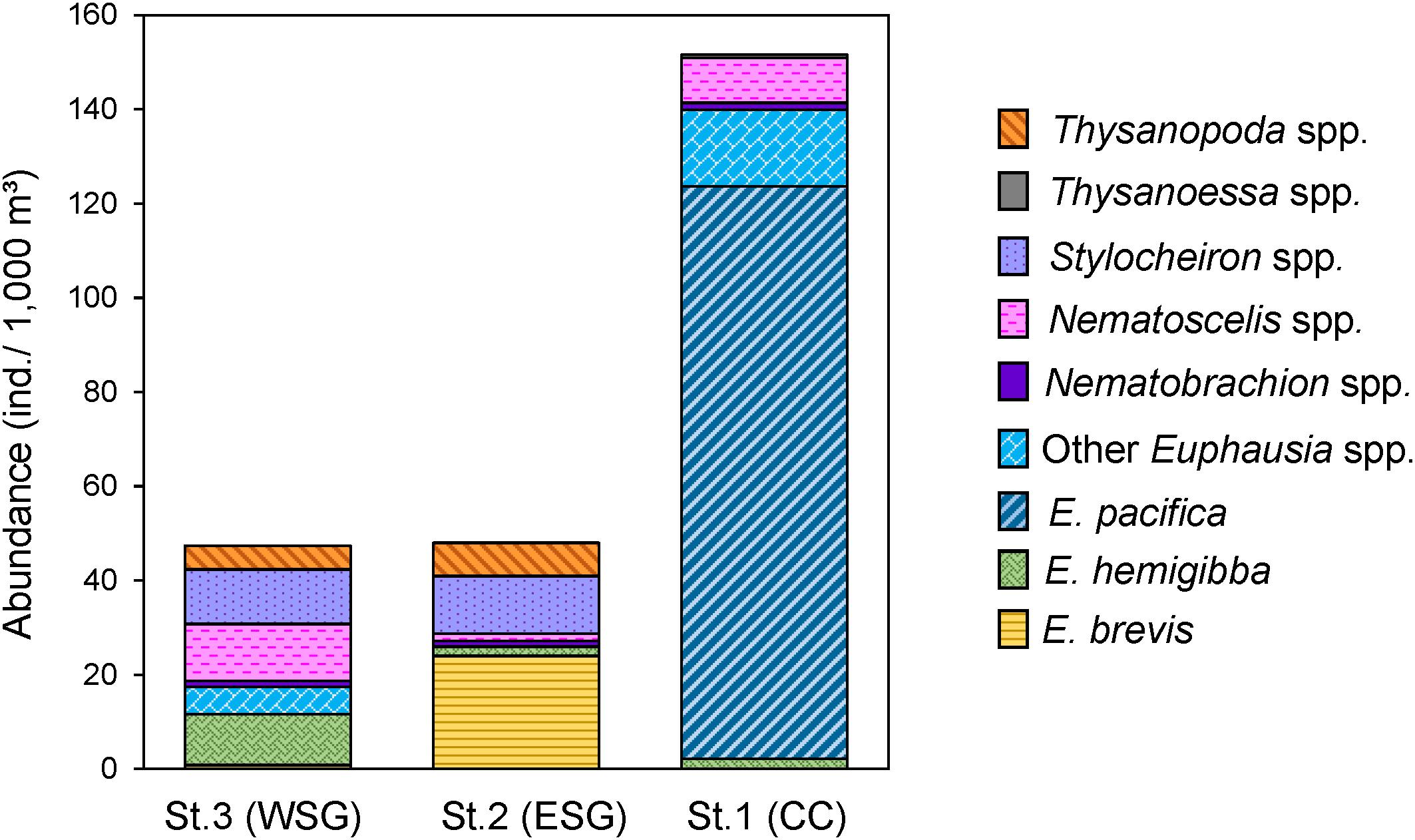
Figure 2. Community structure and abundance (individuals/1,000 m3) of euphausiids at station 1, 2 and 3. Dominant euphausiid species and other species in different genera are indicated by different colors, minor Euphausia species other than the dominant ones are grouped under Other Euphausia spp.
18S V9 Metabarcoding Analysis of Gut Contents
Totally, 79 adult euphausiid individuals (21 E. pacifica, 27 E. brevis, and 31 E. hemigibba) were selected for the feeding ecology study. The total length of euphausiids was 7.6–15 mm for E. pacifica (mean ± standard deviation (SD): 10.0 ± 2.1), 7.0–9.1 mm for E. brevis (mean ± SD: 8.2 ± 0.5), and 9.4–13.2 mm for E. hemigibba (mean ± SD: 11.4 ± 1.1). An overall number of 384,698 sequence reads from eight environmental samples in CC and ESG provinces and 262,758 reads from euphausiid gut samples were obtained. However, 20,533 reads from three negative-control (eye) samples still remained after removing the host and contaminant sequences. Furthermore, >98% of the remaining sequences of negative controls as well as 47.8% of the gut content sequences were classified into “unclassified Crustacea.” We, therefore, double-checked all the OTUs in the negative controls and found that ∼80% of the unclassified OTUs in negative controls also appeared in the gut contents, taking up 95–97% of the unclassified sequences in gut contents. The unclassified OTUs were eventually verified as euphausiid species and other arthropods or remained unclassified based on the BLASTN search against the “nt” database (Supplementary Figure 1). The unclassified sequences discovered in the gut contents were highly likely attributed to the host sequences; thus, all the unclassified sequences in the gut were removed manually. In addition, for each euphausiid species, gut samples that contained insufficient reads (<0.5% of the total gut content sequence reads) were excluded. At last, a total of 12 E. pacifica, 27 E. brevis, and 28 E. hemigibba were used, and sequence reads for final data analysis were 112 to 46,697 for euphausiid guts, and 32,381 to 61,099 for environmental samples.
Gut contents of all three euphausiid species were mainly dominated by the phytoplankton taxonomic groups including Dinophyta, Stramenopiles, and Archaeplastida (Figure 3). Within the phytoplanktonic supergroups, the Kruskal-Wallis test suggested that Dinophyta sequences had significantly higher (p < 0.05) relative abundances than other prey taxa in the guts of all three euphausiid species and were detected in every individual, accounting for 3.3–87.8% (average 45.8%) in E. pacifica, 1.6–97.6% (average 40.5%) in E. brevis, and 4.2–97.6% (average 56.3%) in E. hemigibba. Dinophyta was also abundant in the environmental communities of both CC and ESG regions, averagely representing 29.1% at CC (station 1) and 36.8% at ESG (station 2) in sequence proportions. Stramenopiles was the second most abundant in sequence proportions across the phytoplankton found in the guts, averaging ∼10% in all three species (8.7% in E. pacifica, 8.4% in E. brevis, and 11.7% in E. hemigibba), but comprised a smaller proportion in the water columns (average 4.2% in CC and 1.6% in ESG). Archaeplastida showed relatively small proportions of sequence reads in the water columns, with greater abundance in CC (average 1.8%) than ESG (average 0.2%) (Figures 3B,C), however, Archaeplastida displayed higher percentages in gut contents of euphausiids, making up on average 2.8% in E. brevis, 6.7% in E. hemigibba, and 8.4% in E. pacifica. Kruskal-Wallis test indicated that the relative abundance of Archaeplastida was significantly different between E. brevis and the other two species with p < 0.05, and percentage of Archaeplastida in E. pacifica and E. hemigibba were similar with each other. Moreover, Archaeplastida displayed a significant lower (p < 0.05) relative abundance than Dinophyta and Stramenopiles within E. brevis. Except of phytoplankton, Copepoda sequences represented a similar minor portion in all three species of euphausiids (average 5.7% in E. pacifica and E. brevis, 4.7% in E. hemigibba), but was the most abundant in the environmental community sequence reads. Sequence reads from Rhizaria were relatively abundant in ESG environmental communities (average 7.0% in ESG, 4.8% in CC), and significantly higher percentage (p < 0.05) of Rhizaria was discovered in guts of E. brevis (average 6.3%) than in guts of E. hemigibba (average 1.4%). Rhizaria was found to present a significantly lower percentage than other preys within E. hemigibba. Additionally, Hydrozoa sequences, with a trace amount in the water column (<0.5%), was strikingly discovered in several euphausiid individuals, resulting in a prominent proportion of the gut contents of all three euphausiid species (average 16.6% in E. pacifica, 25.6% in E. brevis, and 8.4% in E. hemigibba). PERMDISP showed no significant differences (p > 0.05) in dispersions among three species, and PERMANOVA indicated a significant difference (p < 0.05) in the gut content composition between E. brevis and E. hemigibba (Supplementary Table 1). The dissimilarity mainly attributed to Dinophyta, Stramenopiles, and Rhizaria with relative greater average dissimilarities and distributed evenly (larger dissimilarity/standard deviation ratios) among euphausiid samples. Hydrozoa also largely contributed to the dissimilarity between two species, but the contributions were inconsistent among all the samples (lower Diss/SD ratio; Supplementary Table 2).
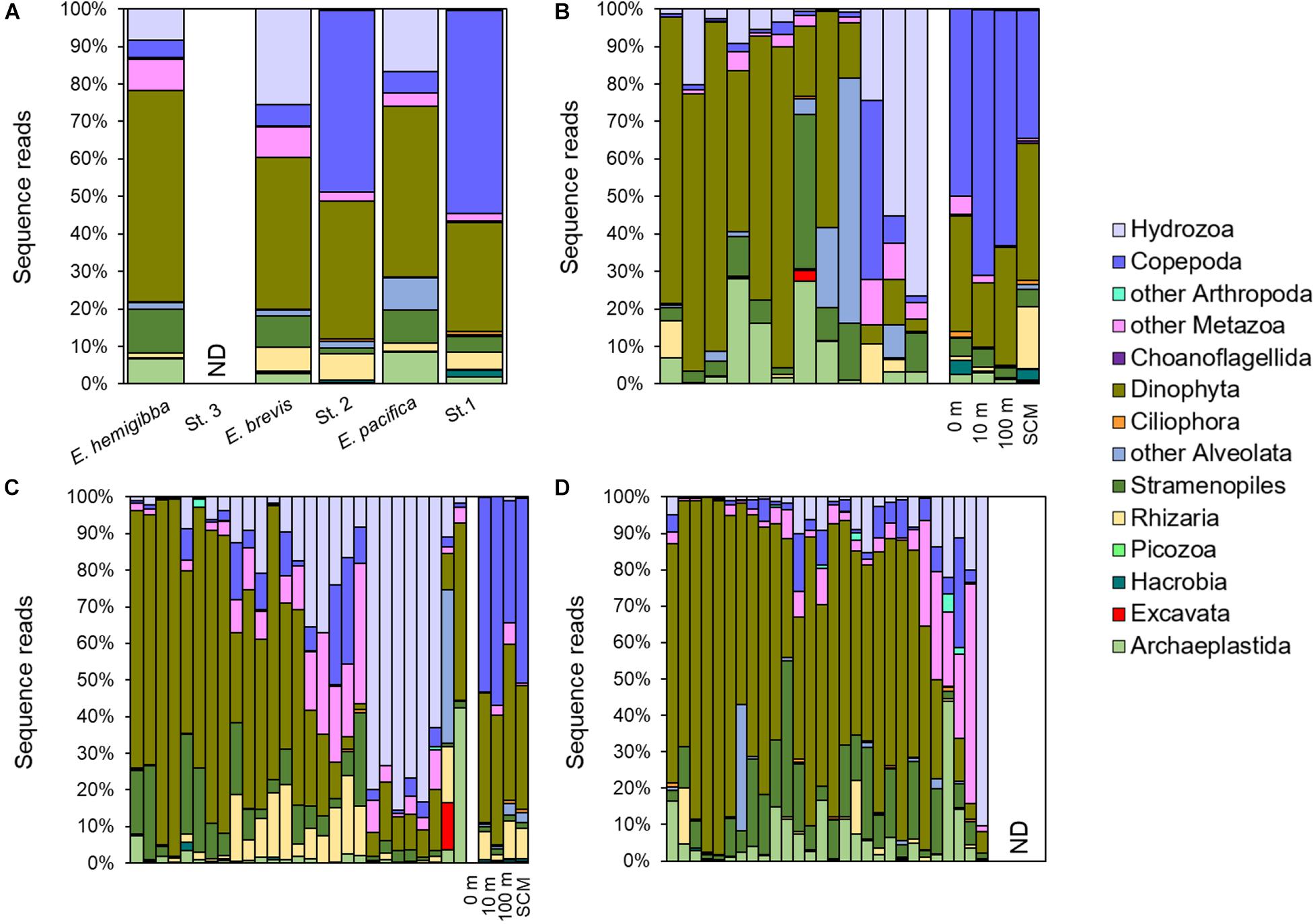
Figure 3. Prey composition of euphausiid species and environmental community structure revealed by 18S V9 metabarcoding. Contribution of different taxonomic groups to the overall diet/community was presented by the proportion of the sequence reads of different supergroups. (A) Average prey composition of three target euphausiid species and environmental communities at station 1, 2, and 3. (B) Individual prey composition of E. pacifica (n = 12) and environmental communities at 0 m, 10 m, 100 m, and SCM of station 1. (C) Individual prey composition of E. brevis (n = 27) and environmental communities at 0 m, 10 m, 100 m, and SCM of station 2. (D) Individual prey composition of E. hemigibba (n = 28), ND: no data.
Dominant Prey Operational Taxonomic Units
A total of 19 OTUs were verified as primary prey of three euphausiid species, including phytoplankton (dinoflagellates, pelagophytes, and green algae) and zooplankton (copepods, hydrozoans, and rhizarians) (Table 3). Hydrozoan OTUs, generally identified as siphonophores, were one of the most dominant OTUs in all three euphausiid species, present in >90% of the euphausiid individuals. Among phytoplankton OTUs, dinoflagellates were the uppermost food items in all three euphausiid species (average of 4.6–14.2% sequence reads), with higher occurrence in E. hemigibba (71%) than E. pacifica (50–58%) and E. brevis (52%). Kareniaceae dinoflagellates were predominant dinoflagellate prey for E. brevis and E. hemigibba. Pelagophytes OTUs were also identified as prime prey for E. pacifica (2.0–4.1% average reads) of CC region and E. hemigibba (3.9% average reads) of WSG region. Pelagomonadales OTUs were common prey of both E. pacifica and E. hemigibba; however, Coccoid pelagophyte OTU was detected only in 58% of the E. pacifica individuals. Green algae OTUs were only listed as dominant in the E. pacifica with a high frequency (58–75%). As for zooplankton prey, Pontellidae copepods for E. brevis, Clausocalanidae copepods for E. hemigibba and E. pacifica were prime food of three species of euphausiids. These copepod OTUs played more important roles in the gut contents of E. brevis and E. hemigibba in the subtropical areas (2.1–2.4% sequence reads; 74–77% occurrence frequency) than E. pacifica (1.7% sequence reads; 67% occurrence frequency). Sphaerozoidae rhizarian was uniquely dominant prey for E. brevis in ESG.
Stable Isotope Analysis
The highest mean nitrogen isotope ratio was in E. pacifica (10.9 ± 0.8‰) and the lowest in E. brevis (3.6 ± 0.2‰), which was consistently 5–5.5‰ higher than that of POM (Figure 4A). The mean δ15N values of the euphausiid species were higher than those of the small or large net-plankton and were lower than those of the myctophid fish. This relationship was also verified between the mean δ13C values of POM, net-plankton, euphausiids, and myctophid fish in both ESG (station 2) and WSG (station 3) regions, however, the mean δ13C value of the E. pacifica was higher than that of the myctophid fish in CC region. The highest value of δ13C was observed in E. brevis (−20.3 ± 0.2‰), and the lowest was observed in E. pacifica (−21.9 ± 0.5‰). The mean trophic levels of E. pacifica, E. brevis, and E. hemigibba were 2.32, 2.48, and 2.57 (Figure 4B), respectively, indicating that the three euphausiid species were between primary and secondary consumers. The trophic levels of E. brevis and E. hemigibba were significantly higher than those of E. pacifica; however, no significant difference was found between E. brevis and E. hemigibba. A relatively wider trophic level spectrum was observed in E. hemigibba (2.07–2.77) in WSG province, compared with the other subtropical species E. brevis (2.37–2.61) in ESG province.
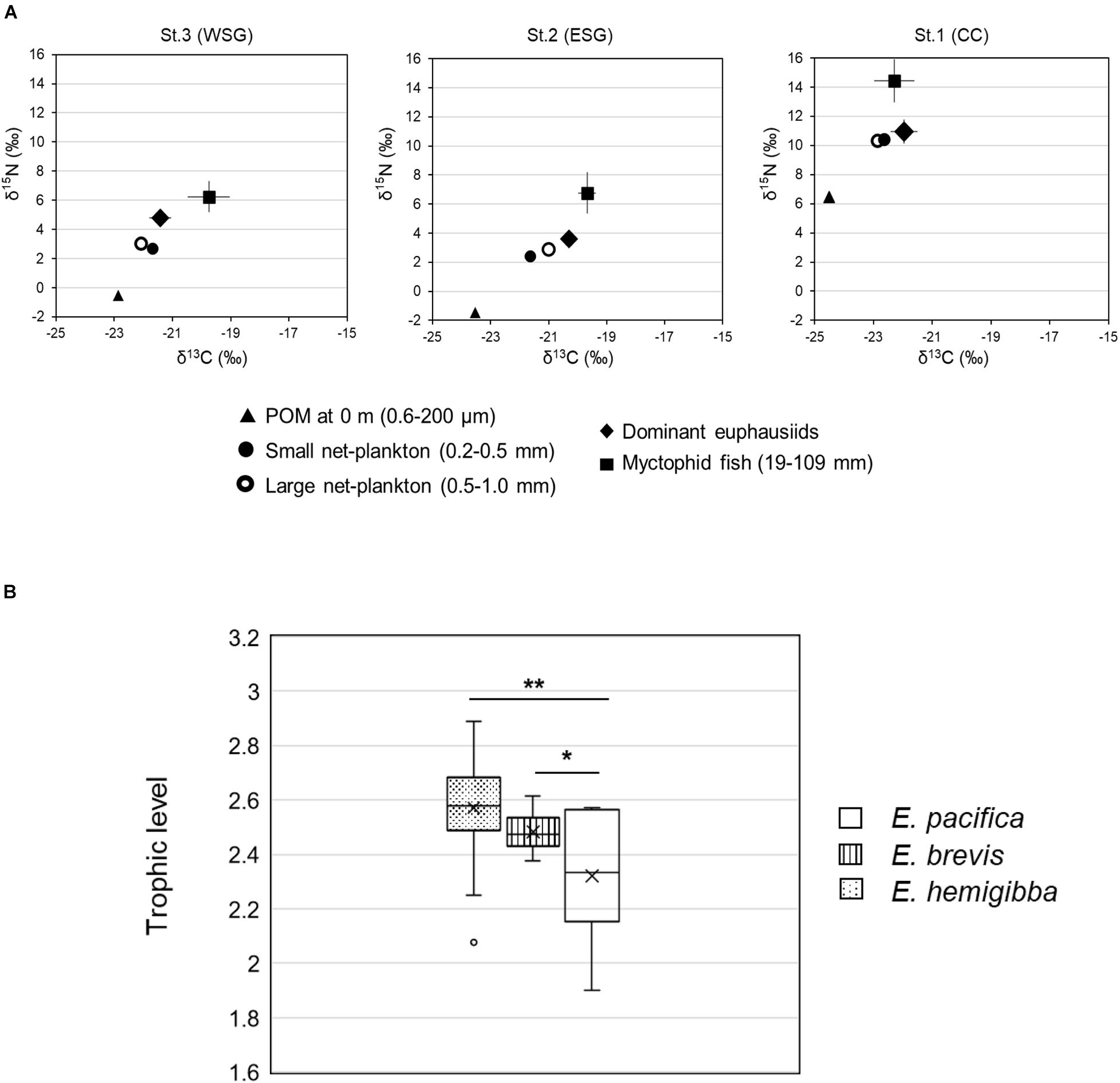
Figure 4. Trophic positions of target euphausiid species in the food web investigated by stable isotope analysis. (A) Trophic positions of E. pacifica, E. brevis, and E. hemigibba in related to Particulate Organic Matter (POM), net plankton, and Myctophid fish at CC, ESG, and WSG region. Different biotic components are indicated by different shapes. Error bars indicate standard deviation. (B) Trophic levels of three euphausiid species. Error bars indicate standard deviation, and cross-mark lines out average trophic level of each euphausiid species. (*: p < 0.05, **: p < 0.01).
Discussion
Euphausiid Community Structure in Relation to Hydrography
This study aimed to reveal the diets of dominant species of euphausiids in the CC and subtropical areas, and the community structures of euphausiids in three water areas were firstly investigated. As reported by Mauchline and Fisher (1969), the hydrographic characteristics of water masses were important factors governing the zonation pattern of euphausiids species in our study, and the euphausiid community in the CC region was distinct from that in the subtropical area. Furthermore, the sea surface temperature is known to be an important driver of diversity in pelagic taxa (Letessier et al., 2009, 2011; Tittensor et al., 2010), agreeing with the diversity patterns of euphausiids in our study. Station 1 (CC area) was in a transition zone with the merging of subarctic water, subtropical water, and equatorial water of the North Pacific, and E. pacifica were highly dominant. A preferred water temperature of E. pacifica ranged between 5 and 16°C, and their abundances were centered in the CC region (Taki, 2007; Lara-Lopez et al., 2012; Tao et al., 2015). Because the warm temperature in the subtropical region around 20–25°N is unfavorable for E. pacifica (Brinton, 1962), this species was not found in both WSG and ESG in our study. On the other hand, most of the predominant euphausiid species of station 2 and 3 were absent in CC province. Roden (1991) noted that the southern boundary of the transition zone (32°N) plays a role as a geographical barrier preventing subtropical species, such as E. brevis from northward extending their distribution. The subtropical species E. brevis and E. hemigibba usually co-occur in the oligotrophic subtropical gyre together with N. atlantica and Stylocheiron suhmi (Moore, 1951; Fager and McGowan, 1963; Wiebe and D’abramo, 1972; Lindley, 1977). However, there were environmental differences in water temperature and chl a concentration within the North Pacific subtropical gyre, possibly leading to different dominant species between ESG and WSG. Based on the results of the present study, more sampling stations in the subtropical regions should be explored for further investigation of euphausiid community structure in related to the hydrographic gradients, and more information about trophic interaction between dominant and minor euphausiid species would provide more insights. The community structures and dominant species, whose diets are further discussed in the following, observed in this study were thus typical in the North Pacific.
Omnivorous Feeding of Dominant Euphausia Species
All the three Euphausia species in this study presented an evident omnivorous feeding. Both phytoplankton (dinoflagellates, pelagophytes, and green algae) and zooplankton (copepods and hydrozoan) were frequently observed from the gut contents by 18S V9 metabarcoding, and omnivorous feeding was also confirmed by the stable isotope analysis. These results also indicated that the feeding pattern of euphausiids was largely influenced by the food resource availabilities of phytoplankton in the environment. An omnivore feeding has been well studied in E. pacifica. In addition to major prey items such as diatoms and ciliates, E. pacifica has been verified to feed on dinoflagellates (Gyrodinium sp. and Protoperidinium sp.) and small copepods (Pseudocalanus sp.) using microscopic observation, stable isotope analysis, and rearing experiments (Dilling et al., 1998; Nakagawa et al., 2001, 2004; Park et al., 2011; Du and Peterson, 2014; Okazaki et al., 2020). In addition, a similar trophic level of 2.5 for E. pacifica in the Oyashio Current was reported by Sogawa et al. (2017) using stable isotope analysis. However, feeding of subtropical euphausiid species have been poorly reported, and only a few studies found foraminifera and unclassified copepods in E. brevis (Mauchline, 1967). Thus, our study was the first to report detailed feeding habits of the dominant species of E. brevis and E. hemigibba in the North Pacific subtropical gyre.
Dinoflagellates are widely recognized as important prey items of pelagic taxa (McClatchie, 1988; Turner et al., 2001; Pethybridge et al., 2014), and as an essential energy supply of all developmental stages of E. pacifica using dinoflagellate-specific fatty acid biomarkers 18:4n-3 and 22:6n-3 (Kohlbach et al., 2019; Fisher et al., 2020). Although some species of Kareniaceae (e.g., Karlodinium sp.), which were detected as dominant OTUs in the present study, could deter grazing from predators by producing karlotoxins, zooplankton such as Acartia tonsa would potentially graze on dinoflagellate strains with lower toxin levels (Waggett et al., 2008). Furthermore, pelagophytes and green algae have not yet been reported in euphausiid diets possibly due to their small size, however, our results showed their importance as food sources of dominant euphausiids. Copepods were also detected as a major prey, even though proportions of copepods were much lower in gut contents of euphausiids than environmental communities in metabarcoding analysis. Cleary et al. (2018) similarly discovered copepod sequence reads in the stomach contents of E. superba, indicating a carnivory supplementary diet of this species. The importance of copepods was verified by Nakagawa et al. (2002) based on carbon content analysis, and E. superba was supposed to feed on Ctenocalanus copepods (Clausocalanidae) according to their overlapping vertical distribution (Marrari et al., 2011).
In the present study, all dominant species belonged to the genus Euphausia. According to comparative morphological studies, the thoracic legs of Euphausia species have a nearly uniform length and form a well-developed feeding basket for omnivorous filter-feeding (Mauchline, 1967), in comparison with other euphausiid species with one or more pairs of elongated thoracic legs (Berkes, 1975; Dalley and McClatchie, 1989; Suh and Choi, 1998). For example, N. difficilis and Nematobrachion boopis displayed higher δ15N values indicating more carnivorous feeding than Euphausia species in the same water area (Kinsey and Hopkins, 1994; Sogawa et al., 2017). Moreover, although the feeding basket structure of Thysanopoda spp. is similar to that of Euphausia spp., mesopelagic Thysanopoda species are more carnivorous as they keep away from the primary producers (Nemoto et al., 1977). Thus, the diet spectrum of the minor euphausiid species was bottlenecked by the inconvenient limbs for filter feeding and their mesopelagic habitat; Euphausia species, however, have their nutritional needs satisfied by an omnivorous dietary ranging from picophytoplankton to microzooplankton. Omnivorous feeding habit enables Euphausia species to adapt to various prey availabilities and become the dominant species of euphausiid communities.
Differences of Feeding Patterns Between Three Euphausiid Species
Although all the three species are omnivorous, and similar gut content compositions were captured for E. pacifica and subtropical euphausiid species, E. pacifica in CC displayed a different feeding habit from E. brevis and E. hemigibba reflecting by lower trophic levels verified by stable isotope analysis. In addition, DNA metabarcoding detected more autotrophic preys (green algae) with greater abundance and occurrence frequency in E. pacifica by comparison with subtropical E. brevis. All the three dominant species conduct diel vertical migration: surface migrant E. pacifica in the northwestern Pacific were most abundant at 5–25 m for feedings during nighttime (Nakagawa et al., 2003; Endo and Yamano, 2006; Sogawa et al., 2016), and E. brevis were most abundant at upper 50 m in the Mediterranean Sea and Bermuda (Wiebe and D’abramo, 1972; Youngbluth, 1975). Despite a similar vertical distribution at night between E. pacifica and E. brevis, higher prey availability of pelagophyte (Stramenopiles) was observed in CC (0 m: 4.8%; 10 m: 4.7%) compared to that in ESG (0 m: 1.5%; 10 m: 1.6%), in addition to more green algae (Archaeplastida) prey (CC: ∼2.5%; ESG: ∼0.2%) (Figures 3B,C). Thus, E. pacifica was allowed access to more autotrophic prey. Moreover, E. pacifica would display a high growth rate when feeding on phytoplankton, such as diatoms, rather than copepods (Lasker, 1966; Ohman, 1984). Autotrophic pelagophyte and green algae are supposed to be the optimal prey of E. pacifica in the CC area, and E. pacifica would depend more on these prey that probably provides them with the optimum ratio of energy gain per energy expenditure (Schoener, 1971).
A lower chl a concentration in oligotrophic environments would lead to an increasing heterotroph (e.g., dinoflagellates, zooplankton) contribution to the whole diet of euphausiids, such as E. lucens, E. spinifera, E. superba, and E. pacifica. (Stuart and Pillar, 1990; Gibbons et al., 1991; Perissinotto et al., 2000; Nakagawa et al., 2002; Henschke et al., 2015). Non-autotrophic prey (copepods and mixotrophic/heterotrophic dinoflagellates) were important foods of E. hemigibba and E. brevis due to a potential poor autotrophic prey availability in the environment. In addition, the lower chl a level in ESG than that in CC (Figure 1B) triggered the lesser uptake of green algae of E. brevis in the present study. Furthermore, the gut content composition of two subtropical euphausiid species was significantly different (Supplementary Table 1): metabarcoding verified a relatively higher contribution of Dinophyta and Stramenopiles in E. hemigibba than E. brevis, while E. brevis consumed significantly more Rhizaria (Supplementary Table 2 and Figure 3). These differences might be attributed to a higher chl a concentration at the subsurface layer in WSG than ESG. Because E. hemigibba were found more frequently in deeper layers (100–150 m) in northeastern Japan and the Mediterranean Sea (Wiebe and D’abramo, 1972; Taki, 2008), they might utilize phytoplankton at the subsurface layer effectively in WSG. On the other hand, trophic level by stable isotope analysis was higher in E. hemigibba than E. brevis, with high variations and no significant difference. This discrepancy in metabarcoding and stable isotope analysis implies that feeding patterns of E. hemigibba and E. brevis change temporally according to food availability in the environments, which also underlines the need of the combining of DNA metabarcoding and stable isotope analysis in feeding habit investigations.
High Proportion and Occurrence of Hydrozoan Sequences
The high proportion and occurrence of hydrozoan sequences detected in the gut contents of the euphausiids were unexpected since hydrozoa (jellyfishes) have been known as predators of copepods, euphausiids, and fish larvae (D’Ambra et al., 2014; Fleming et al., 2015). Likewise, hydrozoan sequences have been recently detected in the gut contents of myctophid fishes of the Southern Ocean, eel larvae of the Northern Atlantic Ocean, and calanoid copepods especially in a low-phytoplankton environment (Yi et al., 2017; Ayala et al., 2018; Hirai et al., 2018; Chow et al., 2019; Yeh et al., 2020), suggesting a previously overlooked importance of gelatinous plankton in pelagic ecosystems. However, hydrozoan as natural prey of euphausiids is contradicted to the intermediate trophic positions of euphausiids uncovered by stable isotope analysis and the minute proportion of Hydrozoa sequences detected by 18S metabarcoding in the environment in our research. One possibility is that euphausiids feed on the early life stages of Hydrozoa or marine snow aggregates combining hydrozoa particles (Dilling et al., 1998; Turner, 2015; Yi et al., 2017; Ayala et al., 2018). Thus, marine snow, as another food source of euphausiids, is suggested to add to the sample collection and be analyzed in the same way as environmental samples in future studies. On the other hand, euphausiids are likely to take hydrozoa atypically in the cod-end during sampling. The feeding pattern difference between two subtropical euphausiid species were unevenly contributed by Hydrozoa among euphausiid individuals indicated by SIMPER, which actually supported this theory. Nicol (1984) noted that Meganyctiphanes euphausiids collected by towed nets within 10 min at sea surface would contain more copepods in their “food baskets” when compared with those collected by hand-held dip nets because potential food items become highly concentrated in towed nets; Judkins and Fleminger (1972) also found greater food diversity in sergestids caught by nets in comparison of those obtained from fish stomachs. Given that the sampling duration was relatively long in the present study, although the mouthpart and stomach of euphausiids were excluded during the dissection process, cod-end feeding of euphausiids might have happened and impacted the gut content analysis. Gut content analysis of euphausiids collected by shorter tows from the surface swarms and additional stable carbon and nitrogen isotope analyses dealing with hydrozoans inhabit in each study site are assured to improve the accuracy of euphausiid diet and trophic linkage study.
DNA Metabarcoding as a Tool of Feeding Ecology Investigation
DNA metabarcoding has been successfully applied to gut contents or feces of a series of organisms ranging from copepods and fish larvae to marine mammals to unveil their dietary diversity, dietary composition, and the contribution of different prey items to their overall diet (Hardy et al., 2017; Zamora-Terol et al., 2020; Kume et al., 2021). The most accepted advantage of DNA metabarcoding over traditional visual analysis is the lesser dependency on the digestion state of prey items. A stomach content analysis of catfish by DNA metabarcoding recovered 91.6% of prey items to species level compared to morphological identification (9.4% to species level, 12.1% to family level), and little difference was shown between the major composition of lightly digested piscine prey items and moderately digested ones (Aguilar et al., 2017). The firstly reported pelagophyte and green algae with evident larger contribution and higher occurrence frequencies in euphausiid diet of our study, once again highlighted the high efficiency and sensitivity of DNA metabarcoding. However, even though potential prey could be easily explored, the natural dietary composition might be biased due to the high sensitivity. Similar to cod-end feeding impacts, cross-contamination derived from body surface during dissection might be left in the gut content sequences (De Barba et al., 2014; Chow et al., 2019); fungi, symbiotic, and parasitic organisms originating from food or digestive tract are difficult to be verified as a true prey due to insufficient parasite knowledge and the limited registered sequences in 18S databases (Hirai et al., 2017; Kodama et al., 2017; Cleary et al., 2018). Furthermore, although V9 hypervariable region of 18S rDNA could reveal the extant diversity of eukaryotes, unclassified OTUs and prey OTUs with lower identify (<85%) during classification ought to be cross-checked by multiple genetic markers and supplementary databases (Da Silva et al., 2019; Choi and Park, 2020). Additionally, trace signals of secondary-prey are likely to remain because of the usage of universal primers, however, these items were generally in low occurrence, thus could be excluded (Hardy et al., 2017; Clarke et al., 2020).
A final challenge of not only feeding ecology study but all the research regarding DNA metabarcoding is how to interpret the sequence reads of different taxonomic groups in relation to their practical biomass (Pompanon et al., 2012; Bucklin et al., 2016). Despite sample size, PCR biases, amplicon efficiency, and different copy number of selected barcodes in different prey items are known to cause different degrees of recovery biases (Deagle et al., 2013). An additional weighing on samples based on gut content mass or fullness, importing blocking primers to inhibit host-sequence amplification, and excluding empty guts like the present study are supposed to contribute to the prey item quantification. By far, the stable isotope and fatty acid analysis are more committed methods for food source quantification, whereas DNA metabarcoding is more suitable to pilot diet investigation of organisms without prior knowledge (Stewart et al., 2017; Kohlbach et al., 2019). Combining the approach of DNA metabarcoding-based gut content analysis and stable isotope analysis is supposed to be the most appropriate way to mirror a convinced feeding ecology of studied consumers. Given all the inadequacies, in the future, feeding ecology study of euphausiids via DNA metabarcoding, we strongly recommend: (1) using disposable dissection tools on different individual specimens to avoid contaminations; (2) employing empty guts and other organs of euphausiids as negative controls to minimize the noises of fungi, parasites, and unclassified sequences; (3) importing additional marine snow samples to provide a more complete food resource availability; (4) using multiple metabarcoding genetic markers and additional reference databases to advance the integration of dietary data; (5) considering the frequency of occurrence as a criterion and relative read abundance for true-prey assessment instead of judging based on a presence/absence data alone.
Data Availability Statement
Raw Illumina Miseq data are available from the NCBI under BioProject accession PRJNA695066.
Author Contributions
FZ, JH, and AT contributed to the research design. JH carried out field sampling. KH contributed to the sampling of environmental samples. SH contributed to the collection and analysis of stable isotope data. FZ contributed to laboratory work, data analyses, and manuscript writing. All authors contributed to the discussion of results and improved the manuscript.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (grant number 18K14519) and the Ministry of Education, Culture, Sports, Science and Technology (grant number 1036979).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the captain and all members of the KH-17-4 cruise on board the RV Hakuho-Maru for their assistance with sample collection. We are also grateful to Sayaka Sogawa for her guidance on euphausiid morphological identification and Takuya Sato for his cooperation in the stable isotope analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.756067/full#supplementary-material
References
Aguilar, R., Ogburn, M. B., Driskell, A. C., Weigt, L. A., Groves, M. C., and Hines, A. H. (2017). Gutsy genetics: identification of digested piscine prey items in the stomach contents of sympatric native and introduced warmwater catfishes via DNA barcoding. Environ. Biol. Fish. 100, 325–336. doi: 10.1007/s10641-016-0523-8
Albaina, A., Aguirre, M., Abad, D., Santos, M., and Estonba, A. (2016). 18S rRNA V9 metabarcoding for diet characterization: a critical evaluation with two sympatric zooplanktivorous fish species. Ecol. Evol. 6, 1809–1824. doi: 10.1002/ece3.1986
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. doi: 10.1371/journal.pone.0006372
Anderson, M. J., Gorley, R. N., and Clarke, K. R. (2008). Permanova+ for Primer: Guide to Software and Statistical Methods. Plymouth: University of Auckland and PRIMER-E Ltd.
Ayala, D. J., Munk, P., Lundgreen, R. B., Traving, S. J., Jaspers, C., Jørgensen, T. S., et al. (2018). Gelatinous plankton is important in the diet of European eel (Anguilla anguilla) larvae in the Sargasso Sea. Sci. Rep. 8:6156. doi: 10.1038/s41598-018-24388-x
Baker, A. D. C., Boden, B. P., and Brinton, E. (1990). A Practical Guide to the Euphausiids of the World. London: British Museum (Natural History).
Berkes, F. (1975). Some aspects of feeding mechanisms of euphausiid crustaceans. Crustaceana 29, 266–270. doi: 10.1163/156854075X00315
Berry, O., Bulman, C., Bunce, M., Coghlan, M., Murray, D. C., and Ward, R. D. (2015). Comparison of morphological and DNA metabarcoding analyses of diets in exploited marine fishes. Mar. Ecol. Prog. Ser. 540, 167–181. doi: 10.3354/meps11524
Boden, B. P., Johnson, M. W., and Brinton, E. (1955). Euphausiacea (Crustacea) of the North Pacific. San Diego, CA: Library–Scripps Digital Collection.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brinton, E. (1962). The Distribution of Pacific Euphausiids. San Diego, CA: Library–Scripps Digital Collection.
Brinton, E. (1967). Vertical migration and avoidance capability of euphausiids in the California current. Limnol. Oceanogr. 12, 451–483. doi: 10.4319/lo.1967.12.3.0451
Brinton, E. (1979). Parameters relating to the distributions of planktonic organisms, especially euphausiids in the eastern tropical Pacific. Prog. Oceanogr. 8, 125–189. doi: 10.1016/0079-6611(79)90001-6
Bucklin, A., Lindeque, P. K., Rodriguez-Ezpeleta, N., Albaina, A., and Lehtiniemi, M. (2016). Metabarcoding of marine zooplankton: prospects, progress and pitfalls. J. Plankton Res. 38, 393–400. doi: 10.1093/plankt/fbw023
Choi, J., and Park, J. S. (2020). Comparative analyses of the V4 and V9 regions of 18S rDNA for the extant eukaryotic community using the Illumina platform. Sci. Rep. 10:6519. doi: 10.1038/s41598-020-63561-z
Chow, S., Inaba, N., Nagai, S., Kurogi, H., Nakamura, Y., Yanagimoto, T., et al. (2019). Molecular diet analysis of Anguilliformes leptocephalus larvae collected in the western North Pacific. PLoS One 14:e0225610. doi: 10.1371/journal.pone.0225610
Clarke, L. J., Trebilco, R., Walters, A., Polanowski, A. M., and Deagle, B. E. (2020). DNA-based diet analysis of mesopelagic fish from the southern Kerguelen Axis. Deep Sea Res. 174:104494. doi: 10.1016/j.dsr2.2018.09.001
Cleary, A. C., and Durbin, E. G. (2016). Unexpected prevalence of parasite 18S rDNA sequences in winter among Antarctic marine protists. J. Plankton Res. 38, 401–417. doi: 10.1093/plankt/fbw005
Cleary, A. C., Durbin, E. G., and Casas, M. C. (2018). Feeding by Antarctic krill Euphausia superba in the West Antarctic Peninsula: differences between fjords and open waters. Mar. Ecol. Prog. Ser. 595, 39–54. doi: 10.3354/meps12568
D’Ambra, I., Carmichael, R. H., and Graham, W. M. (2014). Determination of δ 13 C and δ 15 N and trophic fractionation in jellyfish: implications for food web ecology. Mar. Biol. 161, 473–480. doi: 10.1007/s00227-013-2345-y
Da Silva, L. P., Mata, V. A., Lopes, P. B., Pereira, P., Jarman, S. N., Lopes, R. J., et al. (2019). Advancing the integration of multi-marker metabarcoding data in dietary analysis of trophic generalists. Mol. Ecol. Resour. 19, 1420–1432. doi: 10.1111/1755-0998.13060
Dalley, D. D., and McClatchie, S. (1989). Functional feeding morphology of the euphausiid Nyctiphanes australis. Mar. Biol. 101, 195–203. doi: 10.1007/BF00391458
De Barba, M., Miquel, C., Boyer, F., Mercier, C., Rioux, D., Coissac, E., et al. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323. doi: 10.1111/1755-0998.12188
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 34:1261605. doi: 10.1126/science.1261605
Deagle, B. E., Thomas, A. C., McInnes, J. C., Clarke, L. J., Vesterinen, E. J., Clare, E. L., et al. (2019). Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mol. Ecol. 28, 391–406. doi: 10.1111/mec.14734
Deagle, B. E., Thomas, A. C., Shaffer, A. K., Trites, A. W., and Jarman, S. N. (2013). Quantifying sequence proportions in a DNA-based diet study using Ion Torrent amplicon sequencing: which counts count? Mol. Ecol. Resour. 13, 620–633. doi: 10.1111/1755-0998.12103
DeNiro, M. J., and Epstein, S. (1981). Influence of diet on the distribution of nitrogen isotopes in animals. Geochim. Cosmochim. Acta 45, 341–351. doi: 10.1016/0016-7037(81)90244-1
Dilling, L., Wilson, J., Steinberg, D., and Alldredge, A. (1998). Feeding by the euphausiid Euphausia pacifica and the copepod Calanus pacificus on marine snow. Mar. Ecol. Prog. Ser. 170, 189–201. doi: 10.3354/meps170189
Du, X., and Peterson, W. (2014). Feeding rates and selectivity of adult Euphausia pacifica on natural particle assemblages in the coastal upwelling zone off Oregon, USA, 2010. J. Plankton Res. 36, 1031–1046. doi: 10.1093/plankt/fbu027
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Endo, Y., and Yamano, F. (2006). Diel vertical migration of Euphausia pacifica (Crustacea, Euphausiacea) in relation to molt and reproductive processes, and feeding activity. J. Oceanogr. 62, 693–703. doi: 10.1007/s10872-006-0087-x
Fager, E. W., and McGowan, J. A. (1963). Zooplankton species groups in the North Pacific: co-occurrences of species can be used to derive groups whose members react similarly to water-mass types. Science 140, 453–460. doi: 10.1126/science.140.3566.453
Fisher, J. L., Menkel, J., Copeman, L., Shaw, C. T., Feinberg, L. R., and Peterson, W. T. (2020). Comparison of condition metrics and lipid content between Euphausia pacifica and Thysanoessa spinifera in the northern California Current, USA. Prog. Oceanogr. 188:102417. doi: 10.1016/j.pocean.2020.102417
Fleming, N. E., Harrod, C., Newton, J., and Houghton, J. D. (2015). Not all jellyfish are equal: isotopic evidence for inter-and intraspecific variation in jellyfish trophic ecology. PeerJ 3:e1110. doi: 10.7717/peerj.1110
Folch, J., Lees, M., and Stanley, G. S. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509.
Fry, B. (1988). Food web structure on Georges Bank from stable C, N, and S isotopic compositions. Limnol. Oceanogr. 33, 1182–1190. doi: 10.4319/lo.1988.33.5.1182
Gibbons, M. J., Barange, M., and Pillar, S. C. (1991). Vertical migration and feeding of Euphausia lucens (Euphausiacea) in the Southern Benguela. J. Plankton Res. 13, 473–486. doi: 10.1093/plankt/13.3.473
Gurney, L. J., Froneman, P. W., Pakhomov, E. A., and McQuaid, C. D. (2001). Trophic positions of three euphausiid species from the Prince Edward Islands (Southern Ocean): implications for the pelagic food web structure. Mar. Ecol. Prog. Ser. 217, 167–174. doi: 10.3354/meps217167
Hardy, N., Berry, T., Kelaher, B. P., Goldsworthy, S. D., Bunce, M., Coleman, M. A., et al. (2017). Assessing the trophic ecology of top predators across a recolonisation frontier using DNA metabarcoding of diets. Mar. Ecol. Prog. Ser. 573, 237–254. doi: 10.3354/meps12165
Harms-Tuohy, C. A., Schizas, N. V., and Appeldoorn, R. S. (2016). Use of DNA metabarcoding for stomach content analysis in the invasive lionfish Pterois volitans in Puerto Rico. Mar. Ecol. Prog. Ser. 558, 181–191. doi: 10.3354/meps11738
Henschke, N., Everett, J. D., Suthers, I. M., Smith, J. A., Hunt, B. P., Doblin, M. A., et al. (2015). Zooplankton trophic niches respond to different water types of the western Tasman Sea: a stable isotope analysis. Deep Sea Res. Part I Oceanogr. Res. Pap. 104, 1–8. doi: 10.1016/j.dsr.2015.06.010
Hipfner, J. M. (2009). Euphausiids in the diet of a North Pacific seabird: annual and seasonal variation and the role of ocean climate. Mar. Ecol. Prog. Ser. 390, 277–289. doi: 10.3354/meps08209
Hirai, J., Hamamoto, Y., Honda, D., and Hidaka, K. (2018). Possible aplanochytrid (Labyrinthulea) prey detected using 18S metagenetic diet analysis in the key copepod species Calanus sinicus in the coastal waters of the subtropical western North Pacific. Plankton Benthos Res. 13, 75–82. doi: 10.3800/pbr.13.75
Hirai, J., Hidaka, K., Nagai, S., and Ichikawa, T. (2017). Molecular-based diet analysis of the early post-larvae of Japanese sardine Sardinops melanostictus and Pacific round herring Etrumeus teres. Mar. Ecol. Prog. Ser. 564, 99–113. doi: 10.3354/meps12008
Judkins, D. C., and Fleminger, A. (1972). Comparison of foregut contents of Sergestes similis obtained from net collections and albacore stomachs. Fish. Bull. U.S. 70, 217–223.
Kim, H. S., Yamaguchi, A., and Ikeda, T. (2010). Metabolism and elemental composition of the euphausiids Euphausia pacifica and Thysanoessa inspinata during the phytoplankton bloom season in the Oyashio region, western subarctic Pacific Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 57, 1733–1741. doi: 10.1016/j.dsr2.2010.03.017
Kim, H., Ju, S. J., Kang, J. H., and Shin, K. H. (2019). Diet source of Euphausia pacifica revealed using carbon-and nitrogen-stable isotopes in the Yellow Sea Cold Water Mass in summer. J. Oceanogr. 75, 51–59. doi: 10.1007/s10872-018-0483-z
Kinsey, S. T., and Hopkins, T. L. (1994). Trophic strategies of euphausiids in a low-latitude ecosystem. Mar. Biol. 118, 651–661. doi: 10.1007/BF00347513
Kodama, T., Hirai, J., Tamura, S., Takahashi, T., Tanaka, Y., Ishihara, T., et al. (2017). Diet composition and feeding habits of larval Pacific bluefin tuna Thunnus orientalis in the Sea of Japan: integrated morphological and metagenetic analysis. Mar. Ecol. Prog. Ser. 583, 211–226. doi: 10.3354/meps12341
Kohlbach, D., Lange, B. A., Graeve, M., Vortkamp, M., and Flores, H. (2019). Varying dependency of Antarctic euphausiids on ice algae-and phytoplankton-derived carbon sources during summer. Mar. Biol. 166, 1–17. doi: 10.1007/s00227-019-3527-z
Kume, G., Kobari, T., Hirai, J., Kuroda, H., Takeda, T., Ichinomiya, M., et al. (2021). Diet niche segregation of co-occurring larval stages of mesopelagic and commercially important fishes in the Osumi Strait assessed through morphological, DNA metabarcoding, and stable isotope analyses. Mar. Biol. 168, 1–14. doi: 10.1007/s00227-020-03810-x
Lara-Lopez, A. L., Davison, P., and Koslow, J. A. (2012). Abundance and community composition of micronekton across a front off Southern California. J. Plankton Res. 34, 828–848. doi: 10.1093/plankt/fbs016
Lasker, R. (1966). Feeding, growth, respiration, and carbon utilization of a euphausiid crustacean. J. Fish. Res. Board Can. 23, 1291–1317. doi: 10.1139/f66-121
Letessier, T. B., Cox, M. J., and Brierley, A. S. (2009). Drivers of euphausiid species abundance and numerical abundance in the Atlantic Ocean. Mar. Biol. 156, 2539–2553. doi: 10.1007/s00227-009-1278-y
Letessier, T. B., Cox, M. J., and Brierley, A. S. (2011). Drivers of variability in euphausiid species abundance throughout the Pacific Ocean. J. Plankton Res. 33, 1342–1357. doi: 10.1093/plankt/fbr033
Lindley, J. A. (1977). Continuous plankton records: the distribution of the Euphausiacea (Crustacea: Malacostraca) in the North Atlantic and the North Sea, 1966–1967. J. Biogeogr. 4, 121–133. doi: 10.2307/3038157
Marrari, M., Daly, K. L., Timonin, A., and Semenova, T. (2011). The zooplankton of Marguerite Bay, western Antarctic Peninsula—part II: vertical distributions and habitat partitioning. Deep Sea Res. Part II Top. Stud. Oceanogr. 58, 1614–1629. doi: 10.1016/j.dsr2.2010.12.006
Mauchline, J. (1967). Feeding appendages of the Euphausiacea (Crustacea). J. Zool. 153, 1–43. doi: 10.1111/j.1469-7998.1967.tb05029.x
McClatchie, S. (1988). Functional response of the euphausiid Thysanoessa raschii grazing on small diatoms and toxic dinoflagellates. J. Mar. Res. 46, 631–646. doi: 10.1357/002224088785113496
Minagawa, M., and Wada, E. (1984). Stepwise enrichment of 15N along food chains: further evidence and the relation between δ15N and animal age. Geochim. Cosmochim. Acta 48, 1135–1140. doi: 10.1016/0016-7037(84)90204-7
Moore, H. B. (1951). Physical factors affecting the distribution of euphausiids in the North Atlantic. Bull. Mar. Sci. 1, 278–305.
Nagai, S., Yamamoto, K., Hata, N., and Itakura, S. (2012). Study of DNA extraction methods for use in loop-mediated isothermal amplification detection of single resting cysts in the toxic dinoflagellates Alexandrium tamarense and A. catenella. Mar. Genomics 7, 51–56. doi: 10.1016/j.margen.2012.03.002
Nakagawa, Y., Endo, Y., and Sugisaki, H. (2003). Feeding rhythm and vertical migration of the euphausiid Euphausia pacifica in coastal waters of north-eastern Japan during fall. J. Plankton Res. 25, 633–644. doi: 10.1093/plankt/25.6.633
Nakagawa, Y., Endo, Y., and Taki, K. (2001). Diet of Euphausia pacifica Hansen in Sanriku waters off northeastern Japan. Plankton Biol. Ecol. 48, 68–77.
Nakagawa, Y., Endo, Y., and Taki, K. (2002). Contributions of heterotrophic and autotrophic prey to the diet of euphausiid, Euphausia pacifica in the coastal waters off northeastern Japan. Polar Biosci. 15, 52–65. doi: 10.15094/00006183
Nakagawa, Y., Ota, T., Endo, Y., Taki, K., and Sugisaki, H. (2004). Importance of ciliates as prey of the euphausiid Euphausia pacifica in the NW North Pacific. Mar. Ecol. Prog. Ser. 271, 261–266. doi: 10.3354/meps271261
Nakamura, Y., Tuji, A., Makino, W., Nagata, N., Nakagawa, M., and Takamura, N. (2020). Feeding ecology of a mysid species, Neomysis awatschensis in the Lake Kasumigaura: combining approach with microscopy, stable isotope analysis and DNA metabarcoding. Plankton Benthos Res. 15, 44–54. doi: 10.3800/pbr.15.44
Nemoto, T., Kamada, K., and Brinton, E. (1977). Reproduction and growth of deep sea Thysanopoda euphausiids. Bull. Plankton Soc. Japan 24, 36–43.
Nicol, S. (1984). Cod end feeding by the euphausiid Meganyctiphanes norvegica. Mar. Biol. 80, 29–33. doi: 10.1007/BF00393123
Nicol, S. (2003). Living krill, zooplankton, and experimental investigations: a discourse on the role of krill and their experimental study in marine ecology. Mar. Fresh. Behav. Physiol. 36, 191–205. doi: 10.1080/10236240310001614420
Ohman, M. D. (1984). Omnivory by Euphausia pacifica: the role of copepod prey. Mar. Ecol. Prog. Ser. 19, 125–131. doi: 10.3354/meps019125
Okazaki, Y., Takahashi, K., Nishibe, Y., and Ichinomiya, M. (2020). Post-spring-bloom feeding rates of krill Euphausia pacifica (Euphausiacea) in the Oyashio region, western North Pacific. Prog. Oceanogr. 181:102245. doi: 10.1016/j.pocean.2019.102245
Oozeki, Y., Takasuka, A., Kubota, H., and Barange, M. (2007). Characterizing spawning habitats of Japanese sardine, Sardinops melanostictus, Japanese anchovy, Engraulis japonicus, and Pacific round herring, Etrumeus teres, in the Northwestern Pacific. CCOFI Rep. 48, 191–203.
Park, J. I., Kang, C. K., and Suh, H. L. (2011). Ontogenetic diet shift in the euphausiid Euphausia pacifica quantified using stable isotope analysis. Mar. Ecol. Prog. Ser. 429, 103–109. doi: 10.3354/meps09091
Perissinotto, R., Gurney, L., and Pakhomov, E. A. (2000). Contribution of heterotrophic material to diet and energy budget of Antarctic krill, Euphausia superba. Mar. Biol. 136, 129–135. doi: 10.1007/s002270050015
Pethybridge, H., Bodin, N., Arsenault-Pernet, E. J., Bourdeix, J. H., Brisset, B., Bigot, J. L., et al. (2014). Temporal and inter-specific variations in forage fish feeding conditions in the NW Mediterranean: lipid content and fatty acid compositional changes. Mar. Ecol. Prog. Ser. 512, 39–54. doi: 10.3354/meps10864
Pompanon, F., Deagle, B. E., Symondson, W. O., Brown, D. S., Jarman, S. N., and Taberlet, P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Roden, G. I. (1991). Subarctic-subtropical transition zone of the North Pacific: large-scale aspects and mesoscale structure. NOAA Tech. Rep. NMFS 105, 1–38.
Sarda, F., and Valladares, F. J. (1990). Gastric evacuation of different foods by Nephrops norvegicus (Crustacea: Decapoda) and estimation of soft tissue ingested, maximum food intake and cannibalism in captivity. Mar. Biol. 104, 25–30. doi: 10.1007/BF01313153
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schoener, T. W. (1971). Theory of feeding strategies. Annu. Rev. Ecol. Syst. 2, 369–404. doi: 10.1146/annurev.es.02.110171.002101
Shannon, C. E. (1948). A mathematical theory of communication. Bell. Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Sogawa, S., Sugisaki, H., Saito, H., Okazaki, Y., Ono, T., Shimode, S., et al. (2016). Seasonal and regional change in vertical distribution and diel vertical migration of four euphausiid species (Euphausia pacifica, Thysanoessa inspinata, T. longipes, and Tessarabrachion oculatum) in the northwestern Pacific. Deep Sea Res. Part I Oceanogr. Res. Pap. 109, 1–9. doi: 10.1016/j.dsr.2015.12.010
Sogawa, S., Sugisaki, H., Tadokoro, K., Ono, T., Sato, E., Shimode, S., et al. (2017). Feeding habits of six species of euphausiids (Decapoda: Euphausiacea) in the northwestern Pacific Ocean determined by carbon and nitrogen stable isotope ratios. J. Crustac. Biol. 37, 29–36. doi: 10.1093/jcbiol/ruw014
Stewart, J. D., Rohner, C. A., Araujo, G., Avila, J., Fernando, D., Forsberg, K., et al. (2017). Trophic overlap in mobulid rays: insights from stable isotope analysis. Mar. Ecol. Prog. Ser. 580, 131–151. doi: 10.3354/meps12304
Stuart, V., and Pillar, S. C. (1990). Diel grazing patterns of all ontogenetic stages of Euphausia lucens and in situ predation rates on copepods in the southern Benguela upwelling region. Mar. Ecol. Prog. Ser. 64, 227–241. doi: 10.3354/meps064227
Suh, H. L., and Choi, S. D. (1998). Comparative morphology of the feeding basket of five species of Euphausia (Crustacea, Euphausiacea) in the western North Pacific, with some ecological considerations. Hydrobiologia 385, 107–112.
Sverdrup, H. U., Johnson, M. W., and Fleming, R. H. (1942). The Oceans: Their Physics, Chemistry, and General Biology. New York: Prentice-Hall.
Taki, K. (2007). Seasonal changes in distribution and abundance of euphausiids in the coastal area of north-eastern Japan. Fish. Sci. 73, 522–533. doi: 10.1111/j.1444-2906.2007.01364.x
Taki, K. (2008). Vertical distribution and diel migration of euphausiids from Oyashio Current to Kuroshio area off northeastern Japan. J. Plankton Res. 3, 27–35. doi: 10.3800/pbr.3.27
Tao, Z., Li, C., and Sun, S. (2015). Grazing and metabolism of Euphausia pacifica in the Yellow Sea. PLoS One 10:e0115825. doi: 10.1371/journal.pone.0115825
Tittensor, D. P., Mora, C., Jetz, W., Lotze, H. K., Ricard, D., Berghe, E. V., et al. (2010). Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–1101. doi: 10.1038/nature09329
Turner, J. T. (2015). Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog. Oceanogr. 130, 205–248. doi: 10.1016/j.pocean.2014.08.005
Turner, J. T., Ianora, A., Miralto, A., Laabir, M., and Esposito, F. (2001). Decoupling of copepod grazing rates, fecundity and egg-hatching success on mixed and alternating diatom and dinoflagellate diets. Mar. Ecol. Prog. Ser. 220, 187–199. doi: 10.3354/meps220187
Tverin, M., Esparza-Salas, R., Strömberg, A., Tang, P., Kokkonen, I., Herrero, A., et al. (2019). Complementary methods assessing short and long-term prey of a marine top predator-application to the grey seal-fishery conflict in the Baltic Sea. PLoS One 14:e0208694. doi: 10.1371/journal.pone.0208694
Waggett, R. J., Tester, P. A., and Place, A. R. (2008). Anti-grazing properties of the toxic dinoflagellate Karlodinium veneficum during predator–prey interactions with the copepod Acartia tonsa. Mar. Ecol. Prog. Ser. 366, 31–42. doi: 10.3354/meps07518
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Werner, T., and Buchholz, F. (2013). Diel vertical migration behavior in euphausiids of the northern Benguela current: seasonal adaptations to food availability and strong gradients of temperature and oxygen. J. Plankton Res. 35, 792–812. doi: 10.1093/plankt/fbt030
Whitaker, M. R., Baker, C. C., Salzman, S. M., Martins, D. J., and Pierce, N. E. (2019). Combining stable isotope analysis with DNA metabarcoding improves inferences of trophic ecology. PLoS One 14:e0219070. doi: 10.1371/journal.pone.0219070
Wiebe, P. H., and D’abramo, L. (1972). Distribution of euphausiid assemblages in the Mediterranean Sea. Mar. Biol. 15, 139–149. doi: 10.1007/BF00353642
Yeh, H. D., Questel, J. M., Maas, K. R., and Bucklin, A. (2020). Metabarcoding analysis of regional variation in gut contents of the copepod Calanus finmarchicus in the North Atlantic Ocean. Deep Sea Res. II Top. Stud. Oceanogr. 180:104738. doi: 10.1016/j.dsr2.2020.104738
Yi, X., Huang, Y., Zhuang, Y., Chen, H., Yang, F., Wang, W., et al. (2017). In situ diet of the copepod Calanus sinicus in coastal waters of the South Yellow Sea and the Bohai Sea. Acta Oceanol. Sin. 36, 68–79. doi: 10.1007/s13131-017-0974-6
Youngbluth, M. J. (1975). The vertical distribution and diel migration of euphausiids in the central waters of the eastern South Pacific. Deep Sea Res. 22, 519–536. doi: 10.1016/0011-7471(75)90033-9
Zamora-Terol, S., Novotny, A., and Winder, M. (2020). Reconstructing marine plankton food web interactions using DNA metabarcoding. Mol. Ecol. 29, 3380–3395. doi: 10.1111/mec.15555
Keywords: euphausiids, feeding ecology, 18S V9 metabarcoding, stable isotope analysis, community structure, North Pacific
Citation: Zhou F, Hirai J, Hamasaki K, Horii S and Tsuda A (2021) Feeding Ecology of Three Euphausiid Species in the North Pacific Ocean Inferred From 18S V9 Metabarcoding and Stable Isotope Analysis. Front. Mar. Sci. 8:756067. doi: 10.3389/fmars.2021.756067
Received: 10 August 2021; Accepted: 29 September 2021;
Published: 20 October 2021.
Edited by:
Ciro Rico, Institute of Marine Sciences of Andalusia (ICMAN), SpainReviewed by:
Israel Arreola, Universidad de Guadalajara, MexicoOctavio R. Salazar, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2021 Zhou, Hirai, Hamasaki, Horii and Tsuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fanyu Zhou, emhvdWZhbnl1MDMyMkBnLmVjYy51LXRva3lvLmFjLmpw
 Fanyu Zhou
Fanyu Zhou Junya Hirai
Junya Hirai Koji Hamasaki
Koji Hamasaki Sachiko Horii
Sachiko Horii Atsushi Tsuda1
Atsushi Tsuda1