
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 November 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.754588
Berberine hydrochloride is the main active ingredient of Coptis chinensis, which has demonstrated significant antibacterial activity against bacteria. However, the molecular mechanism underlying its effects is unclear in crabs. In this study, we used transcriptome sequencing to investigate the molecules involved in the therapeutic and defensive responses of Charybdis japonica infected with 105 colony-forming units (CFU)/L of Aeromonas hydrophila following treatment with berberine hydrochloride. A total of 5,409 differentially expressed genes were observed after exposure of C. japonicus for 72 h to 100 mg/L berberine hydrochloride, 100 mg/L berberine hydrochloride plus injection of 105 CFU/L A. hydrophila, and a control group injected with equal amount of physiological saline solution. Enrichment analysis revealed that these genes were involved in metabolism, cellular processes, signal transduction, and immune function. The transcriptomic results indicated that exposure to berberine hydrochloride activated glutathione metabolism, oxidative phosphorylation, fatty acid degradation, retinol metabolism, pyruvate metabolism, and the hypoxia-inducible factor 1 signaling pathway. Transcriptomic analysis and relative mRNA level analysis also identified acid hydrolase genes and phagosomal genes as key factors regulating the adaptation of acid berberine-infused C. japonicus to infection with A. hydrophila, Based on the ability of berberine hydrochloride to induce acid hydrolase and phagosomal gene expression, which can in turn remove bacteria, immersion in berberine hydrochloride may remove A. hydrophila and thus improve the survival of C. japonica. The results of this study provided a new scientific basis for the potential role of berberine hydrochloride in the immune mechanisms of crabs.
Berberine hydrochloride is an isoquinoline alkaloid found mainly in the stems and roots of the genus Berberis, and is the main pharmacological component of the Chinese herbal medicines Coptis chinensis and Phellodendron chinensis (Habtemariam, 2016). As a botanical medicine, berberine hydrochloride has been used to treat intestinal infections, especially bacterial diarrhea, for more than 2,000 years (Tang et al., 2009). Coptis coptidis is used as a prescription drug to control bacterial fish diseases (Ya-Nan et al., 2014). Berberine hydrochloride has demonstrated bacteriostatic and bactericidal effects against Vibrio cholerae, Aeromonas hydrophila, Salmonella typhi, Staphylococcus aureus, and Escherichia coli (Zhang et al., 2009). Its main pharmacological activities include antioxidant and anti-inflammatory activities (Xu et al., 2017). Some studies have also shown that berberine hydrochloride improved growth performance, enhanced skin mucolytic enzyme activity, and increased the growth rate and disease resistance of fish in aquaculture. However, the exact mechanisms of berberine hydrochloride are not yet fully understood. In addition, most studies have focused on fish, with little research on crustaceans. We therefore explored the effect of berberine hydrochloride in the crab, Charybdis japonica.
Charybdis japonica is widely distributed in the coastal areas of Southeast Asia, and its rapid growth, high nutritional value (Lou et al., 2018) have made it one of the most economically important species of crabs. C. japonica has been widely cultivated, but its high culture density has led to outbreaks of infections caused by various bacterial and viral pathogens (Johnson, 1983). One of the most harmful pathogens widely present in crabs is A. hydrophila, which is responsible for diseases that cause major economic losses in crab farming worldwide (González-Escalona et al., 2006; Igbinosa et al., 2012). We therefore tested the effects of berberine hydrochloride against this bacterium in the current study.
Transcriptome sequencing can be analyzed in relation to different environmental stresses, viral and bacterial invasion, and gene expression patterns across tissue types (Czesny et al., 2012). Advances in high-throughput RNA-Seq technology now offer the opportunity to explore the transcriptome and gene expression patterns associated with crab immunity (Xie et al., 2014; Jia et al., 2018). This sequencing technology has been used to identify many immune-related genes in various immune-related organs of crabs, including swimming crabs (Liu et al., 2011), Scyla paramamosain (Liu et al., 2017), Eriocheir sinensis (Zhang et al., 2018), and Sinopotamon henanense (Sun et al., 2016). It provides an efficient technique for further gene expression analysis, novel transcript discovery, and differentially expressed gene identification in crab transcriptome sequencing (Garber et al., 2011).
Improving the immunity and disease resistance of crustaceans is one of the basic measures for preventing and controlling diseases. Immune defense in crabs is entirely dependent on an innate, non-adaptive mechanism to resist environmental stress, and pathogen invasion (Kurata et al., 2006). The crab’s innate immune system plays an important role in early pathogen recognition and relies on a limited number of pattern recognition receptors to recognize evolutionarily conserved pathogen-associated molecular patterns (Neves et al., 2009). The gills are an essential organ for regulating osmotic pressure in crustaceans and play an important role in innate immunity. The gills are directly exposed to the external water environment containing a large number of microorganisms (Bao et al., 2019), and are thus the first organ to be exposed to pathogenic bacteria, including members of the Vibrio family. We therefore investigated the gills of C. japonica as the study organ in these experiments.
The application of big amounts of antibiotics has been depended by the treatment of bacterial diseases in aquatic animals, which has resulted in negative effects such as drug resistance, drug residues, and cross-infection of pathogenic bacteria, endangering human health and contaminating soil and water sources when released into the environment (DuPont, 2007; Martínez, 2008; Seal et al., 2013). The aquaculture department is responsible for producing edible crabs for human consumption. Therefore, chemical drugs are generally not preferred in aquaculture for safety reasons. Secondary metabolites isolated from different plants have been shown to be beneficial for both humans and animals (Lazreg Aref et al., 2011). In this study, we examined the use of berberine hydrochloride as an alternative to antibiotics and an immune-stimulant for the prevention and treatment of A. hydrophila-infection in C. japonica. This study aimed to investigate the effects of berberine hydrochloride on the metabolic pathways of C. japonica based on transcriptomic data, to validate the corresponding immune genes in the transcriptomic metabolic pathways in berberine hydrochloride-treated P. aeruginosa-infected C. japonica using the RT-polymerase chain reaction (RT-PCR), and to examine changes in the metabolic pathways and map the complete pathway system.
The C. japonica individuals used in the study weighed 82 ± 2.6 g, were 9 ± 0.7 cm in length and 6 ± 3.4 cm in width, with a total of 240 samples. C. japonica were purchased from the waters off Malaysia, 3 treatment conditions and a control group, each with 6 replicate aquariums, and each aquarium had 10 C. japonica all with the same net and PVC pipe added as a shelter for the animals to prevent cannibalism. The acclimation period was 2 weeks. During the acclimation period, all pairs of crabs were fed a commercial diet (9812; Shanghai Harmony Feed Co., Ltd., China) at 8:00 and 18:00 daily at 5% of the pair’s body weight. The crabs were exposed to seawater using an artificial sea salt cycle (salinity 28 psu) with water temperature controlled at 25 ± 1°C, pH 8.l ± 0.2, dissolved oxygen concentration of 5.0 mg/L, and a light/dark cycle of 12 h.
Berberine hydrochloride is purchased from Vickie Biotechnology Co. LTD, Purity (HPLC ≥ 98%). After 2 weeks of C. japonica domestication, water quality was maintained at the same level as during the domestication period. C. japonica were not fed for 24 h prior to the test. The pre-test found that 100 mg/L berberine hydrochloride was not harmful to the C. japonica itself. One hundred and twenty C. japonica were soaked with 100 mg/L berberine hydrochloride. 48 h later, 105 CFU/L of A. hydrophila was injected into the fourth leg of the C. japonica in six of the aquaria to which 100 mg/L berberine hydrochloride had been added. The pre-experiment concluded that 105 CFU/L of A. hydrophila can cause death to C. japonica itself after 24 h. C. japonica that were not soaked in berberine hydrochloride in the aquarium were selected for injection of 105 CFU/L of A. hydrophila. After 72 h, a total of 240 crabs were divided into 3 treated groups and a control group with 6 replicates per group. A total of 10 crabs were in per aquarium. Tissue samples from each group of 10 crabs were mixed into one sample, and the sample was firstly anesthetized on ice, then the gill tissue were collected and RNA was isolated on ice. Among them, 100 mg/L berberine hydrochloride (BH), A. hydrophila (AH), 100 mg/L berberine hydrochloride and injected with A. hydrophila (BHAH), and the control group injected with equal amounts of physiological saline solution (CK) were used for RNA-Seq experiments.
The flow of this RNA-seq experiment (total RNA extraction, cDNA library construction, sequencing, and transcriptome analysis) is shown in Figure 1. These processes will be discussed below. Overall RNA was taken from every sample by using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), based on the manufacturer’s conduction. Equal amounts of total RNA (1 μg) from samples in each group were pooled for RNA sequencing using a HiSeq 2000 Illumina Platform. RNA integrity was assessed using a 2100 Bioanalyzer RNA 6000 Nano assay with a minimum RNA integrity number value of 8.0. Ribosomal RNA was then removed. A NETBNext Ultra RNA Library preP Kit from Illumina (New England Biolabs, Ipswich, MA, United States) was used for preparing the cDNA libraries and 150 base pair paired-end reads were sequenced.
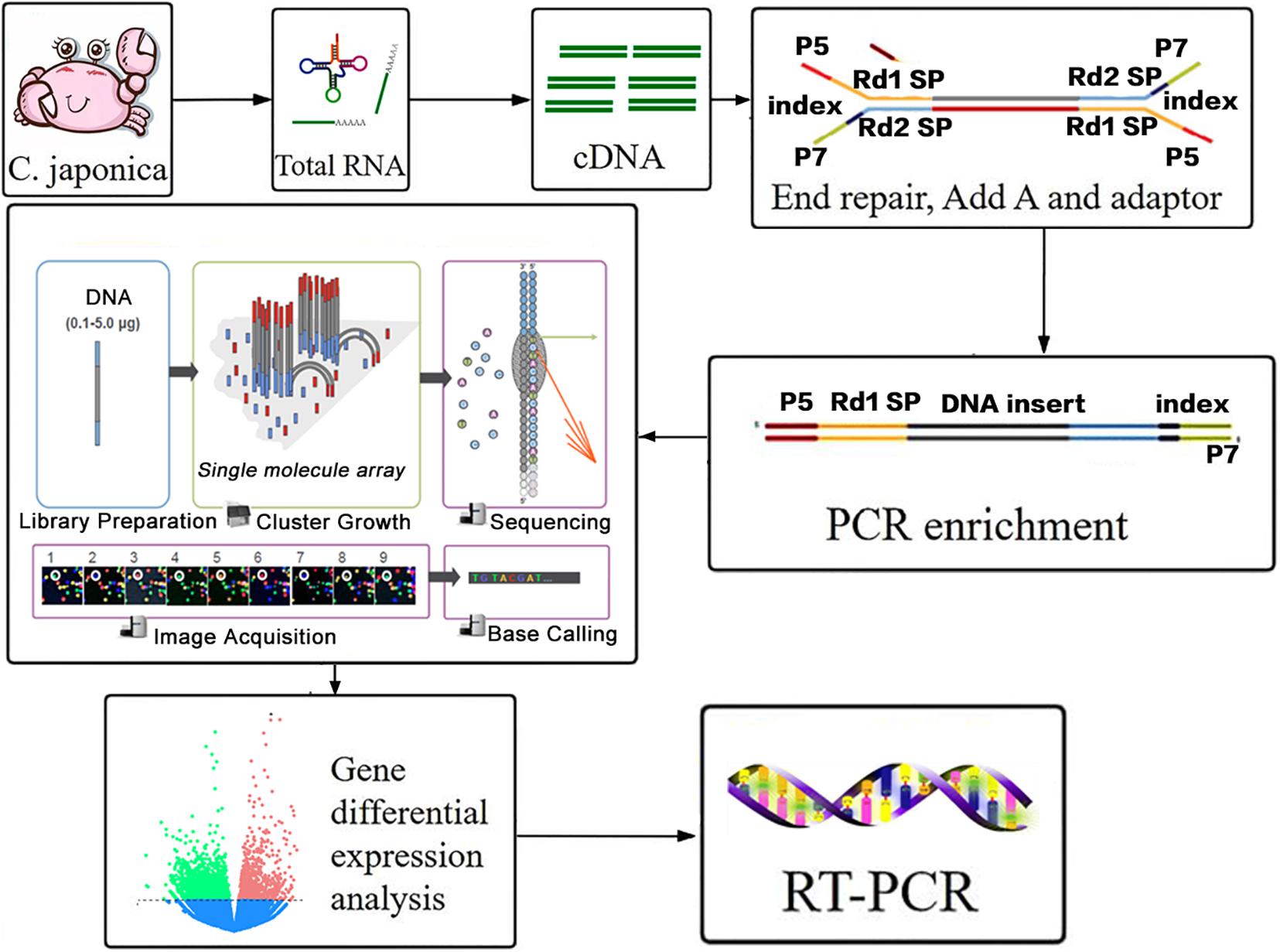
Figure 1. Workflow of transcriptome data analysis in response to berberine hydrochloride immersion infection of Aeromonas hydrophila C. japonica.
The quality of the raw data was checked using FastQC. Low quality bases and adaptors were then removed from each RNA library using Trimmomatic v0.32 (Bolger et al., 2014). A minimum length of 50 bases was chosen as a cut-off for each raw read. All cleaned reads were then aligned and mapped using Tophat v2.0.11 (Trapnell et al., 2009) to provide genomic location information, and only reads that mapped to a single gene were used for further analysis. The genome sequence and annotation data downloaded from http://www.ncgr.ac.cn/grasscarp/were used as a reference for mapping.
The mapped data were used to generate count data using HTSeq v0.5.3 (Love et al., 2014). We then applied DESeq to identify significantly differentially expressed genes (DEGs). DESeq models read counts using a negative binomial distribution with a false discovery rate adjusted P 2 or ≤ 2 and adjusted P) (Conesa et al., 2005). Gene ontology (GO) assignment of the DEGs was performed by Blast2Go measured against all available nucleotide databases, with an e-value cutoff of 10–6. Functional classification of the DEGS was further conducted using the program WEGO (Love et al., 2014; Ye et al., 2018) with default parameters. GO-term enrichment analysis was conducted using the pantherdb database1. Signaling pathways associated with the enriched genes were identified by annotation against the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa & Goto) and KEGG pathway enrichment was analyzed using the Clusterprofiler package. Including non-redundant protein sequences (NR), the Swiss Therapeutic Protein Sequence Database (Swiss-Prot), the Protein Family Database (PFAM), Clusters of Orthologous Groups of Proteins (COG), BlastX similarity search was performed on all unigenes for annotation A corrected p < 0.05 was used as the cut-off criterion.
FPKM and count of Unigene were obtained using Bowtie2 and Express software analysis (Love et al., 2014). The number of Unigene reads corresponding to each sample was obtained by Express software, and the estimateSizeFactors function of Deseq R package was used to standardize the data (Love et al., 2014). Use the nbinomTest function to calculate the p value and fold change values for the different comparison. The GO and KEGG of differential Unigenes with P values less than 0.05 and a difference of more than two times were selected for enrichment analysis to determine the biological functions or pathways mainly affected by differential Unigenes. Meanwhile, unsupervised hierarchical clustering was performed for differential Unigene, and the expression patterns of differential Unigene among different samples were shown in the form of heat maps.
The reliability of the RNA-Seq results was validated by subjecting some of the candidate genes related to immune response to qRT-PCR, using primers designed by Primer Premier 5 (Table 1). Total RNA was extracted from the samples and first-strand cDNA was synthesized using a SYBR Premix Ex TaqTM kit (Takara). PCR was performed in a 20-L reaction mixture in 96-well plates using an Applied Biosystems Prism 7500 sequence detection system (United States), including 10 L of SYBR Premix Ex Taq main mixture and 2 L of cDNA. Relative gene expression was determined according to the 2–ΔΔCT method for 0.2 m of each primer.
All data were presented as the mean ± standard error. Differences between control and treatment groups were analyzed by one-way analysis of variance (ANOVA) and Tukey’s test. A value of p < 0.05 was used to indicate the significance of the data.
The average number of original reads after sequencing was 51,816,020 and the average number of clean reads after filtering was 51,091,279. The mean value of effective base percentage was 97.6%. The average percentage of raw bases with a Phred value >30 was 93.3%. G and C accounted for an average of 46.9% of all clean bases (Table 2).
A total of six databases were used to annotate the sequenced transcriptome including 79,539 genes, of which 34,337 genes were annotated to the databases (Figure 1). A total of 332 genes were annotated to the excretory system and 1,292 to the immune system according to the Organizational Mal Systems database. A total of 1,271 genes were annotated to the folding, sorting, and degradation pathway, 223 to the replication and repair pathway, and 505 to the transcription pathway according to the Genetic Information Processing database. A total of 2,303 genes were annotated to signaling molecules and interaction, 196 to signal transduction, and 72 to membrane transport in the Environmental Information Processing database. In addition, 1,956 genes were associated with transport and catabolism, 942 with cellular community, 409 with cell motility, and 1,453 with cell growth and death in the Cellular Processes database. Totals of 390 and 247 genes were annotated to the nucleotide metabolic pathway and glycan biosynthesis and metabolism, respectively, 271 were annotated to energy metabolism and 328 to metabolism of cofactors and vitamins in the Metabolic Database (Figure 2). Berberine hydrochloride significantly altered the clustering matrix distance of C. japonica. The treatment groups BH (BH1-1, BH1-2, BH1-3, and BH1-4), BHAH (BHAH1-1, BHAH1-2, BHAH1-3, and BHAH1-4) and the control group had a greater separation (Figure 3). Berberine hydrochloride significantly altered the clustering matrix distance of Japanese sturgeon. Treatment groups BH (BH1-1, BH1-2, BH1-3, and BH1-4), BHAH (BHAH1-1, BHAH1-2, BHAH1-3, and BHAH1-4) and control groups had a greater separation (Figure 3).
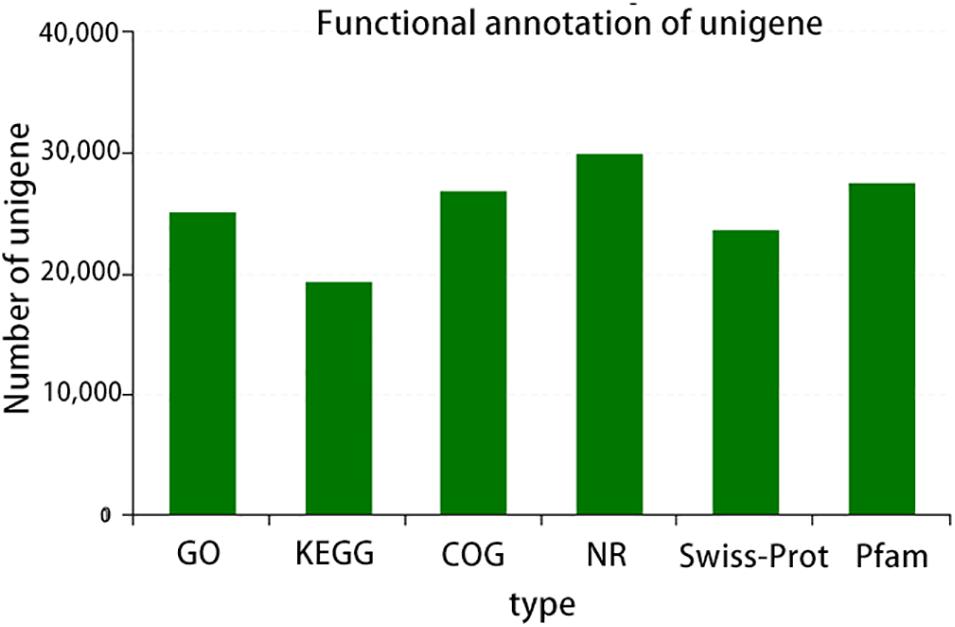
Figure 2. GO, number of transcripts/unigenes annotated to the GO library; KEGG, number of transcripts/unigenes annotated in the KEGG library; COG, number of transcripts/unigenes annotated to the COG library; NR, number of transcripts/unigenes annotated to the NR library; Swiss-Prot, number of transcripts/unigenes annotated to the Swiss-Prot library; PFAM, number of transcripts/unigenes annotated to the PFAM library.
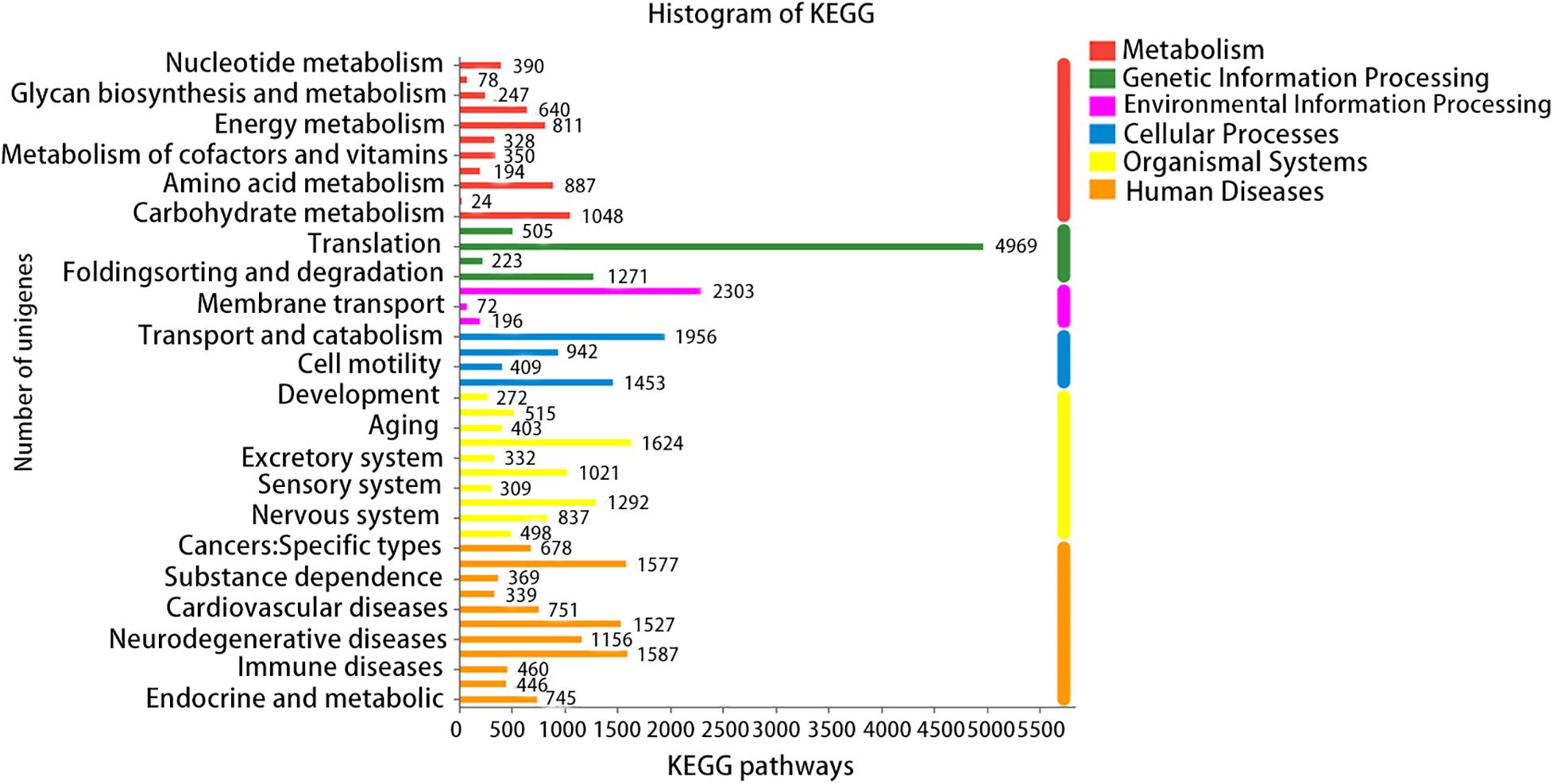
Figure 3. KEGG metabolic pathways. The X-axis indicates the number of genes/transcripts annotated to the respective pathways. KEGG metabolic pathways can be divided into seven categories: metabolism, genetic information processing, environmental information processing, cellular processes, organizational systems, human diseases, and drug development.
A total of 5,409 unigenes were significantly differentially expressed between the control (CK) and BH1 groups, including 3,546 up-regulated and 1,863 down-regulated genes (Figure 4). In addition, 4,535 unigenes were significantly differentially expressed between the CK and BHAH1 groups, including 3,101 up-regulated and 1,434 down-regulated unigenes (Figure 4). The total number of significantly expressed genes for (BH1) vs. (BHAH1) for C. japonica was 4,535, of which the number of up-regulated genes was 3,101 and the number of down-regulated genes was 1,434. Similarly, the total number of significantly expressed genes for (CK) vs. (BHAH1) was 5,409, of which the number of up-regulated genes was 3,546 and the number of down-regulated genes was 1,863 (BH1) vs. (CK) the total number of genes significantly different between samples was 2,116, of which the number of significantly different upregulated genes was 499 and the number of downregulated genes was 1,617. The total number of differentially expressed unigenes was 2,615 (Figure 4). These results indicated that berberine hydrochloride affected glutathione metabolism, oxidative phosphorylation, fatty acid degradation, retinol metabolism, pyruvate metabolism, hypoxia-inducible factor 1 (HIF-1) signaling, and the lysosome and phagosome in C. japonica (Figure 5).
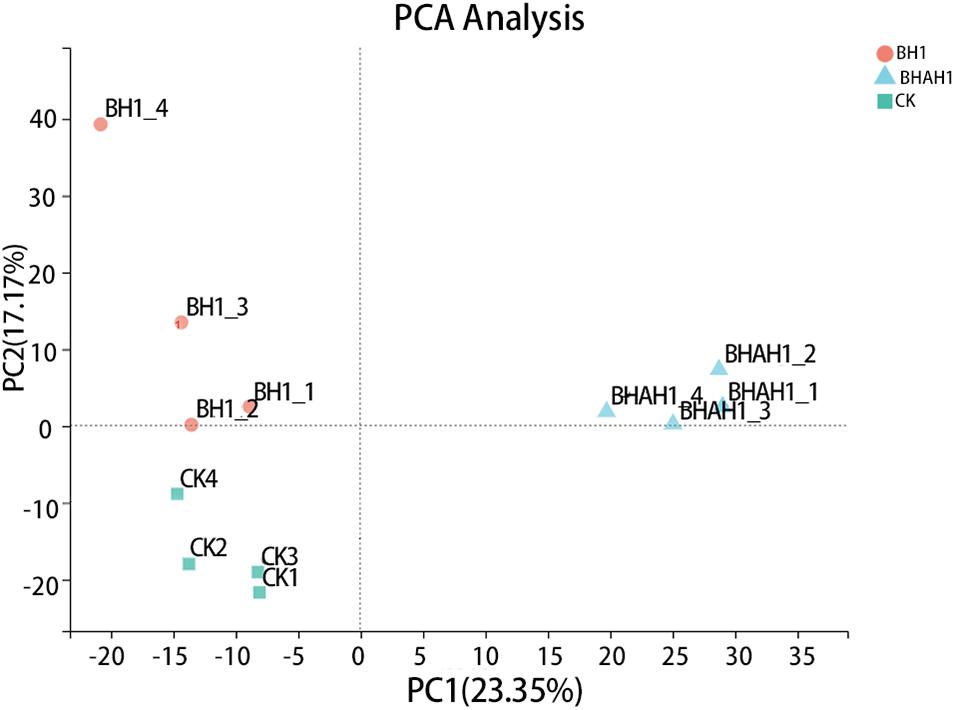
Figure 4. Principal component analysis was performed using the expression levels of unigenes. The horizontal and vertical axes represent the contributions of principal components N (PCN) and M (PCM), respectively, in the two-dimensional graph to the discrimination of samples. Orange dots, C. japonica immersed in berberine hydrochloride; green squares, control group; blue triangles, C. japonica immersed in berberine hydrochloride and injected with A. hydrophila.
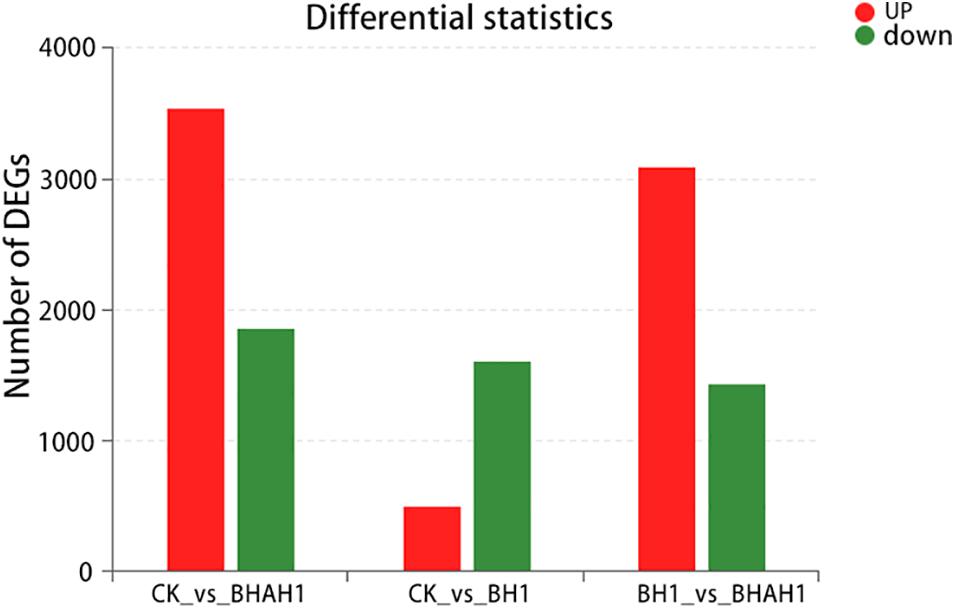
Figure 5. Differentially down-regulated gene expression in each group. Red, up-regulated genes; green, down-regulated genes.
The combination of transcriptomic gene expression and Relative mRNA Level reveals that it contains Antigen processing and presentation (LGMN), V-type H + -transporting ATPase subunit a (ATP6), alpha-N-acetylgalactosaminidase (NAGA), lysosome membrane protein 2 (LIMP2), Saposin (PSAP), glyceraldehyde 3-phosphate dehydrogenase (GAP), and GMP. acetylgalactosaminidase (NAGA), lysosome membrane protein 2 (LIMP2), saposin (PSAP), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), enolase (ENO), nitric-oxide synthase (NOS1) and cathepsin L (CTSL) genes in control (CK), C. japonica dipped in berberine hydrochloride 100 mg/L (BH) and C. japonica dipped in berberine hydrochloride 100 mg/L and injected with 105 CFU/L of Aeromonas hydrophila (BHAH1) genes expression trends were consistent (Figures 6, 7). LGMN, ATP6, NAGA, LIMP2, and PSAP are acid hydrolases contained within the lysosome. Berberine hydrochloride down-regulated the LGMN gene while up-regulating the other acid hydrolases. Berberine hydrochloride also up-regulated some genes in phagocytes, including NOS1 and CTSL. The HIF-1 signaling pathway in C. japonica reduces GAPDH and ENO in anaerobic respiration. These genes are all related to innate immunity in C. japonica, and are also closely related to the autophagy lysosome system. Berberine hydrochloride played a regulatory role in the autolysosomal system in C. japonica infected with A. hydrophila. In terms of energy supply, ATP6 was upregulated in the order CK < BH < AH < BHAH. NOS1 was expressed and induced in the order CK < BH < AH < BHAH on the gill mucosal surface of C. japonica. Berberine hydrochloride also regulated the acidic hydrolases LGMN (BH < BHAH < CK < AH), NAGA (CK < AH < BHAH < BH), LIMP2 (CK < AH < BHAH < BH), and PSAP (CK < BH < AH < BHAH < BHAH) (Figure 8). The molecular mechanism of berberine hydrochloride in C. japonica injected with A. hydrophila was thus determined based on gene expression levels of the transcriptome and relative mRNA levels (Figure 9). Among them, berberine hydrochloride regulates ND1 gene in Oxidative phosphorylation metabolism (CK < AH < BHAH < BH), CPT1A gene in Fatty acid degradation metabolism (AH < CK < BHAH < BH), RDH12 gene in Retinol metabolism and PCCA in Pyruvate metabolism (CK < AH < BHAH < BH). RDH12 gene upregulation and PCCA upregulation in pyruvate metabolism (CK < AH < BHAH < BH) (Figure 10).
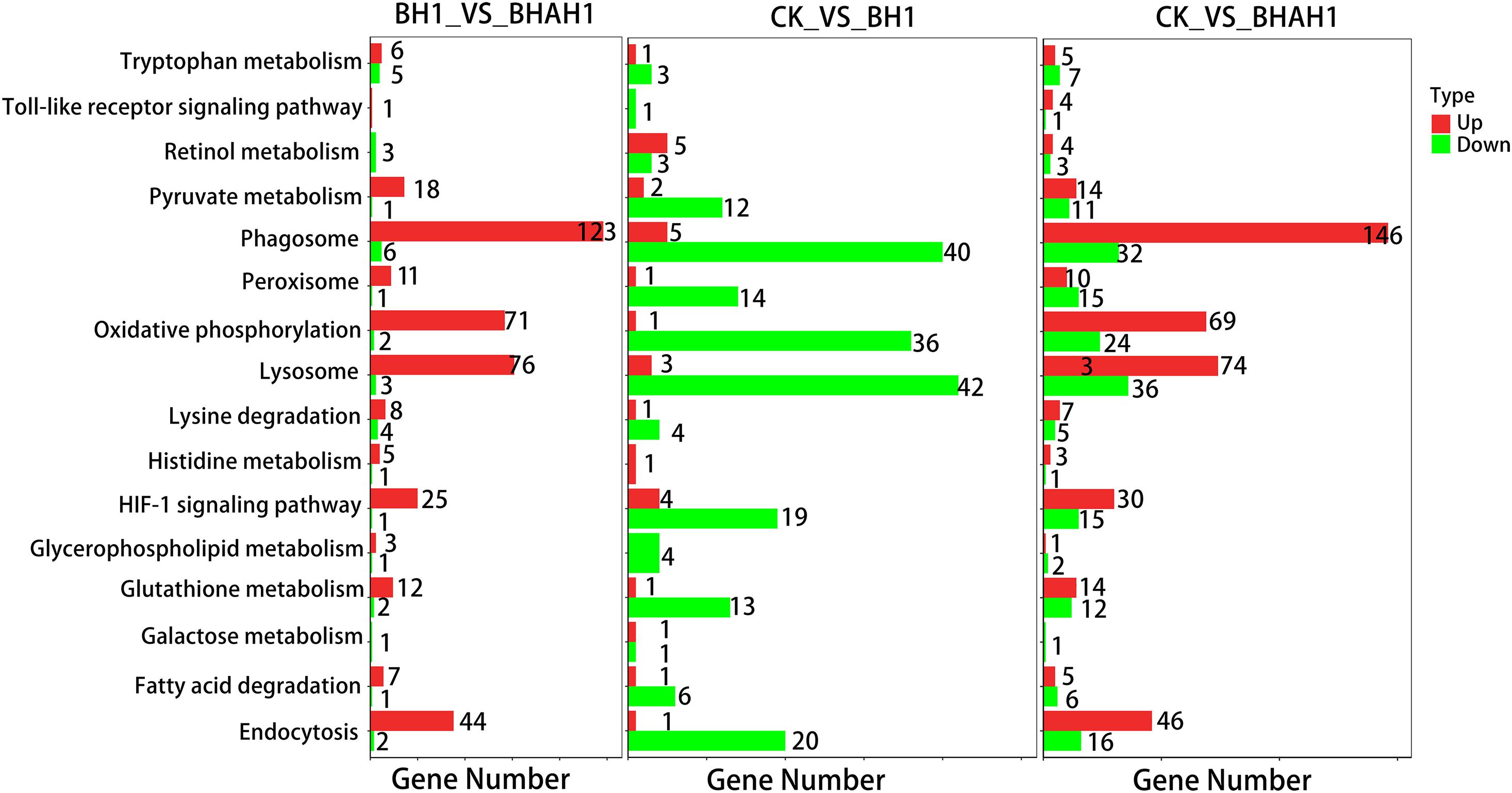
Figure 6. KEGG metabolic pathways based on transcriptome gene expression in gill tissues of C. japonica. Red, up-regulated genes; green, down-regulated genes; CK, control group; BH, C. japonica immersed in berberine hydrochloride; BHAH1, C. japonica immersed in berberine hydrochloride and injected with A. hydrophila.
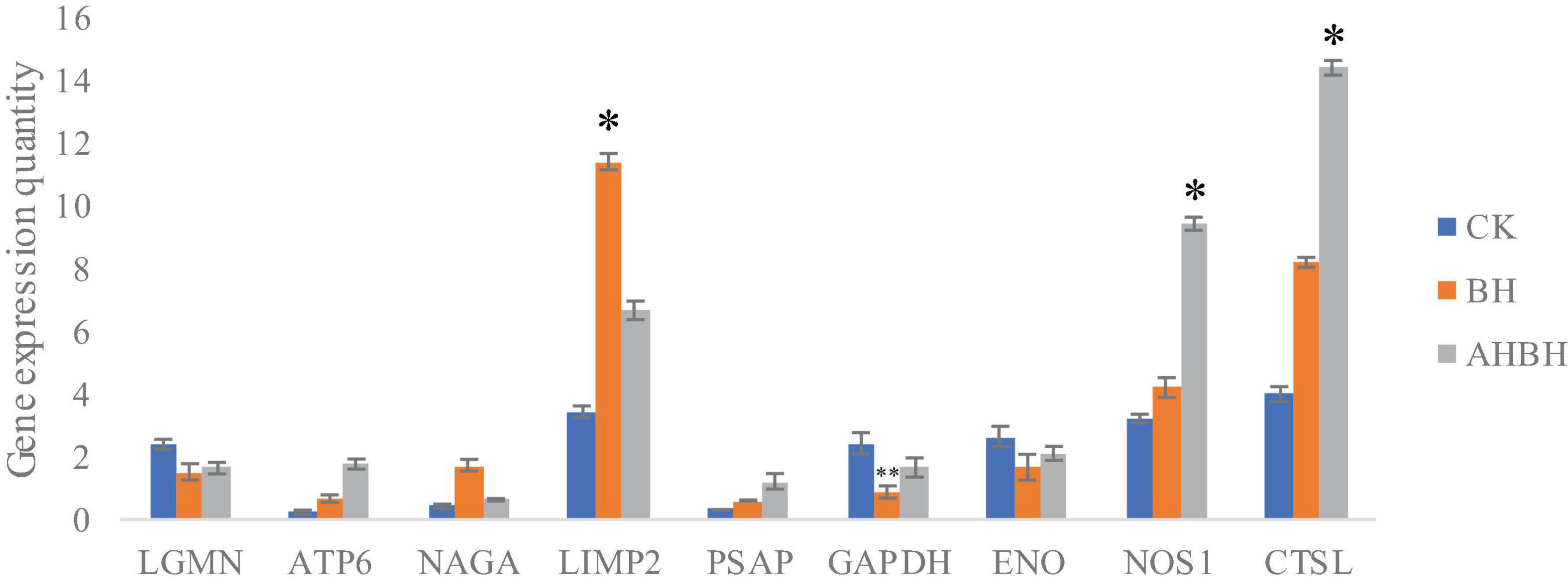
Figure 7. Transcriptome gene expression in gill tissues from C. japonica. CK, control group; BH, C. japonica immersed in berberine hydrochloride; BHAH1, C. japonica immersed in berberine hydrochloride and injected with A. hydrophila. Error bars represent standard deviations of three independent repeats. ∗p < 0.05, ∗∗p < 0.01 analyzed by t-tests. Four crabs were used in each group.
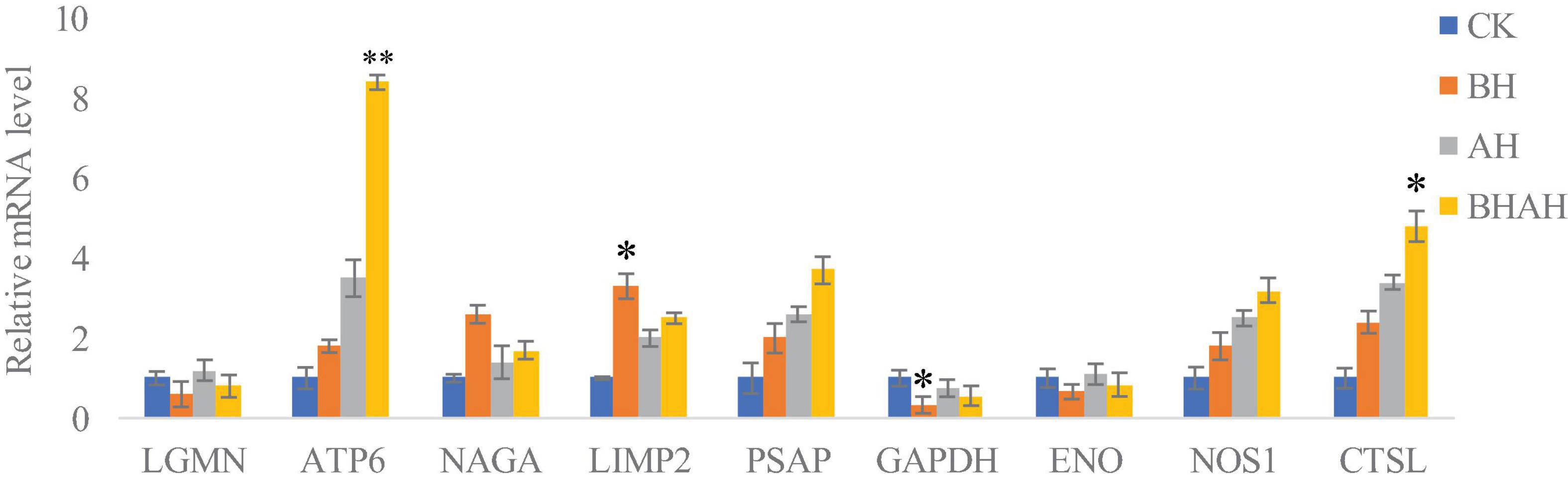
Figure 8. Relative mRNA levels in gill samples from C. japonica. CK, control group; BH, C. japonica immersed in berberine hydrochloride; AH, C. japonica infected with A. hydrophila; BHAH1, C. japonica infected with A. hydrophila and treated with berberine hydrochloride. Error bars represent SD of three independent repeats. ∗p < 0.05, ∗∗p < 0.01 analyzed by t-tests. Four crabs were used in each group.
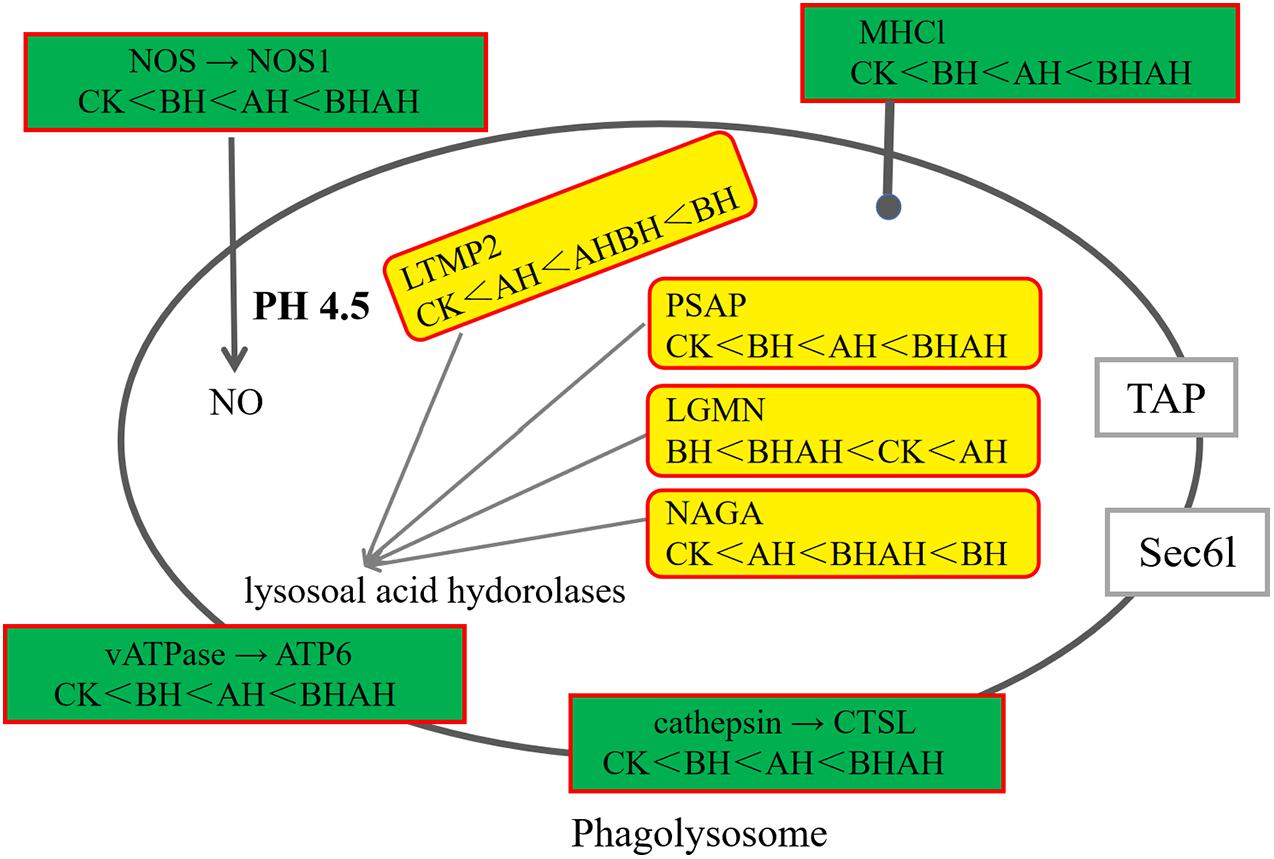
Figure 9. Changes in pathways in C. japonica immersed in 100 mg/L berberine hydrochloride and injected with 105 CFU/L A. hydrophila.
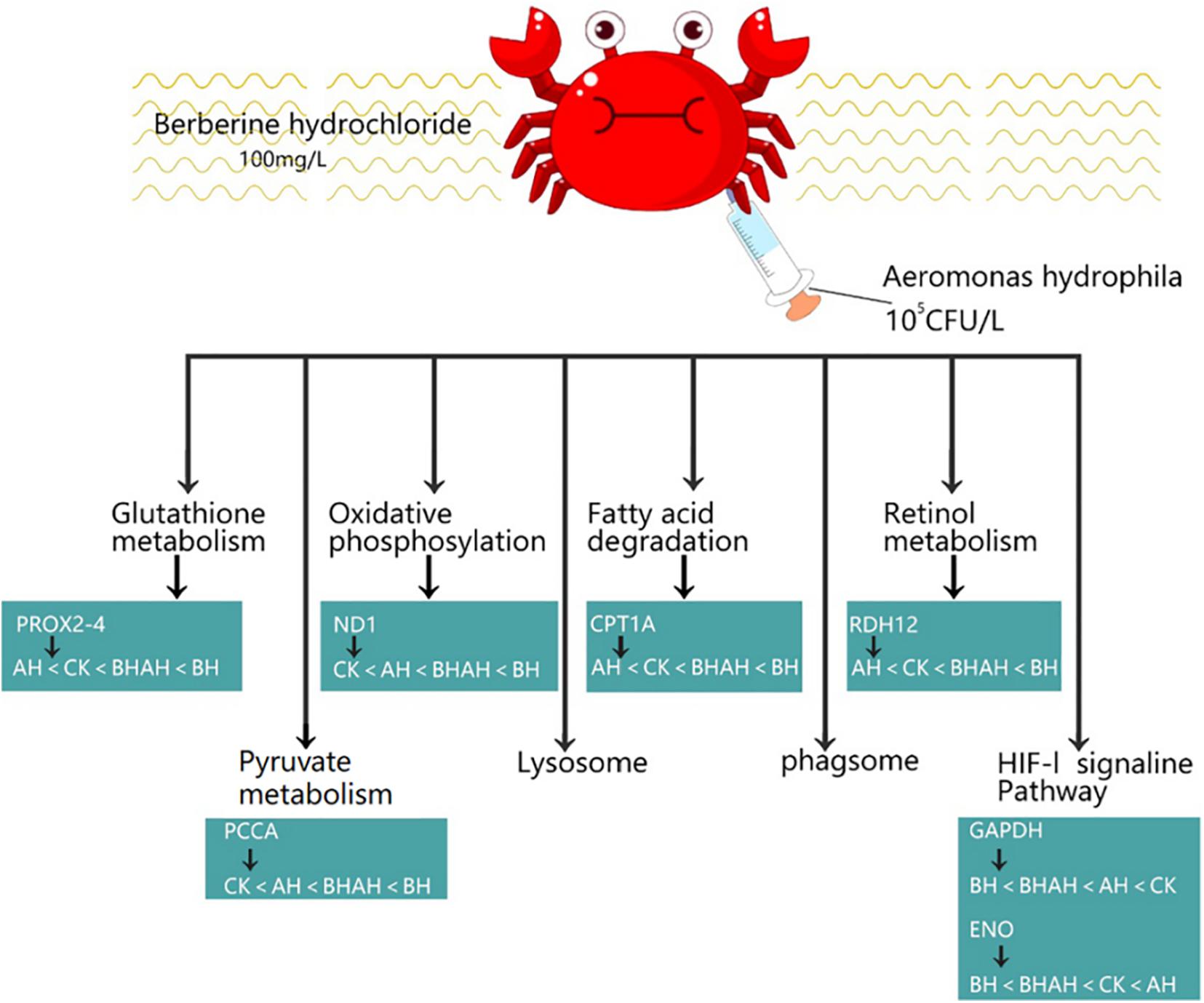
Figure 10. Molecular mechanism of berberine hydrochloride in C. japonica injected with 105 CFU/L A. hydrophila.
Some genes of Glutathione metabolism were also altered in Berberine hydrochloride-soaked C. japonica, such as PRDX2-4 (AH < CK < BHAH < BH). And this experiment also found that berberine hydrochloride could regulate the ND1 gene in Oxidative phosphorylation metabolism (CK < AH < BHAH < BH). In the present experiment there were some changes in genes in Pyruvate metabolism under the action of berberine hydrochloride, such as upregulation of PCCA (CK < AH < BHAH < BH).
In this study, 5,409 transcripts were significantly up-regulated or down-regulated by berberine hydrochloride, and these genes were distributed in various metabolic pathways. Immersion in berberine altered pathways related to glutathione metabolism, oxidative phosphorylation, fatty acid degradation, retinol metabolism, pyruvate metabolism, HIF-1 signaling, and lysosomes and phagosomes. Glutathione metabolism is one of the main components of the oxidative defense system in C. japonica (Pan and Zhang, 2006) and some genes involved in glutathione metabolism, such as PRDX2-4 (AH < CK < BHAH < BH), were altered by berberine hydrochloride immersion. The oxidative phosphorylation pathway is responsible for the production of ATP in mitochondria (Pagliarini and Rutter, 2013). Upregulation of the lipolytic gene CPT1A was accompanied by an increase in energetic ATP during catabolism (Cheng et al., 2016). In this experiment, berberine hydrochloride regulated the CPT1A gene involved in fatty acid degradation metabolism (AH < CK < BHAH < BH), which may ameliorate the damage caused by Vibrio spp. and A. hydrophila infection in crabs. Vitamin A deficiency impairs retinol metabolism and reduces growth and impairs intestinal immune function in fish (Zhang et al., 2017). Vitamin A deficiency also impairs the physical barrier function associated with impaired antioxidant capacity and exacerbates apoptosis (Jiang et al., 2019). However, berberine hydrochloride can affect retinol metabolism and can up-regulate the RDH12 gene in flavanol metabolism (AH < CK < BHAH < BH). The pyruvate metabolism network, including up-regulation of pyruvate kinase, is involved in the pathogenesis of A. hydrophila in fish, thus enhancing the immunity of zebrafish infected with A. hydrophila (Lin et al., 2019). In the current study, berberine hydrochloride up-regulated genes involved in pyruvate metabolism, including PCCA (CK < AH < BHAH < BH), which may be help to resist infection by A. hydrophila.
Lysosomes are digestive organelles in the endocytic pathway, and increased lysosomal enzyme activity may contribute to resistance to pathogens (Habtemariam, 2016). Lysosomes contain acid hydrolytic enzymes, including LGMN, ATP6, NAGA, LIMP-2, and PSAP. Previous studies found that the LGMN gene played a role in both primary and adaptive immunity, including the generation of major histocompatibility complex II-presented antigens, which have effective enhancement of kinetic energy changes between heat absorption and exotherm during contraction in scleractinian fish (Marra et al., 2017). The oxidative phosphorylation pathway is responsible for the production of intracellular ATP in mitochondria, including ATP6, which can supply energy to other metabolic pathways (Hatefi, 1985; Pagliarini and Rutter, 2013). LIMP-2 belongs to a subfamily of scavenger receptors, known as lysosomal transmembrane glycosylation receptors (Tan et al., 2019), and plays an important role in the immune system in response to bacterial infection in fish. LIMP-2 was significantly up-regulated in all mucosal tissues (skin, gills, and intestine) in Scophthalmus maximus following infection by the Gram-negative bacterium Vibrio anguillarum (Tan et al., 2019). PSAP is the precursor of saposin, which is found in lysosomes and secretory proteins. PSAP-1 and PSAP-2 significantly inhibited the growth of various bacteria and killed Miamiensis avidus in Platichthys stellatus, and significantly inhibited the formation of bacterial biofilms (Ghana et al., 2015). A microbial feed additive in Atlantic salmon has been shown to reduce intestinal inflammation by up-regulating the NAGA gene (Yong et al., 2007). The LGMN gene for antigen processing and presentation (BH < BHAH < CK < AH) was down-regulated, the ATP6 gene for the V-type H + transporter ATPase subunit a (CK < BH < AH < BHAH), the NAGA gene for alpha-N-acetylgalactosaminidase (CK < AH < BHAH < BH), the lysosome membrane protein 2 of LIMP2 (CK < AH < BHAH < BH), and PSAP of saposin. SGP1 (CK < BH < AH < BHAH) were all upregulated. Based on the characteristics of these genes, we deduce that berberine hydrochloride improves the immunity of crabs by increasing the levels of some hydrolytic enzyme genes. Berberine hydrochloride was previously shown to induce the expression of acid hydrolase genes that enhanced the degradation and clearance of methicillin-resistant S. aureus and enterosoluble E. coli from macrophages (Yong et al., 2007). Based on the ability of berberine hydrochloride to induce acid hydrolase gene expression and thus remove bacteria, immersion in berberine hydrochloride could remove A. hydrophila, and thus improve survival of C. japonica.
Autophagy is a lysosomal-dependent degradation pathway in eukaryotic cells (Levine et al., 2011). The process of autophagy can be divided into four steps: formation of autophagosomes, fusion of autophagosomes and lysosomes, degradation of autophagosome substrate, and regeneration of autophagosomes. Autophagy can also be induced by conditions such as hypoxia, oxidative stress, and radiation (Radomski et al., 2018). The autophagosome fuses with the lysosome to form the autophagosome and acidic hydrolases within the lysosome lumen degrade the autophagic inner membrane and internal macromolecules to produce small molecules such as amino acids and monosaccharides. In the current study, berberine hydrochloride upregulated some autophagosome genes, including NOS1 and NOS2, which may be related to the microbial-resistance effects of berberine hydrochloride. Nitric oxide is produced by NOS with L-arginine as a substrate, and acts as an antimicrobial host defense effector, which is produced by many immune-associated cells during infection (MacMicking et al., 1997; Bogdan, 1998). The expression and induction of NOS1 has been detected on the mucosal surface of the gills and intestines after challenge, and significant up-regulation of NOS1 was detected in the intestinal tract of turbot following V. anguillarum infection (Pederzoli et al., 2007; Yao et al., 2014). Vibrio has been shown to induce fluid secretion in the intestine, while nitric oxide regulates blood flow and peristalsis and has cytotoxic and antimicrobial activities in the intestine. Vibrio-infected intestinal epithelial cells showed significantly increased NOS activity (Janoff et al., 1997). In addition, the NOS gene also showed high expression in the gut in invertebrates, with the highest expression of the NOS1 gene in the gills (CK < BH < AH < BHAH) following attack by Vibrio parahaemolyticus (Li et al., 2012). This was in accord with the current results in C. japonica infected with A. hydrophila. CTSL is a lysosomal cysteine protease found in rock bream (Whang et al., 2011) and orange spotted groupers (Liang et al., 2012), which acts as a barrier against invading pathogens and is involved in vertebrate immune responses. CTSL acts as a chemical barrier against microbial invasion in the innate immune system (Ellis, 2001). The solubility of CTSL is the main factor leading to the infection of E. sinensis (Li et al., 2012) and Chinese white shrimp (Ren et al., 2010), suggesting its potential role in the immune defense system. In the present study, berberine hydrochloride up-regulated the CTSL (CK < BH < AH < BHAH) in C. japonica to resist A. hydrophila infection.
The HIF-1 signaling pathway plays a central role in the rapid adaptation to an inadequate oxygen supply to prevent cell damage, and the HIF maintenance pathway thus affects many diseases and disease processes (Kim et al., 2017). HIF-1 signaling reduced GAPDH in anaerobic respiration and had a protective effect against Edwardsiella tarda infection (Liu et al., 2005). Fusing GAPDH from pathogenic A. hydrophila, as a putative protective antigen, with four different delivery systems produced a candidate vaccine that induced effective host protection against V. anguillarum, A. hydrophila, and Aspergillus hydrophila (Zhou et al., 2010; Yan et al., 2011). In the current study, berberine hydrochloride also reduced GAPDH gene expression (BH < BHAH < AH < CK), which may have reduced the toxicity of A. hydrophila. Berberine hydrochloride reduced the activity of the eno gene (BH < BHAH < CK < AH), which may have reduced the toxicity of A. hydrophila. The virulence gene eno in Vibrio alginolyticus and A. hydrophila strains has a good toxicological potency on combination with other virulence genes and against the corresponding eno binding of the host (Zuo et al., 2019).
This study revealed the molecular mechanisms following infection of C. japonica by A. hydrophila and immersion in berberine hydrochloride. Exposure to berberine hydrochloride altered the expression of genes involved in glutathione metabolism, oxidative phosphorylation, fatty acid degradation, retinol metabolism, pyruvate metabolism, HIF-1 signaling, and other pathways. Berberine hydrochloride may thus have an adjuvant therapeutic effect against A. hydrophila infection. Transcriptome analysis showed that berberine-soaked C. japonica adapted to A. hydrophila invasion by increasing the expression levels of genes involved in the acid hydrolase and endophagosome, and anaerobic respiration-related genes. These transcriptomic data for the effects of berberine hydrochloride on C. japonica provided strong support for its further development as a therapeutic anti-bacterial agent. The results of this study provided a new scientific basis for the potential role of berberine hydrochloride in protecting crustaceans against bacterial infections in aquaculture. It also provided an additional option for the use of secondary metabolites to replace antibiotics in the treatment of bacterial diseases in aquatic animals.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethical review and approval was not required for the animal study because crabs are crustaceans or arthropods, which do not belong to vertebrates and cephalopods.
MH was responsible for experimental design, experimental operation, and article writing. YL was responsible for data analysis. TZ and DL were responsible for instrument operation. ZZ and QJ were responsible for overall thinking. All authors contributed to the article and approved the submitted version.
This work was supported by National Key R&D Program of China (2020YFD0900305), Science Foundation of Jiangsu (NO. BK20191488) in China, Major Project of Hydrobios Resources in Jiangsu Province (ZYHB16-3), and Agricultural Major New Variety Creation Project in Jiangsu Province (PZCZ201743).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank for Mengling Sun her help of sampling.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.754588/full#supplementary-material
Andrew, S. (2010). Fast QC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Bao, J., Xing, Y.-N., Jiang, H.-B., and Li, X.-D. (2019). Identification of immune-related genes in gills of Chinese mitten crabs (Eriocheir sinensis) during adaptation to air exposure stress. Fish Shellfish Immunol. 84, 885–893.
Bogdan, C. (1998). The multiplex function of nitric oxide in (auto) immunity. J. Exp. Med. 187, 1361–1365. doi: 10.1084/jem.187.9.1361
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cheng, J., Lv, S., Nie, S., Liu, J., Tong, S., Kang, N., et al. (2016). Chronic perfluorooctane sulfonate (PFOS) exposure induces hepatic steatosis in zebrafish. Aquat. Toxicol. 176, 45–52. doi: 10.1016/j.aquatox.2016.04.013
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Czesny, S., Epifanio, J., and Michalak, P. (2012). Genetic divergence between freshwater and marine morphs of alewife (Alosa pseudoharengus): a ‘next-generation’sequencing analysis. PLoS One 7:e31803. doi: 10.1371/journal.pone.0031803
DuPont, H. L. (2007). The growing threat of foodborne bacterial enteropathogens of animal origin. Clin. Infect. Dis. 45, 1353–1361. doi: 10.1086/522662
Ellis, A. (2001). Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 25, 827–839. doi: 10.1016/s0145-305x(01)00038-6
Garber, M., Grabherr, M. G., Guttman, M., and Trapnell, C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477. doi: 10.1038/nmeth.1613
Ghana, V., Viswanath, K., Amod, K., Dalia, D., Jep, L., and Yoichiro, K. (2015). A Microbial Feed Additive Abates Intestinal Inflammation in Atlantic Salmon. Front. Immunol. 6:409. doi: 10.3389/fimmu.2015.00409
González-Escalona, N., Blackstone, G. M., and Depaola, A. (2006). Characterization of a Vibrio alginolyticus strain, isolated from Alaskan oysters, carrying a hemolysin gene similar to the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 72, 7925–7929. doi: 10.1128/aem.01548-06
Habtemariam, S. (2016). Berberine and inflammatory bowel disease: a concise review. Pharmacol. Res. 113, 592–599. doi: 10.1016/j.phrs.2016.09.041
Hatefi, Y. (1985). The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54, 1015–1069. doi: 10.1146/annurev.bi.54.070185.005055
Igbinosa, I. H., Igumbor, E. U., Aghdasi, F., Tom, M., and Okoh, A. I. (2012). Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012:625023.
Janoff, E. N., Hayakawa, H., Taylor, D. N., Fasching, C. E., Kenner, J. R., Jaimes, E., et al. (1997). Nitric oxide production during Vibrio cholerae infection. Am. J. Physiol. Gastrointest. Liver Physiol. 273, G1160–G1167.
Jia, Z., Wang, M., Wang, X., Wang, L., Qiu, L., and Song, L. (2018). Transcriptome sequencing reveals the involvement of reactive oxygen species in the hematopoiesis from Chinese mitten crab Eriocheir sinensis. Dev. Comp. Immunol. 82, 94–103. doi: 10.1016/j.dci.2017.12.030
Jiang, W.-D., Zhou, X.-Q., Zhang, L., Liu, Y., Wu, P., Jiang, J., et al. (2019). Vitamin A deficiency impairs intestinal physical barrier function of fish. Fish Shellfish Immunol. 87, 546–558. doi: 10.1016/j.fsi.2019.01.056
Johnson, P. T. (1983). “Diseases Caused by Viruses, Rickettsiae, Bacteria, and Fungi,” in The Biology of Crusta cea, Vol. 6, ed. A. J. Provenzano (New York, NY: Academic Press), 1–78.
Kim, H., Greenald, D., Vettori, A., Markham, E., Santhakumar, K., Argenton, F., et al. (2017). Zebrafish as a model for von Hippel Lindau and hypoxia-inducible factor signaling. Methods Cell Biol. 138, 497–523.
Kurata, S., Ariki, S., and Kawabata, S.-I. (2006). Recognition of pathogens and activation of immune responses in Drosophila and horseshoe crab innate immunity. Immunobiology 211, 237–249. doi: 10.1016/j.imbio.2005.10.016
Lazreg Aref, H., Gaaliche, B., Fekih, A., Mars, M., Aouni, M., Pierre Chaumon, J., et al. (2011). In vitro cytotoxic and antiviral activities of Ficus carica latex extracts. Nat. Prod. Res. 25, 310–319.
Levine, B., Mizushima, N., and Virgin, H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323–335.
Li, S., Zhang, Z., Li, C., Zhou, L., Liu, W., Li, Y., et al. (2012). Molecular cloning and expression profiles of nitric oxide synthase (NOS) in mud crab Scylla paramamosain. Fish Shellfish Immunol. 32, 503–512.
Liang, J.-Z., Rao, Y.-Z., Lun, Z.-R., and Yang, T.-B. (2012). Cathepsin L in the orange-spotted grouper, Epinephelus coioides: molecular cloning and gene expression after a Vibrio anguillarum challenge. Fish Physiol. Biochem. 38, 1795–1806. doi: 10.1007/s10695-012-9676-3
Lin, Y.-S., Saputra, F., Chen, Y.-C., and Hu, S.-Y. (2019). Dietary administration of Bacillus amyloliquefaciens R8 reduces hepatic oxidative stress and enhances nutrient metabolism and immunity against Aeromonas hydrophila and Streptococcus agalactiae in zebrafish (Danio rerio). Fish Shellfish Immunol. 86, 410–419. doi: 10.1016/j.fsi.2018.11.047
Liu, S., Chen, G., Xu, H., Zou, W., Yan, W., Wang, Q., et al. (2017). Transcriptome analysis of mud crab (Scylla paramamosain) gills in response to Mud crab reovirus (MCRV). Fish Shellfish Immunol. 60, 545–553.
Liu, Y., Cui, Z., Song, C., Wang, S., and Li, Q. (2011). Multiple isoforms of immune-related genes from hemocytes and eyestalk cDNA libraries of swimming crab Portunus trituberculatus. Fish Shellfish Immunol. 31, 29–42. doi: 10.1016/j.fsi.2011.02.016
Liu, Y., Oshima, S. I., Kurohara, K., Ohnishi, K., and Kawai, K. (2005). Vaccine efficacy of recombinant GAPDH of Edwardsiella tarda against edwardsiellosis. Microbiol. Immunol. 49, 605–612. doi: 10.1111/j.1348-0421.2005.tb03652.x
Lou, F., Yang, T., Han, Z., and Gao, T. (2018). Transcriptome analysis for identification of candidate genes related to sex determination and growth in Charybdis japonica. Gene 677, 10–16. doi: 10.1016/j.gene.2018.07.044
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21.
MacMicking, J., Xie, Q.-W., and Nathan, C. (1997). Nitric oxide and macrophage function. Annu. Rev. Immunol. 15, 323–350.
Marra, N. J., Richards, V. P., Early, A., Bogdanowicz, S. M., Bitar, P. D. P., Stanhope, M. J., et al. (2017). Comparative transcriptomics of elasmobranchs and teleosts highlight important processes in adaptive immunity and regional endothermy. BMC Genomics 18:87. doi: 10.1186/s12864-016-3411-x
Martínez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. doi: 10.1126/science.1159483
Neves, J. V., Wilson, J. M., and Rodrigues, P. N. (2009). Transferrin and ferritin response to bacterial infection: the role of the liver and brain in fish. Dev. Comp. Immunol. 33, 848–857. doi: 10.1016/j.dci.2009.02.001
Pagliarini, D. J., and Rutter, J. (2013). Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27, 2615–2627.
Pan, L., and Zhang, H. (2006). Metallothionein, antioxidant enzymes and DNA strand breaks as biomarkers of Cd exposure in a marine crab, Charybdis japonica. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 144, 67–75.
Pederzoli, A., Conte, A., Tagliazucchi, D., Gambarelli, A., and Mola, L. (2007). Occurrence of two NOS isoforms in the developing gut of sea bass Dicentrarchus labrax (L.). Histol. Histopathol. 22, 1057–1064.
Radomski, N., Rebbig, A., Leonhardt, R. M., and Knittler, M. R. (2018). Xenophagic pathways and their bacterial subversion in cellular self-defense–πανταρει–everything is in flux. Int. J. Med. Microbiol. 308, 185–196. doi: 10.1016/j.ijmm.2017.10.012
Ren, Q., Zhang, X.-W., Sun, Y.-D., Sun, S.-S., Zhou, J., Wang, Z.-H., et al. (2010). Two cysteine proteinases respond to bacterial and WSSV challenge in Chinese white shrimp Fenneropenaeus chinensis. Fish Shellfish Immunol. 29, 551–556. doi: 10.1016/j.fsi.2010.03.002
Seal, B. S., Lillehoj, H. S., Donovan, D. M., and Gay, C. G. (2013). Alternatives to antibiotics: a symposium on the challenges and solutions for animal production. Anim. Health Res. Rev. 14, 78–87. doi: 10.1017/s1466252313000030
Sun, M., Li, Y. T., Liu, Y., Lee, S. C., and Wang, L. (2016). Transcriptome assembly and expression profiling of molecular responses to cadmium toxicity in hepatopancreas of the freshwater crab Sinopotamon henanense. Sci. Rep. 6:19405.
Tan, F., Cao, M., Ge, X., Li, C., Tian, M., Zhang, L., et al. (2019). Identification and initial functional characterization of lysosomal integral membrane protein type 2 (LIMP-2) in turbot (Scophthalmus maximus L.). Dev. Comp. Immunol. 99:103412.
Tang, J., Feng, Y., Tsao, S., Wang, N., Curtain, R., and Wang, Y. (2009). Berberine and Coptidis rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. J. Ethnopharmacol. 126, 5–17.
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Whang, I., De Zoysa, M., Nikapitiya, C., Lee, Y., Kim, Y., Lee, S., et al. (2011). Molecular characterization and expression analysis of Cathepsin B and L cysteine proteases from rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 30, 763–772. doi: 10.1016/j.fsi.2010.12.022
Xie, C., Chen, Y., Sun, W., Ding, J., Zhou, L., Wang, S., et al. (2014). Transcriptome and expression profiling analysis of the hemocytes reveals a large number of immune-related genes in mud crab Scylla paramamosain during Vibrio parahaemolyticus infection. PLoS One 9:e114500. doi: 10.1371/journal.pone.0114500
Xu, W. N., Chen, D. H., Chen, Q. Q., and Liu, W. B. (2017). Growth performance, innate immune responses and disease resistance of fingerling blunt snout bream, Megalobrama amblycephala adapted to different berberine-dietary feeding modes. Fish Shellfish Immunol. 68, 458–465. doi: 10.1016/j.fsi.2017.07.051
Ya-Nan, L. U., Fan, C. Q., and Tian, X. Q. (2014). Investigation on the development status and problems of marine Traditional Chinese Medicine based on the “Chinese Pharmacopoeia”. Fish. Inf. Strategy. 029(004): 257–262.
Yan, Z., Liu, Q., Wang, X., Zhou, L., Wang, Q., and Zhang, Y. (2011). Surface Display of Aeromonas hydrophila GAPDH in Attenuated Vibrio anguillarum to Develop a Noval Multivalent Vector Vaccine. Mar. Biotechnol. 13, 963–970. doi: 10.1007/s10126-010-9359-y
Yao, J., Li, C., Zhang, J., Liu, S., Feng, J., Wang, R., et al. (2014). Expression of nitric oxide synthase (NOS) genes in channel catfish is highly regulated and time dependent after bacterial challenges. Dev. Comp. Immunol. 45, 74–86. doi: 10.1016/j.dci.2014.02.005
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nuclc Acids Res. 46, W71–W75.
Yong, Y., Xiao-Li, Y., Xue-Gang, L., Jing, Z., Baoshun, Z., and Lujiang, Y. (2007). Synthesis and antimicrobial activity of 8-alkylberberine derivatives with a long aliphatic chain. Planta Med. 73, 602–604. doi: 10.1055/s-2007-967180
Zhang, D., Li, A., Xie, J., and Ji, C. (2009). In vitro antibacterial effect of berberine hydrochloride and enrofloxacin to fish pathogenic bacteria. Aquac. Res. 41, 1095–1100.
Zhang, C., Pang, Y., Zhang, Q., Huang, G., Xu, M., Tang, B., et al. (2018). Hemolymph transcriptome analysis of Chinese mitten crab (Eriocheir sinensis) with intact, left cheliped autotomy and bilateral eyestalk ablation. Fish Shellfish Immunol. 81, 266–275. doi: 10.1016/j.fsi.2018.07.025
Zhang, L., Feng, L., Jiang, W.-D., Liu, Y., Wu, P., Kuang, S.-Y., et al. (2017). Vitamin A deficiency suppresses fish immune function with differences in different intestinal segments: the role of transcriptional factor NF-κB and p38 mitogen-activated protein kinase signalling pathways. Br. J. Nutr. 117, 67–82. doi: 10.1017/s0007114516003342
Zhou, L., Wang, X., Liu, Q., Wang, Q., Zhao, Y., and Zhang, Y. (2010). A novel multivalent vaccine based on secretary antigen-delivery induces protective immunity against Vibrio anguillarum and Aeromonas hydrophila. J. Biotechnol. 146, 25–30. doi: 10.1016/j.jbiotec.2009.12.010
Keywords: Charybdis japonica, berberine hydrochloride, Aeromonas hydrophila, transcriptome, immune mechanisms
Citation: Han M, Liu Y, Zhang T, Li D, Jiang Q and Zuraini Z (2021) Effect of Berberine Hydrochloride Treatment on Transcriptomic Response in Gill Tissue of Charybdis japonica Infected With Aeromonas hydrophila. Front. Mar. Sci. 8:754588. doi: 10.3389/fmars.2021.754588
Received: 09 August 2021; Accepted: 07 October 2021;
Published: 16 November 2021.
Edited by:
Jianmin Zhao, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Han, Liu, Zhang, Li, Jiang and Zuraini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qichen Jiang, cWljaGVuamlhbmdAbGl2ZS5jbg==; Zakaria Zuraini, enVyYWluaUB1c20ubXk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.