- Laboratory of Fish Nutrition, School of Marine Sciences, Ningbo University, Ningbo, China
In aquatic animals, the light/dark cycle acts as an important biological factor that influences the entire life cycle. Until present, evidence regarding the regulation of physiological metabolic process under different light/dark cycles is limited in Litopenaeus vannamei. In this study, we mainly investigated the effects of different light/dark cycles (12 h light/12 h dark, 0 h light/24 h dark) on the hepatopancreas metabolism and intestinal microbiota homeostasis in L. vannamei using multiomics techniques. One interesting finding was that the body color of L. vannamei became darker after dark treatment for 8 weeks. Further hepatopancreas transcriptome analysis identified down-regulated genes involved in regulating nutrition metabolism, body-color formation, diurnal rhythm, immune function, hormone levels, and posttranslational modifications. The intestinal microbiota analysis showed that dark treatment-induced alterations in intestinal bacterial abundances in L. vannamei, such as decreased (P < 0.05) relative abundance of Formosa, Demequina, Lutimonas and increased (P < 0.05) relative abundance of Ruegeria, Vibrio, Actibacter, Roseovarius, Ilumatobacter, and Kriegella at the genus level. The microbiota functional analysis demonstrated that the dark treatment mainly increased susceptibility of pathogens, decreased nutrition metabolism, and influenced circadian rhythm. This study indicated for the first time that constant darkness treatment darkened the body color and altered hepatopancreas metabolism and intestinal microbiota homeostasis in L. vannamei, which might give potential clues for improving the productive capacities by changing light/dark cycles in shrimp farming.
Introduction
Most species have developed endogenously driven circadian rhythms in physiology and behavior that are attuned to changes in the daily 12 h light/12 h dark cycle (Hussain and Pan, 2015; Schilperoort et al., 2020). In other words, the photoperiod profoundly influences the circadian rhythm of biochemical, physiological, and behavioral processes in almost living organisms (Wright et al., 2013; Dannerfjord et al., 2021). Epidemiological studies and substantial experimental evidence have repeatedly reported that changes in the light/dark cycles have been closely associated with various metabolic disorders (Rutter et al., 2002; Maury et al., 2010). In aquatic animals, the light/dark cycle acts as an important biological factor that influences the entire life cycle from embryonic development to sexual maturation (Downing and Litvak, 2001; Migaud et al., 2010; Arambam et al., 2020). In recent years, growing evidence has shown that the effects of the light/dark cycle on various species are diverse, some have a natural preference for the dark environment while some have an improved physiological state under high-light intensity (Serra et al., 1999; Ruchin, 2006; Li et al., 2021b; Wei et al., 2021). Therefore, it is a promising research direction for further studies.
The circadian nature of physiology and behavior is regulated by a circadian clock that is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (Dannerfjord et al., 2021). Apart from the SCN, the tissues are capable of generating multiple circadian biological rhythms (Hastings et al., 2018; Pett et al., 2018). Earlier studies have demonstrated the mechanism of dark-light-adaptation changes in crustaceans compound eyes (Migaud et al., 2010). Nevertheless, our understanding regarding the influential mechanism underlying the photoperiods on other tissue is unknown in crustaceans and needs to be investigated further. Currently, the relationship between the light/dark cycle and intestinal microbiota has aroused widespread concern (Peyric et al., 2013; Fortes-Silva et al., 2016; Kim et al., 2019). Furthermore, different light/dark cycles could affect growth, digestibility, and physiological metabolism in fish (Leiner and MacKenzie, 2001; Li et al., 2021b). The crustacean hepatopancreas is an important organ constituting the main site for nutrient digestion, absorption, and metabolism (Li et al., 2021a). Therefore, the hepatopancreas and intestine can be the potential targets for studying the responsive mechanism of shrimp in response to different light/dark cycles.
The Pacific white shrimp Litopenaeus vannamei is considered a paramount aquaculture species, accounting for more than 70% of the total global shrimp production worldwide (Jiao et al., 2020). L. vannamei starts benthic life soon after the mysis stage or the early stage of postlarvae. Currently, the effects of different light/dark cycles on larval survival and growth performance have been studied in L. vannamei and Macrobrachium rosenbergii (Sanudin et al., 2014; Wei et al., 2021). Until present, evidence regarding the regulation of physiological metabolic process under different light/dark cycles is limited in L. vannamei. Thus, in this study, we mainly investigated the effects of different light/dark cycles (12 h light/12 h dark, 0 h light/24 h dark) on the hepatopancreas metabolism and intestinal microbiota in L. vannamei using multiomics techniques. Moreover, body color was also measured. This study might provide an important theoretical basis and reference for further understanding the influence of light/dark cycles on the body color, hepatopancreas metabolism, and intestinal microbiota in shrimp.
Materials and Methods
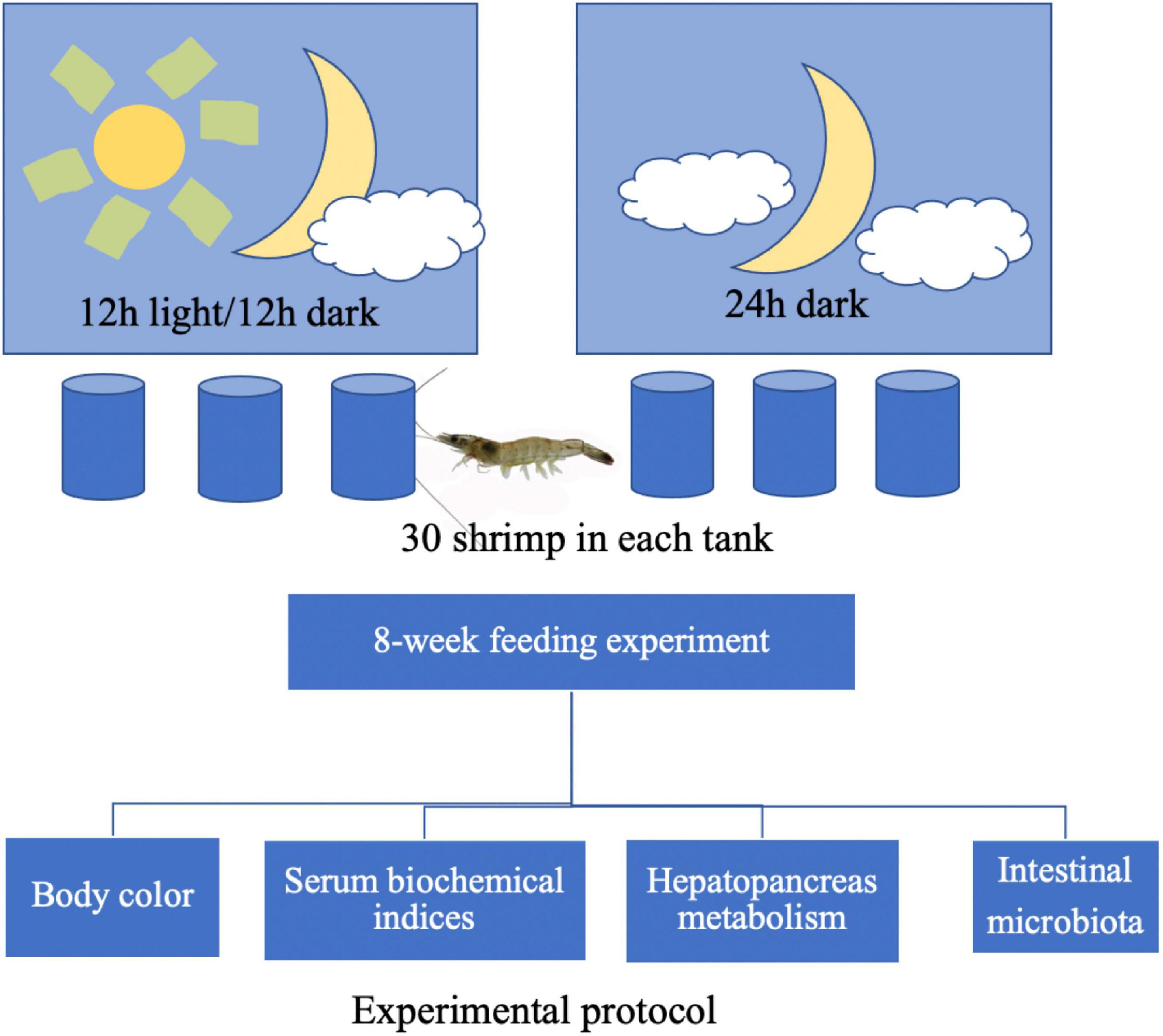
Animals and Design of 8-Week Feeding Experiment
All experimental procedures complied with the Standard Operation Procedures (SOPs) of the Guide for Use of Experimental Animals of Ningbo University.
Litopenaeus vannamei juvenile was obtained from Ningbo Marine Fishery Science and Technology Innovation Base and temporarily reared in a concrete pond with running aerated water (28 ± 2°C, 21.7–23.5 ppt salinity, 4.32–5.5 mg/L dissolved oxygen) prior to the feeding experiment.
Figure 1 shows the experimental protocol. A total of 180 shrimp (approximately 0.72 g per individual) were randomly divided into two groups, namely, the natural light group (12 h light/12 h dark) and dark treatment group (0 h light/24 h dark), with three replicates per group. A total of 30 shrimp were reared in a 100-L cylindrical fiberglass tank, considered as one replicate. After the 8-week feeding trail, body color was captured by digital cameras and then analyzed by the Just Color Picker 5.5 software. Hemolymph samples of six shrimp in each tank were collected from the pericardial cavity and placed into 1.5 ml centrifuge tubes overnight at 4°C before centrifugation (1,811 g, 10 min). The supernatant was mixed as one replicate and collected for the biochemical analysis. The hepatopancreas of six shrimp in each tank was collected rapidly and sterilely, mixed as one sample, and frozen in liquid nitrogen. Similarly, the intestinal content of six shrimp in each tank was collected through a sterile operation, placed into one sterile Eppendorf centrifuge tube, and immediately frozen in liquid nitrogen. Later, intestine and hepatopancreas samples were sent to Hangzhou Mingke Biotechnology Co., Ltd., China for further 16S rDNA-based microbiota and transcriptome analysis, respectively.
Analytical Testing
Hemolymph Biochemical Analysis
Hemolymph triglyceride (TG), total cholesterol (TC), low-density lipoprotein cholesterol (LDL) and high-density lipoprotein cholesterol (HDL), glucose (GLU), glutamic-pyruvic transaminase (ALT), glutamic oxalacetic transaminase (AST), albumin (ALB), and total protein (TP) were determined using Selectra ProM Clinical Chemistry System (ELITech, Sees, France). The parameters were estimated using enzymatic spectroscopic methods on dedicated system packs according to the conventional laboratory protocols. This system has been calibrated, and quality controls were duly maintained before measurement. An independent-sample t-test was used to compare the index. The data was expressed as mean ± SD. P < 0.05 was considered statistically significant.
Hepatopancreas Transcriptome Analysis
Total hepatopancreas RNA was extracted from tissues using TRIzol reagents (Invitrogen, Waltham, MA, United States) and then determined its quality and quantity as previously described (Jiao et al., 2020). Subsequently, mRNA was isolated and fragmented for cDNA synthesis. The sequencing library was constructed and sequenced by HiSeq 2500 platform (Illumina, San Diego, CA, United States).
The raw paired-end reads were trimmed and quality controlled for obtaining clean data. Later, clean data were assembled de novo with Trinity and searched to identify proteins to retrieve their functional annotations. The expression value of each transcript was calculated using fragments per kilobase of exon per million mapped reads method, while the software package RSEM was used for quantifying gene and isoform abundances. P-value < 0.05 and | log2(fold change)| ≥ 1 were identified as the threshold for significance of gene expression differences. In addition, the functional-enrichment analysis including GO and KEGG were performed by Goatools and KOBAS online analysis to identify differential expression genes (DEGs) at Bonferroni-corrected P-value ntial.
Intestinal Microbiome Analysis
Total intestinal genome DNA was extracted using CTAB/SDS method and measured its quality using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The V4-V5 region of the 16S rRNA gene was amplified using primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′) with the unique barcode for PCRs. Sequencing libraries were generated on an Illumina MiSeq platform. Paired-end reads were later harvested for further bioinformatics analysis as previously described (Jiao et al., 2021). Briefly, alpha diversity indexes, Simpson, Chao, Shannon, and Goods coverage were calculated with QIIME (Version 1.7.0), and rarefaction curves were displayed with GraphPad Prism 5. The microbial community distribution of all samples was visualized based on community composition information at the taxonomic levels. An independent-sample t-test was used to compare the microbial community distribution between two groups. The functional analysis of 16S rRNA genebased microbial community samples was predicted using the PICRUSt algorithm and further analyzed statistically by metagenomic profiles (STAMP) (version 2.1.3) package. Linear discriminant analysis Effect Size (LEfSe) analysis shows differentially abundant genera as biomarkers, which was determined using Kruskal-Wallis test (P < 0.05) with a linear discriminant analysis (LDA) score > 3.5.
Results
Body Color of Litopenaeus vannamei
Figure 2 shows the effects of different light/dark cycles on body color in L. vannamei. As shown in Figure 2, compared with the natural light group, the body color of L. vannamei became darker after dark treatment for 8 weeks. The result of the measurement of RGB color value showed that each parameter (i.e., red, green, and blue) value was decreased (P < 0.05) in constant dark treatment compared with the natural light group.
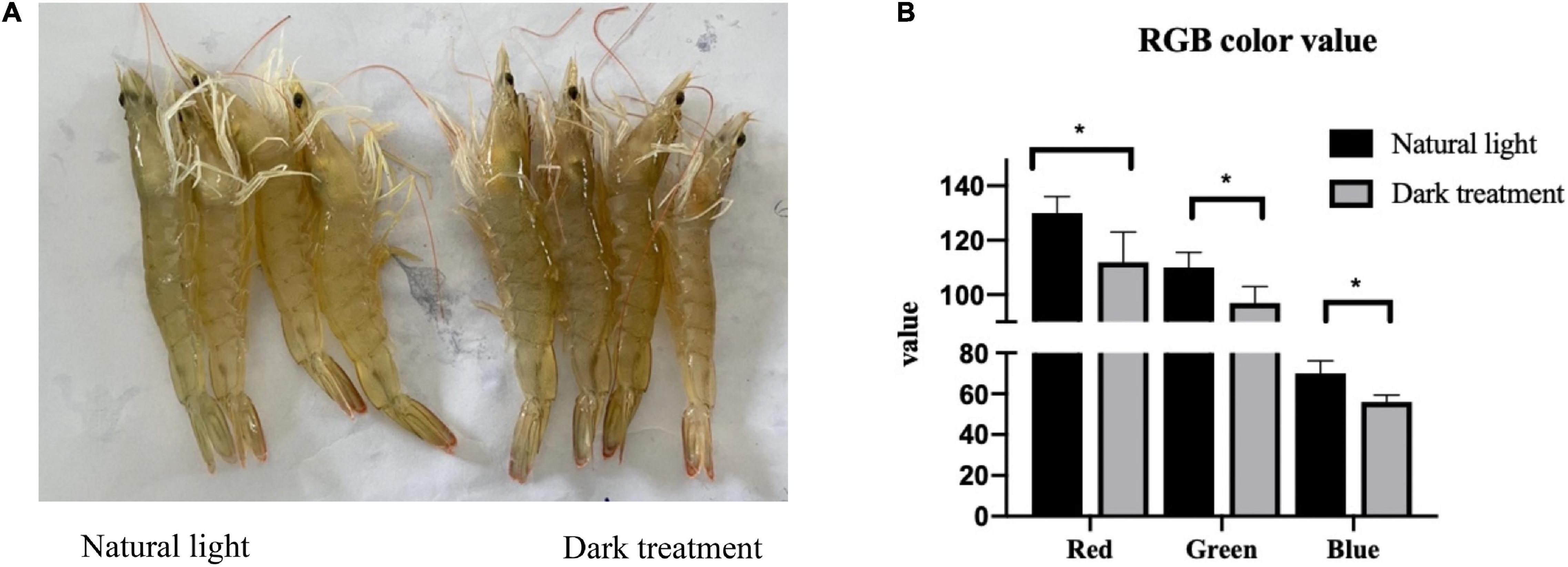
Figure 2. The body color of Litopenaeus vannamei. (A) Image captured by digital cameras. (B) Analysis of red, green, and blue (RGB) color values based on images. *indicated significantly difference among tow groups (P < 0.05).
Serum Biochemical Indexes
Figure 3 shows the effects of different light/dark cycles on serum biochemical indexes in L. vannamei. As shown in Figure 3, compared with the natural light group, constant dark treatment significantly increased (P < 0.05) the serum levels of ALT, AST, LDL, and ALB and decreased (P < 0.05) serum glucose levels in L. vannamei. The serum levels of TP, HDL, TG, and TC were not influenced (P > 0.05) by the constant dark treatment.
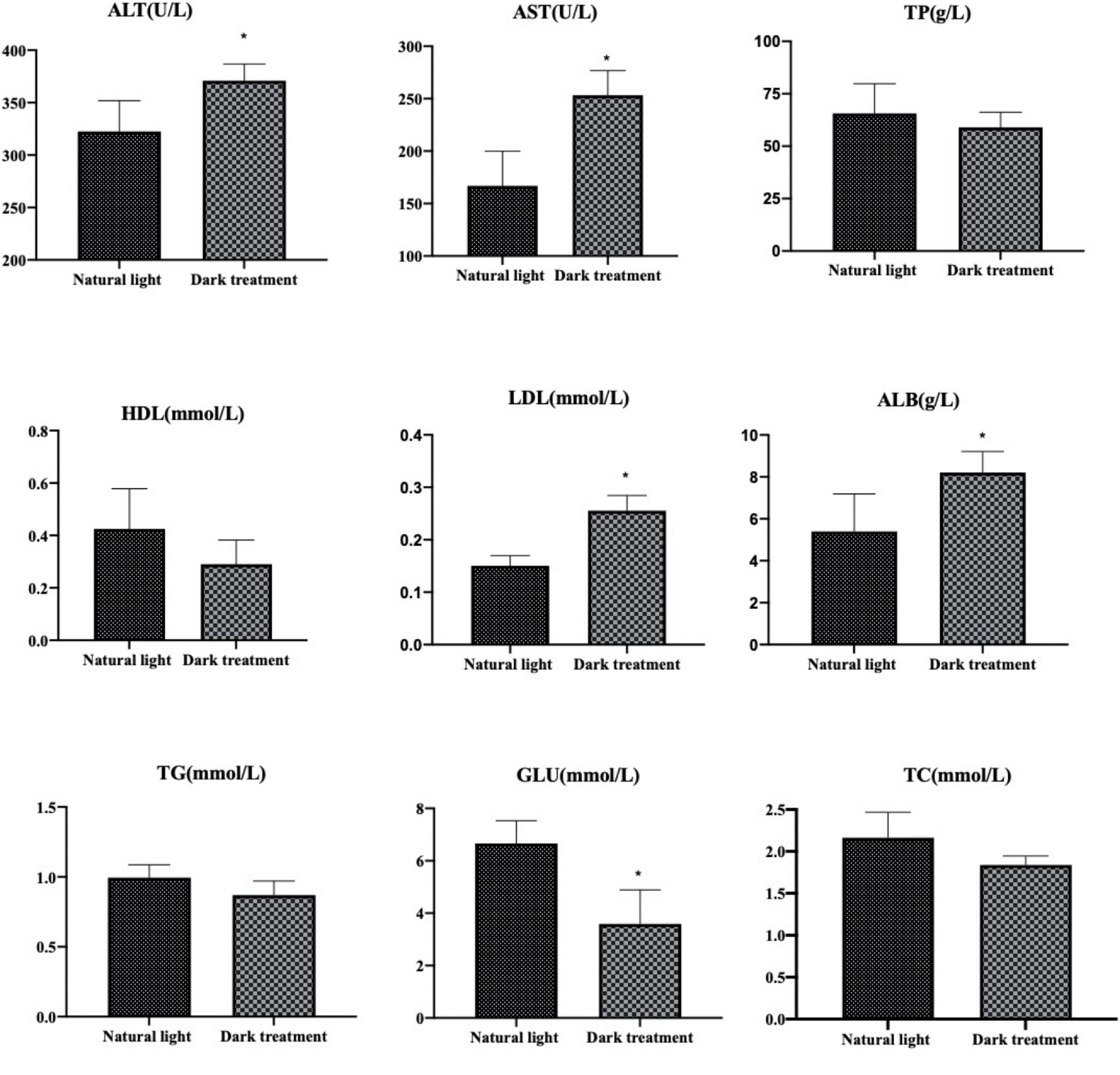
Figure 3. Serum biochemical indexes in Litopenaeus vannamei. *indicated significantly difference among two groups (P < 0.05).
Transcriptome Analysis
Summary of RNA Sequencing
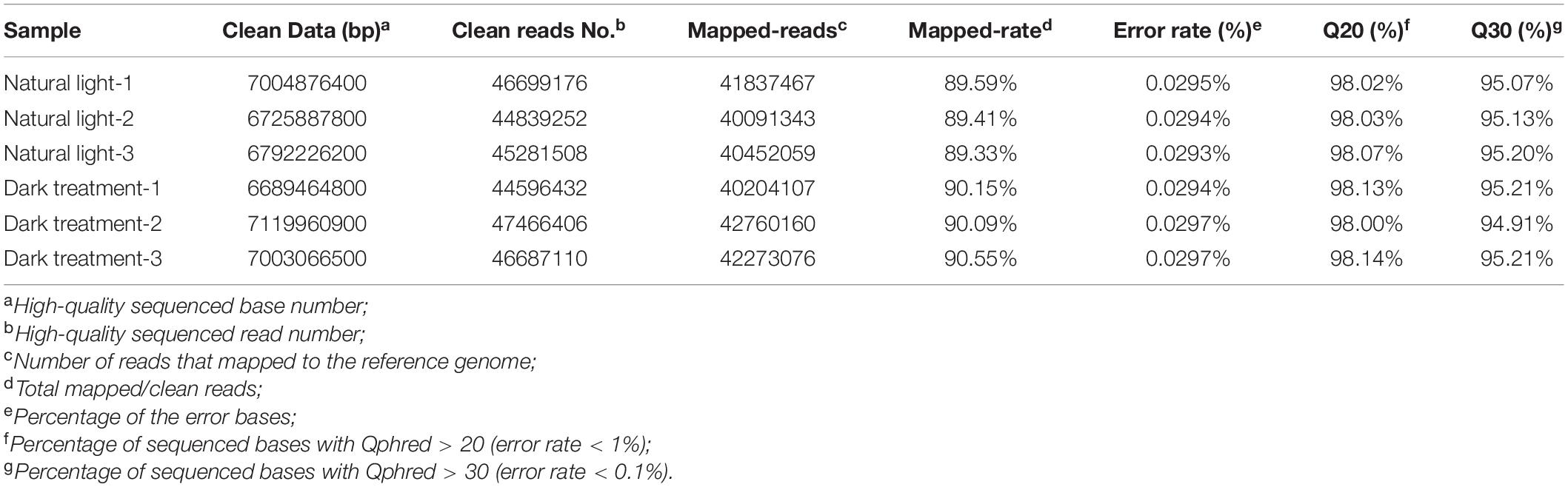
Table 1 shows the two cDNA libraries that were constructed using the hepatopancreas from the natural light group and dark treatment group. In this experiment, 136,819,936 and 138,749,948 clean reads were obtained from the hepatopancreas transcriptome sequencing in the natural light group and dark treatment group, respectively. Approximately, 89.33–90.55% of the reads were mapped onto the reference genome. The percentage of sequenced bases with Qphred > 30 is higher than 94.91%, and the average error rate was 0.0293–0.0297%. The above summary of RNA sequencing showed that good-quality sequence data were harvested for further bioinformatics analysis.
Identification of Differentially Expressed Genes
As shown in Figure 4, the heatmap plot (A) and a volcano plot (B) present the difference analysis of expressed genes between the natural light group and dark treatment group in the hepatopancreas of L. vannamei. In this study, we can see that three replicates in the natural light group (i.e., natural light-1, natural light-2, and natural light-3) were clustered together and three replicates in the dark treatment group (i.e., dark treatment-1, dark treatment-2, and dark treatment-3) were clustered together, indicating that gene expression in the hepatopancreas of L. vannamei was significantly different between the two groups. Furthermore, the volcano plot identified 483 DEGs, among which 223 genes were upregulated and 260 genes were downregulated.
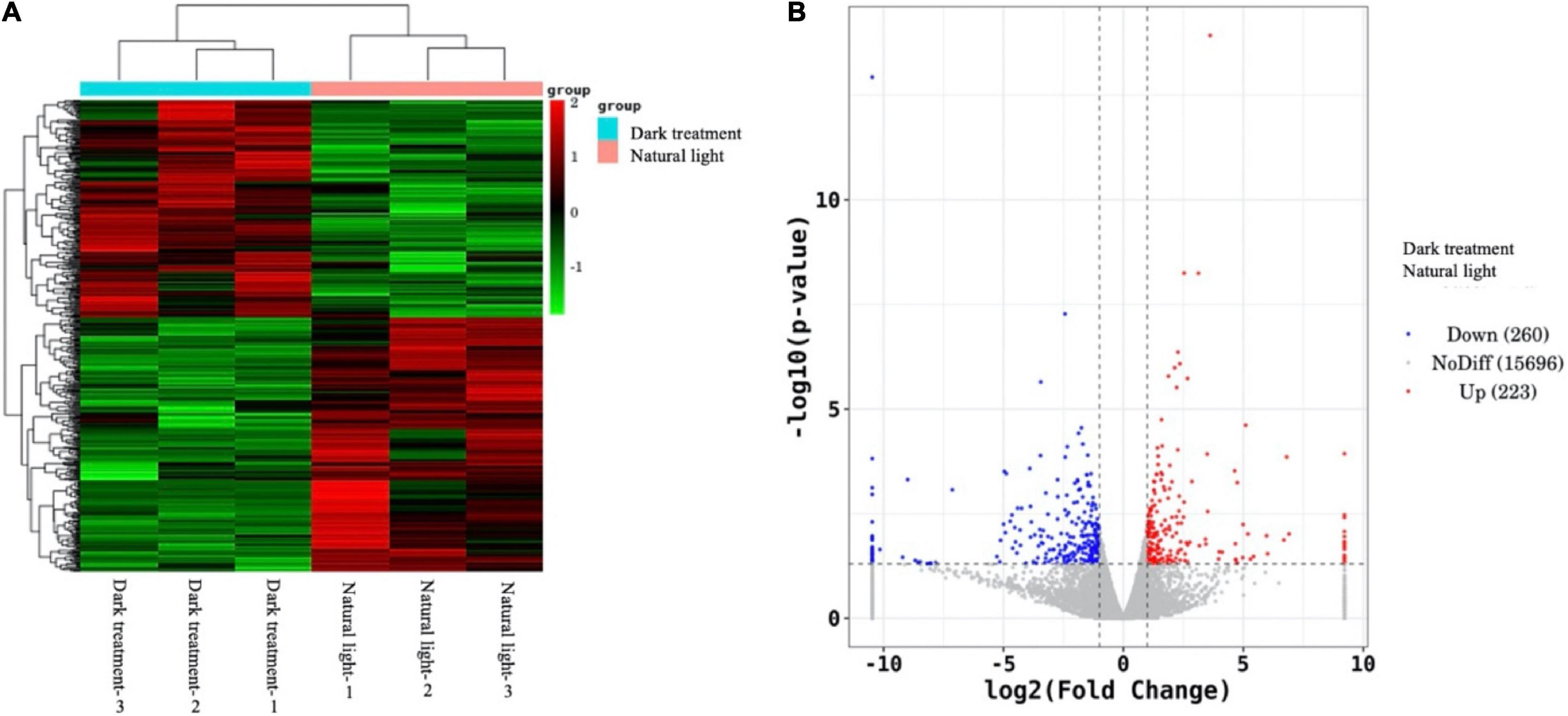
Figure 4. Difference analysis of expressed genes between the natural light group and dark treatment group in the hepatopancreas of Litopenaeus vannamei. The heatmap plot (A) denotes clusters of samples, while the volcano plot (B) represents statistics of the total expressed genes that include upregulated genes (indicated as red dots), no difference genes (indicated as gray dots), and downregulated genes (indicated as blue dots).
KEGG Enrichment Analysis
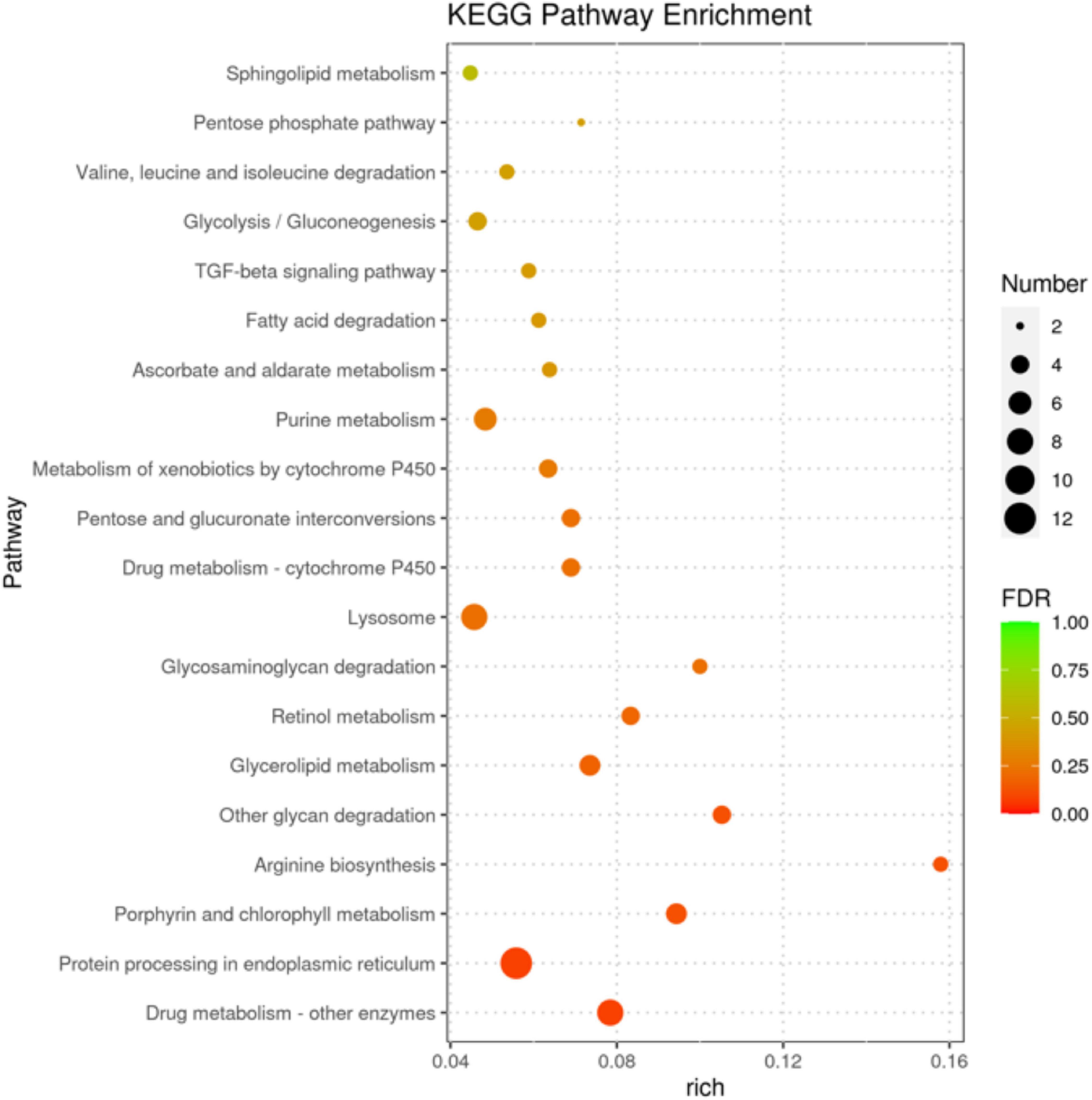
In Figure 5, the senior bubble chart showed the top 20 enrichment pathways, namely, protein processing in the endoplasmic reticulum, drug metabolism-other enzymes, carbohydrate metabolism (i.e., glycolysis/gluconeogenesis, pentose phosphate pathway, ascorbate and aldarate metabolism, pentose phosphate pathway, and amino sugar and nucleotide sugar metabolism), metabolism of cofactors and vitamins (i.e., porphyrin and chlorophyll metabolism), amino acid metabolism (i.e., arginine biosynthesis and valine, leucine, and isoleucine degradation), lipid metabolism (i.e., glycerolipid metabolism, fatty acid degradation, and sphingolipid metabolism), transport and catabolism (i.e., lysosome and phagosome), and metabolism of cofactors and vitamins (i.e., porphyrin and chlorophyll metabolism and retinol metabolism).
Identification of Key Differentially Expressed Genes
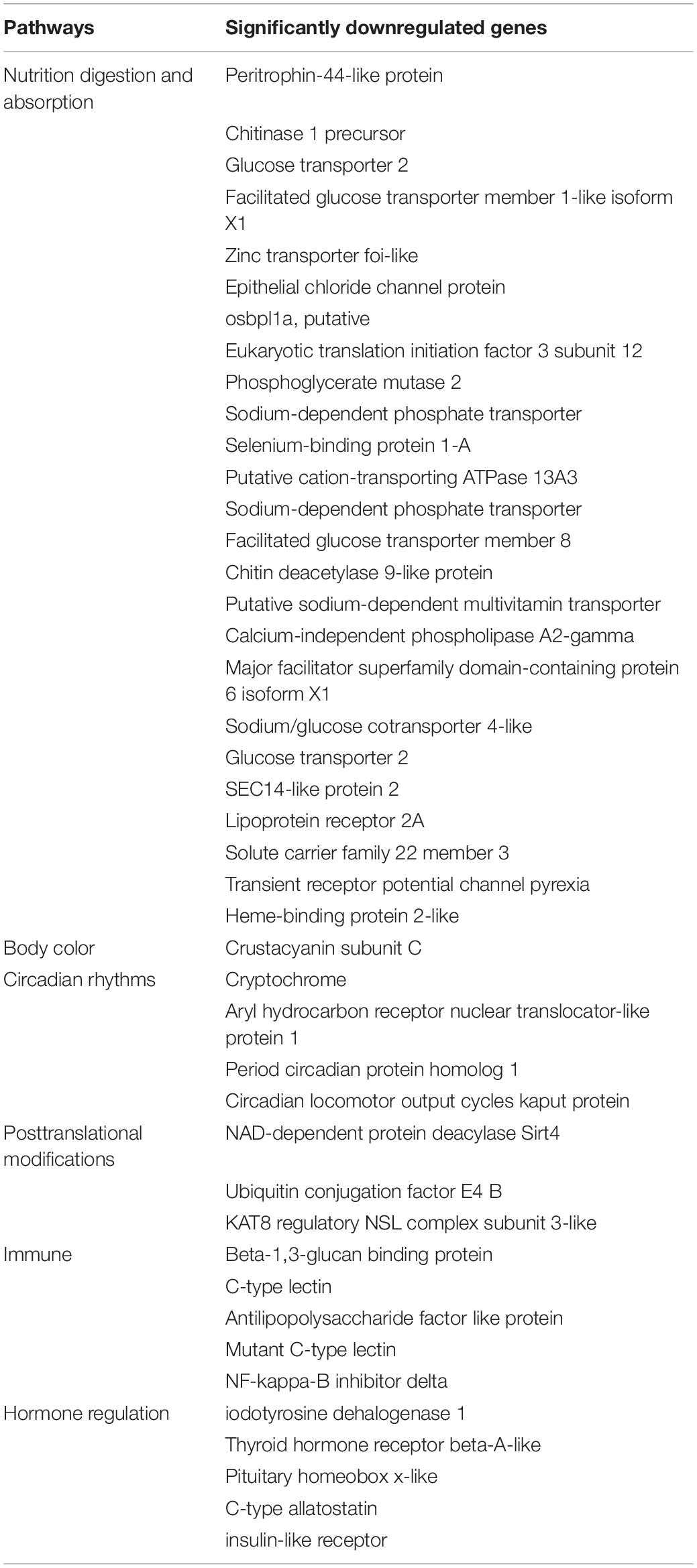
As shown in Table 2, key genes regulating nutrition metabolism, body color formation, diurnal rhythm, immune function, hormone levels, and posttranslational modifications were downregulated after constant darkness for 8 weeks.
Intestinal Microbiota Analysis
Characteristics of Sequencing Results
As Figure 6A shown, 2,61,195 high-quality reads were produced, with an average of 38,086 ± 1,499 in the natural light group and 48,979 ± 9,345 in the dark treatment group. The rarefaction curves tended toward the saturation plateau (Figure 6B), while the coverage estimations of Good revealed that the amounts of obtained bacterial species in the natural light group and dark treatment group were 0.9993 and 0.9997, respectively, which indicates an adequate sequencing coverage that was able to capture the diversity of the bacterial communities. OTUs, the diversity index (Shannon and Simpson), and the estimators of the community richness index (Chao) were summarized. No significant differences (P > 0.05) were observed in OTUs, richness index (Chao), and diversity index (Shannon and Simpson) between the natural light group and dark treatment group.
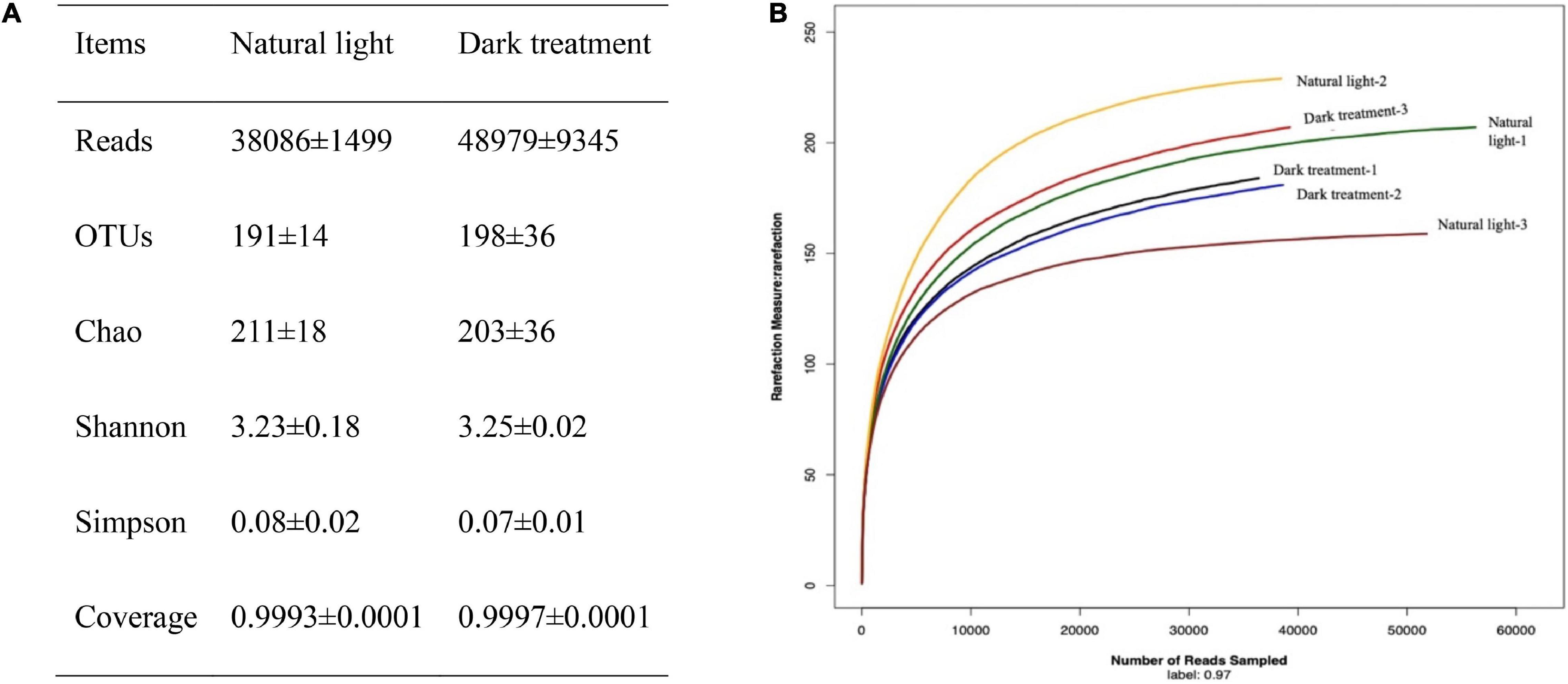
Figure 6. Characteristics of sequencing results. (A) Summary of sequencing data and (B) rarefaction analysis. Rarefaction curves of OTUs clustered at 97% sequence identity across the sample. The samples labeled with natural light-1, natural light-2, and natural light-3 corresponded to three replicates of the natural light group; the samples labeled with dark treatment-1, dark treatment-2, and dark treatment-3 corresponded to three replicates of the dark treatment group.
Comparison of the Intestinal Microbiota Composition
The taxonomic assignment of all sequences was used to assess the microbial composition at the phylum and genus level as summarized in Figures 7A,C. At the phylum level, the intestinal microbiota is mainly composed of Bacteroidetes and Proteobacteria. Compared with the natural light group, dark treatment significantly decreased (P < 0.05) the relative abundance of intestinal Bacteroidetes and increased (P < 0.05) the relative abundance of Proteobacteria at the phylum level of L. vannamei (Figure 7B). At the genus level, dark treatment significantly decreased (P < 0.05) the relative abundance of Formosa, Demequina, and Lutimonas and increased (P < 0.05) the relative abundance of Ruegeria, Vibrio, Actibacter, Roseovarius, Ilumatobacter, and Kriegella at the intestine of L. vannamei (Figure 7D).
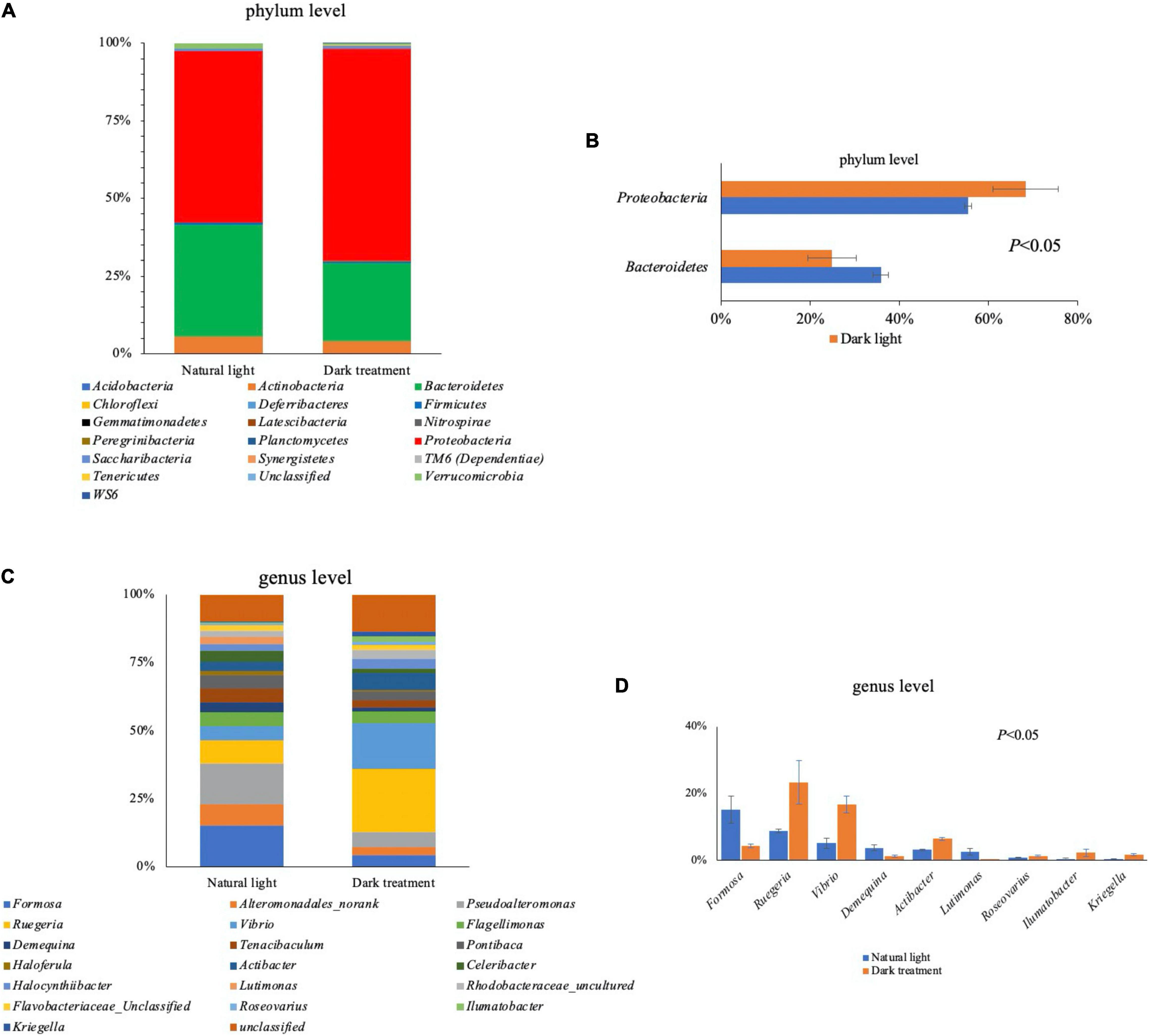
Figure 7. Intestinal microbiota in Litopenaeus vannamei. Overall bacteriome composition at the phylum (A) and genus (C) level; significant different microbiota composition between the natural light group and dark treatment group at the phylum (B) and genus (D) levels.
In Figure 8, we applied the LEfSe to distinguish the specific bacterial taxa that varied in relative abundance by different treatments. As more bacteria were identified from phyla to species with LDA scores > 2.0, we raised the criteria and selected specific bacterial taxa with LDA scores > 3.5. It could be observed that Ruegeria, Vibrio, Actibacter, Simiduia, Ilumatobacter, Cellvibrionales, Kriegella, Tamlana, Muricauda, and Iamia were biomarkers in the dark treatment group, while Formosa, Pseudoalteromonas, Lutimonas, Celeribacter, Demequina, Bacteroidales, and Halieaceae were biomarkers in the natural light group.
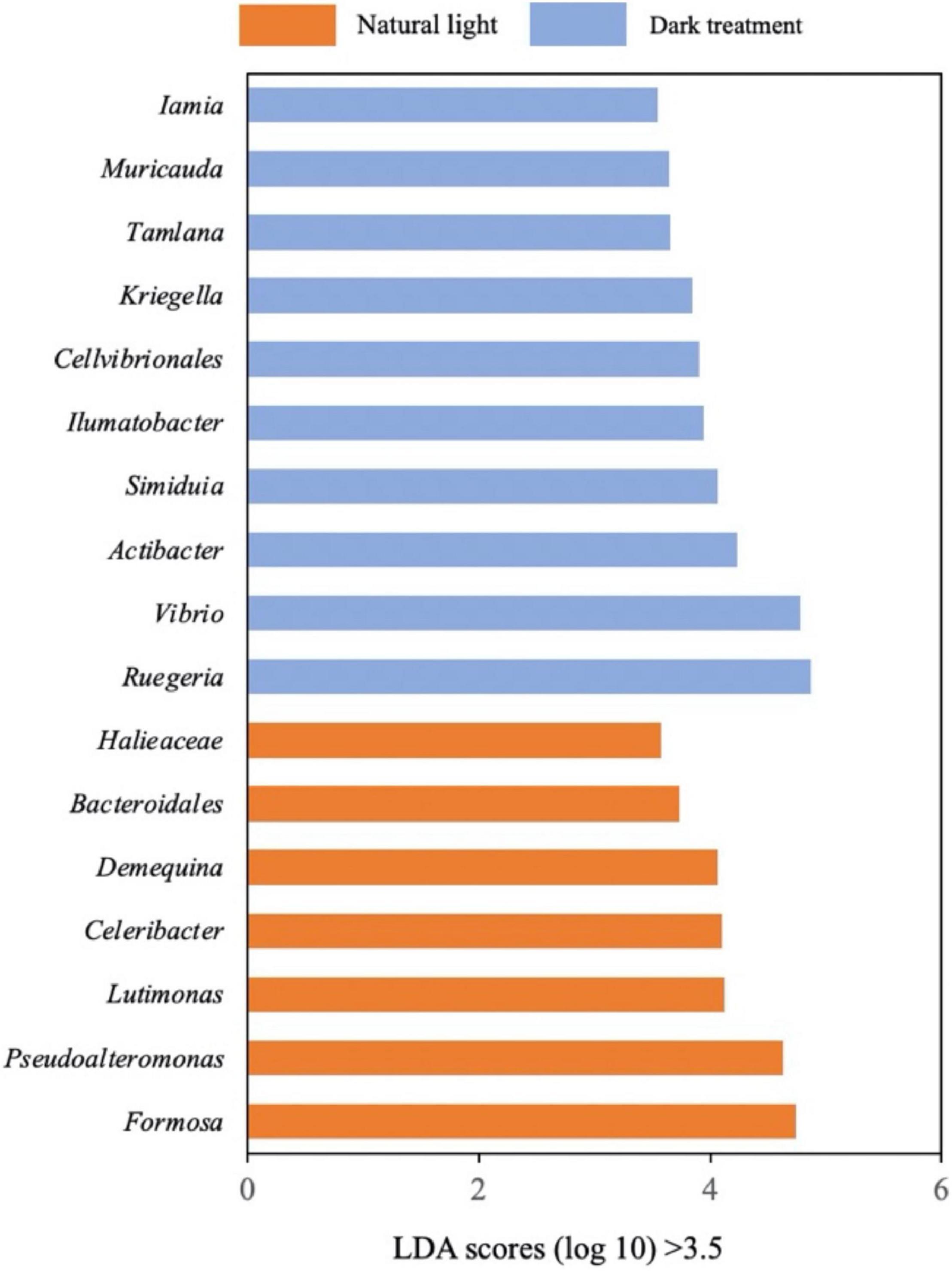
Figure 8. Linear discriminant analysis Effect Size (LEfSe). Differentially abundant bacteria were identified as biomarkers using the Kruskal–Wallis test (P < 0.05) with a linear discriminant analysis (LDA) score of > 3.5.
Functional Prediction of the Intestinal Microbial Community
In Figure 9, significant differences were observed in the KEGG pathways between the natural light group and the dark treatment group. Compared with the natural light group, the dark treatment group significantly increased (P < 0.05) KEGG pathways involving mainly in the two-component system, bacterial chemotaxis, phosphotransferase system (PTS), Vibrio cholerae infection, Staphylococcus aureus infection, and biofilm formation of Vibrio cholerae. The dark treatment group significantly decreased (P < 0.05) KEGG pathways involving mainly metabolism (e.g., nitrogen metabolism, sulfur metabolism, and galactose metabolism), circadian rhythm, and biosynthesis of the siderophore group non-ribosomal peptides.
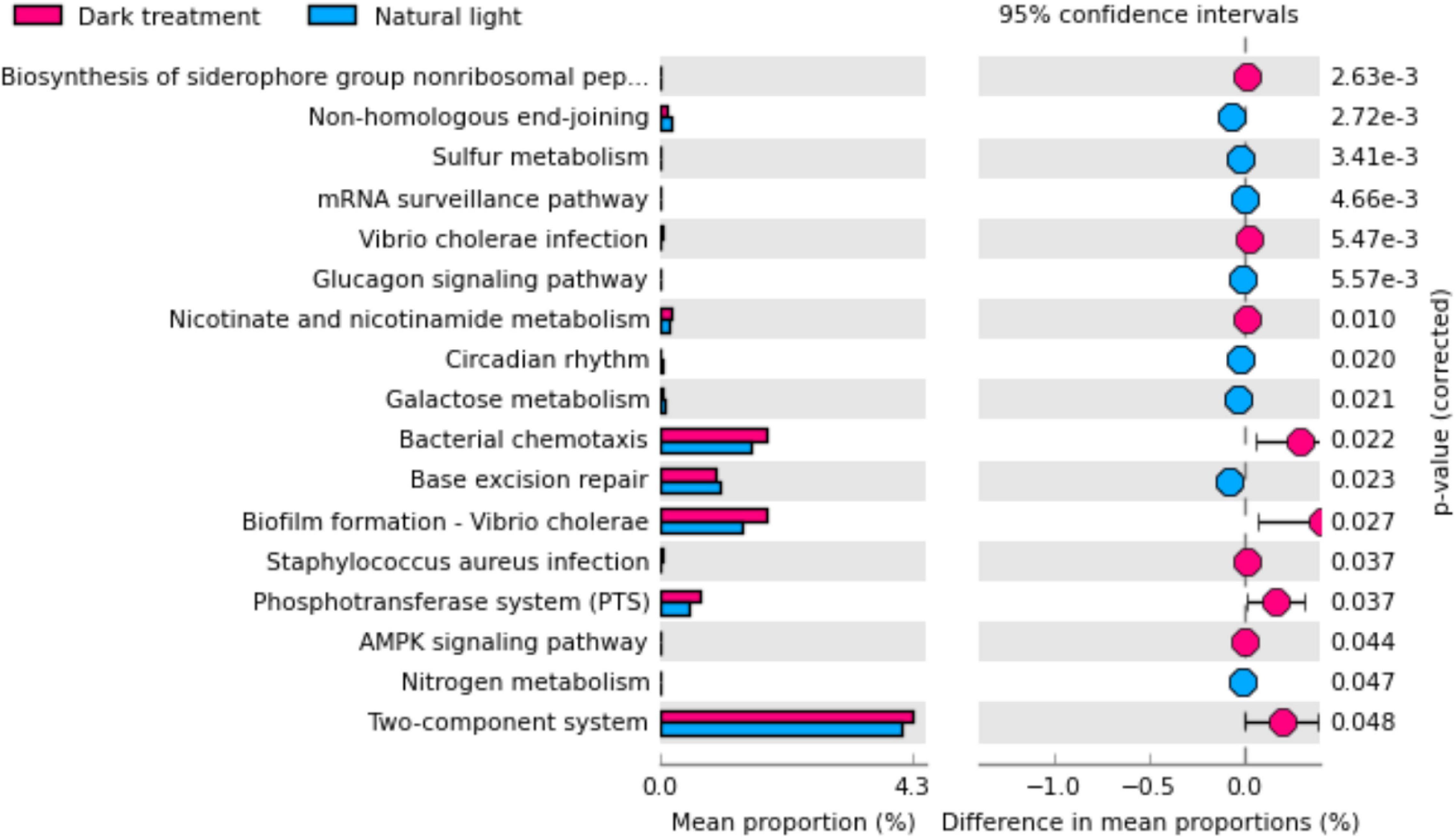
Figure 9. The functional analysis was predicted from 16S rRNA gene-based microbial compositions. Differences were considered significant at P < 0.05 using an independent t-test.
Discussion
The change in somatic color in crustacea has received a great deal of attention as a conspicuous and quantifiable phenomenon related to physiological and ecological factors. Crustacean showed an ability to change coloration in response to photoperiod, which may play roles including photoprotection and enhancing camouflage in a unique marine environment (Russell and Dierssen, 2018). In our study, one interesting finding was that the body color of L. vannamei became darker after dark treatment for an 8-week feeding trail, which may be related to the decreased gene expression of crustacyanin subunit C (from transcriptome data). Although the molecular mechanism of the crustacean body color change is not clear, several studies have indicated that the regulation of body color can be associated with gene expression of crustacyanin (Zhao et al., 2021). Astaxanthin, one of the carotenoids found in nature, appears to be the main pigment responsible for color in crustaceans, accounting for approximately 65–98% of all the carotenoids found in shrimp species (Wade et al., 2005; Ertl et al., 2013). The stability of the highly reactive astaxanthin pigment is provided by crustacyanin, a protein that binds to the pigment to form a carotenoprotein complex, which stabilizes the pigment as well as the tertiary and quaternary structures of proteins (Wade et al., 2009). Crustacyanin subunit A and C expression levels were lower in dark shrimp than that in light shrimp (Ertl et al., 2013). In addition, it has been reported that L. vannamei infected with Vibrio parahaemolyticus, V. fluvialis, V. mimicus, V. alginolyticus, and Vibrio sp. found to take less feed and body color tended to be darker (Mastan, 2015). Later, the intestinal microbiota analysis confirmed that the relative number of Vibrio has been increased after the dark treatment. Taken together, we suspected that dark treatment decreased the gene expression of crustacyanin subunit C and increased intestinal Vibrio number and, therefore, changed body color in L. vannamei. However, currently, the molecular mechanism remains unclear, which allows us to dig deeper into the possible mechanisms in the future.
Serum enzymes such as AST and ALT are generally used in assessing liver function (Senior, 2012). We found that dark treatment elevated serum levels of AST and ALT, which are usually considered as a sign of hepatocellular damage (Panteghini et al., 1983). The transcriptomics data further revealed the influence of different photoperiods (12 h light/12 h dark, 0 h light/24 h dark) on the metabolism in L. vannamei. We observed that DEGs mostly involved in nutrient digestion, absorption, and transport were significantly suppressed after dark treatment for the 8-week feeding trail, which might partially explain the changed serum levels of ALB, LDL, and glucose. In detail, digestive enzymes (i.e., chitinase and chitin deacetylase 9-like protein), nutrition transport regulation (i.e., glucose transporters, ion/metal transporters, putative sodium-dependent multivitamin transporter, transient receptor potential channel pyrexia, solute carrier family 22 members, and selenium-binding protein 1-A), and lipoprotein binding (i.e., lipoprotein receptor 2A, SEC14-like protein 2, and osbpl1a) genes were downregulated. Similarly, Sanudin et al. (2014) reported that the ingestion rate of frozen Artemia nauplii under dark conditions (24 h dark) was lower than that under light conditions (24 h light) in the smallest shrimp (total length of 0.5 cm) (Sanudin et al., 2014).
Most organisms have evolved an internal circadian clock that drives circadian rhythms in metabolism, physiology, and behavior (Ko and Takahashi, 2006). The circadian clocks use a 24 h light-dark cycle as the environmental signal (zeitgeber) to establish endogenous circadian timing systems that synchronize biological functions (Roenneberg et al., 2013). In our study, we observed that circadian clock genes including cryptochrome, aryl hydrocarbon receptor nuclear translocator-like protein 1 (BMAL1), period circadian protein homolog 1, and circadian locomotor output cycles kaput protein were downregulated under constant darkness treatment in the hepatopancreas of L. vannamei. In similar to our data, continuous darkness decreased the mRNA levels of clock genes cryptochrome 1 (Cry1) and period 2 (Per2) in the eyestalk of L. amboinensis (Choi et al., 2019). In mammals, darkness exposure for 8 weeks decreased the mRNA levels of BMAL1 in the brain and muscle and decreased the mRNA levels of Per1 and Per2 in the liver (Li et al., 2020).
Molecular insights into the mechanisms of circadian rhythms have provided clues that posttranslational modifications control the circadian clock function (Gallego and Virshup, 2007). Posttranslational modifications regulate the localization, degradation, and activity of these clock genes by methylation, phosphorylation, acetylation, sumoylation, ubiquitination, etc. We observed that darkness exposure downregulated genes, which were associated with protein ubiquitination (i.e., ubiquitin conjugation factor E4 B), acetylation (i.e., KAT8 regulatory NSL complex subunit 3 and NAD-dependent protein deacylase Sirt4), and methylation (i.e., thiopurine S-methyltransferase-like). Ubiquitin conjugation factor E4 B functions as an E4 ligase that mediates the assembly of polyubiquitin chains on substrates ubiquitinated by another E3 ubiquitin ligase (Hatakeyama et al., 2001). NAD-dependent protein deacylase SIRT4 is a lysine deacetylase and acts as a crucial player in maintaining insulin secretion and glucose homeostasis (Anderson et al., 2017). As part of the NSL complex, KAT8 regulatory NSL complex subunit 3 is involved in the acetylation of nucleosomal histone H4 on several lysine residues and may be involved in the regulation of transcription (Cai et al., 2010). Thiopurine S-methyltransferase-like drives methylation by directing thiopurines [6-mercaptopurine (6MP)] toward methylated entities [6-methylmercaptopurine (6MMP)] and, thus, decreases levels of active 6-thioguanine nucleotides (Urbancic et al., 2019).
A marked oscillation in hormone exhibited daily rhythms under light/dark cycles (Bayarri et al., 2004; Oliveira et al., 2013). Merging evidence suggests that disturbance of the circadian rhythm of the body increases endocrine disorder risk (Gamble et al., 2014; Zimmet et al., 2019). We found that constant darkness exposure exhibited decreased several gene expressions, which was involved in hormone regulation, including insulin-like receptor, C-type allatostatin, iodotyrosine dehalogenase 1, pituitary homeobox x-like, and thyroid hormone receptor beta-A-like in the hepatopancreas of L. vannamei. In invertebrates, the role of the insulin pathways includes not only glucose homeostasis but also the regulation of a variety of fundamental processes, such as growth, aging, and reproduction (Claeys et al., 2002). As the pivotal protein in any insulin family-based signaling pathway, the insulin-like receptor is responsible for mediating the signal carried by insulin-like peptides (ILPs) from the intercellular to the intracellular environment (Sharabi et al., 2016). A similar result was reported that constant darkness exposure exhibited decreased BMAL1 expression and inhibition of the NAMPT/NAD + /SIRT1 pathway, further suppressed GLUT4 expression, and lead to insulin resistance in the liver cells (Li et al., 2020). C-type allatostatin is named because of its potent inhibitory effect on juvenile hormone biosynthesis by the corpora allata in insects and also modulates the immune response in the hepatopancreas of mud crab Scylla paramamosain (Xu et al., 2020). The thyroid hormone receptors are members of the steroid receptor superfamily that mediate the action of thyroid hormone signaling in numerous tissues to regulate important physiological and developmental processes (Anyetei-Anum et al., 2018). Iodotyrosine dehalogenase 1 is a transmembrane protein involved in the encoding iodotyrosine deiodinase, and it controls the reuse of iodide for thyroid hormone synthesis (Gnidehou et al., 2004). Pituitary homeobox plays an important role in regulating the transcription of enzymes involved in steroidogenesis (Hiroi et al., 2003). Our results provide the first evidence that constant dark treatment could influence hormone regulation in the hepatopancreas of L. vannamei.
The involvement of the light-dark cycle as an important regulator of immune functions has been extensively described in mammals, but there is a paucity of information on the influence of this biological phenomenon in aquatic animals. Limited studies have reported that the innate immune system had a circadian rhythm based on the light-dark cycle in fish (Esteban et al., 2006; Ceballos-Francisco et al., 2020). Serum-mediated bacterial killing activities and enzyme activities associated with immune defense (i.e., lysozyme, alkaline phosphatase, and myeloperoxidase) reached the highest during the light phase in rainbow trout (Oncorhynchus mykiss) (Lazado et al., 2018). High-light intensity (16D:8L and 0D:24L dark/light cycles) increased lymphocyte count and improved the overall physiological state of carp (Cyprinus carpio L.) yearlings, while zero illumination had an inverse effect (Ruchin, 2006). We found that dark treatment significantly suppressed immune-related gene expression in the hepatopancreas of L. vannamei, including beta-1,3-glucan binding protein, C-type lectin, antilipopolysaccharide factor-like protein, and NF-kappa-B inhibitor delta. Beta-1,3-glucan binding protein might act as a pathogenic recognition protein to activate shrimp immune defense against invasive pathogens by the agglutination, binding, and enhancing encapsulation and phenoloxidase activity of the hemocytes (Phupet et al., 2018). C-type lectin could inhibit the proliferation of hemolymph microbiota by maintaining the expression of antimicrobial peptides in shrimp (Wang et al., 2014). An antilipopolysaccharide factor is a small protein that has been verified responsive to different pathogen challenges and with broad-spectrum antimicrobial activities (Li et al., 2015). NF-kappa-B inhibitor delta functions in the regulation of inflammatory responses and regulates the expression of IL-2, IL-6, and other cytokines through regulation of the NF-kappa-B activity (Arnold et al., 2012). These results indicated that constant dark treatment impaired the immune function in the hepatopancreas of L. vannamei.
Intestinal microbiota modulates host physiological processes and plays an important role in promoting and maintaining the health of the host. Dark treatment significantly increased the relative abundance of genus Ruegeria, Vibrio, Actibacter, Roseovarius, Ilumatobacter, and Kriegella at the intestine of L. vannamei. It is worth noting that dark treatment promoted the proliferation of pathogenic bacteria. Vibrio spp. is the most typical and well-known pathogen causing vibriosis infections in aquatic animals (Arunkumar et al., 2020). As a member of the Roseobacter clade, Roseovarius crassostreae is a pathogenic bacteria of Roseovarius Oyster Disease and has resulted in mass mortalities of cultured Crassostrea virginica in the northeastern United States (Maloy et al., 2007). Furthermore, microbiota KEGG functional analysis demonstrated that dark treatment increased the abundance of pathways in Vibrio cholerae infection, Staphylococcus aureus infection, and biofilm formation of Vibrio cholerae. Besides, the relative abundance of Ruegeria was increased after the dark treatment at the intestine of L. vannamei. Ruegeria is a globally distributed gram-negative marine bacterium, which could produce the antibacterial compound tropodithietic acid and biofilm formation against several marine pathogens (Garcia et al., 2014). Ilumatobacter genome includes two type I polyketide synthases. The type I polyketide synthases can catalyze the synthesis of polyketides, which are highly effective antibiotics (Zhao et al., 2019). The above findings suggested that increased Ruegeria and Ilumatobacter might help combat pathogenic bacteria. Moreover, the dark treatment significantly decreased the relative abundance of Formosa, Demequina, and Lutimonas. Demequina can help its host absorb and utilize carbohydrates by producing α-amylase, xylanase, and cellulase (Al-Naamani et al., 2015). Lutimonas is a strictly aerobic heterotrophic nitrifying bacterium for the degradation of ammonia (Fu et al., 2009). To date, less is known about the physiological function of Actibacter, Kriegella, and Formosa, which needs to be further clarified in the future.
Conclusion
The overall result generated from our findings indicated that constant darkness darkened body color and altered hepatopancreas metabolism and intestinal microbiota homeostasis in L. vannamei. Genes involved in regulating nutrition metabolism, body color formation, diurnal rhythm, immune function, hormone levels, and posttranslational modifications were downregulated after constant darkness for 8 weeks. Further intestinal microbiota analysis showed that dark treatment-induced alterations in intestinal bacterial abundances and circadian rhythm increased susceptibility of pathogens, and decreased nutrition metabolism. The results of this study would provide an important theoretical basis and reference for further understanding of the impact of different light/dark cycles in shrimp physiological changes, which might give clues for improving the productive capacities by changing light/dark cycles in shrimp farming.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA761846.
Ethics Statement
The animal study was reviewed and approved by Ethics-Scientific Committee for Experiments on Animals of Ningbo University.
Author Contributions
LFJ and QCZ designed and performed the experiment, analyzed the data, and drafted the manuscript. TMD helped to perform the experiment and collected samples. XYT and JJL helped to revise the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by National Key R&D Program of China (2018YFD0900400), Ningbo Public Welfare Science and Technology Project (202002N3041), Industrial Chain Collaborative Innovation Project of the Demonstration Work on Innovative Development of the Marine Economy of the State Oceanic Administration (NBHY-2017-S2), and KC Wong Magna Fund in Ningbo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Naamani, L., Dobretsov, S., Al-Sabahi, J., and Soussi, B. (2015). Identification and characterization of two amylase producing bacteria Cellulosimicrobium sp. and Demequina sp. isolated from marine organisms. J. Agricult. Mar. Sci. 20, 8–15. doi: 10.24200/jams.vol20iss0pp8-15
Anderson, K. A., Huynh, F. K., Fisher-Wellman, K., Stuart, J. D., Peterson, B. S., Douros, J. D., et al. (2017). SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab, 83:e15. doi: 10.1016/j.cmet.2017.03.003
Anyetei-Anum, C. S., Roggero, V. R., and Allison, L. A. (2018). Thyroid hormone receptor localization in target tissues. J. Endocrinol. 237, R19–R34.
Arambam, K., Singh, S. K., Biswas, P., Patel, A. B., Jena, A. K., and Pandey, P. K. (2020). Influence of light intensity and photoperiod on embryonic development, survival and growth of threatened catfish, Ompok bimaculatus early larvae. J. Fish Biol. 97, 740–752. doi: 10.1111/jfb.14428
Arnold, C. N., Pirie, E., Dosenovic, P., McInerney, G. M., Xia, Y., Wang, N., et al. (2012). A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc. Natl. Acad. Sci. U.S.A. 109, 12286–12293. doi: 10.1073/pnas.1209134109
Arunkumar, M., LewisOscar, F., Thajuddin, N., Pugazhendhi, A., and Nithya, C. (2020). In vitro and in vivo biofilm forming Vibrio spp: A significant threat in aquaculture. Proc. Biochem. 94, 213–223. doi: 10.1016/j.procbio.2020.04.029
Bayarri, M. J., Rodriguez, L., Zanuy, S., Madrid, J. A., Sanchez-Vazquez, F. J., Kagawa, H., et al. (2004). Effect of photoperiod manipulation on the daily rhythms of melatonin and reproductive hormones in caged European sea bass (Dicentrarchus labrax). Gen Comp. Endocrinol. 136, 72–81. doi: 10.1016/j.ygcen.2003.12.004
Cai, Y., Jin, J., Swanson, S. K., Cole, M. D., Choi, S. H., Florens, L., et al. (2010). Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 285, 4268–4272. doi: 10.1074/jbc.C109.087981
Ceballos-Francisco, D., Cuesta, A., and Esteban, M. A. (2020). Effect of light-dark cycle on skin mucosal immune activities of gilthead seabream (Sparus aurata) and European sea bass (Dicentrarchus labrax). Fishes Basel 5:10. doi: 10.3390/fishes5010010
Choi, J. Y., Choi, Y. U., Kho, J., and Choi, C. Y. (2019). Effects of various photoperiods and specific wavelengths on circadian rhythm in ornamental cleaner shrimp Lysmata amboinensis. Biol. Rhythm. Res. 50, 897–907. doi: 10.1080/09291016.2018.1502237
Claeys, I., Simonet, G., Poels, J., Van Loy, T., Vercammen, L., De Loof, A., et al. (2002). Insulin-related peptides and their conserved signal transduction pathway. Peptides 23, 807–816. doi: 10.1016/s0196-9781(01)00666-0
Dannerfjord, A. A., Brown, L. A., Foster, R. G., and Peirson, S. N. (2021). Light input to the mammalian circadian clock. Methods Mol. Biol. 2130, 233–247. doi: 10.1007/978-1-0716-0381-9_18
Downing, G., and Litvak, M. K. (2001). The effect of light intensity and spectrum on the incidence of first feeding by larval haddock. J. Fish. Biol. 59, 1566–1578. doi: 10.1111/j.1095-8649.2001.tb00221.x
Ertl, N. G., Elizur, A., Brooks, P., Kuballa, A. V., Anderson, T. A., and Knibb, W. R. (2013). Molecular characterisation of colour formation in the prawn Fenneropenaeus merguiensis. PLoS One 8:e56920. doi: 10.1371/journal.pone.0056920
Esteban, M. A., Cuesta, A., Rodriguez, A., and Meseguer, J. (2006). Effect of photoperiod on the fish innate immune system: a link between fish pineal gland and the immune system. J. Pineal Res. 41, 261–266. doi: 10.1111/j.1600-079X.2006.00362.x
Fortes-Silva, R., Oliveira, I. E., Vieira, V. P., Winkaler, E. U., Guerra-Santos, B., and Cerqueira, R. B. (2016). Daily rhythms of locomotor activity and the influence of a light and dark cycle on gut microbiota species in tambaqui (Colossoma macropomum). Biol. Rhythm Res. 47, 183–190. doi: 10.1080/09291016.2015.1094972
Fu, S., Fan, H., Liu, S., Liu, Y., and Liu, Z. (2009). A bioaugmentation failure caused by phage infection and weak biofilm formation ability. J. Environ. Sci. 21, 1153–1161. doi: 10.1016/s1001-0742(08)62396-7
Gallego, M., and Virshup, D. M. (2007). Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148. doi: 10.1038/nrm2106
Gamble, K. L., Berry, R., Frank, S. J., and Young, M. E. (2014). Circadian clock control of endocrine factors. Nat. Rev. Endocrinol. 10, 466–475. doi: 10.1038/nrendo.2014.78
Garcia, M. J. P., D’Alvise, P. W., Rygaard, A. M., and Gram, L. (2014). Biofilm formation is not a prerequisite for production of the antibacterial compound tropodithietic acid in Phaeobacter inhibens DSM17395. J. Appl. Microbiol. 117, 1592–1600. doi: 10.1111/jam.12659
Gnidehou, S., Caillou, B., Talbot, M., Ohayon, R., Kaniewski, J., Noël-Hudson, M. S., et al. (2004). Iodotyrosine dehalogenase 1 (DEHAL1) is a transmembrane protein involved in the recycling of iodide close to the thyroglobulin iodination site. FASEB J. 18, 1574–1576. doi: 10.1096/fj.04-2023fje
Hastings, M. H., Maywood, E. S., and Brancaccio, M. (2018). Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469.
Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K. I. (2001). U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120. doi: 10.1074/jbc.m102755200
Hiroi, N., Kino, T., Bassett, M., Rainey, W. E., Phung, M., Abu-Asab, M., et al. (2003). Pituitary homeobox factor 1, a novel transcription factor in the adrenal regulating steroid 11beta-hydroxylase. Horm. Metab. Res. 35, 273–278. doi: 10.1055/s-2003-41301
Hussain, M. M., and Pan, X. Y. (2015). Circadian regulators of intestinal lipid absorption. J. Lipid Res. 56, 761–770. doi: 10.1194/jlr.r051573
Jiao, L. F., Dai, T. M., Cao, T. L., Jin, M., Sun, P., and Zhou, Q. C. (2020). New insight into the molecular basis of chromium exposure of Litopenaeus vannamei by transcriptome analysis. Mar. Pollut. Bull. 160:111673. doi: 10.1016/j.marpolbul.2020.111673
Jiao, L. F., Dai, T. M., Lu, J. J., Tao, X. Y., Jin, M., Sun, P., et al. (2021). Effects of Dietary Manganese Supplementation on Growth Performance, Antioxidant Capacity, Immune Function and Intestinal Microbiota in Pacific White Shrimp Litopenaeus Vannamei. New jersey, NJ: Wiley.
Kim, Y. M., Snijders, A. M., Brislawn, C. J., Stratton, K. G., Zink, E. M., Fansler, S. J., et al. (2019). Light-Stress influences the composition of the murine gut microbiome, memory function, and plasma metabolome. Front. Mol. Biosci. 6:108. doi: 10.3389/fmolb.2019.00108
Ko, C. H., and Takahashi, J. S. (2006). Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277.
Lazado, C. C., Gesto, M., Madsen, L., and Jokumsen, A. (2018). Interplay between daily rhythmic serum-mediated bacterial killing activity and immune defence factors in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immun. 72, 418–425. doi: 10.1016/j.fsi.2017.11.025
Leiner, K. A., and MacKenzie, D. S. (2001). The effects of photoperiod on growth rate and circulating thyroid hormone levels in the red drum, Sciaenops ocellatus: evidence for a free-running circadian rhythm of T(4) secretion. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130, 141–149. doi: 10.1016/s1095-6433(01)00373-7
Li, S., Guo, S., Li, F., and Xiang, J. (2015). Functional diversity of anti-lipopolysaccharide factor isoforms in shrimp and their characters related to antiviral activity. Mar. Drugs 13, 2602–2616. doi: 10.3390/md13052602
Li, S., Zhai, J. Y., Chu, W. W., Geng, X. Y., Chen, Z. J., and Du, Y. Z. (2020). Altered circadian clock as a novel therapeutic target for constant darkness-induced insulin resistance and hyperandrogenism of polycystic ovary syndrome. Transl. Res. 218, 13–29. doi: 10.1016/j.trsl.2020.02.003
Li, X., Han, T., Zheng, S., and Wu, G. (2021a). Nutrition and functions of amino acids in aquatic crustaceans. Adv. Exp. Med. Biol. 1285, 169–198. doi: 10.1007/978-3-030-54462-1_9
Li, X., Wei, P., Liu, S., Tian, Y., Ma, H., and Liu, Y. (2021b). Photoperiods affect growth, food intake and physiological metabolism of juvenile European Sea Bass (Dicentrachus labrax L.). Aquacult. Rep. 20:100656. doi: 10.1016/j.aqrep.2021.100656
Maloy, A. P., Ford, S. E., Karney, R. C., and Boettcher, K. J. (2007). Roseovarius crassostreae, the etiological agent of Juvenile Oyster Disease (now to be known as Roseovarius Oyster Disease) in Crassostrea virginica. Aquaculture 269, 71–83. doi: 10.1016/j.aquaculture.2007.04.008
Mastan, S. A. (2015). Incidences of white feces syndrome (WFS) in farm-reared shrimp, Litopenaeus vannamei, andhra pradesh. Indo Am. J. Pharmaceut. Res. 5:9.
Maury, E., Ramsey, K. M., and Bass, J. (2010). Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ. Res. 106, 447–462. doi: 10.1161/circresaha.109.208355
Migaud, H., Davie, A., and Taylor, J. F. (2010). Current knowledge on the photoneuroendocrine regulation of reproduction in temperate fish species. J. Fish Biol. 76, 27–68. doi: 10.1111/j.1095-8649.2009.02500.x
Oliveira, C. C., Aparicio, R., Blanco-Vives, B., Chereguini, O., Martin, I., and Javier Sanchez-Vazquez, F. (2013). Endocrine (plasma cortisol and glucose) and behavioral (locomotor and self-feeding activity) circadian rhythms in Senegalese sole (Solea senegalensis Kaup 1858) exposed to light/dark cycles or constant light. Fish Physiol. Biochem. 39, 479–487. doi: 10.1007/s10695-012-9713-2
Panteghini, M., Falsetti, F., Chiari, E., and Malchiodi, A. (1983). Determination of aspartate aminotransferase isoenzymes in hepatic diseases–preliminary findings. Clin. Chim. Acta 128, 133–140. doi: 10.1016/0009-8981(83)90063-3
Pett, J. P., Kondoff, M., Bordyugov, G., Kramer, A., and Herzel, H. (2018). Co-existing feedback loops generate tissue-specific circadian rhythms. Life Sci. Alliance 1:e201800078. doi: 10.26508/lsa.201800078
Peyric, E., Moore, H. A., and Whitmore, D. (2013). Circadian clock regulation of the cell cycle in the zebrafish intestine. PLoS One 8:e73209. doi: 10.1371/journal.pone.0073209
Phupet, B., Pitakpornpreecha, T., Baowubon, N., Runsaeng, P., and Utarabhand, P. (2018). Lipopolysaccharide- and beta-1,3-glucan-binding protein from Litopenaeus vannamei: Purification, cloning and contribution in shrimp defense immunity via phenoloxidase activation. Dev. Comp. Immunol. 81, 167–179. doi: 10.1016/j.dci.2017.11.016
Roenneberg, T., Kantermann, T., Juda, M., Vetter, C., and Allebrandt, K. V. (2013). Light and the human circadian clock. Handb. Exp. Pharmacol. 217, 311–331. doi: 10.1007/978-3-642-25950-0_13
Ruchin, A. B. (2006). Effect of light on white blood cell count in carp Cyprinus carpio L. Izv Akad Nauk Ser. Biol. 5, 634–637.
Russell, B. J., and Dierssen, H. M. (2018). Color change in the Sargassum crab, Portunus sayi: response to diel illumination cycle and background albedo. Mar. Biol. 165:28.
Rutter, J., Reick, M., and McKnight, S. L. (2002). Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 71, 307–331.
Sanudin, N., Tuzan, A. D., and Yong, A. S. K. (2014). Feeding activity and growth performance of shrimp post larvae Litopenaeus vannamei under light and dark condition. J. Agricult. Sci. 6:103.
Schilperoort, M., van den Berg, R., Bosmans, L. A., van Os, B. W., Dolle, M. E. T., and Smits, N. A. M. (2020). Disruption of circadian rhythm by alternating light-dark cycles aggravates atherosclerosis development in APOE∗3-Leiden.CETP mice. J. Pineal Res. 68:e12614. doi: 10.1111/jpi.12614
Senior, J. R. (2012). Alanine aminotransferase: a clinical and regulatory tool for detecting liver injury-past, present, and future. Clin. Pharmacol. Ther. 92, 332–339. doi: 10.1038/clpt.2012.108
Serra, E. L., Medalha, C. C., and Mattioli, R. (1999). Natural preference of zebrafish (Danio rerio) for a dark environment. Braz. J. Med. Biol. Res. 32, 1551–1553. doi: 10.1590/s0100-879x1999001200016
Sharabi, O., Manor, R., Weil, S., Aflalo, E. D., Lezer, Y., Levy, T., et al. (2016). Identification and characterization of an insulin-Like receptor involved in crustacean reproduction. Endocrinology 157, 928–941. doi: 10.1210/en.2015-1391
Urbancic, D., Kotar, A., Smid, A., Jukic, M., Gobec, S., Martensson, L. G., et al. (2019). Methylation of selenocysteine catalysed by thiopurine S-methyltransferase. Biochim. Biophys. Acta Gen. SUBJ 1863, 182–190. doi: 10.1016/j.bbagen.2018.10.002
Wade, N., Goulter, K. C., Wilson, K. J., Hall, M. R., and Degnan, B. M. (2005). Esterified astaxanthin levels in lobster epithelia correlate with shell colour intensity: potential role in crustacean shell colour formation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 141, 307–313. doi: 10.1016/j.cbpc.2005.04.004
Wade, N. M., Tollenaere, A., Hall, M. R., and Degnan, B. M. (2009). Evolution of a novel carotenoid-binding protein responsible for crustacean shell color. Mol. Biol. Evol. 26, 1851–1864. doi: 10.1093/molbev/msp092
Wang, X. W., Xu, J. D., Zhao, X. F., Vasta, G. R., and Wang, J. X. (2014). A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J. Biol. Chem. 289, 11779–11790. doi: 10.1074/jbc.M114.552307
Wei, J., Tian, L., Wang, Y. K., Yu, L. Y., and Zhu, X. P. (2021). Effects of salinity, photoperiod, and light spectrum on larval survival, growth, and related enzyme activities in the giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 530:735794. doi: 10.1016/j.aquaculture.2020.735794
Wright, K. P. Jr., McHill, A. W., Birks, B. R., Griffin, B. R., Rusterholz, T., and Chinoy, E. D. (2013). Entrainment of the human circadian clock to the natural light-dark cycle. Curr. Biol. 23, 1554–1558. doi: 10.1016/j.cub.2013.06.039
Xu, Z., Wei, Y., Guo, S., Lin, D., and Ye, H. (2020). B-type allatostatin modulates immune response in hepatopancreas of the mud crab Scylla paramamosain. Dev. Comp. Immunol. 110:103725. doi: 10.1016/j.dci.2020.103725
Zhao, F. Y., Zhang, Y. Y., Dong, W. G., Zhang, Y. Q., Zhang, G. X., Sun, Z. P., et al. (2019). Vermicompost can suppress Fusarium oxysporum f. sp. lycopersici via generation of beneficial bacteria in a long-term tomato monoculture soil. Plant Soil 440, 491–505. doi: 10.1007/s11104-019-04104-y
Zhao, J. C., Liu, X. T., Liao, X. Z., He, Z. H., Chen, X. Y., Sun, C. B., et al. (2021). Transcriptome analysis reveals the effects of sand substrate removal and body colour change of kuruma shrimp, Marsupenaeus japonicus. Aquacult. Res. 52, 577–588. doi: 10.1111/are.14915
Keywords: light/dark cycles, multi-omics techniques, microbiota homeostasis, circadian biological rhythm, crustacean, physiological metabolism
Citation: Jiao LF, Dai TM, Tao XY, Lu JJ and Zhou QC (2021) Influence of Light/Dark Cycles on Body Color, Hepatopancreas Metabolism, and Intestinal Microbiota Homeostasis in Litopenaeus vannamei. Front. Mar. Sci. 8:750384. doi: 10.3389/fmars.2021.750384
Received: 30 July 2021; Accepted: 31 October 2021;
Published: 29 November 2021.
Edited by:
Vincenzo Parrino, University of Messina, ItalyReviewed by:
Chunxiao Zhang, Jimei University, ChinaPindaro Alvarez-Ruiz, Instituto Politécnico Nacional (IPN), Mexico
Sehrish Taj, Hainan University, China
Copyright © 2021 Jiao, Dai, Tao, Lu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qicun Zhou, emhvdXFpY3VuQG5idS5lZHUuY24=
 Lefei Jiao
Lefei Jiao Tianmeng Dai
Tianmeng Dai Qicun Zhou
Qicun Zhou