- 1Coastal Marine Field Station, School of Science, University of Waikato, Tauranga, New Zealand
- 2Environmental Research Institute, University of Waikato, Tauranga, New Zealand
The fast expansion of the global seaweed aquaculture industry has created an interest in translocating seedlings cultivated from wild type brood stock. However, such translocations must be applied with caution as introduced cultivars can reduce genetic structure and diversity of wild populations. An understanding of the genetic structure and connectivity of target species is required to guide decision making around aquaculture translocation activities. In this study we used 14 microsatellite loci in a three-level hierarchical sampling design to analyze the genetic structure and connectivity of the native kelp Ecklonia radiata across 12 sites among four geographic regions (Northland, Bay of Plenty, Gisborne, and Wellington) in the North Island of New Zealand. Our aim was to provide guidance for translocation of cultivars to prevent the introduction of locally absent genotypes of E. radiata. Strong genetic structure and low geneflow were observed at all hierarchical levels, indicating the presence of multiple genetically distinct sub-populations. On a regional scale, high genetic differentiation was found between the Wellington region and the other three regions (FST = 0.407–0.545), and within regions most sites were significantly different (measured by pairwise FST) with high relatedness found between individuals within sites (mean 28.2% ± 0.7 SE). Bayesian modeling and redundancy analysis showed a high degree of genetic clustering and indicate that ocean currents and other factors that have resulted in biogeographical breaks along the coast are likely to be the main factors shaping genetic structure and connectivity of E. radiata on the North Island, rather than isolation by distance. Based on these findings, we recommend that that cultivars of E. radiata should not be translocated outside their area of origin to avoid introducing locally absent genotypes to local sub-populations.
Introduction
Seaweed aquaculture has grown exponentially during the last 50 years and now has a global annual yield of 32 million tons and a commercial value of US$13.3 billion (FAO, 2020). The vast expansion of the sector has created an interest in translocating strains (i.e., movement outside area of origin), as intraspecific differentiation in key commercial traits, such as growth rate and biochemical content, is common within seaweeds due to genotypic variation and local adaptation (Oliveira et al., 2013; Gosch et al., 2015; Mata et al., 2017). However, translocation of seaweed cultivars (i.e., strains selected for cultivation) must be approached with caution. Introduced cultivars can reduce genetic differentiation between wild populations (i.e., reduce genetic structure) by outcompeting unique local genotypes and reduce genetic variation within wild populations (i.e., reduce genetic diversity) by over-dominating the local gene pool (Ellstrand et al., 2013; Bolstad et al., 2017; Valero et al., 2017; Hu et al., 2021). Such genetic changes may have negative ecological and economic consequences, as loss of genetic structure can lead to the extinction of valuable genetic traits adapted to specific ecological niches (Evankow et al., 2019; Shan et al., 2019). Furthermore, the loss of genetic diversity within wild macrophyte populations may decrease productivity and tolerance to environmental stress in these communities (Hughes and Stachowicz, 2004; Reusch et al., 2005; Wernberg et al., 2018).
The extent of gene flow from farmed seaweed to wild populations remains poorly quantified (Valero et al., 2017; Campbell et al., 2019) and may vary between species and with cultivation method (Shan et al., 2019). However, evidence to date suggests that gene flow from farmed to wild seaweed populations is low (Guzinski et al., 2018; Li et al., 2020; Graf et al., 2021). For instance, gene flow from cultivars to wild populations is typically low for Saccharina japonica (Liu et al., 2012; Zhang et al., 2017) because selective breeding of cultivars has delayed spore release to late summer when most cultivars have already been harvested and any offspring of remaining cultivars are unlikely to survive high summer temperatures (Shan et al., 2019). However, gene flow from cultivars to wild populations has been documented extensively for animal aquaculture and agriculture (Ellstrand et al., 2013; Glover et al., 2013), with effects varying from possibly advantageous (e.g., increased fitness in wild lettuce; Hooftman et al., 2009; Hartman et al., 2013) to devastating (e.g., wide-spread changes in age and size at maturation in wild stock of Norwegian Atlantic salmon; Bolstad et al., 2017). Consequently, the introduction of cultivated organisms to wild populations is recognized as one of the main risks associated with aquaculture of any species (Naylor et al., 2001) and the cultivation of locally absent seaweed genotypes is often restricted (Loureiro et al., 2015). Therefore, mapping genetic structure and connectivity of wild populations is crucial to ensure that seedlings cultivated from wild type broodstock can be translocated without introducing locally absent genotypes (Barbier et al., 2019; Evankow et al., 2019).
The native kelp Ecklonia radiata (C.Agardh.) J.Agardh is a target for aquaculture in New Zealand because of its wide distribution (Shears and Babcock, 2007) and commercial applications as biostimulants, sea vegetables and food additives (White and White, 2020). However, the genetic structure and connectivity of E. radiata has not been investigated in New Zealand. Consequently, the geographic extent that seedlings cultivated from wild type brood stock can be translocated without changing local genetic structure remains unknown. In Australia, the genetic structure of E. radiata is largely region specific, with high genetic connectivity among populations on the west coast (Coleman et al., 2011a; Coleman, 2013) and low genetic connectivity among populations on the south coast (Coleman et al., 2009). These regional differences in genetic structure are largely attributed to variations in oceanography as the gene flow of E. radiata is correlated to the strength of ocean currents (Coleman et al., 2011b). In New Zealand, ocean currents are known to play a key role in long distance connectivity among populations of seagrass (Jones et al., 2008) and several species of seaweeds (Collins et al., 2010; Macaya and Zuccarello, 2010; Buchanan and Zuccarello, 2012; Fraser et al., 2013). Accordingly, the genetic structure of E. radiata in New Zealand may also be influenced by local oceanographic conditions. Within the North Island of New Zealand, distinct genetic sub-populations exist for multiple taxonomic groups including seaweeds (Buchanan and Zuccarello, 2012; Muangmai et al., 2015; Huanel et al., 2020), seagrasses (Jones et al., 2008), anemones (Veale and Lavery, 2012), mollusks (Ross et al., 2012), and crustaceans (Stevens, 1990). Furthermore, five bioregions have been proposed within the North Island, based on the distribution E. radiata and 150 other species of seaweed and invertebrates, indicating that there may be distinct sub-populations within each of these bioregions, and potential breaks in connectivity between bioregions (Shears et al., 2008). This general high level of genetic and distributional subdivision within seaweeds and benthic invertebrates on the North Island suggest that E. radiata is also likely to be divided into genetically distinct sub-populations.
In this study, we used 14 polymorphic microsatellite loci in a three-level hierarchical sampling design to analyze the genetic structure and connectivity of E. radiata among four regions of the North Island of New Zealand. The aims were to (1) assess patterns of genetic structure at a local (<300 km) and regional (150–1,200 km) level; and (2) estimate levels of genetic connectivity between local and regional populations; in order to (3) provide guidance for translocation of cultivars to prevent the introduction of locally absent genotypes of E. radiata within the North Island of New Zealand.
Materials and Methods
Sample Collection
Thirty sporophytes of E. radiata were sampled from each of 12 sites on the North Island of New Zealand between September 2019 and March 2020 (Figure 1A), conducted under Special Permit 724 issued by the Ministry for Primary Industries, Manatū Ahu Matua to the University of Waikato. A hierarchical design was used with three sites sampled within each of four regions (Northland, Bay of Plenty, Gisborne, and Wellington). At each site, samples of tissue from secondary blades were collected haphazardly by hand from adult (stage 3) plants (Mann and Kirkman, 1981) while snorkeling on rocky reefs within 2–5 m depth and with>1 m between each sampled individual (sample area ≈ 1,000 m2). After collection, samples were stored in separate polyethylene bags and stored on ice for transport back to the laboratory. Within 24 h of collection, samples were cleaned using a scalpel and lint free tissue paper to remove epiphytes and a 2 cm2 blade section was cut and dried in silica gel for storage for a maximum of 12 months until further processing (Coleman et al., 2009). Blade tissue was homogenized using a Precellys Evolution Homogenizer (Bertin Instruments, France) and genomic DNA was extracted using a DNeasy® Plant Pro Kit (Qiagen, Germany) following the manufacturer’s instructions with the addition of 80–100 μL of PS solution to each sample.
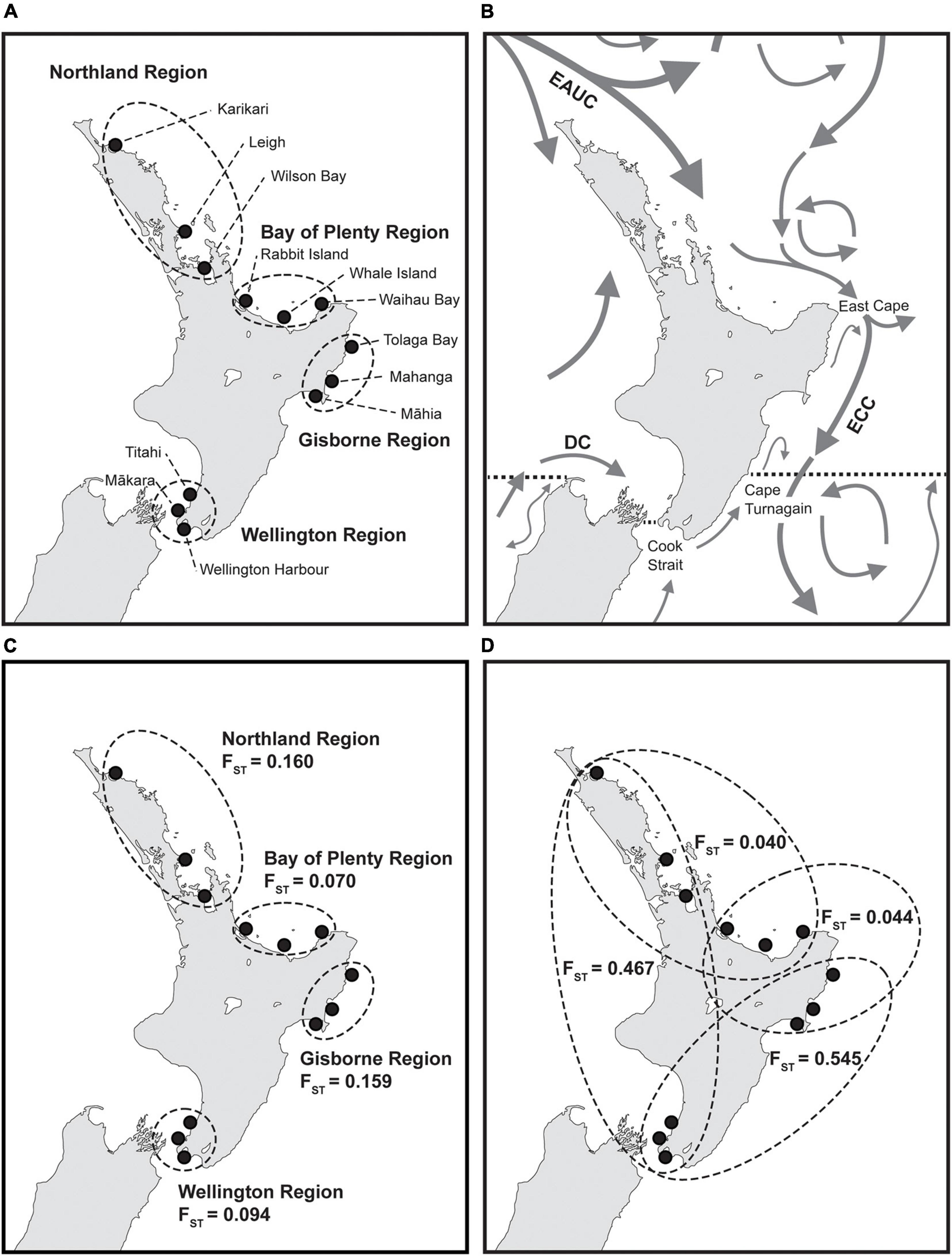
Figure 1. Map of New Zealand with (A) sampling sites (filled black circles) and sampling regions (dashed circles), (B) major ocean currents (Heath, 1985), proposed biogeographic breaks (dashed black lines, Shears et al., 2008) and locations referred to in the text, (C) genetic differentiation (FST) of E. radiata within each region and (D) FST between each neighboring region (FST Northland vs. Gisborne = 0.043, FST Bay of Plenty vs. Wellington = 0.407). EAUC: East Auckland Current; ECC: East Cape Current; DC: D’Urville Current. (B) Modified with permission from Buchanan and Zuccarello (2012).
Primer Selection
Twenty five microsatellite loci developed for three other species of Ecklonia (Itou et al., 2012; Akita et al., 2018, 2020) were tested to determine their suitability for use in E. radiata (Supplementary Table 1). All loci were initially tested on 8 individuals from five different sites within the four sampling regions. DNA was amplified in 25 μL singleplex PCR reactions containing 12.5 μL MyTag HS Red Mix (MilliporeSigma, Germany), 0.4 μM non-labeled forward primer, 0.4 μM fluorescent-labeled reverse primer, and 1 μL of genomic DNA. PCRs were run on an Applied Biosystems SimplyAmp Thermal Cycler (Thermo Fisher Scientific, United States) and comprised of an initial denaturation phase at 95°C for 1 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 7 min. Five microliter of PCR product was run on a 2% agarose gel and amplicon size was determined using a size standard marker (Bioline HyperLadder™ 25 bp). Loci showing strong evidence of primer dimer on gels were discarded. Loci that produced an amplicon within 150 bp of the expected size range in at least 6 of the 8 tested individuals were selected for genotyping. Loci showing no or inconsistent amplification on gels were further tested on six new individuals, using the same parameters described above. Again, loci were amplified, visualized and selected for genotyping as described earlier and loci that did not consistently produce an amplicon within 150 bp of the expected size range were omitted from further testing. Following this process, 20 loci were selected and subsequently genotyped across all 360 samples (30 samples from each of the 12 sites). Loci were amplified in PCR reactions using the same parameters described above. Multiplex genotyping was carried out by Genetic Analysis Service, Department of Anatomy, University of Otago using an ABI 3730xl DNA Analyzer (Thermo Fisher Scientific, United States) and allele sizes were scored using GeneMarker® (SOFTGENETICS, United States). Three of the 20 loci that were genotyped produced electropherograms too variable for allele sizes to be scored consistently across the 360 samples and were discarded (Supplementary Table 1).
Data Analyses
Data (Supplementary Data 1) was checked for deviations from Hardy-Weinberg equilibrium and presence of linkage disequilibrium using Genepop on the Web v4.7 (Raymond and Rousset, 1995; Rousset, 2008). Three of the 17 genotyped loci were too monomorphic to enable testing of Hardy-Weinberg equilibrium at any site and were therefore excluded from further statistical analysis (Supplementary Tables 1, 2). None of the remaining 14 loci deviated significantly from Hardy-Weinberg equilibrium when data was analyzed collectively across all sites. However, it is likely that some of these loci were not in Hardy-Weinberg equilibrium as 12 of the 14 loci were monomorphic (or almost monomorphic) at one or more sites. Consequently, it was not possible to test for Hardy-Weinberg equilibrium at these sites and they therefore could not be included in the collective assessment of Hardy-Weinberg equilibrium. To determine the influence of this high level of monomorphism on results, a sensitivity analysis was run where all analyses described below were conducted with all 14 loci, and then were repeated excluding all loci where more than half the sites were too monomorphic to calculate test Hardy-Weinberg equilibrium. This sensitivity analysis showed that excluding the 8 loci with the highest monomorphism had no major influence on the main results and all 14 selected loci where therefore retained for analysis (Supplementary Tables 3, 4). Linkage disequilibrium was tested across all sites and loci, and P-values were adjusted using the false discovery rate (fdr) correction in the R-studio STATS package (Team RStudio, 2021). No significant linkage disequilibrium was found between any loci but an average 58% ± 15 SD of the loci combinations could not be checked for linkage disequilibrium due to monomorphism. However, as this uncertainty was evenly spread across all loci with no consistent patterns, all remaining loci were retained for analysis.
Allele frequencies and total number of alleles were calculated for each locus using GeneAlEx v6.5 (Peakall and Smouse, 2006, 2012). Arlequin v.3.5.2.2 (Excoffier and Lischer, 2010) was used to perform global analysis of molecular variance (AMOVA) as weighted average over all 14 loci within the three hierarchical sampling levels with 1,000 permutations. The number of genetic clusters (K) was analyzed using a Bayesian modeling approach in STRUCTURE v2.3.4 (Pritchard et al., 2000), running 100,000 burn-in and 300,000 Monte Carlo Marco chain repeats after burn-in with 20 iterations for all analyzed levels of K (1–14). Clustering was analyzed using an admixture model with correlated allele frequencies and no prior information about the data. STRUCTURE HARVESTER (Earl and vonHoldt, 2012) was used to find the number of clusters best fitting the data based on highest delta K and to find the iteration with the highest likelihood (Evanno et al., 2005). The clustering analysis was run multiple times to successively identify all hierarchical layers of structuring within the data. All samples were included in the first analysis and for each subsequent analysis, the samples within each of the previously identified clusters were analyzed in separate simulations. FST is a measure of genetic differentiation ranging from 0 to 1, with 0 showing no genetic difference between samples and 1 showing maximum genetic difference between samples. FST was calculated in Arlequin. Within region FST was calculated as the average pairwise FST between each pair of sites within each region. Between regions FST was calculated by treating each region as one single population and calculating a single pairwise FST value for each pair of regions. Pairwise FST between sites was visualized using R-studio (script sourced from The Banta Lab).1
Relatedness between the individuals collected from each site was quantified using ML-Relate (Kalinowski et al., 2006) which includes a null allele correction for any loci at any site that showed significant heterozygote deficiency (α = 0.05). Relatedness was further quantified using maximum likelihood estimates, counting all estimates of half sibling, full sibling, and parent/offspring (i.e., not unrelated) as related. Individuals with missing values at more than 2/3 of the loci showed abnormally high values of relatedness and were therefore deleted and not used in any analysis (13 samples out of 360). High levels of relatedness were found between individuals at all sites (mean 28.2% ± 0.7 SE) which can bias genetic analysis (Waples and Anderson, 2017). To determine the influence of this high level of relatedness on results, another sensitivity analysis was run where all analyses described below were repeated excluding all samples that were related to more than 33.3% of the other samples within their sites (thereby lowing mean relatedness to 21.1% ± 0.6 SE). This sensitivity analysis showed that excluding the 97 most related samples had no major effect on the main results and all 347 samples were therefore retained for analysis (Supplementary Tables 3, 4). To determine the influence of geographic distance on genetic structure, a redundancy analysis was performed in R-studio using the vegan package (Oksanen et al., 2020) and visualized using ggplot (Wickham, 2016). Site coordinates (longitude and latitude) were set as independent variables and site-specific allele frequencies (72 different alleles in total across the 14 selected loci) as constrained dependent variables (Meirmans, 2015).
Results
Genetic diversity was highest within northern sites (sites within the Northland and Bay of Plenty regions) compared to southern sites (sites within the Gisborne and Wellington regions, Table 1). The mean total number of alleles and mean number of unique alleles were 34.83 and 2.8, respectively, for northern sites, compared to 26.2 and 0.7, respectively, for southern sites. Relatedness between individuals was high at all sites (mean 28.2% ± 0.7 SE), with individuals within southern sites being slightly more related than samples within the northern sites (32.0% ± 1.0 SE and 24.3% ± 0.8 SE, respectively).
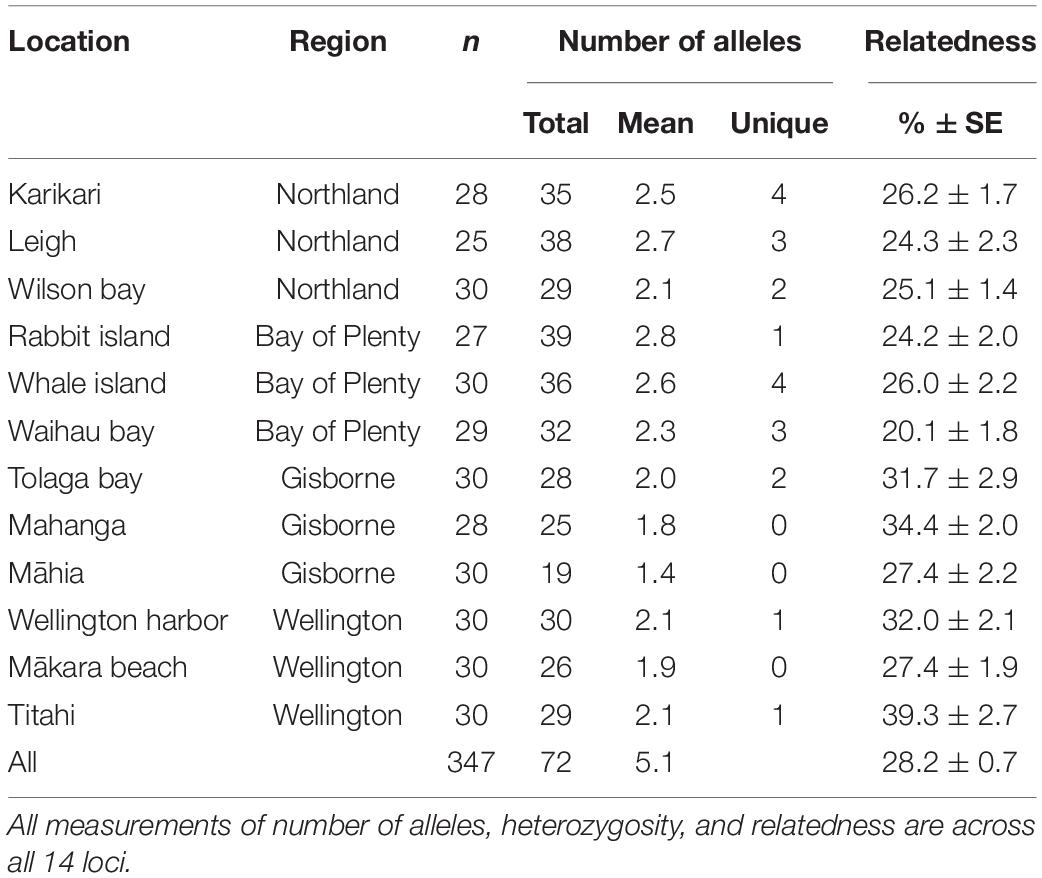
Table 1. Number of individuals genotyped (n); total and mean number of alleles, and number of unique alleles; and% relatedness between individuals ± standard error.
Analysis of molecular variance showed high genetic variation among regions (27.41%) and low genetic variation among sites within regions (7.10%), with all levels of variation being significant (p < 0.001, Table 2). The same pattern was evident for genetic differentiation (FST, Supplementary Tables 5, 6). FST values were low within regions (FST = 0.070–0.160, Figure 1C), indicating low genetic differentiation, but were generally higher between regions (FST = 0.040–0.545, Figure 1D), indicating comparatively higher genetic differentiation. Between region FST was high between the Wellington region and the three other regions (FST = 0.407–0.545) and low between the Northland, Bay of Plenty and Gisborne regions (FST = 0.040–0.044). Pairwise FST between sites showed significant genetic differentiation between all sites except for 2 pairs (Figure 2). Highest pairwise FST was again found between the Wellington sites and sites from the other regions, with Titahi Bay and Mākara Beach showing the highest differentiation from other sites and Wellington Harbor showing only intermediate differentiation.
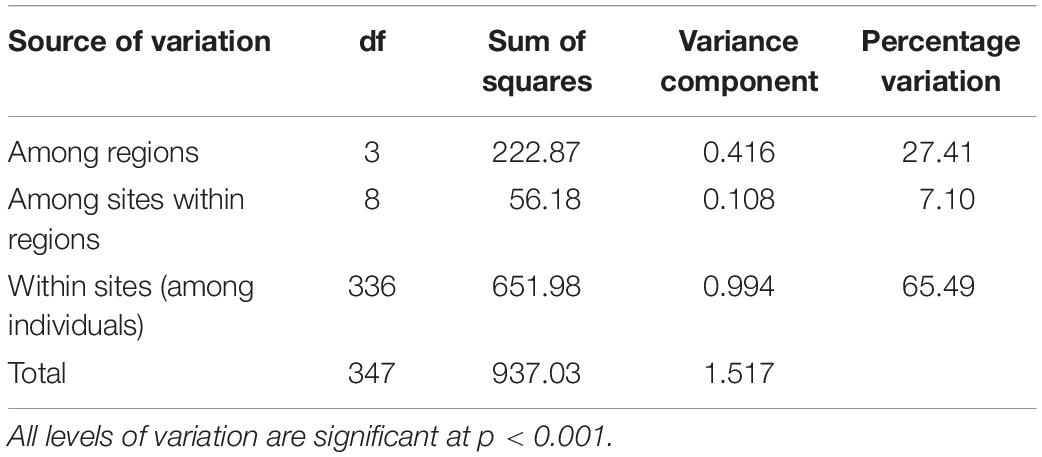
Table 2. Global analysis of molecular variance (AMOVA) for E. radiata among the four regions, among the three sites within each region, and among the individuals (25–30) within each site.
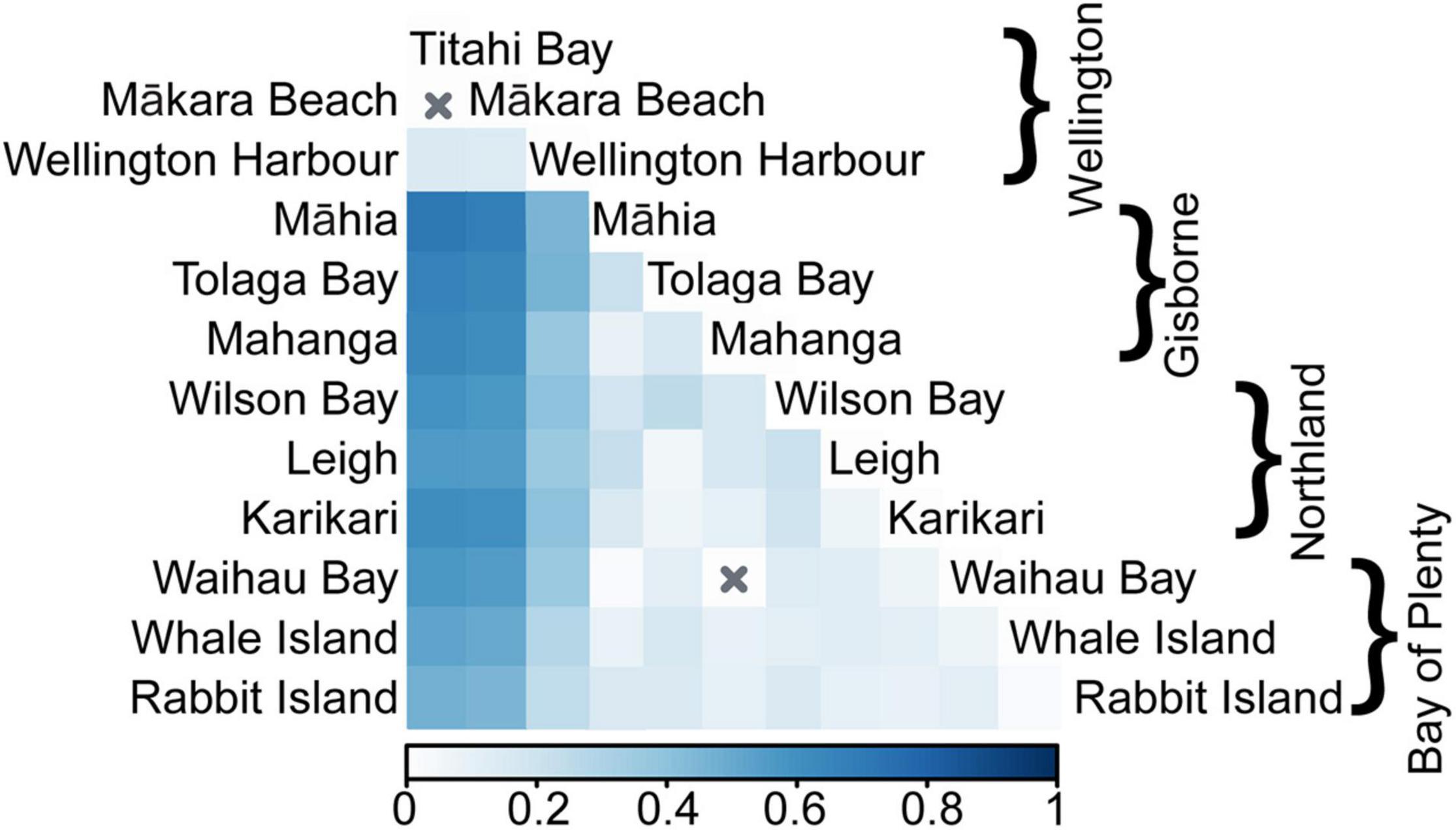
Figure 2. Pairwise genetic differentiation (FST) between each site (FST values indicated by color gradient) with X indicating that sites are not significantly different.
Bayesian modeling of population structure predicted that the most likely number of genetic clusters (inferred from delta K) for any of the STRUCTURE simulations was two (K = 2, Figure 3). When all samples were included in the analysis, the three Wellington sites were all grouped into one distinct cluster and the nine sites from the other three regions were grouped into a second distinct cluster. At the lower hierarchical levels of clustering (1) the Northland region separated from the Bay of Plenty and Gisborne region, (2) the Bay of Plenty region separated slightly from the Gisborne region, and (3) Wellington Harbor separated sharply from the Wellington open coast sites, Mākara Beach and Titahi Bay. There was no evidence of genetic structure among sites within each of the Northland, Bay of Plenty, and Gisborne regions.
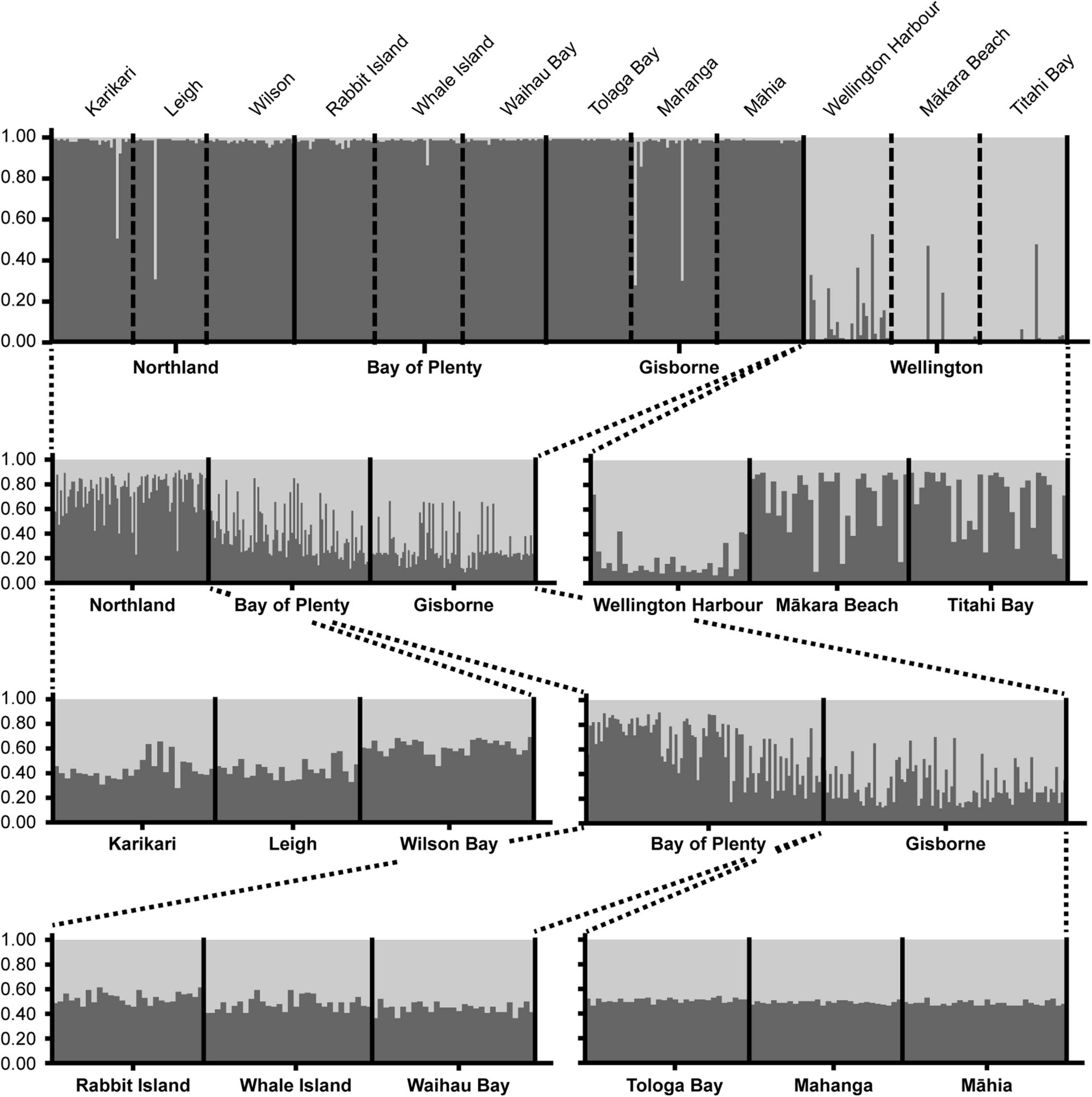
Figure 3. Bayesian modeling of population structure for E. radiata obtained using STRUCTURE (K = 2 for all plots). Top plot was run with all samples included and each subplot underneath was run with a subset of the samples indicated by the dashed lines. The x-axes show all individuals included in the simulations, with each individual represented by a column and grouped within their sites and regions. The y-axes show the genetic membership of each individual to the two clusters.
Redundancy analysis showing the effect of geographic location (latitude and longitude) on allele frequency was highly significant (P = 0.001, 74% constrained variance and 26% unconstrained variance), with latitude and longitude explaining 74% of the genetic variation in the data (Figure 4). The redundancy analysis showed the same pattern of population structure as the clustering analysis, with strong genetic separation between the wellington sites and the other sites along the RDA 1 axis (67% of the total variation) and a weaker genetic separation between the Northland sites and the other sites along the RDA 2 axis (7% of total variation). Most of the 72 alleles analyzed across the 14 loci were plotted in the center of the graph and only 8 alleles were plotted outside the center, indicating that most of the variation in the data was provided by these 8 alleles.
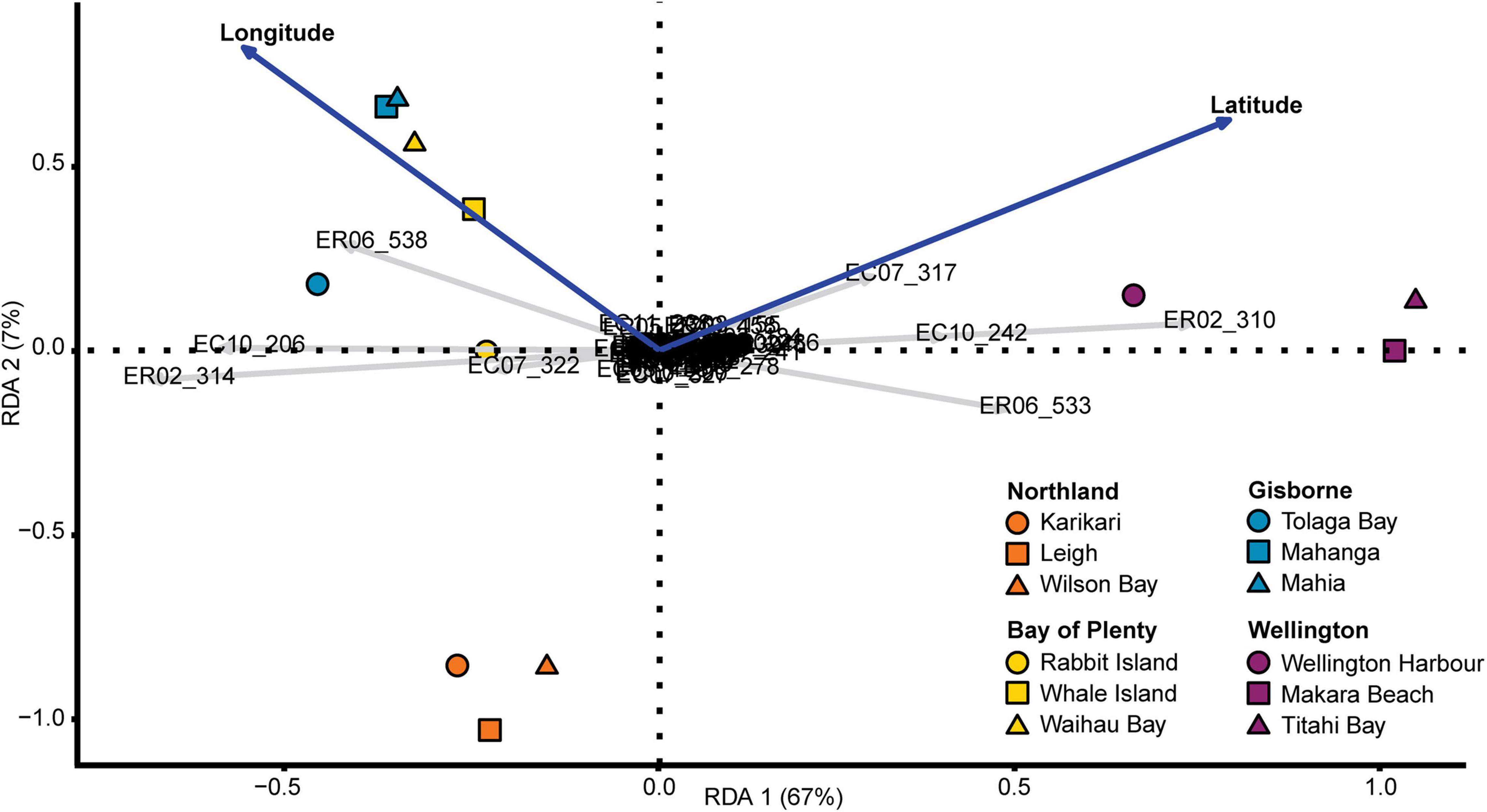
Figure 4. Redundancy analysis (RDA) of allele frequencies explained by geographical location (longitude and latitude). Gray vectors with allele names indicate magnitude and direction of the variation associated with each identified allele where the vector length indicates the amount of variation and the vector direction indicates the correlation of the variation with the two RDA axes (parallel lines being highly correlated). Similarly, the blue vectors indicate the magnitude and direction of the variation associated with the latitude and longitude coordinates of the sites. Sites are plotted according to region (color) and position within region (shape). The percent variation explained by the two RDA components are shown on the two axes.
Discussion
Genetic Structure and Connectivity of Ecklonia radiata
Genetic analysis of microsatellite loci showed strong population structure and low genetic connectivity of E. radiata within the North Island of New Zealand. Clustering analysis showed structural differences on all hierarchical levels, with the Wellington region separating from the other regions at the highest level, the Northland region separating from the Bay of plenty and Gisborne regions at the intermediate level, and Wellington Harbor separating from Mākara Beach and Titahi Bay at the within region level (Figure 3). This same pattern of hierarchical genetic structuring was found through analysis of pairwise genetic differentiation (FST) and redundancy analysis, which showed clear distinction between the Wellington region and the other regions and between Wellington Harbor and the other sites within the Wellington region (Figures 1, 4). These findings demonstrate high levels of genetic structure both across long and short geographic distances. The high level of relatedness between individuals within sites and the large number of sites that were significantly different from one another (all but two site pairs through Pairwise FST), suggest that geneflow is generally low. However, regional FST showed that gene flow was higher among the Northland, Bay of Plenty and Gisborne regions (FST = 0.040–0.044) than between these regions and Wellington (FST = 0.407–0.545). Further support for the notion of reduced connectivity in the Wellington region was provided by the higher levels of relatedness recorded for southern sites (32.0% ± 1.0 SE) compared to those in the north (24.3% ± 0.8 SE). The redundancy analysis showed high correlation between genetic structure and geographic location (74% constrained variance), but also showed a high degree of clustering in agreement with the groupings identified using STRUCTURE, indicating that gene flow in E. radiata in New Zealand is primarily shaped by dispersal barriers rather than by isolation by distance. In combination, these analyses suggest that multiple sub-populations of E. radiata are present within the locations sampled in this study and that genetic connectivity is limited.
E. radiata has a life cycle of two alternate generations where macroscopic diploid sporophytes produce spores that develop into microscopic haploid gametophytes (males and females) which then produce eggs and sperm that fuse and grow into sporophytes. The pelagic phase of E. radiata zoospores is relatively short (usually 1–2 h, Wernberg et al., 2019) and based on studies of related species, gametophyte sperm dispersal is expected to occur over the scale of centimeters, whereas spore dispersal has an approximate maximum dispersal range of 1 km (Reed, 1990; Gaylord et al., 2006). Because E. radiata is negatively buoyant, long-distance dispersal of mature plants is unlikely unless rafting with buoyant species (Fraser et al., 2013). Consequently, the dispersal potential of E. radiata is likely to be limited. These expectations were generally met as evidenced by significant levels of genetic differentiation between most sub-populations. Furthermore, this low dispersal life history may also explain the high levels of relatedness recorded for individuals within sites. High levels of relatedness have previously been reported for the related species E. cava (Itou et al., 2019), and these results indicate inbreeding, which may have contributed to the high FST values.
Strong genetic differentiation between Wellington and the three northern regions matches patterns of genetic structure reported for other seaweed and invertebrate species. For these species, populations in the Wellington region have been shown to be distinct from northern populations, and more closely aligned to a South Island sub-population, indicating a major break in connectivity on the North Island for these species (Ross et al., 2009, 2012; Buchanan and Zuccarello, 2012; Veale and Lavery, 2012). Shears et al. (2008) found major biogeographic breaks based on distributional data covering E. radiata and 150 species of seaweed and invertebrates. In this case, the location of the genetic breaks recorded for E. radiata (between Wellington and northern regions and between the sites within the Wellington region), coincide with the biogeographic boundaries reported by Shears et al. (2008). This suggests that the sharp breaks in E. radiata connectivity between and within regions are most likely generated by the same processes (oceanography and climate) that influence the distribution of a numerous other marine species in the area.
Our detection of regional differences in genetic connectivity is similar to that reported for E. radiata in Australia, where this species displays high genetic connectivity along the west coast, but low connectivity along the south coast (Coleman et al., 2009, 2011a; Coleman, 2013). Ocean currents are a strong driver of differences in genetic connectivity among E. radiata populations in Australia (Coleman et al., 2011b) and may also explain the patterns of genetic connectivity observed in this study. Ocean currents in New Zealand are thought to shape long distance connectivity among other species of seaweeds (Collins et al., 2010; Macaya and Zuccarello, 2010; Buchanan and Zuccarello, 2012; Fraser et al., 2013) and seagrass (Jones et al., 2008). The two main ocean currents shaping the oceanography along the north-east coast of the North Island of New Zealand are the East Auckland Current and the East Cape Current, which travel south-eastward through the Northland, Bay of Plenty and Gisborne region, passing Cape Turnagain where the East Cape Current turns east with the Chatham Rise into the Pacific (Figure 1B; Heath, 1985). The higher genetic connectivity of E. radiata found along the north-eastern coast of the North Island compared to low connectivity found between Wellington and the other regions may therefore be facilitated by these major ocean currents. In the Wellington region, water movement northwards is prevented by the D’Urville Current, which pushes water south-eastwards through Cook Straight.
Implications for Aquaculture
In commercial kelp cultivation, new broodstock is cultivated and seeded onto ropes in on-shore hatcheries, and then, following a nursery period, the ropes are deployed off-shore for the juvenile kelp to mature (Zhang, 2018; Kim et al., 2019). This process creates high potential for spread of locally absent cultivar alleles, which may affect the genetic structure of wild populations and leave them more vulnerable to environmental stress (Ellstrand et al., 2013; Bolstad et al., 2017; Wernberg et al., 2018). The spread of locally absent cultivar alleles can be avoided by developing local cultivars for local use only (Barbier et al., 2019). Our analysis showed clear distinction between the Wellington region and the three other regions but also revealed strong genetic structure and significant differentiation within and between individual sites. We therefore recommend that cultivars of E. radiata should not be translocated outside their area of origin to avoid introducing locally absent genotypes to local sub-populations. Specific guidelines for the scale of translocation of kelp seed-stock have already been developed in some locations (e.g., Gulf of Alaska, United States, Kim et al., 2019), and based on the results of this study and future genetic research, similar guidelines could be developed for E. radiata in New Zealand.
Microsatellite markers have proven to be a powerful tool for the assessment of genetic structure and connectivity of natural populations (Guichoux et al., 2011). However, results provided by microsatellites may be less detailed than what might have been revealed through full genome sequencing (e.g., Graf et al., 2021). Therefore, it is possible that further genetic differentiation among E. radiata sub-populations in New Zealand could be revealed through detailed genomic analysis. Still, given the large number of samples genotyped (360) and the high number of loci (14) included in our analyses, we believe that the results presented here are representative of the general genetic structure and connectivity of E. radiata in New Zealand. The patterns of low geneflow presented here suggest that the genetic structure of E. radiata in the South Island of New Zealand is also likely to be strong. Therefore, we recommend a similarly conservative approach to the translocation of kelp cultivars in this region.
Selective breeding is an integral part of the global seaweed aquaculture industry and has made it possible to improve key cultivar traits such as growth rate, morphology, reproductive timing, biochemical content, stress tolerance, texture, and taste (Hwang et al., 2019). However, selective breeding tends to reduce the genetic diversity of cultivars as it often leads to inbreeding, whether intentional or not (Valero et al., 2017). Reducing the genetic diversity of cultivars is not only a threat to the genepool of local wild populations (Haygood et al., 2003; Bolstad et al., 2017), but may also leave cultivars more susceptible to pathogens (Hwang et al., 2019). For instance, low genetic diversity caused by inbreeding was the main cause of the heavy ice-ice disease outbreak that lead to the sharp decline of the eucheumatoid seaweed farming in the Philippines (Trono and Largo, 2019). It is therefore important in aquaculture not only to use local cultivars, but also to maintain a large effective population size and frequently introduce new genotypes from wild type broodstock to ensure high genetic diversity among cultivars (Table 1; Barbier et al., 2019; Graf et al., 2021; Hu et al., 2021). As has previously been recorded in other New Zealand seaweed species, substantially higher genetic diversity was found among the northern sub-population of E. radiata compared to the southern sub-populations (Fraser et al., 2009; Buchanan and Zuccarello, 2012; Muangmai et al., 2015). The northern regions seem therefore to be the best suited areas for E. radiata cultivation with regards to maintaining high genetic diversity among cultivars.
Conclusion
The strong genetic structure found at all hierarchical levels for E. radiata in the North Island of New Zealand underlines the general need for population genetic analyses to be conducted before initiating translocation of broodstock for seaweed aquaculture. Bayesian analysis of population structure, redundancy analysis and measurements of genetic differentiation (FST) all showed high levels of genetic variation and indicated that genetically distinct sub-populations are likely to be present both between and within regions. From a genetic perspective, we therefore recommend that cultivars of E. radiata should not be translocated outside their area of origin to avoid introducing locally absent genotypes to local sub-populations. The high relatedness between samples found here highlights the importance of increasing the distance between collected individuals of Ecklonia sp. for genetic analysis (preferably to more than 4 m, Itou et al., 2019) to minimize the impact of non-random mating and increase the importance of migration and drift within the signal. The microsatellite data collected in this study can serve as a reference set for future analysis of the genetic structure of E. radiata by comparing the genetic membership of individuals from sites outside the range of this study with the sub-populations sampled here.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JN-D: investigation, data curation, data analysis, visualization, and writing—original draft, review, and editing. MM: conceptualization, design, supervision, funding, and writing—review, and editing. CG: conceptualization, supervision, writing—review, and editing. PR: writing—review, and editing. RL: conceptualization, design, supervision, data analysis, and writing—review, and editing. All authors contributed to the article and approved the submitted version.
Funding
This research is part of the Entrepreneurial Universities Macroalgal Biotechnologies Program, jointly funded by the University of Waikato and the Tertiary Education Commission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Peter Randrup and Alisa Mihaila for sample collection and sample processing assistance. We also thank the two reviewers for very constructive feedback and suggestions for data analysis and literature.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.749154/full#supplementary-material
Footnotes
References
Akita, S., Hashimoto, K., Hanyuda, T., and Kawai, H. (2020). Molecular phylogeny and biogeography of Ecklonia spp. (Laminariales, Phaeophyceae) in Japan revealed taxonomic revision of E. kurome and E. stolonifera. Phycologia 59, 330–339. doi: 10.1080/00318884.2020.1756123
Akita, S., Koiwai, K., Hanyuda, T., Kato, S., Nozaki, R., Uchino, T., et al. (2018). Development of 11 Ecklonia radicosa (Phaeophyceae, Laminariales) SSRs markers using next-generation sequencing and intra-genus amplification analysis. J. Appl. Phycol. 30, 2111–2115. doi: 10.1007/s10811-018-1406-5
Barbier, M., Charrier, B., Araujo, R., Holdt, S. L., Jacquemin, B., and Rebours, C. (2019). in Pegasus - Phycomorph European Guidelines for a Sustainable Aquaculture of Seaweeds COST Action FA1406, eds M. Barbier and B. Charrier (Roscoff: European Commission), doi: 10.21411/2c3w-yc73
Bolstad, G. H., Robertsen, G., Jonsson, B., Sægrov, H., Diserud, O. H., Fiske, P., et al. (2017). Gene flow from domesticated escapes alters the life history of wild Atlantic salmon. Nat. Ecol. Evol. 1:0124. doi: 10.1038/s41559-017-0124
Buchanan, J., and Zuccarello, G. C. (2012). Decoupling of short- and long-distance dispersal pathways in the endemic New Zealand seaweed Carpophyllum maschalocarpum (Phaeophyceae, Fucales). J. Phycol. 48, 518–529. doi: 10.1111/j.1529-8817.2012.01167.x
Campbell, I., Macleod, A., Sahlmann, C., Neves, L., Funderud, J., Øverland, M., et al. (2019). The environmental risks associated with the development of seaweed farming in Europe – prioritizing key knowledge gaps. Front. Mar. Sci. 6:107. doi: 10.3389/fmars.2019.00107
Coleman, M. A. (2013). Connectivity of the habitat-forming kelp, Ecklonia radiata within and among estuaries and open coast. PLoS One 8:e64667. doi: 10.1371/journal.pone.0064667
Coleman, M. A., Chambers, J., Knott, N. A., Malcolm, H. A., Harasti, D., Jordan, A., et al. (2011a). Connectivity within and among a network of temperate marine reserves. PLoS One 6:e20168. doi: 10.1371/journal.pone.0020168
Coleman, M. A., Roughan, M., Macdonald, H. S., Connell, S. D., Gillanders, B. M., Kelaher, B. P., et al. (2011b). Variation in the strength of continental boundary currents determines continent-wide connectivity in kelp. J. Ecol. 99, 1026–1032. doi: 10.1111/j.1365-2745.2011.01822.x
Coleman, M. A., Gillanders, B. M., and Connell, S. D. (2009). Dispersal and gene flow in the habitat-forming kelp, Ecklonia radiata: relative degrees of isolation across an eastwest coastline. Mar. Freshw. Res. 60, 802–809. doi: 10.1071/MF08268
Collins, C. J., Fraser, C. I., Ashcroft, A., and Waters, J. M. (2010). Asymmetric dispersal of southern bull-kelp (Durvillaea antarctica) adults in coastal New Zealand: testing an oceanographic hypothesis. Mol. Ecol. 19, 4572–4580. doi: 10.1111/j.1365-294X.2010.04842.x
Earl, D. A., and vonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Ellstrand, N. C., Meirmans, P., Rong, J., Bartsch, D., Ghosh, A., de Jong, T. J., et al. (2013). Introgression of crop alleles into wild or weedy populations. Annu. Rev. Ecol. Evol. Syst. 44, 325–345. doi: 10.1146/annurev-ecolsys-110512-135840
Evankow, A., Christie, H., Hancke, K., Brysting, A. K., Junge, C., Fredriksen, S., et al. (2019). Genetic heterogeneity of two bioeconomically important kelp species along the Norwegian coast. Conserv. Genet. 20, 615–628. doi: 10.1007/s10592-019-01162-8
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
FAO (2020). The State of World Fisheries and Aquaculture 2020. Sustainability in Action. Rome: FAO, doi: 10.4060/ca9229en
Fraser, C. I., Nikula, R., Spencer, H. G., and Waters, J. M. (2009). Kelp genes reveal effects of subantarctic sea ice during the Last Glacial Maximum. Proc. Natl. Acad. Sci. U.S.A. 106, 3249–3253. doi: 10.1073/pnas.0810635106
Fraser, C. I., Zuccarello, G. C., Spencer, H. G., Salvatore, L. C., Garcia, G. R., and Waters, J. M. (2013). Genetic affinities between trans-oceanic populations of non-buoyant macroalgae in the high latitudes of the southern hemisphere. PLoS One 8:e69138. doi: 10.1371/journal.pone.0069138
Gaylord, B., Reed, C. D., Raimondi, T. P., and Washburn, L. (2006). Macroalgal spore dispersal in coastal environments: mechanistic insights revealed by theory and experiment. Ecol. Monogr. 76, 481–502. doi: 10.1890/0012-96152006076[0481:MSDICE]2.0.CO;2
Glover, K. A., Pertoldi, C., Besnier, F., Wennevik, V., Kent, M., and Skaala, Ø (2013). Atlantic salmon populations invaded by farmed escapees: quantifying genetic introgression with a Bayesian approach and SNPs. BMC Genet. 14:74. doi: 10.1186/1471-2156-14-74
Gosch, B. J., Paul, N. A., de Nys, R., and Magnusson, M. (2015). Spatial, seasonal, and within-plant variation in total fatty acid content and composition in the brown seaweeds Dictyota bartayresii and Dictyopteris australis (Dictyotales, Phaeophyceae). J. Appl. Phycol. 27, 1607–1622. doi: 10.1007/s10811-014-0474-4
Graf, L., Shin, Y., Yang, J. H., Choi, J. W., Hwang, I. K., Nelson, W., et al. (2021). A genome-wide investigation of the effect of farming and human-mediated introduction on the ubiquitous seaweed Undaria pinnatifida. Nat. Ecol. Evol. 5, 360–368. doi: 10.1038/s41559-020-01378-9
Guichoux, E., Lagache, L., Wagner, S., Chaumeil, P., Léger, P., Lepais, O., et al. (2011). Current trends in microsatellite genotyping. Mol. Ecol. Resour. 11, 591–611. doi: 10.1111/j.1755-0998.2011.03014.x
Guzinski, J., Ballenghien, M., Daguin-Thiébaut, C., Lévêque, L., and Viard, F. (2018). Population genomics of the introduced and cultivated Pacific kelp Undaria pinnatifida: marinas—not farms—drive regional connectivity and establishment in natural rocky reefs. Evol. Appl. 11, 1582–1597. doi: 10.1111/eva.12647
Hartman, Y., Uwimana, B., Hooftman, D. A. P., Schranz, M. E., van de Wiel, C. C. M., Smulders, M. J. M., et al. (2013). Genomic and environmental selection patterns in two distinct lettuce crop-wild hybrid crosses. Evol. Appl. 6, 569–584. doi: 10.1111/eva.12043
Haygood, R., Ives, A. R., and Andow, D. A. (2003). Consequences of recurrent gene flow from crops to wild relatives. R. Soc. 270, 1879–1886. doi: 10.1098/rspb.2003.2426
Heath, R. A. (1985). A review of the physical oceanography of the seas around New Zealand — 1982. New Zeal. J. Mar. Freshw. Res. 19, 79–124. doi: 10.1080/00288330.1985.9516077
Hooftman, D. A. P., Hartman, Y., Oostermeijer, J. G. B., and Den Nijs, H. C. M. (2009). Existence of vigorous lineages of crop-wild hybrids in lettuce under field conditions. Environ. Biosafety Res. 8, 203–217. doi: 10.1051/ebr/2010001
Hu, Z. M., Shan, T. F., Zhang, J., Zhang, Q. S., Critchley, A. T., Choi, H. G., et al. (2021). Kelp aquaculture in China: a retrospective and future prospects. Rev. Aquac. 13, 1324–1351. doi: 10.1111/raq.12524
Huanel, O. R., Nelson, W. A., Robitzch, V., Mauger, S., Faugeron, S., Preuss, M., et al. (2020). Comparative phylogeography of two Agarophyton species in the New Zealand archipelago. J. Phycol. 56, 1575–1590. doi: 10.1111/jpy.13046
Hughes, A. R., and Stachowicz, J. J. (2004). Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl. Acad. Sci. U.S.A. 101, 8998–9002. doi: 10.1073/pnas.0402642101
Hwang, E. K., Yotsukura, N., Pang, S. J., Su, L., and Shan, T. F. (2019). Seaweed breeding programs and progress in eastern Asian countries. Phycologia 58, 484–495. doi: 10.1080/00318884.2019.1639436
Itou, T., Kanno, M., Suyama, Y., Inaba, K., and Aoki, M. N. (2019). Opening the black box: microspatial patterns of zoospore dispersal, parentage, and selfing in the kelp Ecklonia cava as revealed by microsatellite markers. J. Appl. Phycol. 31, 3283–3294. doi: 10.1007/s12686-011-9574-5
Itou, T., Kanno, M., Suyama, Y., Kamiyama, A., Sakamoto, S. X., Kijima, A., et al. (2012). Development of 12 polymorphic microsatellite DNA markers for the kelp Ecklonia cava (Phaeophyceae, Laminariales). Conserv. Genet. Resour. 4, 459–461.
Jones, T. C., Gemmill, C. E. C., and Pilditch, C. A. (2008). Genetic variability of New Zealand seagrass (Zostera muelleri) assessed at multiple spatial scales. Aquat. Bot. 88, 39–46. doi: 10.1016/j.aquabot.2007.08.017
Kalinowski, S. T., Wagner, A. P., and Taper, M. L. (2006). ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579. doi: 10.1111/j.1471-8286.2006.01256.x
Kim, J., Stekoll, M., and Yarish, C. (2019). Opportunities, challenges and future directions of open-water seaweed aquaculture in the United States. Phycologia 58, 446–461. doi: 10.1080/00318884.2019.1625611
Li, Q., Shan, T., Wang, X., Su, L., and Pang, S. (2020). Evaluation of the genetic relationship between the farmed populations on a typical kelp farm and the adjacent subtidal spontaneous population of Undaria pinnatifida (Phaeophyceae, Laminariales) in China. J. Appl. Phycol. 32, 653–659. doi: 10.1007/s10811-019-01917-3
Liu, F., Yao, J., Wang, X., Repnikova, A., Galanin, D. A., and Duan, D. (2012). Genetic diversity and structure within and between wild and cultivated Saccharina japonica (Laminariales, Phaeophyta) revealed by SSR markers. Aquaculture 35, 139–145. doi: 10.1016/j.aquaculture.2012.06.022
Loureiro, R., Gachon, C. M. M., and Rebours, C. (2015). Seaweed cultivation: potential and challenges of crop domestication at an unprecedented pace. New Phytol. 206, 489–492. doi: 10.1111/nph.13278
Macaya, E. C., and Zuccarello, G. C. (2010). DNA barcoding and genetic divergence in the giant kelp Macrocystis (laminariales). J. Phycol. 46, 736–742. doi: 10.1111/j.1529-8817.2010.00845.x
Mann, E. H., and Kirkman, H. (1981). Biomass method for measuring productivity of Ecklonia radiata, with the potential for adaptation to other large brown algae. Aust. J. Mar. Freshw. Res. 32, 297–304.
Mata, L., Lawton, R. J., Magnusson, M., Andreakis, N., De Nys, R., Paul, N. A., et al. (2017). Within-species and temperature-related variation in the growth and natural products of the red alga Asparagopsis taxiformis. J. Appl. Phycol. 29, 1437–1447. doi: 10.1007/s10811-016-1017-y
Meirmans, P. G. (2015). Seven common mistakes in population genetics and how to avoid them. Mol. Ecol. 24, 3223–3231. doi: 10.1111/mec.13243
Muangmai, N., Fraser, C. I., and Zuccarello, G. C. (2015). Contrasting patterns of population structure and demographic history in cryptic species of Bostrychia intricata (Rhodomelaceae, Rhodophyta) from New Zealand. J. Phycol. 51, 574–585. doi: 10.1111/jpy.12305
Naylor, R. L., Williams, S. L., and Strong, D. R. (2001). Aquaculture: a gateway for exotic species. Sci. New Ser. 294, 1655–1656. doi: 10.1126/science.1064875
Oksanen, J., Guillaume Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). Vegan: Community Ecology Package. R Package Version 2.5-7. Available online at: https://cran.r-project.org/package=vegan (accessed November 28, 2020).
Oliveira, A. S., Sudatti, D. B., Fujii, M. T., Rodrigues, V., and Pereira, R. C. (2013). Inter- and intrapopulation variation in the defensive chemistry of the red seaweed Laurencia dendroidea (Ceramiales, Rhodophyta). Phycologia 52, 130–136. doi: 10.2216/12-058.1
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard, J. K., Stephens, M., and Donelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Raymond, M., and Rousset, F. (1995). Genpop 1.2 Population genetics software for exact test and ecumenicism. J. Hered. 86, 248–249. doi: 10.1093/oxfordjournals.jhered.a111573
Reed, D. C. (1990). The effects of variable settlement and early competition on patterns of kelp recruitment. Ecology 71, 776–787.
Reusch, T. B. H., Ehlers, A., Hämmerli, A., and Worm, B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl. Acad. Sci. U.S.A. 102, 2826–2831. doi: 10.1073/pnas.0500008102
Ross, P. M., Hogg, I. D., Pilditch, C. A., and Lundquist, C. J. (2009). Phylogeography of New Zealand’s coastal benthos. New Zeal. J. Mar. Freshw. Res. 43, 1009–1027. doi: 10.1080/00288330.2009.9626525
Ross, P. M., Hogg, I. D., Pilditch, C. A., Lundquist, C. J., and Wilkins, R. J. (2012). Population genetic structure of the New Zealand estuarine clam Austrovenus stutchburyi (Bivalvia: Veneridae) reveals population subdivision and partial congruence with biogeographic boundaries. Estuar. Coast. 35, 143–154. doi: 10.1007/s12237-011-9429-z
Rousset, F. (2008). GENEPOP’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Shan, T., Li, Q., Wang, X., Su, L., and Pang, S. (2019). Assessment of the genetic connectivity between farmed populations on a typical kelp farm and adjacent spontaneous populations of Saccharina japonica (Phaeophyceae, Laminariales) in China. Front. Mar. Sci. 6:494. doi: 10.3389/fmars.2019.00494
Shears, N. T., and Babcock, R. C. (2007). Quantitative Description Mainland New Zealand’s Shallow Subtidal Reef Communities. Wellington: Department of Conservation.
Shears, N. T., Smith, F., Babcock, R. C., Duffy, C. A. J., and Villouta, E. (2008). Evaluation of biogeographic classification schemes for conservation planning: application to New Zealand’s coastal marine environment. Conserv. Biol. 22, 467–481. doi: 10.1111/j.1523-1739.2008.00882.x
Stevens, P. M. (1990). A genetic analysis of the pea crabs (Decapoda: Pinnotheridae) of New Zealand. I. Patterns of spatial and host-associated genetic structuring in Pinnotheres novaezelandiae Filhol. J. Exp. Mar. Biol. Ecol. 141, 195–212. doi: 10.1016/0022-0981(90)90224-Z
Trono, G. C., and Largo, D. B. (2019). The seaweed resources of the Philippines. Bot. Mar. 62, 483–498. doi: 10.1515/bot-2018-0069
Valero, M., Guillemin, M.-L., Destombe, C., Jacquemin, B., Gachon, C. M. M., Badis, Y., et al. (2017). Perspectives on domestication research for sustainable seaweed aquaculture. Perspect. Phycol. 4, 33–46. doi: 10.1127/pip/2017/0066
Veale, A. J., and Lavery, S. D. (2012). The population genetic structure of the waratah anemone (Actinia tenebrosa) around New Zealand. New Zeal. J. Mar. Freshw. Res. 46, 523–536. doi: 10.1080/00288330.2012.730053
Waples, R. S., and Anderson, E. C. (2017). Purging putative siblings from population genetic data sets: a cautionary view. Mol. Ecol. 26, 1211–1224. doi: 10.1111/mec.14022
Wernberg, T., Coleman, M. A., Babcock, R. C., Bell, S. Y., Bolton, J. J., Connell, S. D., et al. (2019). Biology and ecology of the globally significant kelp Ecklonia radiata. Oceanogr. Mar. Biol. 57, 265–324. doi: 10.1201/9780429026379-6
Wernberg, T., Coleman, M. A., Bennett, S., Thomsen, M. S., Tuya, F., and Kelaher, B. P. (2018). Genetic diversity and kelp forest vulnerability to climatic stress. Sci. Rep. 8:1851. doi: 10.1038/s41598-018-20009-9
White, L. N., and White, W. L. (2020). Seaweed utilisation in New Zealand. Bot. Mar. 63, 303–313. doi: 10.1515/bot-2019-0089
Keywords: seaweed, marine, farming, genetic differentiation, sub-populations, microsatellites, translocation
Citation: Nepper-Davidsen J, Magnusson M, Glasson CRK, Ross PM and Lawton RJ (2021) Implications of Genetic Structure for Aquaculture and Cultivar Translocation of the Kelp Ecklonia radiata in Northern New Zealand. Front. Mar. Sci. 8:749154. doi: 10.3389/fmars.2021.749154
Received: 29 July 2021; Accepted: 01 November 2021;
Published: 22 November 2021.
Edited by:
Matthias Schmid, Trinity College Dublin, IrelandReviewed by:
Trevor Bringloe, The University of Melbourne, AustraliaDavid Seth Portnoy, Texas College, United States
Copyright © 2021 Nepper-Davidsen, Magnusson, Glasson, Ross and Lawton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacob Nepper-Davidsen, Sm4xMDBAc3R1ZGVudHMud2Fpa2F0by5hYy5ueg==
 Jacob Nepper-Davidsen
Jacob Nepper-Davidsen Marie Magnusson
Marie Magnusson Christopher R. K. Glasson1,2
Christopher R. K. Glasson1,2 Philip M. Ross
Philip M. Ross Rebecca J. Lawton
Rebecca J. Lawton