- Key Laboratory for Feed Quality Testing and Safety, Fisheries College, Jimei University, Xiamen, China
This study aimed to investigate the effects of quercetin (QUE) on fat deposition and the underlying mechanism. Fish were fed four test diets: normal fat diet (NFD), high-fat diet (HFD), and HFD supplemented with 0.5 or 1.0 g/kg quercetin (QUE0.5 or QUE1.0). The results showed that HFD feeding resulted in poor growth and feed utilization while QUE treatment reversed this. The fat contents of serum and liver were increased by HFD and QUE supplementation significantly decreased fat content. Furthermore, gene expressions and ultrastructure observation showed that mitochondrial biogenesis and mitophagy were inhibited and endoplasmic reticulum stress (ERS) in the HFD group. QUE can activate the biogenesis and autophagy of mitochondria and suppress ERS, which is related to its fat-lowering effect.
Introduction
Fat plays a key role in metabolism as it supplies energy, essential fatty acids, and phospholipids for animals (Watanabe, 1982). It is reported that the increase of dietary lipid level moderately can improve feed efficiency and protein deposition (Du et al., 2006). Therefore, high-fat feed is widely used in fish farming. However, recent studies have indicated that the intake of high fat diet (HFD) often induces excess fat deposition, metabolic disorders, and immune dysfunction (Jin et al., 2013; Lu et al., 2014; Xie et al., 2021a; Yin et al., 2021).
It is well-known that fat deposition in the liver represents complex processes, such as uptake, oxidation, transport, and so on (Tilg and Moschen, 2010; Rocha et al., 2020). As the main state of fatty acid oxidation, mitochondrial damage caused by oxidation retardation is the immediate reason for excessive hepatic fat deposition (Rector et al., 2010; Tilg and Moschen, 2010). The lower hepatic fat secretion means an attenuated capacity for transporting lipids out of the liver of fish fed an HFD, which largely contributes to fat deposition in the liver (Lu et al., 2014, 2017). Moreover, the damaged mitochondria often generate excess reactive oxygen species (ROS) and lead to oxidative stress thereby perpetuating the vicious cycle (Wallace, 2007; Shadel and Horvath, 2015). When the endoplasmic reticulum is damaged by ROS, it will generate superfluous abnormal protein folding and then endoplasmic reticulum stress (ERS) (Malhotra and Kaufman, 2007). Further, there is crosstalk and/or interaction between mitochondrial damage and ERS. Based on the above, more researchers suggest that the therapies acting on mitochondria and ER are novel strategies for the prevention and protection against excess fat deposition (Wang et al., 2018; Dong et al., 2020).
Quercetin (QUE), a flavonoid, is widely present in various plants, such as tea, fruitage, and vegetables (Formica and Regelson, 1995; Boots et al., 2008). In mammals, many studies evidenced the regulatory effects of QUE on mitochondria and ER under metabolism diseases (Porras et al., 2017; Liu et al., 2018). In addition, the previous studies showed the antioxidant effects of QUE in fish (Pês et al., 2016, 2018). Furthermore, QUE could alleviate HFD-induced fat accumulation in blunt snout bream (Megalobrama amblycephala) (Jia et al., 2019). However, the molecular mechanism of QUE on fat deposition still needs further study.
Spotted seabass (Lateolabrax maculatus) is a carnivorous species that has been largely cultured in East Asia, particularly in China. The lipid requirement of this fish is ~10%, while the lipid content is often >15% in commercial feeds (Xu et al., 2011; Xie et al., 2021b). Compared to other cultured fish species, its artificial rearing is often associated with fatty liver probably due to its carnivorous instinct and high dietary lipid level. Hence, this fish model was used to investigate the effects of QUE on fat deposition, mitochondria, and endoplasmic reticulum status.
Materials and Methods
Ethics Statement
The feeding trial was conducted according to the principles of the Basel Declaration and Recommendations of Animal Research Institute Committee guidelines, Jimei University, China.
Diets, Feeding Test, and Sampling
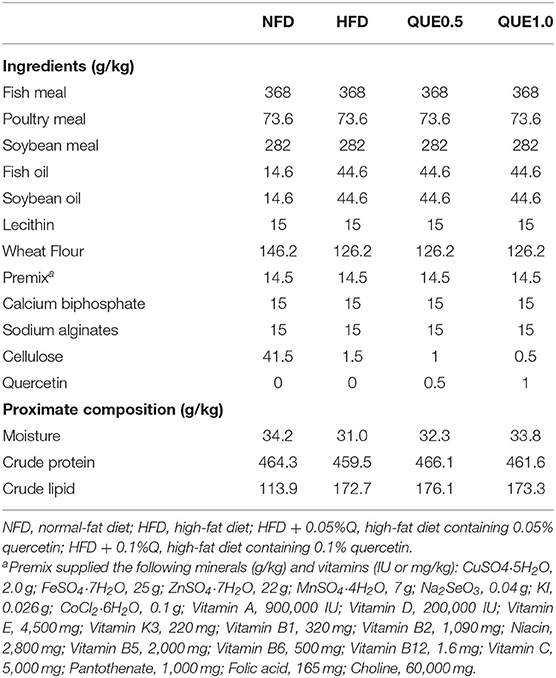
According to the nutrition requirements of spotted seabass, a normal fat diet (NFD) with 11% crude lipid and an HFD with 17% crude lipid were formulated, named as NFD and HFD, respectively. QUE treatments were prepared by the supplementary of 0.5 or 1.0 g/kg QUE (Shanghai Aladdin Biochemical Technology Co., Ltd., China) to HFD. The formulation and proximate composition of the experimental diets are presented in Table 1. The method of feed manufacturing referred to the previous study (Cai et al., 2020).
For the feeding test, fish were obtained from a fish hatchery in Zhangzhou (Fujian, China). First, the fish were kept in a 1,500-L tank to adapt to the experimental facilities and conditions. During this period, fish were fed a commercial diet two times a day (8:00 and 17:00). At the end of the acclimation period, 240 healthy fish averaging 12.02 ± 0.17 g were randomly distributed into 12 circular fiberglass tanks (200 L) in a recirculating aquaculture system. The experiment was conducted using three replicates for each dietary treatment, and fish were fed the diets to apparent satiation two times a day (8:00 and 17:00) for 8 weeks. During the feeding test, the water conditions were maintained as following: water temperature 26–27°C; pH 6.9–7.2; dissolved oxygen > 5.5 mg/L.
After 8 weeks of feeding, fish were starved 24 h and weighed for calculation of growth parameters. Six fish per tank were anesthetized with 100 mg/L MS-222 (Sigma, Ronkonkoma, NY, USA), randomly selected, and sampled for analysis. Blood was collected from the caudal vein and centrifuged to gain serum. The liver was sampled on ice, flash frozen in liquid nitrogen, and then stored at −80°C. Moreover, liver samples for transmission electron microscope examination were fixed in the 2.5% glutaraldehyde solution.
Measures of Biochemical Parameters
The activities of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) and the contents of triacylglycerols (TG), total cholesterol (T-CHO), and non-esterified-free fatty acids (NEFA) were measured by commercial kits (Nanjing JianCheng Bioengineering Institute, Nanjing, China).
Total lipid level in the liver was detected by Folch's method (Folch, 1957). Malondialdehyde (MDA) content was measured by the thiobarbituric acid method and superoxide dismutase (SOD) activity was determined by the hydroxylamine method, according to our previous study (Zhou et al., 2019). The reduced glutathione (GSH) content, catalase (CAT) activity, and total antioxidant capacity (T-AOC) were measured by commercial kits (Nanjing JianCheng Bioengineering Institute, Nanjing, China). The protein level in the liver homogenate was determined by the BCA protein assay kit (LABLEAD Inc., Beijing, China).
Analysis of Mitochondrial Status
Mitochondria were isolated from a fresh liver sample by a tissue mitochondria isolation kit (C3606, Beyotime Biotechnology, China). Then, the T-AOC and MDA contents of isolation mitochondria were determined. Another fresh liver was used to made single-cell suspension by mechanical method for mitochondrial ROS measurement. The hepatocyte was incubated with 5-μM MitoSOX Red (M36008, Invitrogen, Waltham, MA, USA) at 28°C for 10 min, and the fluorescence was determined by a microplate reader (Thermo Scientific, Waltham, MA, USA).
Histology Examination
For transmission electron microscopy analysis, liver samples were fixed in cold 2.5% glutaraldehyde solution for 24 h and post-fixed in 1% OsO4 at 4°C for 2 h. Then, the ultrathin slices were cut into 60-nm thickness after being embedded in epoxy resin, stained with uranyl acetate and lead citrate and observed under a transmission electron microscope (Hitachi H-7650, Tokyo, Japan).
Real-Time Quantitative PCR
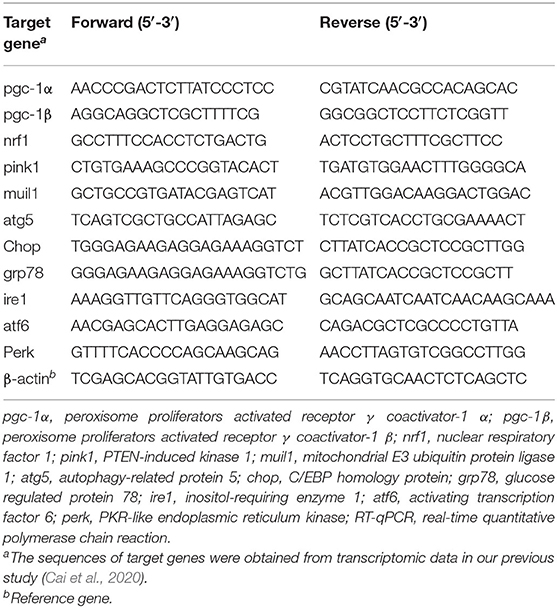
Total RNA was isolated using the FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme Biotech Co., Ltd, China). RNA purity was determined by a spectrophotometer (Thermo Scientific, United States) at 260/280 nm, and RNA integrity was investigated by using an agarose gel electrophoresis assay. Complementary DNA (cDNA) was synthesized by a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd, China). After eliminating genomic DNA by gDNA wiper, cDNA was used to measure specific the relative expression of genes in a quantitative PCR system (ABI, New York, NY, USA). ChamQ™ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd, China) was used for real-time PCR by the protocol of the manufacturer. The expressions of genes were normalized by β-actin and calculated by 2−ΔΔCt method according to the study of Xie et al. (2020). The sequences of primers used in this study were listed in Table 2.
Statistical Analysis
One-way ANOVA was used by the IBM SPSS 20 program. Tukey's test was used in significant difference analysis and P < 0.05 was considered as significant level. All data were shown as means ± SE with three replicates.
Results
Growth Performance and Fat Deposition
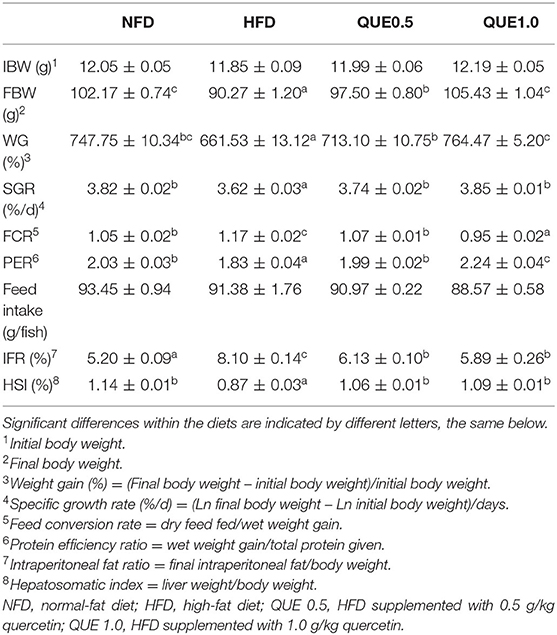
Fish-fed HFD showed the lowest weight gain (WG) and specific growth rate (SGR; P < 0.05). WG and SGR were significantly increased in the two QUE groups (P < 0.05), and fish in the QUE1.0 group exhibited the highest WG. The feed conversion rate (FCR) in the HFD group was higher than that of the NFD group, and the tendency of protein efficiency ratio (PER) was the opposite. QUE supplementation decreased FCR and increased PER, and the QUE1.0 group exhibited the lowest FCR and the highest PER among all groups (Table 3).
Fish-fed HFD showed excess fat deposition in the liver comparing to other groups. The contents of TG, T-CHO, and NEFA in the liver of fish-fed HFD were significantly higher than those of fish-fed NFD. The supplementation of 1 g/kg QUE significantly decreased the contents of total lipid, TG, T-CHO, and NEFA (Figure 1).
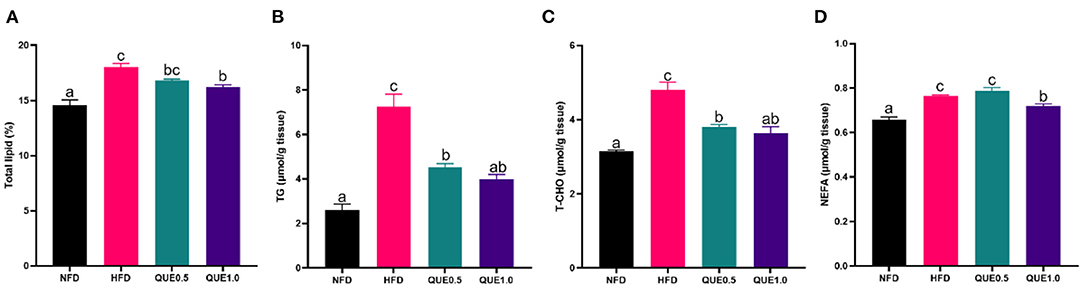
Figure 1. Fat levels in the liver of fish fed with experimental diets. (A) Total lipid content. (B) TG content. (C) T-CHO content. (D) NEFA content. TG, triacylglycerols; T-CHO, total cholesterol; NEFA, non-esterified-free fatty acids. Different letters indicate a significant difference among groups (P < 0.05).
Blood Chemistry
Both ALT and AST activities of the NFD group were significantly higher than the HFD group, while the QUE application decreased these two transaminase activities. For fat deposition, TG, T-CHO, and NEFA contents were all remarkably higher in fish-fed NFD compared to the NFD group. Meanwhile, the QUE application decreased these contents (Figure 2).
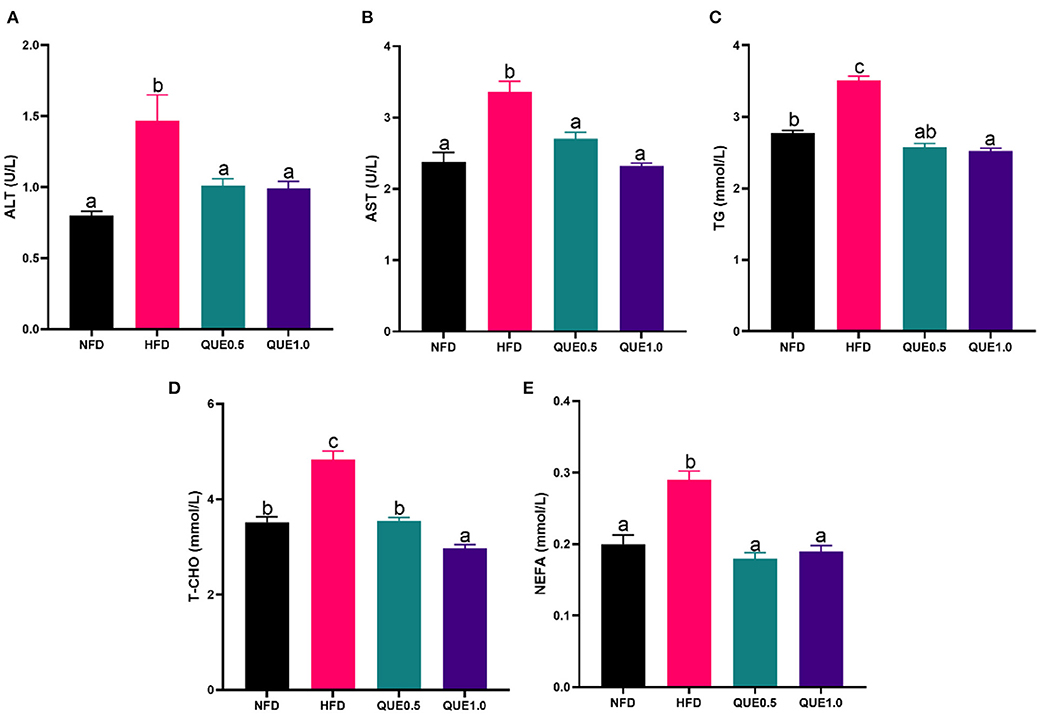
Figure 2. Blood biochemistry of fish fed with experimental diets. (A) ALT activity; (B) AST activity. (C) TG content. (D) T-CHO content. (E) NEFA content. AST, aspartate aminotransferase; ALT, alanine aminotransferase; TG, triacylglycerols; T-CHO, total cholesterol; NEFA, non-esterified-free fatty acids. Different letters indicate a significant difference among groups (P < 0.05).
Oxidative Status in Liver and Mitochondrion
In the liver, the activities/levels of CAT, SOD, GSH, and T-AOC of fish-fed HFD were significantly decreased, while MDA content was significantly increased. Both 0.5 and 1 g/kg supplementary of QUE dramatically elevated SOD activity, GSH content, and T-AOC and reduced MDA content (Figure 3).
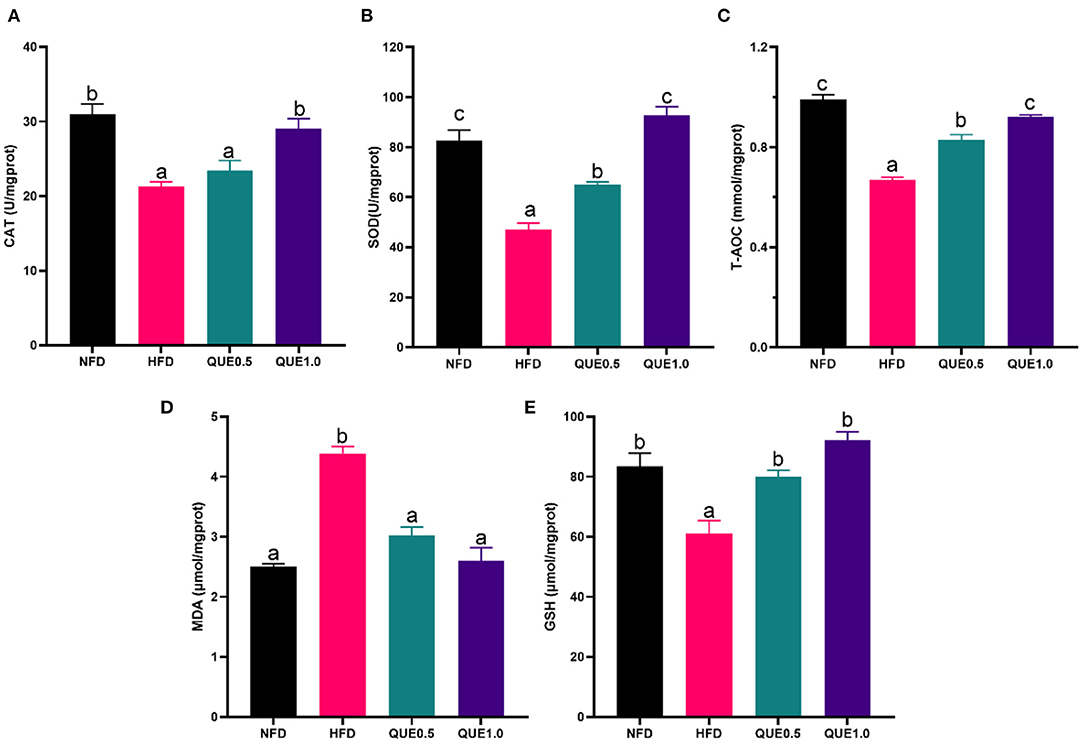
Figure 3. Oxidative status in the liver of fish fed with experimental diets. (A) CAT activity. (B) SOD activity. (C) GSH content. (D) T-AOC. (E) MDA content. SOD, superoxide dismutase; GSH, glutathione; CAT, catalase; T-AOC, total antioxidant capacity; MDA, malondialdehyde. Different letters indicate a significant difference among groups (P < 0.05).
In mitochondrion, fish-fed HFD showed the lowest T-AOC and the highest contents of MDA and ROS. Meanwhile, the supplementation of 0.1% QUE significantly enhanced T-AOC and reduced the contents of MDA and ROS (Figure 4).
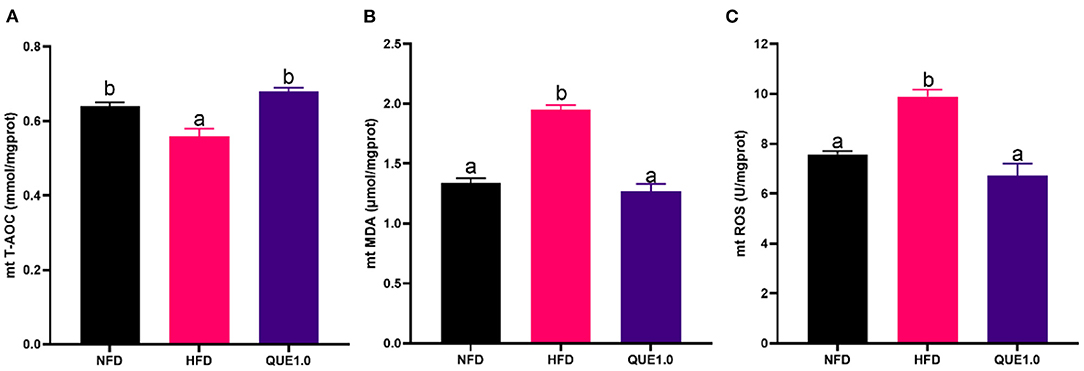
Figure 4. Oxidative status in hepatic mitochondria of fish fed with experimental diets. (A) T-AOC. (B) MDA content. (C) ROS content. T-AOC, total antioxidant capacity; MDA, malondialdehyde. Different letters indicate a significant difference among groups (P < 0.05).
Mitochondrial Status
The liver of fish-fed NFD showed normal ultrastructure. In these fish, the nucleus was round and the nucleolus was visible. Hepatocytes displayed dark and slender mitochondria. However, abnormal ultrastructure was found in mitochondria of fish-fed HFD, such as swell and nucleus atrophy. Further, the HFD-induced abnormal of mitochondria can be attenuated in QUE-fed fish (Figure 5).
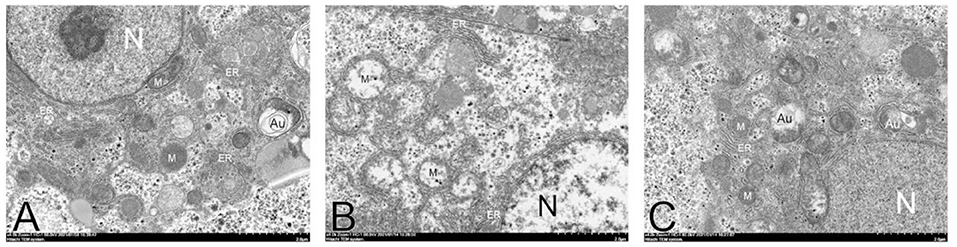
Figure 5. Transmission electron microscope images of hepatocytes in fish fed with experimental diets. N, nucleolus; M, mitochondria; Au, autophagosome; ER, endoplasmic reticulum. (A) NFD group. (B) HFD group. (C) QUE1.0 group. NHD, normal fat diet; HFD, high-fat diet; QUE, quercetin.
The expressions of mitochondria-related genes (pgc1-α, pgc1-β, and nrf1) were significantly downregulated by HFD. While, QUE can upregulate these expressions. In addition, HFD decreased the expression of mitophagy-related genes (pink1, atg5, and muil1; Figure 6).
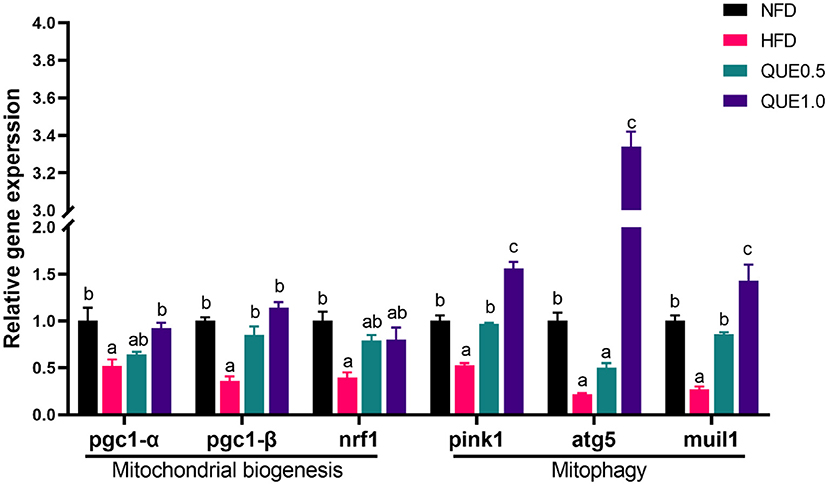
Figure 6. The expressions of mitochondrion-related genes in the liver of fish fed with experimental diets. Different letters indicate a significant difference among groups (P < 0.05).
Endoplasmic Reticulum Status
The expressions of ERS-related genes (chop, grp78, ire1, atf6, and perk) were significantly increased in the HFD group, and the application of QUE significantly downregulated the expressions of these genes (Figure 7).
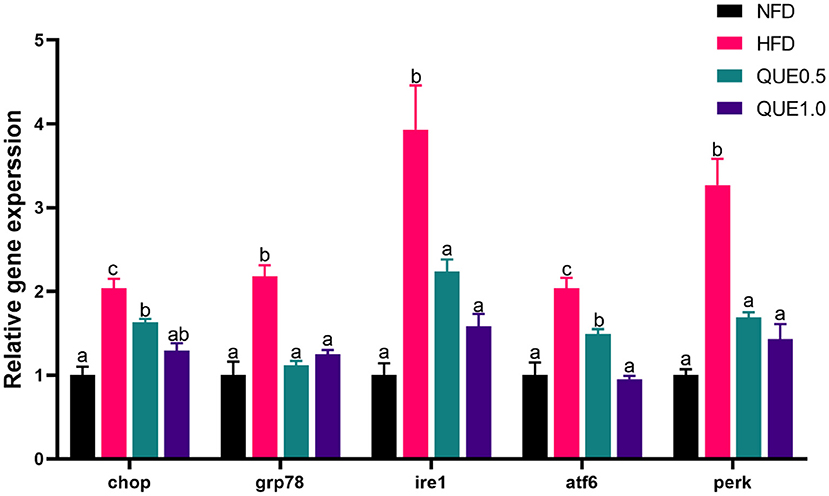
Figure 7. The expressions of ERS-related genes in the liver of fish fed with experimental diets. ERS, endoplasmic reticulum stress. Different letters indicate a significant difference among groups (P < 0.05).
Discussion
Similar to some previous studies, fish-fed HFD showed poor growth, excess fat deposition, and increased transaminase activities (Dong et al., 2020; Yin et al., 2021). QUE supplementation mitigated the decreasing growth performance and excess fat deposition, which is in agreement with previous studies (Jia et al., 2019). NEFA in the liver is a primary cause of lipotoxicity (Brent, 2010), while QUE can decrease its level and thus attenuate lipotoxicity. Excess fat often induces more ROS in mitochondria and causes oxidative stress (Marí et al., 2006). It is reported that QUE can be served as an antioxidant due to its free radical scavenging capacity (Boots et al., 2008; Pês et al., 2016, 2018). Similarly, our study also showed the antioxidant ability of QUE. It is considered that oxidative stress is often associated with the dysfunction of the mitochondrion (Lu et al., 2014, 2017). This was evidenced that the over ROS production of mitochondria in fish-fed HFD. Further, the HFD-induced abnormality of mitochondria and oxidative stress can be attenuated in QUE application. These findings indicate the regulatory effects of QUE on mitochondrion.
The mitochondria network in metabolism tissues is a dynamic state (Auger et al., 2015). Under normal conditions, new mitochondria can be constantly produced and the aberrant ones should be removed by mitophagy (Palikaras and Tavernarakis, 2014). The suppression of biogenesis and/or elimination will cause the dysfunction of the mitochondrion (Greene et al., 2012). PGC-1 is a key factor for mitochondrial biogenesis due to its promotion for downstream genes transcription, such as nrf1 (Rector et al., 2010). PINK1 is an activator in the classical mitophagy pathway stimulating E3 ubiquitin ligases to ubiquitinate membrane proteins of dysfunctional mitochondria (Tolkovsky, 2009). This study indicated that HFD feeding arrested mitochondrial biogenesis and mitophagy by the downregulation of the genes. QUE can protect mitochondria through activation of the gene expressions. In addition, it was reported that QUE can play a key role in the mitochondrial homeostasis of mice (Liu et al., 2018).
It is considered that there is a crosstalk between the endoplasmic reticulum and mitochondrion (Daw et al., 2020). Previous studies have indicated that ERS can be caused by the dysfunction of the mitochondrion (Malhotra and Kaufman, 2007; Hafiz et al., 2016). The damaged endoplasmic reticulum induces ERS through unfolded proteins response by IRE1, ATF6, and PERK pathways (Walter and Ron, 2011). These unfolded proteins induce the release of molecular chaperones and then apoptosis and inflammation (Szegezdi et al., 2006; Kaplowitz et al., 2007; Urra et al., 2013). Many evidence indicate that ERS contributed to the liver damage induced by HFD (Cao et al., 2018; Jia et al., 2020). Our results also found that HFD upregulated the expression of key genes in ERS pathway. Furthermore, the application of QUE suppressed the activation of ERS by downregulating the expressions of related genes.
Conclusion
The supplementary of quercetin can attenuate excess fat deposition in fish tissues induced by the intake of HFD. Quercetin can activate the biogenesis and autophagy of mitochondria and suppress ERS, which is related with its fat-lowering effect.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This animal study was reviewed and approved by the Animal Research Institute Committee of Jimei University.
Author Contributions
Y-ZD and C-XZ designed the study. Y-ZD and TX conducted the experiments. Y-ZD wrote the manuscript. J-BL corrected the writing style. LW and KS provided technical support. All authors read and approved the final version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (32072984), the Natural Science Foundation of Fujian Province (2020J01664), and the China Agriculture Research System (CARS-47).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors genuinely appreciate Xiaoqi Li, Youmei Jin, and Yuxi Jiang for their valuable assistance in feeding trials and lab works.
References
Auger, C., Alhasawi, A., Contavadoo, M., and Appanna, V. D. (2015). Dysfunctional mitochondrial bioenergetics and the pathogenesis of hepatic disorders. Front. Cell Dev. Biol. 3:40. doi: 10.3389/fcell.2015.00040
Boots, A. W., Haenen, G. R. M. M., and Bast, A. (2008). Health effects of quercetin: From antioxidant to nutraceutical. Eur. J. Pharmacol. 585, 325–337. doi: 10.1016/j.ejphar.2008.03.008
Brent, A. N. (2010). Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology 52, 774–788. doi: 10.1002/hep.23719
Cai, L., Wang, L., Song, K., Lu, K., Zhang, C., and Rahimnejad, S. (2020). Evaluation of protein requirement of spotted seabass (Lateolabrax maculatus) under two temperatures, and the liver transcriptome response to thermal stress. Aquaculture 516:734615. doi: 10.1016/j.aquaculture.2019.734615
Cao, X. F., Dai, Y. J., Liu, M. Y., Yuan, X. Y., and Wang, C. C. (2018). High-fat diet induces aberrant hepatic lipid secretion in blunt snout bream by activating endoplasmic reticulum stress-associated IRE1/XBP1 pathway. BBA Mol. Cell Biol. L. 3:1864. doi: 10.1016/j.bbalip.2018.12.005
Daw, C. C., Ramachandran, K., Enslow, B. T., Maity, S., Bursic, B., Novello, M. J., et al. (2020). Lactate elicits ER-Mitochondrial mg2+ dynamics to integrate cellular metabolism. Cell 183, 474–489. doi: 10.1016/j.cell.2020.08.049
Dong, Y., Li, L., Espe, M., Lu, K., and Rahimnejad, S. (2020). Hydroxytyrosol attenuates hepatic fat accumulation via activating mitochondrial biogenesis and autophagy through the AMPK pathway. J. Agr. Food Chem. 68, 9377–9386. doi: 10.1021/acs.jafc.0c03310
Du, Z. Y., Clouet, P., Zheng, W. H., Degrace, P., and Liu, Y. J. (2006). Biochemical hepatic alterations and body lipid composition in the herbivorous grass carp (Ctenopharyngodon idella) fed high-fat diets. Brit. J. Nutr. 95, 905–915. doi: 10.1079/BJN20061733
Folch, J. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Bid. Chem. 226, 497–509. doi: 10.1016/S0021-9258(18)64849-5
Formica, J. V., and Regelson, W. (1995). Review of the biology of Quercetin and related bioflavonoids. Food Chem. Toxicol. 33, 1061–1080. doi: 10.1016/0278-6915(95)00077-1
Greene, A. W., Grenier, K., Aguileta, M. A., Muise, S., Farazifard, R., Haque, M. E., et al. (2012). Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13:378. doi: 10.1038/embor.2012.14
Hafiz, M., Geum, H. G., Hyung-Ryong, K., and Han-Jung, C. (2016). Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 17:327. doi: 10.3390/ijms17030327
Jia, E., Yan, Y., Zhou, M., Li, X., Jiang, G., Liu, W., et al. (2019). Combined effects of dietary quercetin and resveratrol on growth performance, antioxidant capability and innate immunity of blunt snout bream (Megalobrama amblycephala). Anim. Feed Sci. Tech. 256: 114268. doi: 10.1016/j.anifeedsci.2019.114268
Jia, R., Cao, L., Du, J., He, Q., Gu, Z., Jeney, G., et al. (2020). Effects of high-fat diet on steatosis, endoplasmic reticulum stress and autophagy in liver of tilapia (Oreochromis niloticus). Front. Mar. Sci. 7:363. doi: 10.3389/fmars.2020.00363
Jin, Y., Tian, L., Zeng, S., Xie, S., Yang, H., Liang, G., et al. (2013). Dietary lipid requirement on non-specific immune responses in juvenile grass carp (Ctenopharyngodon idella). Fish Shellfish Immun. 34, 1202–1208. doi: 10.1016/j.fsi.2013.01.008
Kaplowitz, N., Than, T. A., Shinohara, M., and Ji, C. (2007). Endoplasmic reticulum stress and liver injury. Semin. Liver Dis. 27, 367–377. doi: 10.1055/s-2007-991513
Liu, P., Lin, H., Xu, Y., Feng, Z., Jing, W., Liu, J., et al. (2018). Frataxin-Mediated PINK1-Parkin-Dependent mitophagy in hepatic steatosis: the protective effects of quercetin. Mol. Nutr. Food Res. 62:1800164. doi: 10.1002/mnfr.201800164
Lu, K., Lu, K., Wang, L., Wang, L., Zhang, D., Zhang, D., et al. (2017). Berberine attenuates oxidative stress and hepatocytes apoptosis via protecting mitochondria in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fish Physiol. Biochem. 43, 65–76. doi: 10.1007/s10695-016-0268-5
Lu, K. L., Xu, W. N., Liu, W. B., Wang, L. N., Zhang, C. N., and Li, X. F. (2014). Association of mitochondrial dysfunction with oxidative stress and immune suppression in blunt snout bream Megalobrama amblycephala fed a high-fat diet. J. Aquat. Anim. Health. 26, 100–112. doi: 10.1080/08997659.2014.893460
Malhotra, J. D., and Kaufman, R. J. (2007). Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a Double-Edged sword? Antioxid. Redox Sign. 9, 2277–2294. doi: 10.1089/ars.2007.1782
Marí, M., Caballero, F., Colell, A., Morales, A., Caballeria, J., Fe Rnandez, A., et al. (2006). Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. 4, 185–198. doi: 10.1016/j.cmet.2006.07.006
Palikaras, K., and Tavernarakis, N. (2014). Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Exp. Gerontol. 56, 182–188. doi: 10.1016/j.exger.2014.01.021
Pês, T. S., Saccol, E. M. H., Londero, É. P., Bressan, C. A., Ourique, G. M., Rizzetti, T. M., et al. (2018). Protective effect of quercetin against oxidative stress induced by oxytetracycline in muscle of silver catfish. Aquaculture 484, 120–125. doi: 10.1016/j.aquaculture.2017.10.043
Pês, T. S., Saccol, E. M. H., Ourique, G. M., Londero, É. P., Gressler, L. T., Golombieski, J. I., et al. (2016). Quercetin in the diet of silver catfish: effects on antioxidant status, blood parameters and pituitary hormone expression. Aquaculture 458, 100–106. doi: 10.1016/j.aquaculture.2016.02.020
Porras, D., Nistal, E., Martínez-Flórez, S., Pisonero-Vaquero, S., Olcoz, J. L., Jover, R., et al. (2017). Protective effect of quercetin on high-fat diet-induced non-alcoholic fatty liver disease in mice is mediated by modulating intestinal microbiota imbalance and related gut-liver axis activation. Free Radical Bio. Med. 102, 188–202. doi: 10.1016/j.freeradbiomed.2016.11.037
Rector, R. S., Thyfault, J. P., Uptergrove, G. M., Morris, E. M., Naples, S. P., Borengasser, S. J., et al. (2010). Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J. Hepatol. 52, 727–736. doi: 10.1016/j.jhep.2009.11.030
Rocha, M., Apostolova, N., Diaz-Rua, R., Muntane, J., and Victor, V. M. (2020). Mitochondria and T2D: role of autophagy, ER stress, and inflammasome. Trends Endocrin. Met. 31, 725–741. doi: 10.1016/j.tem.2020.03.004
Shadel, G. S., and Horvath, T. L. (2015). Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. doi: 10.1016/j.cell.2015.10.001
Szegezdi, E., Logue, S. E., Gorman, A. M., and Samali, A. (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885. doi: 10.1038/sj.embor.7400779
Tilg, H., and Moschen, A. R. (2010). Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 52, 1836–1846. doi: 10.1002/hep.24001
Tolkovsky, A. M. (2009). Mitophagy. BBA-Mol. Cell Res. 1793, 1508–1515. doi: 10.1016/j.bbamcr.2009.03.002
Urra, H., Dufey, E., Lisbona, F., Rojas-Rivera, D., and Hetz, C. (2013). When ER stress reaches a dead end. BBA Mol. Cell Res. 1833, 3507–3517. doi: 10.1016/j.bbamcr.2013.07.024
Wallace, D. C. (2007). Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76, 781–821. doi: 10.1146/annurev.biochem.76.081205.150955
Walter, P., and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Wang, N., Liu, Y., Ma, Y., and Wen, D. (2018). Hydroxytyrosol ameliorates insulin resistance by modulating endoplasmic reticulum stress and prevents hepatic steatosis in diet-induced obesity mice. J. Nutr. Biochem. 57, 180–188. doi: 10.1016/j.jnutbio.2018.03.018
Watanabe, T. (1982). Lipid nutrition in fish. Comp. Biochem. Phys. B. 1, 3–15. doi: 10.1016/0305-0491(82)90196-1
Xie, N., Wen, H., Xie, S., Jiang, M., Yu, L., Wu, F., et al. (2021a). Adaptations of hepatic lipid and glucose metabolism in response to high-macronutrient diets in juvenile grass carp. Aquacult. Nutr. 00, 1–12. doi: 10.1111/anu.13311
Xie, S., Lin, Y., Wu, T., Tian, L., Liang, J., and Tan, B. (2021b). Dietary lipid levels affected growth performance, lipid accumulation, inflammatory response and apoptosis of Japanese seabass (Lateolabrax japonicus). Aquacult. Nutr. 27, 807–816. doi: 10.1111/anu.13225
Xie, S., Yin, P., Tian, L., Liu, Y., and Niu, J. (2020). Lipid metabolism and plasma metabolomics of juvenile largemouth bass Micropterus salmoides were affected by dietary oxidized fish oil. Aquaculture 522:735158. doi: 10.1016/j.aquaculture.2020.735158
Xu, J., Qin, J., Yan, B., Zhu, M., and Luo, G. (2011). Effects of dietary lipid levels on growth performance, feed utilization and fatty acid composition of juvenile Japanese seabass (Lateolabrax japonicus) reared in seawater. Aquacult. Int. 19, 79–89. doi: 10.1007/s10499-010-9342-7
Yin, P., Xie, S., Zhuang, Z., He, X., Tang, X., Tian, L., et al. (2021). Dietary supplementation of bile acid attenuate adverse effects of high-fat diet on growth performance, antioxidant ability, lipid accumulation and intestinal health in juvenile largemouth bass (Micropterus salmoides). Aquaculture 531:735864. doi: 10.1016/j.aquaculture.2020.735864
Keywords: Lateolabrax maculatus, fatty liver, mitochondrial biogenesis, mitophagy, endoplasmic reticulum stress
Citation: Dong Y-Z, Xia T, Lin J-B, Wang L, Song K and Zhang C-X (2021) Quercetin Attenuates High-Fat Diet-Induced Excessive Fat Deposition of Spotted Seabass (Lateolabrax maculatus) Through the Regulatory for Mitochondria and Endoplasmic Reticulum. Front. Mar. Sci. 8:746811. doi: 10.3389/fmars.2021.746811
Received: 24 July 2021; Accepted: 23 August 2021;
Published: 20 September 2021.
Edited by:
Chunnuan Zhang, Henan University of Science and Technology, ChinaReviewed by:
Shiwei Xie, Guangdong Ocean University, ChinaSonglin Li, Shanghai Ocean University, China
Copyright © 2021 Dong, Xia, Lin, Wang, Song and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Xiao Zhang, Y3h6aGFuZ0BqbXUuZWR1LmNu
 Yan-Zou Dong
Yan-Zou Dong Tian Xia
Tian Xia Ling Wang
Ling Wang Chun-Xiao Zhang
Chun-Xiao Zhang