
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 09 September 2021
Sec. Marine Molecular Biology and Ecology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.746598
The complete mitochondrial genome of Eleutheronema rhadinum was determined for the first time. The E. rhadinum mitogenome was 16,718 bp in length, comprising 13 protein coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), 1 control region (CR), and 1 L-strand replication origin (OL). The genome organization, and base composition were similar to other teleosts. An additional non-coding region detected in the HSL cluster, was with significant similarity to the nearby tRNA-Leu (CUN), which could be a useful marker for identifying E. rhadinum and Eleutheronema tetradactylum. Furthermore, phylogenetic analysis showed species in the family Polynemidae were well clustered together. But sister relationships between Polynemidae and Sciaenidae, Menidae plus Lactariidae were refused by present study.
The East Asian fourfinger threadfin, Eleutheronema rhadinum, is endemic to East Asia region, widely distributed in coastal areas of China (Zhang et al., 1955; Motomura et al., 2002; Zhuang et al., 2006), Japan and northern Vietnam (Motomura et al., 2001, 2007). The species is an important commercial fish in China, with the annual catch of more than 1,100 tons from 1970 to 1999 (Motomura, 2004). Due to its relatively high price, recently, it has become an important aquaculture species in southern China (You et al., 2014; Ou et al., 2017). Because of the high similarity, it is difficult to identify Eleutheronema species by visual check. Especially, the distribution of E. rhadinum and E. tetradactylum are overlapped in the Northern continental shelf of South China Sea. For a long time in the past, E. rhadinum had long been recognized as a junior synonym of E. tetradactylum (Kagwade, 1970; Motomura et al., 2002).
Mitochondrial genome is a closed-circular molecule, ranging from 15 to 18 kb in length. It comprises a light and a heavy chain, encoding 13 protein-coding genes, 2 rRNAs, 22 tRNAs, an origin of light strand replication (OL), and a control region (Taanman, 1999). Due to its small size, maternal inheritance, low level of recombination, and fast rate of evolution (Moritz et al., 1987), mtDNA has been widely used as a tool for taxonomy, molecular ecology, and population genetics studies (Wilson et al., 1985; Satoh et al., 2016; Zhong et al., 2016, 2018a,b). However, to date, there have been no reports about the mitochondrial genome of E. rhadinum.
The genera Eleutheronema belongs to the Family Polynemidae, which had been previously placed in the Order Polynemiformes (Wang, 1958) and Mugiliformes (Rosen, 1964). Subsequently, Nelson gave subordinal status to Polynemidae and placed it in the Order Perciformes based on morphological analyses (Nelson, 1984), having a sister relationship with Sphyraenidae and Sciaenidae (Johnson, 1993). However, a recently phylogenetic hypothesis grouped the diversity of Percomorphs into nine supraordinal series. The Family Polynemidae was placed in the Series Carangaria, which had a close relation with Menidae and Lactariidae (Sanciangco et al., 2016), but the Order-level was still incertae sedis (Betancur-R et al., 2017).
In the present study, the first mitochondrial genome of E. rhadinum was annotated. Its organization and characterization were fully analyzed. These results would not only provide better understanding of the systematics of E. rhadinum, but also facilitate future studies of species identification and population genetics.
During the ichthyological survey in Yangzhong section of the Yangtze River, China, a specimen of E. rhadinum was caught. The individual was preserved in 70% ethanol and deposited in the collection of Freshwater Fisheries Research Institute of Jiangsu Province with the reference number JSFFRI-18010. The specimen was identified as E. rhadinum due to its 4 pectoral filaments, 95 pored lateral-line scales, and 13/16 scale rows above/below lateral line by morphological identification (Zhong et al., 2019). Total genomic DNA from the pectoral fin was extracted using an Ezup Column Animal Genomic DNA kit (Sangon, Shanghai, China) and evaluated by 1% agarose gel electrophoresis.
To amplify the entire mitogenome, 30 pairs of fish-universal primers (Miya and Nishida, 1999) and 17 pairs of specific primers designed based on mtDNA sequence of E. tetradactylum (accession no. NC_021620) were employed. PCR amplification was performed as described previously (Zhong et al., 2018b). The PCR products were evaluated by 1.5% agarose gel electrophoresis and Sanger-sequenced by Sangon Biotech Co., Ltd. (Shanghai, China) with the same primers.
Raw sequences were screened and aligned into contigs using BioEdit (Hall, 2013) then spliced the complete mitogenome. The mitogenome was preliminarily annotated by MitoAnnotator in the MitoFish database (Iwasaki et al., 2013). The software tRNAscan-SE version 2.0 was used to identify tRNA genes and predict the secondary structures (Lowe and Chan, 2016). Non-coding regions were identified by sequence homology analysis. Tandem repeats in the control region were searched by Tandem Repeats Finder (Benson, 1999). L-strand replication origin (OL) was predicted with the Mfold web server (Zuker, 2003). The base composition was calculated by MEGA 6 (Tamura et al., 2013). Composition skewness was performed by AT skew and GC skew (Perna and Kocher, 1995).
To discuss the phylogenetic relationships of Polynemidae, E. rhadinum and 20 other related mitogenomes downloaded from GenBank were used. The concatenated sequences of the 13 protein coding genes (PCGs) were aligned in BioEdit excluding the stop codons. Because of its high heterogeneity and consistently poor phylogenetic performance, the ND6 gene was deleted (Miya and Nishida, 2000). The final alignment length was 3,633 amino acids. The Maximum Likelihood (ML) phylogenetic tree was constructed by MEGA 6 based on the mtREV24 + G + I + F model with 1,000 bootstrap replicates.
The newly assembled mitogenome of E. rhadinum was 16,718 bp in length (Figure 1), which was longer than that of E. tetradactylum (16,470 bp). The variability in mitogenome length can be attributed to a new additional intergenic non-coding region between tRNA-Ser and tRNA-Leu (Zhang et al., 2014). The gene content was typical for teleost, compromising 13 PCGs, 22 tRNA genes, 2 rRNA genes, 1 control region (CR), and 1 OL (Supplementary Table 1). Most genes (12 PCGs, 14 tRNAs, 2 rRNAs and CR) were encoded on the heavy strand (H-strand), while the ND6 gene, 8 tRNAs and OL were harbored on the light strand (L-strand).
The nucleotide composition of E. rhadinum mitogenome is slightly biased toward AT (55.2%). The overall base composition was estimates as 28.2% A, 27.0% T, 28.8% C, and 16.0% G (Table 1). Control region (CR) sequence had the strongest AT biases (67.4%), while PCGs, tRNAs and rRNAs showed similar AT biases to the whole mitogenome. Similarly to other teleosts, positive AT-skew (0.022) and negative GC-skew (−0.286) were found for E. rhadinum mitogenome, indicating a strong excess of C over G nucleotides and a slight excess of A over T nucleotides. In PCGs, there were no deflections of the base composition in the first codon positions. However, markedly G-skewed (−0.328) and T-skewed (−0.382) were detected in the second codon positions. Meanwhile, in the third codon positions, a strong anti-G bias (−0.647) was detected as noted in typical teleosts (Satoh et al., 2016). In RNA genes and CR, positive AT-skews (0.074 to 0.155) and negative GC-skews (−0.092 to −0.149) were also found, respectively.
The 13 PCGs in the E. rhadinum mitogenome were similar to those of other teleosts. The total length of the PCGs was 11,449 bp, comprising 68.30% of the entire mitochondrial length. Most of the PCGs used ATG as the initiation codon, except the COI gene, which started with GTG like in many other Osteichthyes mitogenomes. TAR (TAA and TAG) was used as termination codons. Nevertheless, incomplete termination codons (TA- and T-) were detected in ND2, COII, ATPase6, ND3, and ND4, which could add A or AA to the 3′ region of the transcripts by post-transcriptional polyadenylation (Ojala et al., 1981). Three overlapping regions were identified between ATPase8 and ATPase6 (10 bp), ND4 and ND4L (7 bp), and ND5 and ND6 (4 bp), which were conventional in teleosts.
Like other teleosts, twenty-two tRNA genes were predicted in the E. rhadinum mitogenome, ranging from 64 to 75 bp (Supplementary Figure 1). Arrangements of 14 H-strand-encoded and 8 L-strand-encoded tRNAs was identical to the typical one in vertebrate mitogenomes. Two kinds of leucine (tRNA-Leu UUR and CUN) and serine (tRNA-Ser UCN and AGN) were found. Secondary structures predicted by tRNAscan-SE suggested that all tRNAs could be folded as a typical cloverleaf structure except for tRNA-Ser (AGN). Due to the lack of the dihydrouridine arm (DHU), tRNA-Ser (AGN) could not form a stable secondary structure, which is common in vertebrates (Garey and Wolstenholme, 1989).
Two rRNA genes, 12S rRNA and 16S rRNA, were identified between tRNA-Phe and tRNA-Leu, separating by tRNA-Val (Figure 1). The 12S rRNA was 996 bp in length with 52.7% AT content. The 16S rRNA was 1793 bp in length with 55.1% AT content.
Two typical non-coding regions, control region (CR) and L-strand replication origin (OL), were found in the E. rhadinum mitogenome. The OL was detected in the WANCY cluster between tRNA-Asn and tRNA-Cys with a length of 38 bp (Table 1), containing a 20 bp stem and a 14 bp loop (Figure 2A). Significant asymmetry was displayed in the stem, more pyrimidines were used at the 5′ end. The sequence 5′-GCGGG-3′ but conserved motif 5′-GCCGG-3′ was identified at the base of the stem, which was also noted in other fishes, such as Odontobutis haifengensis (Zhong et al., 2018b) and Paraplagusia japonica (Gong et al., 2013). The conserved motif was likely to be involved in the mechanism of RNA transformation to DNA (Wong and Clayton, 1985).
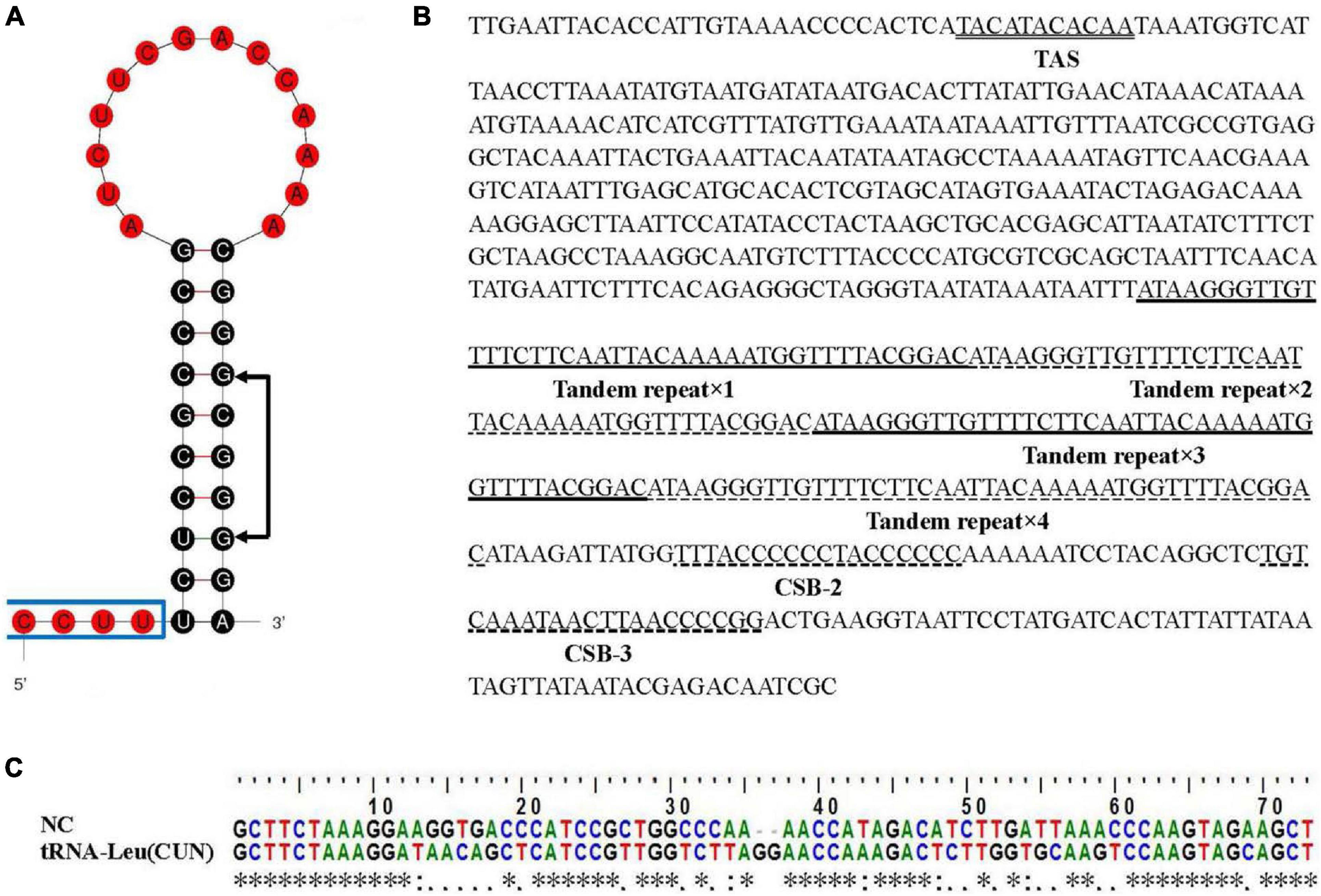
Figure 2. Non-coding regions in the Eleutheronema rhadinum mitogenome. (A) The OL stem-loop secondary structure. The conserved motif 5′-GCCGG-3′ was indicated with arrows. (B) Control region in the E. rhadinum mitogenome. Termination associated sequence (TAS), conserved sequence blocks (CSB-2 and CSB-3) and tandem repeats are underlined. (C) Sequence alignment between NC and tRNA-Leu (CUN). * means the corresponding two bases are the same, : means the corresponding two bases are matched, and . means the corresponding two bases are mismatched.
The control region (CR) was detected between tRNA-Pro and tRNA-Phe with a length of 700 bp. After blasting, several regulatory elements were identified (Figure 2B). A termination associated sequence (TAS) motif (TACATACACAA) was found at the beginning of CR, which was considered to be a signal for terminating of H-strand elongation (Clayton, 1991). However, three typical central conserved sequence blocks (CSB-F, E, and D) and a conserved sequence blocks (CSB-1) were not detected, which were also absent in some gobies belonged to the Perciformes (Kim et al., 2004; Liu et al., 2013). Instead, a perfect repeat region was detected in the middle of CR, which contained a 42 bp sequence tandemly repeated four times. Slipped-strand mispairing was thought to be the most likely mechanism for tandem repeats in mitochondrial genomes (Broughton and Dowling, 1997). Furthermore, two conserved sequence blocks (CSB-2 and 3) were identified in the end of CR, which were involved in RNA polymerase positioning during transcription and priming replication (Liu et al., 2013).
A novel additional non-coding region (NC) was identified on the H-strand in the E. rhadinum mitogenome by comparing to mitogenome sequences of other species in Polynemidae family. The tRNA cluster HSL (tRNAHis-tRNASer-tRNALeu) is located between ND4 and ND5 in typical teleost mitogenome. By contrast, in the mitogenome of E. rhadinum, the HSL tRNA cluster was divided into two parts by the novel intergenic non-coding region (Figure 1). The length of NC was 165 bp in length, while the sequence was only 5–8 bp in other species of Polynemidae family. Homology searches on NC revealed significant similarity to the nearby tRNA-Leu (CUN) (Figure 2C). The novel NC region could be a useful marker for identifying E. rhadinum and E. tetradactylum and for phylogenetic analyses. The duplication/random loss model would be applicable to explain the insertion of the additional non-coding region (Dowton et al., 2003). Tandem duplication occurred in the tRNA-Leu (CUN), following by a random loss of function and becoming pseudogene. With the accumulation of data, CR and three tRNA clusters (IQM, WANCY, and HSL) were found to be frequent of base insertion and deletion, tandem repetitive and gene rearrangement (Zhong et al., 2018b).
Phylogenetic tree revealed that all 21 fishes were grouped well in family level (Figure 3). Six polynemid species formed a monophyletic Polynemidae cluster, which had been also supported by morphological evidence (Kang et al., 2018). Especially, E. rhadinum and E. tetradactylum formed the Eleutheronema genus, and Polynemus dubius and Polynemus paradiseus formed the Polynemus genus. Then, Eleutheronema genus and Polynemus genus clustered with Polydactylus forming Polynemidae family. Furthermore, it has also clearly showed that Polynemidae was not included in the Order Mugiliformes and Order Perciformes. Therefore, the sister relationship of Polynemidae and Sciaenidae confirmed by morphological characters (Kang et al., 2018) was inconsistent with the results in this present study. In addition, the close relation between Polynemidae and Menidae plus Lactariidae suggested by Sanciangco et al. (2016) was not supported as well. Nevertheless, molecular analysis based on mitogenomes supported the monophyly of family Polynemidae and Sphyraenidae in Percomorph fishes. Due to its well-supported monophyletic cluster, the independent Order Polynemiformes might be reset as Wang’s (1958). However, to confirm the order-level status of the Family Polynemidae, and more importantly, to fix the “bush at the top” of the teleost tree (Percomorpha phylogenetic intrarelationships) bit by bit, it is necessary to make further analyses of polynemid species based on more molecular and morphological information and extensive taxon sampling.
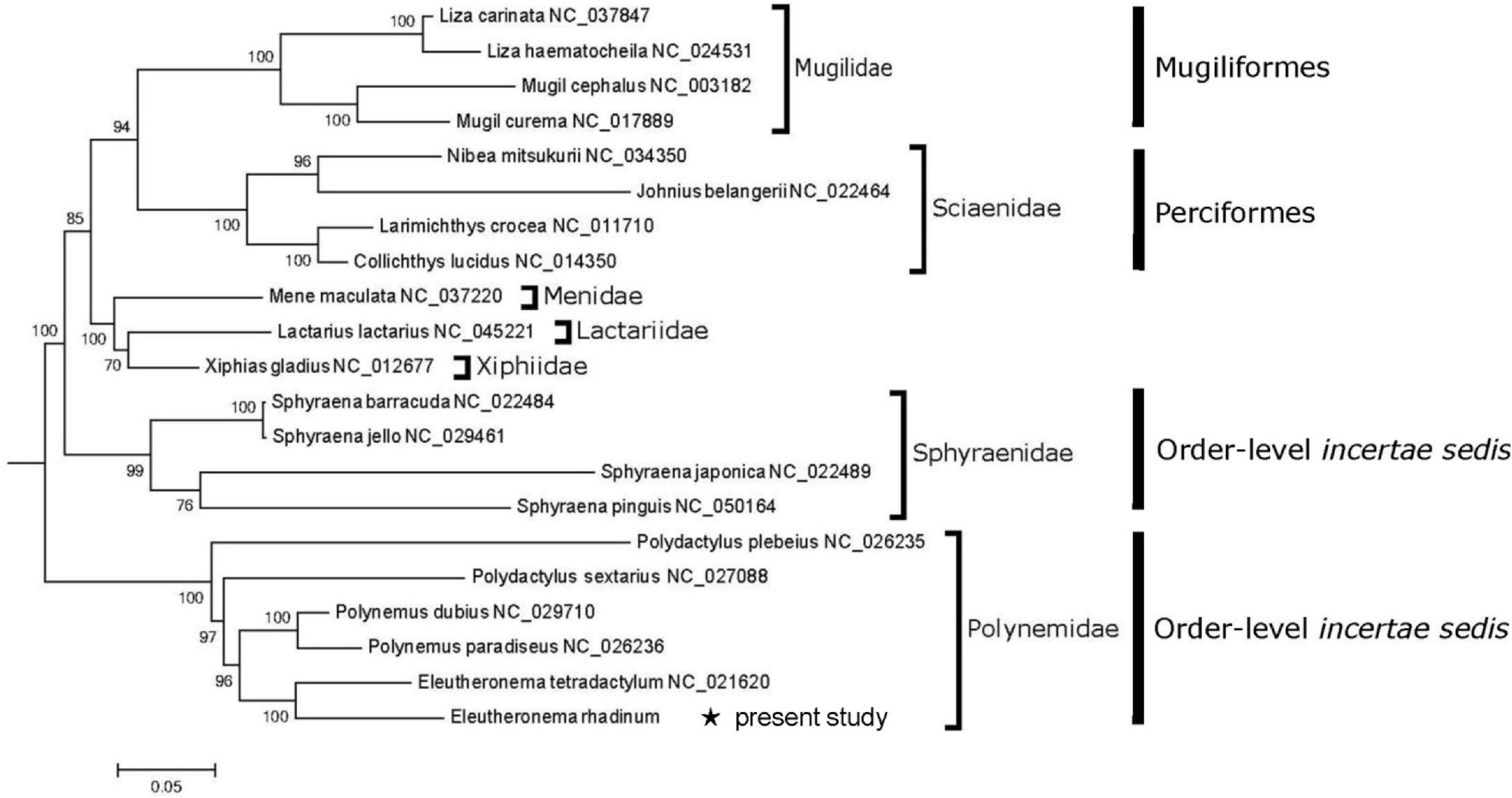
Figure 3. A phylogenetic tree inferred by ML analysis and based on 12 protein-coding genes. Bootstrap values are expressed as percentages and are noted on nodes. * indicates E. rhadinum examined in the present study.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MW845829).
The animal study was reviewed and approved by the Freshwater Fisheries Research Institute of Jiangsu Province.
LZ and XC designed the research and revised the manuscript. DL and ST were involved in the sample collection and preprocessing. MW and LZ conducted the research and drafted the manuscript. LZ and DL conducted the molecular work and data analysis. All authors reviewed and approved the manuscript.
This study was supported by the Earmarked fund for China Agriculture Research System of MOF and MARA (CARS-46), Inland Water Fishery Resources Monitoring Program of Jiangsu Province (2020–2021), and Major Project of Hydrobios Resources in Jiangsu Province (ZYHB16-3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.746598/full#supplementary-material
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Betancur-R, R., Wiley, E. O., Arratia, G., Acero, A., Bailly, N., Miya, M., et al. (2017). Phylogenetic classification of bony fishes. BMC Evol. Biol. 17:162. doi: 10.1186/s12862-017-0958-3
Broughton, R. E., and Dowling, T. E. (1997). Evolutionary dynamics of tandem repeats in the mitochondrial DNA control region of the minnow Cyprinella spiloptera. Mol. Biol. Evol. 14, 1187–1196. doi: 10.1093/oxfordjournals.molbev.a025728
Clayton, D. A. (1991). Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem. Sci. 16, 107–111. doi: 10.1016/0968-0004(91)90043-U
Dowton, M., Castro, L. R., Campbell, S. L., Bargon, S. D., and Austin, A. D. (2003). Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J. Mol. Evol. 56, 517–526. doi: 10.1007/s00239-002-2420-3
Garey, J. R., and Wolstenholme, D. R. (1989). Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNAser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 28, 374–387. doi: 10.1007/BF02603072
Gong, L., Shi, W., Wang, Z. M., Miao, X. G., and Kong, X. Y. (2013). Control region translocation and a tRNA gene inversion in the mitogenome of Paraplagusia japonica (Pleuronectiformes: Cynoglossidae). Mitochondrial DNA 24, 671–673. doi: 10.3109/19401736.2013.773984
Hall, T. (2013). BioEdit. Available online at: http://www.mbio.ncsu.edu/BioEdit/bioedit.html. (accessed September 18, 2013).
Iwasaki, W., Fukunaga, T., Isagozawa, R., Yamada, K., Maeda, Y., Satoh, T. P., et al. (2013). MitoFish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 30, 2531–2540. doi: 10.1093/molbev/mst141
Kang, S. H., Imamura, H., and Kawai, T. (2018). Morphological evidence supporting the monophyly of the family Polynemidae (Teleostei: Perciformes) and its sister relationship with Sciaenidae. Ichthyol. Res. 65, 29–41. doi: 10.1007/s10228-017-0591-6
Kim, I. C., Kweon, H. S., Kim, Y. J., Kim, C. B., Gye, M. C., Lee, W. O., et al. (2004). The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene 336, 147–153. doi: 10.1016/j.gene.2004.04.009
Liu, T. X., Jin, X. X., Wang, R. X., and Xu, T. J. (2013). Complete sequence of the mitochondrial genome of Odontamblyopus rubicundus (Perciformes: Gobiidae): genome characterization and phylogenetic analysis. J. Genet. 92, 423–432. doi: 10.1007/s12041-013-0283-6
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Miya, M., and Nishida, M. (1999). Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar. Biotechnol. 1, 416–426. doi: 10.1007/PL00011798
Miya, M., and Nishida, M. (2000). Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenet. Evol. 17, 437–455. doi: 10.1006/mpev.2000.0839
Moritz, C., Dowling, T. E., and Brown, W. M. (1987). Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 18, 269–292. doi: 10.1146/annurev.es.18.110187.001413
Motomura, H. (2004). Threadfins of the World (Family Polynemidae), An Annotated and Illustrated Catalogue of Polynemid Species Known to Data. FAO Species Catalogue for Fishery Purposes No. 3. Rome: Food and Agriculture Organization.
Motomura, H., Ito, M., Takayama, M., Haraguchi, Y., and Matsunuma, M. (2007). Second Japanese record of a threadfin, Eleutheronema rhadinum (Perciformes, Polynemidae), with distributional implications. Biogeography 9, 7–11.
Motomura, H., Iwatsuki, Y., Kimura, S., and Yoshino, T. (2002). Revision of the Indo-West Pacific polynemid fish genus Eleutheronema (Teleostei: Perciformes). Ichthyol. Res. 49, 47–61. doi: 10.1007/s102280200005
Motomura, H., Senou, H., and Iwatsuki, Y. (2001). A record of a threadfin, Eleutheronema tetradactylum, from aomori prefecture, northern Japan, and description of a newly-recognized diagnostic character for the species (Perciformes: Polynemidae). JPN. J. Ichthyol. 48, 41–47. doi: 10.11369/jji1950.48.41
Ojala, D., Montoya, J., and Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474. doi: 10.1038/290470a0
Ou, Y. J., Xie, M. J., Li, J. E., Wen, J. H., Zhou, H., and Liu, Q. Q. (2017). First maturation and mass seedling propagation of cultured Eleutheronema tetradactylum in Guangdong Province. South China Fish. Sci. 13, 97–104. doi: 10.3969/j.issn.2095-0780.2017.04.012
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358. doi: 10.1007/BF01215182
Rosen, D. E. (1964). The relationships and taxonomic position of the halfbeaks, killifishes, silversides, and their relatives. Bull. Mus. Nat. Hist. 27, 219–267.
Sanciangco, M. D., Carpenter, K. E., and Betancur, R. (2016). Phylogenetic placement of enigmatic percomorph families (Teleostei: Percomorphaceae). Mol. Phylogenet. Evol. 94, 565–576. doi: 10.1016/j.ympev.2015.10.006
Satoh, T. P., Miya, M., Mabuchi, K., and Nishid, M. (2016). Structure and variation of the mitochondrial genome of fishes. BMC Genomics 17:719. doi: 10.1186/s12864-016-3054-y
Taanman, J. W. (1999). The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta Bioenerg. 1410, 103–123. doi: 10.1016/S0005-2728(98)00161-3
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wilson, A. C., Cann, R. L., Carr, S. M., George, M., Gyllensten, U. B., Helm-Bychowski, K. M., et al. (1985). Mitochondrial DNA and two perspectives on evolutionary genetics. Biol. J. Linn. Soci. 26, 375–400. doi: 10.1111/j.1095-8312.1985.tb02048.x
Wong, T. W., and Clayton, D. A. (1985). In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell 42, 951–958. doi: 10.1016/0092-8674(85)90291-0
You, J. J., Liu, M. H., Fu, R. B., Li, W. Y., Zhang, X., and Luo, H. Z. (2014). Characteristics of morphological development and growth of four-finger threadfin Eleutheronema tetradactylum (Shaw) larvae and juveniles. J. Dalian Ocean Univ. 29, 577–581.
Zhang, B., Sun, Y. N., and Shi, G. (2014). The complete mitochondrial genome of the fourfinger threadfin Eleutheronema tetradactylum (Perciforms: Polynemidae) and comparison of light strand replication origin within Percoidei. Mitochondrial DNA 25, 411–413. doi: 10.3109/19401736.2013.809433
Zhang, C. L., Cheng, Q. T., Zheng, B. S., Li, S. Z., Zheng, W. L., and Wang, W. B. (1955). Fishes of the Yellow Sea and Pohai. Beijing: Science Press.
Zhong, L. Q., Wang, M. H., Li, D. M., Tang, S. K., Zhang, T. Q., Bian, W. J., et al. (2018a). Complete mitochondrial genome of freshwater goby Rhinogobius cliffordpopei (Perciformes, Gobiidae): genome characterization and phylogenetic analysis. Genes Genomics 40, 1137–1148. doi: 10.1007/s13258-018-0669-1
Zhong, L. Q., Wang, M. H., Li, D. M., Tang, S. K., and Chen, X. H. (2019). First record of the East Asian fourfinger threadfin, Eleutheronema rhadinum (Jordan & Evermann, 1902), from Zhenjiang, China. Cybium 43, 209–211. doi: 10.26028/cybium/2019-423-010
Zhong, L. Q., Wang, M. H., Li, D. M., Tang, S. K., Zhang, T. Q., Bian, W. J., et al. (2016). Complete mitochondrial genome of Chinese icefish Neosalanx tangkahkeiis (Salmoniformes, Salangidae): comparison reveals Neosalanx taihuensis not a valid name. Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 3303–3305. doi: 10.3109/19401736.2015.1015014
Zhong, L. Q., Wang, M. H., Li, D. M., Tang, S. K., Zhang, T. Q., Bian, W. J., et al. (2018b). Complete mitochondrial genome of Odontobutis haifengensis (Perciformes, Odontobutiae): a unique rearrangement of tRNAs and additional noncoding regions identified in the genus Odontobutis. Genomics 110, 382–388. doi: 10.1016/j.ygeno.2017.12.008
Zhuang, P., Wang, Y. H., Li, S. F., Deng, S. M., Li, C. S., and Ni, Y. (2006). Fishes of the Yangtze Estuary. Shanghai: Shanghai Science and Technology Press.
Keywords: Eleutheronema rhadinum, mitochondrial genome, Polynemidae, non-coding region, phylogeny
Citation: Zhong L, Wang M, Li D, Tang S and Chen X (2021) Mitochondrial Genome of Eleutheronema rhadinum With an Additional Non-coding Region and Novel Insights Into the Phylogenetics. Front. Mar. Sci. 8:746598. doi: 10.3389/fmars.2021.746598
Received: 26 July 2021; Accepted: 23 August 2021;
Published: 09 September 2021.
Edited by:
Xiaoping Huang, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Zhong, Wang, Li, Tang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqiang Zhong, bHF6aG9uZ2ZmcmlAaG90bWFpbC5jb20=; Xiaohui Chen, eGhjaGVuZmZyaUBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.