- 1College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 2Zhejiang Key Laboratory of Aquatic Germplasm Resources, College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
- 3Ninghai Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ningbo, China
The circadian rhythm has a great impact on the growth, metabolism and development of animals, but little is known about the circadian rhythm of marine bivalves. Understanding of the feeding rhythm is of great significance to increase the yield of razor clam Sinonovacula constricta, an economically important bivalve mollusk. The aim of this experiment was to study the effects of circadian rhythm of feeding time on digestive enzymes activities and their gene expression in S. constricta within a light (ZT8-ZT20)/dark (ZT20-ZT8) cycle. The present results showed that circadian rhythm of feeding rate (FR) was highly associated with digestive enzyme activities and relative expression of their genes. The highest values of FR were basically observed in the night from ZT0-ZT2 and ZT6-ZT8, which were significantly higher than those values in the daytime from ZT12-ZT14 and ZT18-ZT20 (P < 0.05). The digestive enzymes activities displayed the highest values at ZT2 and ZT8, and the lowest at ZT14 and ZT20. Among them, cellulase and pepsin were found to have significantly different activities (P < 0.05), rather than amylase and lipase. Notably, the relative expression of digestive enzyme genes shared the similar pattern with the activities of digestive enzymes. The highest values of relative gene expression of amylase (AMY), lipase (LIP), cellulase (CEL), and pepsin (PEP) were found at ZT2 and ZT8 in the night, while the lowest values were found at ZT14 during the day. It is therefore suggested that the biological clock may regulate the process from feeding to digestion. Furthermore, it might be better to feed at night to reduce cultivating cost and increase economic benefits in the farming industry of S. constricta.
Introduction
The circadian rhythm is a 24-h cyclical change in environmental factors such as light and temperature caused by the rotation of the earth, which has an important impact on the physiology and behavior of organisms (Fustin et al., 2013). The circadian rhythms inside and outside the organism are closely related to the rhythm changes of the external environment. When the rhythm of environmental factors is changed, the nervous system and endocrine system of animals will change accordingly, leading to changes in their behaviors, lifestyles, and physiological conditions (Wu et al., 2002).
The feeding rhythm of animals affected by cyclical change factors (e.g., light and tide) is essential for the establishment of scientific feeding mode (Mistlberger, 1994; Sanchezvazquez, 1995; Wang, 2004; Connor and Gracey, 2011). Many activities such as feeding, oxygen consumption and digestion have been used to study rhythmic behavior in European oyster Ostrea edulis, New Zealand cockle Austravenus stutchburyi, clam Saxidomus purpuratus, etc (Morton, 1971; Williams and Pilditch, 1997; Kim et al., 2003). Furthermore, the absorption of food after feeding is closely related to the process of digestion and metabolism. Meanwhile, digestive enzyme activity is an important indicator of digestion and absorption capacity, which determines the ability to digest and absorb nutrients for food (Bobrowska et al., 2011; Wu et al., 2013). The relationship between the activity of digestive enzymes and the feeding pattern has been studied in cockle Cerastoderma edule (Ibarrola et al., 1998), clam Ruditapes decussatus and Venerupis pullastra (Albentosa and Moyano, 2008), and scallop Patinopecten yessoensis (Li et al., 2010). In addition, some previous studies have found that the activities of digestive enzymes such as pepsin, amylase, cellulase, and lipase are served as indicators that can measure the digestion and absorption of protein and carbohydrates in bivalves (Supannapong et al., 2008; Tizon et al., 2013). For animals, digestion is a complex process that requires signal transduction regulation inside and outside the body. The enzyme-related protein precursor mRNA is regulated to stimulate transcription and synthesis after ingestion, so that the enzyme precursors are released to promote digestion, absorption, and growth (Yúfera et al., 2018). For example, amylase (AMY) gene is proved to be involved in the growth of razor clam Sinonovacula constricta and Pacific oyster Crassostrea gigas, while cellulase (CEL) can improve the digestibility of food and synthesize glucose to provide energy for the body (Meenu et al., 2014; Thongsaiklaing et al., 2014; Rong et al., 2015; Liu et al., 2017). Moreover, lipase (LIP) gene expression is correlated with feeding status, whereas pepsin (PEP) has important digestive functions in both vertebrates and invertebrates (Liang et al., 2003). The presence of various forms of pepsin precursors such as pepsin A and pepsin C may be related to food or feeding habits (Carginale et al., 2004).
S. constricta is an economically and ecologically important benthic marine bivalve, which naturally distributes along the western Pacific coasts of China, Japan, and South Korea (Morton, 2010). It has been widely cultivated in the intertidal zone and estuary waters with more than 400 years of cultivating history in the Zhejiang and Fujian provinces, China (Shen et al., 2013). Due to relatively short production cycle and high productive efficiency, it has been become an important marine aquaculture species in China with 852,925 tons of production in 2018 (FAO, 2020). Food intake is one of the important conditions for increasing the yield of industrial aquaculture (Rønnestad et al., 2013). Good ingestion and efficient digestion of nutrients are necessary to meet the high demand of matter and energy, which are able to support the high-growth rate (Navarro-Guillén et al., 2018). In order to determine the optimal feeding time and increase the growth rate of S. constricta, we continuously measured feeding rate, digestive enzyme activity, and relative expression of digestive enzyme genes at different time points in the day and night. The present results will be beneficial for reducing farming cost and increasing farming benefits.
Materials and Methods
Experimental Animals
S. constricta were obtained from Ningbo Ocean and Fishery Science and Technology Innovation Base (Ningbo, Zhejiang province). One-year-old adults (n = 60) with an average shell length of (5.9 ± 0.3) cm were collected to culture for a week in seawater within light and dark (L/D) cycles. The lights in the experimental environment were turned on, and shoot them directly into the tank from ZT (Zeitgeber Time) 8-ZT20 to simulate the daytime, and from ZT20-ZT8 to cover the tank with a black cloth to simulate the night. All clams were not fed before the start of the experiment. During the formal experiment, the clams were fed with the live microalgae of Chaetoceros muelleri with the concentration of (2.5 ± 0.2) × 108 cell/L. The water temperature and salinity were maintained at (25.0 ± 2) °C and (20 ± 1) ppt, respectively. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Zhejiang Wanli University, China.
Feeding Rate Experiment
The clams in similar size were selected and placed into 12 tanks, having five individuals in each tank. The time setting was to divide 24 h into four time periods, including ZT0-ZT2, ZT6-ZT8, ZT12-ZT14, and ZT18-ZT20. For each time point, it corresponded to three parallel tanks. Before the experiment, a preliminary experiment was conducted to determine the concentration of microalgae C. muelleri. No clam was placed in the control tank, and the same concentration of algae for tank was used to calculate the loss rate of algae. During the experiment, the water was exchanged at each time point, and the water temperature was kept constant for 3 days.
Before and after each time point, water samples of the experimental groups and control groups were sampled to count the concentration of algae on a blood cell counter. The feeding rate (FR) was calculated according to the following formula:
Ctd and Ct: the algae concentration in the control group and experimental group at the end of the experiment (cells/L), T: the experimental time (h), V: the volume of the experimental water (L), N: the number of experimental clam, Sd: the control bait variation coefficient.
Analysis of Digestive Enzyme Activities
A total of 240 clams with similar sizes were randomly distributed into three tanks (group A, group B, and group C). Samples were obtained at 6 h intervals (ZT2, ZT8, ZT14, and ZT20) after feeding, having continuous sampling for 3 days. Four individuals were randomly selected from each group, and the visceral mass tissue was dissected and immediately frozen in liquid nitrogen, and stored at −80°C.
The enzyme extracts were prepared to measure the enzyme activities of α-amylase, lipase, cellulase, and pepsin. Each sample was weighed at 100 mg, which was mixed with 0.9 mL of normal saline for mechanically homogenization using an automatic sample rapid grinding machine1. The samples were centrifuged at 3,500 rpm for 10 min at 4°C. All samples were kept in ice in order to avoid enzymes denaturation or damage. Enzyme extracts were kept at −20°C until analysis within 24 h.
All enzymatic activity analysis was conducted by using the commercial kits from Nanjing Jiancheng Bioengineering Institute2. α-amylase activity as protein per mg of visceral mass reacted with the substrate at 37°C for 30 min to hydrolyze 10 mg amylon and recorded at OD600 nm. The unit of lipase activity was per g tissue protein reacted with methyl resorufin substrate at 37°C and recorded at OD580nm. The cellulase activity as tissue per g catalyzed to 1 μg glucose per minute and recorded at OD550nm. One unit of pepsin activity was defined as tissue protein per mg decomposed into 1 μg amino acid at 37°C per minute and recorded at OD660nm. The protein concentration extracted from the tissue uses the BSA kit3. The enzyme activity of α-amylase, lipase, cellulase, and pepsin were all measured according to the standard protocol.
RNA Extraction and Real-Time Quantitative Expression Analysis of Digestive Enzyme Genes
The total RNA was extracted from the visceral mass by using the Trizol reagent following the manufacturer’s instructions4. The RNA quality was tested by electrophoresis, and the concentration was measured with a nucleic acid detector NanoVue Plus. Total RNA was reverse transcribed to cDNA by RT-PCR kits5.
The whole predicted coding sequence (CDS) sequences of AMP, LPS, CEL, and PEP were obtained from the genome of S. constricta (WSYO00000000.1). The primer sequences were designed following the CDS sequences and listed in Table 1. The mRNA expression levels per each gene were assessed by qRT-PCR using ChamQ SYBR qPCR Master Mix6. The 20 μL reaction volume for amplification contained 10 μL of SYBR qPCR Master Mix, 1 μL of each primer (10 μM), and 8 μL of cDNA sample (10 ng/μL). Initial denaturation was conducted at 95°C for 10 s, followed by 40 cycles at 95°C for 5 s and at 60°C for 30 s. 18S rRNA gene was selected as the housekeeping gene, and the expression levels of AMP, LPS, CEL, and PEP gene were normalized to that of 18S rRNA by using the 2–ΔΔCT method.
Statistical Analysis
Data were presented as the means ± standard deviation, and one-way ANOVA analysis were used to compare the difference in food intake ratio, digestive enzyme activity and digestive enzyme gene expression at different time points in the day and night. GraphPad prism 8 software was used to statistically analysis. P < 0.05 was considered statistically as the significant difference, and P < 0.01 as the extremely significant difference.
Results
Analysis of Diurnal Difference of Feeding Rate
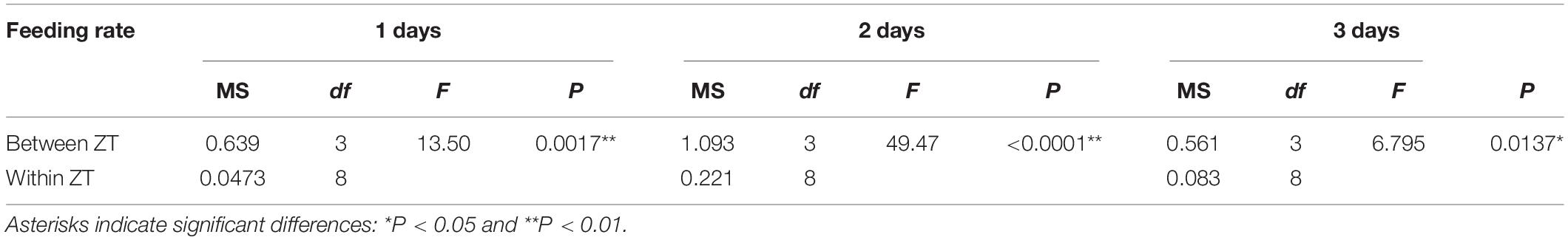
Under the condition of 12 h light/12 h dark (L/D) cycles, the food intake rates at ZT0-ZT2 and ZT6-ZT8 during the night were higher than those at ZT12-ZT14 and ZT18-ZT20 during the day, indicating an obvious circadian rhythm of feeding (Figure 1). For the first day at ZT0-ZT2 and ZT18-ZT20, the food intake rates were 2.15 × 108 (cells/h) and 1.225 × 108 (cells/h). For the second day at ZT6-ZT8 and ZT12-ZT14, the food intake rates were 2.03 × 108 (cells/h) and 0.85 × 108 (cells/h). For the third day at ZT6-ZT8 and ZT18-ZT20, the food intake rates were 2.33 × 108 (cells/h) and 1.38 × 108 (cells/h). In total, the highest and lowest ingestion points were significantly different in the experimental period by one-way ANOVA analysis (P < 0.05), with 1.69–2.39 times between them (Table 2).
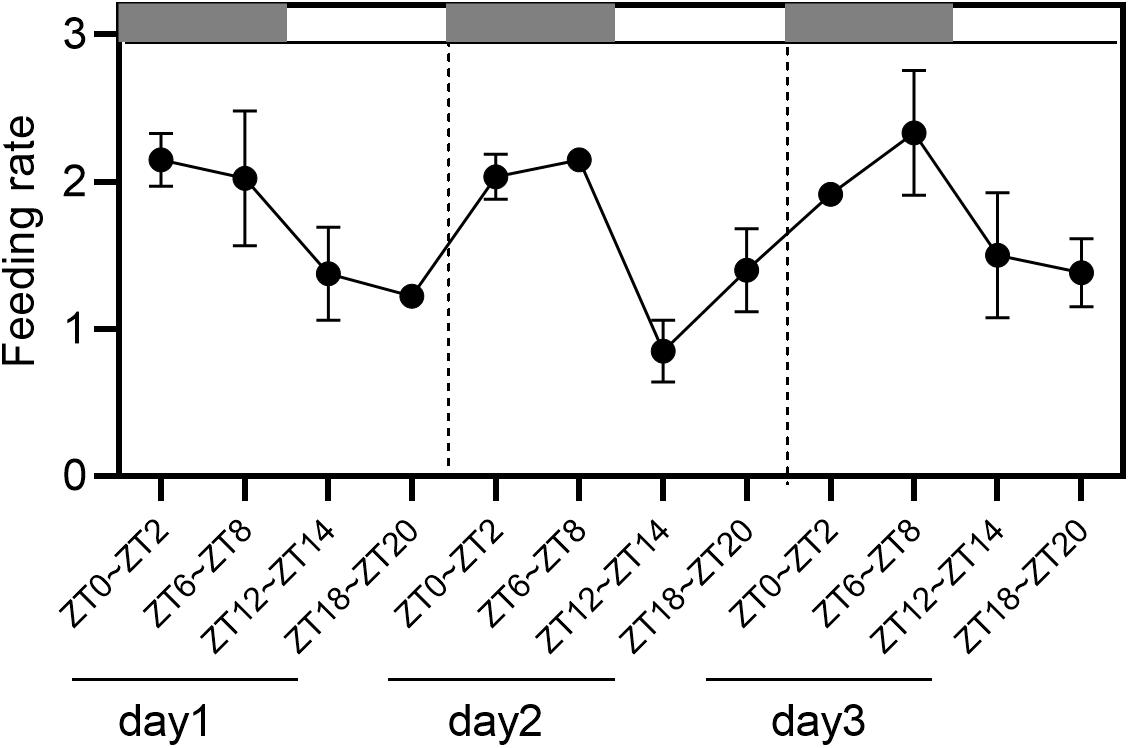
Figure 1. The results of feeding rate in 12 h light/12 h dark (L/D) cycles during 3 days. Values are represented as means ± SD (n = 3). The gray grid represents the dark time point.
The Circadian Rhythm of Four Digestive Enzyme Activities
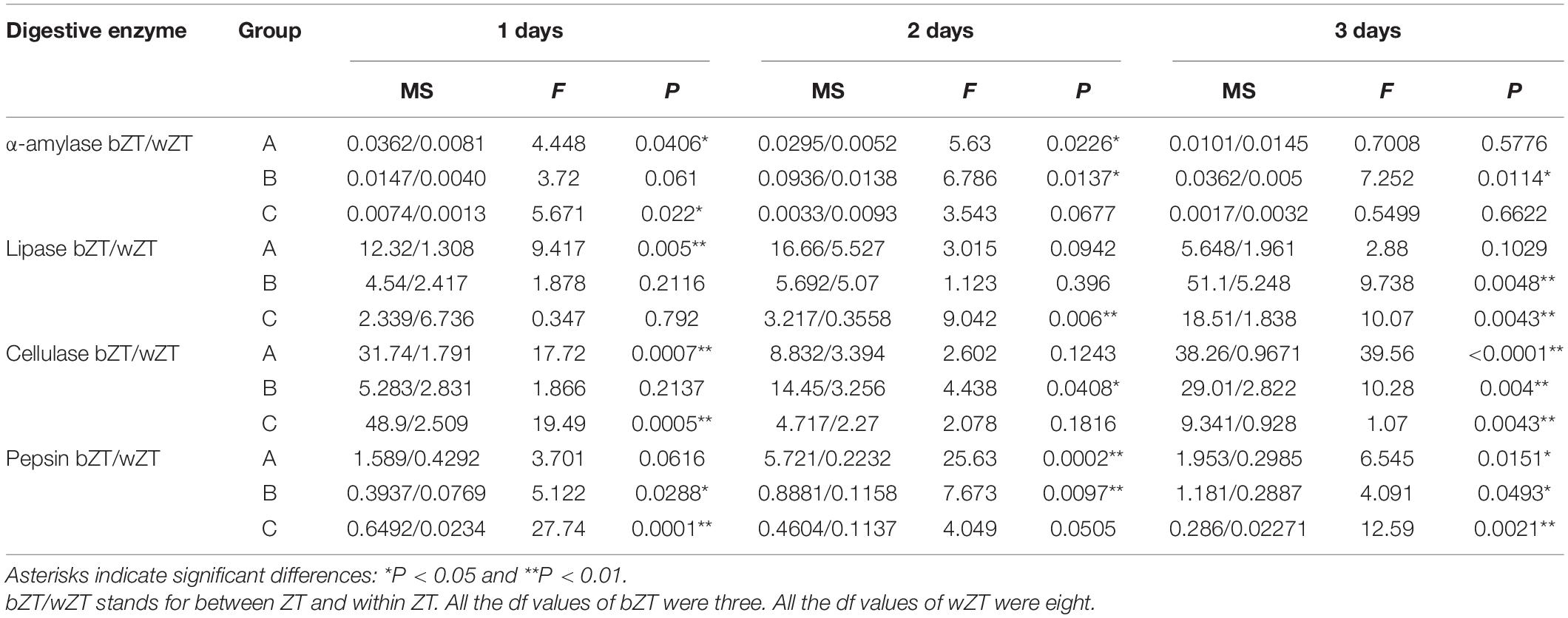
The results of α-amylase, lipase, cellulase, and pepsin activities (expressed as U mg–1) were shown in Figure 2 and Table 3. These four enzymes have a diurnal variation trend in group A, B, C. The activities of lipase reached the highest value at ZT20 and ZT2, followed by low levels of activity between ZT8 and ZT14. Among all these four enzymes, the activities of lipase were significantly lower than those of other enzymes (P < 0.05). However, only part of the results detected significant differences between the highest and lowest points of lipase activities (Table 3 and Figure 2A). For α-amylase, the maximum values of enzyme activities were generally detected at ZT20 and ZT2, while the minimum values were found at ZT8 and ZT14. Nevertheless, α-amylase activities during the whole day and night cycle were statistically different between time periods, but it was only partially present (Table 3 and Figure 2B). As indicated, cellulase and pepsin had the highest activities, displaying the same changing trend (P < 0.05, Table 3 and Figures 2C,D). Basically, the enzyme activities were found to be higher in the dark (ZT20-ZT8) than those in the light (ZT8-ZT20).
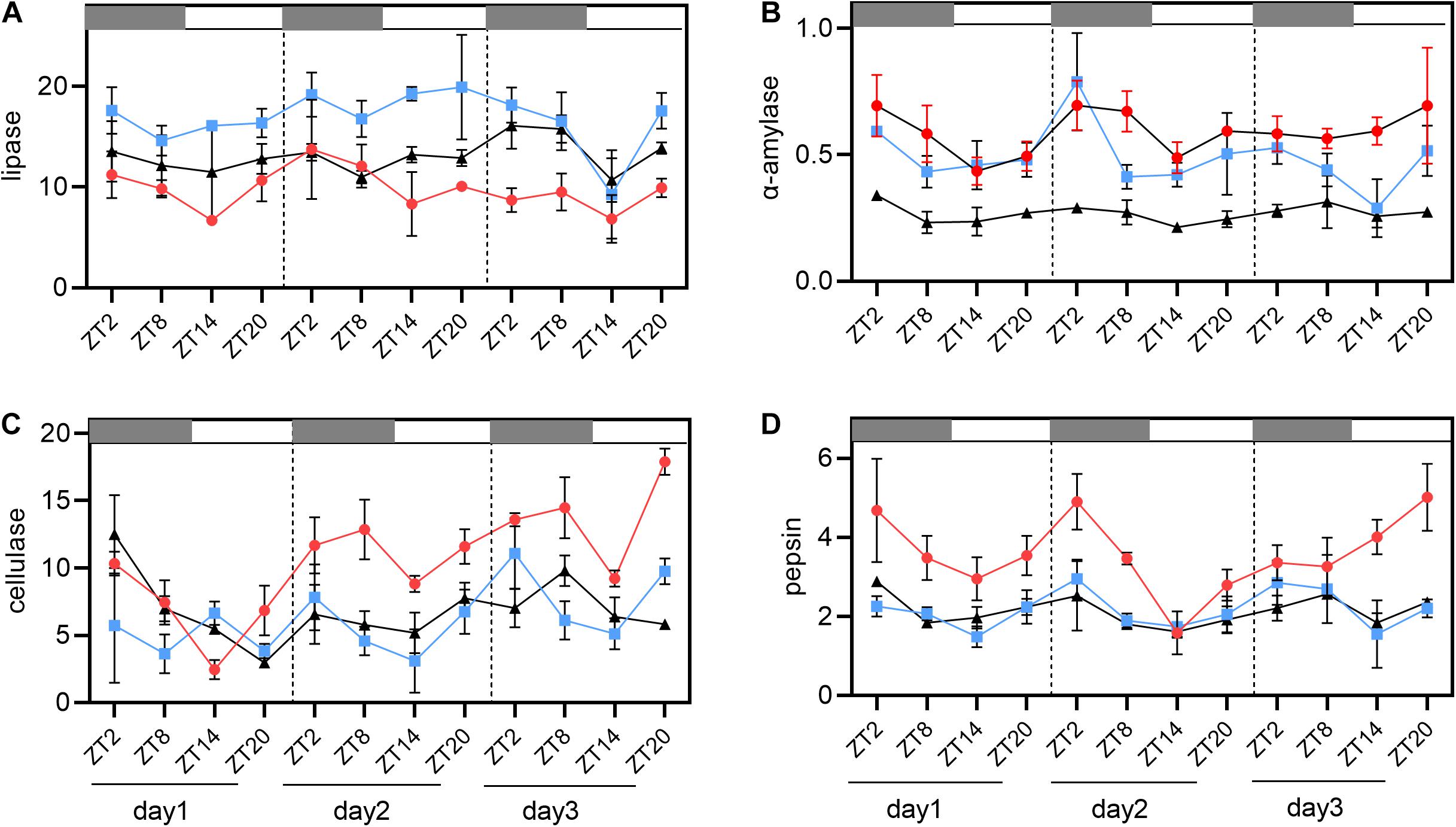
Figure 2. Analysis of four digestive enzyme activities in 12 h light/12 h dark (L/D) cycles during 3 days. (A) Lipase activity diagram. (B) α-amylase activity diagram. (C) Cellulase activity diagram. (D) Pepsin activity diagram. Values are represented as means ± SD (n = 4). Group A, B, C are three parallel tanks with the same cultivating conditions, and the red circle represents Group A, the blue square represents Group B and black triangle represents Group C. The gray grid represents the dark time point.
Relative Expression of Four Digestive Enzyme Genes
The results of relative expression of digestive enzyme genes were shown in Figure 3. The relative expression of AMY, LPS, CEL, and PEP had a regular trend from high to low levels from ZT2 to ZT20 of 3 days, with no significant change in diurnal fluctuations. The relative expression of LPS had the highest expression level at ZT2 and ZT8, the lowest expression at ZT14 (Figure 3A). Meanwhile, AMY was detected to be the highest at ZT2 and the lowest at ZT14 (Figure 3B). Similarly, the gene expression of CEL was found to be the highest at ZT2 and the lowest at ZT14 (Figure 3C). Consistently, PEP showed the highest expression level at ZT2 and the lowest expression at ZT14 and ZT20 (Figure 3D).
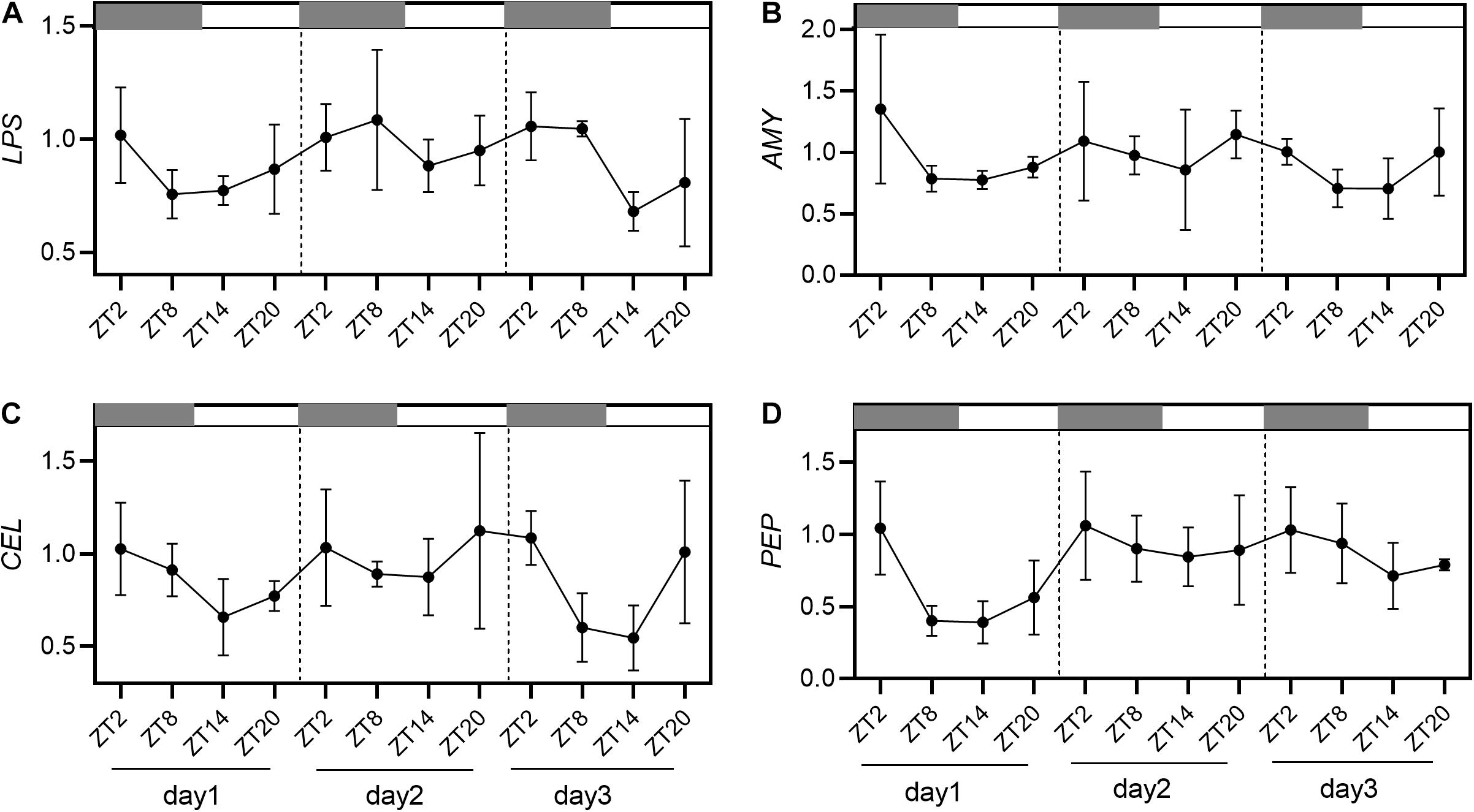
Figure 3. Relative gene expression of LPS (A), AMY (B), CEL (C), and PEP (D) in 12 h light/12 h dark (L/D) cycles during 3 days. Values are represented as means ± SD (n = 4). The gray grid represents the dark time point.
Discussion
For most organisms, circadian rhythm is affected by endogenous factors and external environmental factors (Mata-Sotres et al., 2016). Under natural light conditions, the behavior of most animals is divided into two types: diurnal and nocturnal. Understanding and clarifying the circadian rhythm can provide a scientific basis for revealing accurately the laws and inner mechanisms of many life phenomena. Although the feeding rhythm is of great significance for establishing a scientific feeding mode to farmed animals, there are few studies on the feeding rhythm and their mechanisms in aquatic animals. In fish, as reported, the Gilthead sea bream Sparus aurata showed an feeding rhythm during the day under a light/dark condition (Mata-Sotres et al., 2015). In contrast, the Japanese prawn Penaeus japonicus feeds at night under the same conditions (Reymond and Lagardère, 1990). In mollusks, many species showed similar feeding and metabolic rhythm. For example, Manila clam Ruditapes philippinarum exhibited a 24 h circadian rhythm of feeding rates, having a higher feeding rate at night than that in the day (Jiang et al., 2009). A study on circadian feeding activity and digestive physiology in the abalone Haliotis discus hannai found that the percentages of ingesting food were significantly higher at night than in the daytime, demonstrating their nocturnal characteristics (Gao et al., 2021). An obvious circadian rhythm in water filtration rate was also found in the scallop Chlamys farreri, which had a significantly higher water filtration rate at night than that in daytime (Du et al., 2012). In S. constricta, the obvious diurnal changes in food intake rates were proved by the energy budget study (Li et al., 2006). In the present study, we found that the feeding rate of razor clam in dark was significantly higher than that in light, speculating that there may be a circadian rhythm of feeding, which is indirectly regulated by biological clock genes. Additionally, the highest feeding point of mollusks at night may be related to avoiding being preyed by predators. Moreover, some studies in aquatic animals have found that light is the most important external factor influencing the feeding activity and different modes of light conditions will reduce the intake of food (Li et al., 2010). Especially, for the nocturnal animals that forage at night, light inhibits their feeding behavior. However, the long-term evolutionary internal biological clock can regulate the life patterns of animals, which may control the time and amount of consumed food (Annie et al., 1999). These findings may thus help us optimize the feeding time and frequency in clam aquaculture.
The degree of digestion is derived from the intestinal transport factor and digestive enzyme activity that reflects the digestion and absorption capacities of intestinal tract (Mata-Sotres et al., 2015; Navarro-Guillén et al., 2015). Generally, there is a close relationship between the digestive enzyme activity and feeding behavior, which the increase of digestive enzyme activity is several hours earlier than the time of peak feeding, suggesting that the early enzyme secretion can optimize the percentages of digestion and absorption, and reduce the risk of predation due to shortened feeding time. For nocturnal animals, even under an unnatural light/dark cycle, the digestive enzyme activity is prone to be higher at night than in the daytime. In the current study, the light/dark cycle and feeding rate had a significant impact on four digestive enzyme activities in the visceral mass of S. constricta, including α-amylase, lipase, cellulase, and pepsin. The vitality of digestive enzymes after the increase of food intake in ZT2 and ZT8 at night is significantly higher than that in low food intake ZT14 and ZT20 in the daytime, demonstrating that food digestion has a circadian rhythm behavior. There is also the same finding that the production of digestive enzymes increased significantly after increasing food intake, like in C. edule (Ibarrola et al., 1998) and clam R. philippinarum (Houki and Tomohiko, 2020). However, relatively speaking, lipase activity is significantly lower than the other three. Numerous studies have shown that carnivorous animals have higher lipase content than omnivorous and herbivorous animals, and those with high lipase content are generally seen in animals with mature intestines, lipid hydrolysis mainly occurs in the pyloric caeca and/or proximal intestine (Caruso et al., 2009; Tengjaroenku et al., 2000). The razor clam, as a filter-feeding shellfish, has an incompletely differentiated intestine, which the ability to hydrolyze lipids is weak.
The expression of digestive enzyme genes is strongly correlated to their enzyme activities, which directly reflect the digestive physiology of aquatic animals. Existing research has shown that the increase of enzyme genes expression at night was the regulation of translation process and may be the anticipation of next food intake (José Antonio et al., 2016). A recent research on the feeding and digestion physiology of abalone H. discus hannai under the light/dark cycle found that digestive enzymes and feeding related genes were significantly higher at night than during the day with rhythmic 24 h oscillations (Gao et al., 2021). In this research, the mRNA levels of four key digestive enzyme genes (AMY, LPS, CEL, and PEP) by qRT-PCR during the light/dark cycles were basically consistent with the production of their enzymes. Furthermore, the genes expression showed a periodic pattern of high at night and low during the day for consecutive 3 days, indicating that circadian rhythm may be the internal mechanism that was regulated by circadian clock genes (Paredes et al., 2014; Qin et al., 2020). Then the circadian clock genes may regulate the expression of AMY, LPS, CEL, and PEP, thereby participating indirectly in the feeding and digestion activities of S. constricta. Although the similar trends of gene expression and activities of key digestive enzymes, and feeding rates in this experiment were discovered, deep understanding of their corresponding relationships still need a lot of in-depth and meticulous research.
Conclusion
The feeding rate, digestive enzyme activities and relative expression of their genes all had a circadian rhythm in S. constricta, which it can be basically determined that ZT0-ZT2 will reach the peak at night and ZT12-ZT14 will reach the lowest value during the day from feeding to digestion. Therefore, it can be speculated that razor clams have higher nighttime activities than during the day, suggesting that the feeding time in the industrial farming can be arranged at night to reduce cultivating costs and increase farming benefits.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The adult hard clams Sinonovacula constricta at the age of 1 year were collected from the genetic breeding research center of Zhejiang Wanli University, China. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China.
Author Contributions
HY and YD conceived and designed the project. TZ collected the samples and contributed reagents. YL performed the experiments and data analysis, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31902393), National Key Research and Development Program of China (2018YFD0901405 and 2020YFD0900802), Zhejiang Major Program of Science and Technology (2021C02069-7), Ningbo Major Project of Science and Technology (2019B10005), and National Marine Genetic Resource Center Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.744212/full#supplementary-material
Footnotes
- ^ https://www.roche.com/
- ^ http://www.njjcbio.com/
- ^ https://www.thermofisher.cn/
- ^ https://www.sangon.com/
- ^ https://www.takarabiomed.com.cn/
- ^ http://vazyme.bioon.com.cn/
References
Albentosa, M., and Moyano, F. J. (2008). Influence of nutritional stress on digestive enzyme activities in juveniles of two marine clam species, Ruditapes decussatus and Venerupis pullastra. J. Sea Res. 59, 249–258. doi: 10.1016/j.seares.2008.02.004
Annie, M., Stephen, C. B., and Jean-Franois, H. (1999). Daily burrowing cycle and feeding activity of juvenile sea cucumbers Holothuria scabra in response to environmental factors. J. Exp. Mar. Biol. Ecol. 239, 125–156. doi: 10.1016/S0022-0981(99)00034-9
Bobrowska, B., Tokrz, A., Białek, S., and Seweryn, M. (2011). Effect of dietary supplementation on the prognostic value of urinary and serum 8-isoprostaglandin F2α in chemically-induced mammary carcinogenesis in the rat. Lipids Health Dis. 10:40. doi: 10.1186/1476-511X-10-40
Carginale, V., Trinchella, F., Capasso, C., Scudiero, R., Riggio, M., and Parisi, E. (2004). Adaptive evolution and functional divergence of pepsin gene family. Gene 333, 81–90. doi: 10.1016/j.gene.2004.02.011
Caruso, G., Denaro, M. G., and Genovese, L. (2009). Digestive enzymes in some teleost species of interest for mediterranean aquaculture. Open Fish Sci. J. 2, 74–86. doi: 10.2174/1874401X00902010074
Connor, K. M., and Gracey, A. Y. (2011). Circadian cycles are the dominant transcriptional rhythm in the intertidal mussel Mytilus californianus. Proc. Natl. Acad. Sci. U.S.A. 108, 16110–16115. doi: 10.1073/pnas.1111076108
Du, M. R., Fang, J. G., Mao, Y. Z., Zhang, J. H., Ge, C. Z., Jiang, Z. J., et al. (2012). Diurnal rhythm of filtration rate of Zhikong scallop Chlamys farreri veliger and spat. Prog. Fish. Sci. 33, 73–77.
FAO (2020). Fishery and Aquaculture Statistics 2018. Available online at: http://www.fao.org/fishery/statistics/yearbook/en (accessed September 1, 2021).
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., and Yoshida, M. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. doi: 10.1016/j.cell.2013.10.026
Gao, X. L., Pang, G. W., Luo, X., You, W. W., and Ke, C. H. (2021). Effects of light cycle on circadian feeding activity and digestive physiology in Haliotis discus hannai. Aquaculture 539:736642. doi: 10.1016/j.aquaculture.2021.736642
Houki, S., and Tomohiko, K. (2020). Feeding and digestion periodicity of Manila clam Ruditapes philippinarum in natural intertidal and subtidal zones estimated from the morphological condition of the crystalline style. J. Molluscan Stud. 86, 361–371.
Ibarrola, I., Larretxea, X., Iglesias, J. I. P., Urrutia, M. B., and Navarro, E. (1998). Seasonal variation of digestive enzyme activities in the digestive gland and the crystalline style of the common cockle Cerastoderma edule. Comp. Biochem. Physiol. 121, 25–34. doi: 10.1016/S1095-6433(98)10097-1
Jiang, S. L., Zhao, C. M., Wang, Y. H., Yin, H., Sun, Y., and Shi, X. Y. (2009). Effects of water temperature, shell length and the concentration of algae on the ingestion rate of Ruditapes philippinarum. Prog. Fish. Sci. 30, 78–83.
José Antonio, M. S., Francisco, J. M., and Gonzalo, M. R. (2016). Daily rhythms of digestive enzyme activity and gene expression in gilthead seabream (Sparus aurata) during ontogeny. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 197, 43–51.
Kim, W. S., Huh, H. T., Je, J. G., and Han, K. N. (2003). Evidence of two-clock control of endogenous rhythm in the Washington clam, Saxidomus purpuratus. Mar. Biol. 142, 305–309. doi: 10.1007/s00227-002-0952-0
Li, B., Chen, B., Qi, Z. H., Jiang, Z., Zhang, J., and Fang, J. (2010). Relationship between differential retention of Escherichia coli and Enterococcus faecalis and variations in enzyme activity in the scallop Patinopecten yessoensis. Mar. Pollut. Bull. 60, 1600–1605. doi: 10.1016/j.marpolbul.2010.03.017
Li, D. X., Li, T. W., Su, X. R., Lin, Z. H., Chai, X. L., and Fang, J. (2006). Energy budgets of Sinonovacula constricta. Fish. Sci. 25, 401–404.
Liang, X. F., Bai, J. J., Lao, H. H., Li, G. S., Zhou, T. H., and Hiroshi, Y. O. (2003). Nutritional regulaition of lipoprotein lipase gene experssion and visceral fat deposition in red set bream (Pagrus major). Oceanologia Et Limnologiia Sinica 34, 625–630. doi: 10.3321/j.issn:0029-814X.2003.06.006
Liu, C. S., Dong, Y. H., Xue, Q. G., and Lin, Z. H. (2017). Association of α-amylase gene with growth traits in the razor clam Sinonovacula. Invertebrate Surviv. J. 14, 494–504.
Mata-Sotres, A., Yufera, J., Manuel, J. M., and Martinez-Rodriguez, G. (2016). Daily rhythms of digestive enzyme activity and gene expression in gilthead seabream (Sparus aurata) during ontogeny. Comp. Biochem. Physiol. A 197, 43–51.
Mata-Sotres, J. A., Martínez-Rodríguez, G., Pérez-Sánchez, J., Martínez-Rodríguez, G., Pérez-Sánchez, J., and Sánchez-Vázquez, F. J. (2015). Daily rhythms of clock gene expression and feeding behavior during the larval development in gilthead seabream, Sparus aurata. Chronobiol. Int. 32:1061. doi: 10.3109/07420528.2015.1058271
Meenu, K., Singh, G., and Vishwakarma, R. A. (2014). “Chapter 22-molecular mechanism of cellulase production systems in trichoderma,” in Biotechnology and Biology of Trichoderma, eds A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, M. Schmoll, M. Tuohy, and V. G. Gupta (Philadelphia, PA: Elsevier Science), 319–324. doi: 10.1016/B978-0-444-59576-8.00022-9
Mistlberger, R. E. (1994). Circadian food-anticipatory activity: formal models and physiological mechanisms. Neurosci. Biobehav. Rev. 18, 171–195. doi: 10.1016/0149-7634(94)90023-x
Morton, B. (1971). The diurnal rhythm and tidal rhythm of feeding anddigestion in Ostrea edulis. Biol. J. Linn. Soc. 3, 329–342. doi: 10.1111/j.1095-8312.1971.tb00526.x
Morton, B. (2010). The functional morphology of Sinonovacula constricta with a discussion on the taxonomic status of the Novaculininae (Bivalvia). Proc. Zool. Soc. Lond. 202, 299–325. doi: 10.1111/j.1469-7998.1984.tb05085.x
Navarro-Guillén, C., Conceio, L. E. C., Pinto, W., Siguero, I., and Yúfera, M. (2018). Fast growing greater amberjack post-larvae require a high energy-high protein weaning diet. Aquaculture 499, 195–202. doi: 10.1016/j.aquaculture.2018.09.037
Navarro-Guillén, C., Moyano, F. J., and Yúfera, M. (2015). Diel food intake and digestive enzyme production patterns in Solea senegalensis larvae. Aquaculture 435, 33–42. doi: 10.1016/j.aquaculture.2014.09.017
Paredes, J. F., Vera, L. M., Martinez-Lopez, F. J., Navarro, I., and Sánchez, V. F. J. (2014). Circadian rhythms of gene expression of lipid metabolism in gilthead seabream liver: synchronisation to light and feeding time. Chronobiol. Int. 31, 613–626. doi: 10.3109/07420528.2014.881837
Qin, C. J., Sun, J. X., Wang, J., Han, Y., and Hu, P. (2020). Discovery of differentially expressed genes in the intestines of Pelteobagrus vachellii within a light/dark cycle. Chronobiol. Int. 37, 339–352. doi: 10.1080/07420528.2019.1690498
Reymond, H., and Lagardère, J. P. (1990). Feeding rhythms and food of Penaeus japonicus Bate (Crustacea Penaeidae) in salt marsh ponds. Aquaculture 84, 125–143. doi: 10.1016/0044-8486(90)90343-L
Riisgård, H. U. (1991). Filtration rate and growth in the blue mussel, Mytilus edulis Linneaus, 1758: dependence on algal concentration. J. Shellfish Res. 10, 29–35.
Rong, Y. J., Zhang, L., Chi, Z. M., and Wang, X. H. (2015). A carboxymethyl cellulase from a marine yeast (Aureobasidium pullulans 98): its purification, characterization, gene cloning and carboxymethyl cellulose digestion. J. Ocean Univ. China. 14, 913–921. doi: 10.1007/s11802-015-2574-4
Rønnestad, I., Yúfera, M., Ribeiro, L., and Boglione, C. (2013). Feeding behaviour and digestive physiology in larval fish: current knowledge, and gaps and bottlenecks in research. Rev. in Aquac. 5(Suppl. S1), S59–S98. doi: 10.1111/raq.12010
Sanchezvazquez, E. J. (1995). Light-dark and food restriction cycles in sea bass: effect of conflicting zeitgebers on demand-feeding rhythms. Physiol. Behav. 58:705. doi: 10.1016/0031-9384(95)00116-Z
Shen, H. D., Liu, G., Fang, L., and Li, J. (2013). Effect of microalgae and temperature on absorption efficiency of razor clam (Sinonovacula constricta Lamark, 1818). Aquac. Res. 44, 1524–1530. doi: 10.1111/j.1365-2109.2012.03159.x
Supannapong, P., Pimsalee, T., Komol, T. A., Engkagul, A., Kovitvadhi, U., and Kovitvadhi, S. (2008). Digestive enzymes and in-vitro digestibility of different species of phytoplankton for culture of the freshwater pearl mussel, Hyriopsis bialatus. Aquac. Int. 16, 437–453. doi: 10.1007/s10499-007-9156-4
Tengjaroenku, L. B., Smith, B., Caceci, J., and Smith, S. A. (2000). Distribution of intestinal enzyme activities along the intestinal tract of cultured Nile tilapia, Oreochromis niloticus L. Aquaculture 182, 317–327. doi: 10.1016/S0044-8486(99)00270-7
Thongsaiklaing, T., Sehawong, W., Kubera, A., and Ngernsiri, L. (2014). Analysis of the α-amylase gene sequence and the enzyme activity of indian rock oyster Saccostrea forskali. Fish. Sci. 80, 589–601. doi: 10.1007/s12562-014-0708-z
Tizon, R. U., Serrano, A. E., and Traifalgar, R. F. (2013). Effect of unialgal diets on digestive enzyme activity inthe angelwing clam (Pholas orientalis). Isr. J. Aquac. Bamid. 890, 1–6. doi: 10.1071/MF12240
Wang, J. Y. (2004). Aquaculture on the control measures of ecological environment pollution. J. Beijing Fish. 4, 4–6.
Williams, B. G., and Pilditch, C. A. (1997). The entrainment of persistent tidal rhythmicity in a filter-feeding bivalve using cycles of food availability. J. Biol. Rhythms 12, 173–181. doi: 10.1177/074873049701200208
Wu, G., Chen, P., Hang, R., Yang, S. Y., and Shen, J. L. (2002). Influence of salinity and day and night rhythm on feeding rate(FR) of Philippinarum ruditapes. J. Oceanogr. Tanwan Strait. 21, 73–77. doi: 10.1007/s11769-002-0041-9
Wu, Y., Liu, W. B., Li, H. Y., Xu, W. N., He, J. X., and Li, X. F. (2013). Effects of dietary supplementation of fructooligosaccharide on growth performance, body composition, intestinal enzymes activities and histology of blunt snout bream (Megalobrama amblycephala) fingerlings. Aquac. Nutr. 19, 886–894. doi: 10.1111/anu.12033
Yúfera, M., Moyano, F. J., and Martínez-Rodríguez, G. (2018). “The digestive function in developing fish larvae and fry. From molecular gene expression to enzymatic activity”, in Emerging Issues in Fish Larvae Research ed. M. Yúfera (Cham: Springer International Publishing), 51–86. doi: 10.1007/978-3-319-73244-2_3
Keywords: Sinonovacula constricta, circadian rhythm, feeding rate, digestive enzyme, gene expression
Citation: Liu Y, Yao H, Zhou T, Lin Z and Dong Y (2021) The Discovery of Circadian Rhythm of Feeding Time on Digestive Enzymes Activity and Their Gene Expression in Sinonovacula constricta Within a Light/Dark Cycle. Front. Mar. Sci. 8:744212. doi: 10.3389/fmars.2021.744212
Received: 20 July 2021; Accepted: 20 September 2021;
Published: 14 October 2021.
Edited by:
Weiwei You, Xiamen University, ChinaReviewed by:
Xiaolong Gao, Xiamen University, ChinaKwang-Sik Choi, Jeju National University, South Korea
Copyright © 2021 Liu, Yao, Zhou, Lin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanhan Yao, eWFvaGFuaGFuMTAyMEAxMjYuY29t; Yinghui Dong, ZG9uZ3lpbmdodWkxMThAMTI2LmNvbQ==
 Yanzi Liu1,2
Yanzi Liu1,2 Hanhan Yao
Hanhan Yao Yinghui Dong
Yinghui Dong